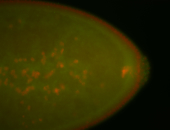
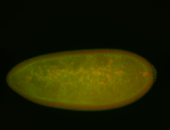
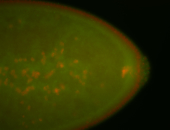
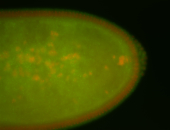
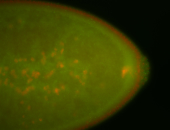
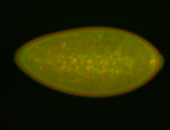
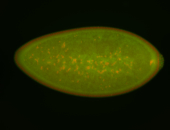
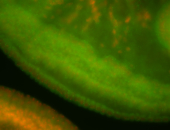
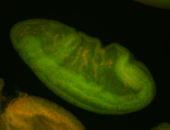
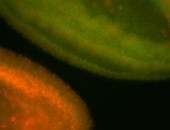
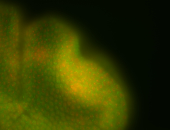
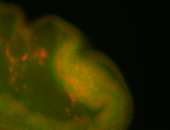
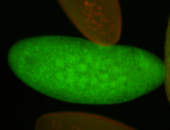
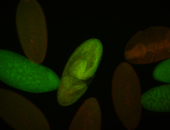
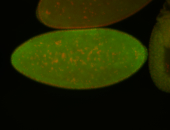
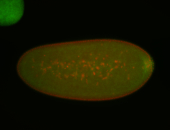
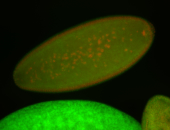
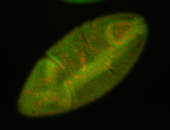
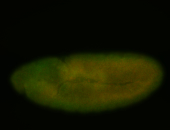
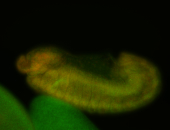
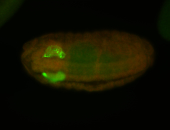
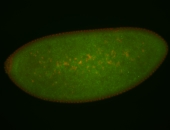
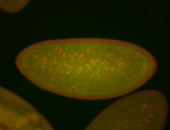
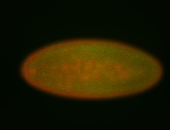
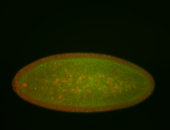
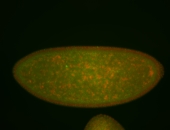
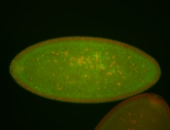
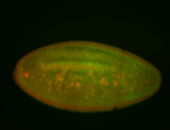
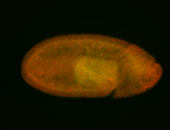
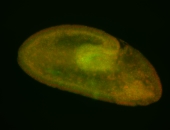
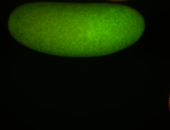
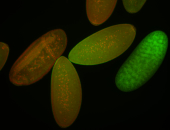
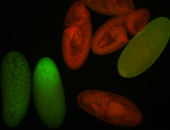
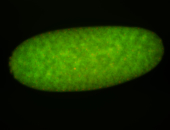
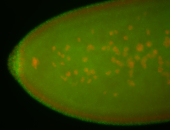
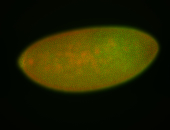
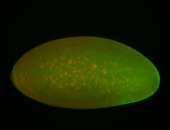
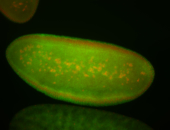
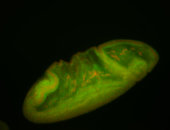
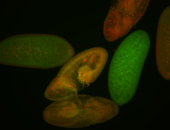
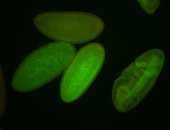
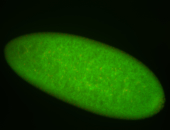
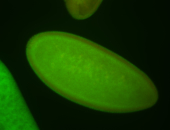
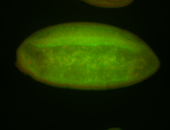
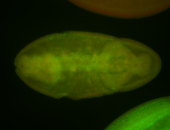
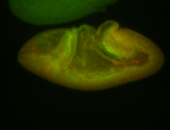
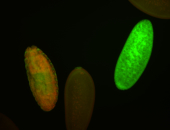
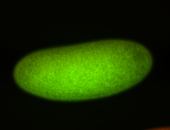
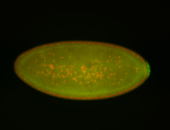
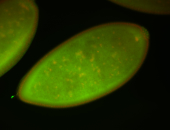
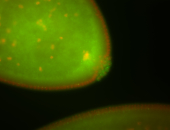
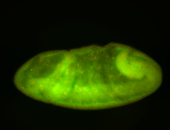
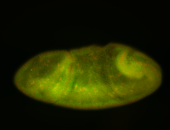
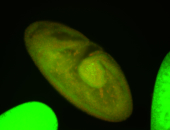
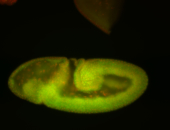
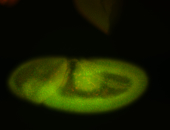
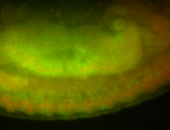
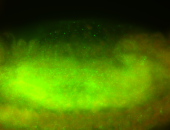
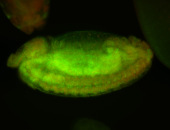
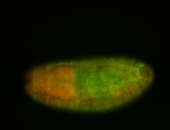
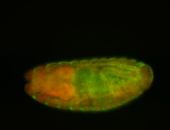
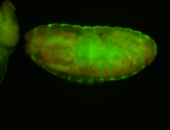
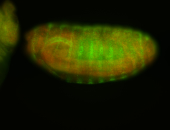
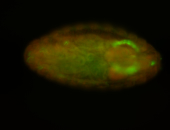
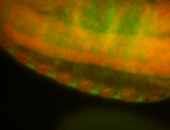
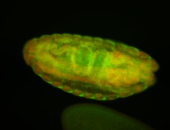
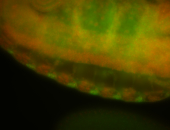
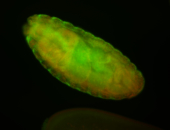
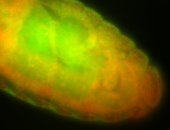
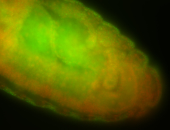
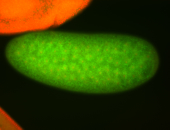
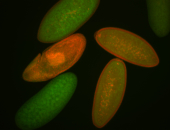
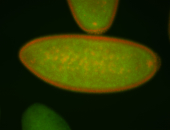
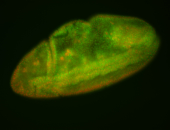
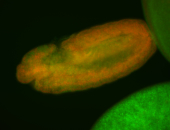
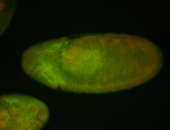
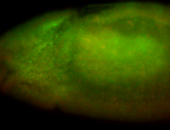
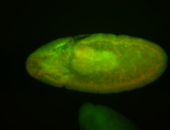
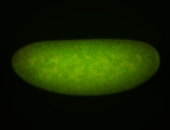
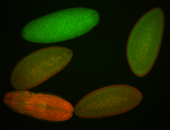
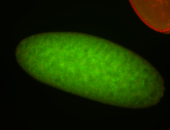
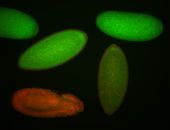
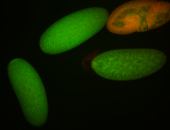
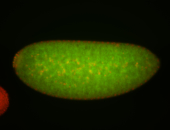
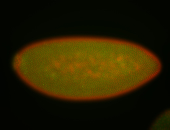
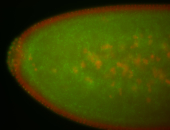
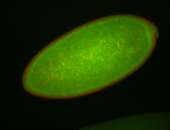
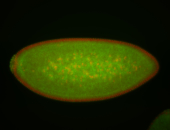
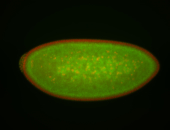
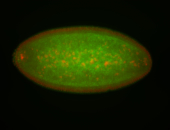
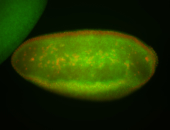
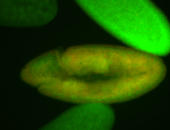
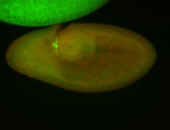
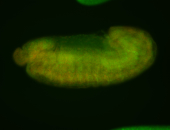
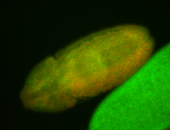
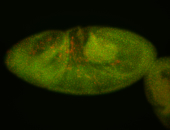
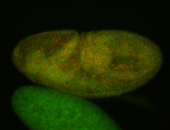
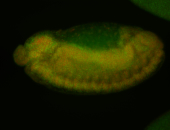
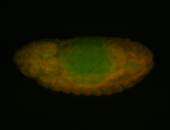
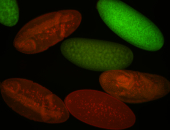
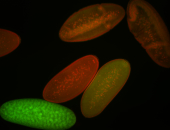
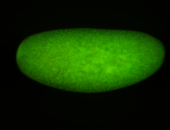
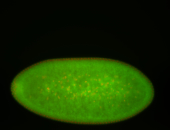
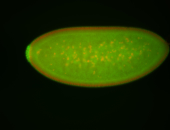
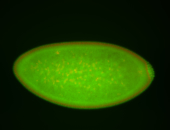
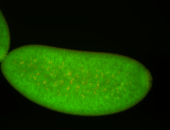
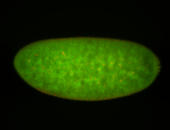
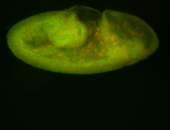
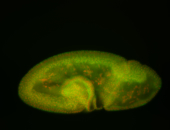
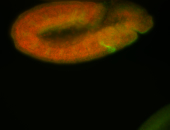
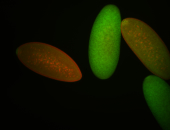
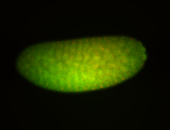
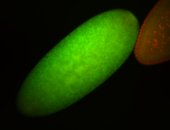
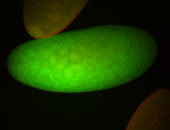
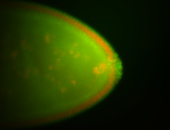
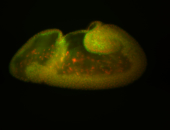
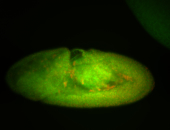
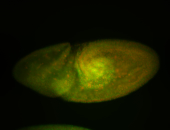
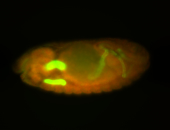
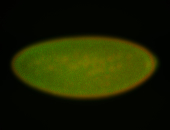
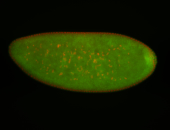
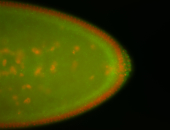
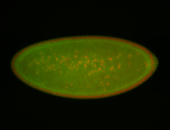
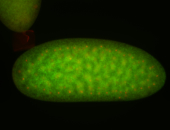
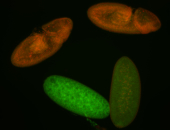
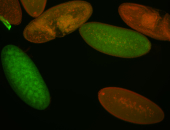
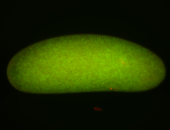
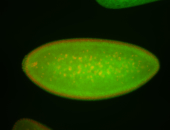
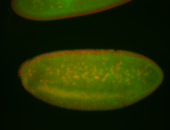
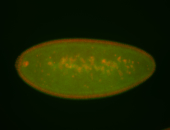
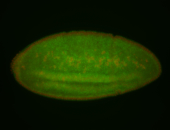
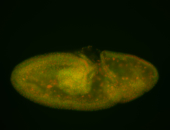
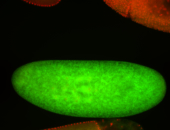
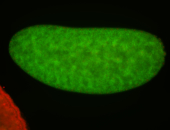
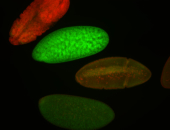
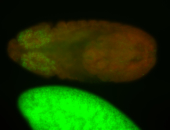
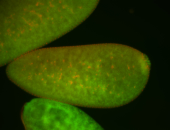
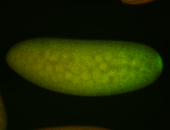
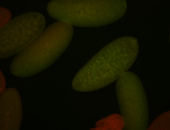
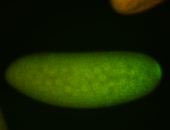
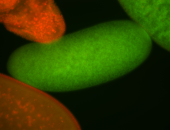
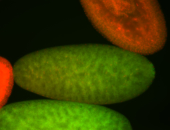
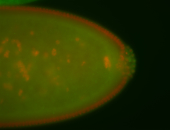
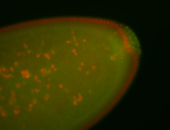
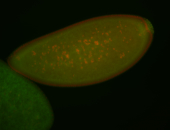
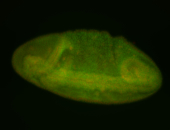
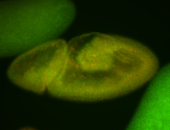
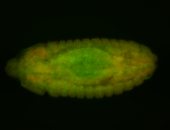
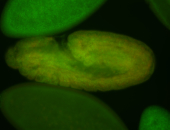
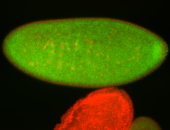
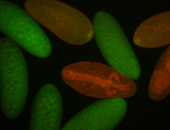
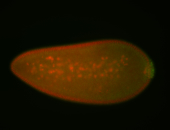
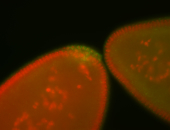
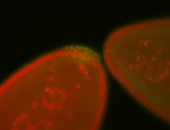
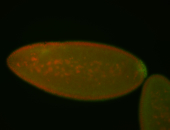
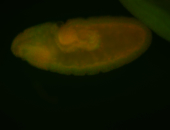
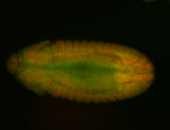
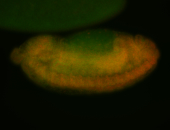
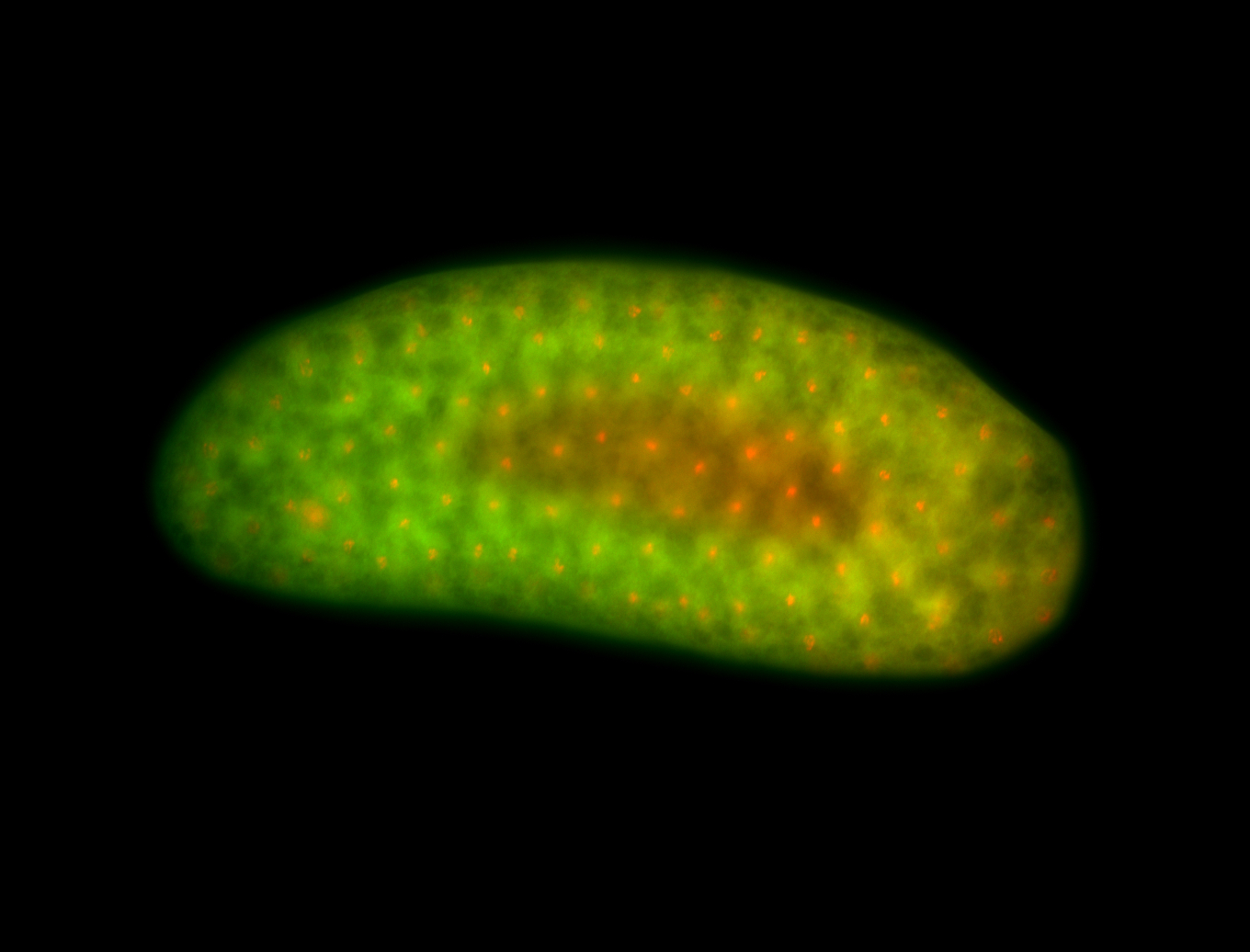
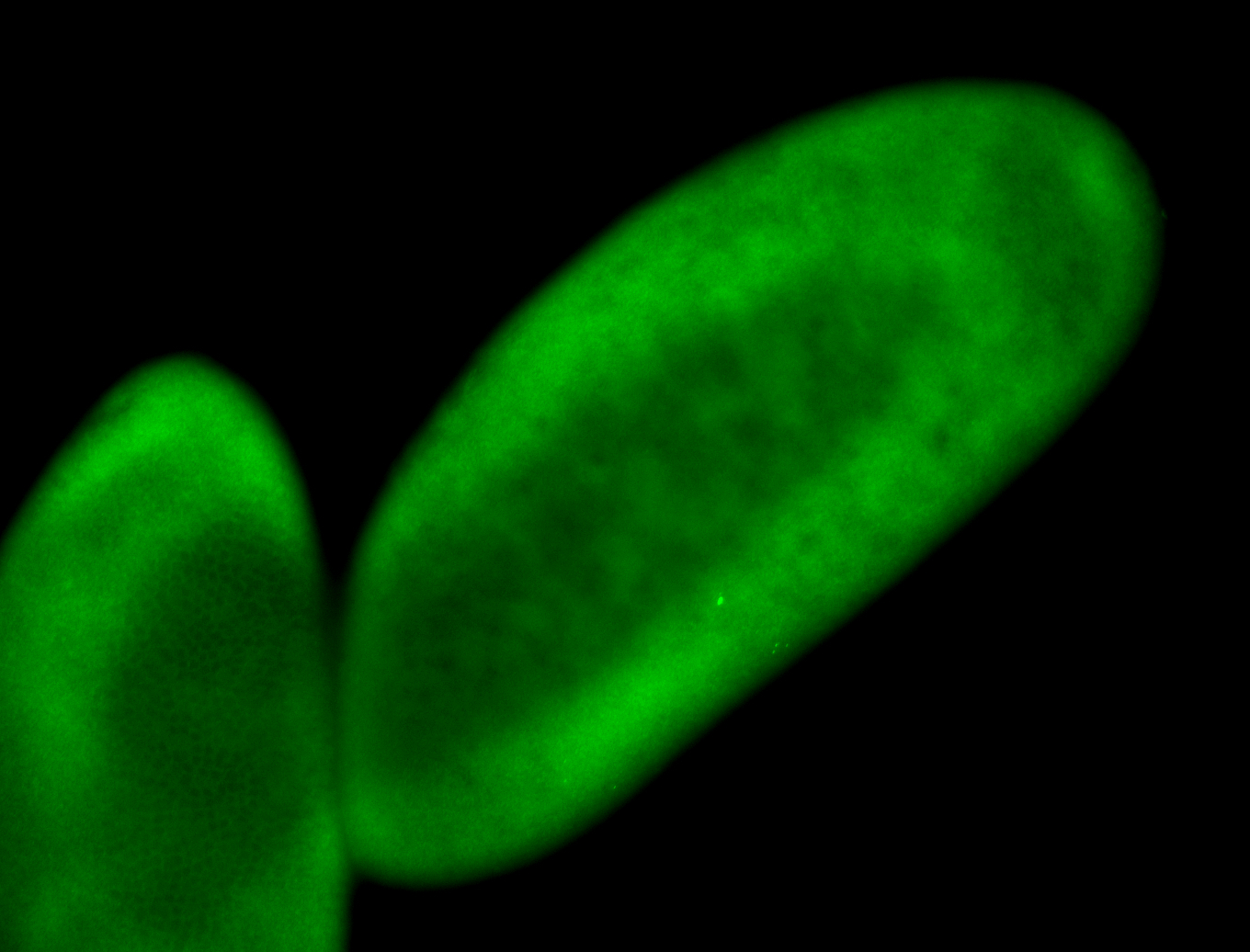
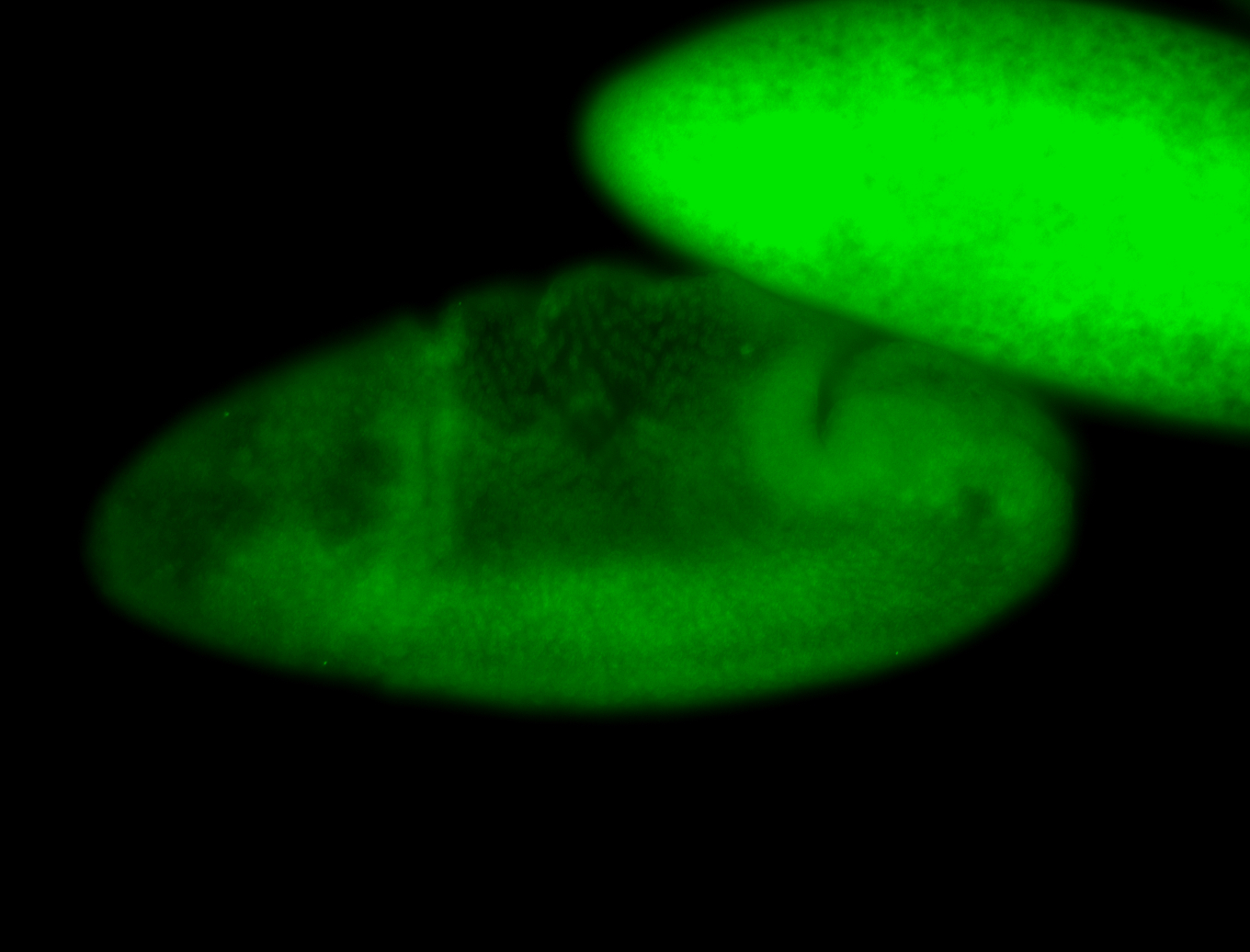
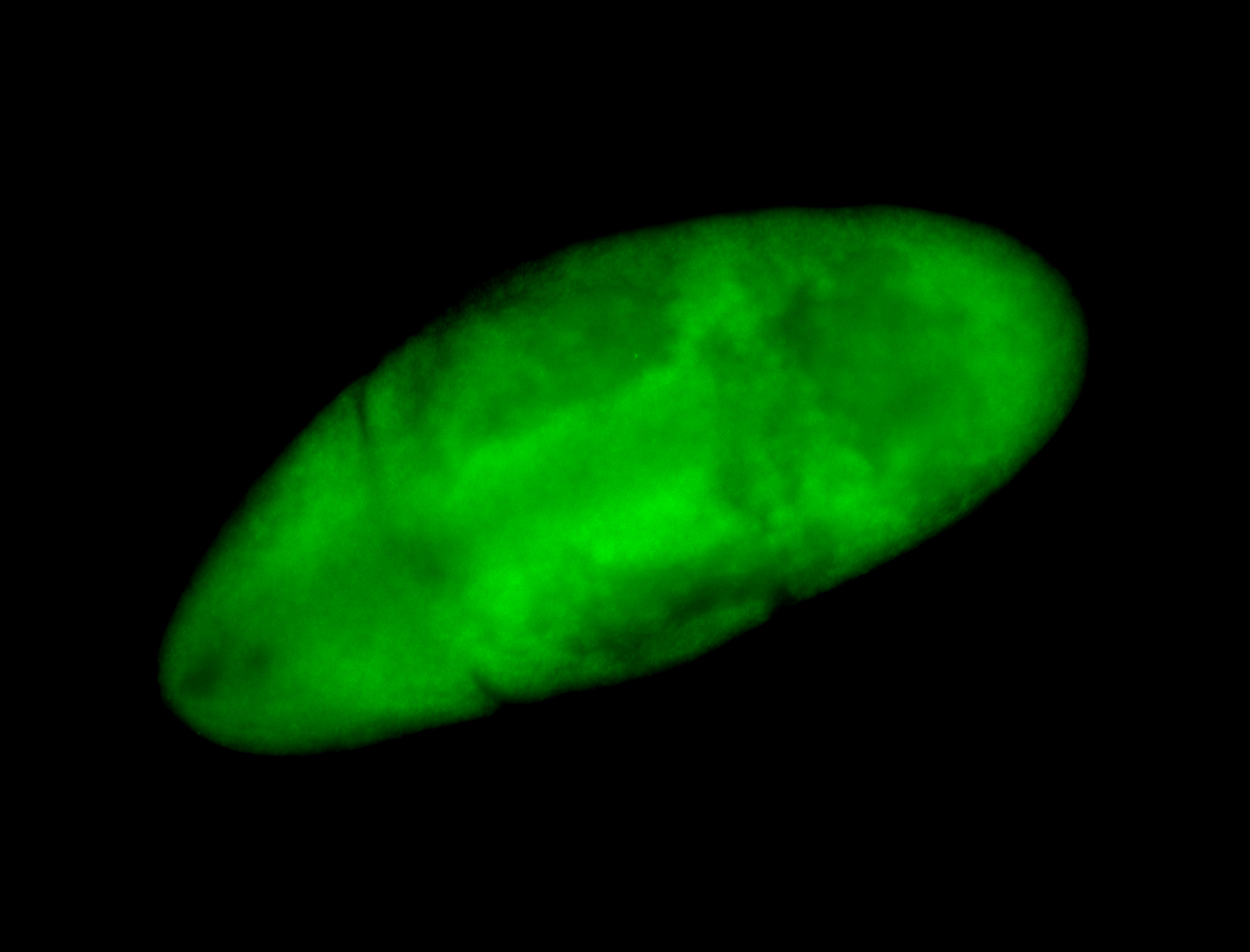
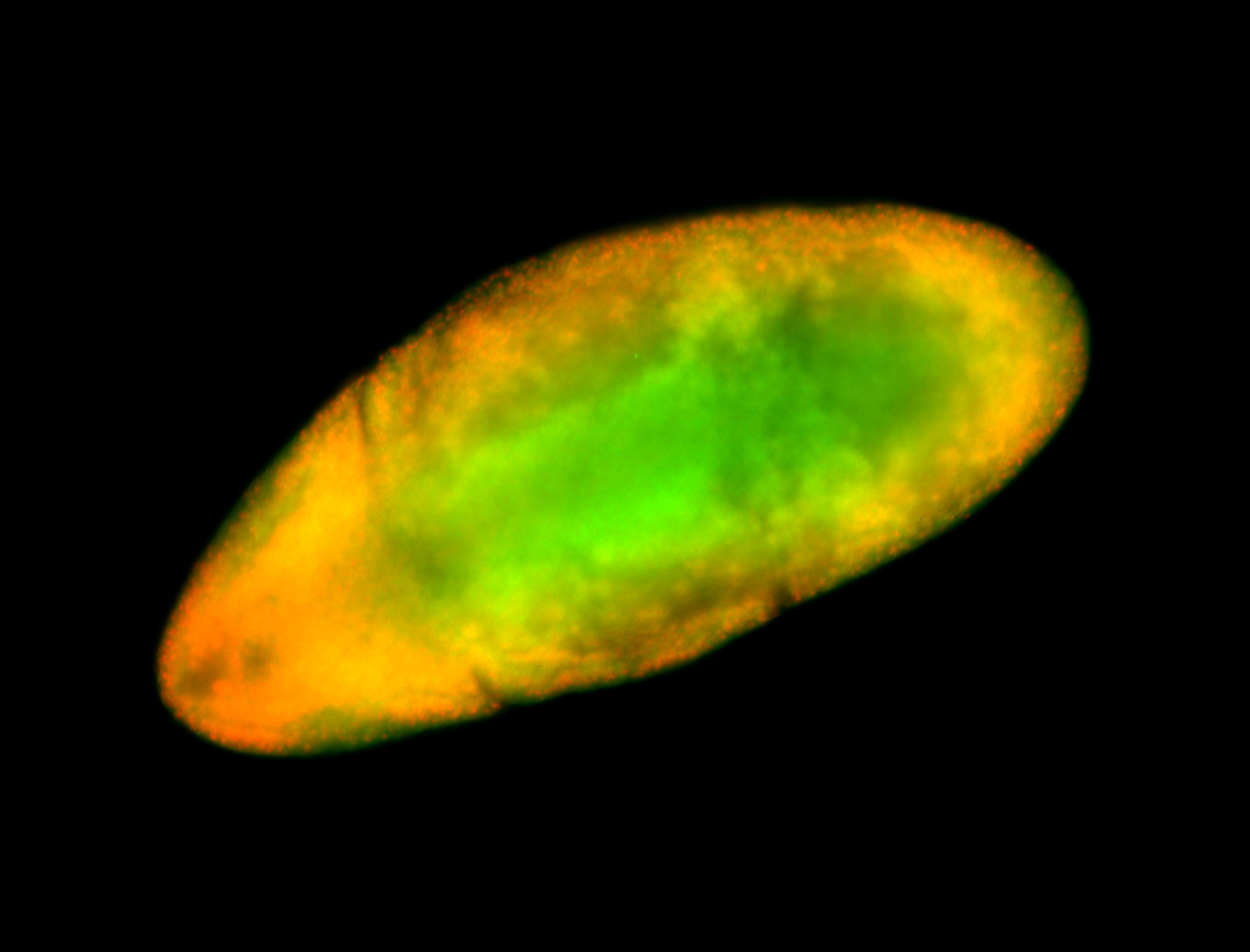
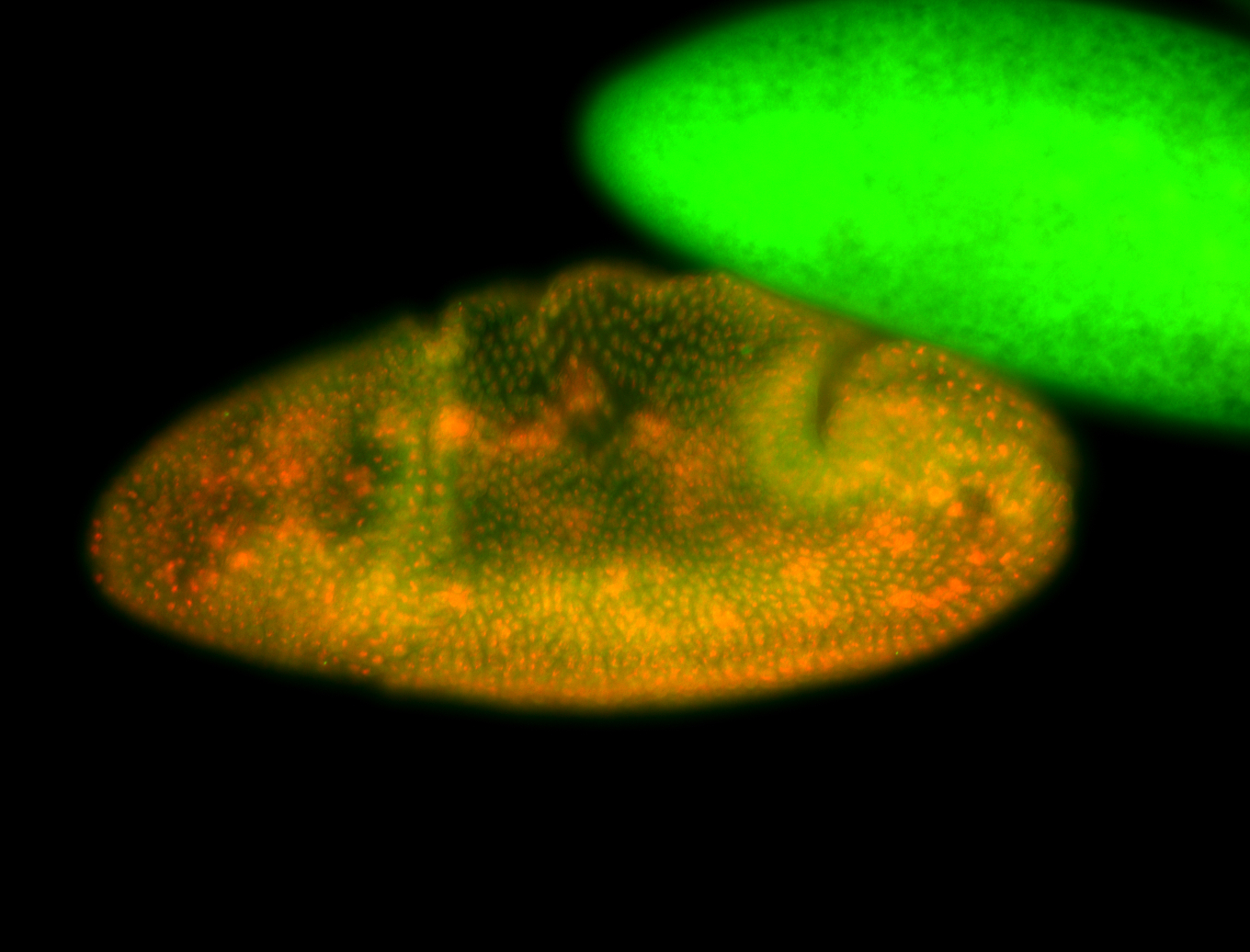
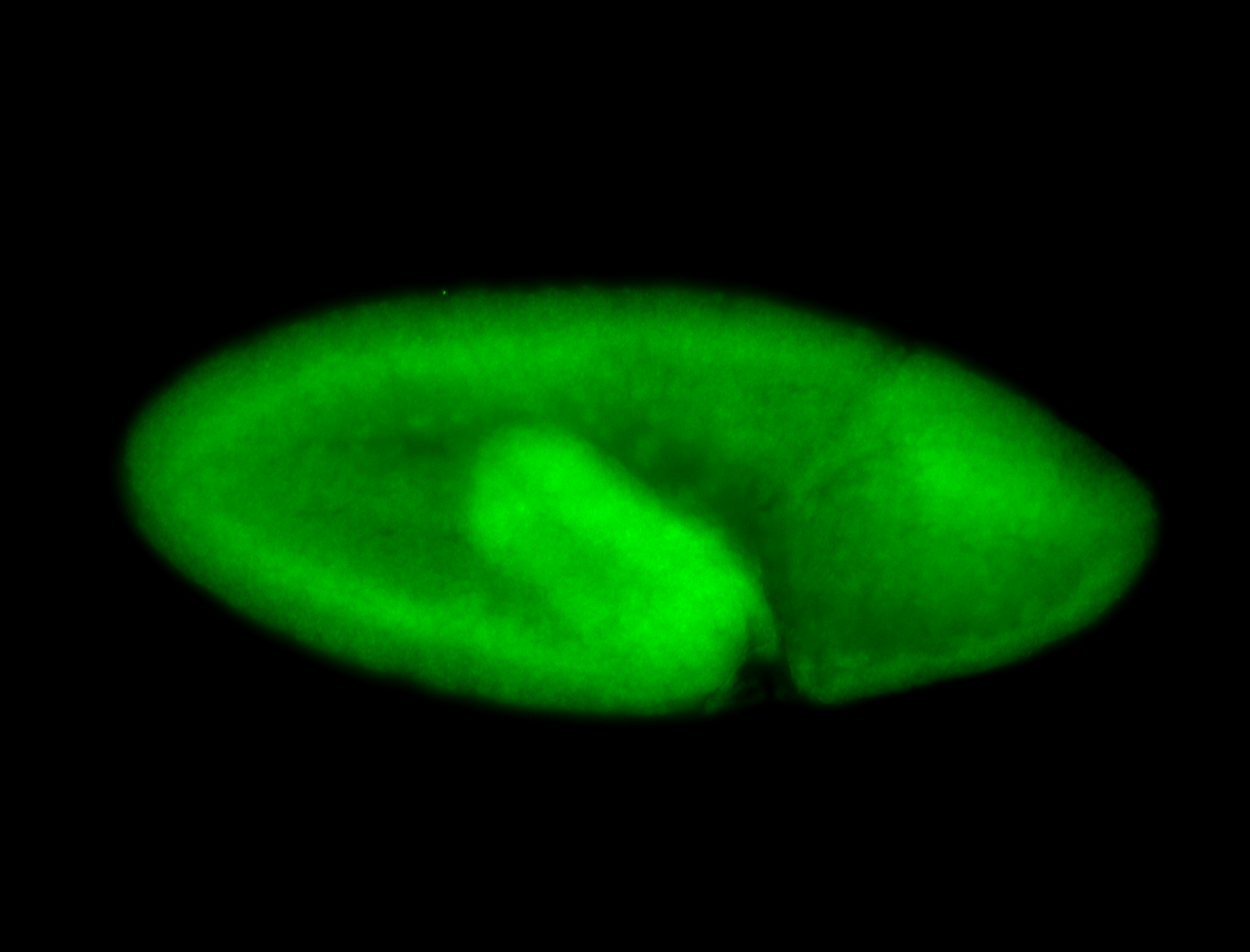
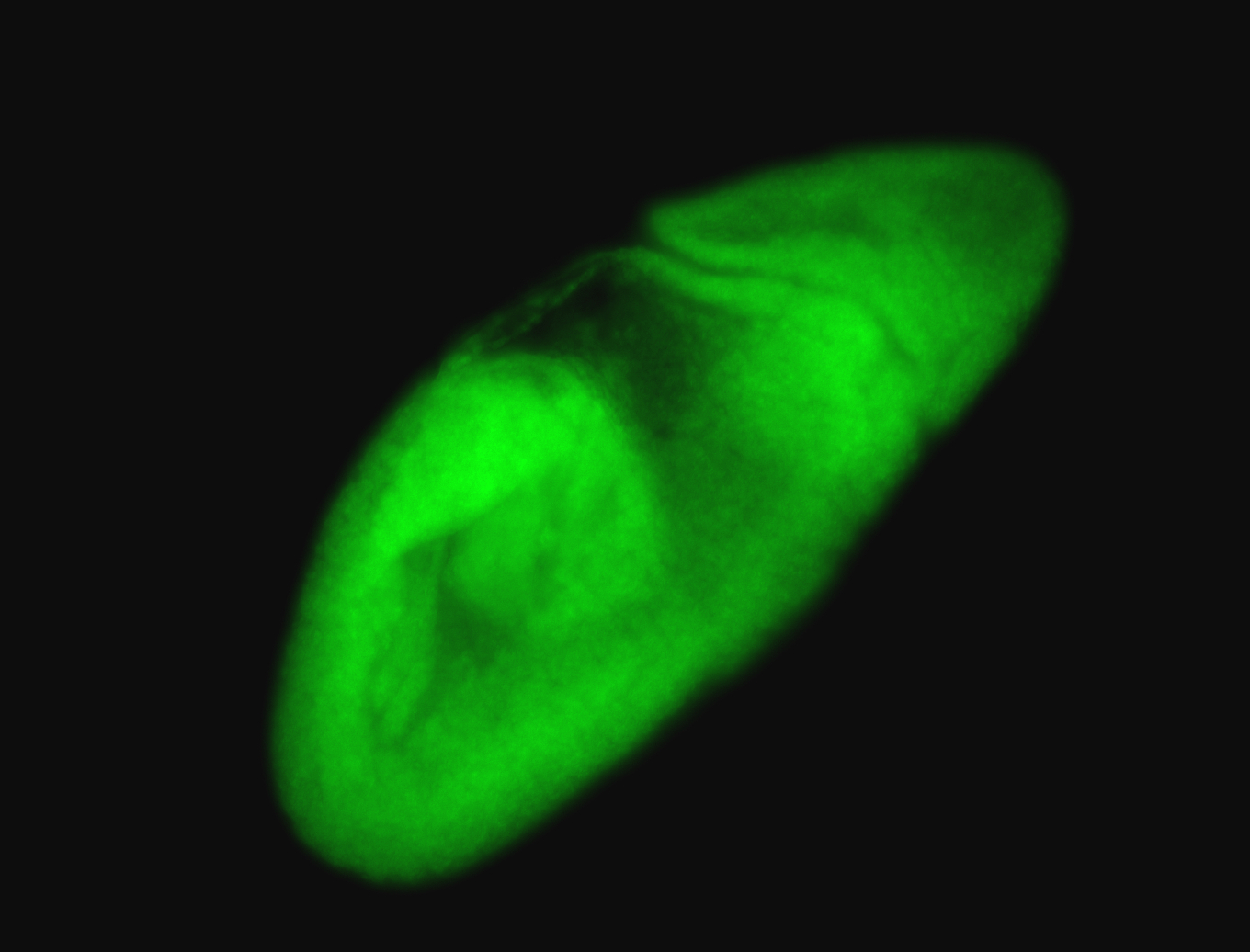
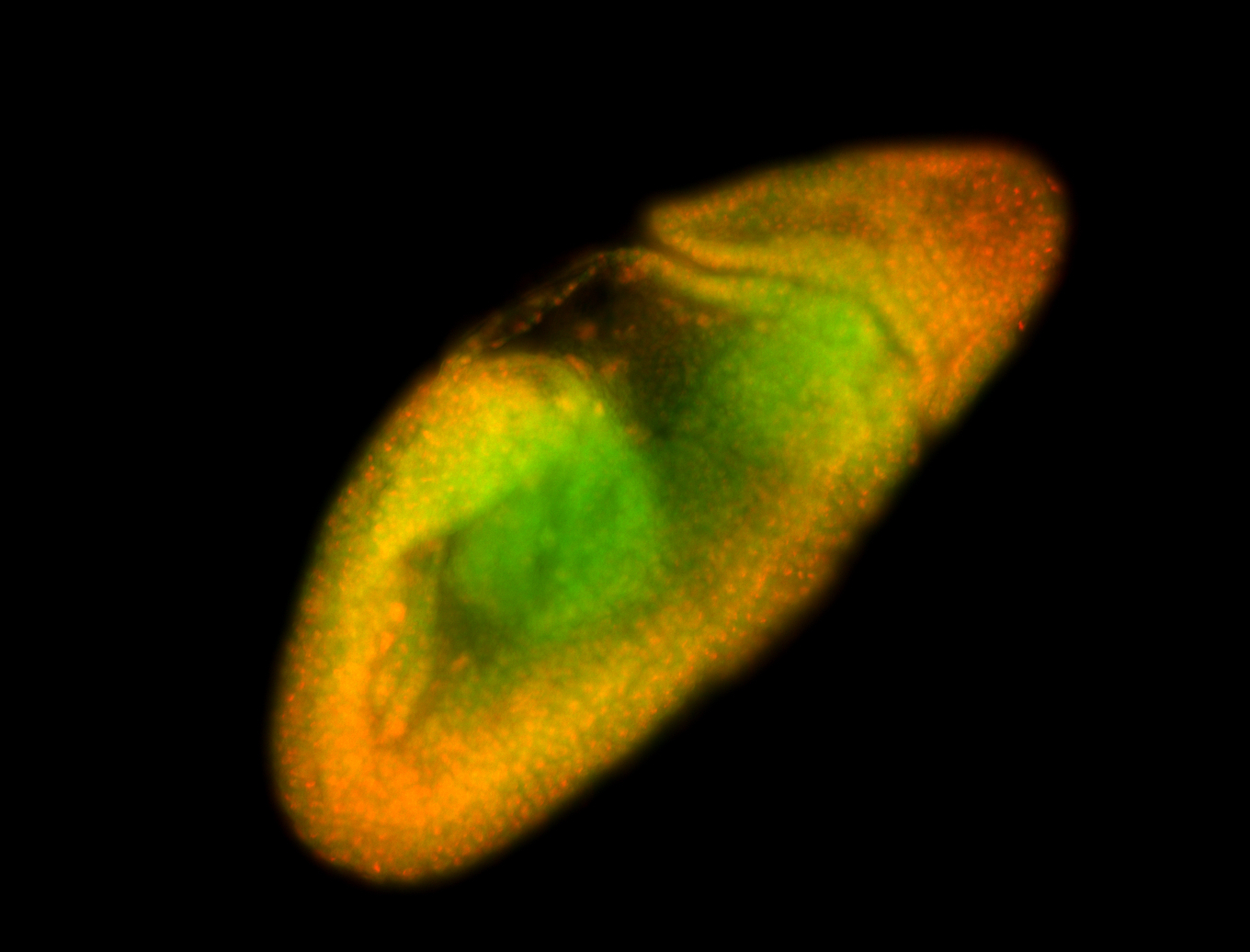
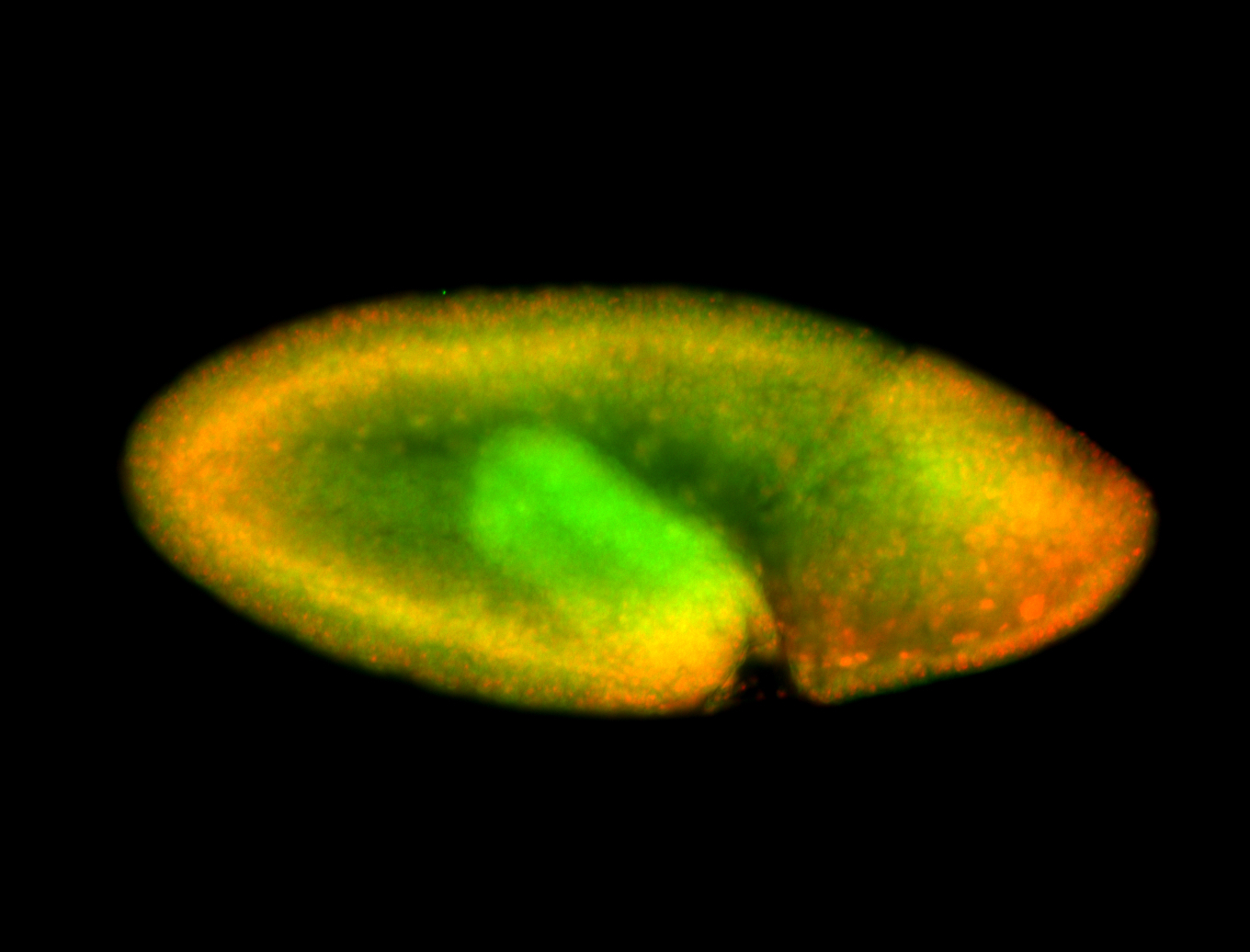
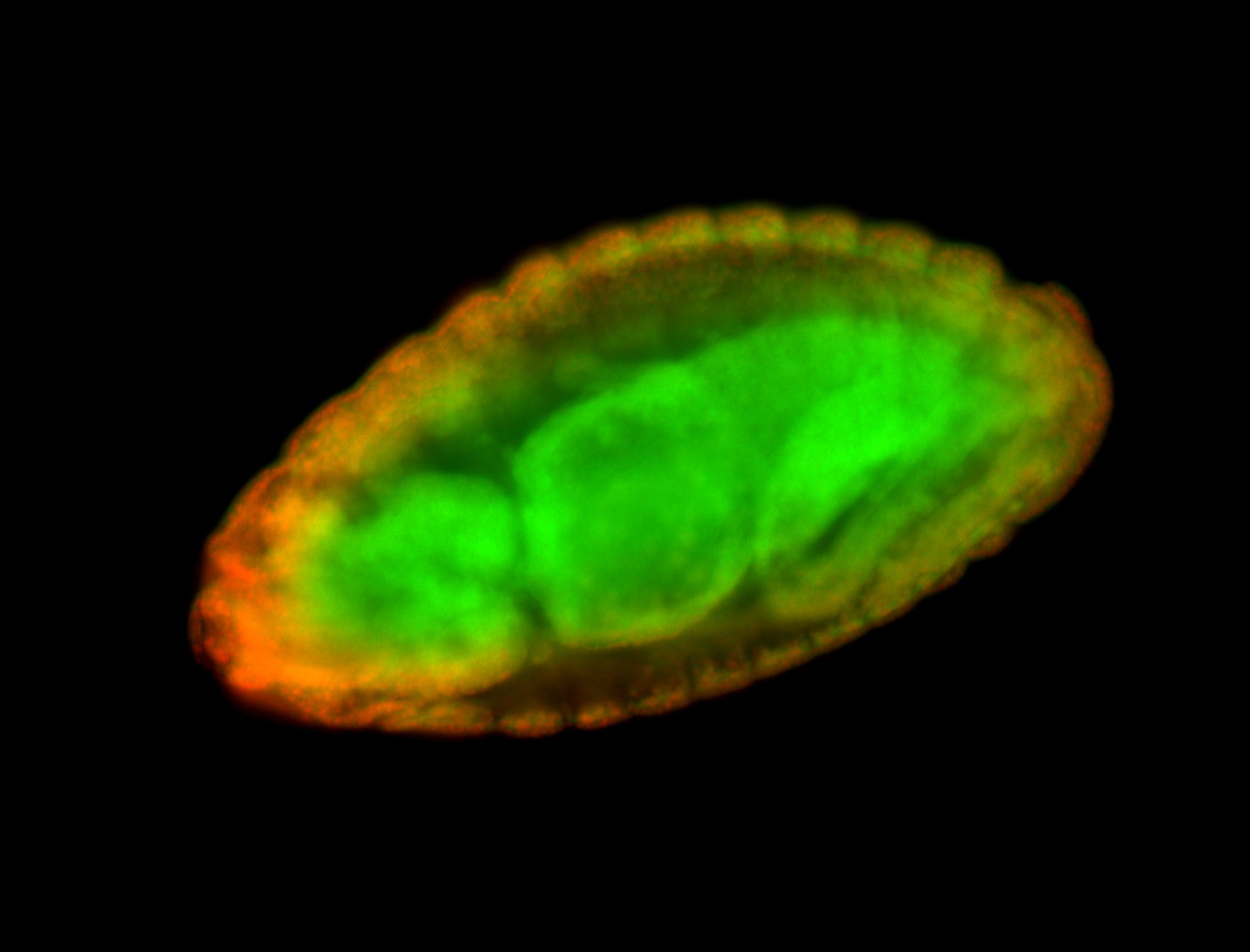
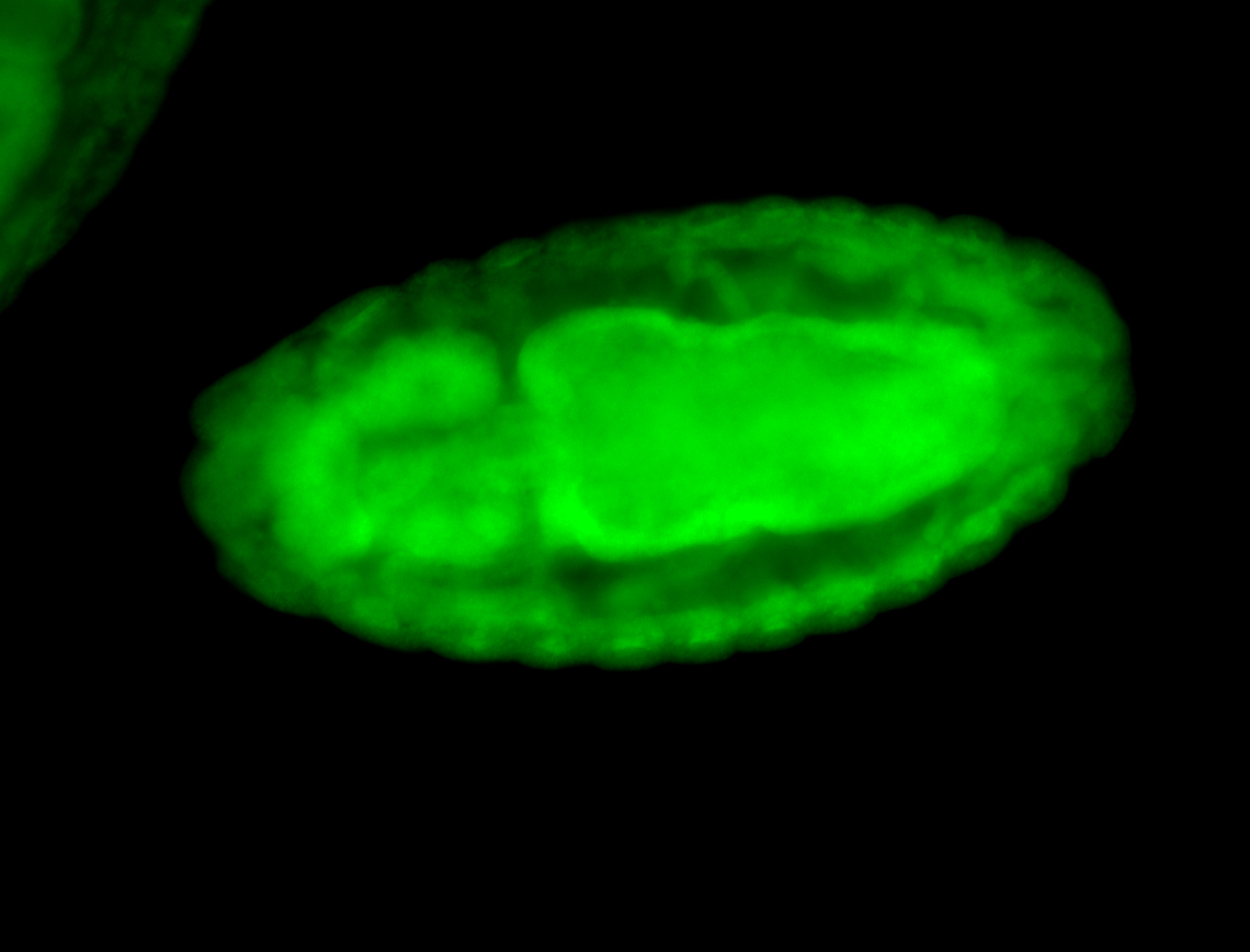
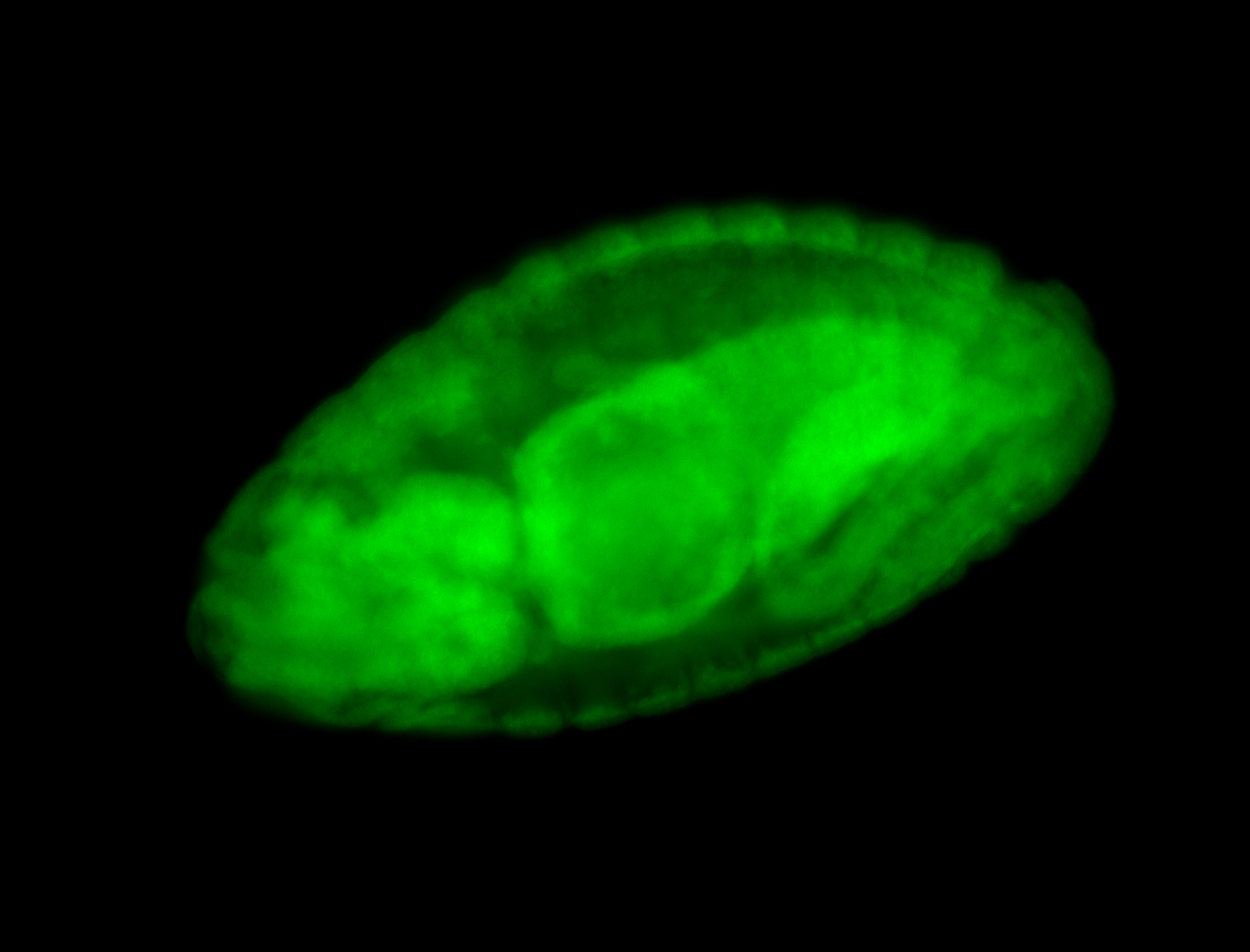
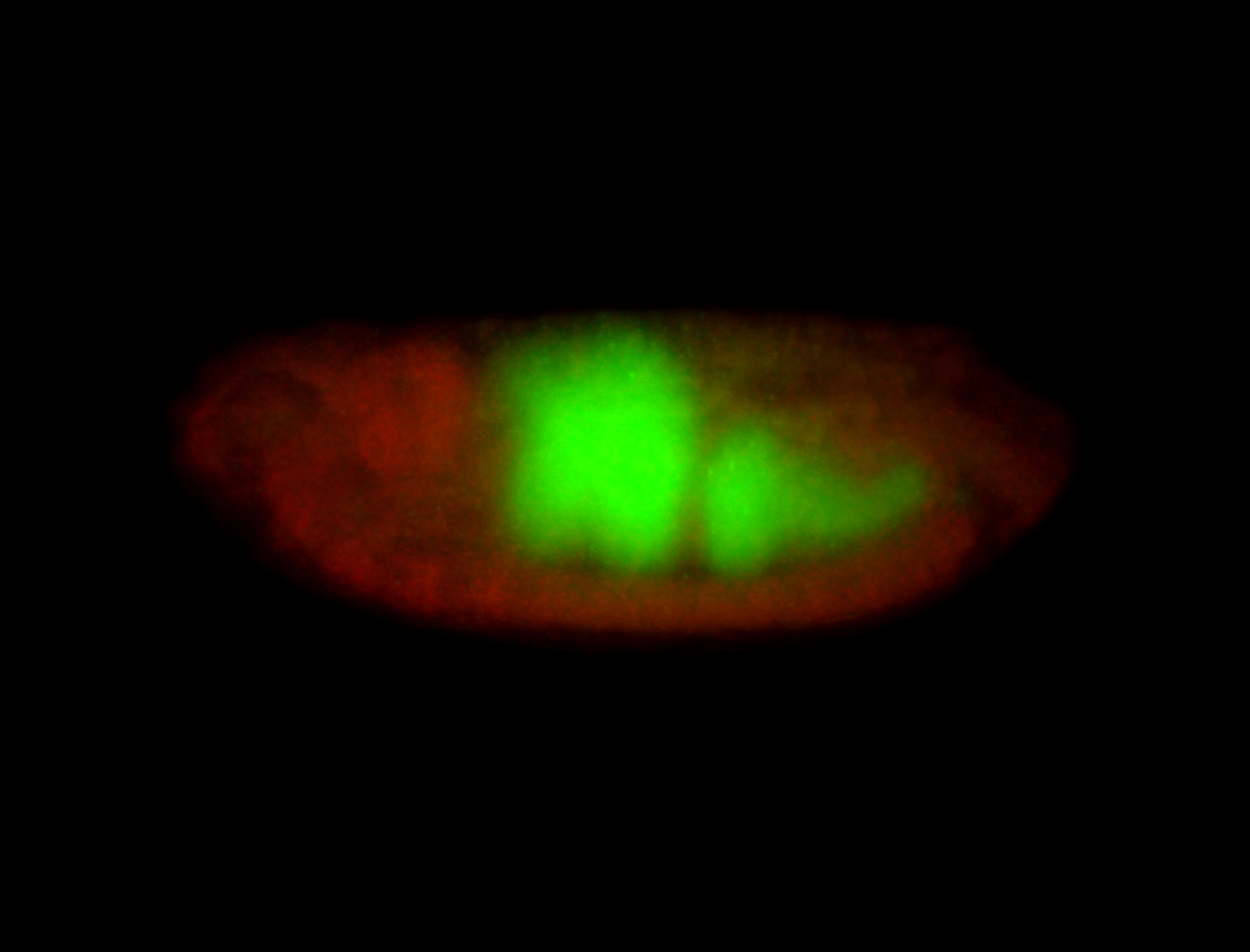
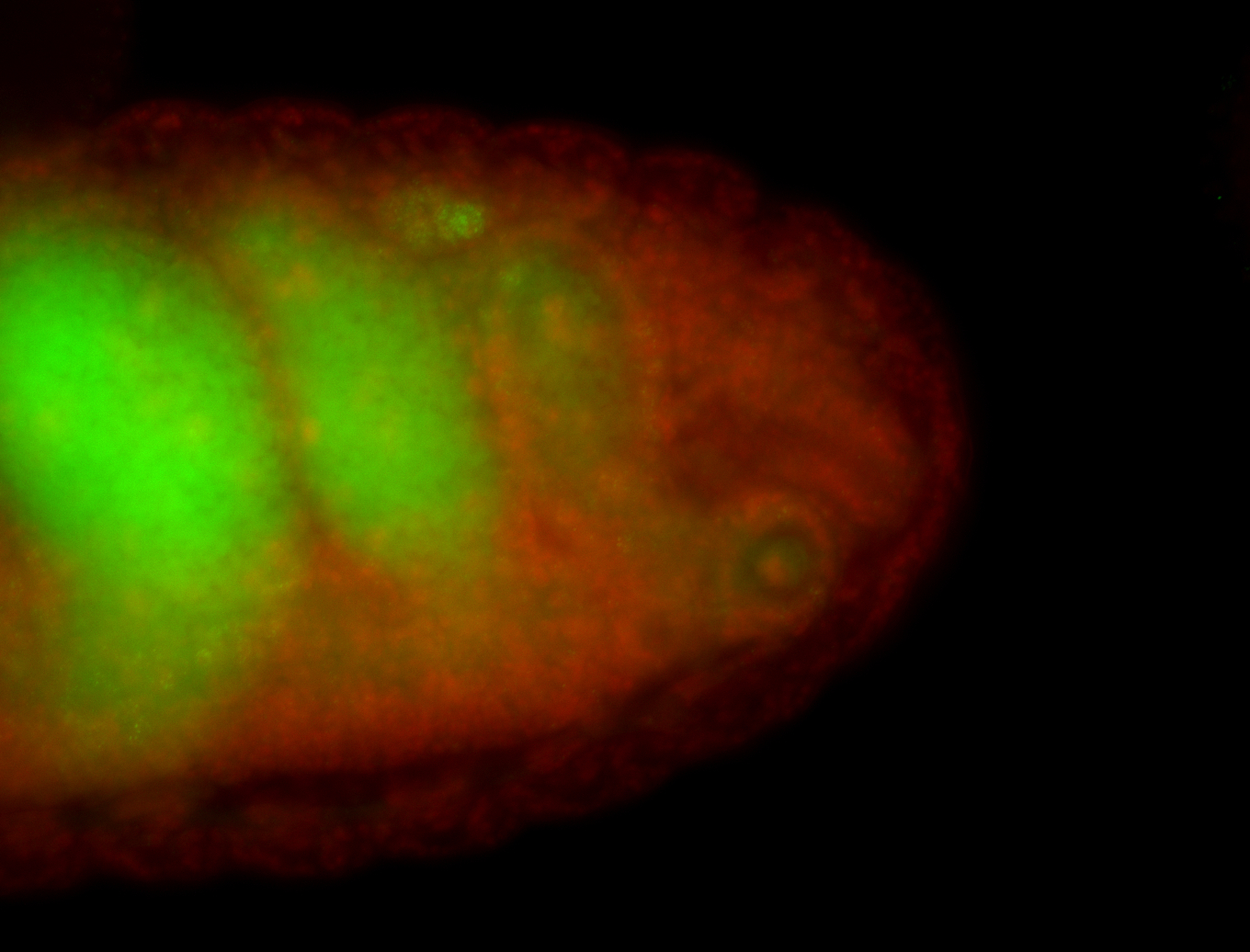
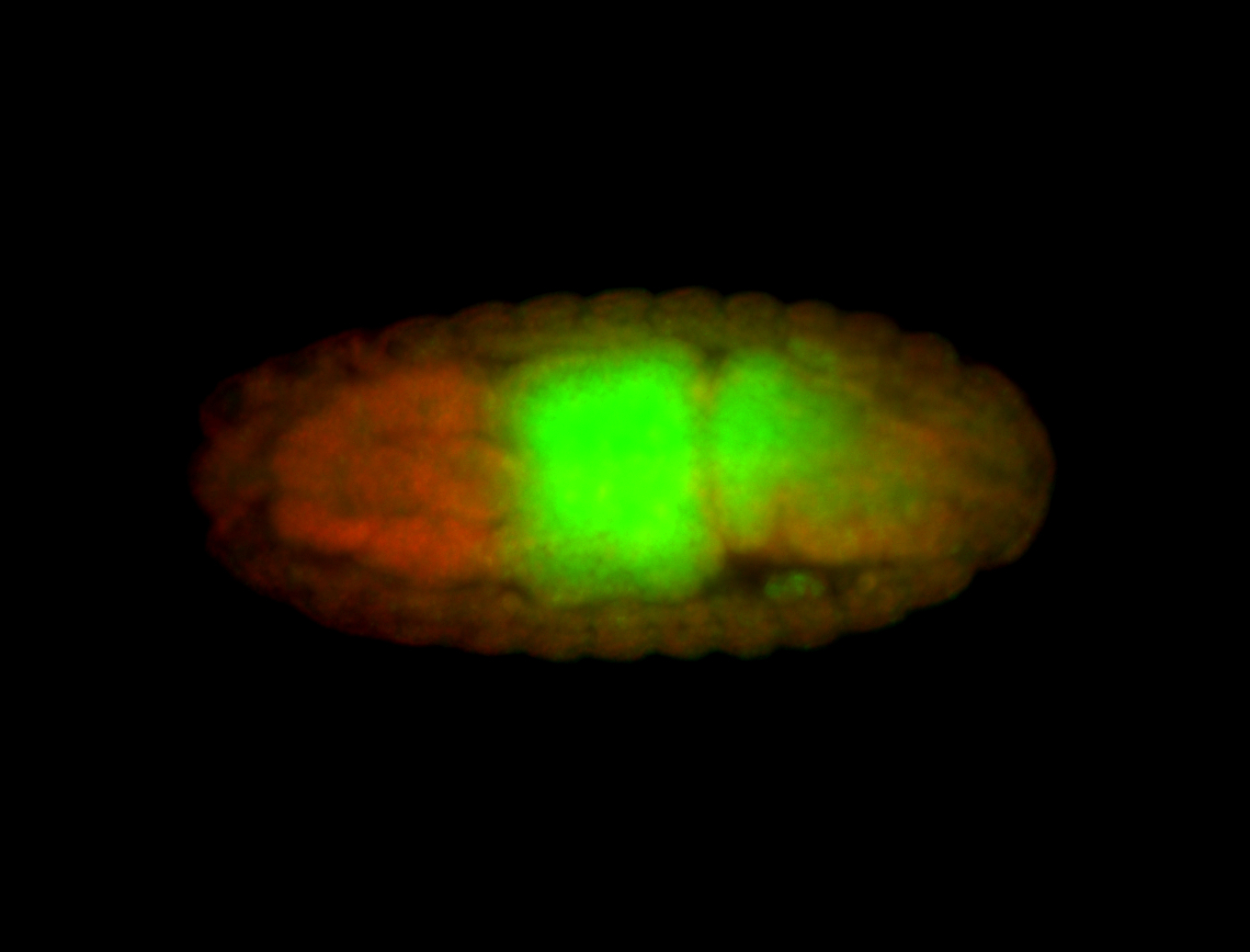
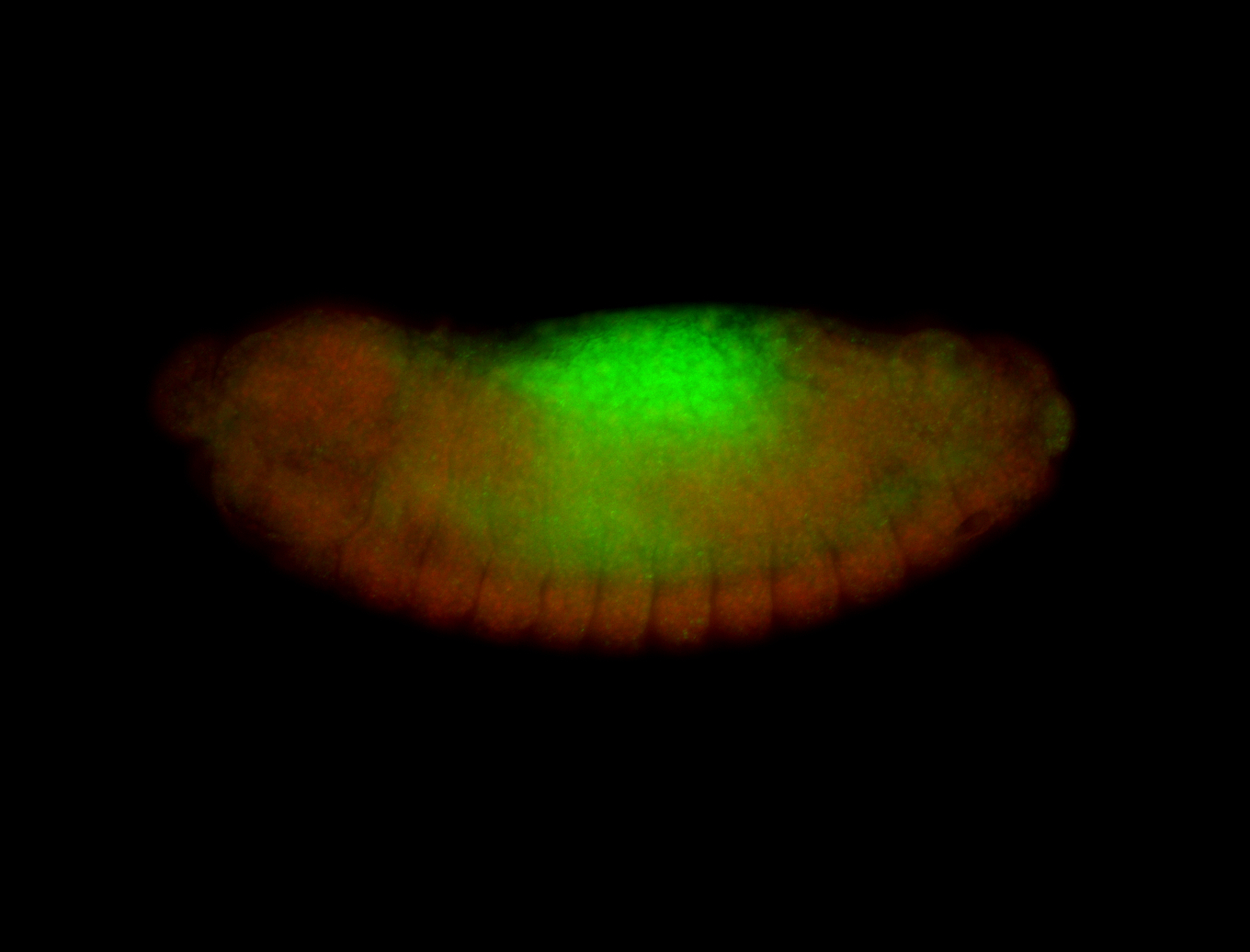
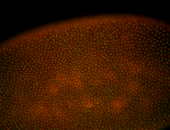
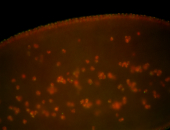
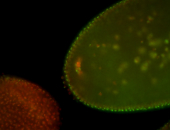
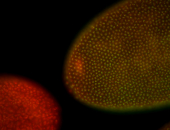
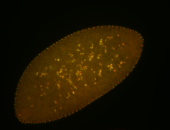
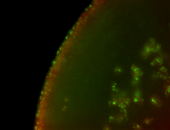
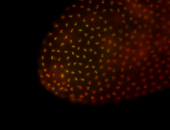
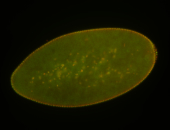
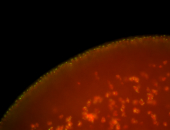
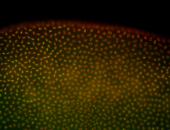
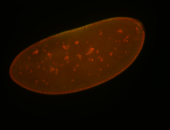
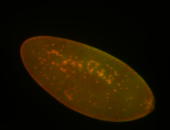
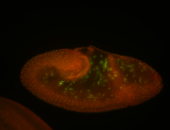
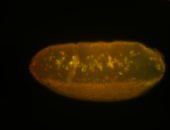
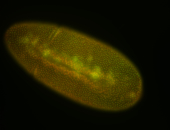
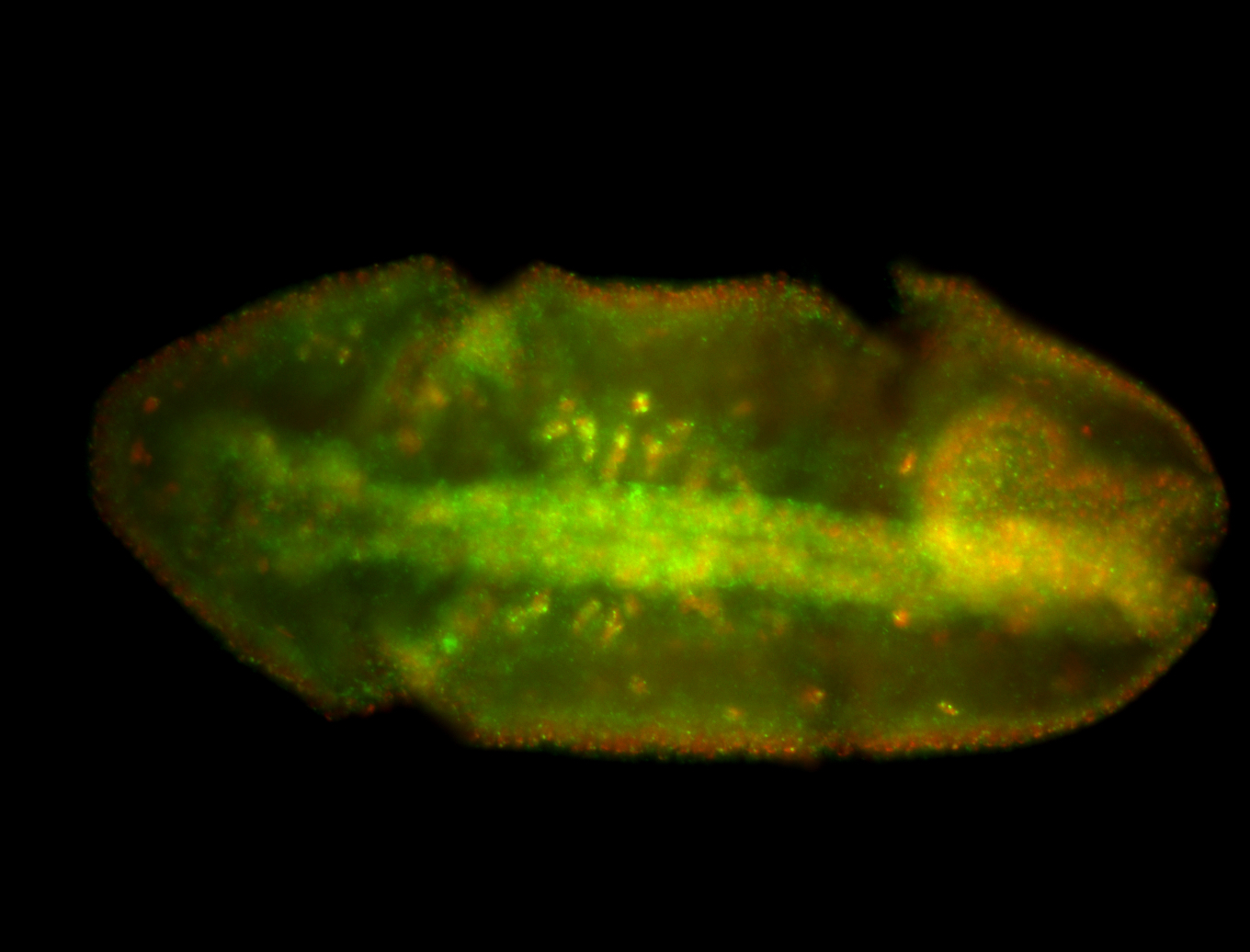
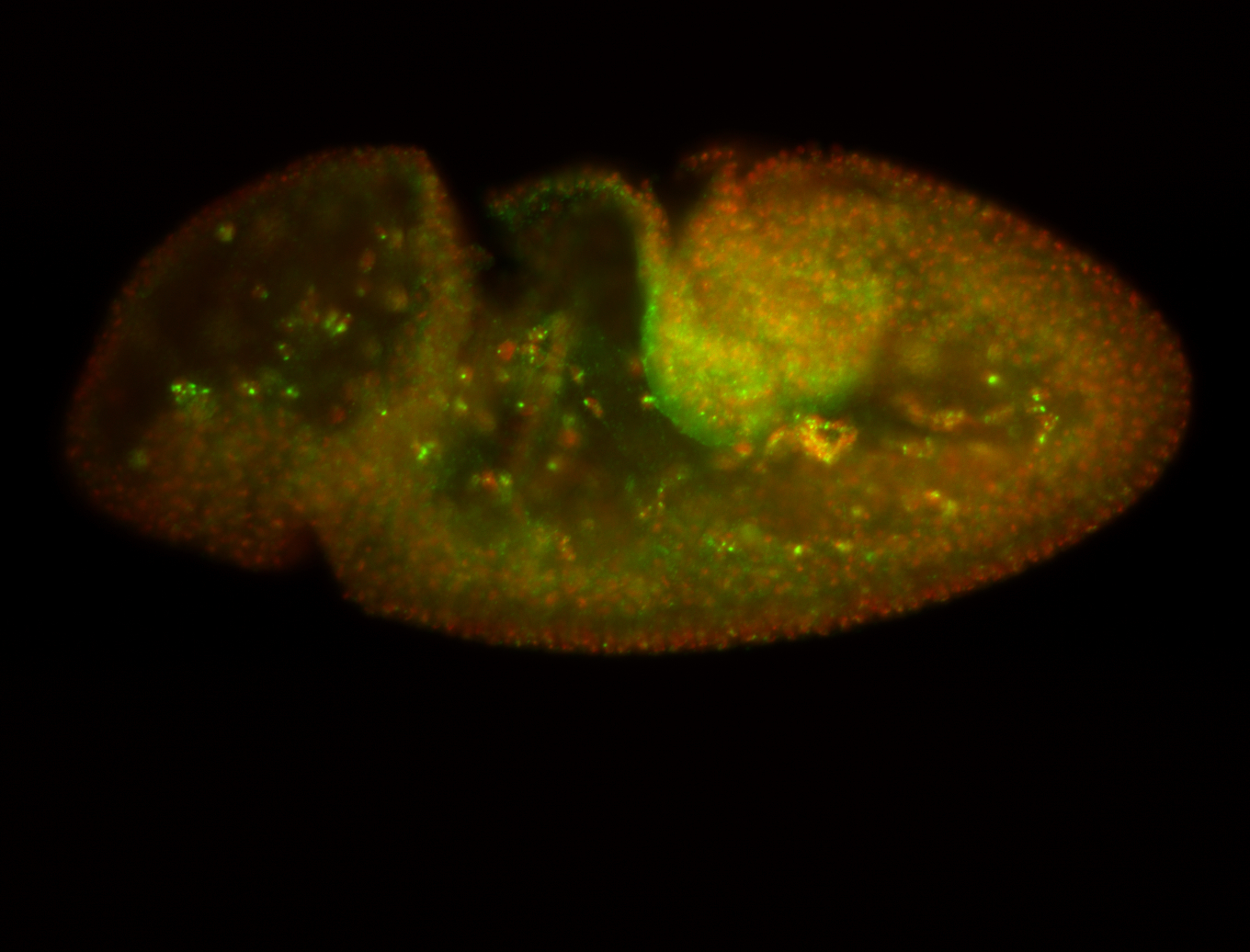
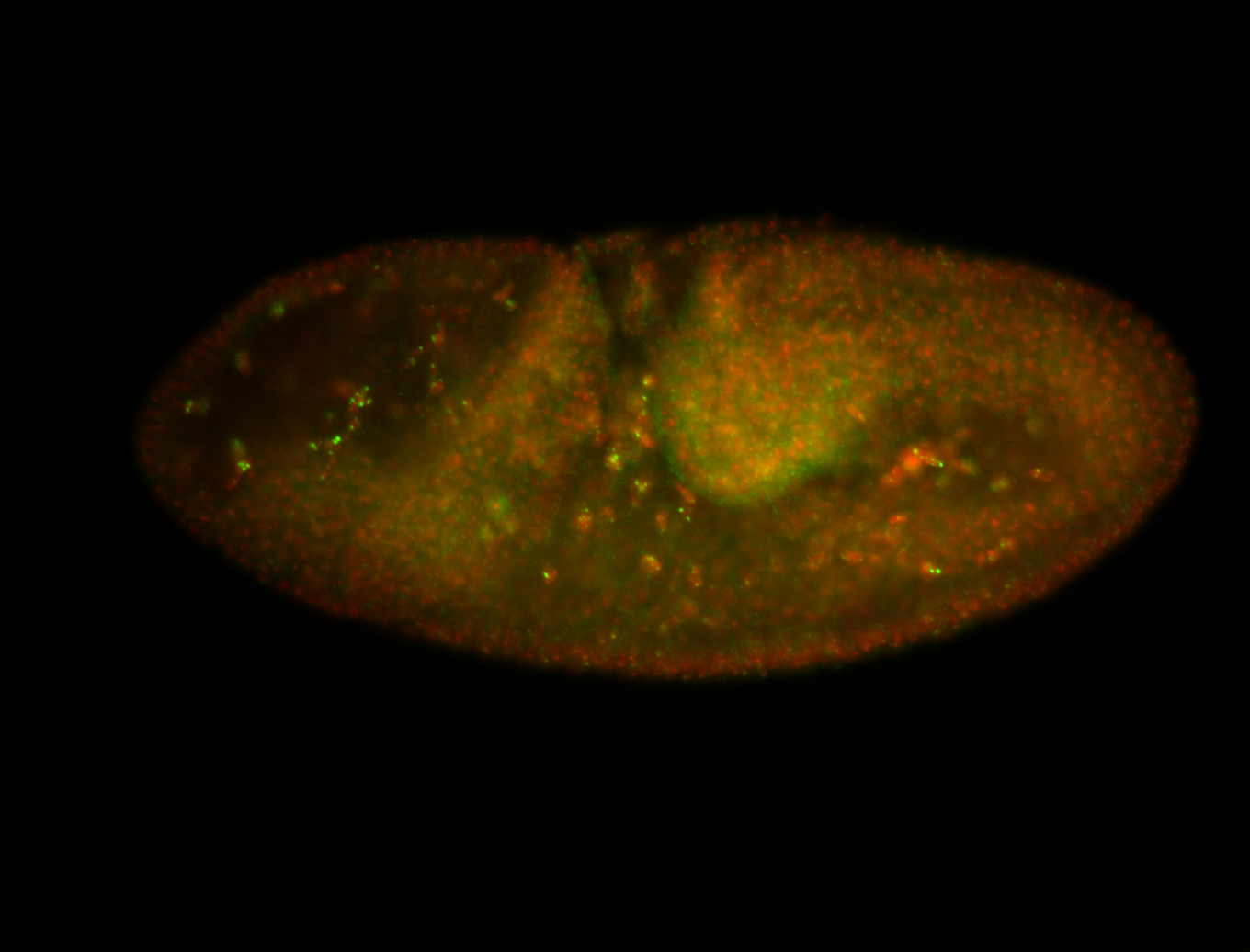
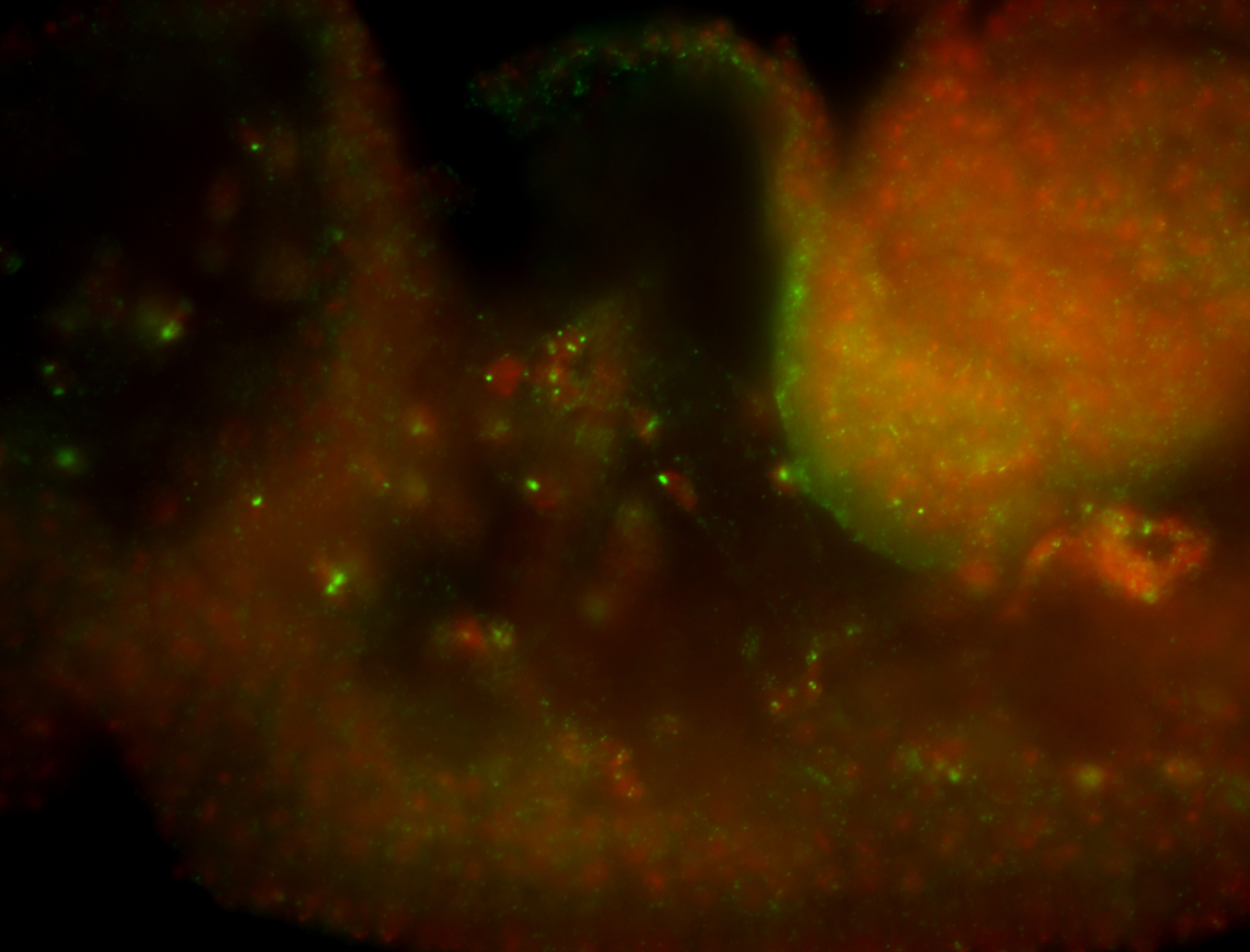
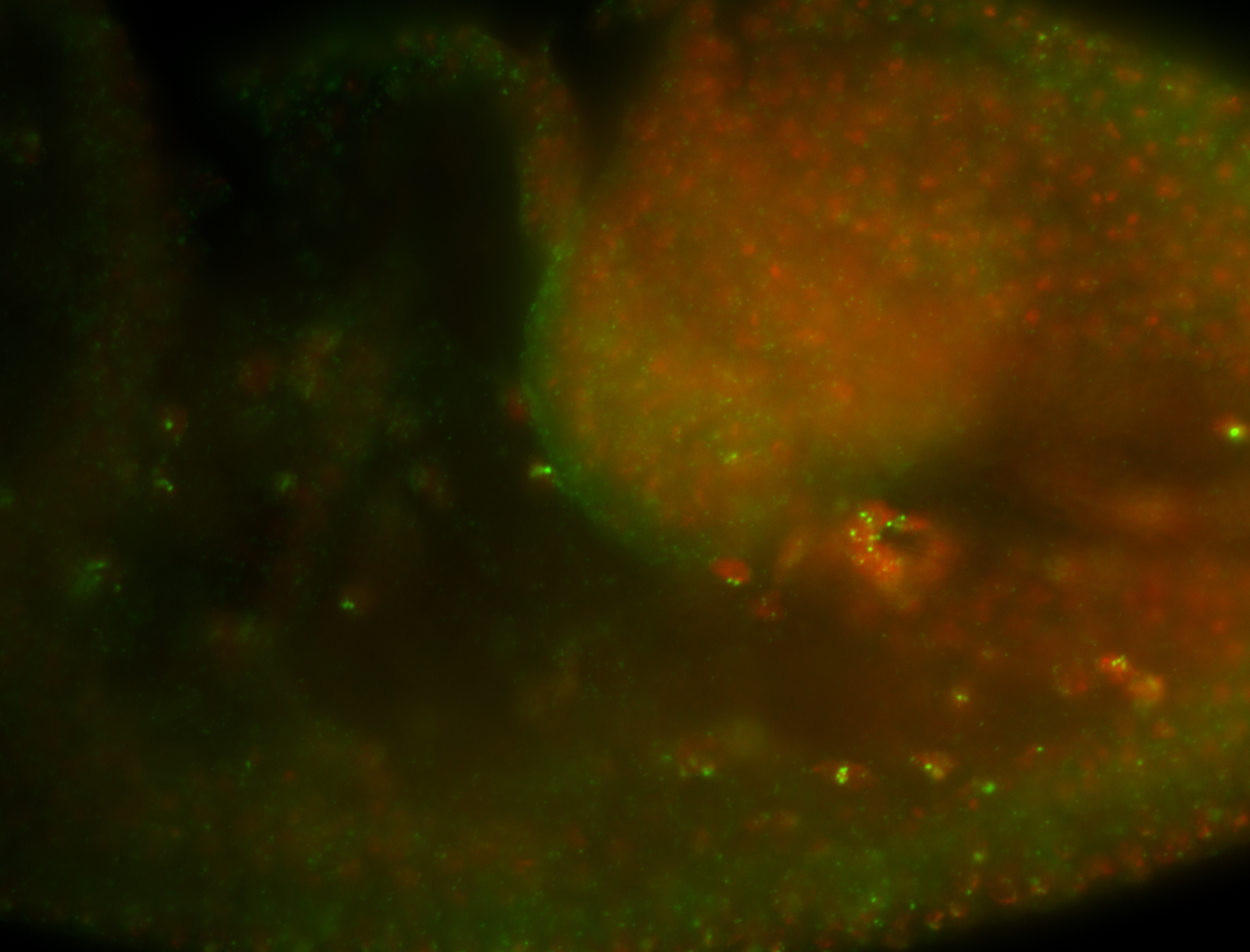
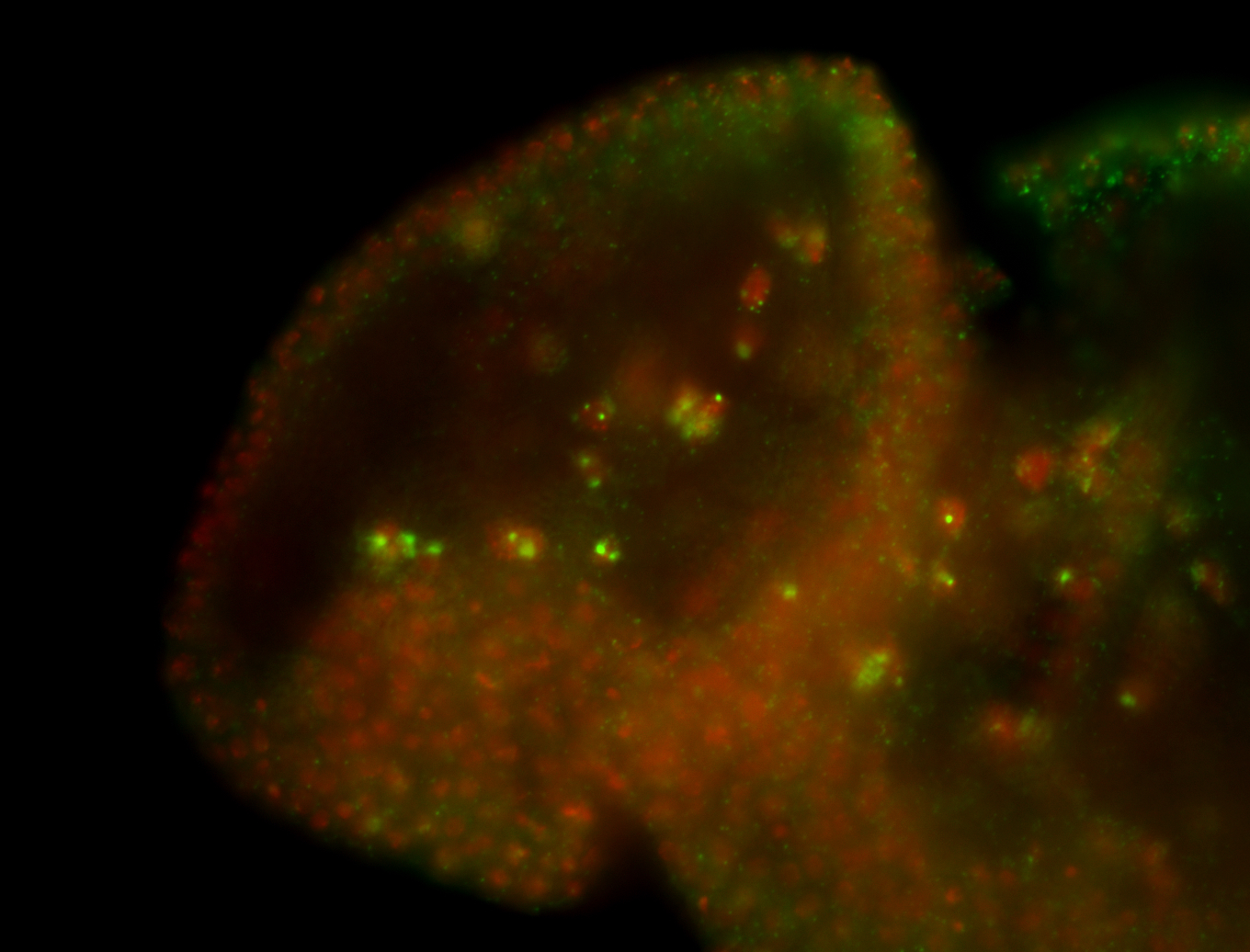
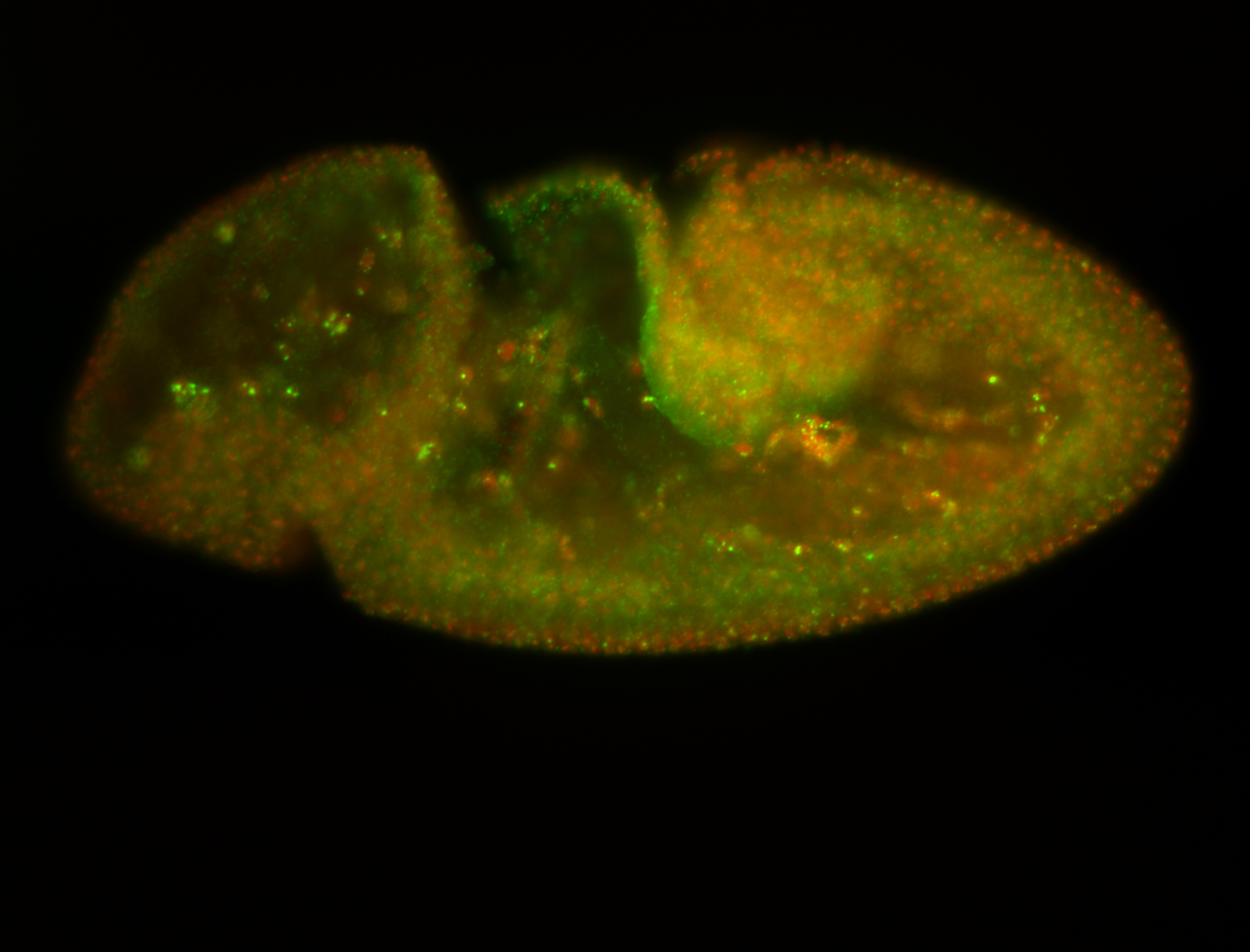
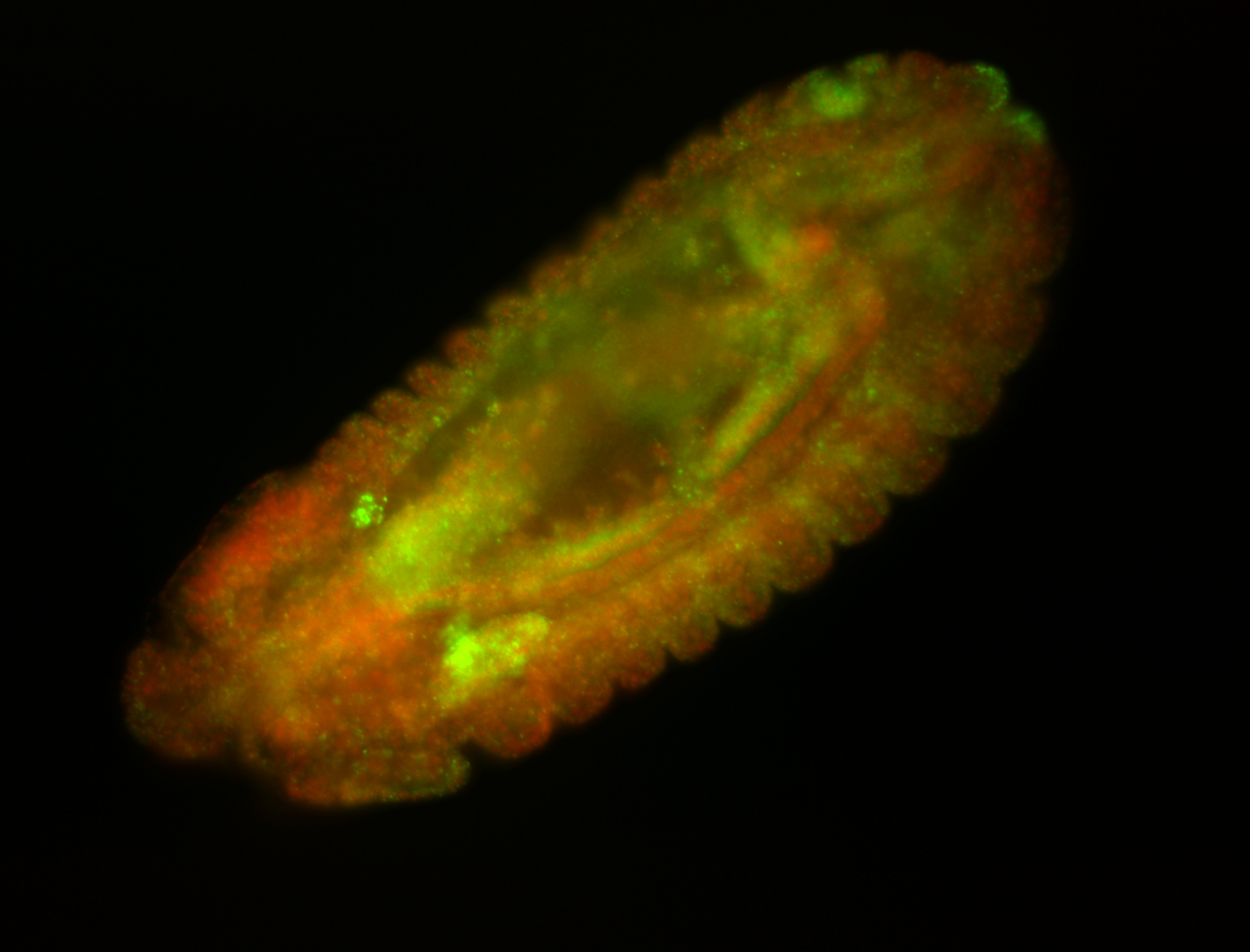
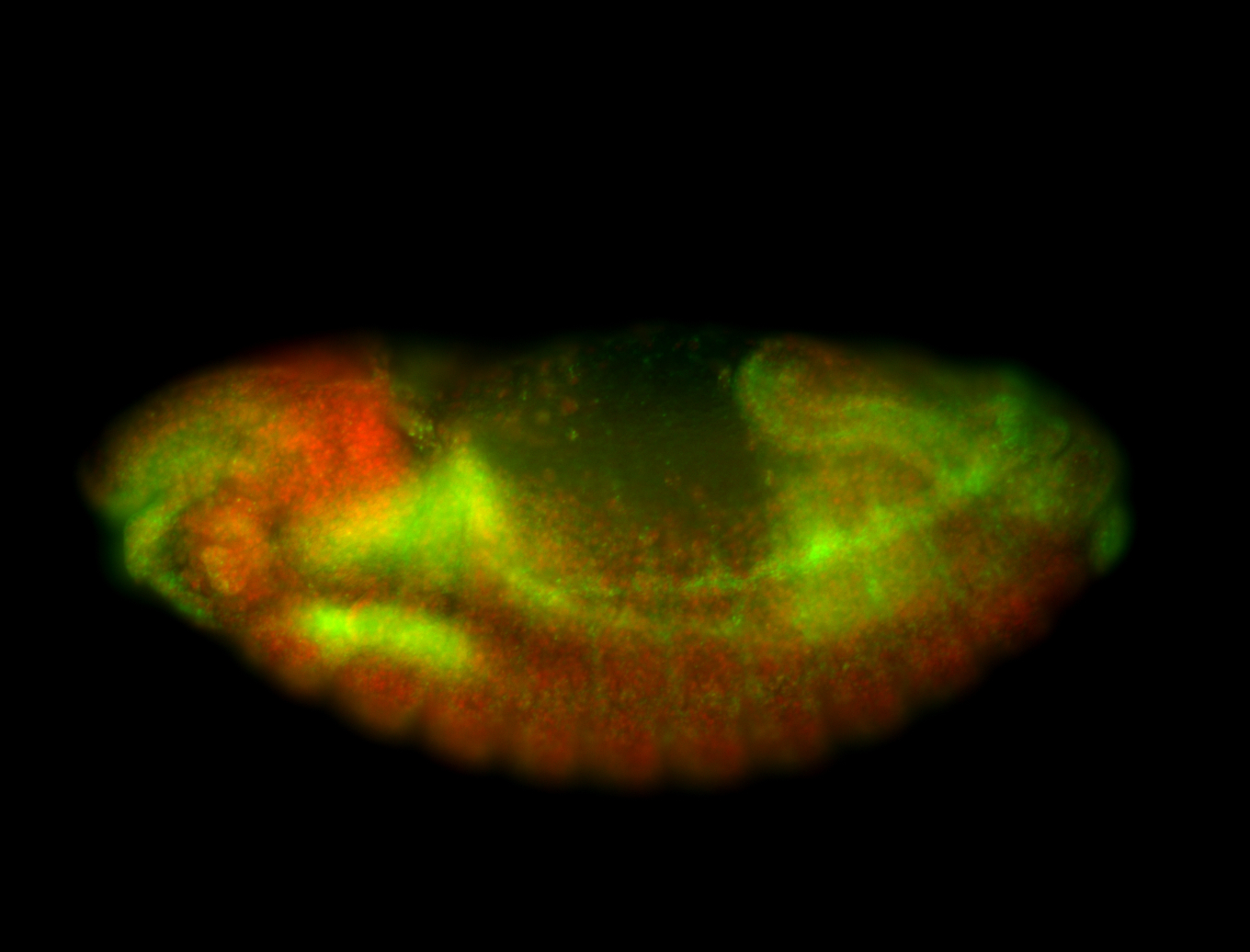
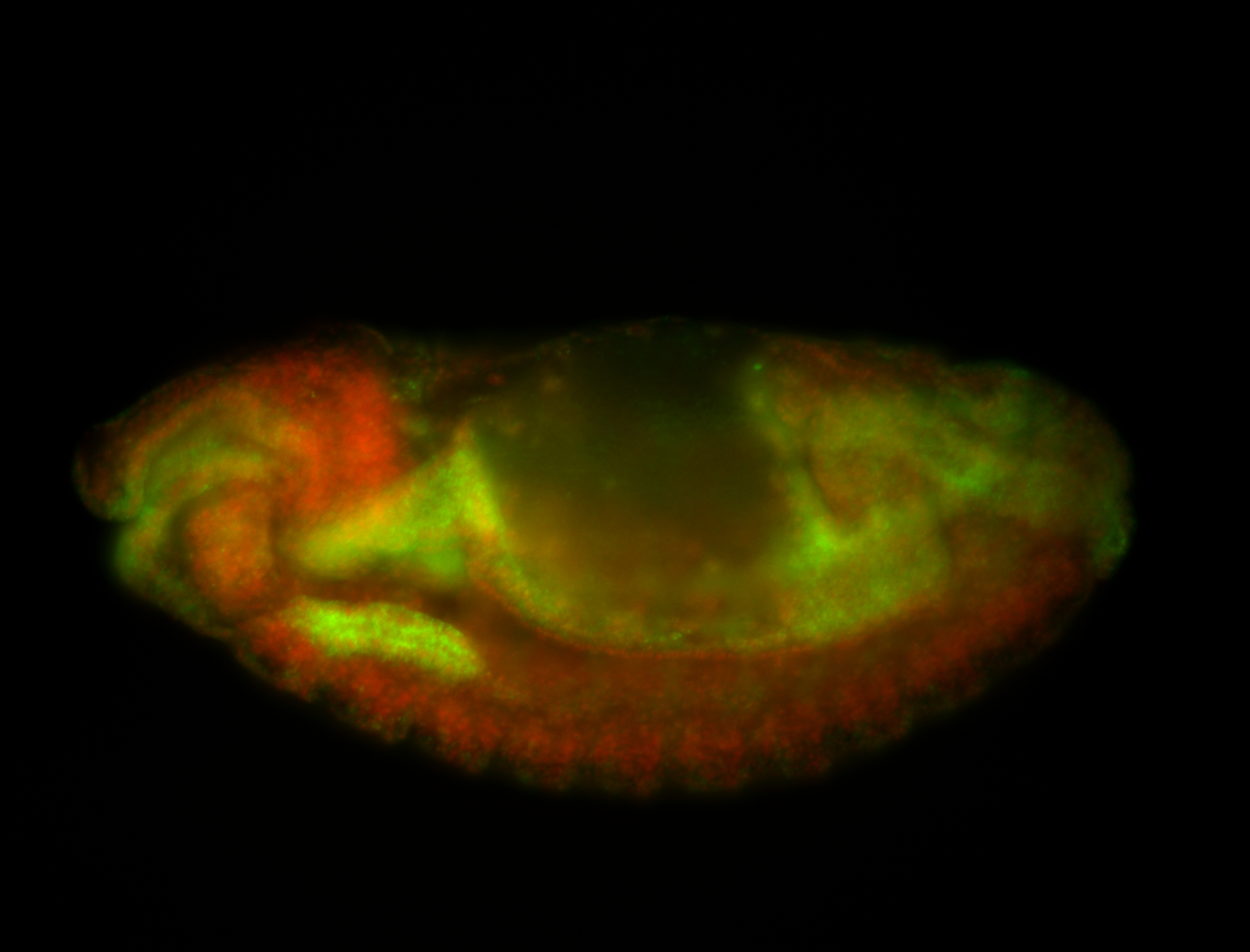
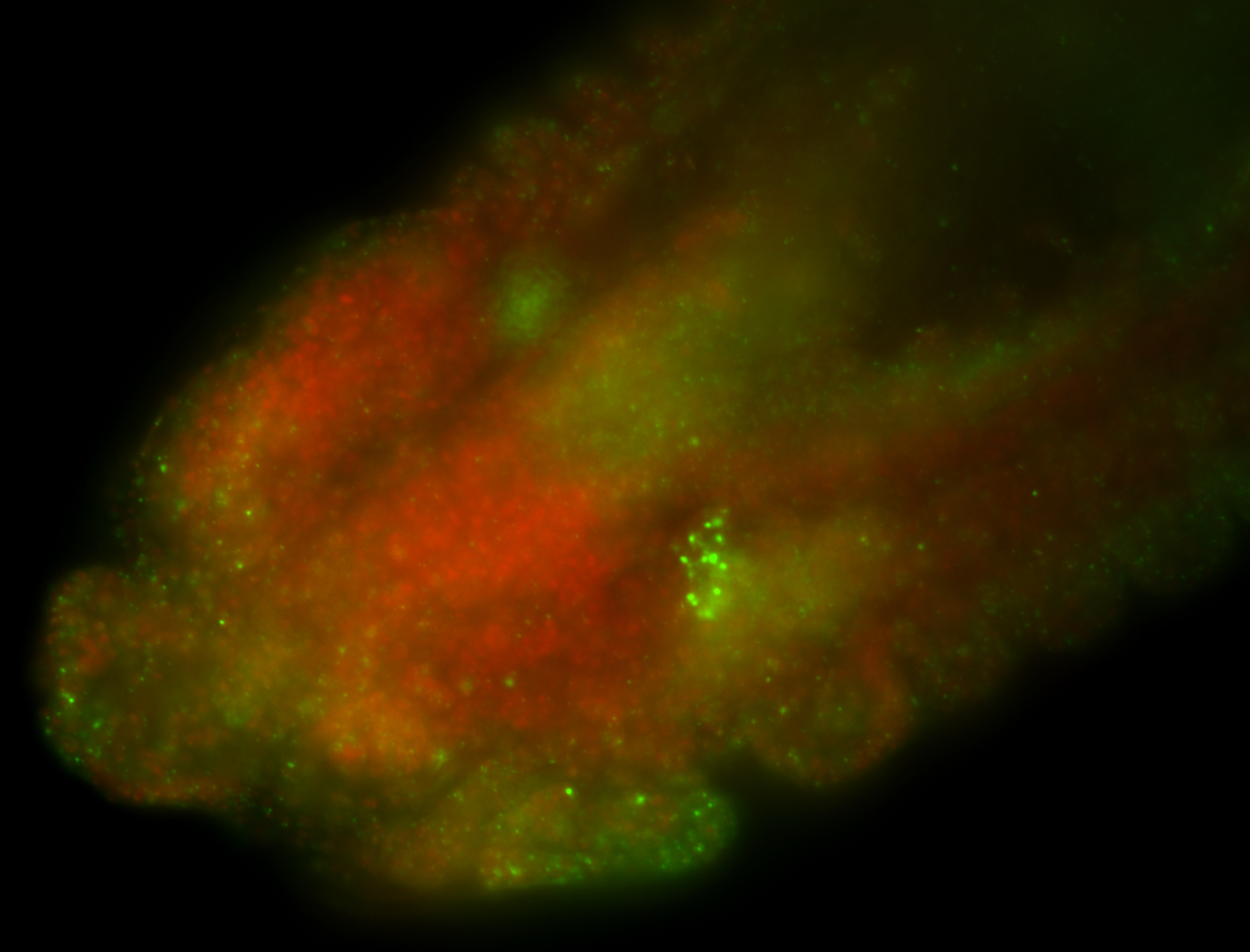
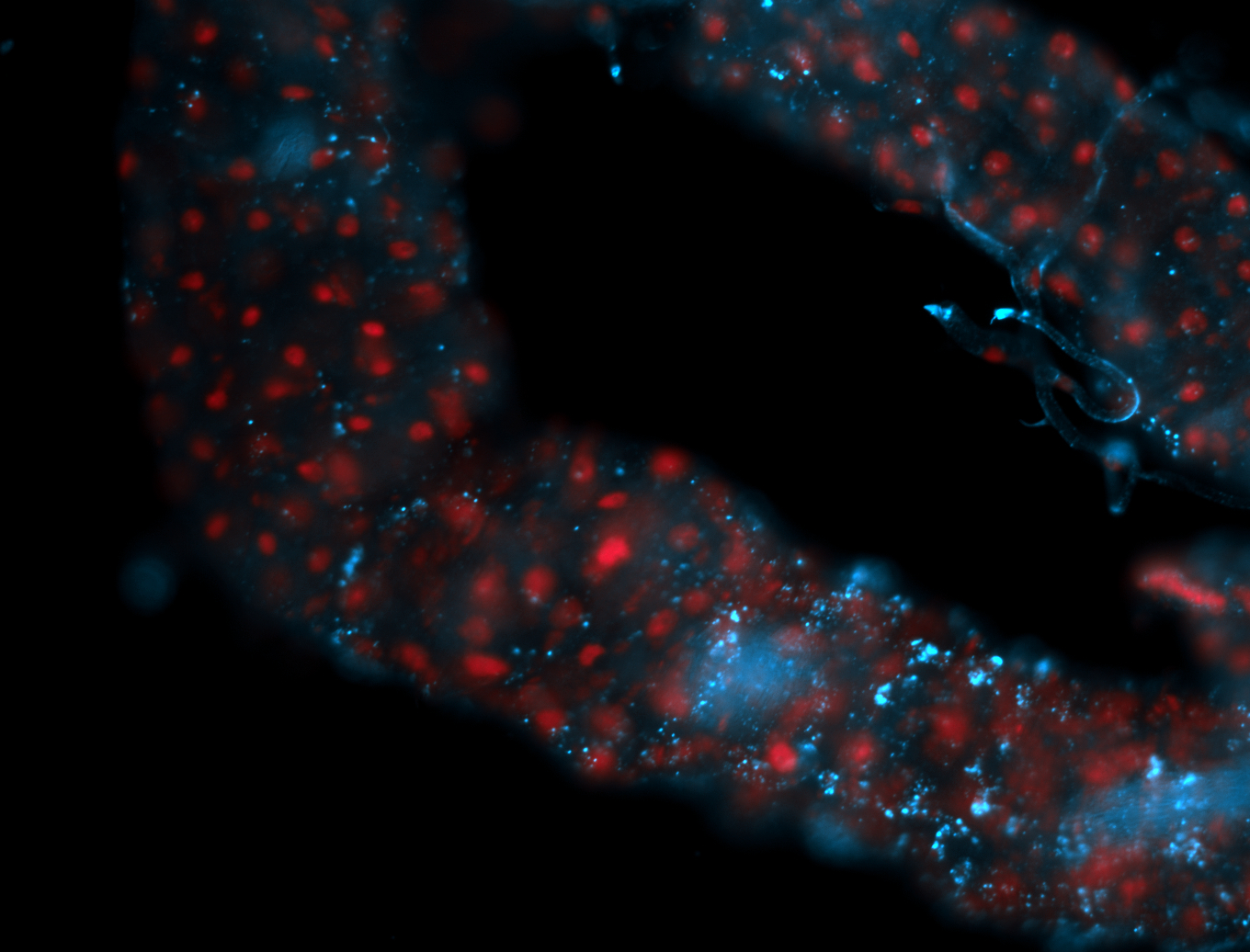
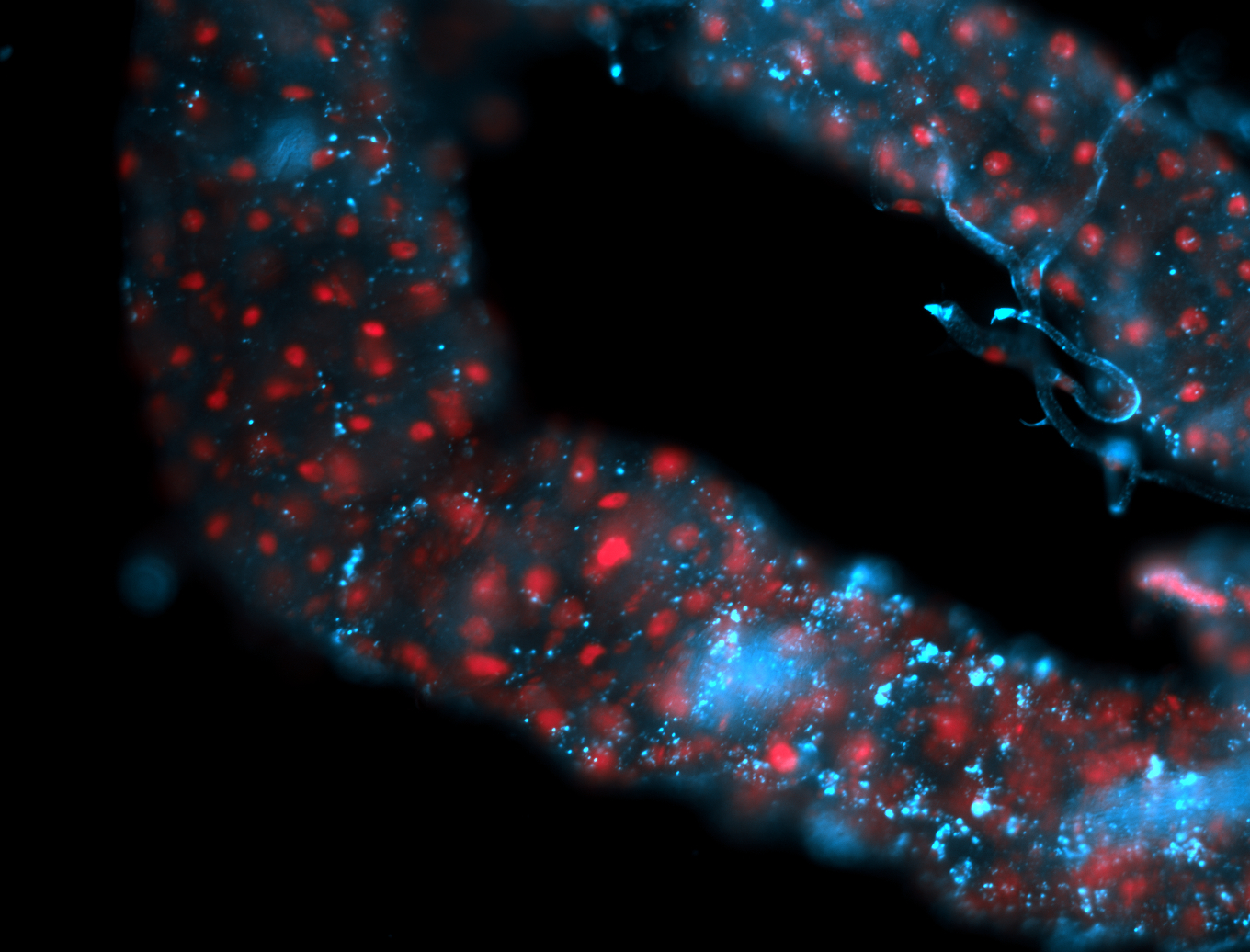
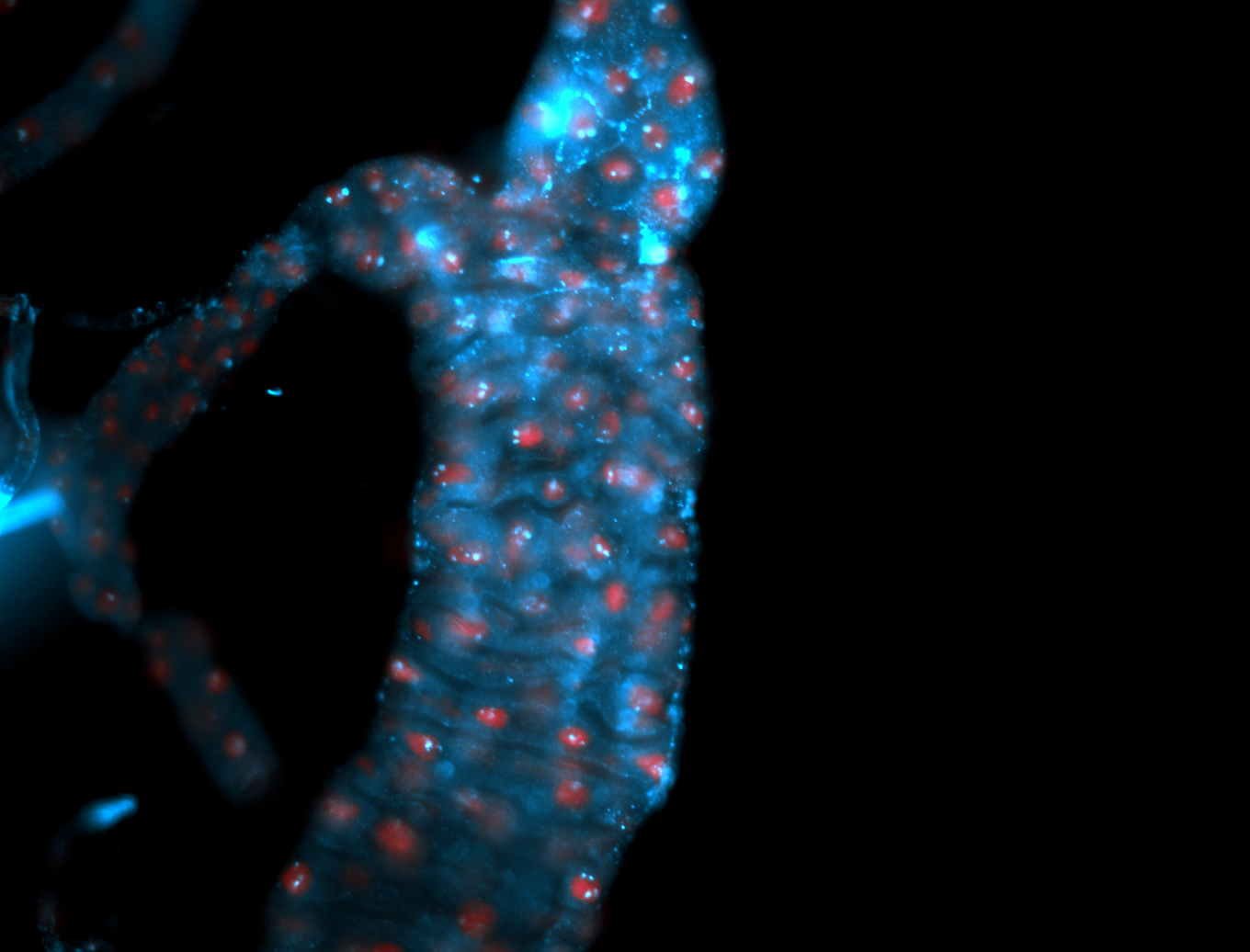
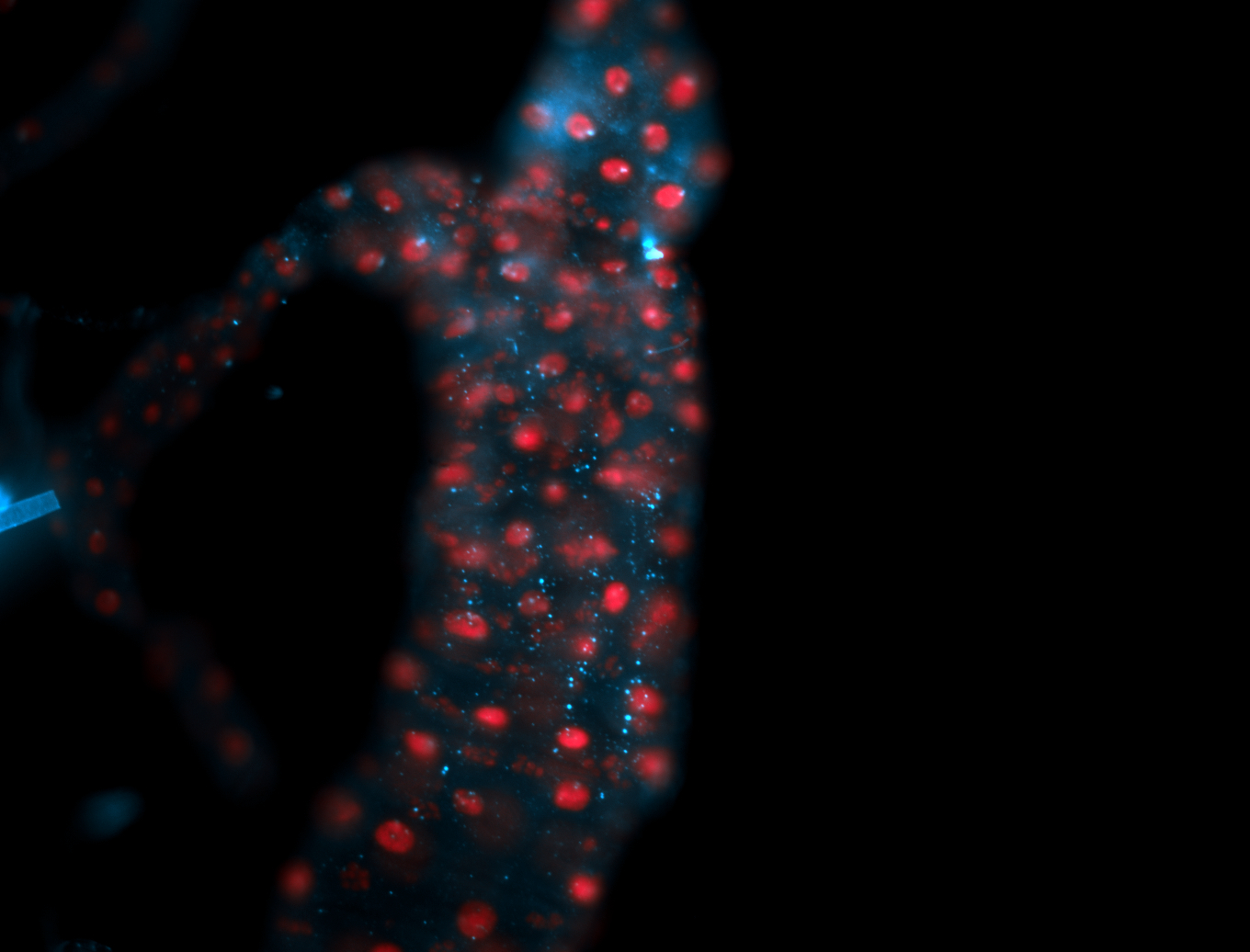
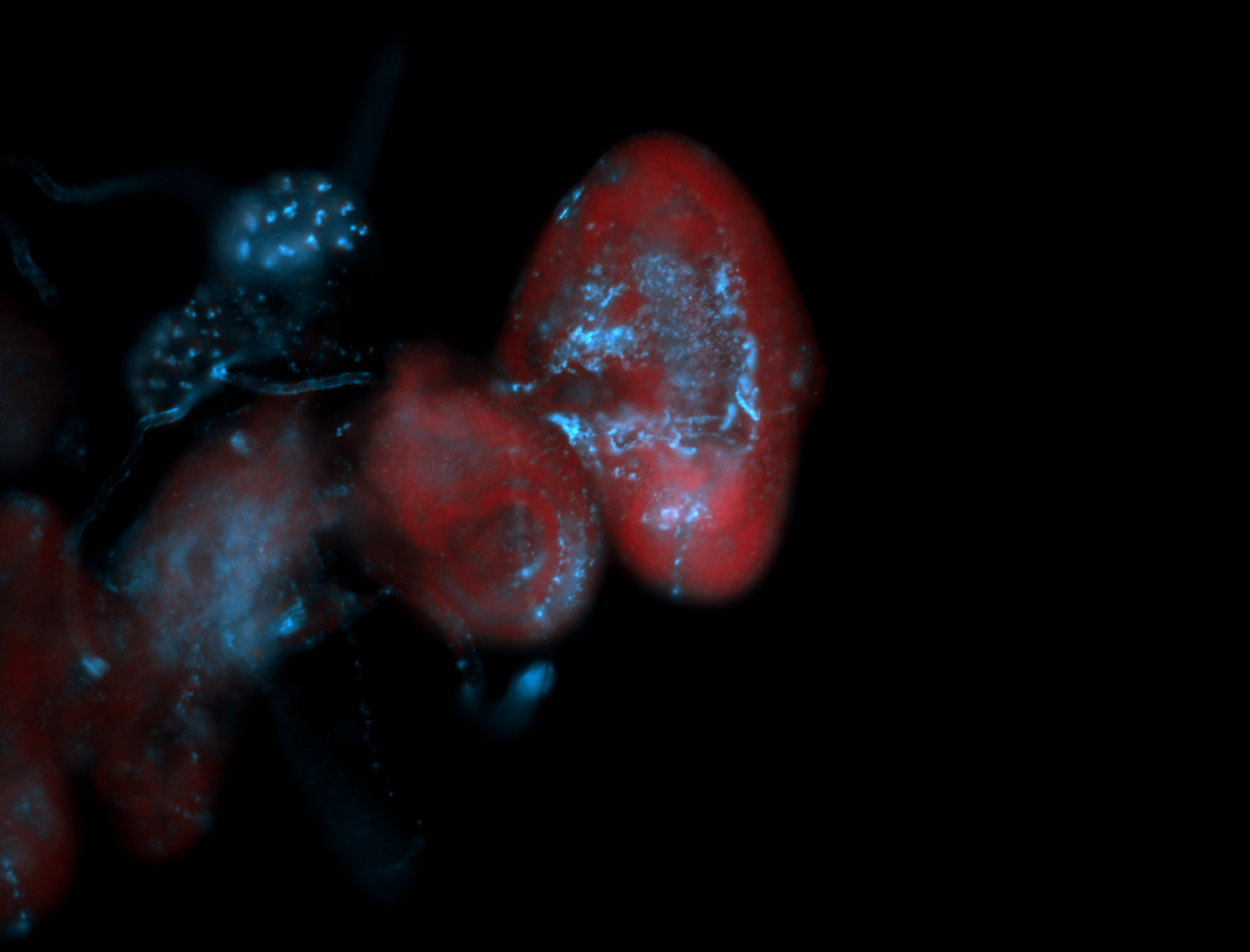
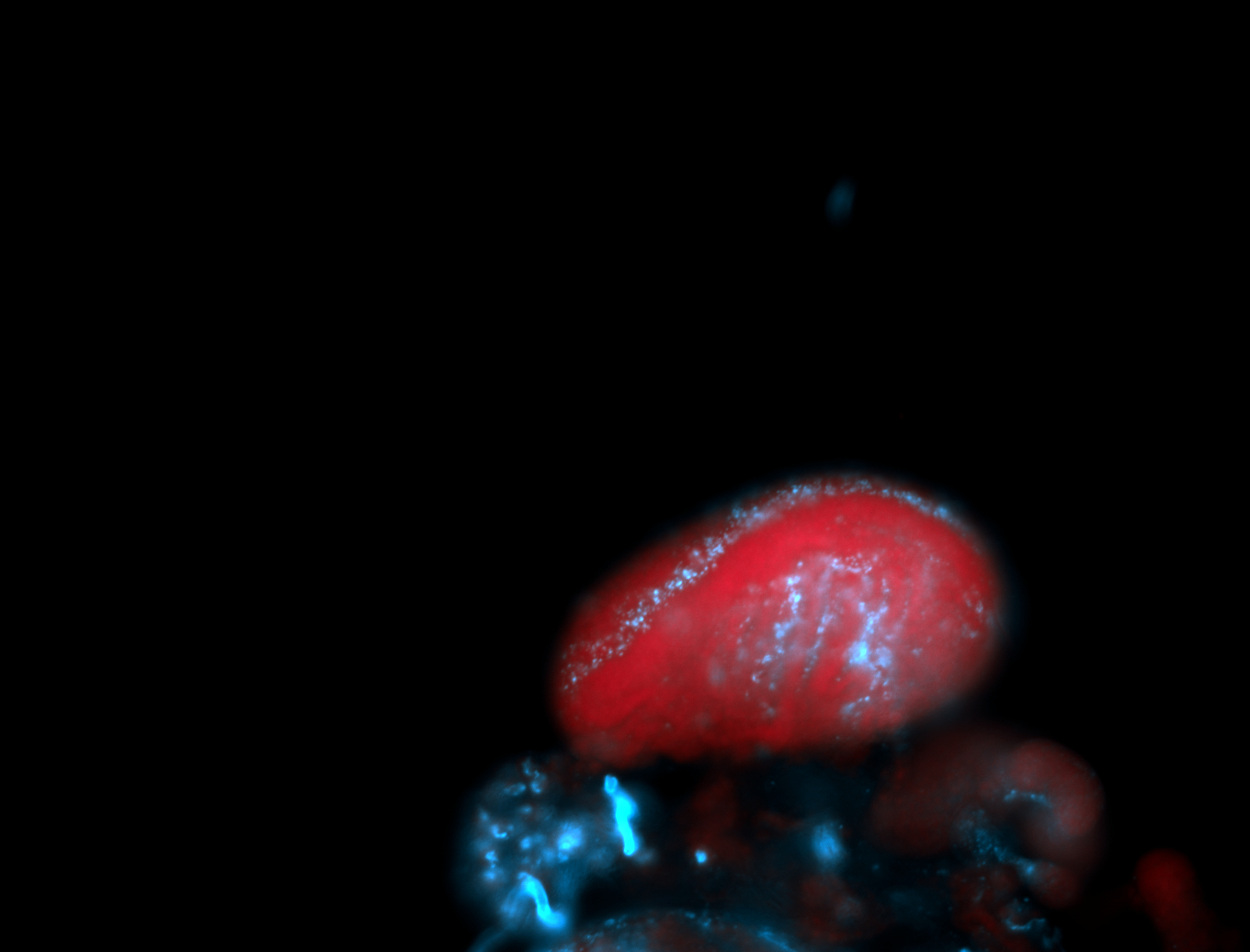
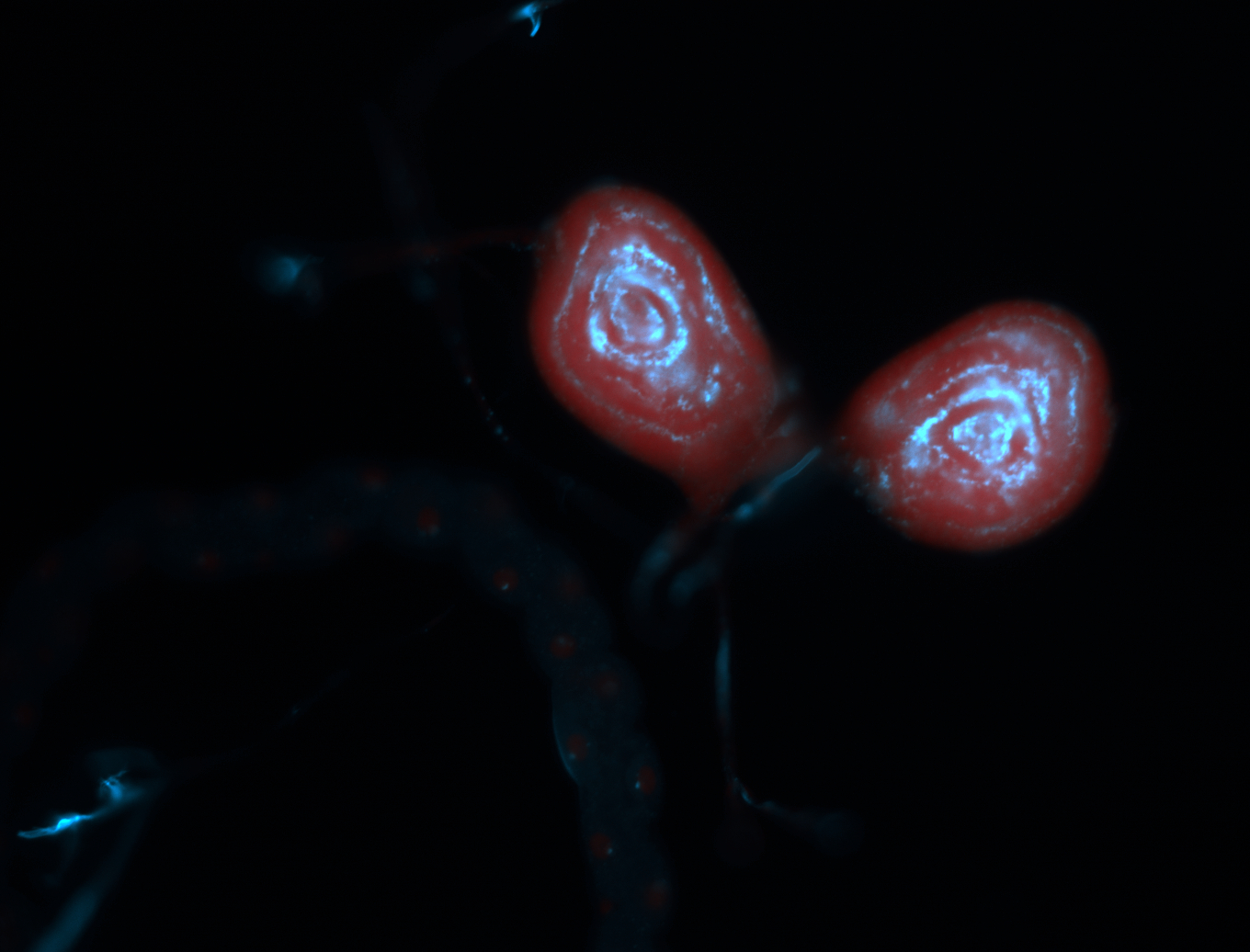
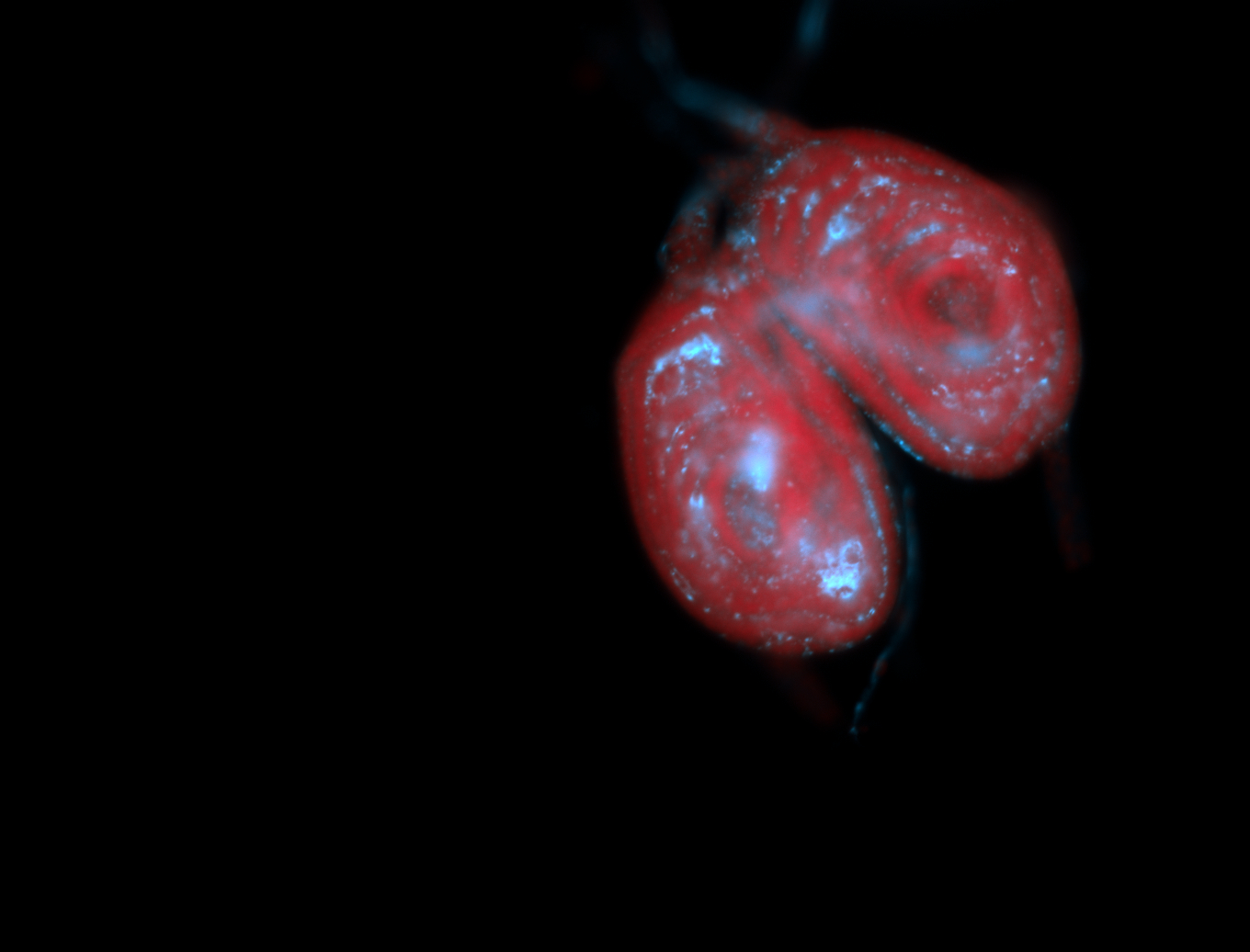
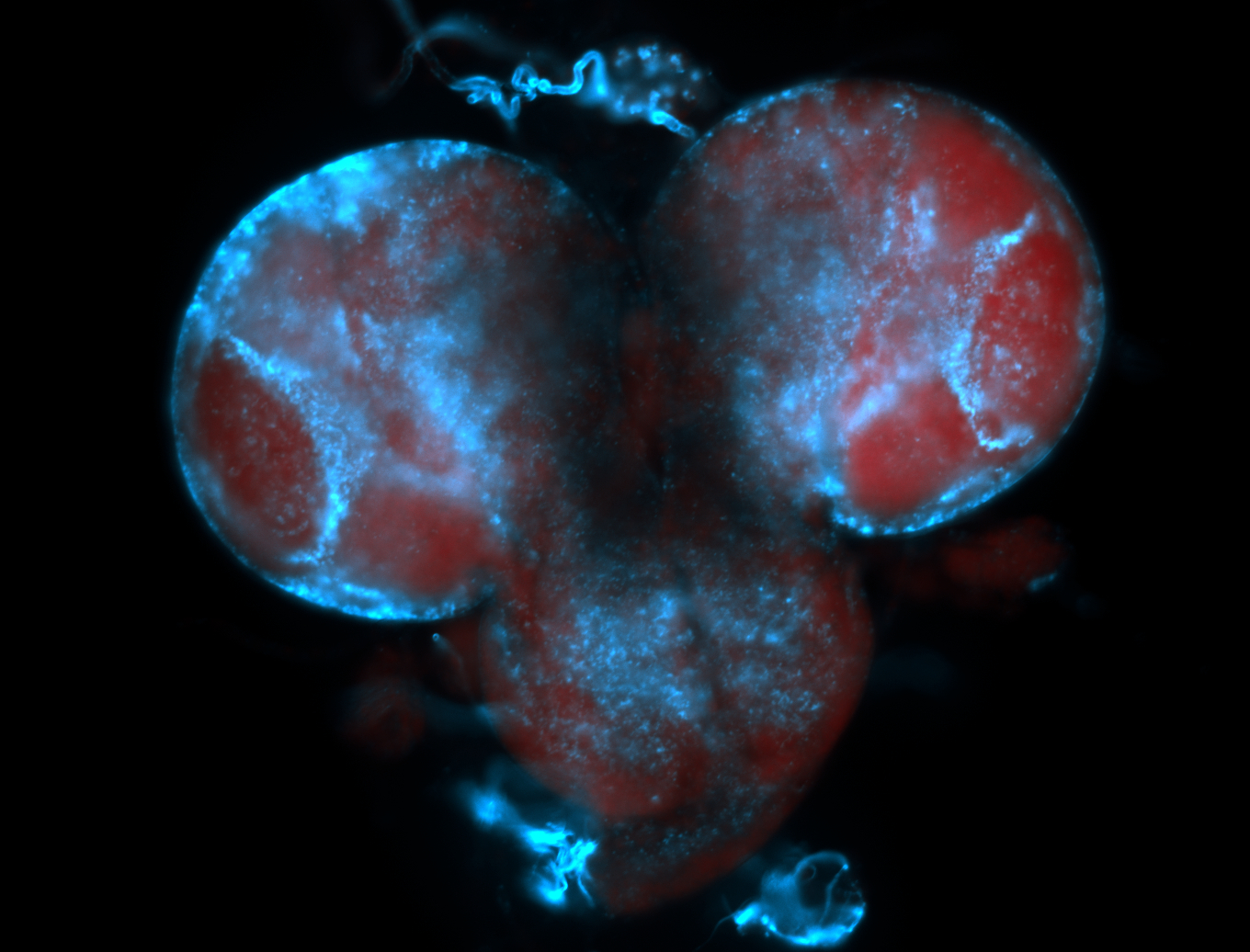
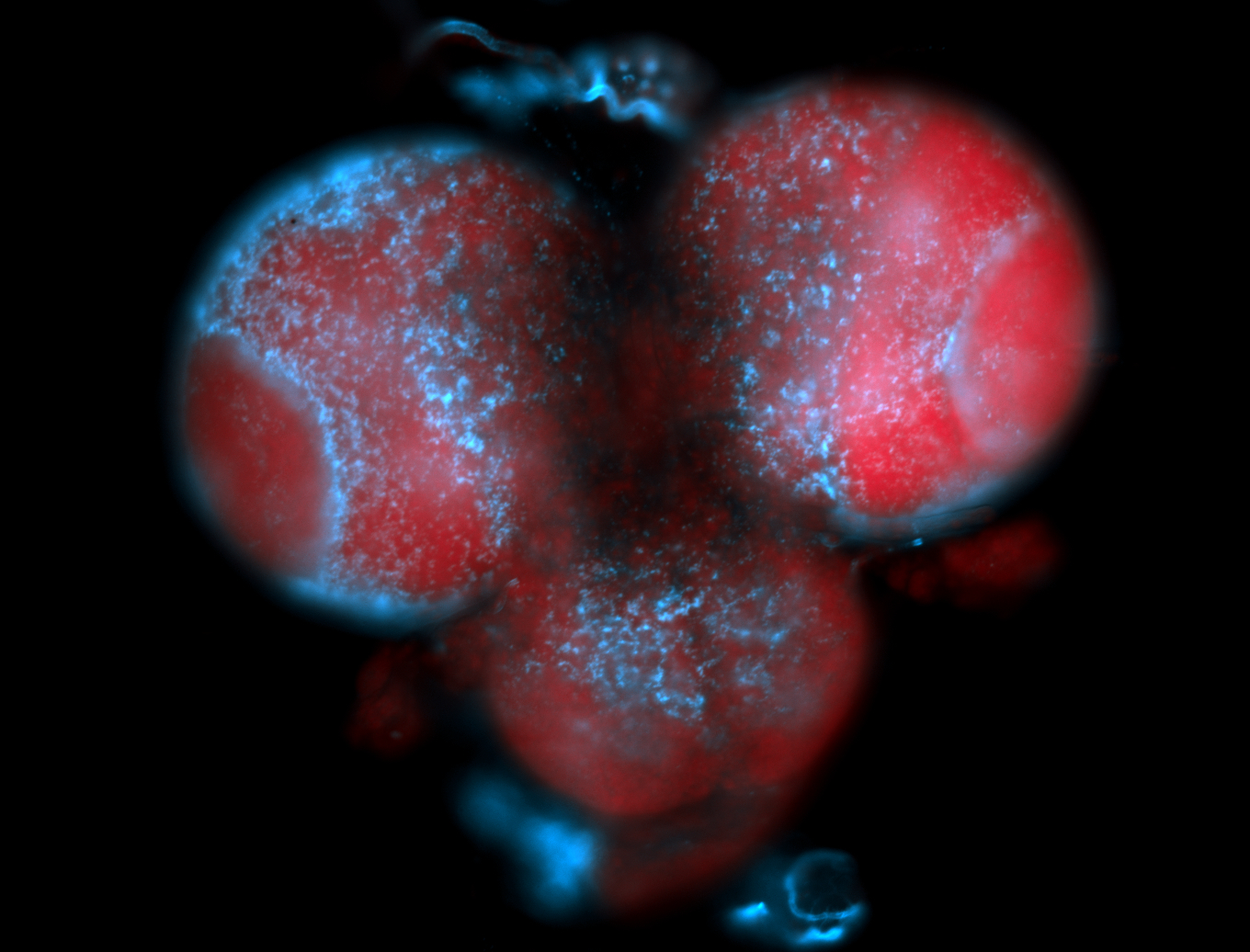
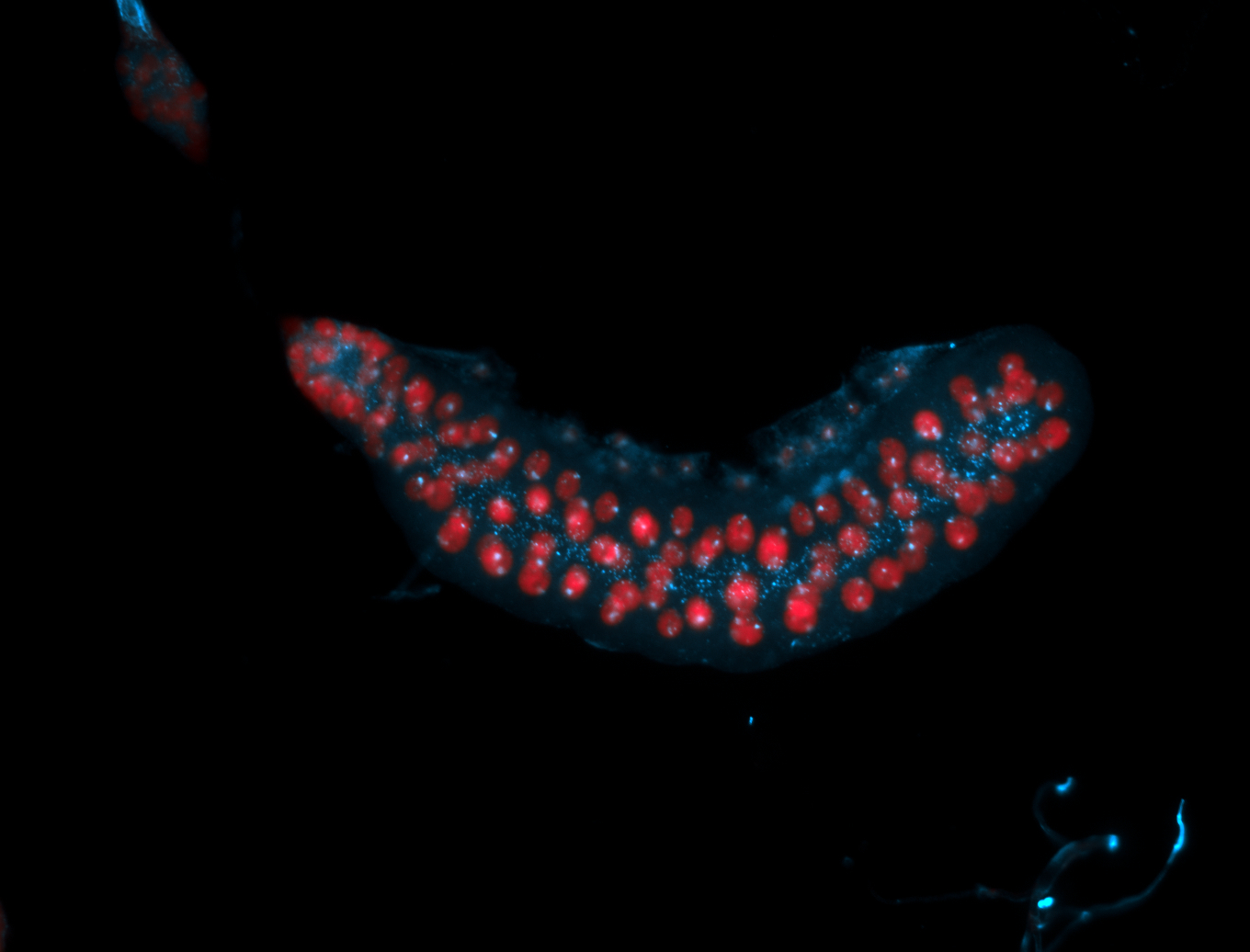
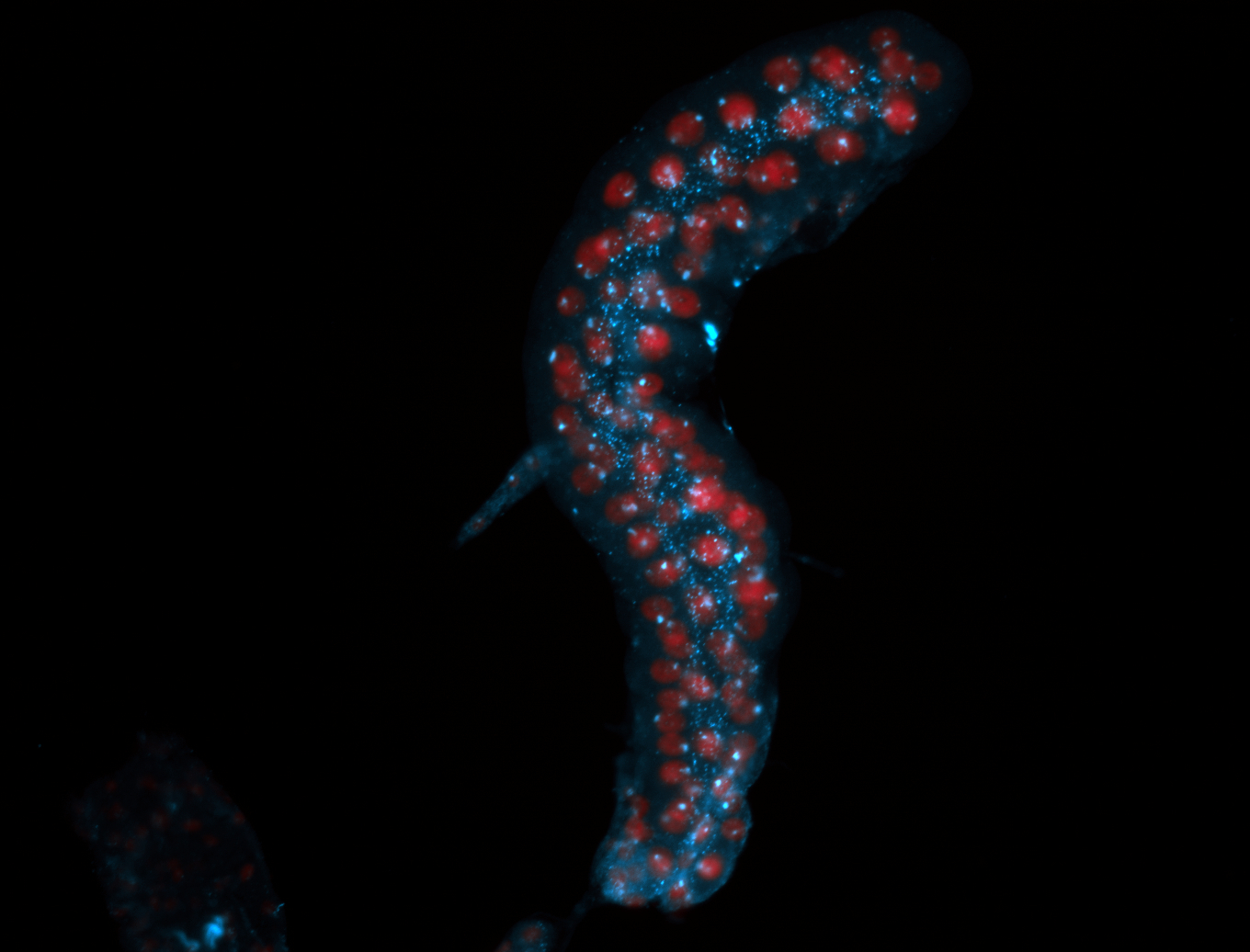
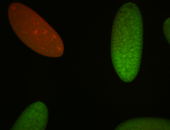
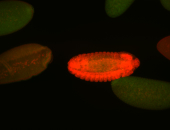
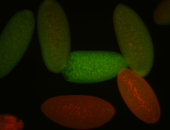
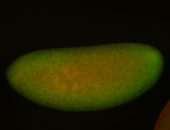
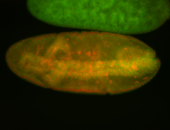
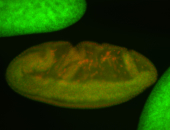
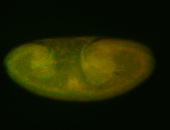
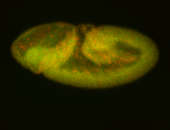
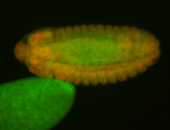
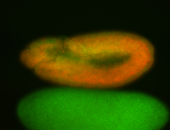
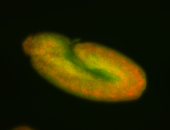
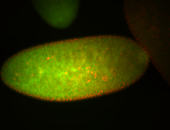
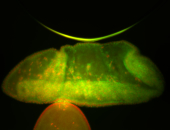
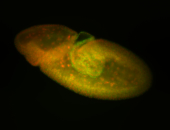
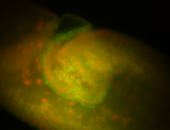
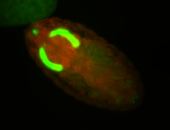
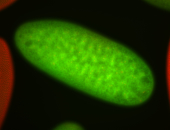
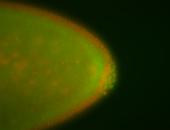
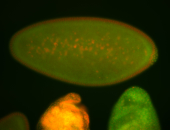
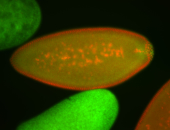
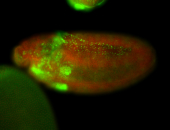
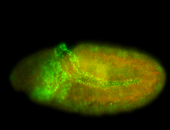
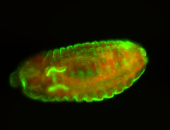
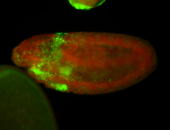
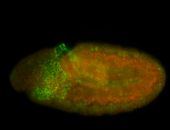
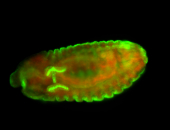
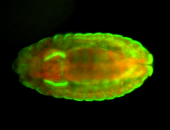
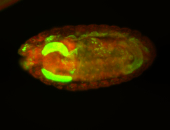
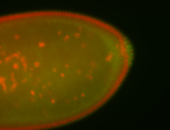
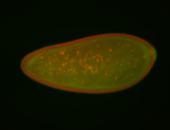
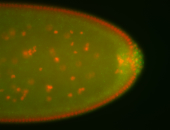
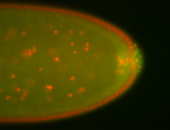
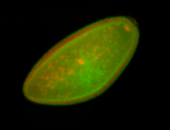
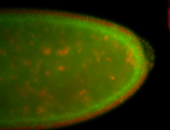
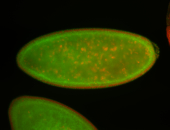
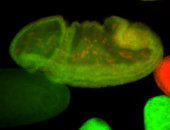
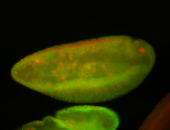
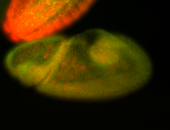
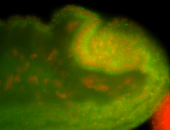
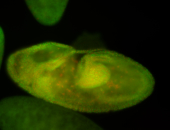
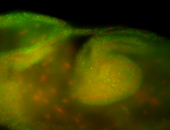
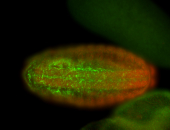
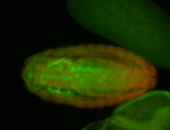
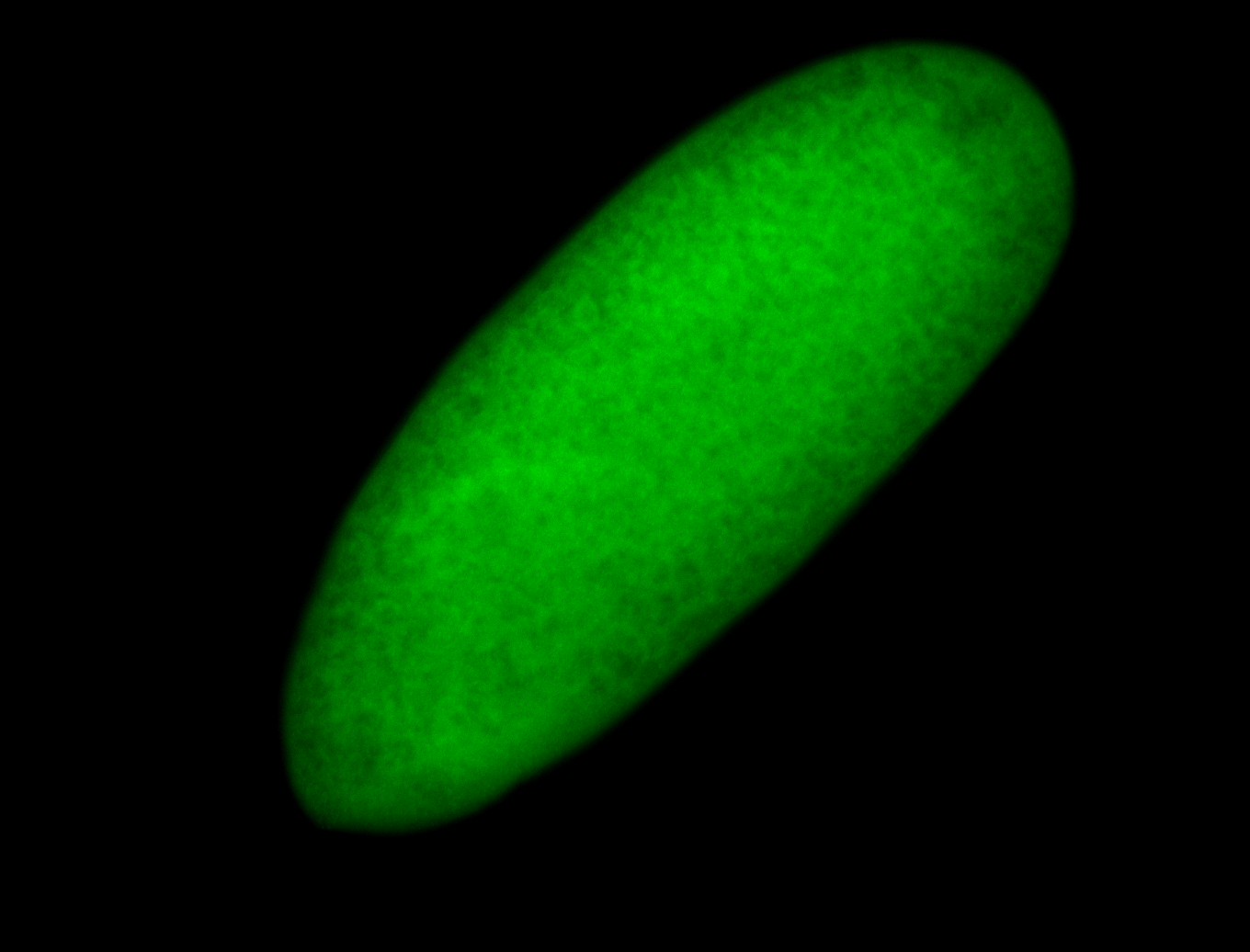
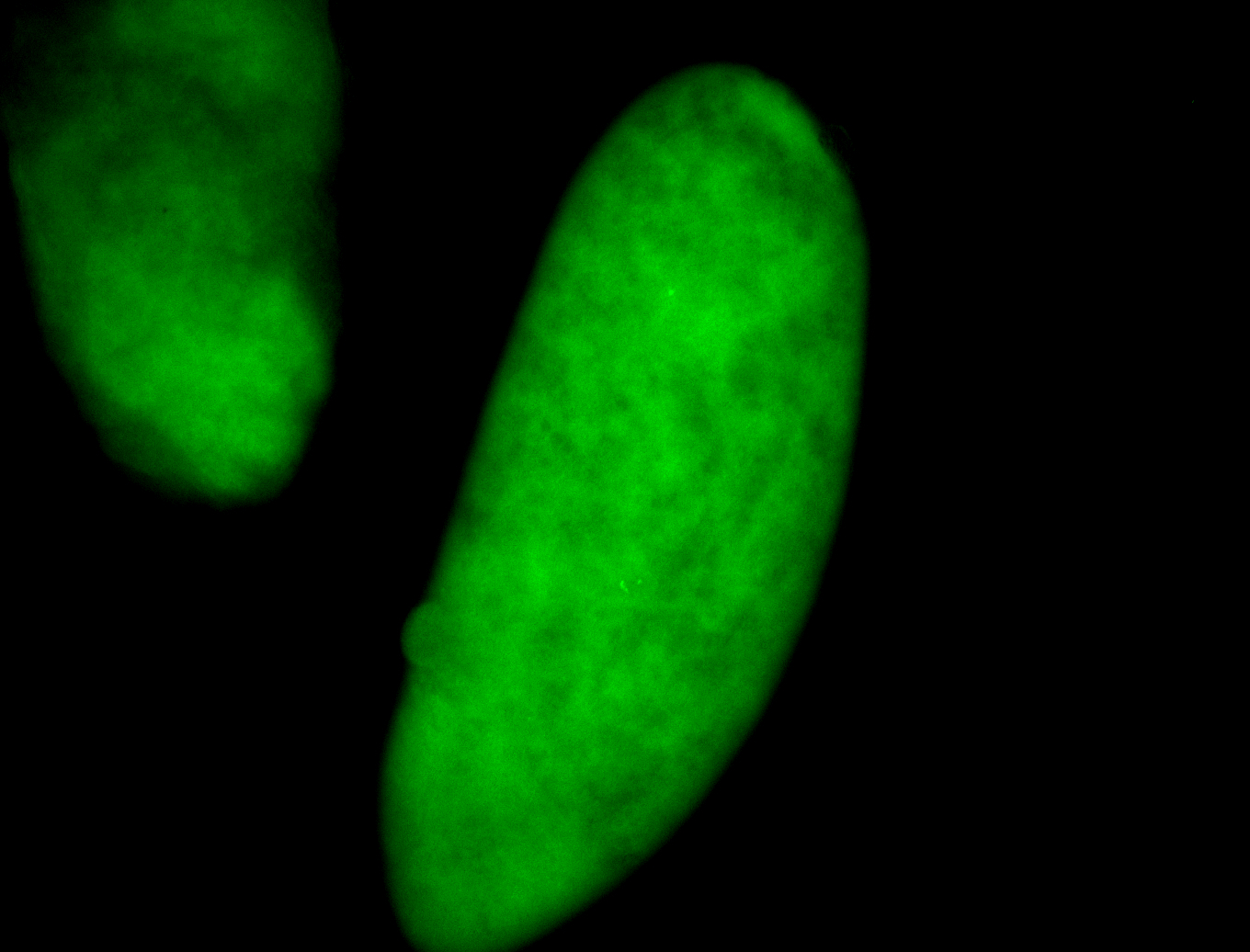
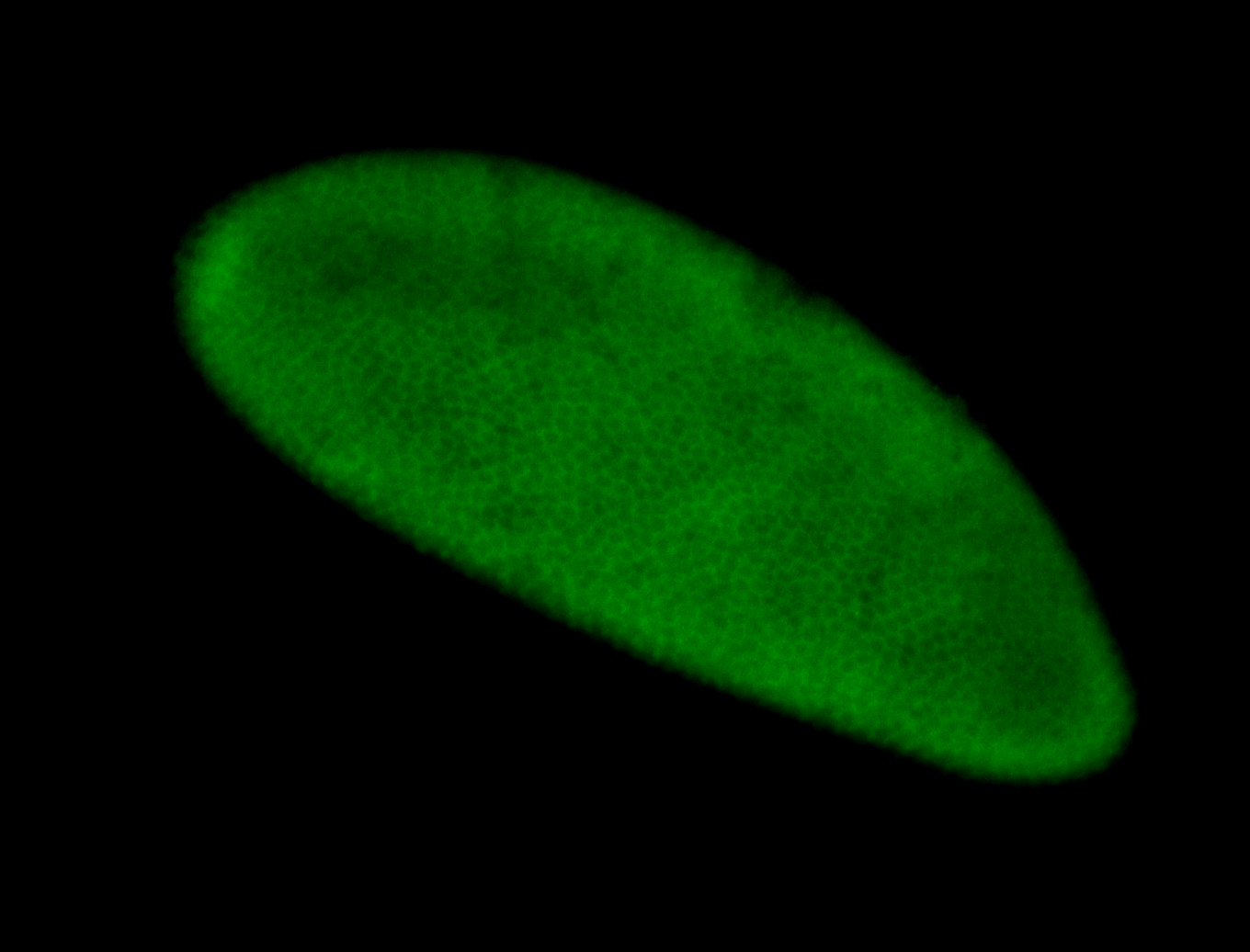
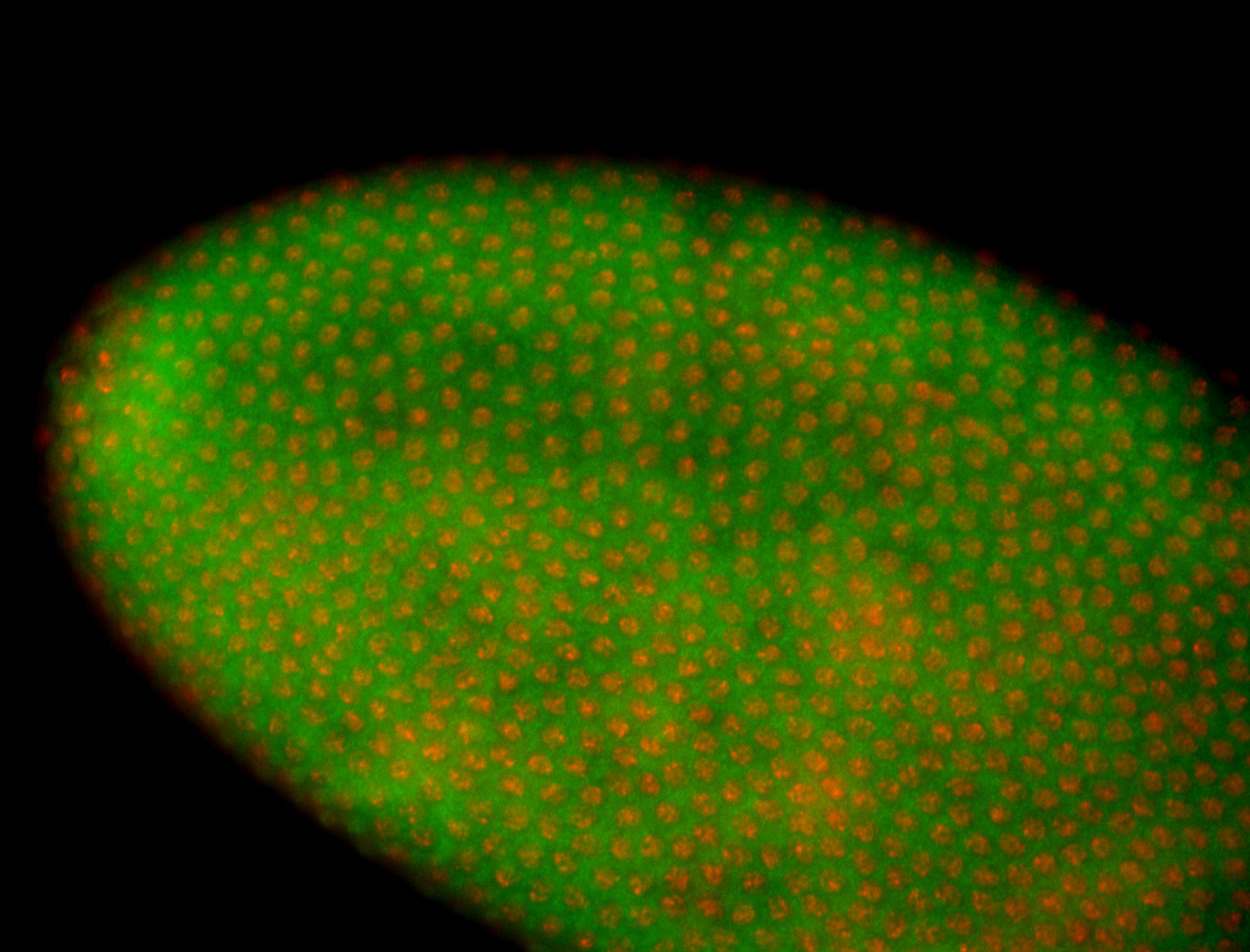
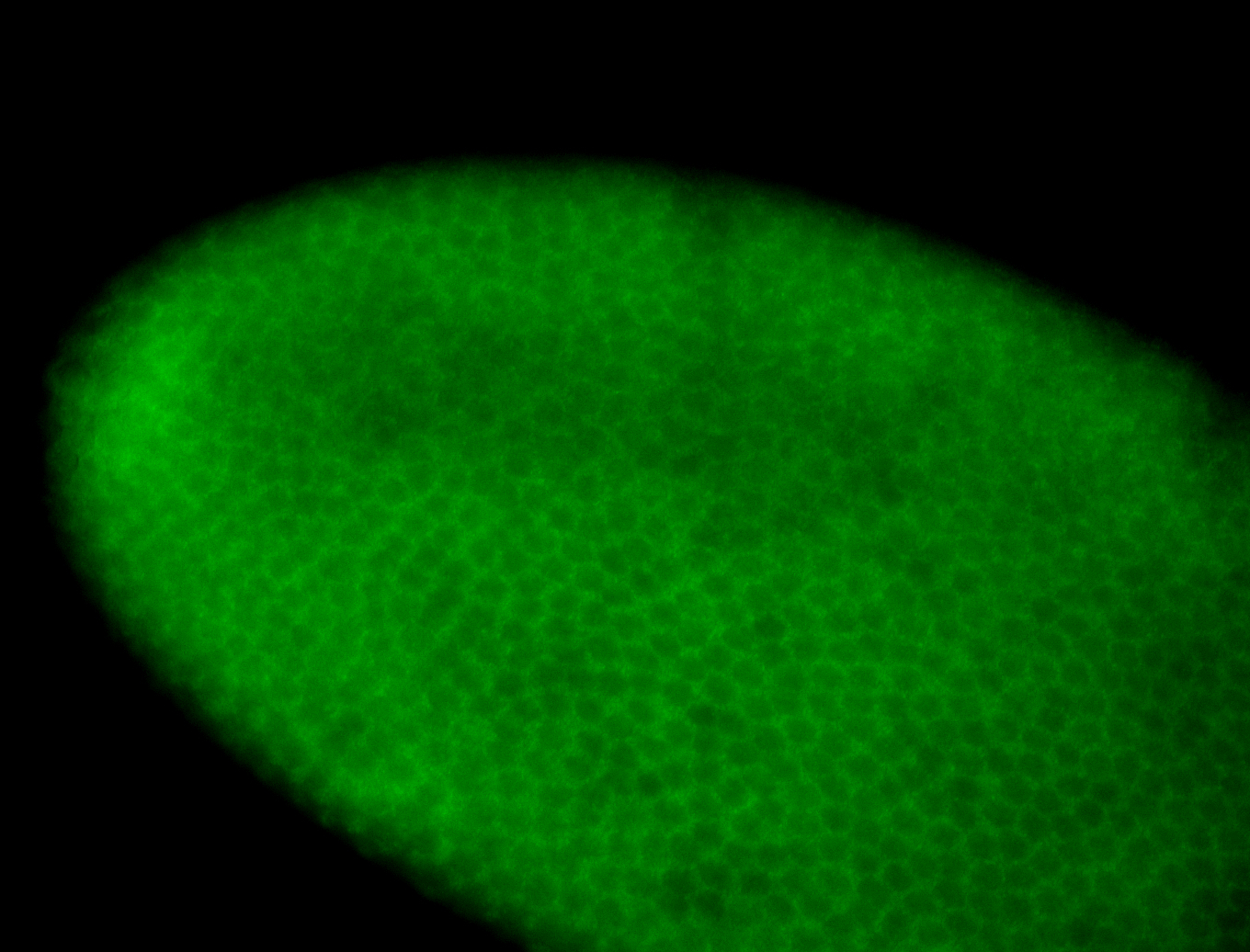
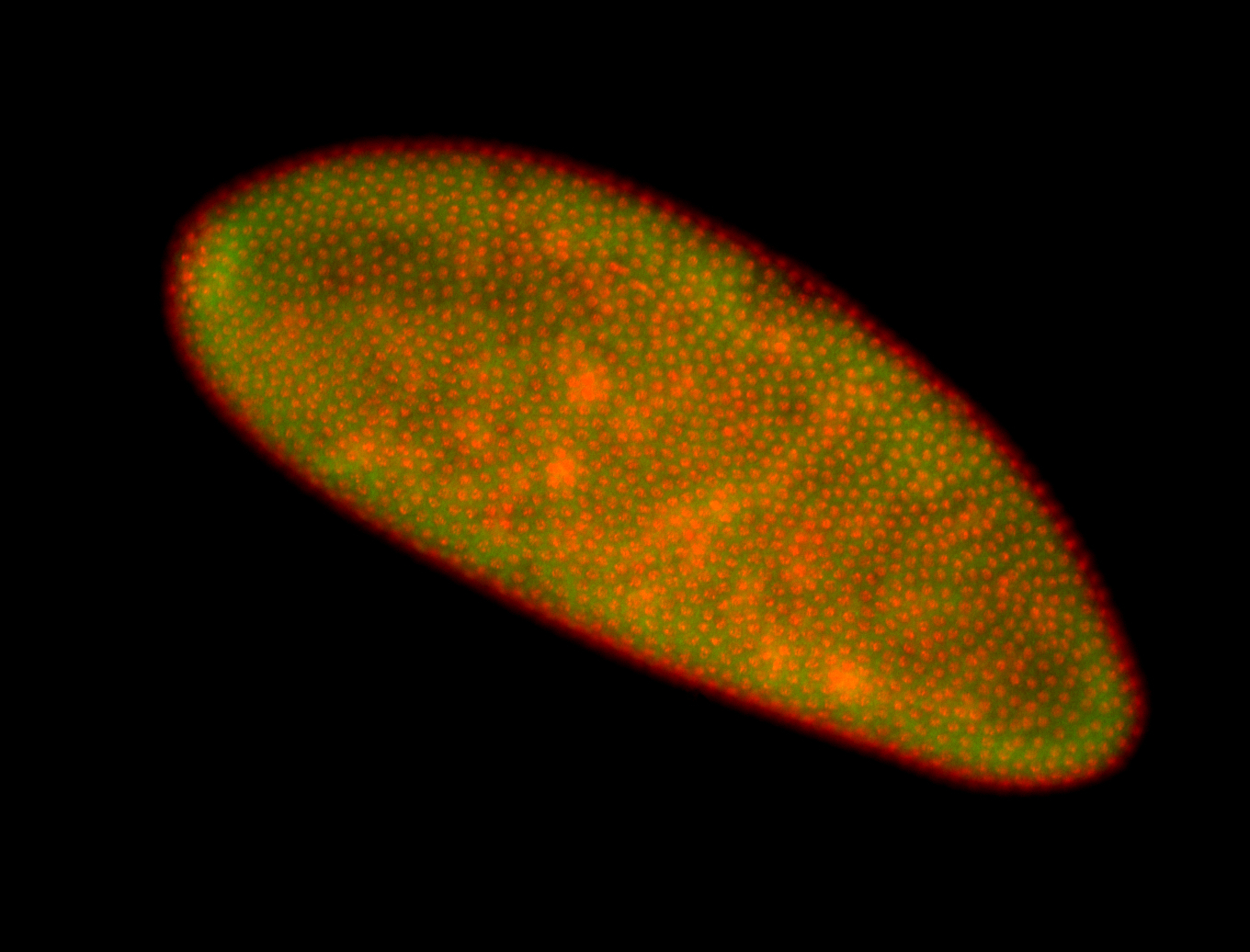
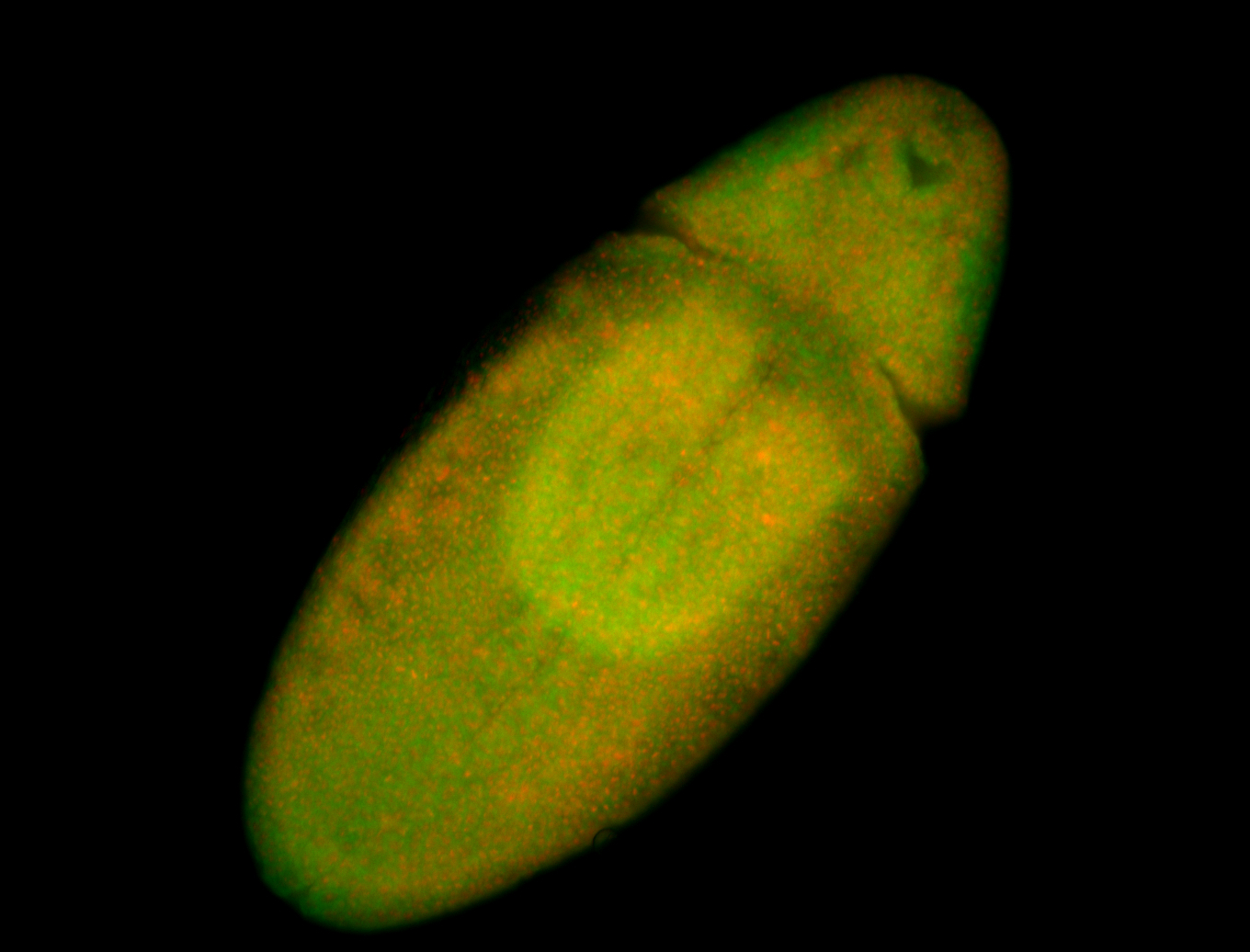
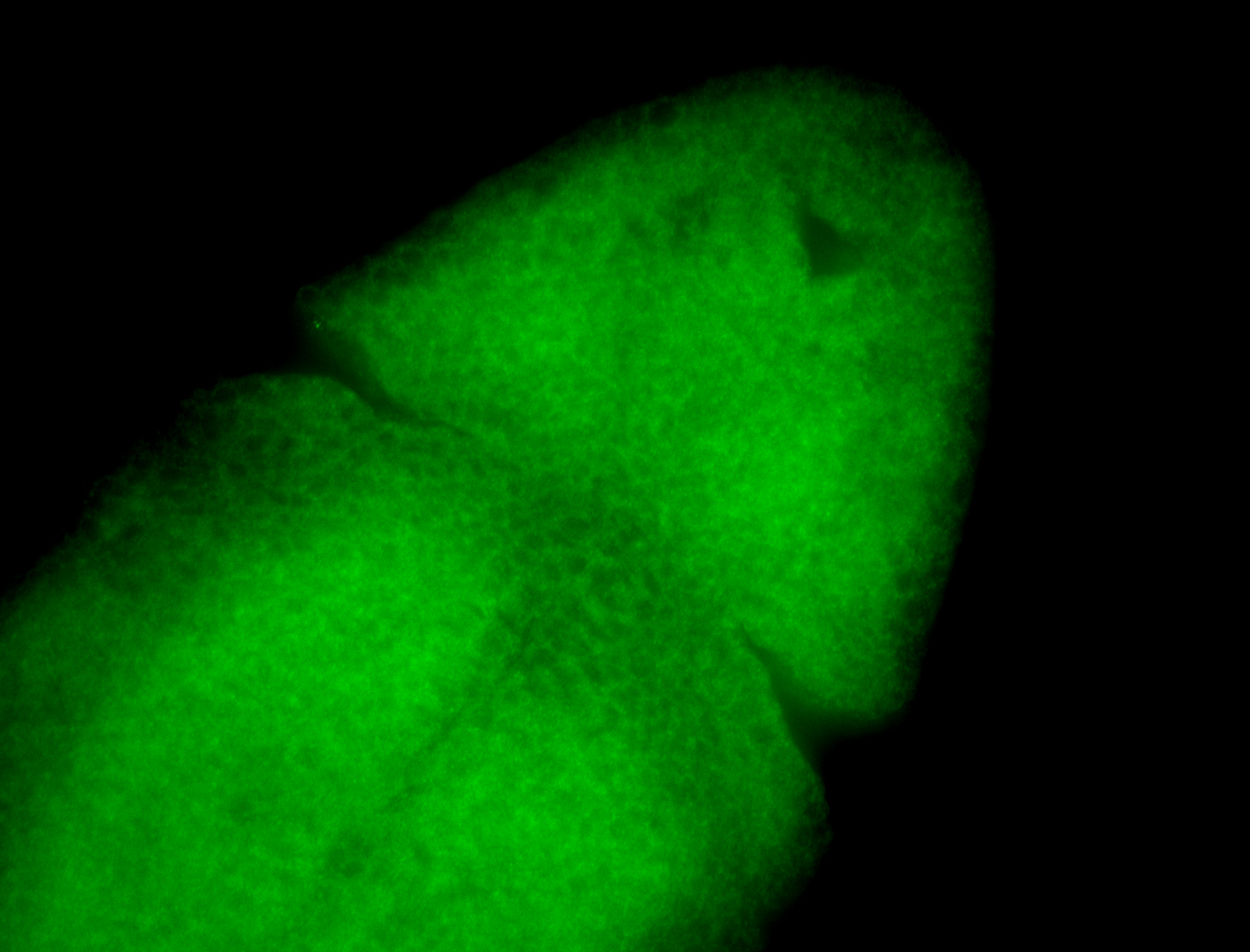
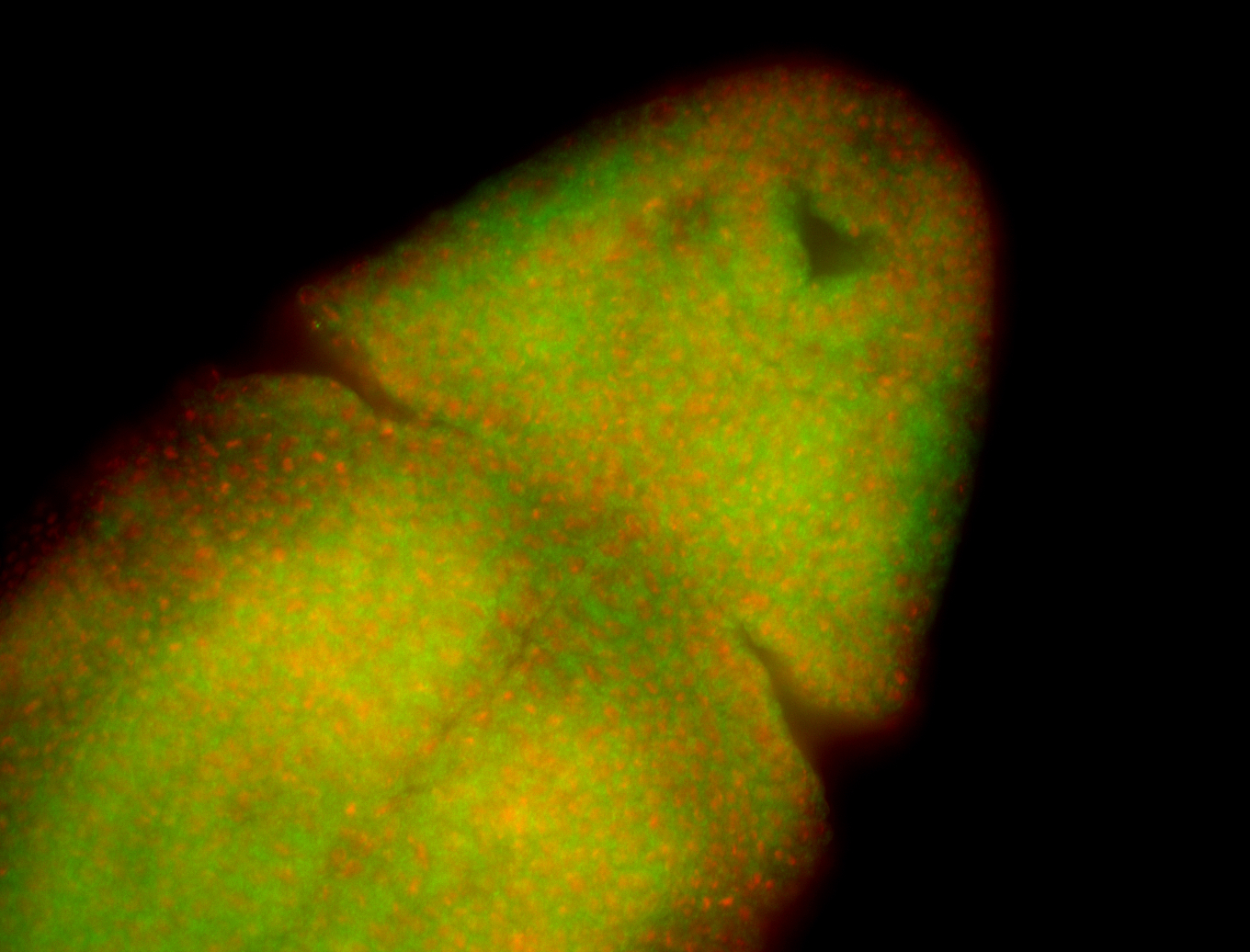
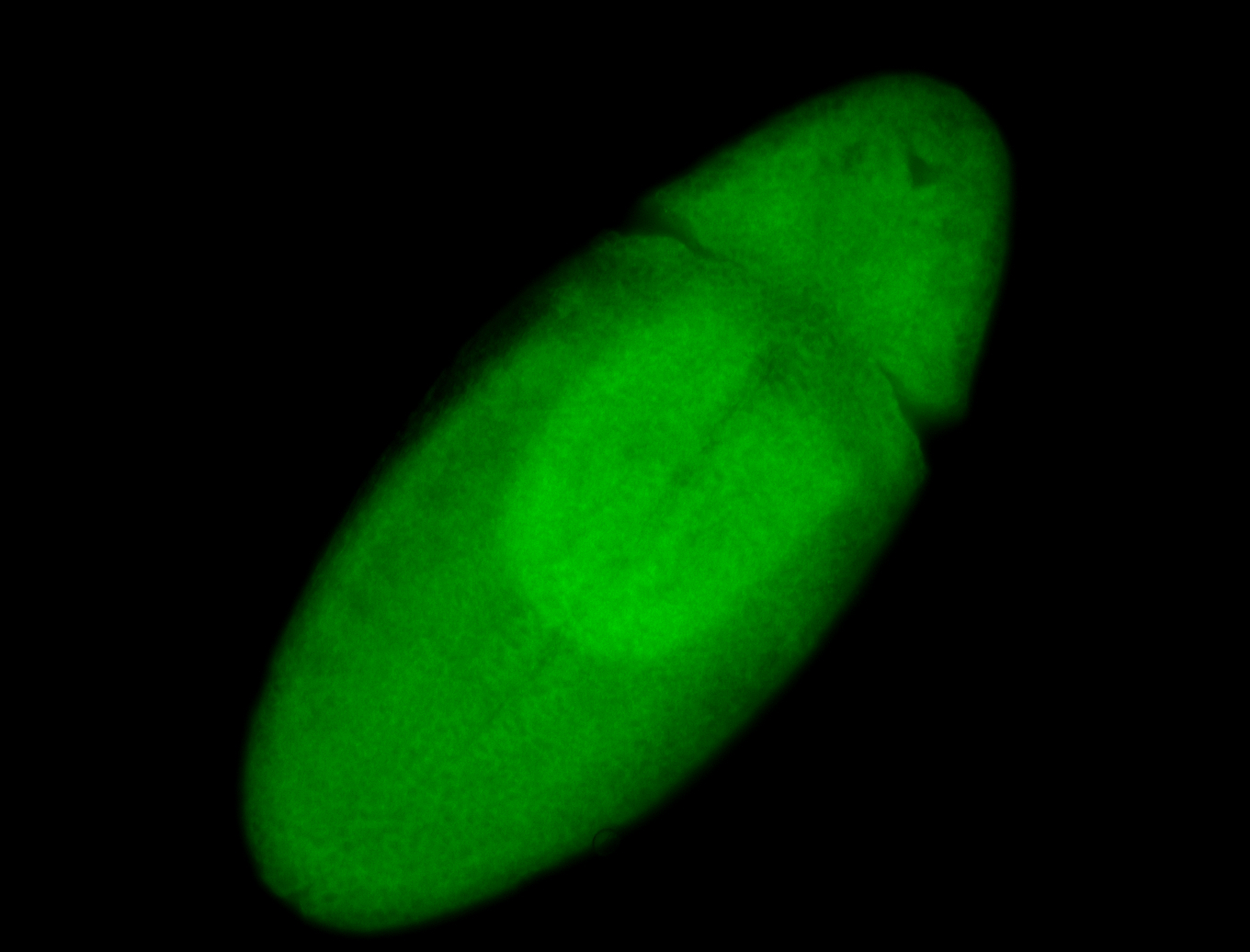
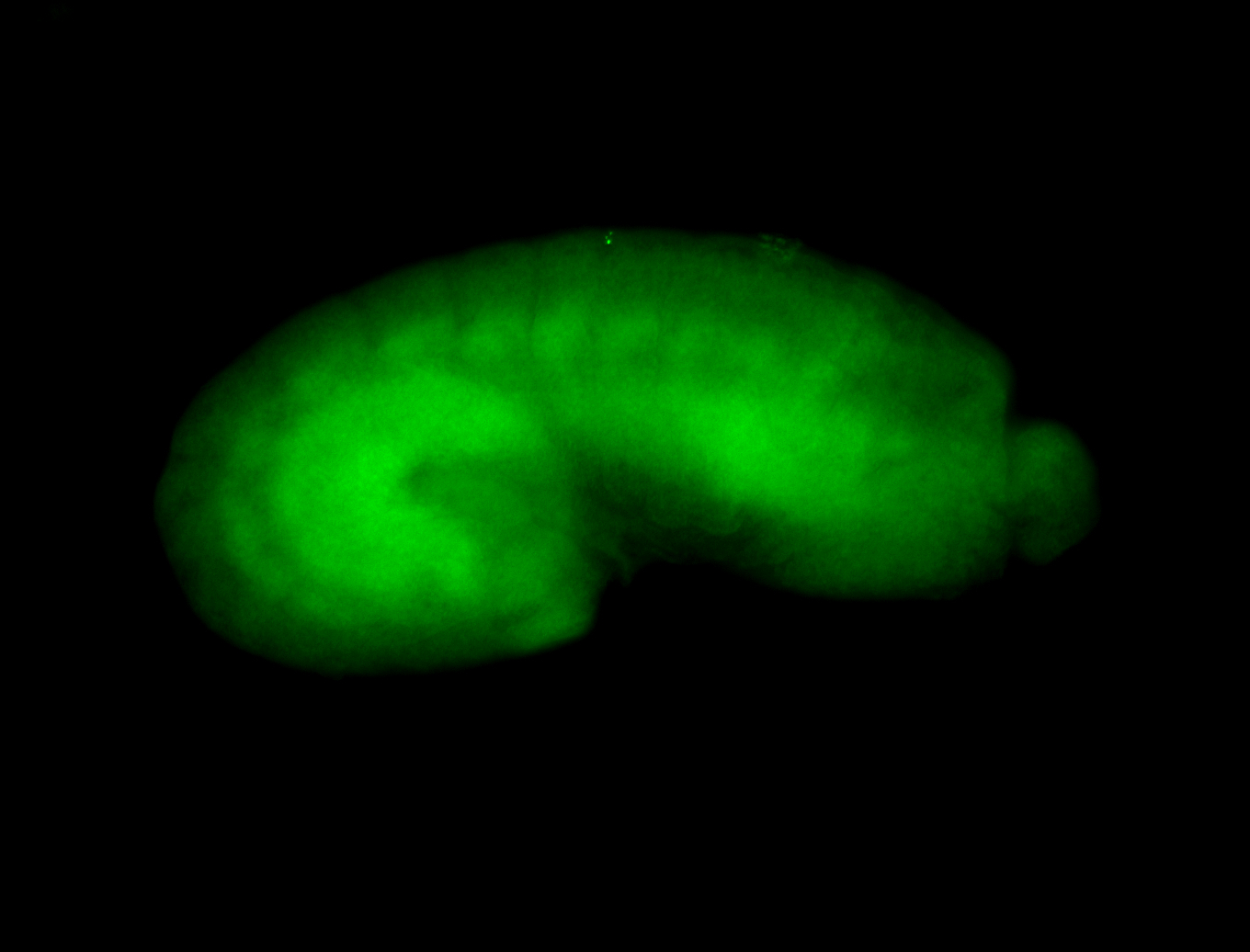
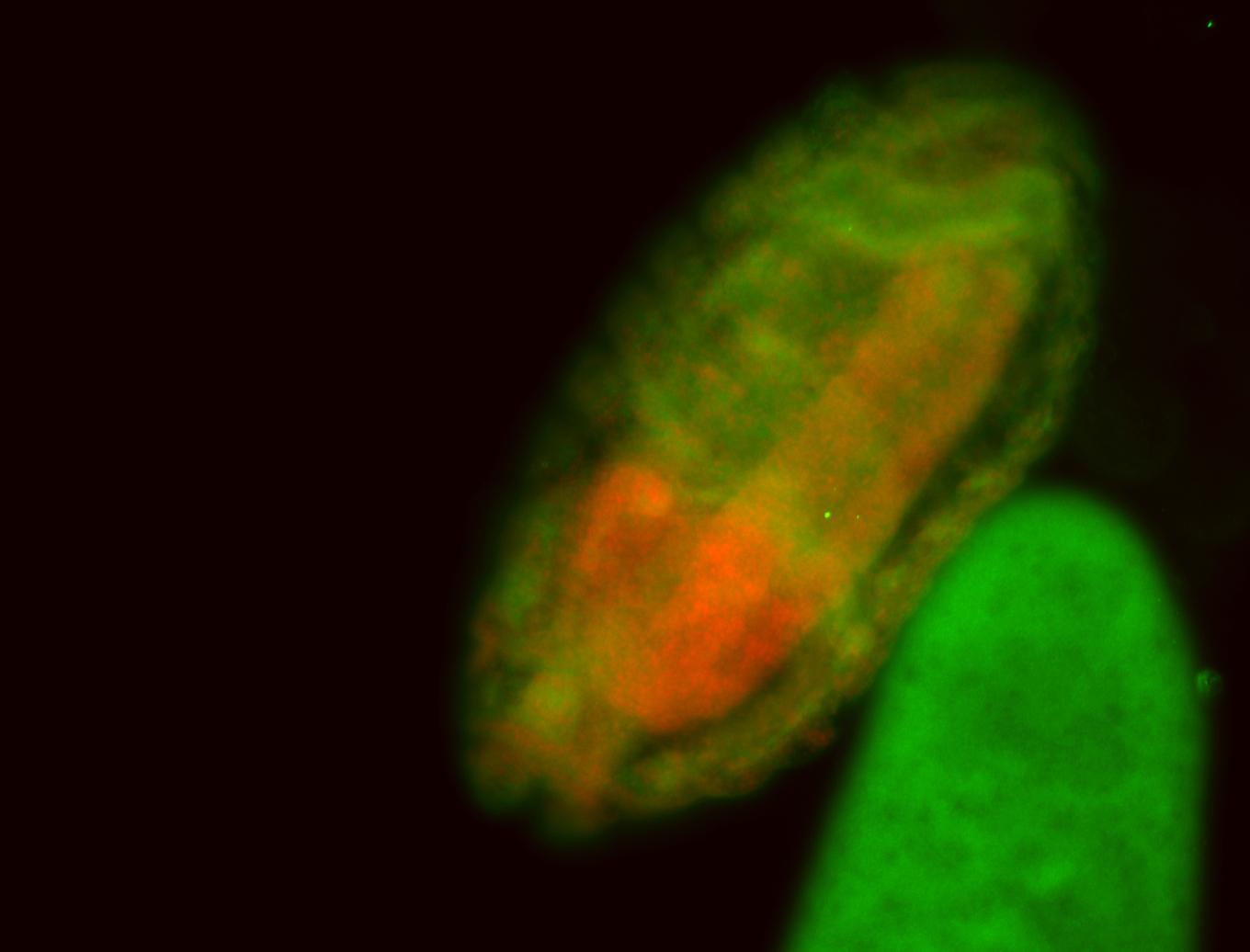
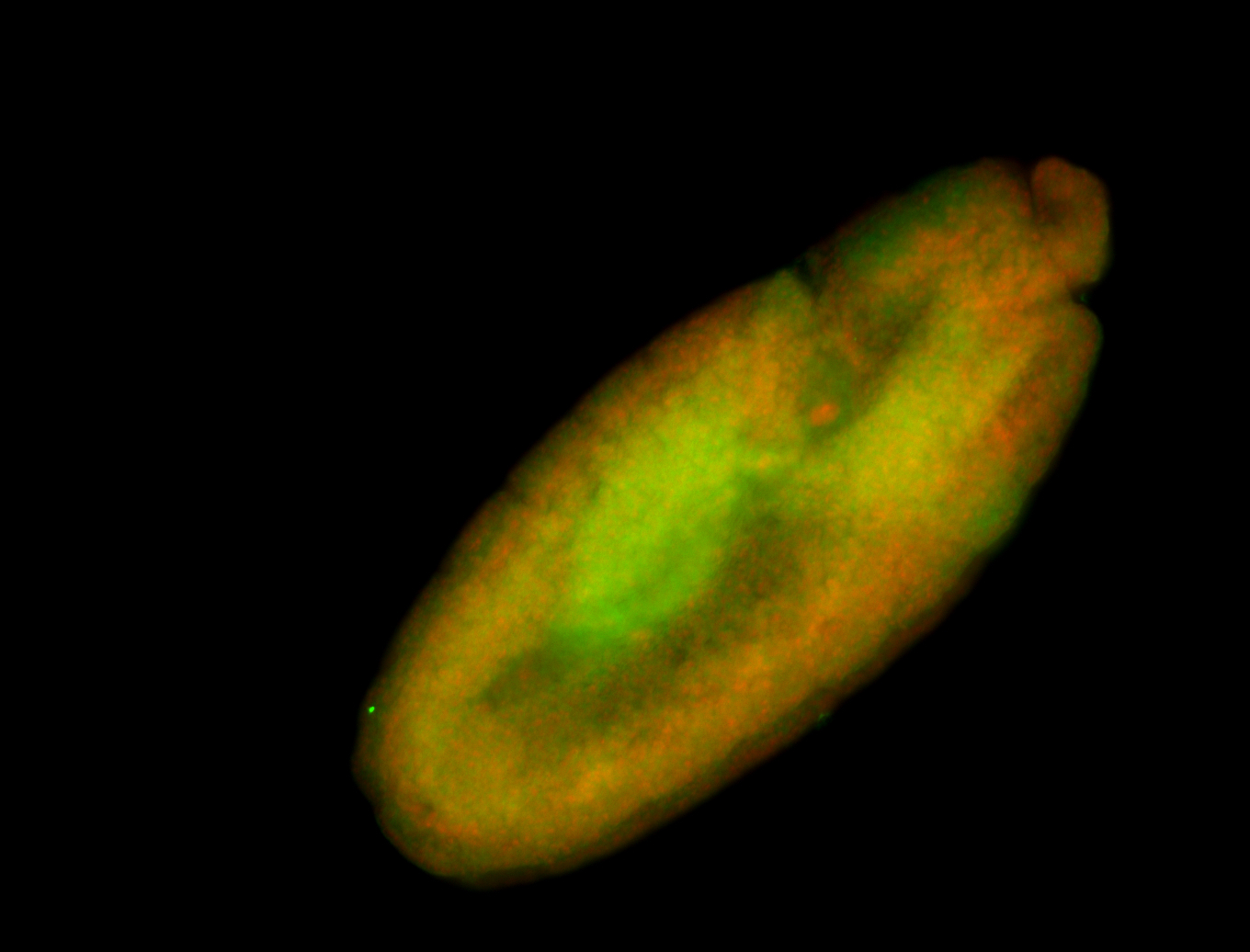
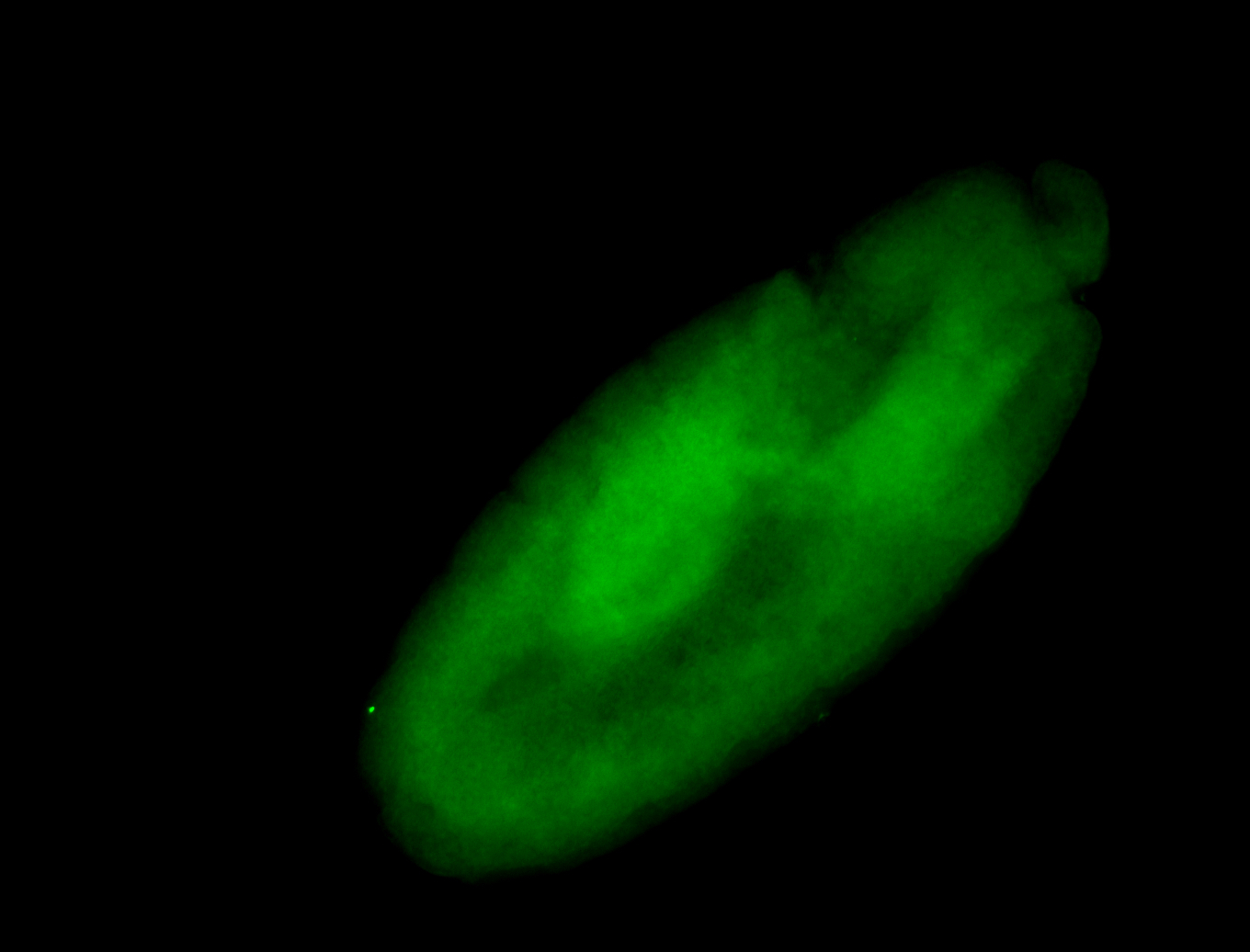
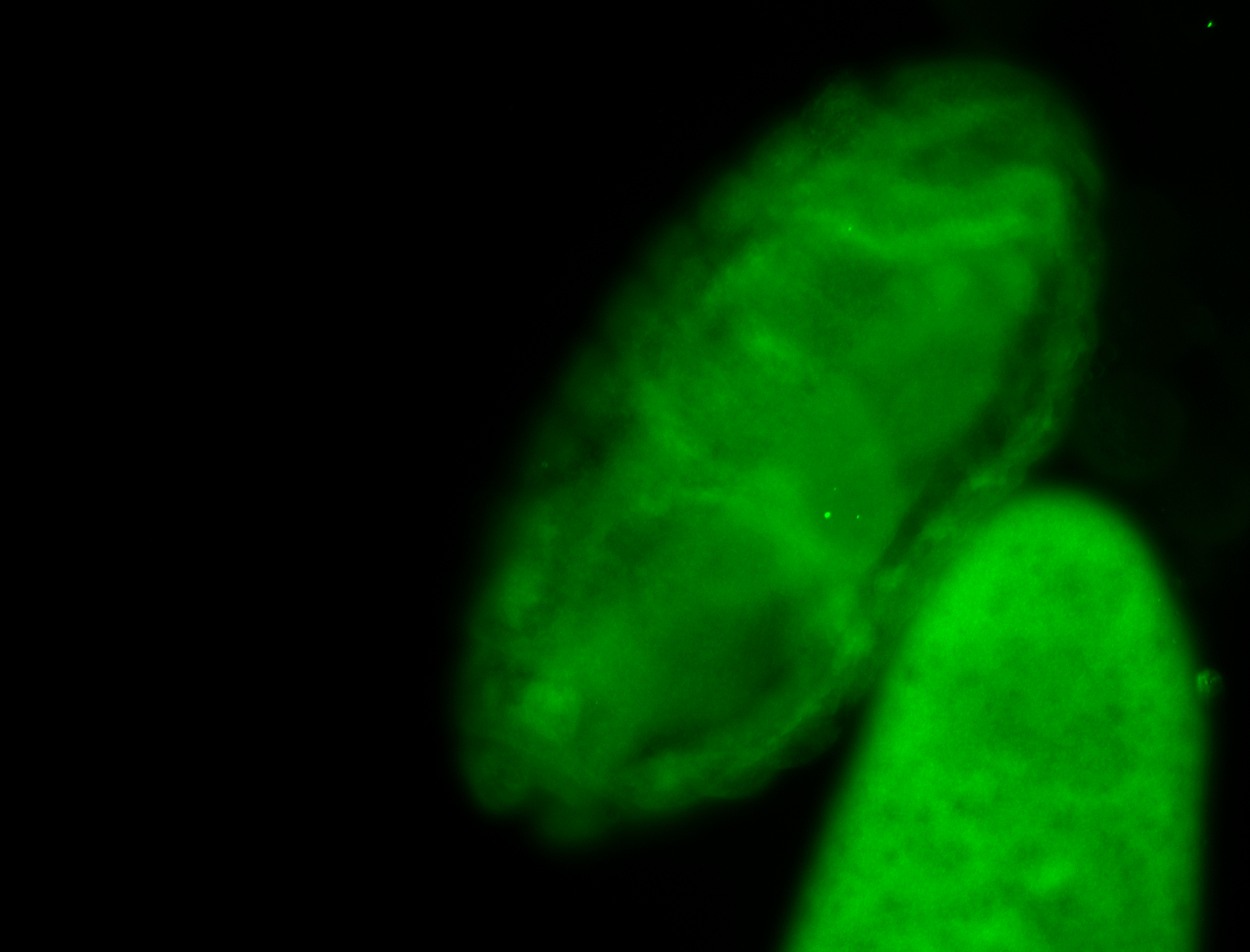
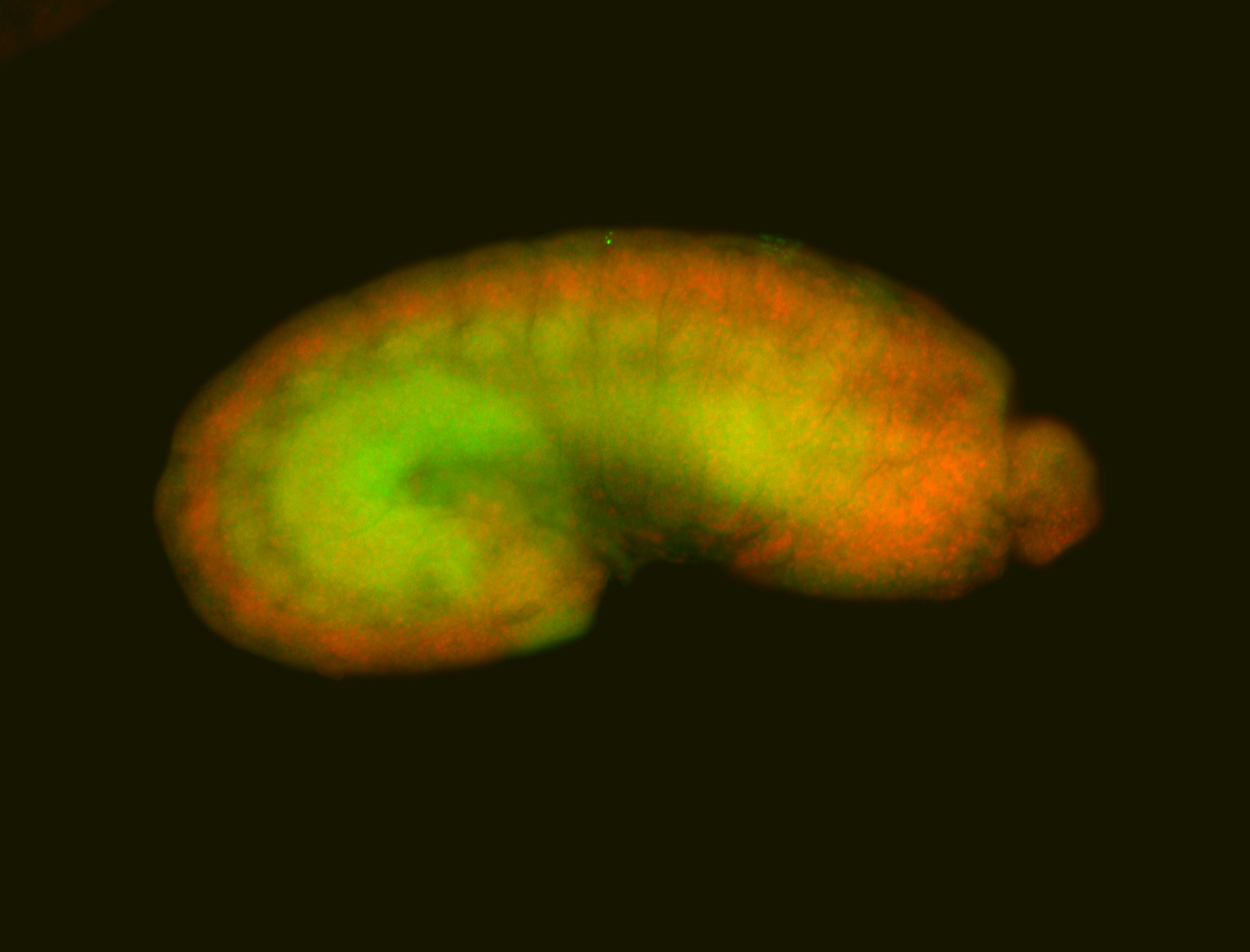
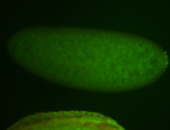
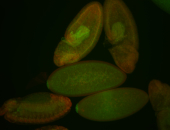
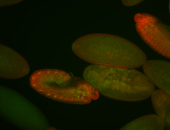
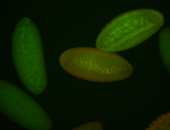
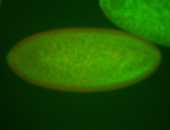
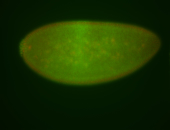
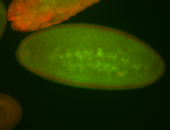
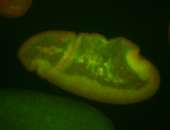
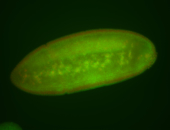
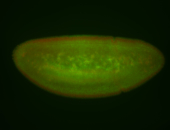
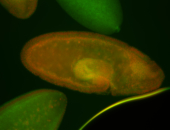
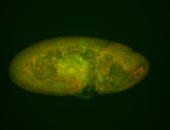
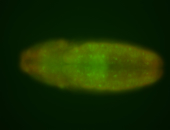
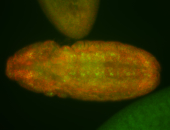
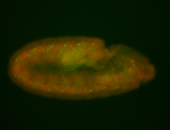
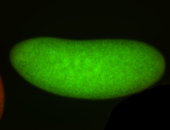
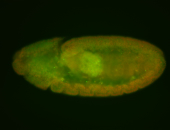
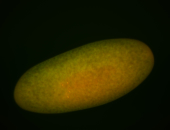
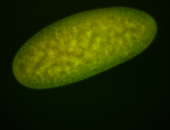
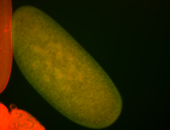
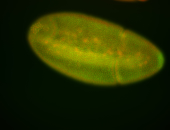
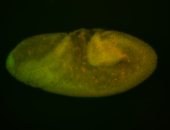
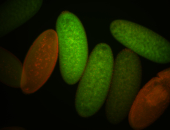
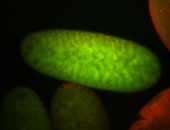
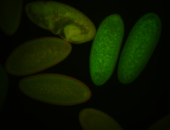
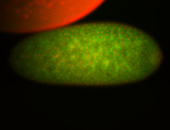
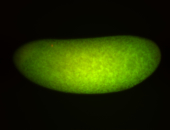
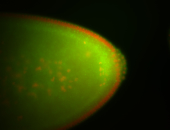
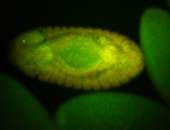
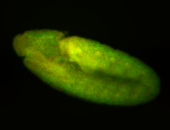
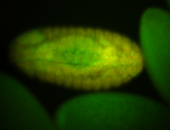
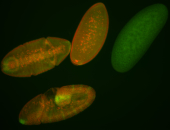
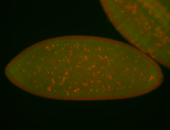
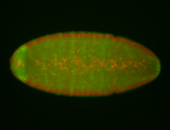
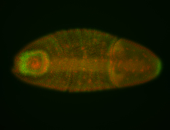
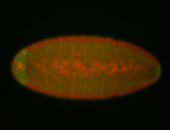
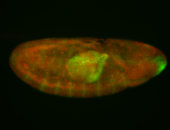
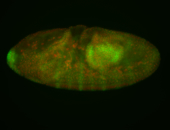
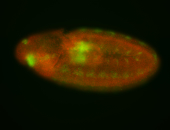
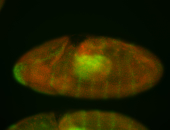
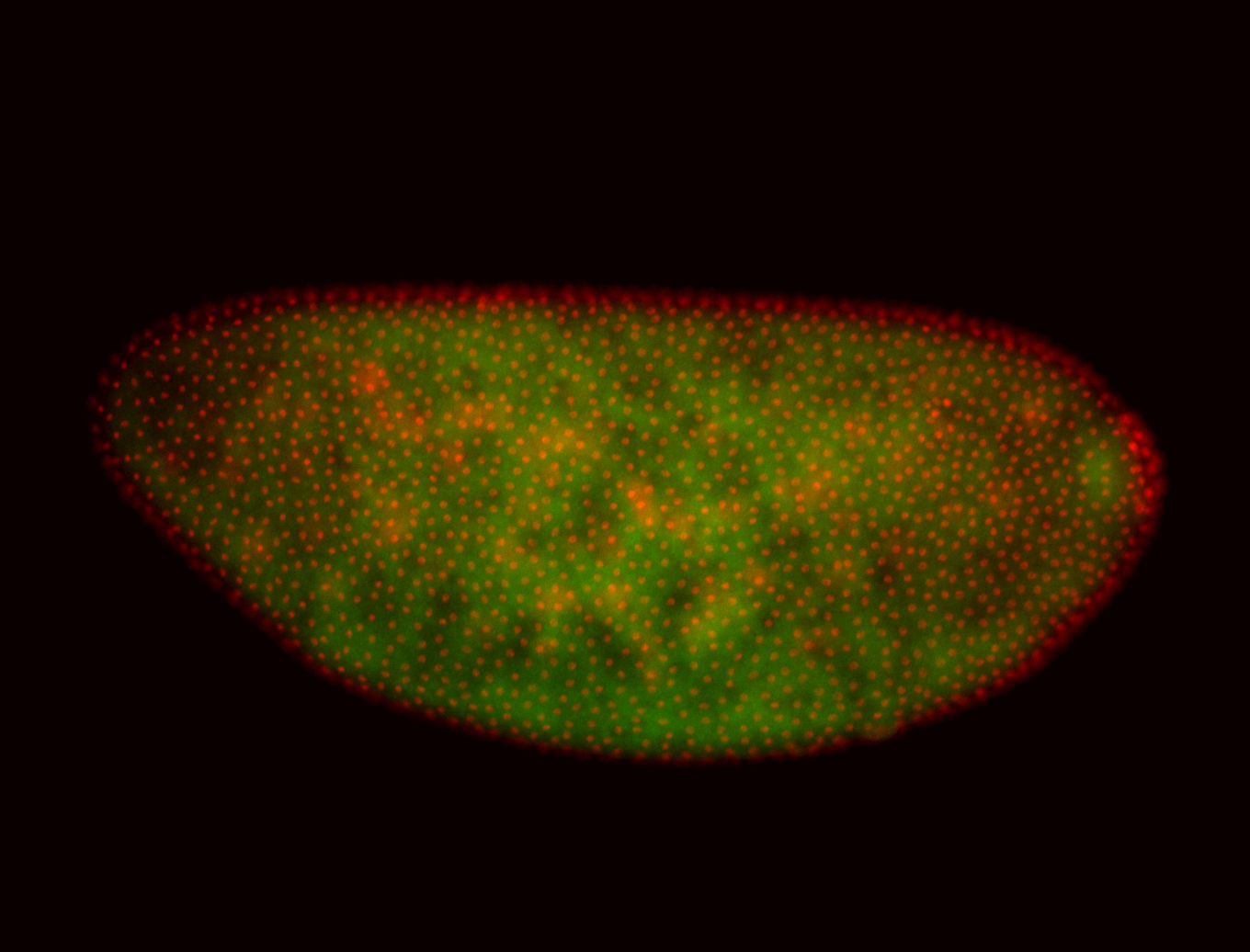
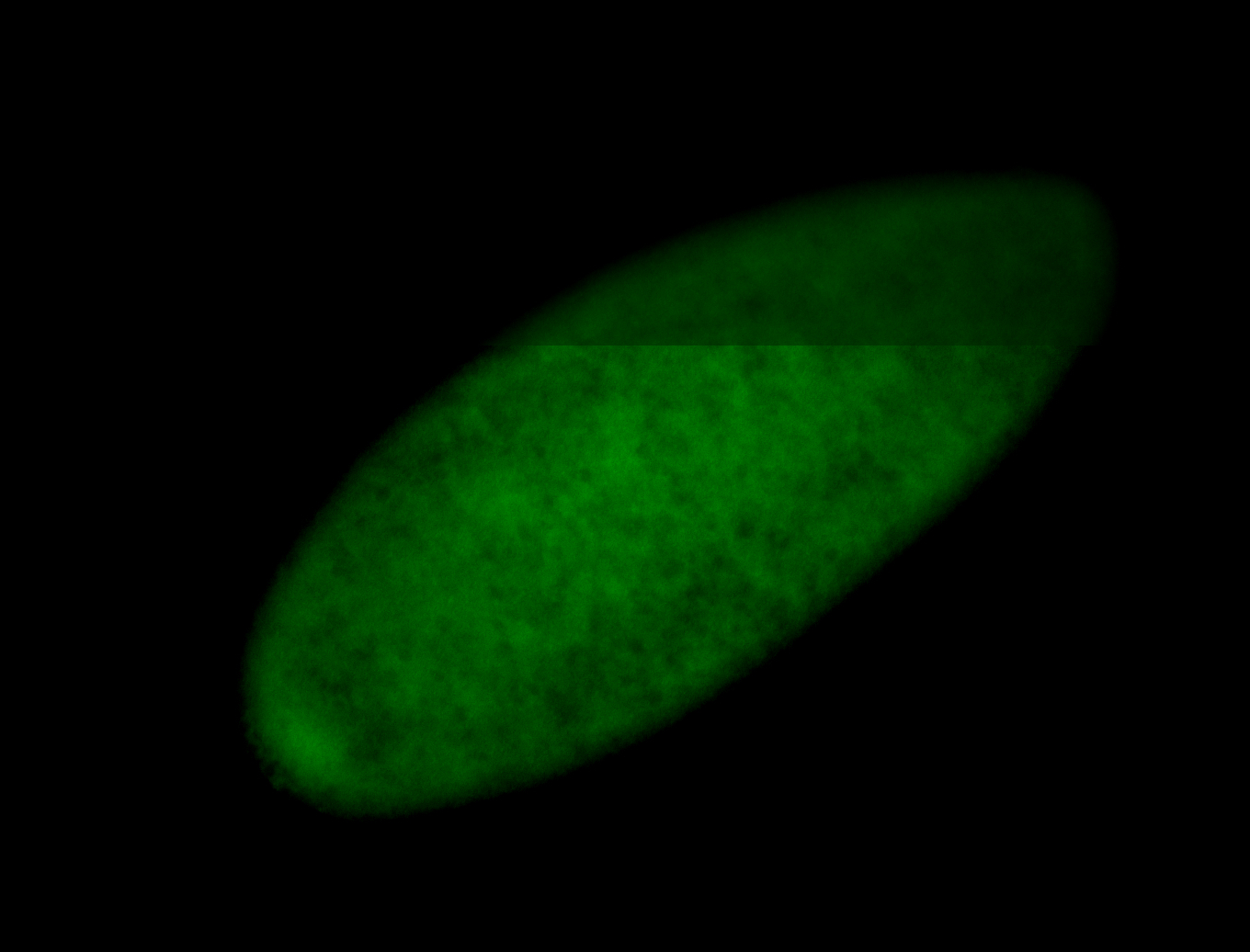
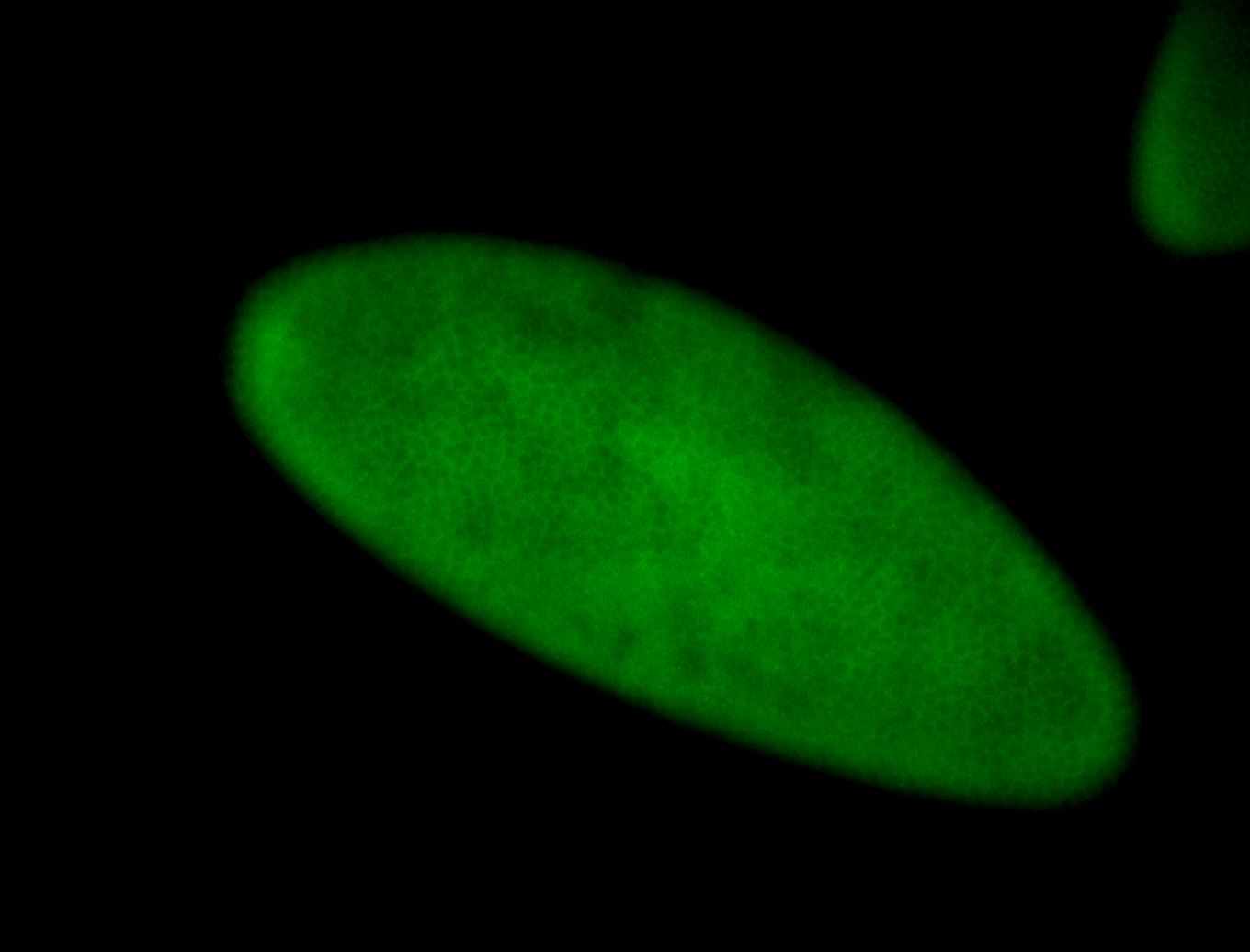
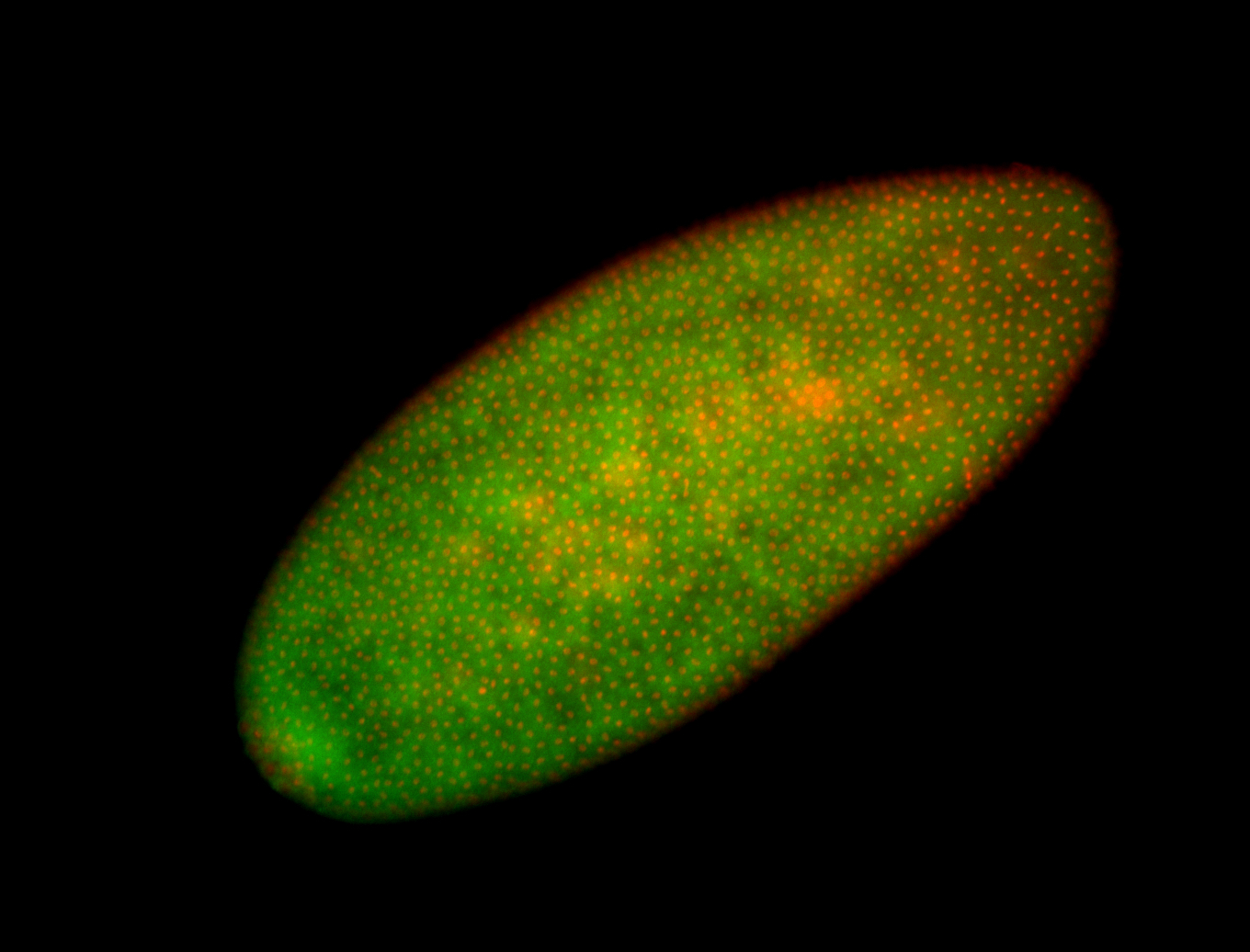
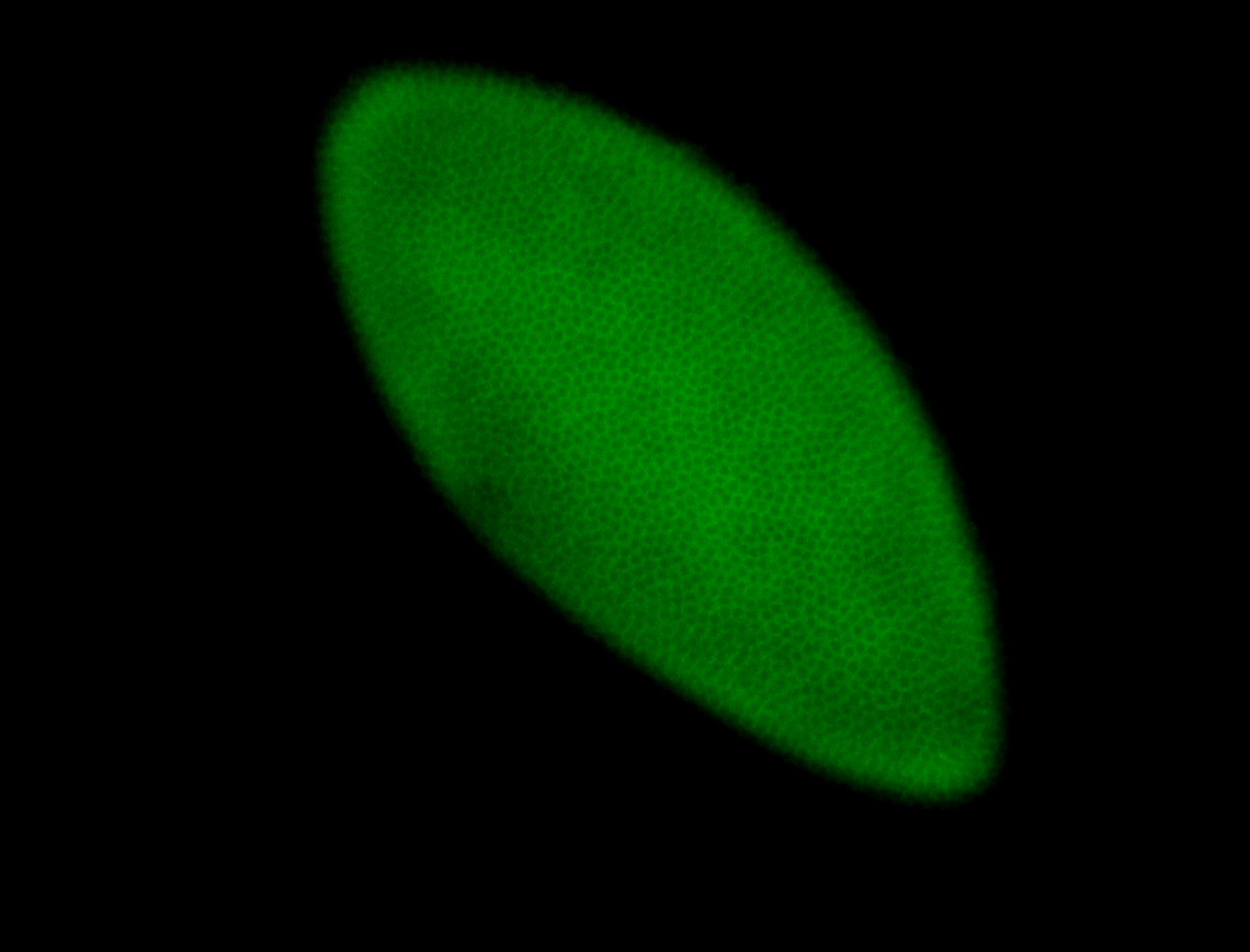
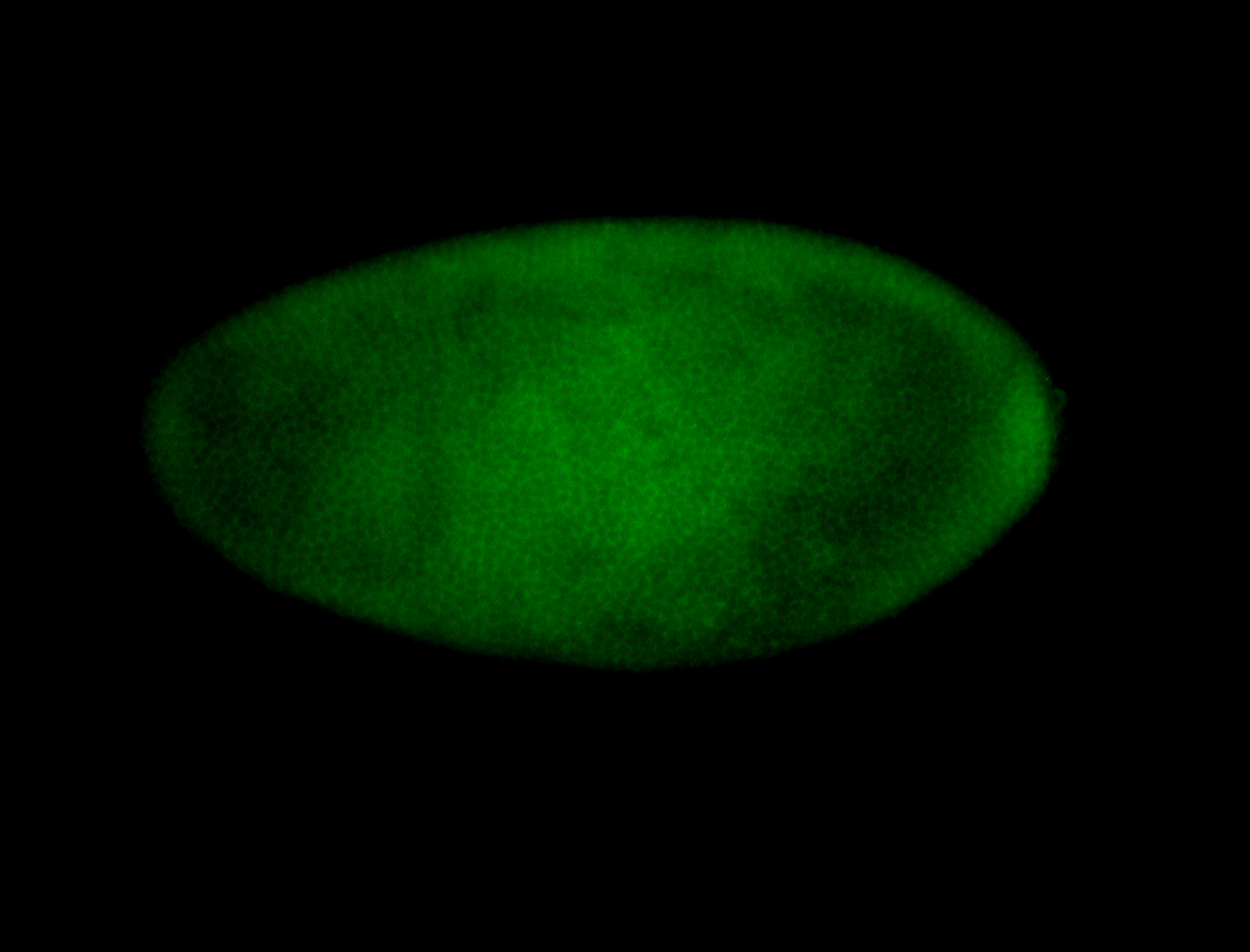
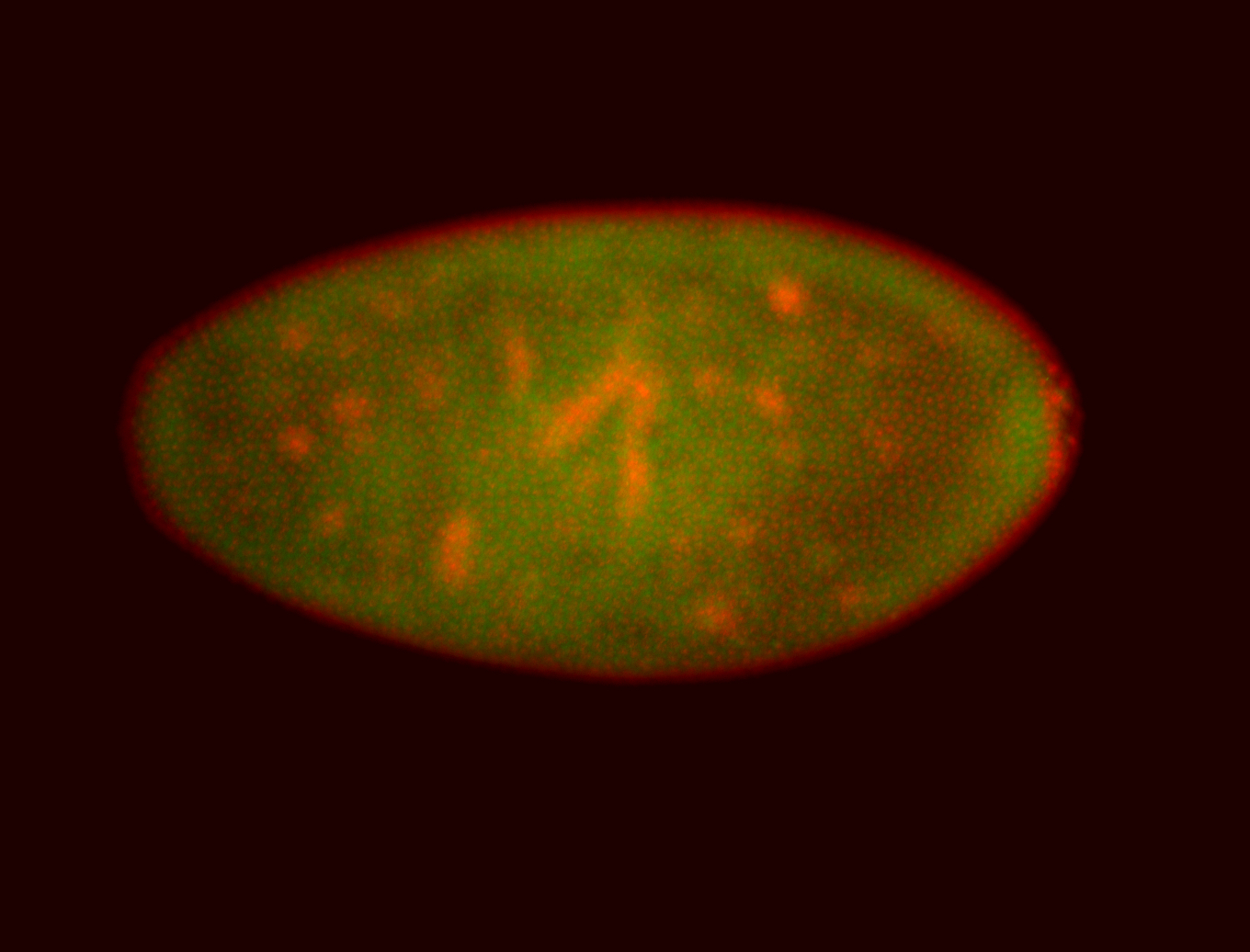
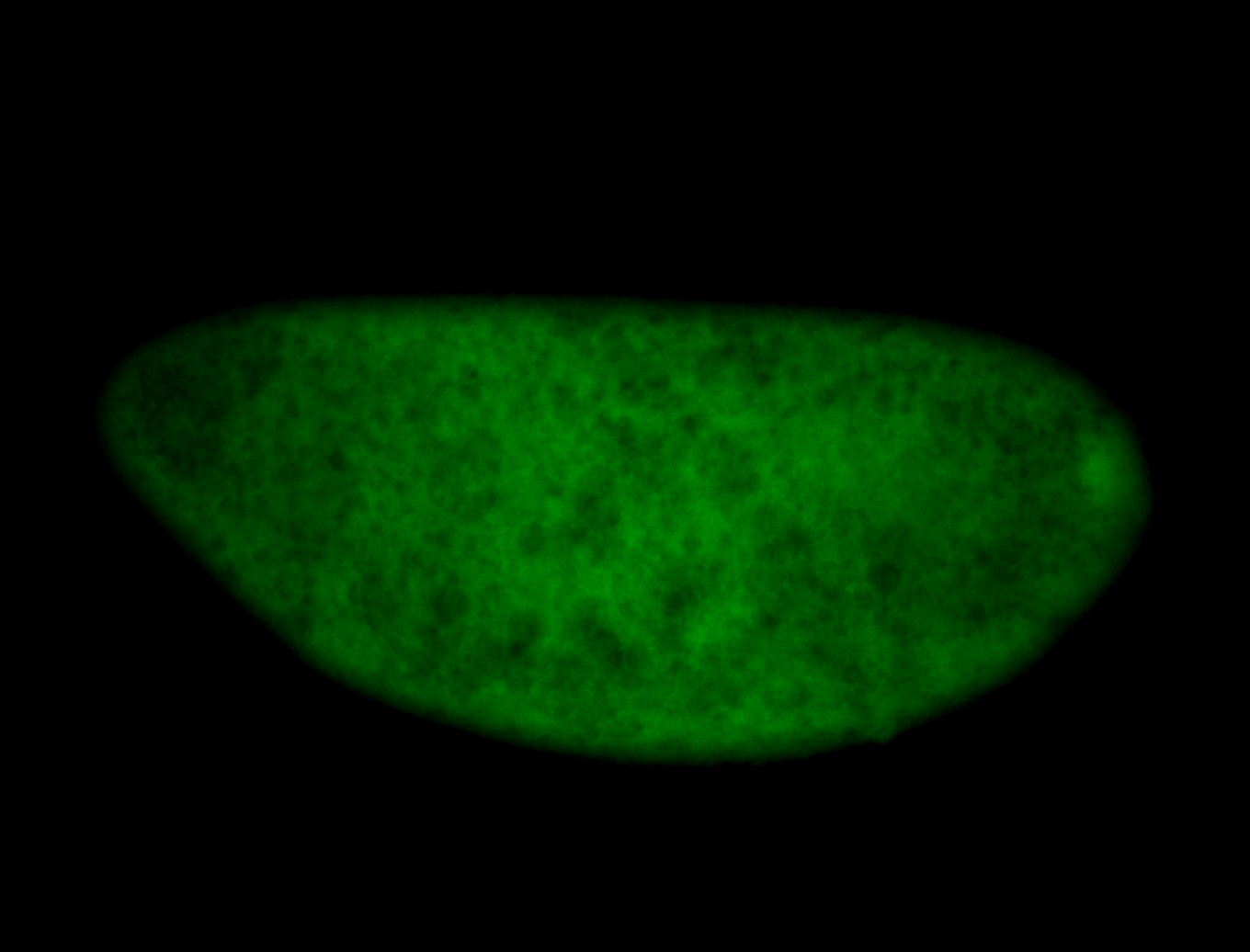
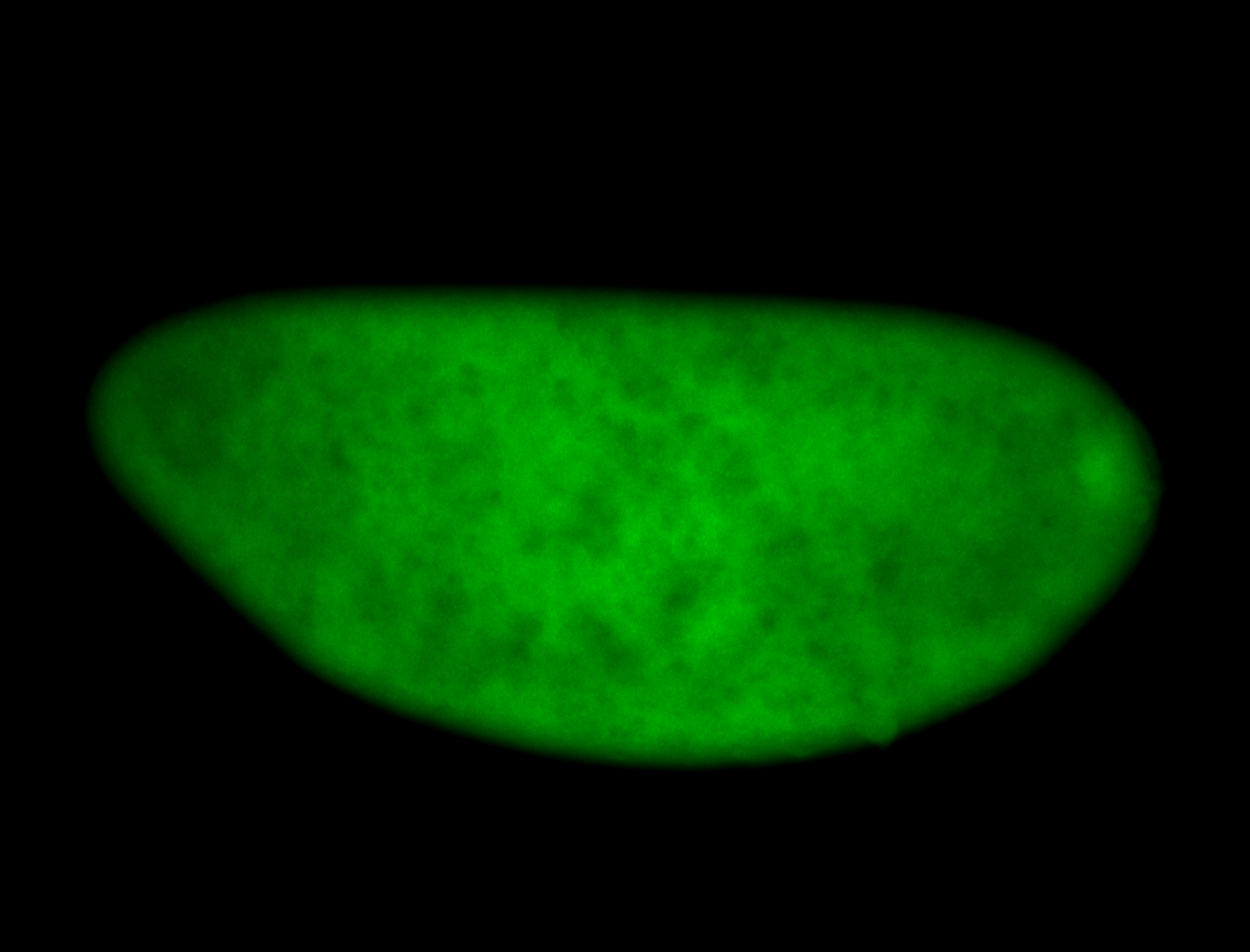
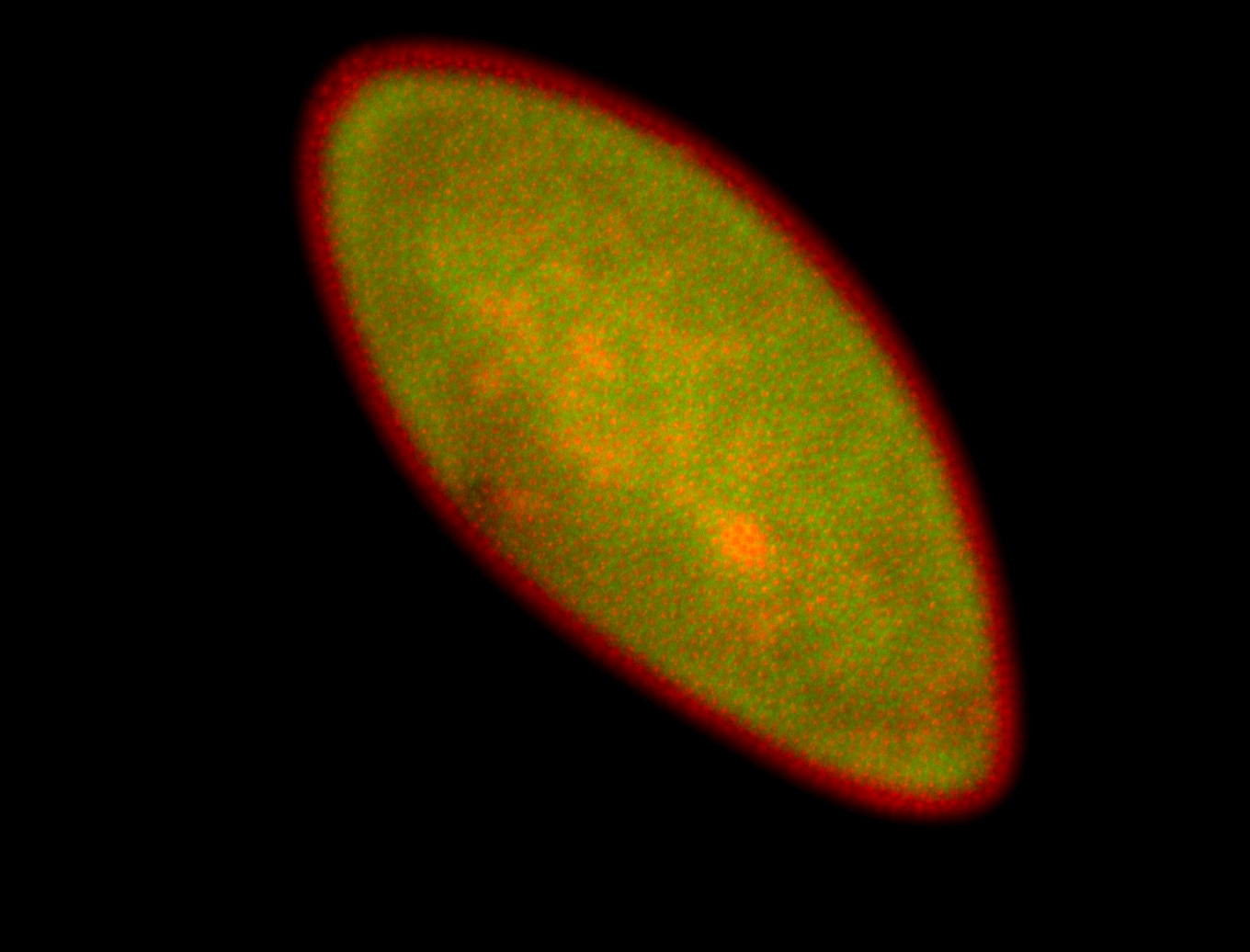
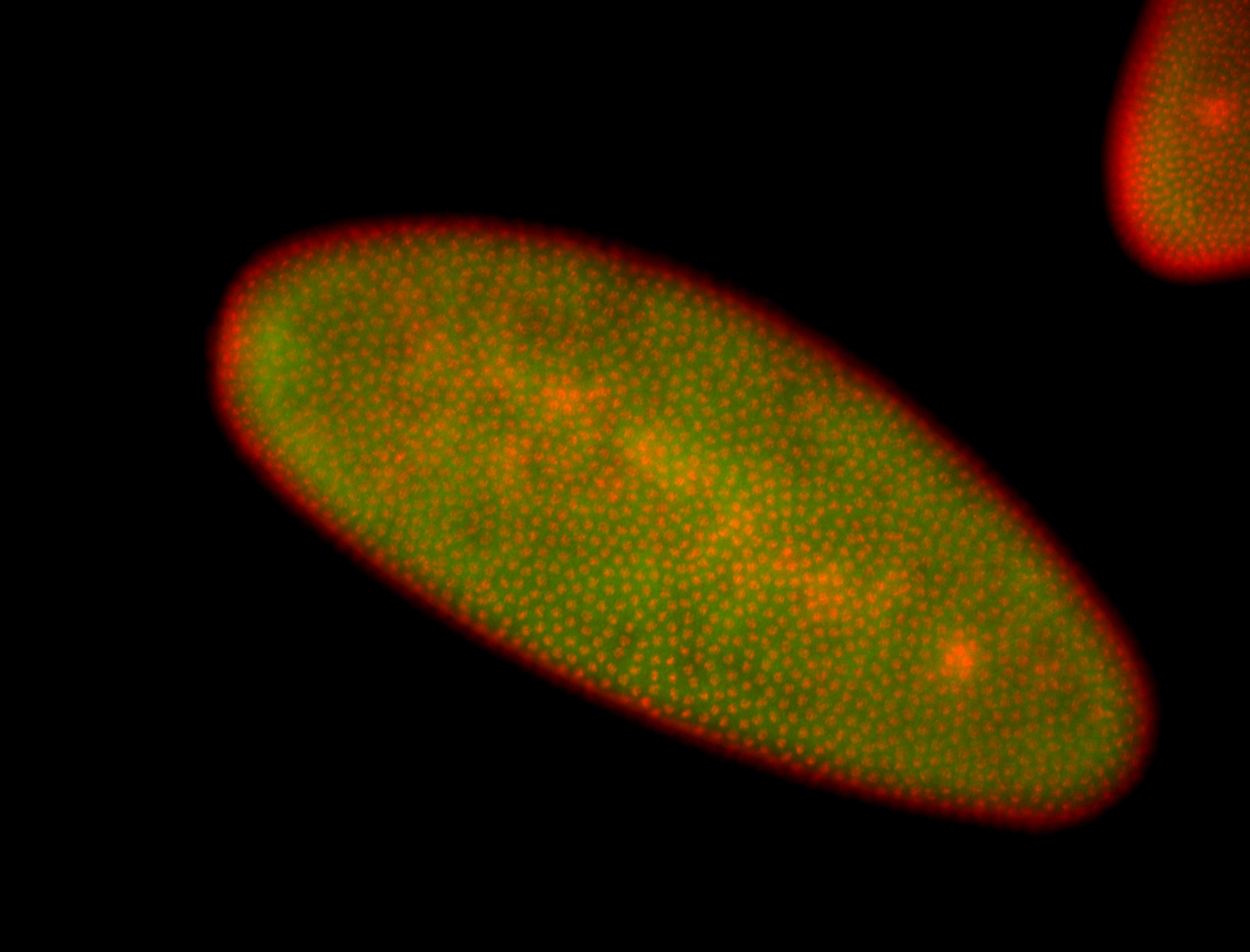
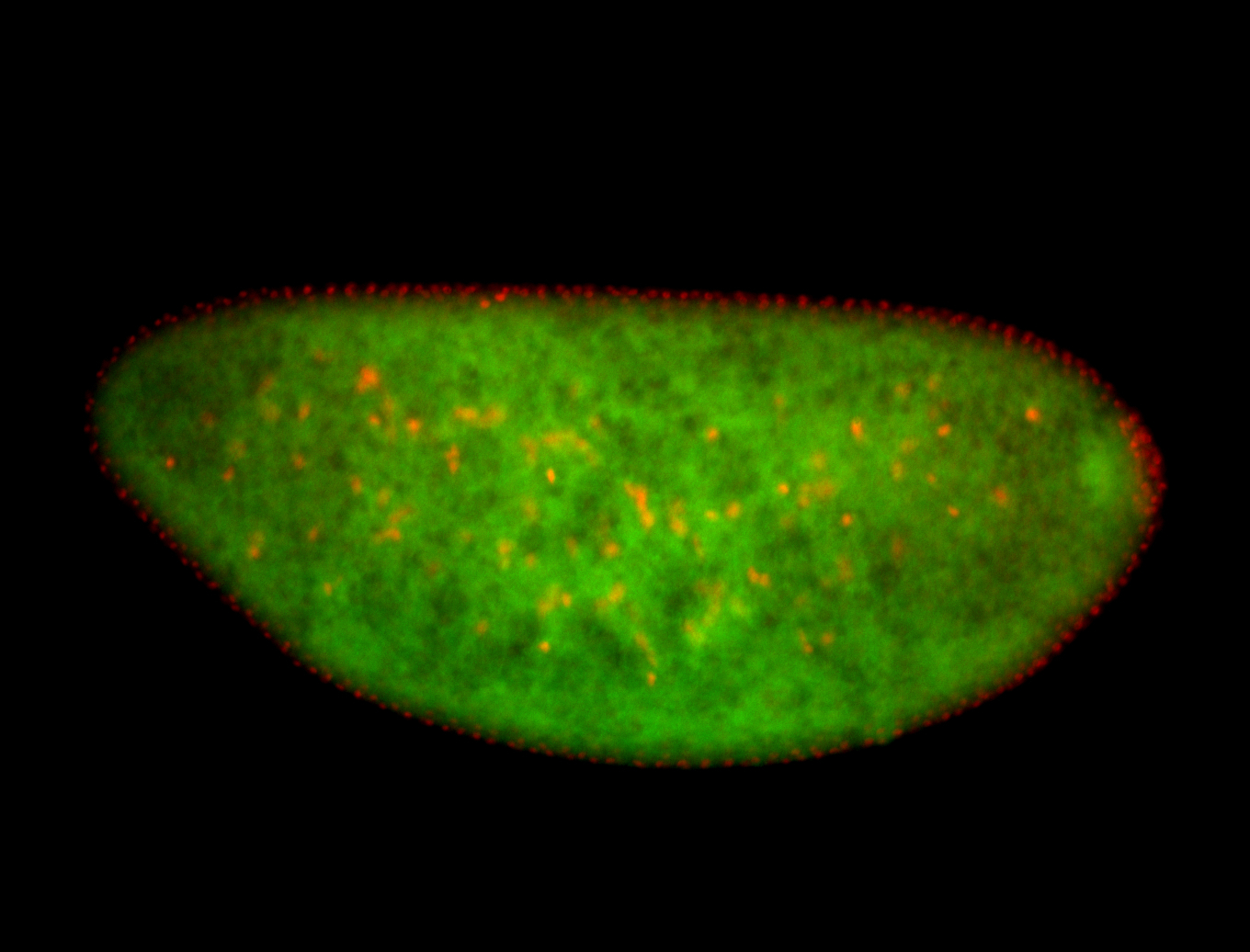
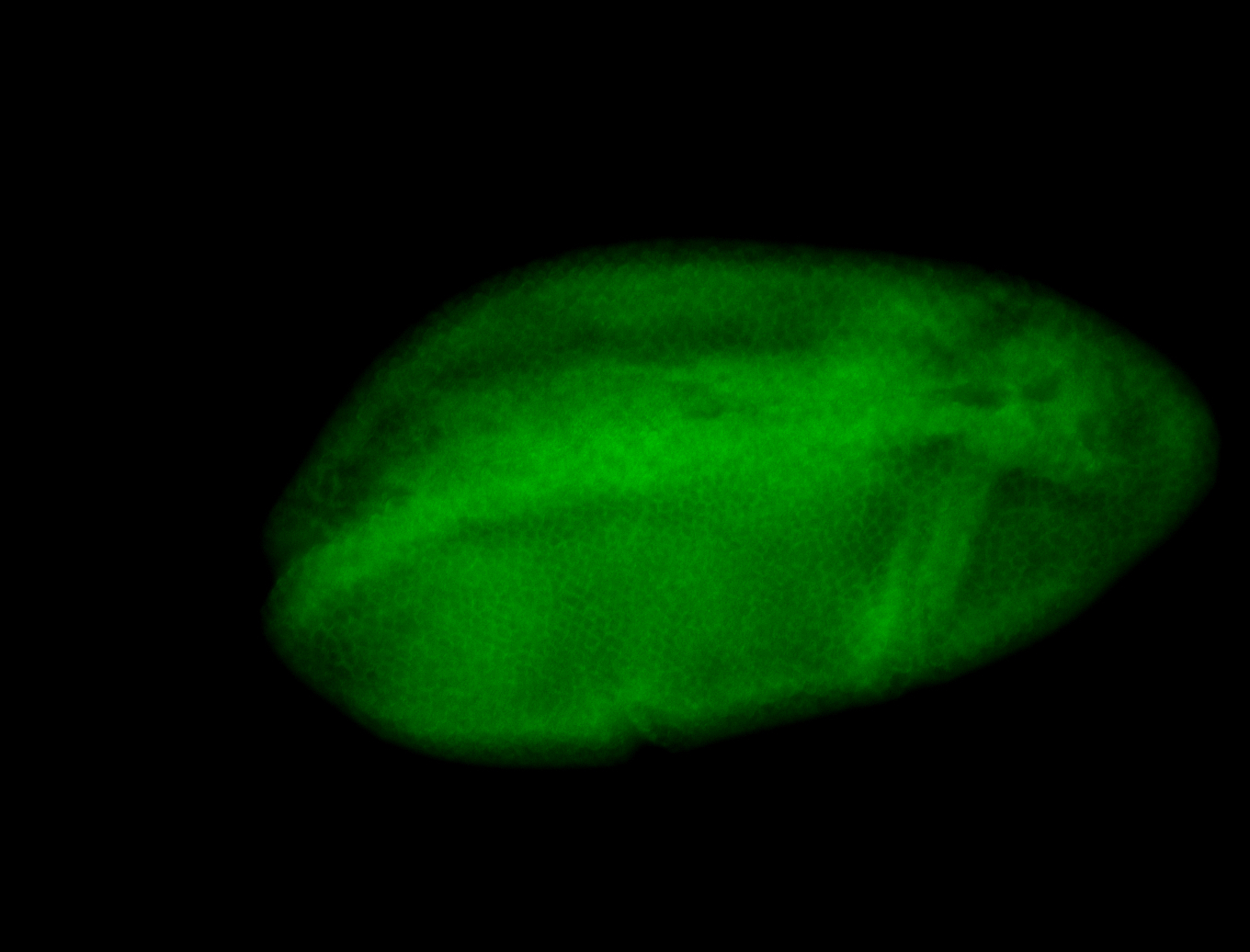
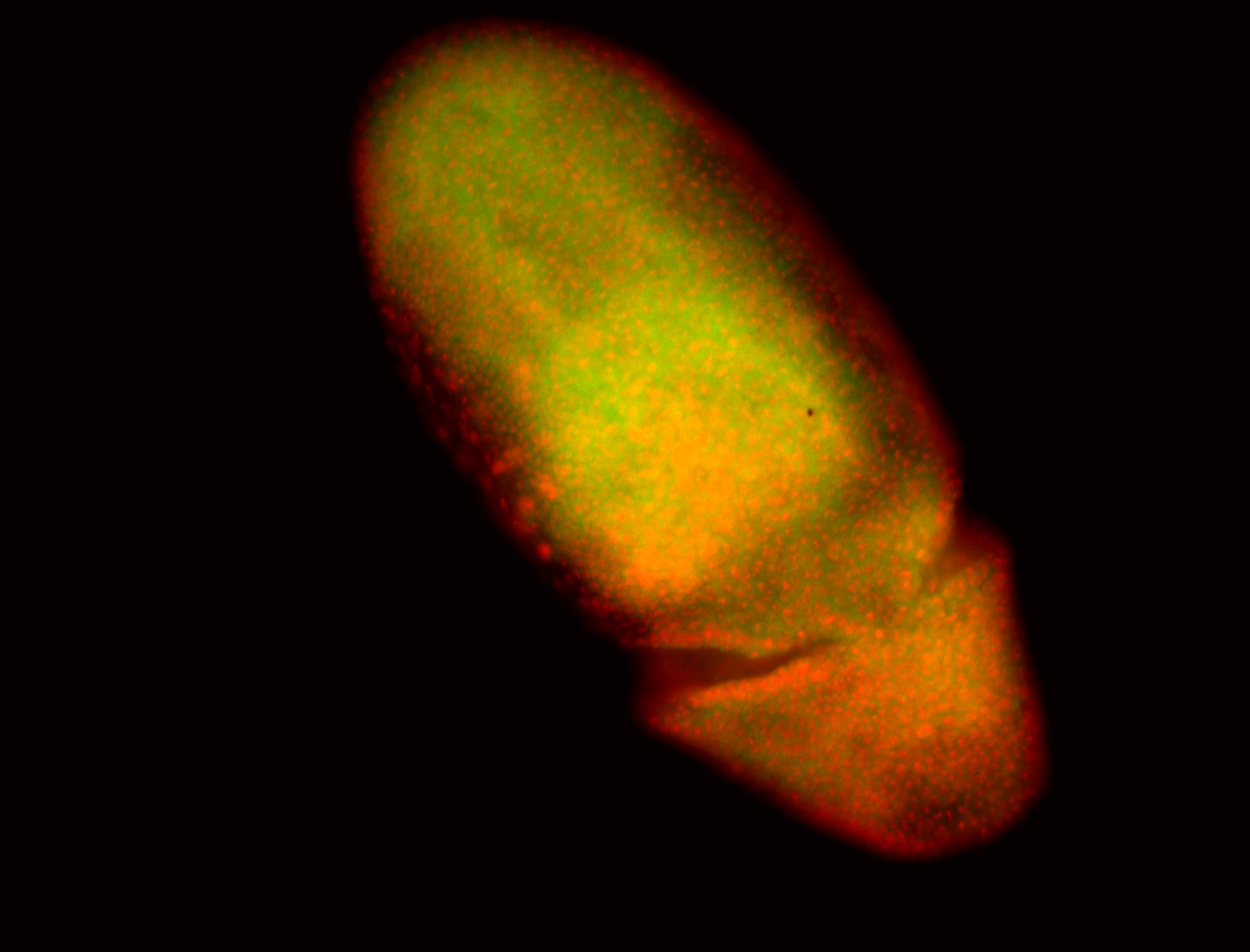
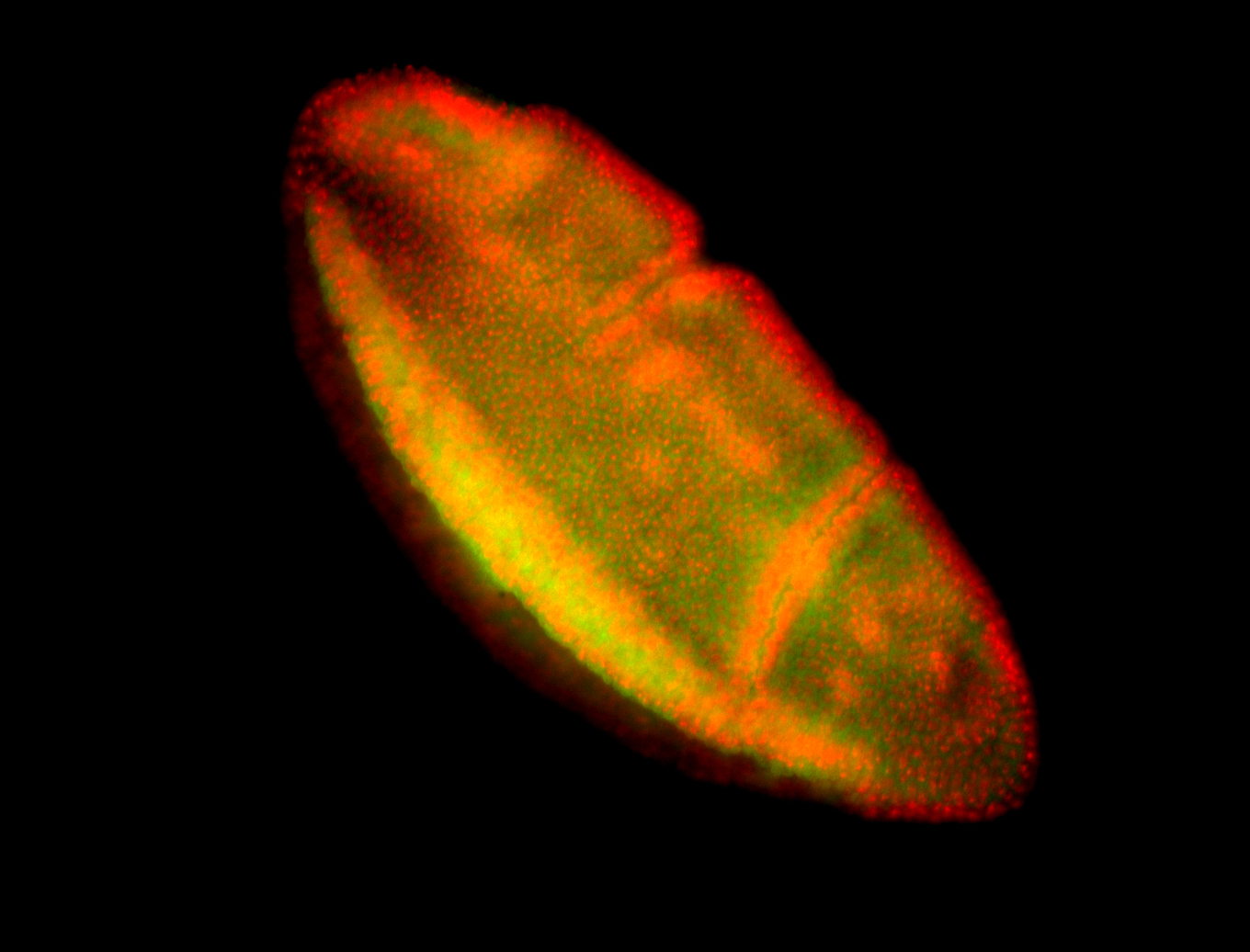
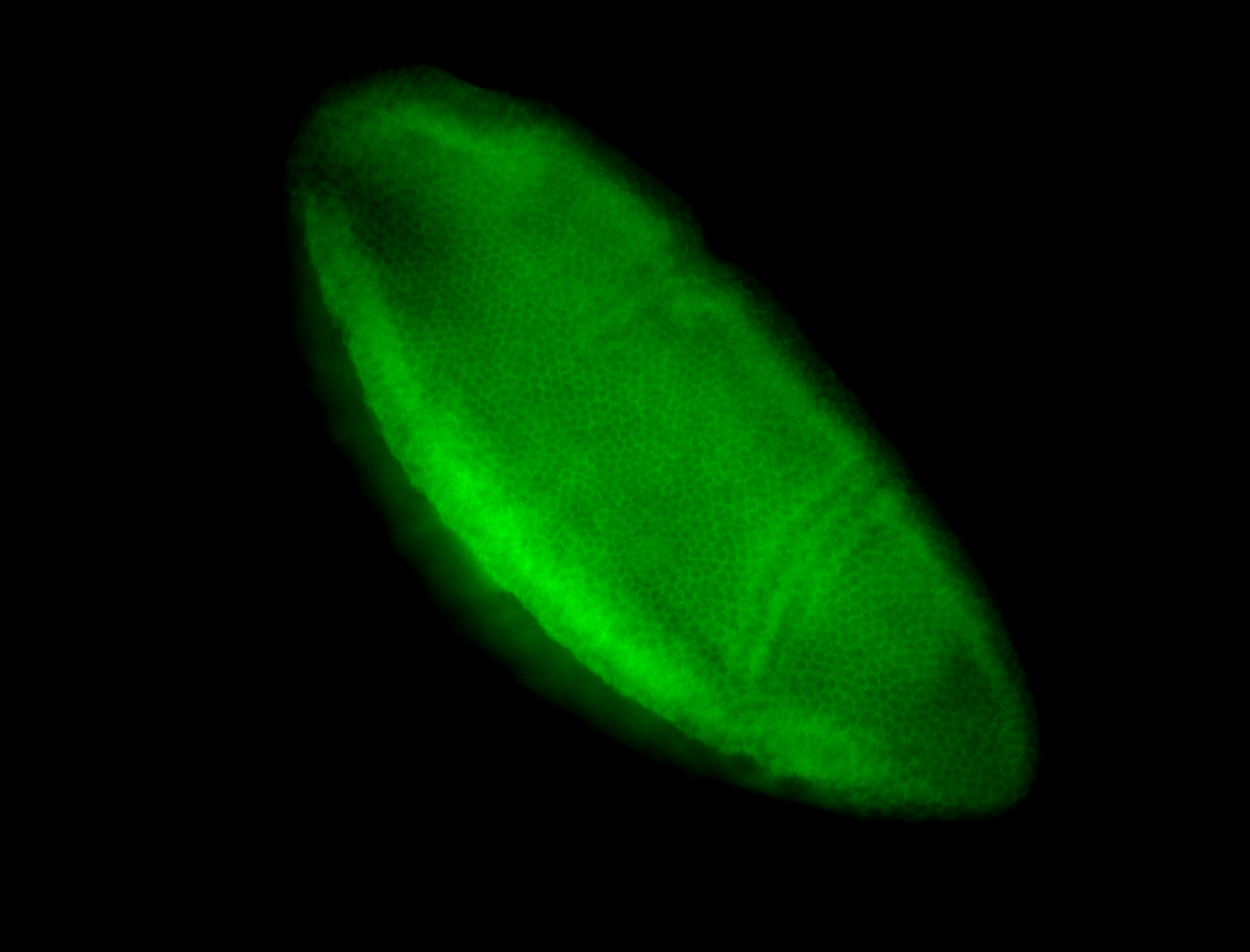
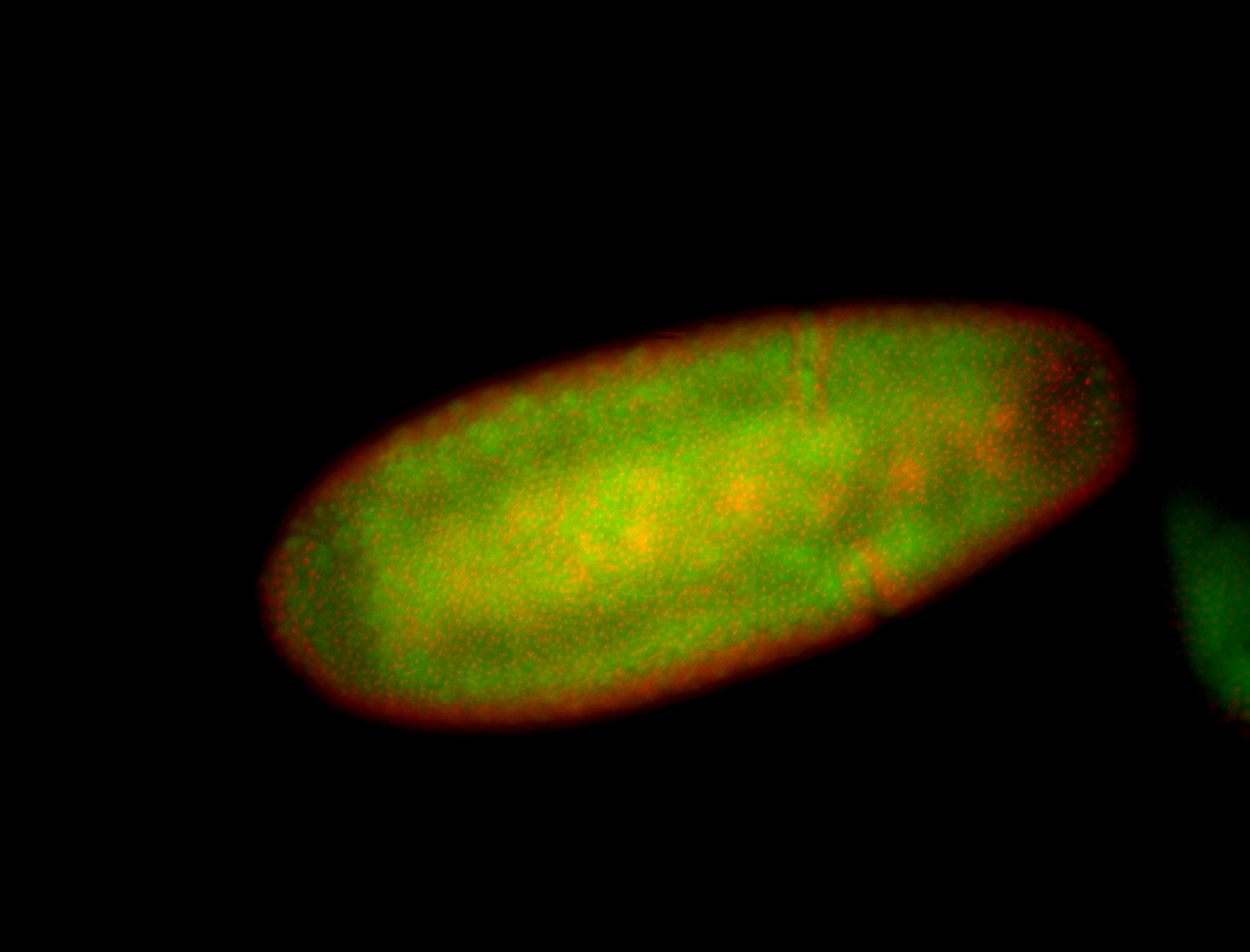
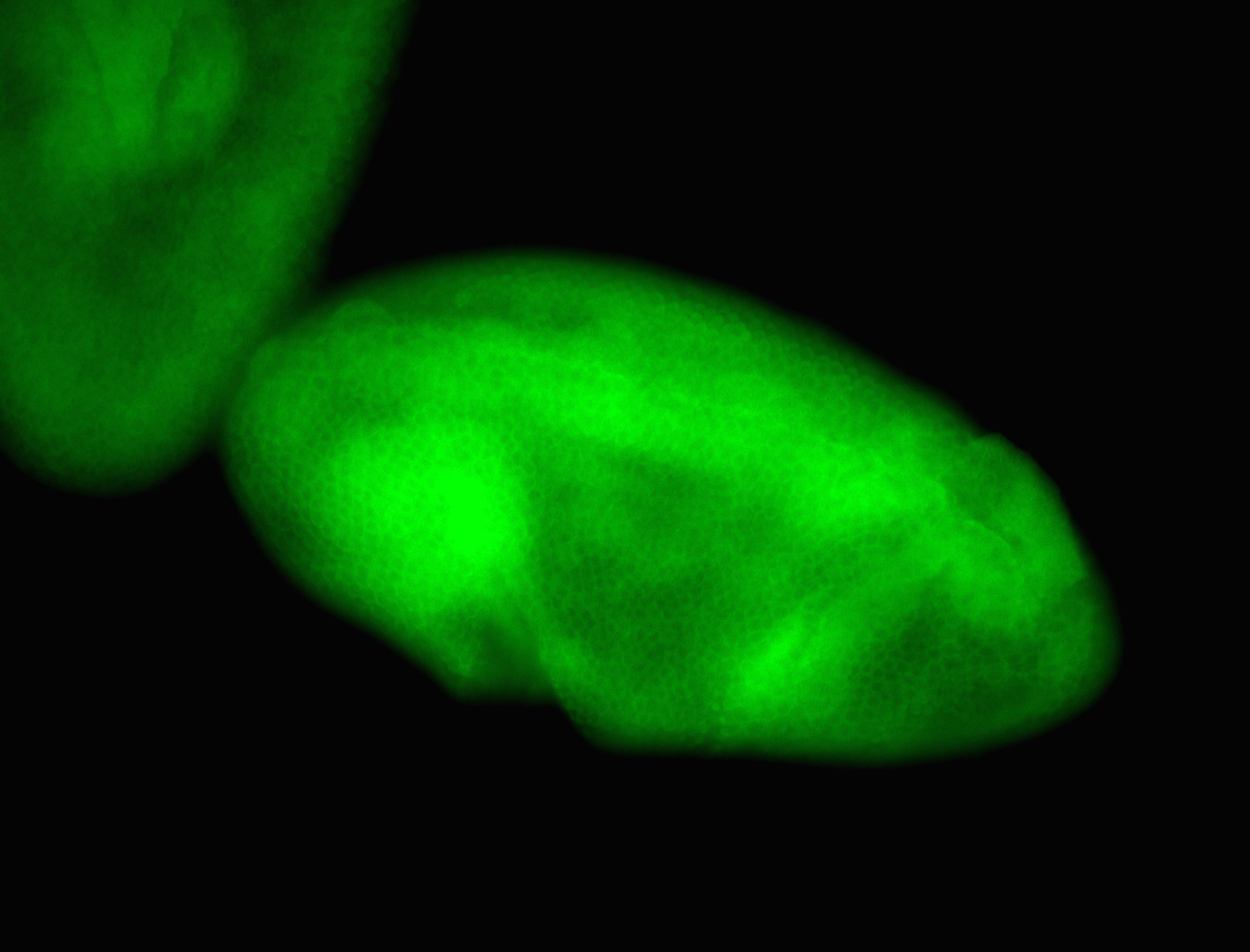
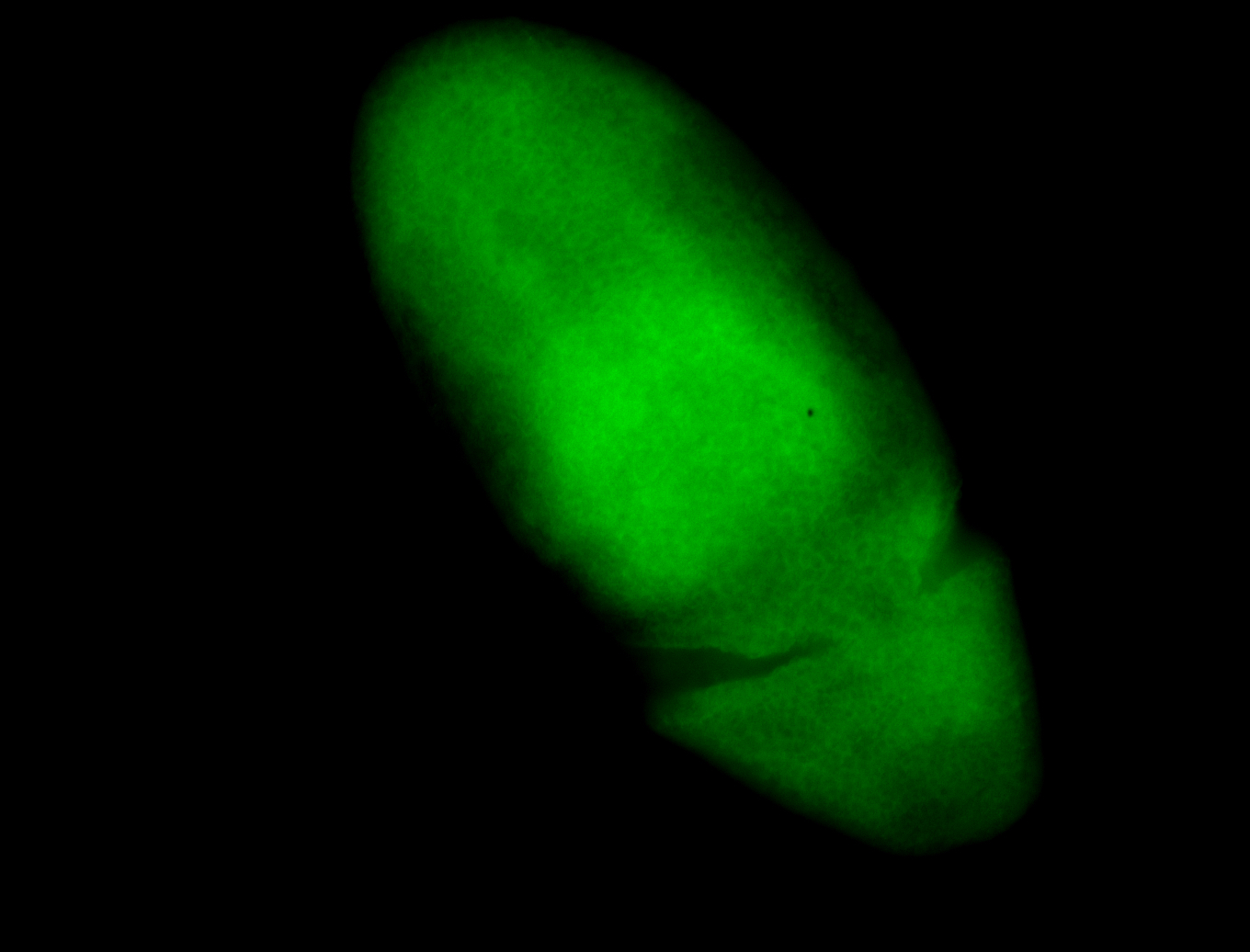
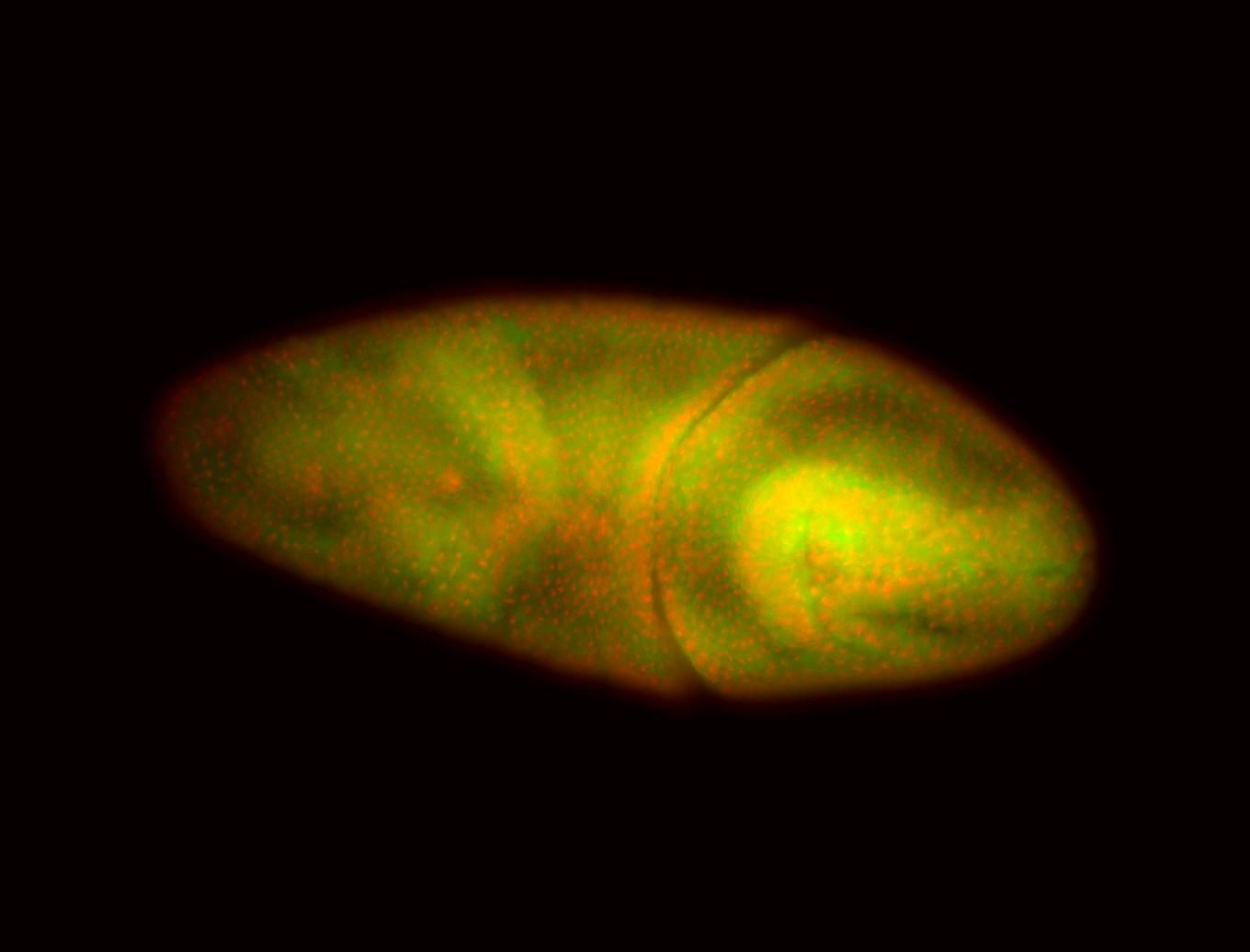
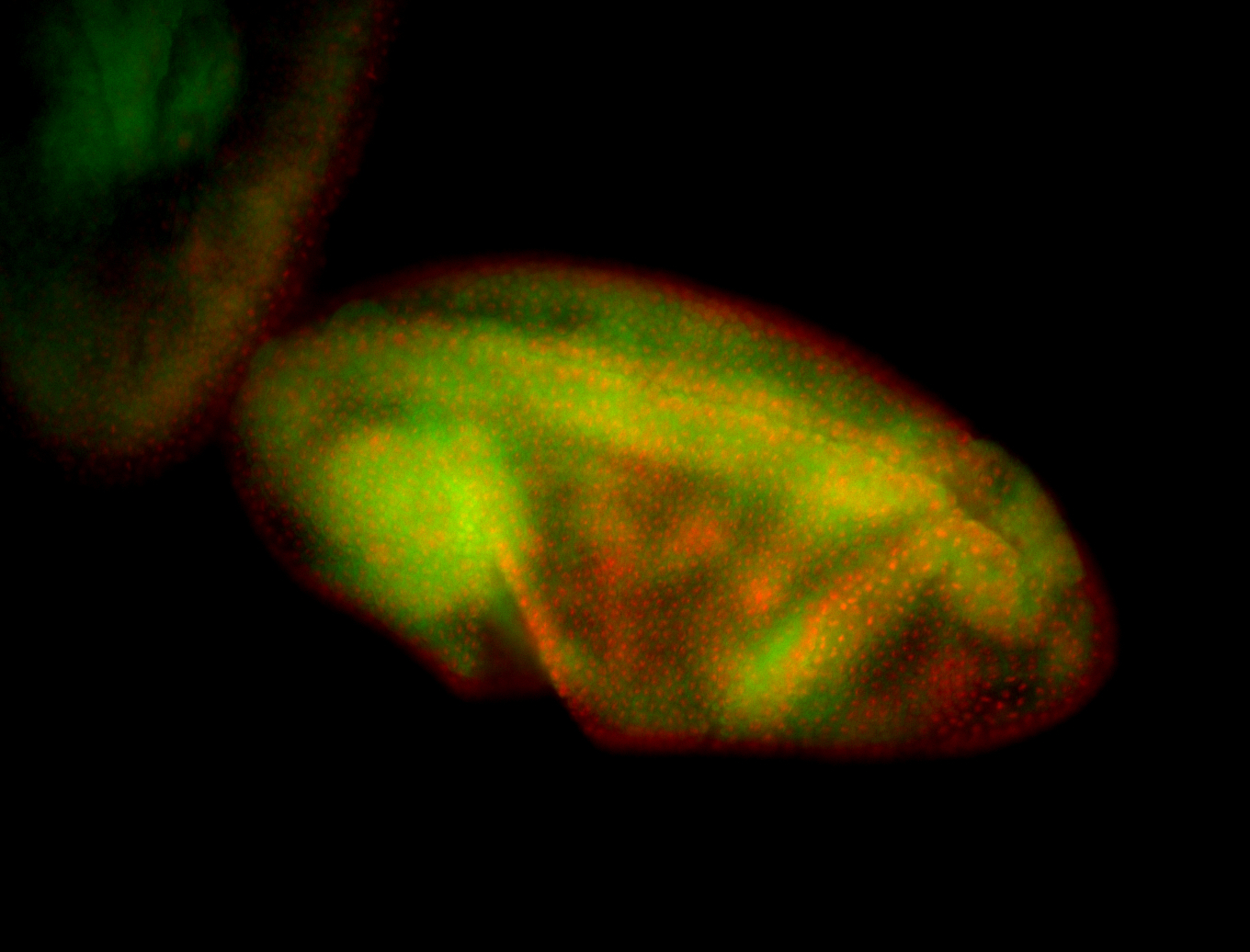
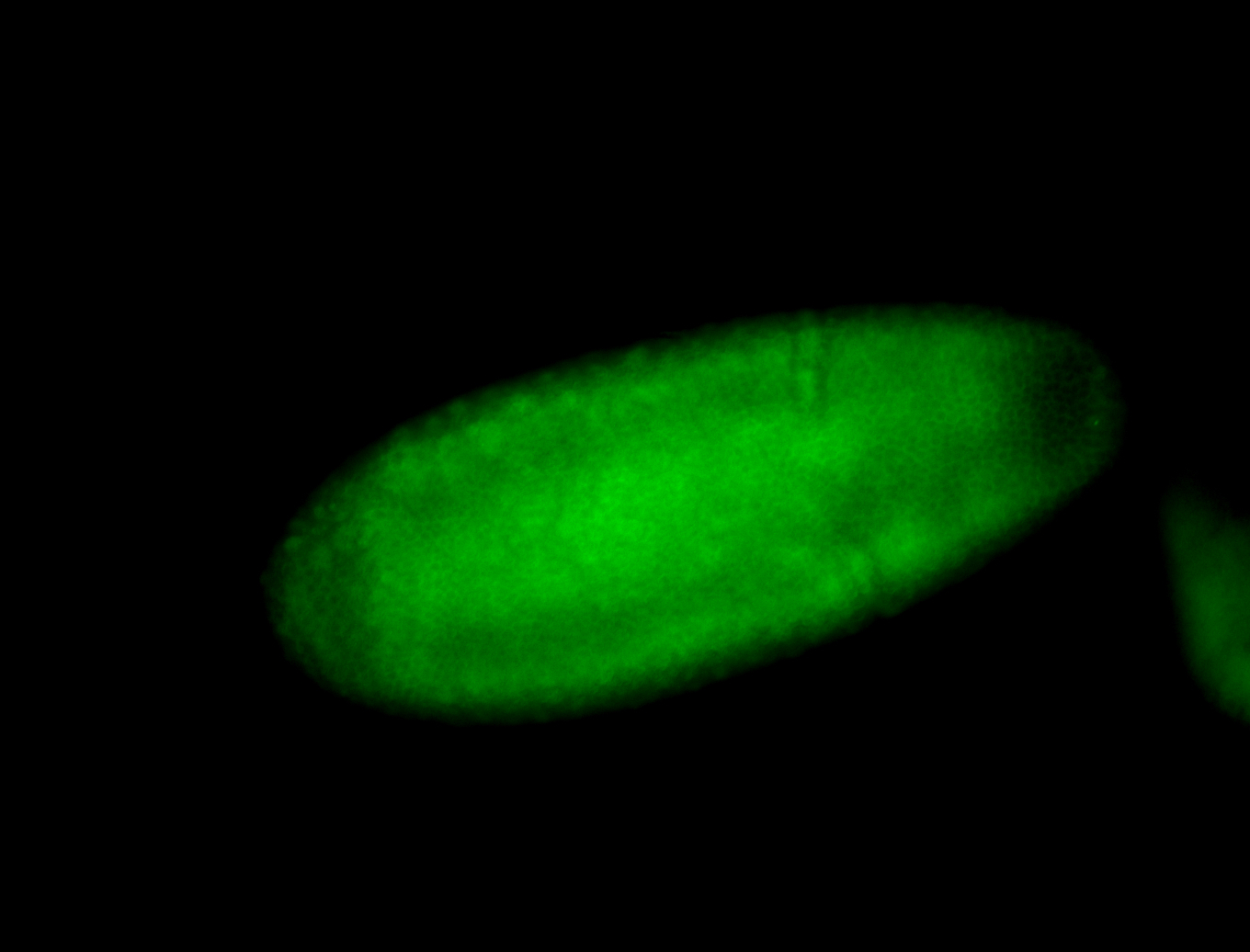
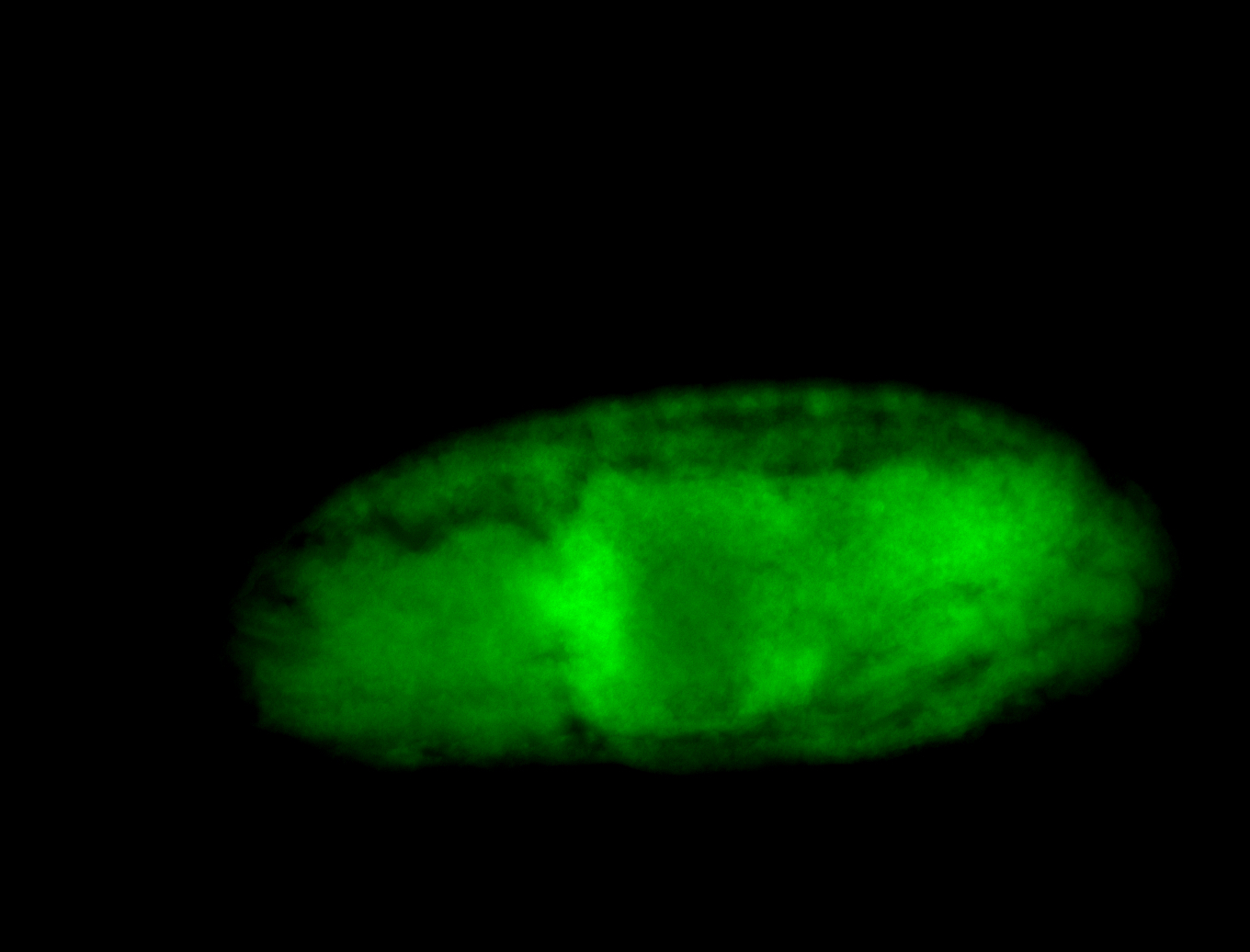
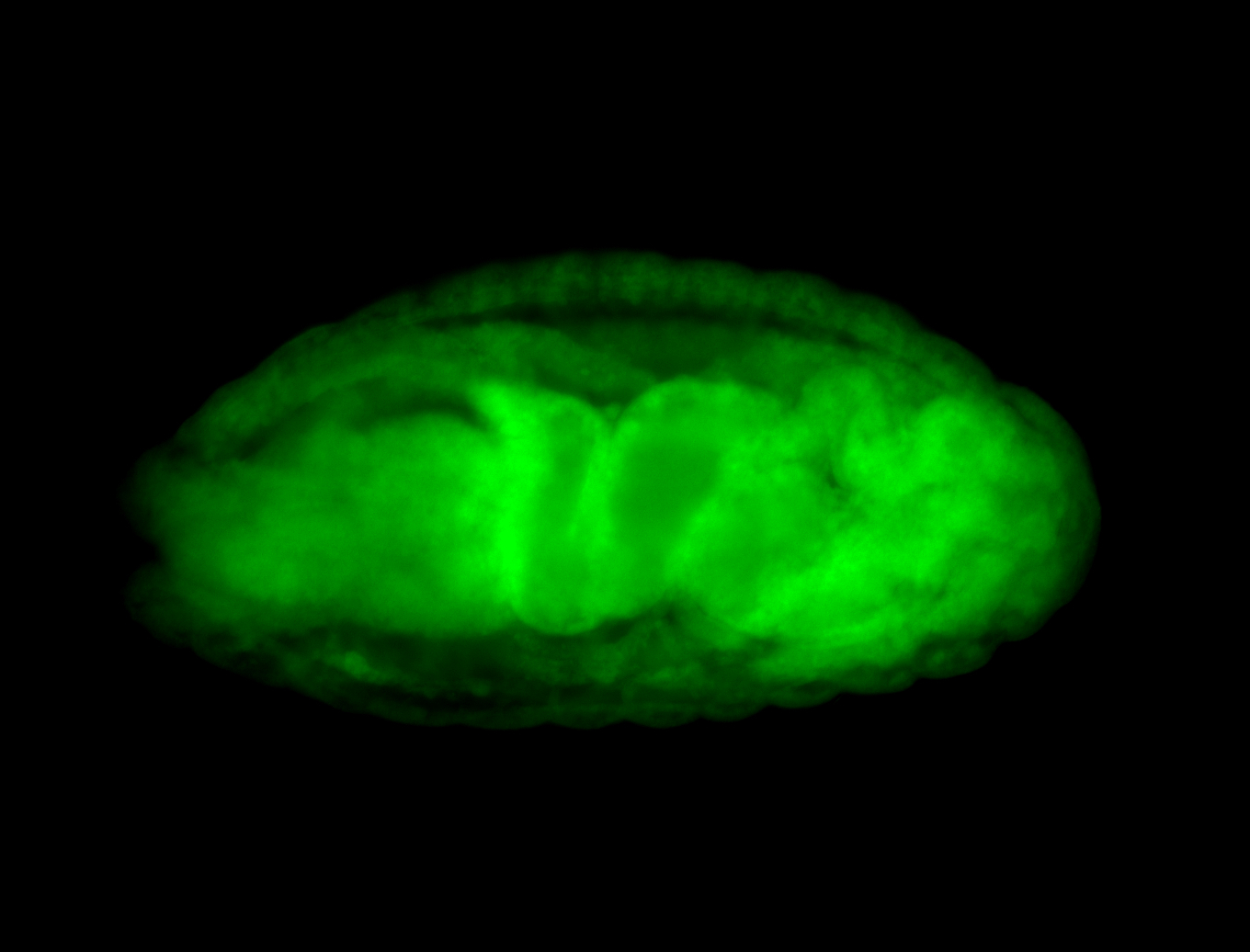
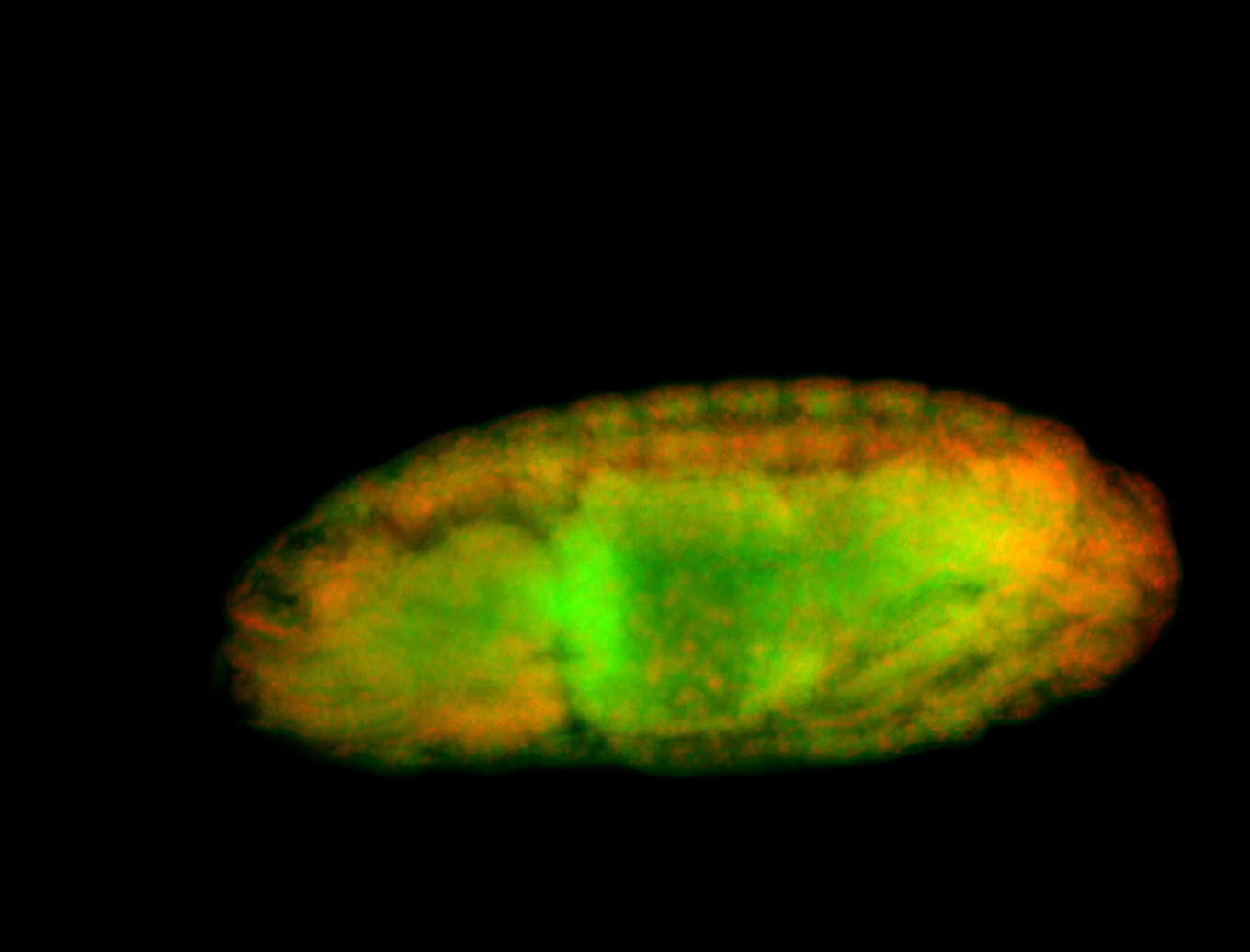
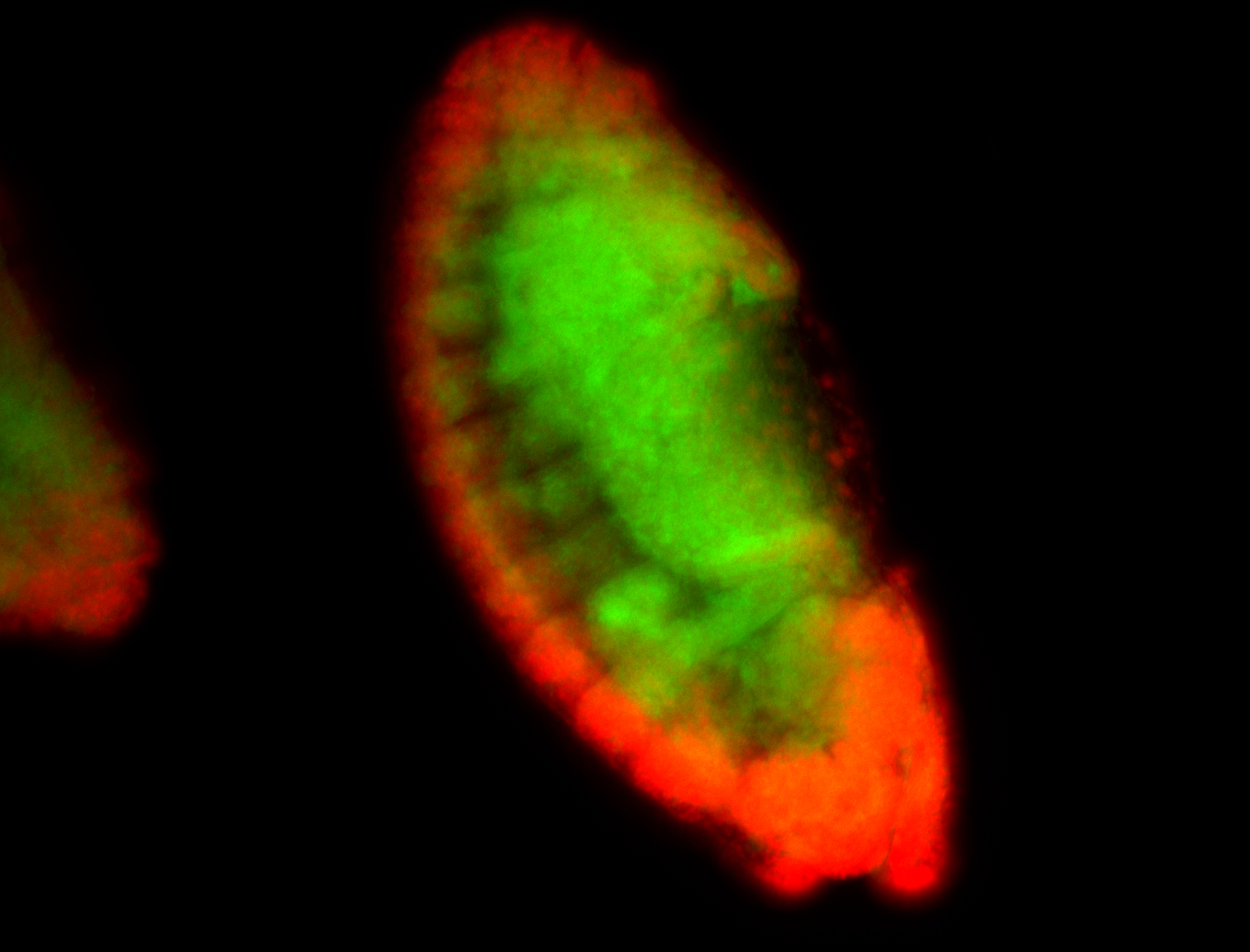
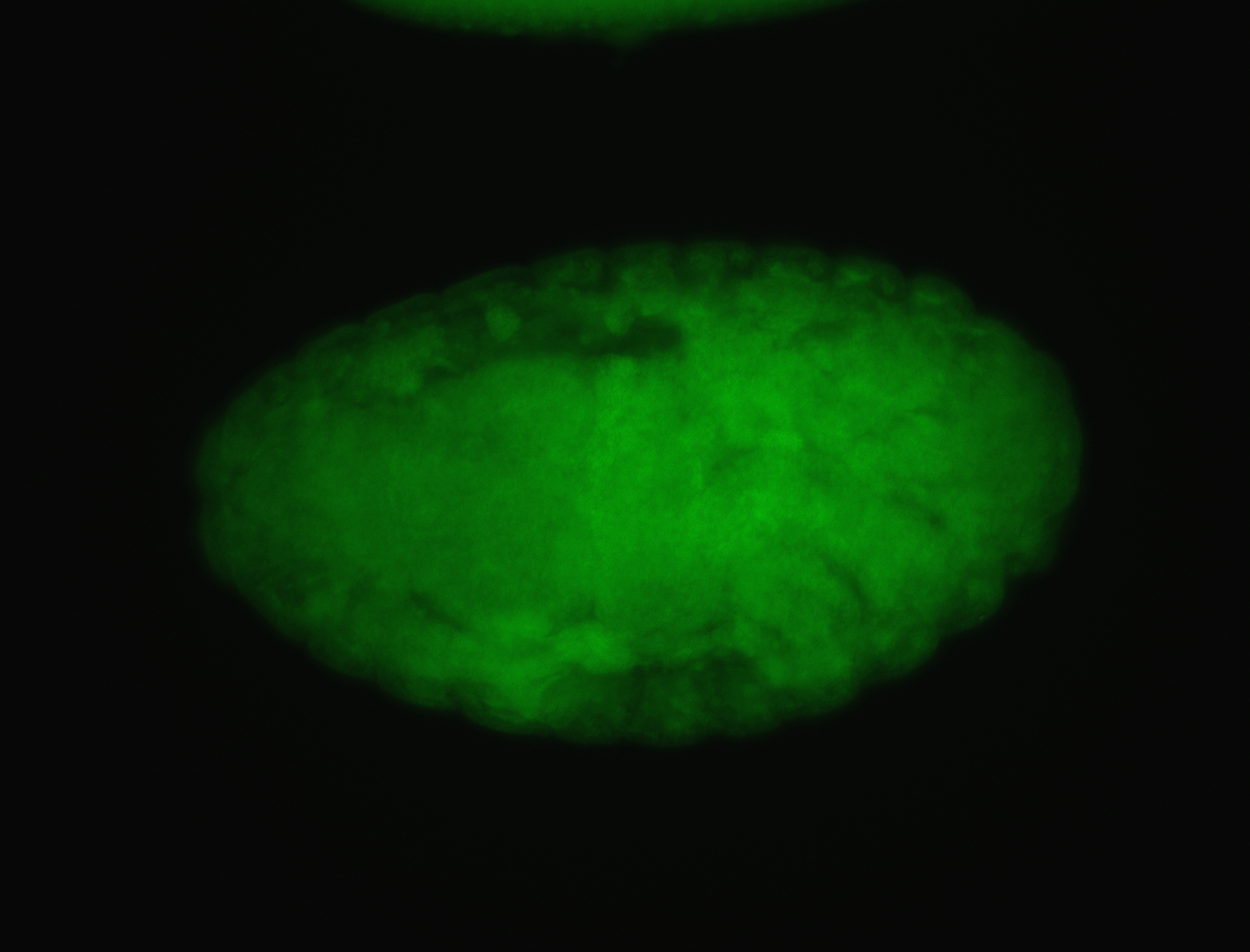
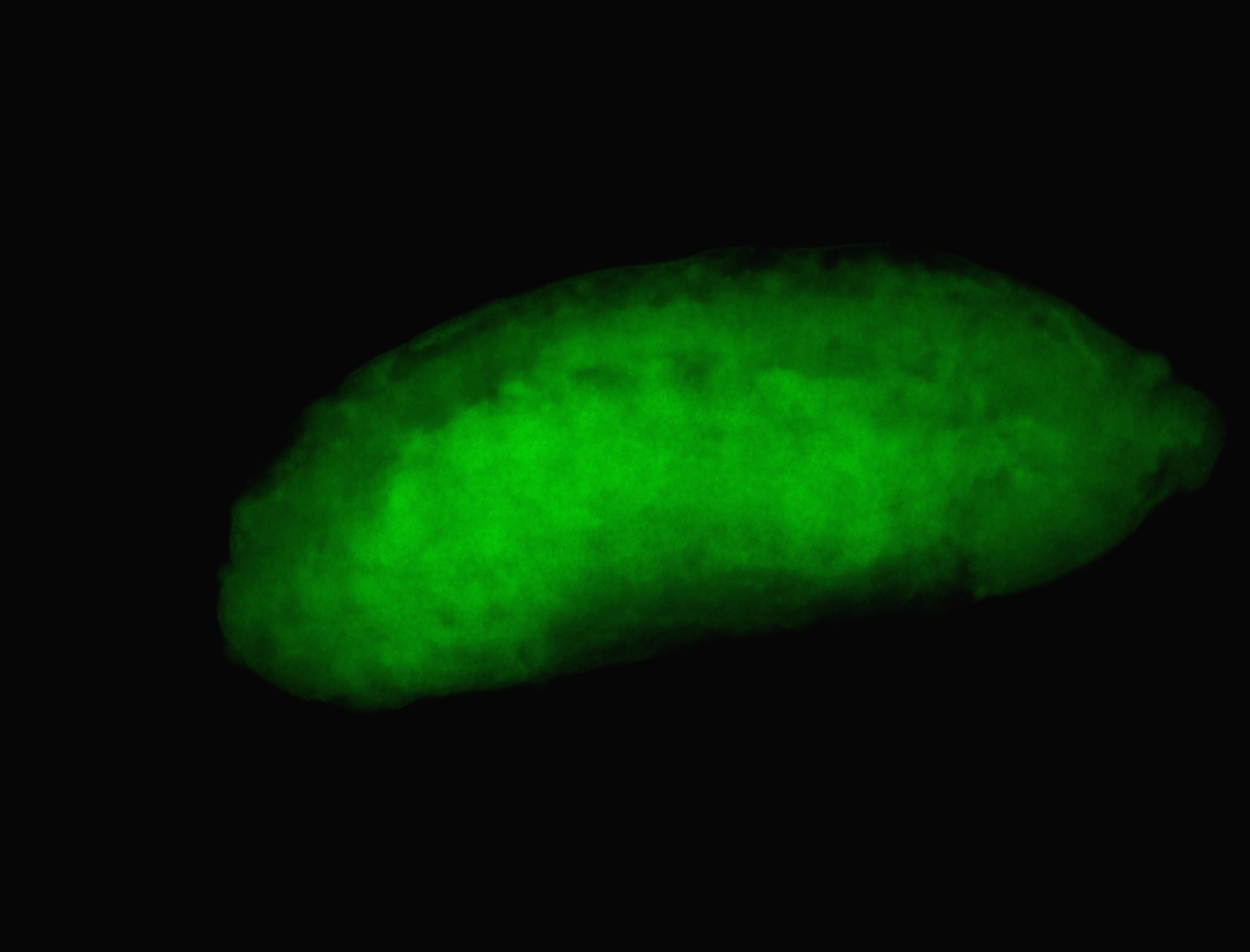
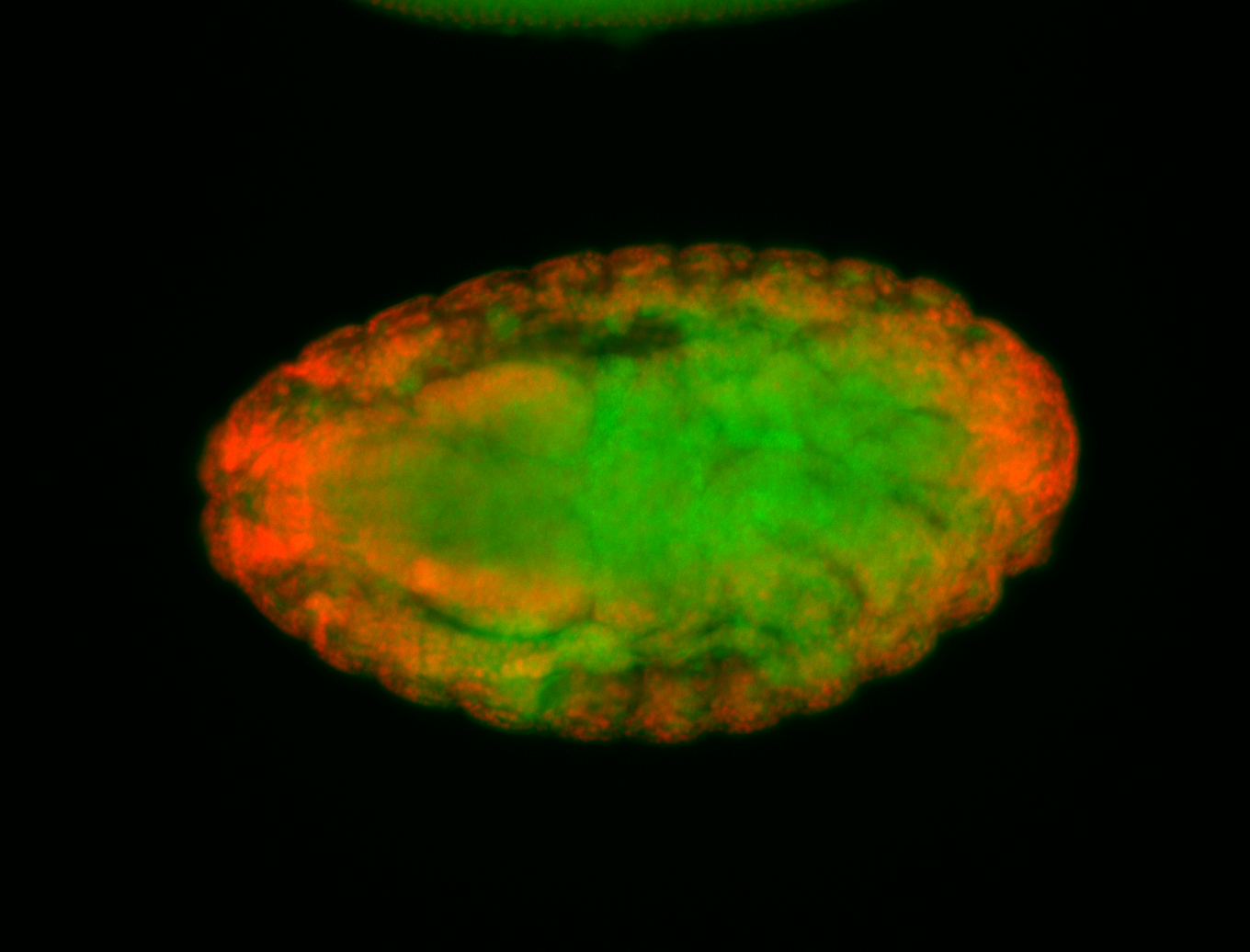
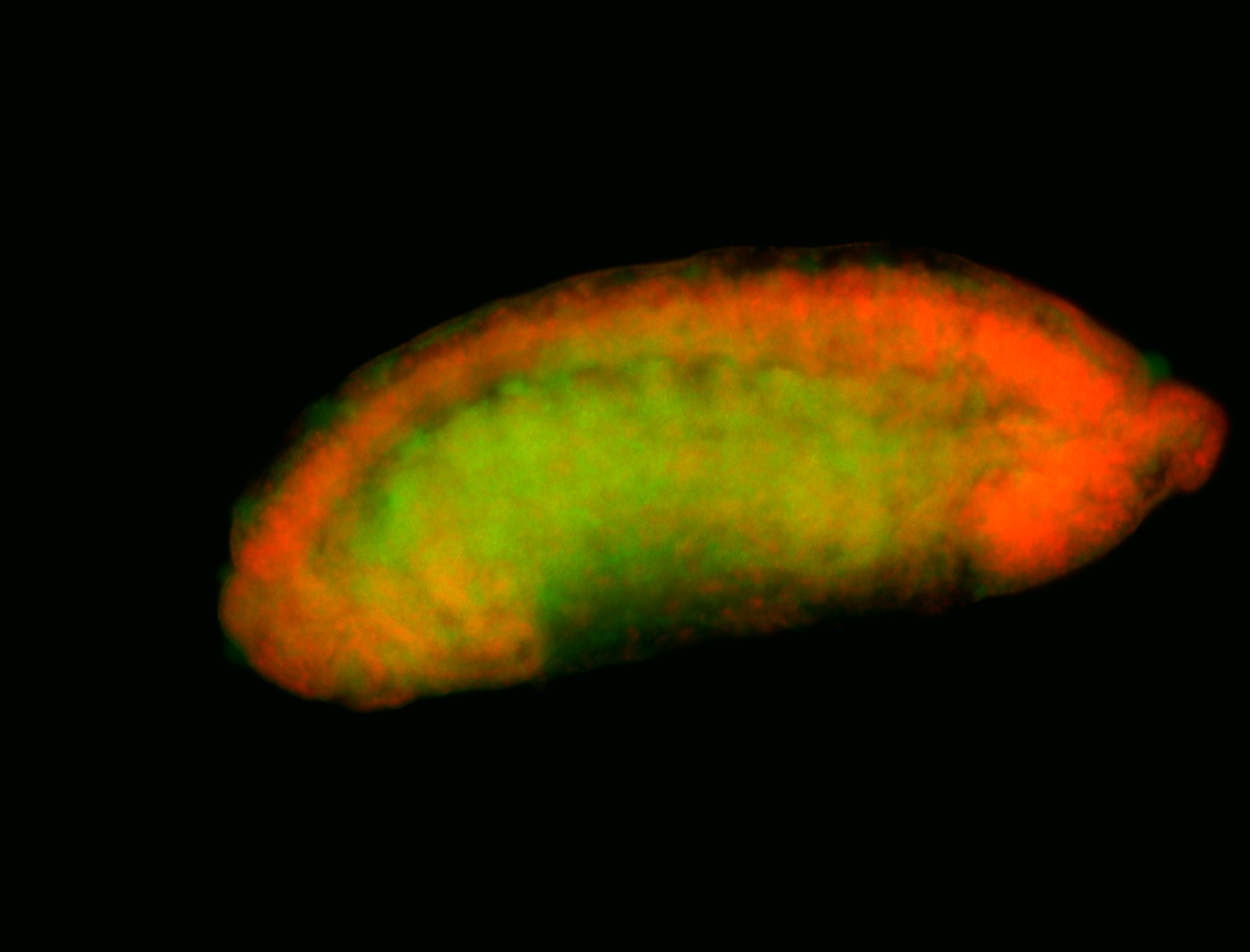
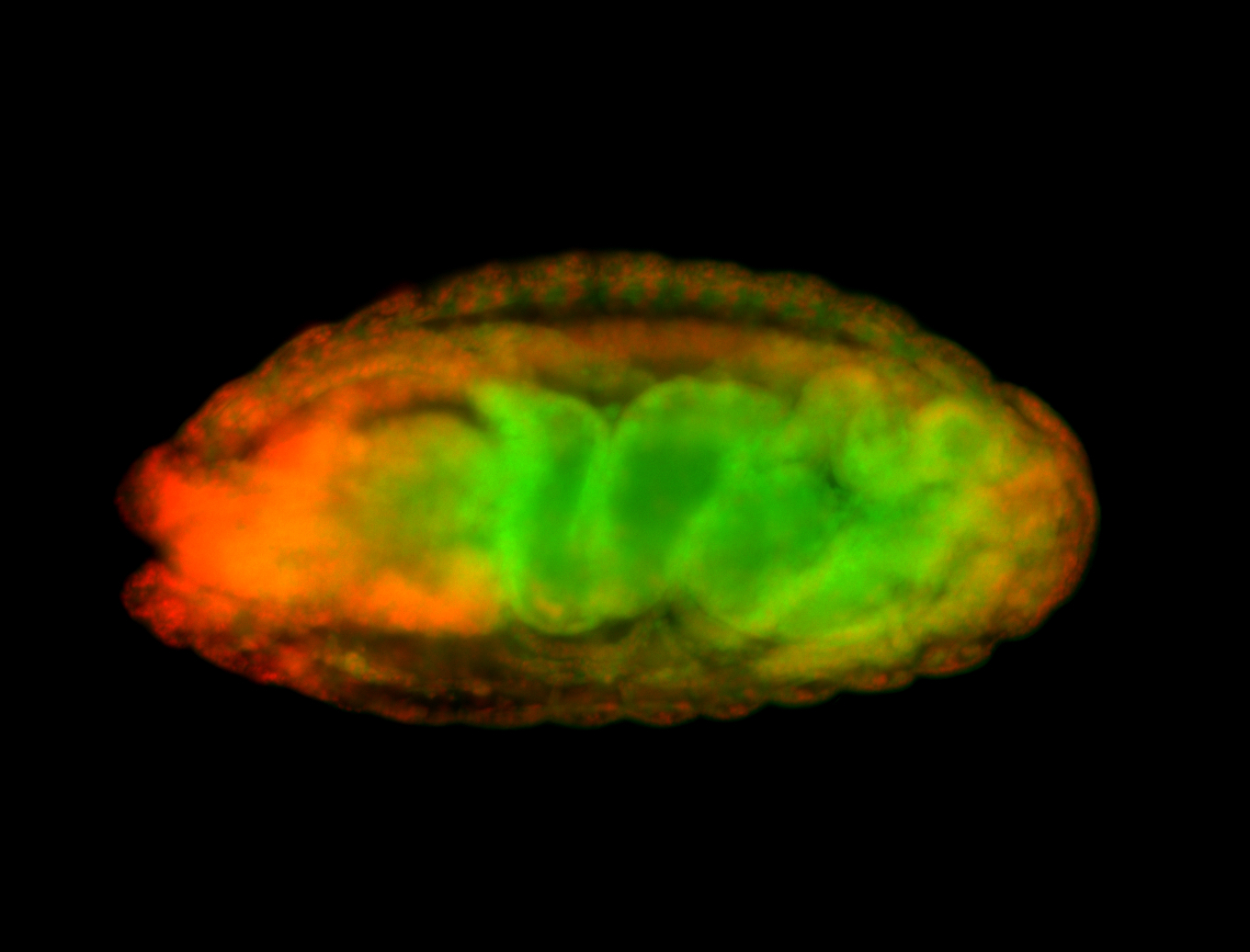
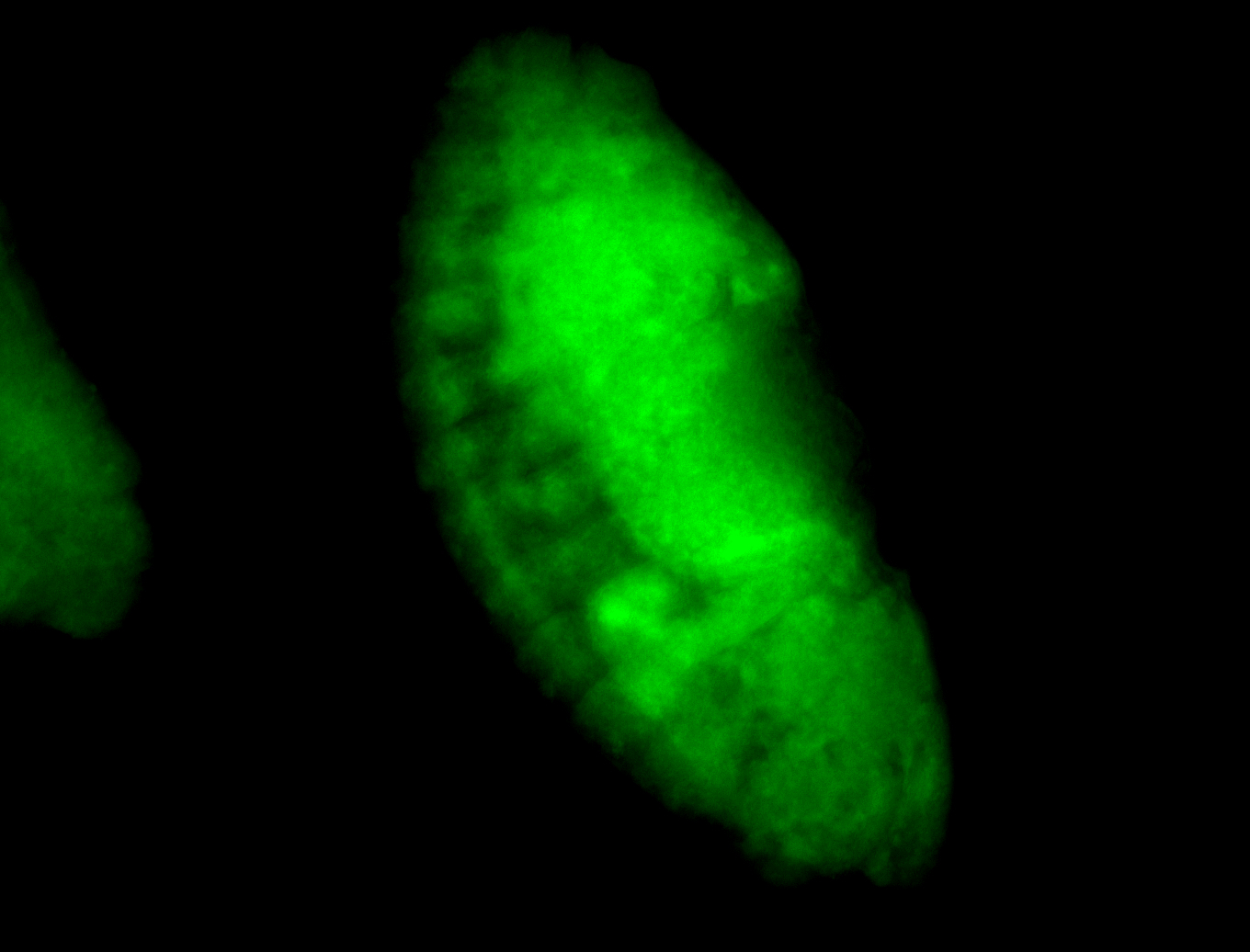
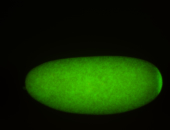
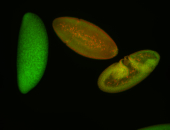
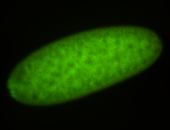
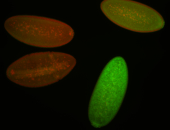
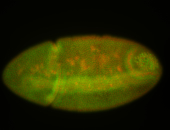
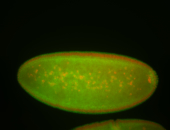
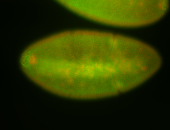
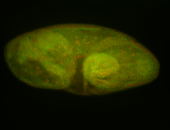
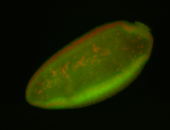
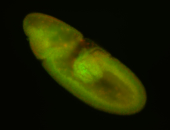
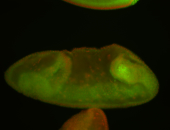
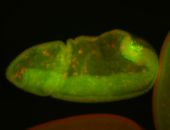
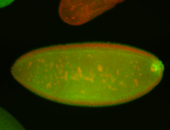
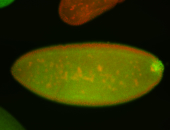
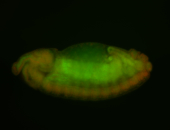
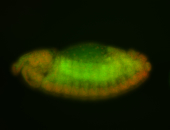
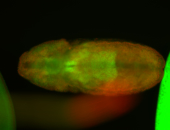
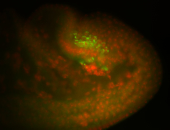
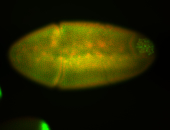
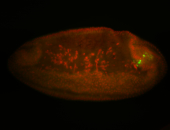
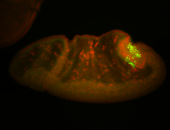
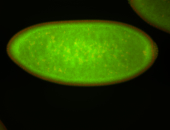
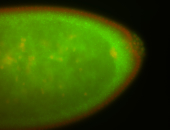
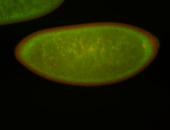
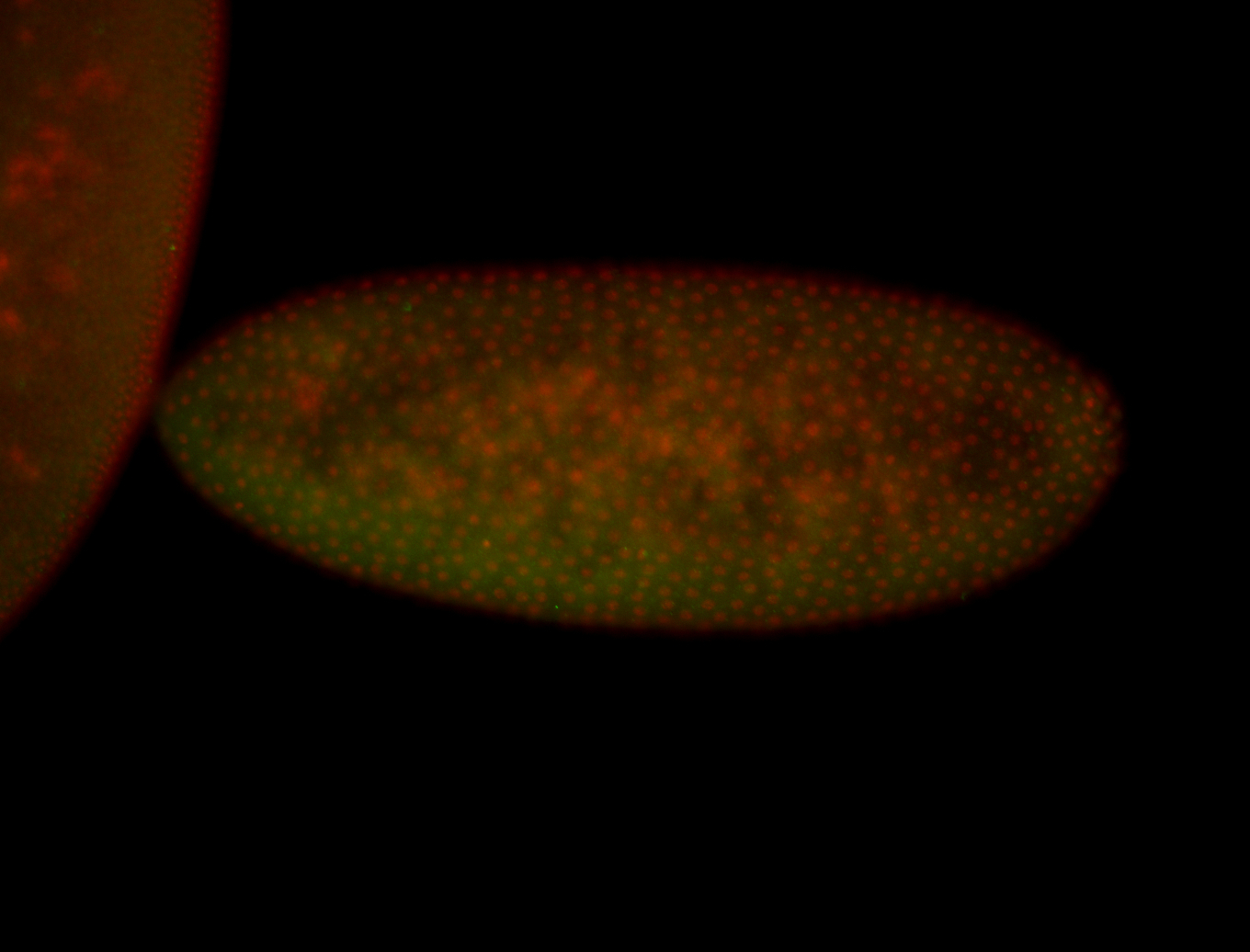
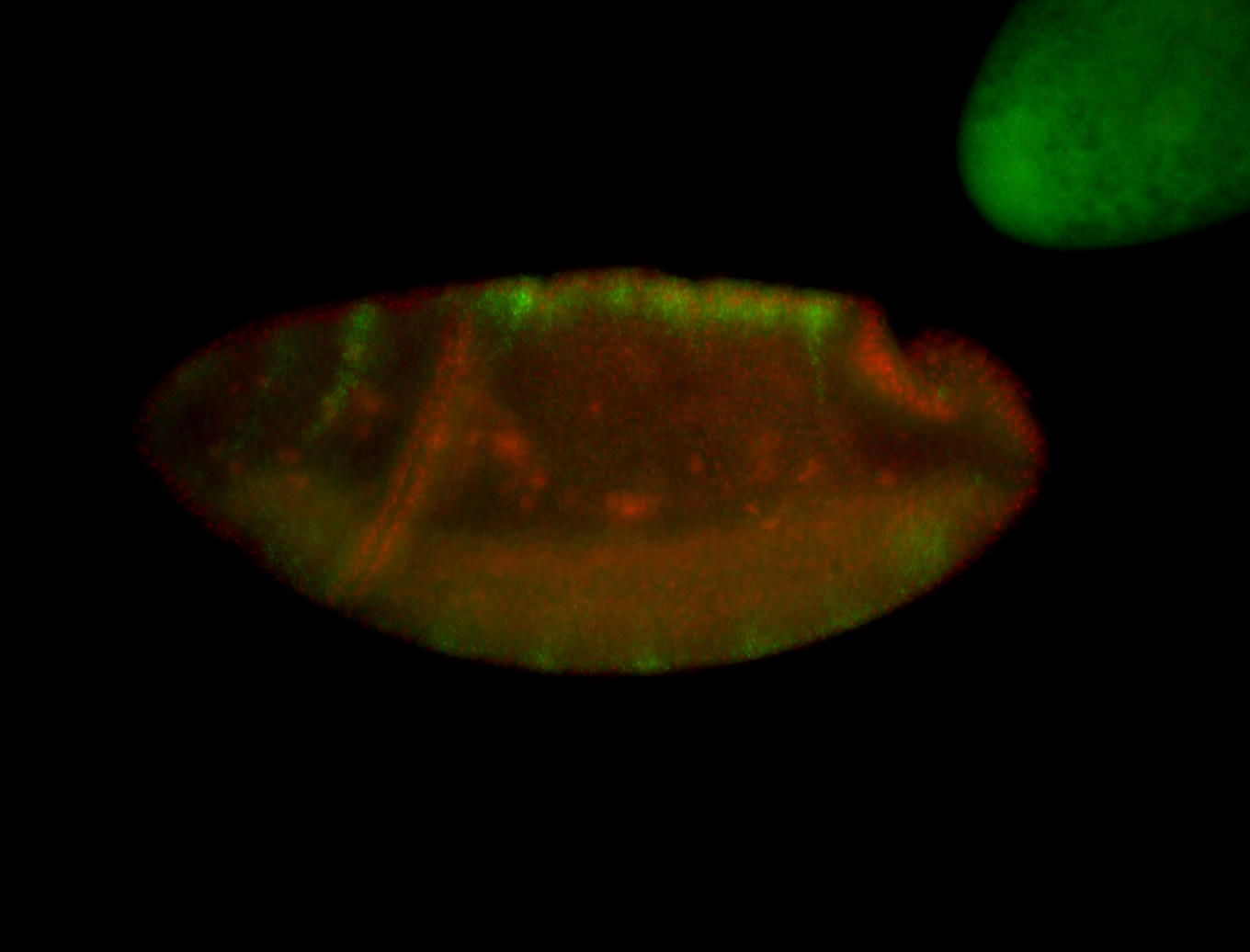
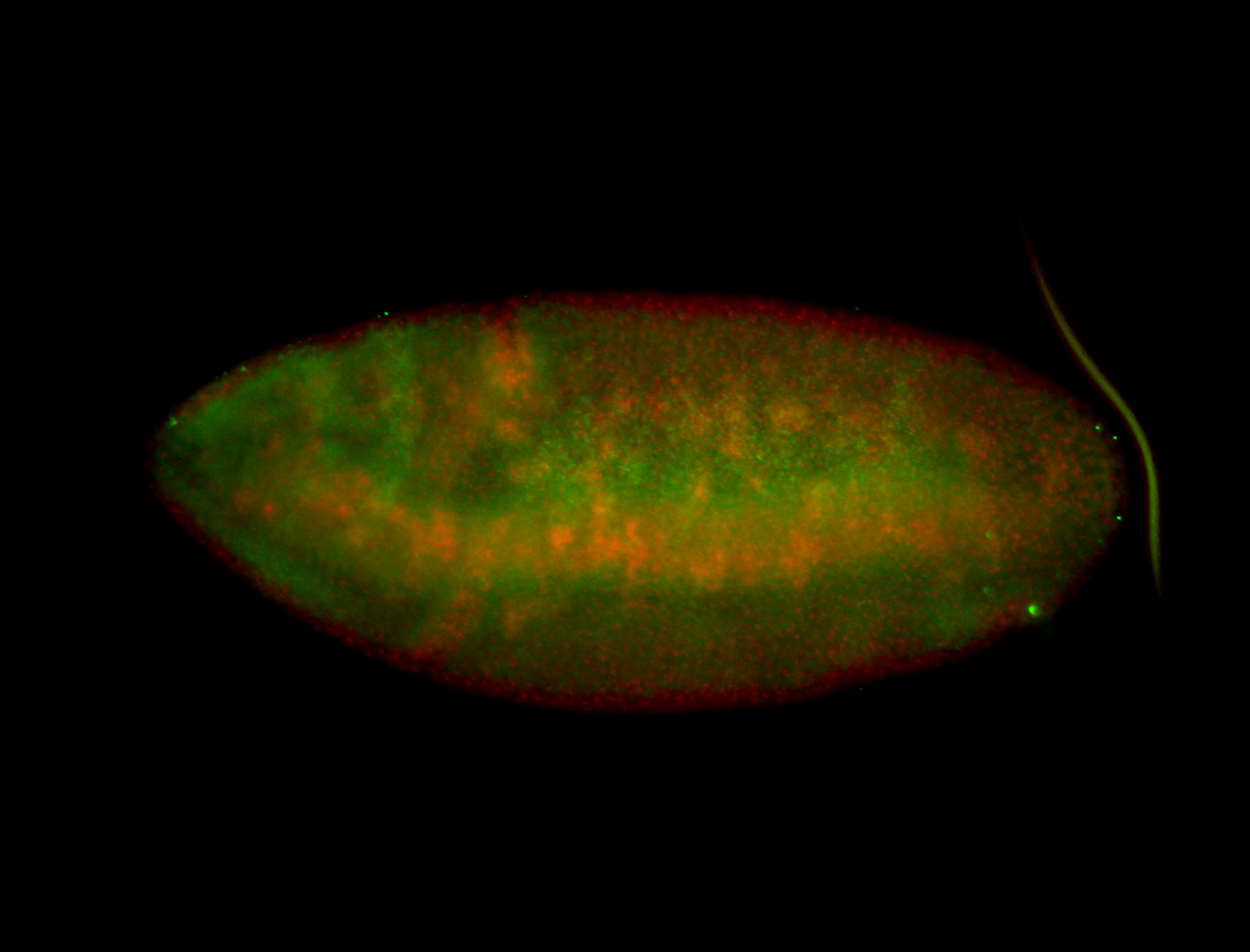
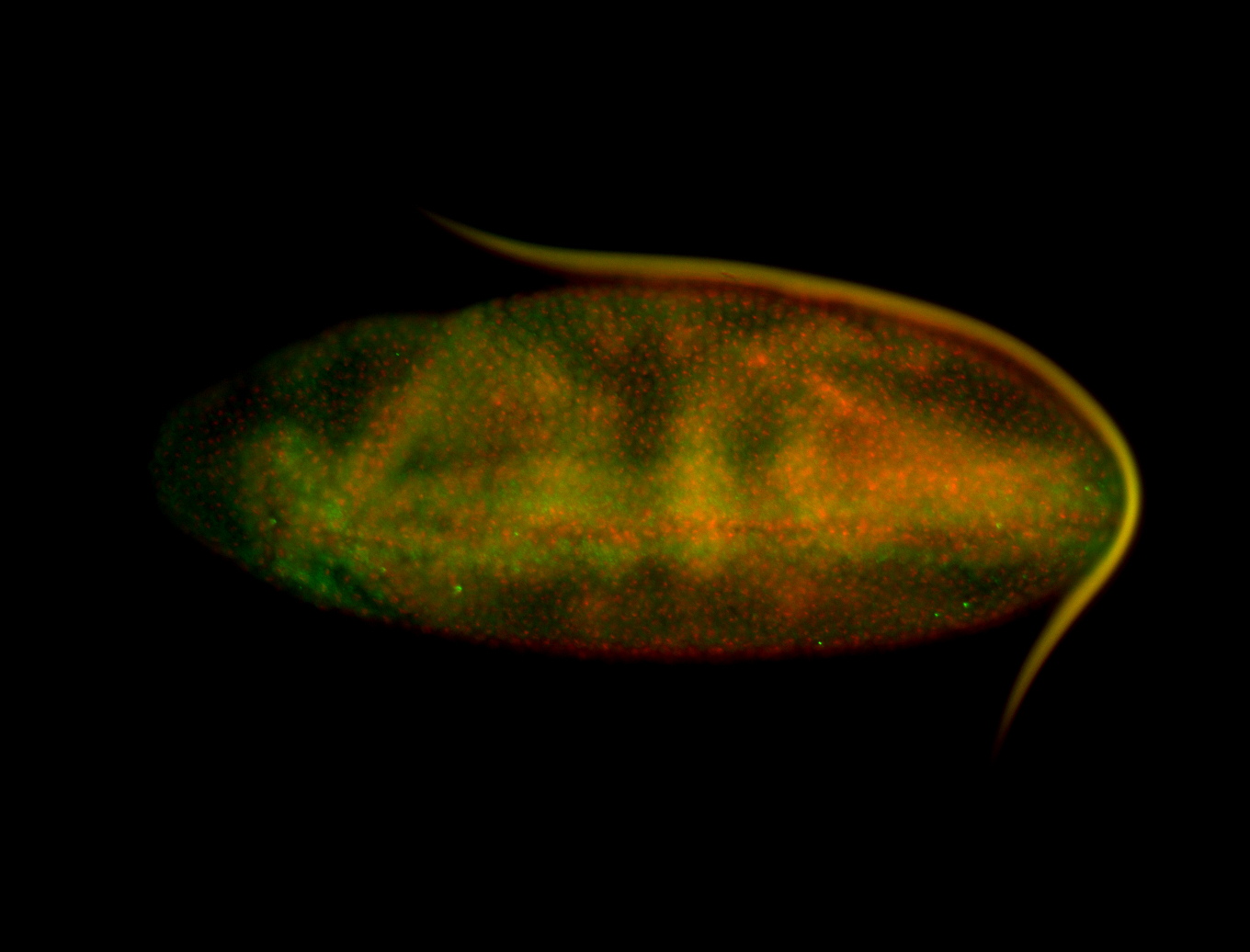
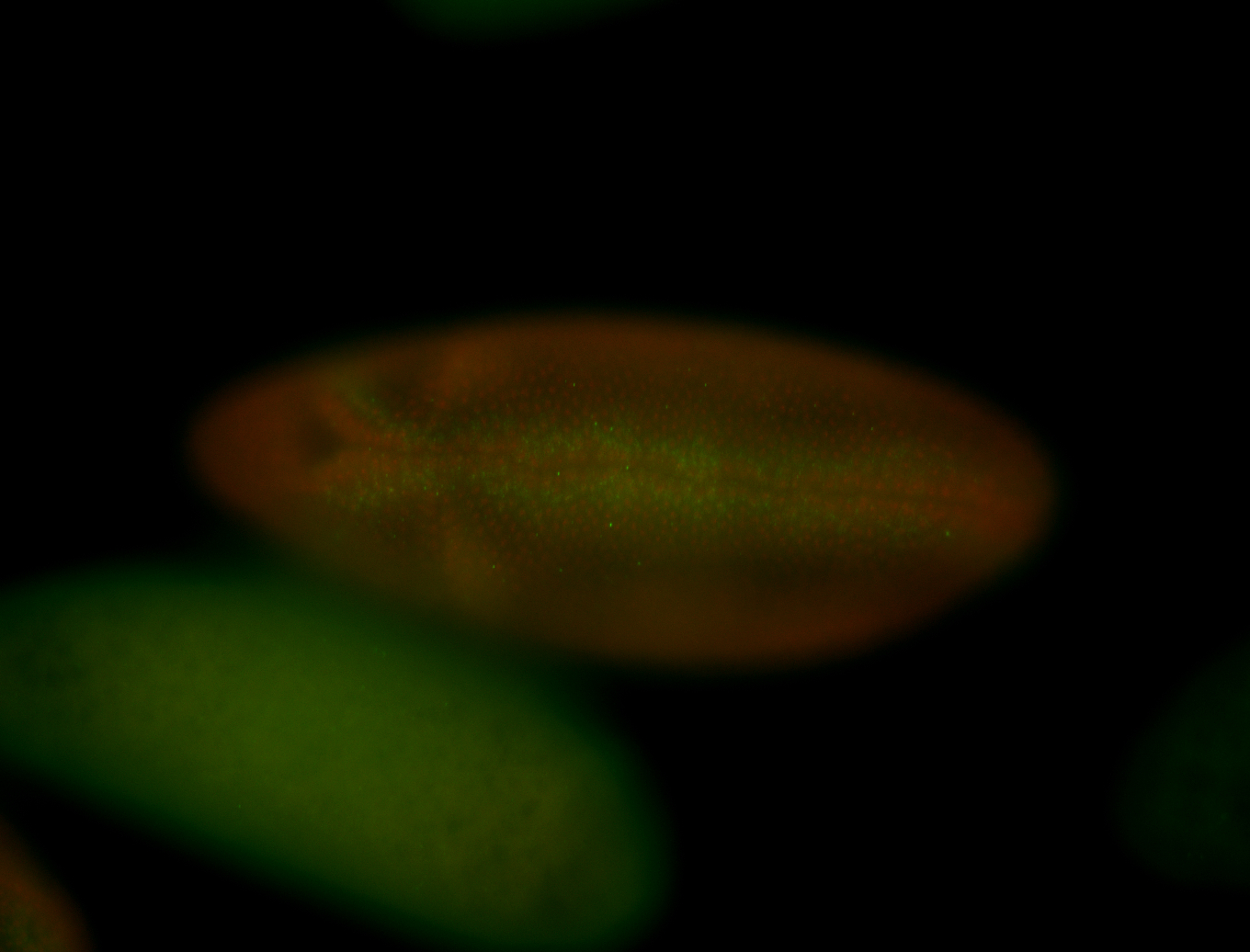
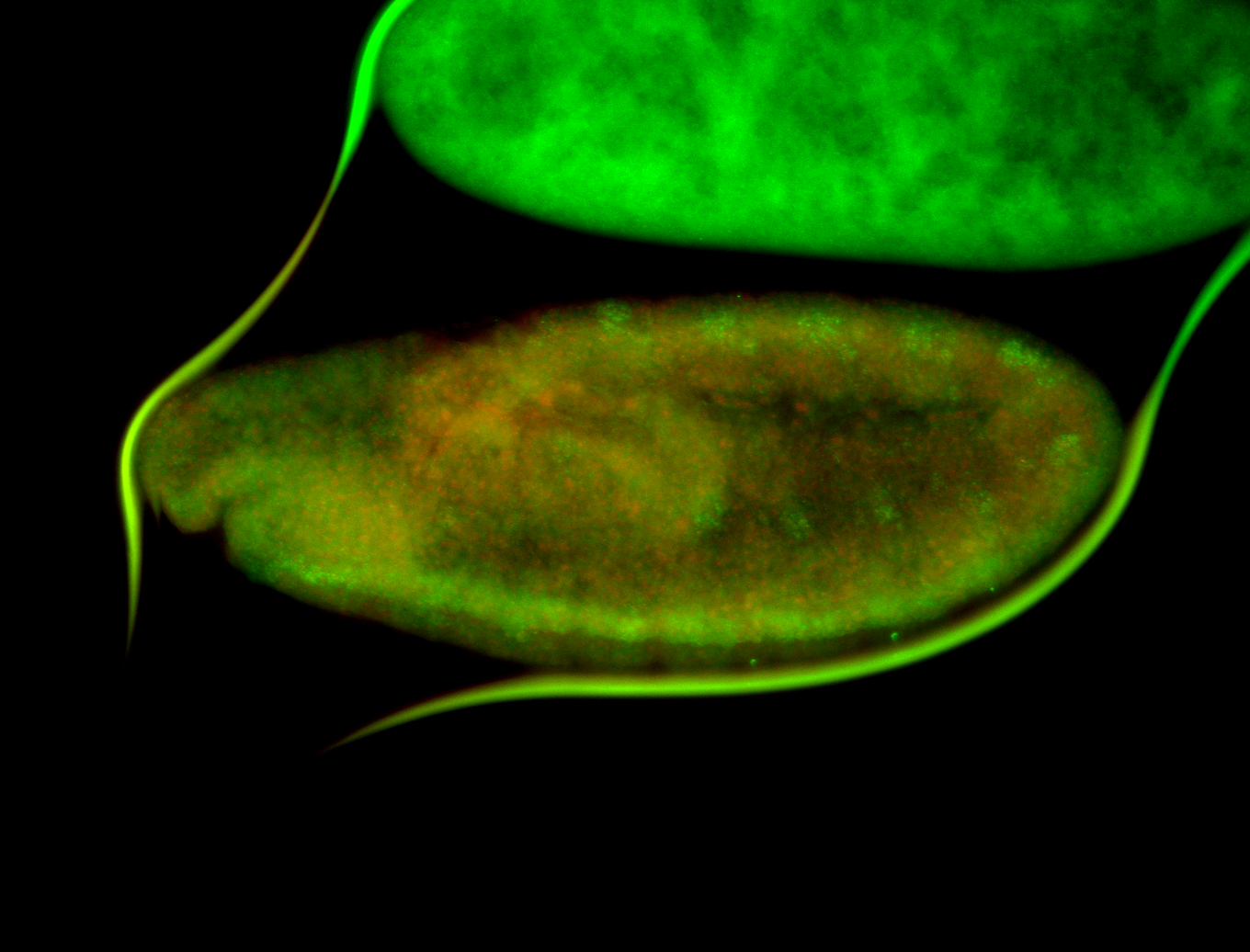
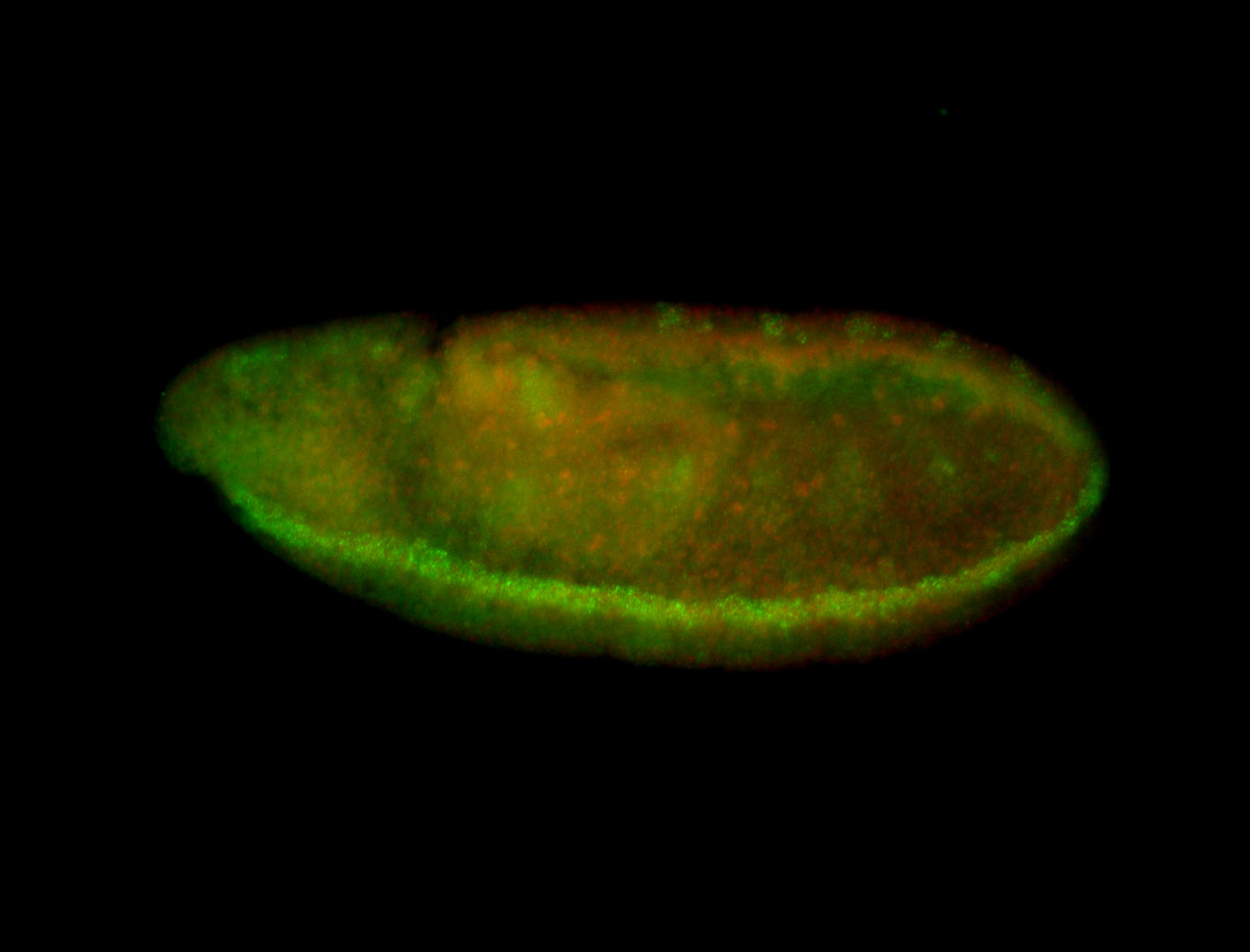
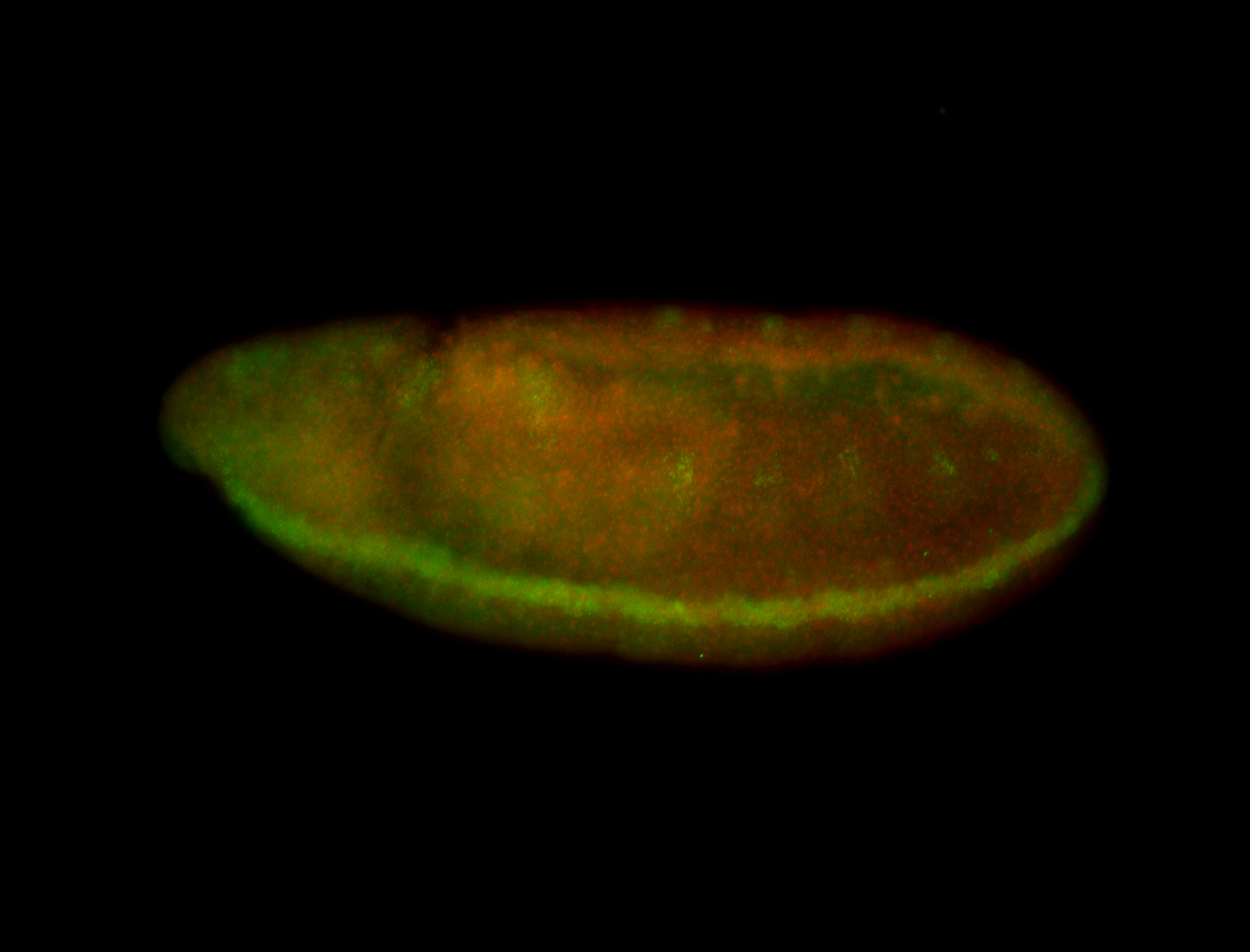
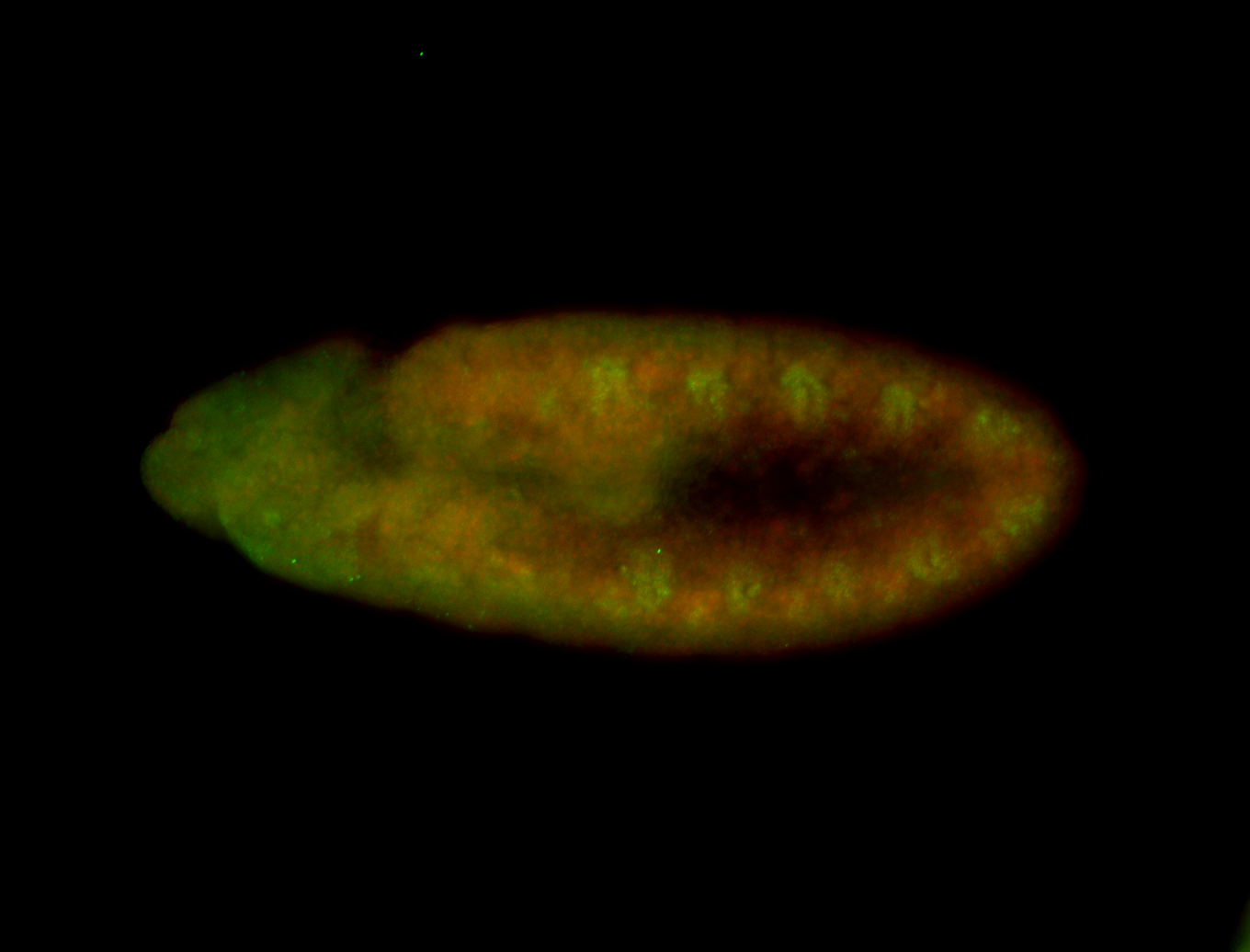
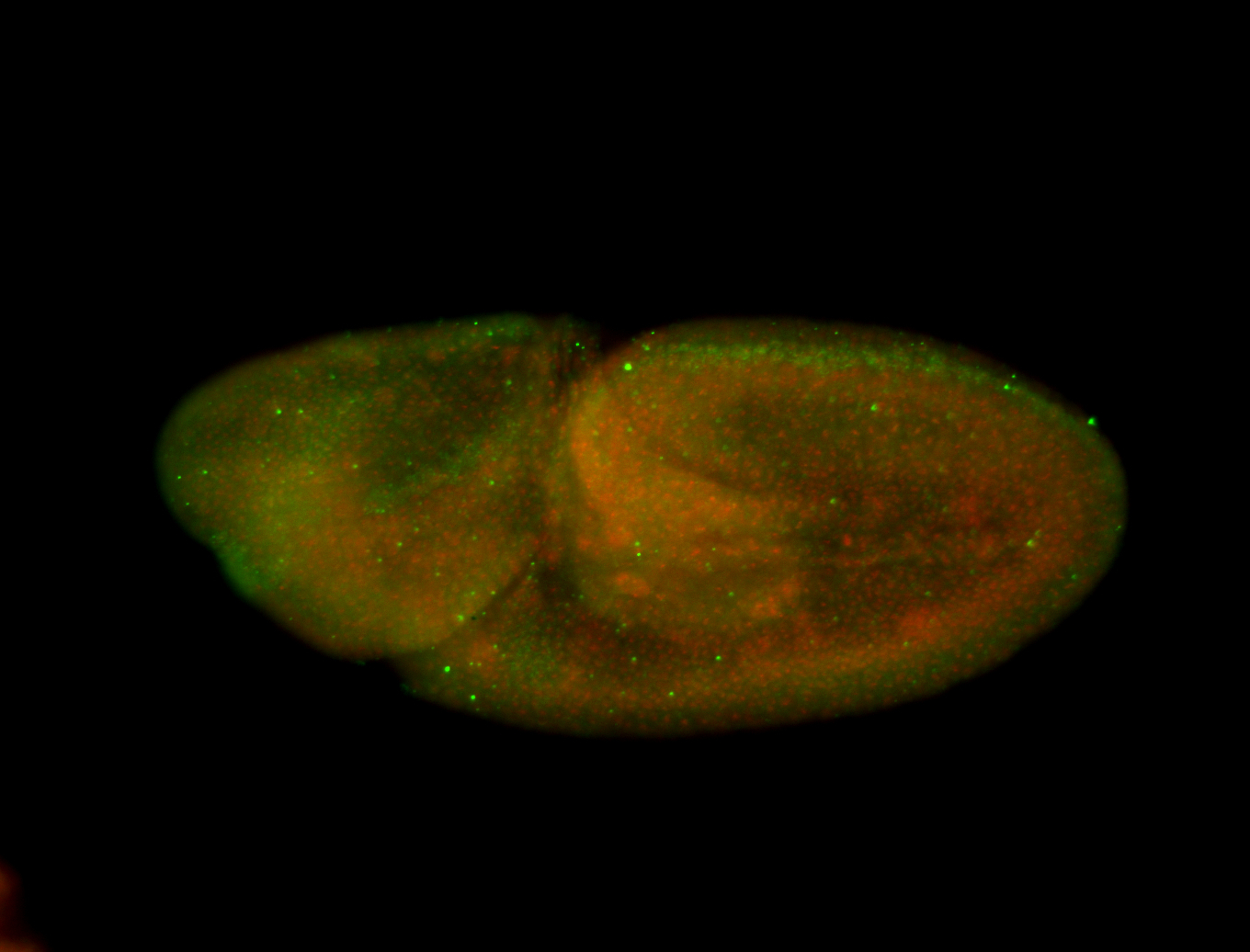
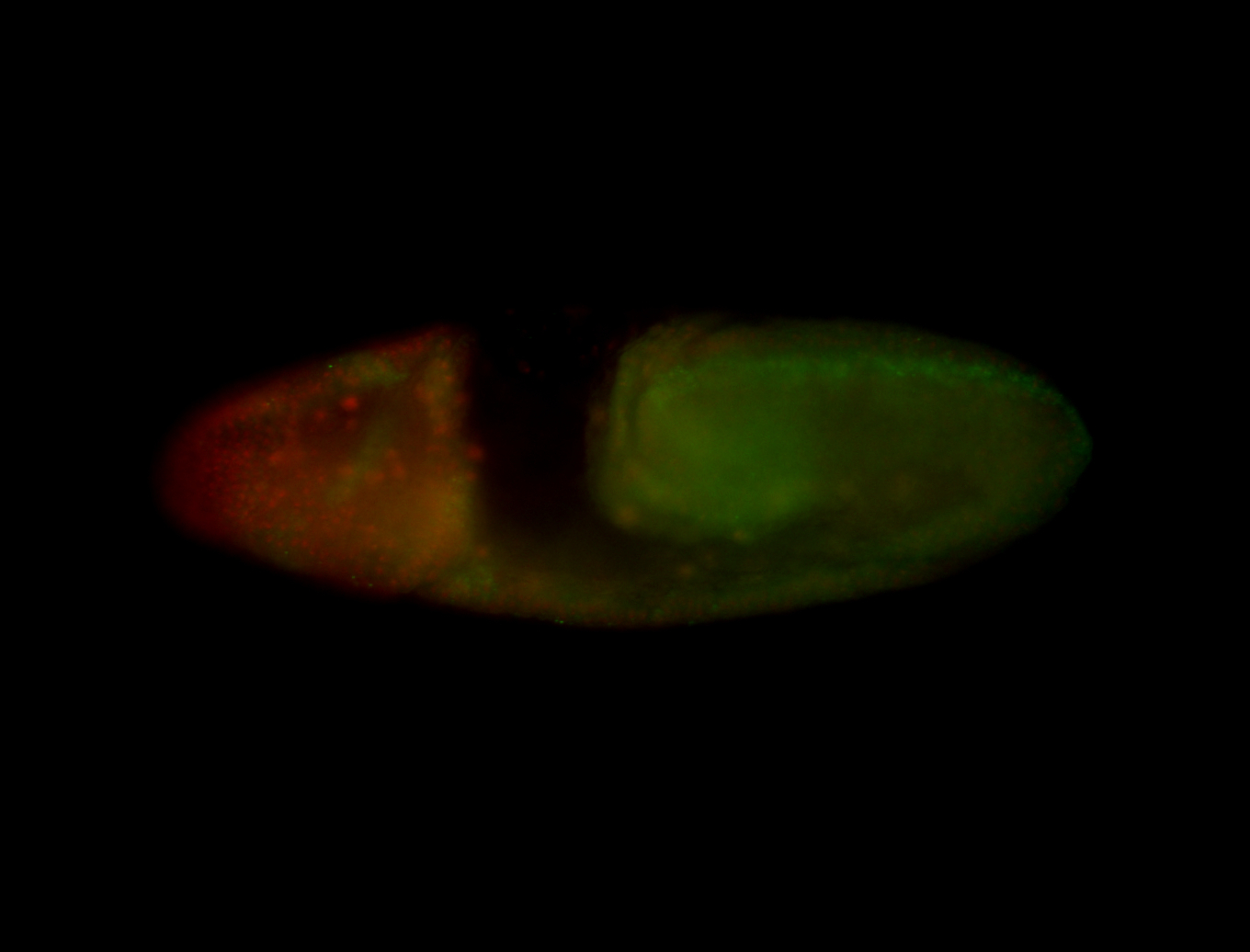
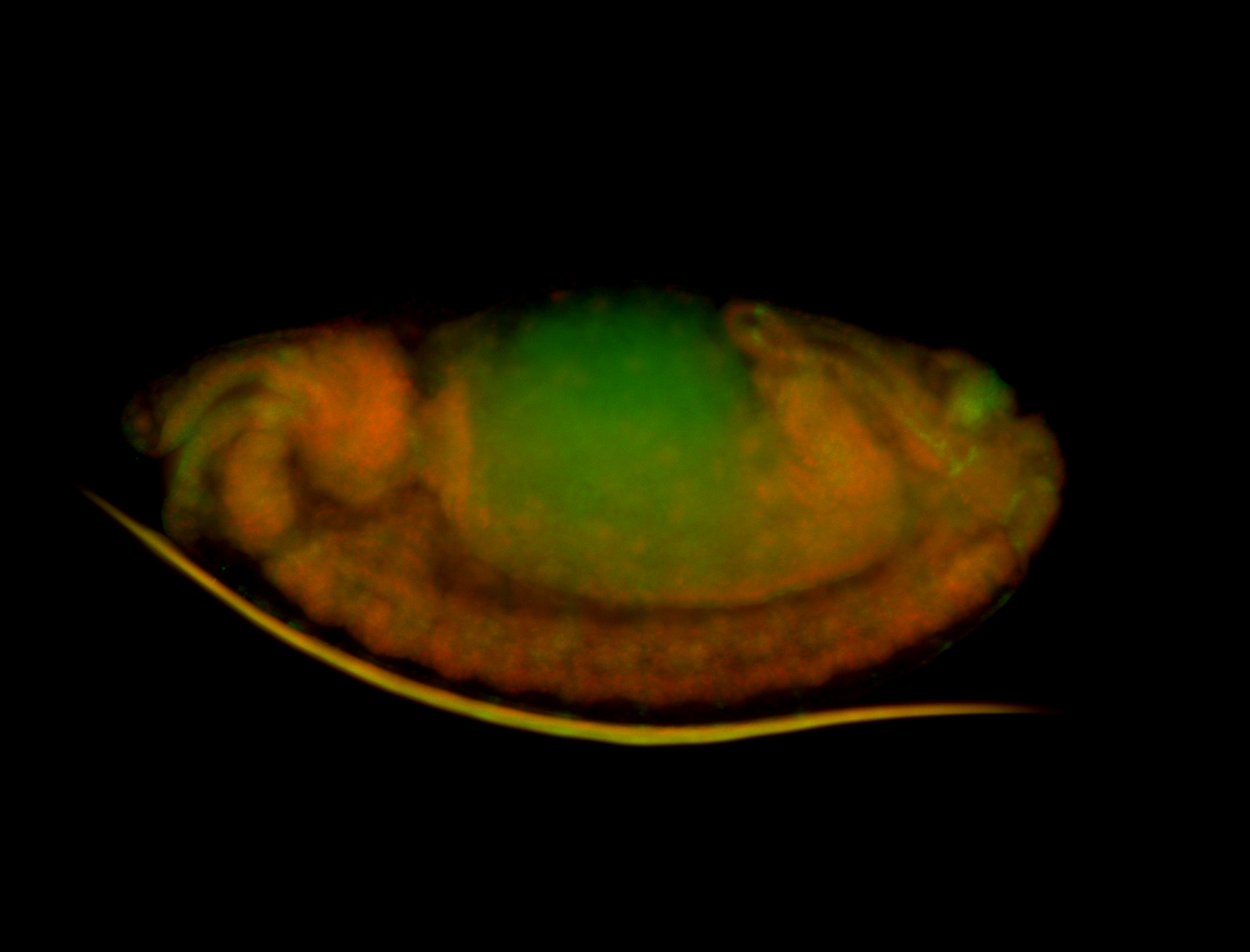
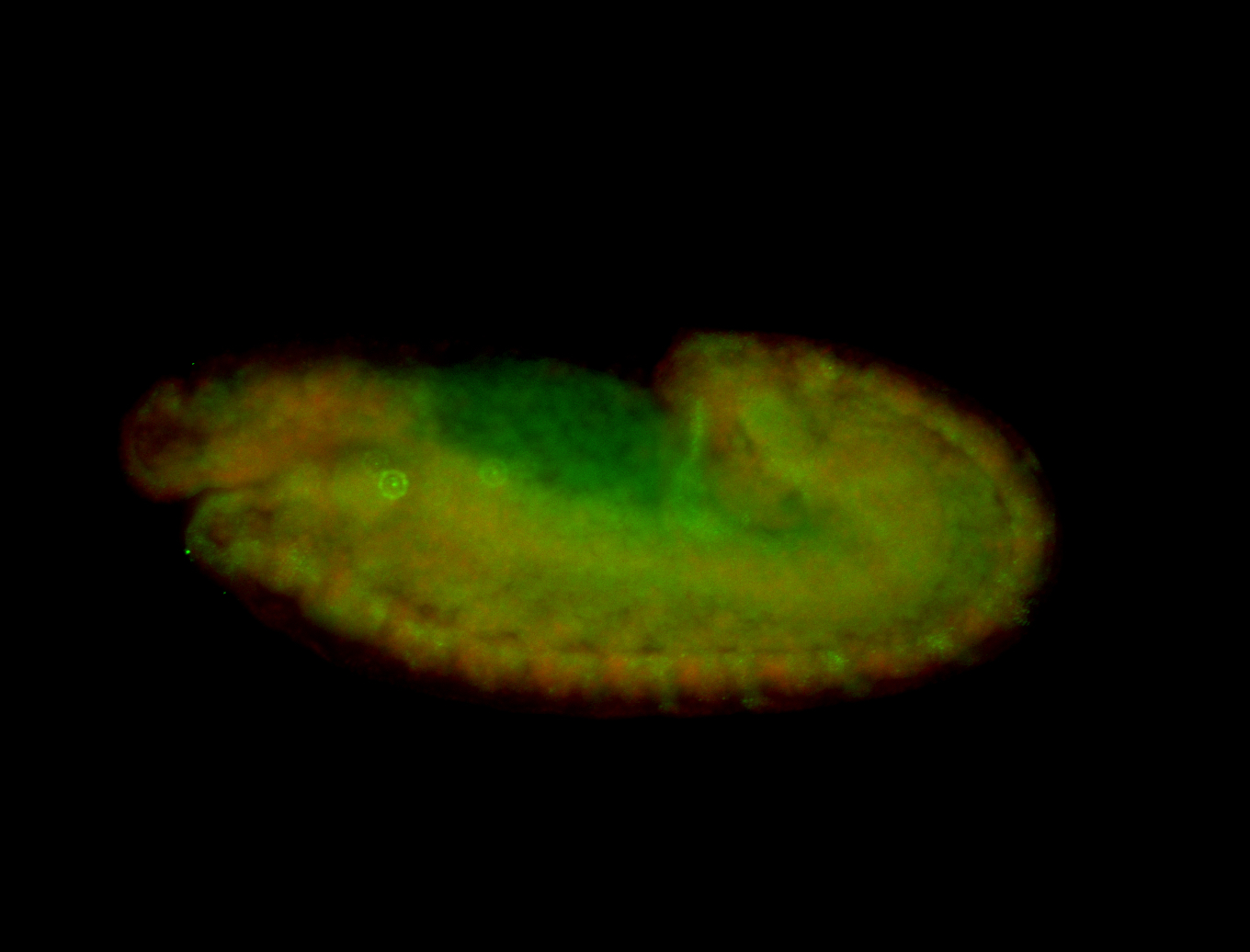
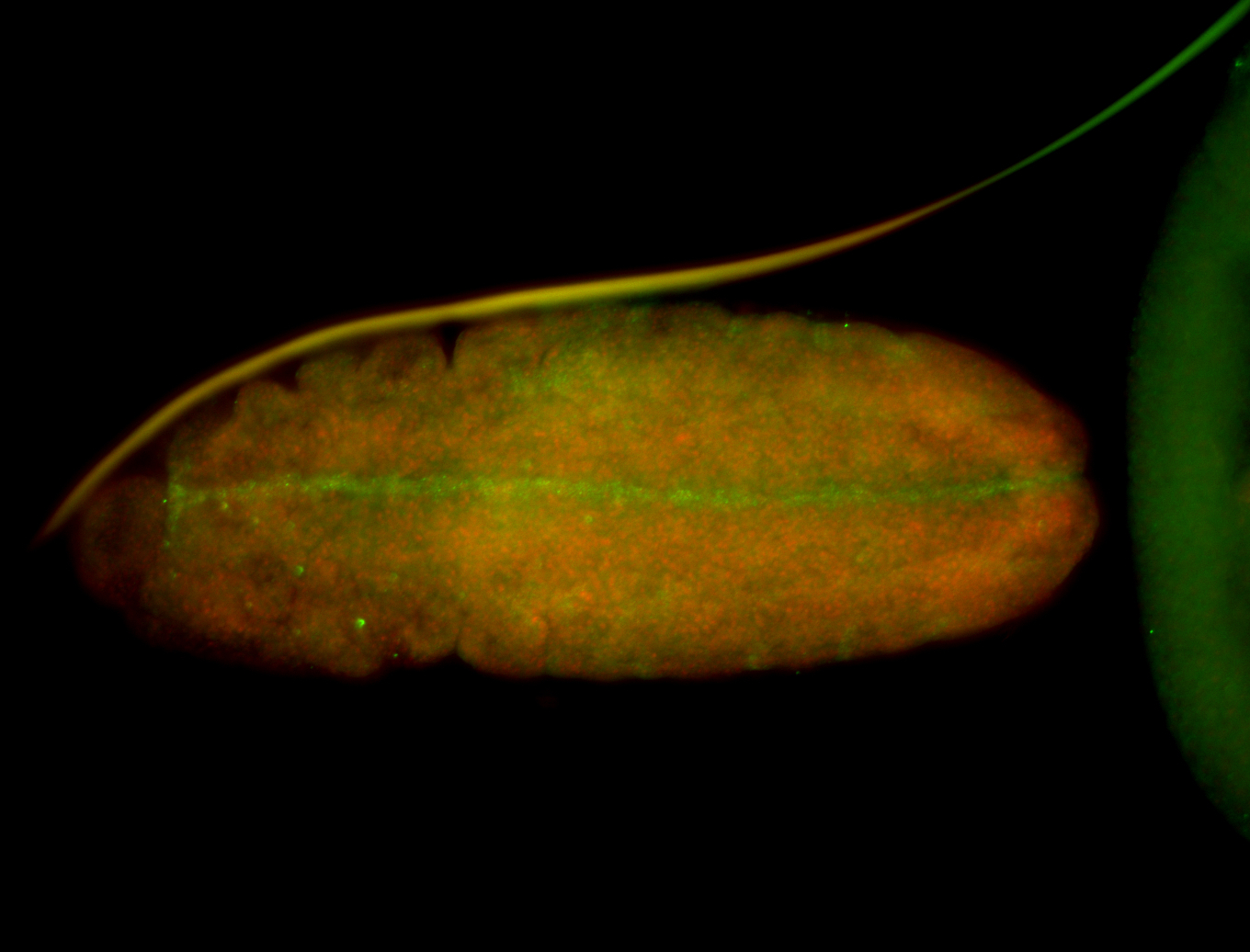
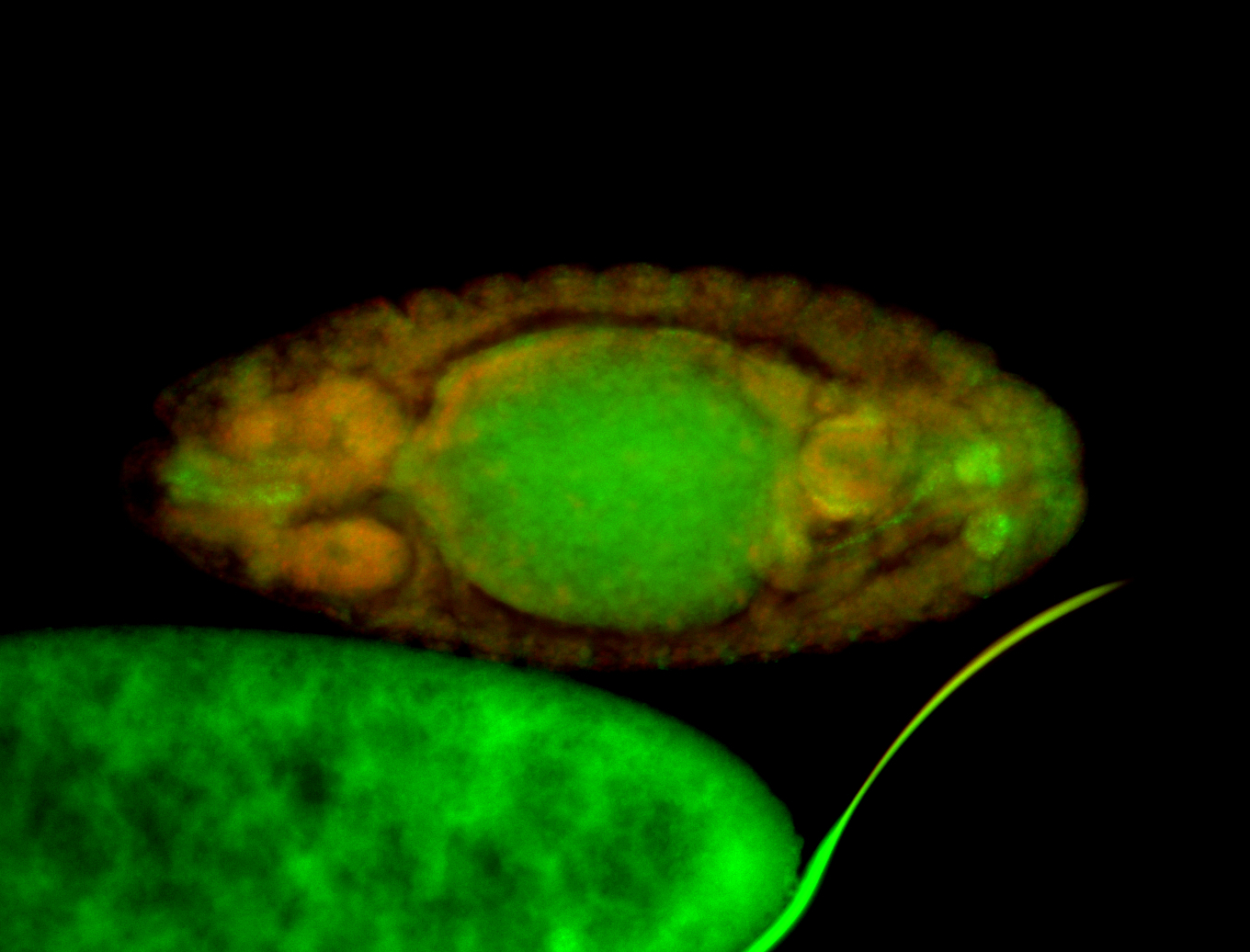
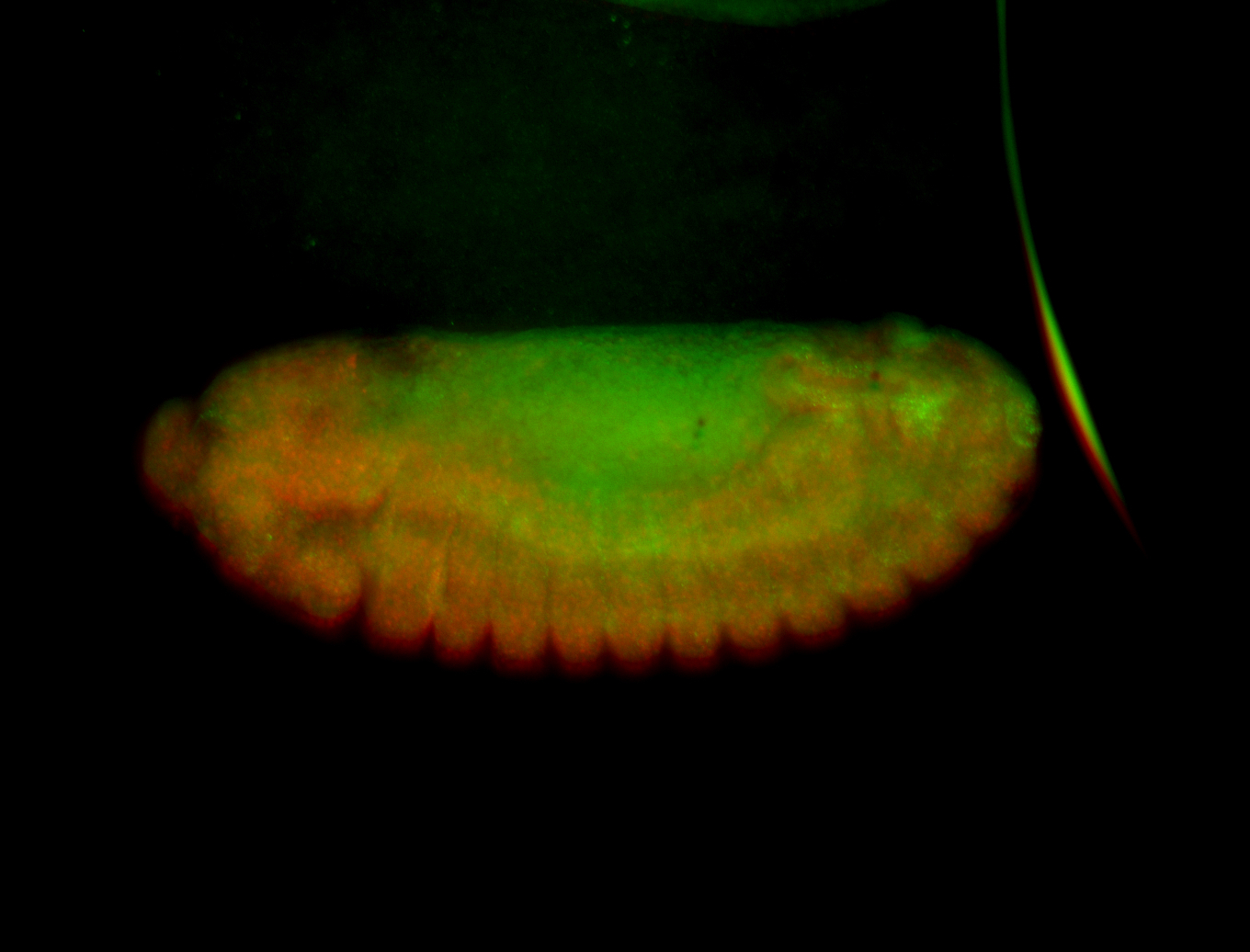
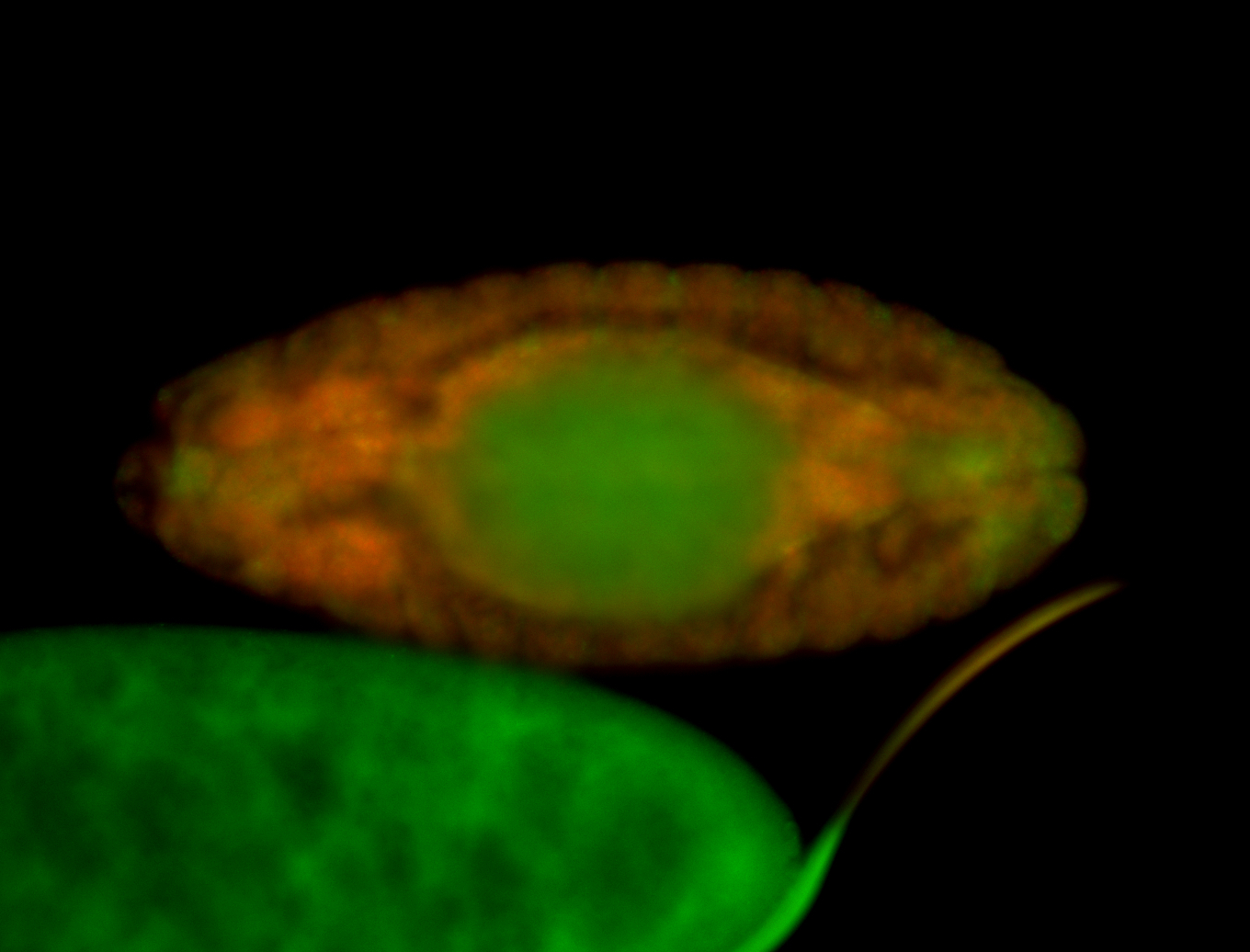
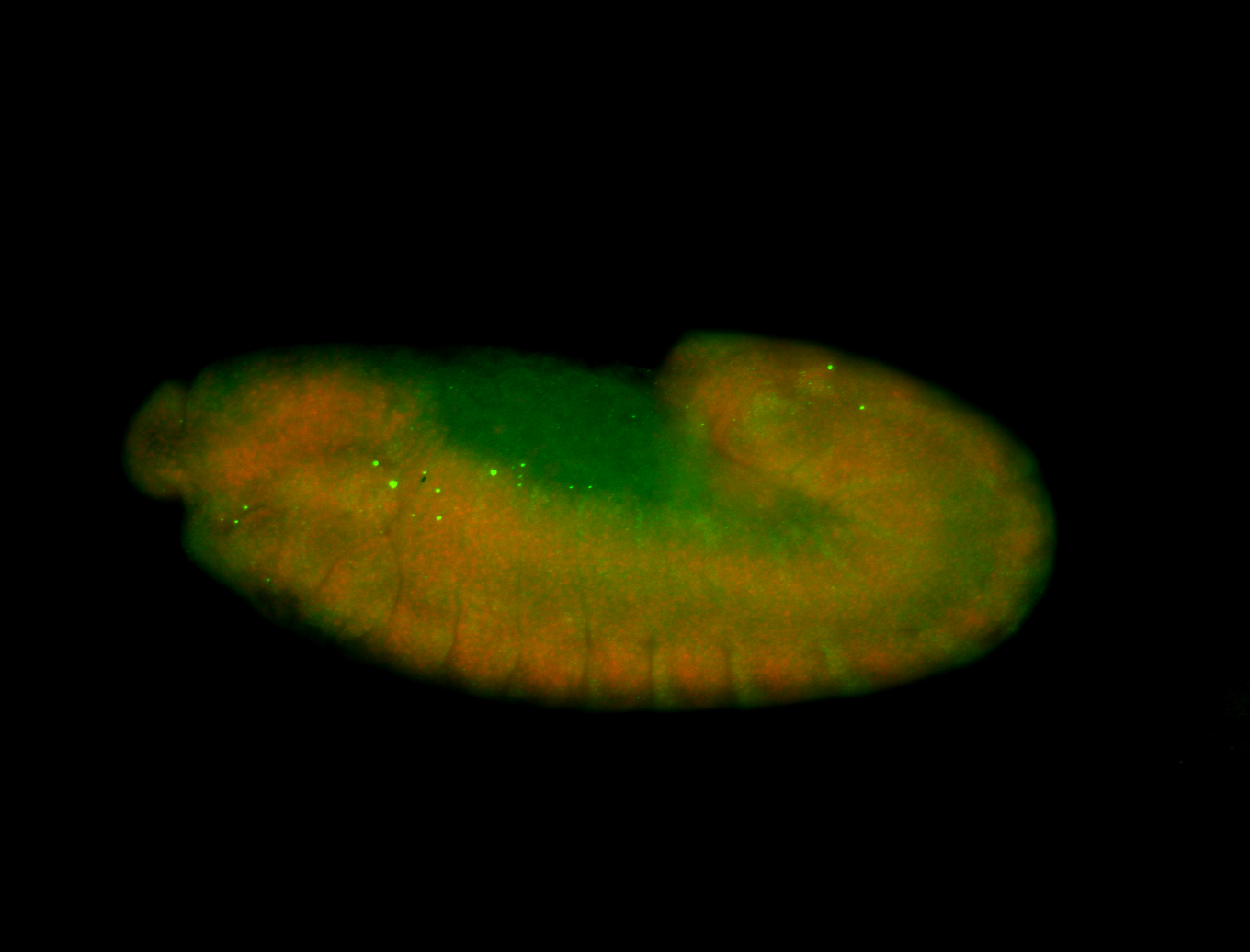
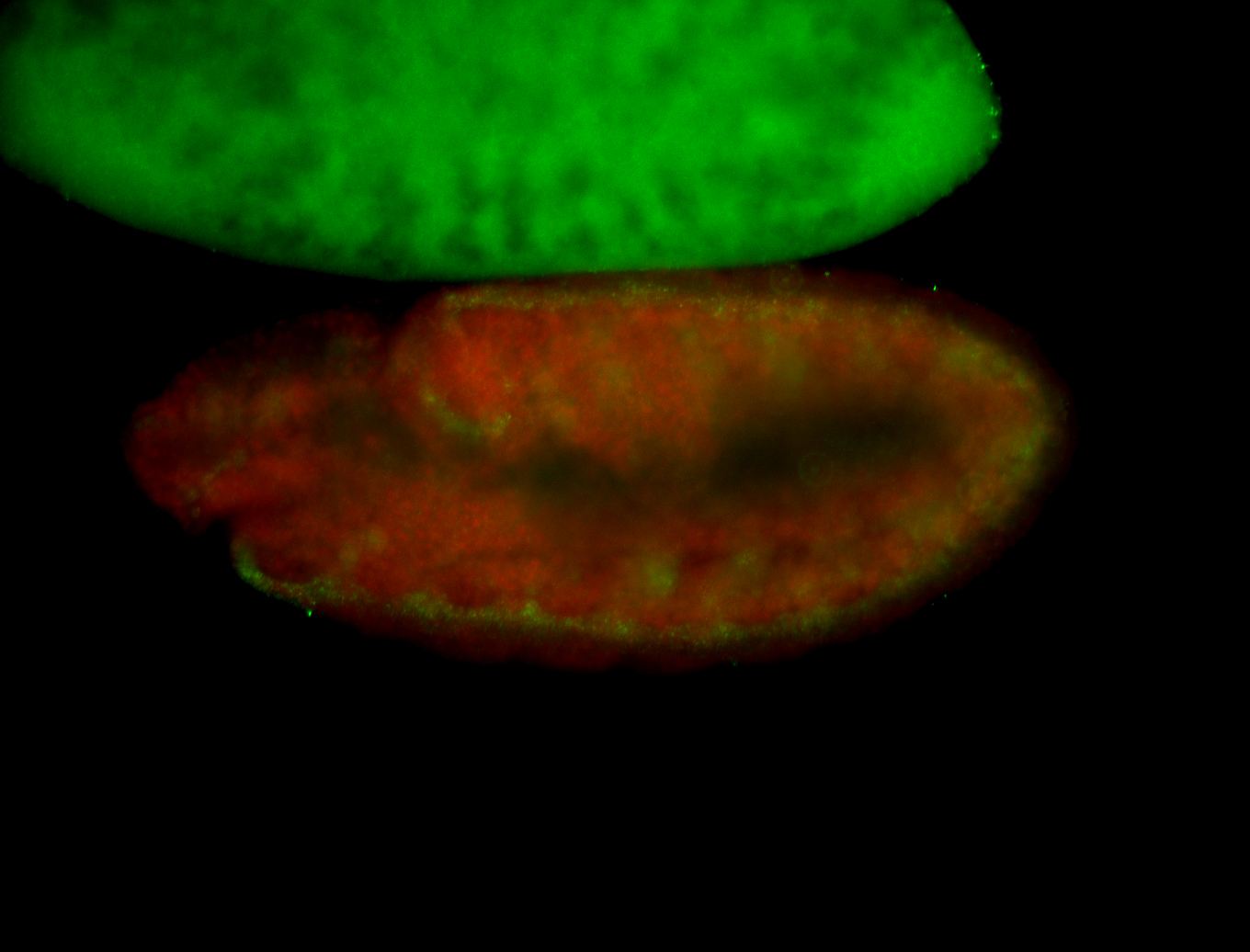
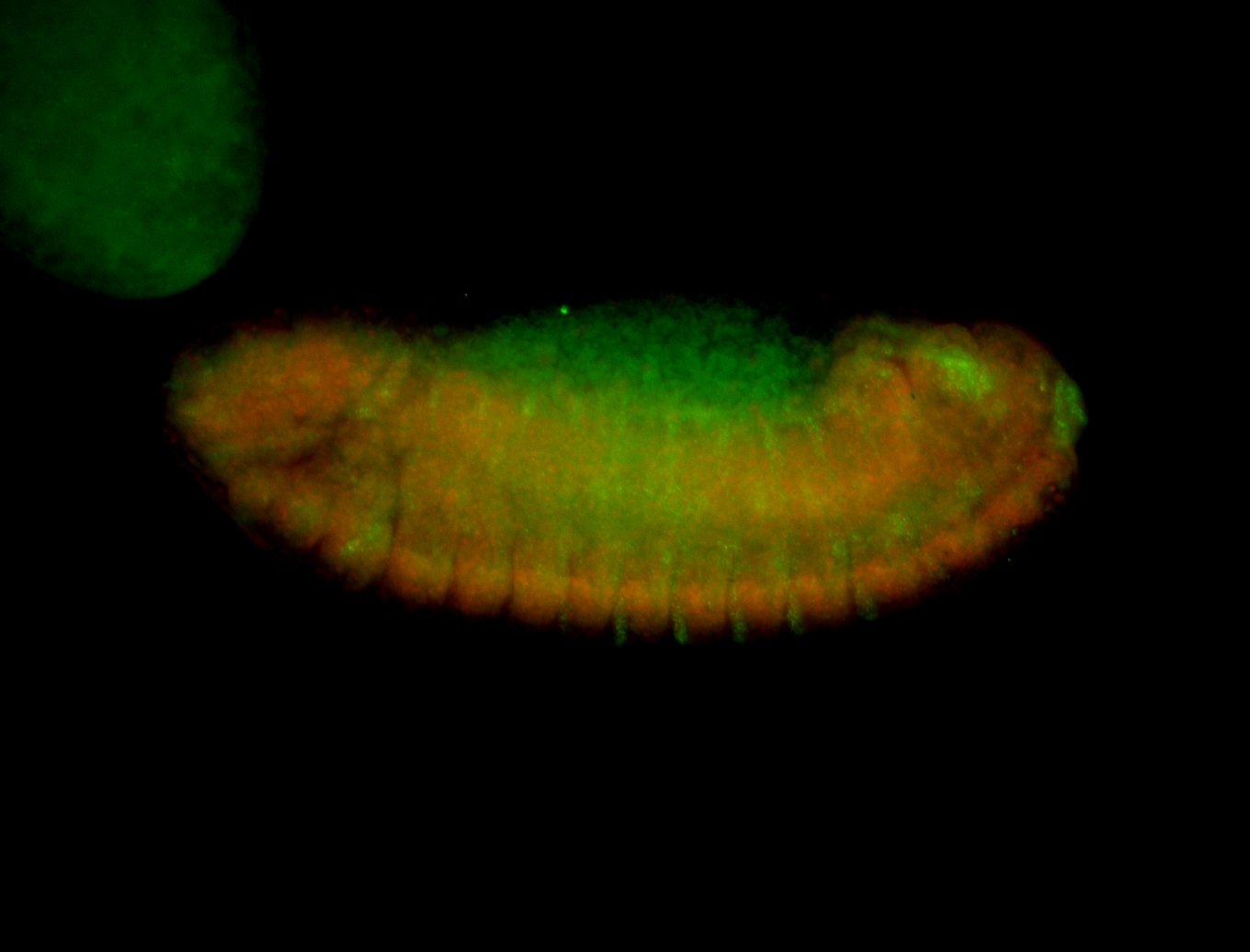
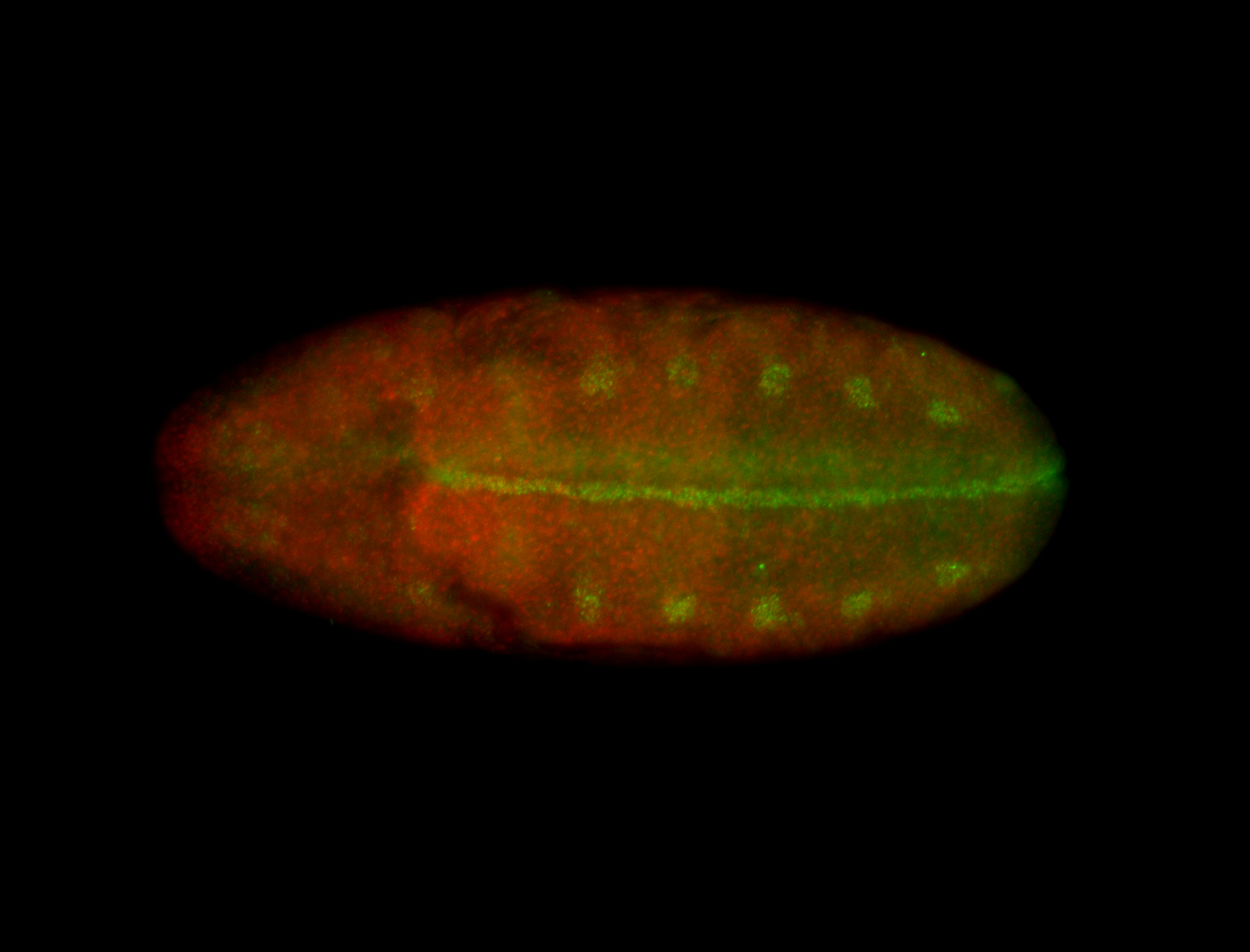
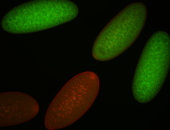
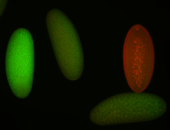
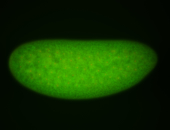
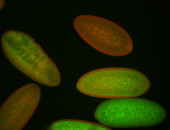
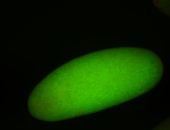
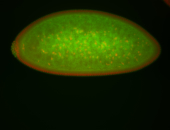
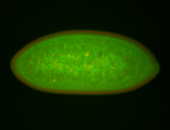
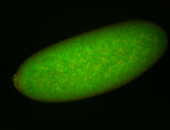
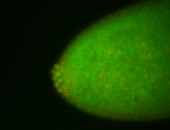
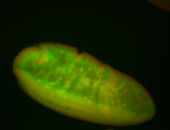
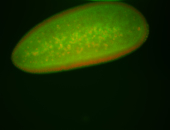
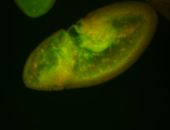
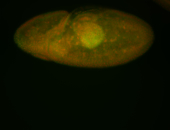
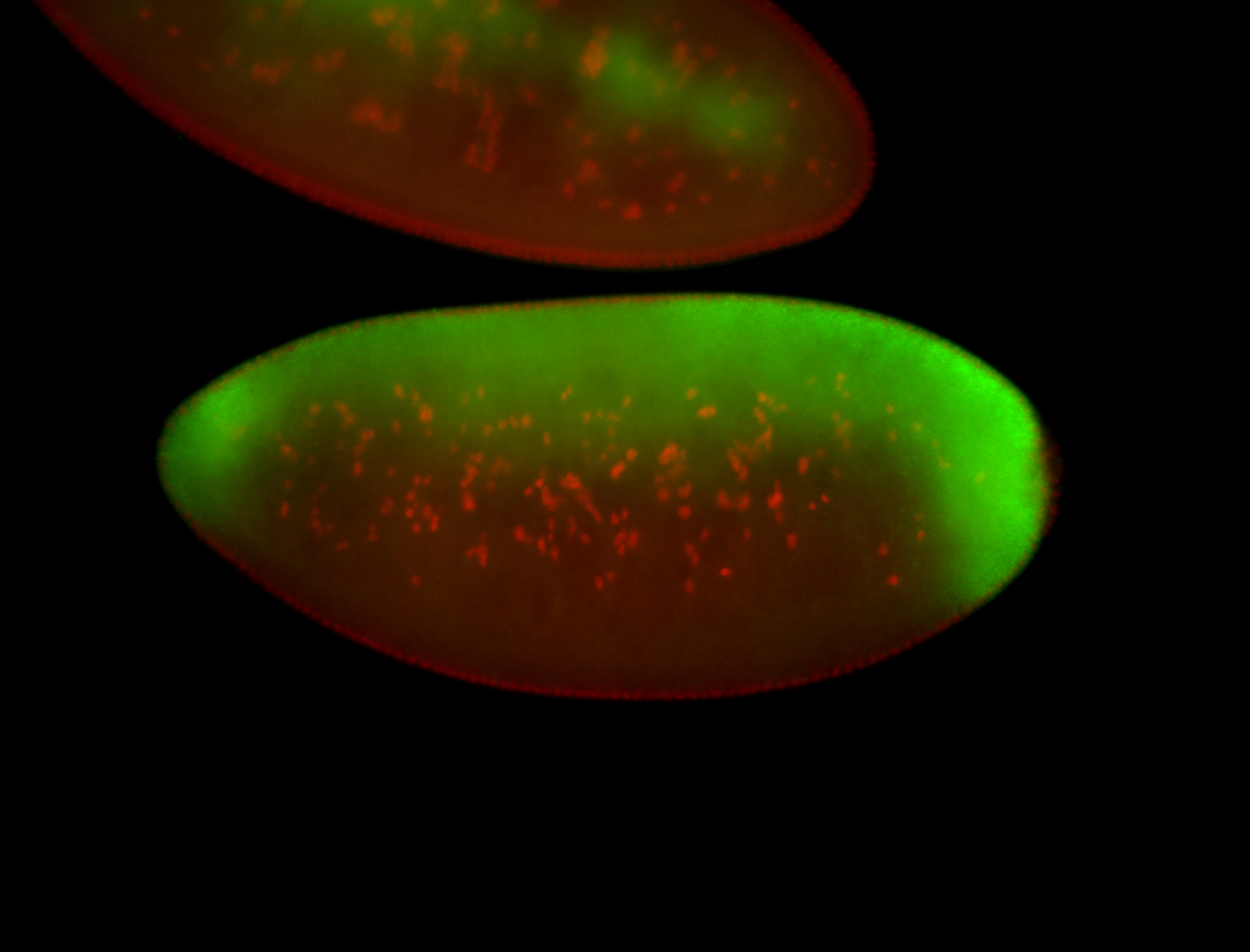
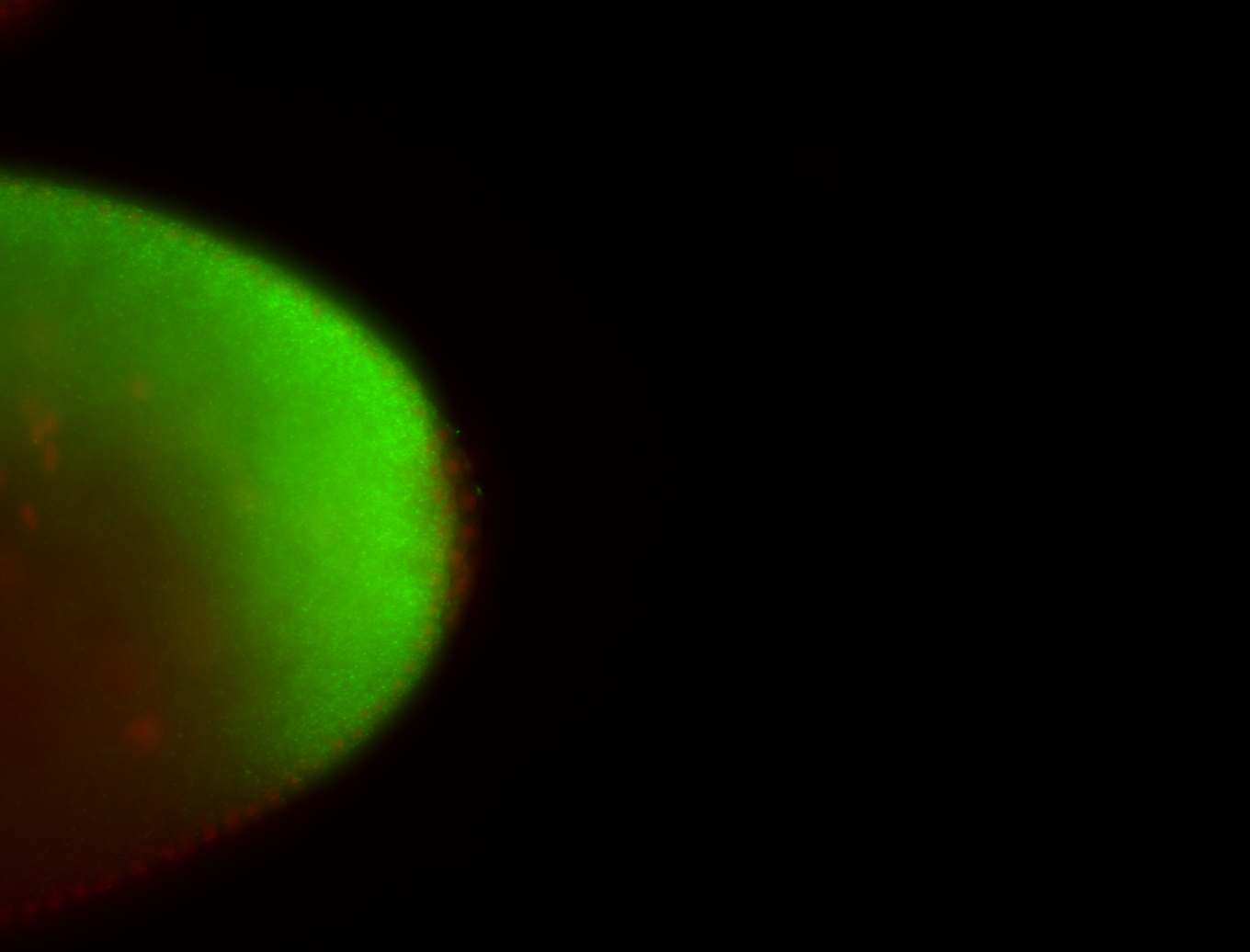
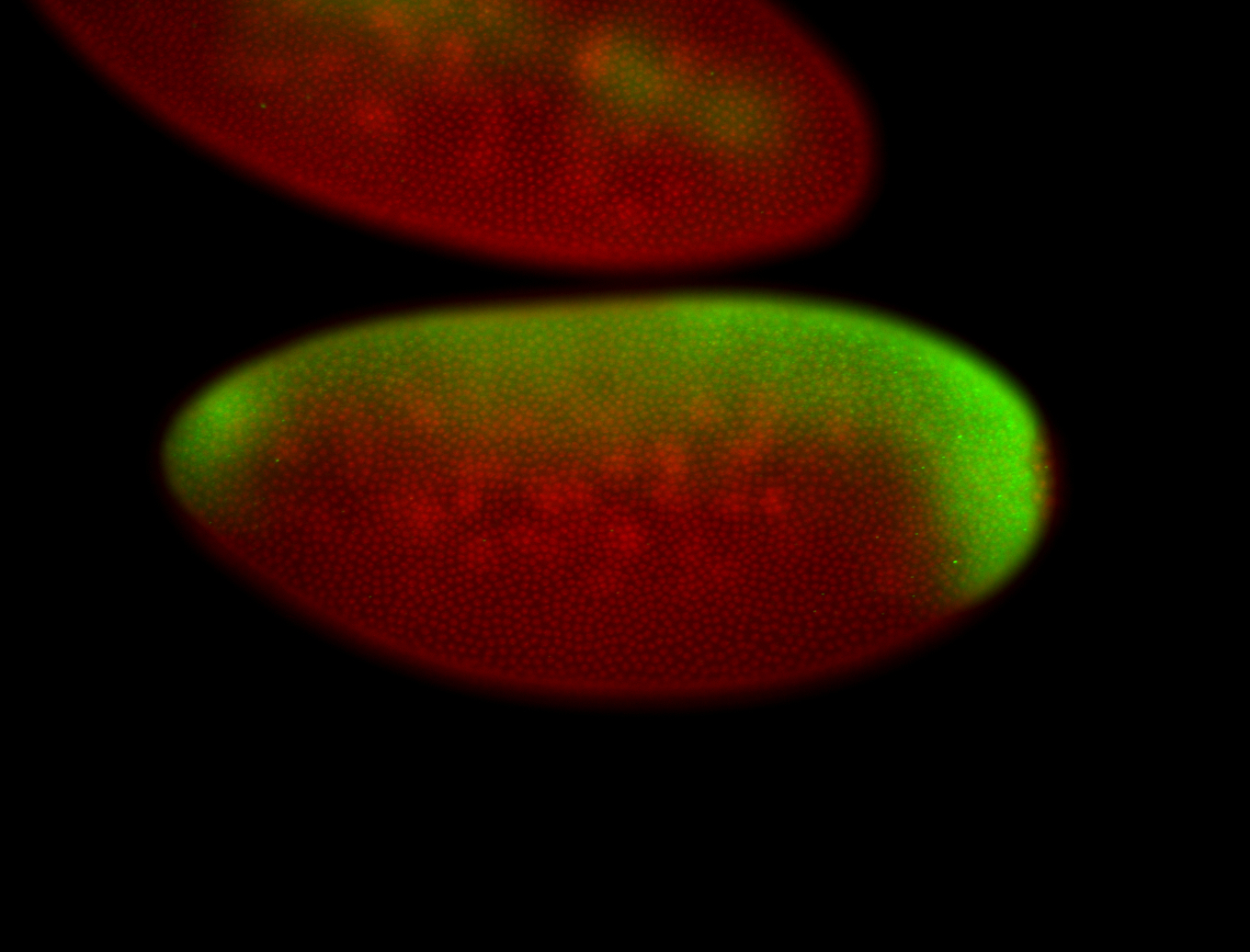
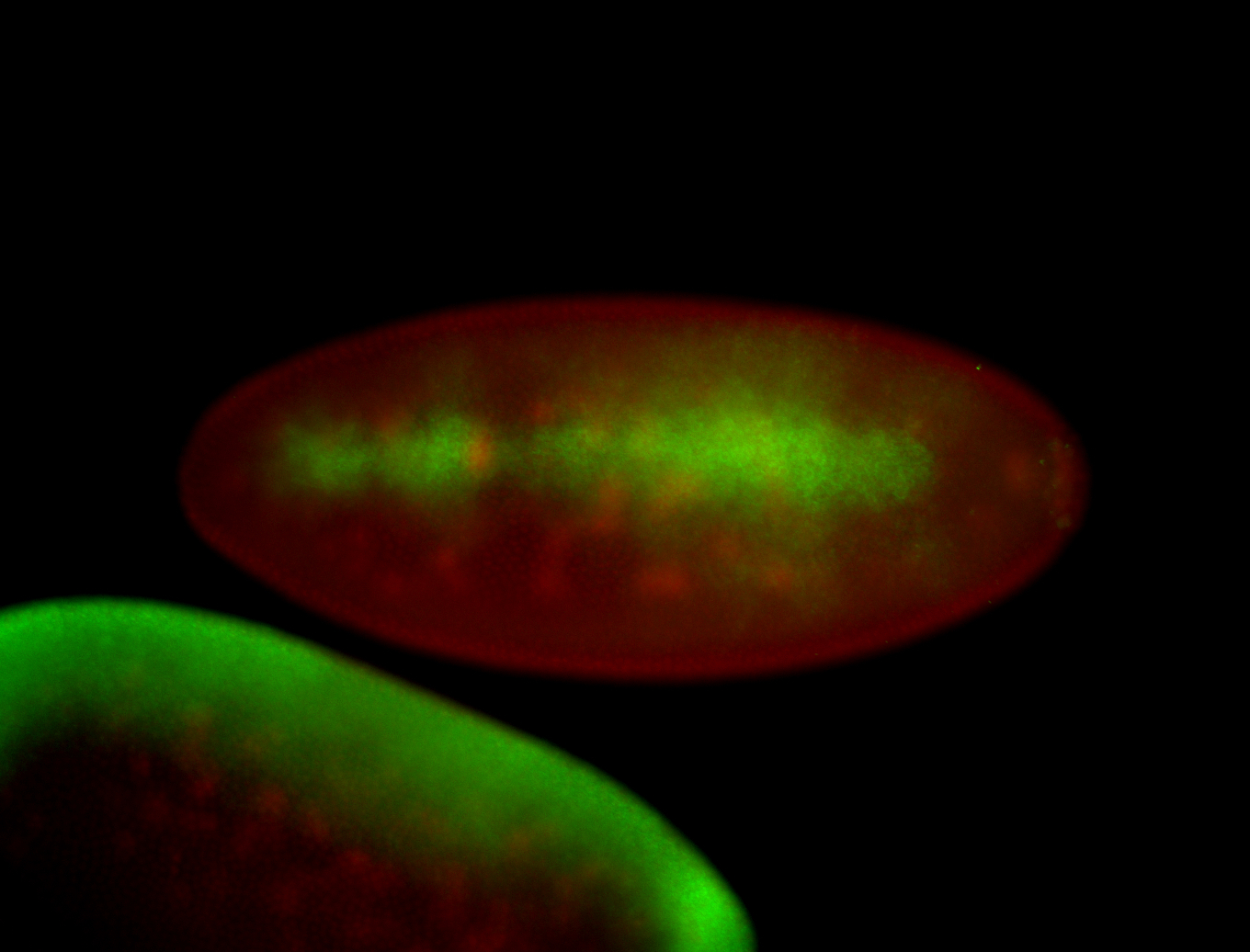
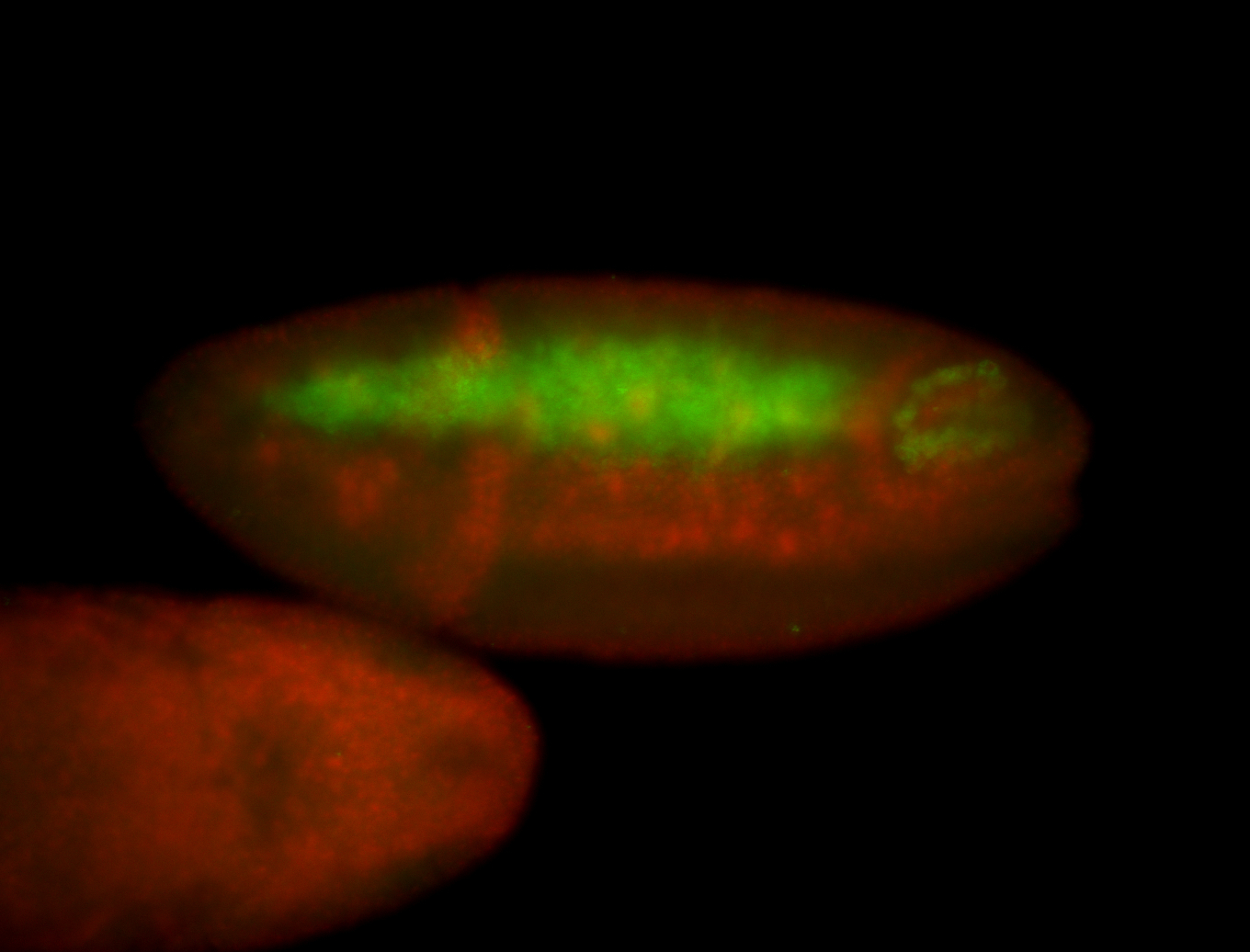
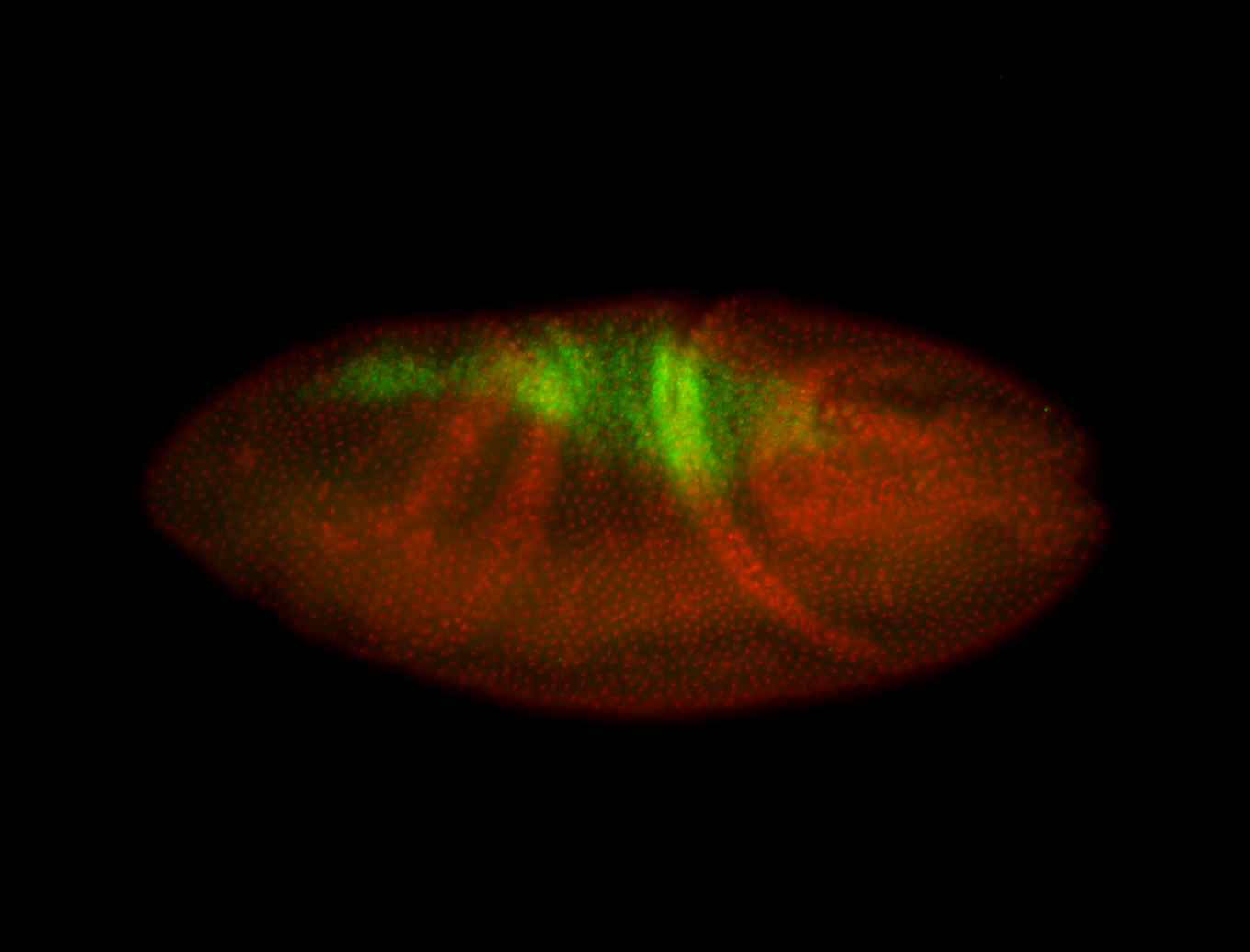
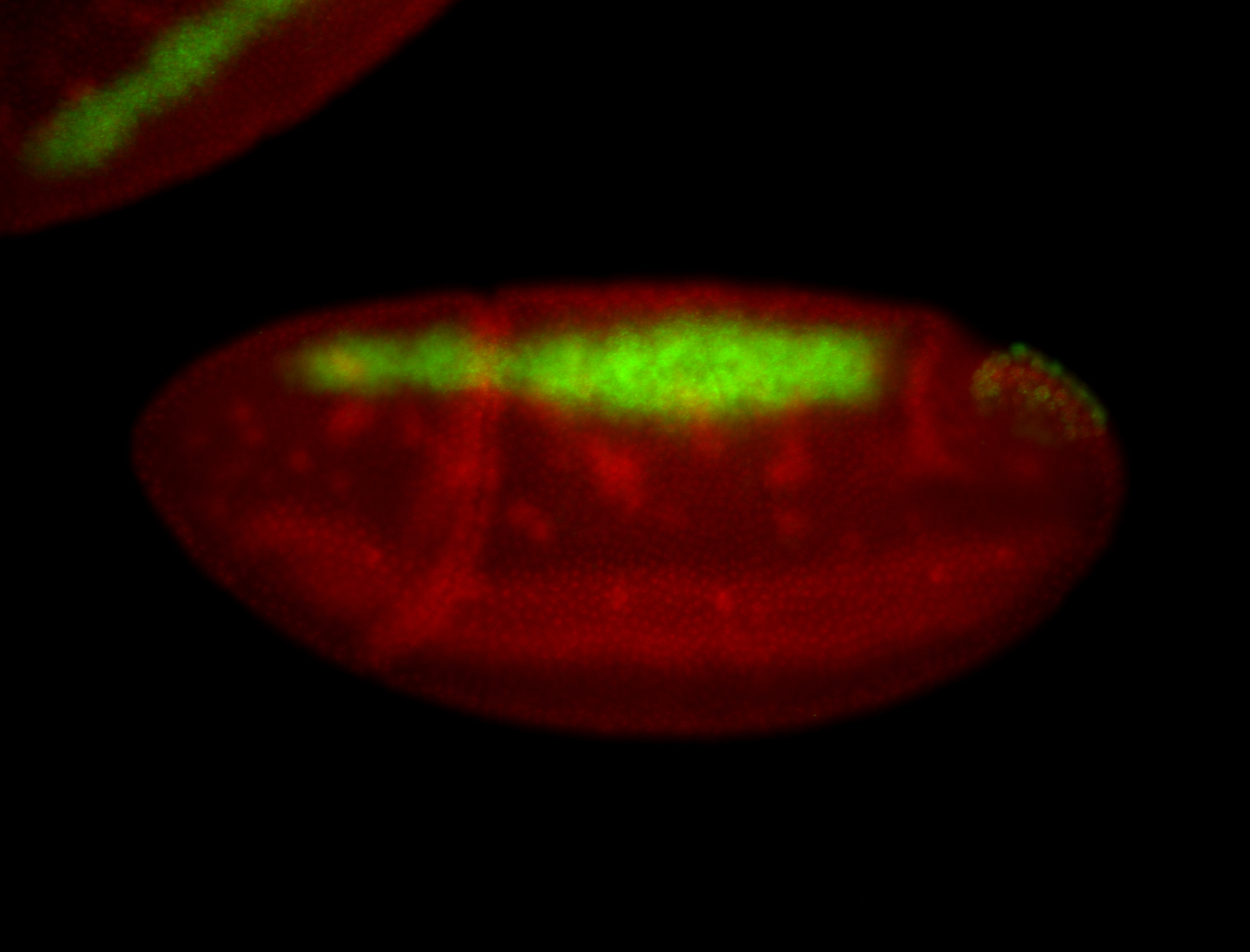
For a full list of searchable terms, please see the term page.
NOTE: It takes a few seconds for the term suggestions to load.
Result Summary
| Number of genes | 45 | |
| Export | ||
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe on gel. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Low probe. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Re-annotated. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| The probe looks good on gel. |
| Few cells have expression... |
| Low zygotic expression. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
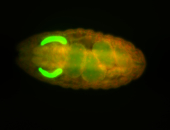
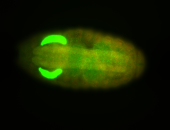
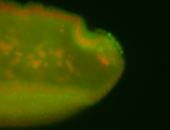
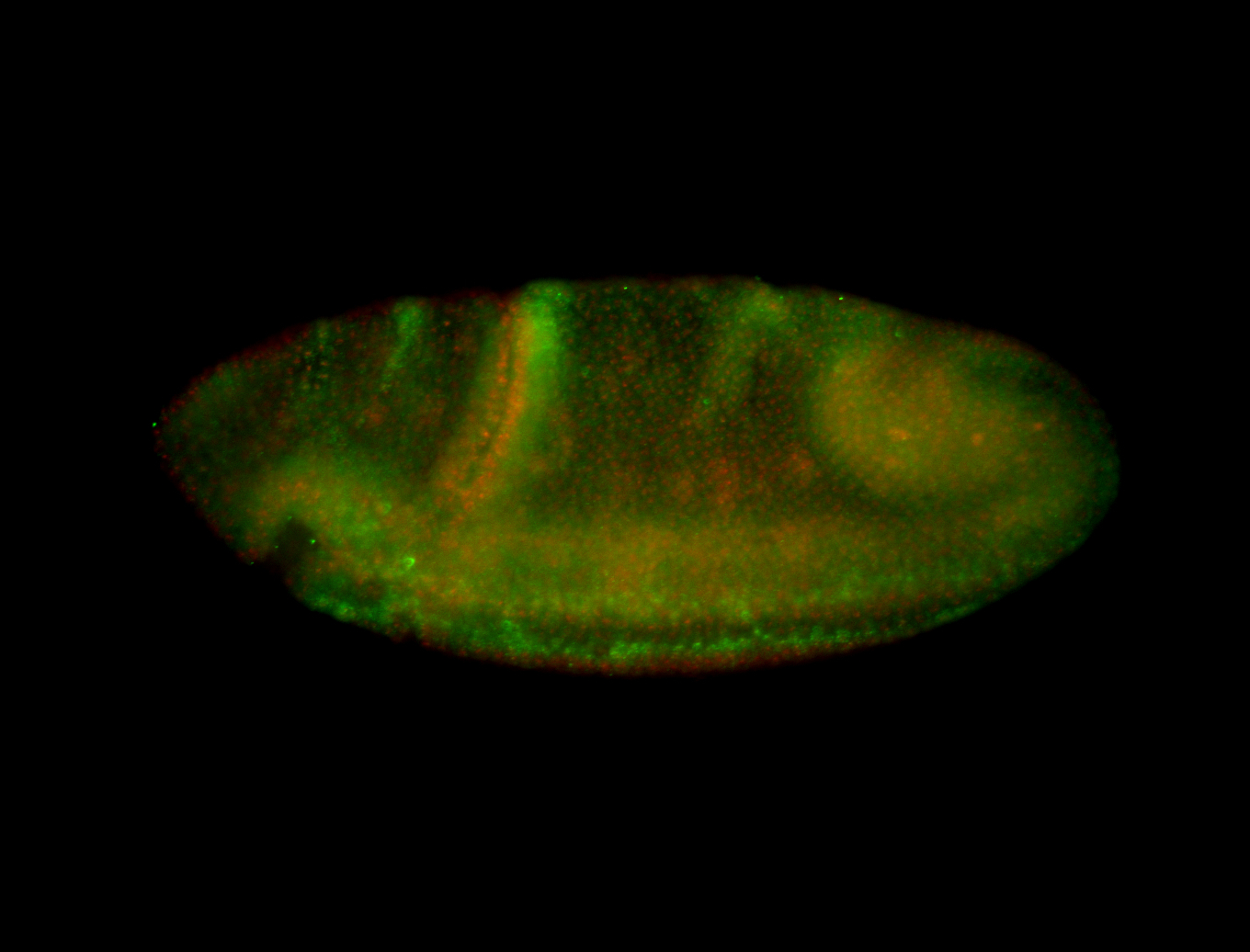
| Tracheal pits. |
| Tracheal pits. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
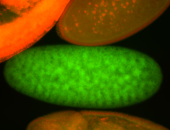
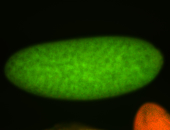
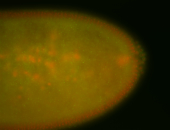
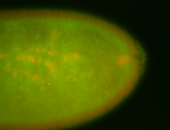
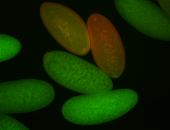
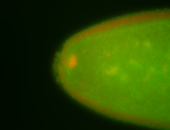
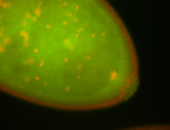
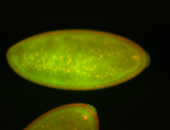
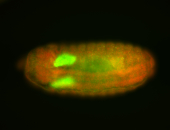
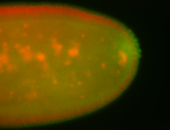
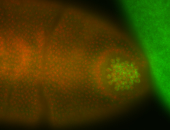
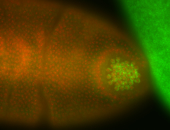
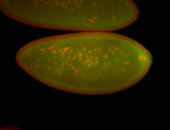
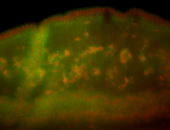
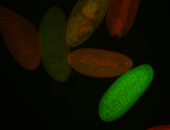
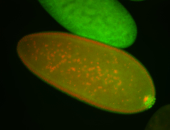
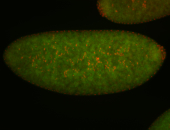
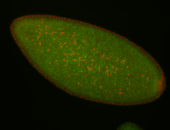
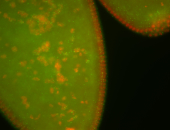
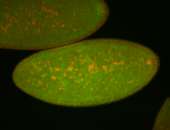
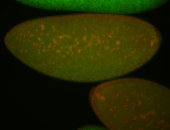
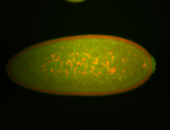
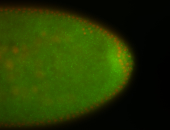
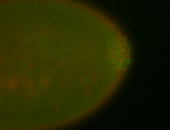
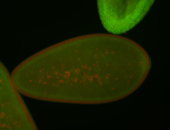
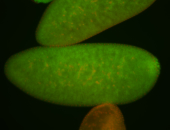
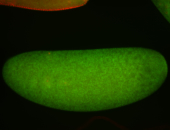
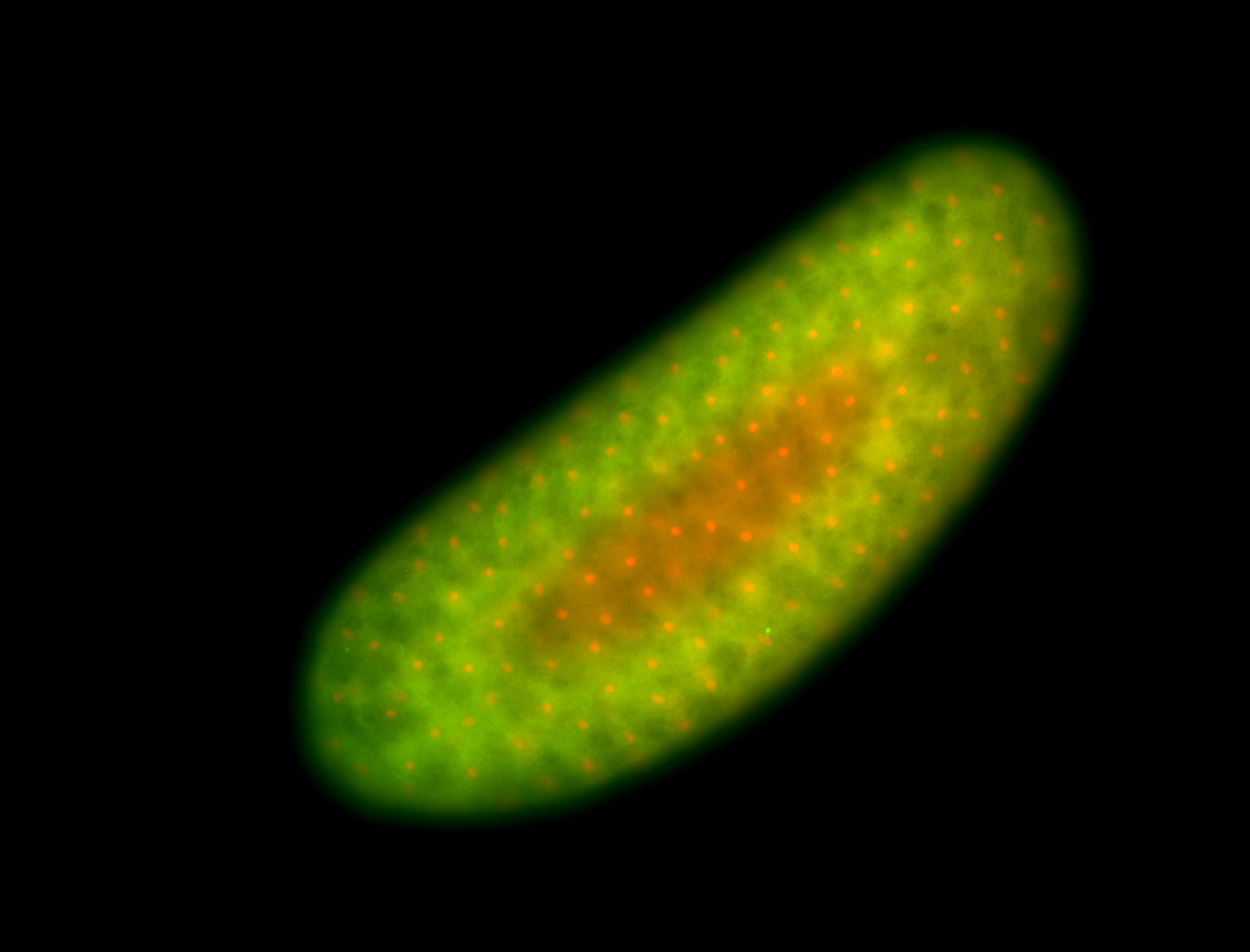
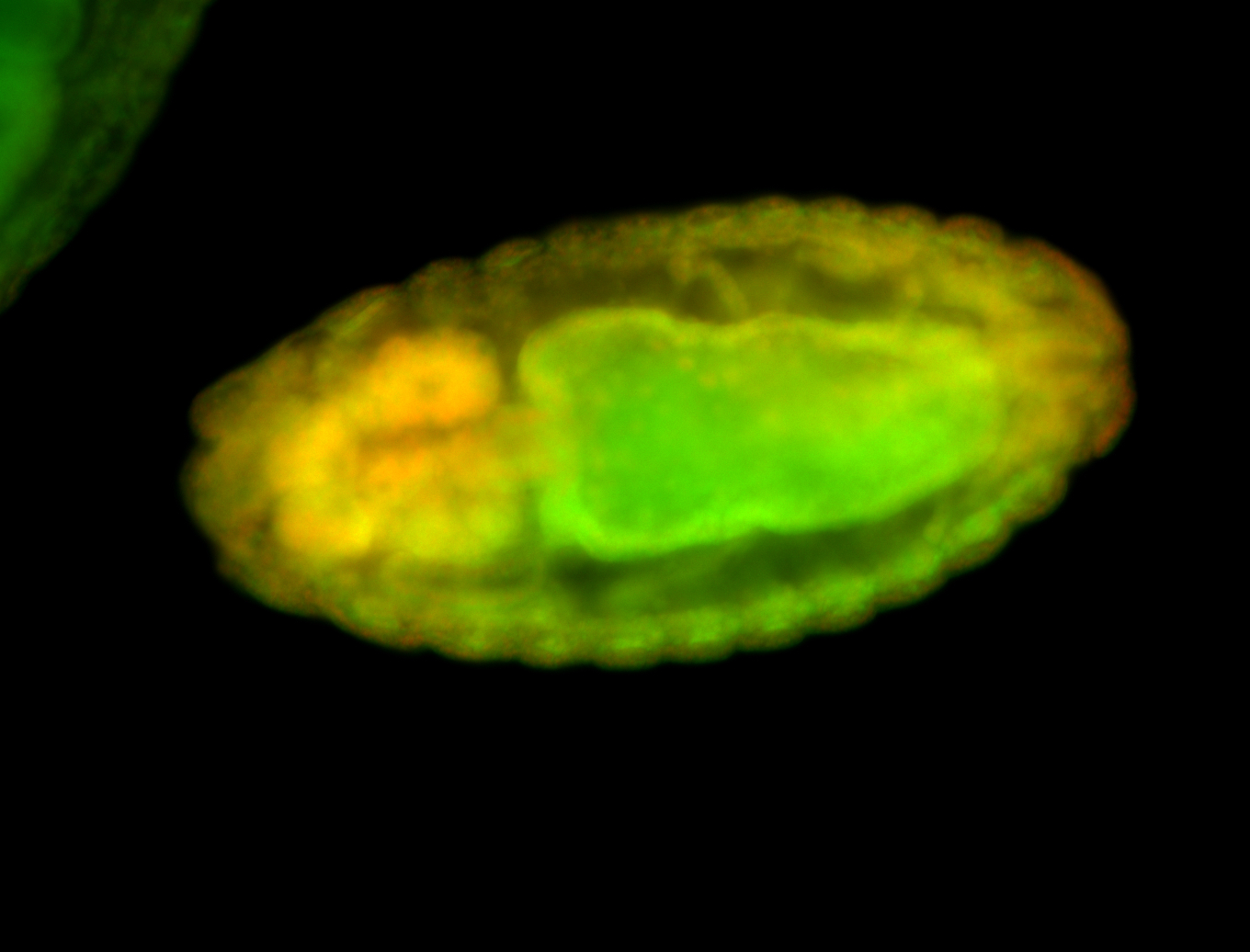
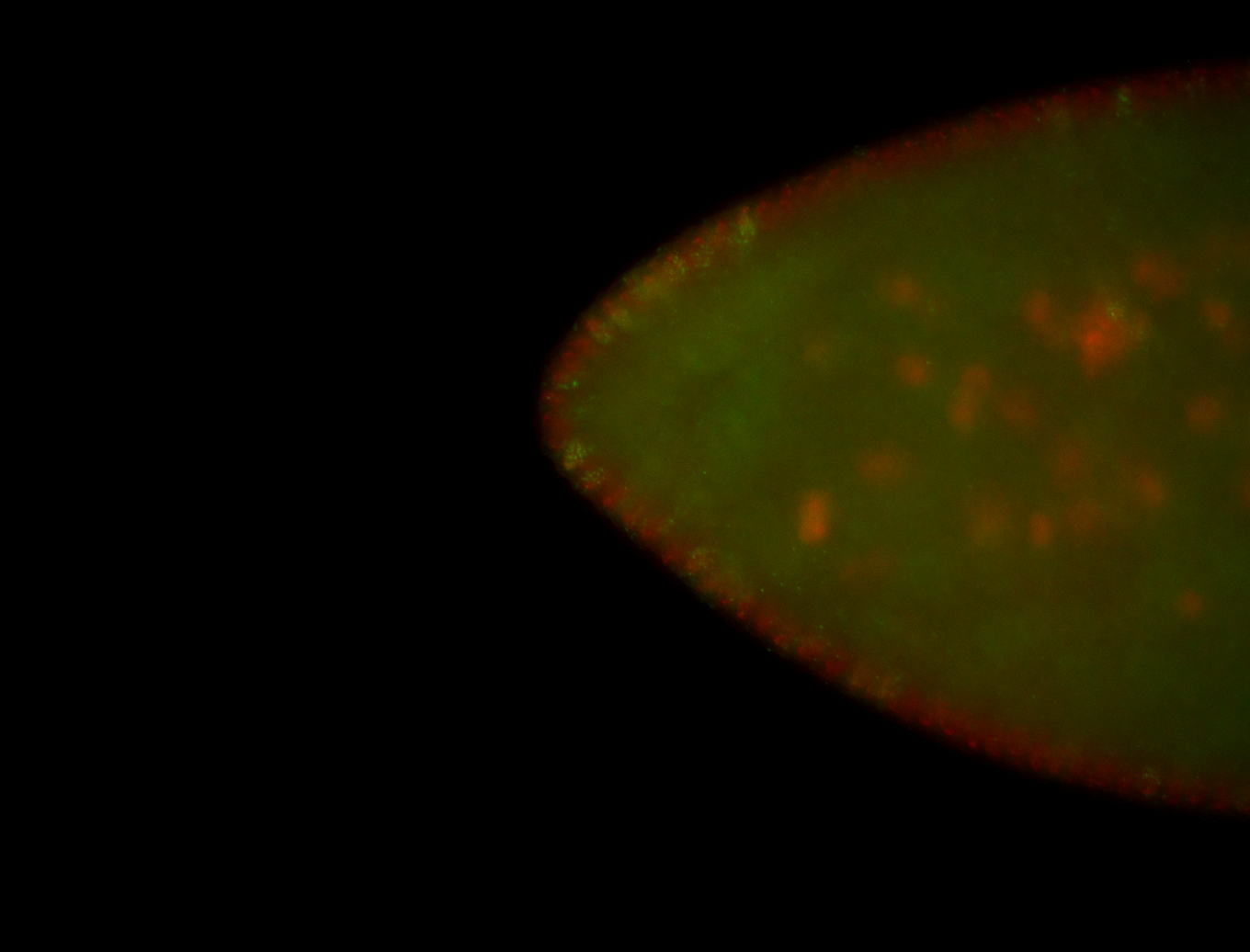
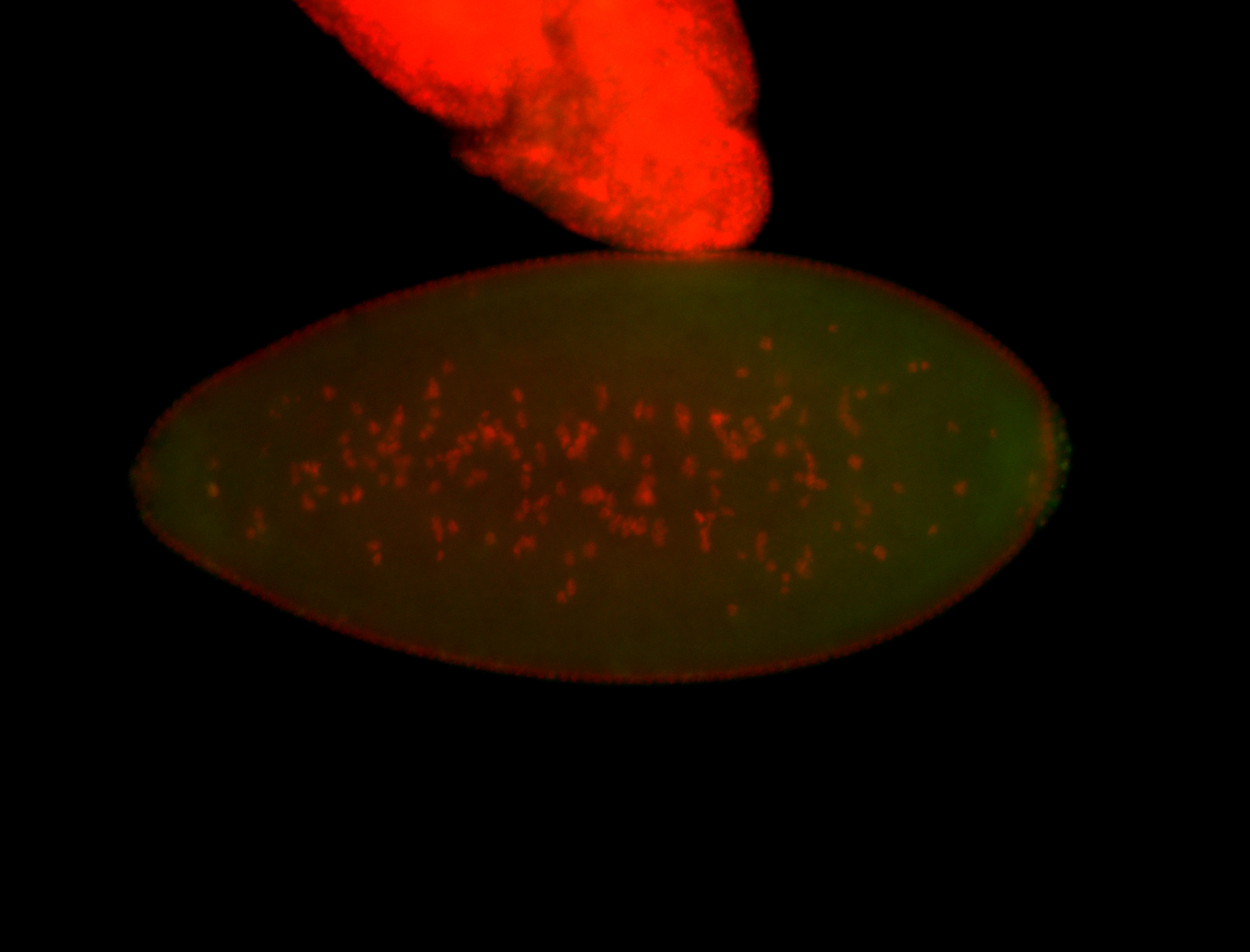
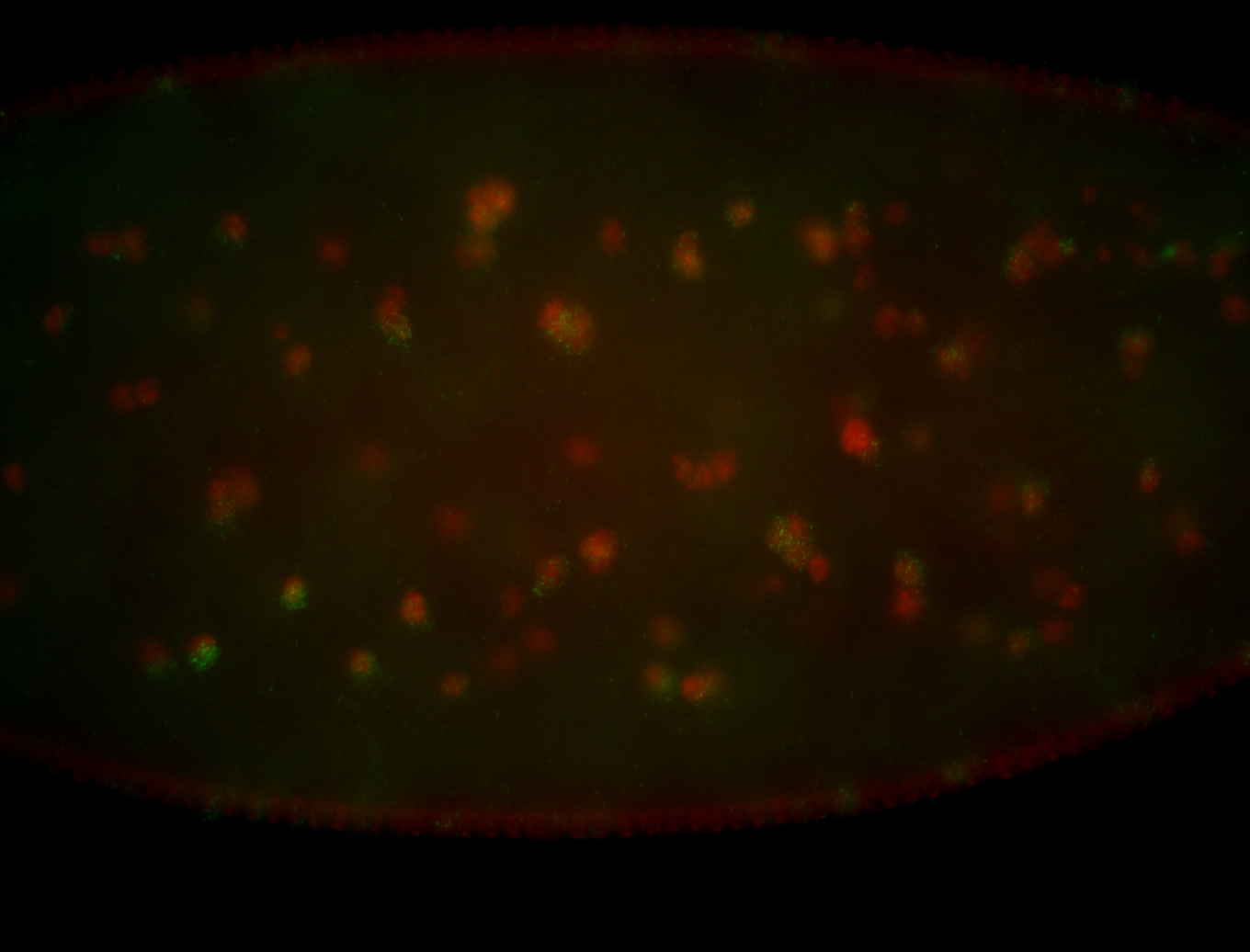
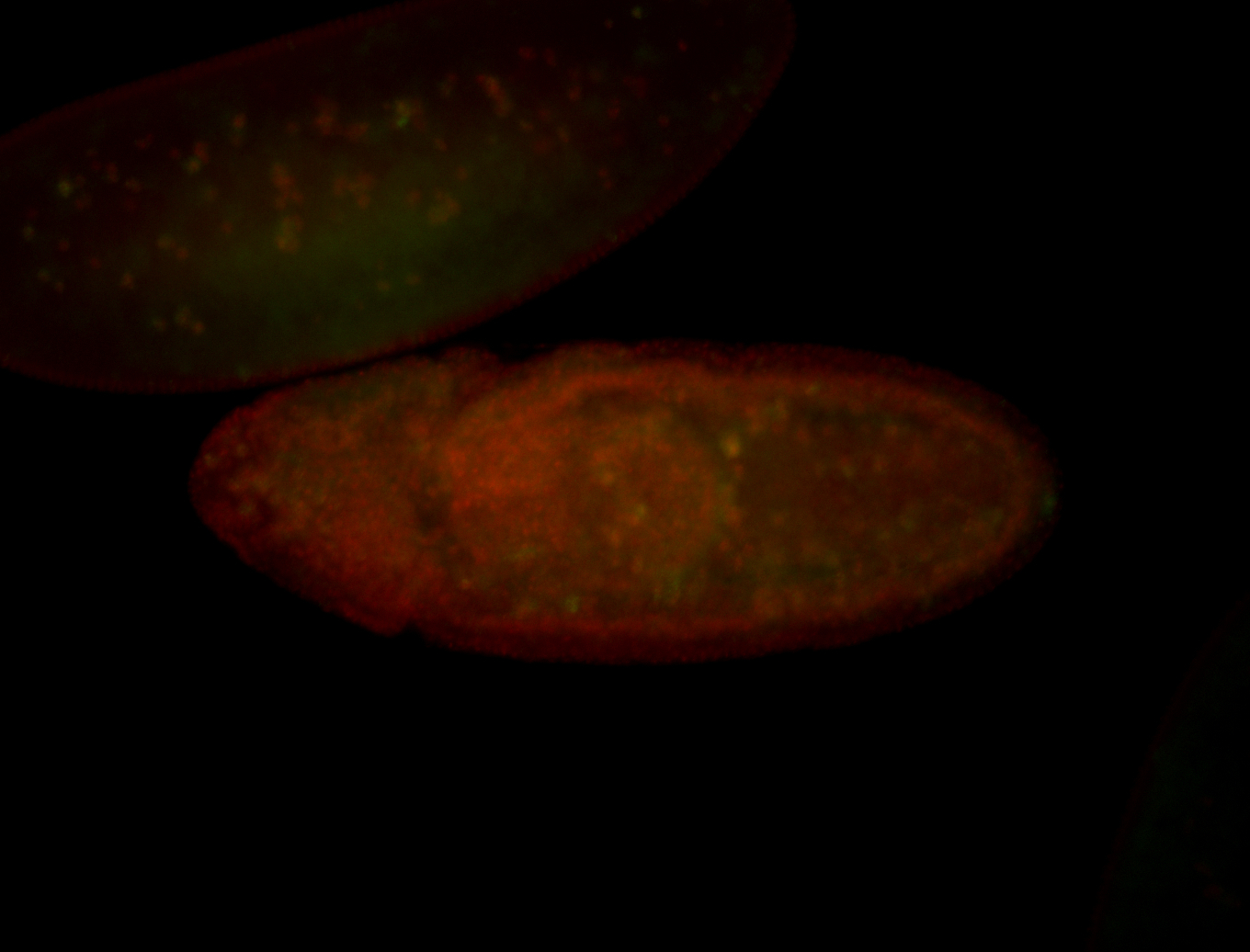
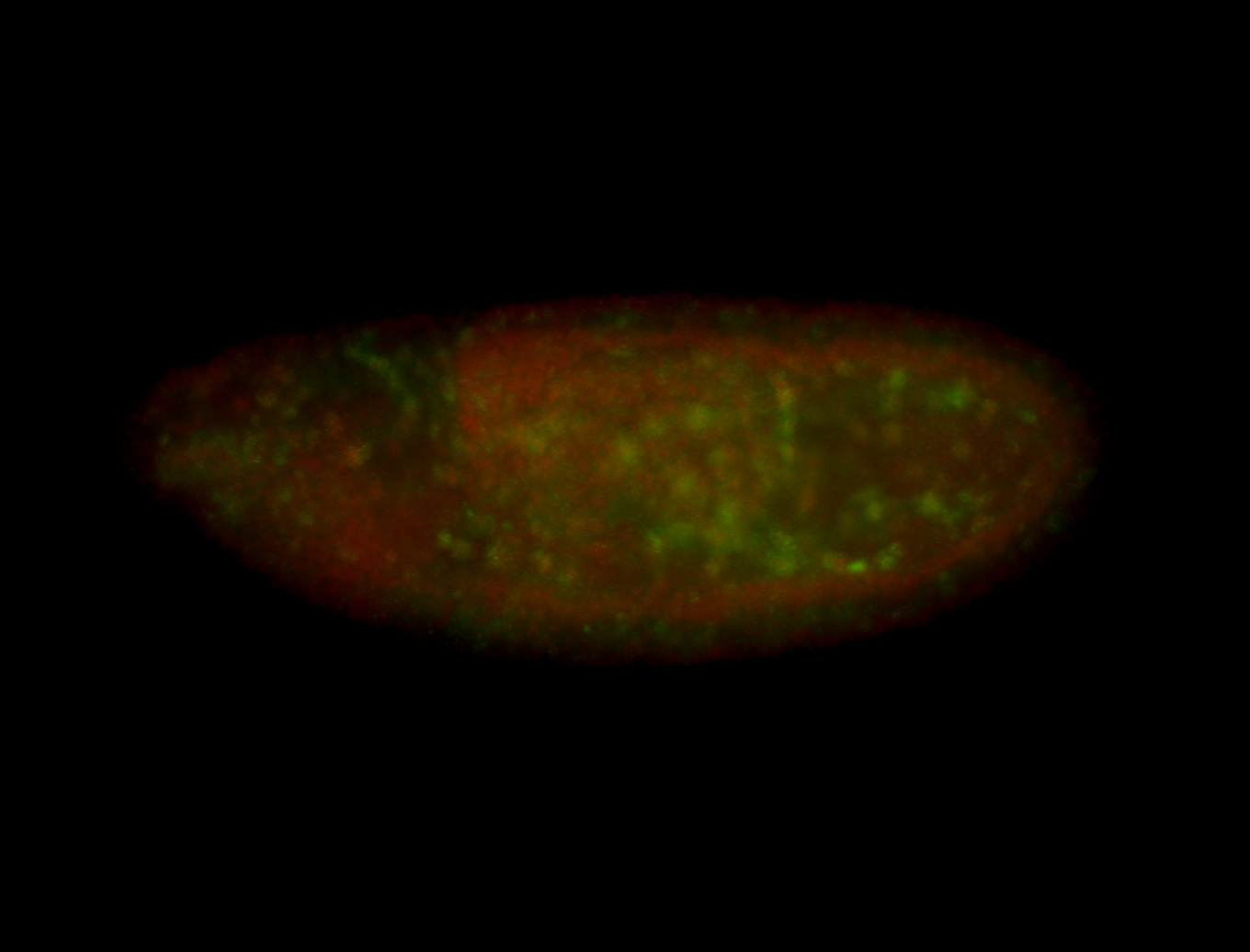
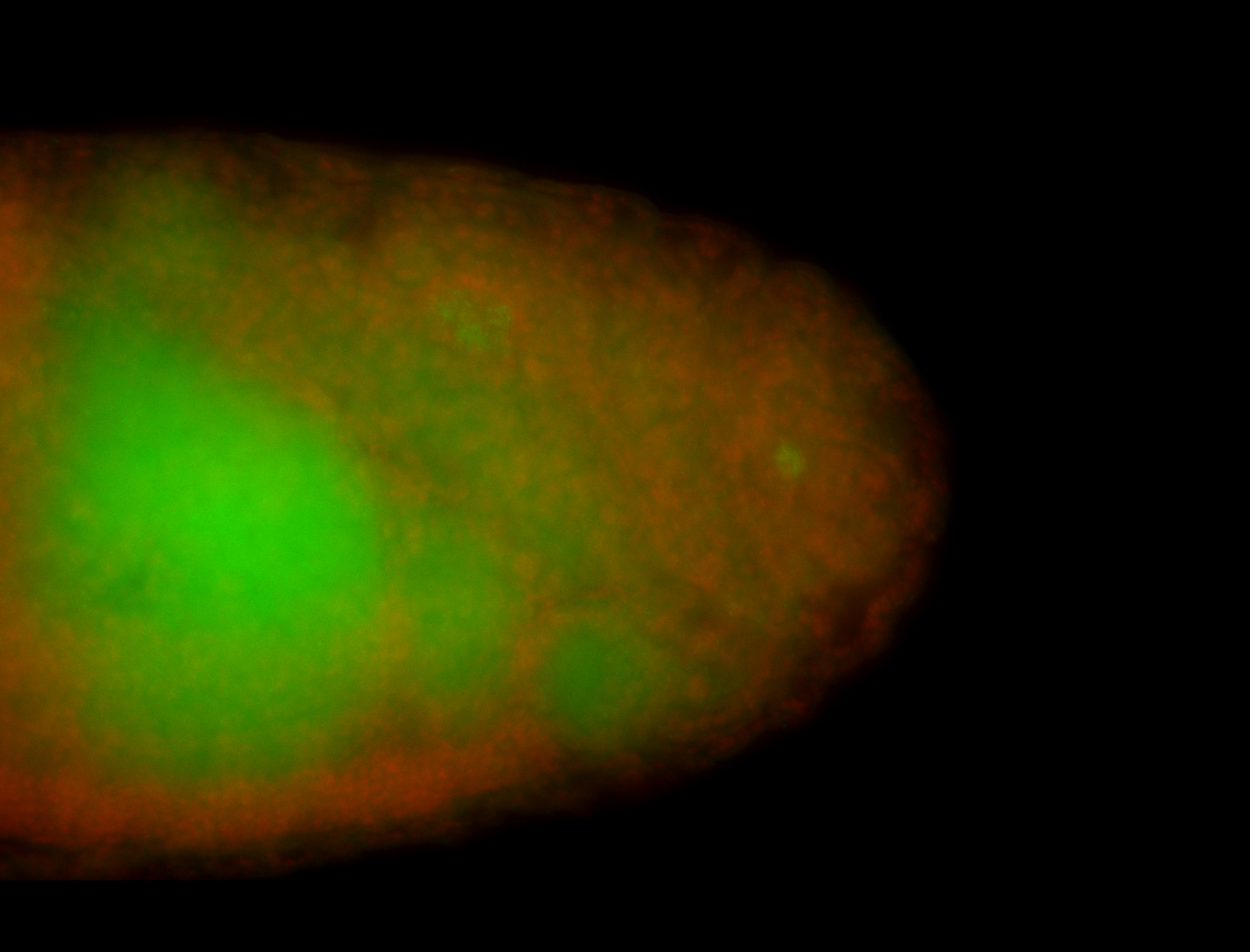
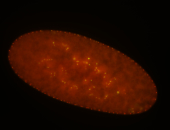
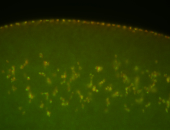
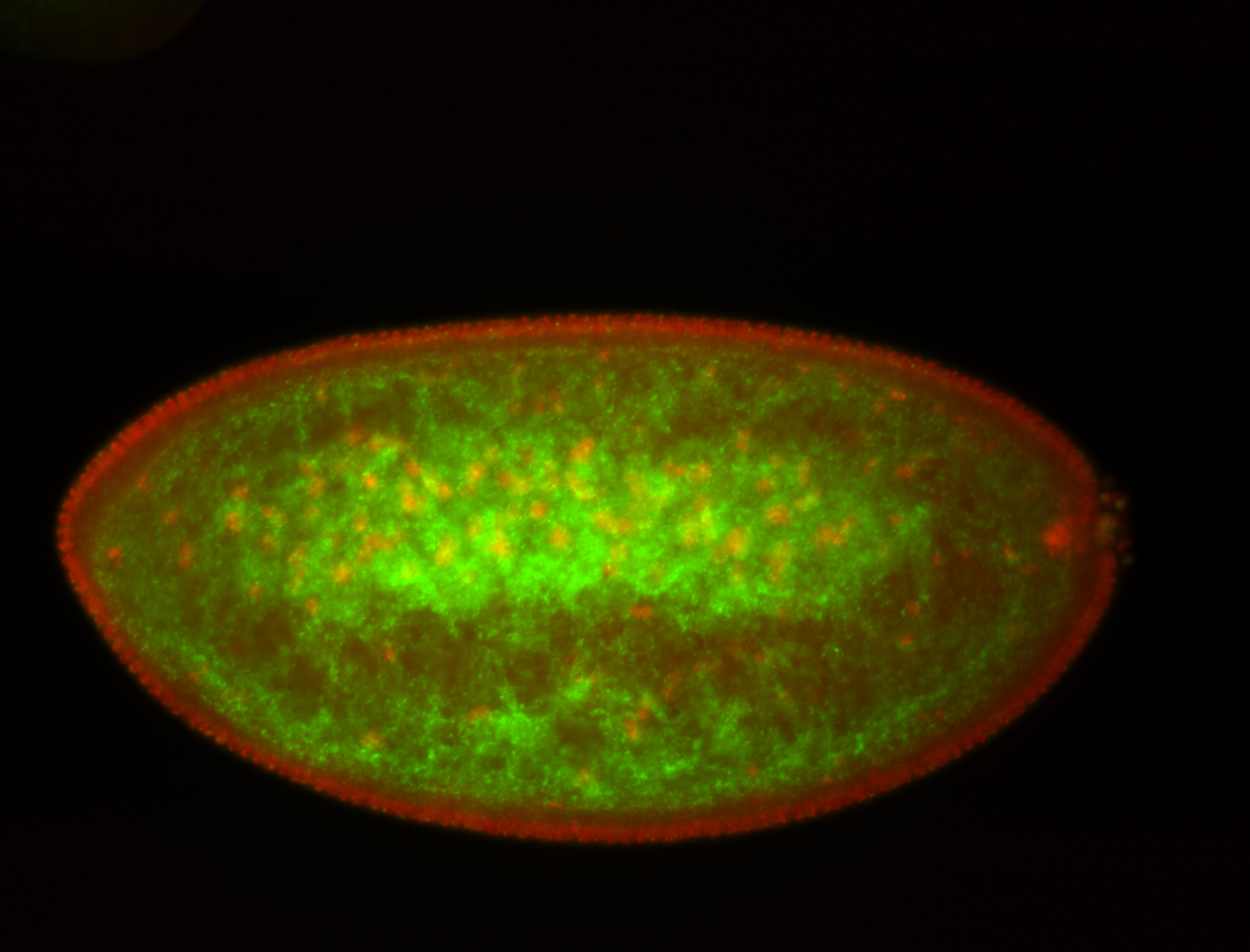
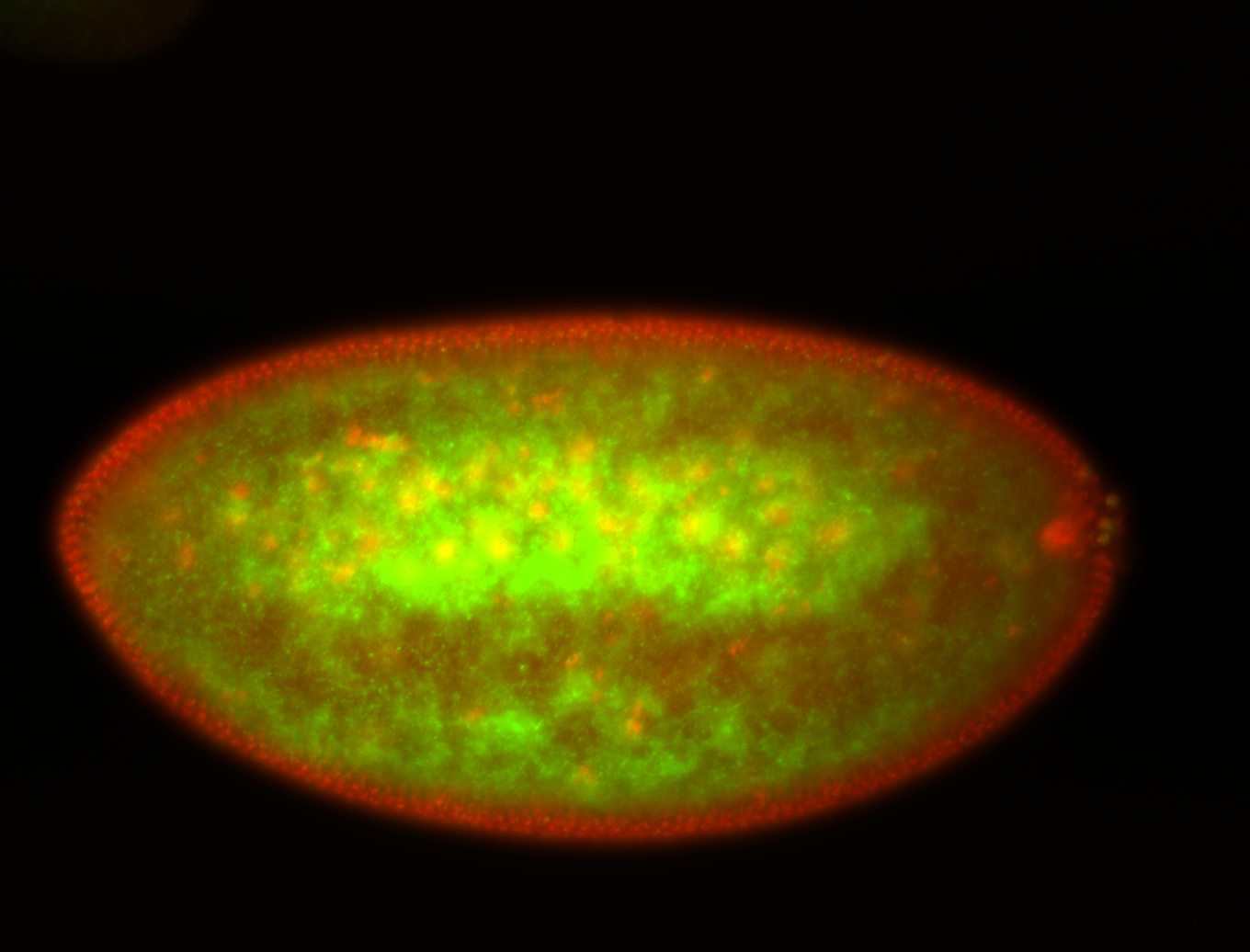
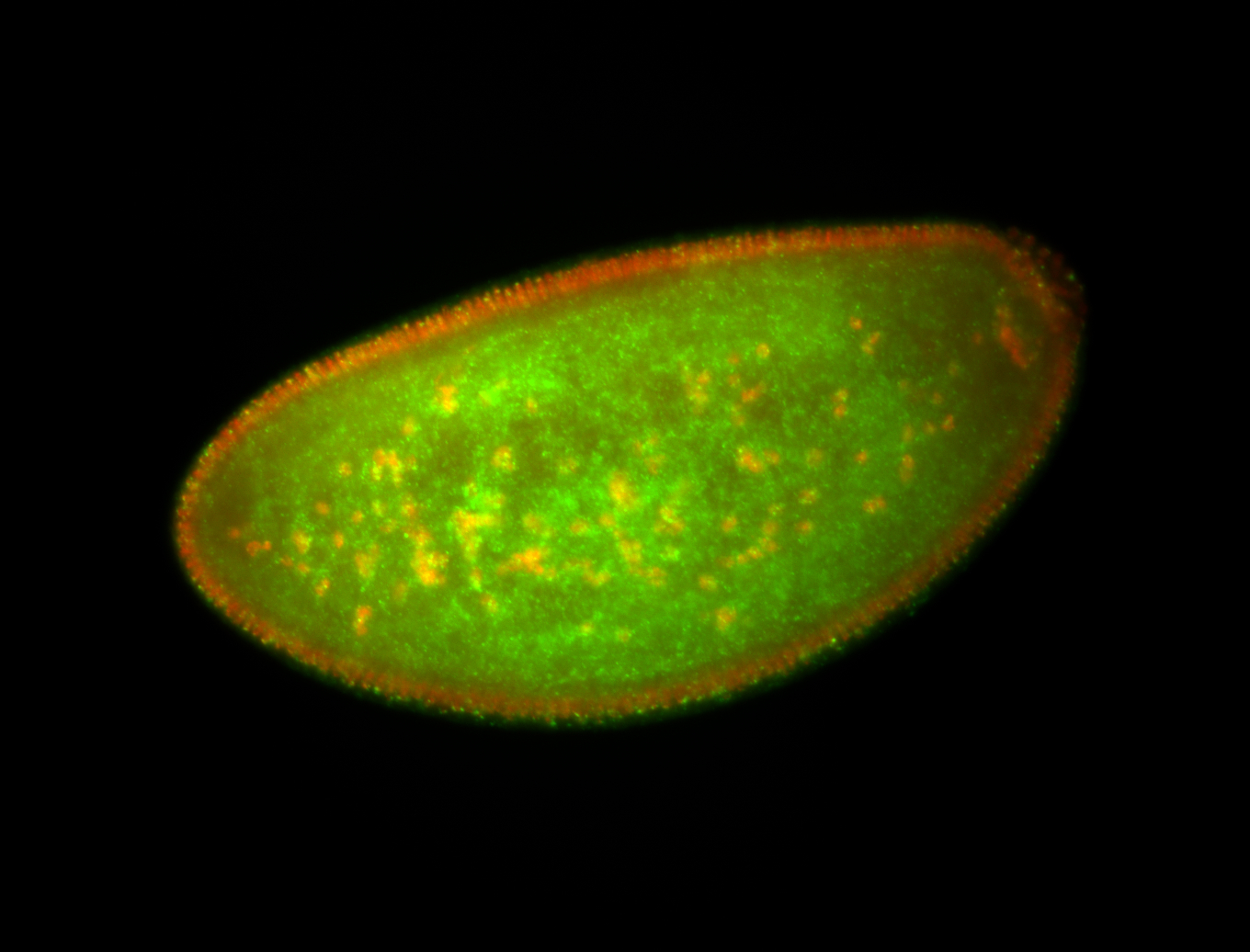
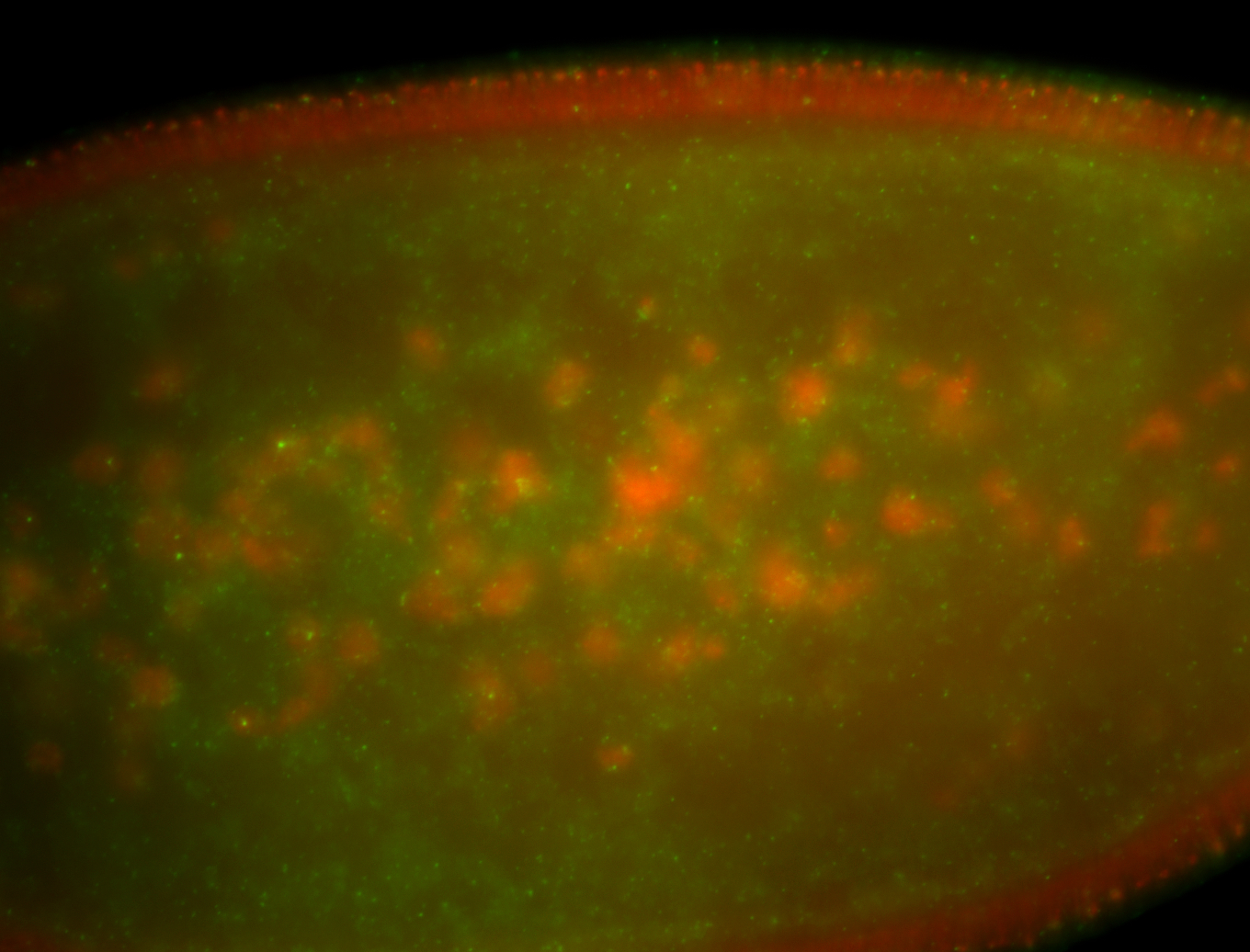
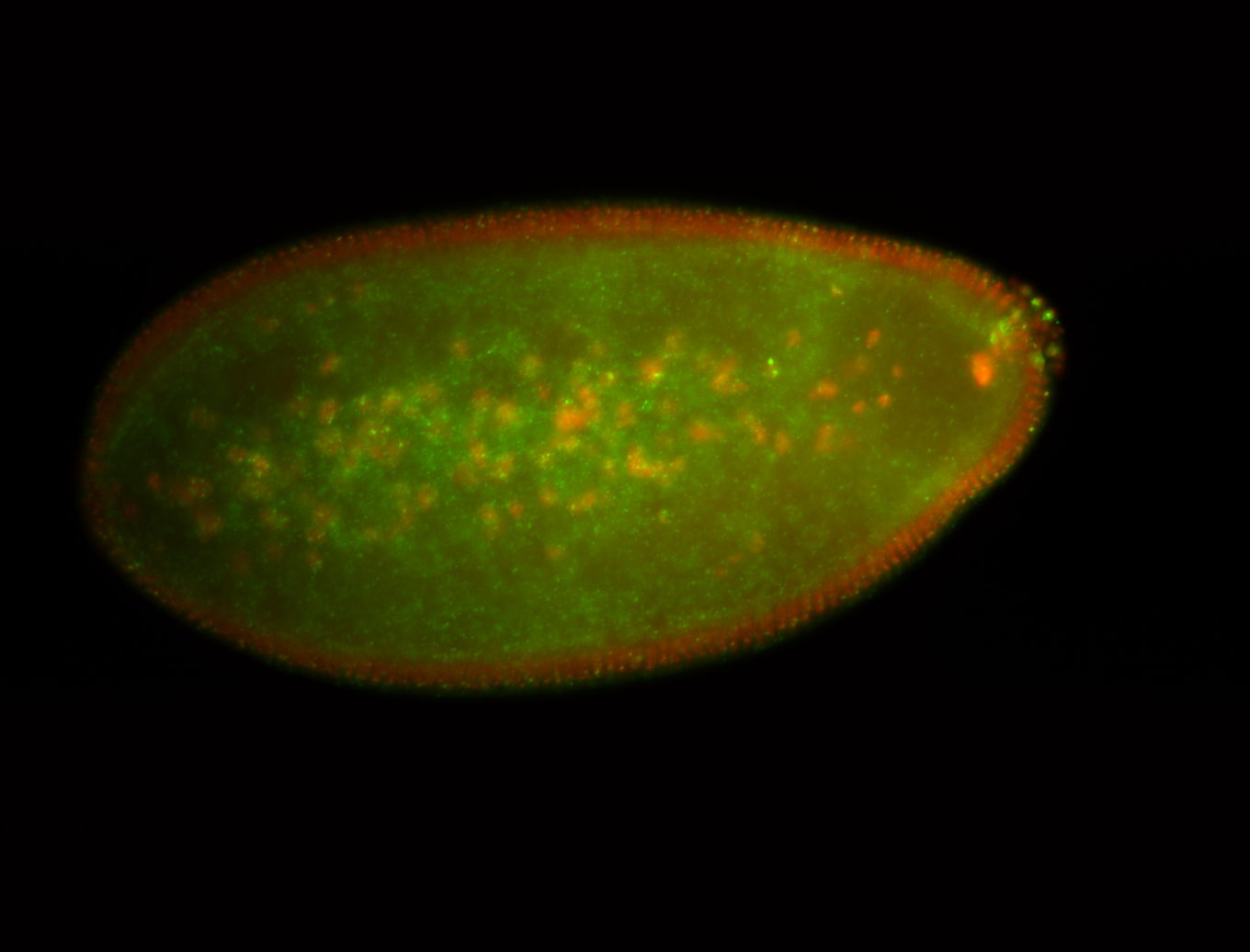
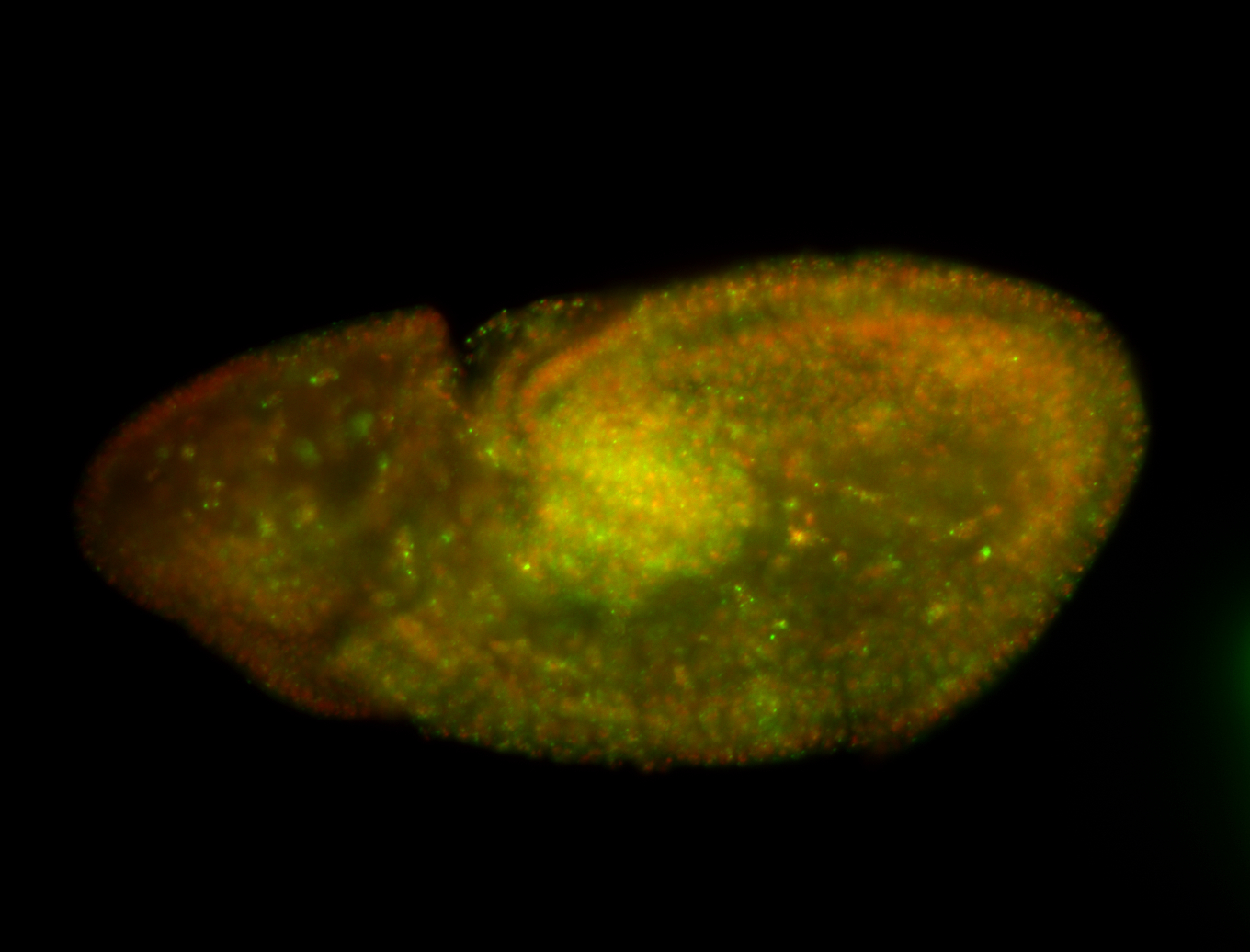
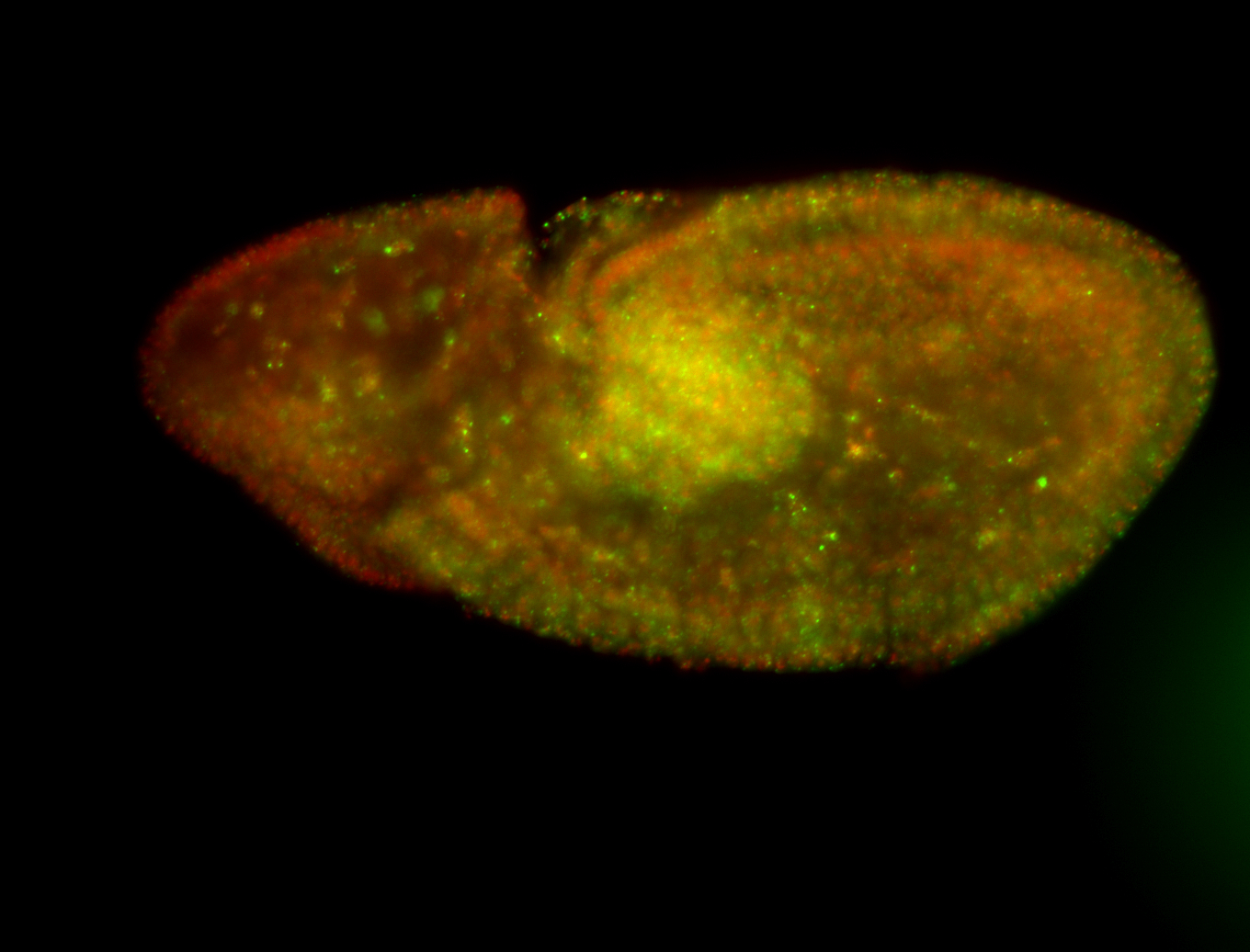
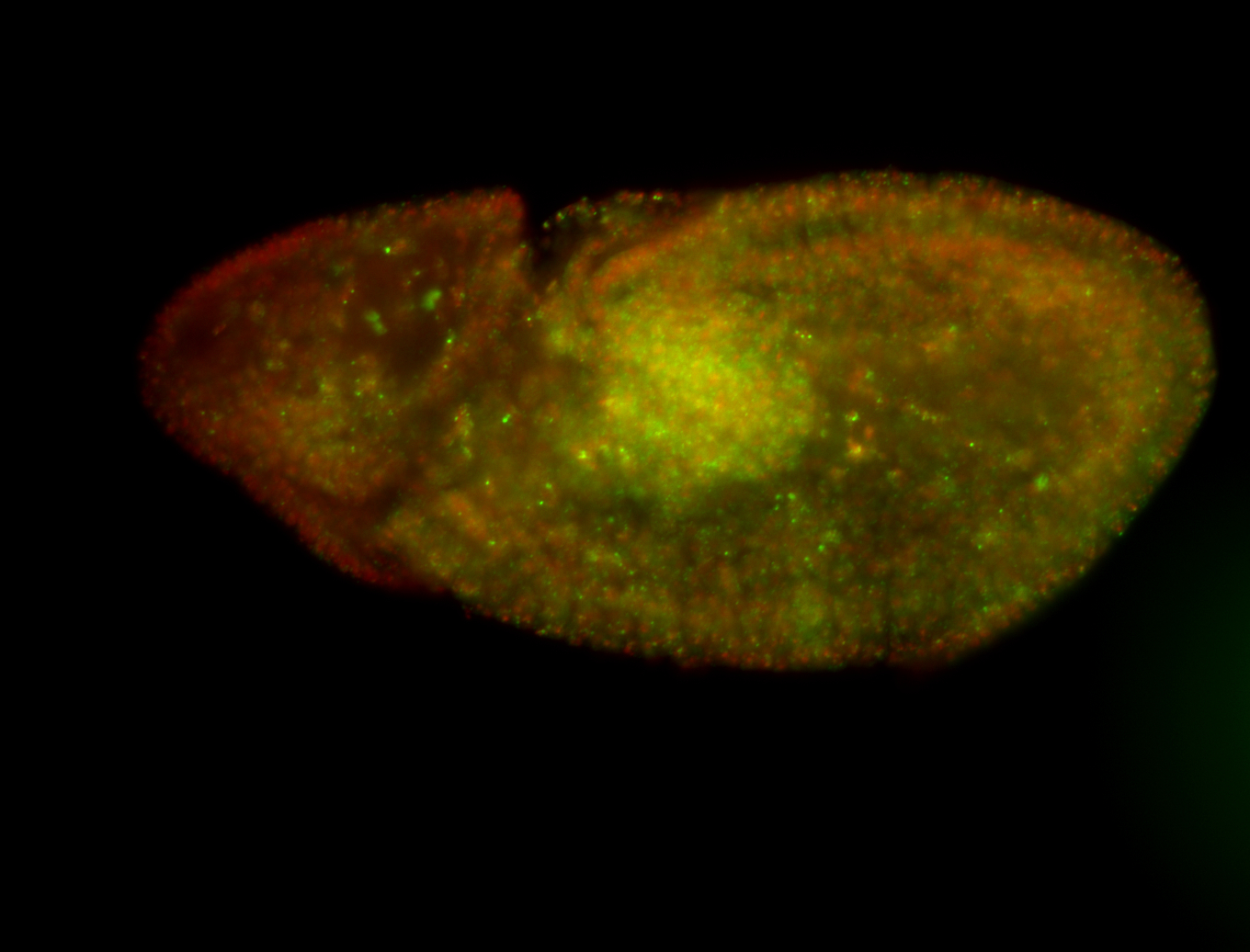
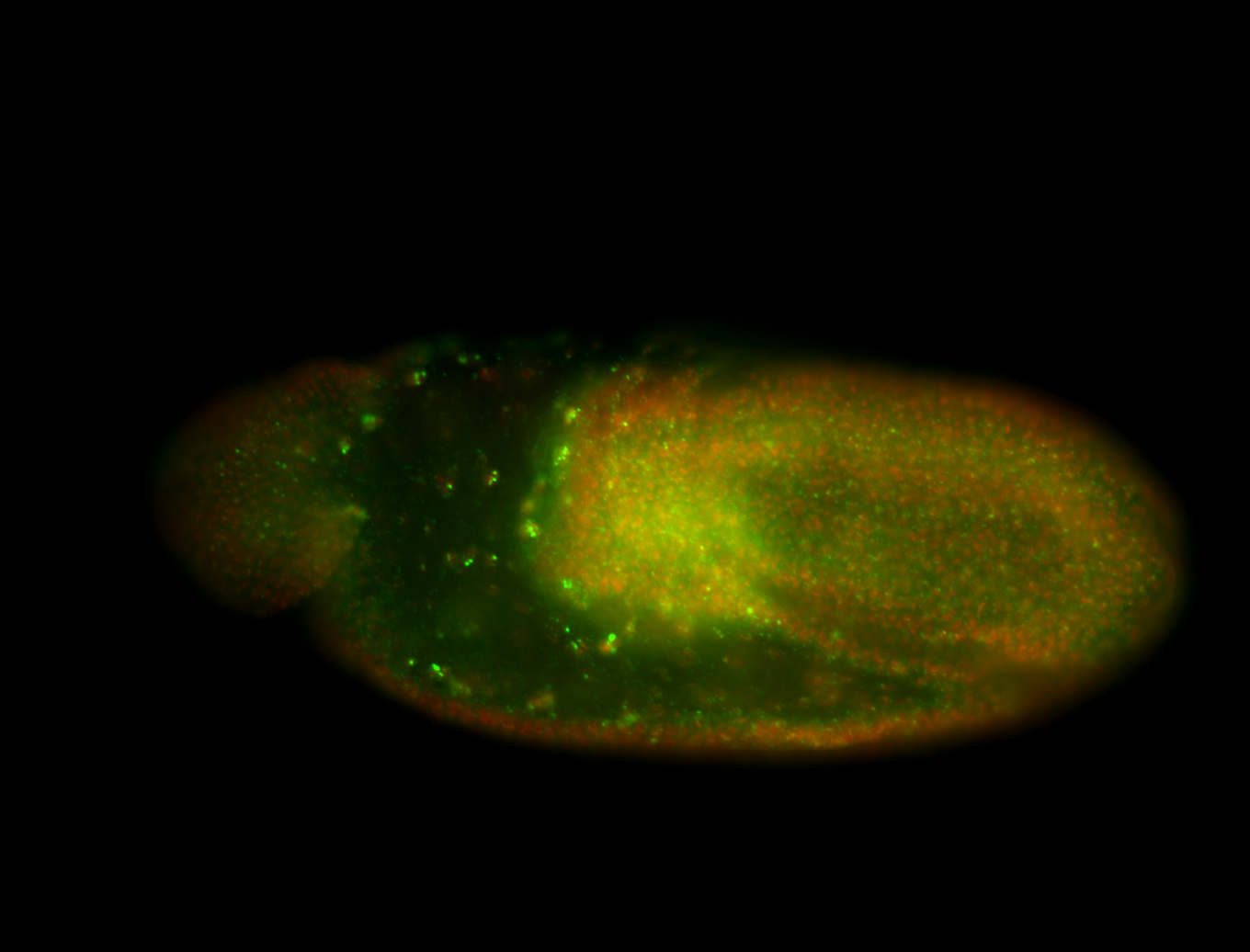
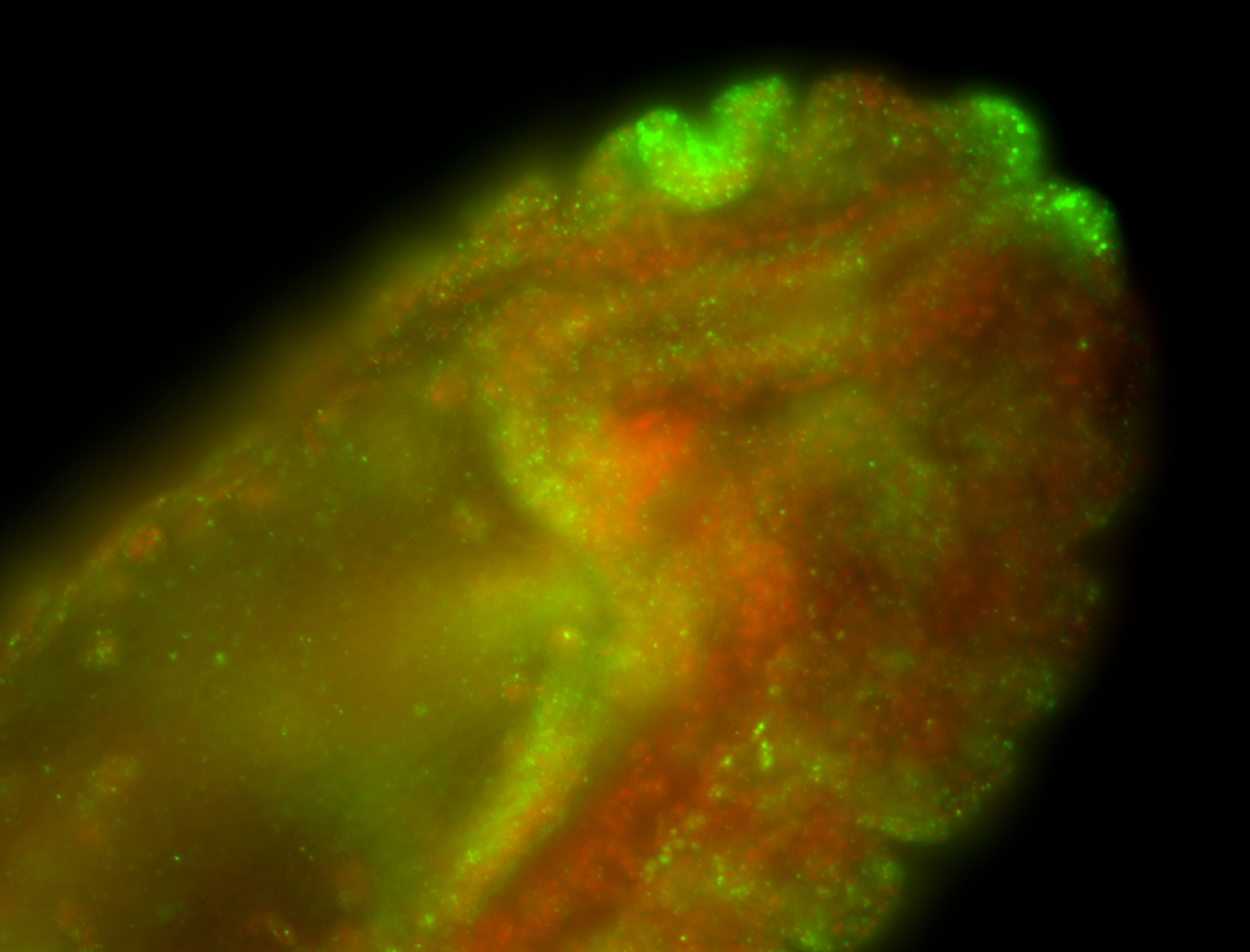
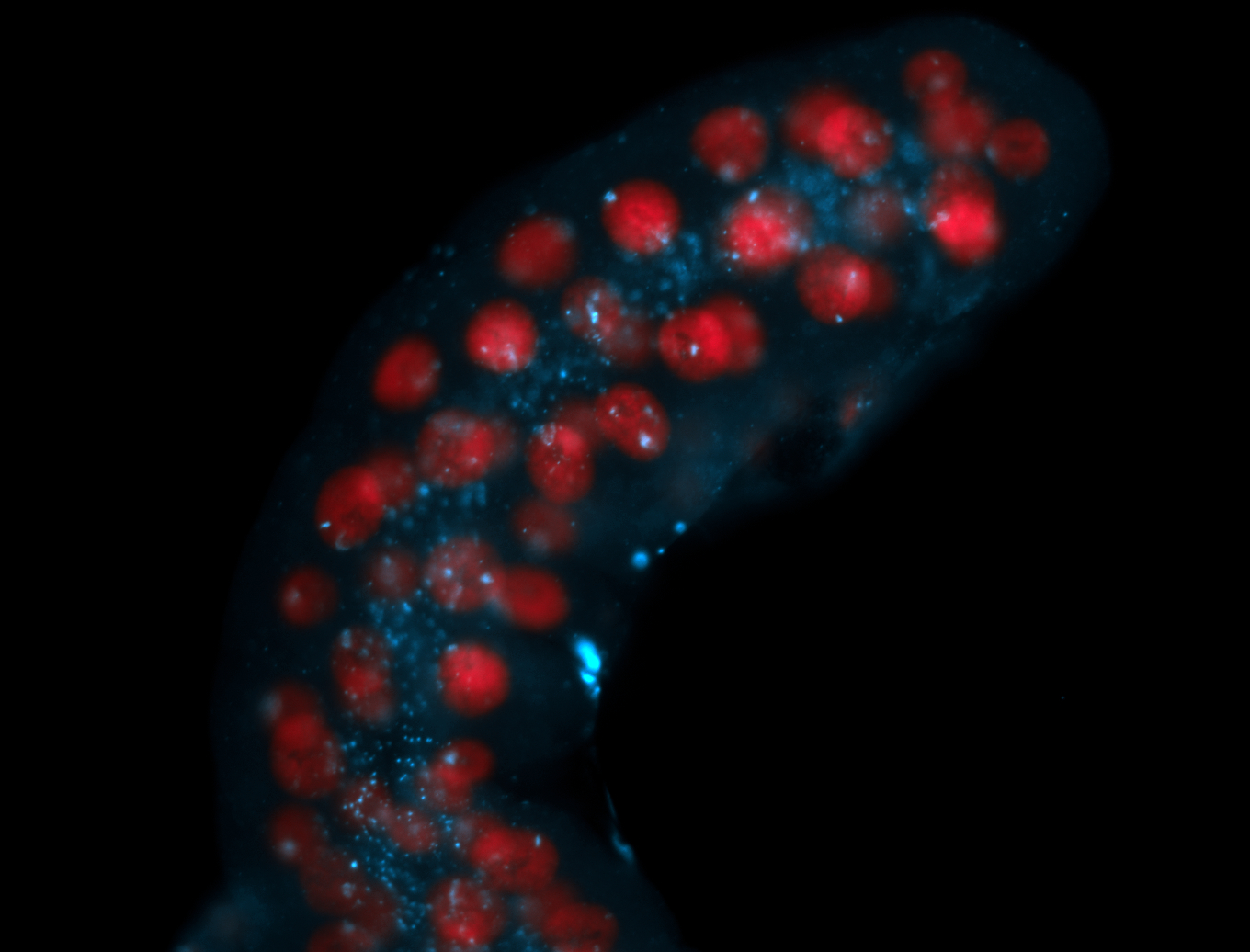
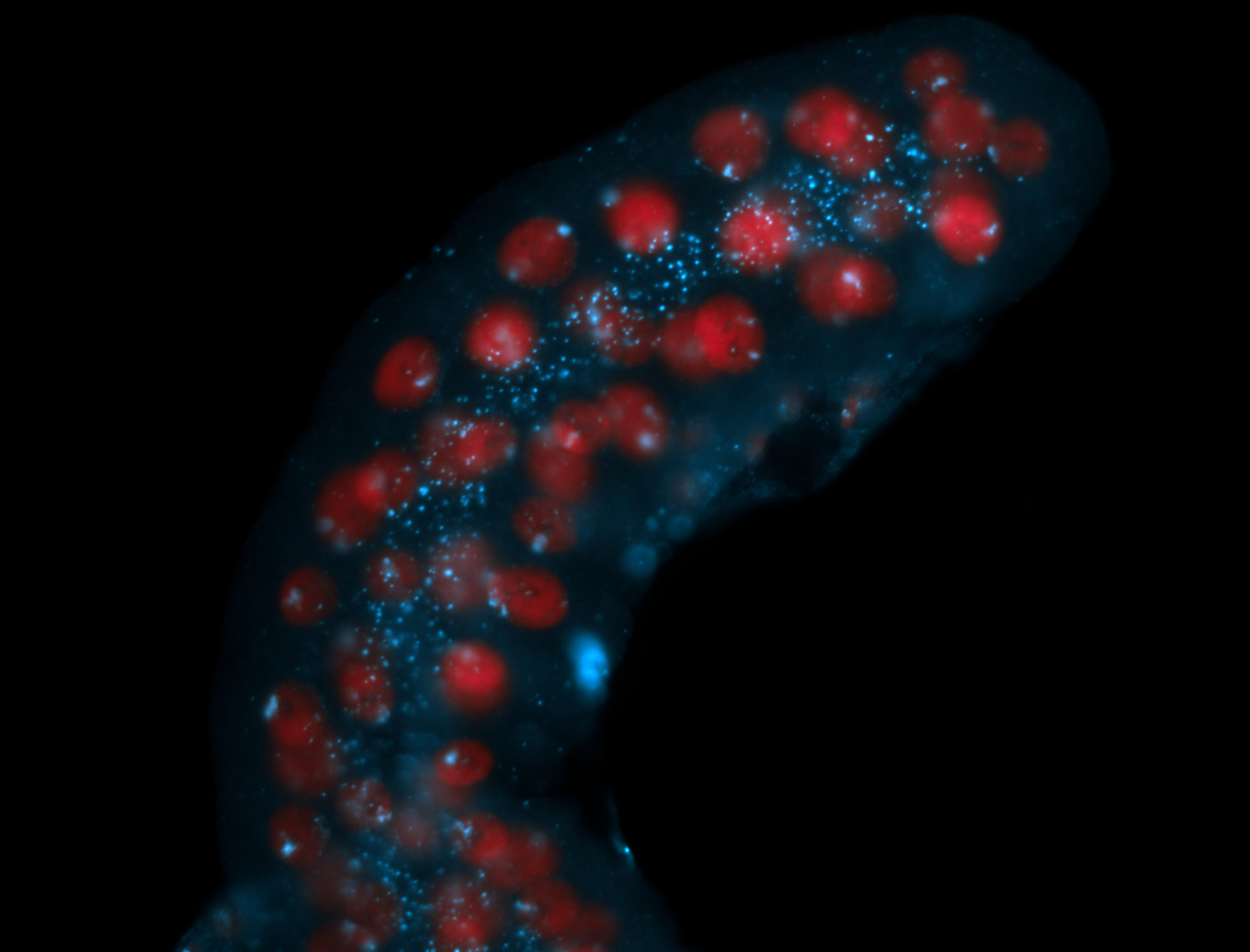
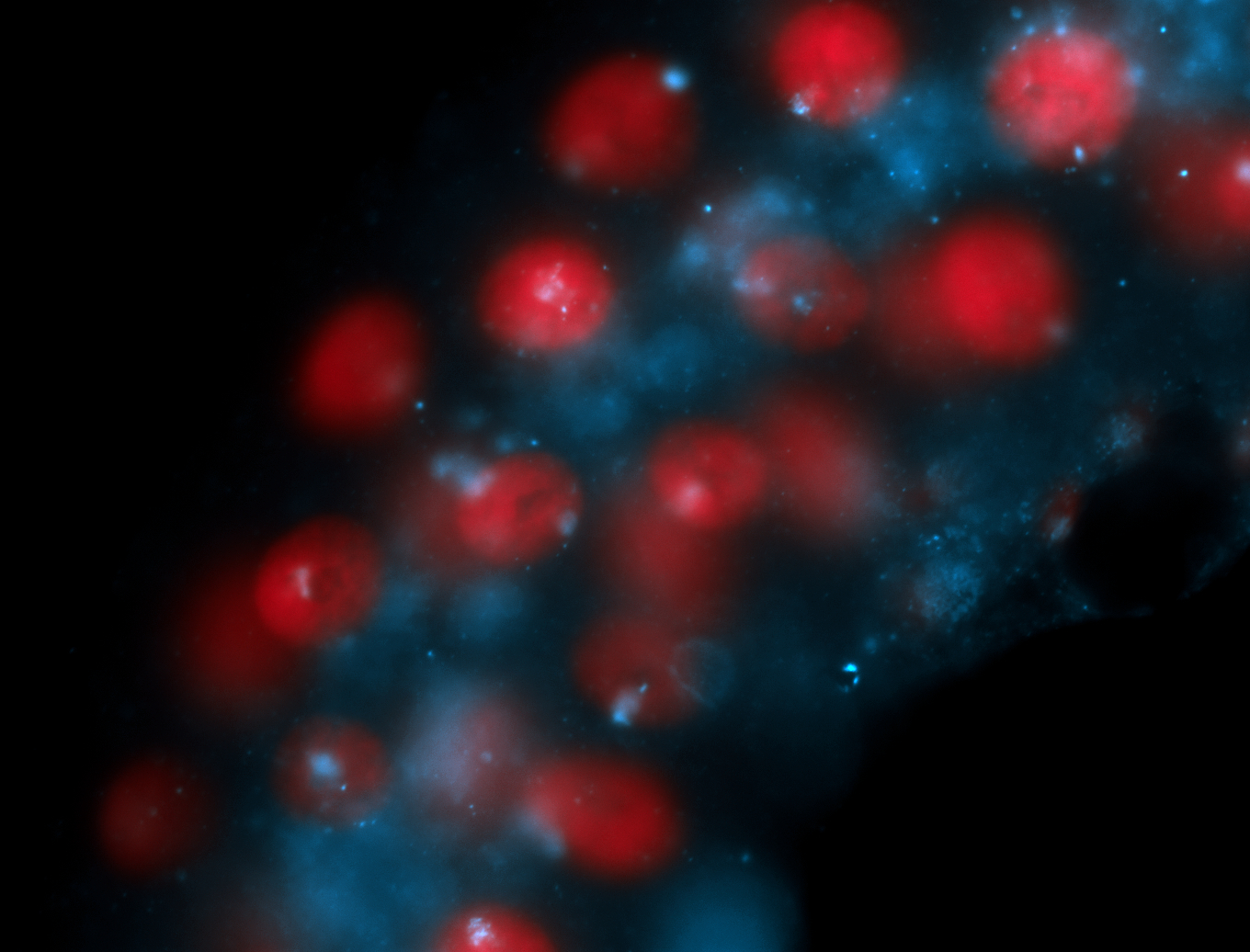
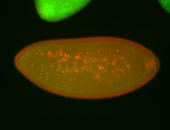
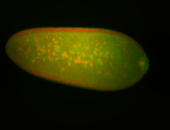
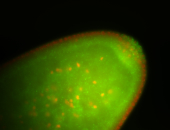
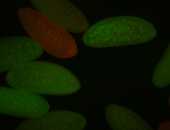
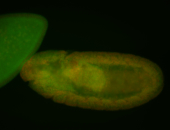
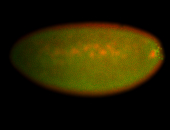
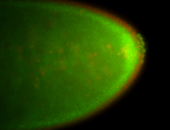
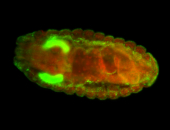
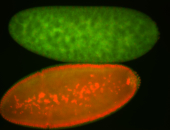
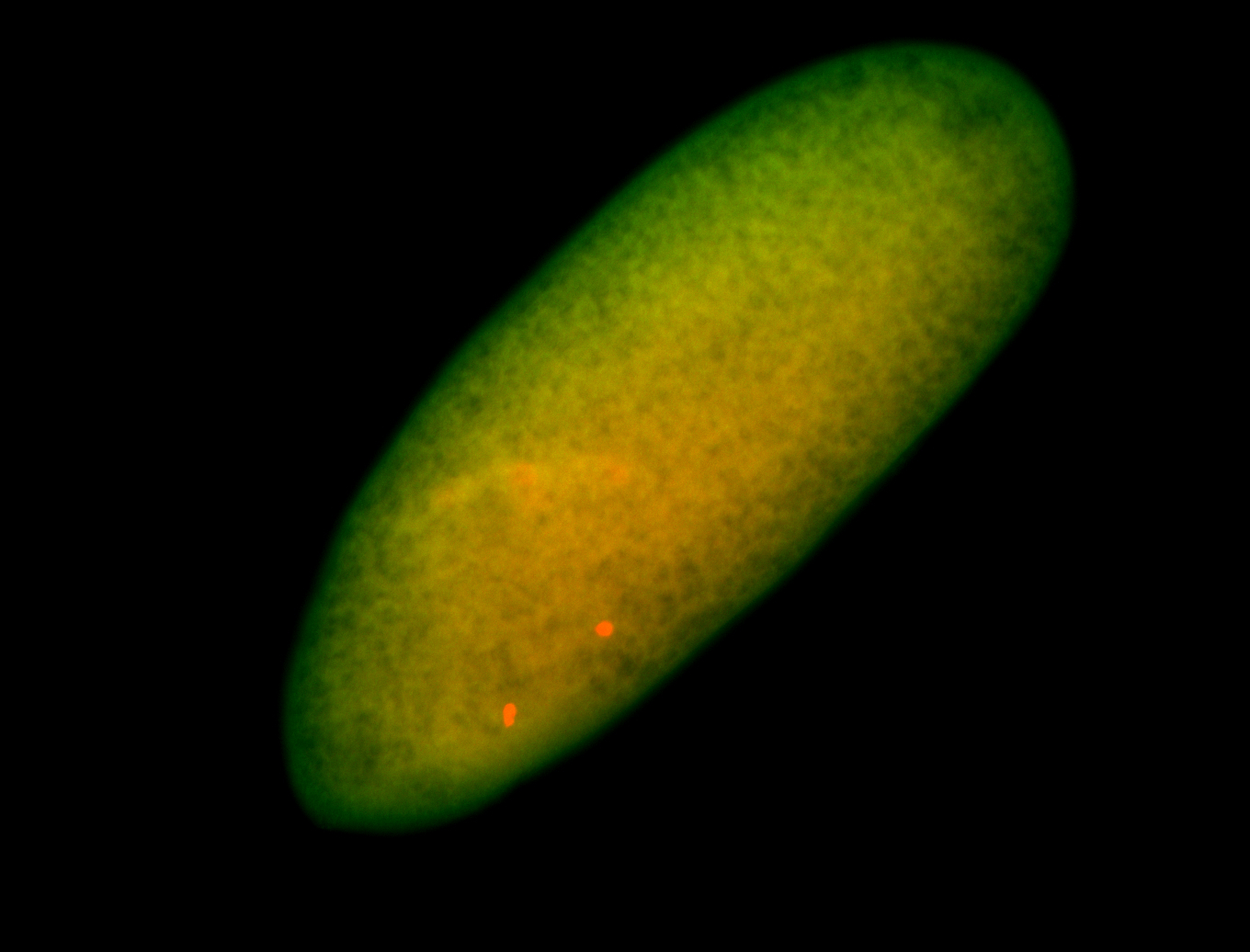
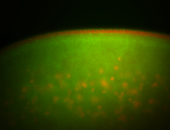
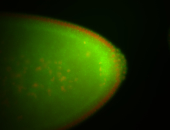
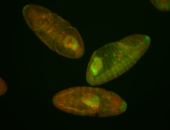
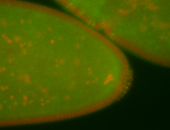
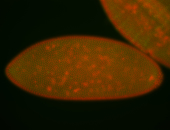
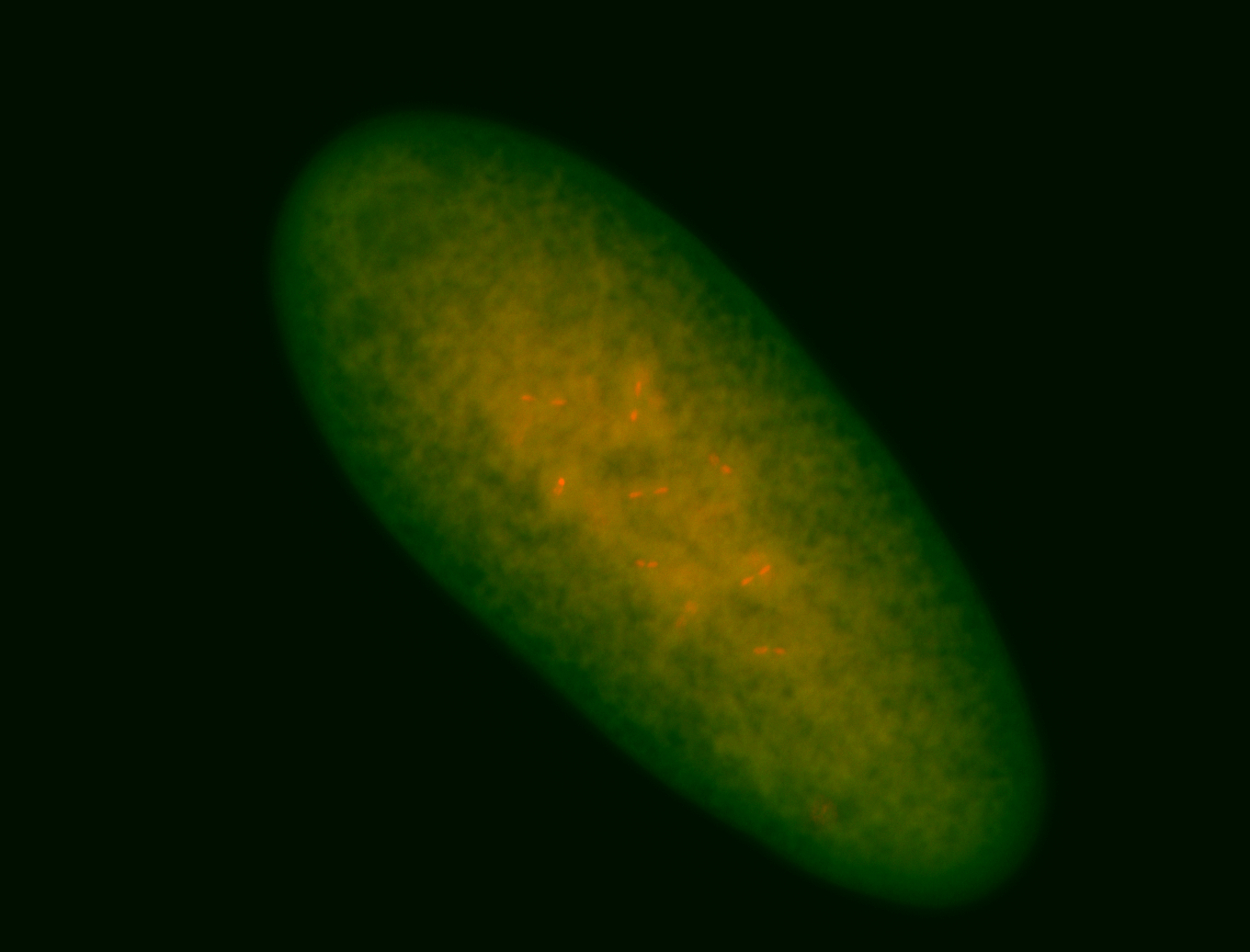
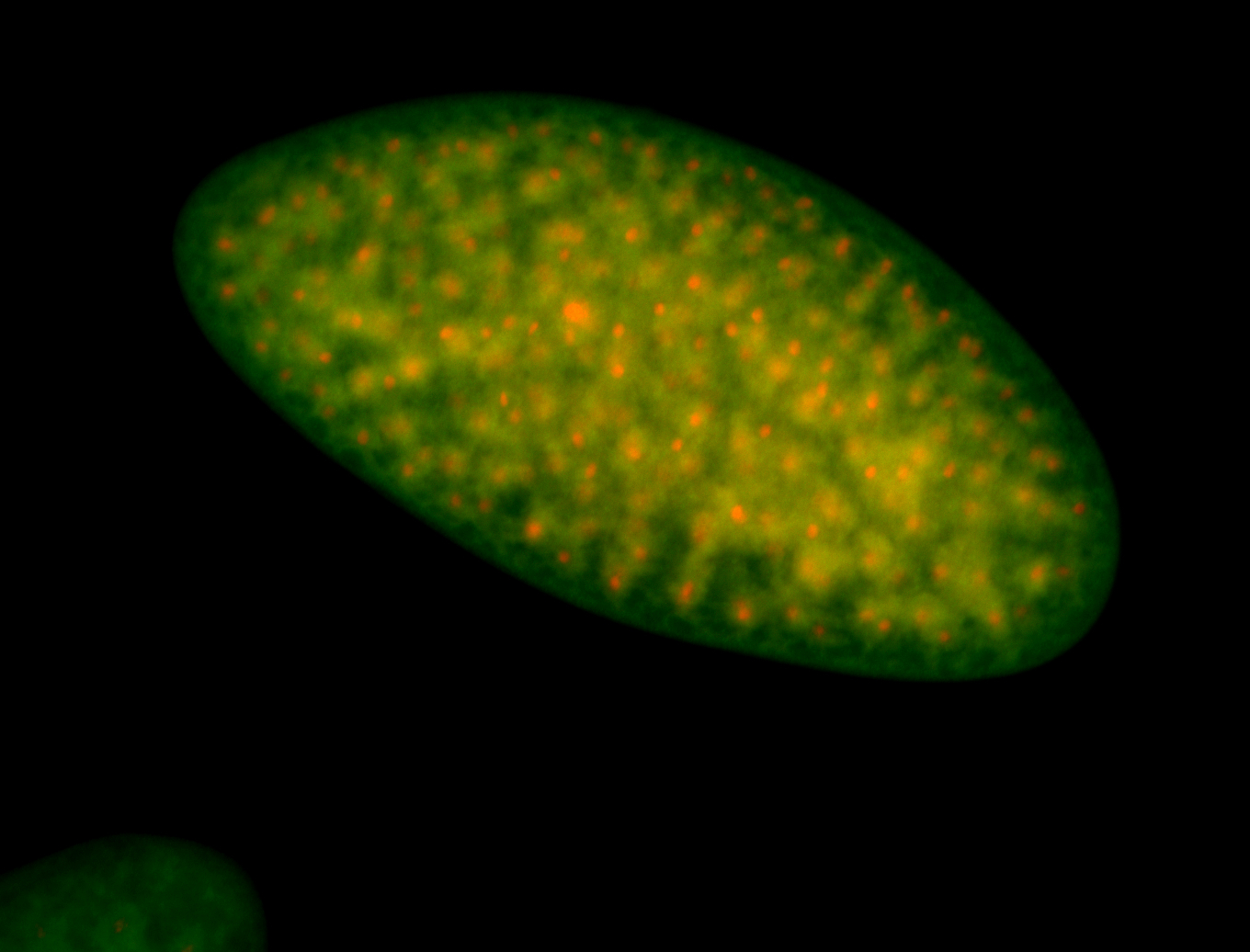
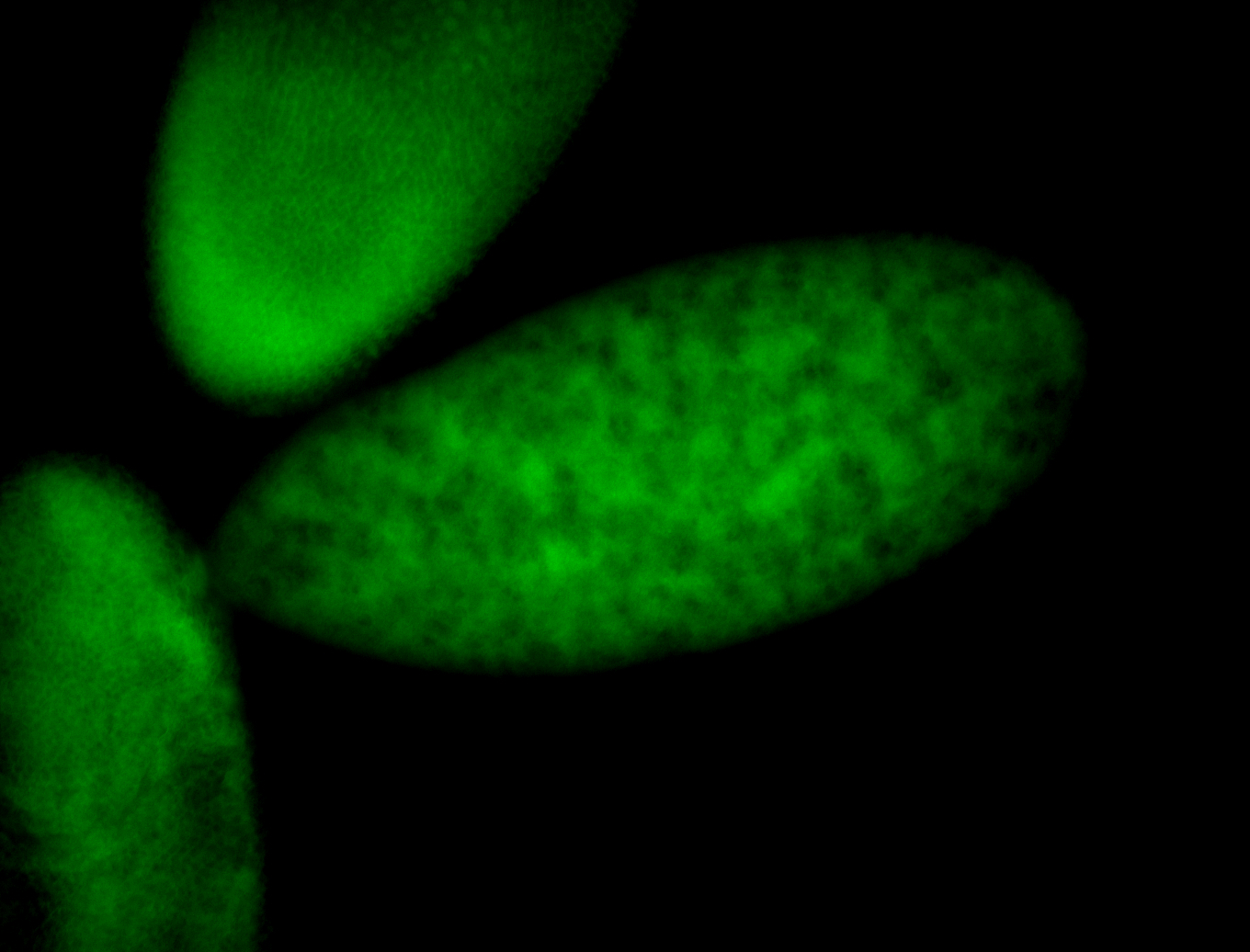
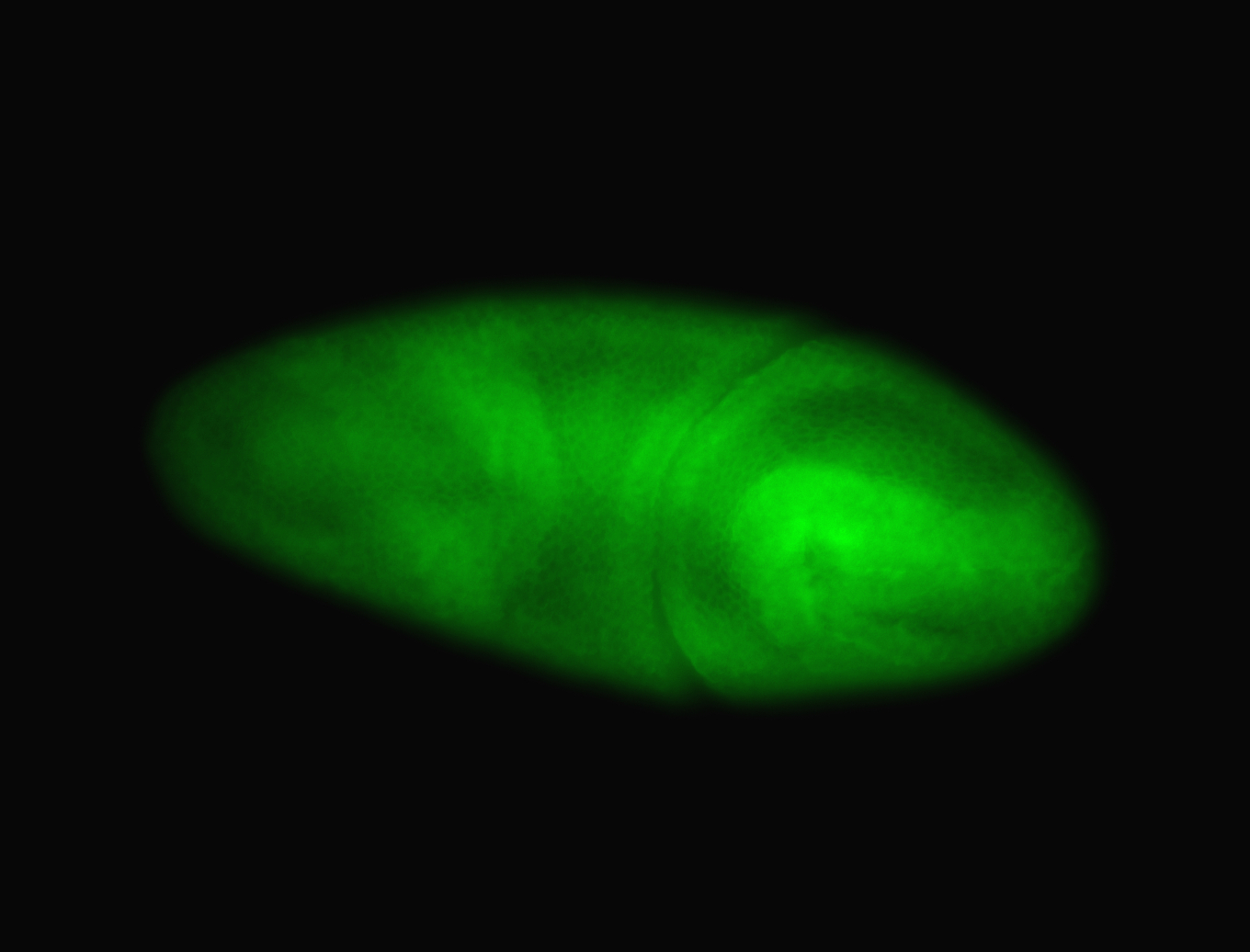
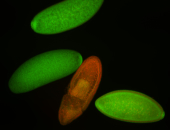
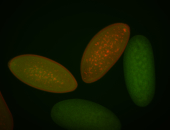
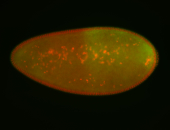
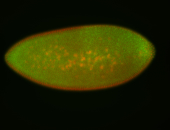
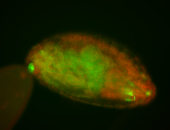
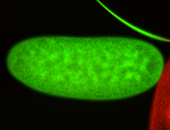
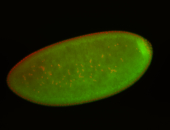
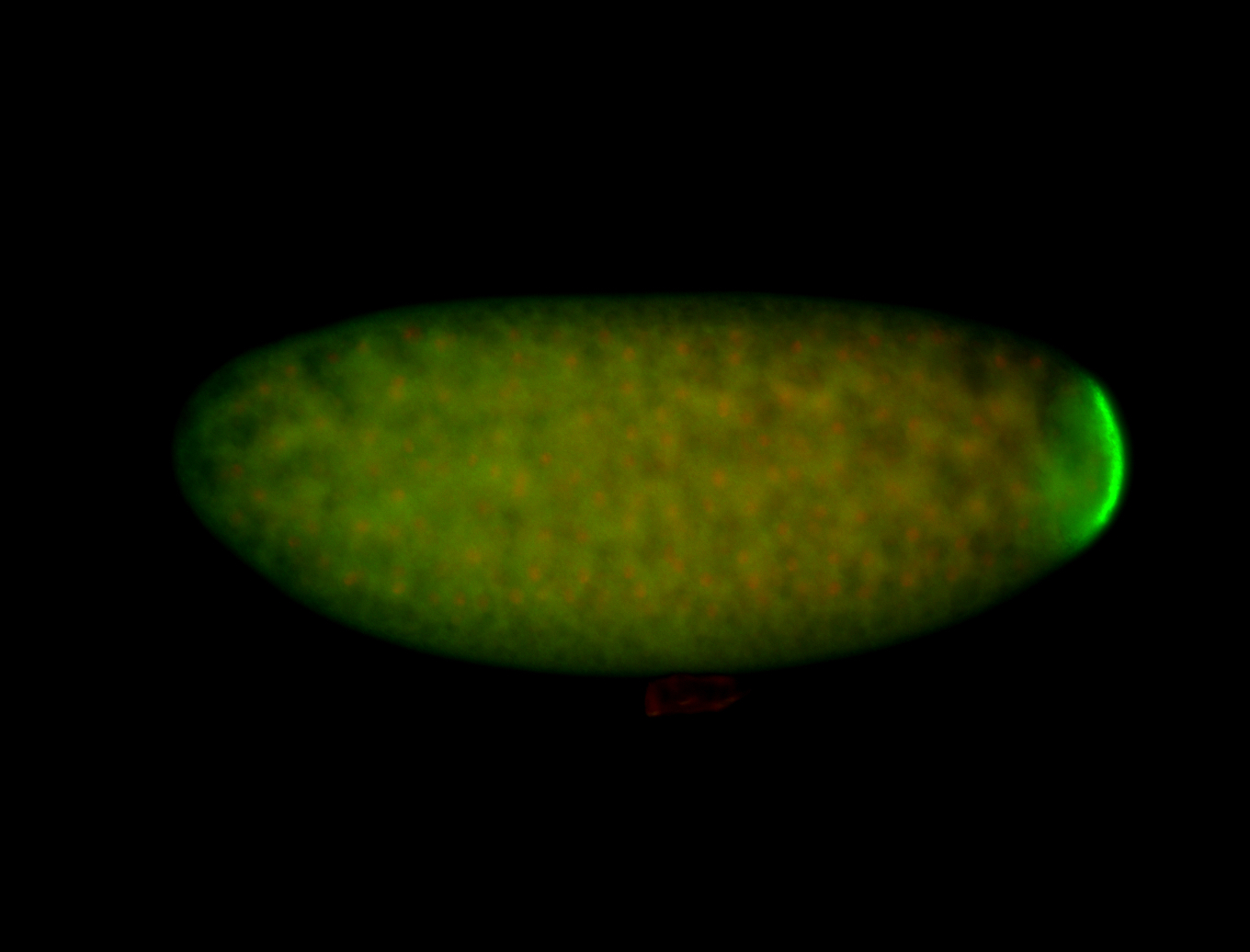
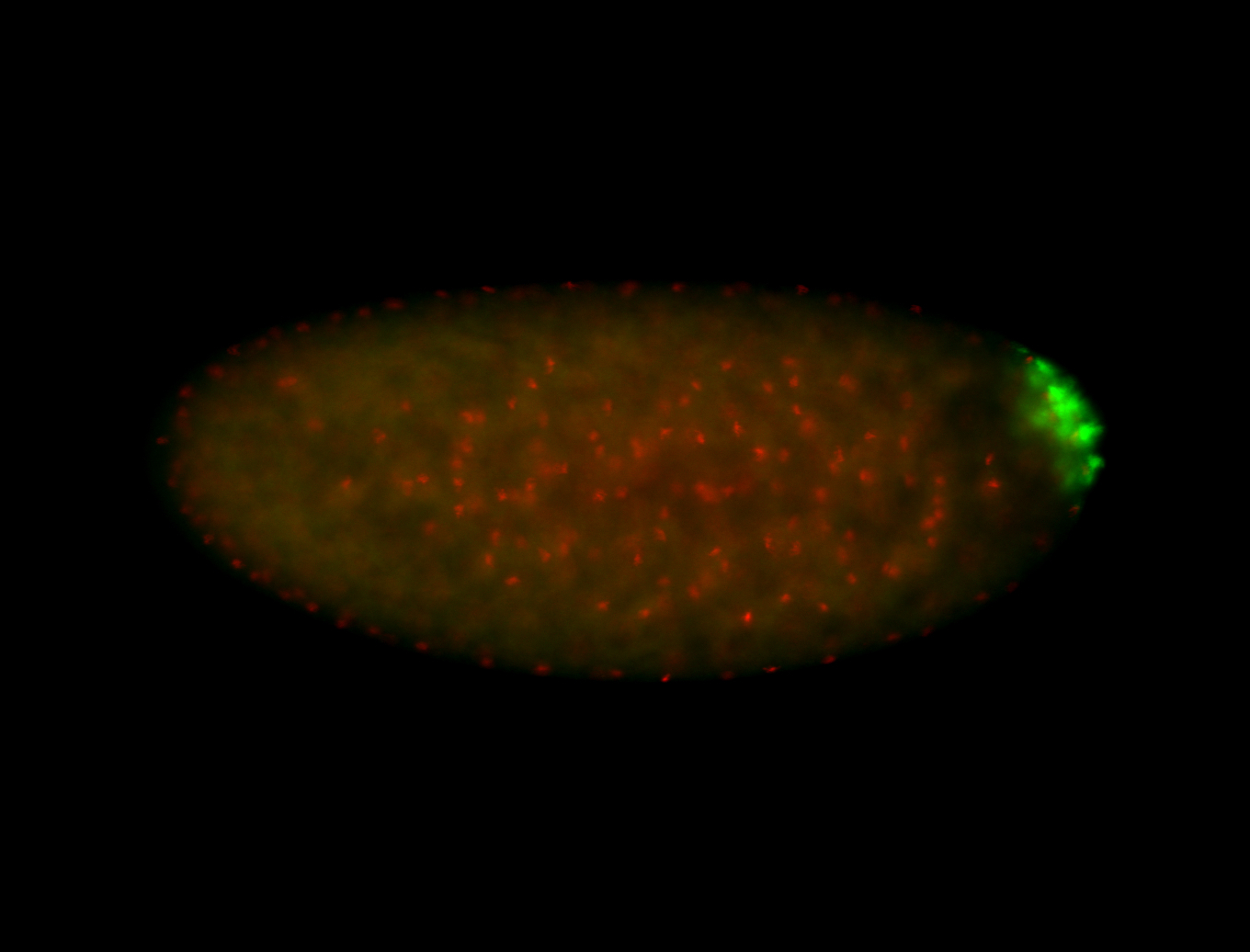
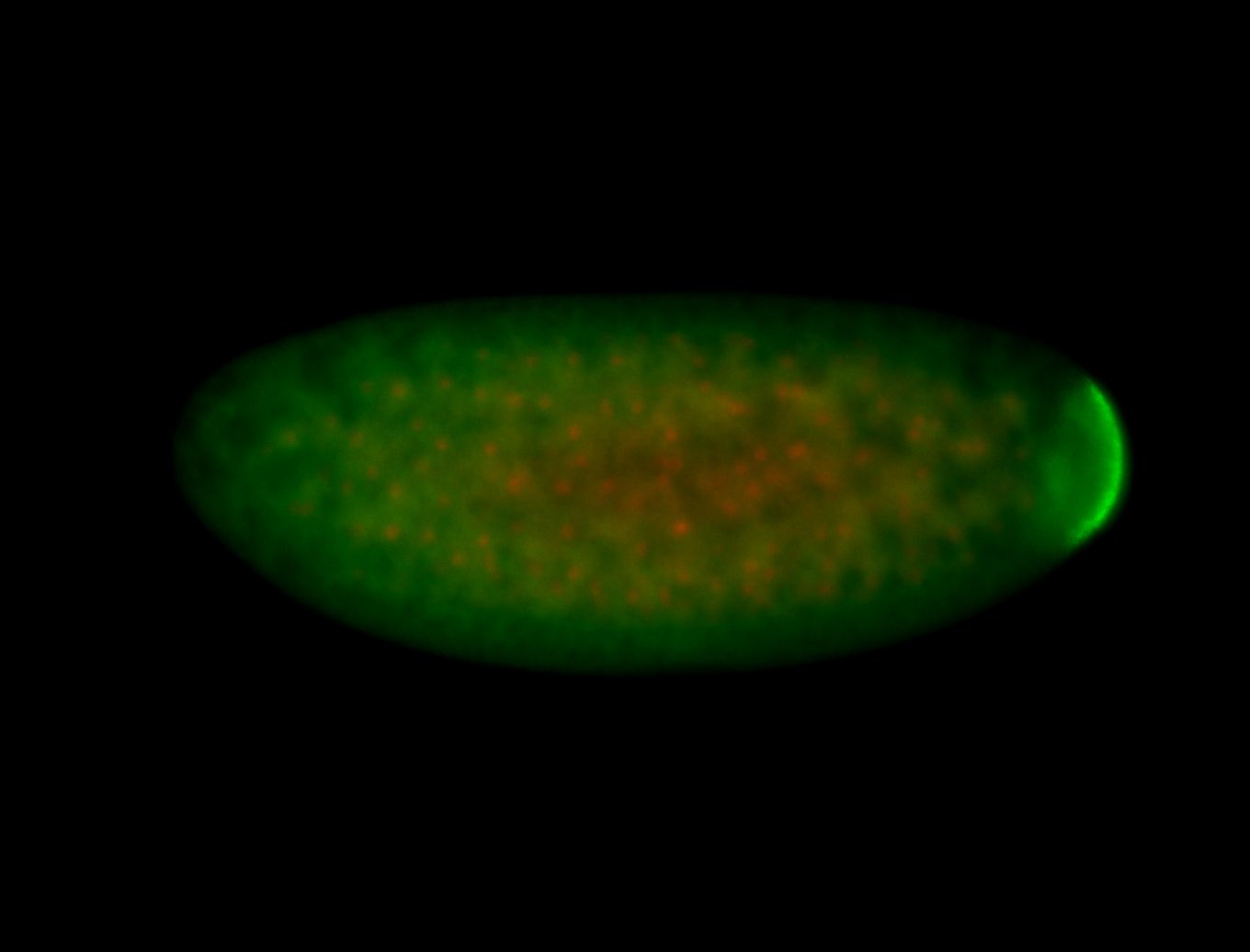
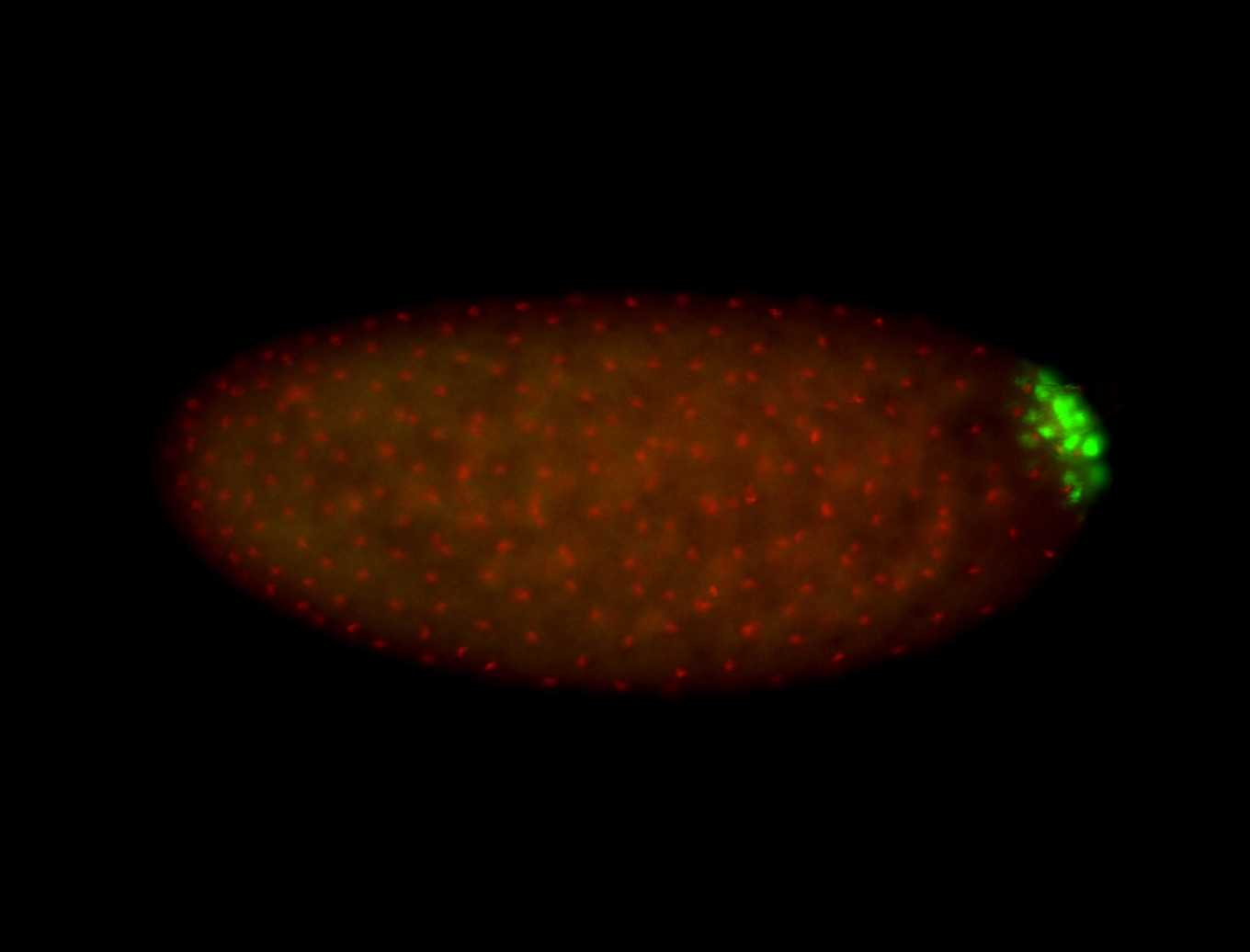
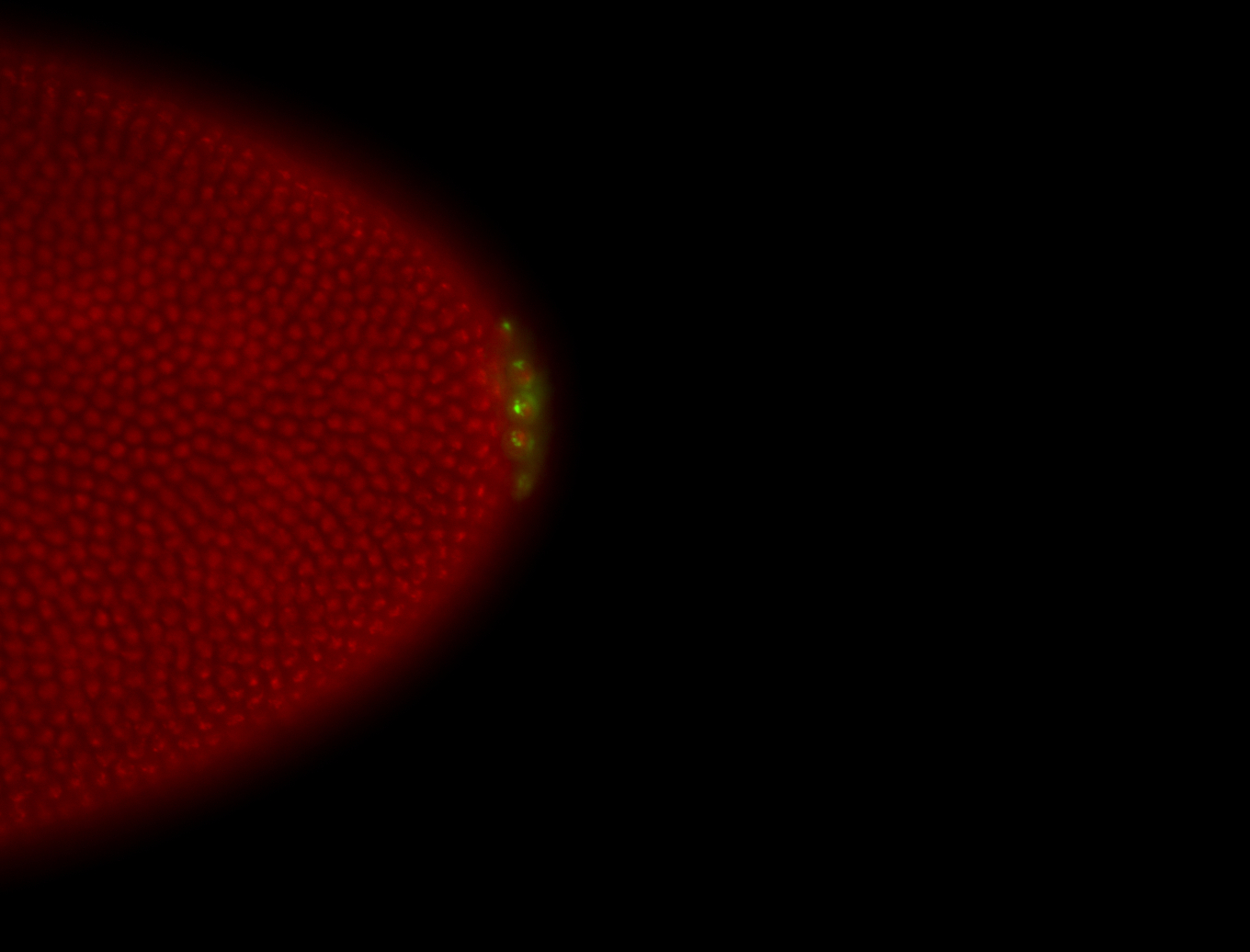
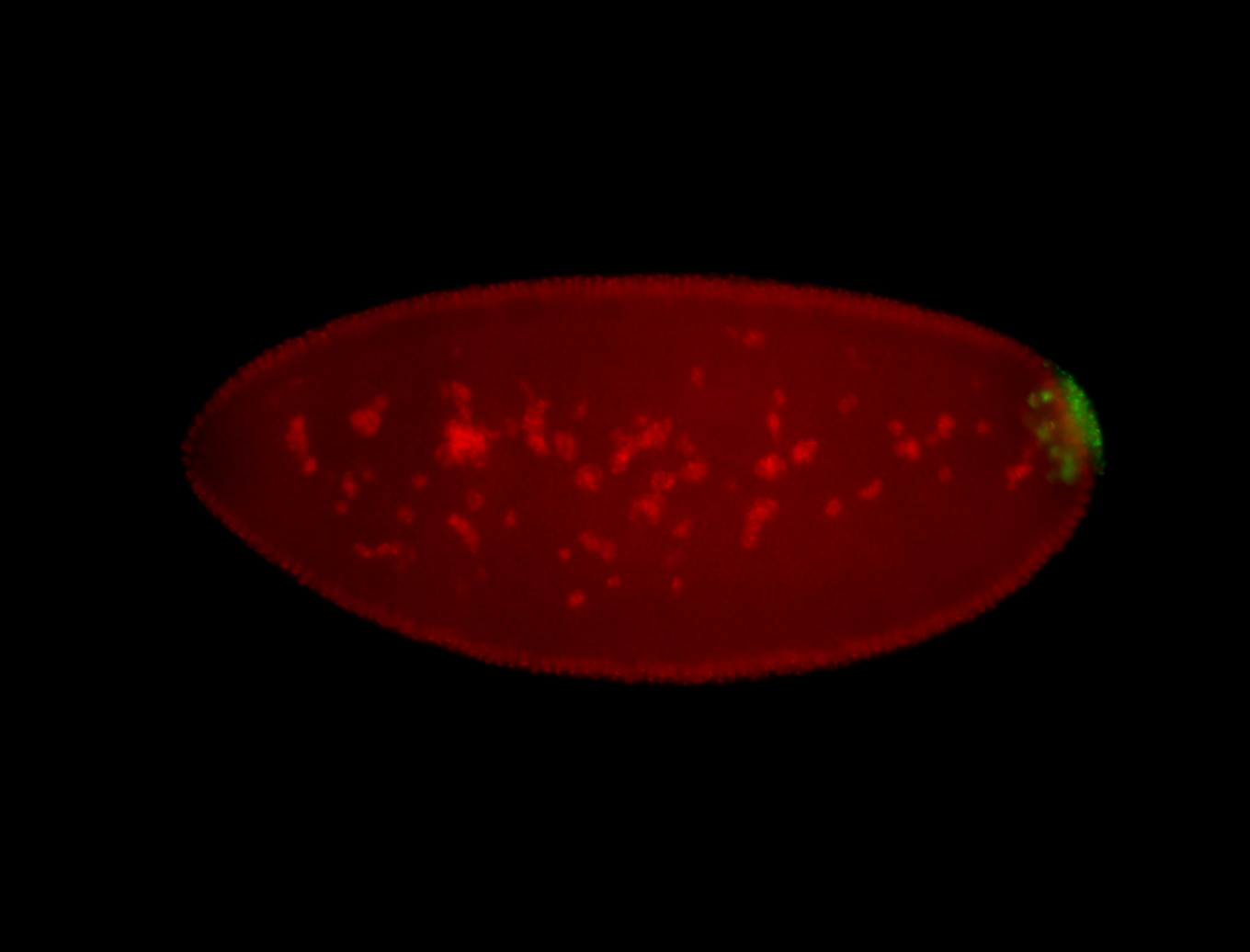
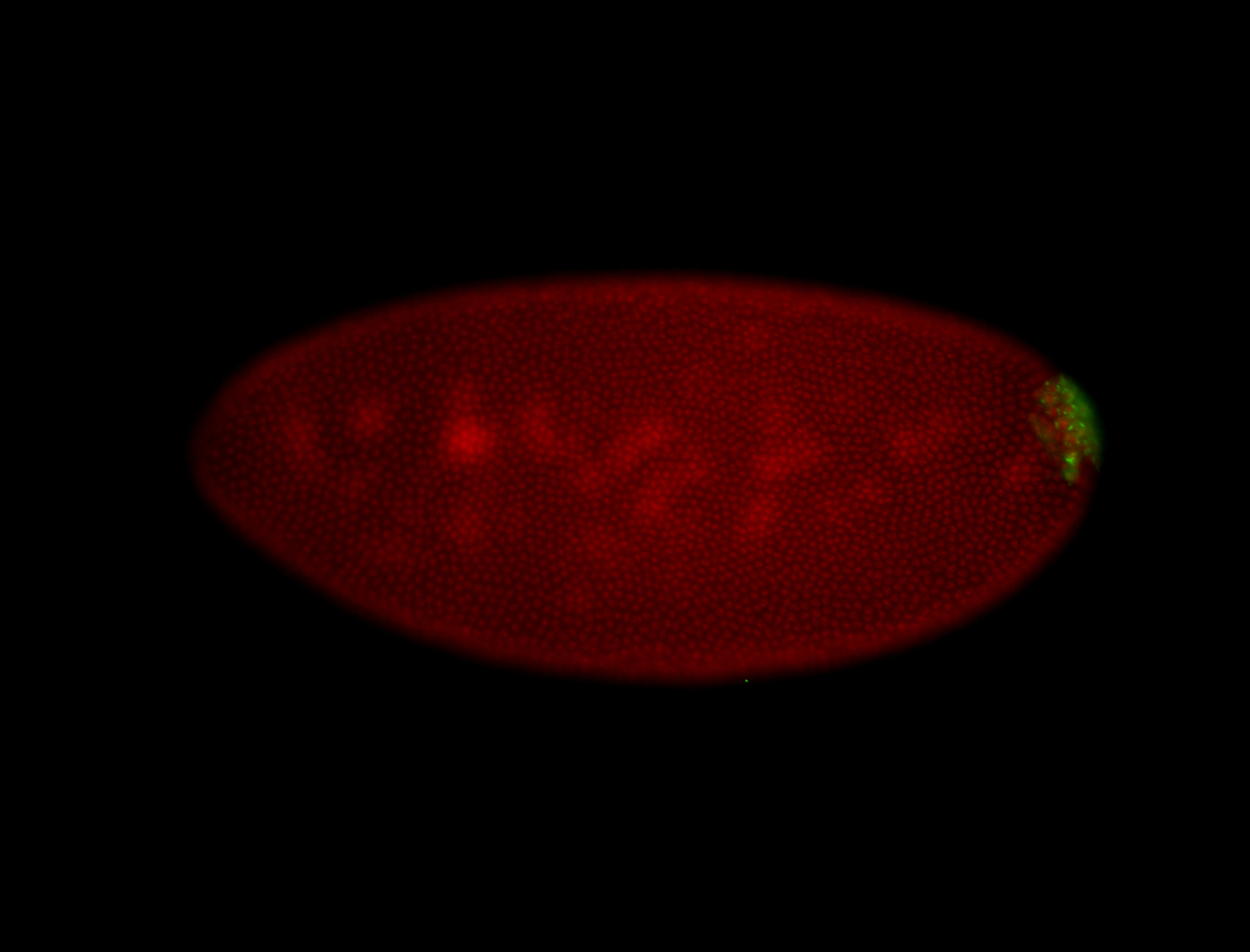
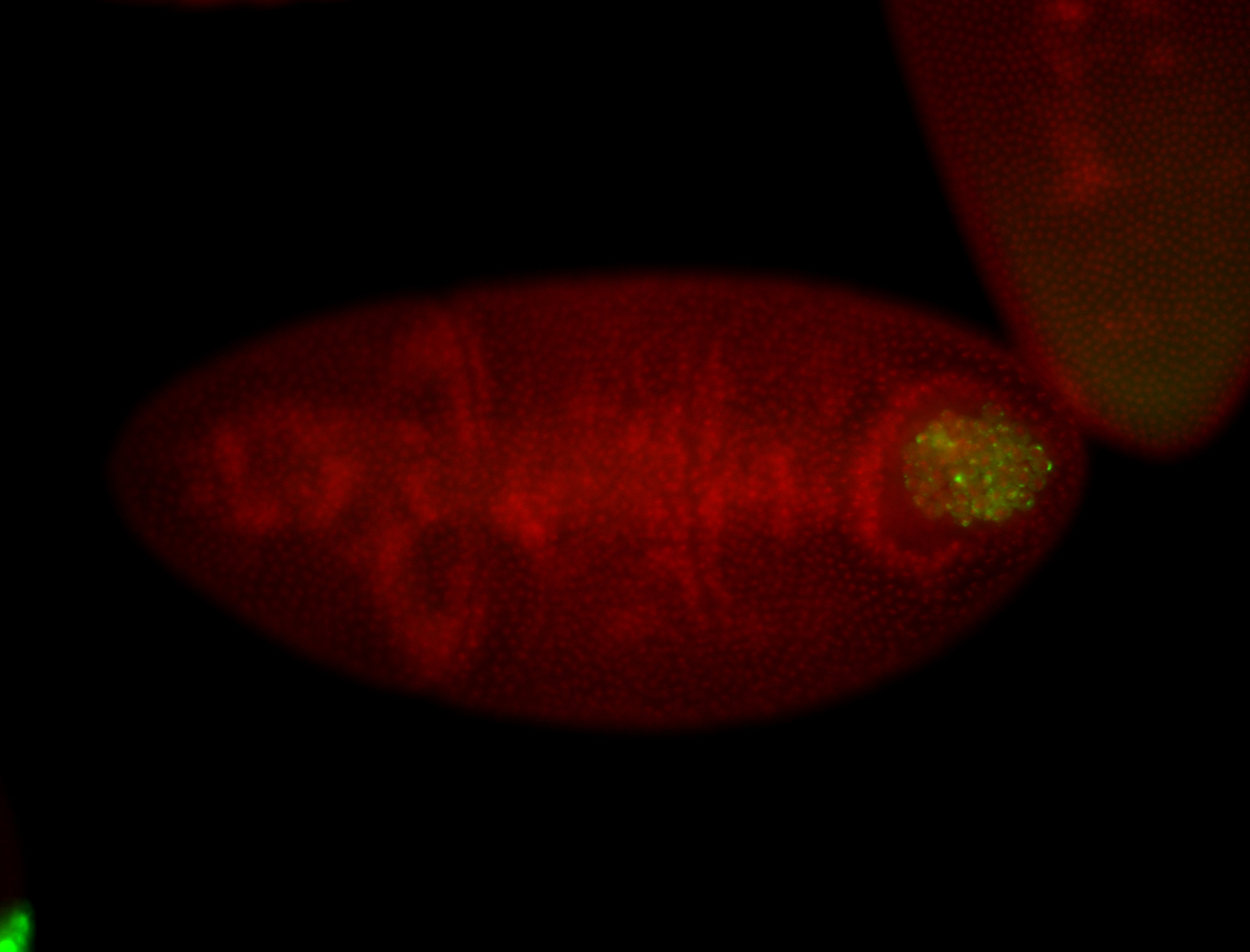
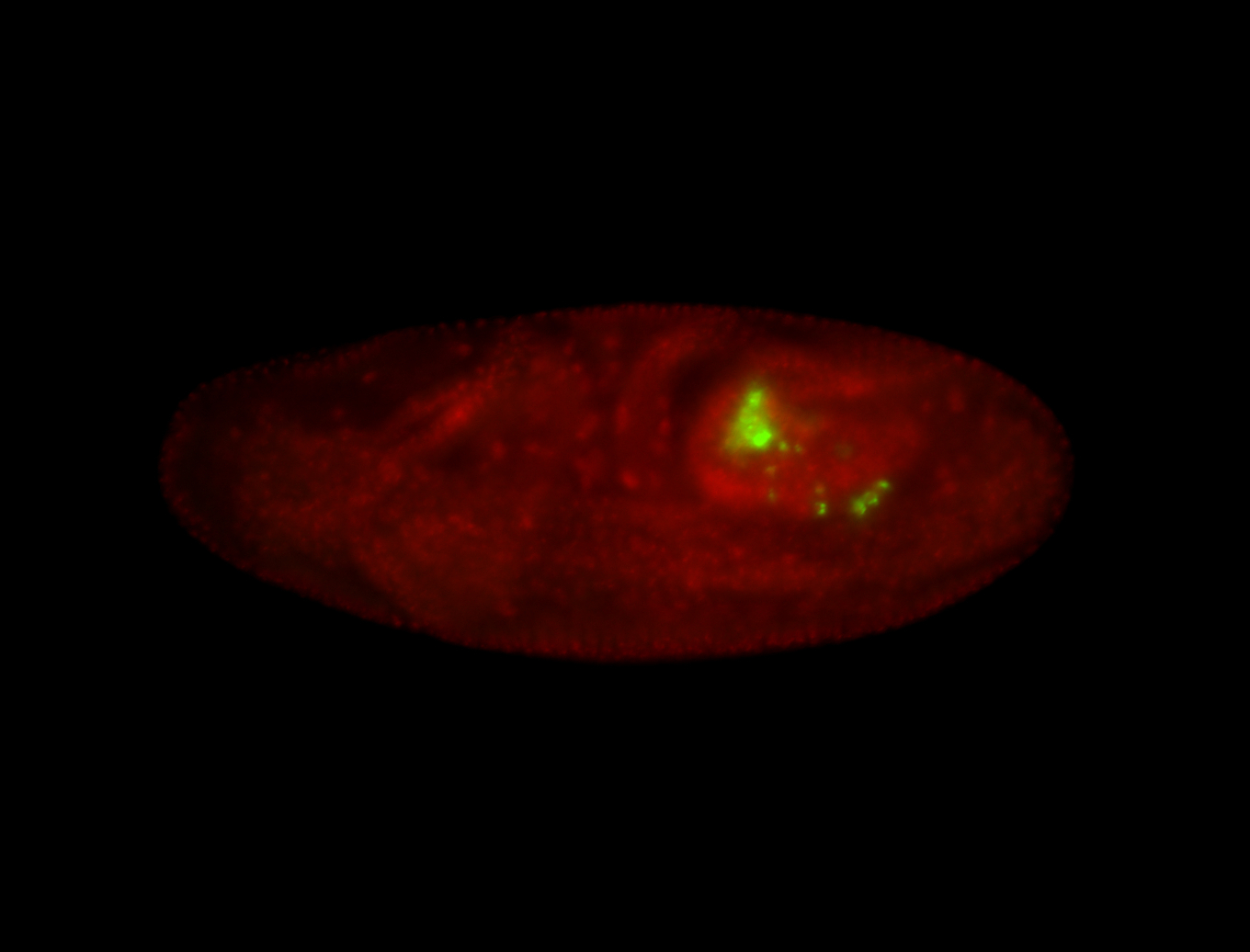
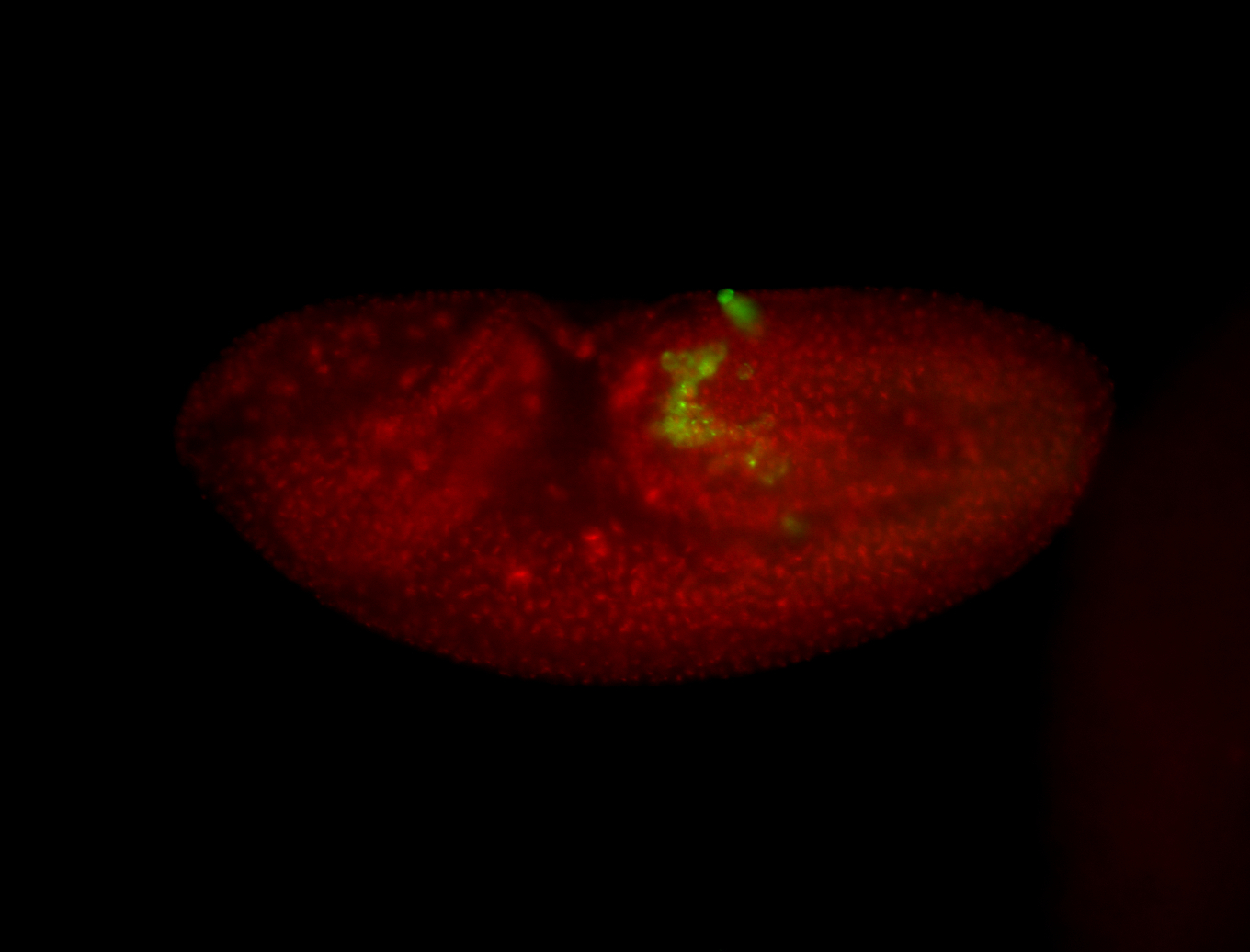
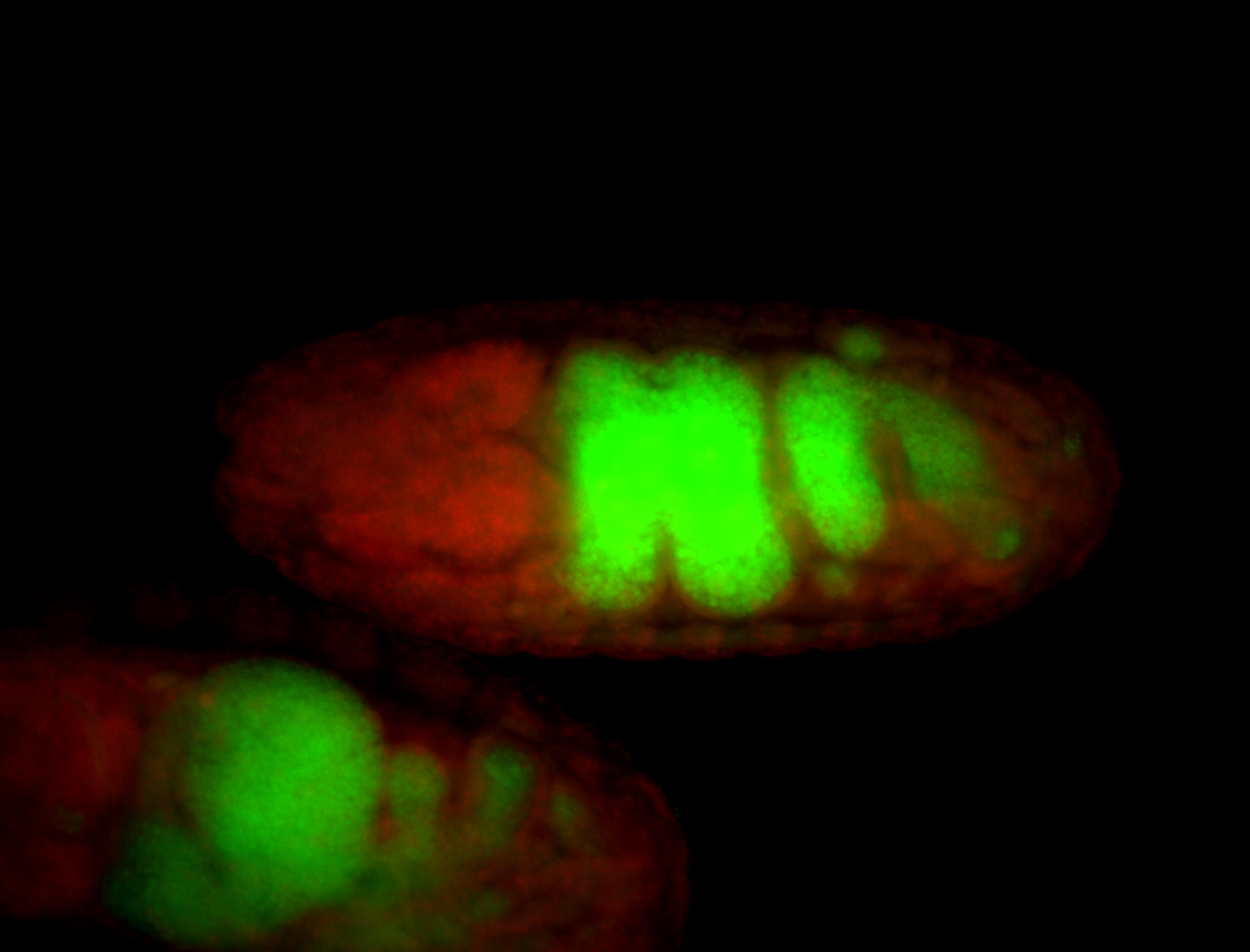
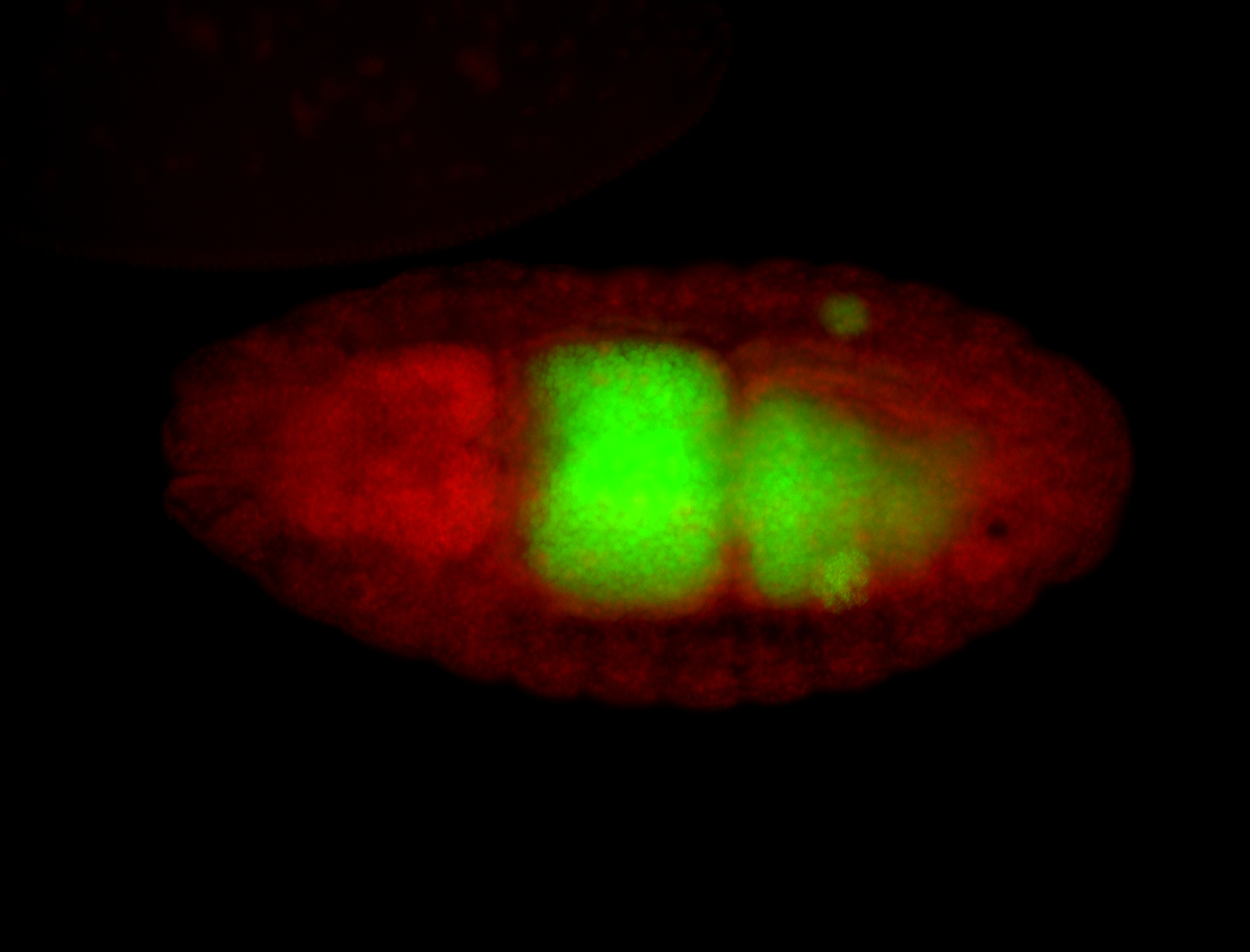
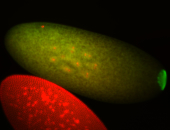
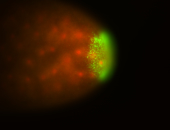
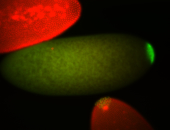
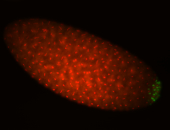
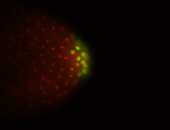
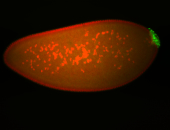
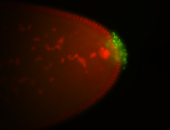
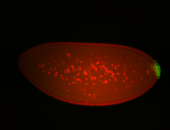
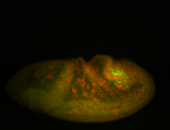
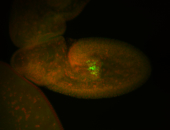
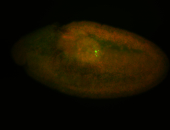
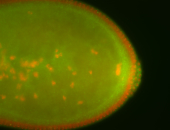
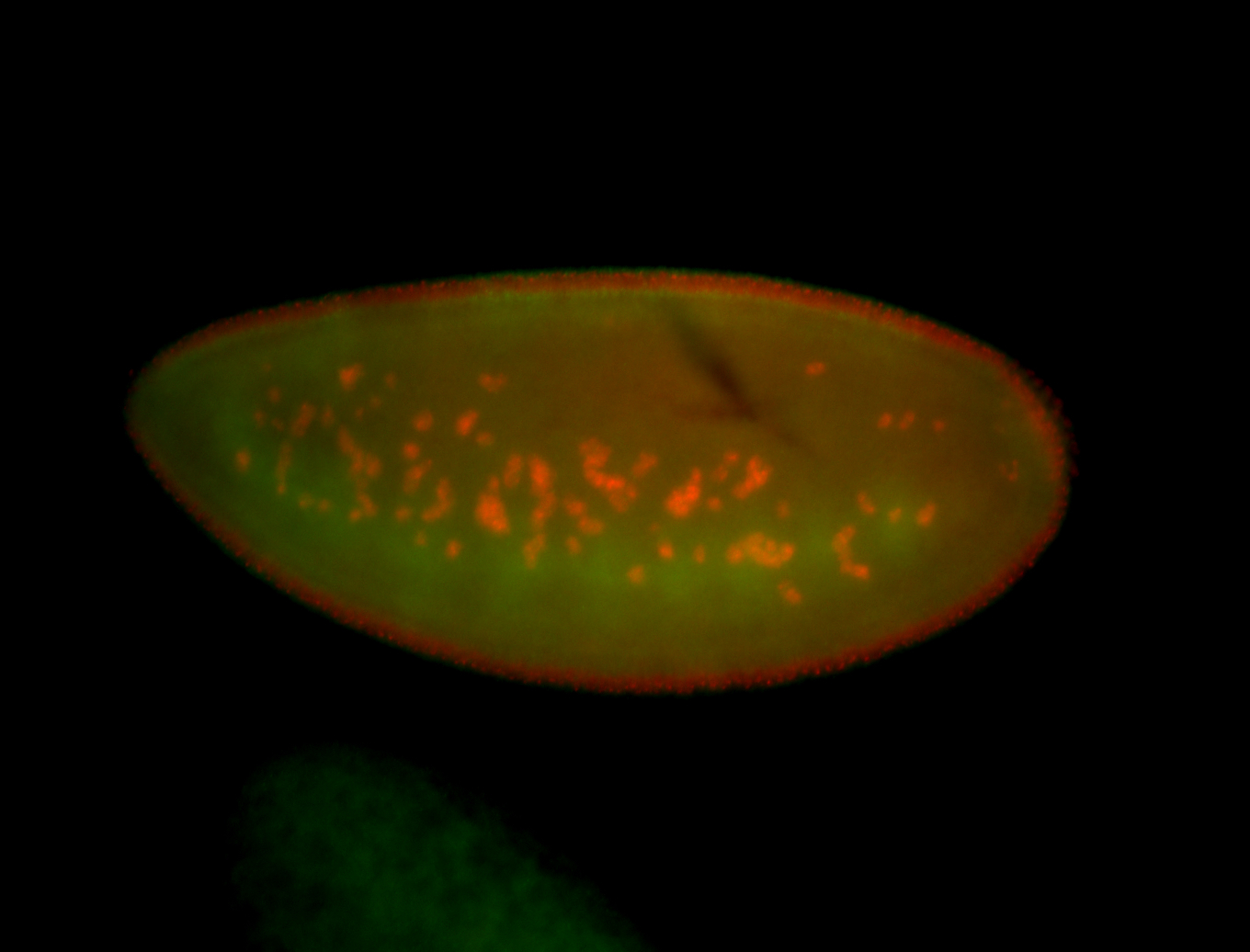
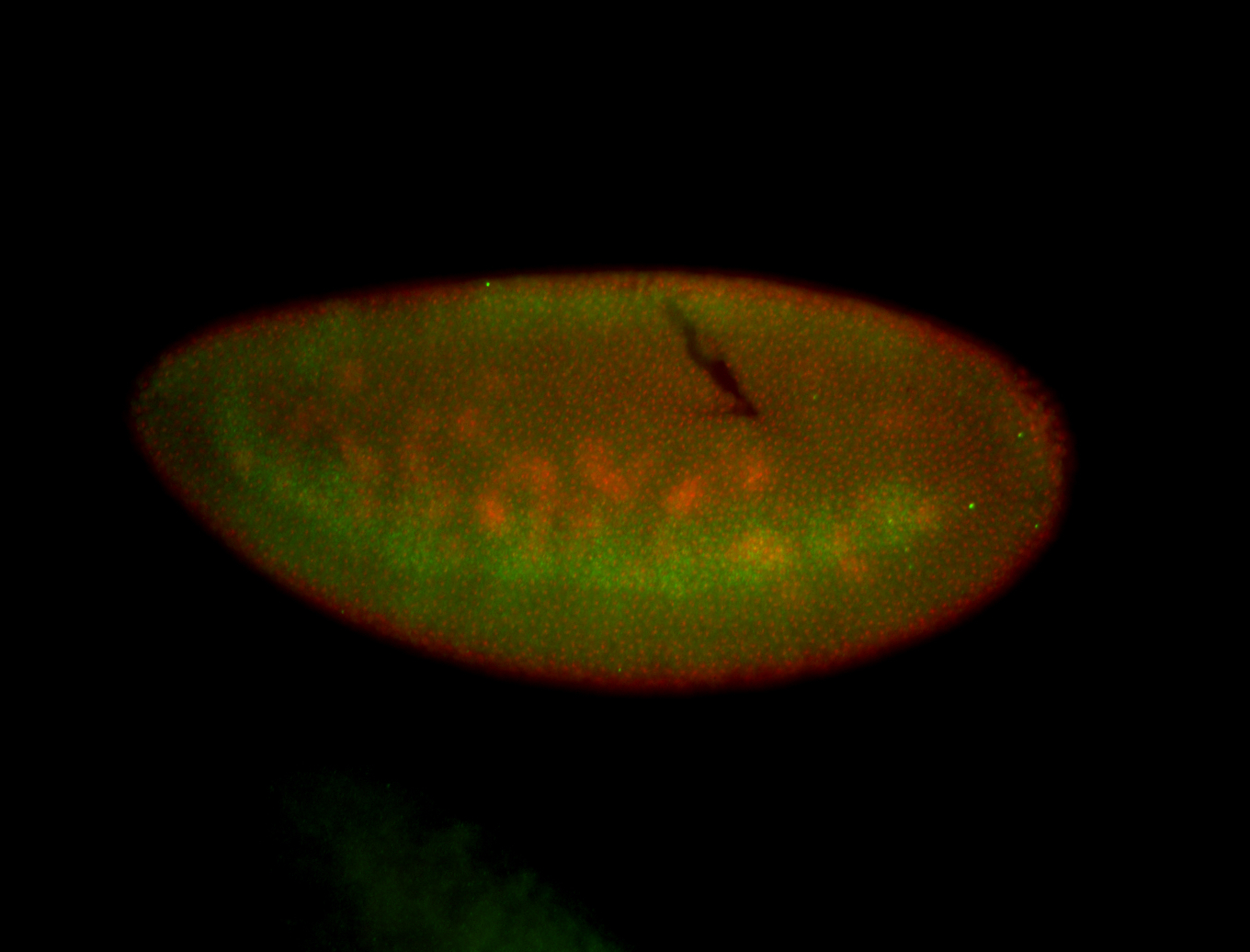
| Foci in pole cells. Could be zygotic expression, not completely obvious. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
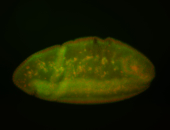
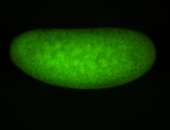
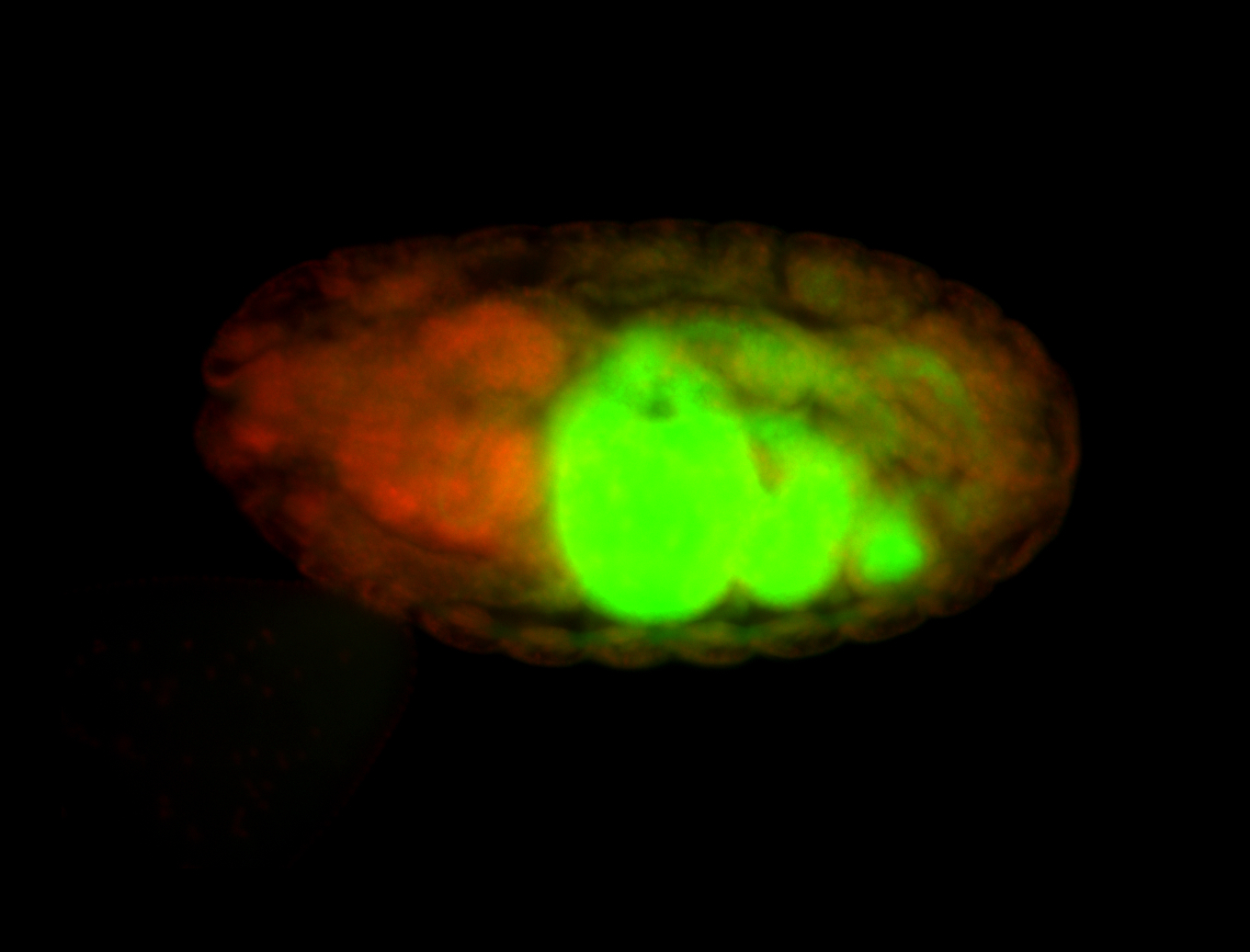
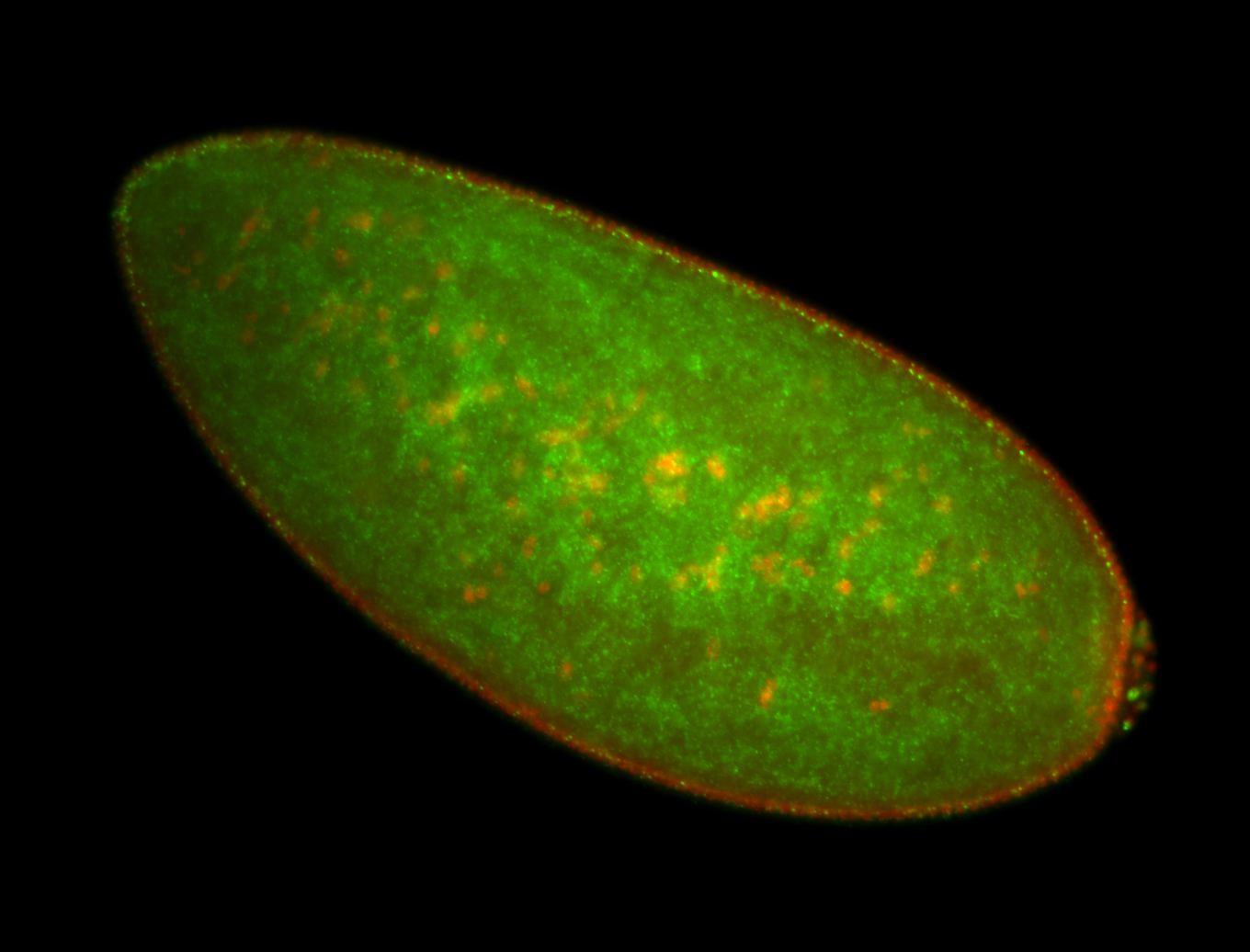
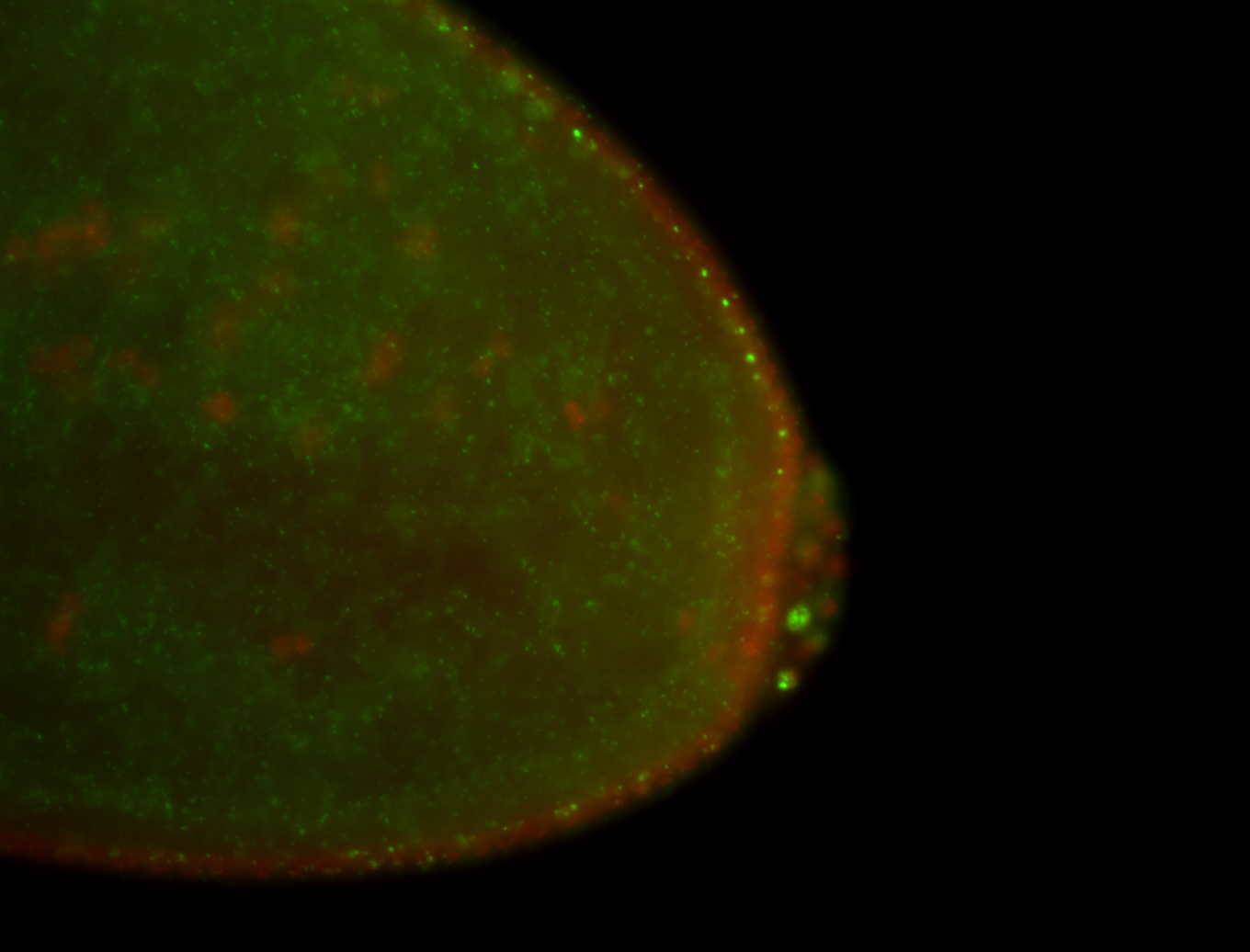
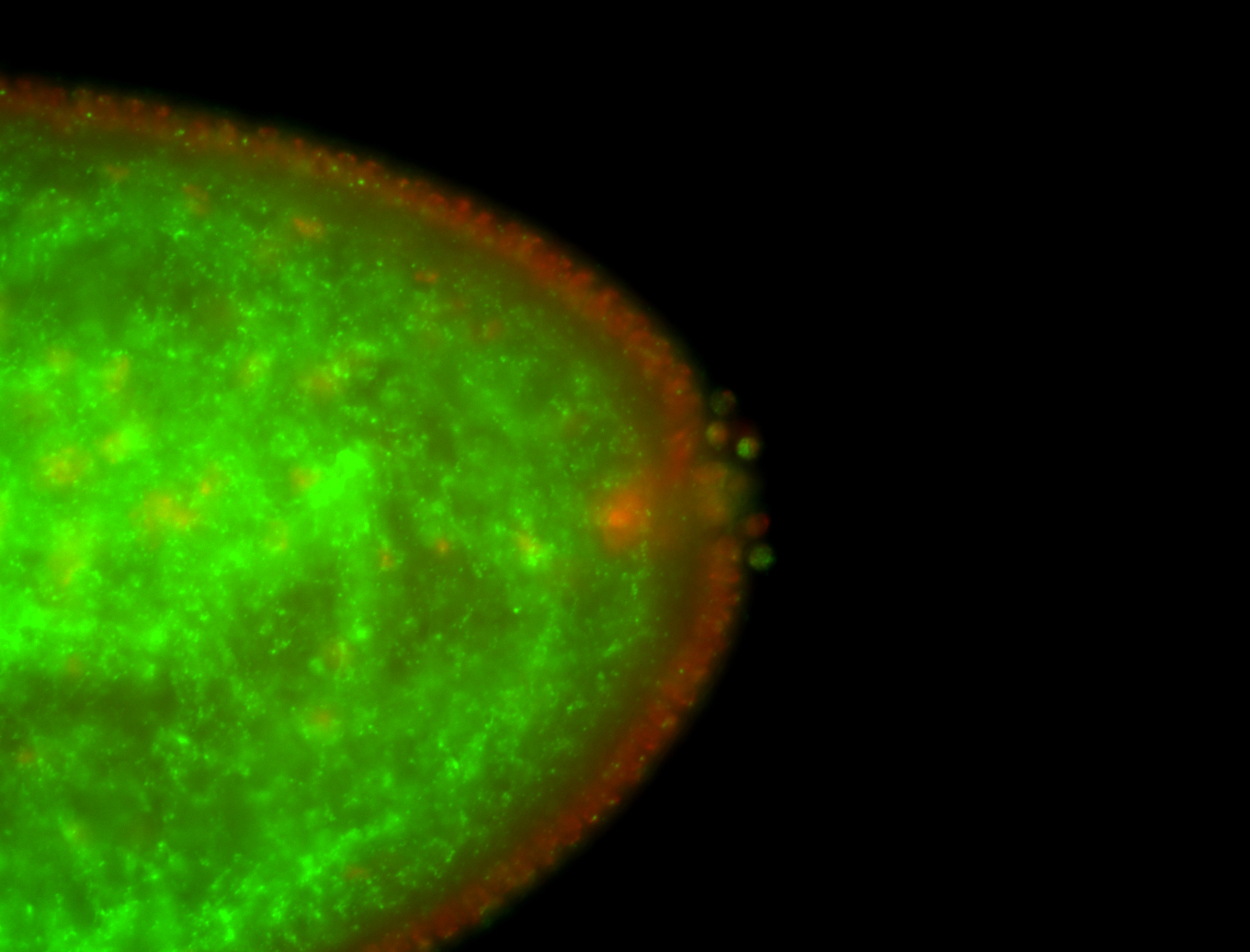
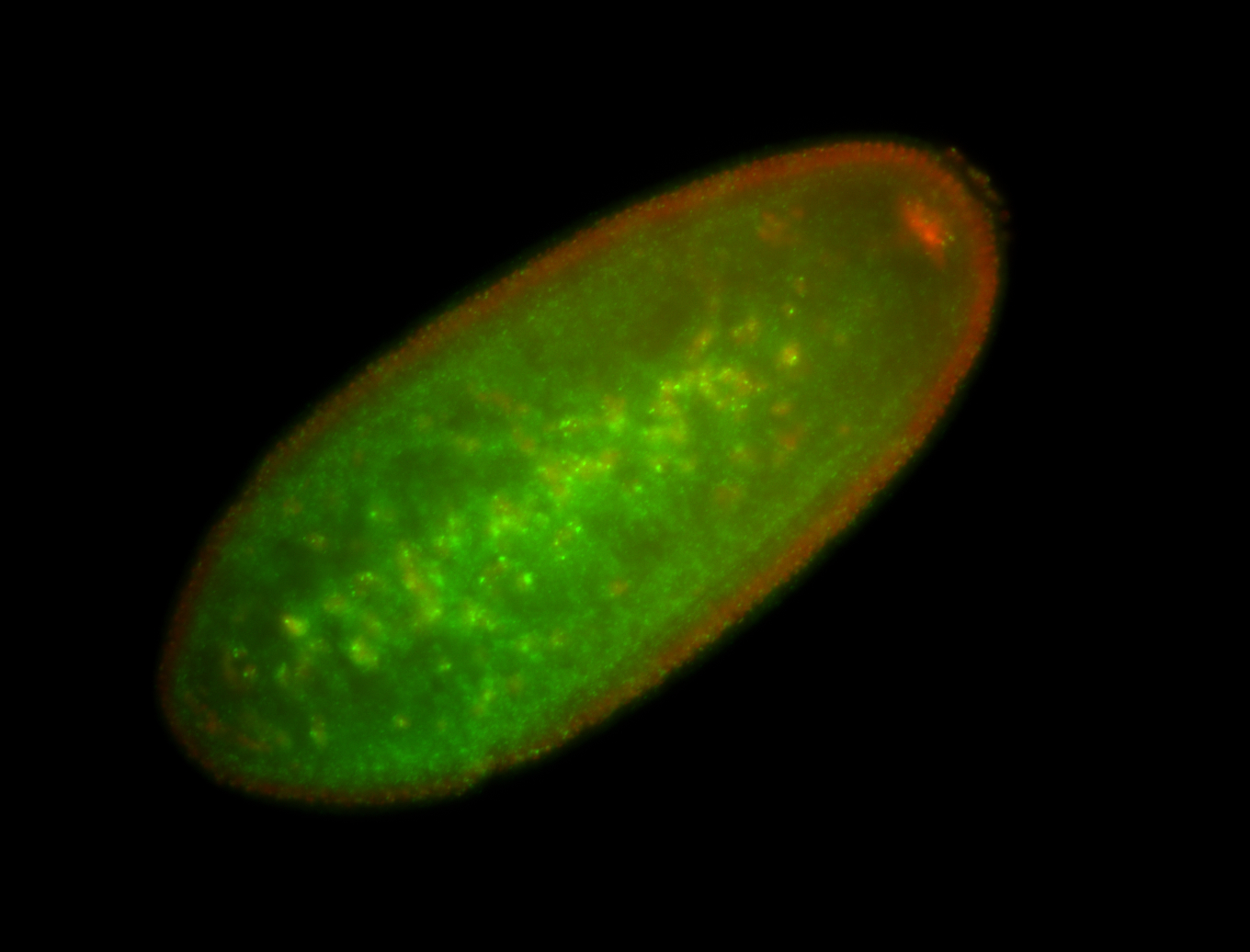
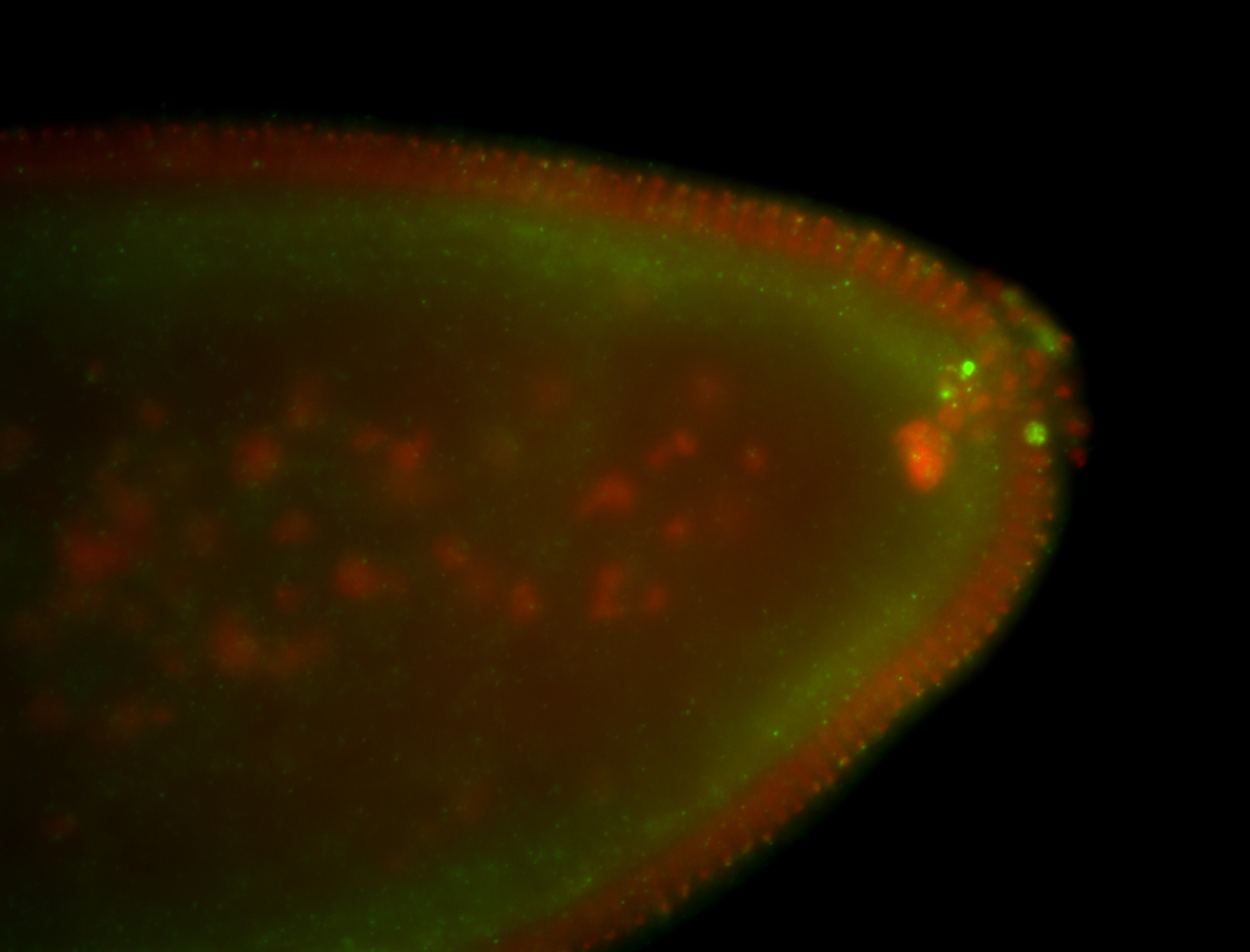
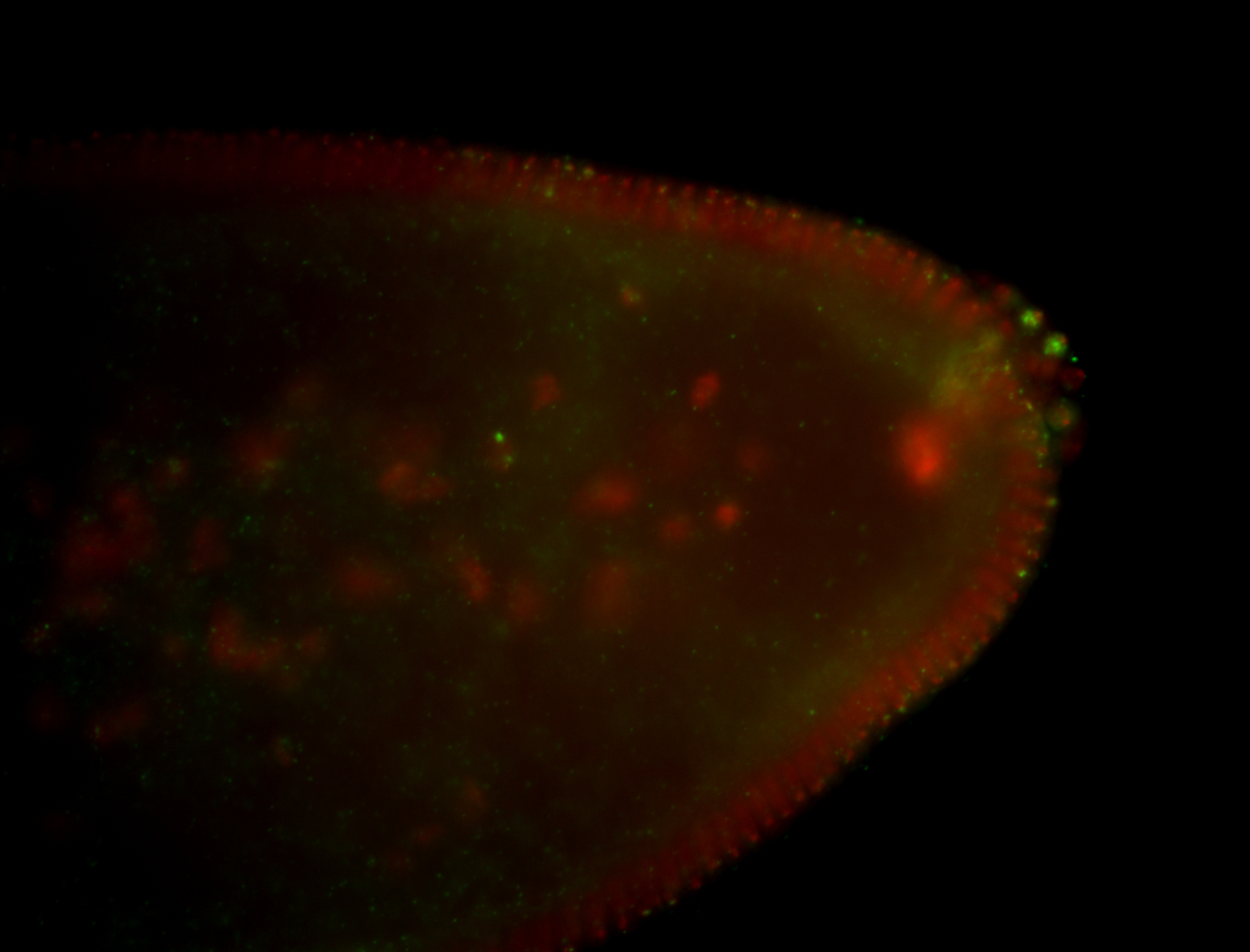
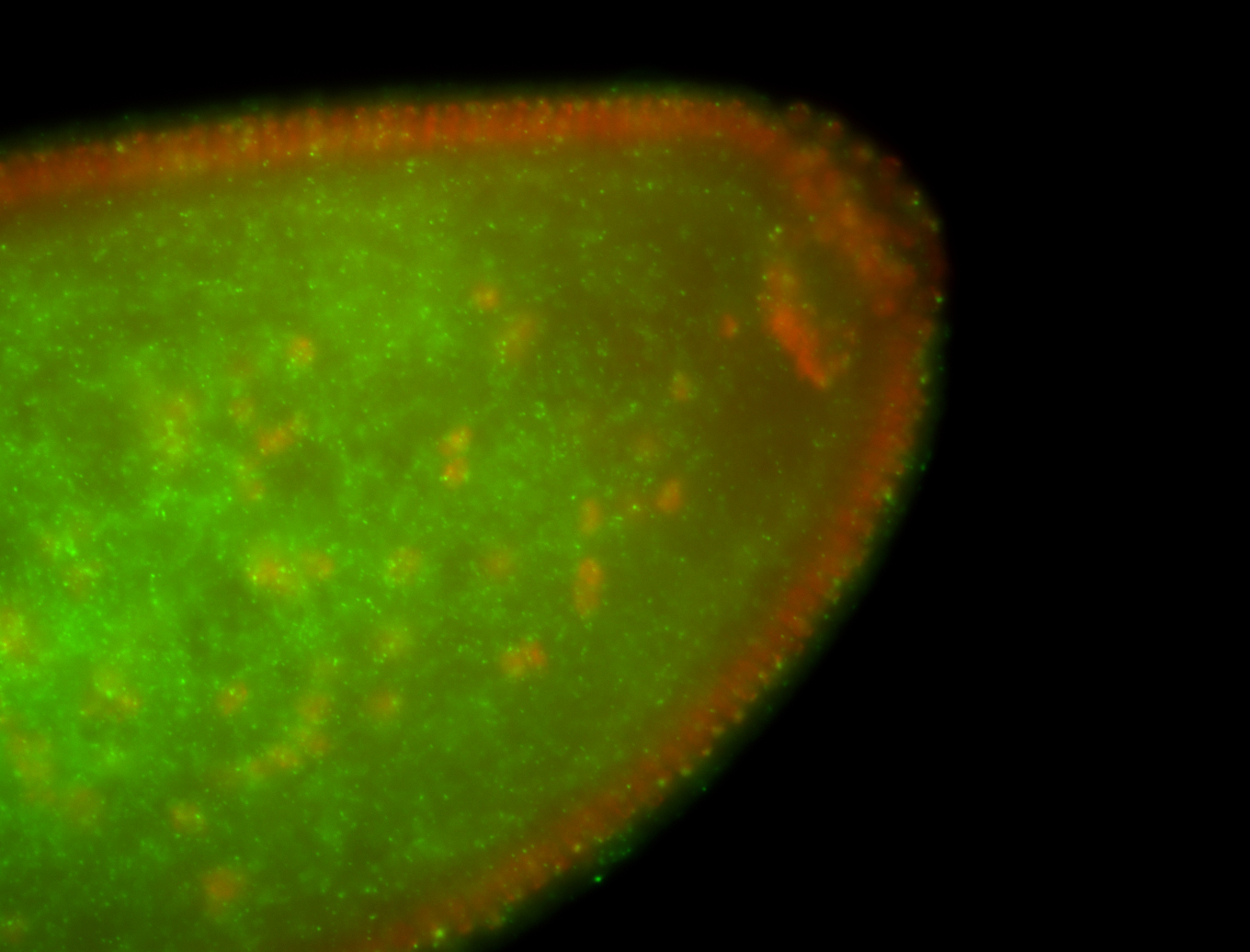
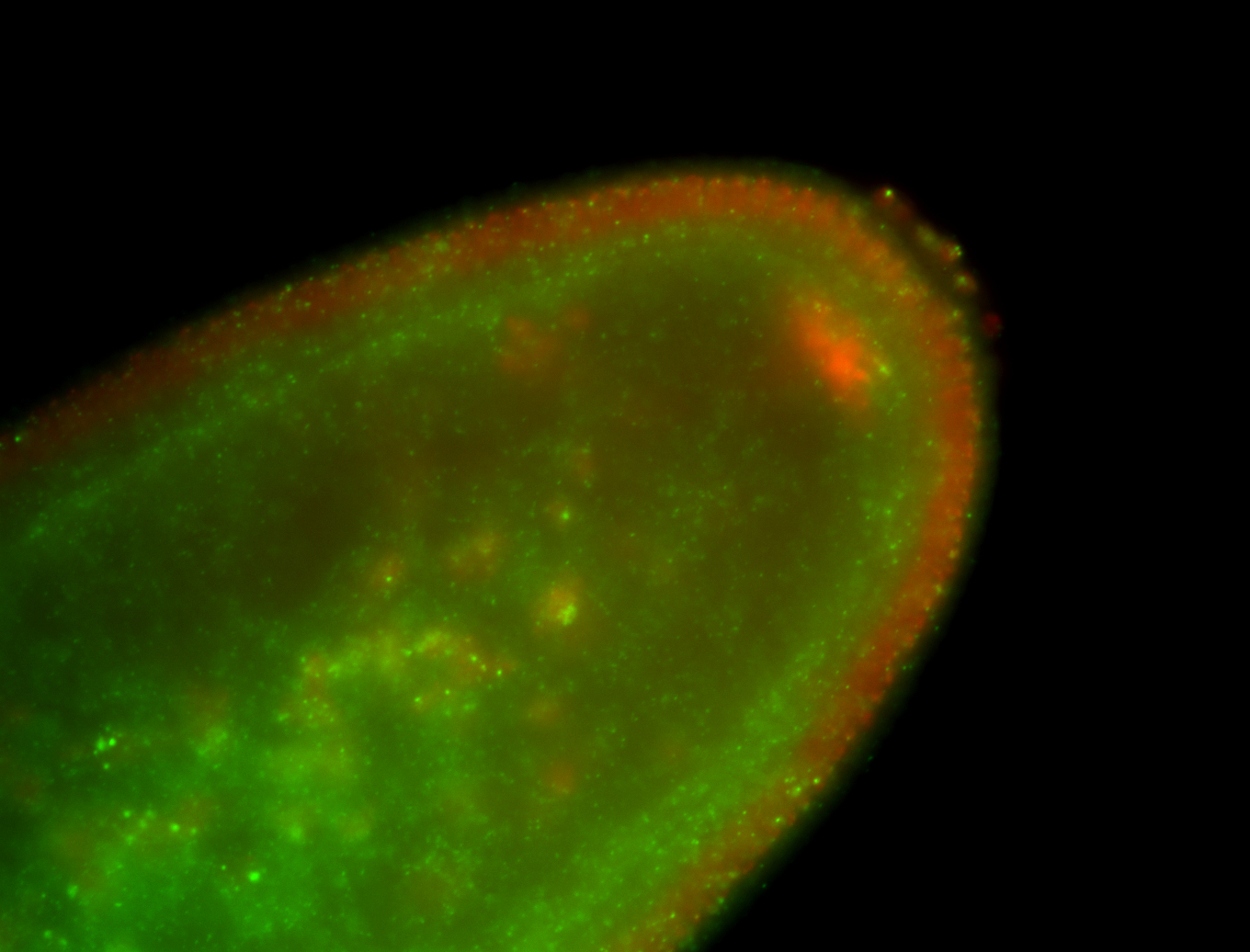
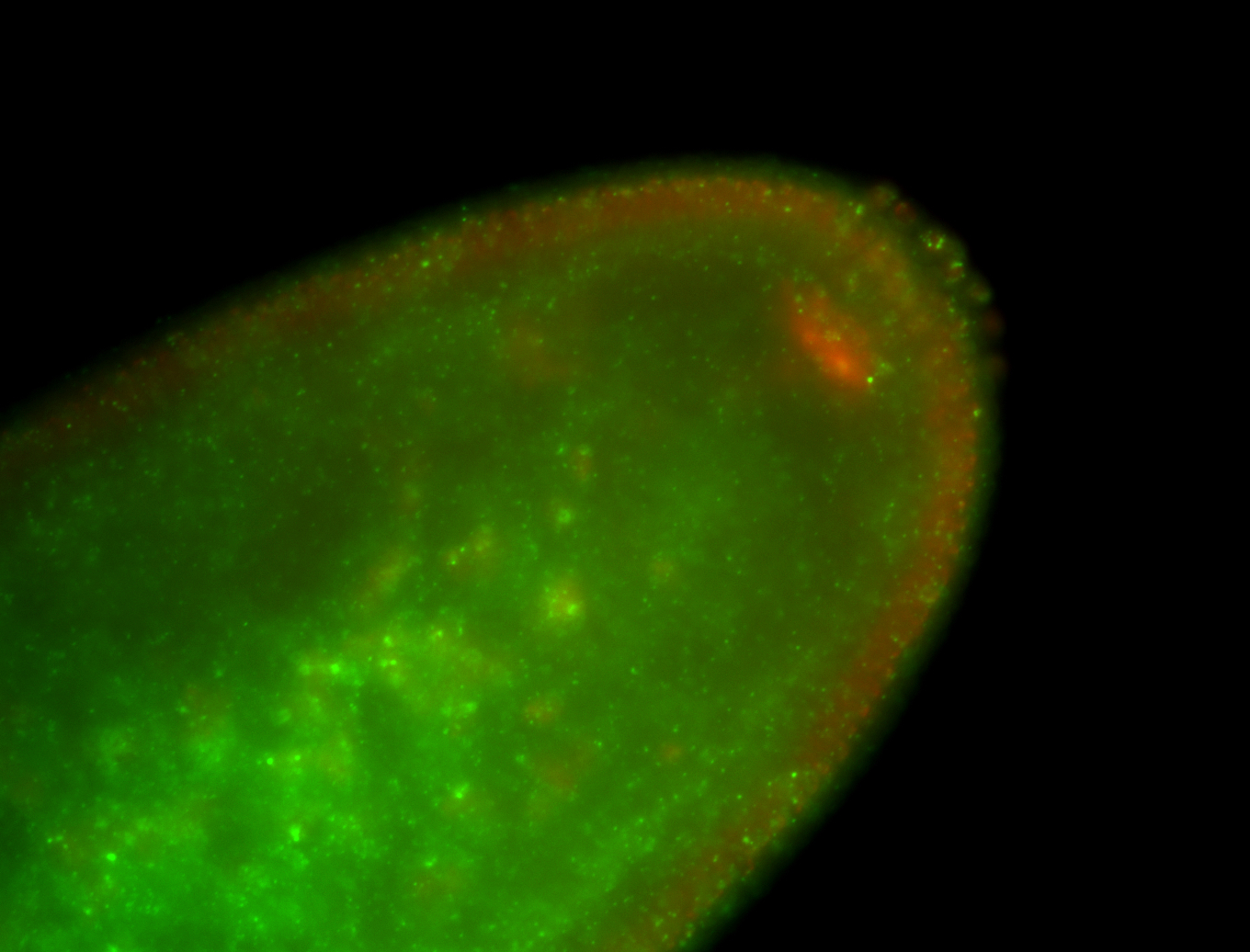
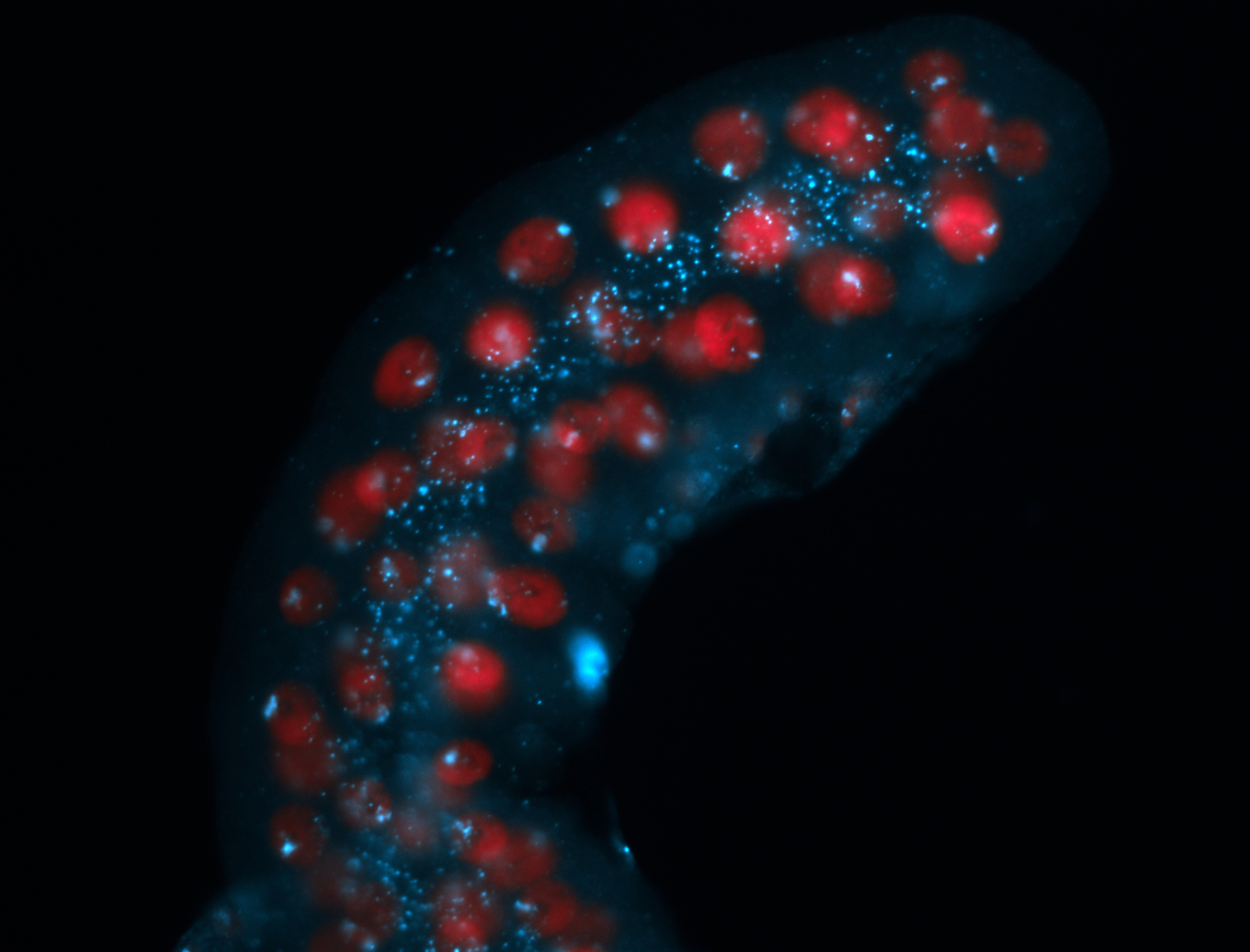
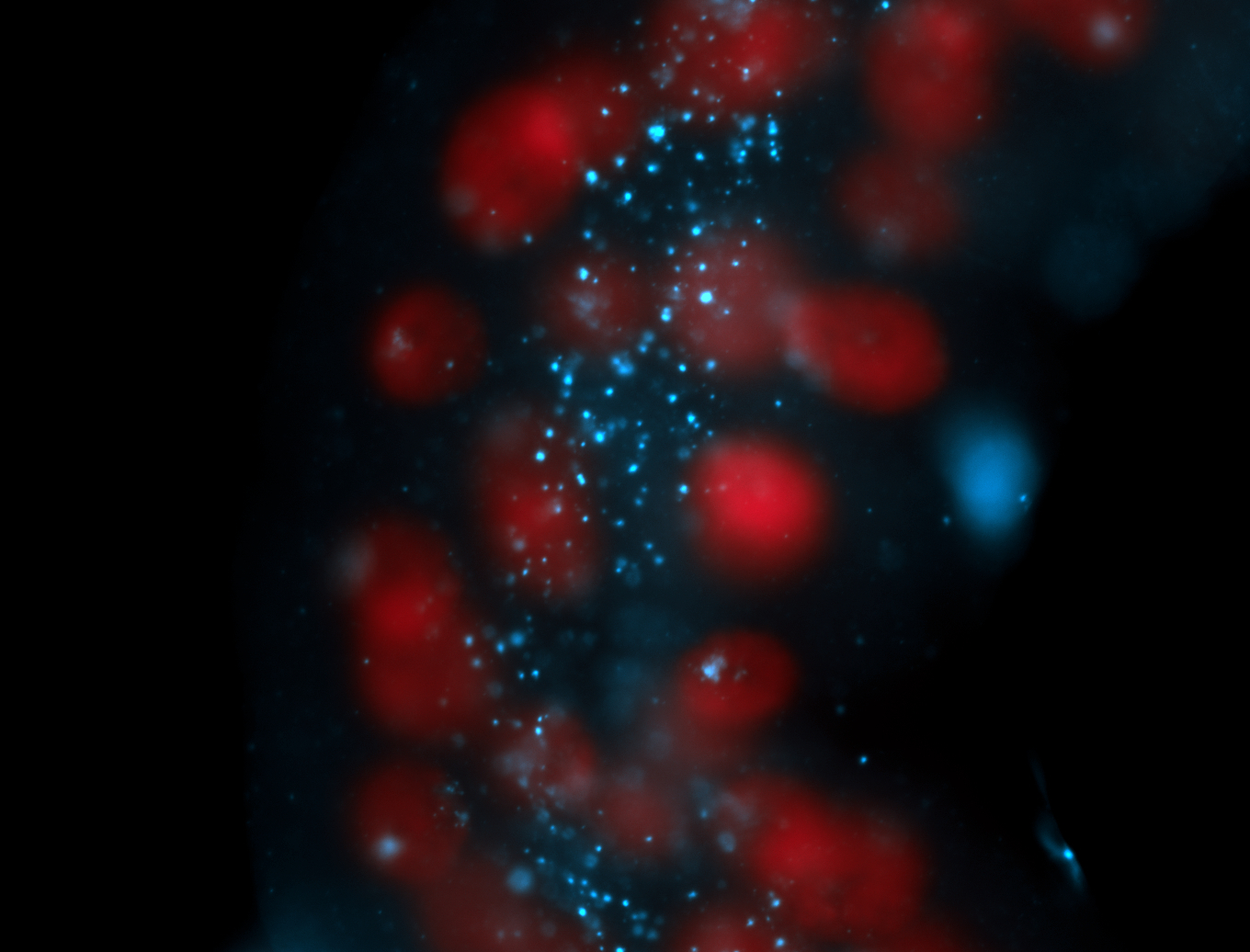
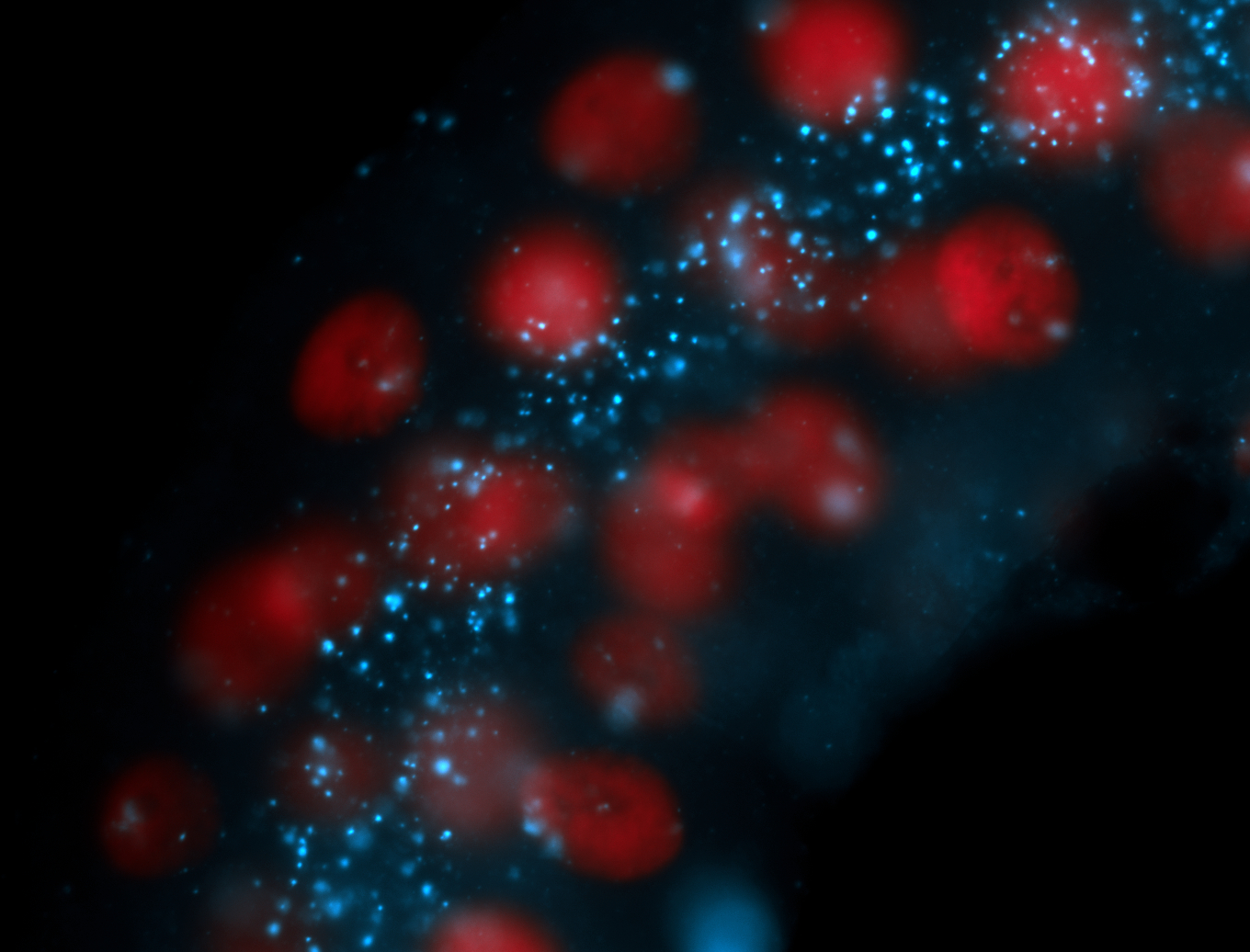
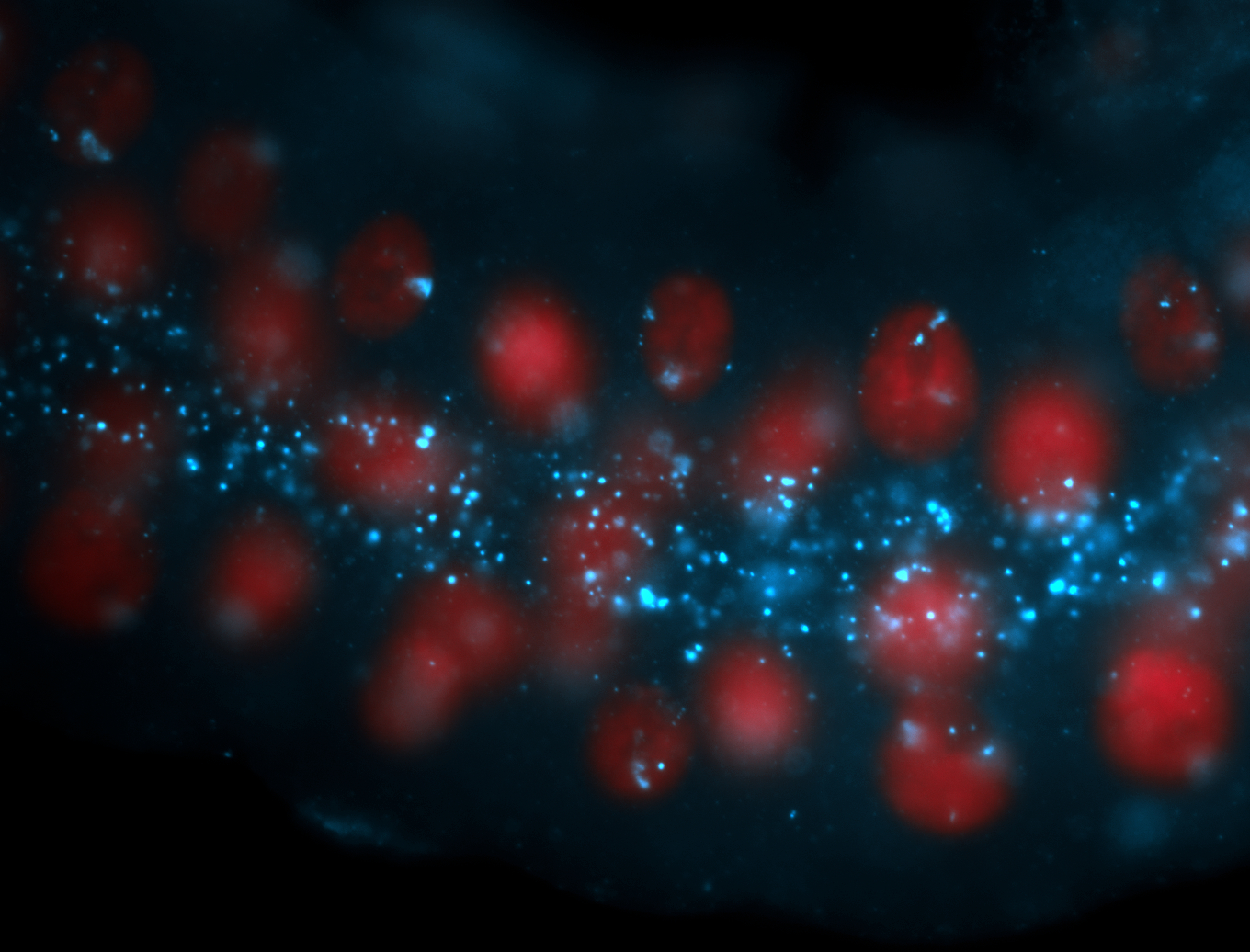
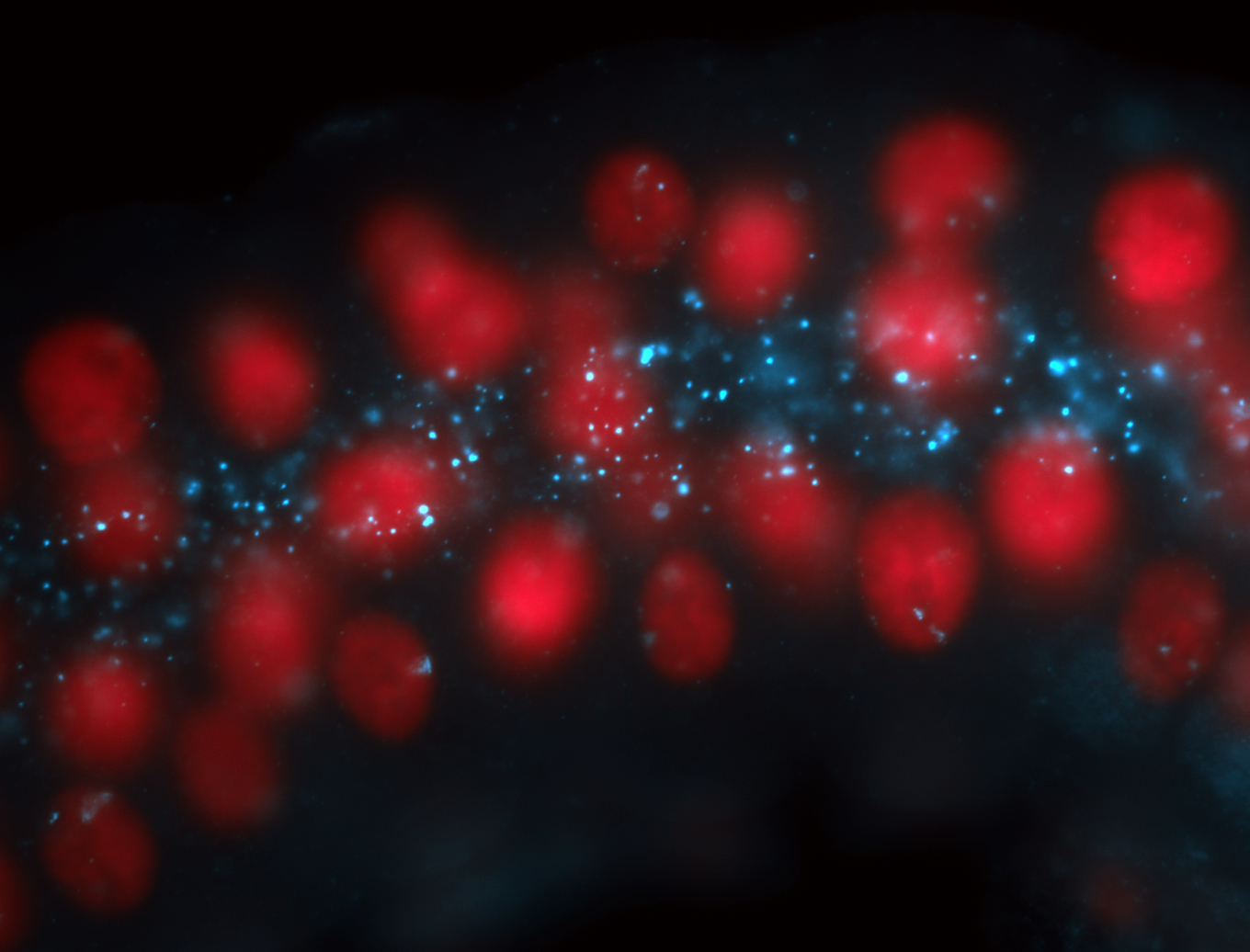
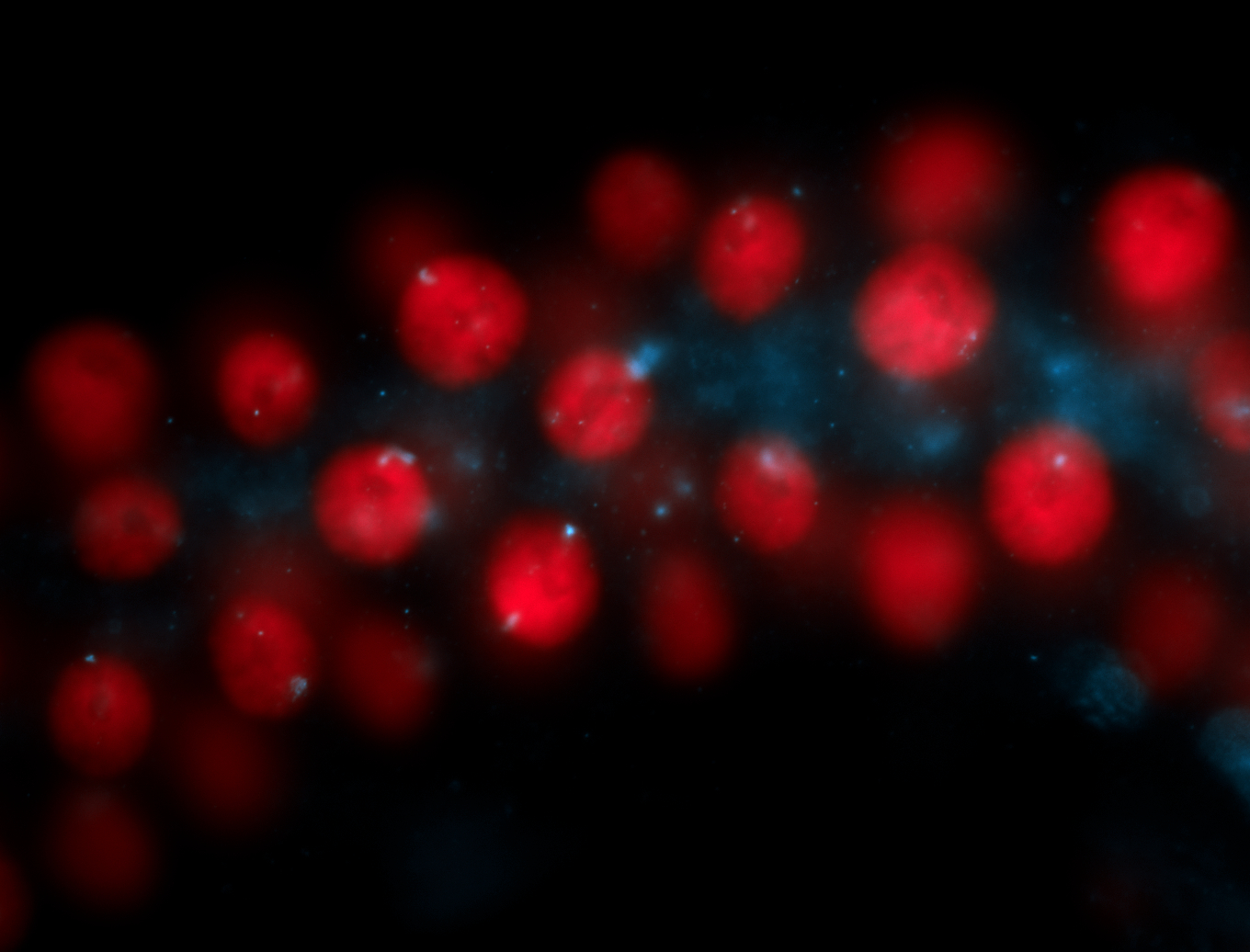
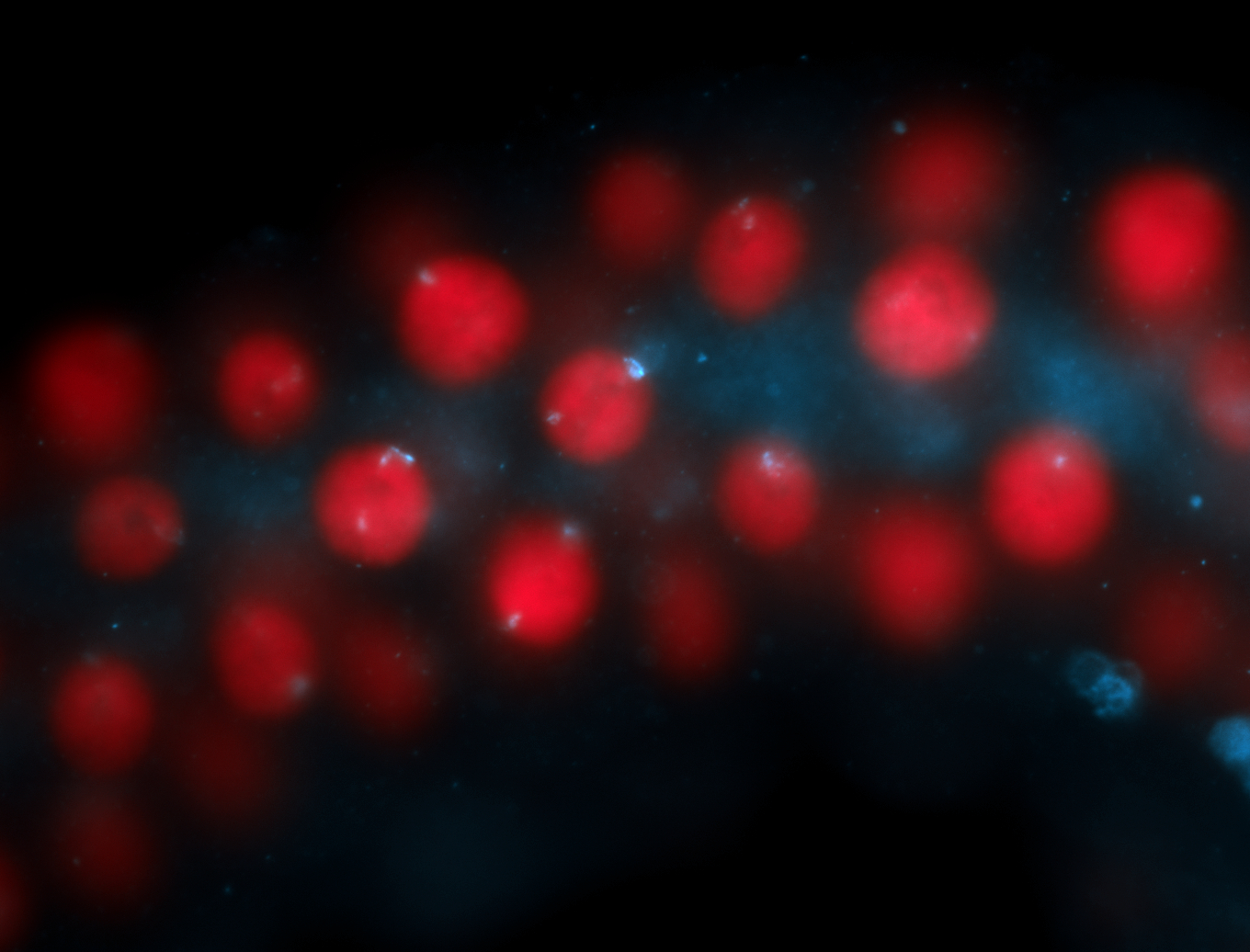
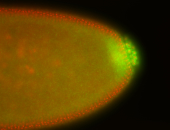
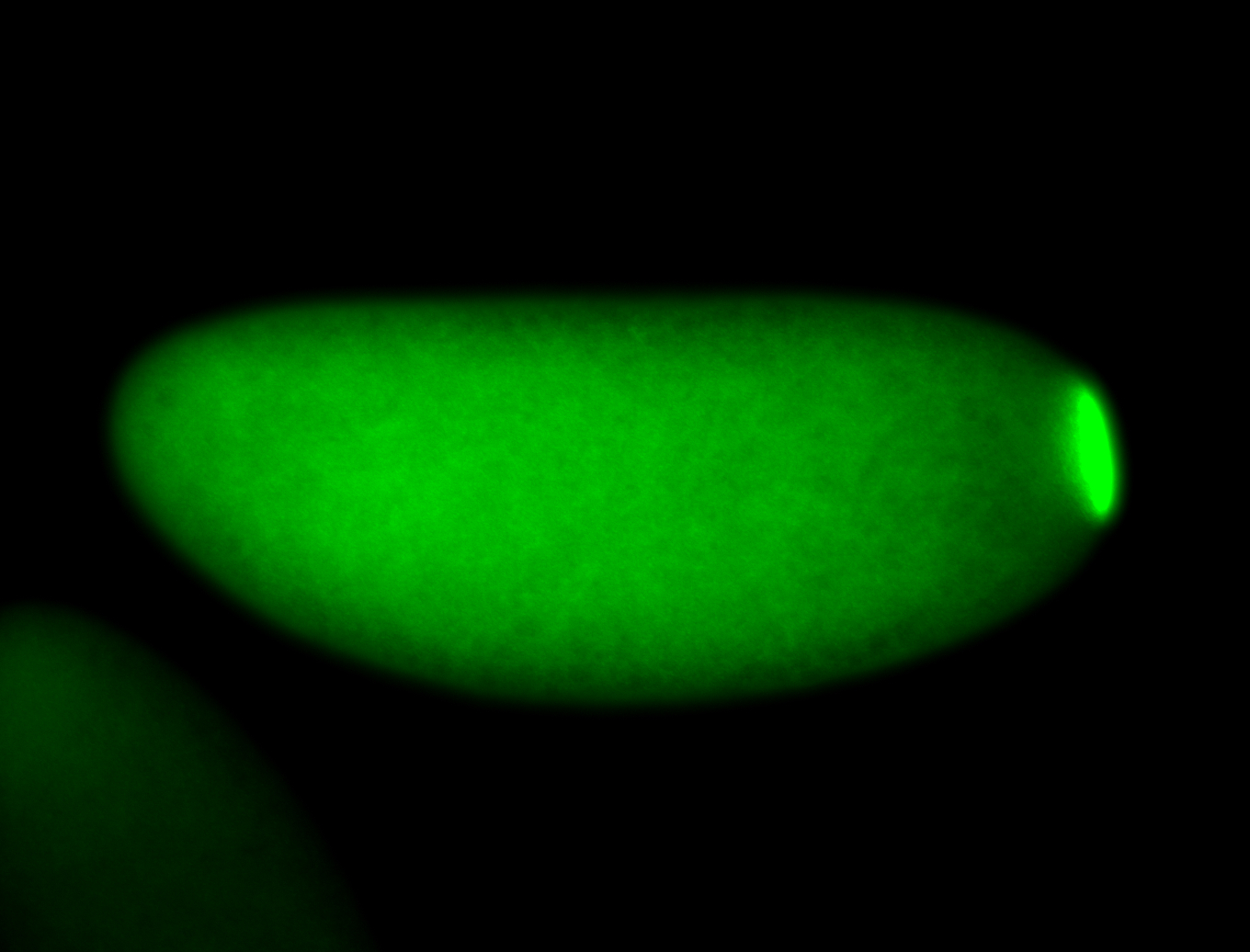
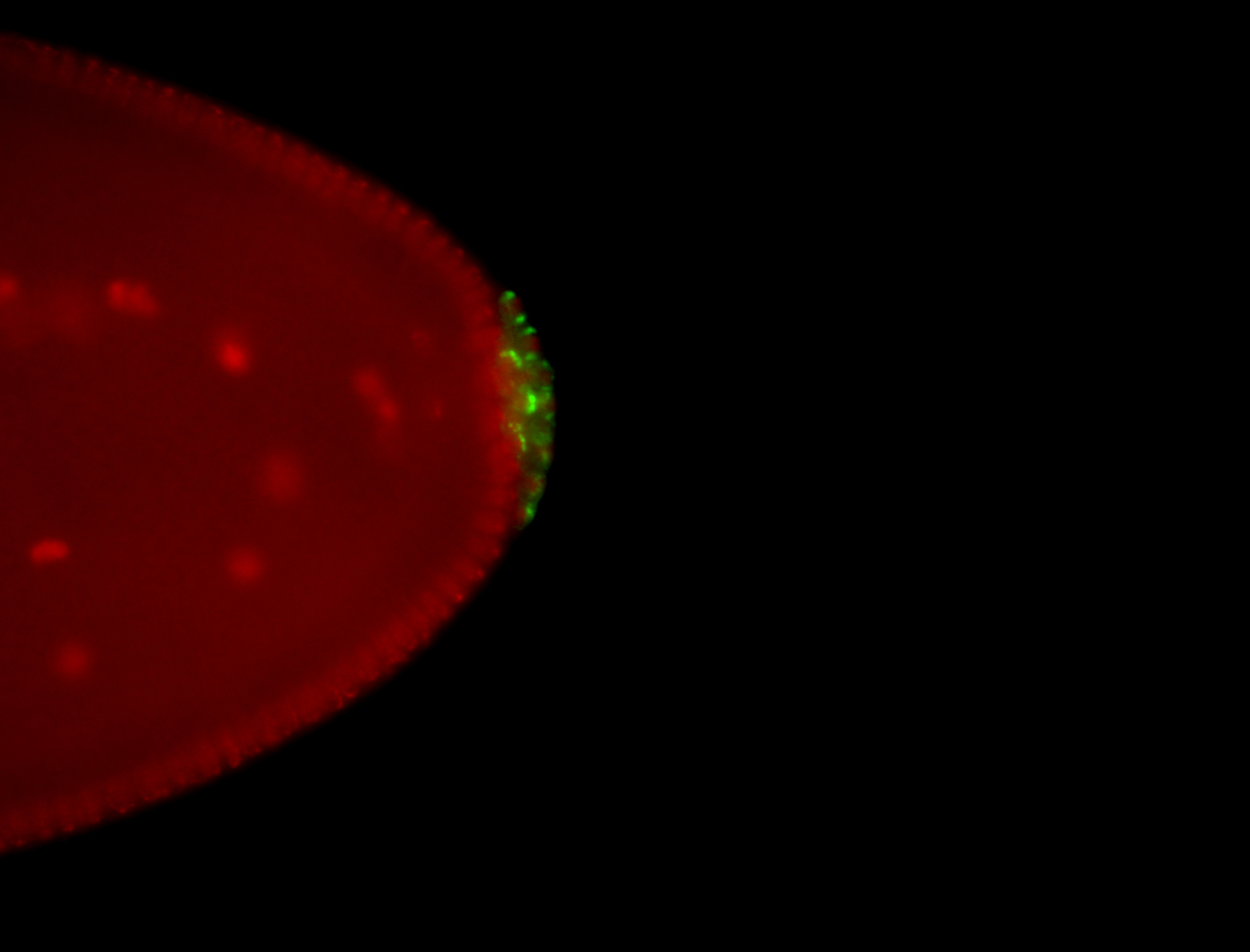
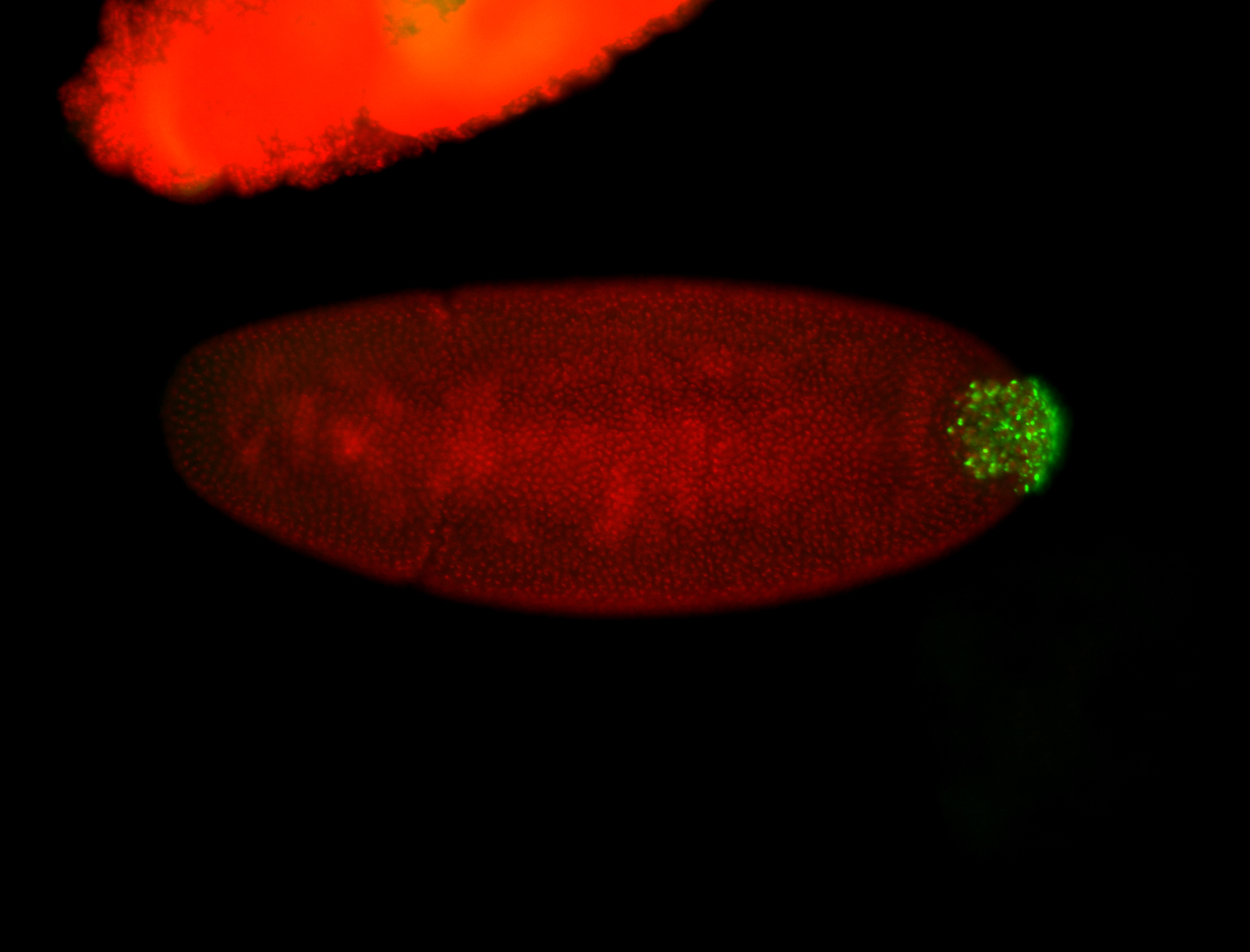
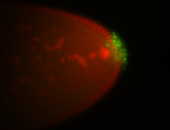
| Localized to pole cells, and pole plasm. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
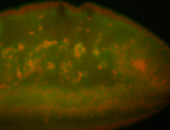
| I am not sure if it has a cell division pattern... |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
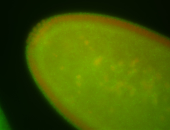
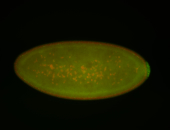
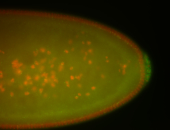
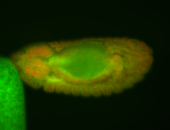
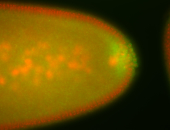
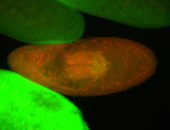
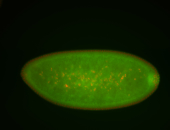
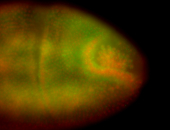
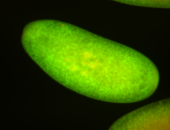
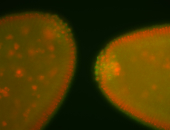
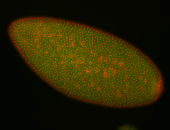
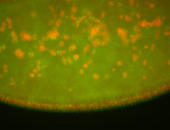
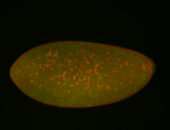
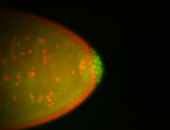
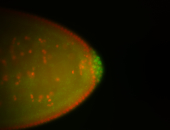
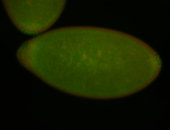
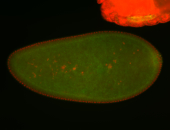
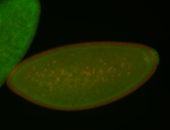
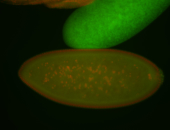
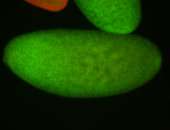
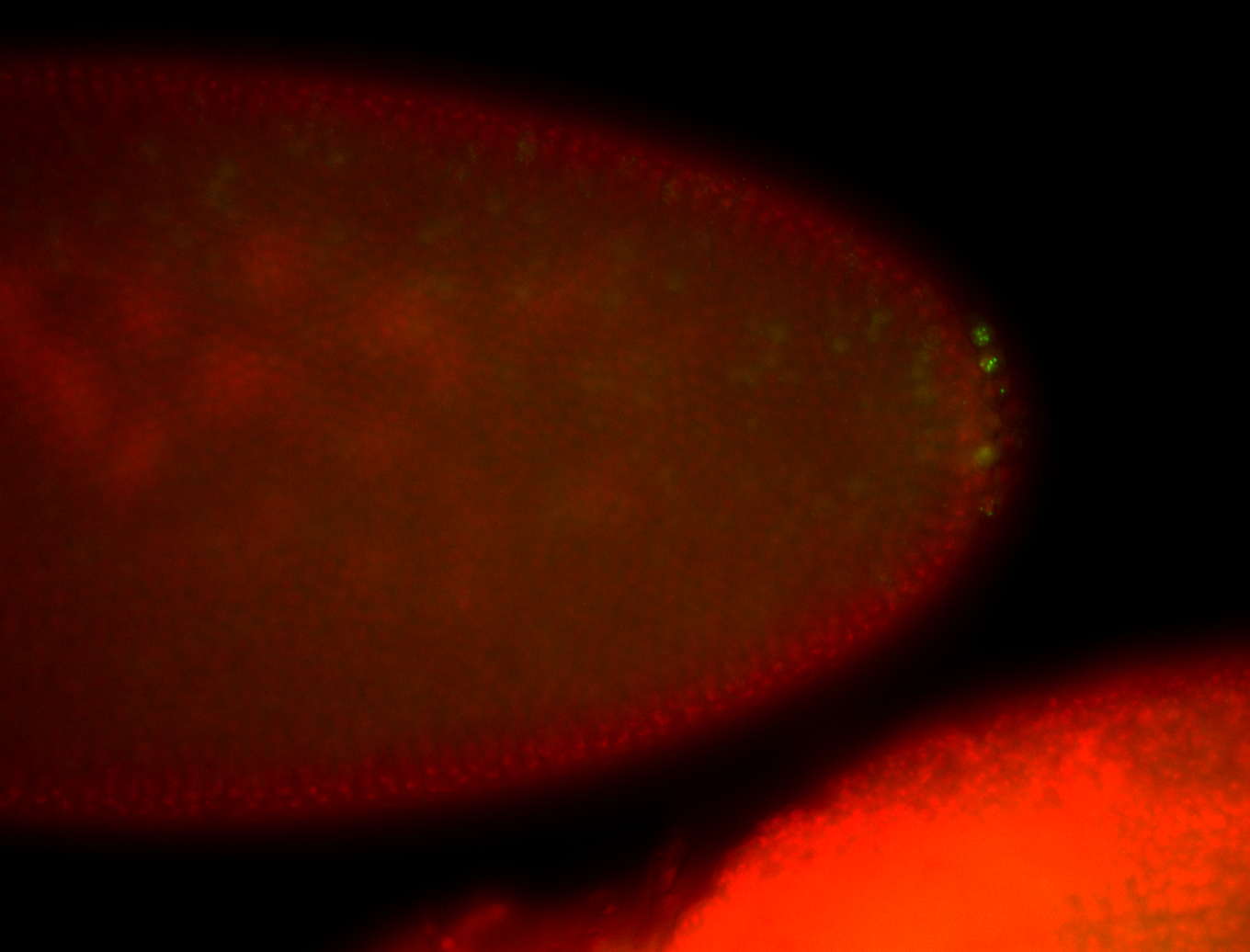
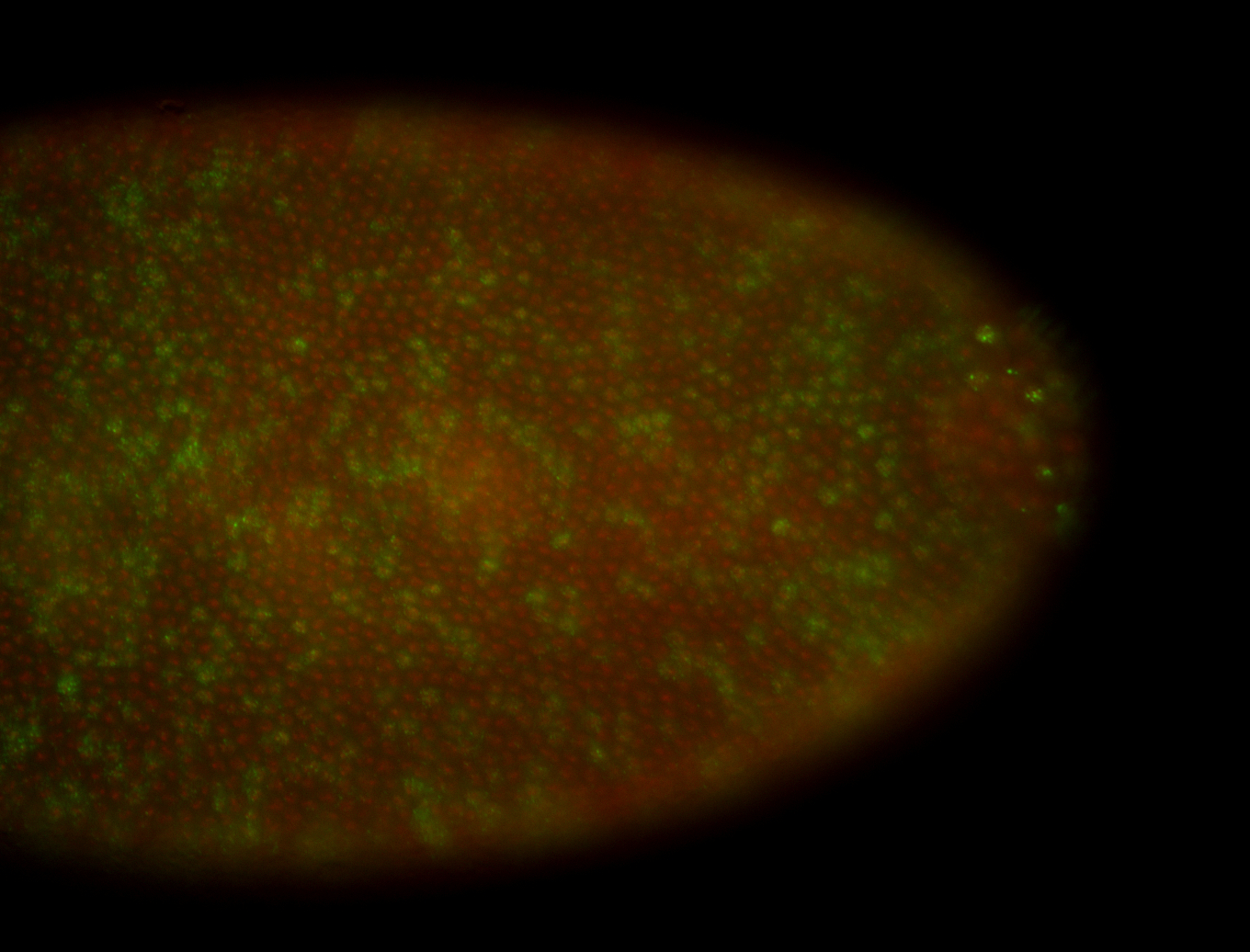
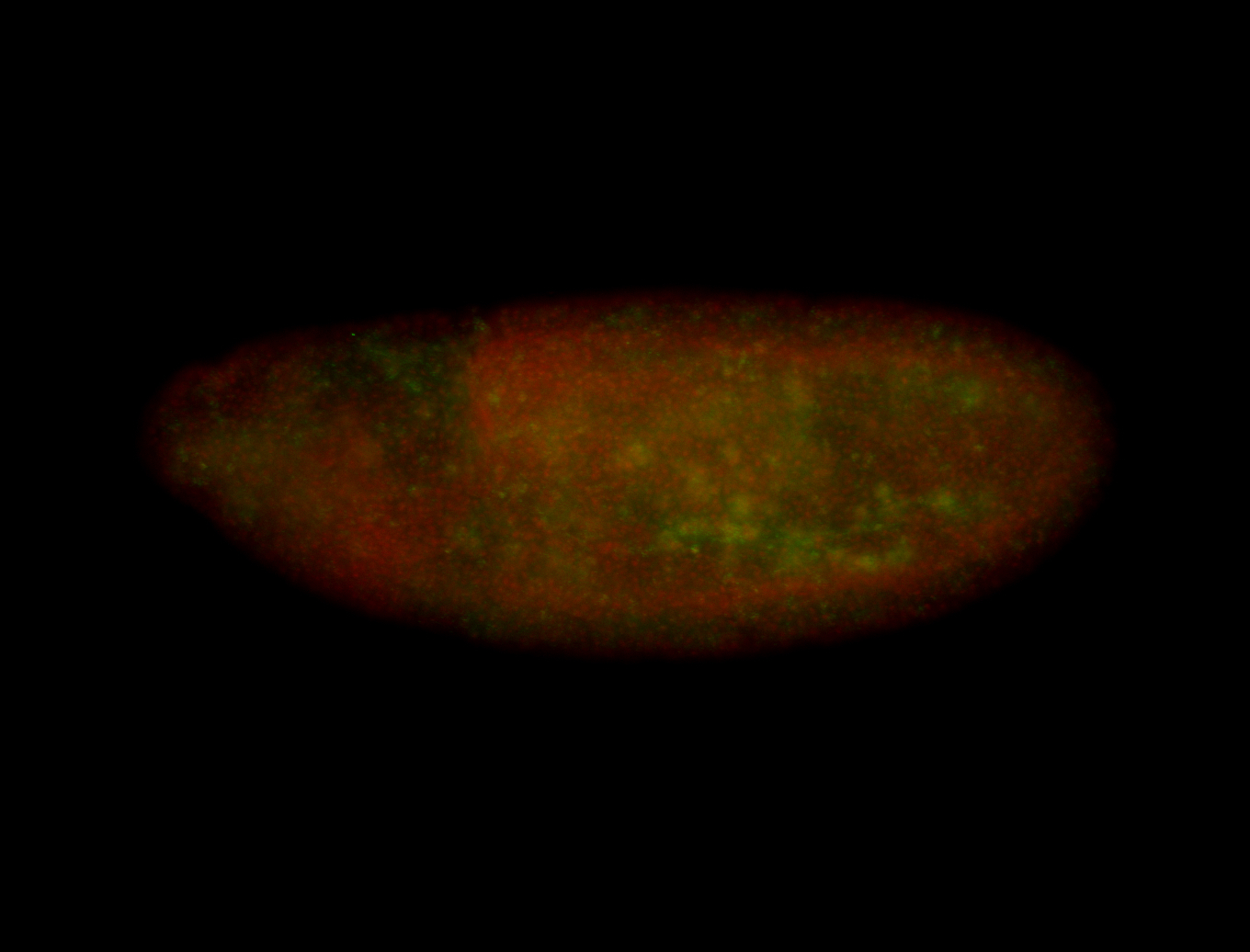
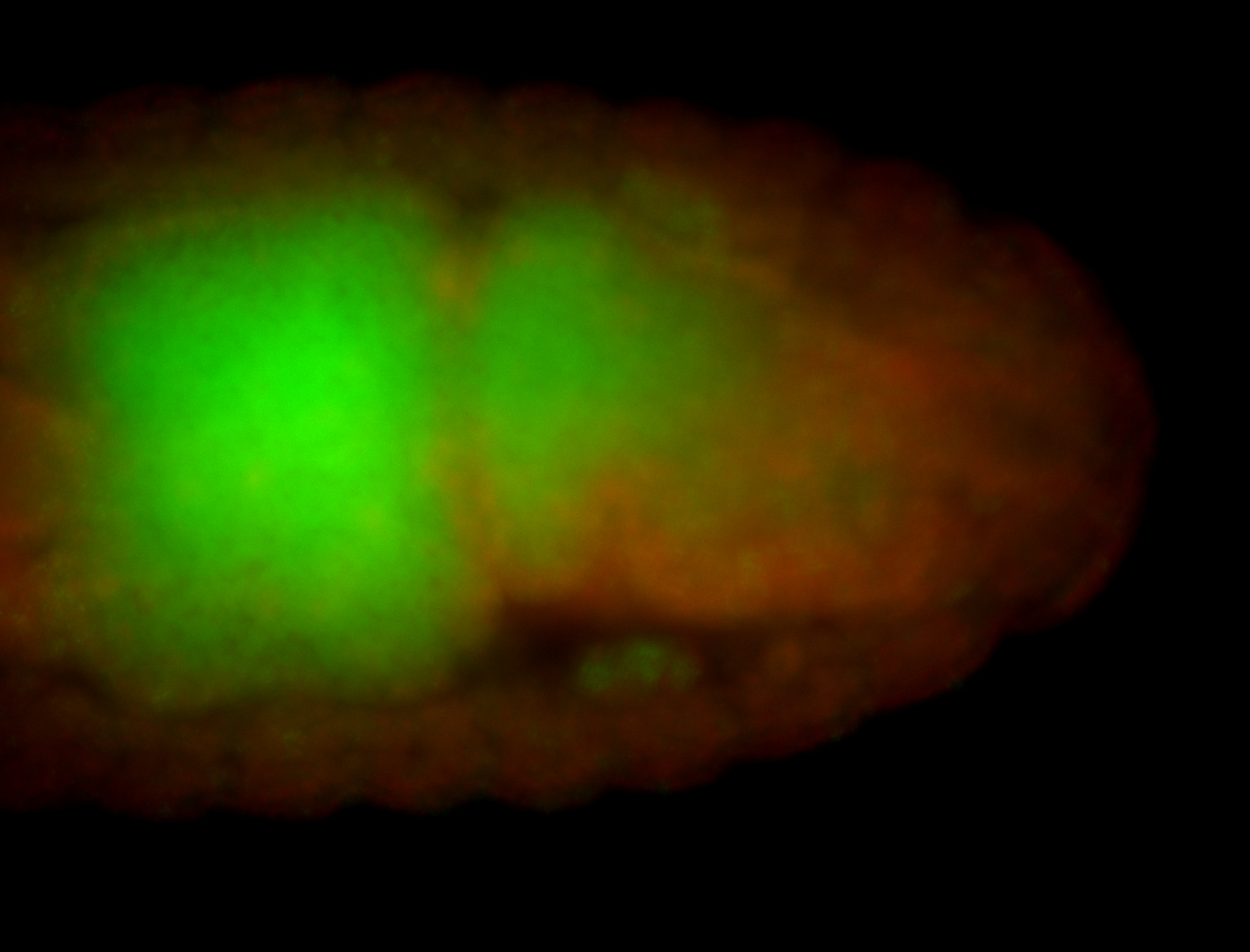
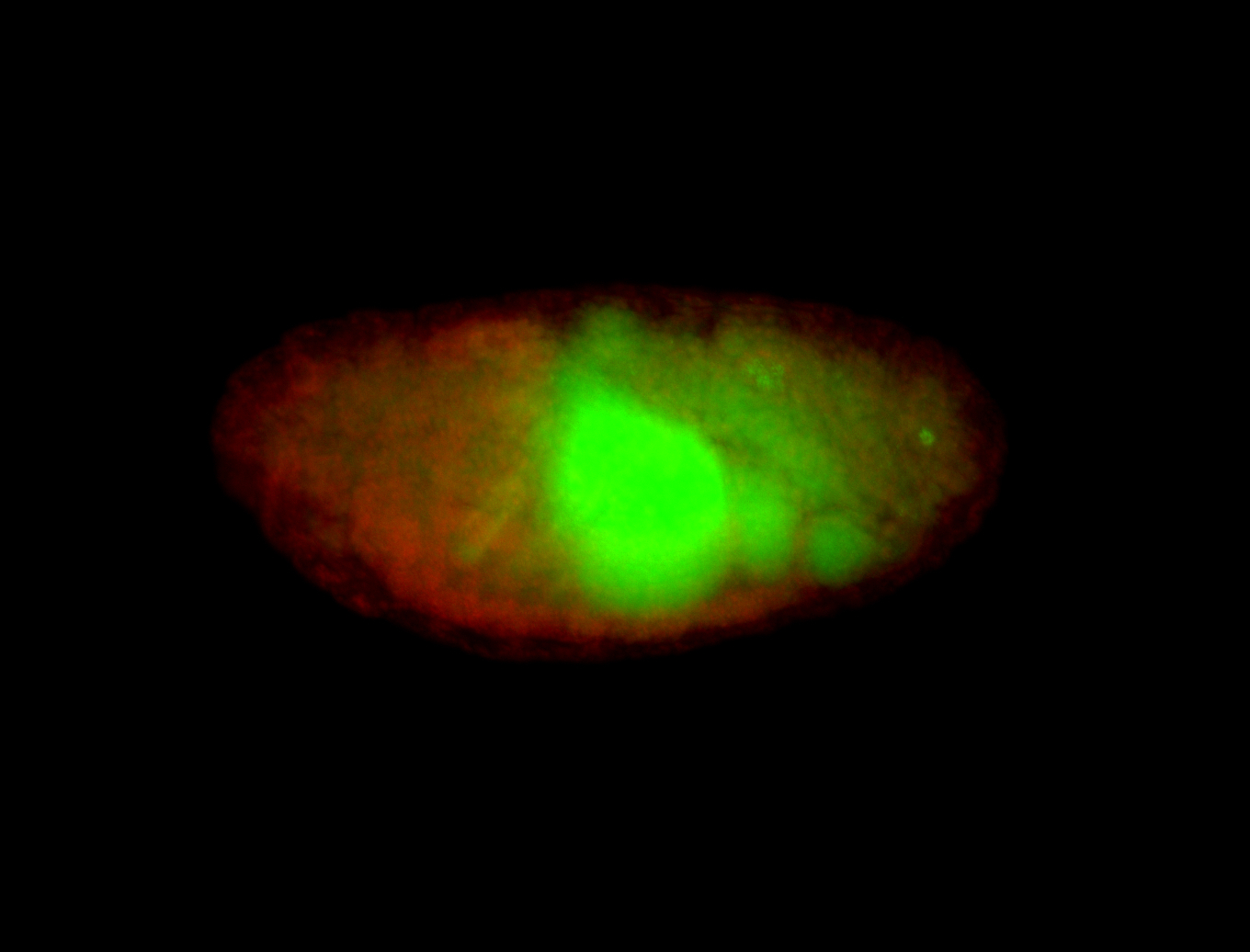
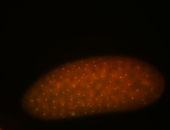
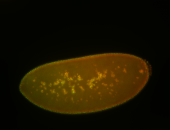
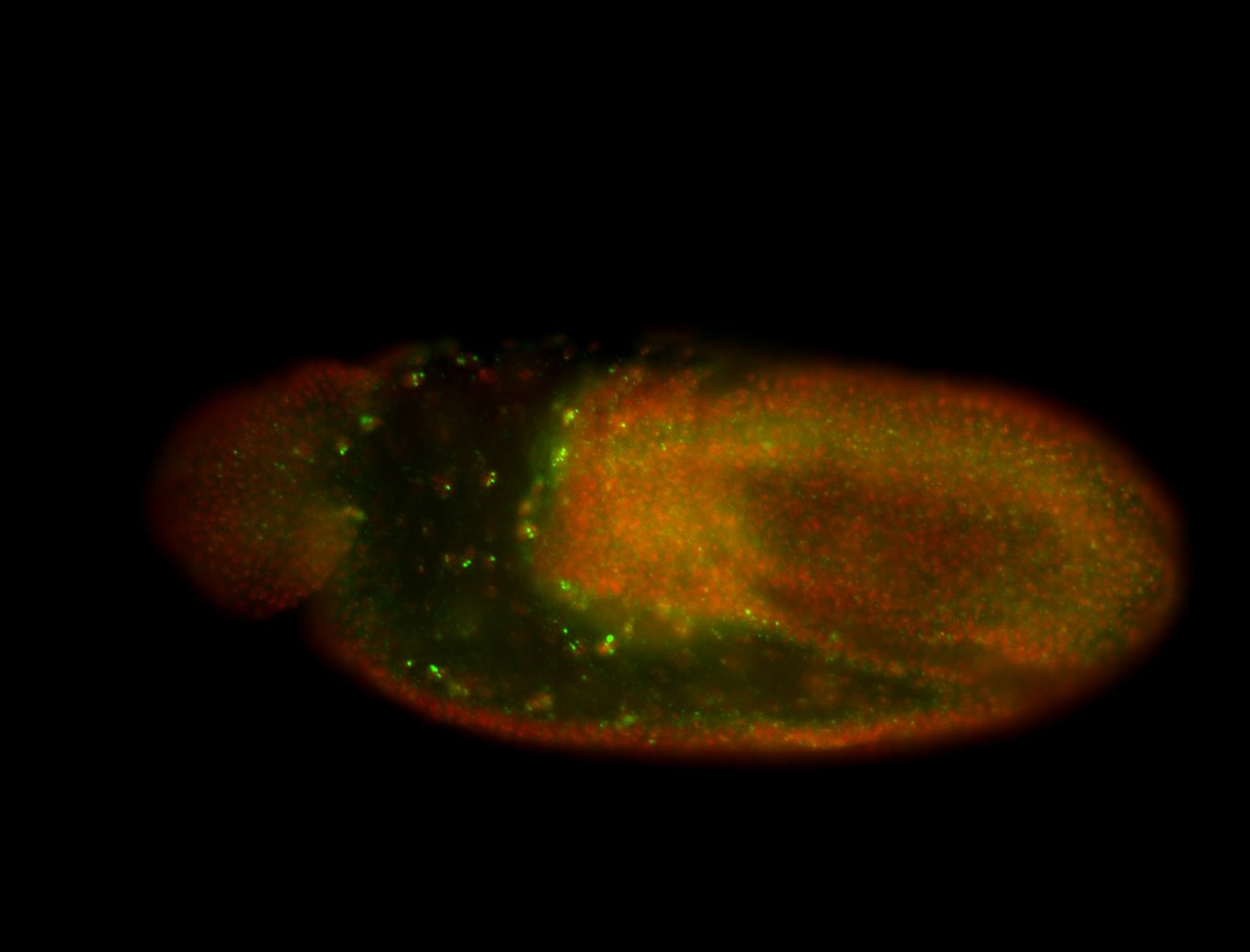
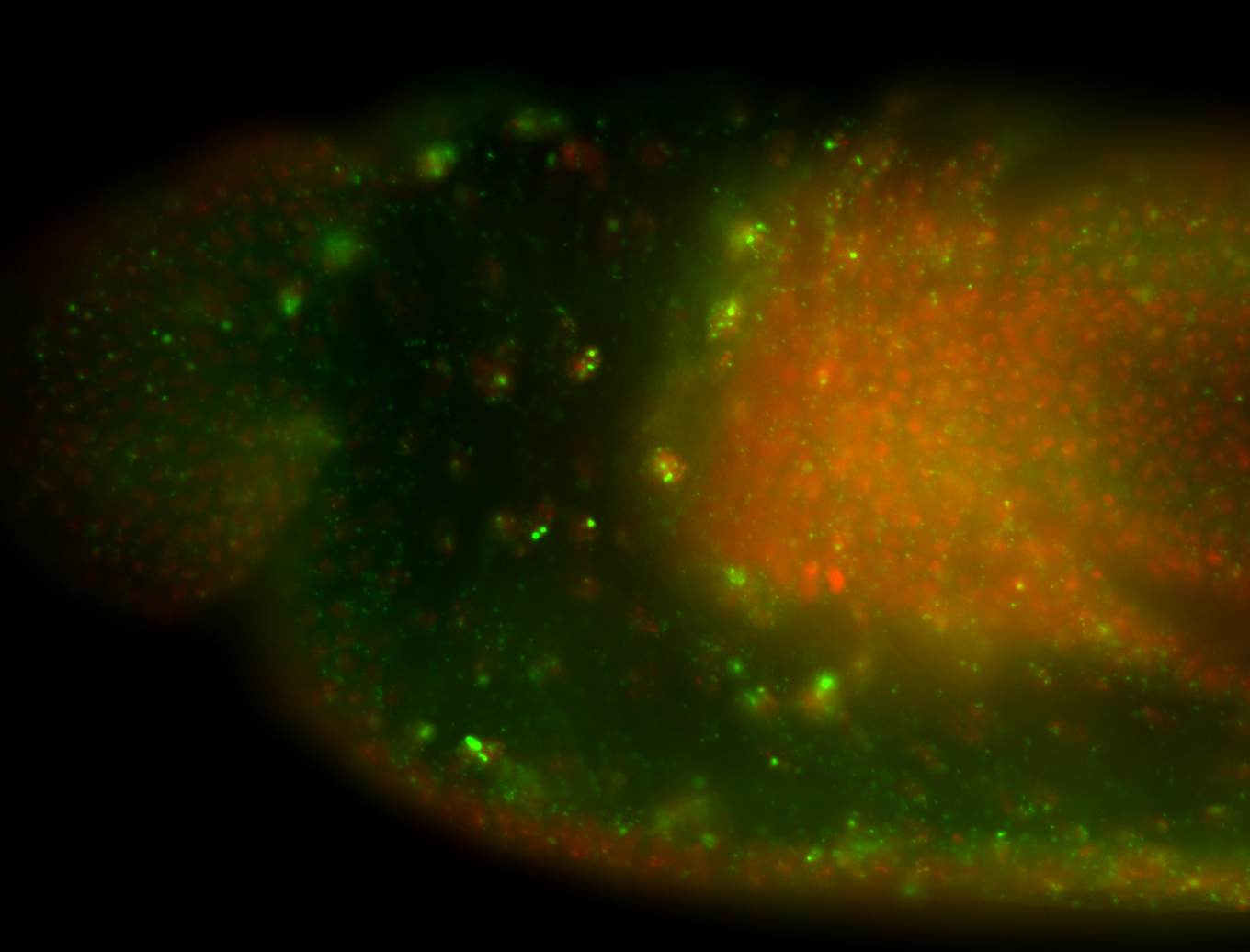
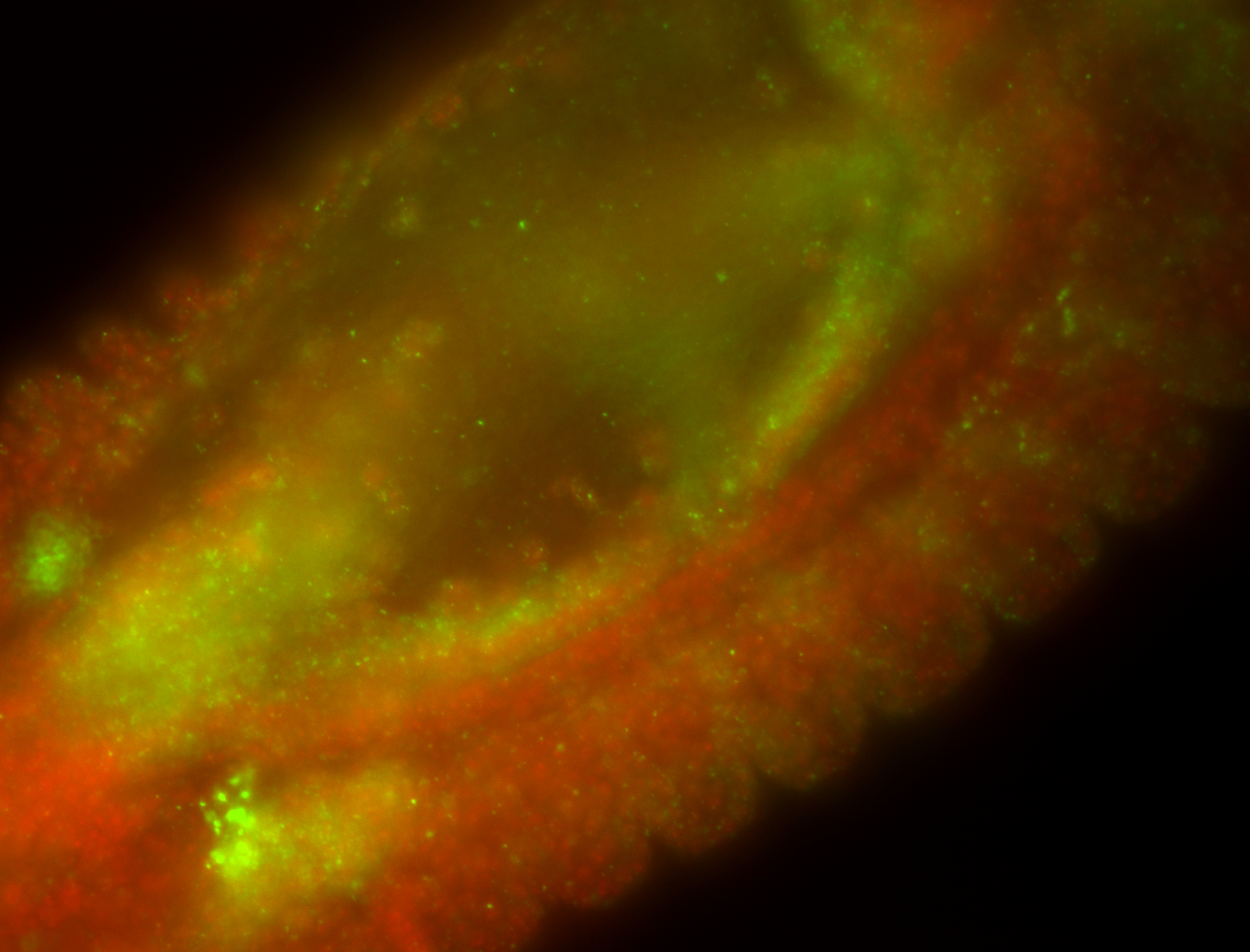
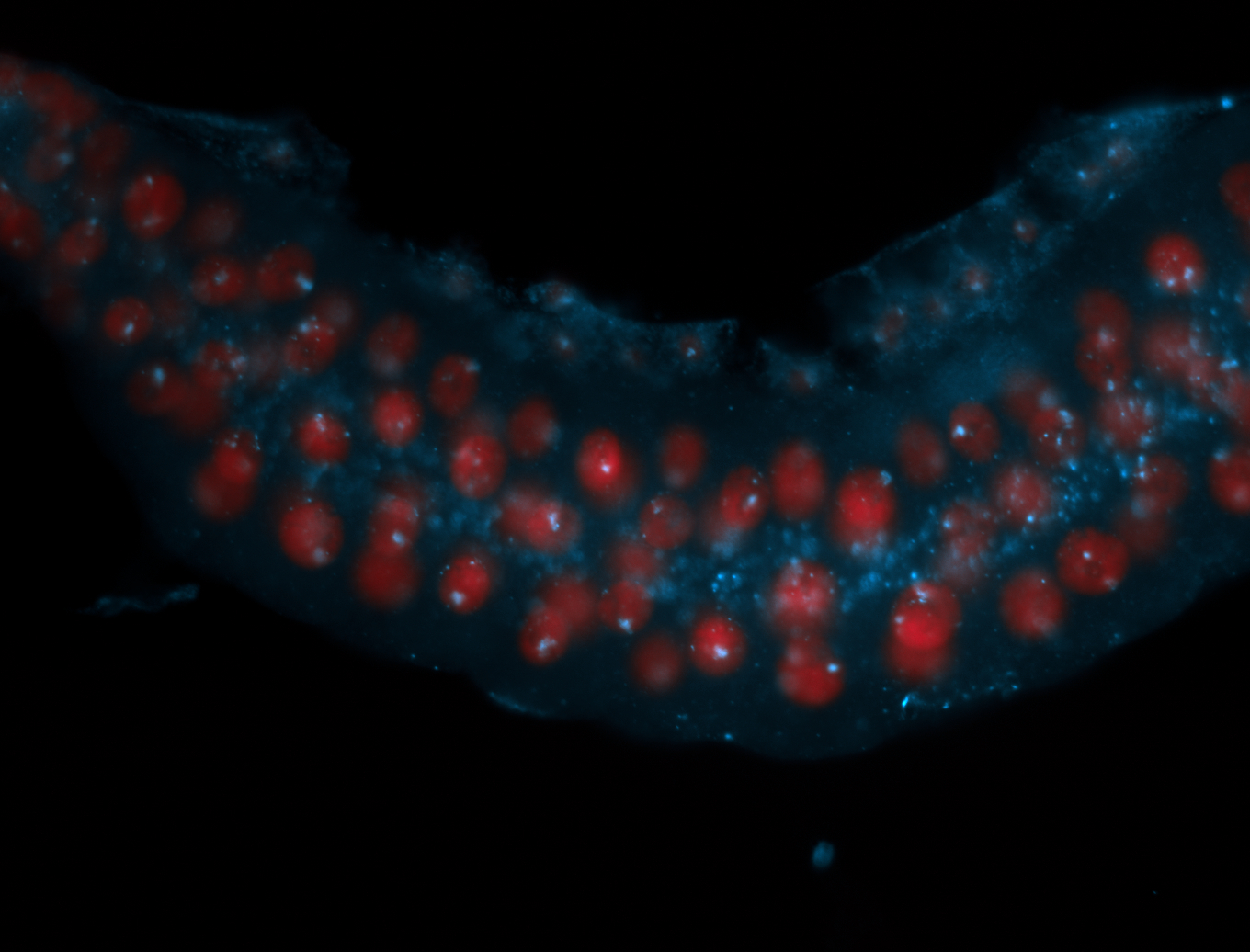
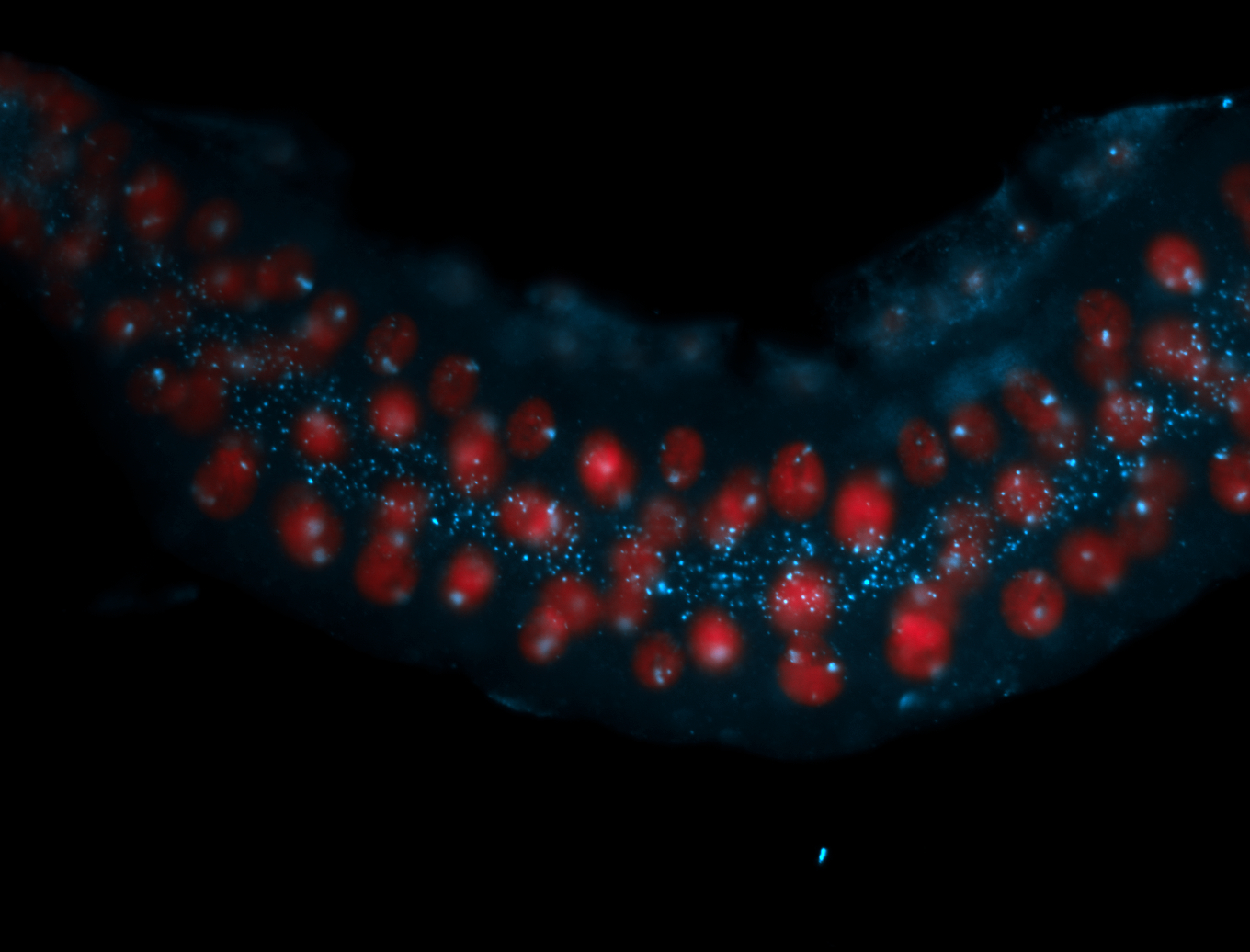
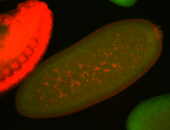
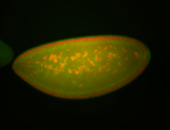
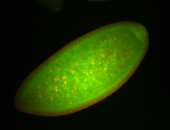
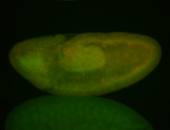
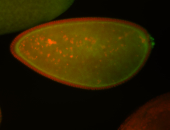
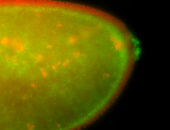
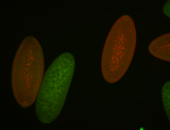
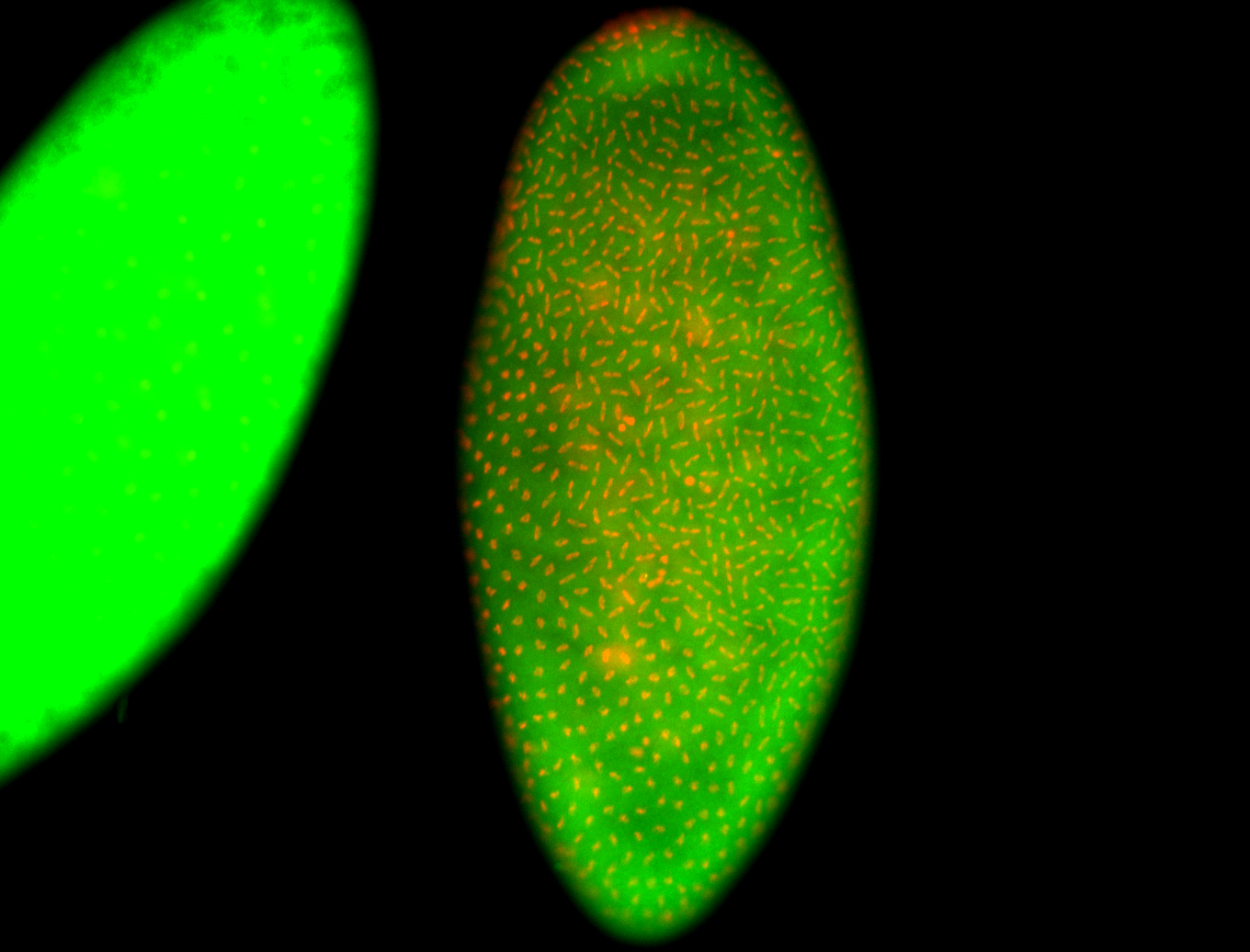
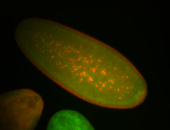
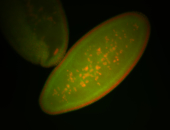
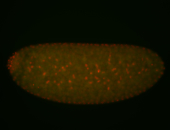
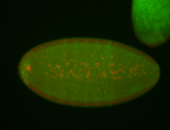
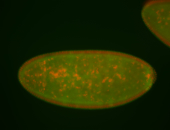
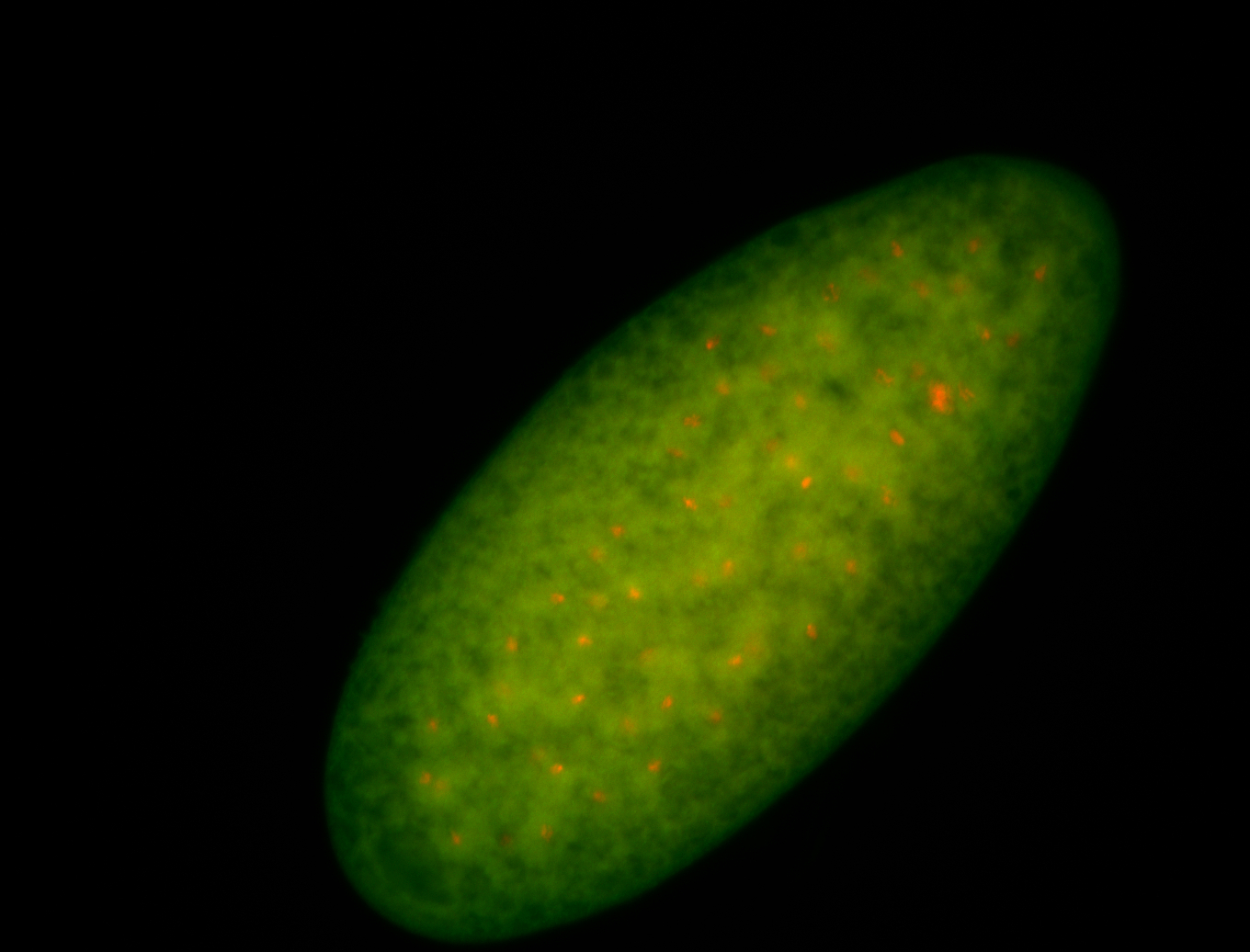
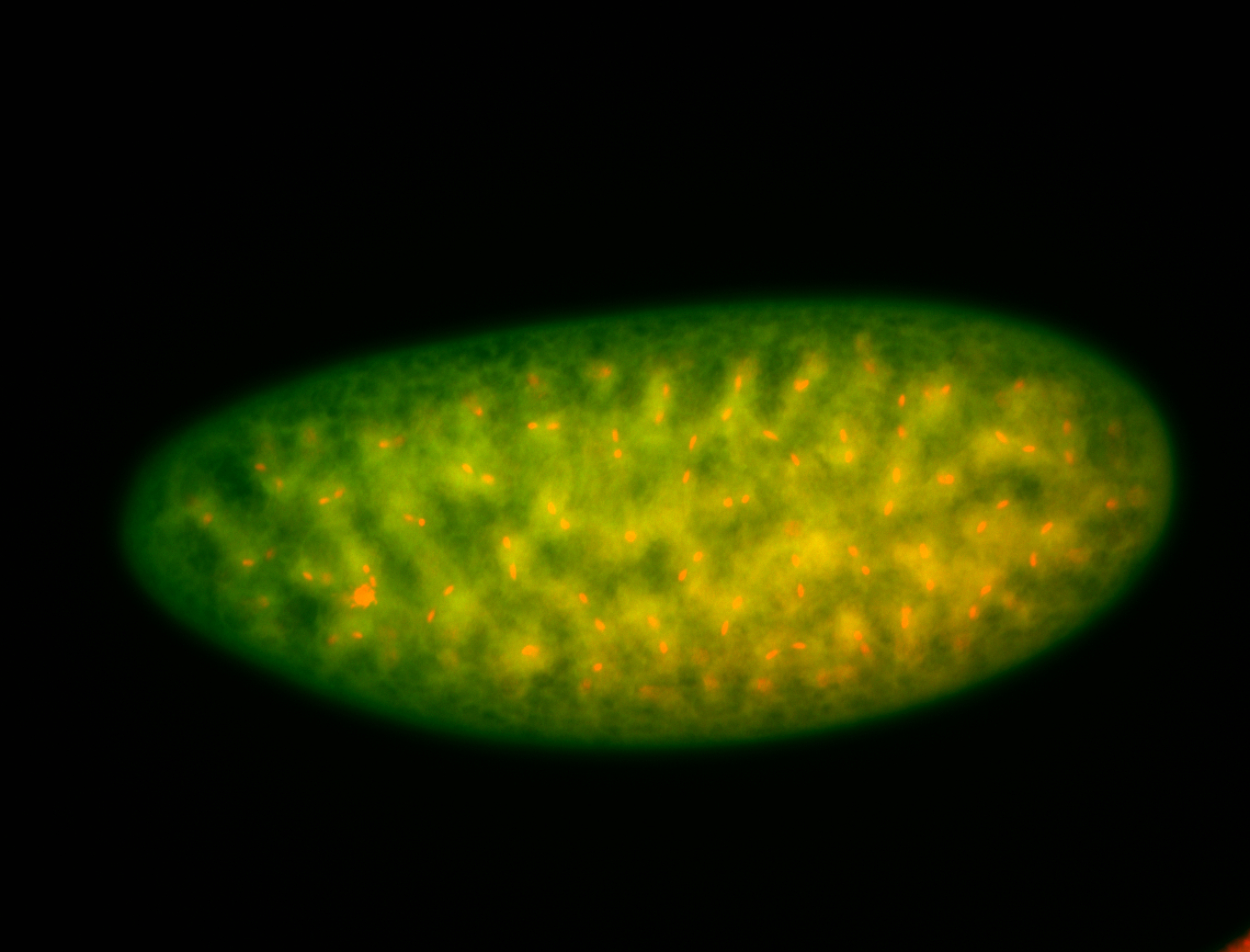
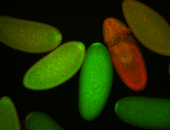
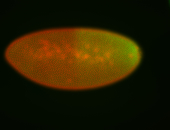
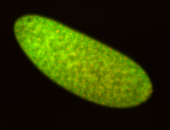
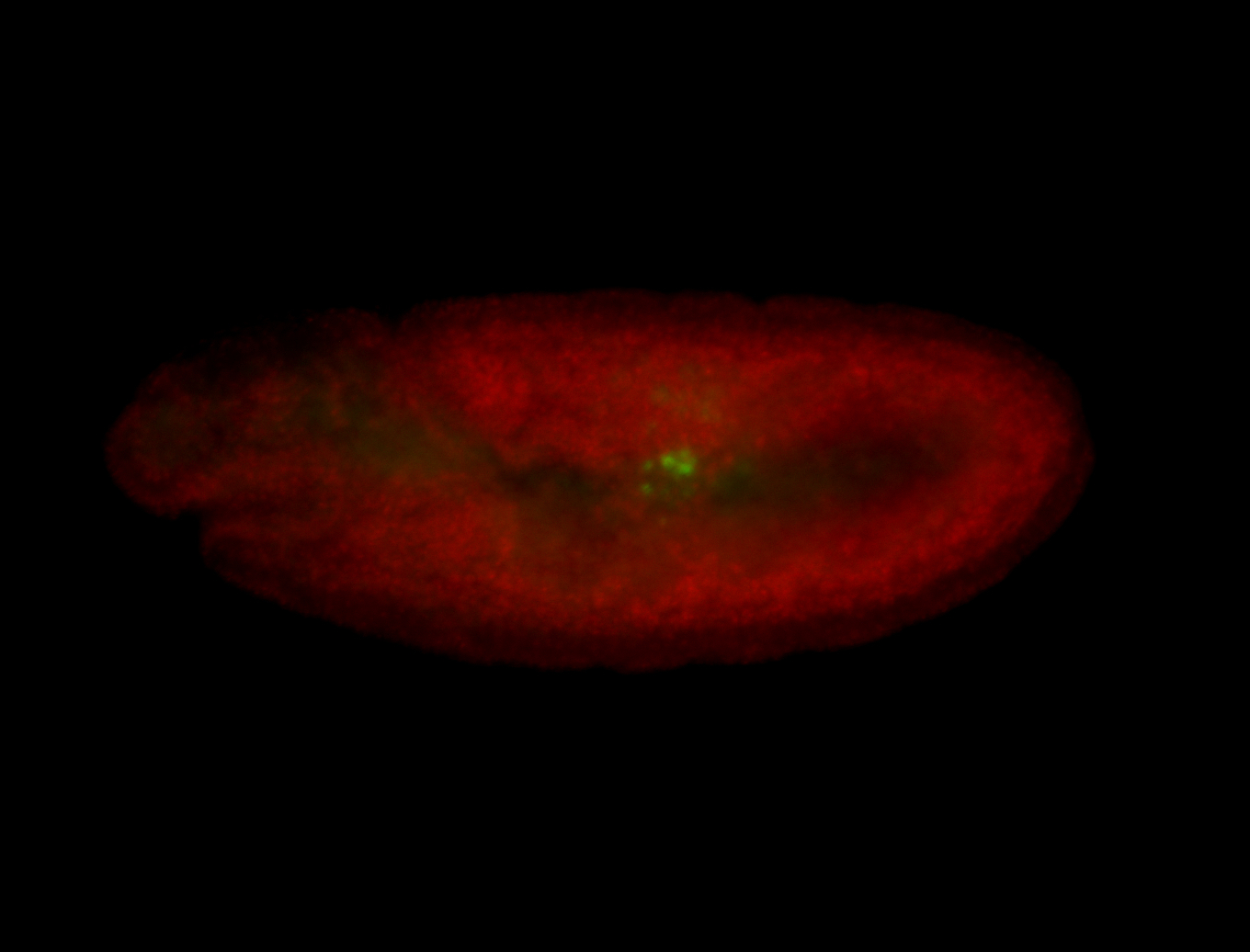
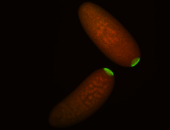
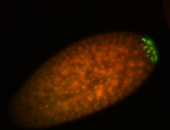
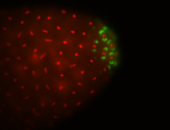
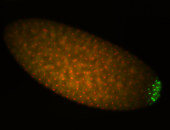
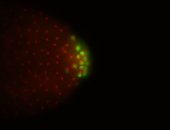
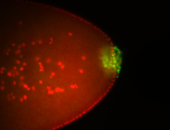
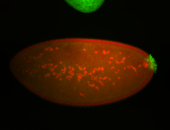
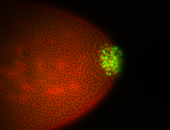
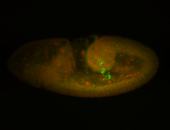
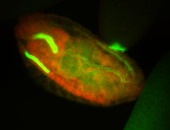
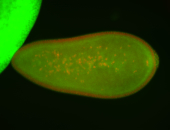
| Cytoplasmic foci around the pole cells. |
| Good probe on gel (a bit low) |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
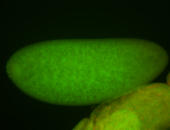
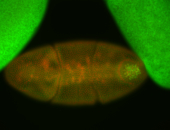
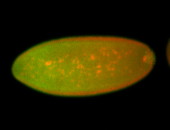
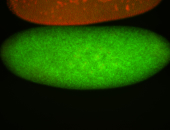
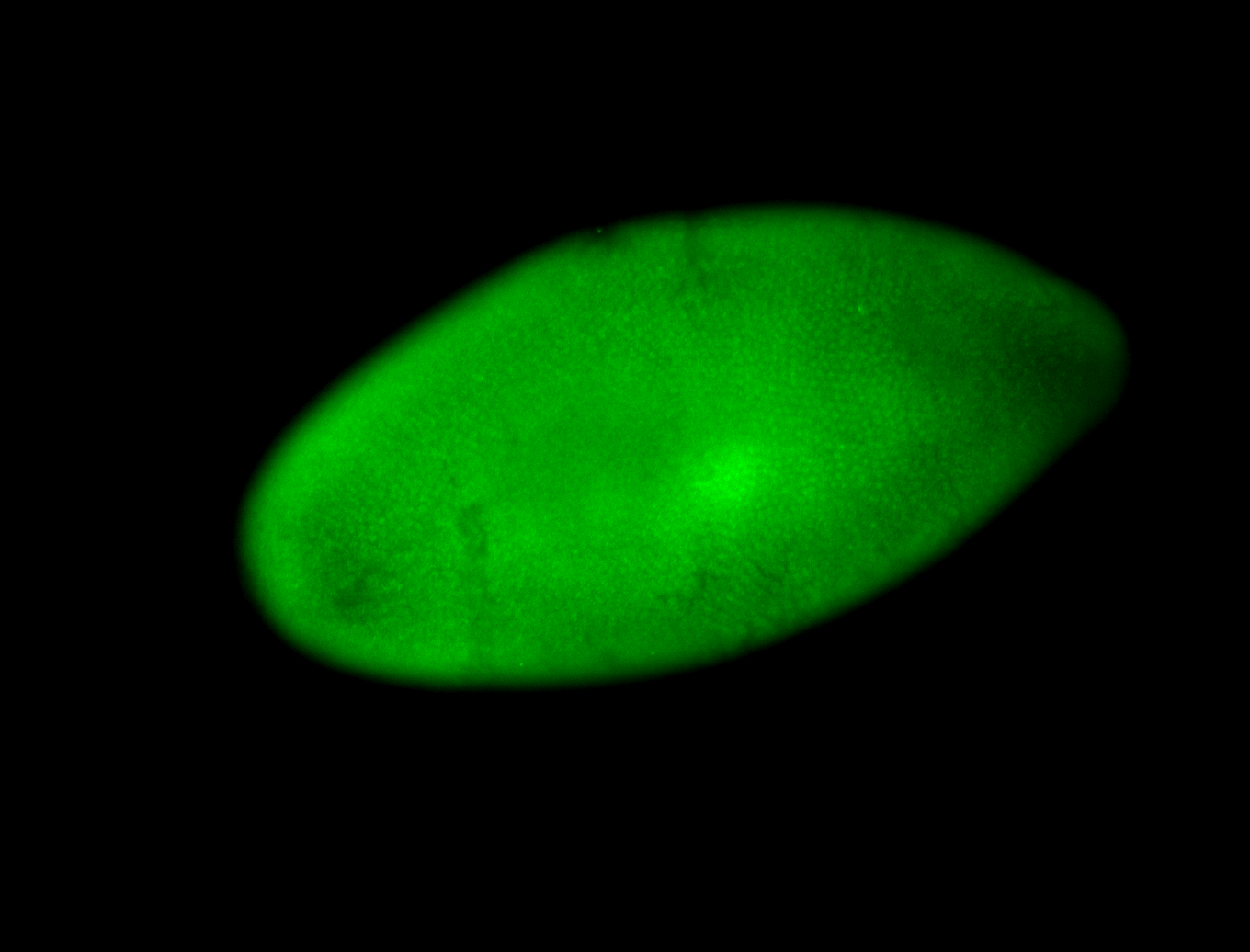
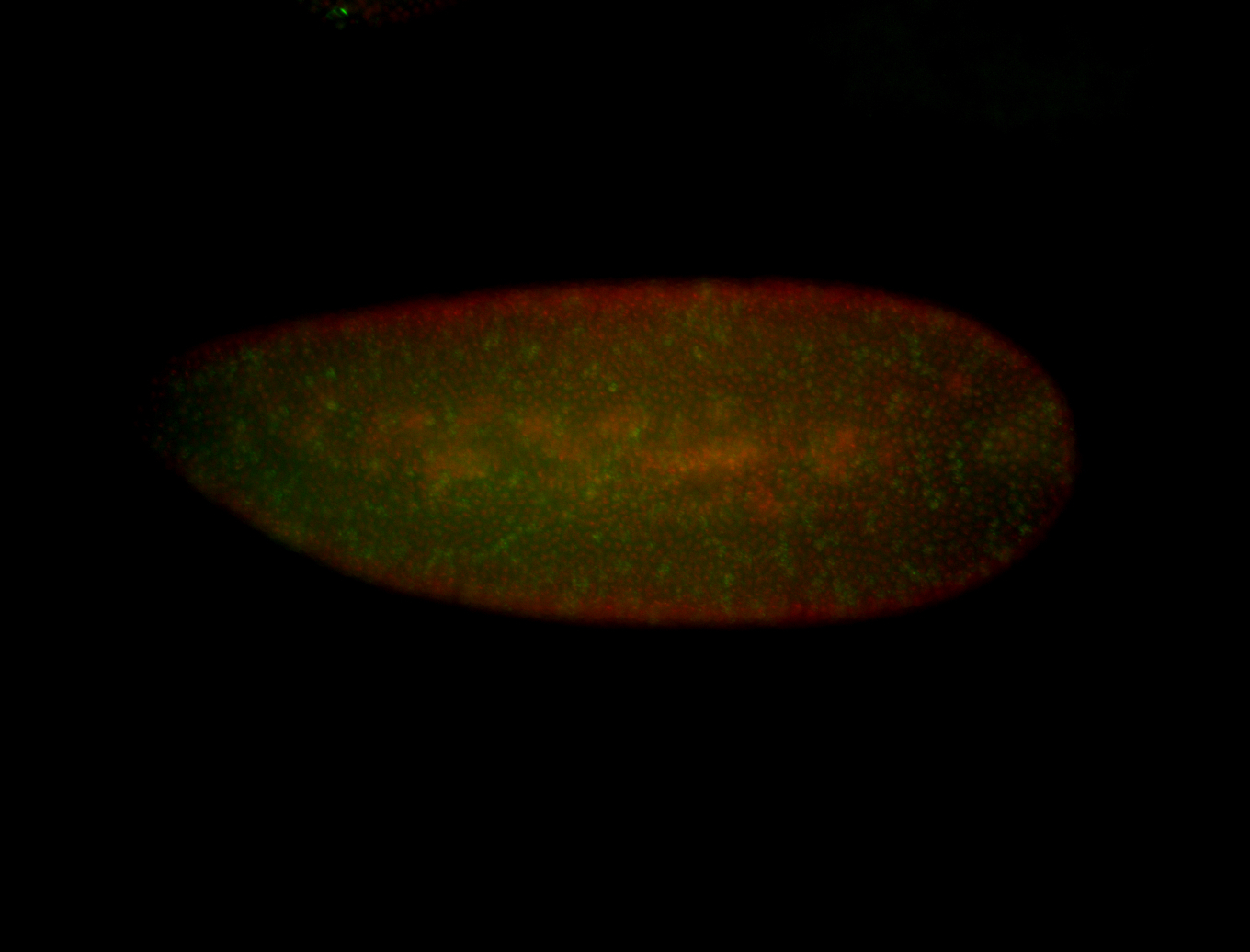
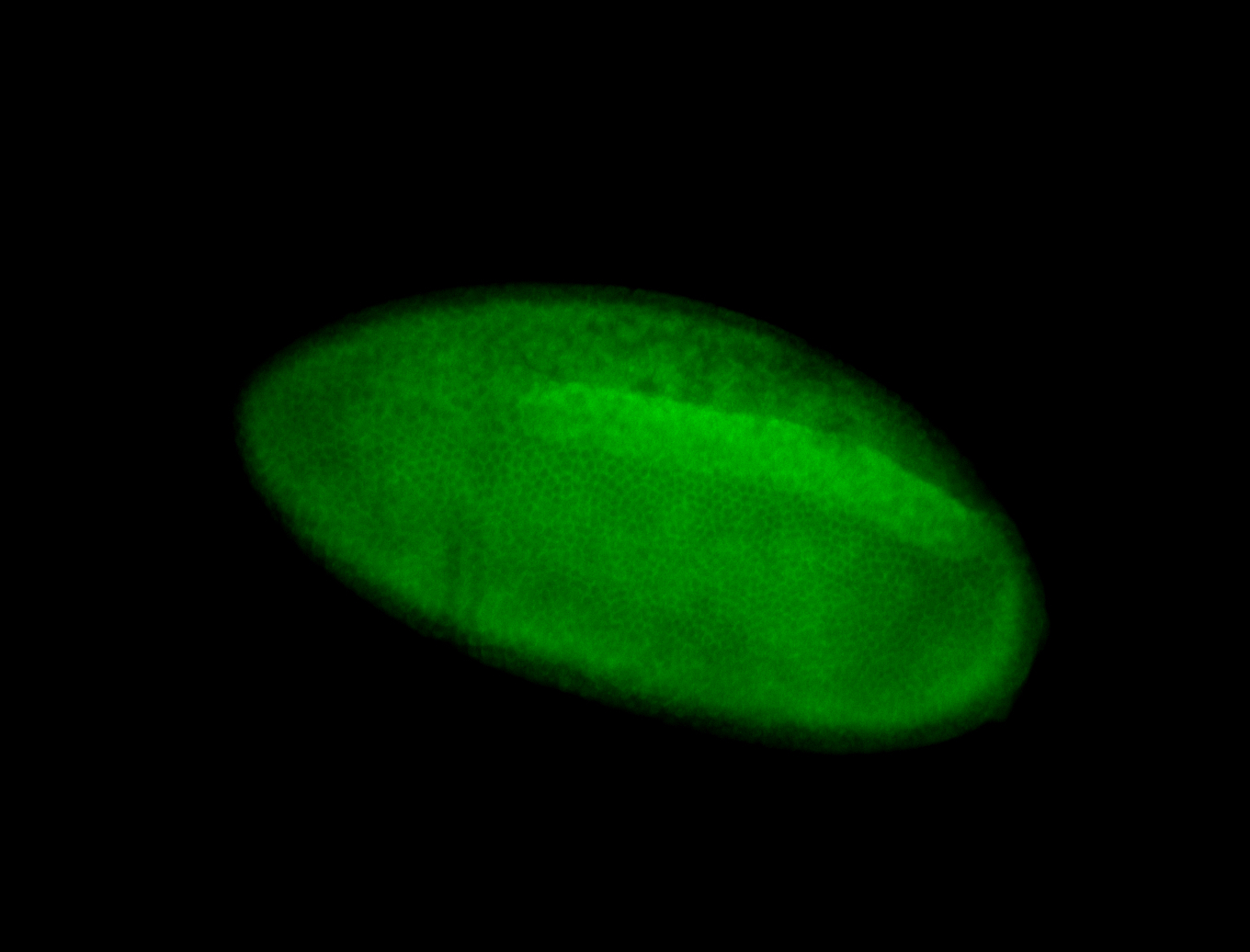
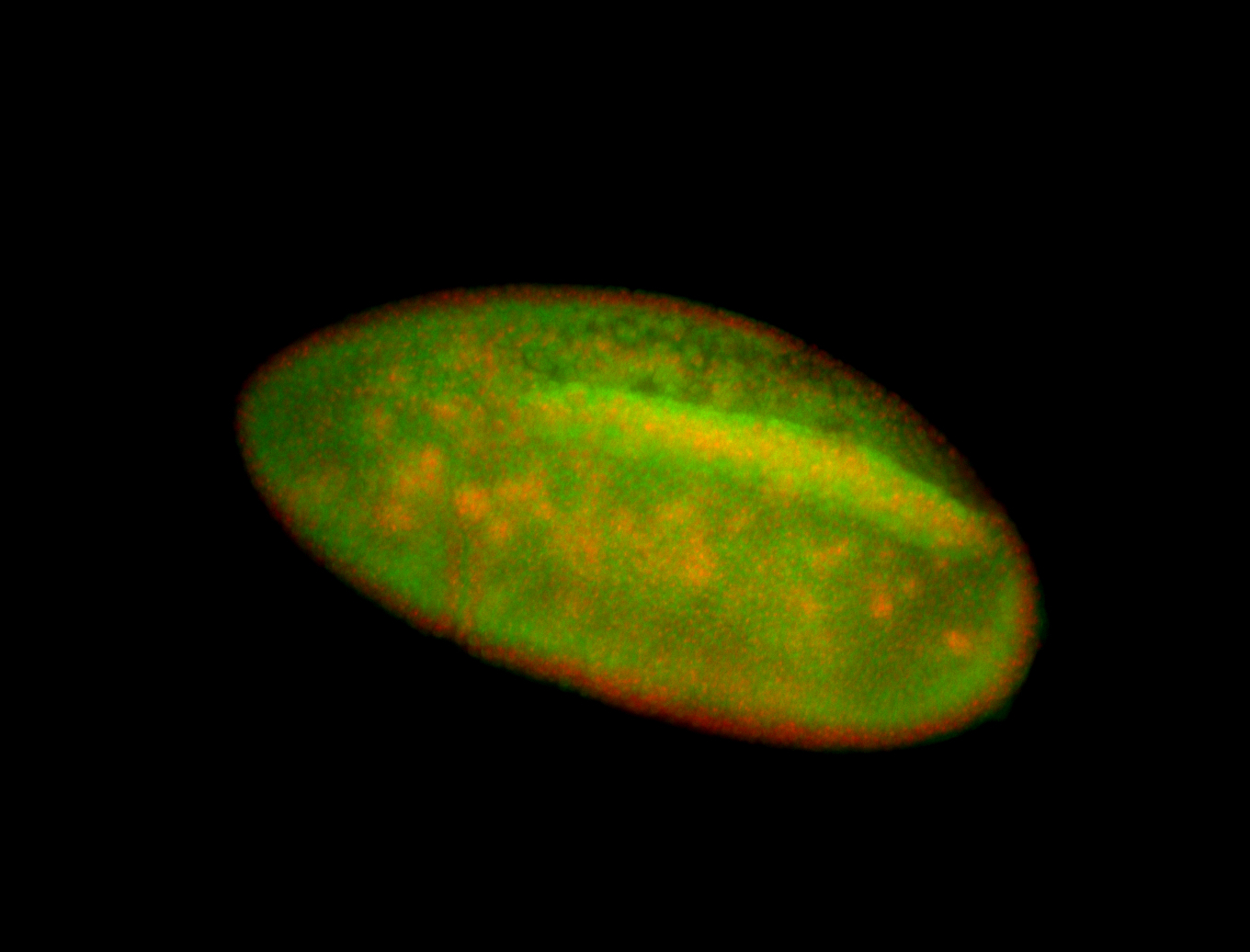
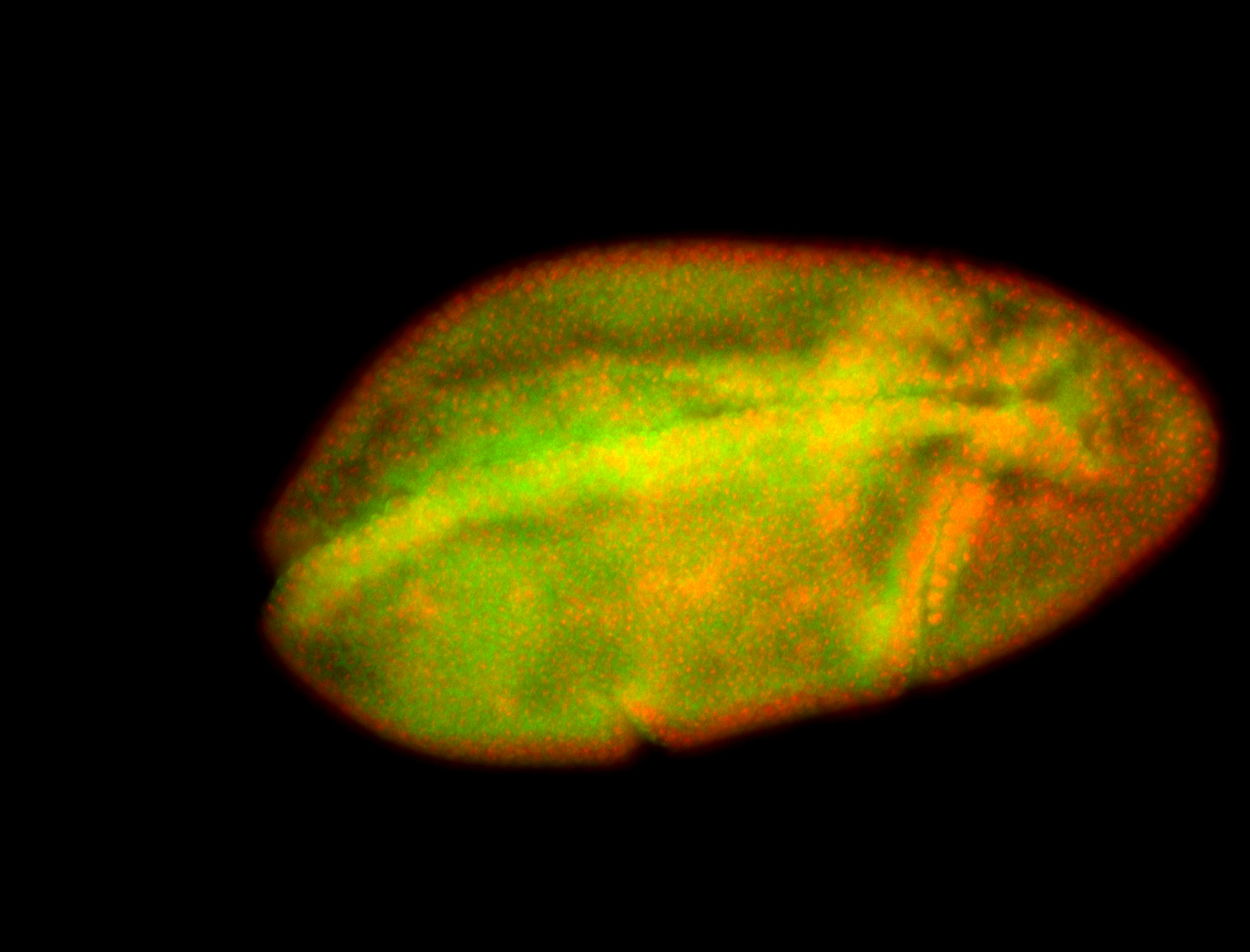
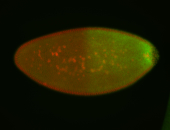
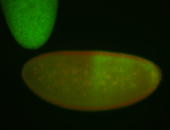
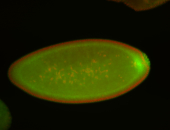
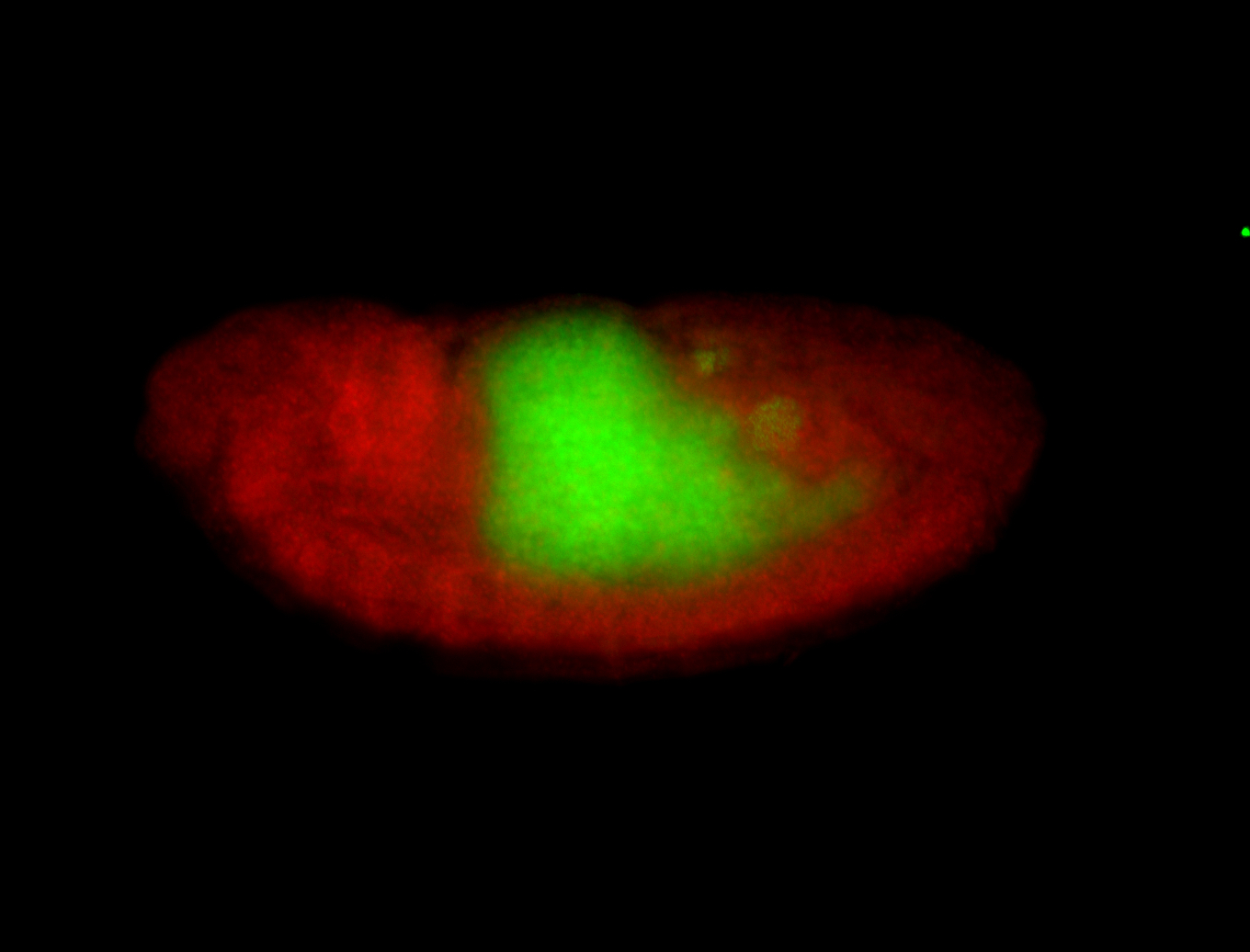
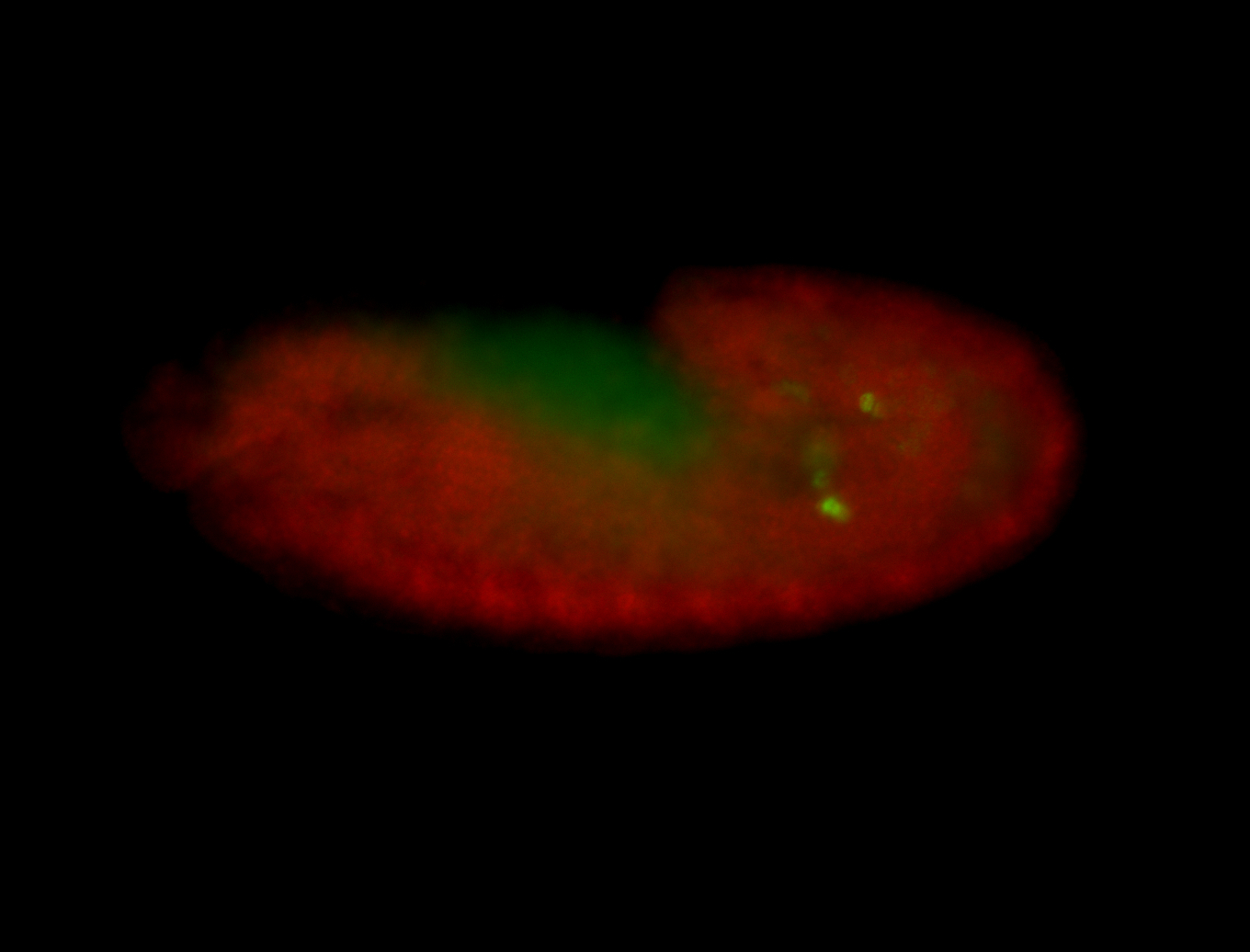
| Posterior enriched, but not posterior localized. |
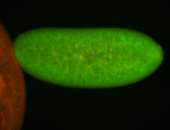
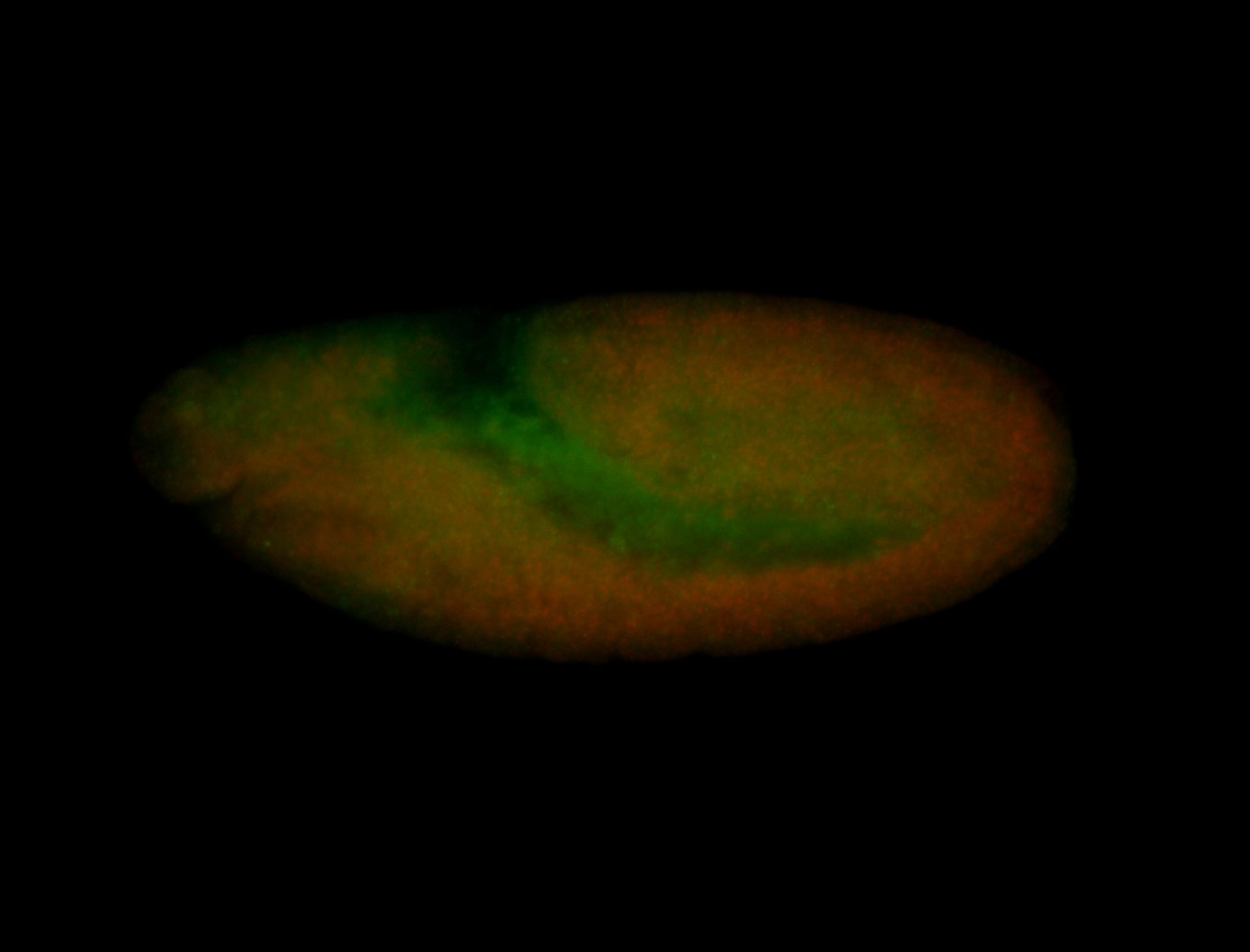
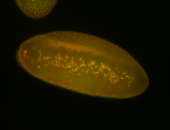
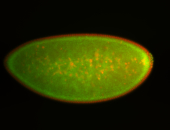
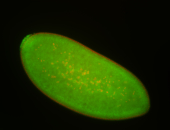
| Anterior and posterior midgut enrichment. |
| Anterior and posterior midgut enrichment. |
| Good probe on gel. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe on gel |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| This data comes from probe 'RE02642' on plate AM01-F8 (RA.300-68). Same CG number. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
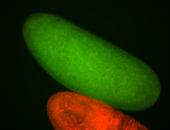
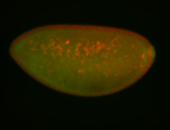
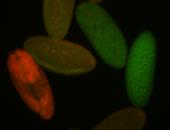
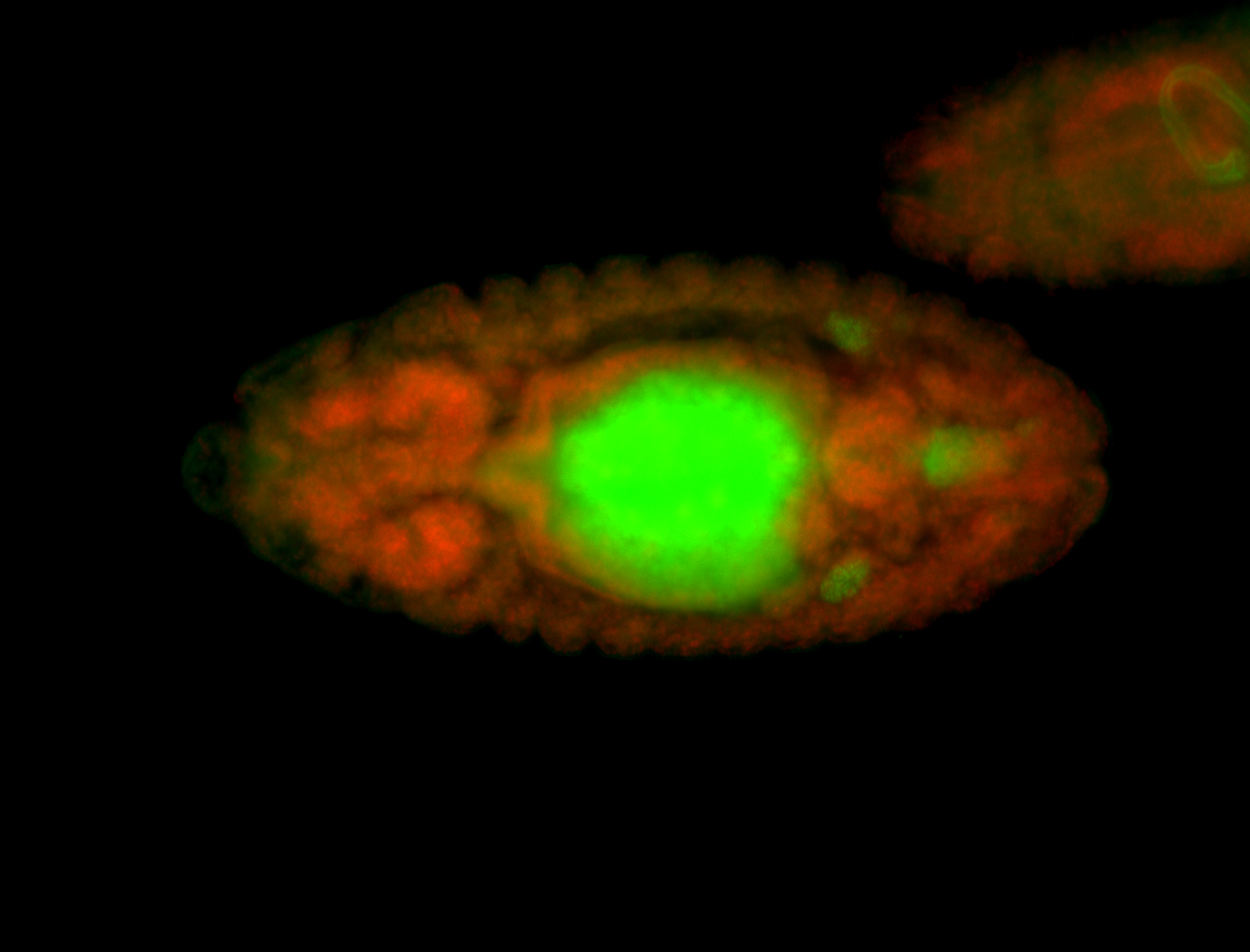
| Perinuclear |
| Perinulclear |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Probe CR43264-RA Links: Permalink
Genes: CR43264
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
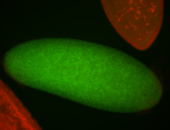
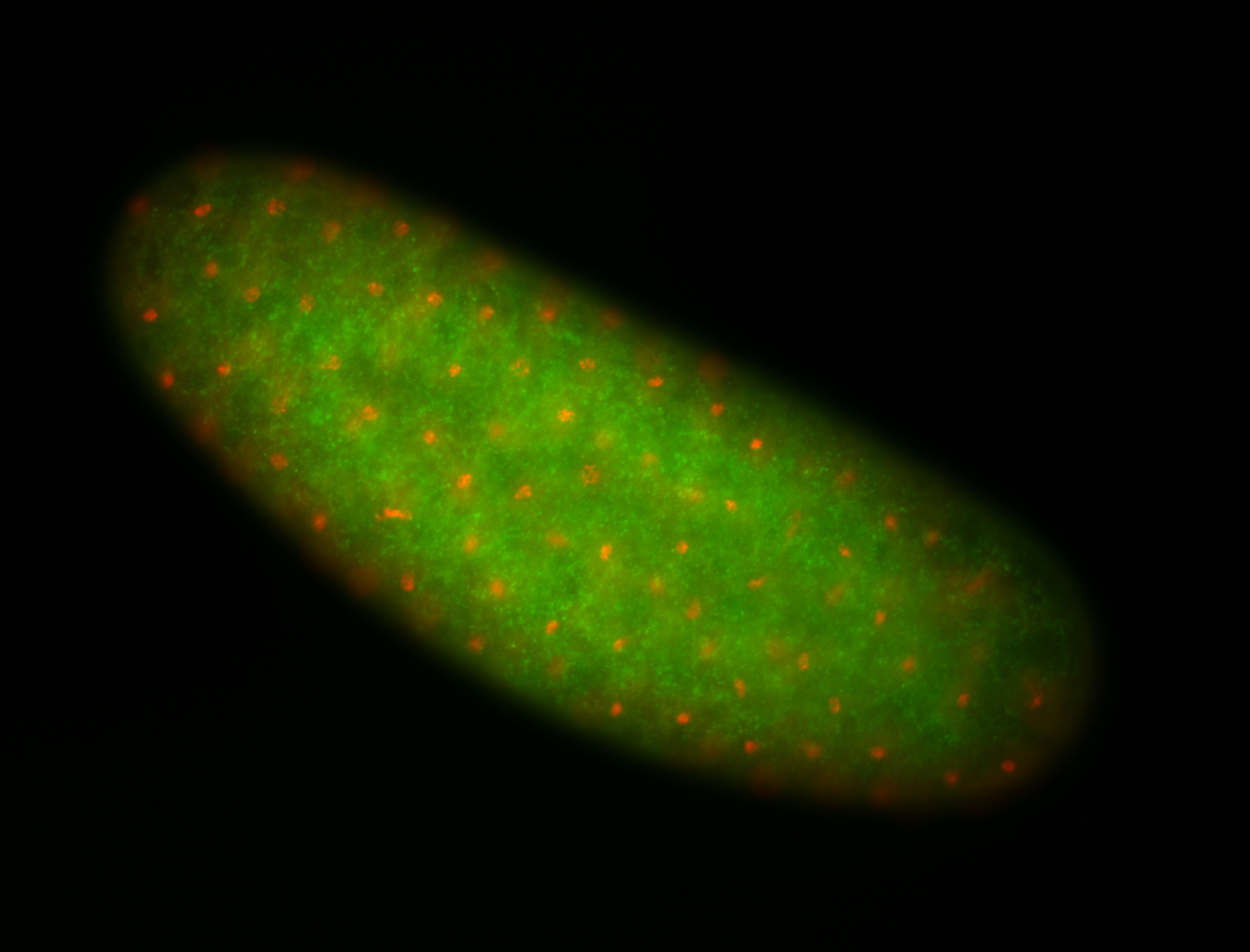
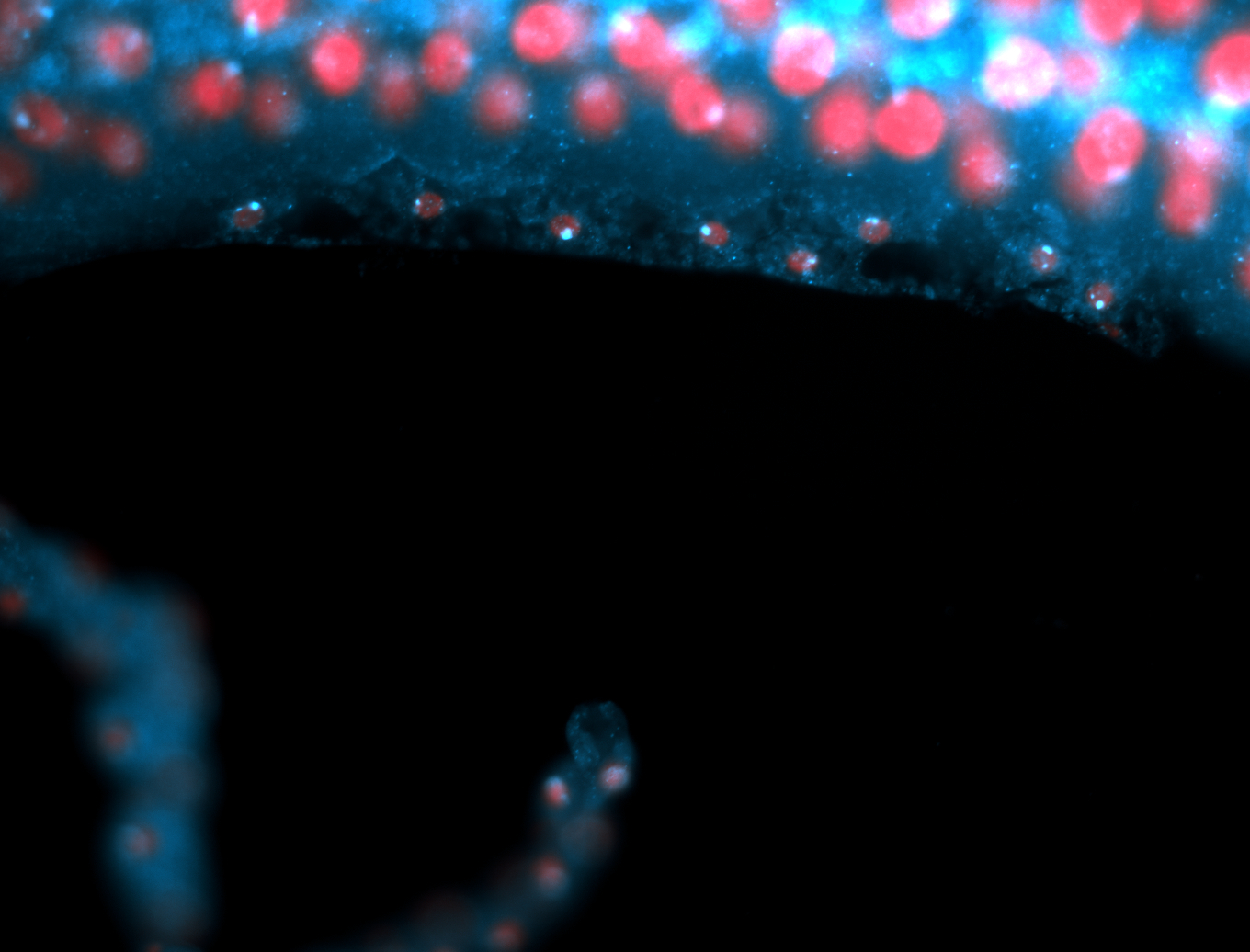
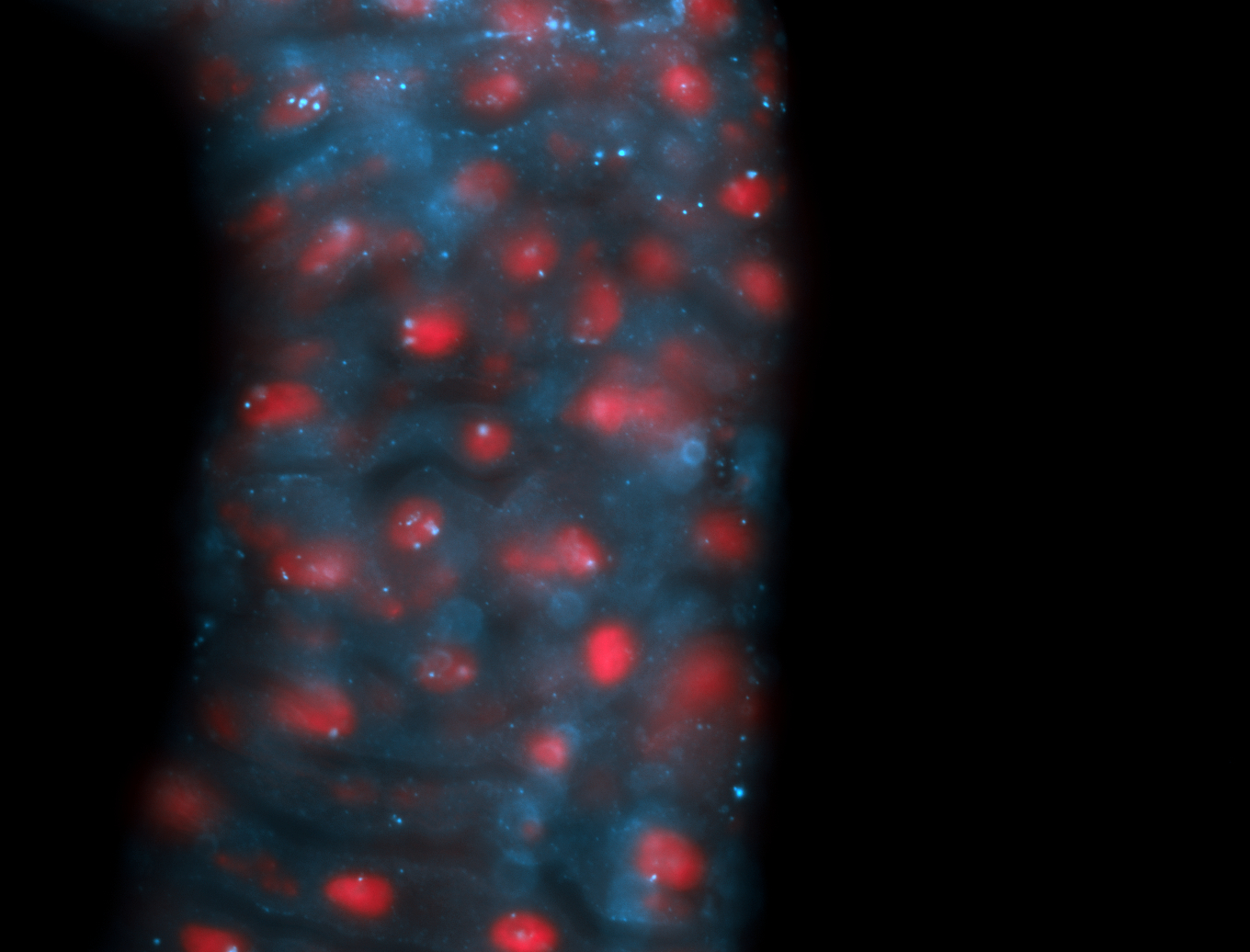
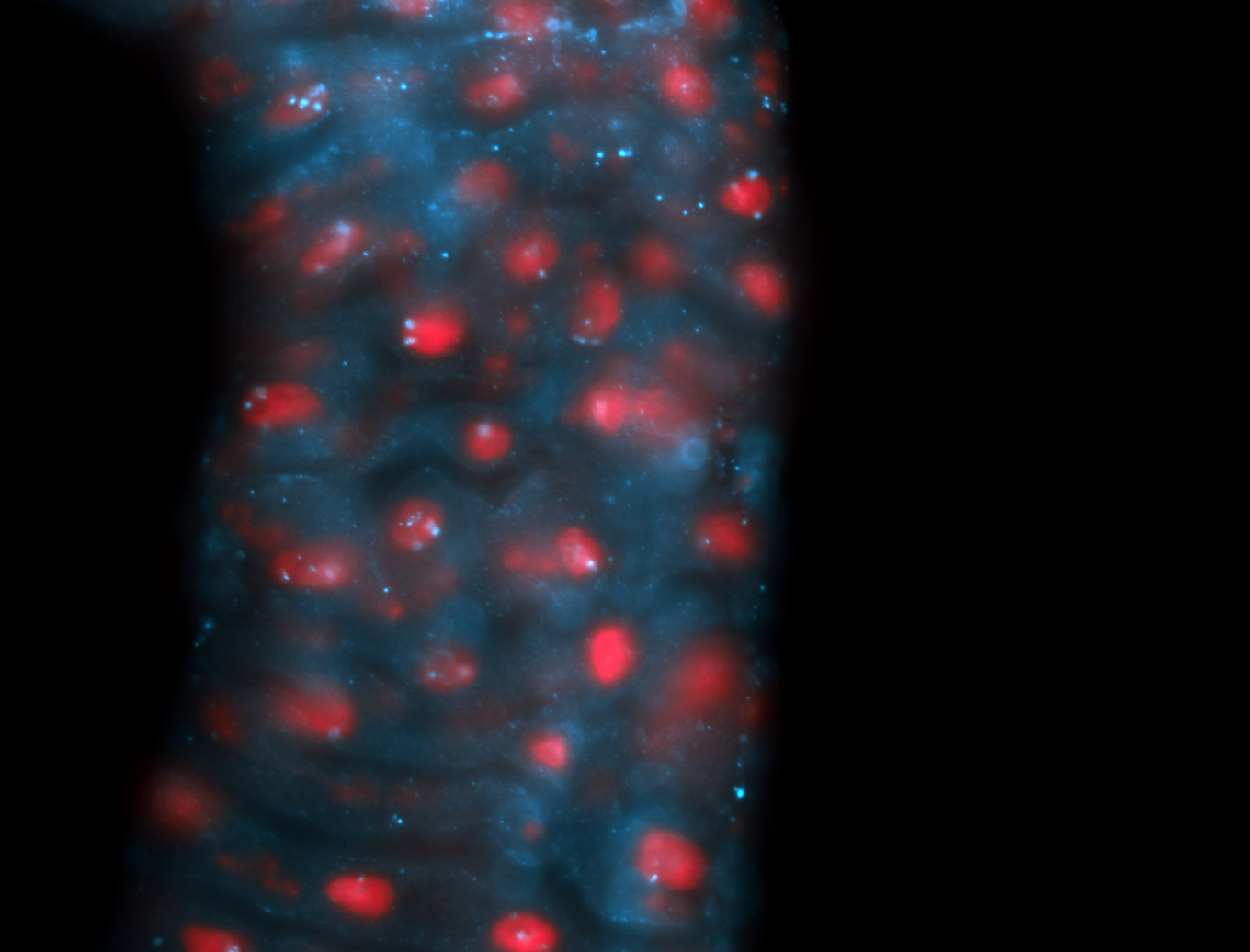
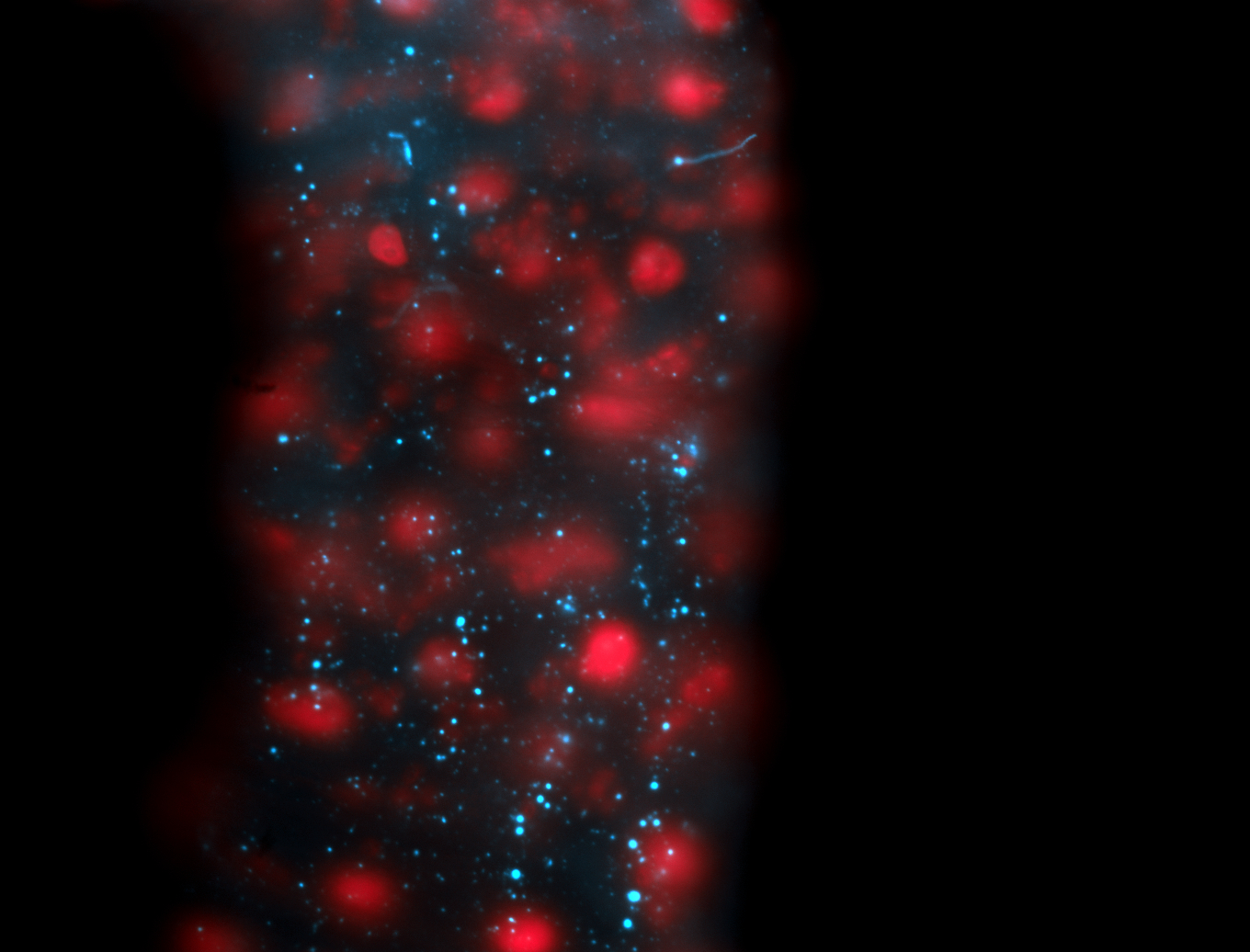
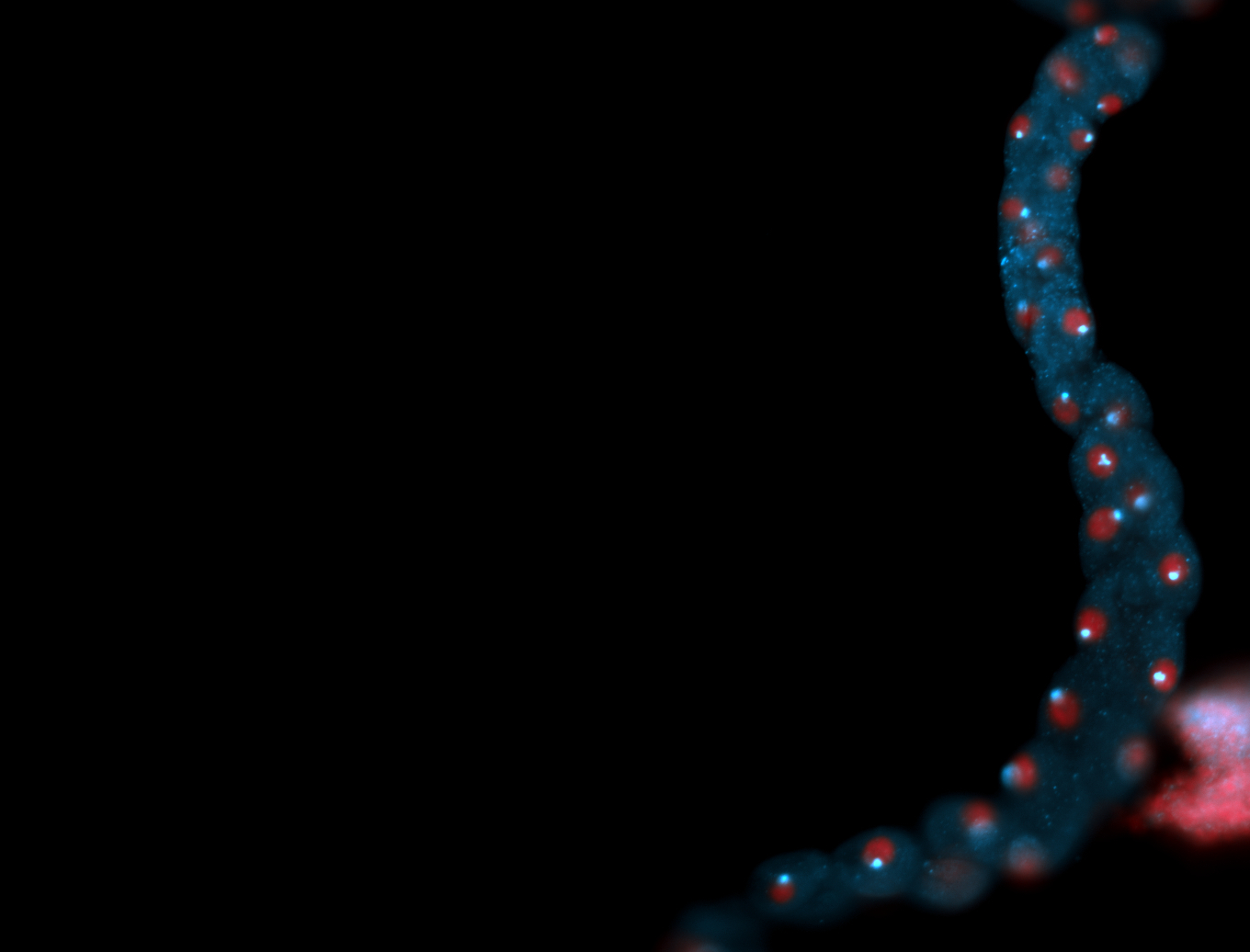
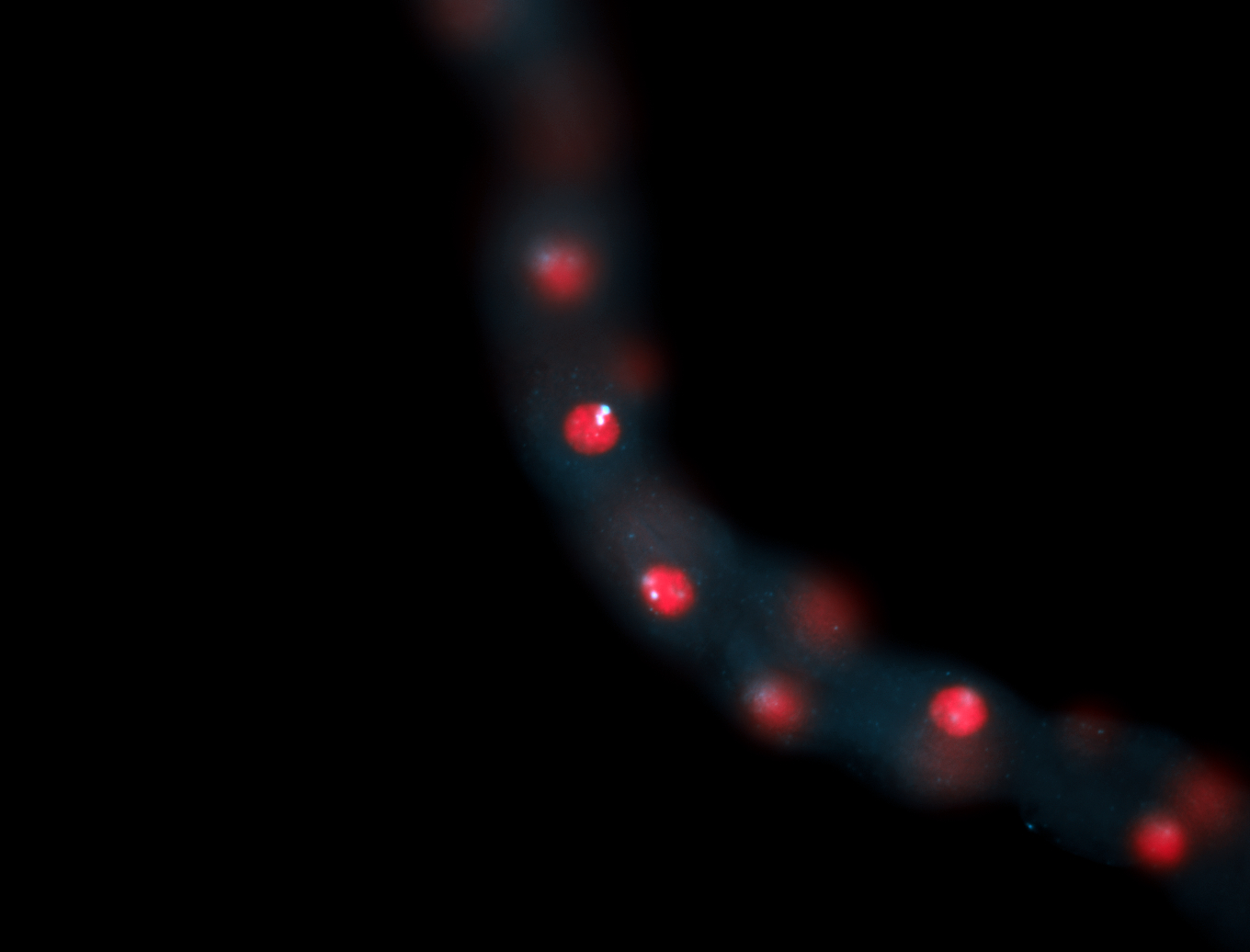
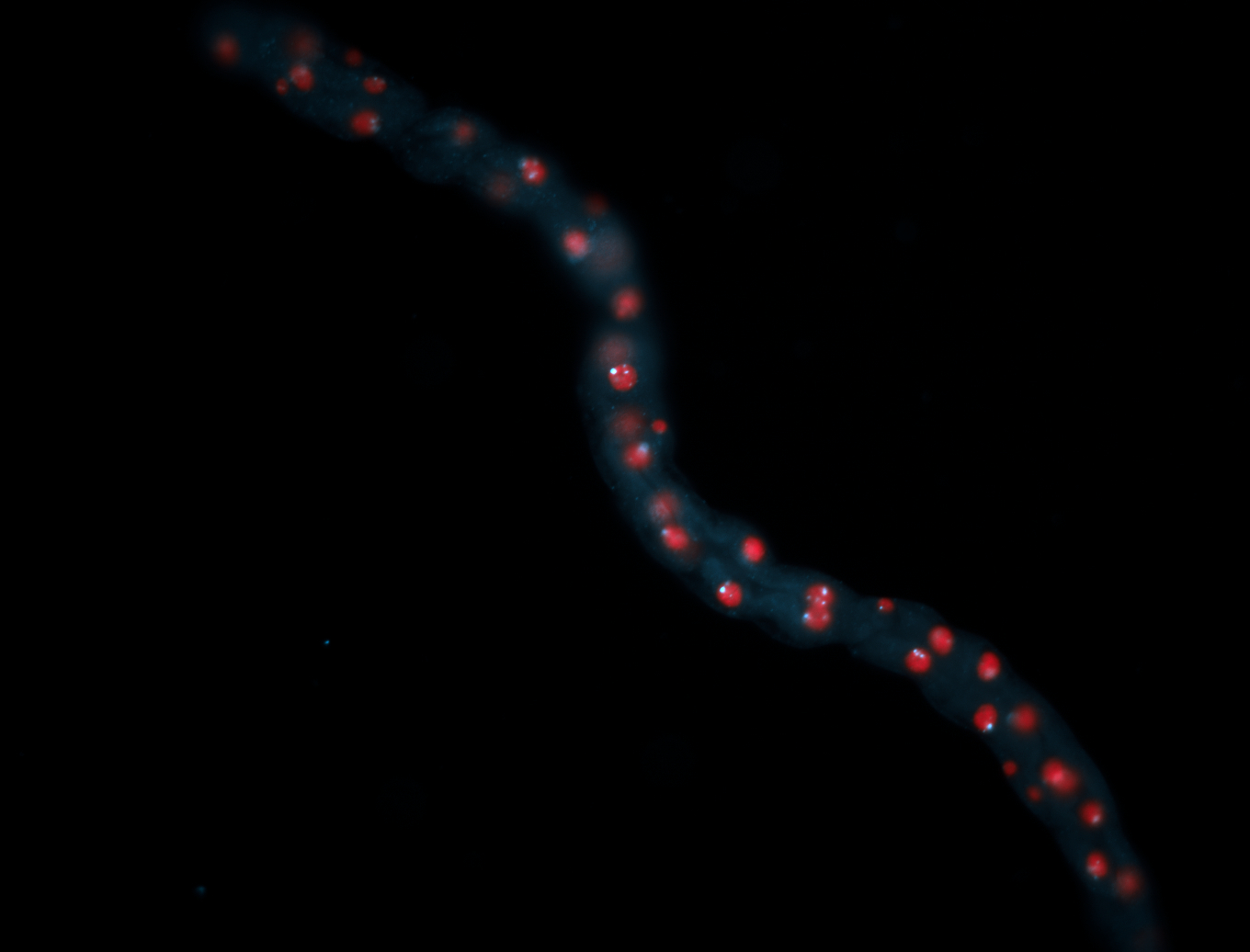
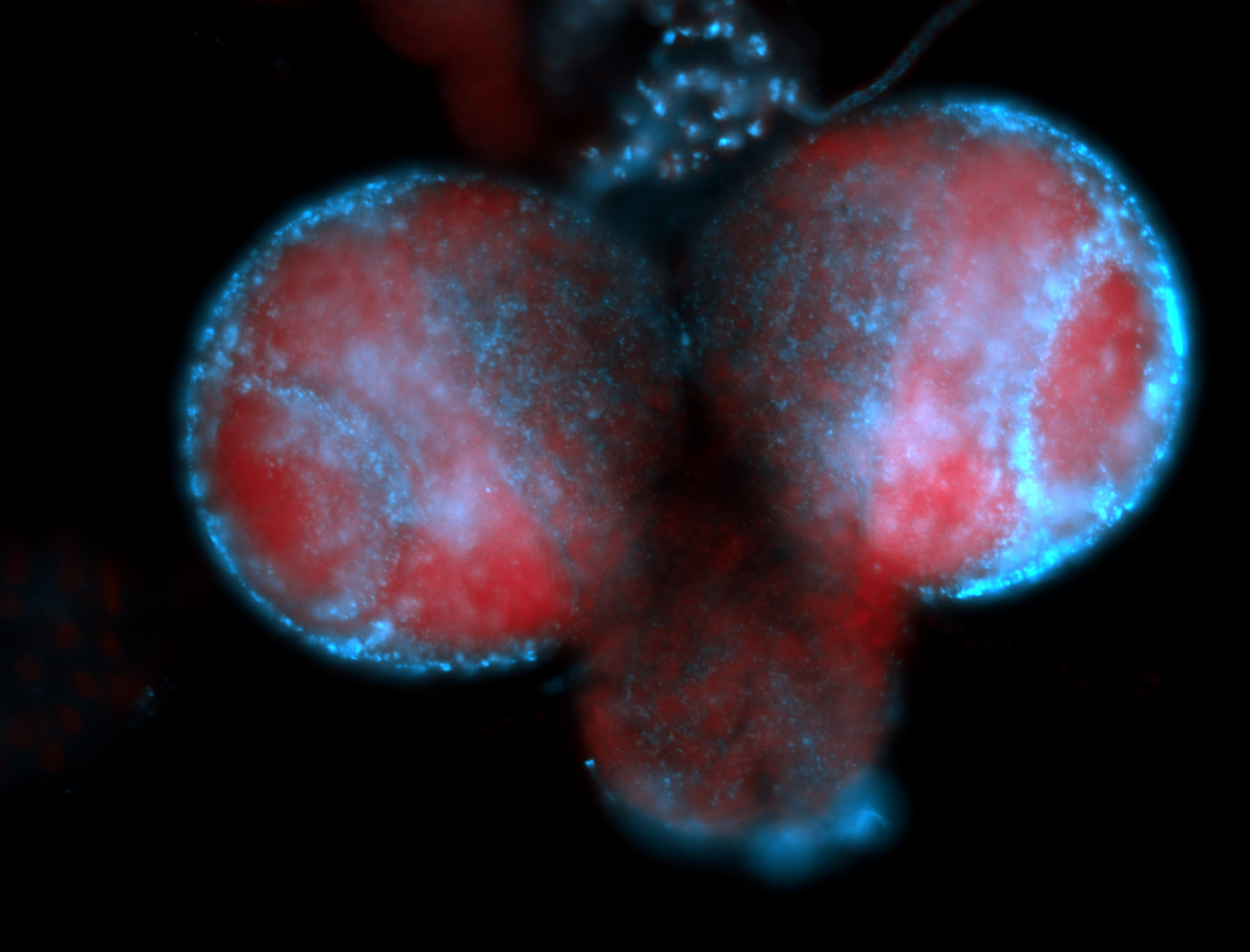
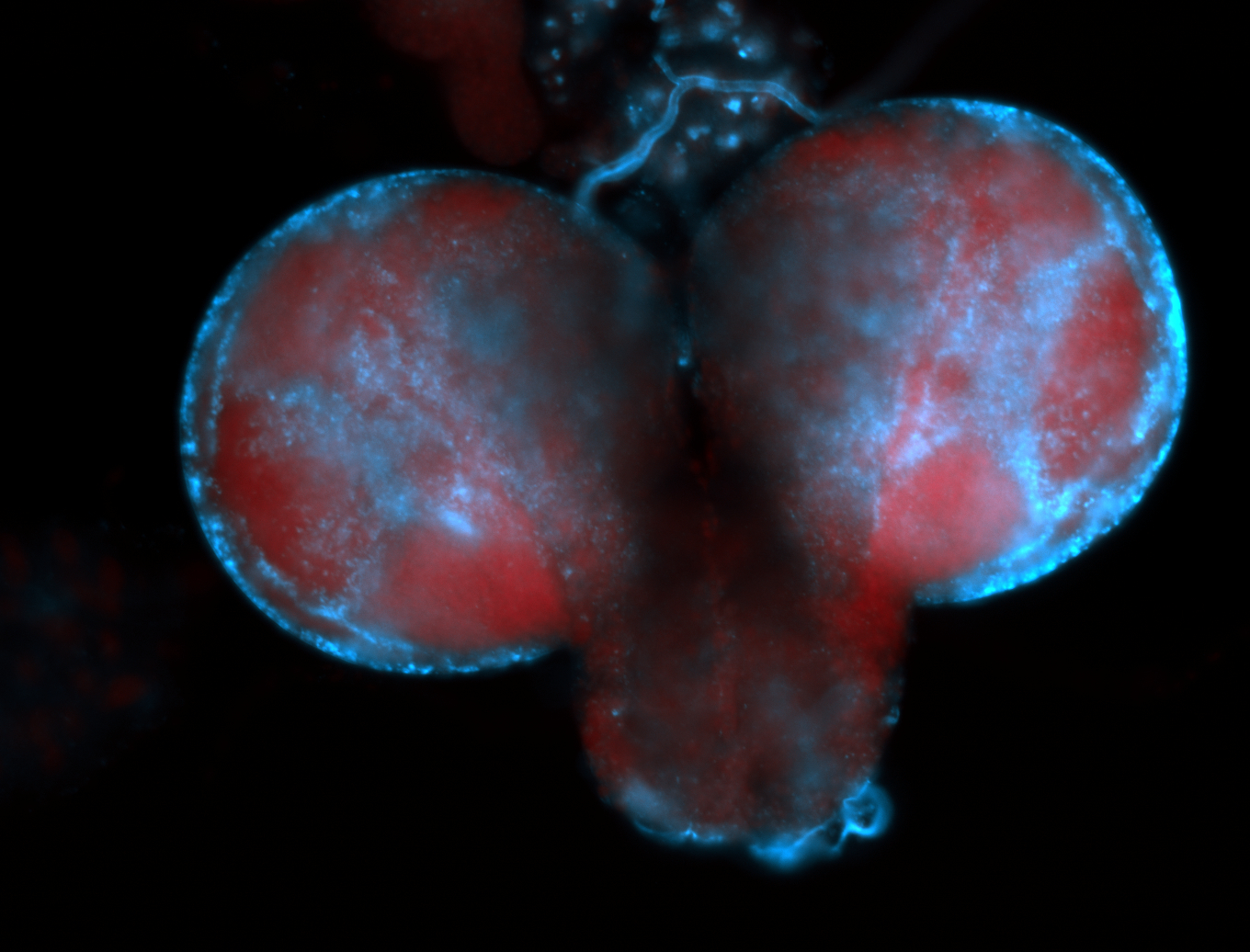
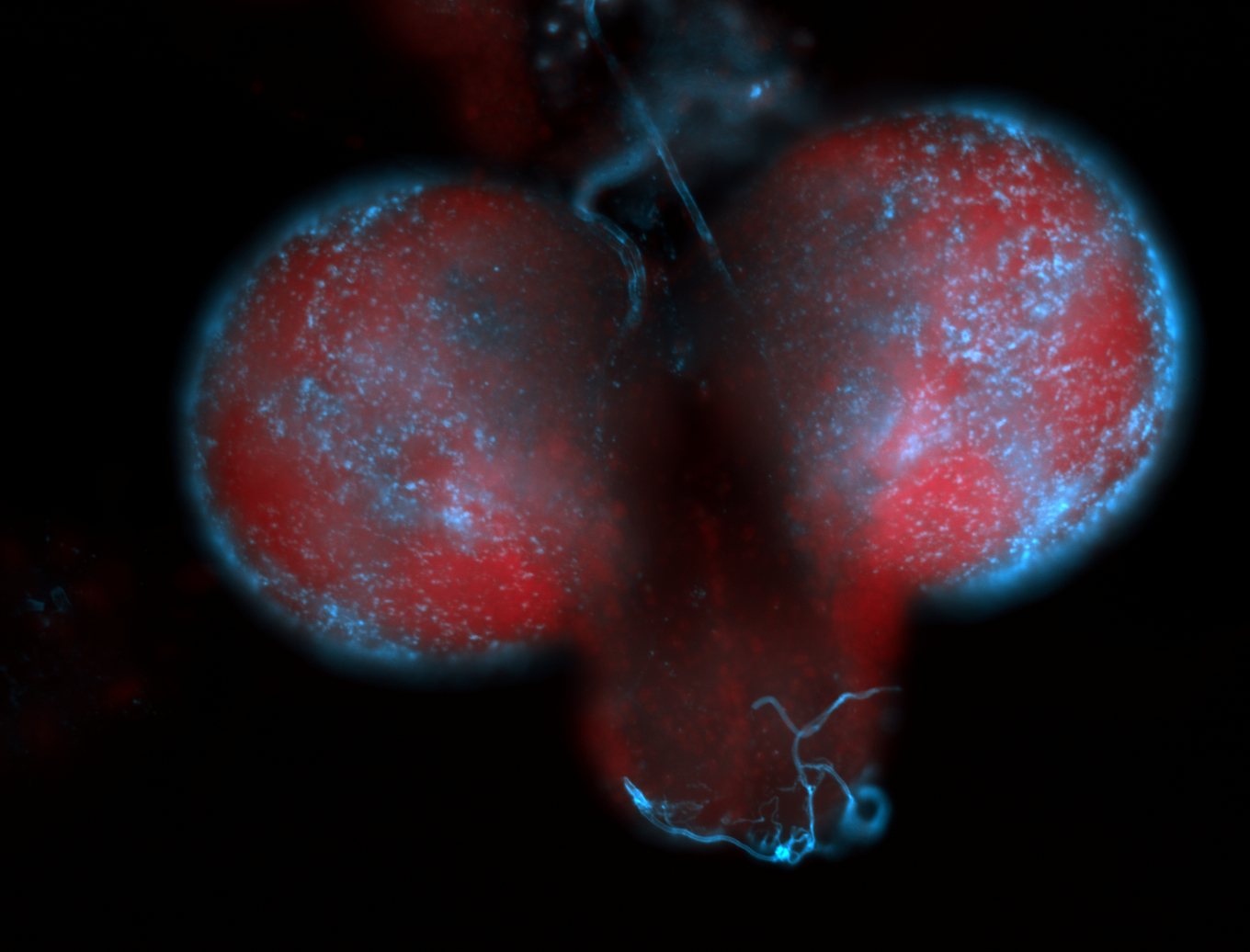
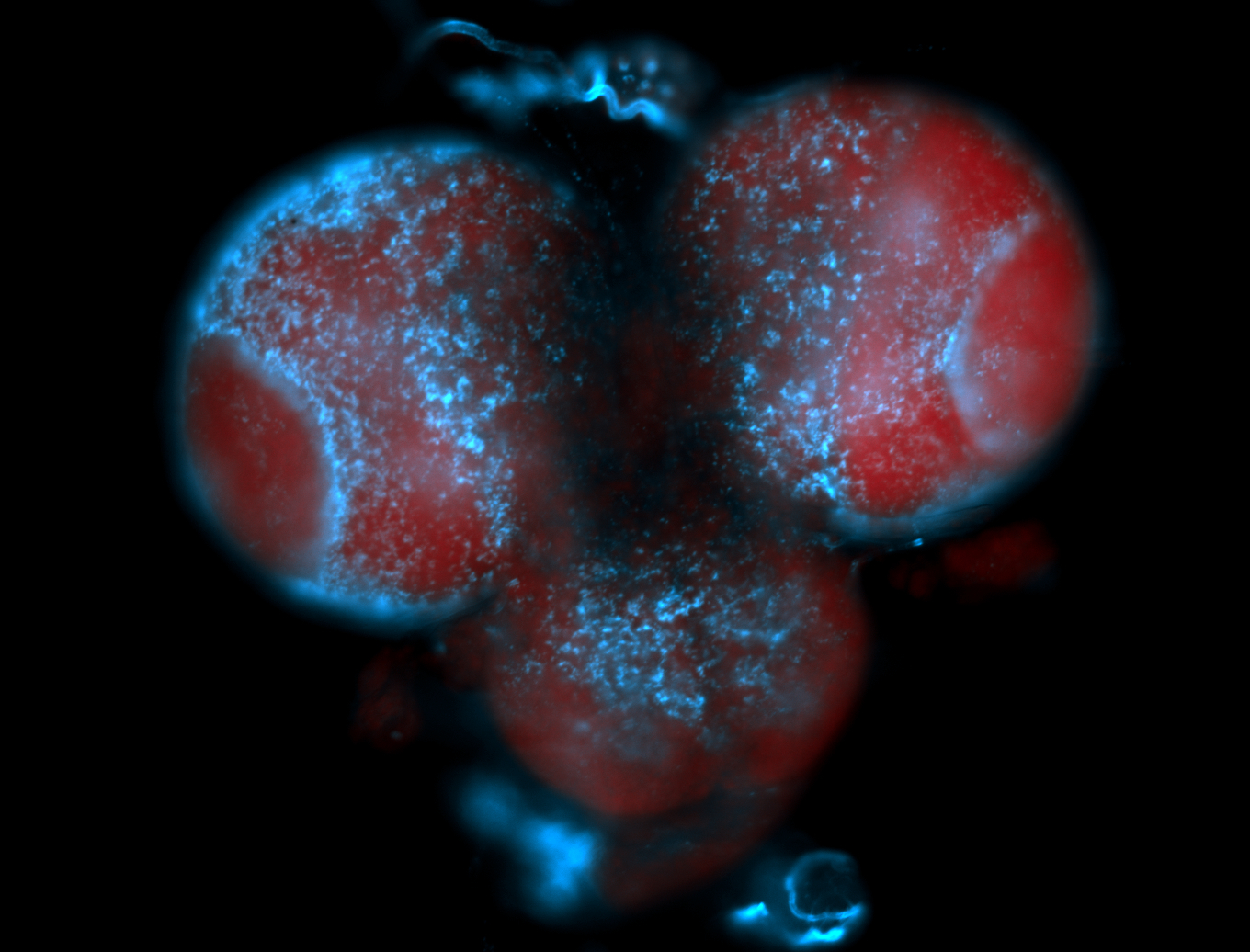
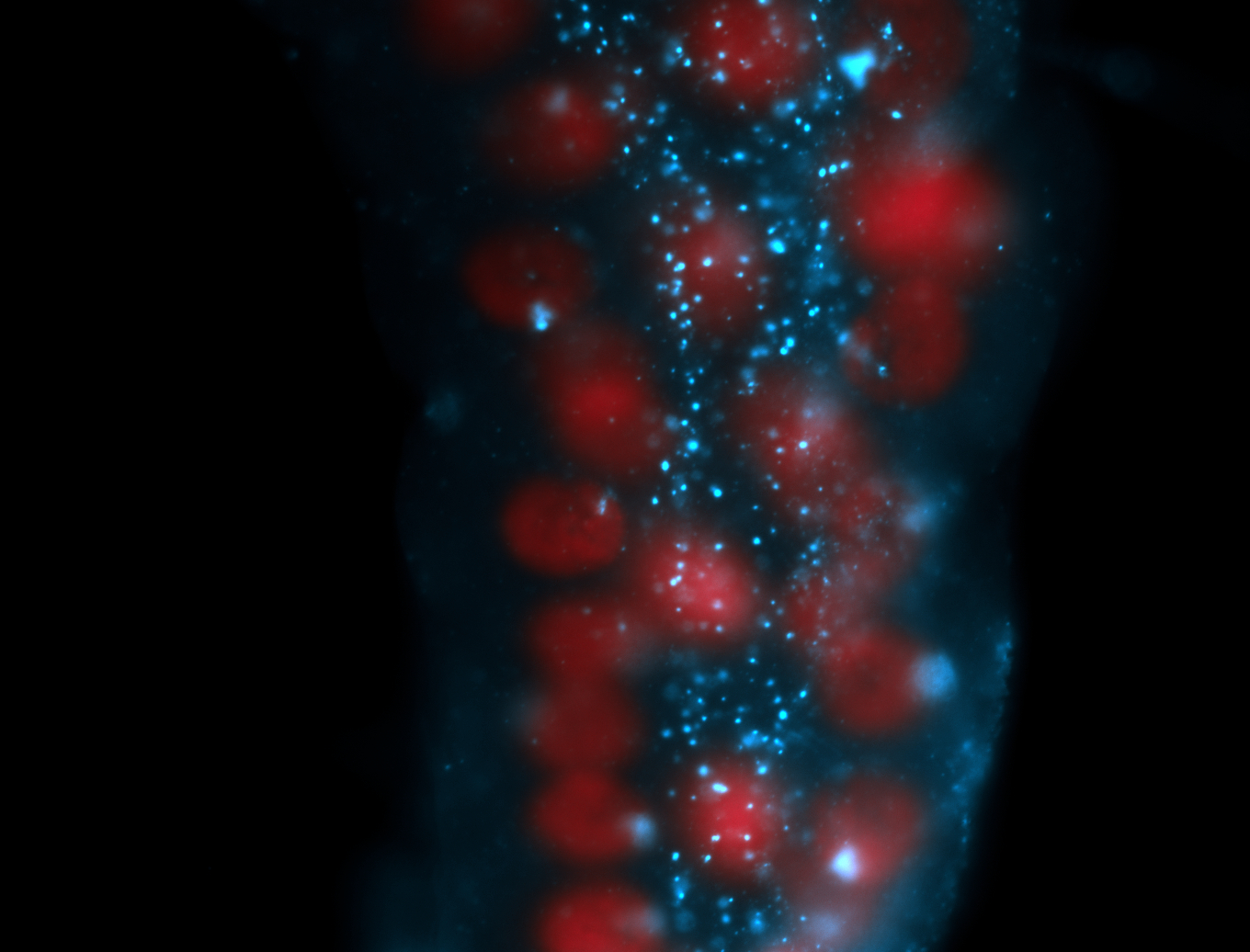
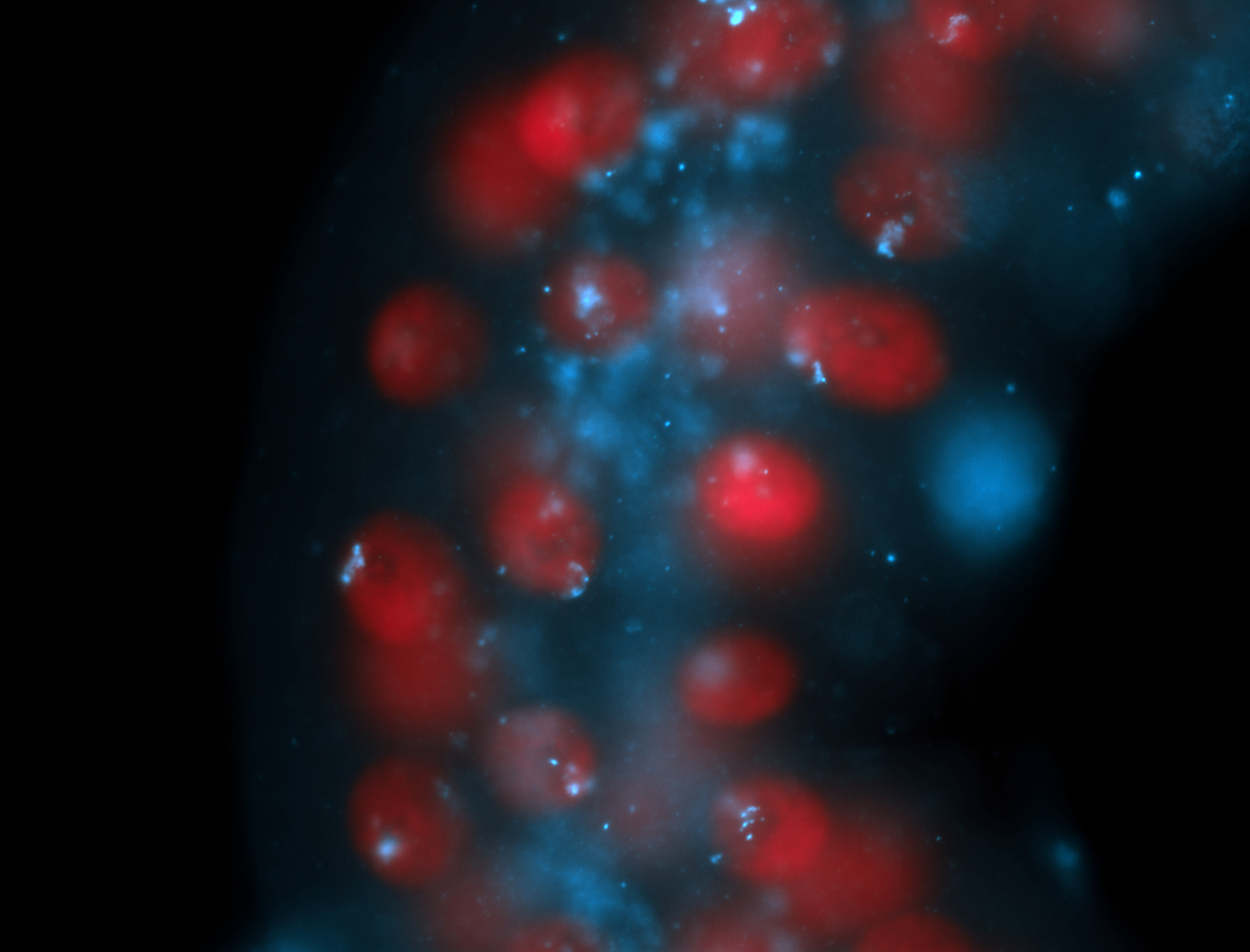
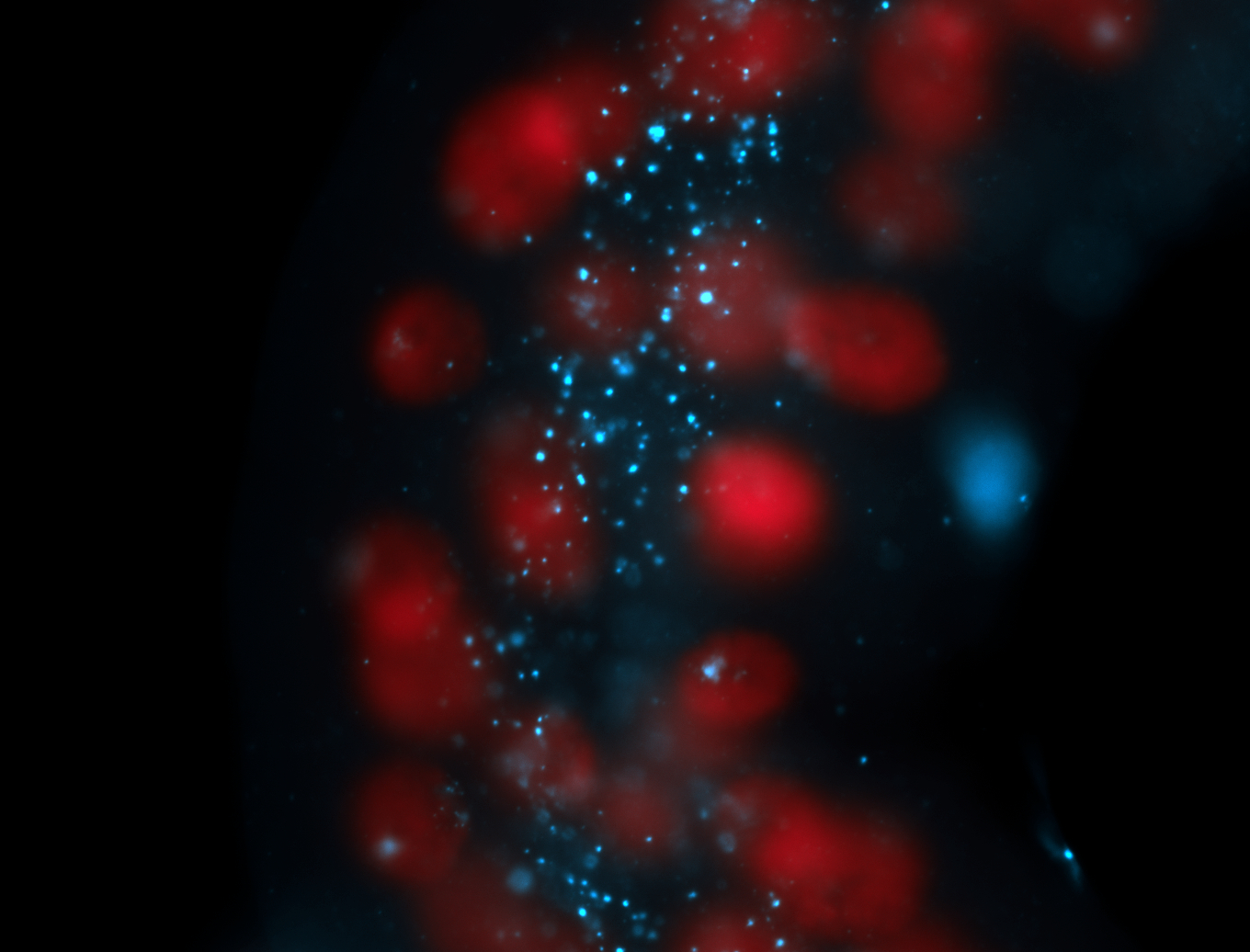
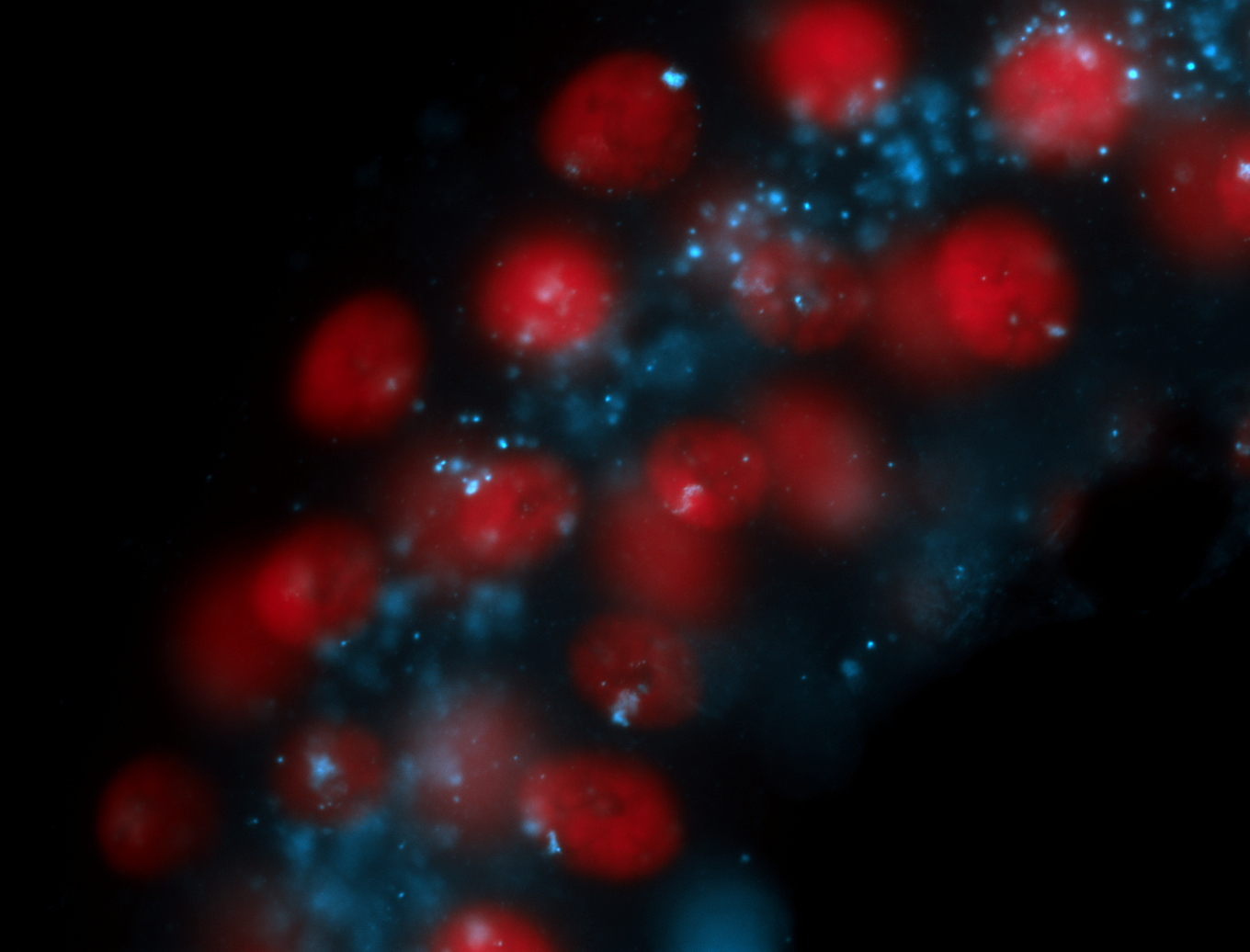
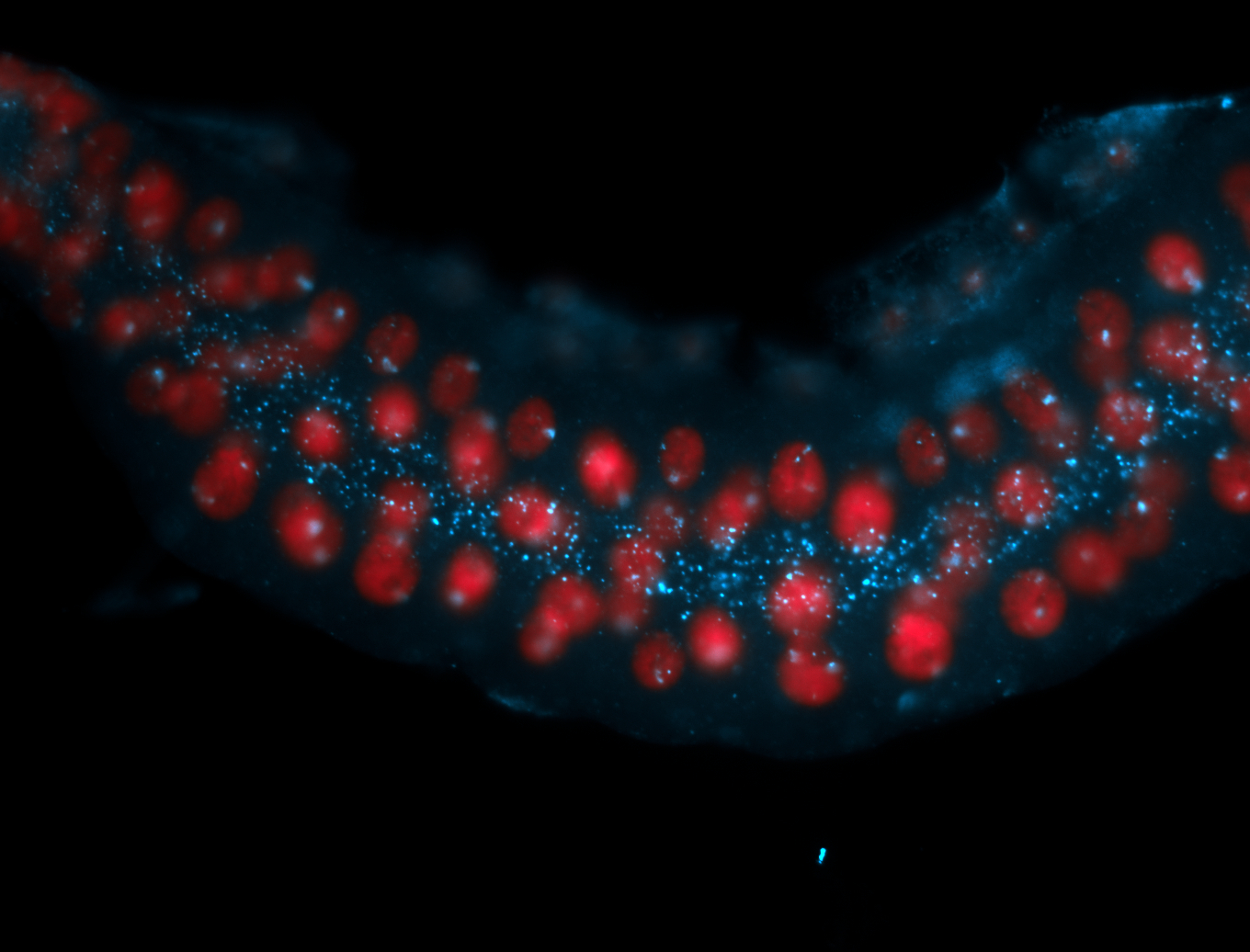
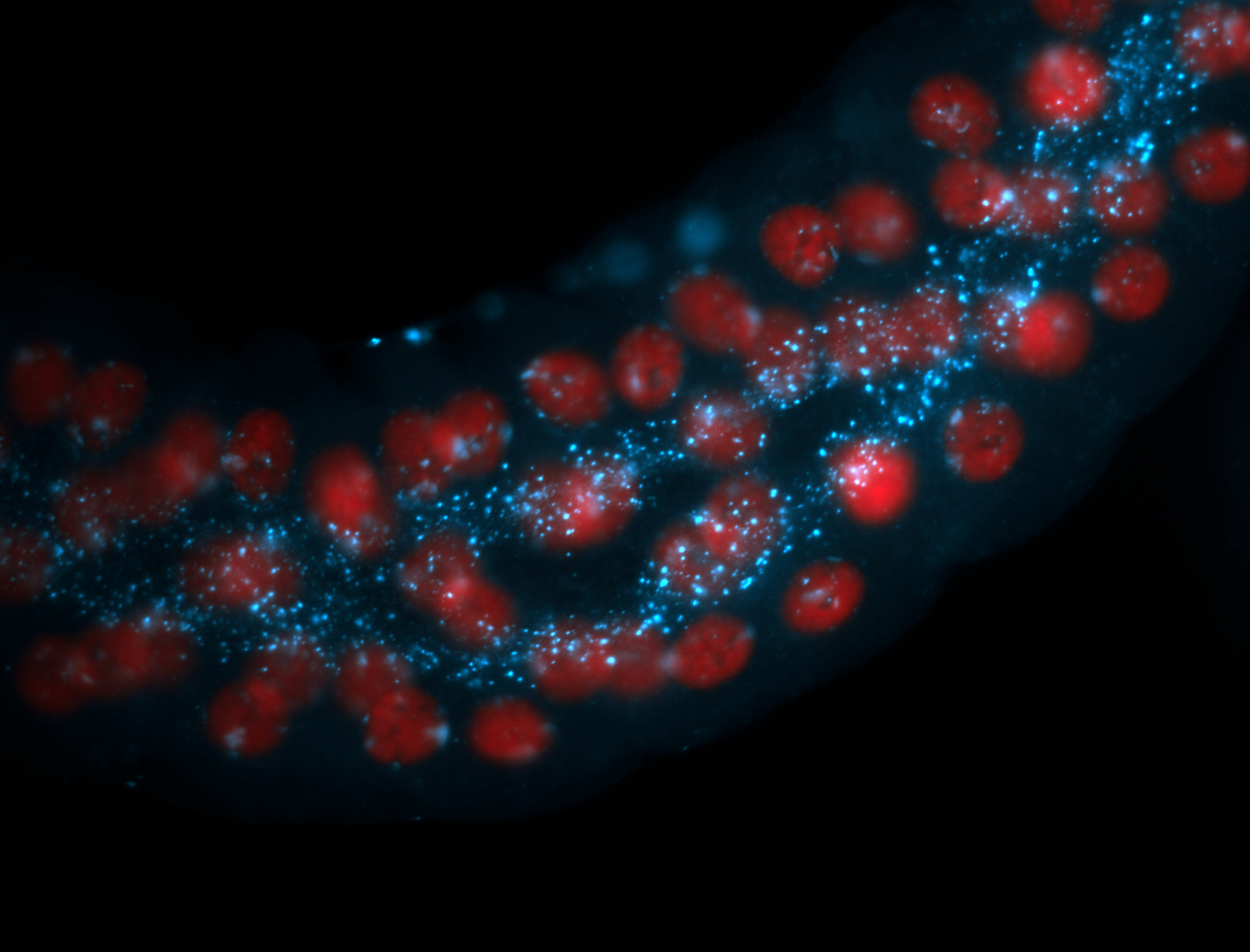
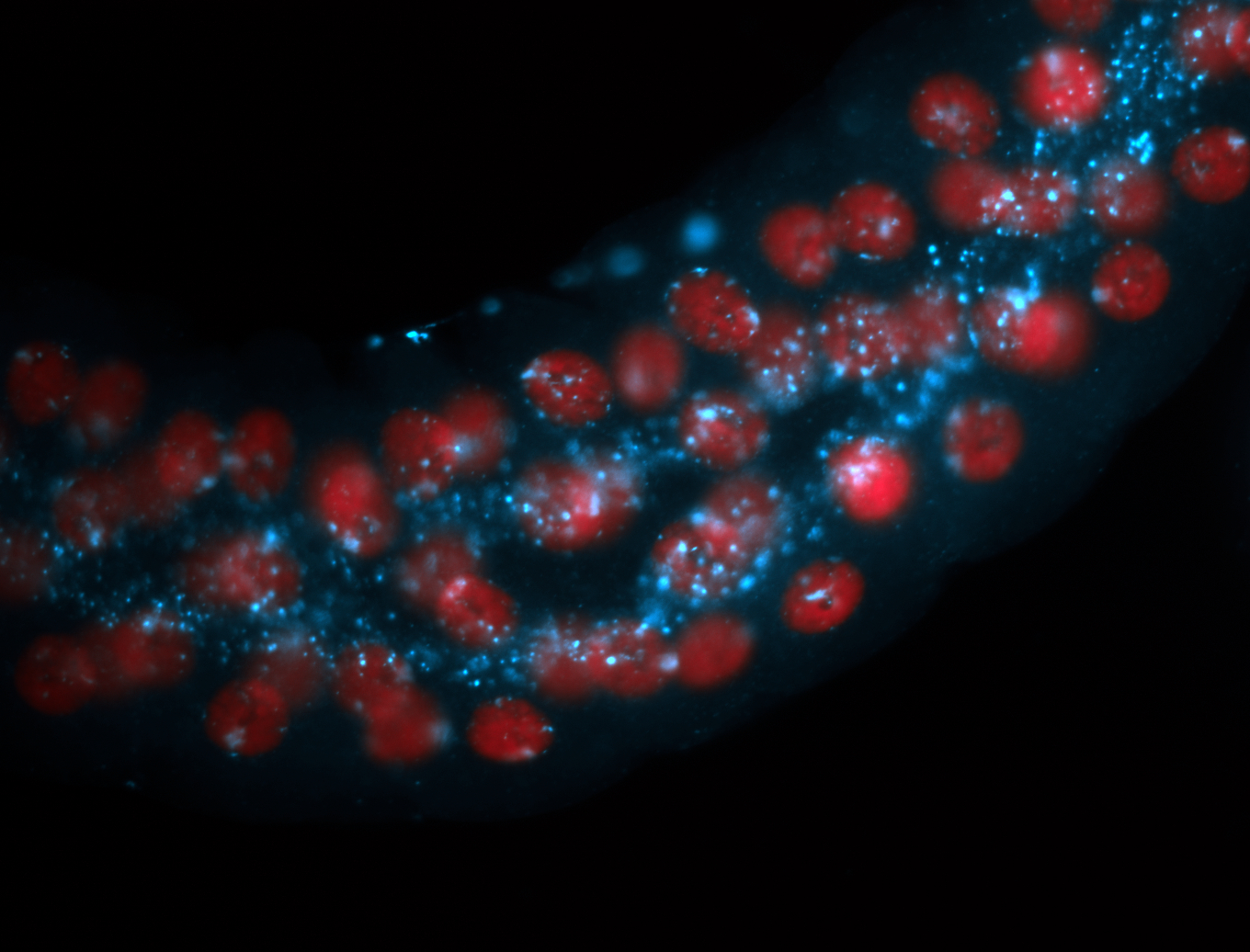
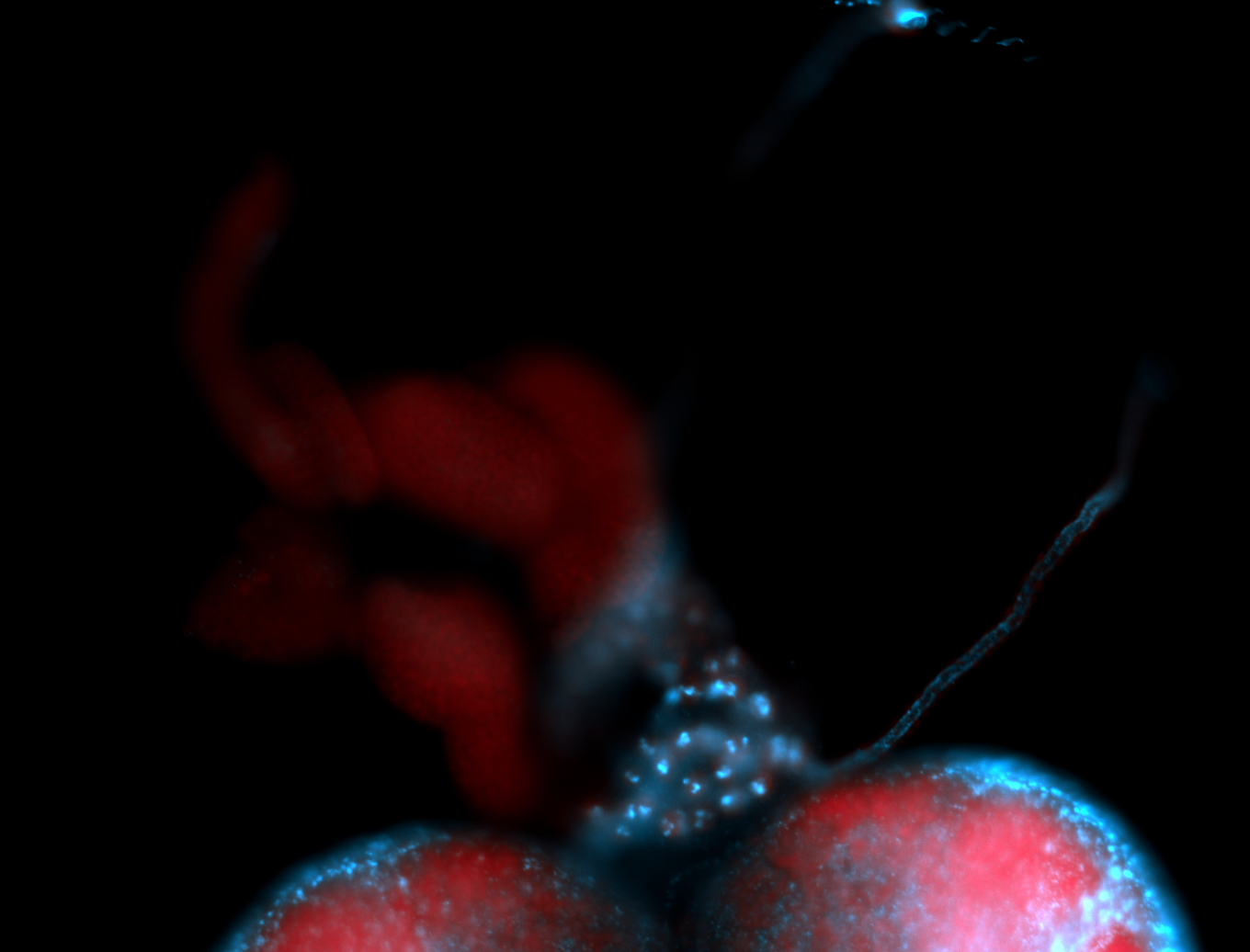
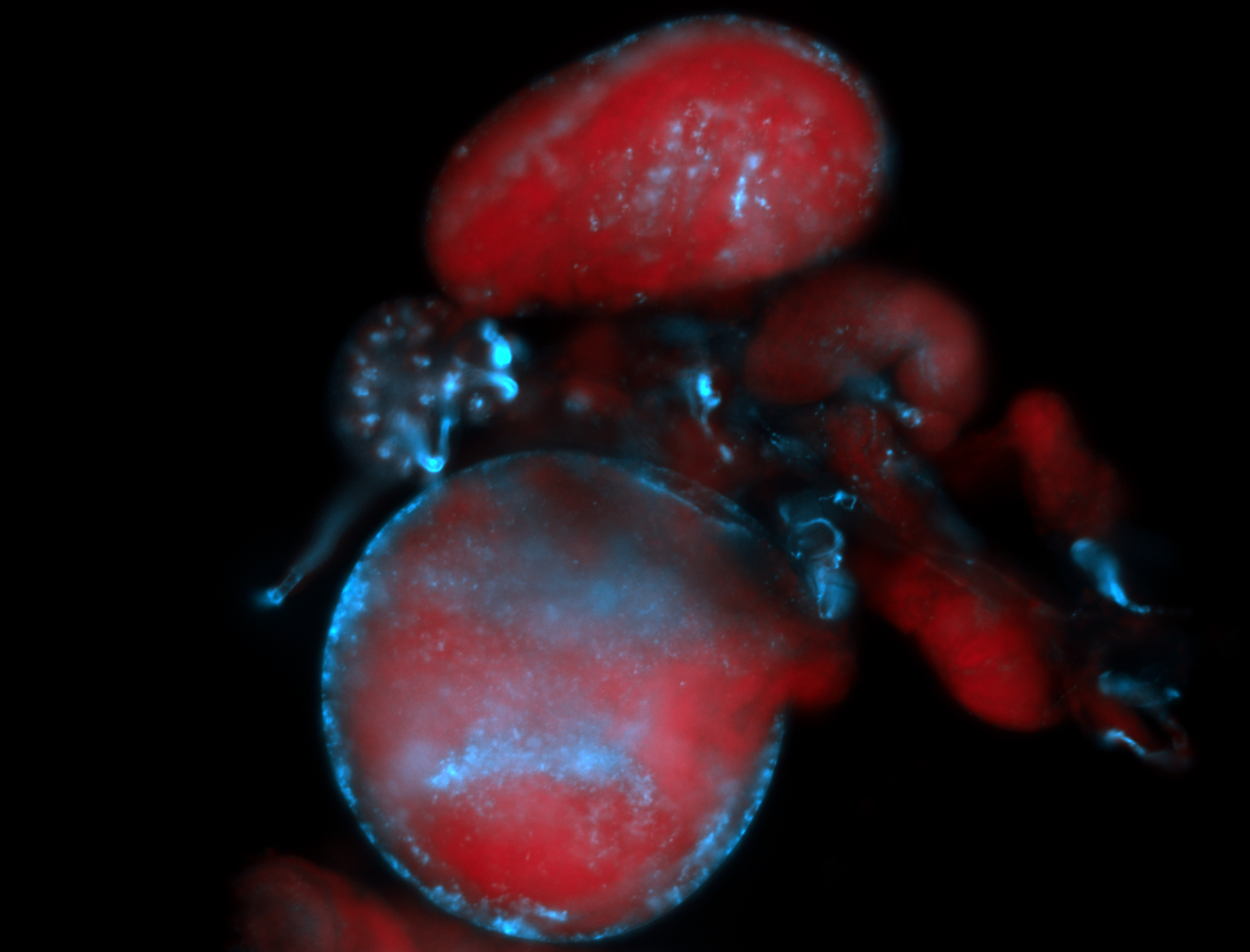
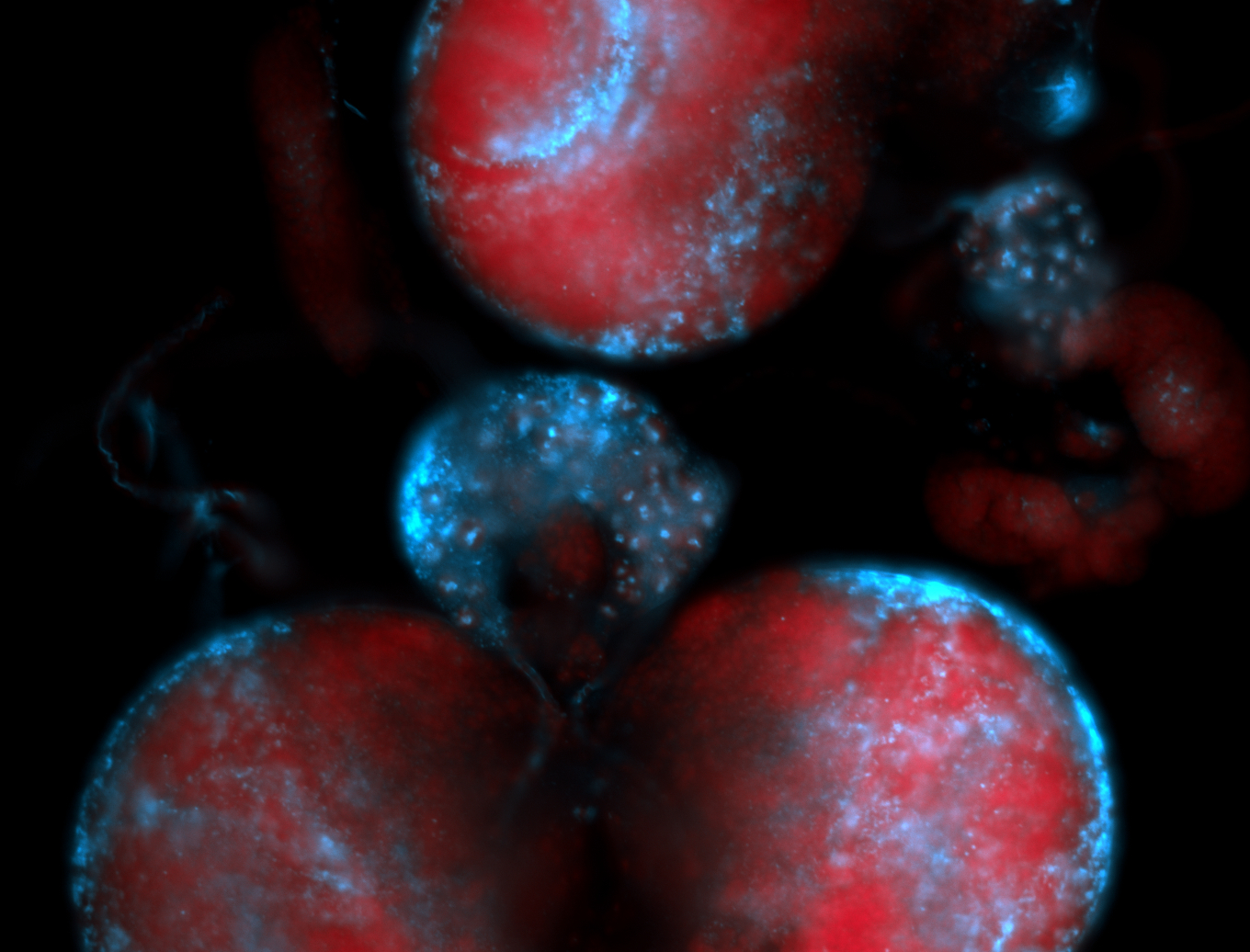
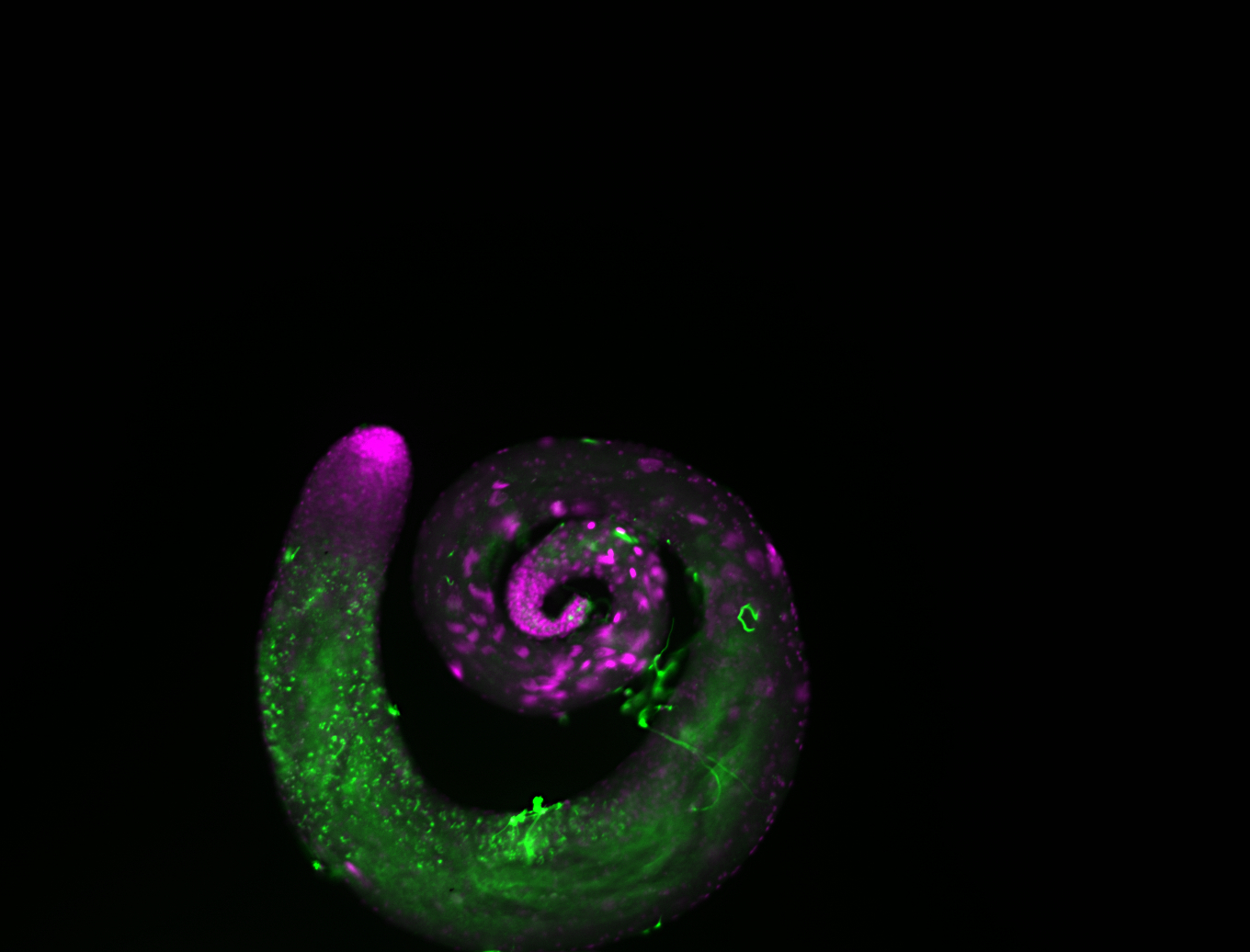
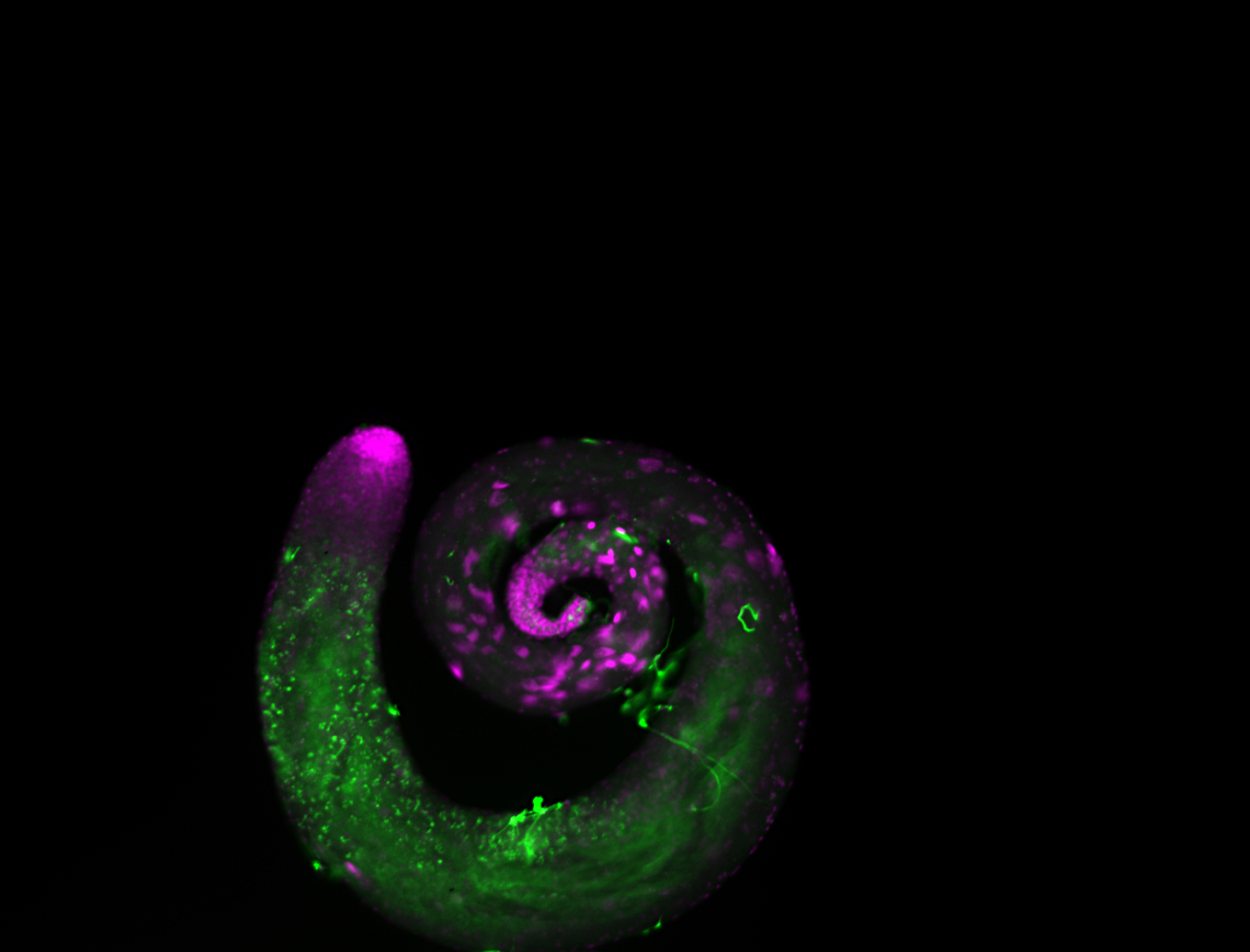
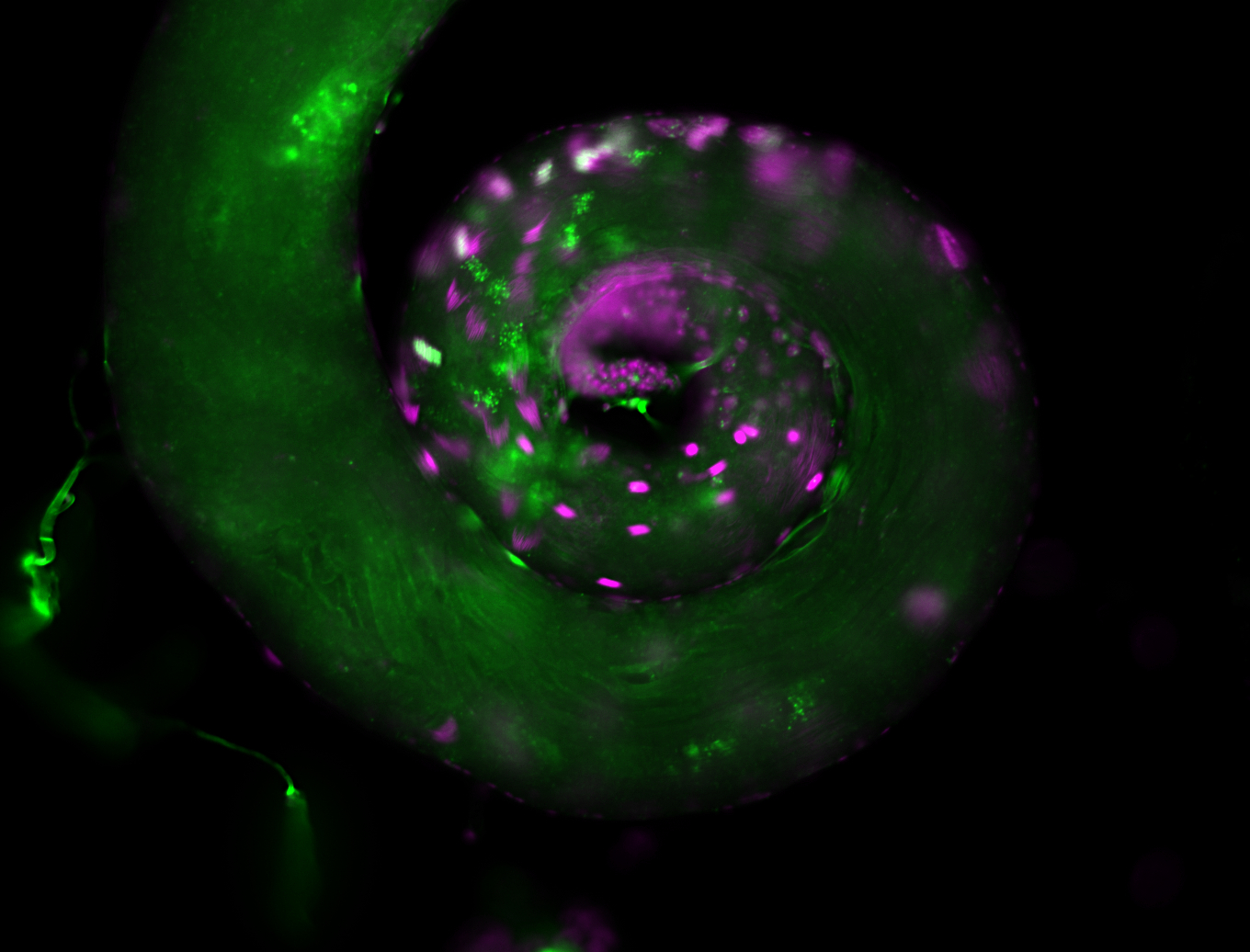
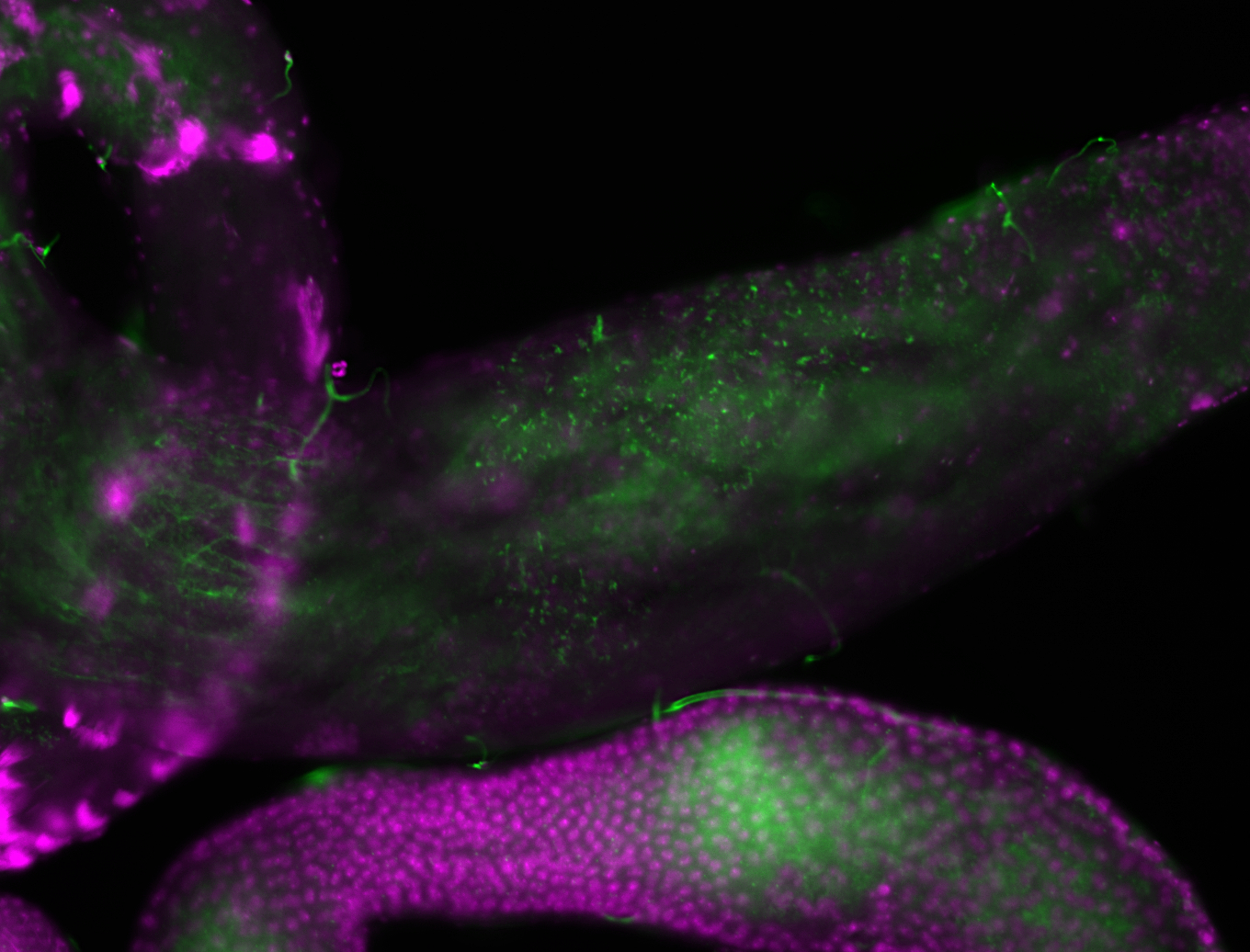
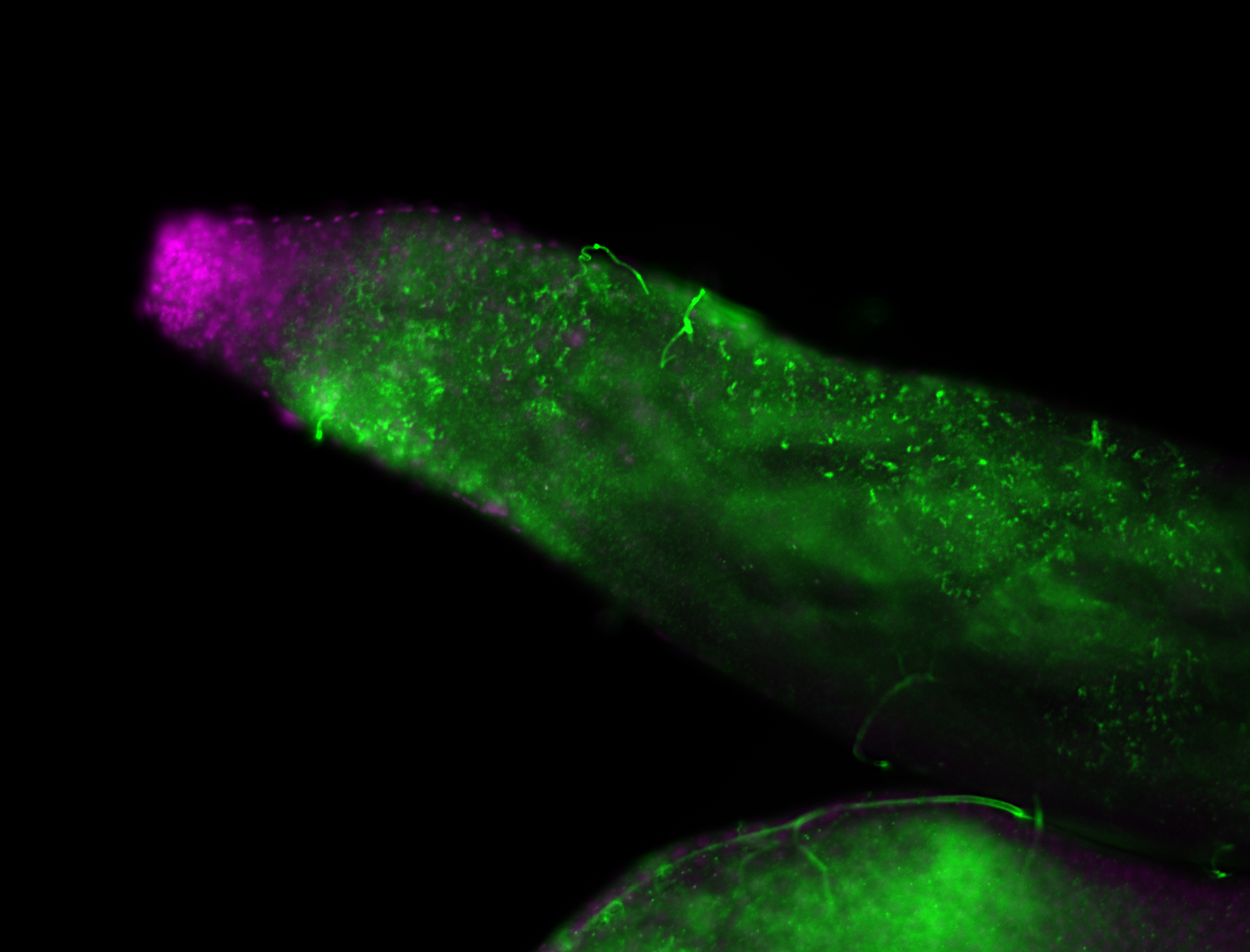
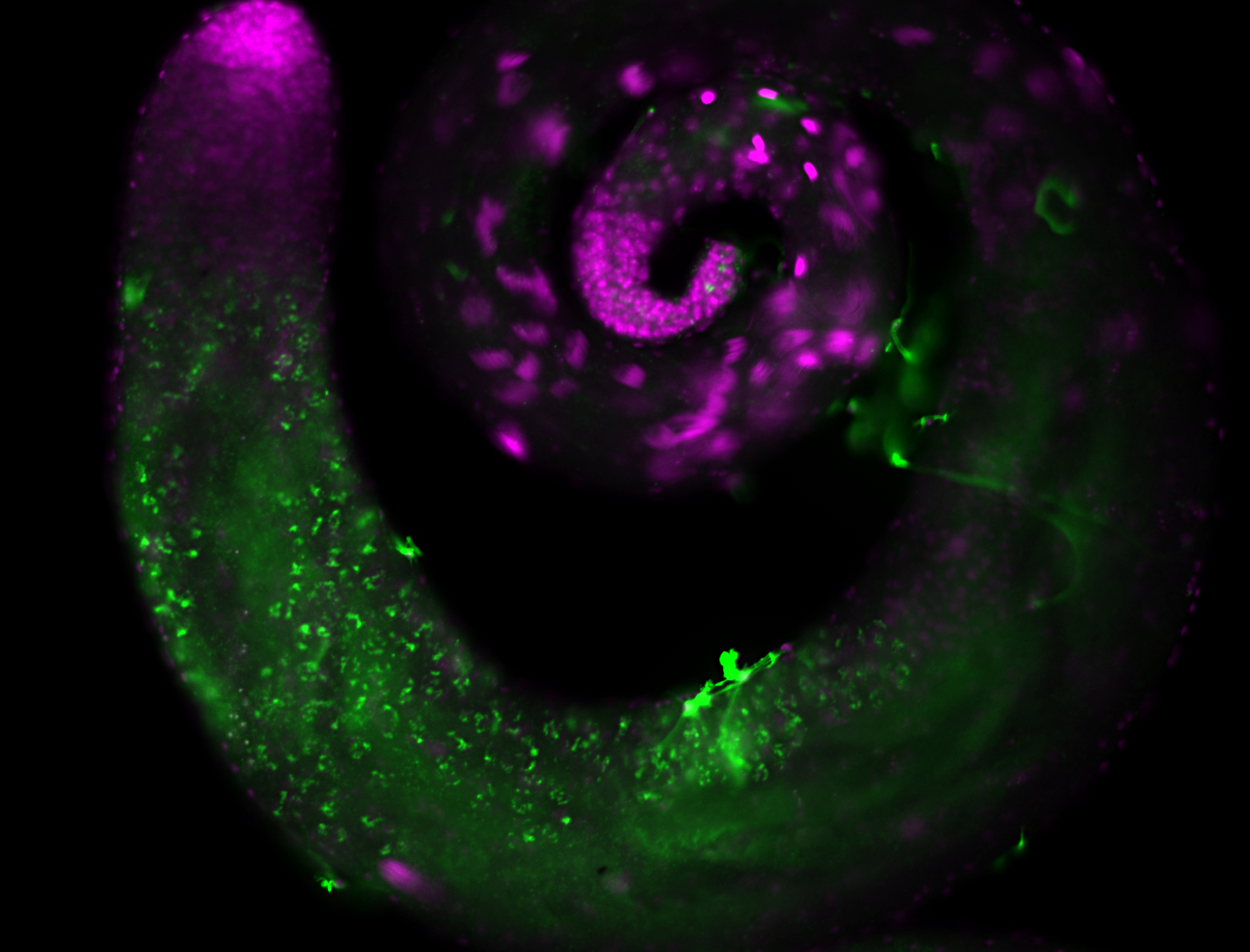
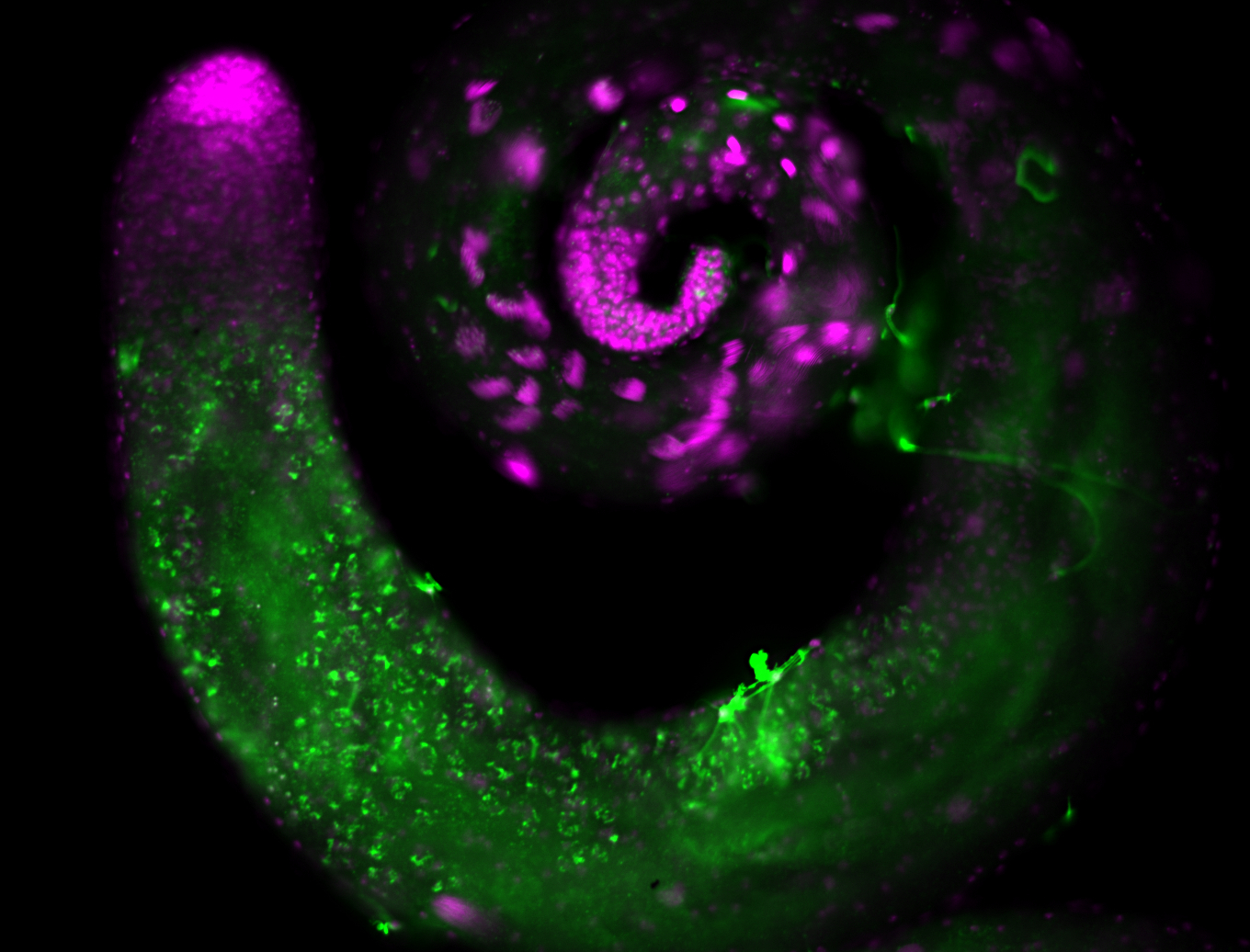
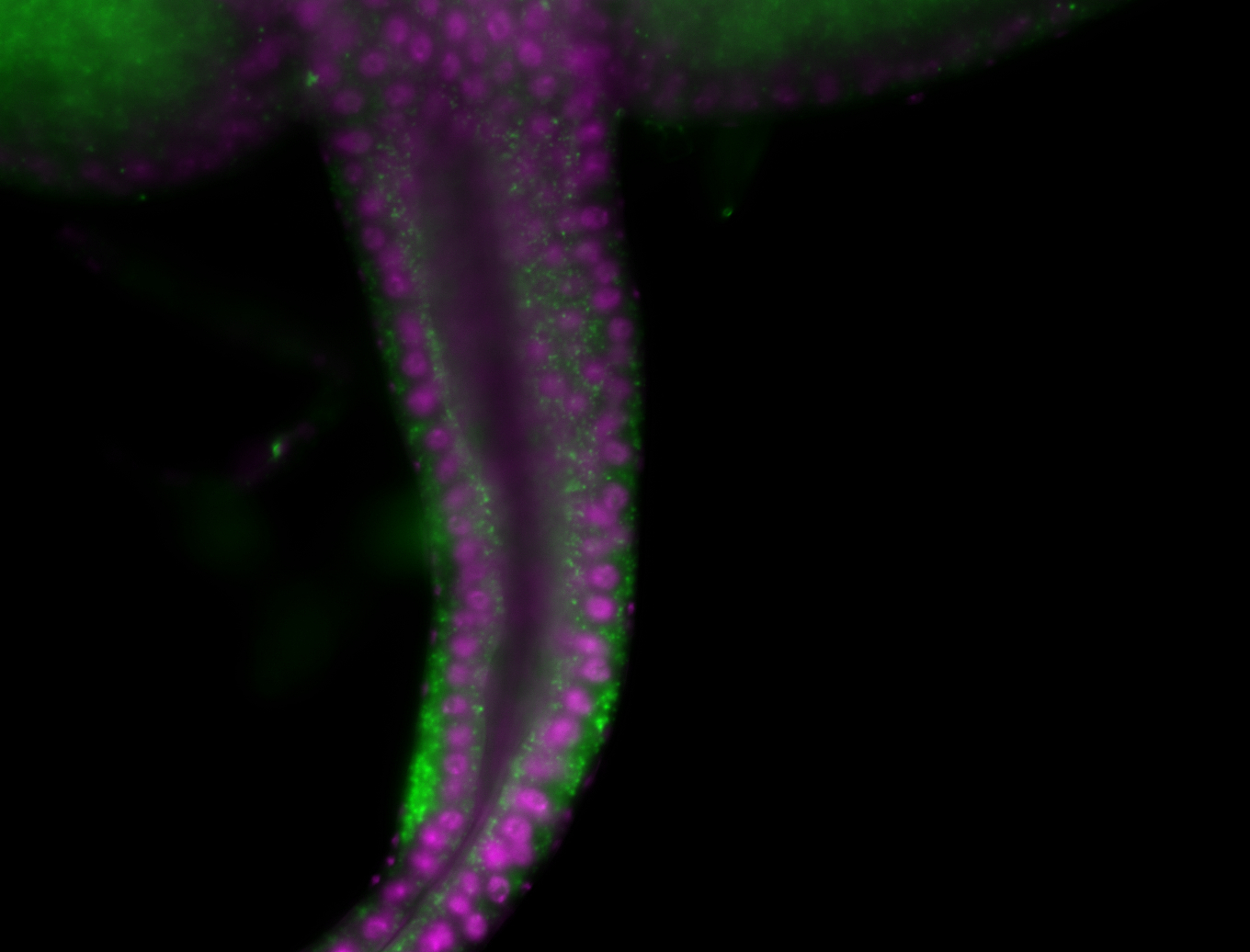
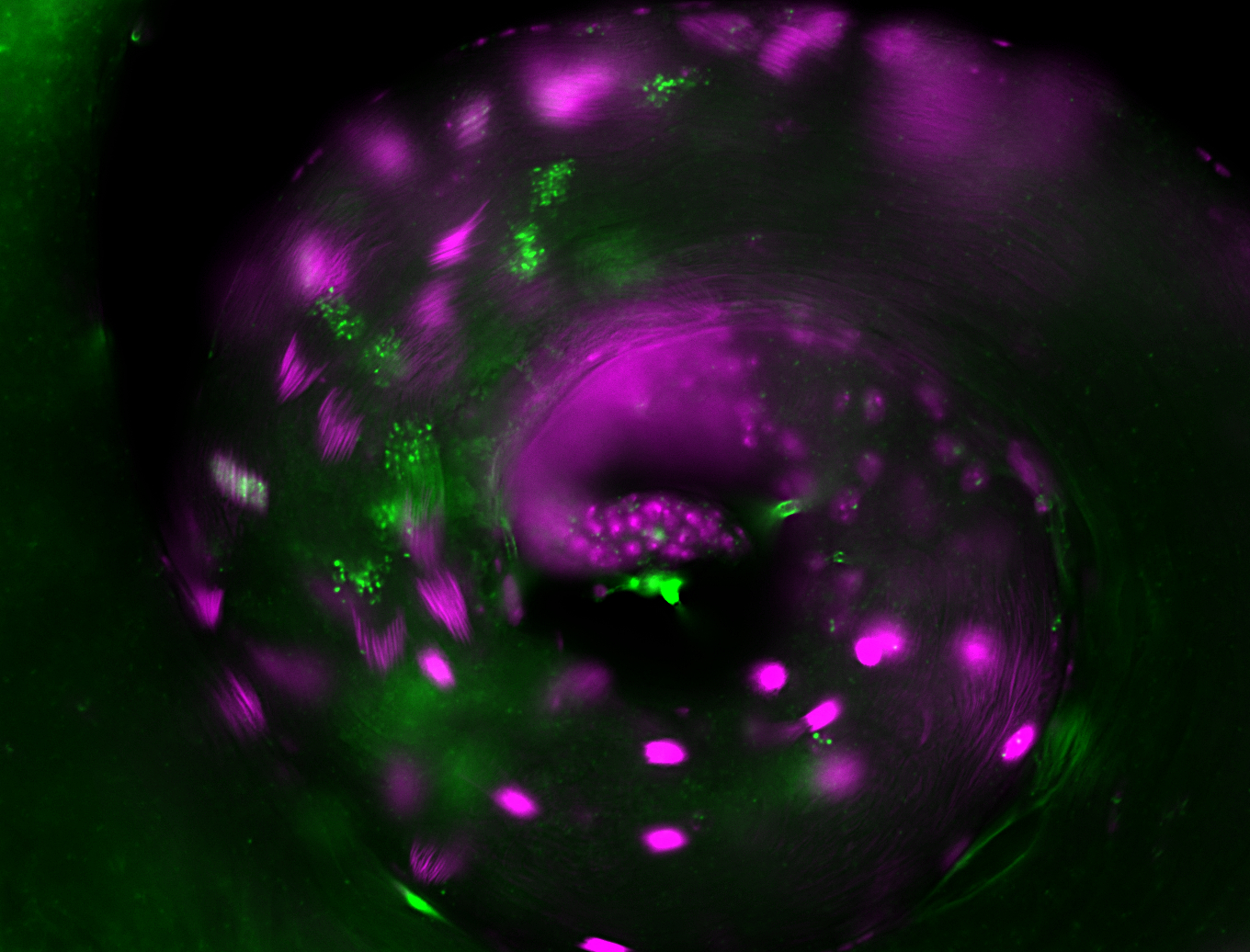
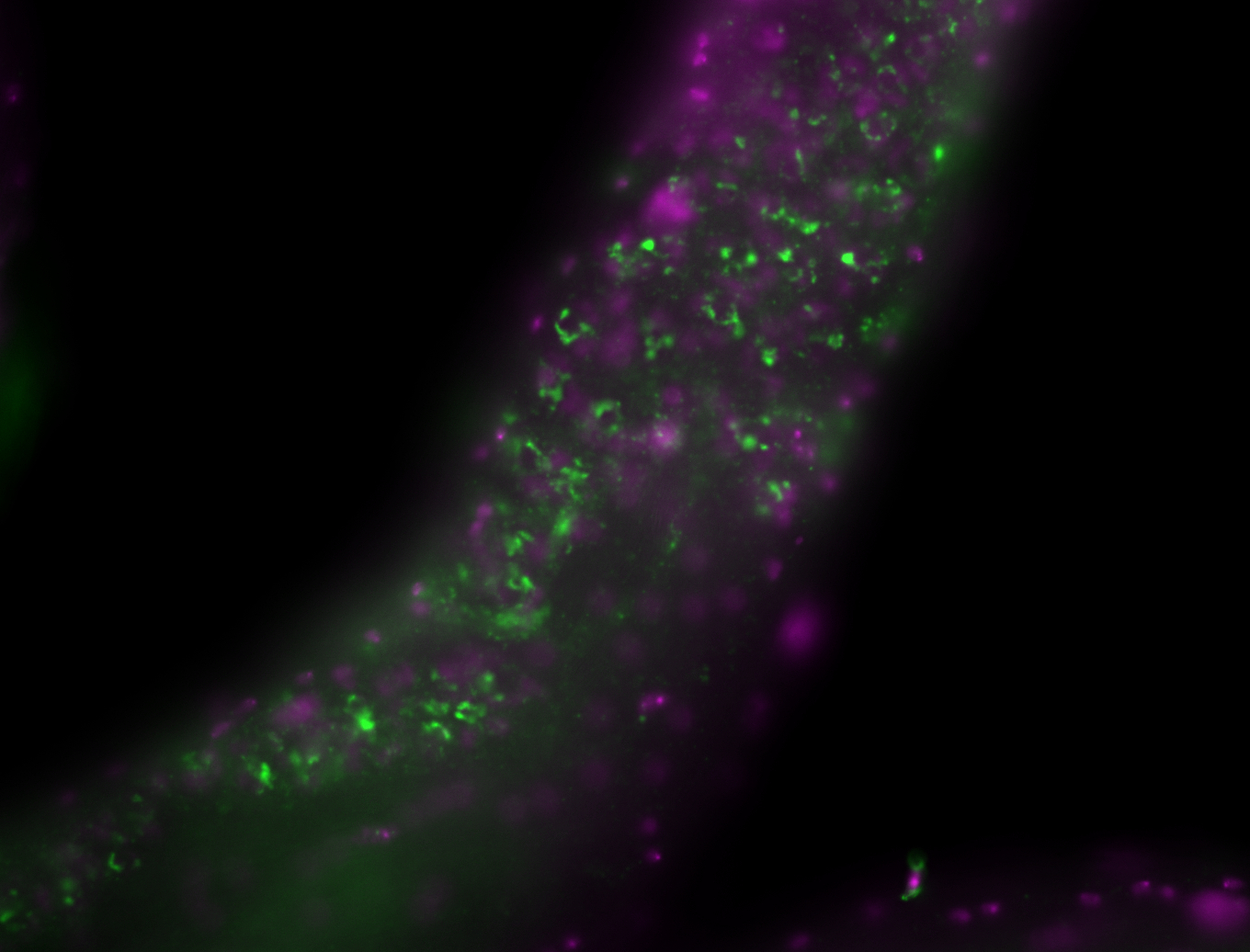
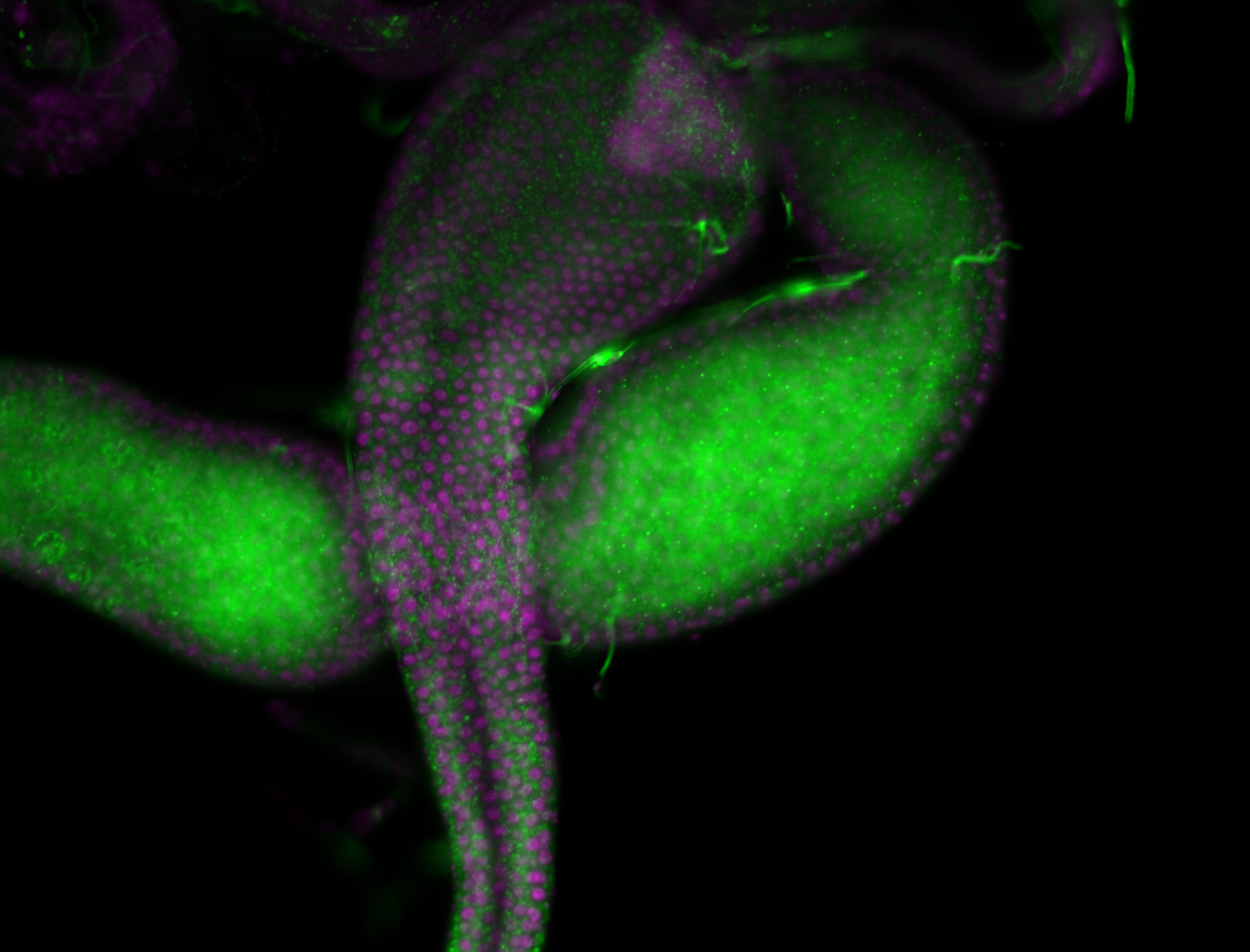
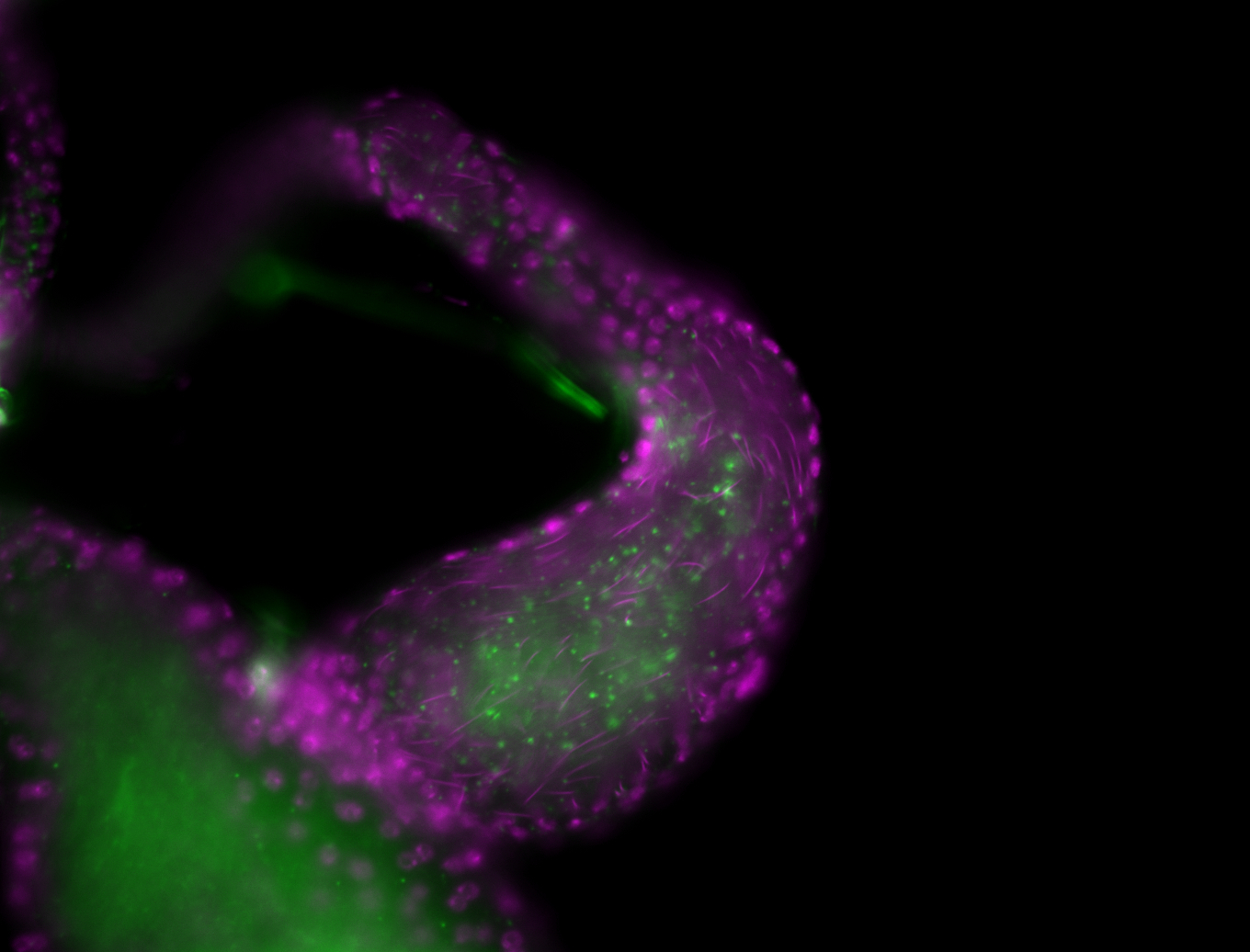
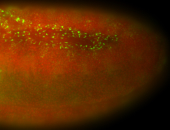
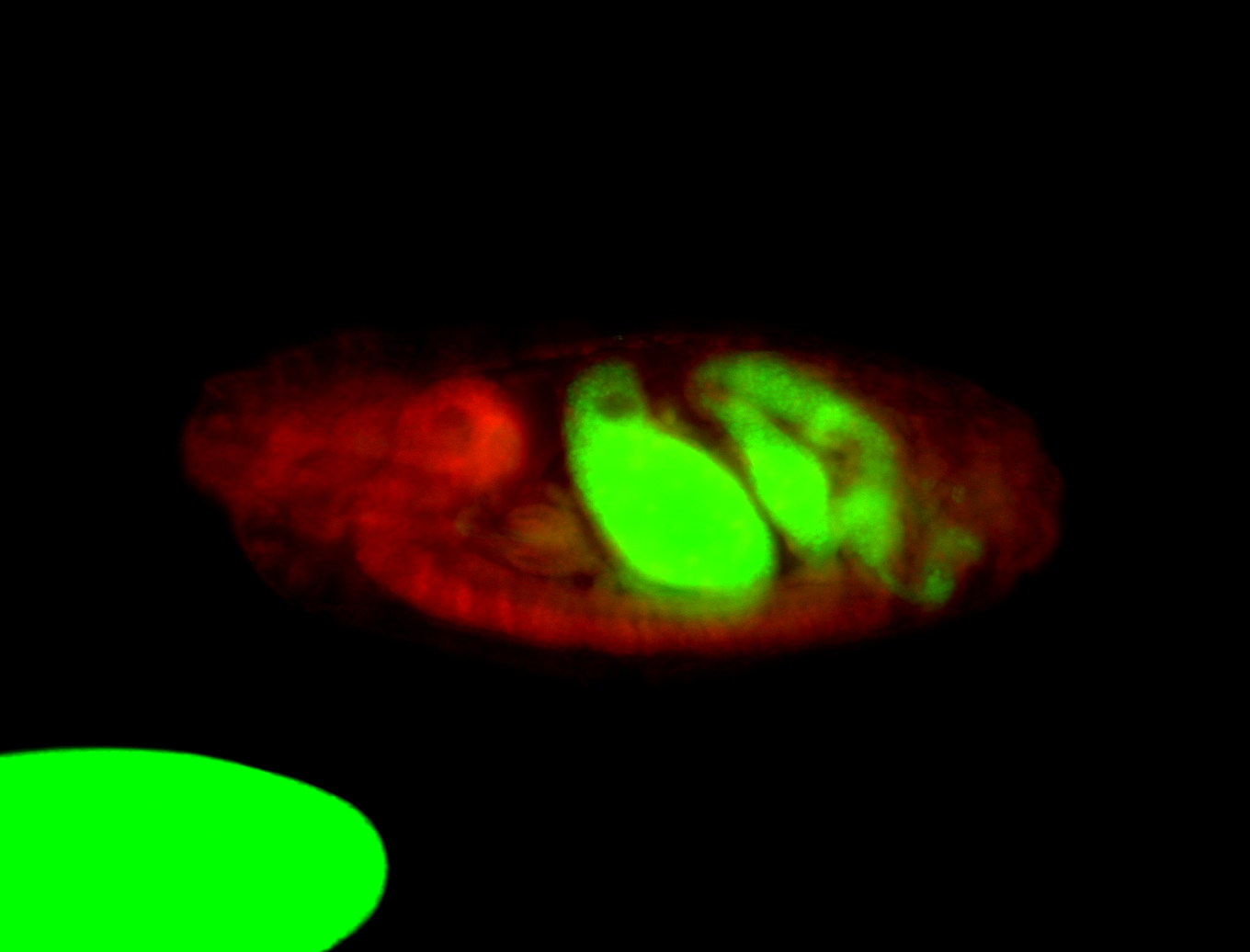
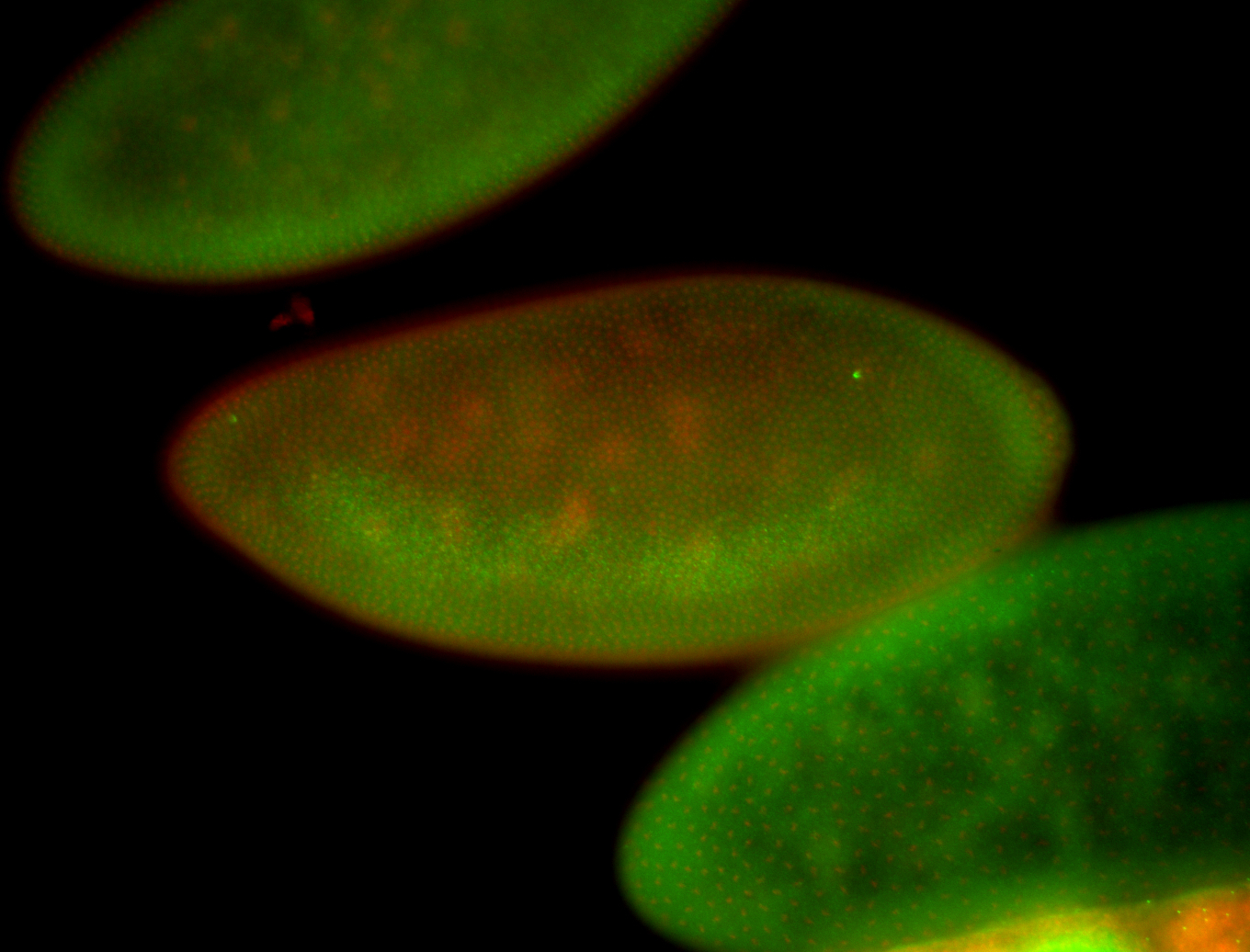
| -performed via HCR-FISH using 2 probe sets targeting the RA isoform transcript |
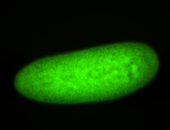
| -this non-coding gene is an extremely highly expressed lncRNA in male testes according to scRNA-seq data, and exhibits a strong nuclear pattern in in embryo yolk, pole cell, and random blastoderm cells |
Probe GM08252 Links: Permalink
Genes: flam flam-intron
Embryo
Larva
Adult
Comments
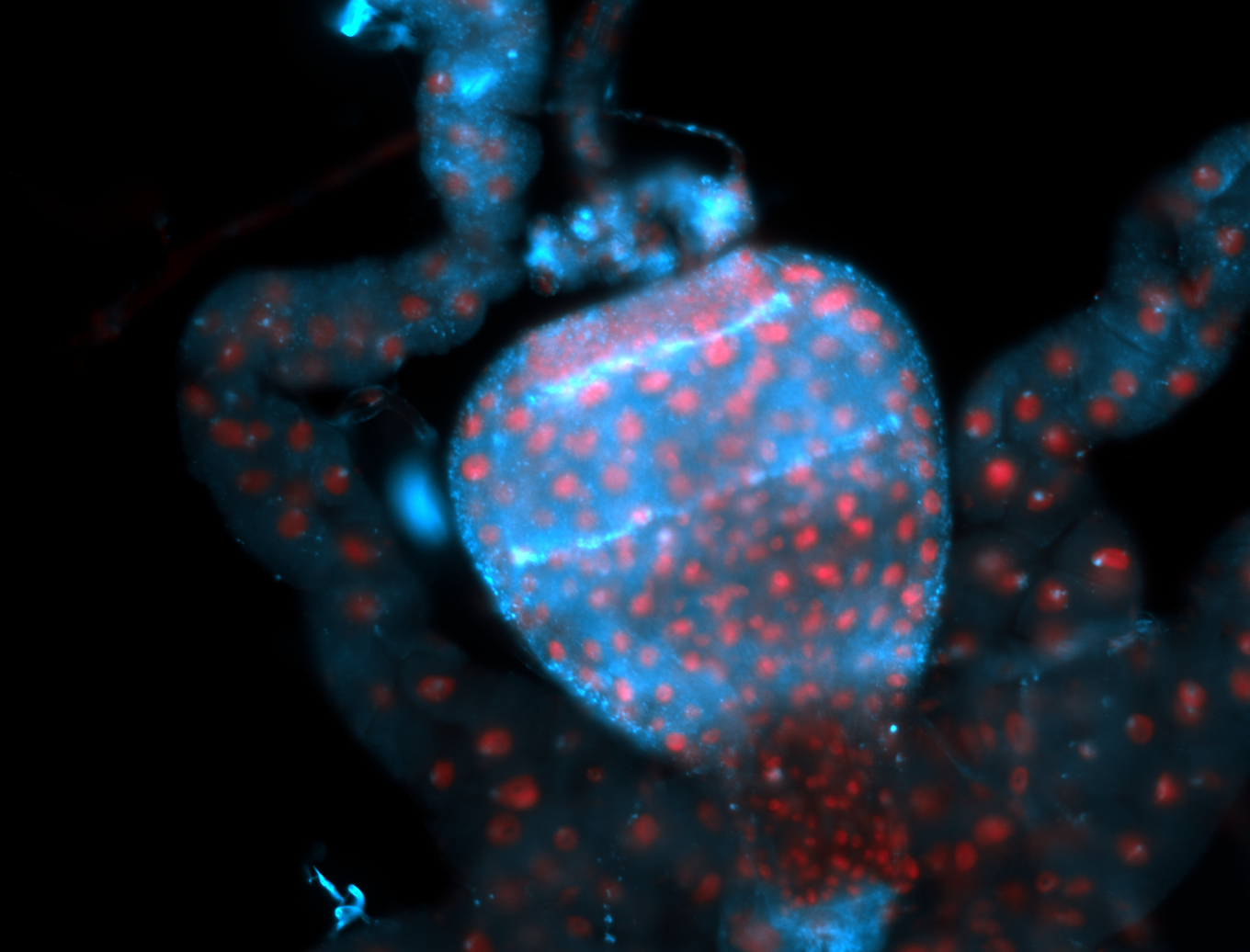
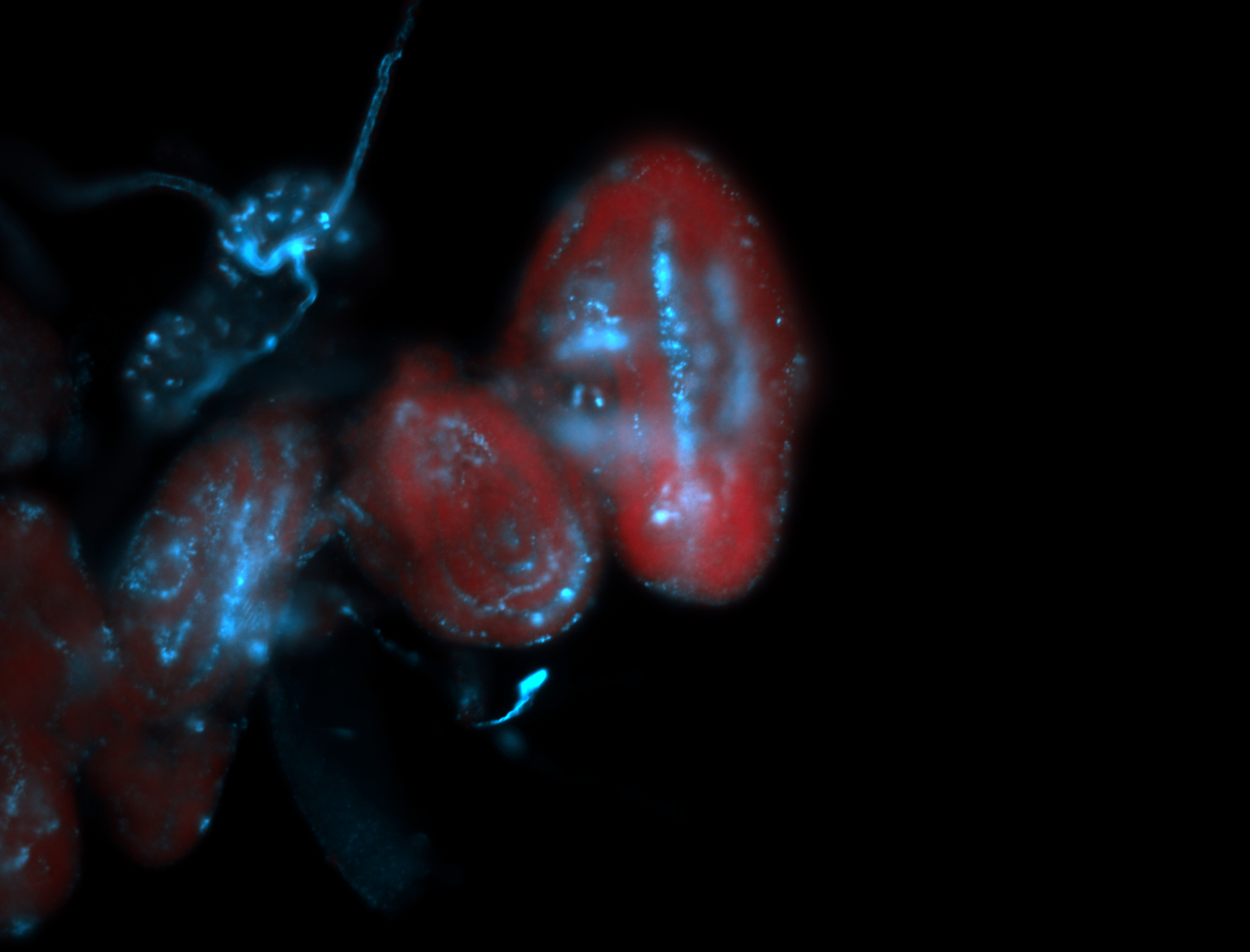
| The probe may be recognizing 16 transcripts - Including flamenco. The probe may recognize some other areas in the genome that contain a highly homologous sequence. |
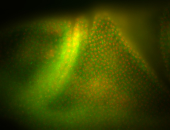
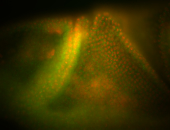
| Amazing patterns! |
| Very interesting patterns in adult testes and associated tissues. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Very interesting late patterns -R |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
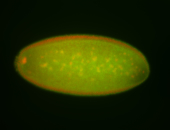
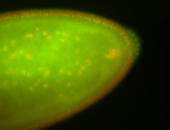
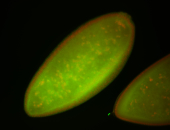
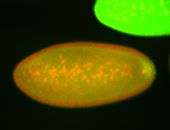
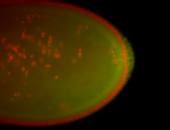
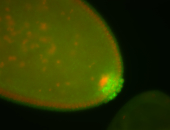
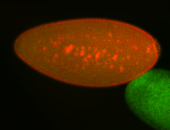
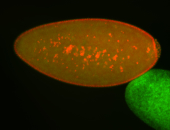
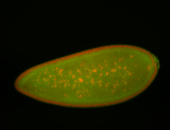
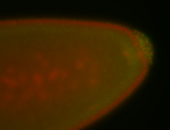
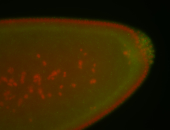
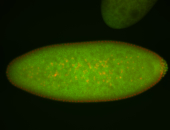
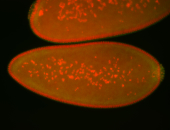
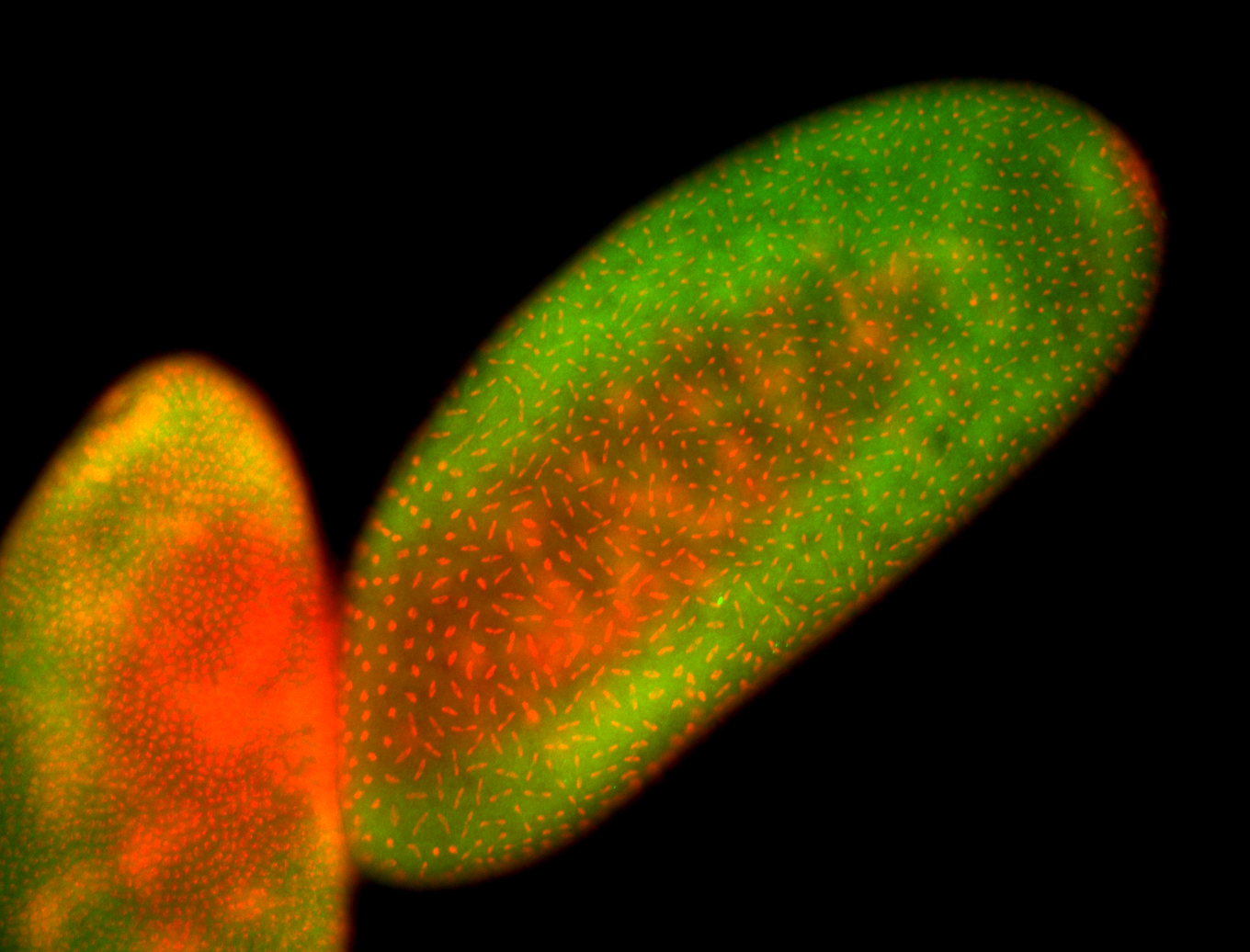
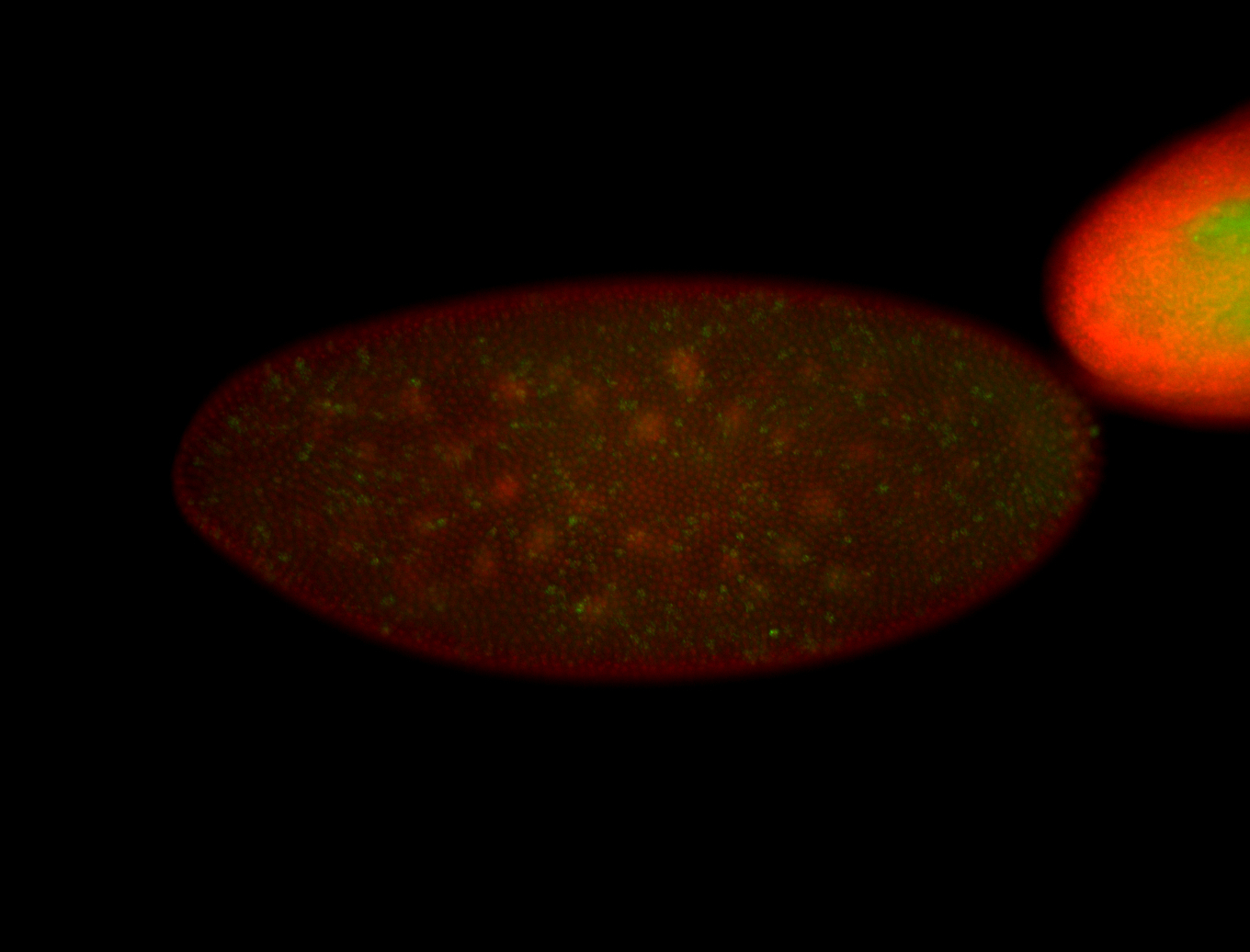
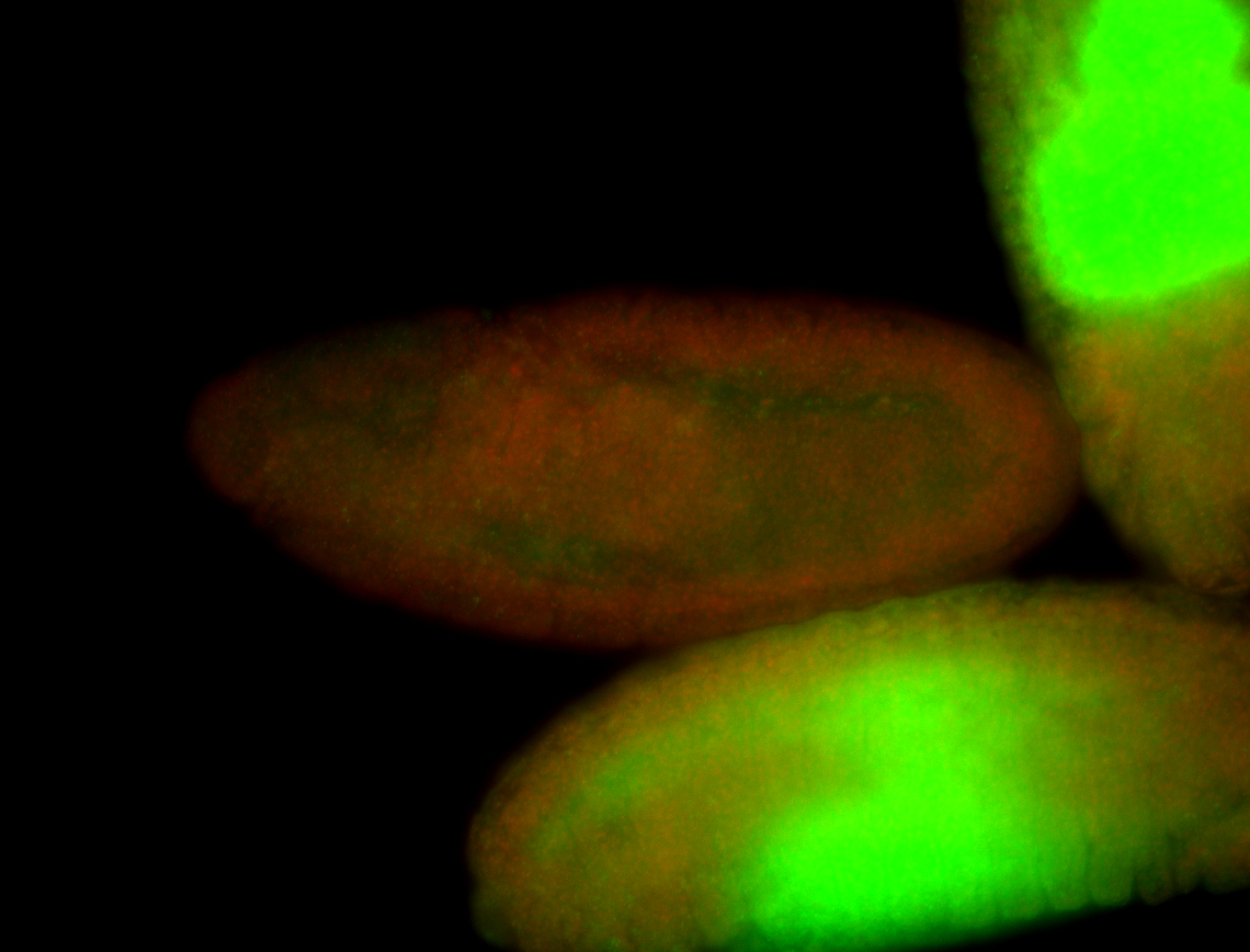
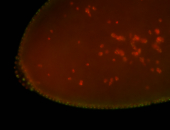
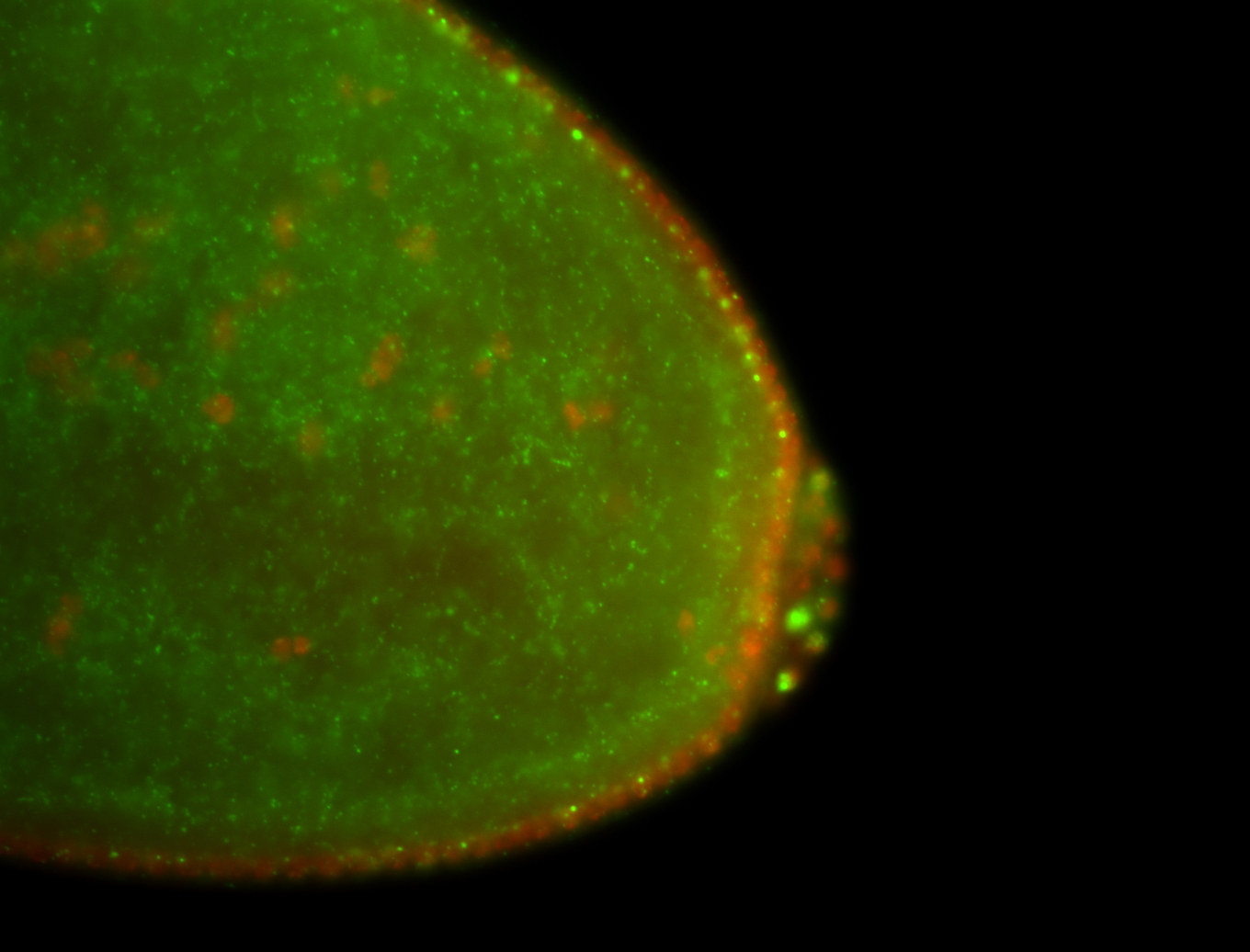
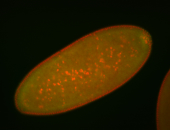
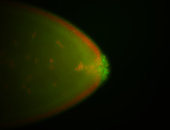
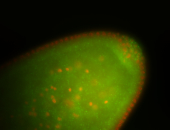
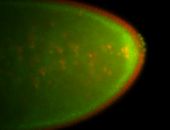
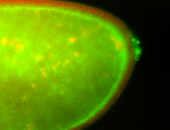
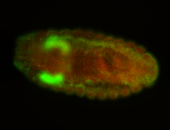
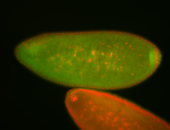
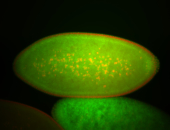
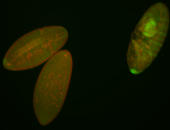
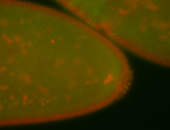
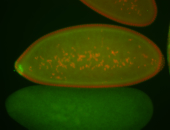
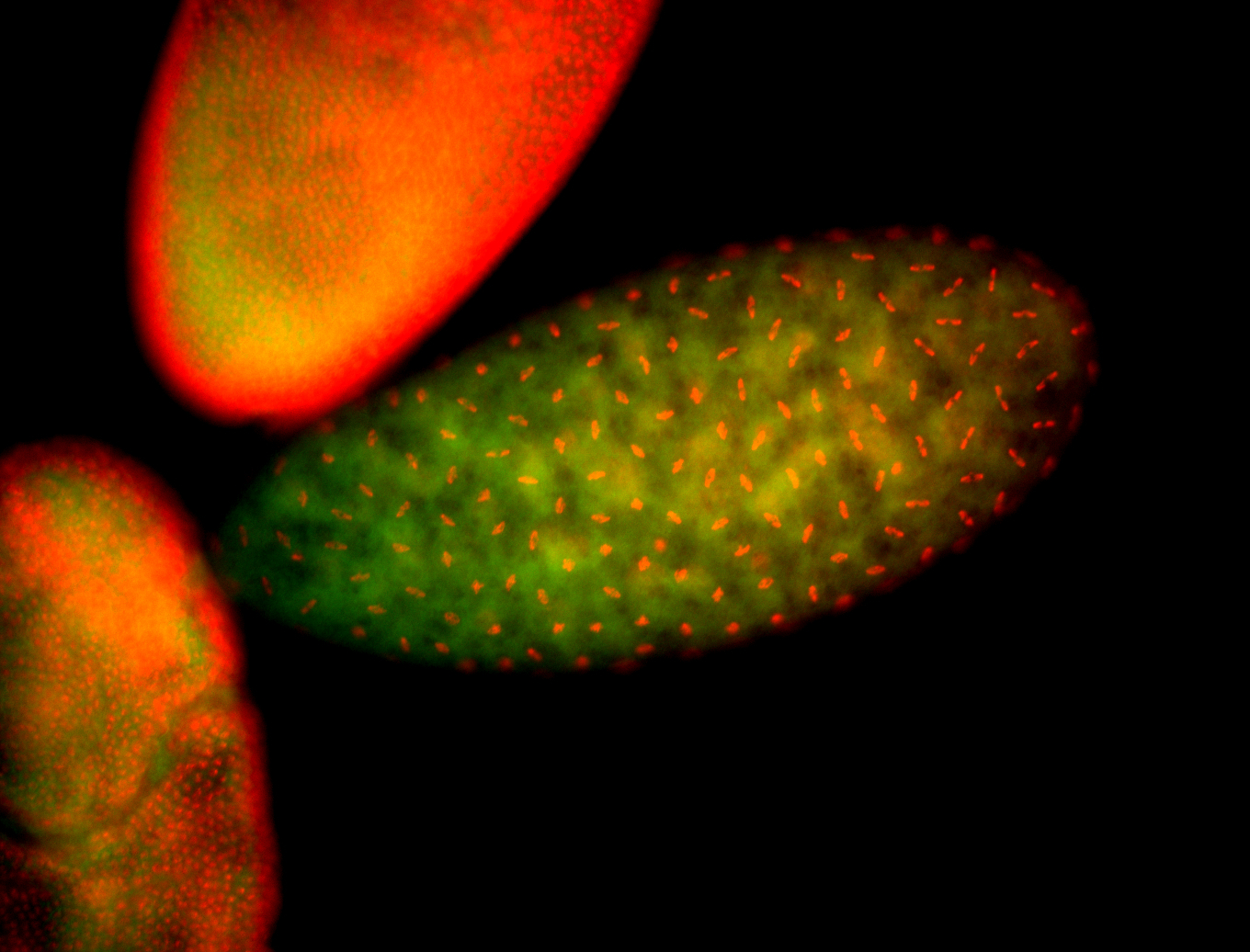
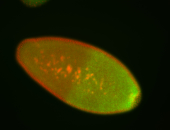
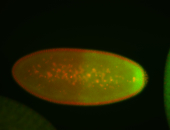
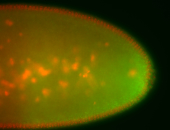
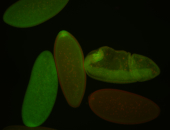
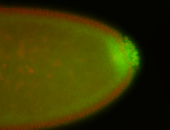
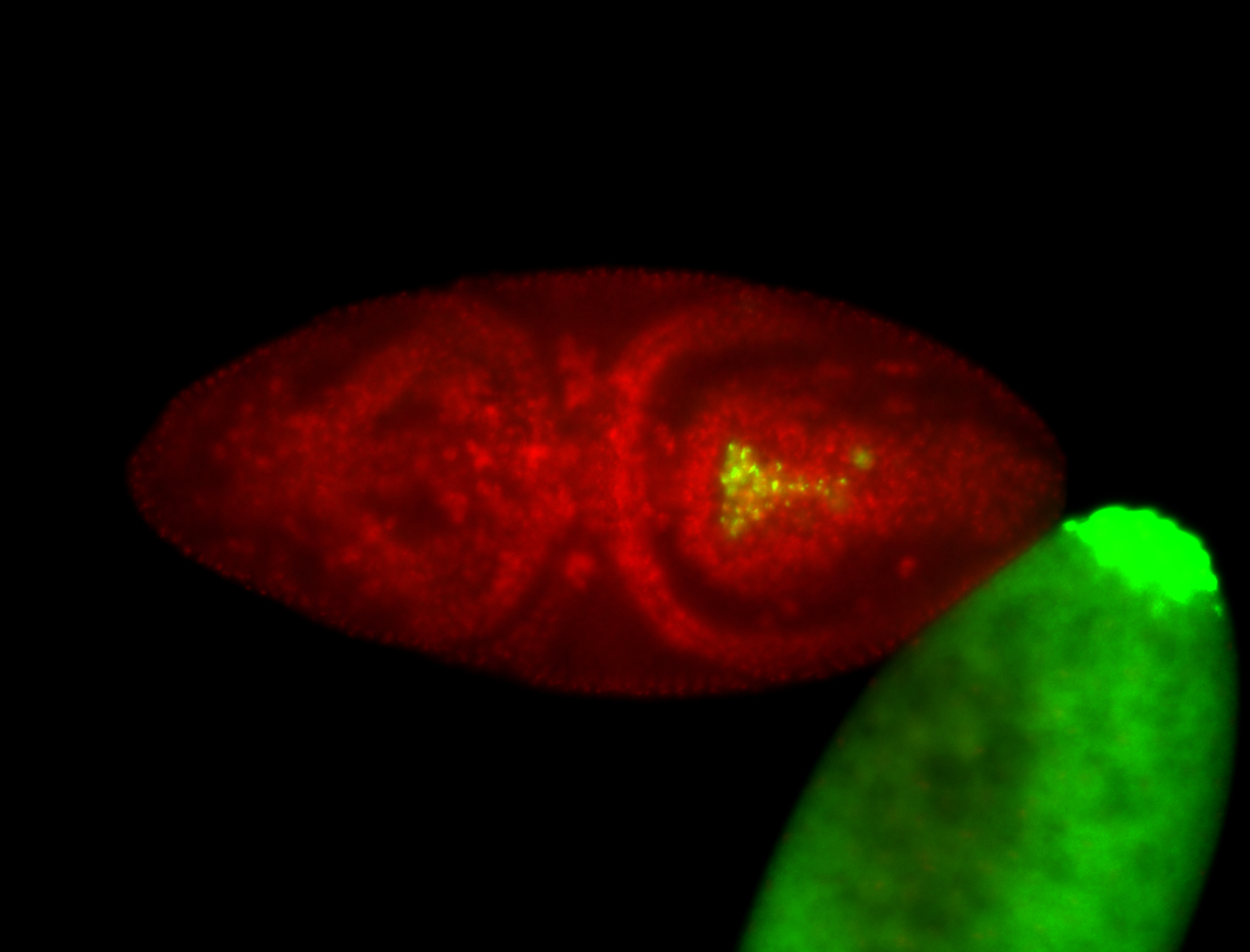
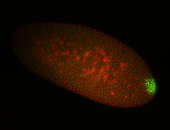
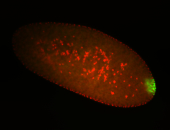
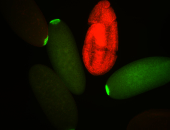
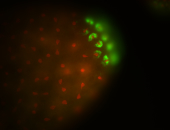
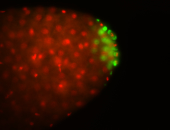
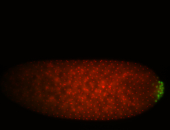
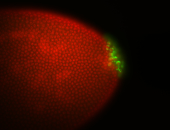
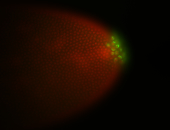
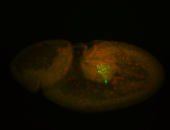
| Cytoplasmic foci around pole cells. |
| Pole plasm and pole buds enriched. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe on gel. Data from RE17418 probe |
| Salivary glands, probably non-specific. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Salivary glands, probably non-specific. |
| The probe looks weak on our gel. |
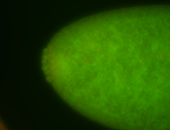
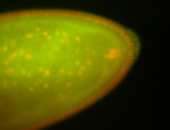
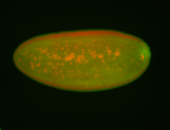
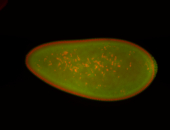
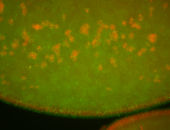
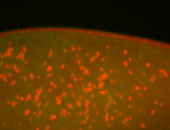
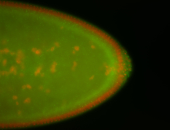
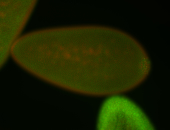
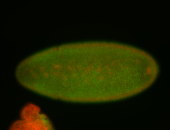
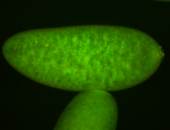
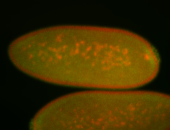
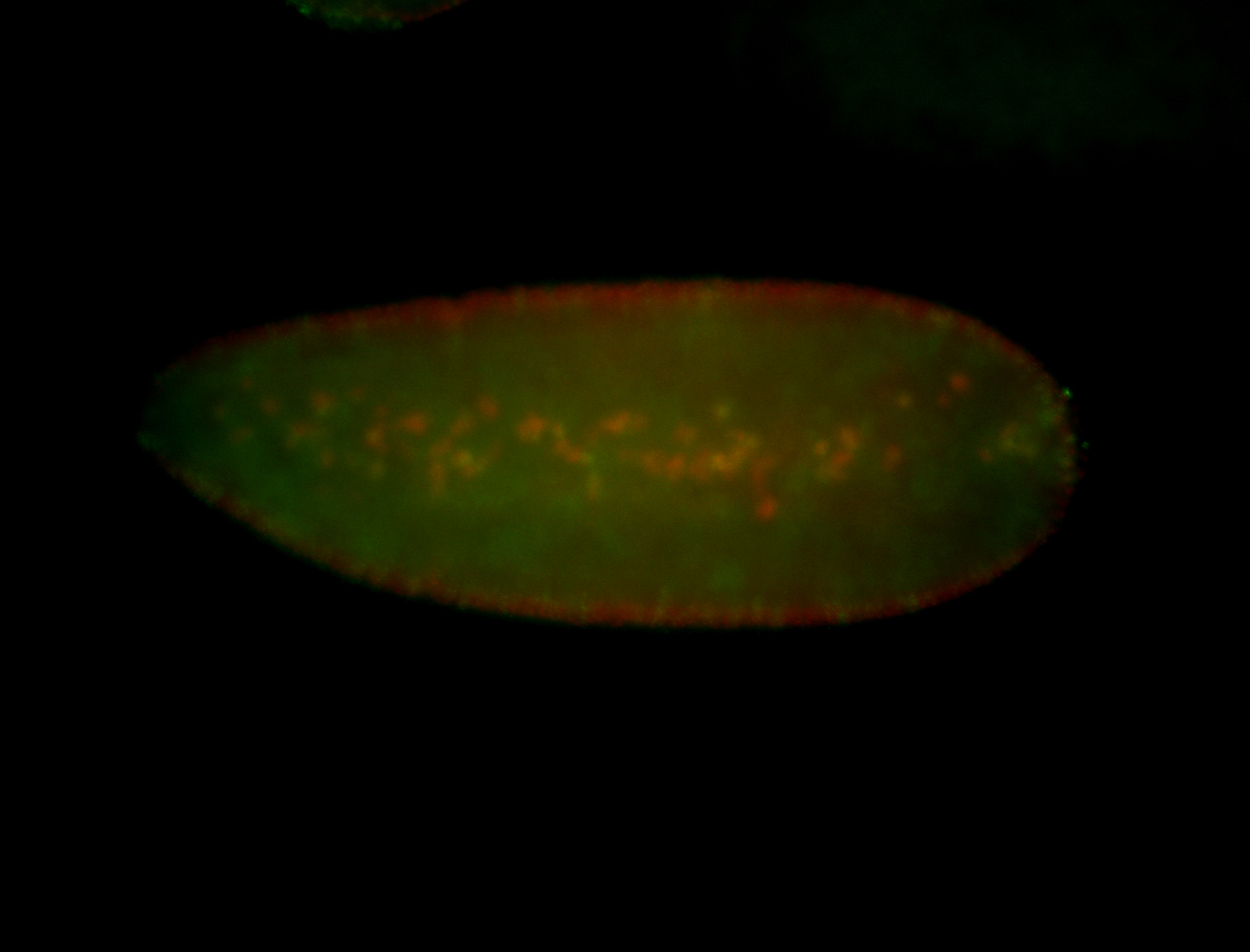
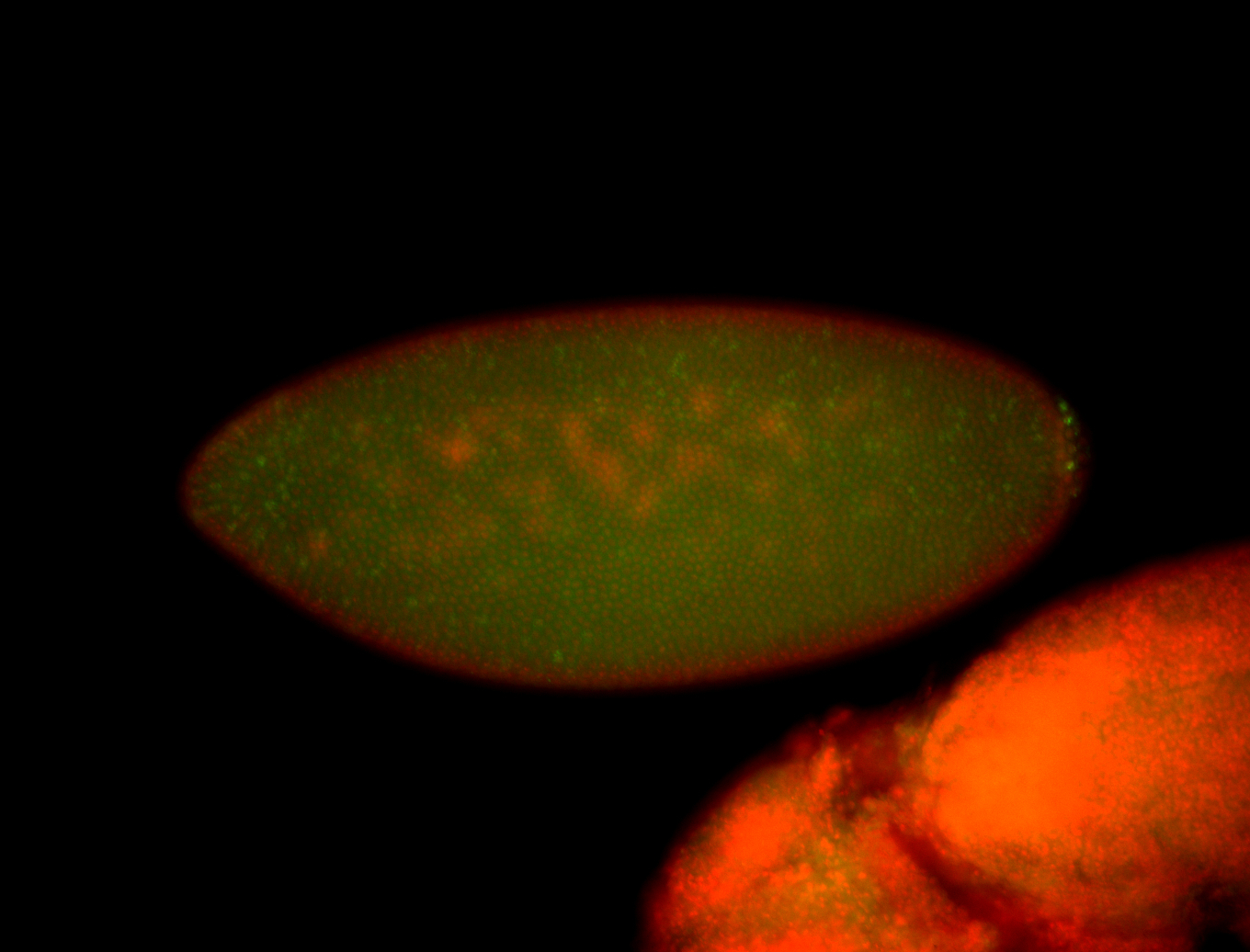
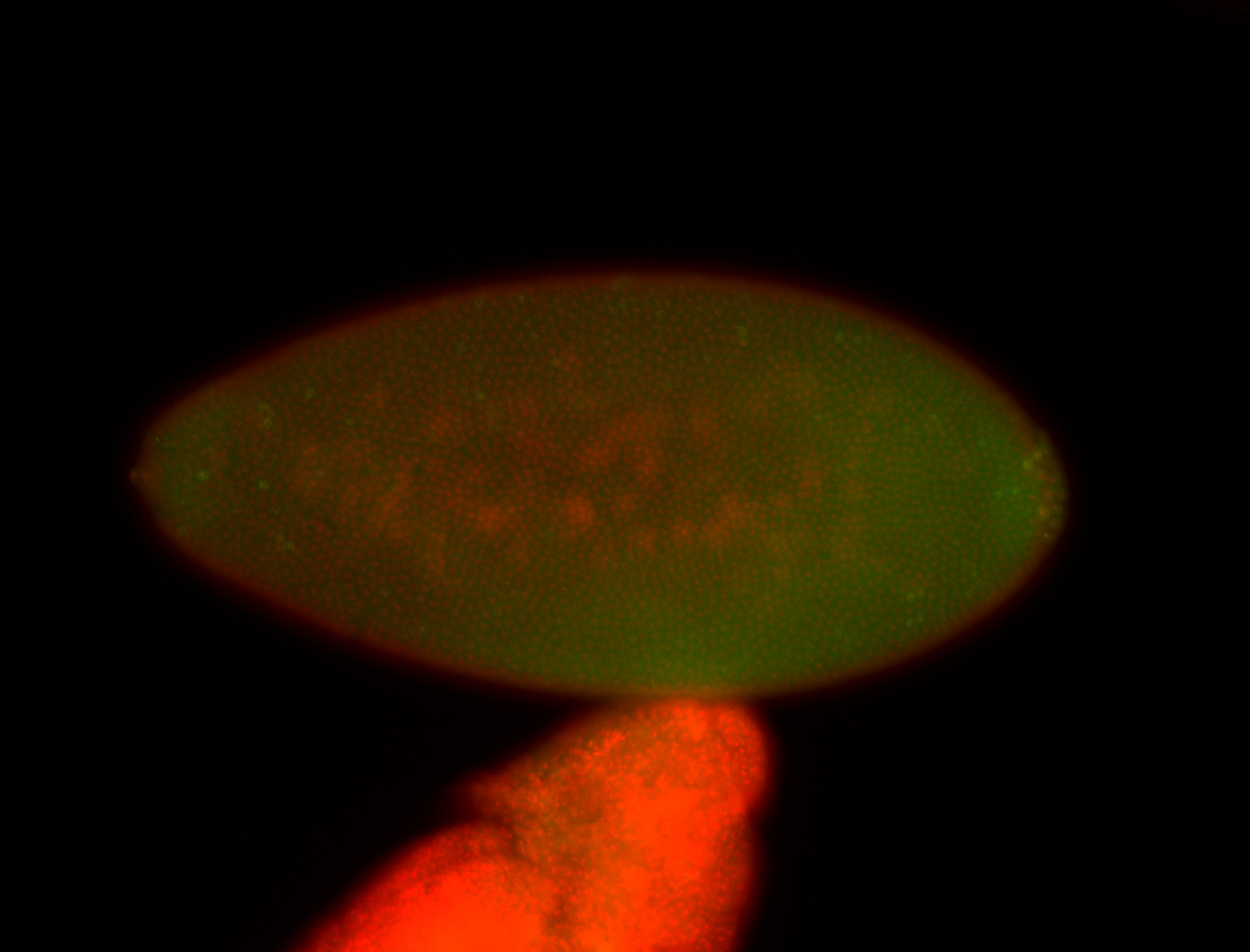
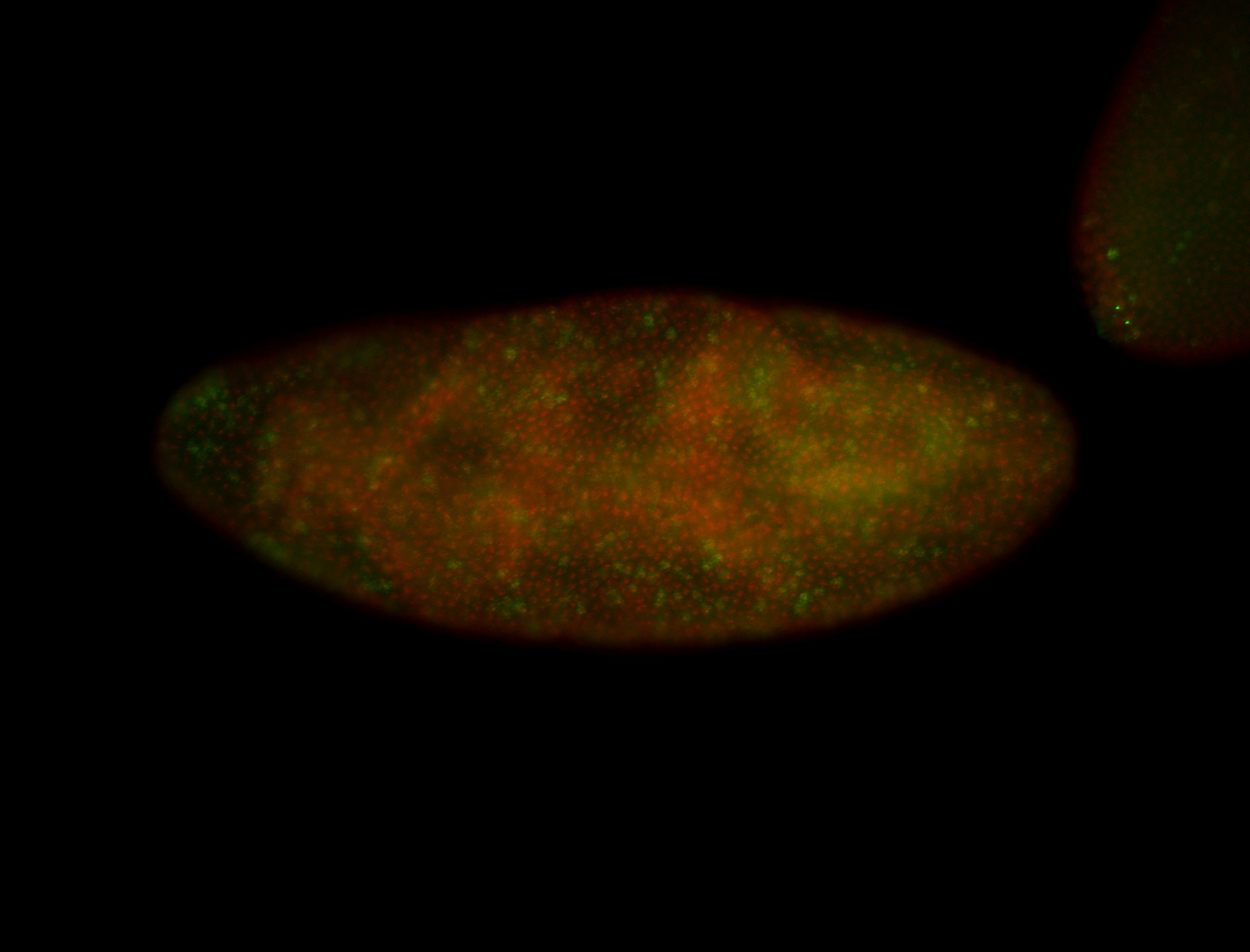
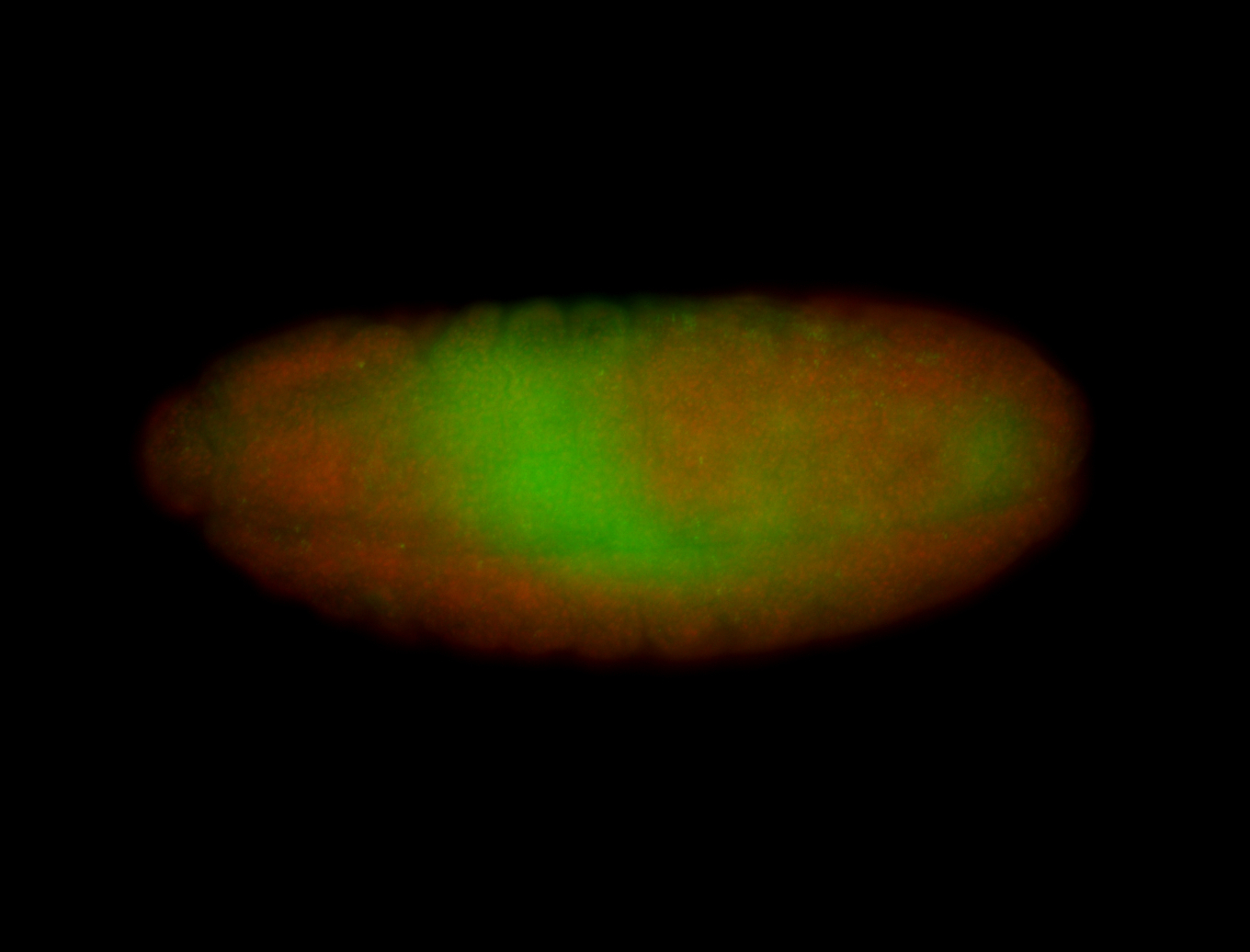
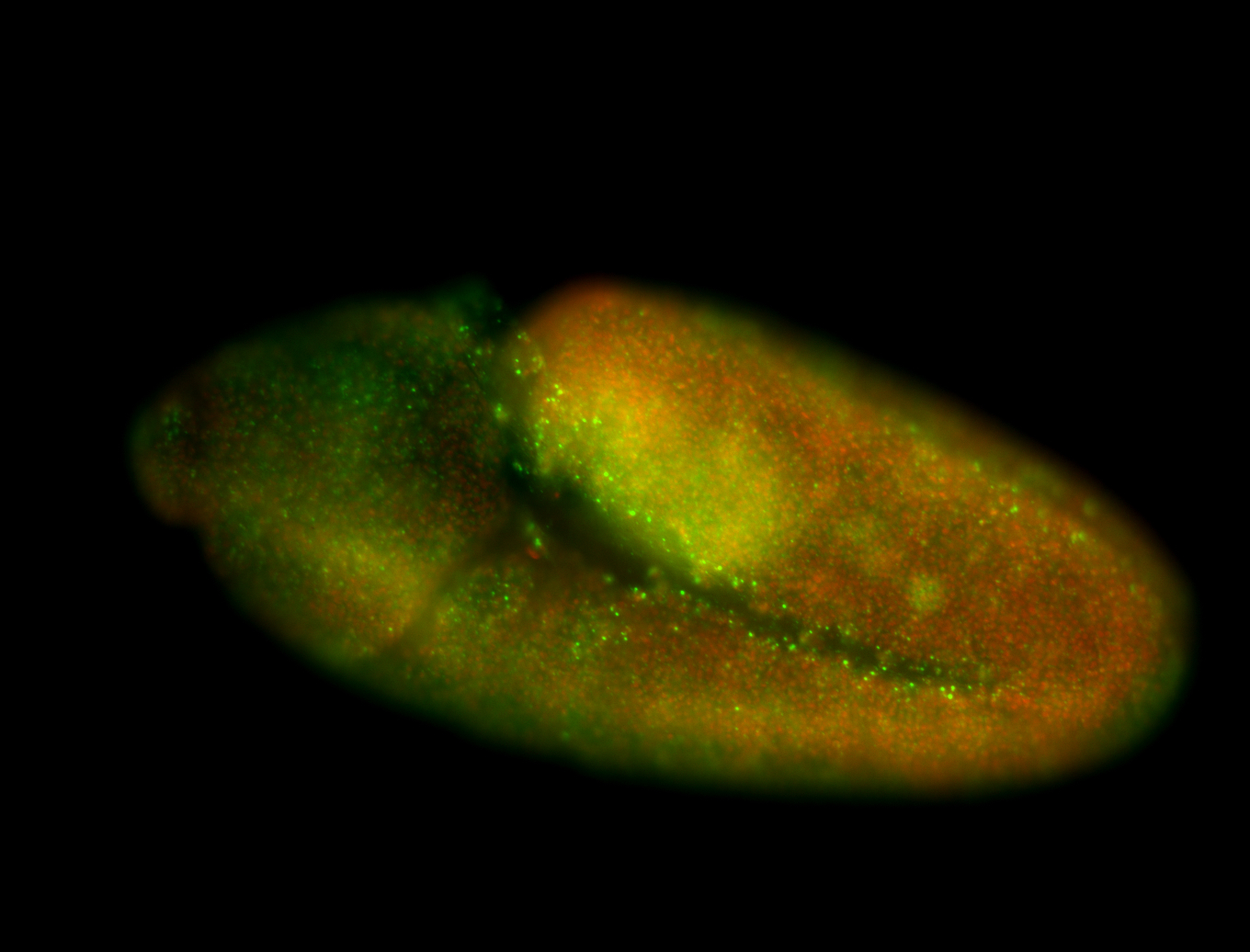
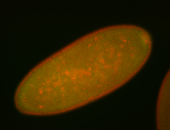
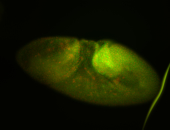
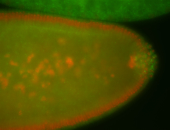
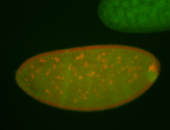
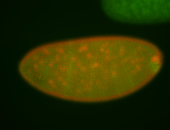
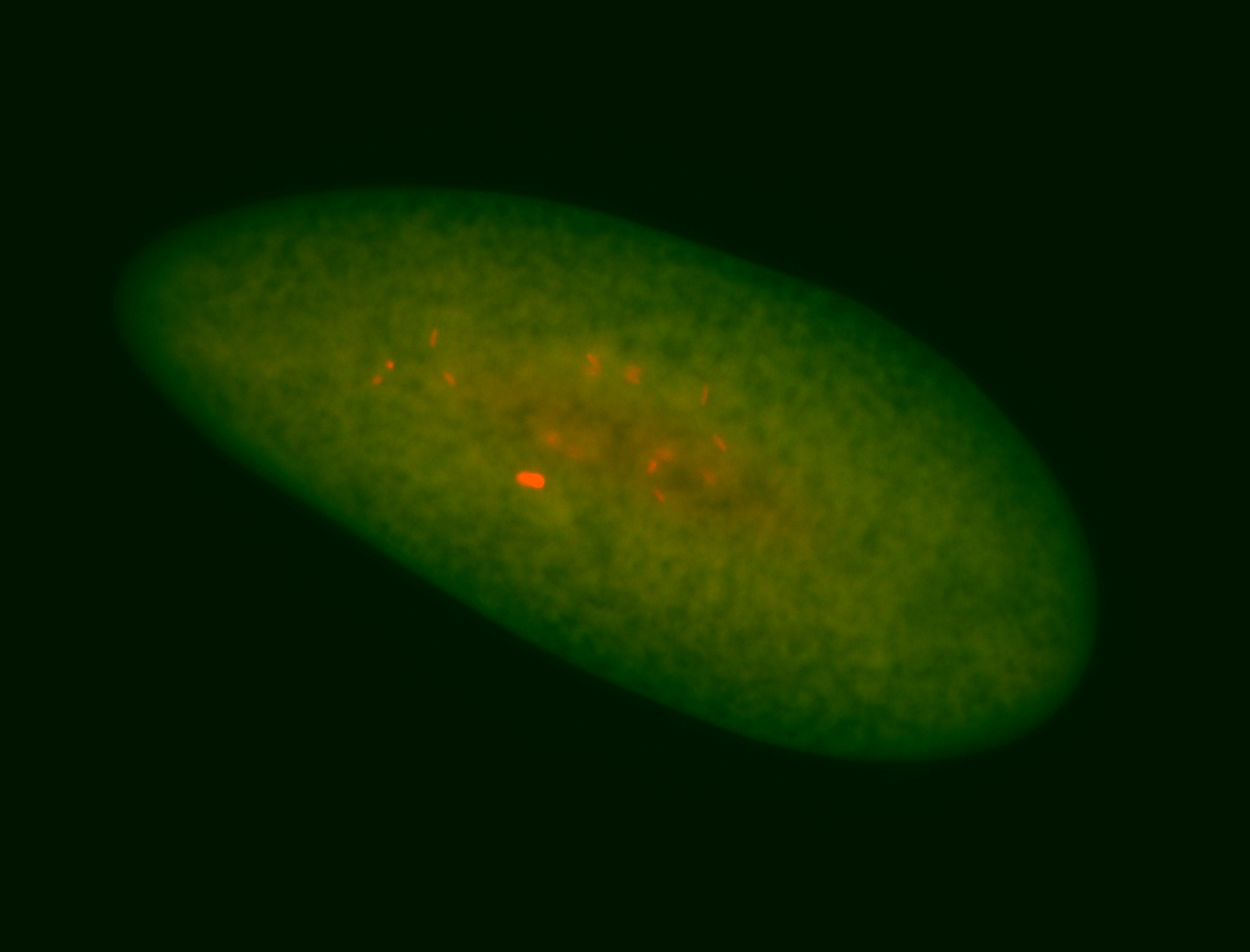
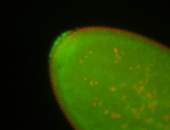
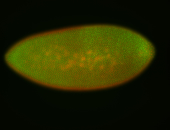
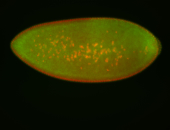
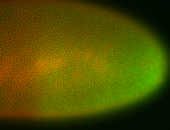
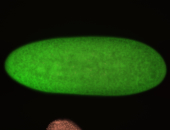
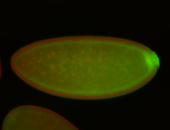
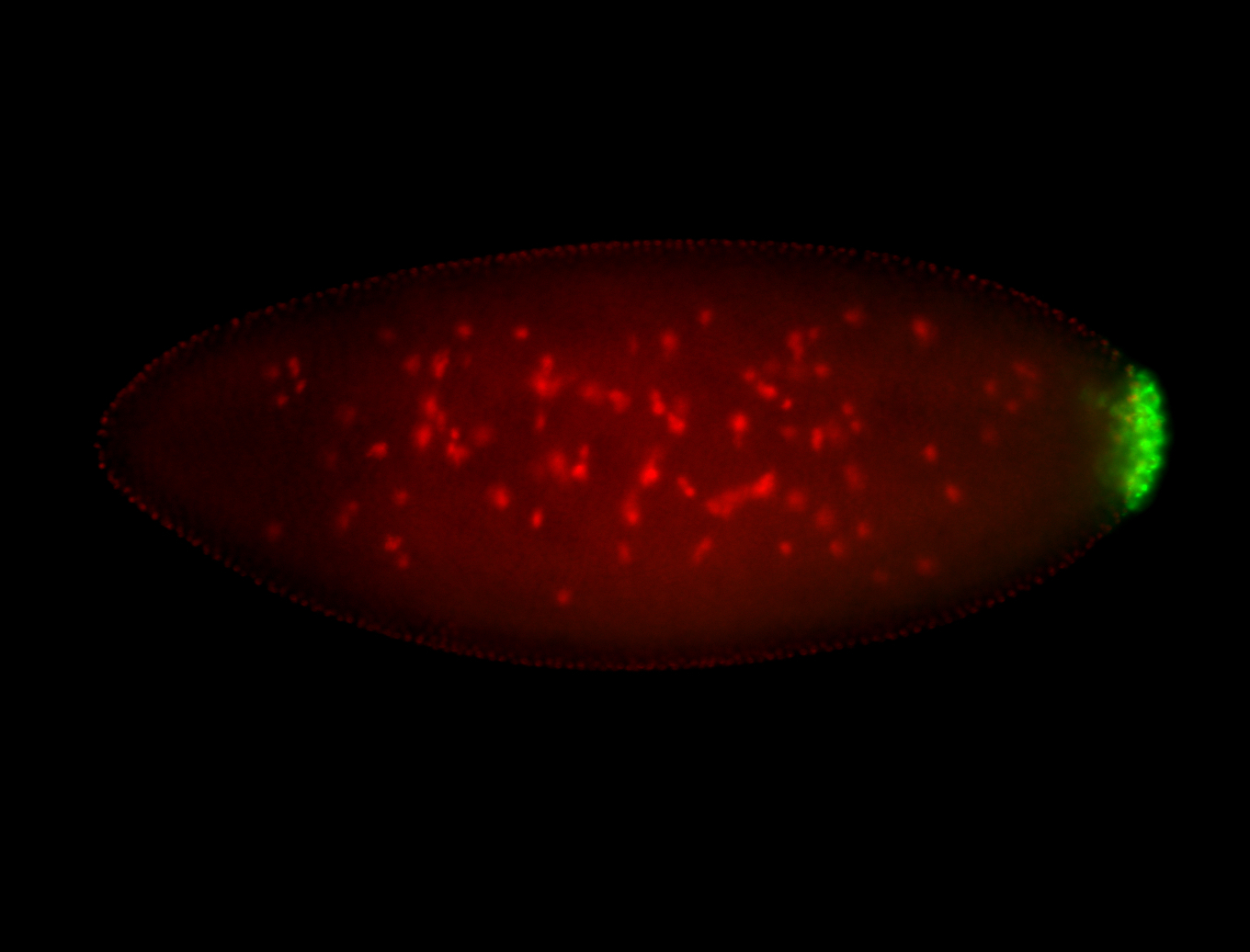
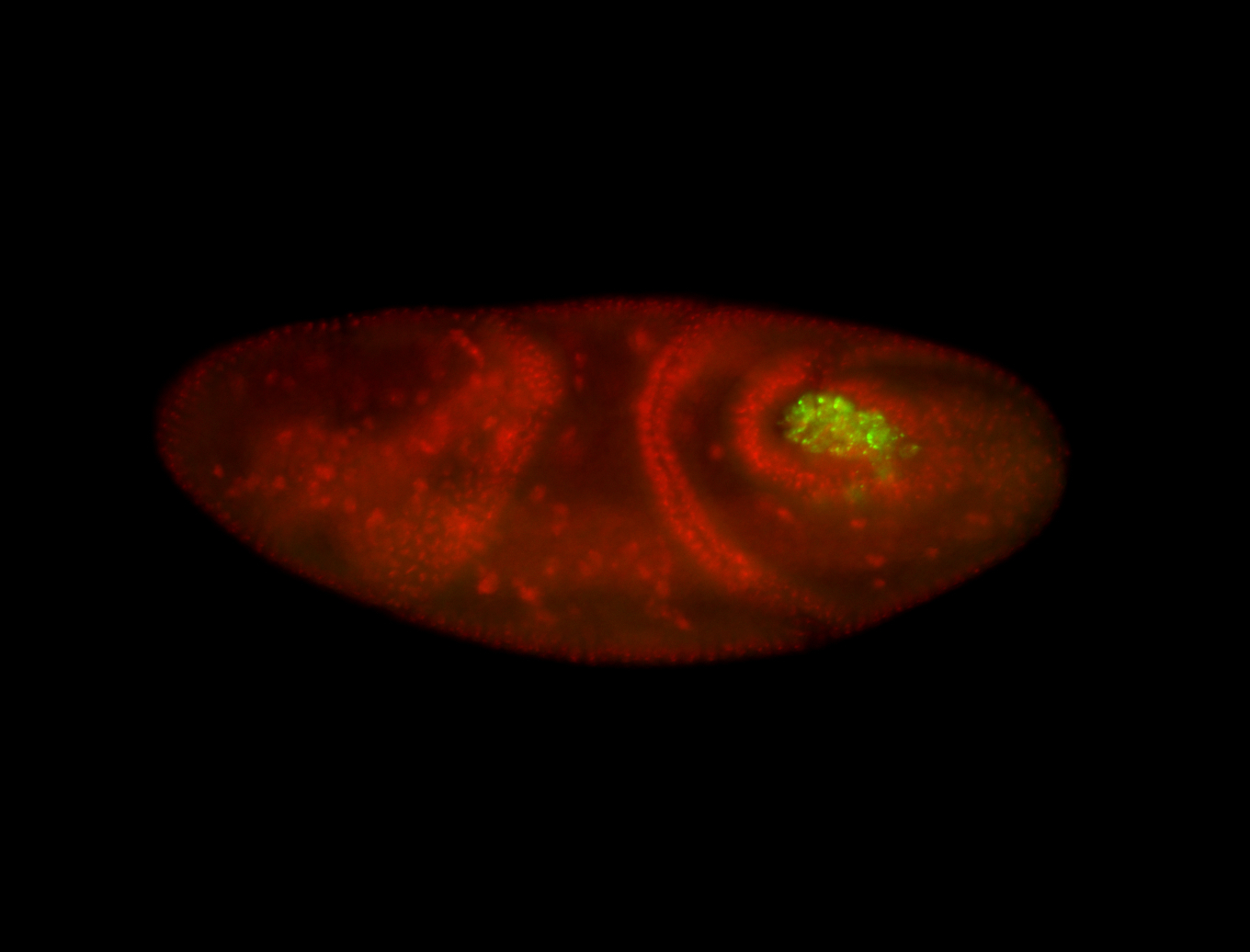
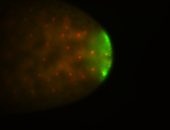
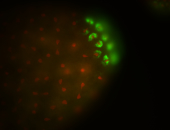
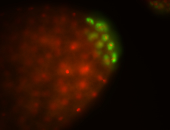
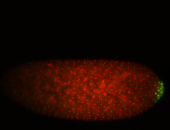
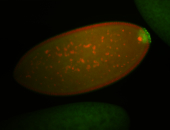
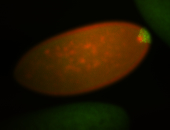
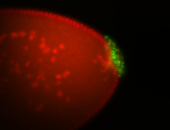
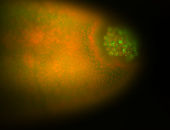
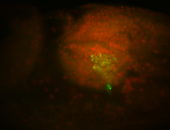
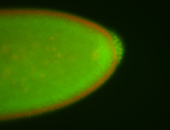
| Pole plasm enriched. Maybe zygotic in pole cells... not sure. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Partly nascent, partly nuclear subregion |
| Partly nascent, partly nuclear subregion. |
| Partly nascent, partly nuclear subregion |
| Salivary glands, probably non-specific. |
| Good probe on gel. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe on gel |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
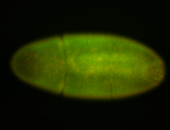
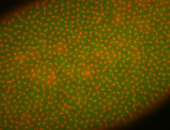
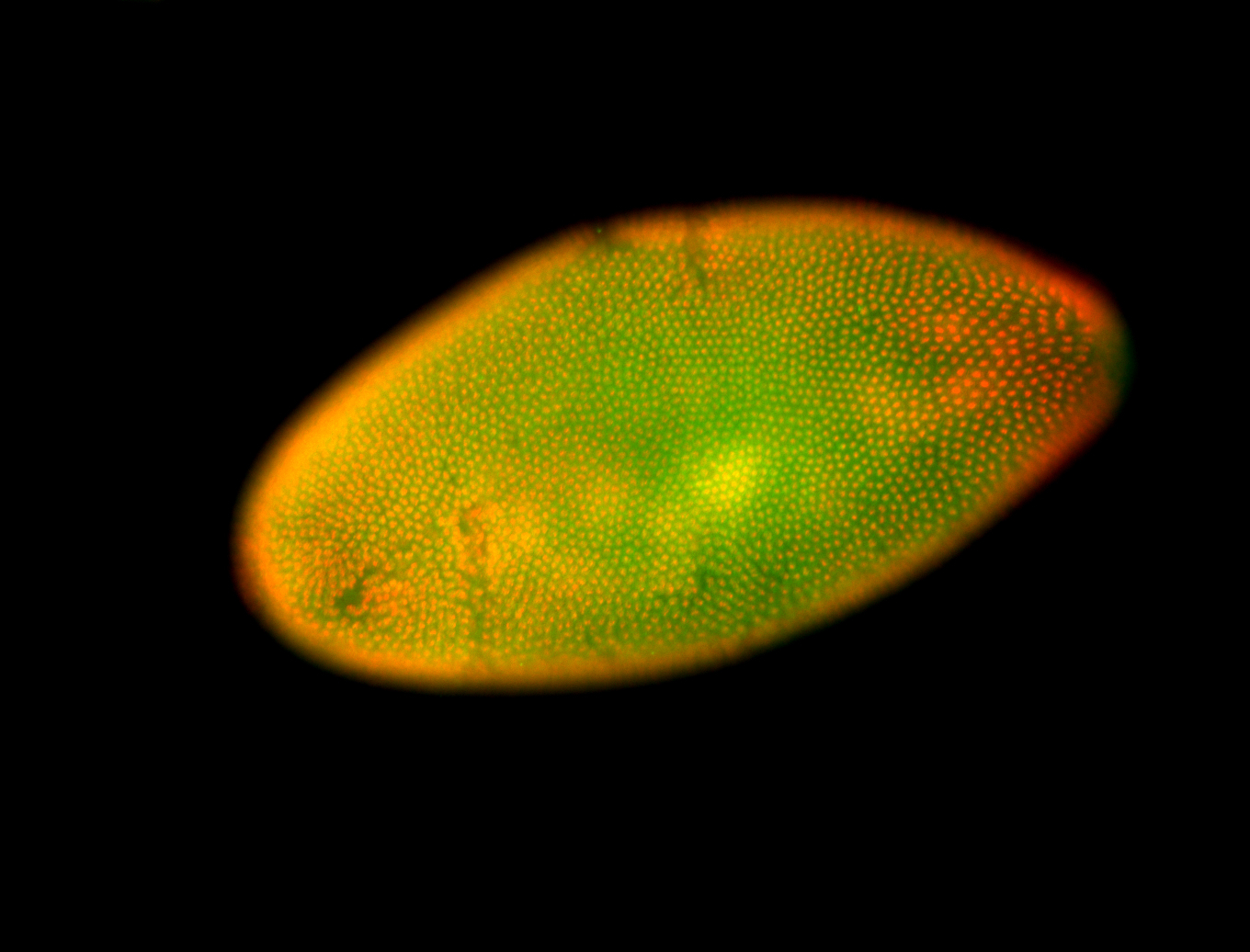
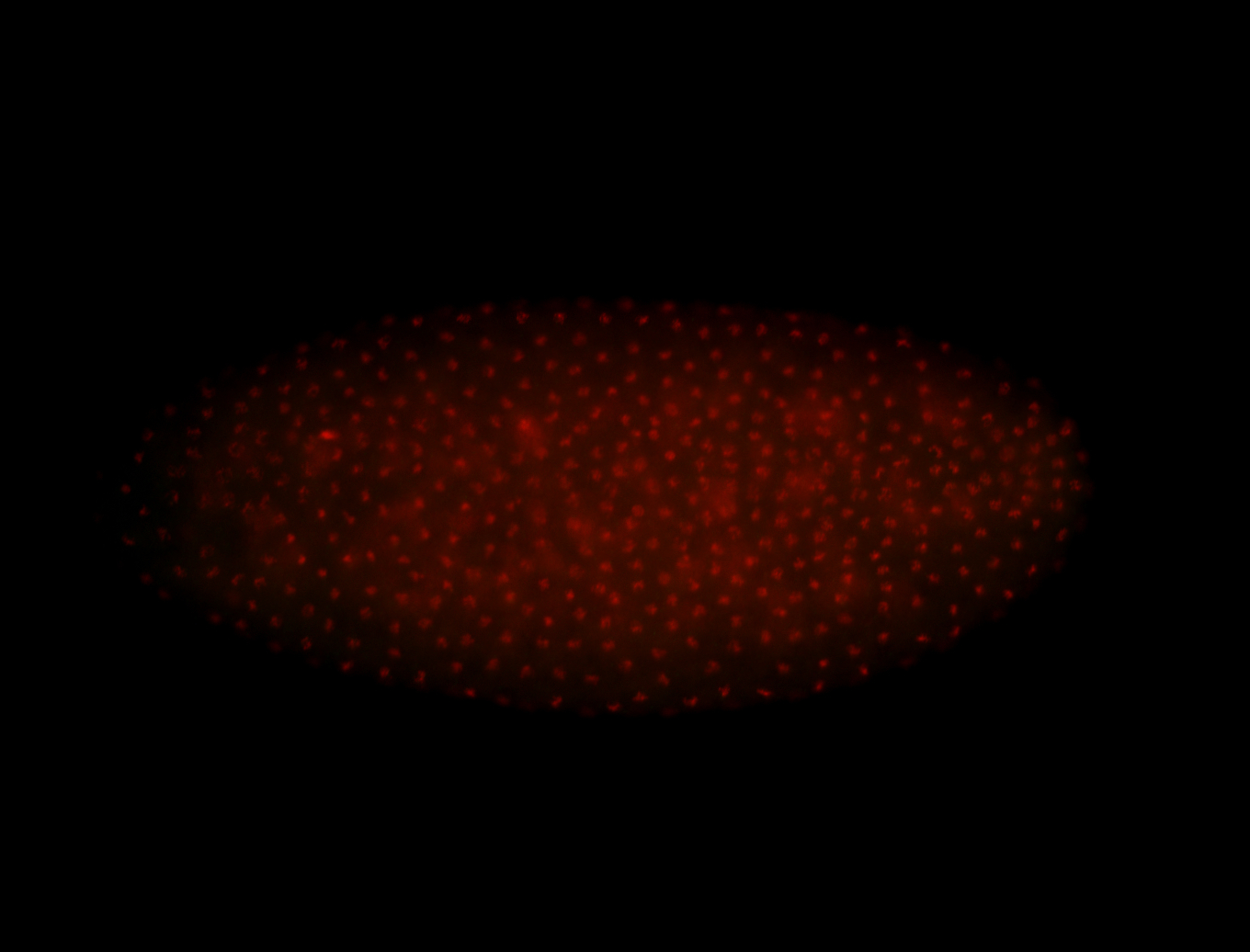
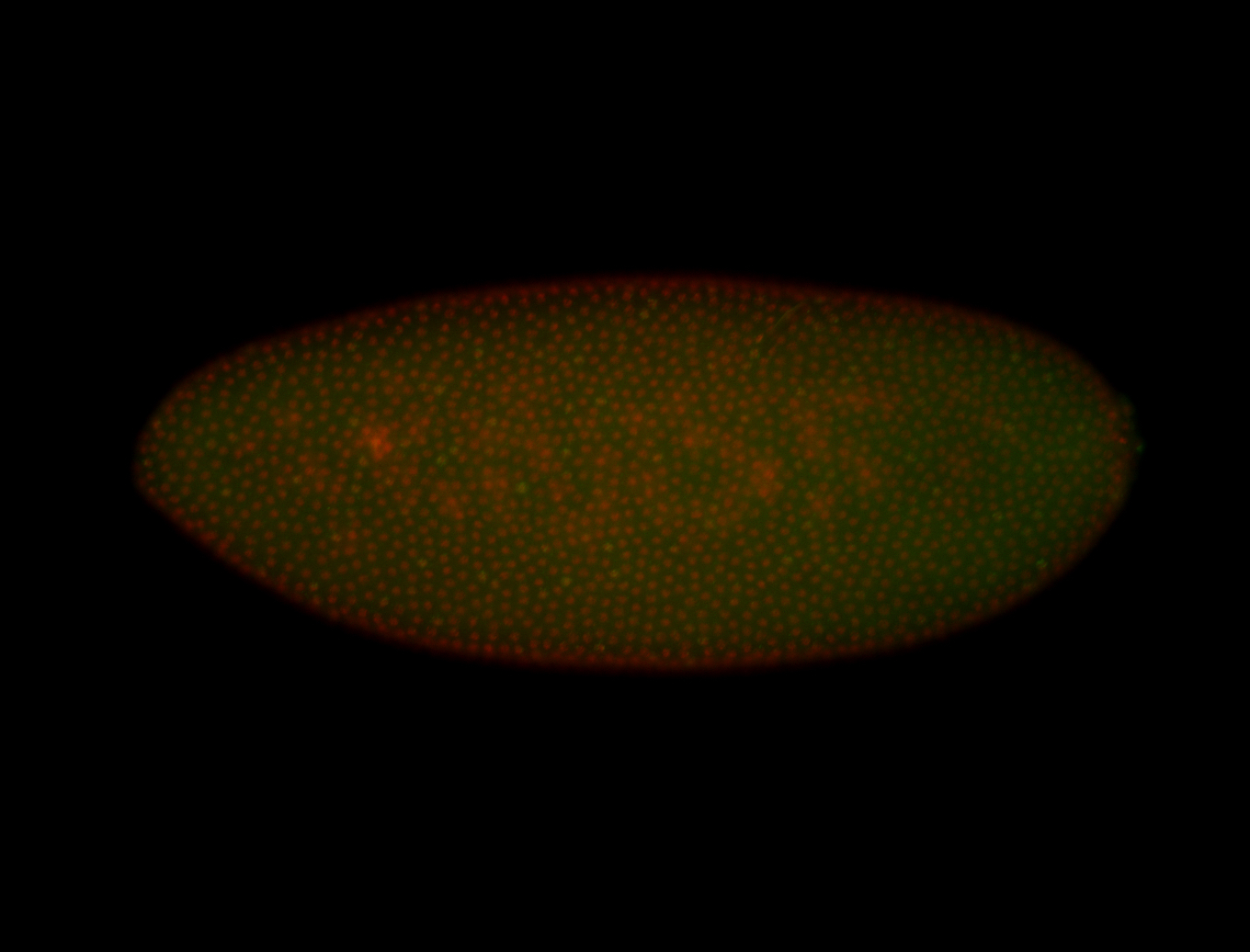
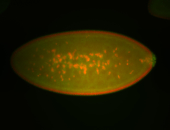
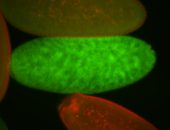
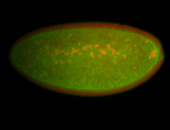
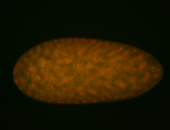
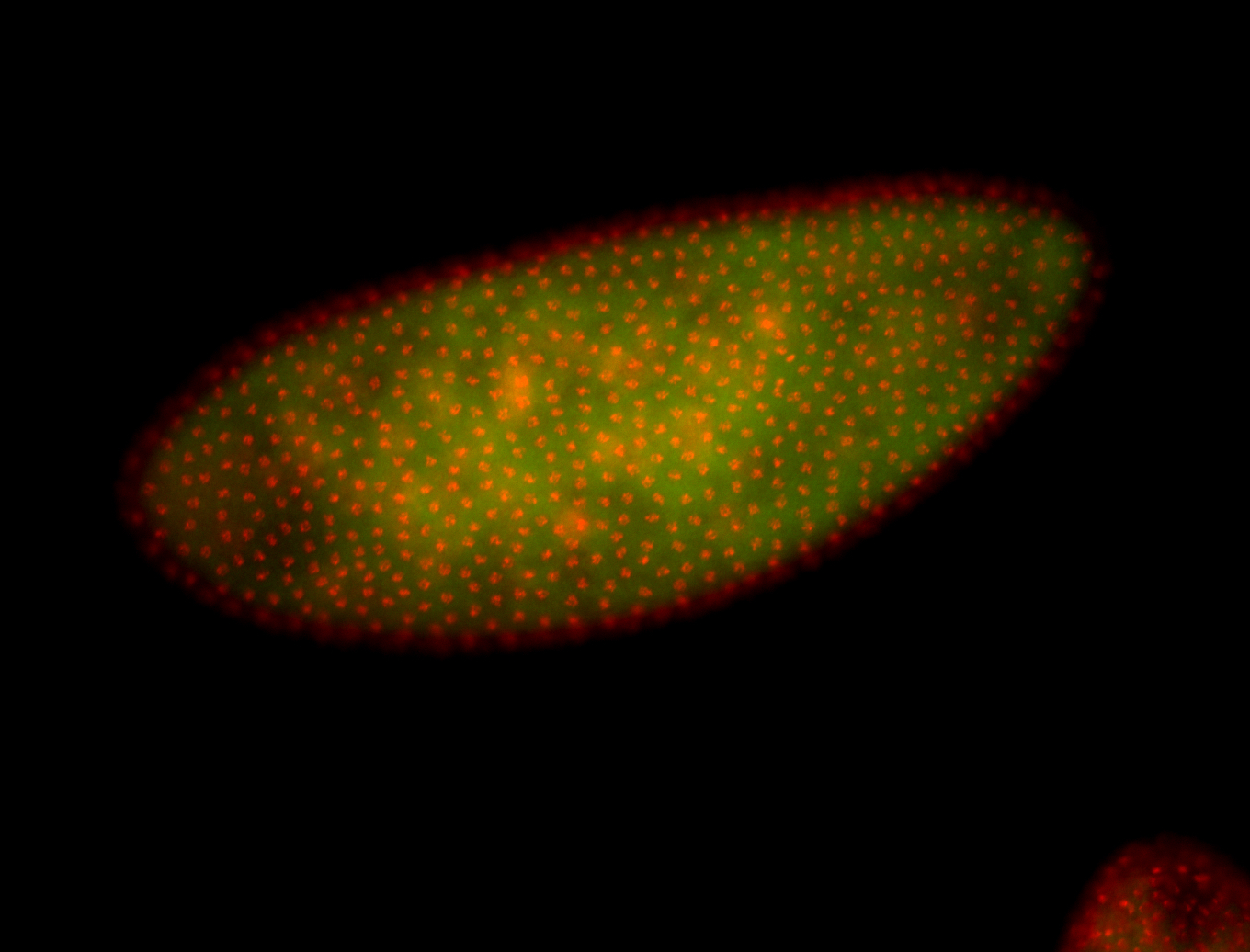
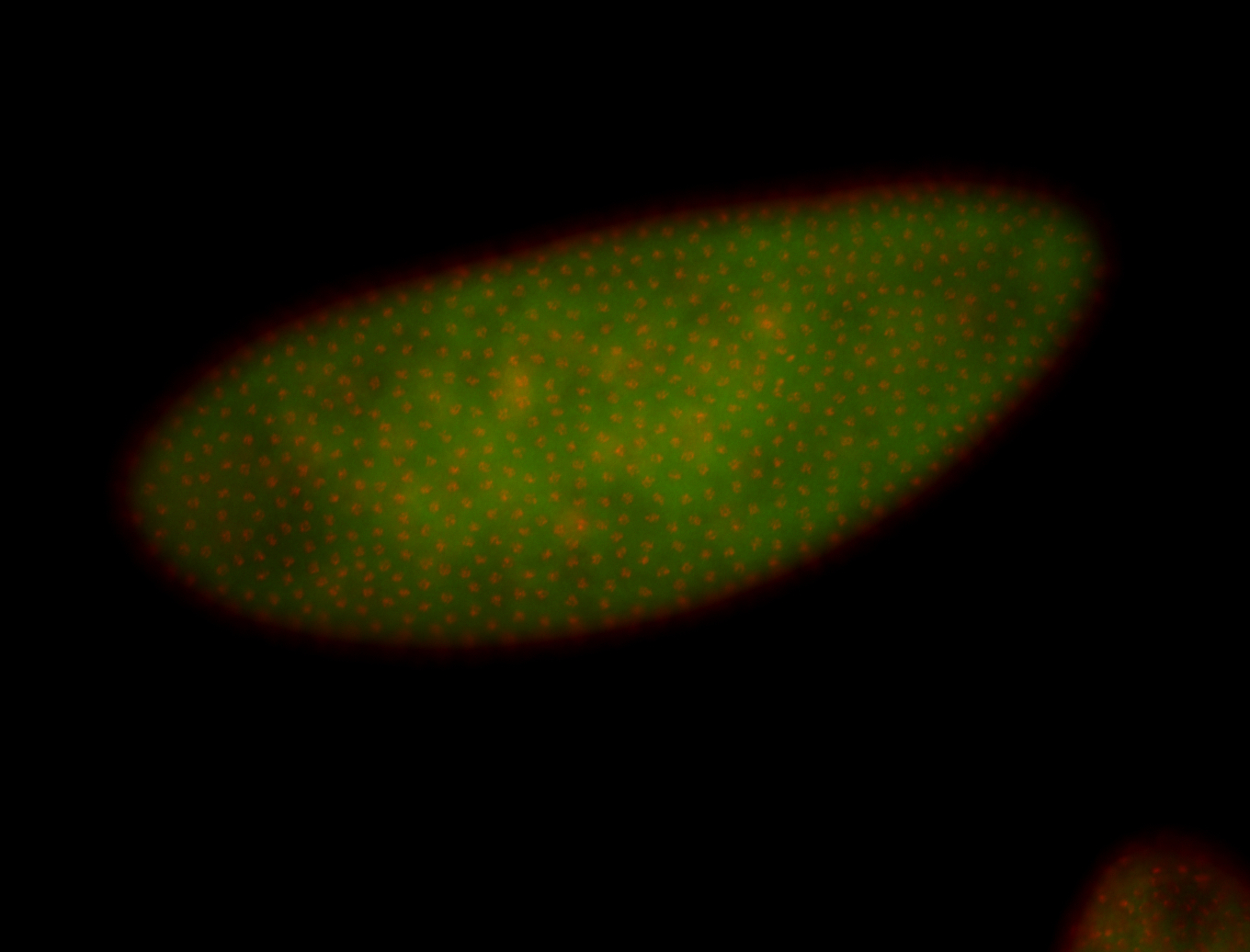
| 84 ms I think it is all zygotic, nothing sub-cellular (?). I wish we had 40x images. |
| 84 ms |
| 84 ms |
| 84 ms Re-annotated by Ronit- 2014. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe on gel. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Re-annotated |
| Need more photos for a better description. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
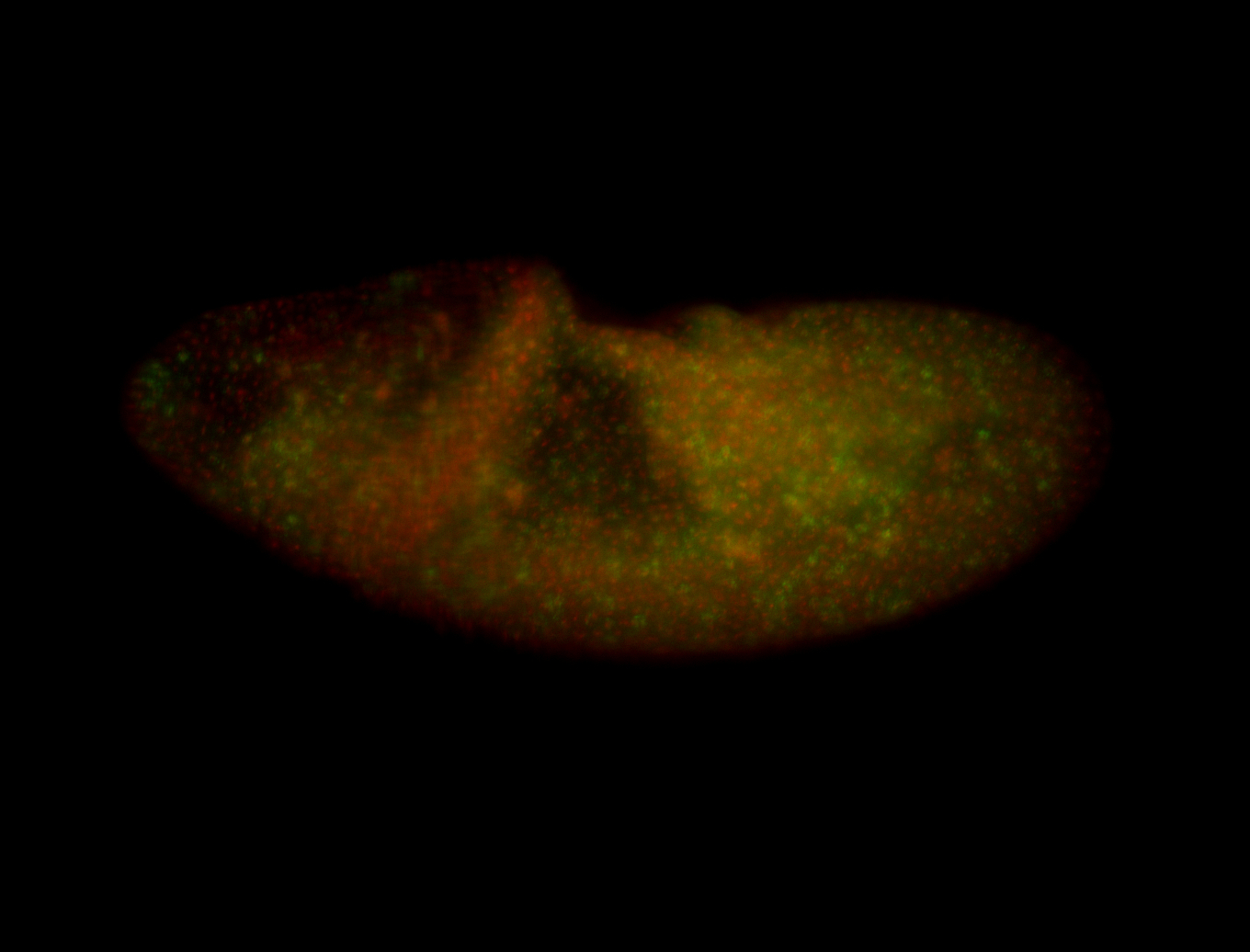
| Posterior 1/2 |
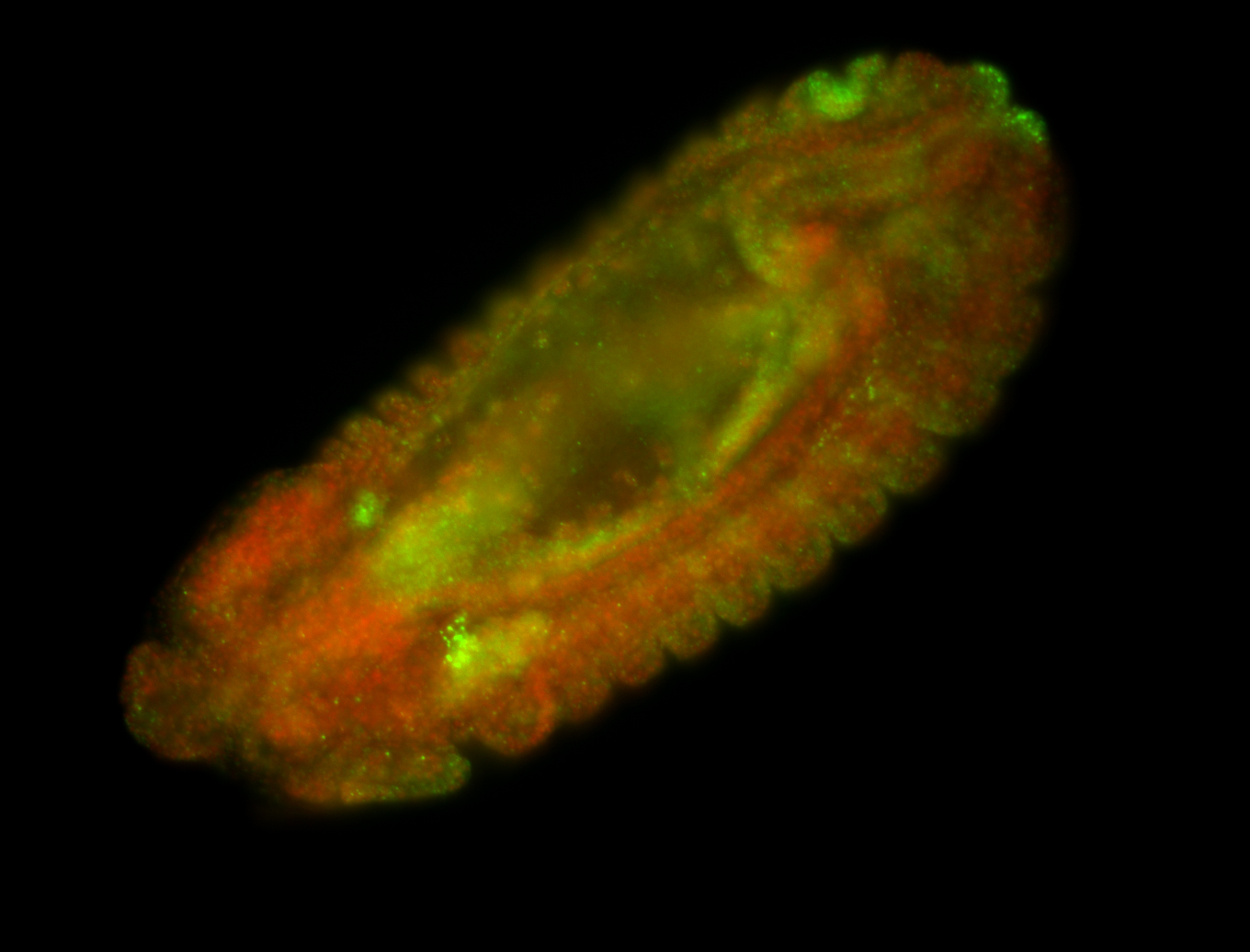
| Tracheal lumen, probably non-specific. Need better fotos to determine sub-cellular localization. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Low probe on gel, good insitu reaction. |
| Anterior and posterior midgut expression. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
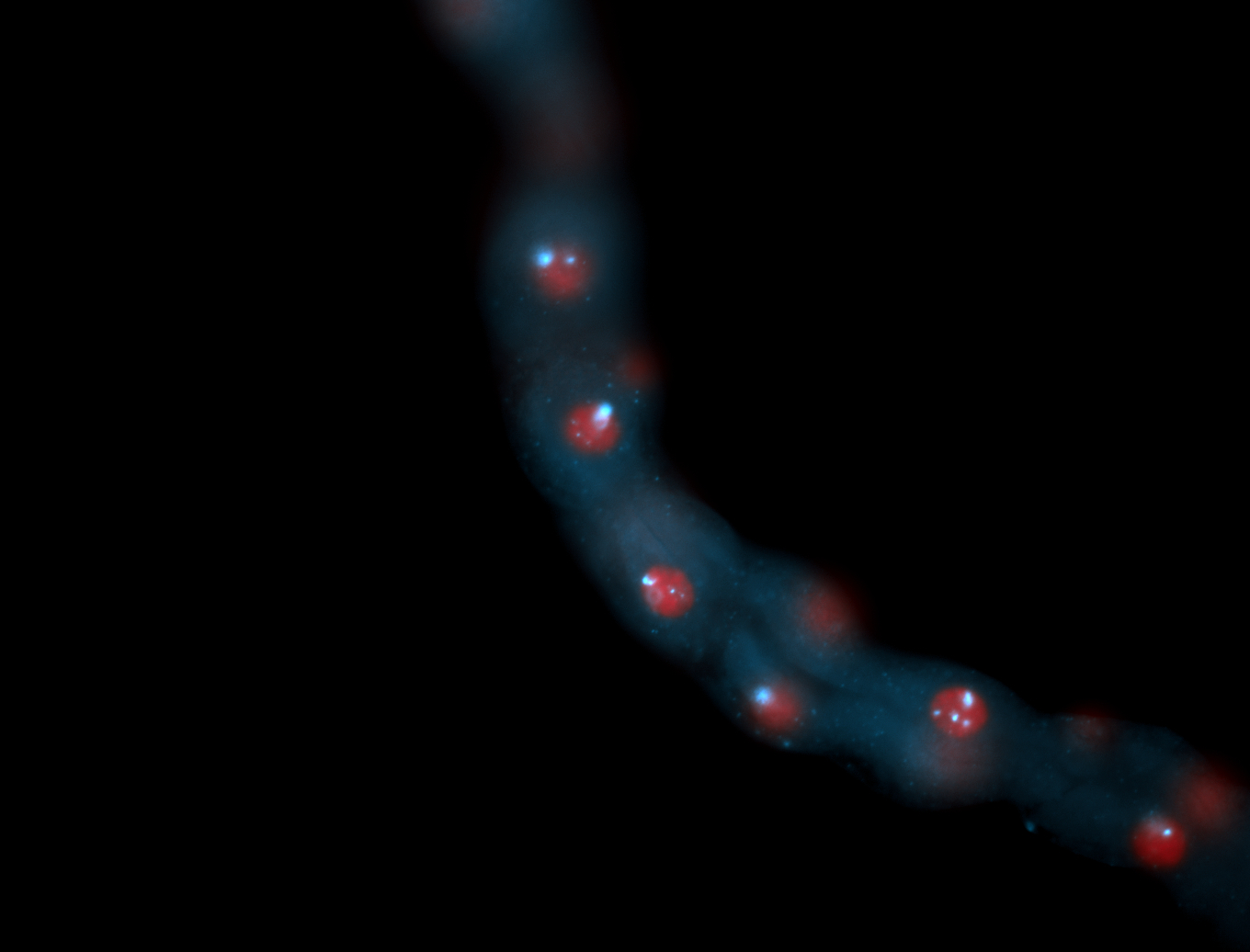
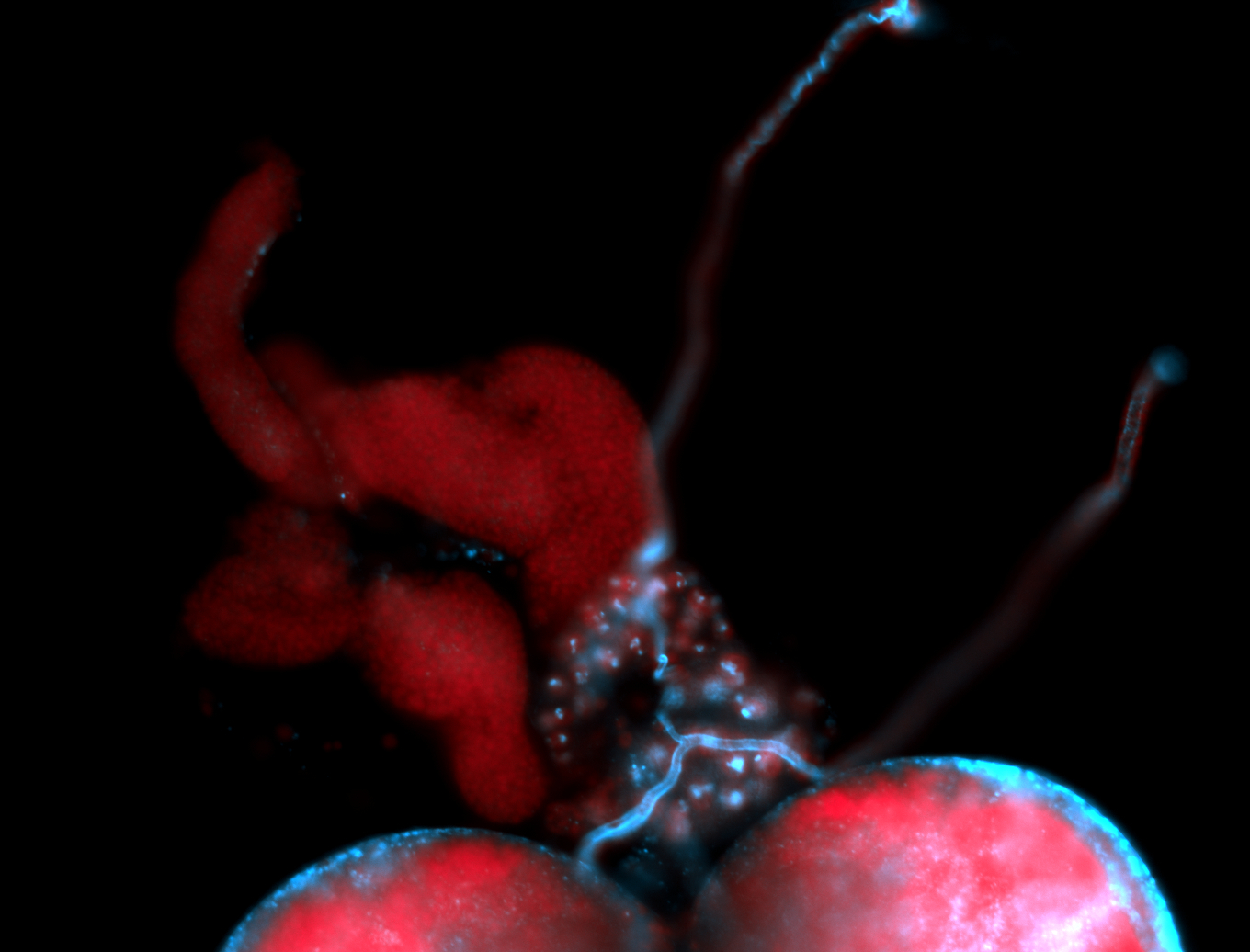
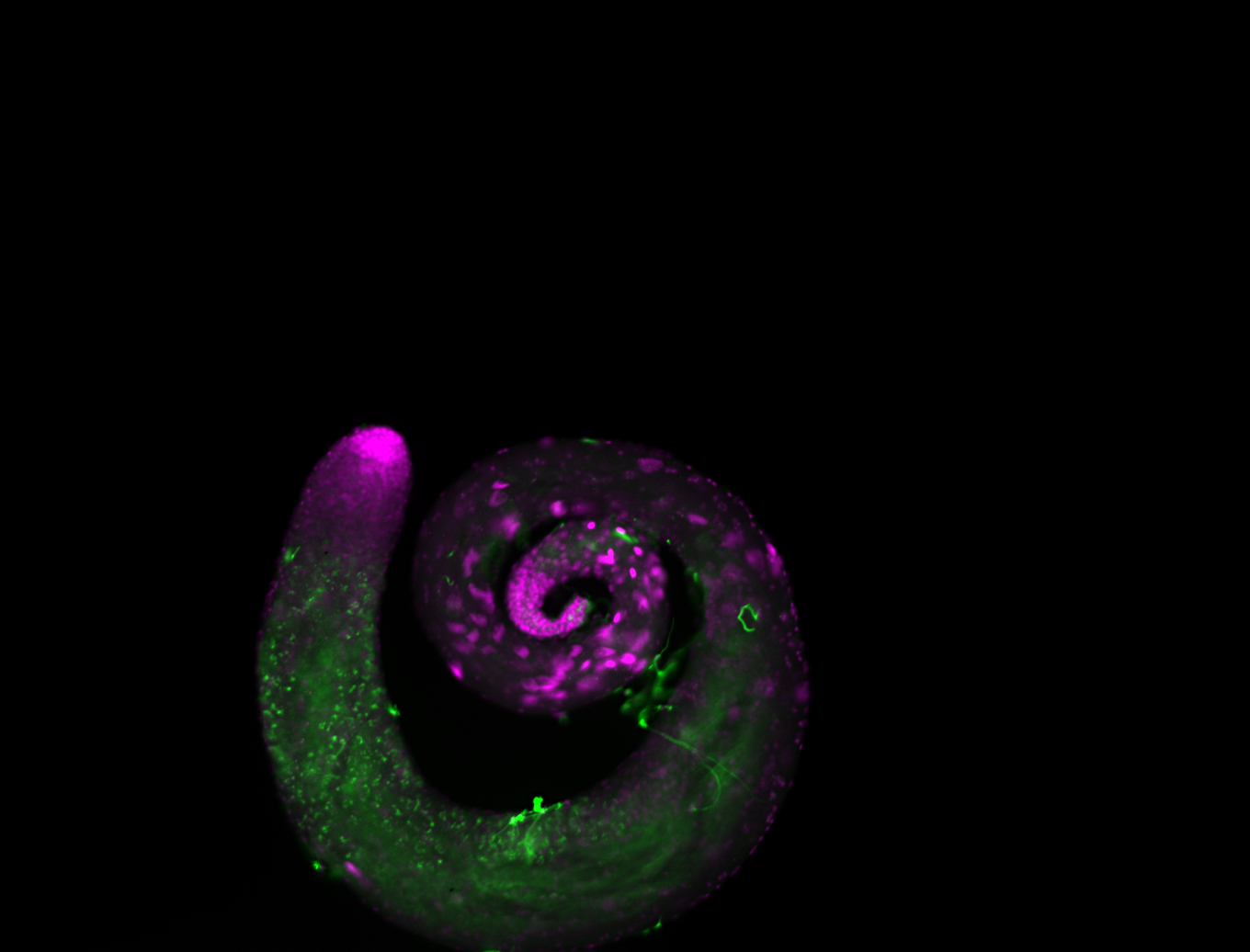
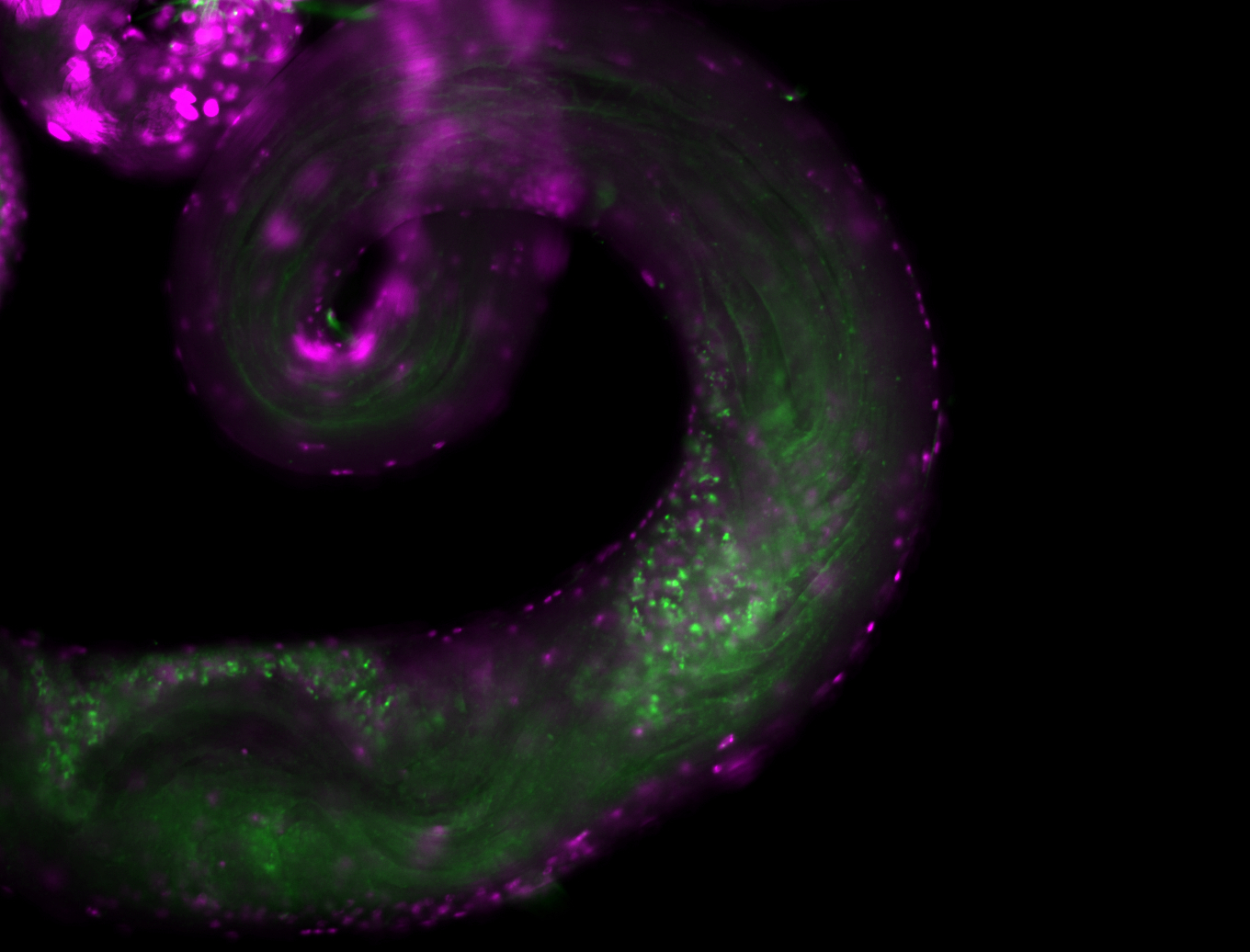
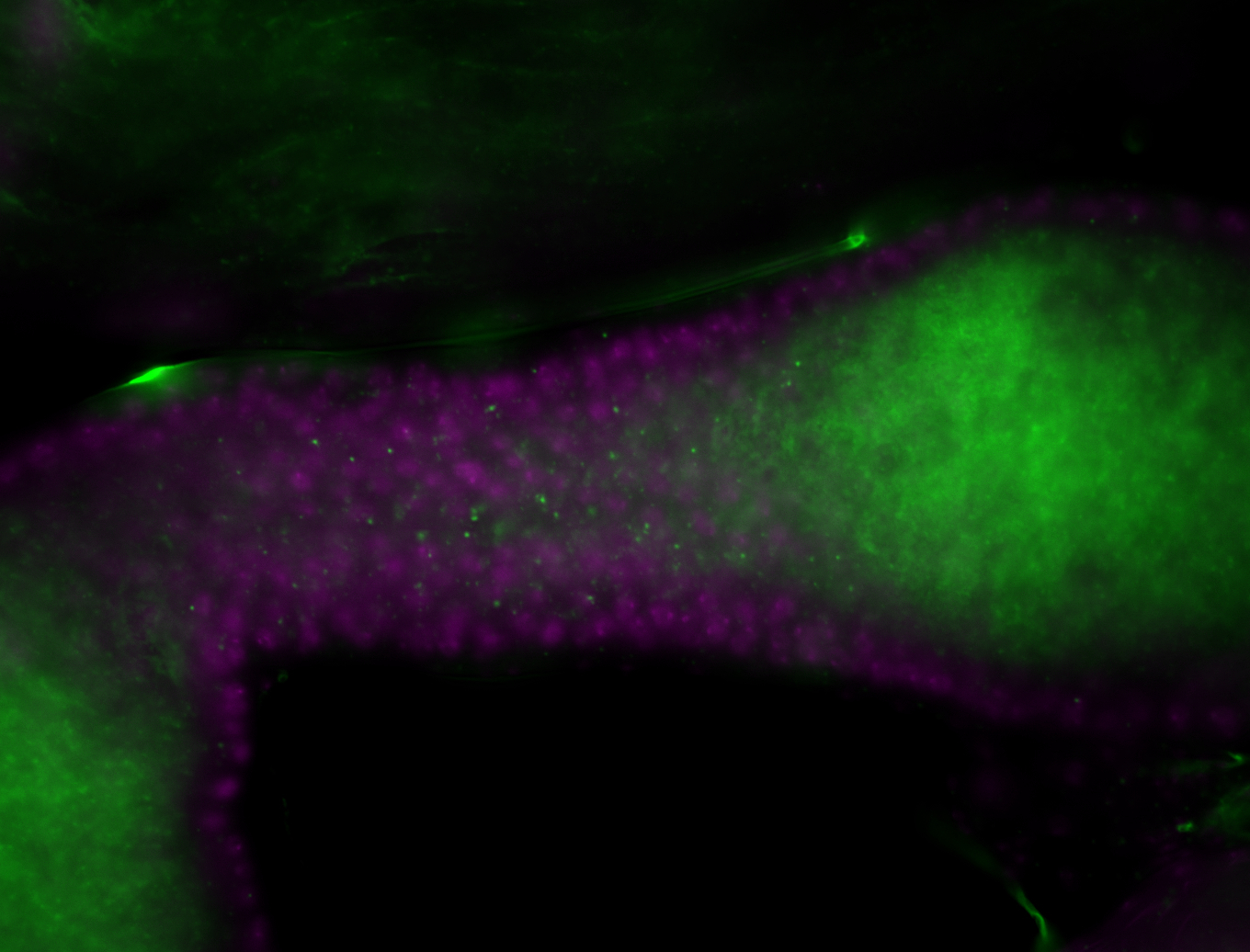
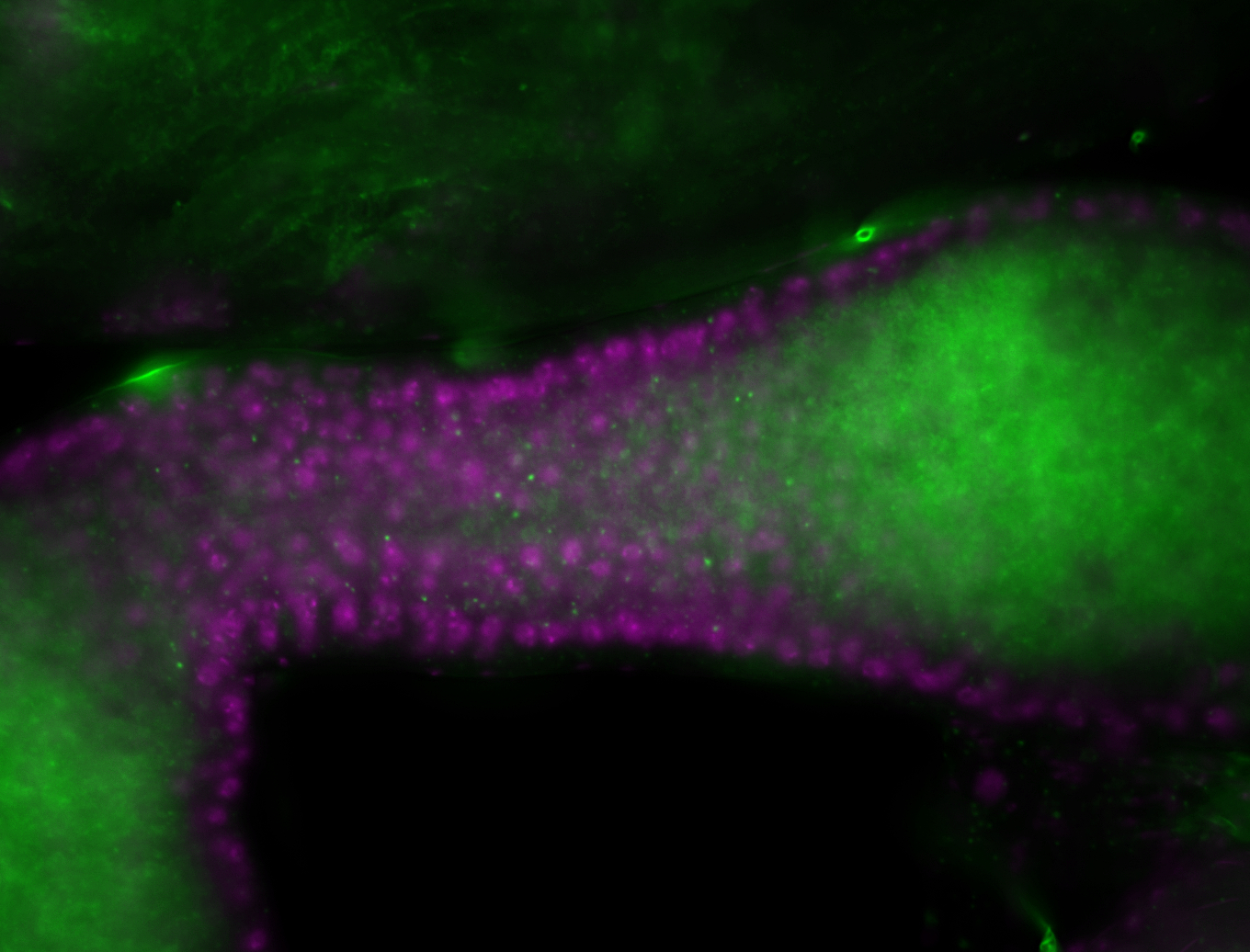
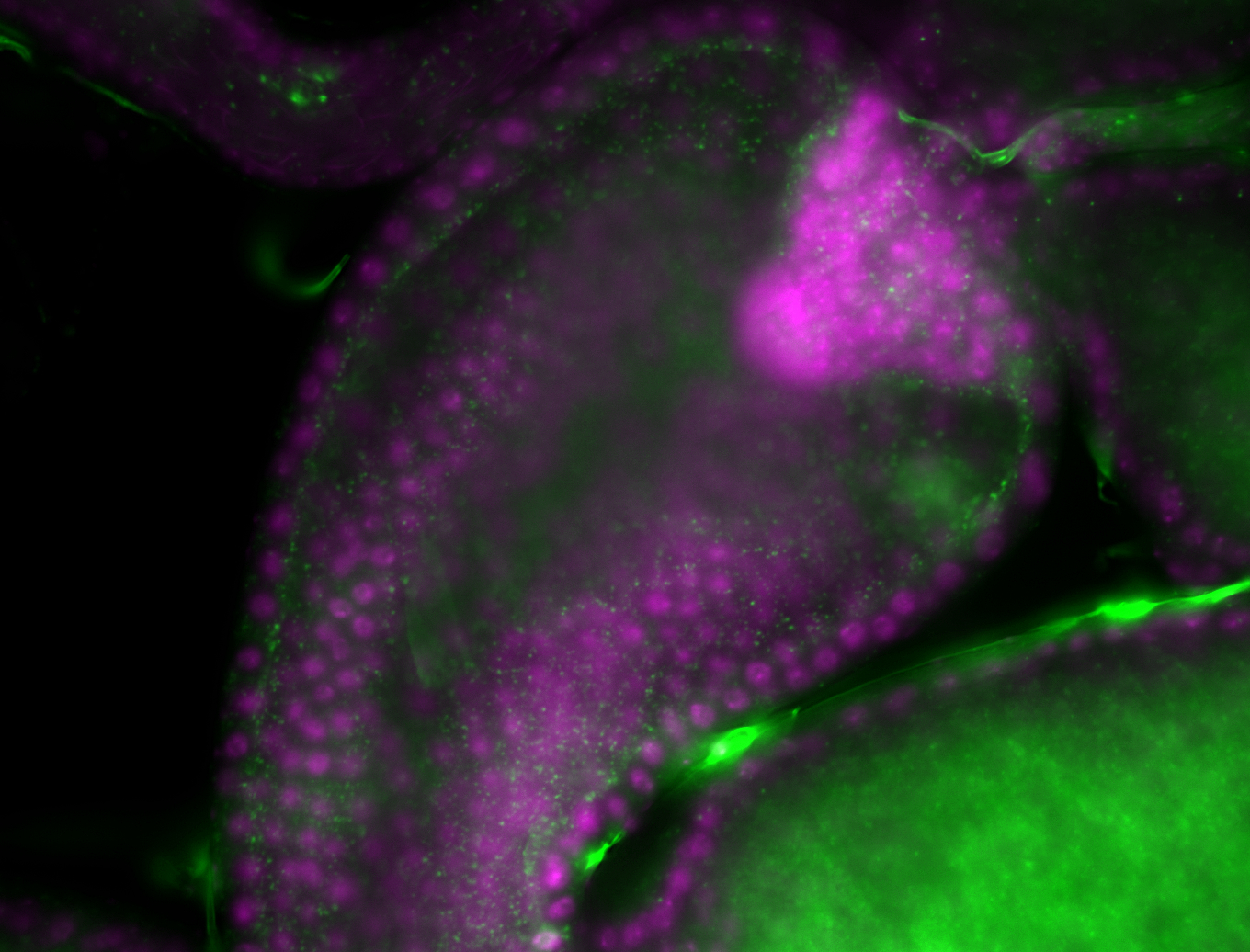
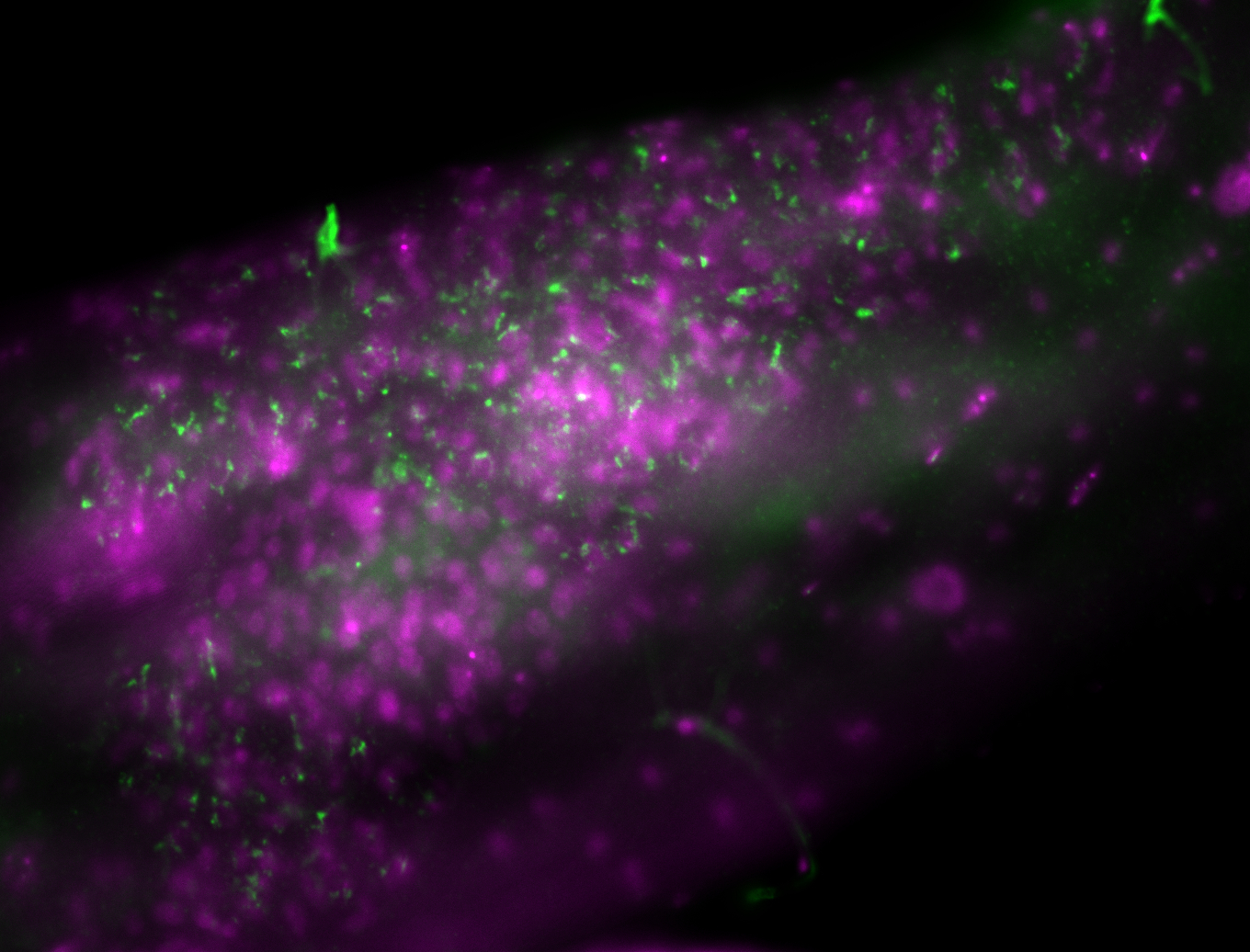
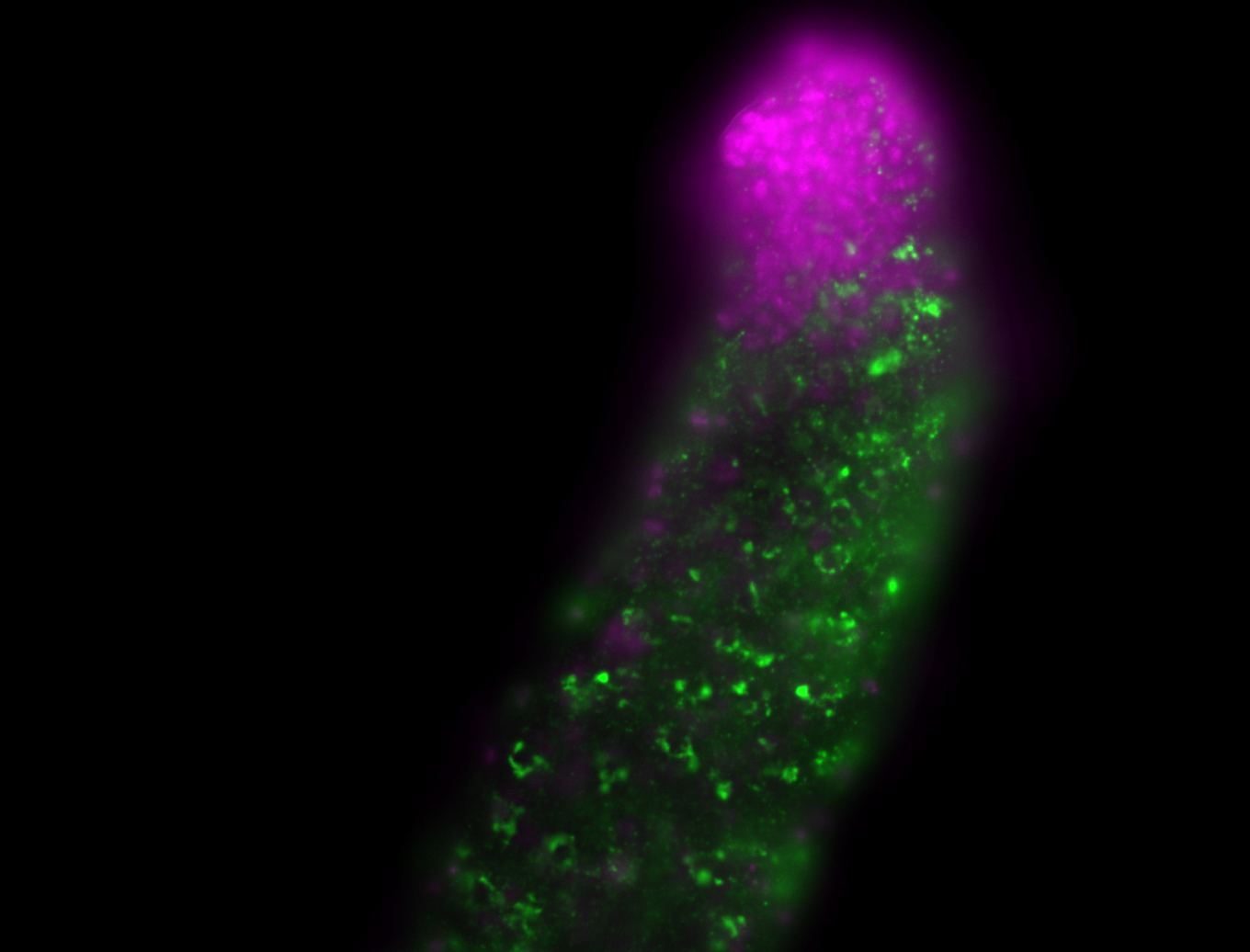
| -performed via HCR-FISH using 3 probe sets targeting the RB transcript isoform |
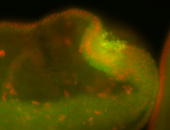
| -interesting subcellular localization patterns in the pole/germ cells; makes a great marker for tracking the development of germ line cells throughout the entirety of embryogenesis |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
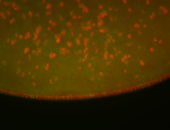
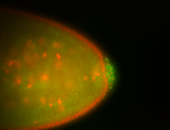
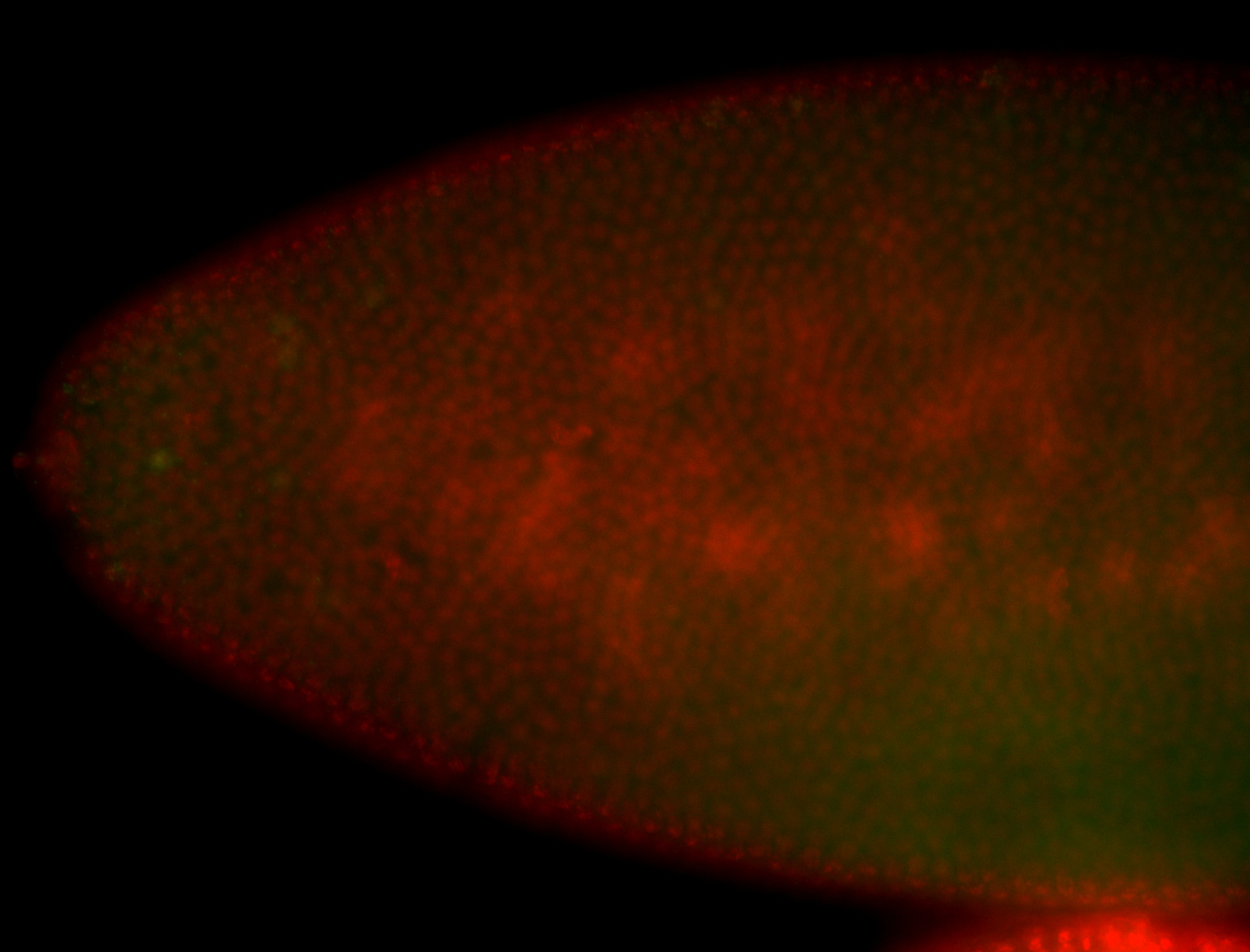
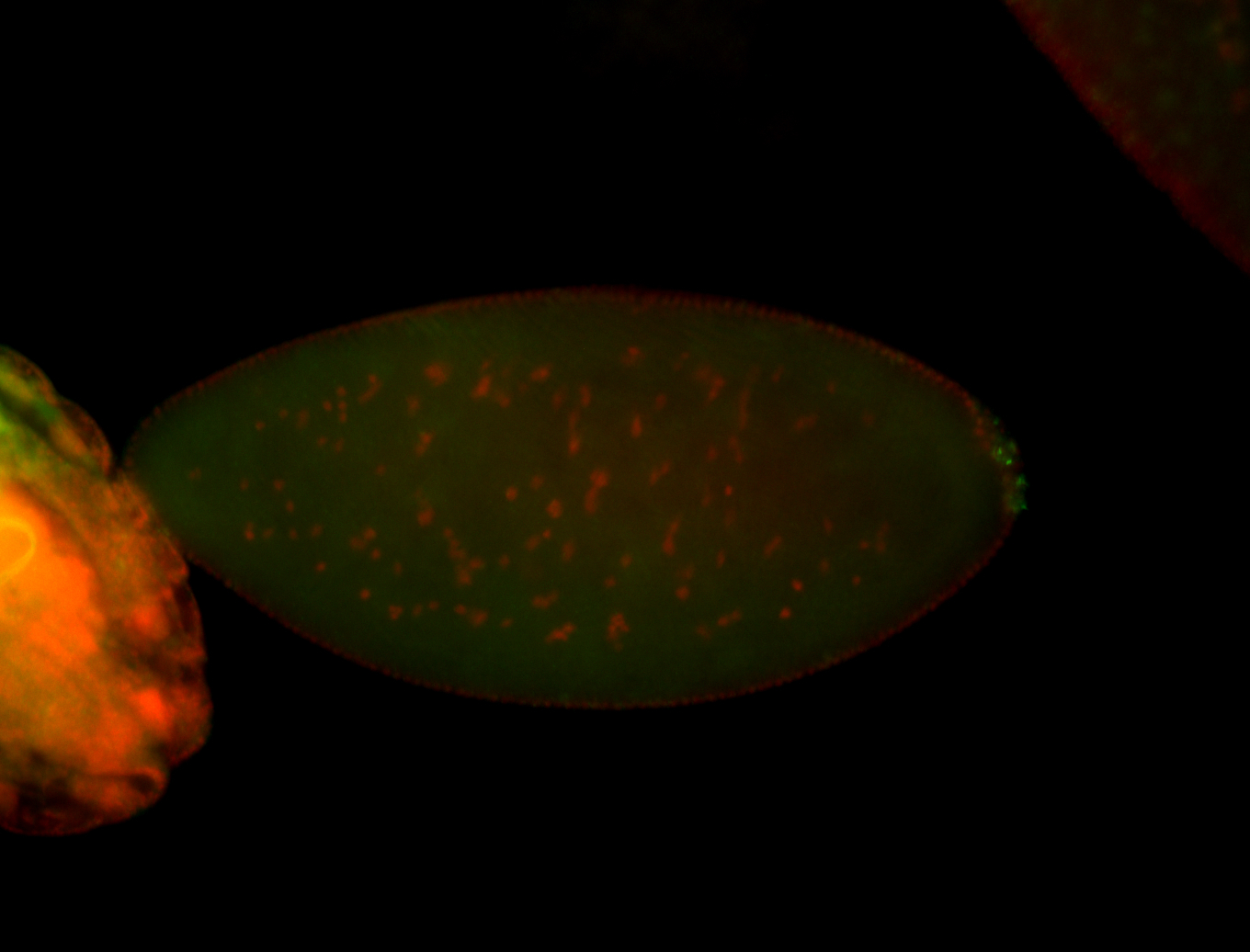
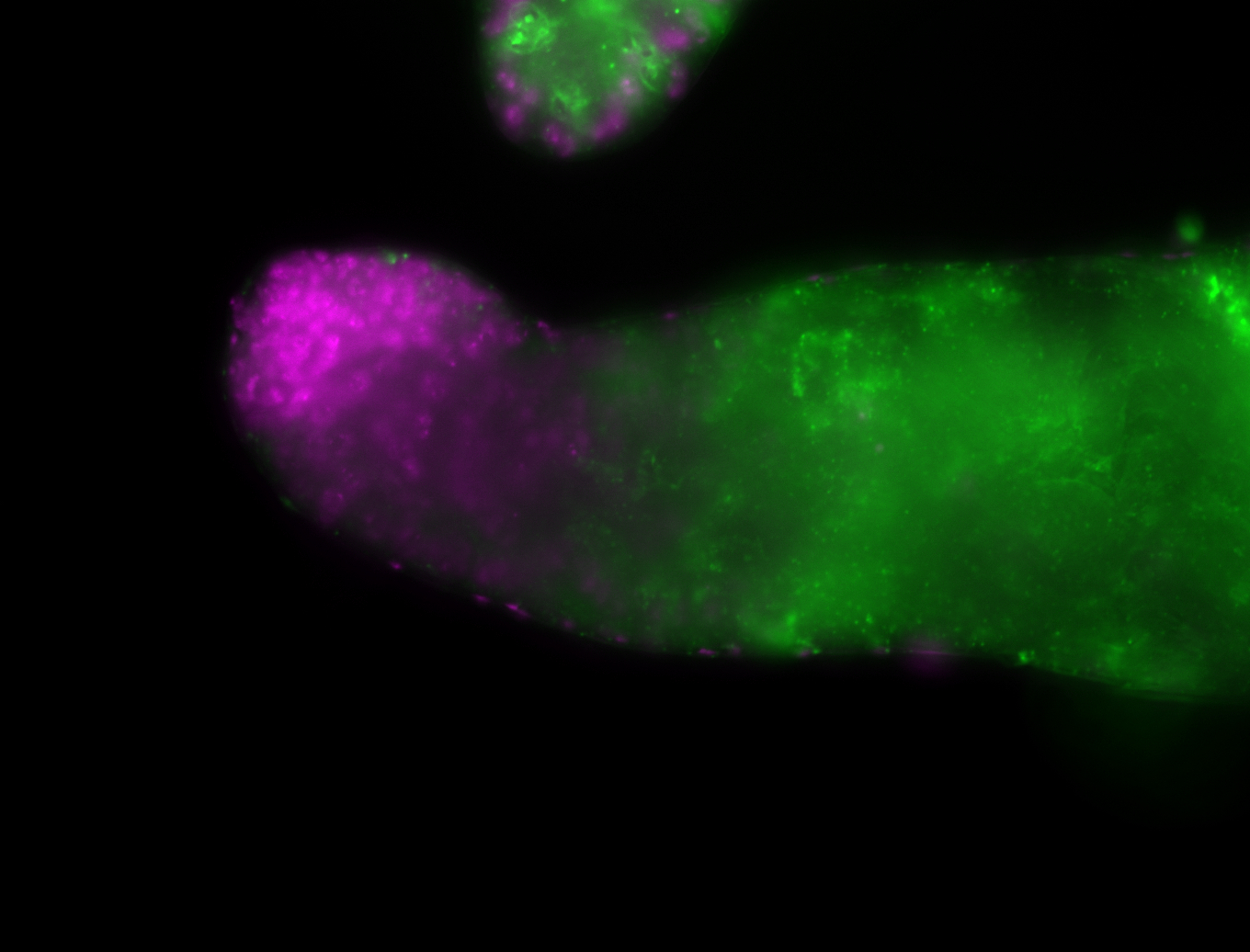
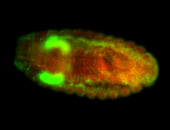
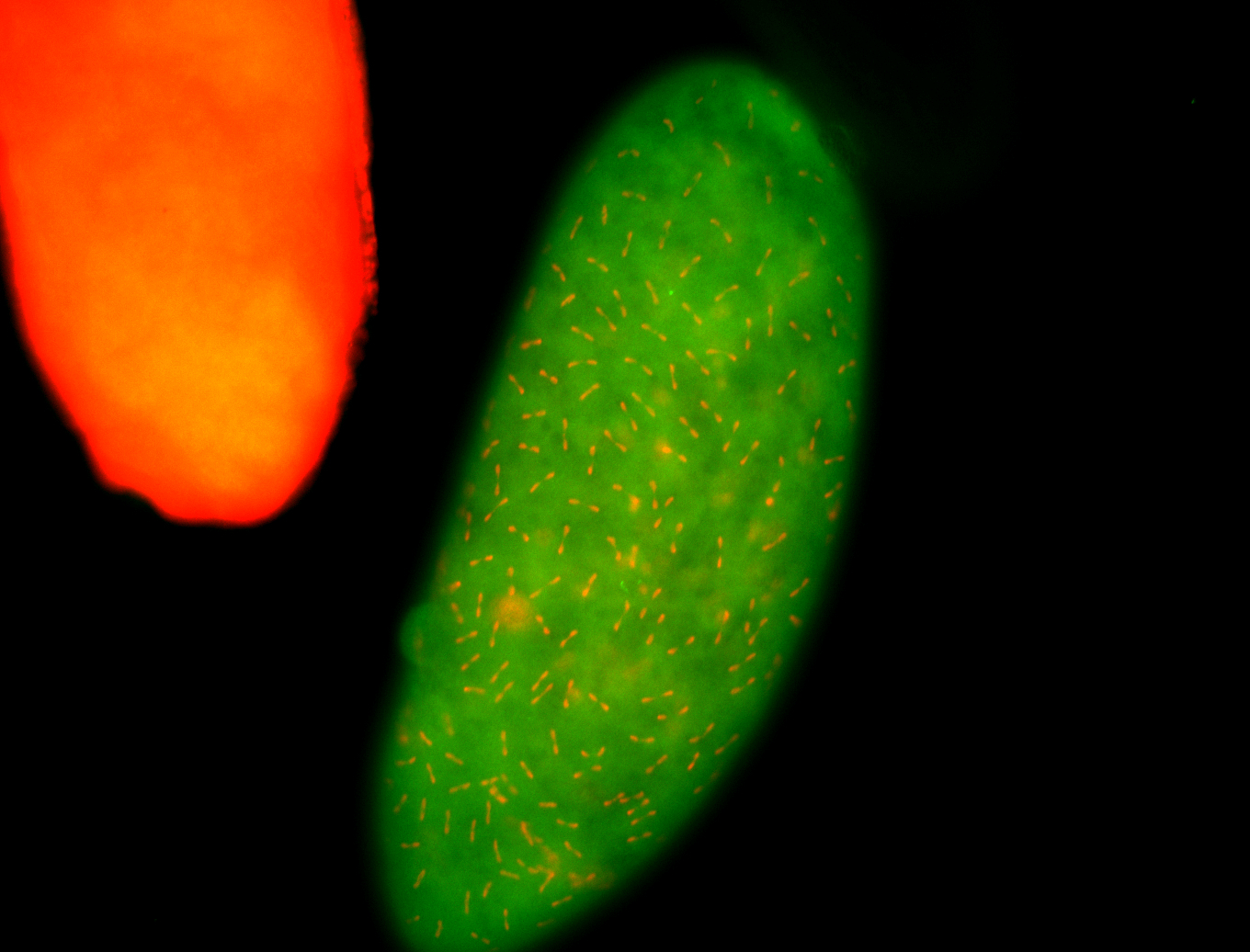
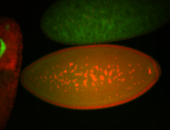
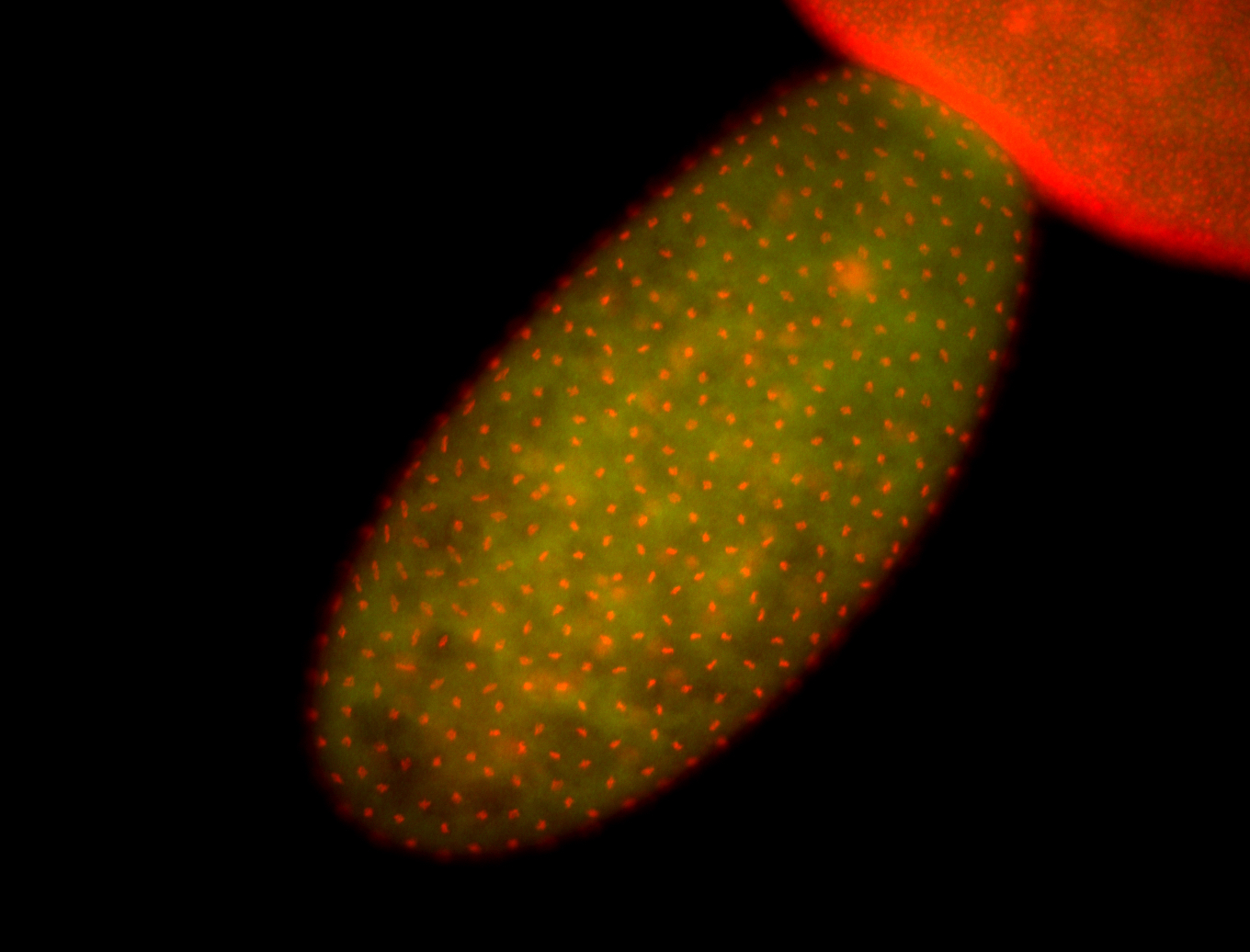
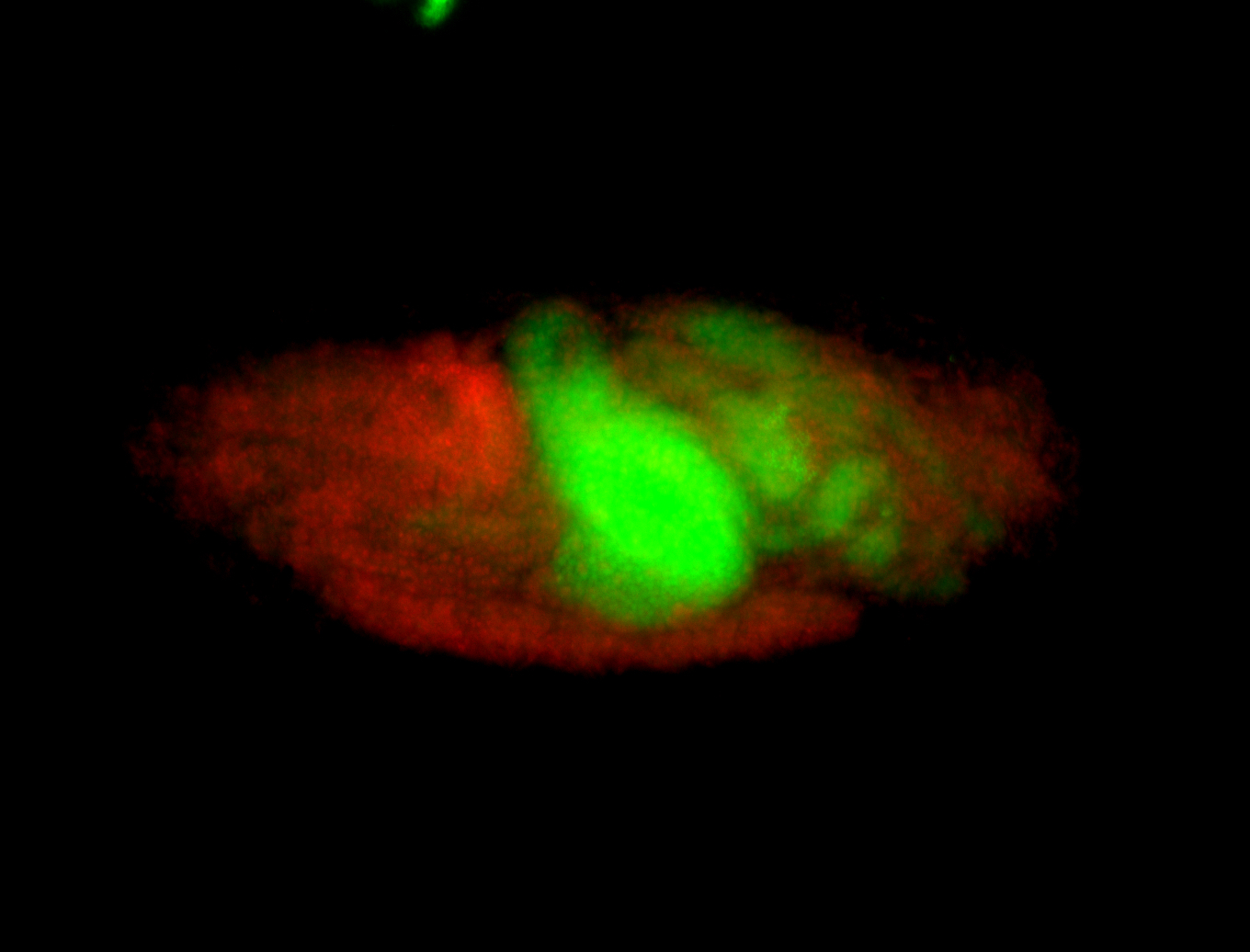
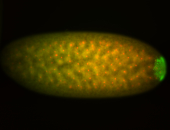
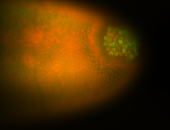
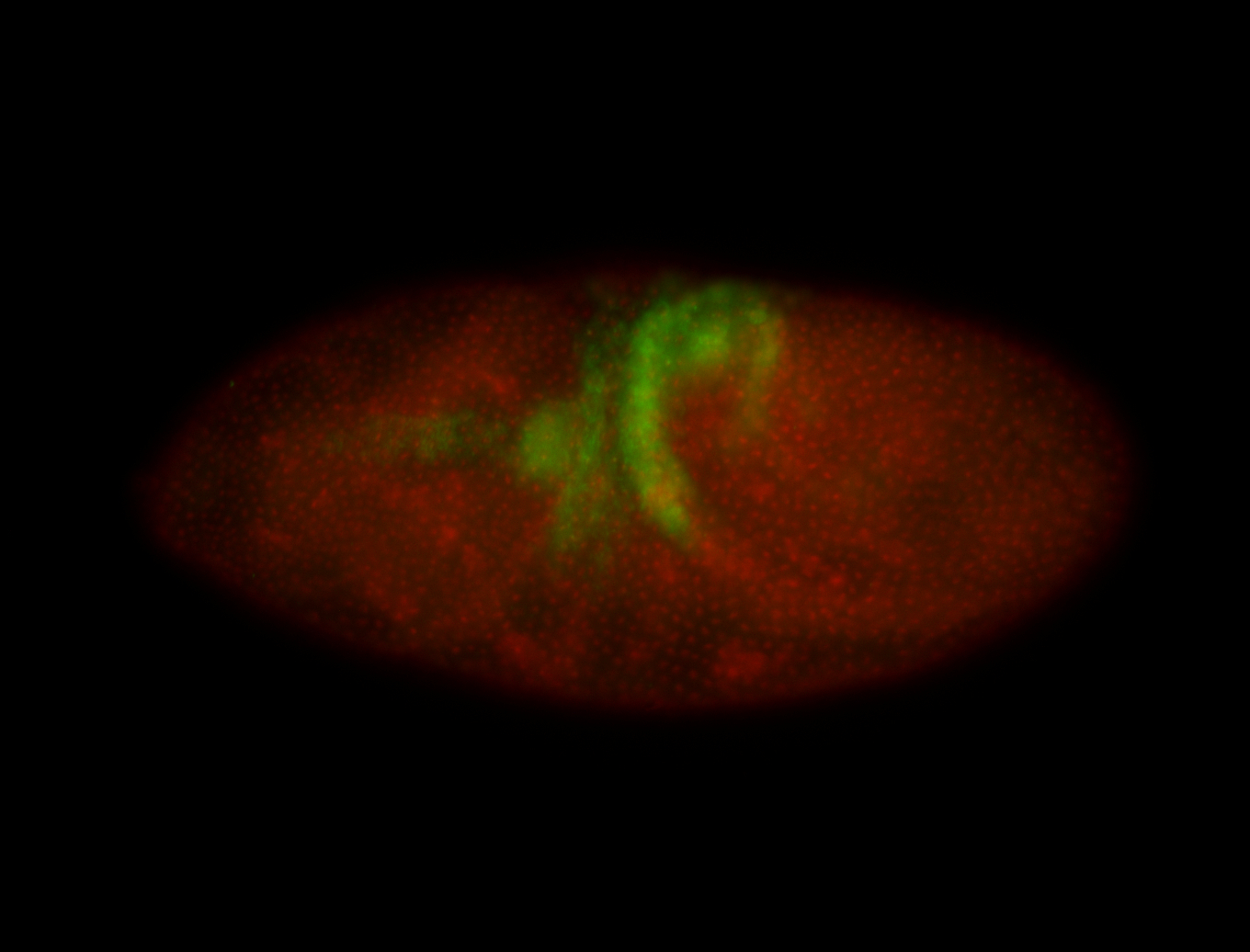
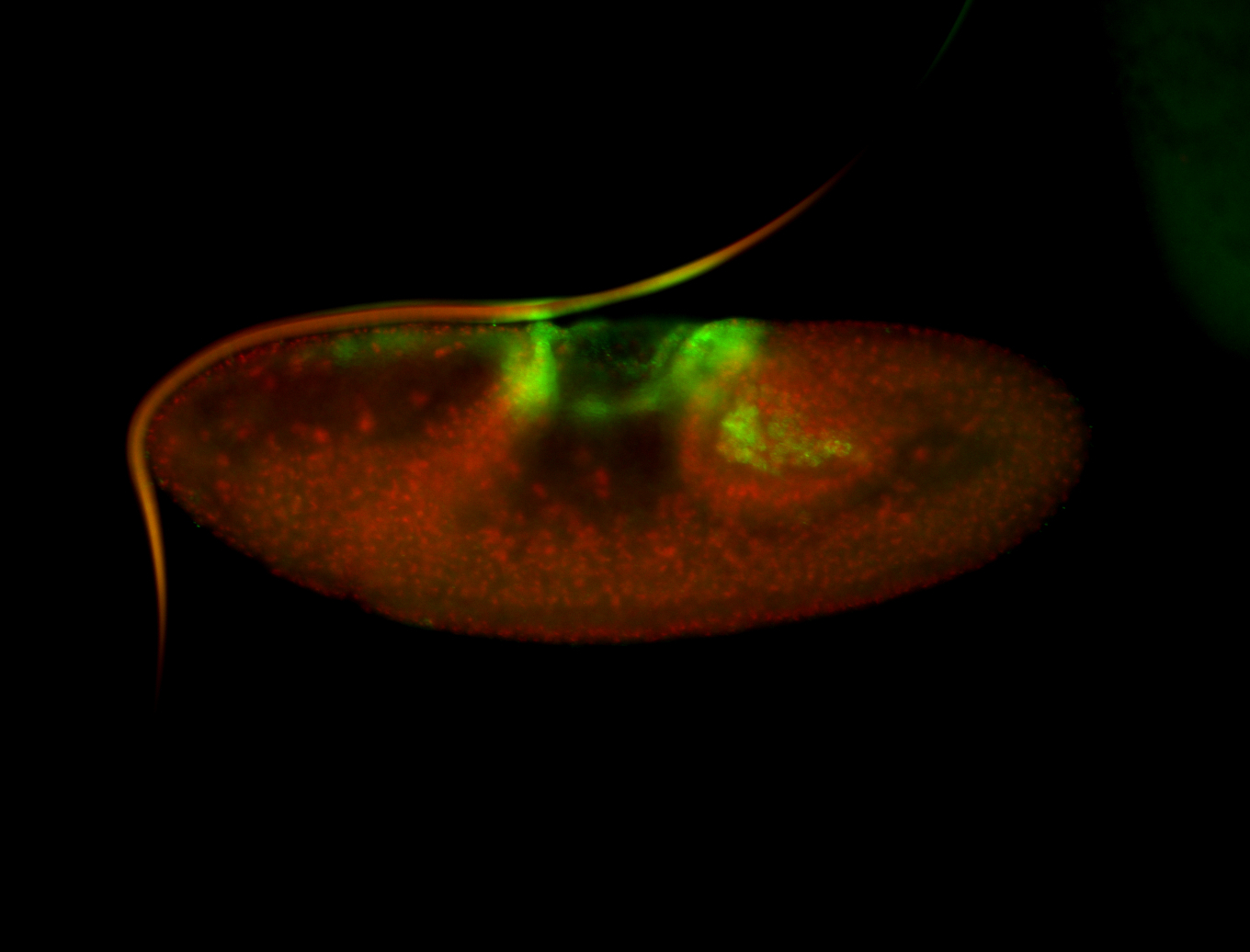
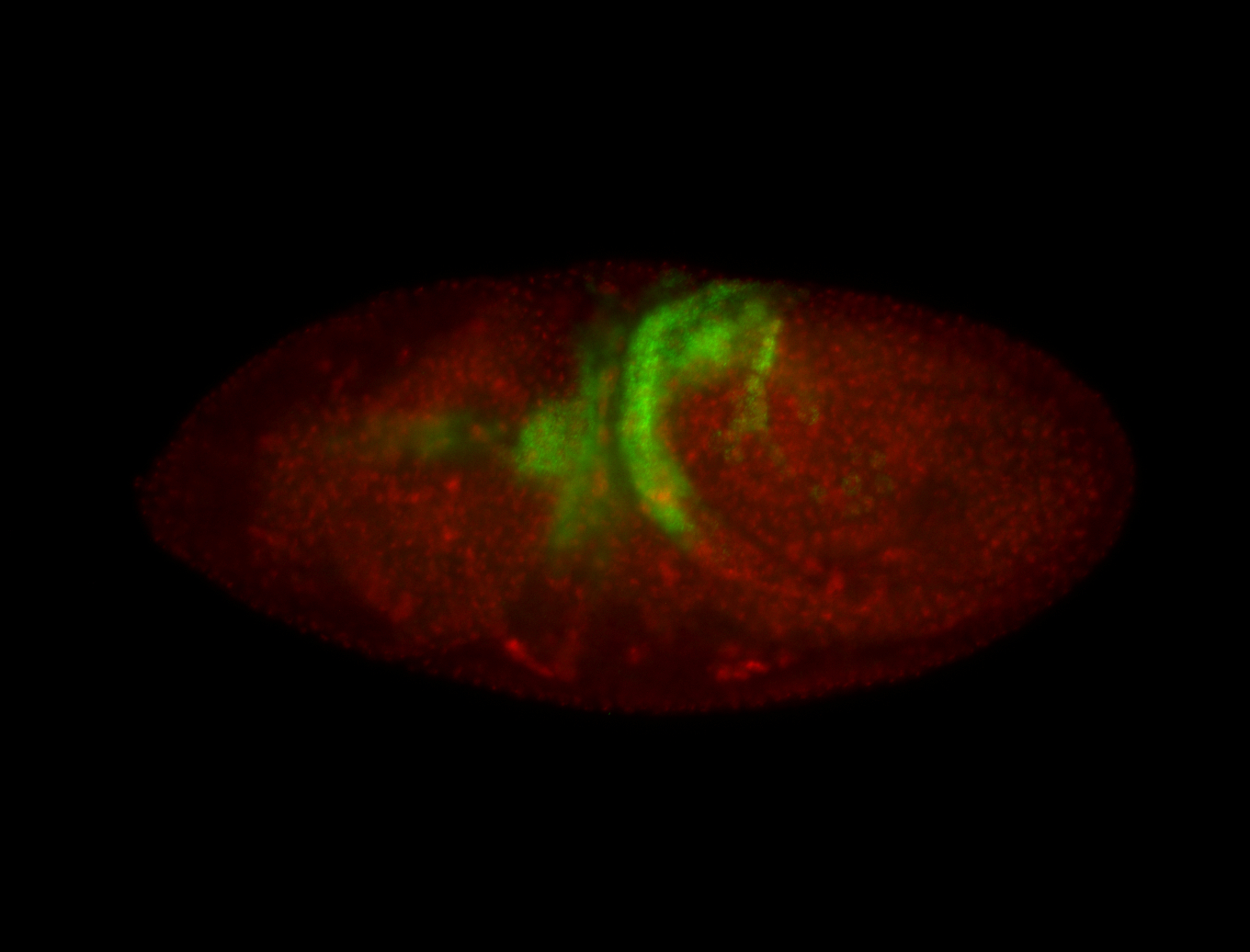
| Again, perinuclear pole cell asymmetric expression. |
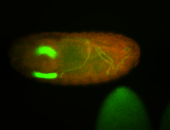
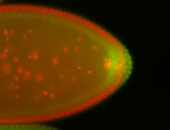
| Pole cell asymmetric expression. I see small 'dots' around the pole cells, stronger in one 1/2 of the pole nuclei. |
| Good probe on gel |
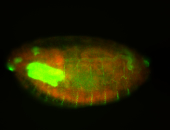
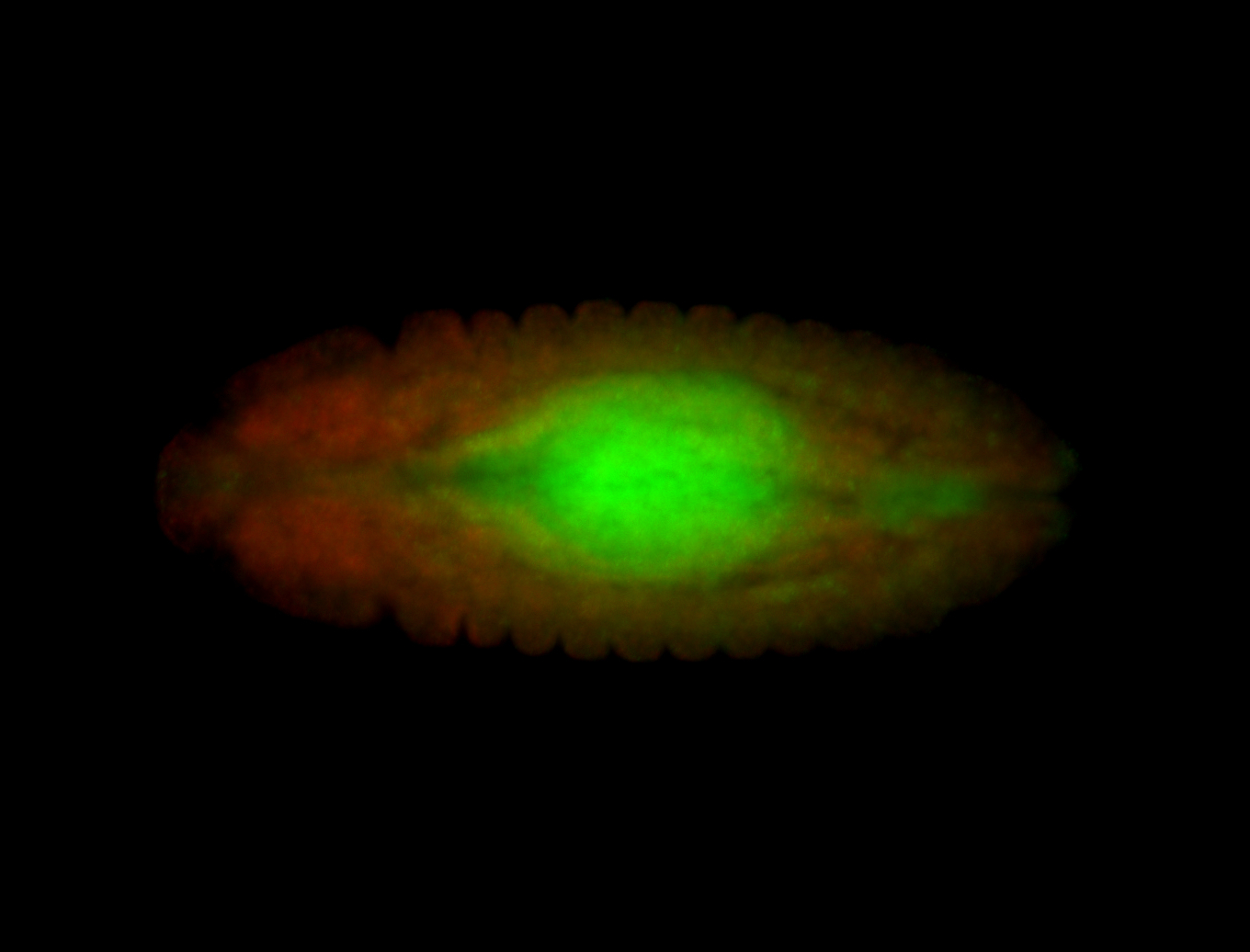
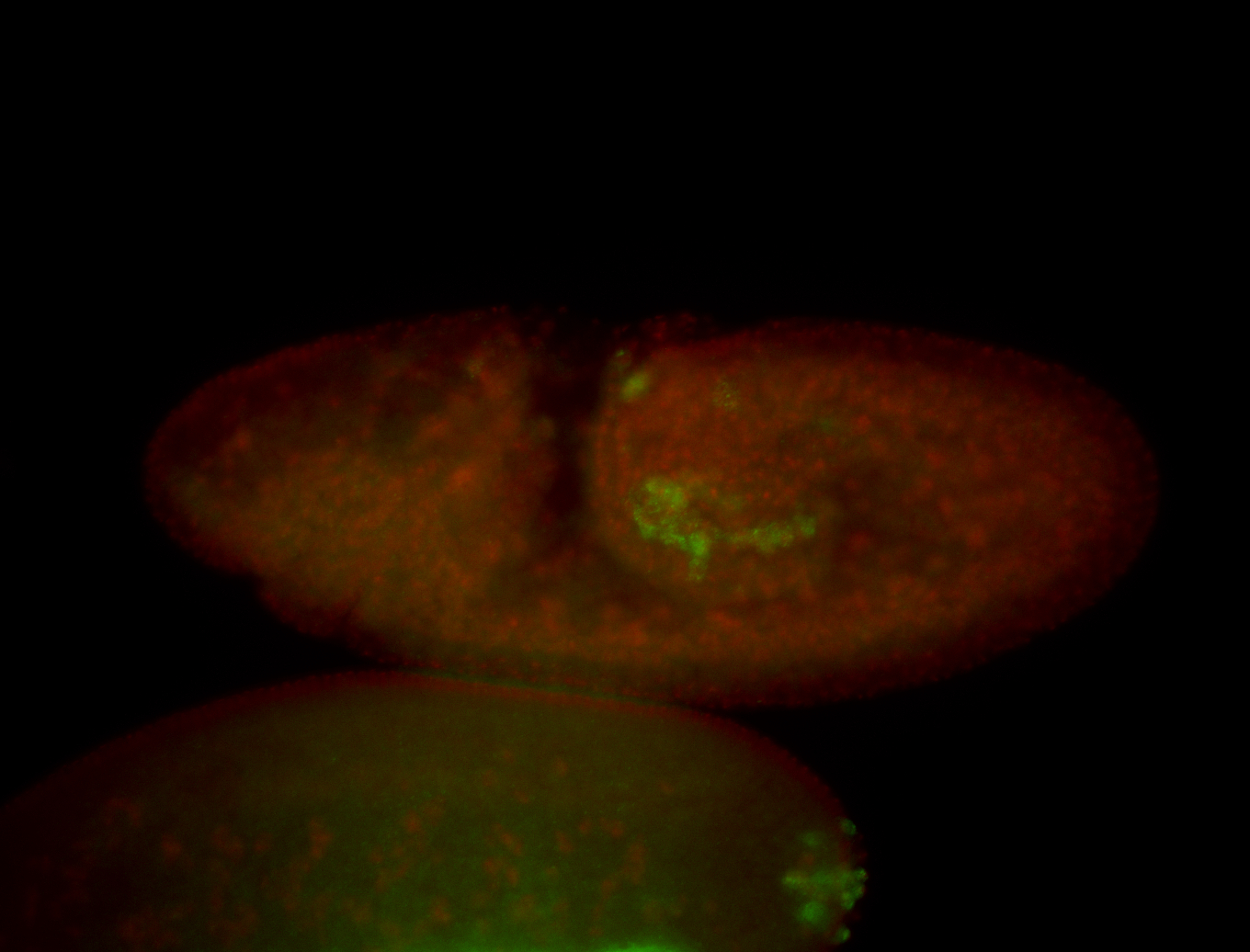
| Posterior localization, even in unfertilized eggs. RNA accumulates asymmetrically around the pole buds with a 'fiber-like' texture. When nuclei divide, the RNA will stay in the nuclei that are closer to the posterior end. Very interesting! |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|---|---|
| Male reproductive tissues |
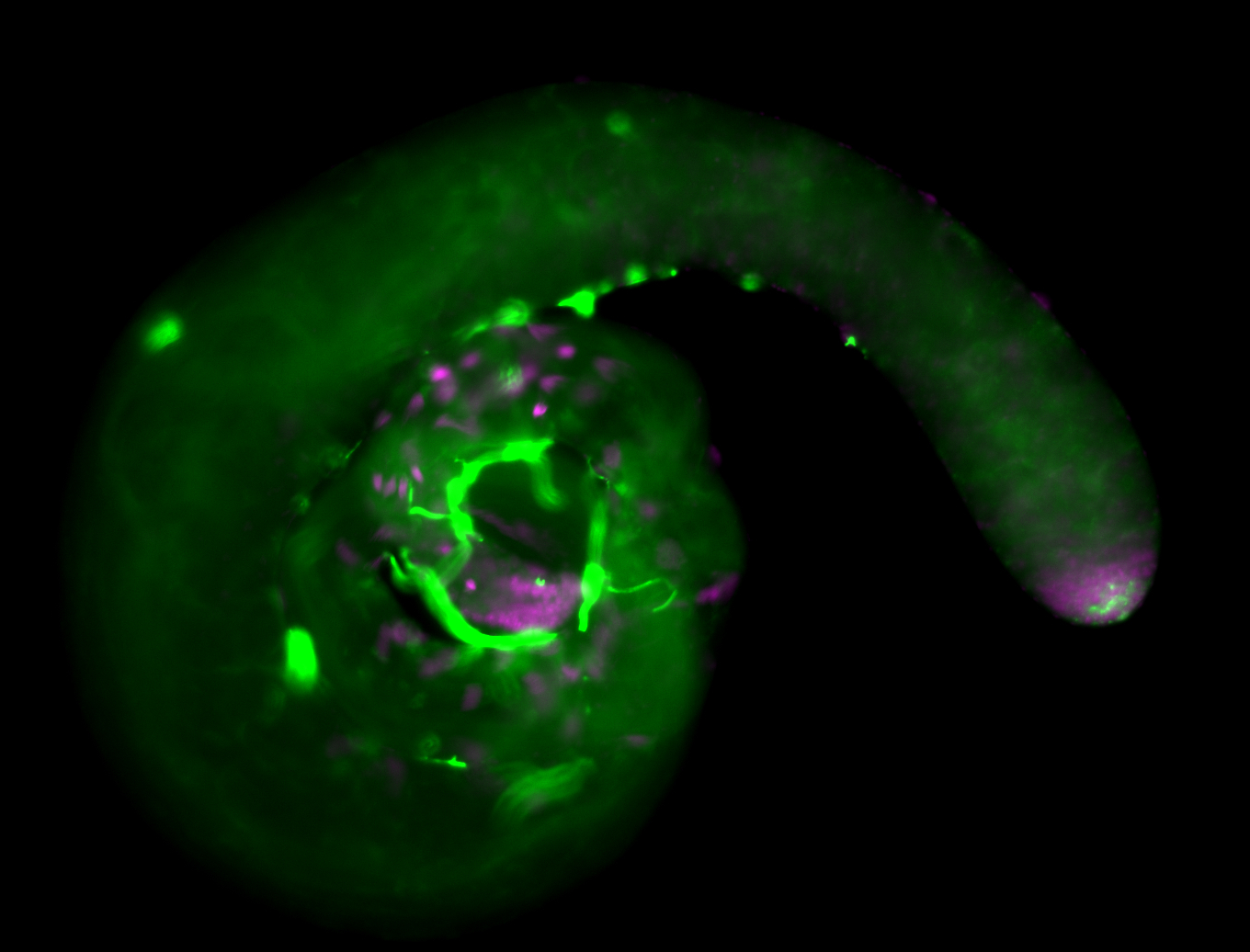
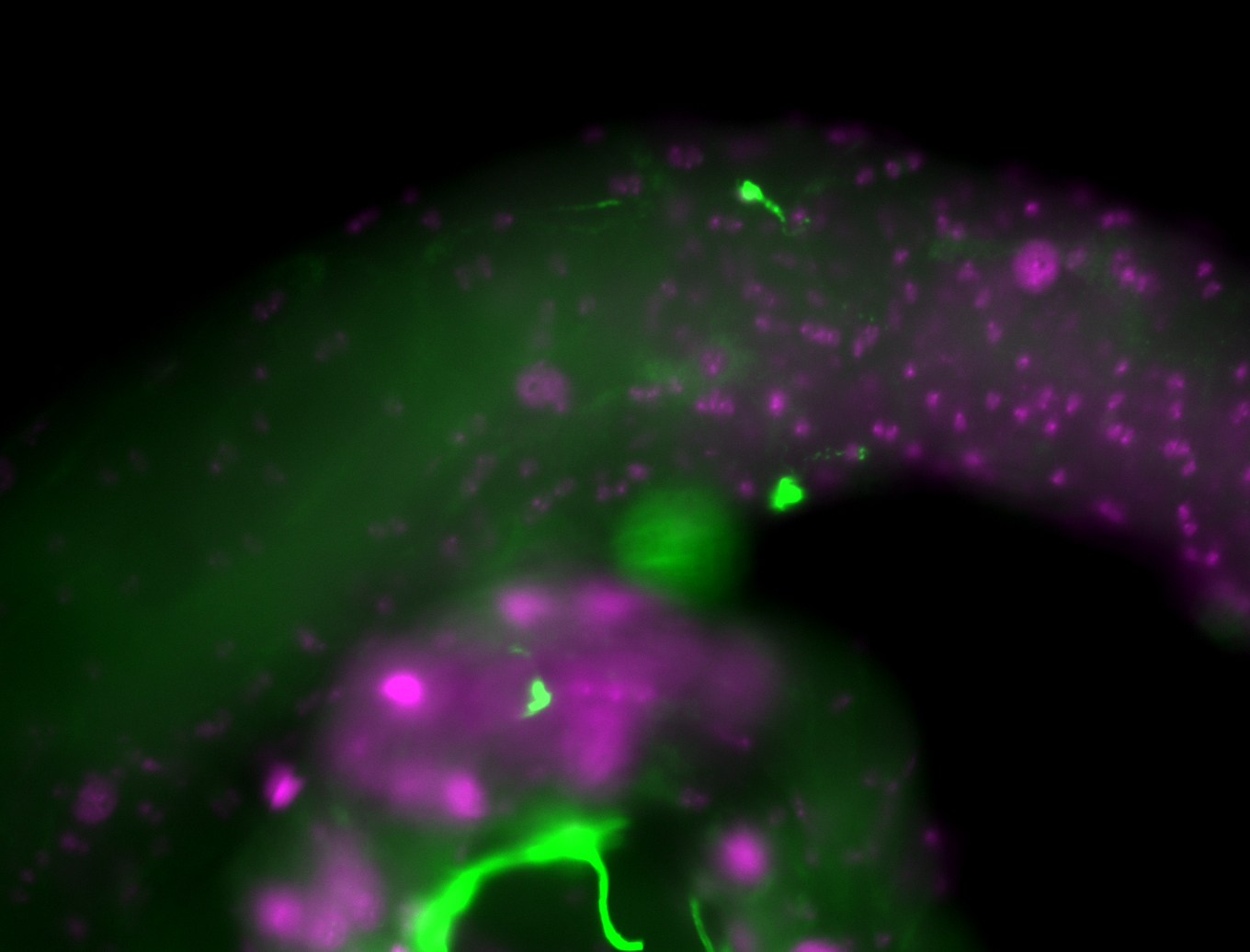
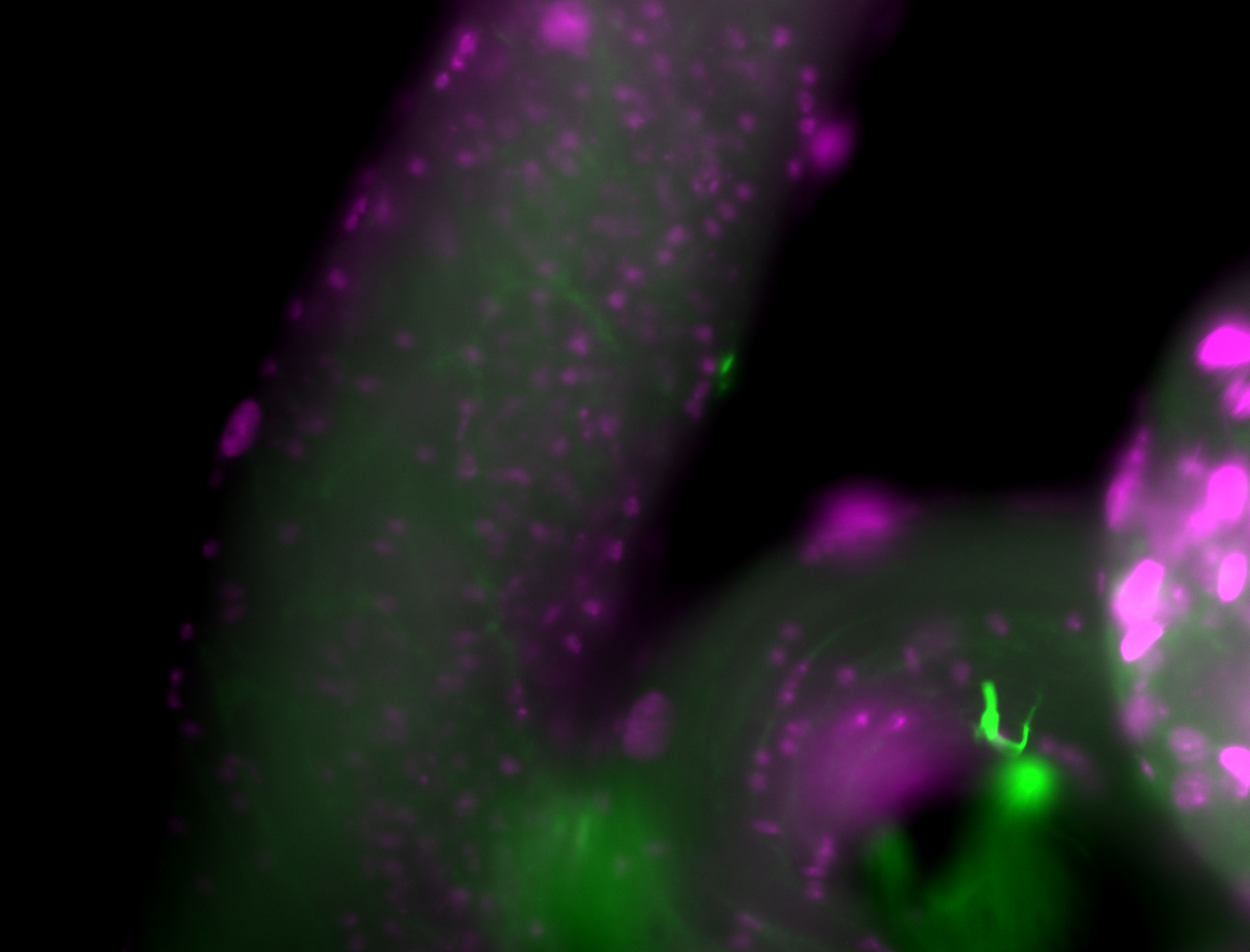
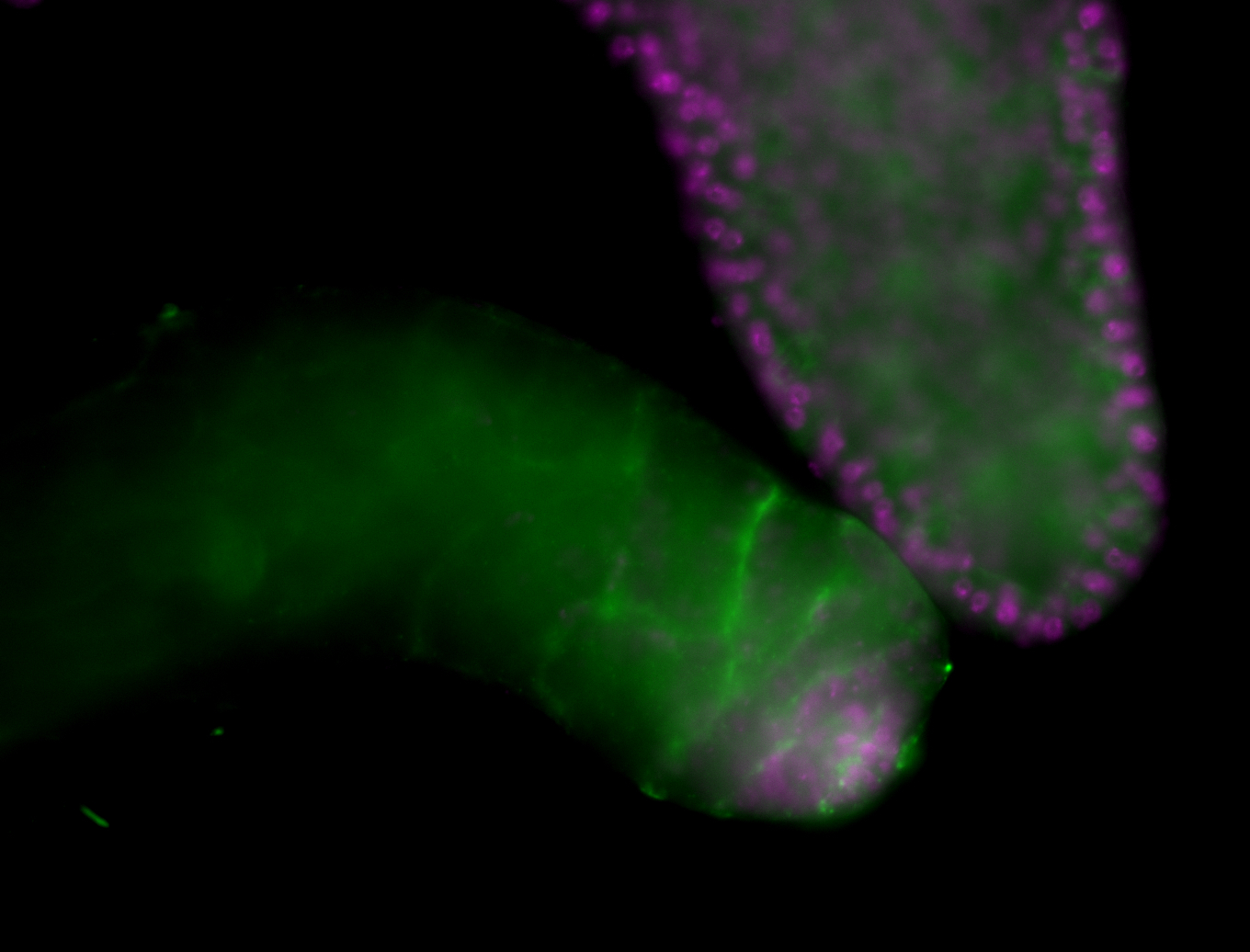
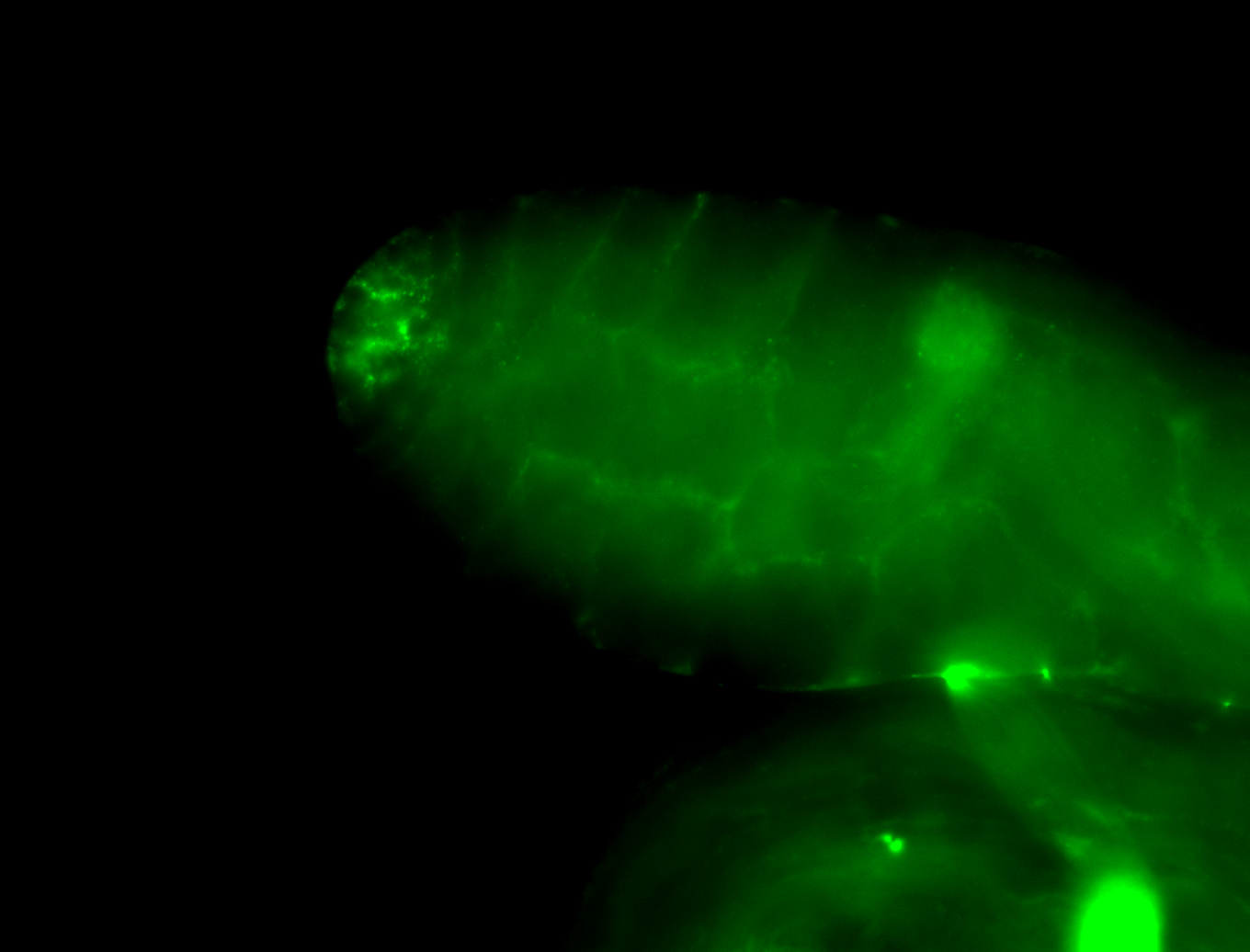
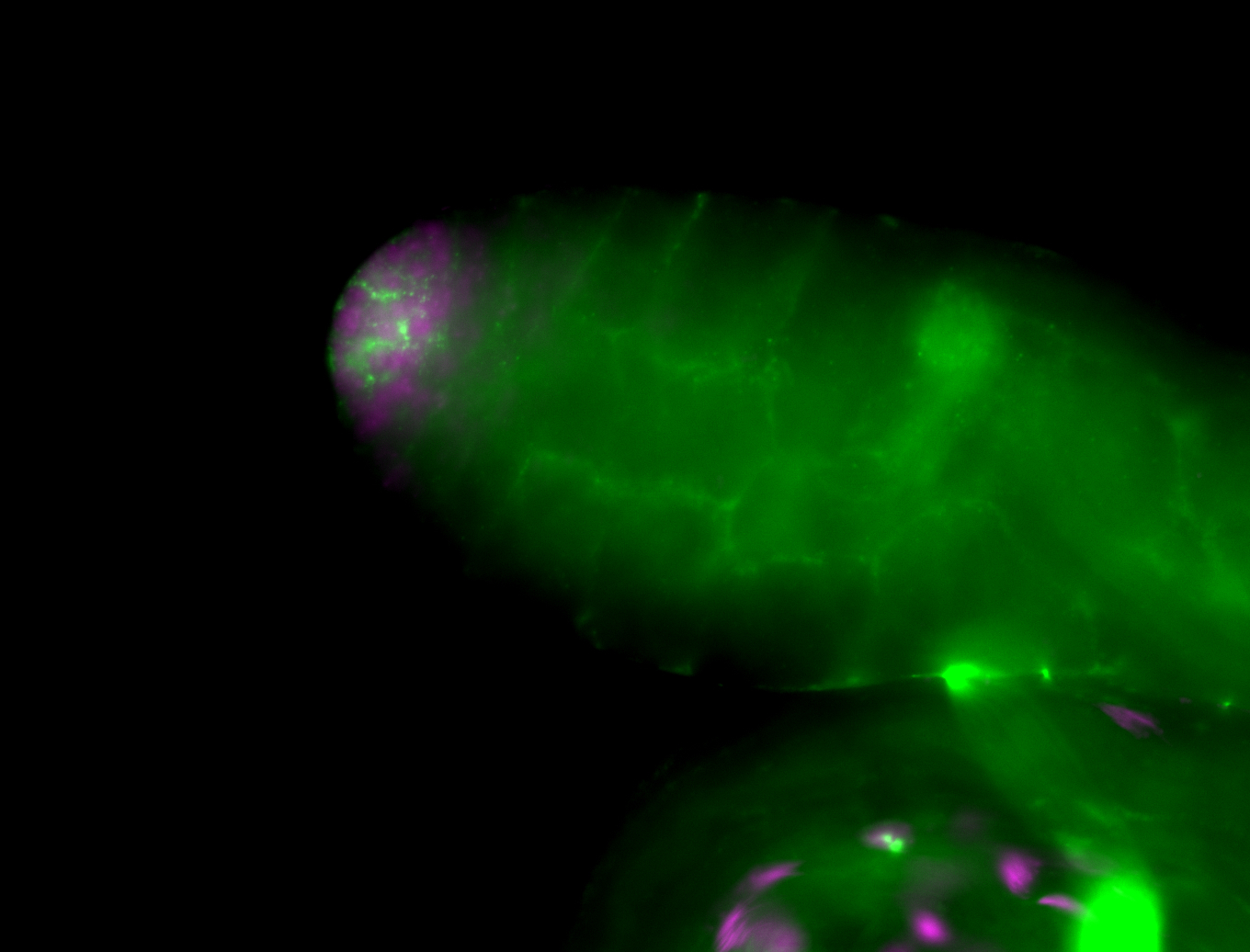
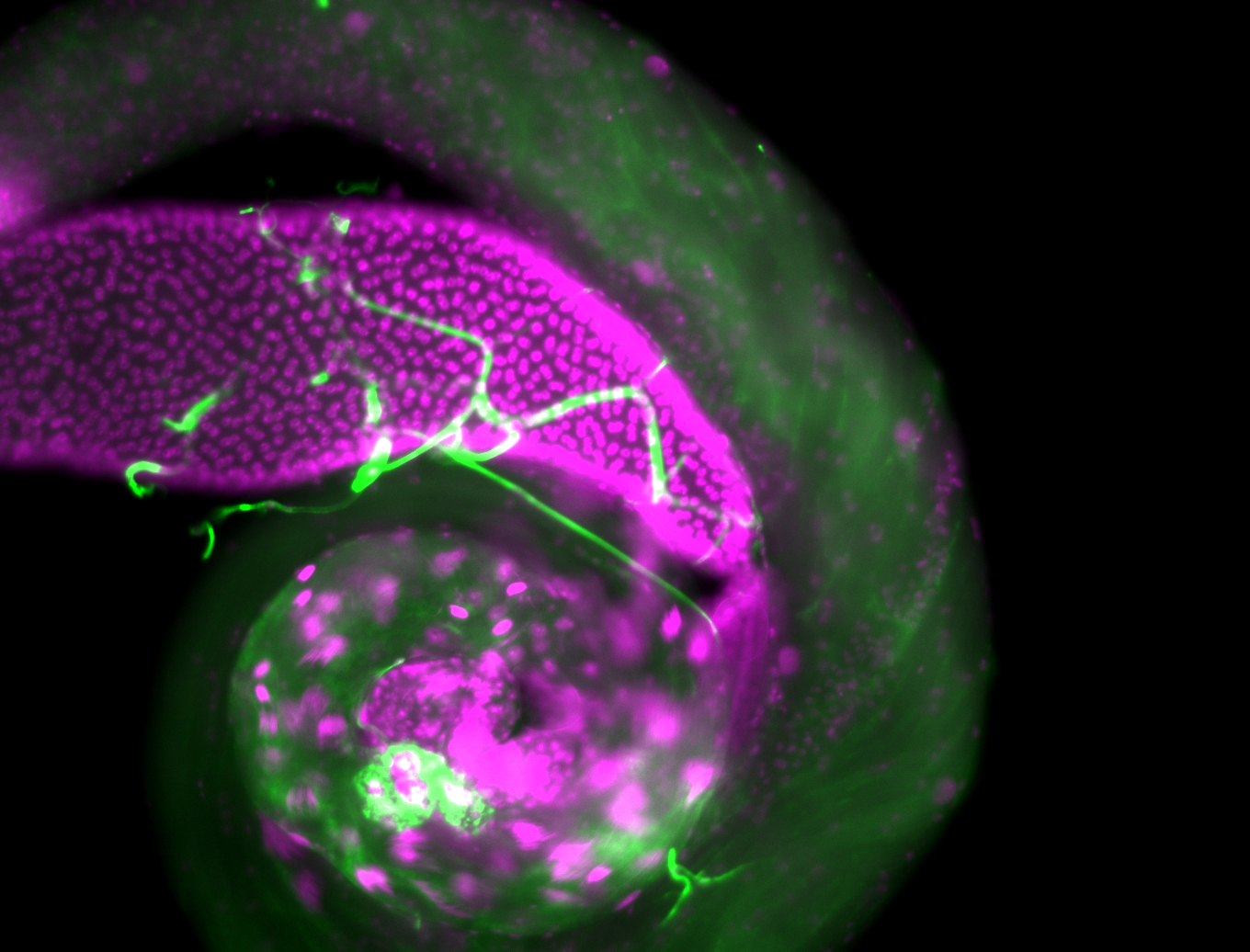
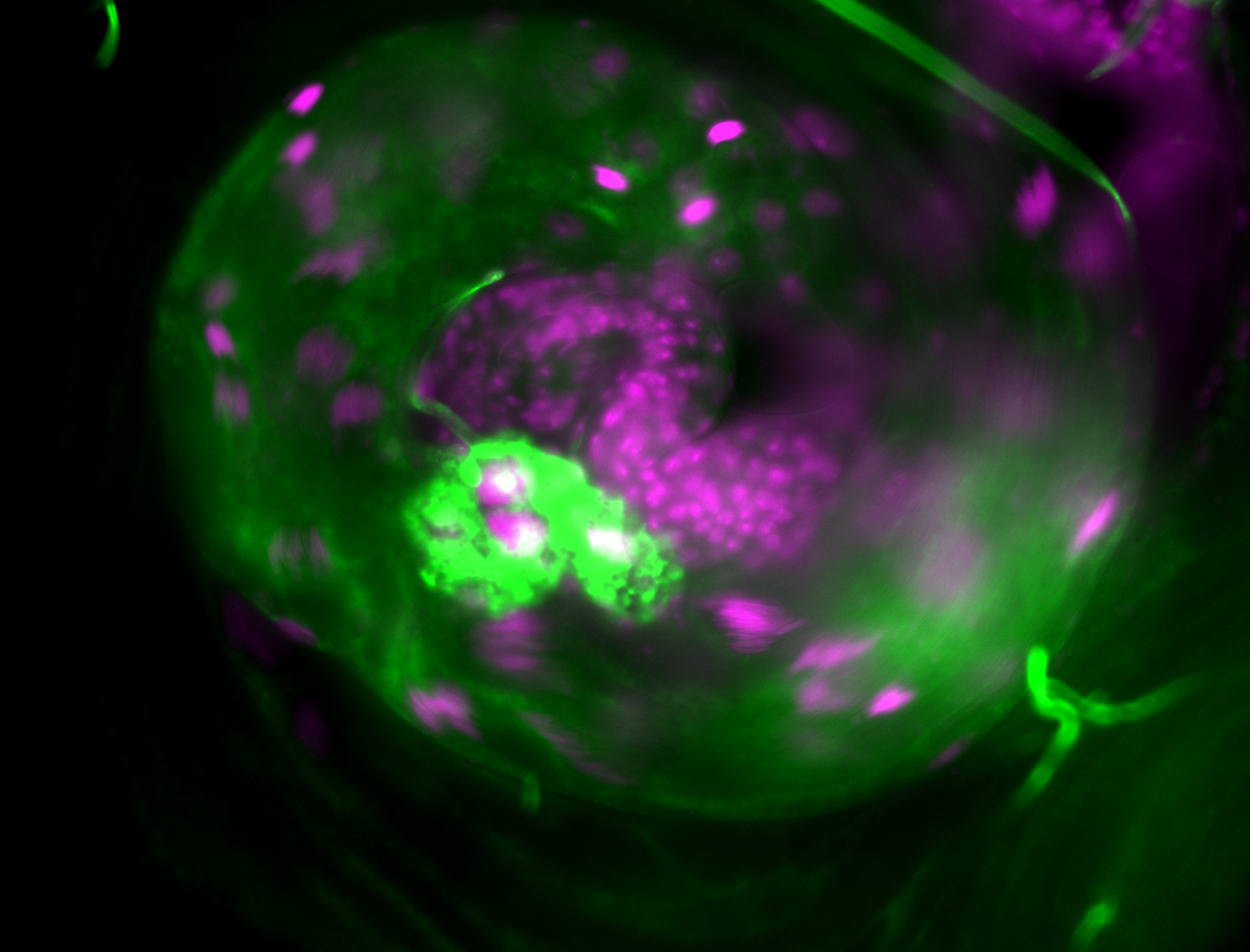
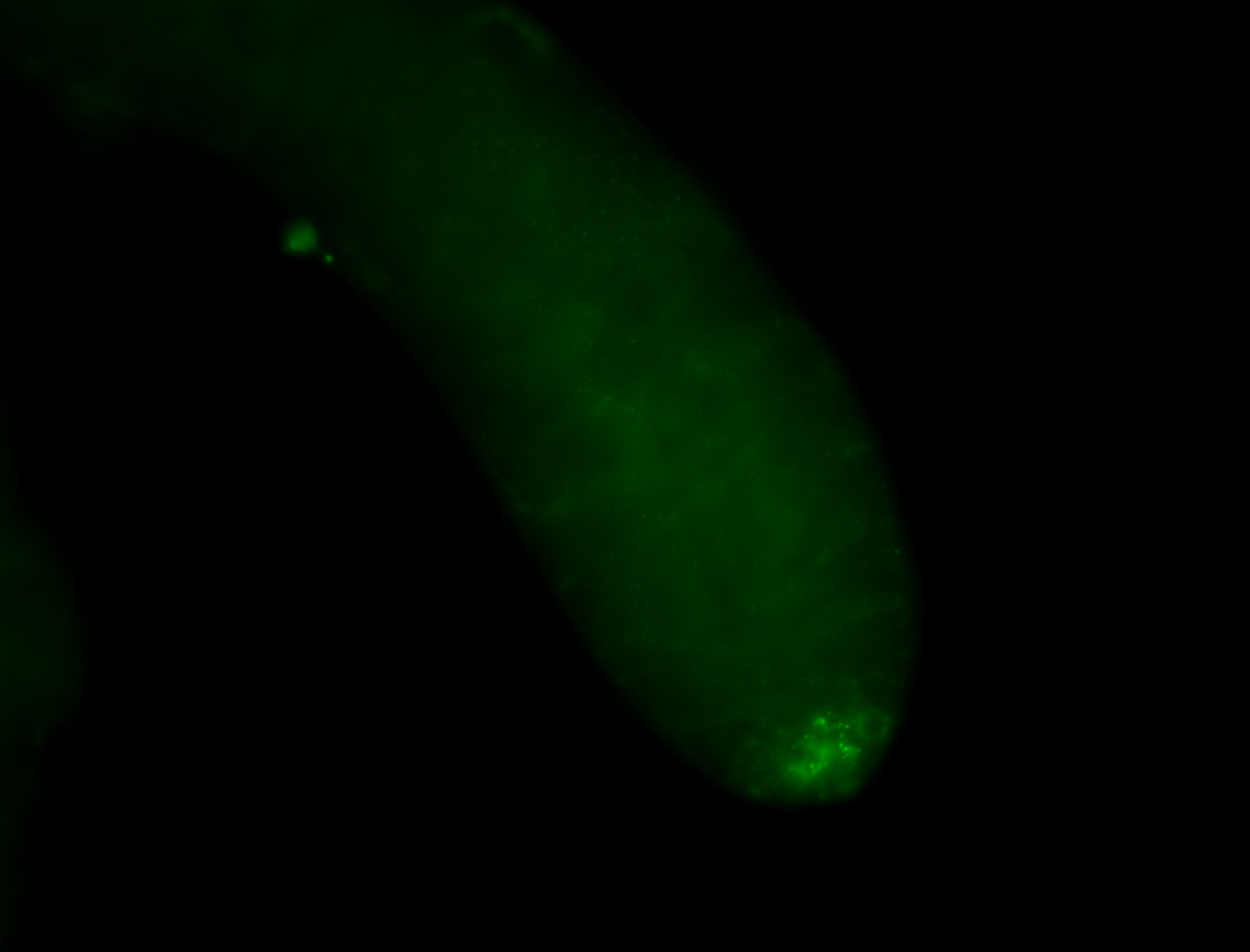
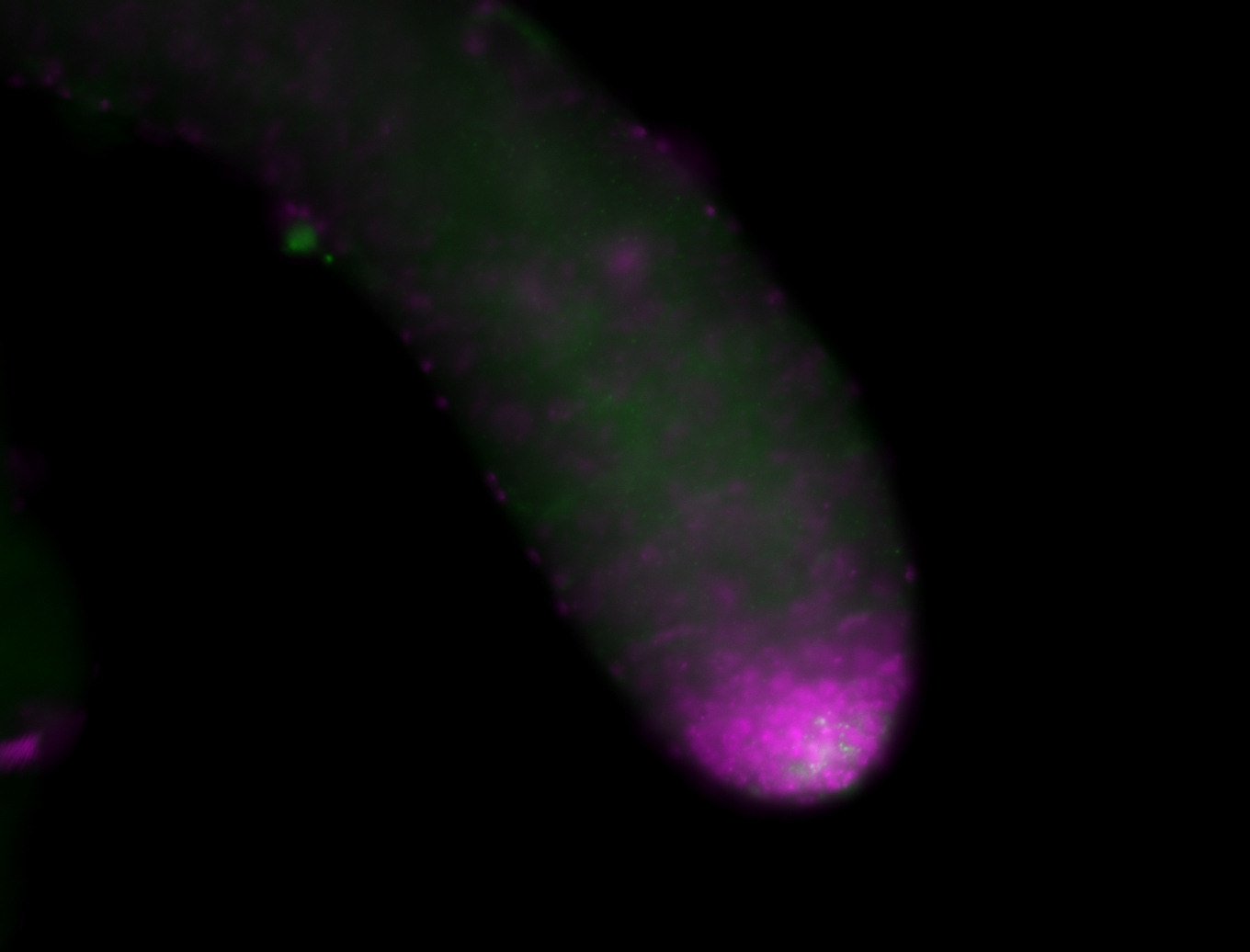
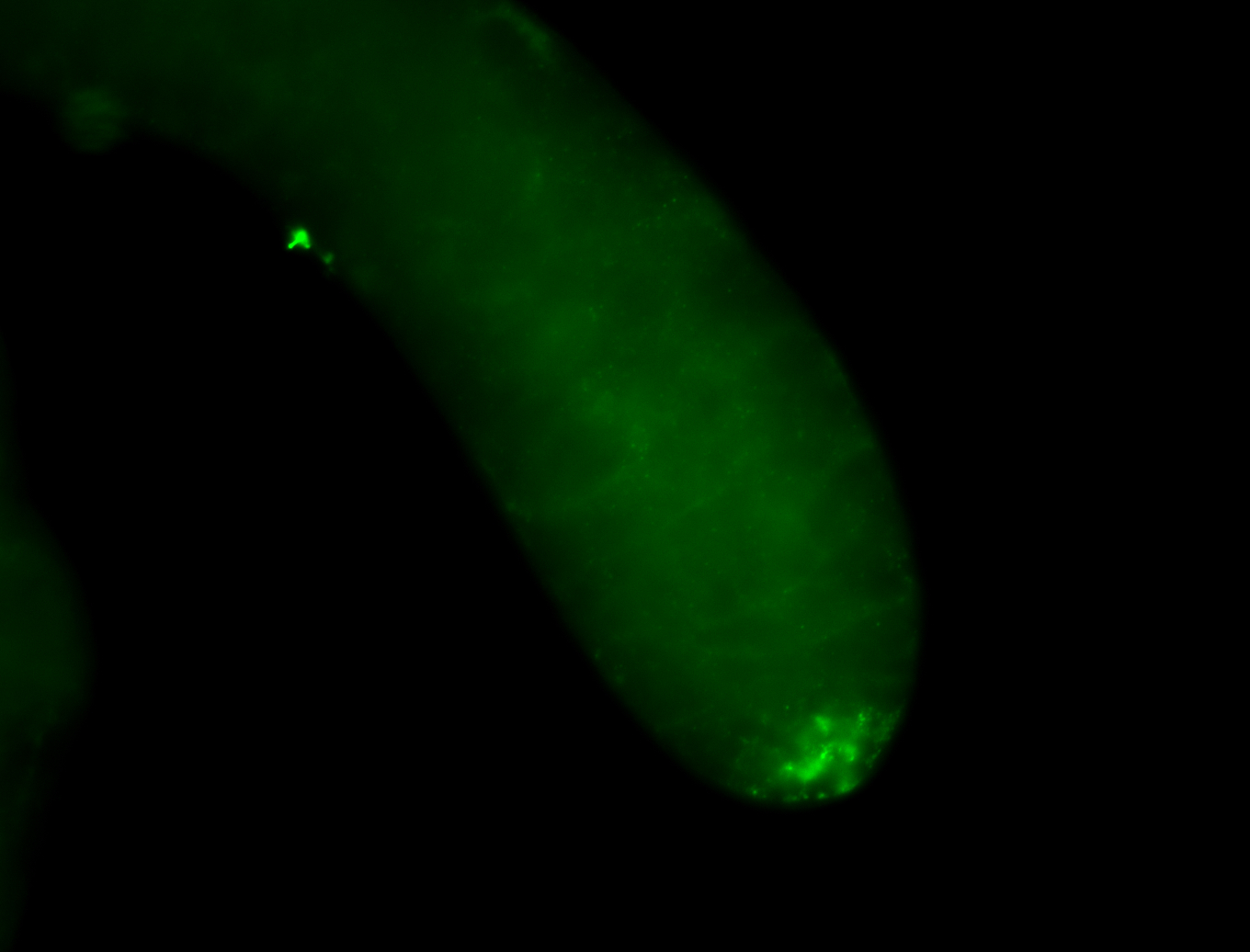
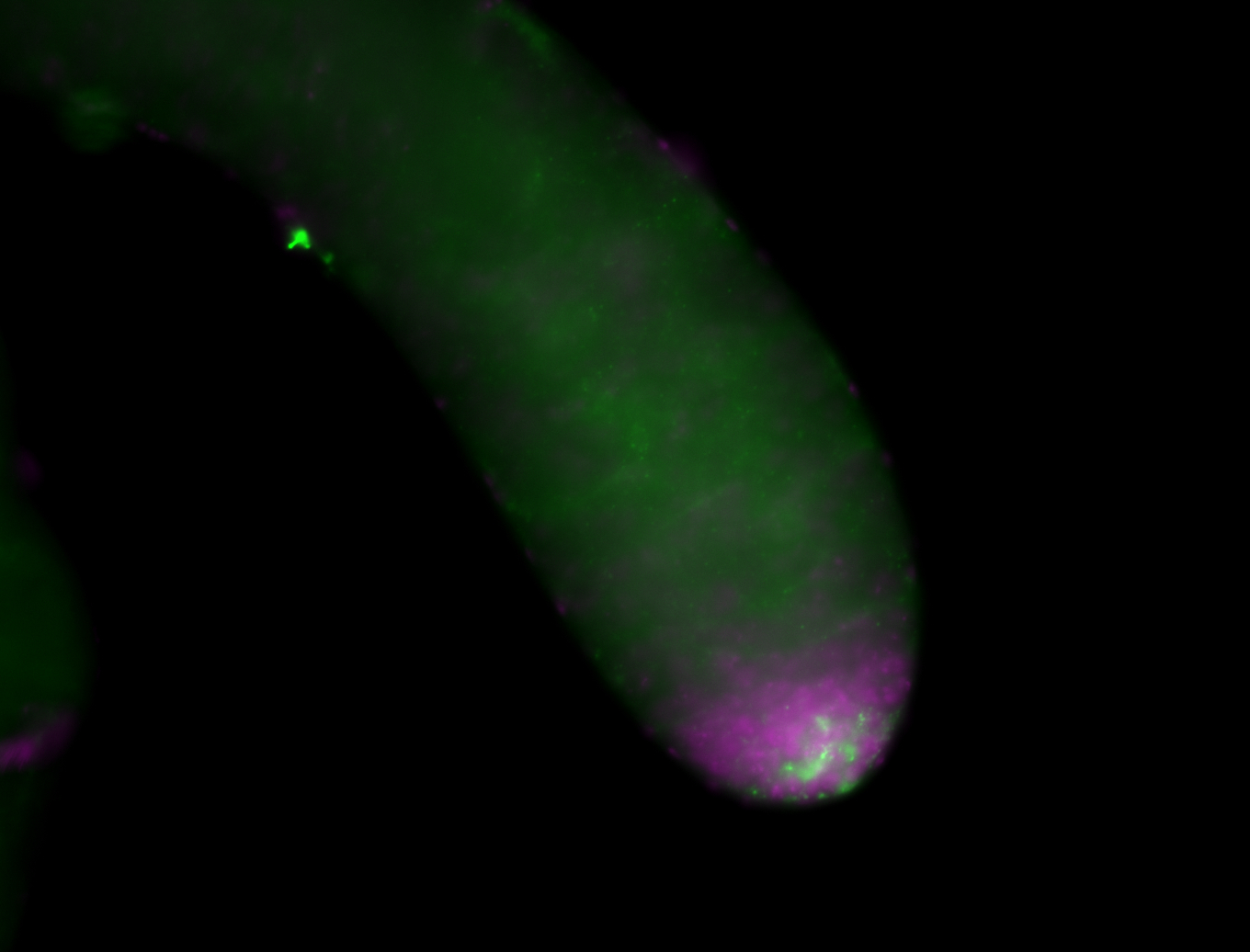
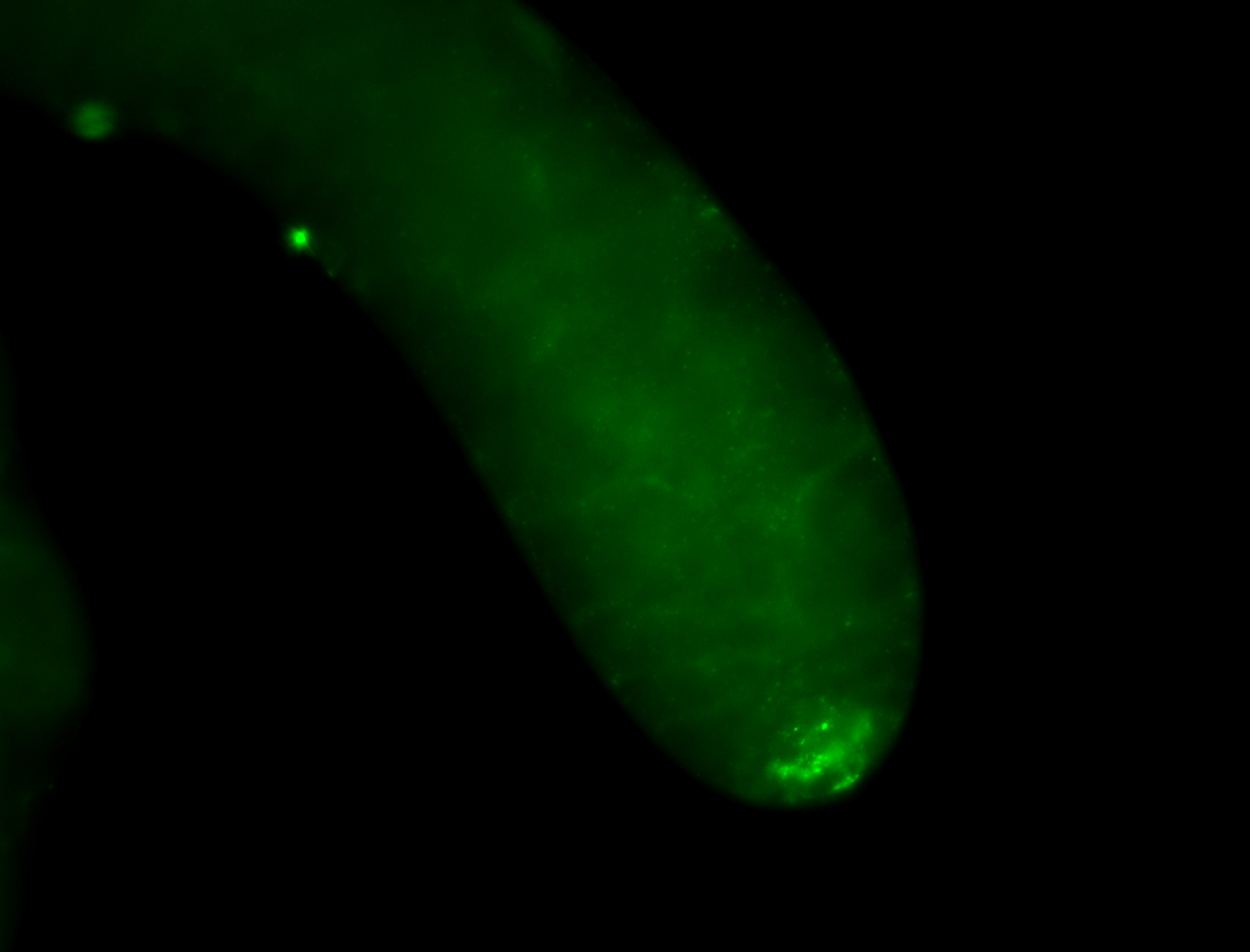
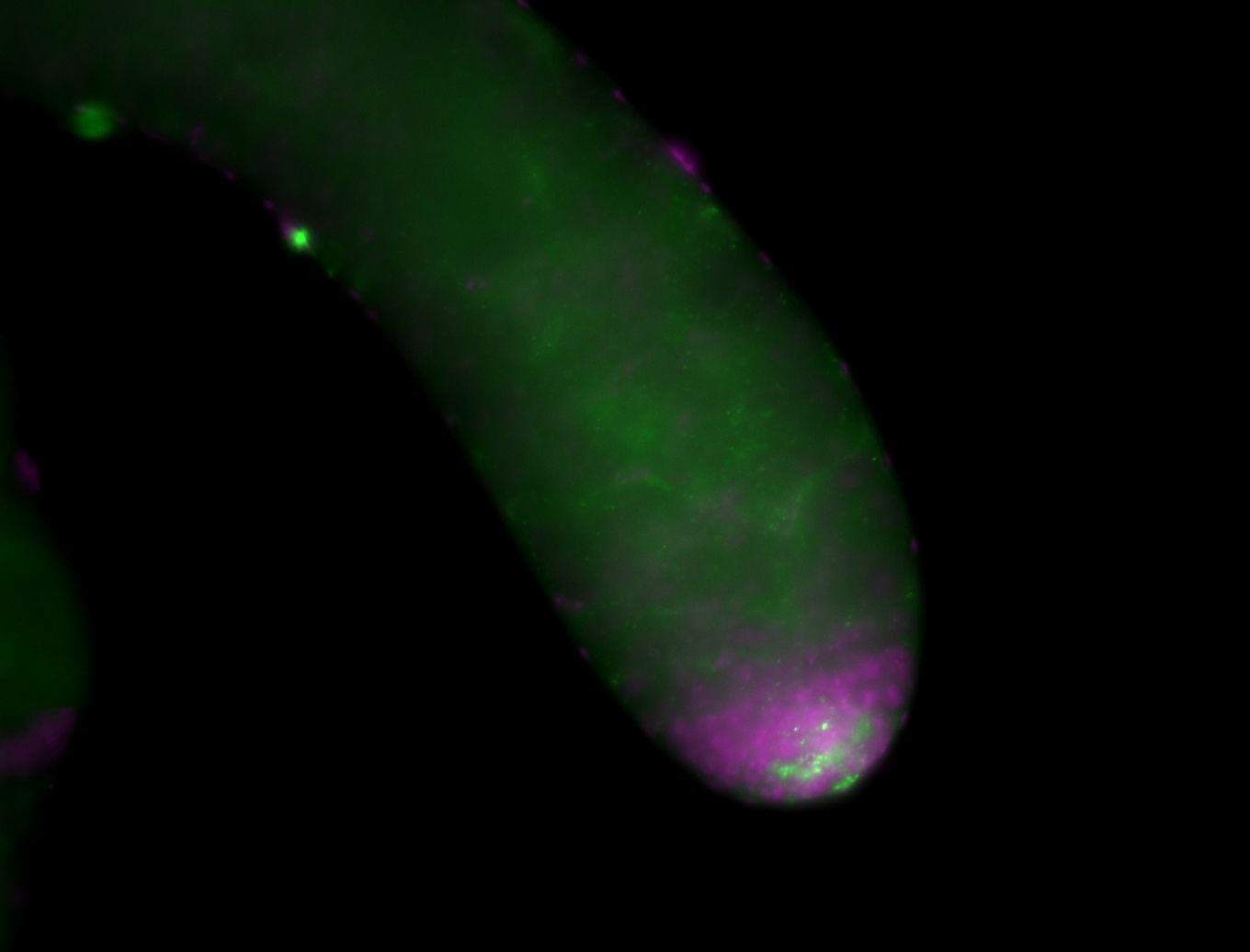
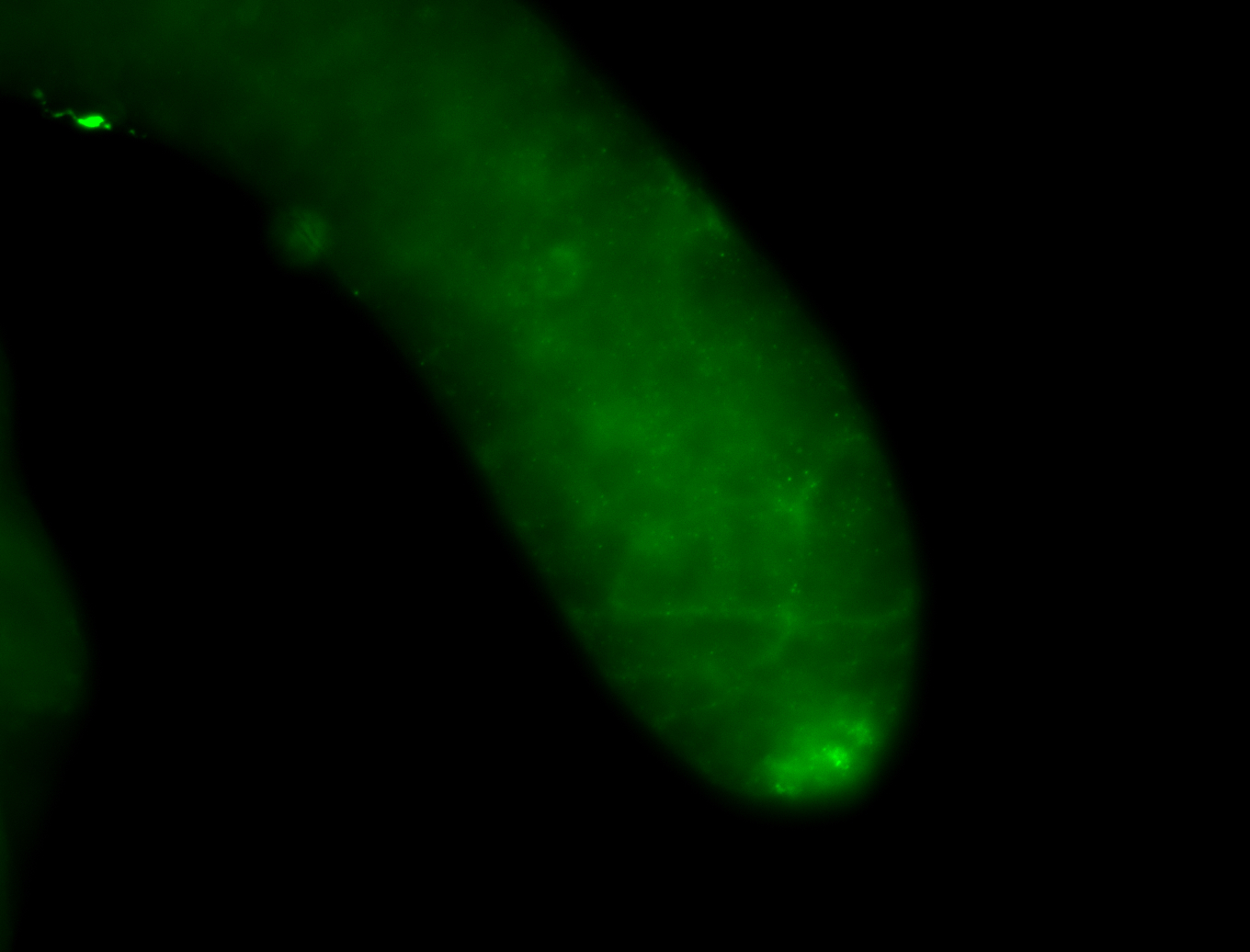
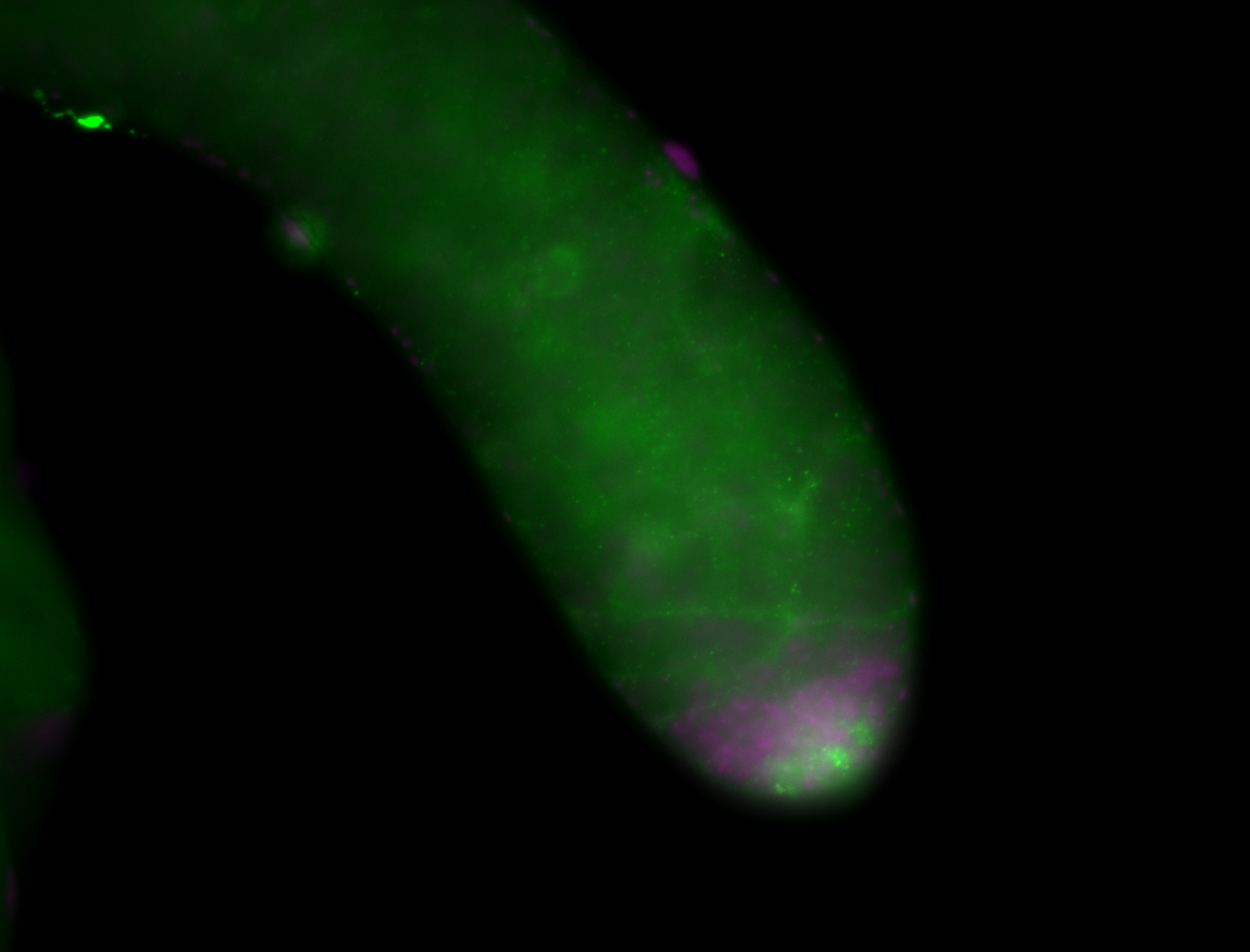
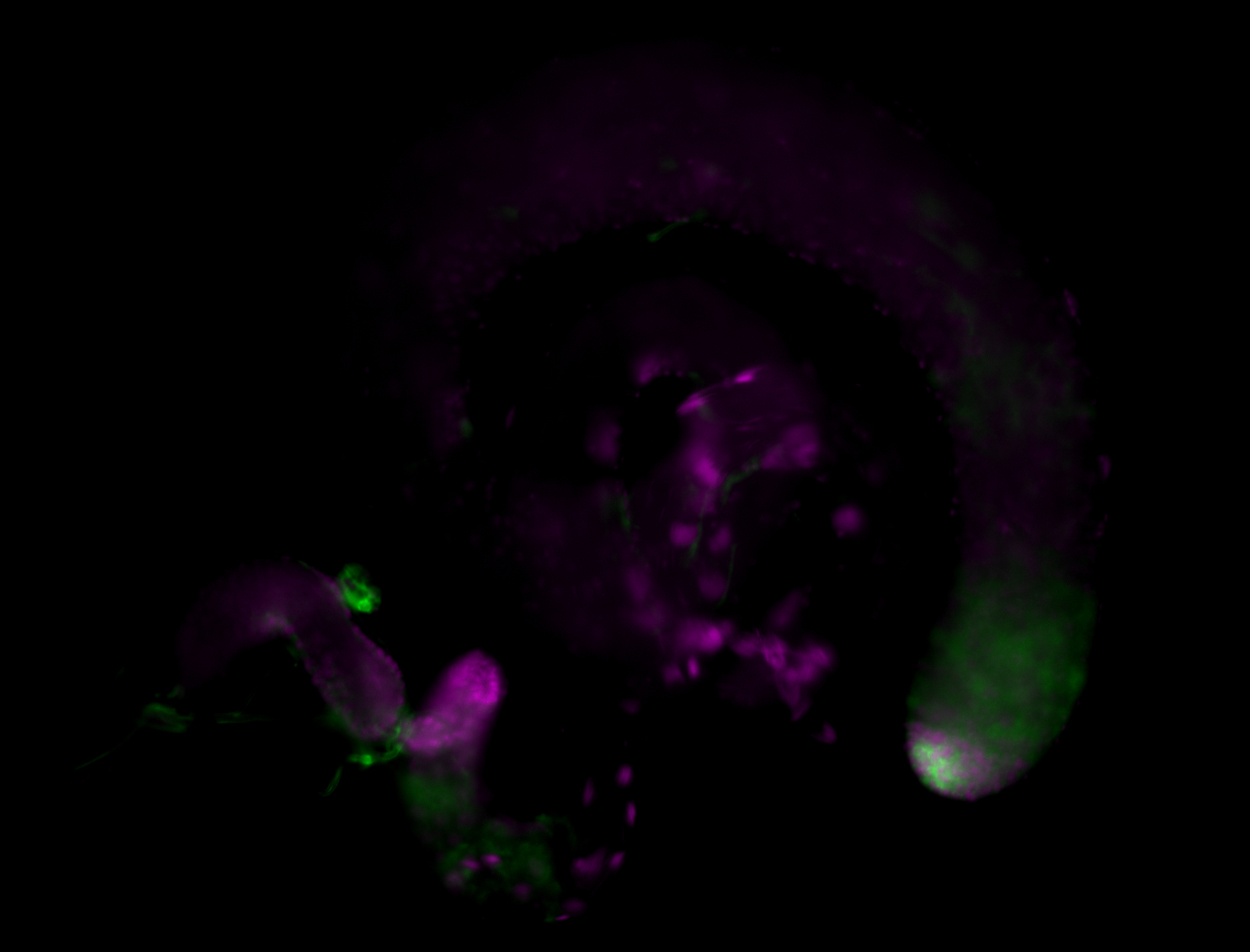
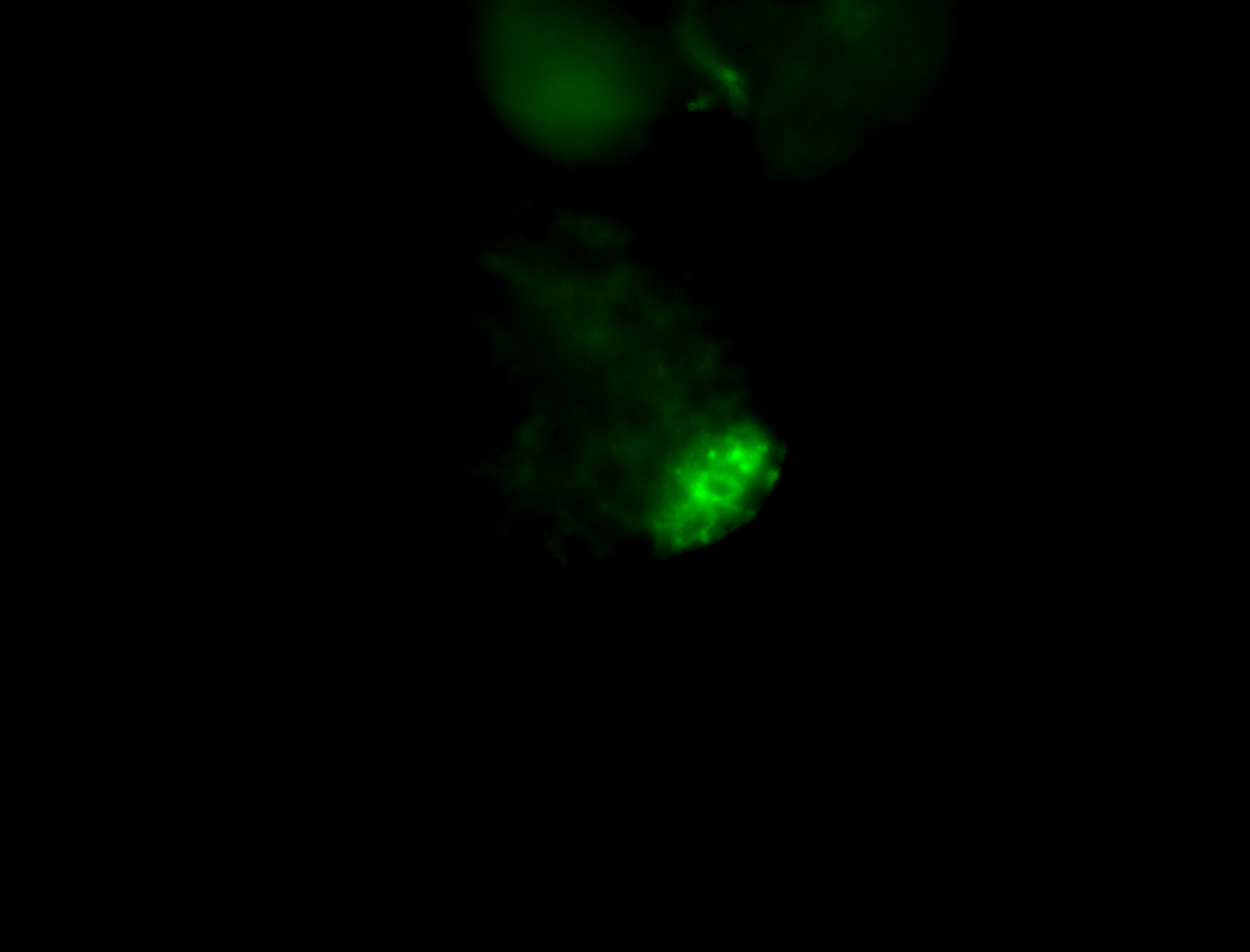
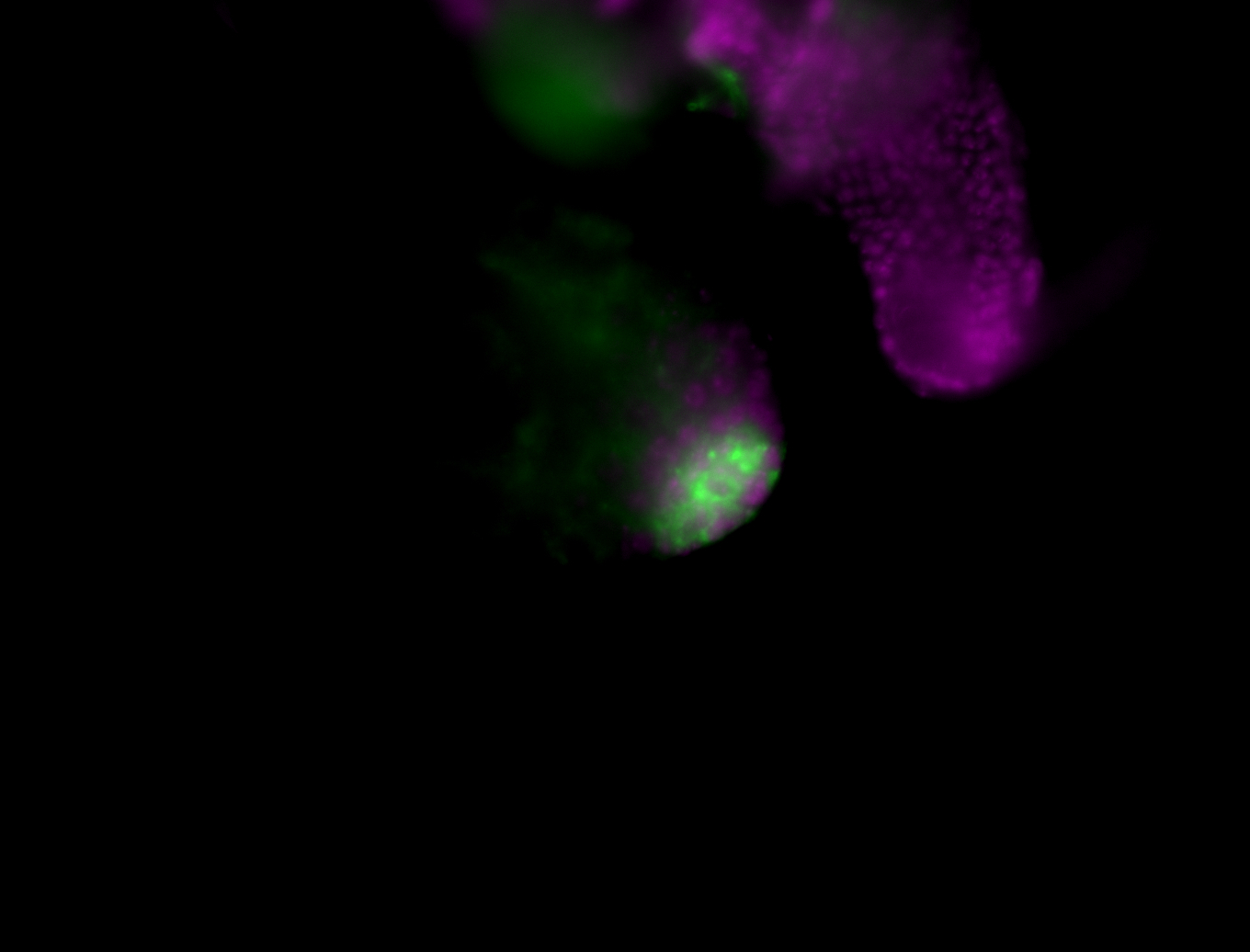
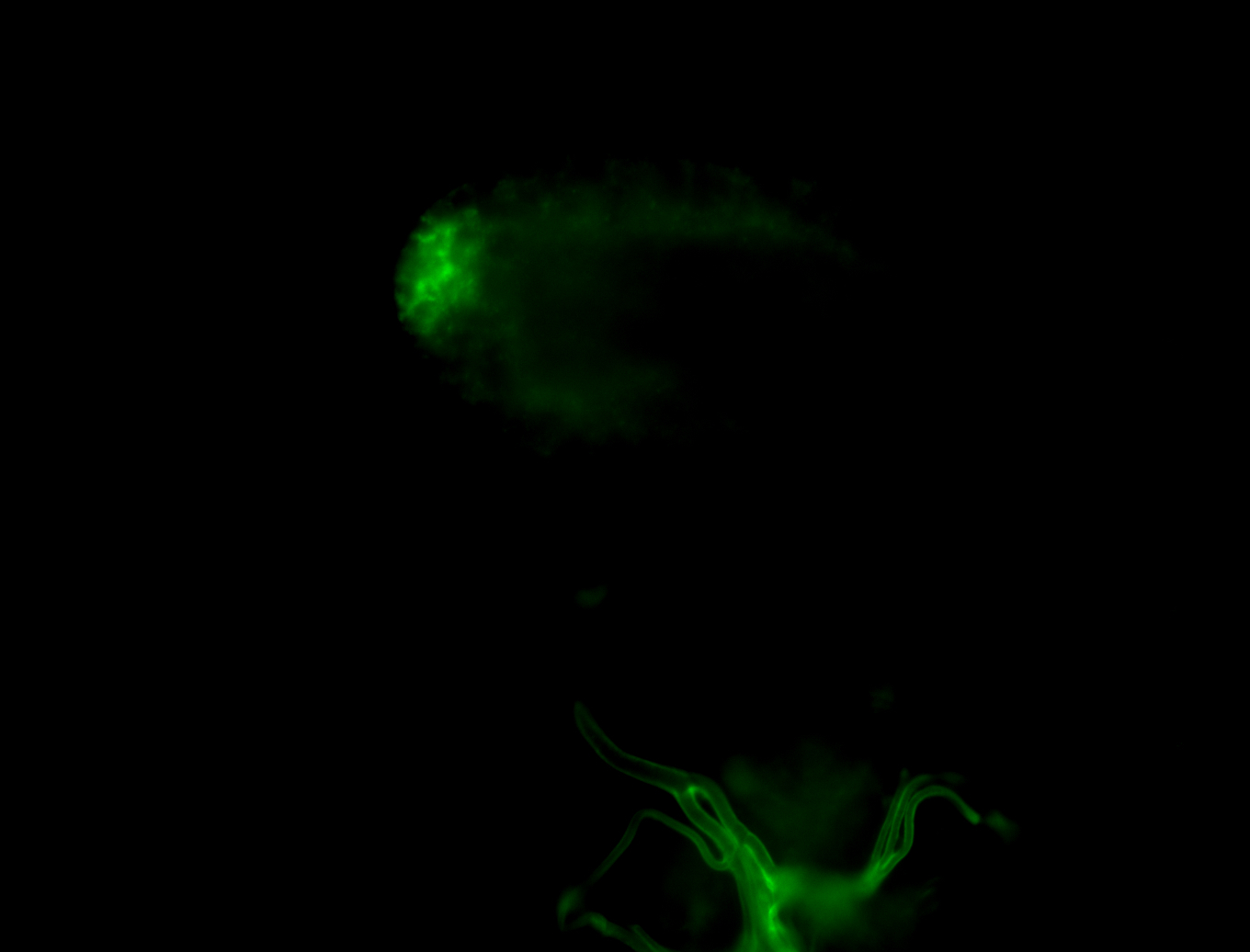
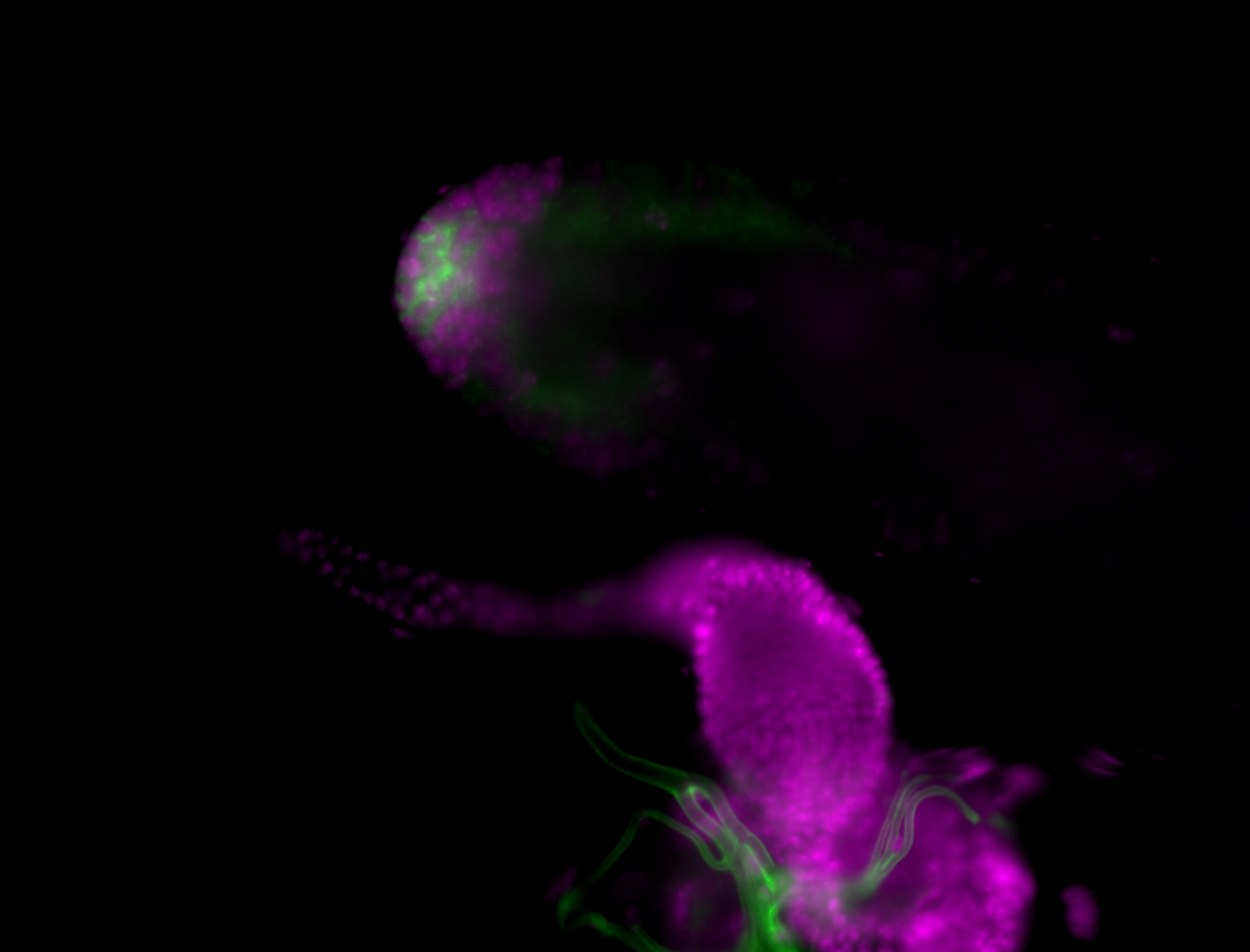
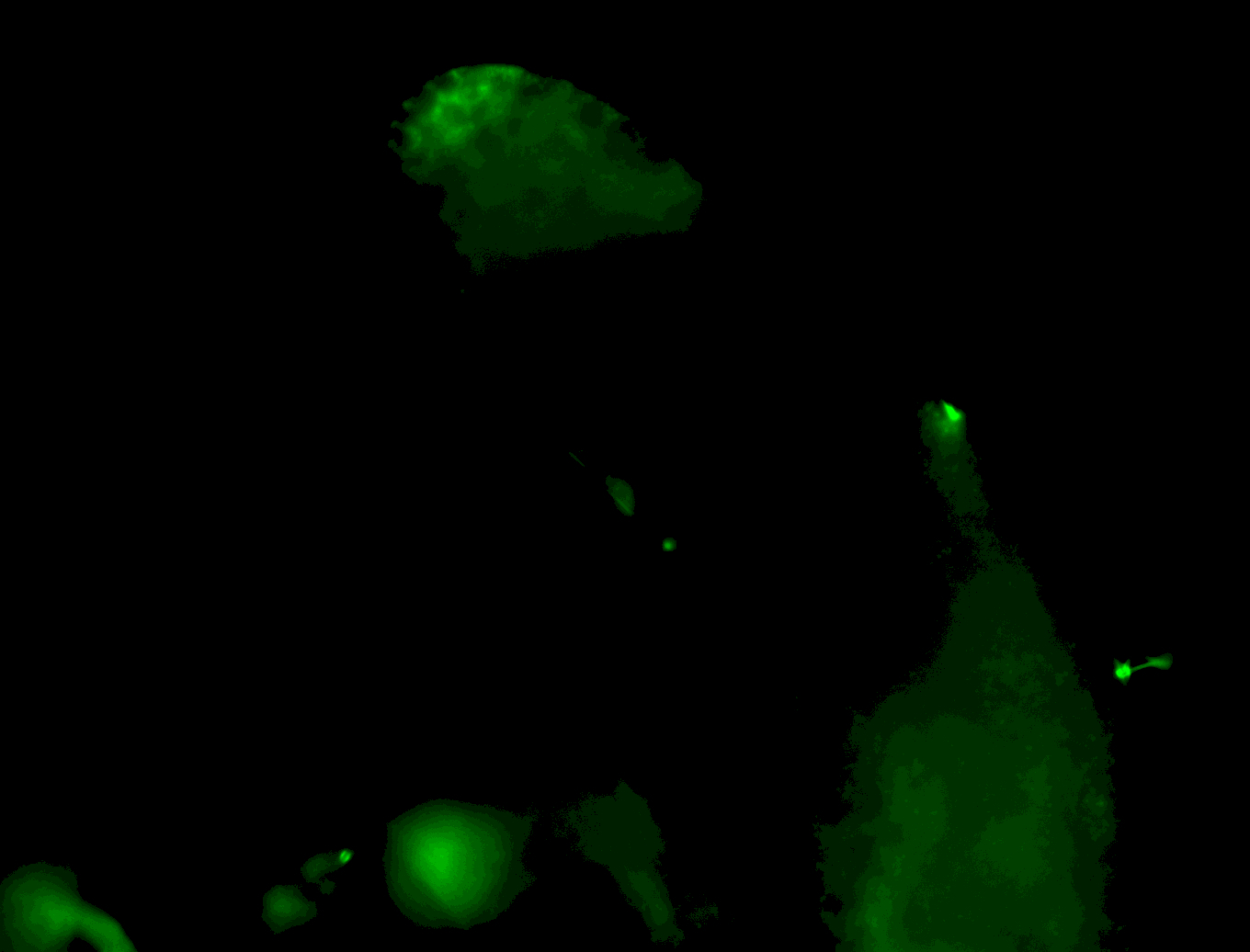
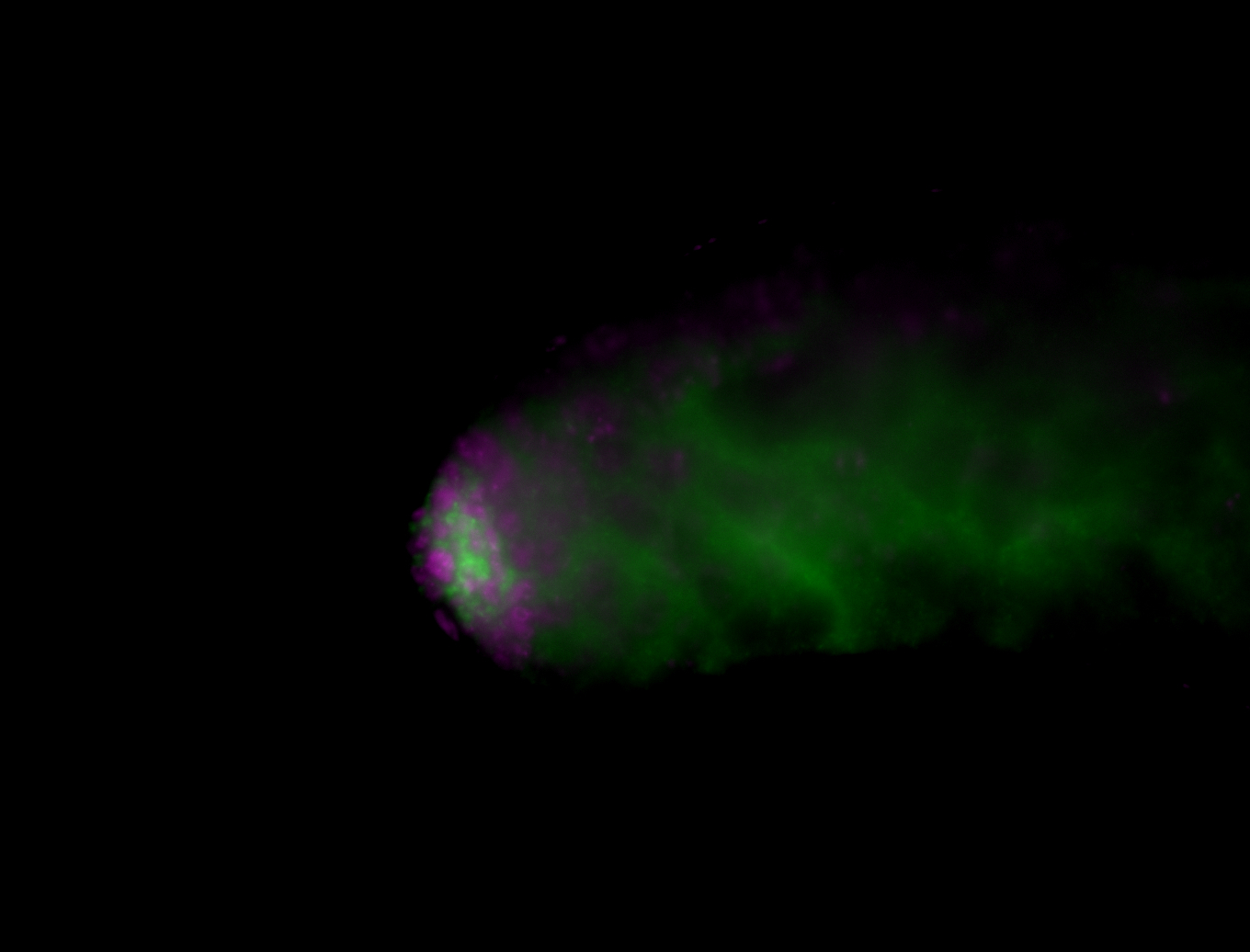
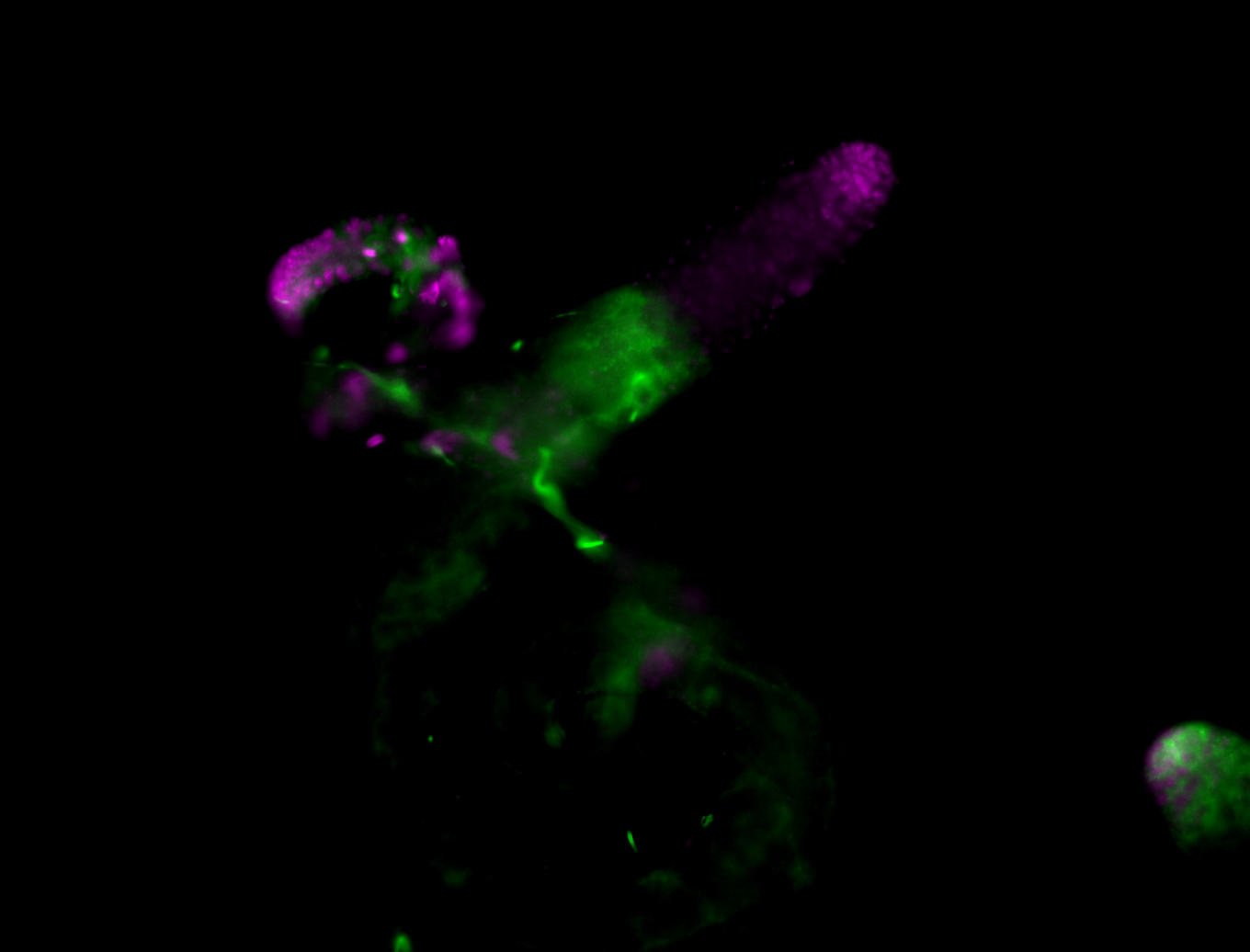
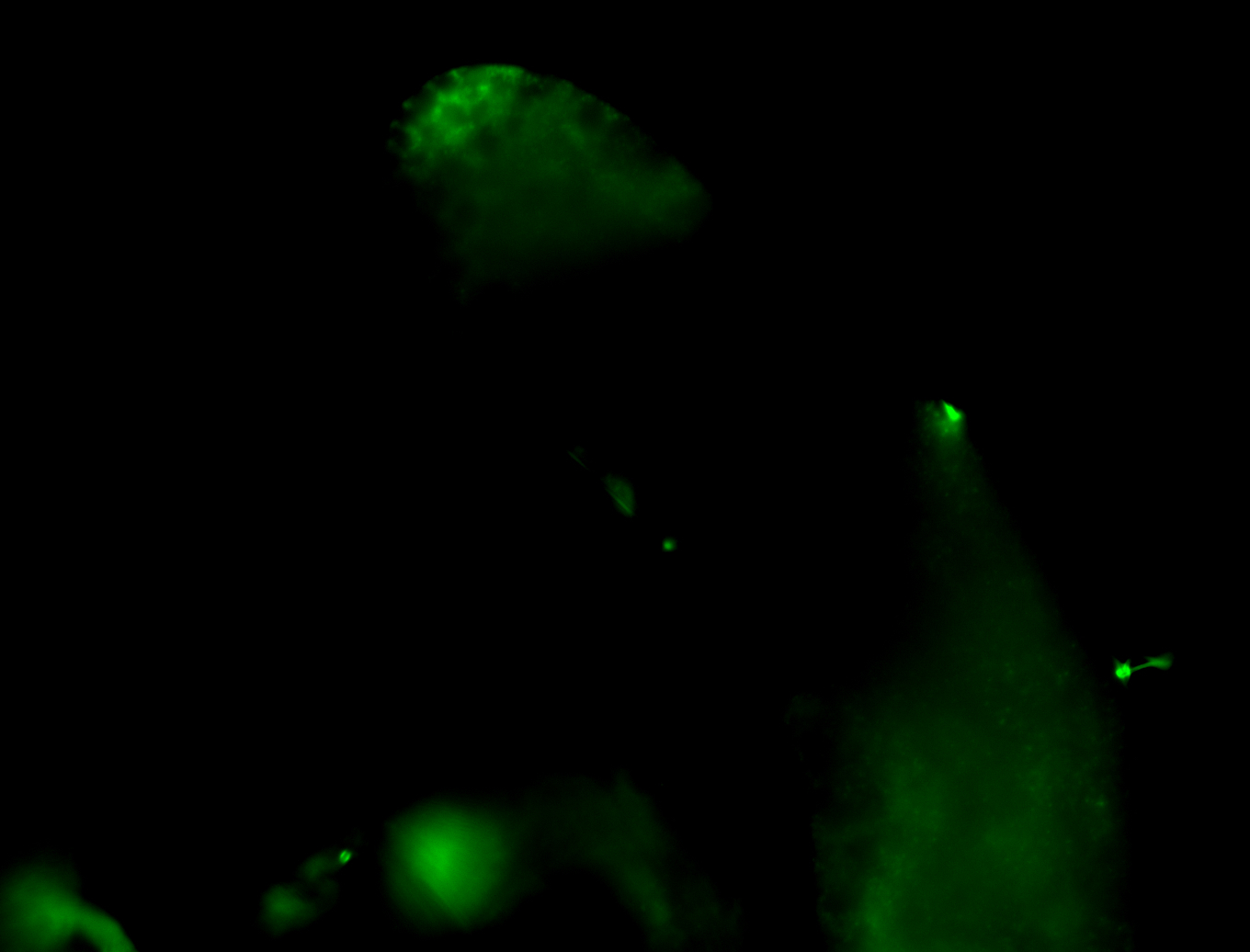
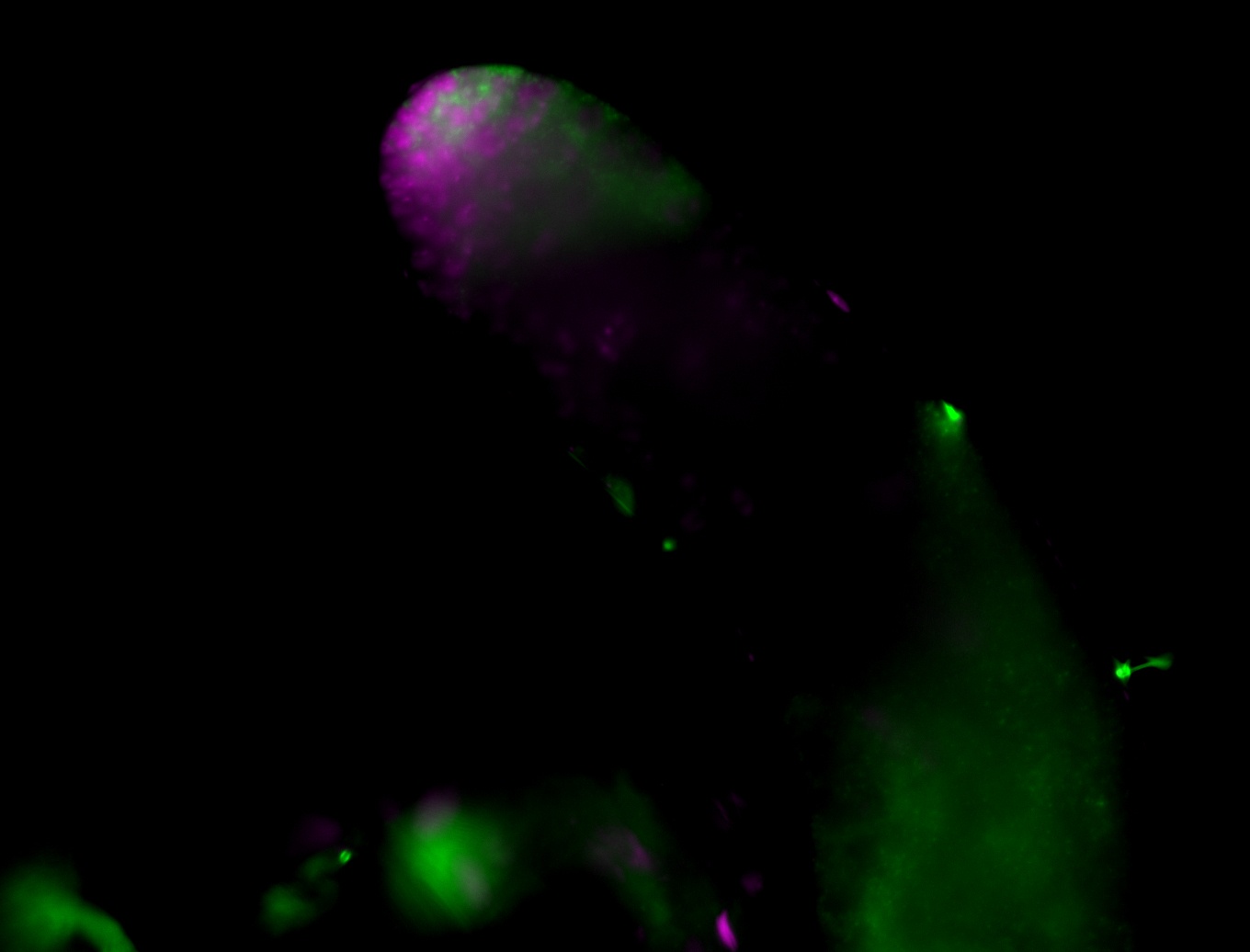
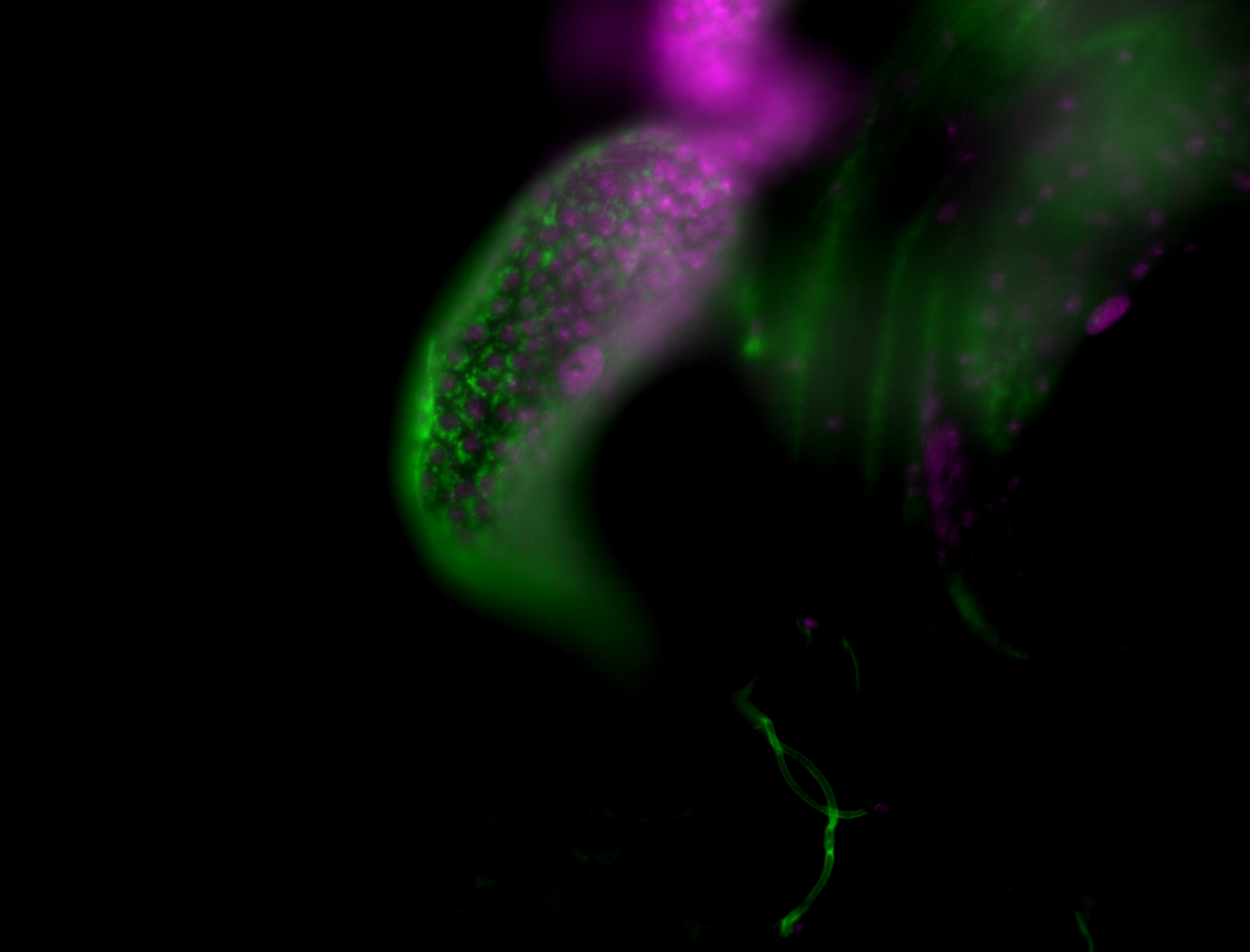
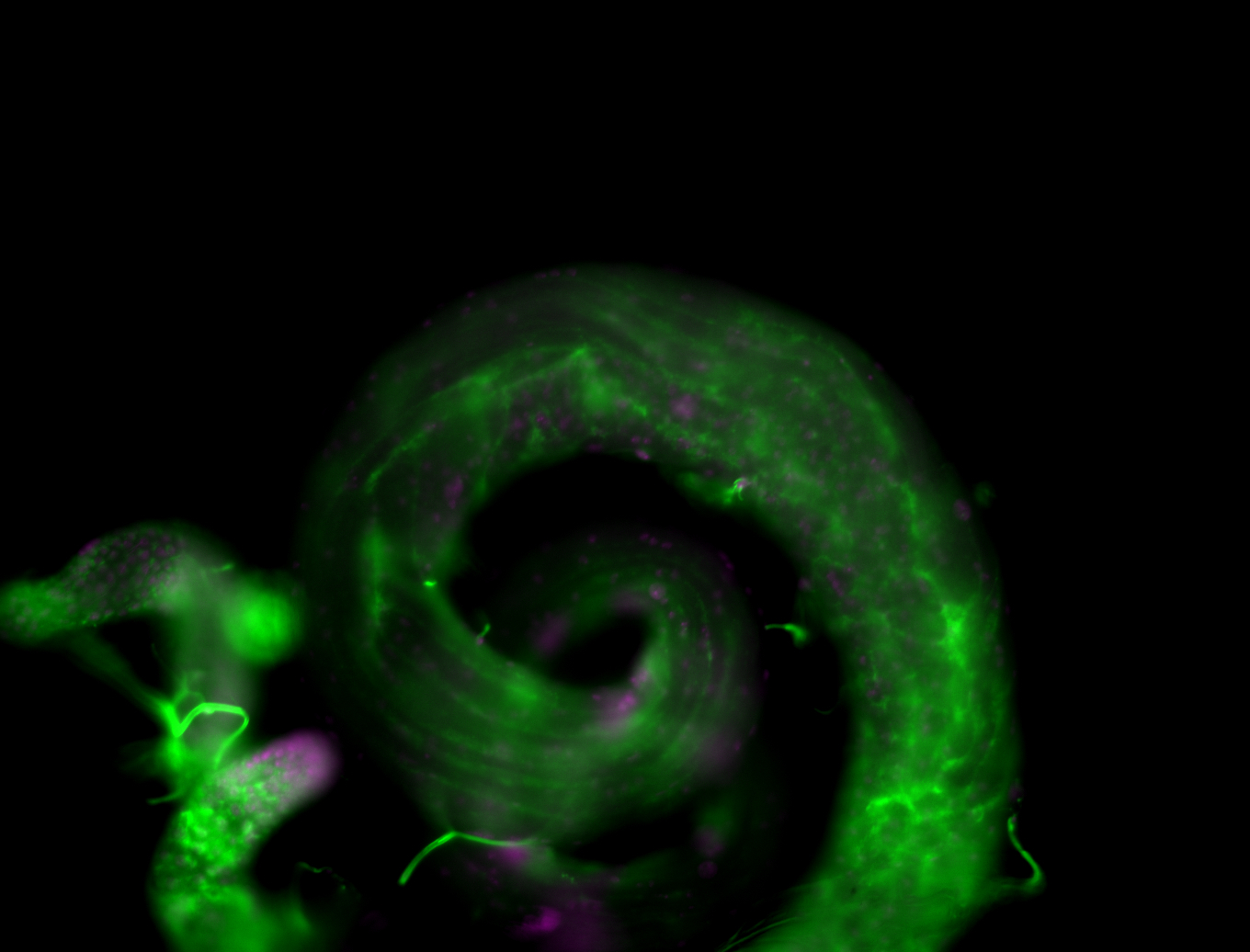
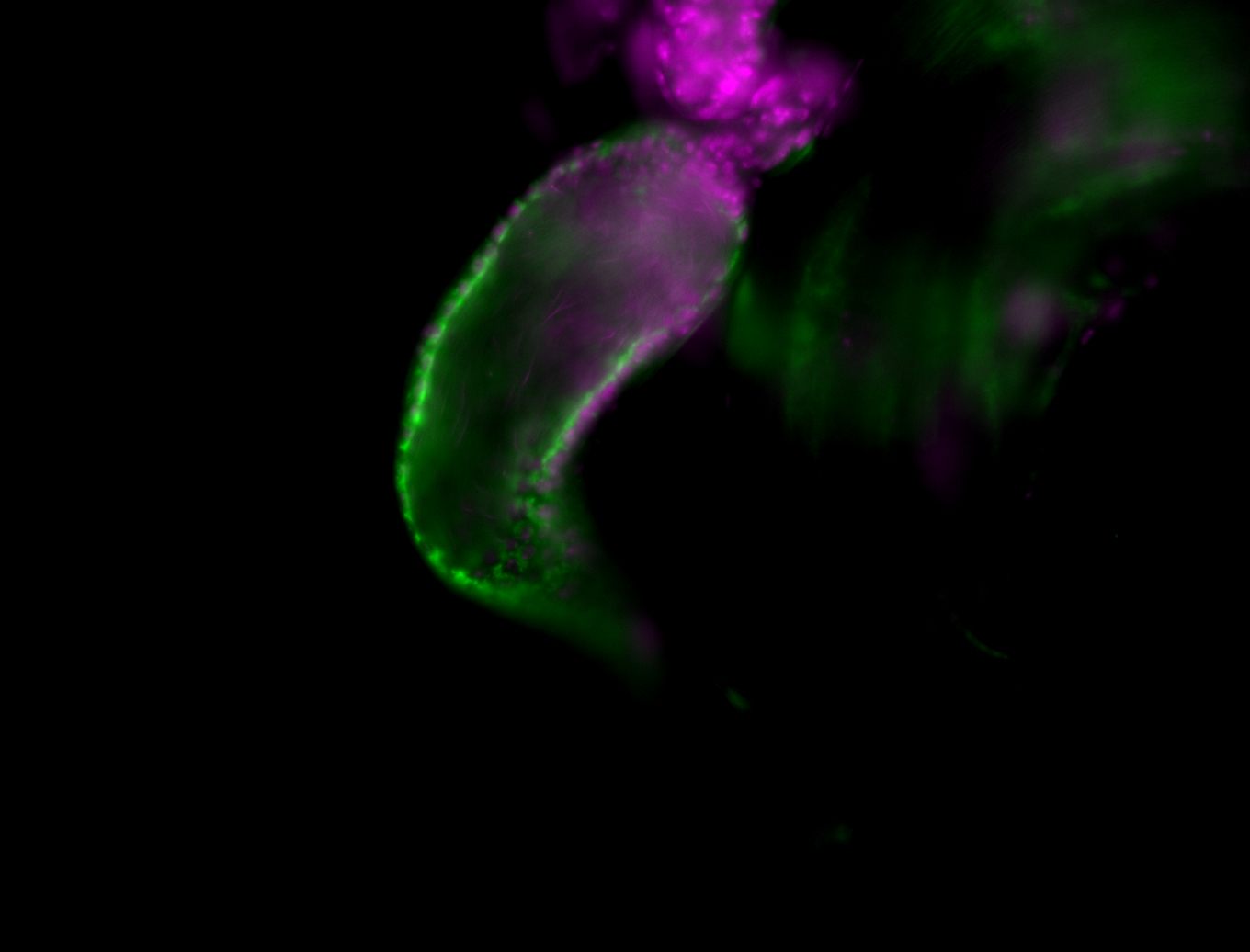
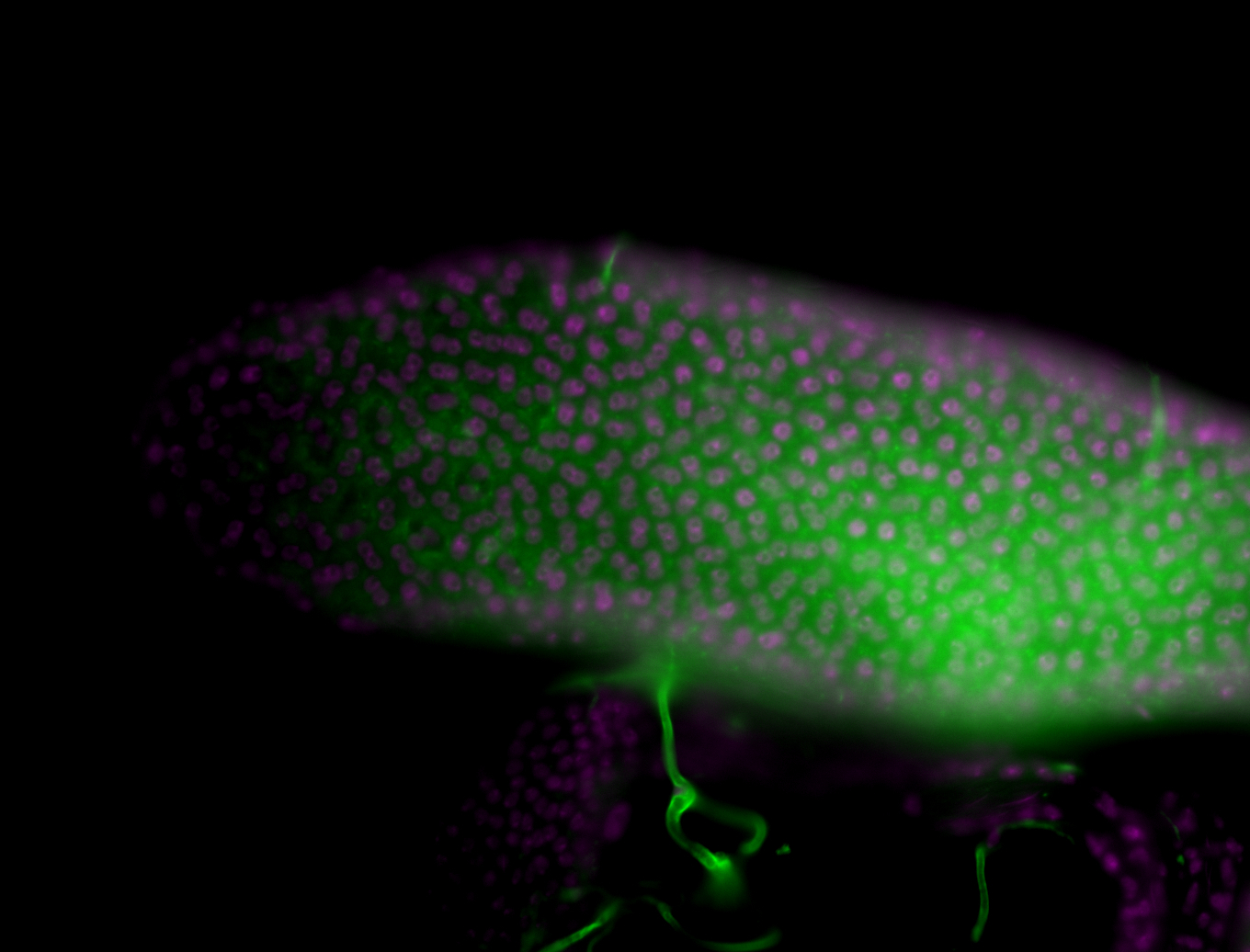
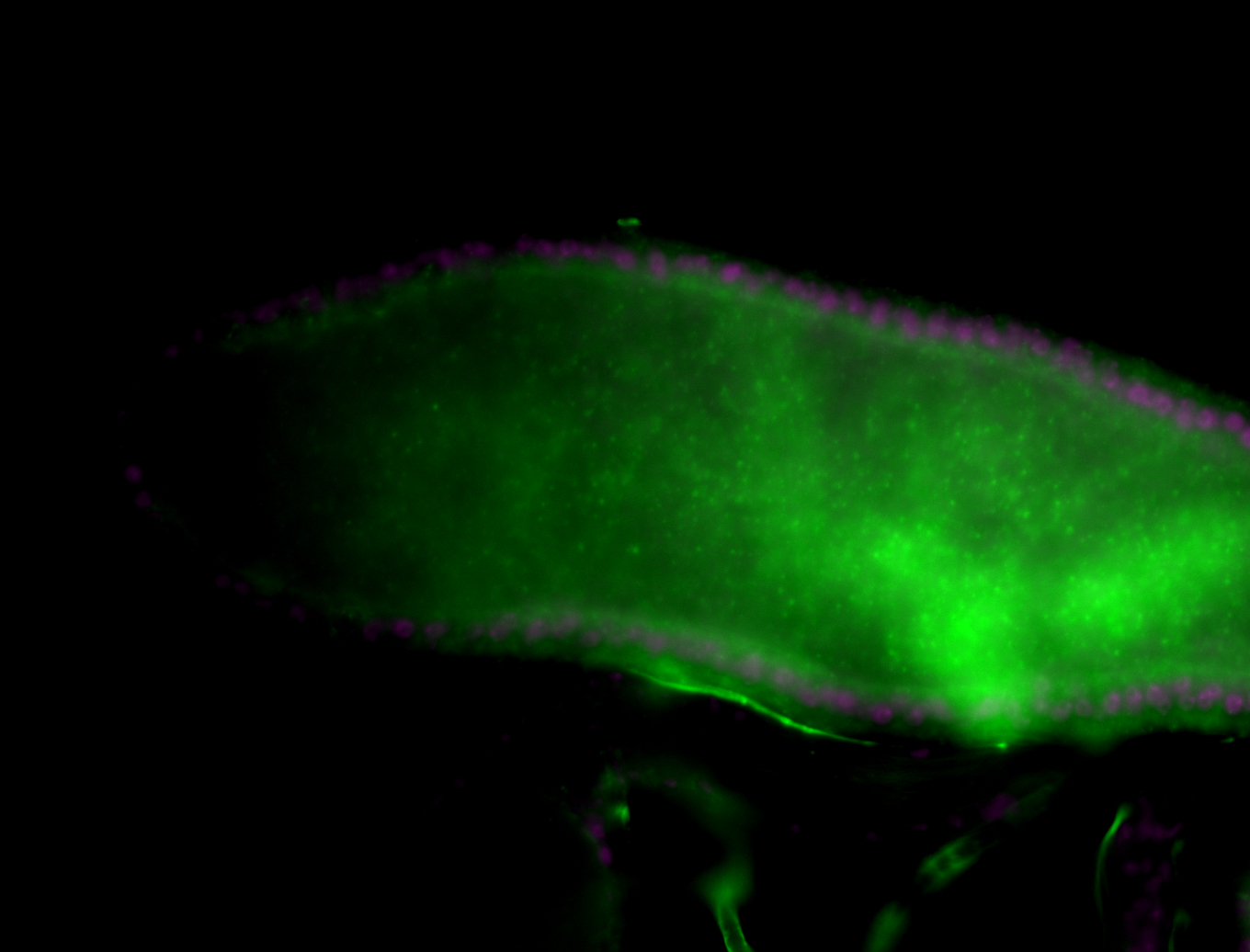
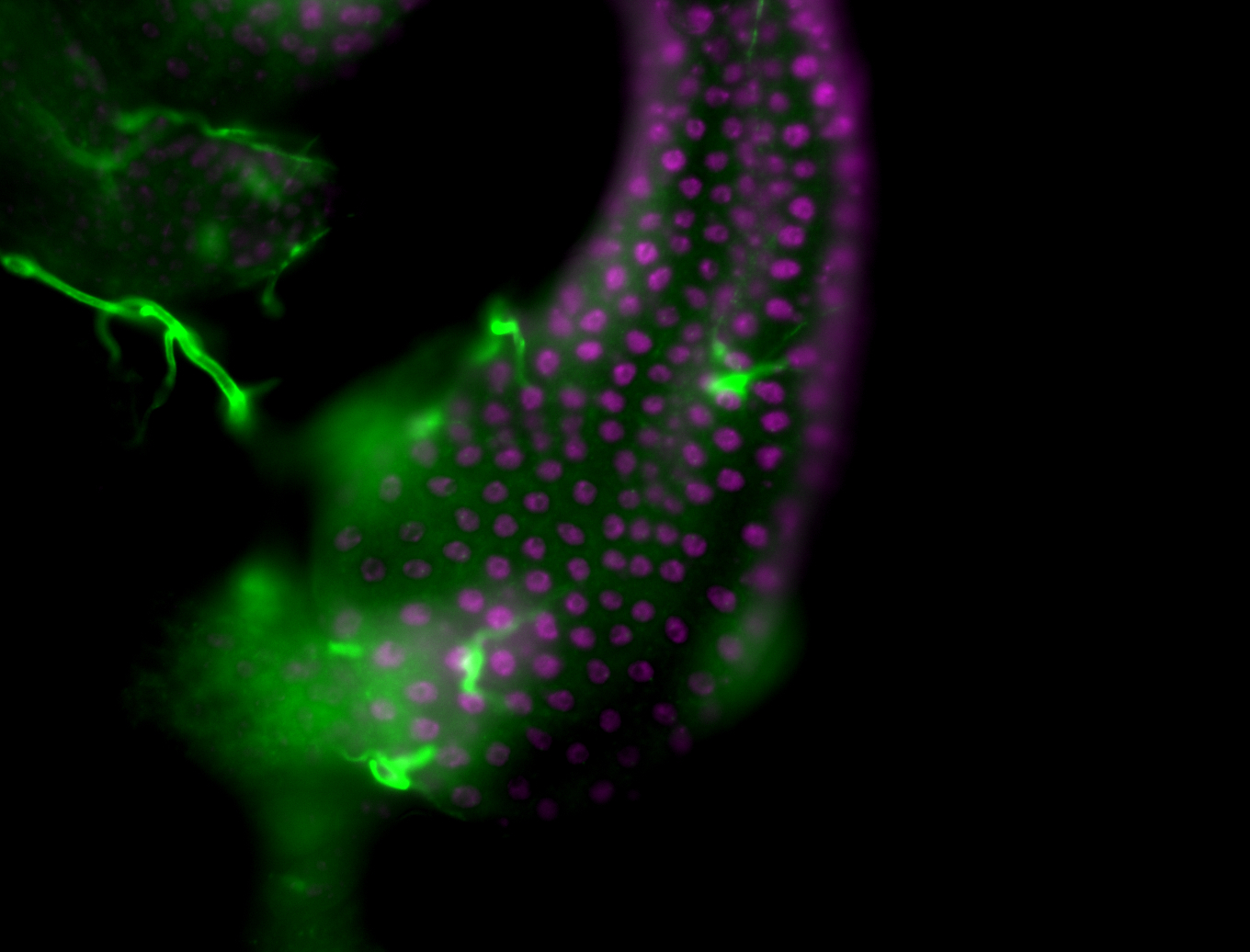
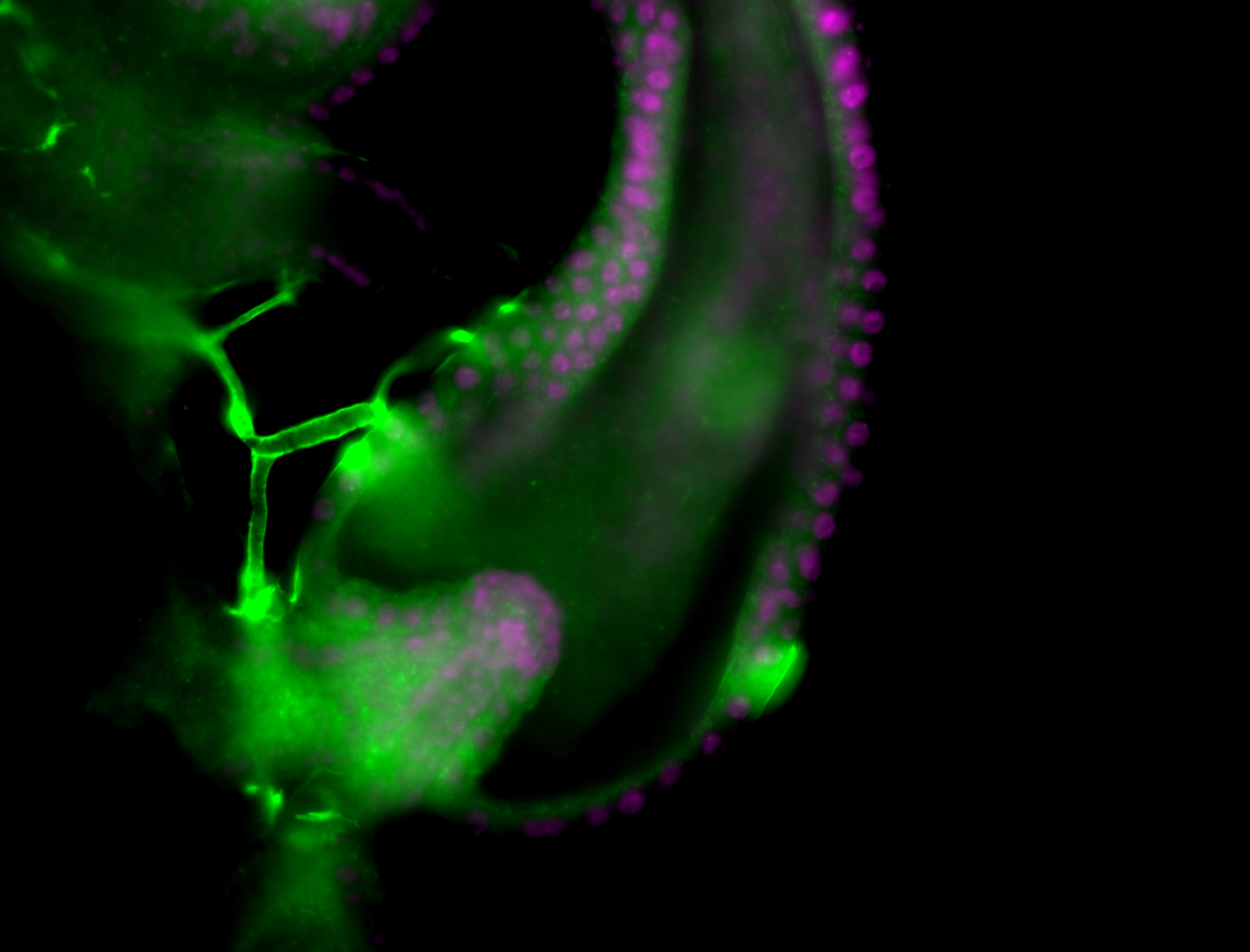
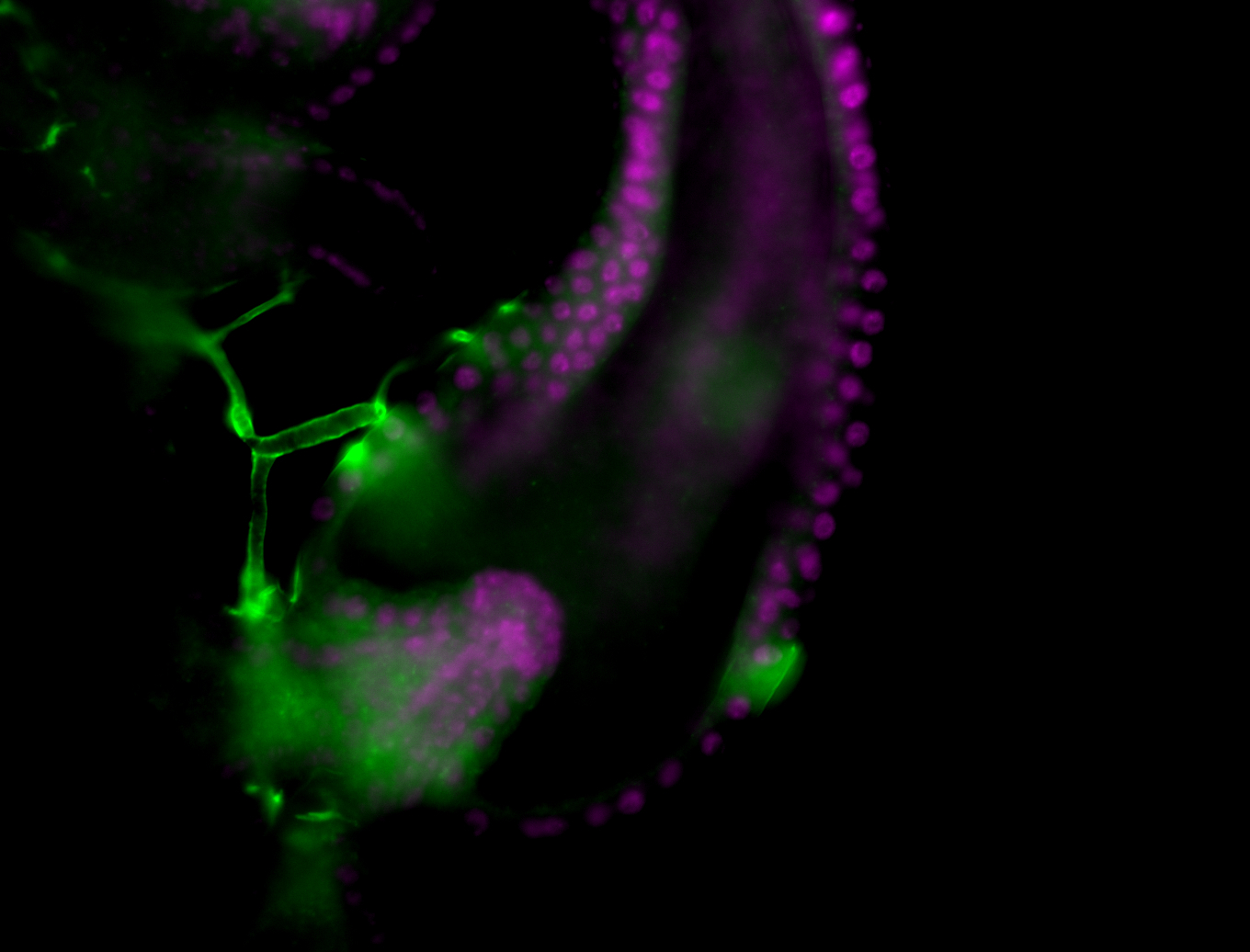
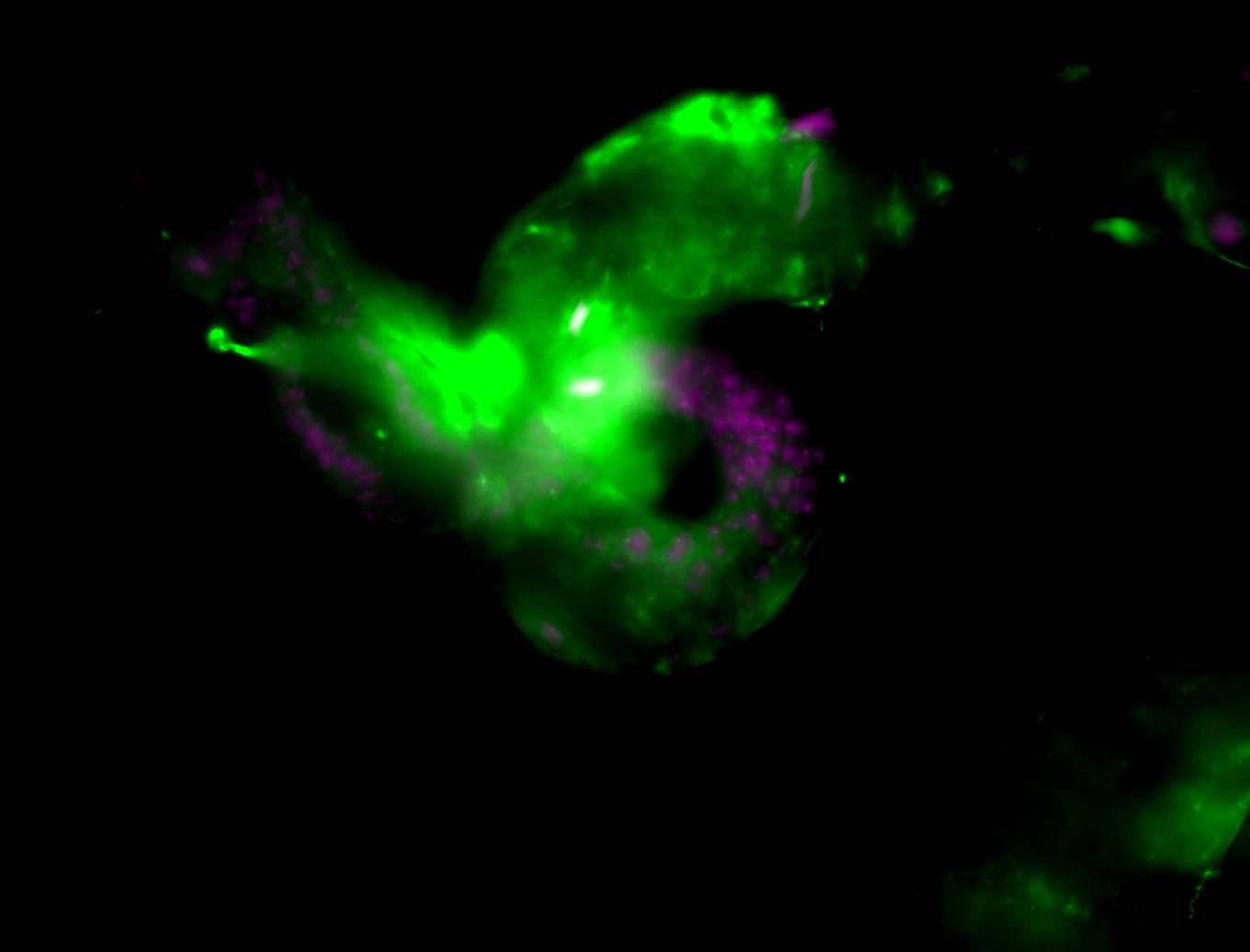
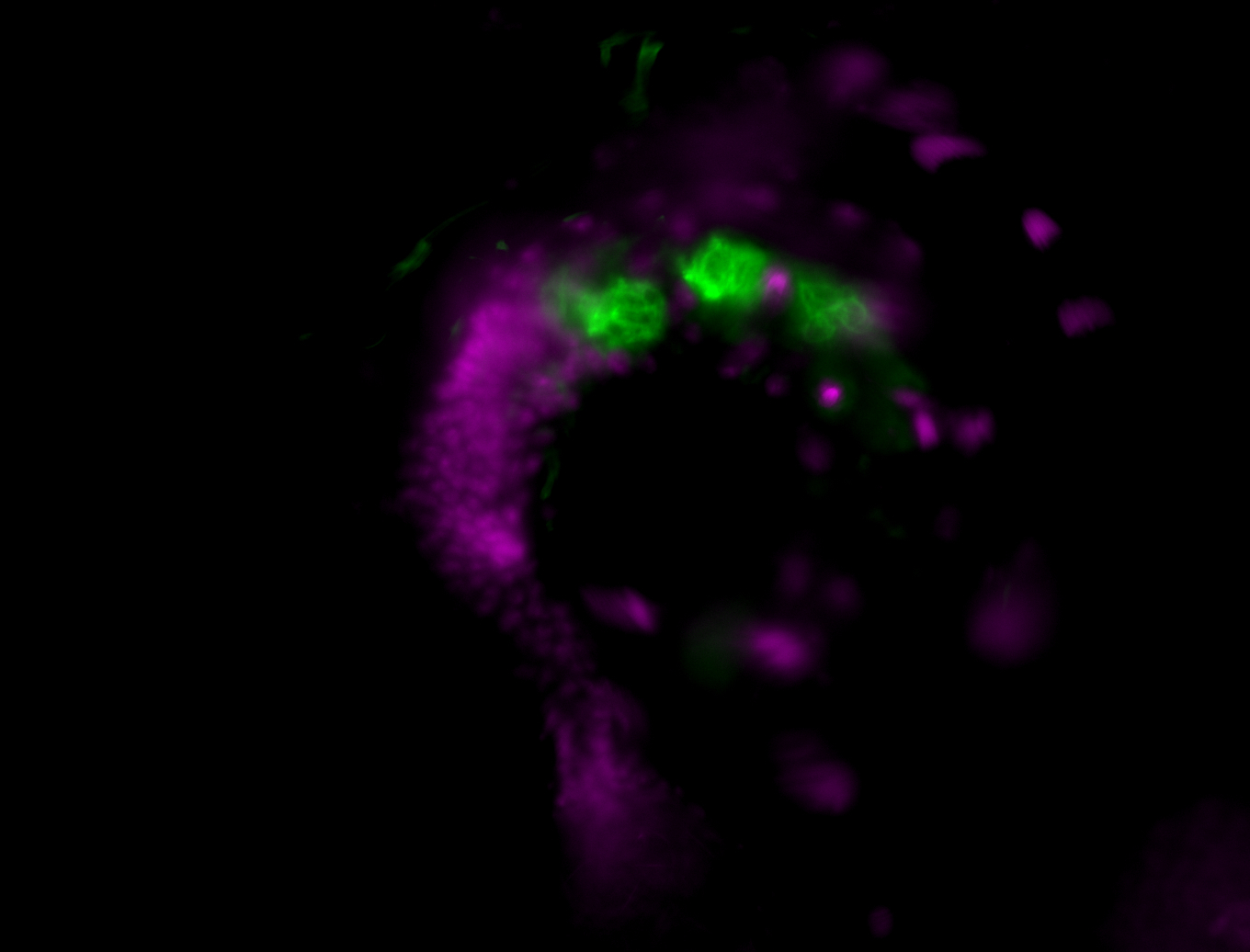
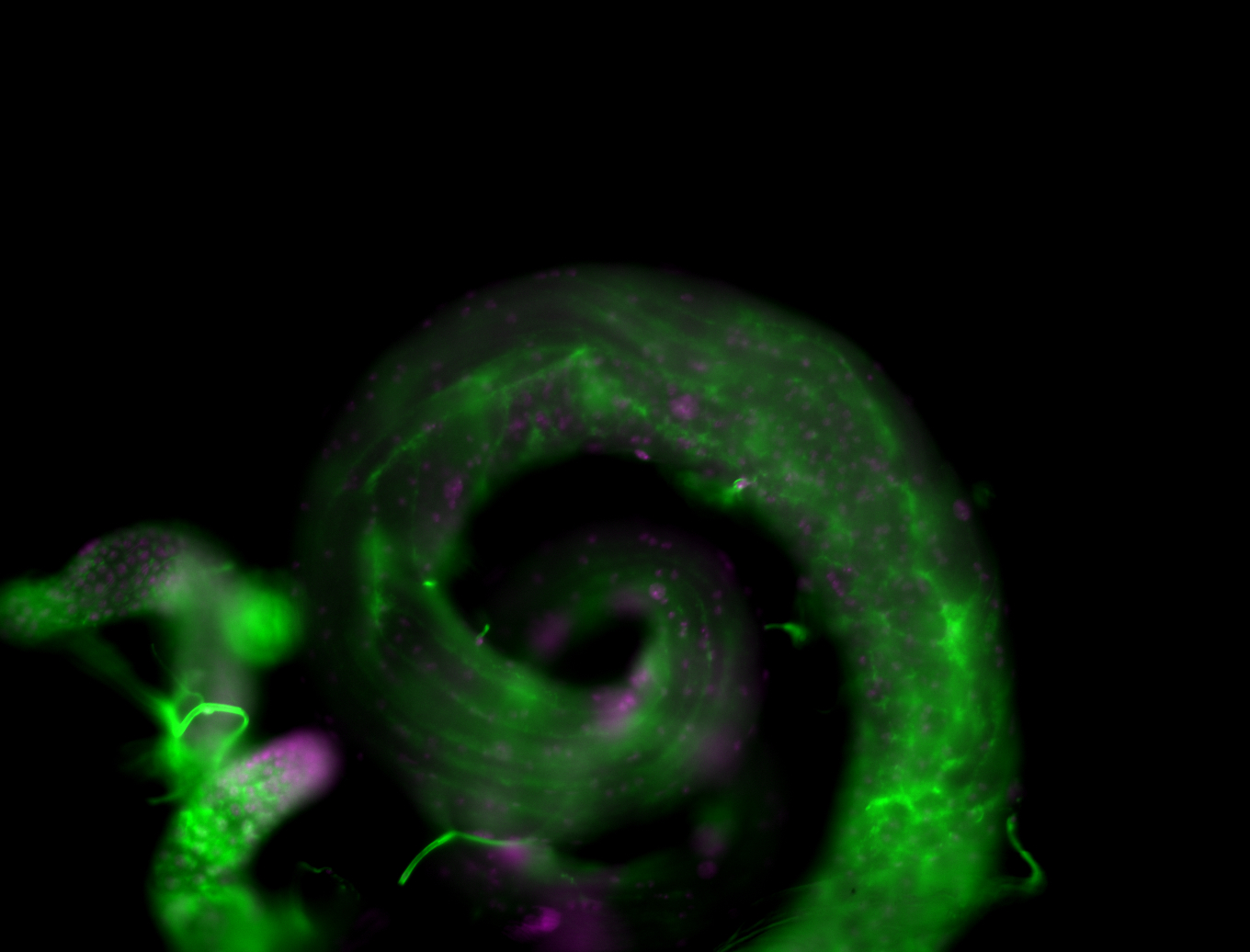
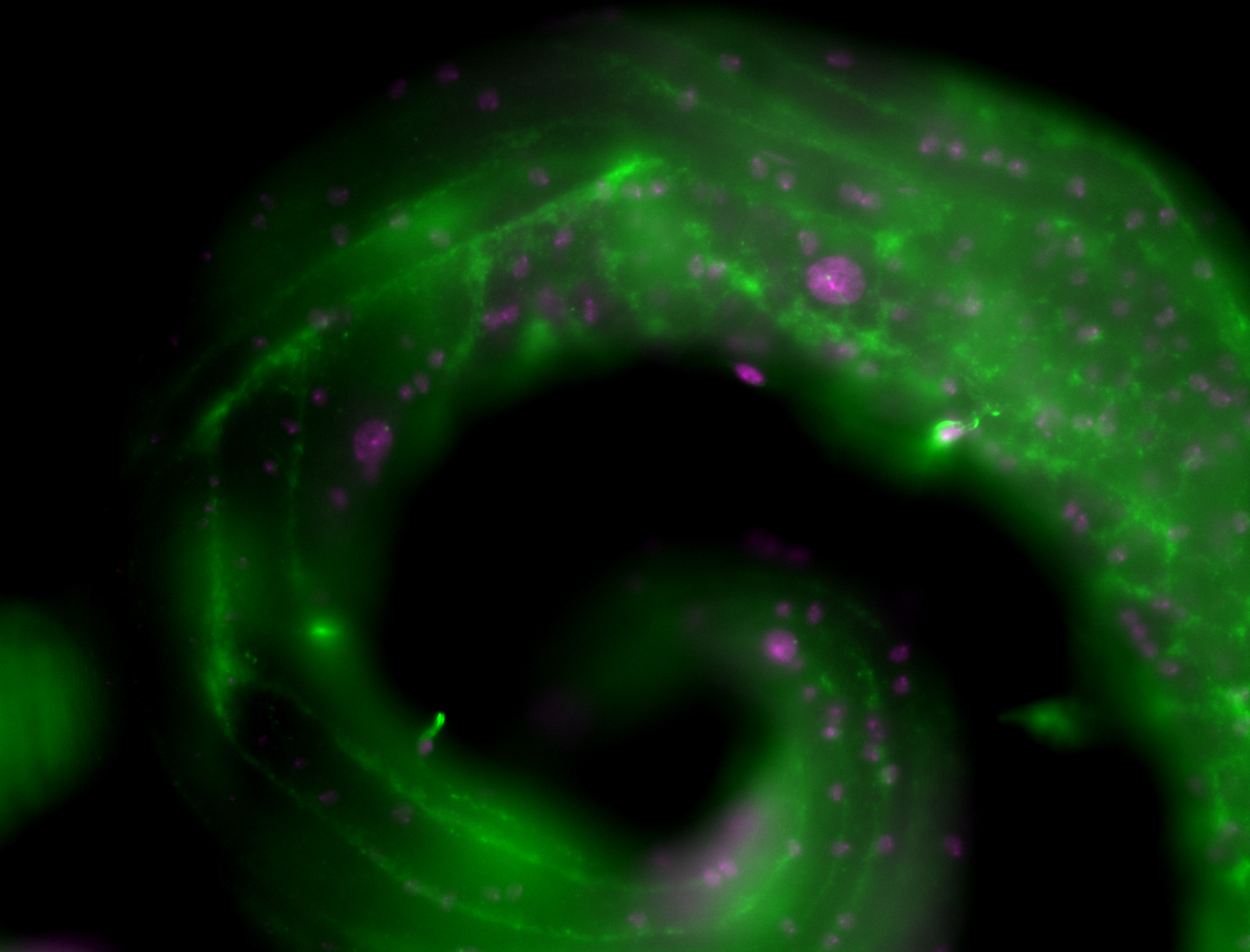
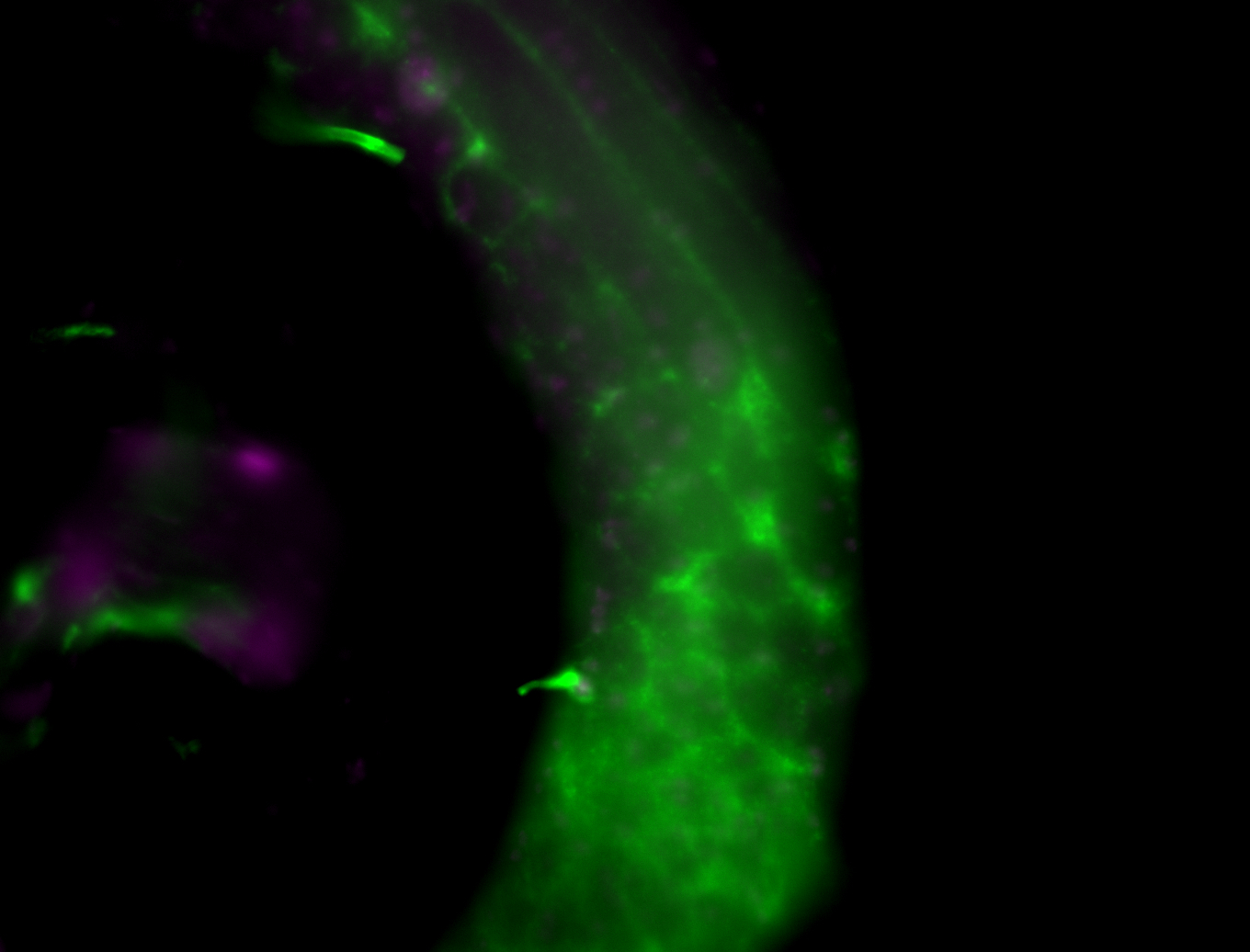
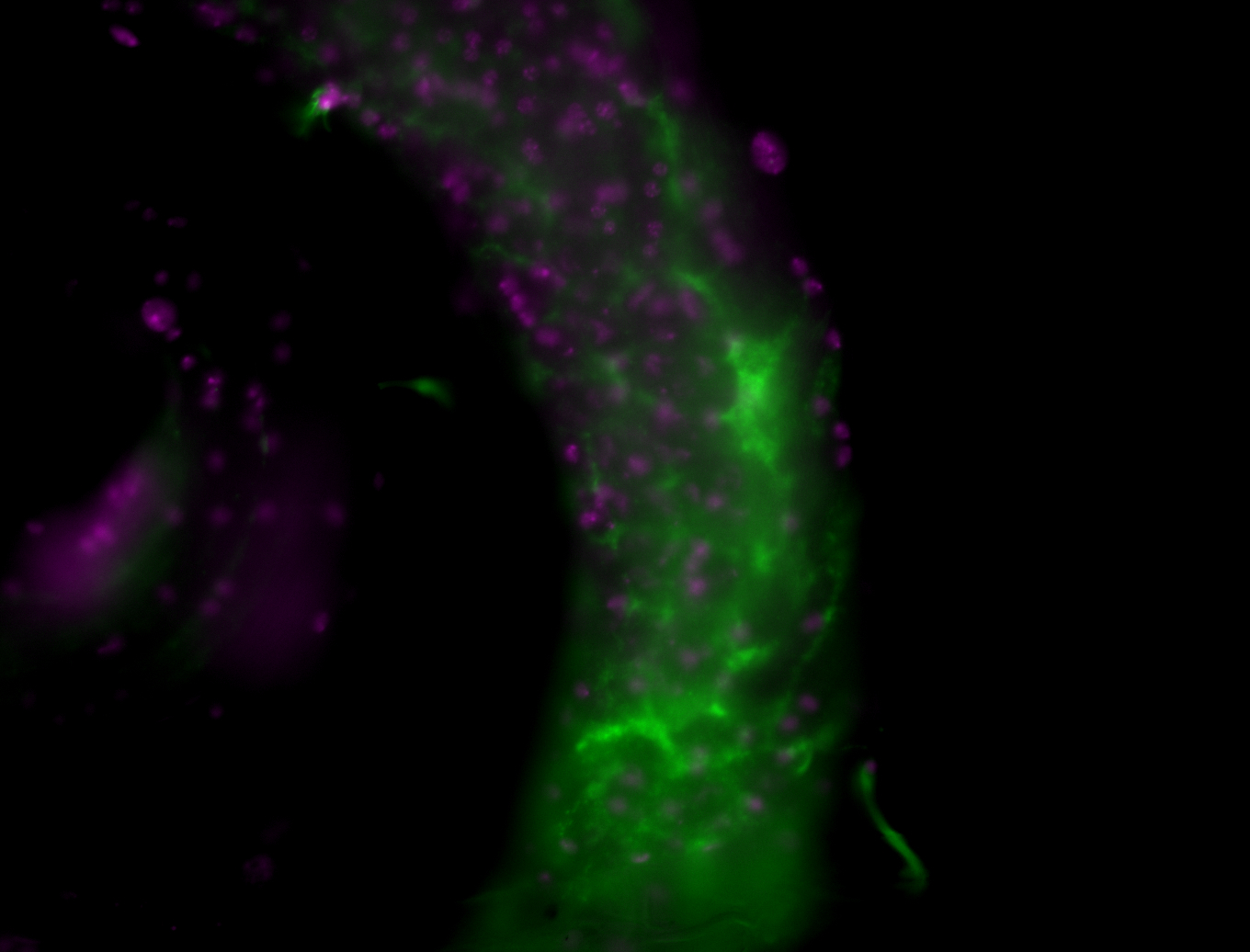
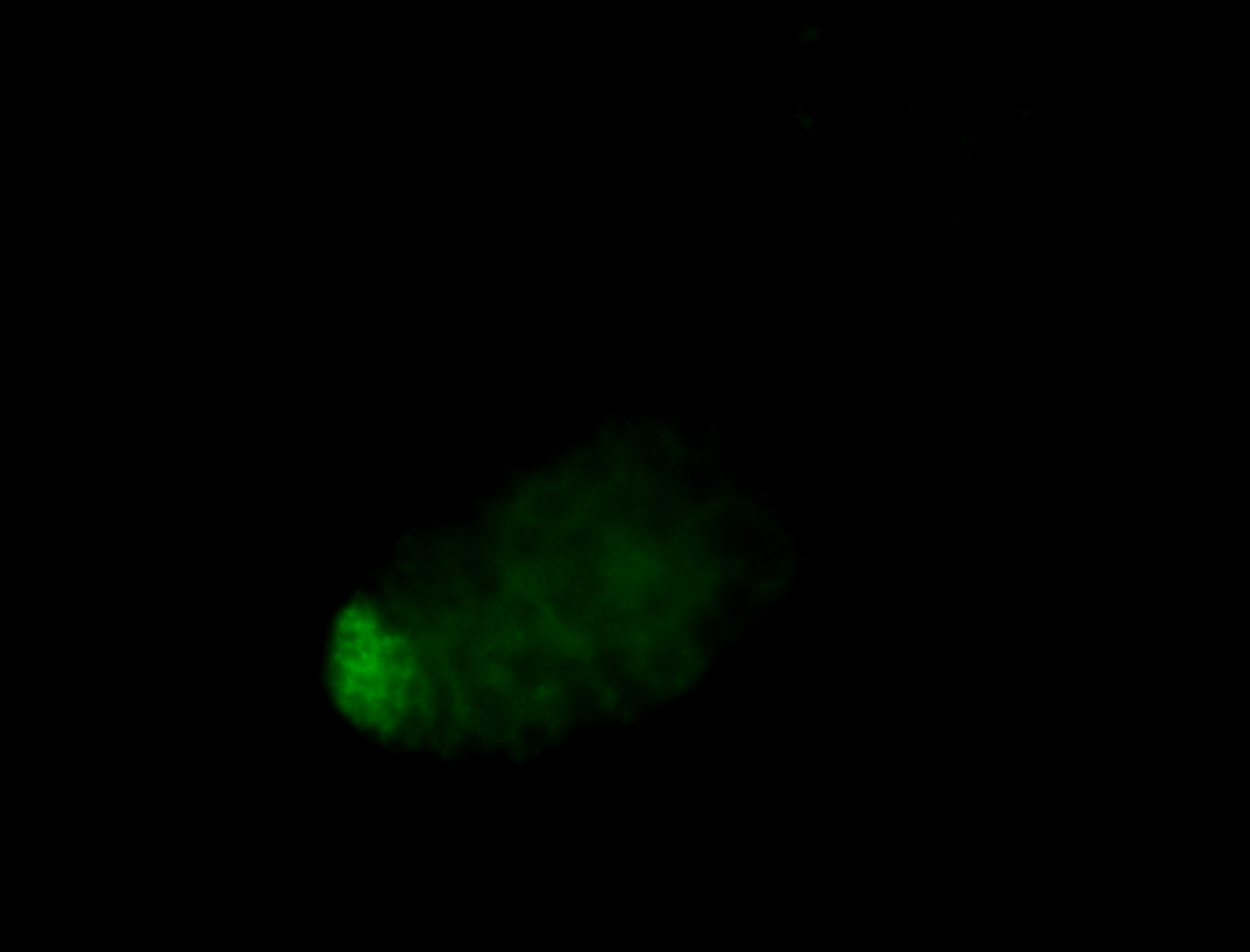
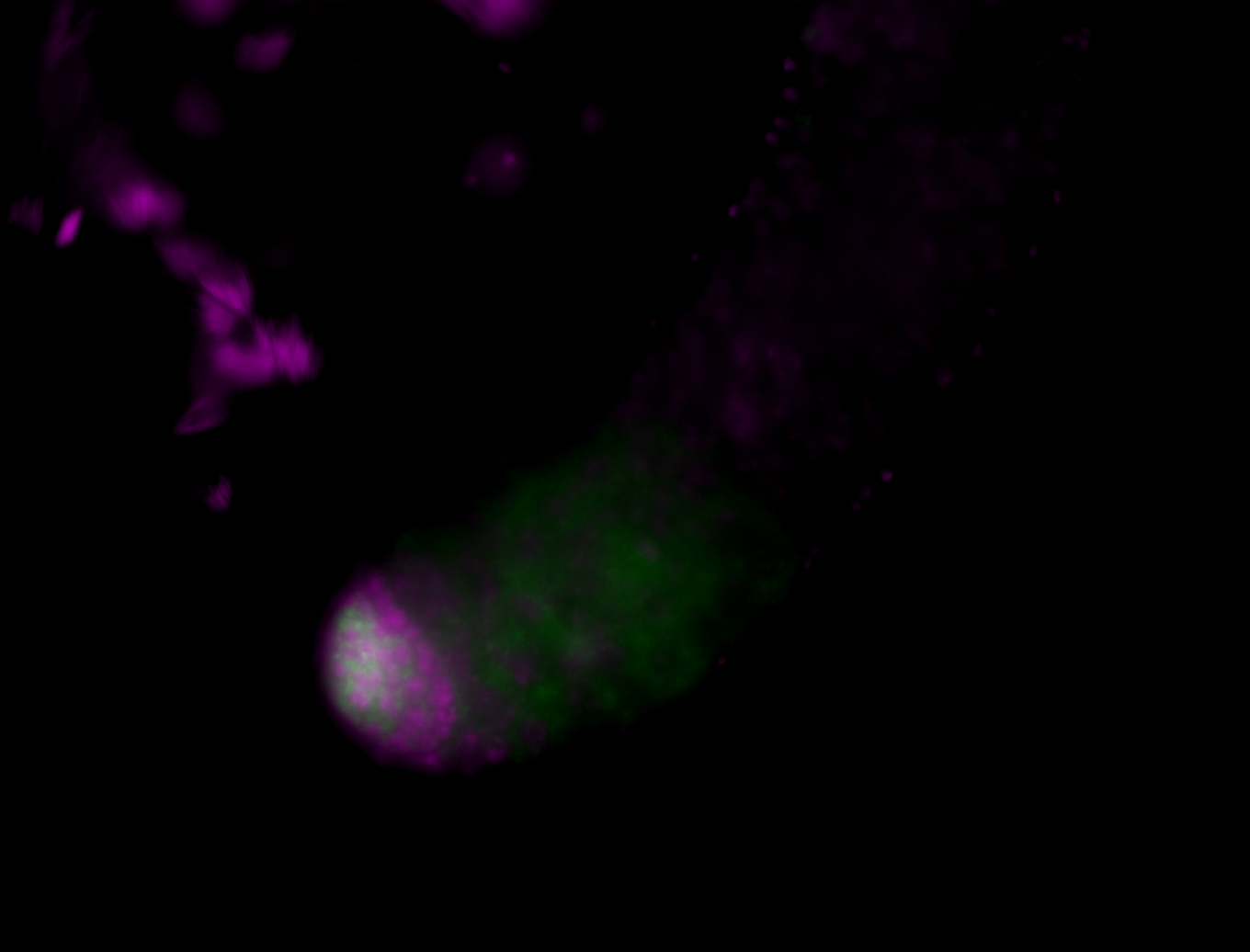
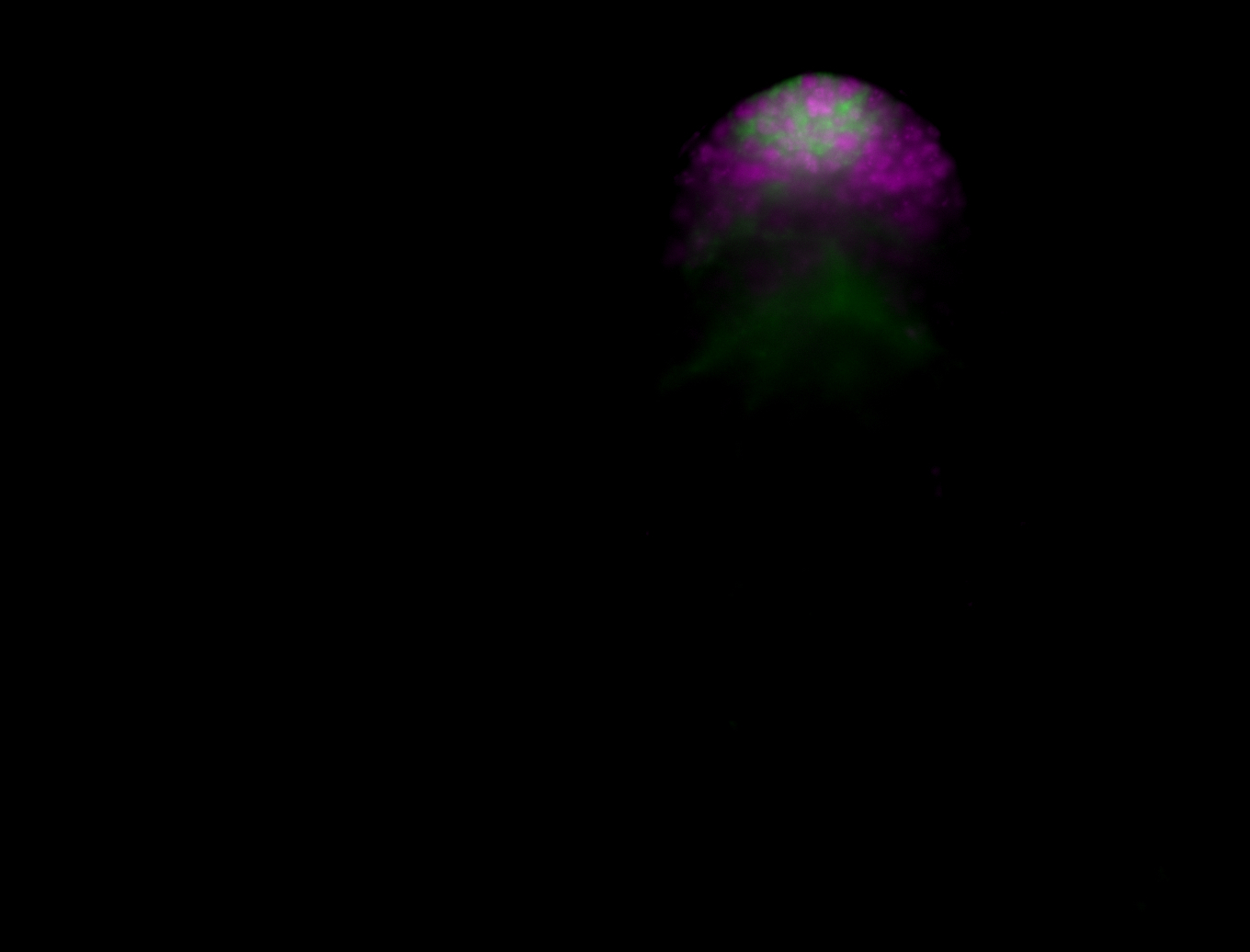
|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
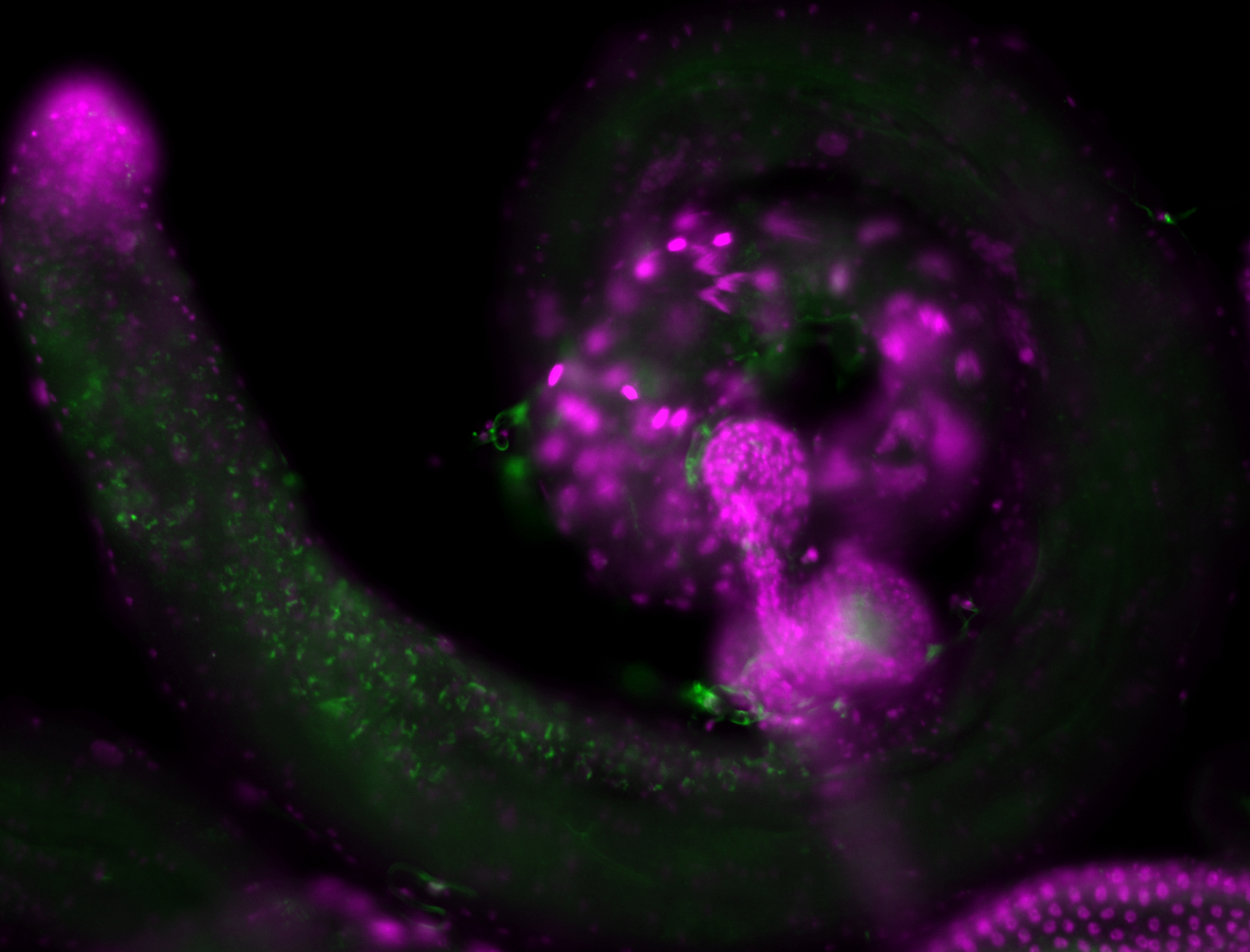
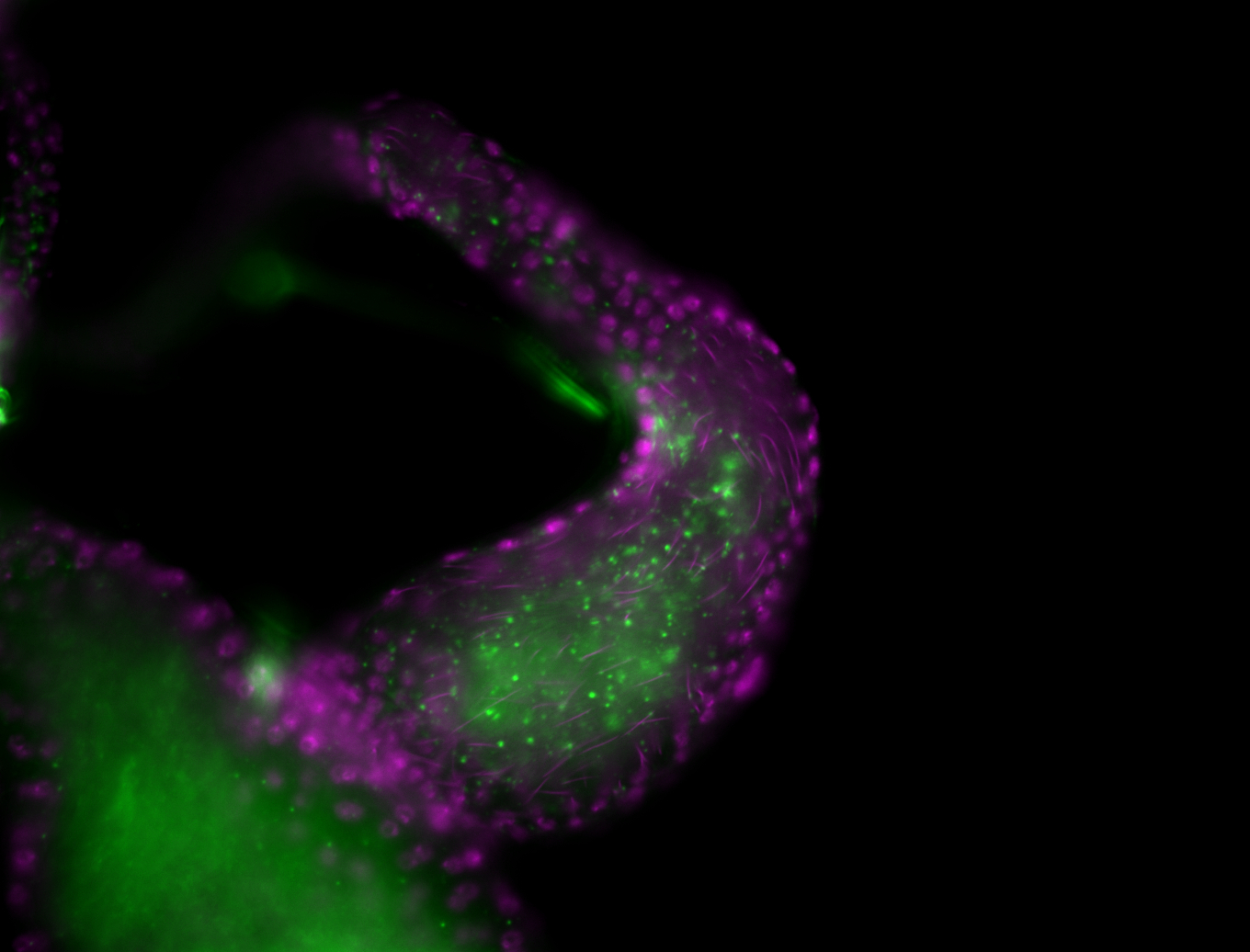
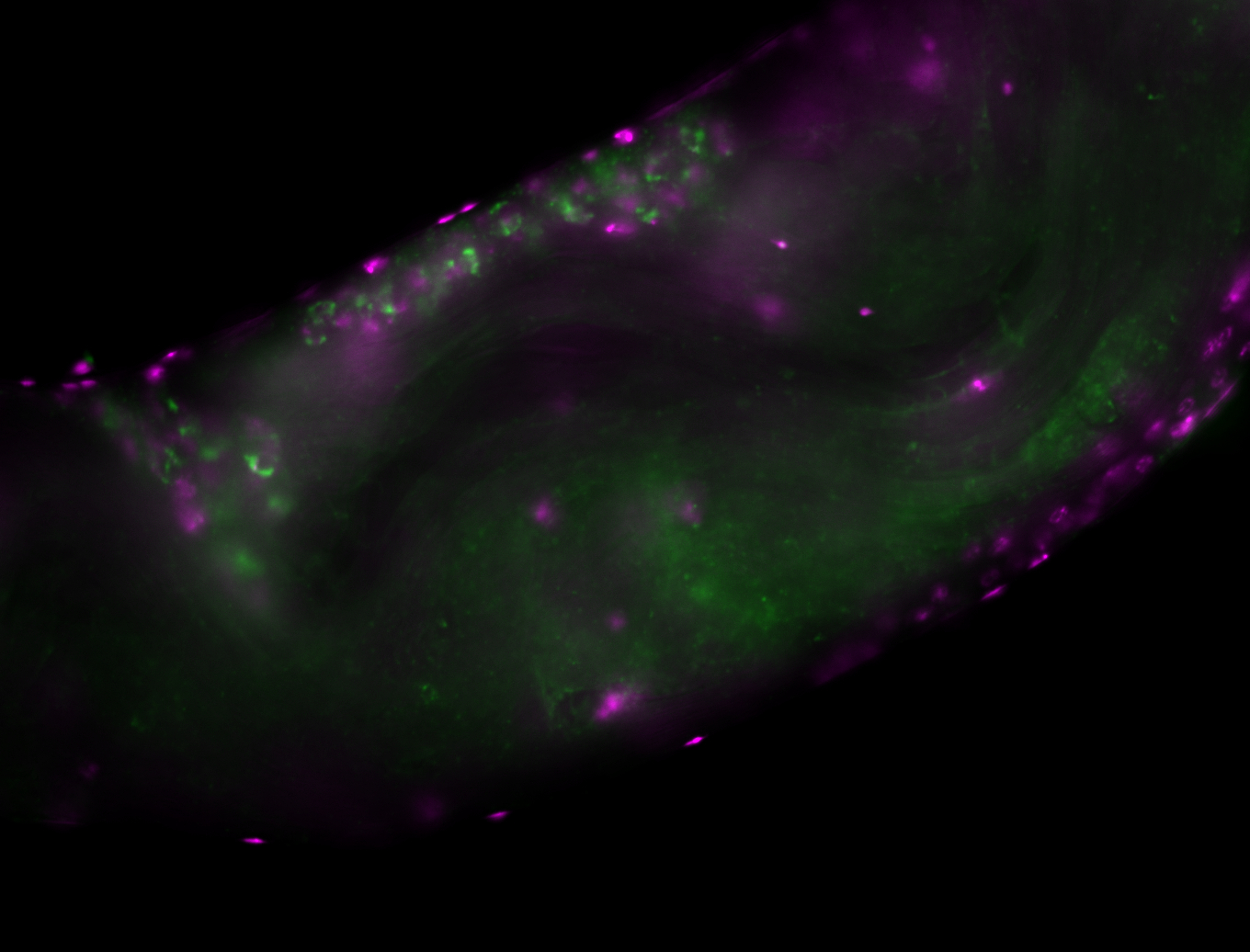
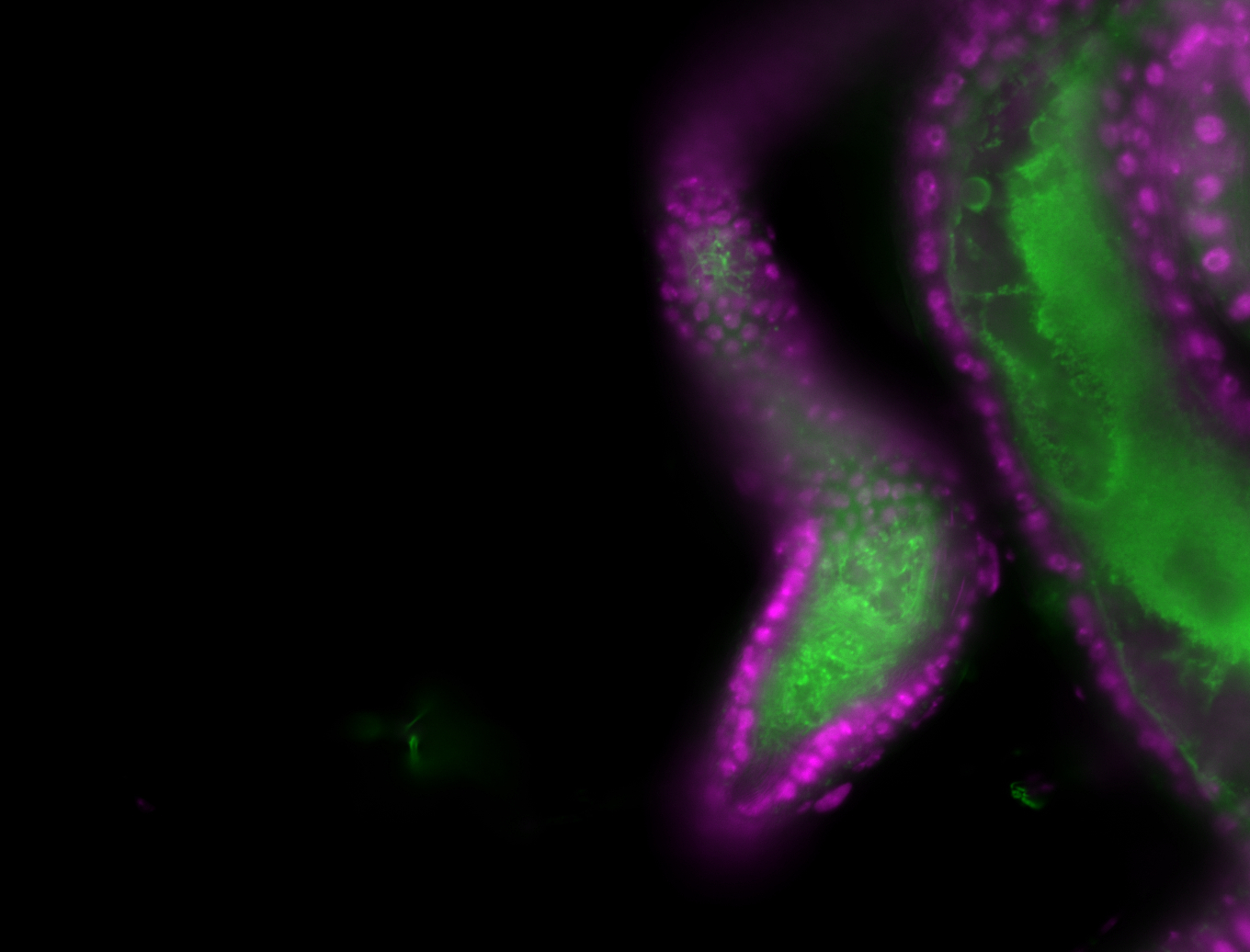
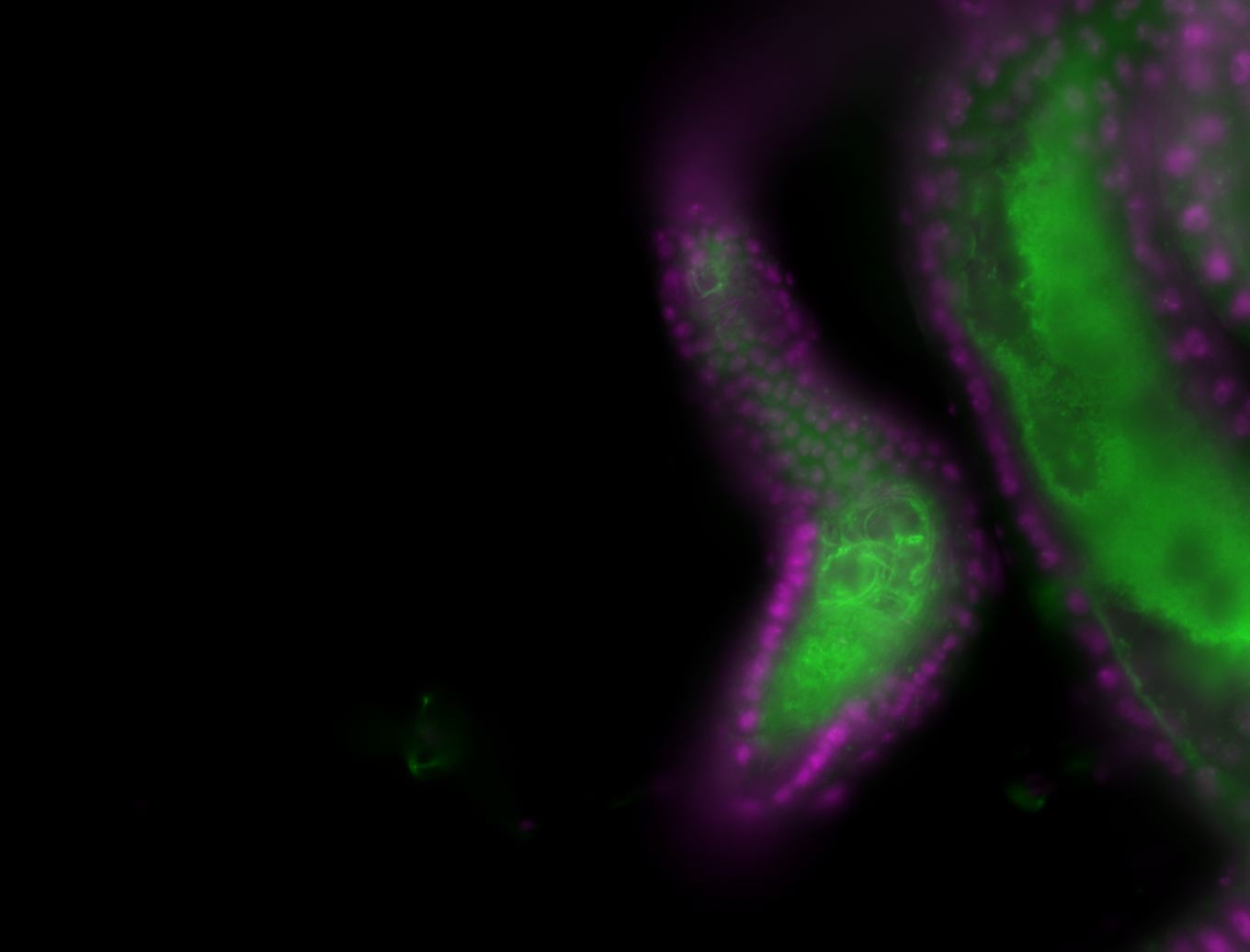
| -performed via HCR-FISH using 3 probe sets targeting the RB transcript isoform |
| -neat and diverse expression patterns throughout development |
| -images are from two separate experiments under identical conditions for better coverage |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
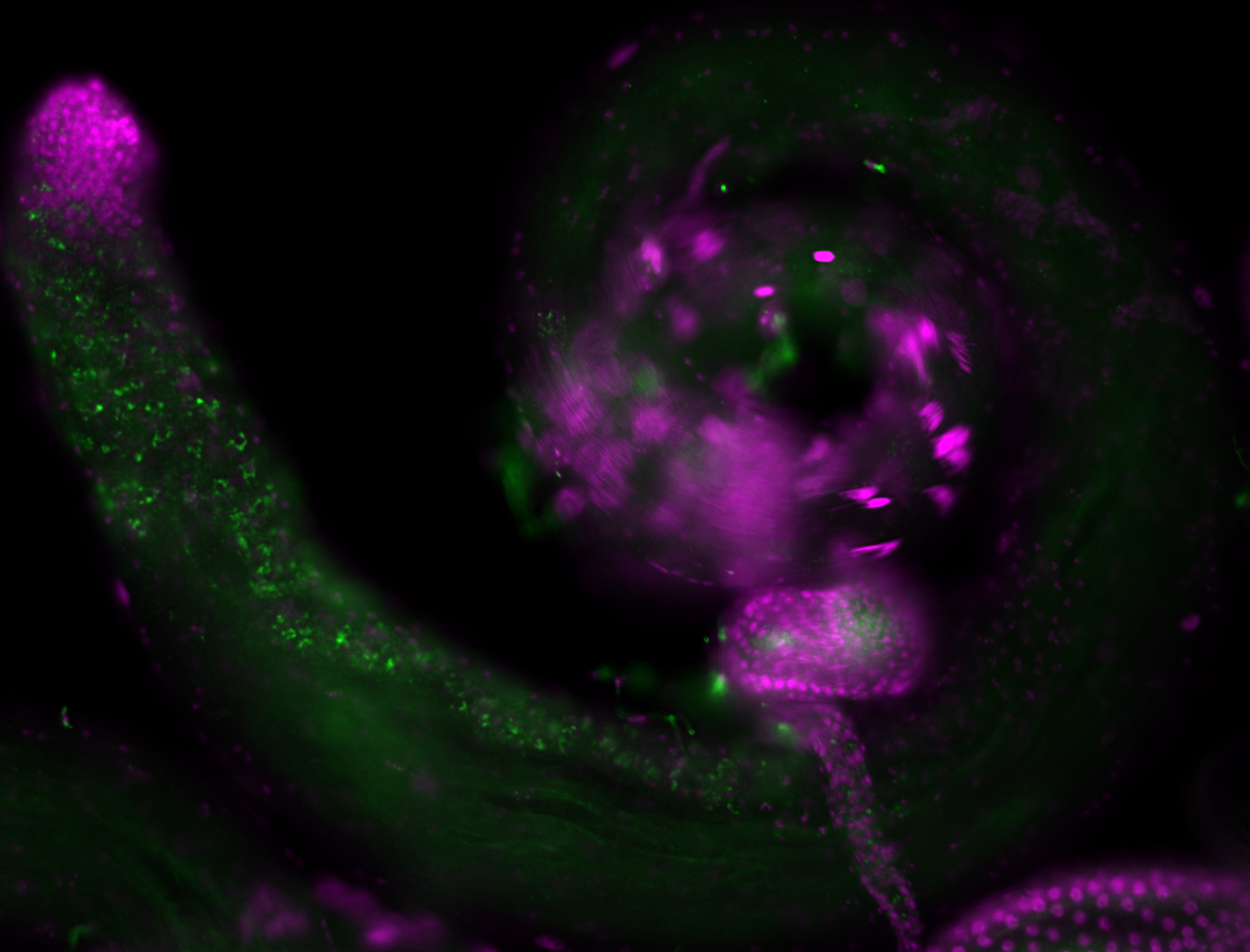
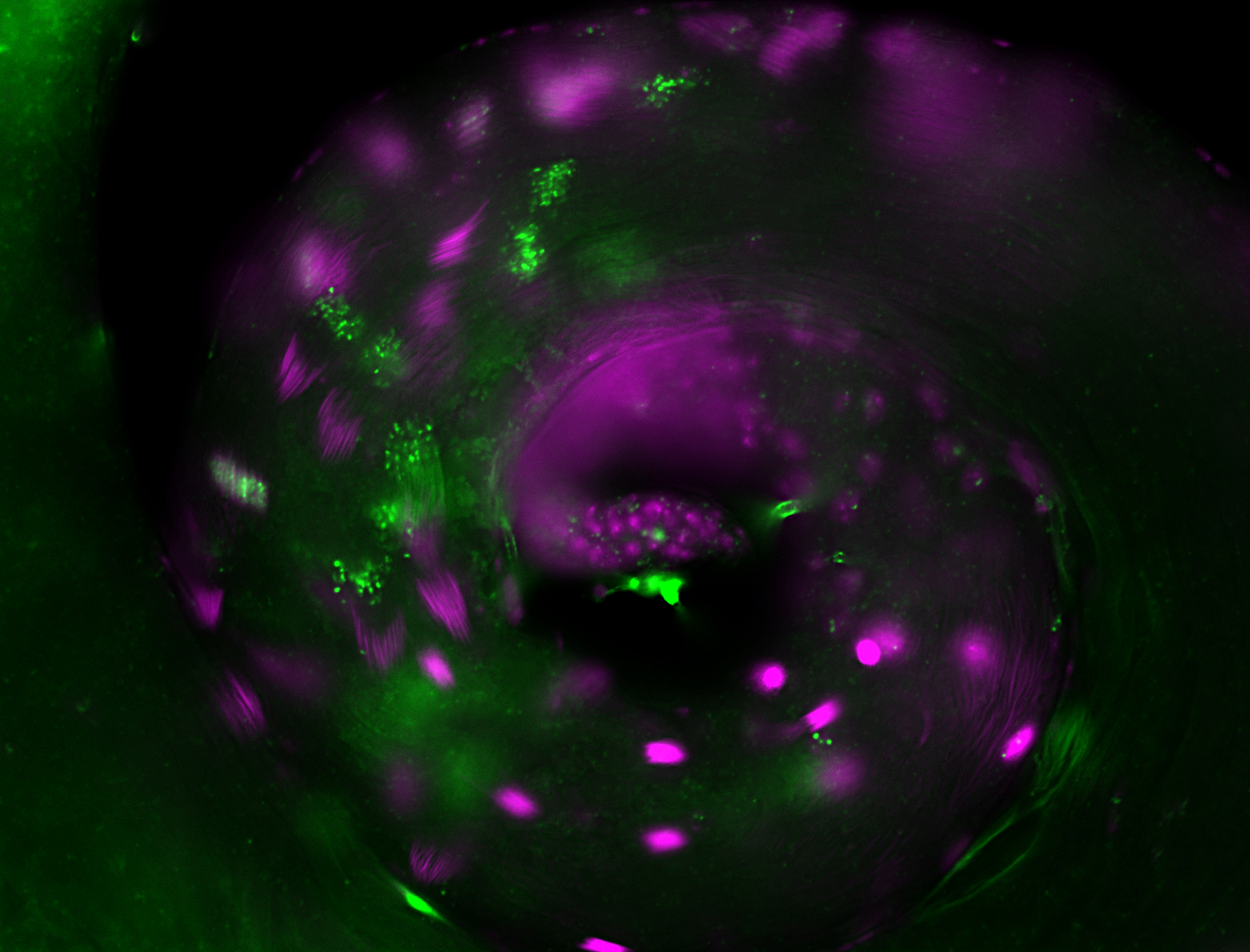
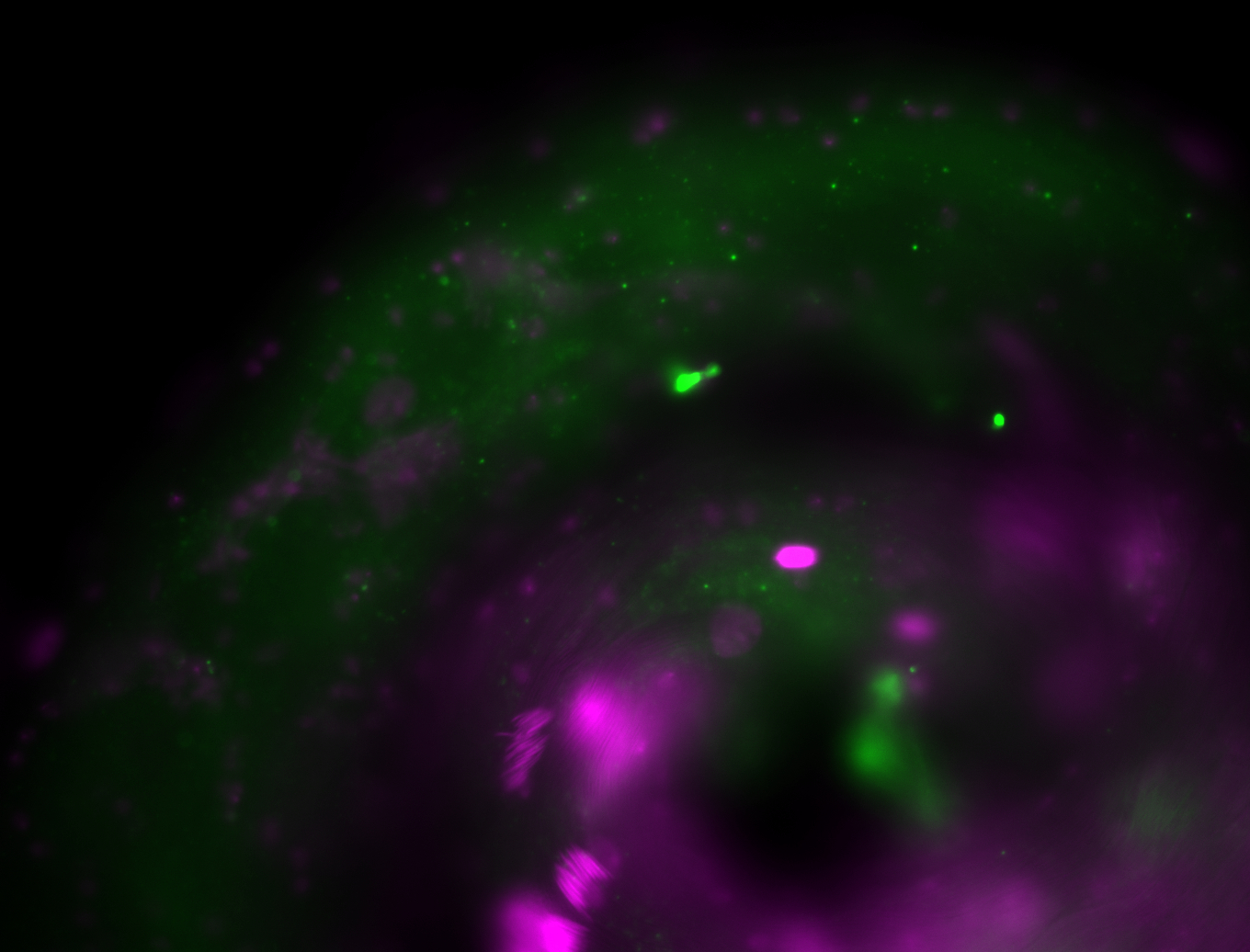
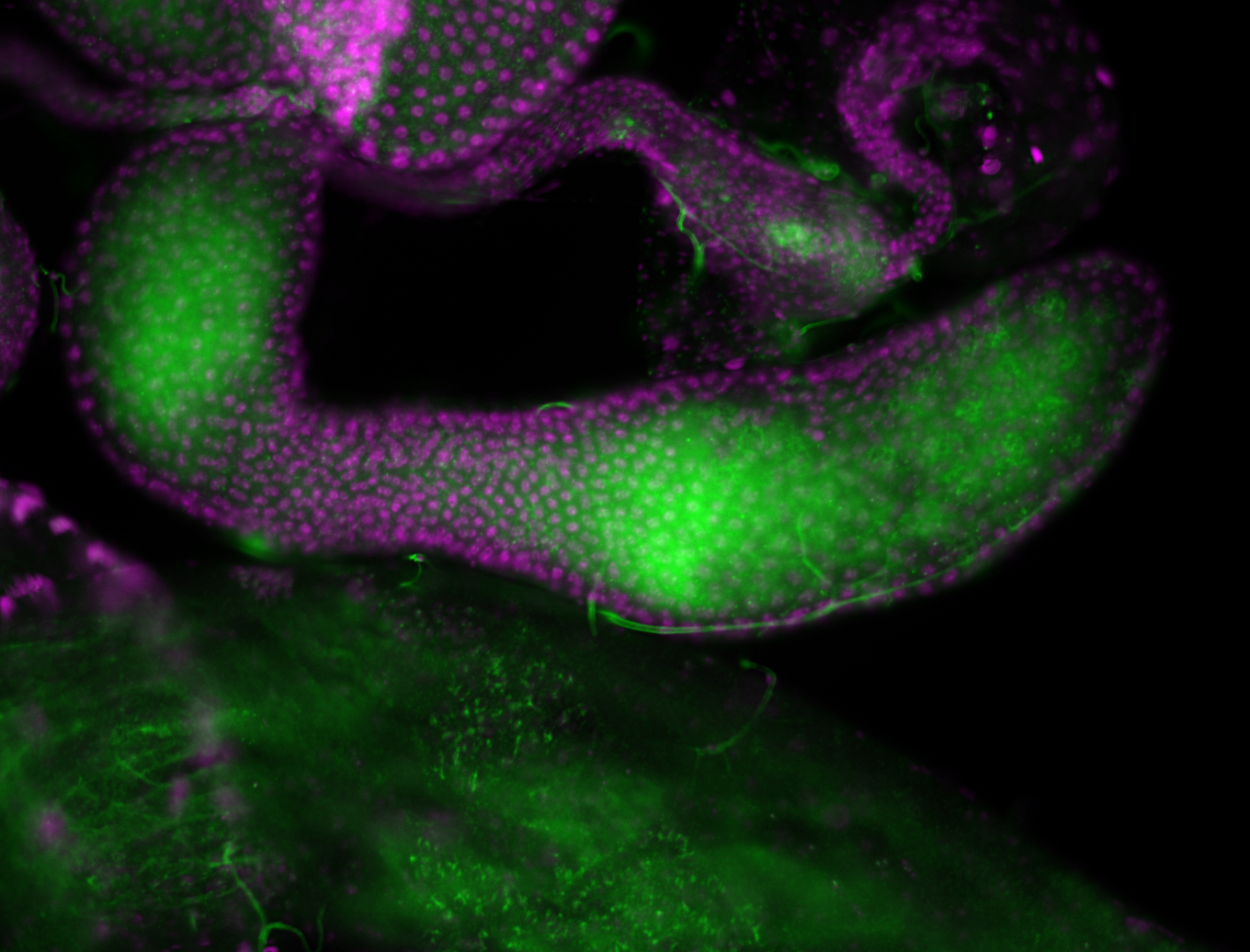
| -performed via HCR-FISH using 3 probe sets targeting the RA transcript isoform |
| -neat expression patterns in early embryos and early germ band elongation stages; expression is no longer detectable after stage 9-10 (fits modENCODE RNA-seq data well) |