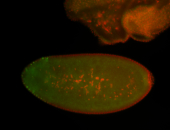
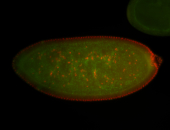
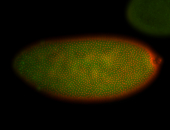
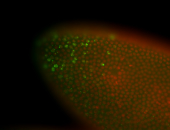
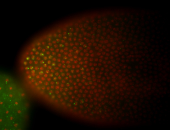
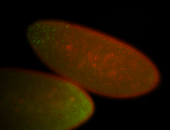
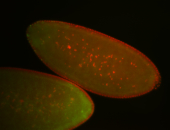
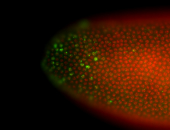
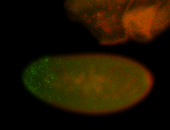
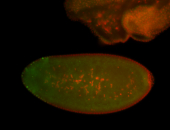
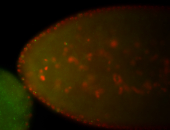
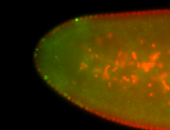
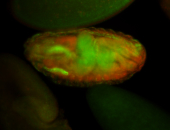
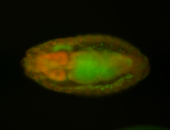
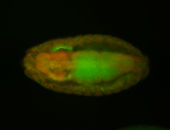
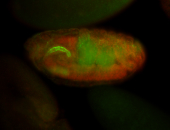
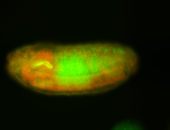
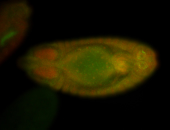
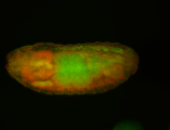
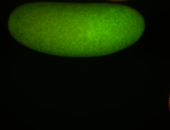
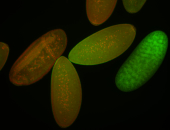
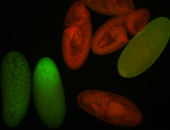
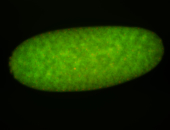
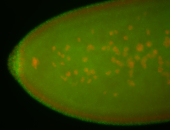
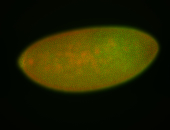
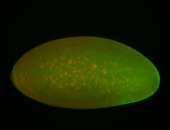
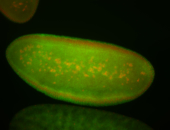
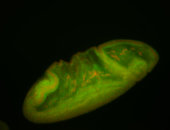
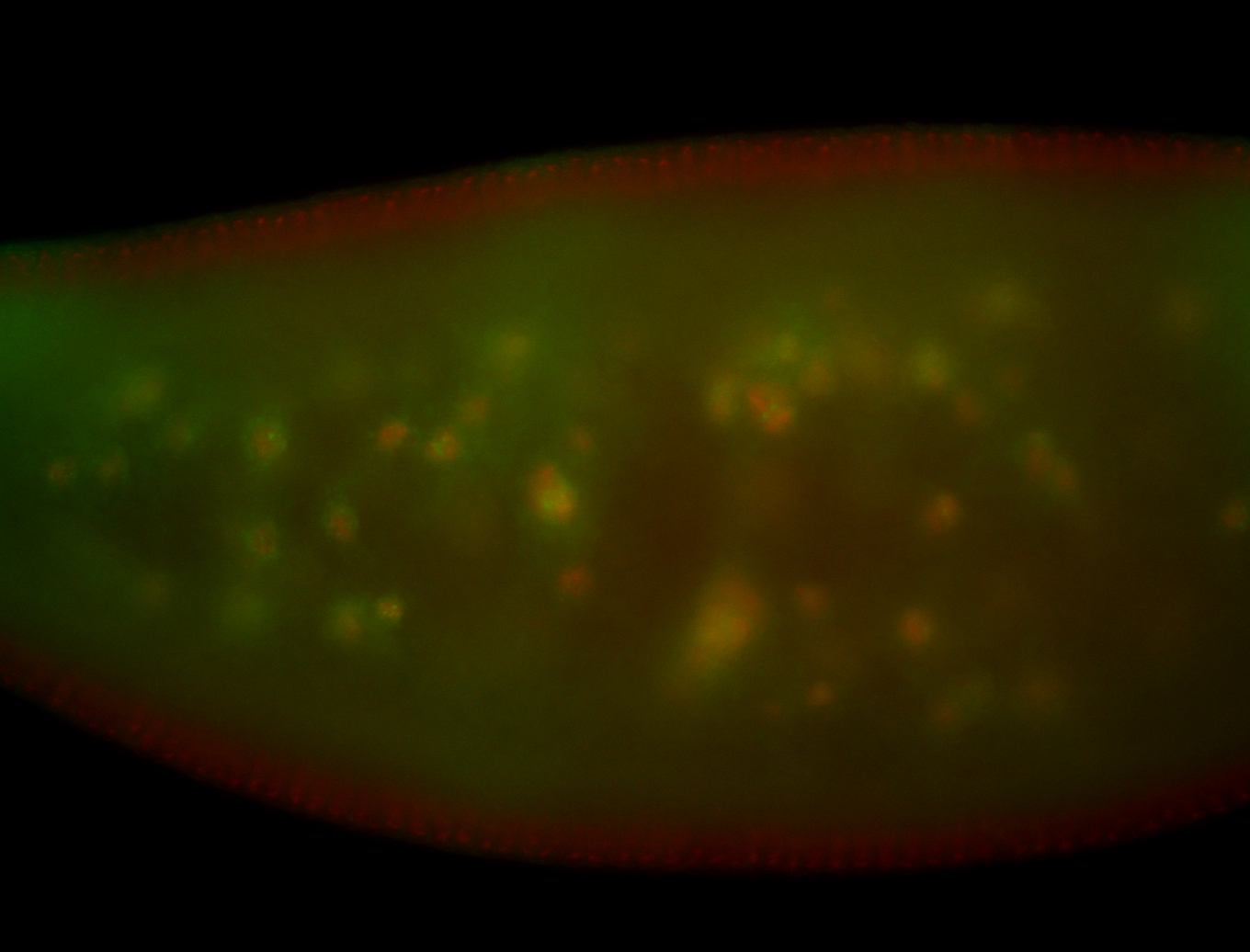
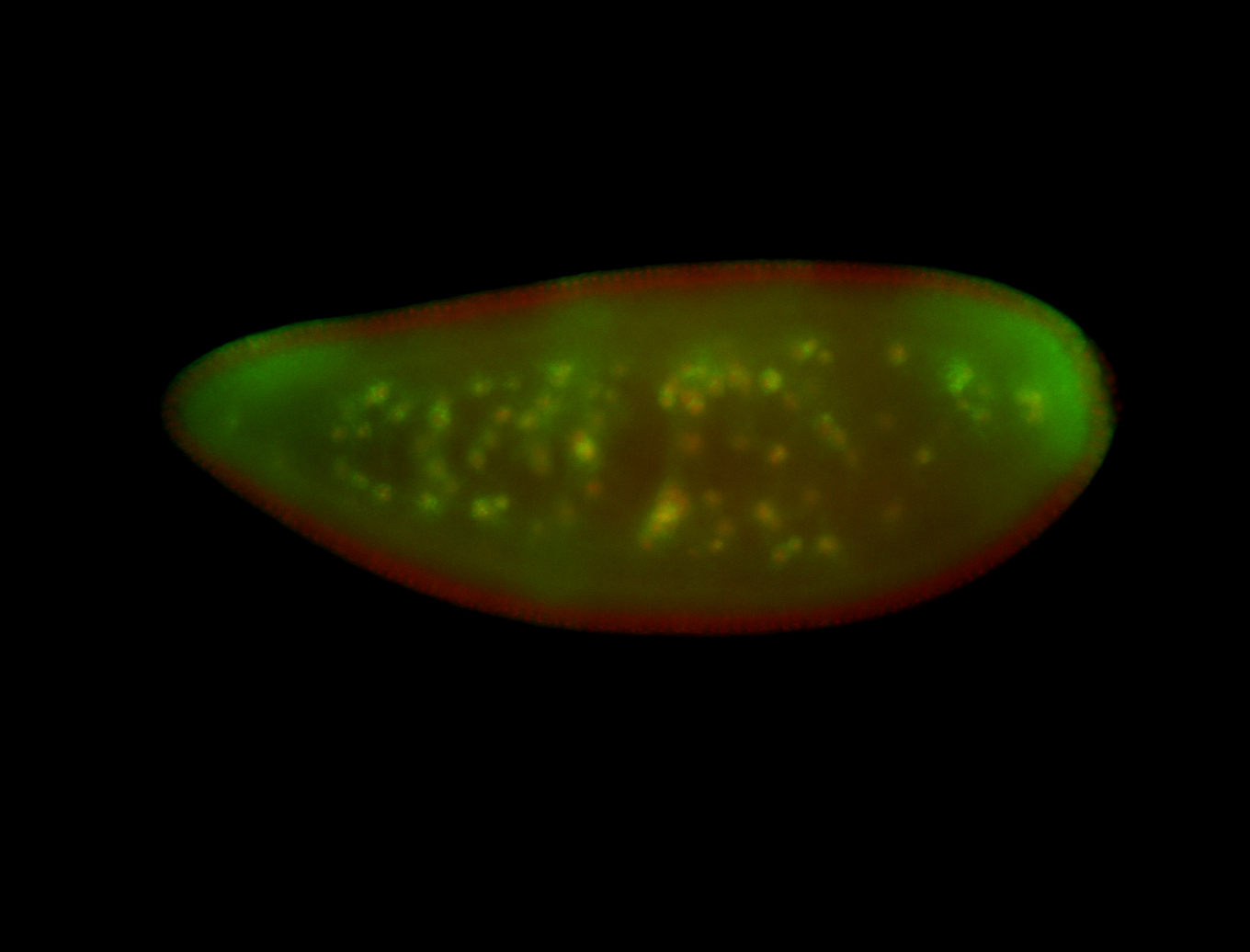
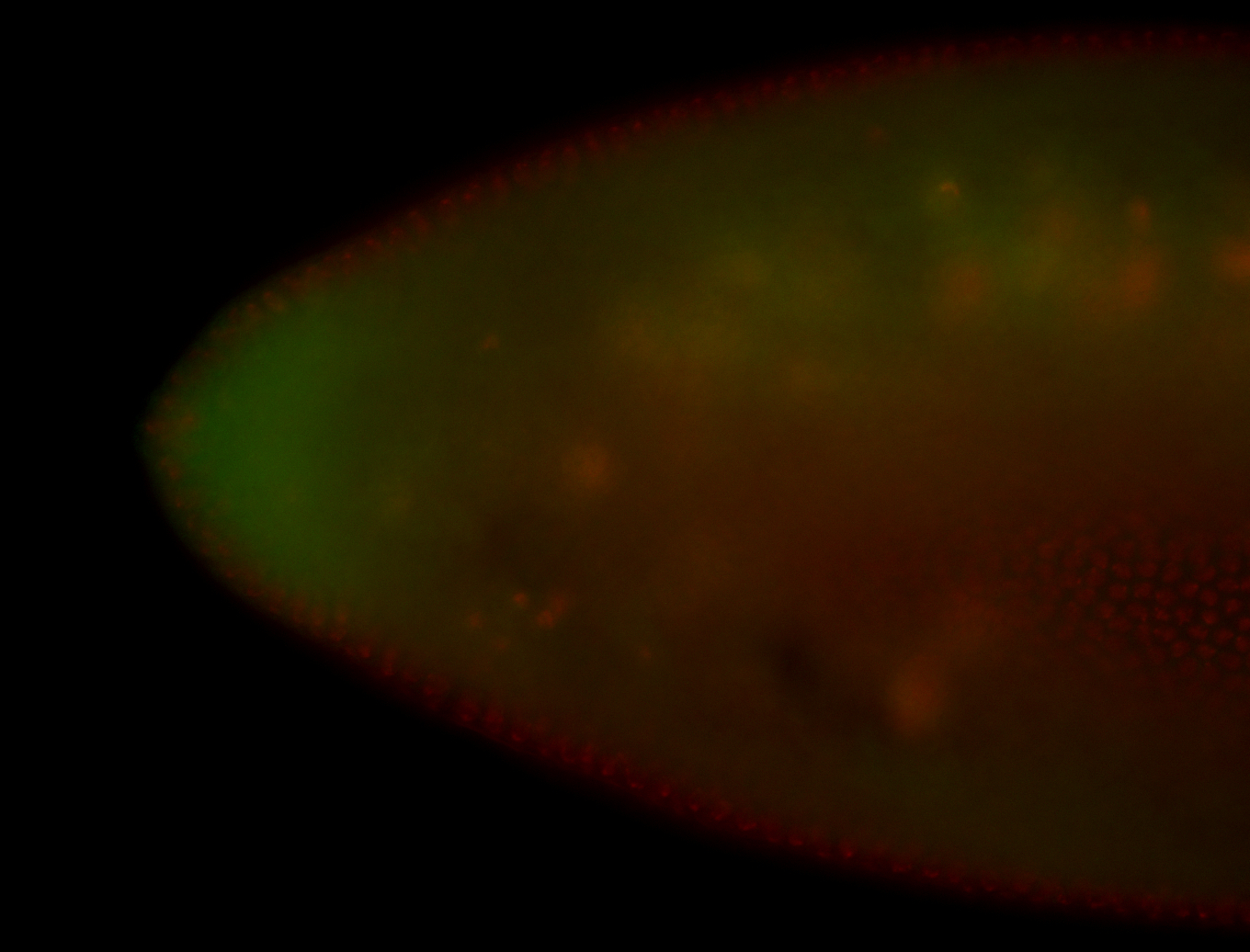
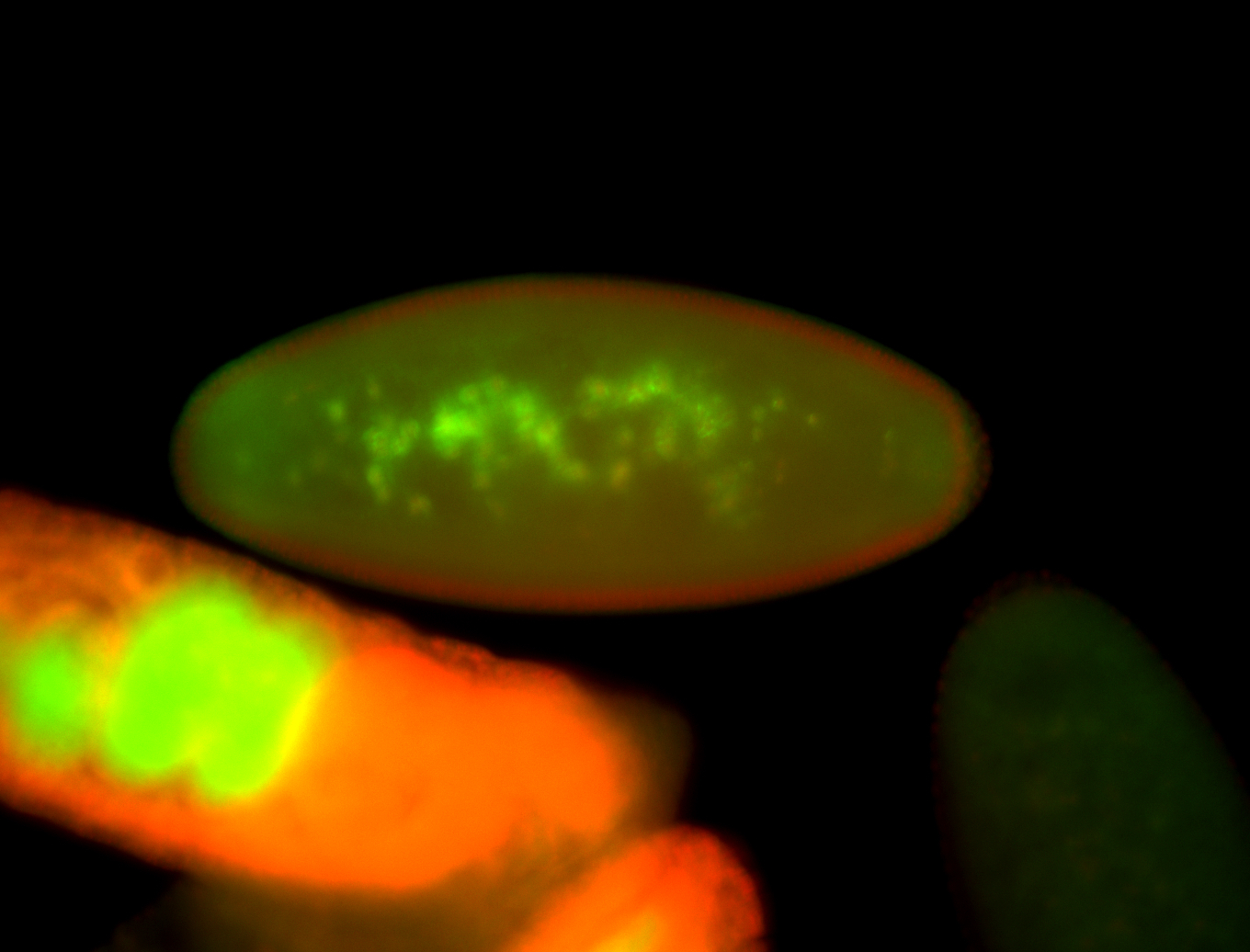
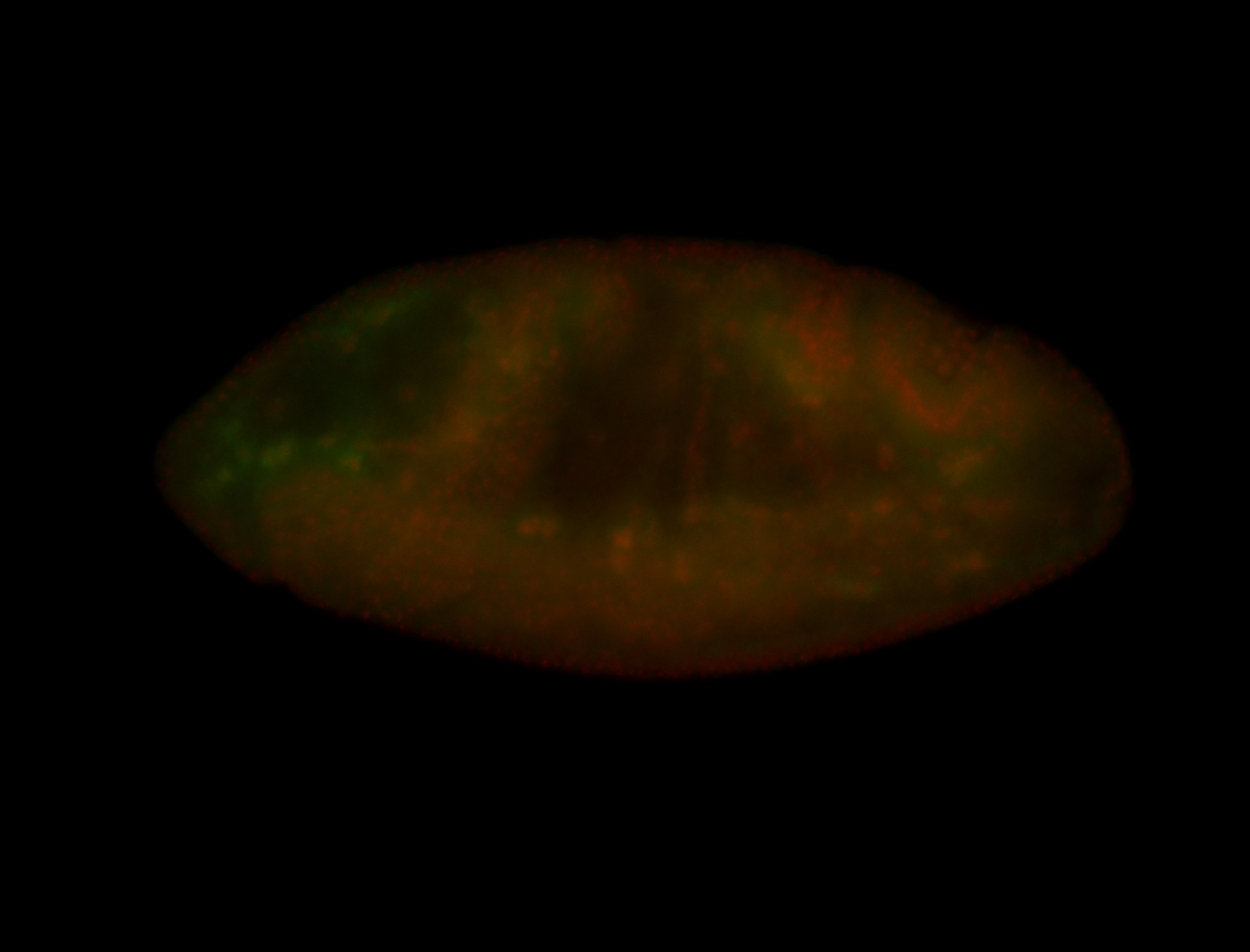
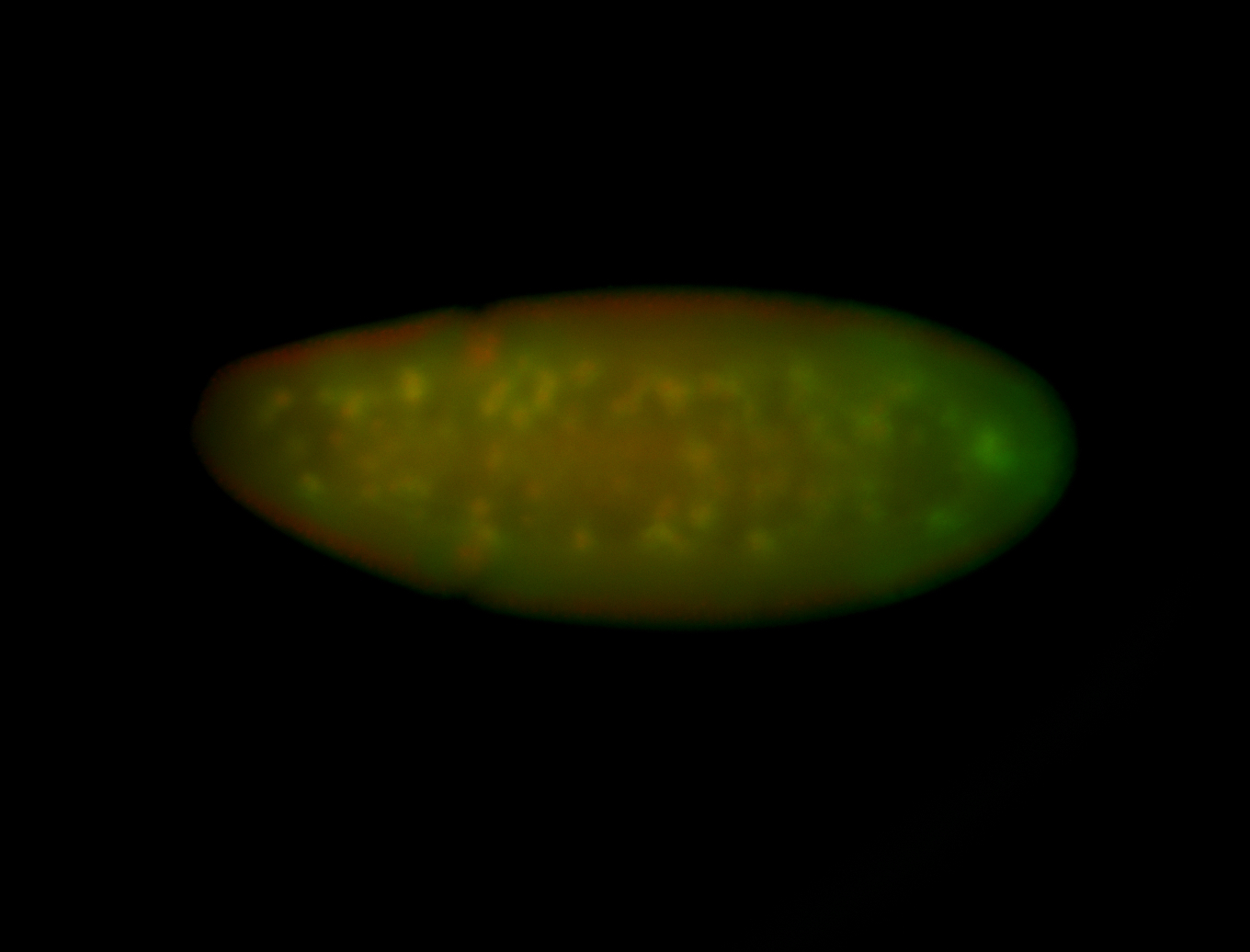
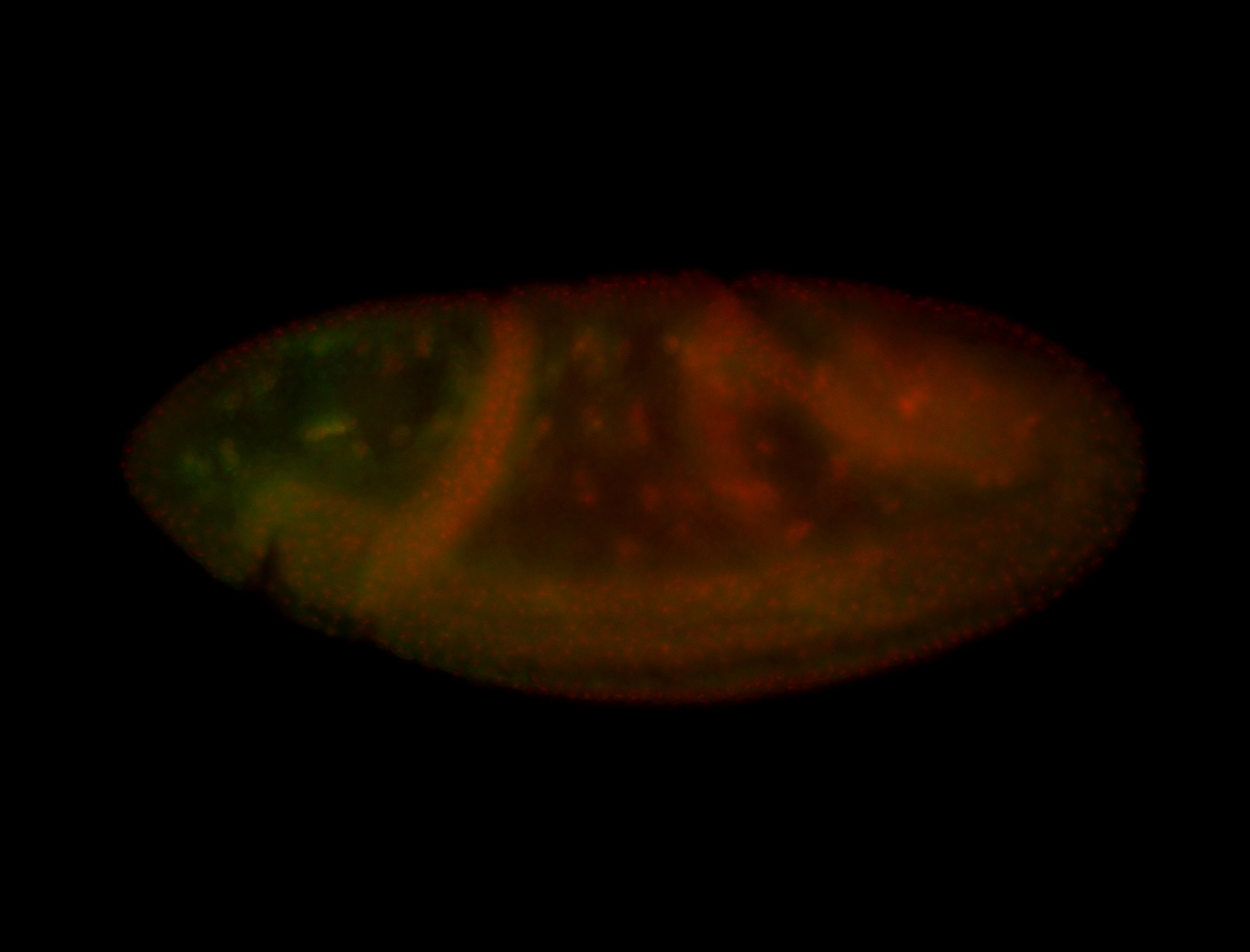
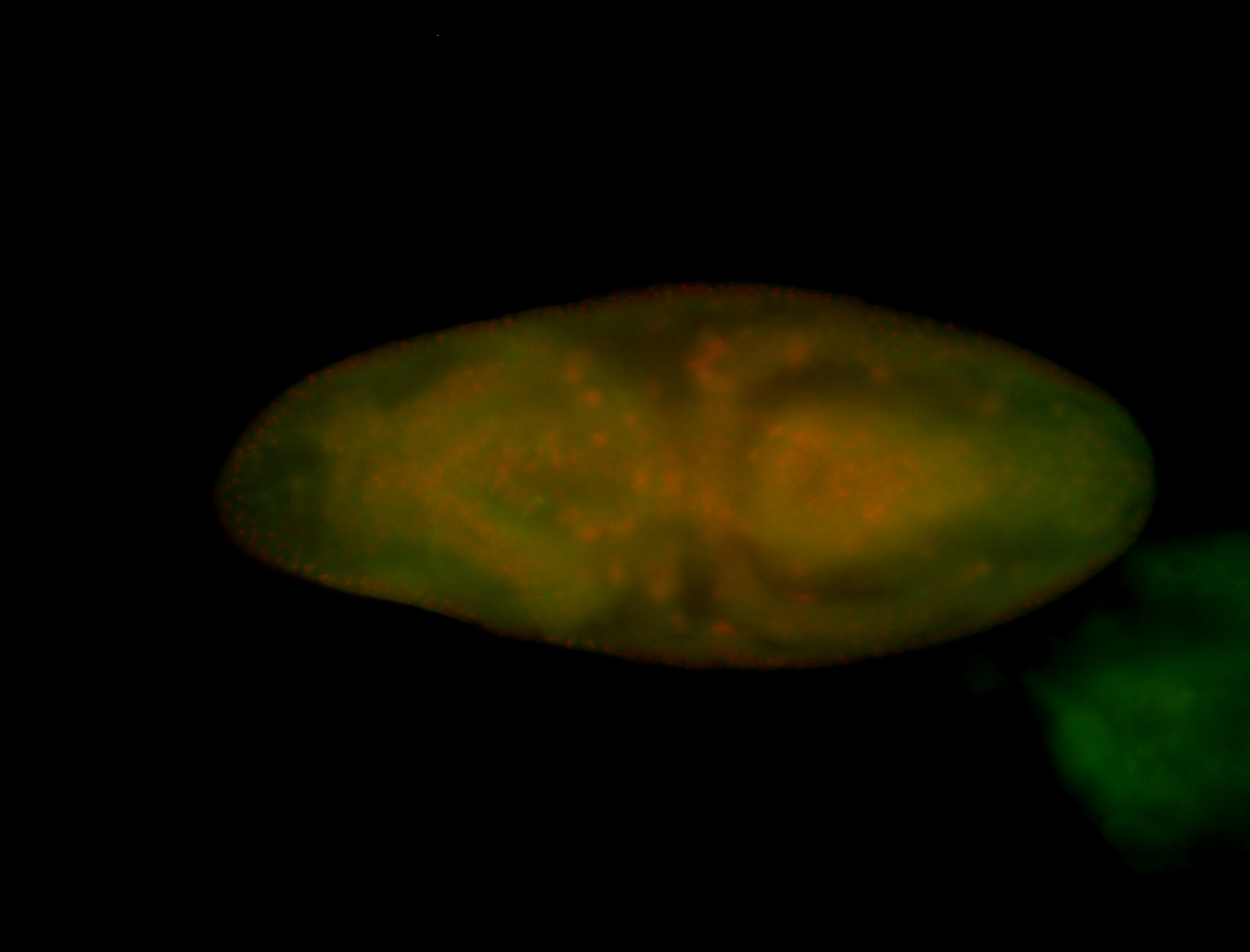
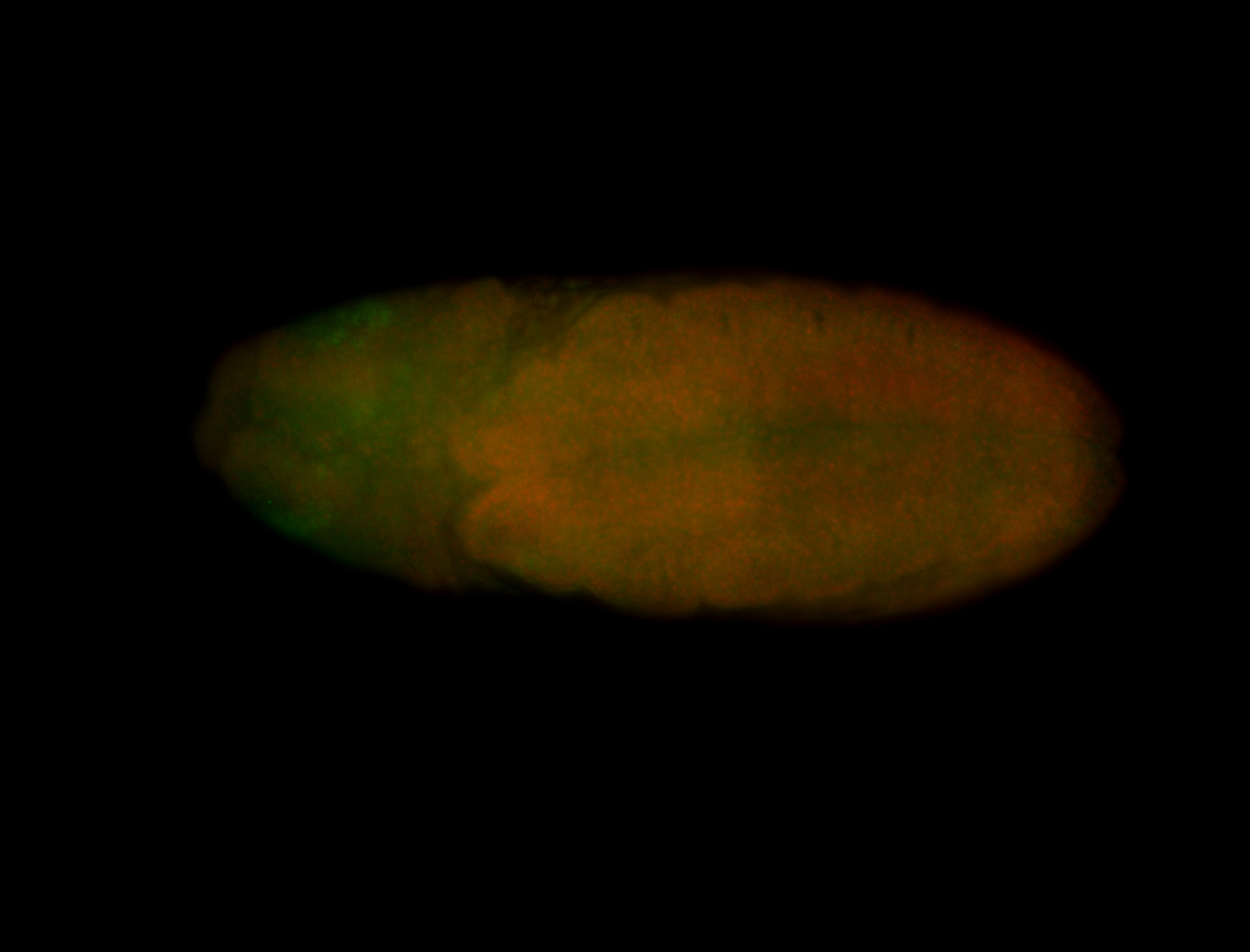
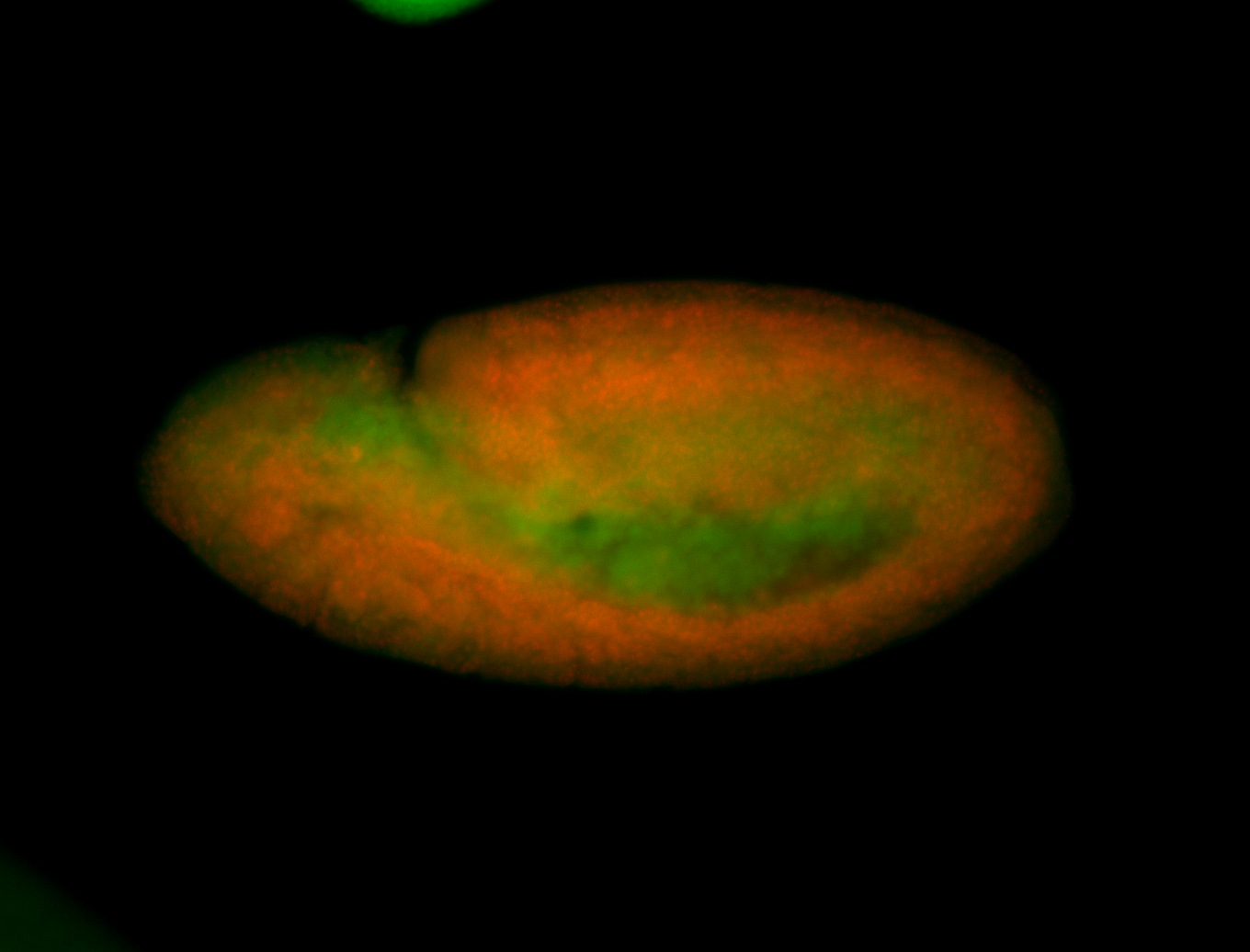
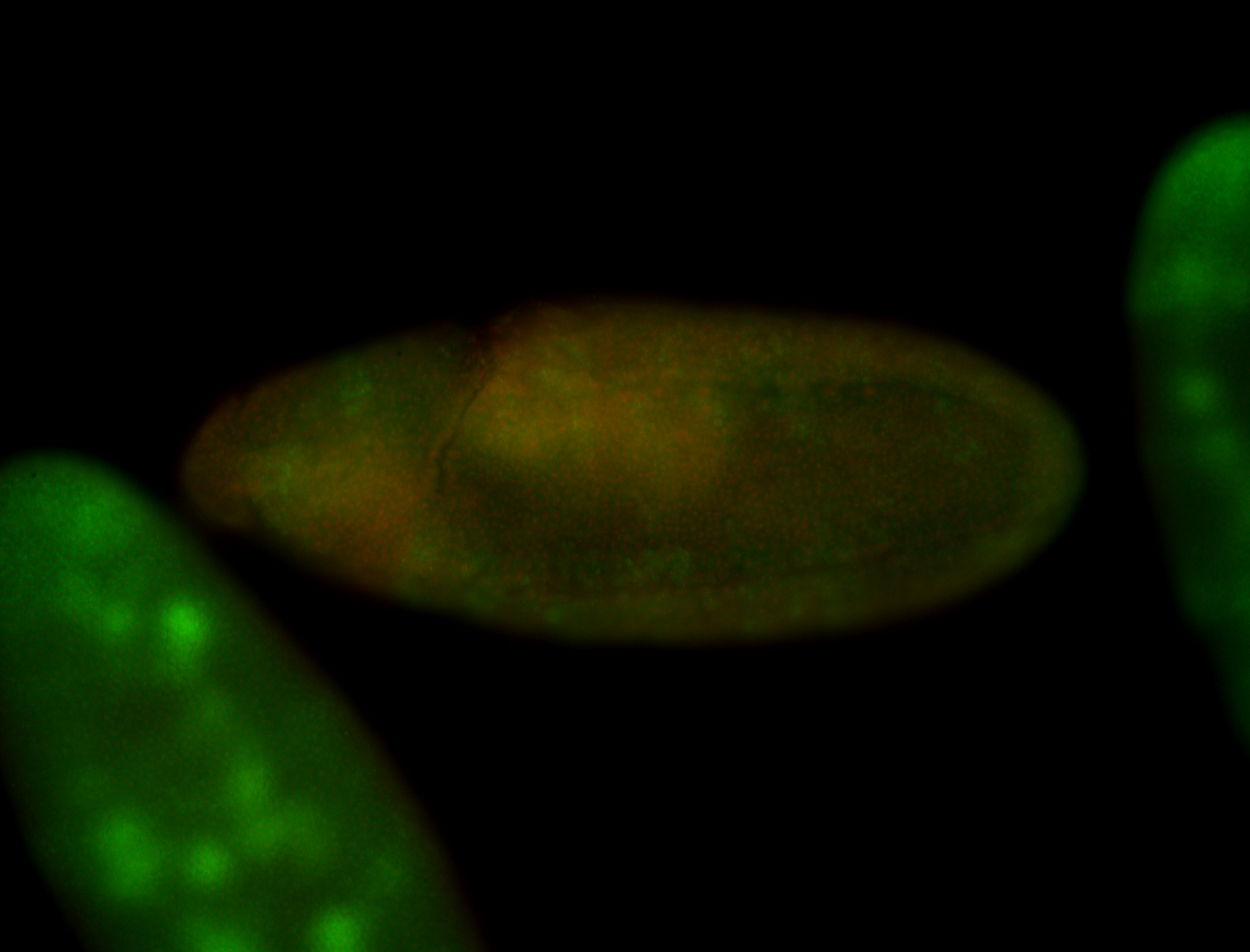
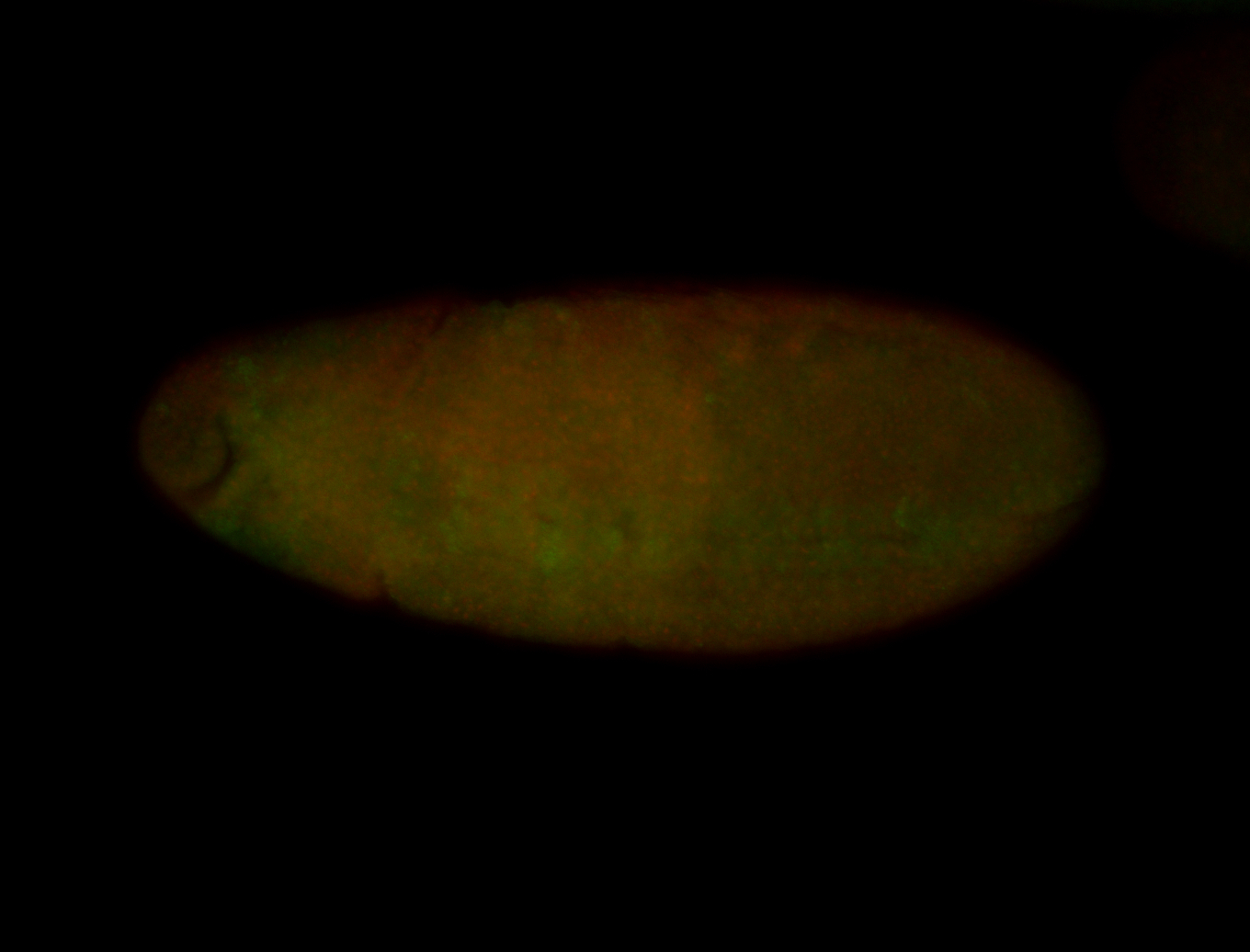
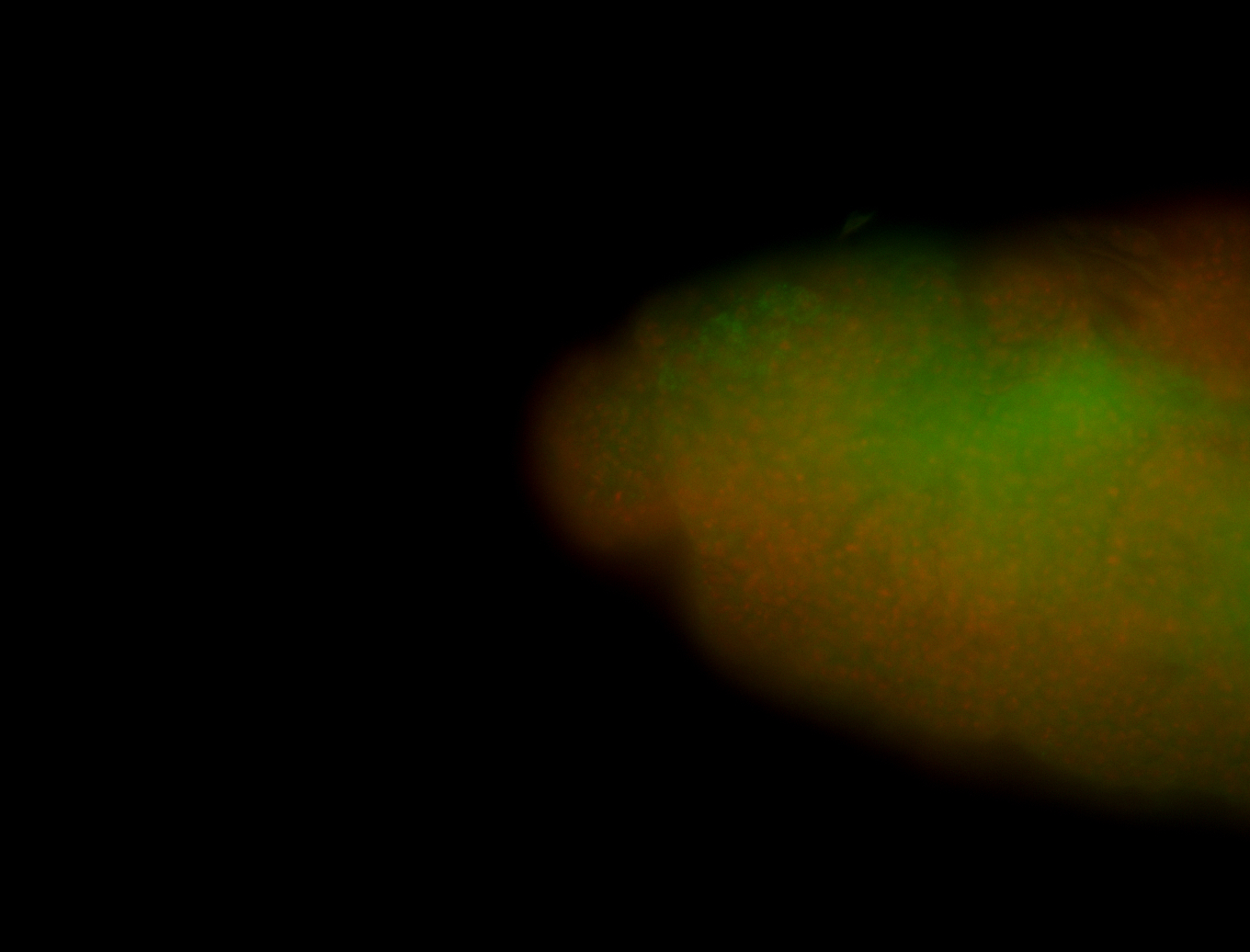
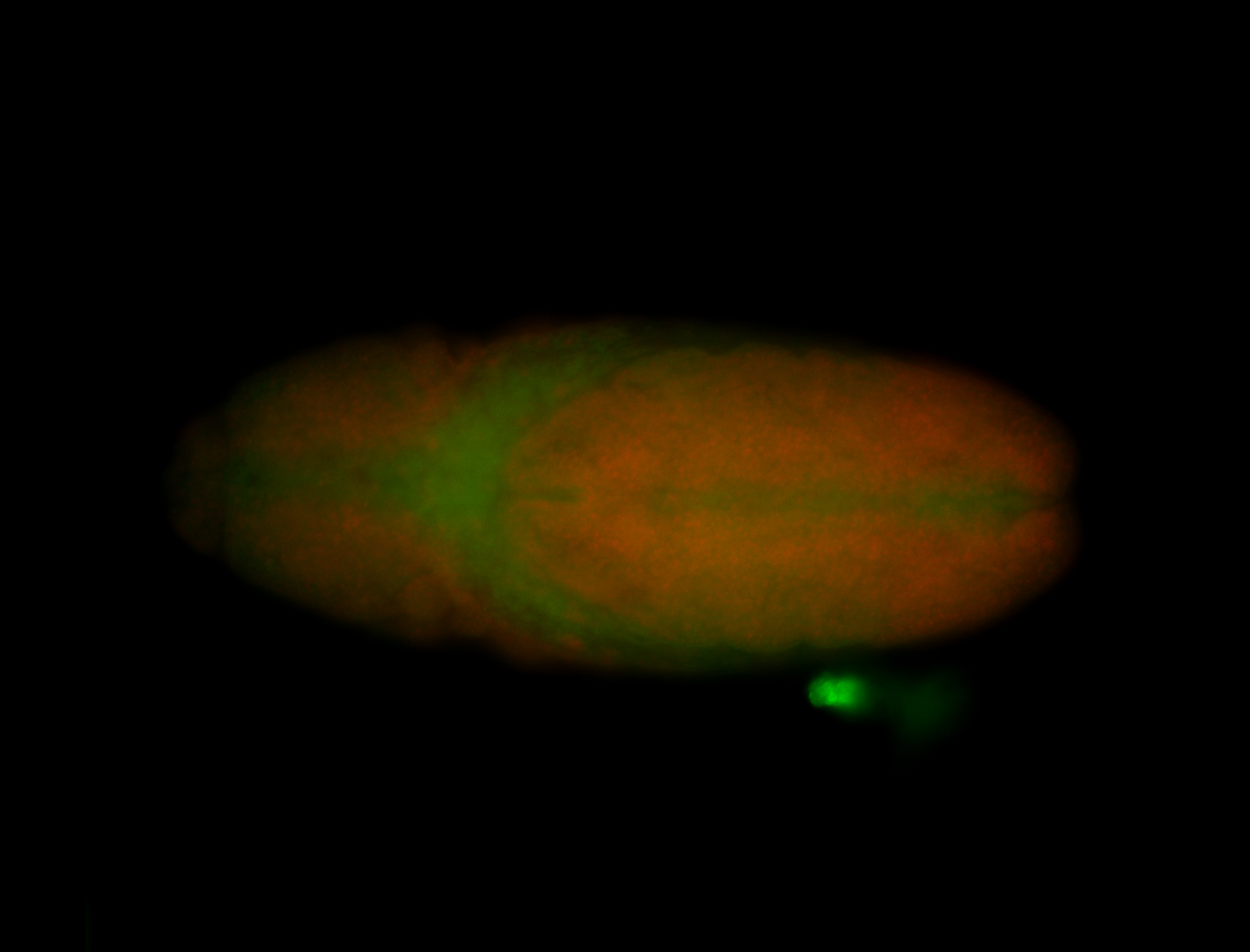
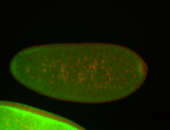
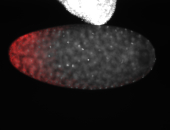
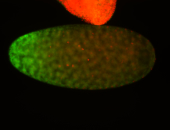
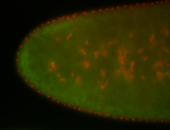
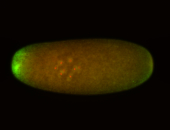
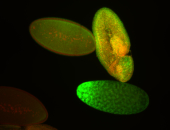
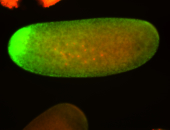
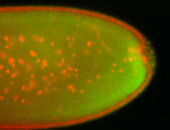
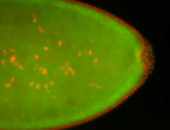
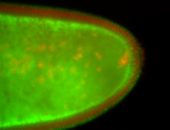
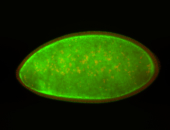
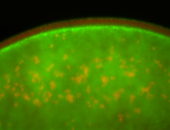
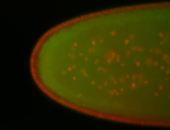
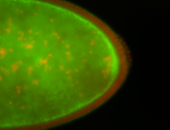
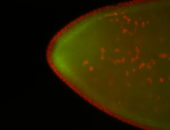
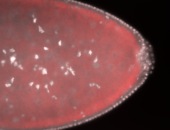
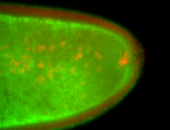
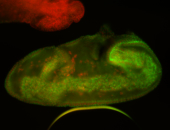
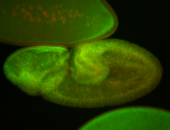
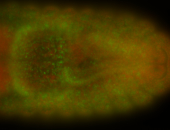
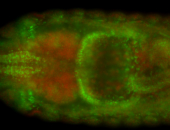
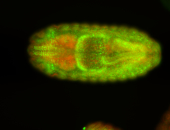
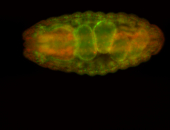
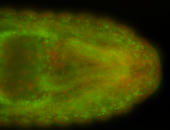
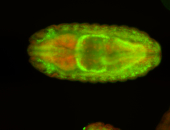
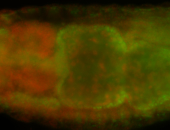
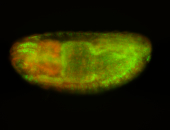
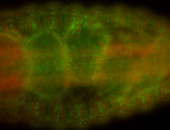
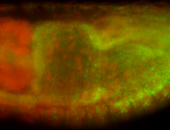
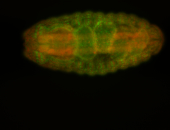
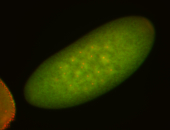
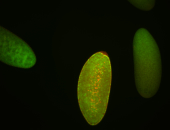
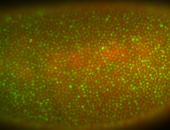
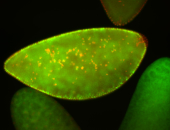
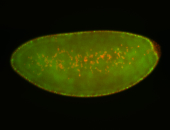
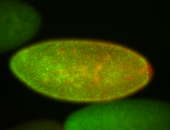
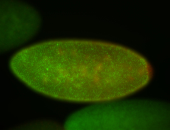
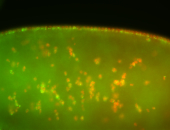
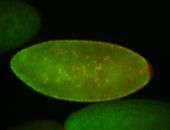
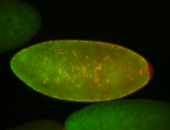
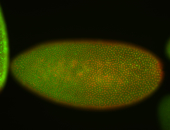
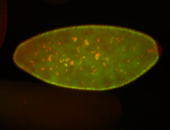
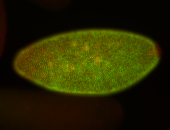
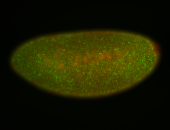
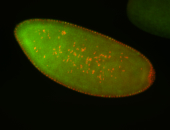
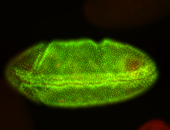
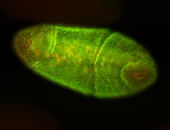
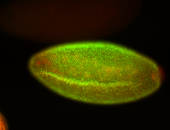
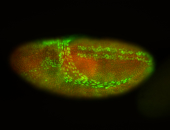
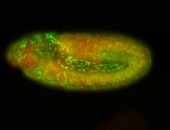
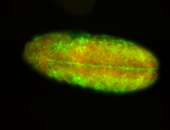
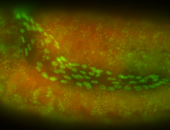
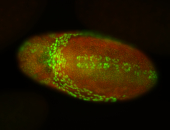
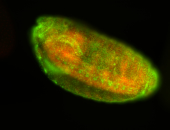
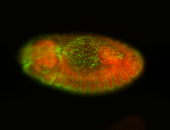
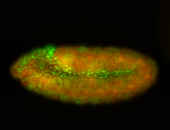
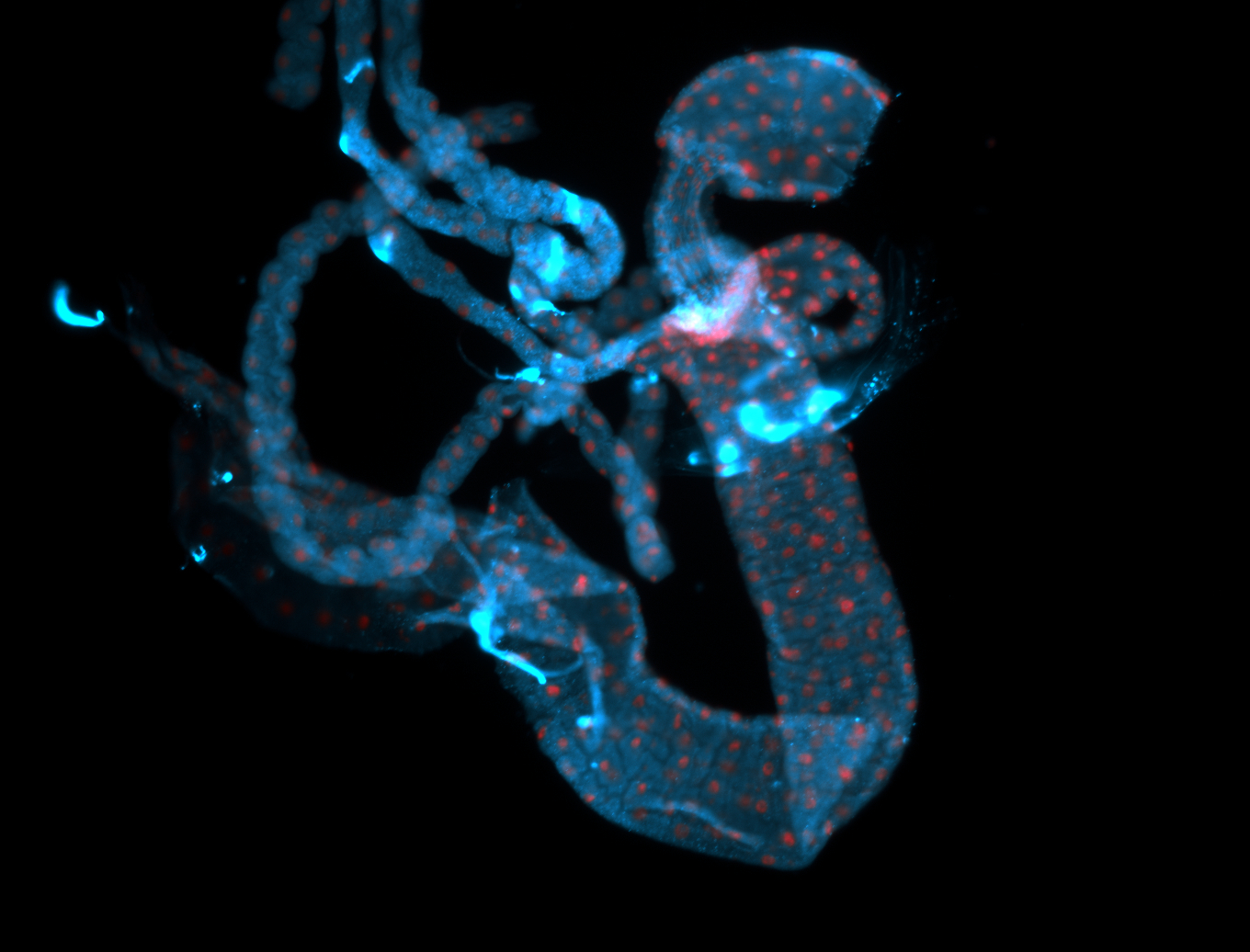
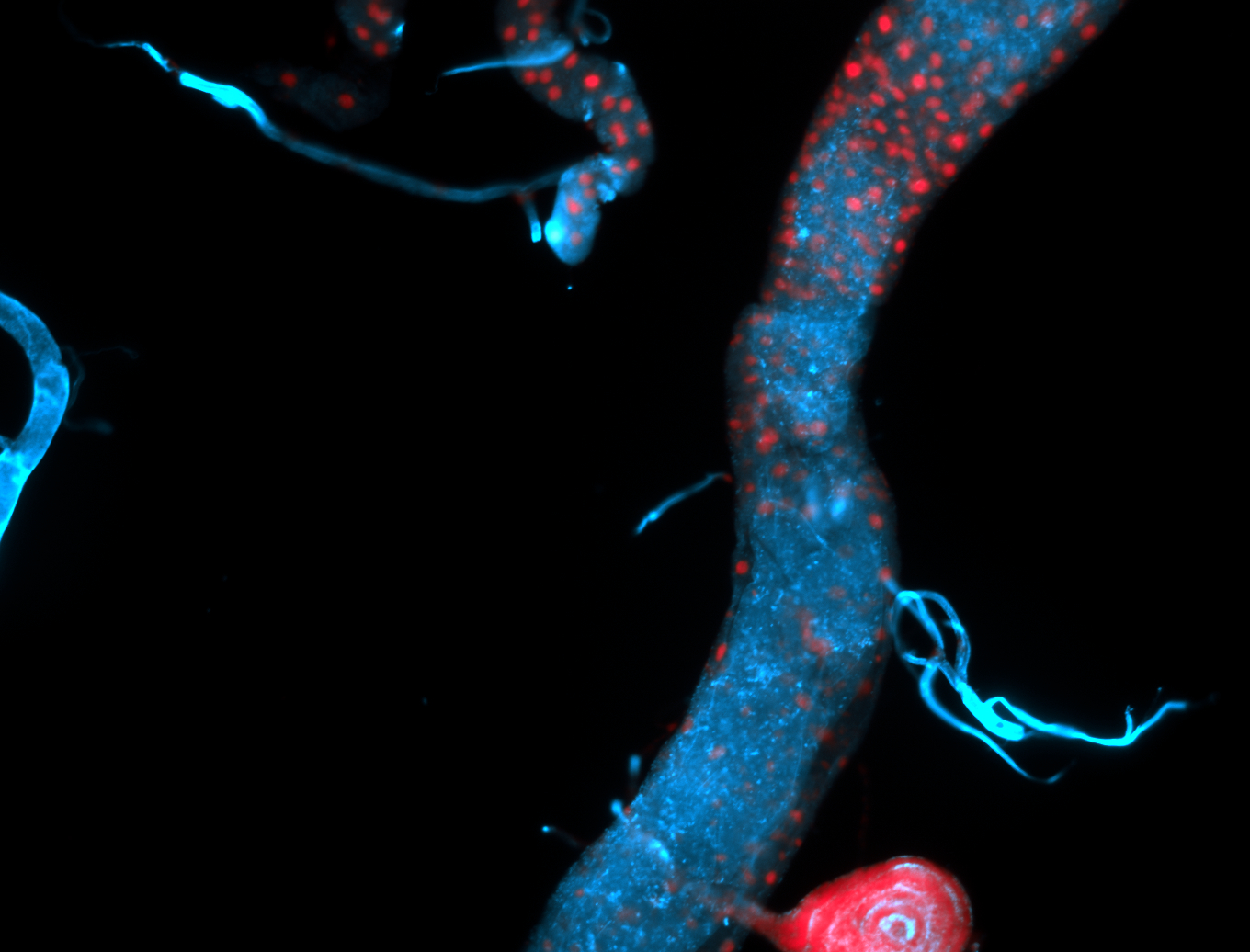
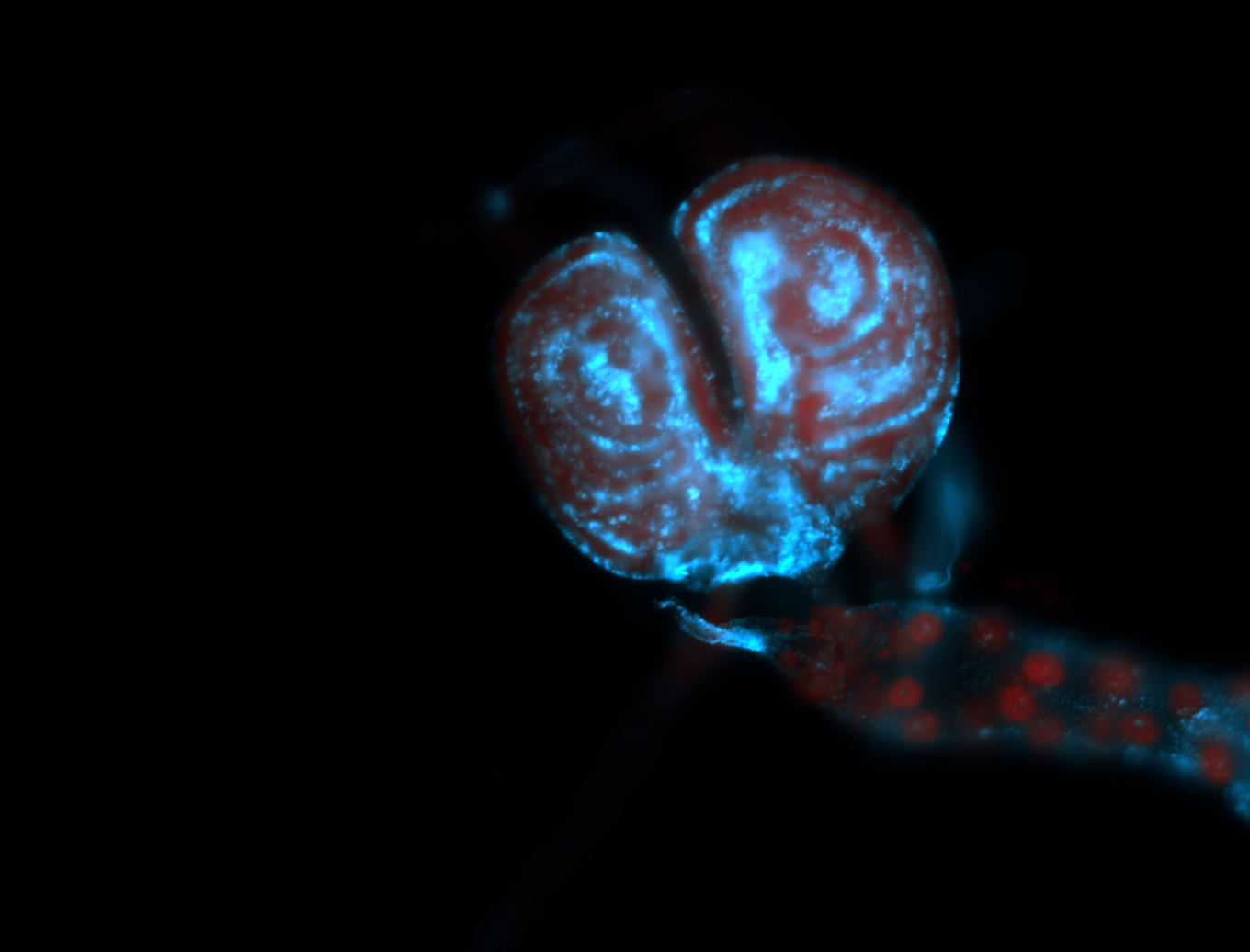
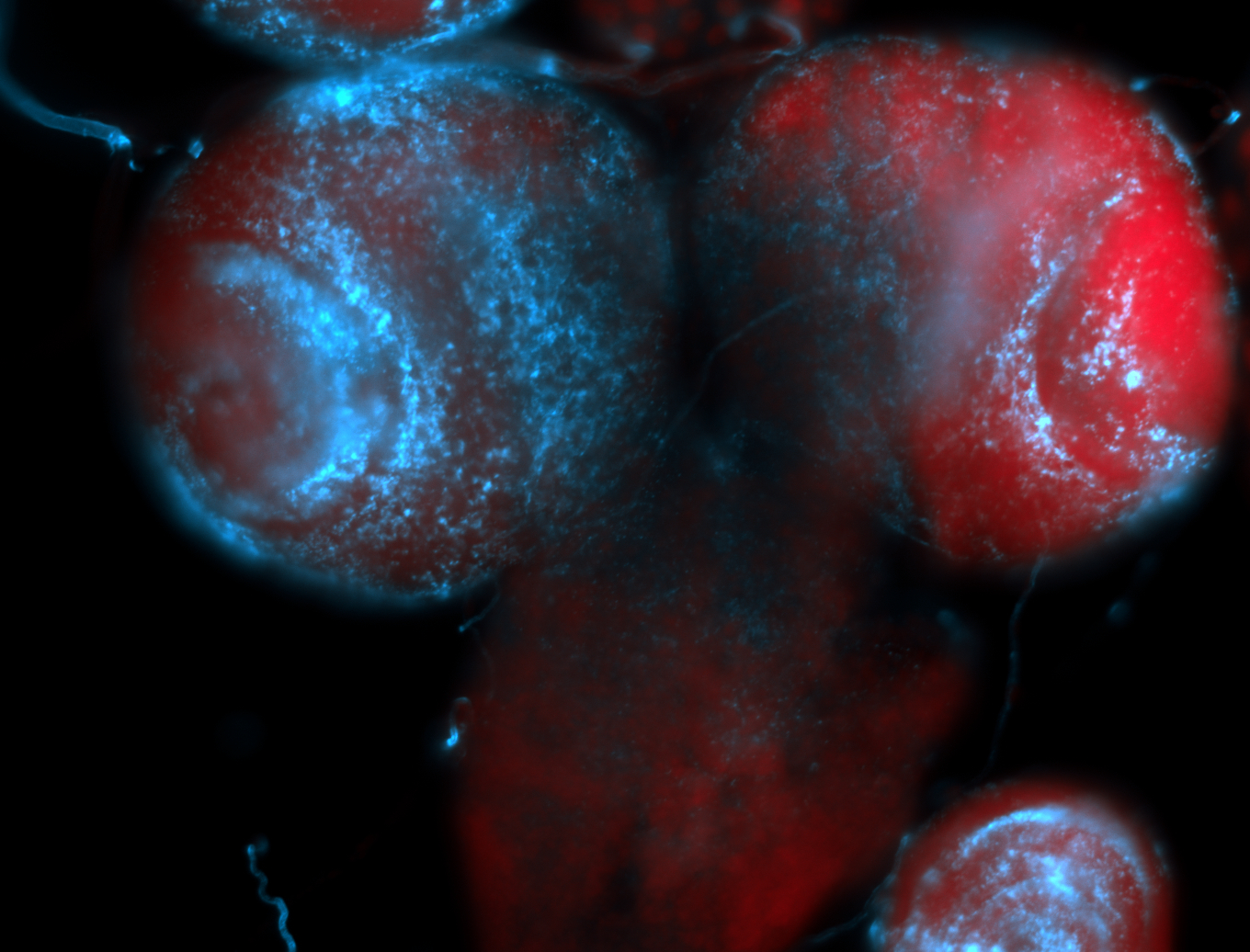
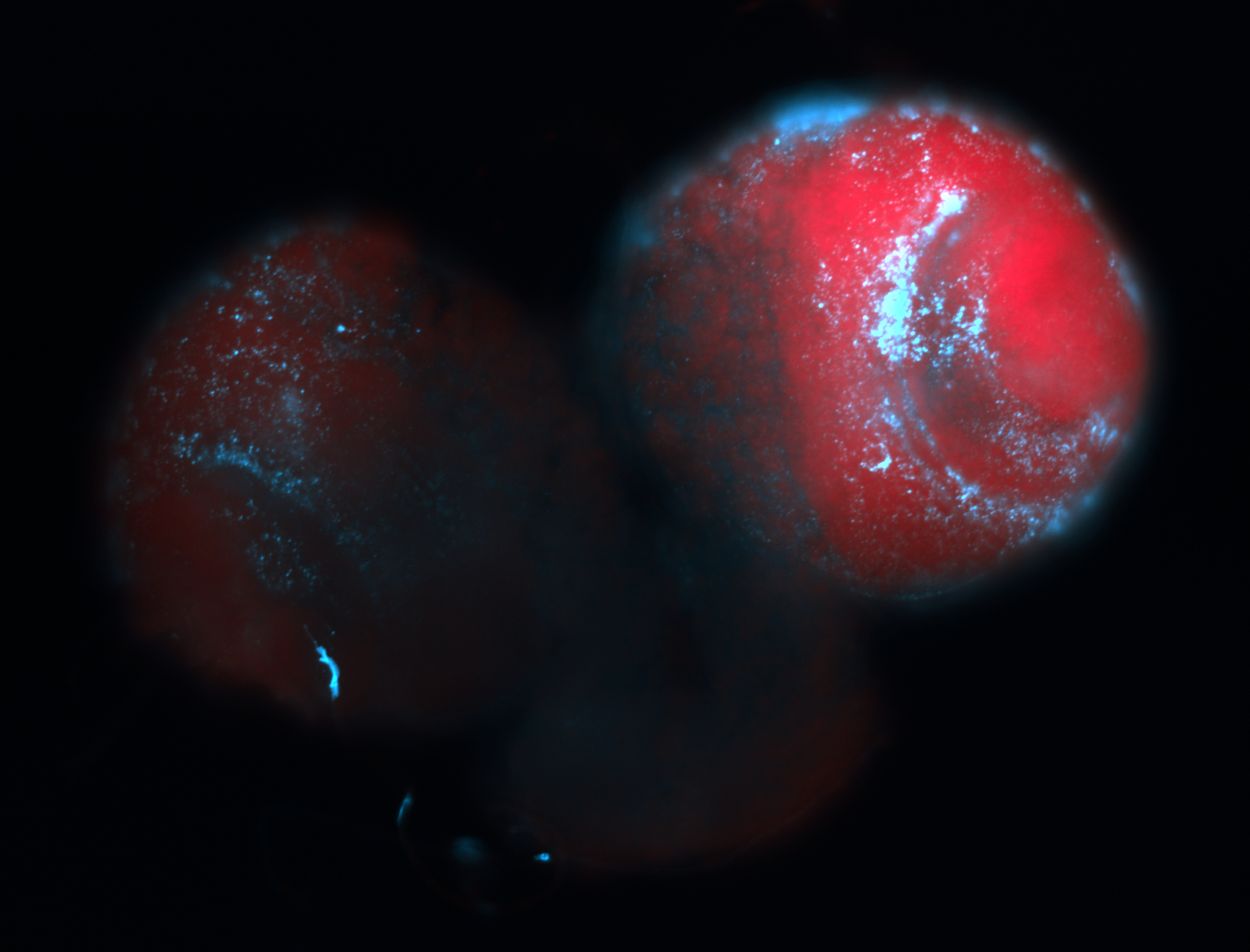
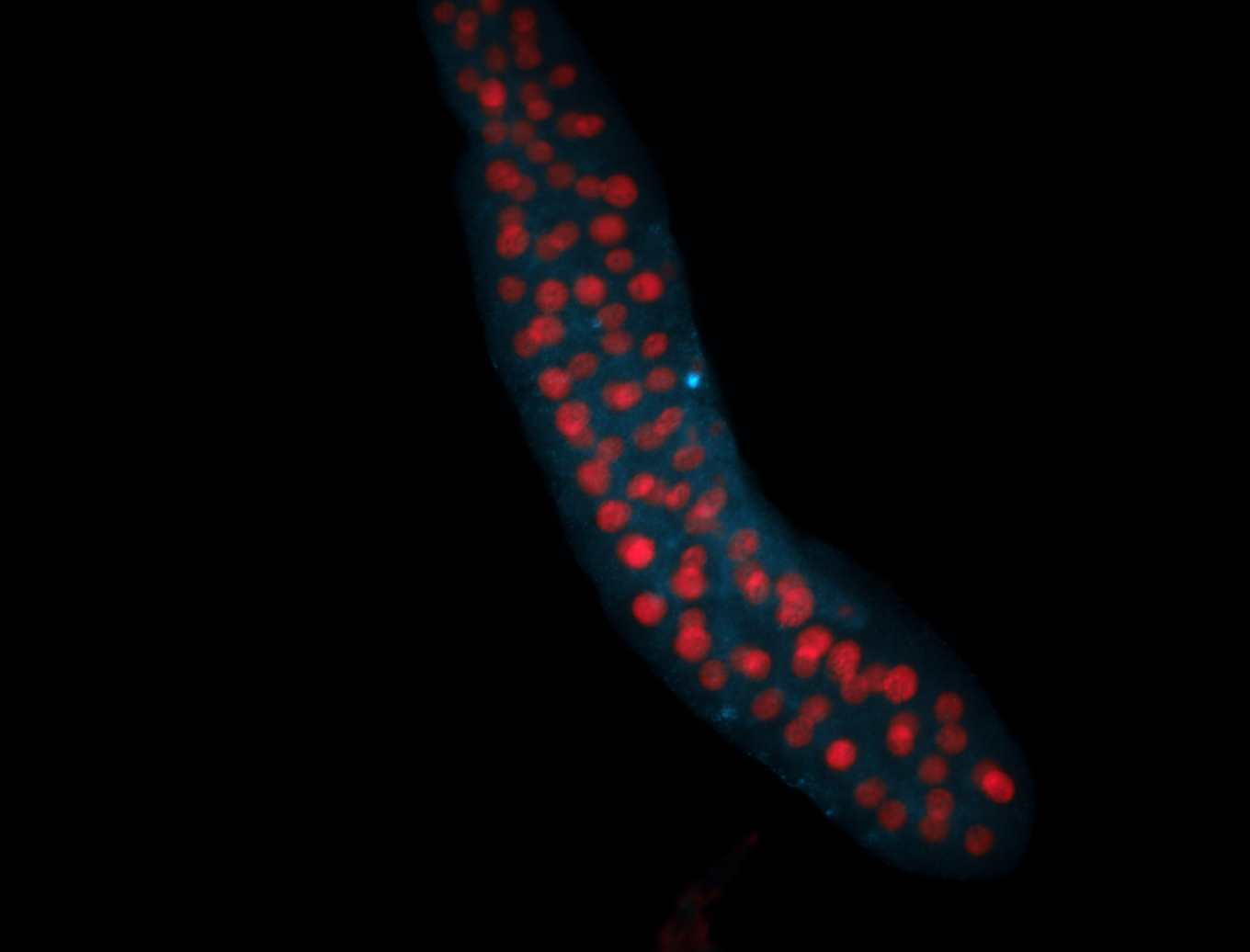
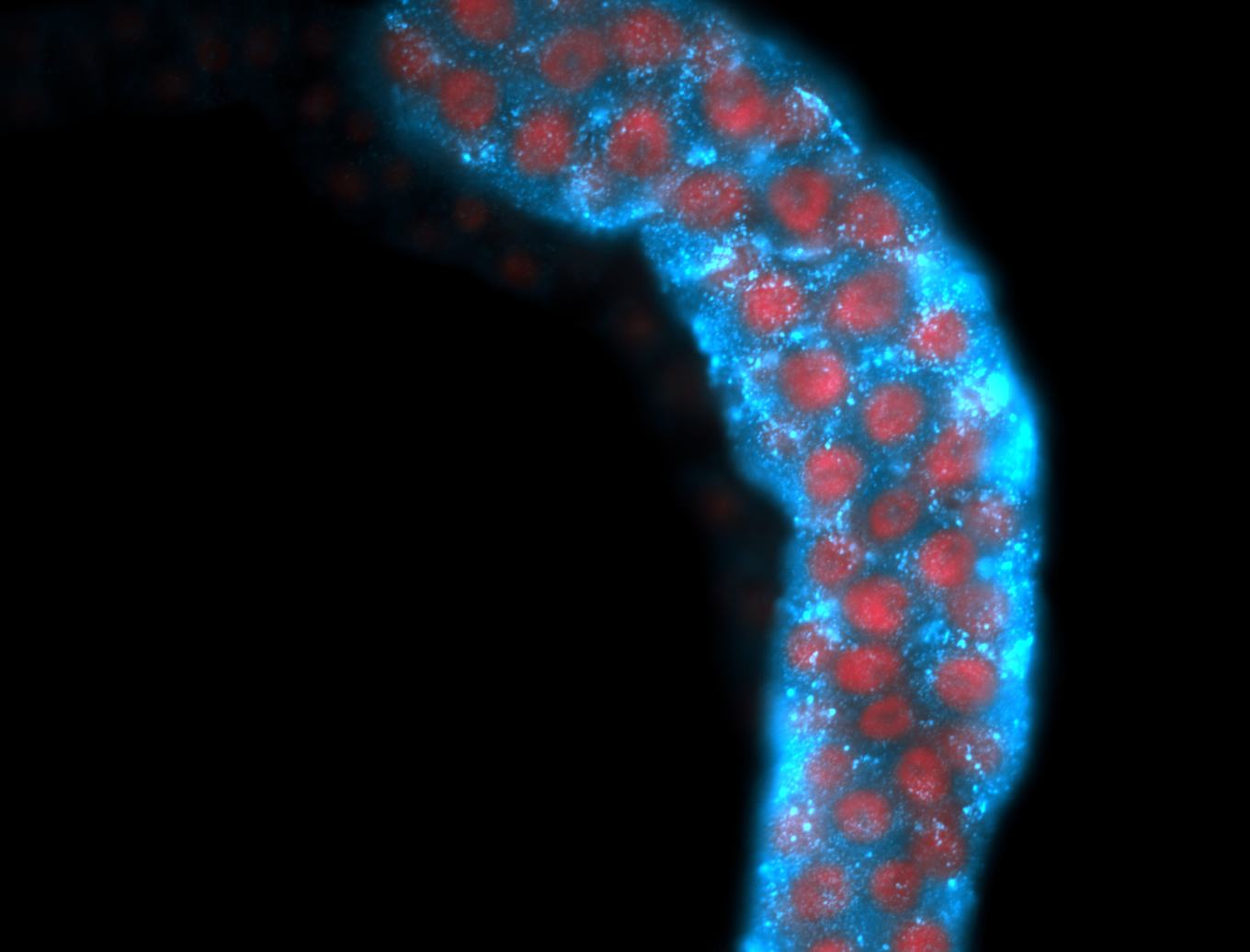
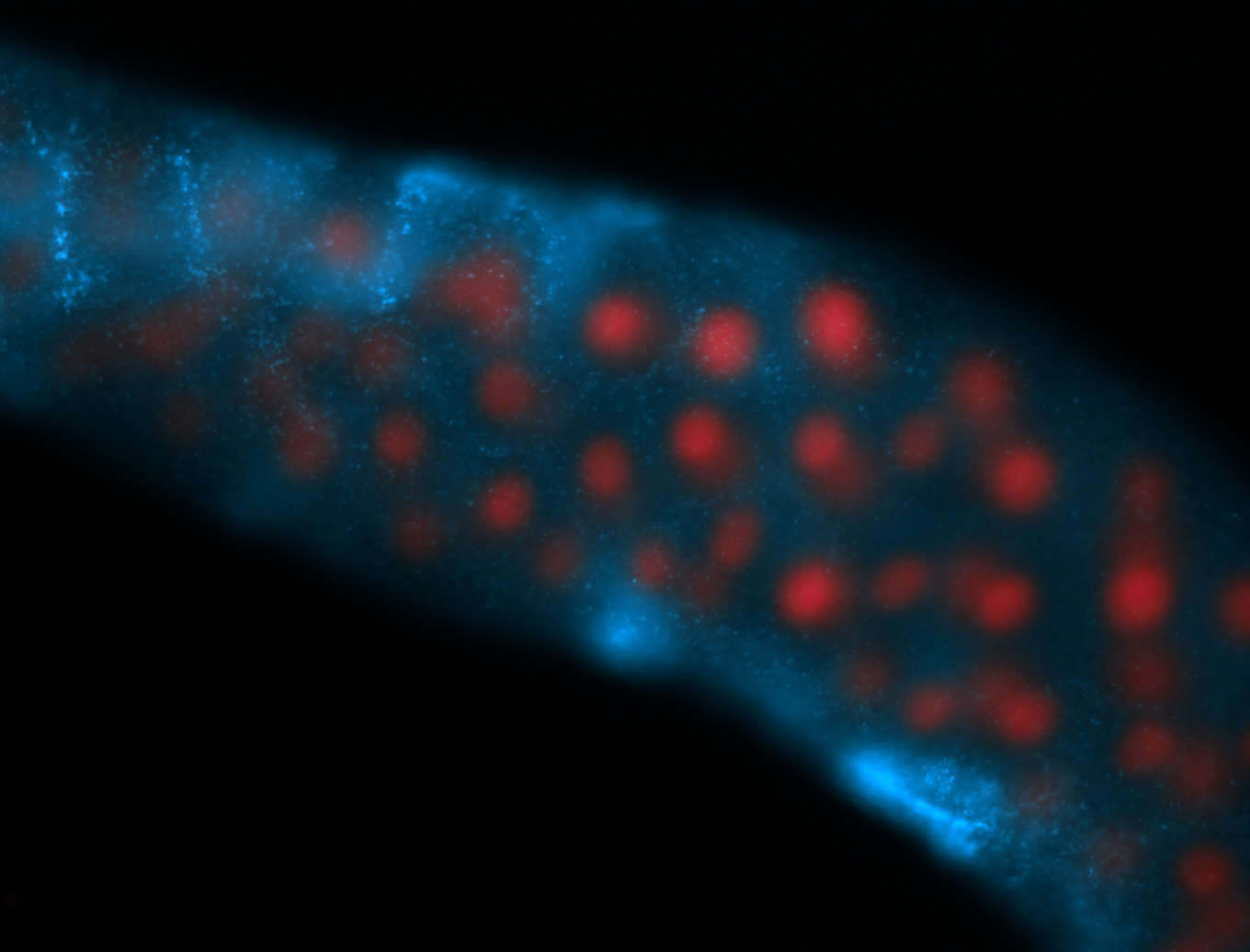
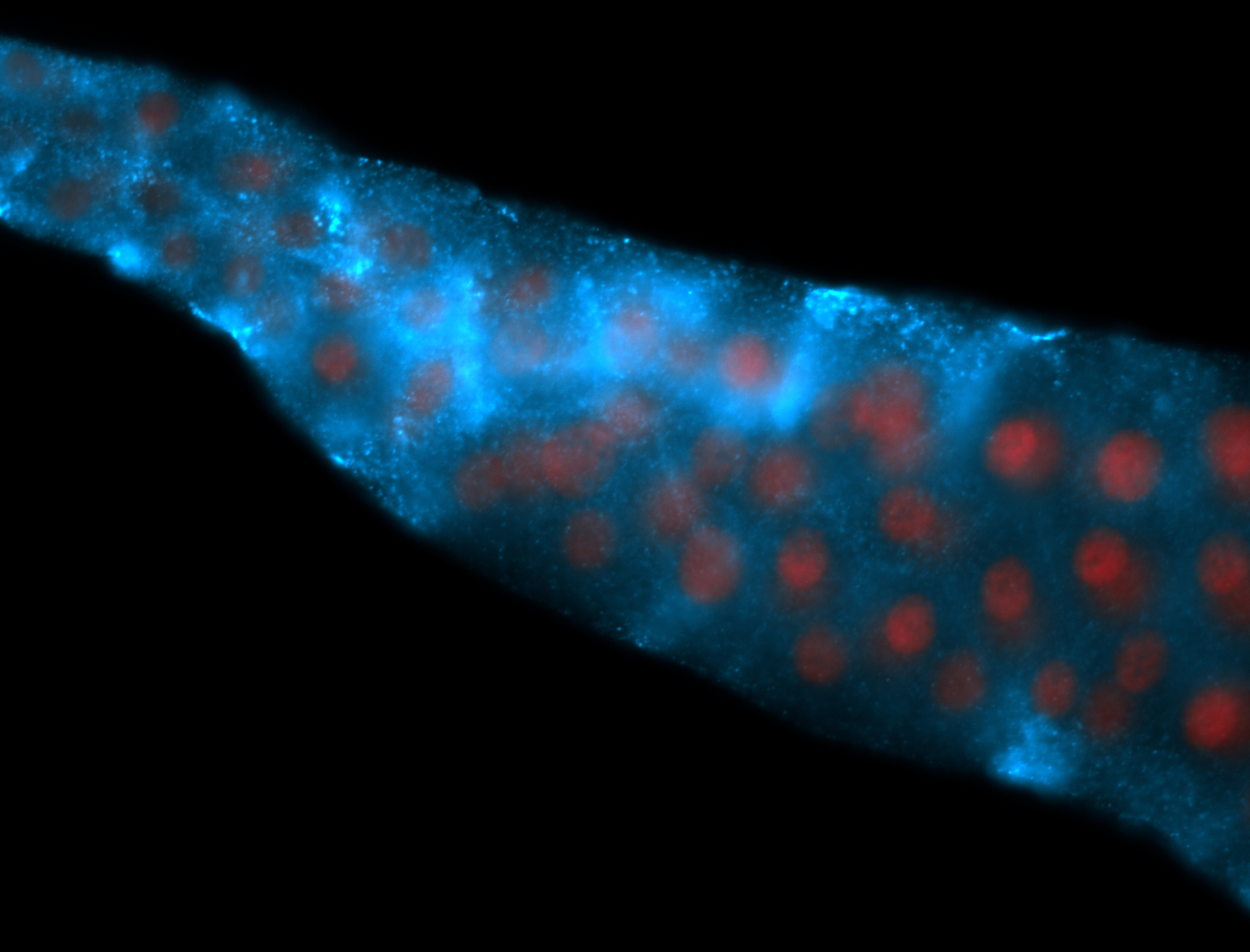
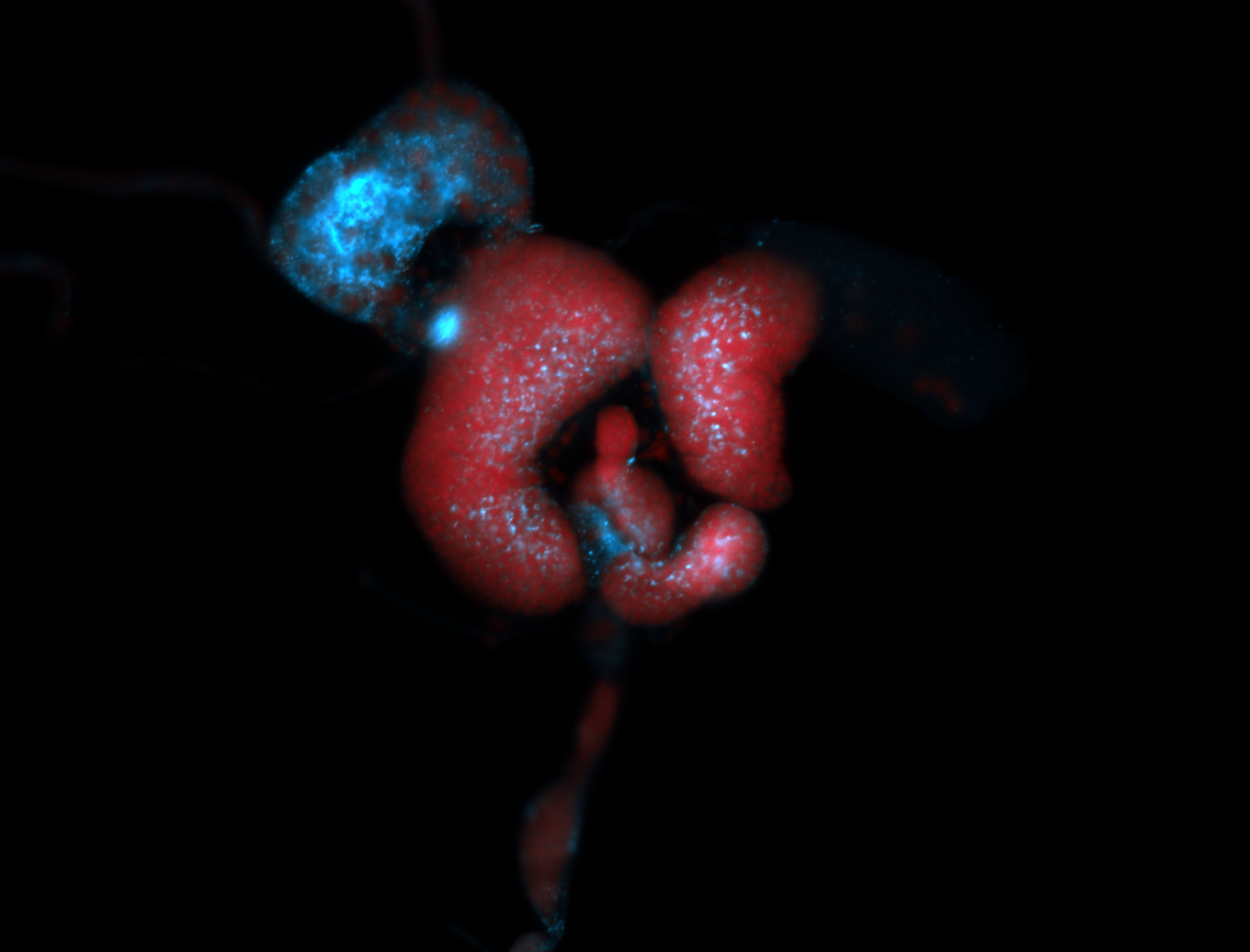
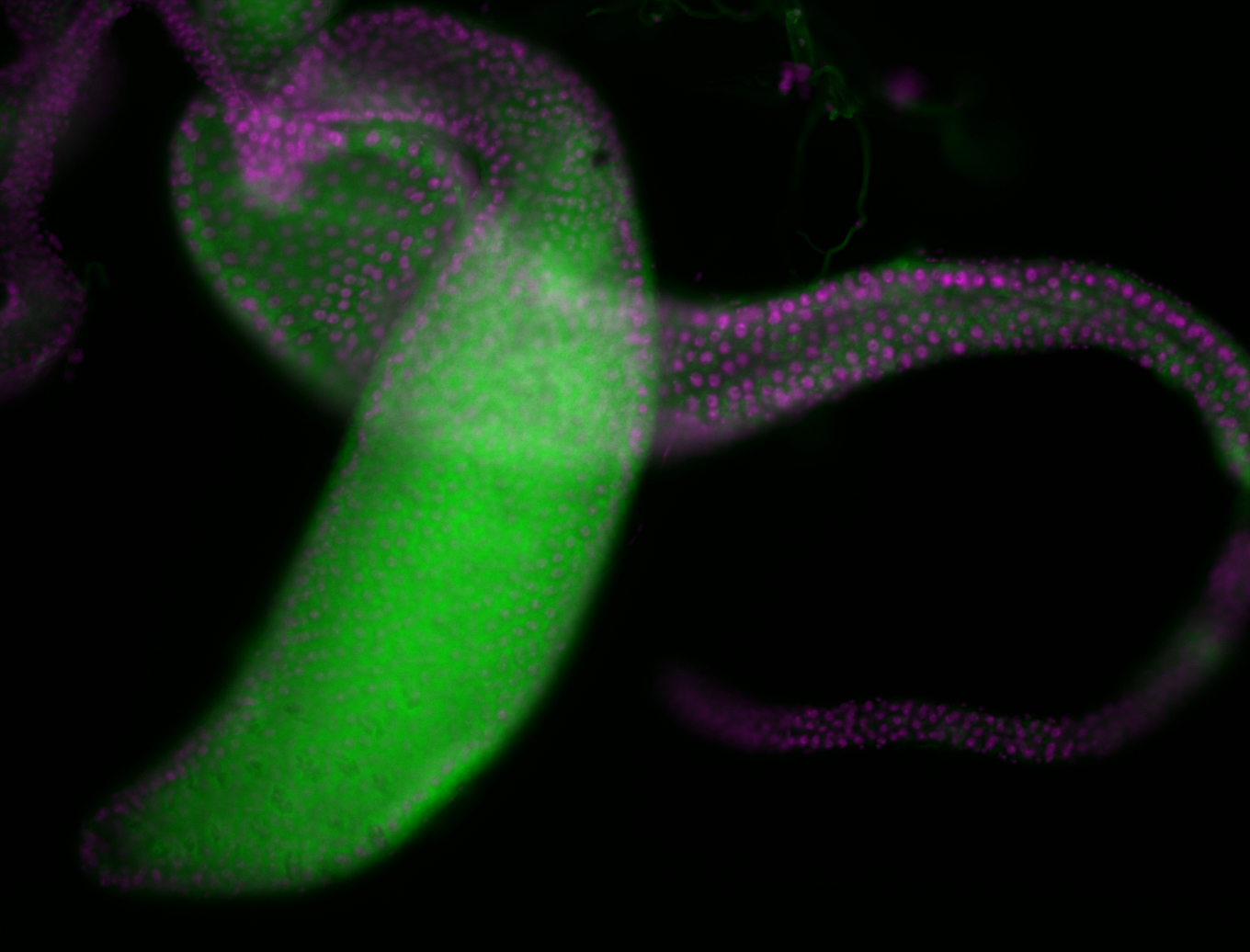
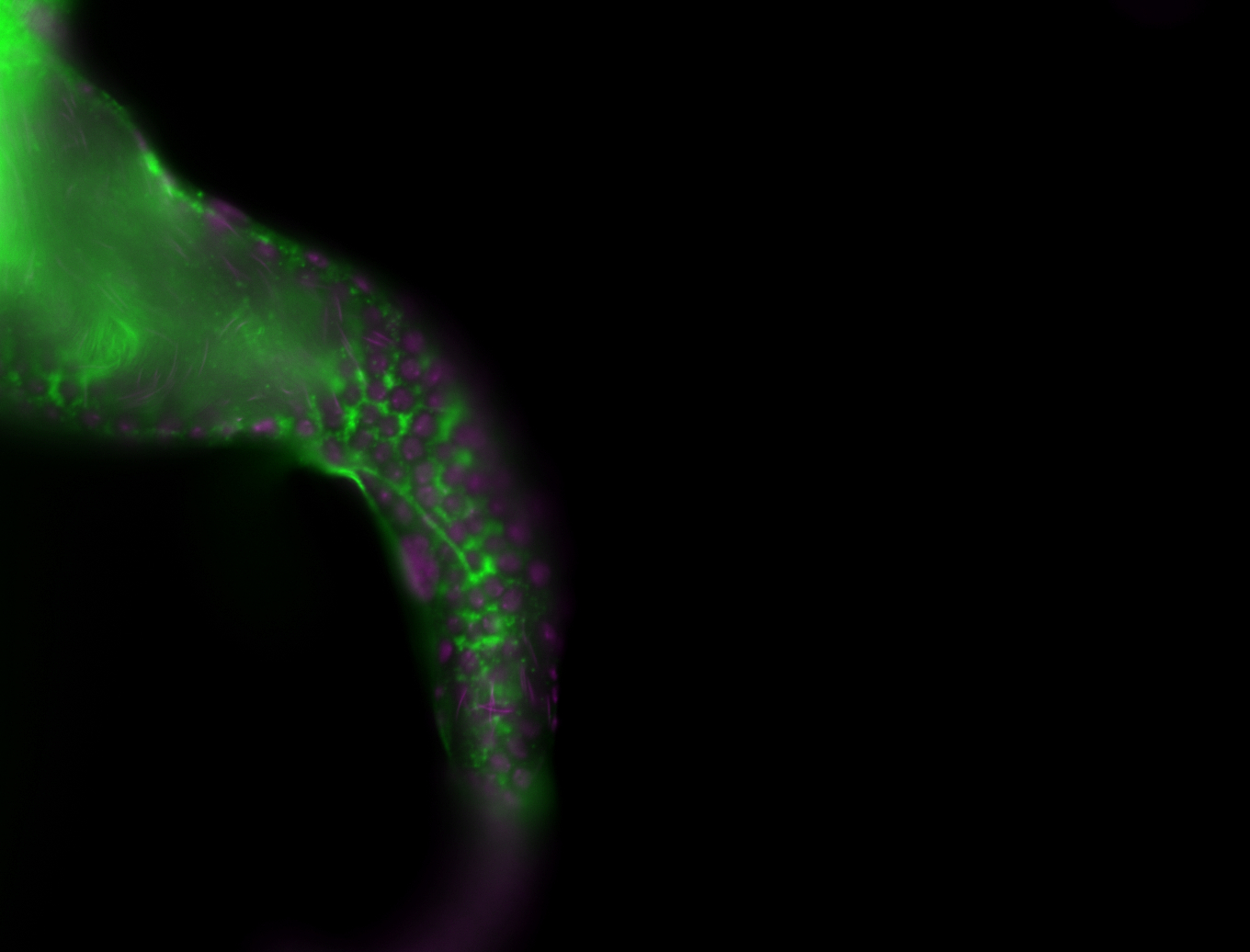
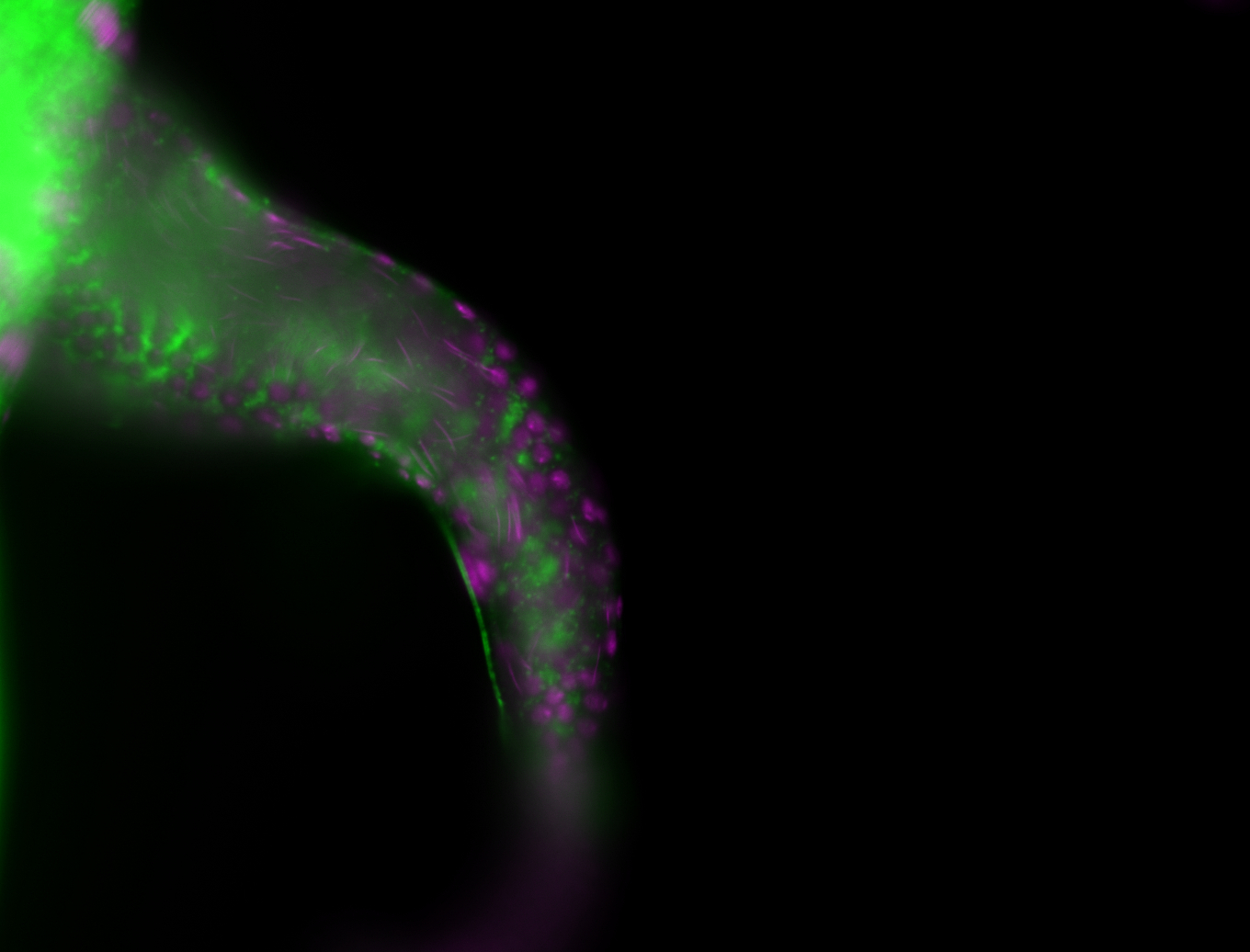
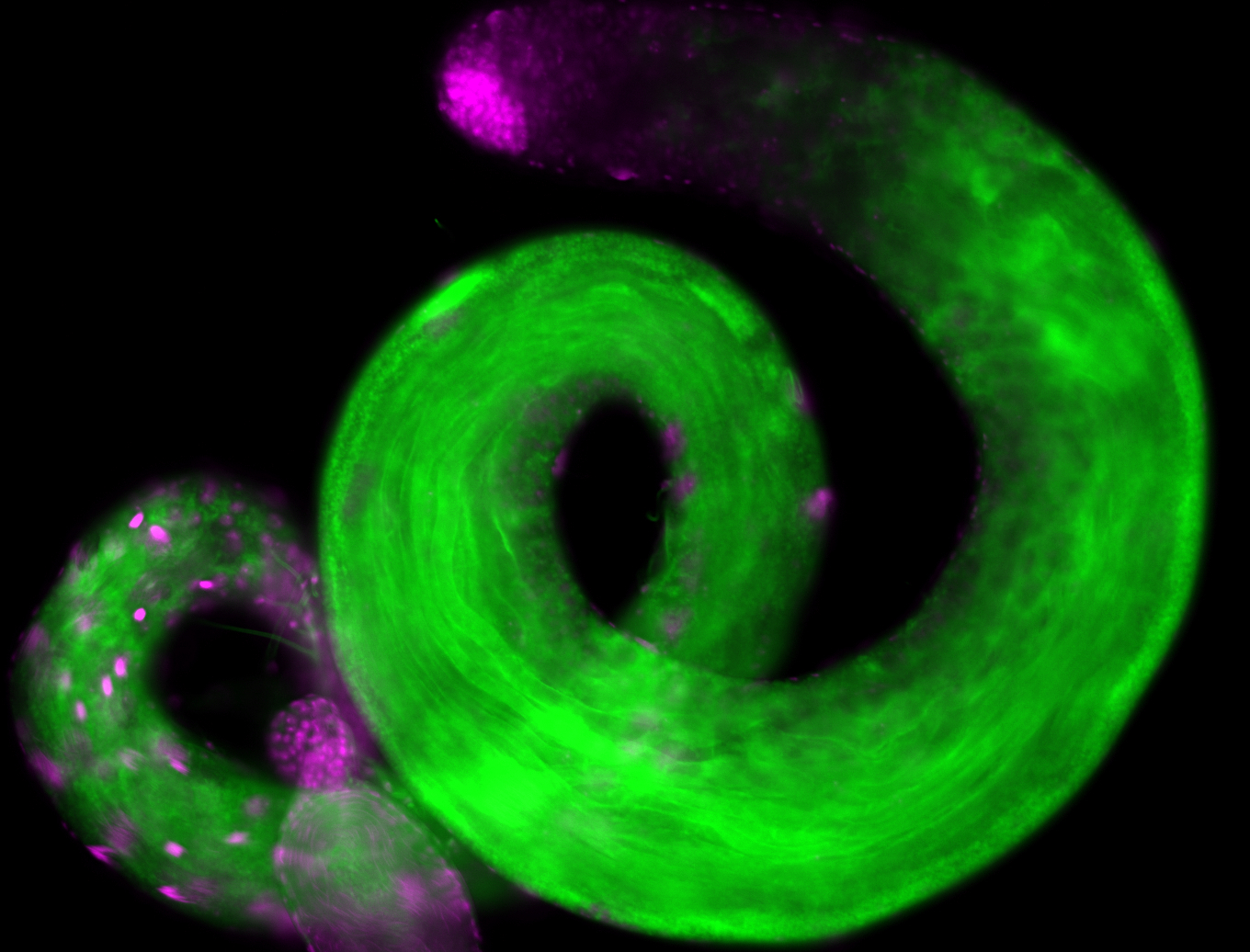
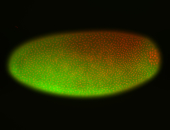
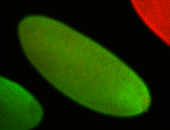
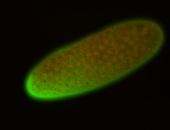
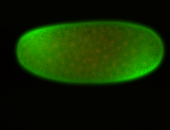
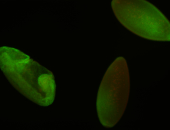
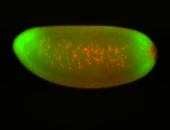
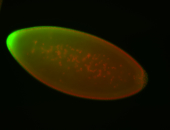
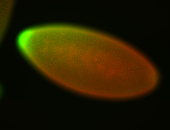
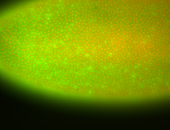
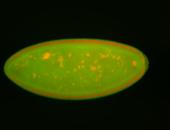
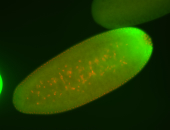
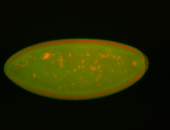
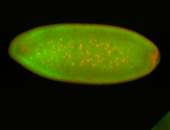
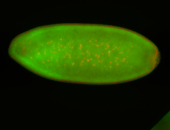
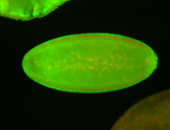
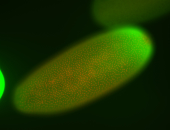
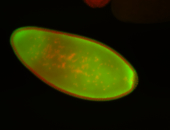
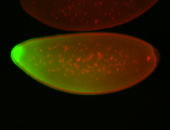
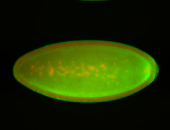
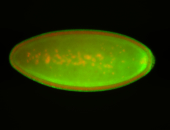
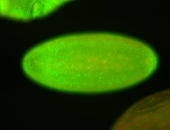
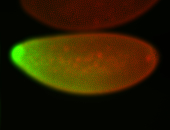
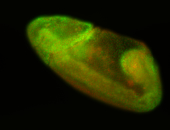
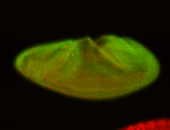
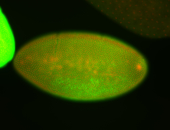
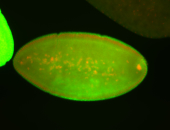
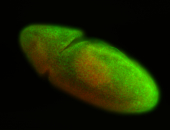
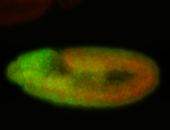
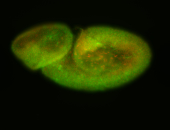
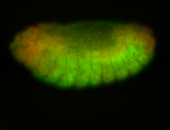
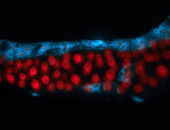
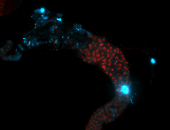
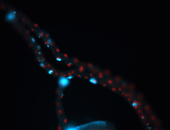
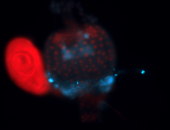
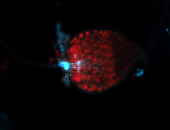
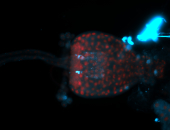
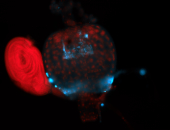
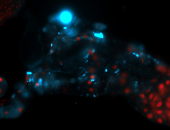
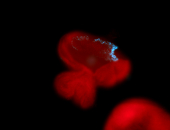
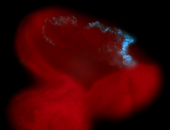
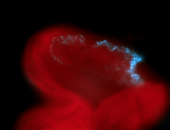
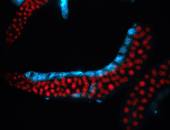
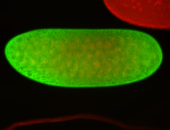
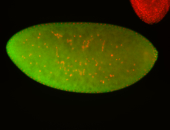
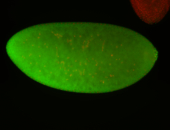
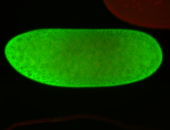
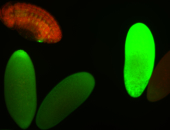
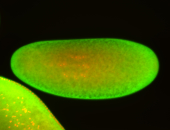
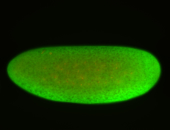
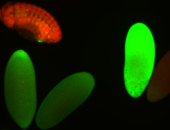
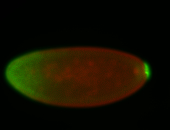
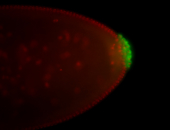
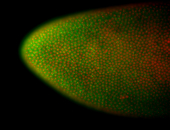
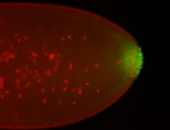
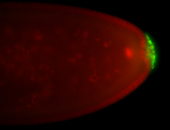
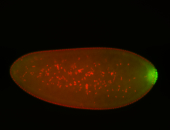
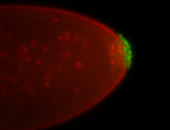
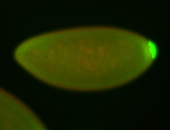
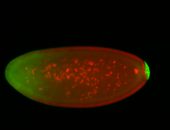
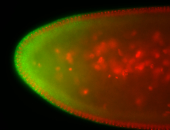
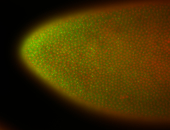
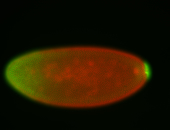
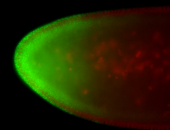
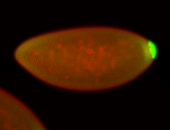
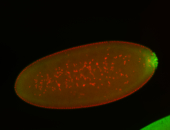
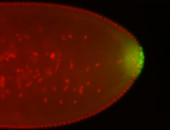
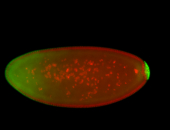
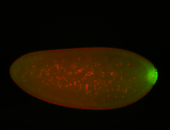
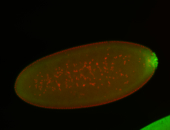
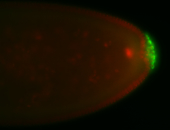
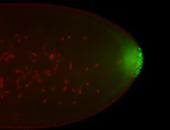
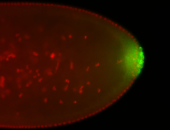
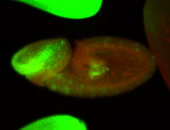
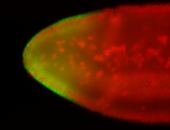
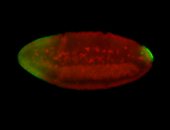
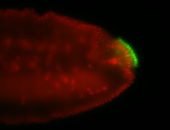
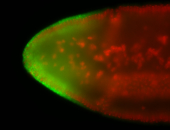
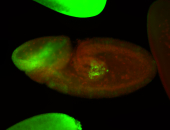
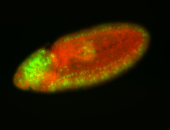
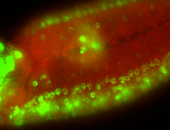
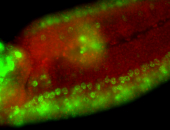
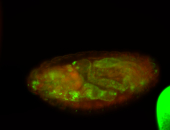
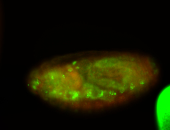
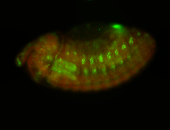
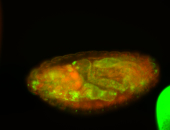
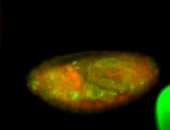
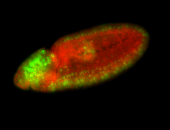
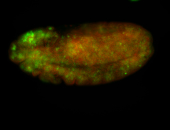
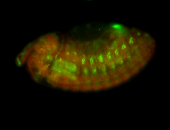
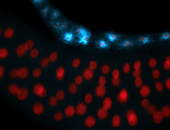
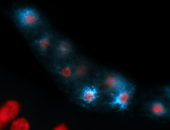
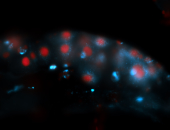
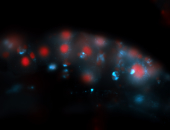
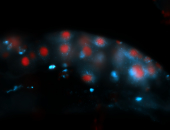
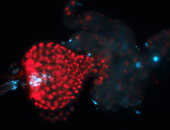
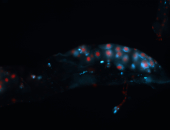
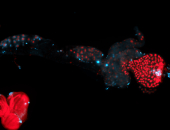
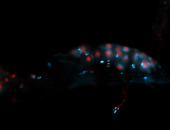
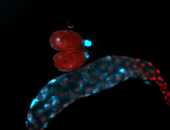
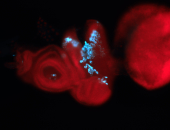
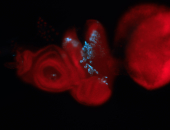
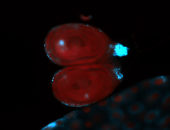
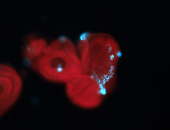
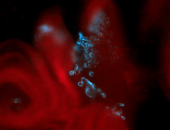
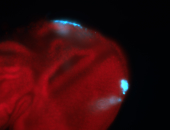
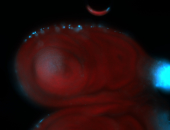
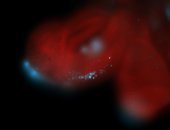
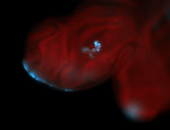
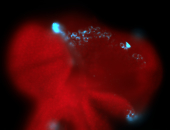
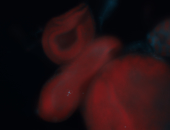
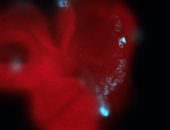
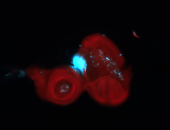
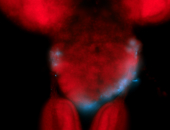
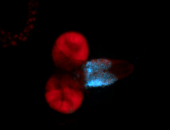
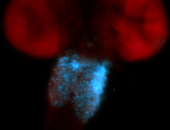
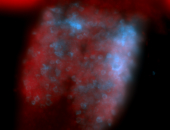
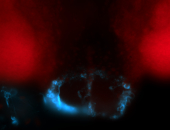
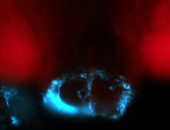
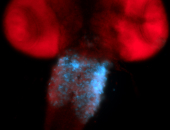
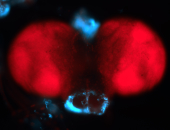
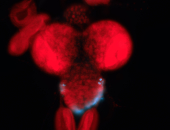
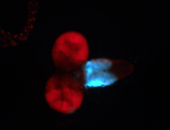
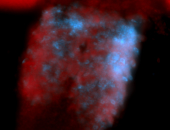
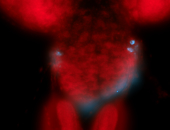
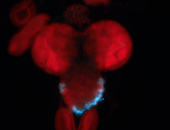
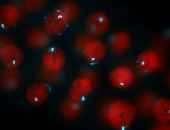
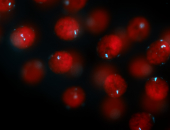
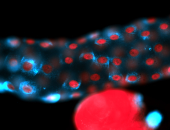
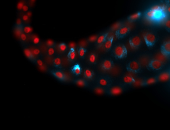
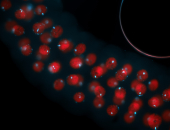
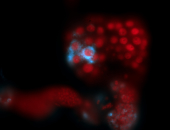
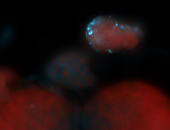
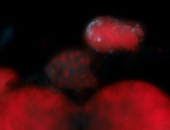
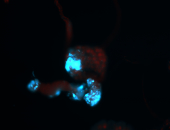
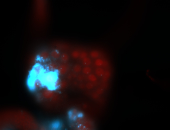
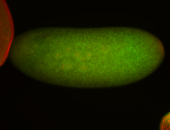
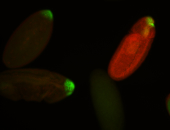
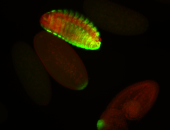
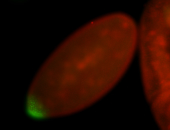
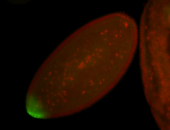
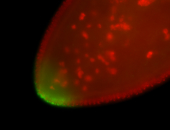
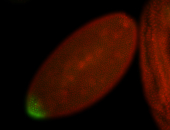
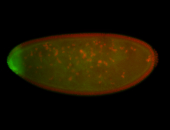
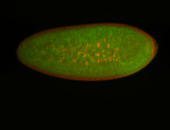
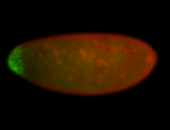
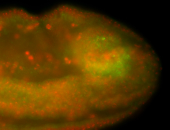
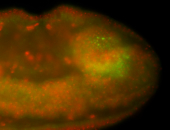
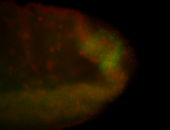
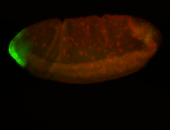
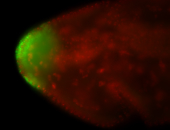
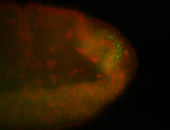
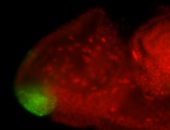
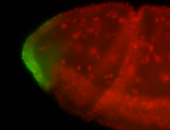
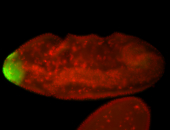
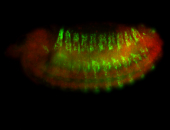
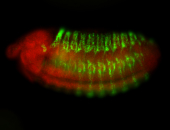
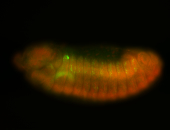
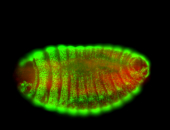
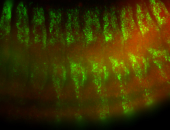
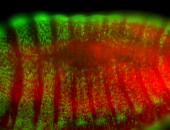
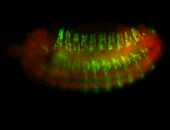
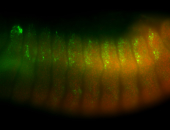
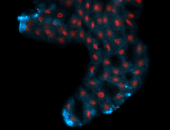
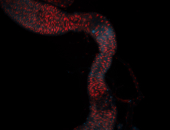
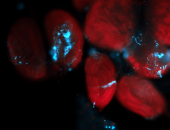
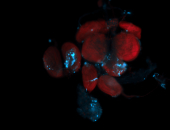
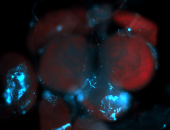
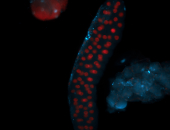
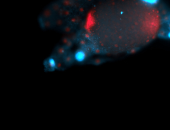
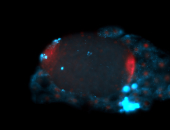
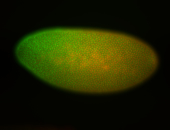
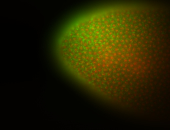
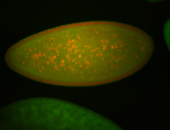
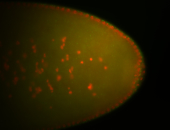
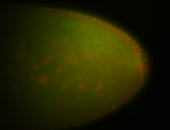
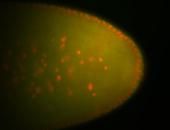
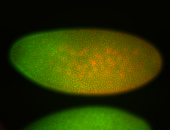
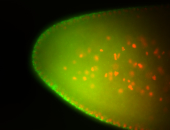
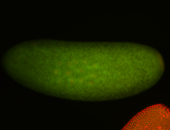
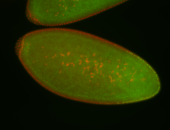
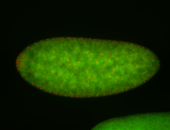
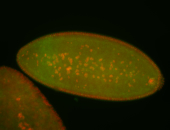
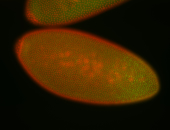
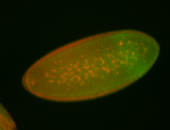
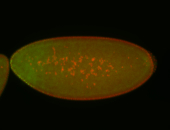
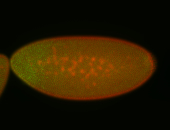
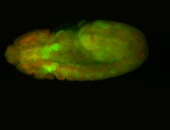
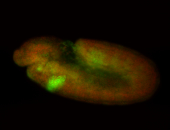
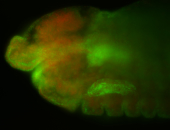
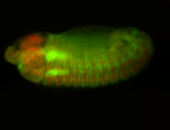
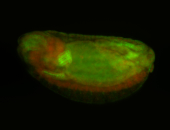
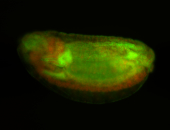
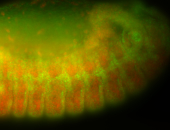
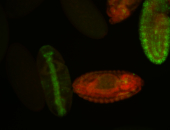
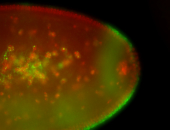
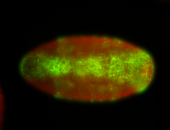
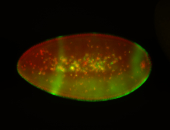
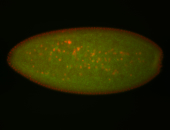
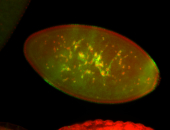
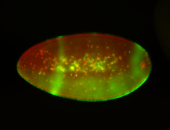
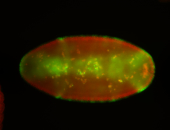
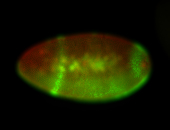
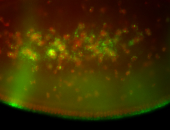
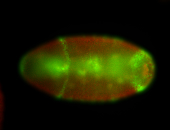
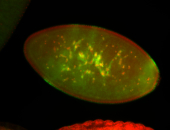
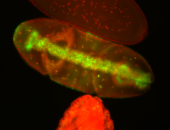
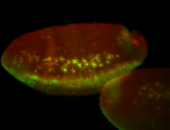
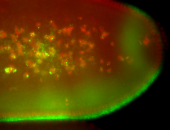
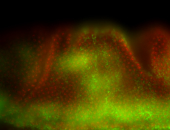
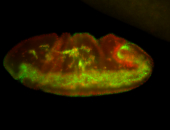
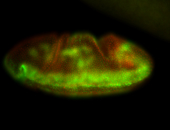
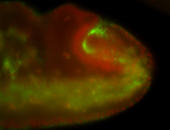
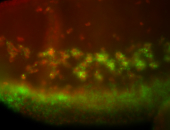
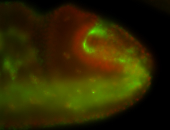
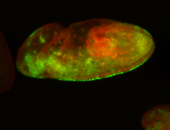
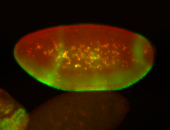
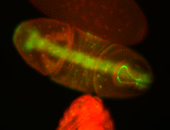
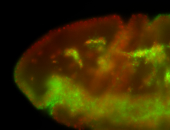
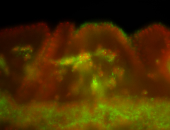
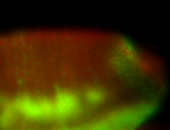
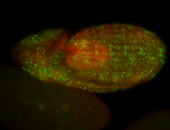
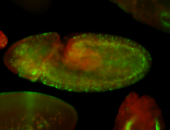
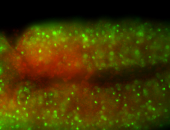
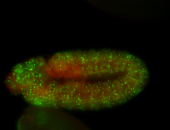
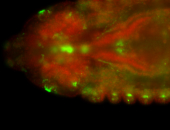
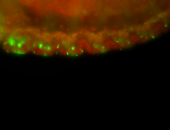
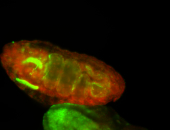
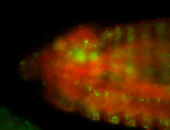
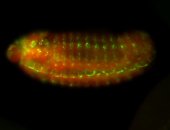
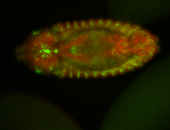
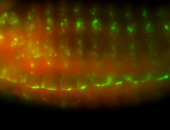
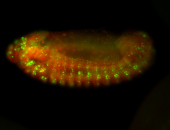
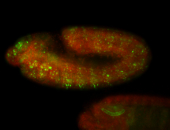
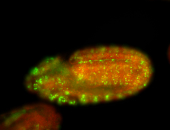
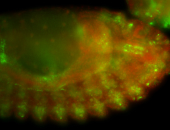
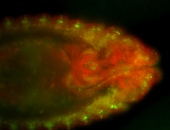
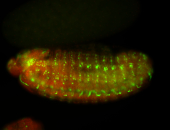
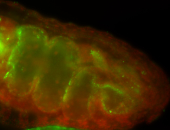
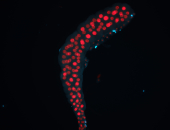
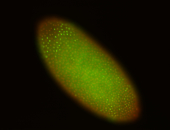
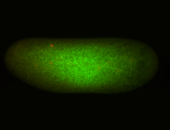
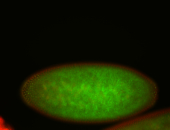
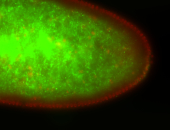
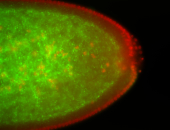
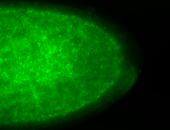
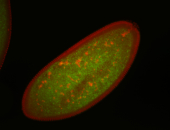
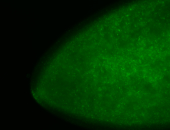
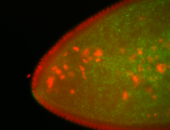
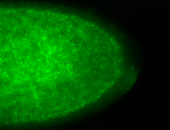
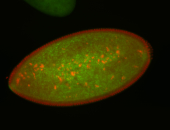
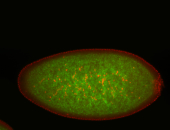
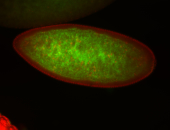
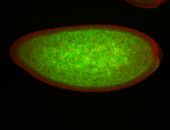
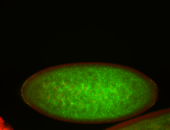
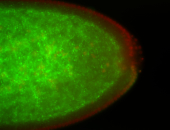
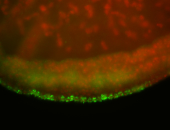
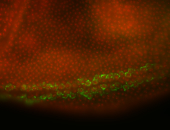
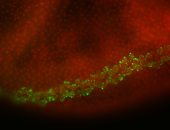
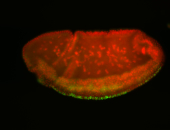
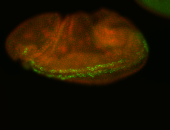
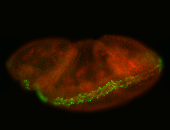
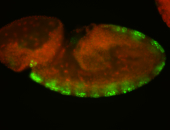
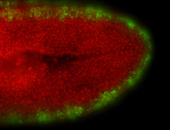
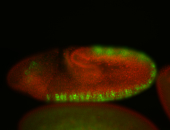
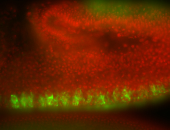
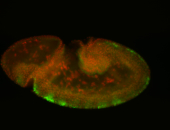
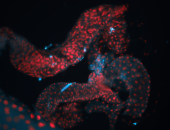
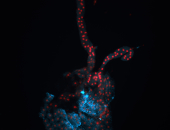
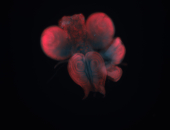
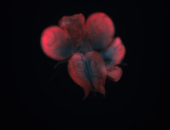
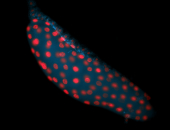
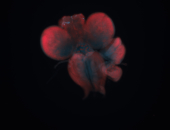
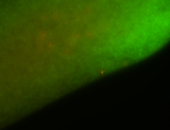
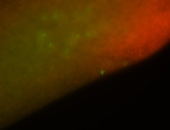
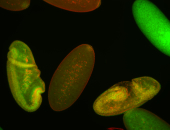
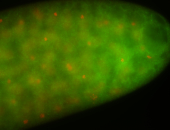
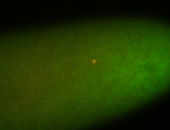
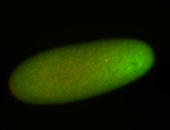
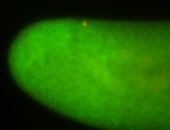
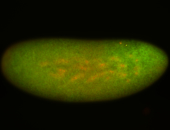
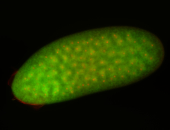
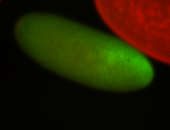
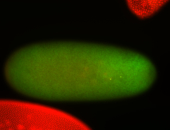
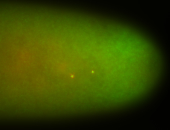
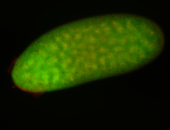
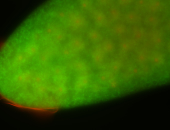
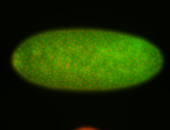
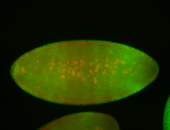
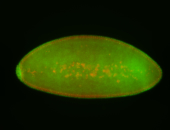
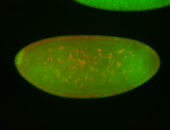
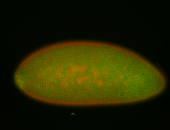
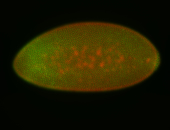
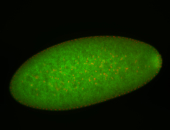
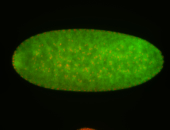
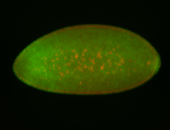
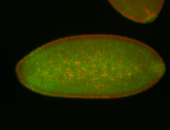
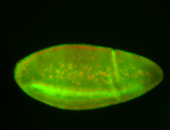
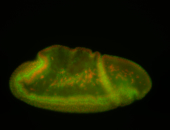
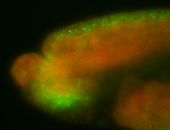
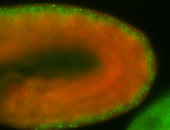
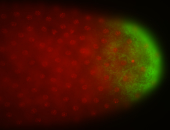
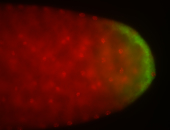
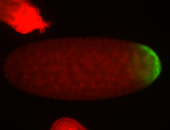
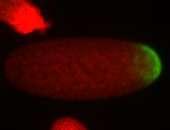
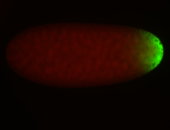
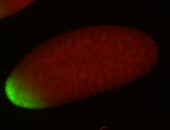
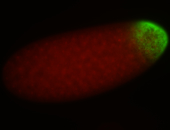
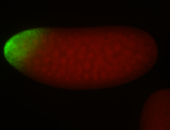
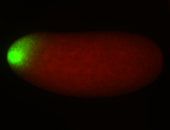
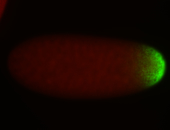
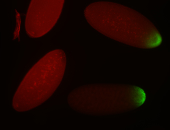
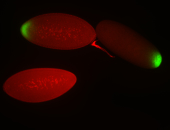
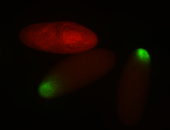
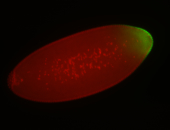
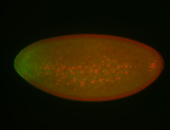
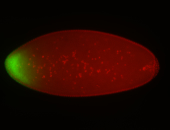
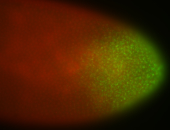
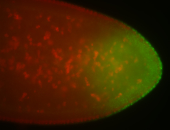
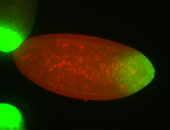
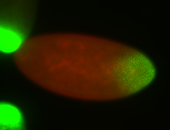
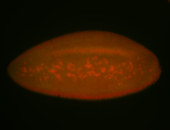
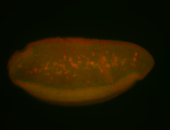
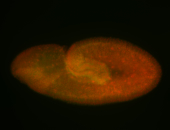
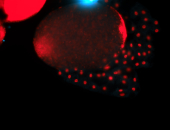
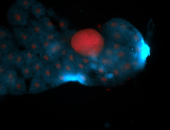
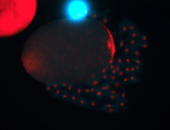
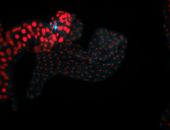
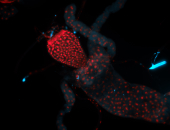
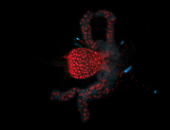
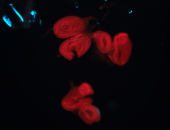
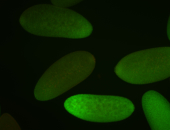
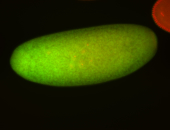
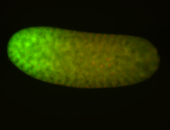
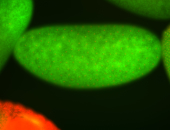
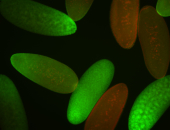
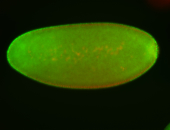
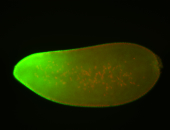
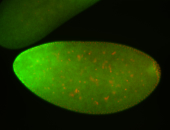
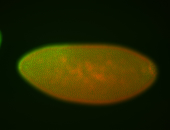
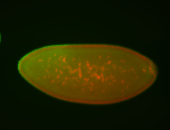
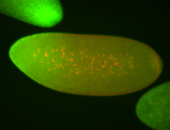
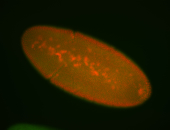
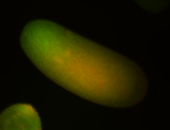
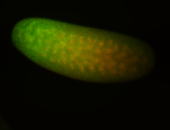
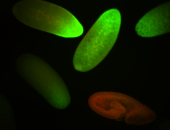
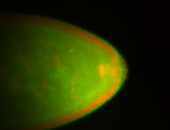
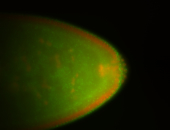
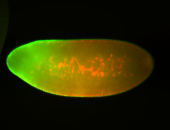
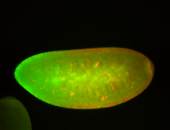
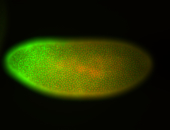
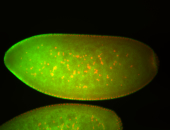
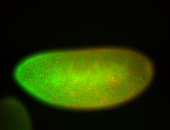
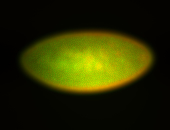
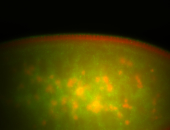
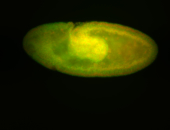
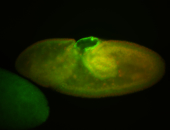
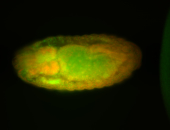
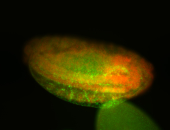
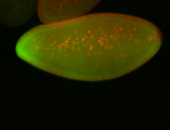
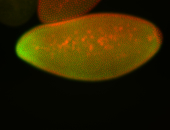
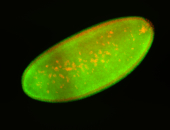
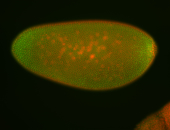
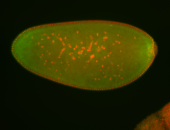
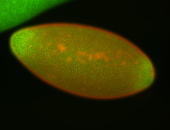
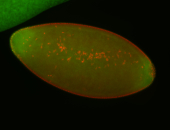
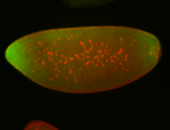
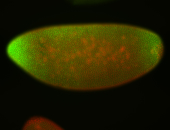
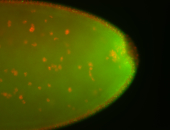
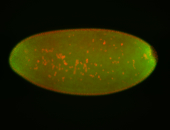
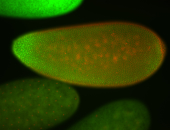
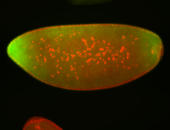
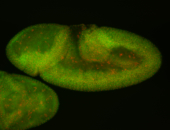
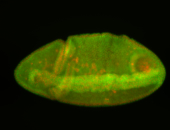
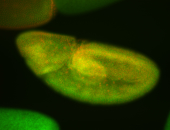
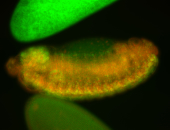
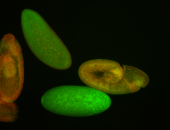
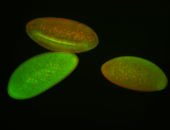
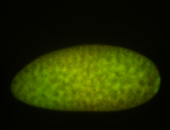
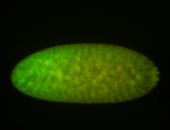
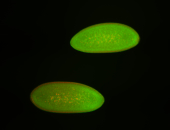
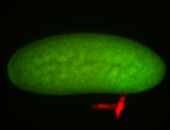
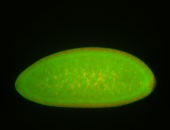
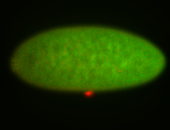
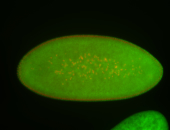
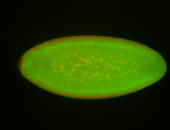
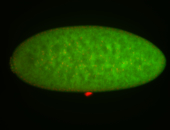
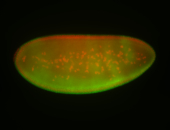
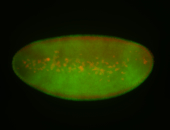
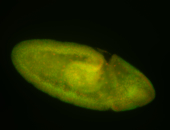
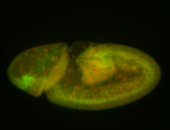
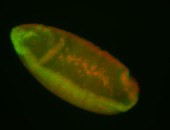
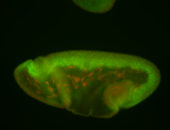
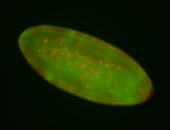
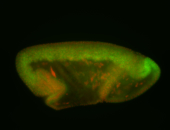
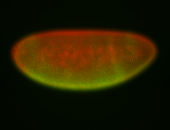
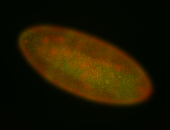
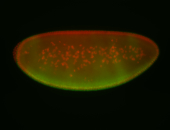
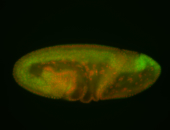
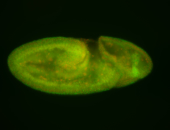
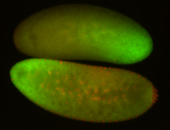
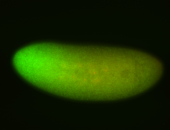
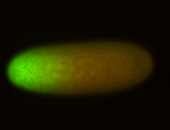
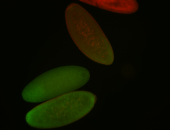
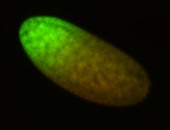
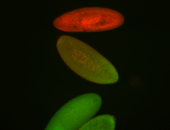
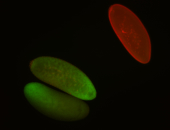
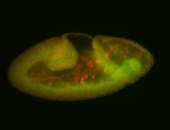
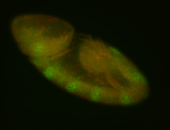
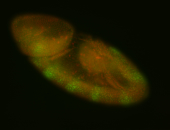
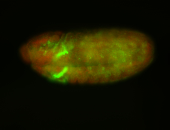
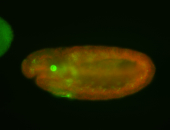
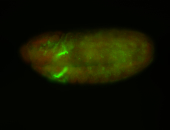
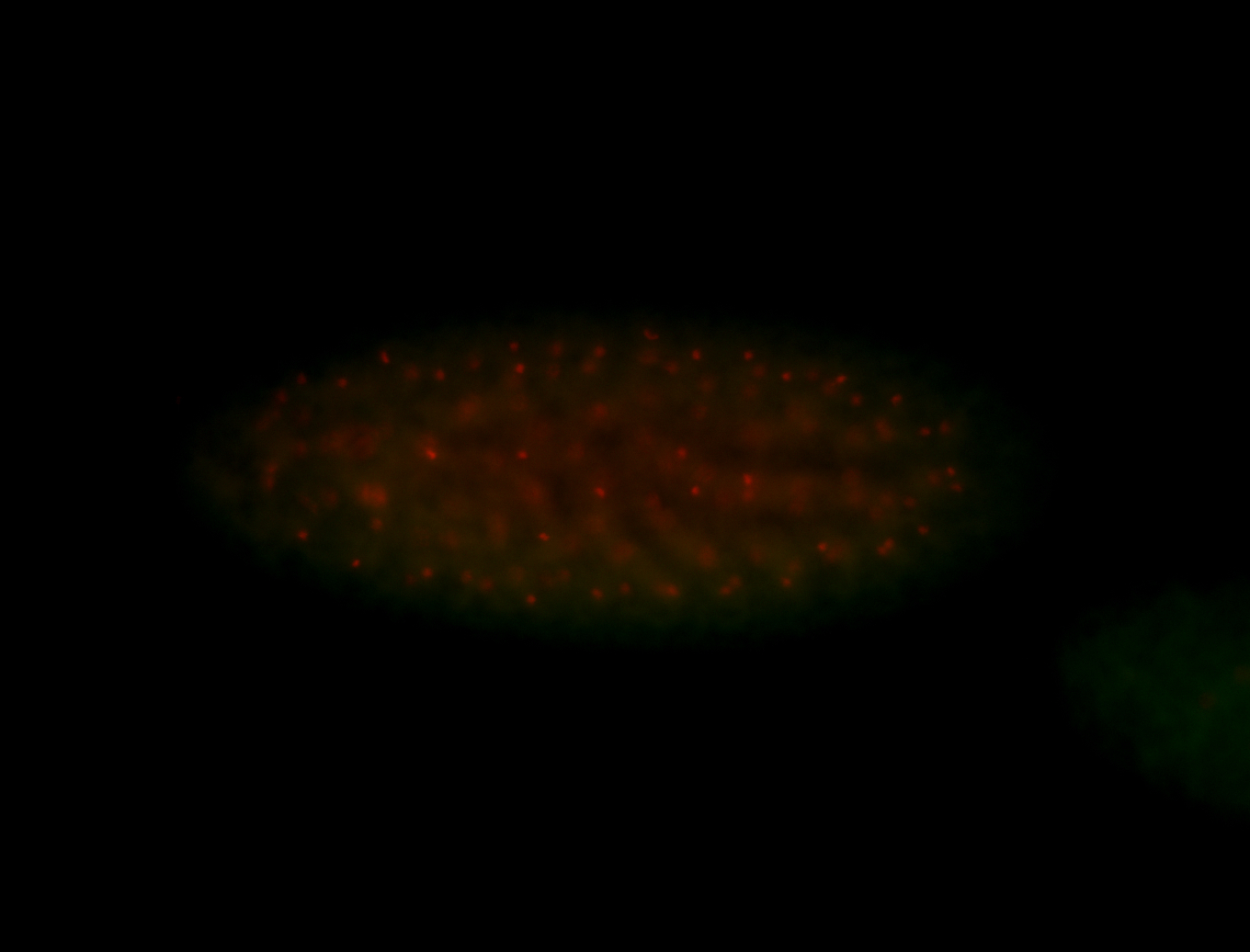
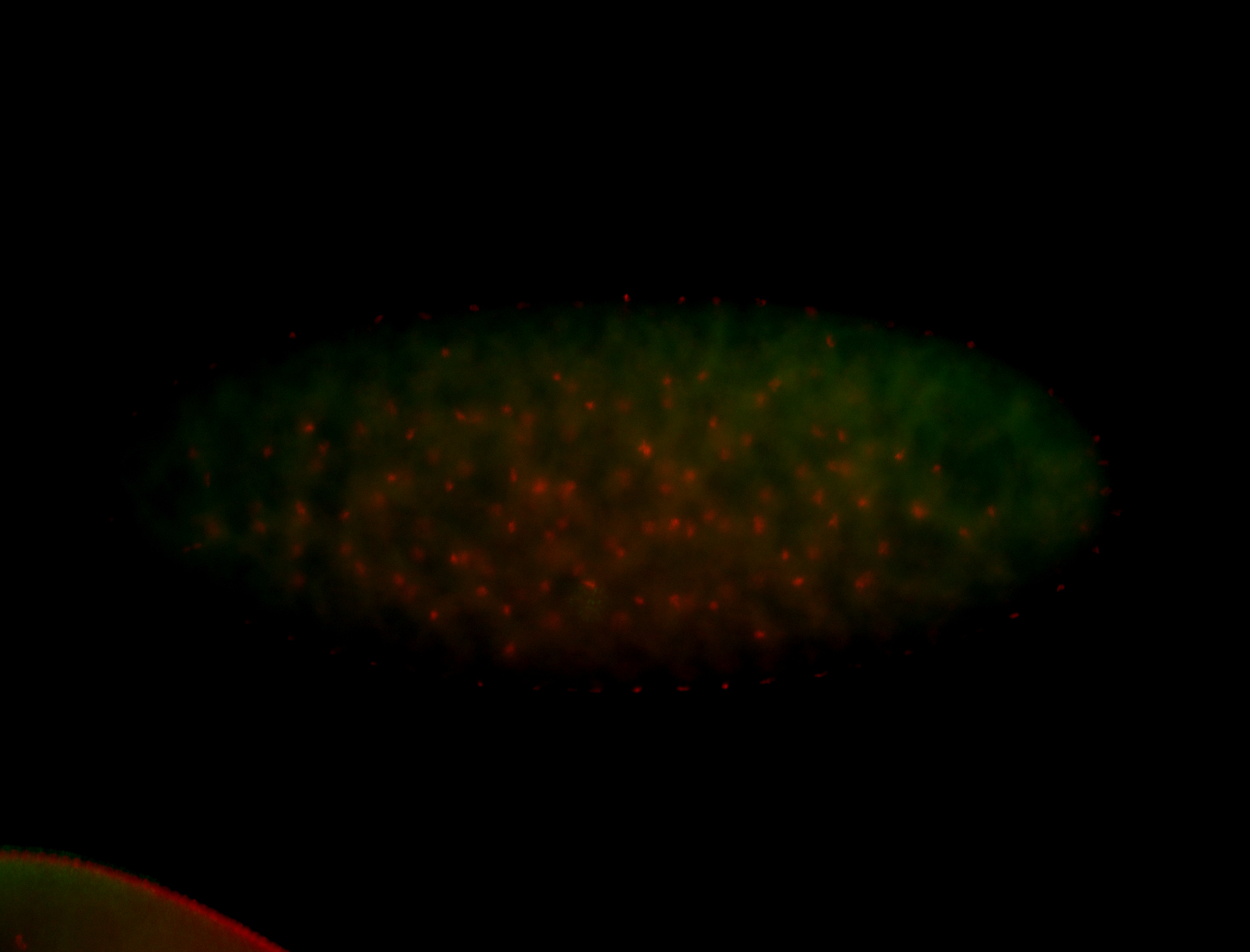
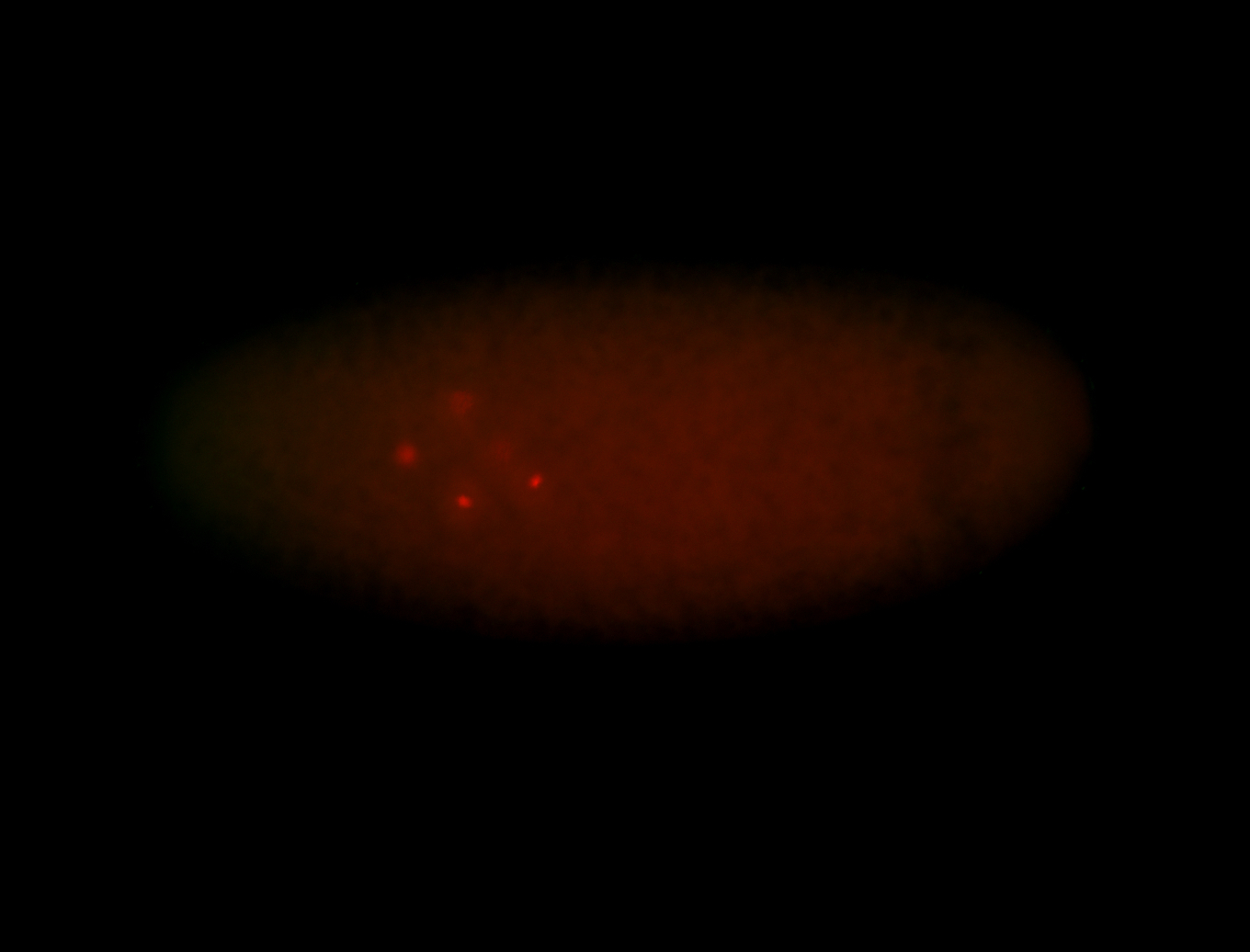
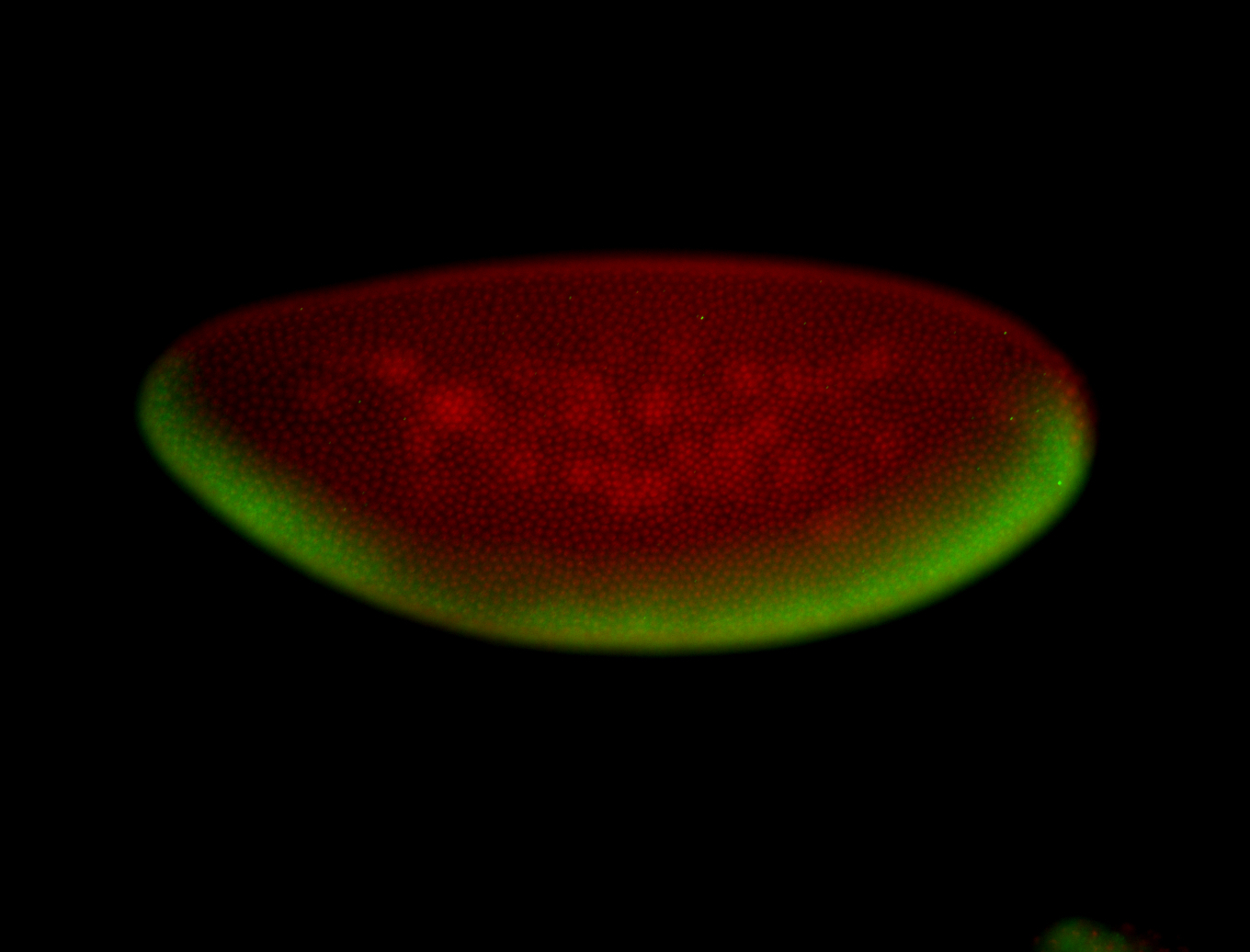
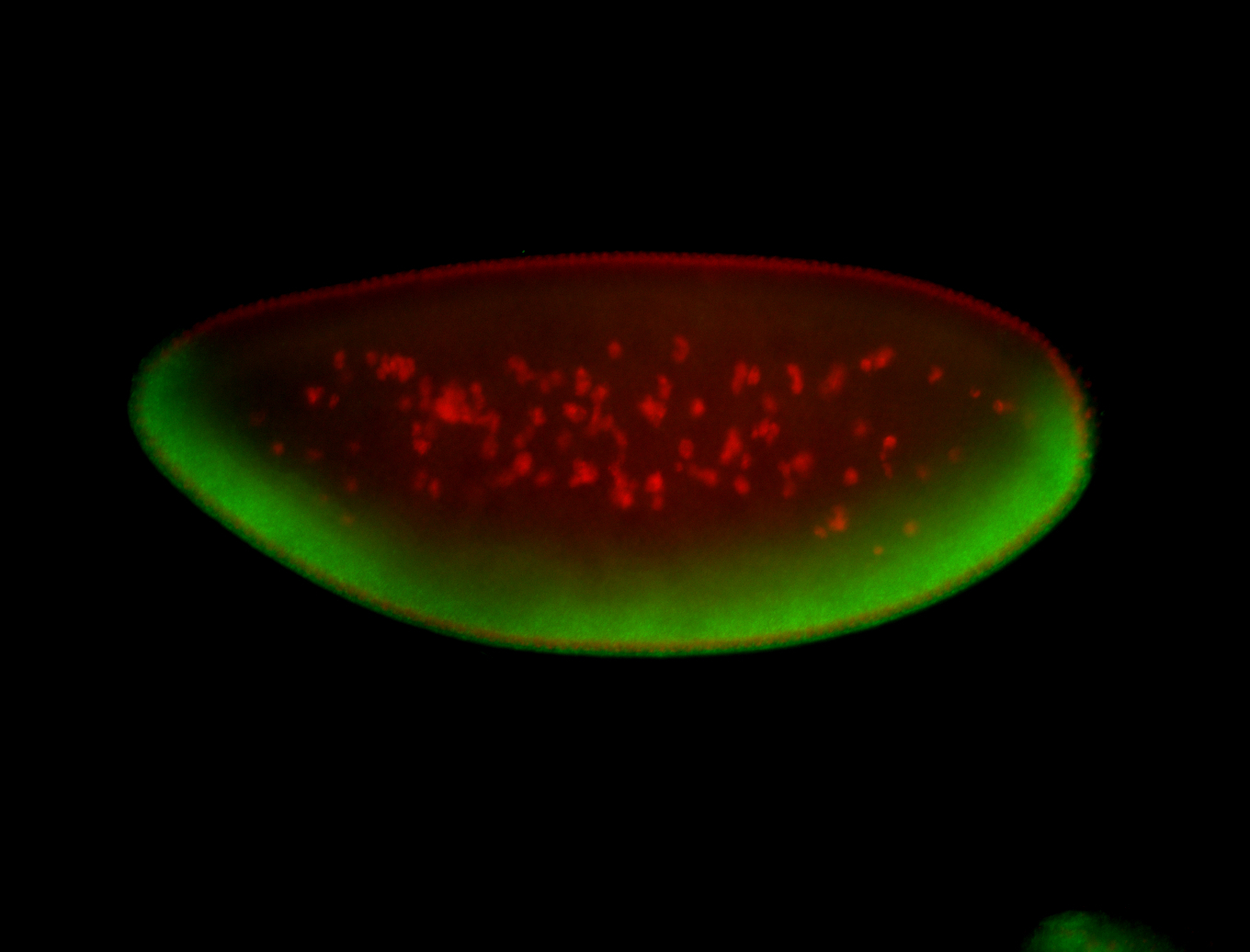
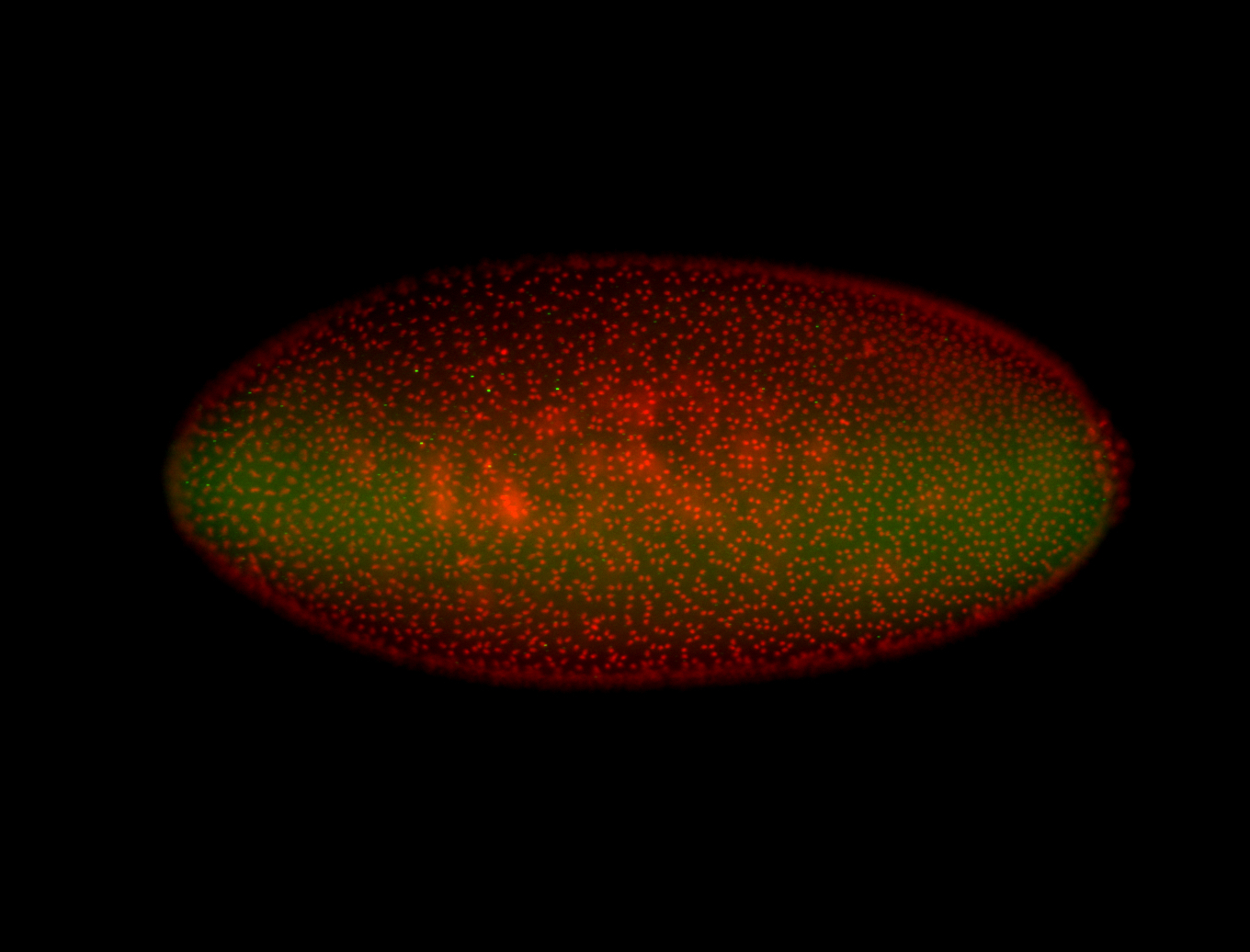
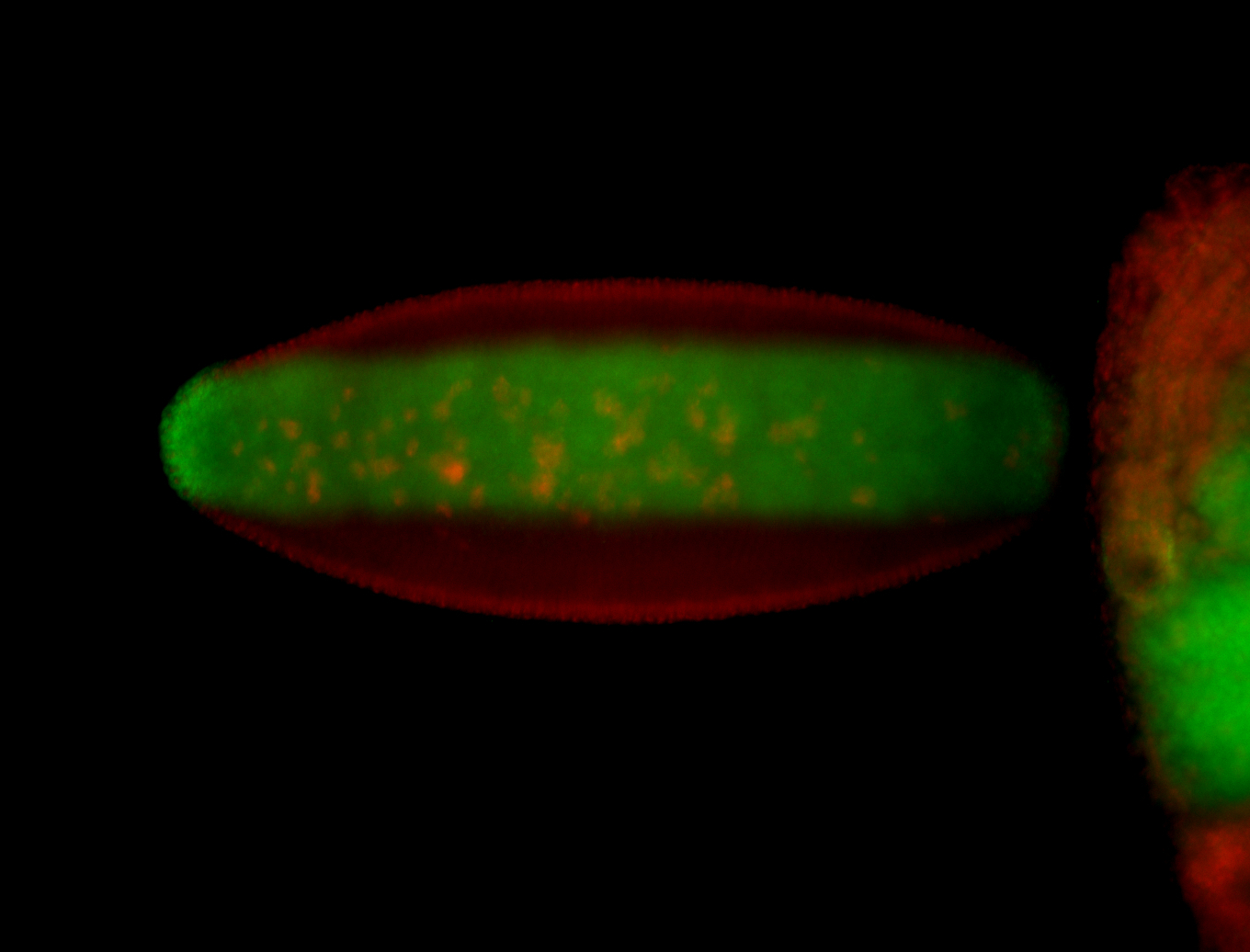
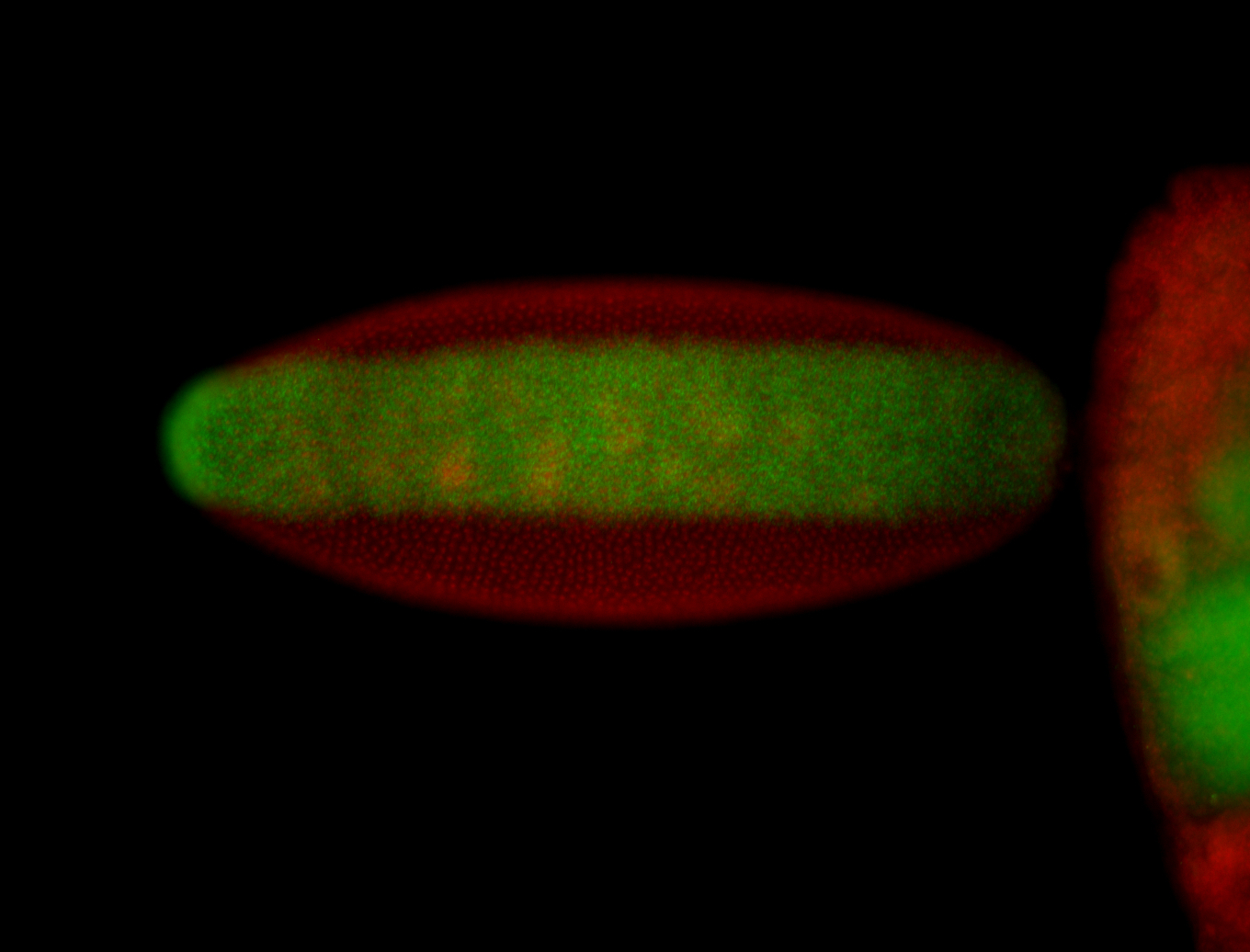
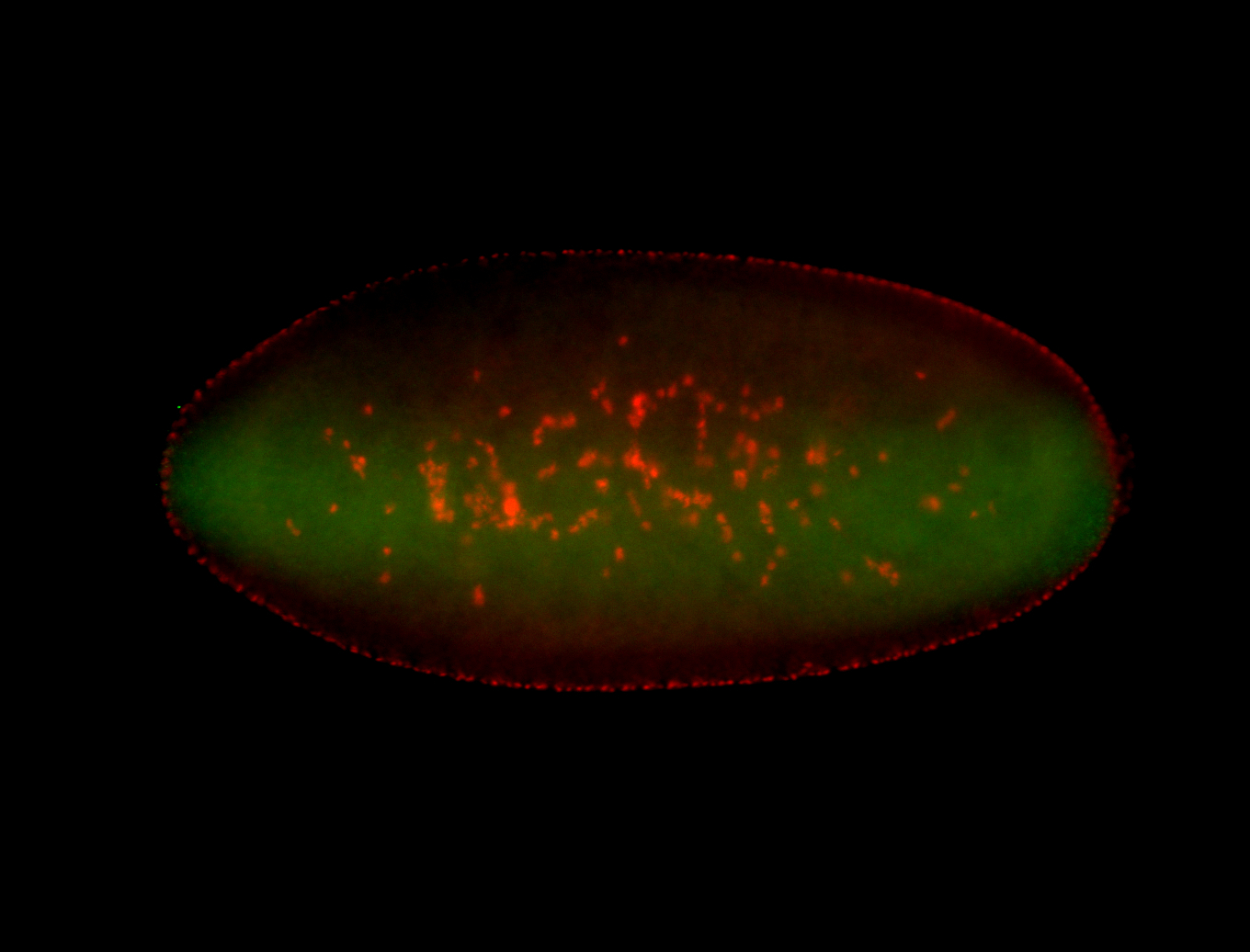
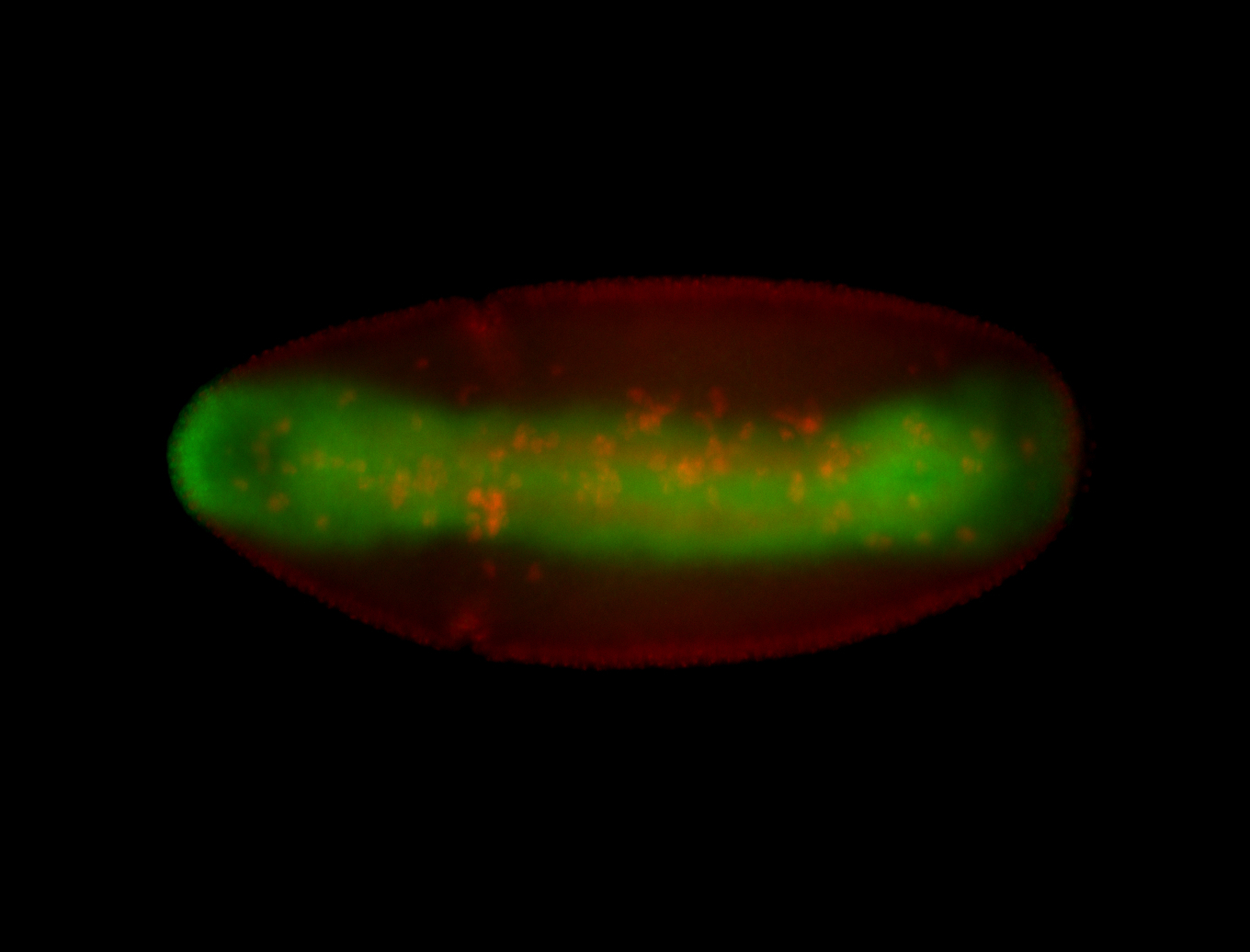
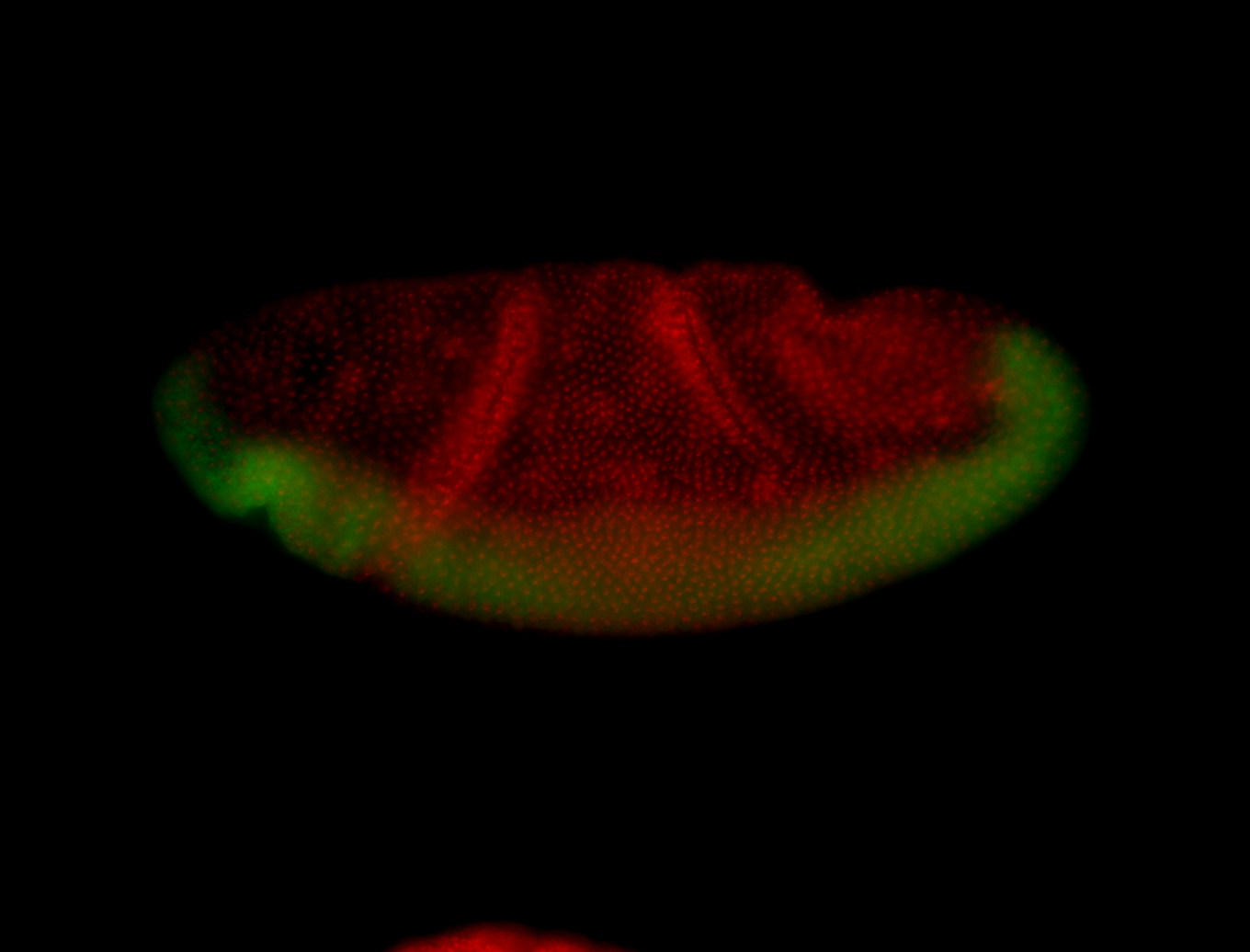
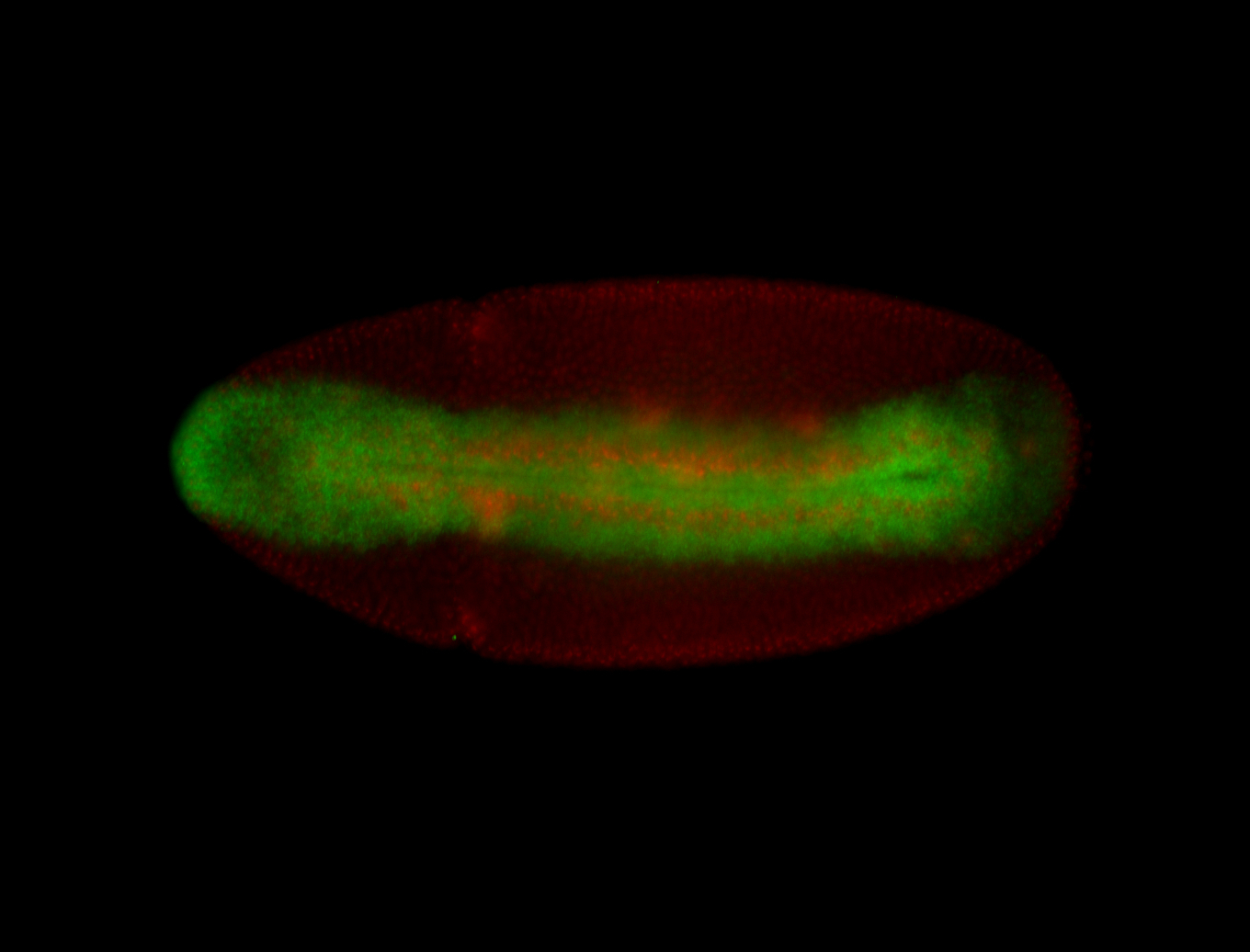
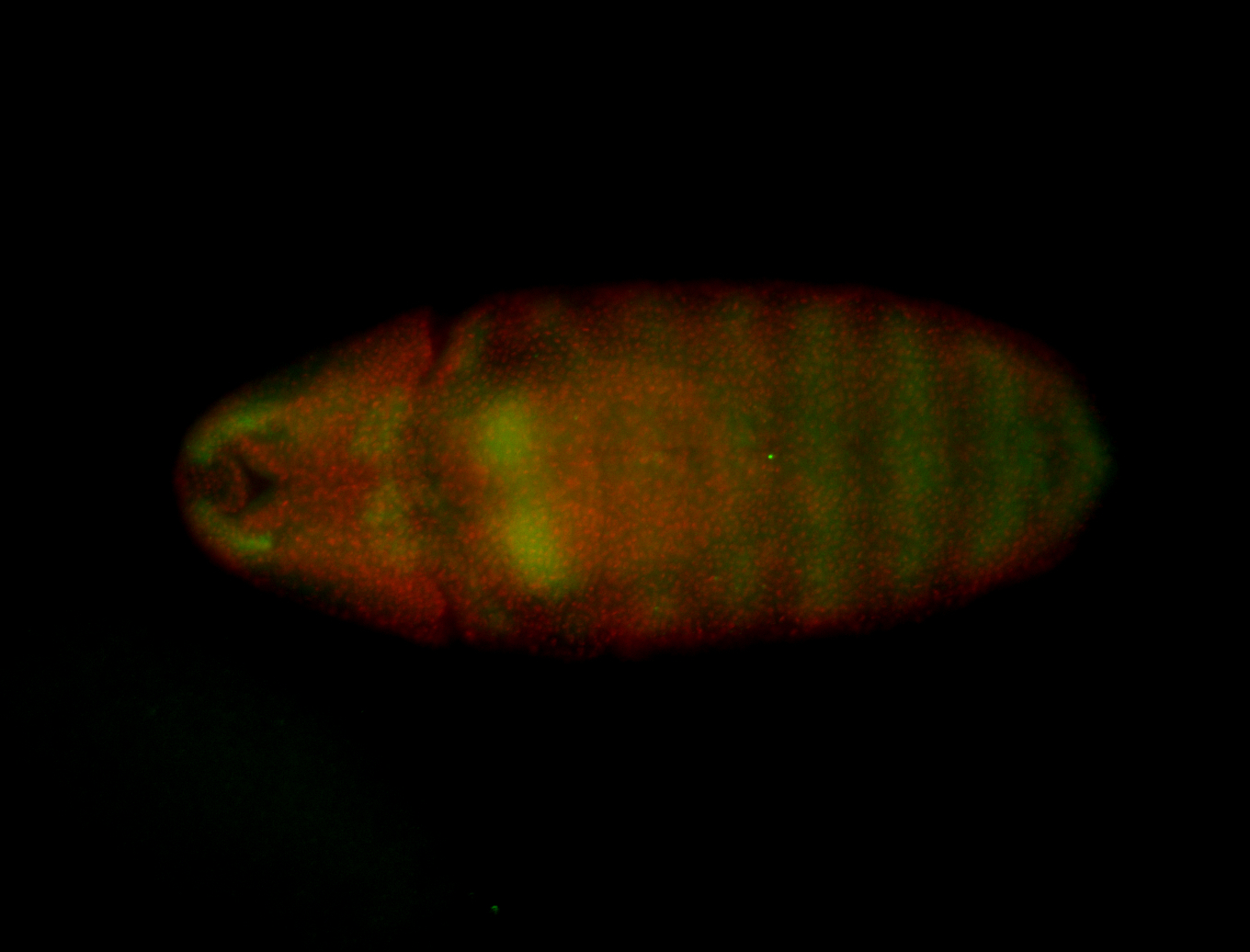
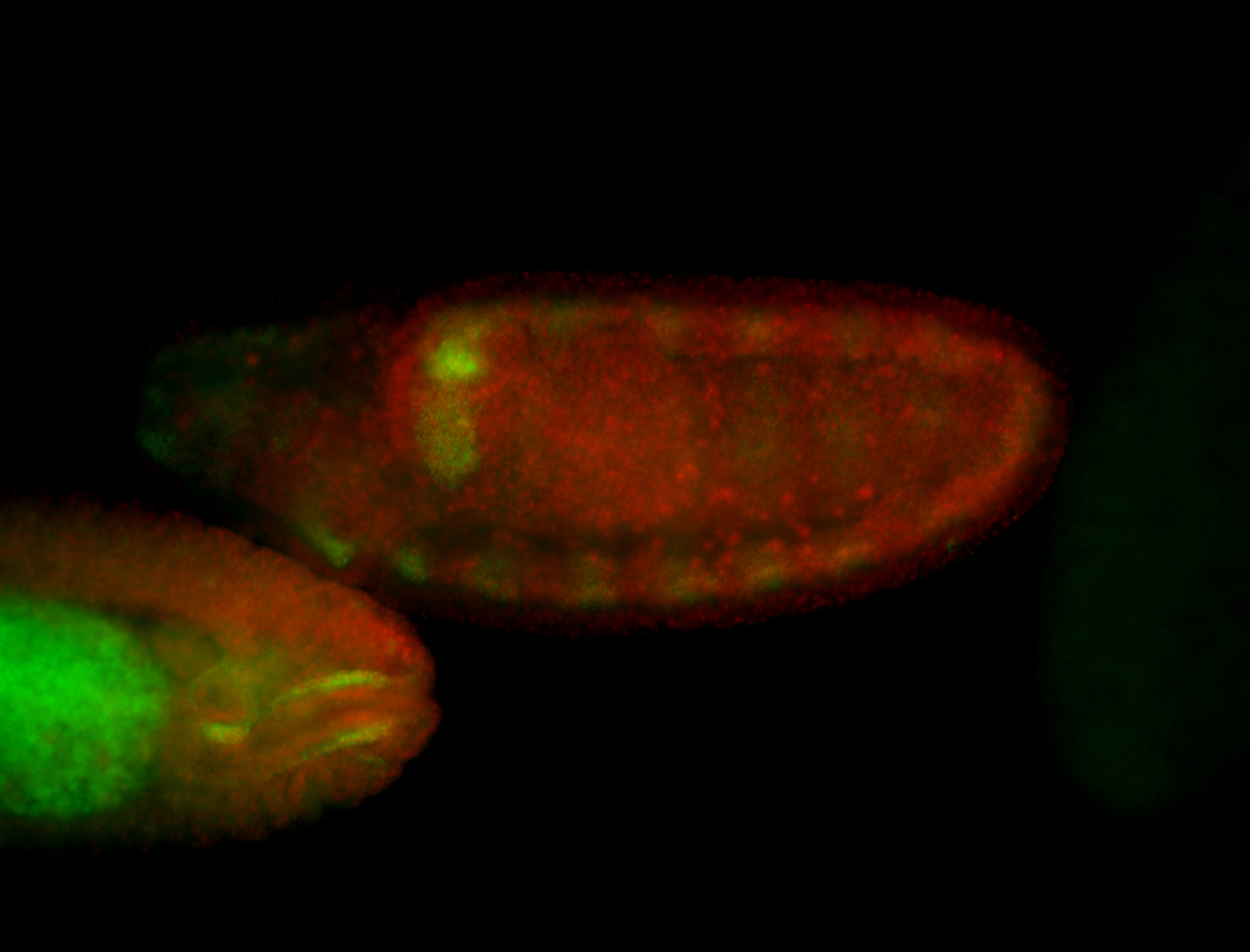
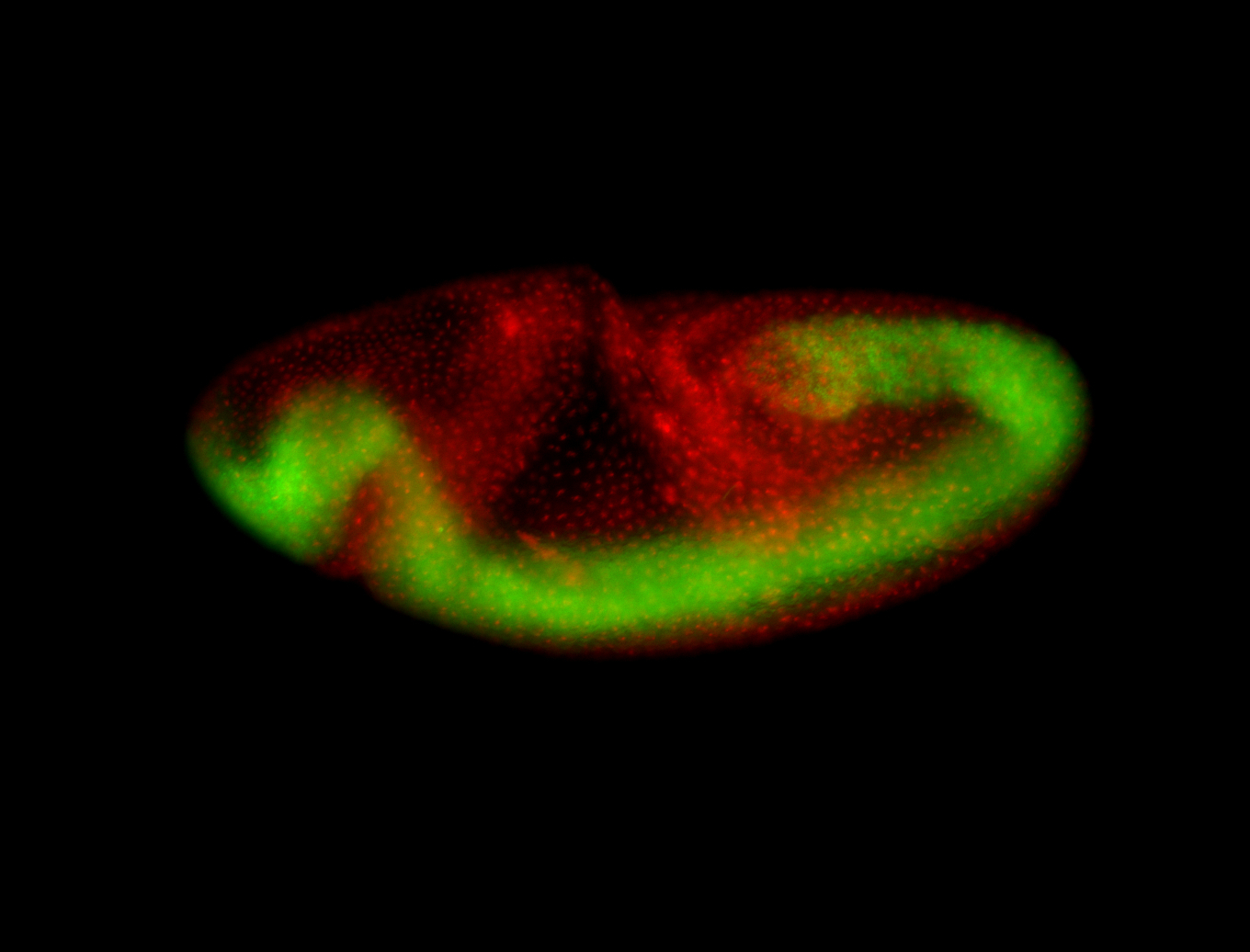
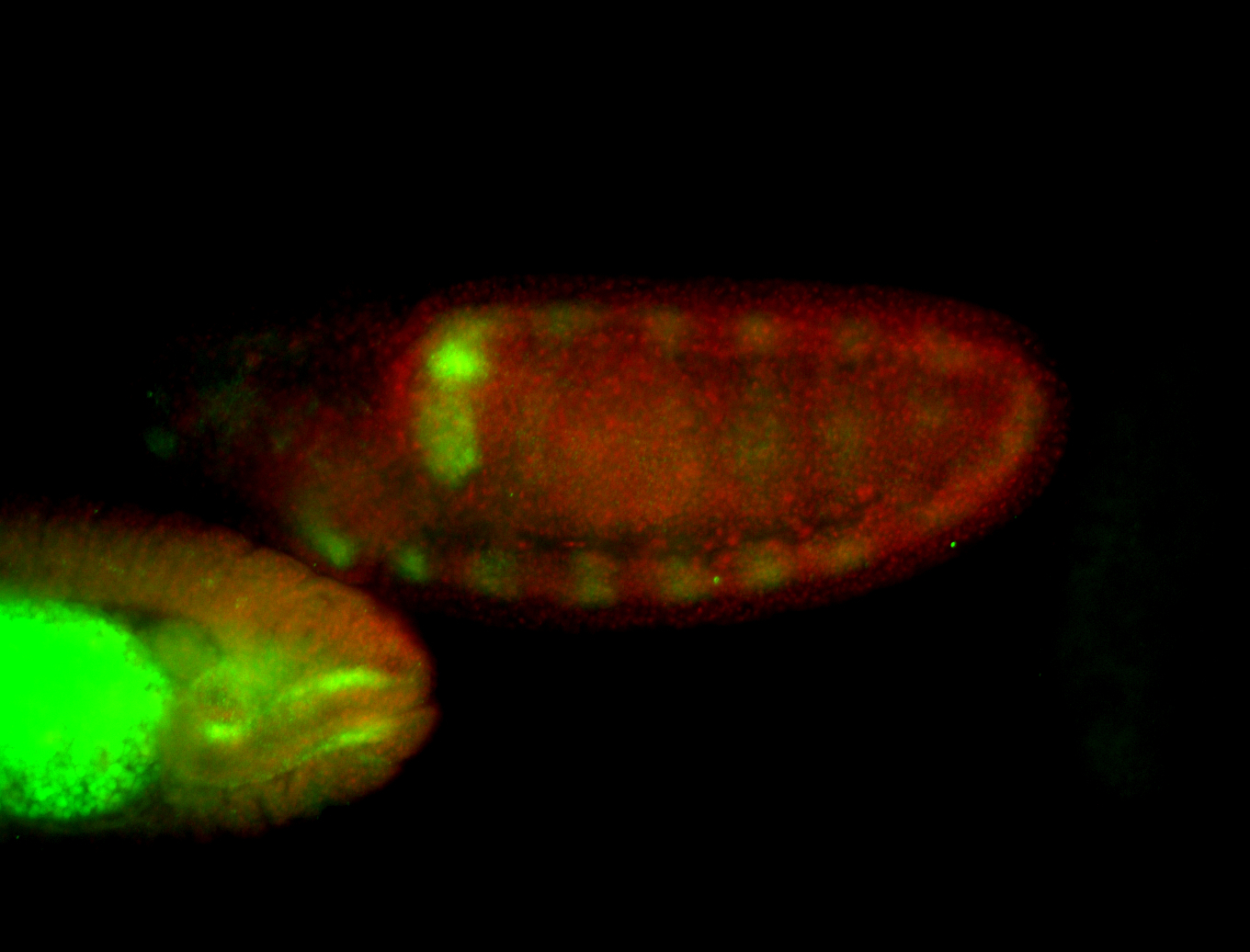
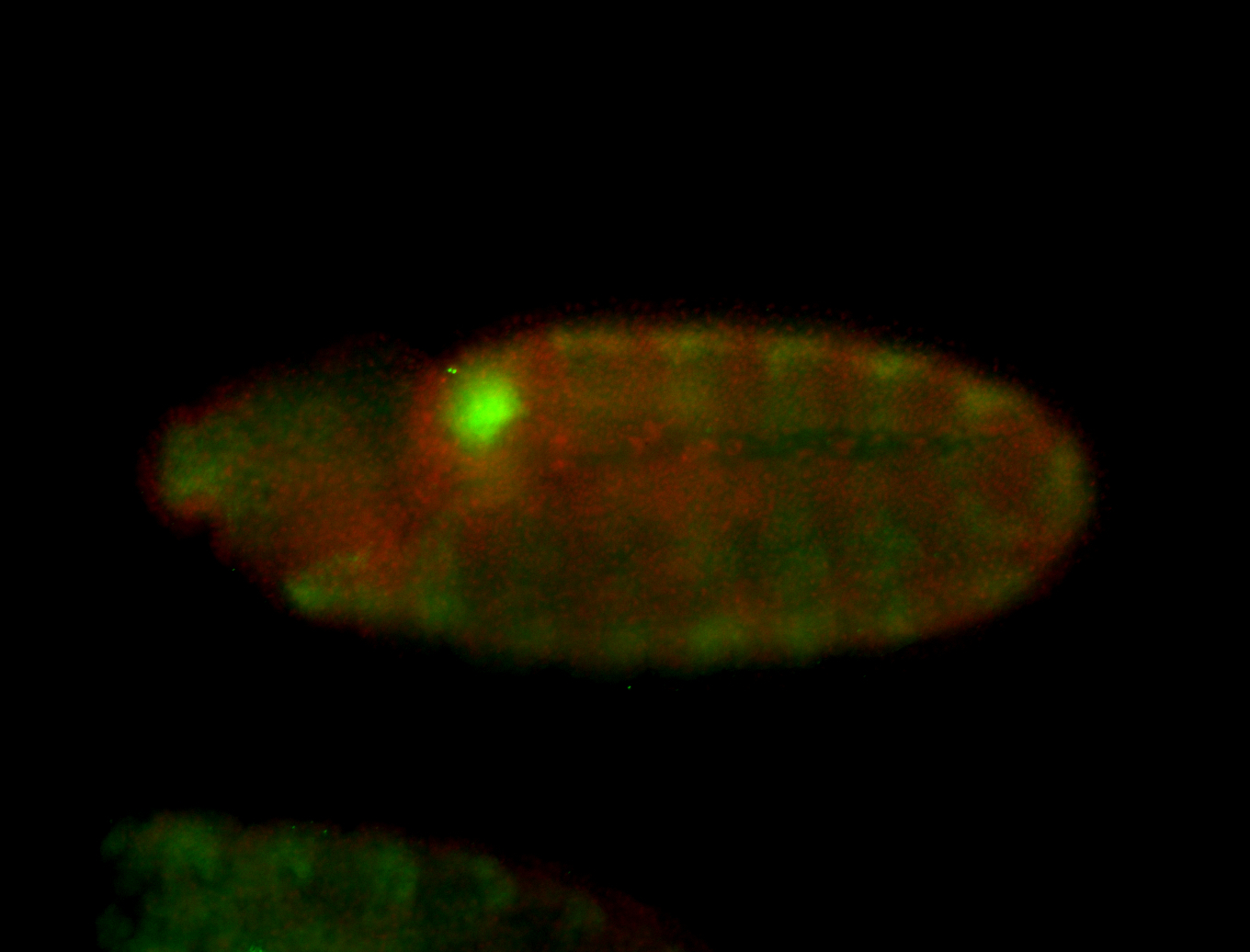
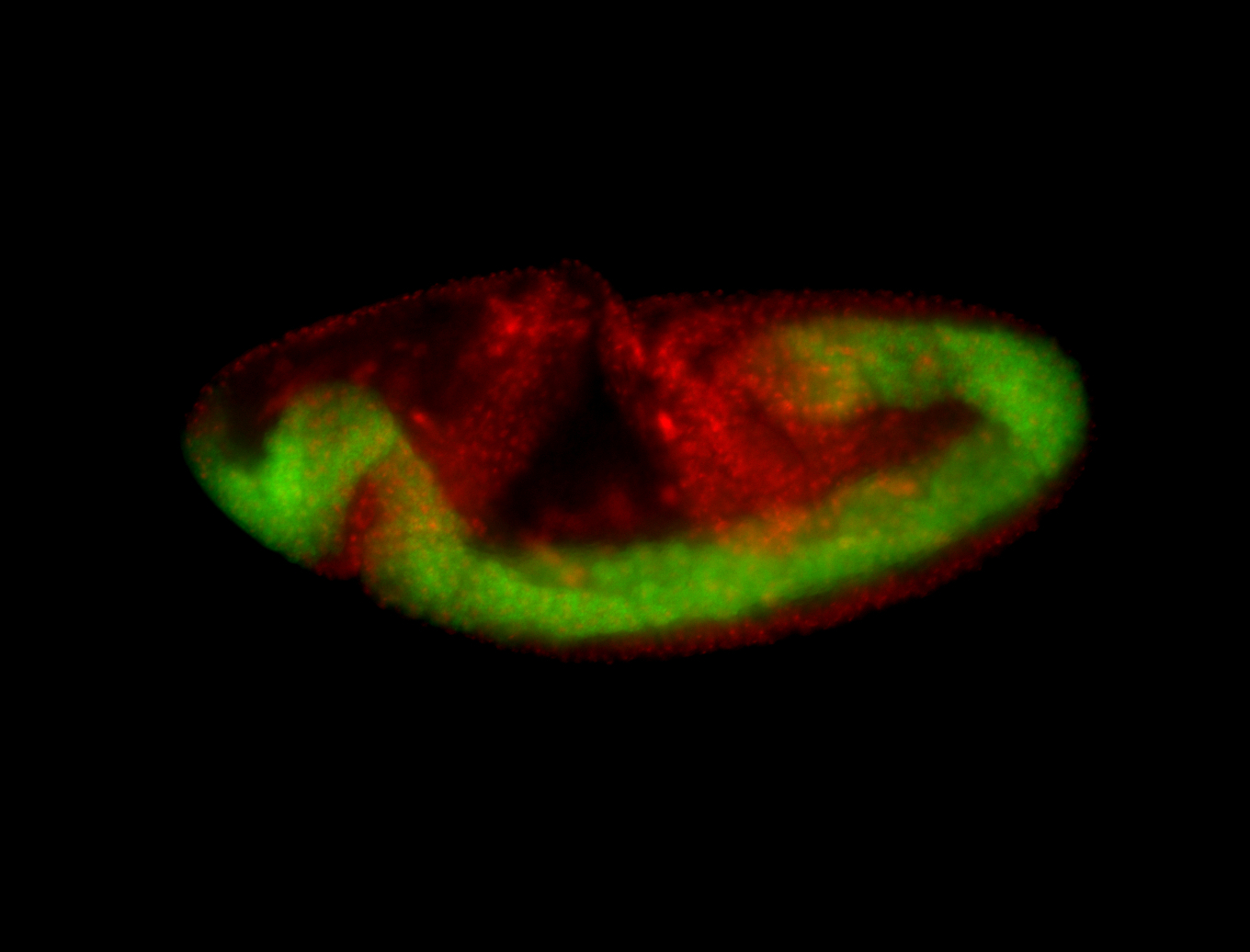
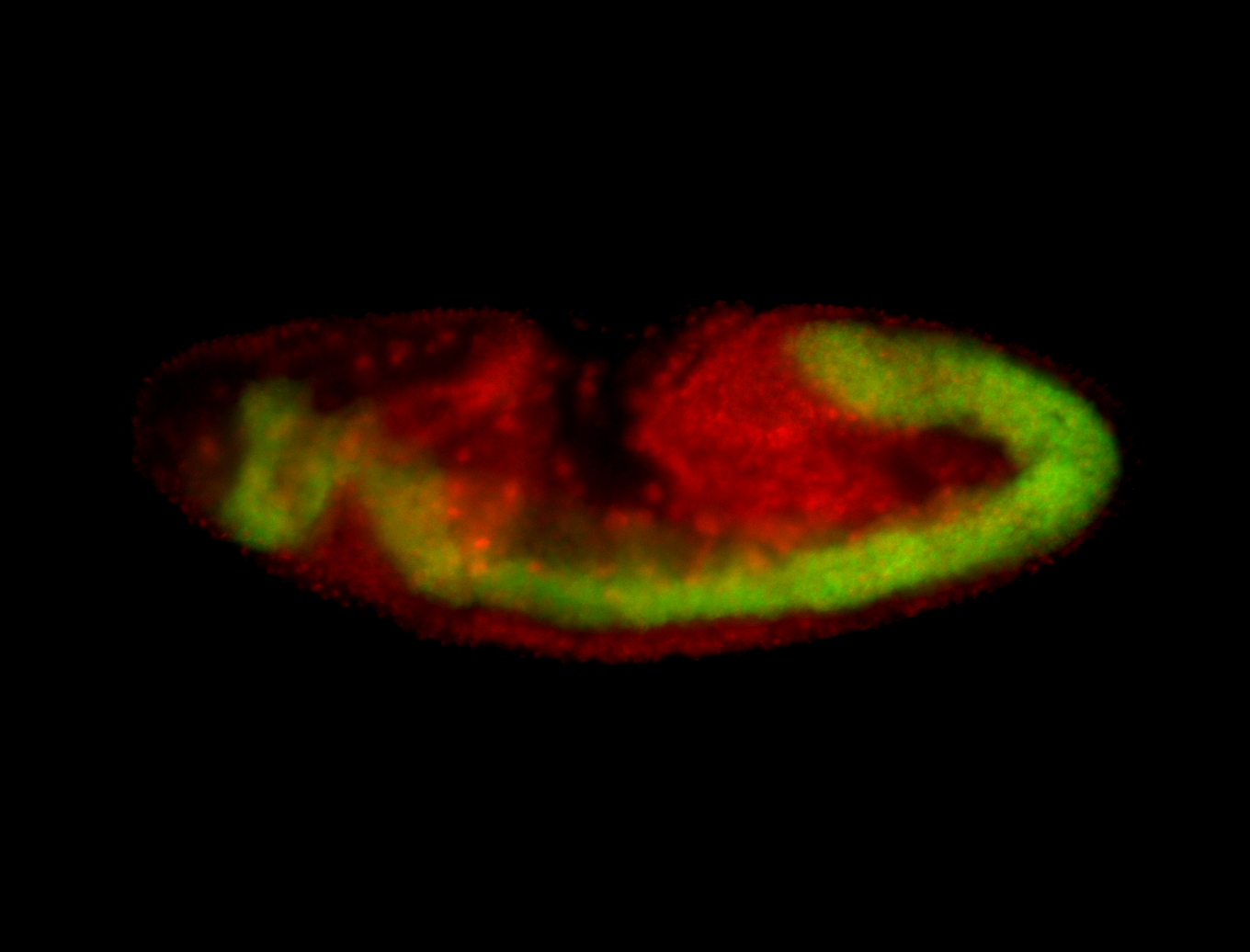
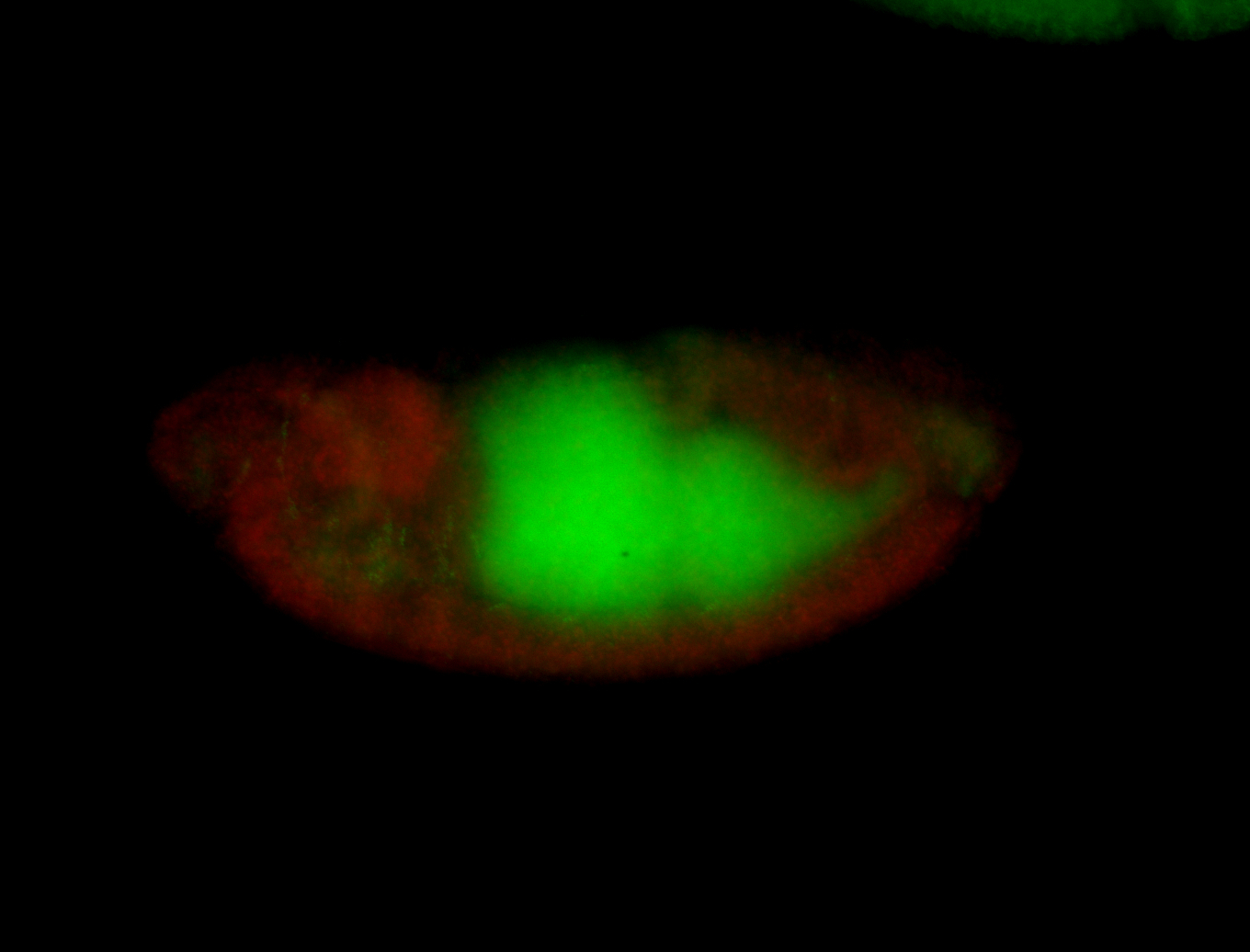
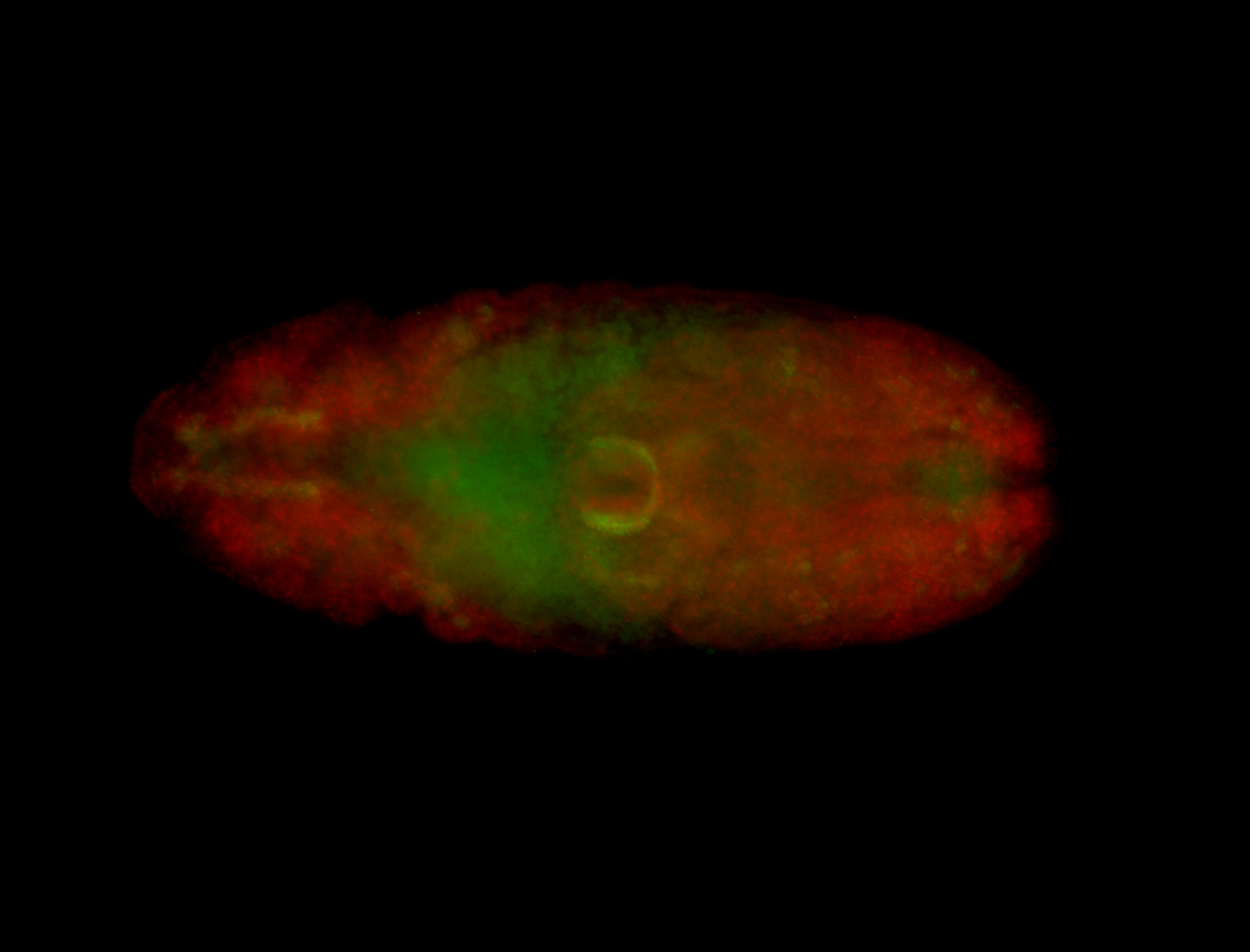
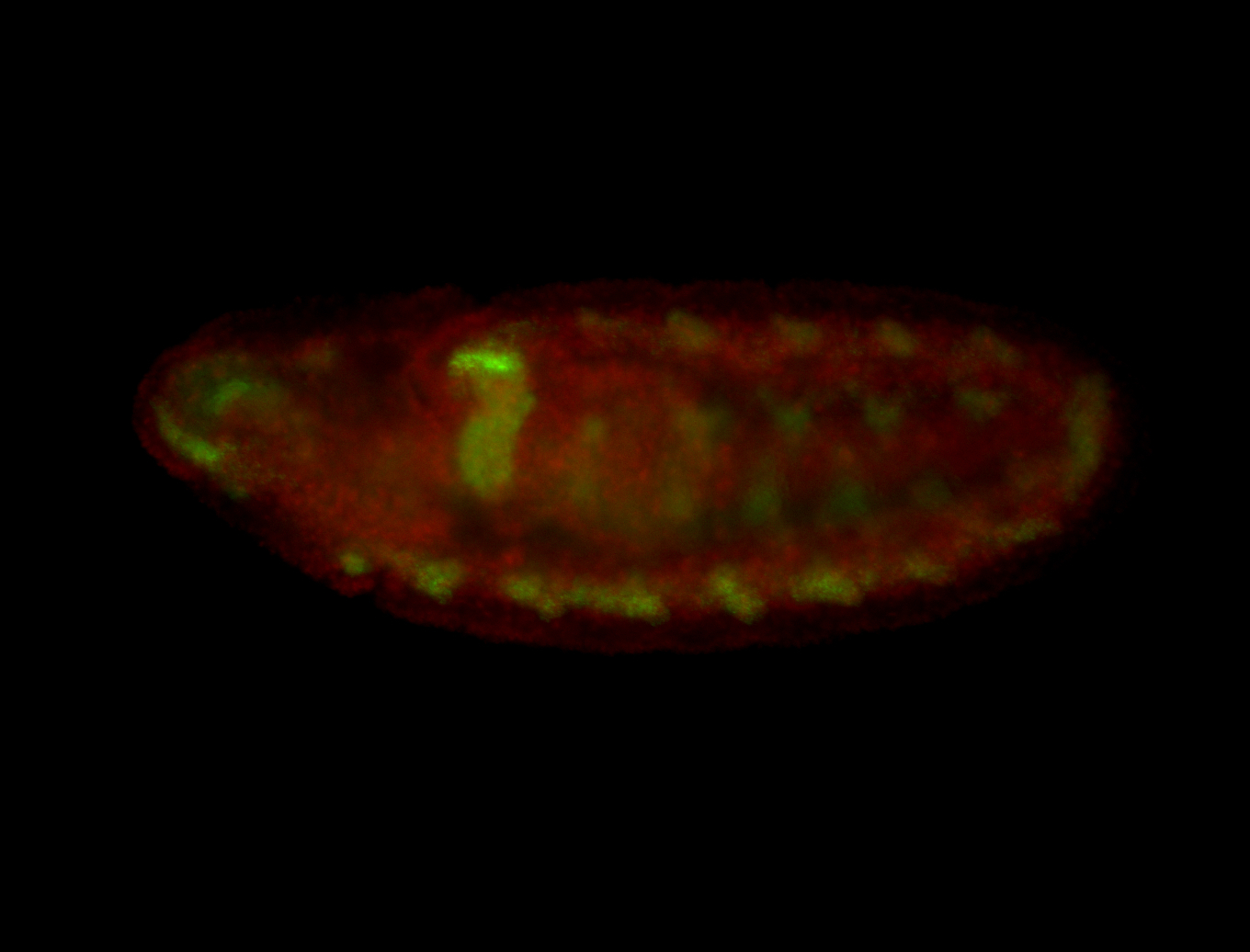
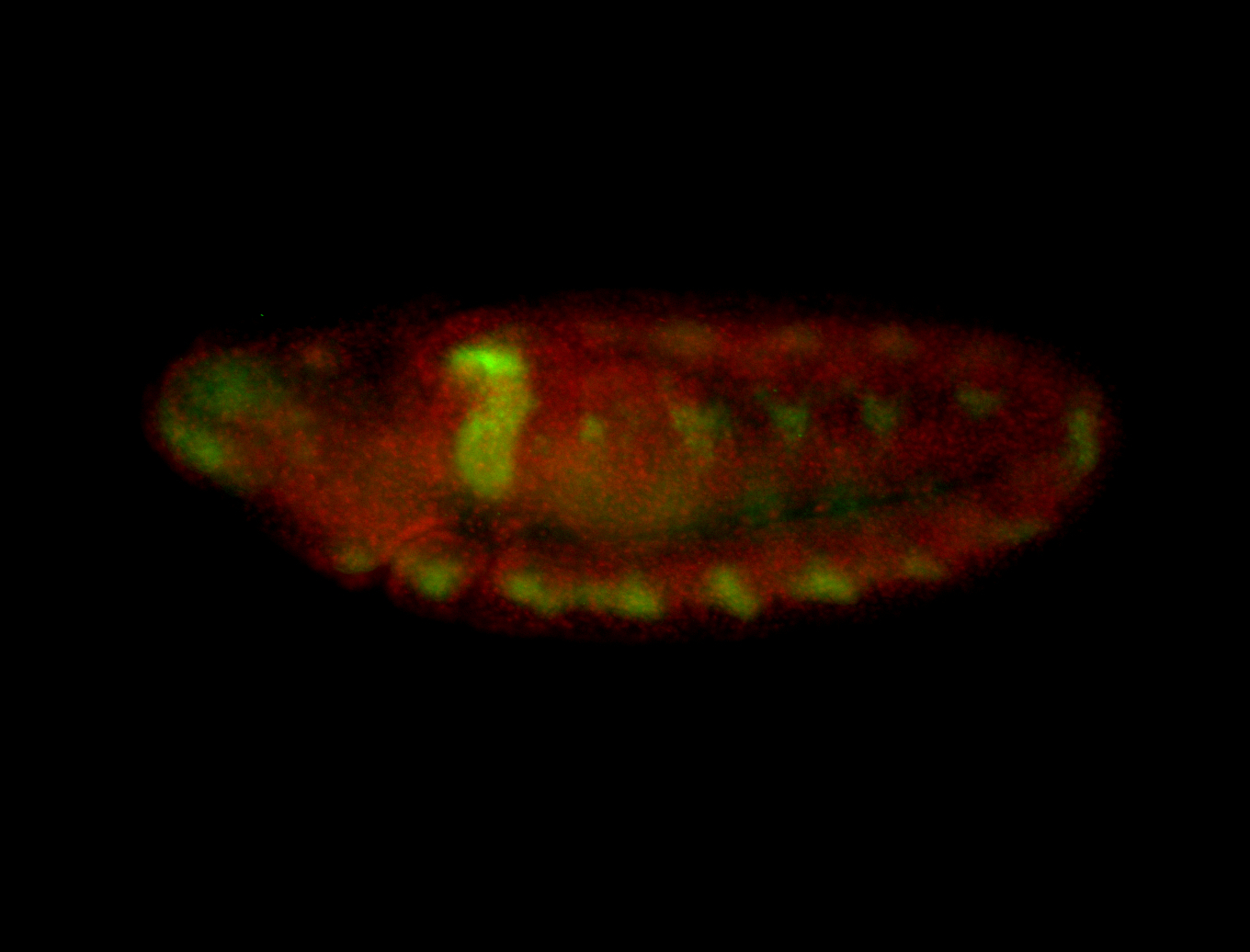
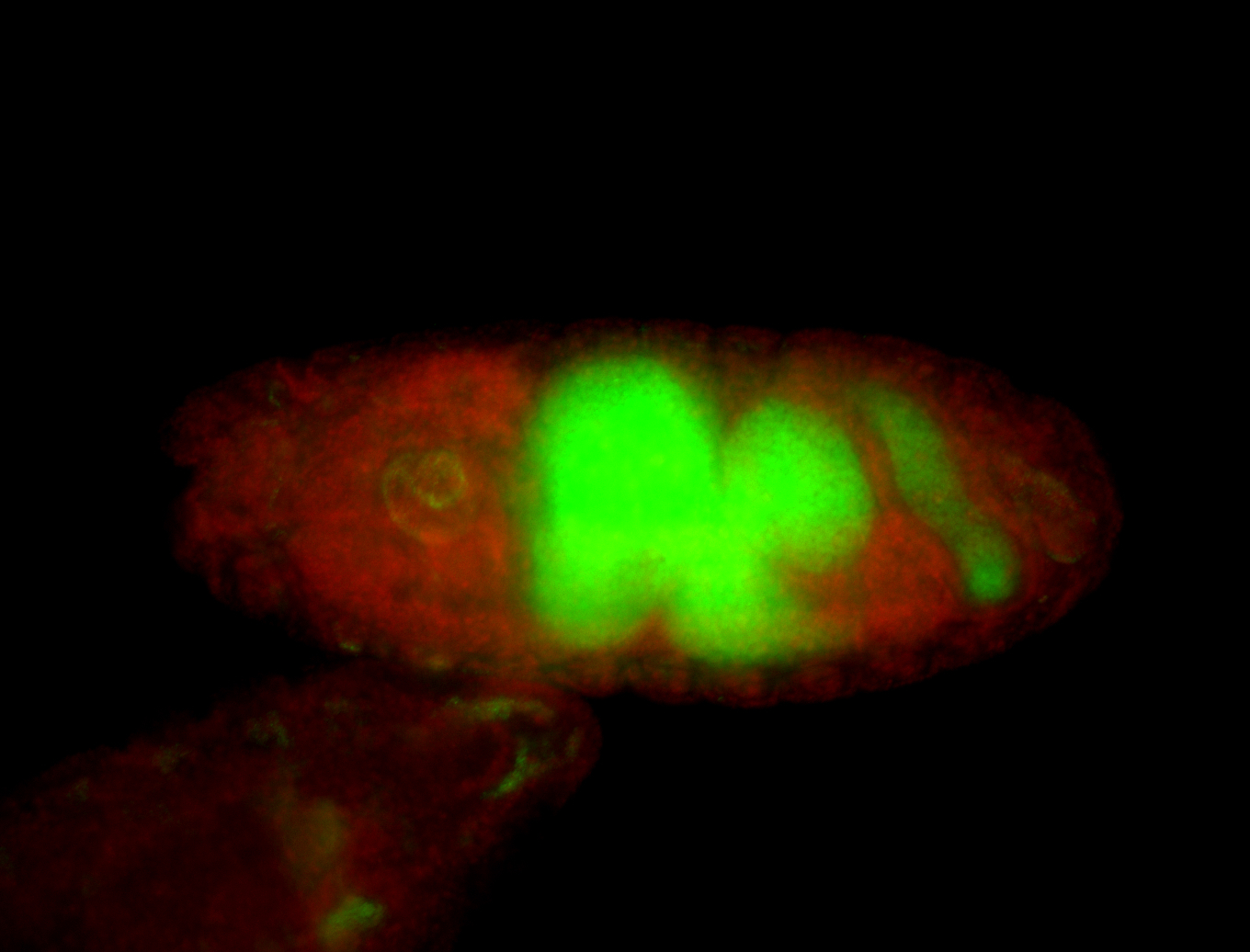
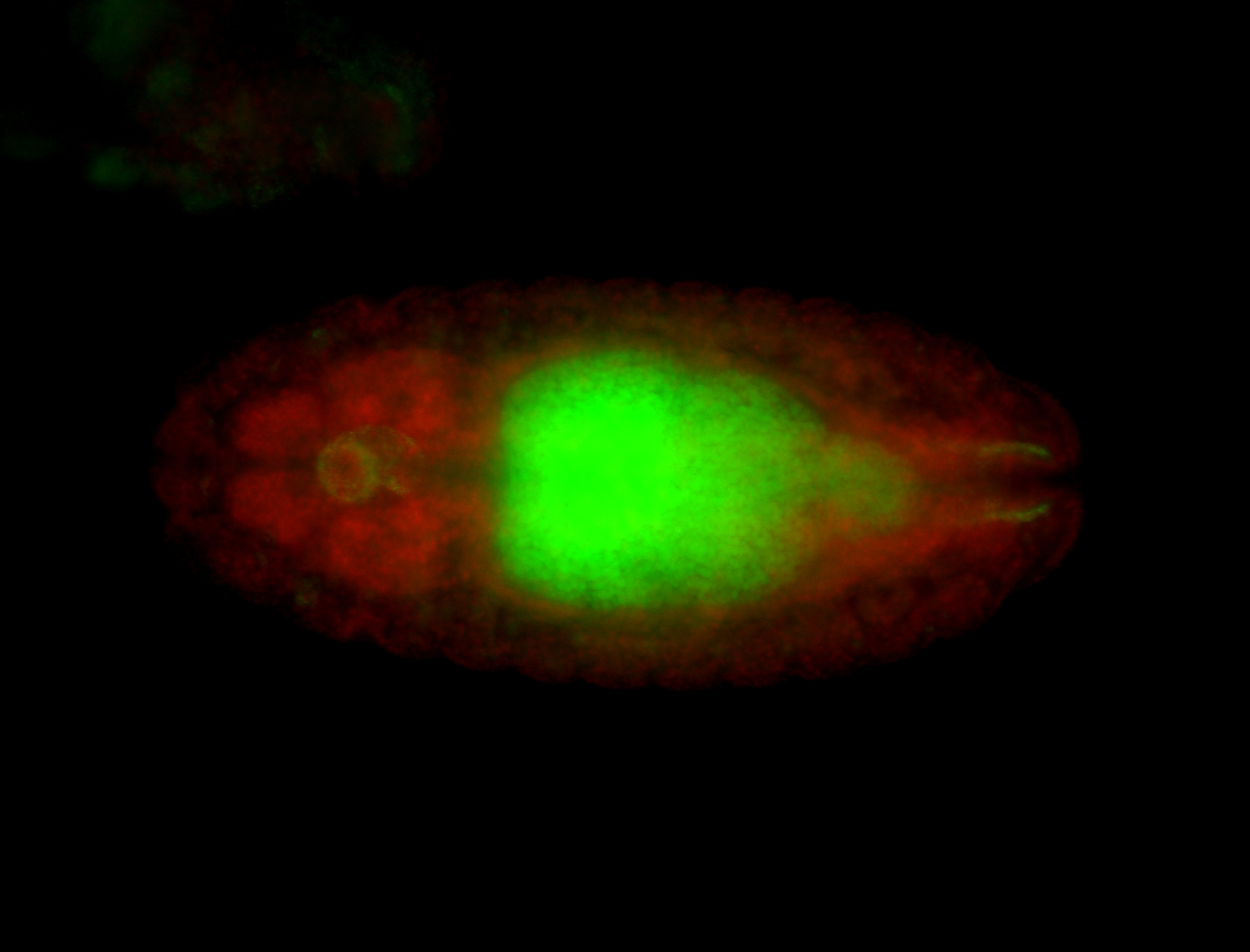
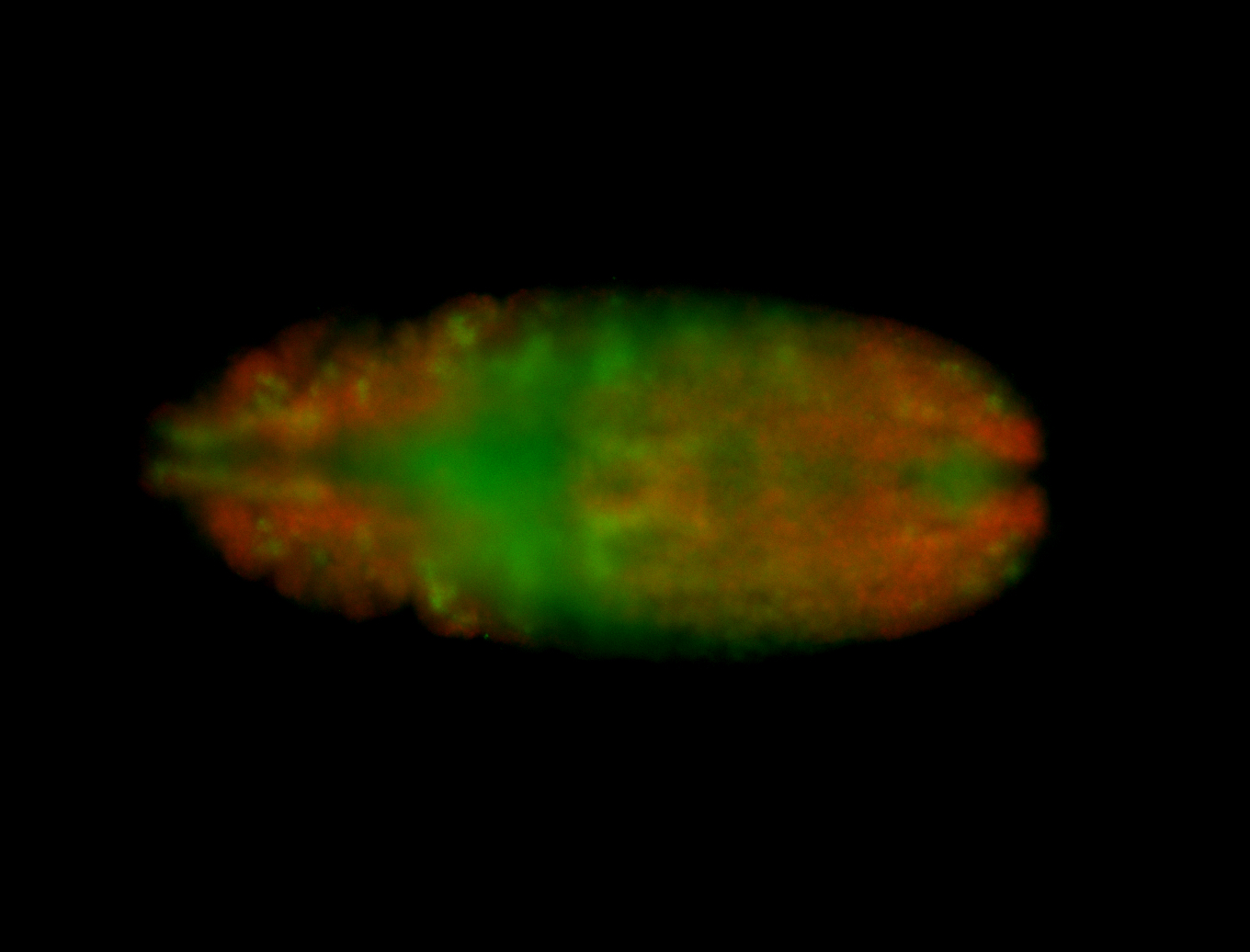
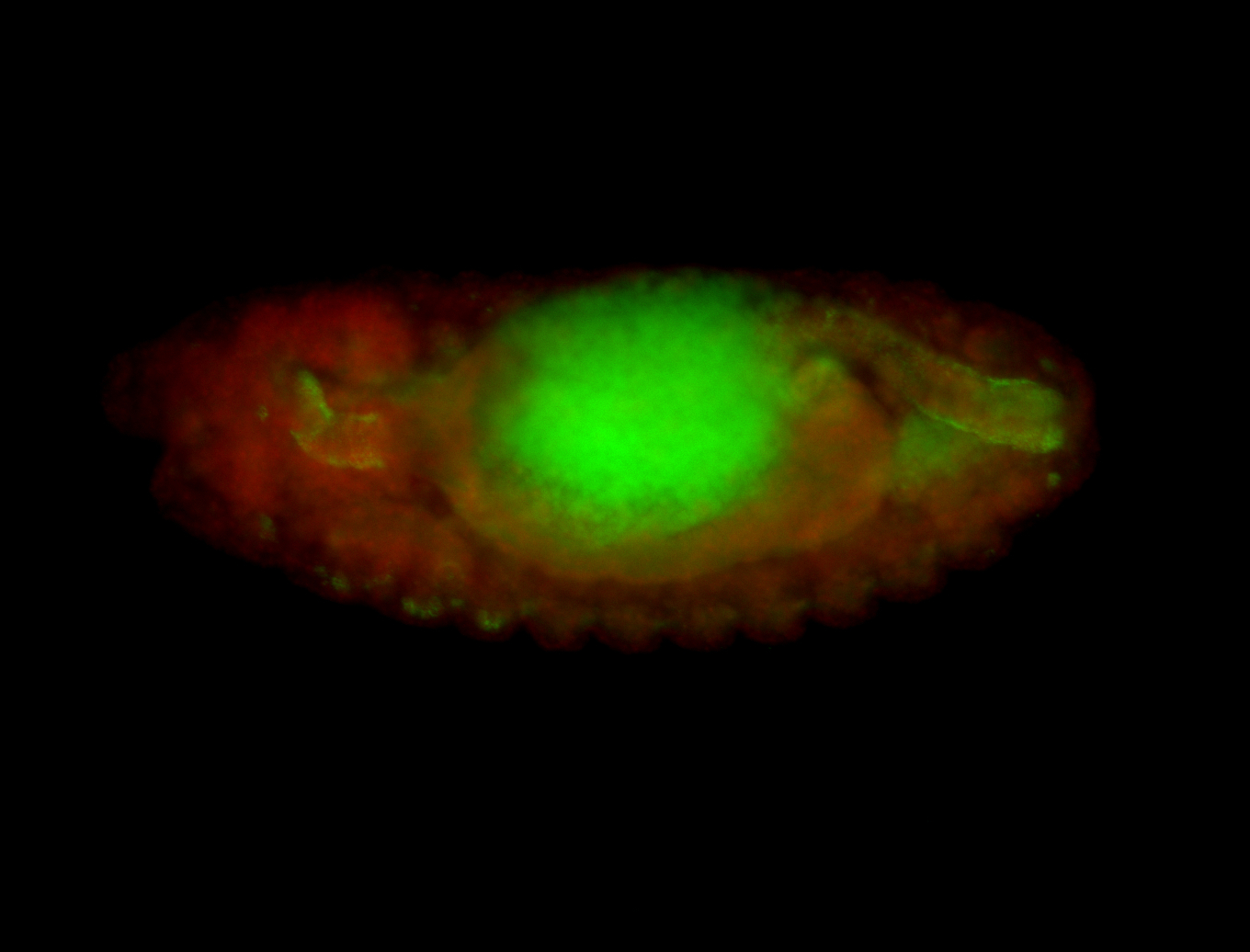
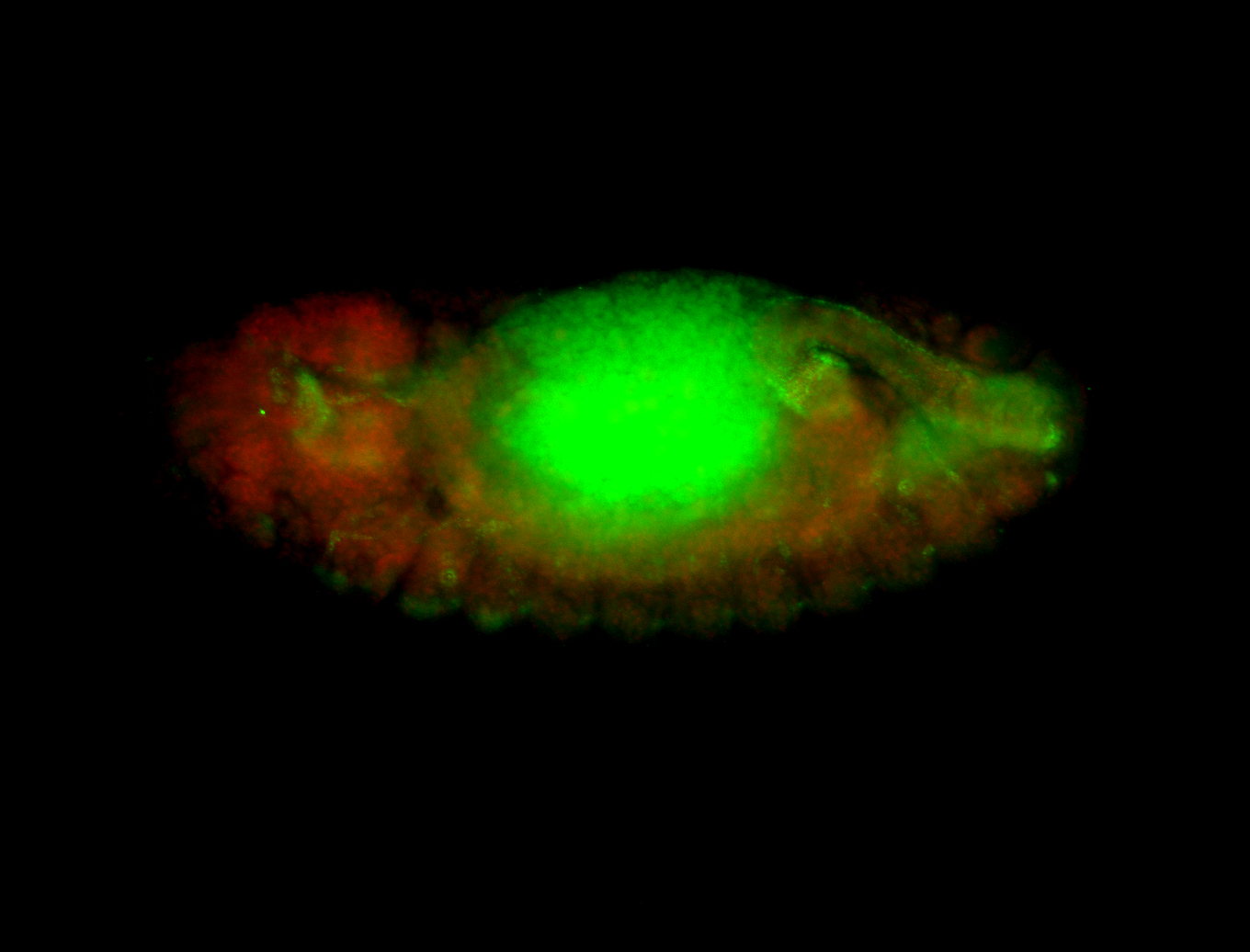
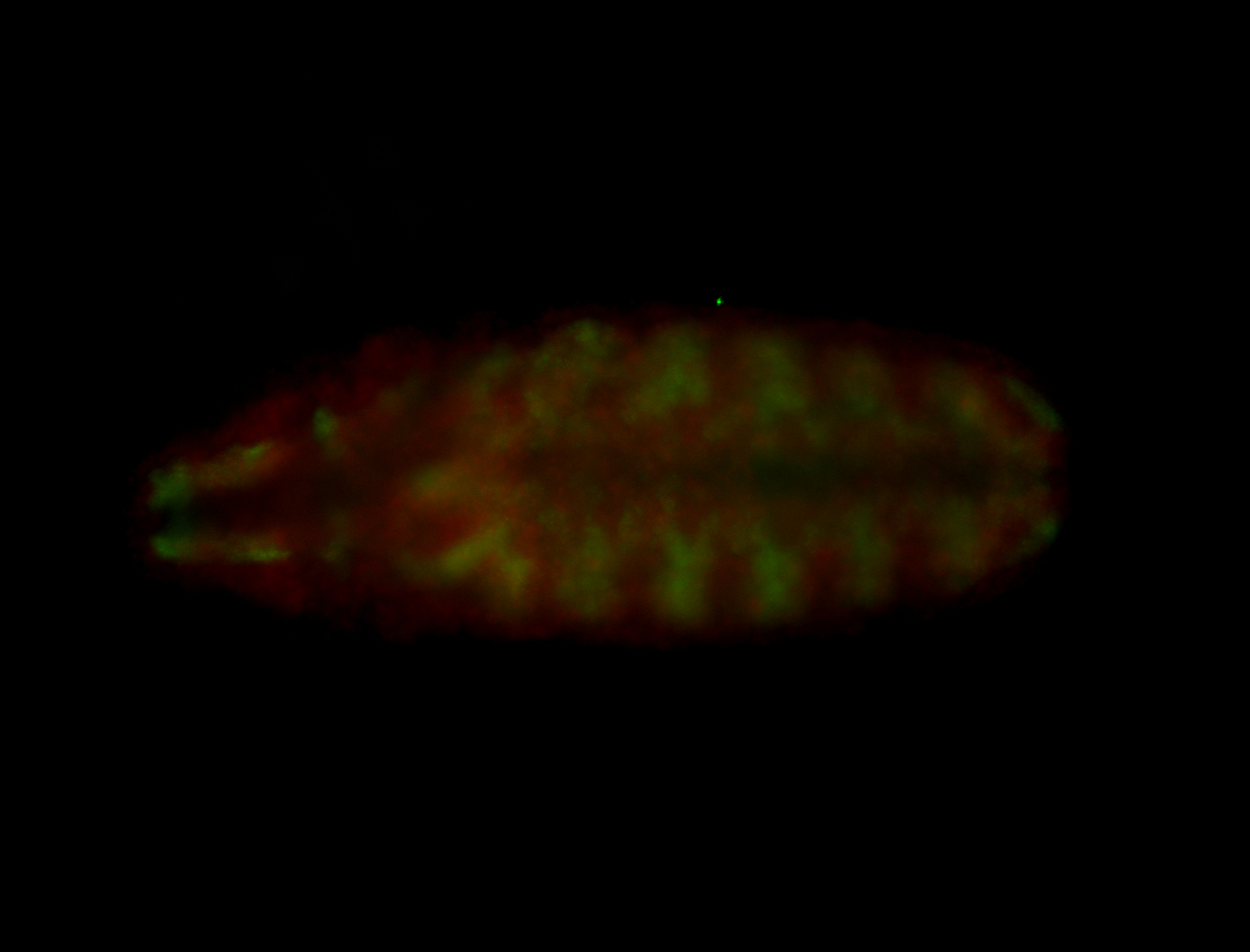
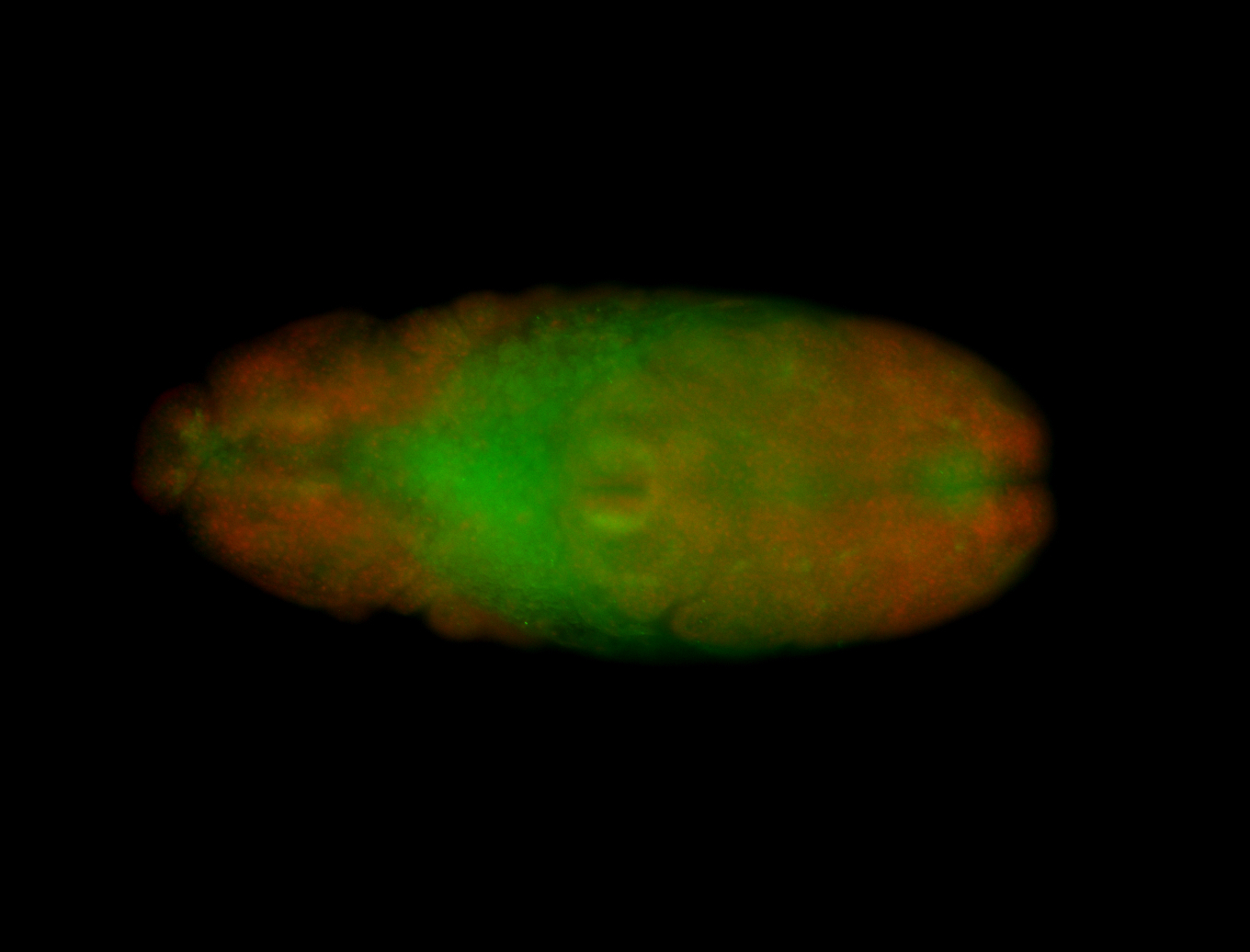
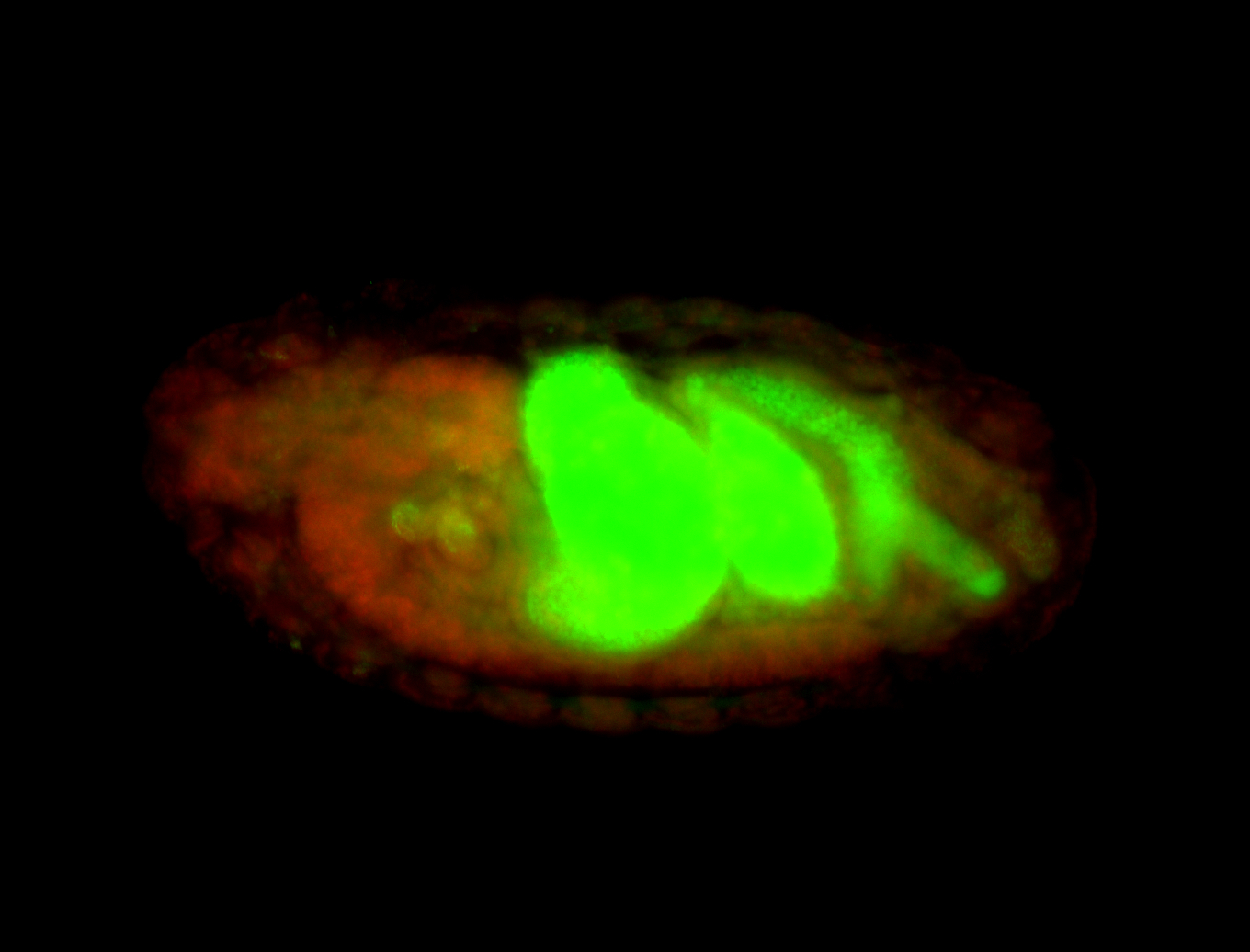
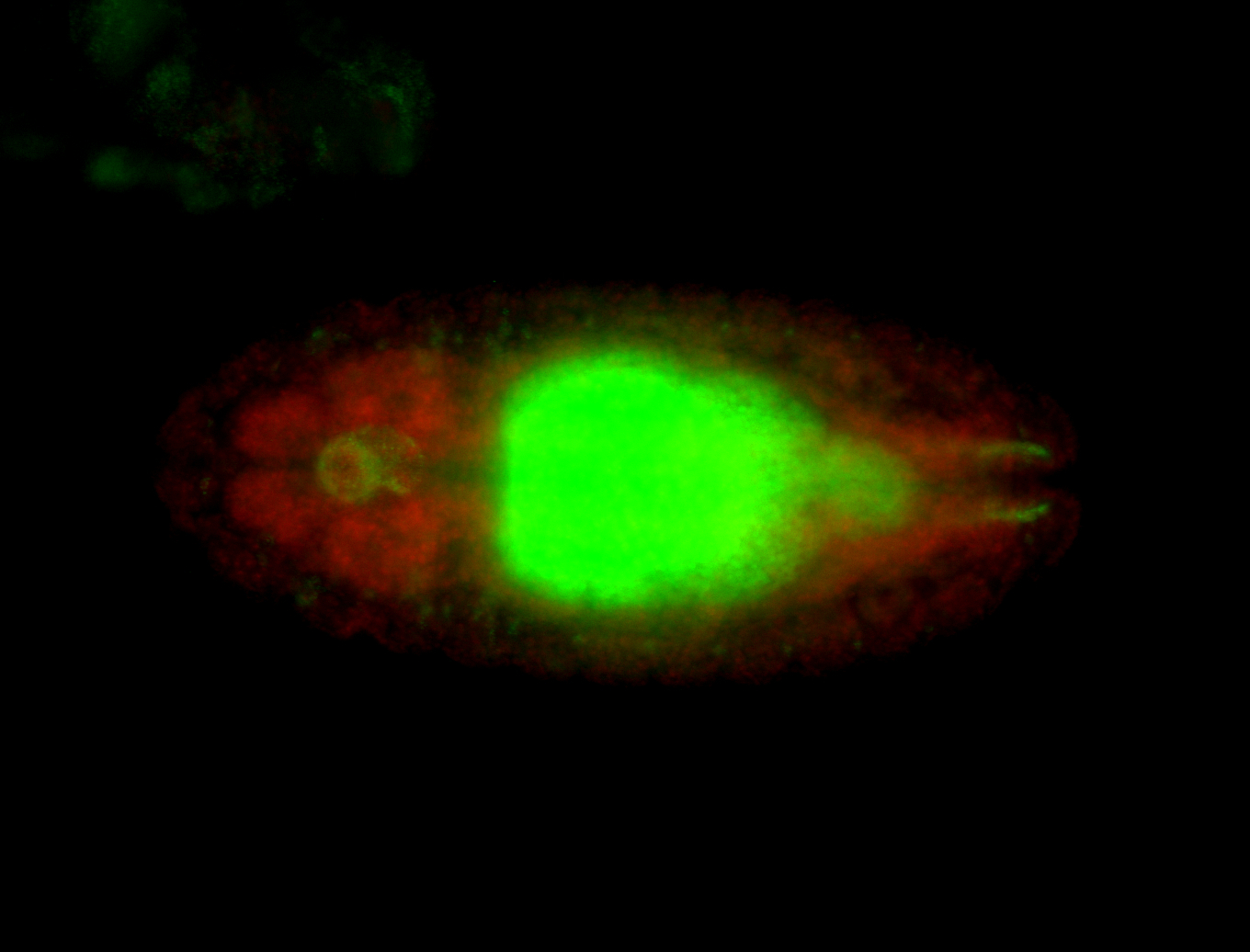
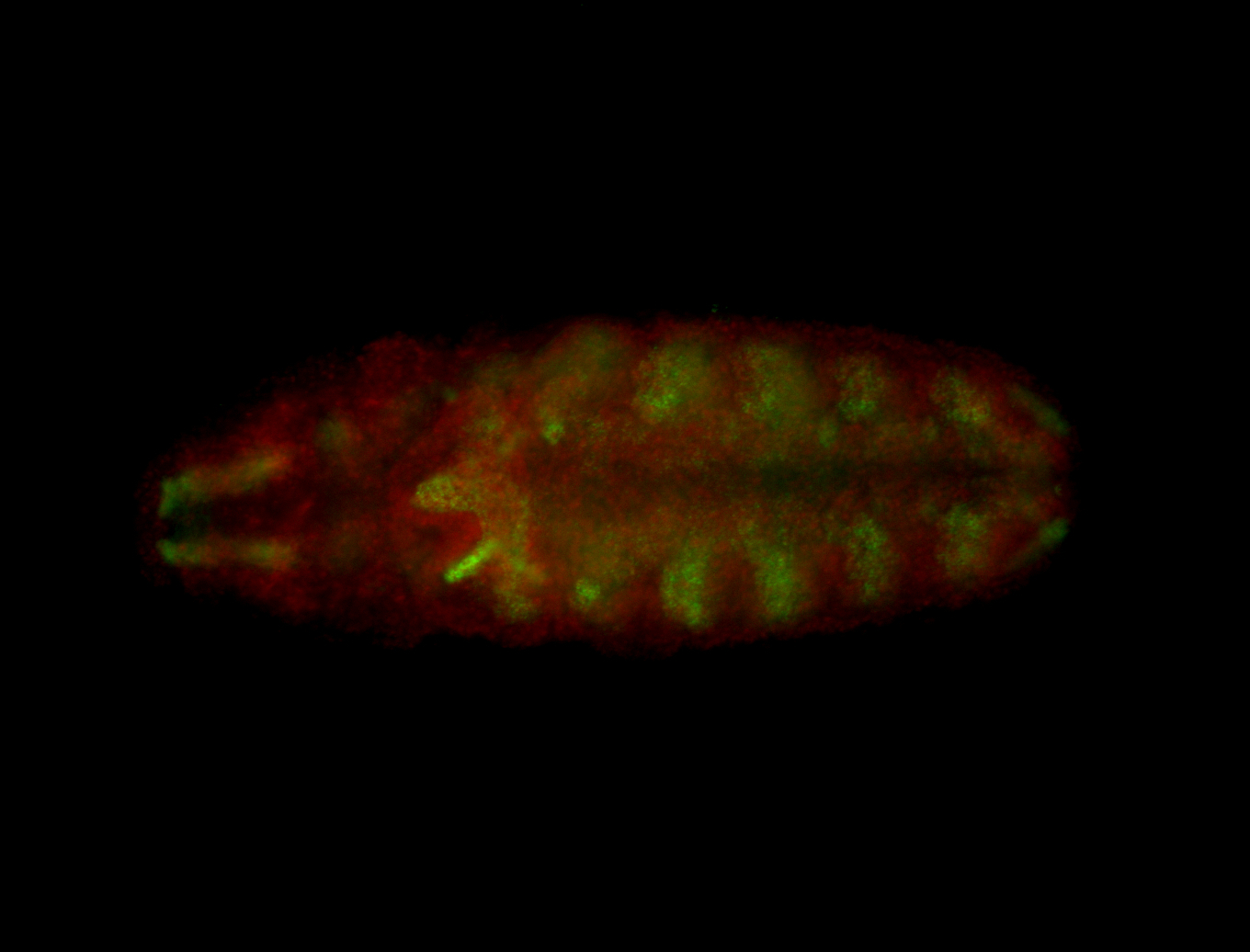
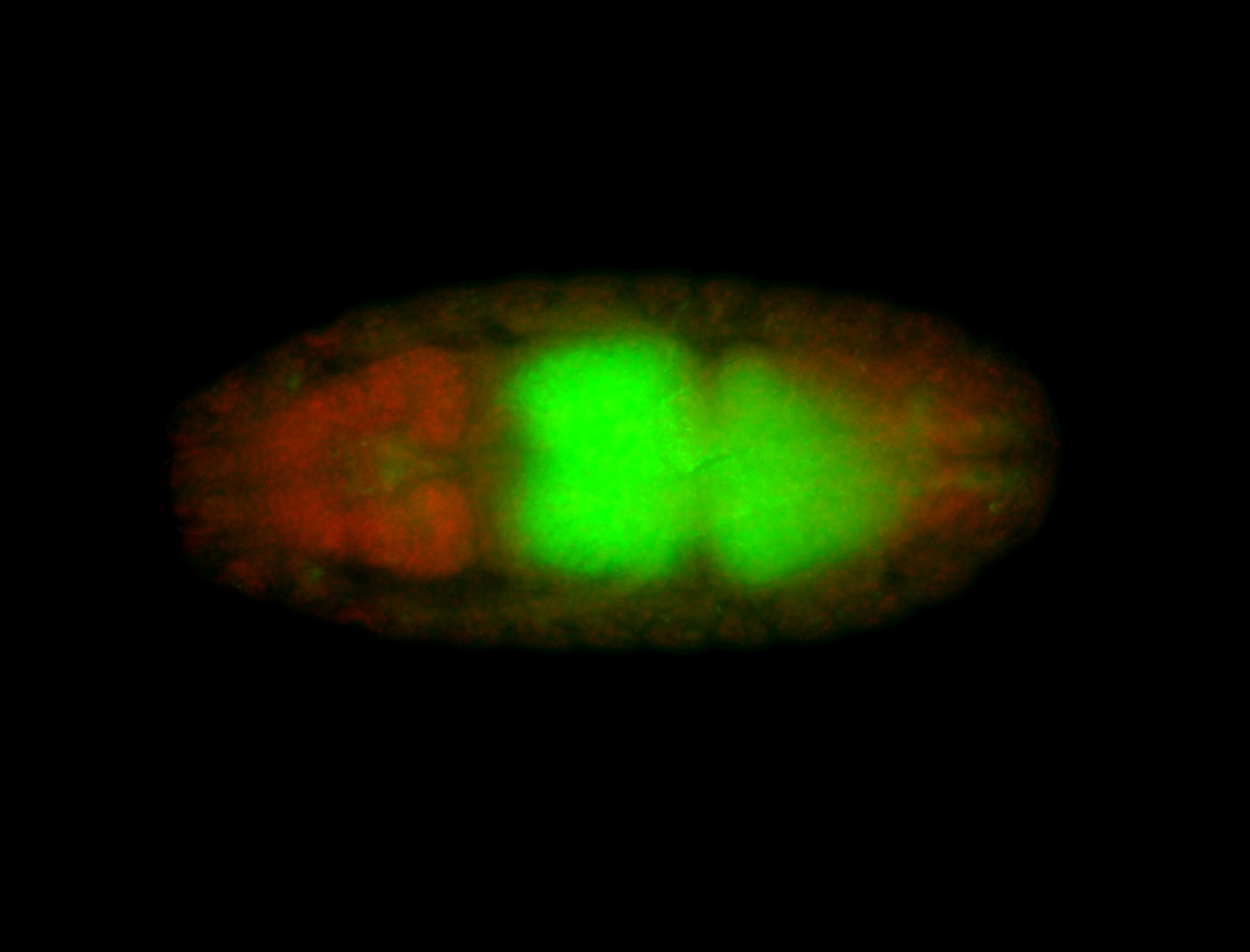
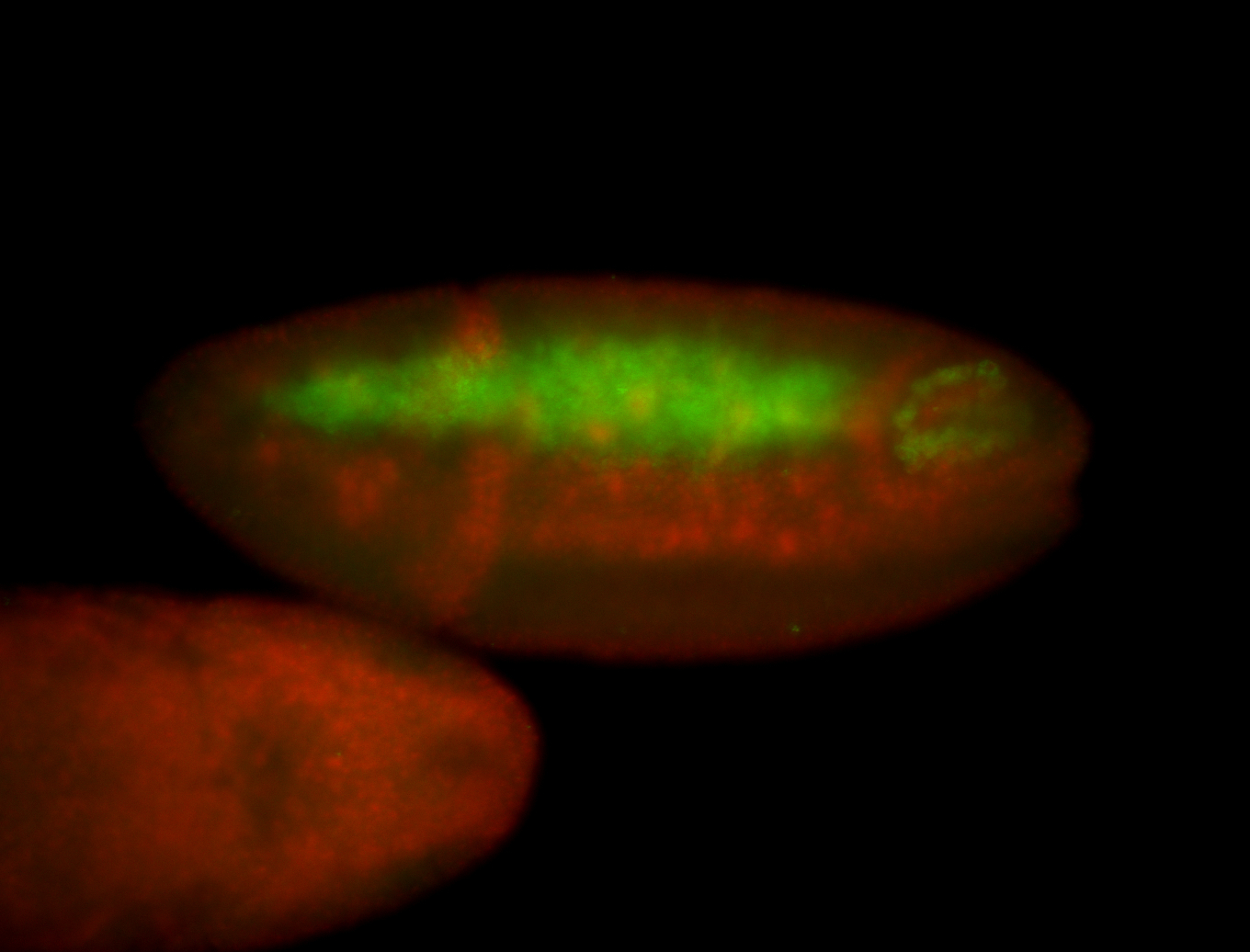
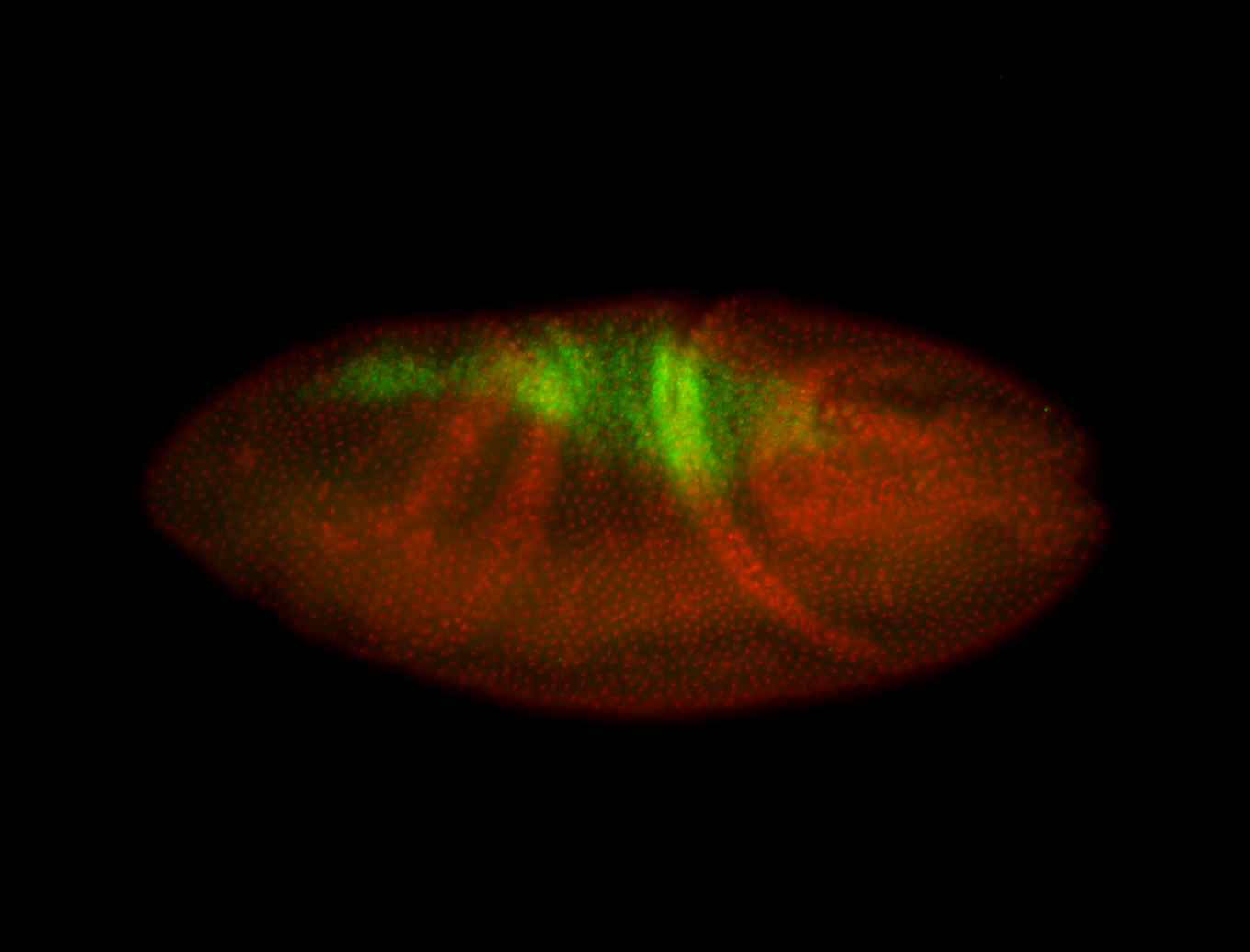
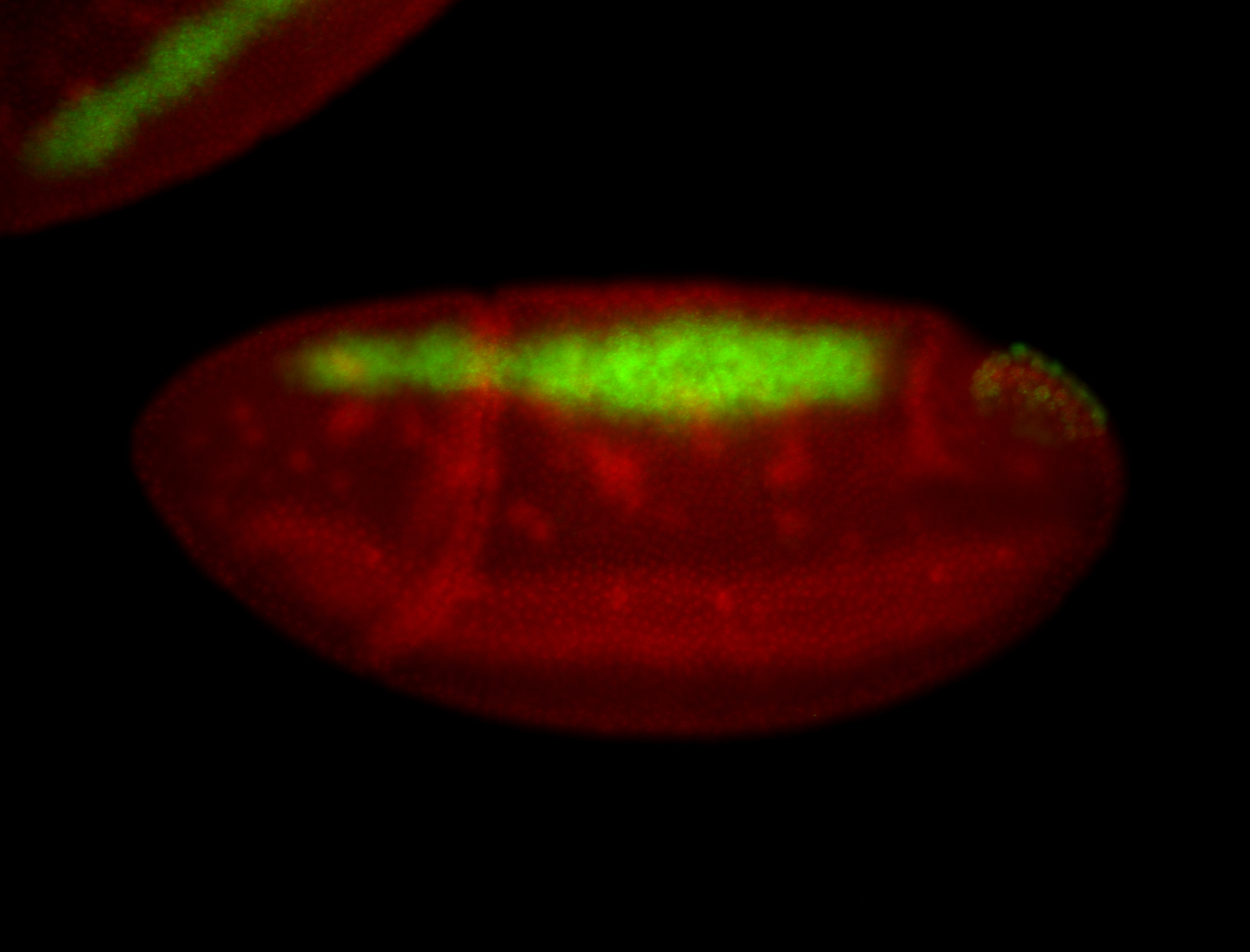
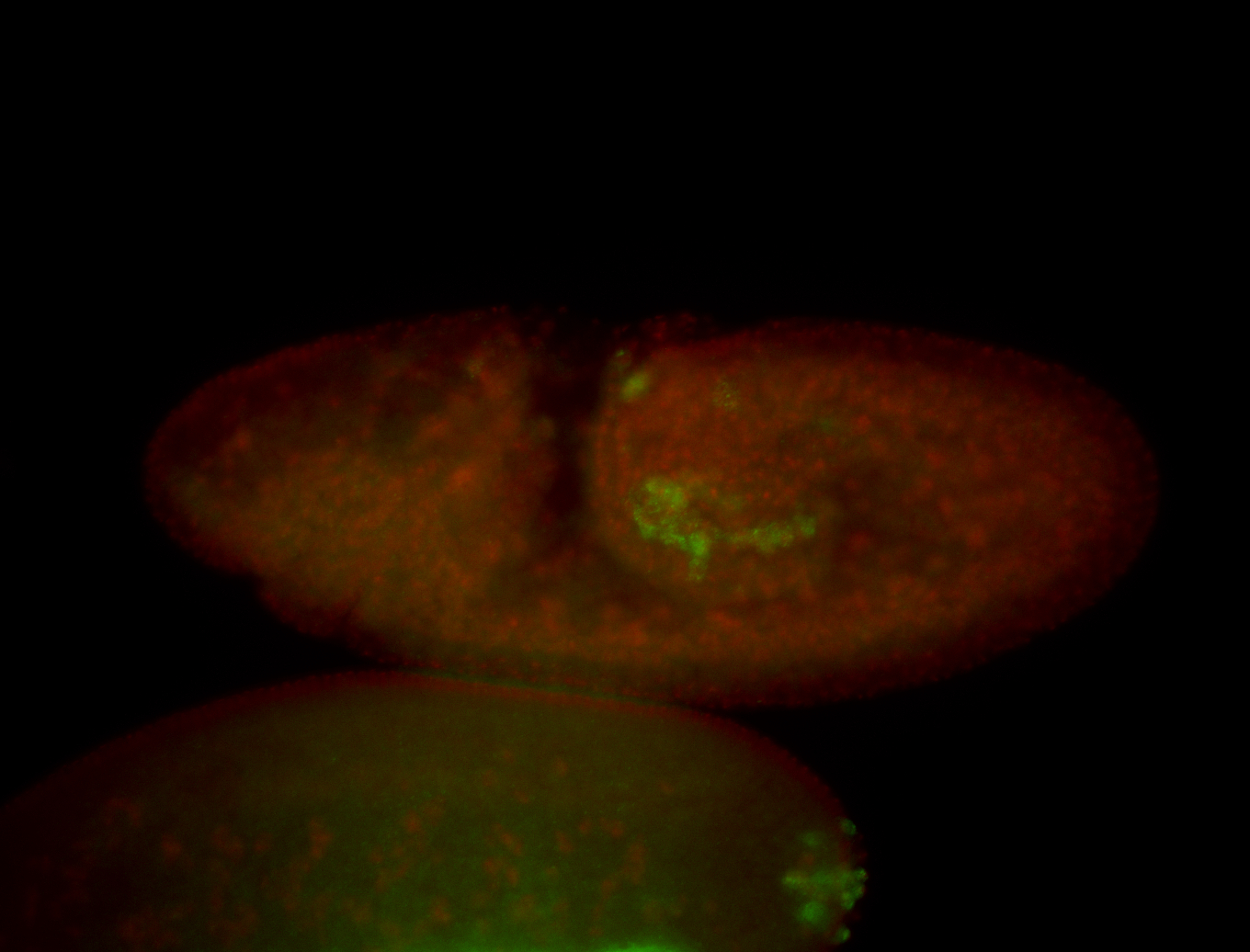
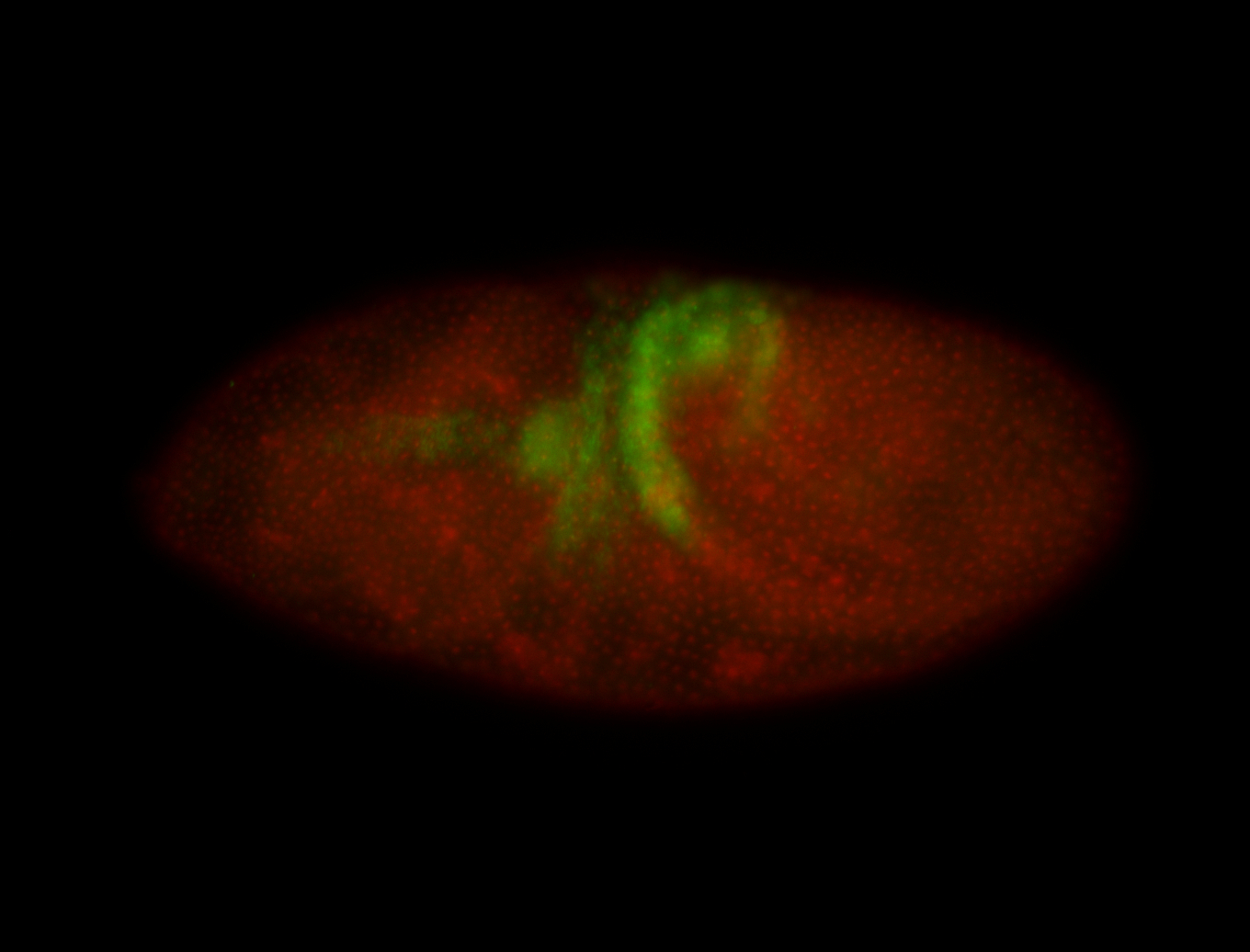
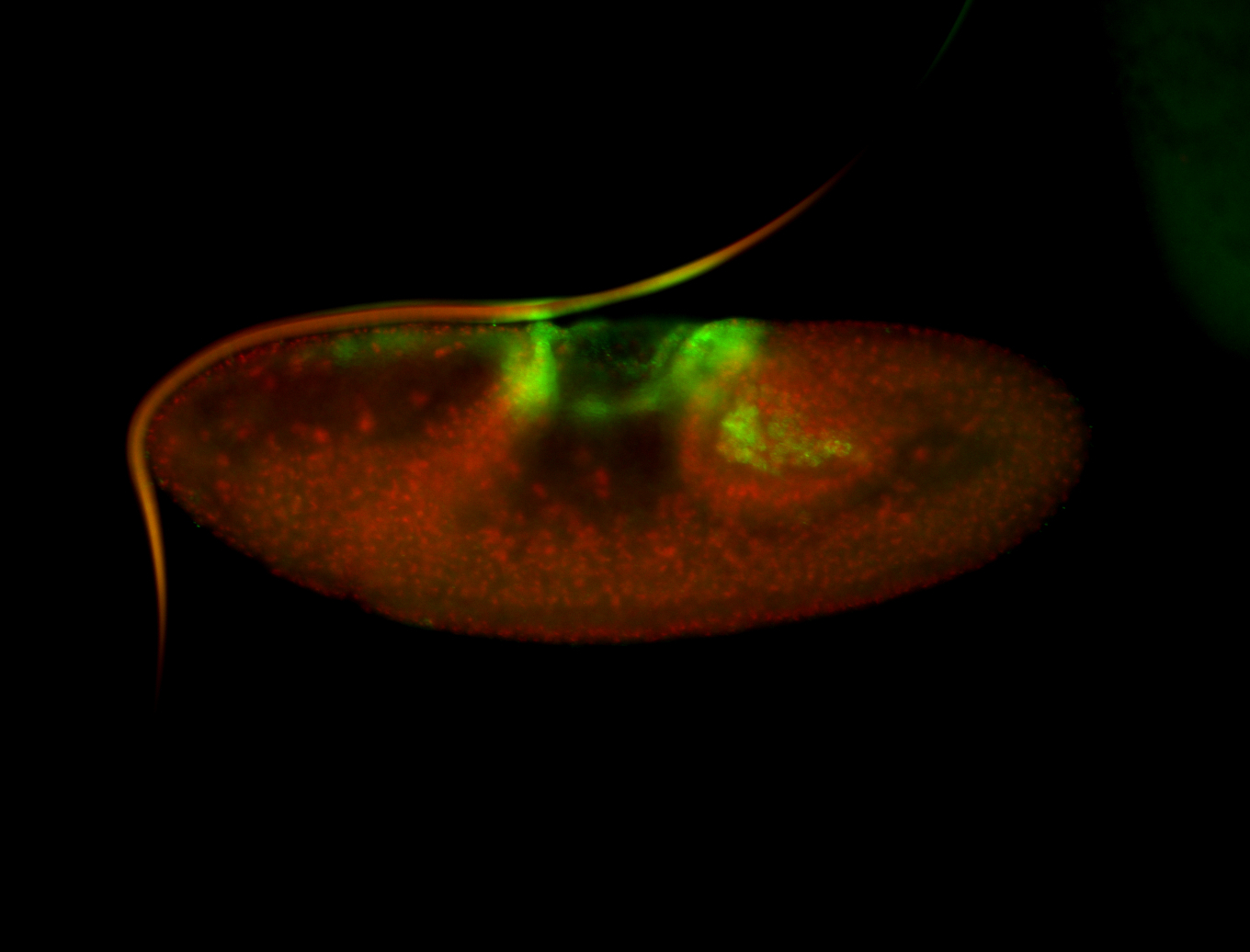
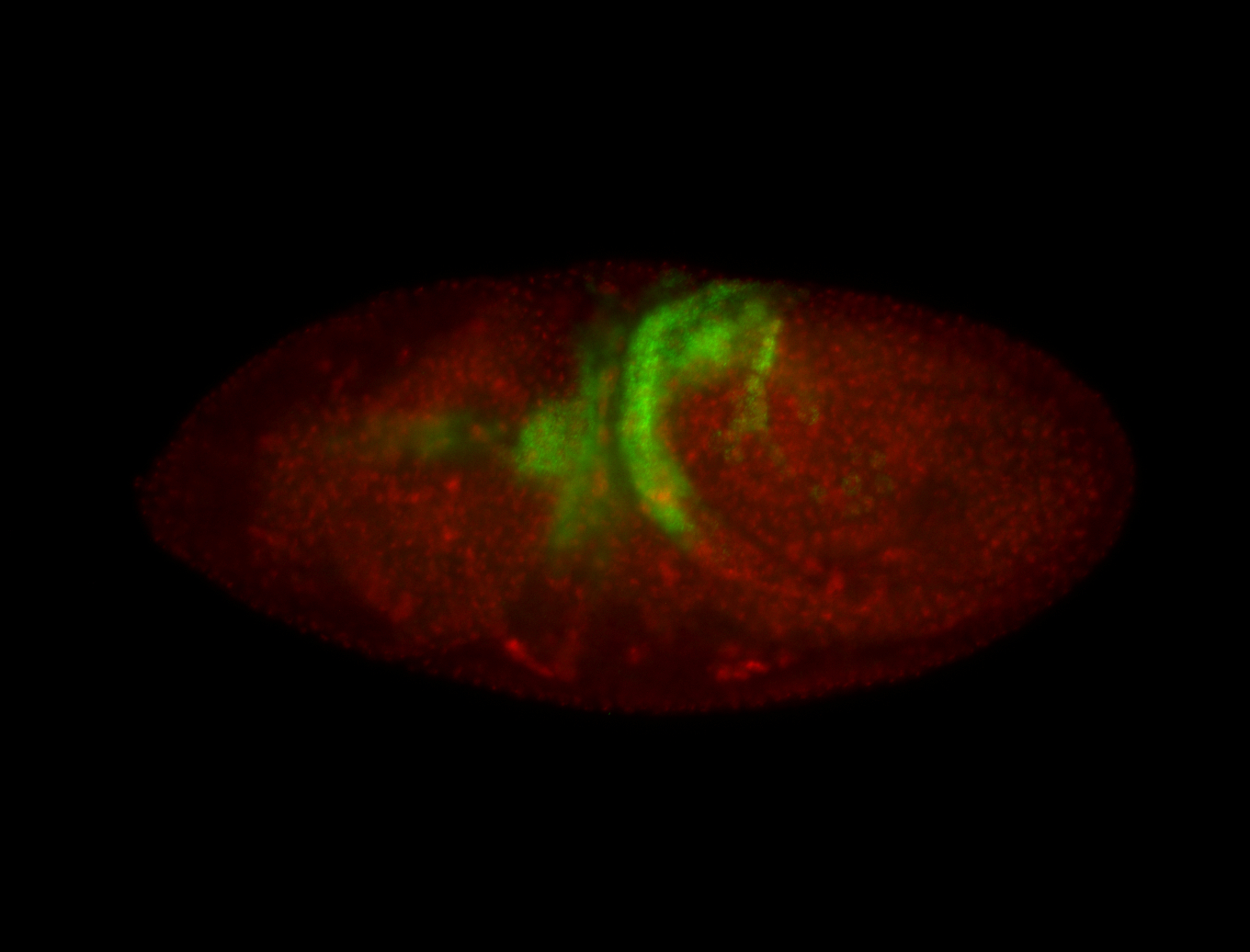
For a full list of searchable terms, please see the term page.
NOTE: It takes a few seconds for the term suggestions to load.
Result Summary
| Number of genes | 28 | |
| Export | ||
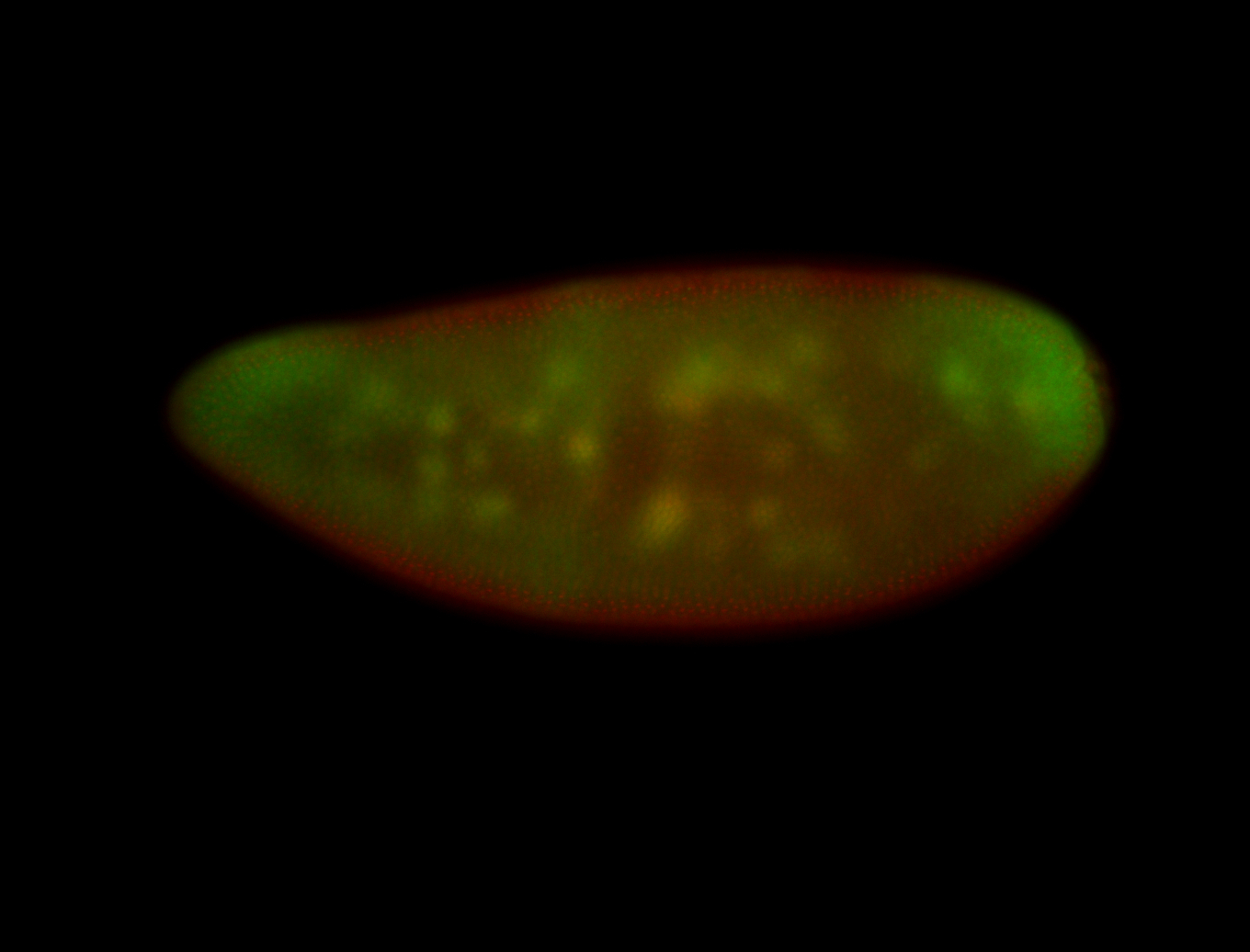
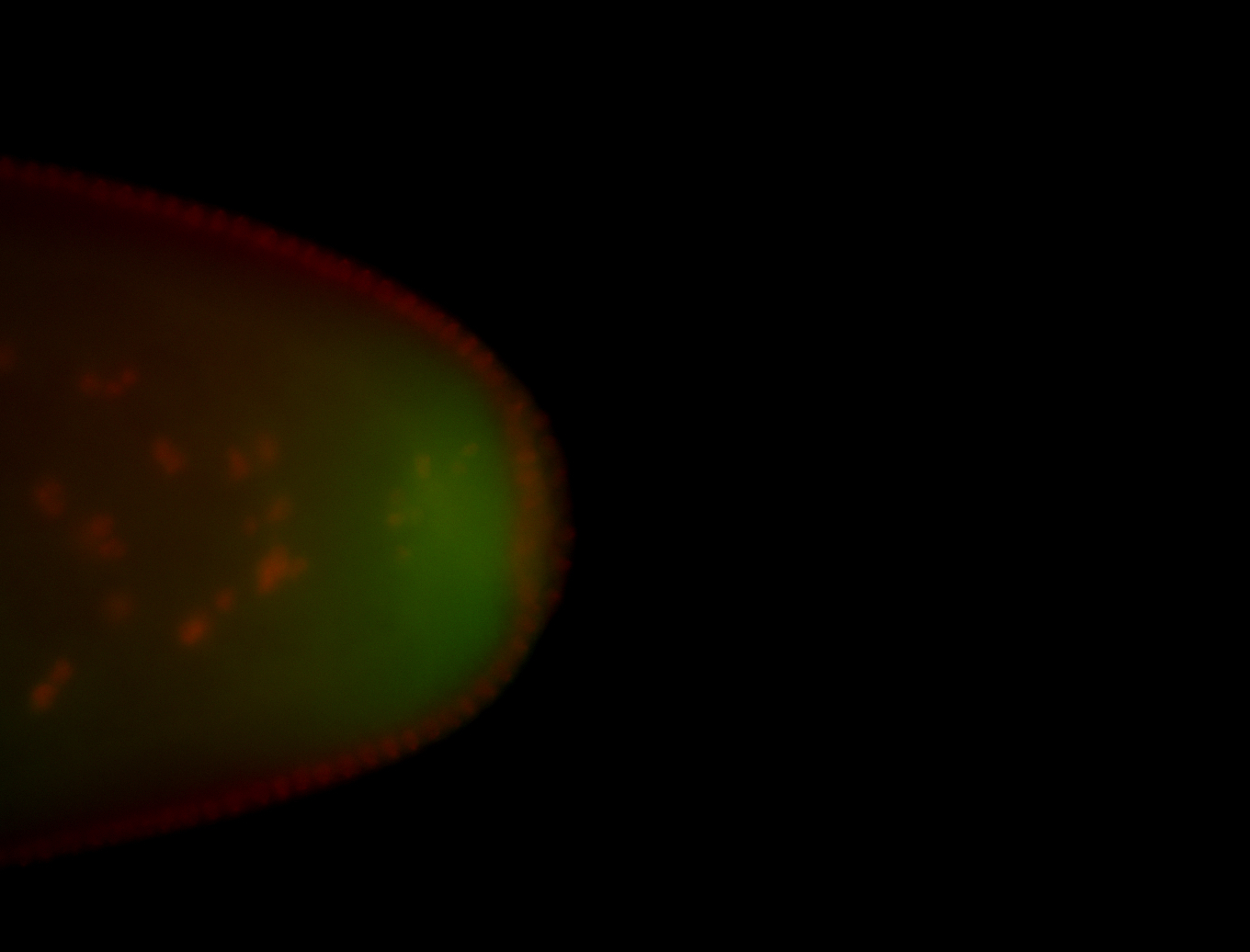
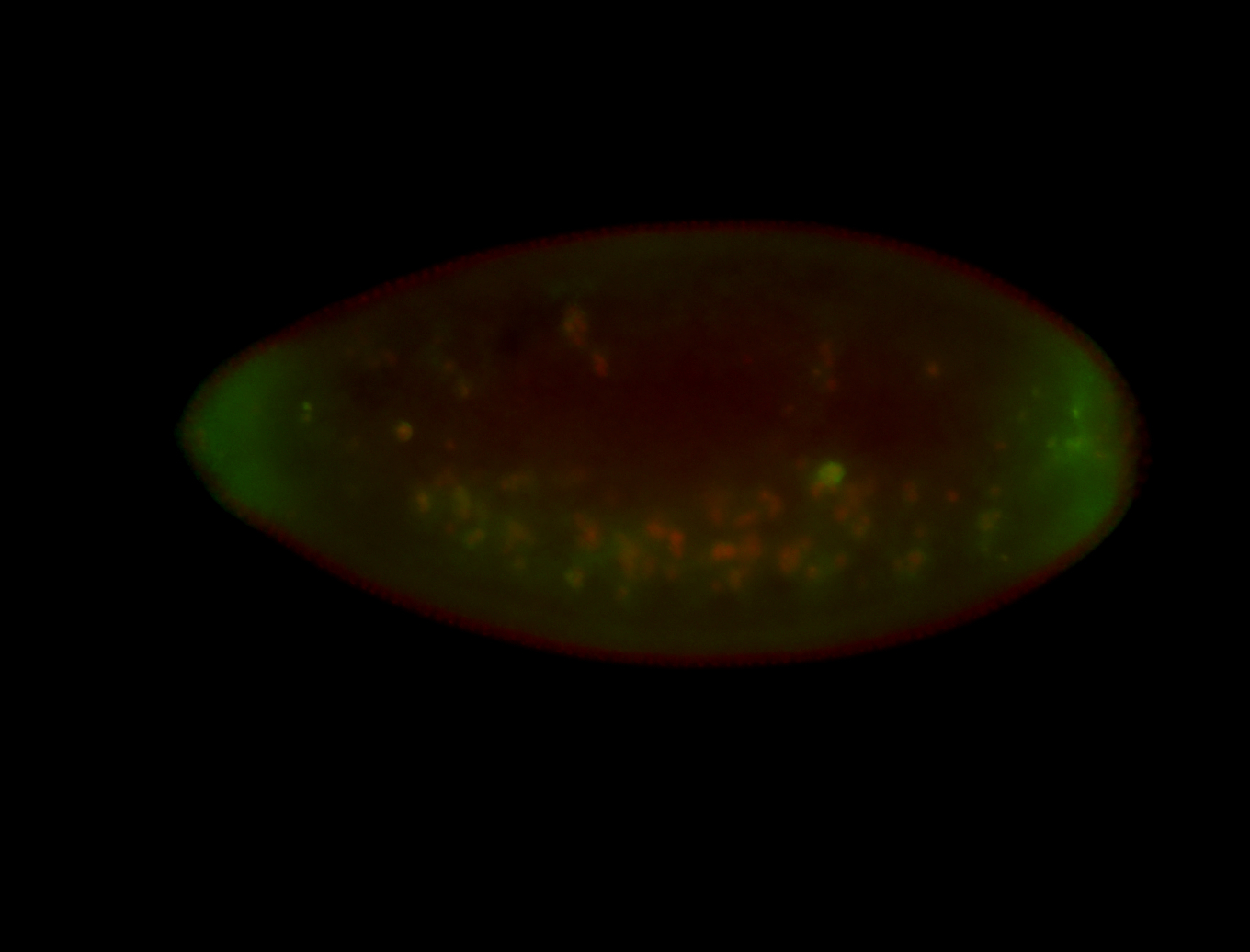
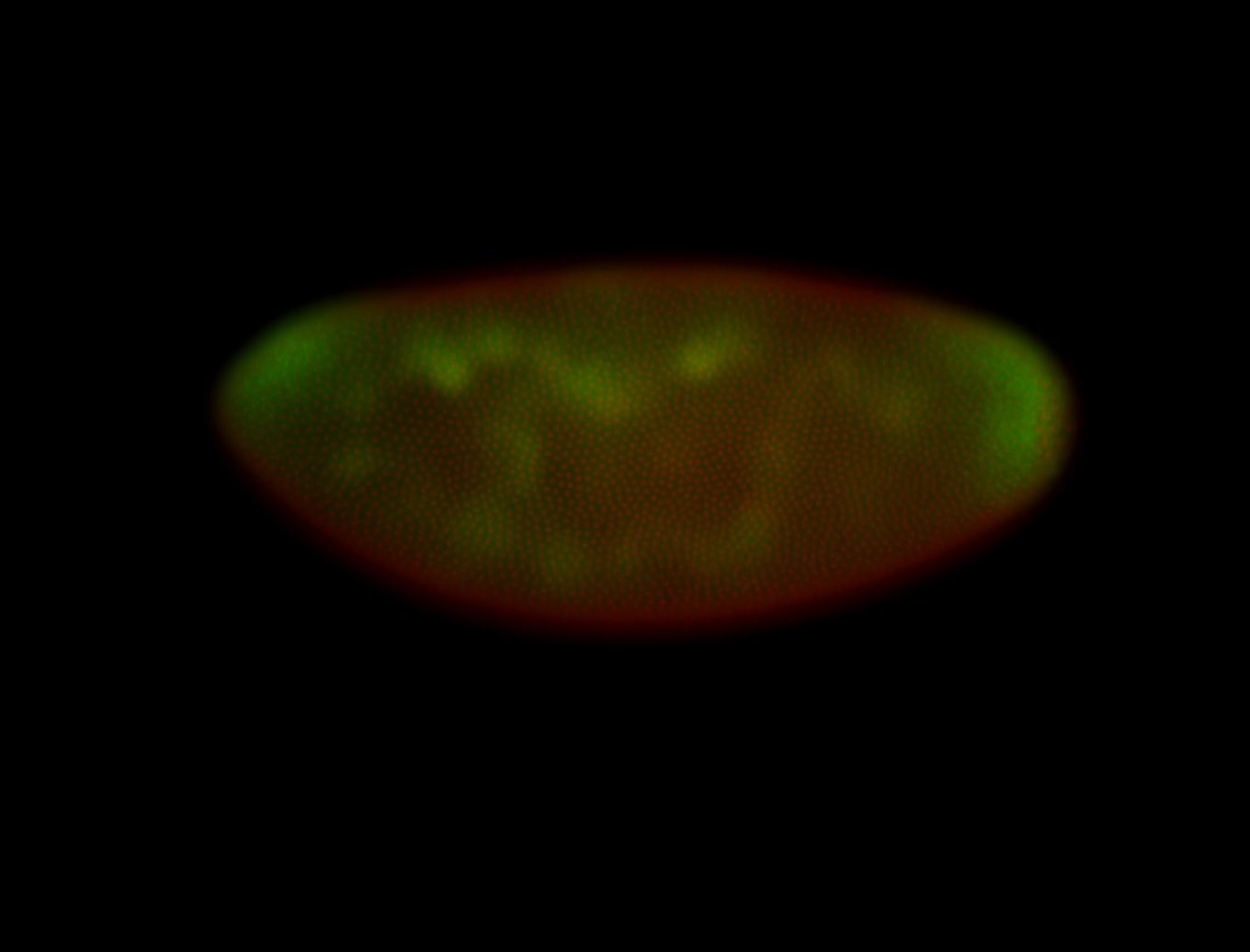
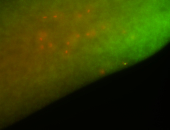
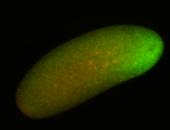
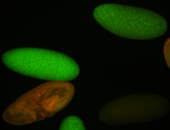
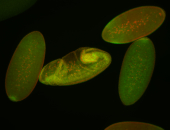
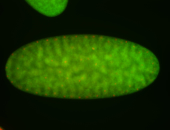
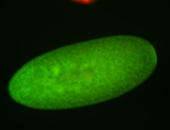
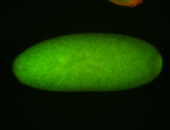
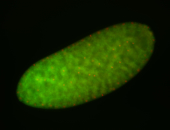
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Very low probe on gel. |
| Very interesting patterns. |
| Salivary glands, probably non-specific. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Probe CG16815-RB Links: Permalink
Genes: CG16815
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RB isoform transcript |
Probe CG31131-ex Links: Permalink
Genes: CG31131
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting common exons |
| -modENCODE RNAseq data on FlyBase shows low/no expression, however, we observed strong expression patterns throughout development |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Weak probe, good insitu. |
Embryo
Larva
Adult
Comments
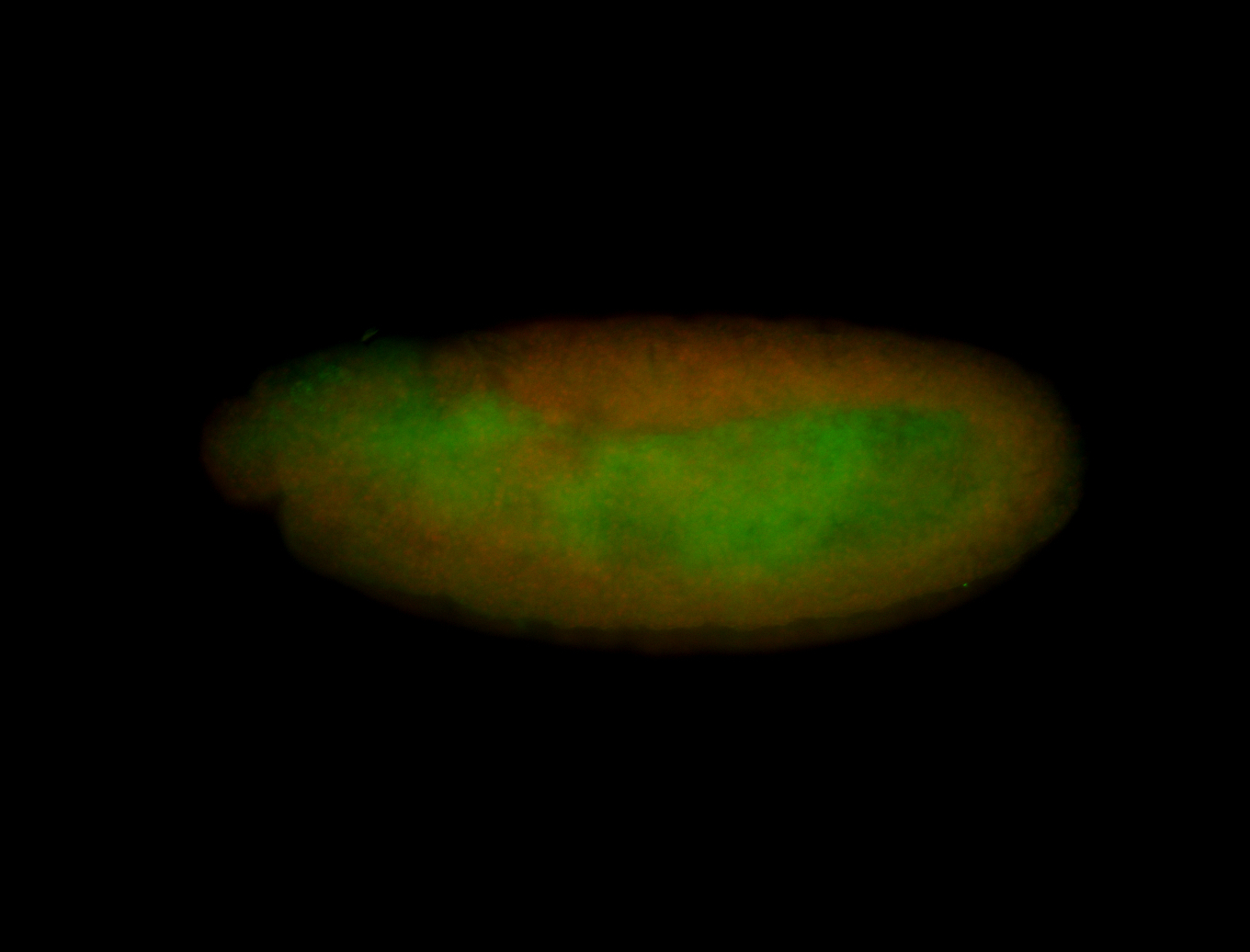
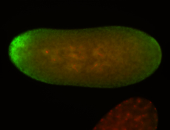
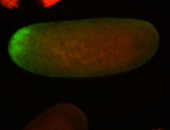
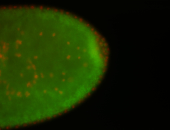
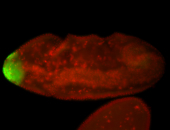
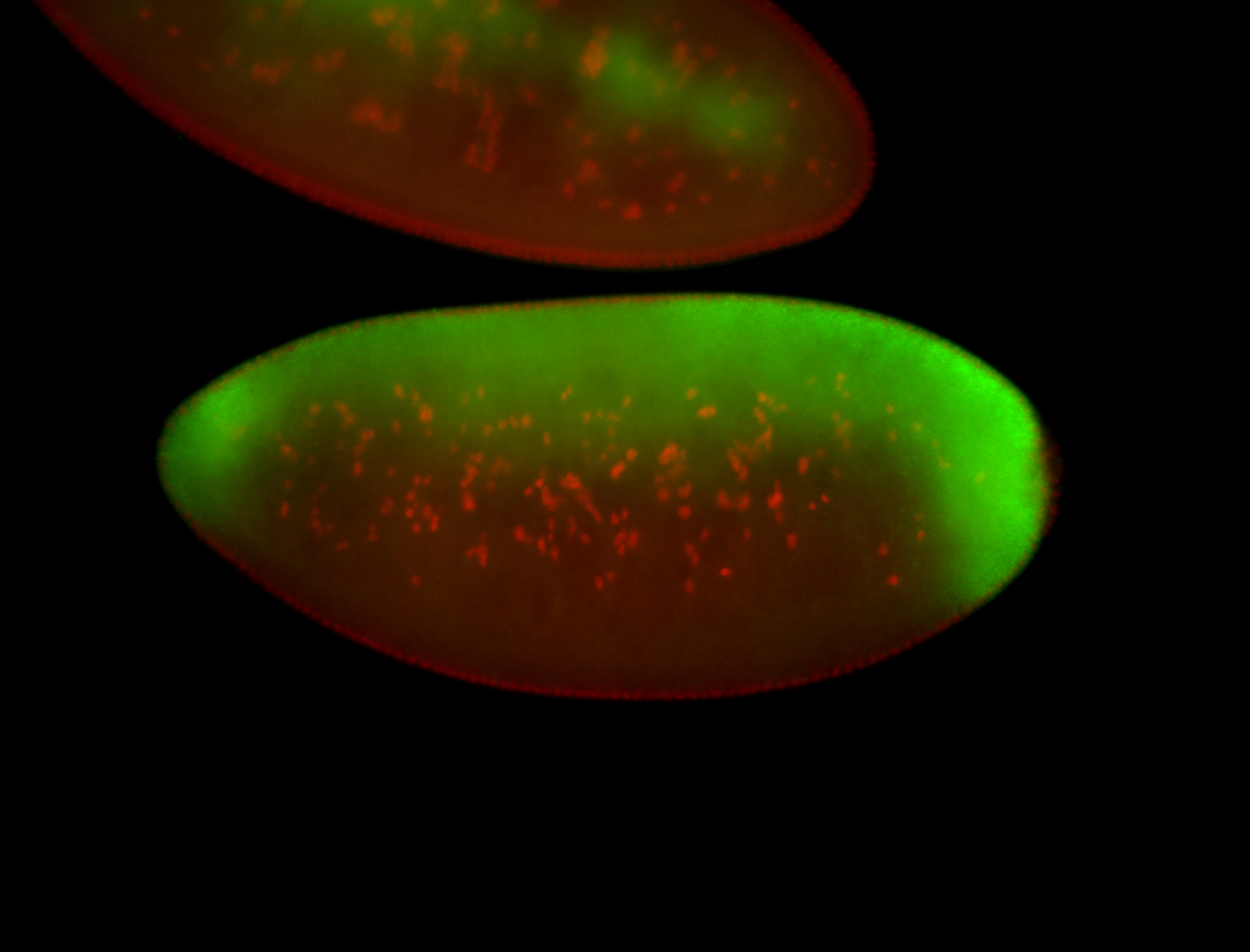
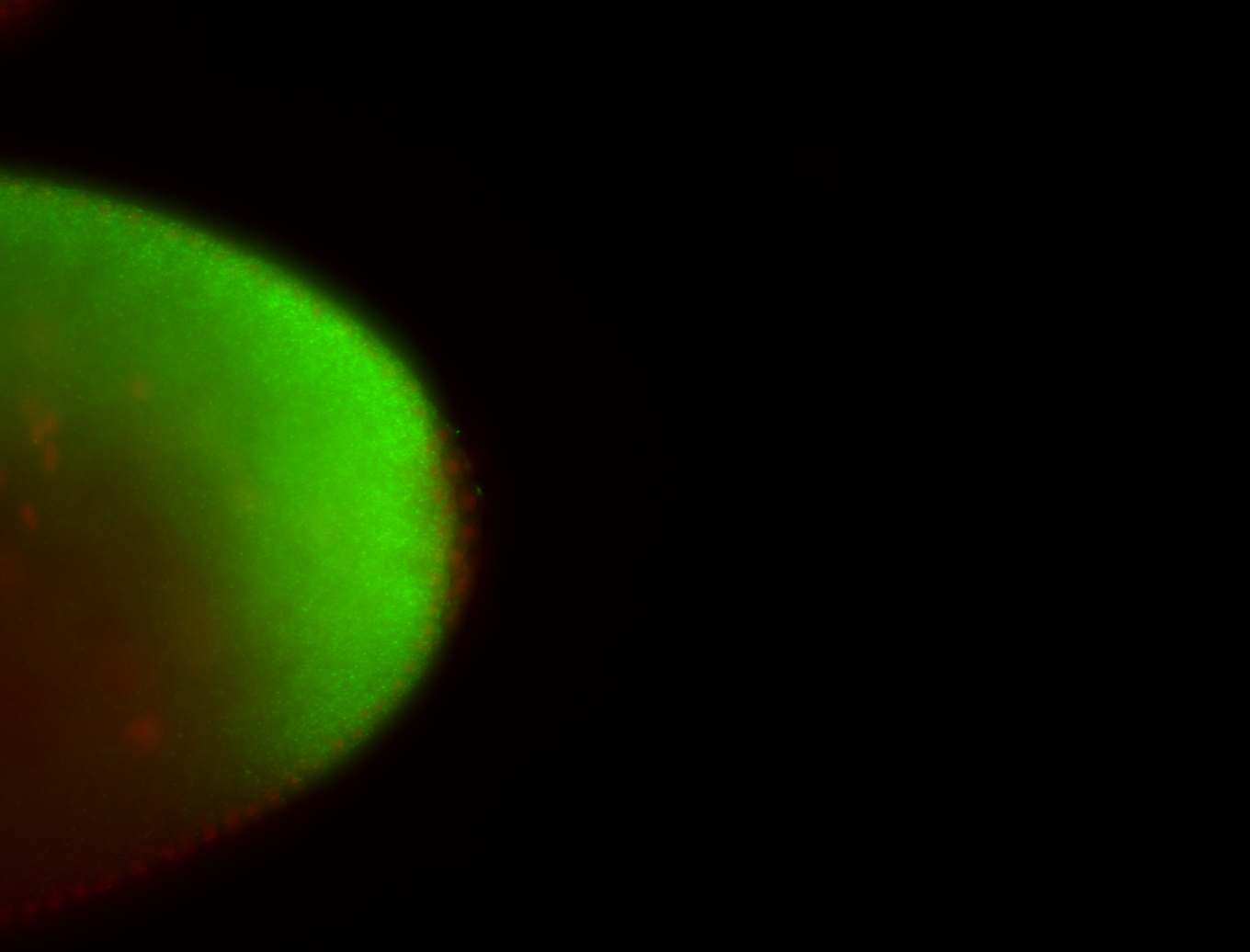
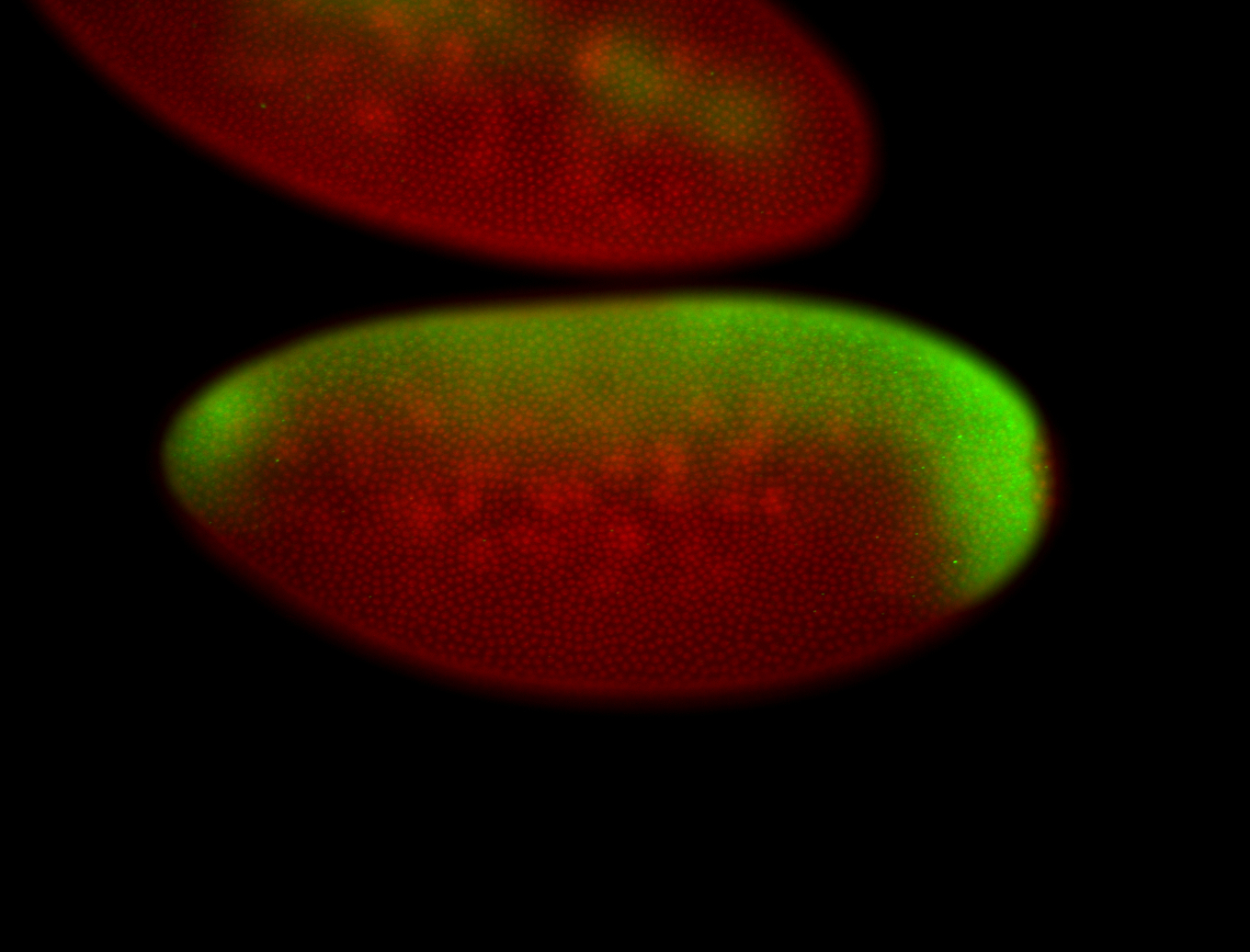
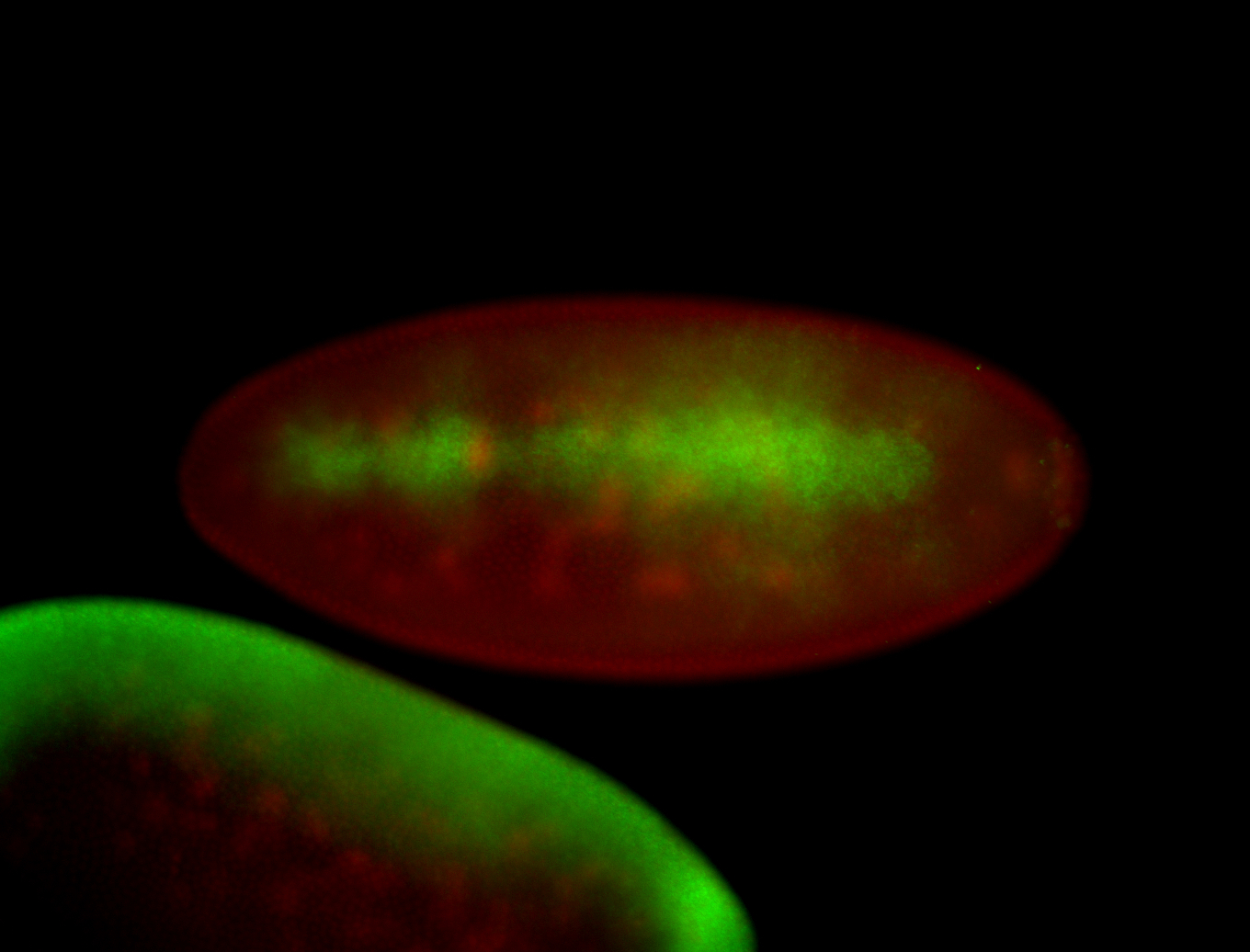
| All nuclei. Strong in the Amnioserosa. |
| Enriched in the amnioserosa, the central nervous system, the ventral midline, and maybe the tracheal pits. |
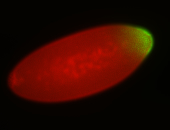
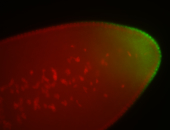
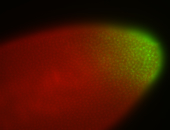
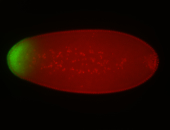
| Anterior to posterior gradient Subnuclear localization. |
| The probe is low, but good. Nice insitu data!!! |
Probe LP10071 Links: Permalink
Genes: Ef1alpha48D
Embryo
Larva
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe on gel. |
| Maybe membranes... need better data. |
Probe Hsromega-ex Links: Permalink
Genes: Hsr-omega
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting common exons |
| -lcnRNA |
Embryo
Larva
Adult
| Area | Images | Terms |
|---|
Comments
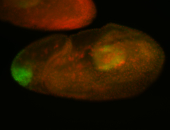
| Good probe on gel Goes from all-over to 'posterior enriched'. |
| Anterior zygotic and posterior maternal expression. |
Embryo
Larva
Adult
| Area | Images | Terms |
|---|
Comments
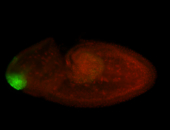
| Good probe on gel. |
| Sub and peri nuclear with apical enrichment. Anterior cells only. Very specific and interesting. |
| It is hard to distinguish if the axons in the larval ventral nerve cord are 'axons' or 'tracheal extensions' |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -There is no entry for this gene on BDGP, and modENCODE RNAseq data on FlyBase indicates that it is not expressed in the embryo. However, we observed obvious patterns during embryogenesis. Additionally, FlyBase does not have any in-depth characterizations of this gene. |
| -Osi21 does not overlap with any other genes, ruling out the possibility of the targeting of alternative transcripts. BLAST also confirms the specificity of our probes to Osi21. Additionally, no other genes for which probes were designed for on this plate show patterns such as those observed here, ruling out the possibility of cross-contamination during probe preparation. |
| -This may be a result of Osi21's poor characterization or incorrect RNAseq data |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
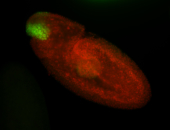
| Note: this in situ was done twice with the same result The next gene on the plate has a similar expression pattern |
| Nice anterior to posterior gradient. Apical expression. Cytoplasmic foci also in a gradient anterior to posterior. Very interesting. |
Probe LD29301 Links: Permalink
Genes: Su(var)2-HP2
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| re-annotated |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe on gel Maybe some small foci. |
Embryo
Larva
Adult
| Area | Images | Terms |
|---|
Comments
| Degrades. |
| Very interesting. |
| Perinuclear around neuroblasts. |
| Anterior dorsal and posterior dorsal enriched and apically enriched on these cells as well. |
| Good probe on gel. Amazing in situ. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Re-annotated |
| Neuroblasts |
Embryo
Larva
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Central nervous system |
| Anterior gradient. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Apical, anterior to posterior gradient. |
| Anterior to posterior gradient! |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe on gel |
| It may be membrane associated... a closer look is required to determine this Interesting zygotic expression. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting common exons |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Re-annotated |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Anterior ot posterior gradient and apical to apical/lateral enrichment in epithelial cells. |
| Anterior to posterior gradient! |
| Segmented zygotic expression. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -retested using HCR-FISH 2025 for full coverage of late stage expression patterns |
| -performed using 3 probe pairs (HCR-FISH) against the RB isoform |
| -really interesting patterns! |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA transcript isoform |
| -neat expression patterns in early embryos and early germ band elongation stages; expression is no longer detectable after stage 9-10 (fits modENCODE RNA-seq data well) |