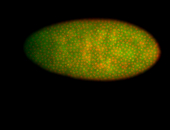
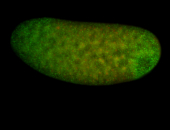
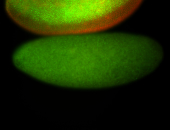
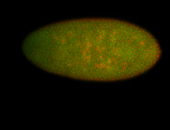
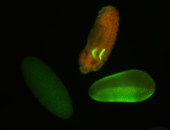
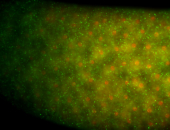
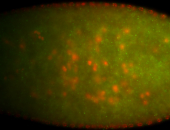
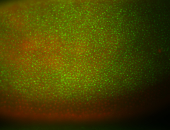
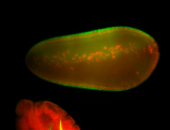
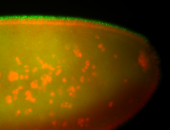
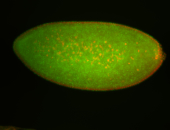
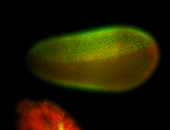
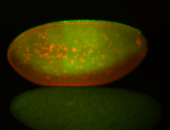
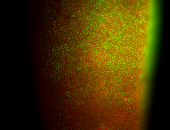
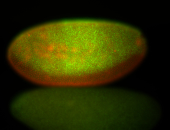
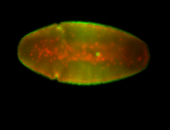
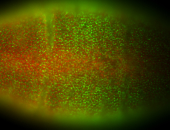
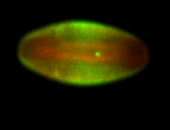
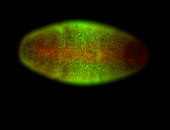
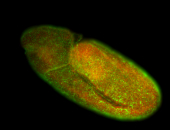
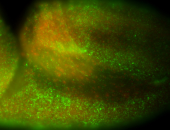
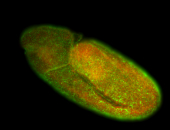
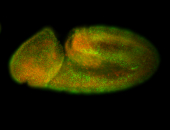
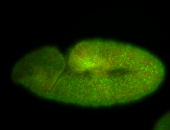
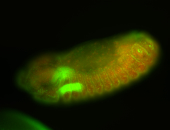
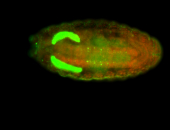
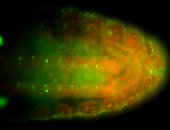
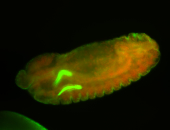
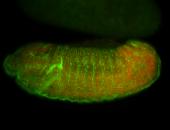
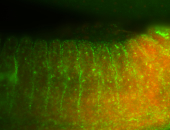
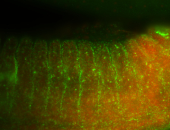
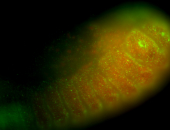
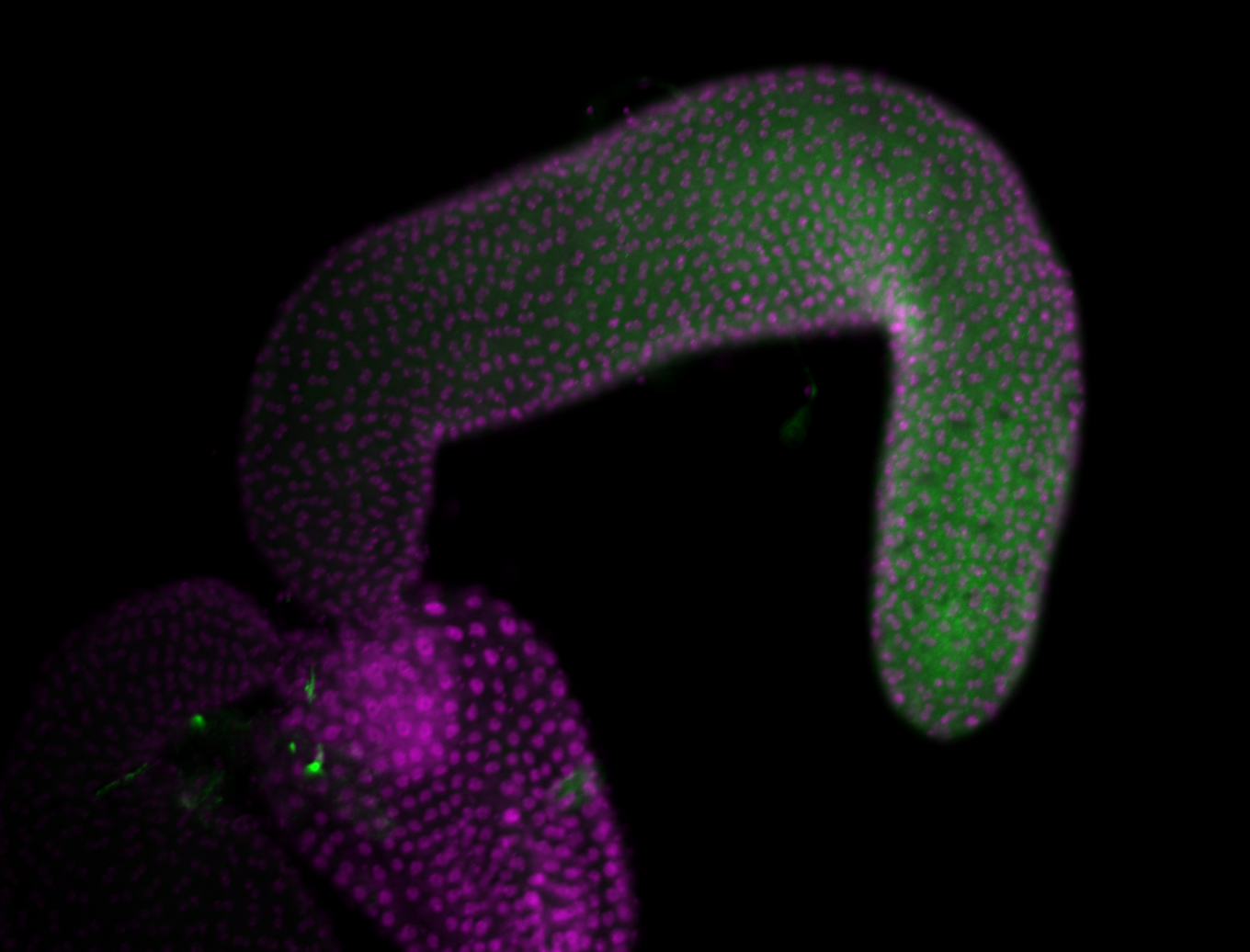
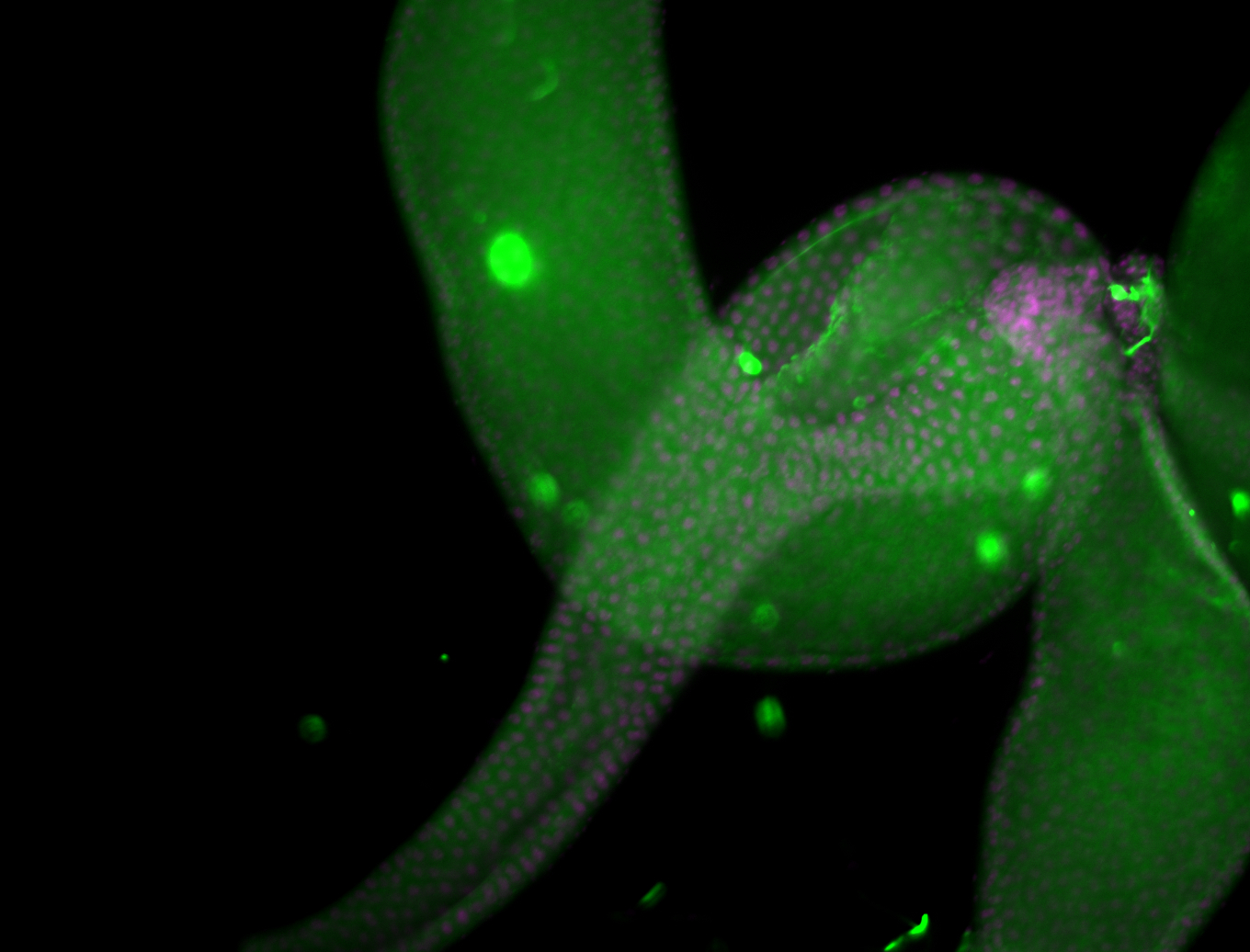
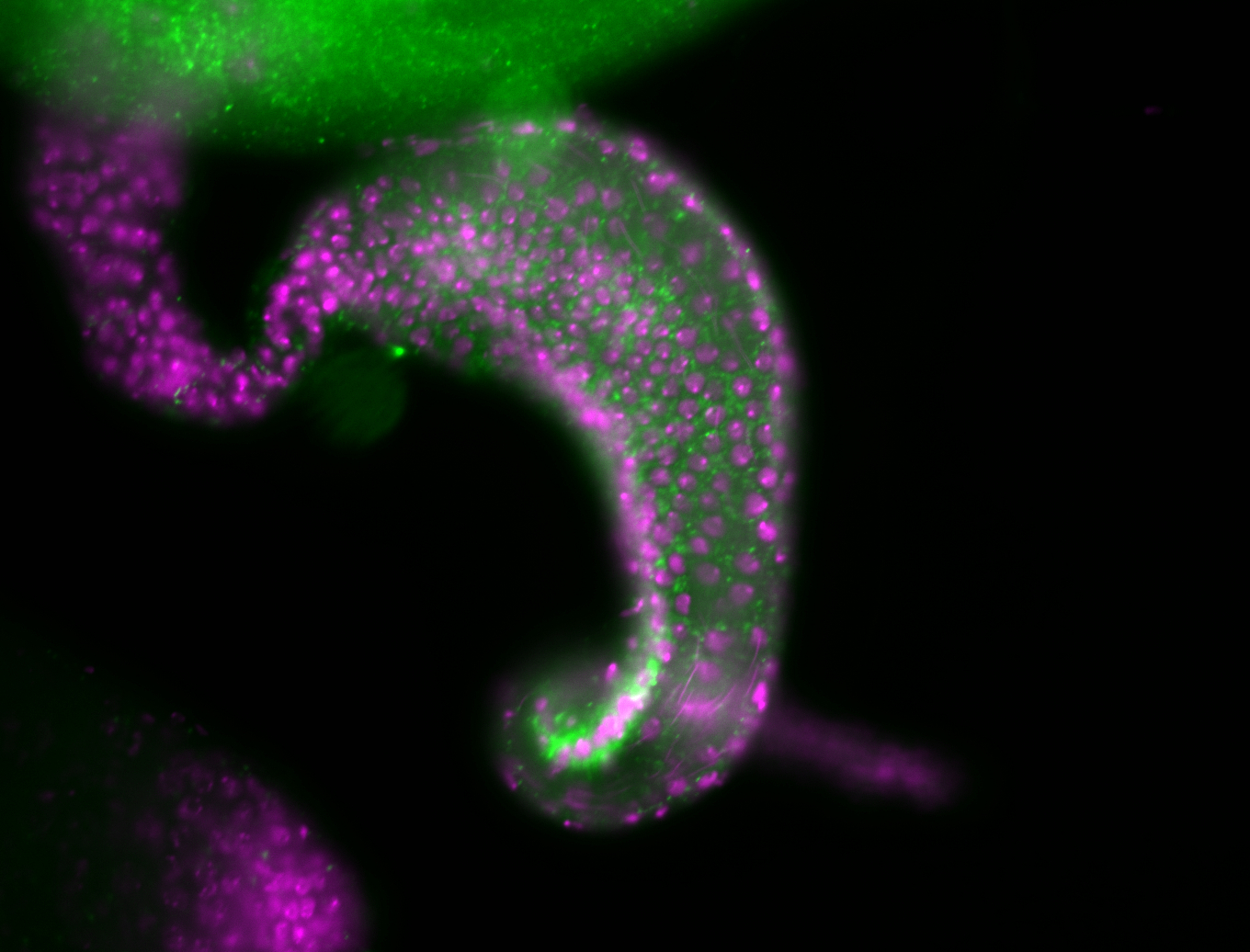
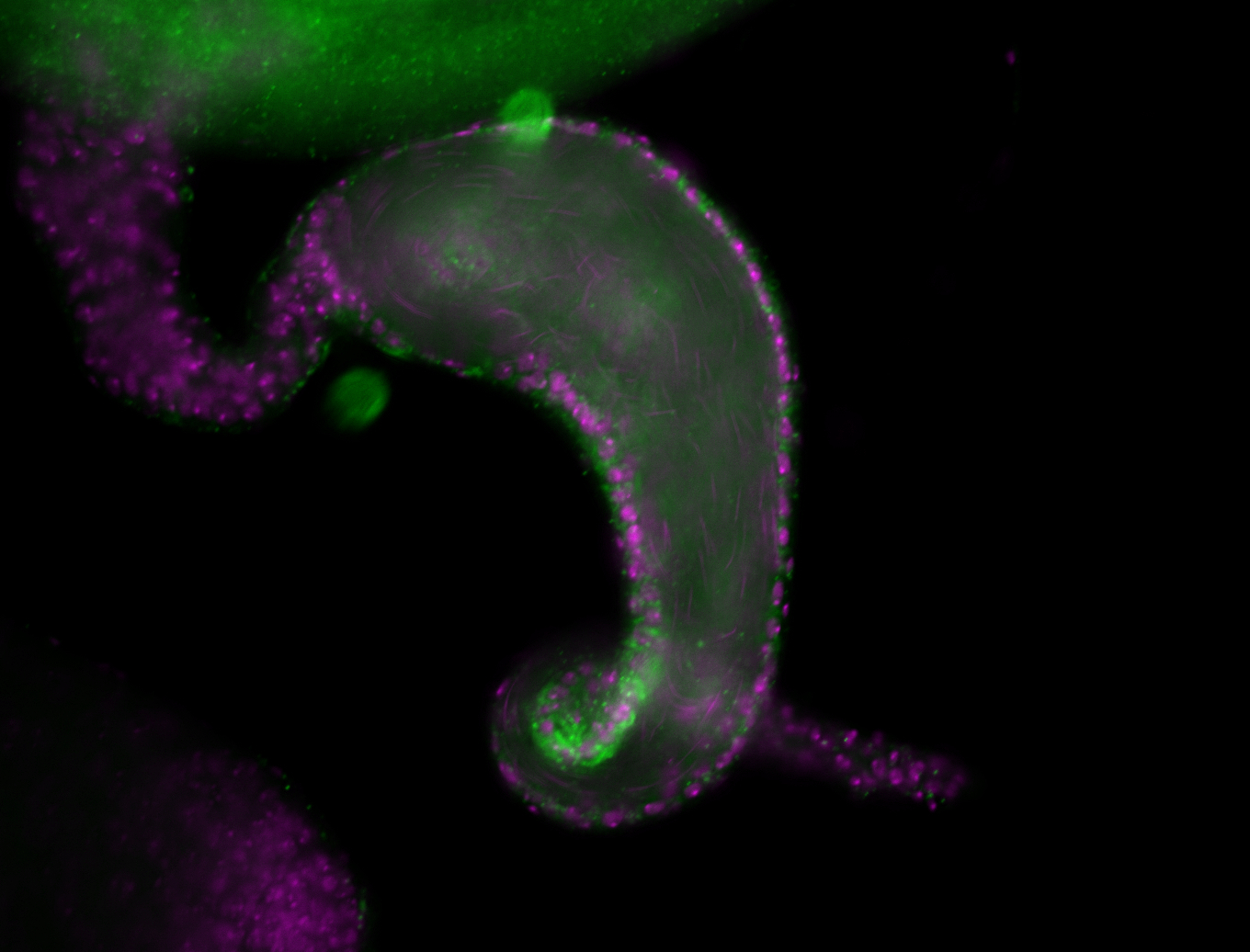
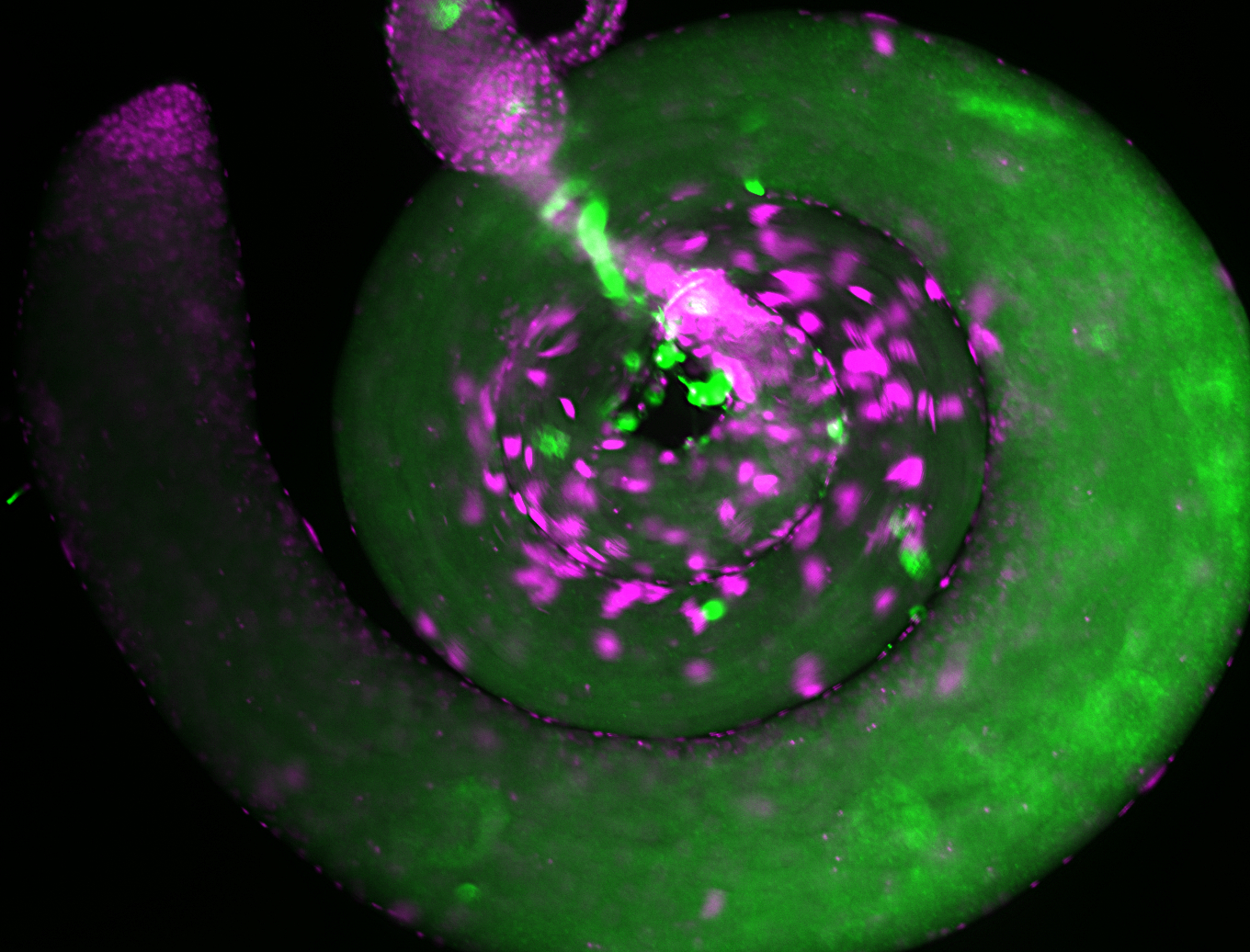
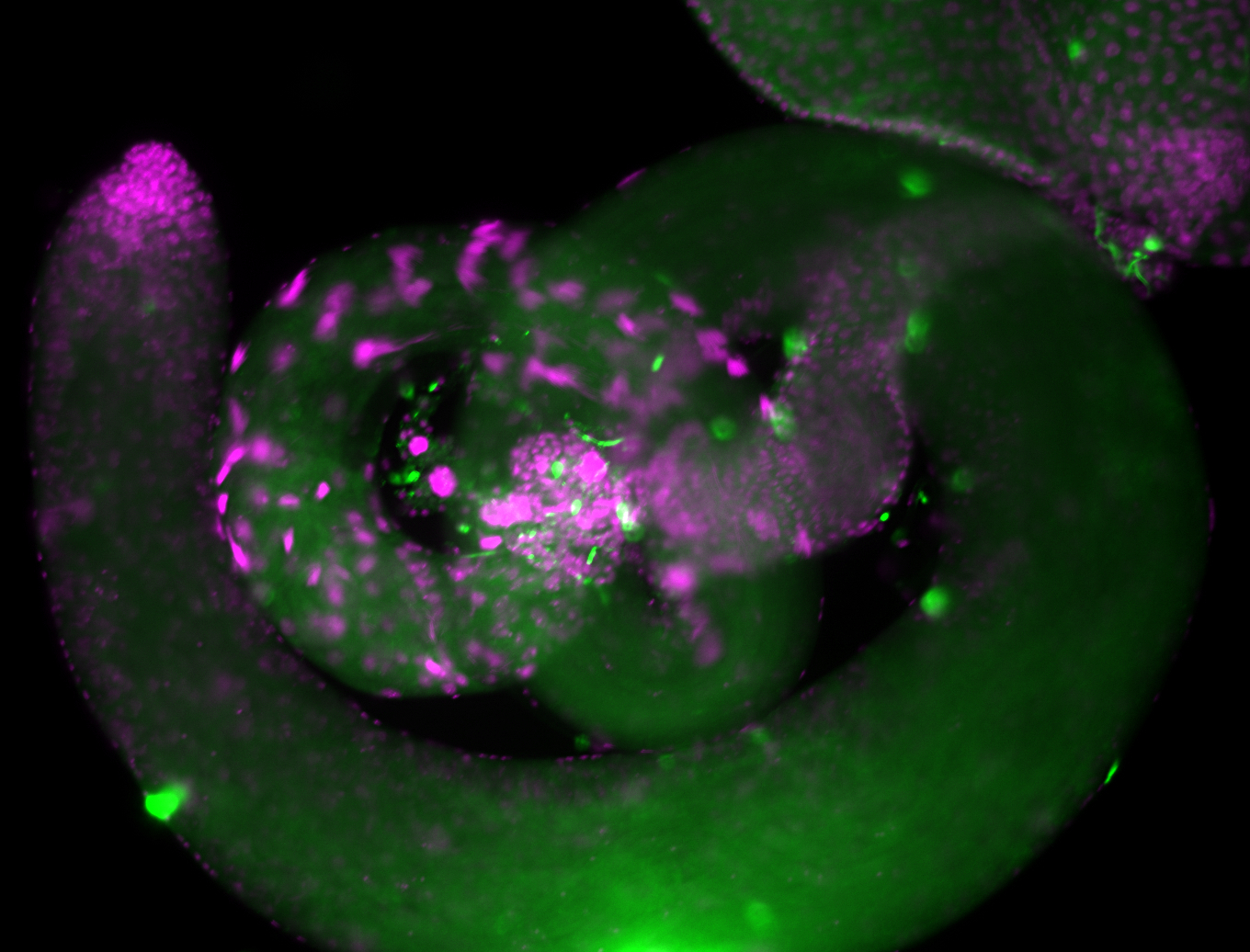
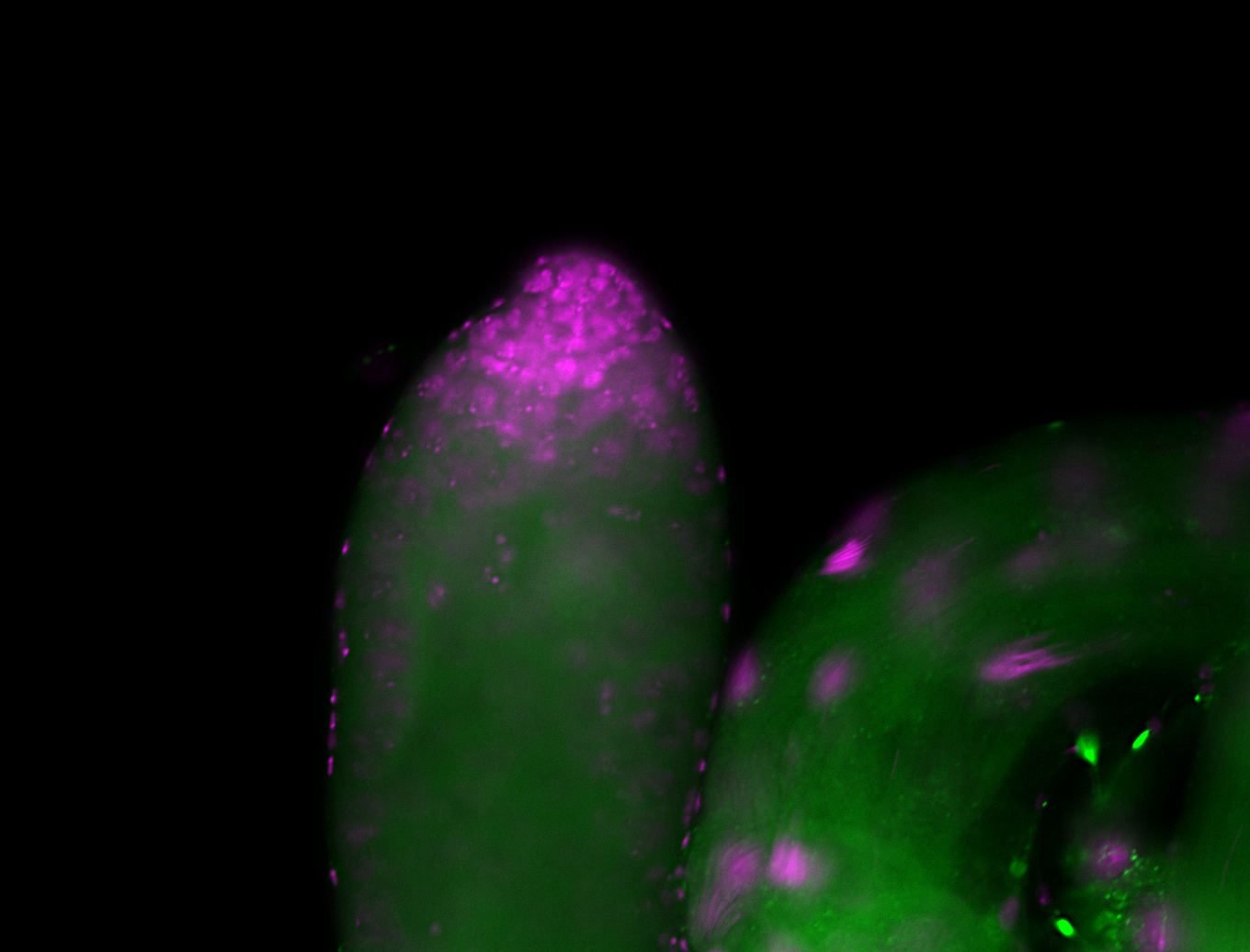
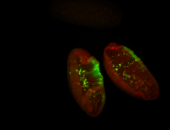
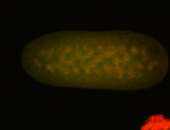
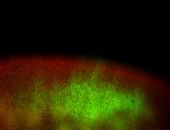
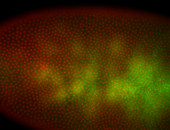
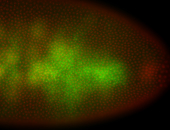
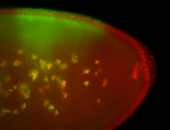
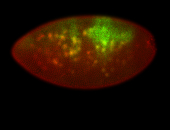
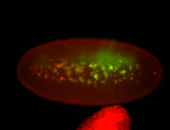
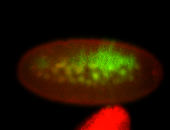
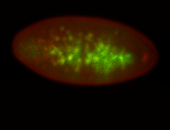
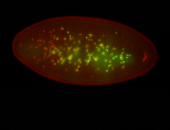
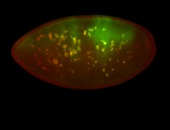
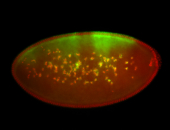
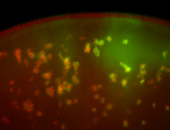
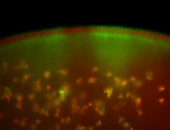
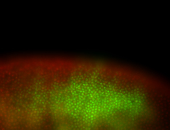
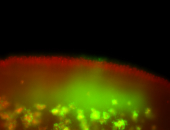
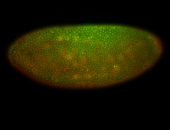
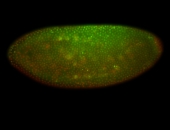
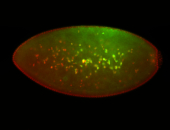
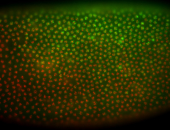
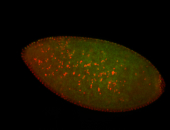
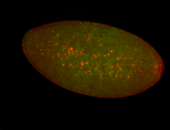
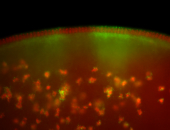
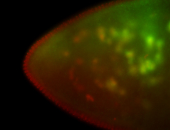
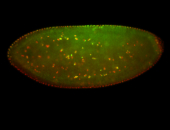
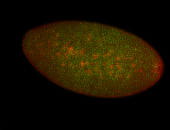
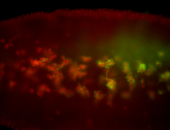
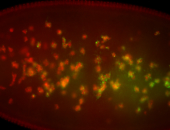
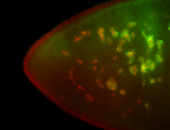
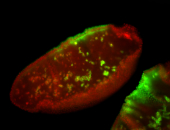
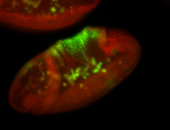
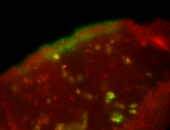
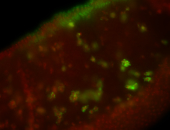
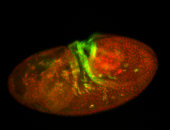
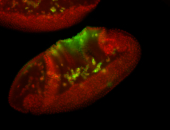
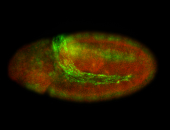
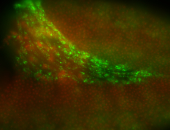
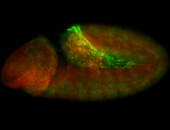
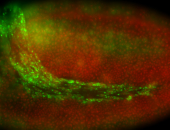
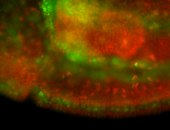
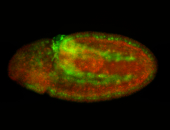
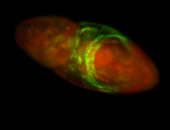
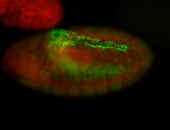
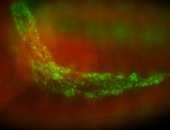
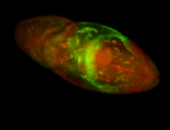
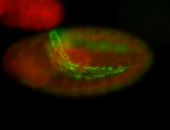
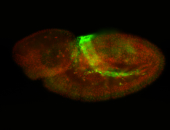
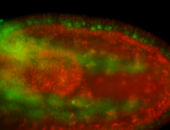
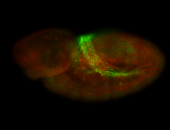
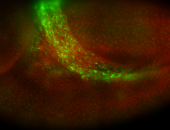
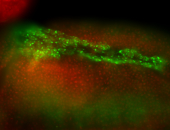
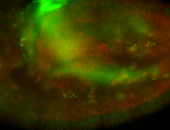
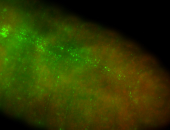
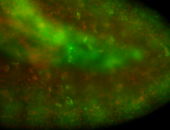
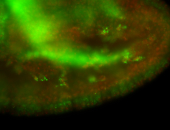
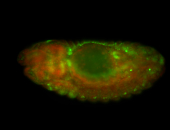
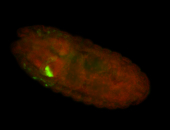
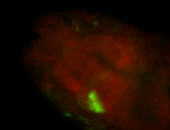
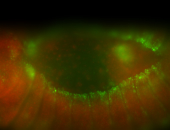
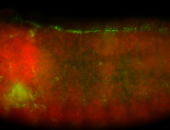
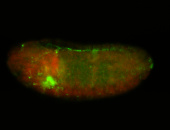
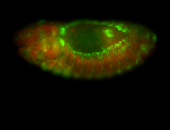
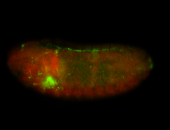
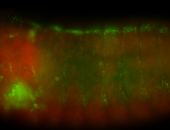
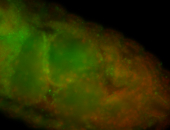
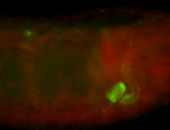
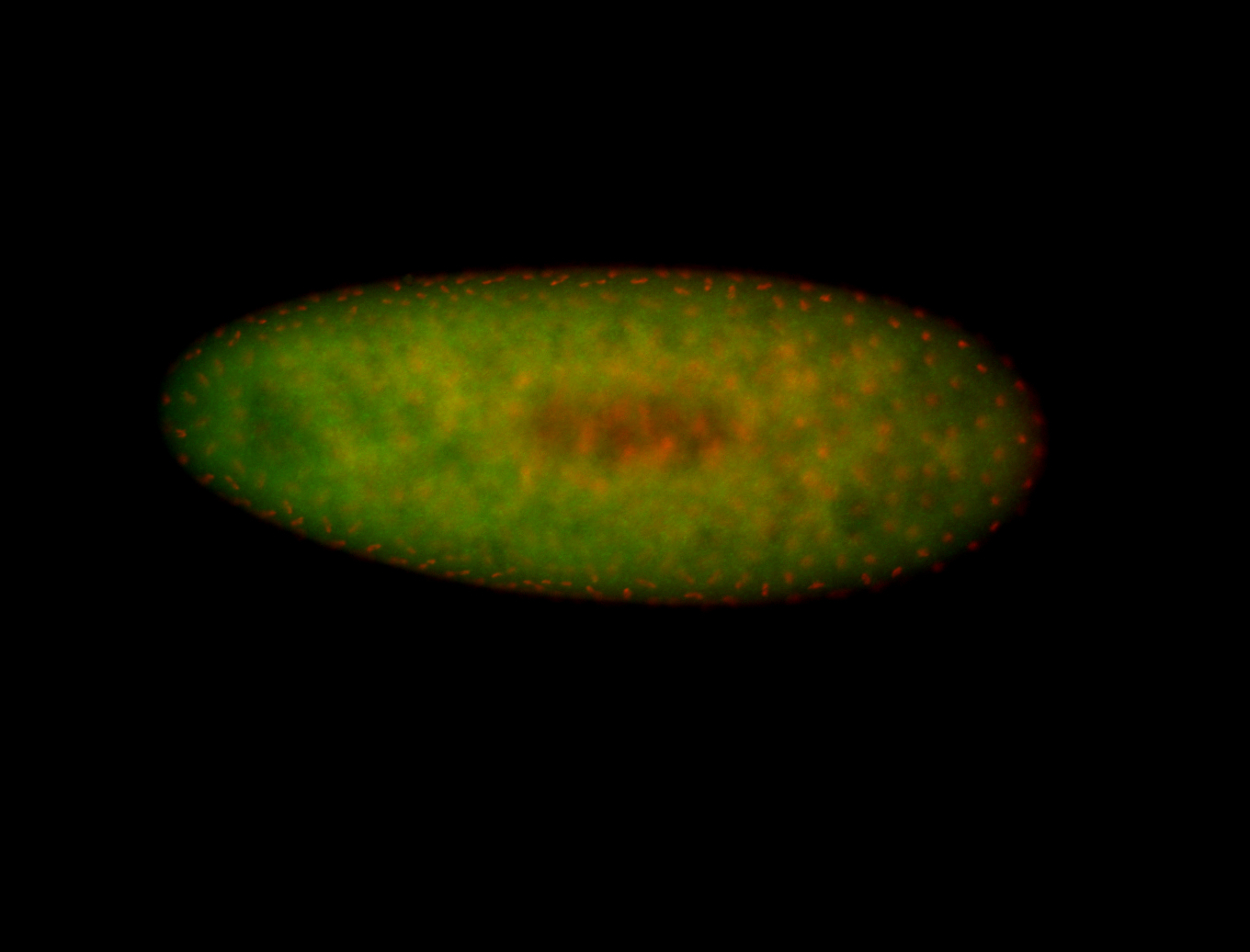
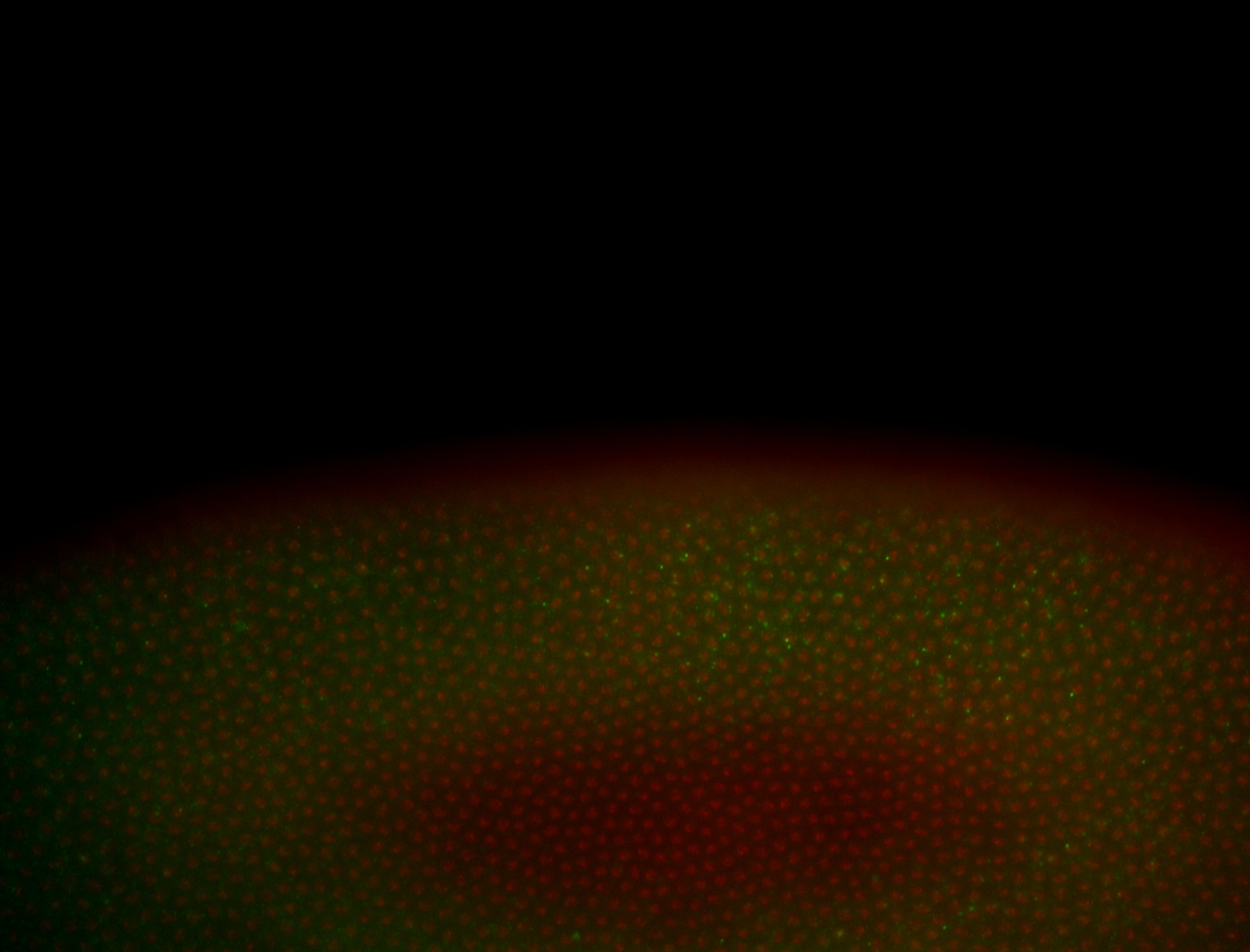
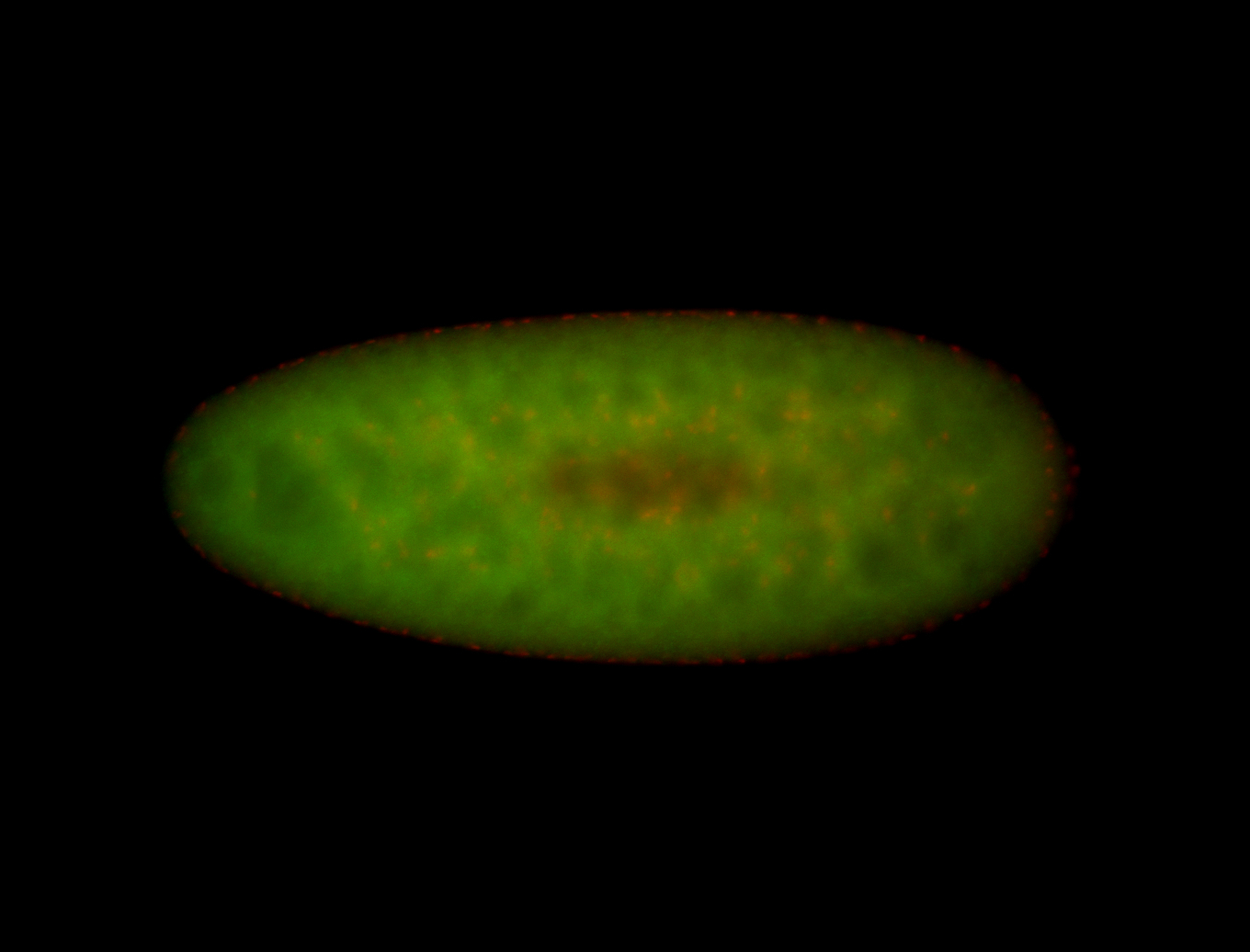
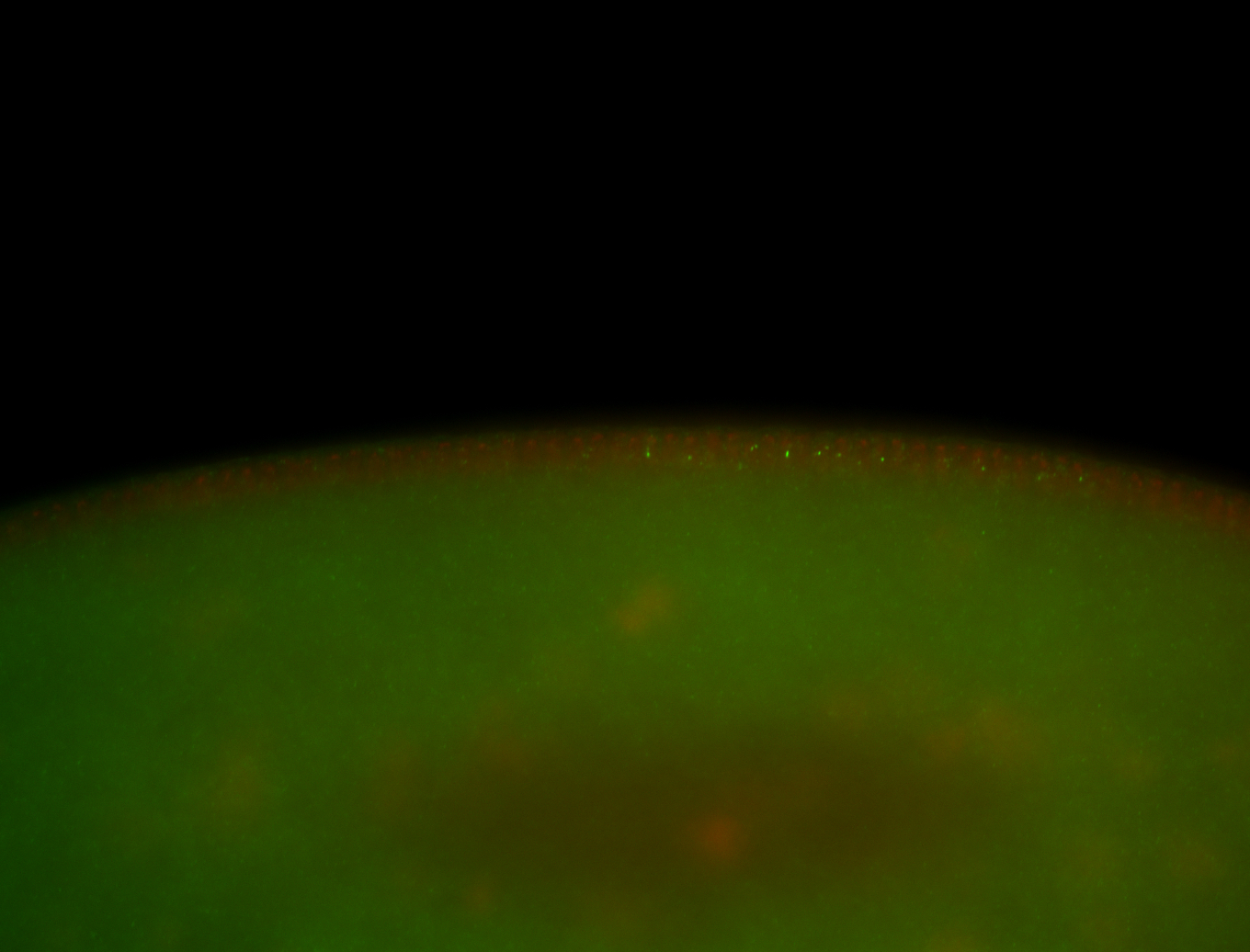
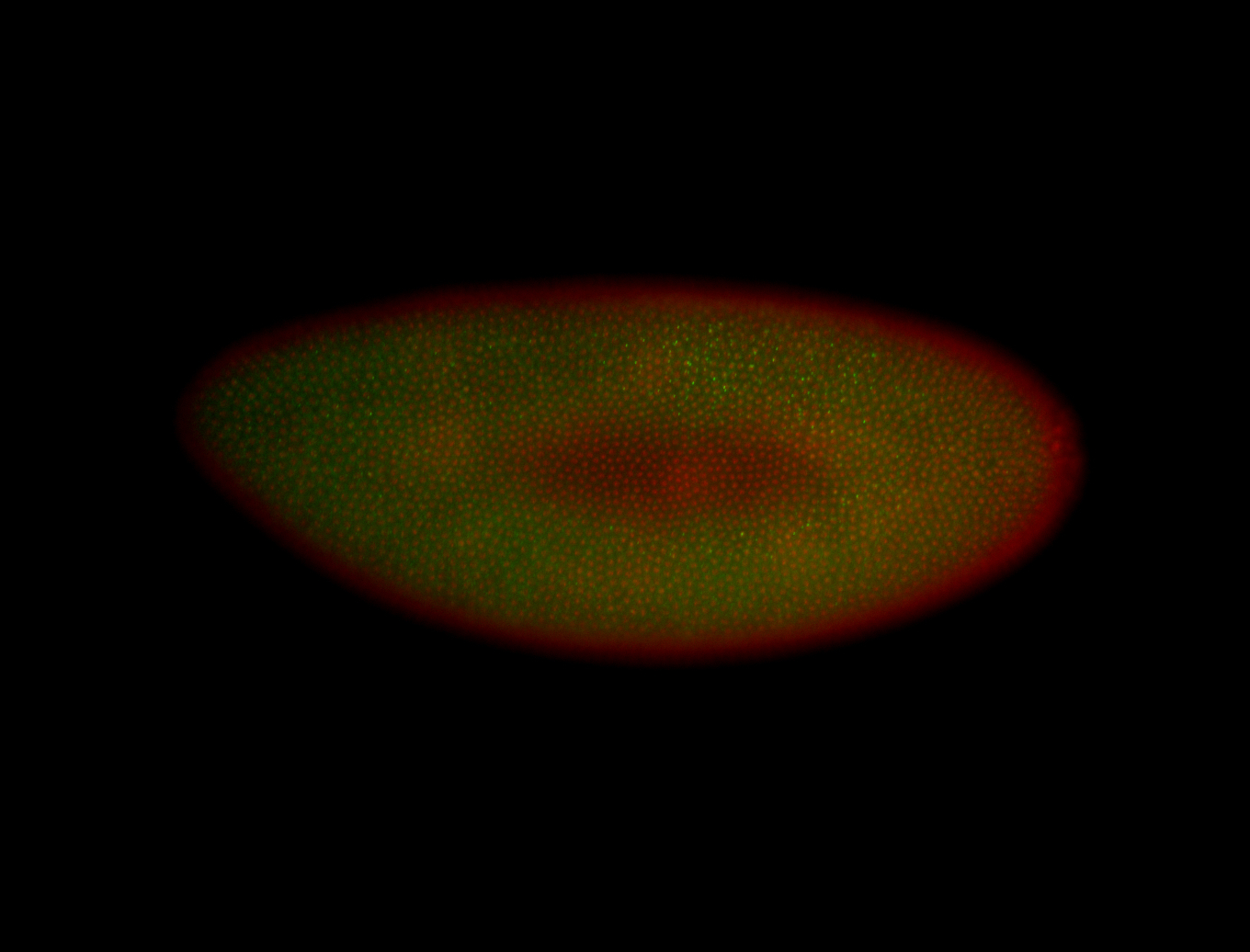
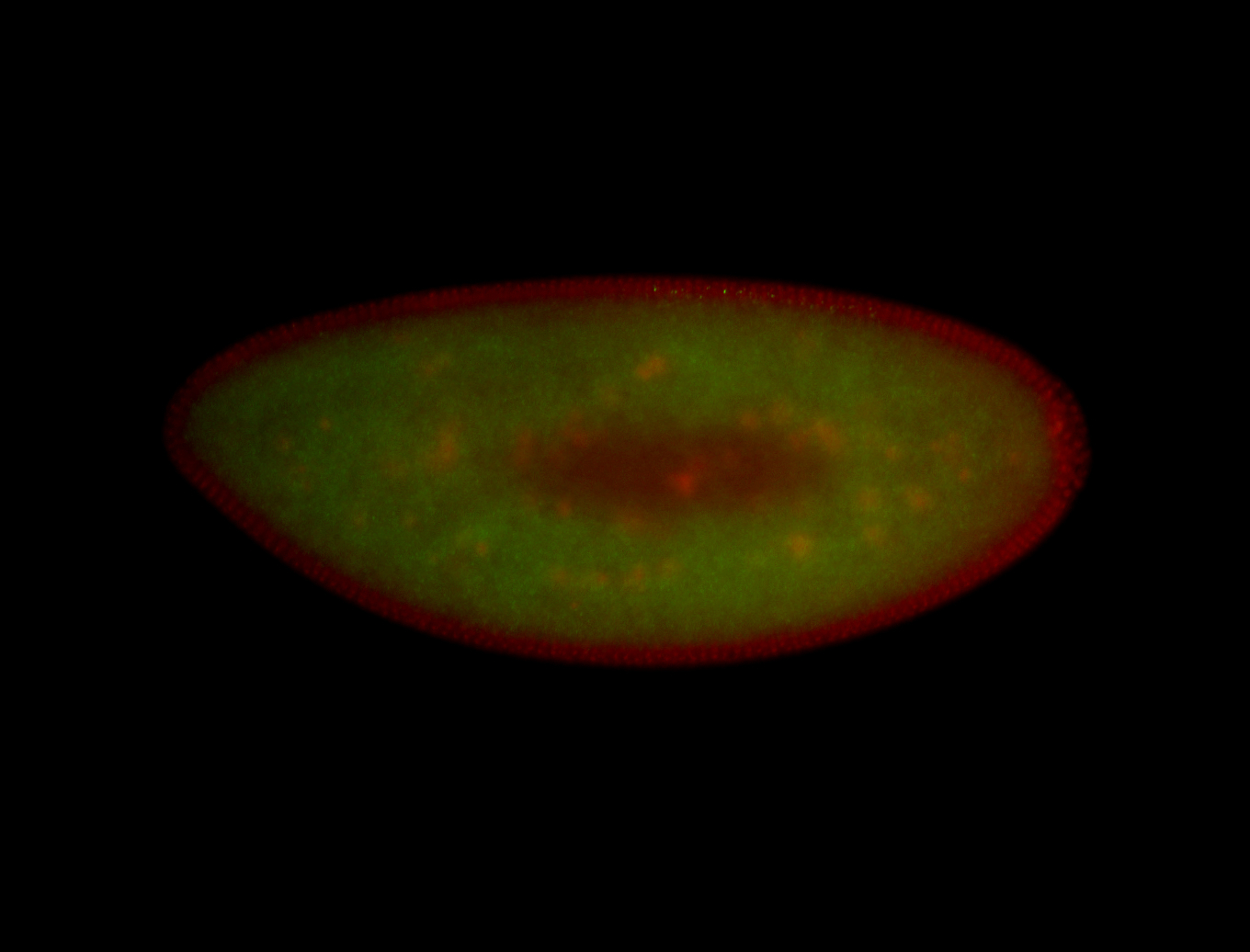
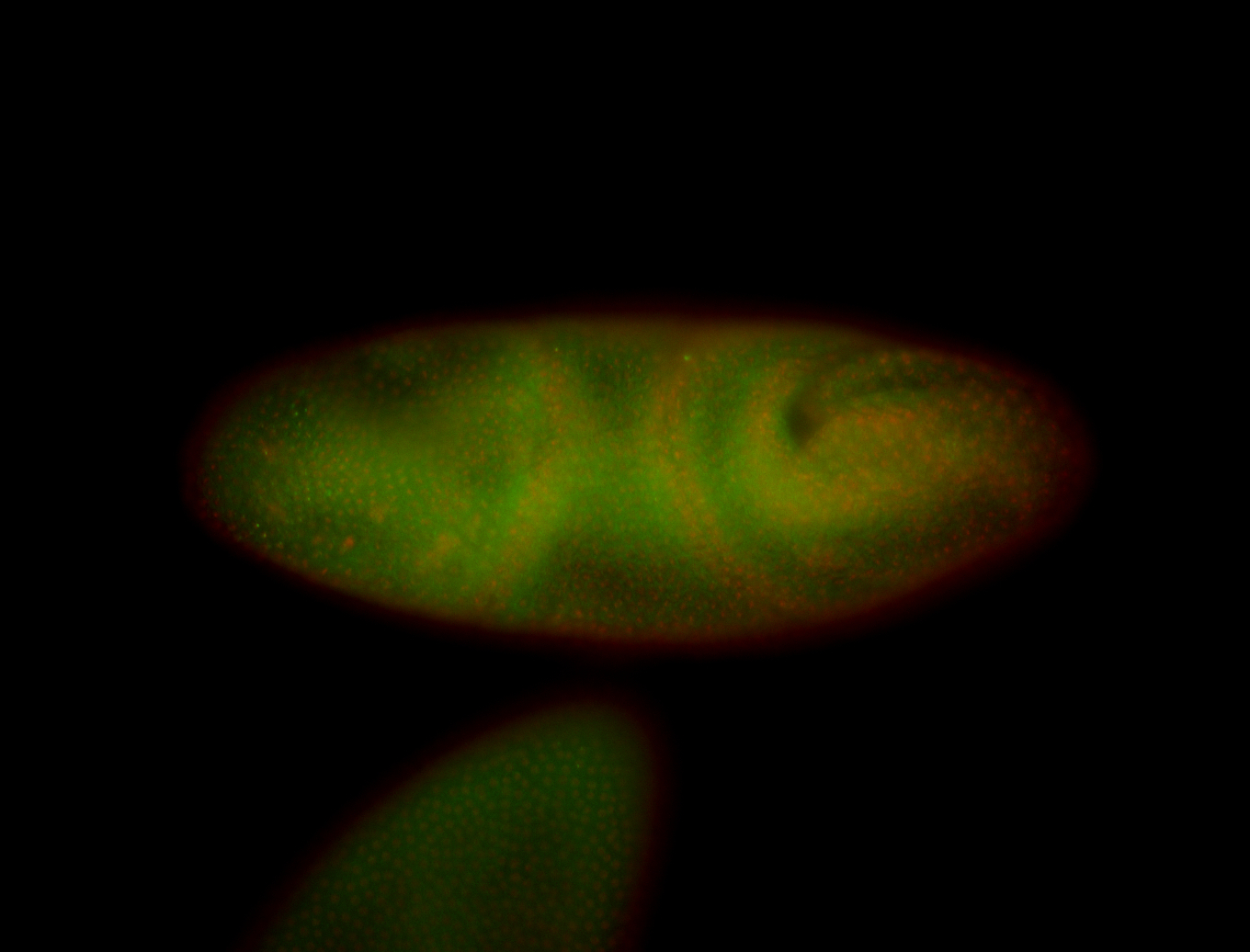
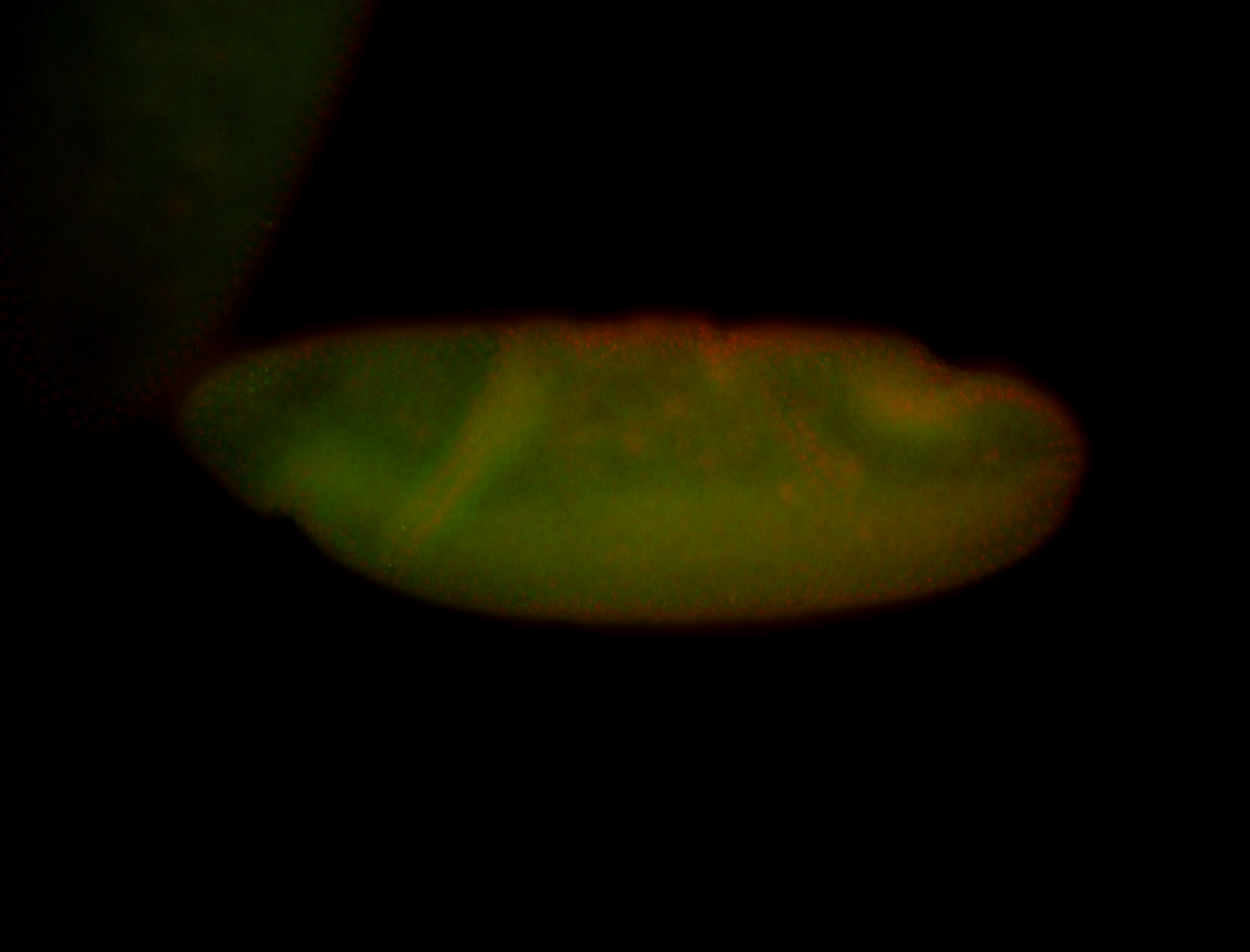
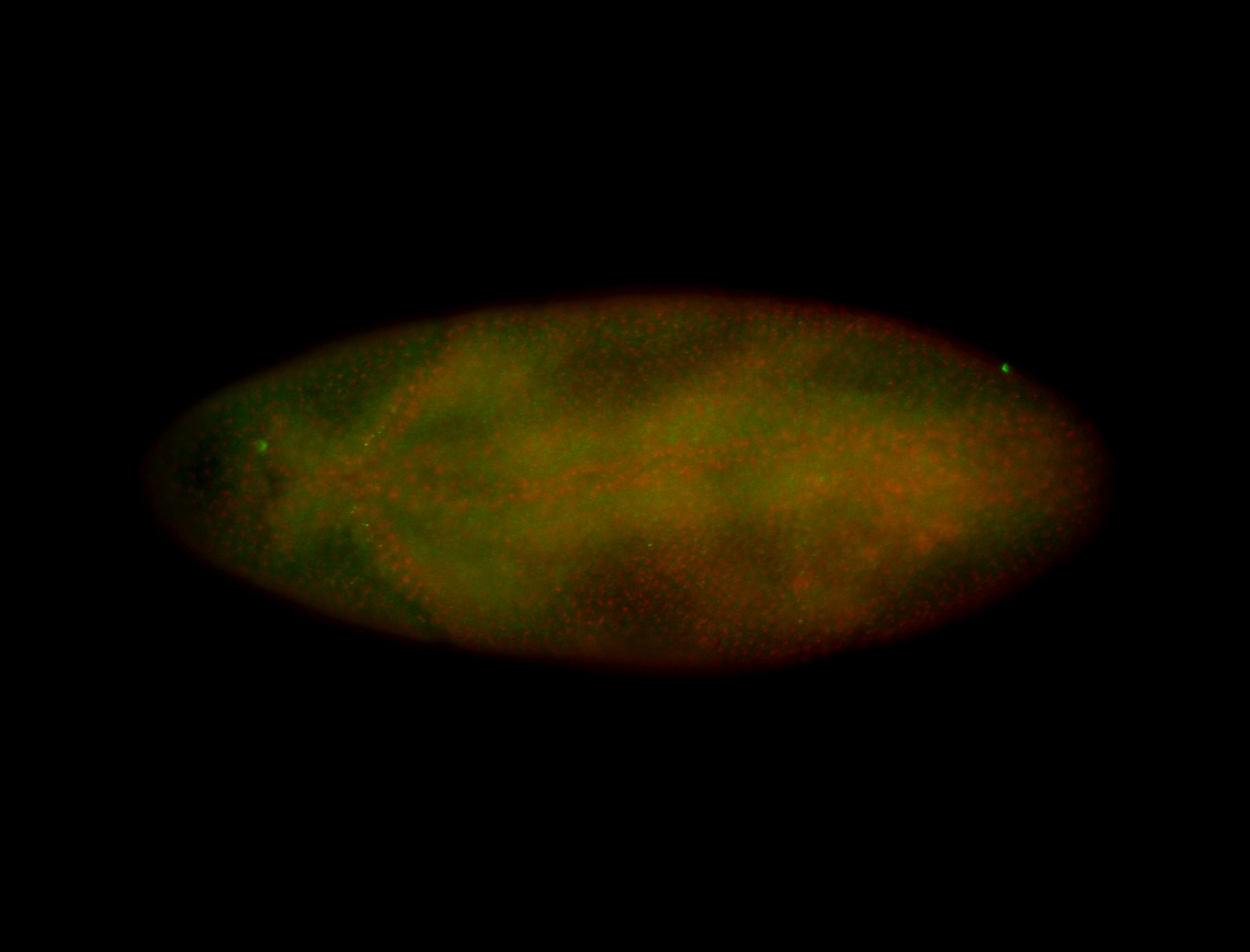
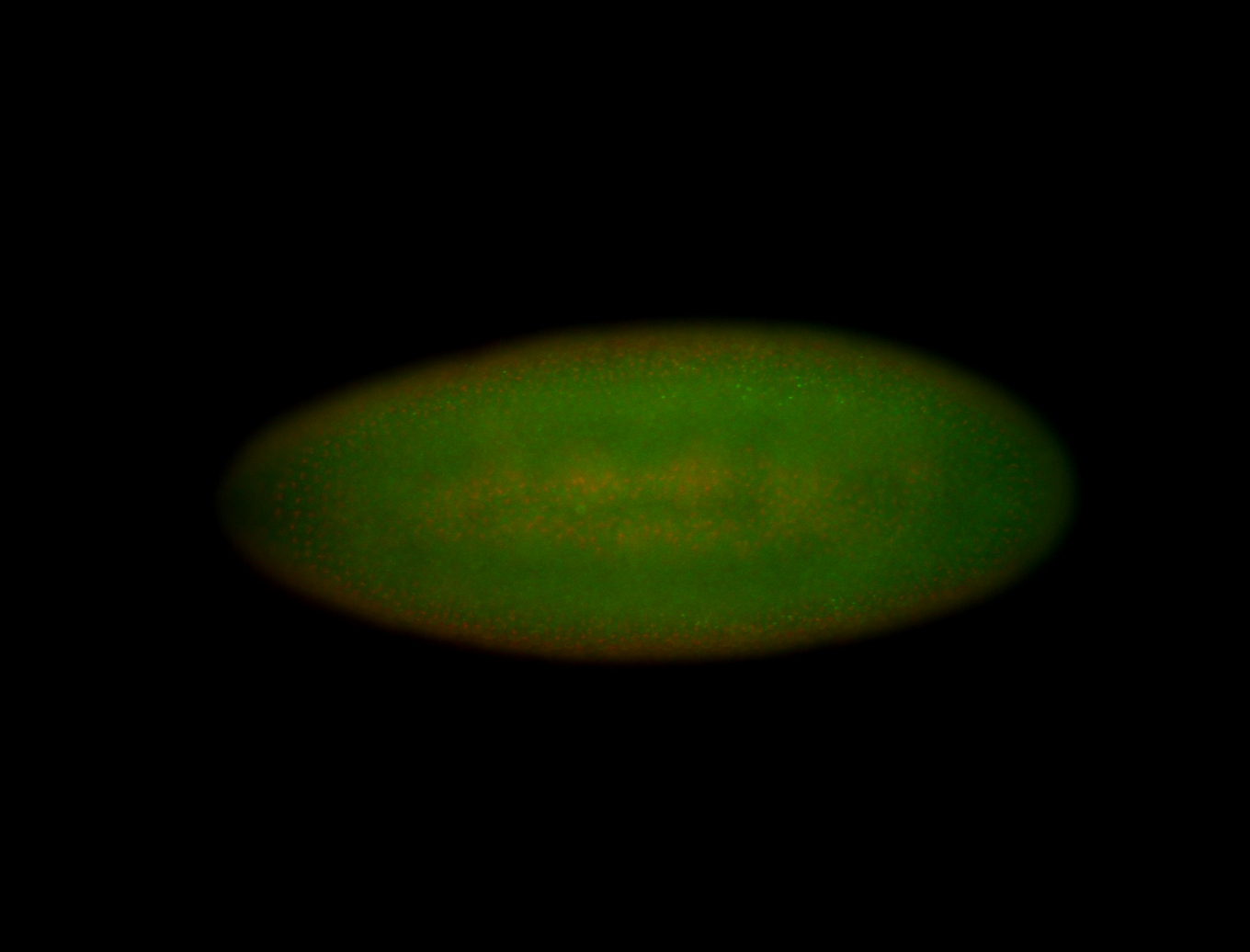
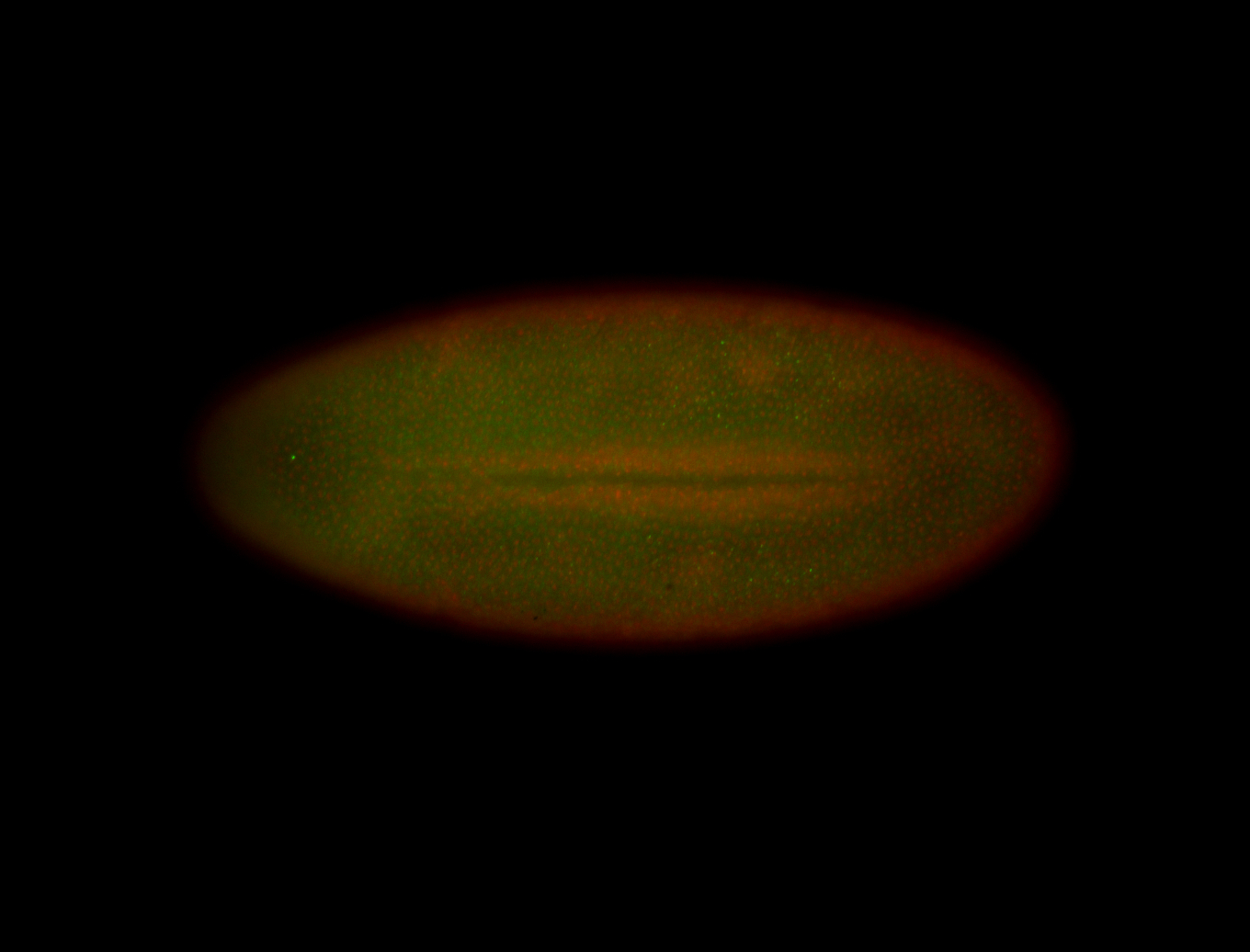
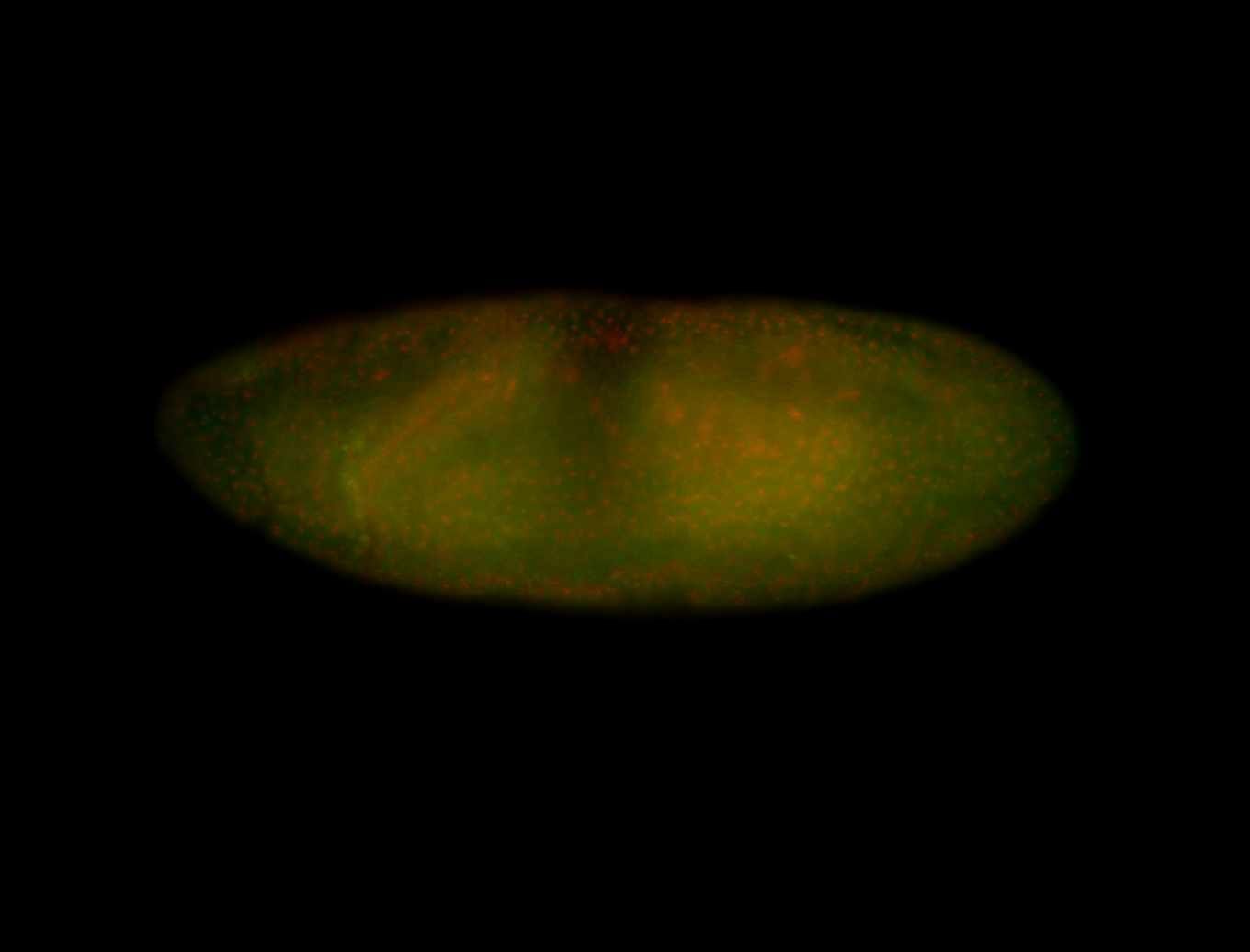
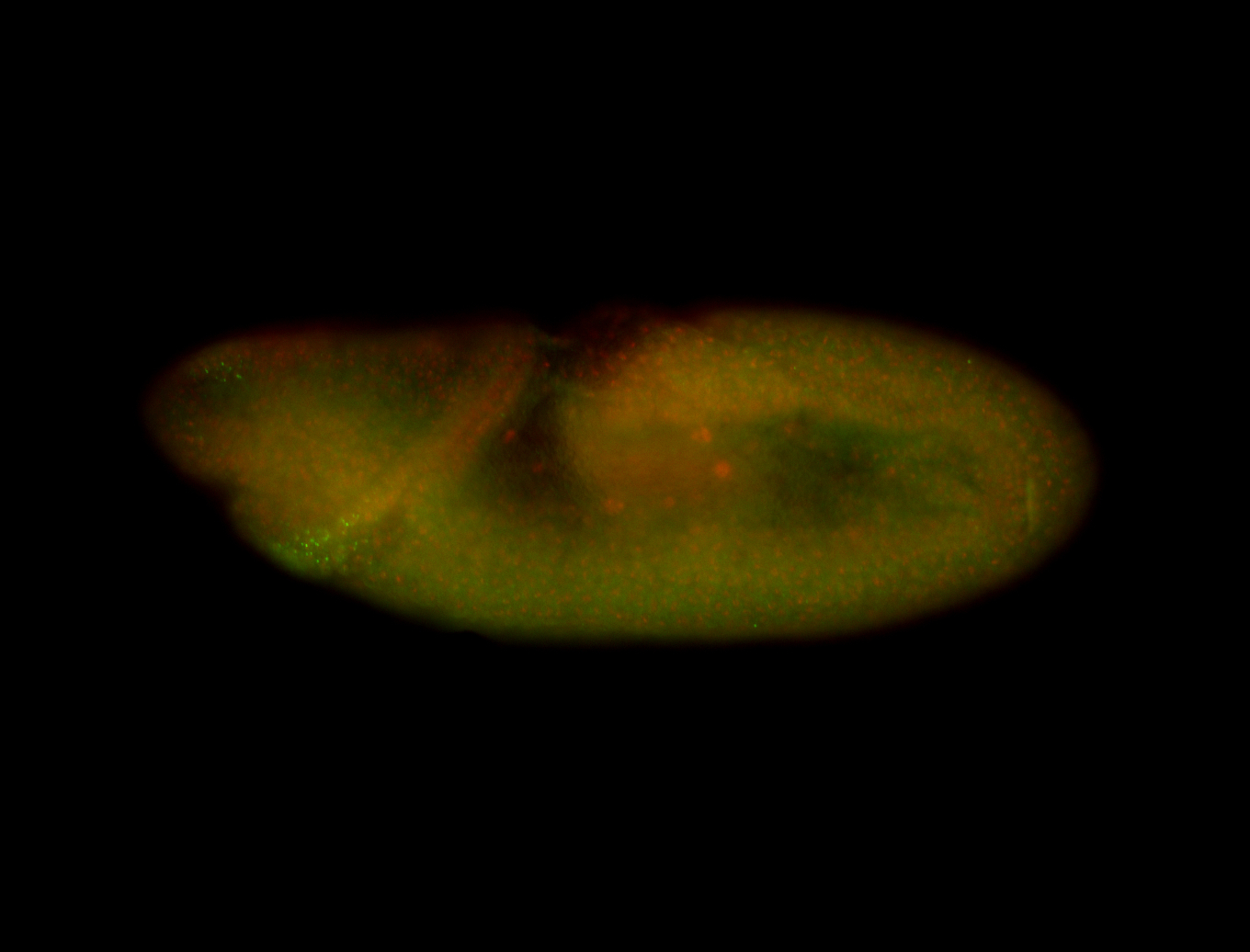
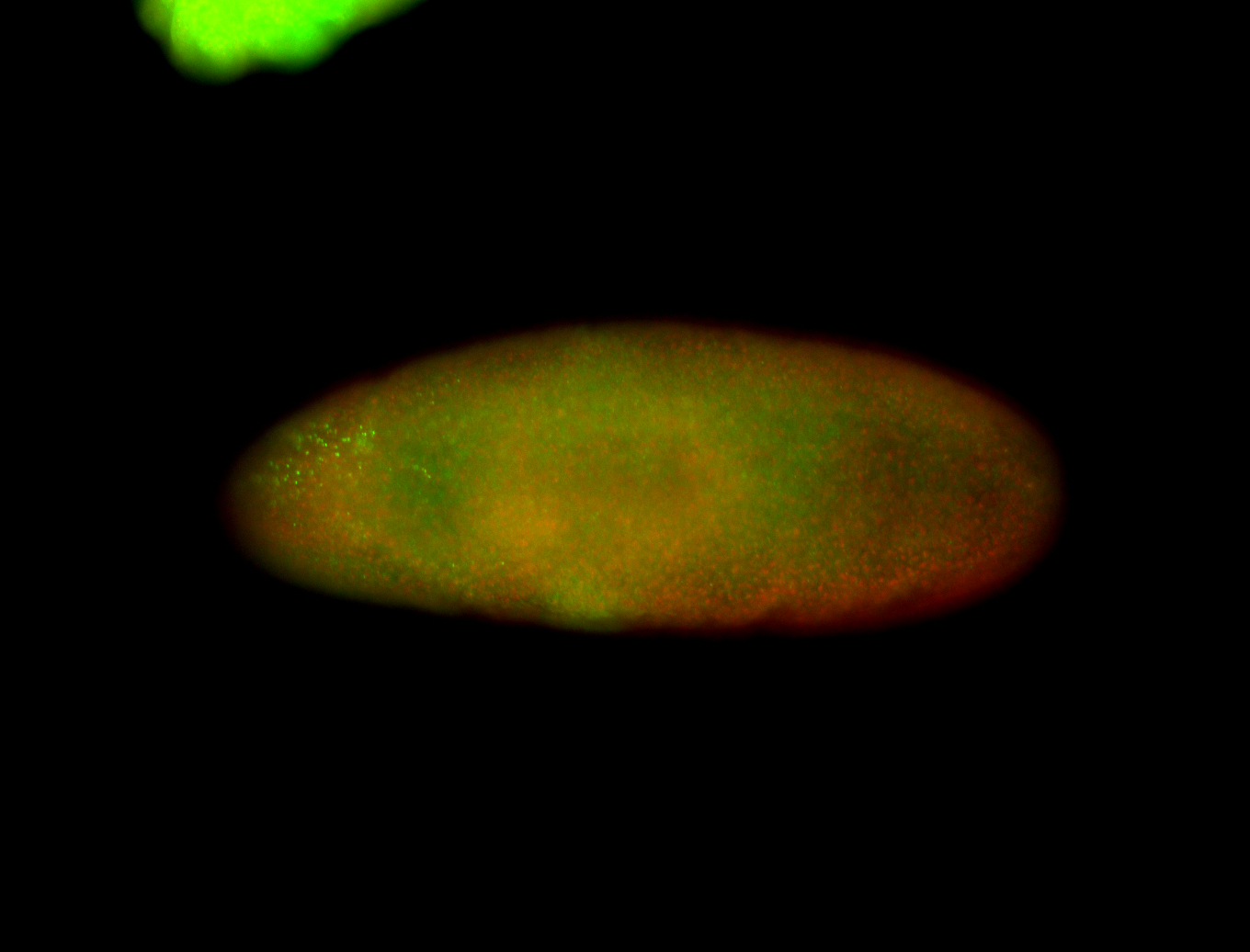
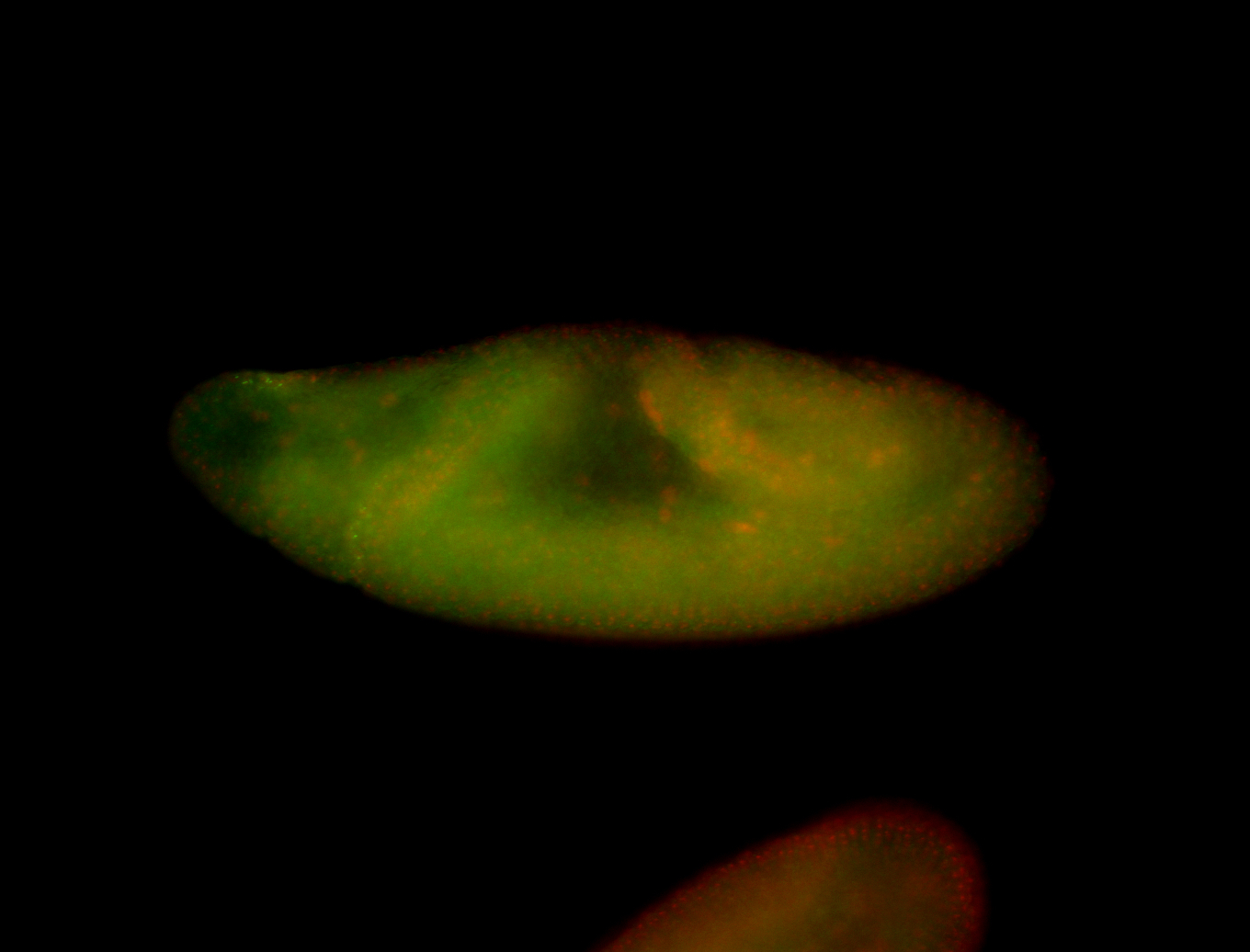
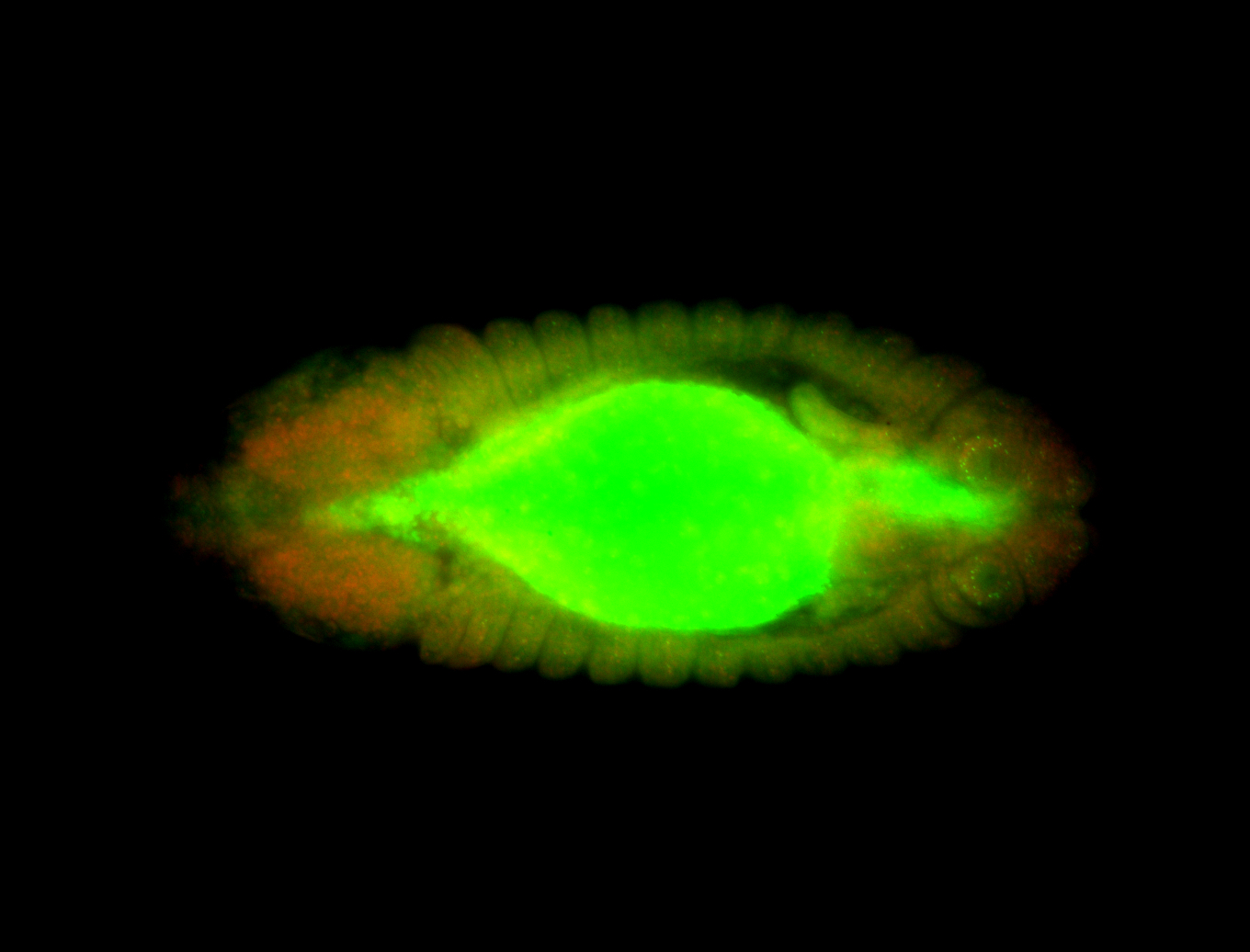
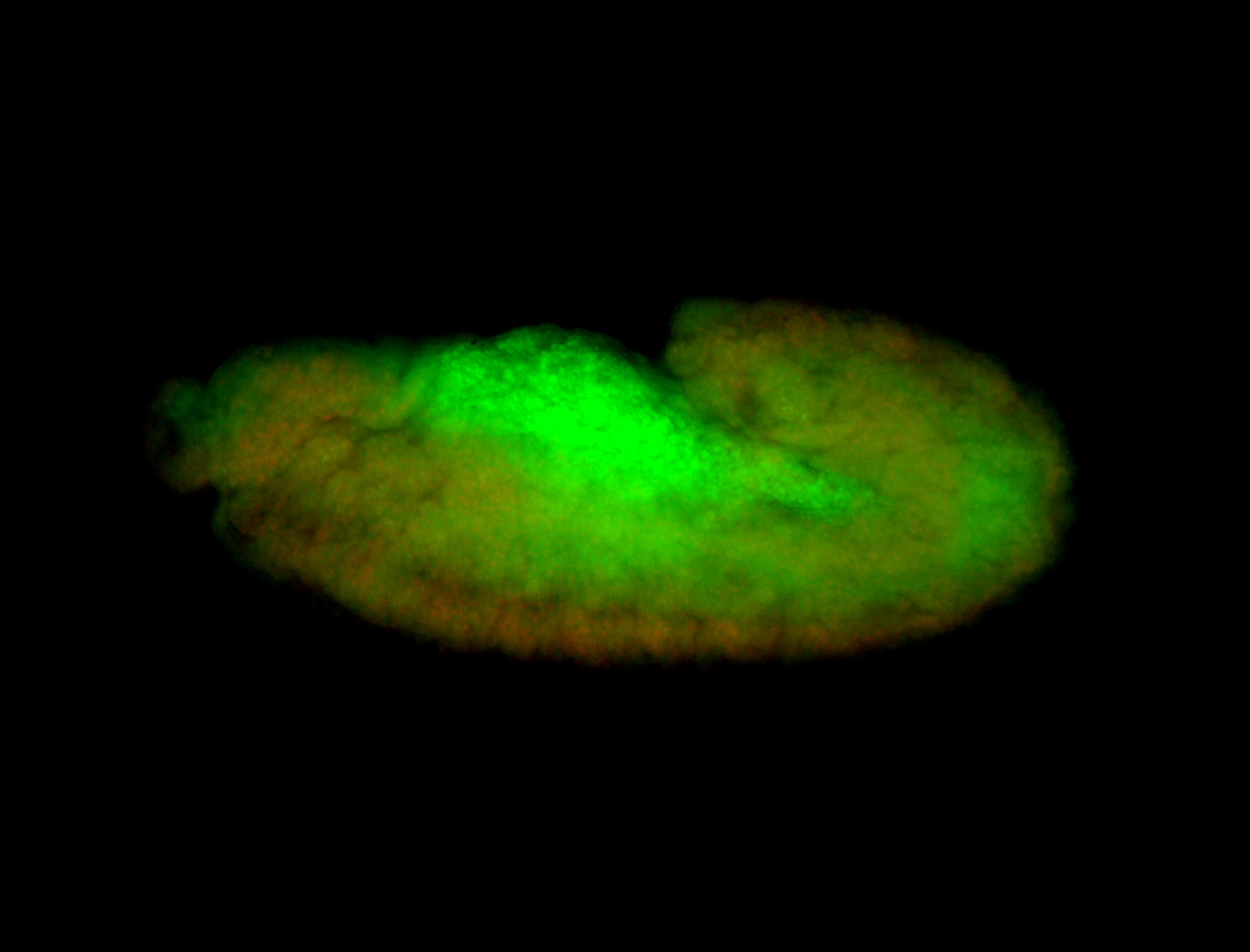
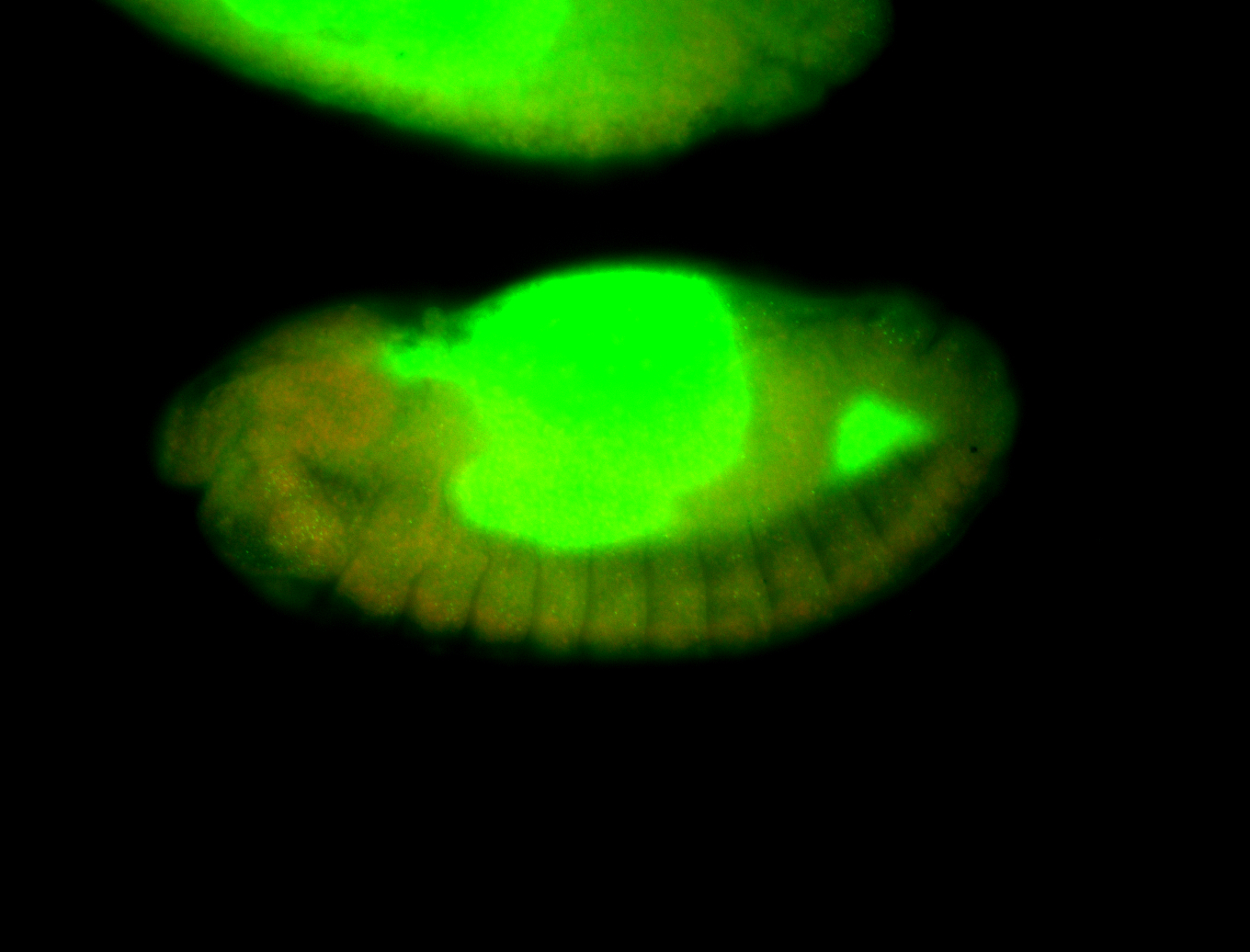
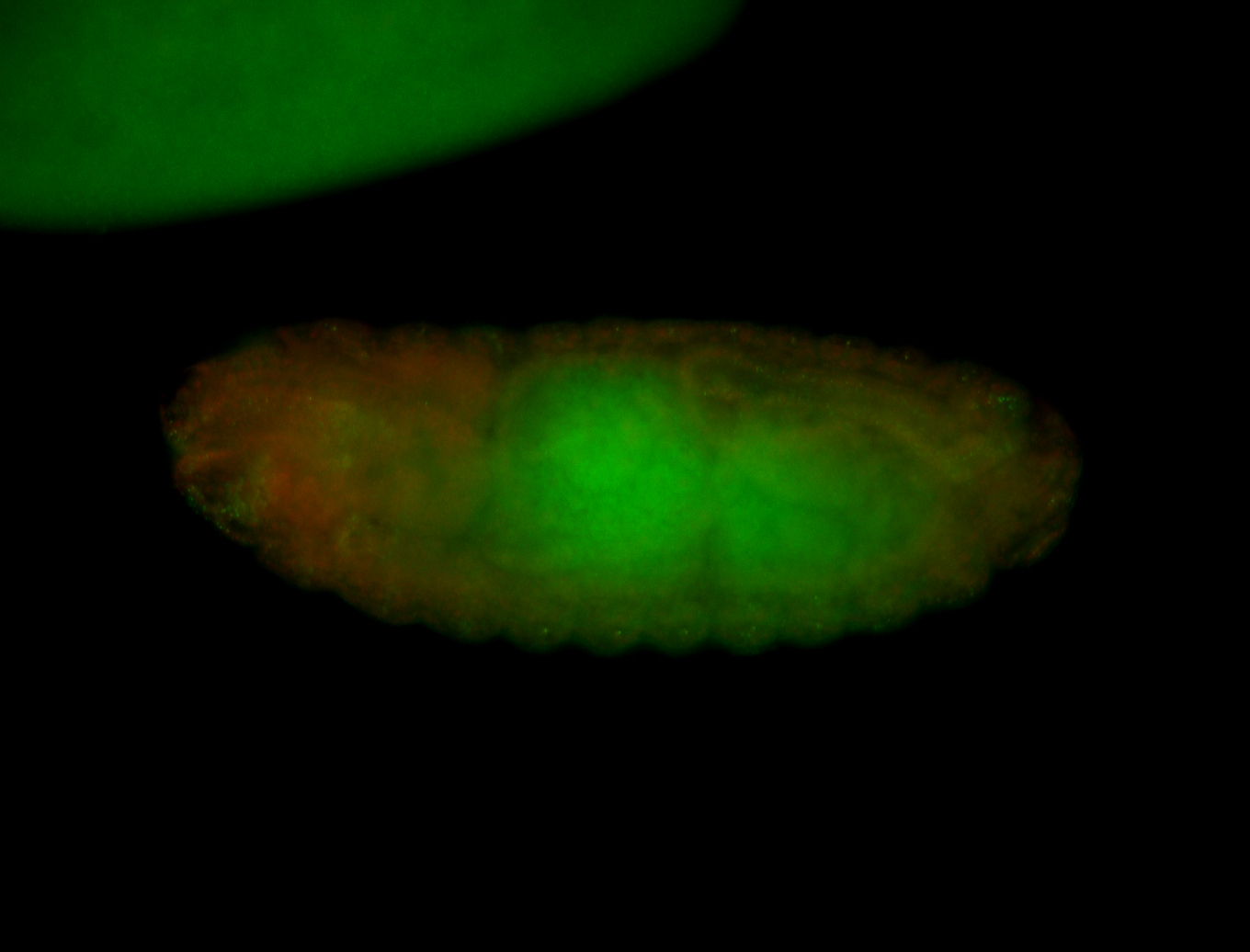
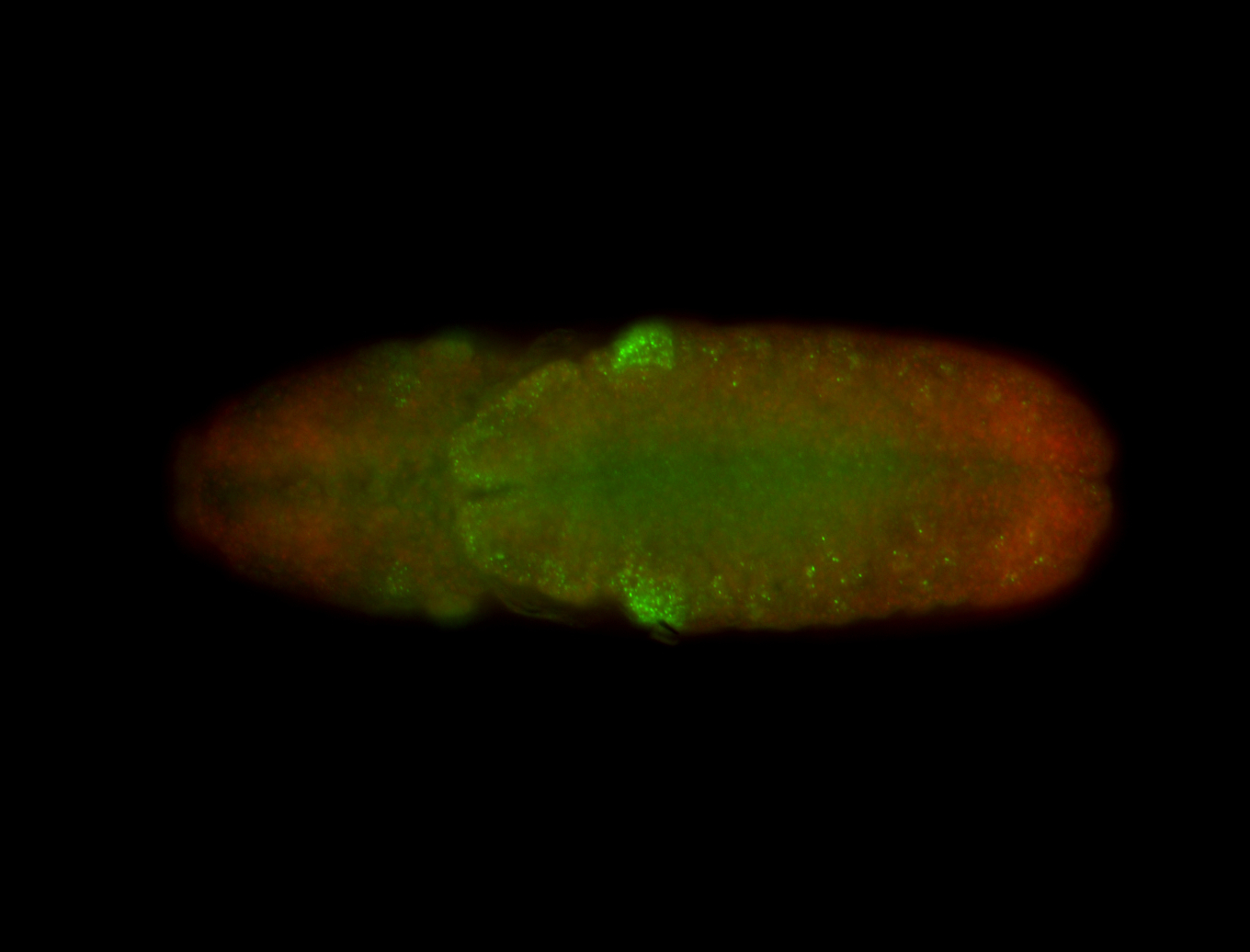
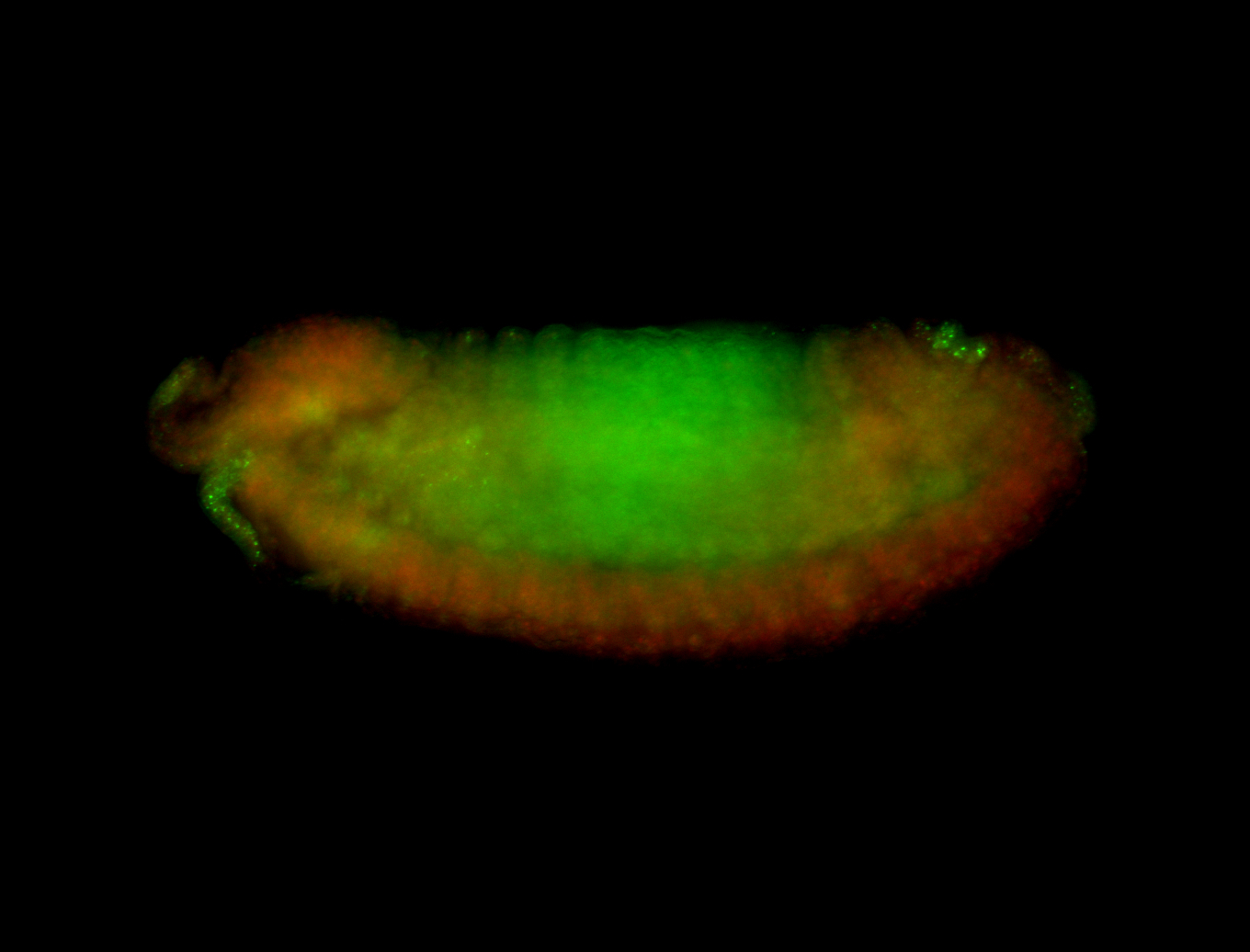
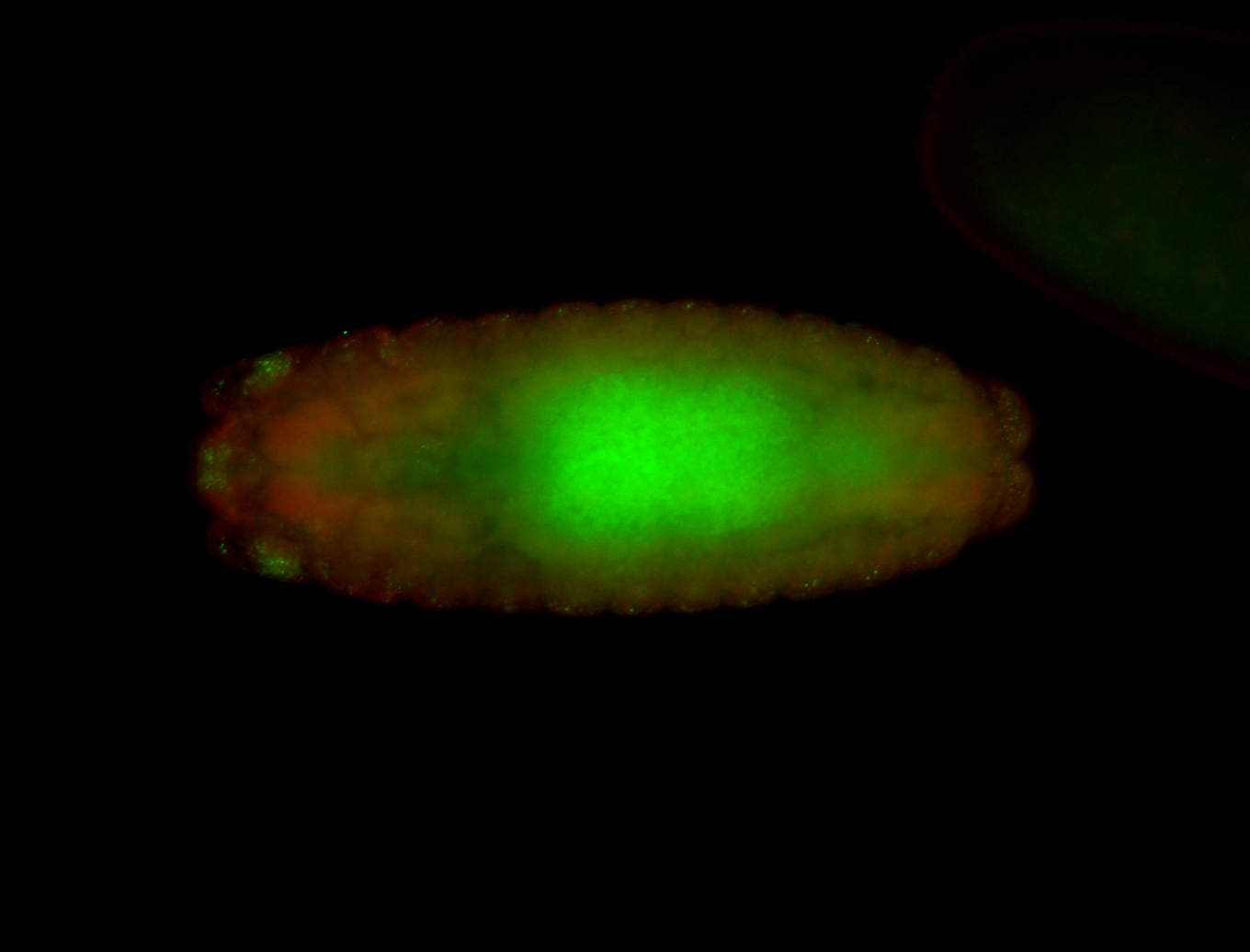
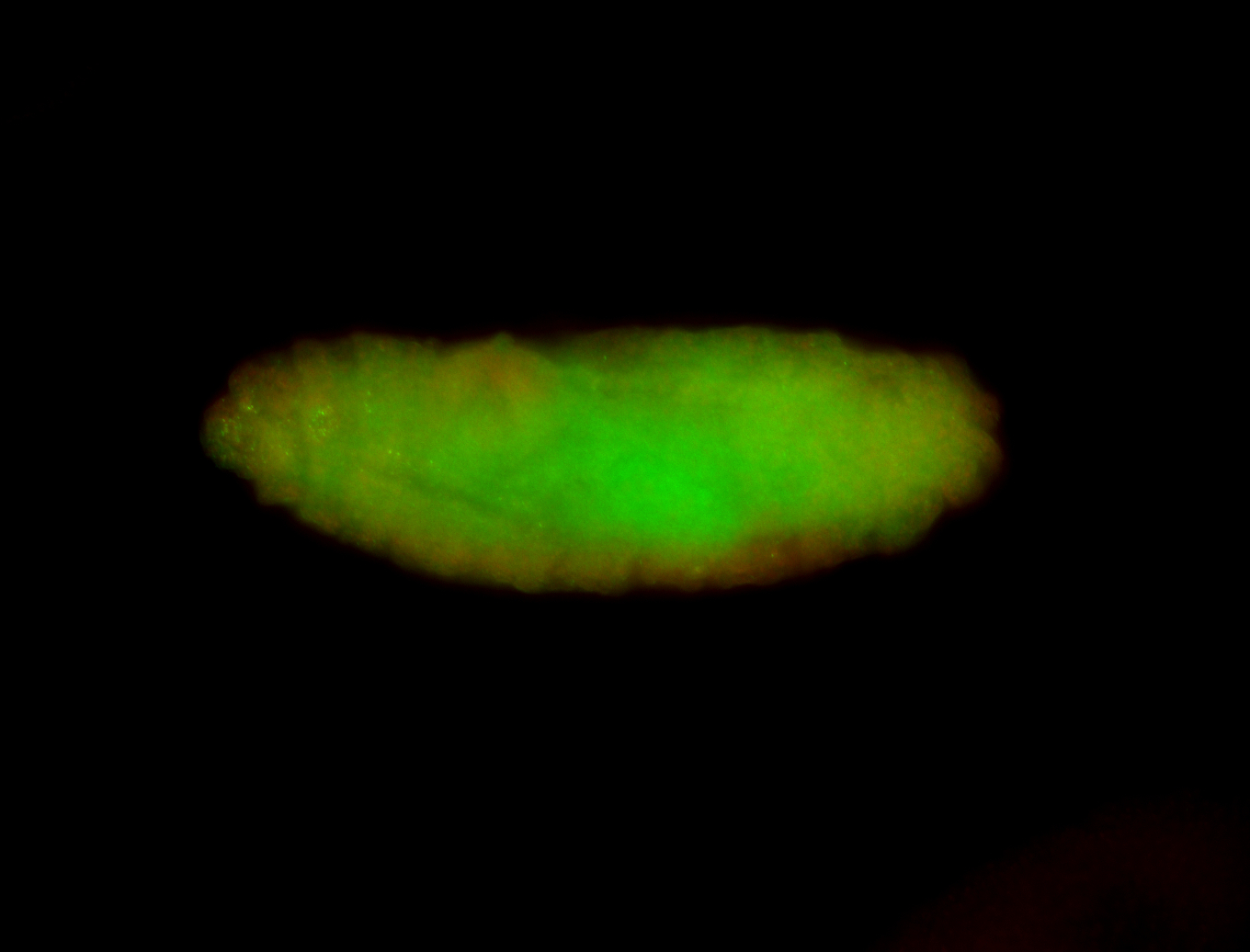
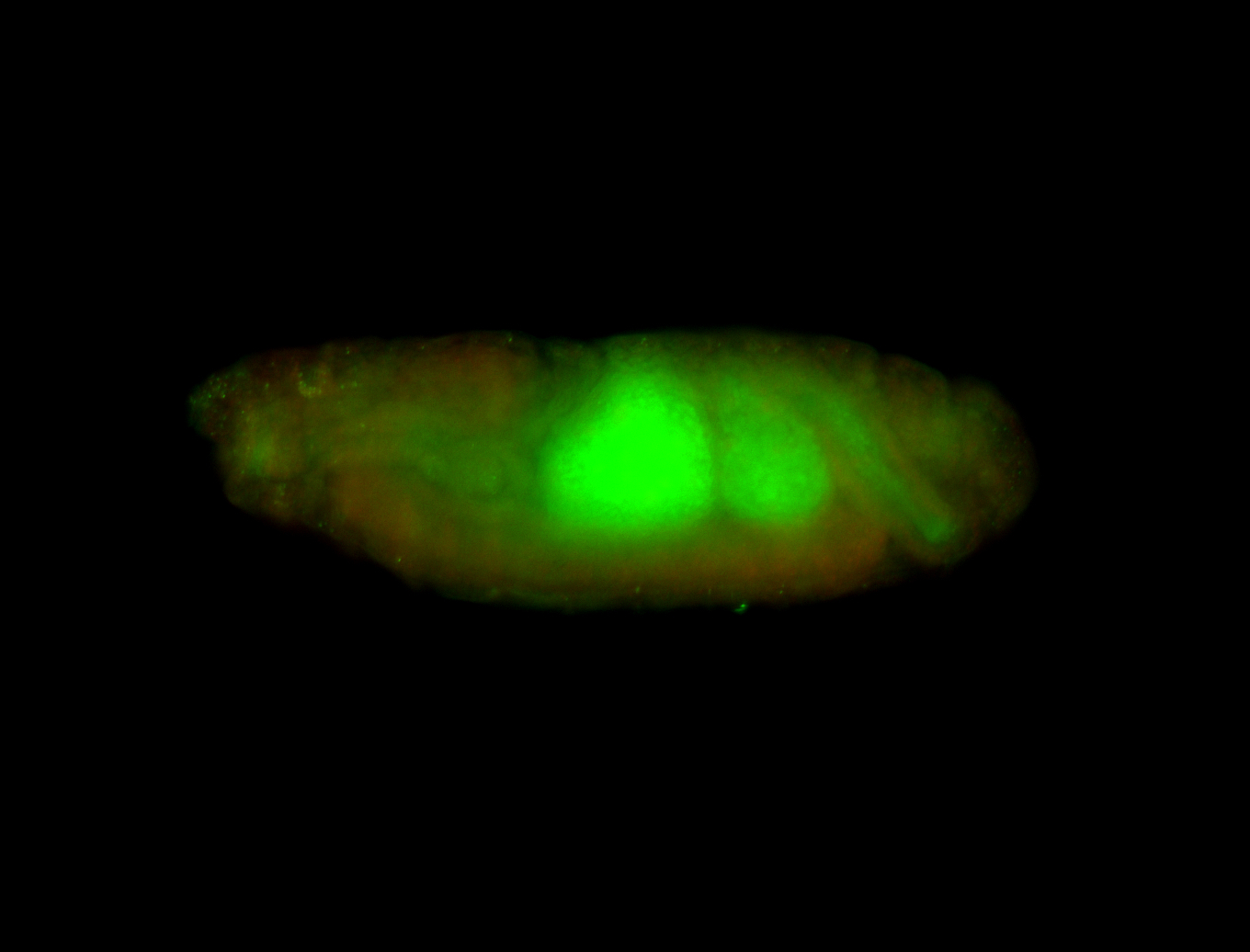
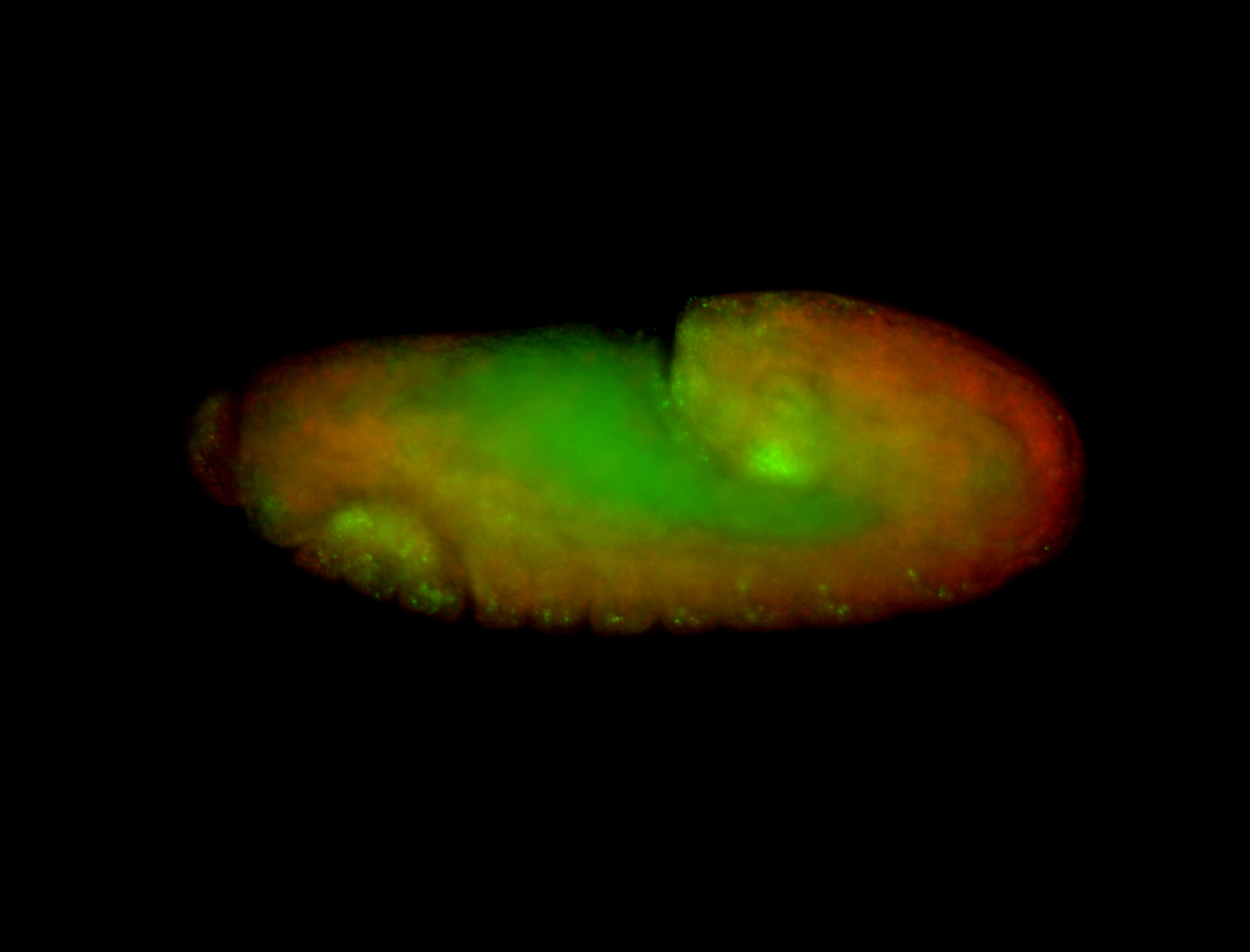
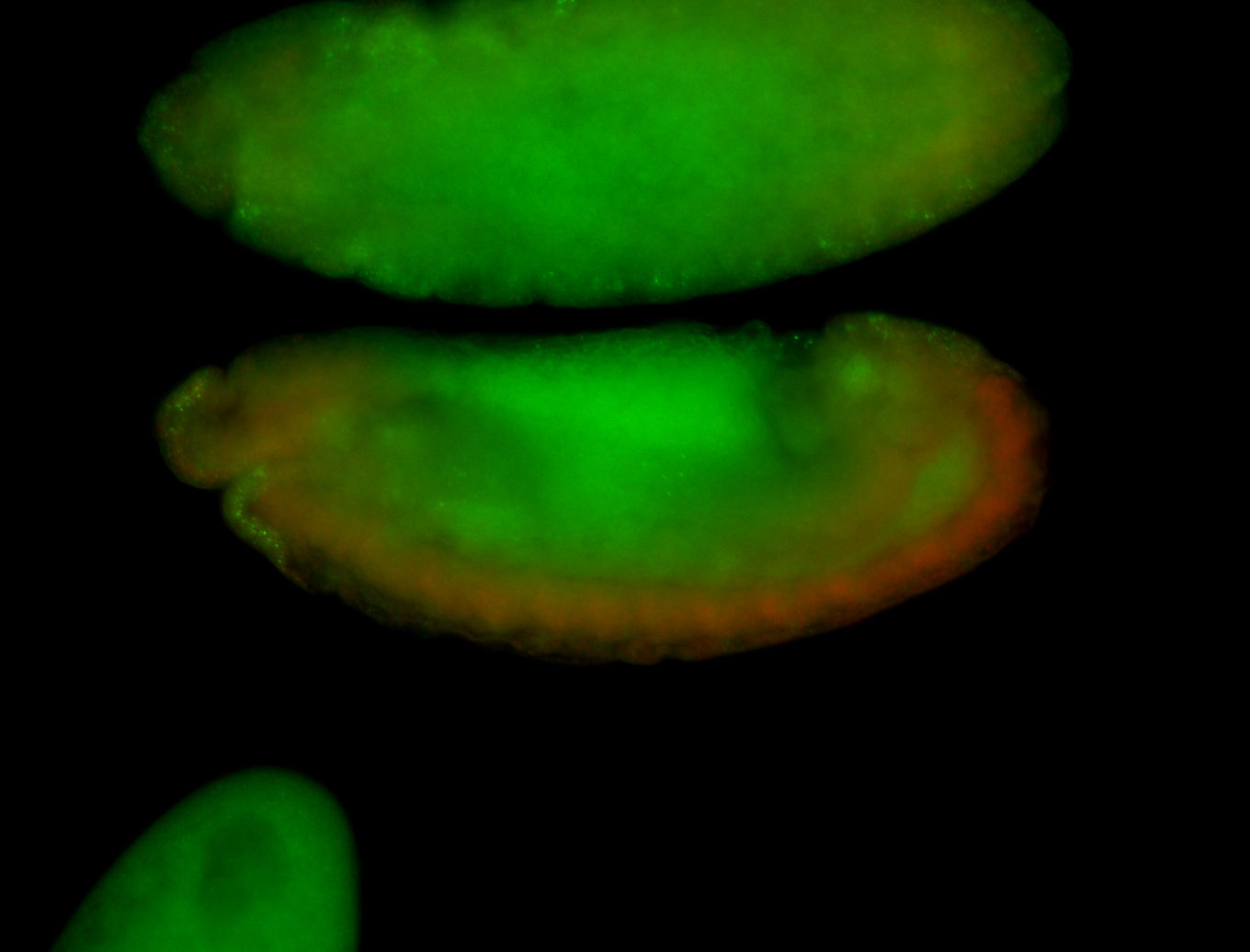
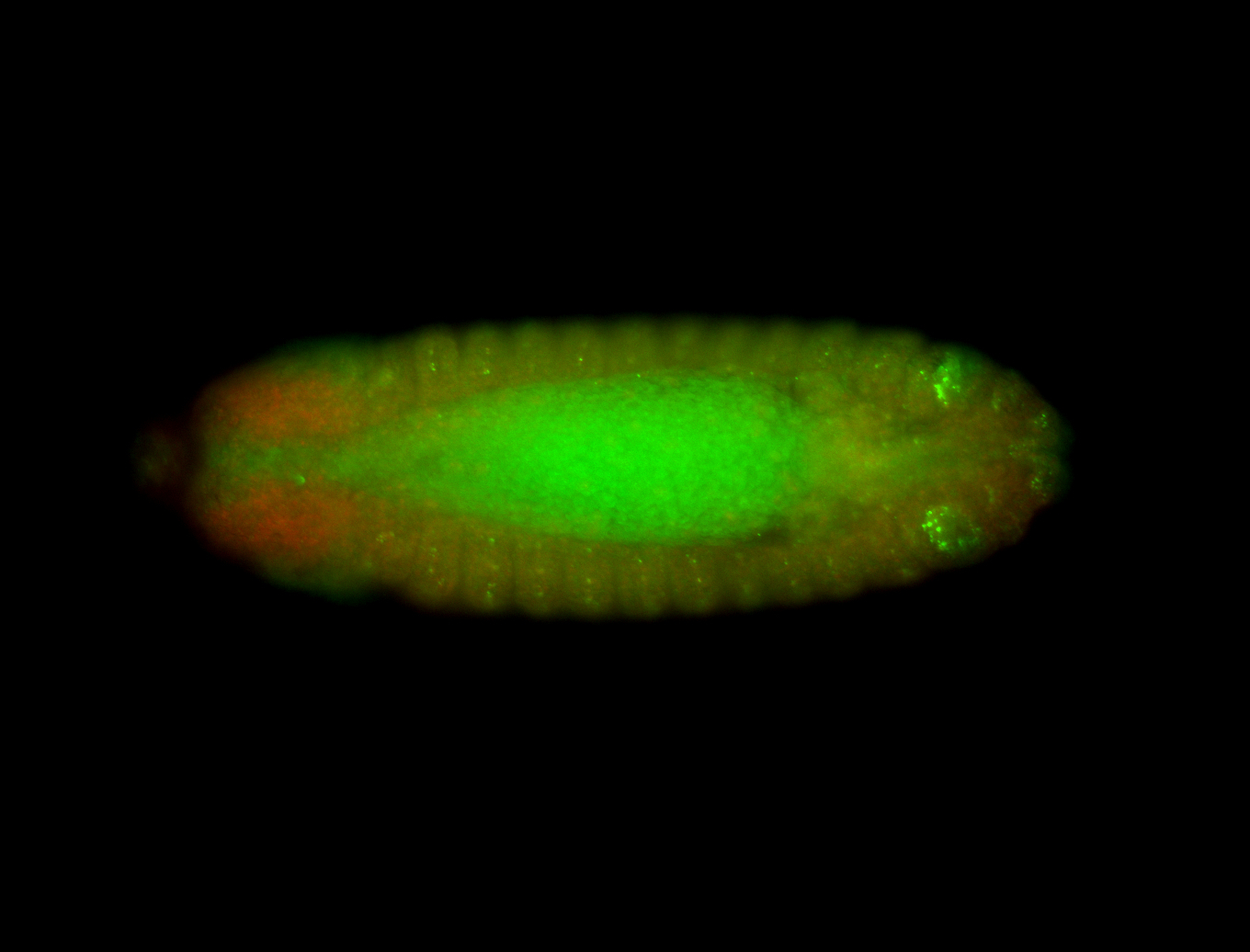
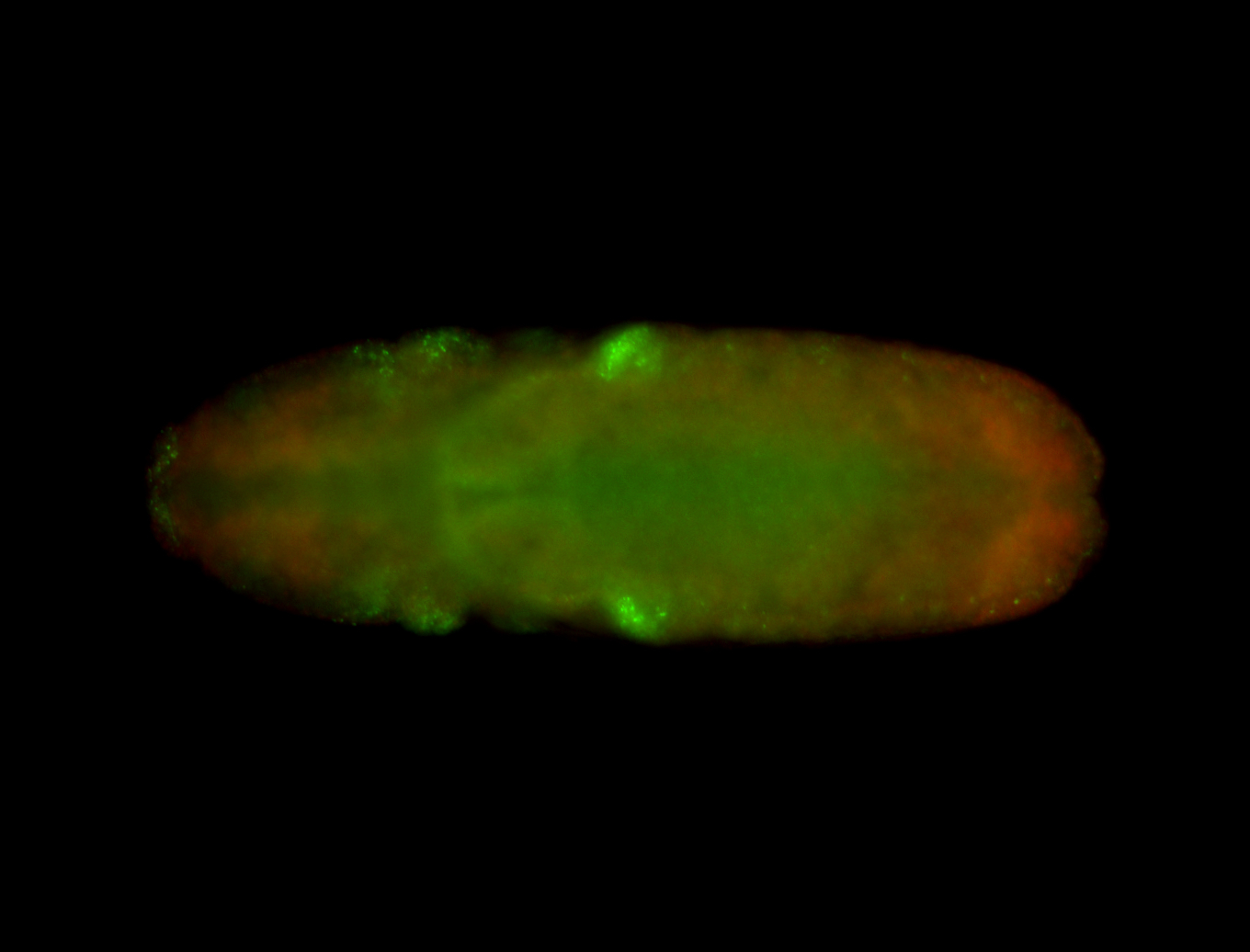
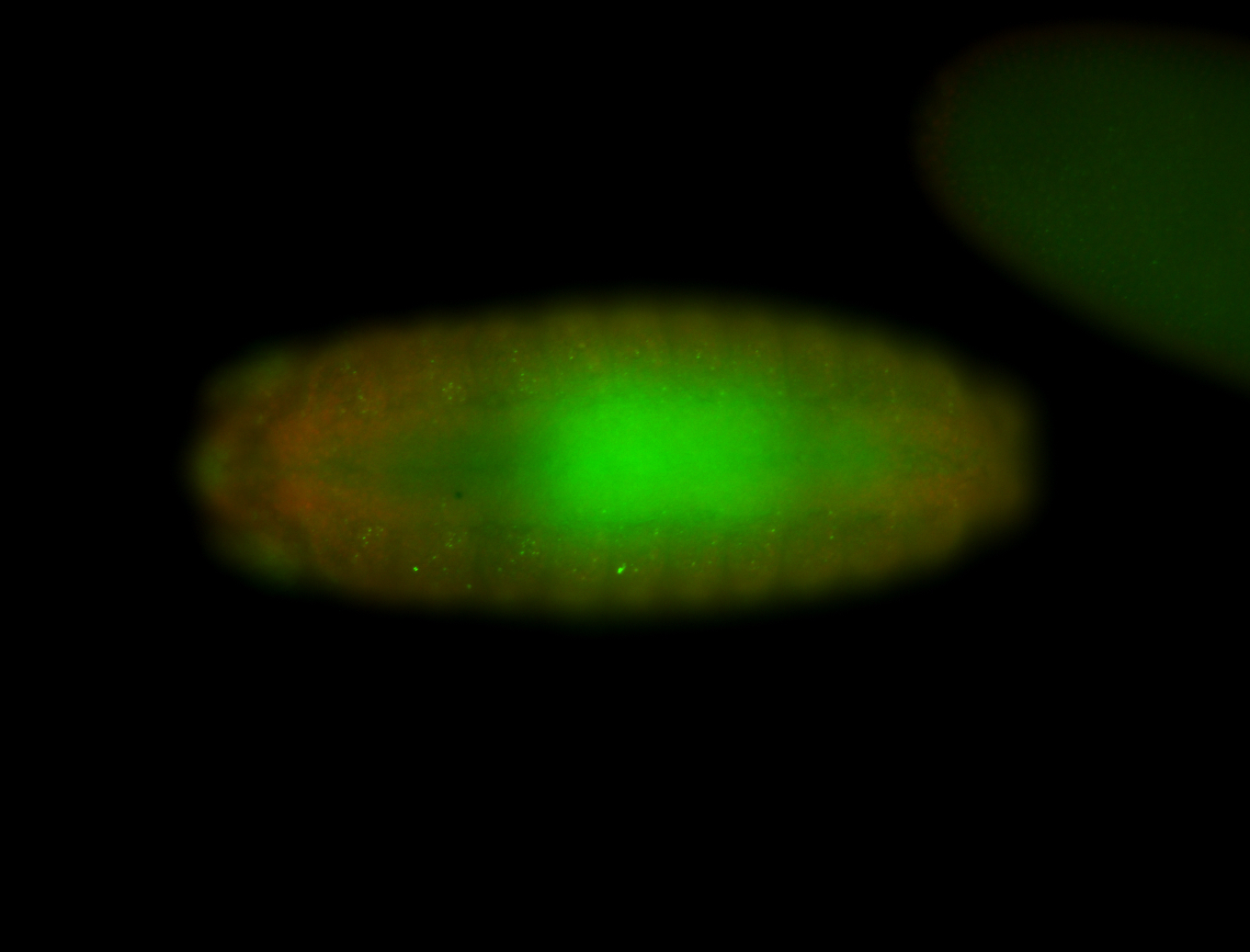
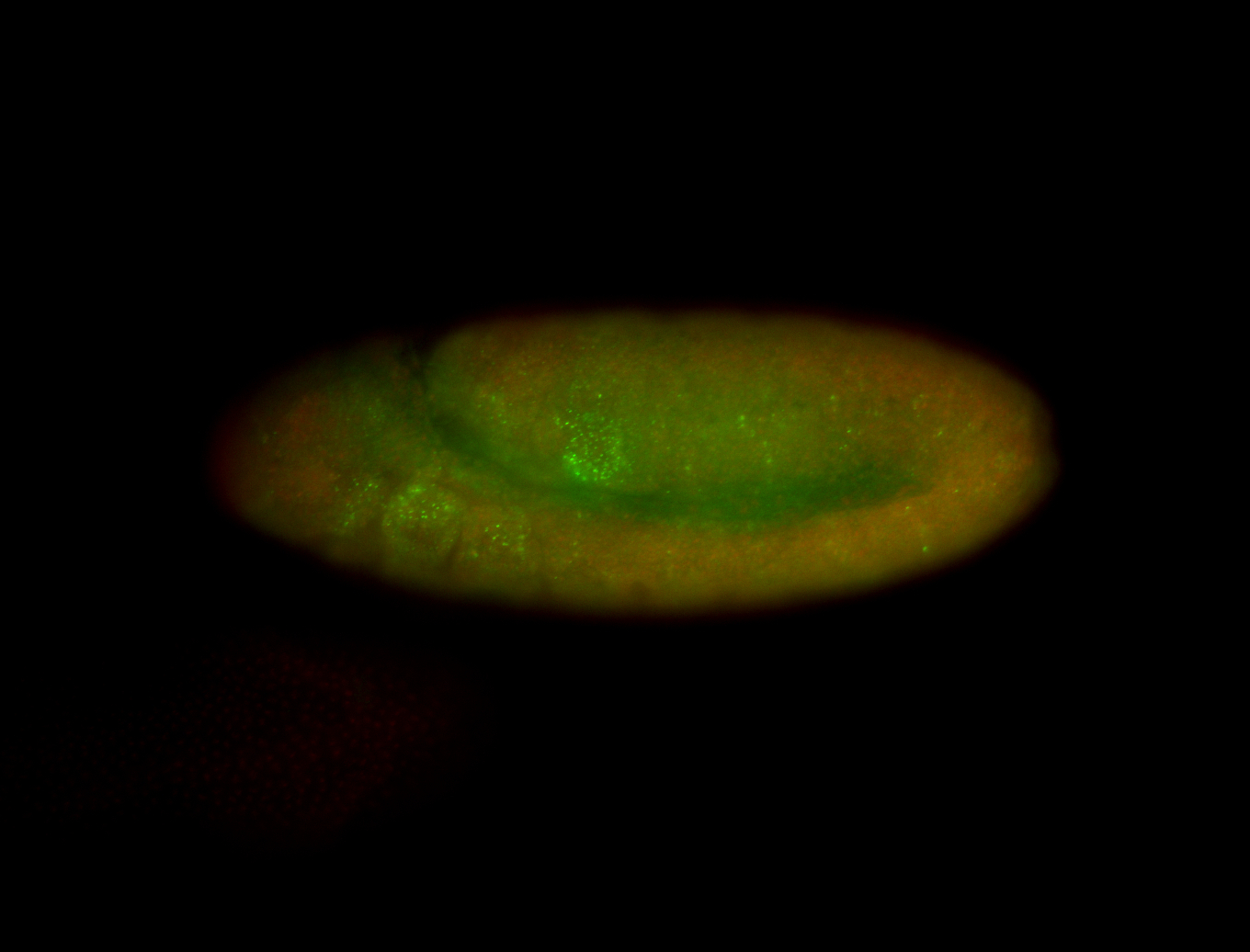
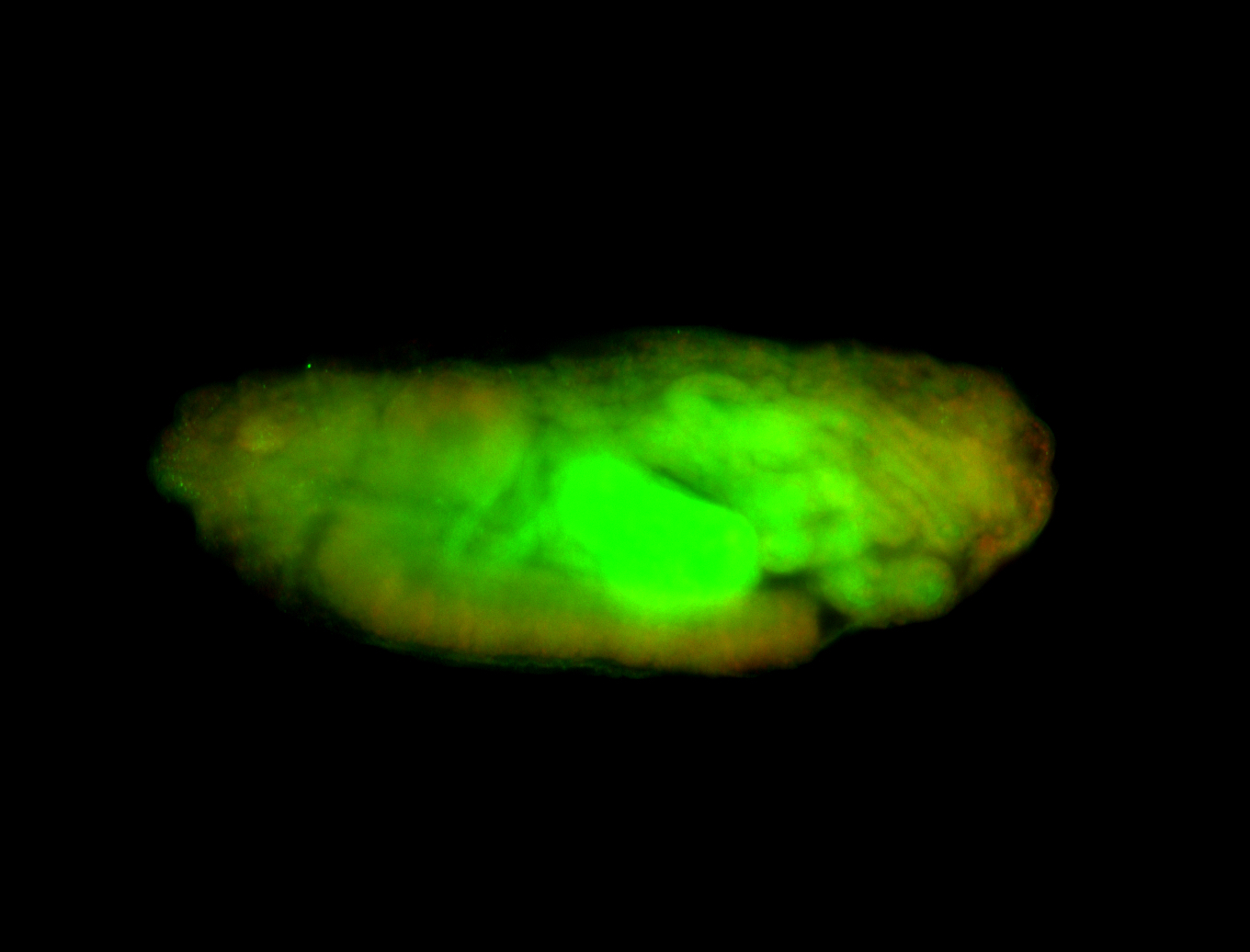
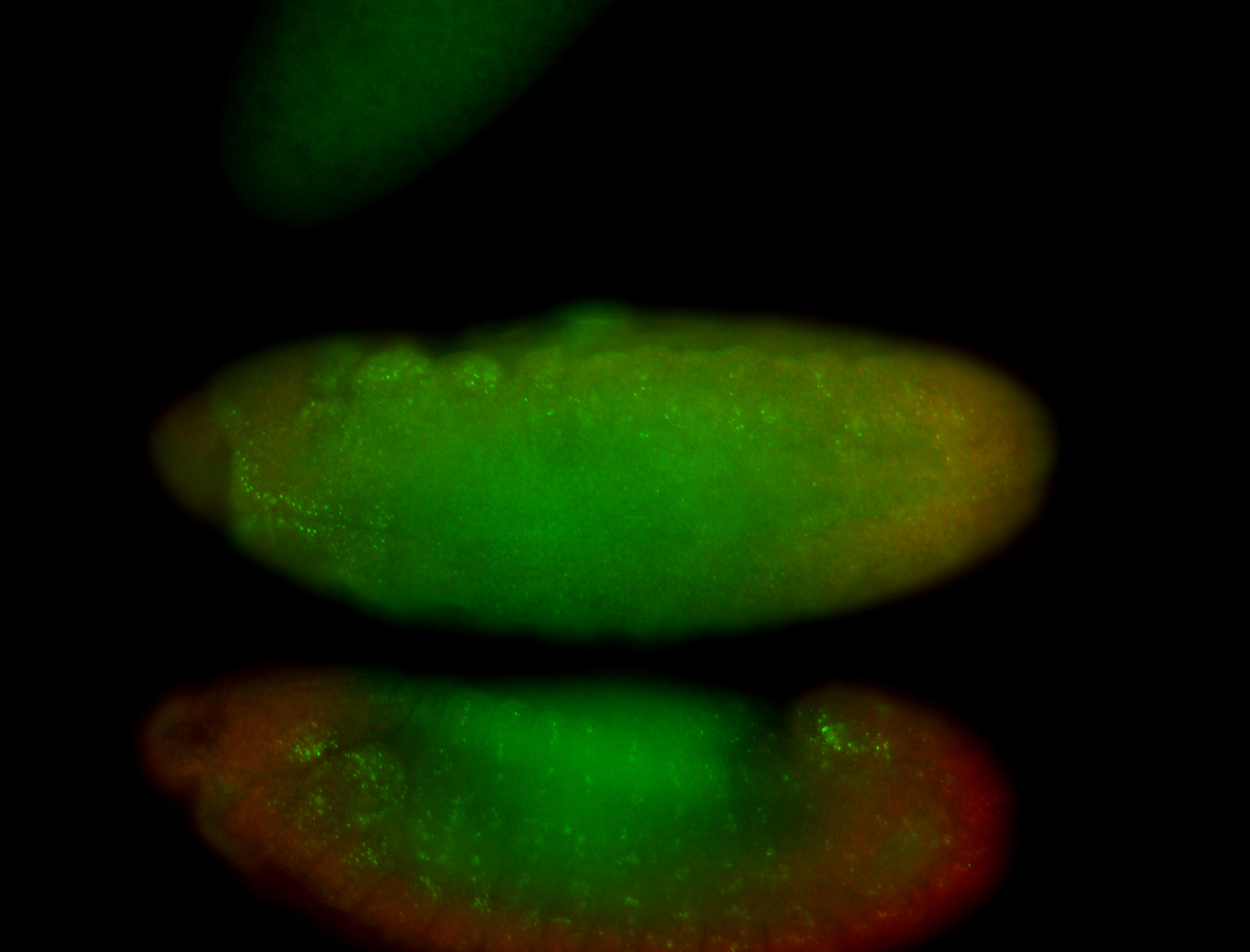
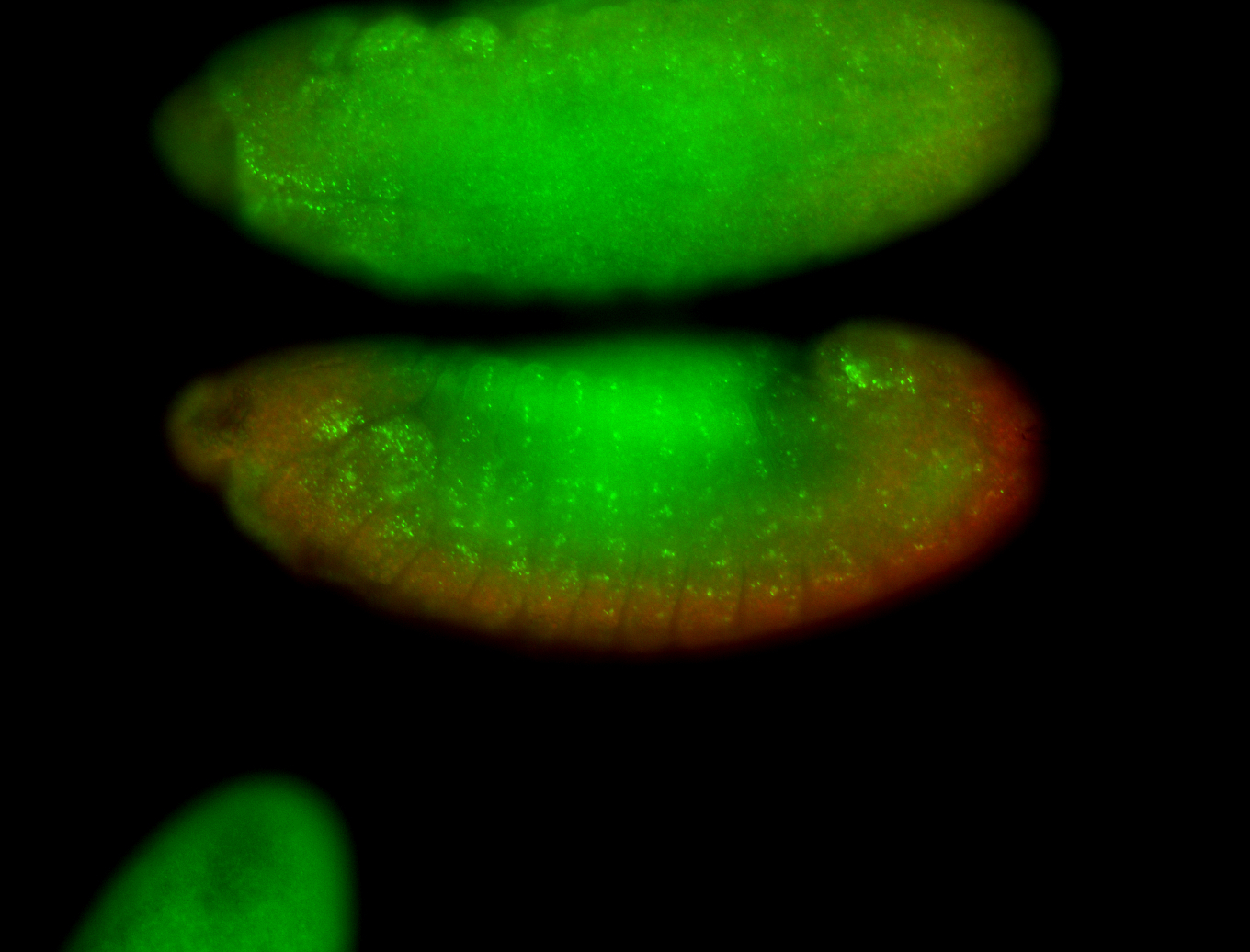
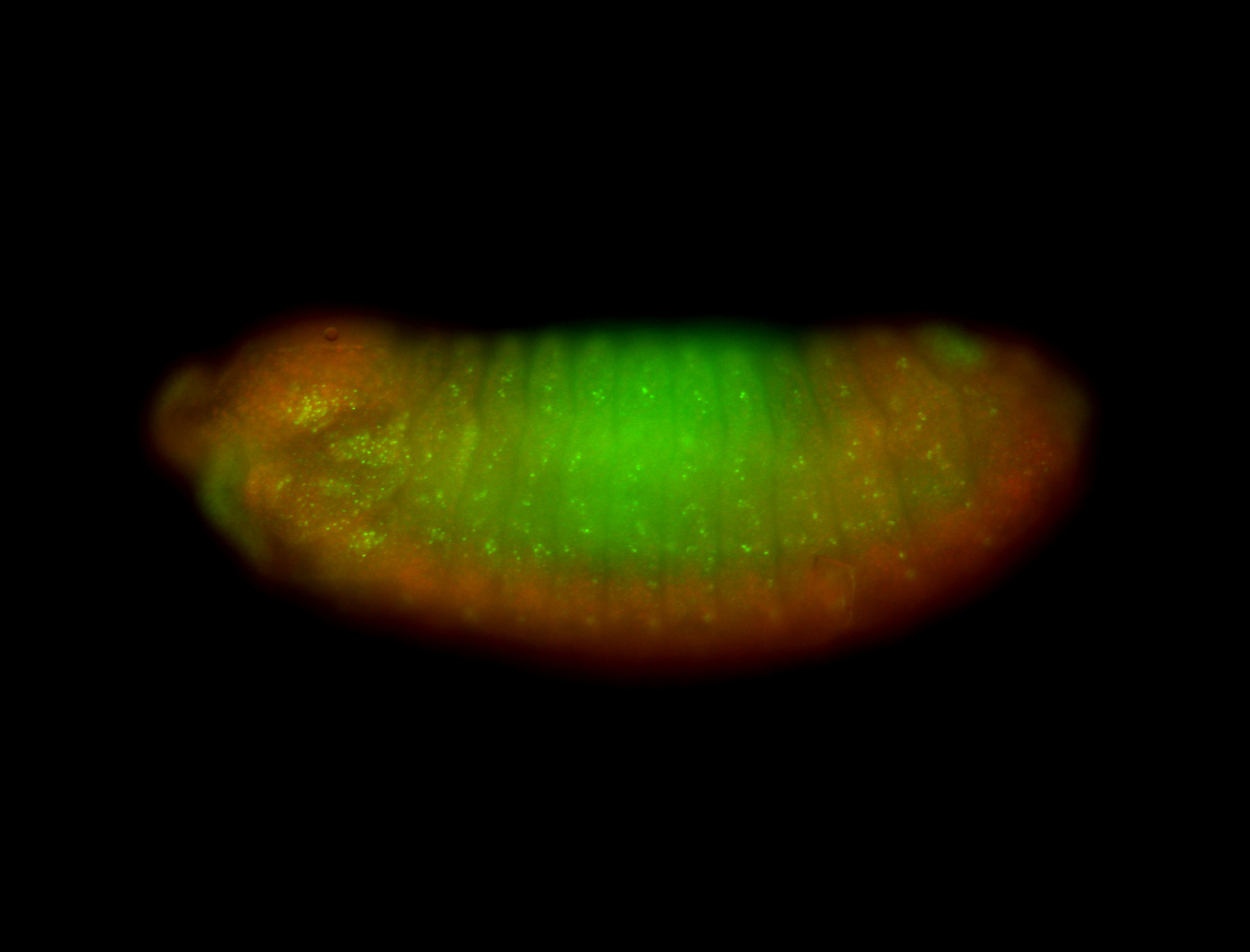
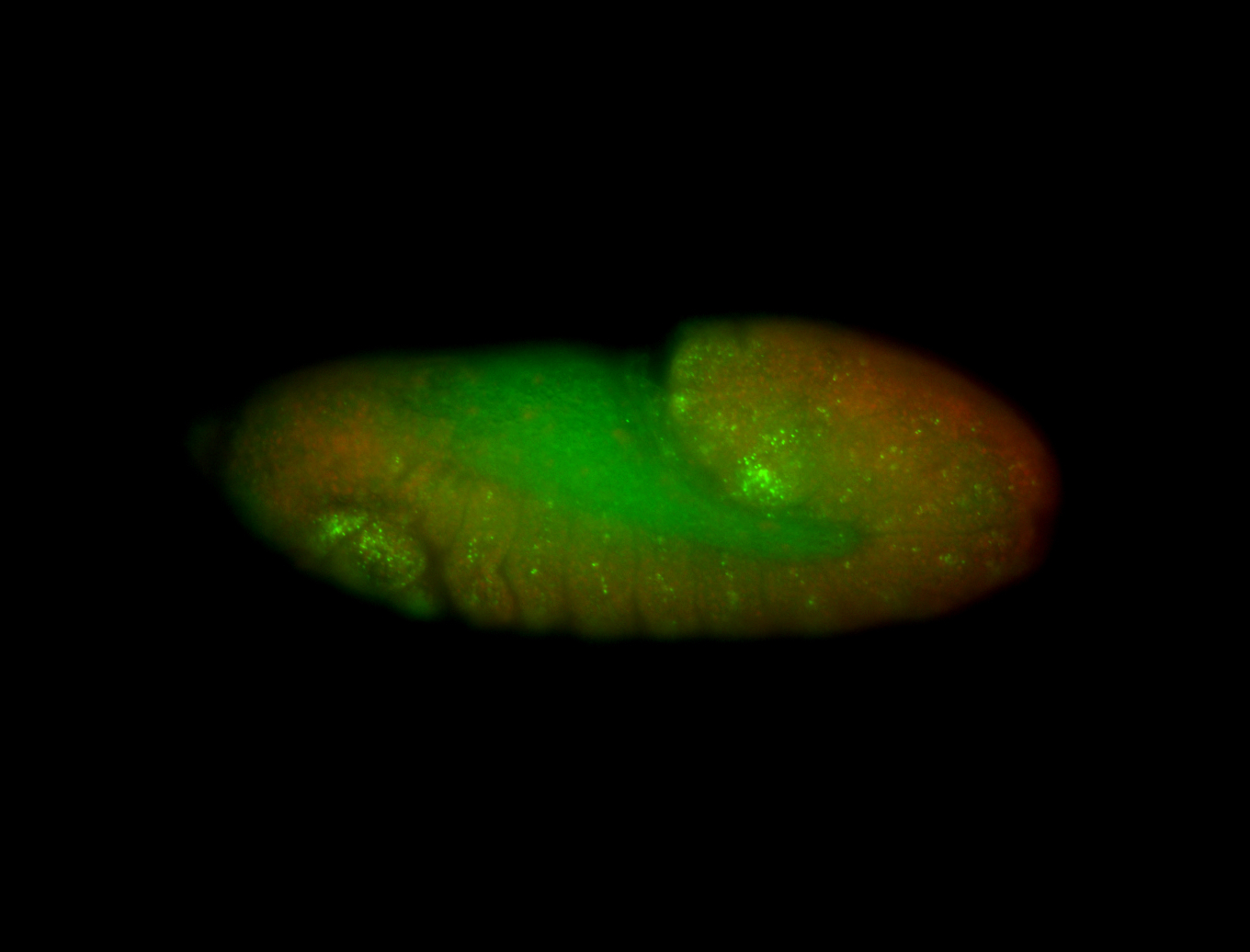
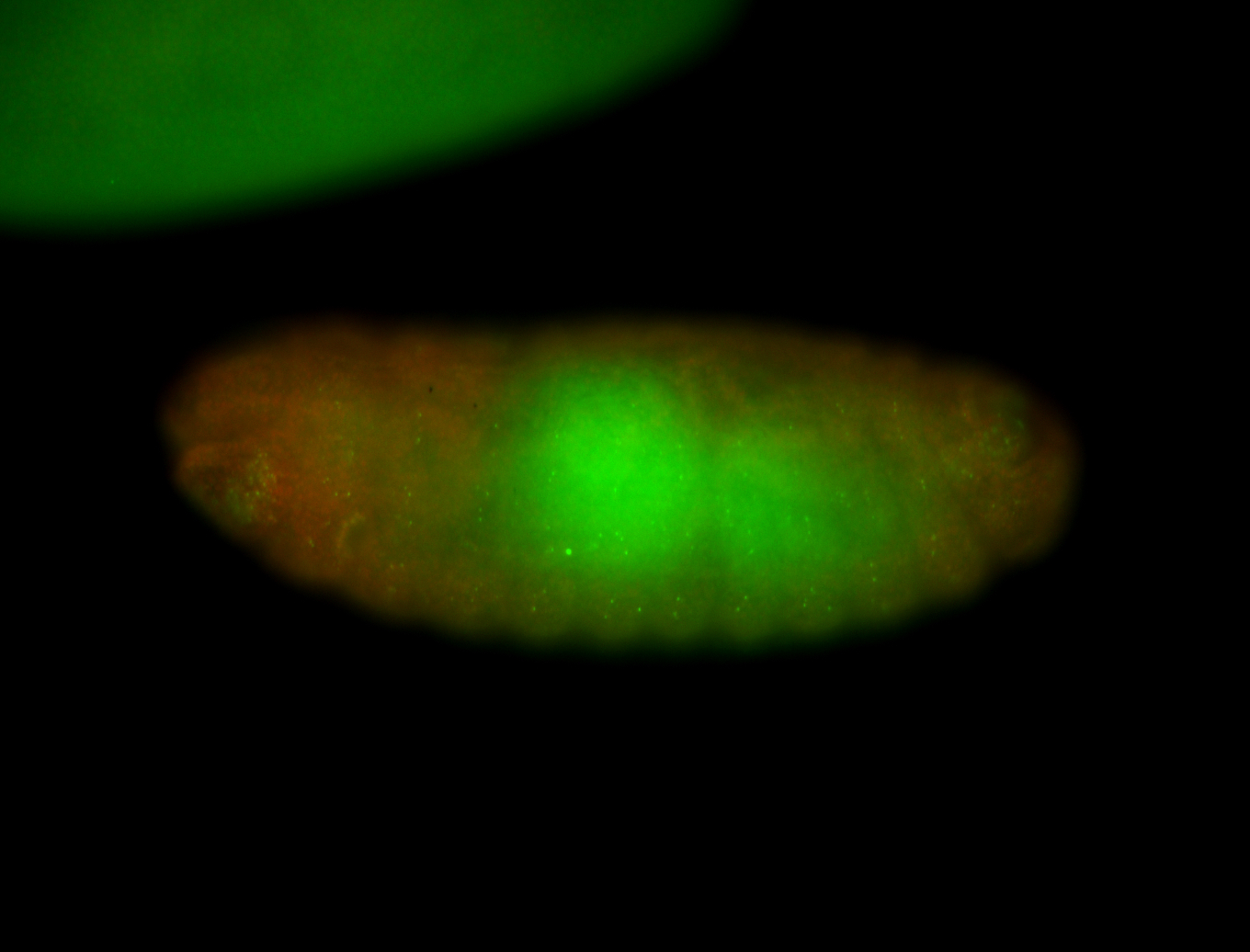
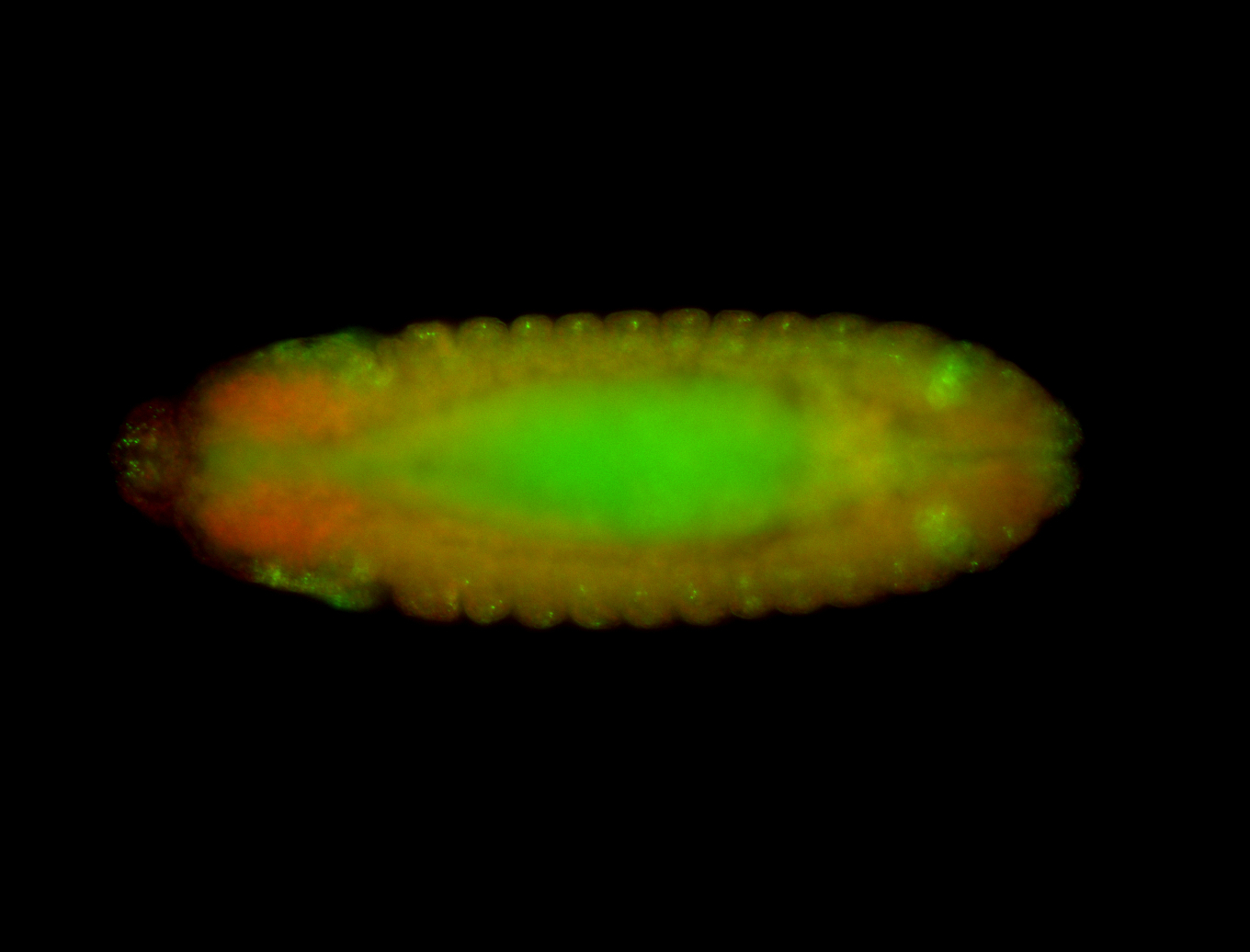
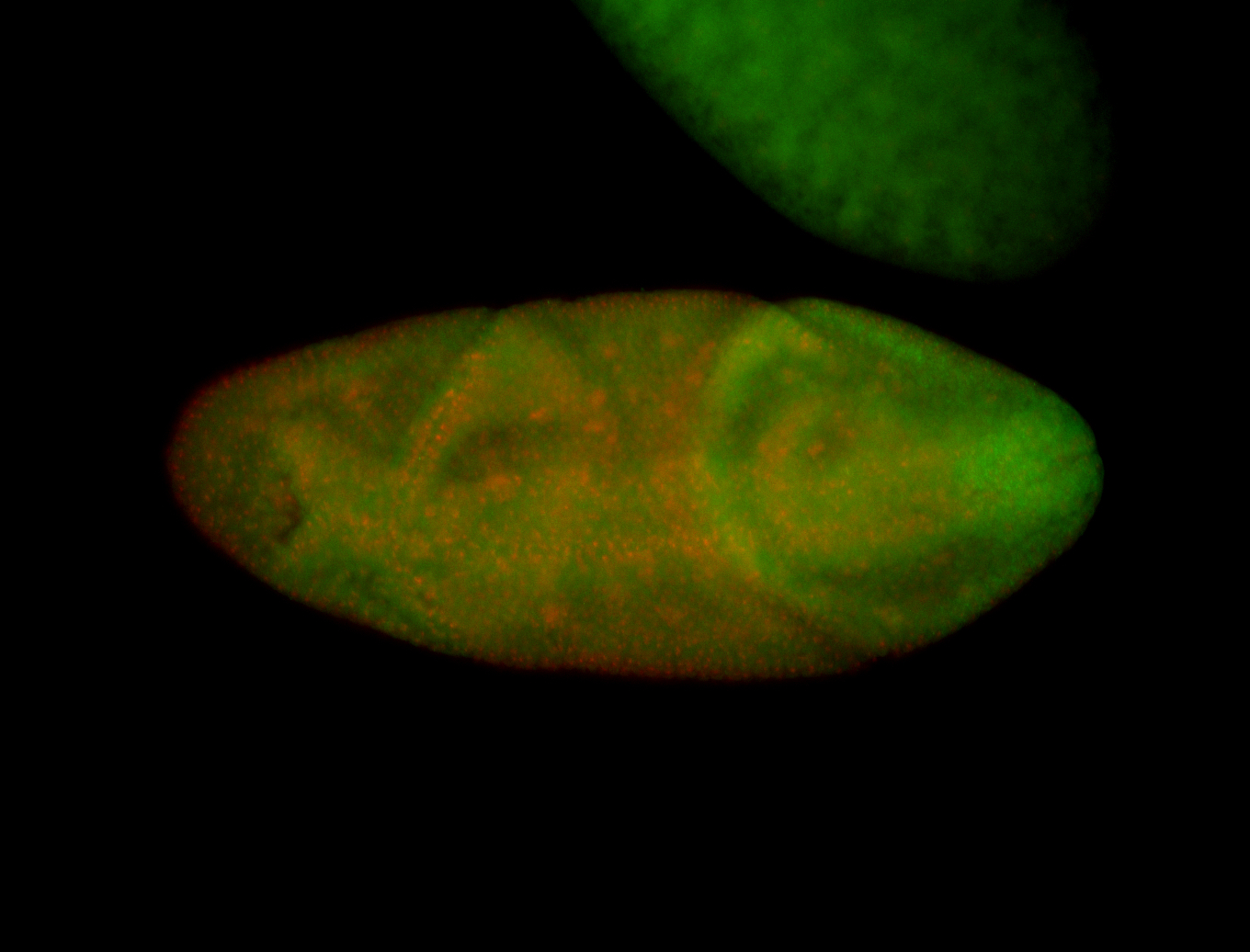
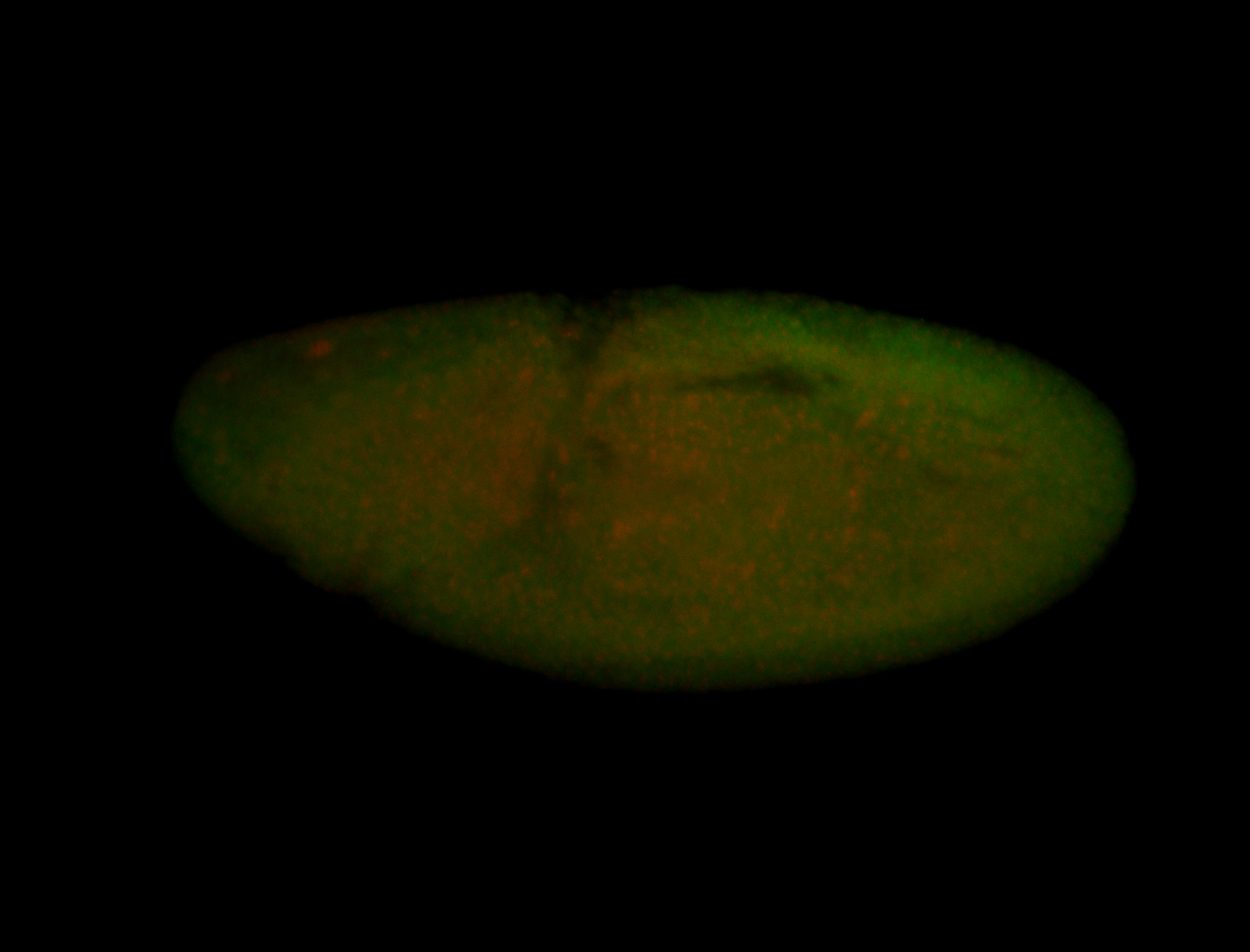
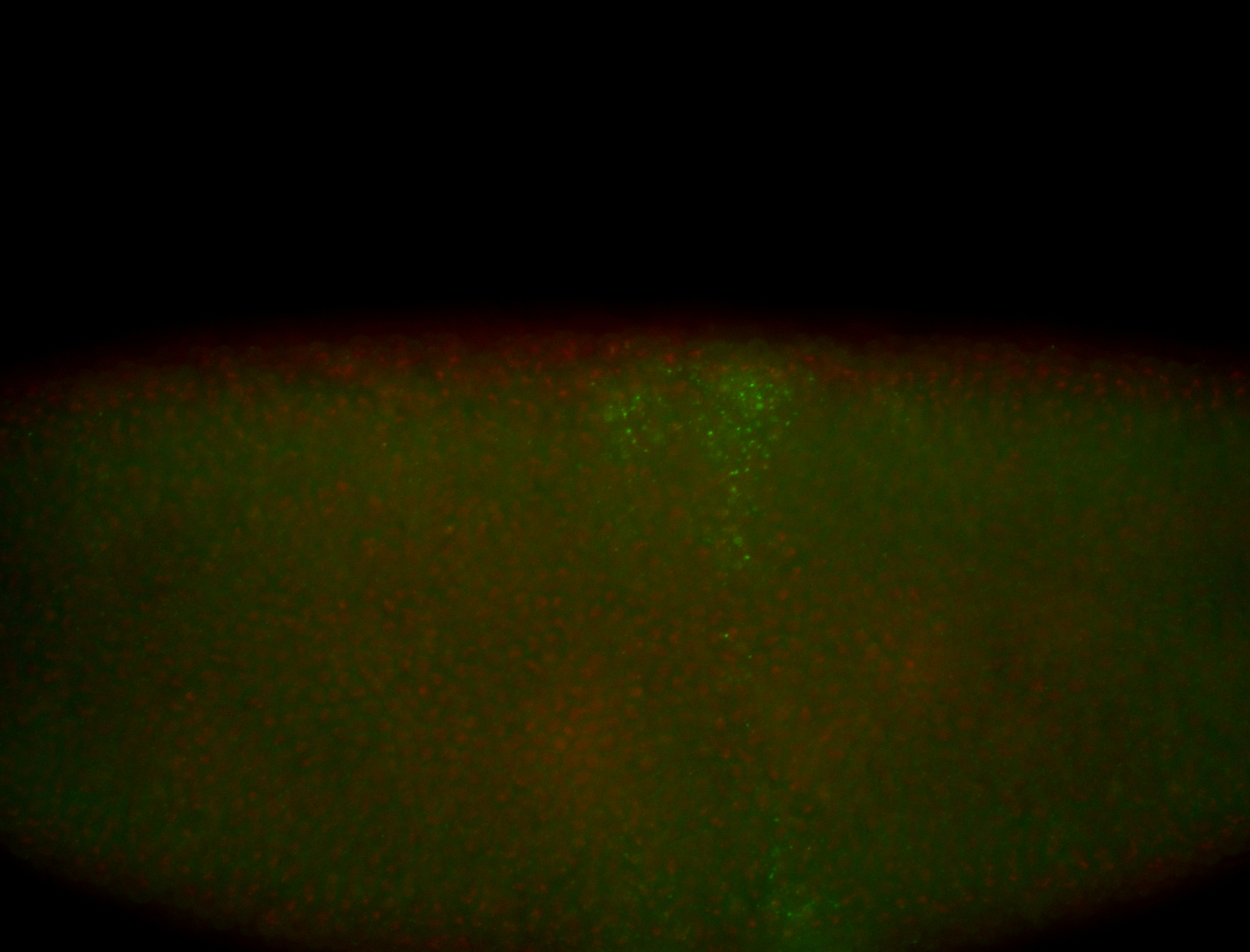
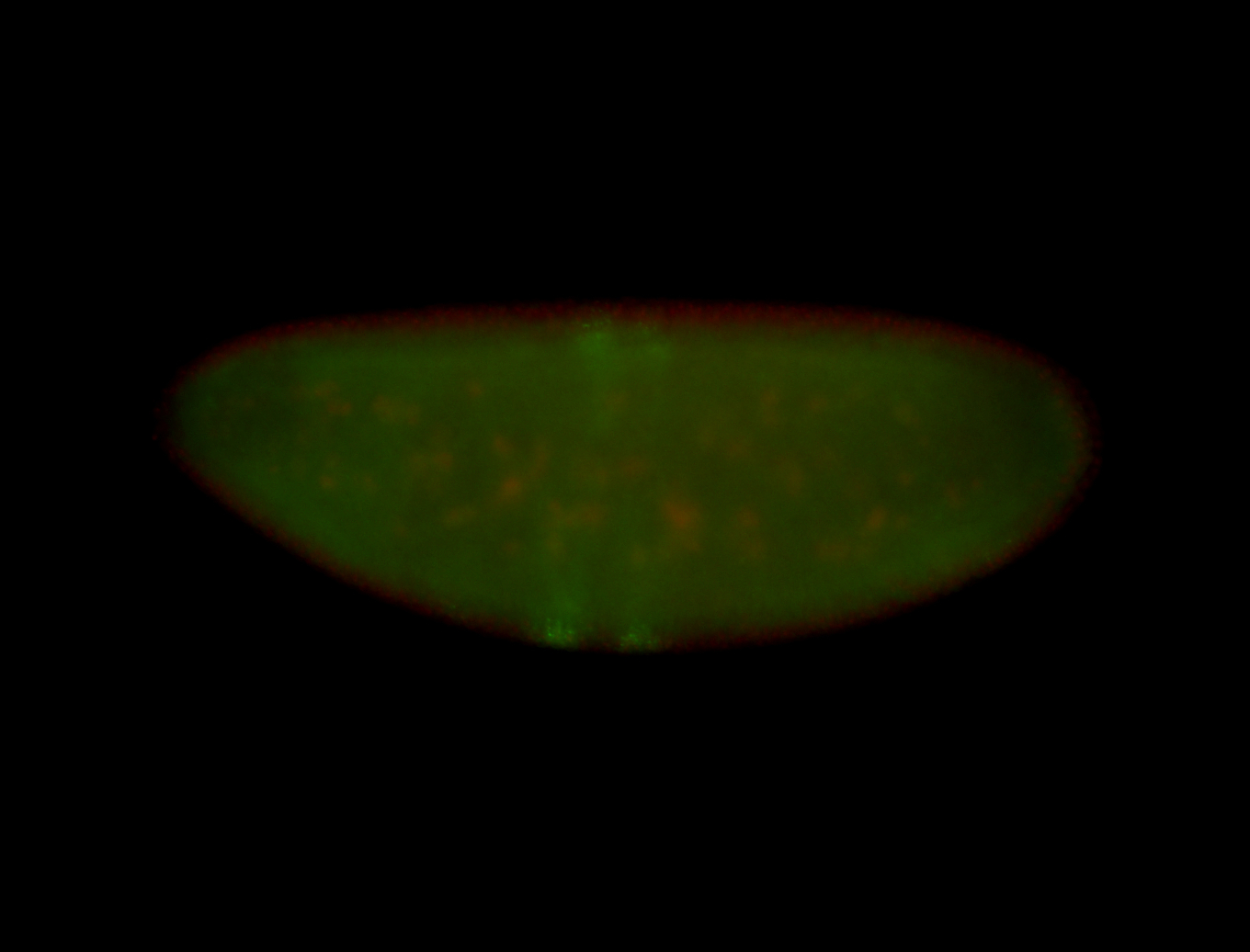
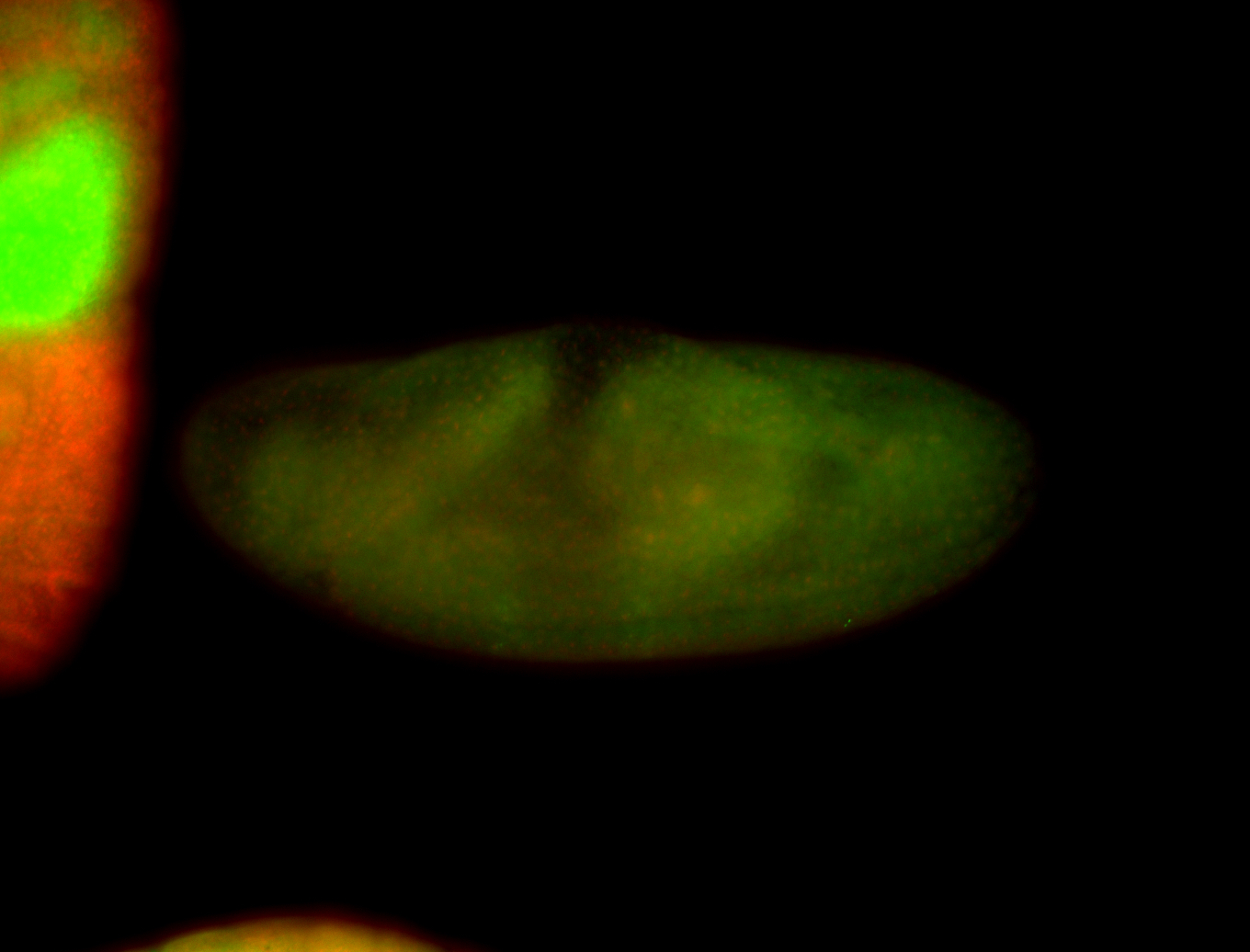
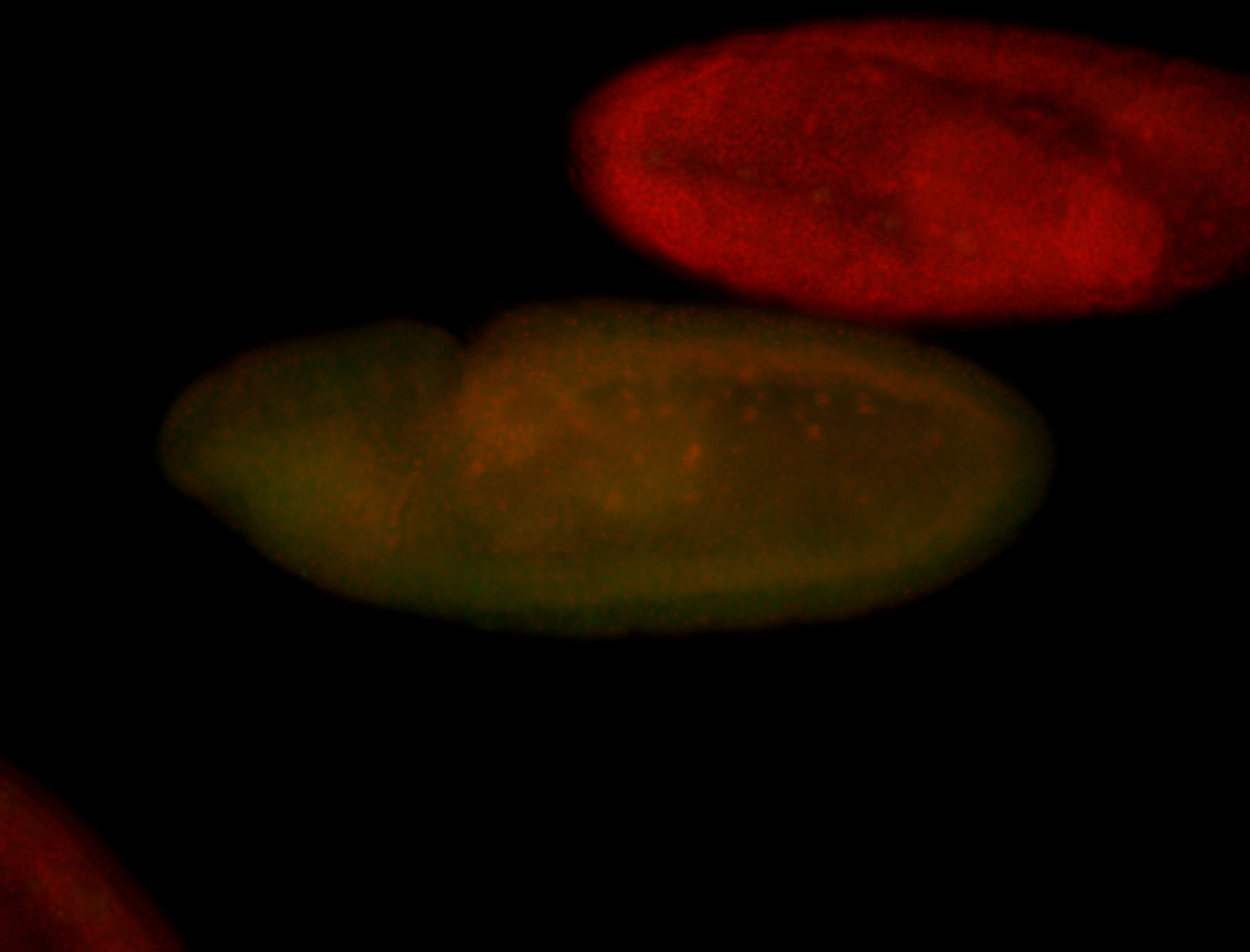
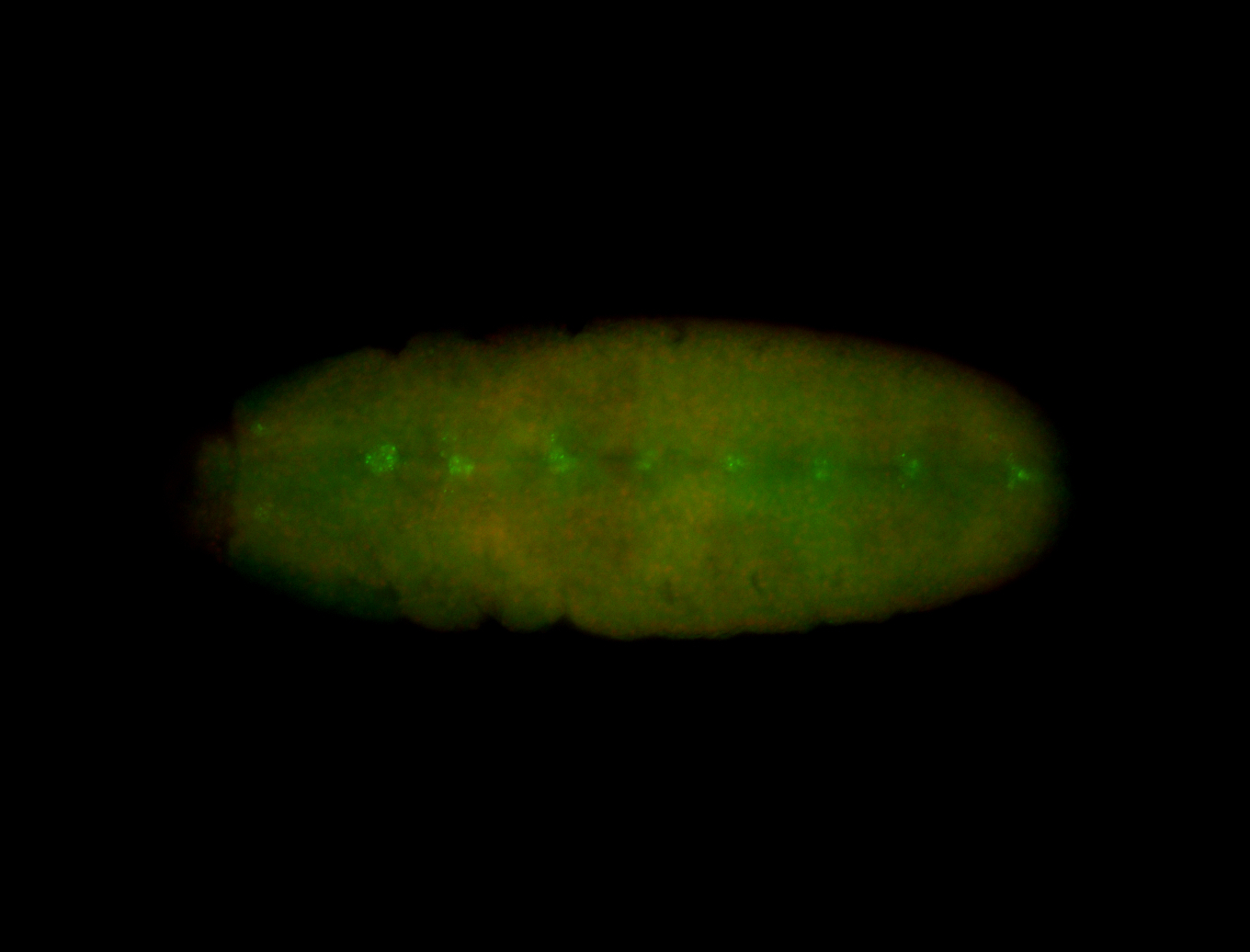
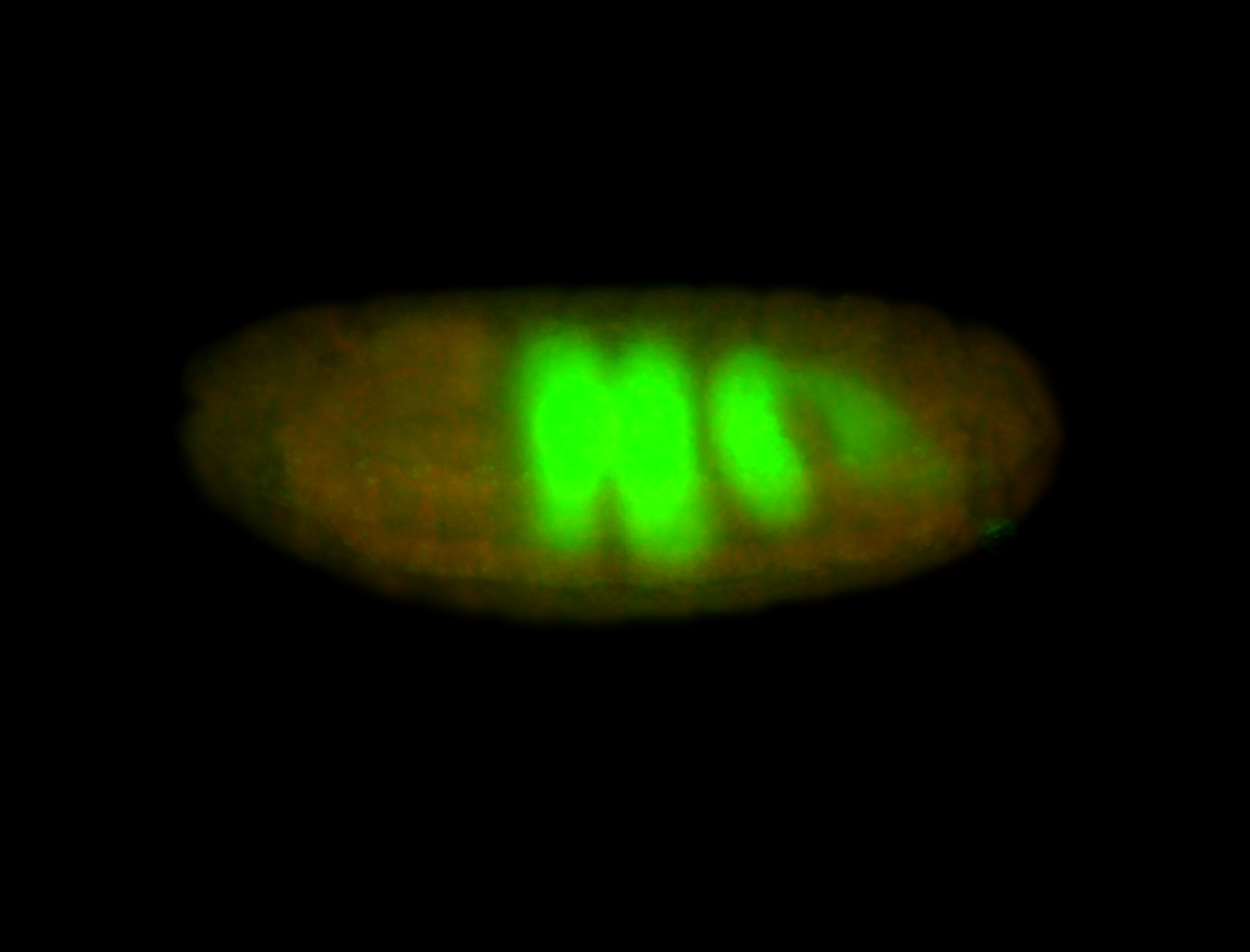
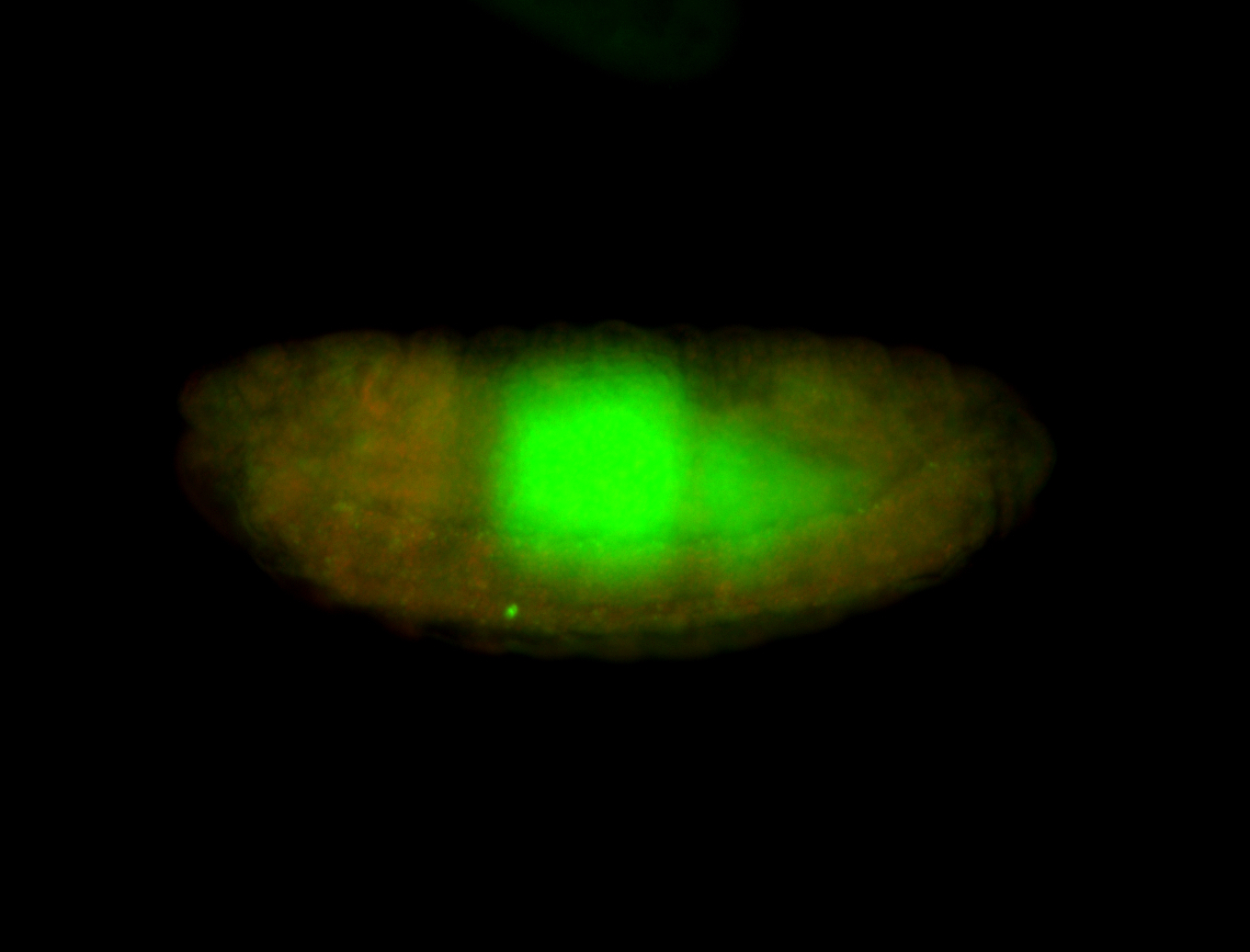
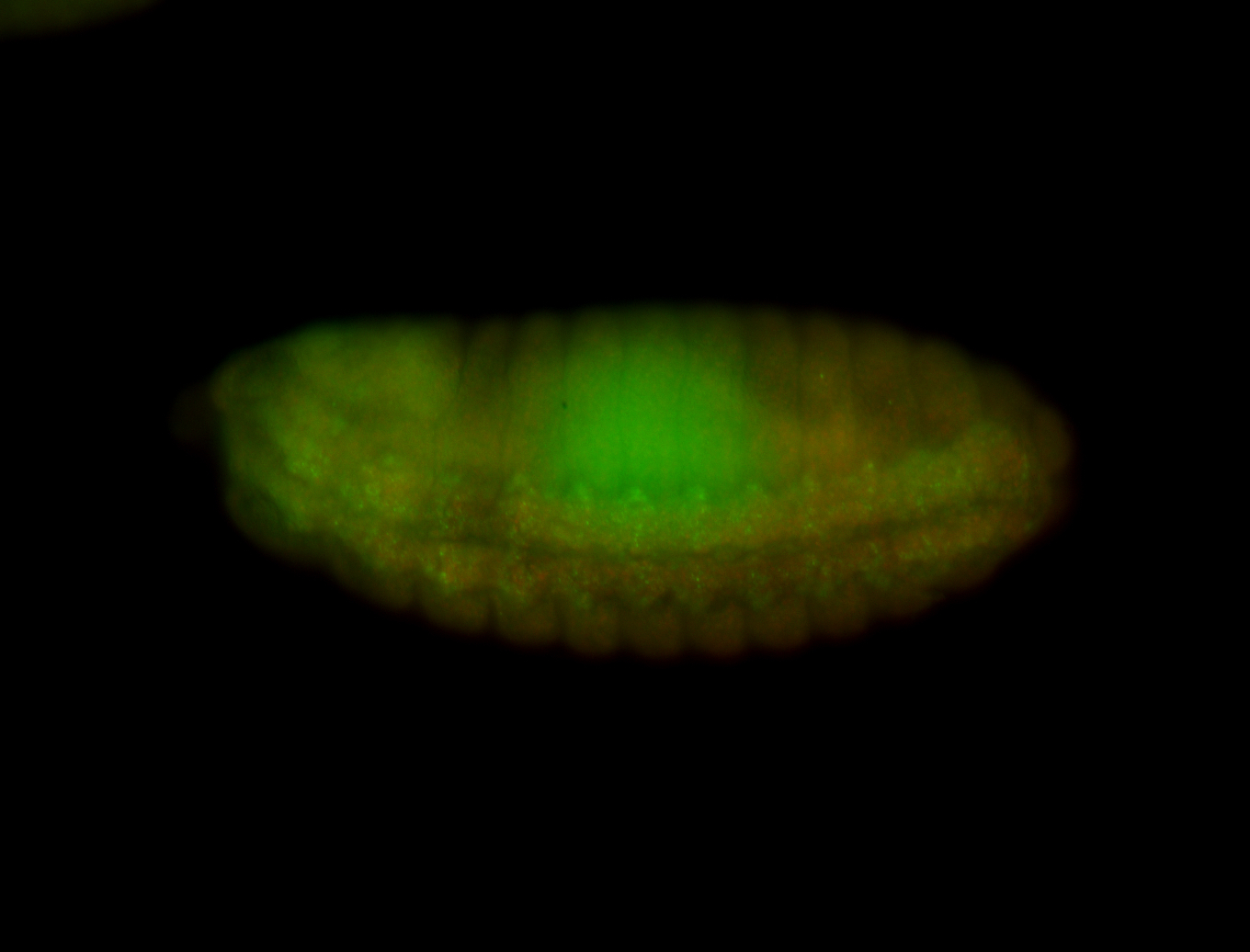
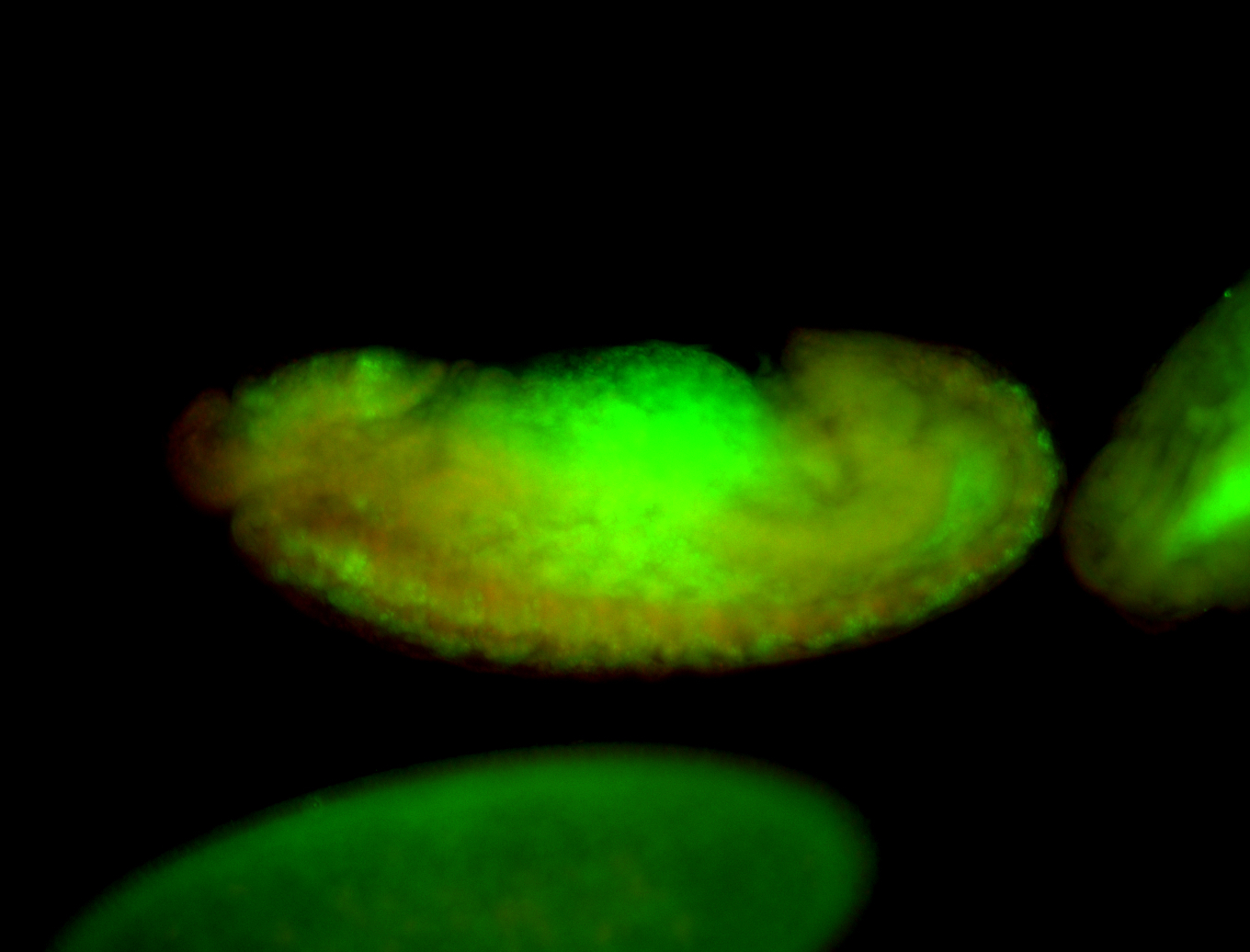
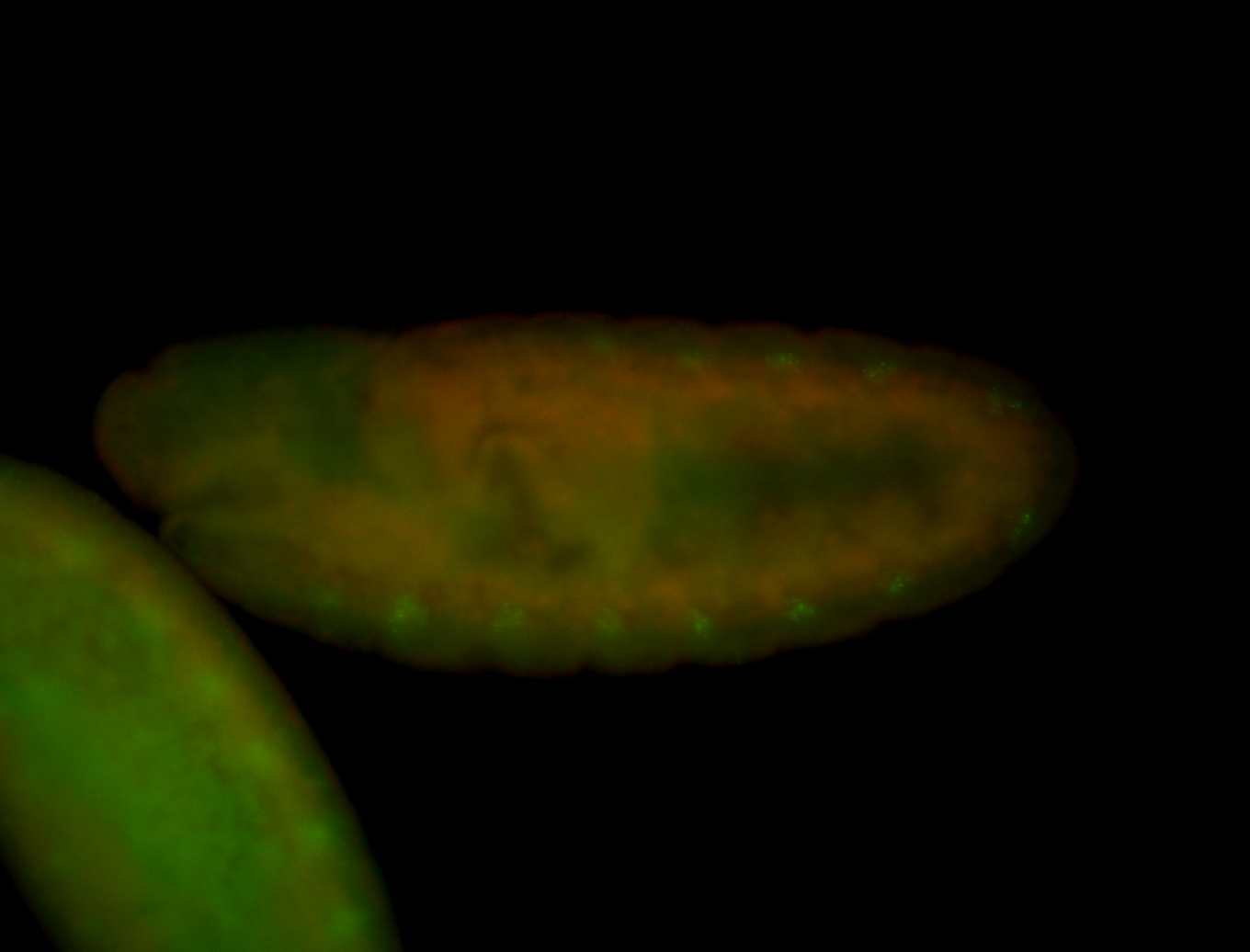
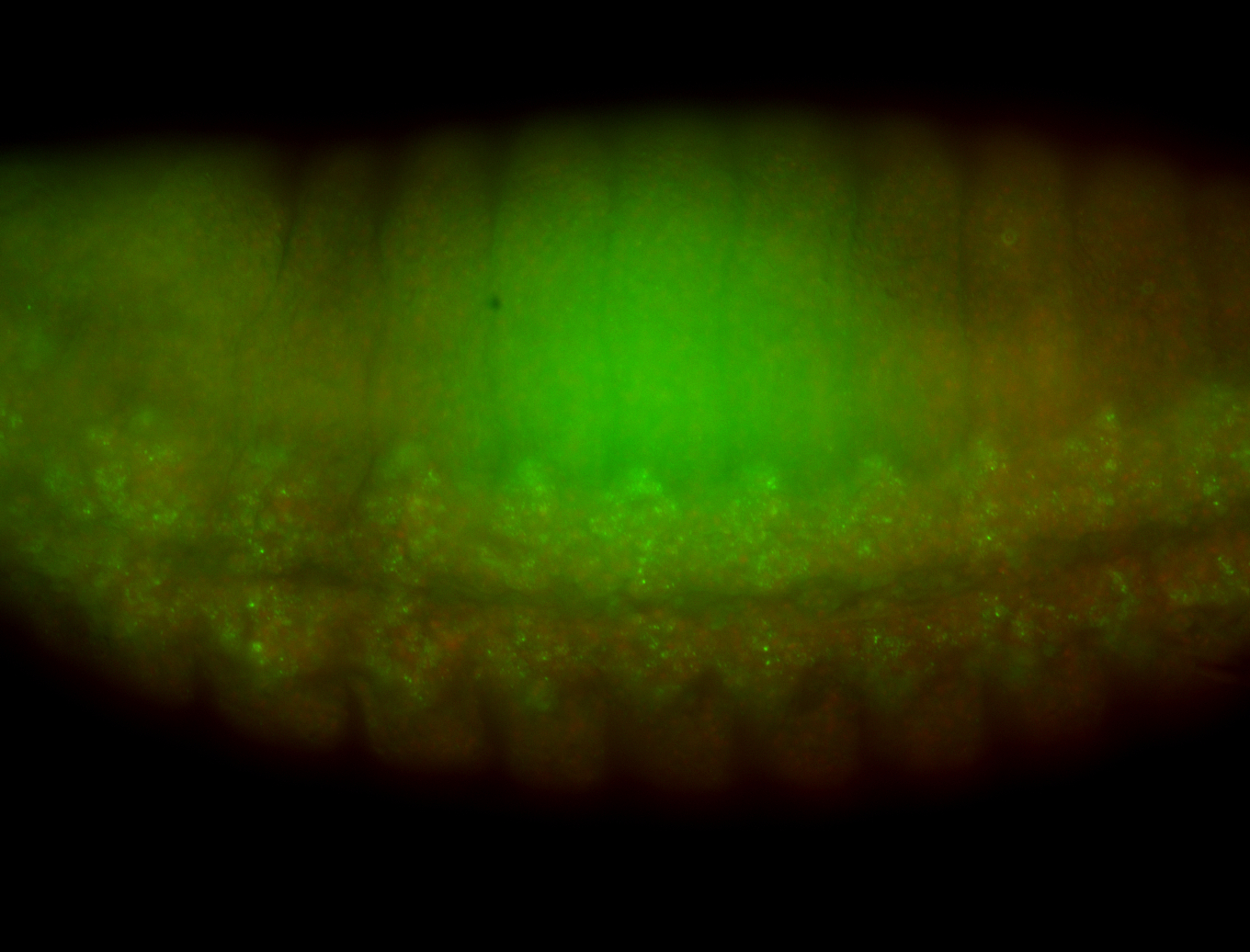
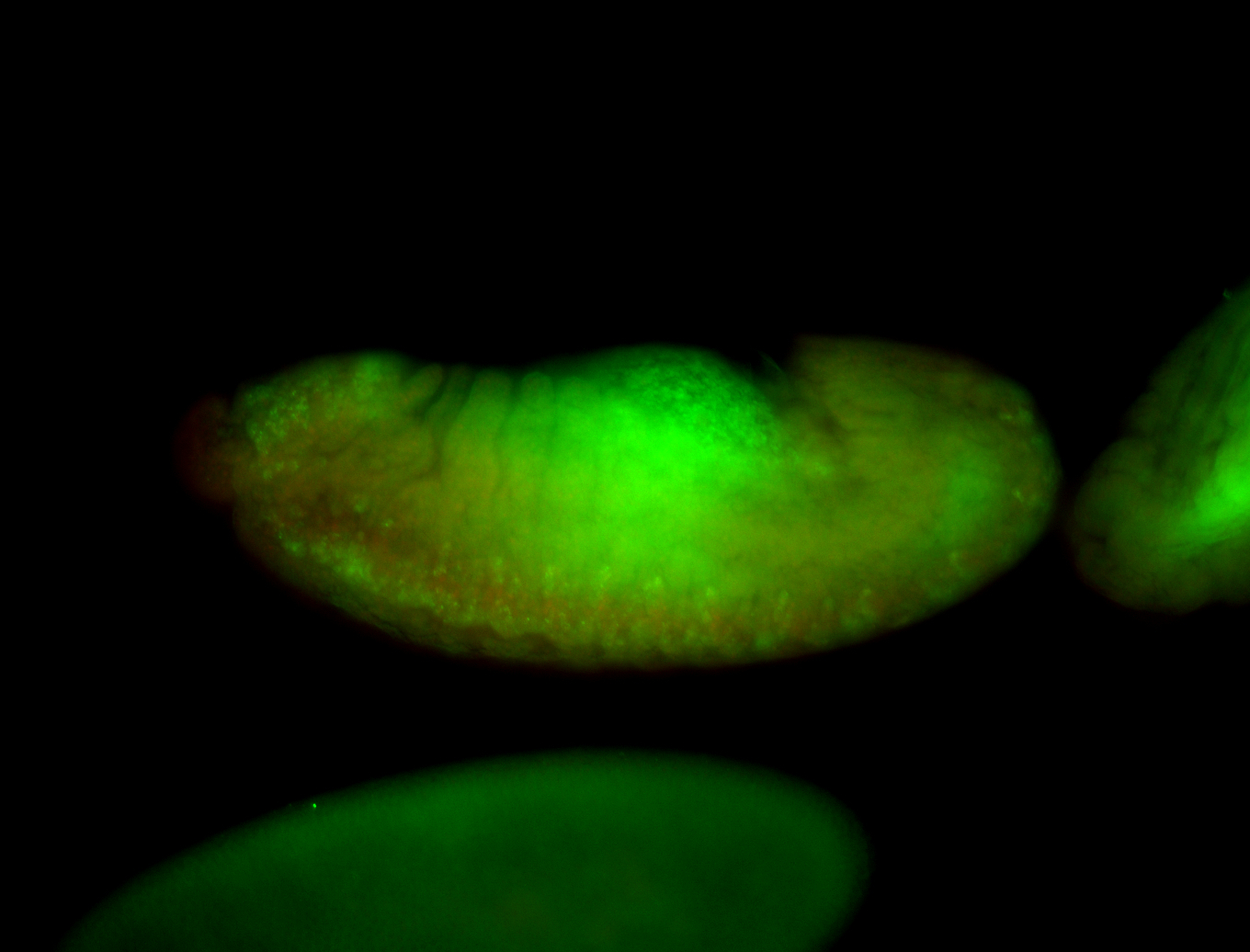
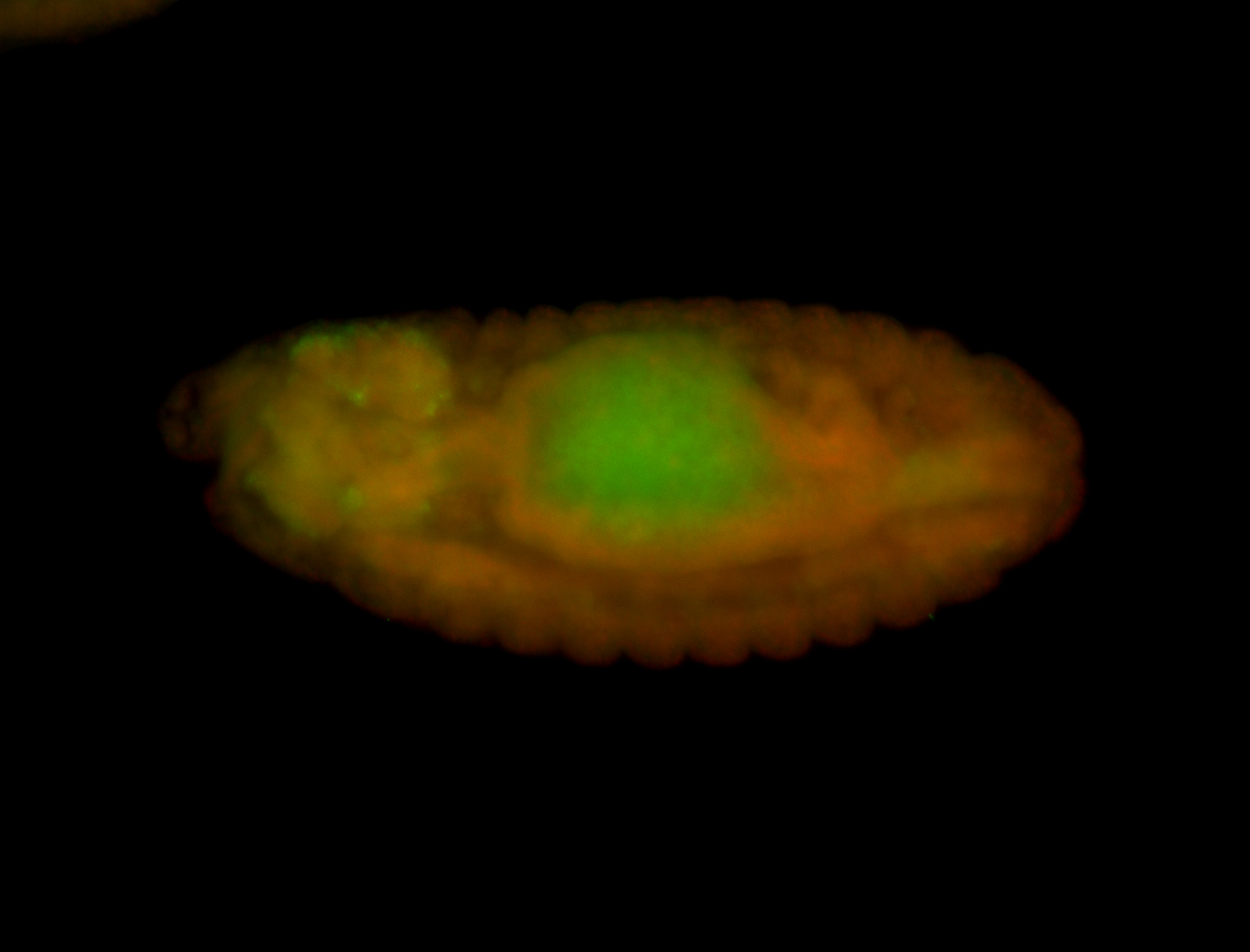
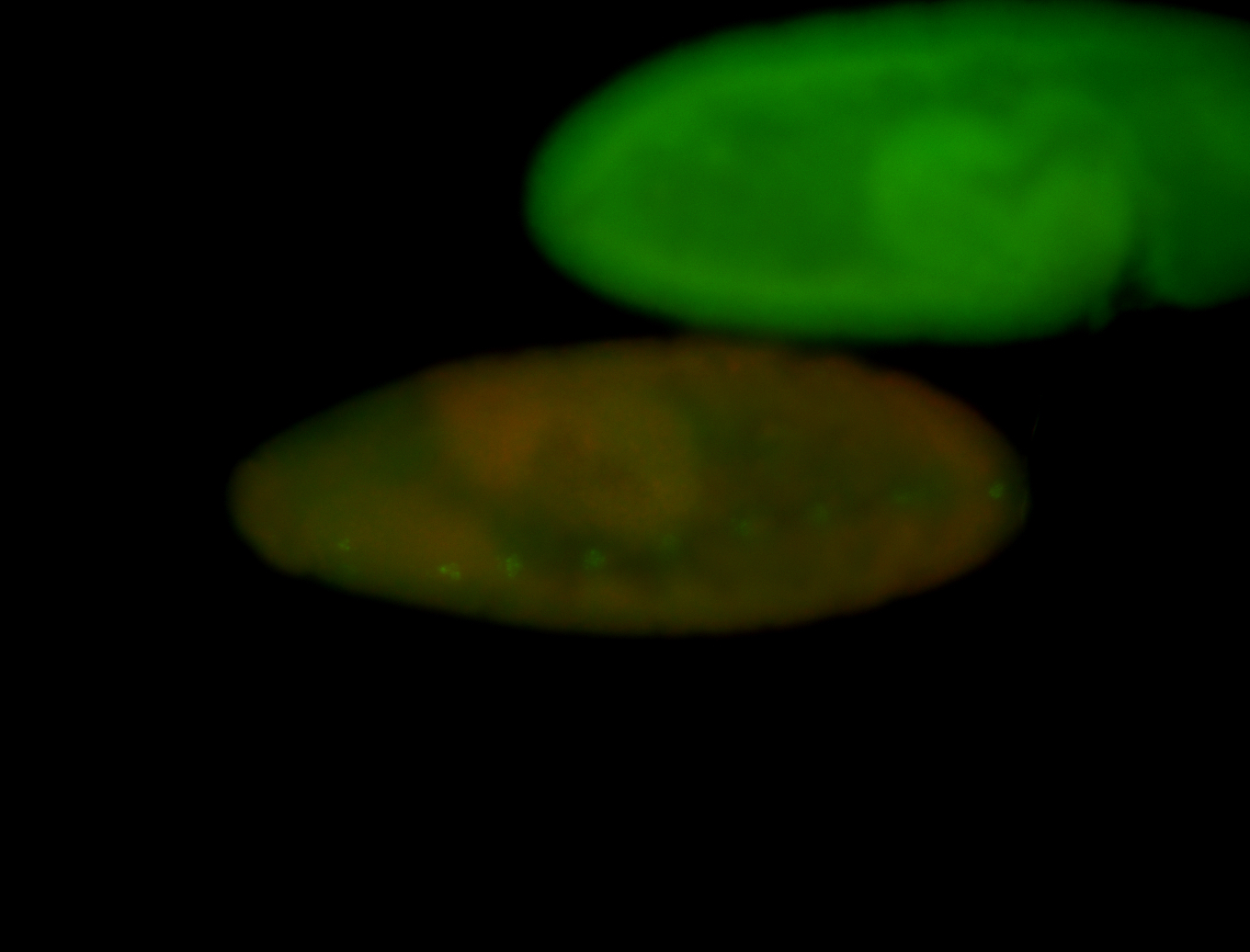
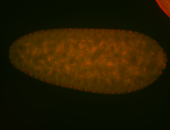
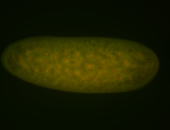
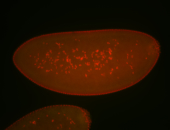
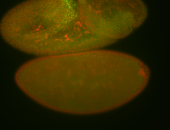
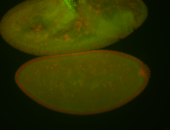
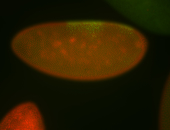
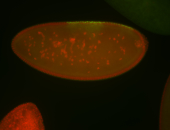
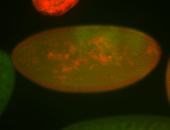
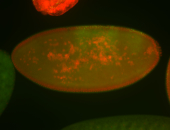
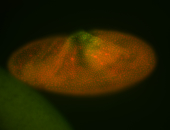
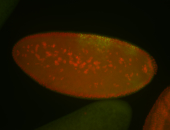
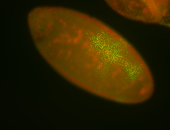
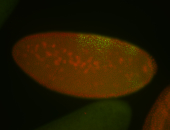
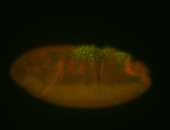
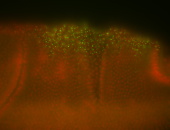
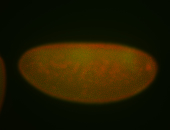
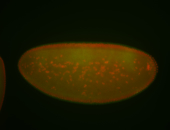
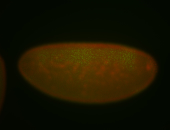
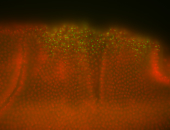
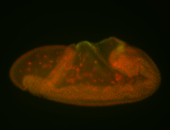
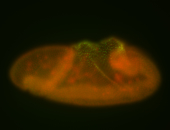
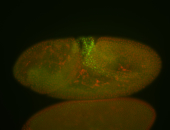
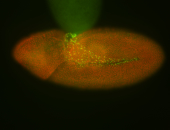
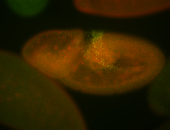
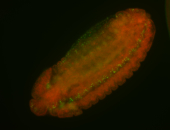
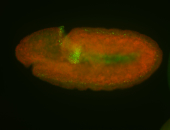
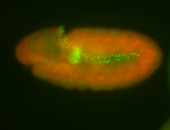
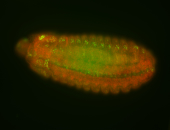
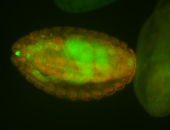
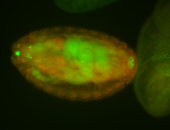
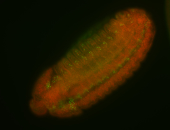
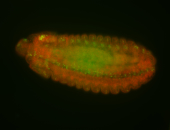
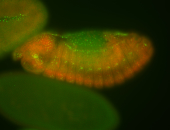
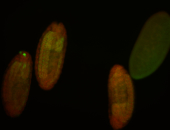
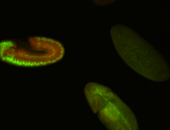
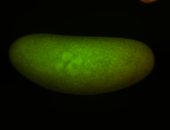
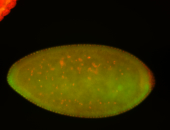
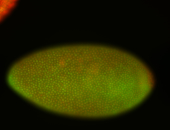
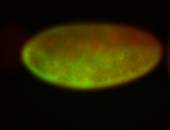
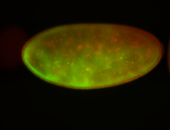
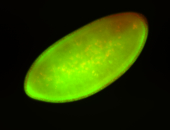
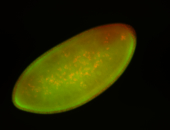
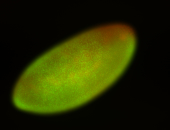
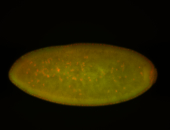
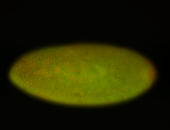
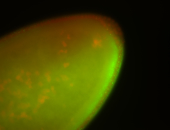
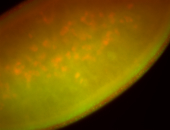
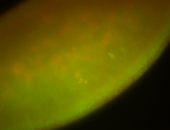
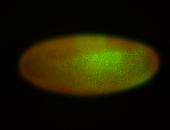
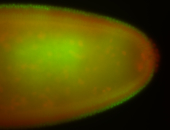
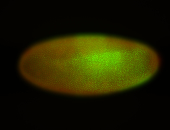
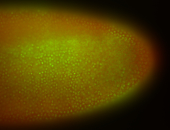
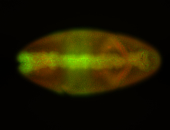
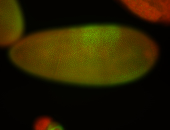
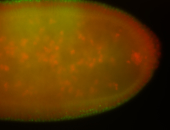
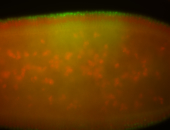
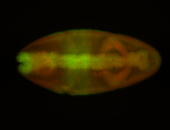
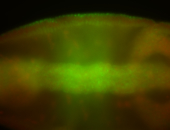
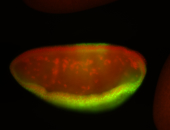
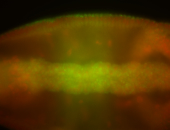
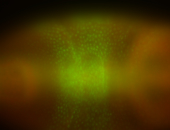
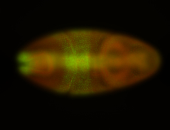
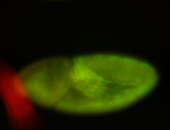
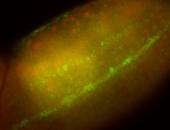
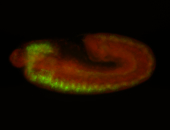
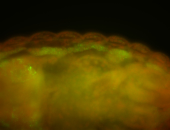
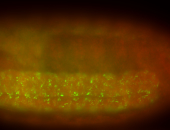
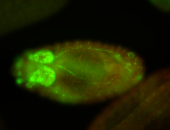
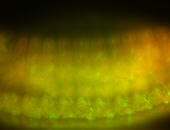
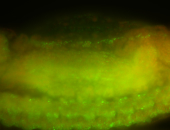
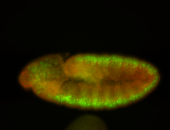
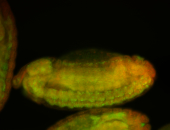
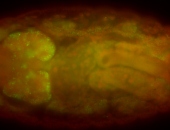
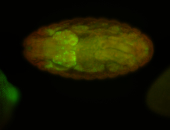
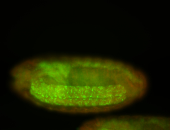
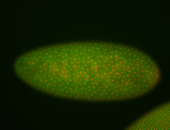
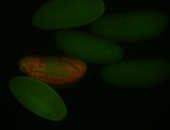
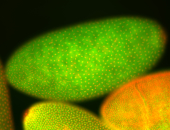
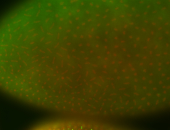
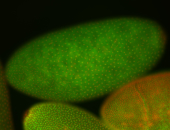
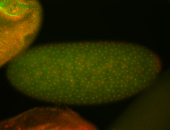
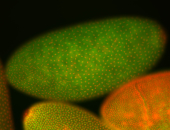
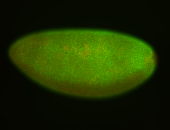
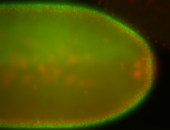
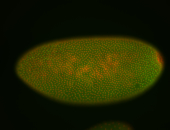
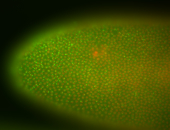
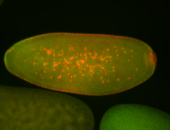
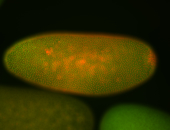
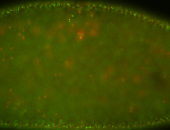
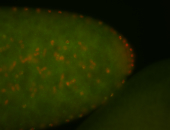
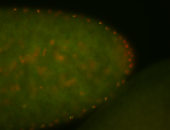
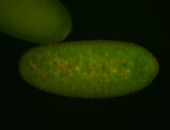
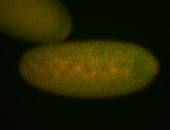
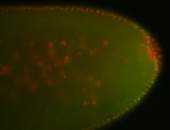
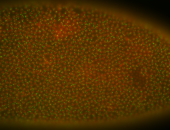
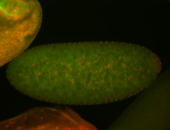
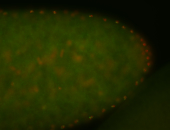
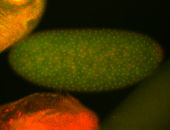
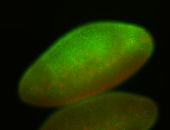
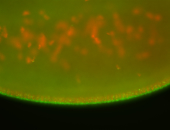
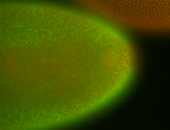
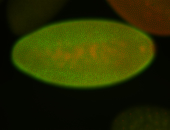
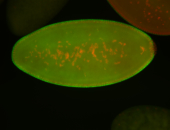
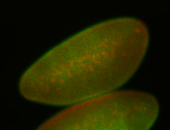
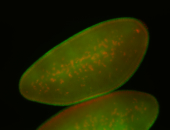
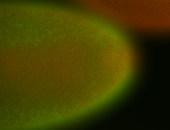
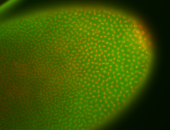
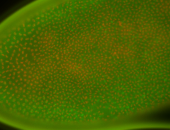
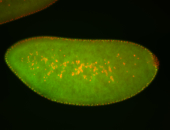
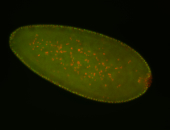
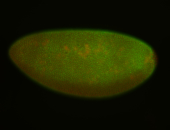
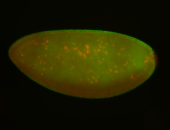
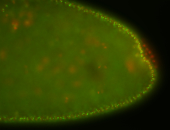
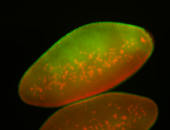
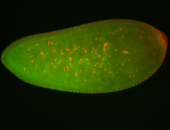
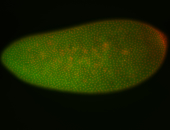
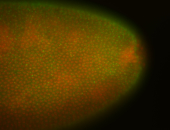
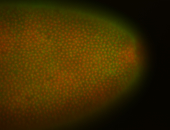
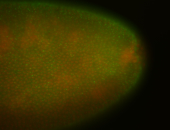
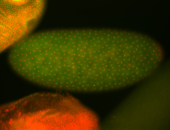
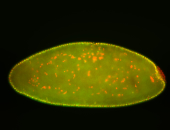
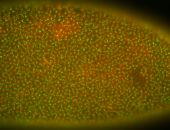
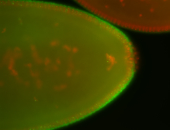
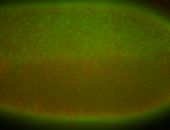
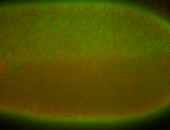
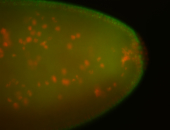
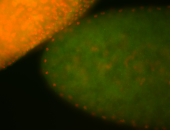
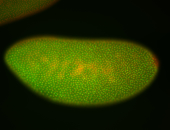
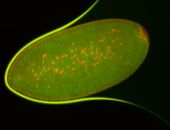
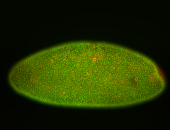
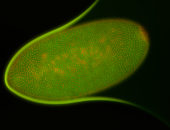
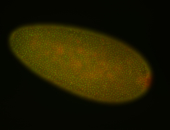
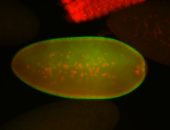
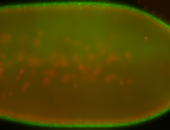
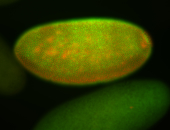
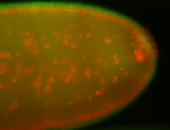
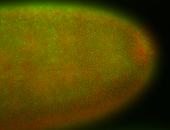
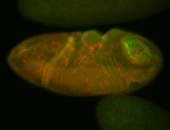
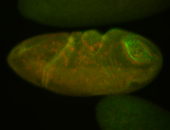
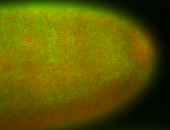
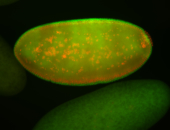
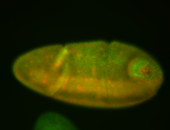
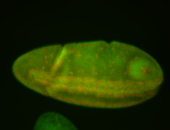
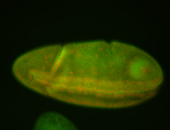
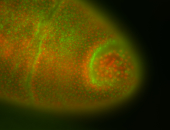
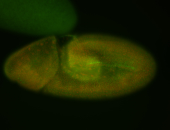
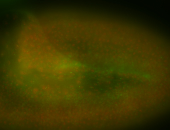
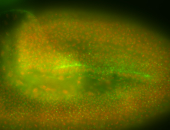
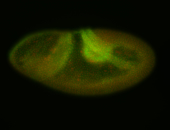
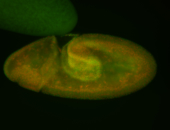
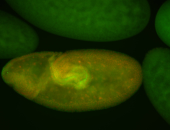
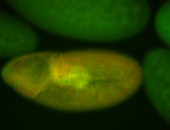
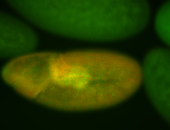
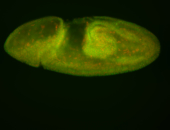
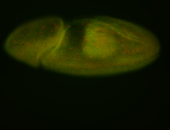
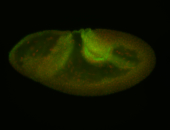
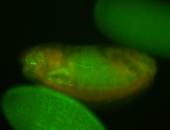
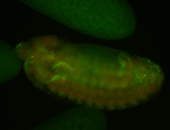
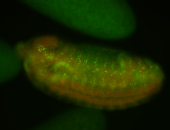
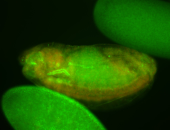
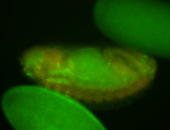
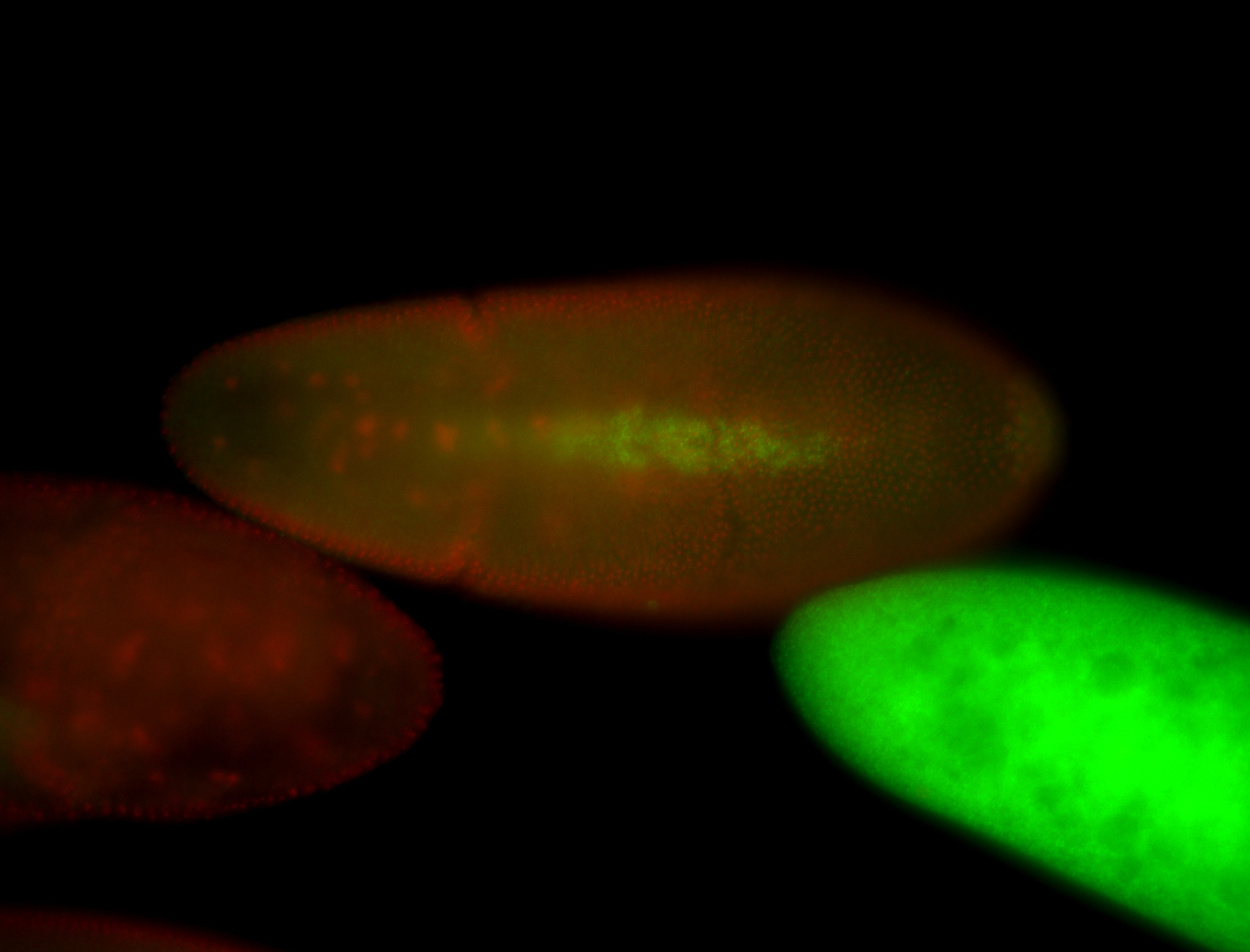
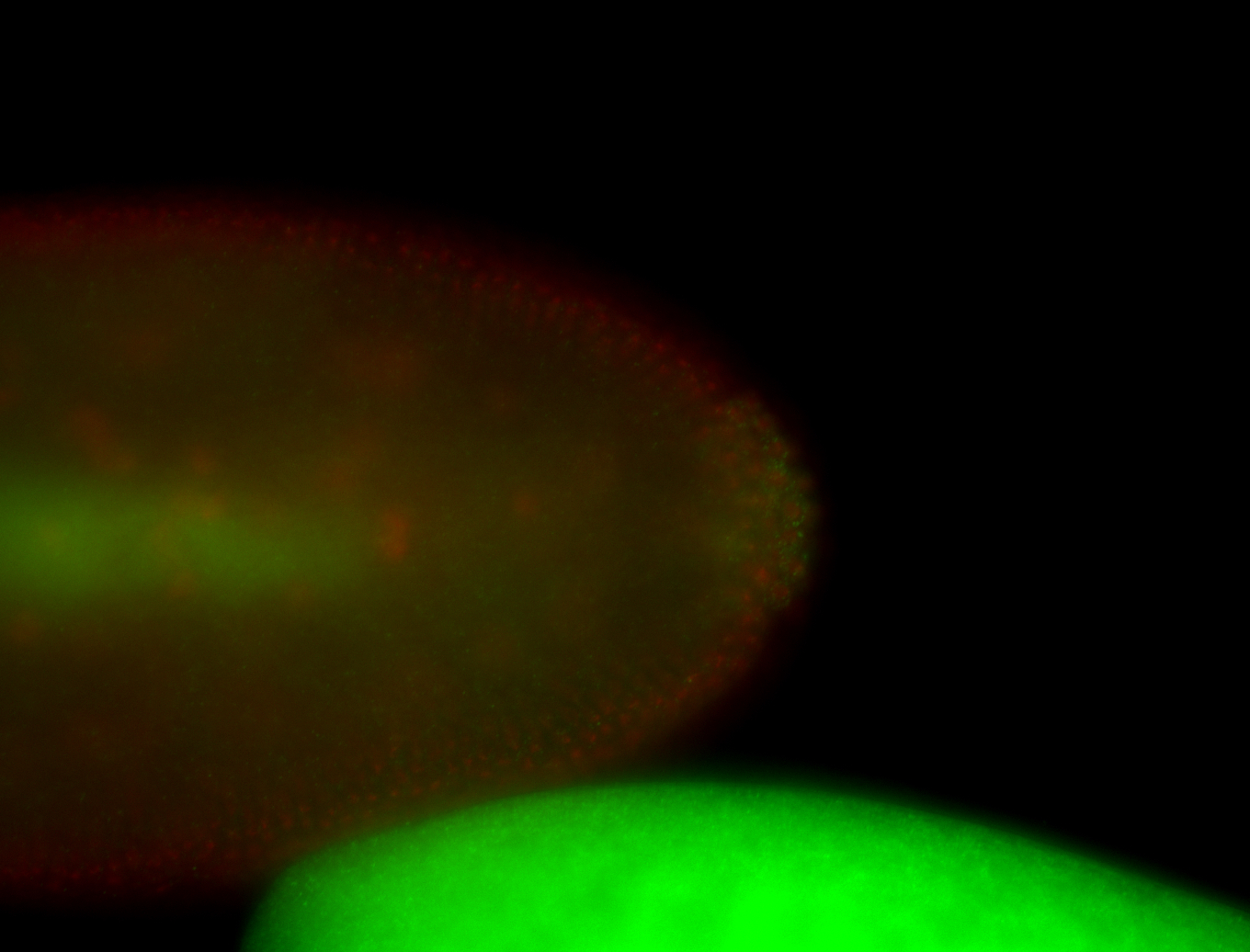
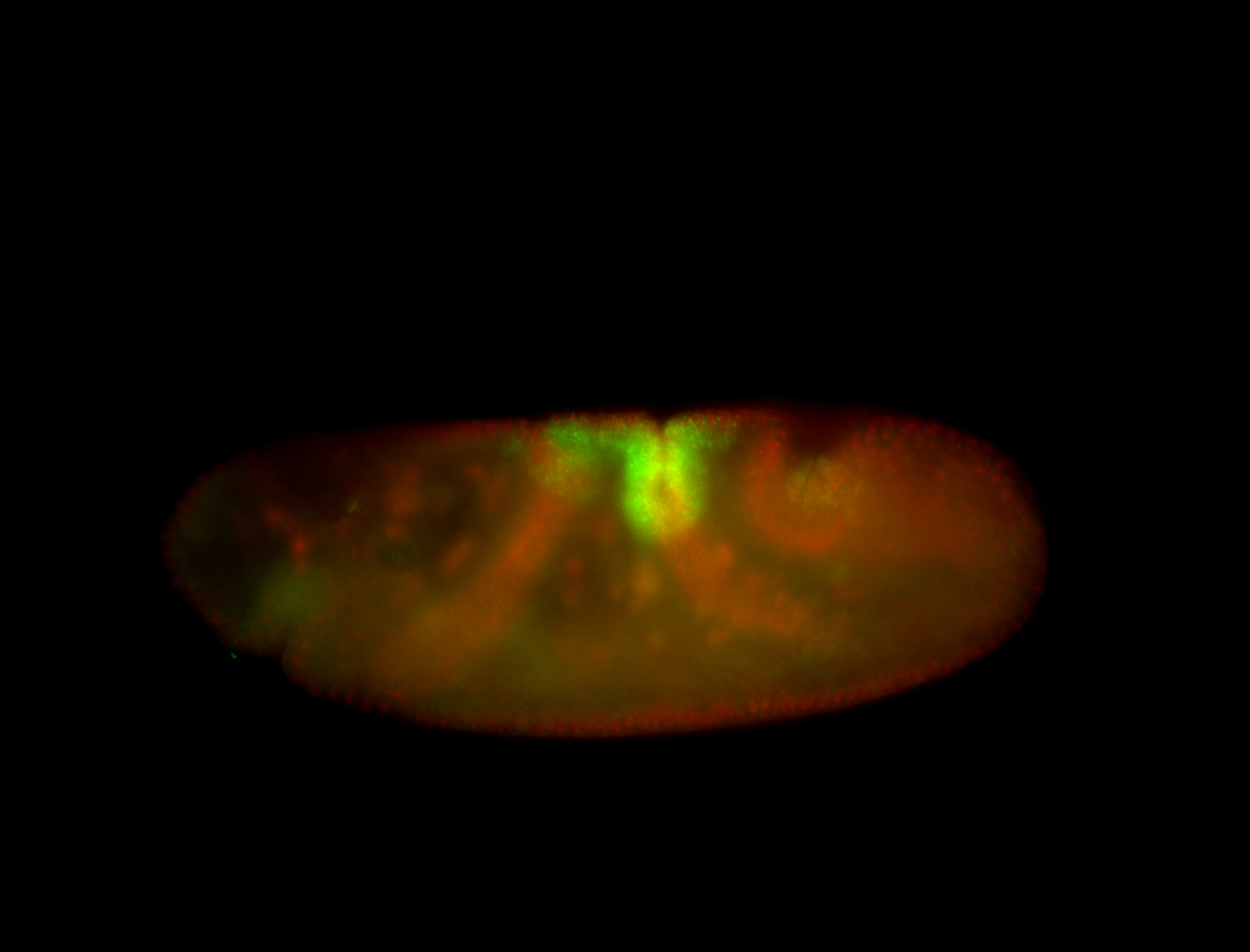
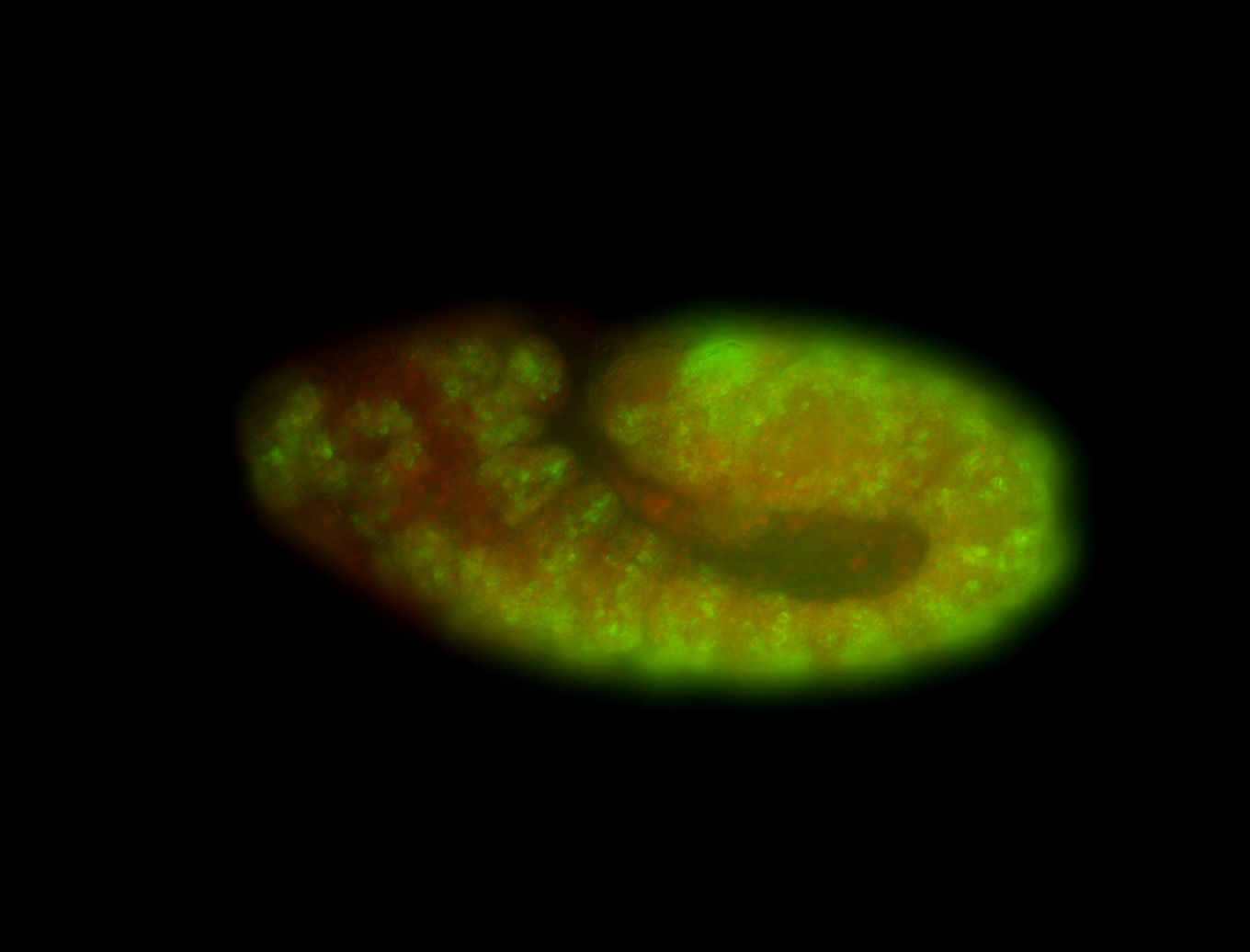
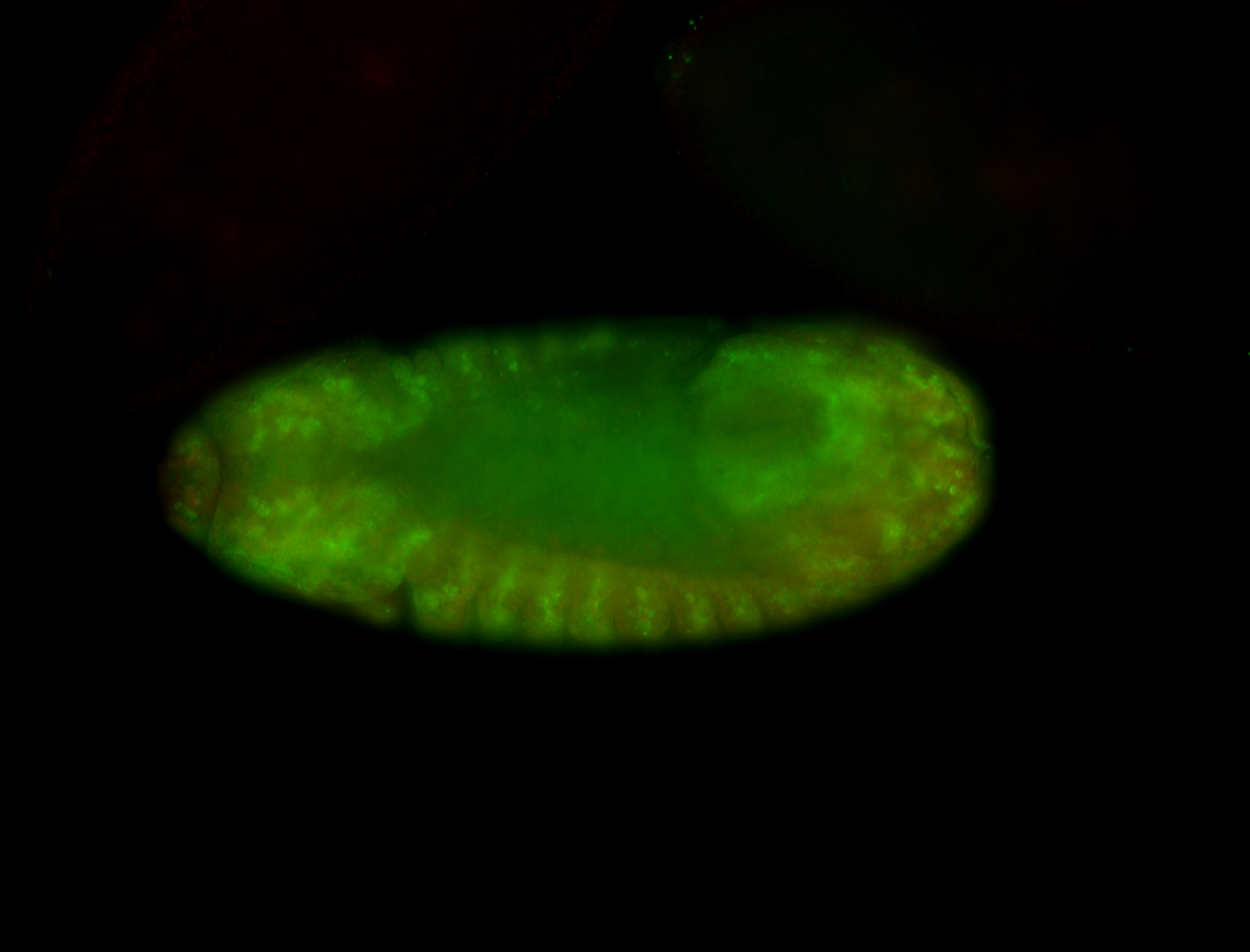
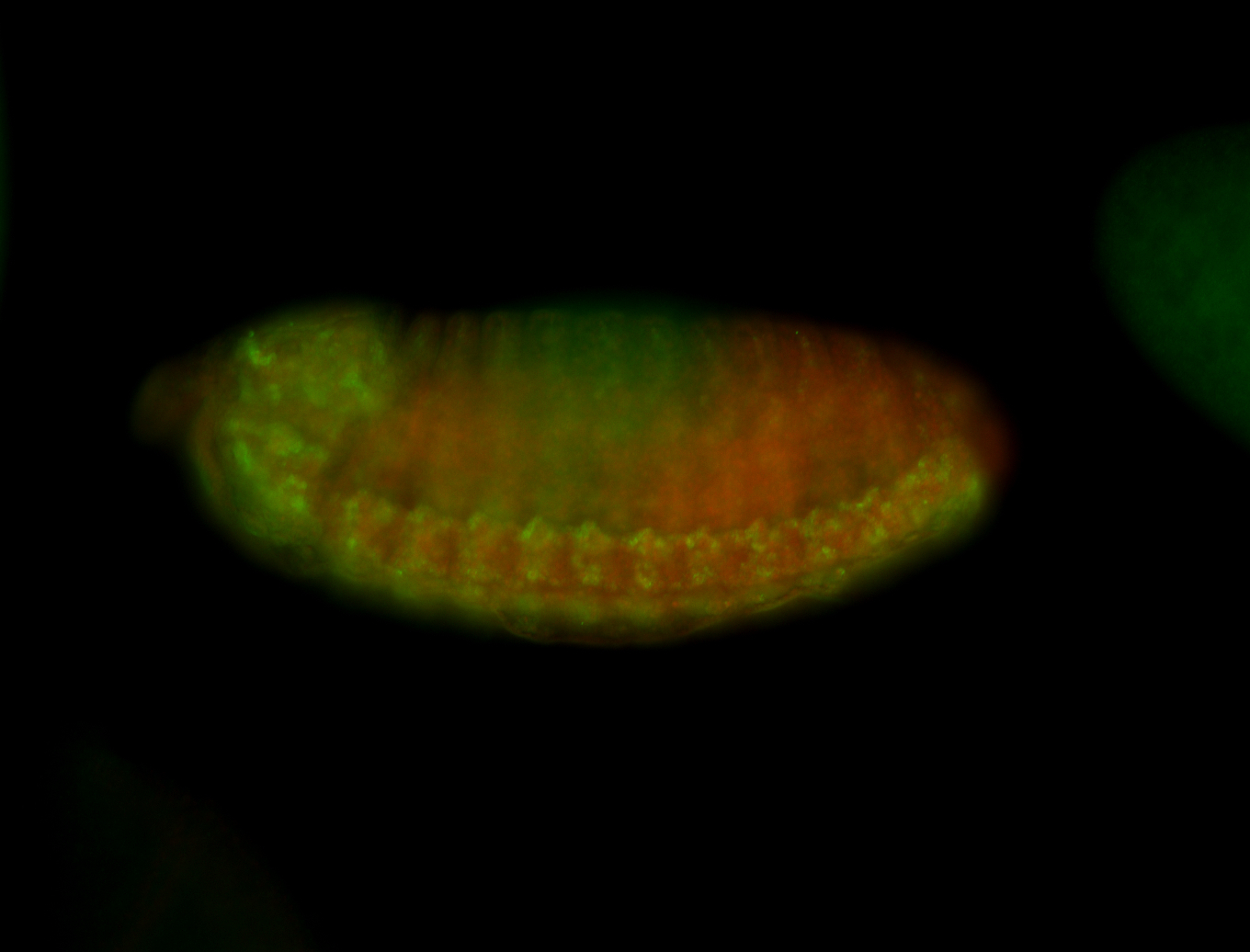
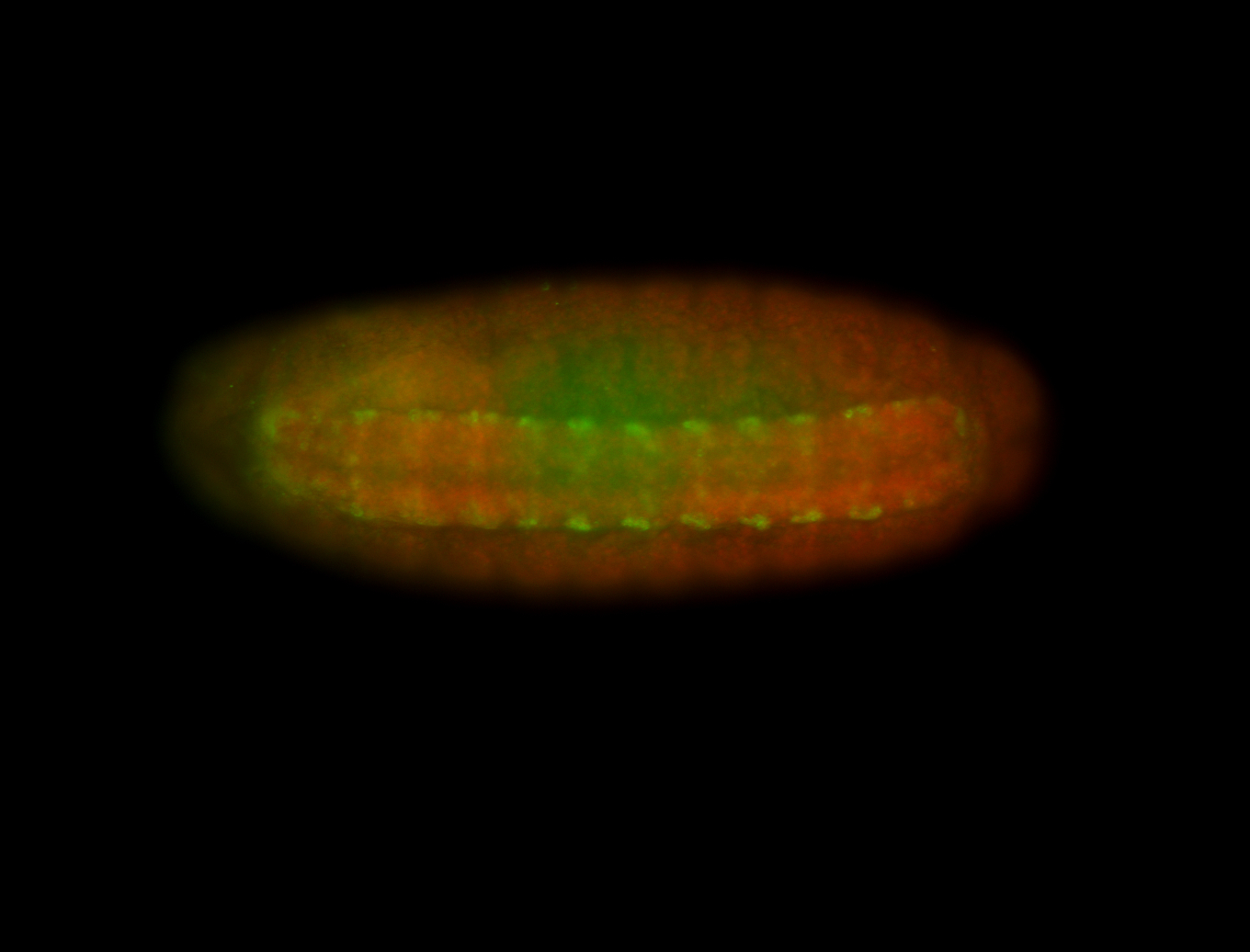
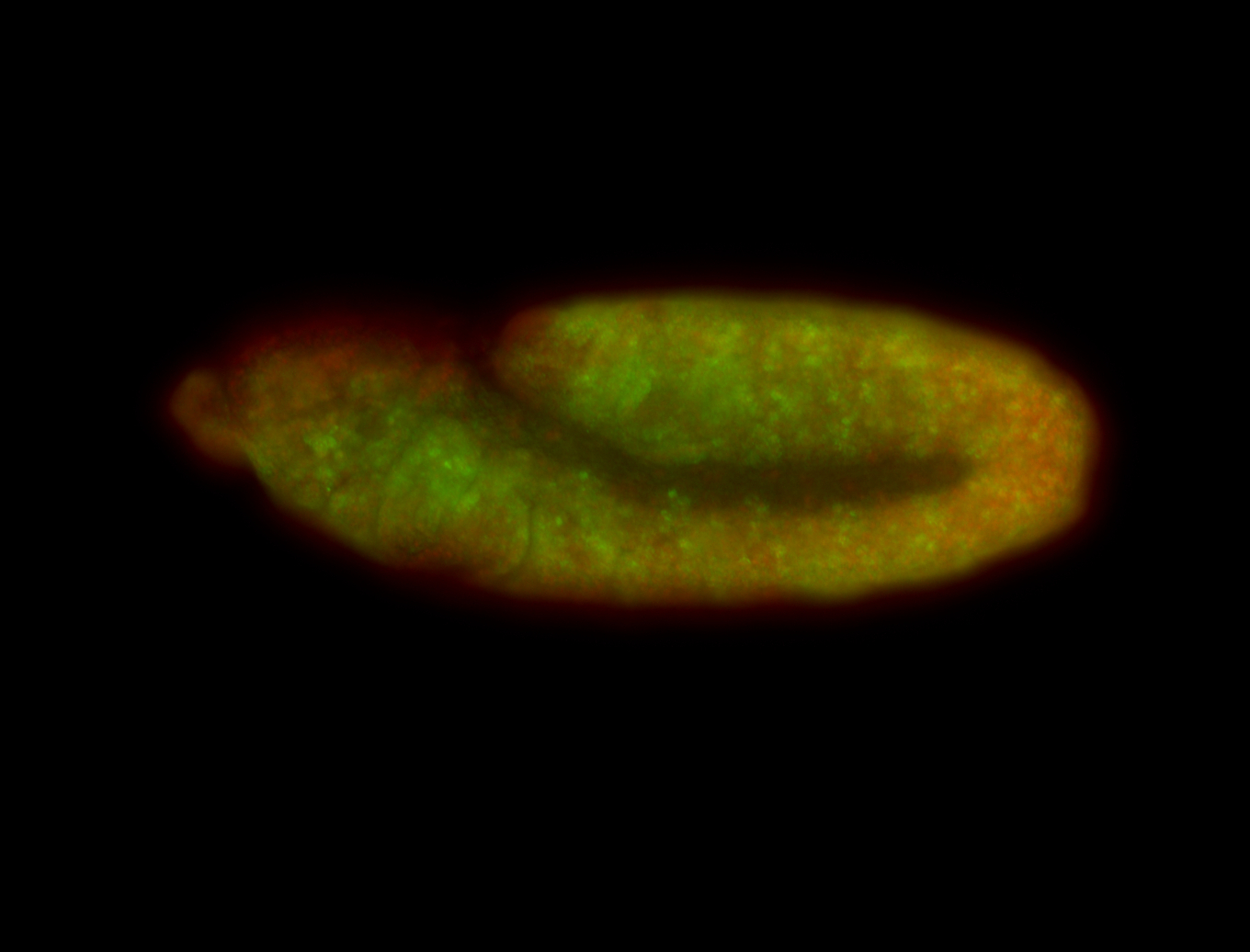
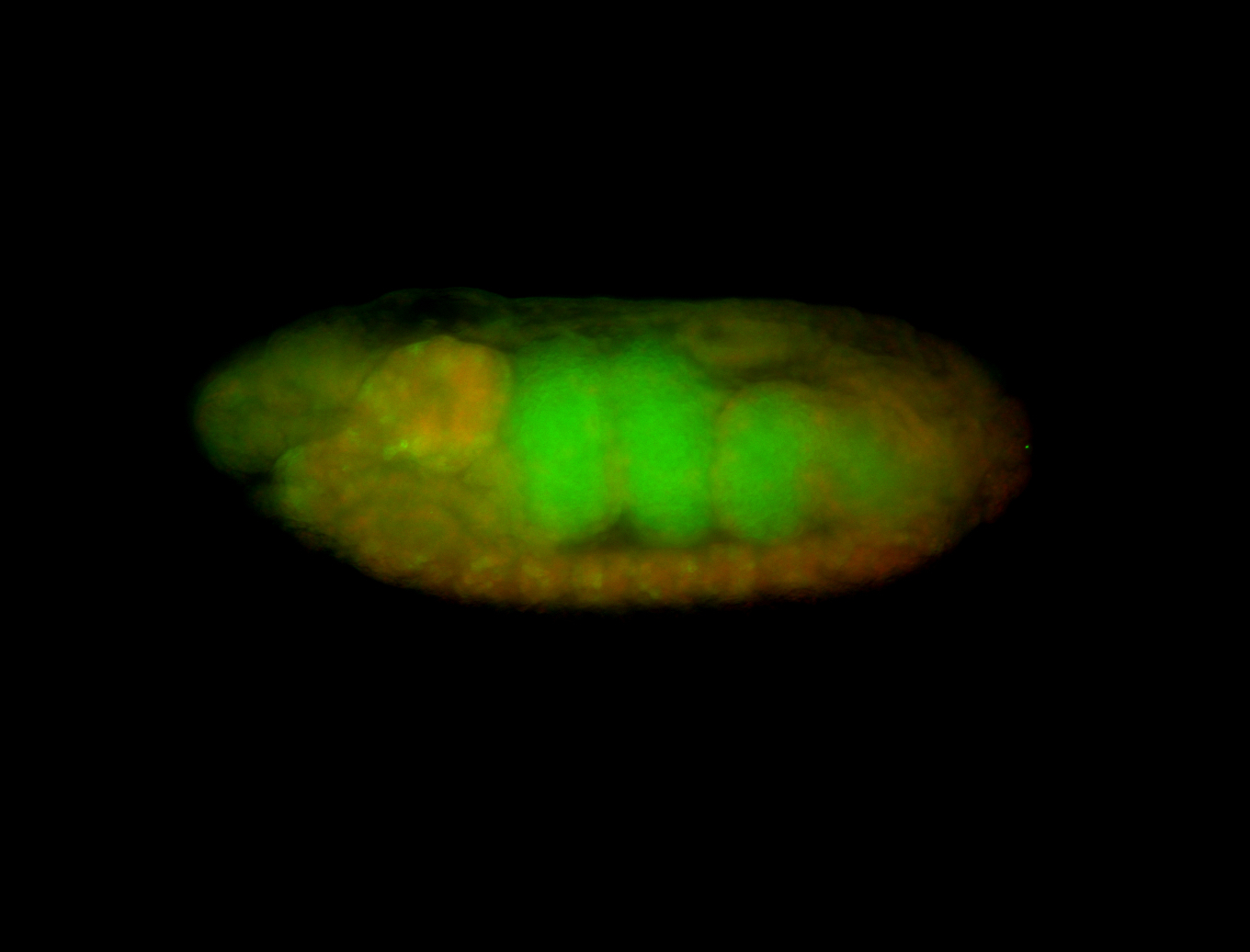
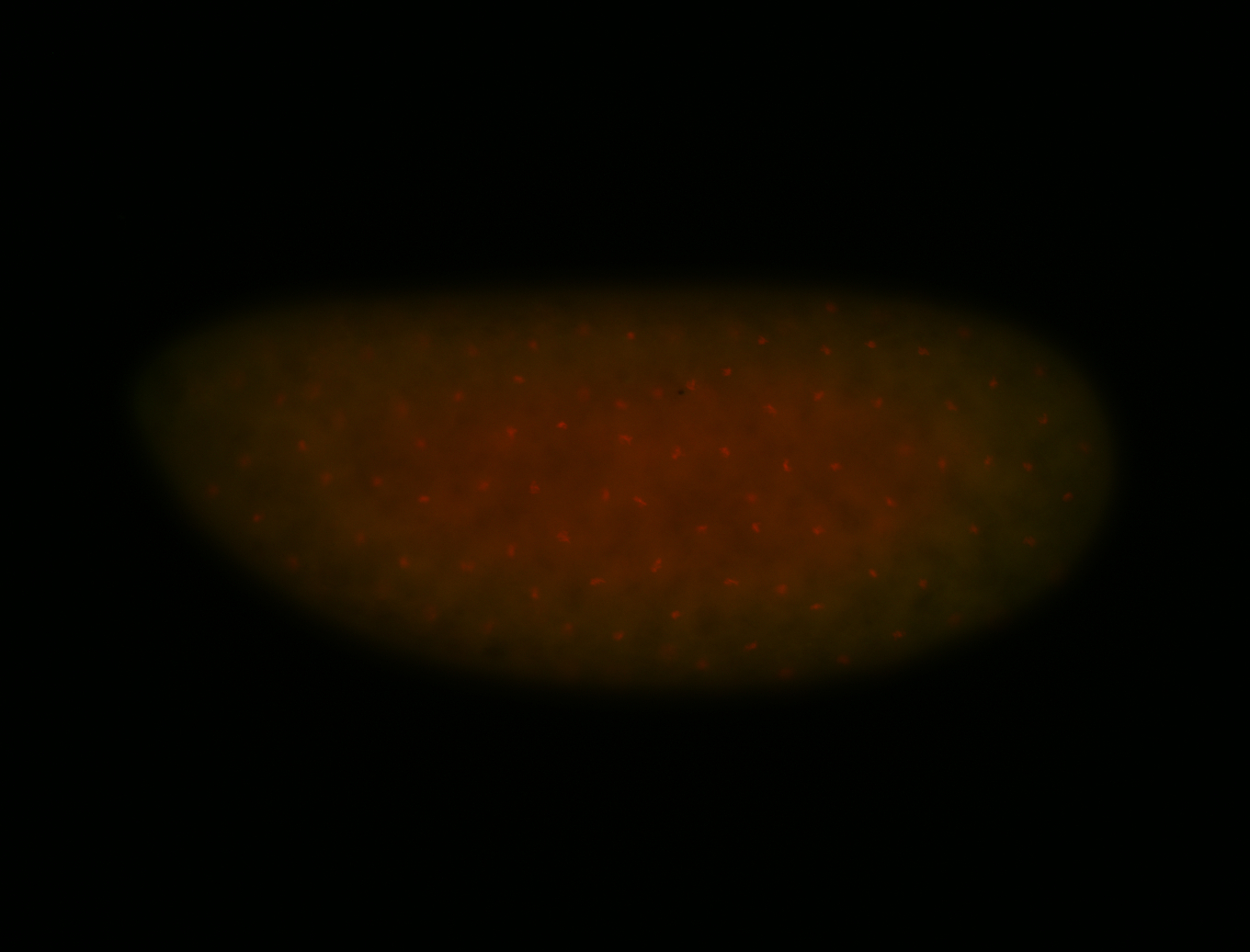
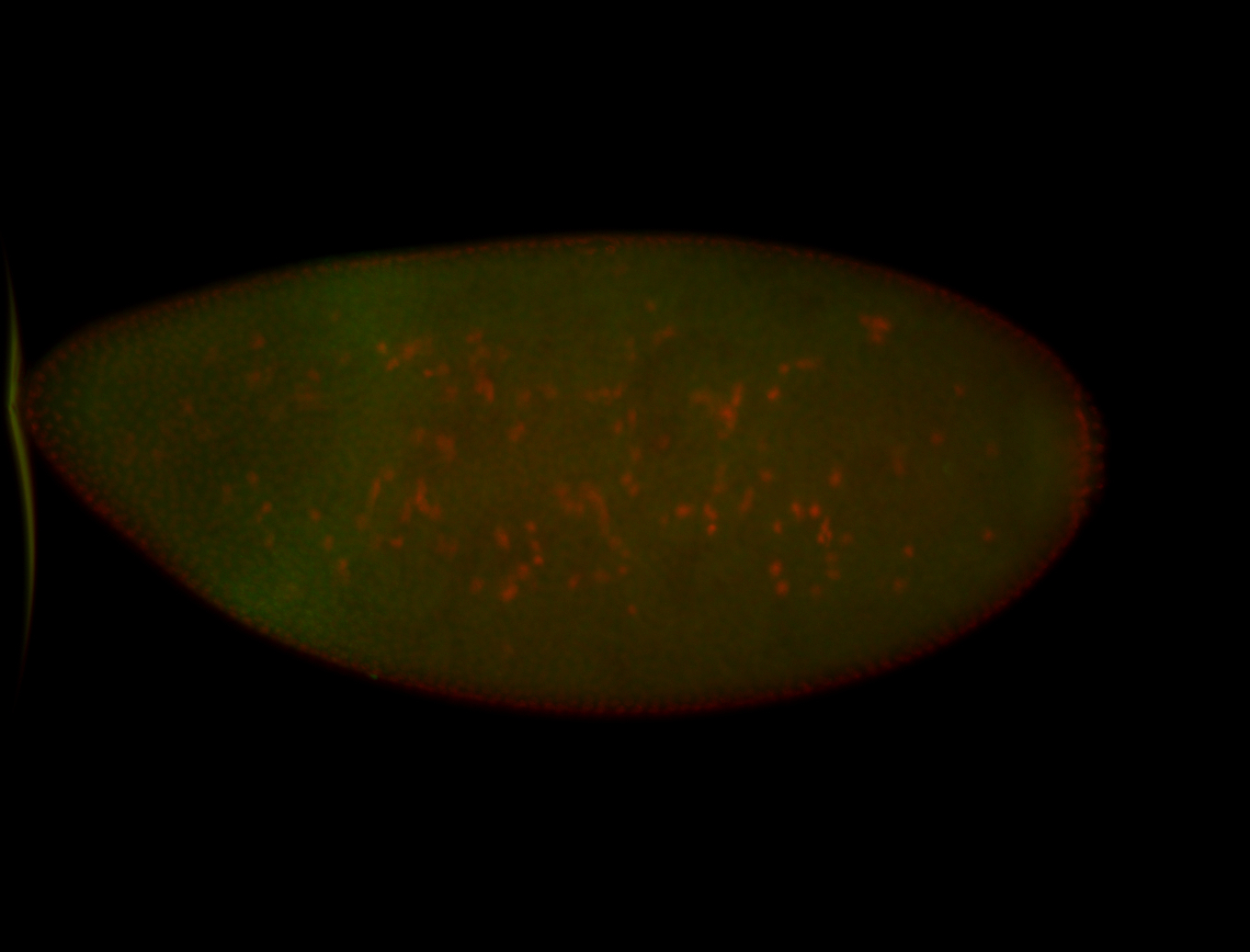
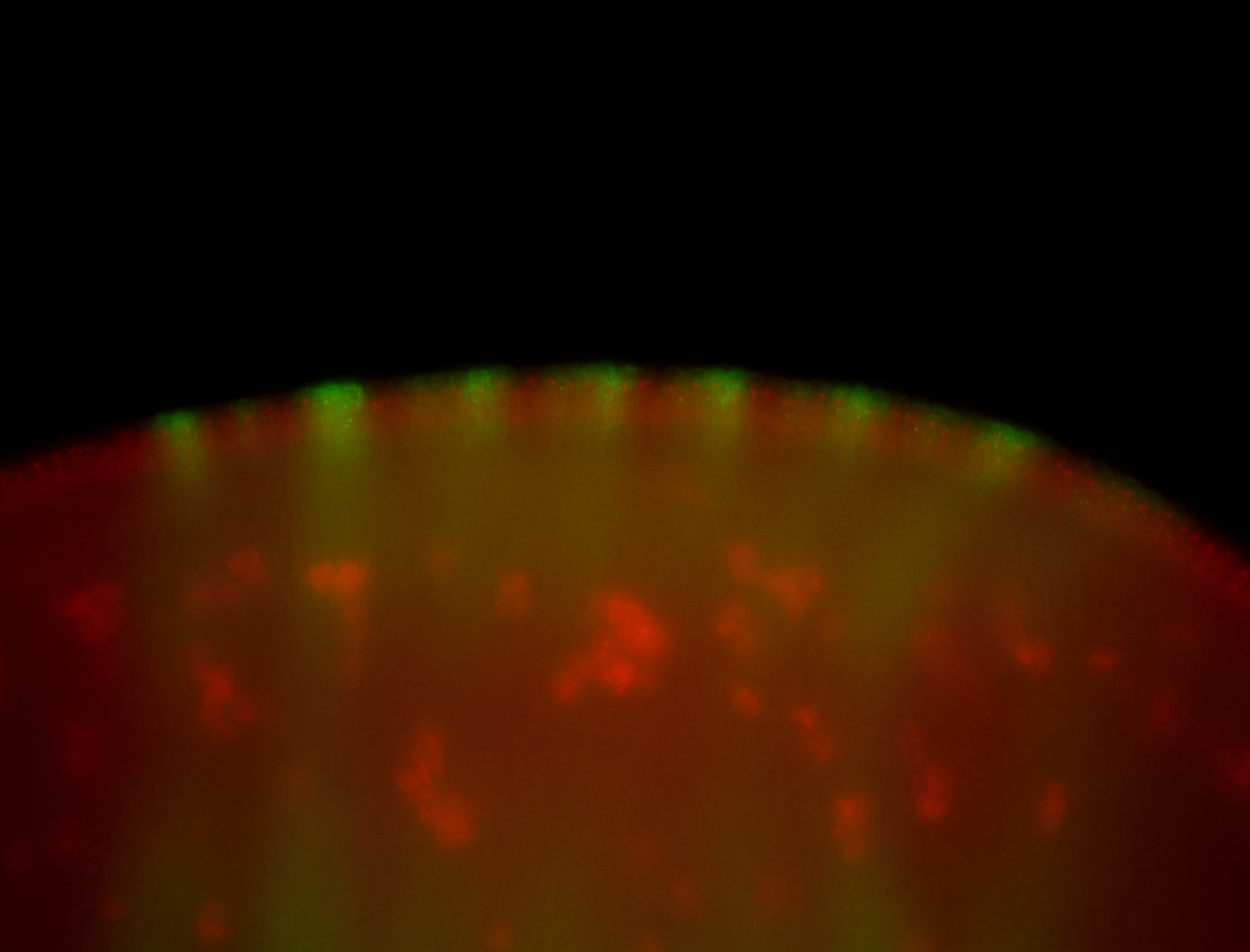
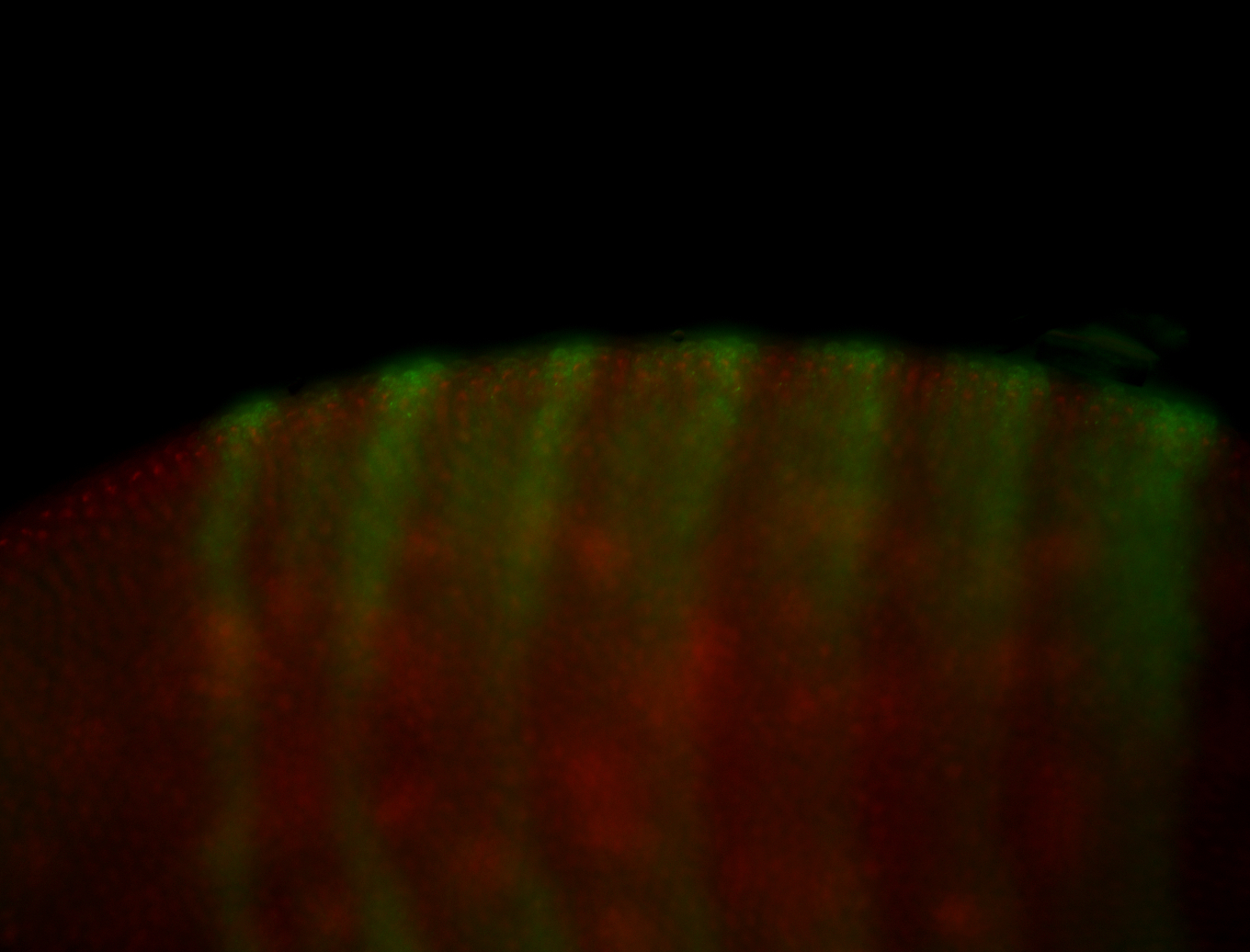
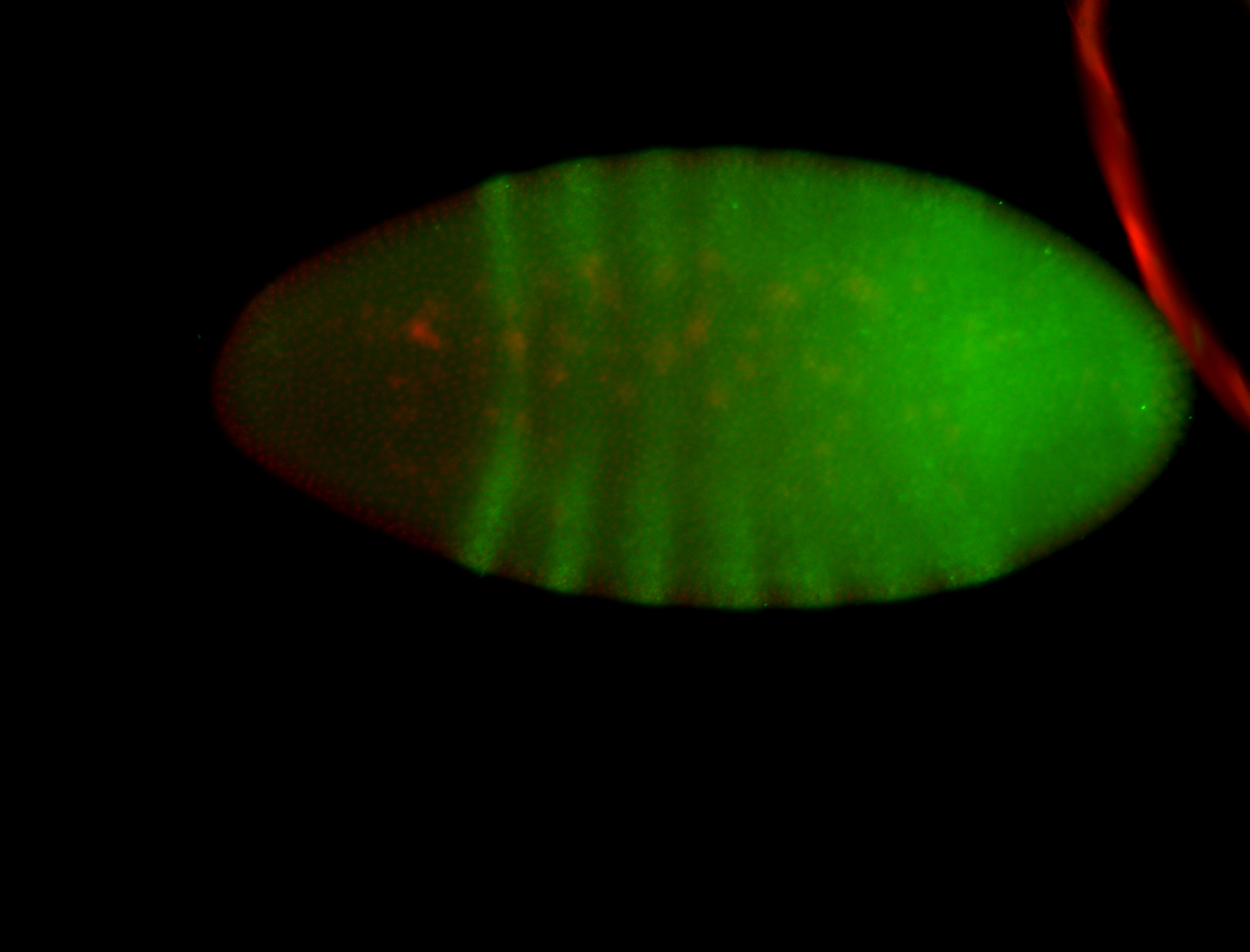
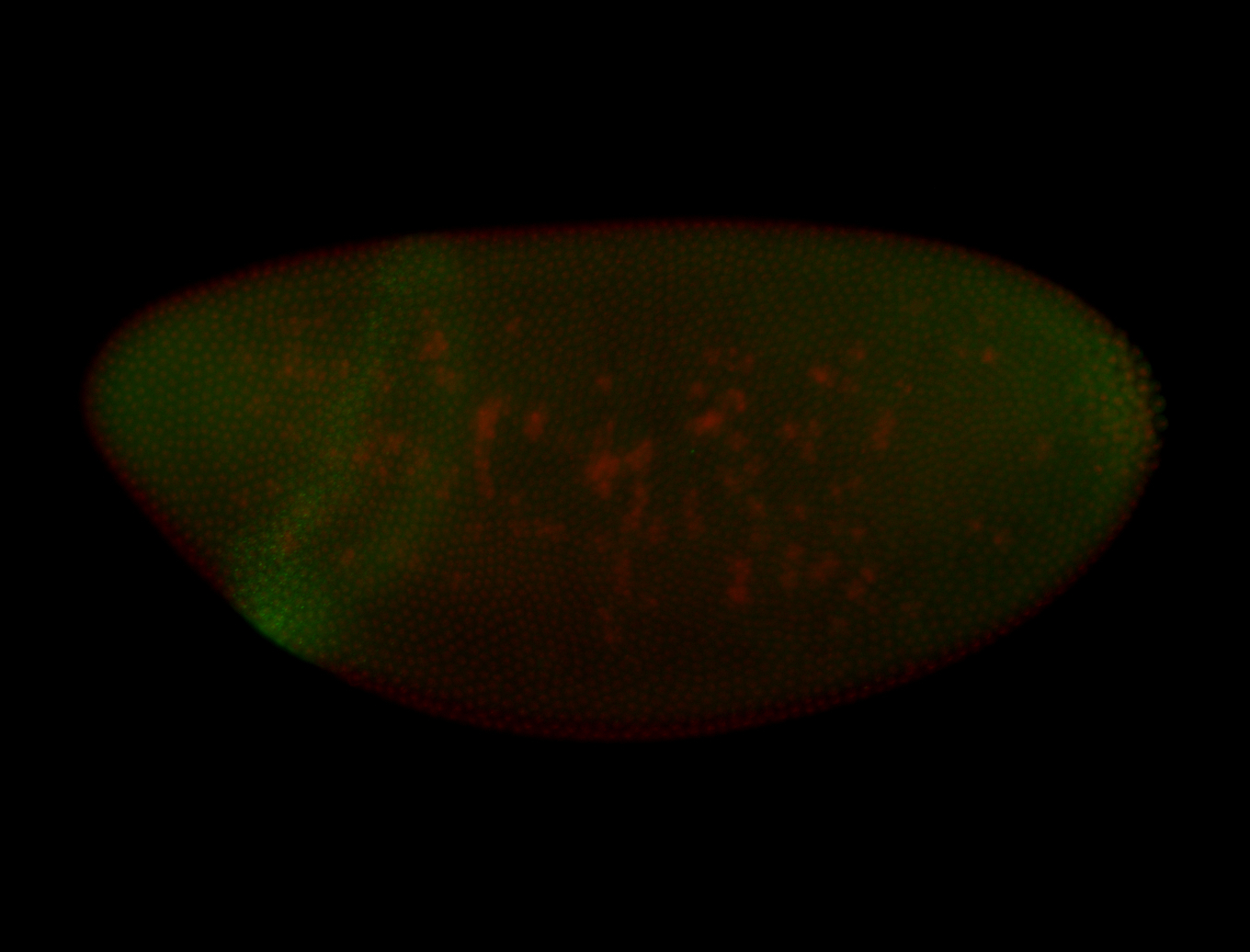
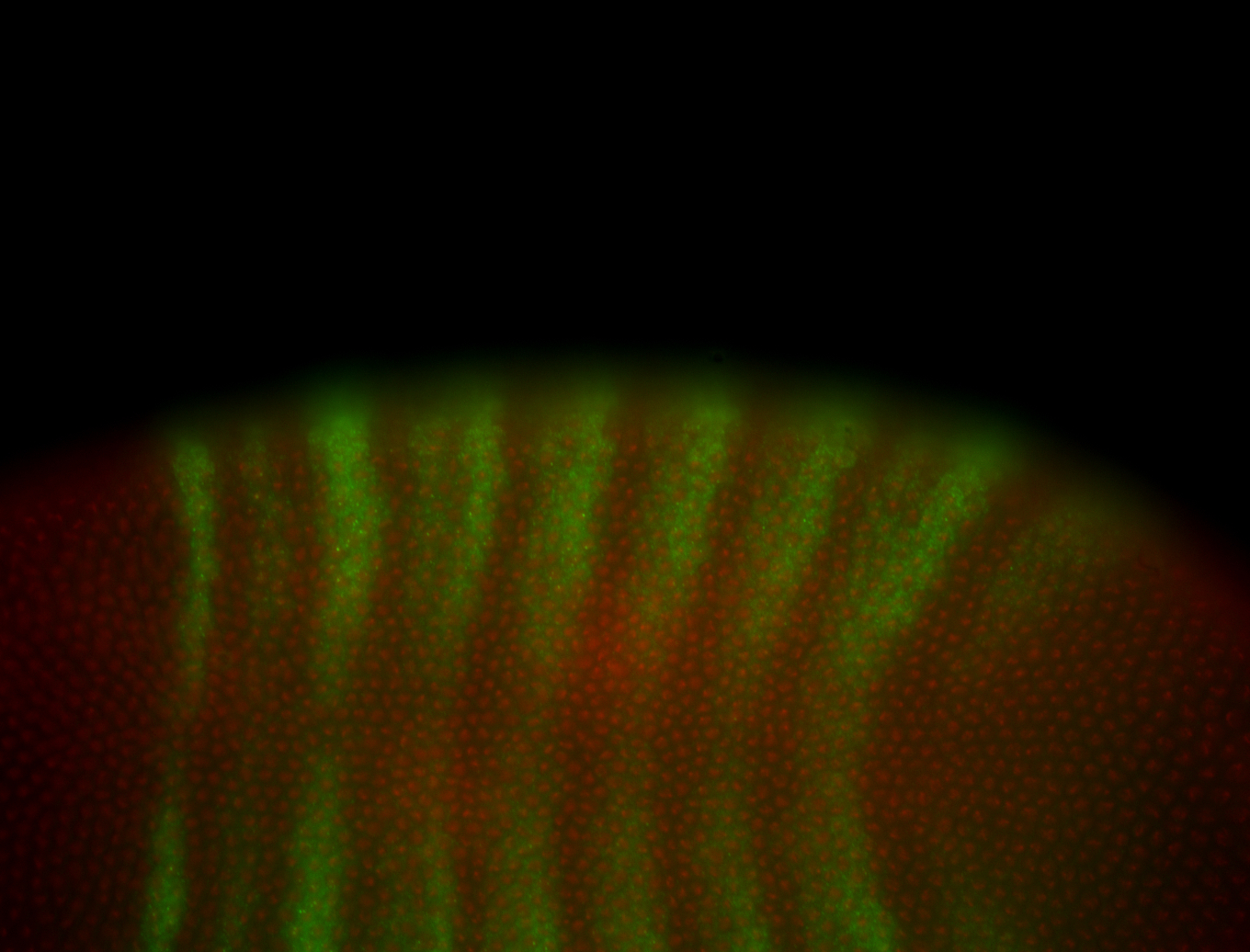
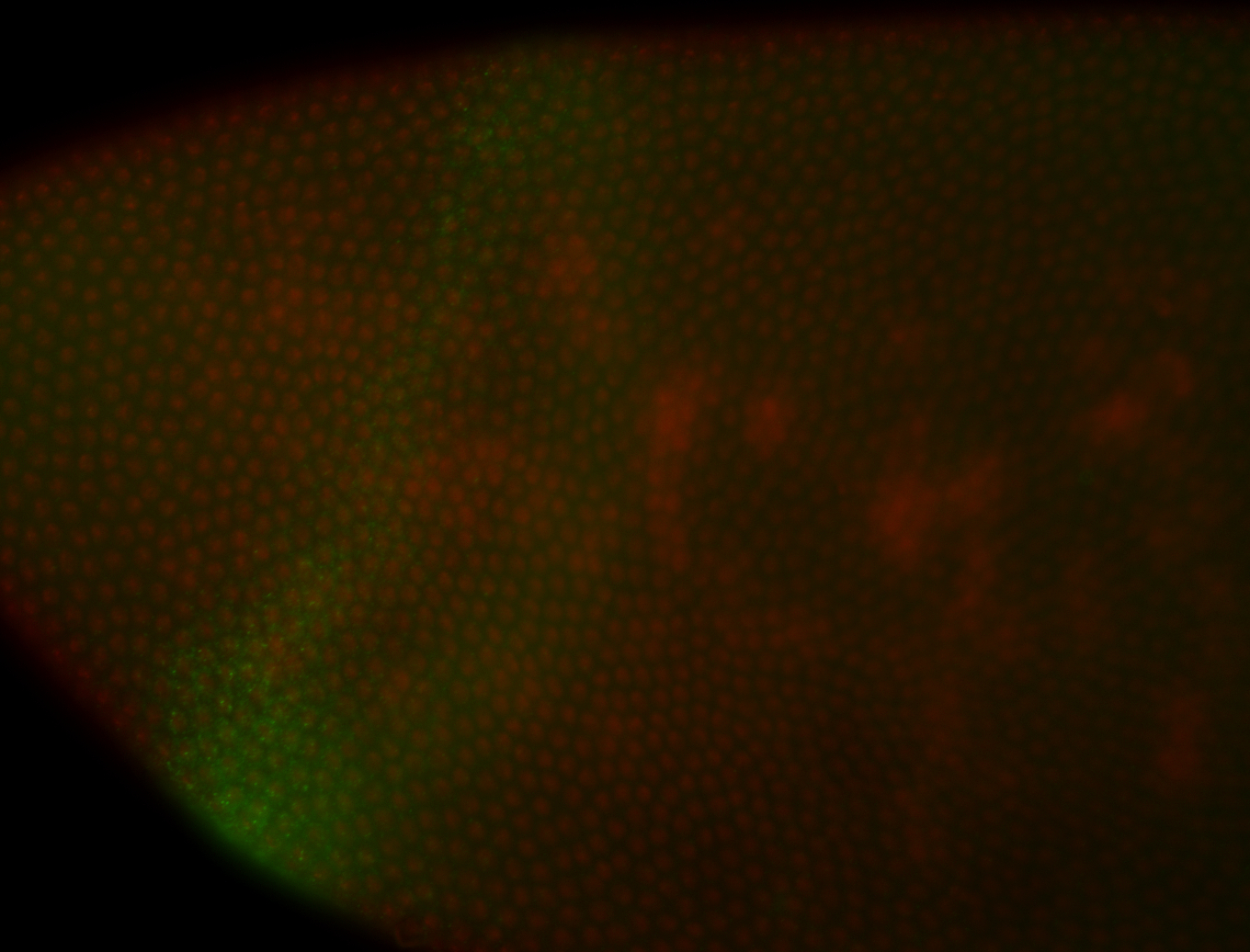
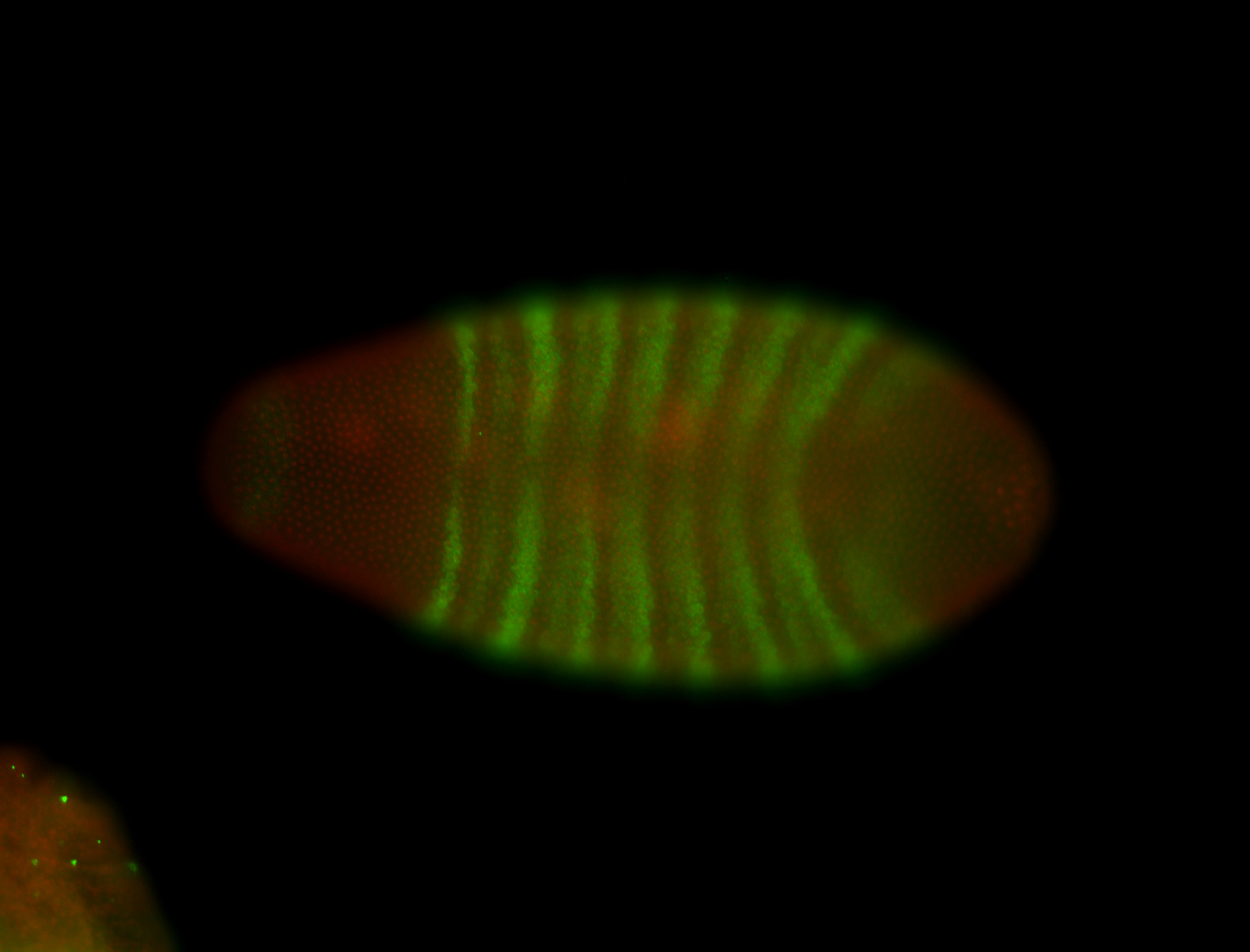
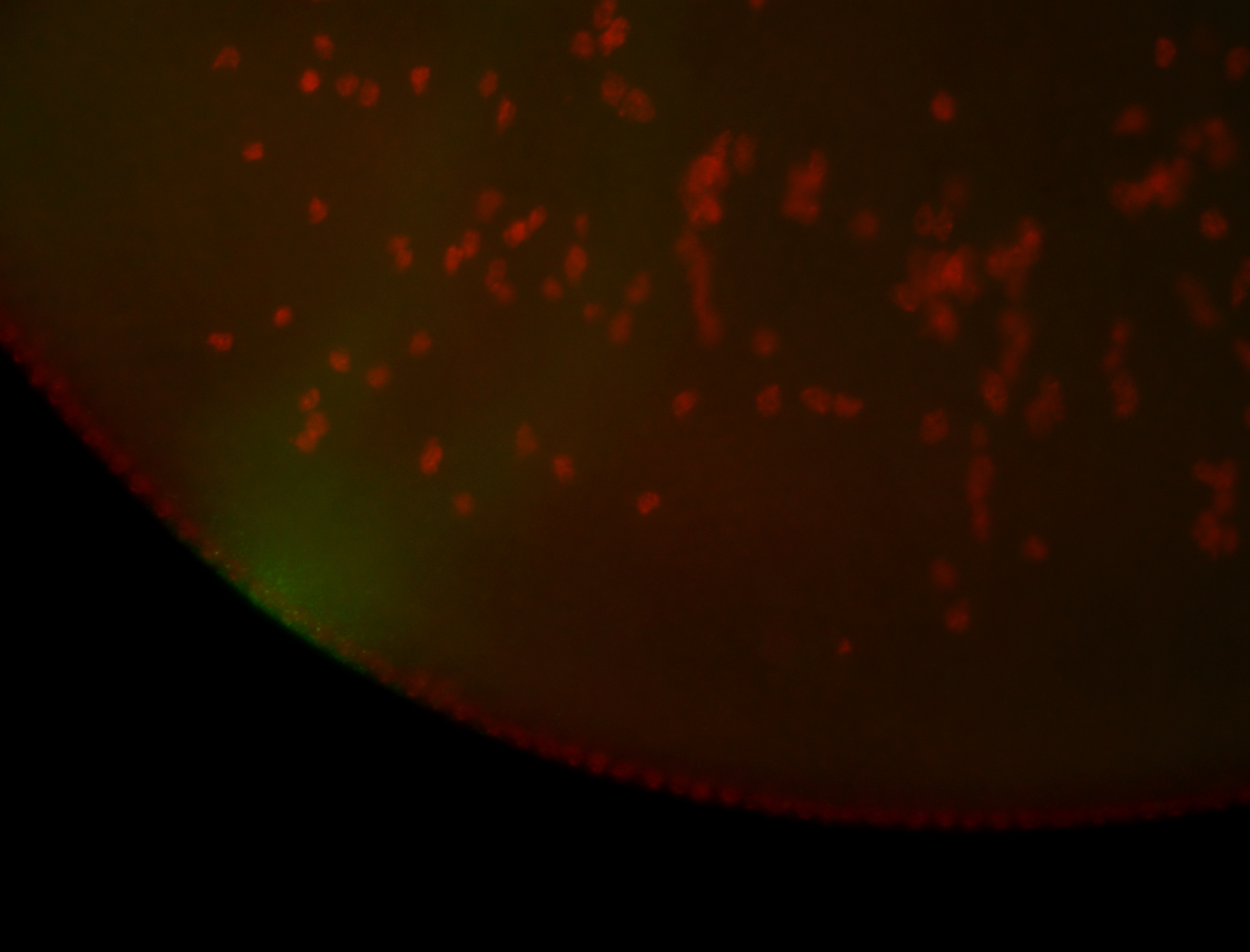
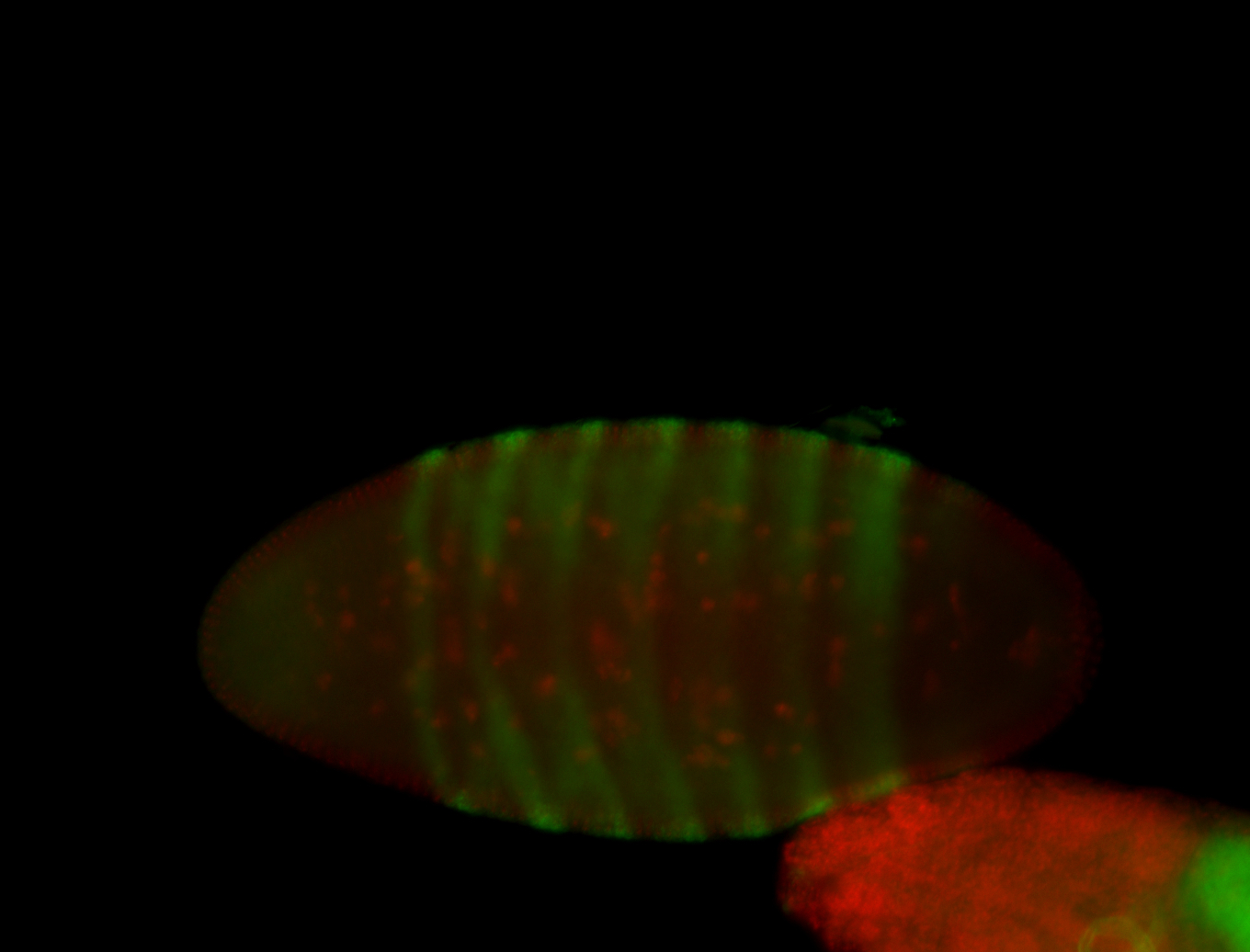
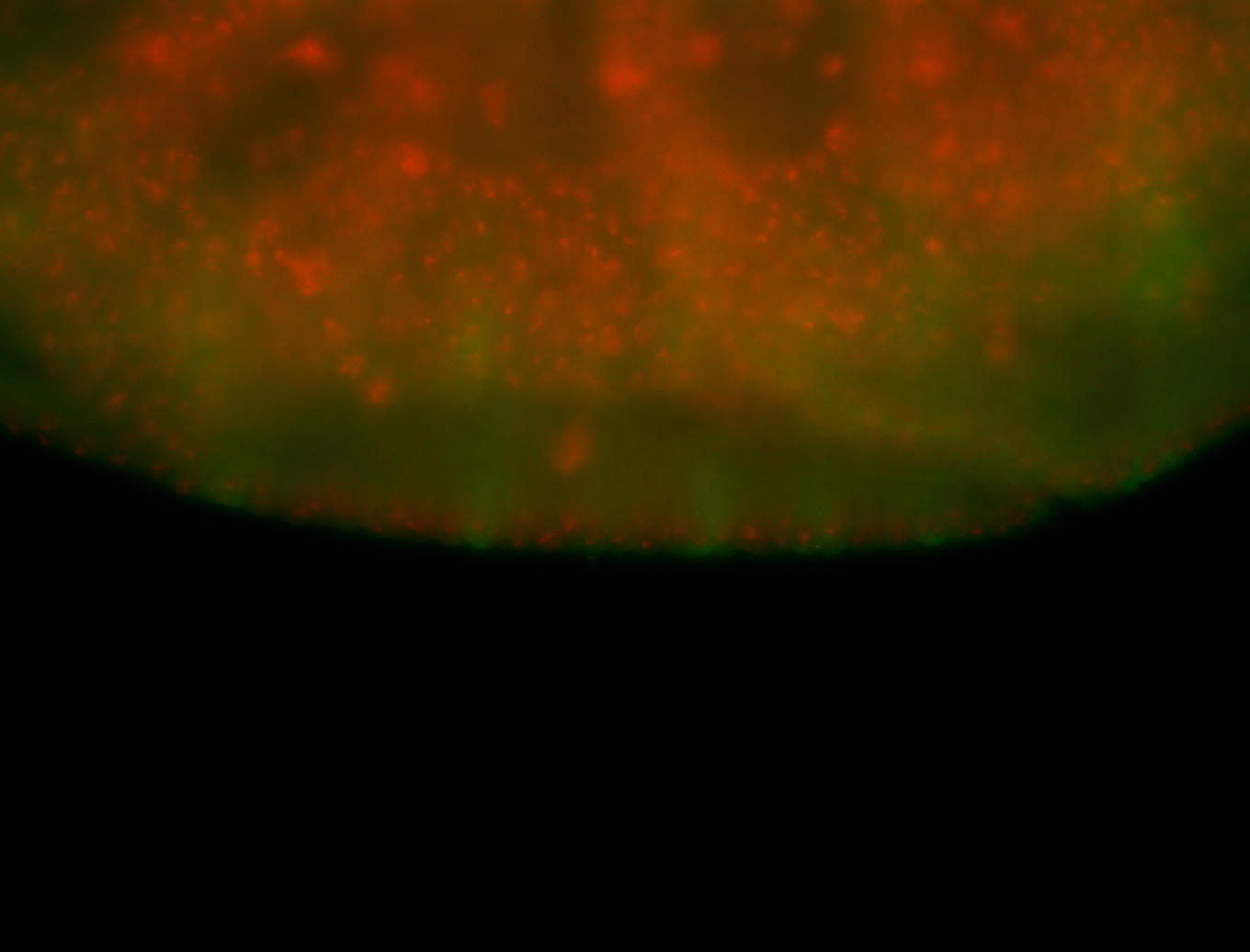
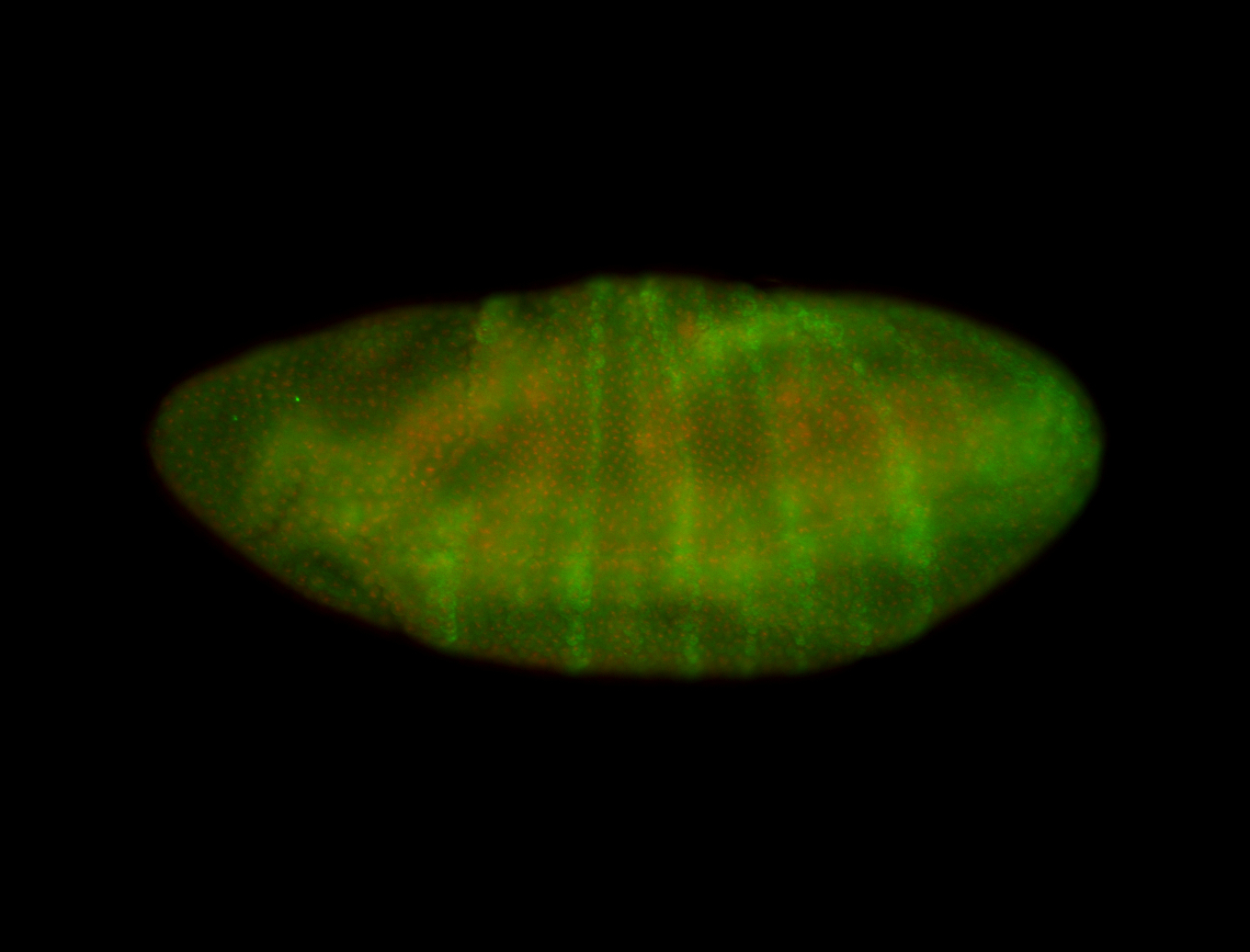
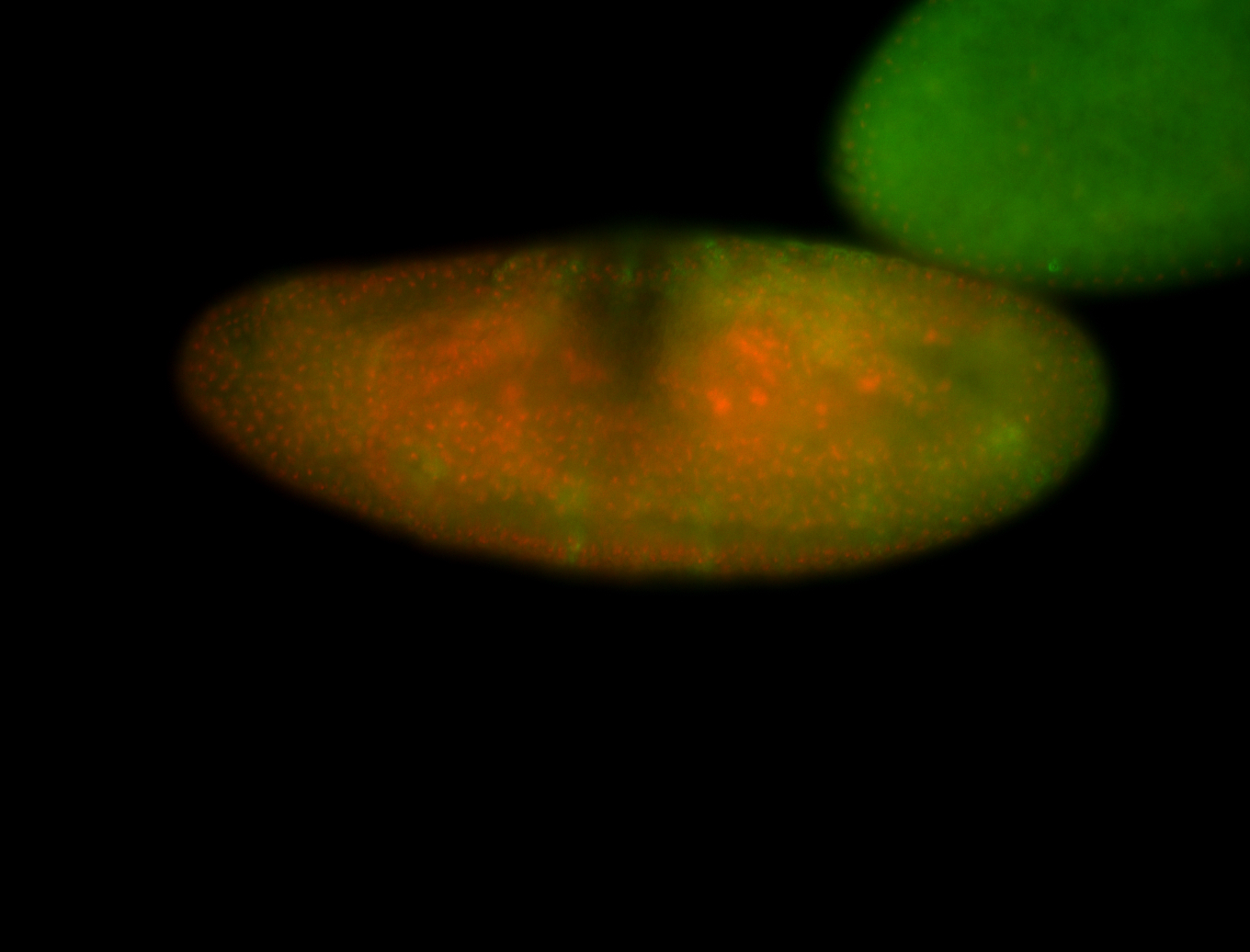
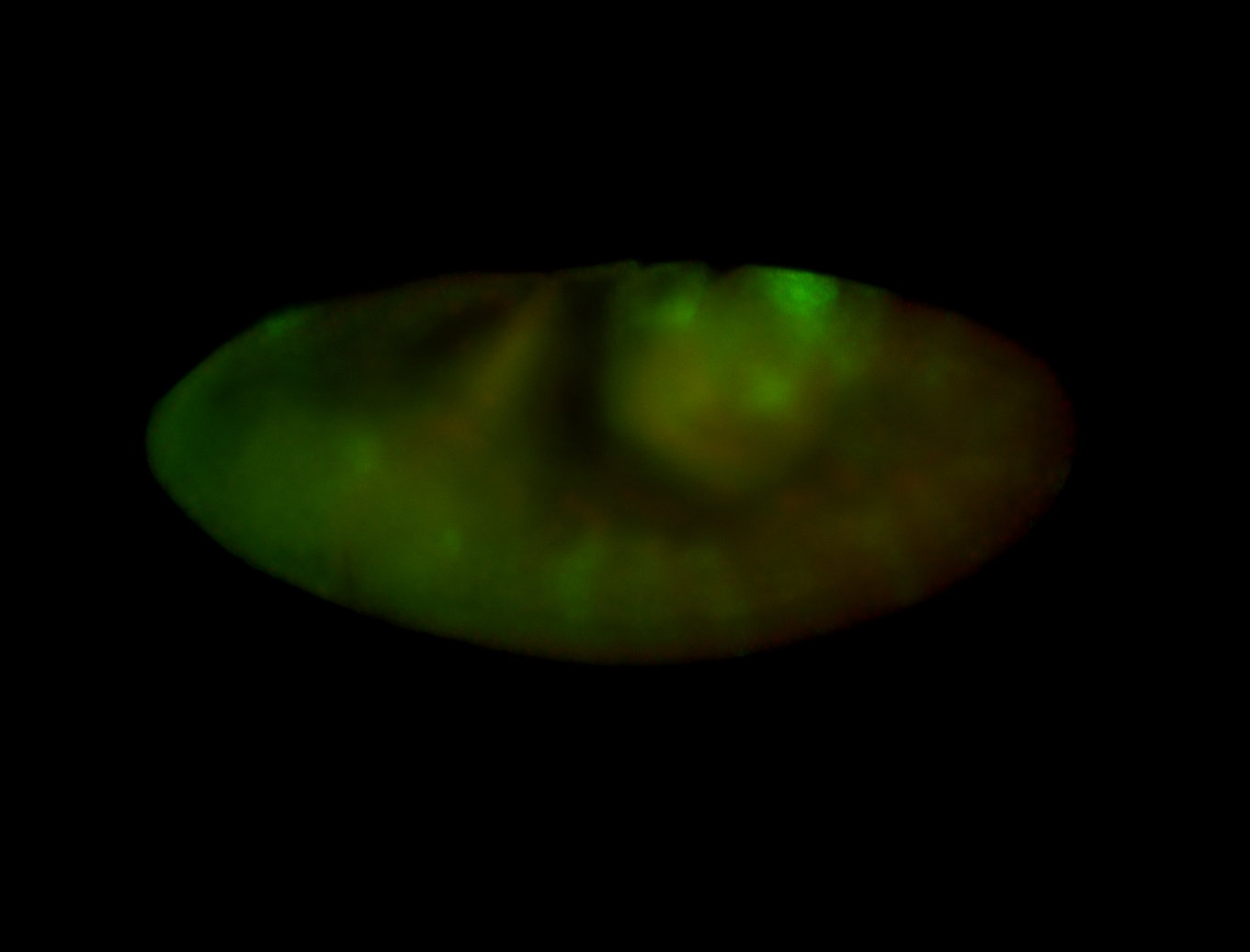
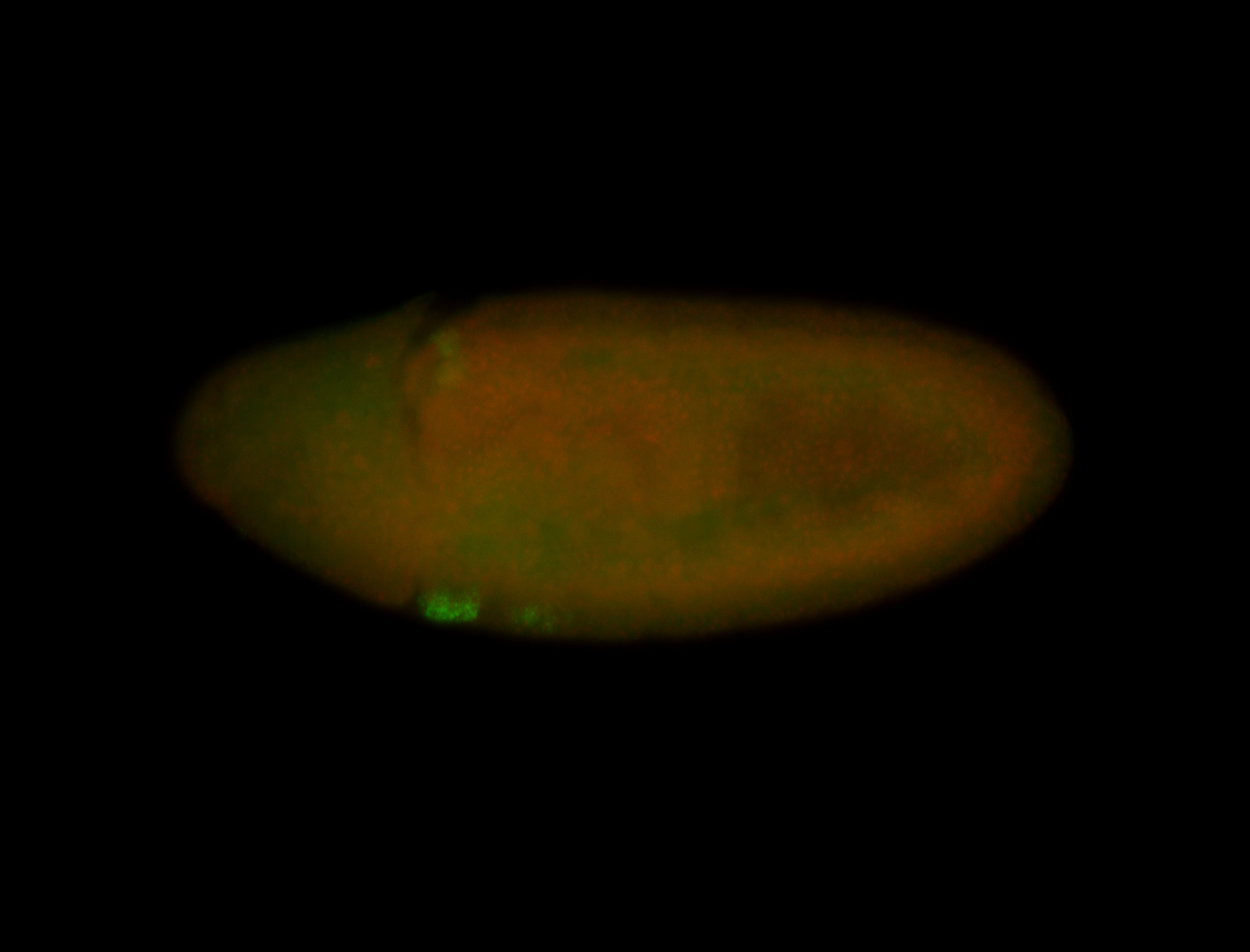
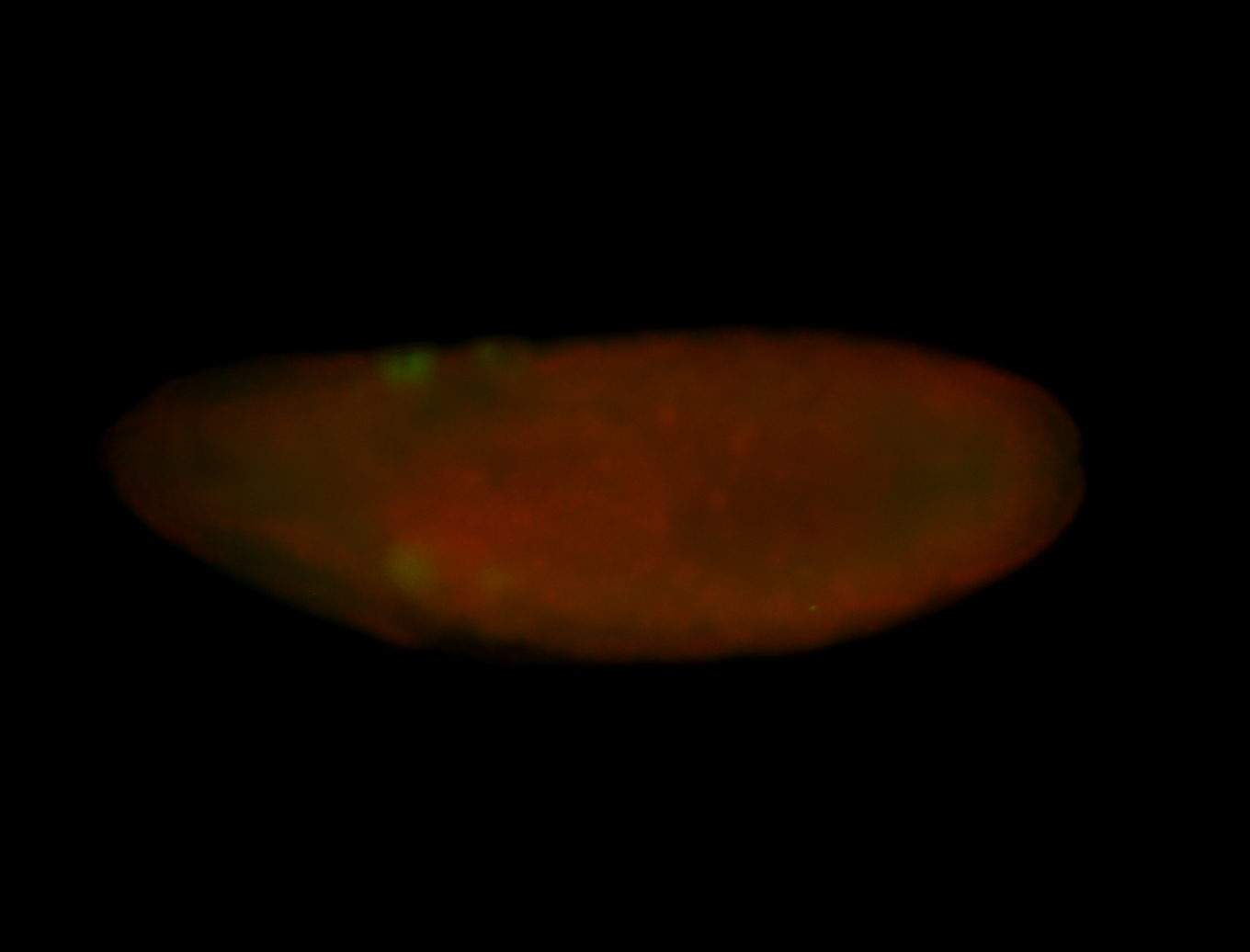
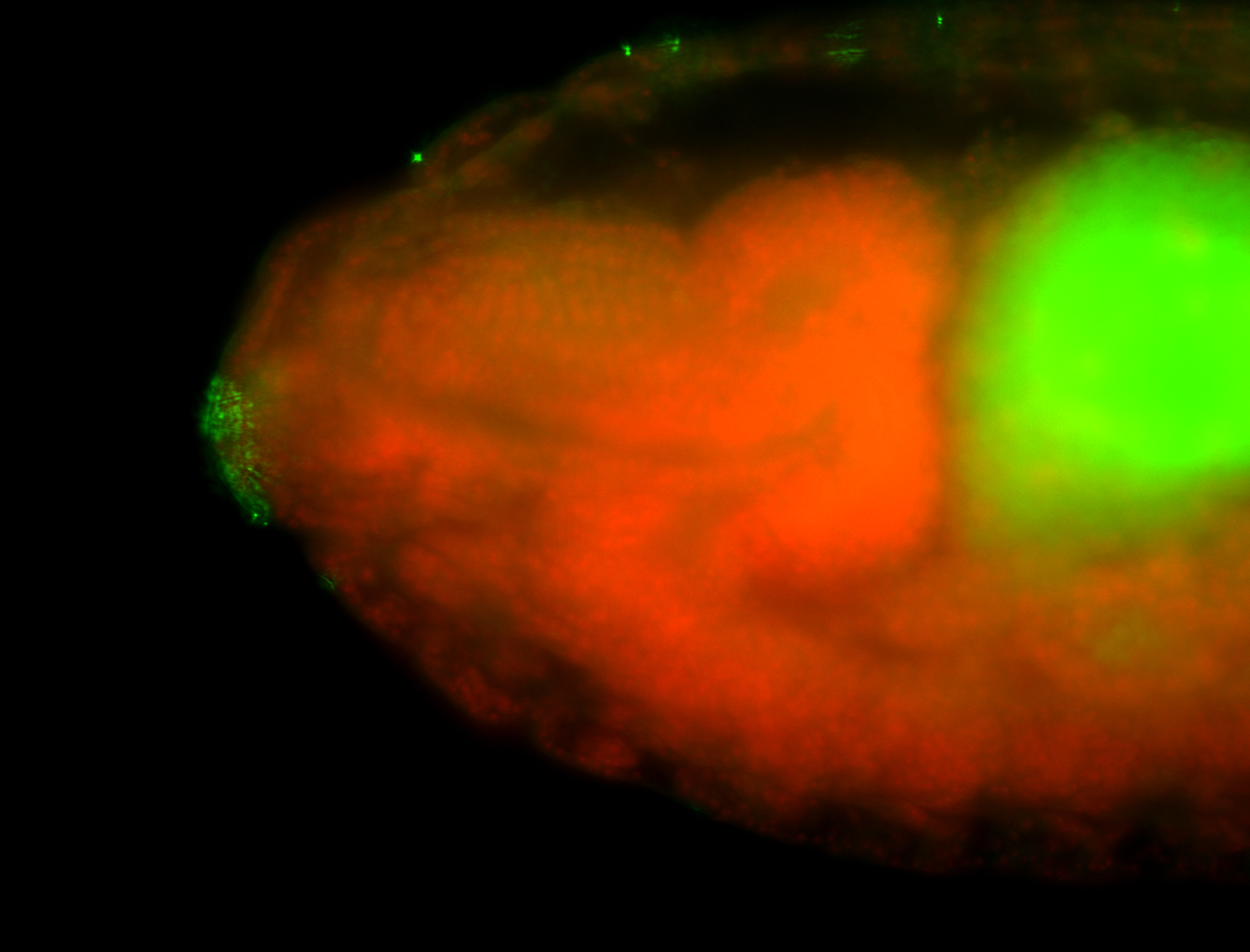
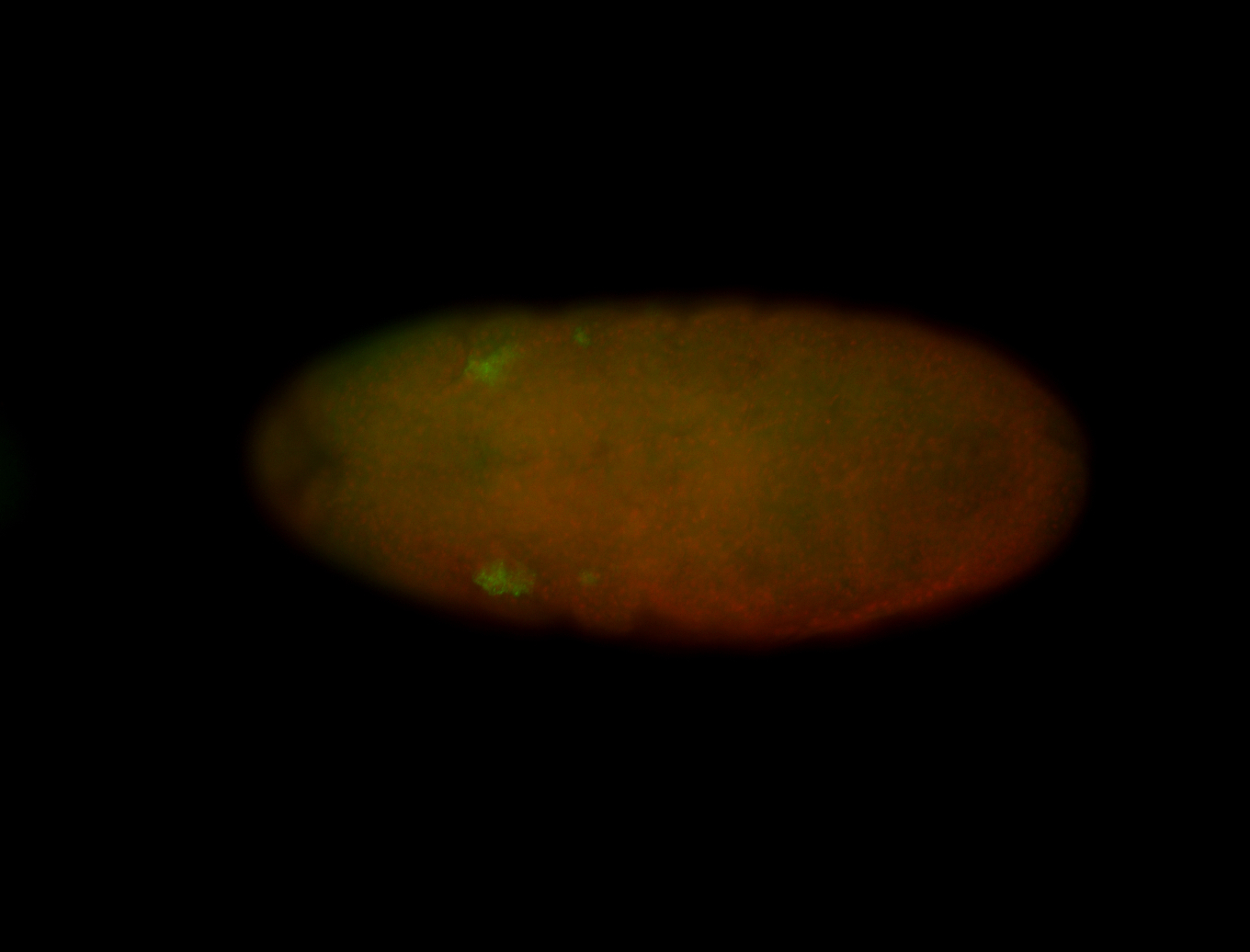
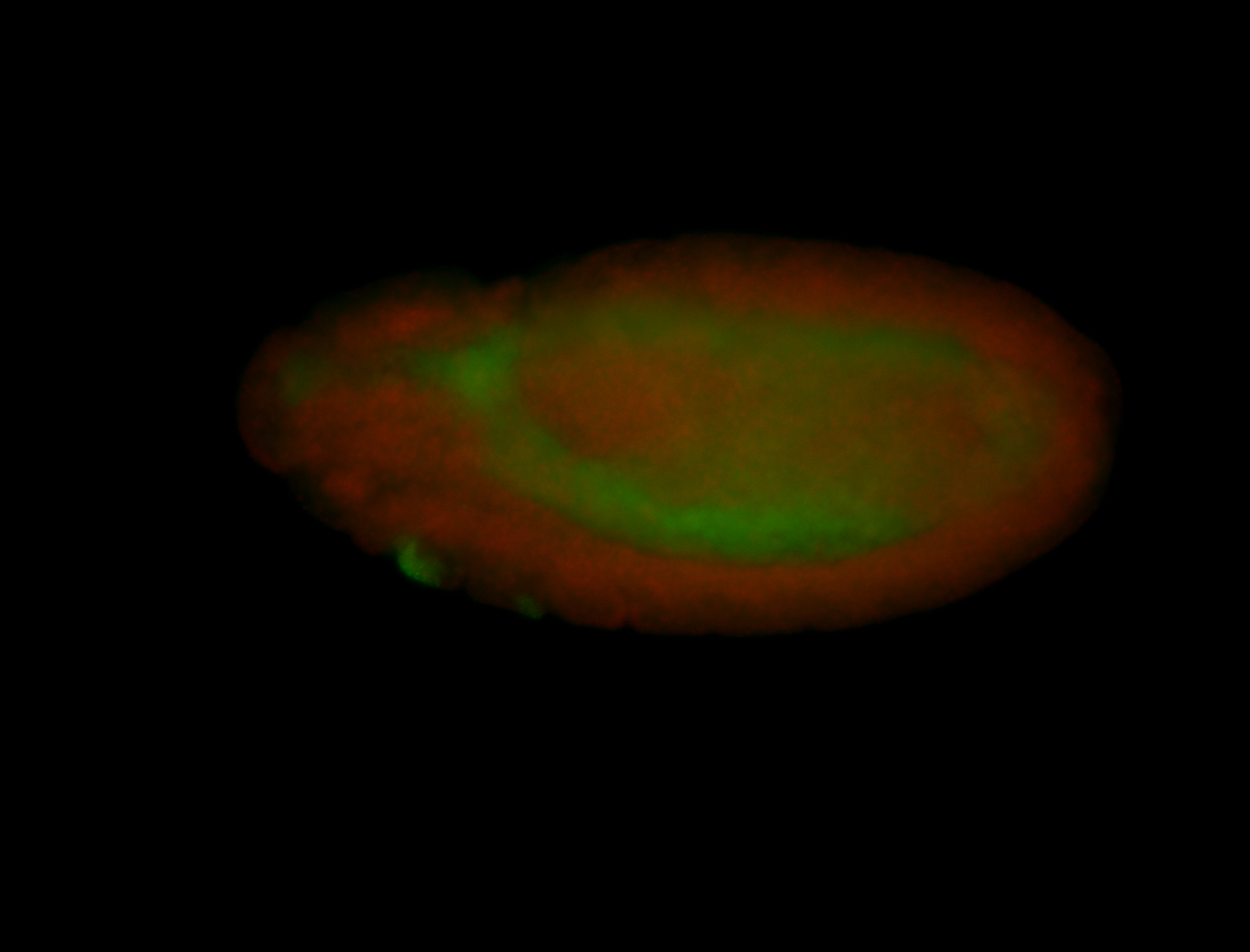
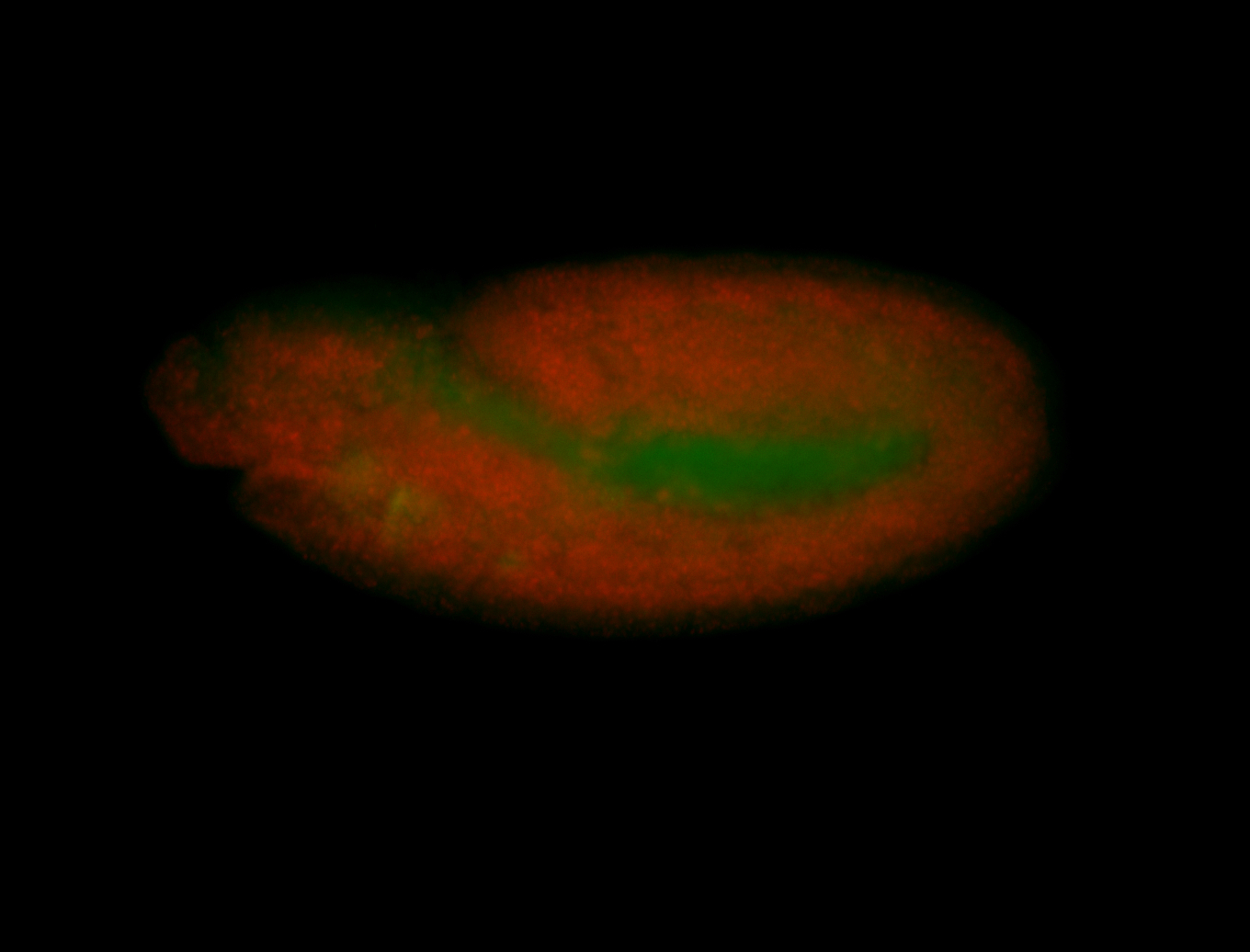
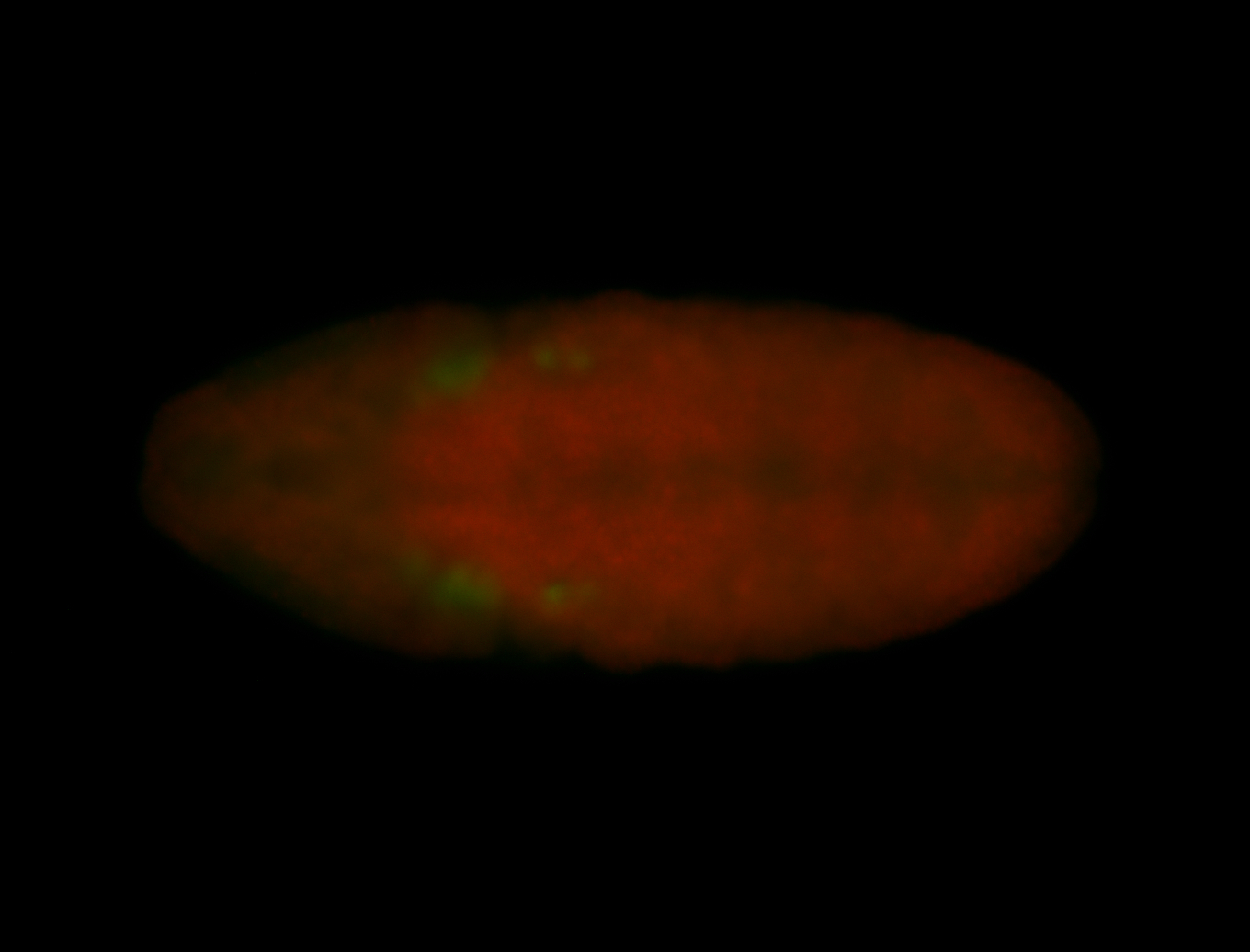
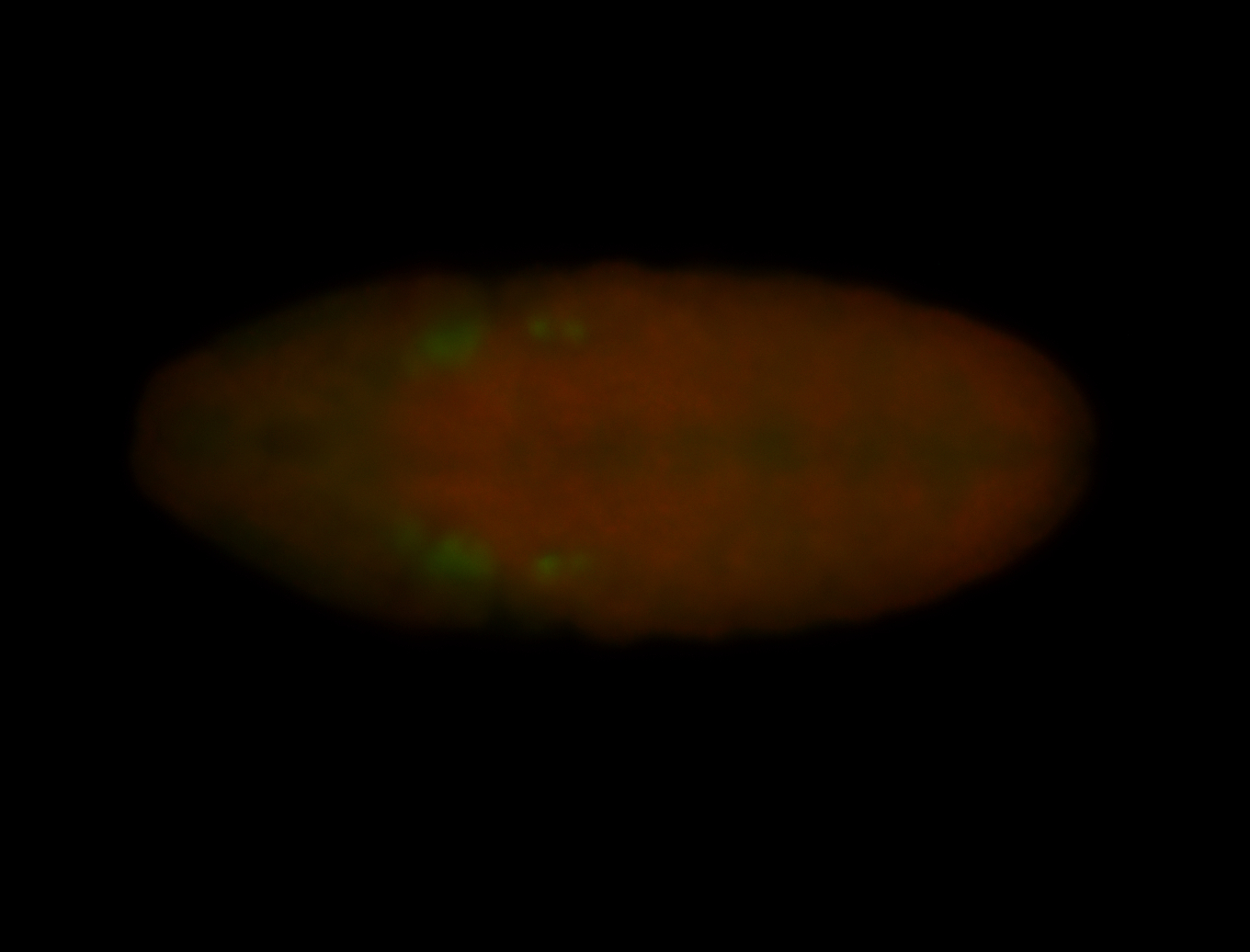
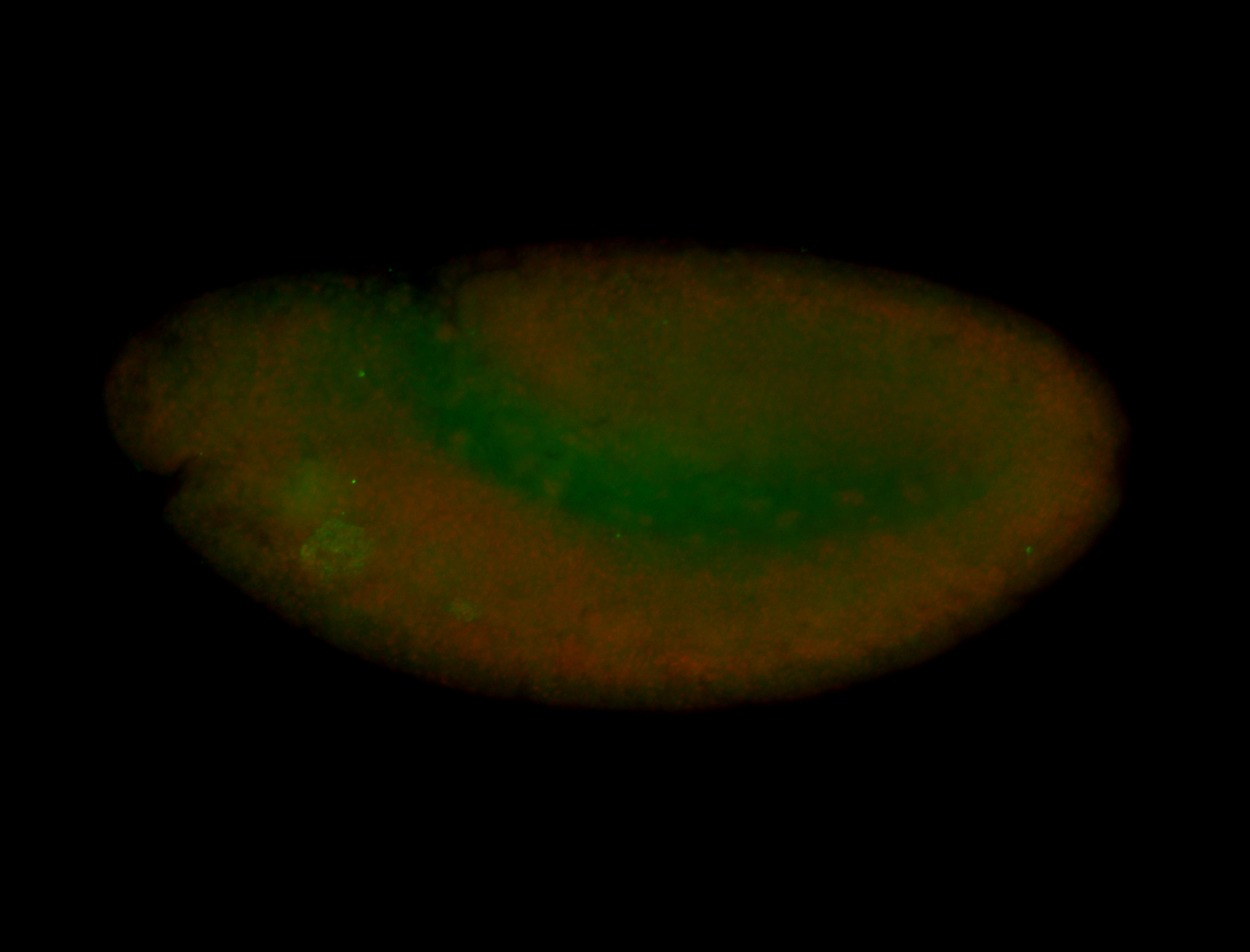
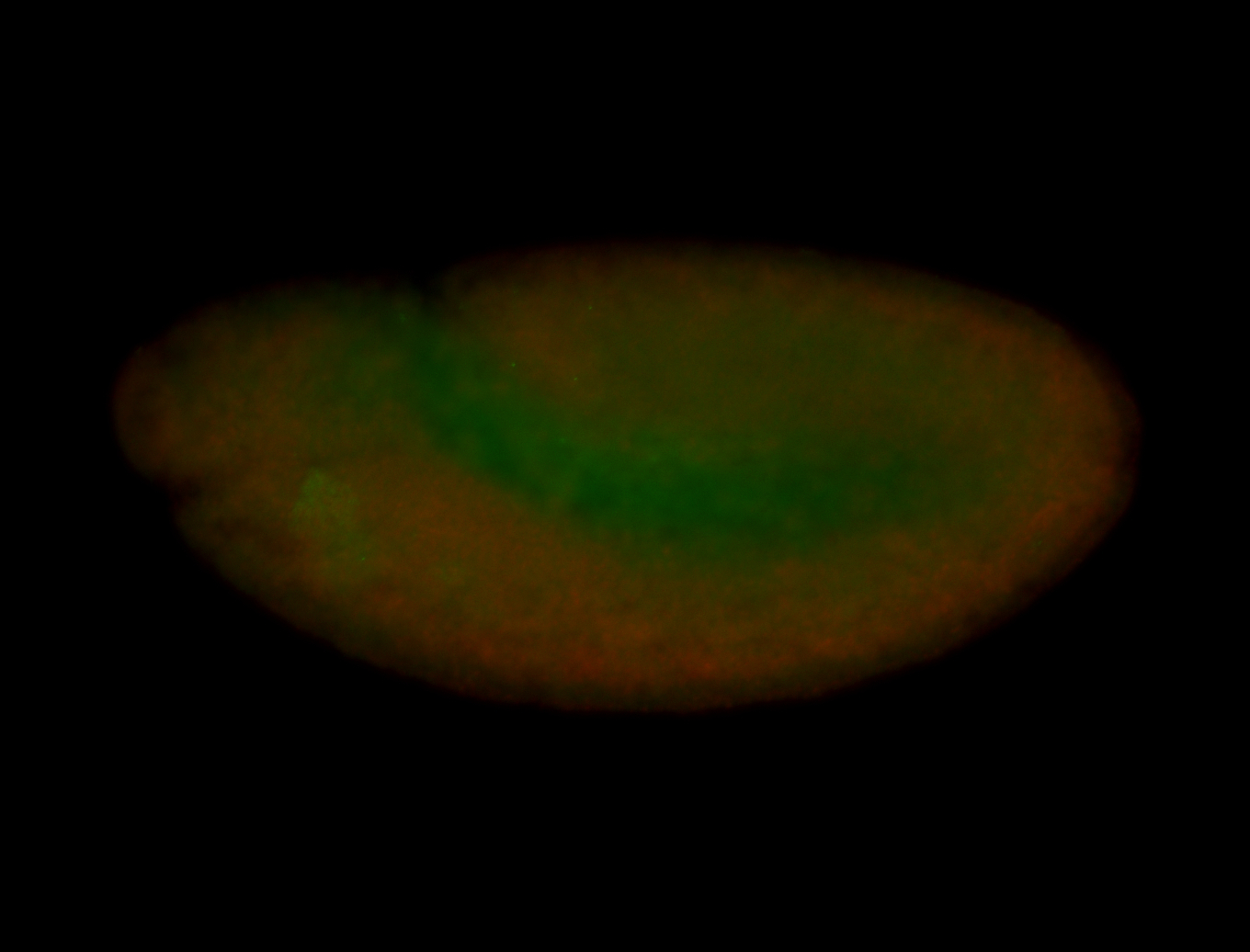
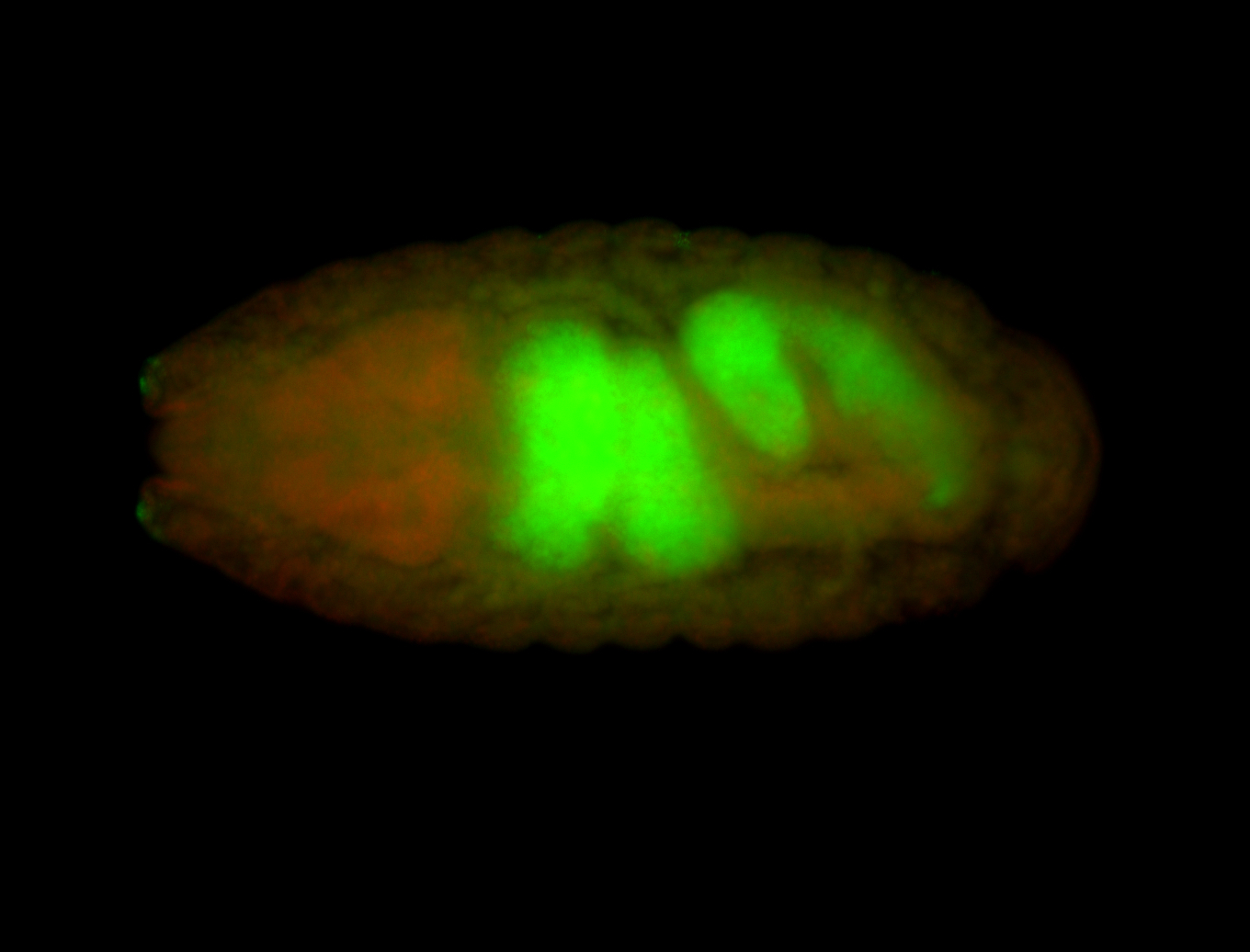
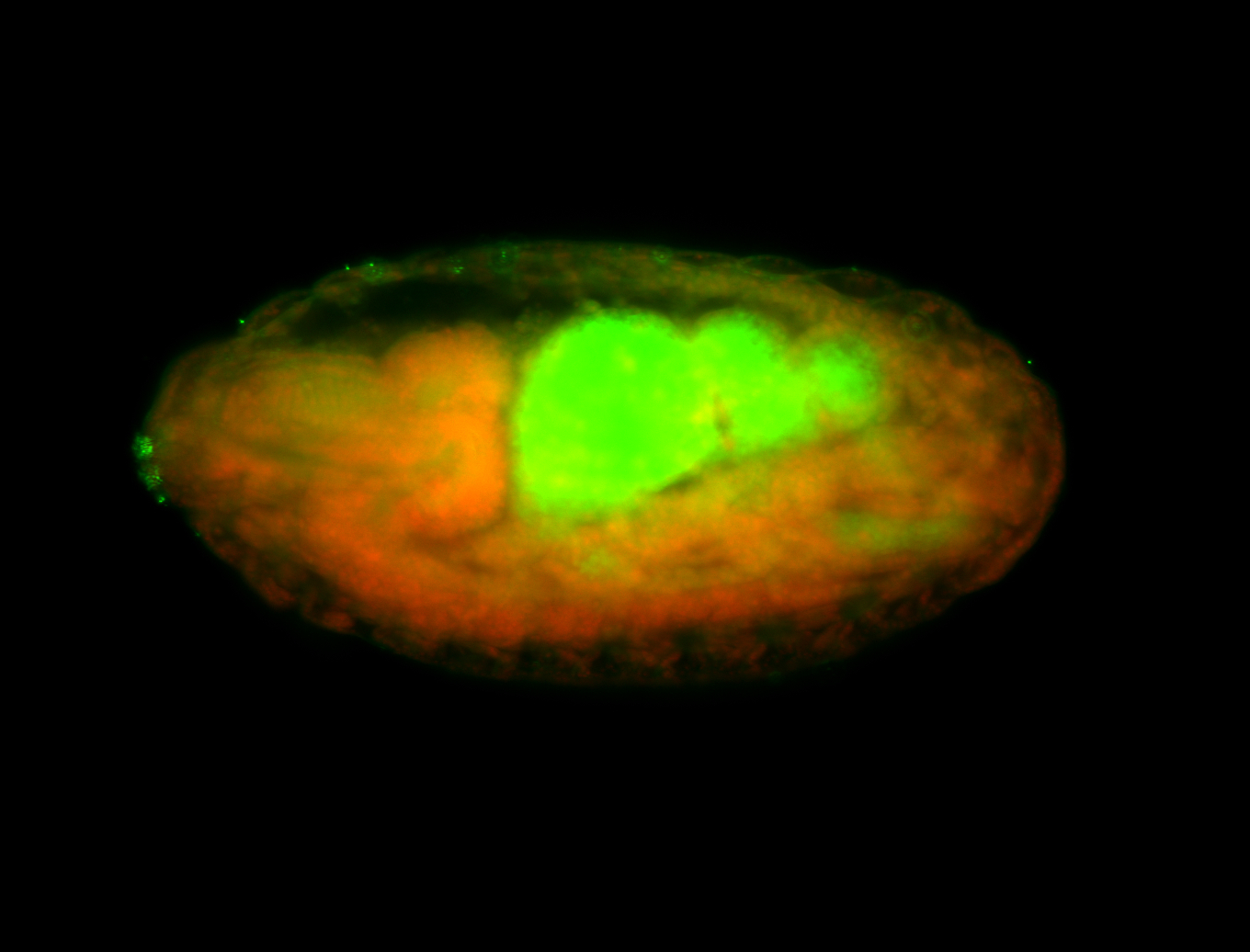
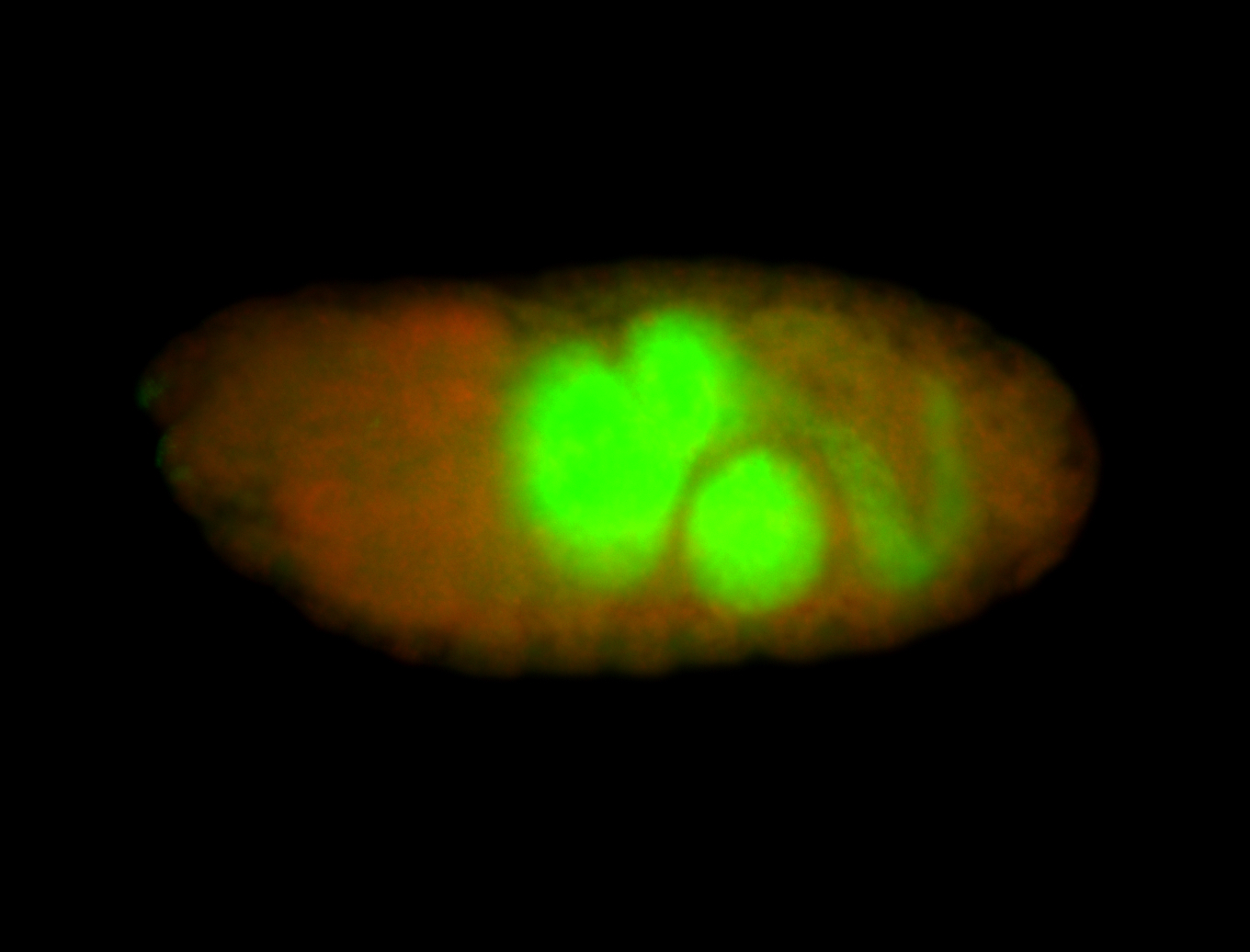
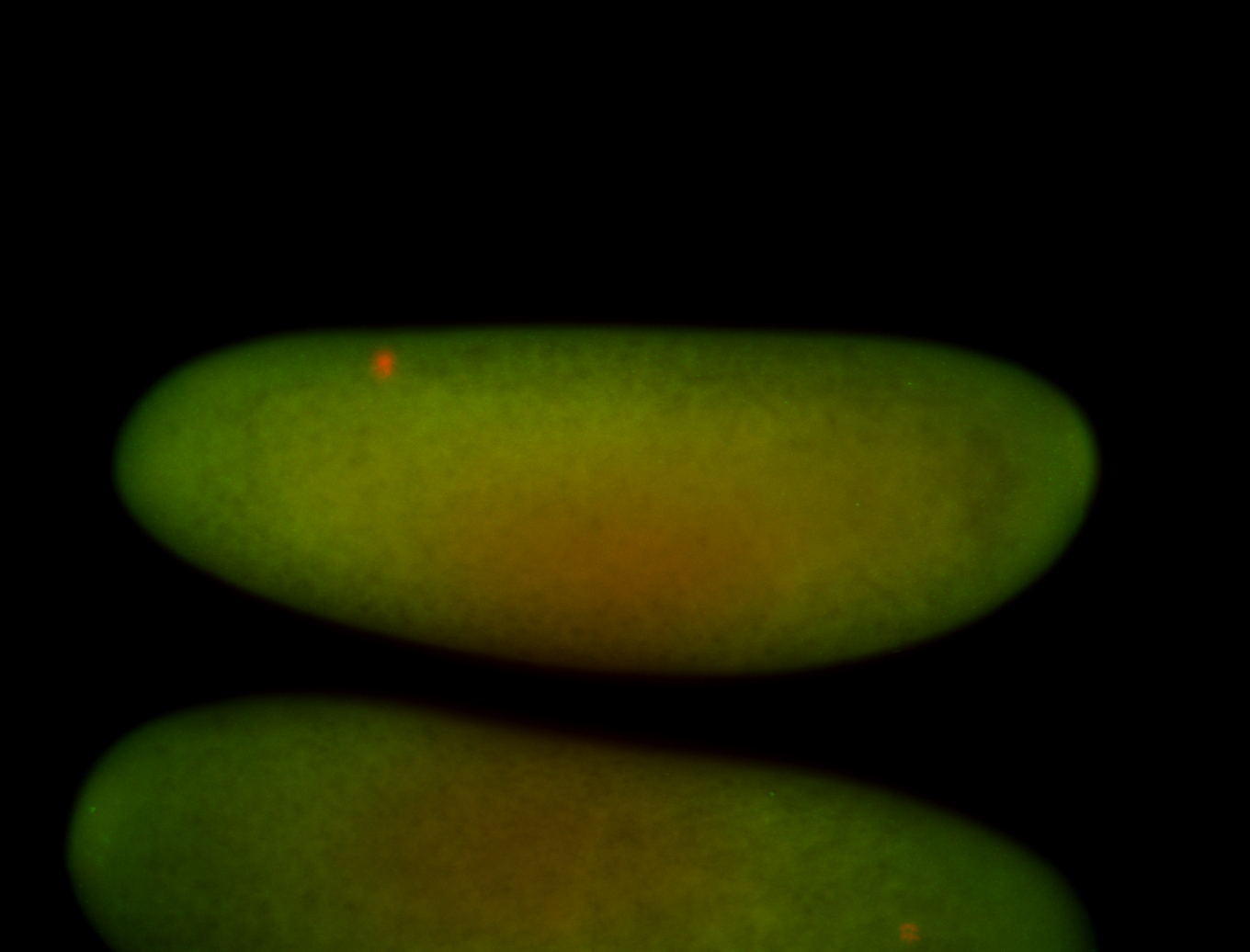
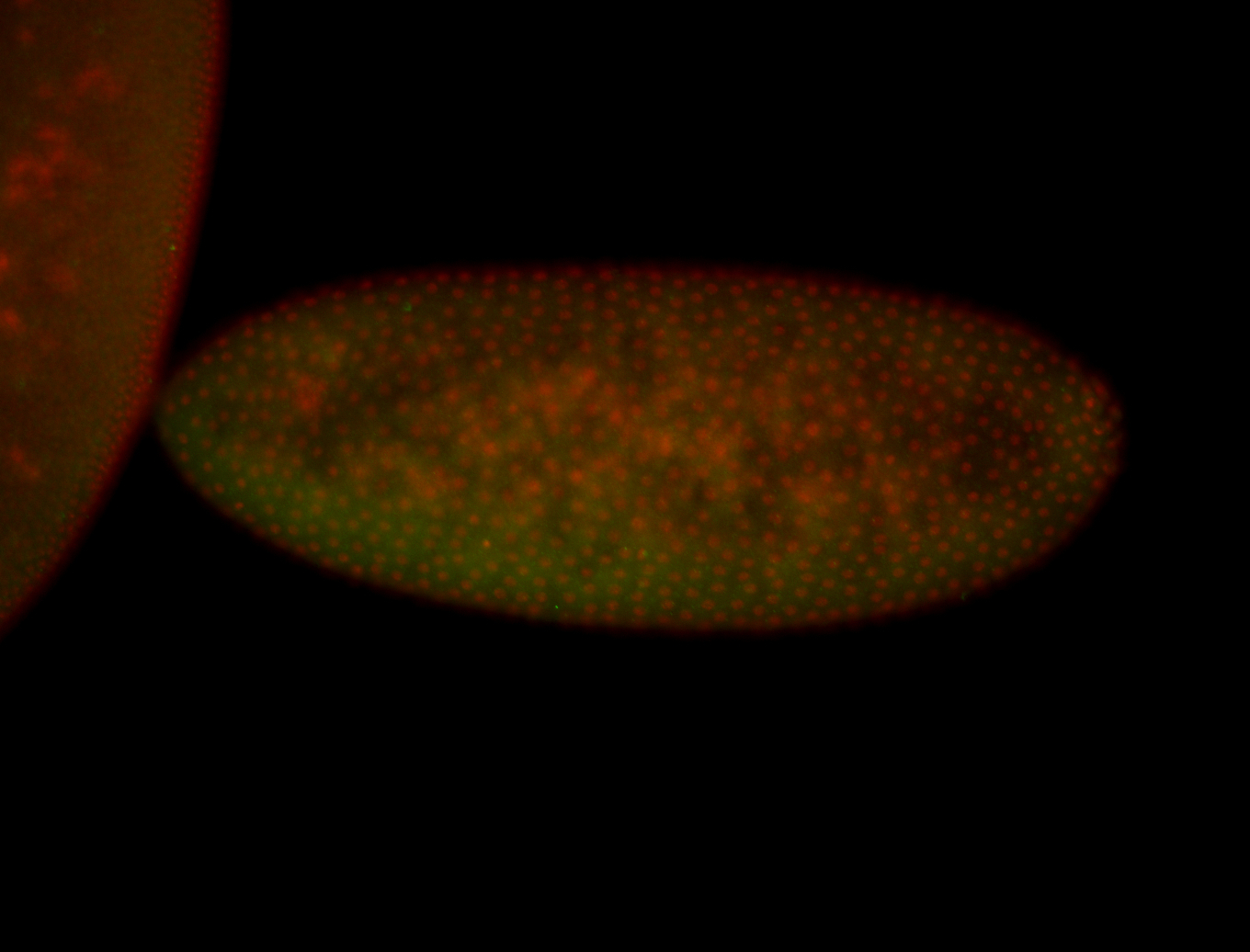
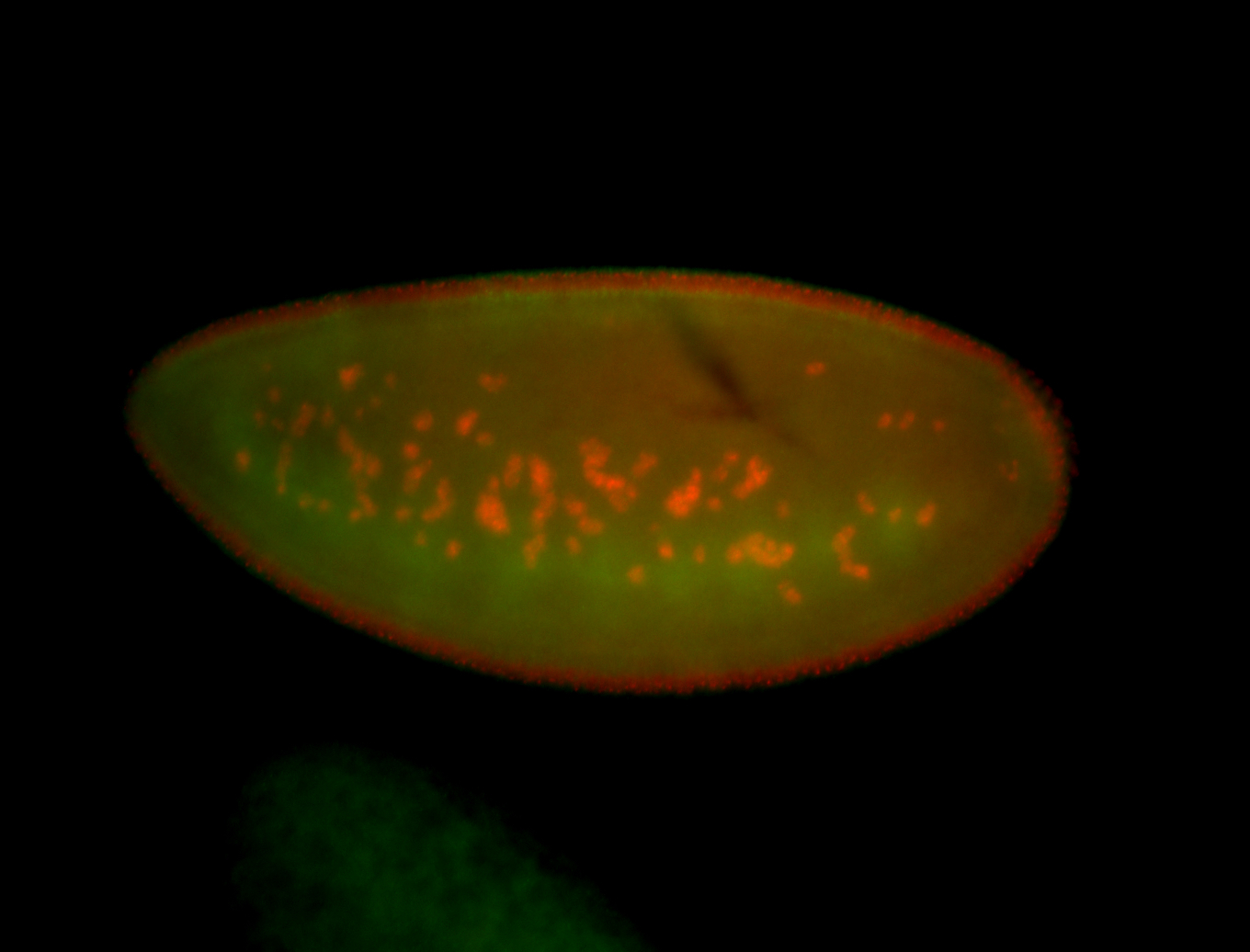
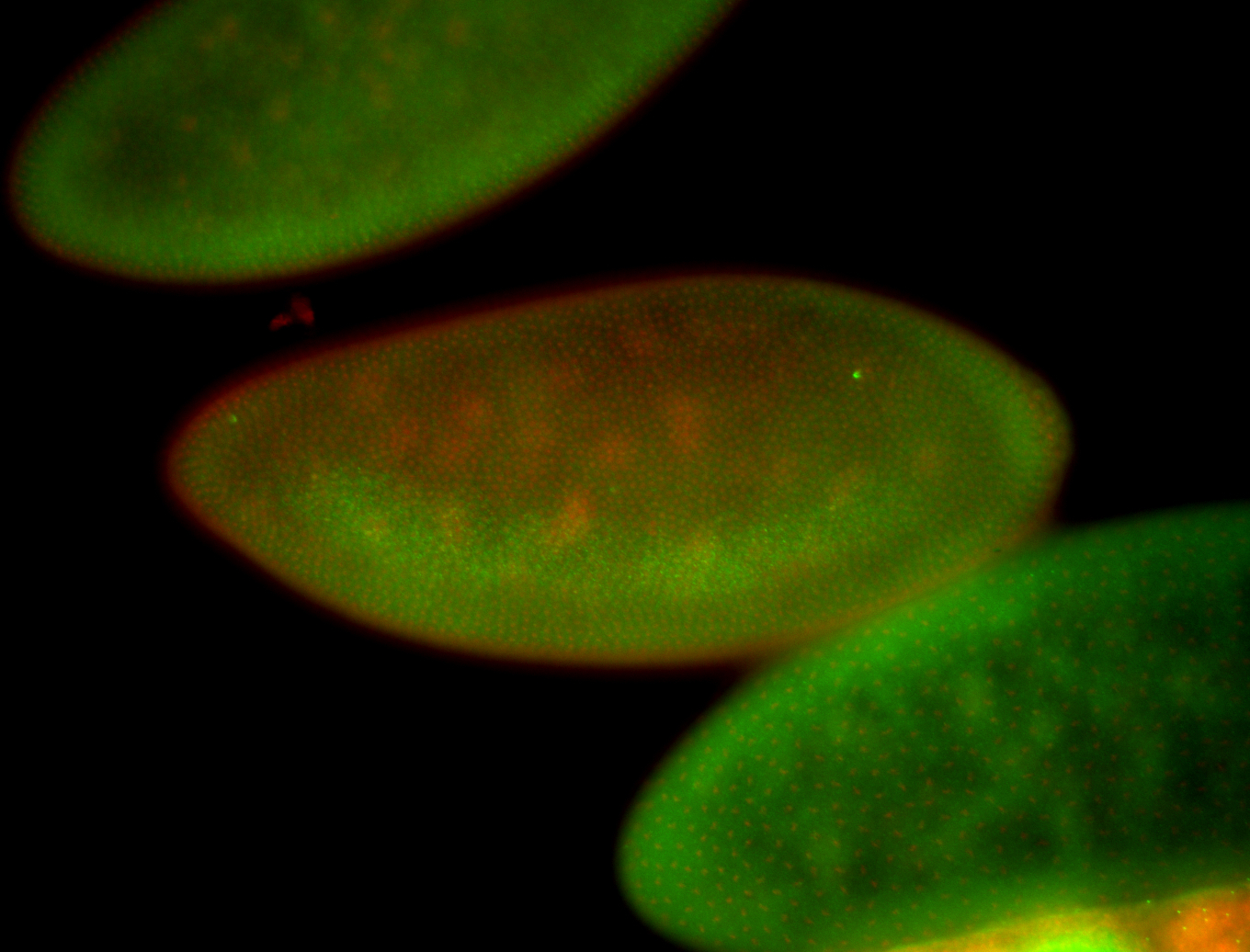
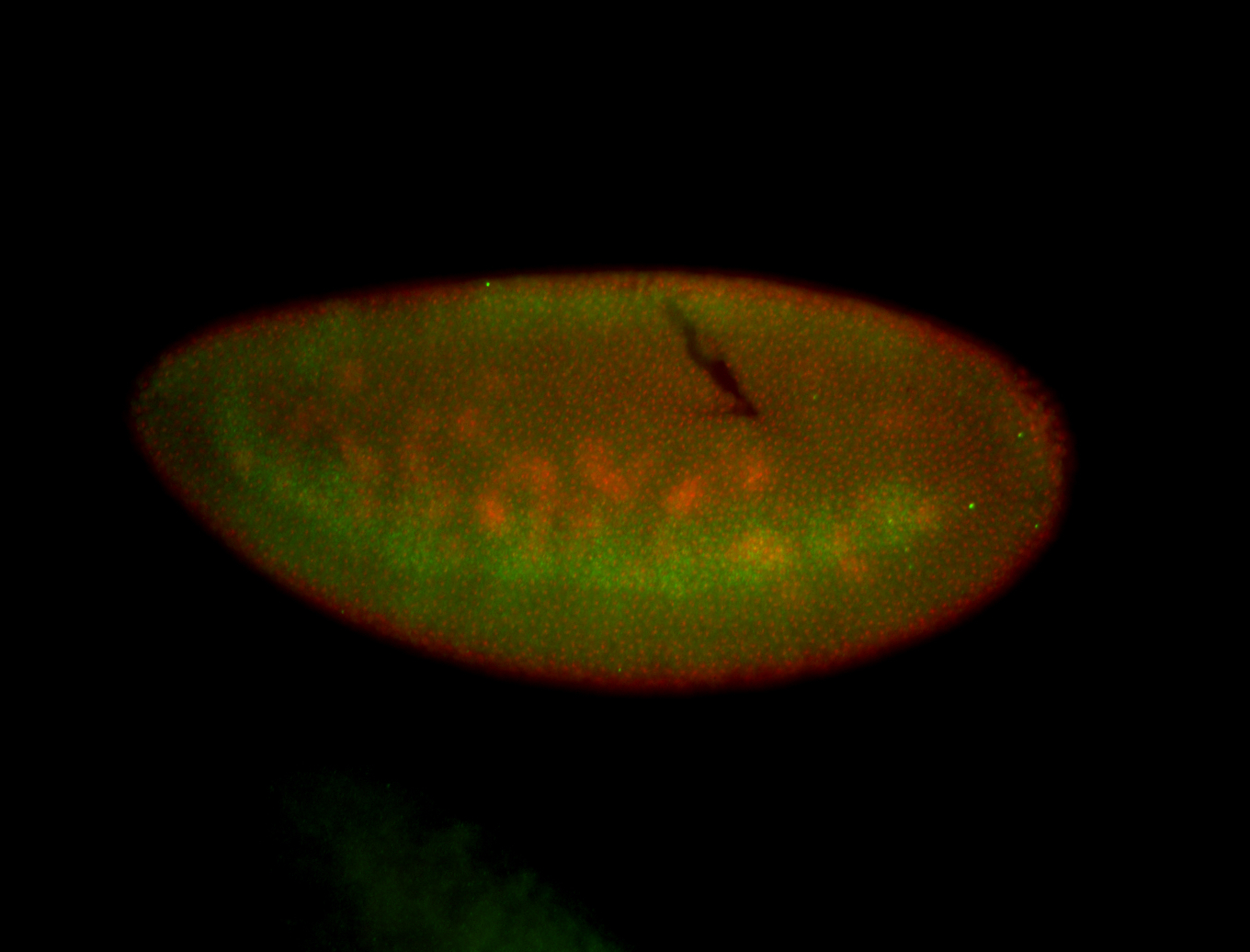
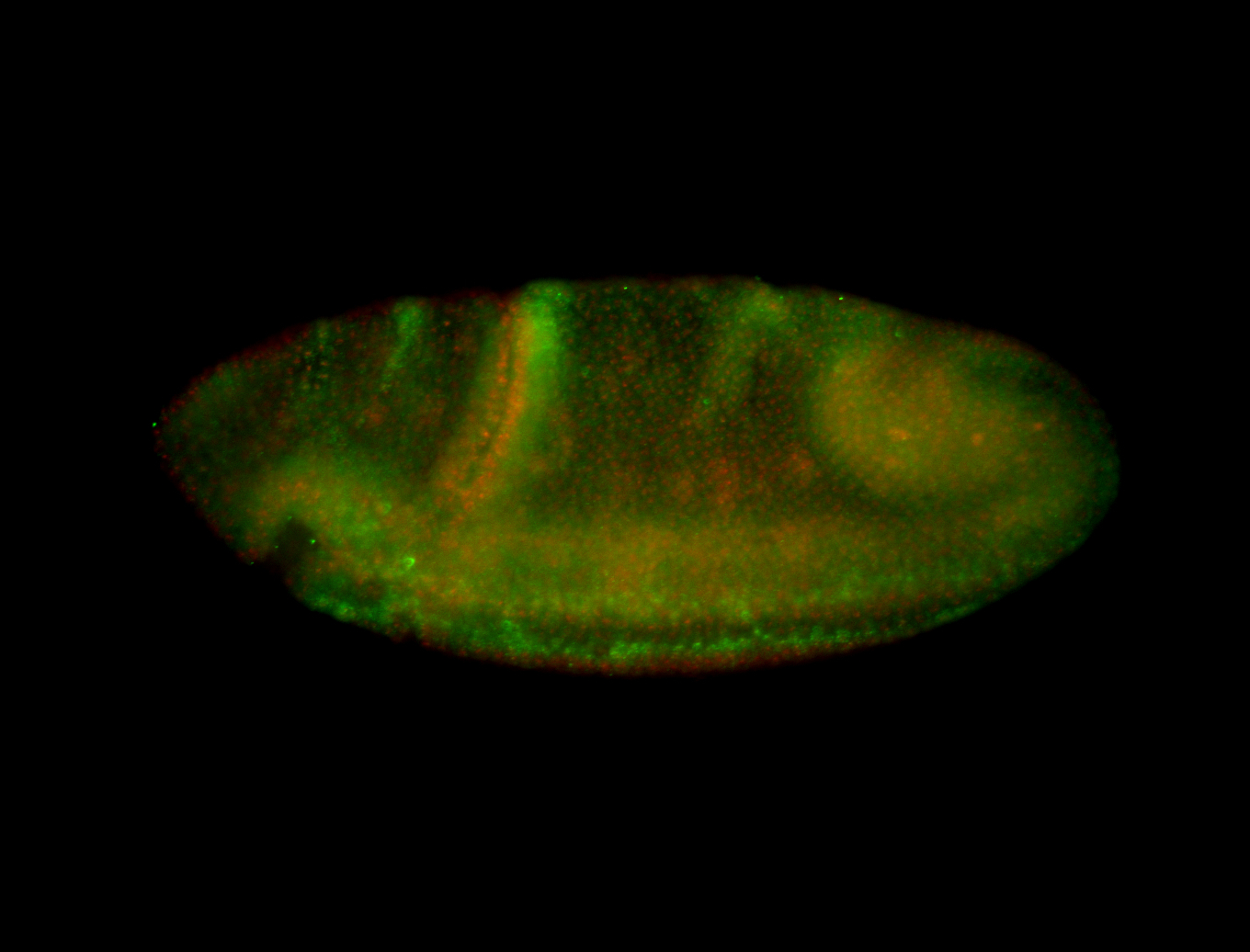
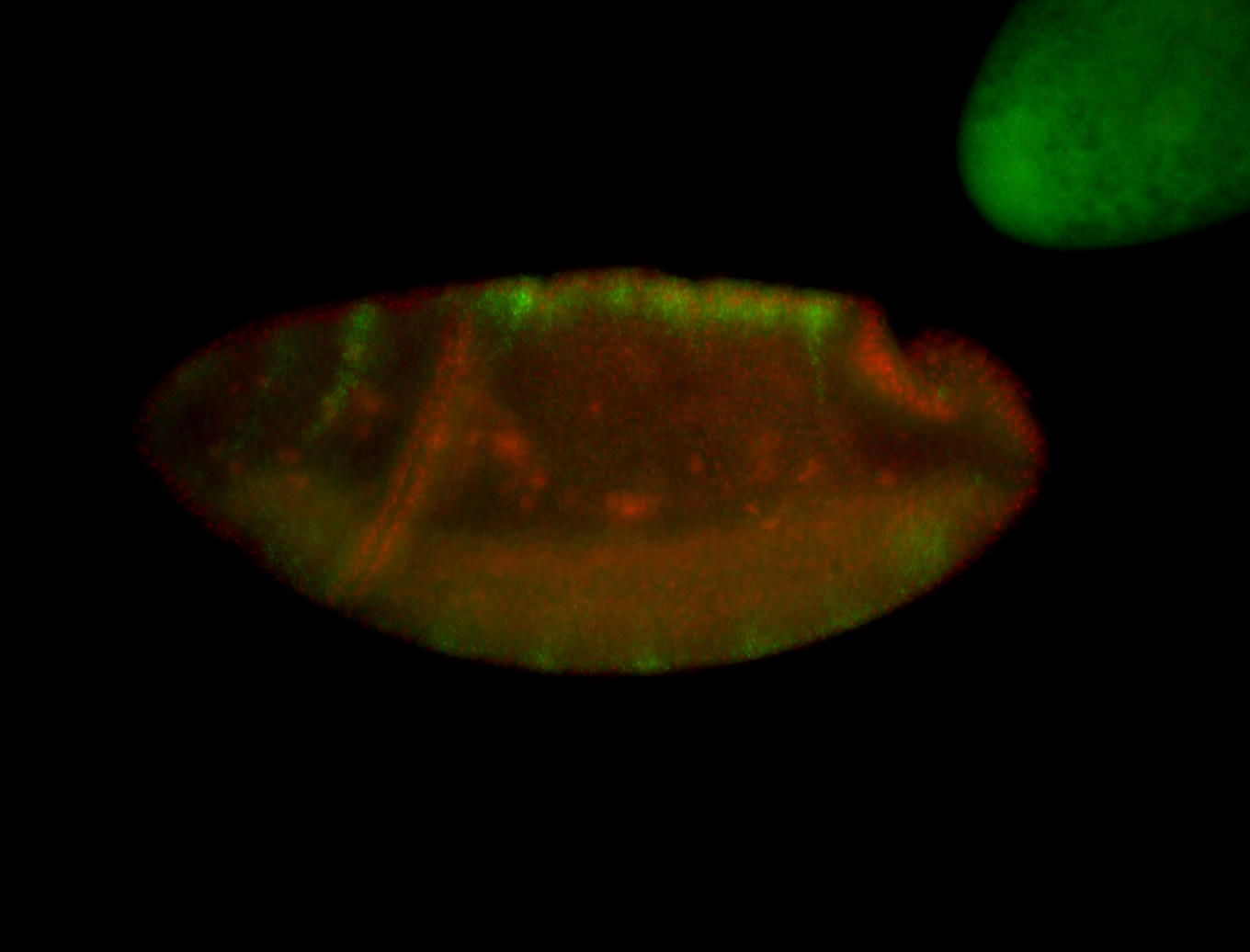
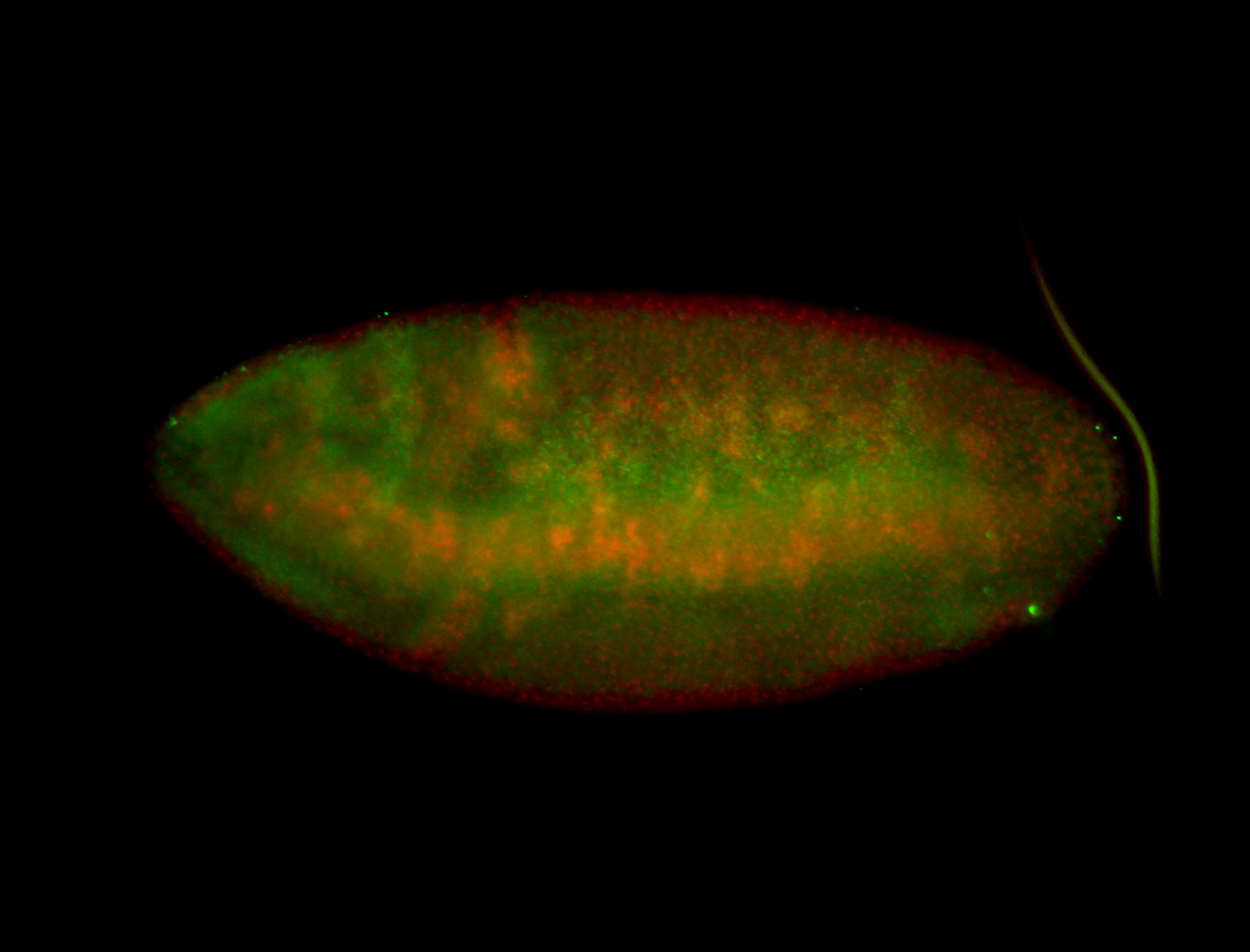
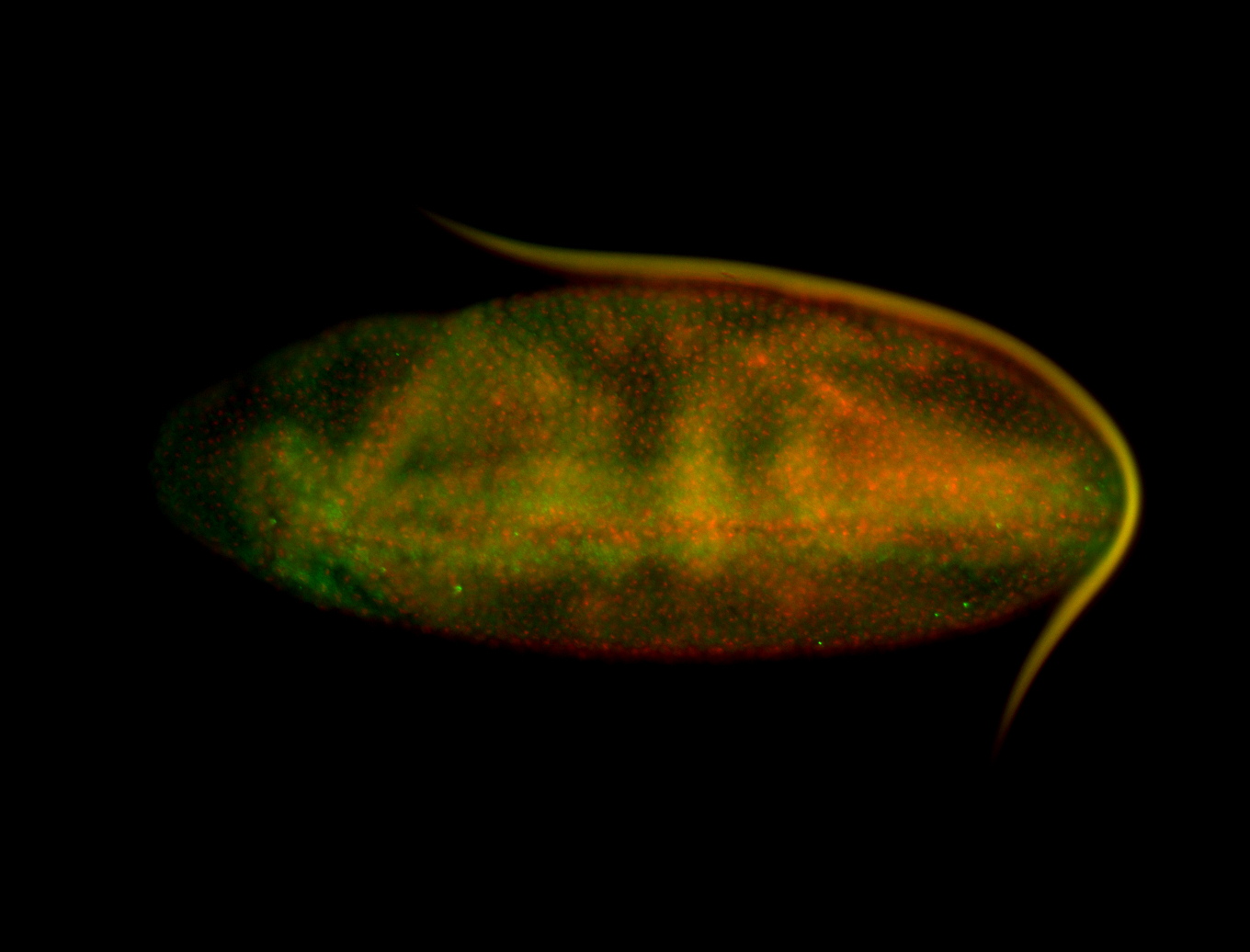
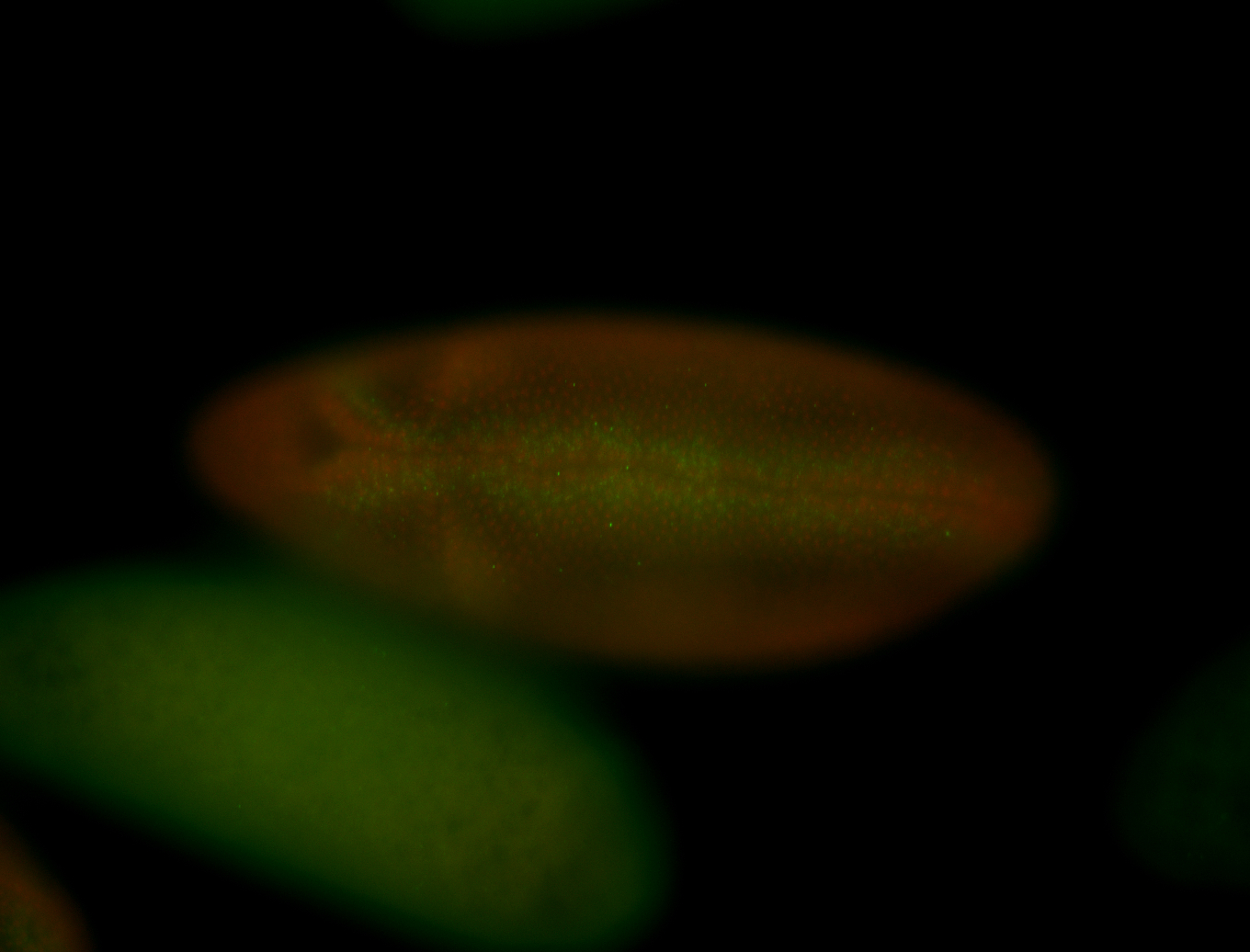
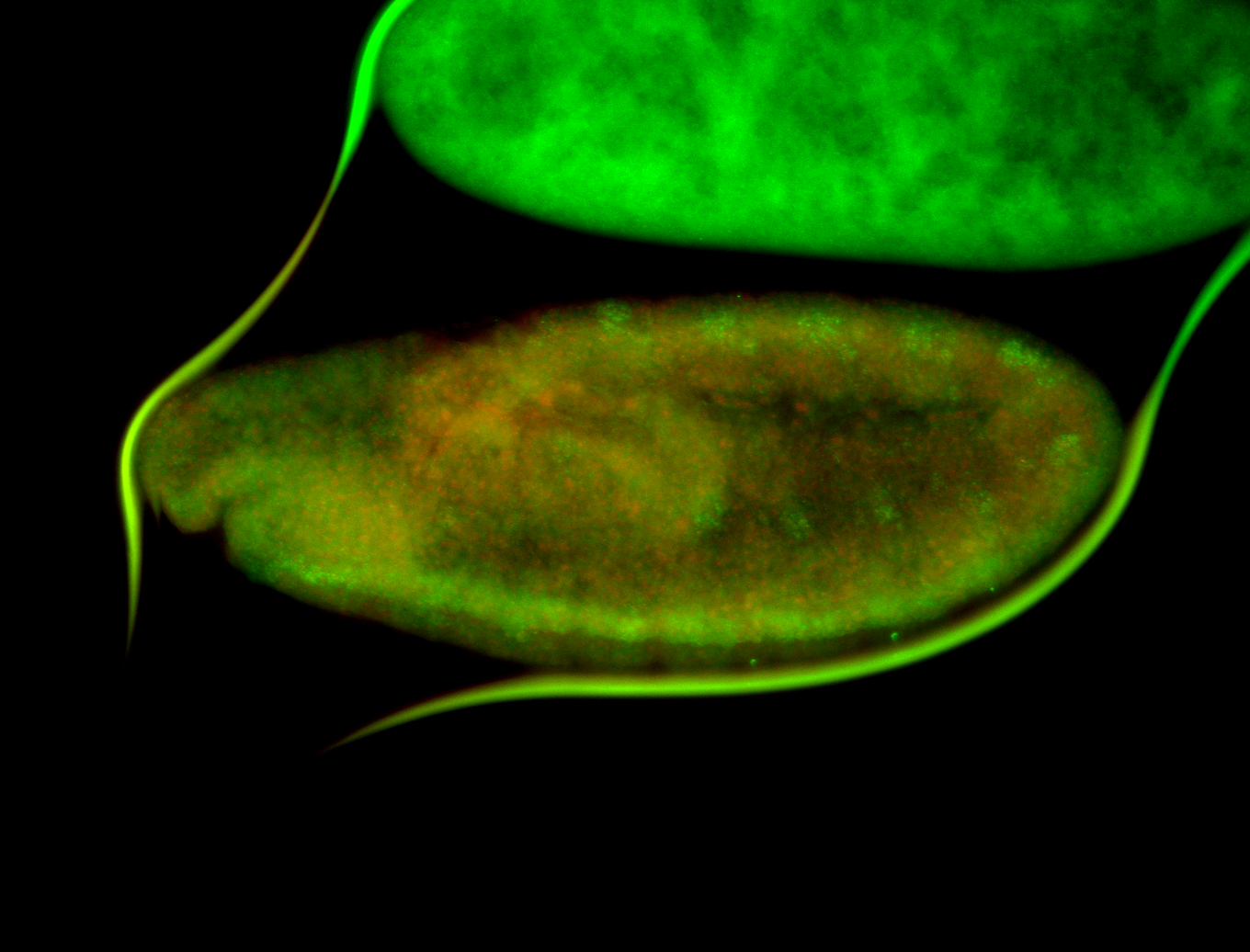
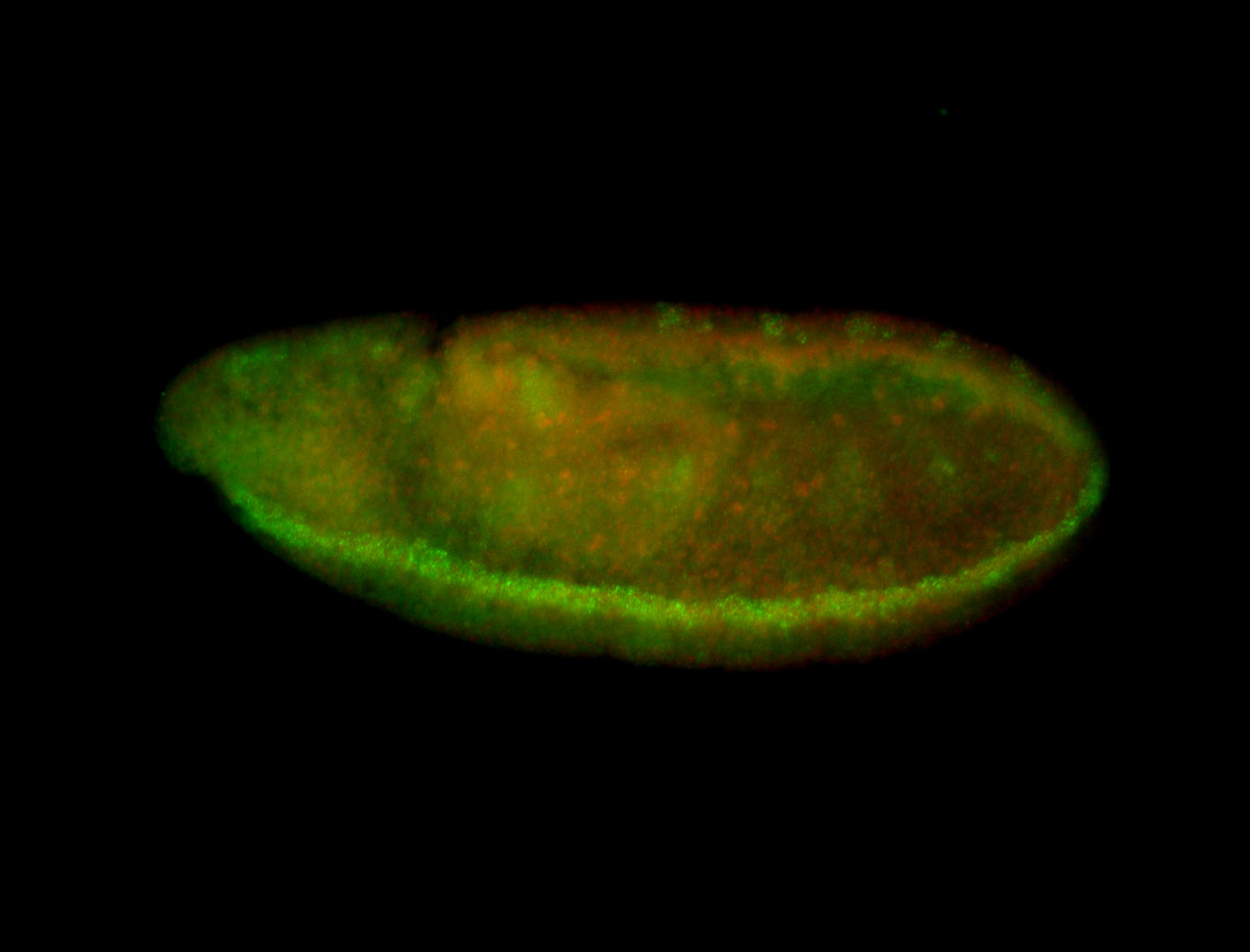
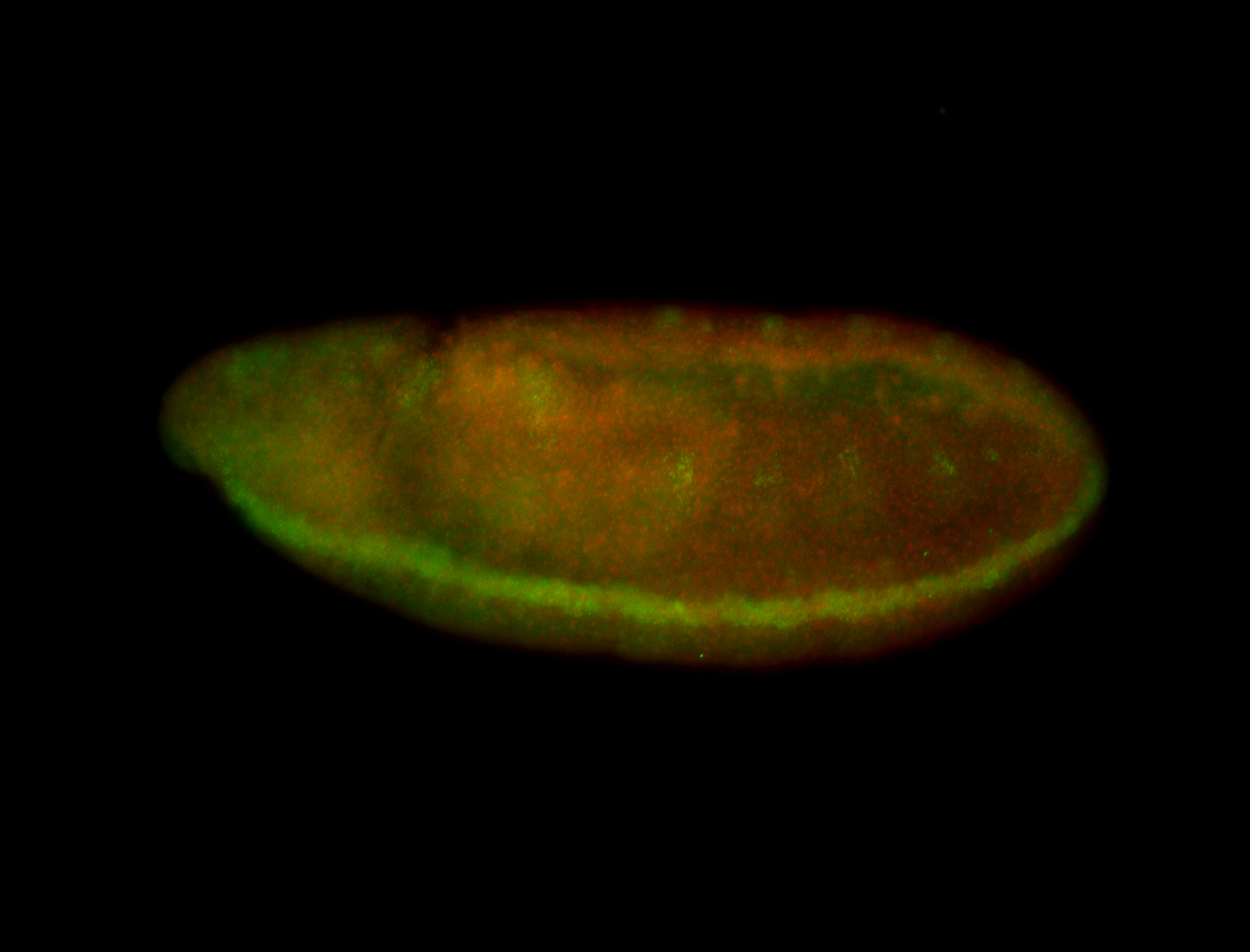
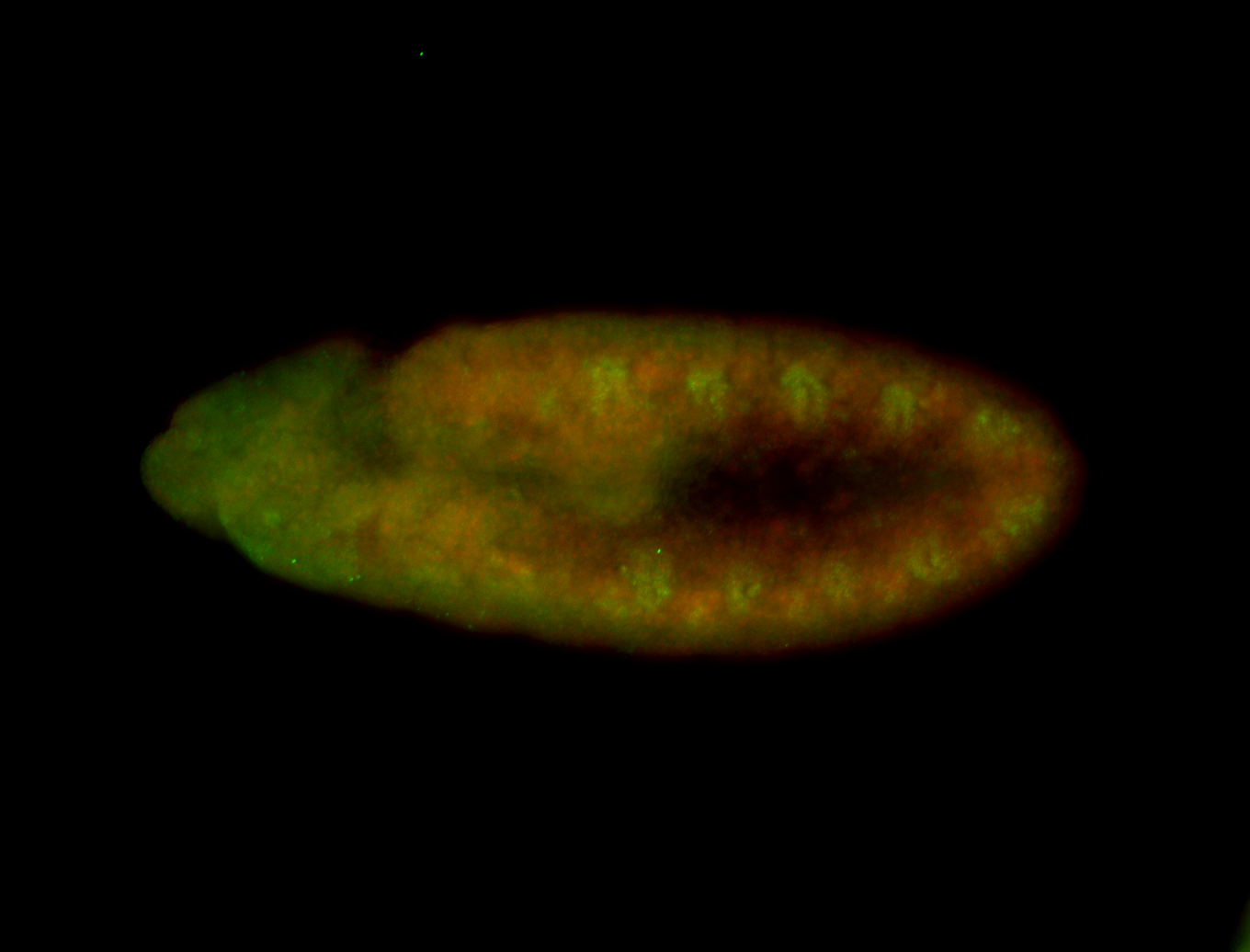
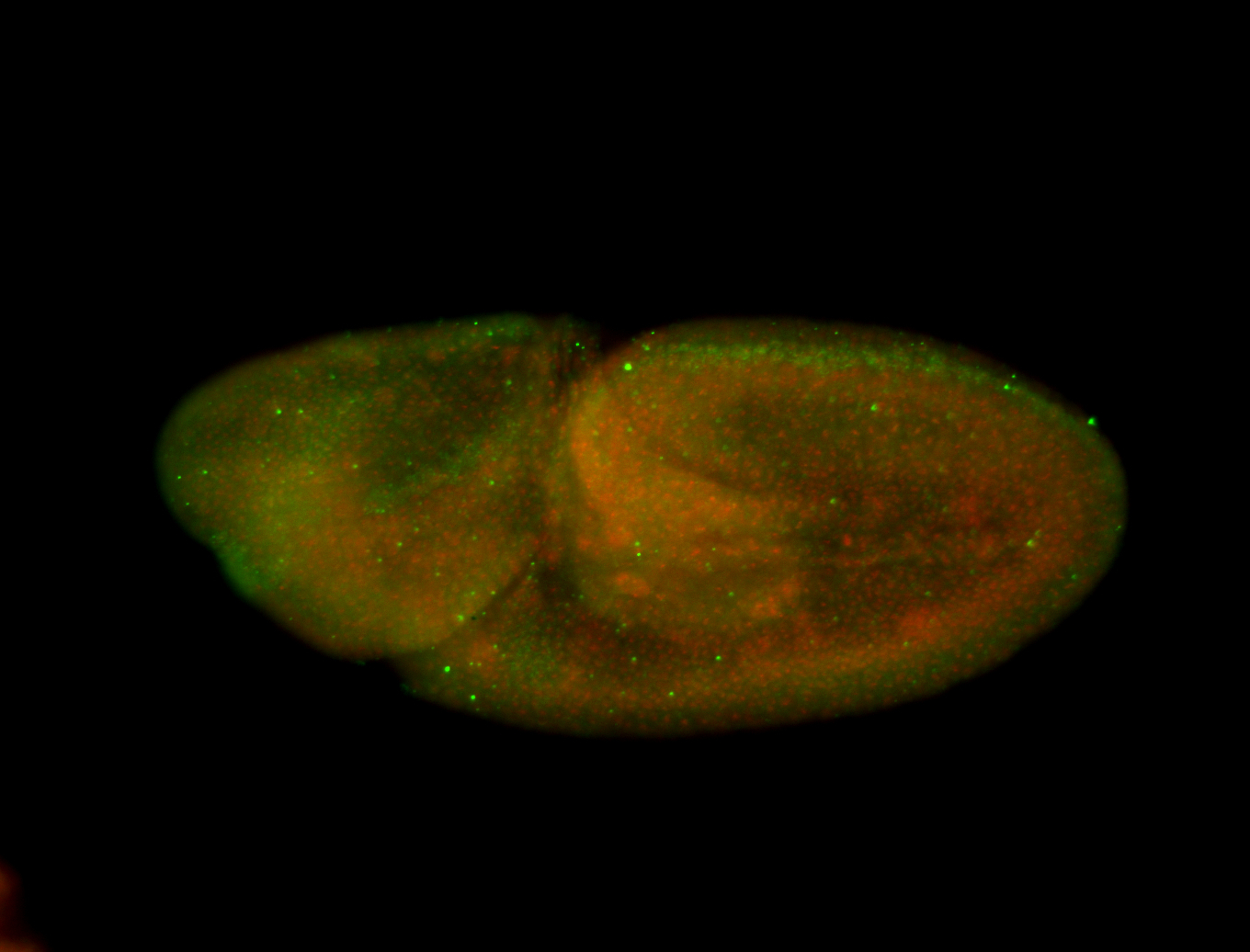
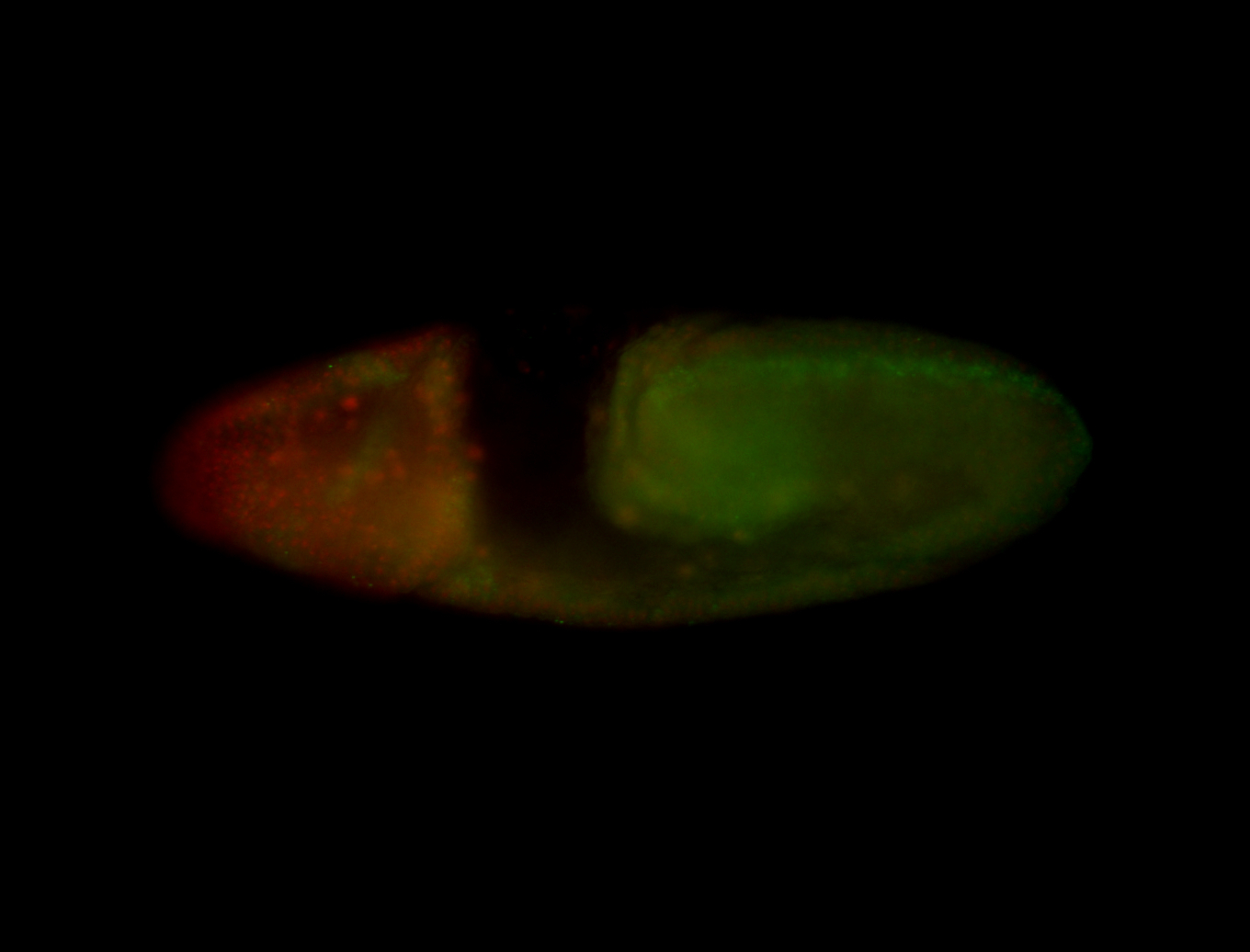
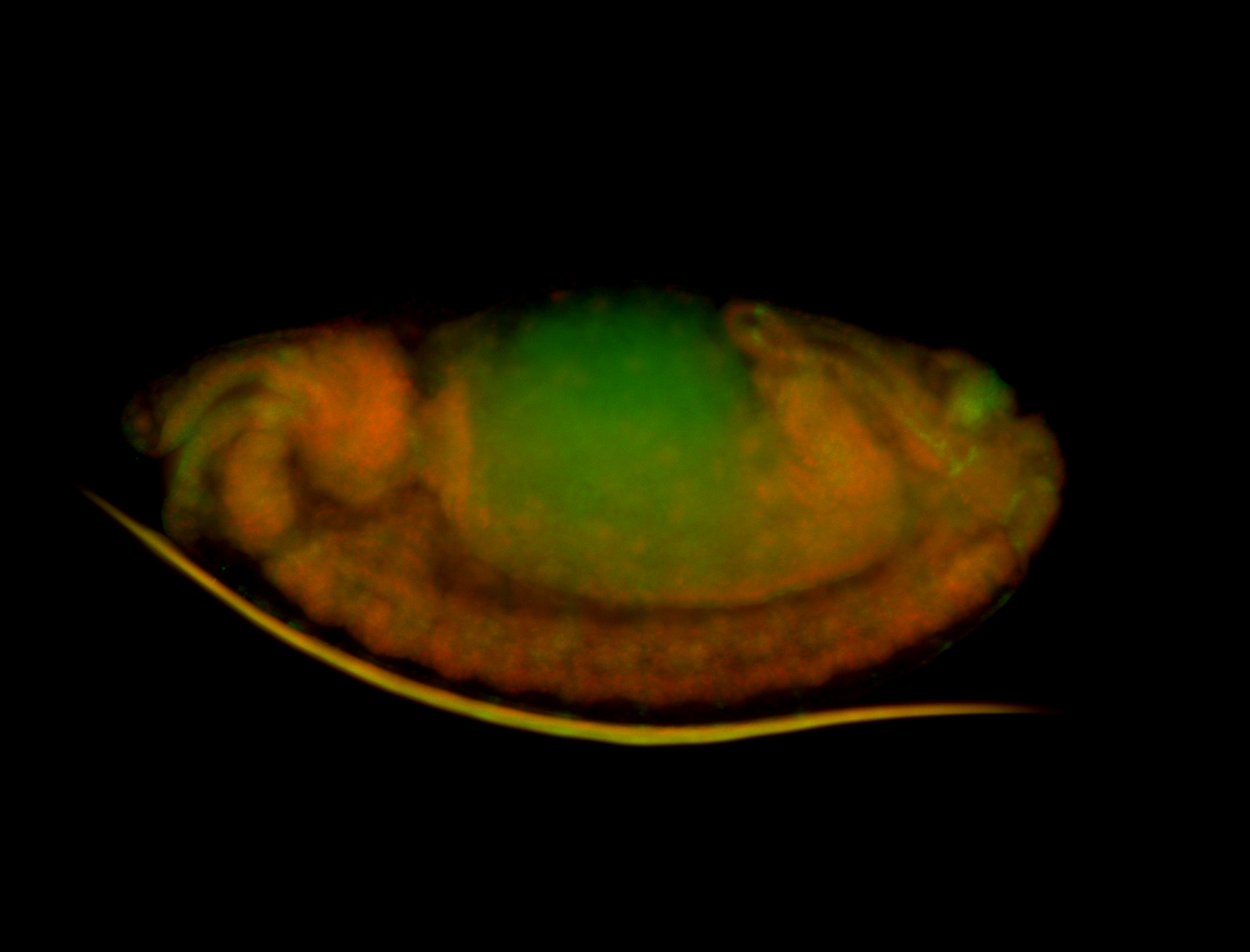
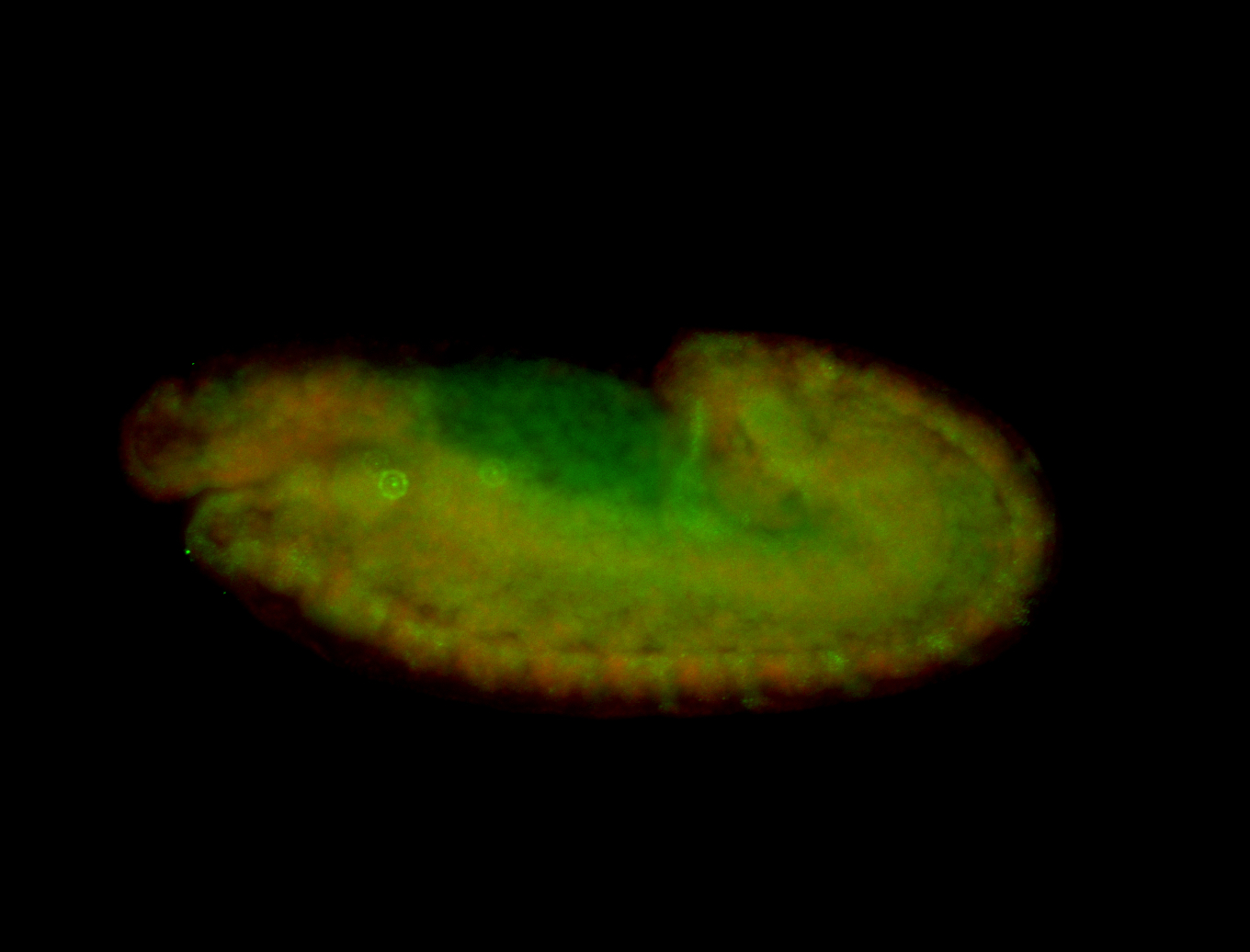
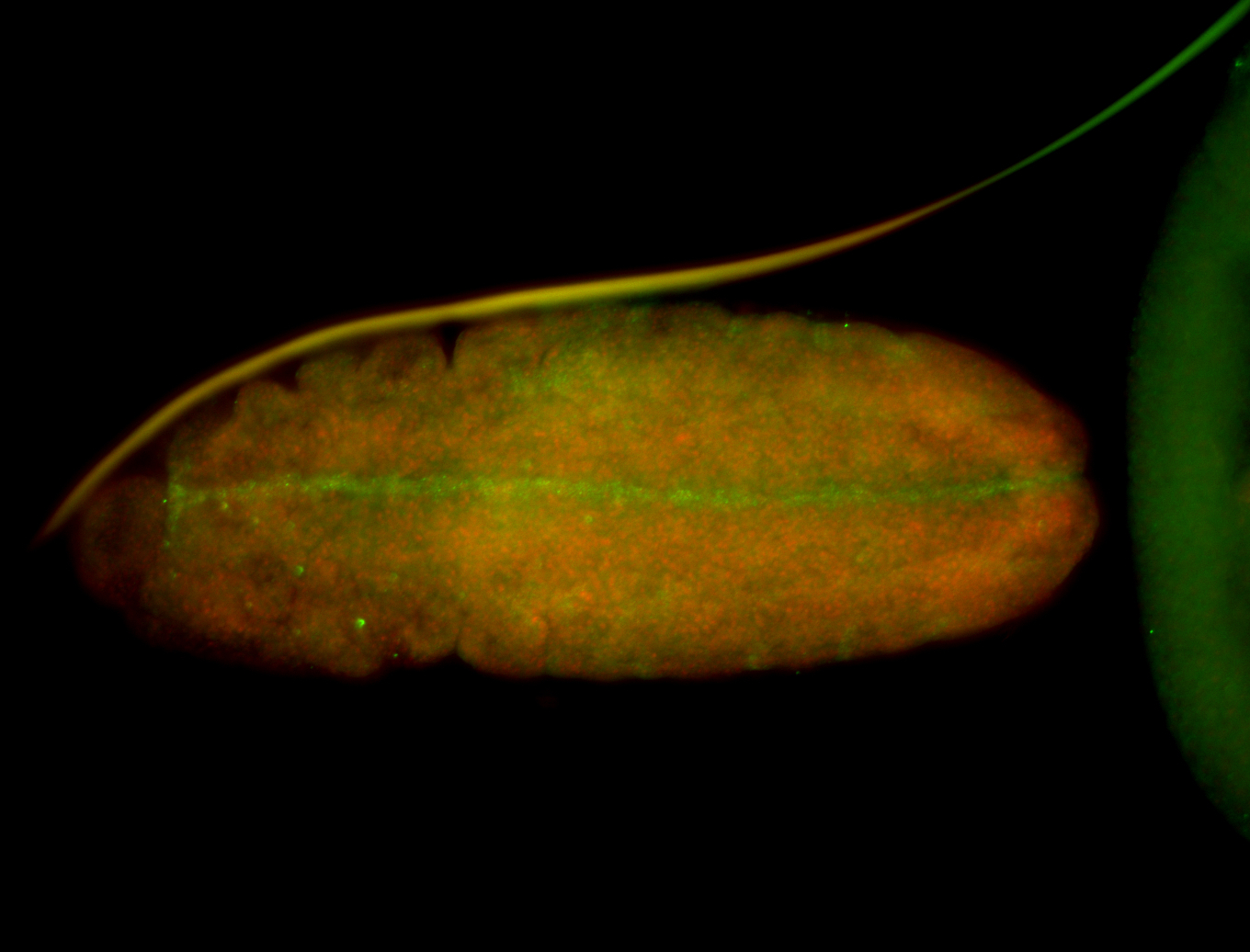
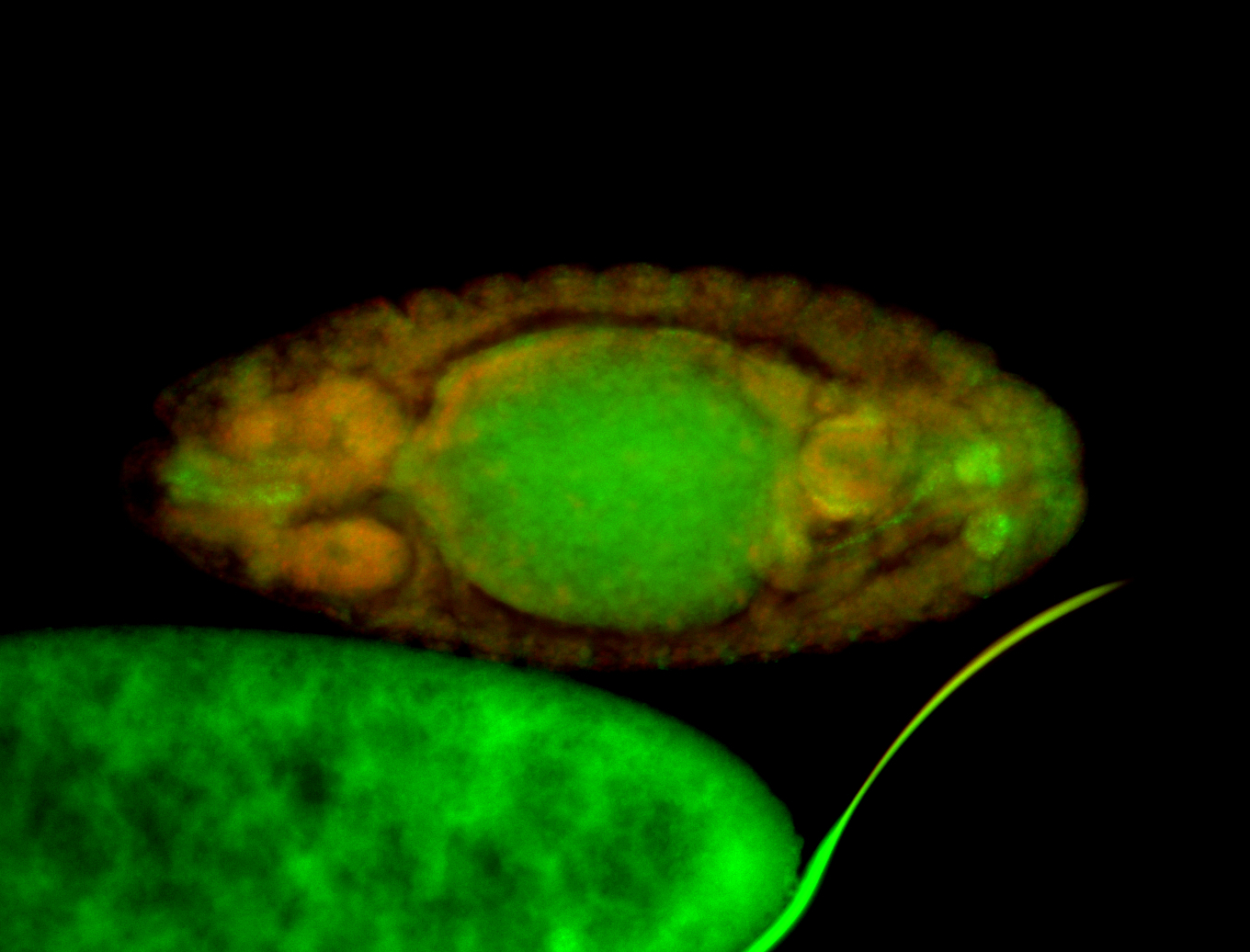
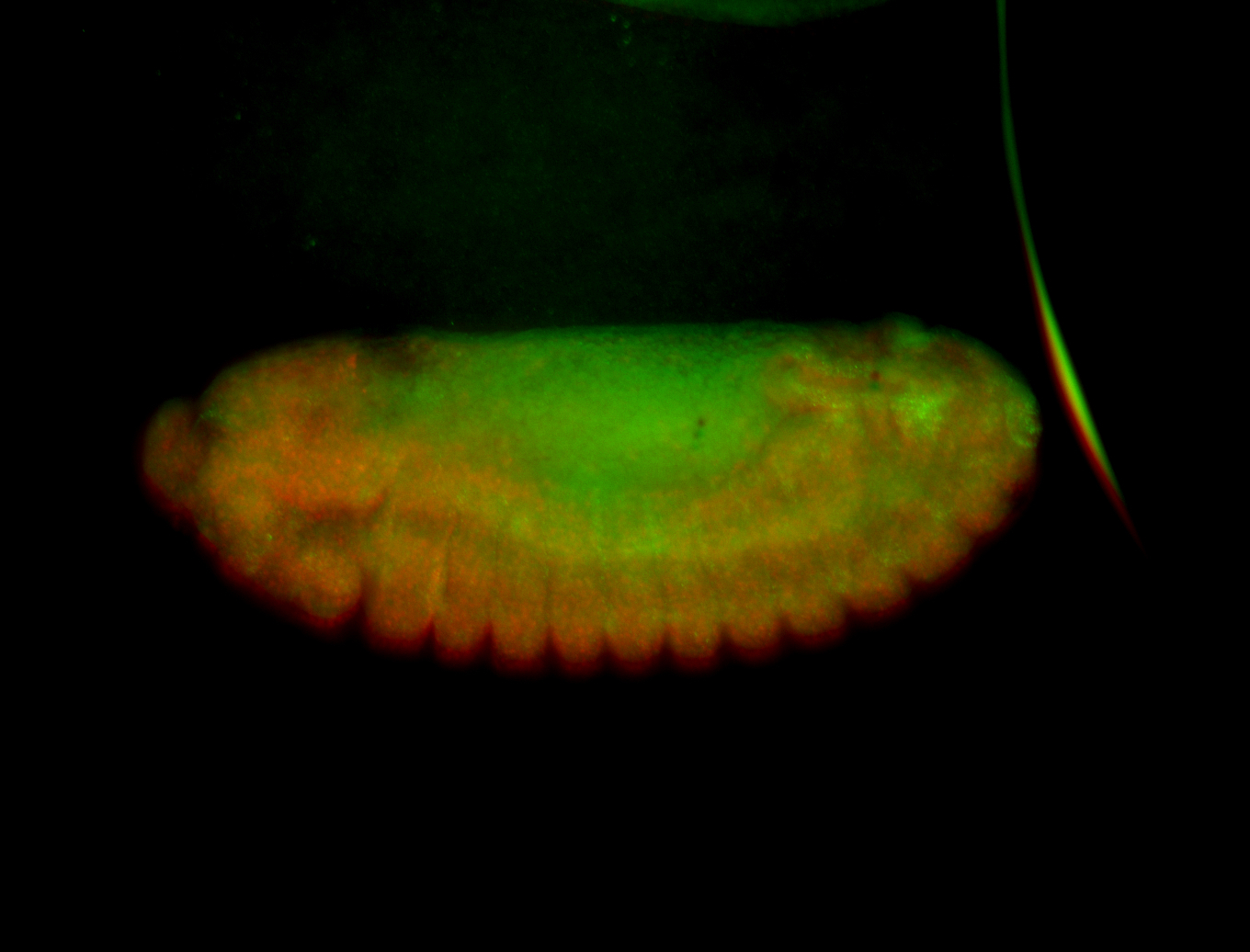
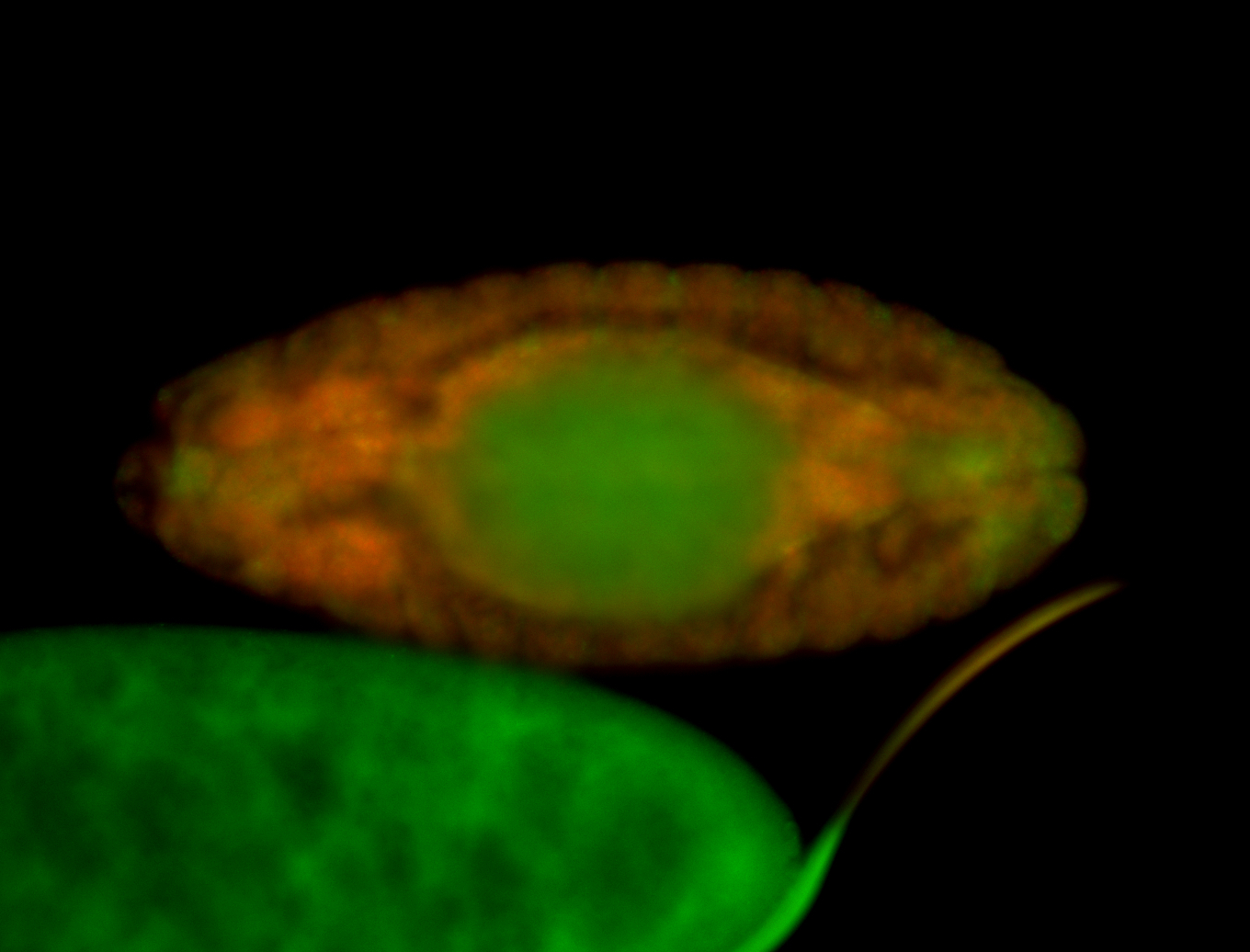
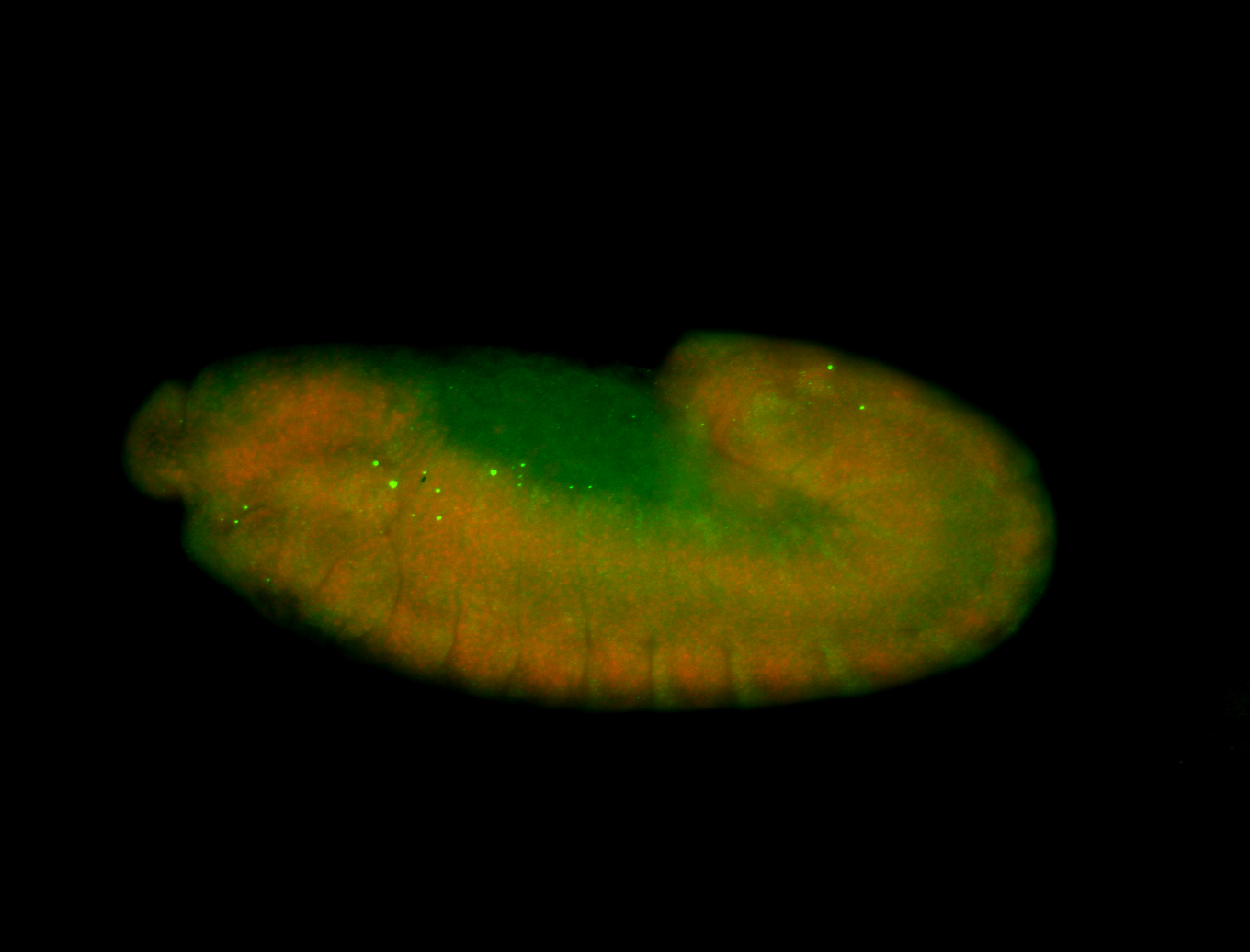
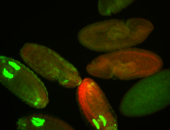
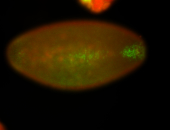
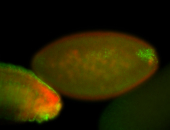
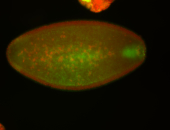
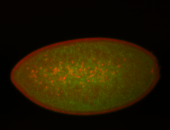
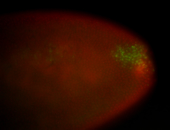
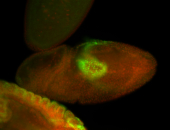
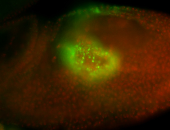
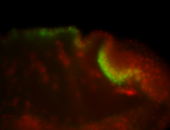
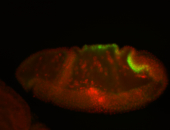
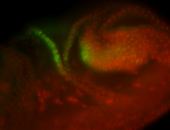
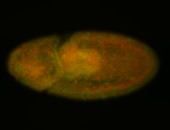
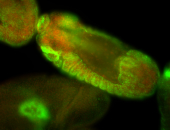
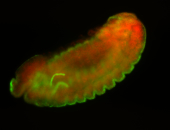
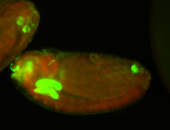
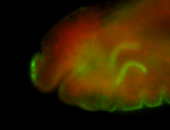
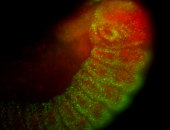
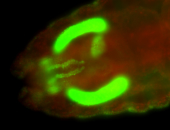
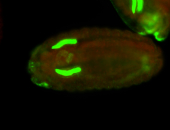
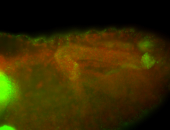
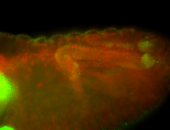
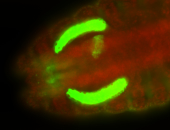
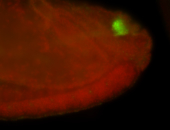
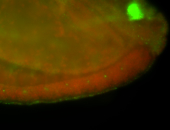
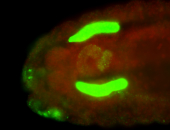
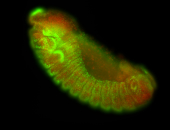
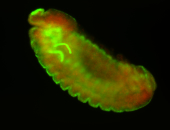
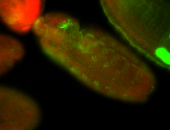
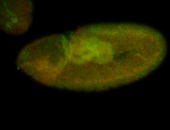
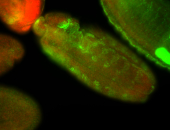
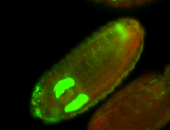
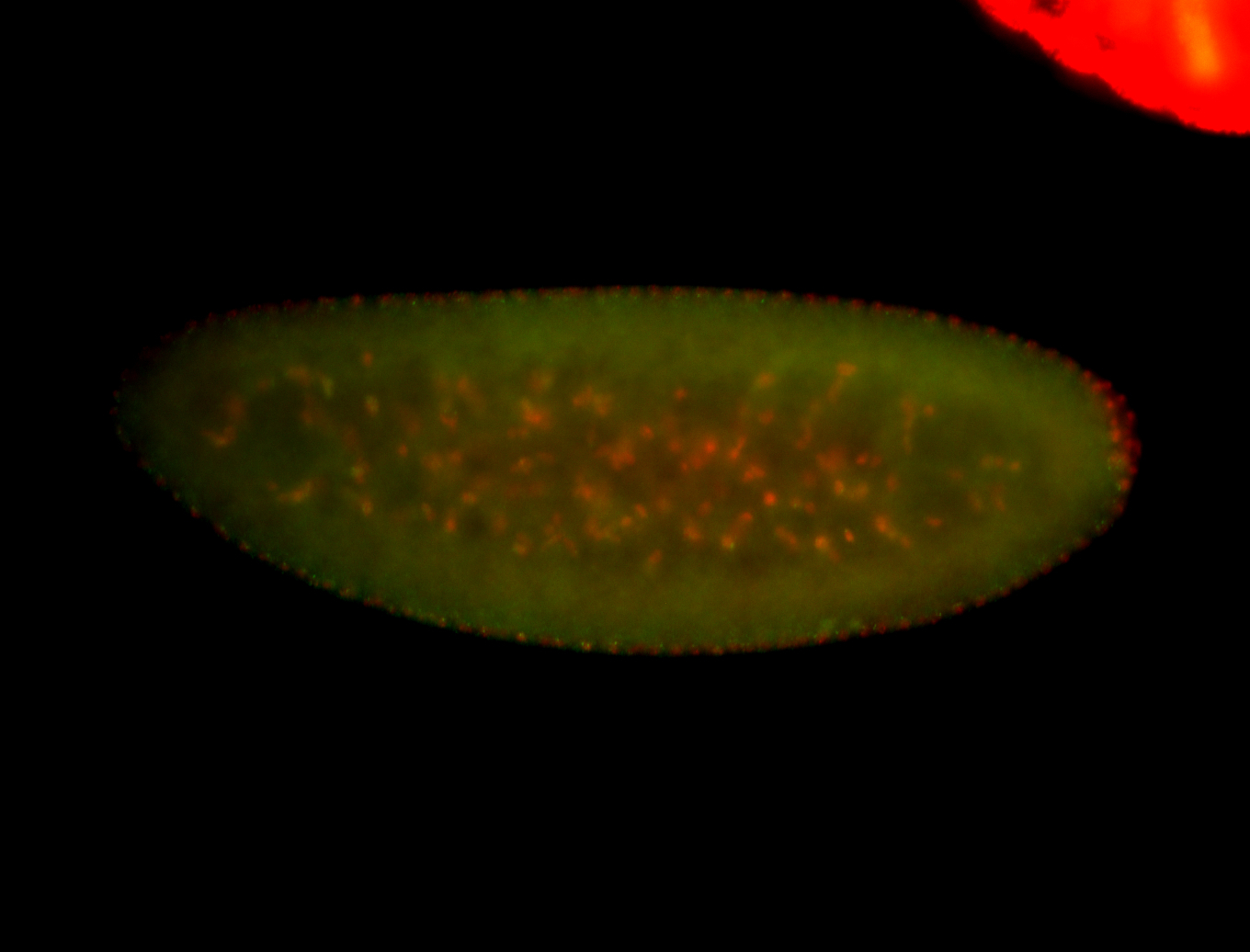
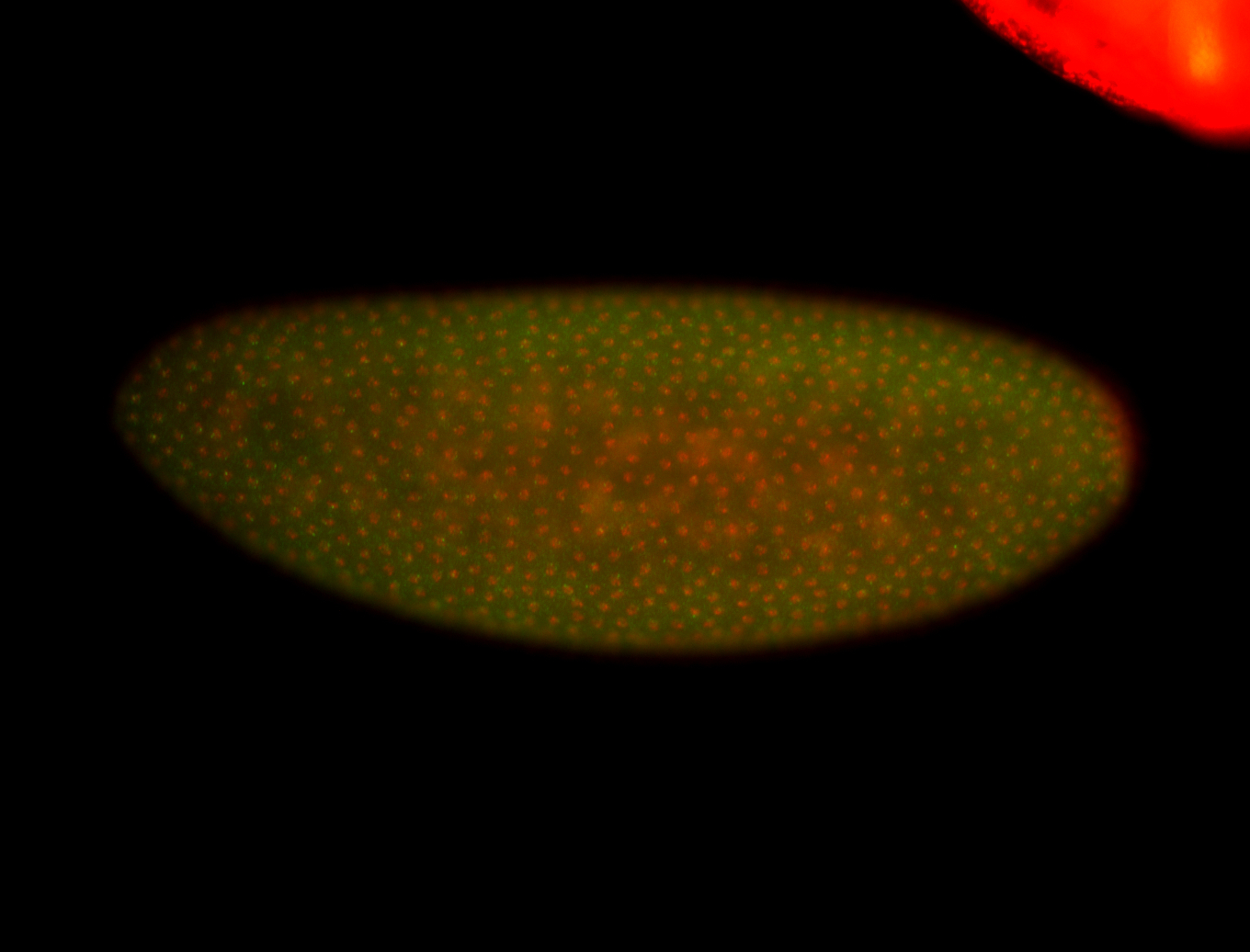
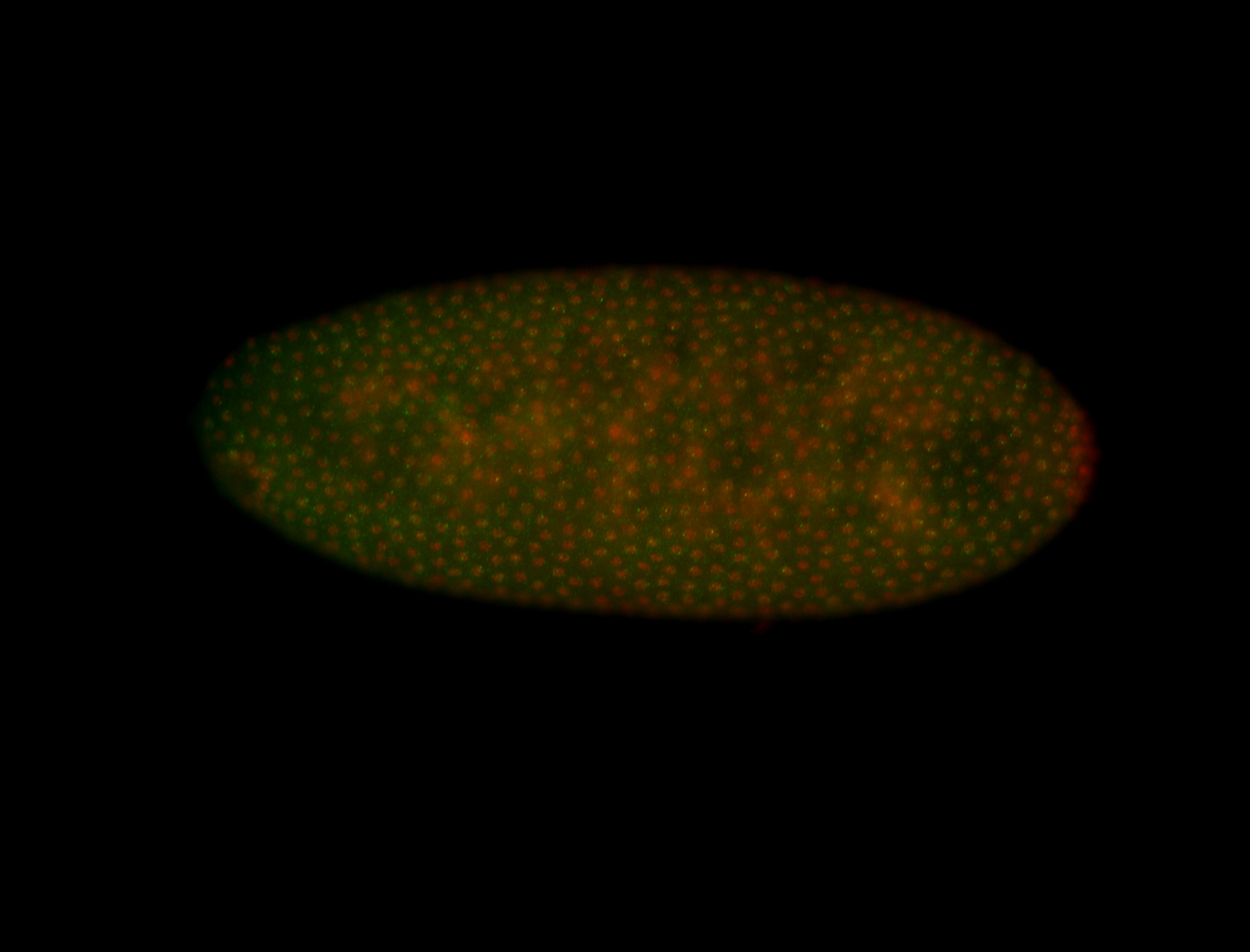
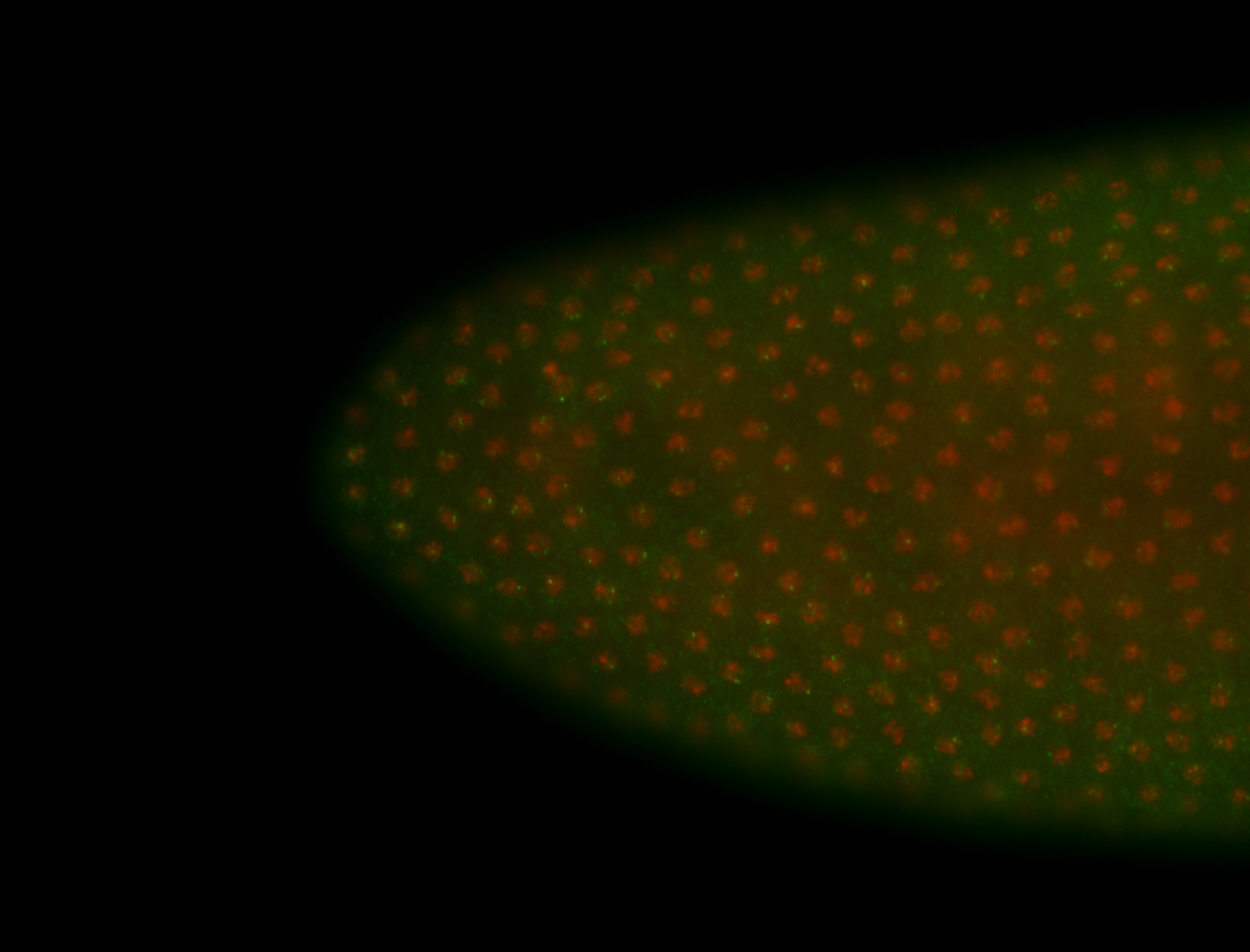
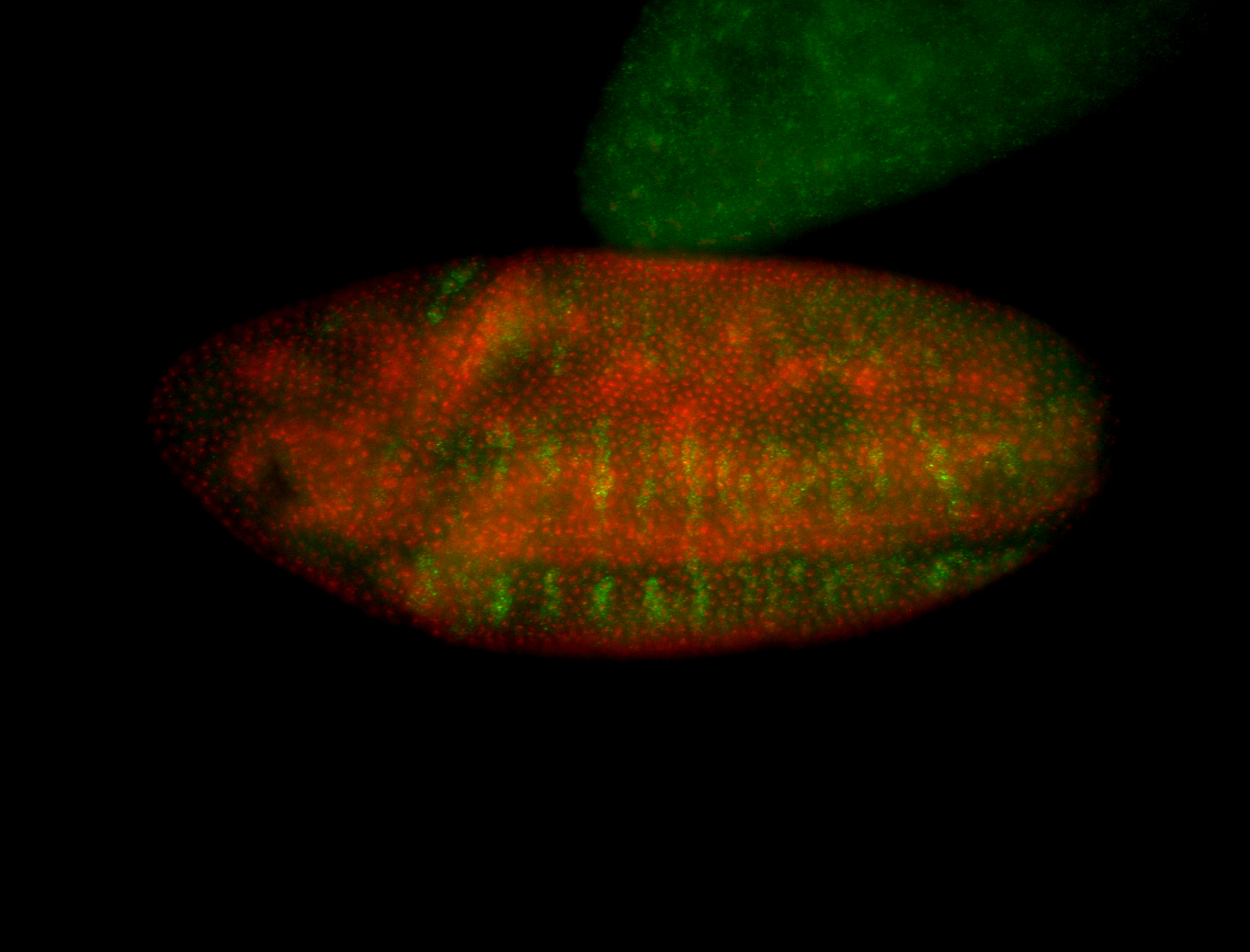
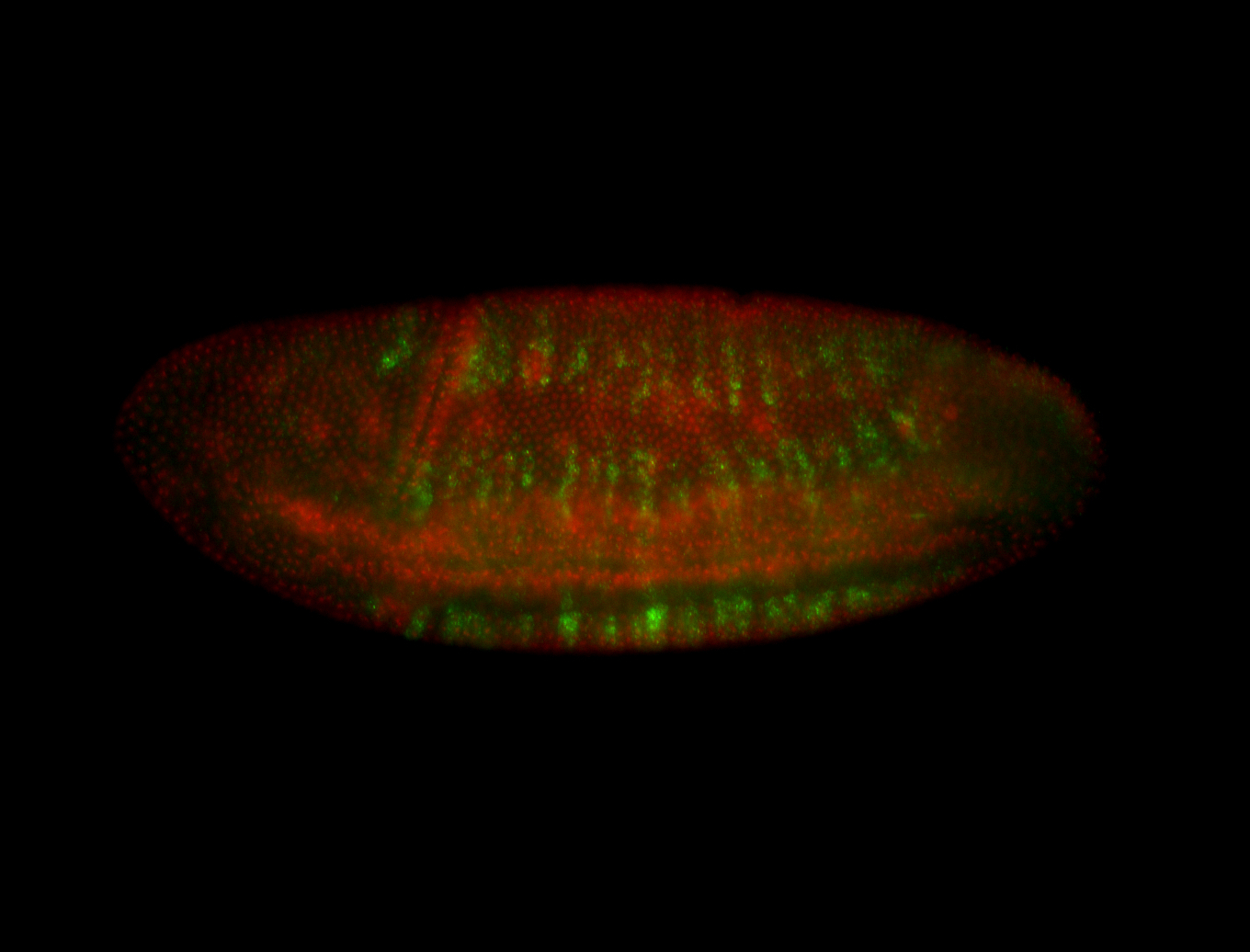
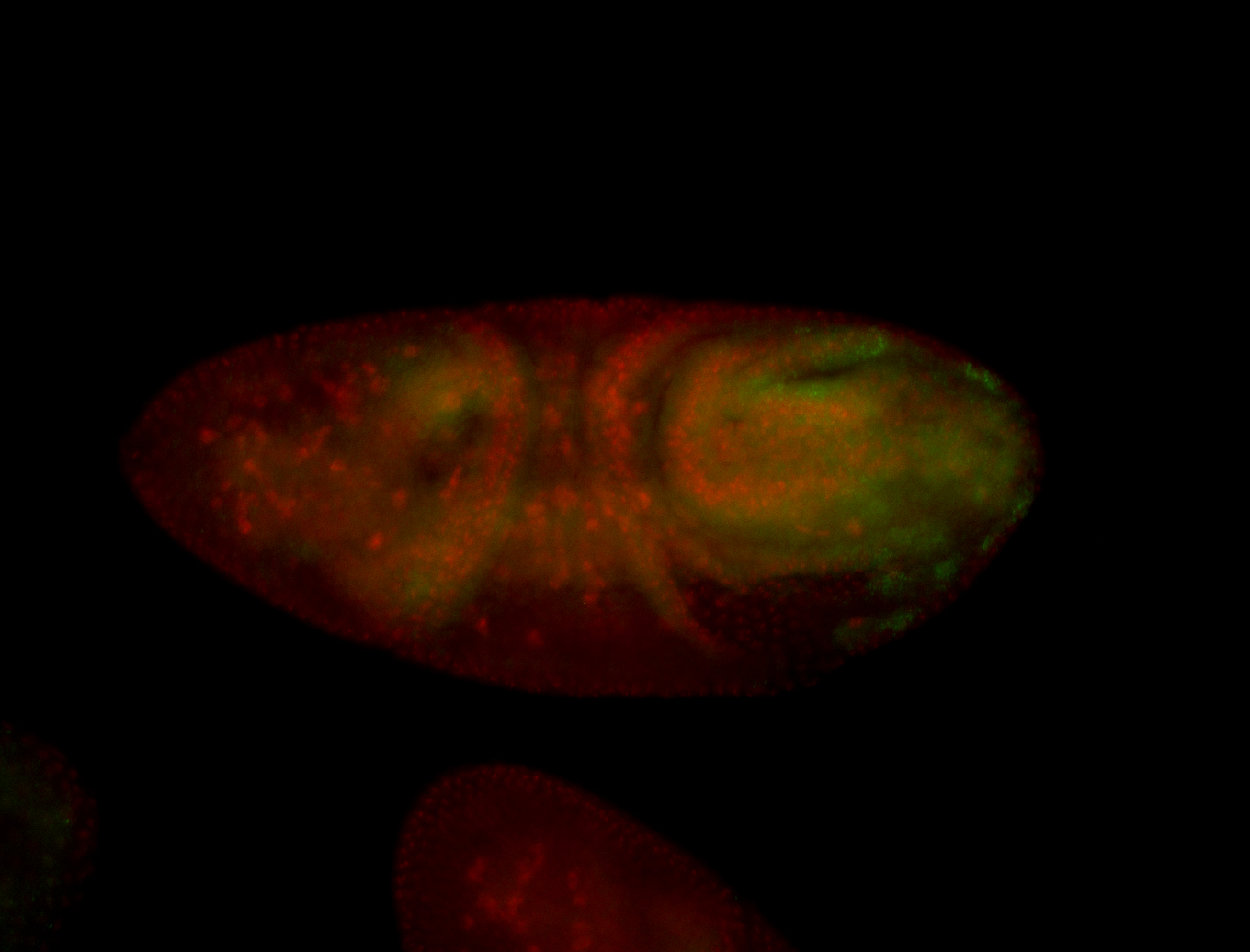
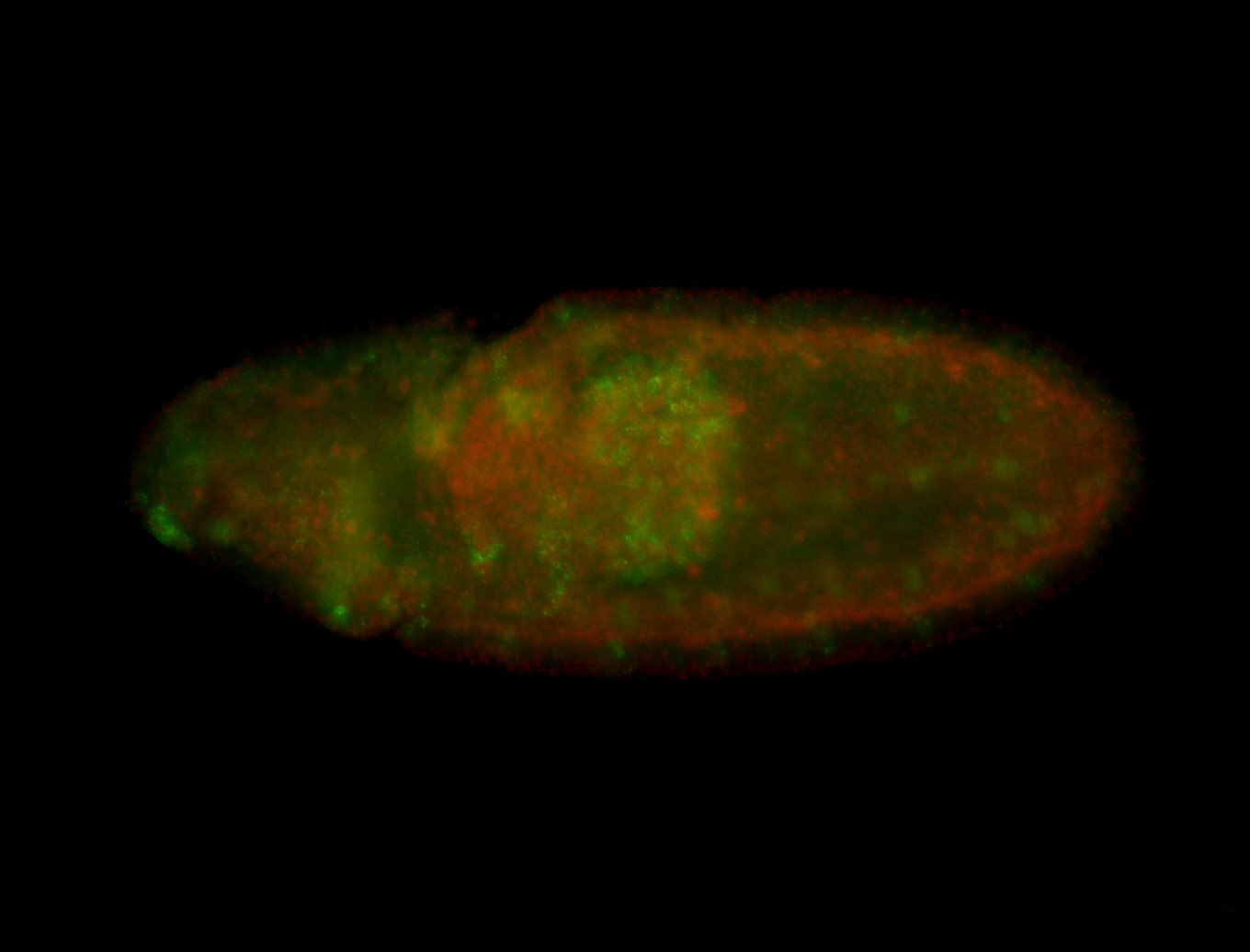
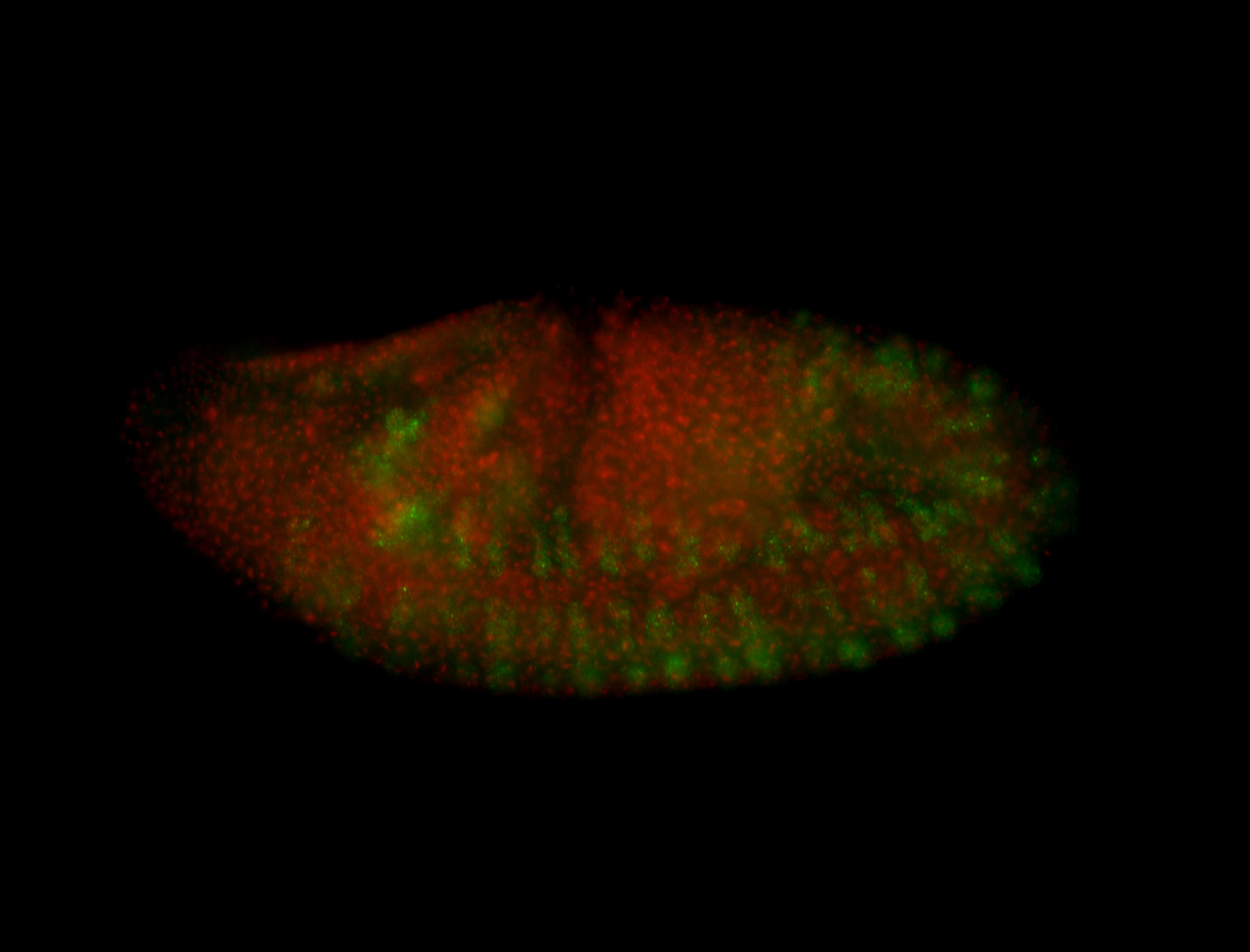
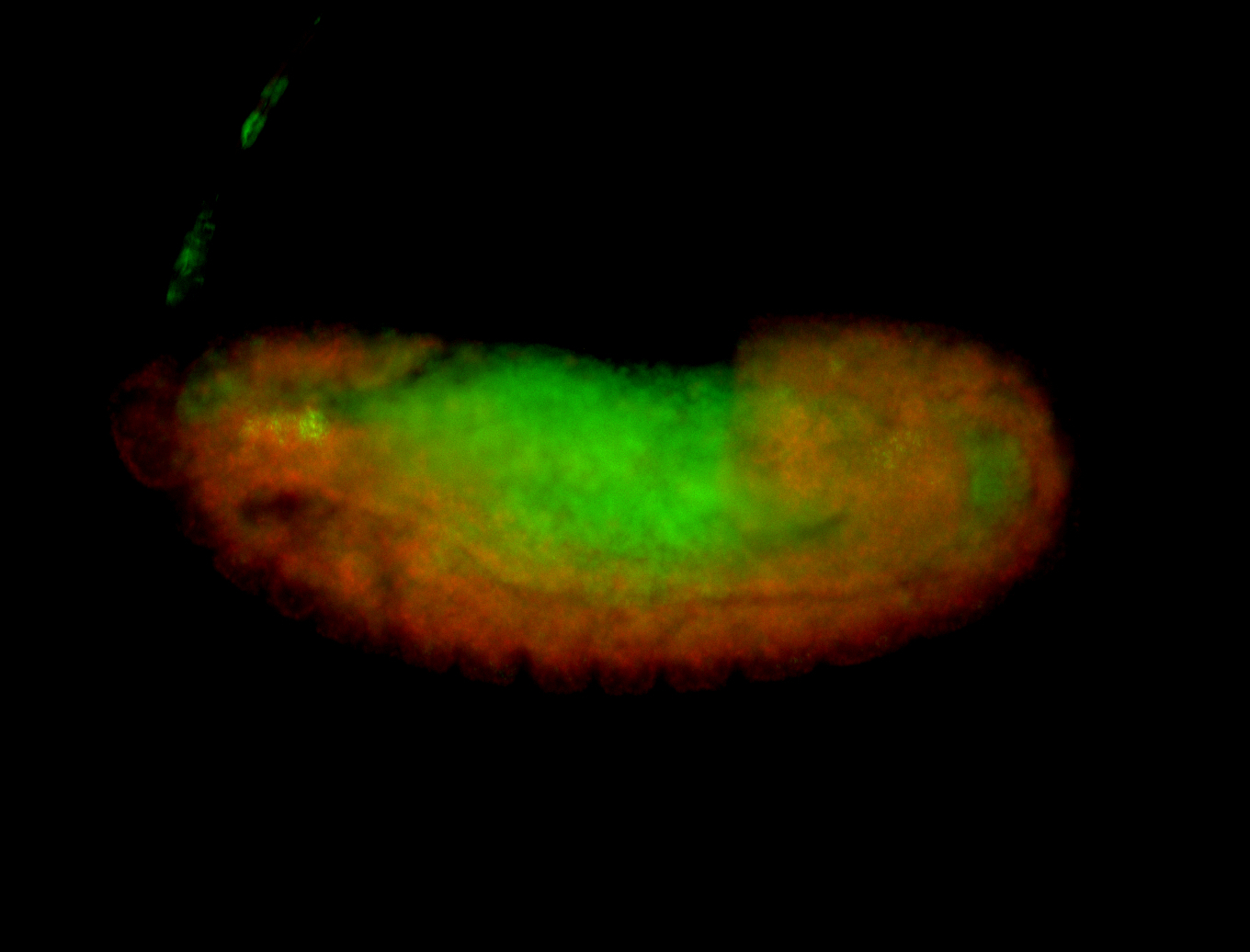
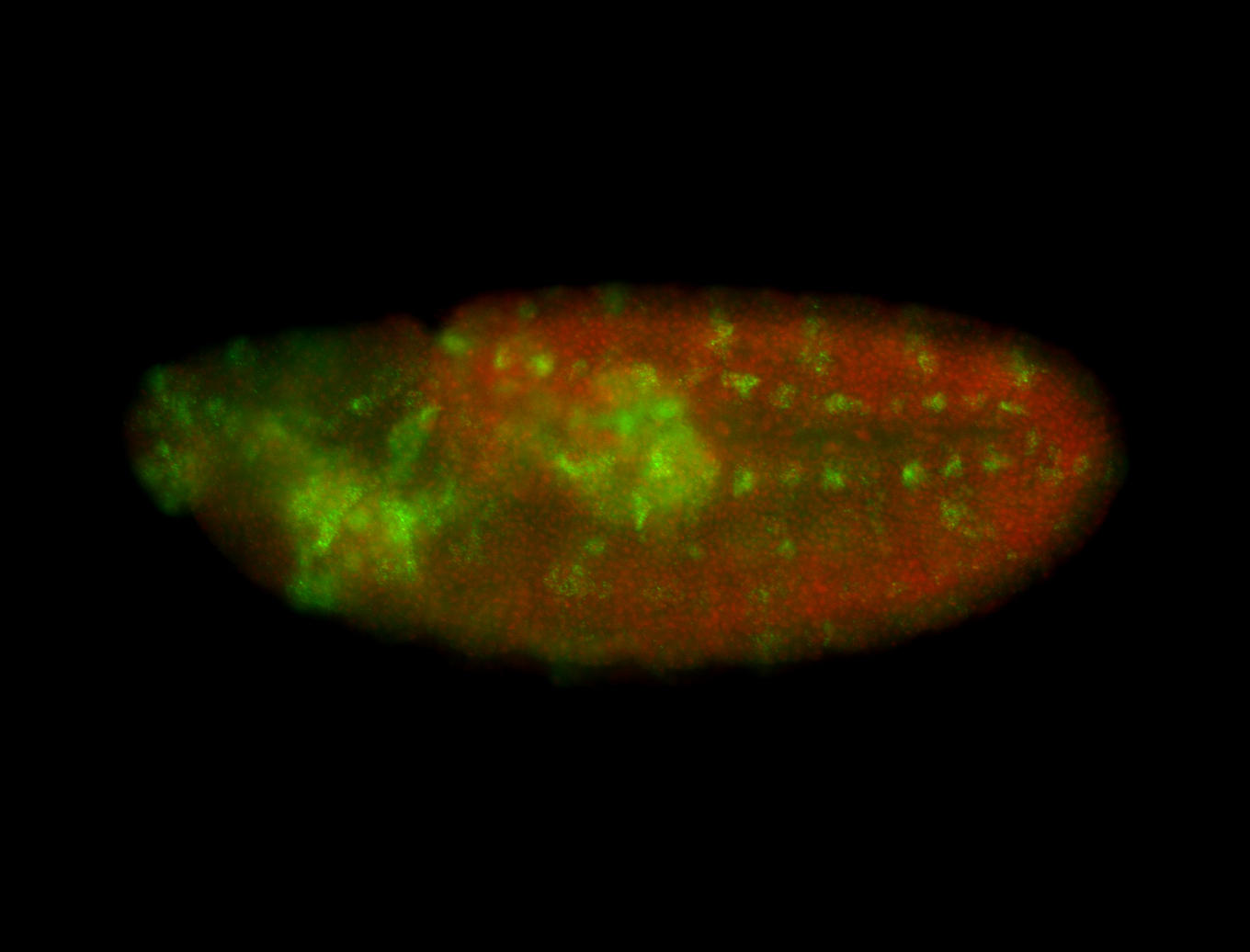
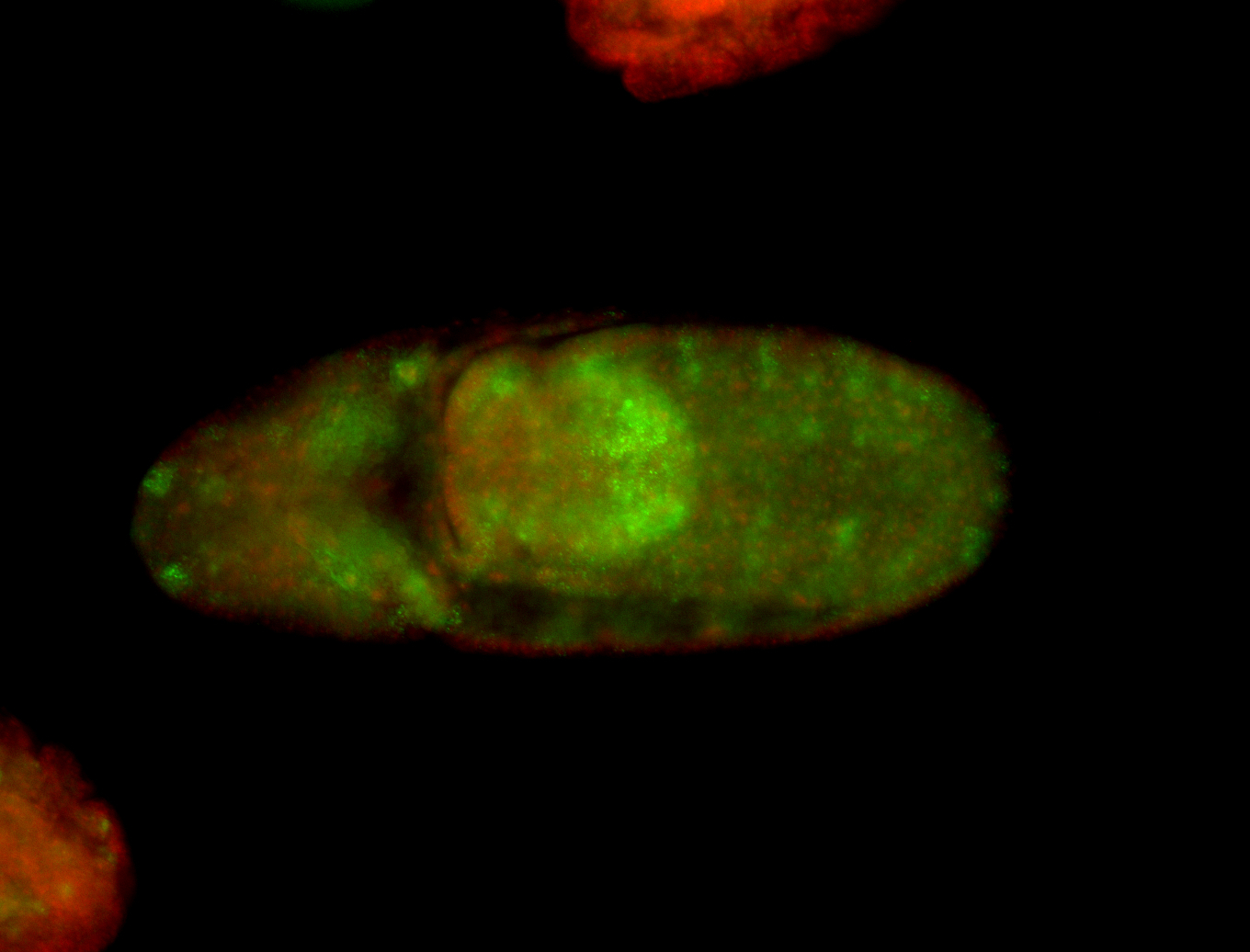
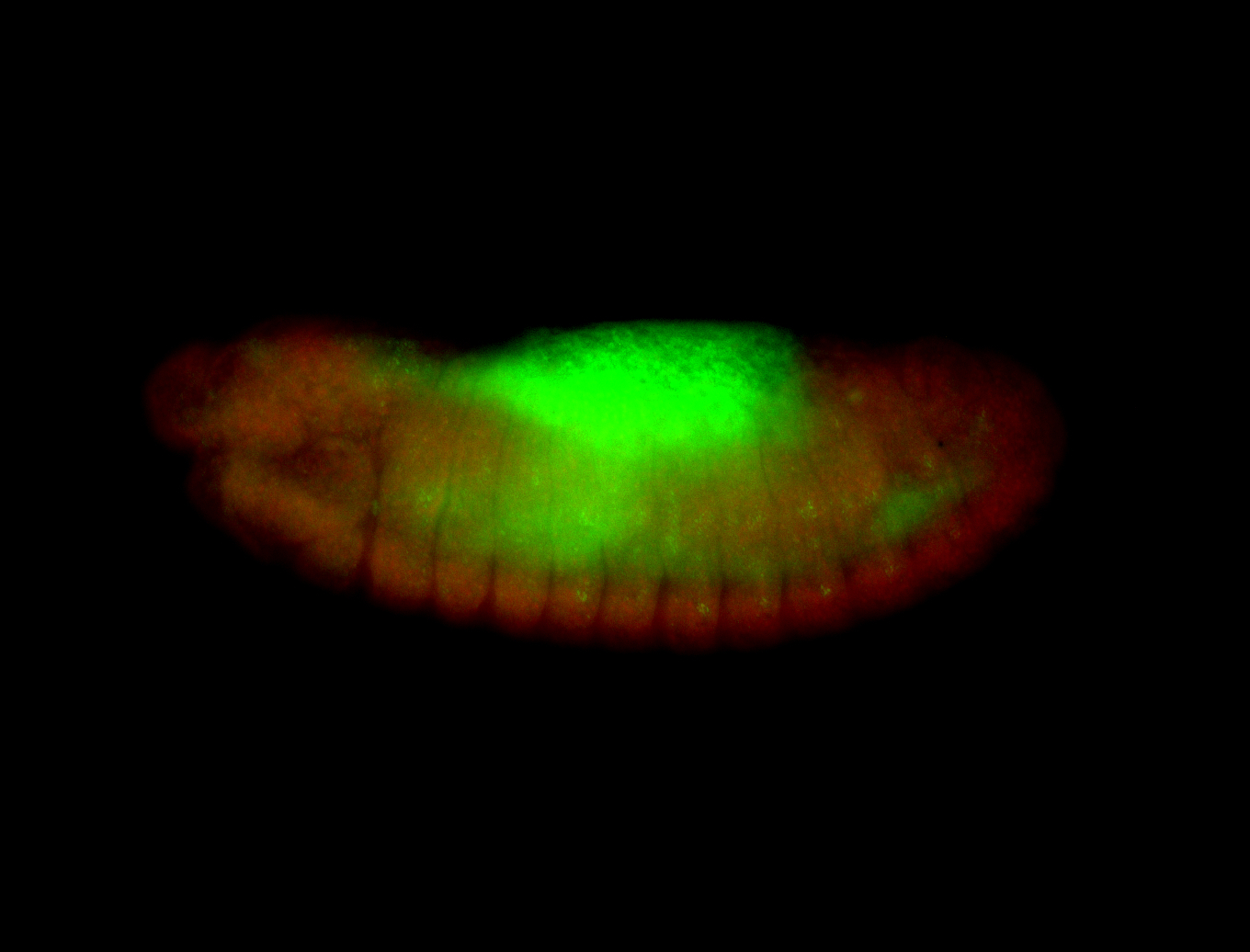
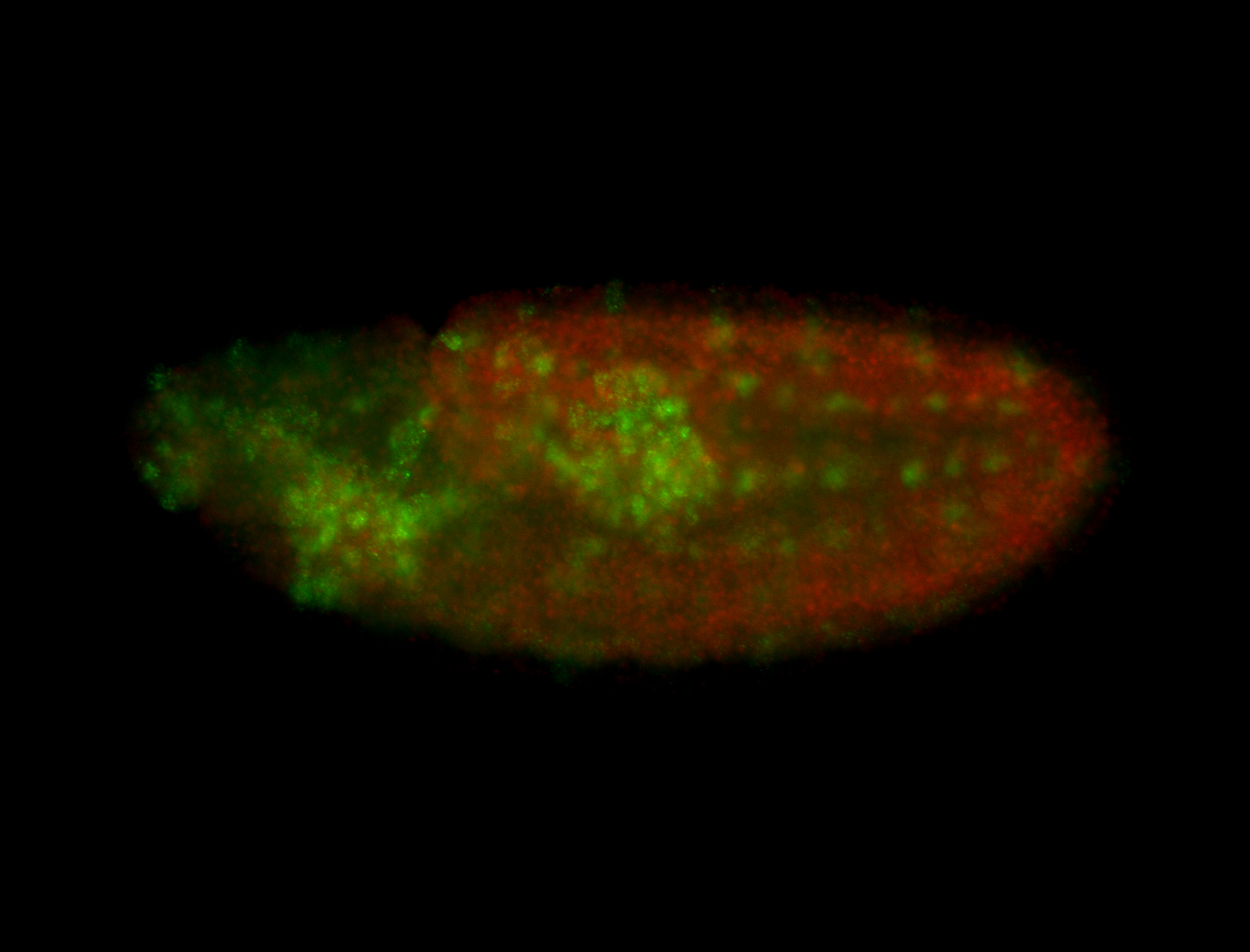
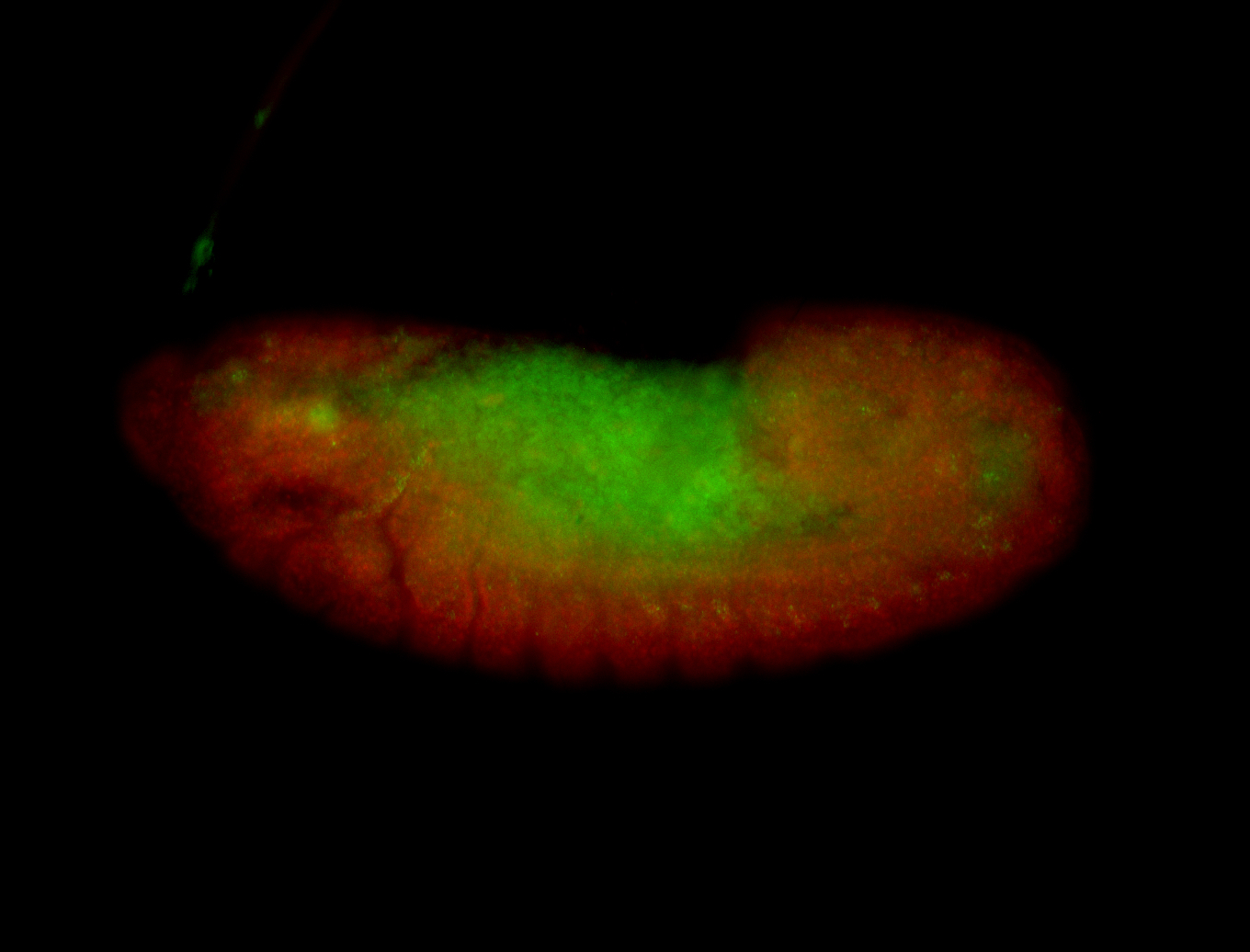
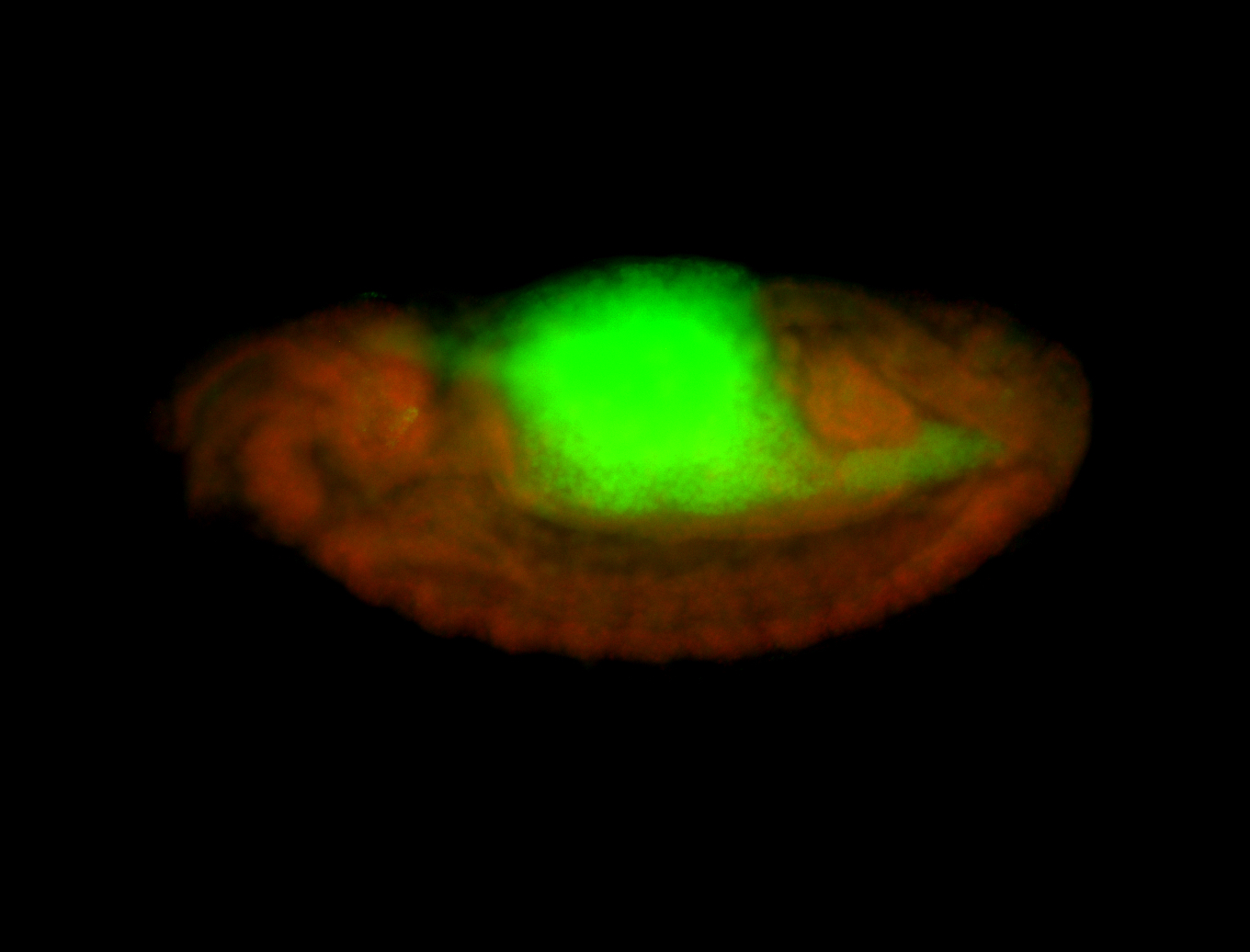
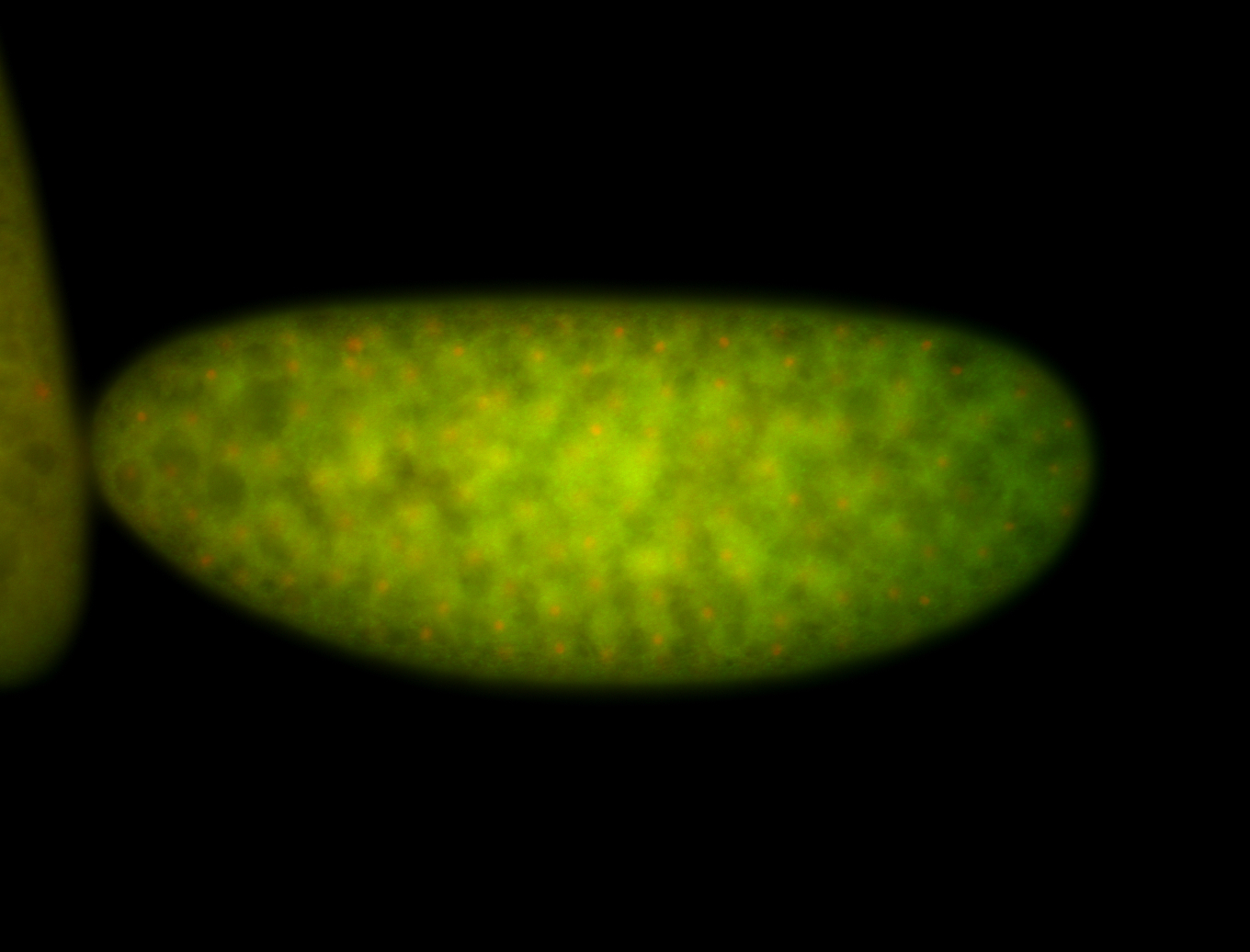
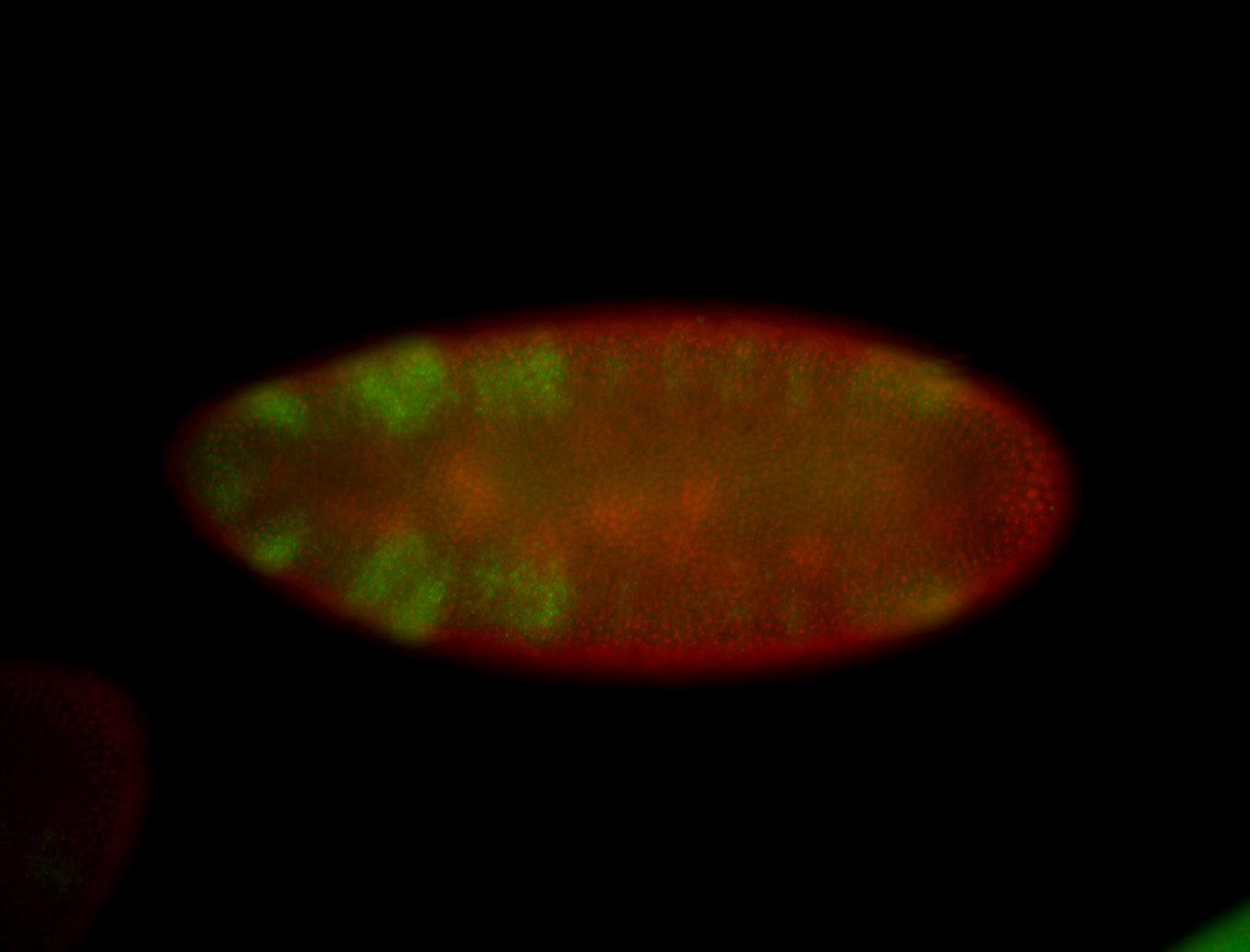
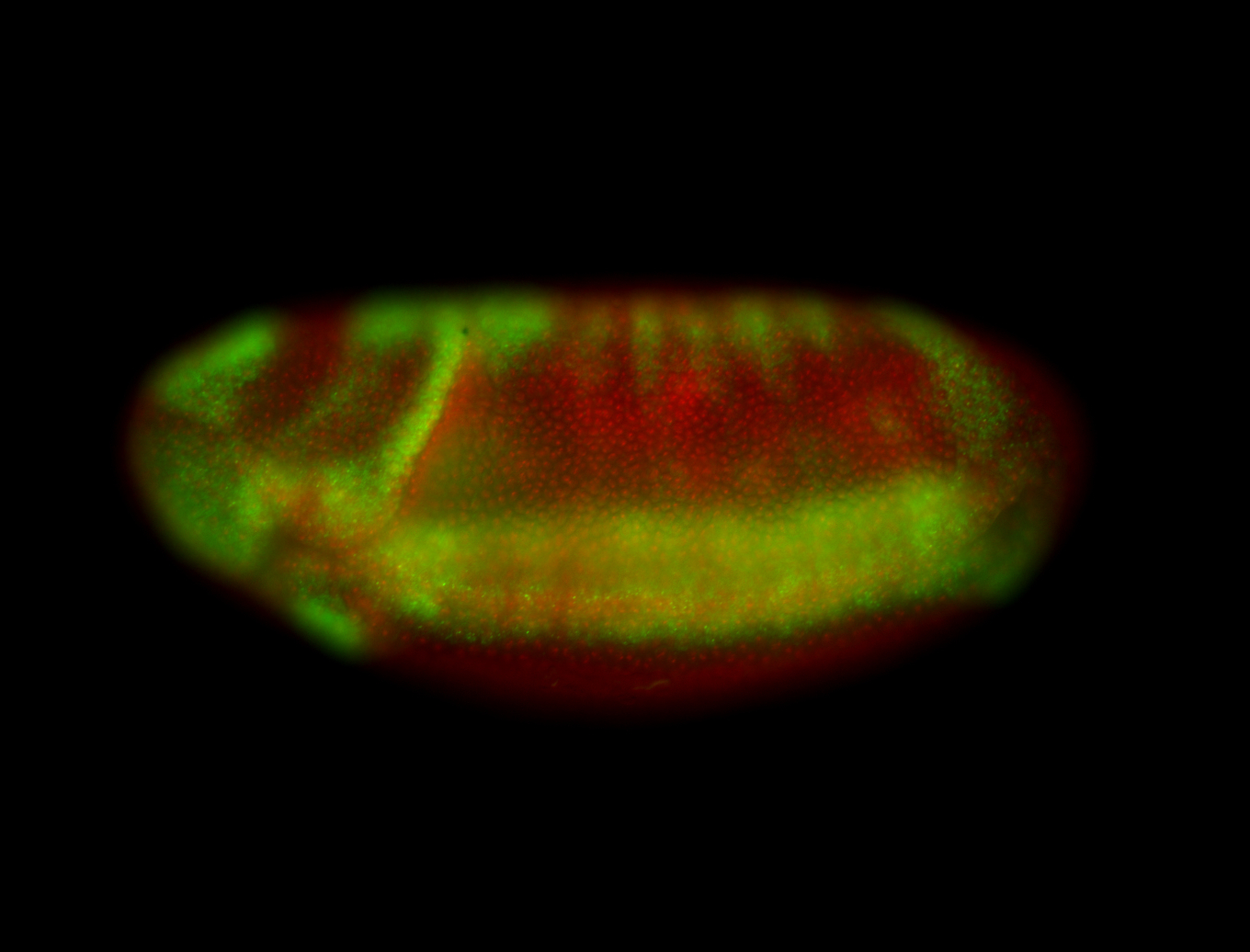
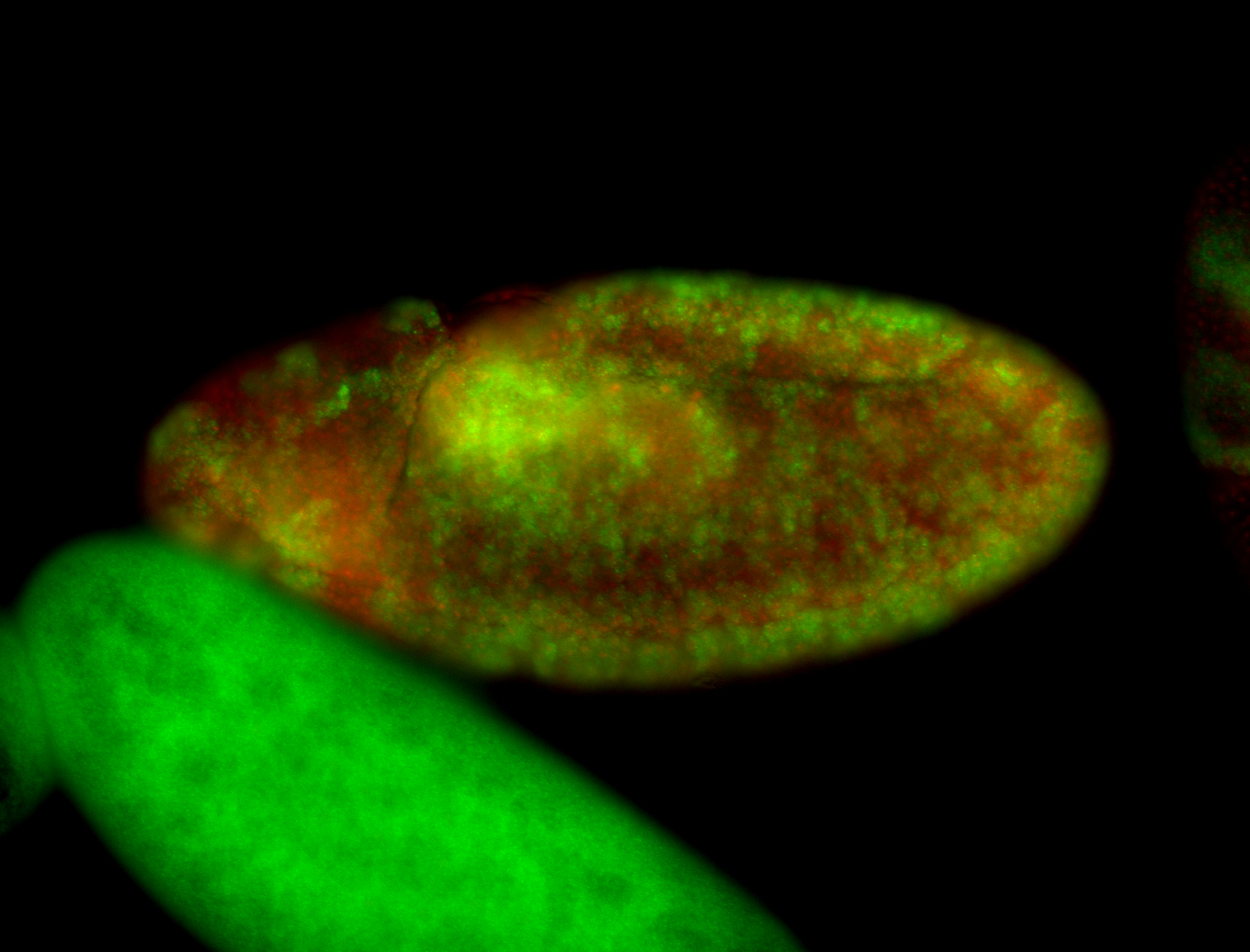
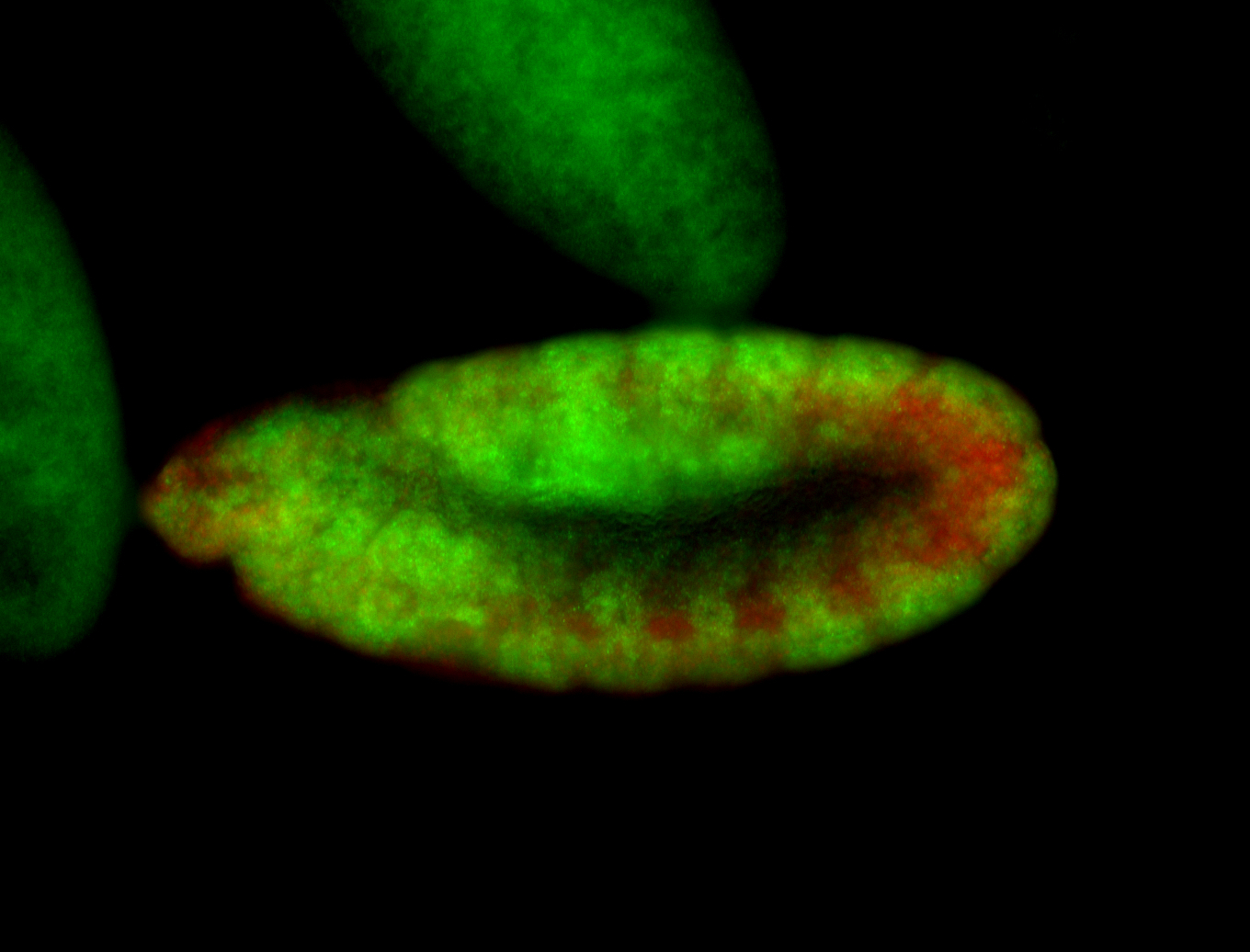
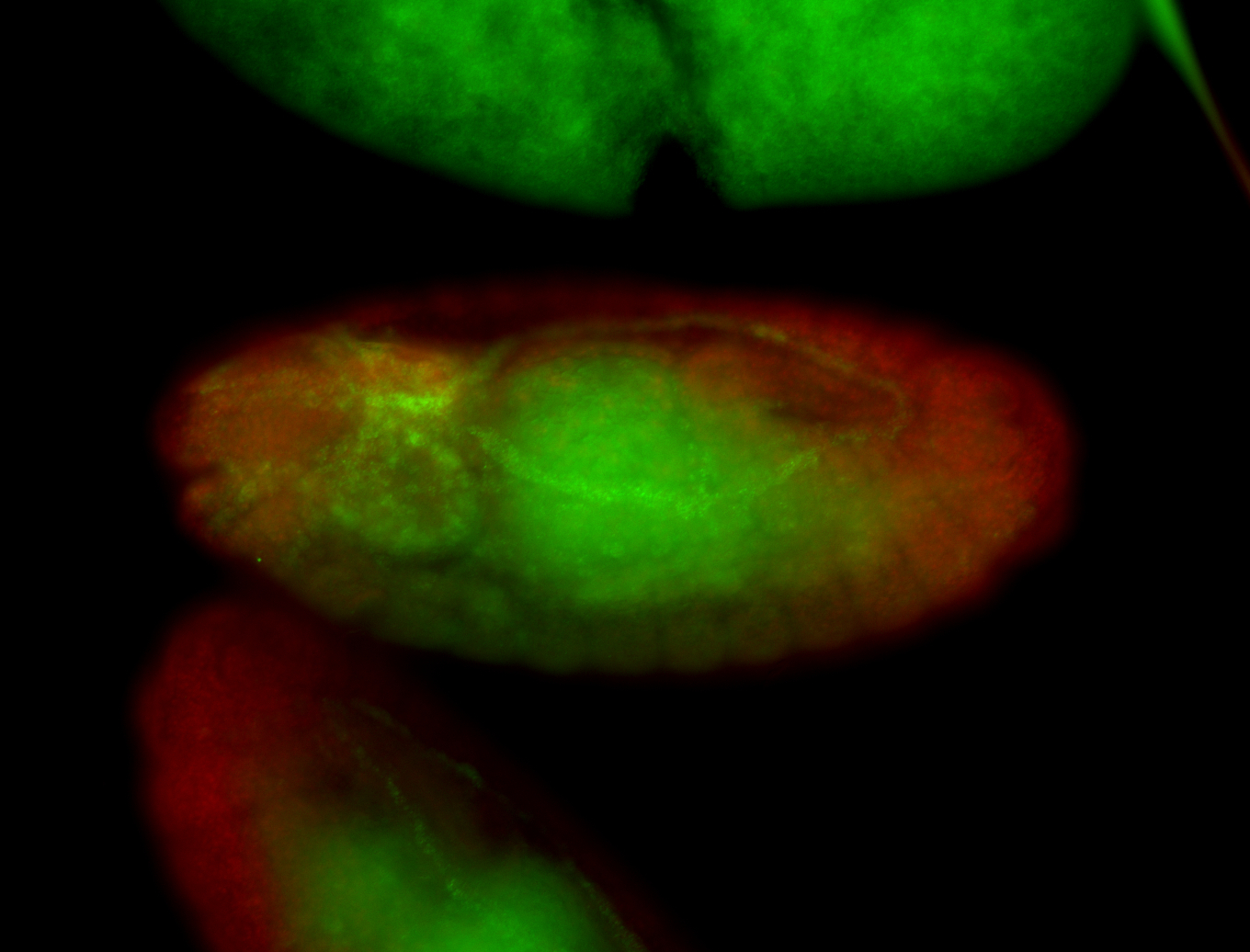
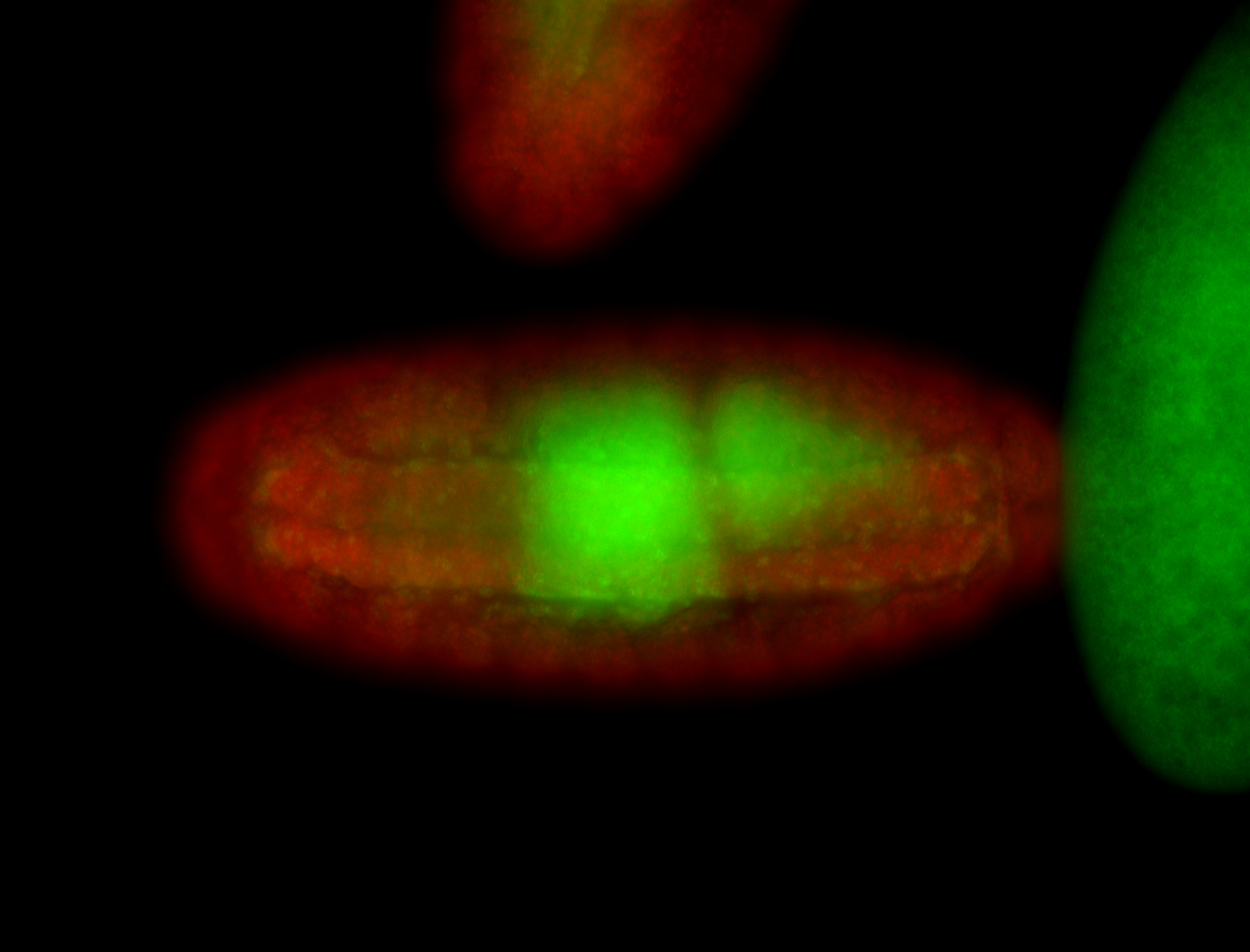
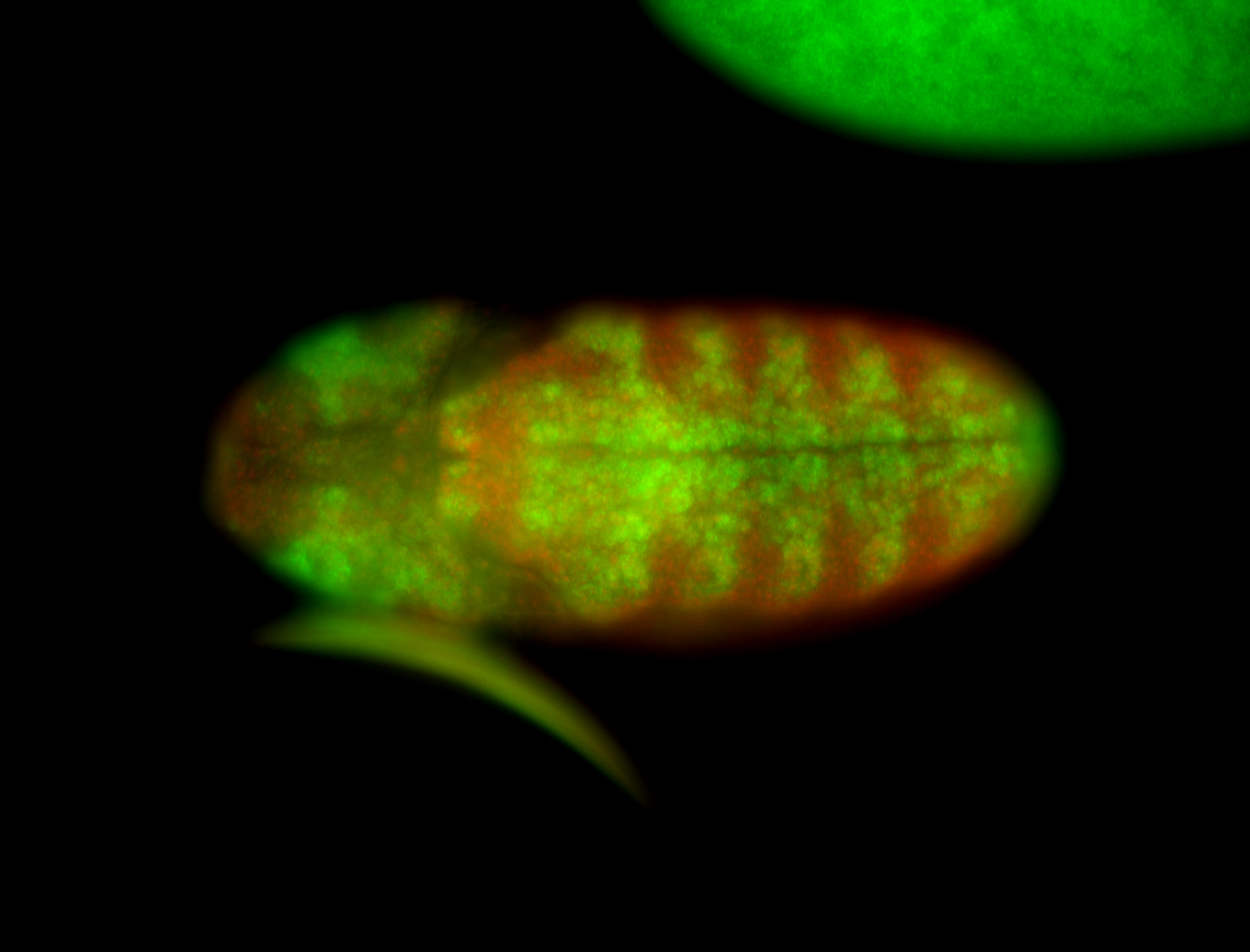
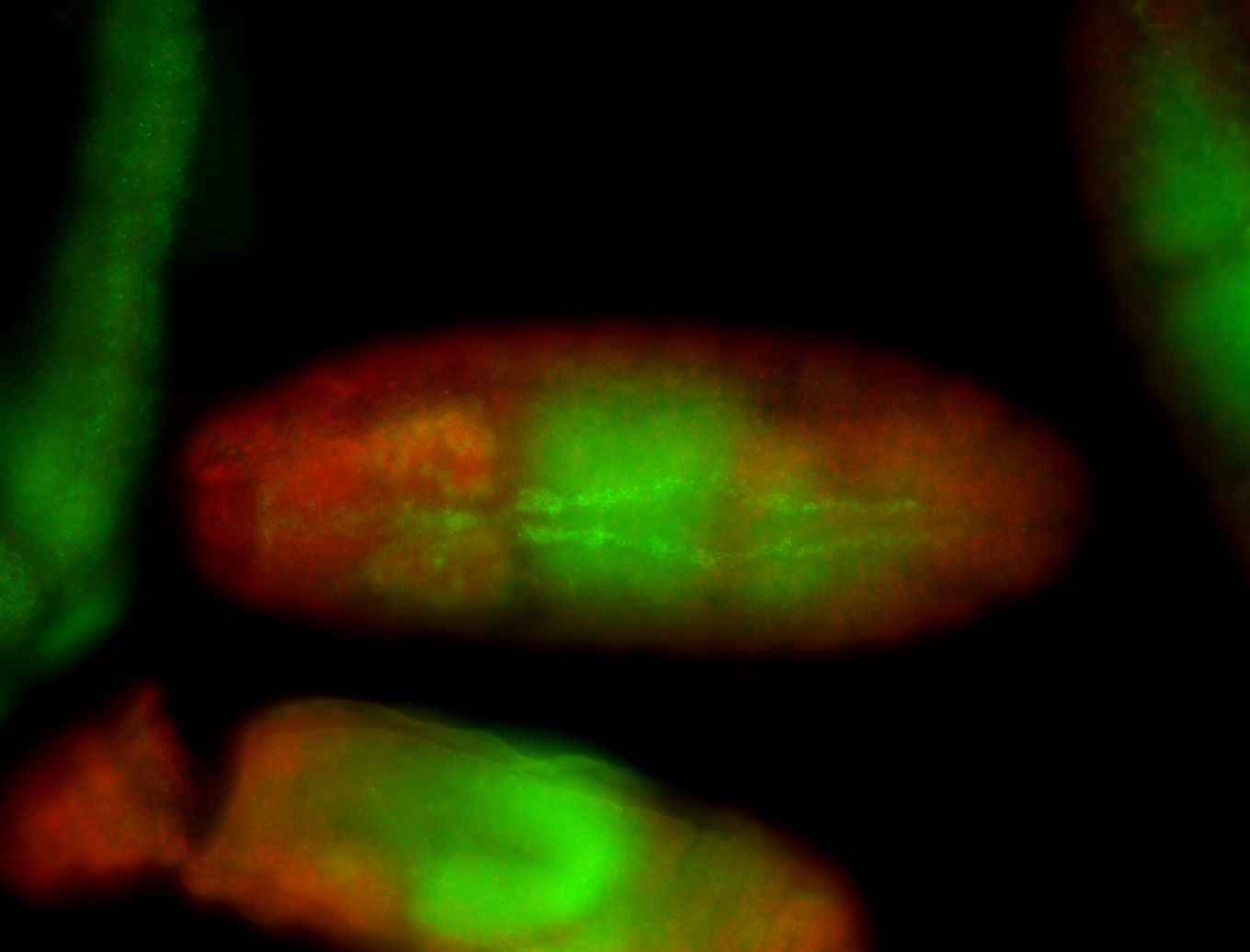
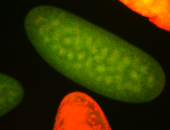
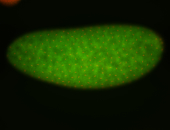
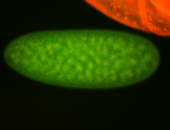
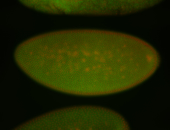
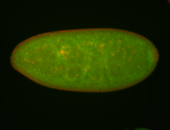
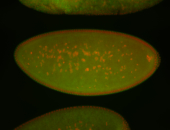
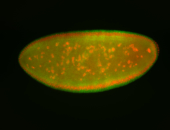
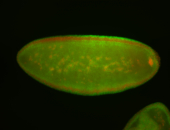
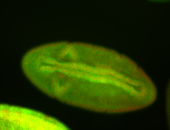
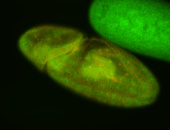
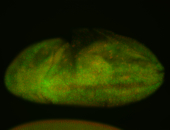
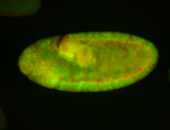
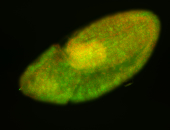
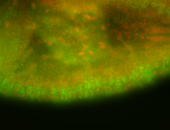
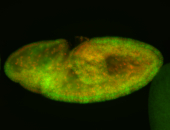
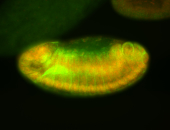
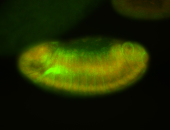
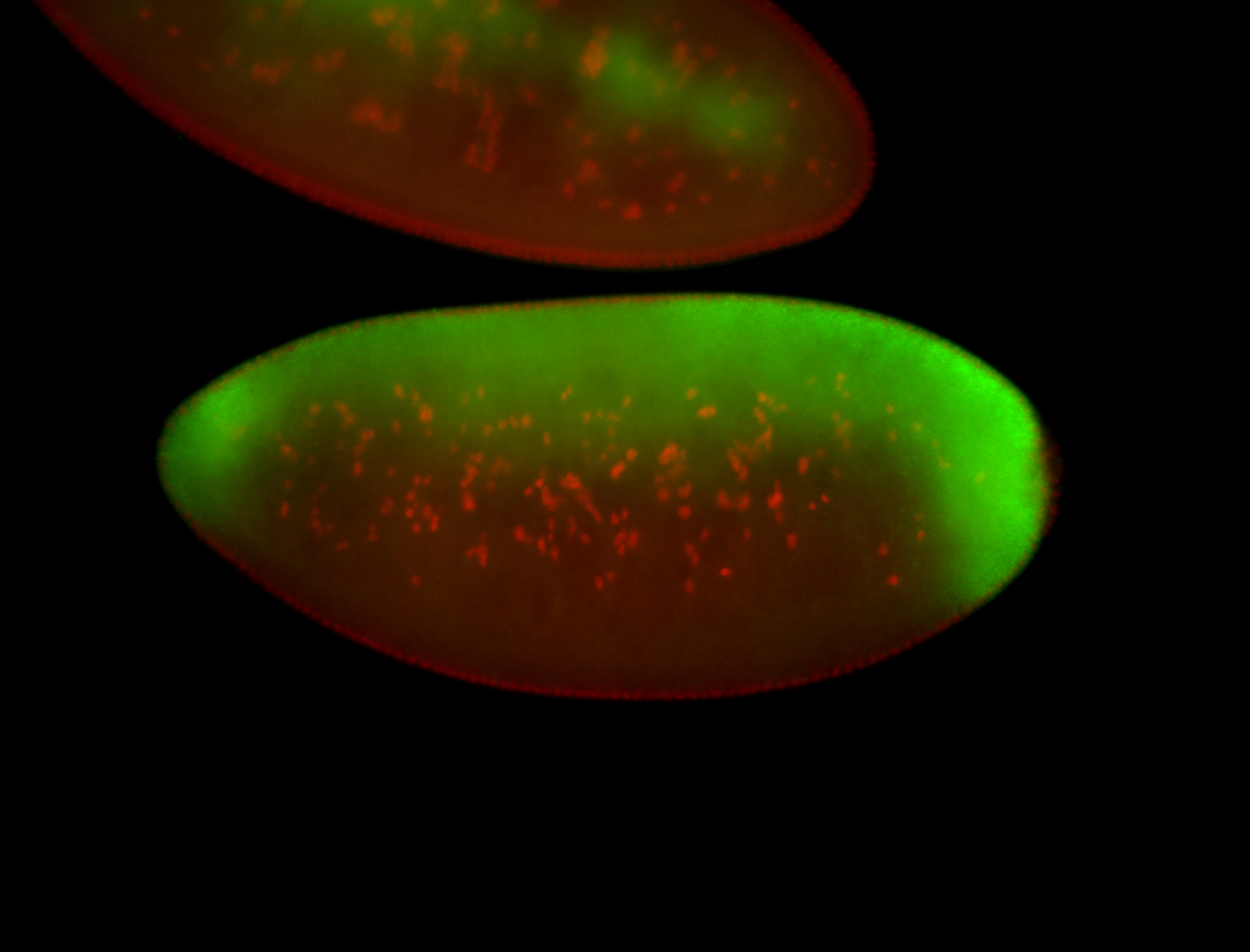
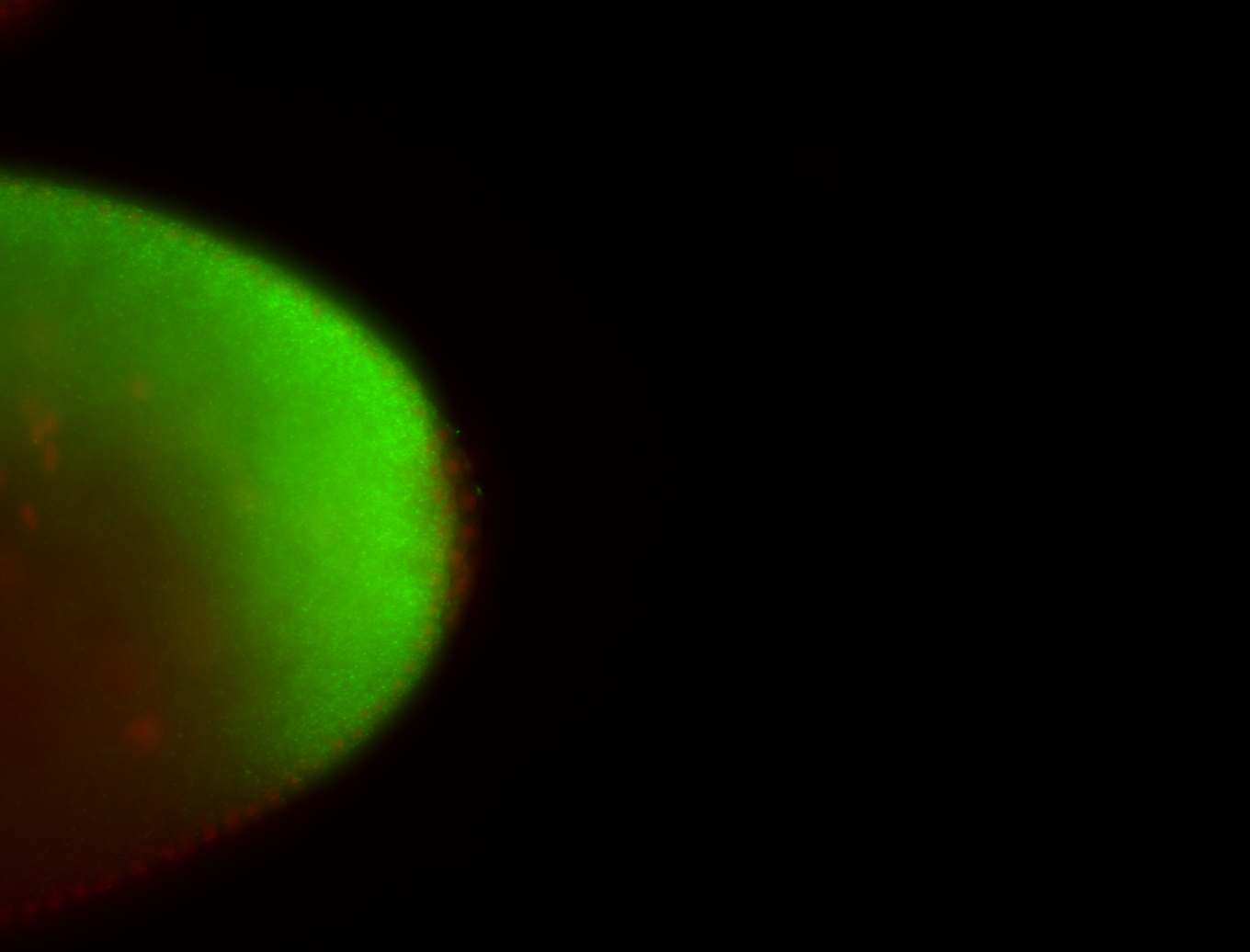
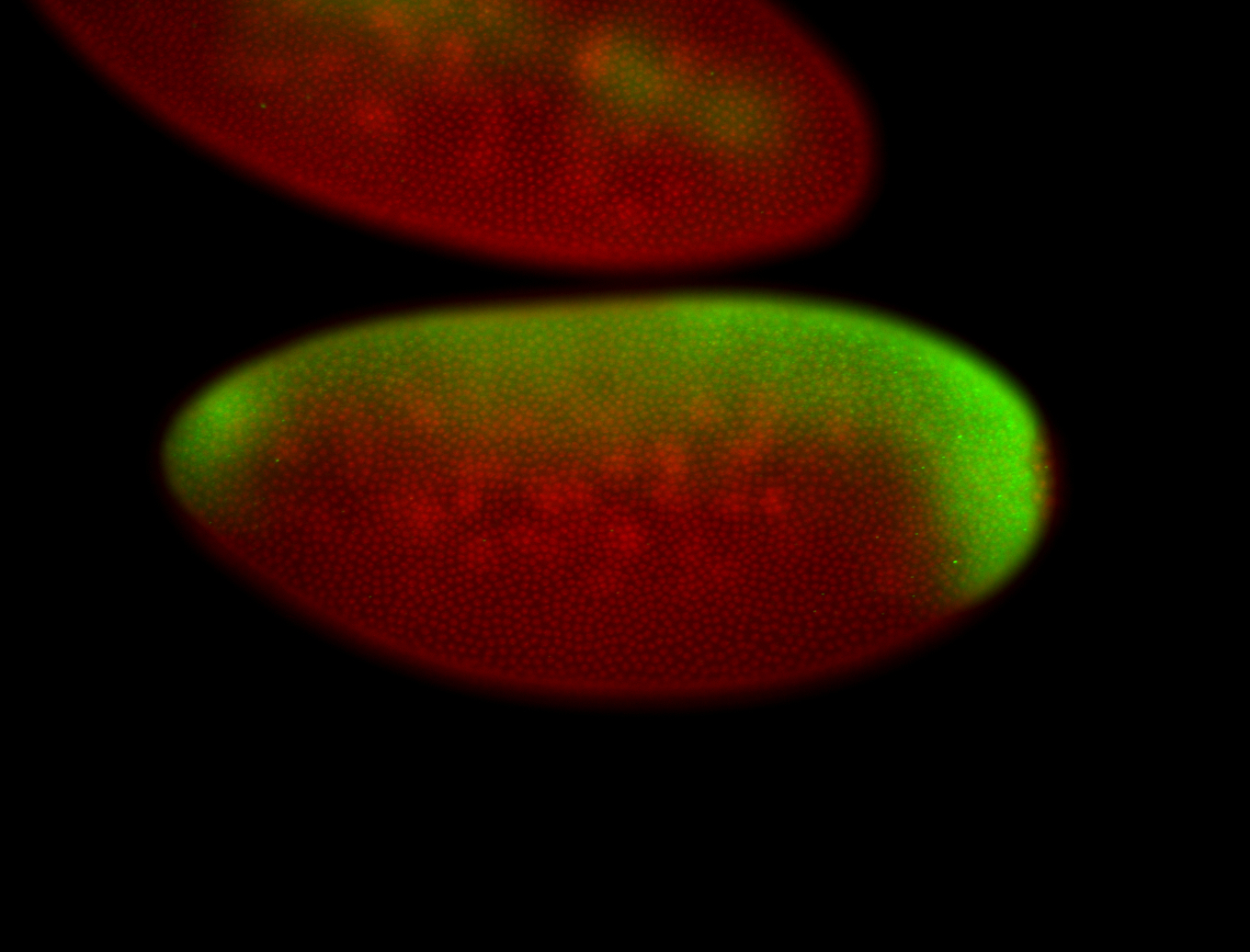
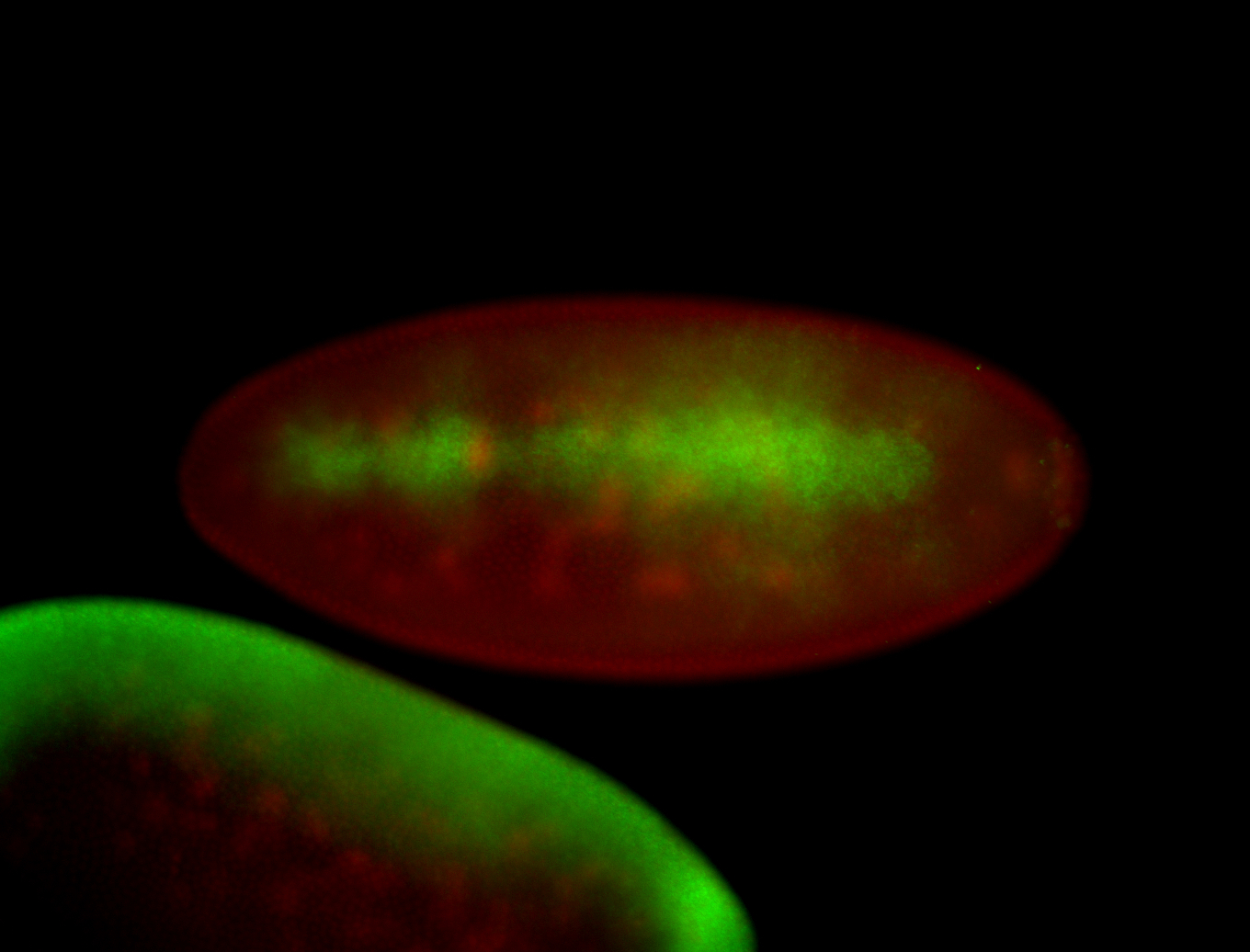
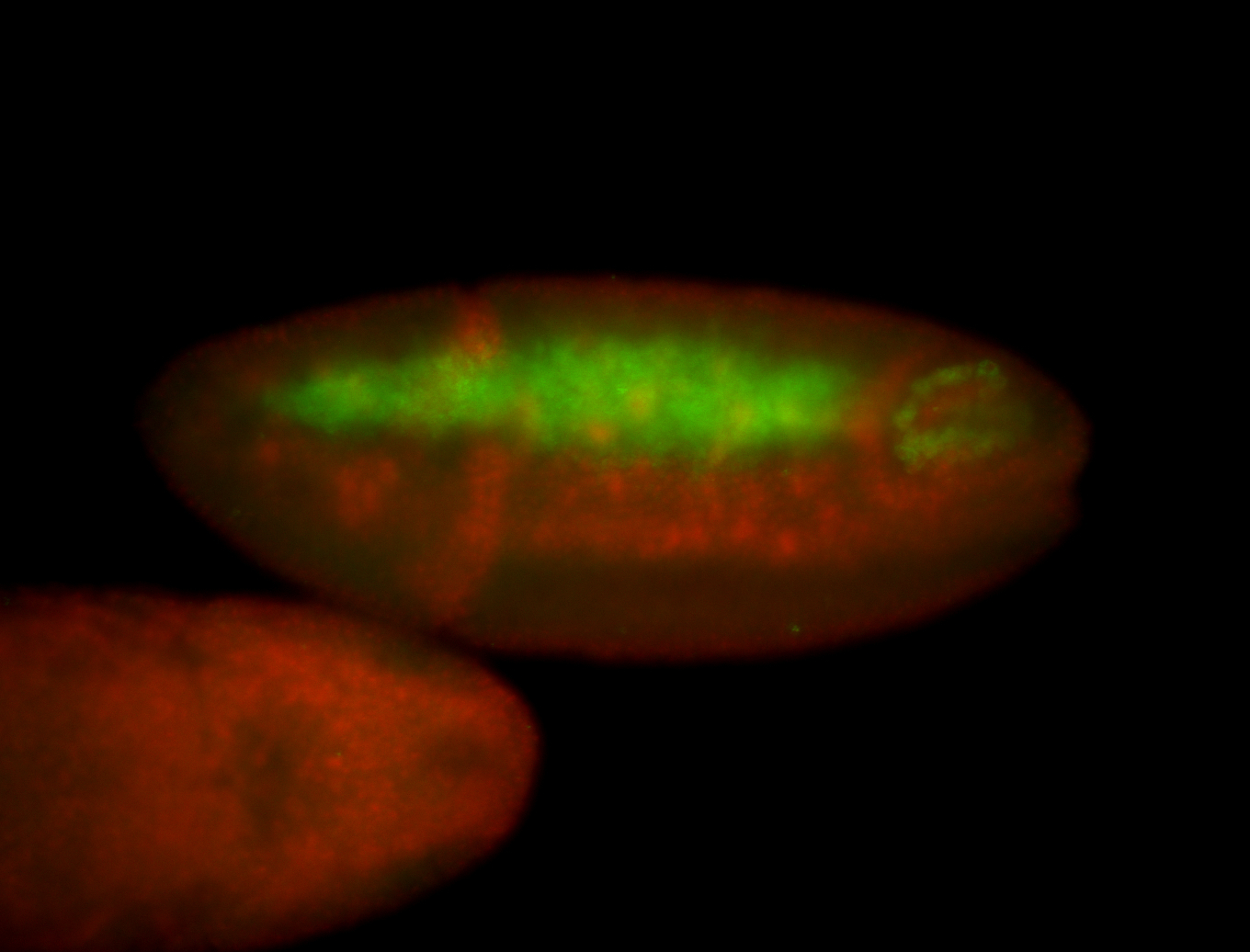
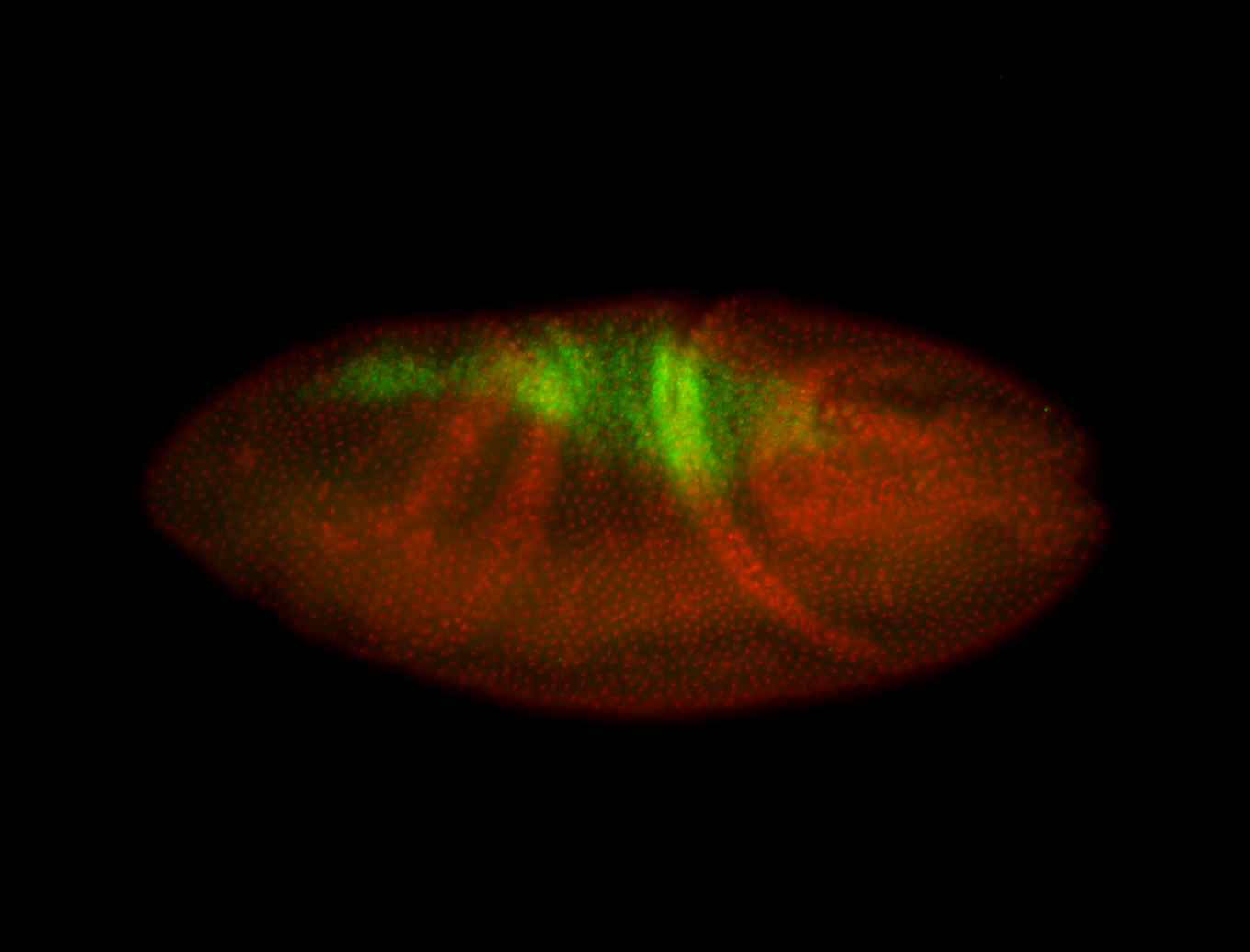
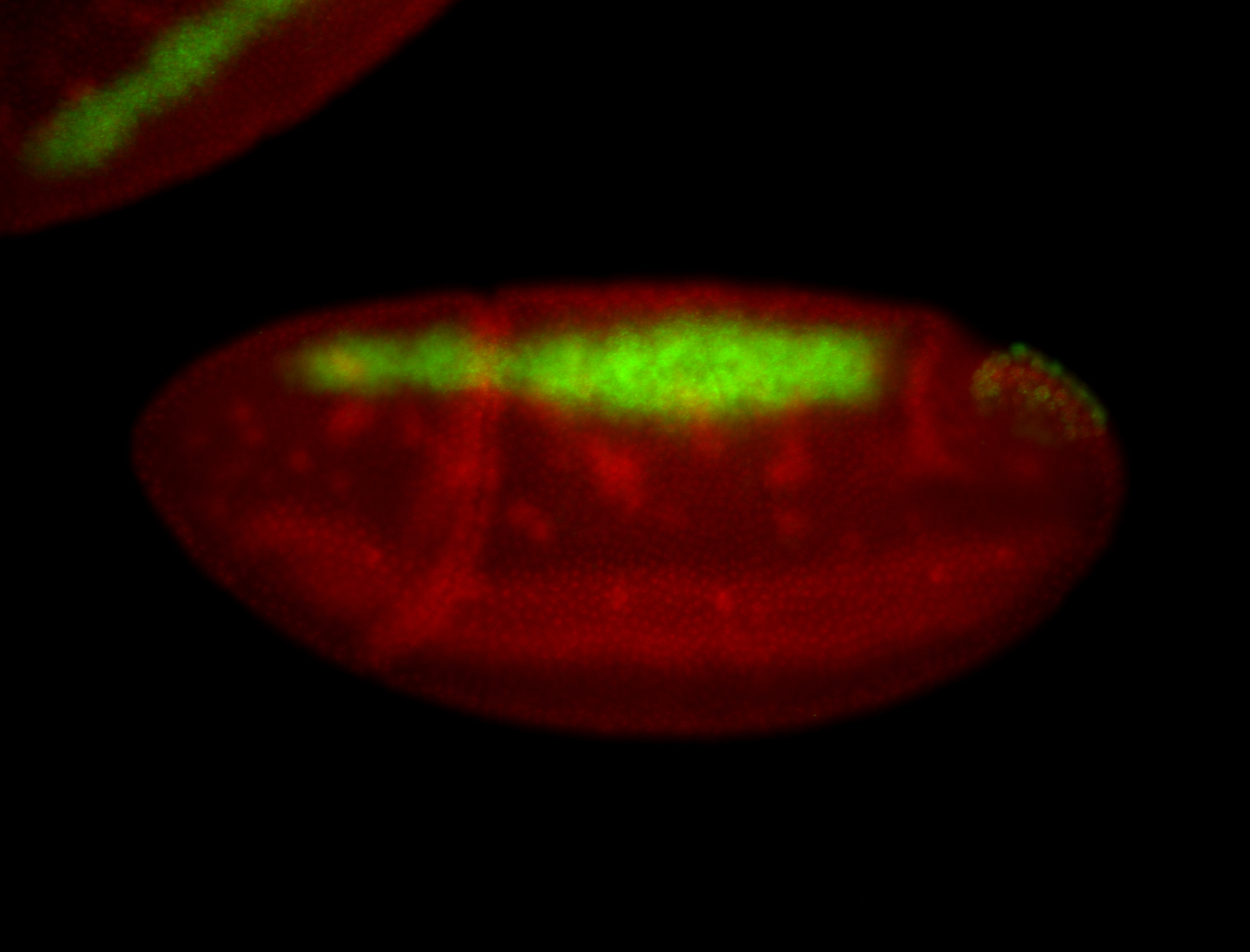
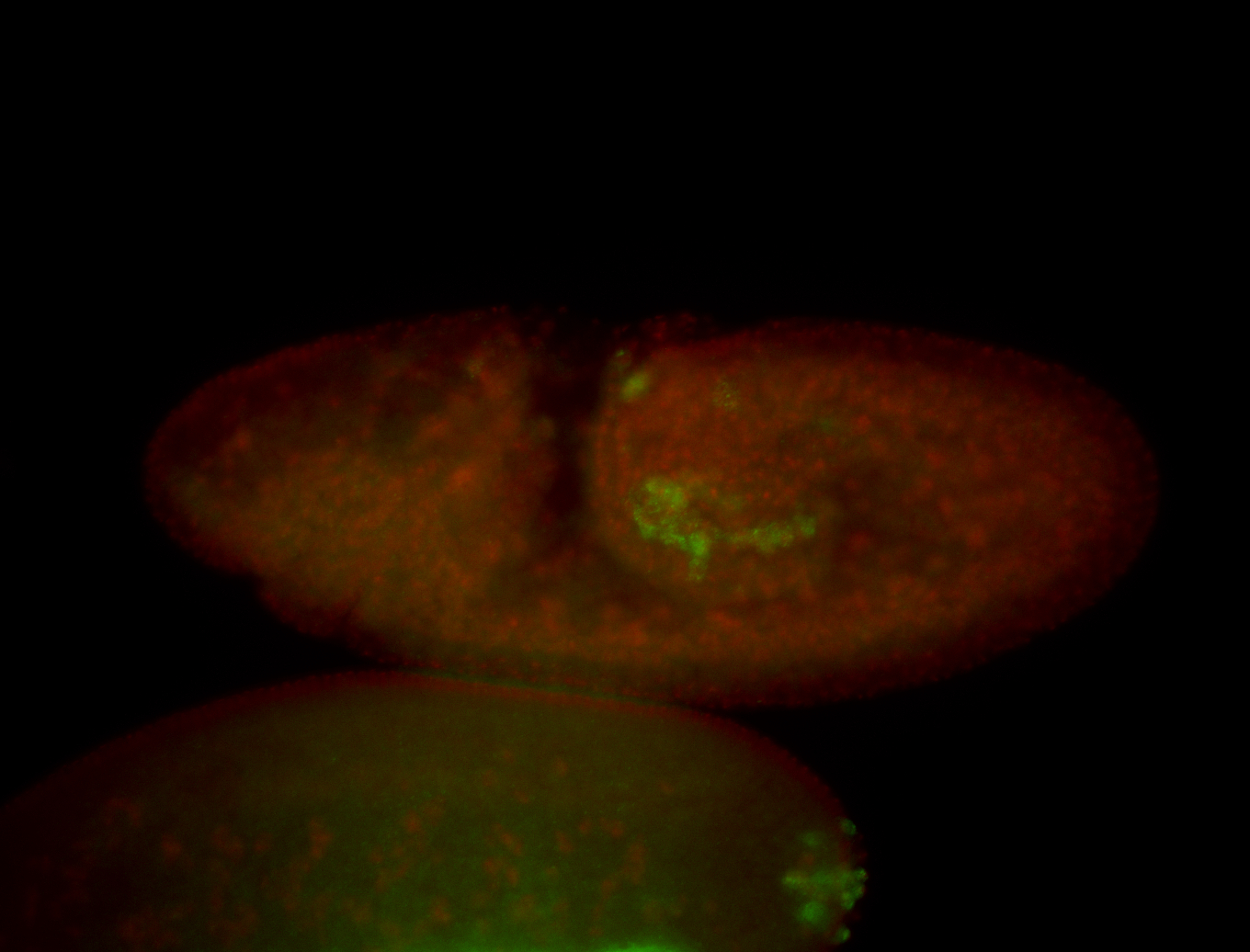
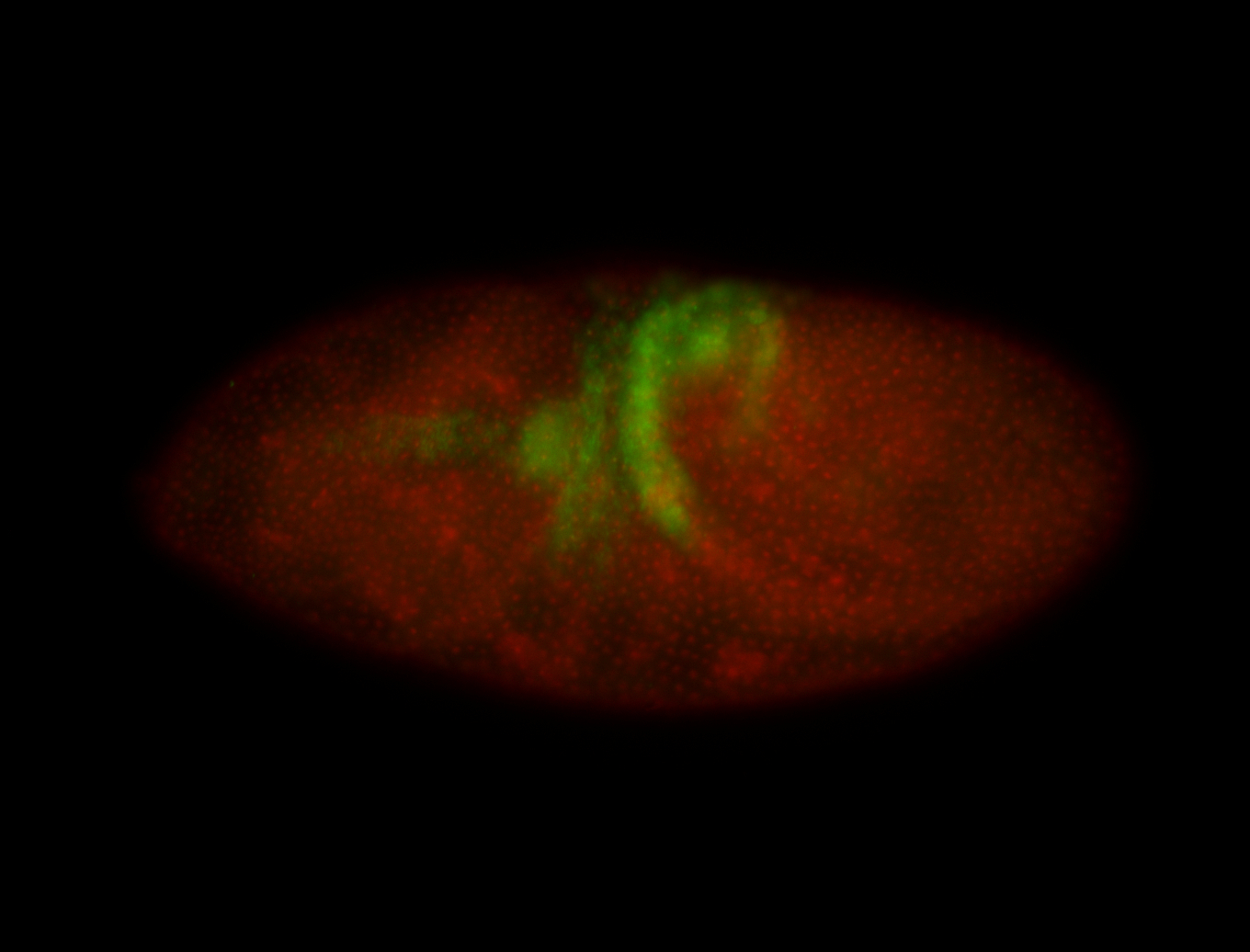
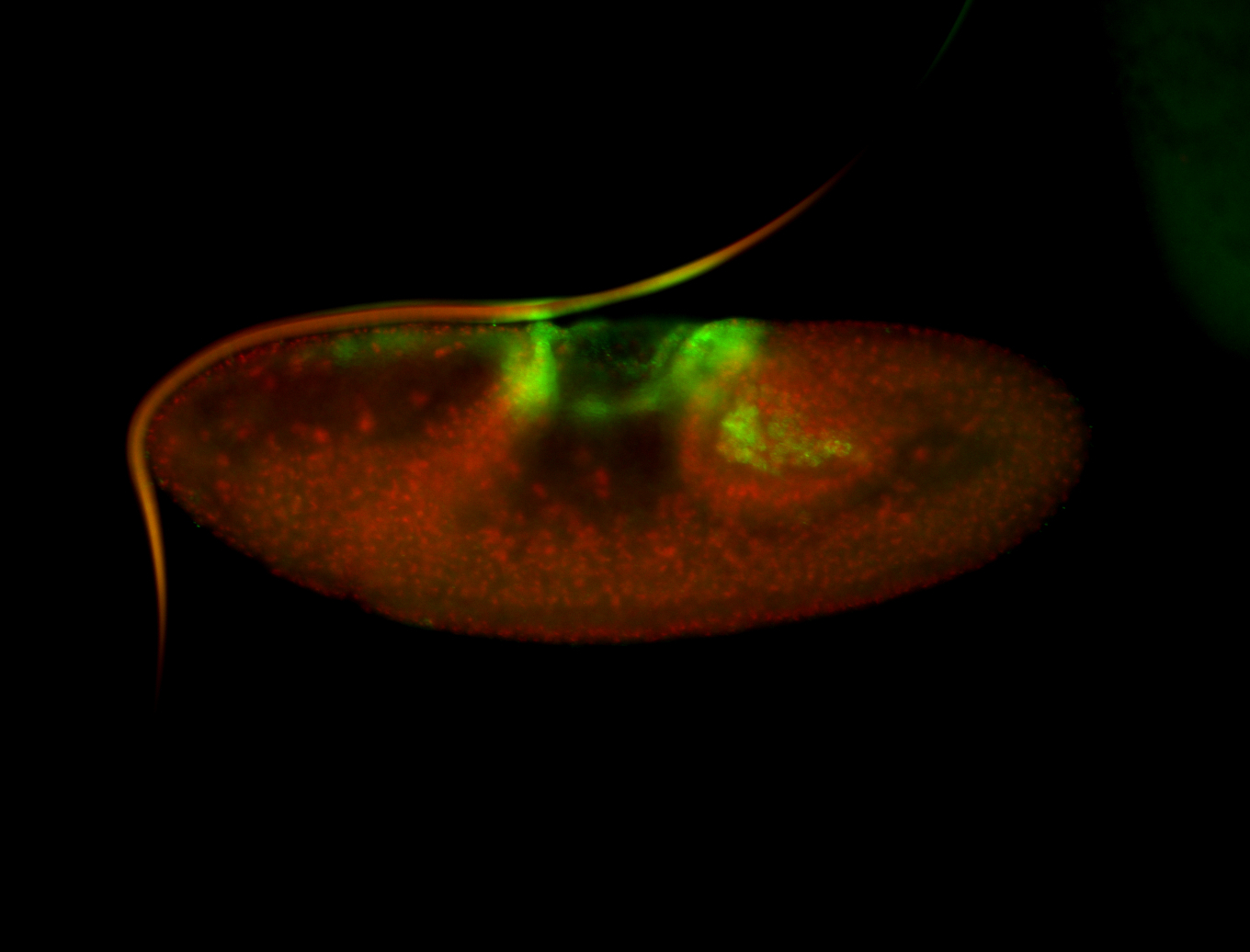
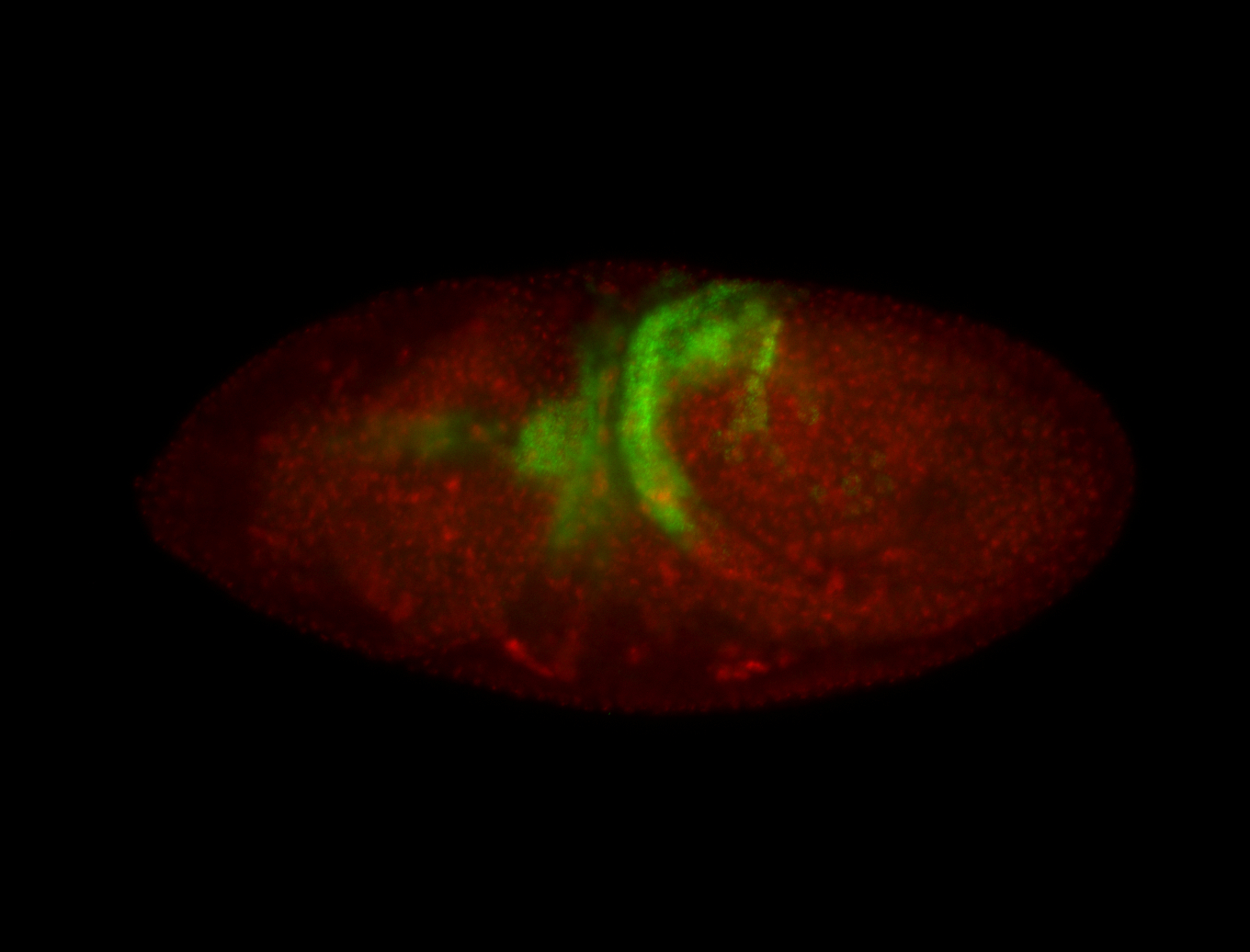
For a full list of searchable terms, please see the term page.
NOTE: It takes a few seconds for the term suggestions to load.
Result Summary
| Number of genes | 27 | |
| Export | ||
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
Comments
| Good probe on gel, a bit low. Very nice insitu... interesting patterns. |
| Membrane localization apically, in small clusters. Very interesting. |
| Membrane localization apically, in clusters. Very interesting. |
| Salivary glands, probably non-specific. |
Probe CG31131-ex Links: Permalink
Genes: CG31131
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting common exons |
| -modENCODE RNAseq data on FlyBase shows low/no expression, however, we observed strong expression patterns throughout development |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe. Amazing insitu! Little maternal. |
| Sub-cellular localization at the edges of amnioserosa cells. |
| Great patterns! |
Probe CR43314-RA Links: Permalink
Genes: CR43314
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -quite unique expression pattern that appear as highly localized and nuclear small foci along a strip of the early blastoderm; and as small highly localized foci in the early anterior-dorsal head, API, ventral cephalic furrow; and lastly in ectodermal regions of late stages representing PNS patterns |
| -images are from two separate experiments using different hairpin-fluorophores for signal amplification but otherwise identical conditions |
Probe CR43617-RA Links: Permalink
Genes: CR43617
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -posterior single stripe; expression is reported to be similar to the lncRNA "iab4" (CR31271) |
Probe CR46003-RC Links: Permalink
Genes: CR46003
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RC isoform transcript |
| -the gene is a lncRNA with quite interesting patterns in the CNS (brain and VNC) |
| -seems to be expressed in the anterior transverse furrow, shown in stage 6-7 images |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting common exons |
| -Lateral and ventral spots in the epidermis of late stage samples are likely imaginal discs |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -There is no entry for this gene on BDGP, and modENCODE RNAseq data on FlyBase indicates that it is not expressed in the embryo. However, we observed obvious patterns during embryogenesis. Additionally, FlyBase does not have any in-depth characterizations of this gene. |
| -Osi21 does not overlap with any other genes, ruling out the possibility of the targeting of alternative transcripts. BLAST also confirms the specificity of our probes to Osi21. Additionally, no other genes for which probes were designed for on this plate show patterns such as those observed here, ruling out the possibility of cross-contamination during probe preparation. |
| -This may be a result of Osi21's poor characterization or incorrect RNAseq data |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe on gel. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -detected using HCR-FISH protocol (3 probe sets) |
| -probes were designed against dap-RA isoform transcript |
| -really neat expression patterns |
| -images are from two separate experiments under identical conditions for better coverage |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -Most signals in the late stages are likely imaginal discs |
| -6 "ring"-like structures on the anterior ventral side are likely the leg discs -2 circles above leg discs on each side are likely wing discs -Band posterior to the VNC is likely the genital disc -small dots located on the late stage midgut may be intestinal stem cells (ISCs) |
| -Other signals in late stages likely correspond to imaginal discs which we cannot identify based on our resolution |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RF isoform transcript |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting common exons |
| -Potential weak ventral nerve cord signal |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -retested recently for late stage coverage (2025); appears to be expressed in a different program in the gnathal elements and sensory regions in late stages |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RB transcript isoform |
| -neat and diverse expression patterns throughout development |
| -images are from two separate experiments under identical conditions for better coverage |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Salivary glands, probably non-specific. |
| Dorsal and posterior. |
| Dorsal and posterior. |
| Low expression. Mild enrichment. Semi-good probe on gel. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA transcript isoform |
| -expression seen in the stomatogastric sensory primordium, and the anterior and posterior midgut primordium; forms a complex along with ac (CG3796) so should colocalize in neuronal regions |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA transcript isoform |
| -very diverse and neat patterns at all stages |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Salivary glands, probably non-specific Need more images to tell the full story. |
| Low probe, nice insitu. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA transcript isoform |
| -neat expression patterns in early embryos and early germ band elongation stages; expression is no longer detectable after stage 9-10 (fits modENCODE RNA-seq data well) |