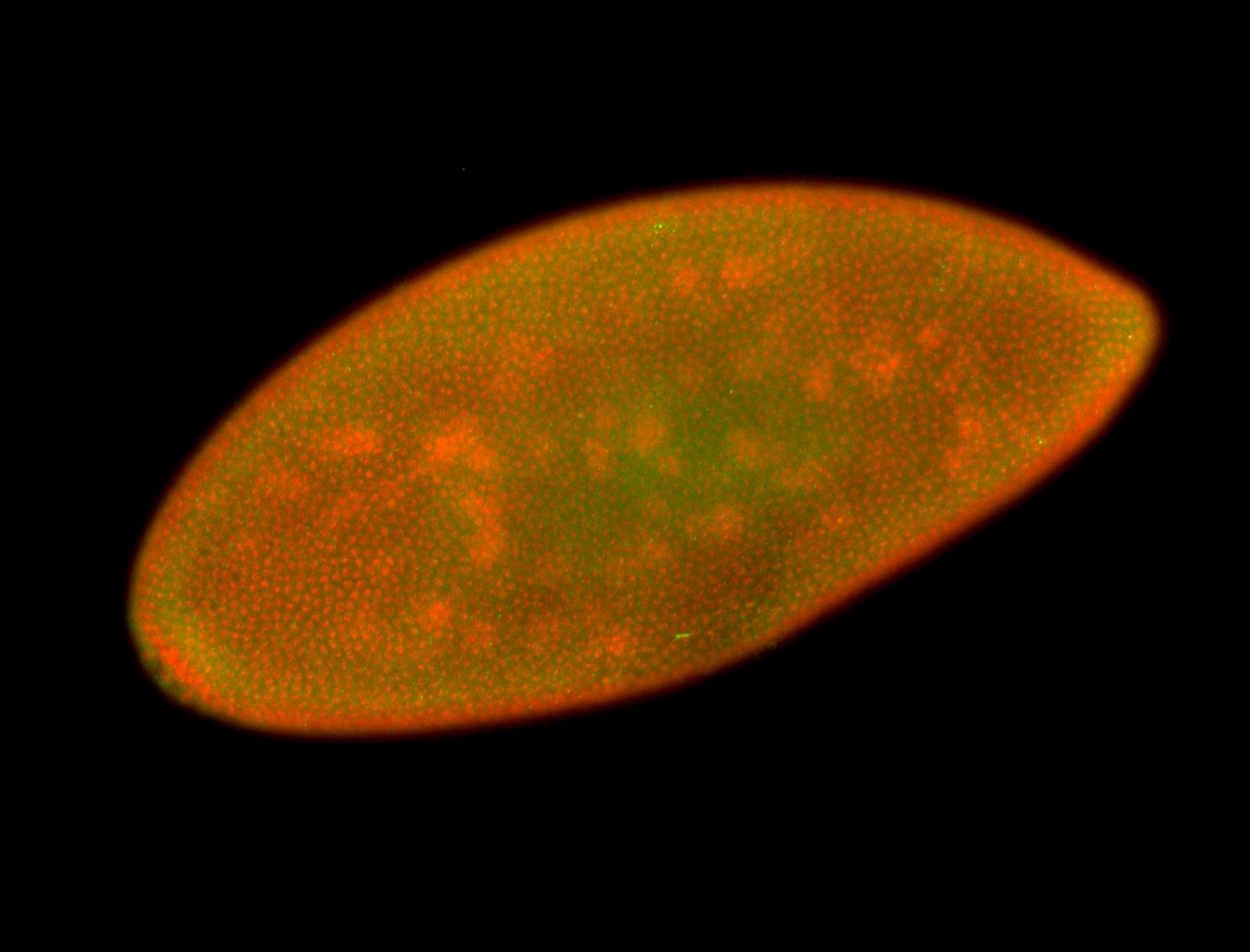
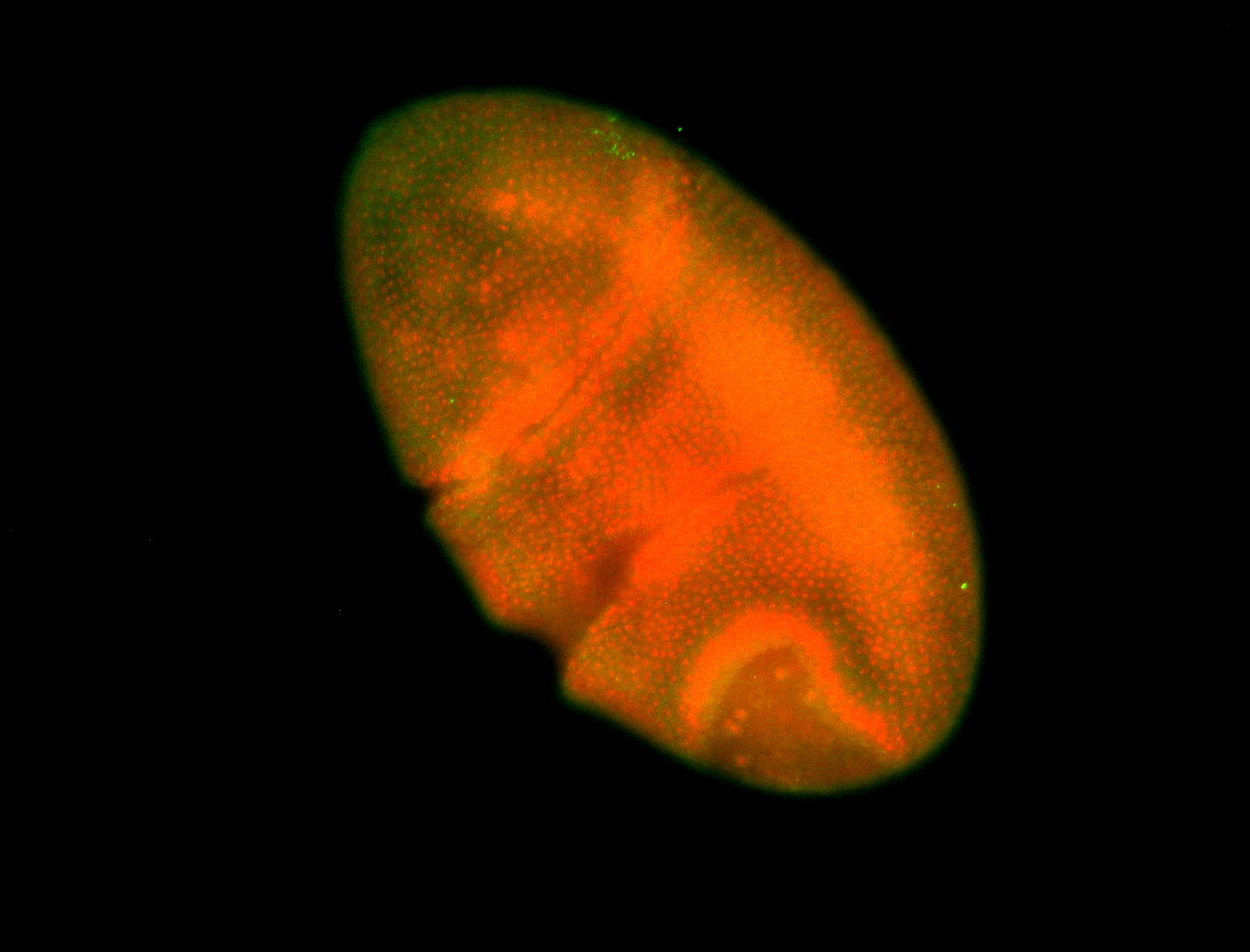
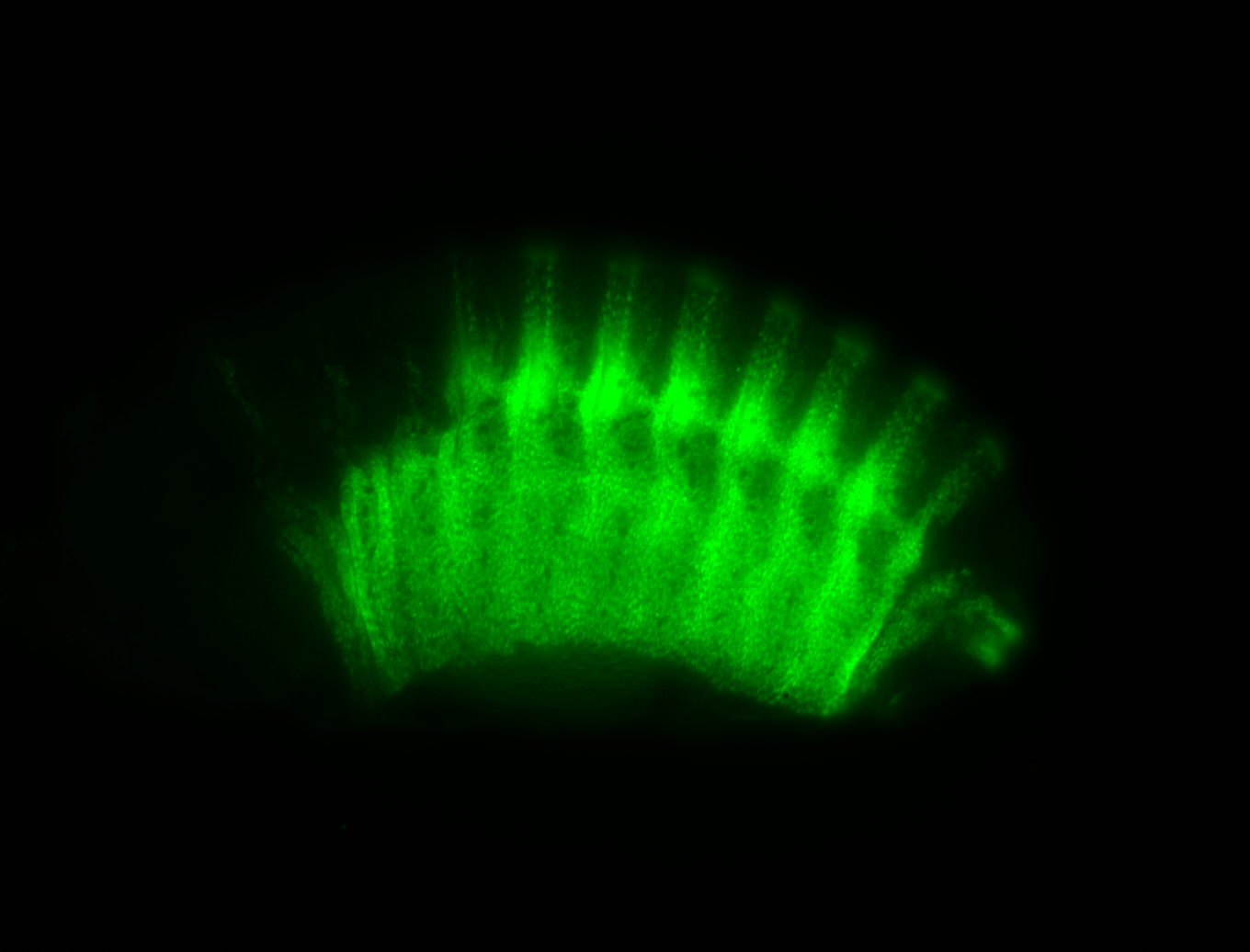
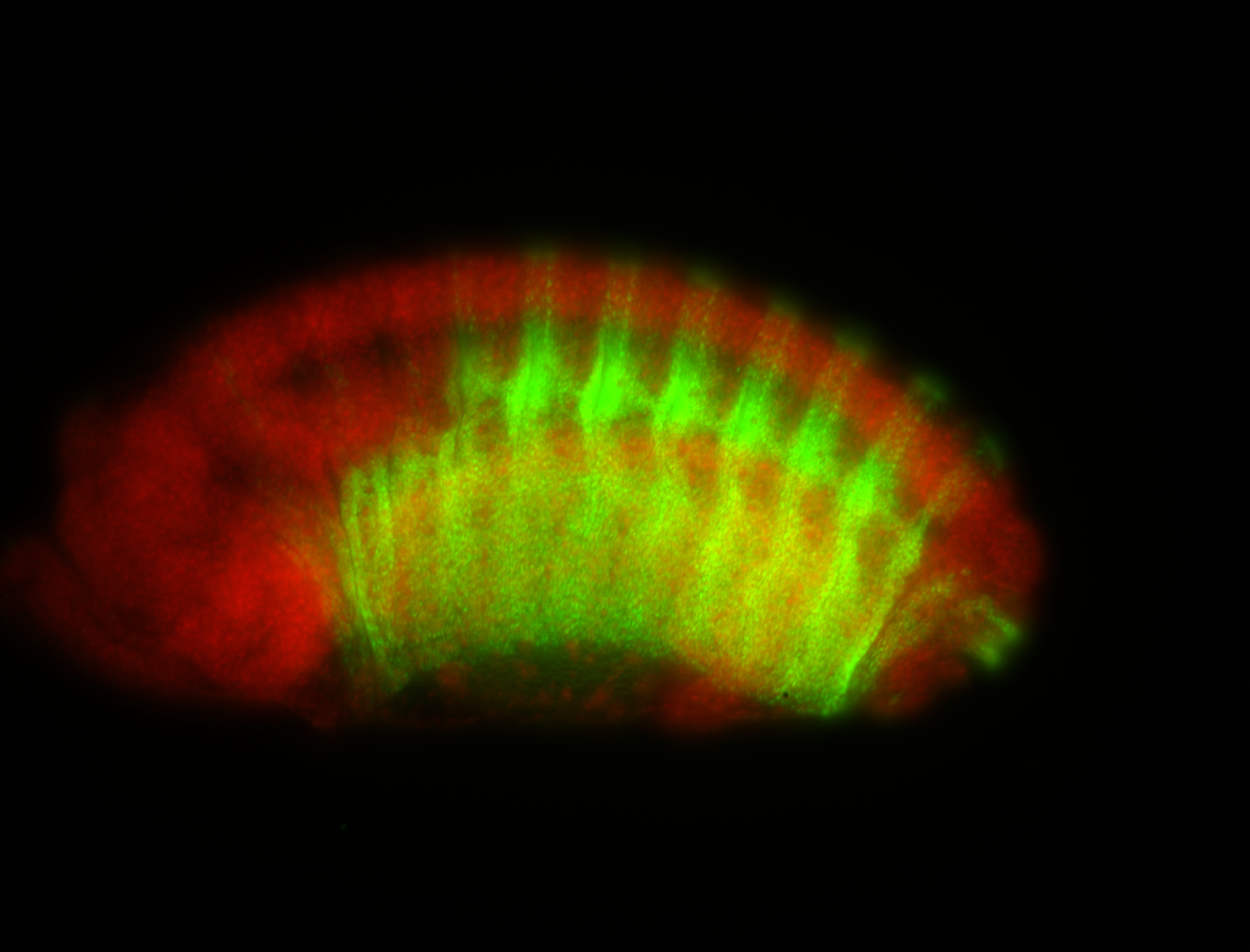
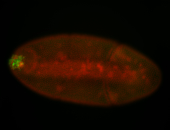
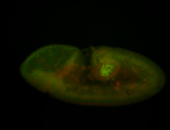
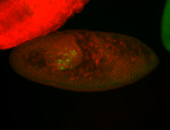
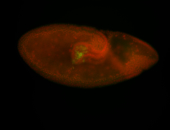
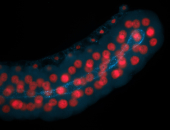
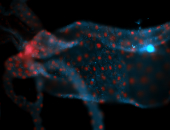
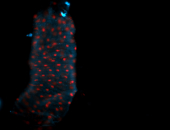
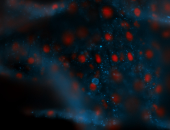
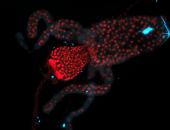
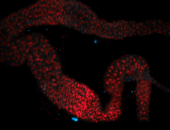
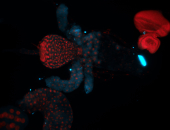
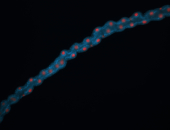
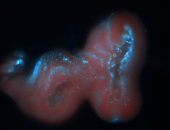
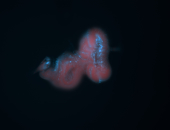
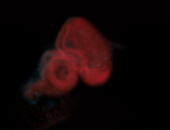
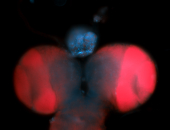
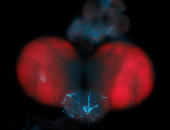
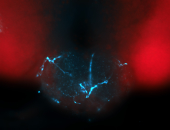
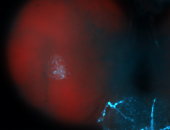
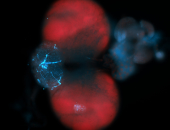
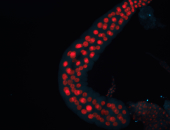
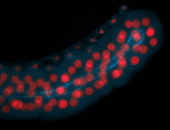
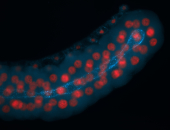
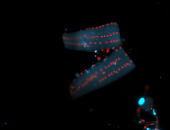
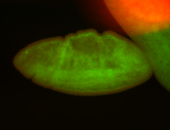
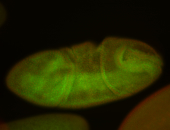
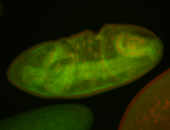
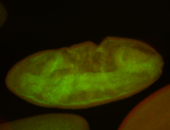
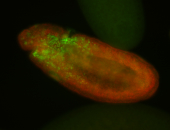
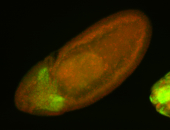
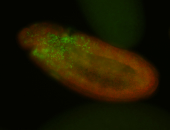
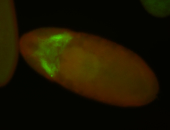
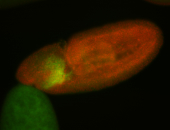
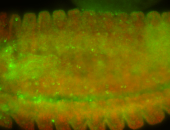
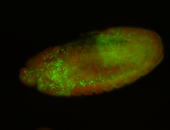
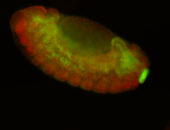
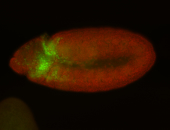
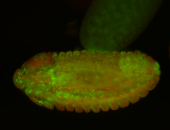
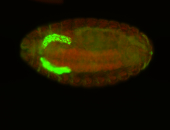
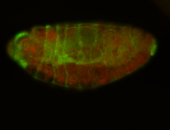
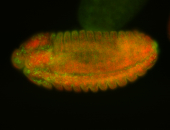
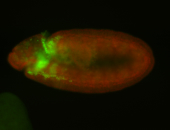
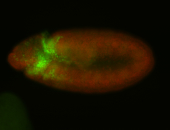
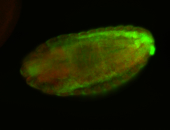
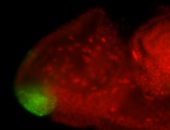
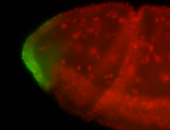
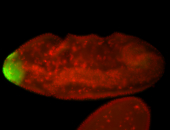
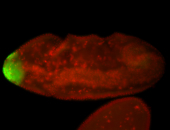
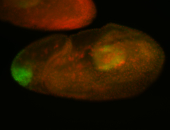
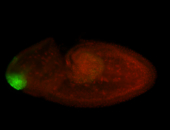
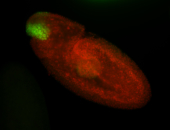
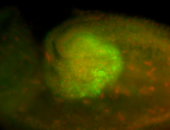
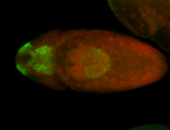
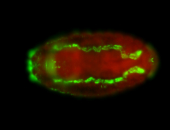
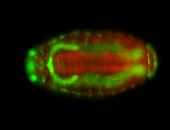
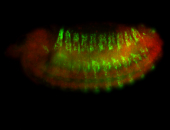
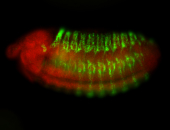
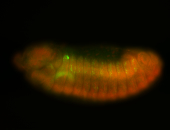
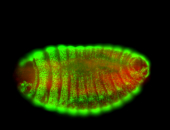
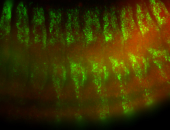
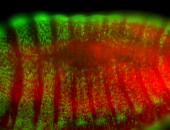
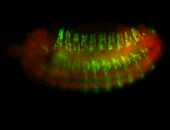
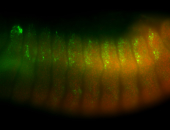
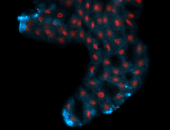
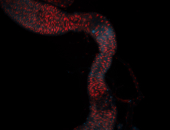
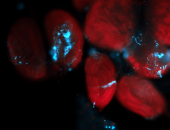
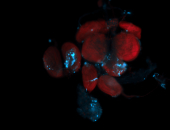
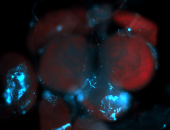
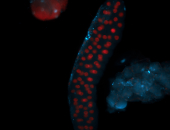
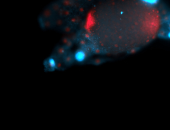
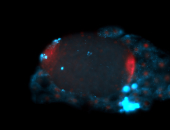
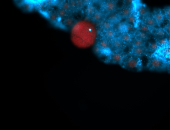
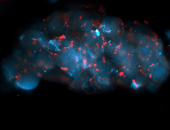
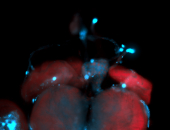
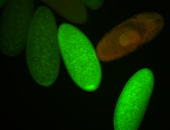
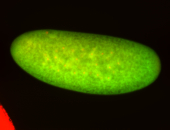
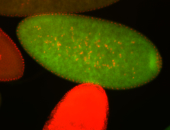
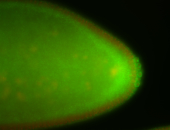
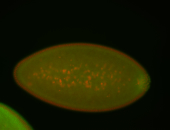
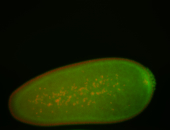
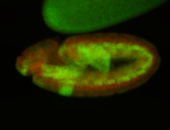
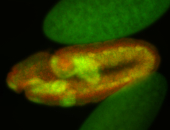
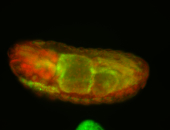
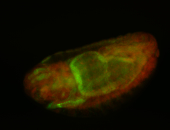
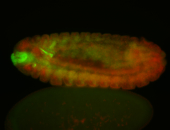
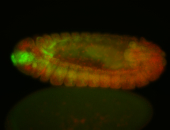
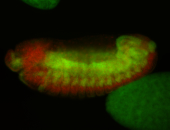
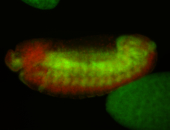
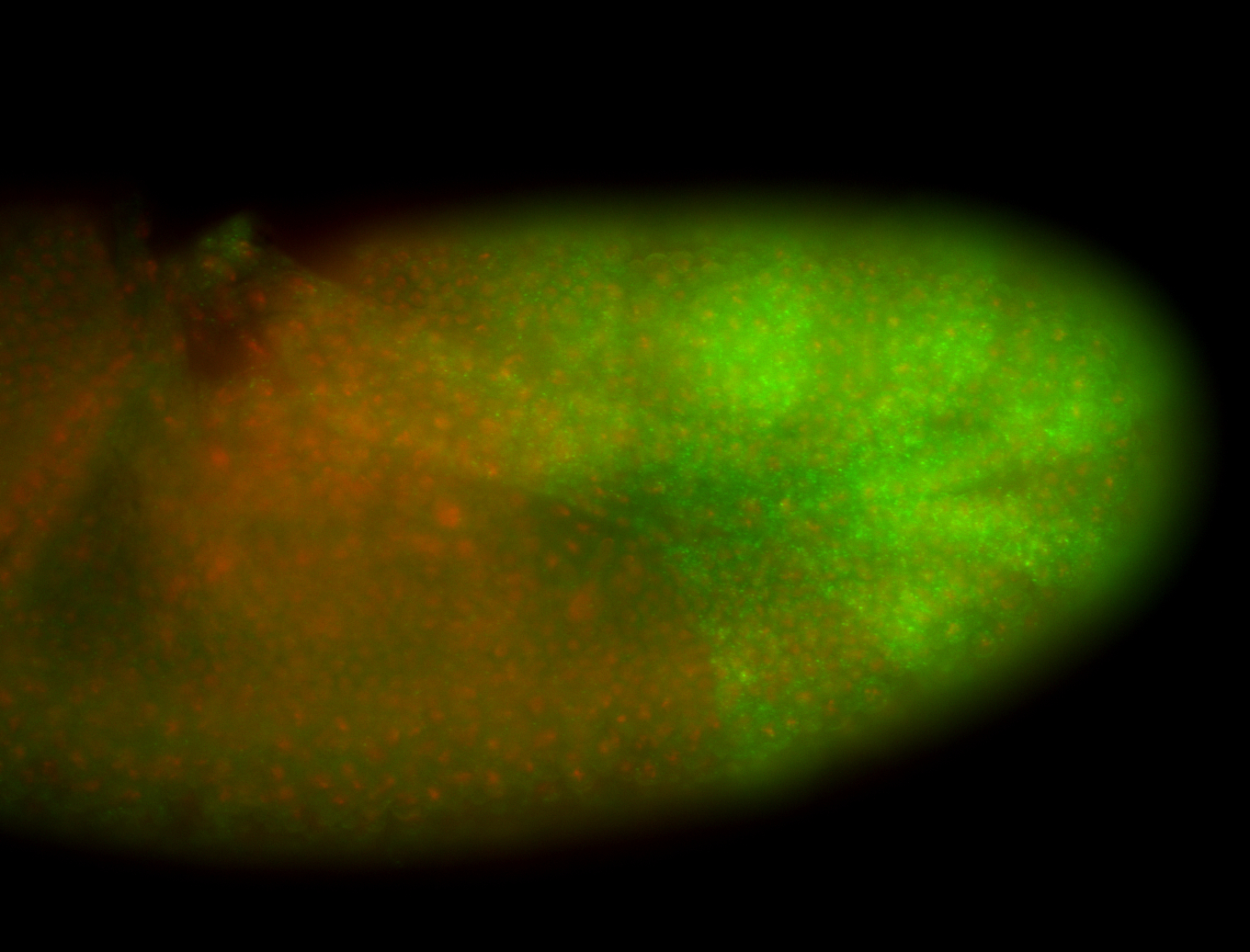
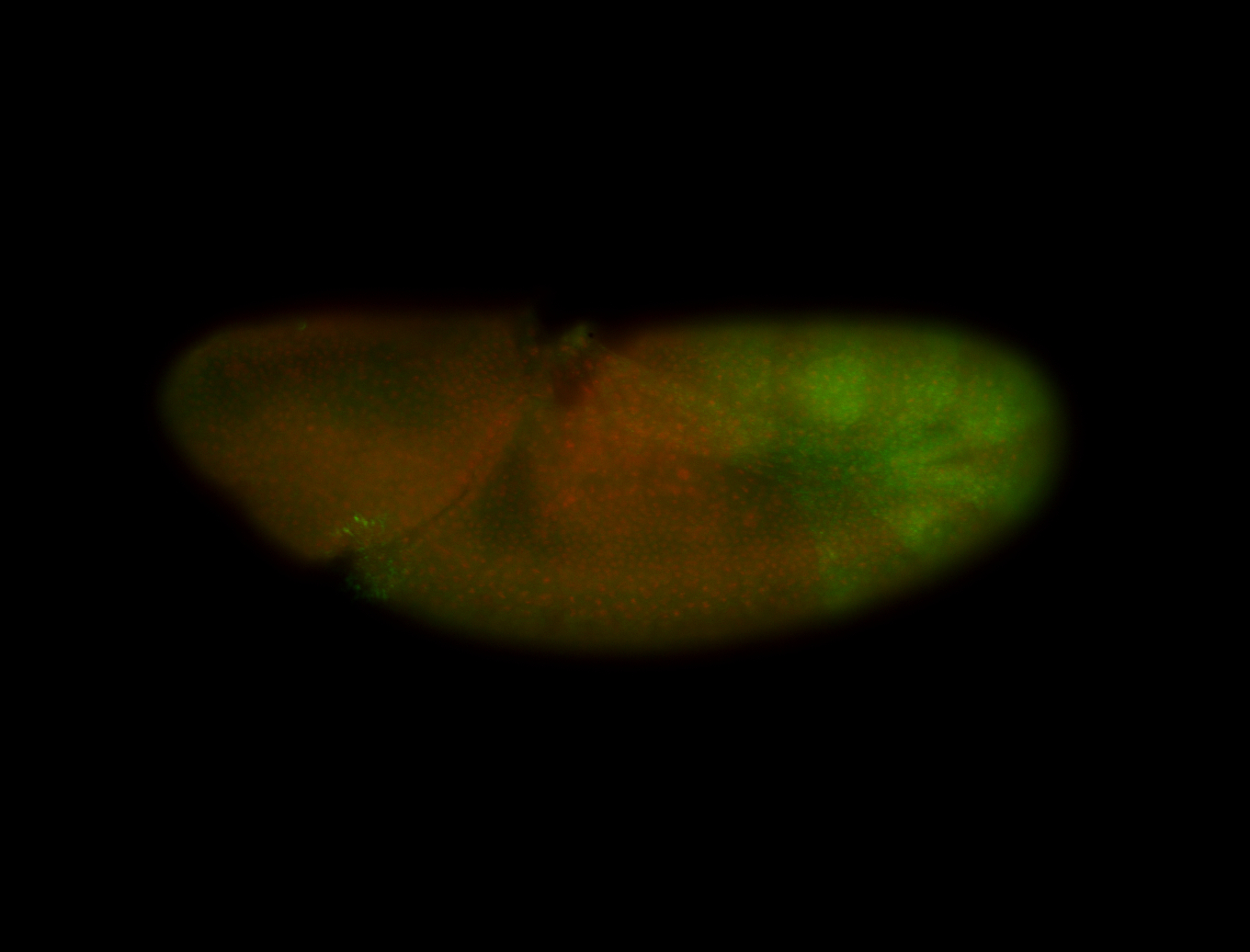
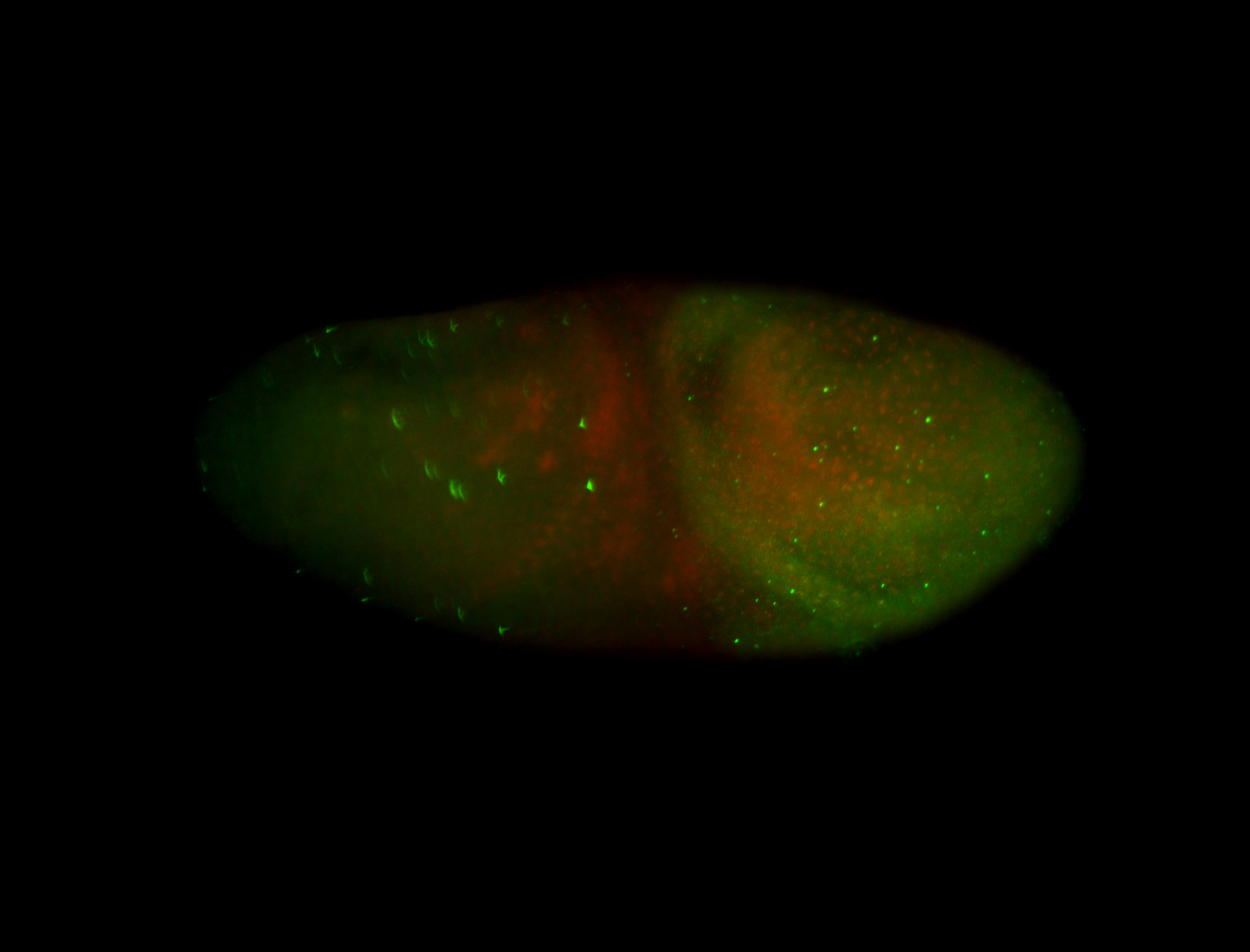
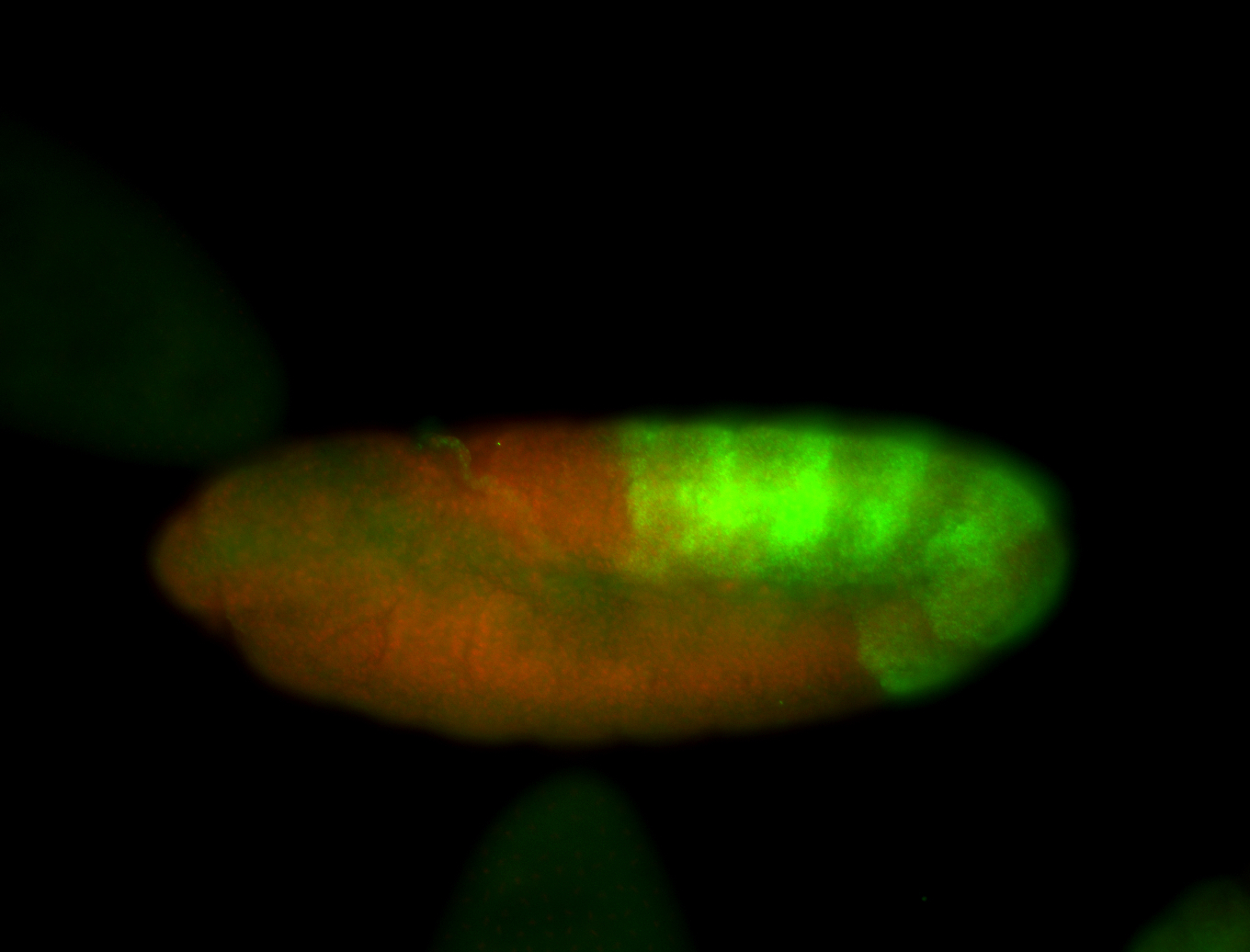
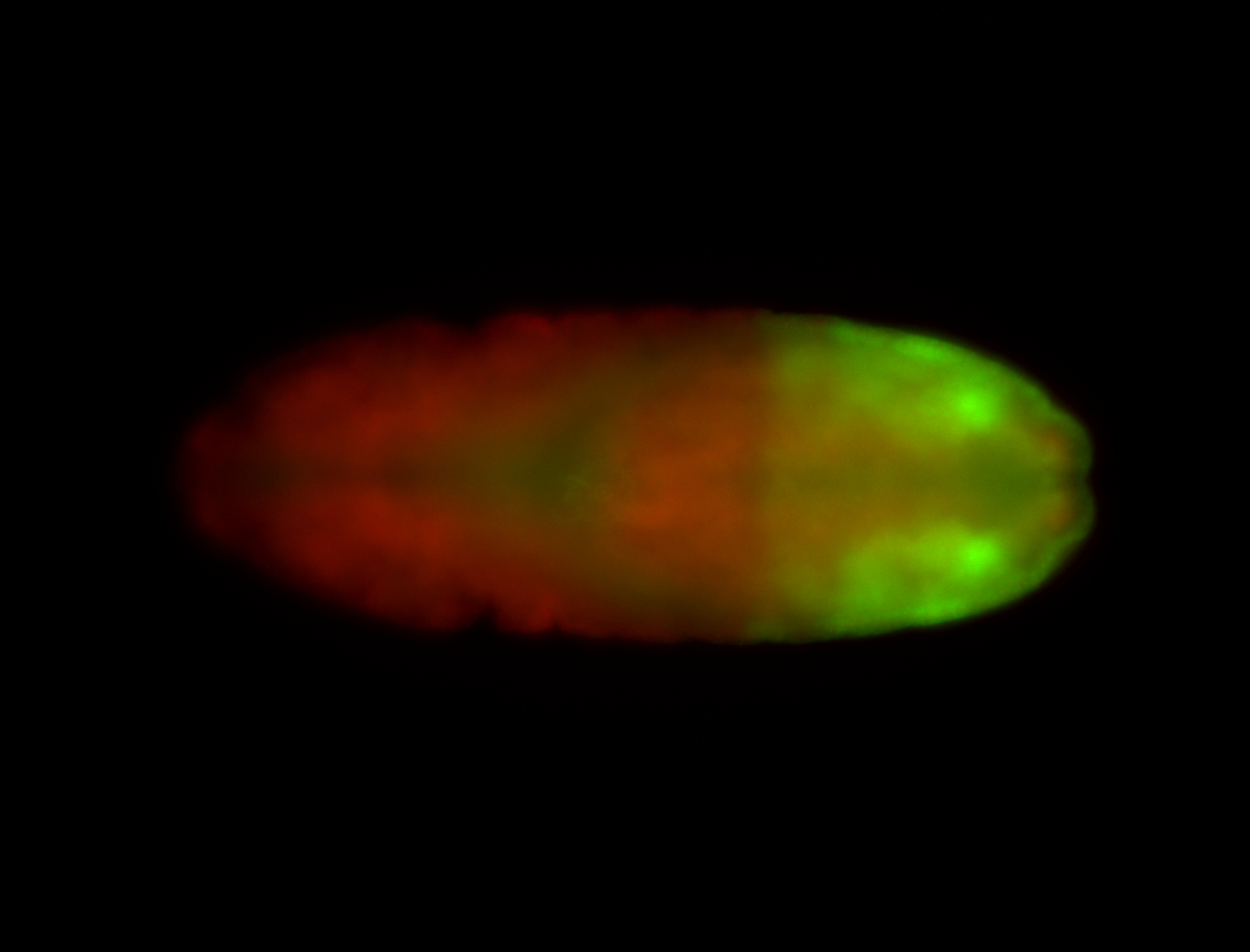
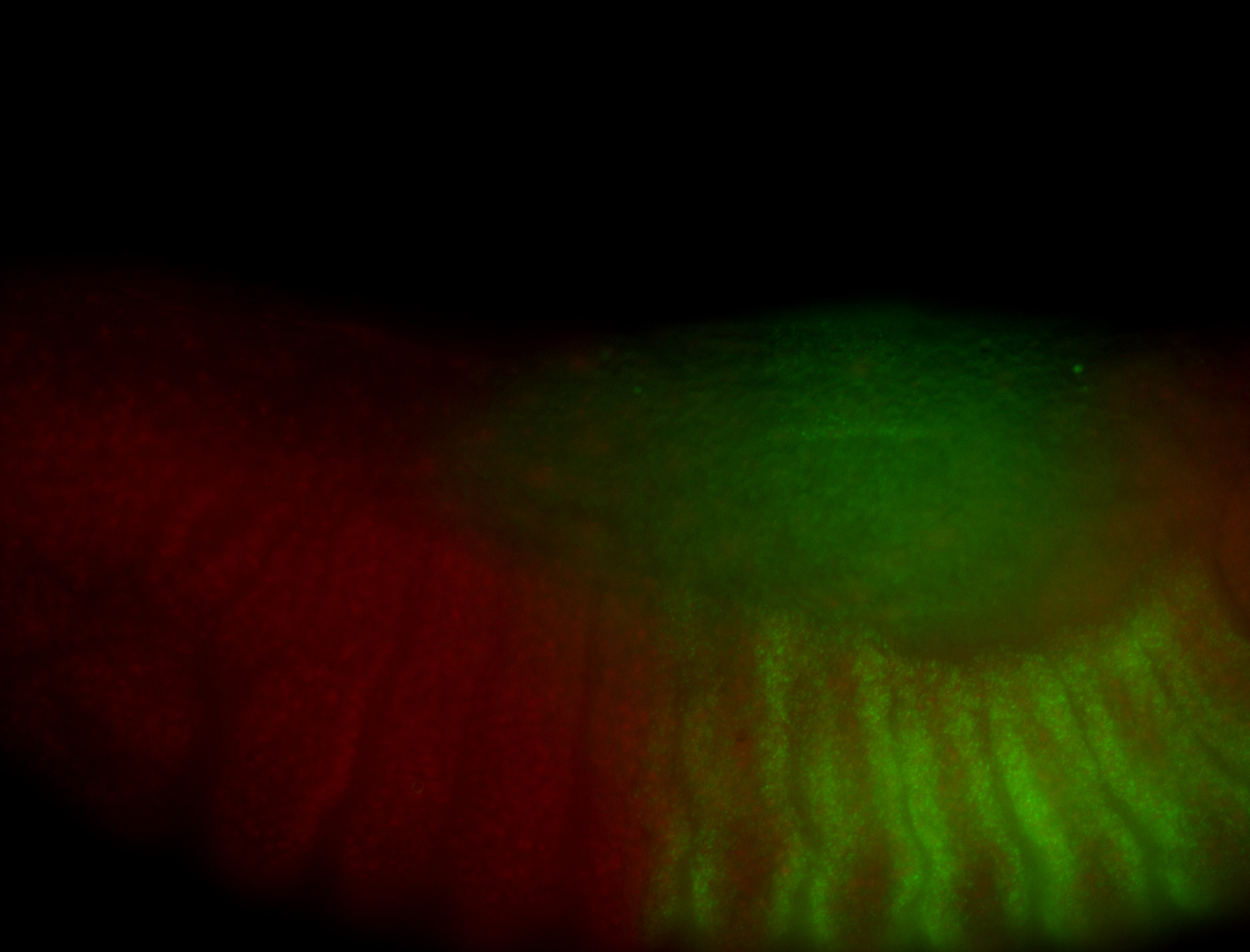
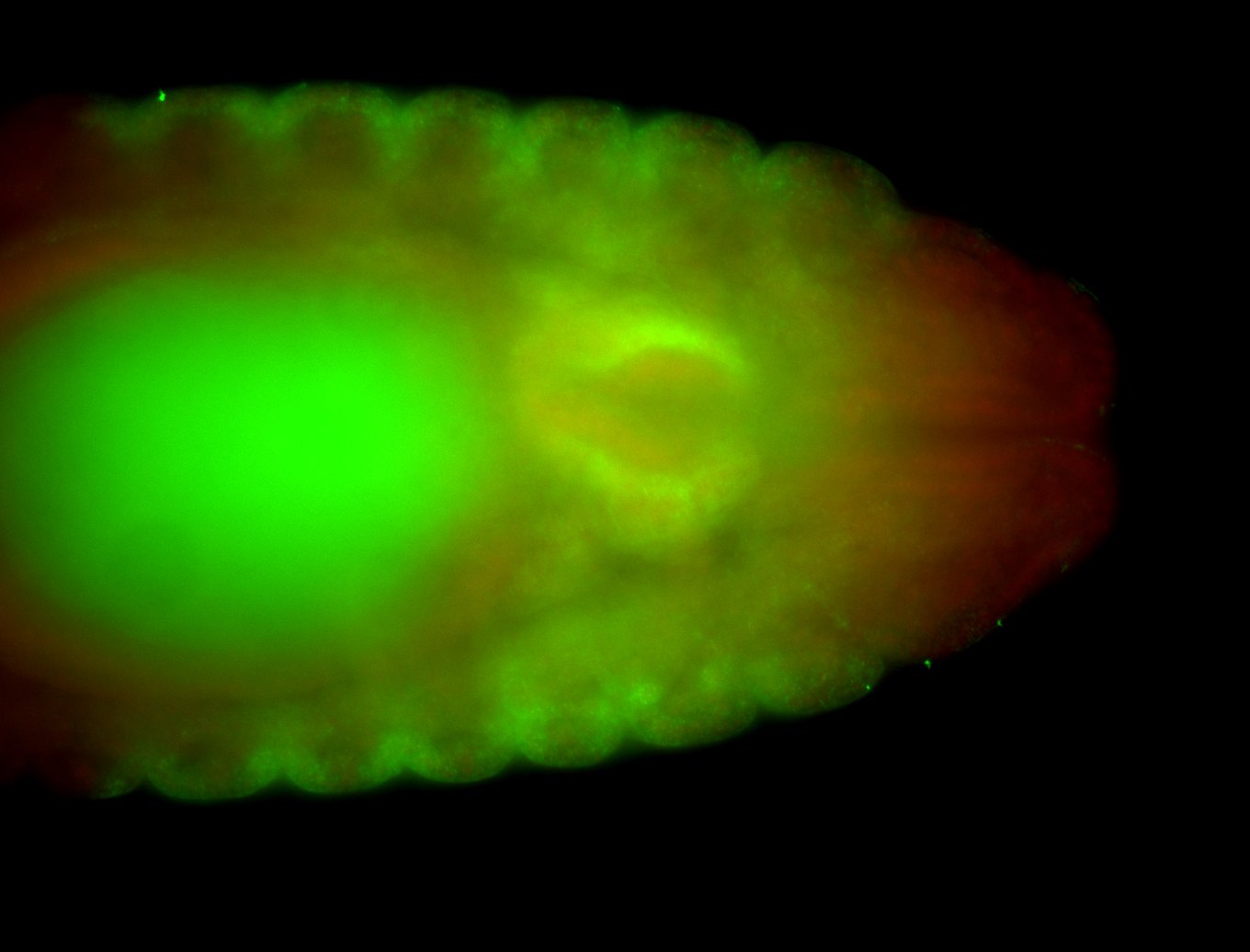
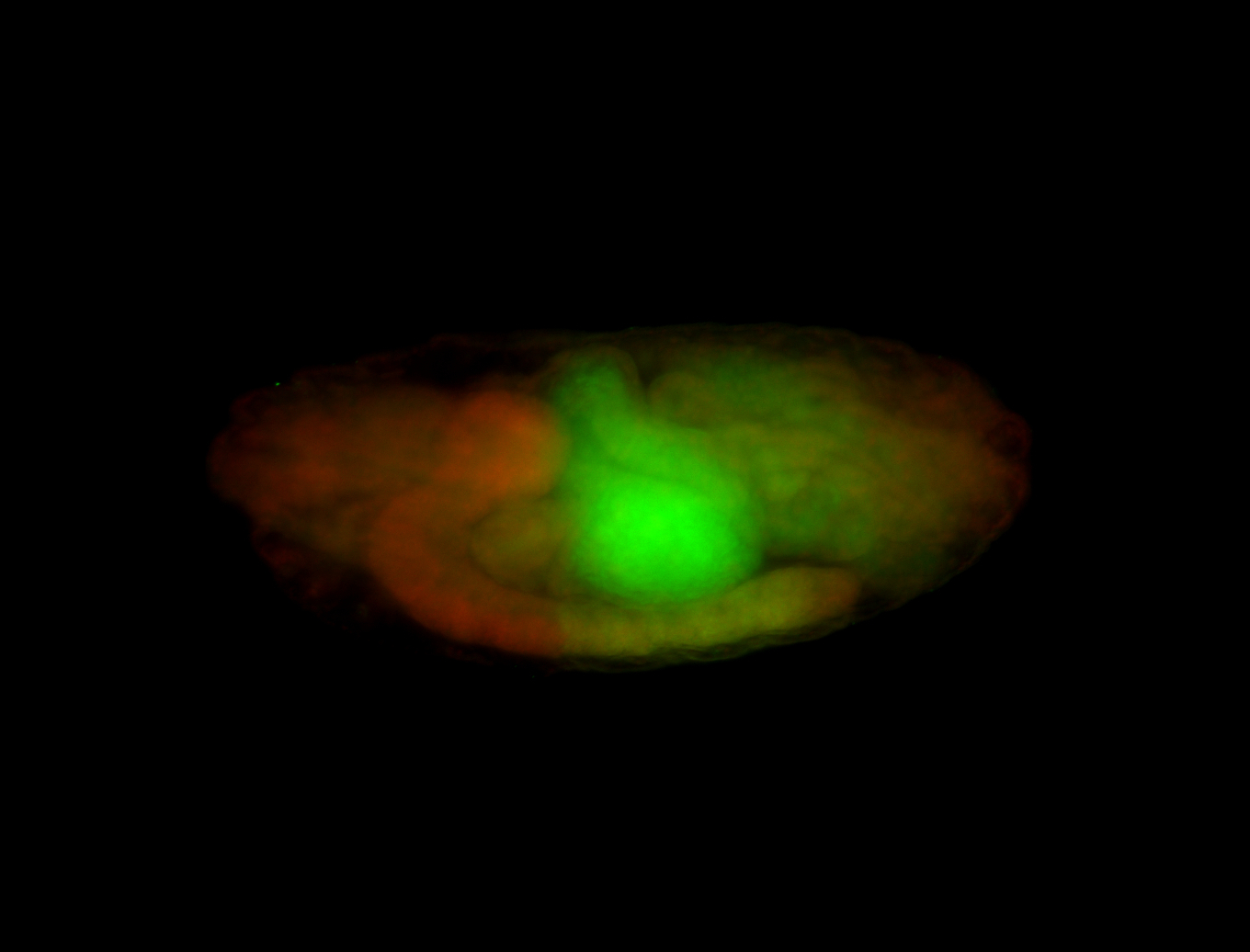
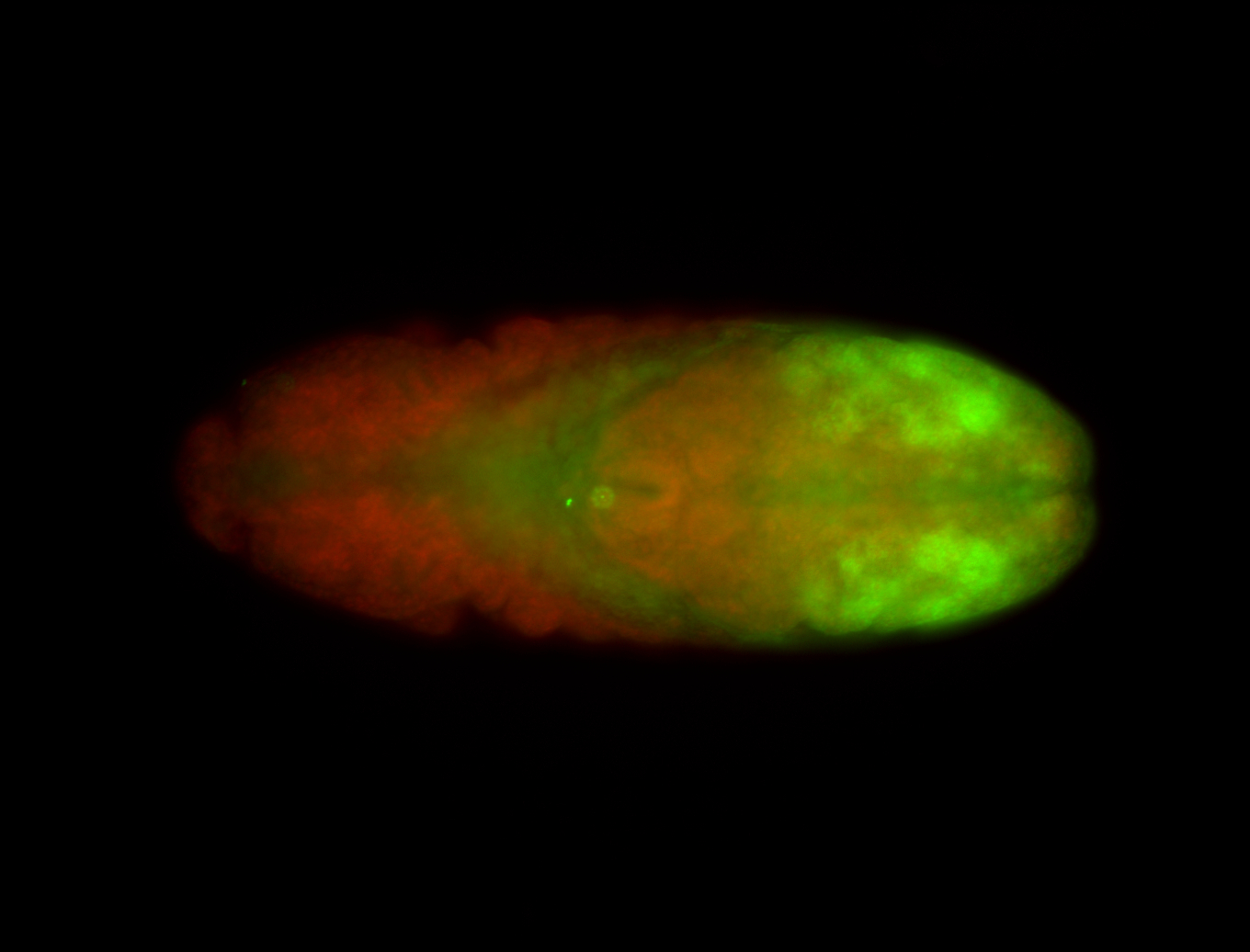
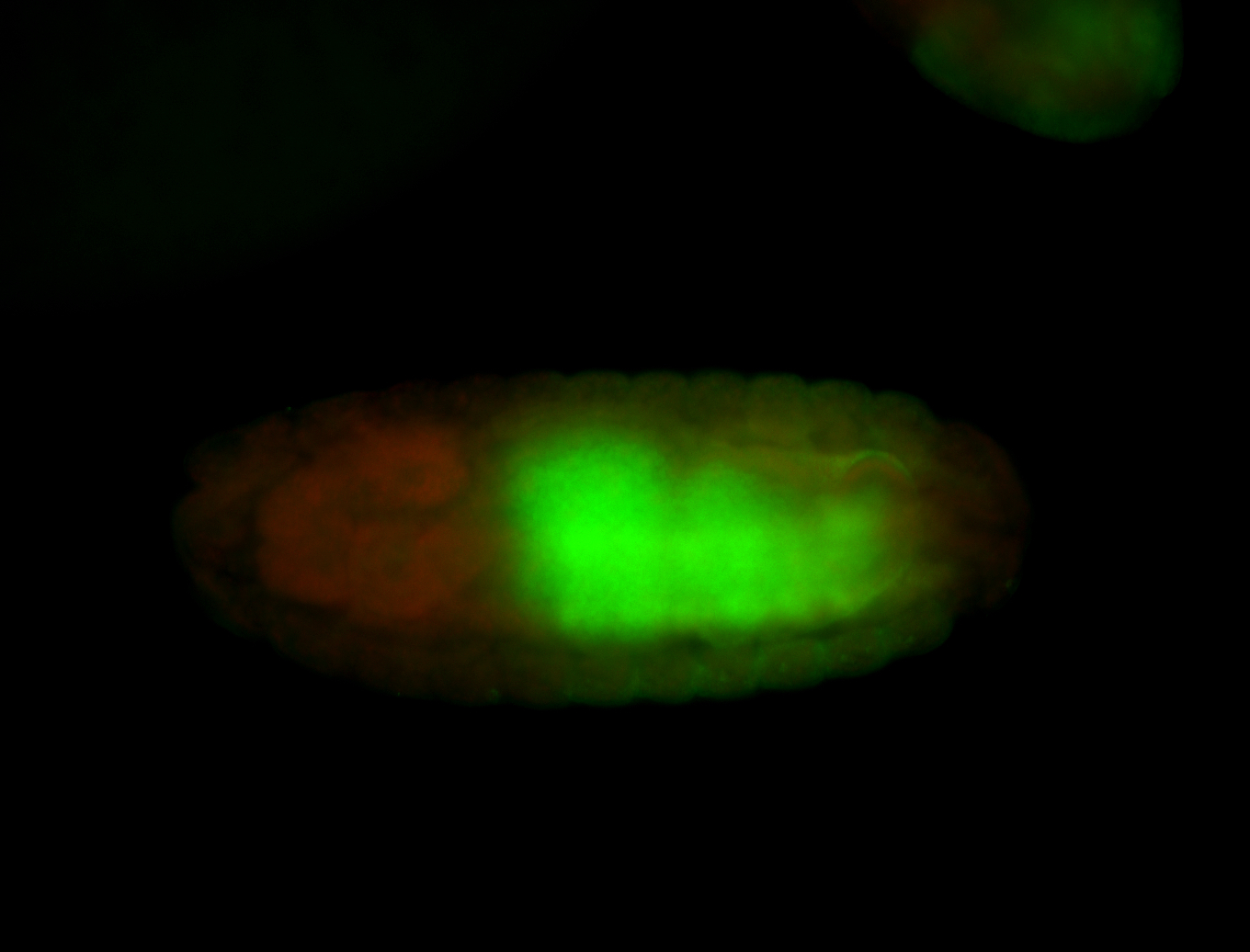
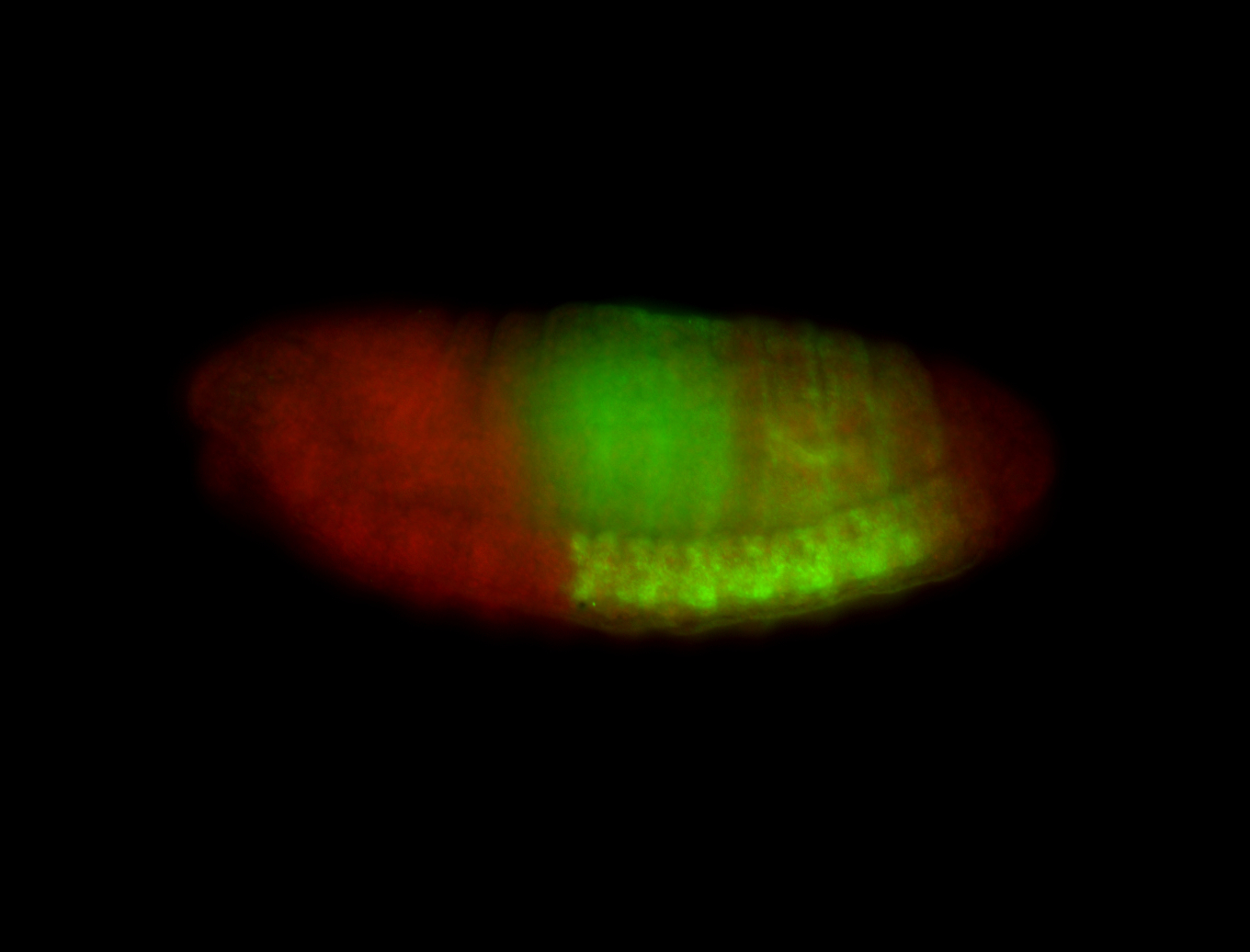
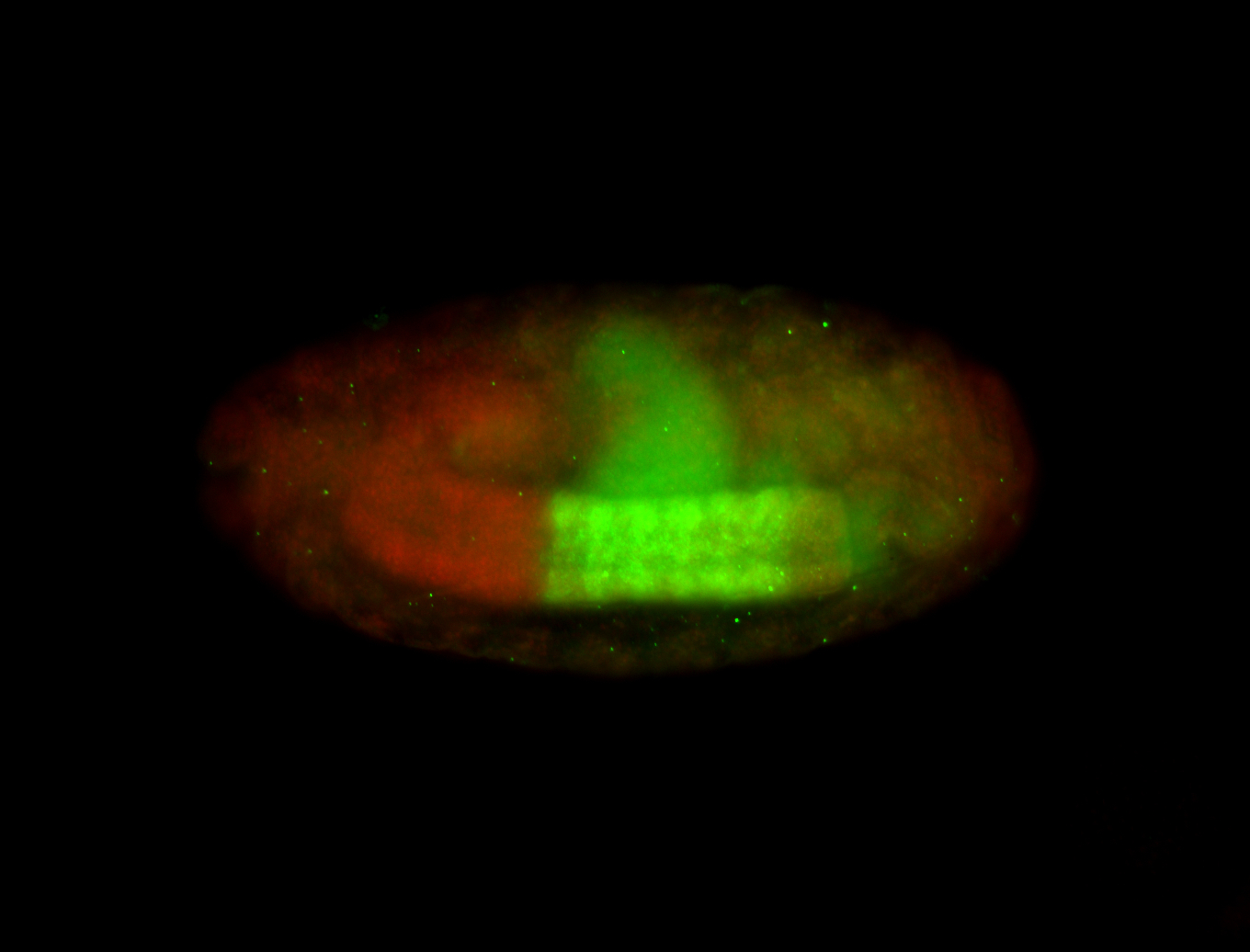
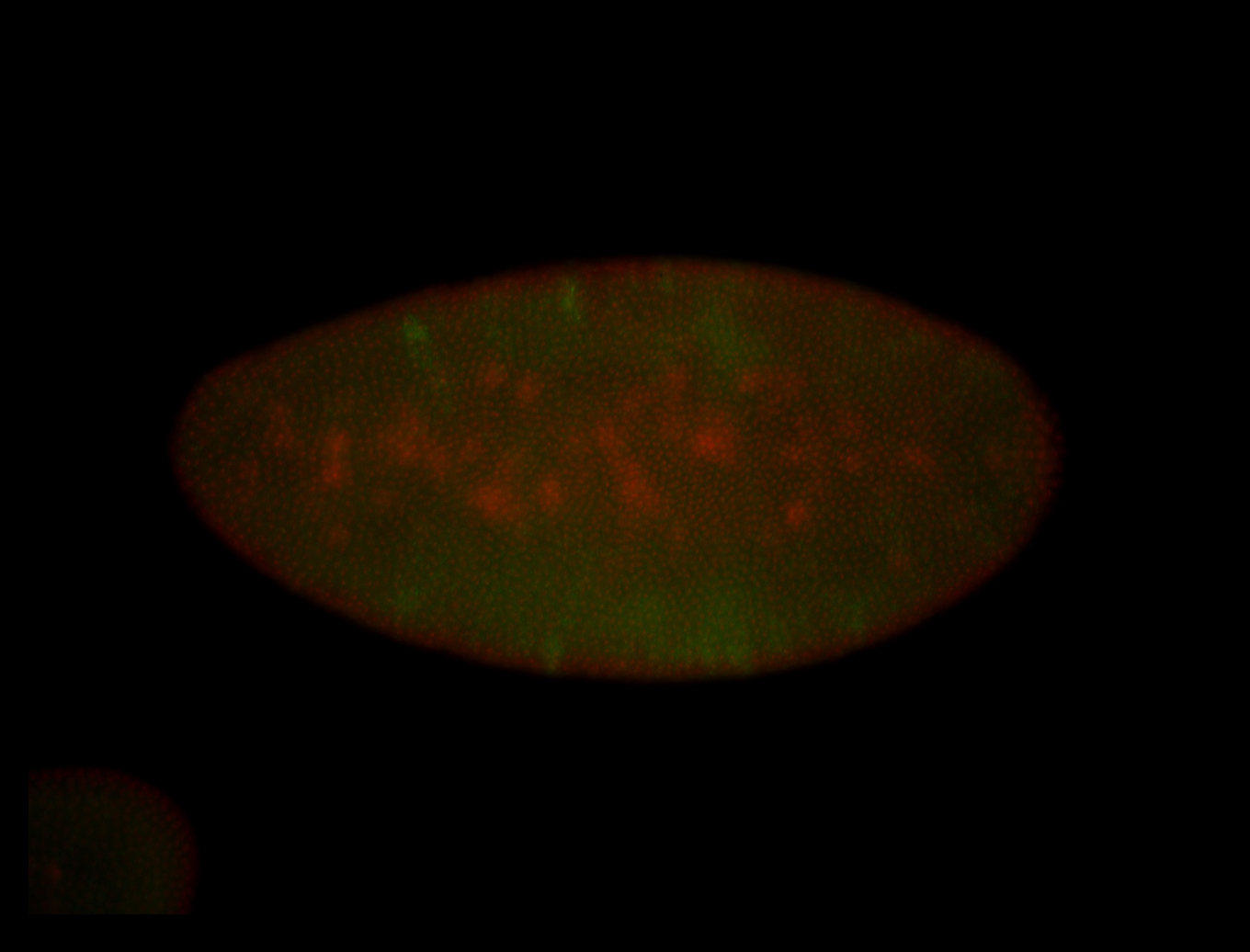
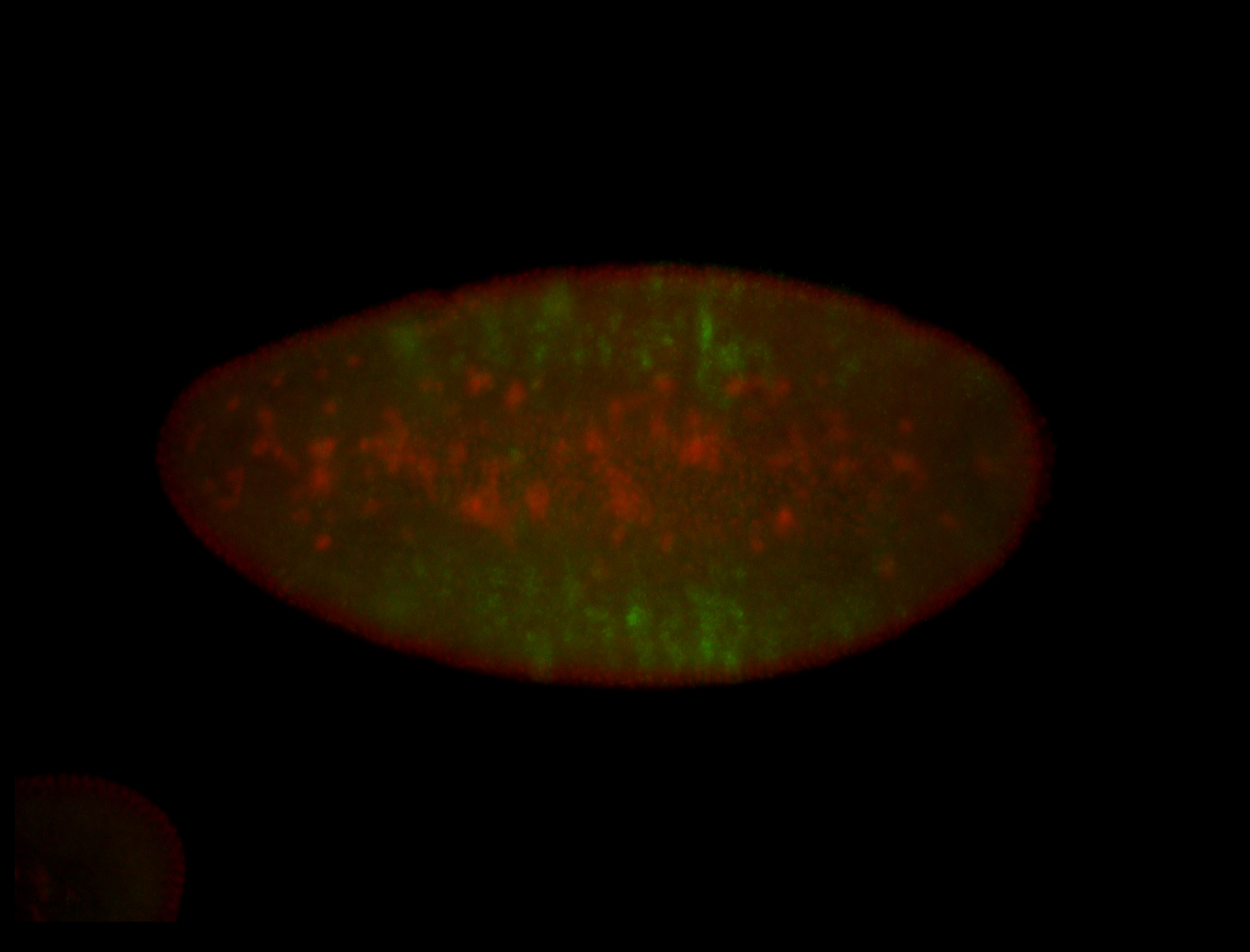
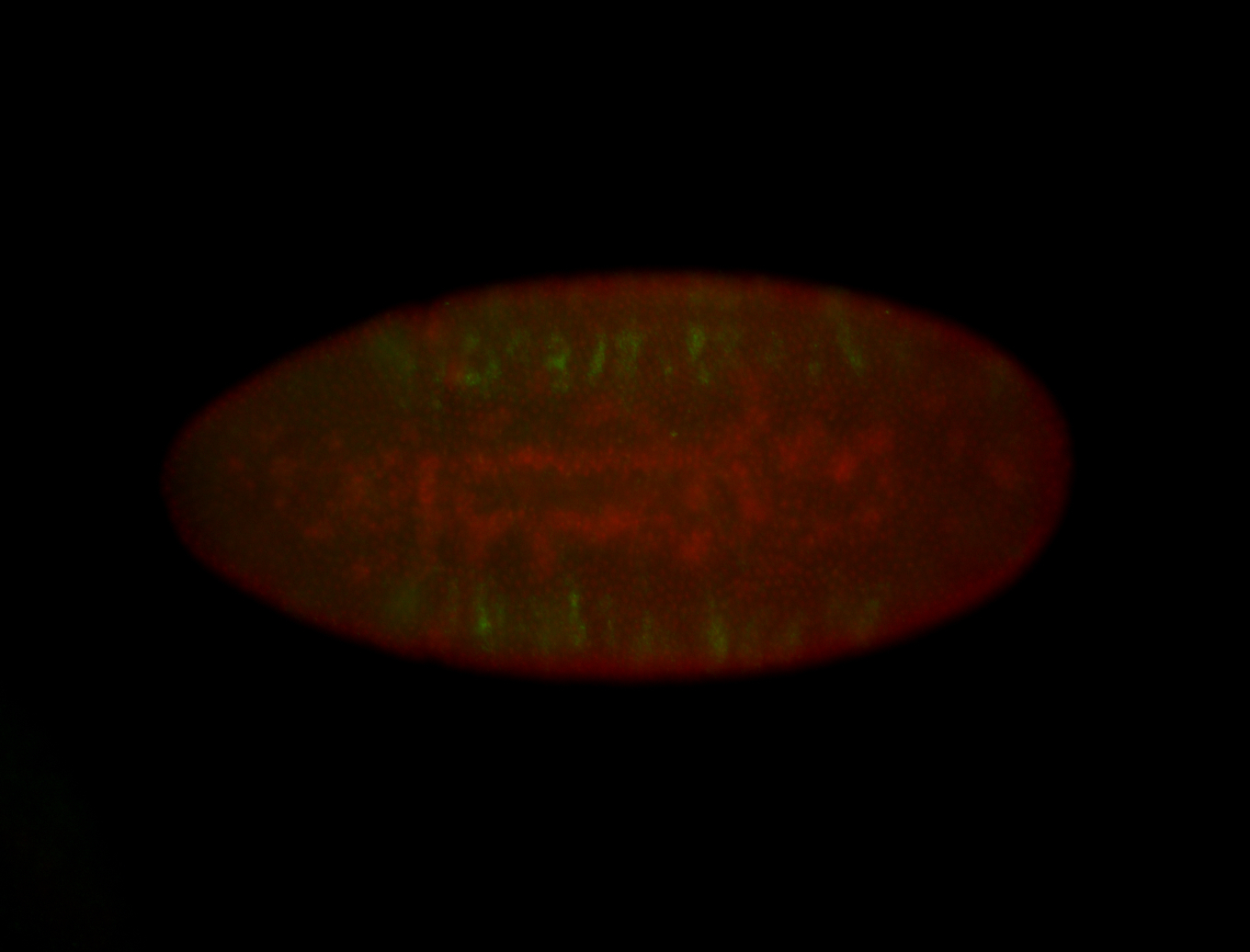
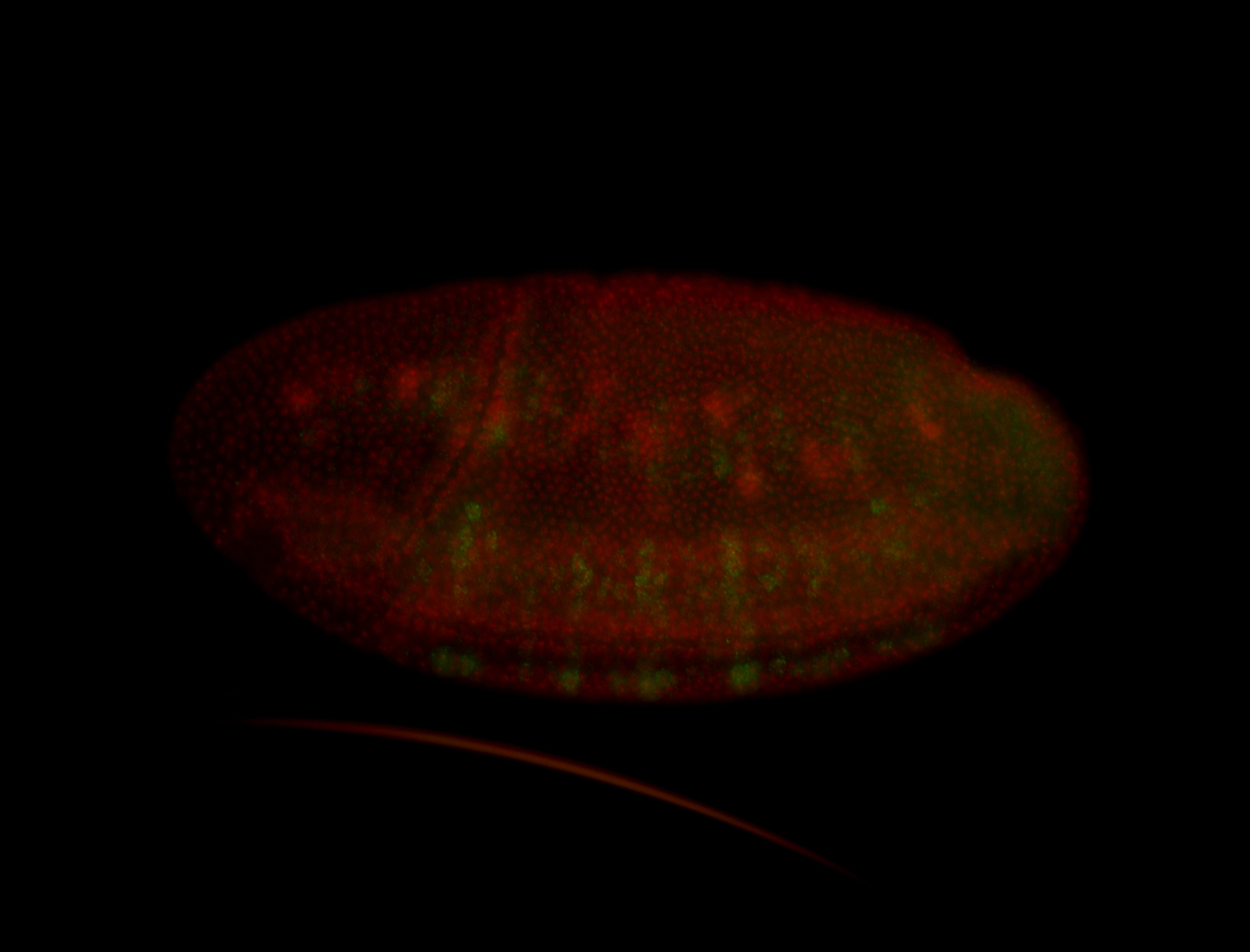
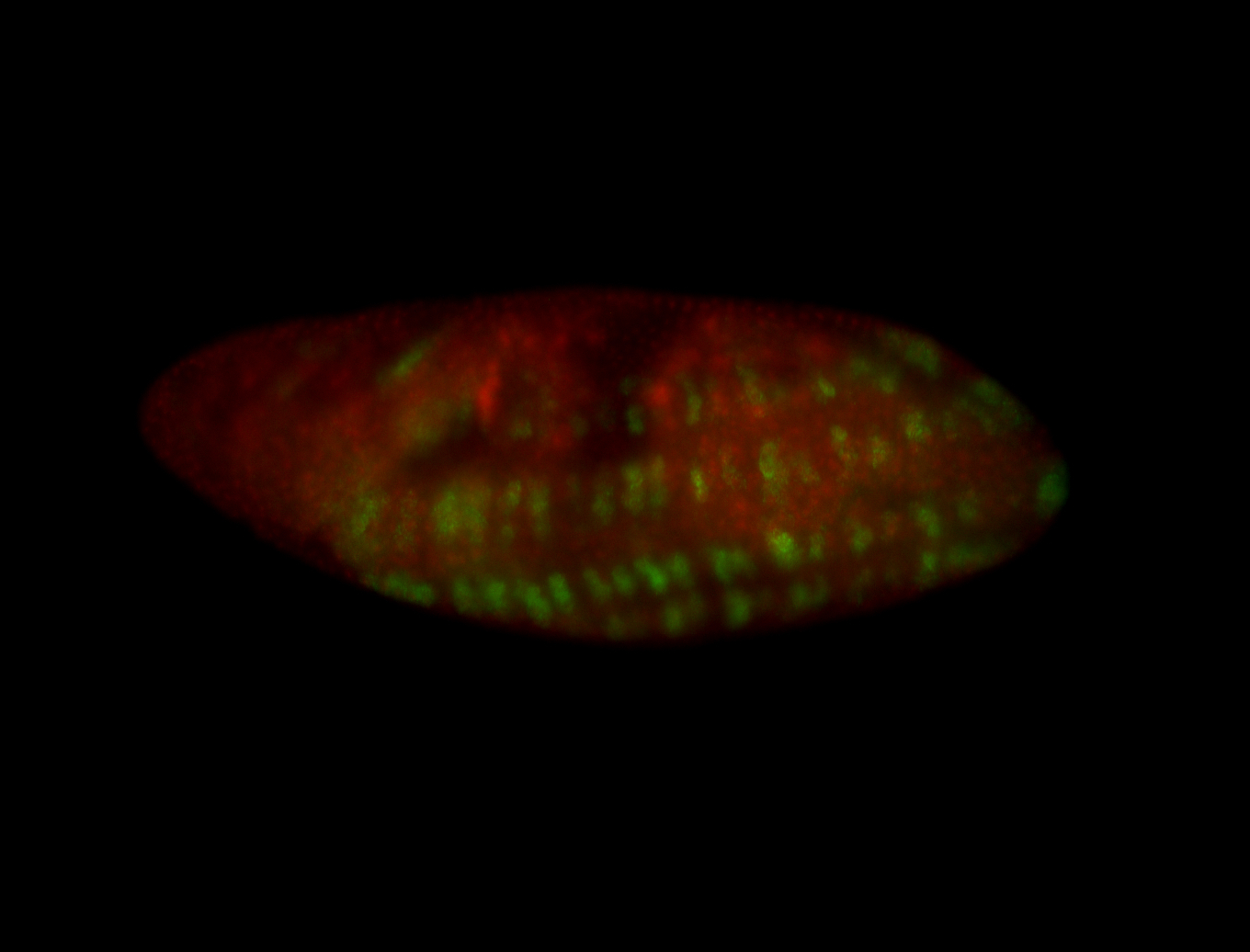
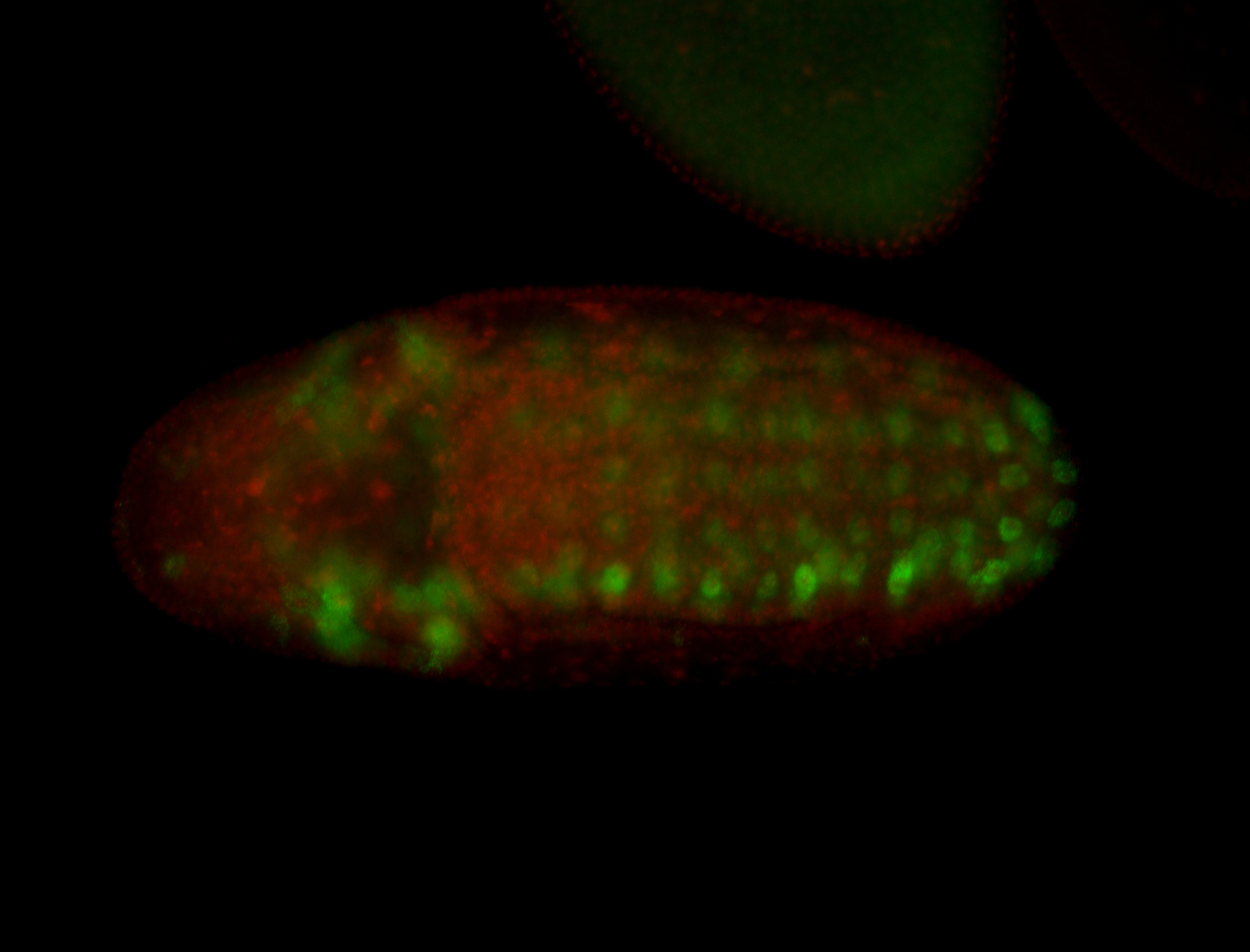
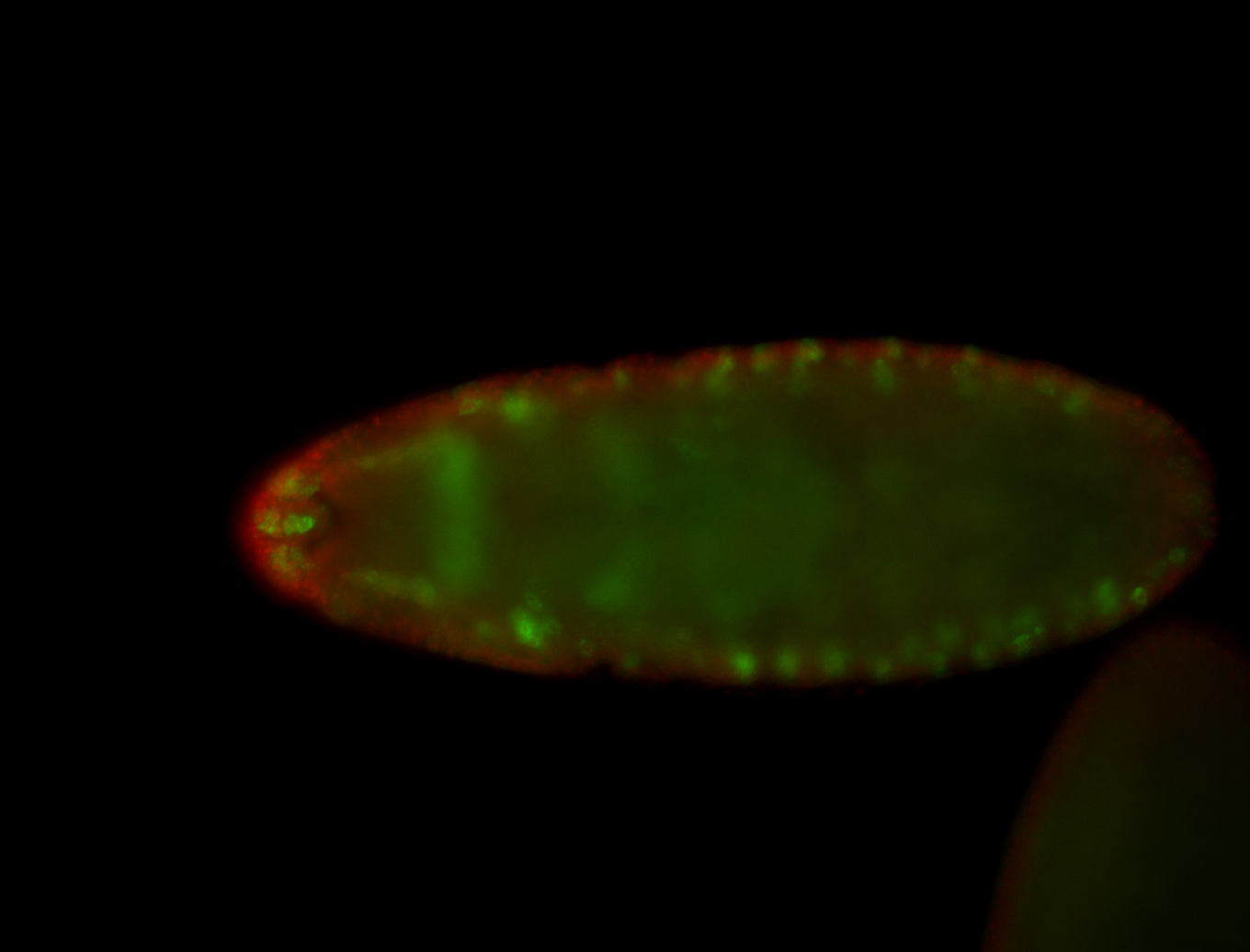
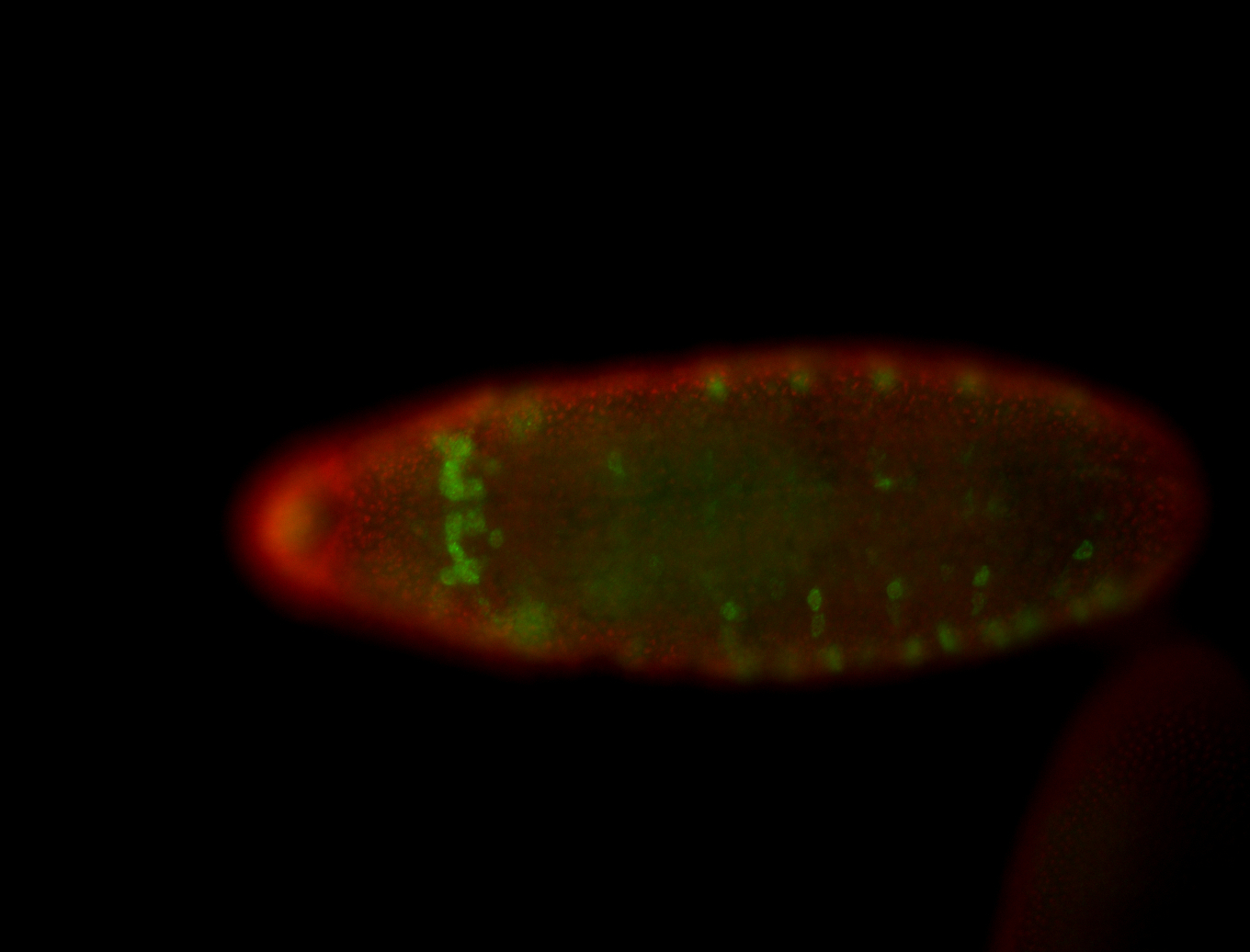
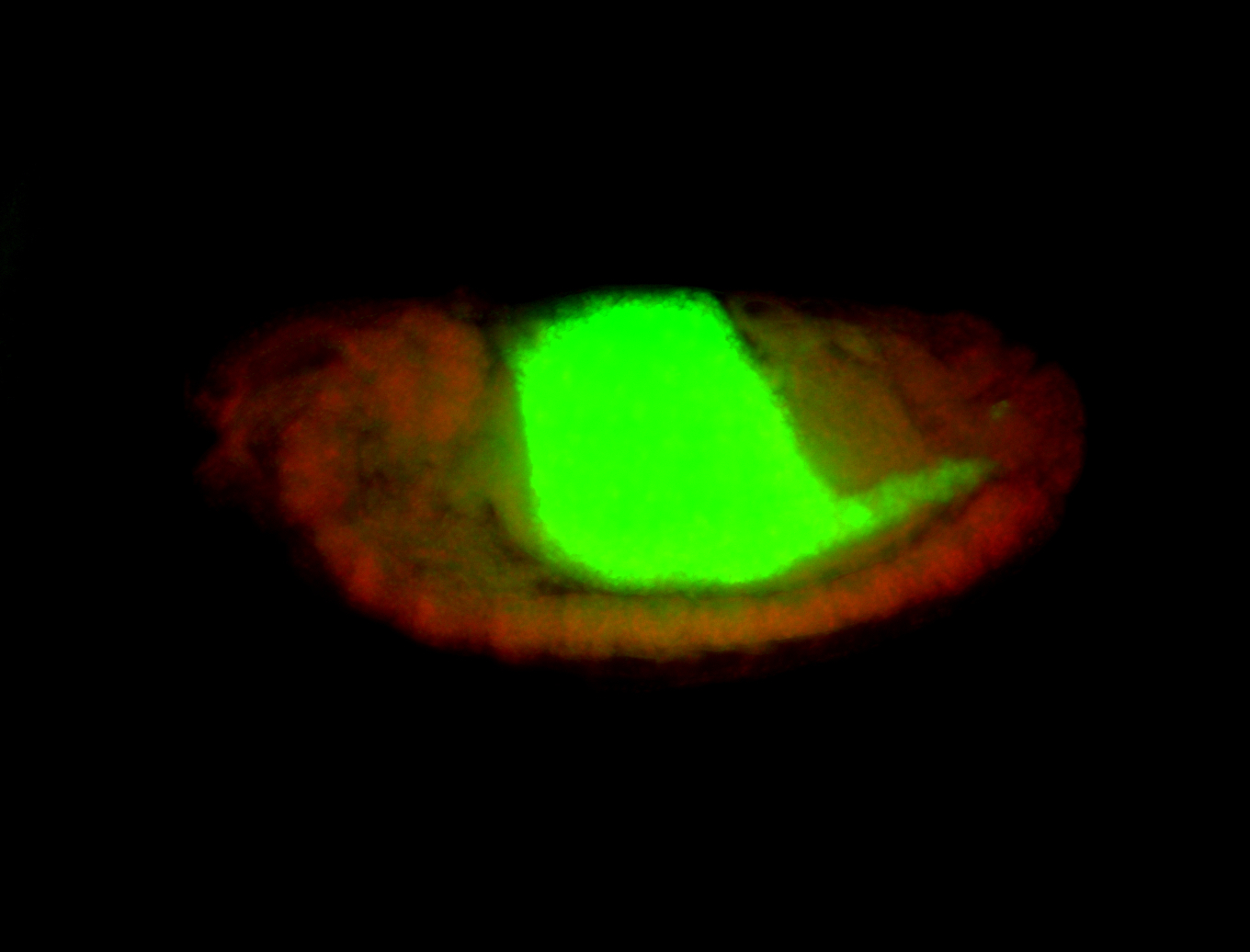
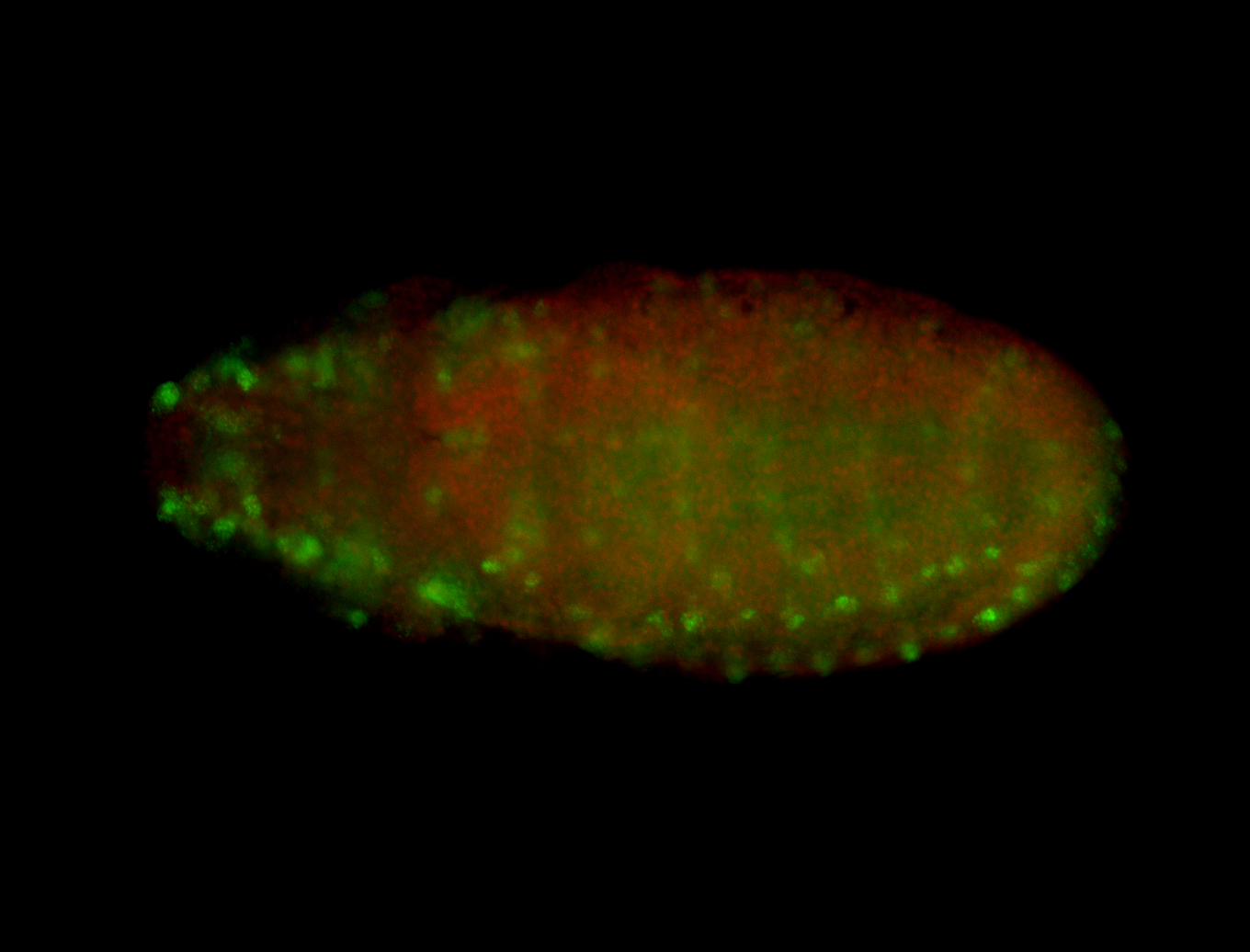
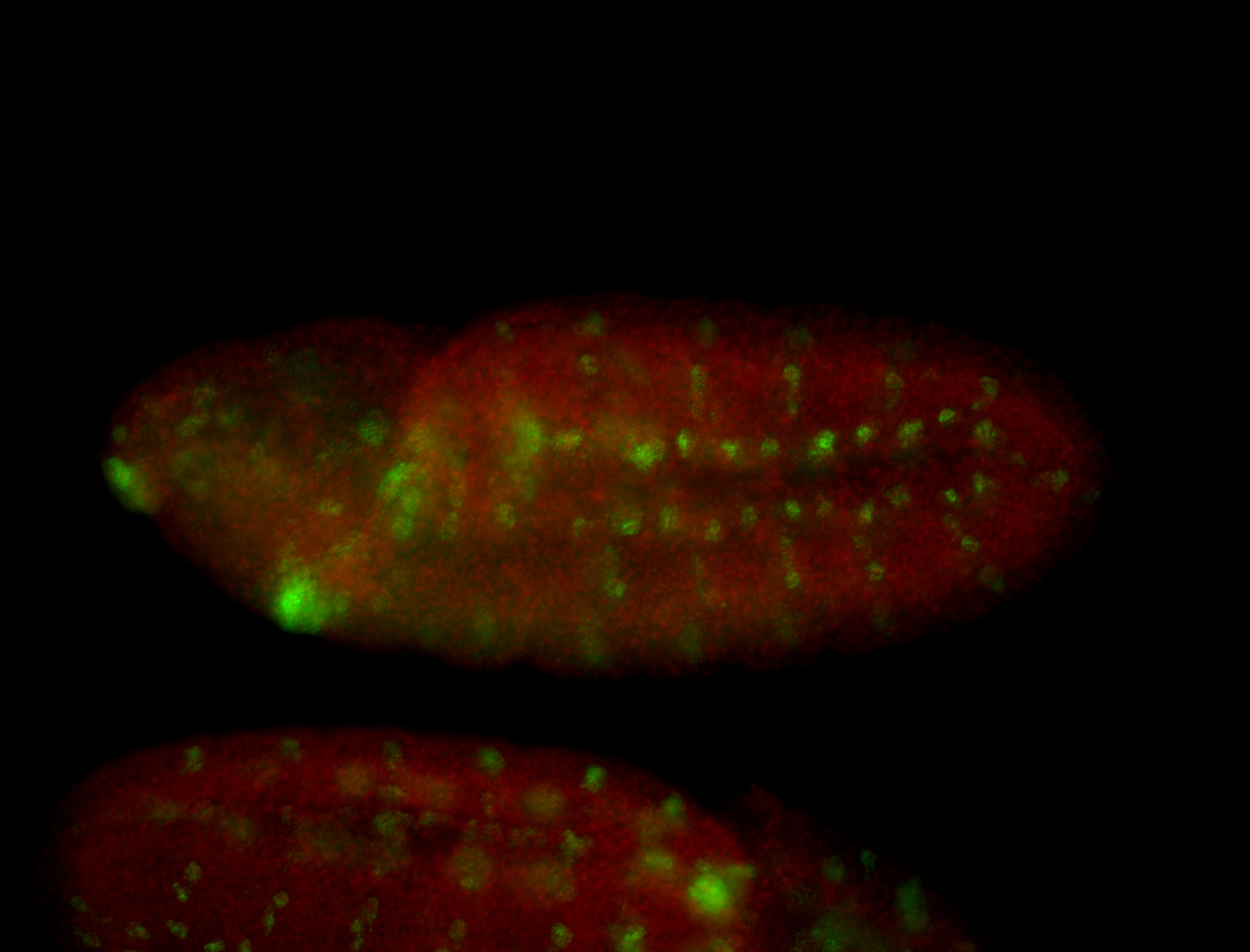
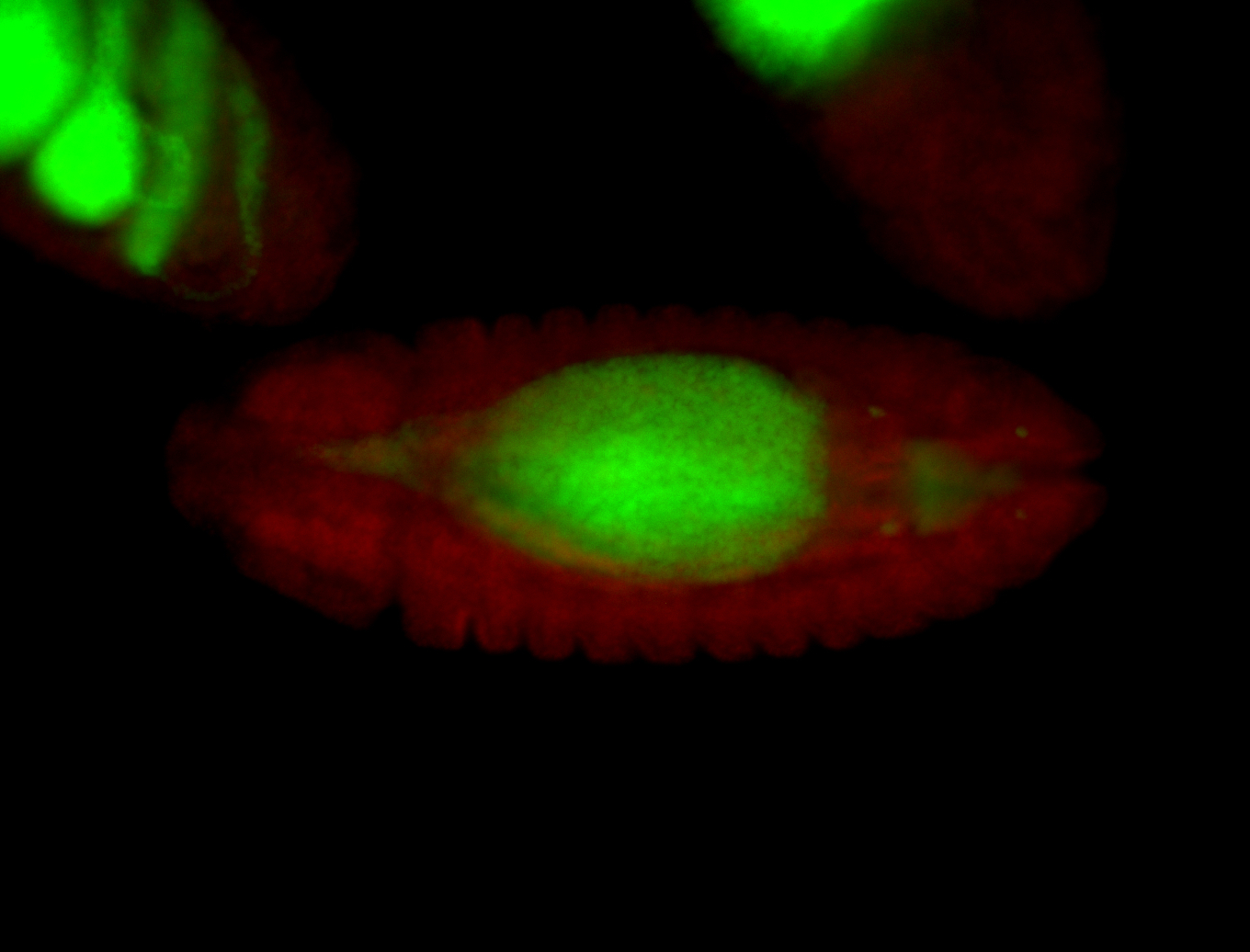
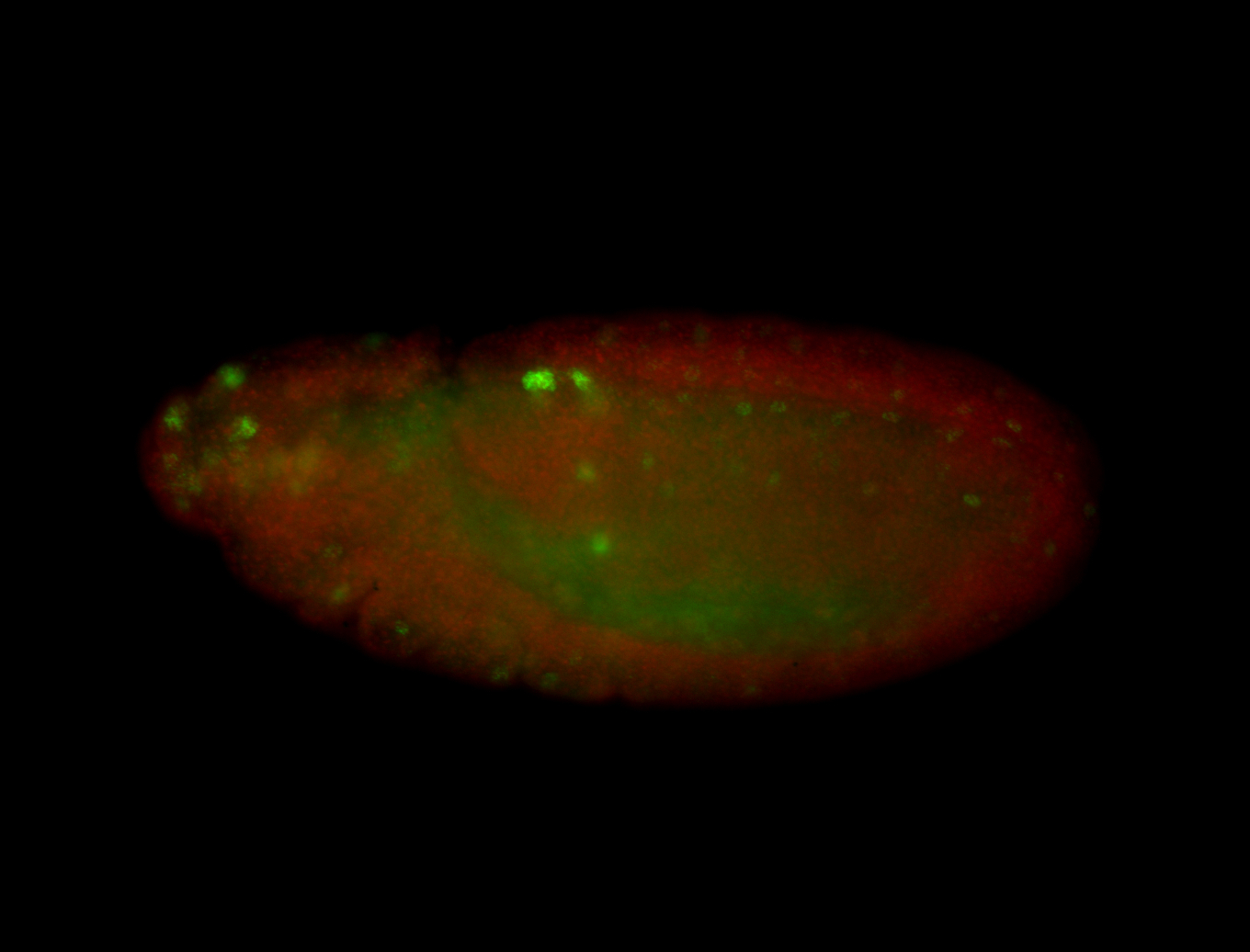
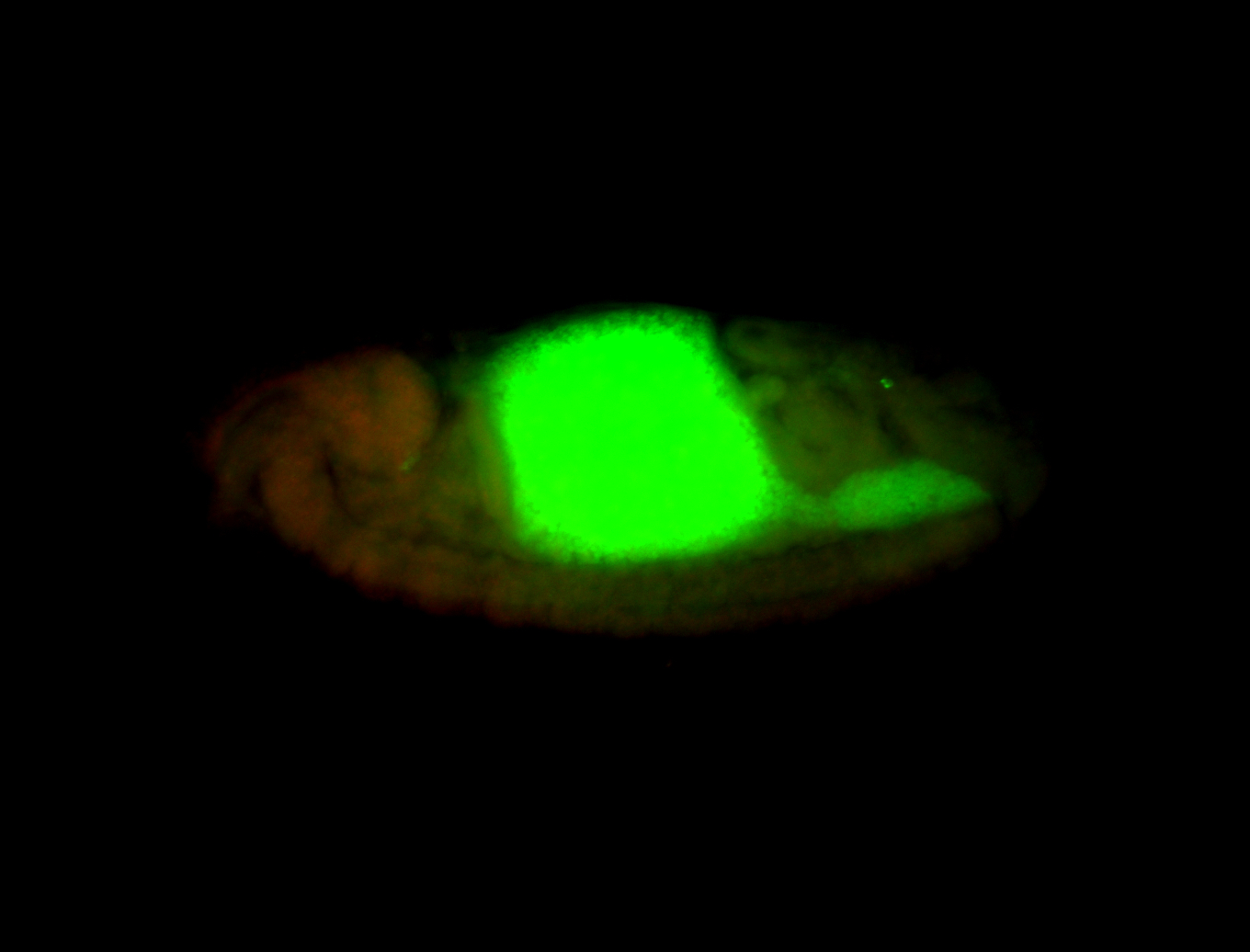
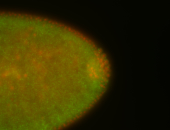
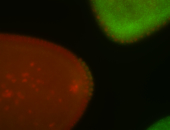
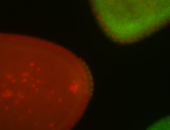
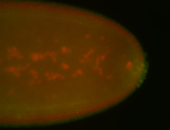
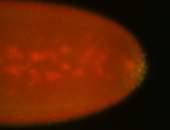
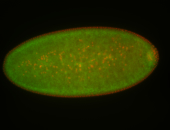
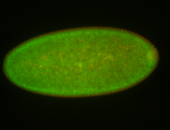
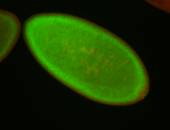
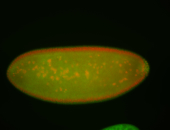
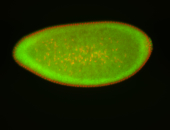
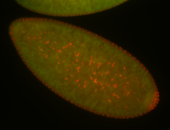
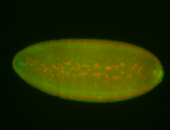
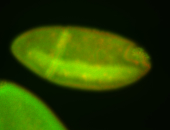
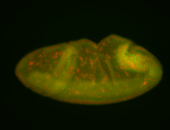
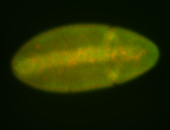
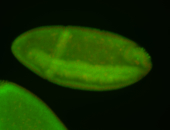
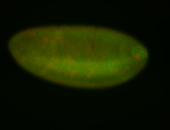
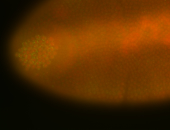
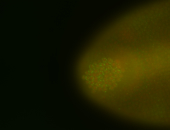
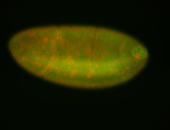
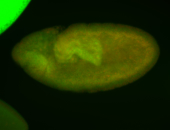
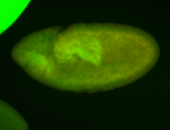
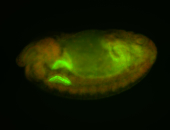
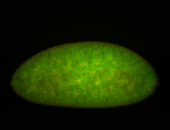
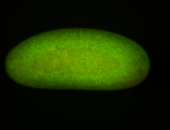
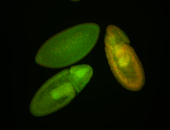
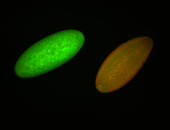
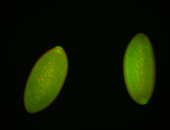
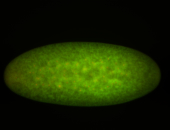
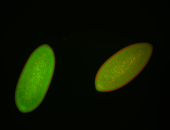
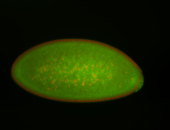
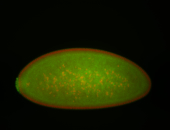
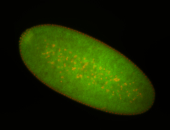
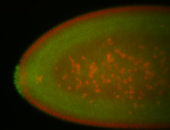
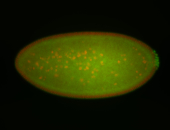
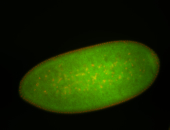
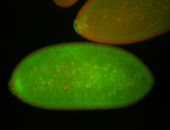
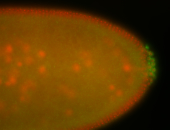
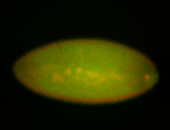
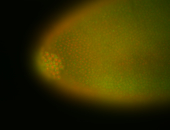
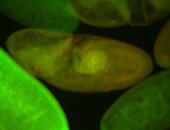
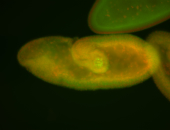
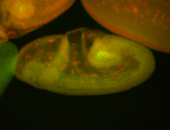
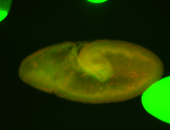
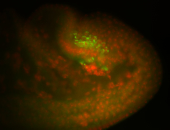
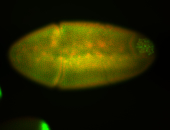
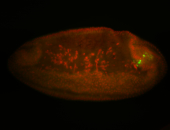
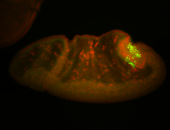
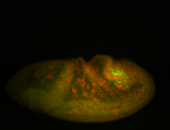
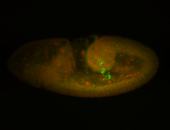
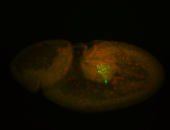
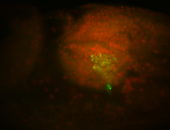
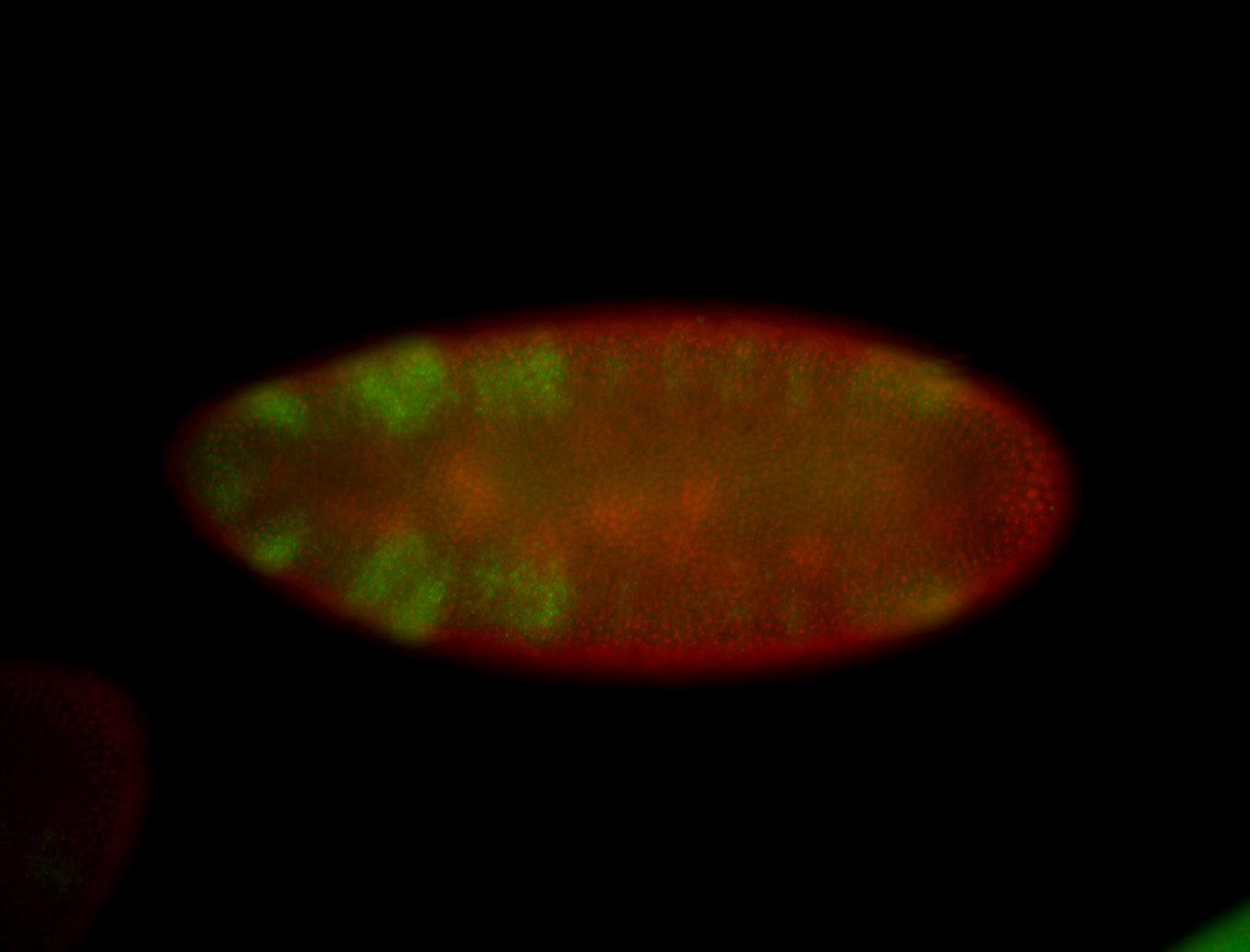
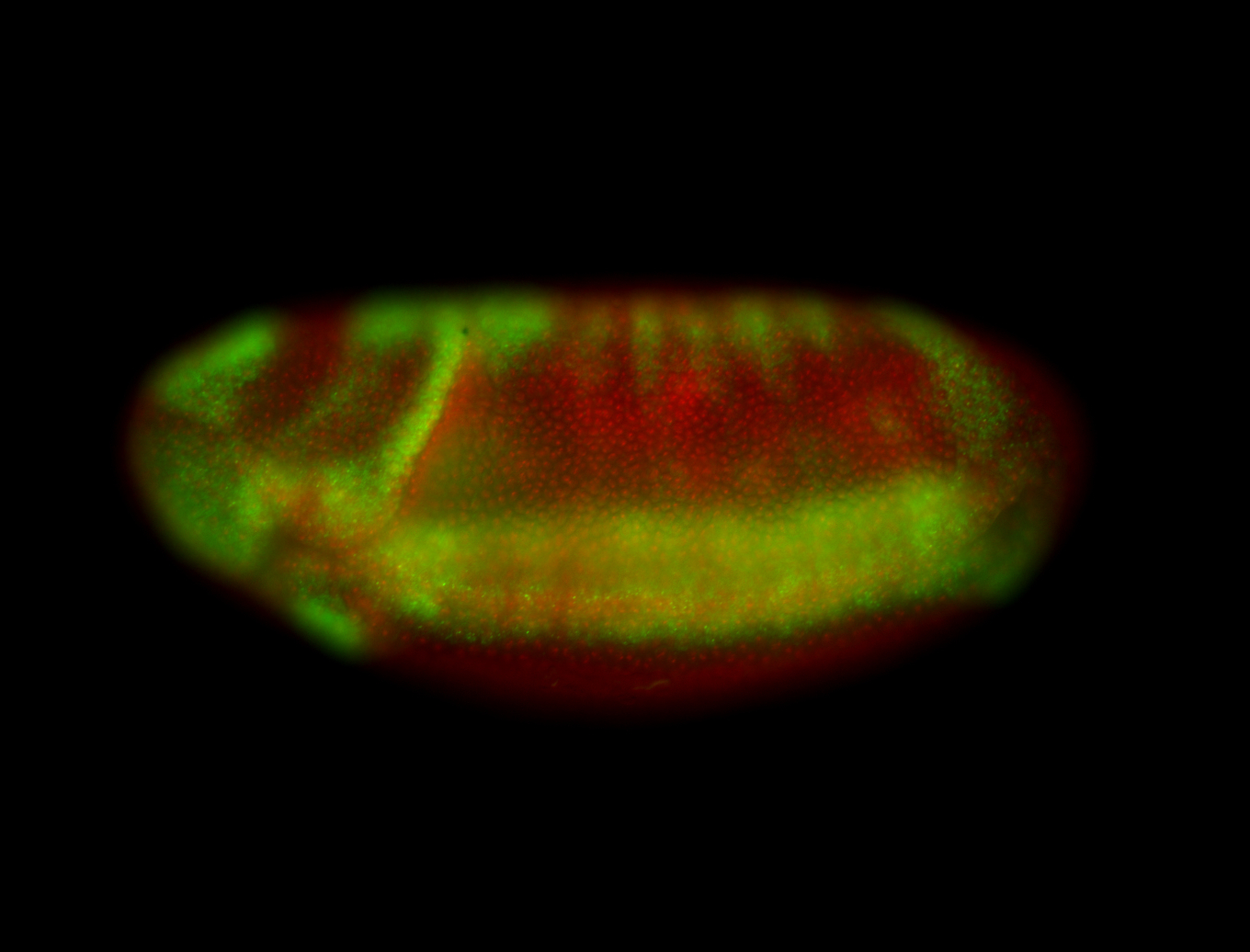
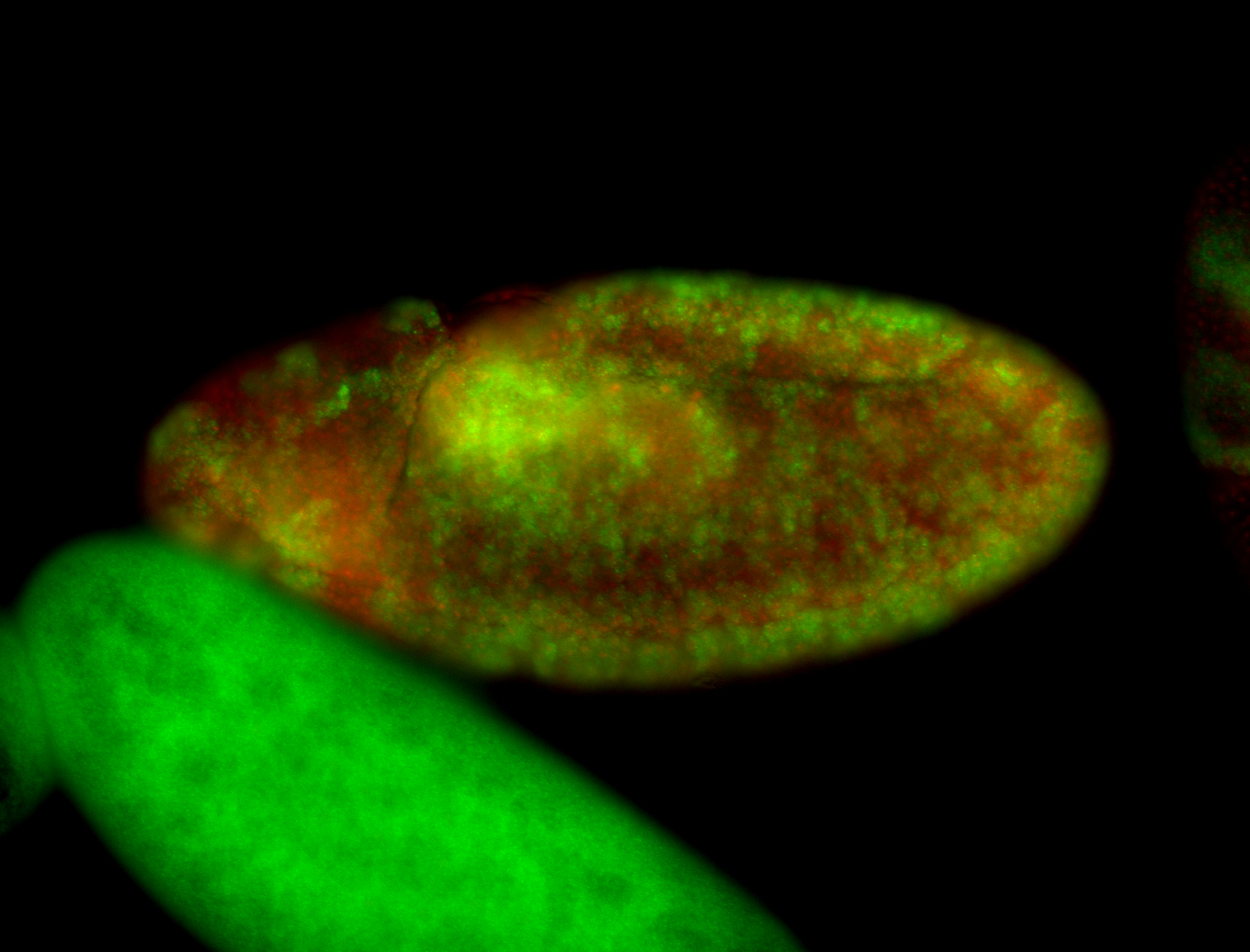
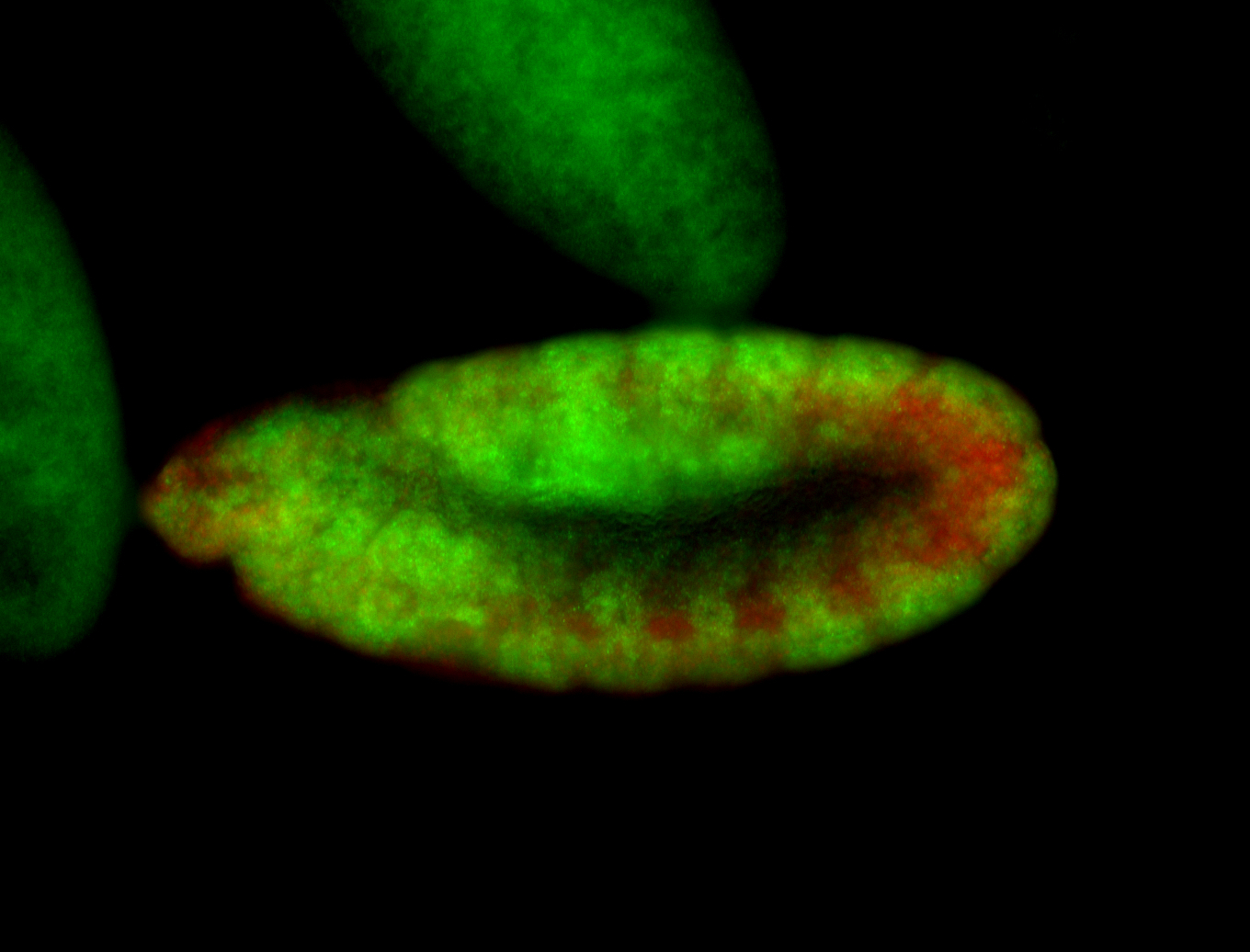
For a full list of searchable terms, please see the term page.
NOTE: It takes a few seconds for the term suggestions to load.
Result Summary
| Number of genes | 22 | |
| Export | ||
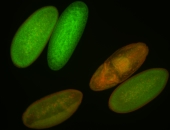
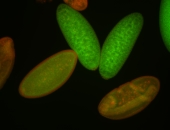
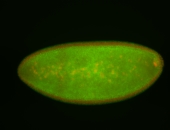
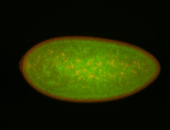
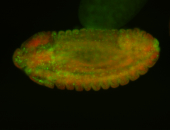
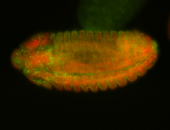
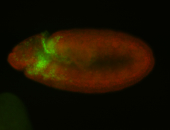
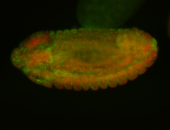
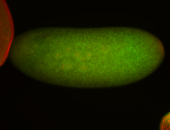
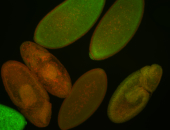
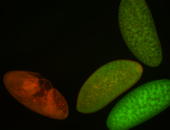
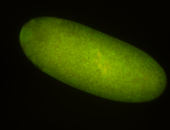
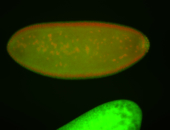
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
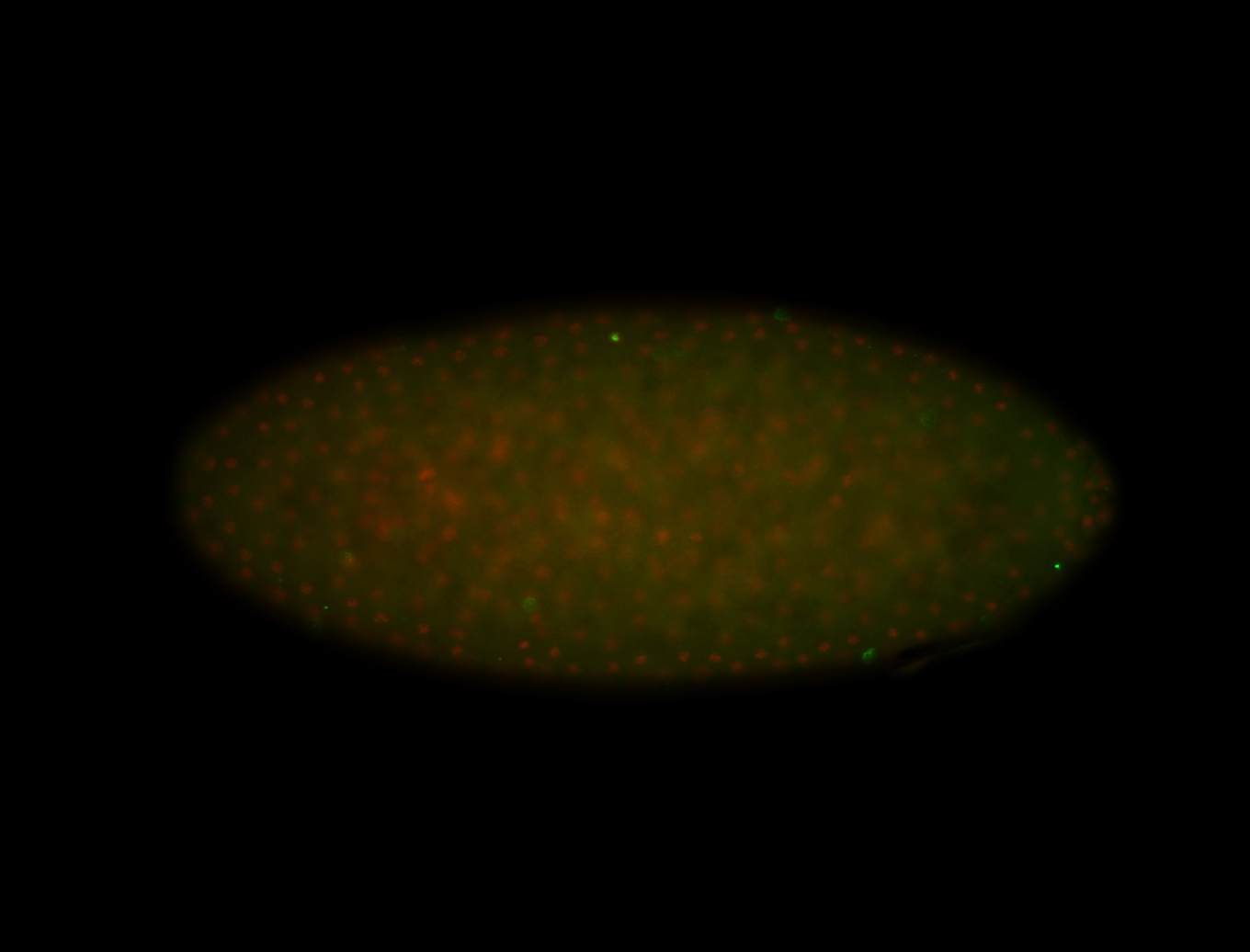
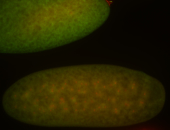
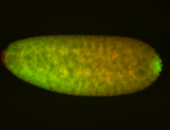
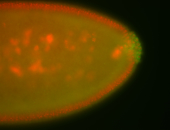
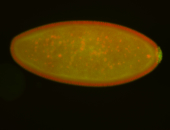
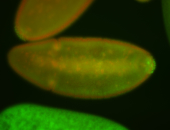
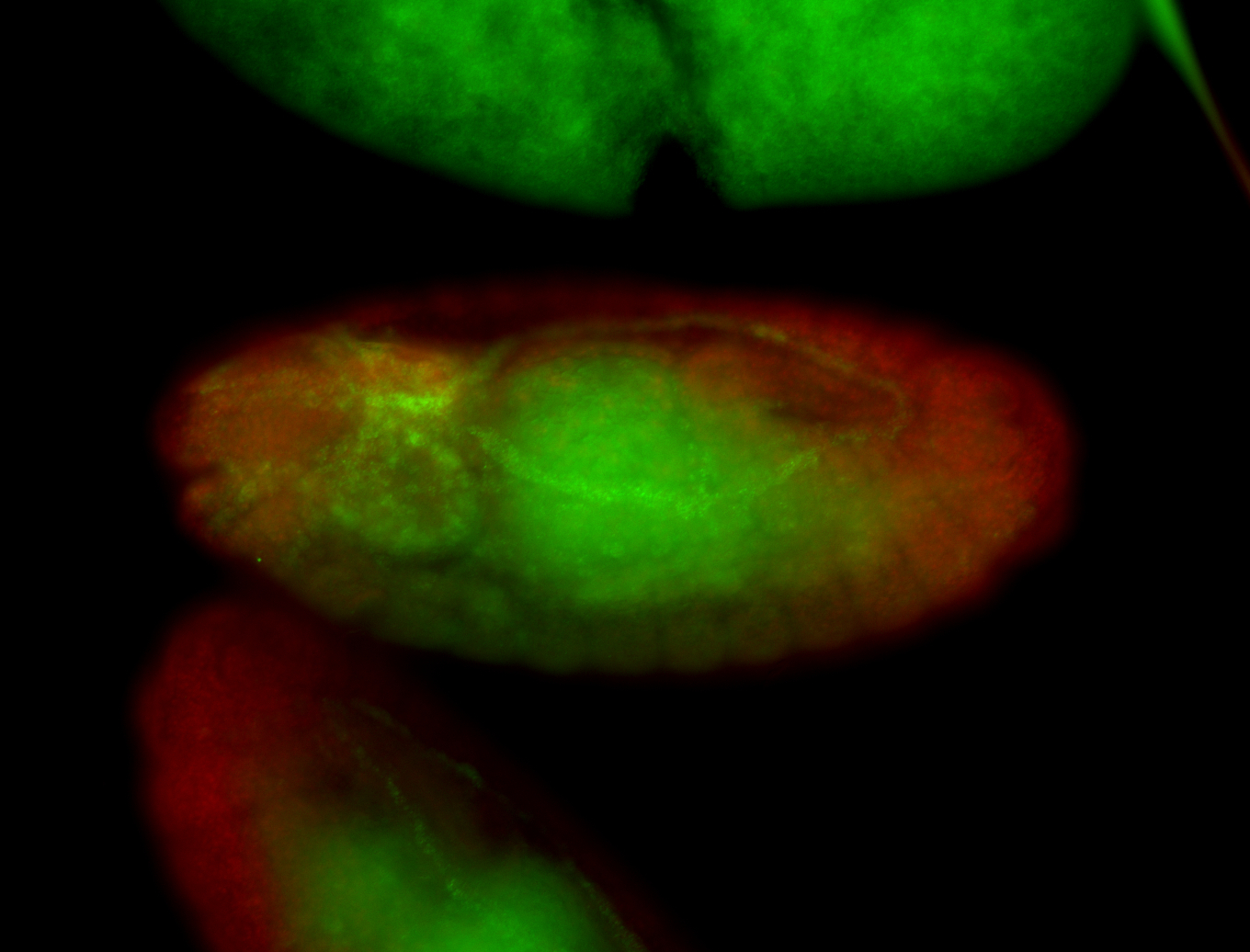
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
Adult
| Area | Images | Terms |
|---|
Comments
| It is hard to distinguish if the axons in the larval ventral nerve cord are 'axons' or 'tracheal extensions' |
Probe CG31131-ex Links: Permalink
Genes: CG31131
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
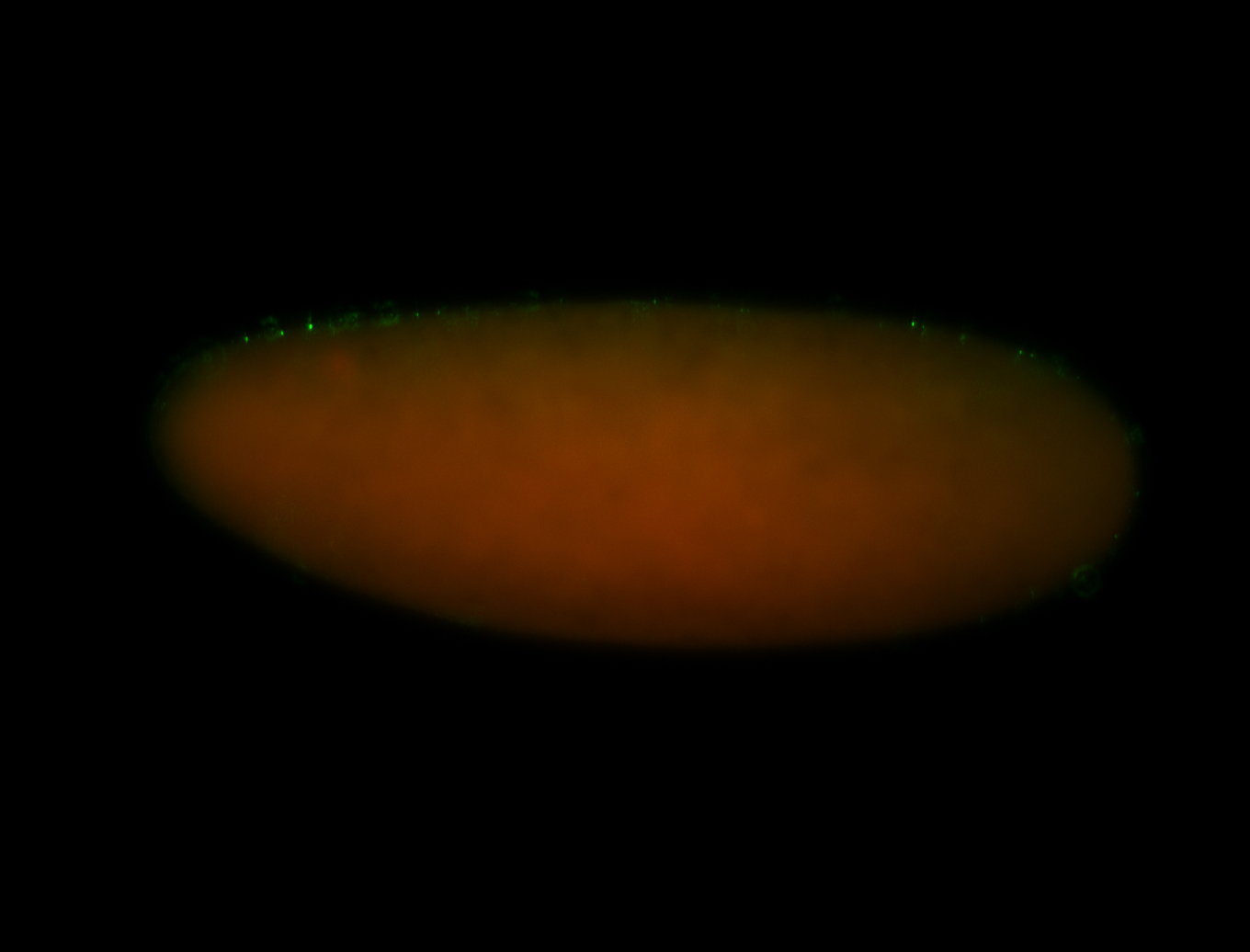
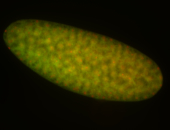
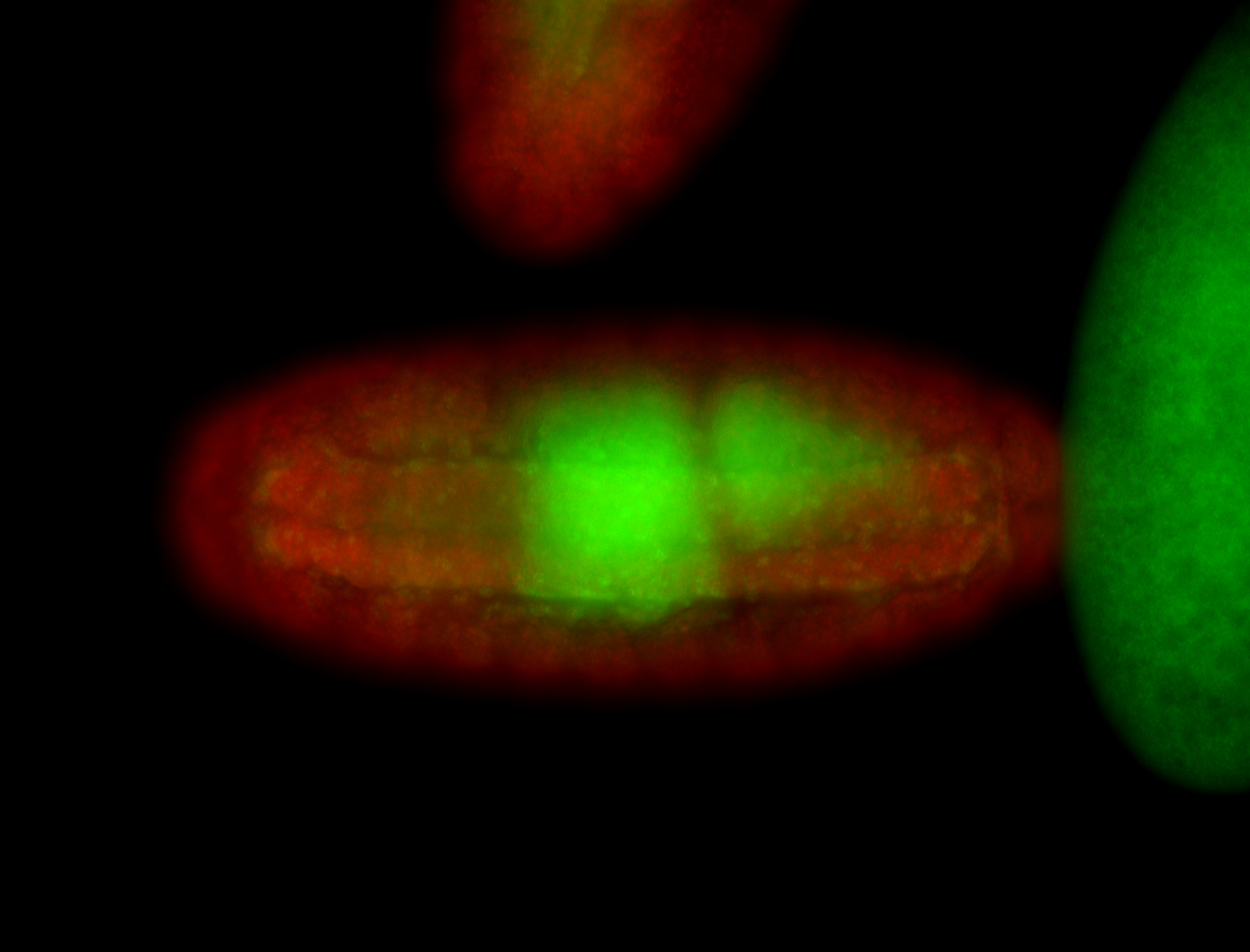
| -performed via HCR-FISH using 3 probe sets targeting common exons |
| -modENCODE RNAseq data on FlyBase shows low/no expression, however, we observed strong expression patterns throughout development |
Probe CG42239-RA Links: Permalink
Genes: CG42239
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
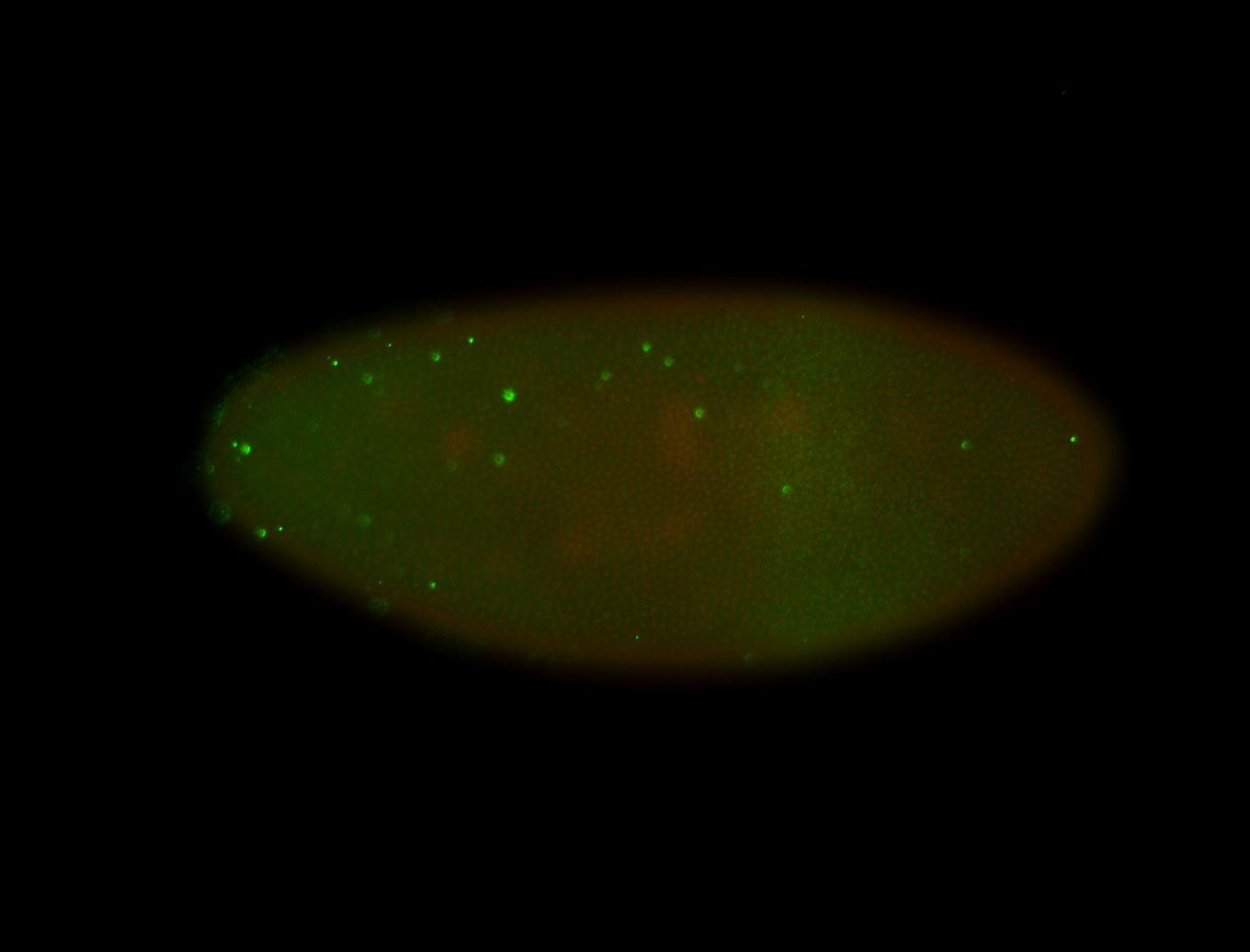
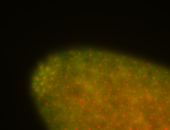
| -becomes very hard to detect after early germband elongation and is rapidly degraded, it may still be weakly ubiquitous in later stages but expression levels border between expressed and non-expressed |
| -performed via HCR-FISH using 3 probe sets targeting the RA transcript isoform |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
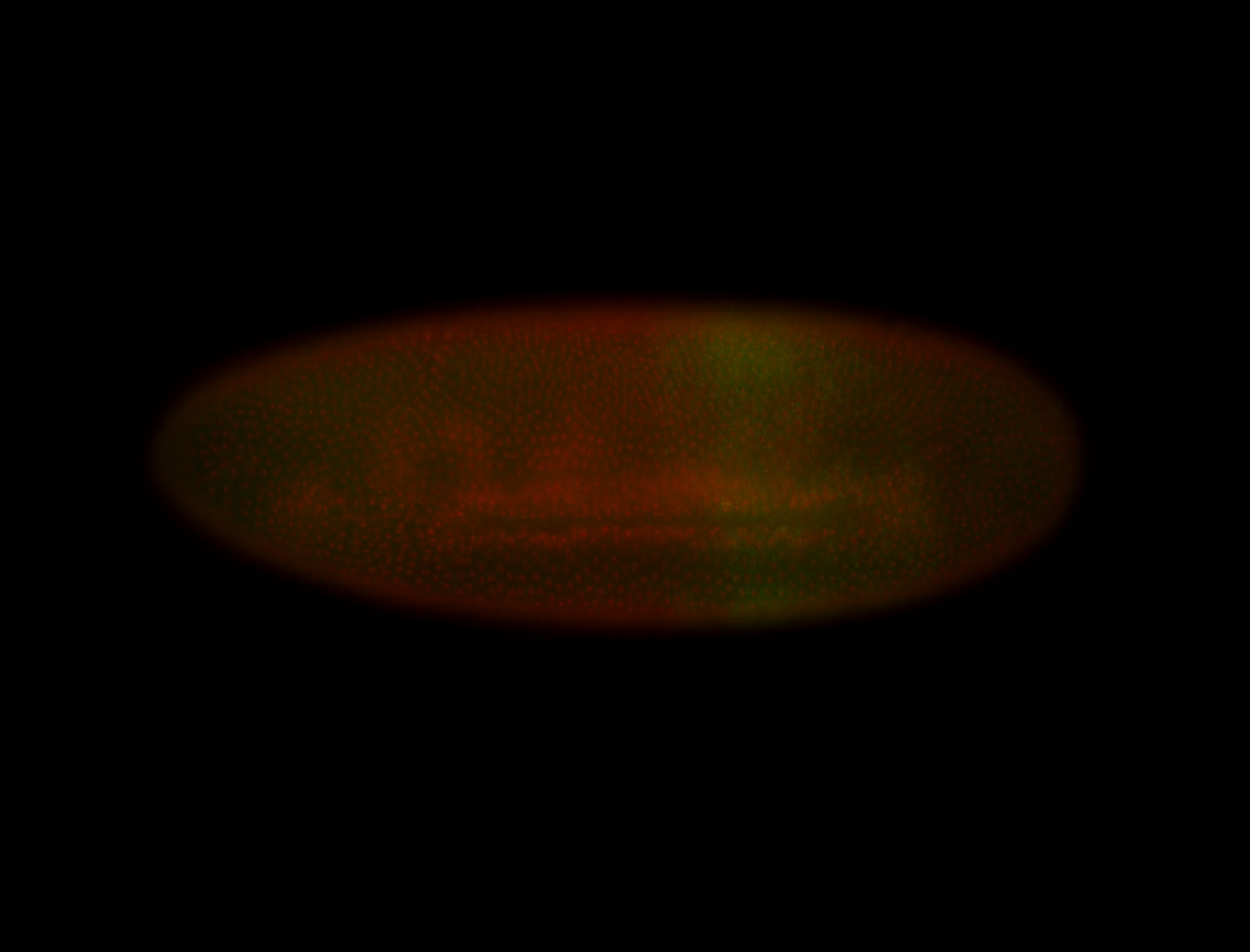
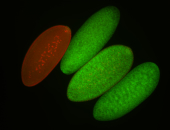
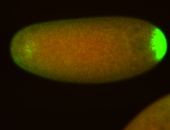
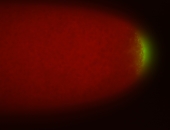
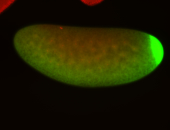
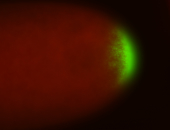
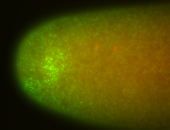
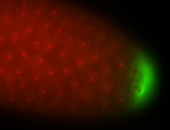
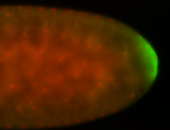
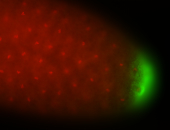
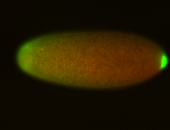
| Posterior enriched |
Probe CR43617-RA Links: Permalink
Genes: CR43617
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
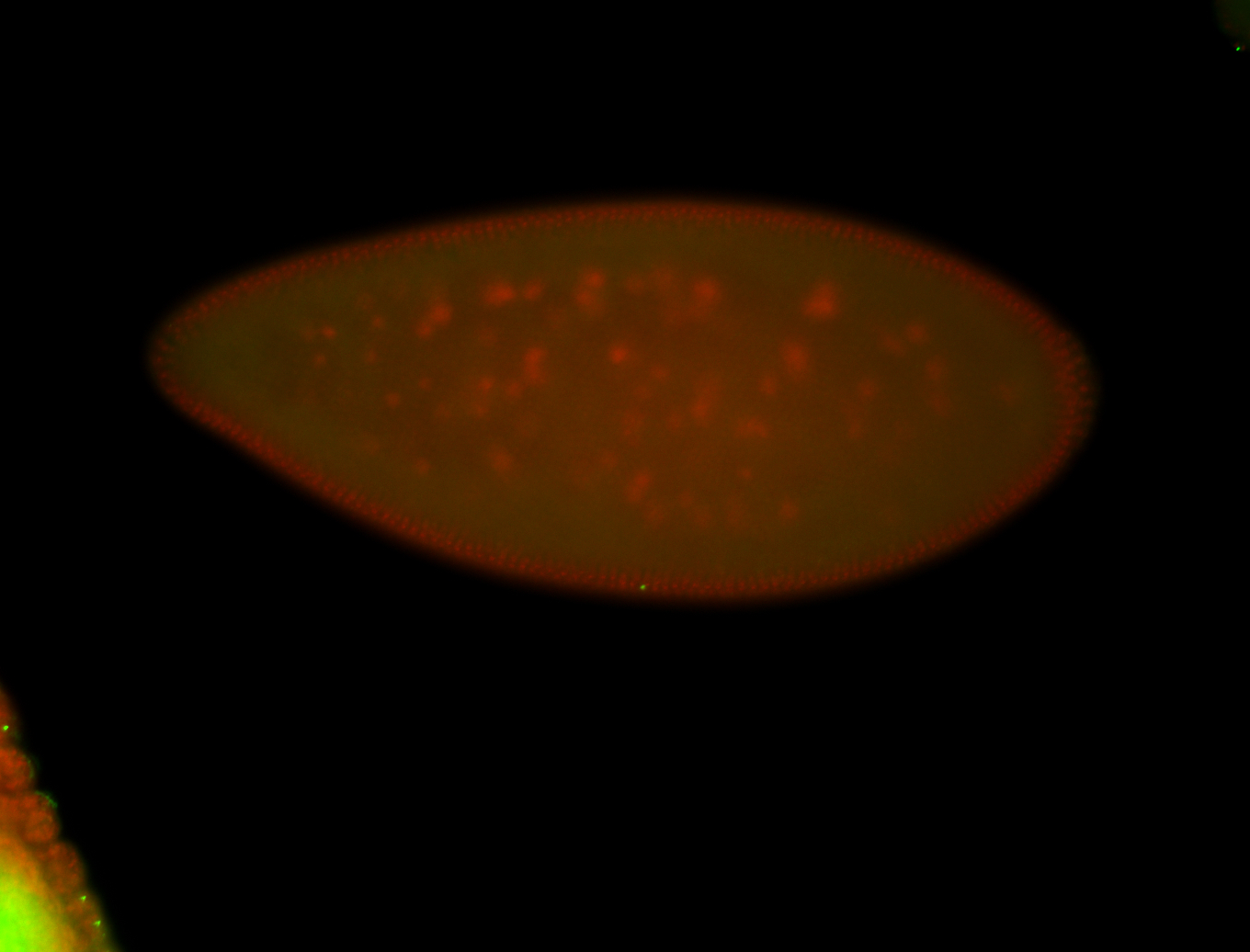
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -posterior single stripe; expression is reported to be similar to the lncRNA "iab4" (CR31271) |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| All over |
Probe GEFmeso-ex Links: Permalink
Genes: GEFmeso
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting common exons |
Embryo
Larva
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe on gel. |
| Sub and peri nuclear with apical enrichment. Anterior cells only. Very specific and interesting. |
| It is hard to distinguish if the axons in the larval ventral nerve cord are 'axons' or 'tracheal extensions' |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
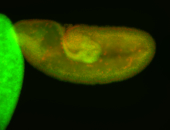
| Anterior midgut, stronger at the posterior midgut. |
| Probe looks good on gel |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -There is no entry for this gene on BDGP, and modENCODE RNAseq data on FlyBase indicates that it is not expressed in the embryo. However, we observed obvious patterns during embryogenesis. Additionally, FlyBase does not have any in-depth characterizations of this gene. |
| -Osi21 does not overlap with any other genes, ruling out the possibility of the targeting of alternative transcripts. BLAST also confirms the specificity of our probes to Osi21. Additionally, no other genes for which probes were designed for on this plate show patterns such as those observed here, ruling out the possibility of cross-contamination during probe preparation. |
| -This may be a result of Osi21's poor characterization or incorrect RNAseq data |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting common exons |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
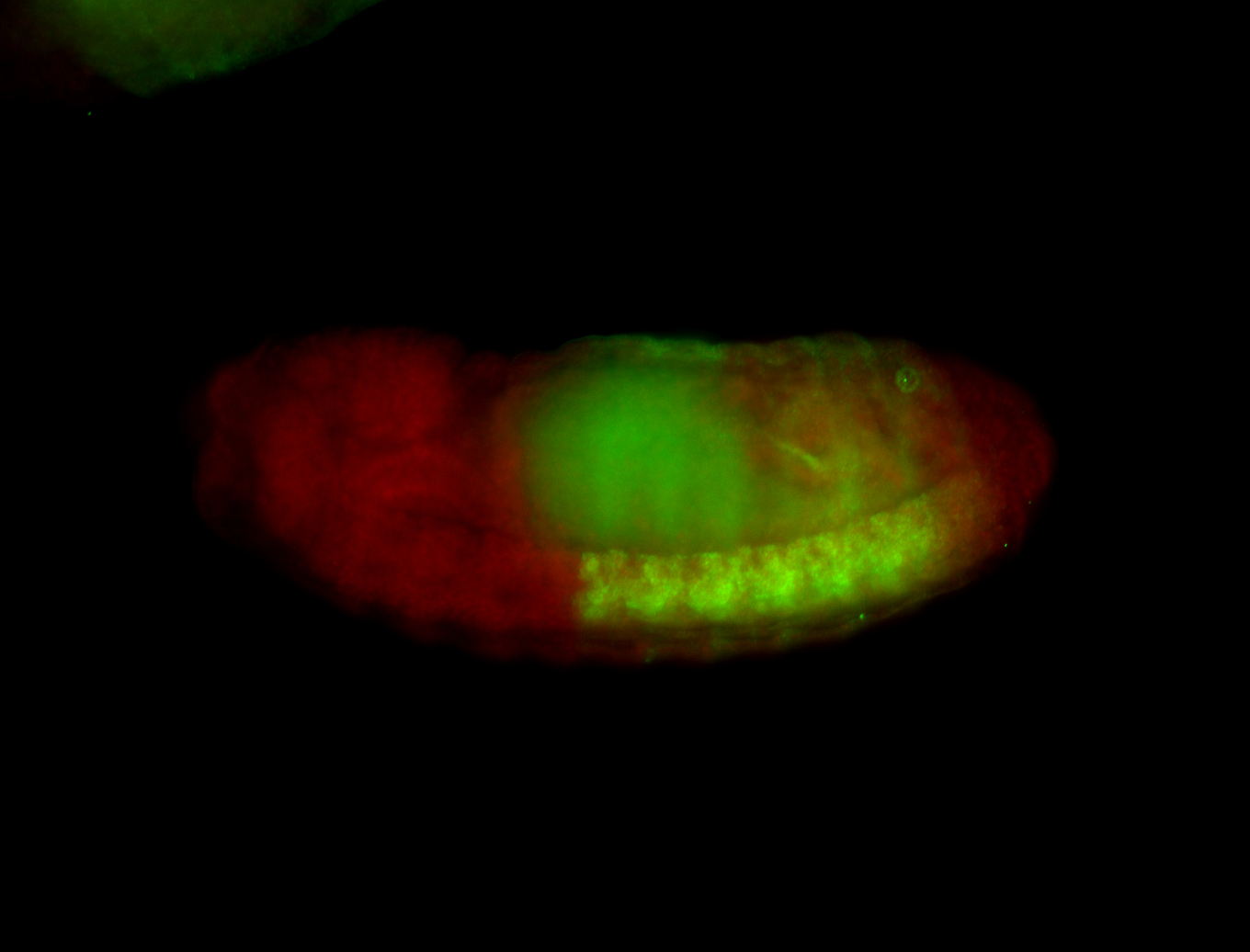
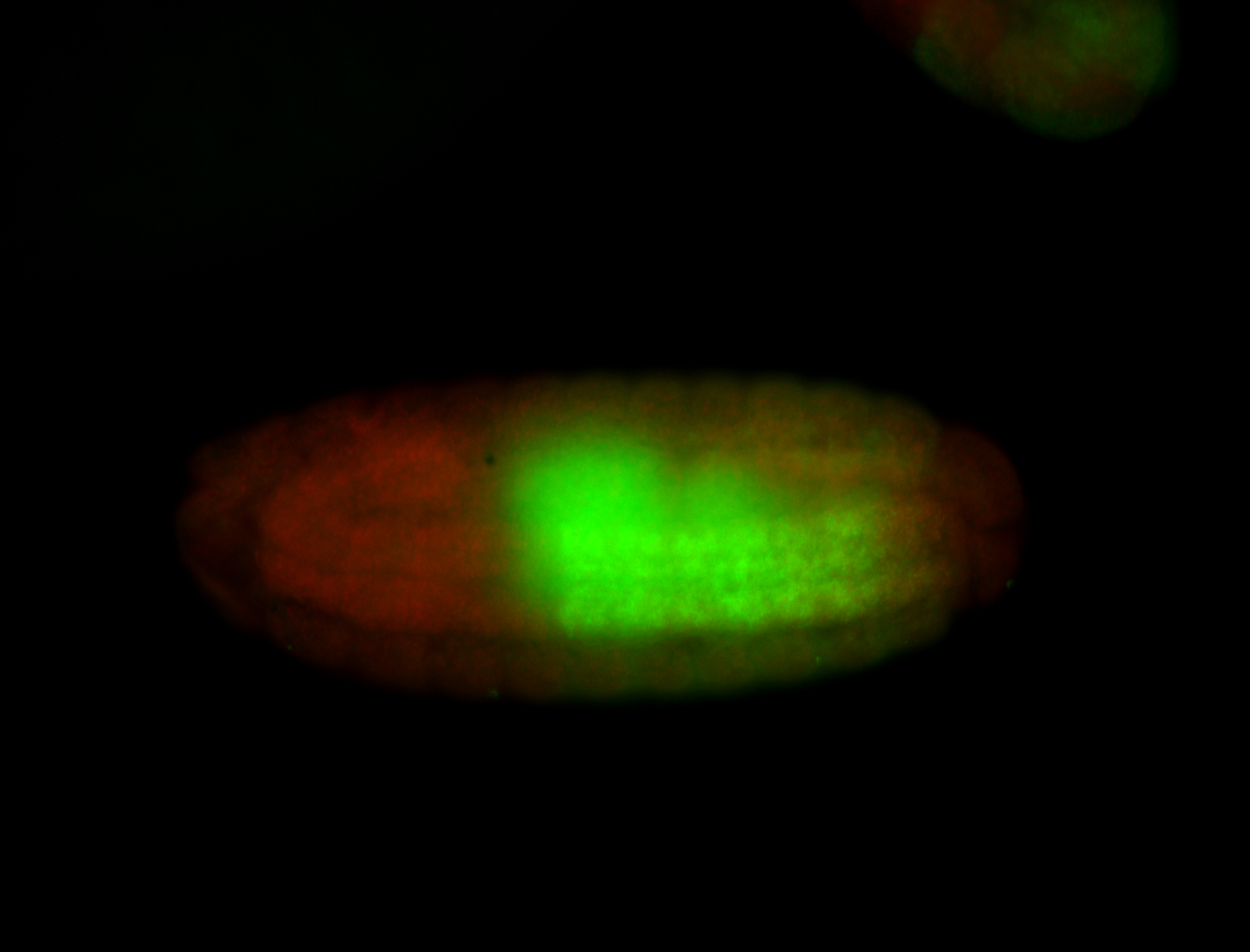
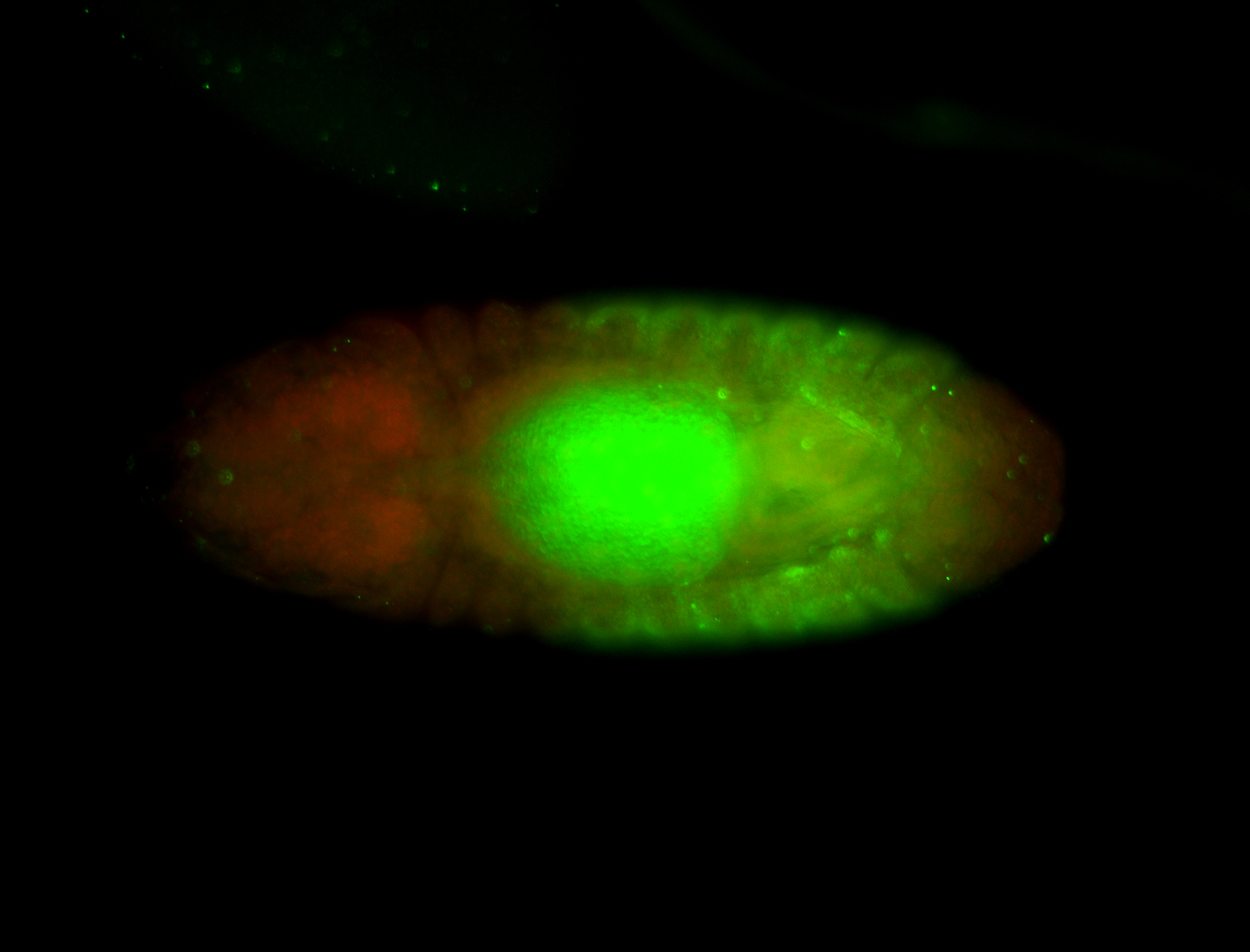
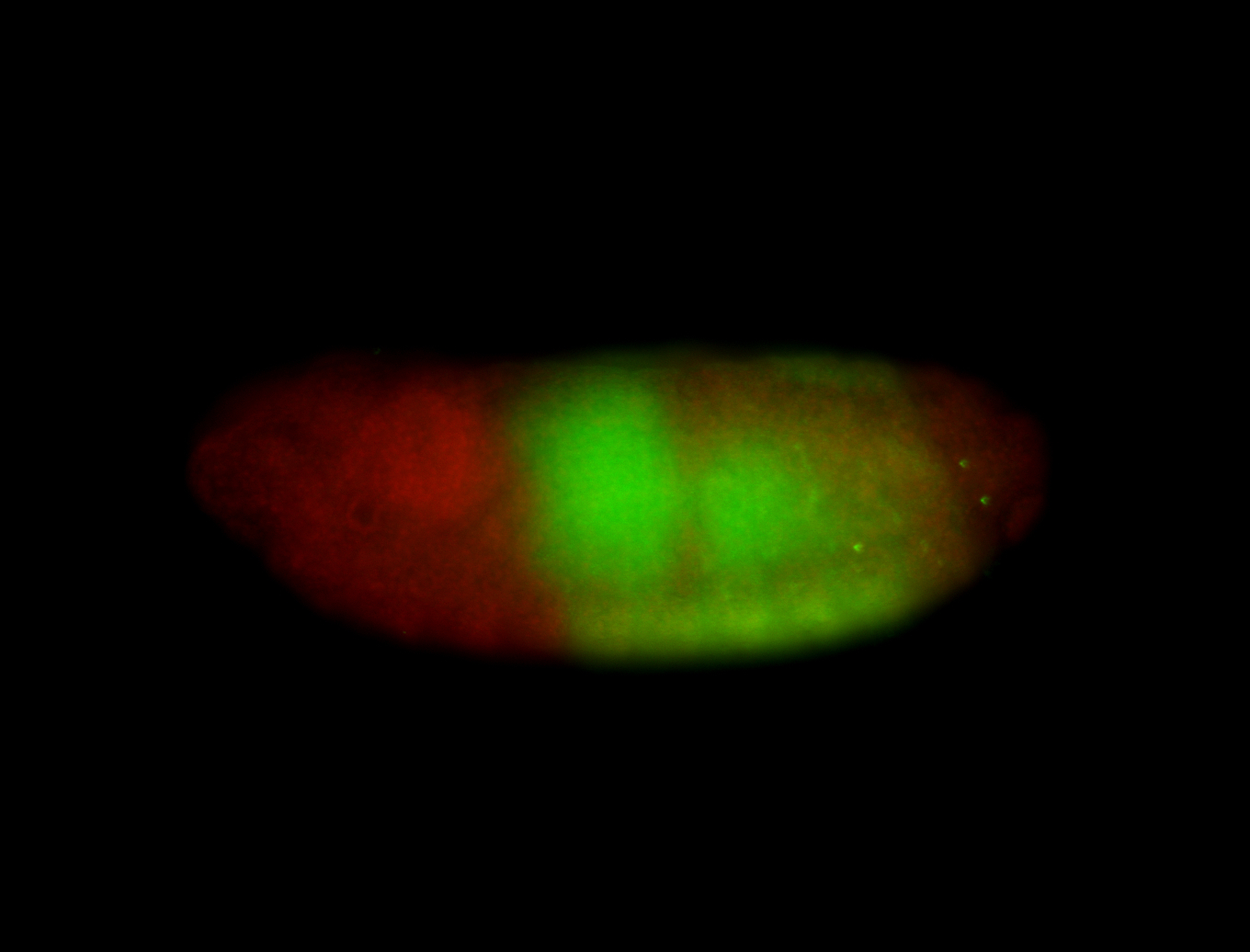
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
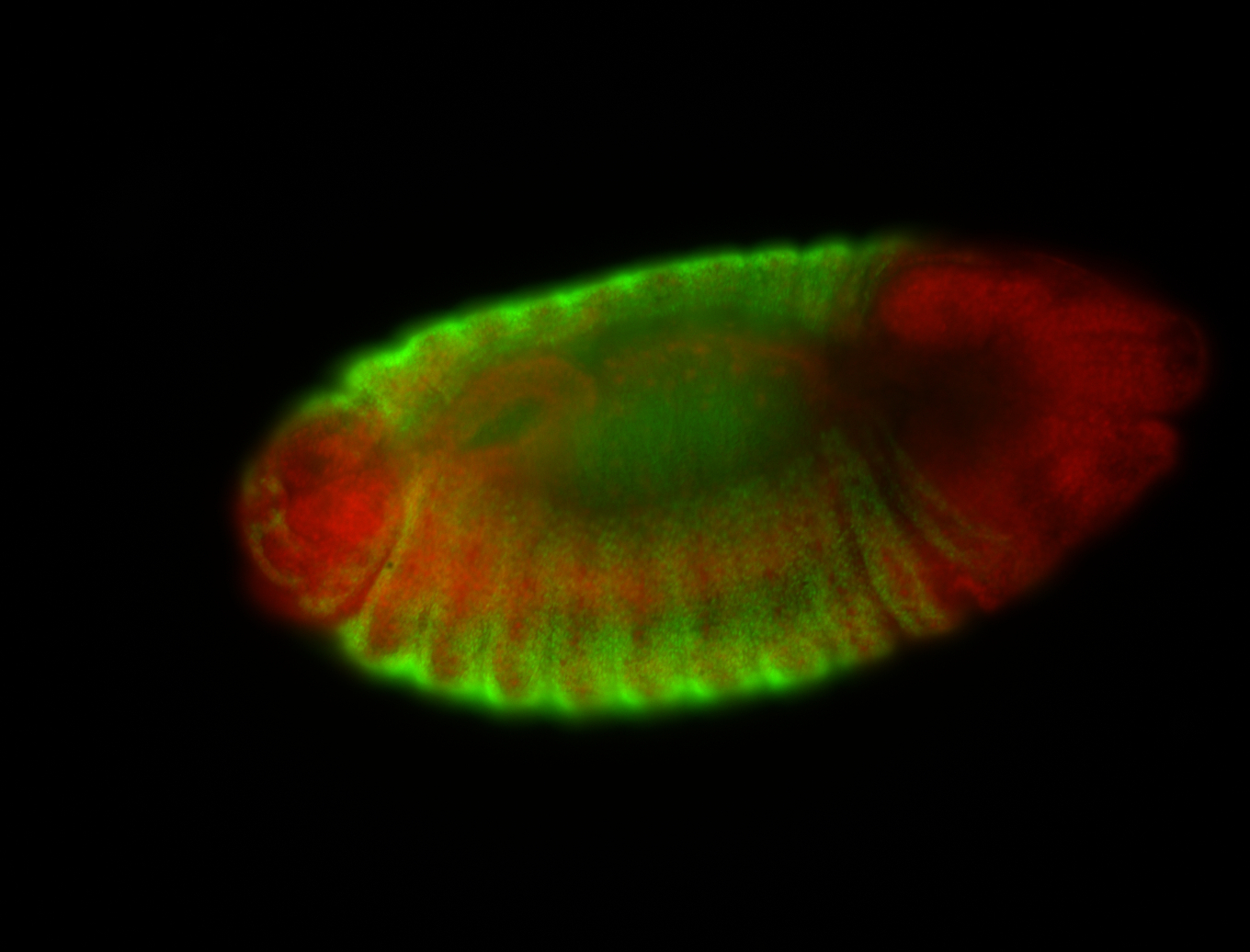
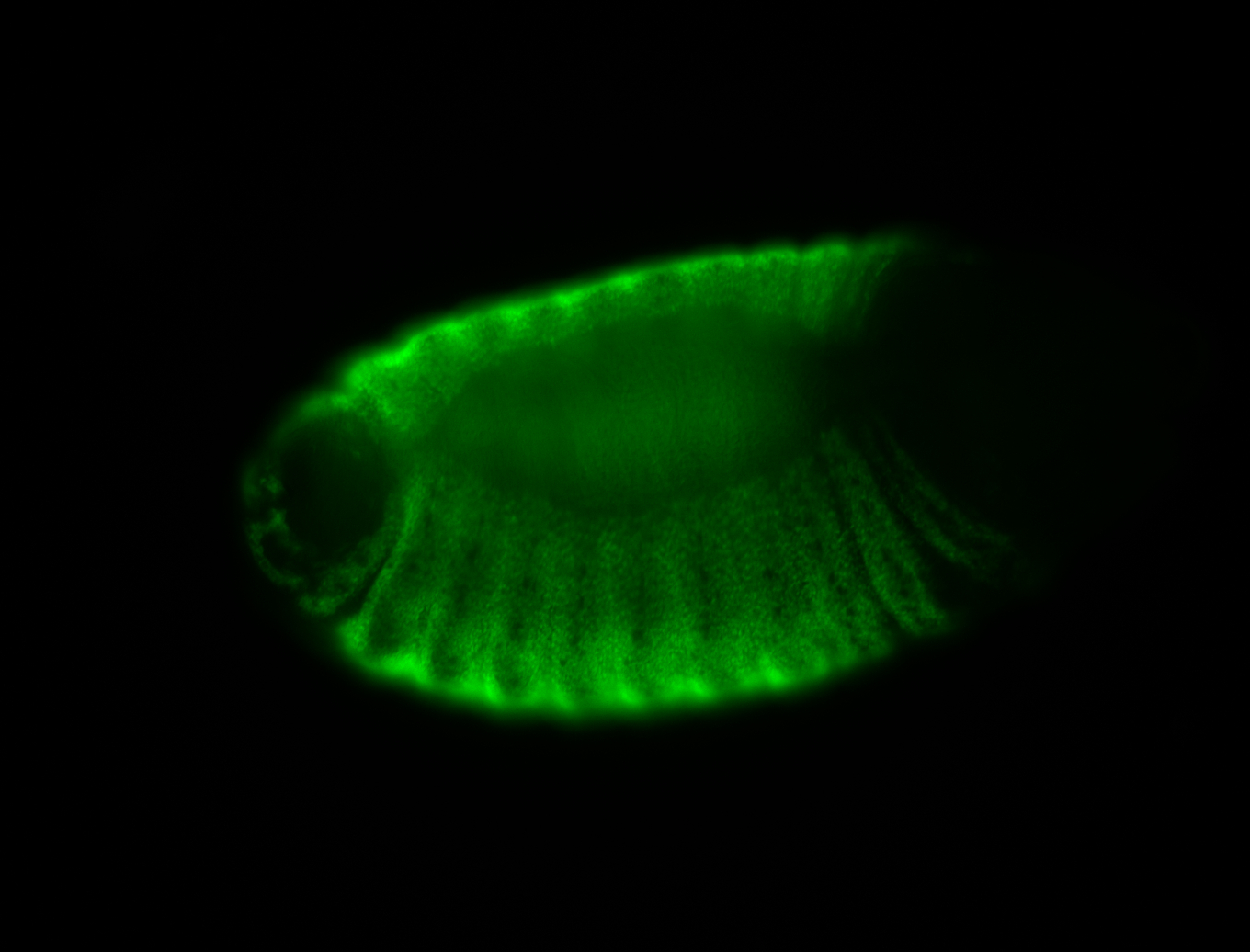
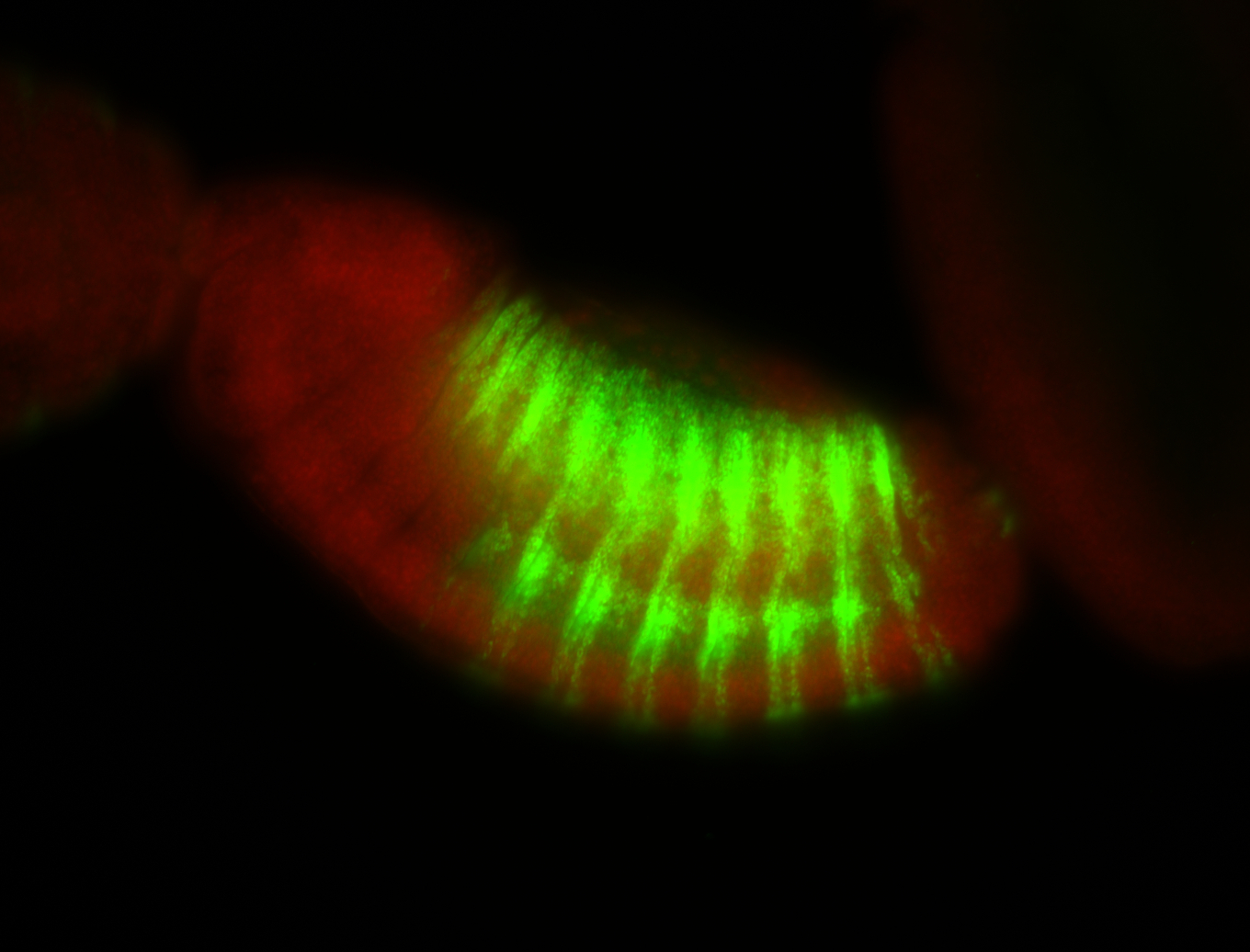
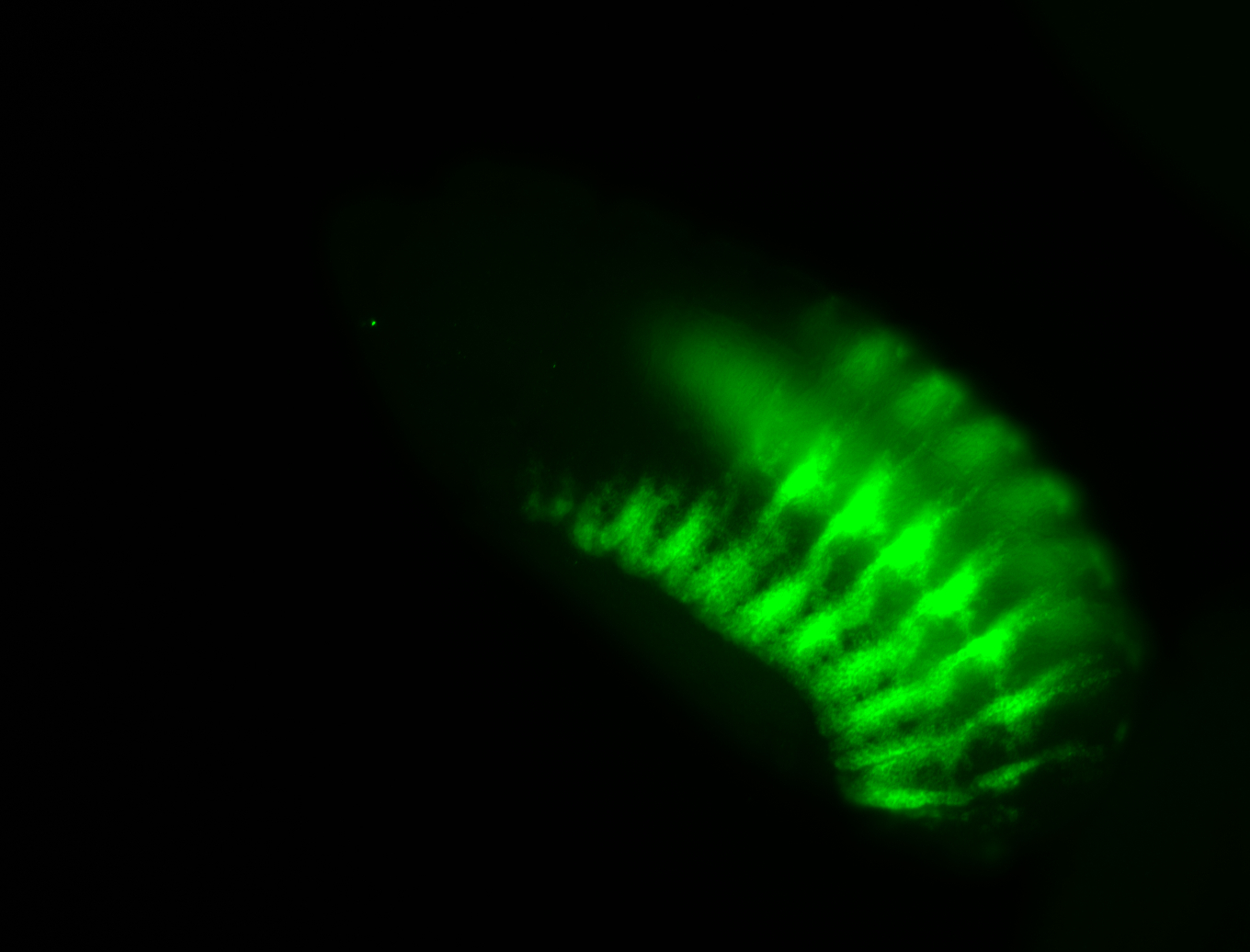
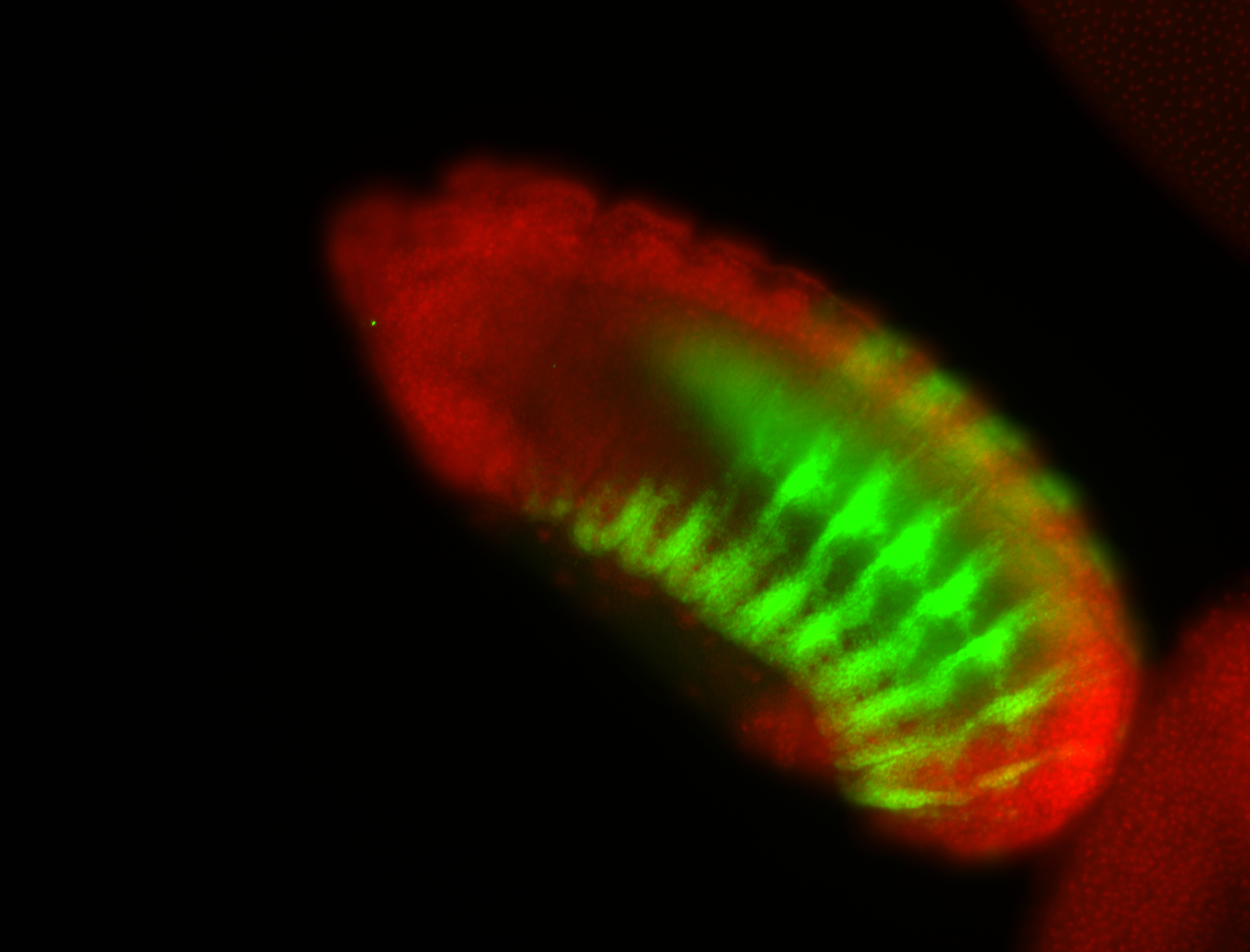
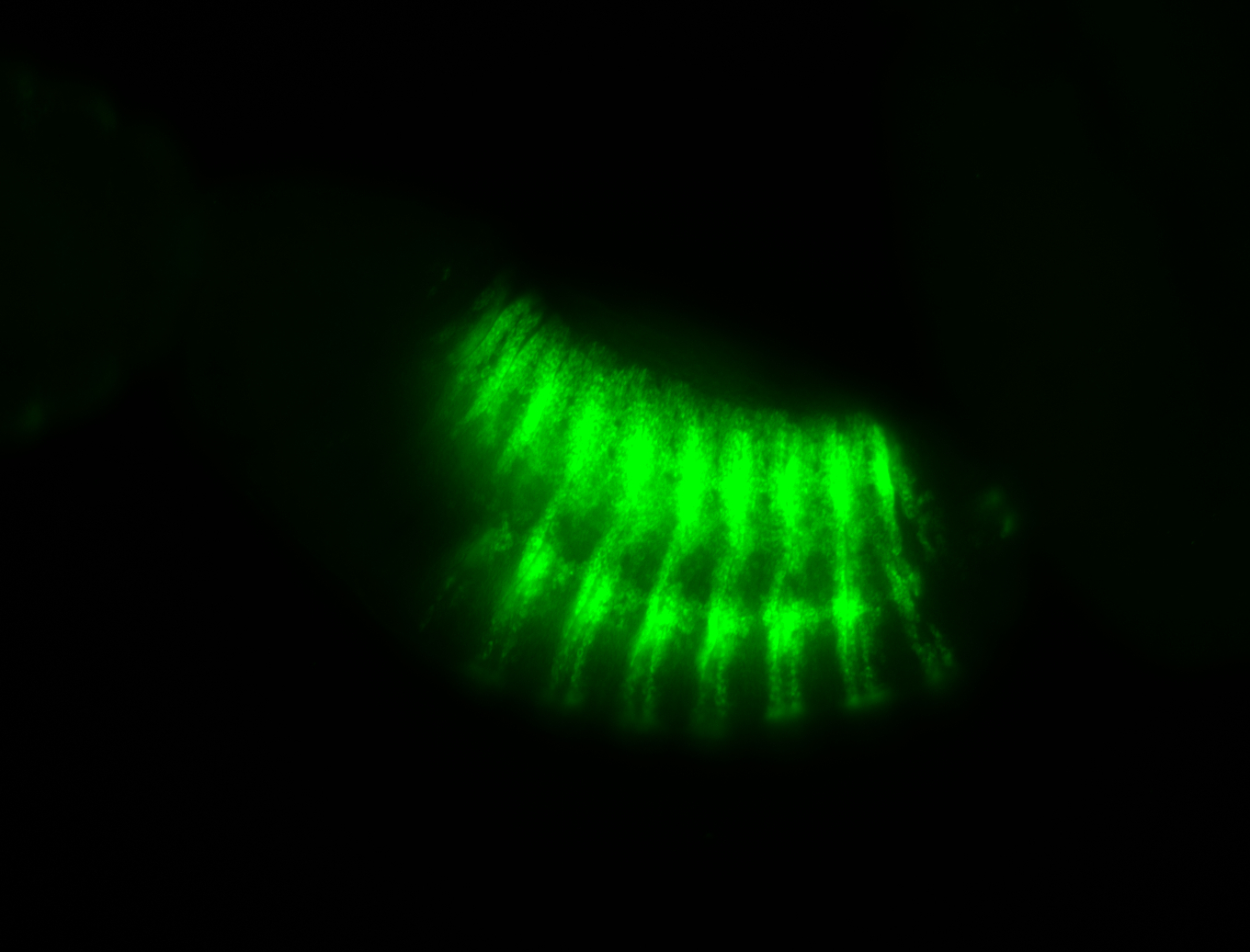
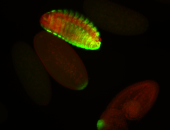
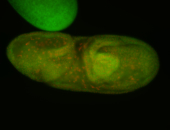
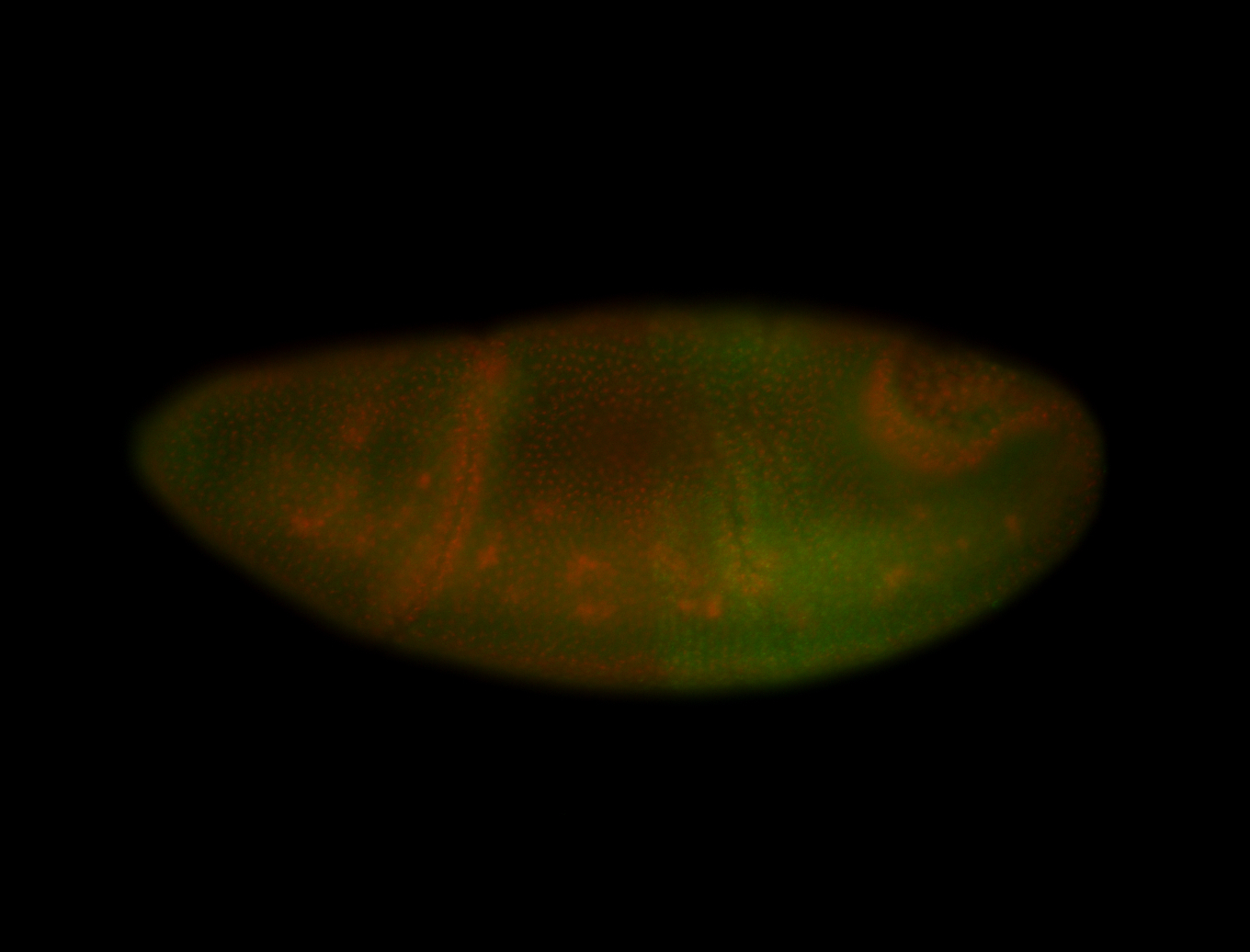
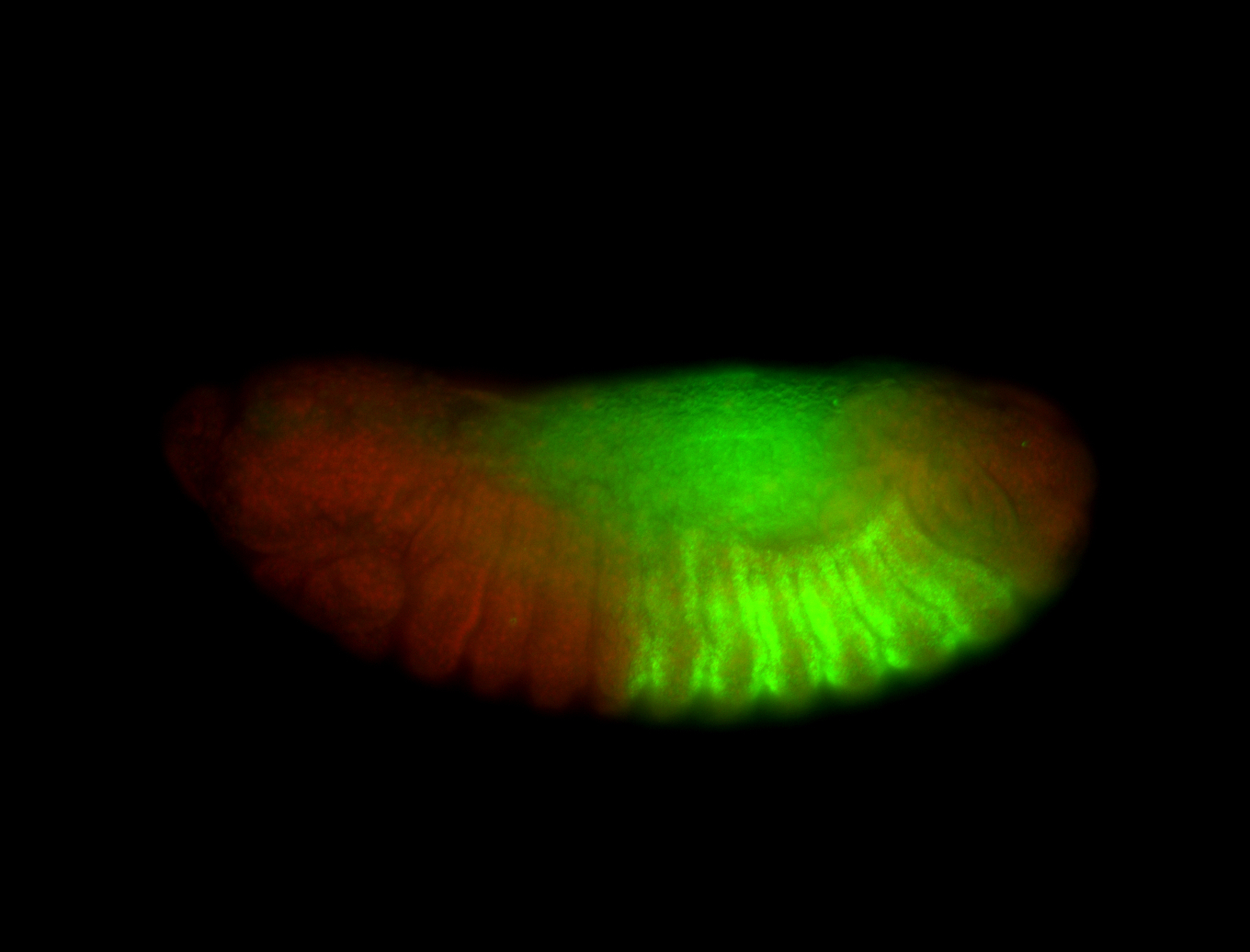
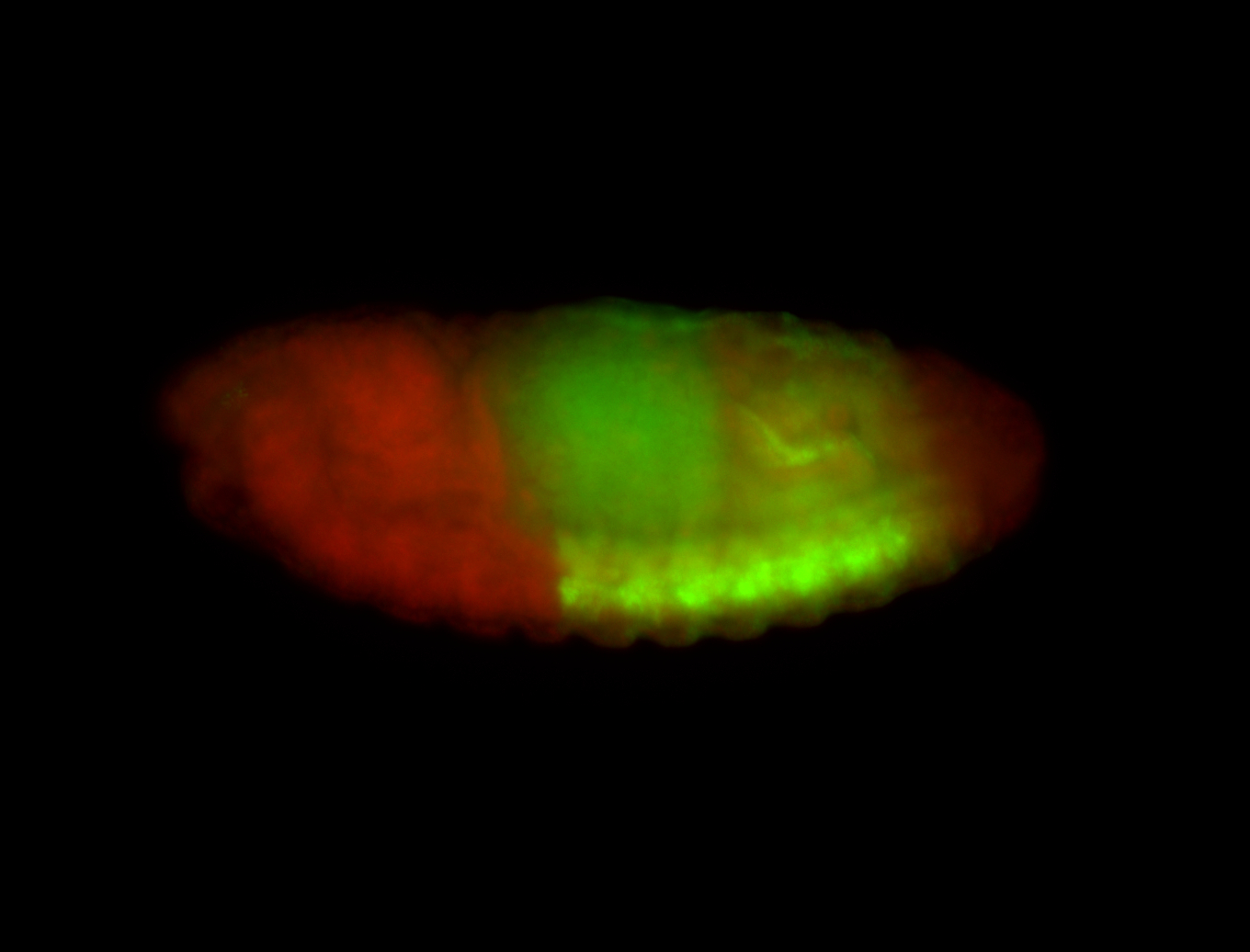
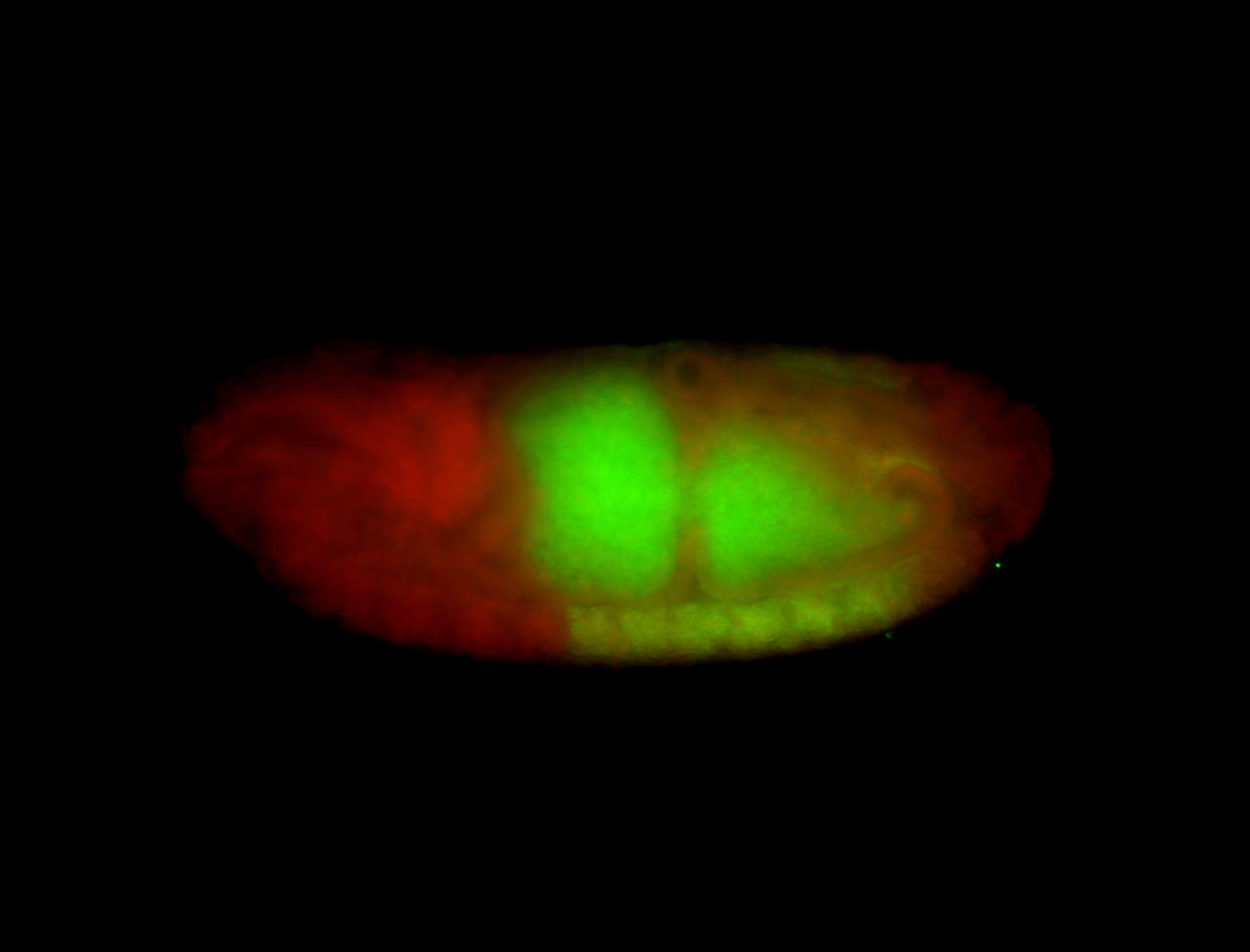
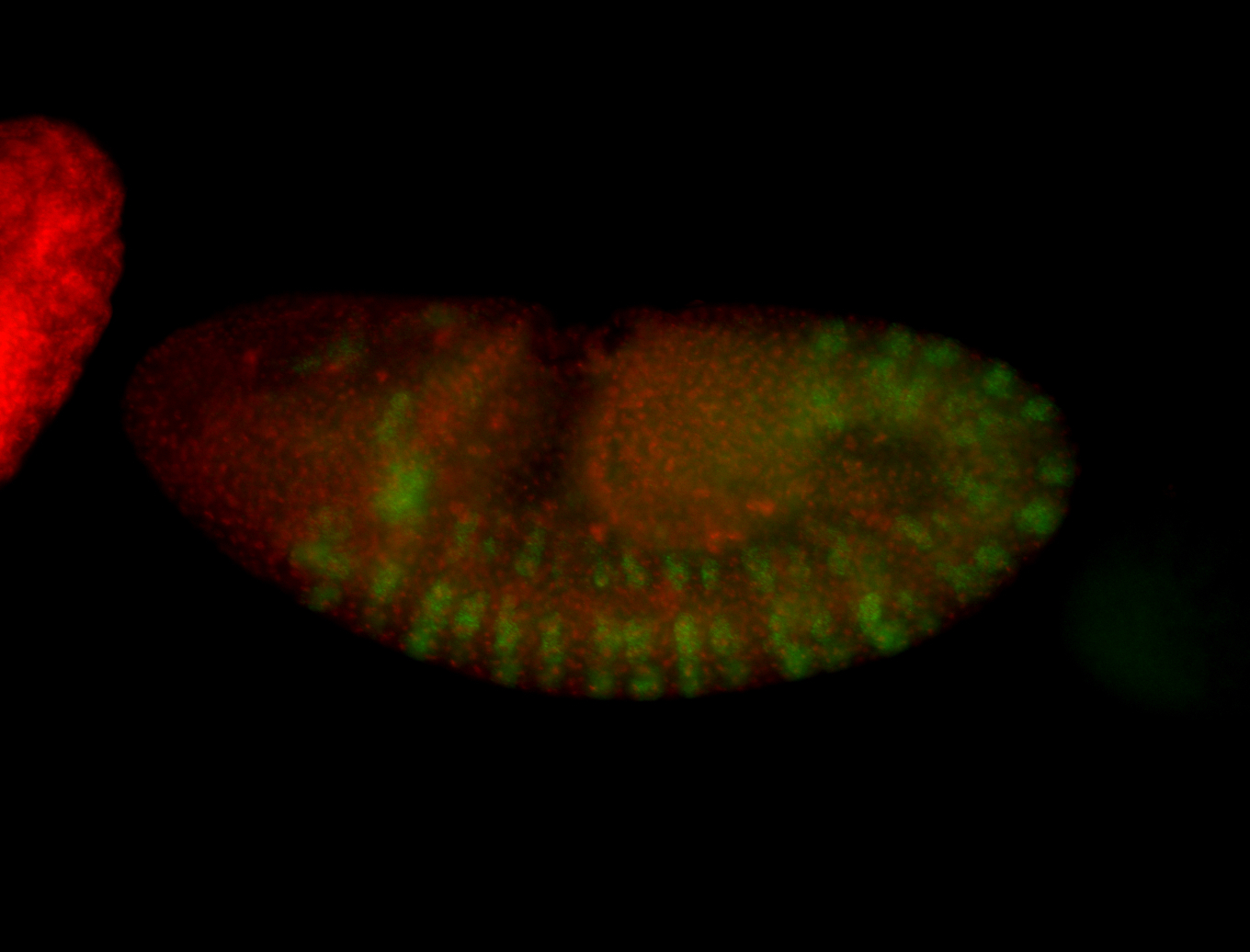
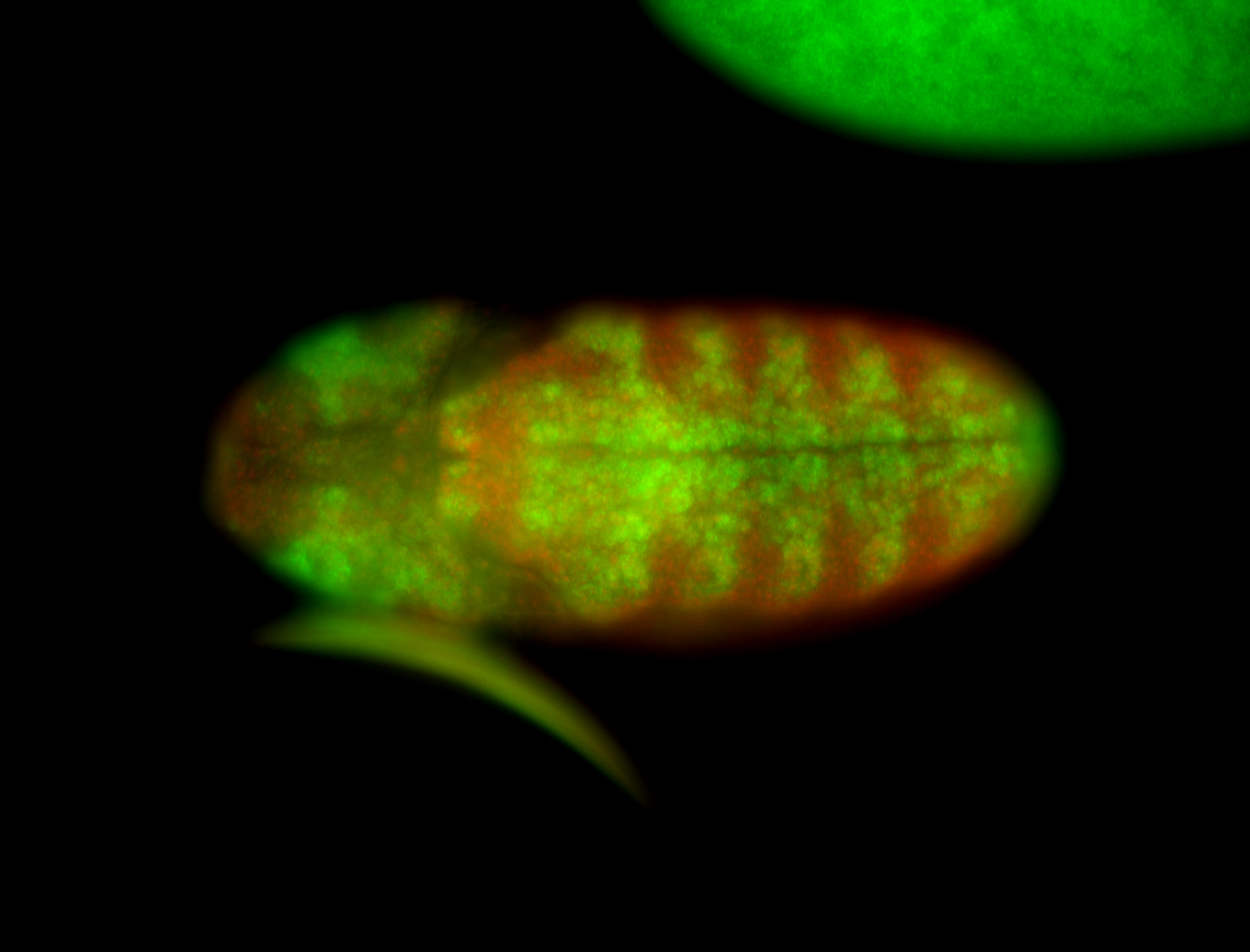
| -possesses a striking segmented pattern in the epidermis, mesoderm, and VNC; reports of expression in the cardiac primordium and dorsal vessel were not clearly observed and could possibly be amnioserosa expression |
| -a classic homeobox gene |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA transcript isoform |
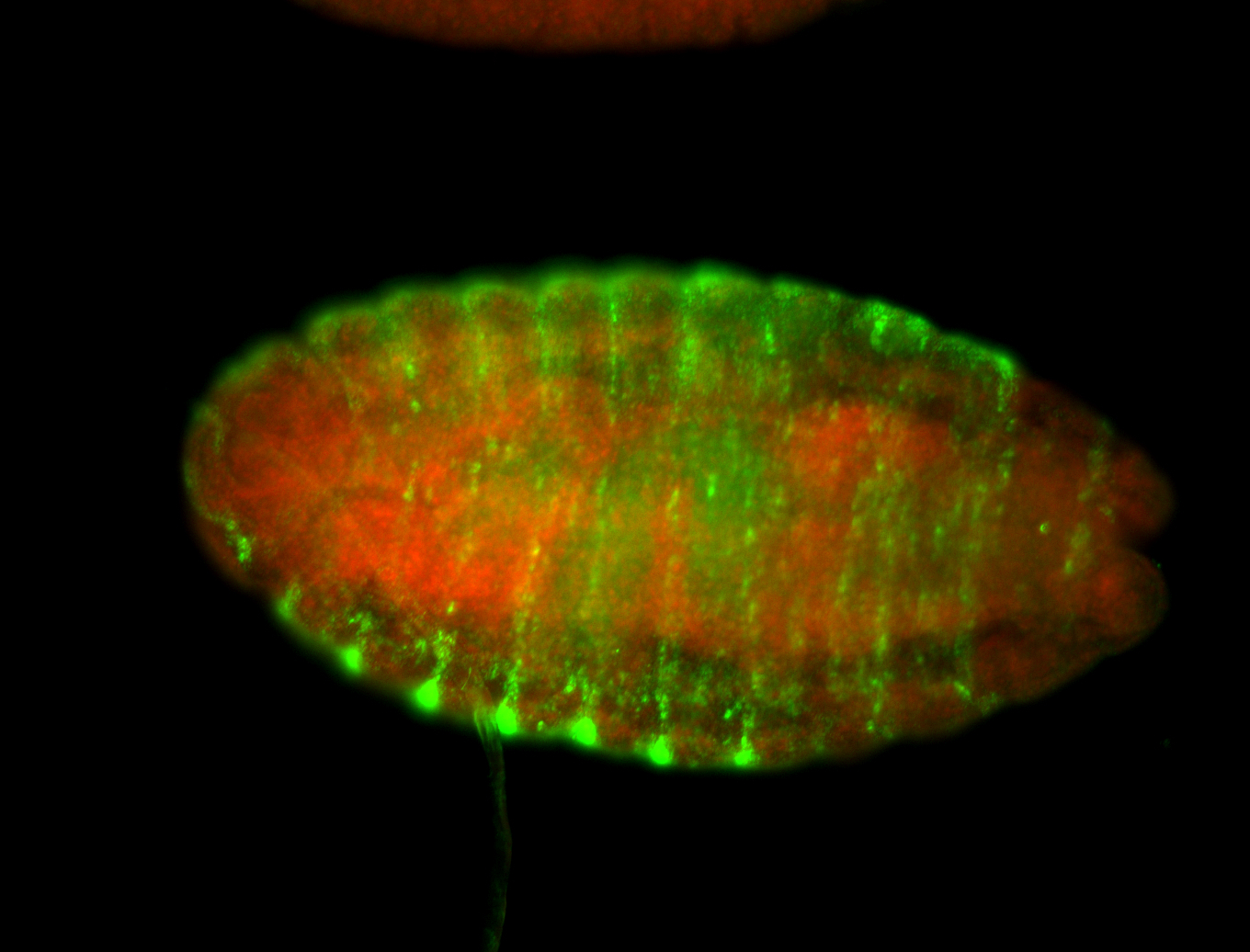
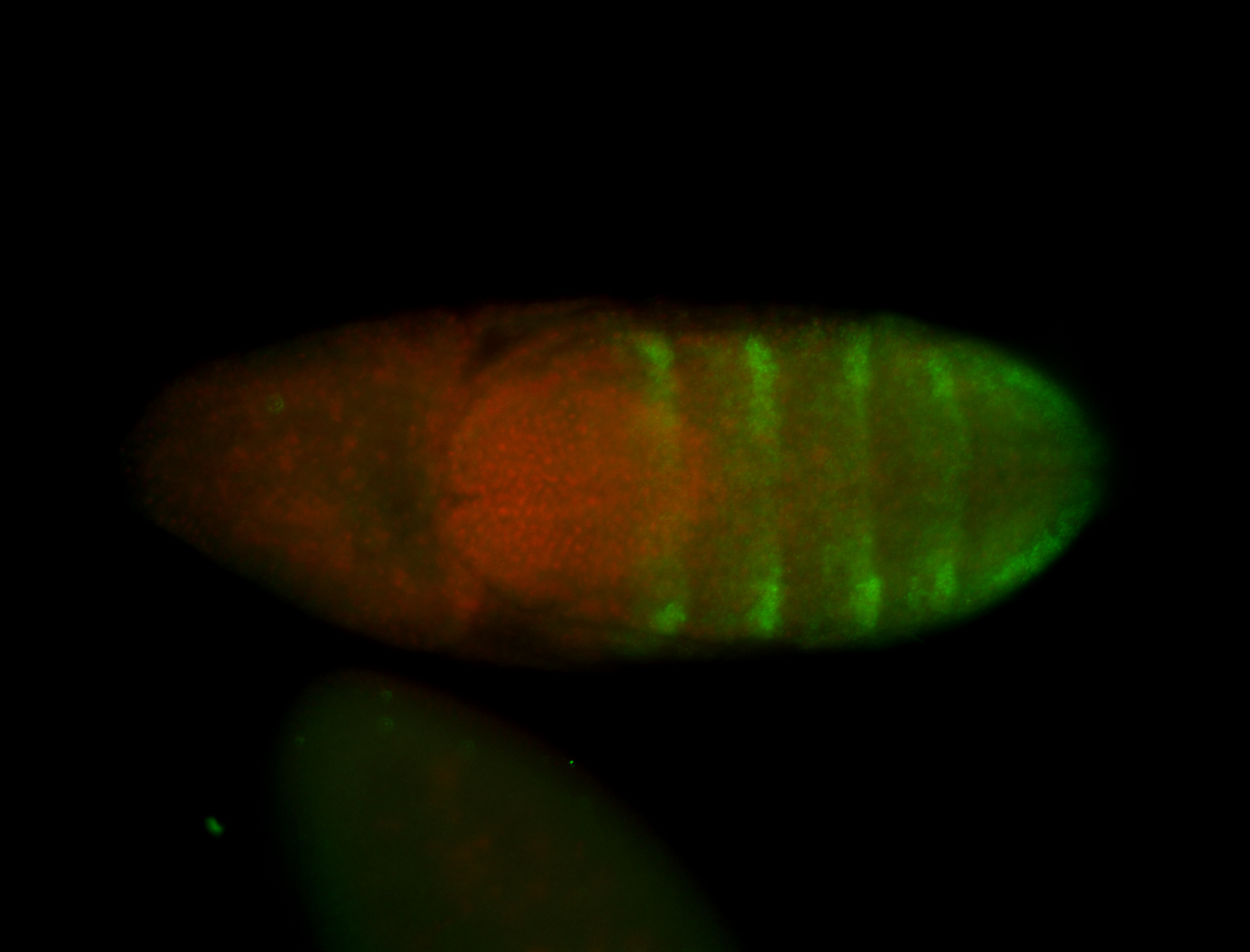
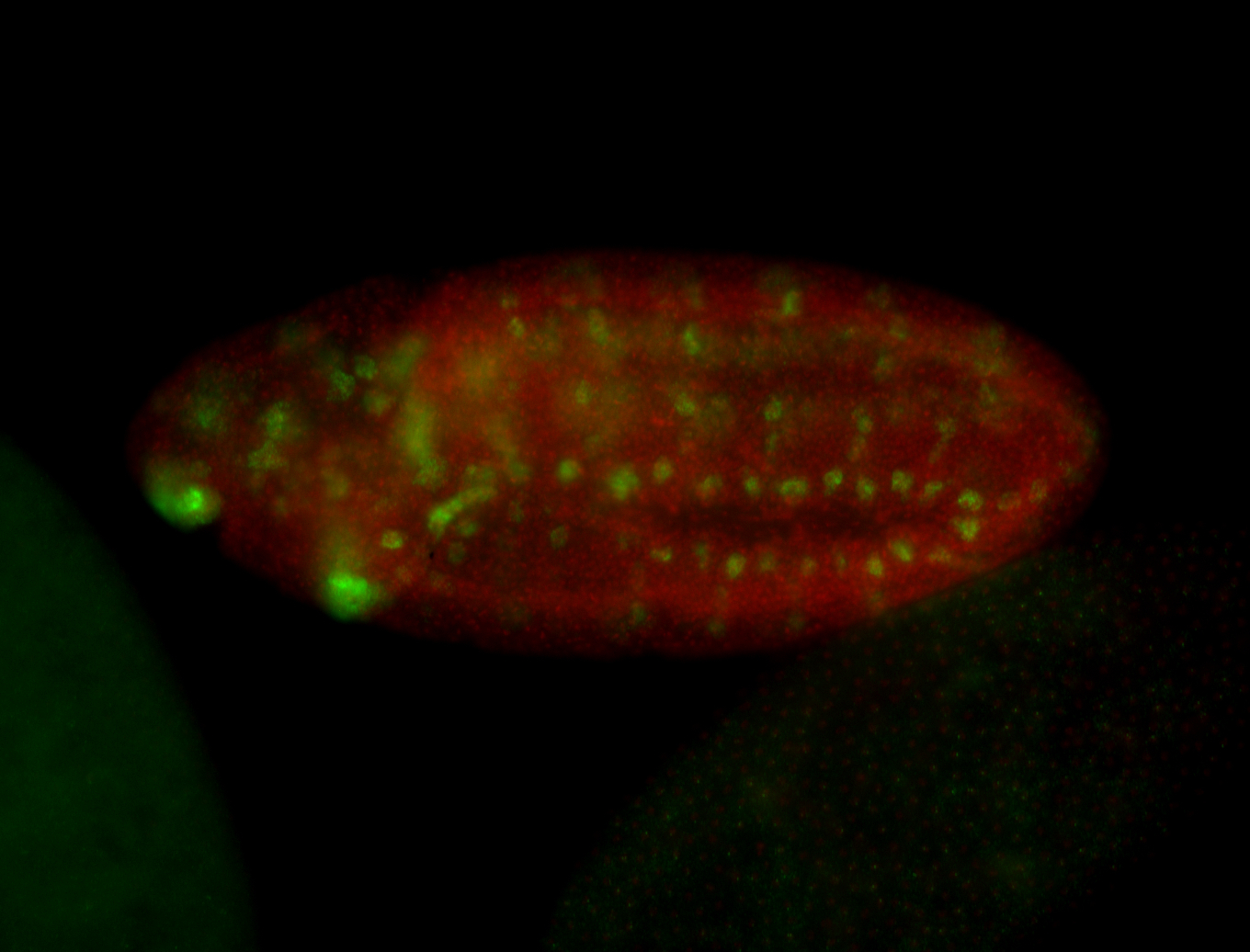
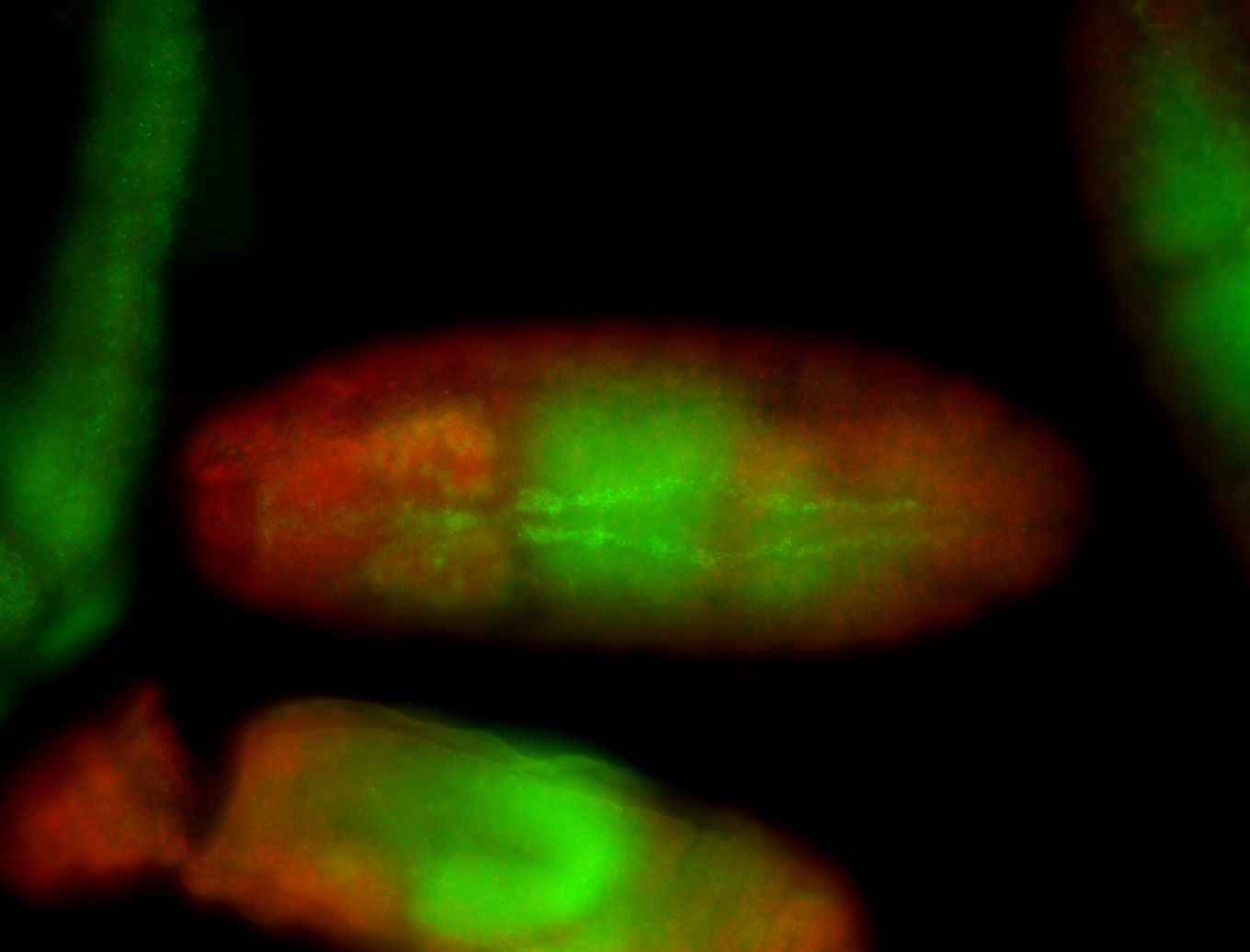
| -neat marker for neural precursor formation, appears to not be expressed past stage 13-14 except for 4 small points near the dorsal-posterior region of the ectoderm (possibly tip cells of the Malpighian tubules); clear stomatogastric nervous system primordium expression |
| -images are from two separate experiments under identical conditions for better coverage |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting common exons |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
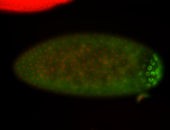
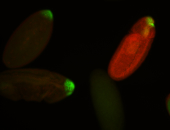
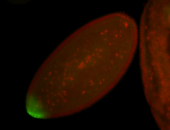
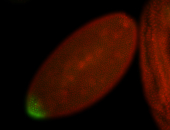
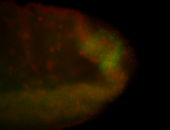
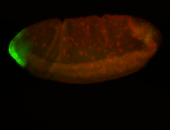
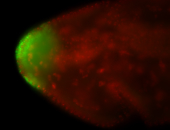
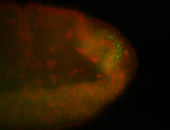
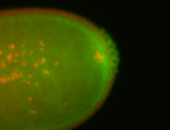
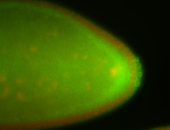
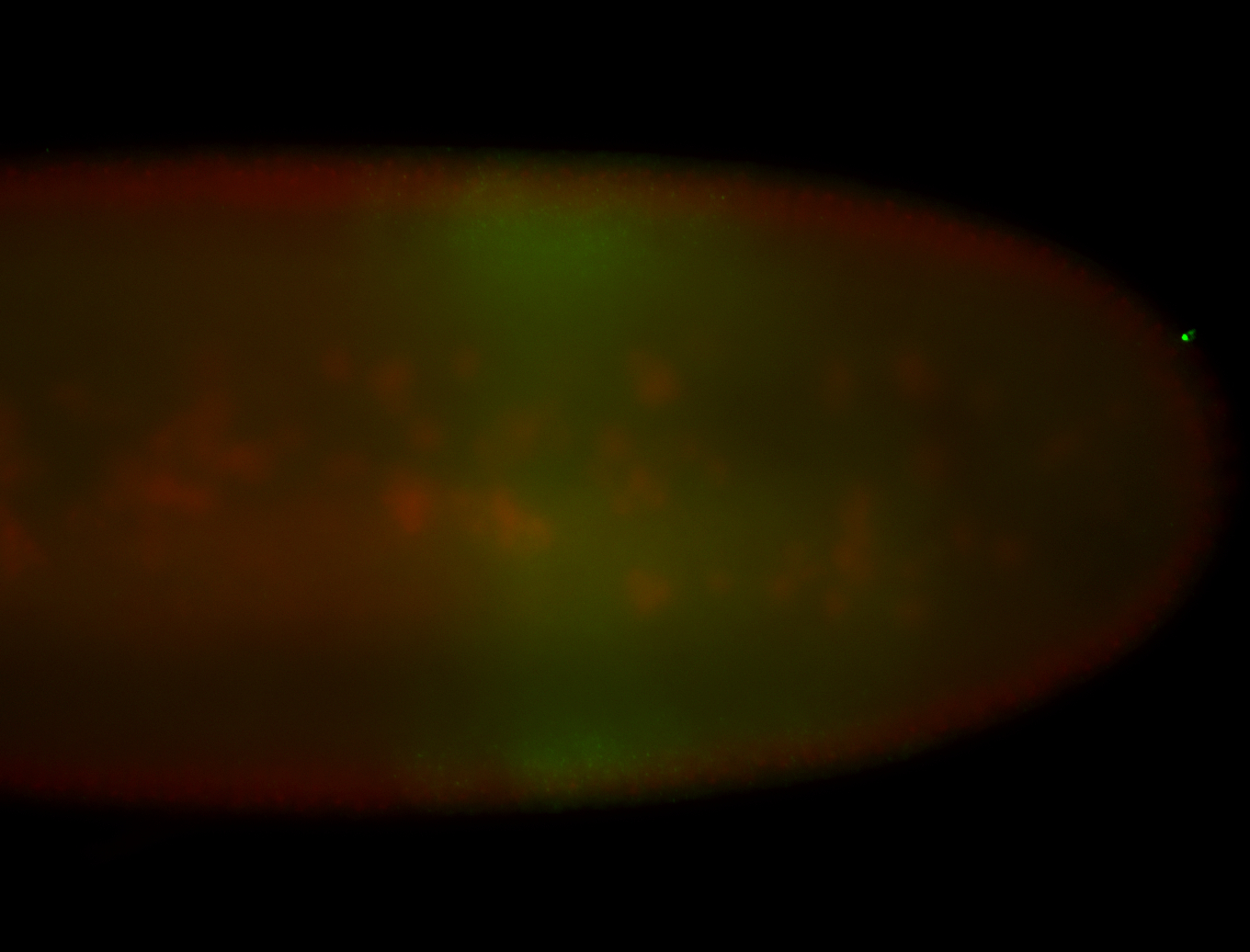
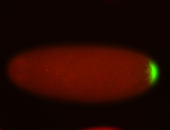
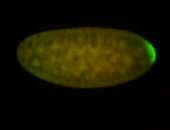
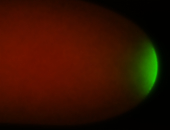
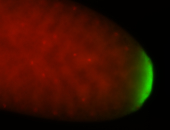
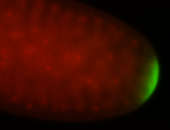
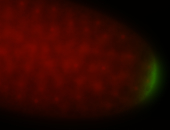
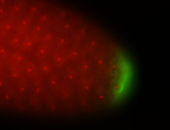
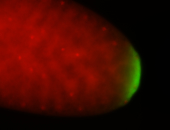
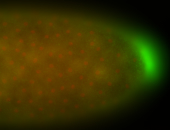
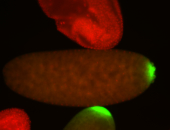
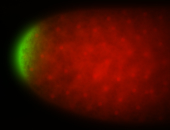
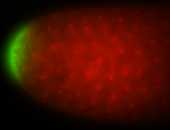
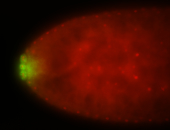
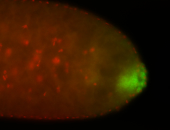
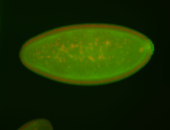
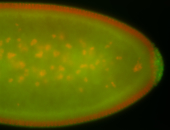
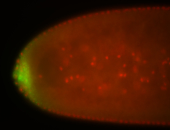
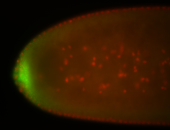
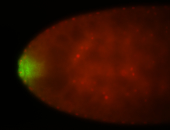
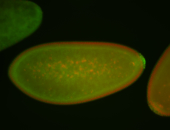
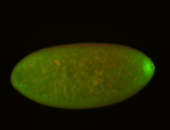
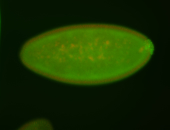
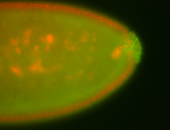
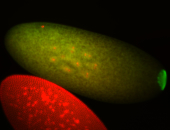
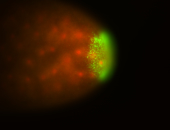
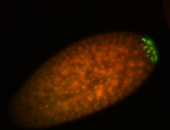
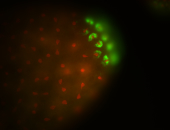
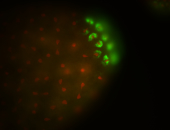
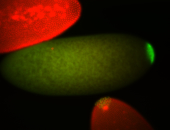
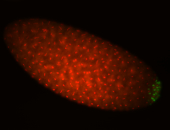
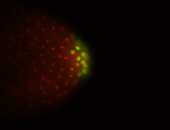
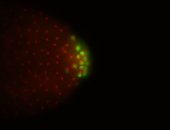
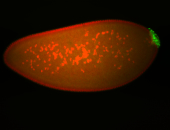
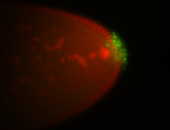
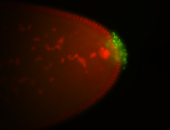
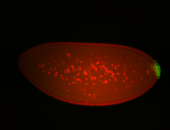
| Maternal RNA is posteriorly localized in yolk plasm and then in pole cells. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
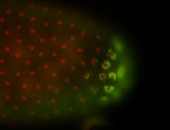
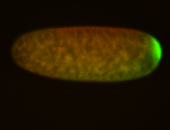
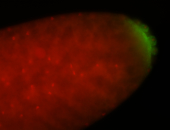
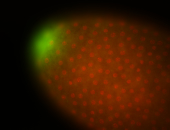
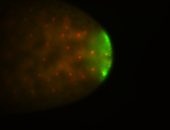
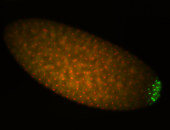
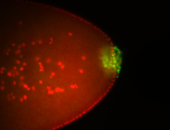
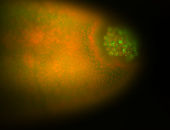
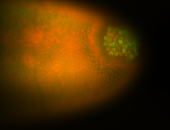
| Good probe on gel In unfertilized eggs, and 2 nuclei embryos: I see osk very strongly expressed posteriorly (as expected) AND anterior expression as well (see 2 nuclei anterior 40x photo). The RNA probe shows anterior 'structures' (interesting and probably novel). |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
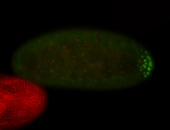
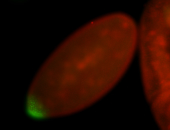
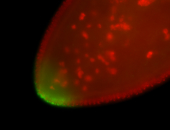
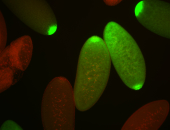
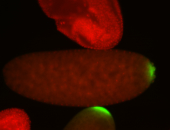
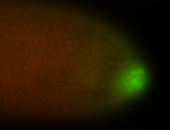
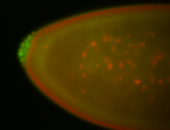
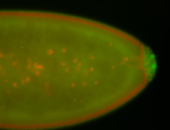
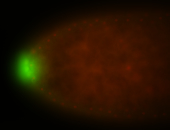
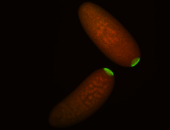
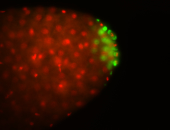
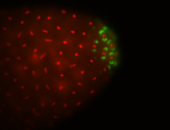
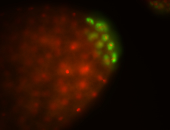
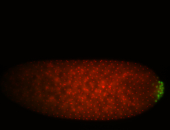
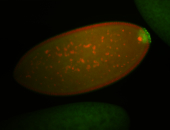
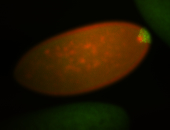
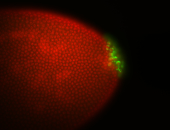
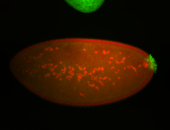
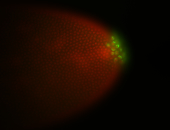
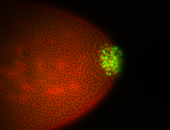
| Again, perinuclear pole cell asymmetric expression. |
| Pole cell asymmetric expression. I see small 'dots' around the pole cells, stronger in one 1/2 of the pole nuclei. |
| Good probe on gel |
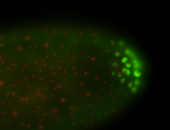
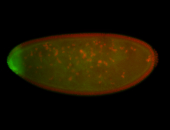
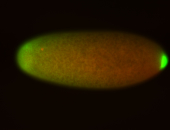
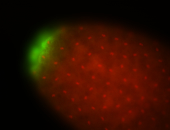
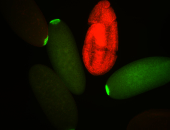
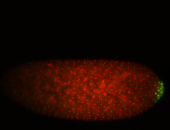
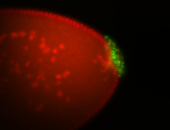
| Posterior localization, even in unfertilized eggs. RNA accumulates asymmetrically around the pole buds with a 'fiber-like' texture. When nuclei divide, the RNA will stay in the nuclei that are closer to the posterior end. Very interesting! |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA transcript isoform |
| -very diverse and neat patterns at all stages |