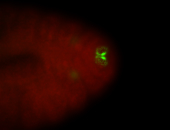
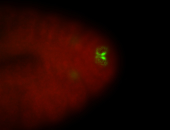
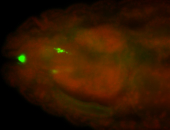
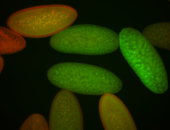
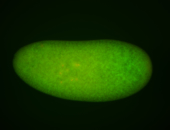
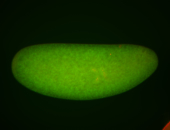
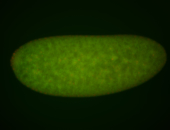
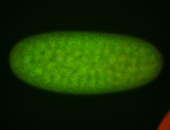
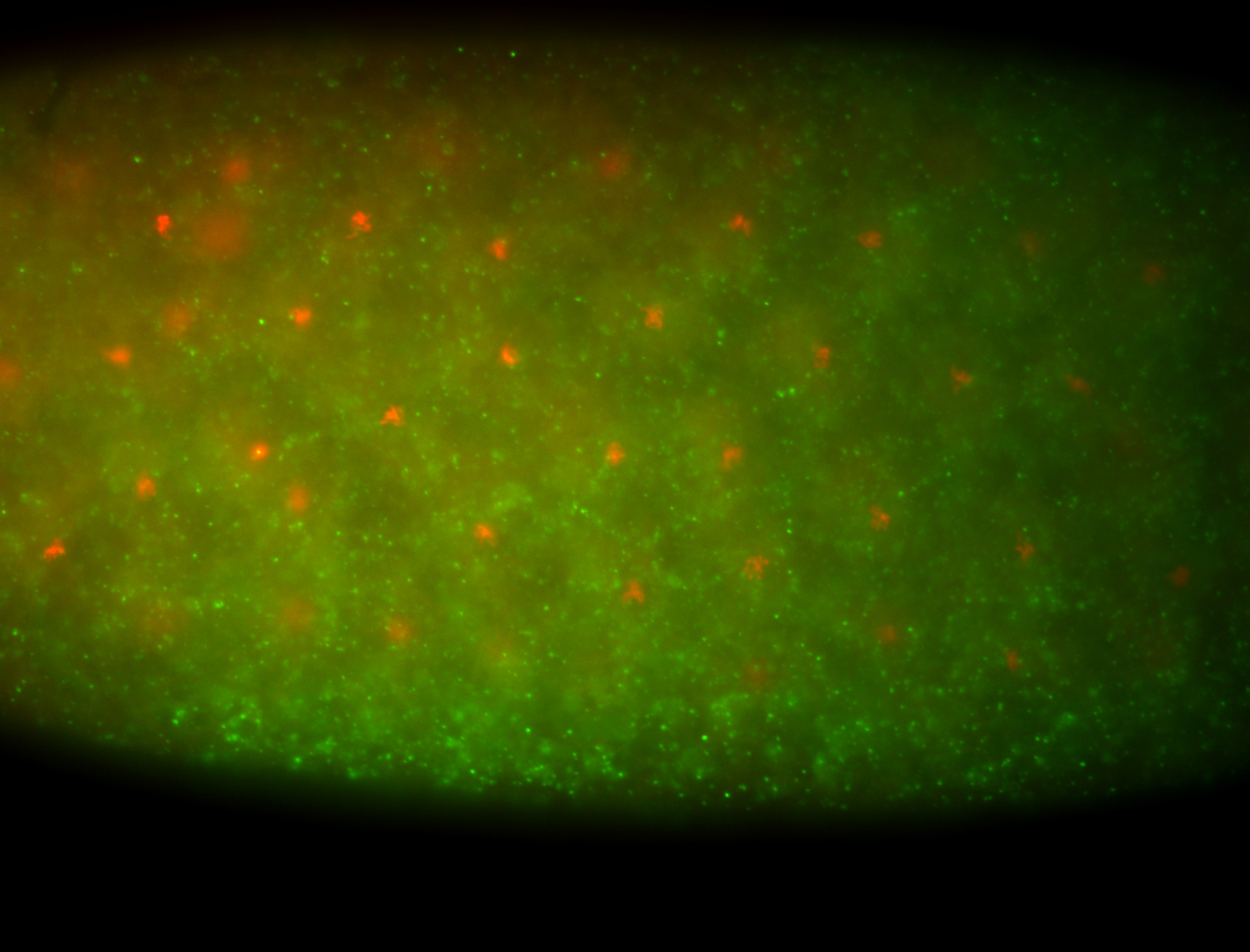
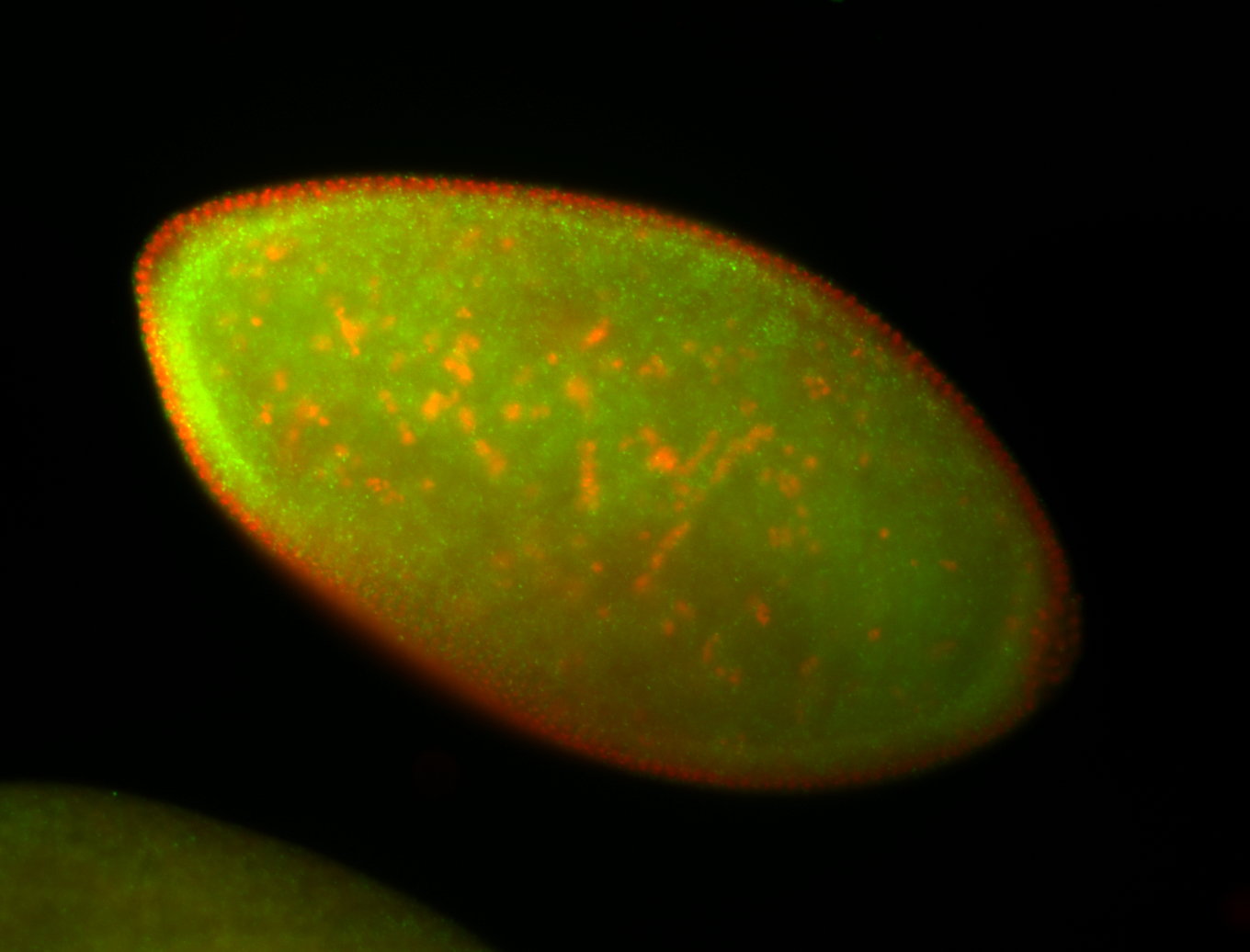
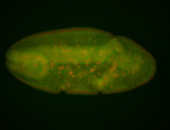
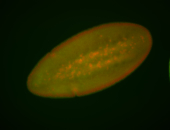
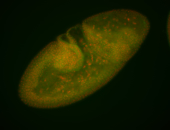
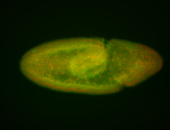
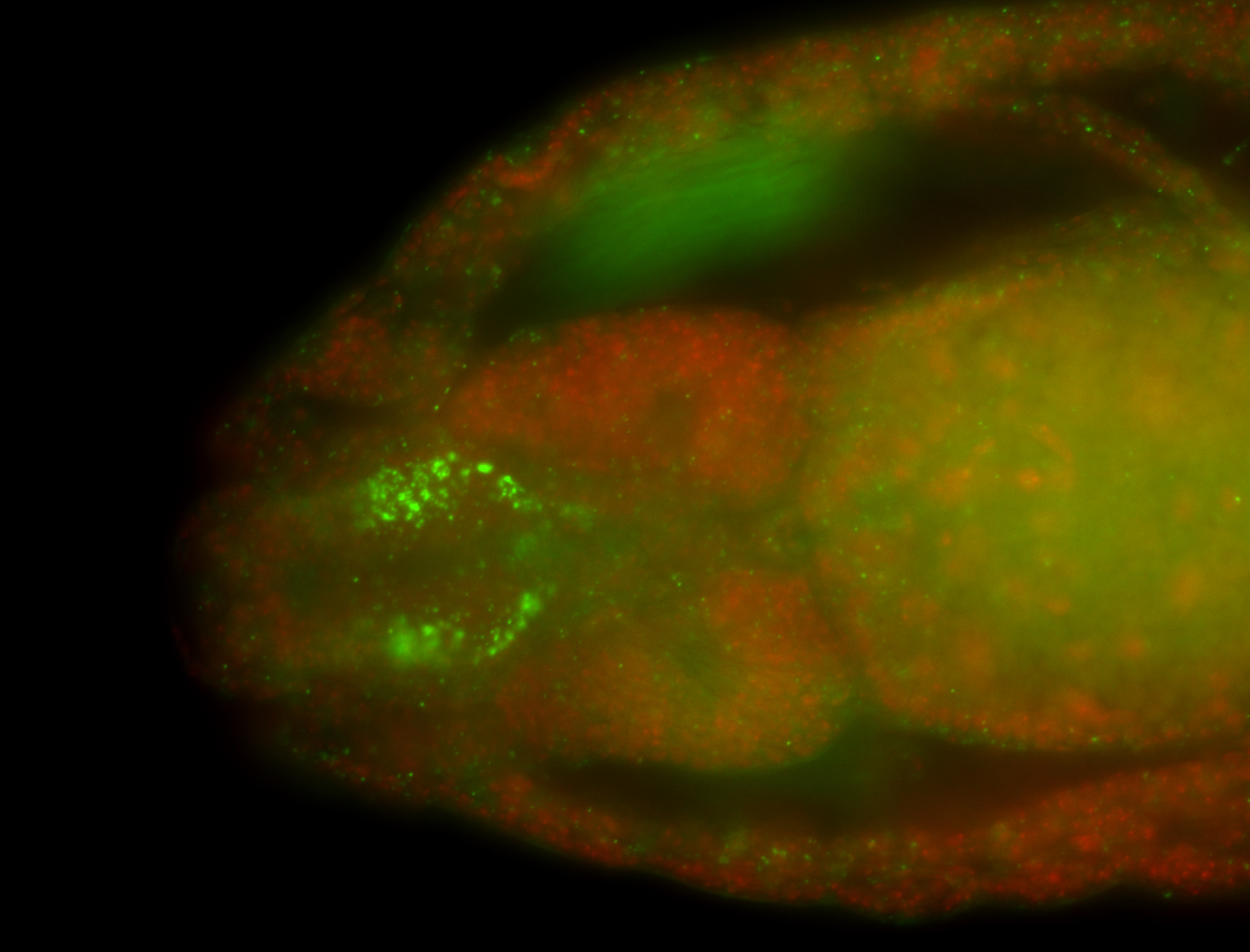
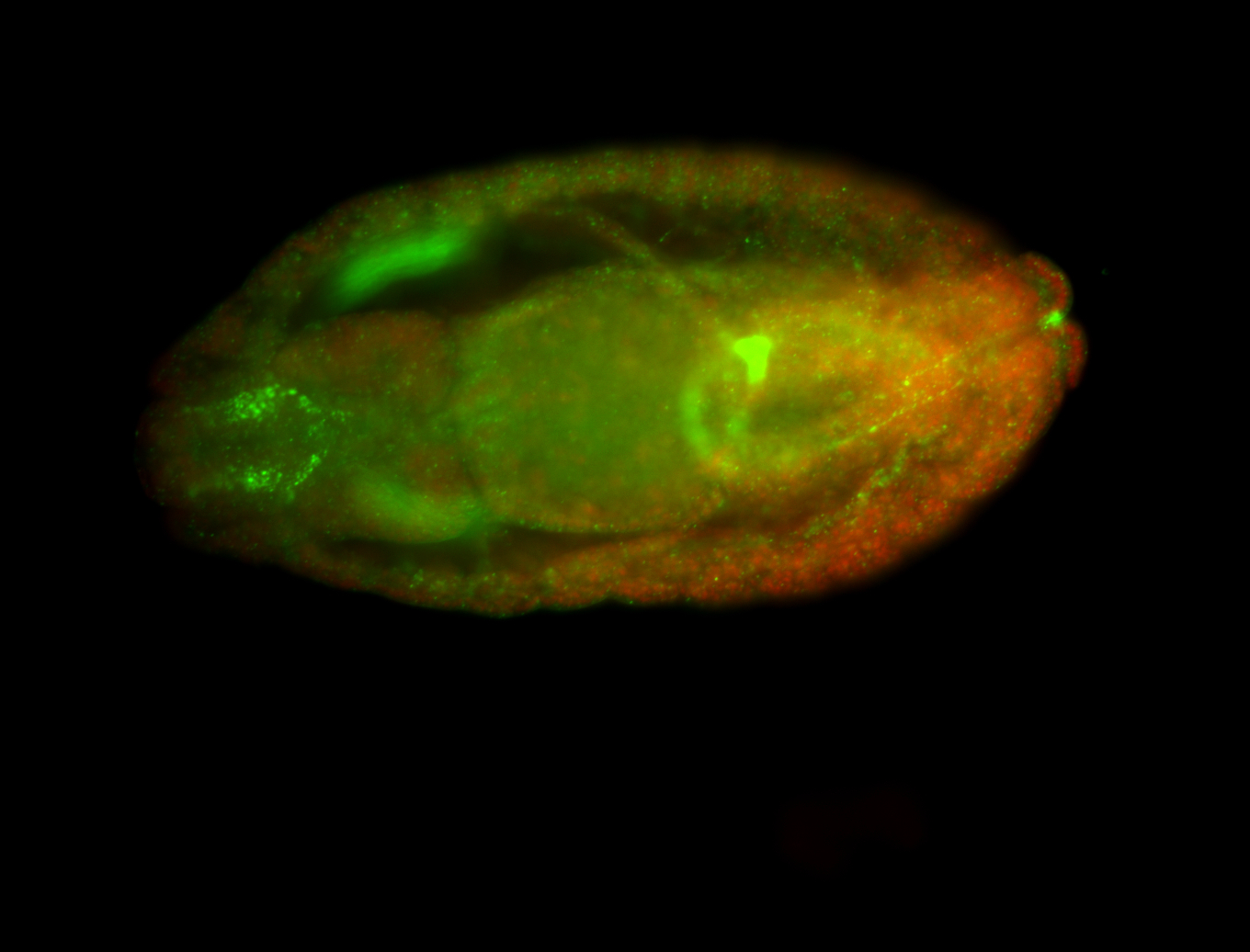
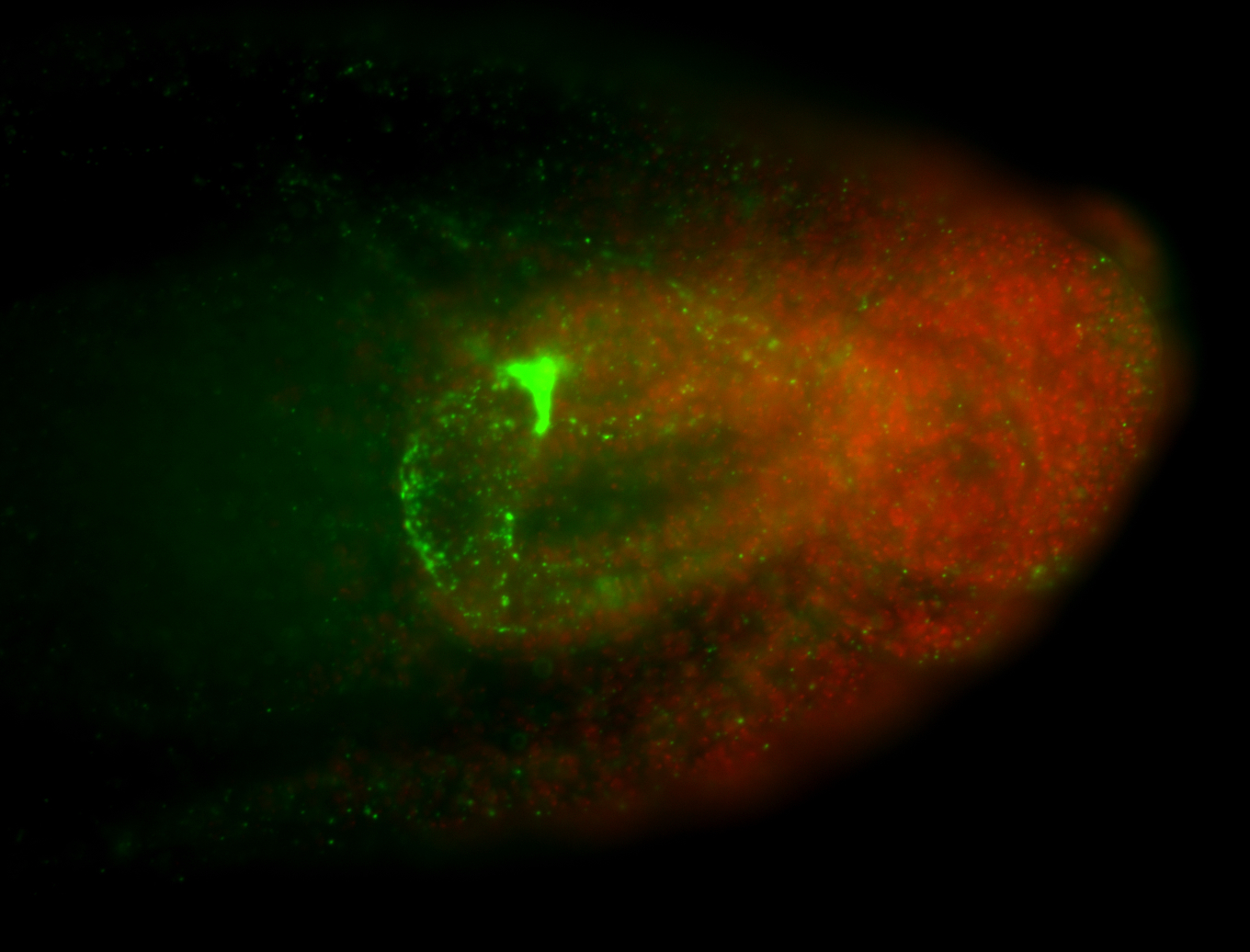
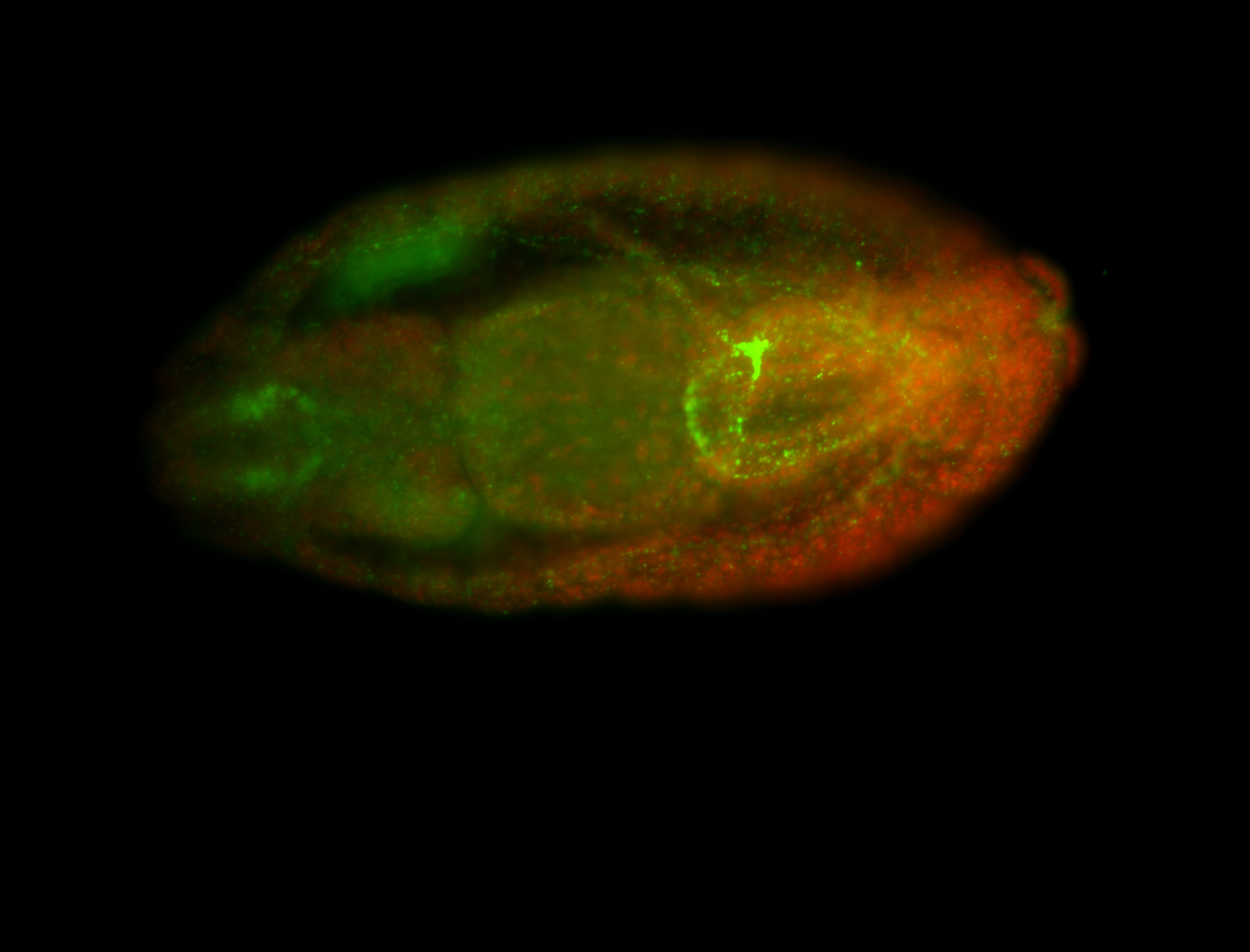
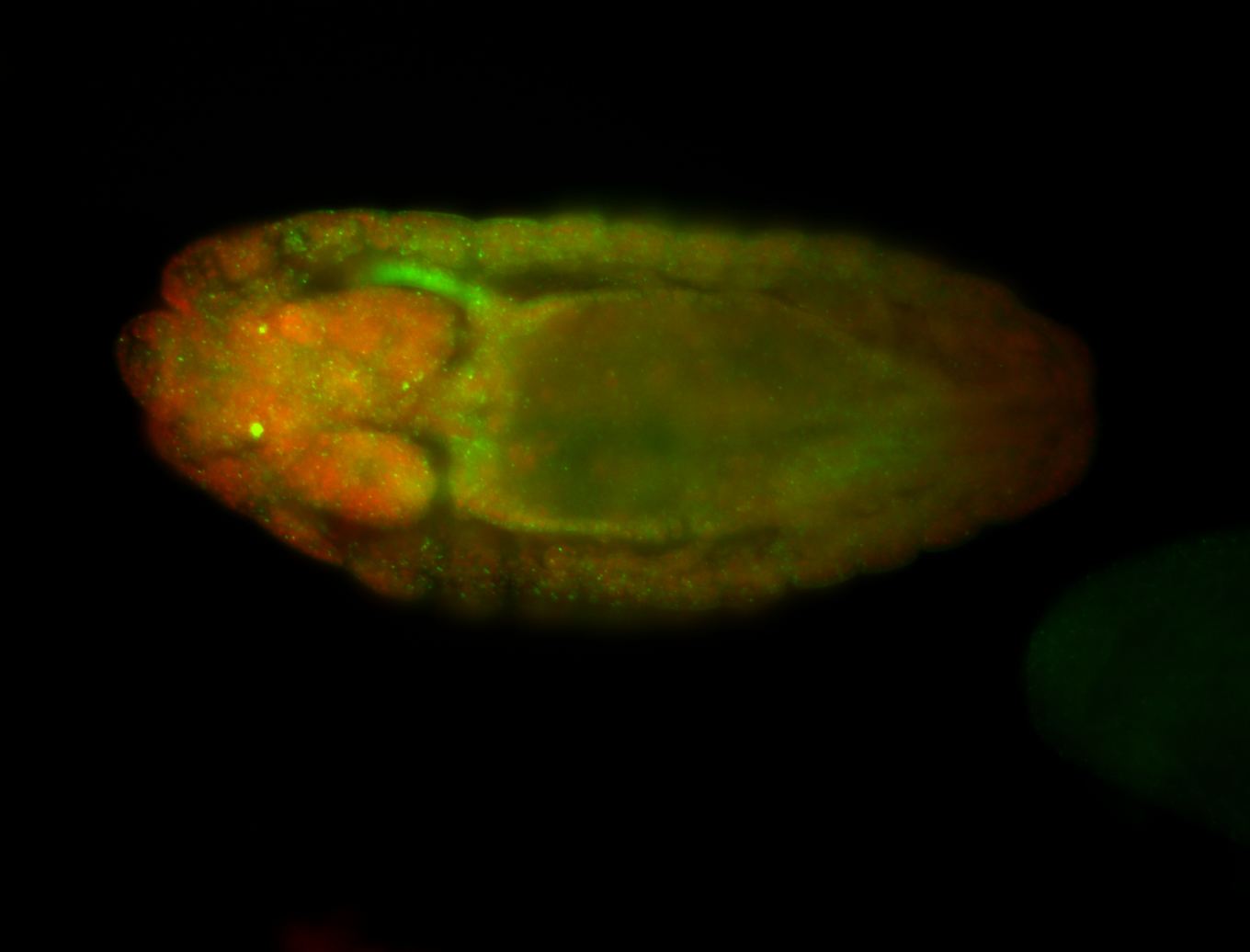
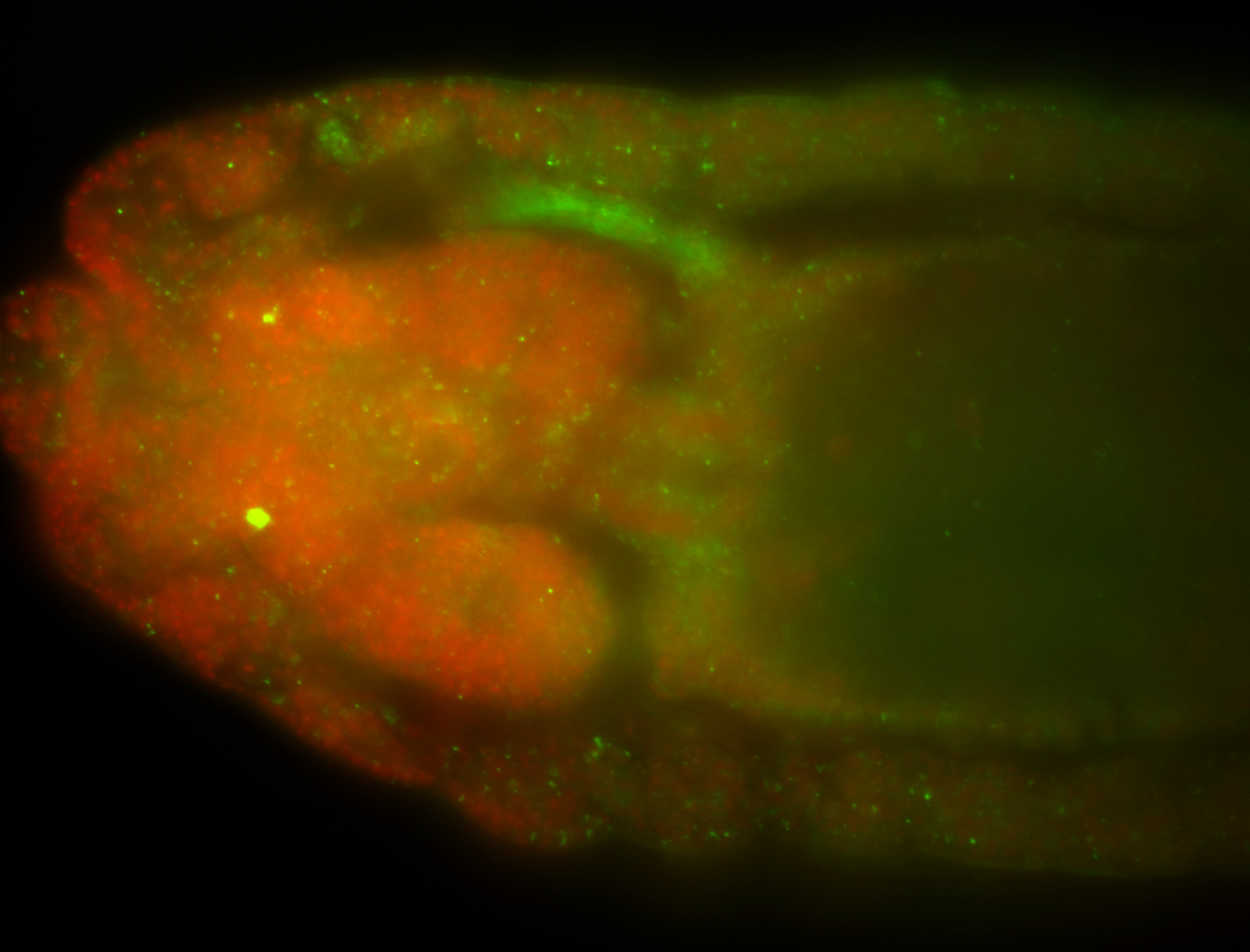
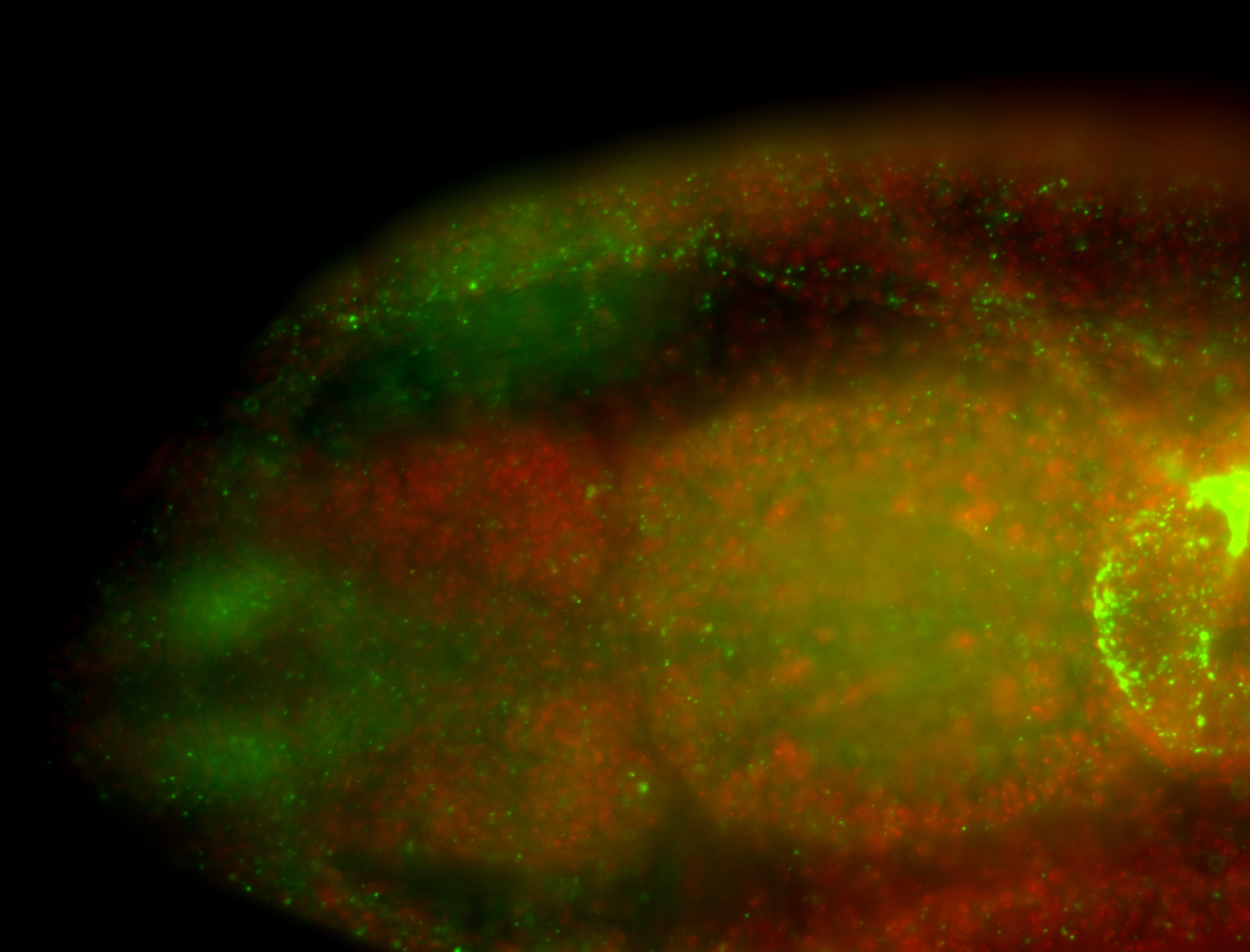
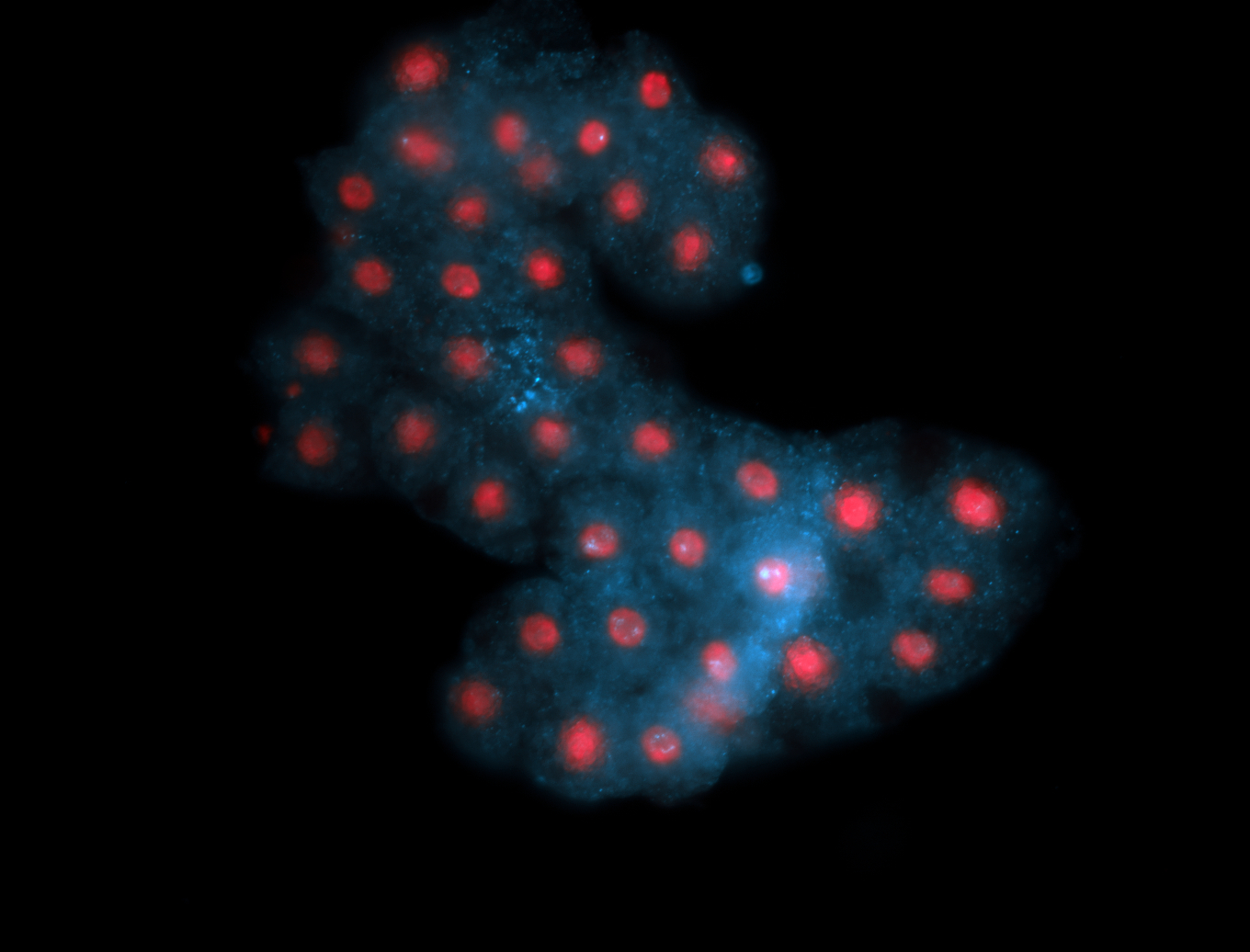
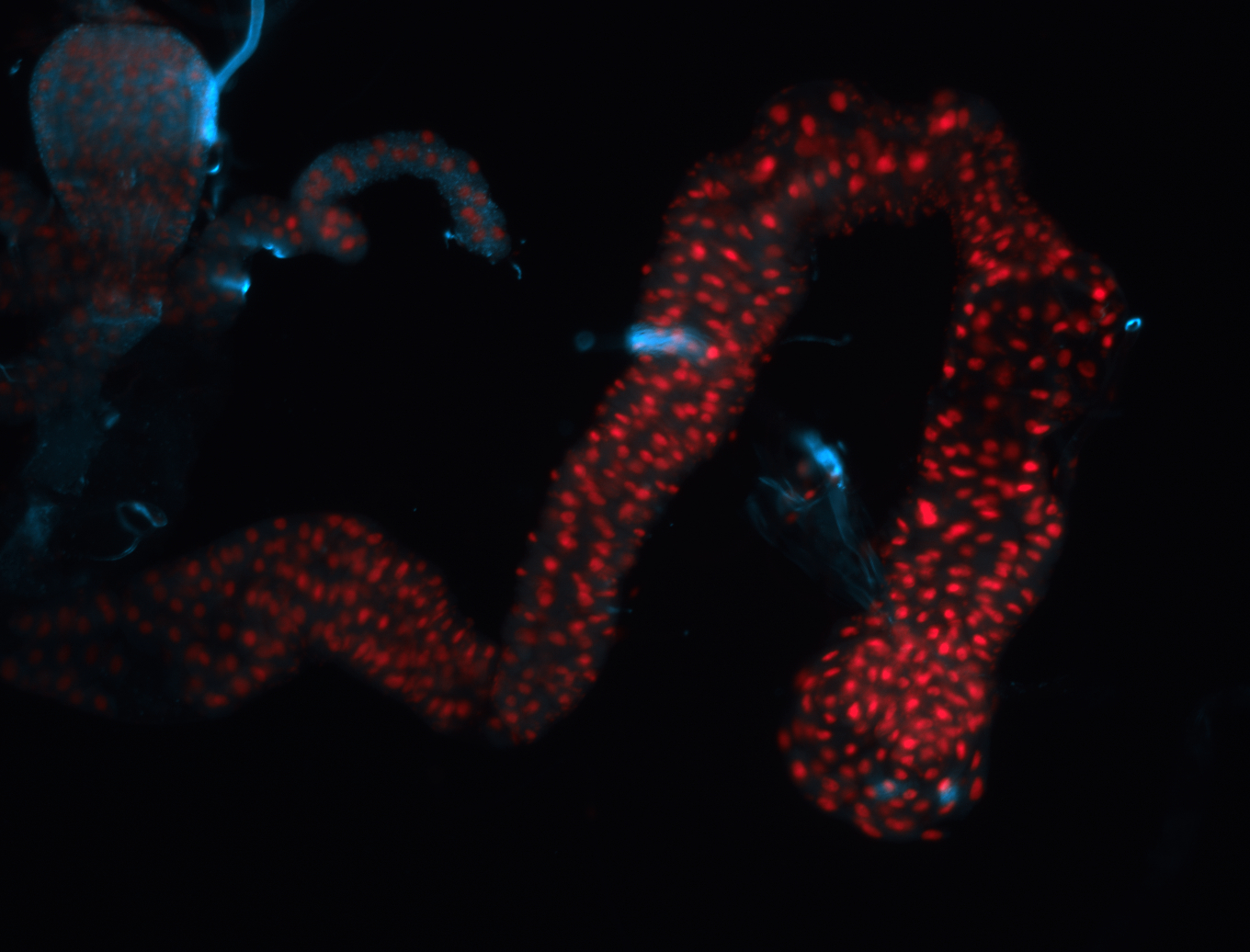
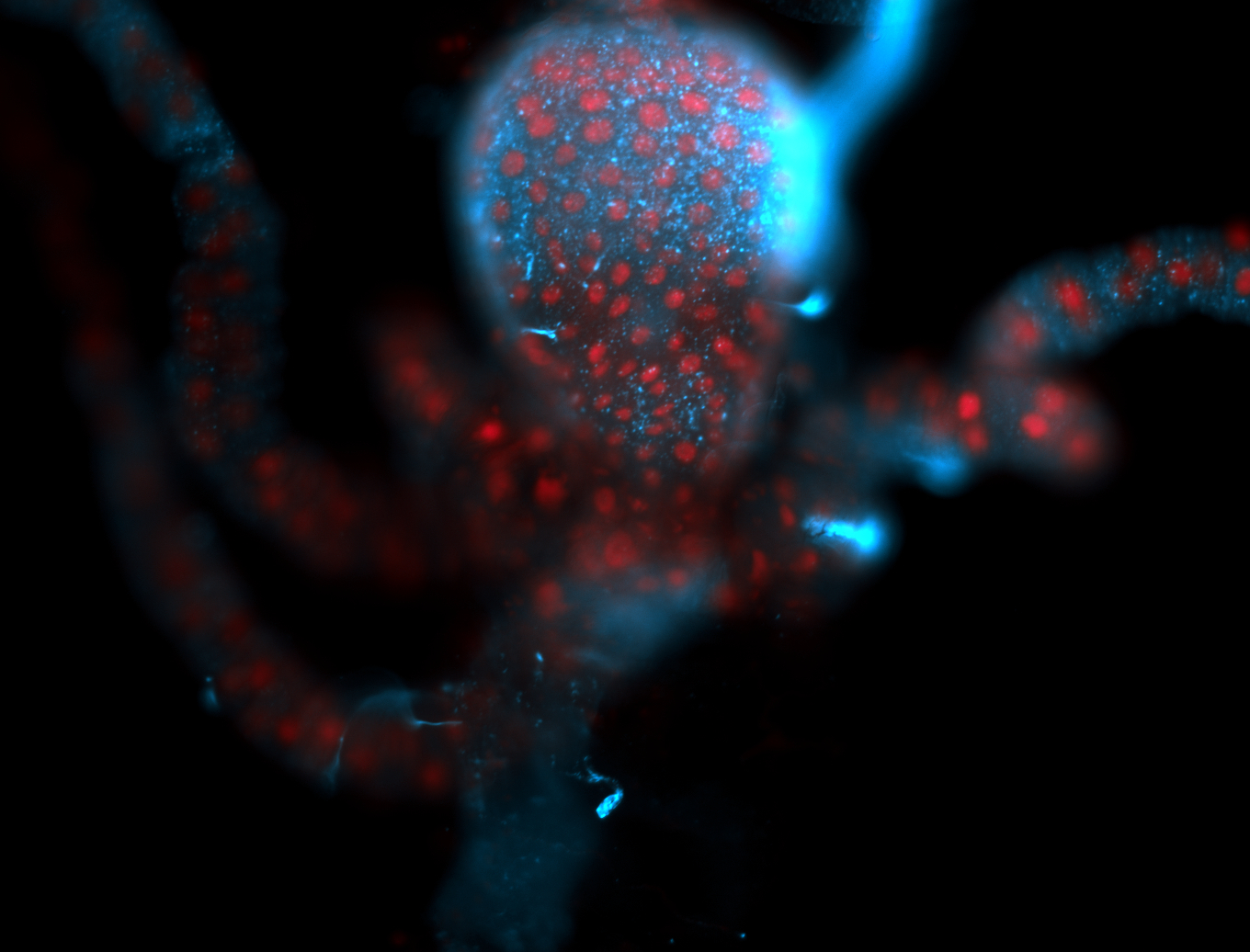
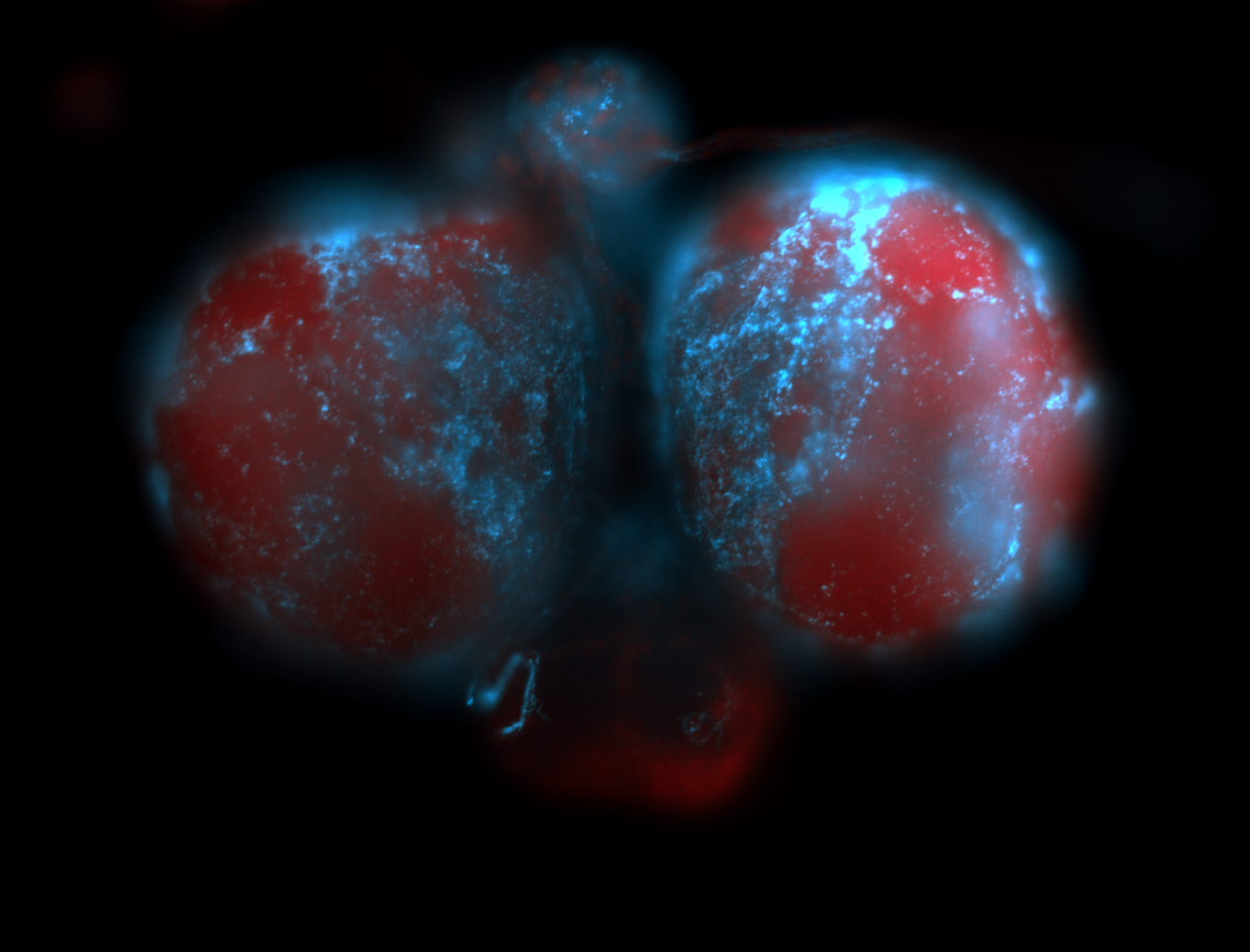
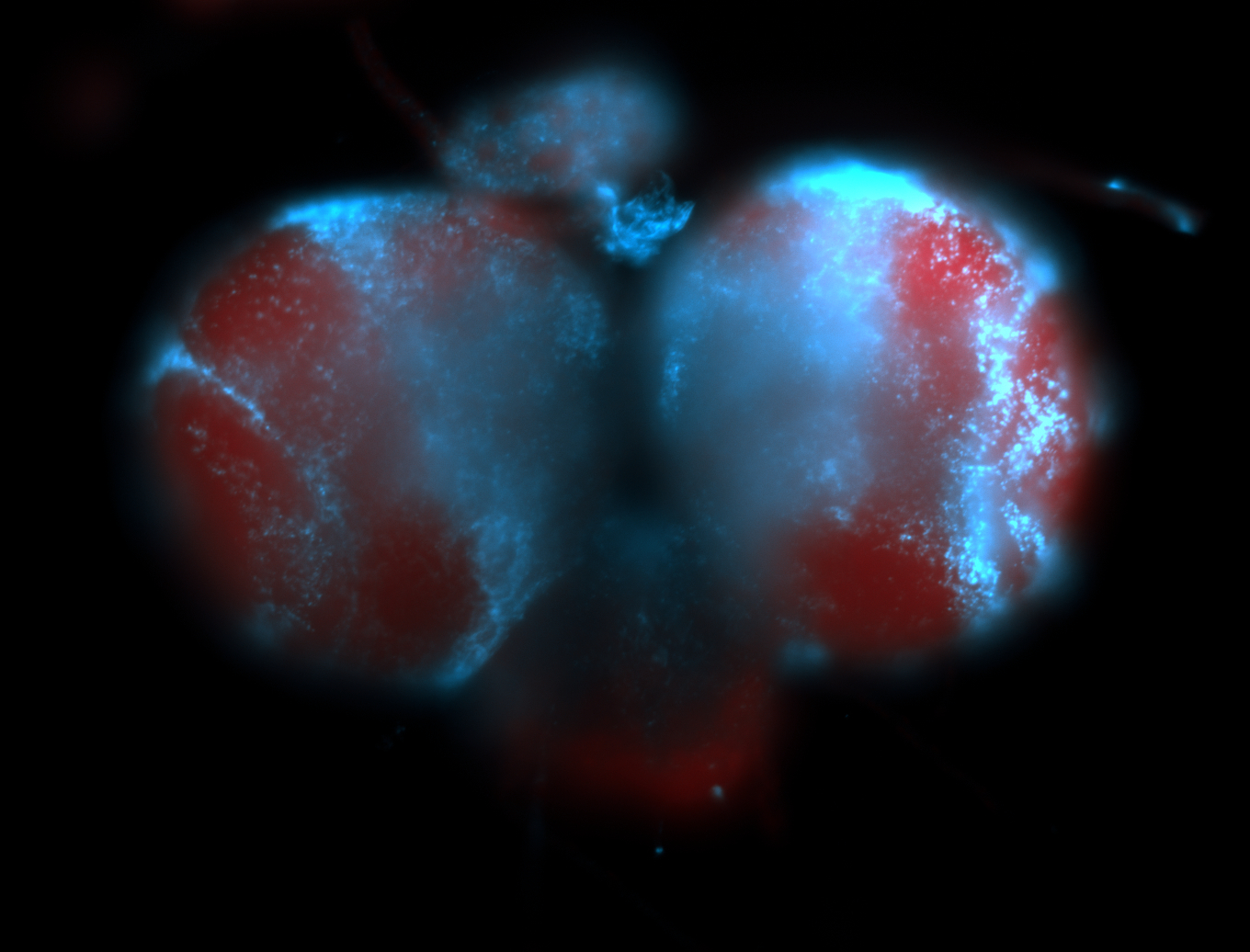
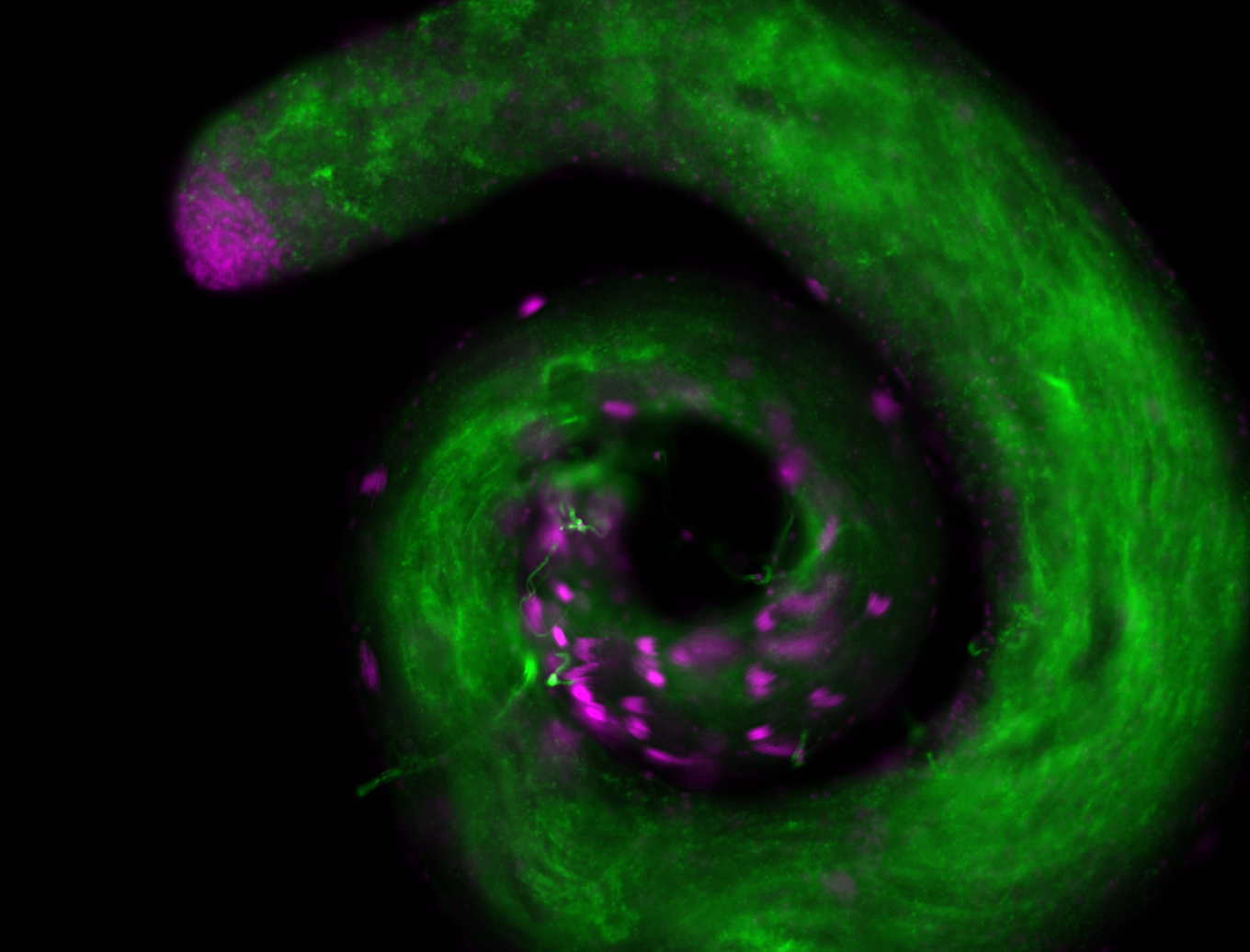
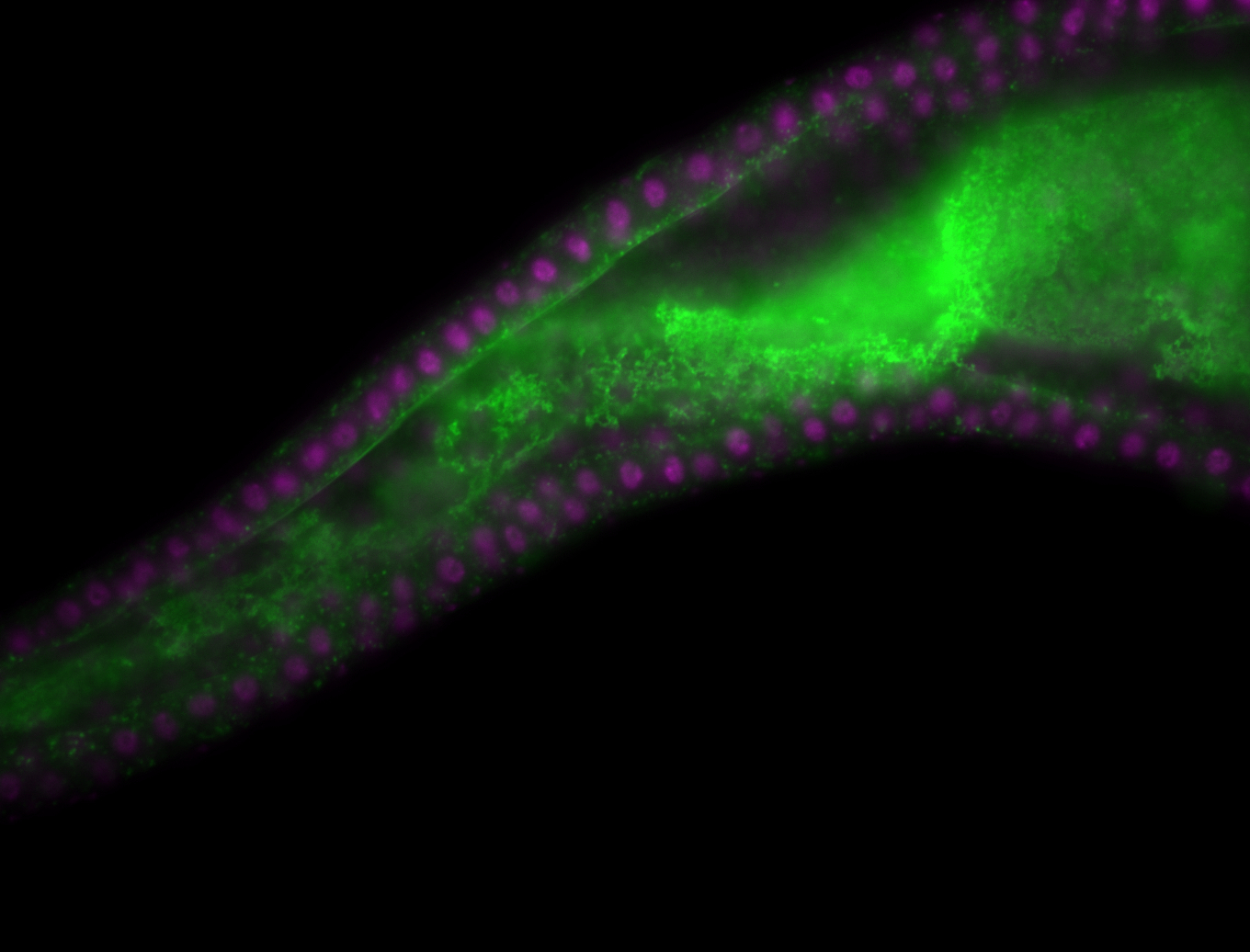
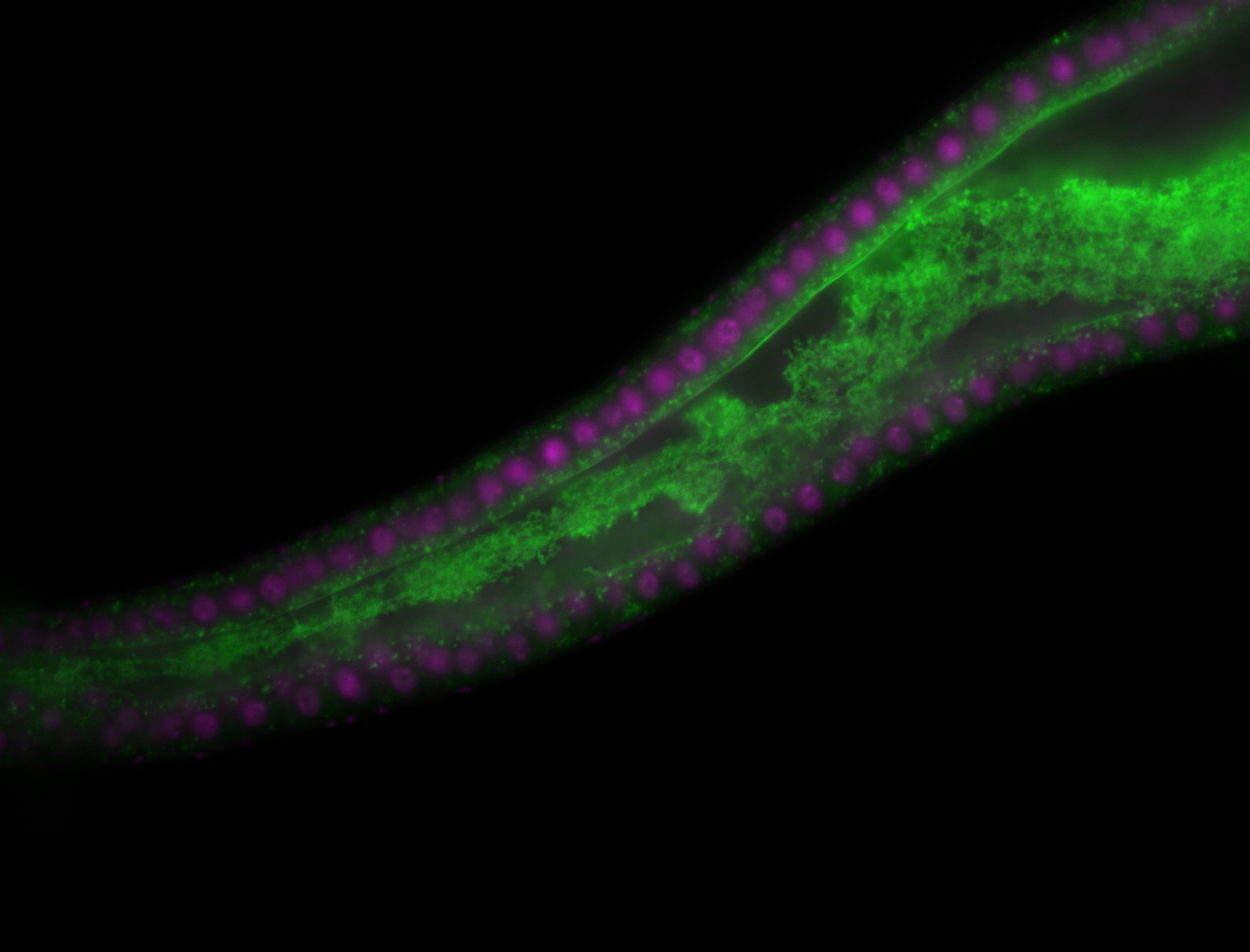
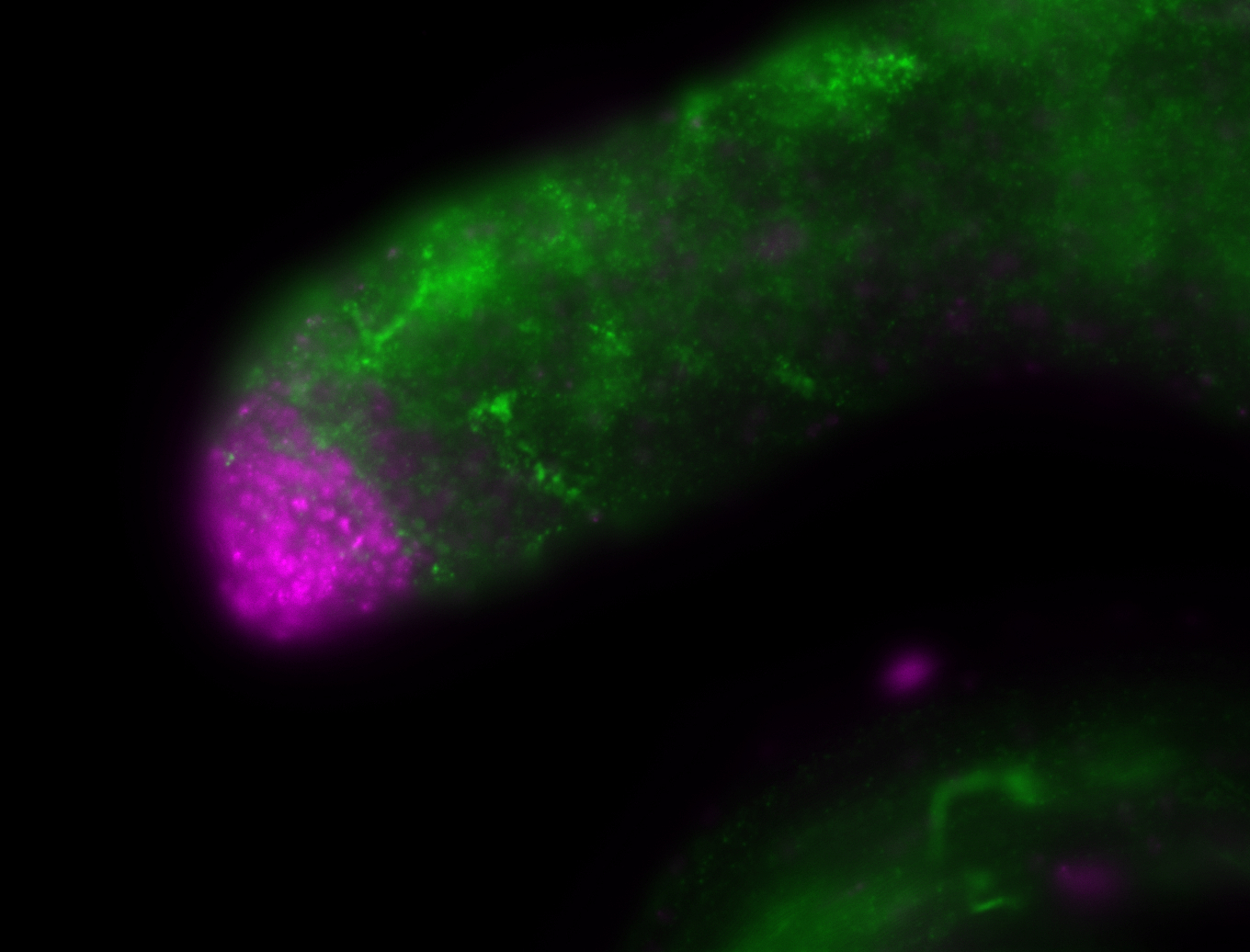
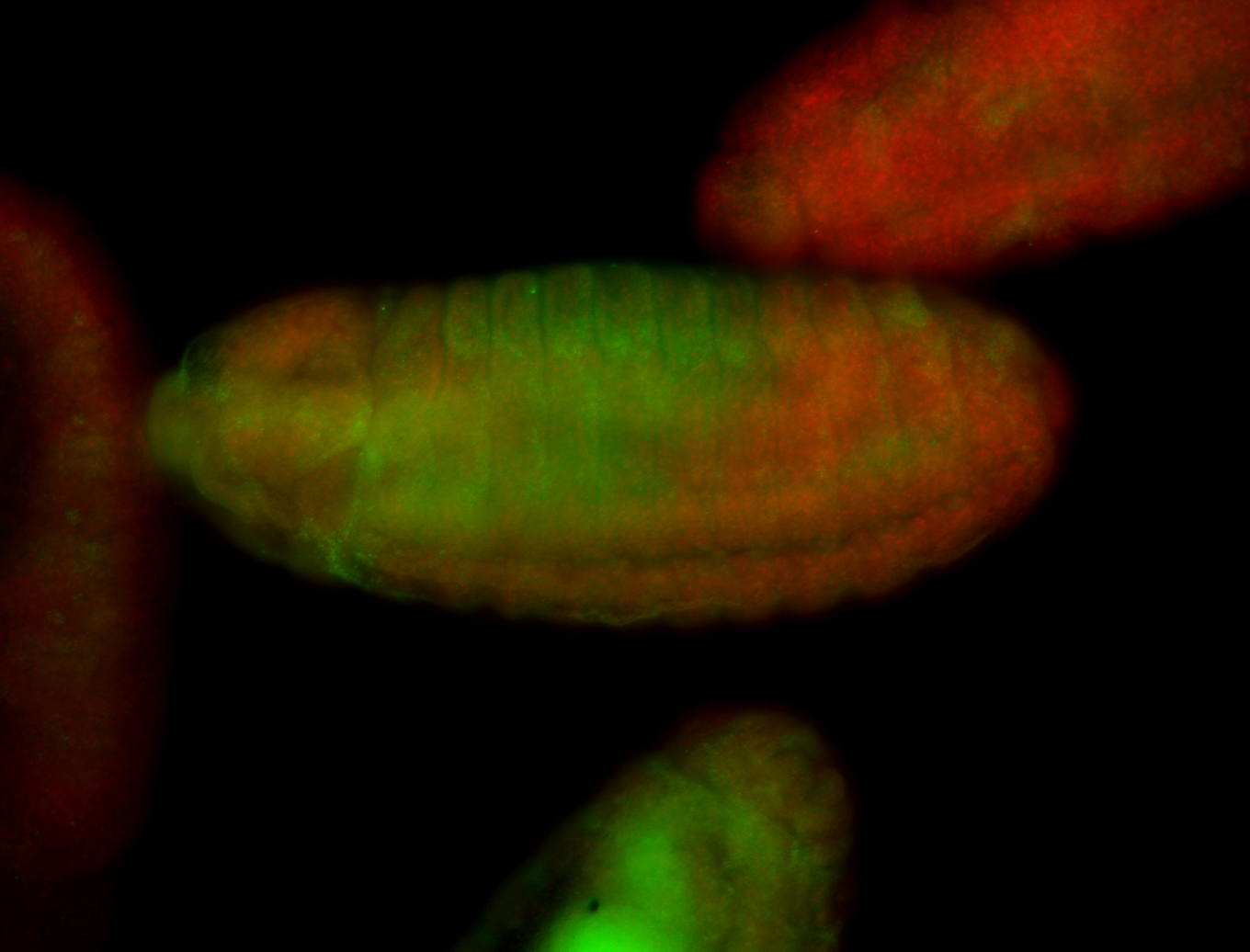
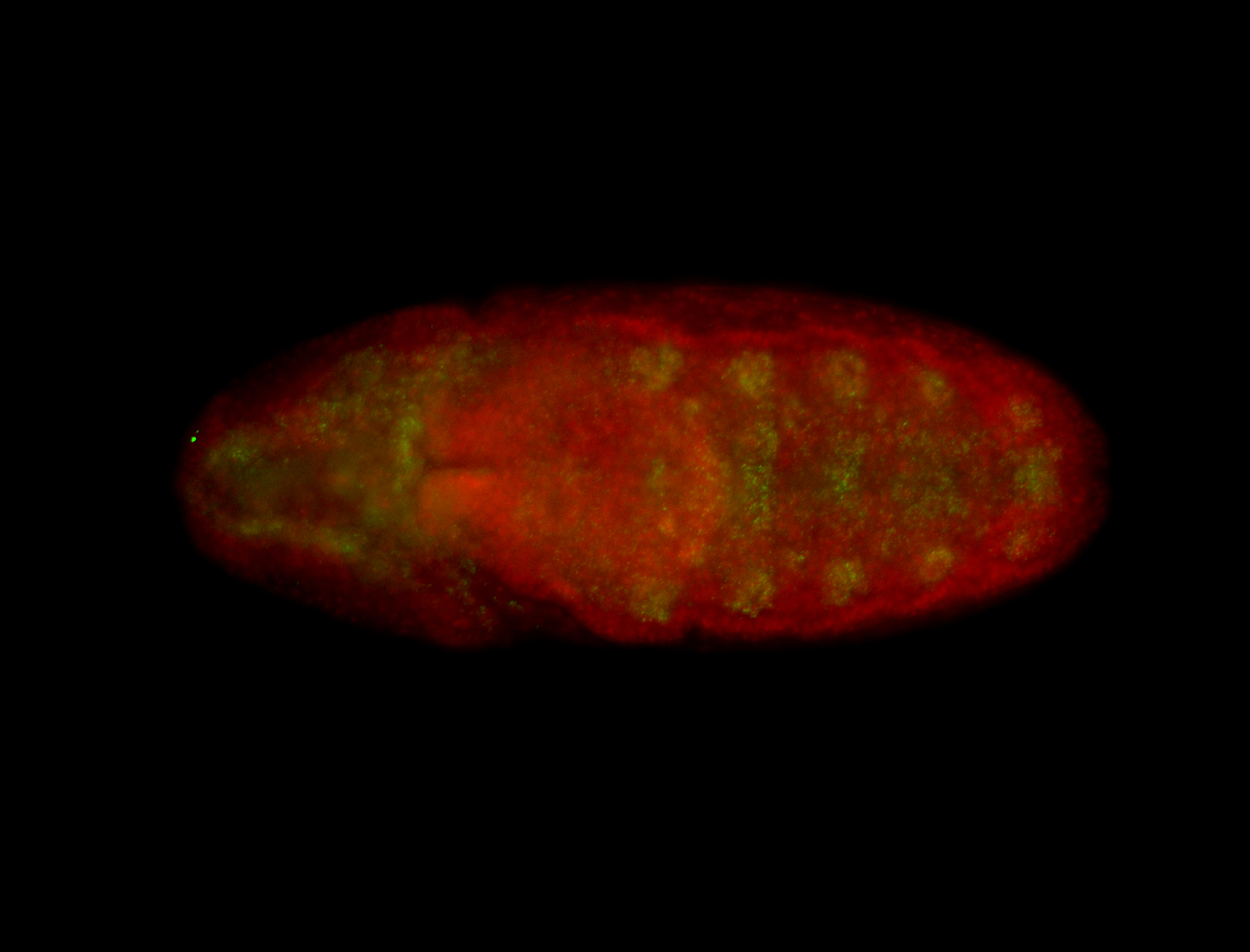
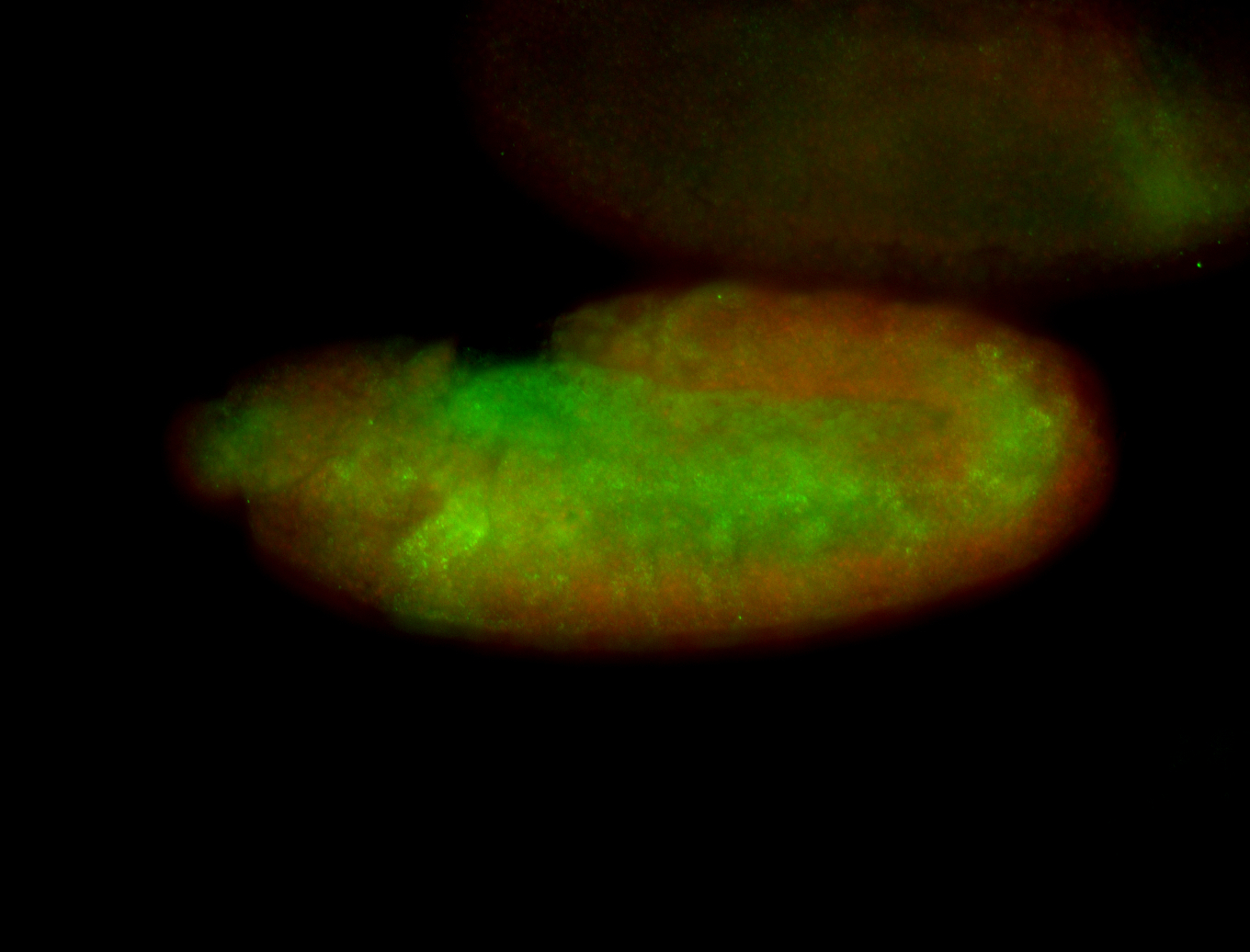
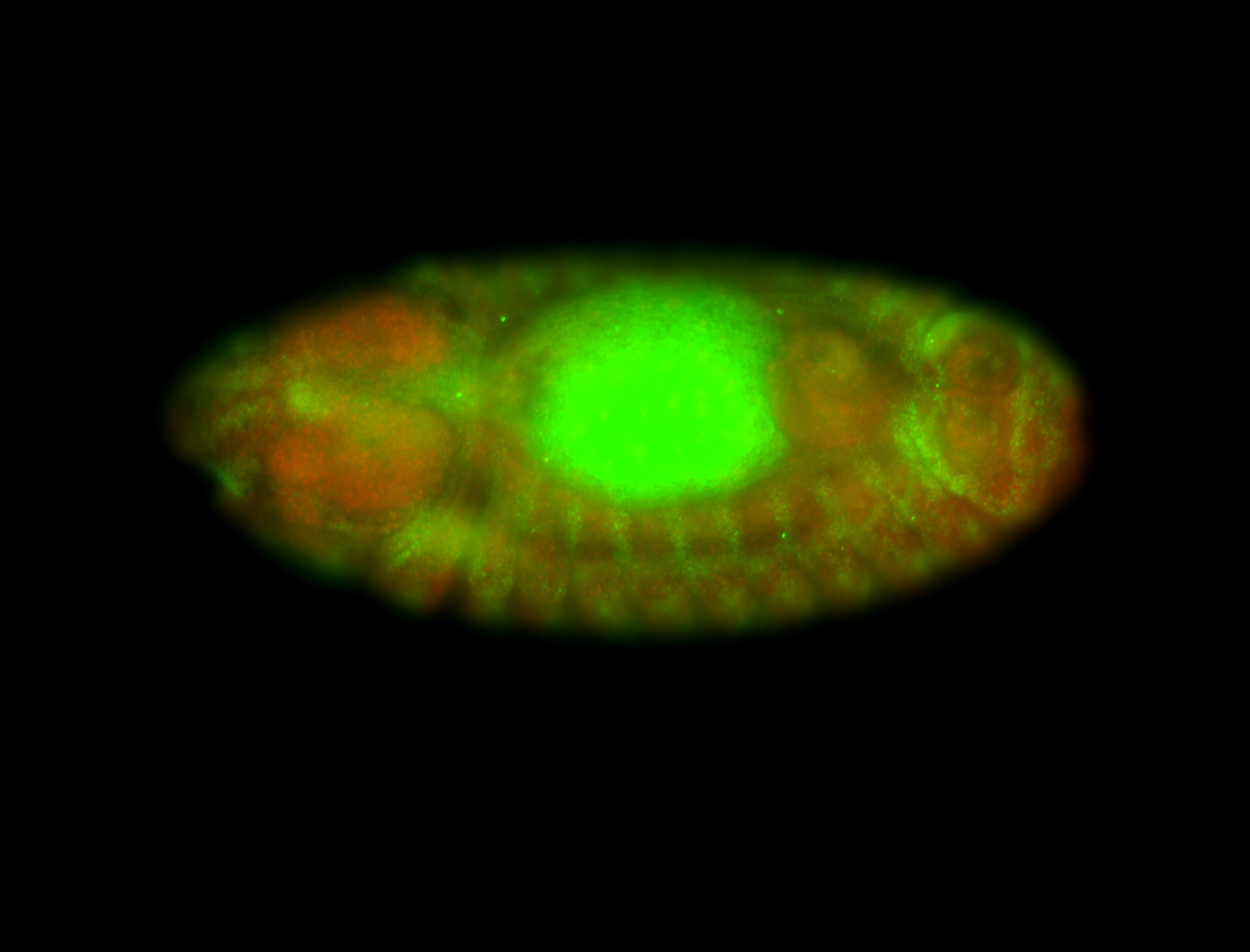
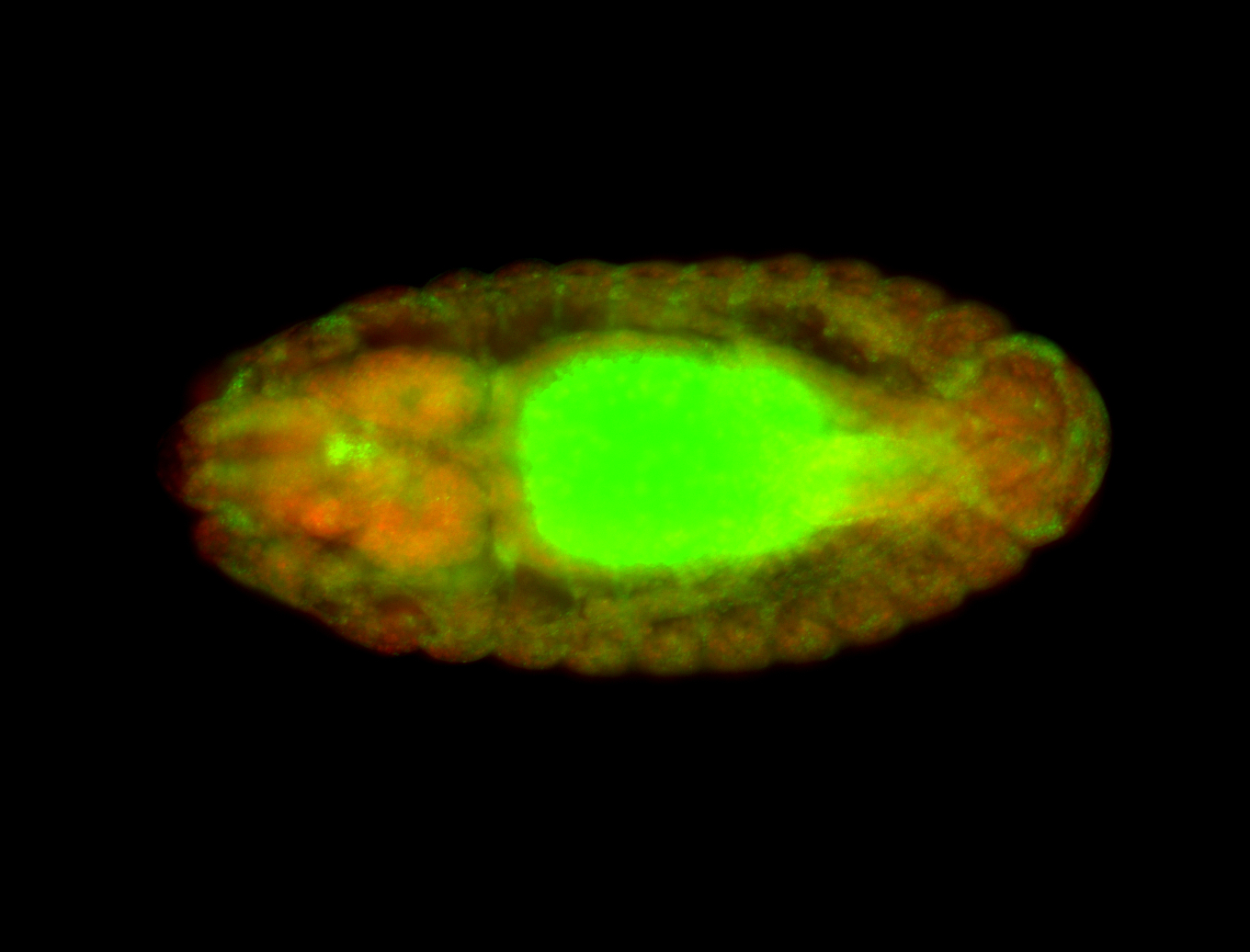
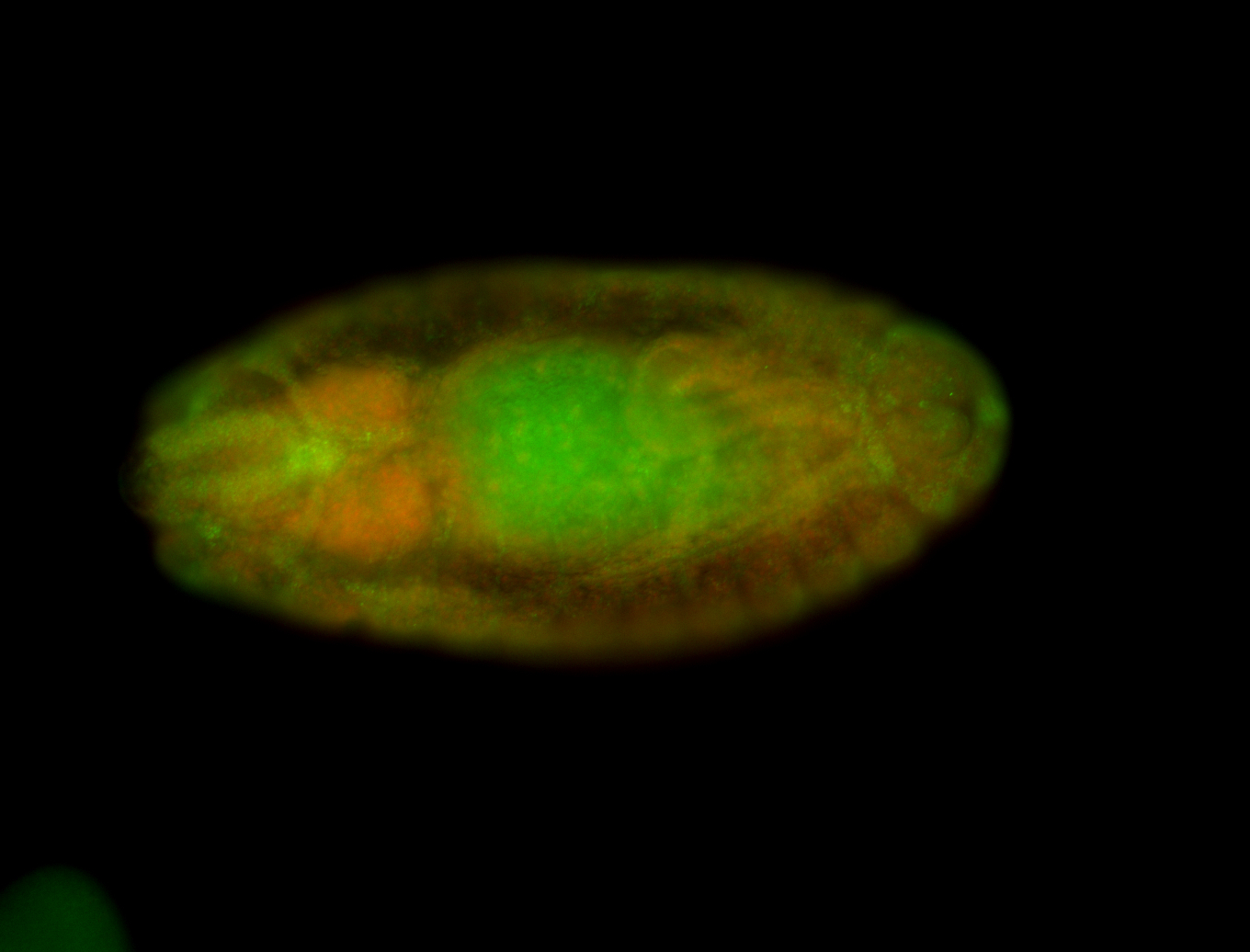
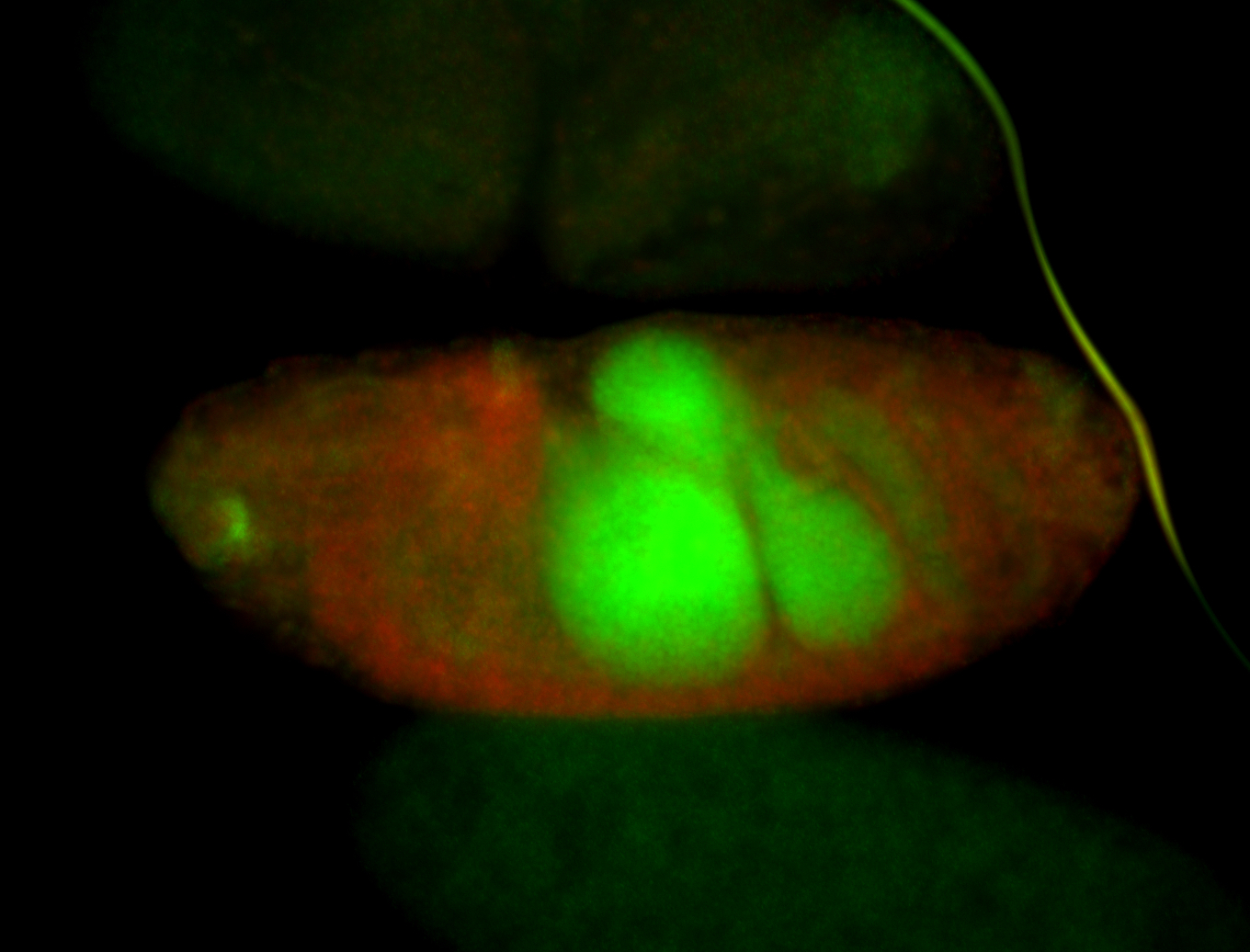
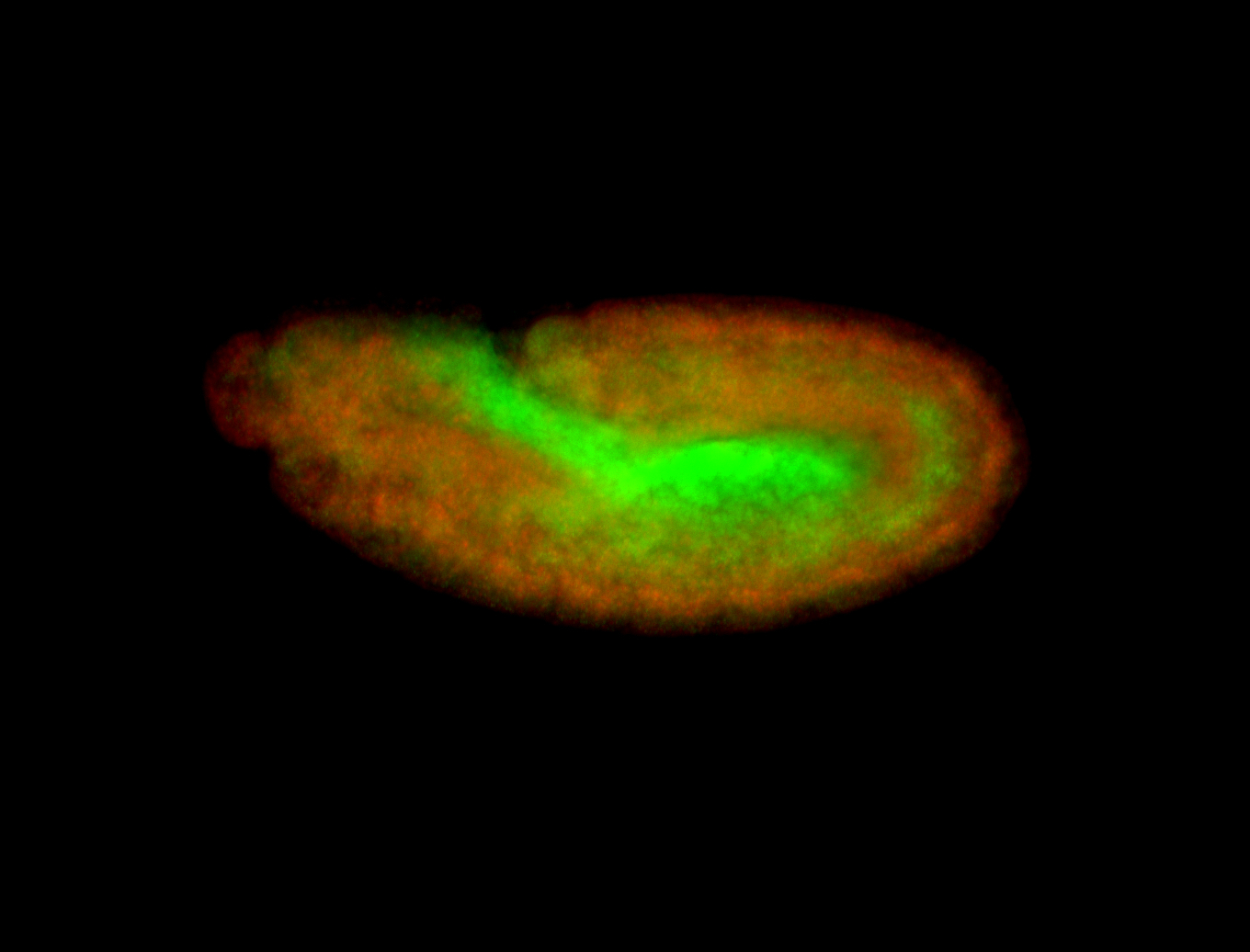
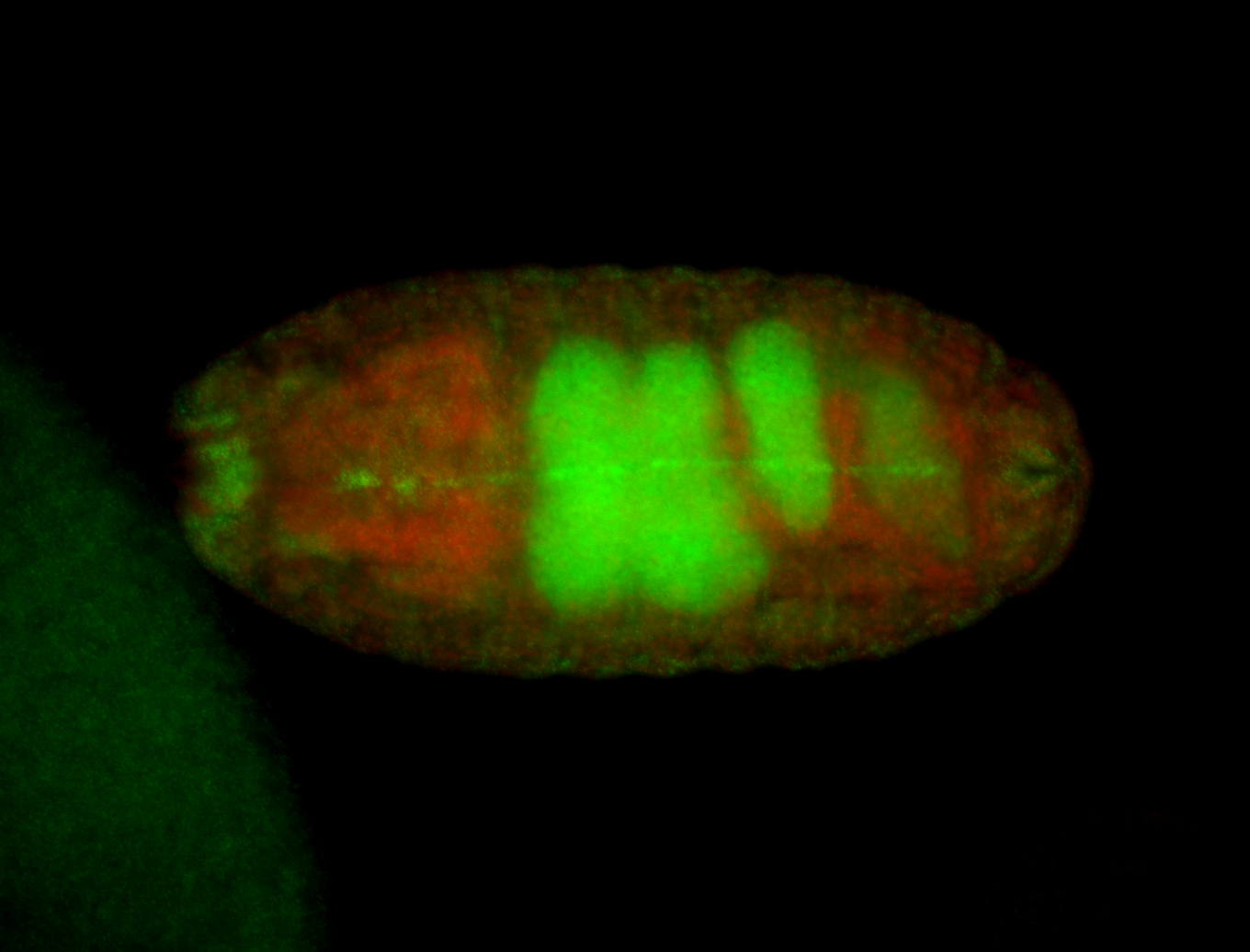
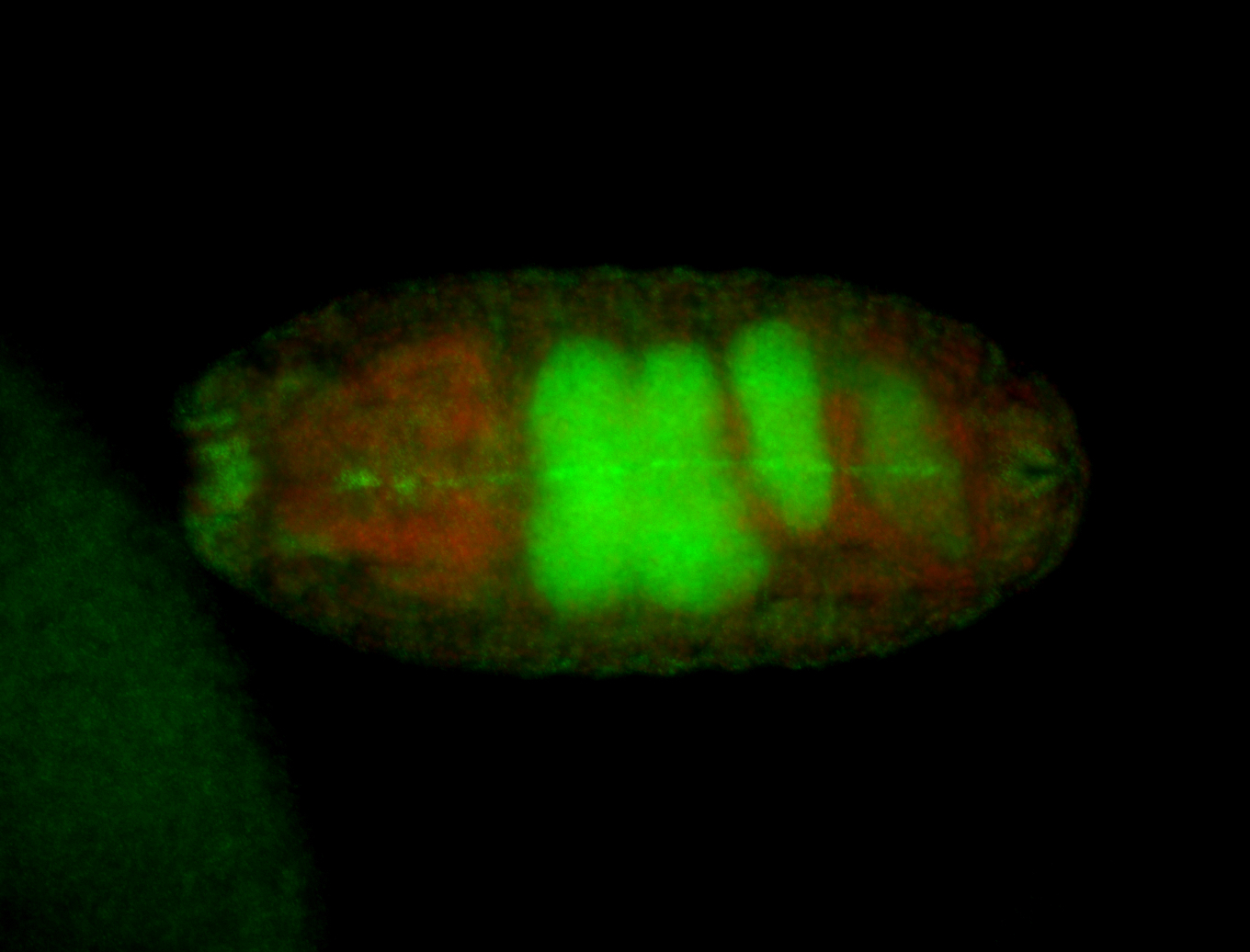
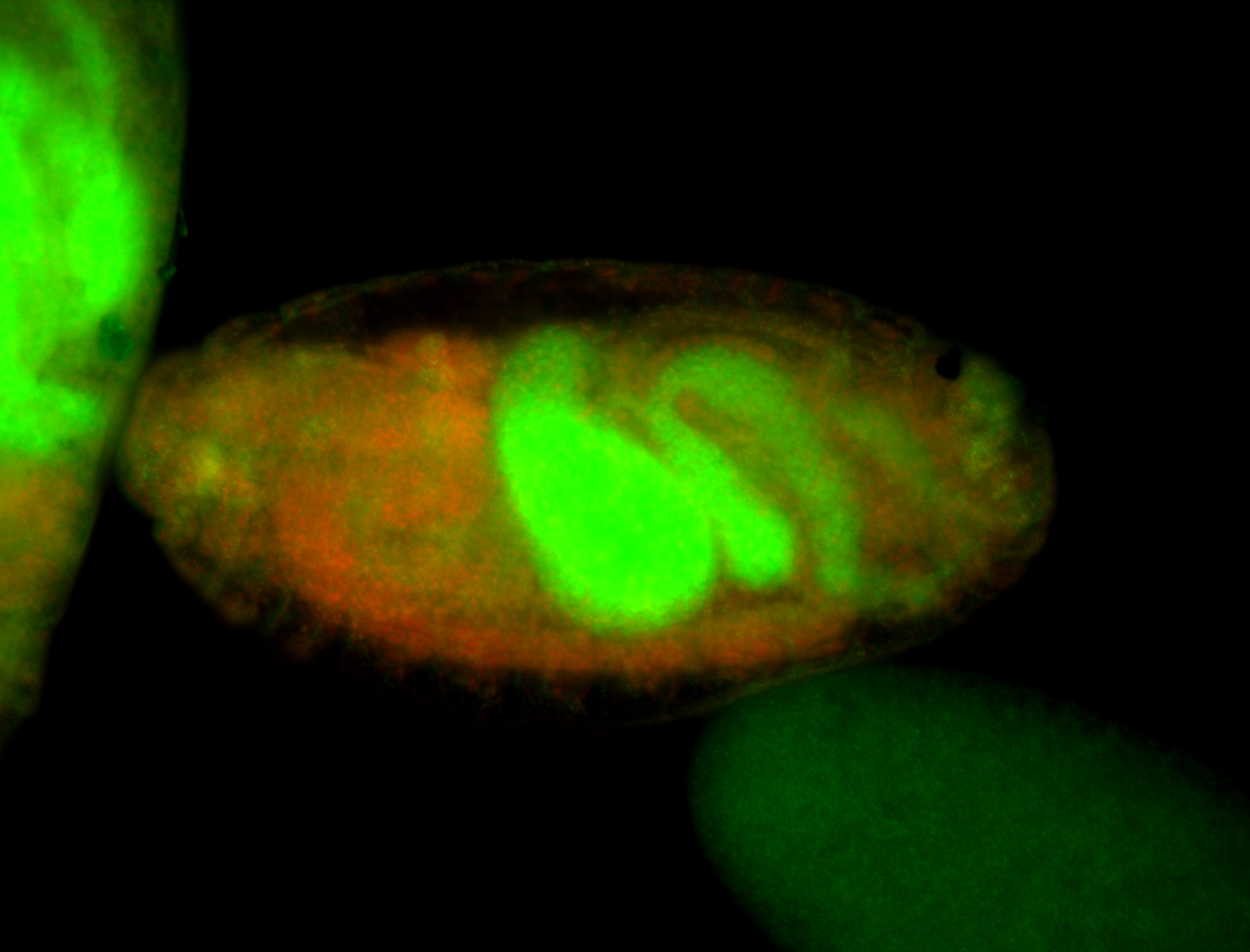
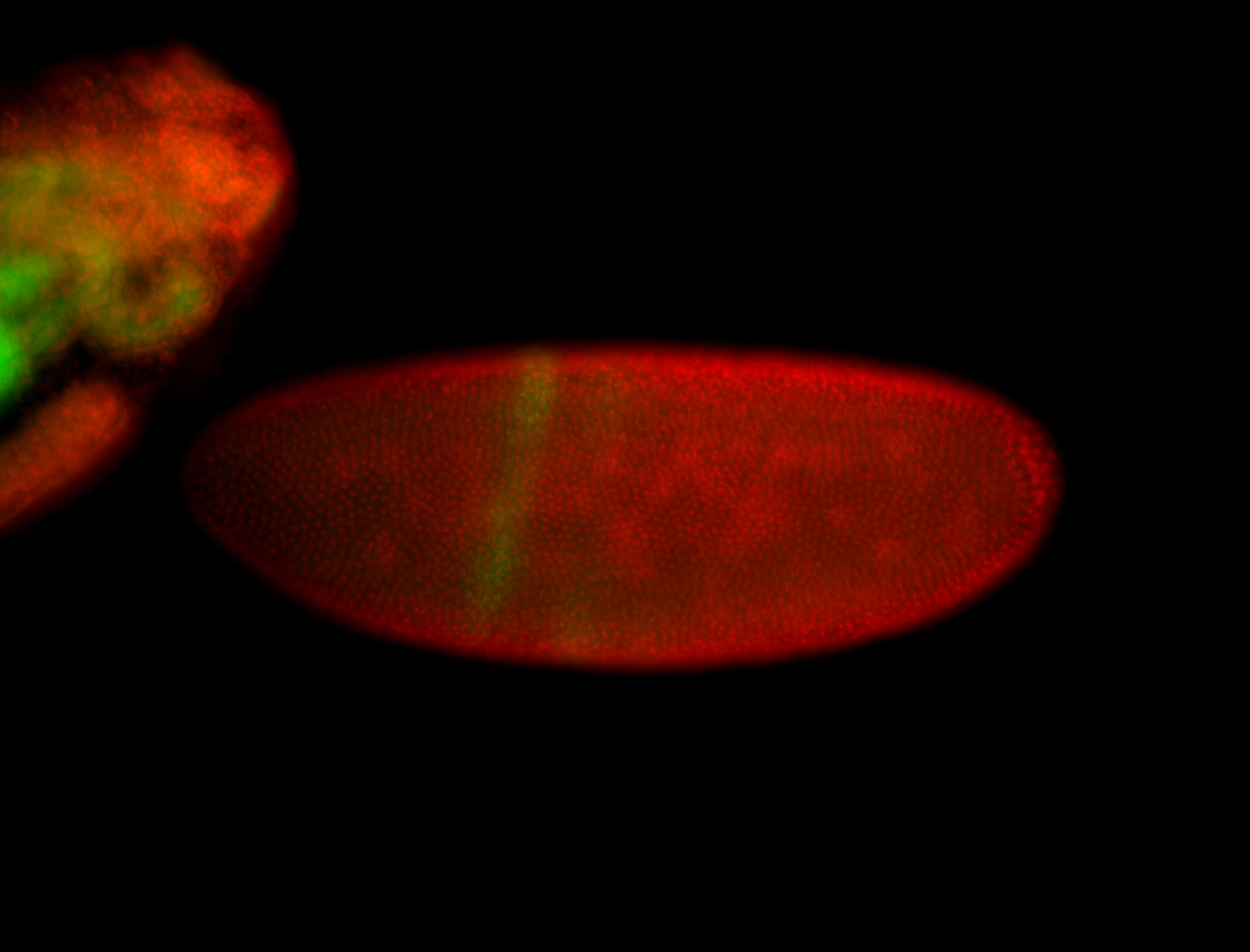
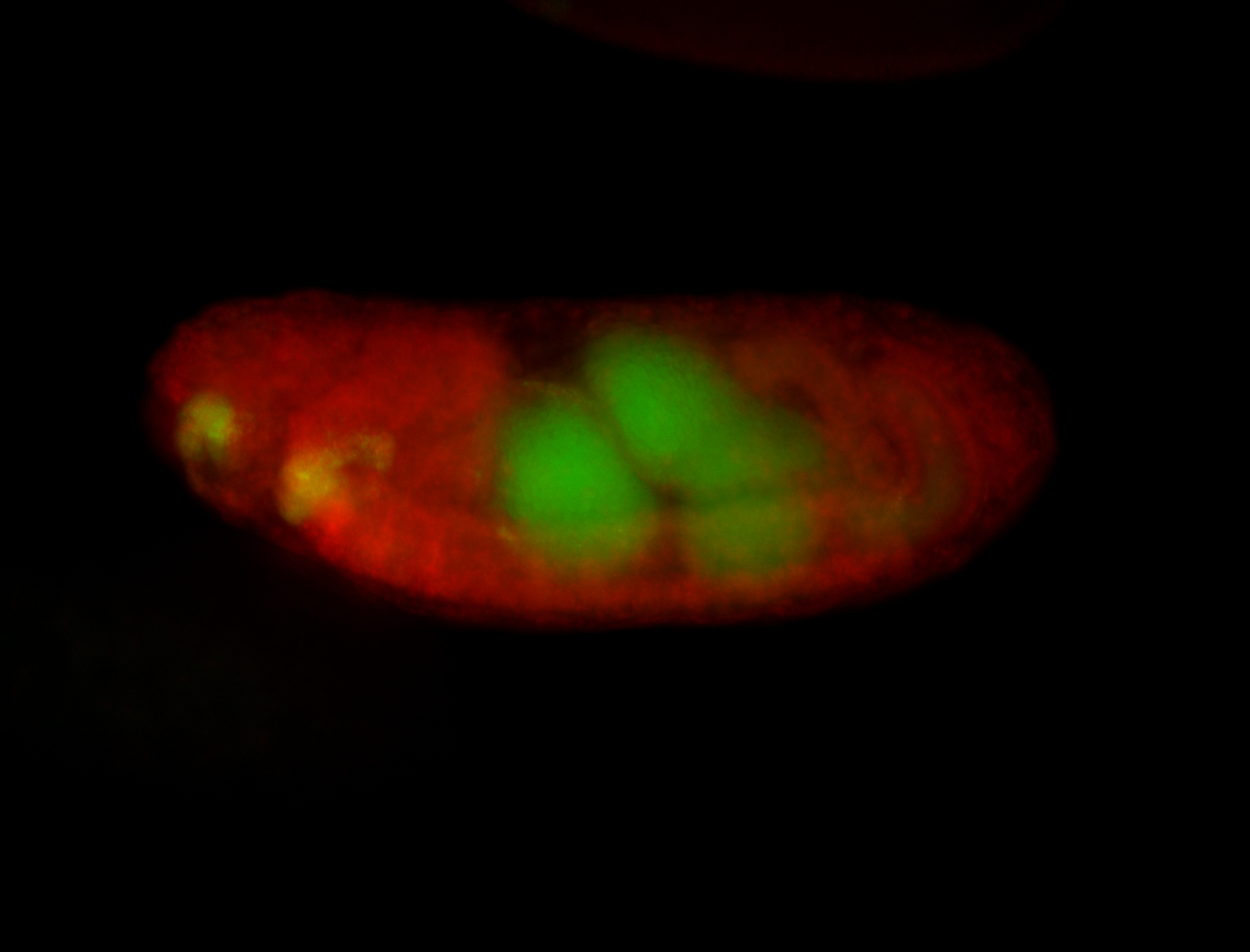
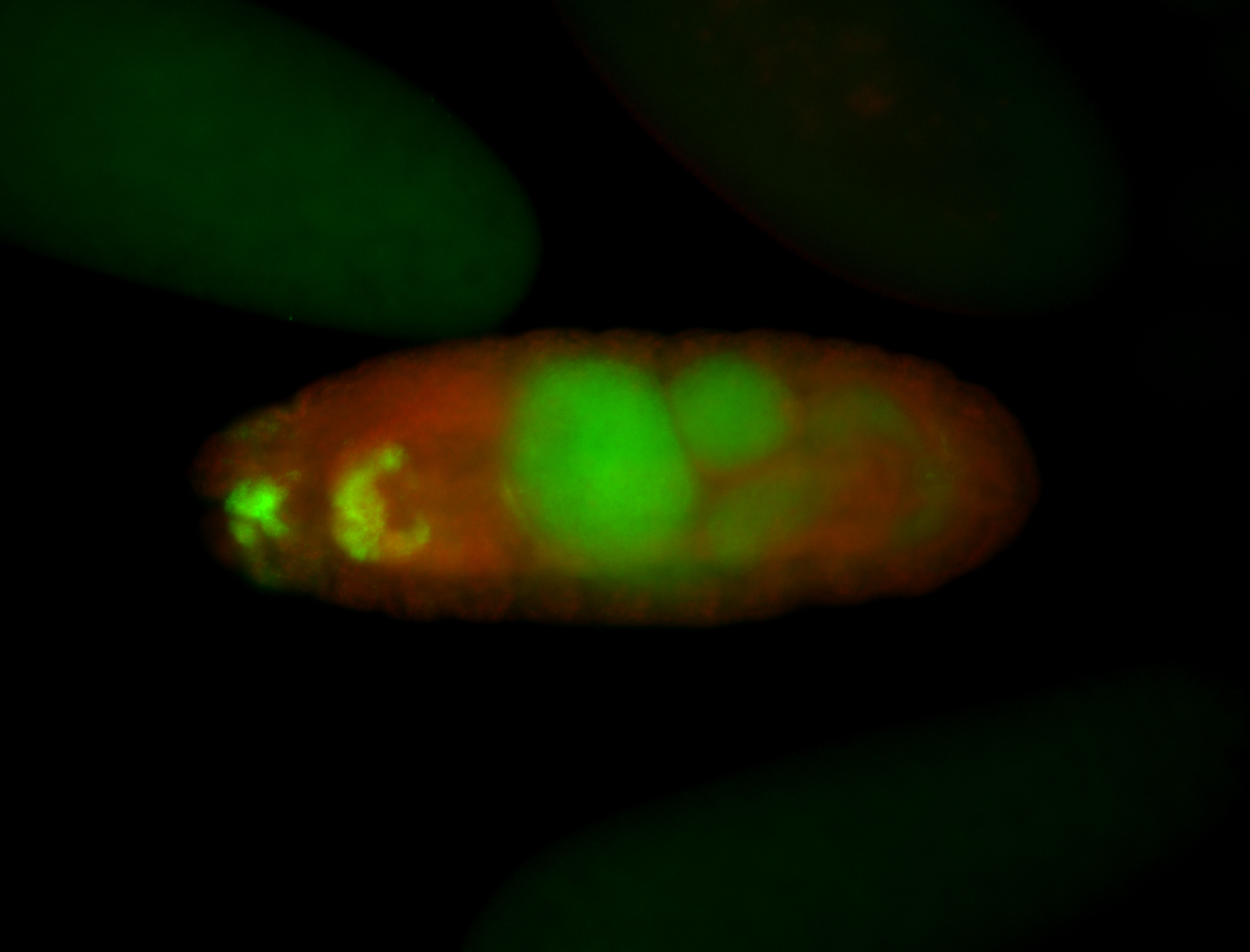
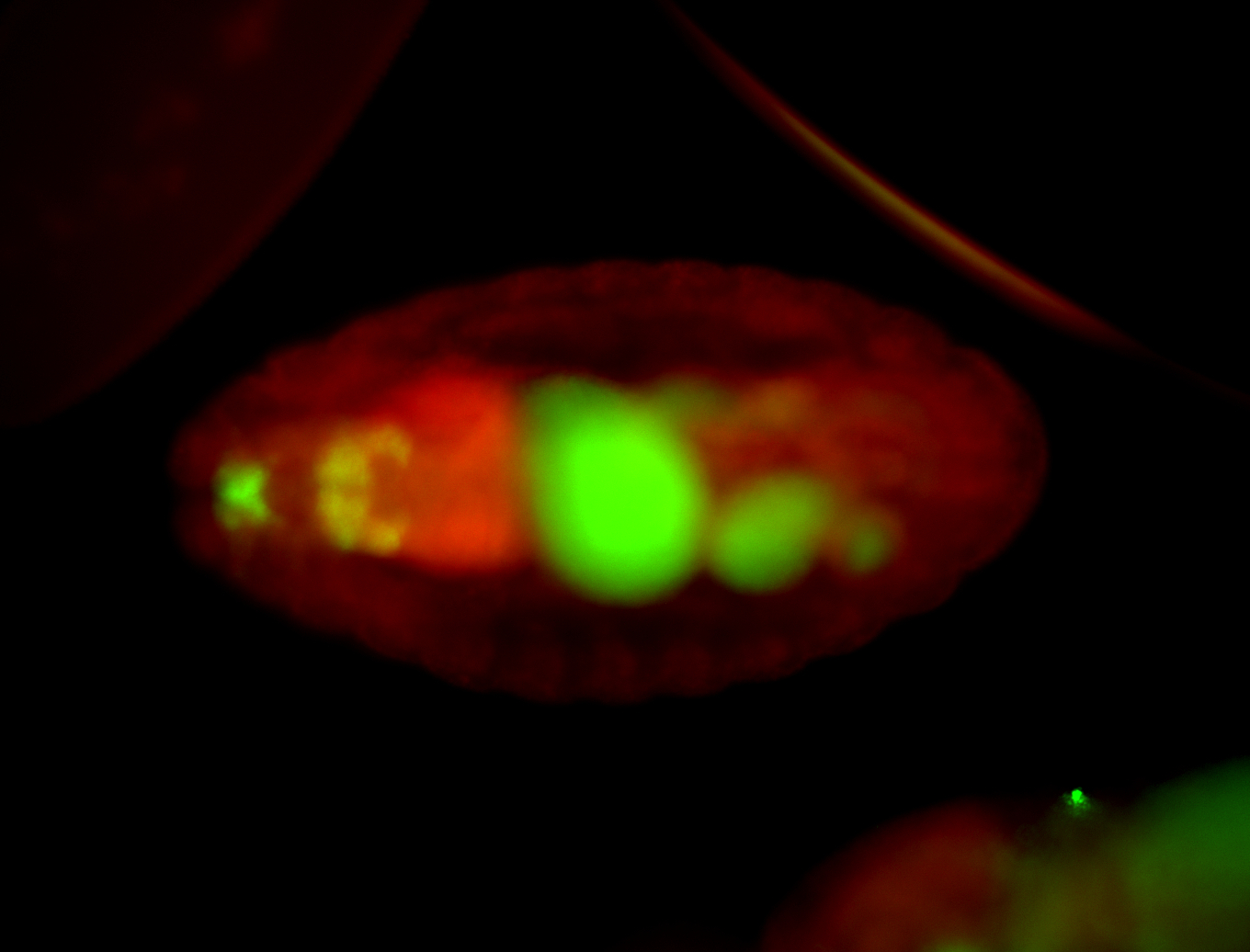
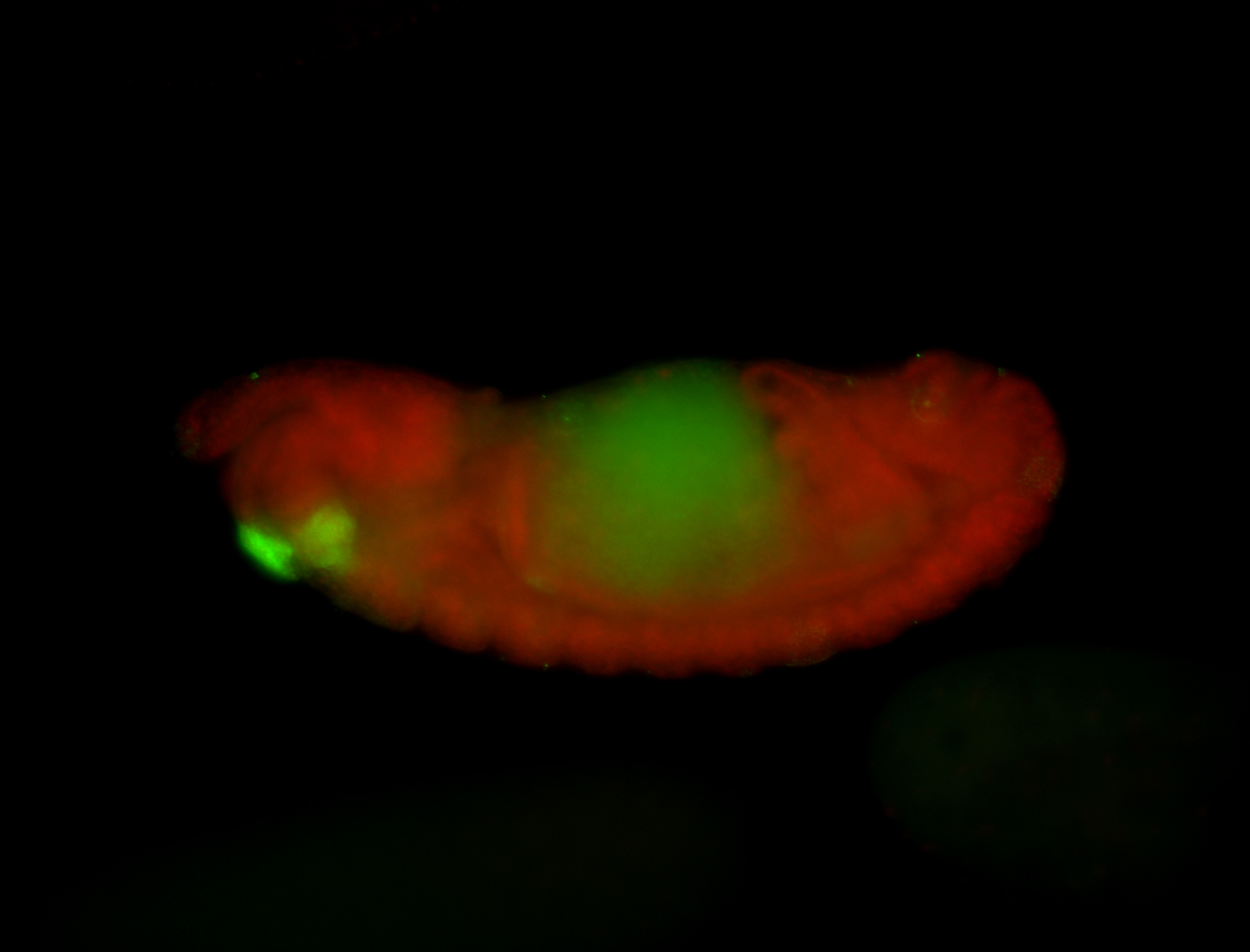
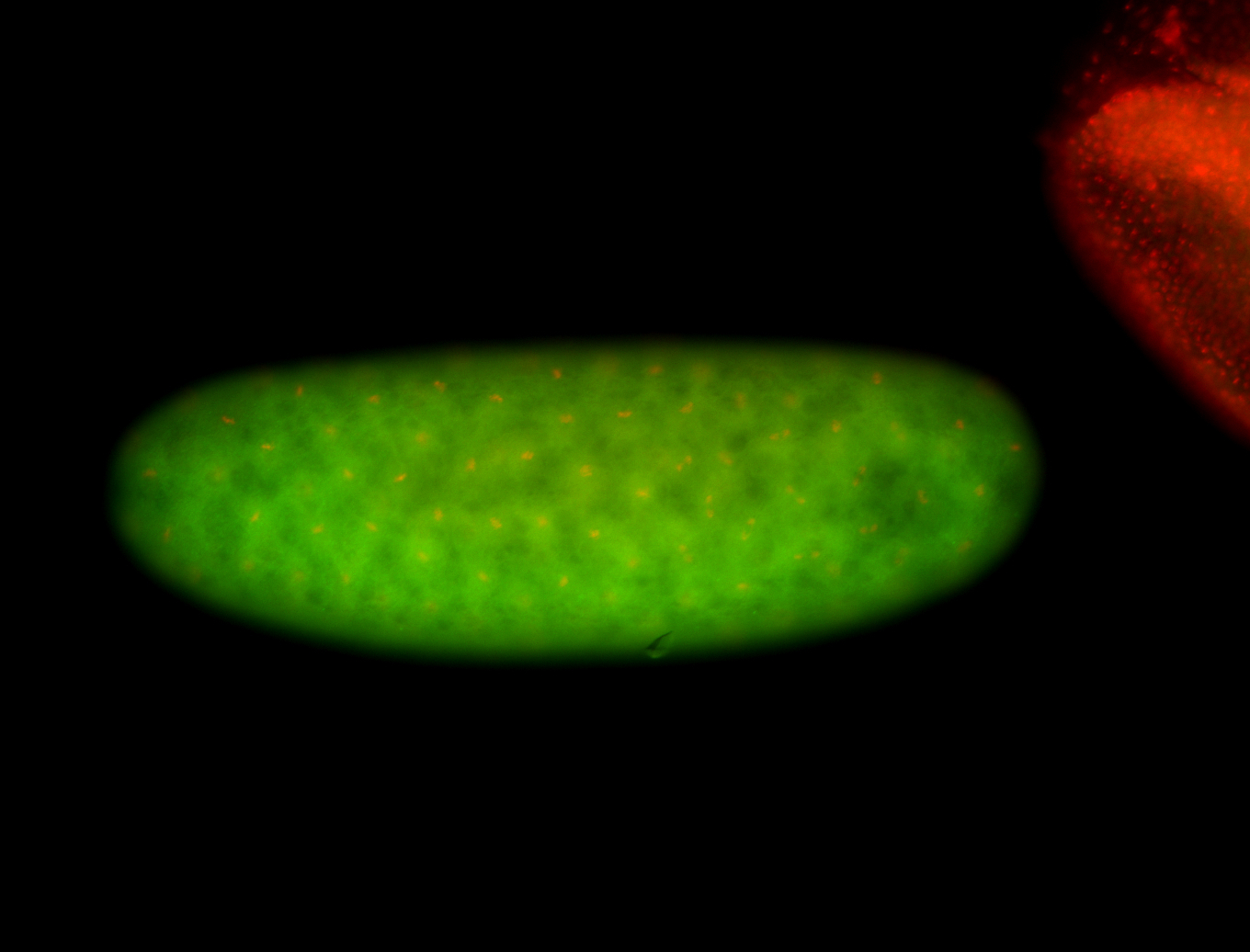
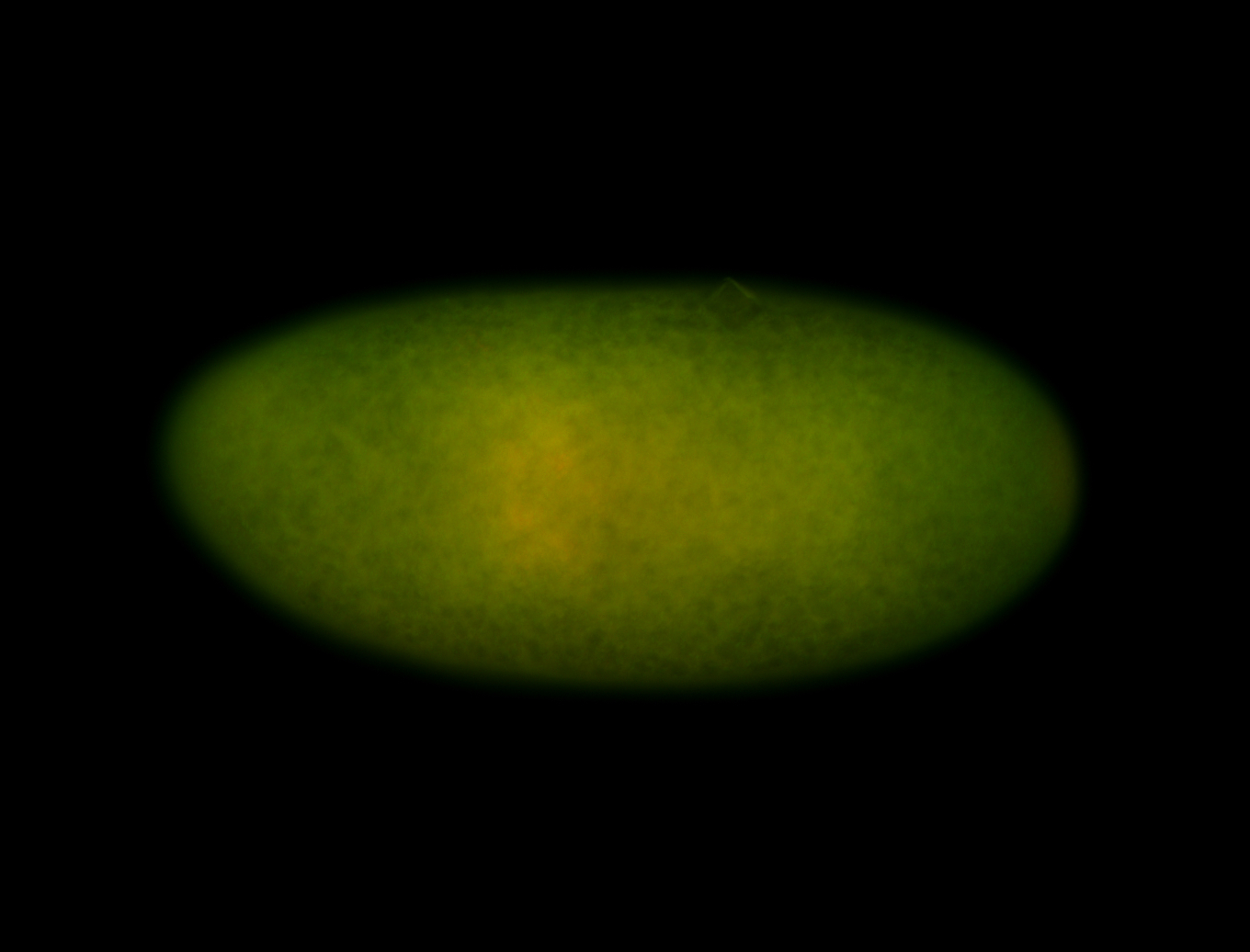
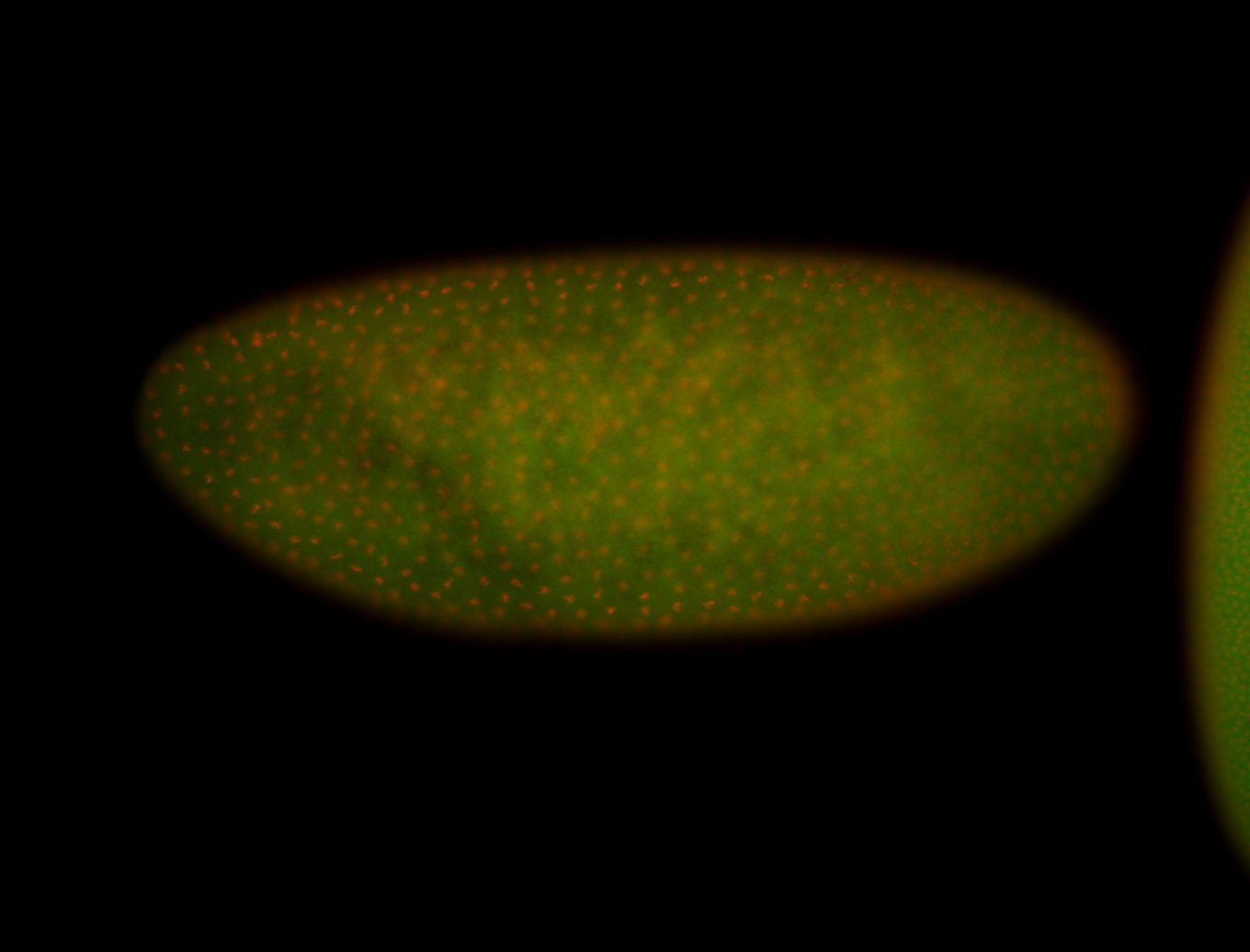
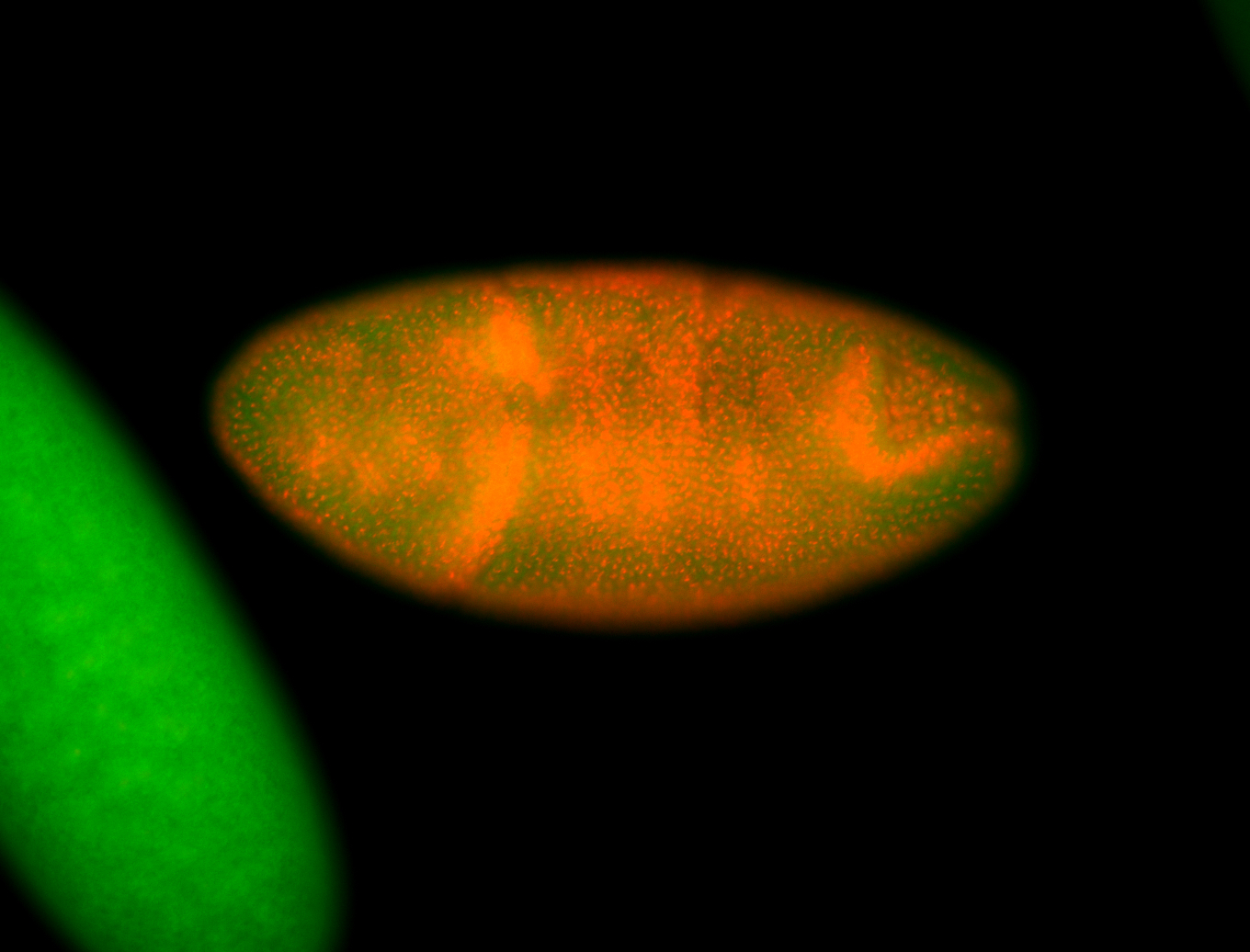
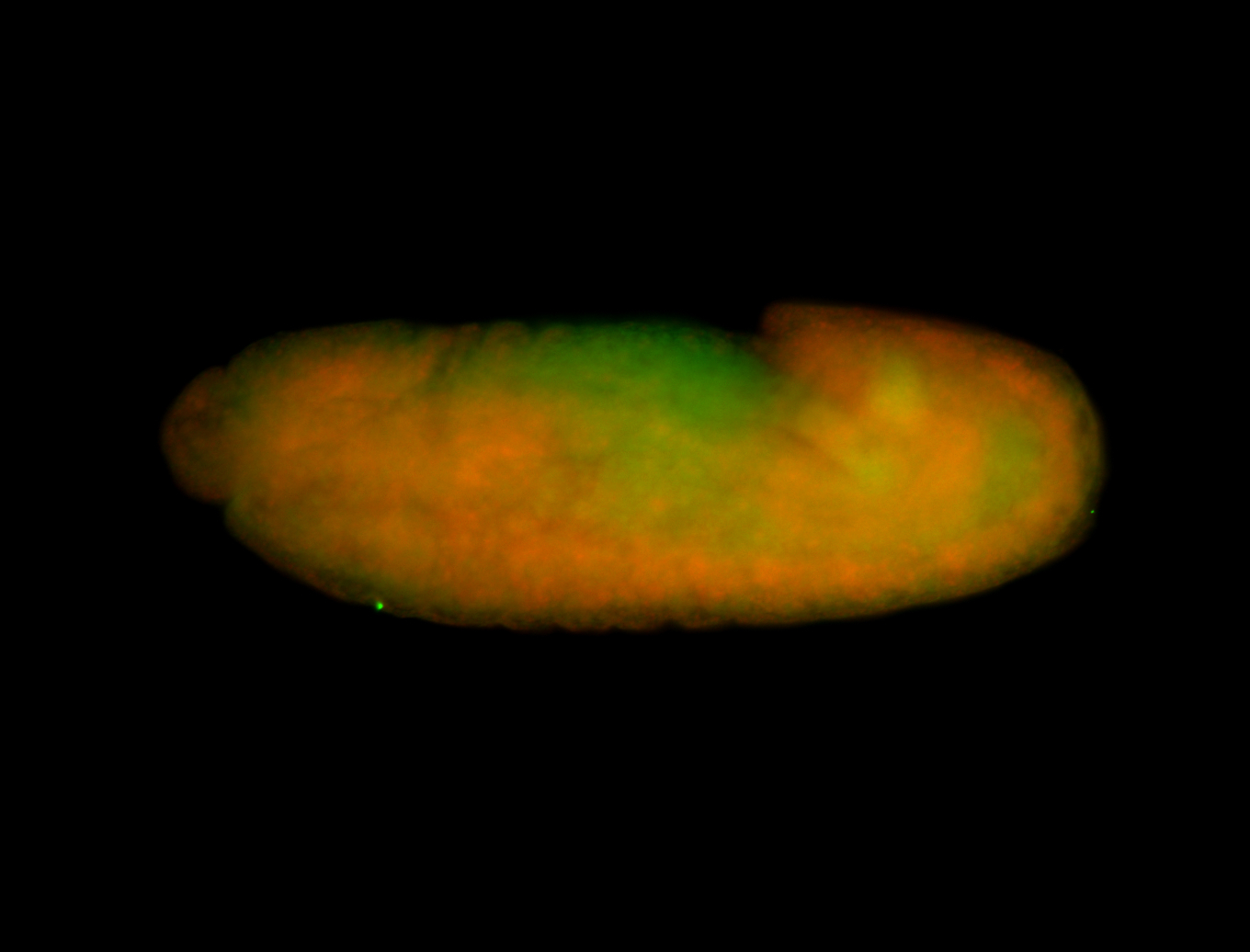
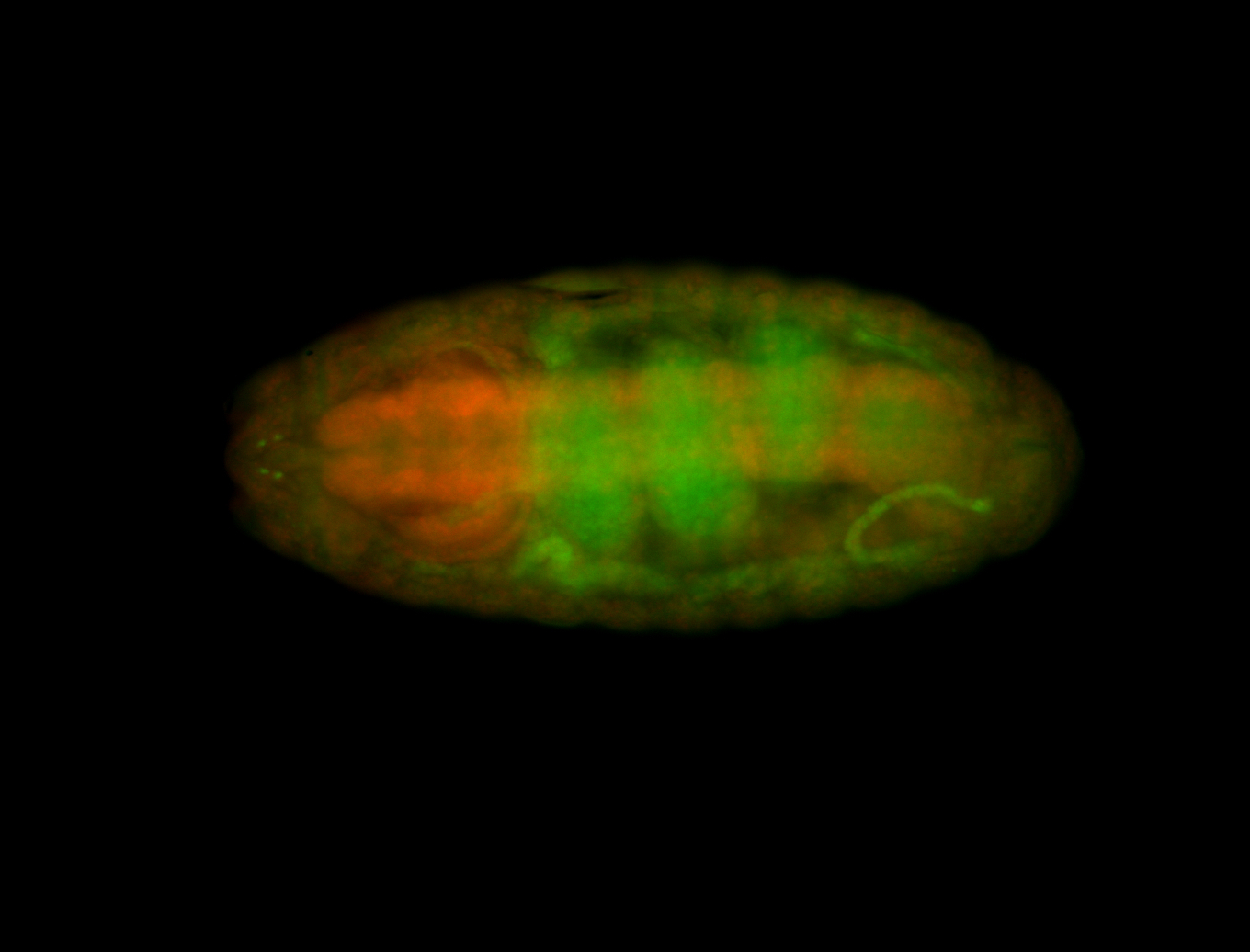
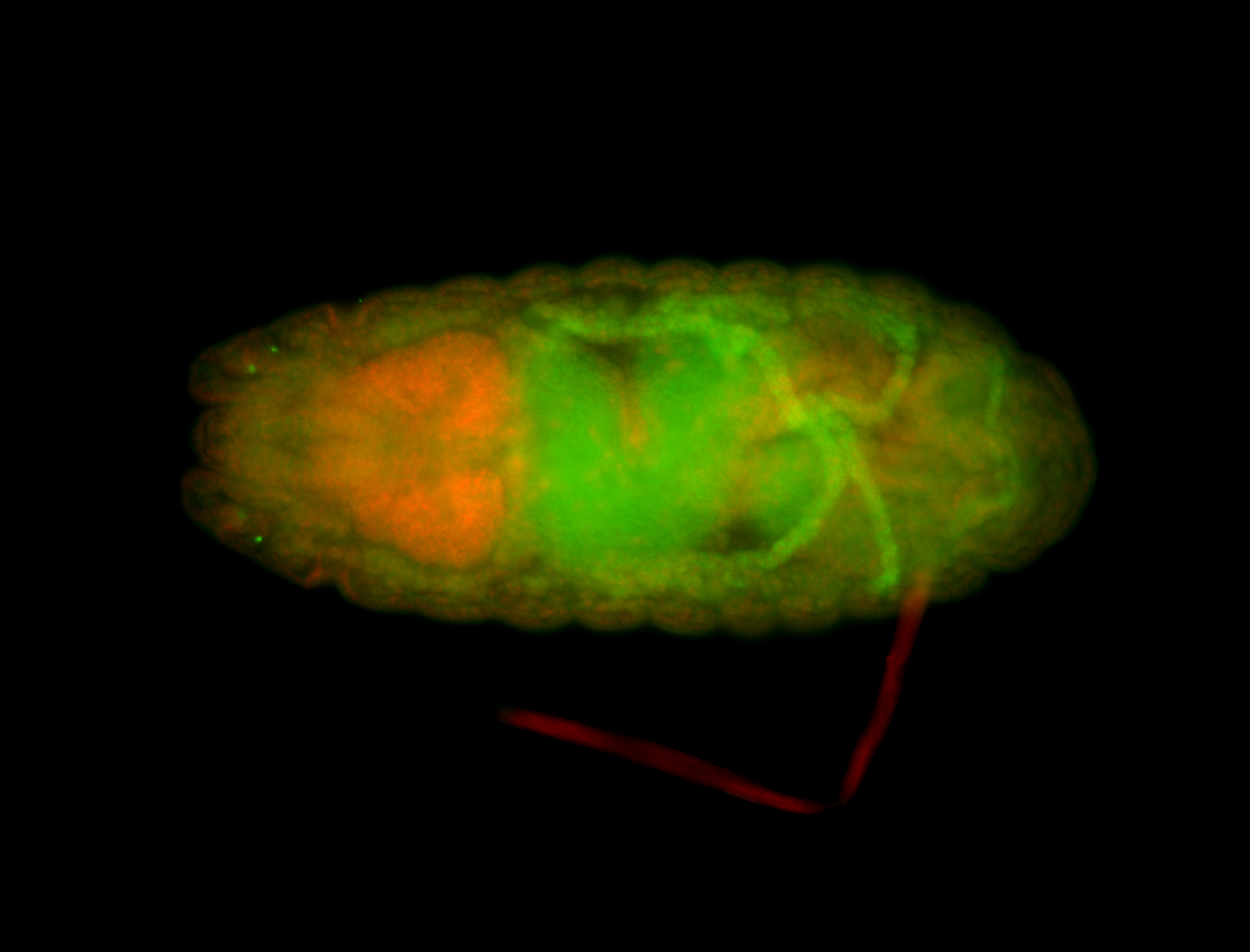
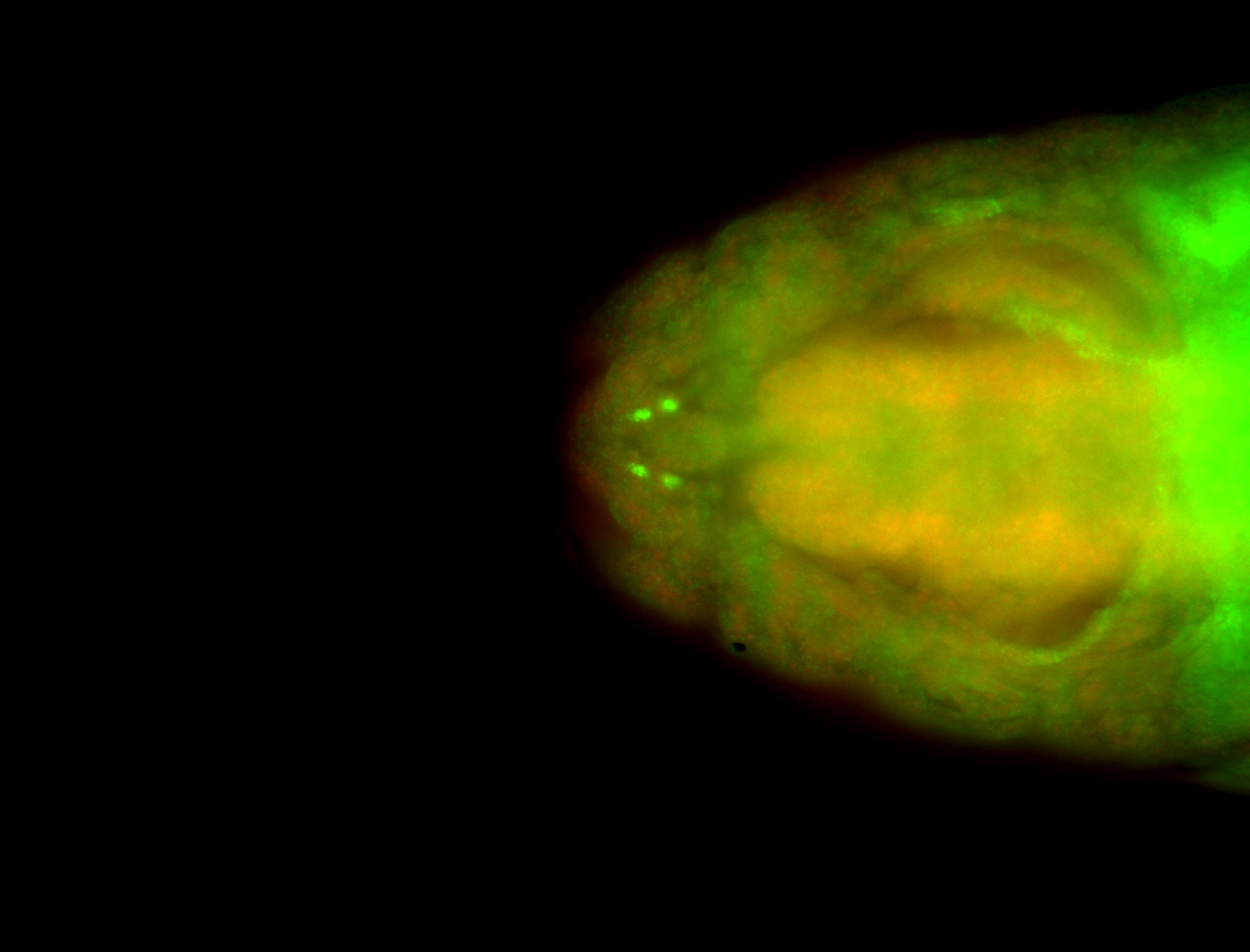
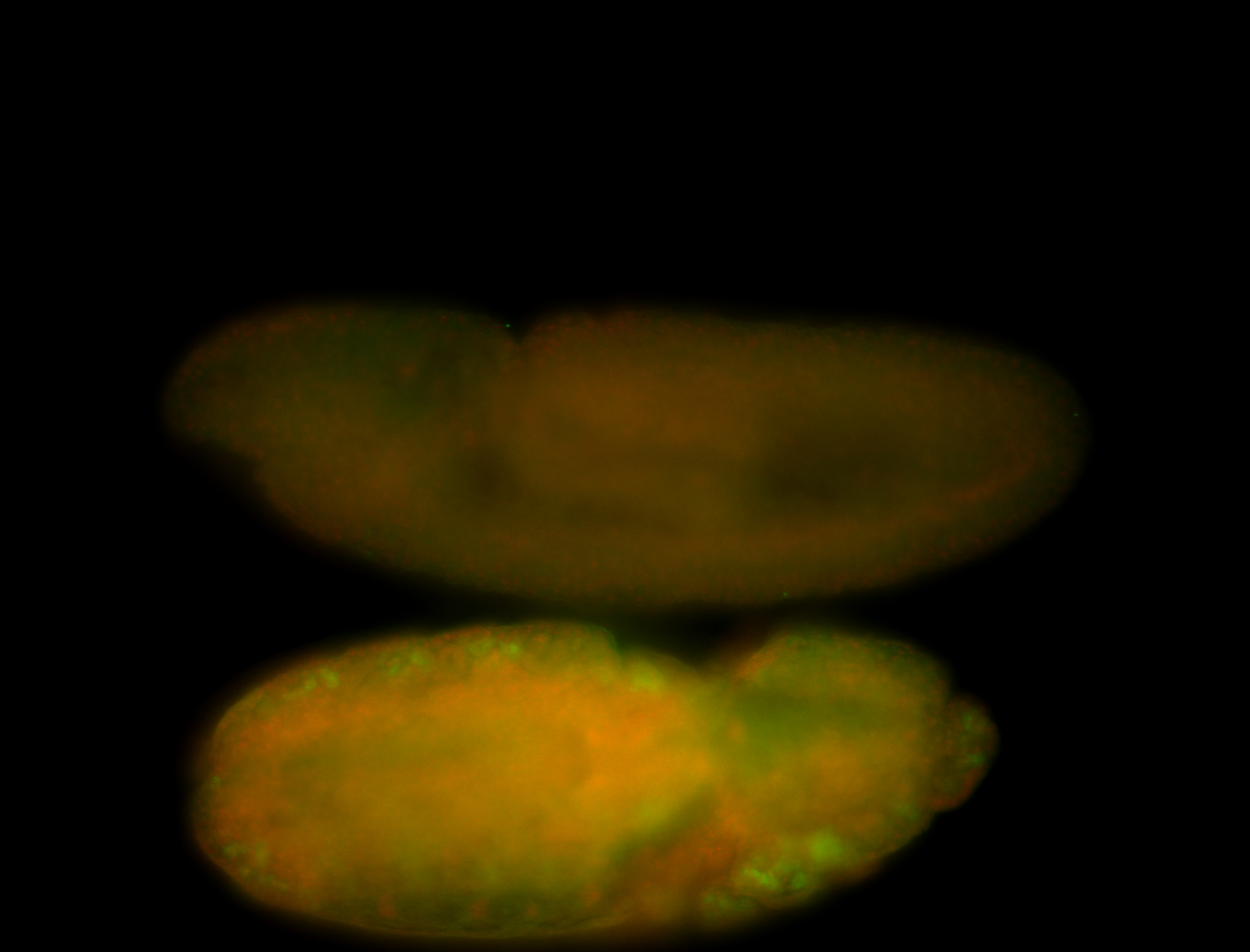
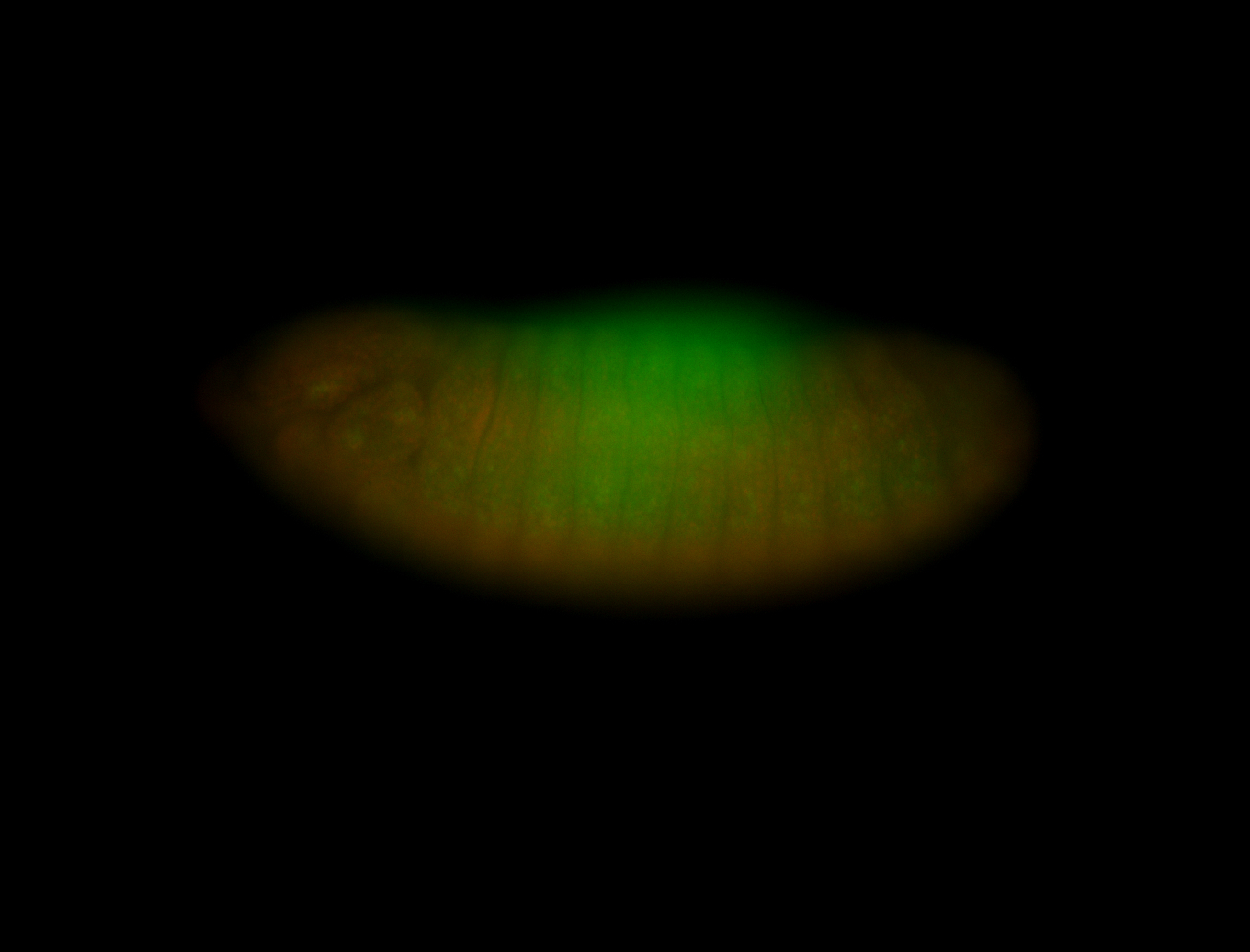
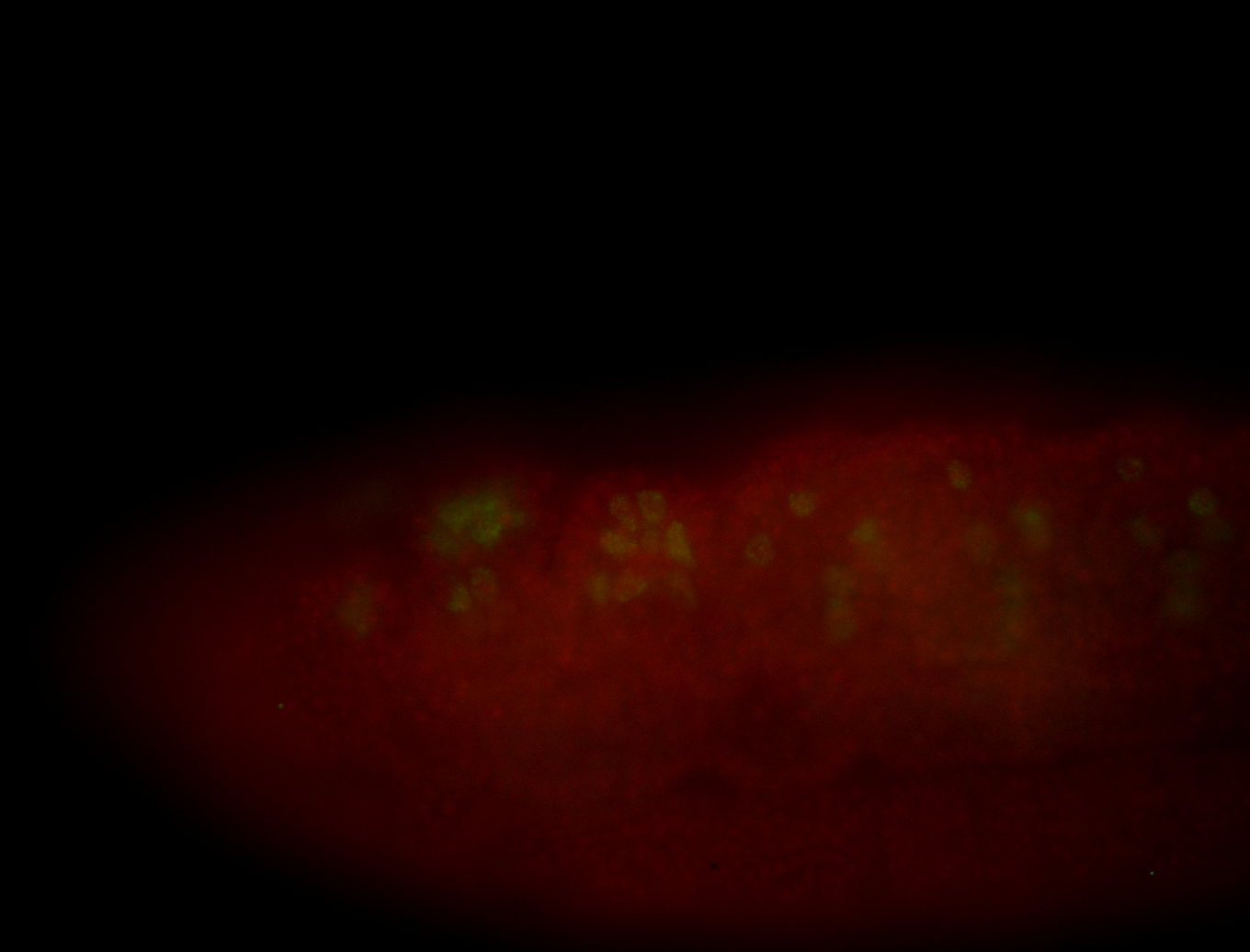
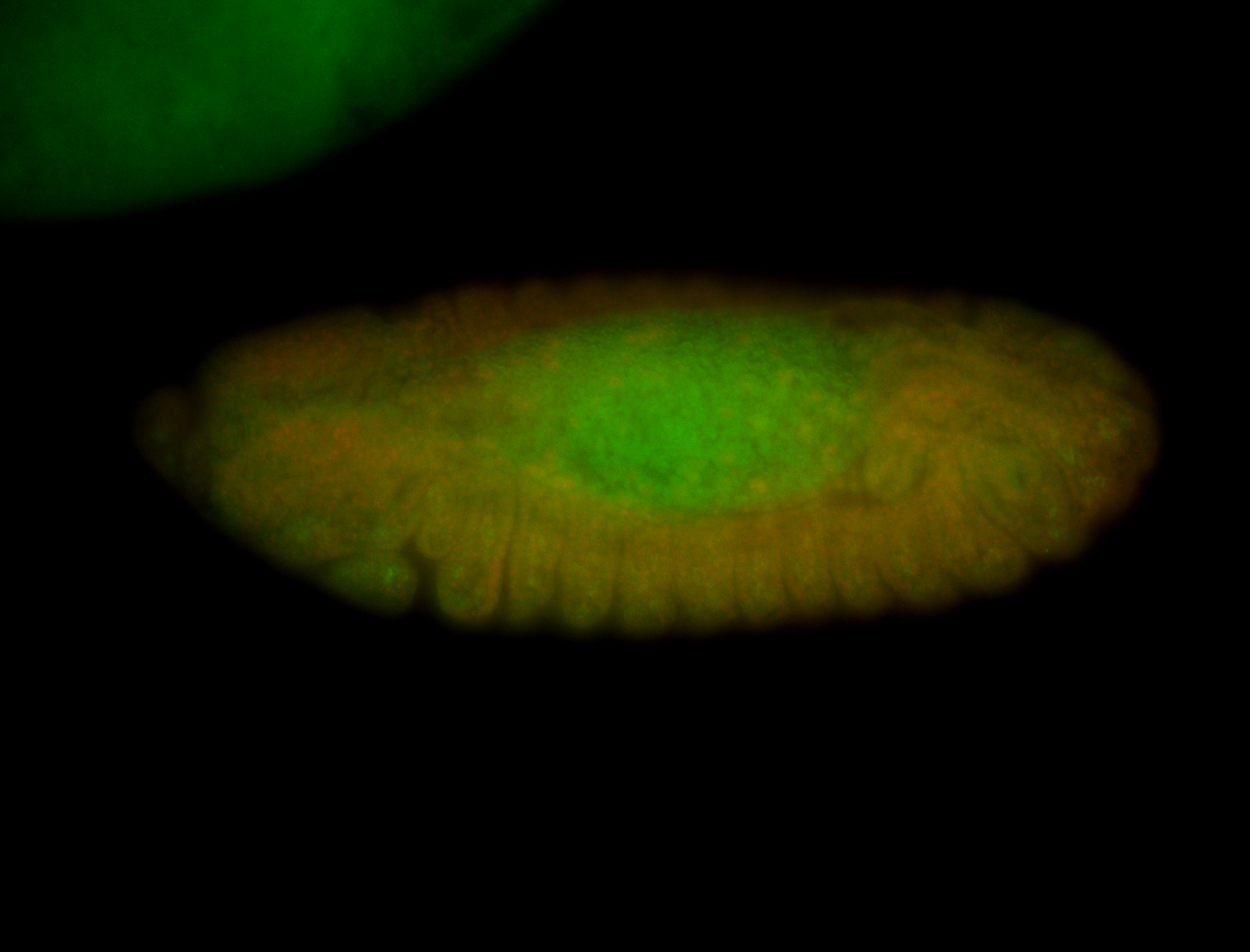
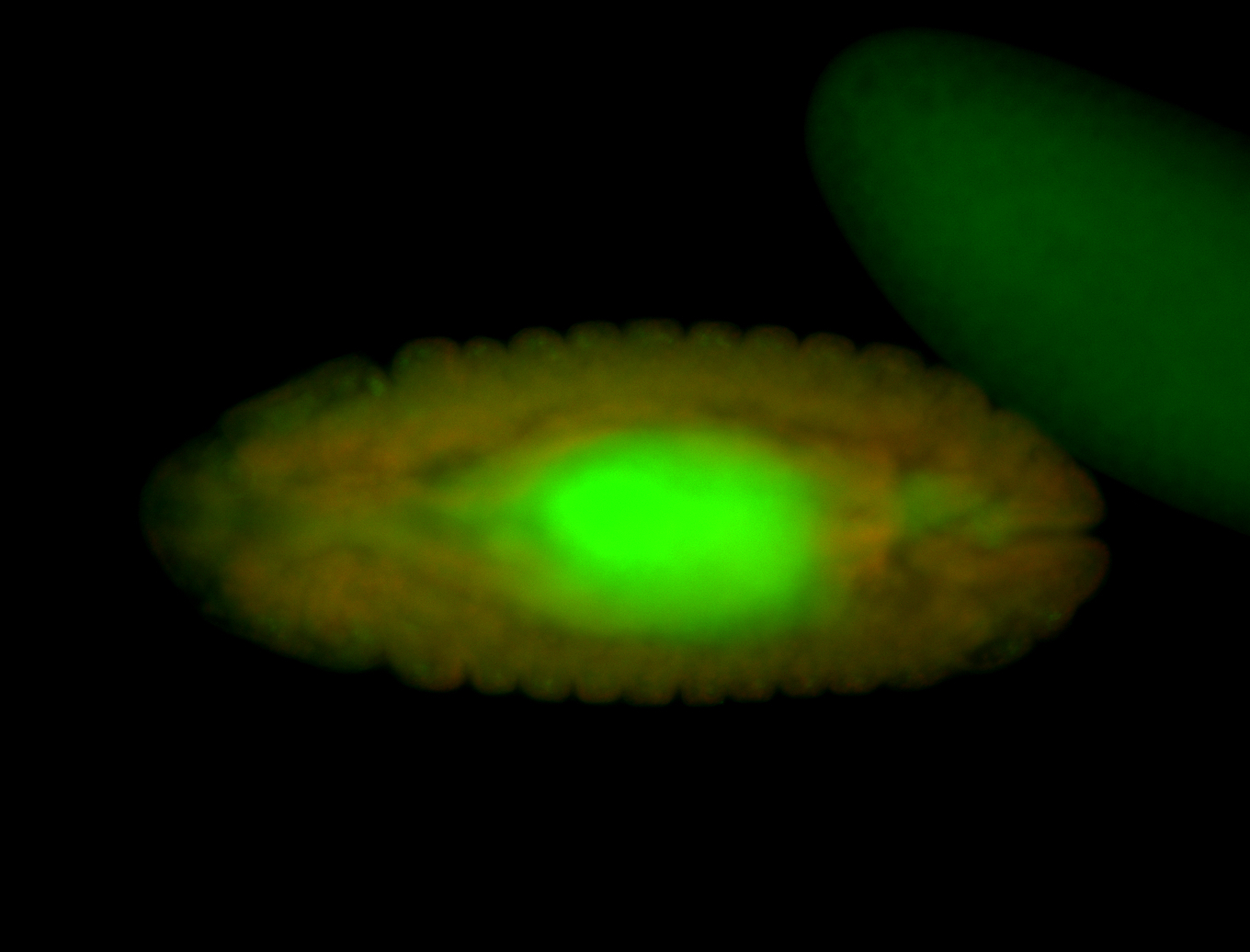
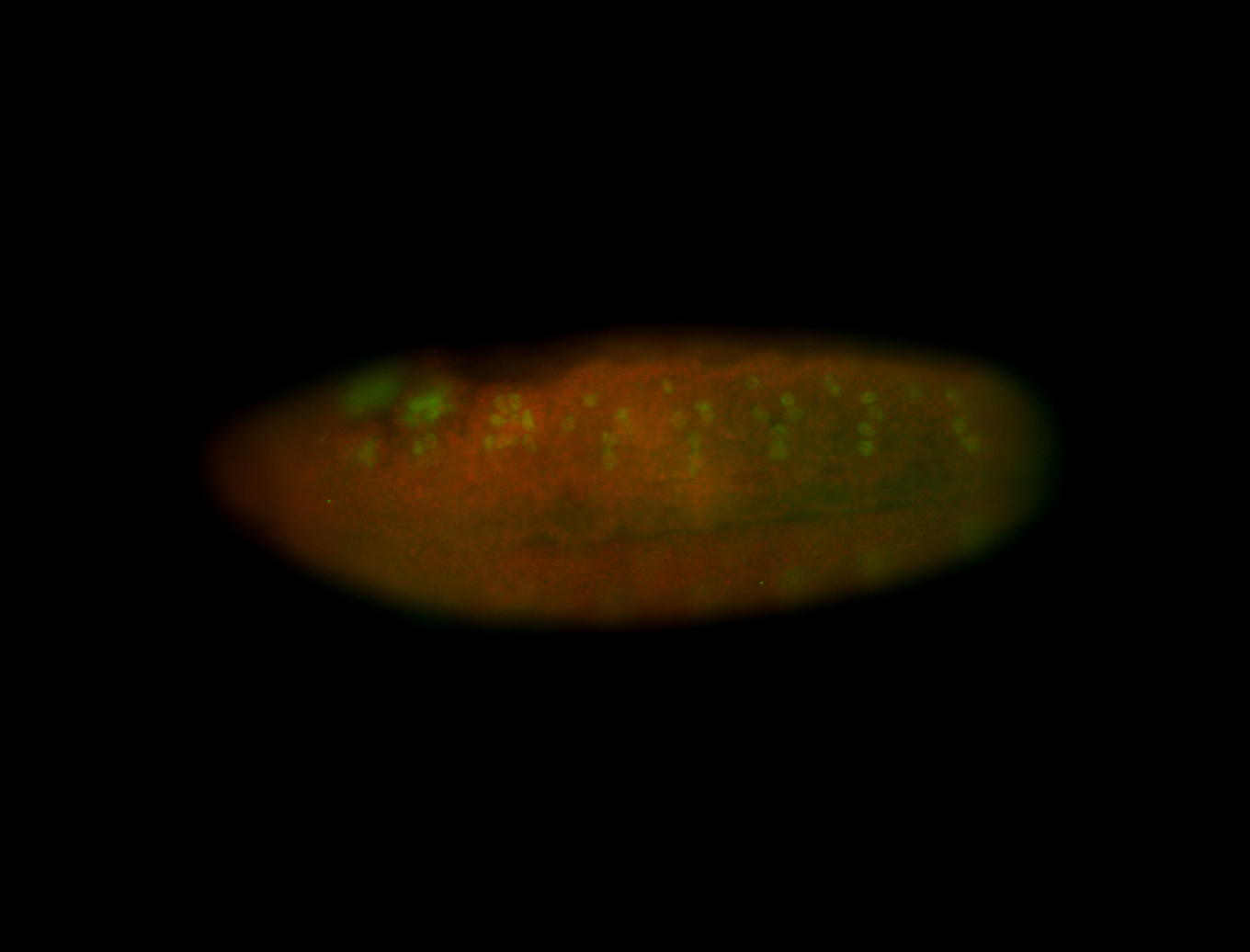
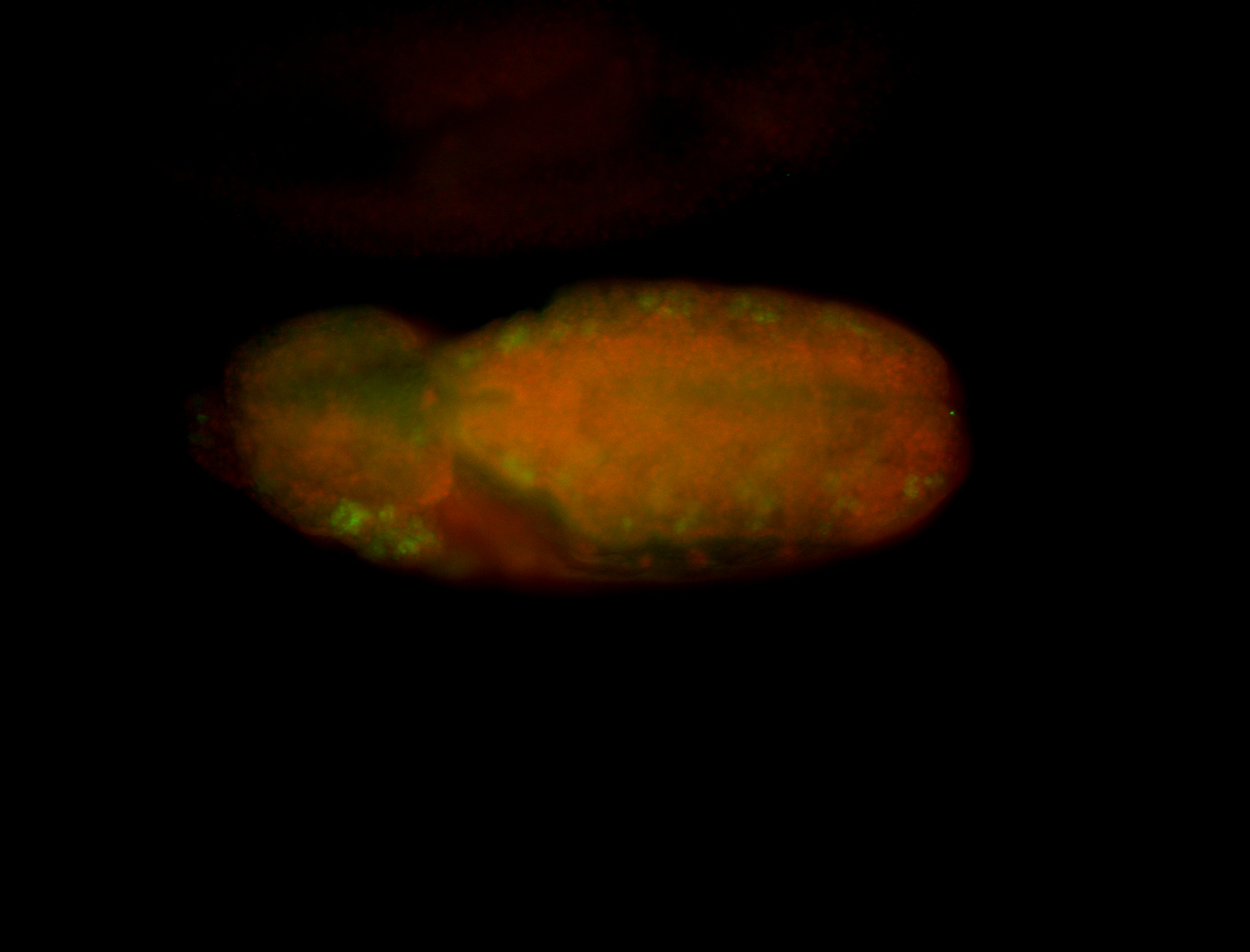
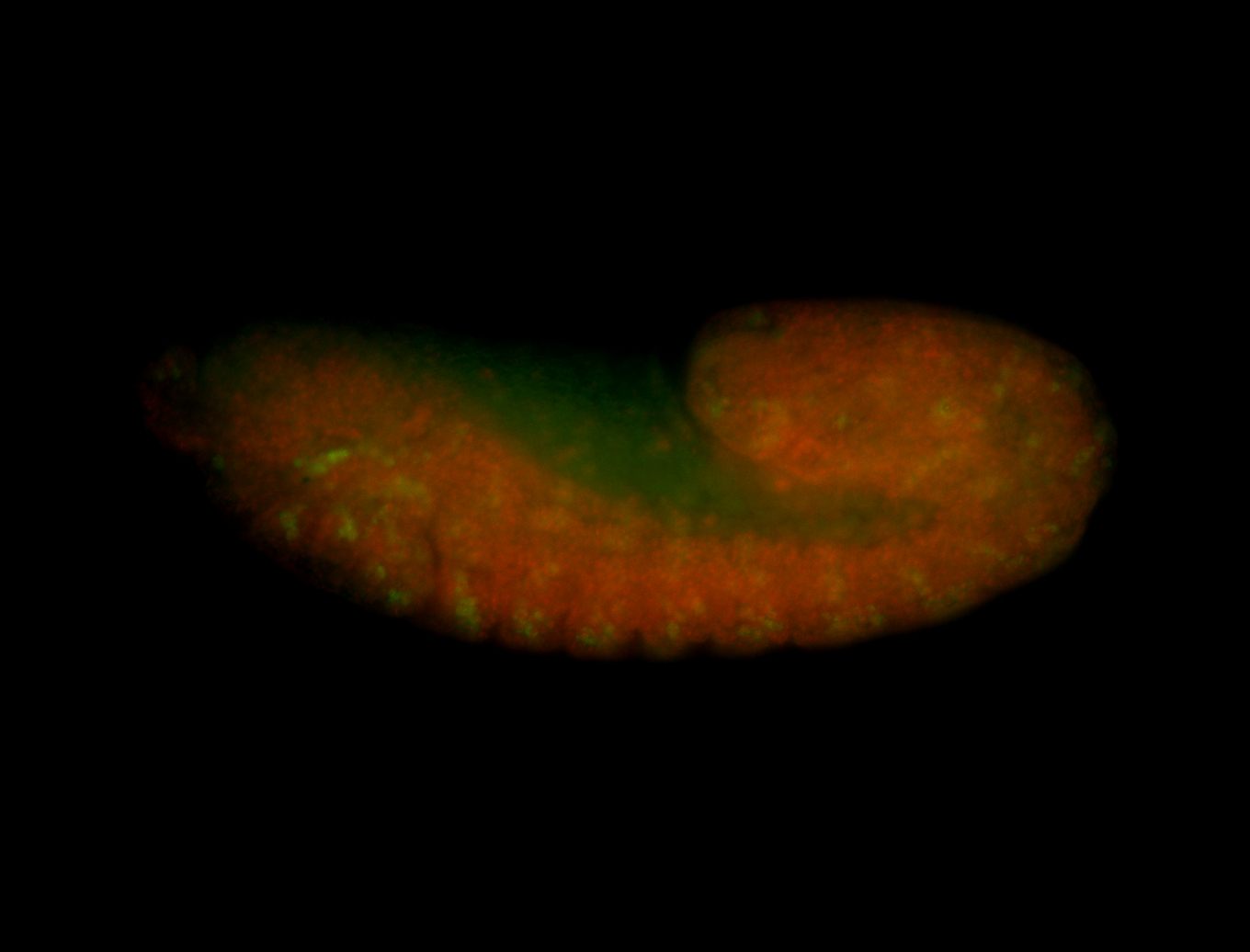
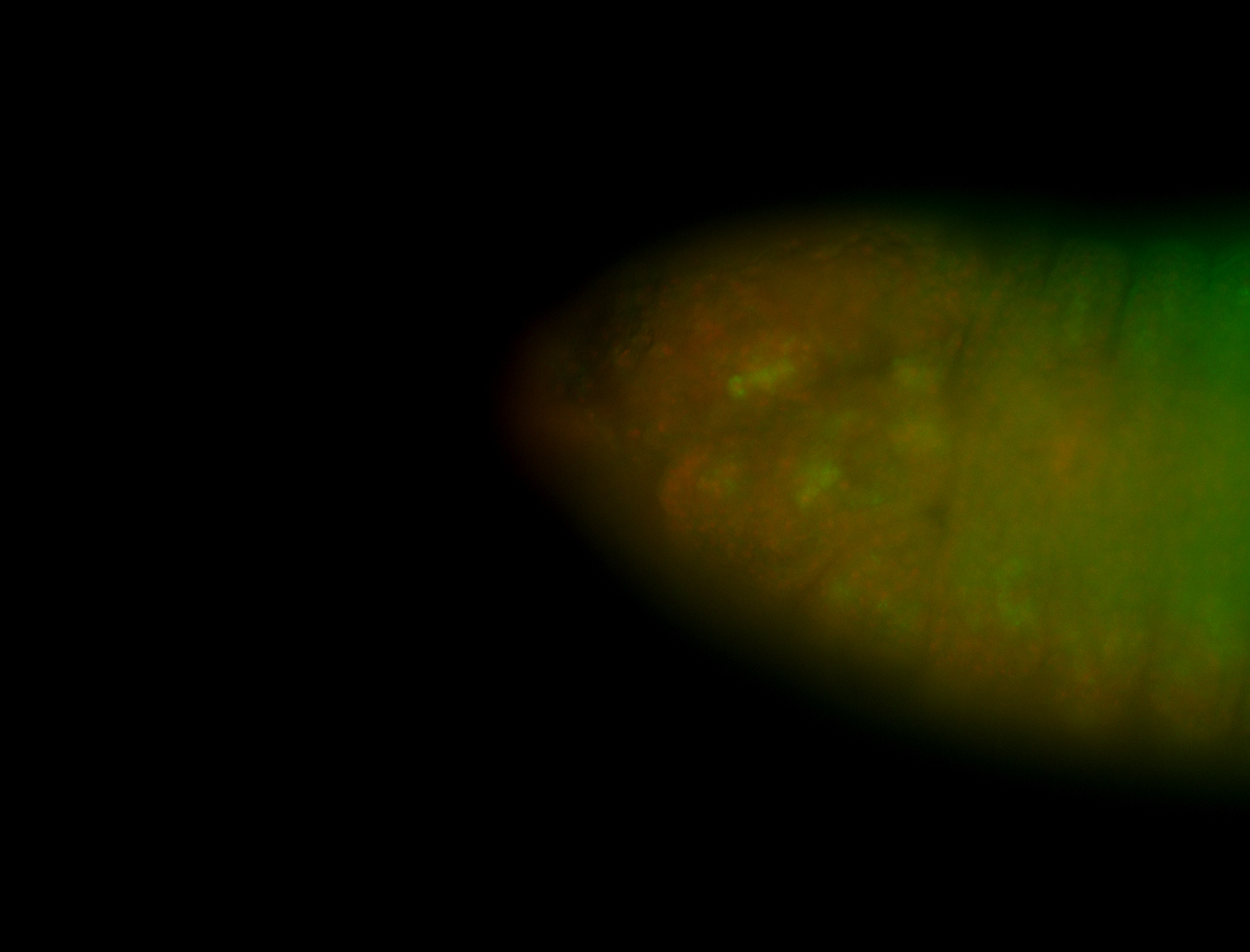
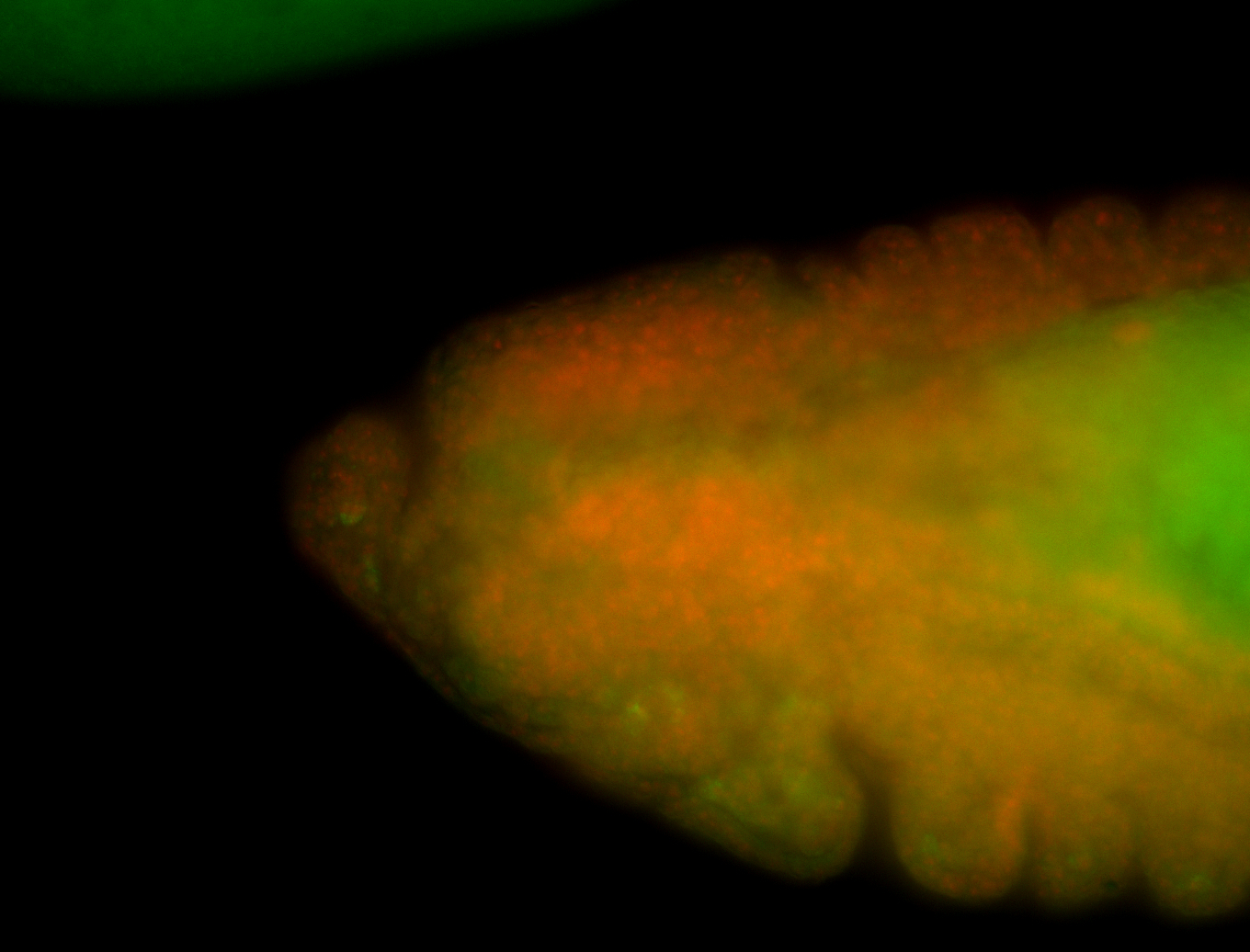
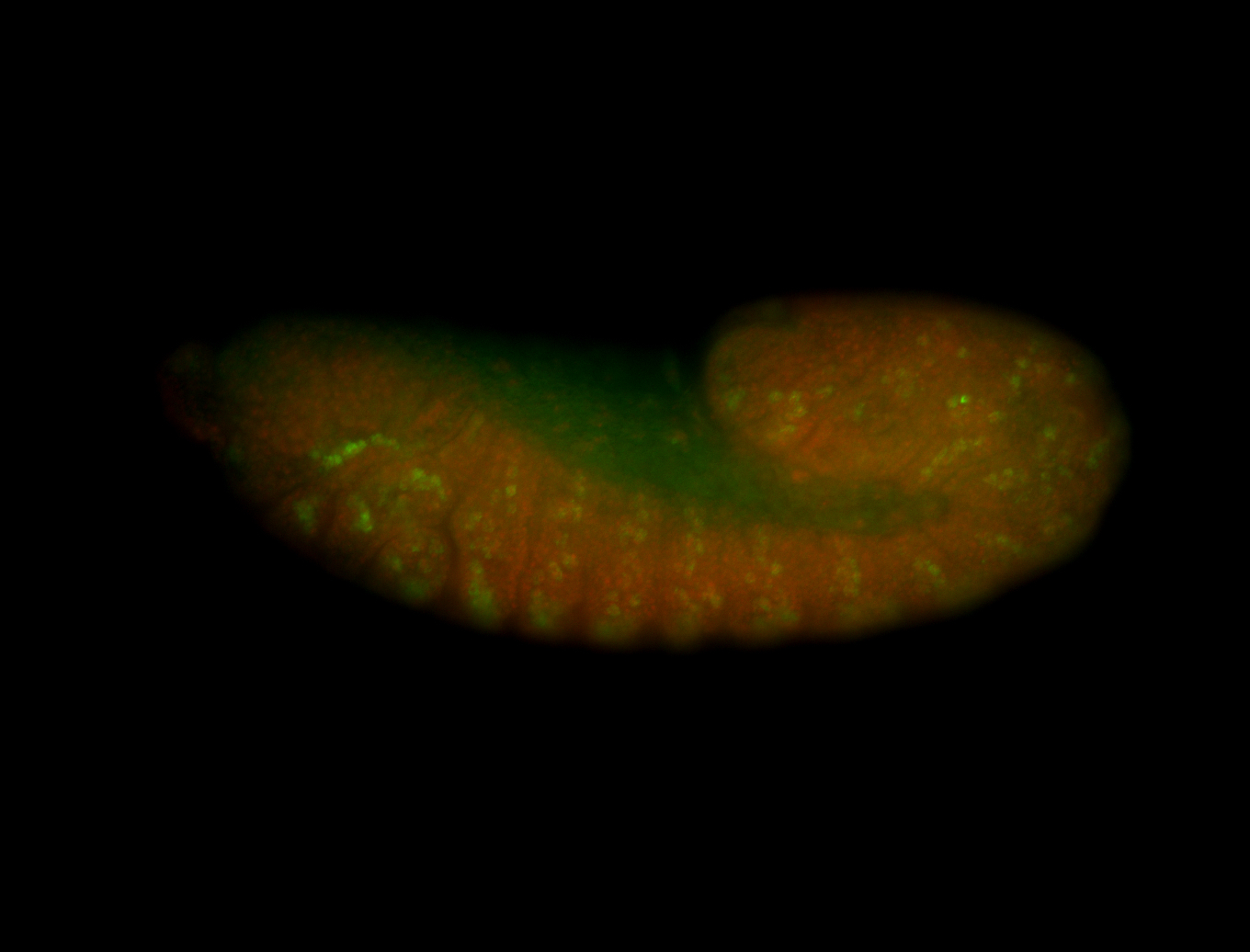
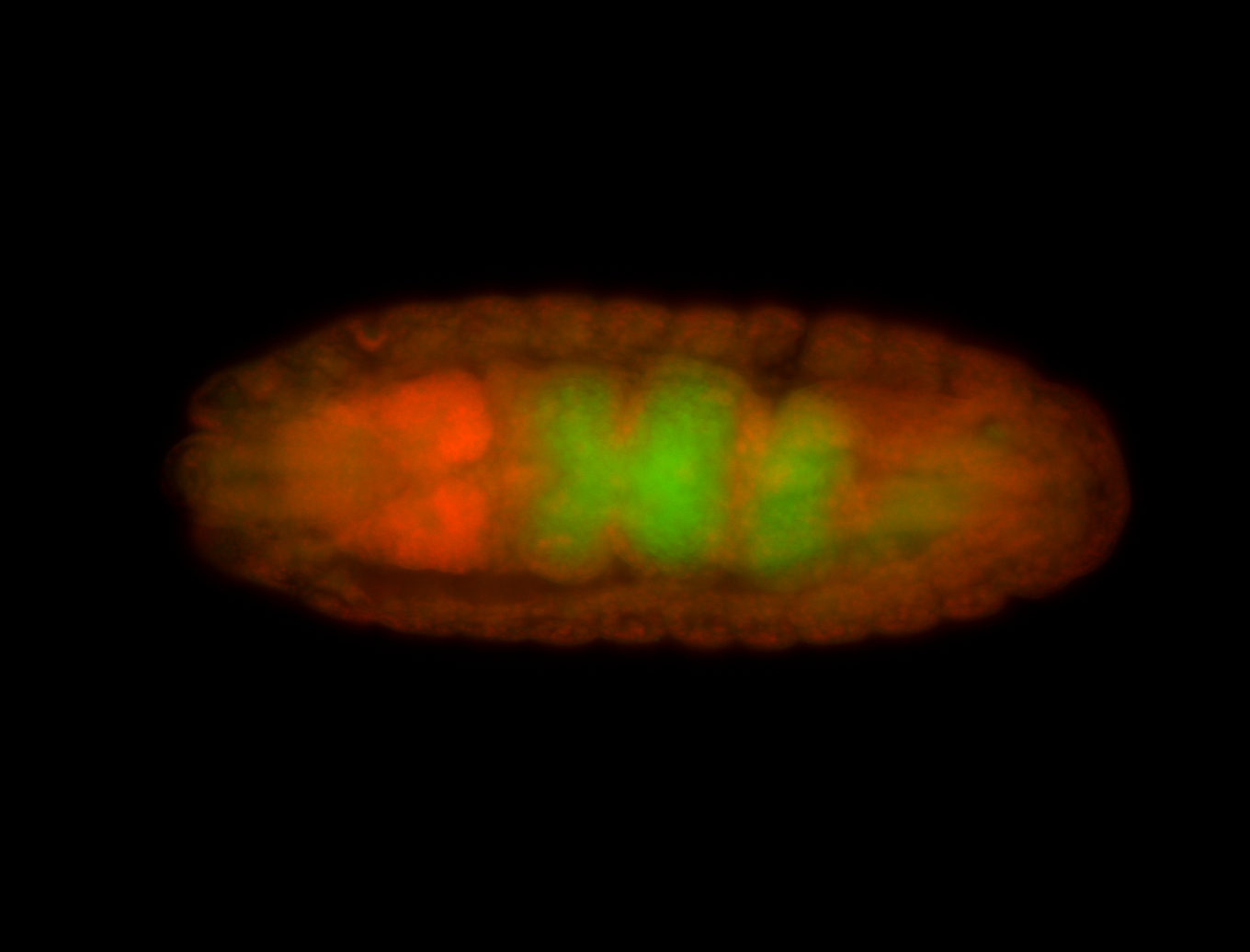
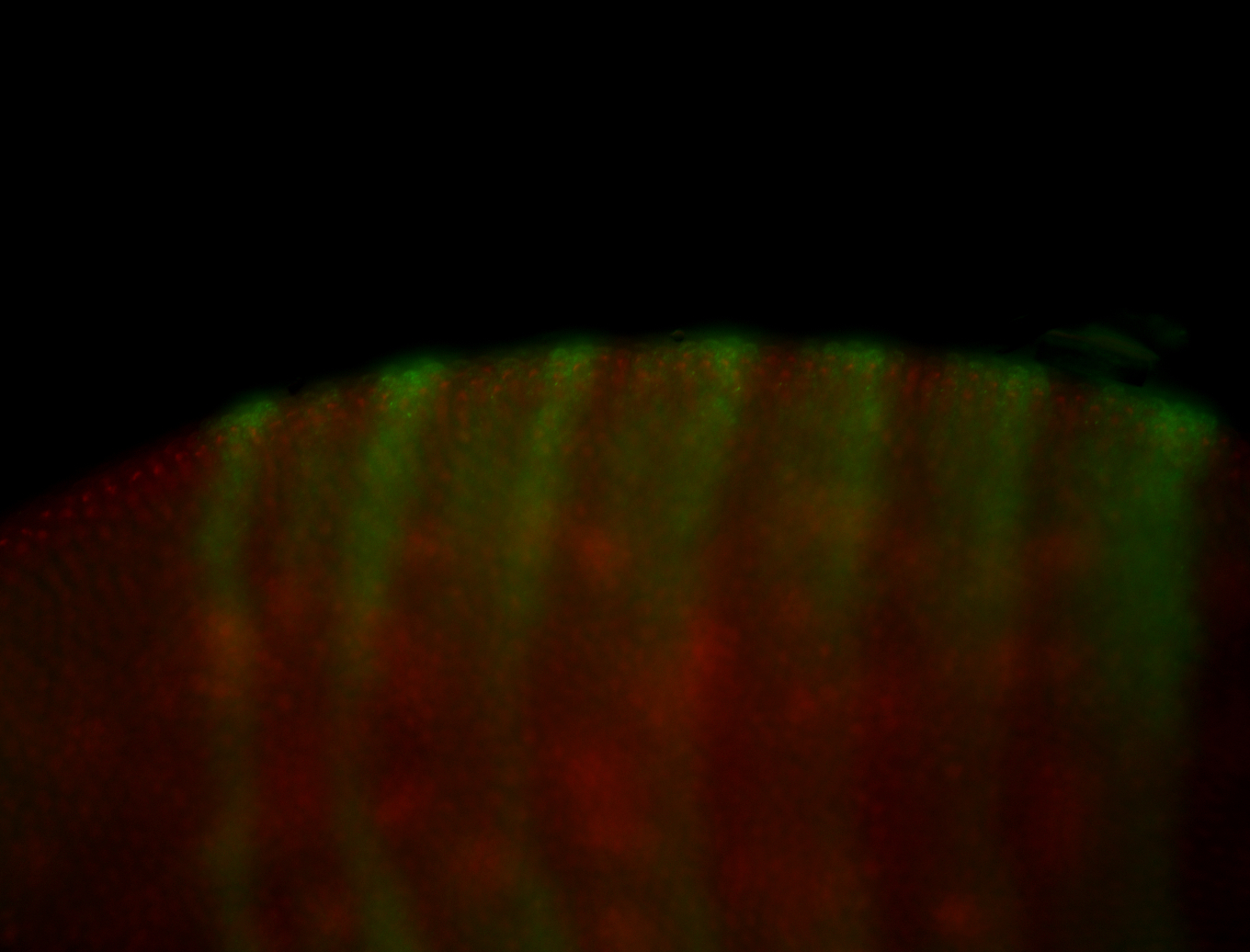
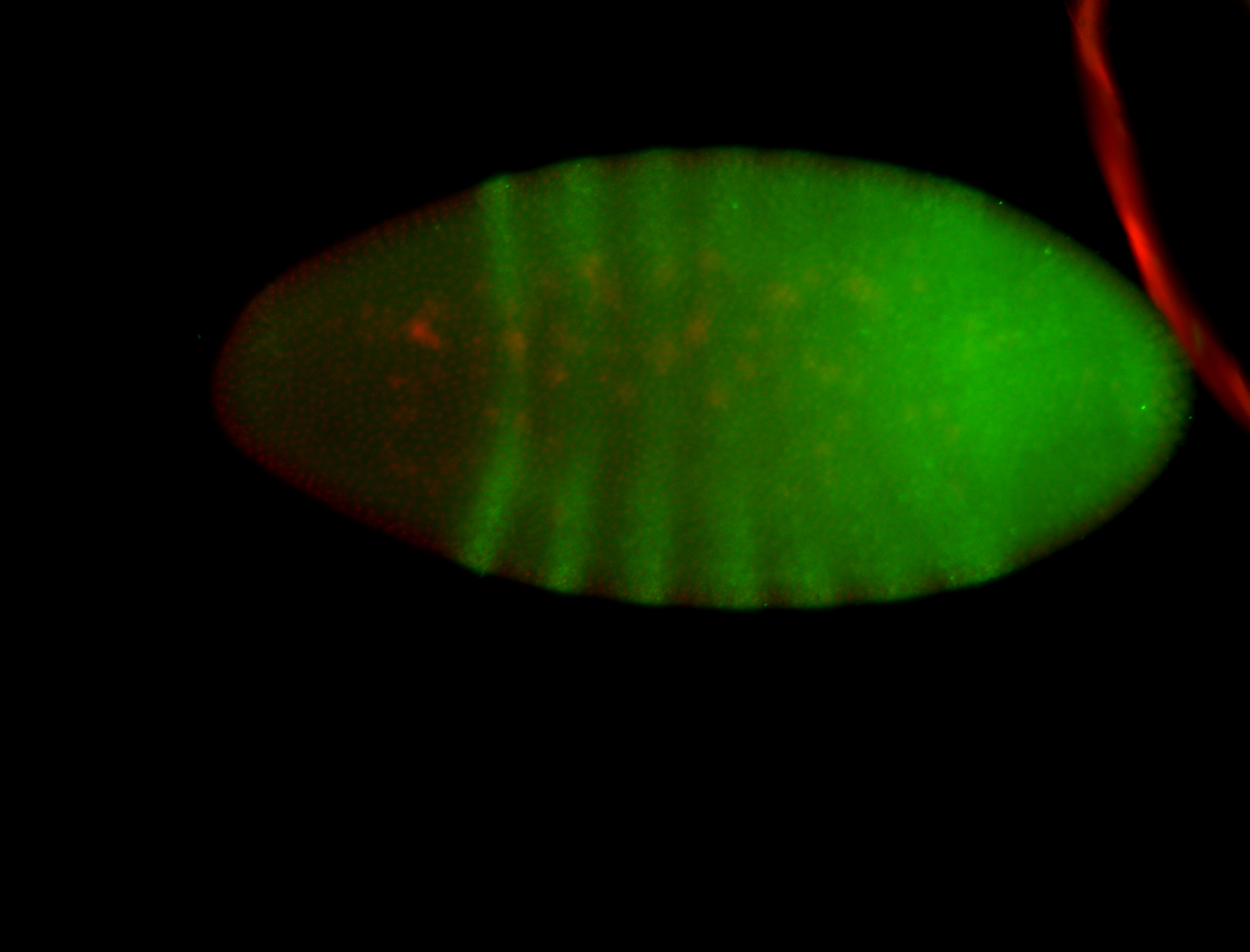
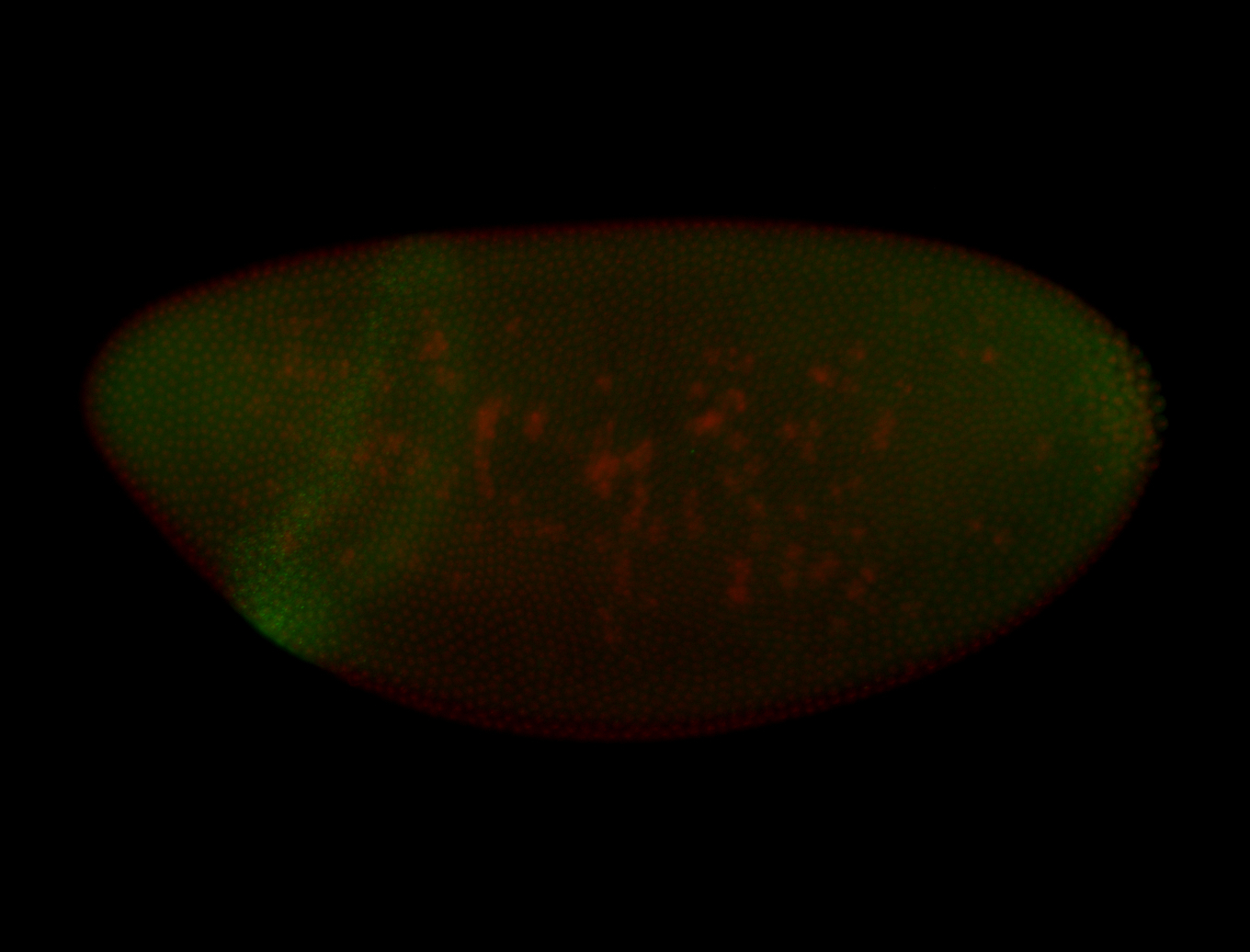
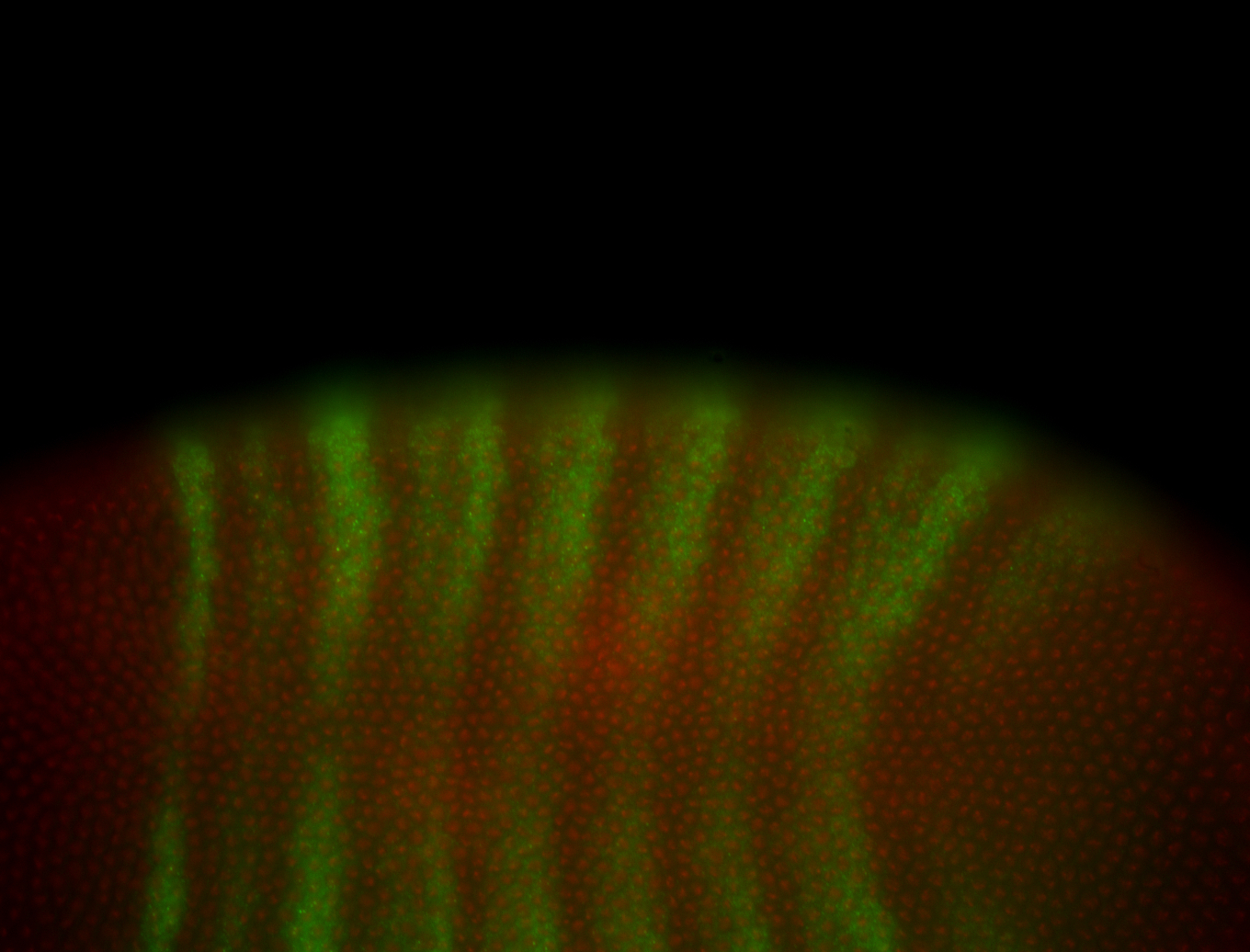
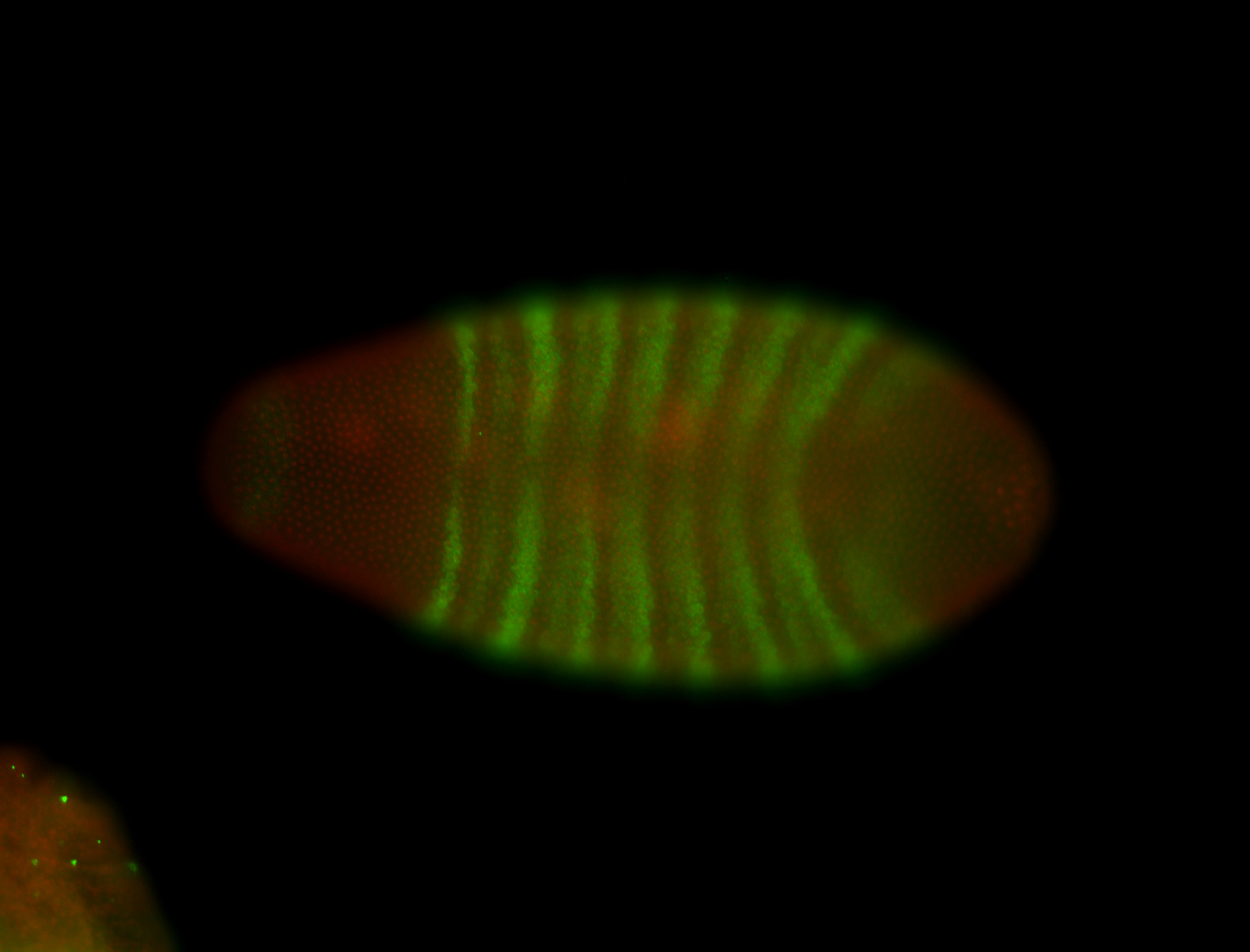
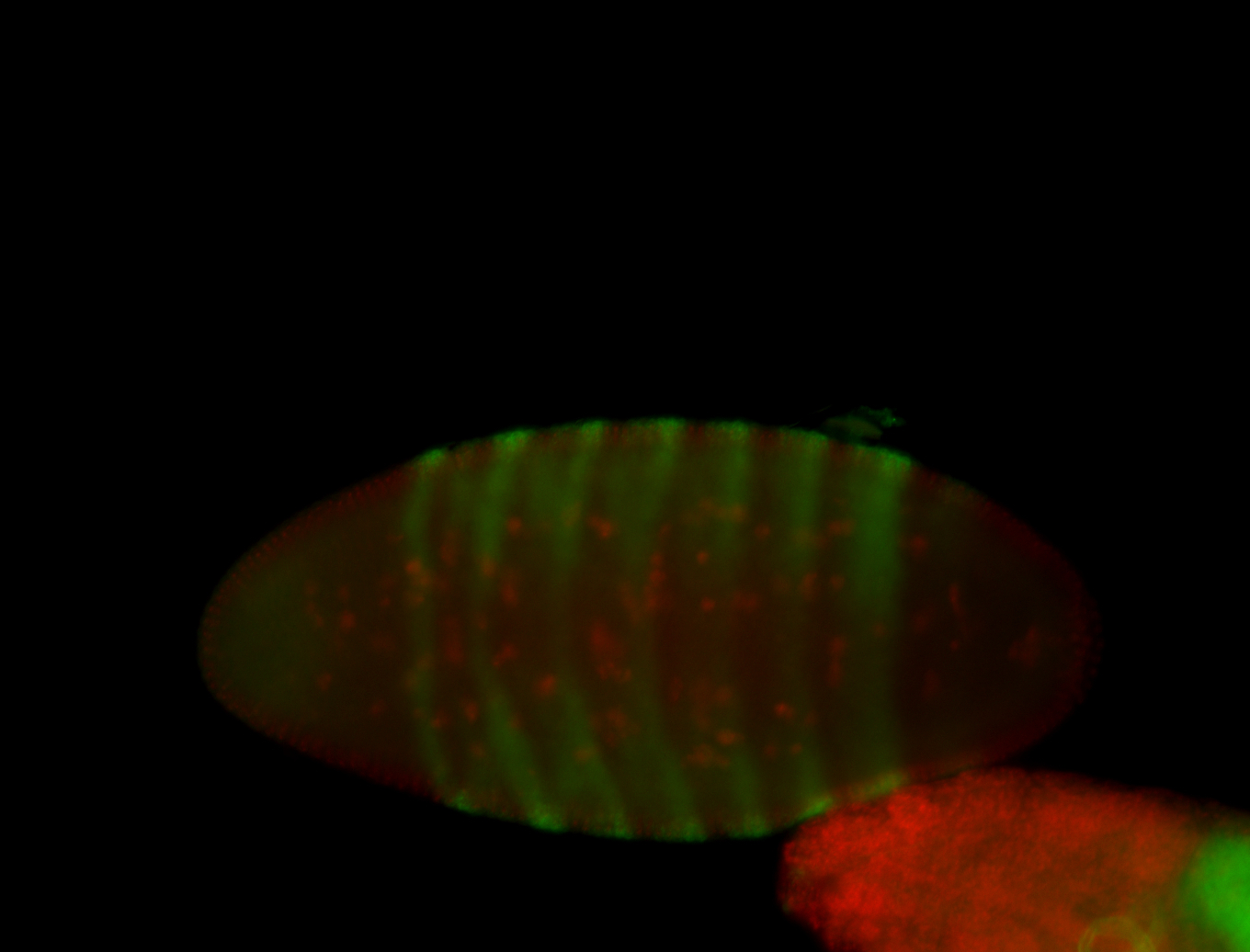
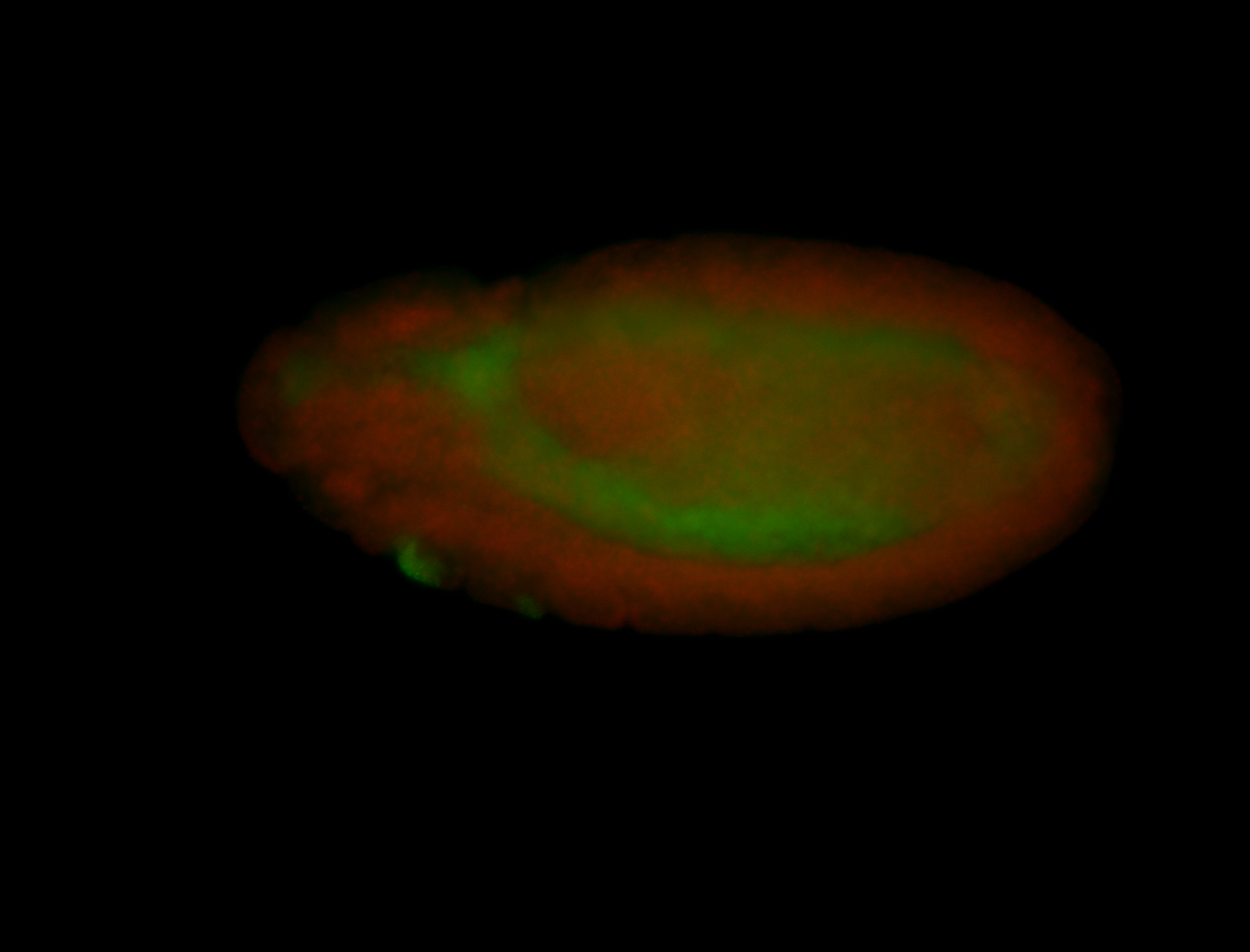
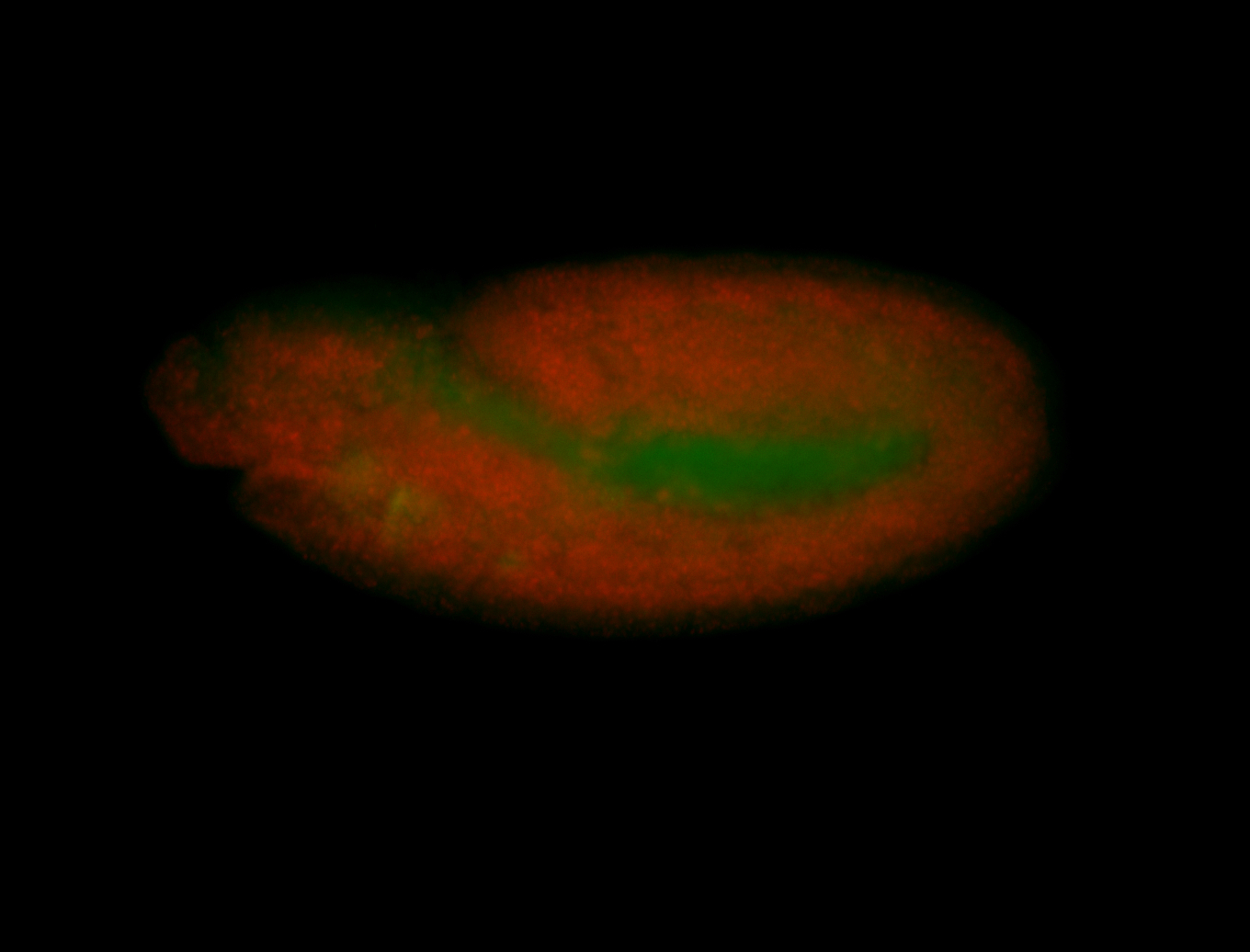
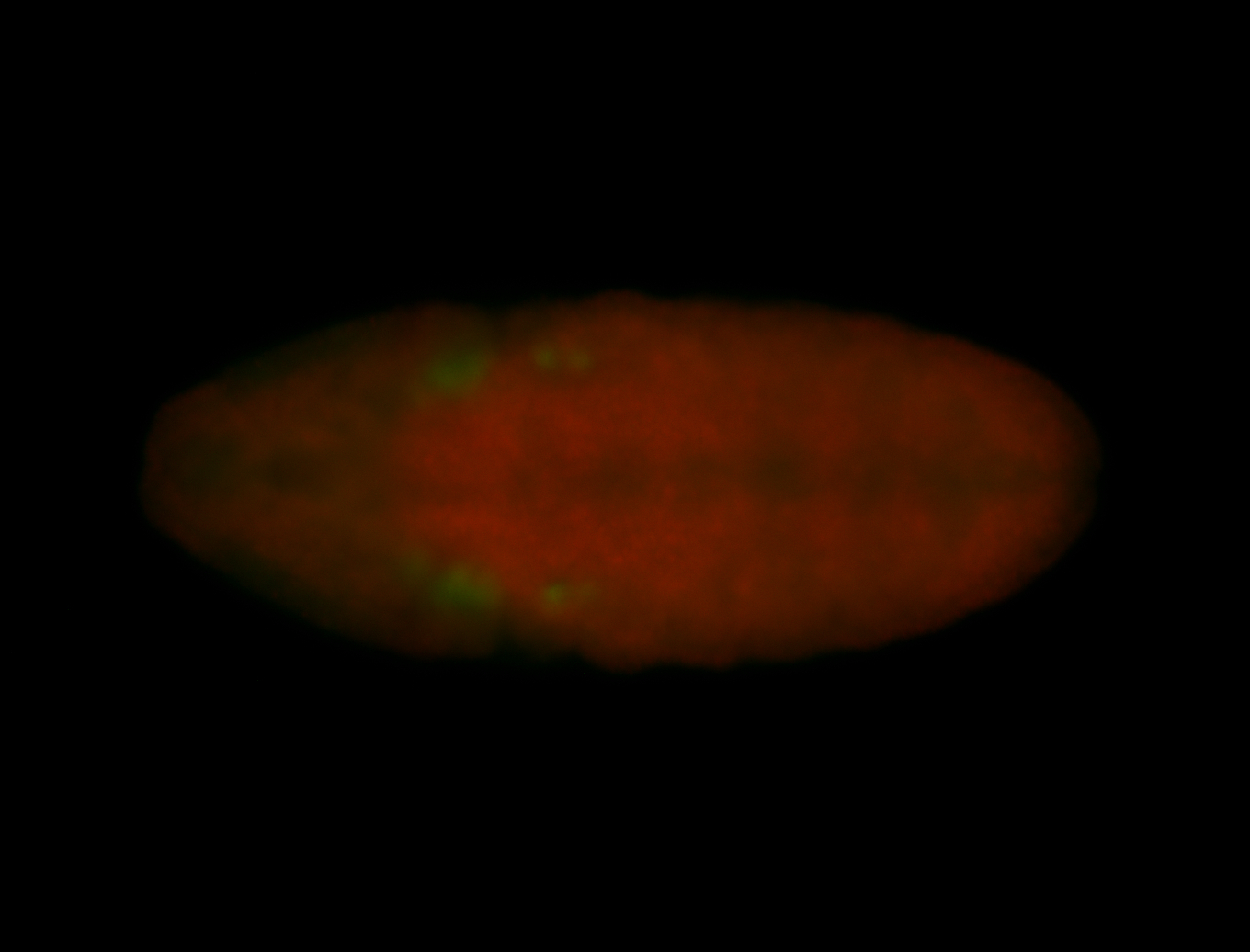
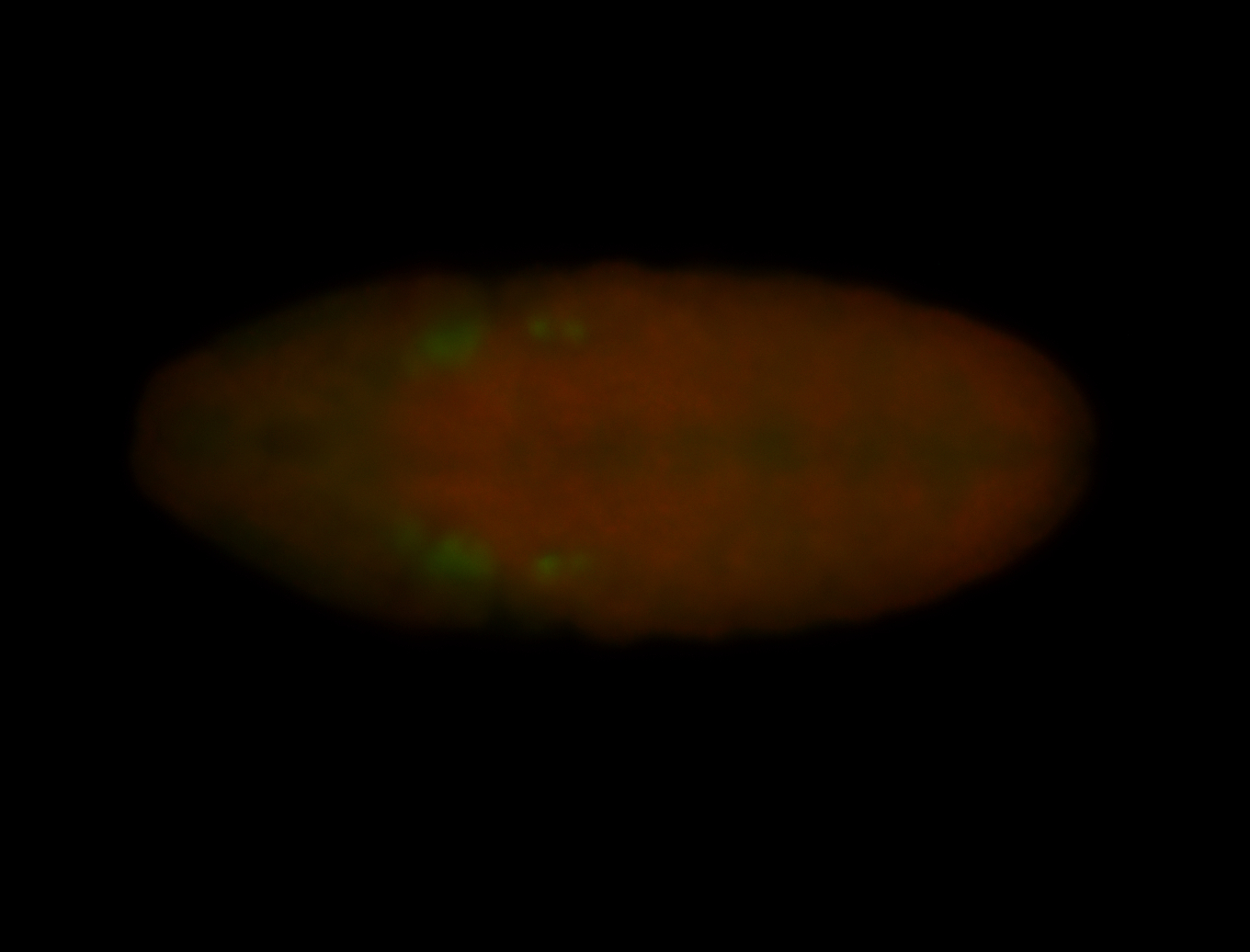
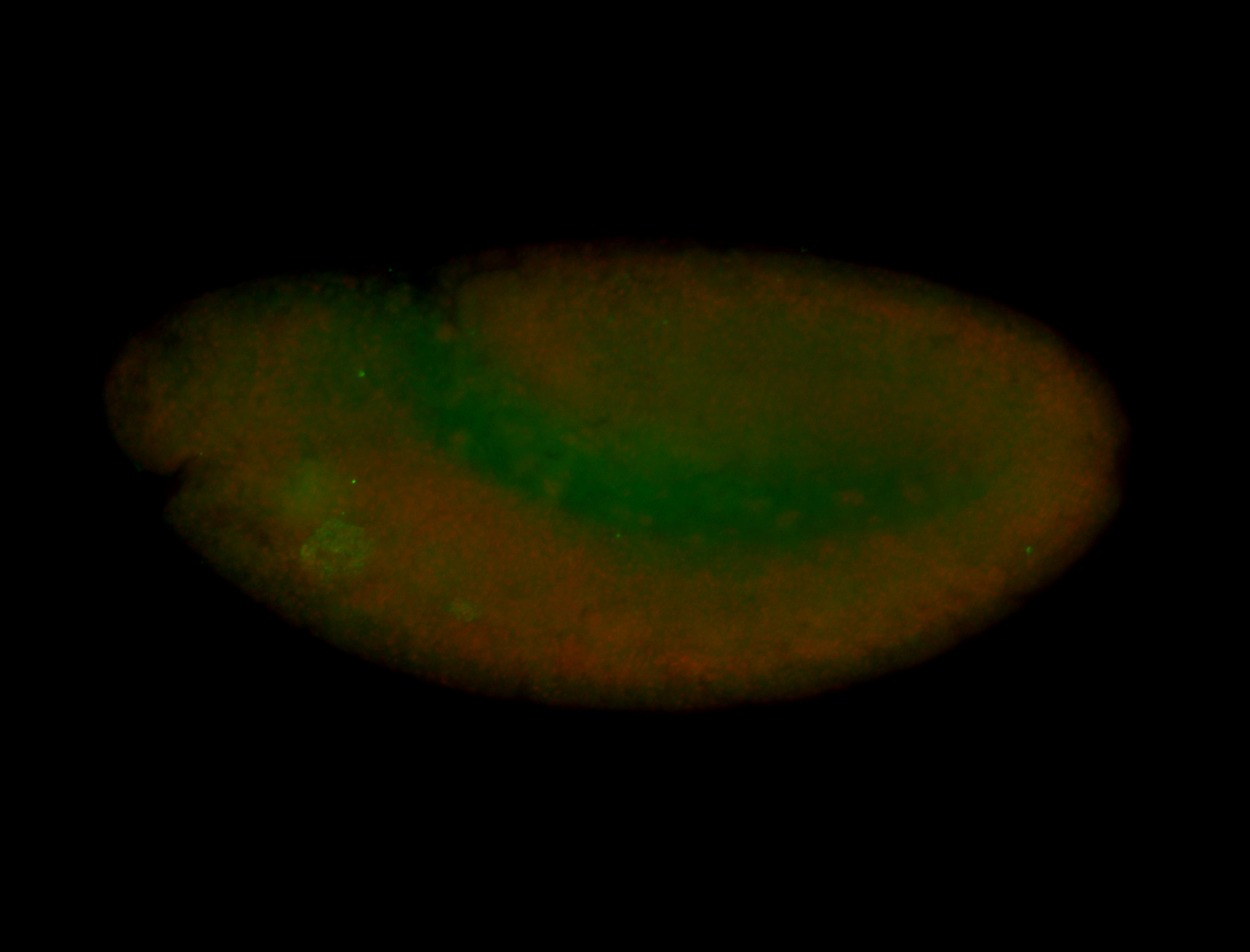
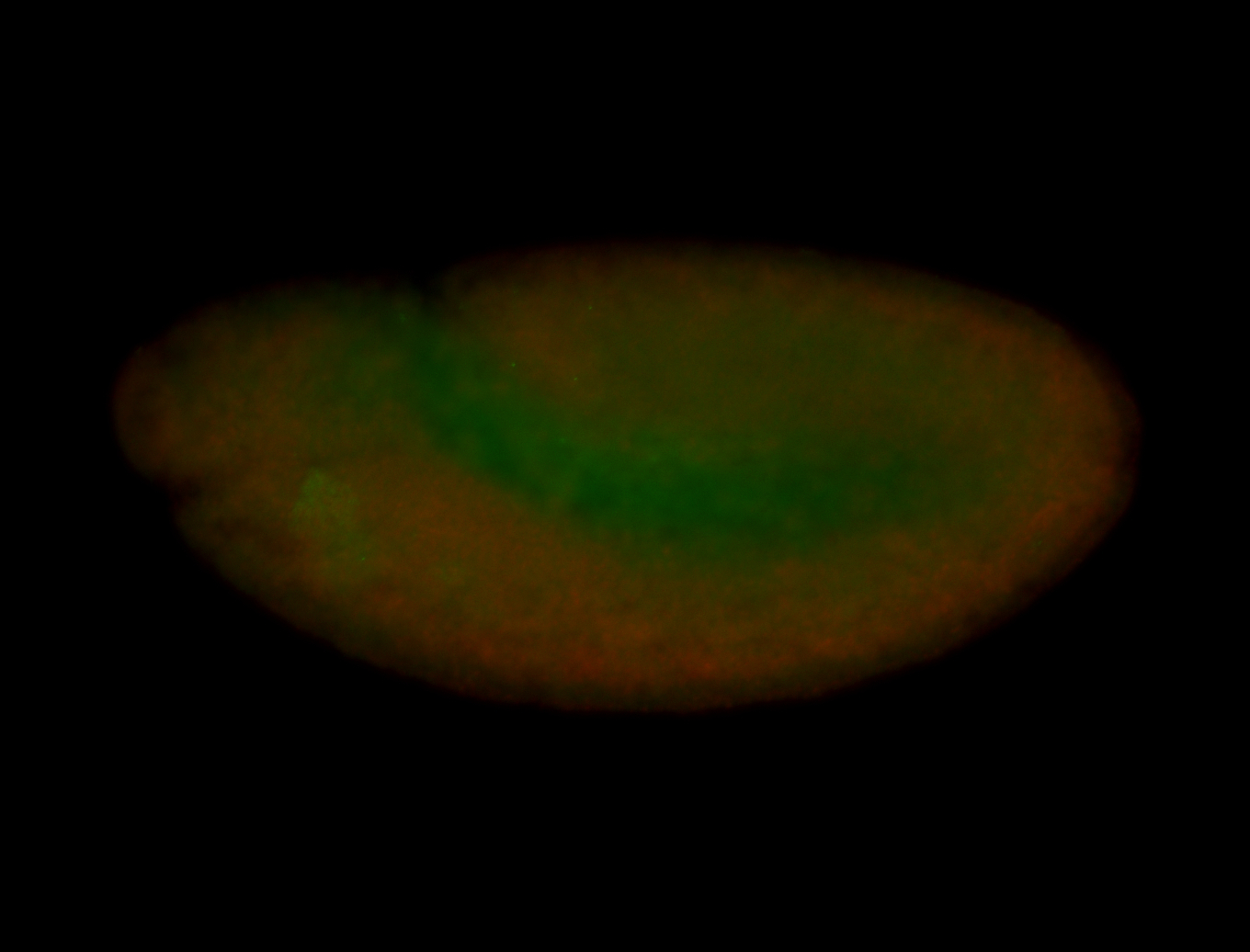
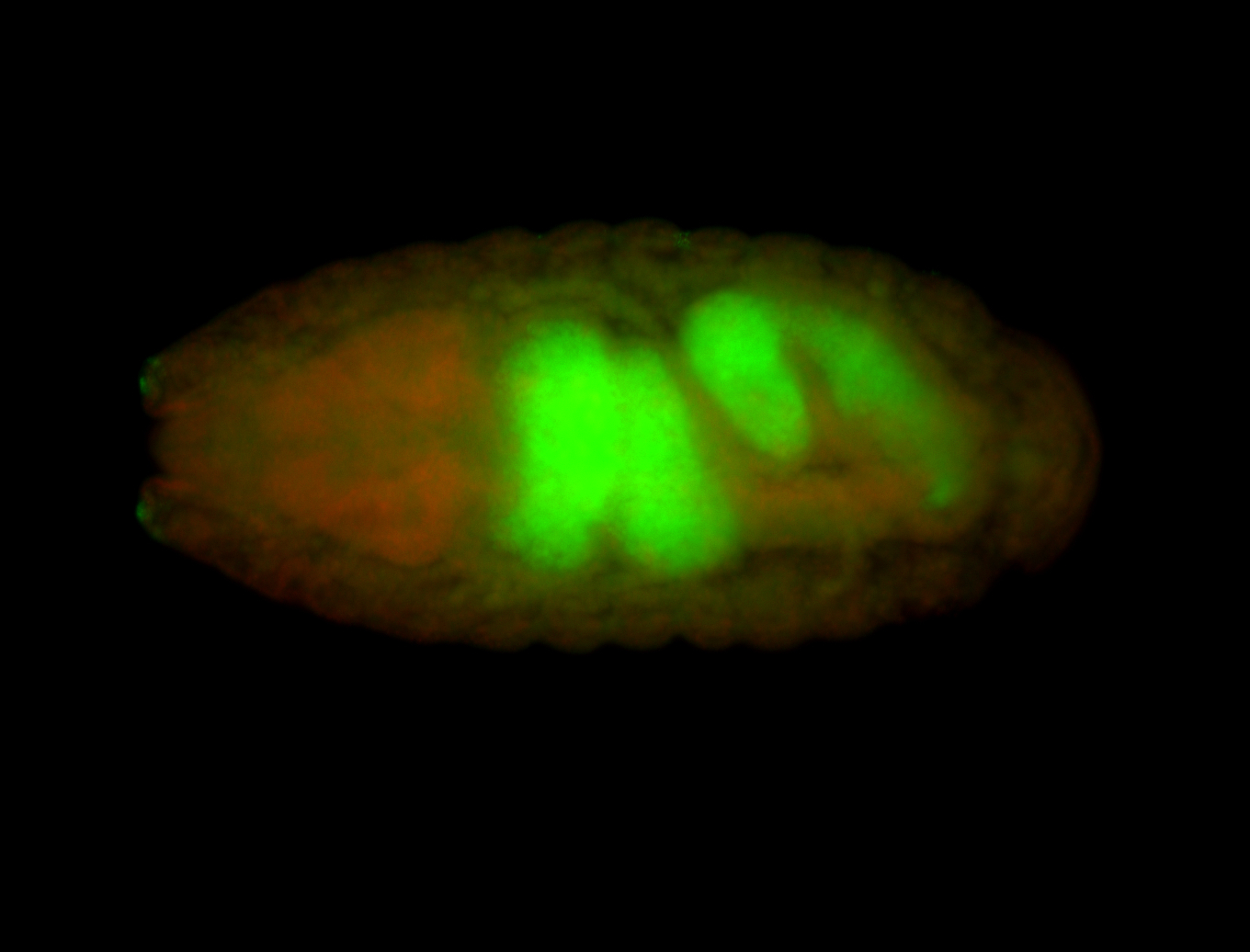
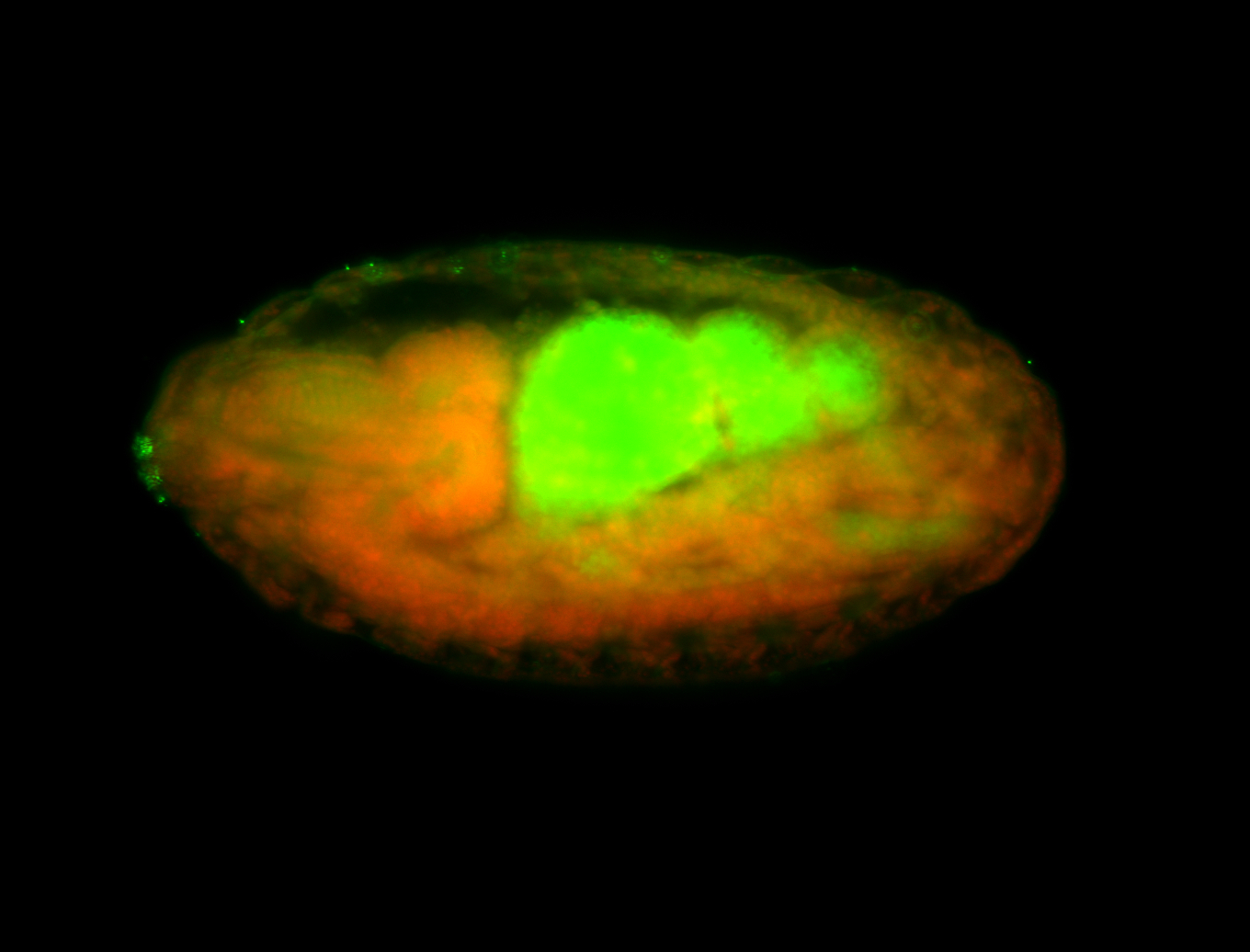
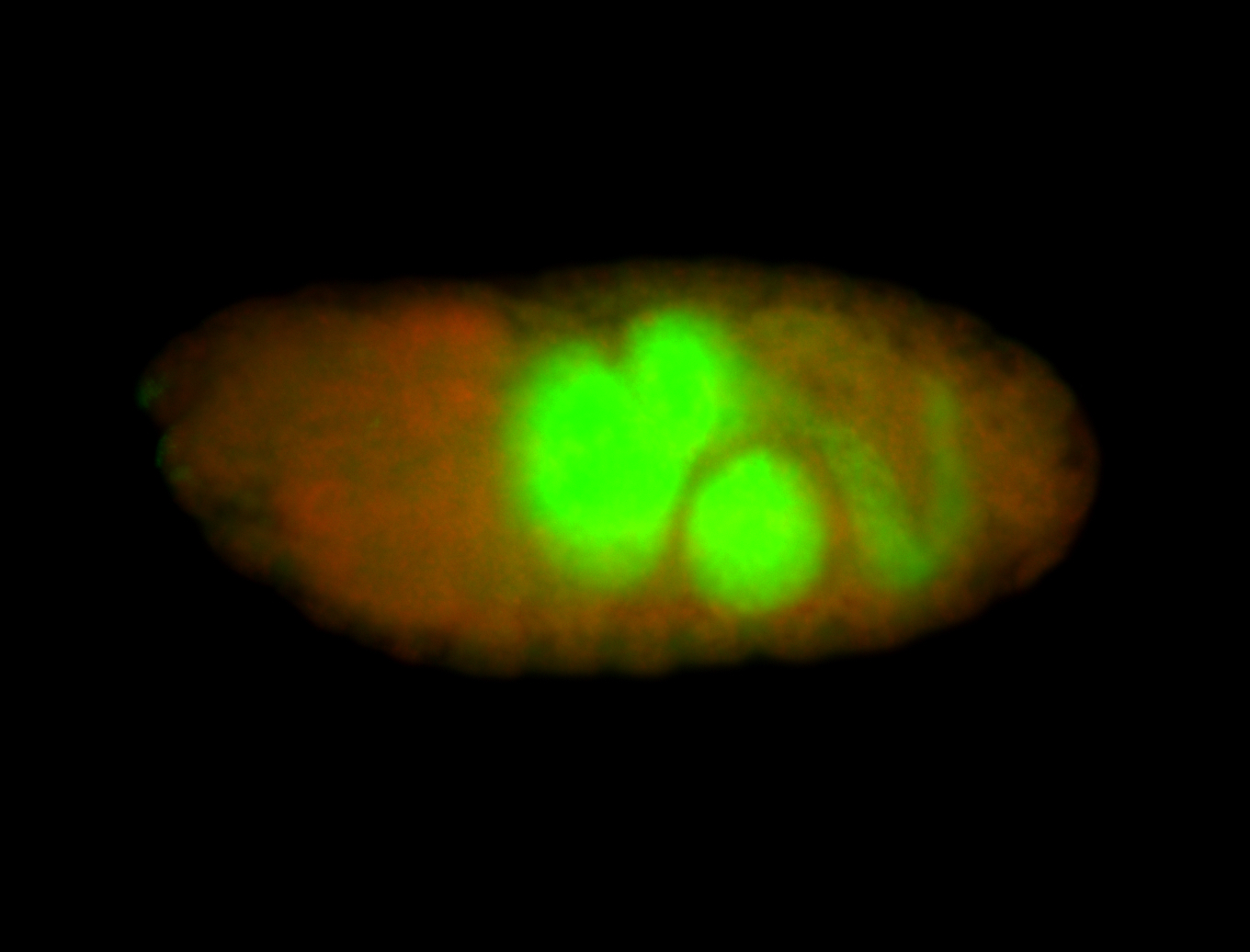
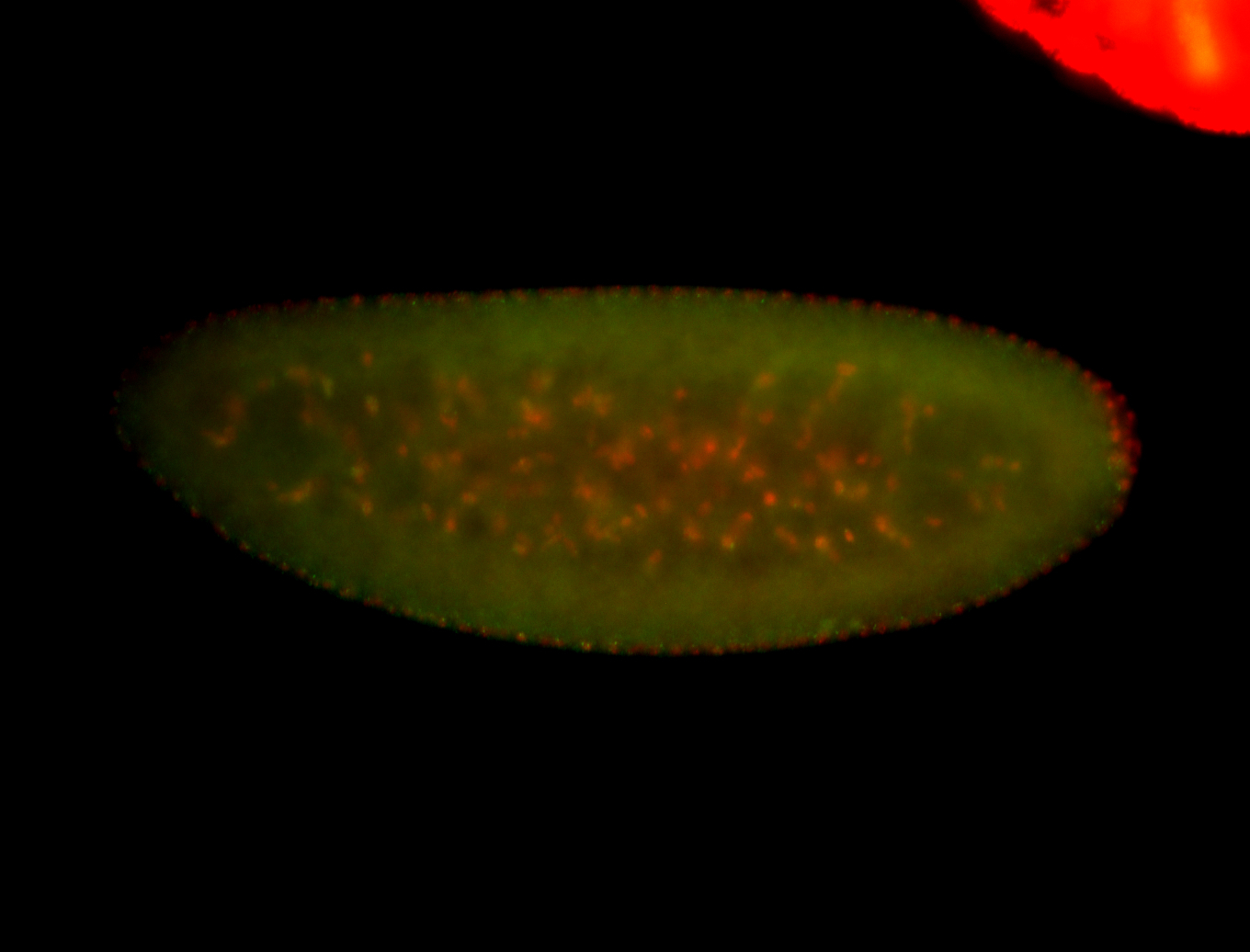
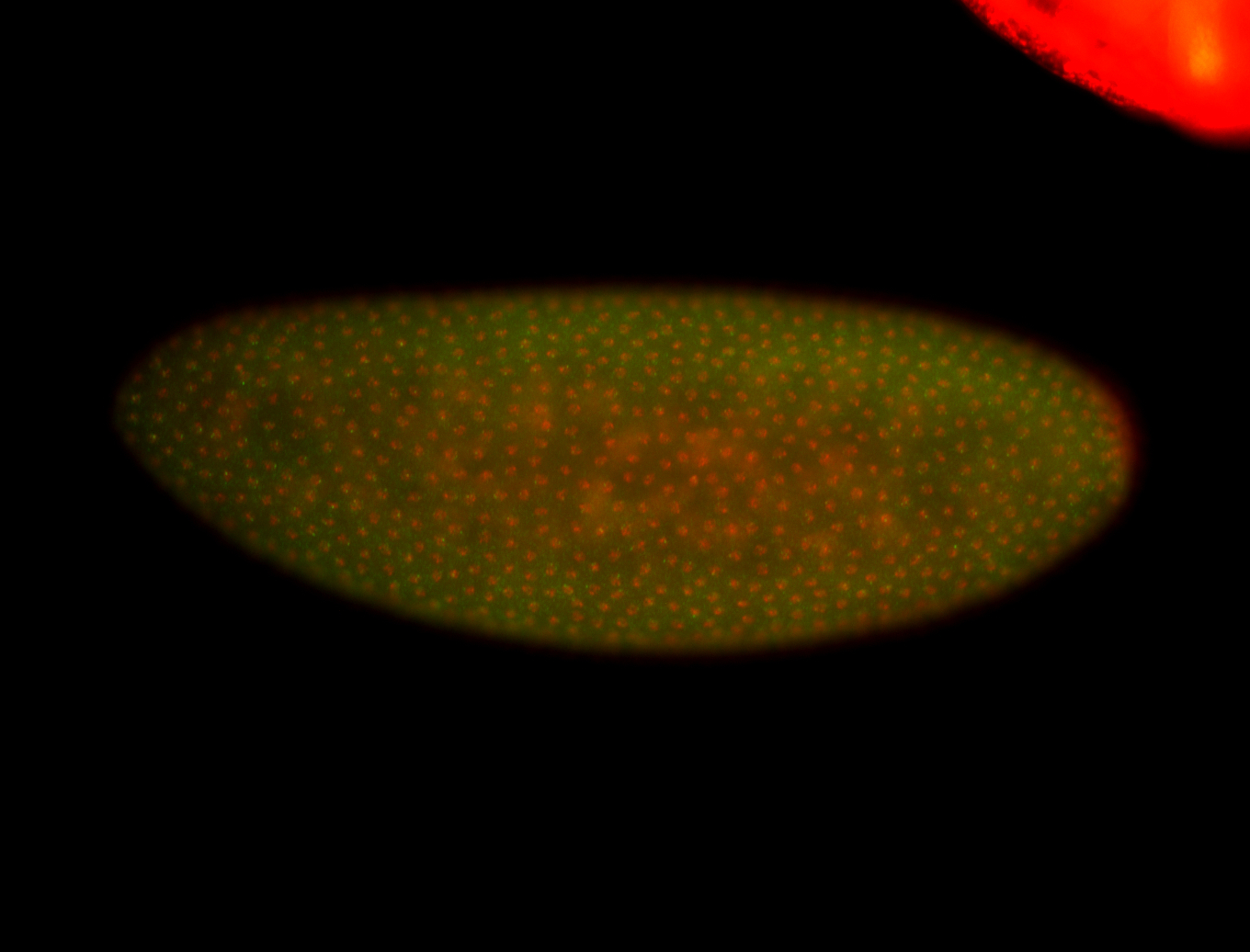
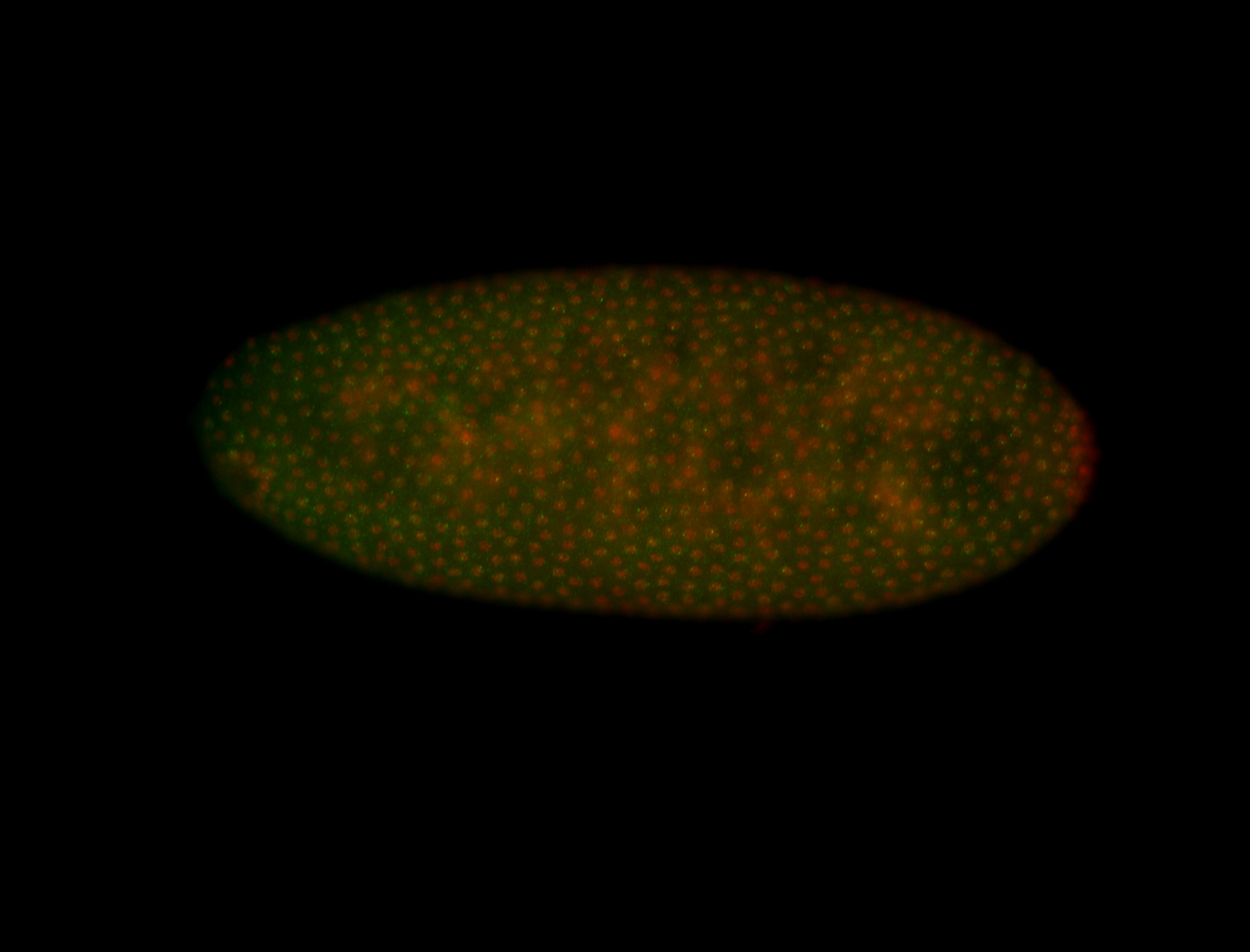
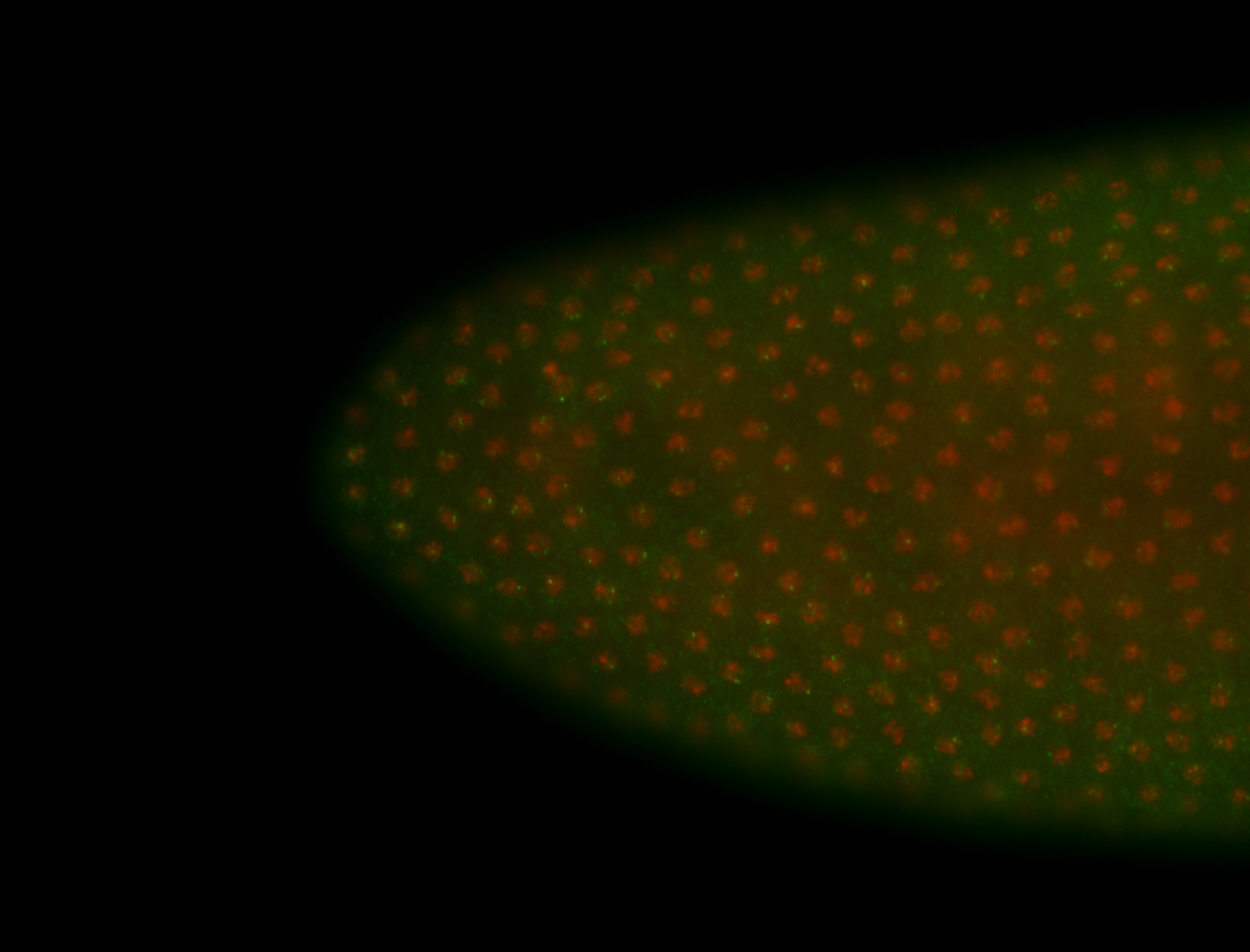
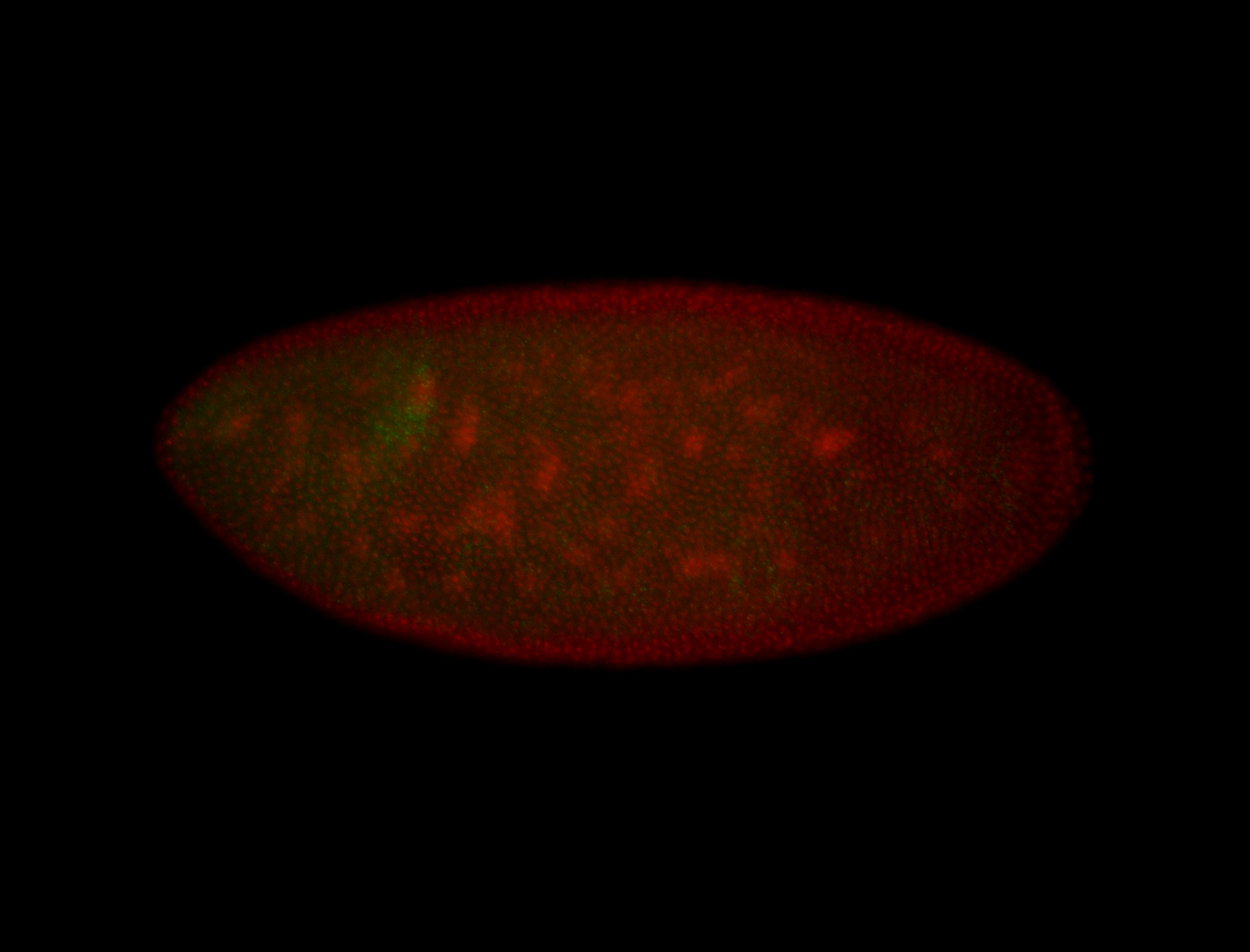
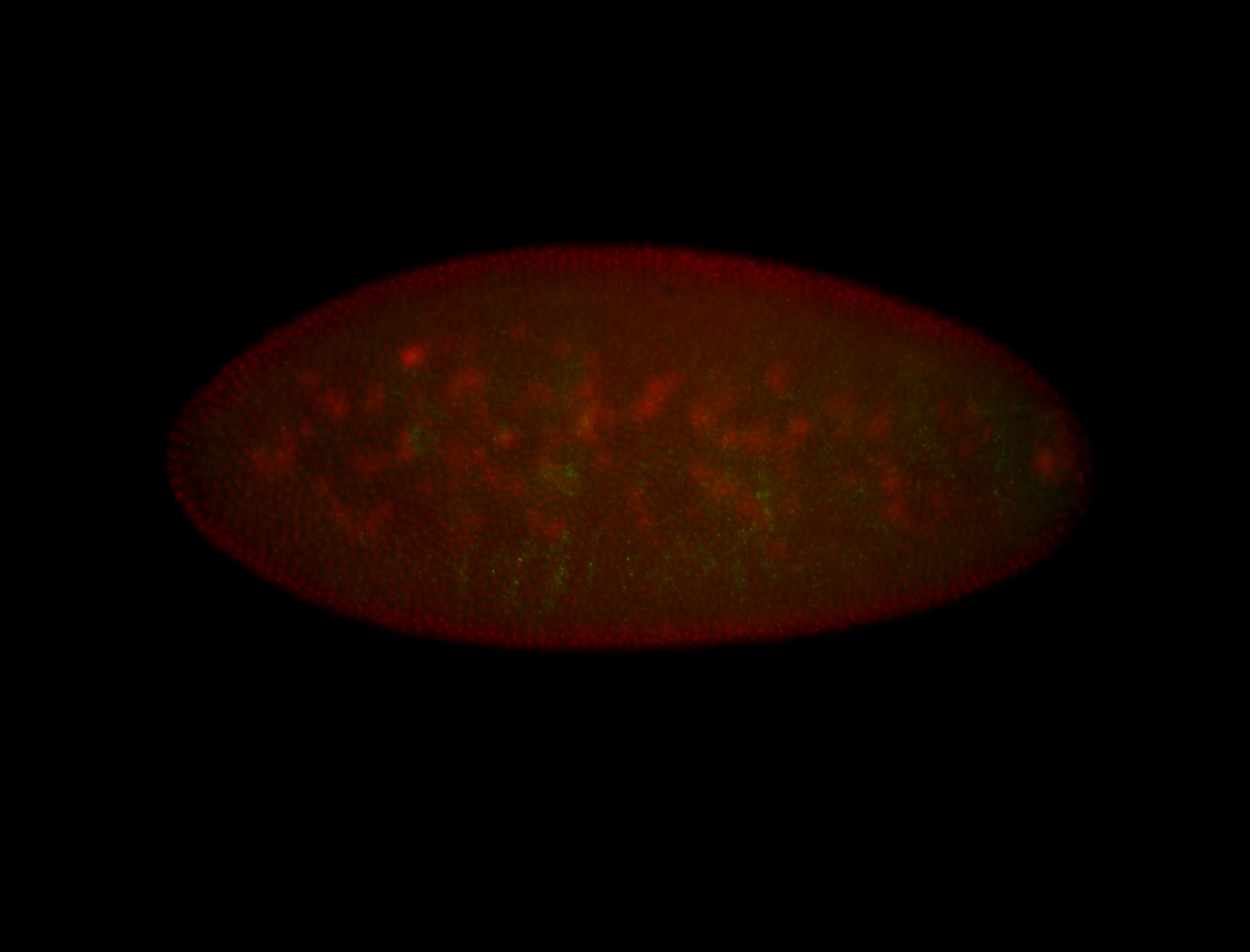
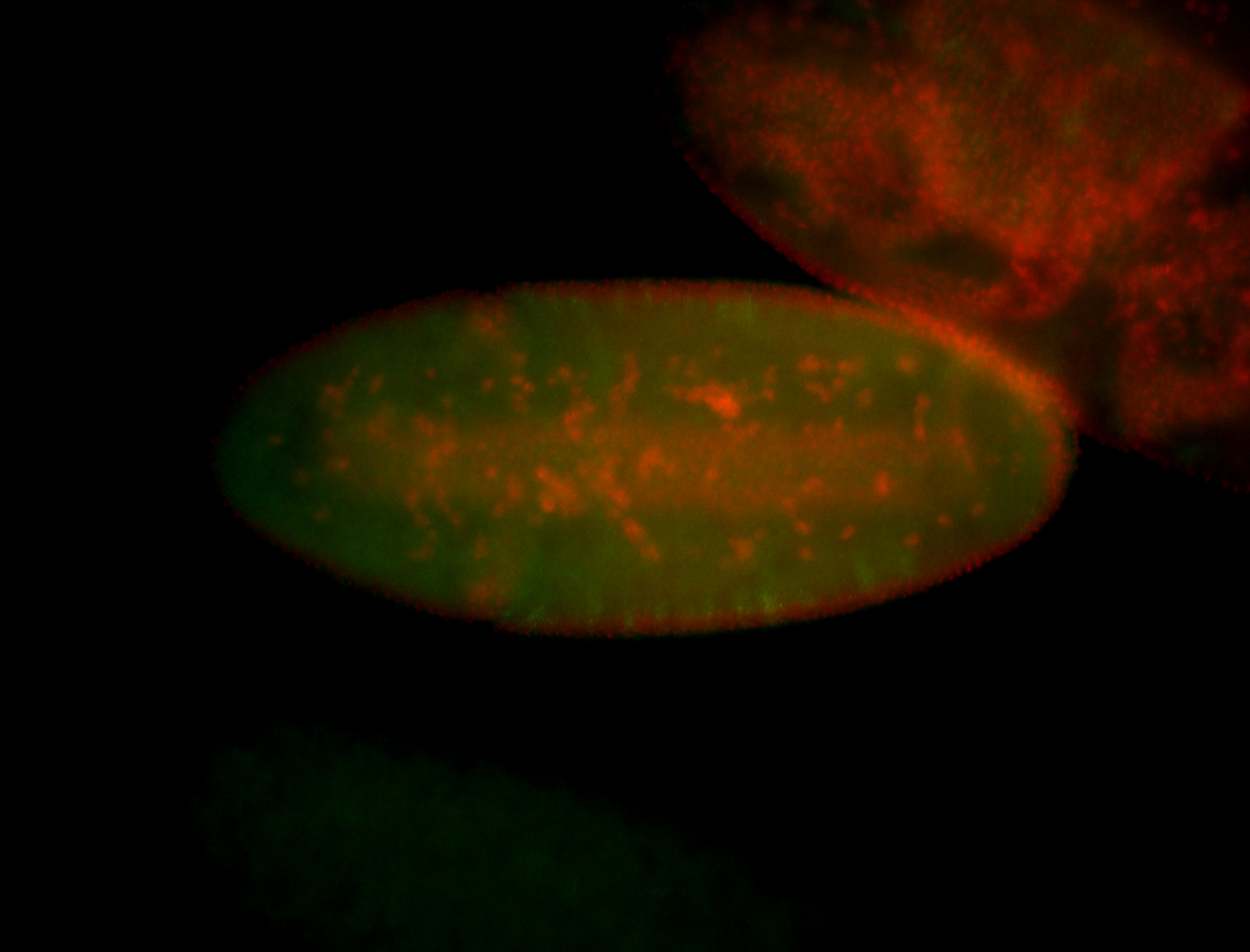
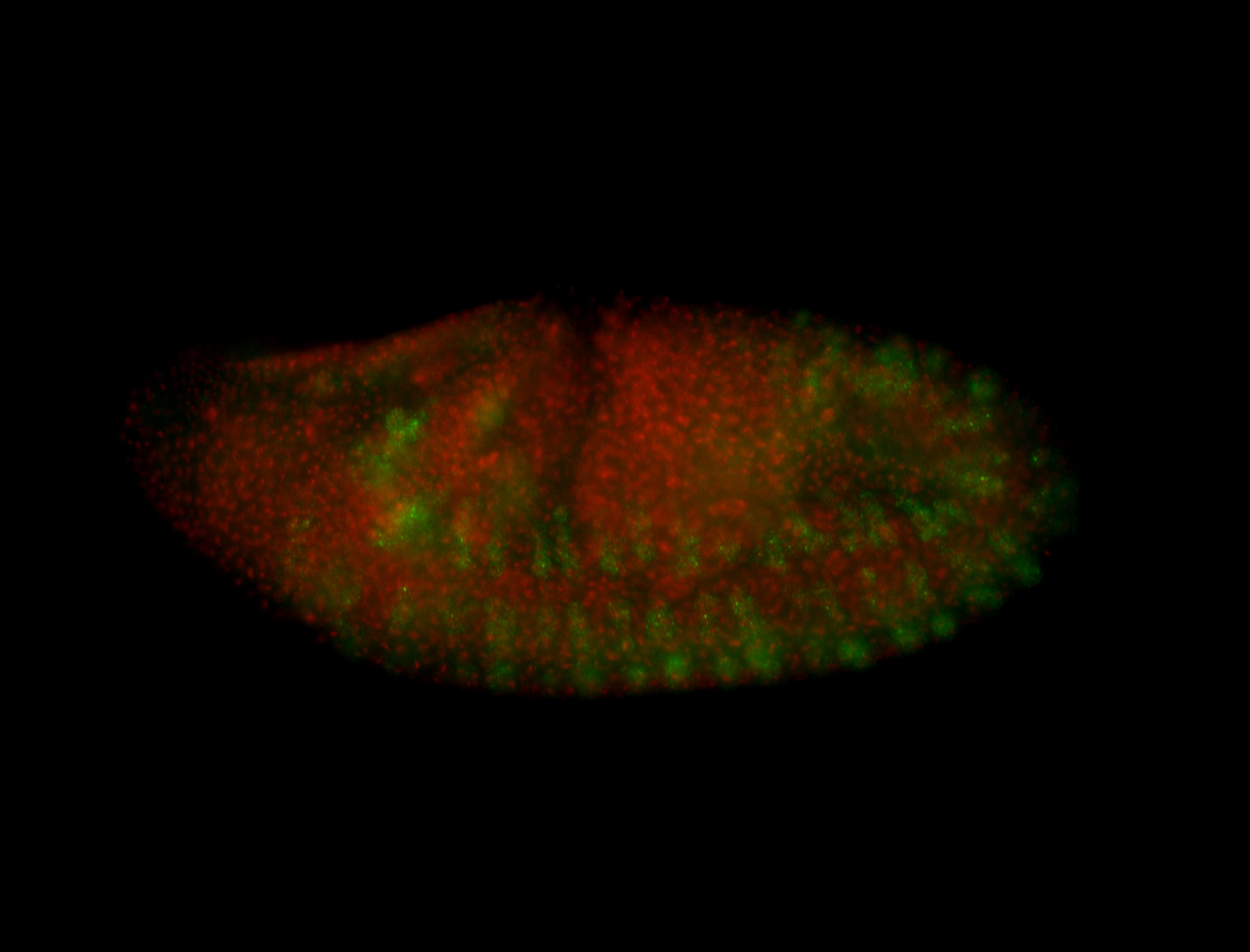
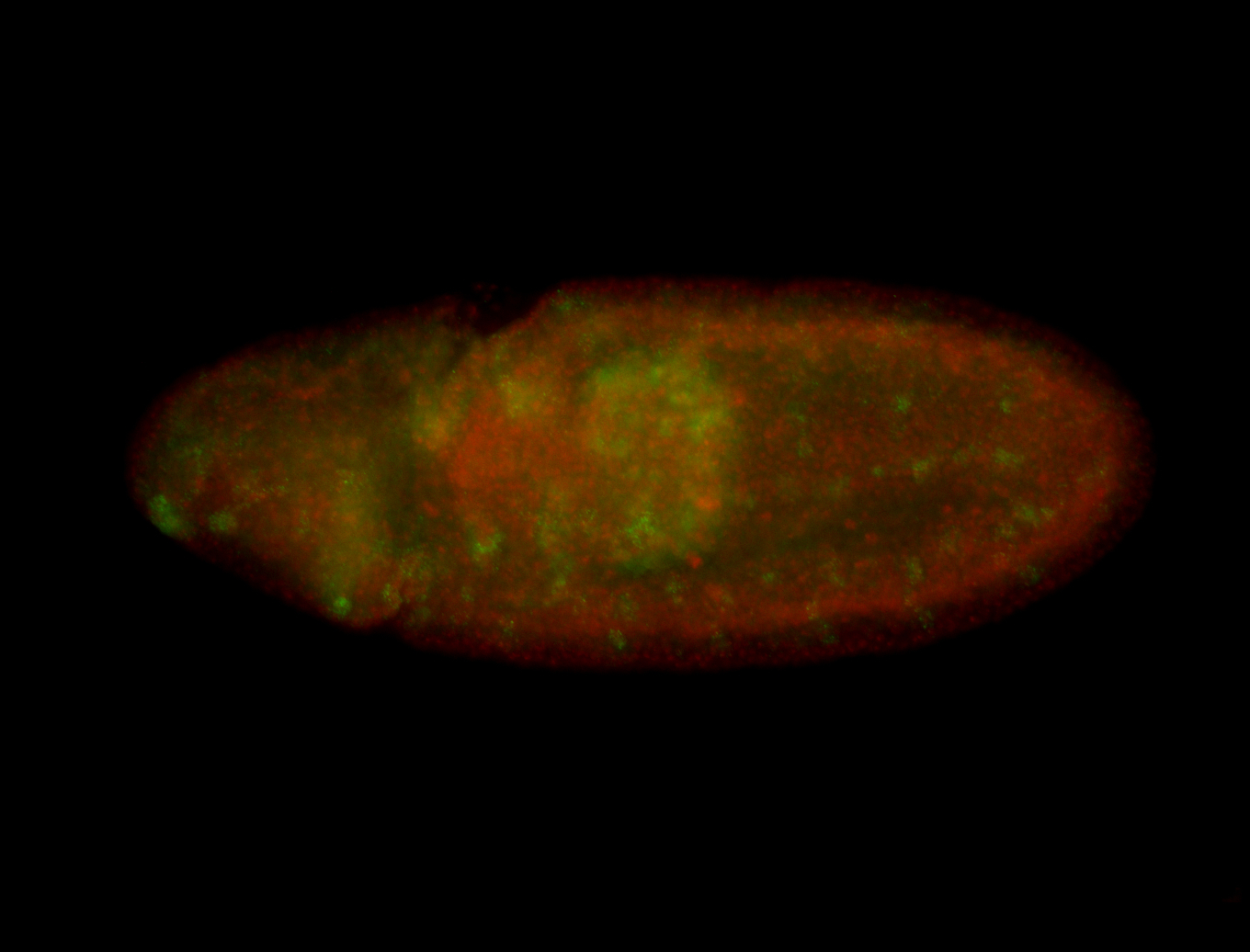
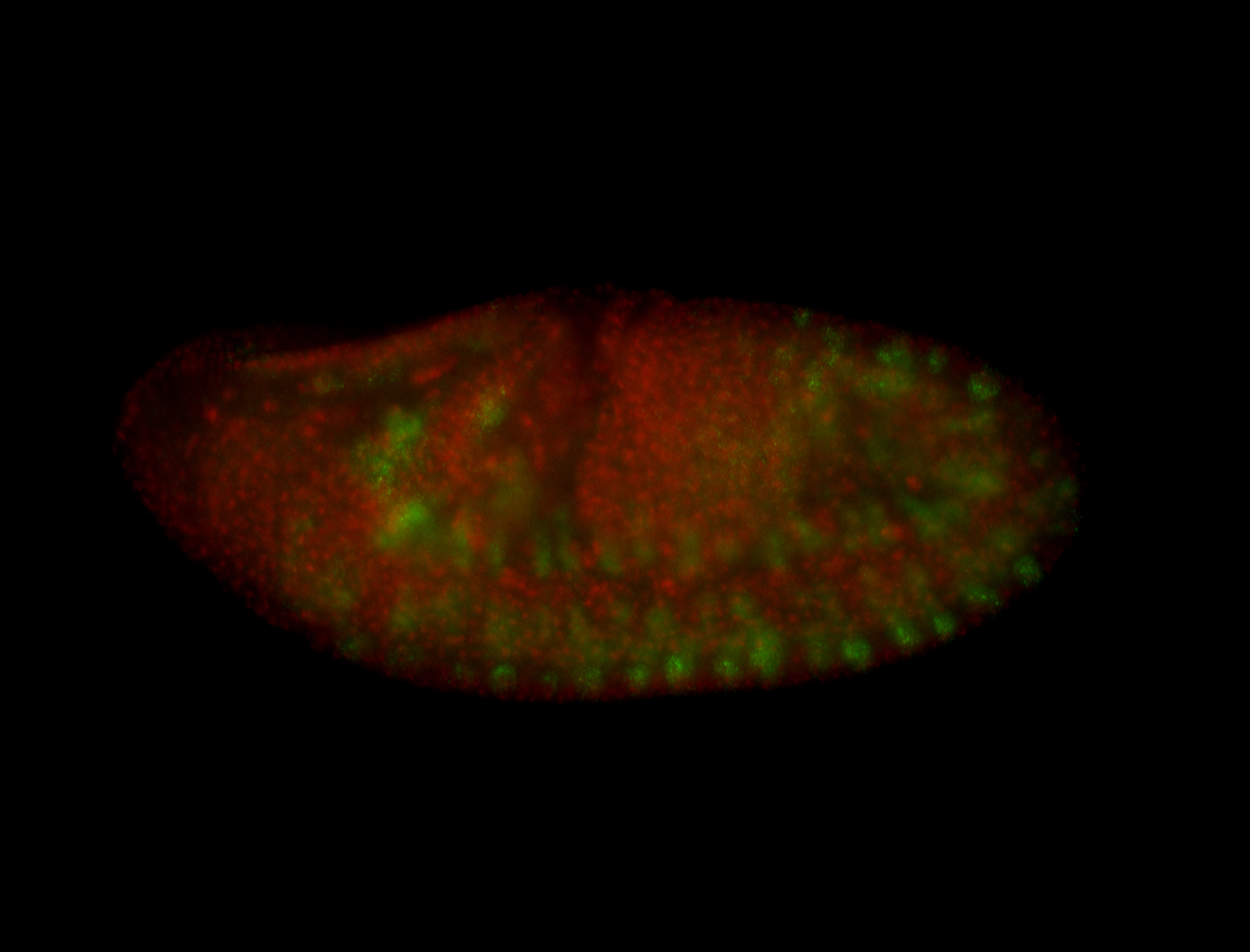
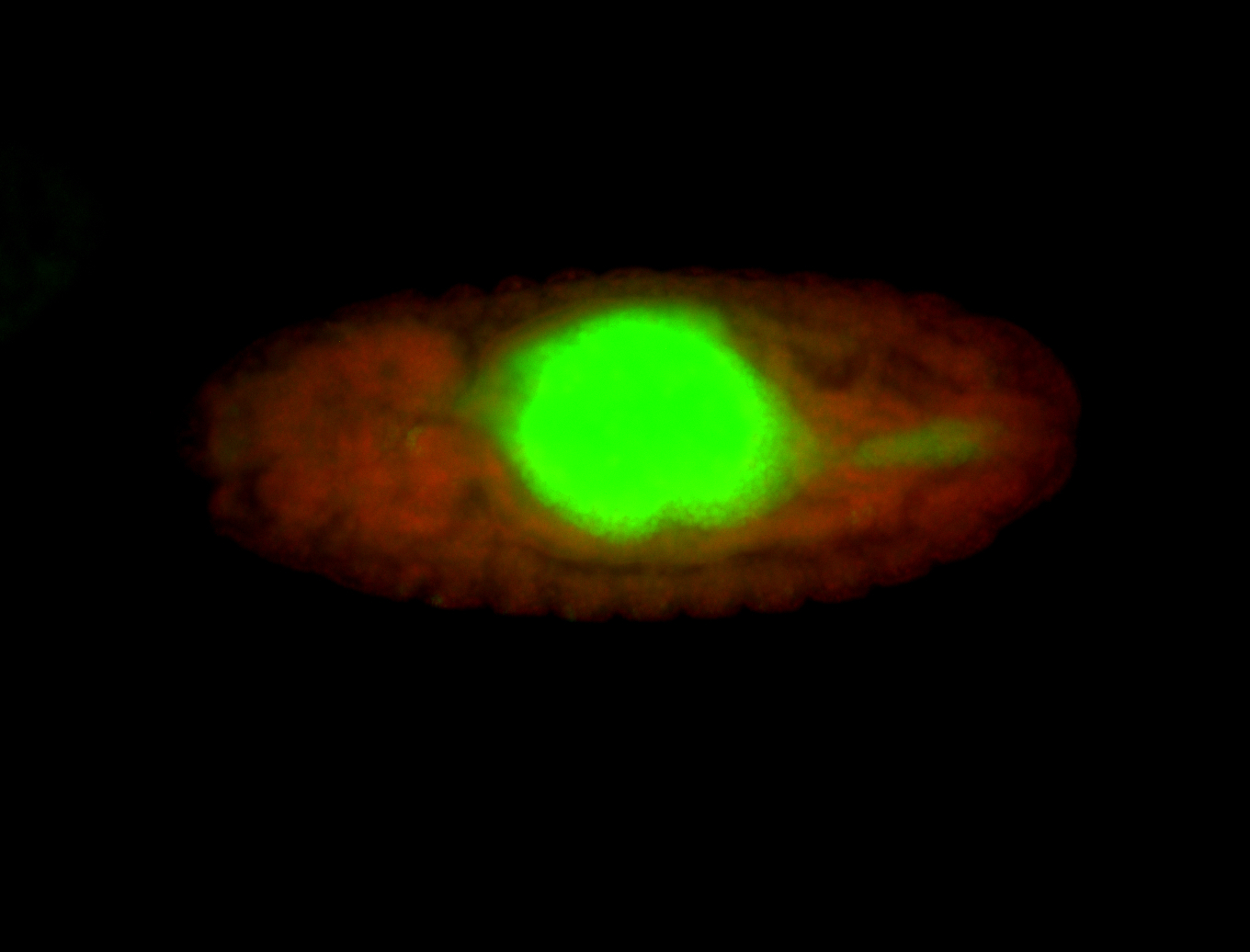
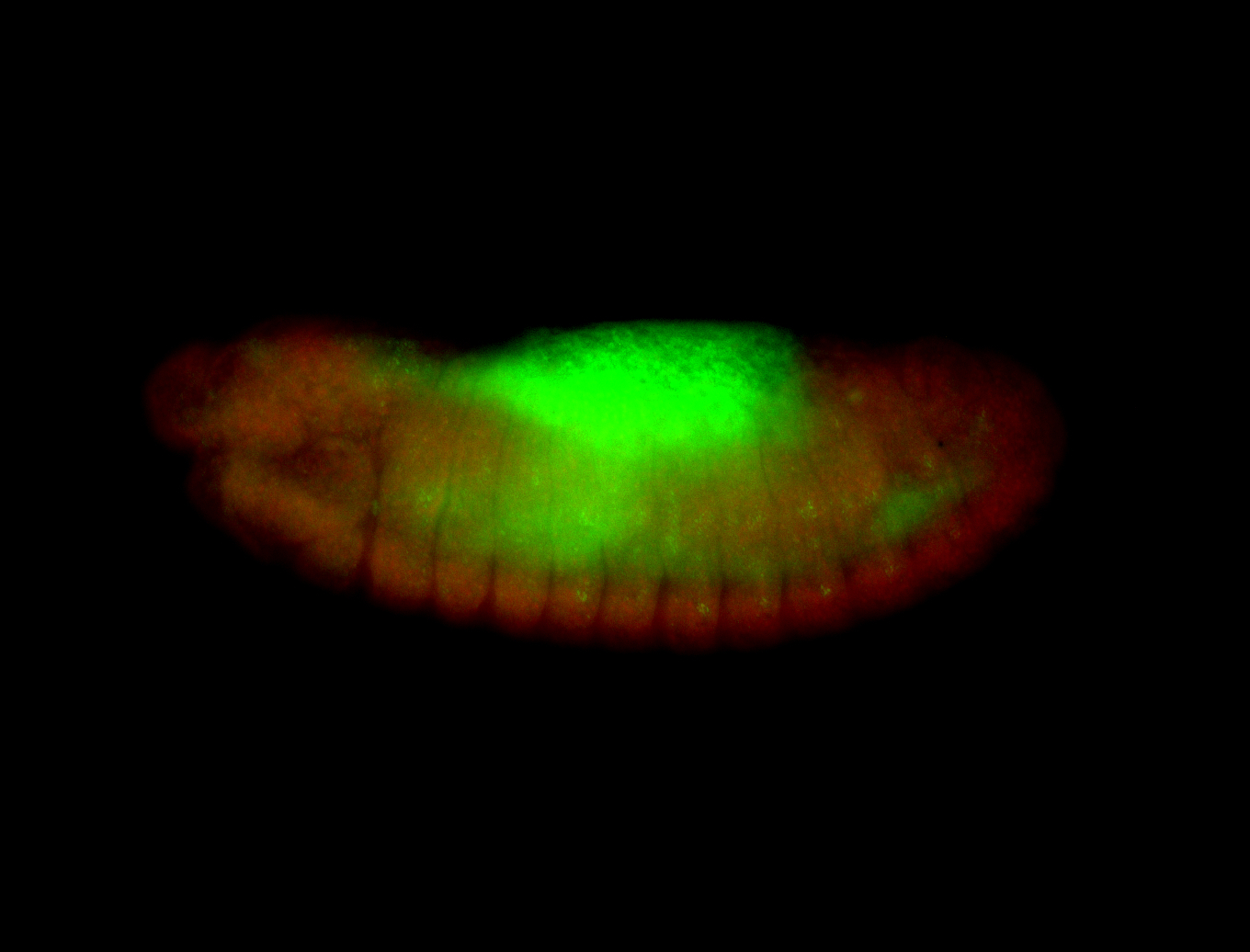
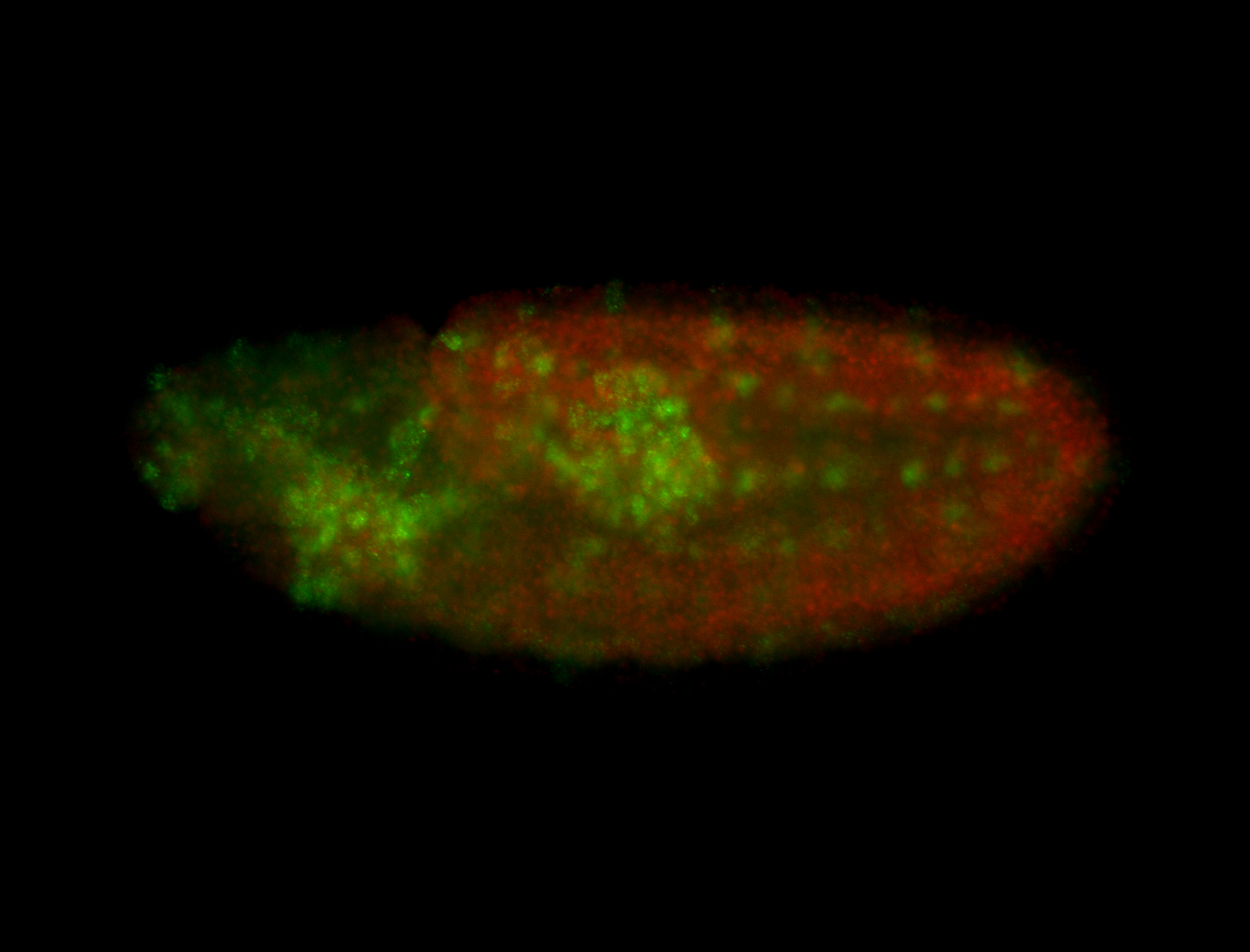
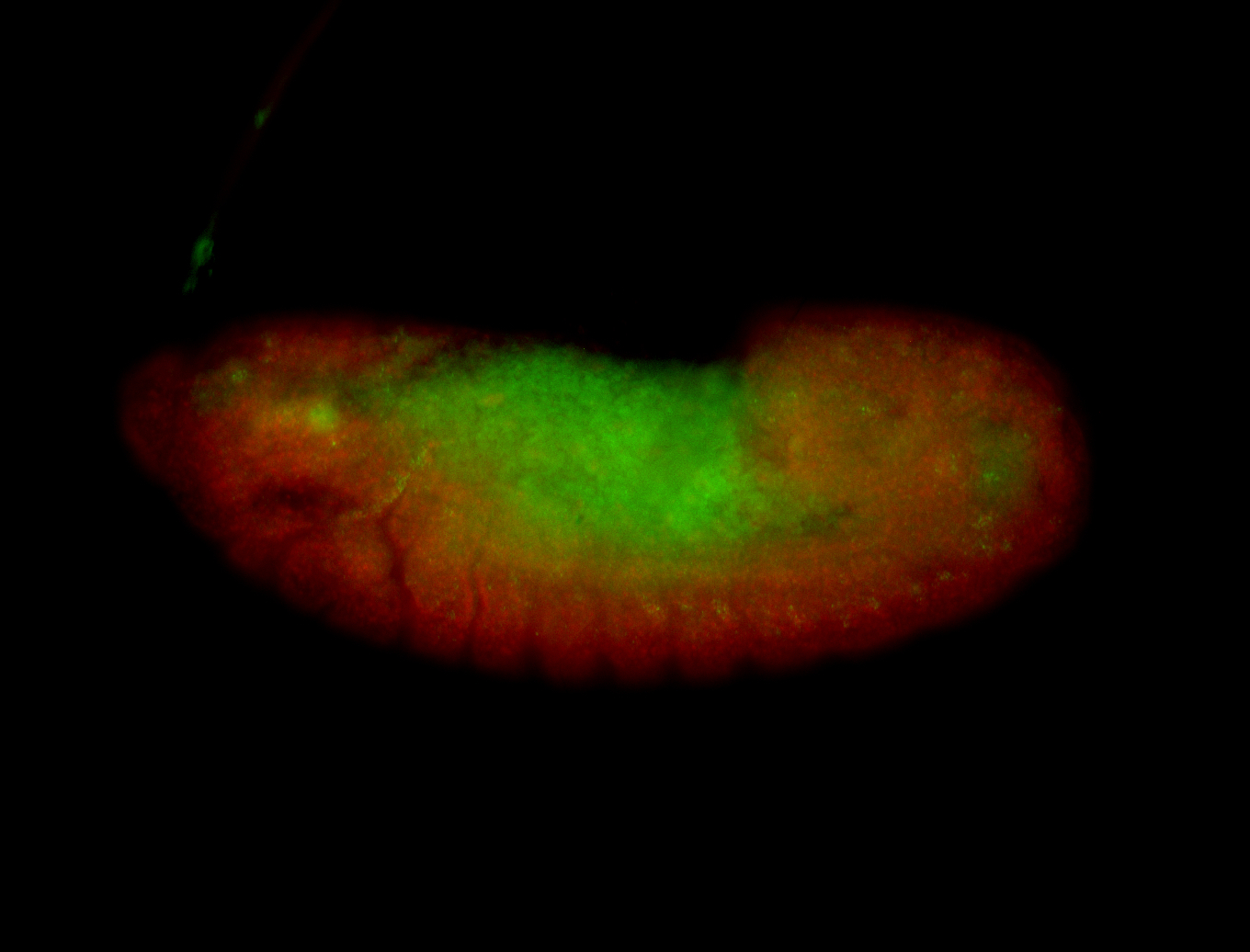
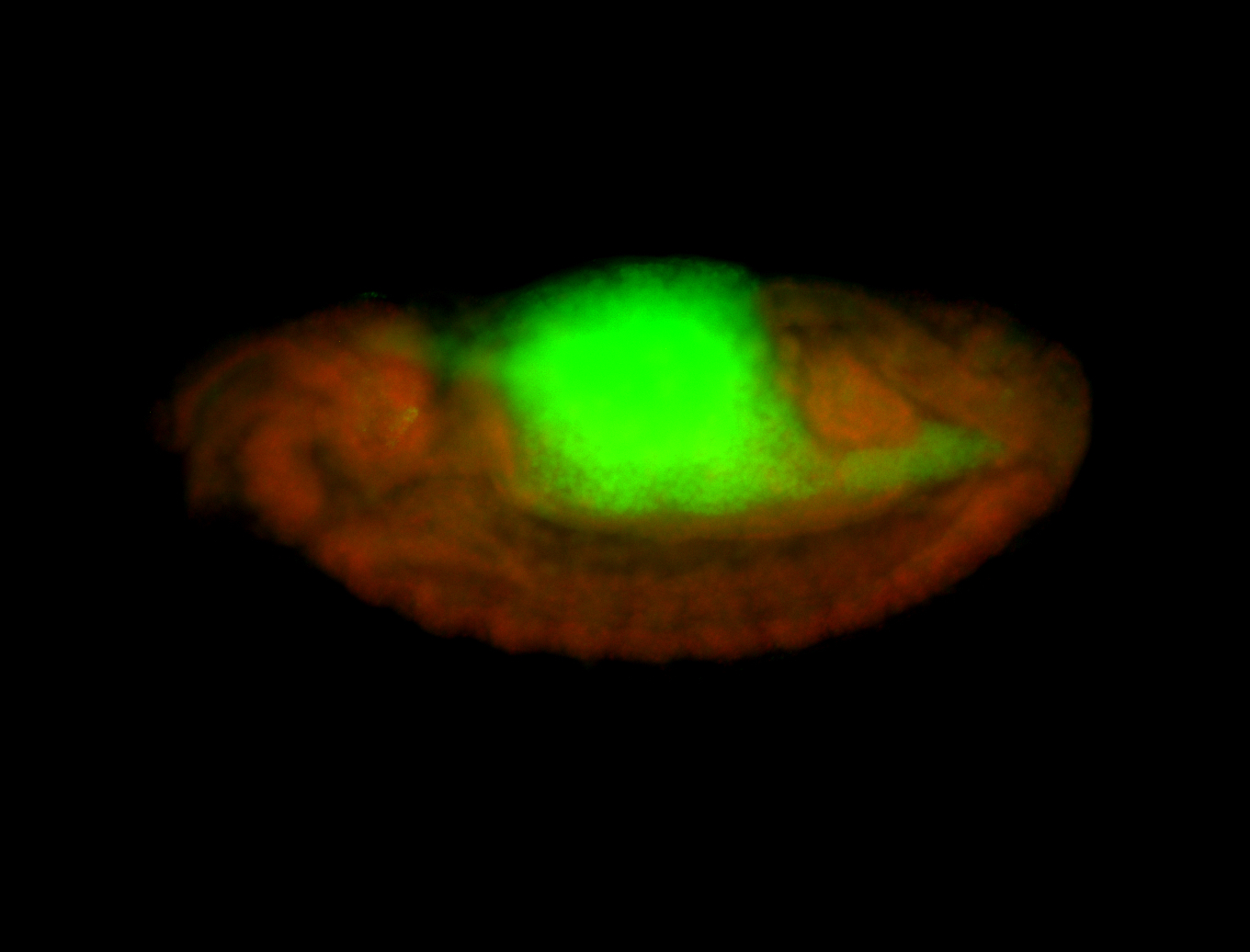
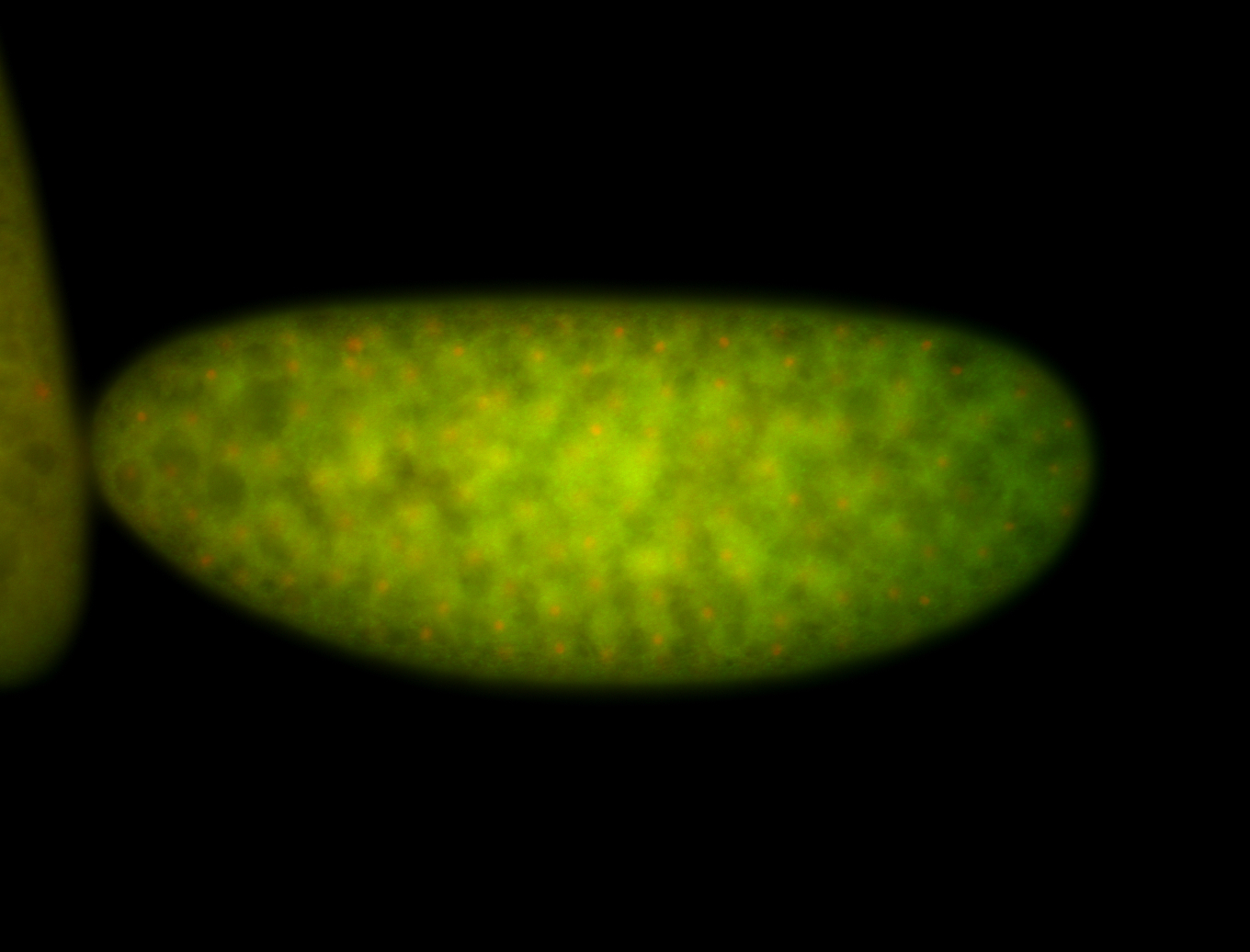
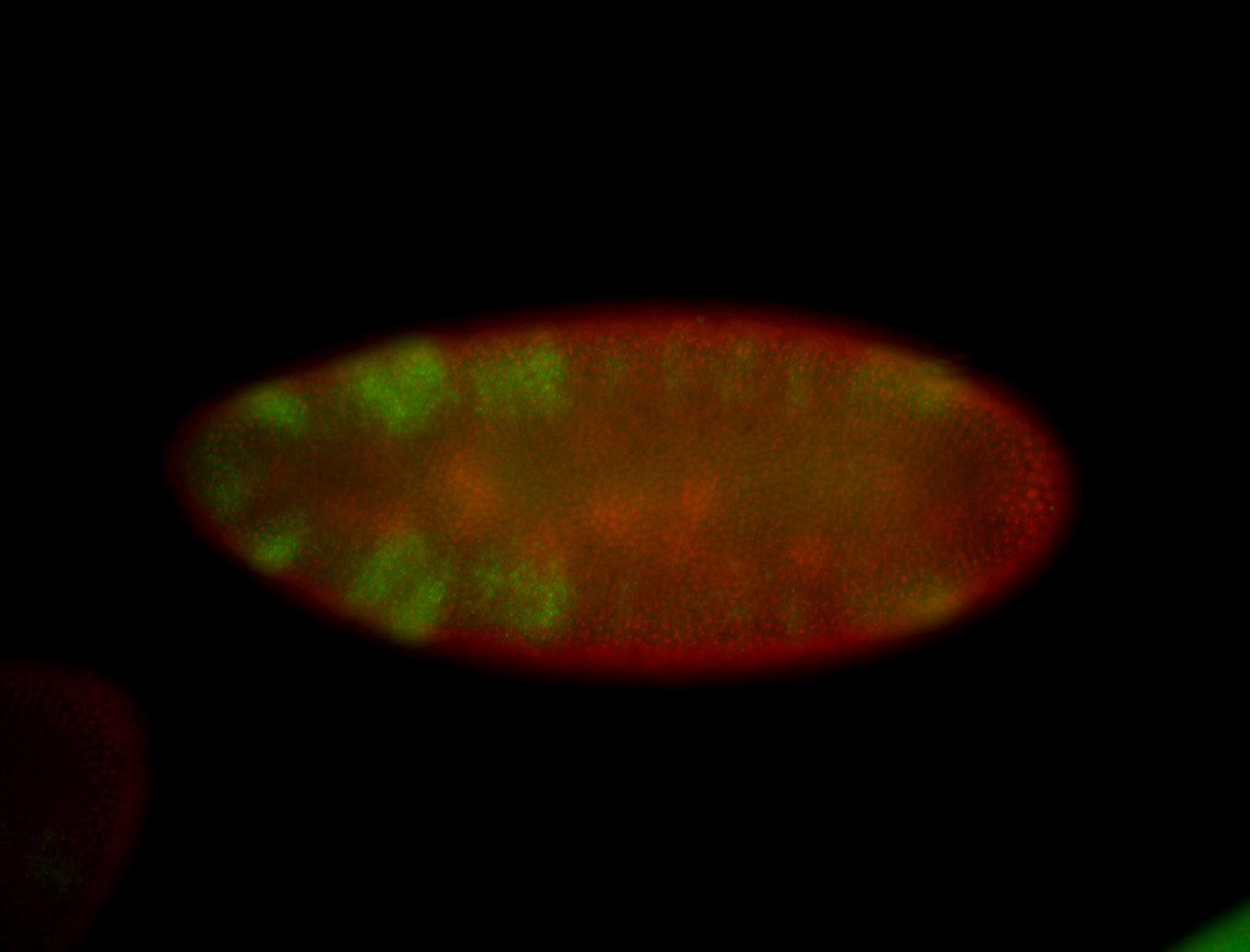
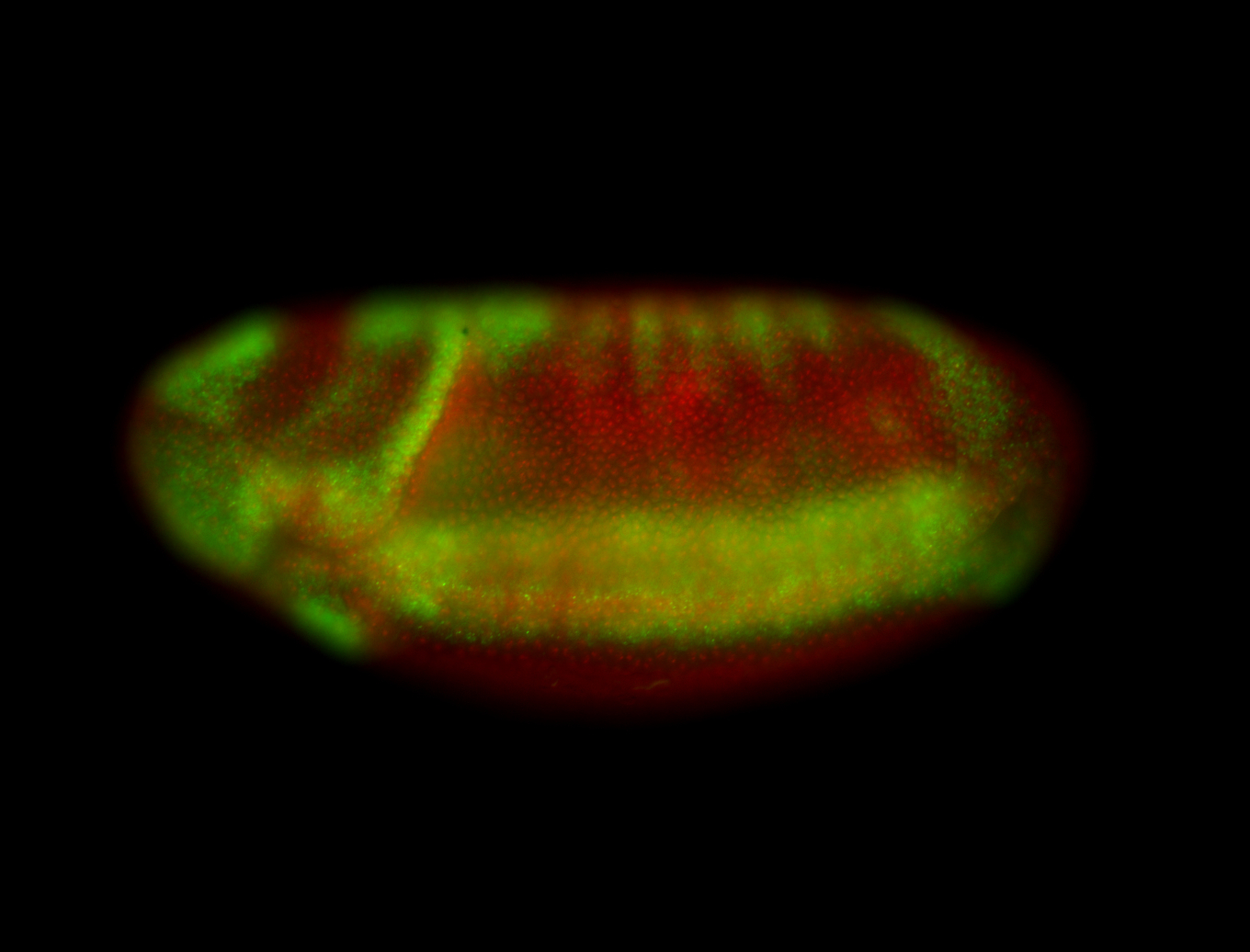
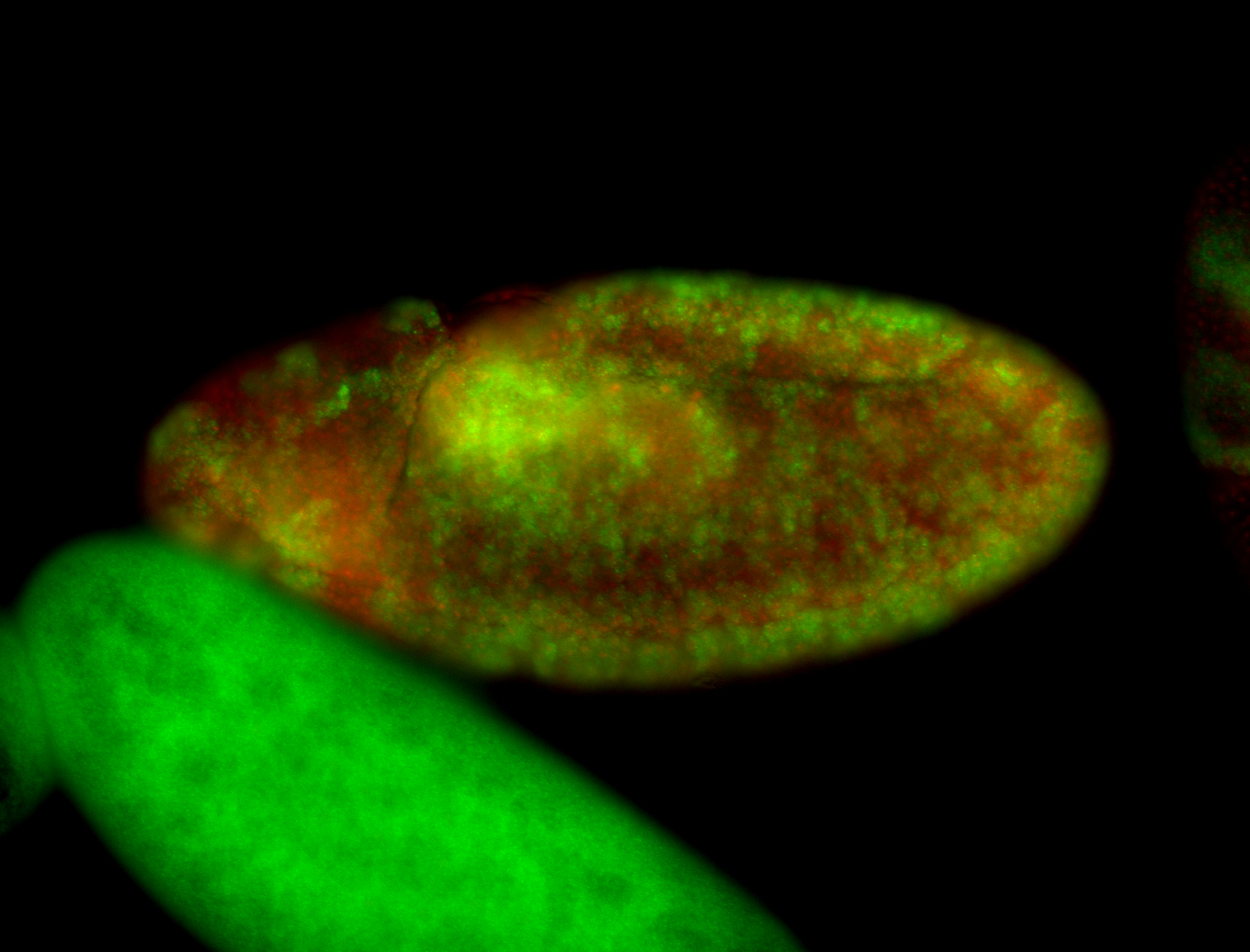
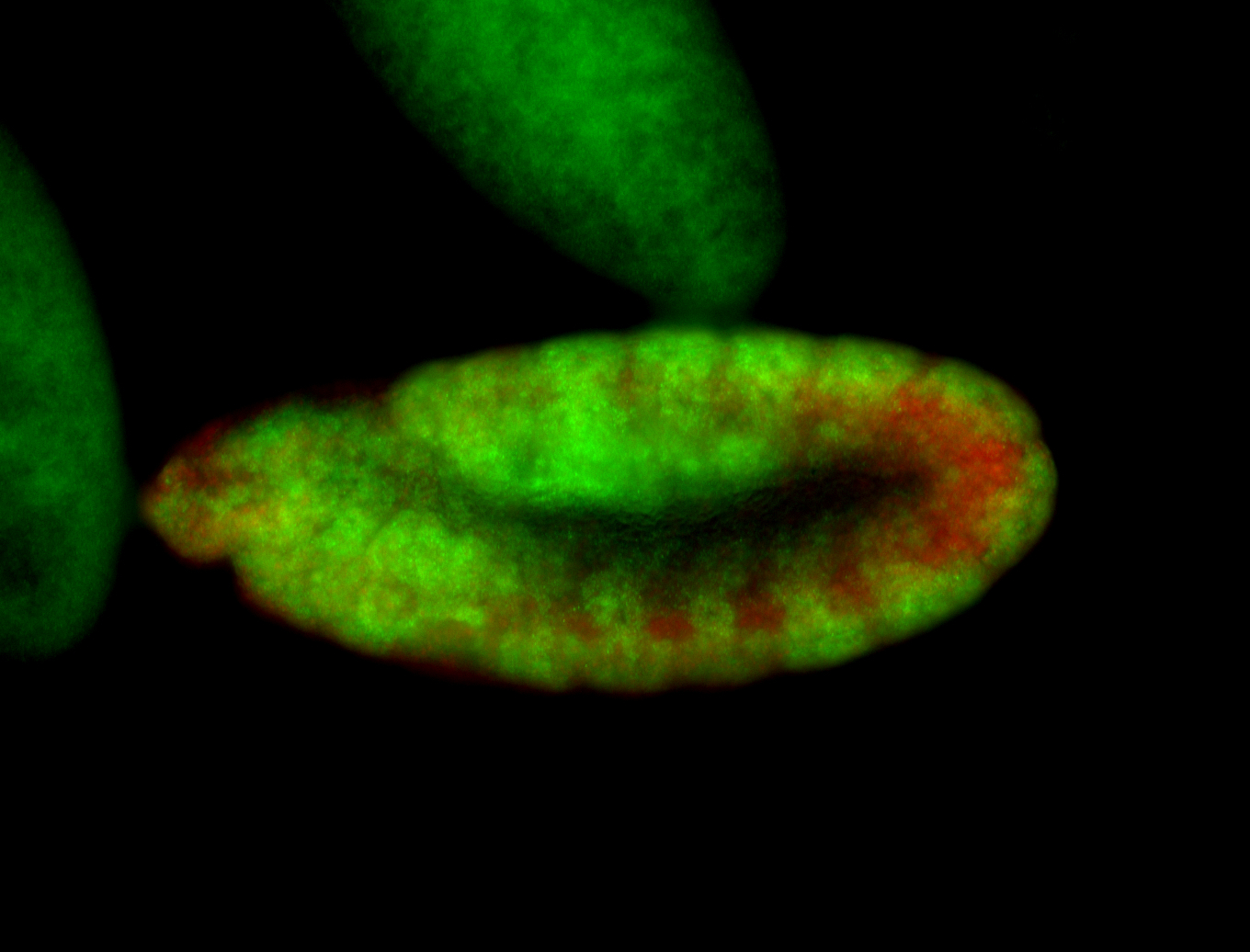
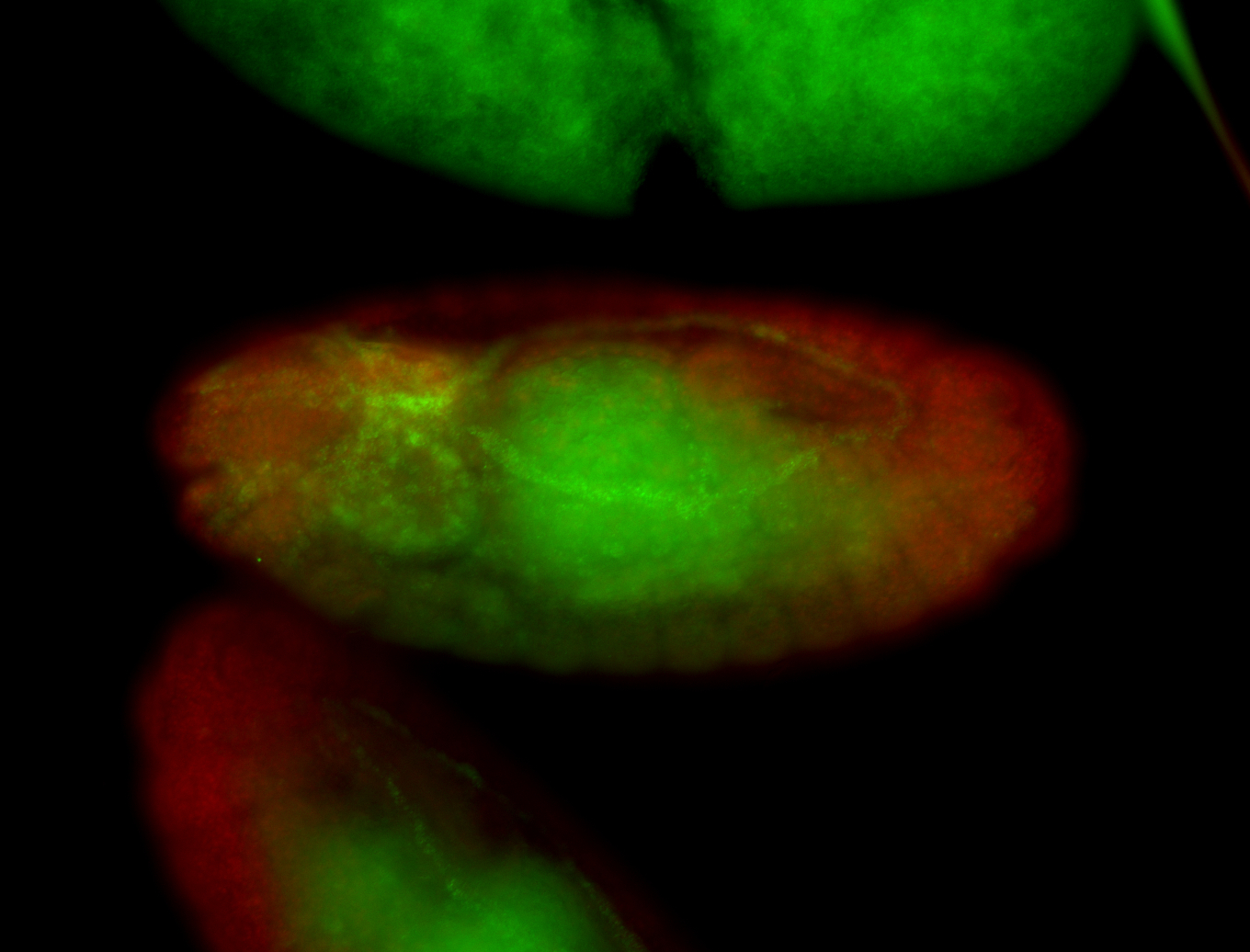
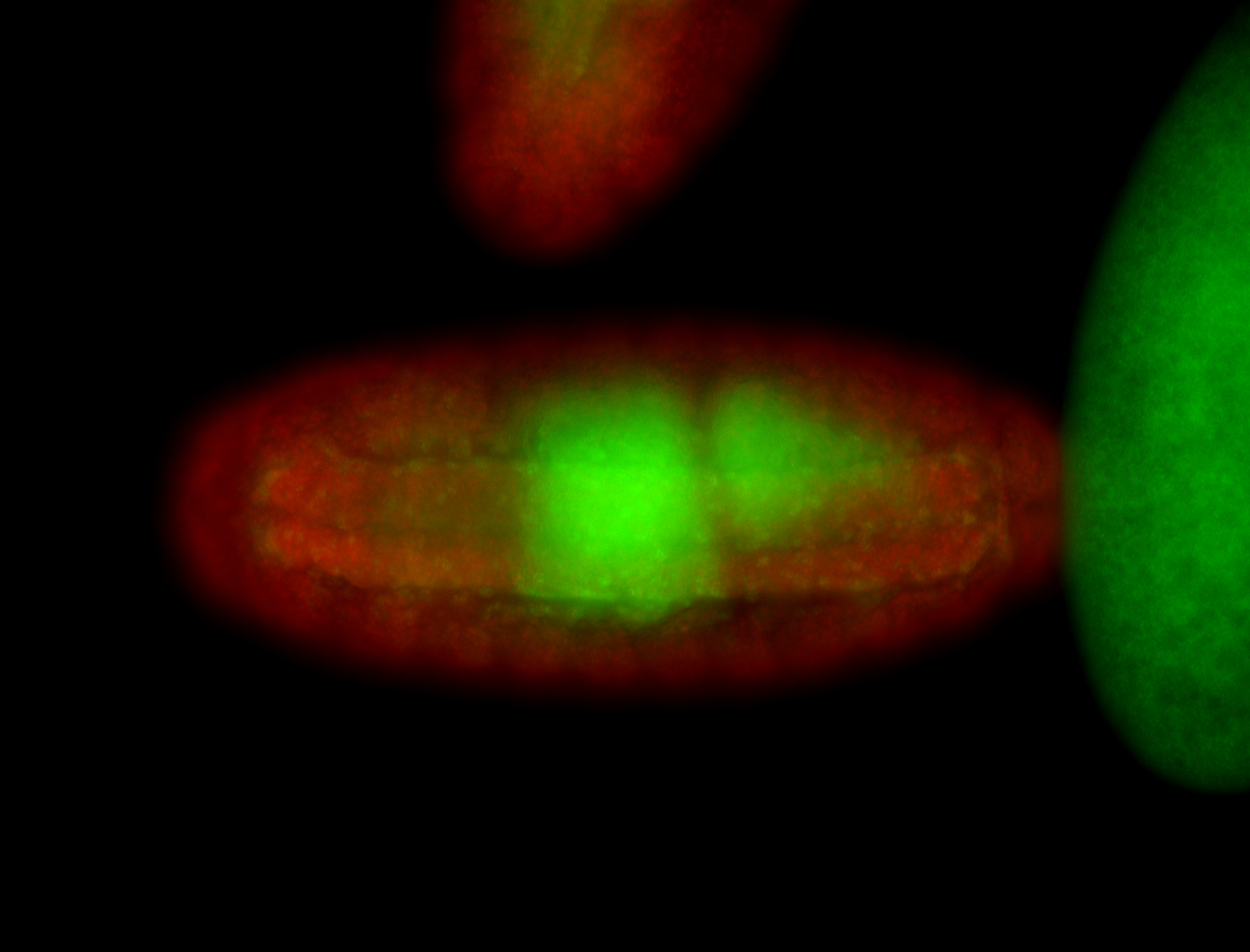
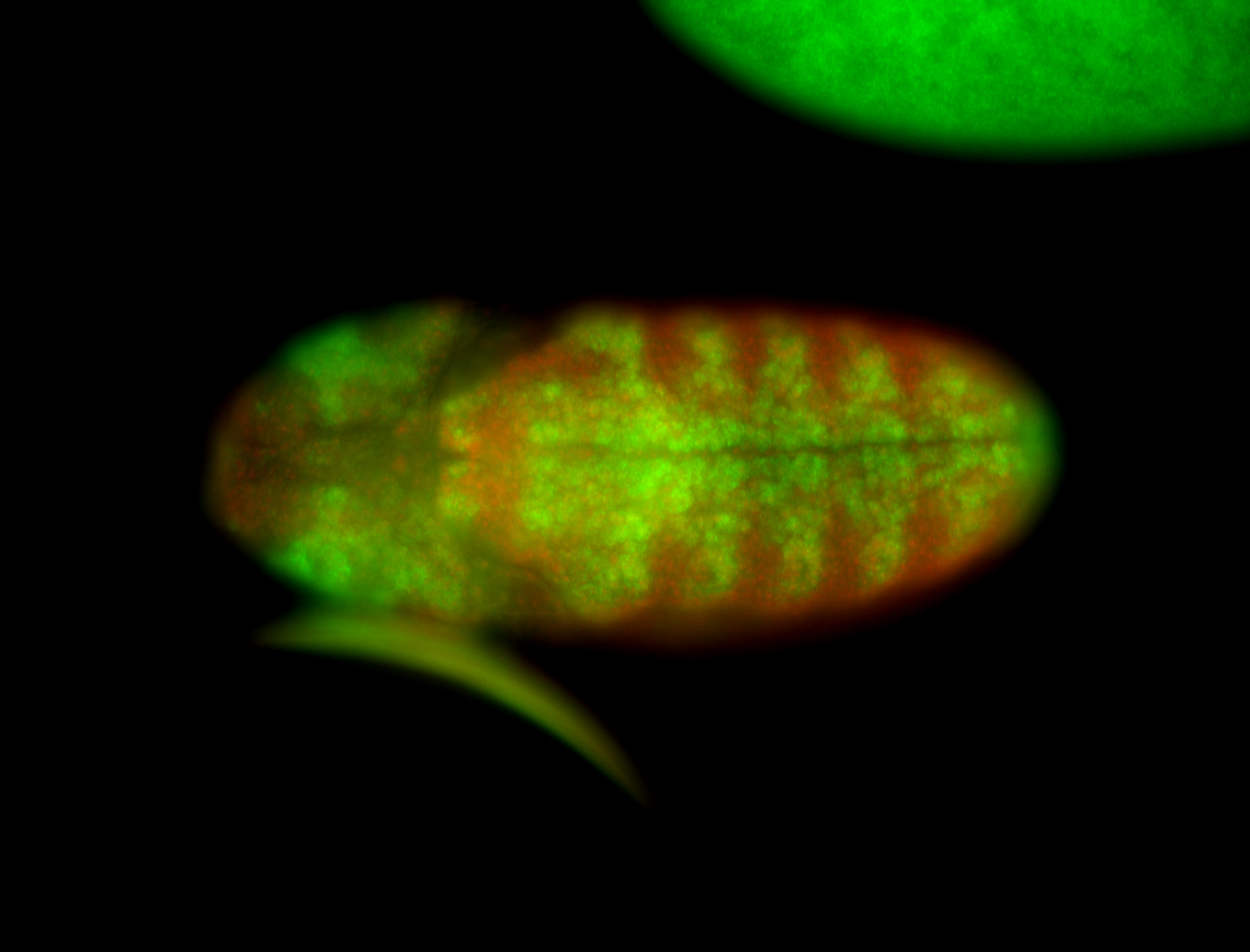
For a full list of searchable terms, please see the term page.
NOTE: It takes a few seconds for the term suggestions to load.
Result Summary
| Number of genes | 12 | |
| Export | ||
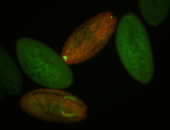
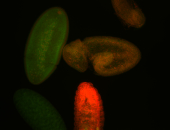
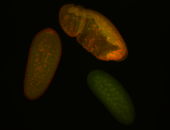
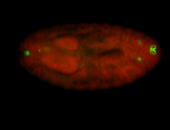
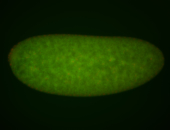
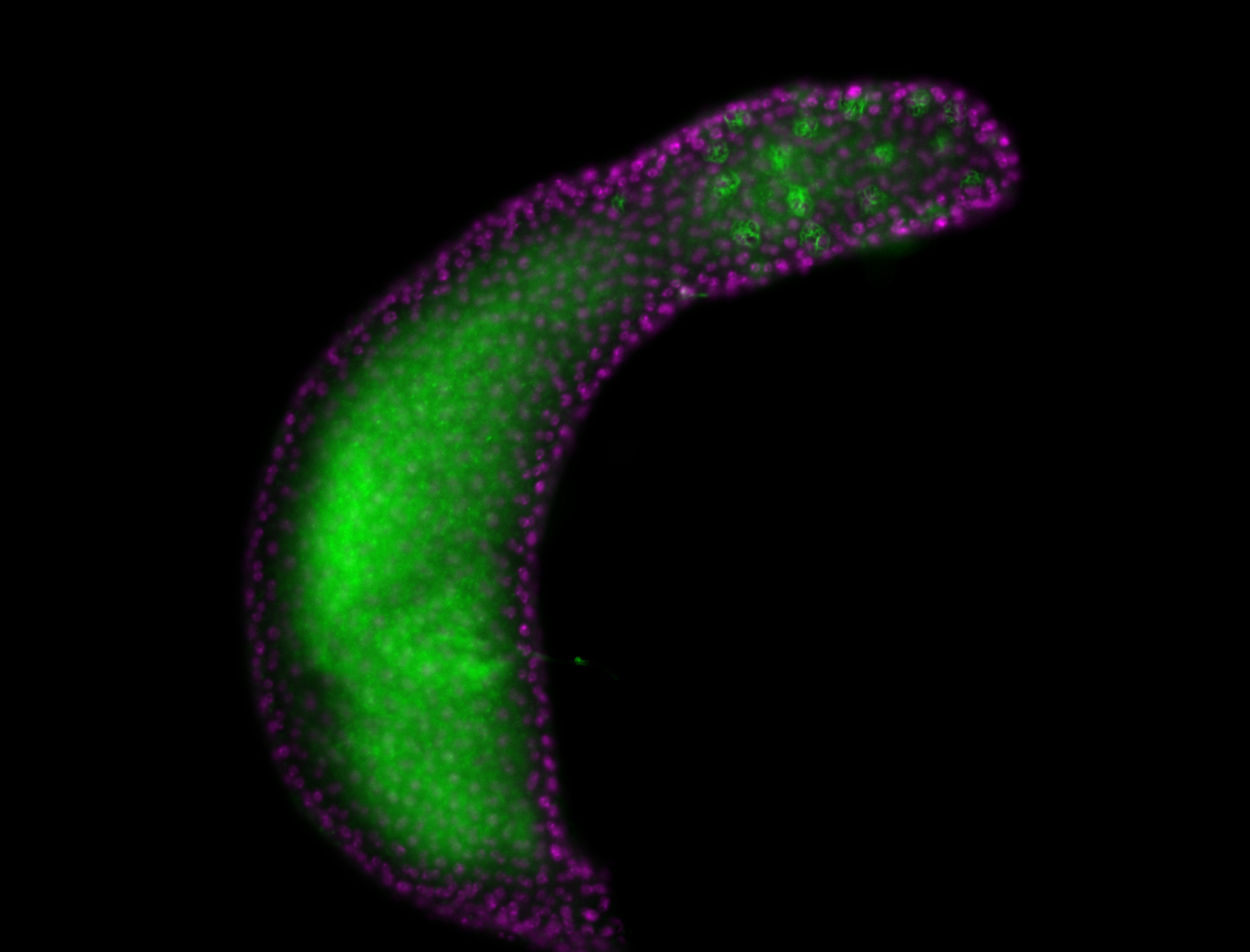
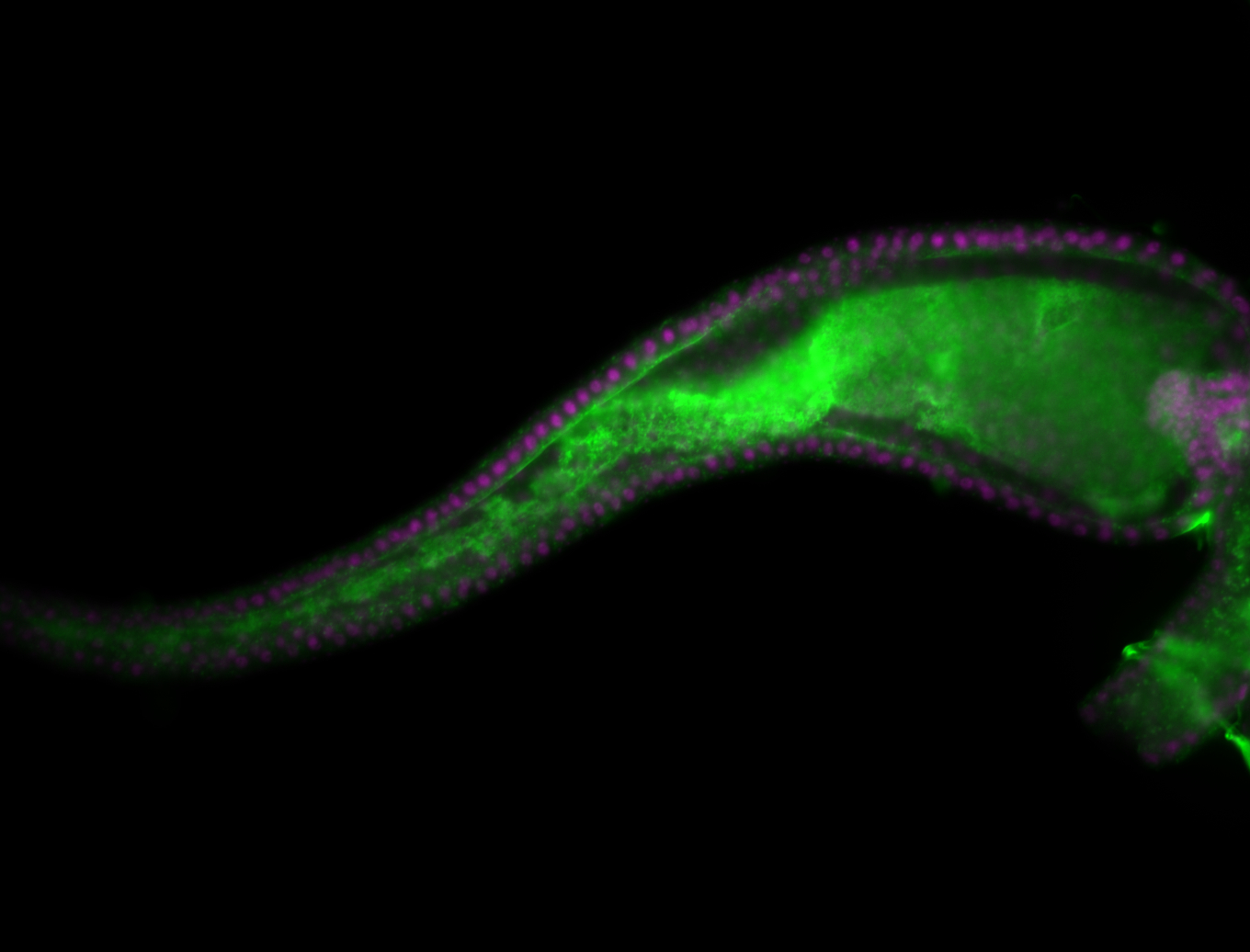
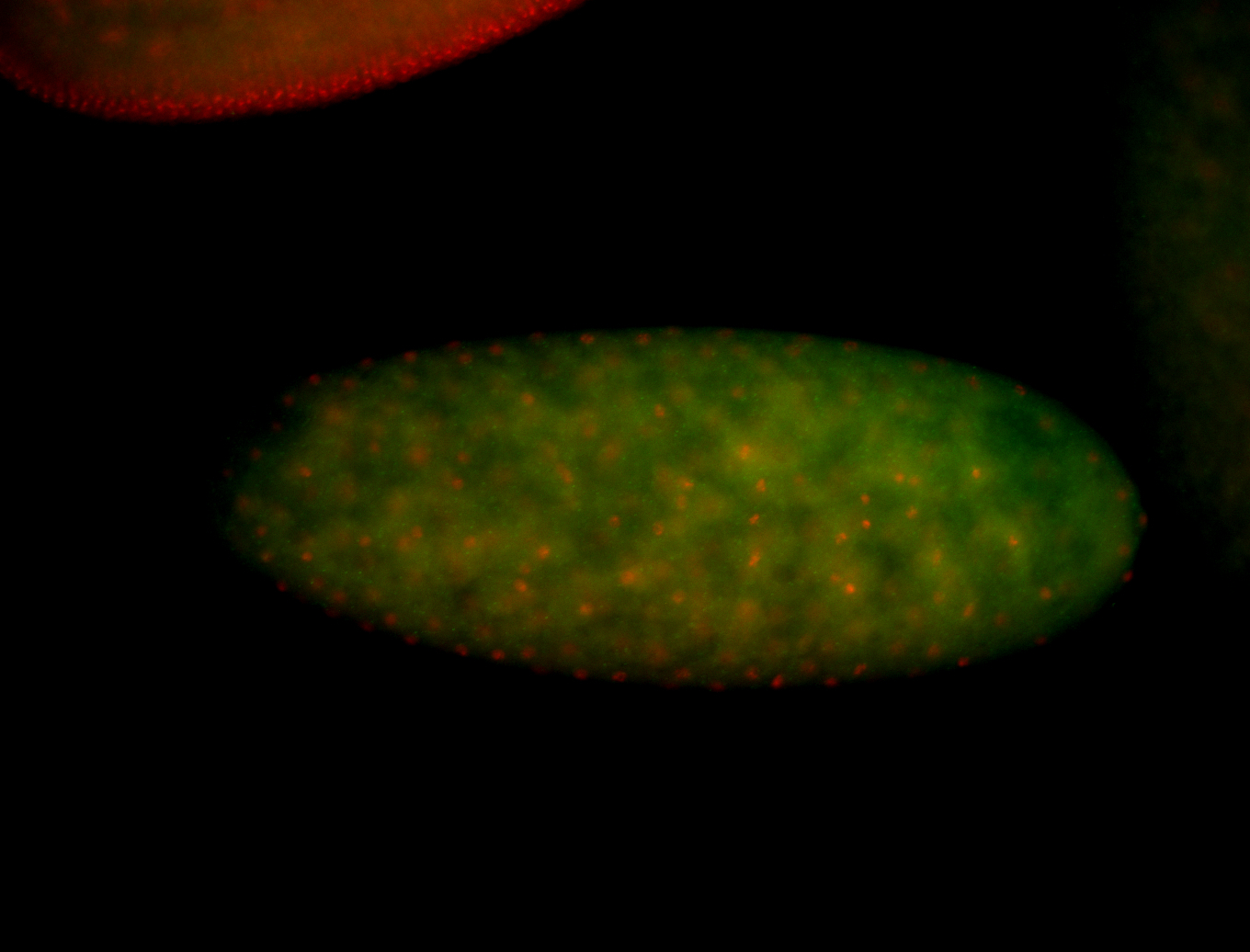
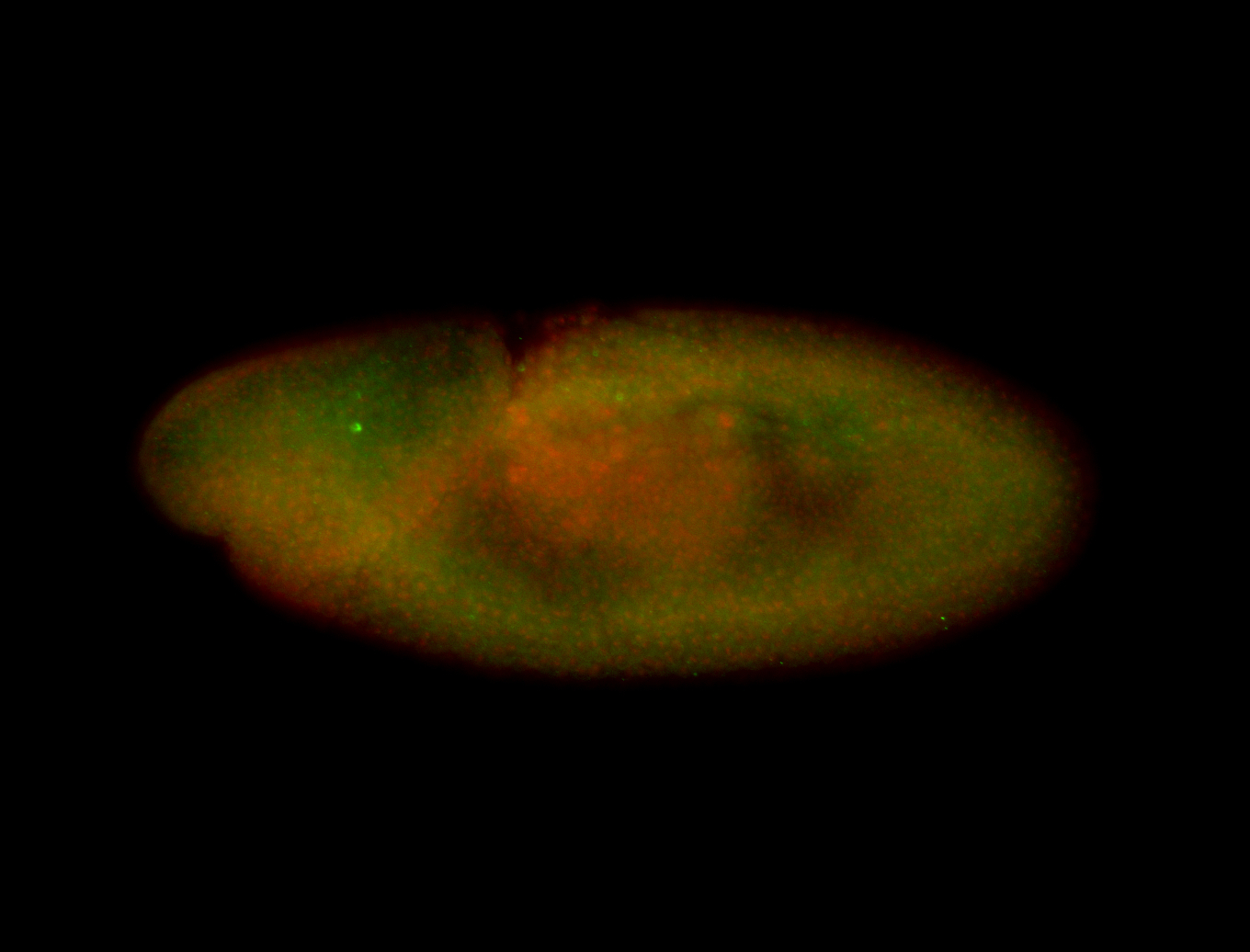
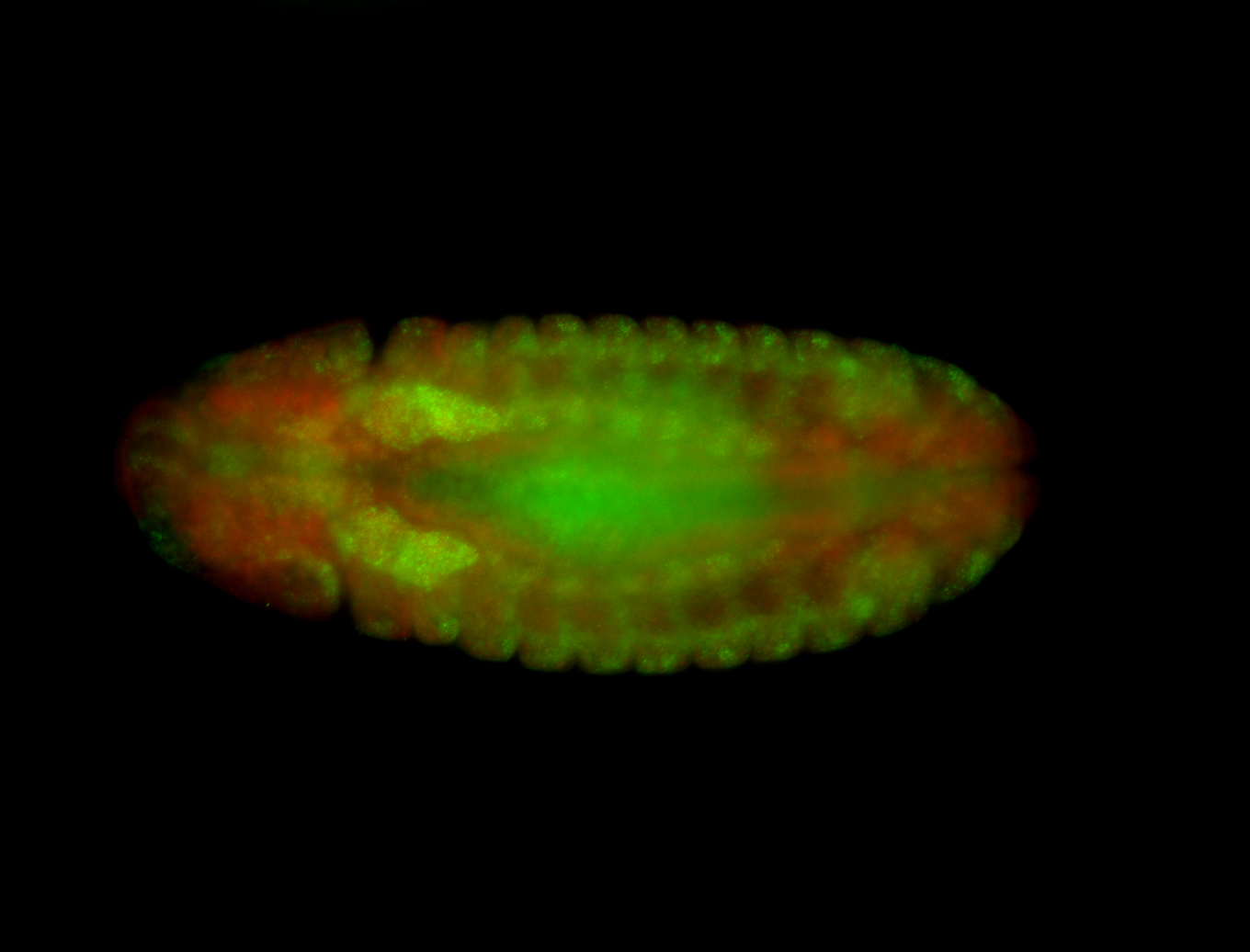
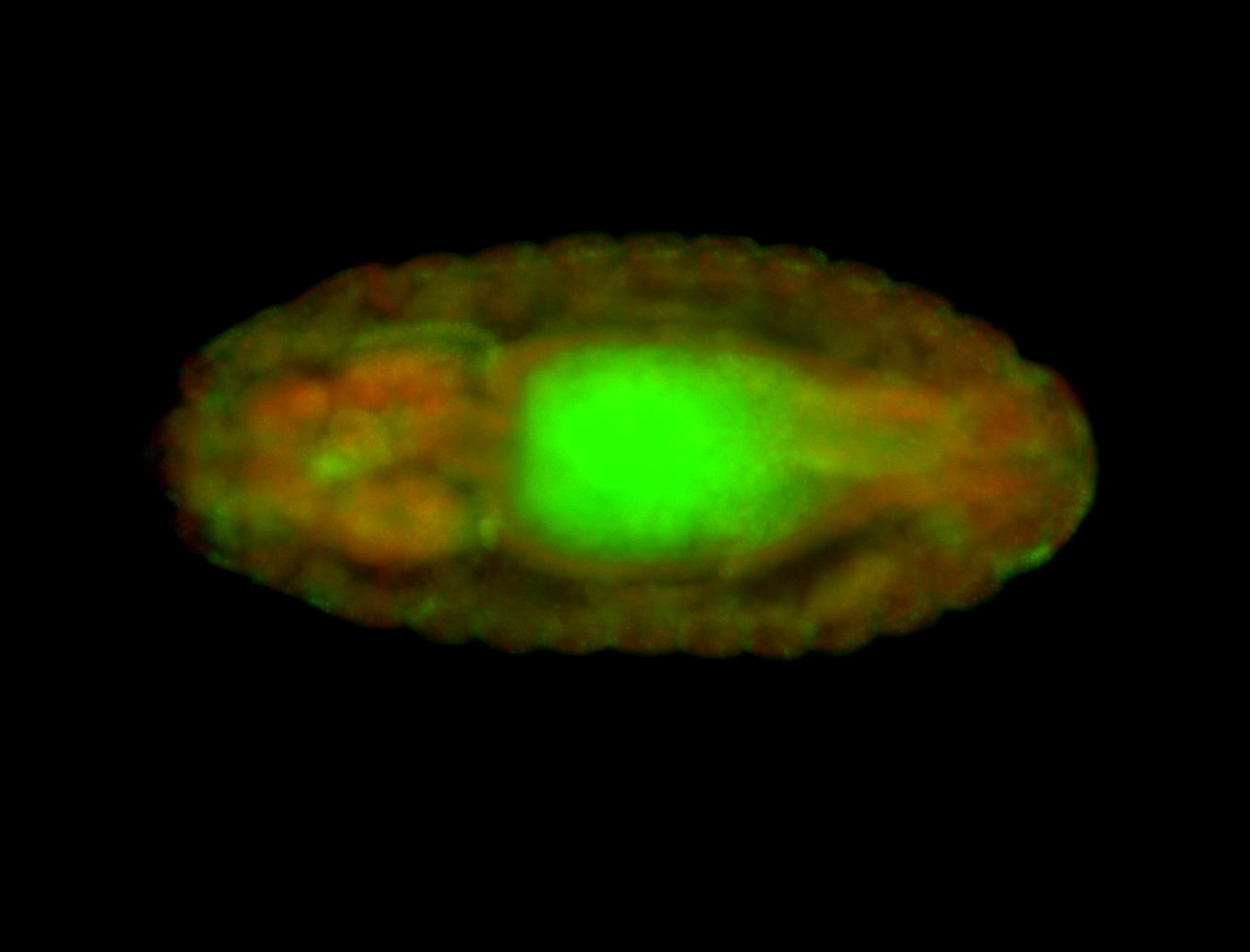
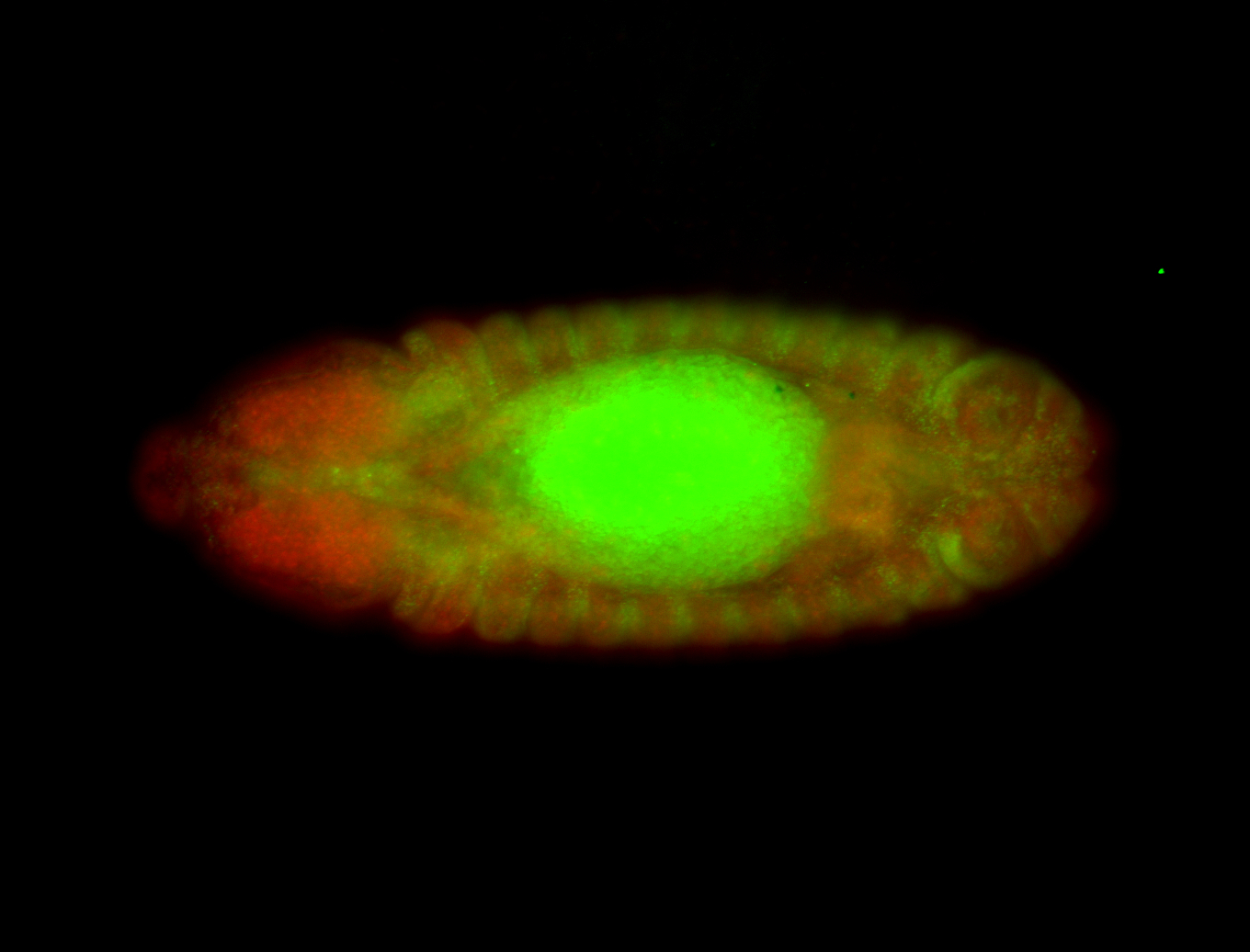
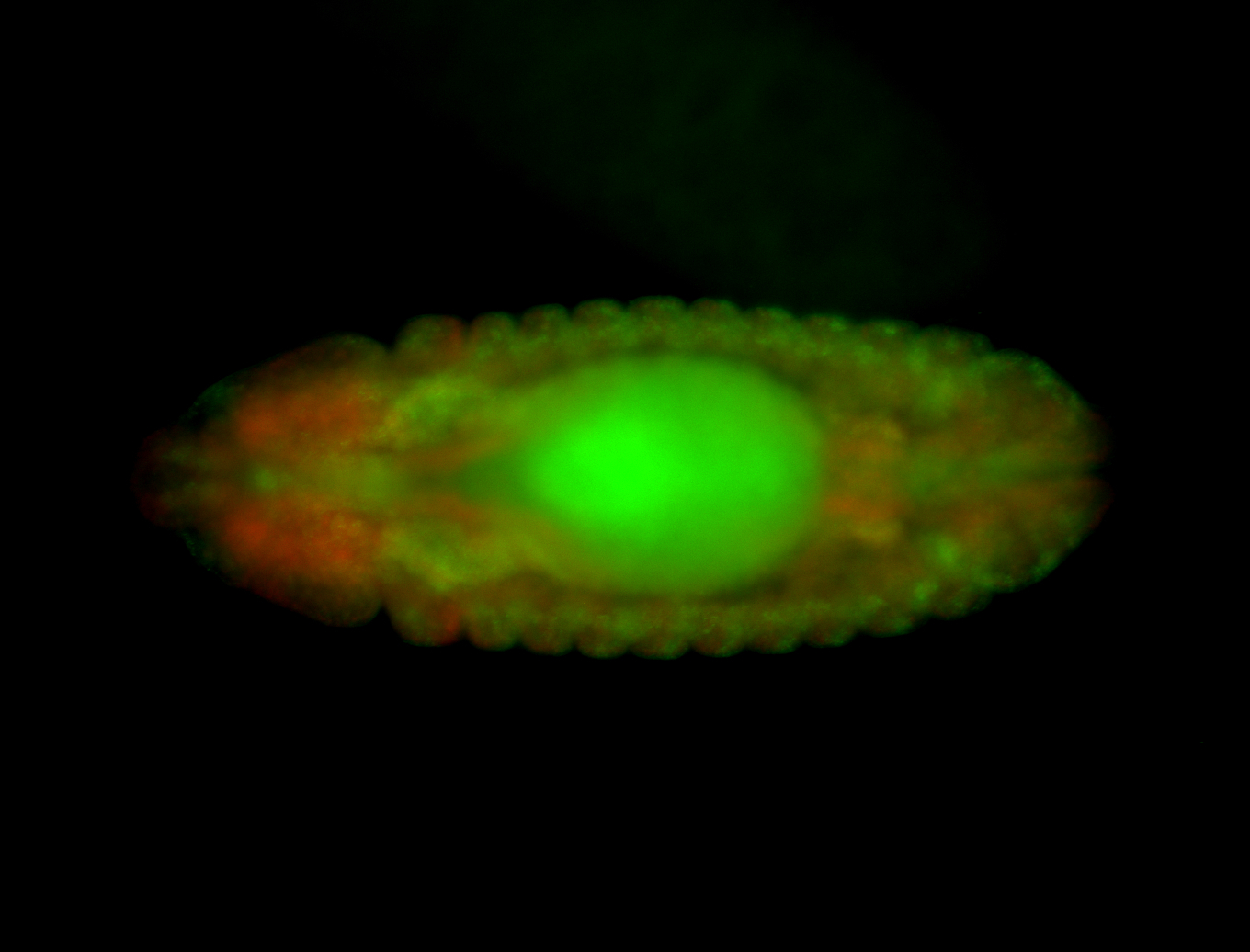
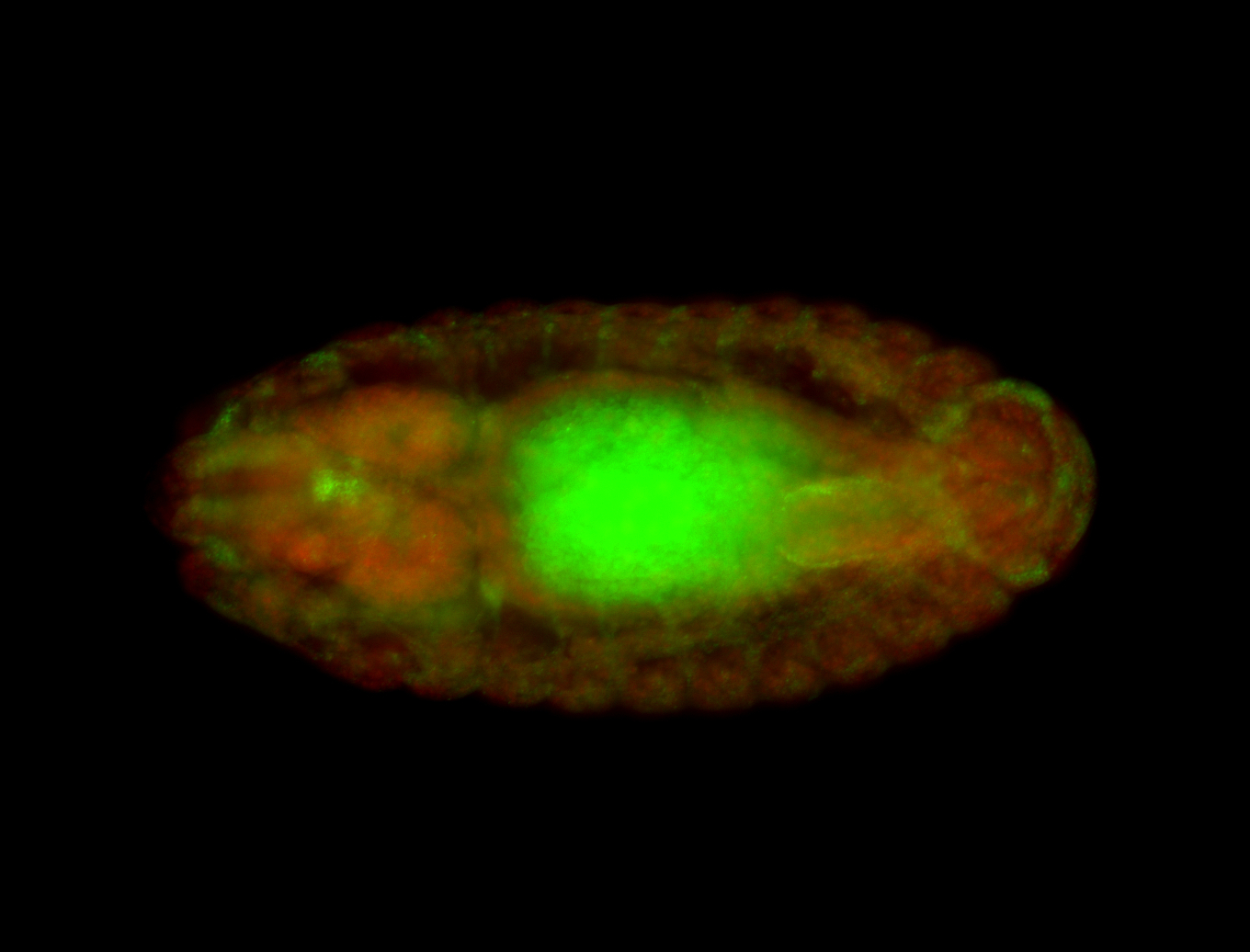
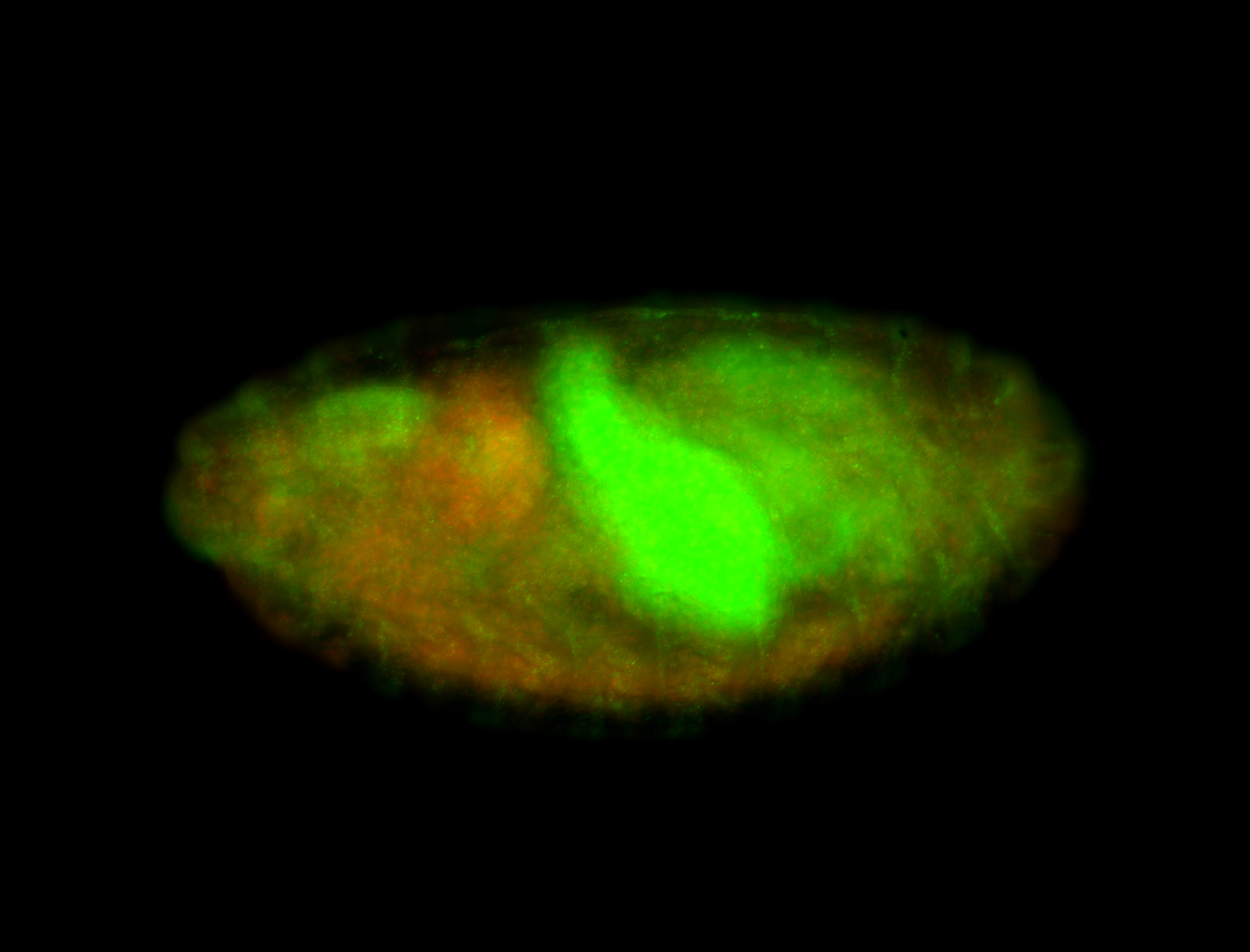
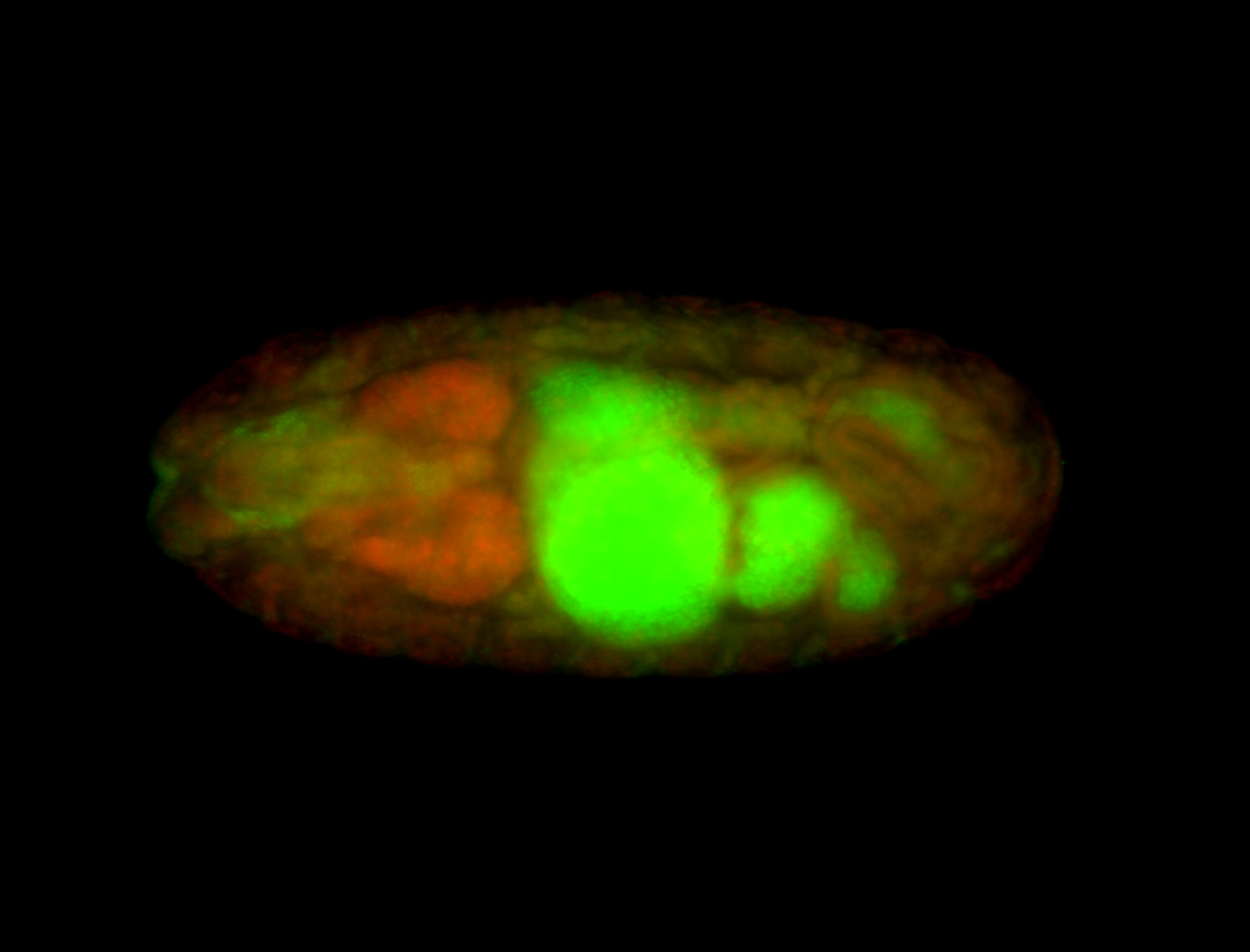
Embryo
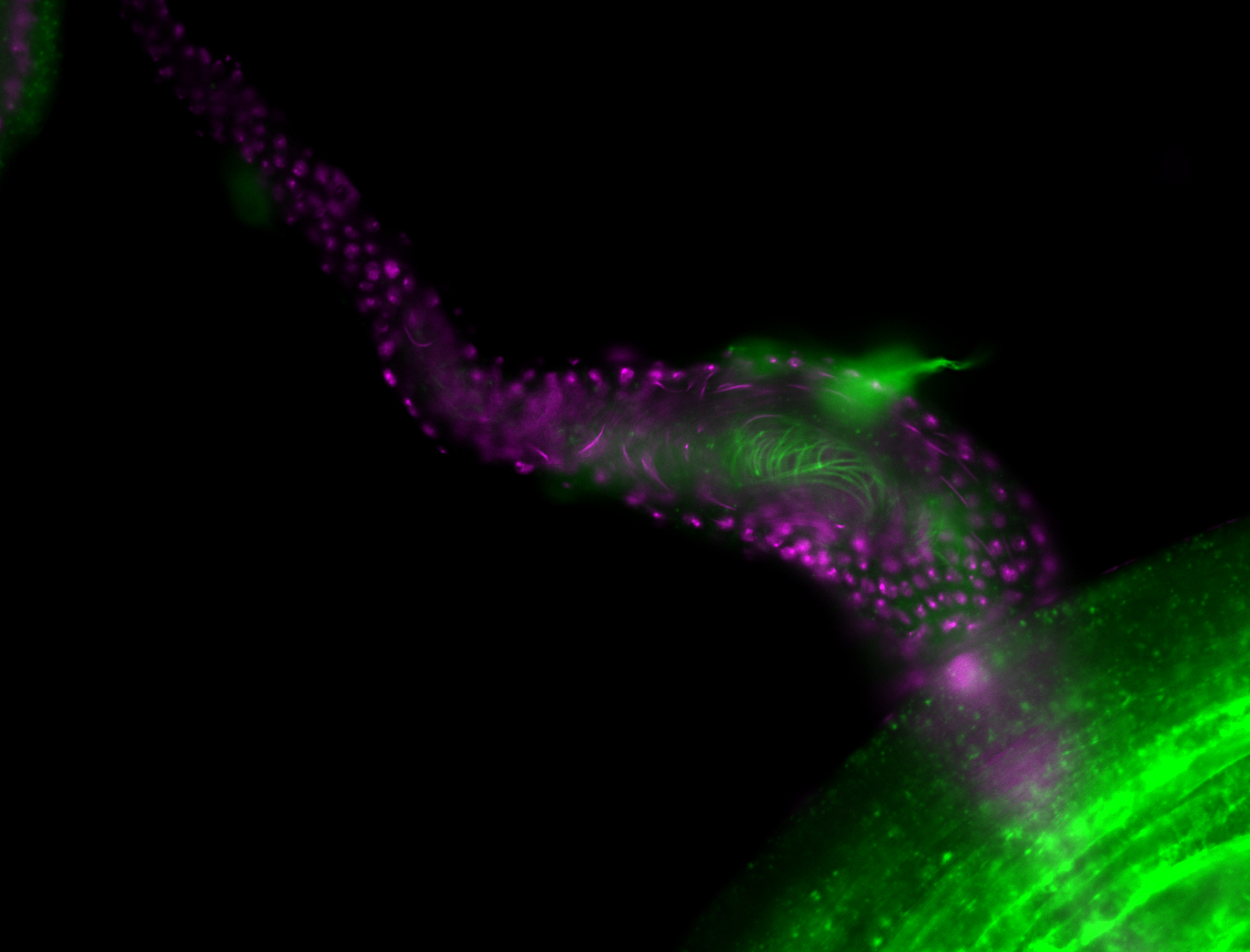
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Rays - posterior spiracles. |
| Good probe on gel. |
Embryo
Larva
Adult
Comments
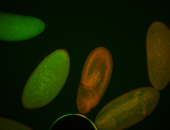
| Re-annotated by Ronit. Maybe some foci. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
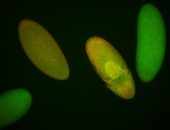
| -performed via HCR-FISH using 3 probe sets targeting the RE transcript isoform |
| -very neat germ band stage and late stage patterns; salivary gland pattern may be real |
| -images are from two separate experiments under identical conditions for better coverage |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
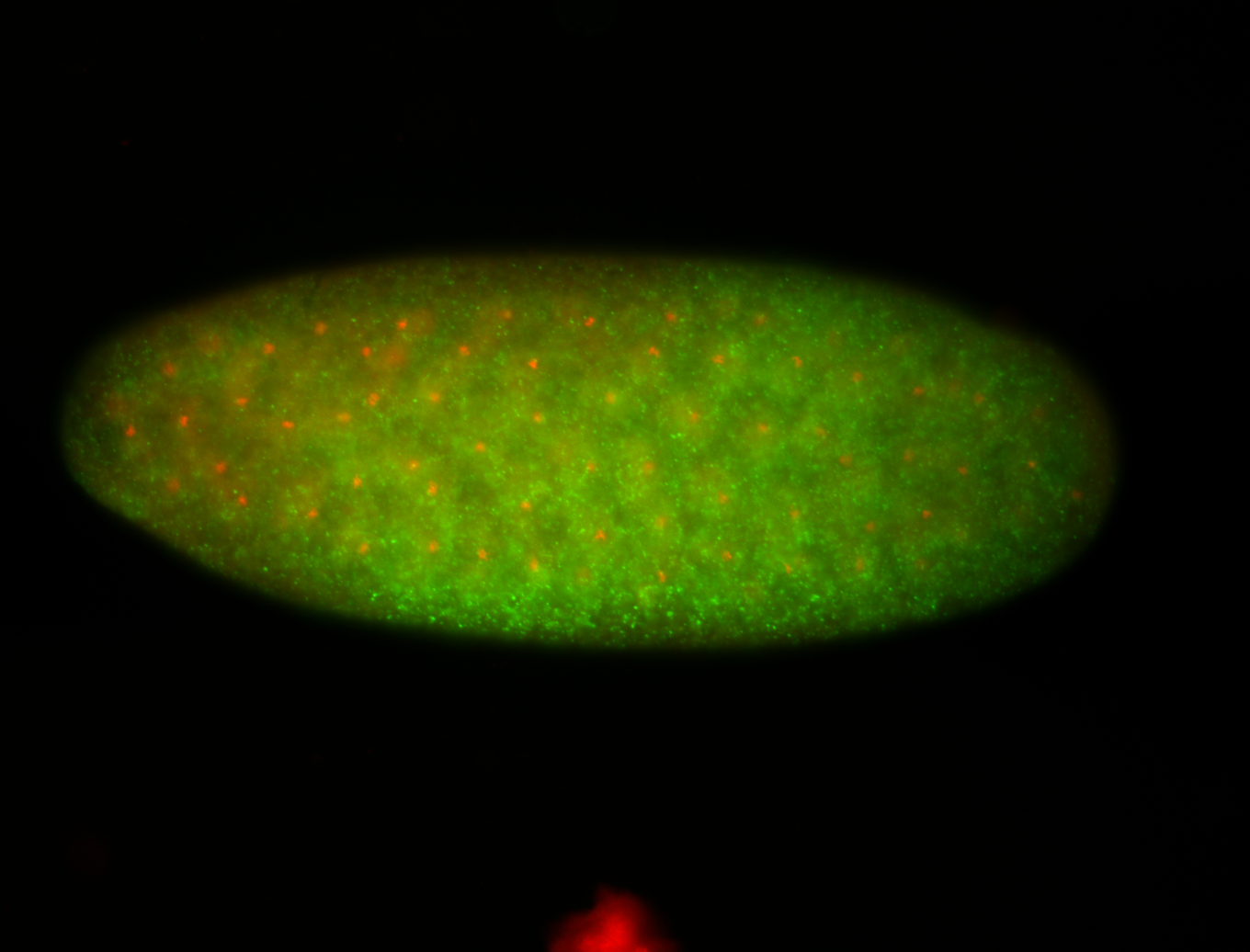
| -performed via HCR-FISH using 3 probe sets targeting the RB transcript isoform |
| -very diverse expression patterns in the late embryos; possibly expressed in the germ cell |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -There is no entry for this gene on BDGP, and modENCODE RNAseq data on FlyBase indicates that it is not expressed in the embryo. However, we observed obvious patterns during embryogenesis. Additionally, FlyBase does not have any in-depth characterizations of this gene. |
| -Osi21 does not overlap with any other genes, ruling out the possibility of the targeting of alternative transcripts. BLAST also confirms the specificity of our probes to Osi21. Additionally, no other genes for which probes were designed for on this plate show patterns such as those observed here, ruling out the possibility of cross-contamination during probe preparation. |
| -This may be a result of Osi21's poor characterization or incorrect RNAseq data |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -tested via HCR-FISH using 3 probe sets |
| -probes designed to target the transcript isoform Scr-RF |
| -a classic homeobox gene |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RD isoform transcript |
| -maternal signal may not persist until stage 8-9 due to possible background (maternal signal may or may not be real but does align with RNAseq data) |
| -no midgut and dorsal vessel signal was observed under our experimental conditions but was reported by BDGP |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed using HCR-FISH (3 probe sets) |
| -probes targeted dac-RB isoform of transcript |
| -very neat patterning in later stages; possibly visceral muscles of the hindgut |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -retested recently for late stage coverage (2025); appears to be expressed in a different program in the gnathal elements and sensory regions in late stages |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA transcript isoform |
| -expression seen in the stomatogastric sensory primordium, and the anterior and posterior midgut primordium; forms a complex along with ac (CG3796) so should colocalize in neuronal regions |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA transcript isoform |
| -very diverse and neat patterns at all stages |