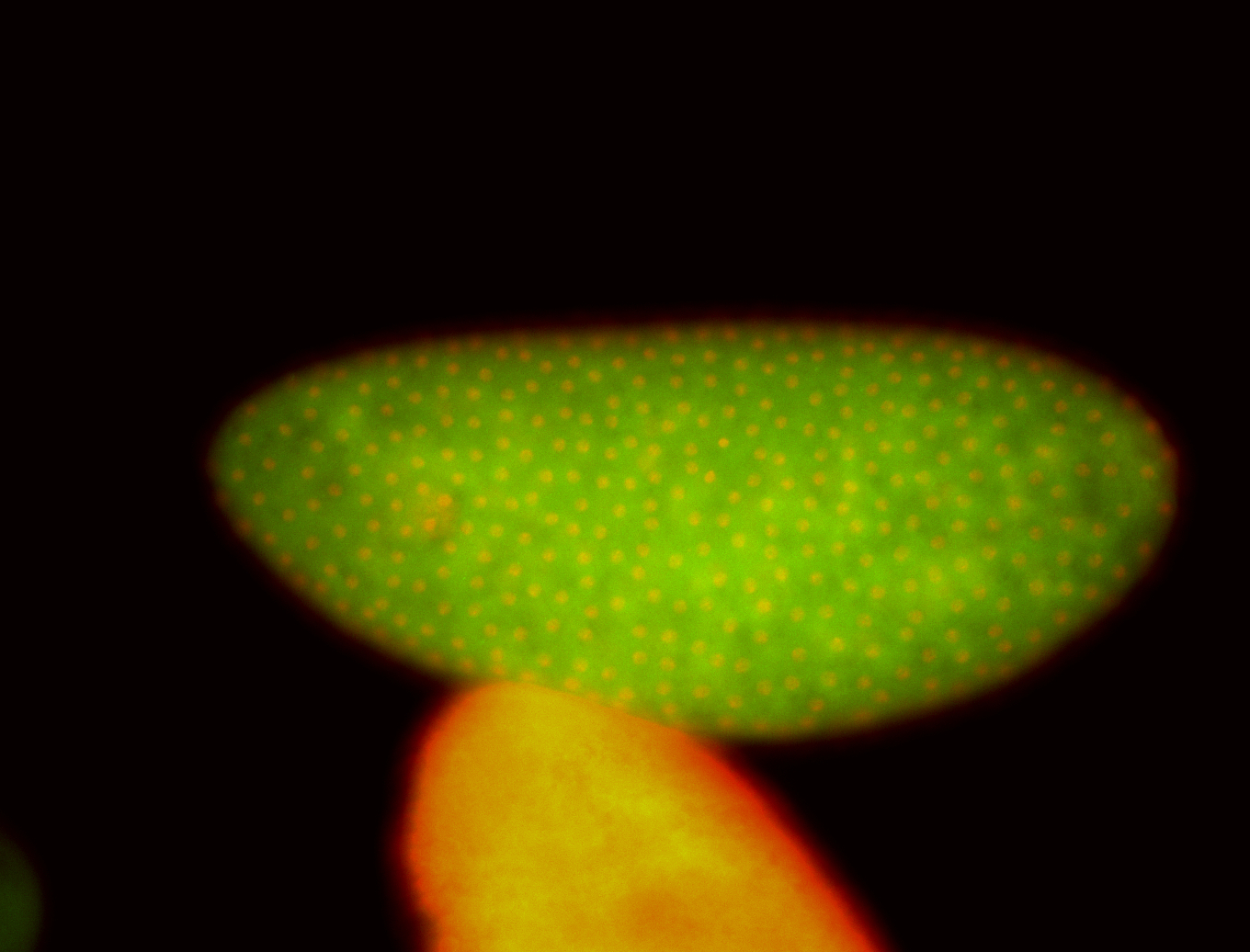
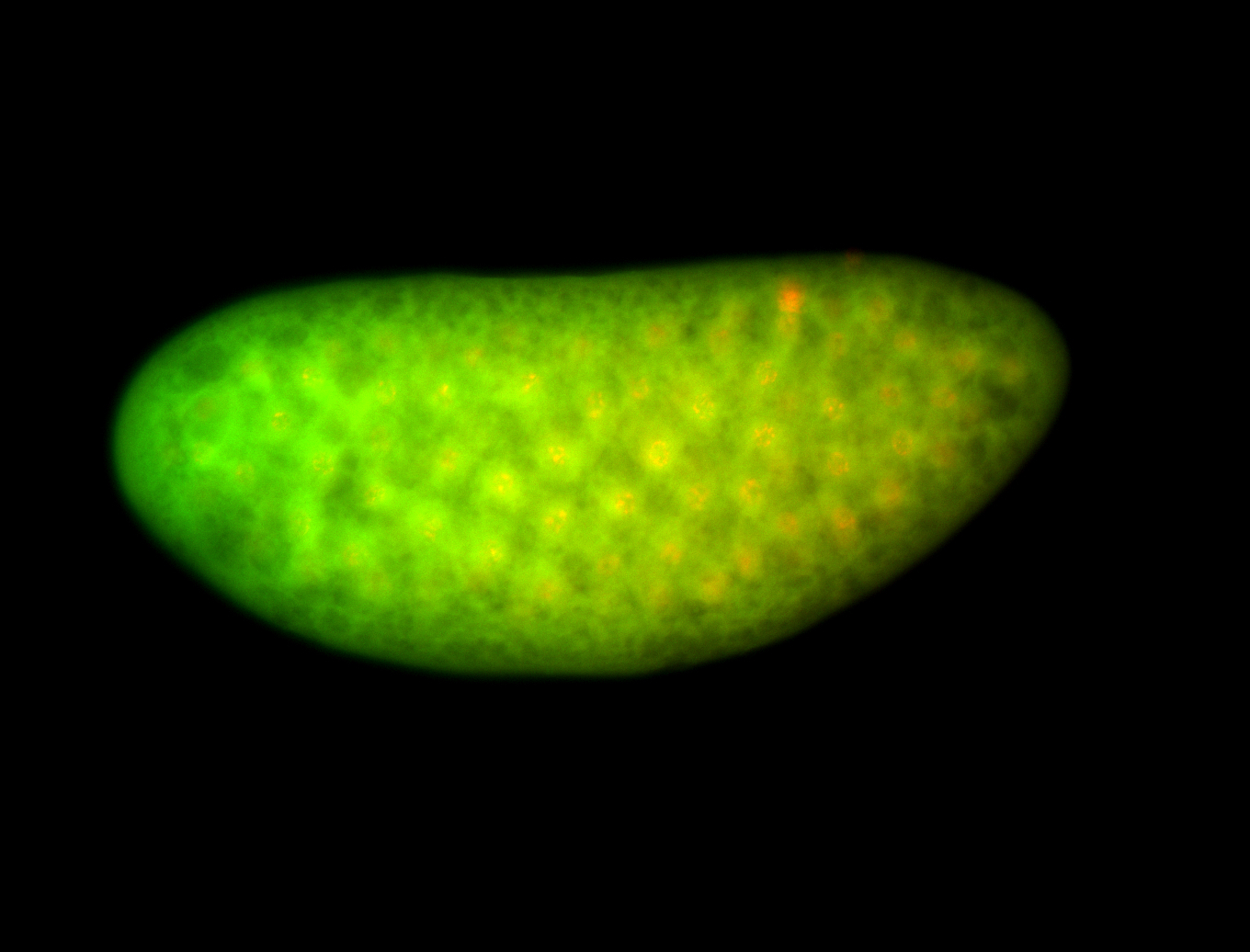
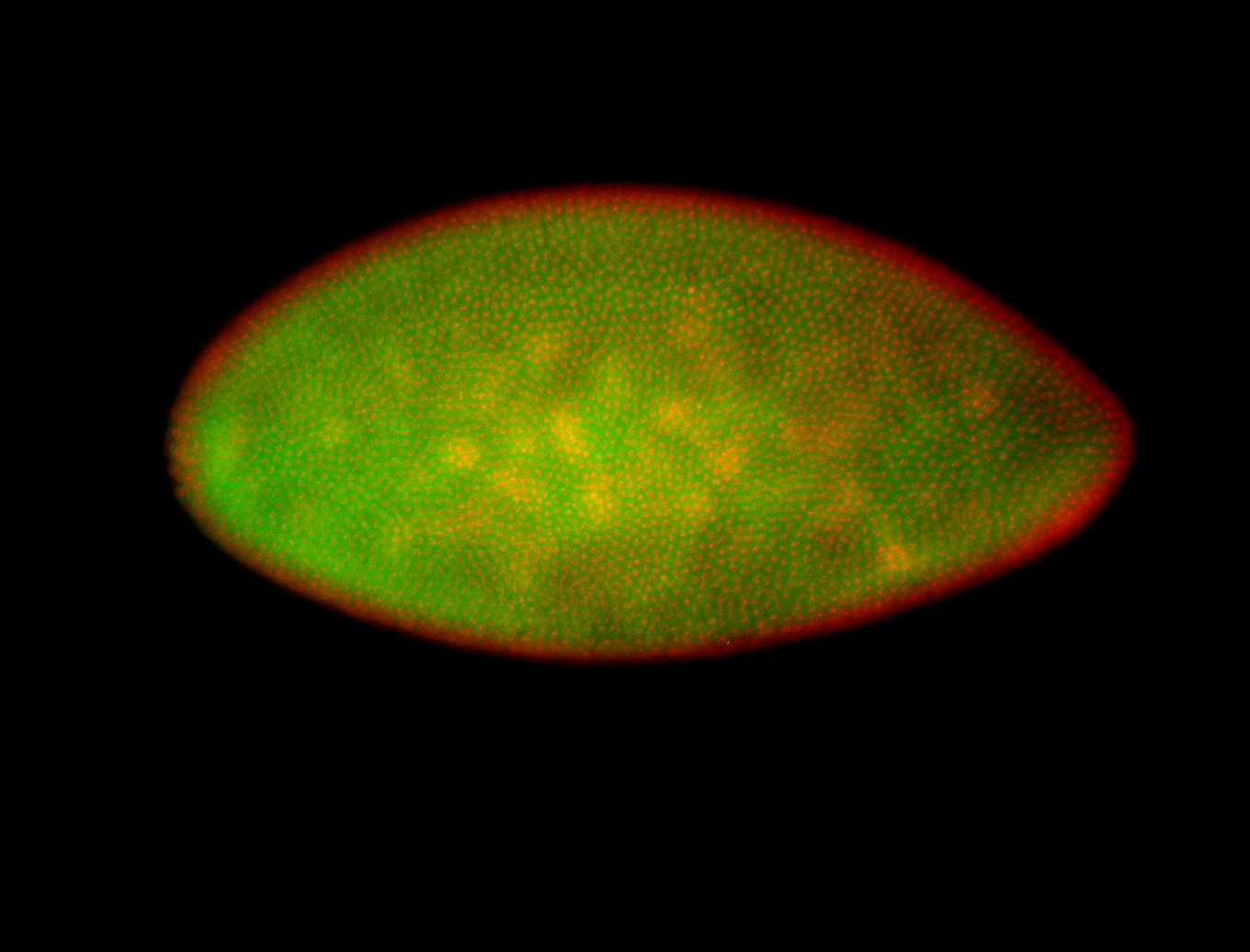
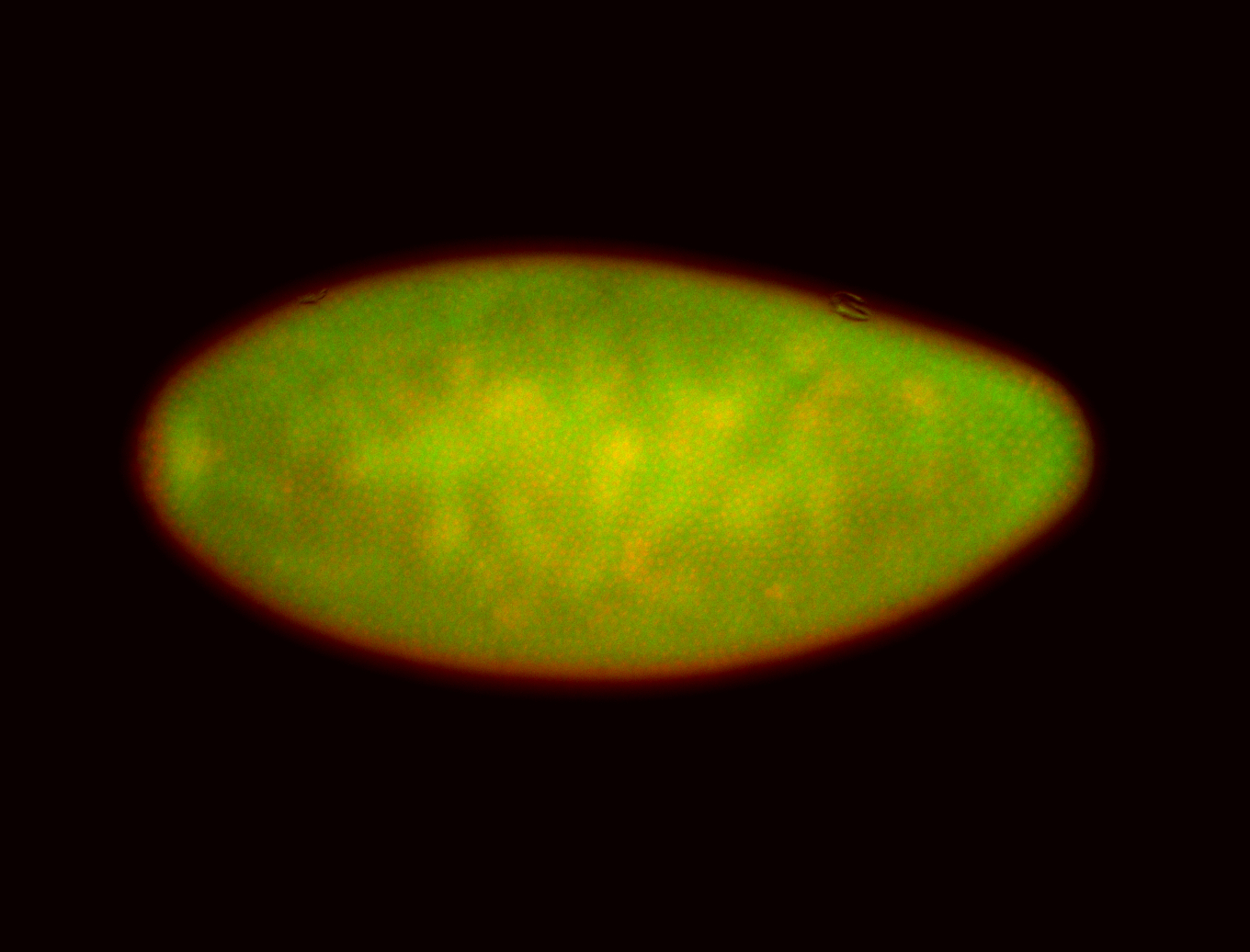
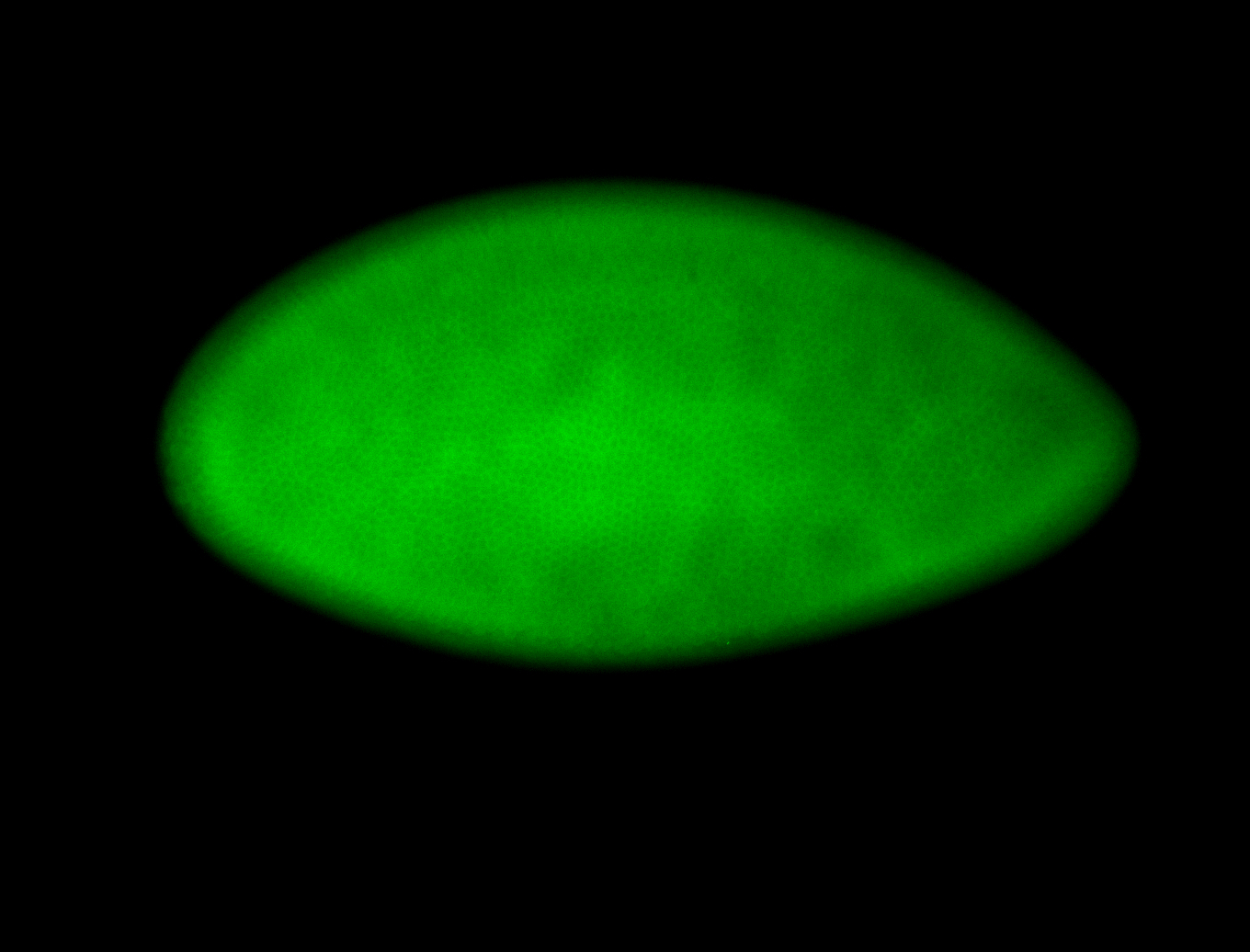
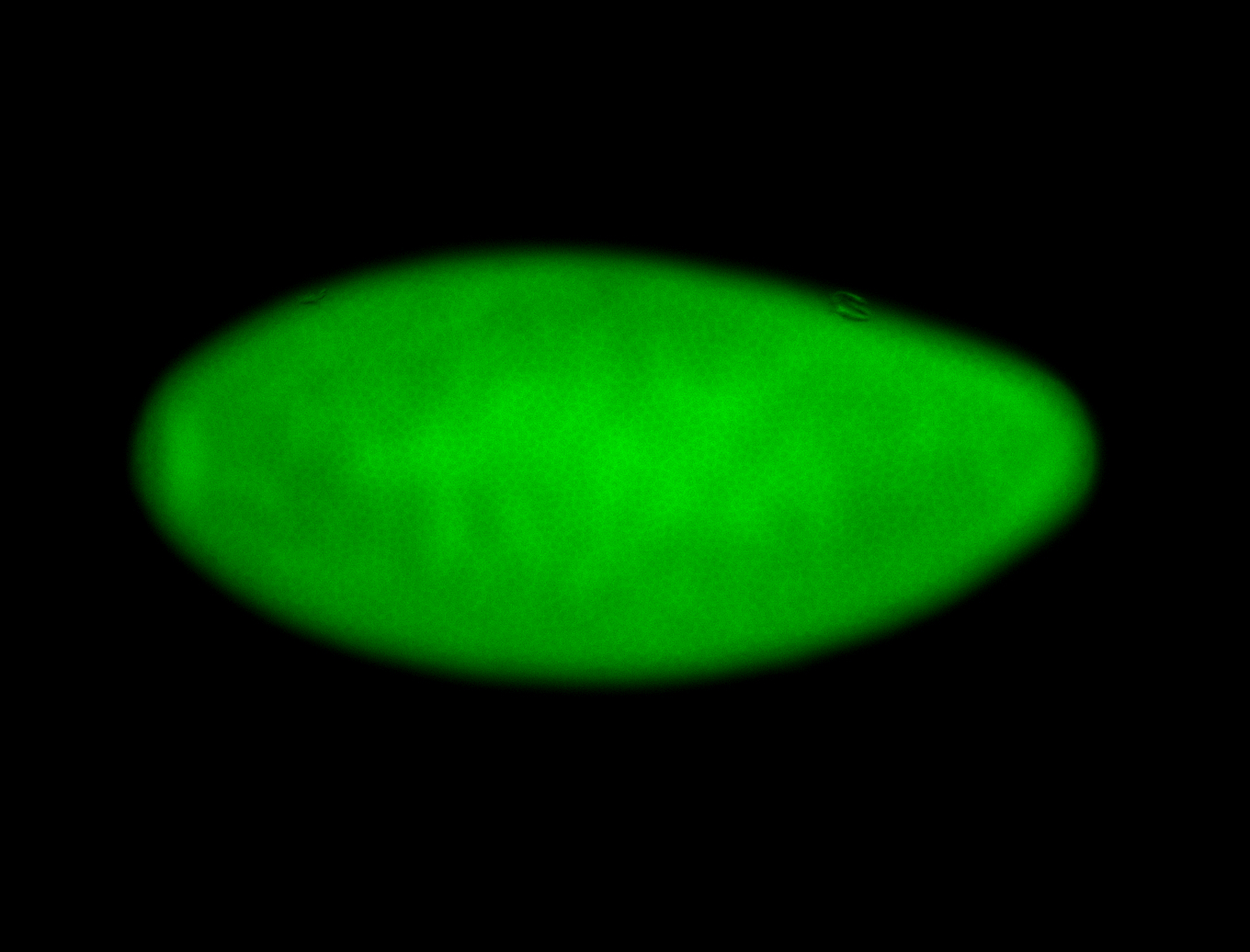
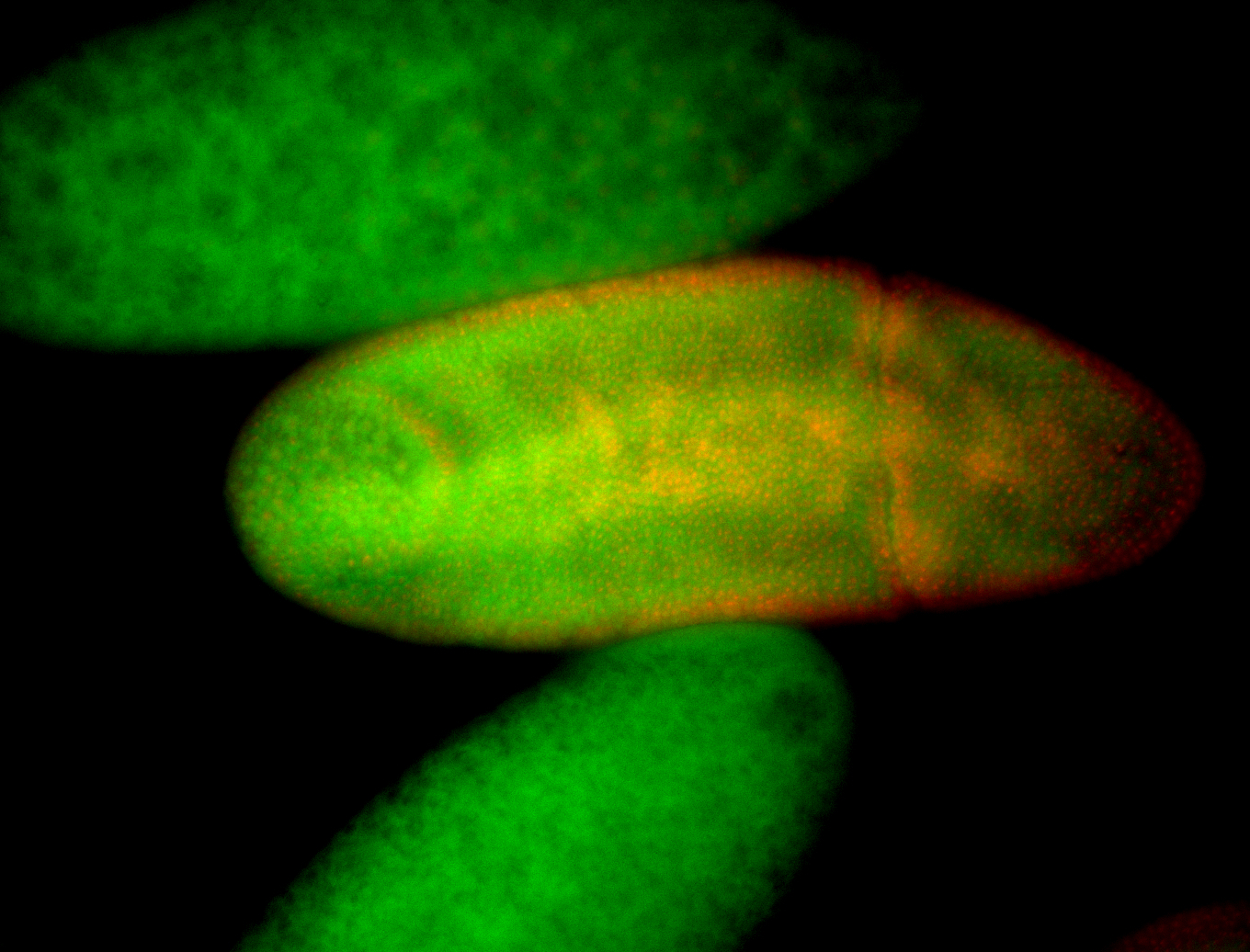
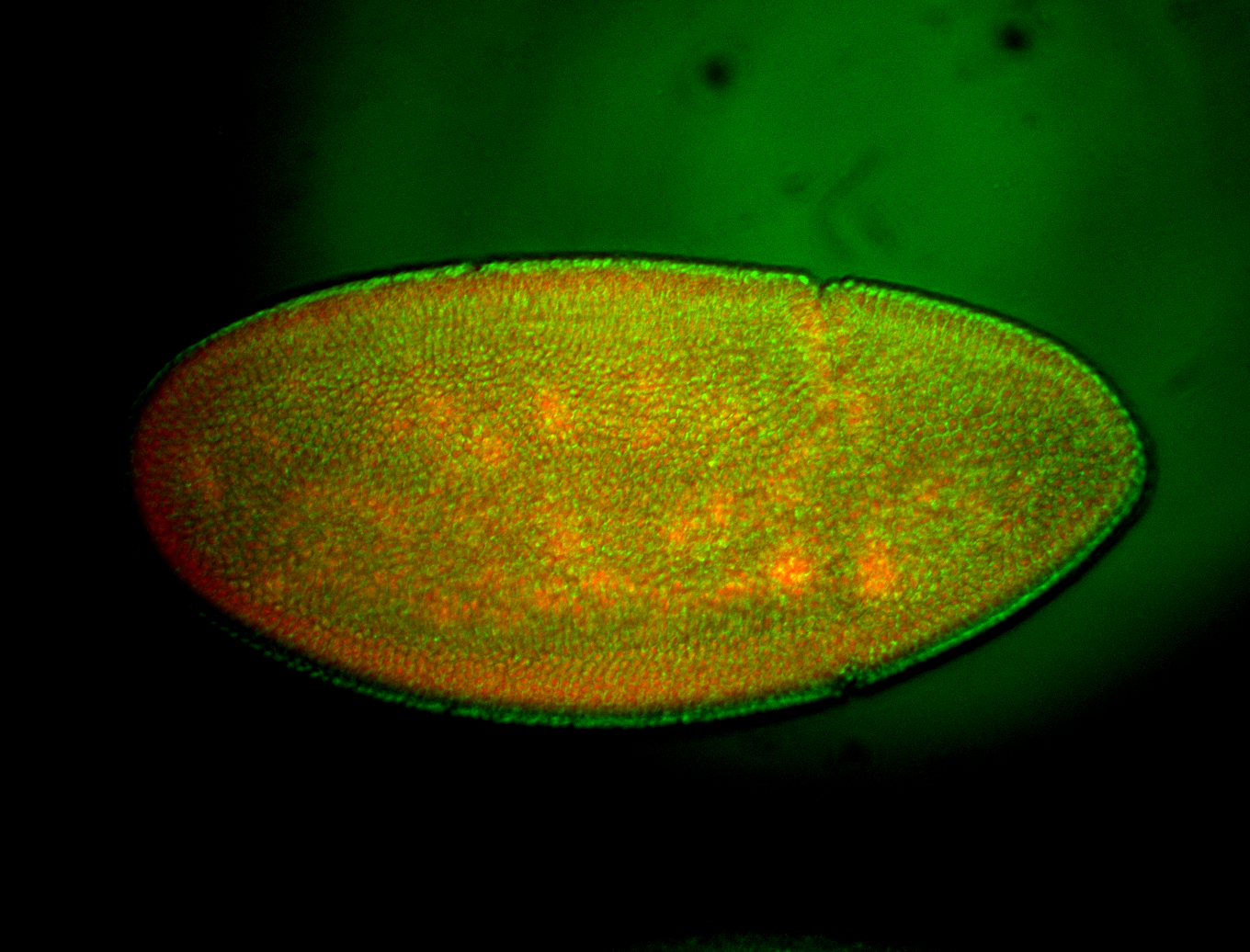
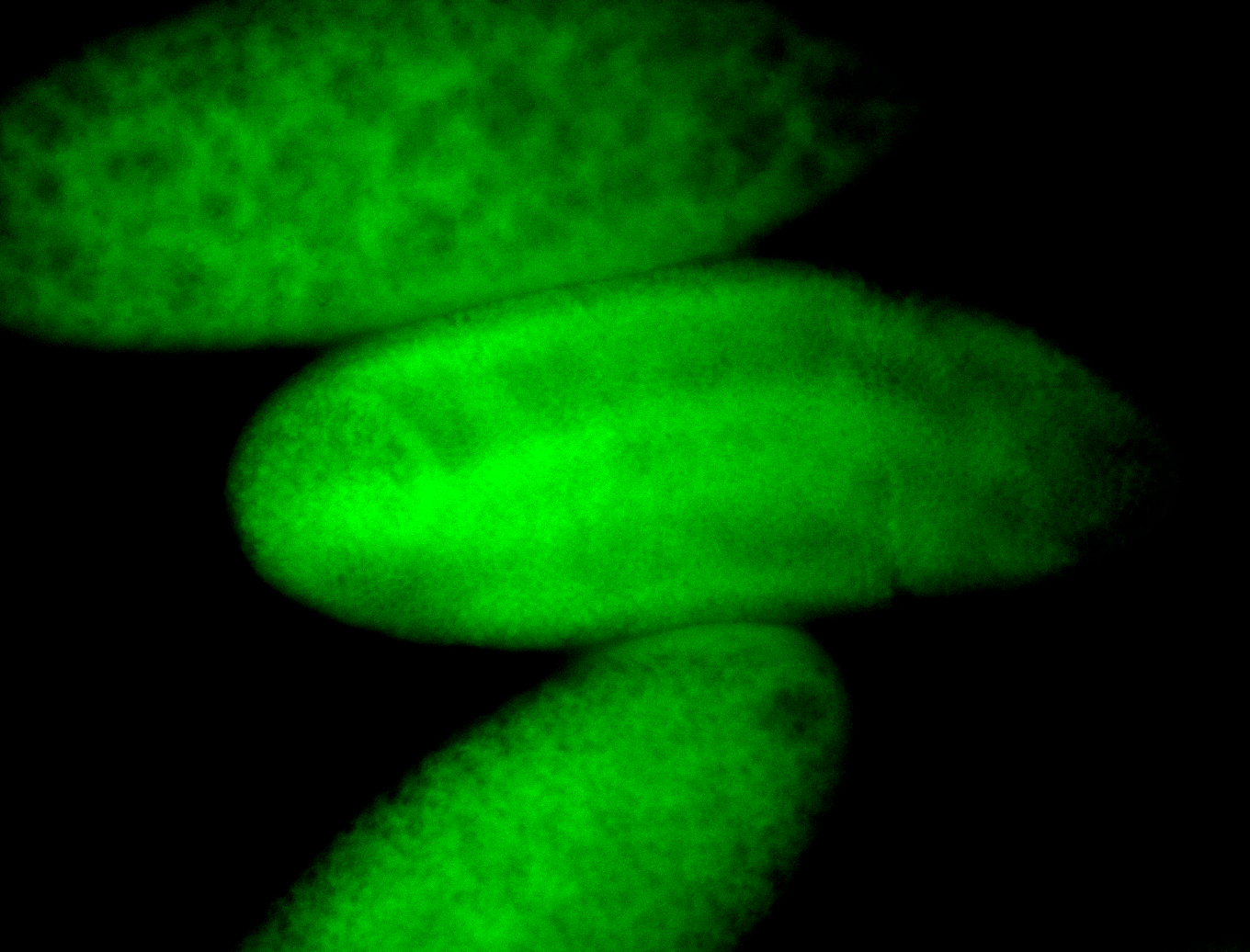
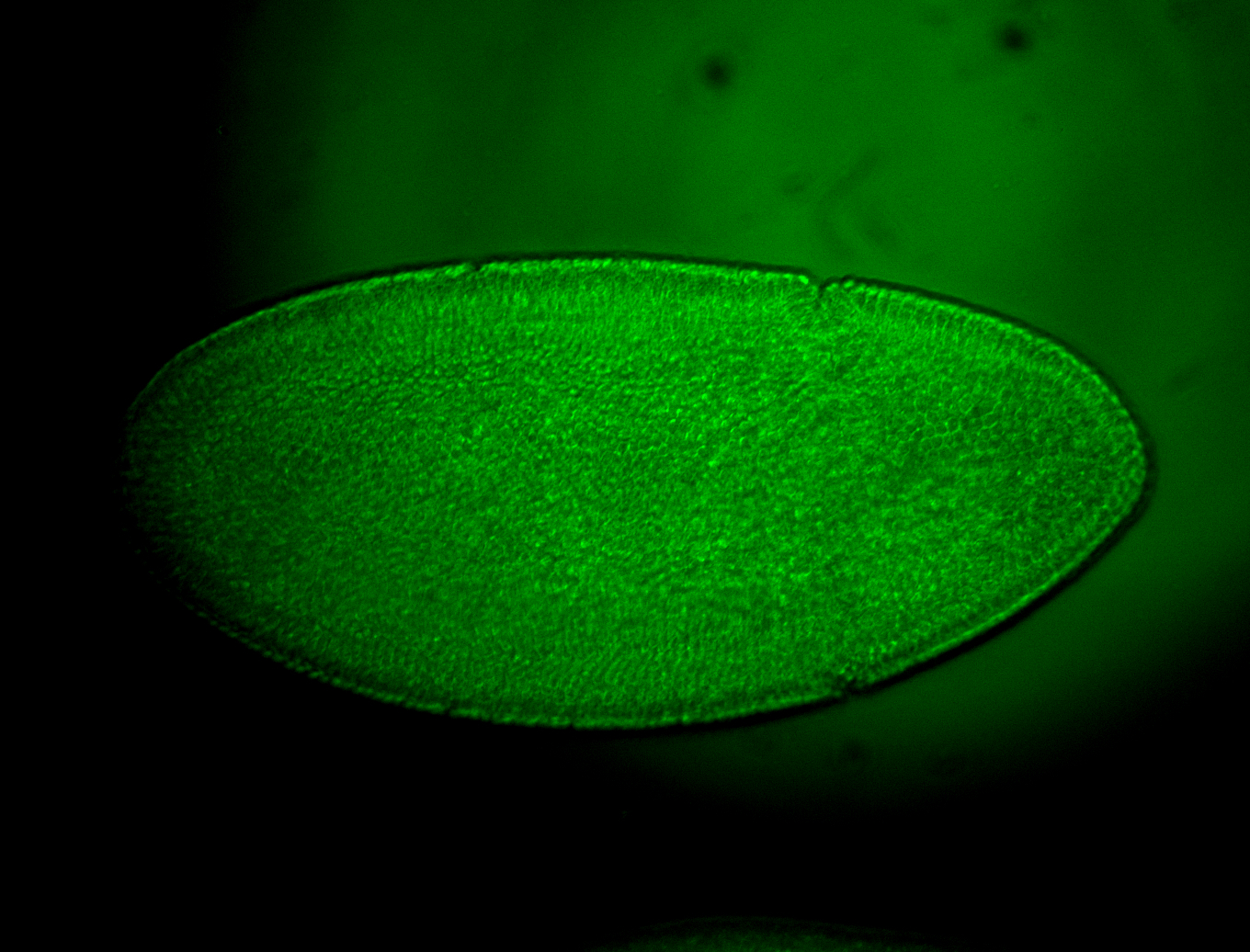
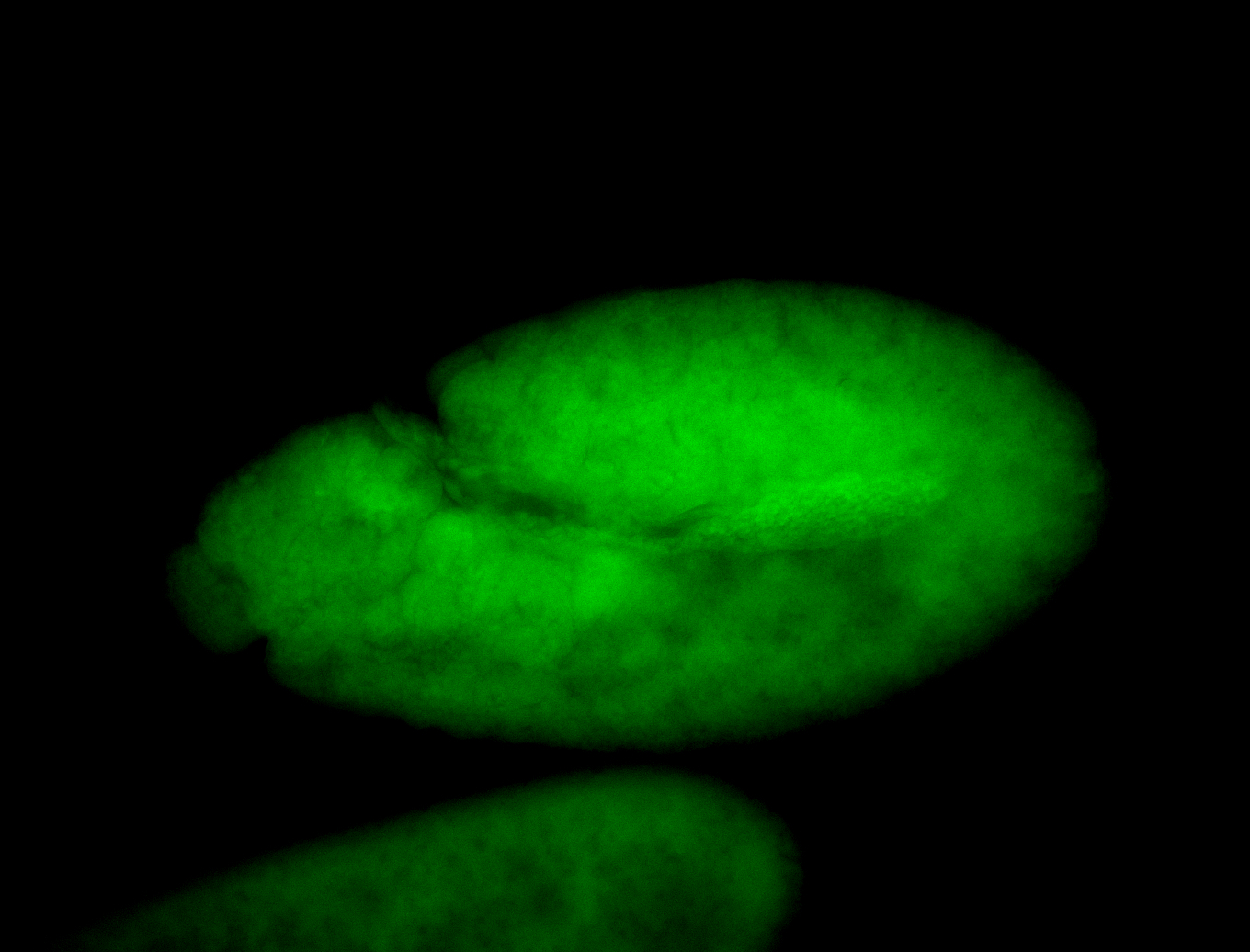
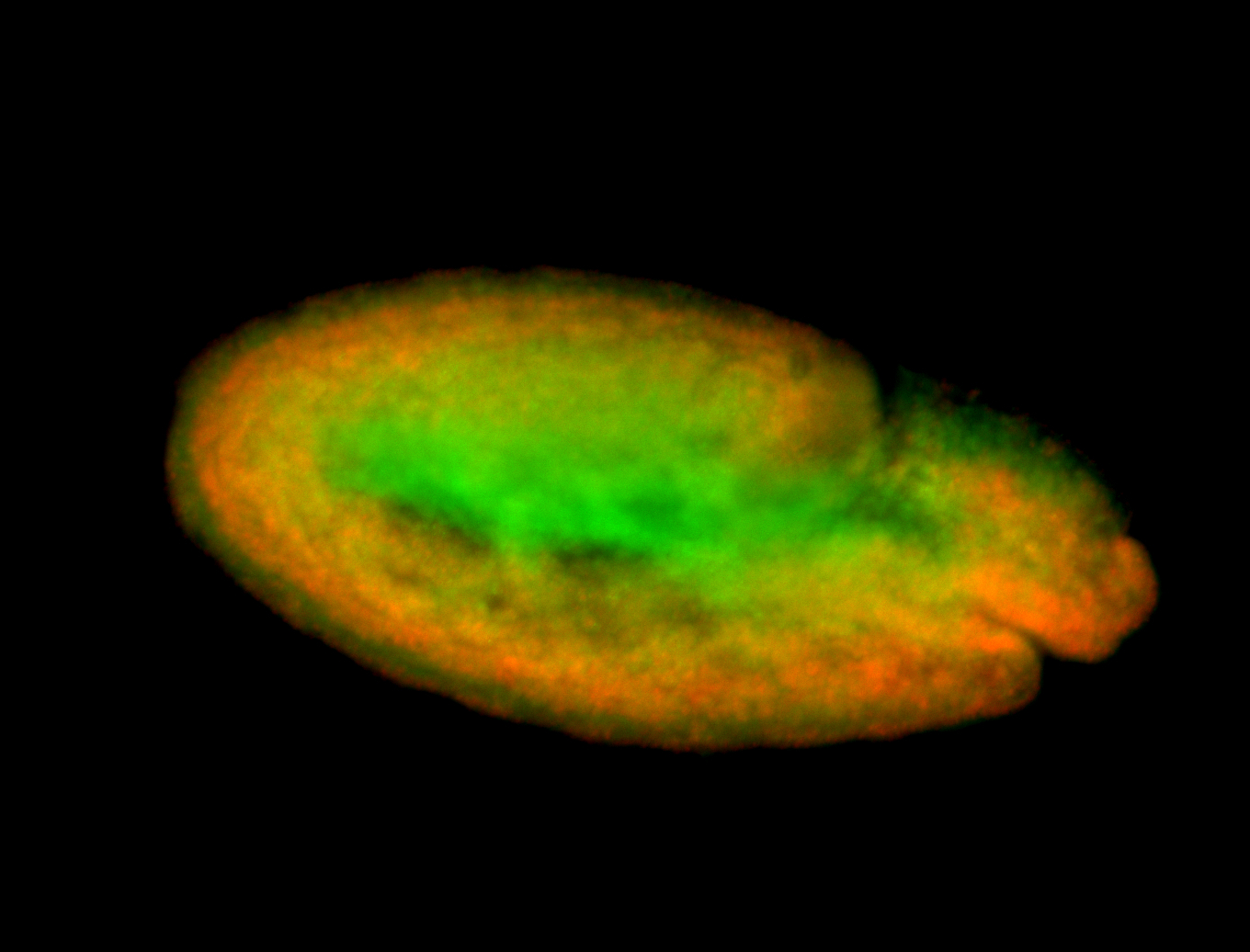
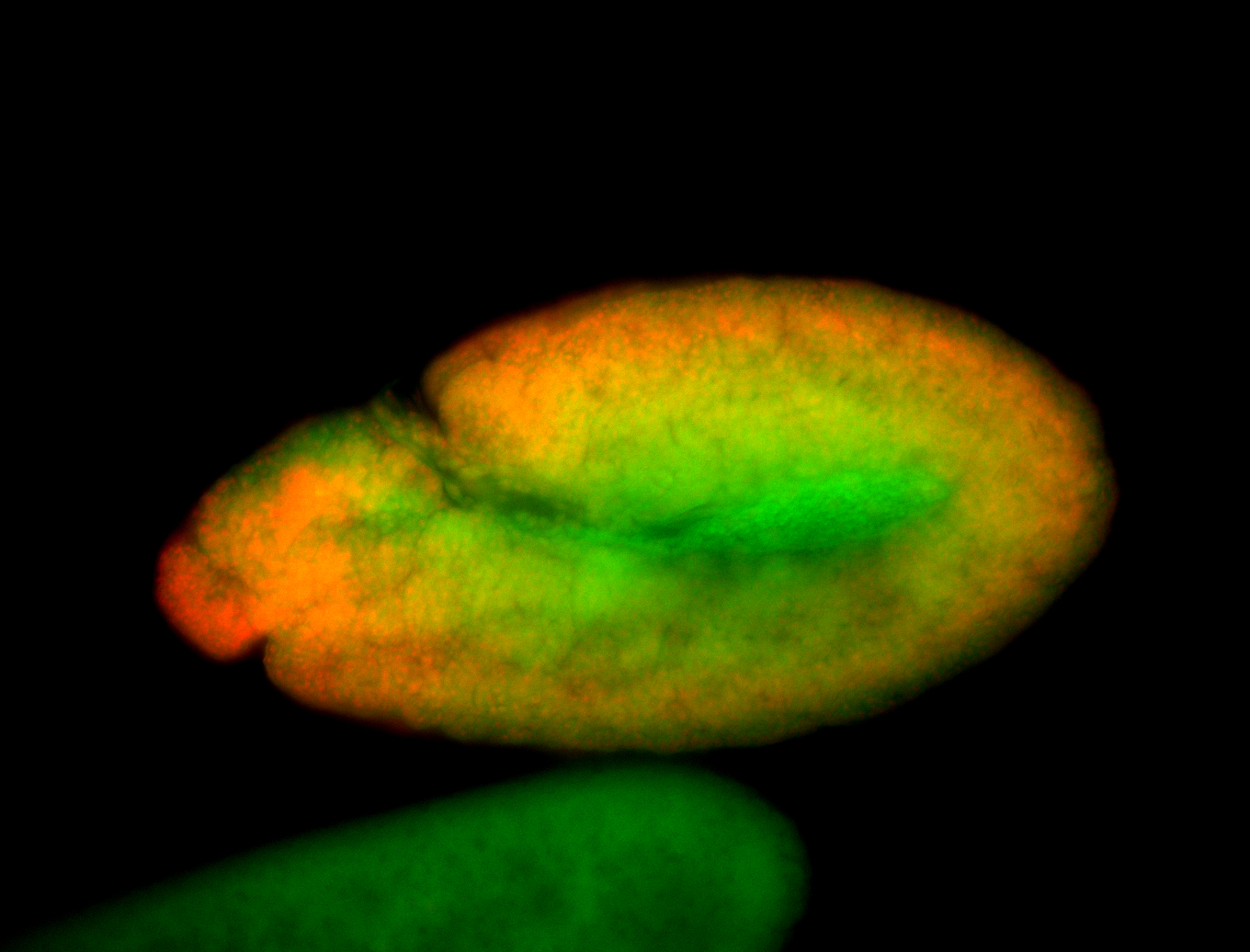
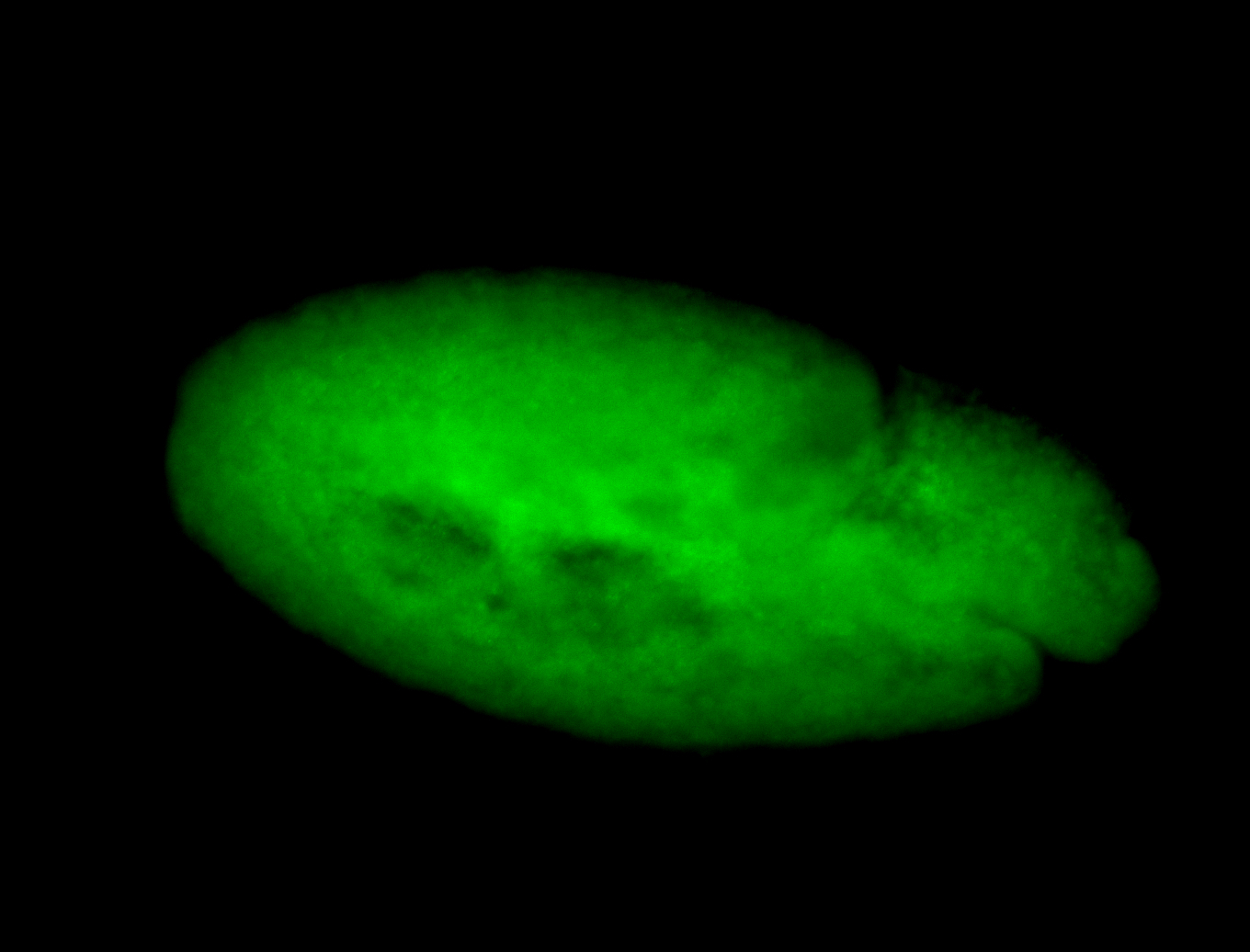
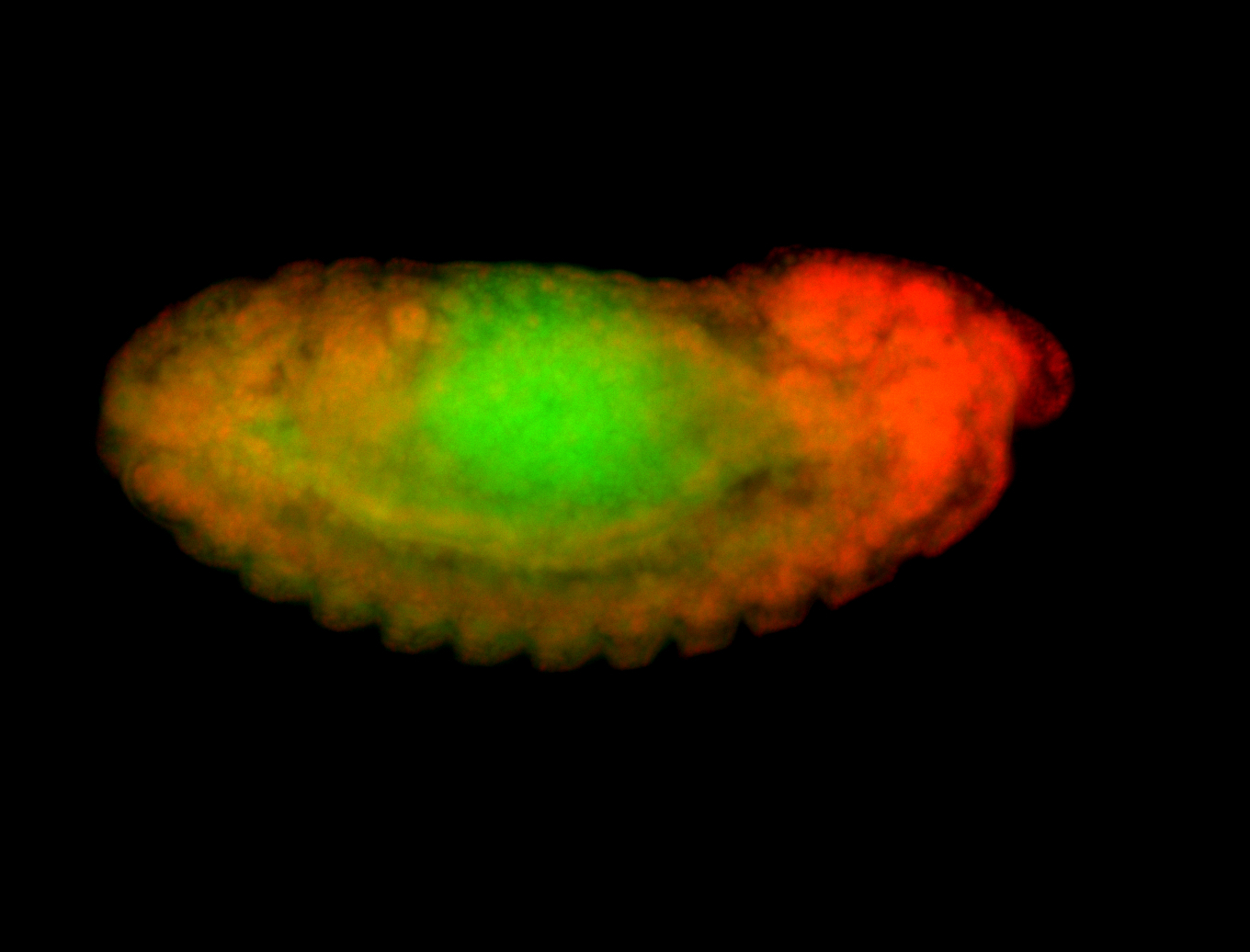
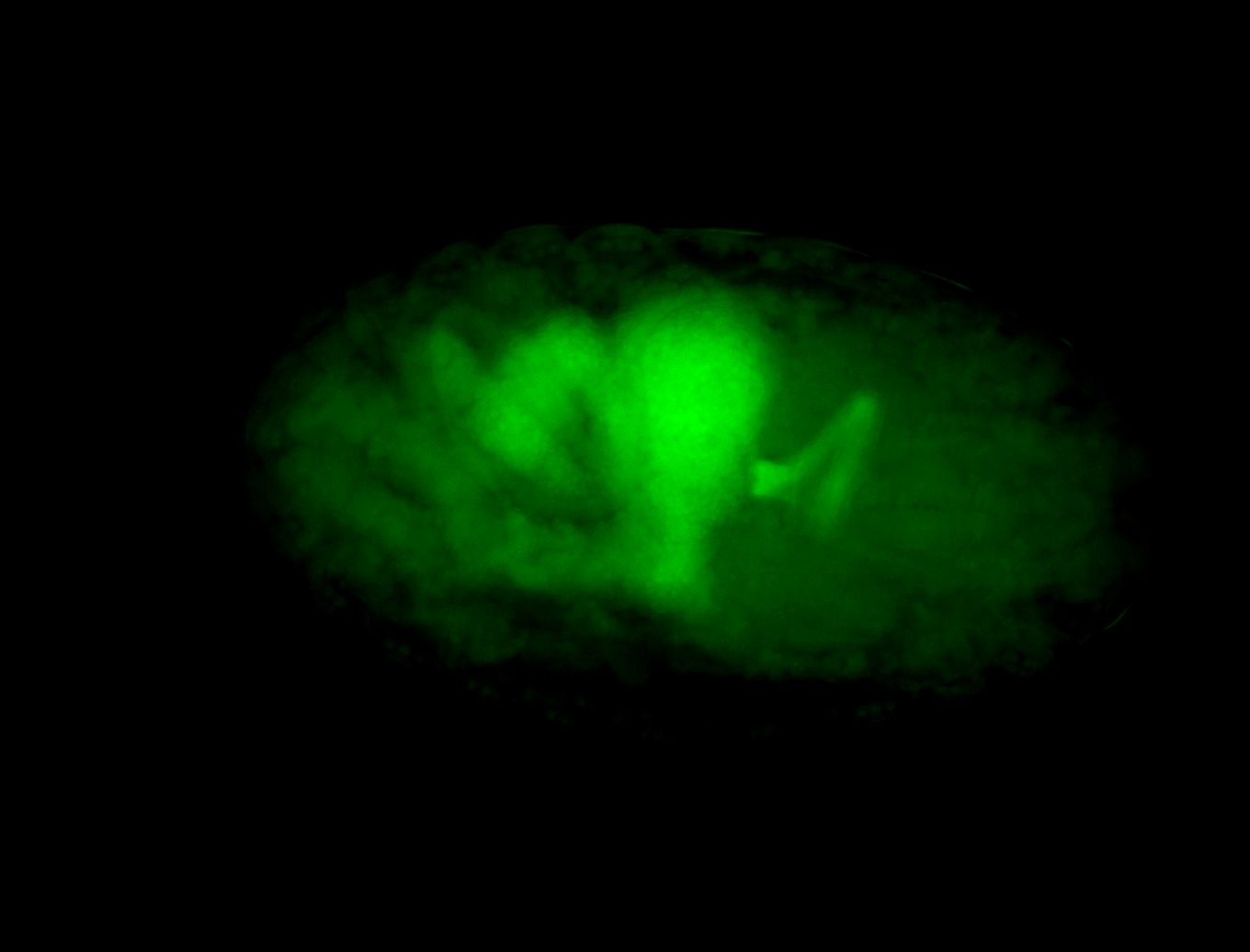
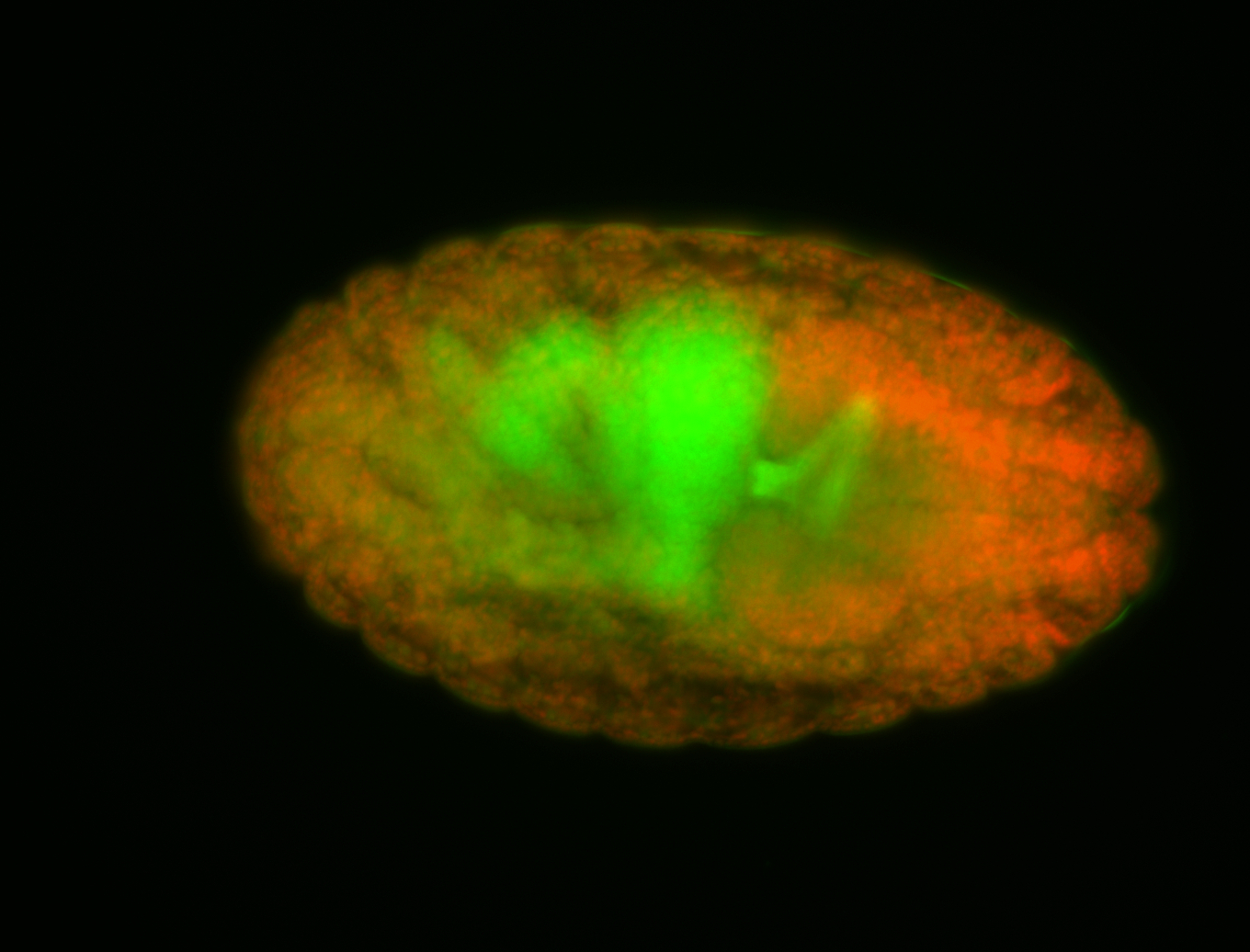
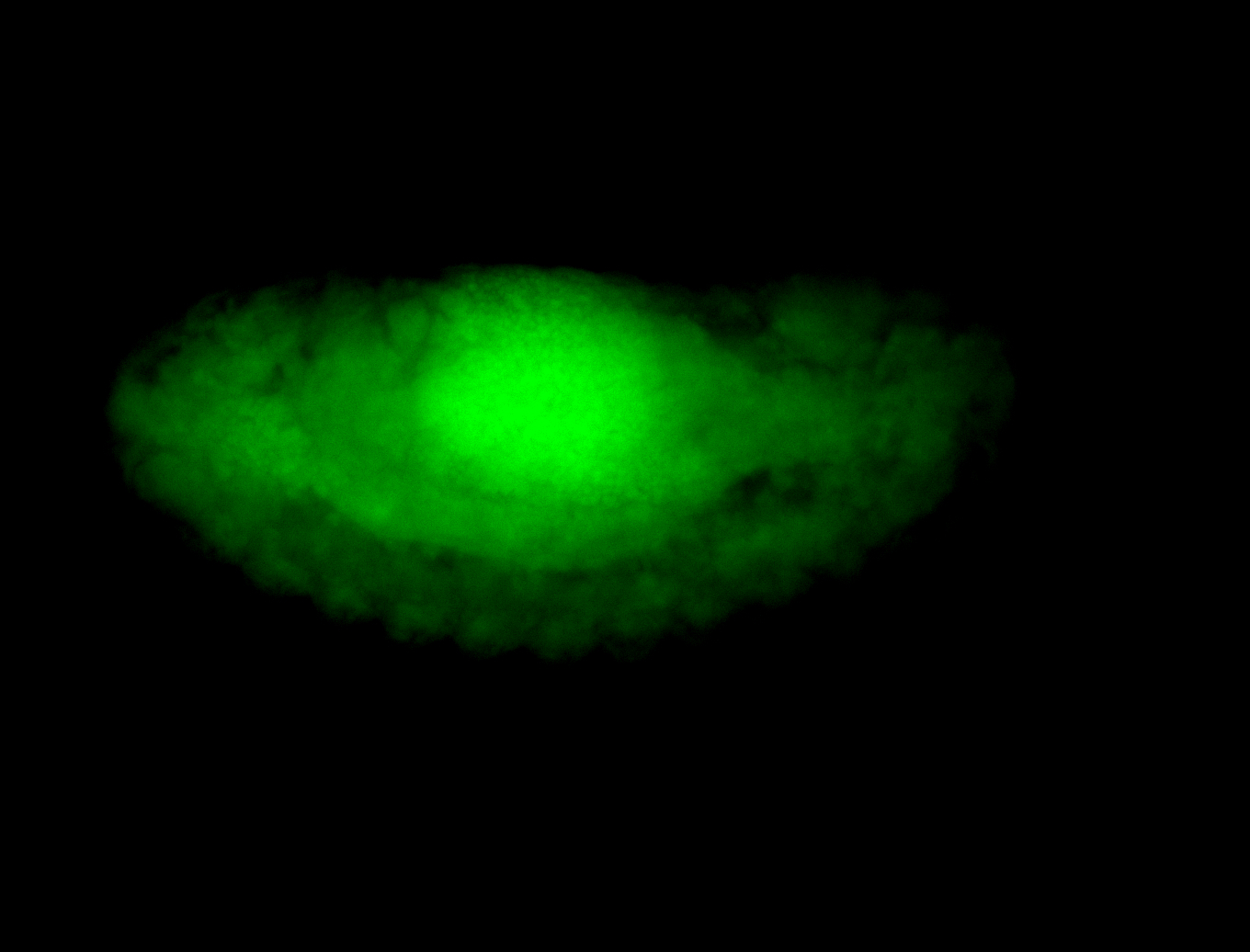
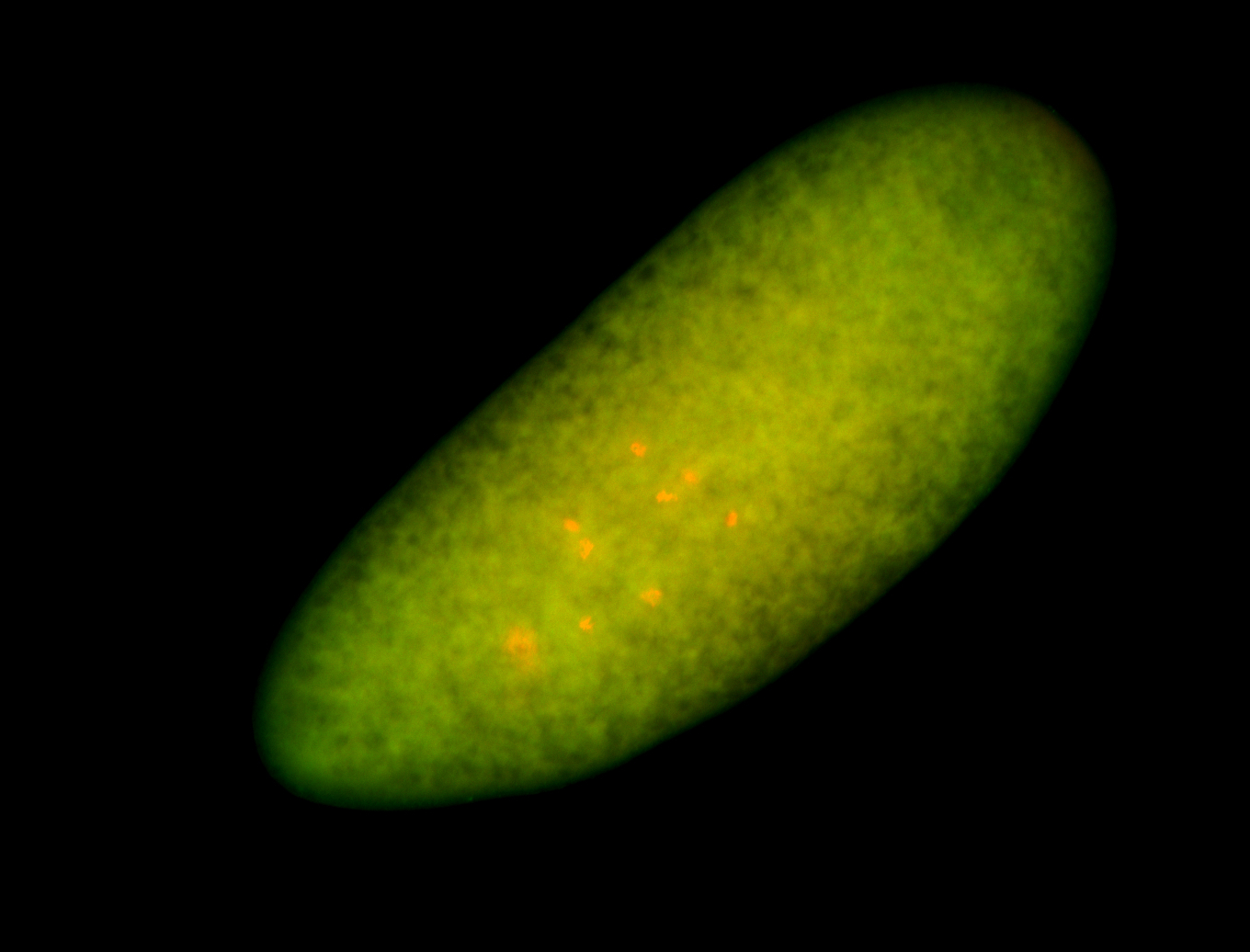
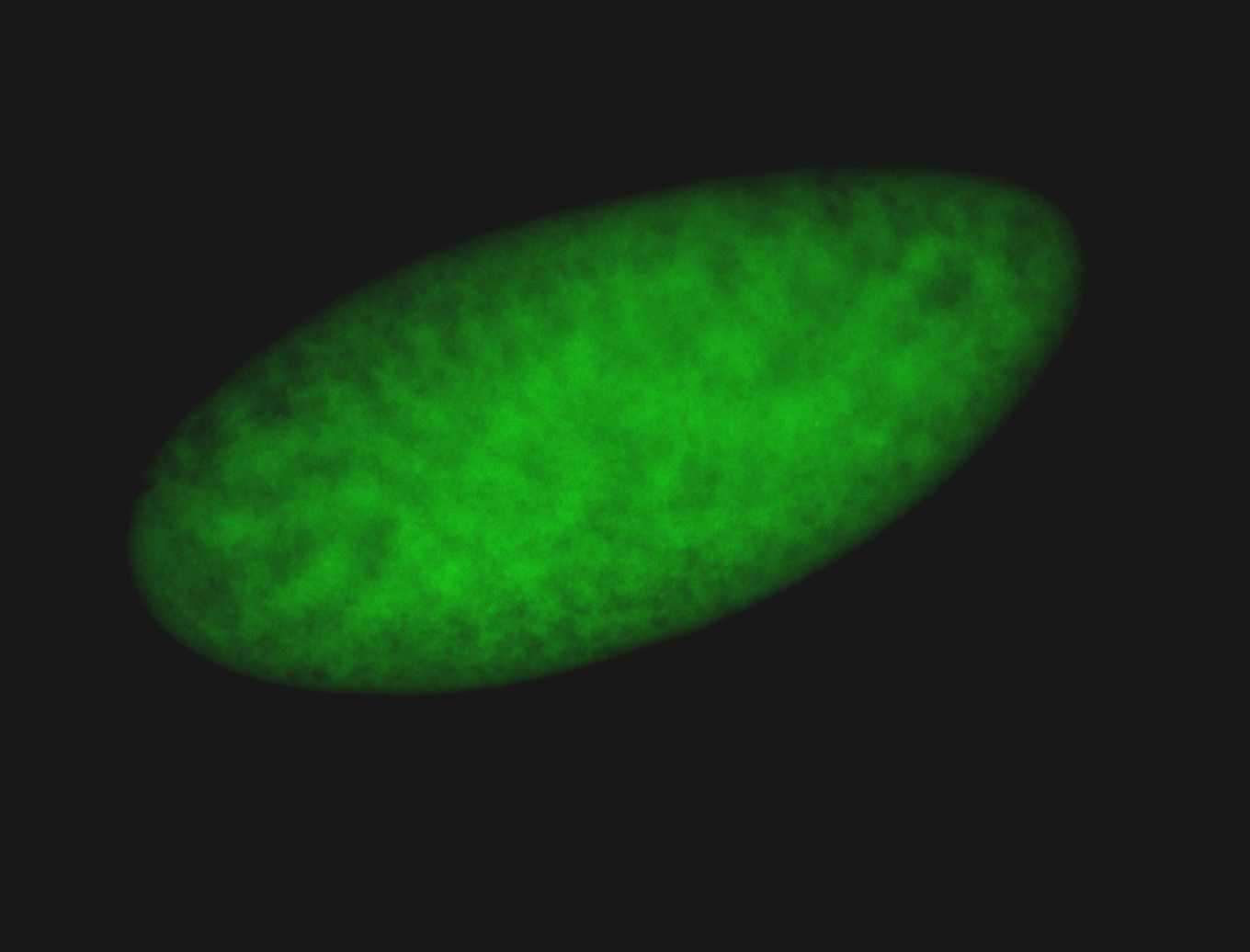
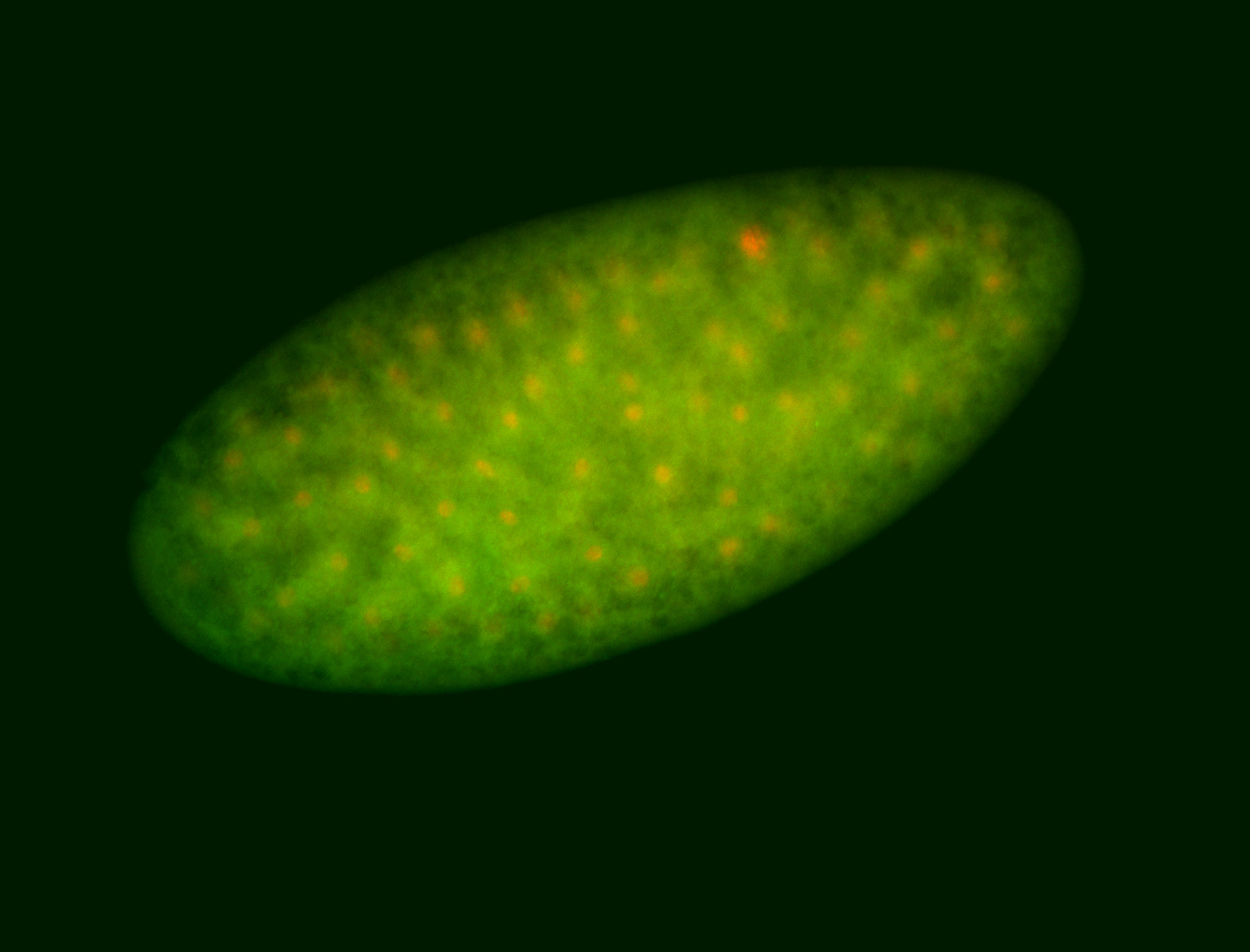
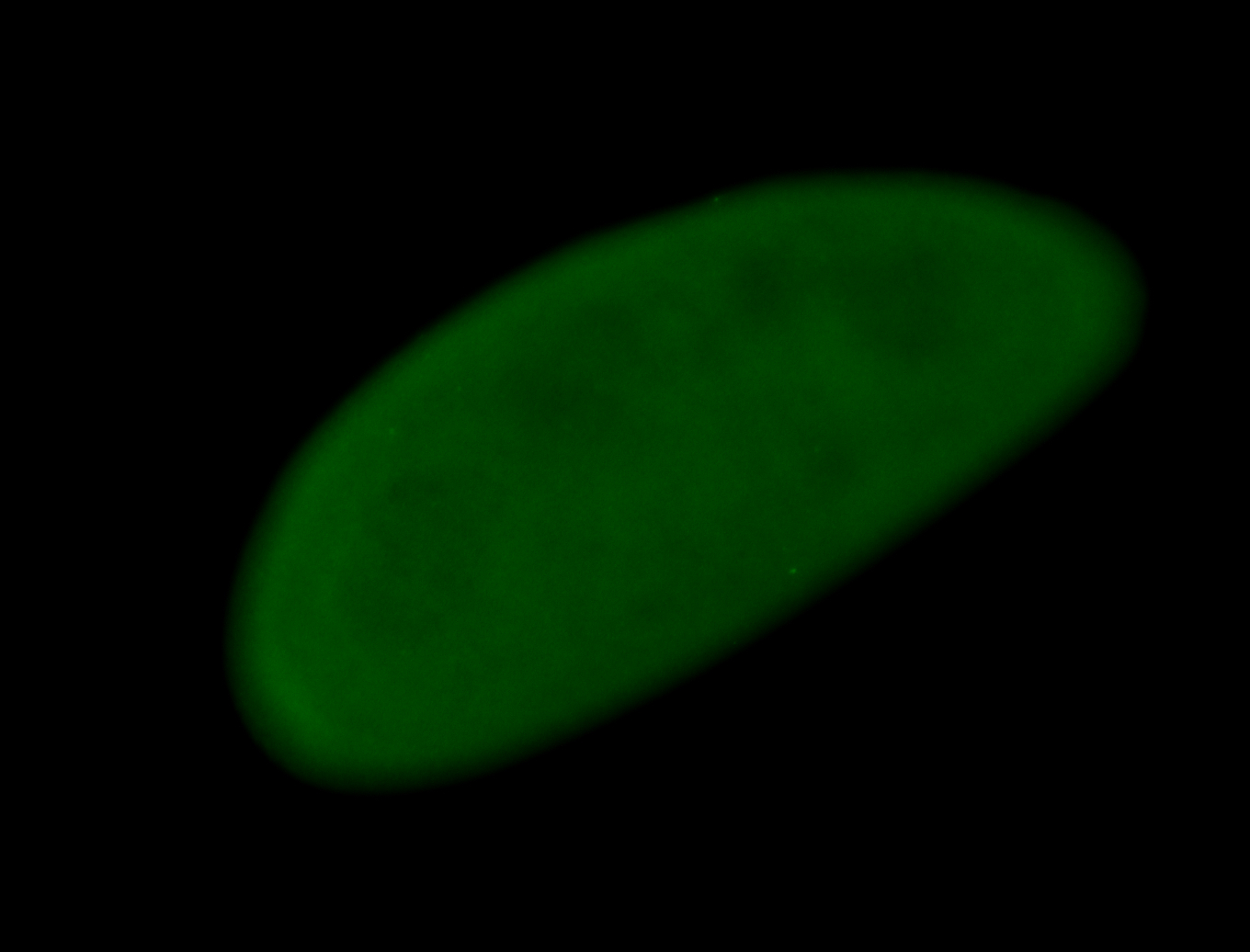
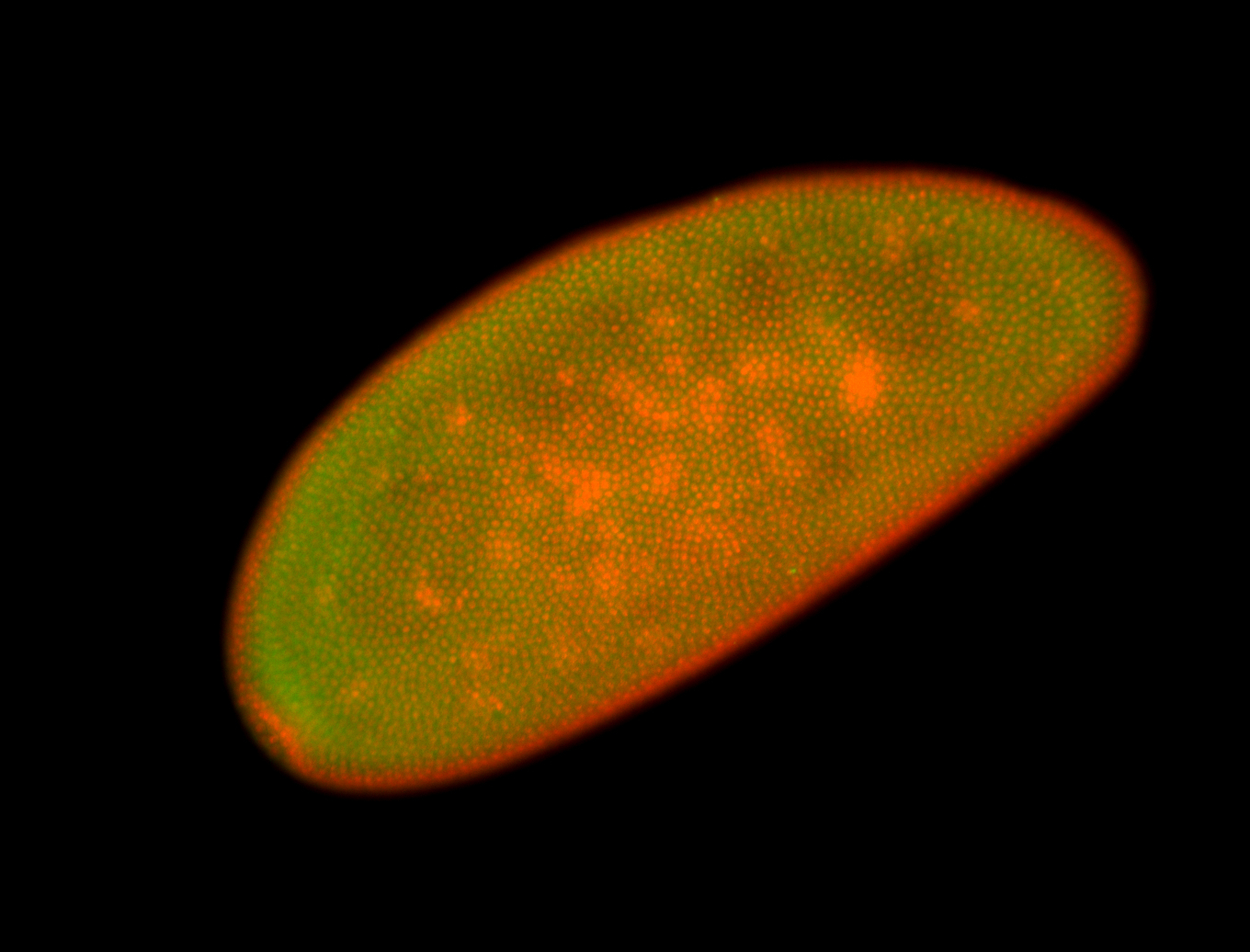
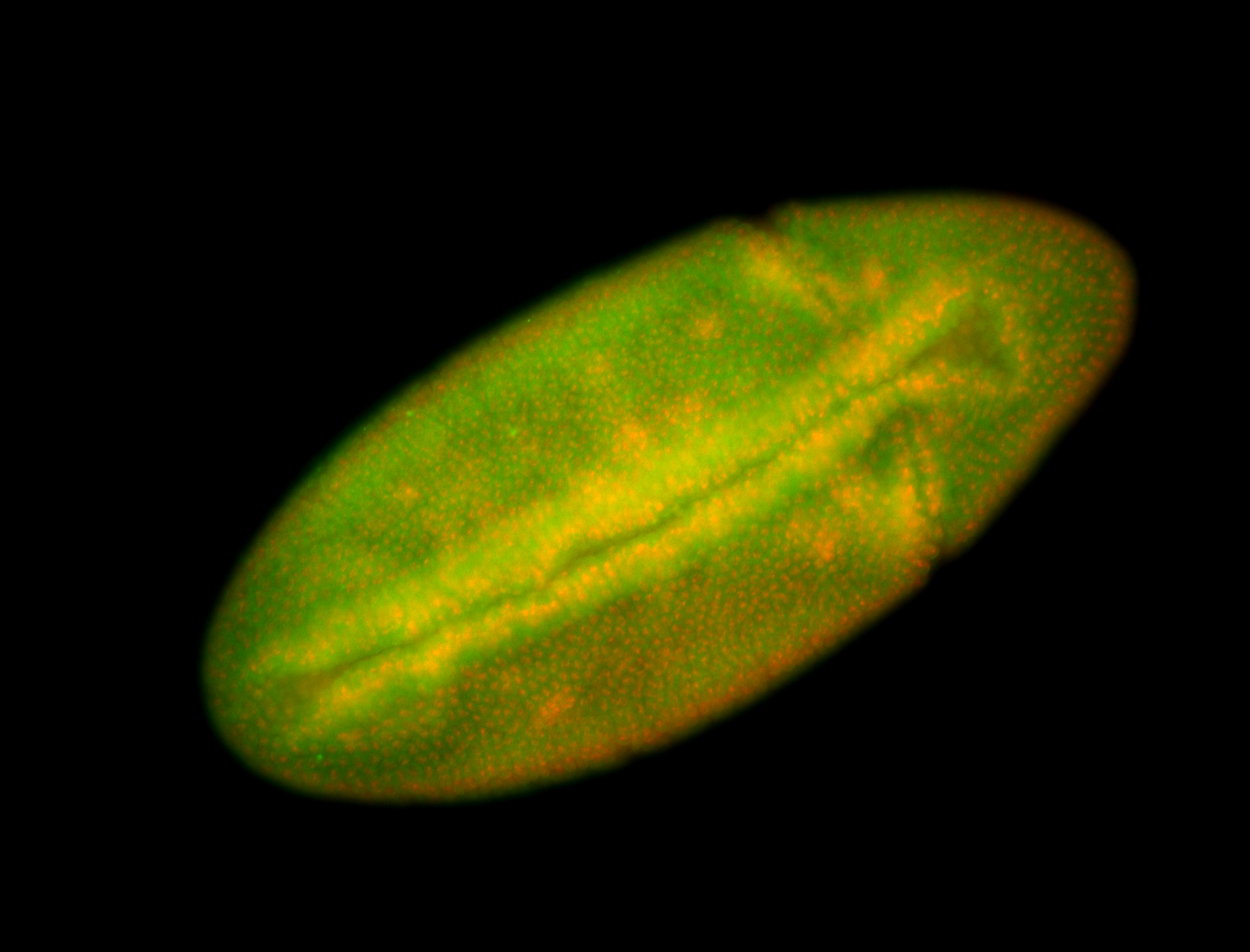
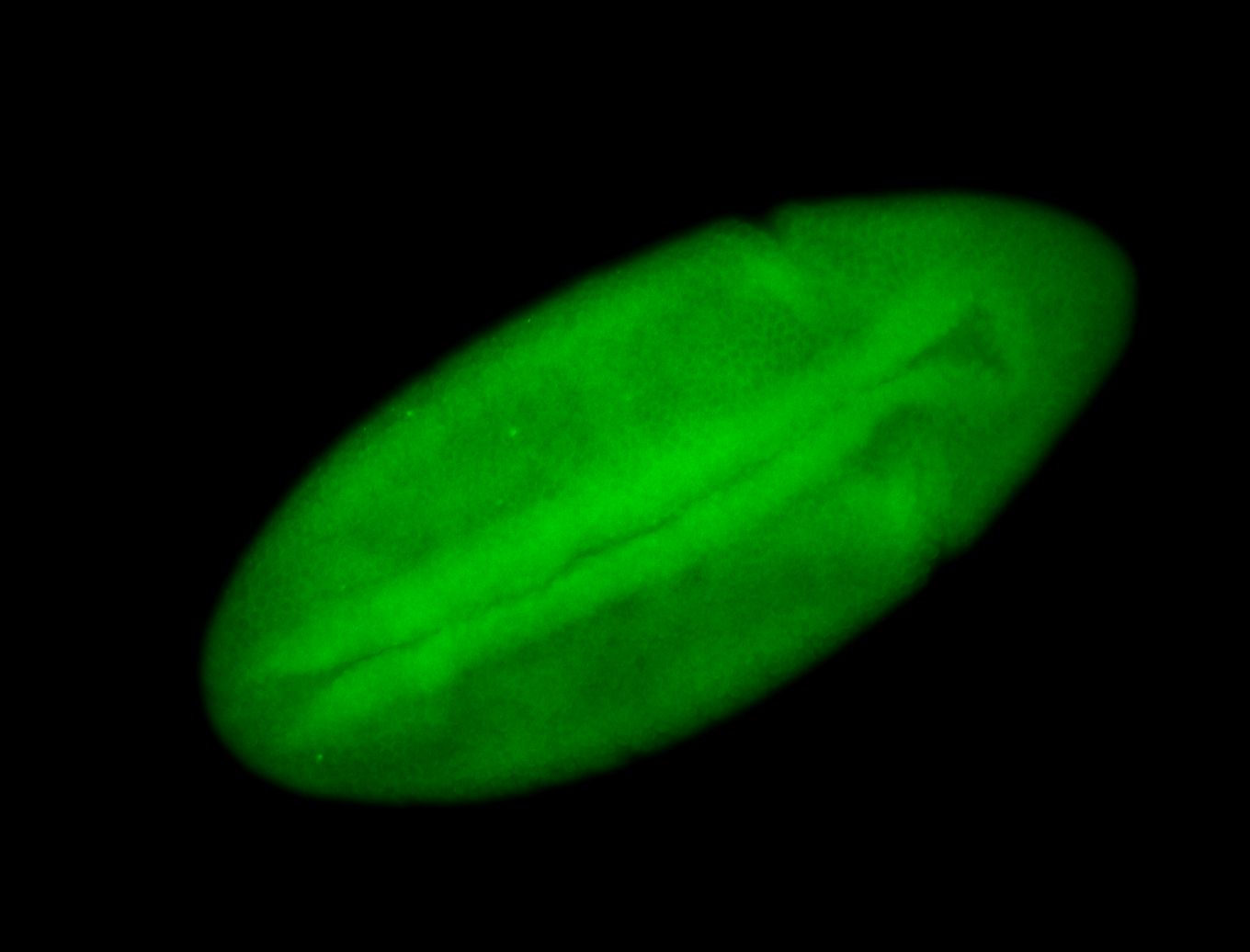
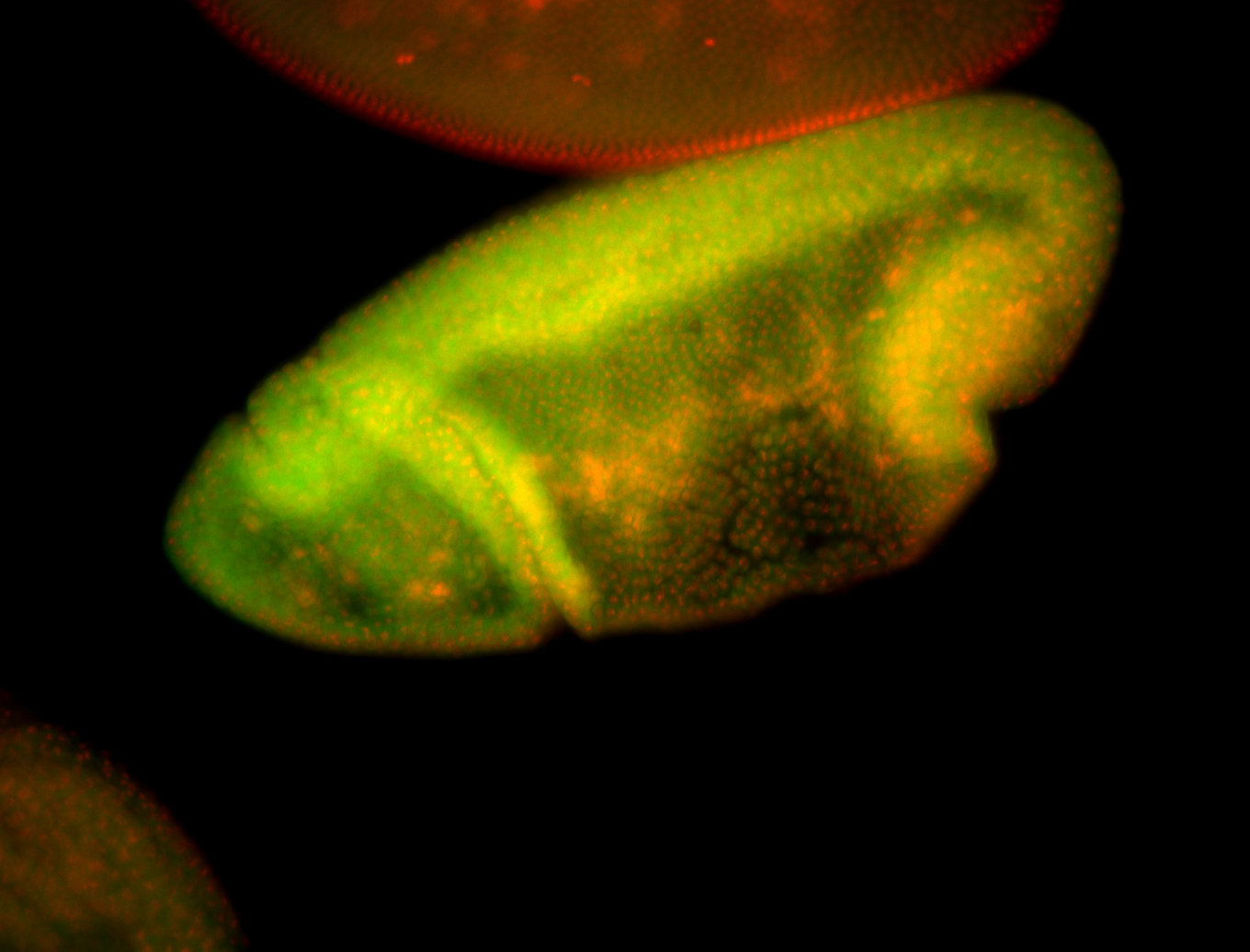
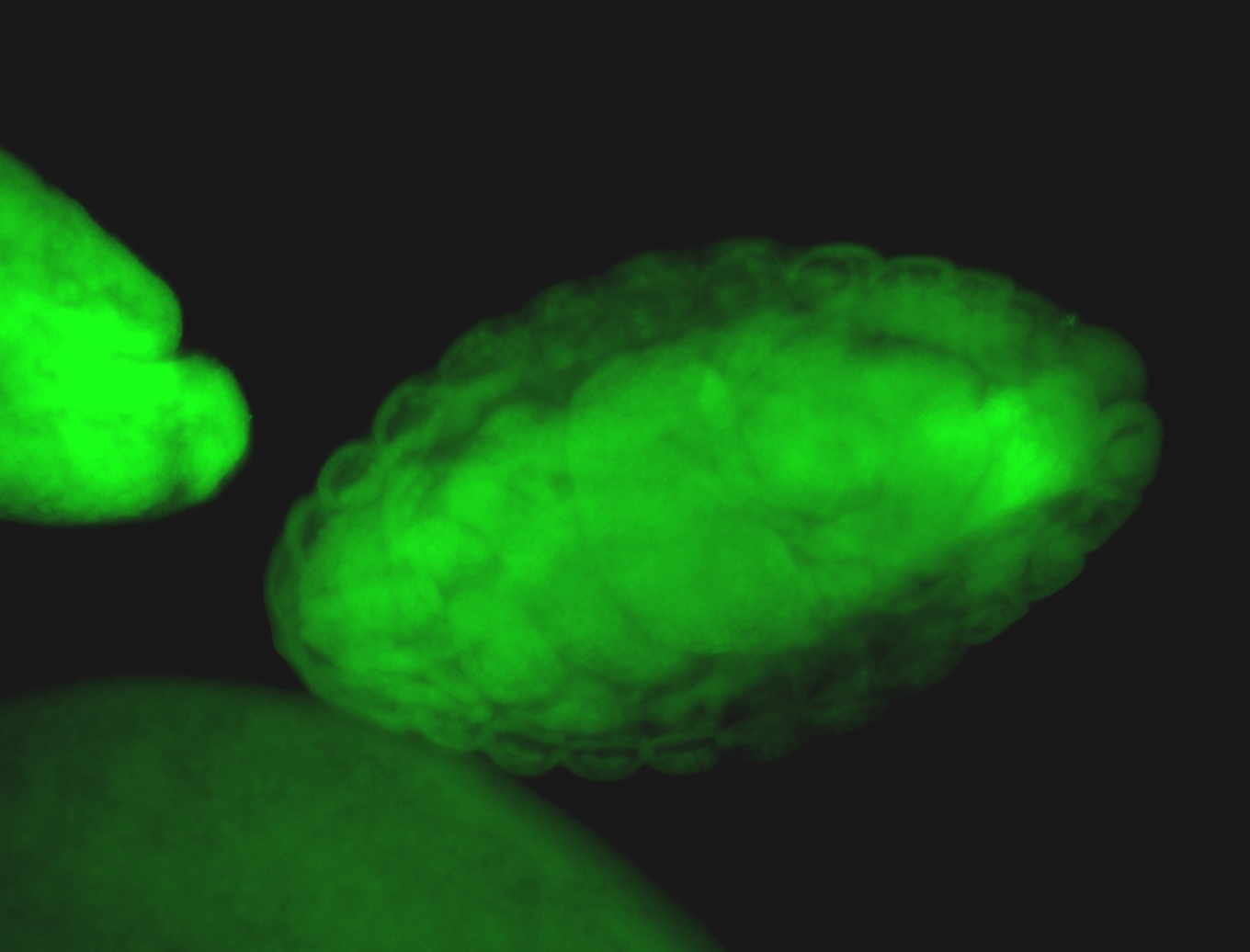
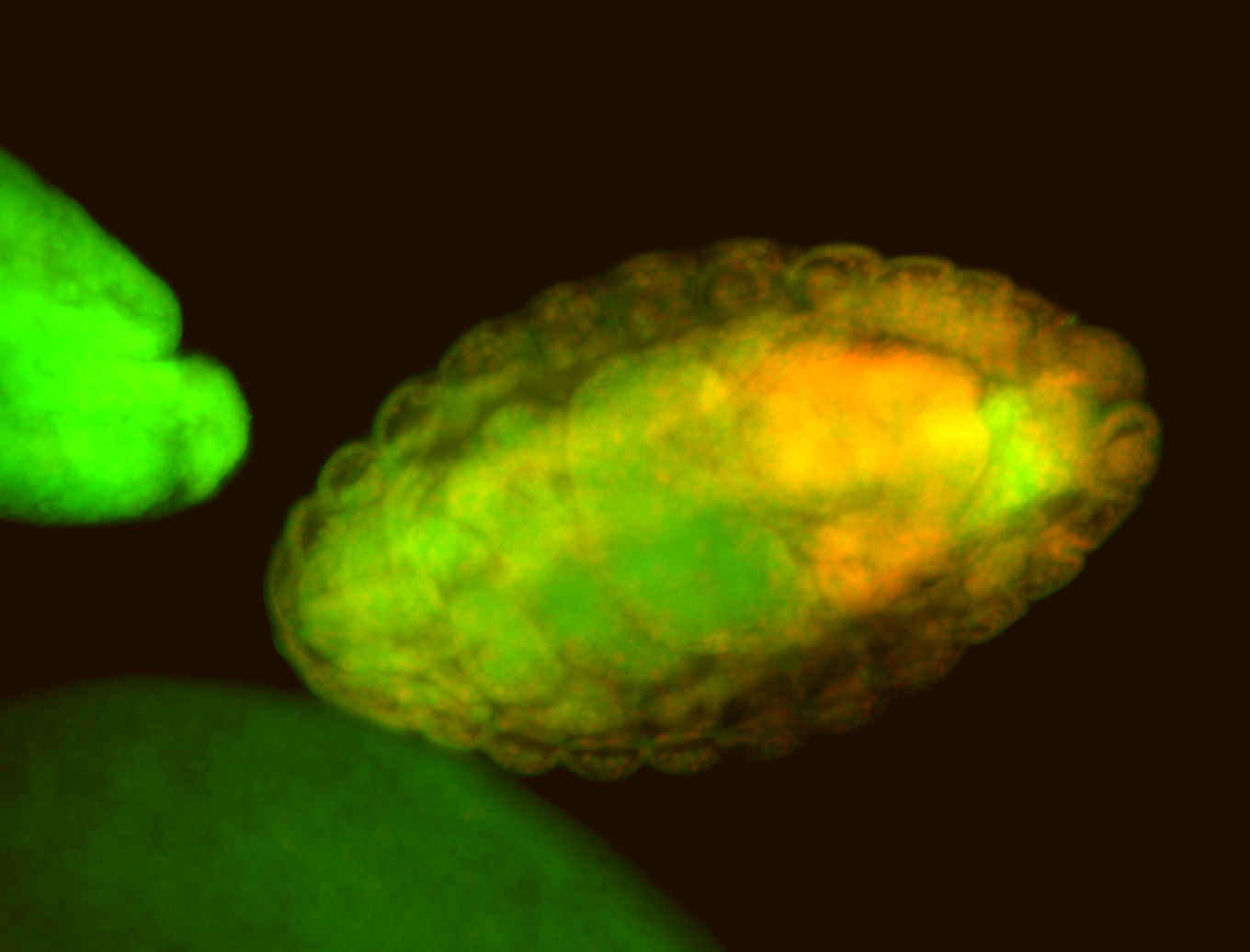
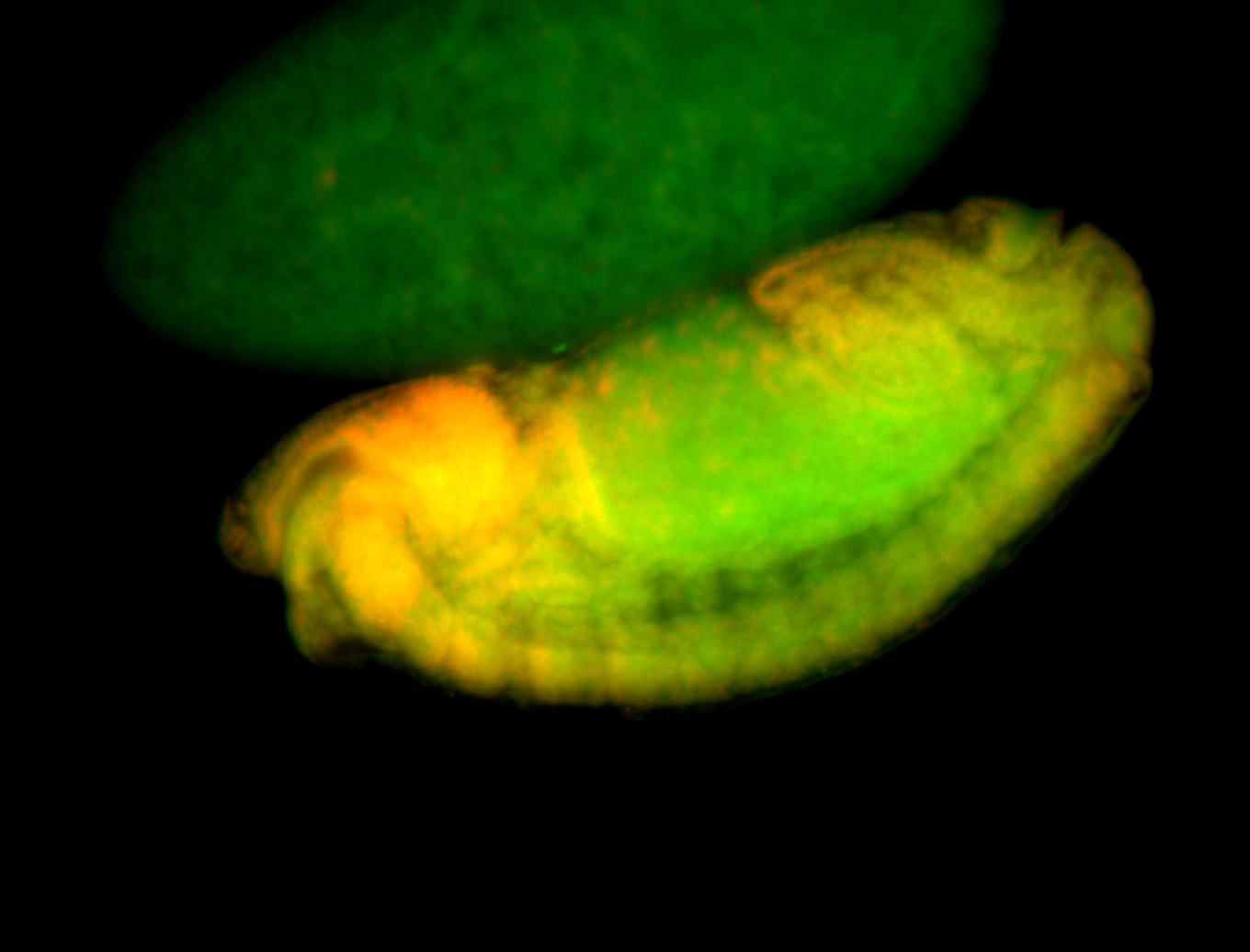
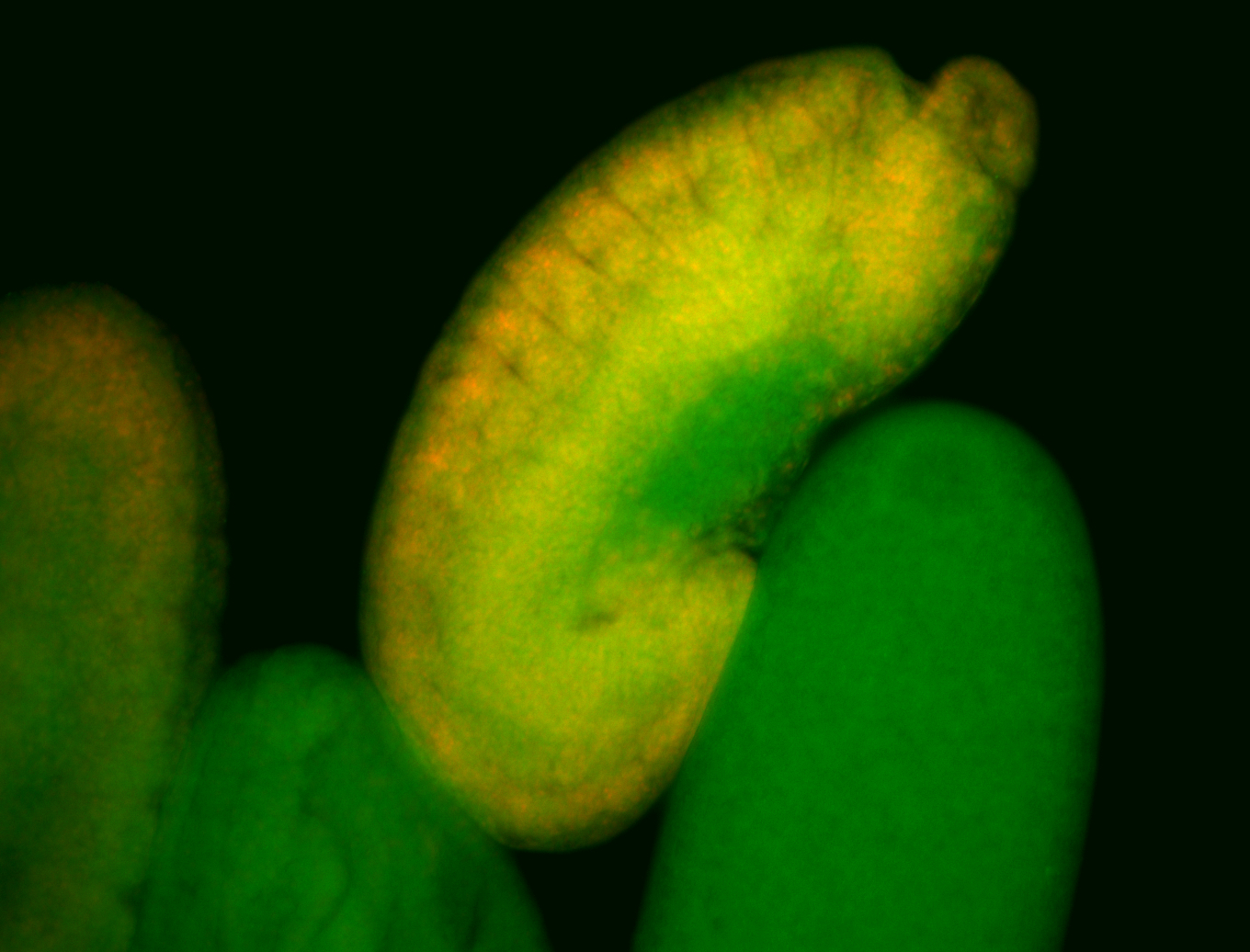
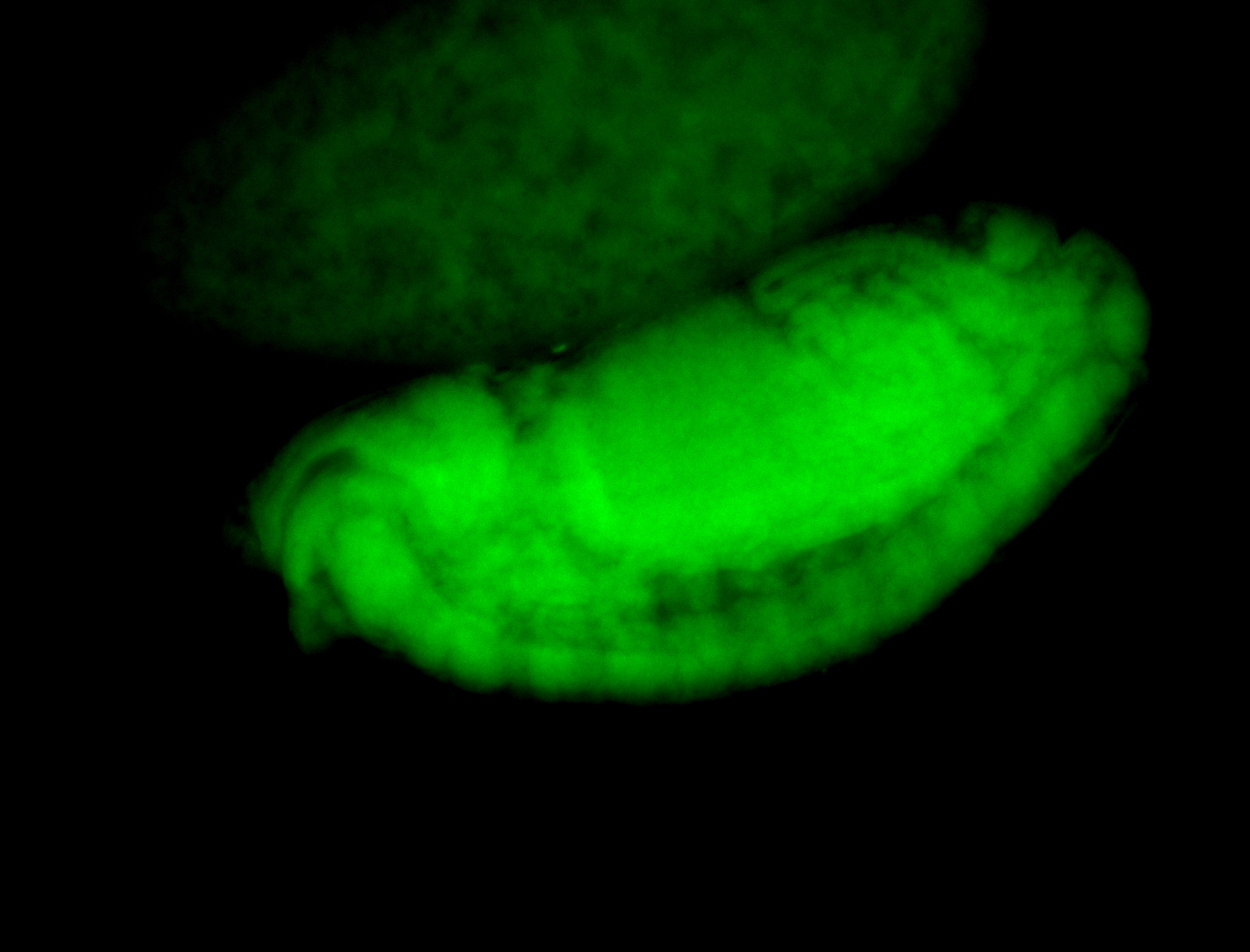
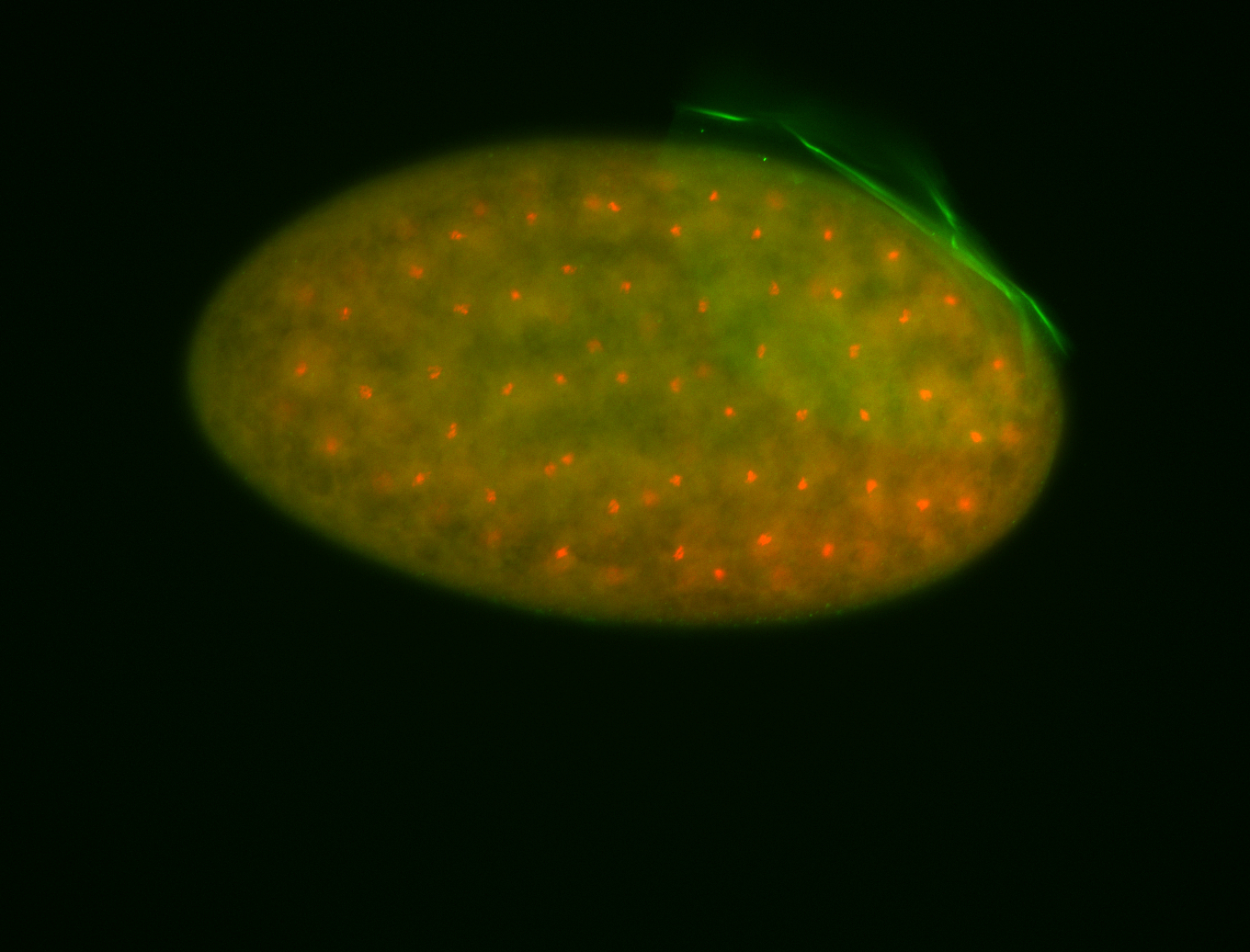
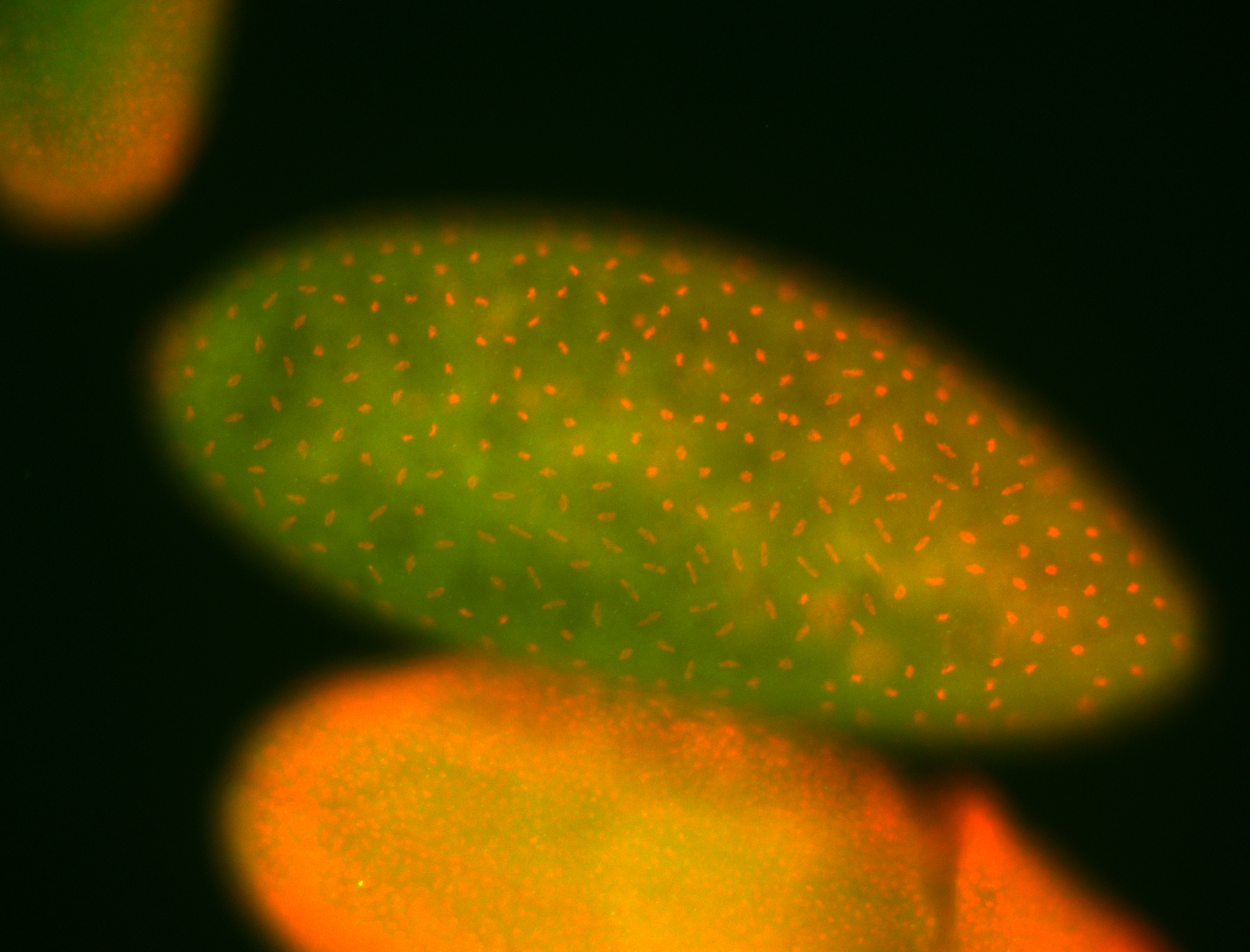
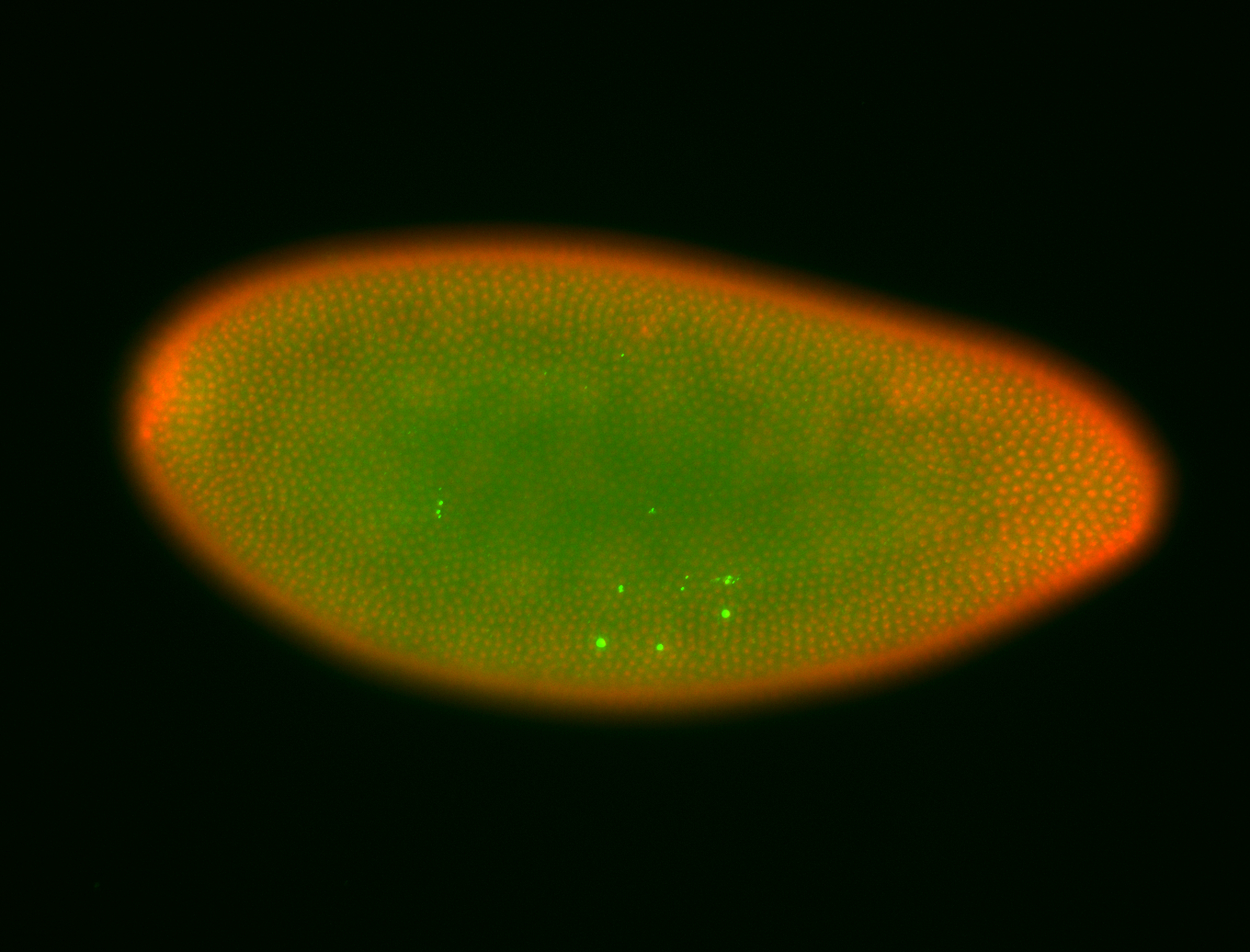
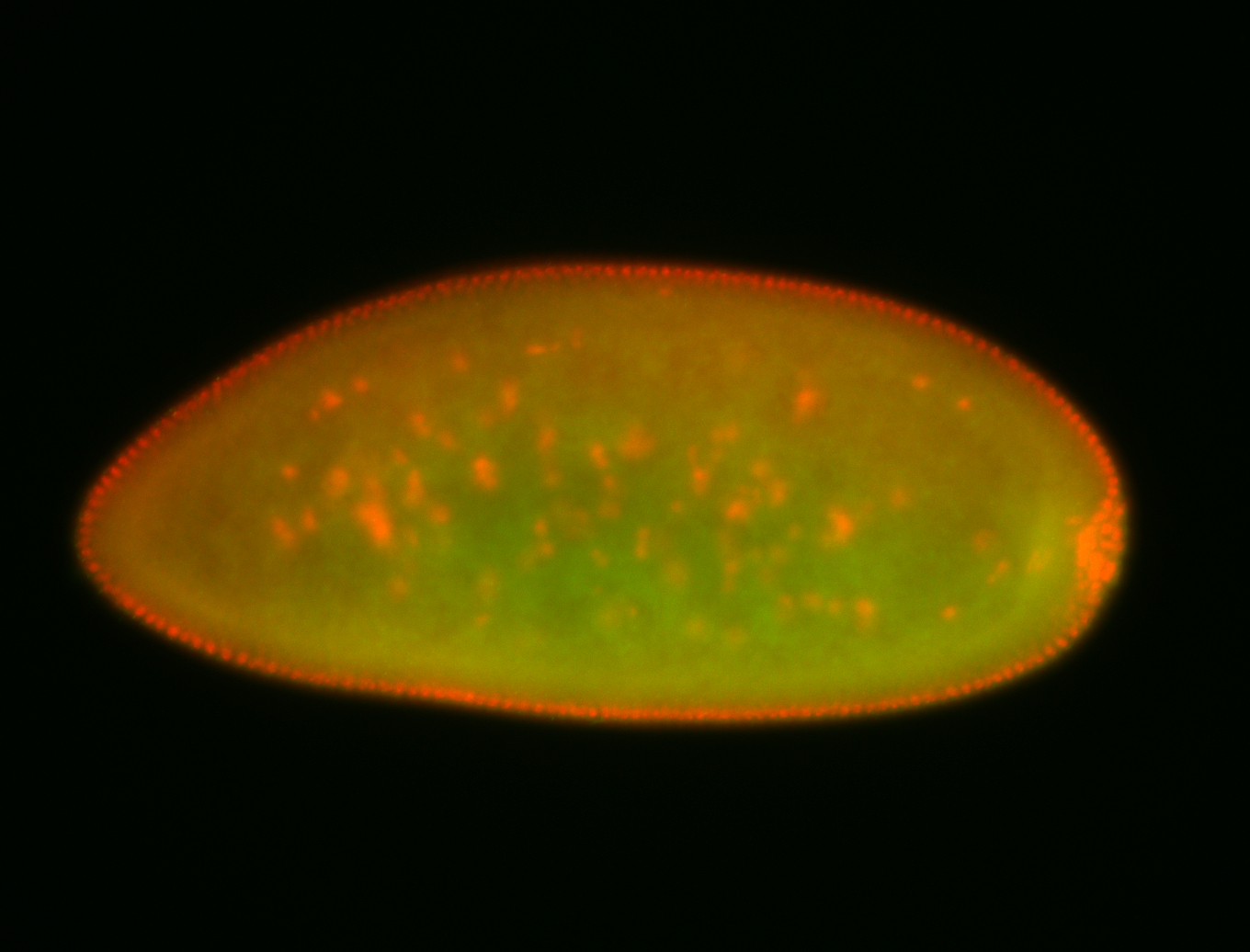
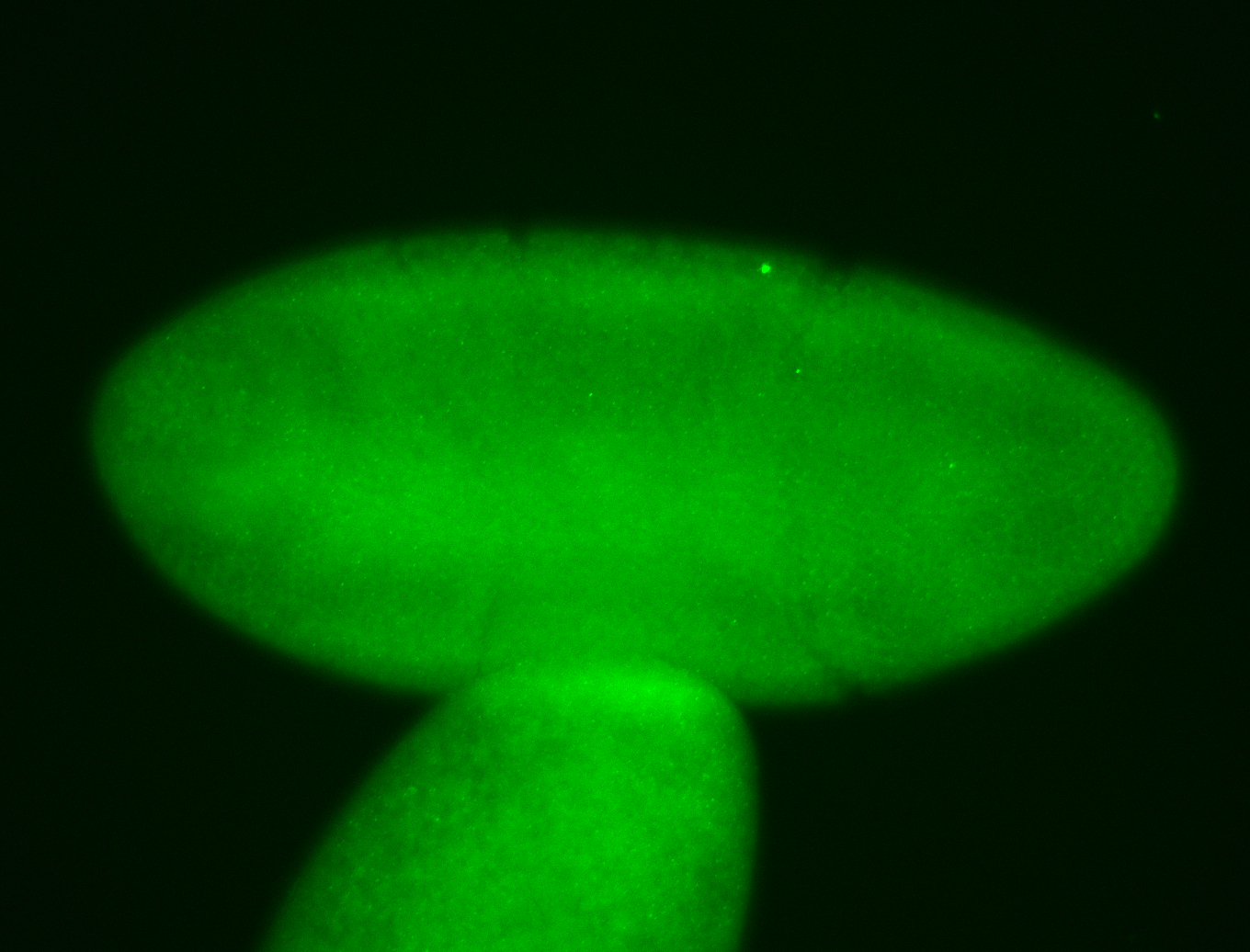
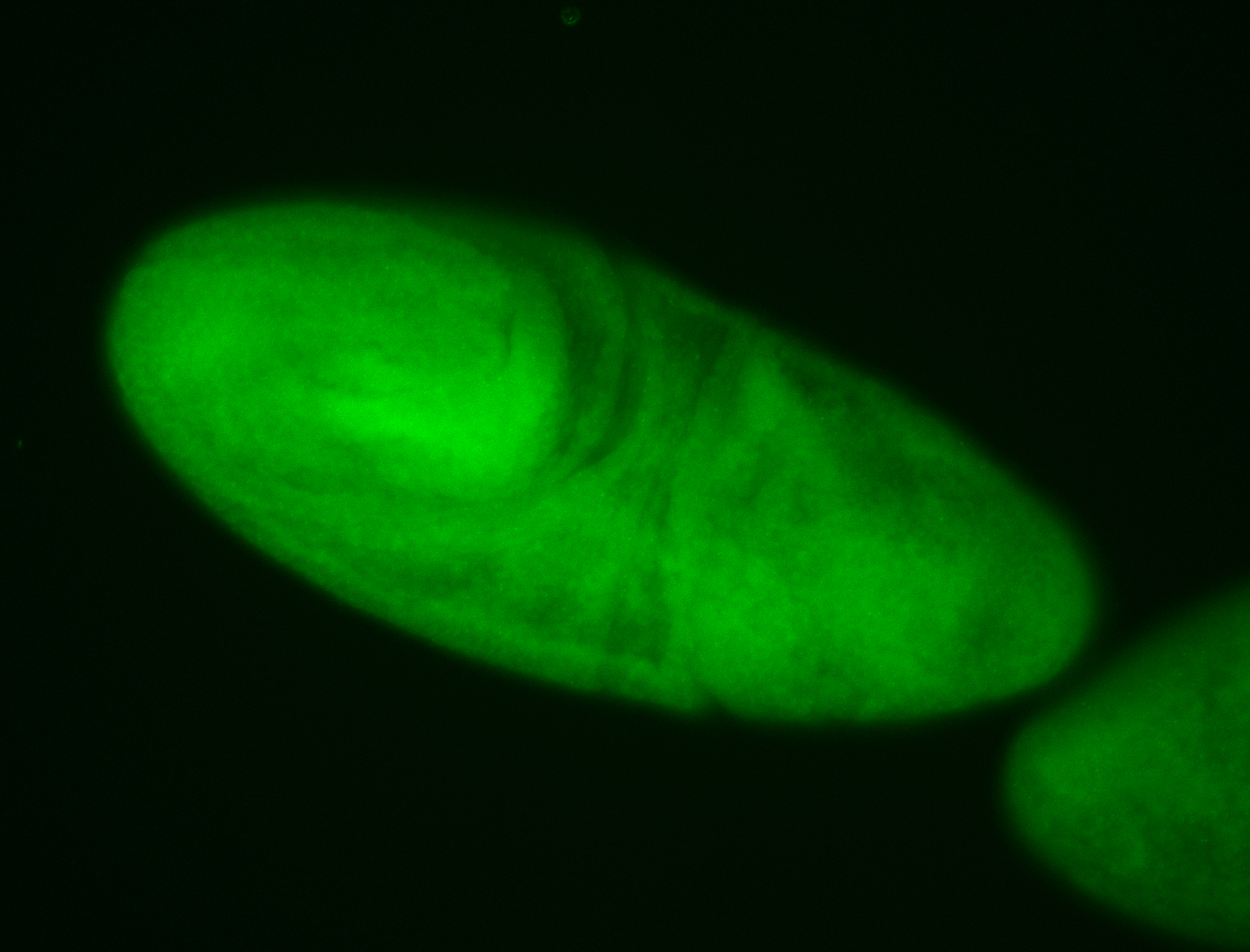
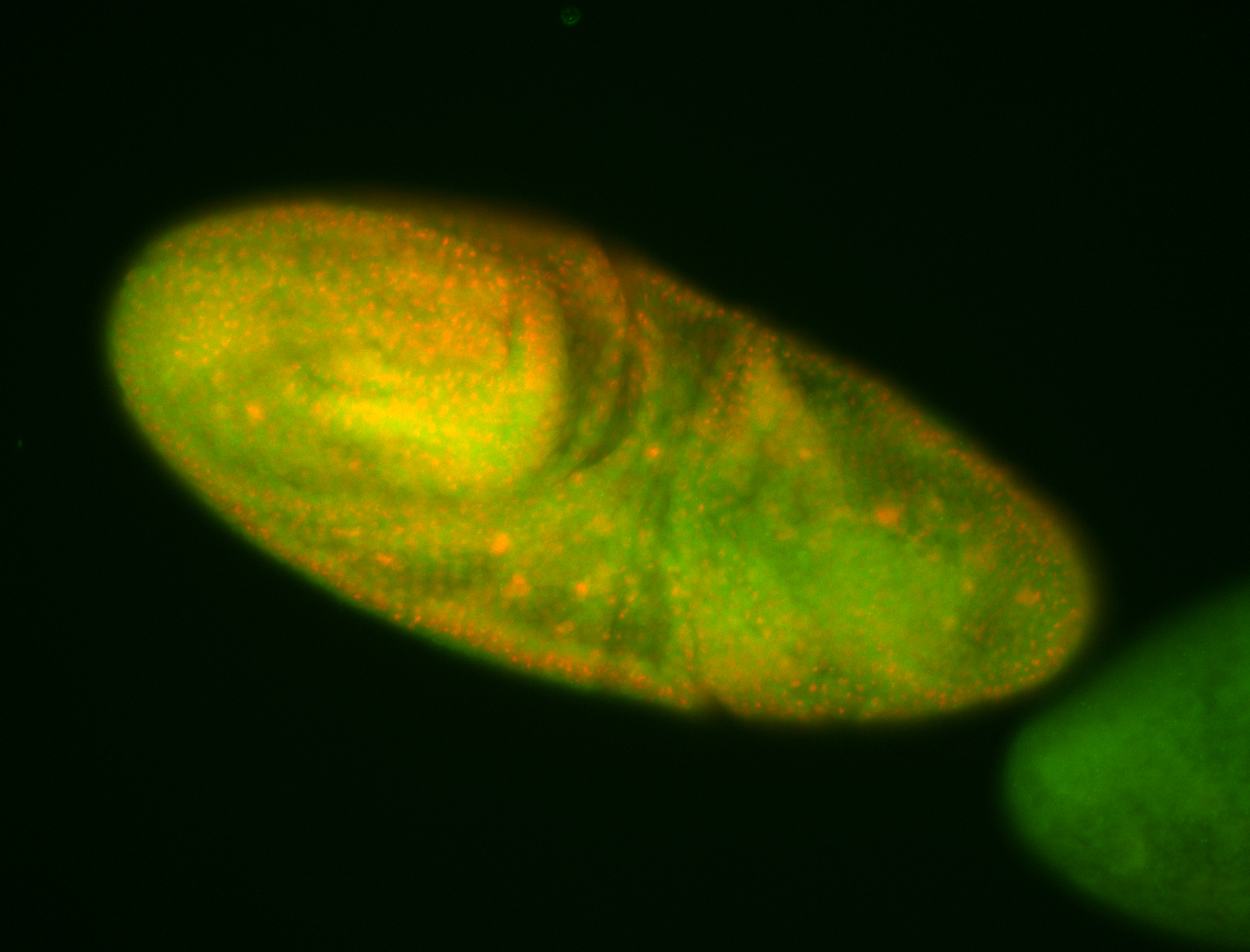
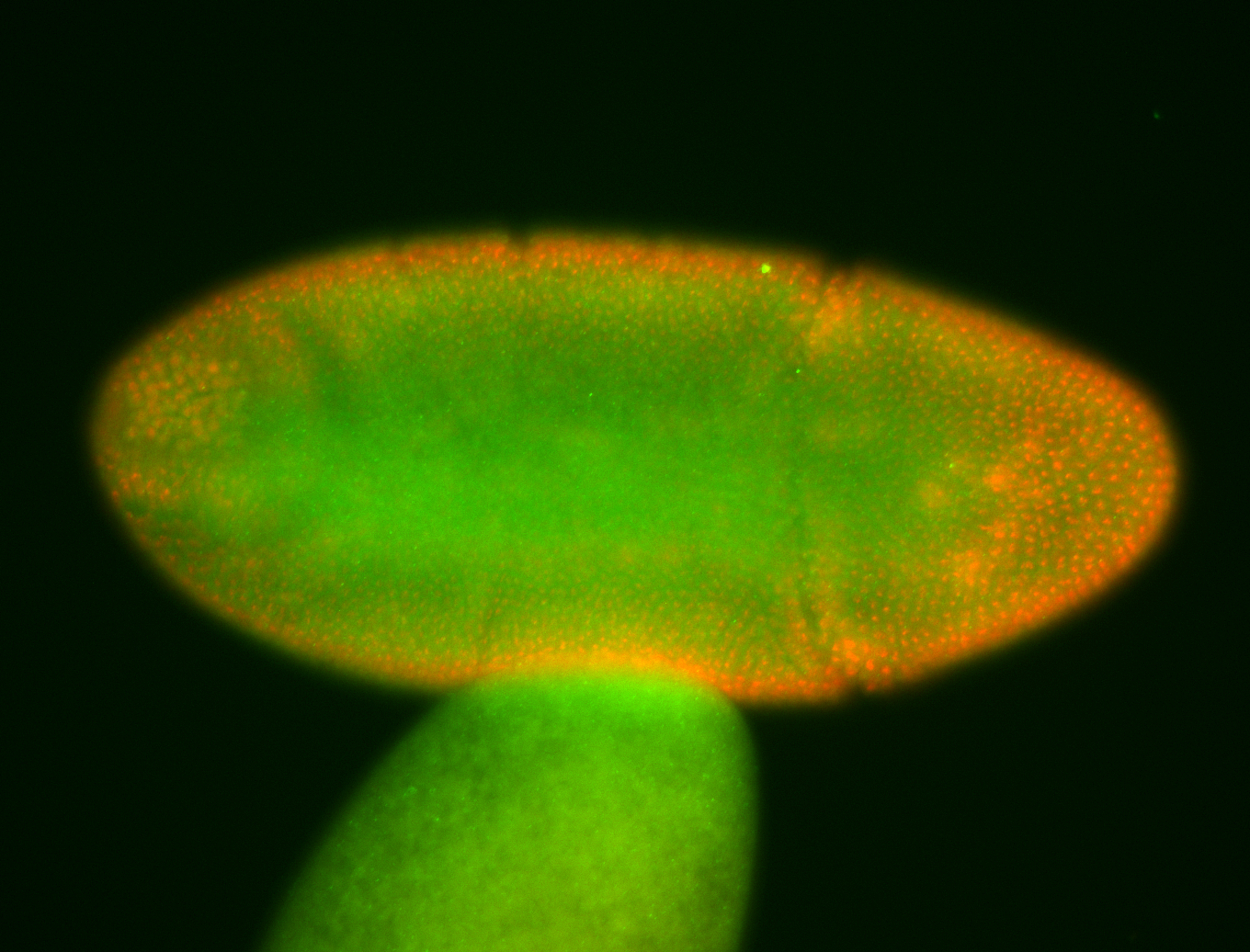
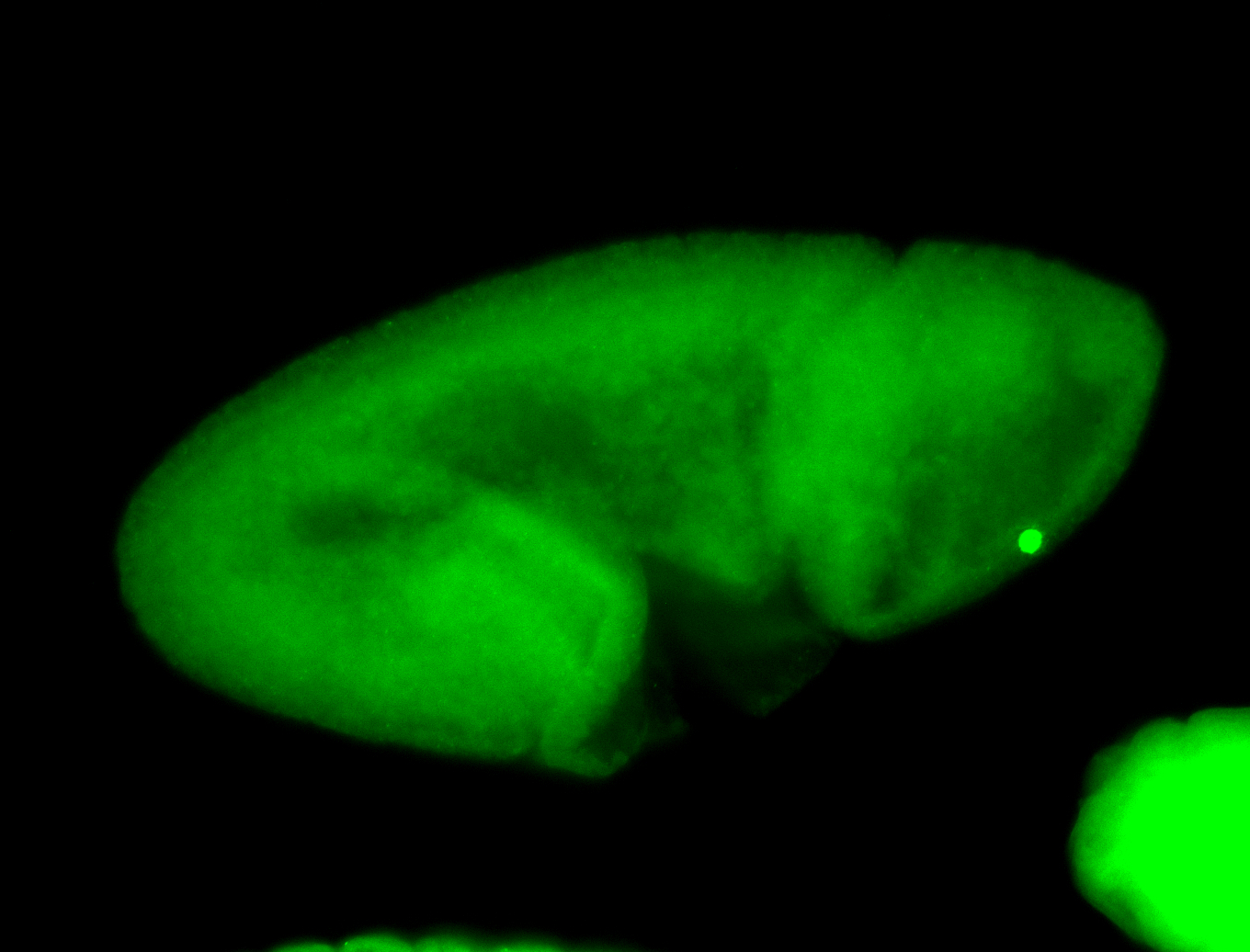
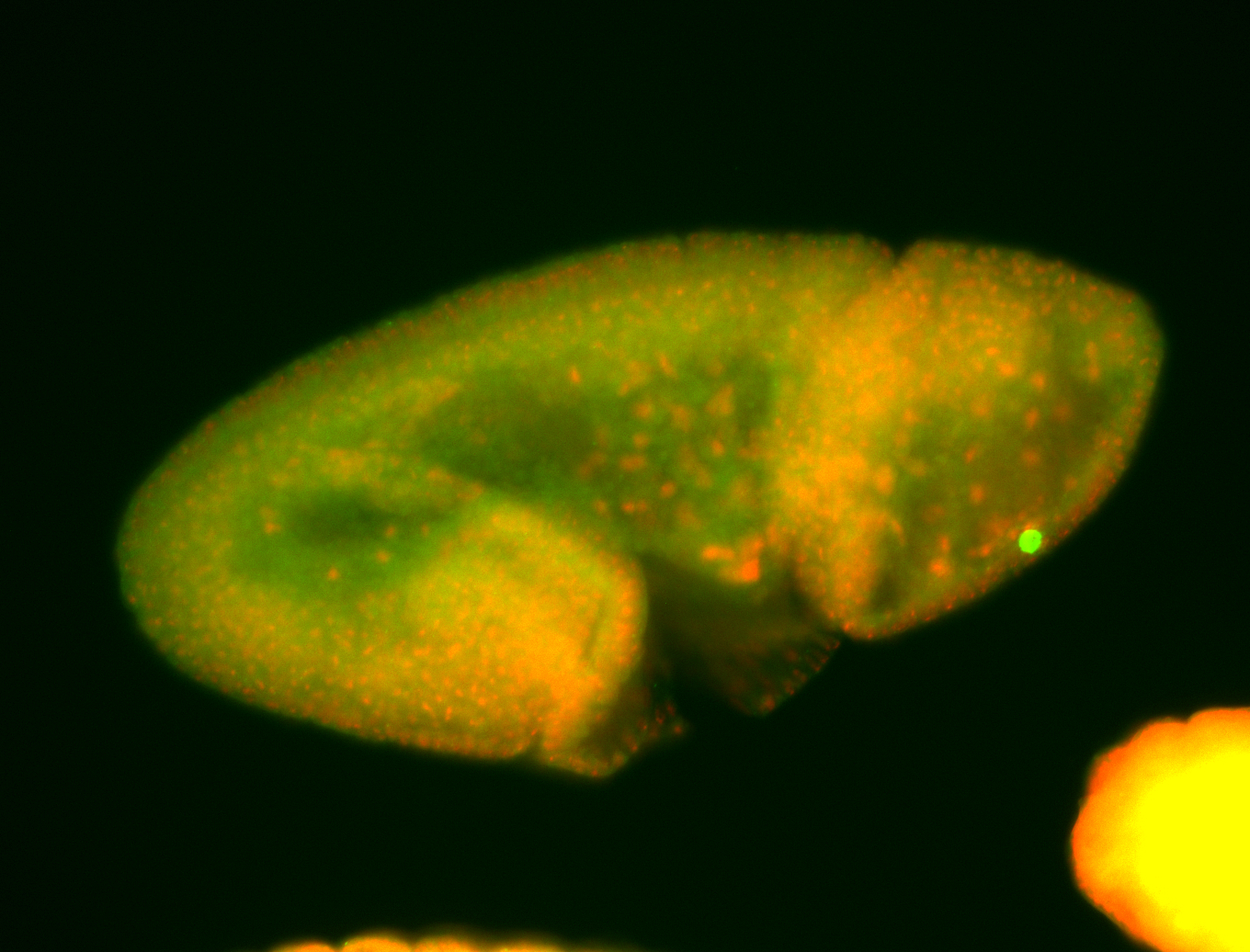
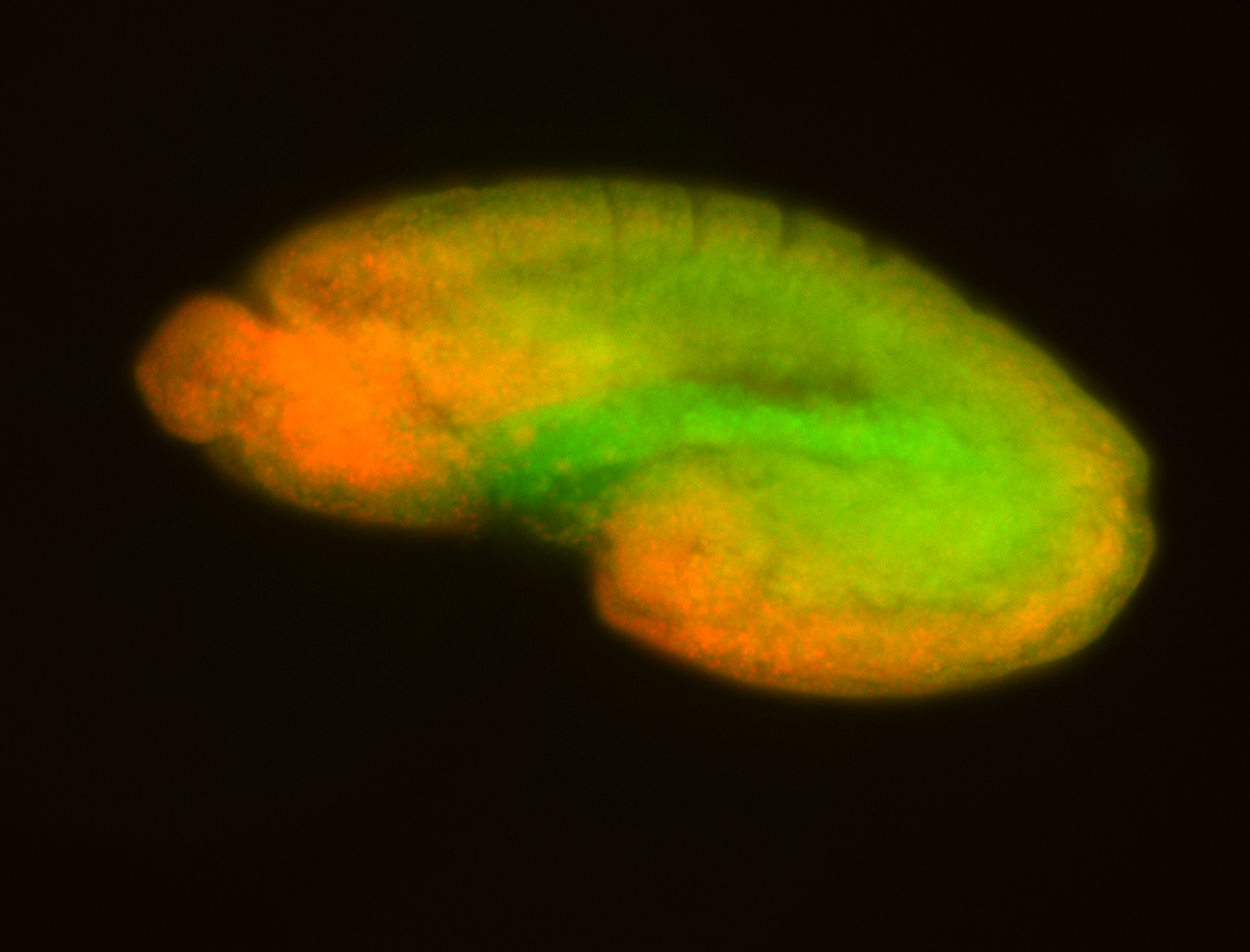
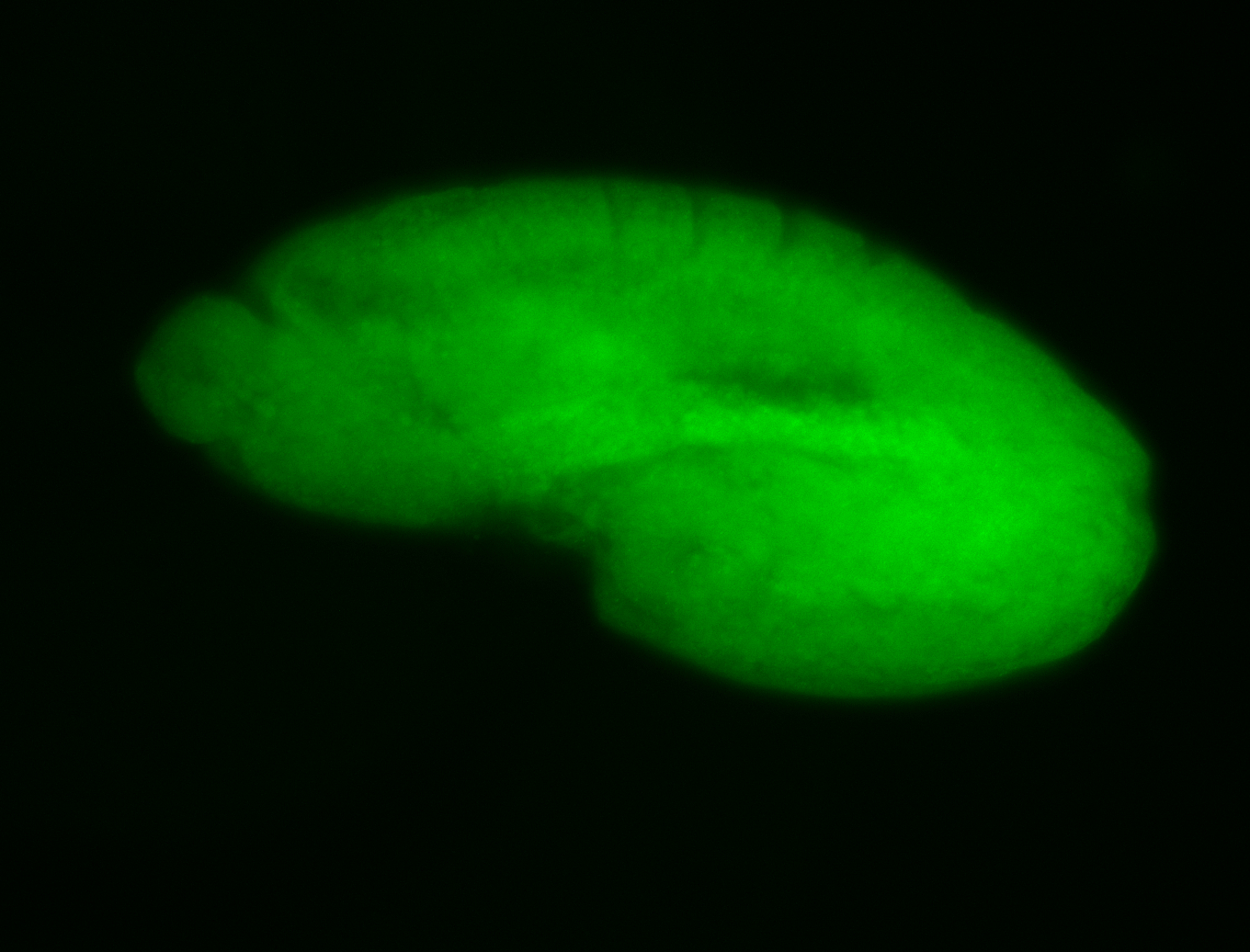
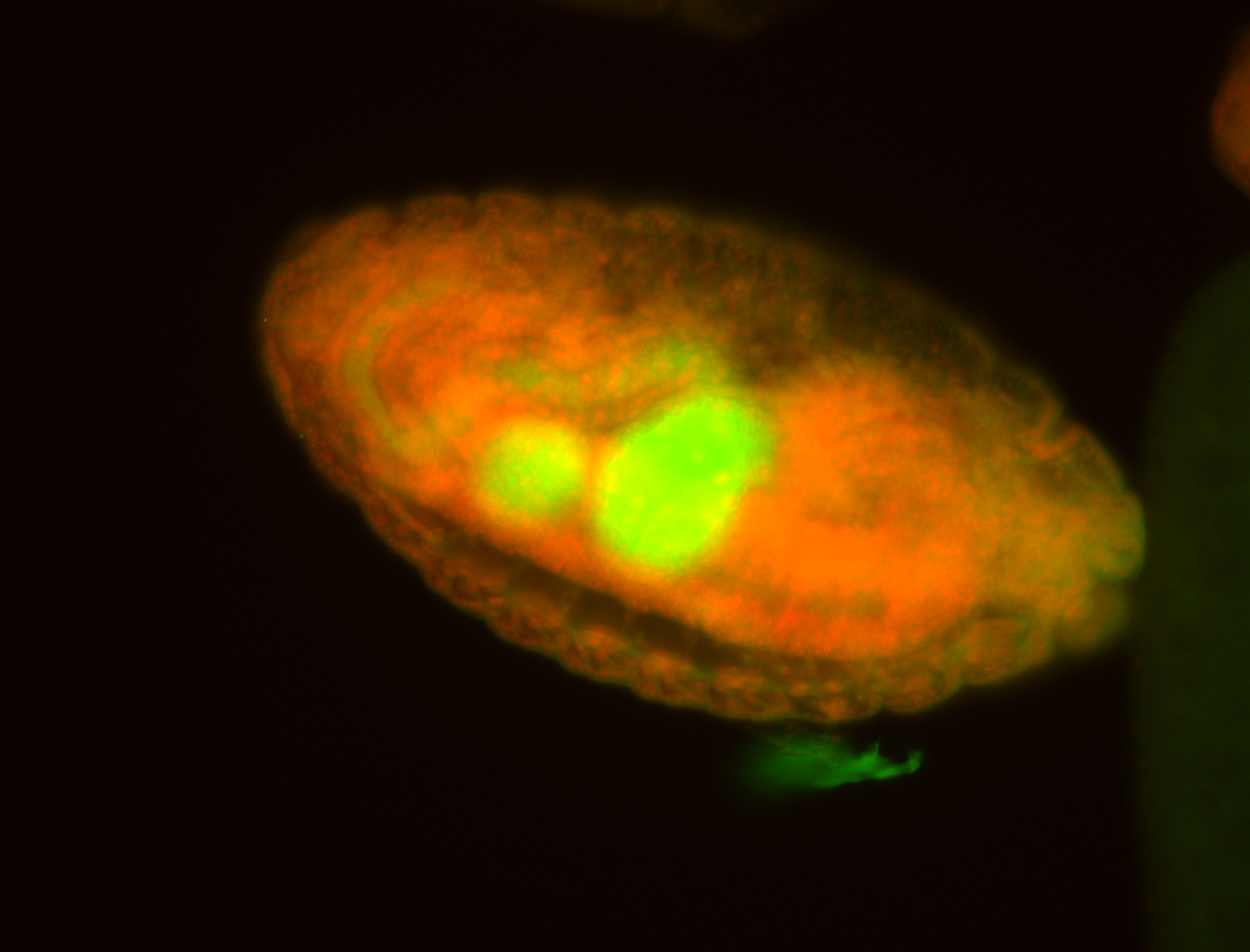
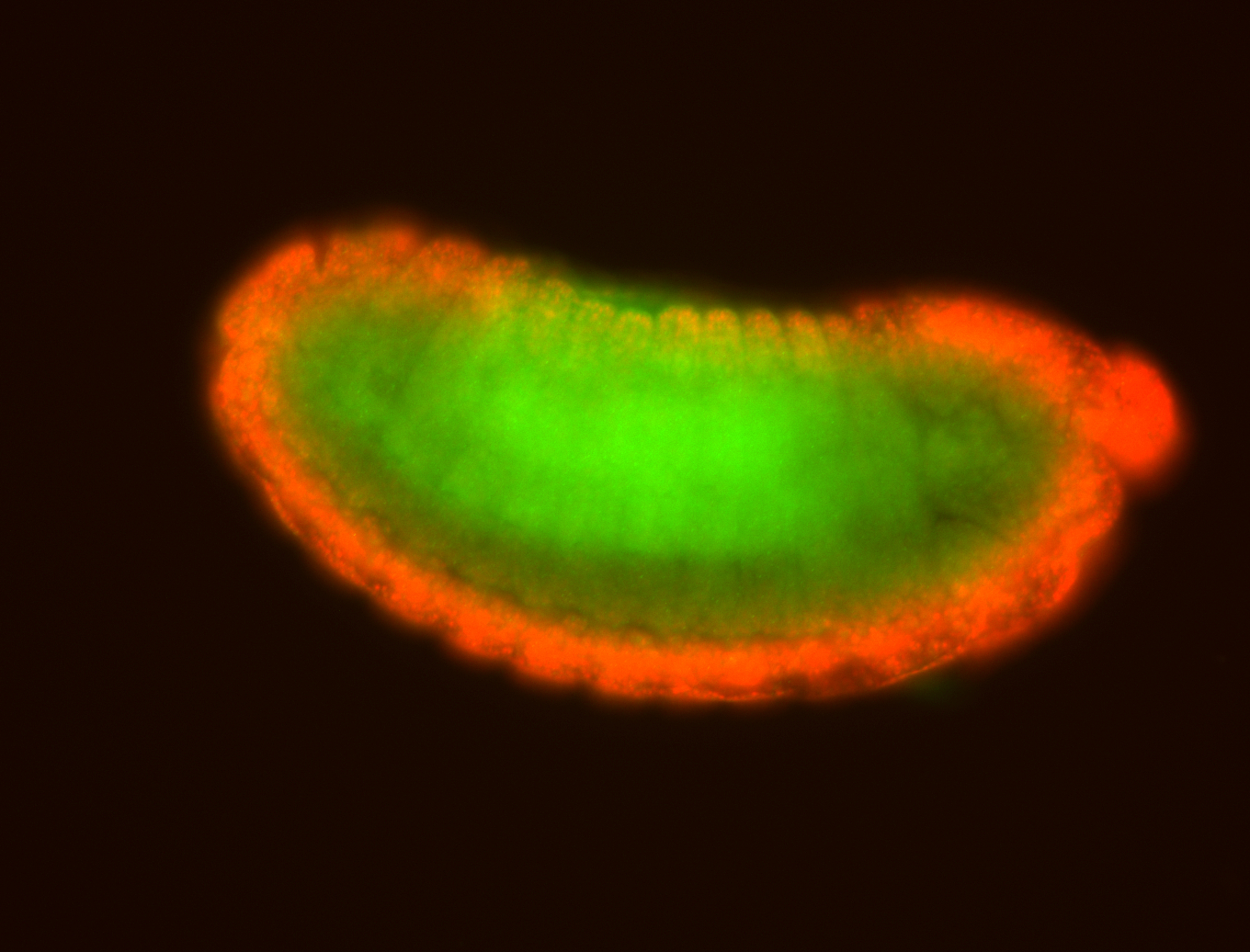
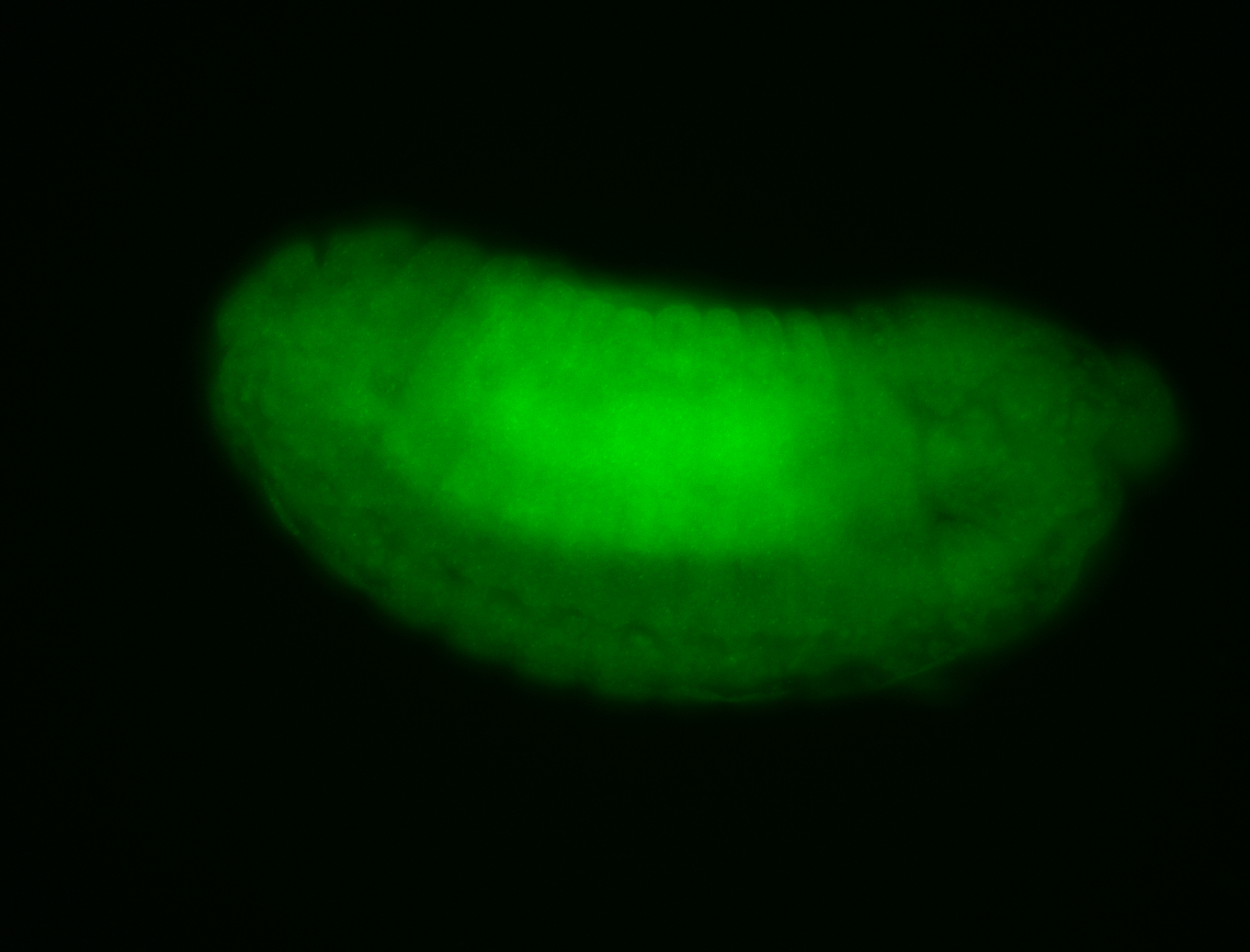
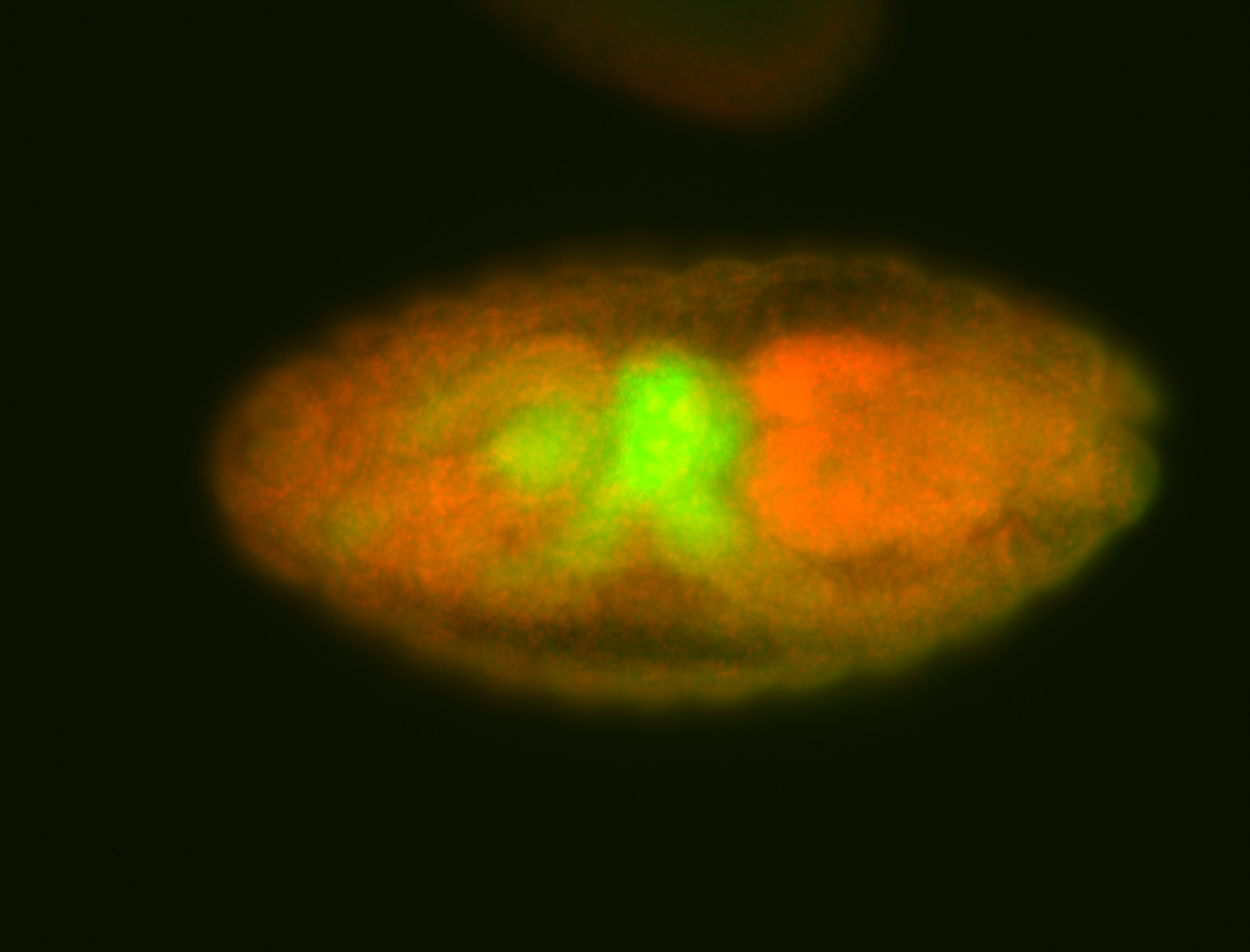
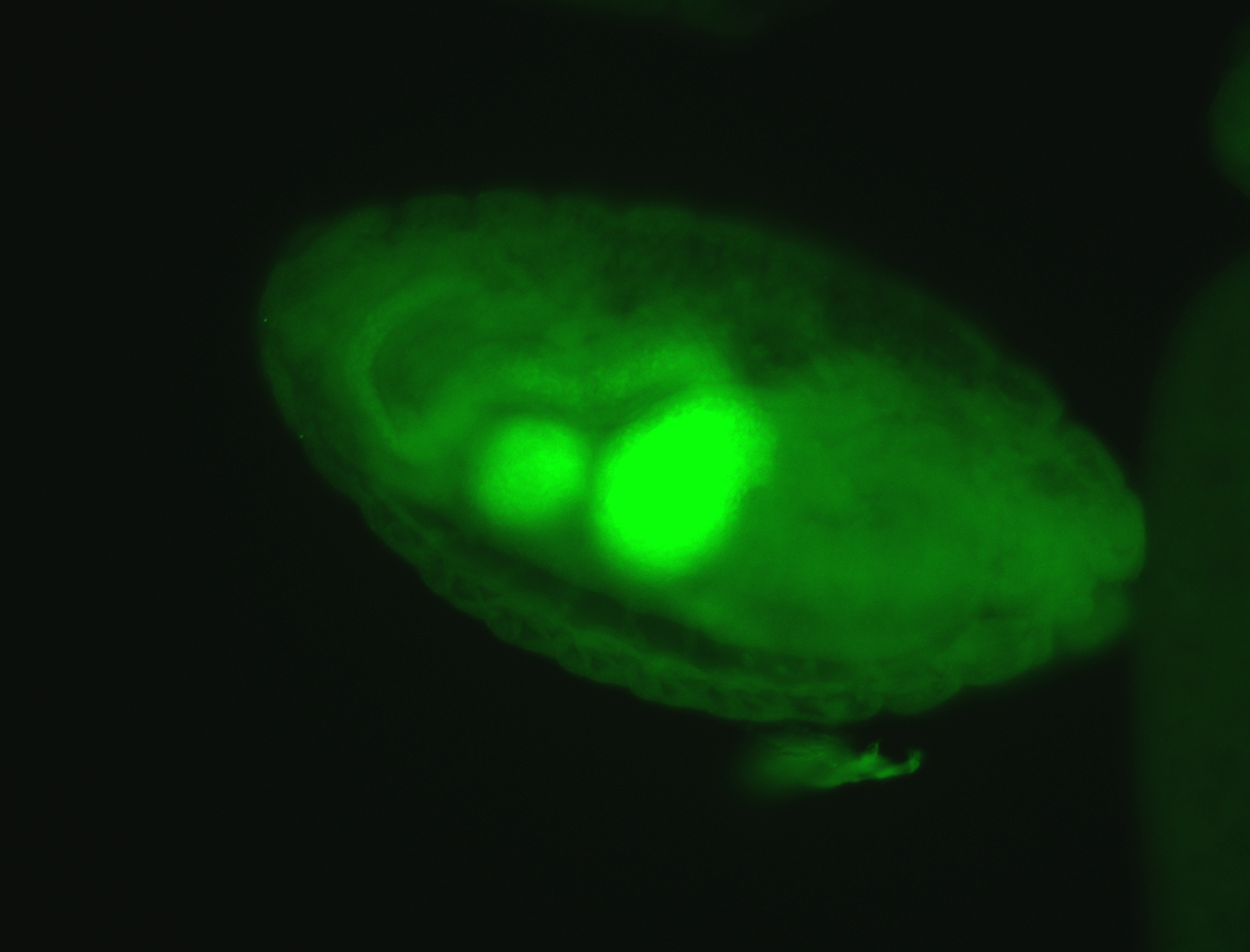
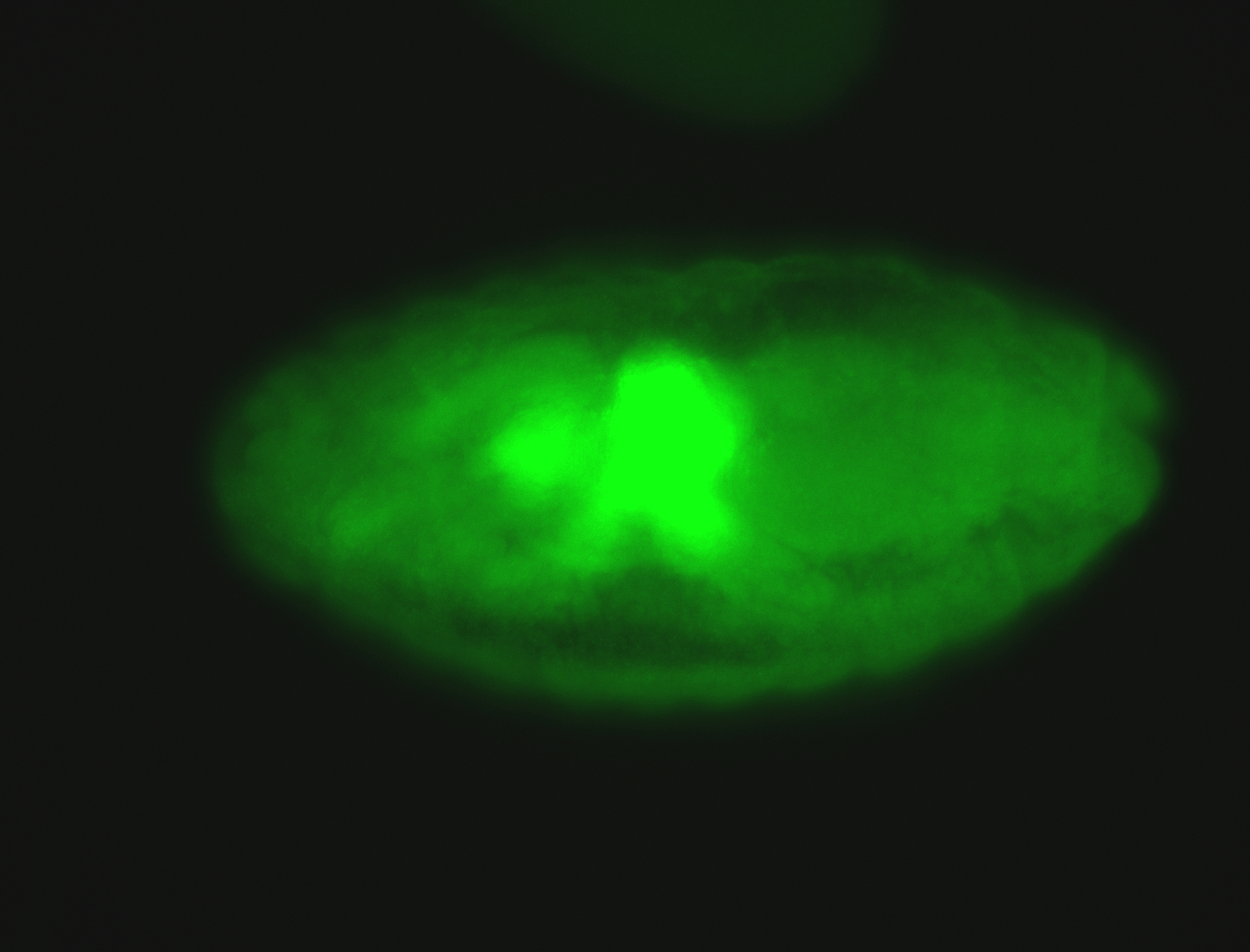
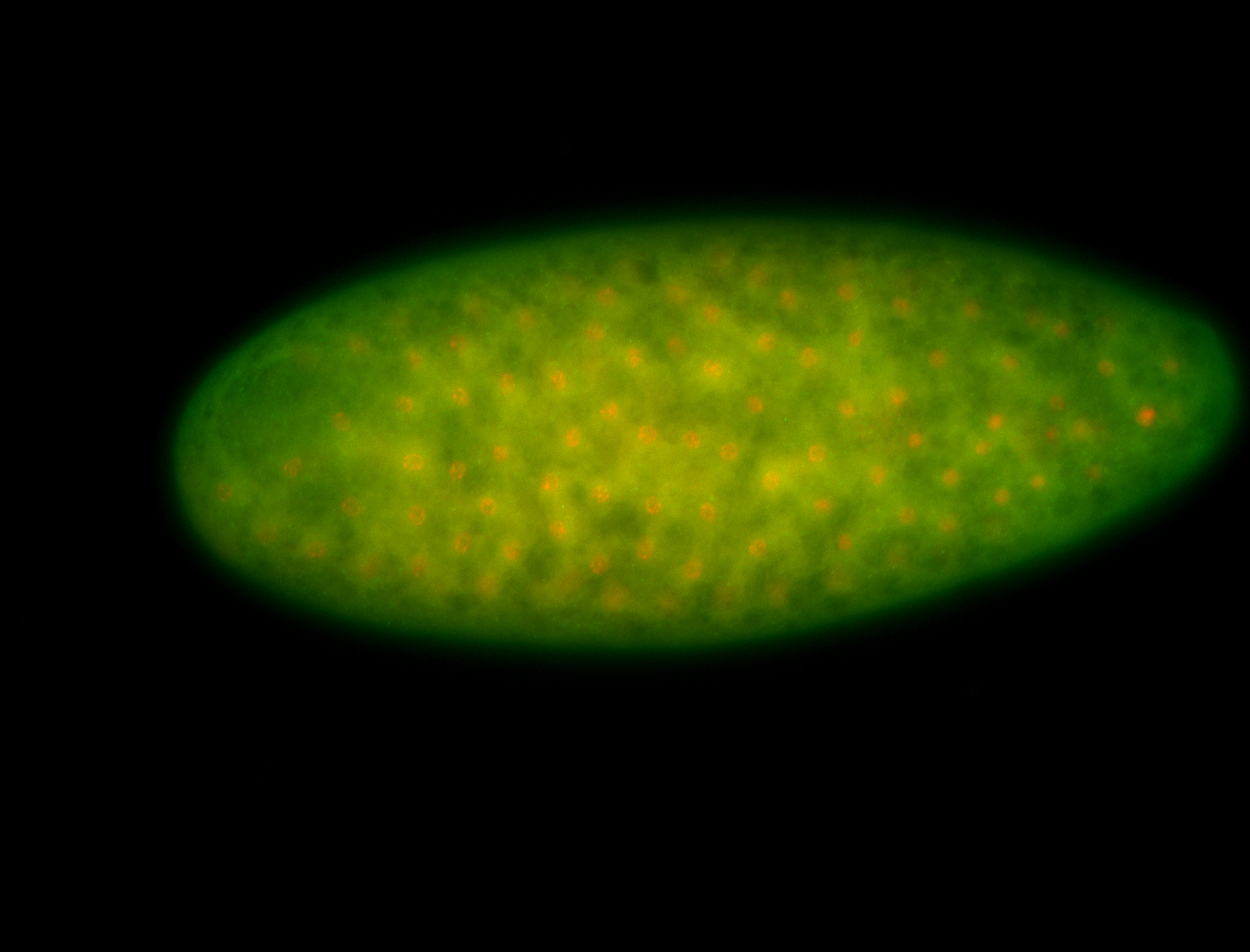
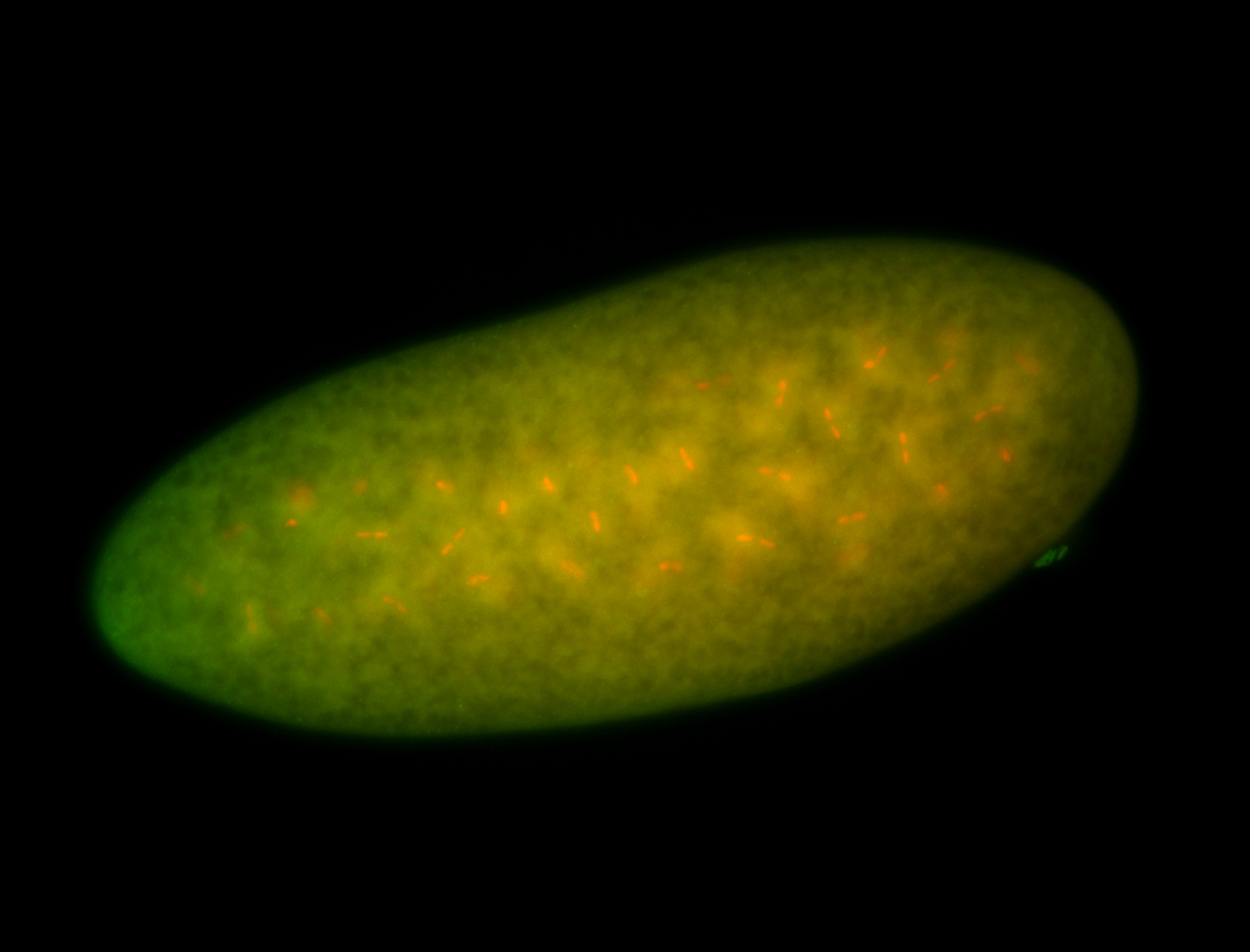
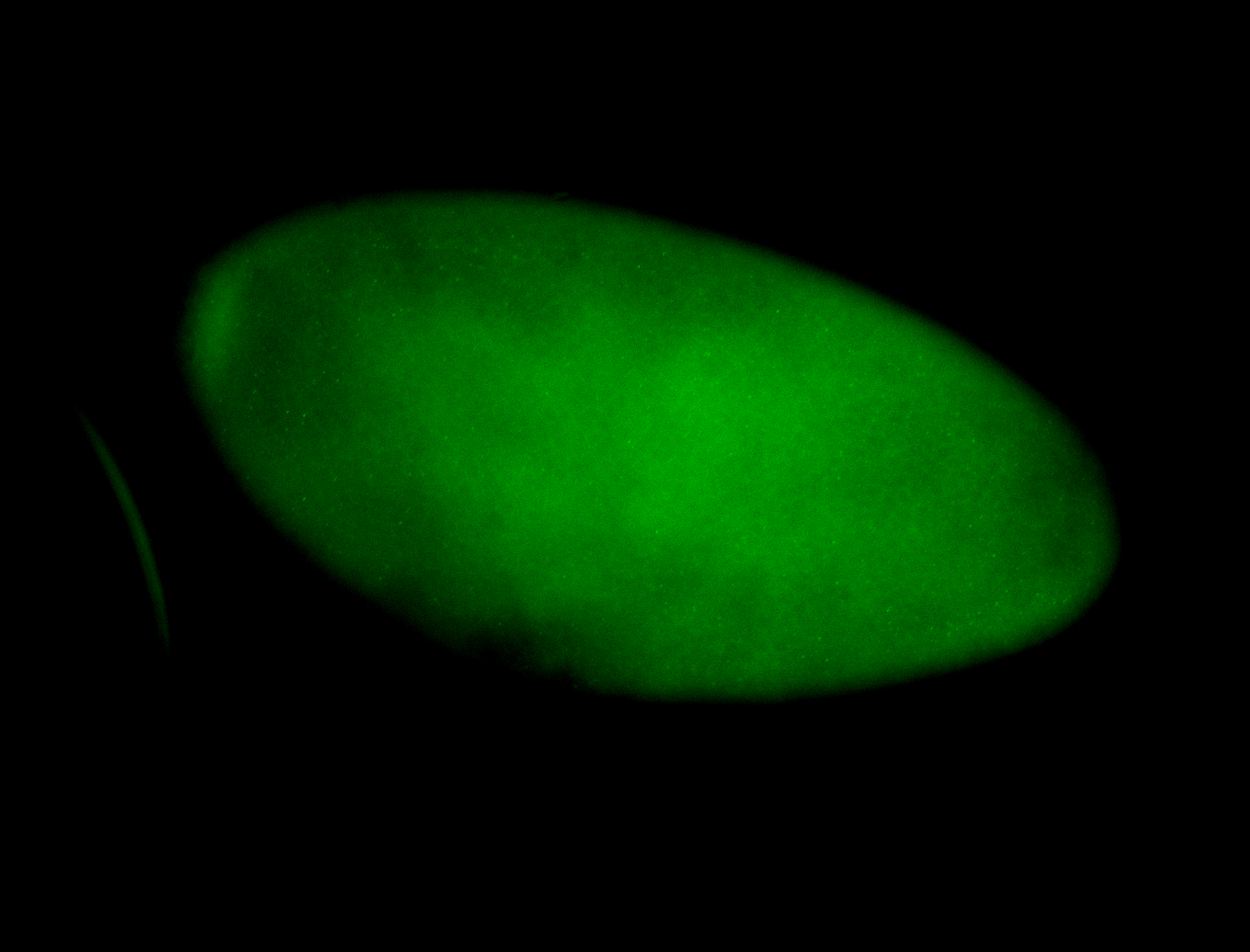
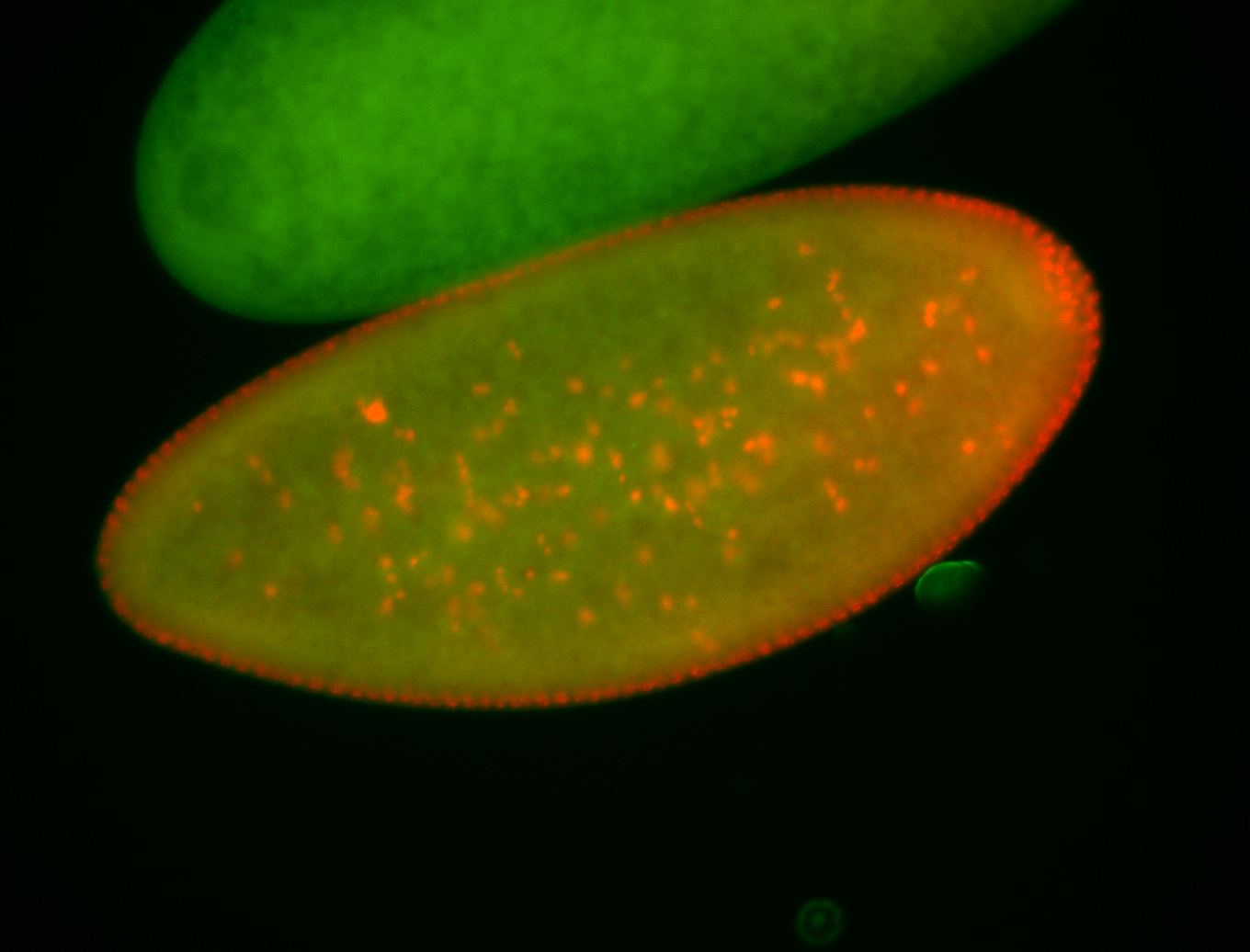
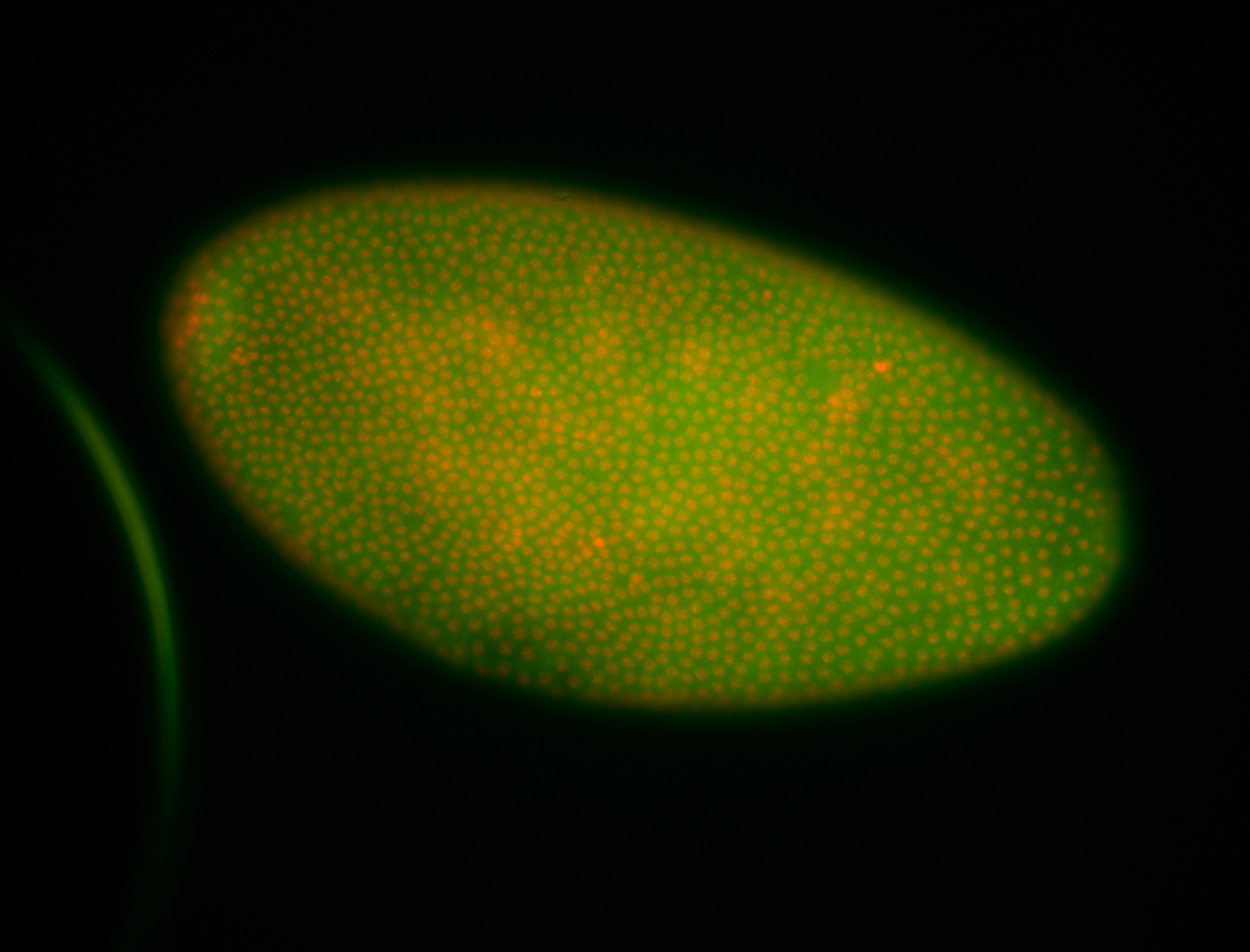
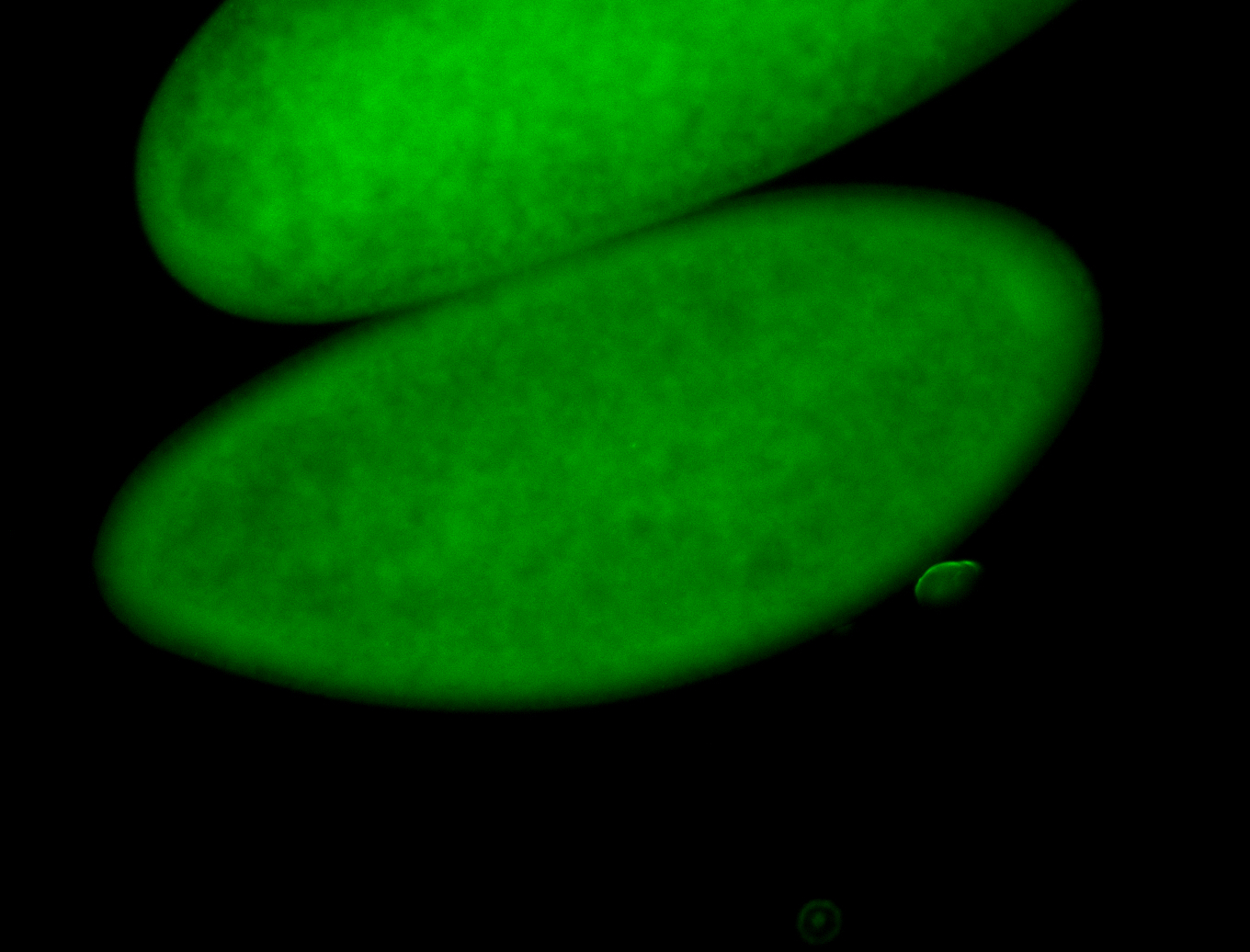
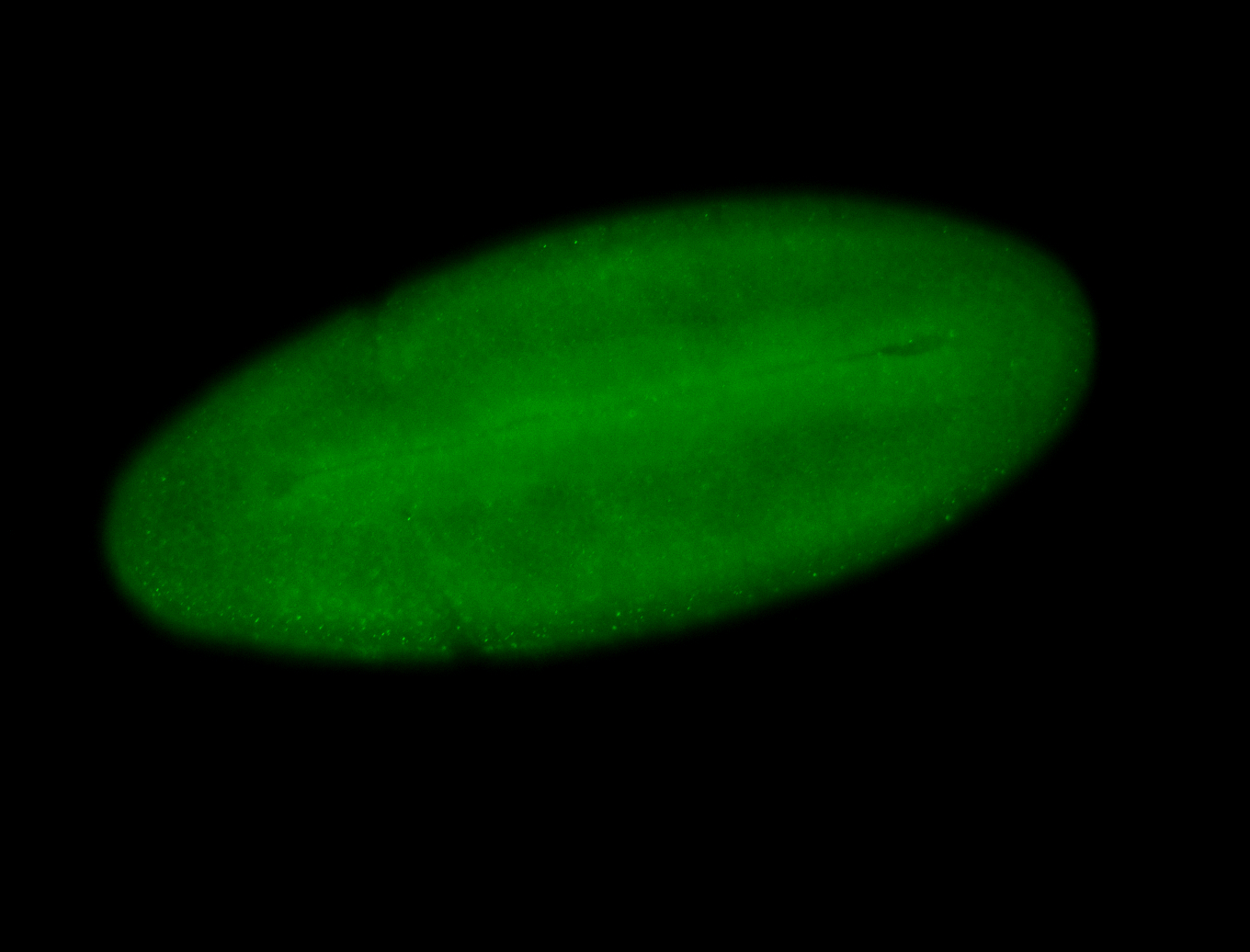
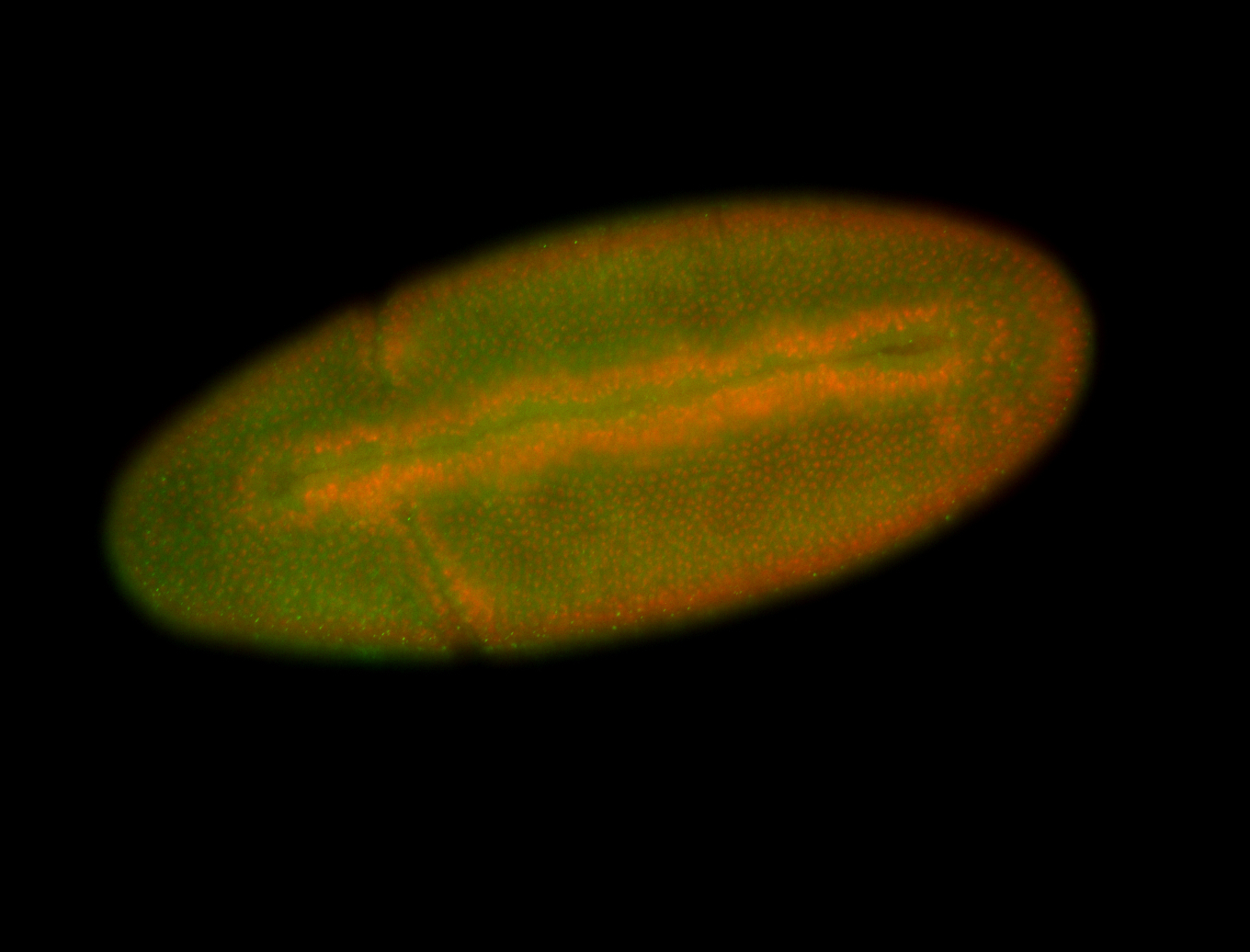
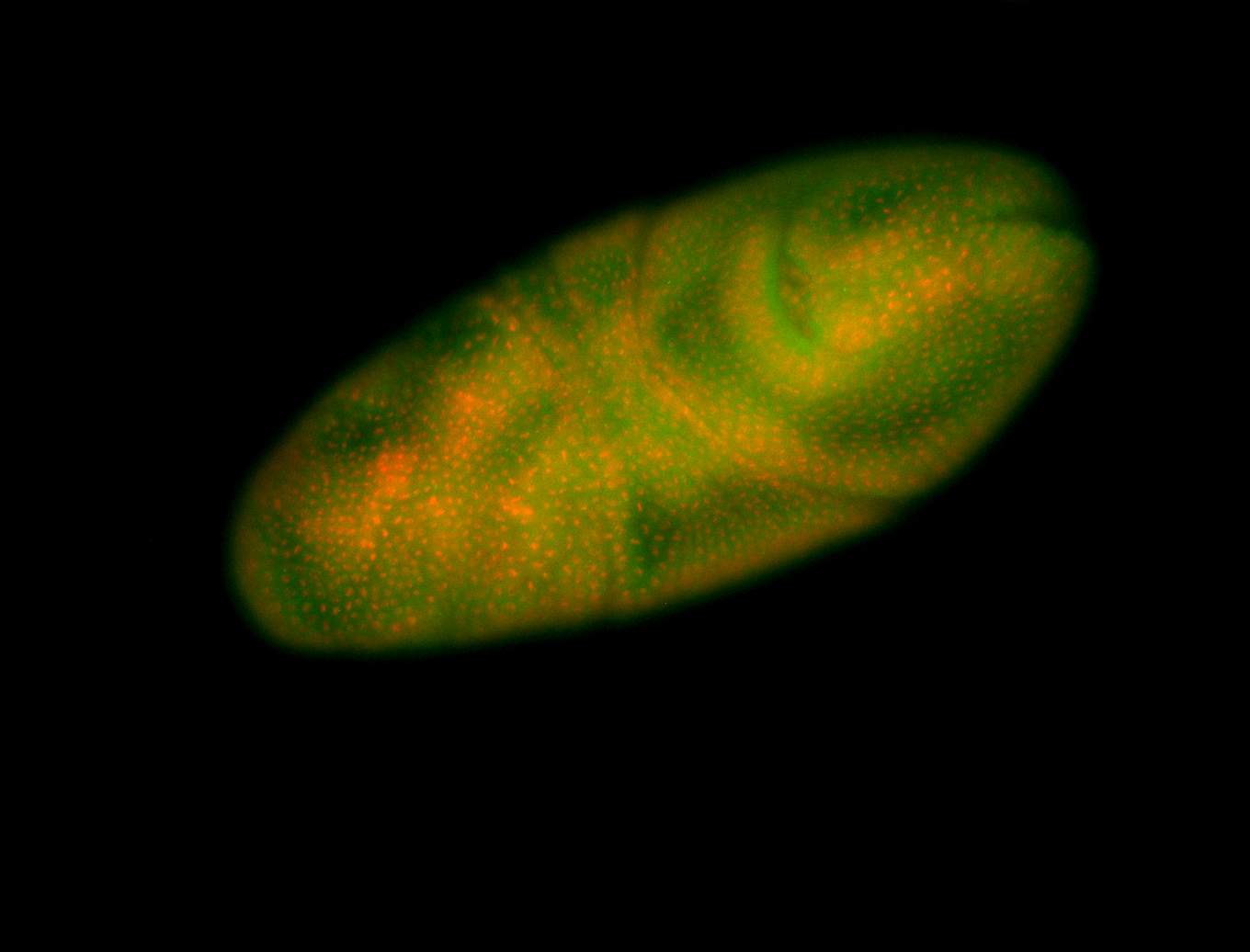
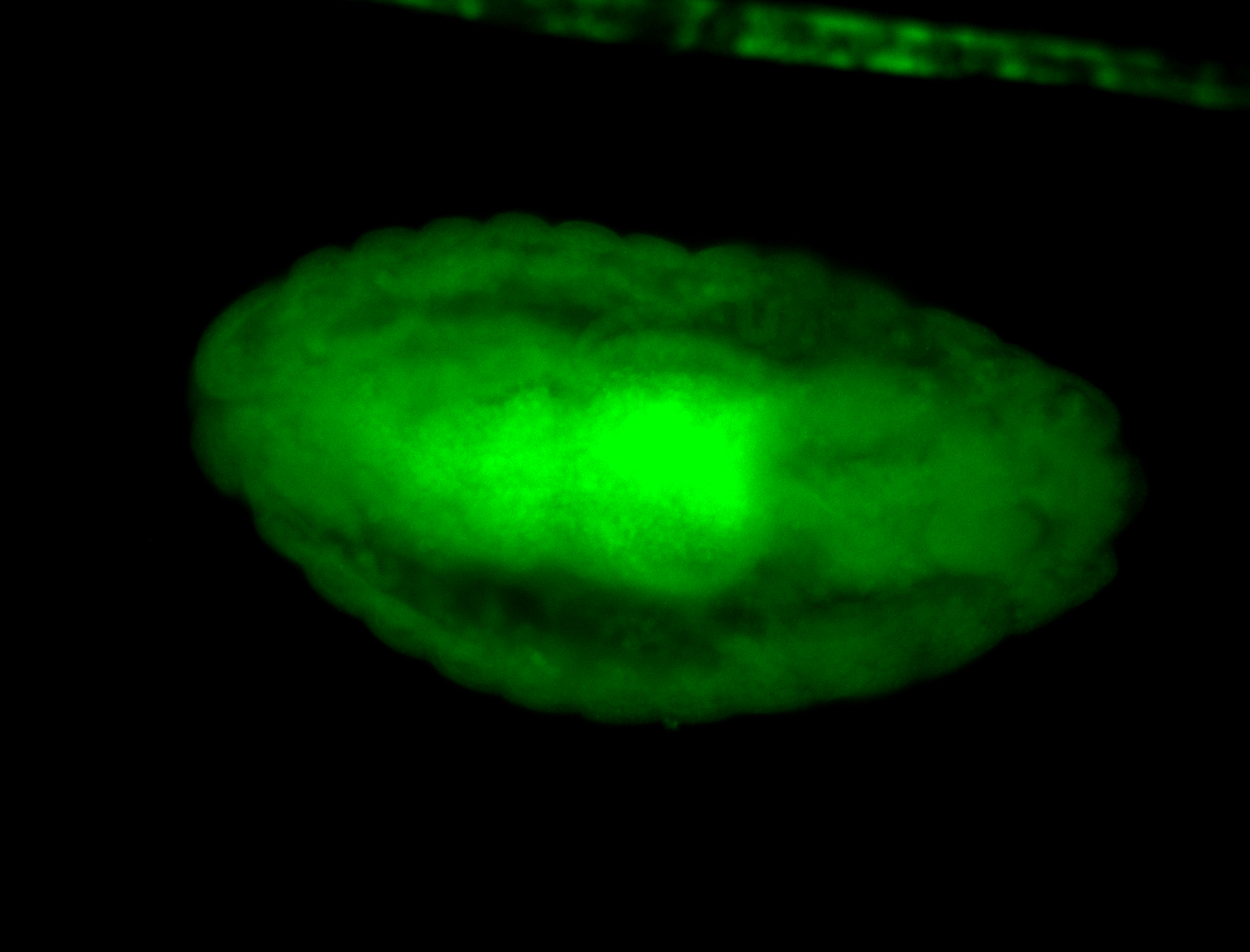
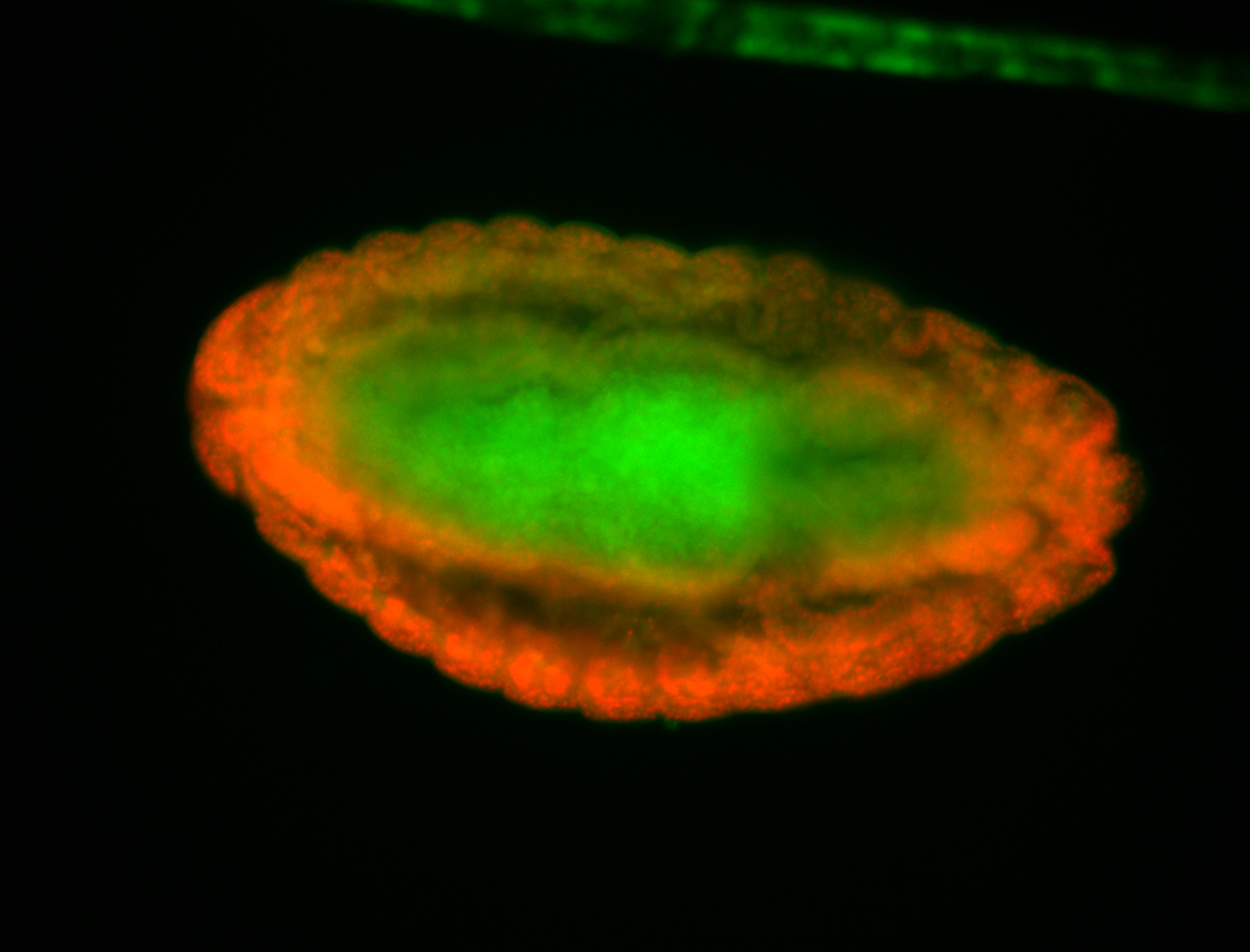
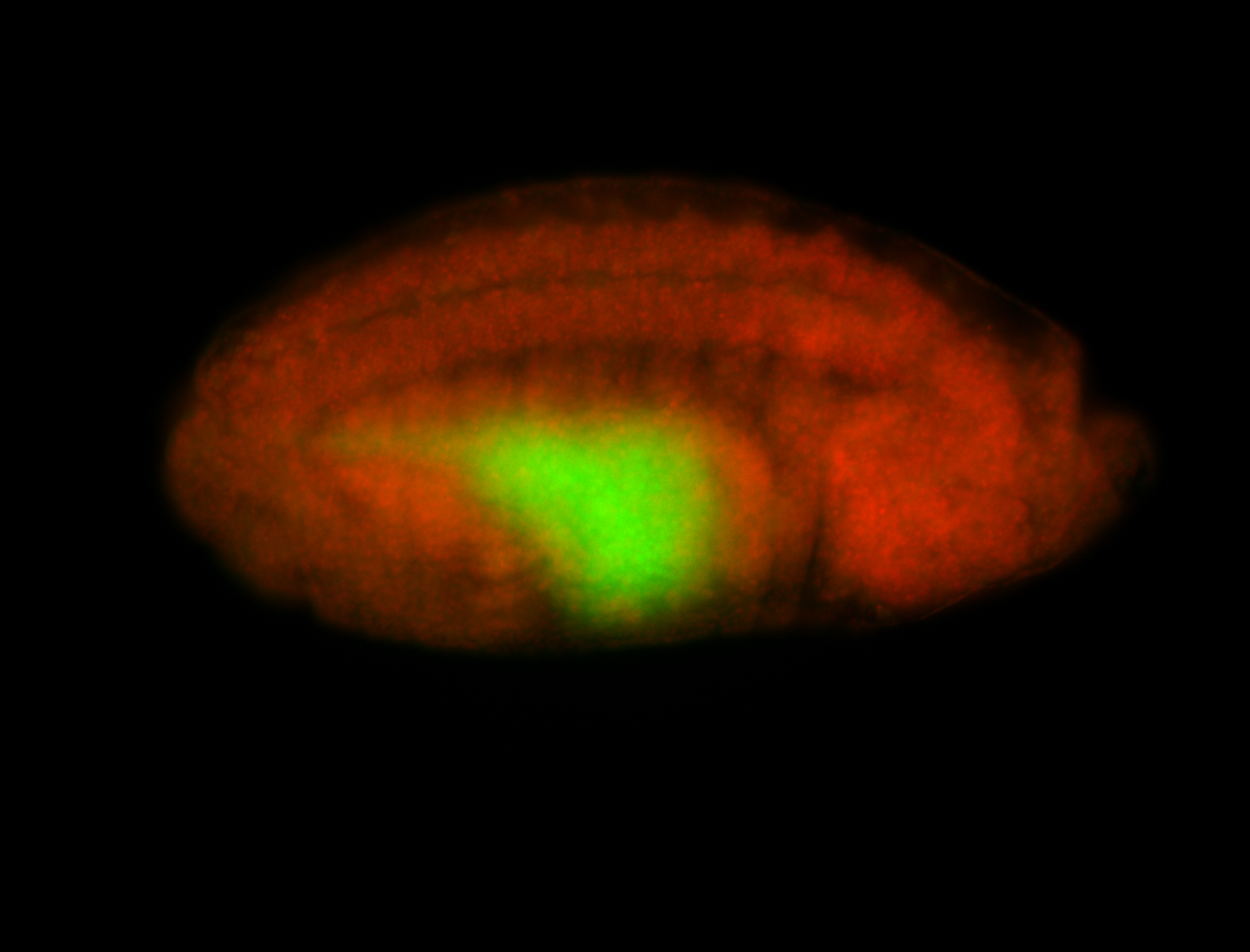
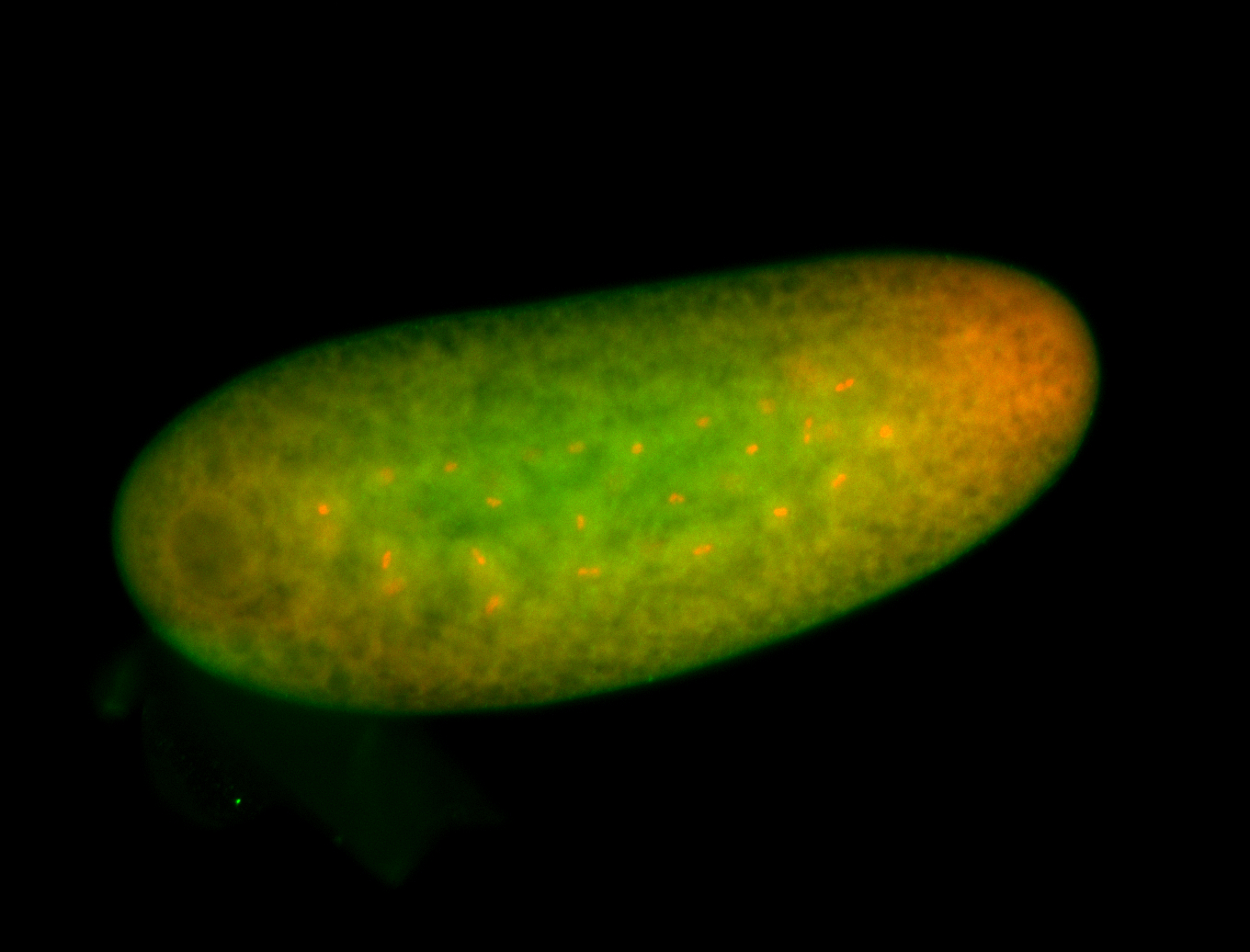
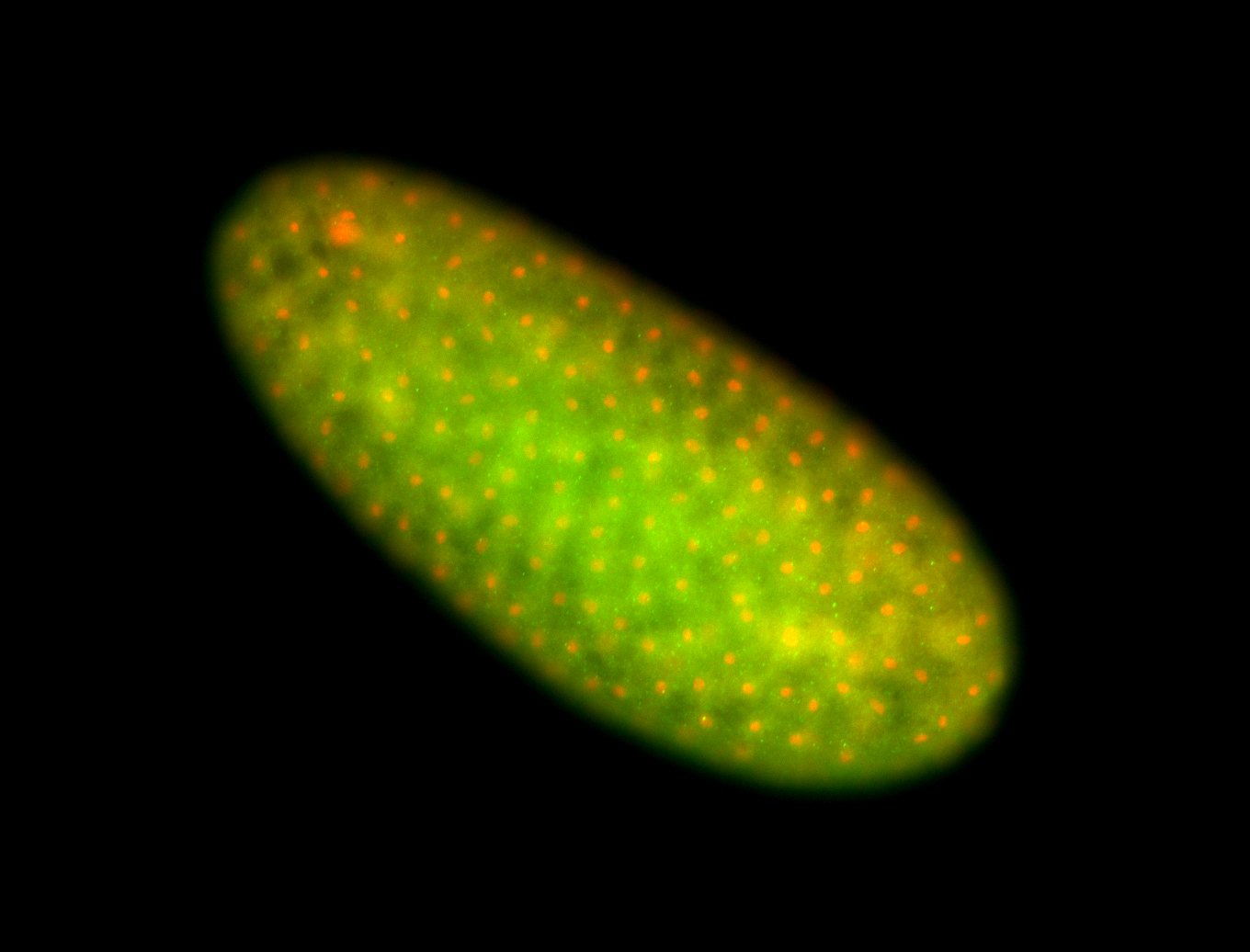
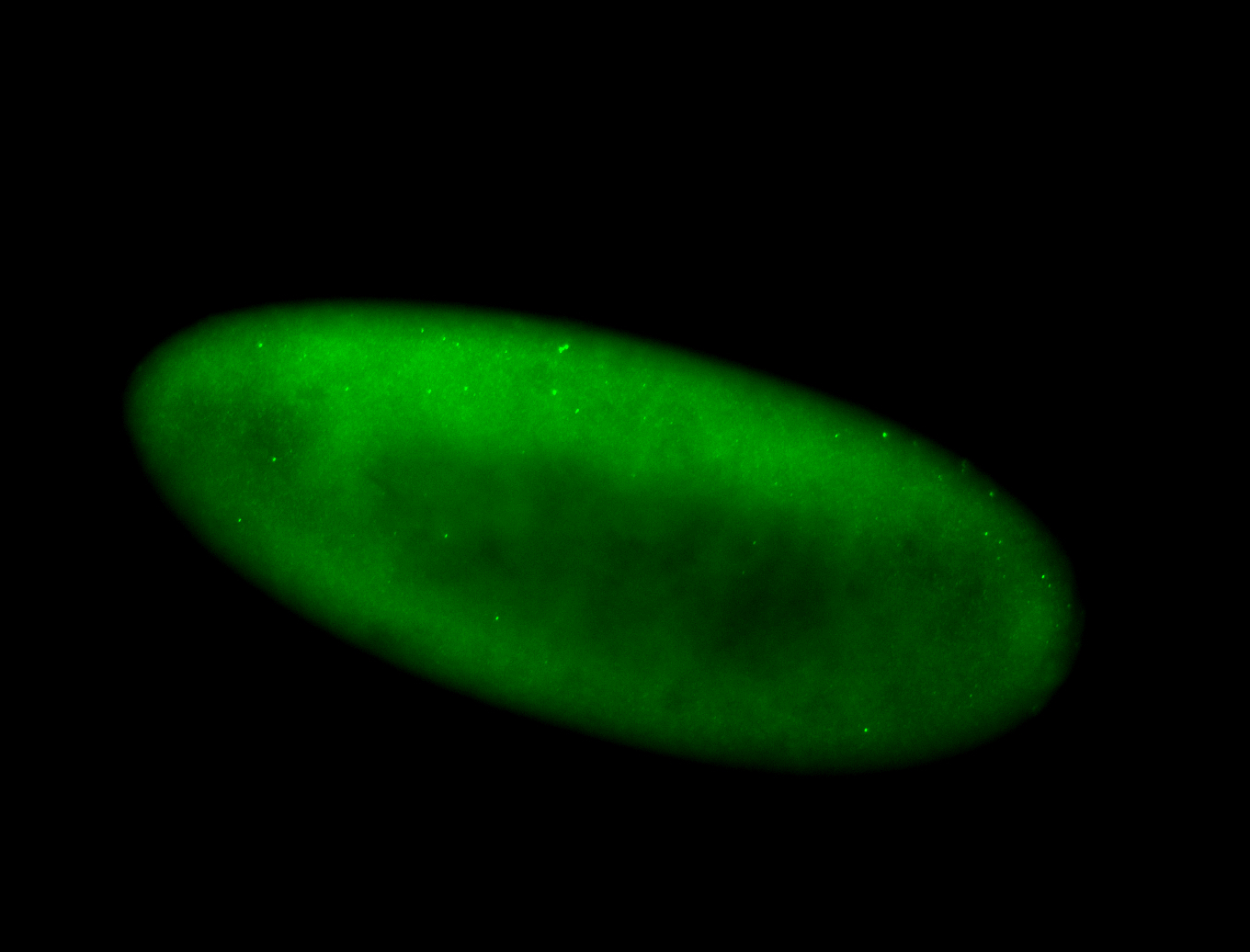
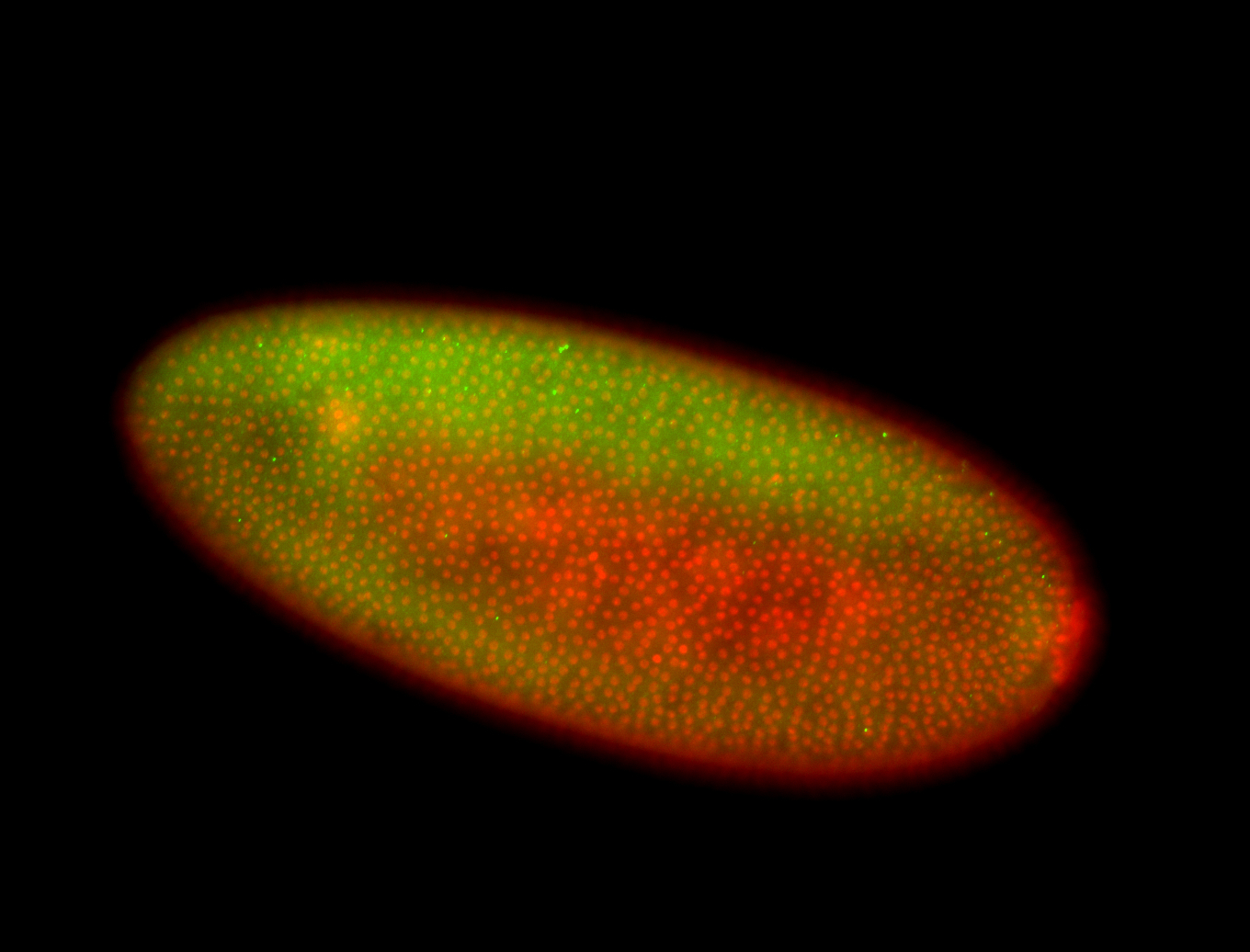
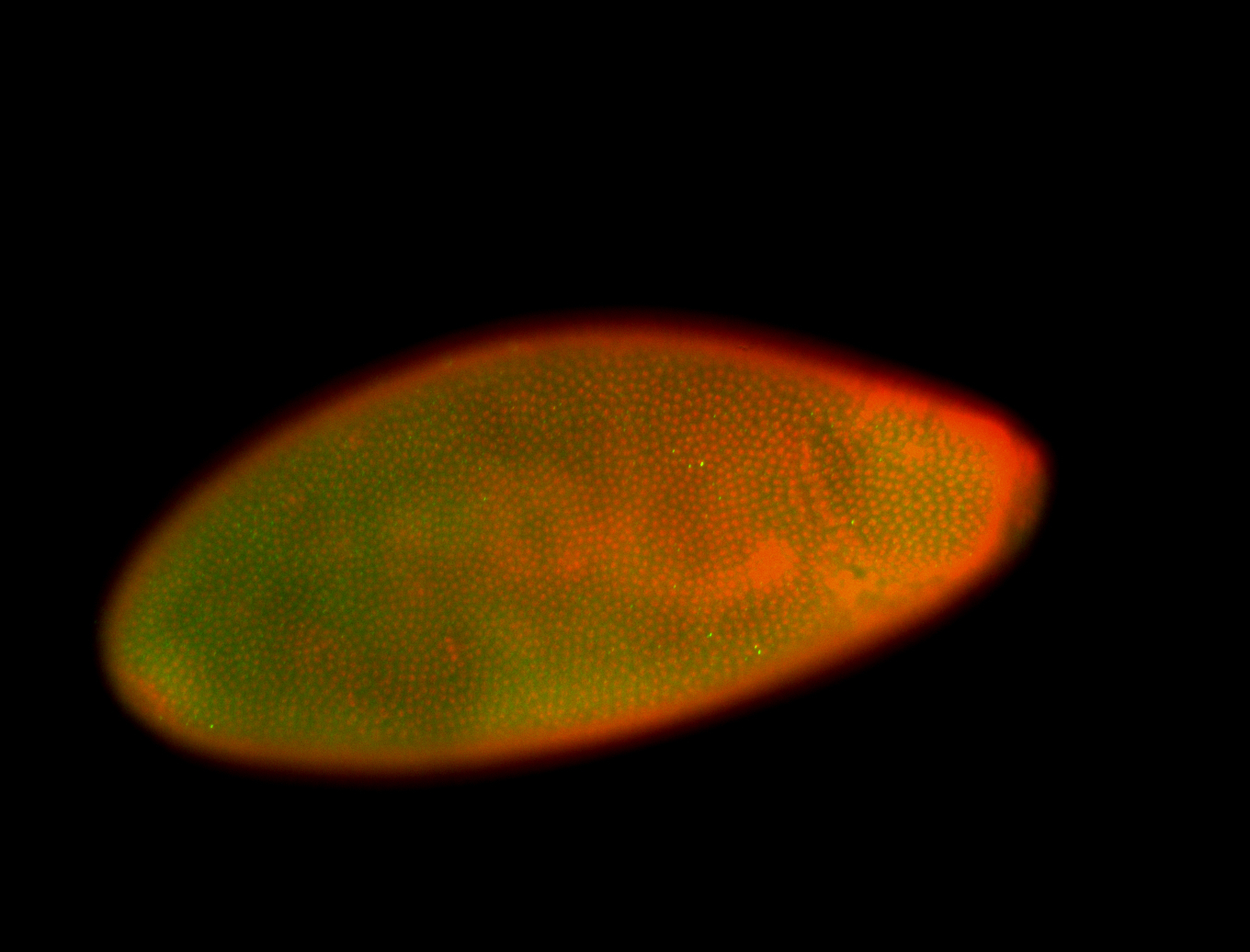
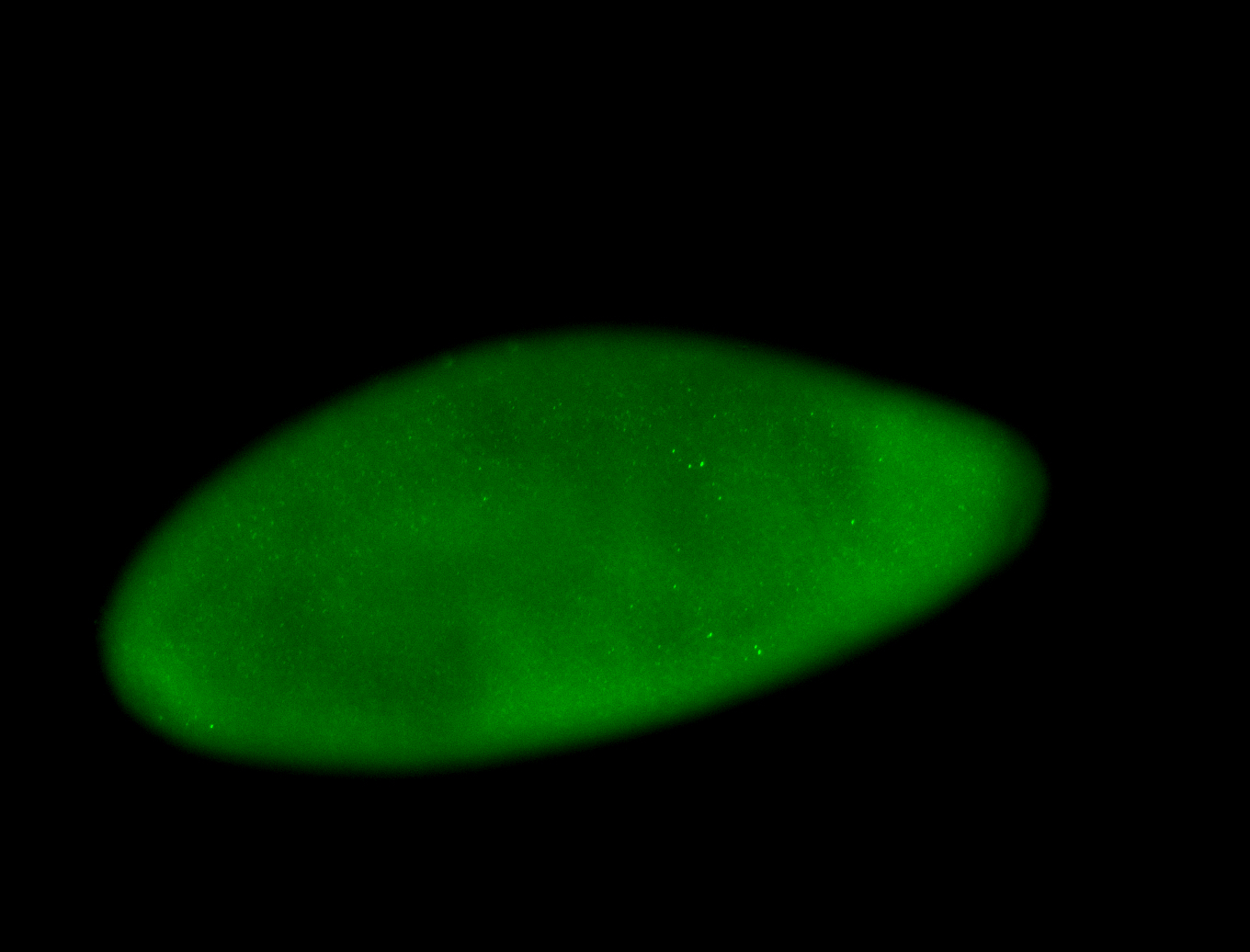
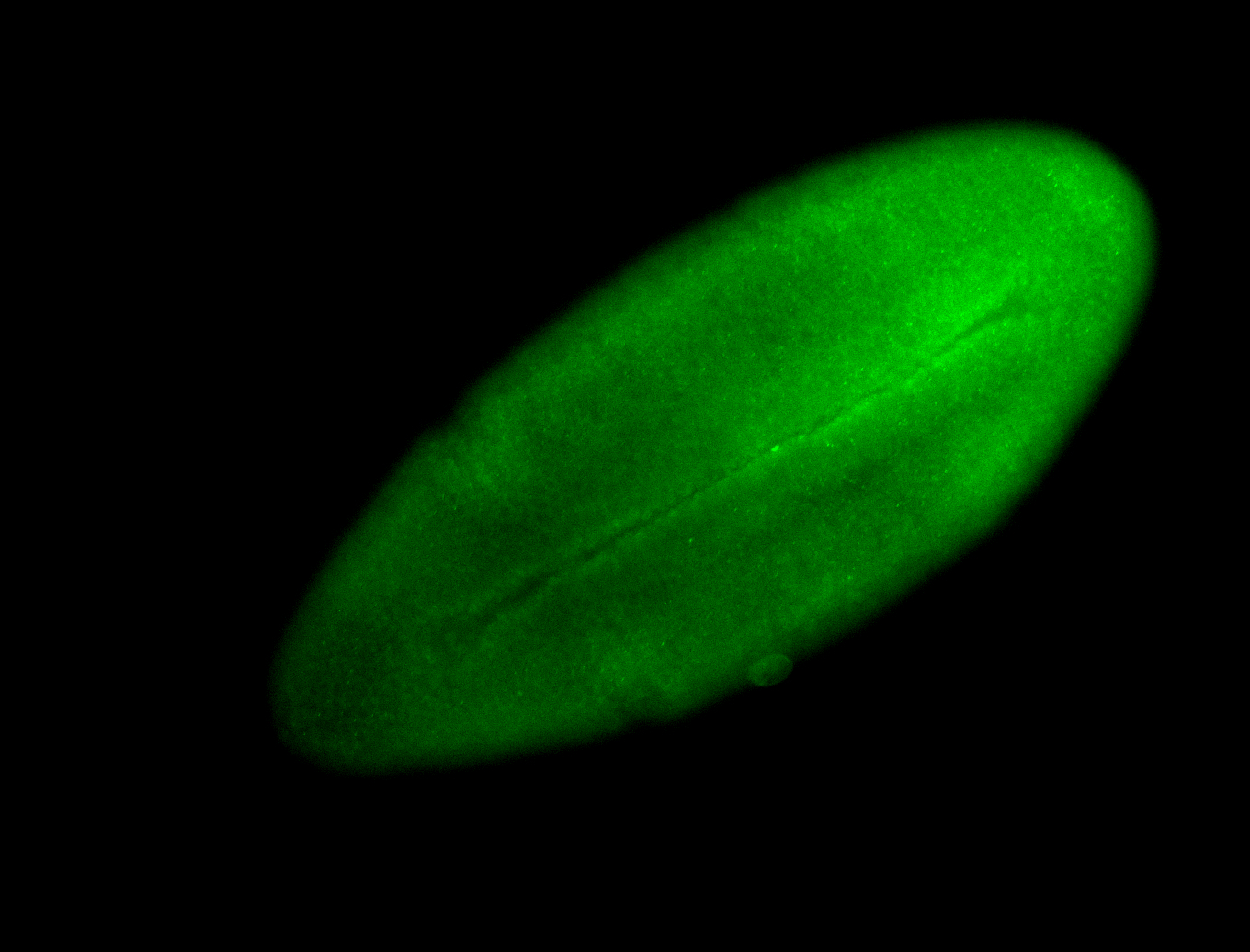
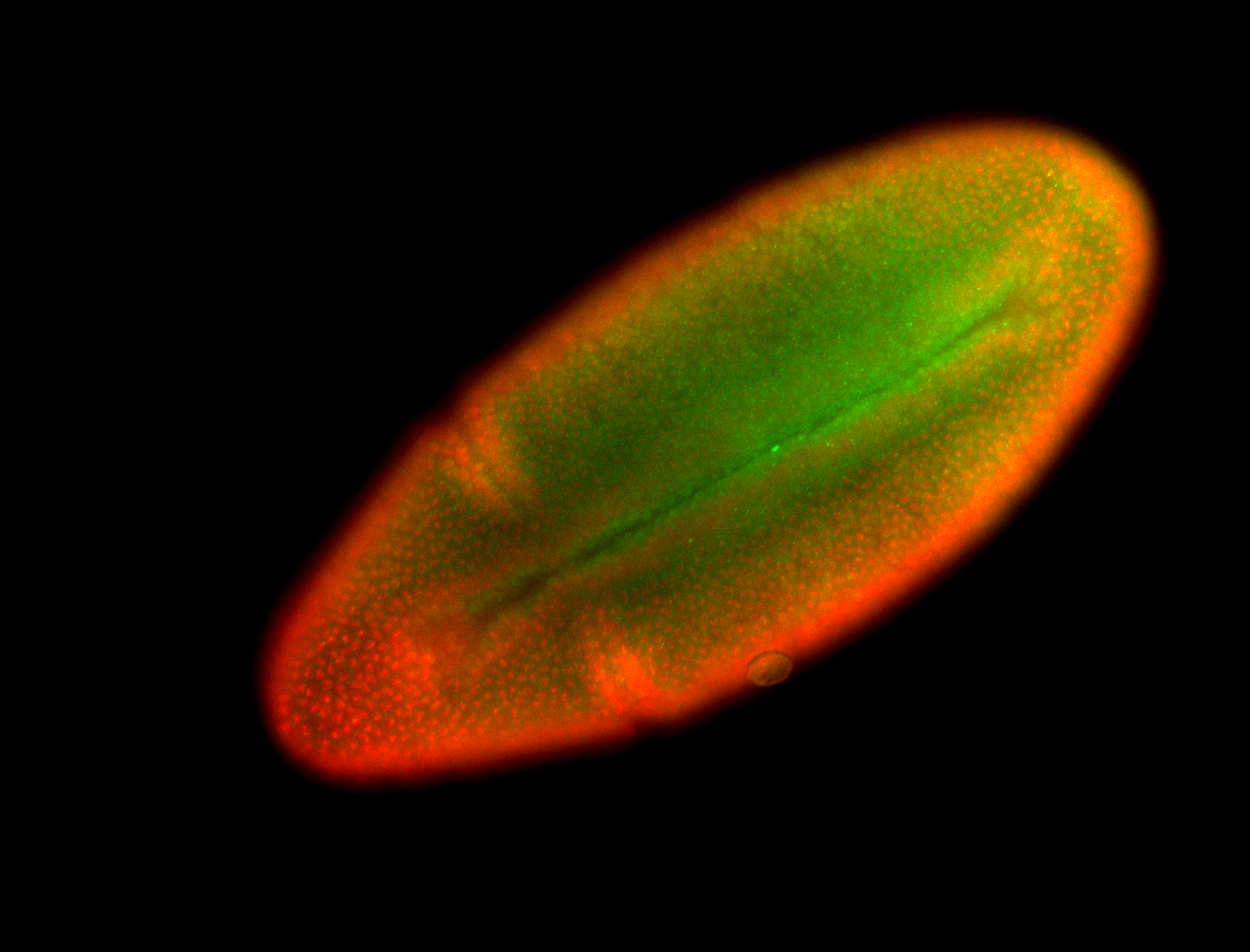
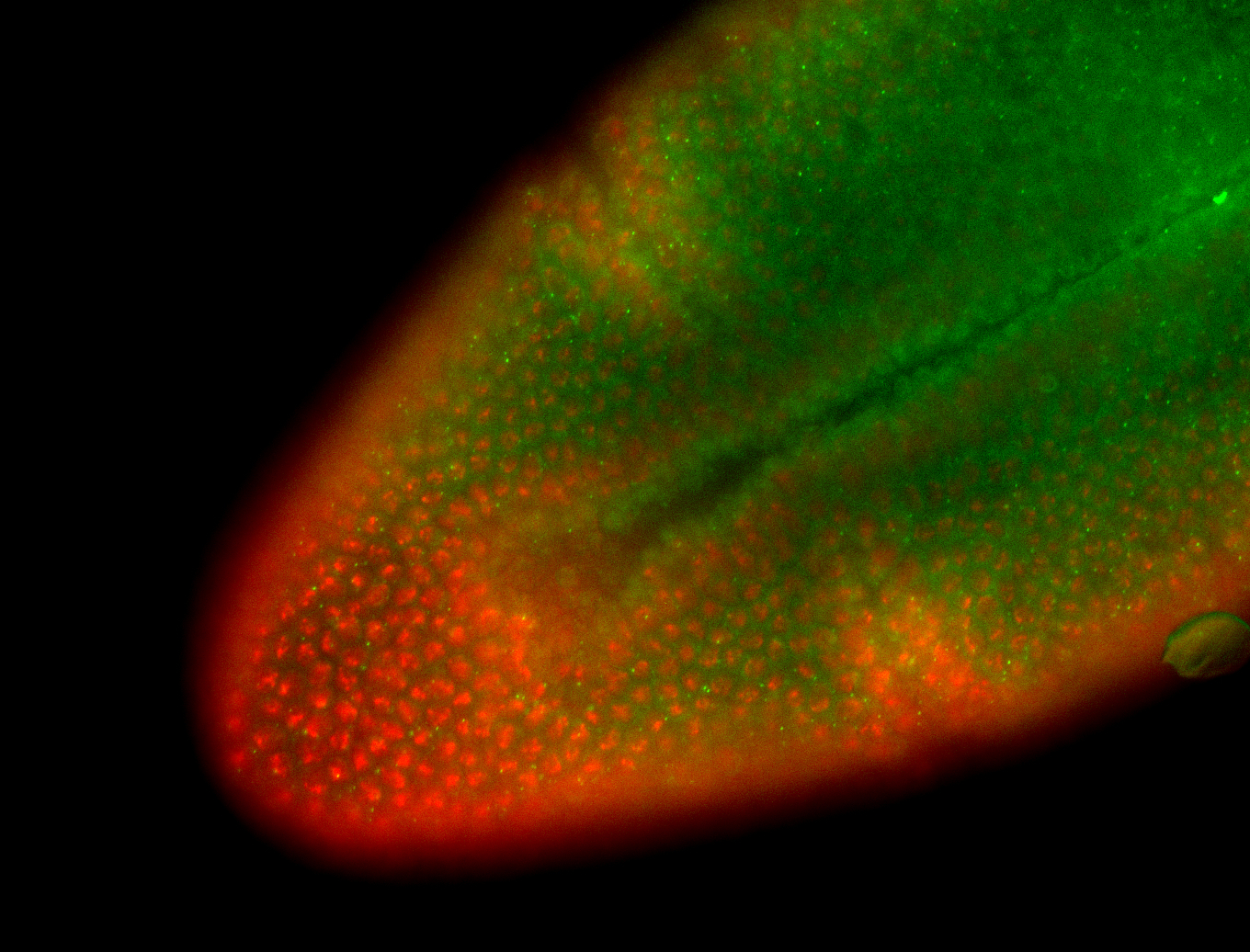
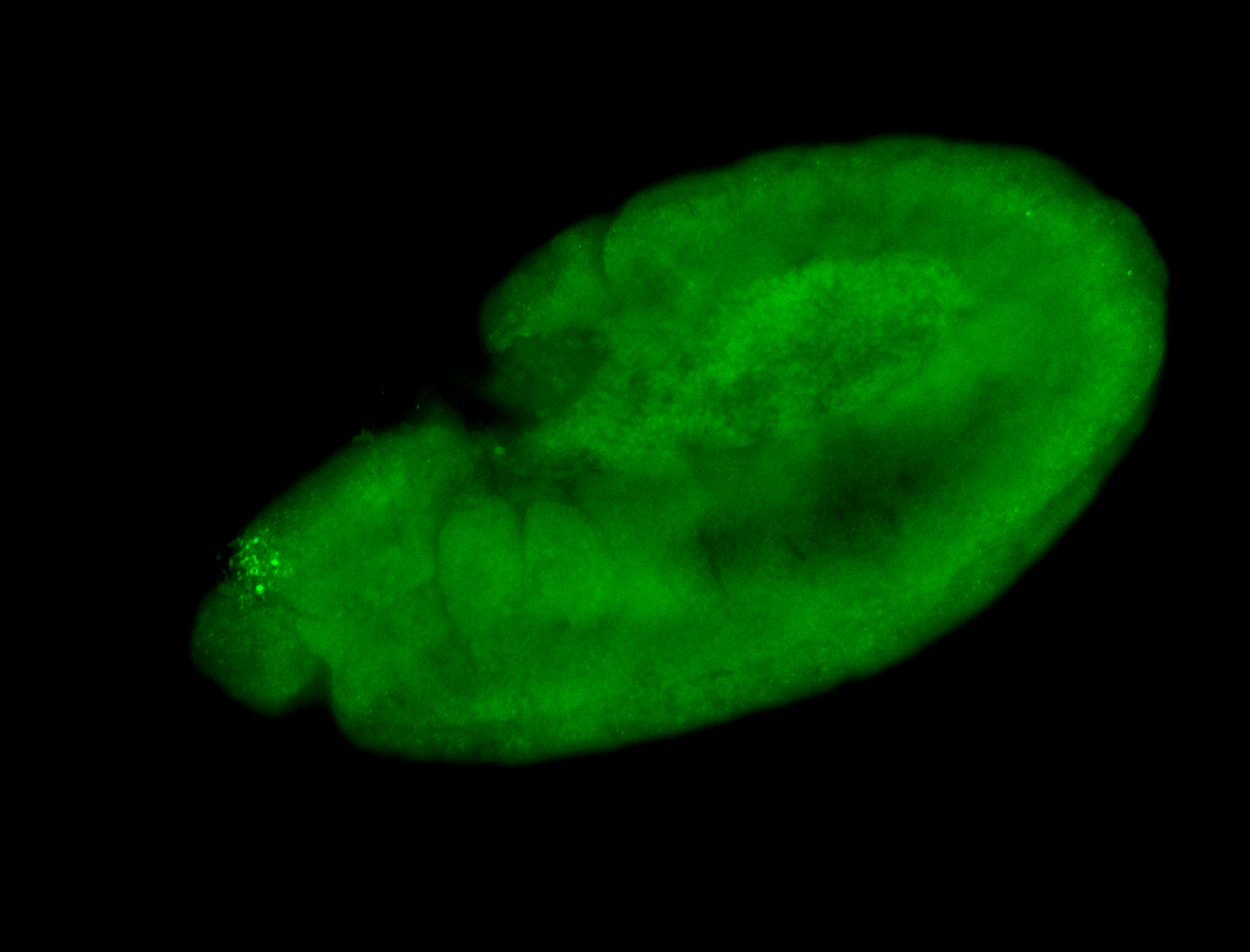
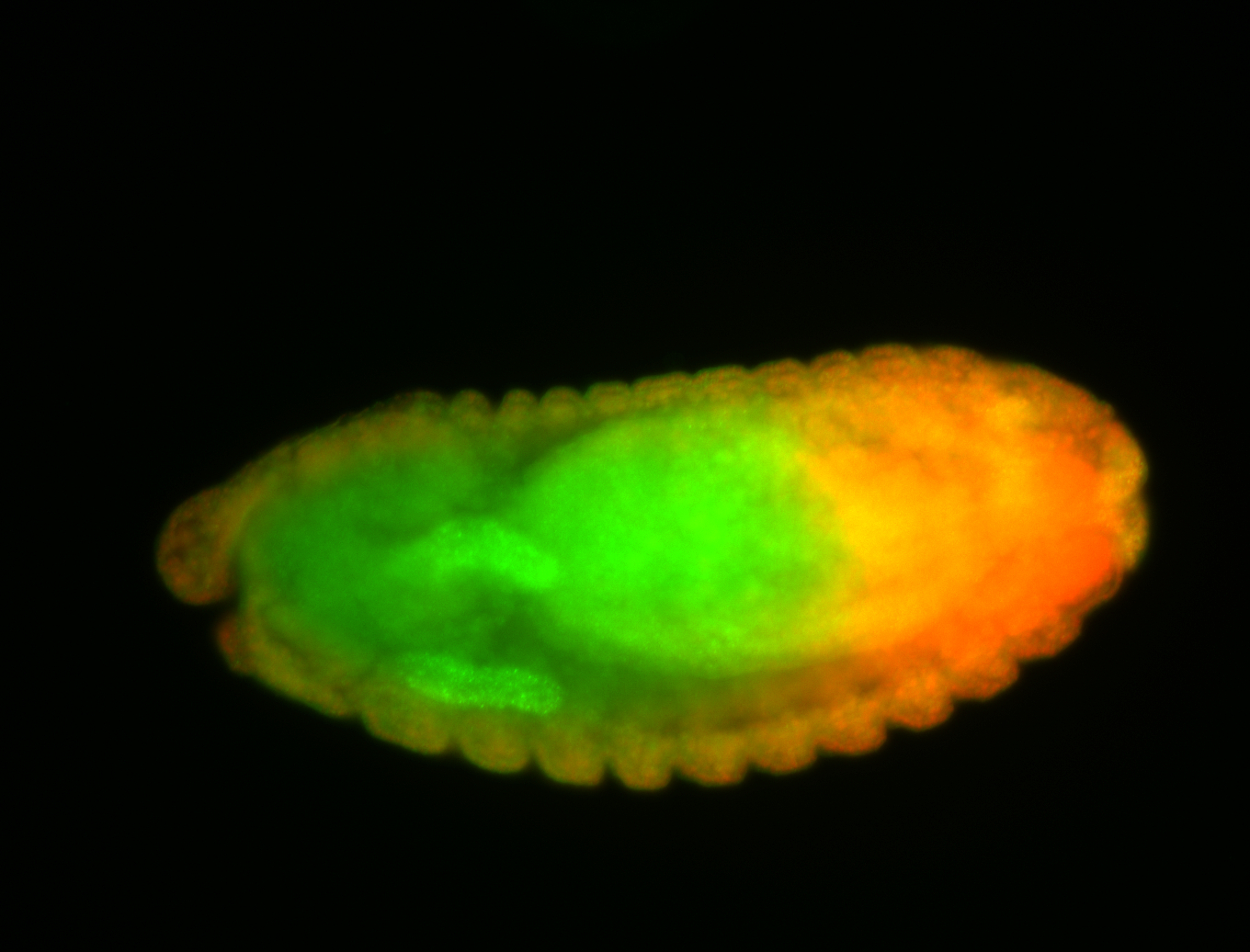
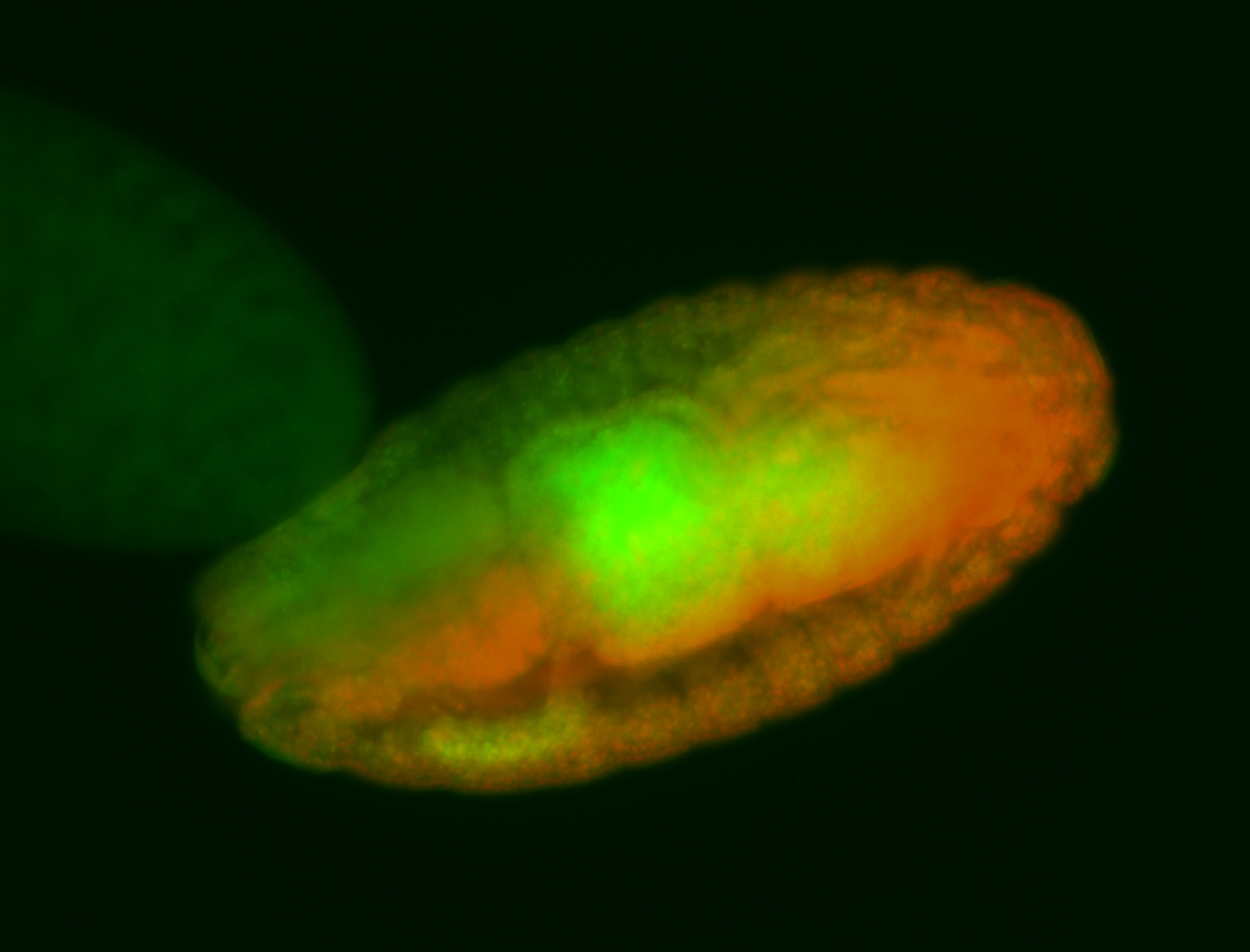
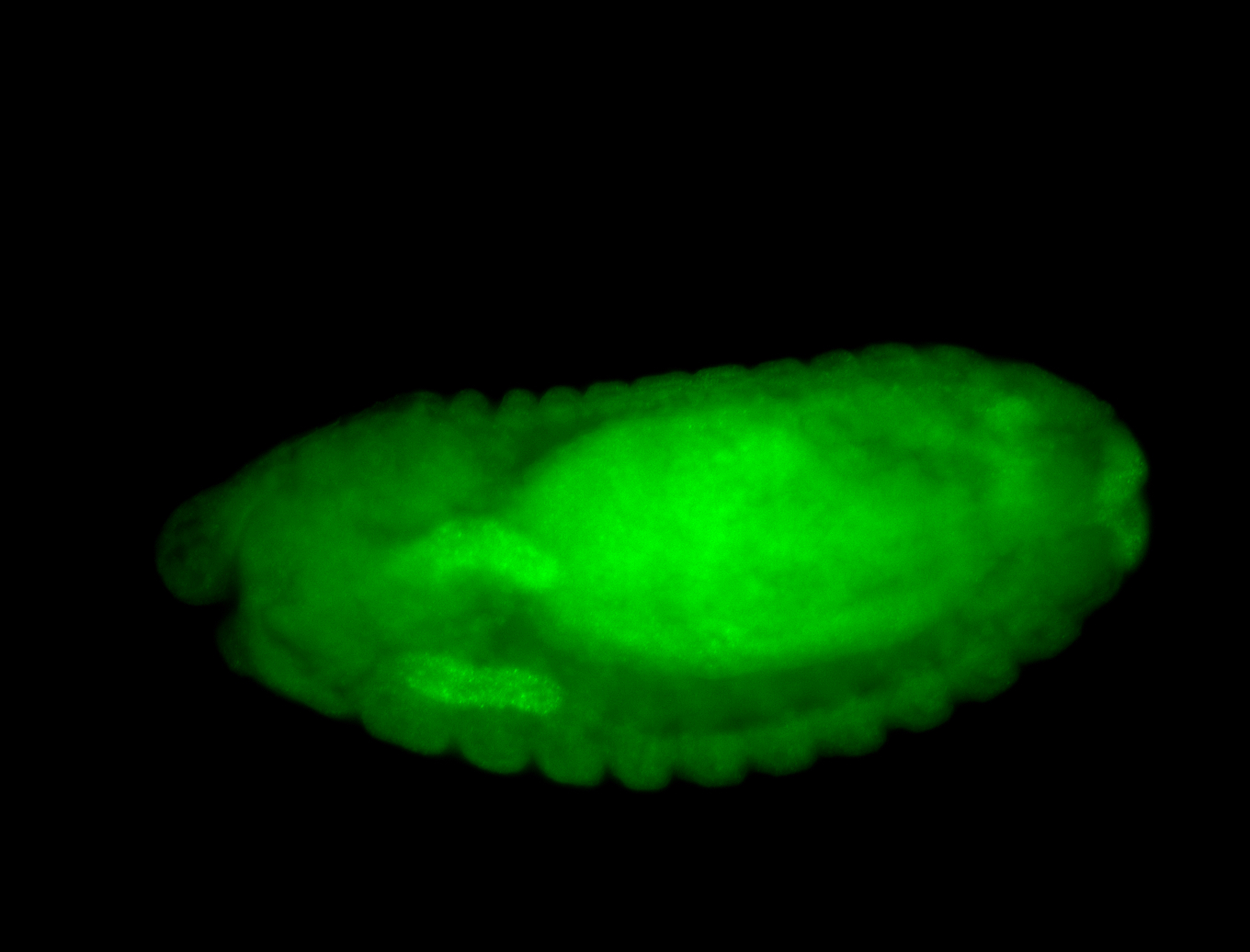
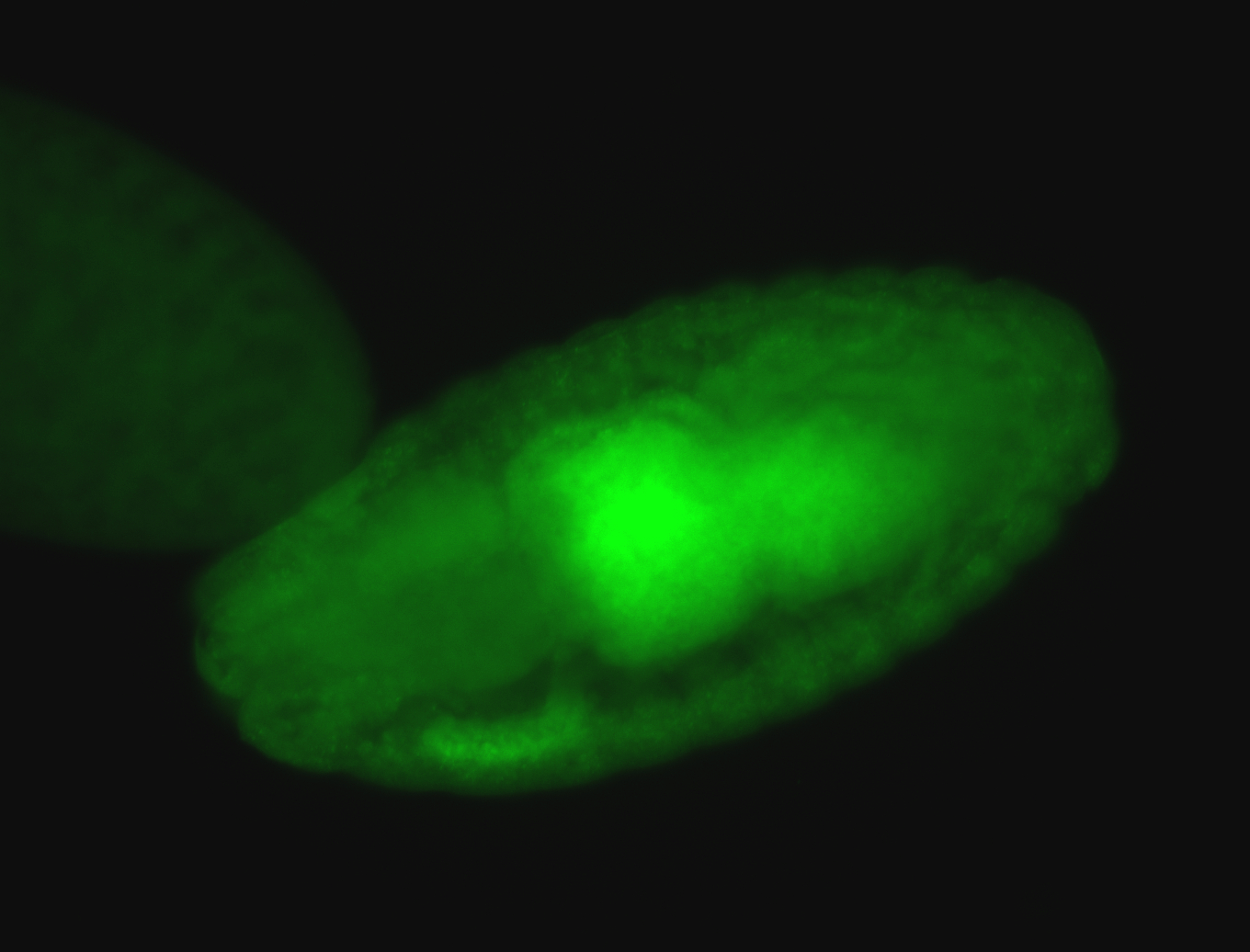
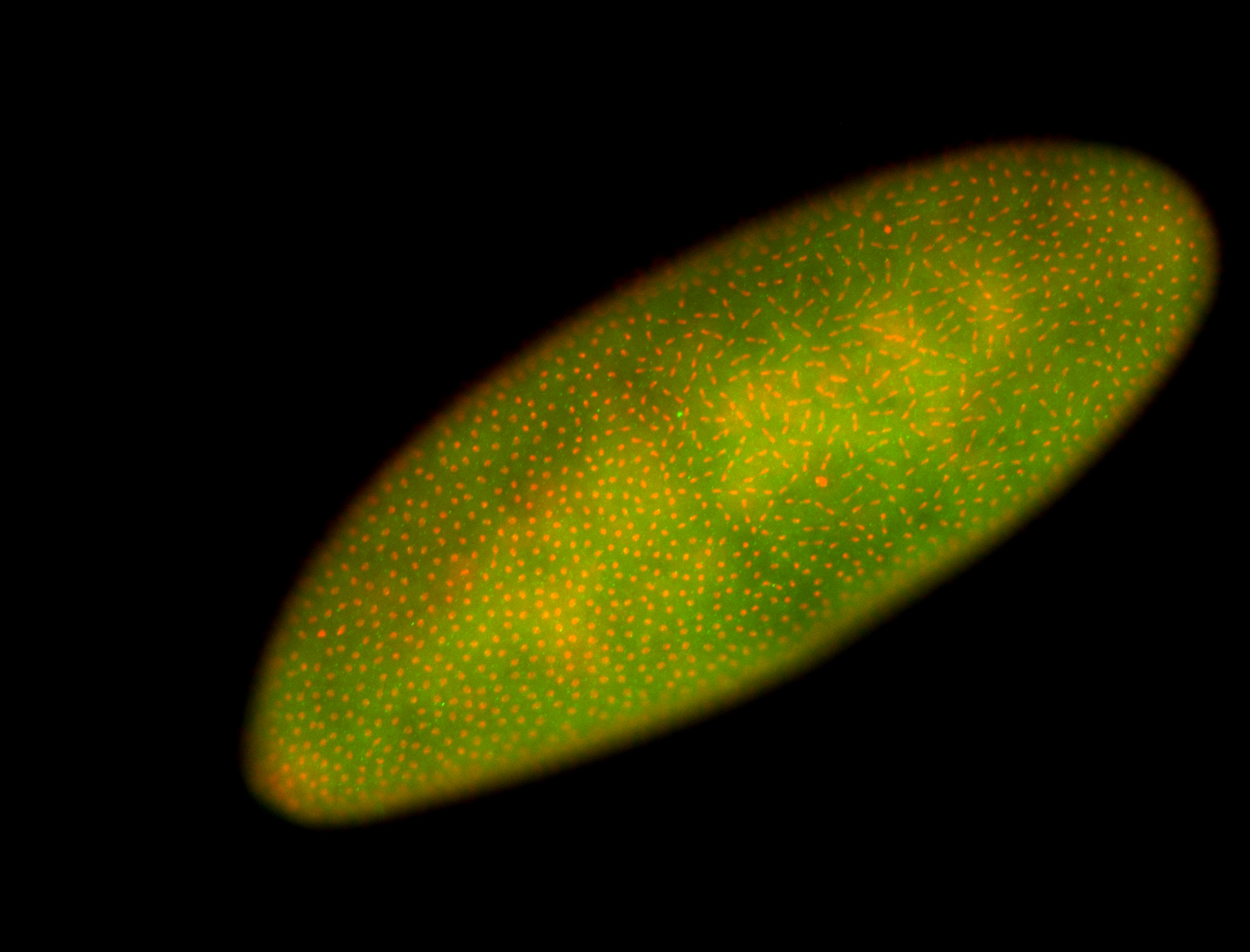
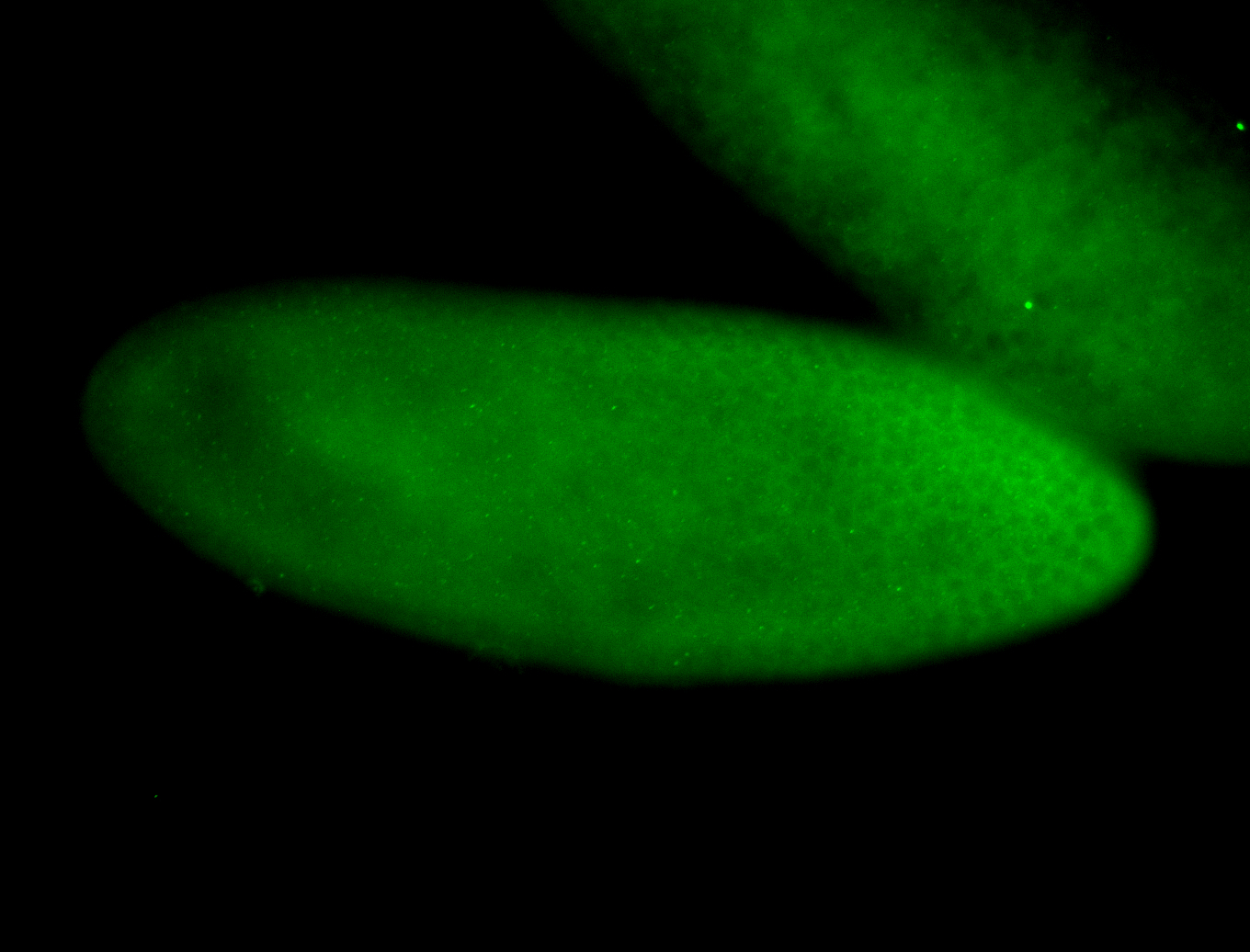
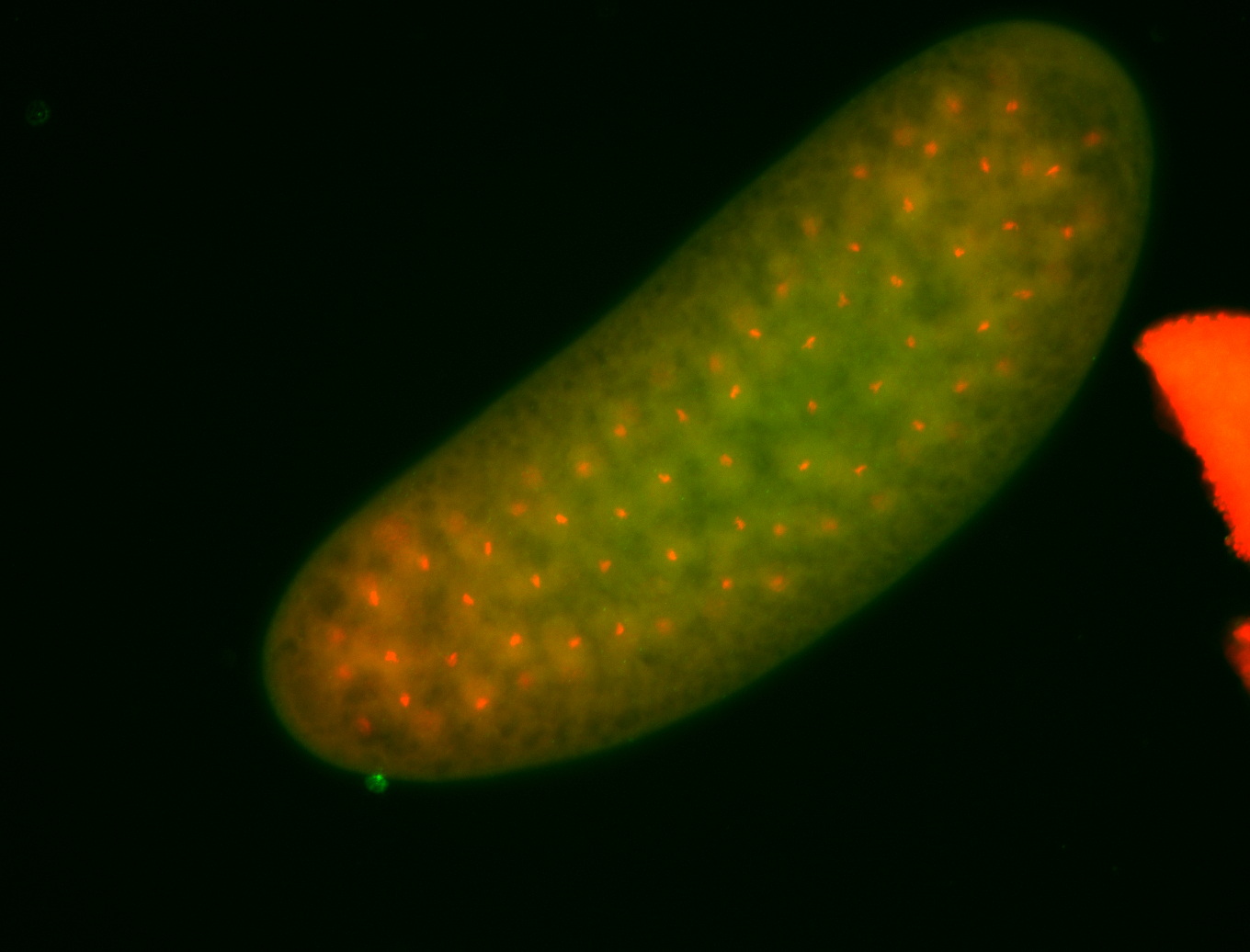
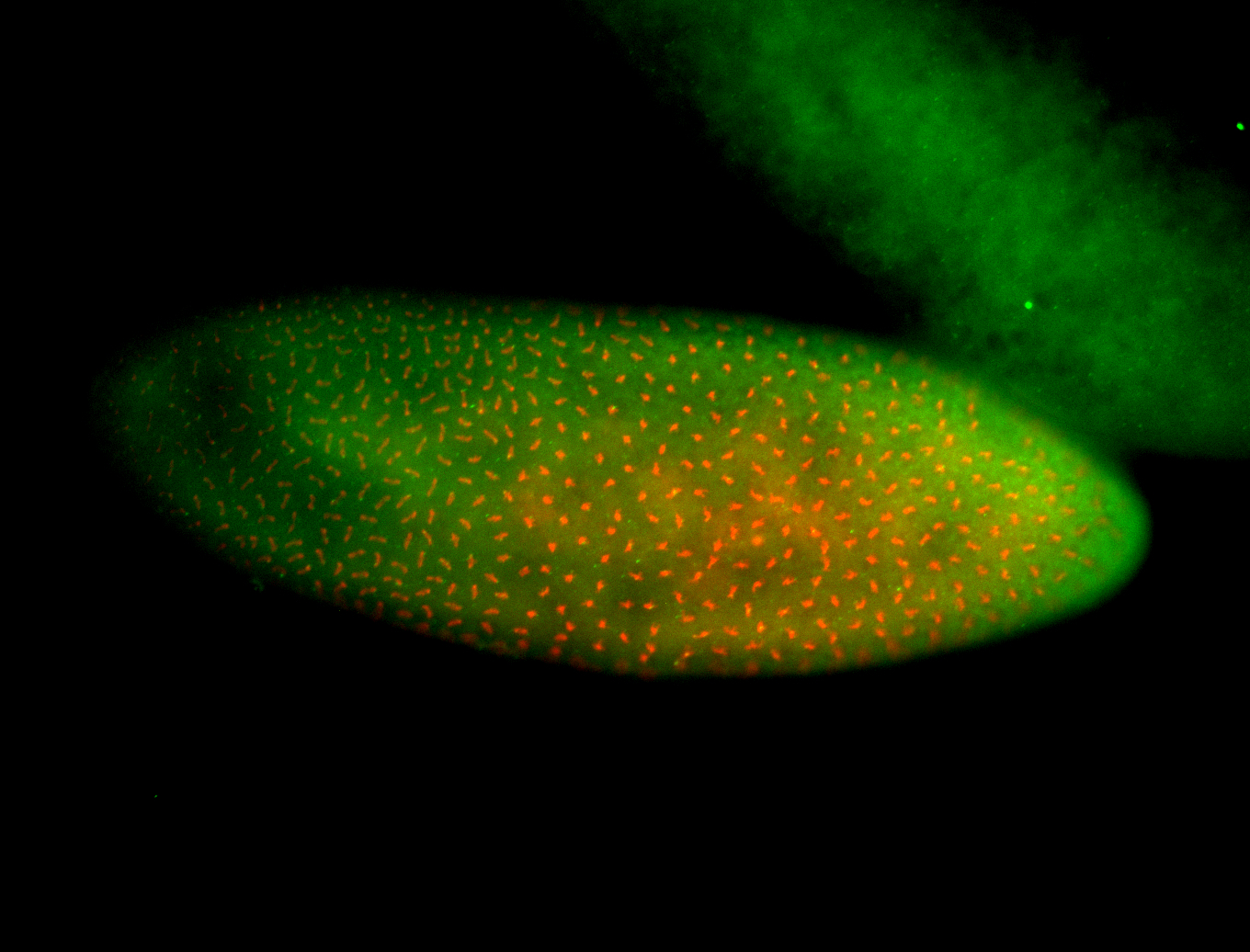
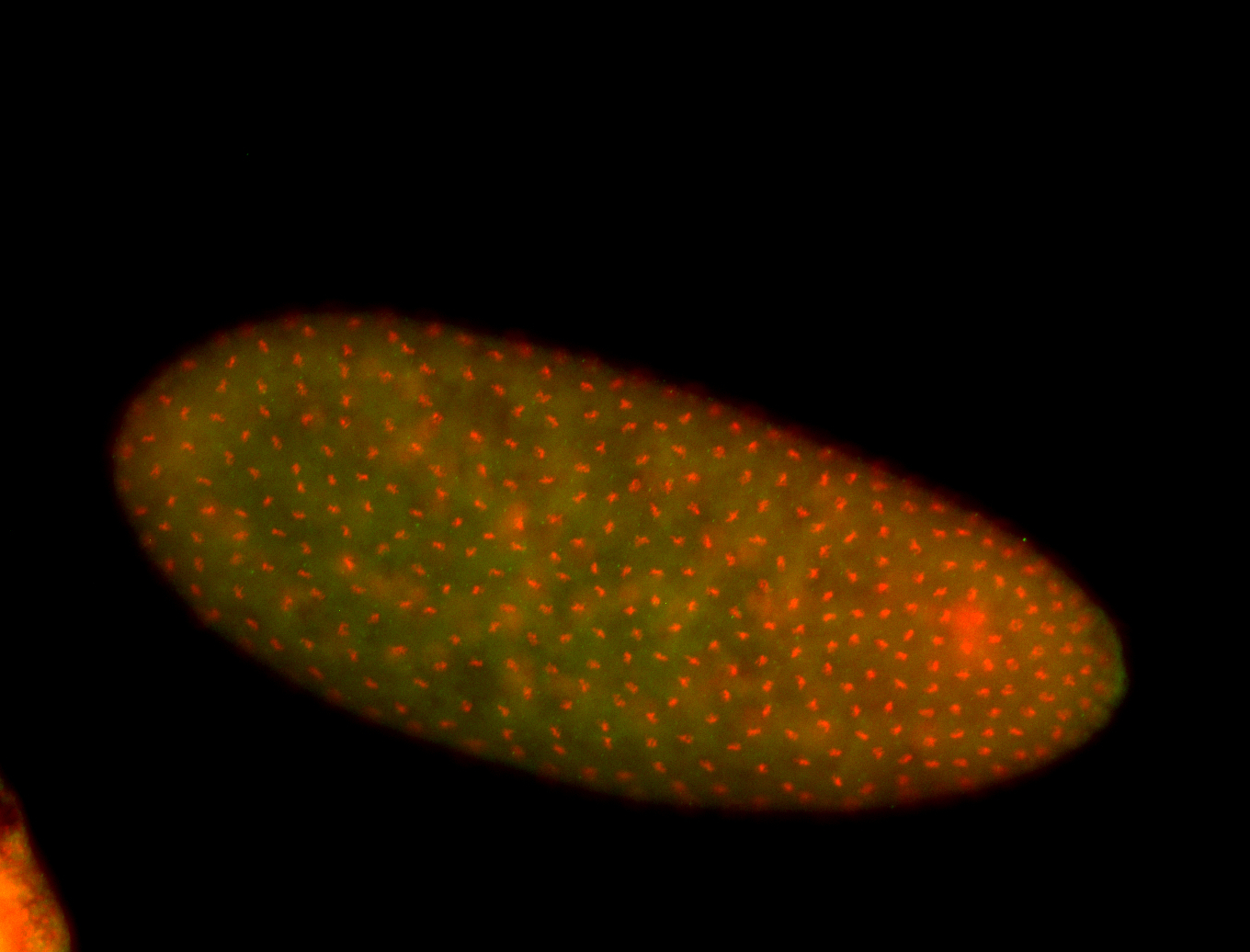
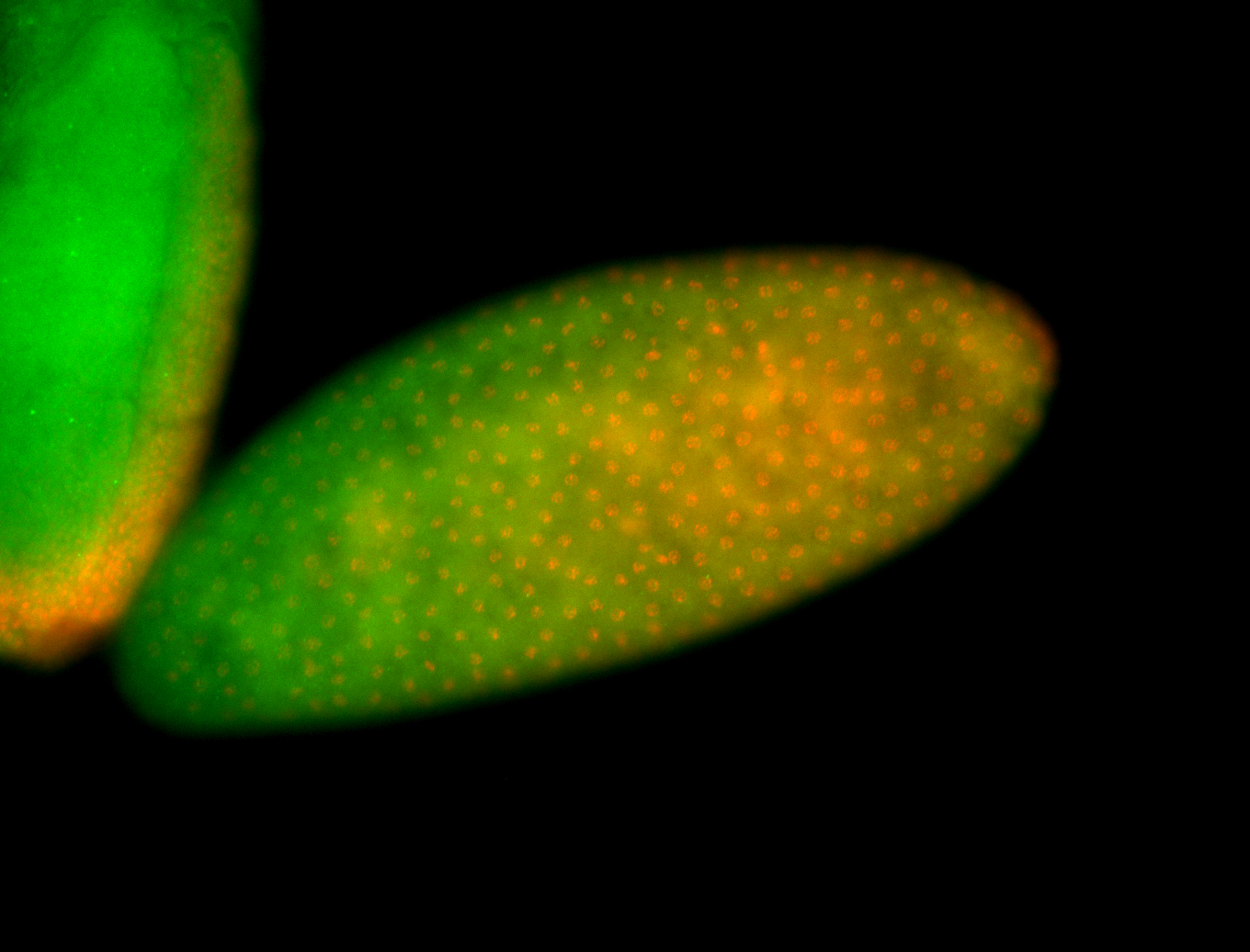
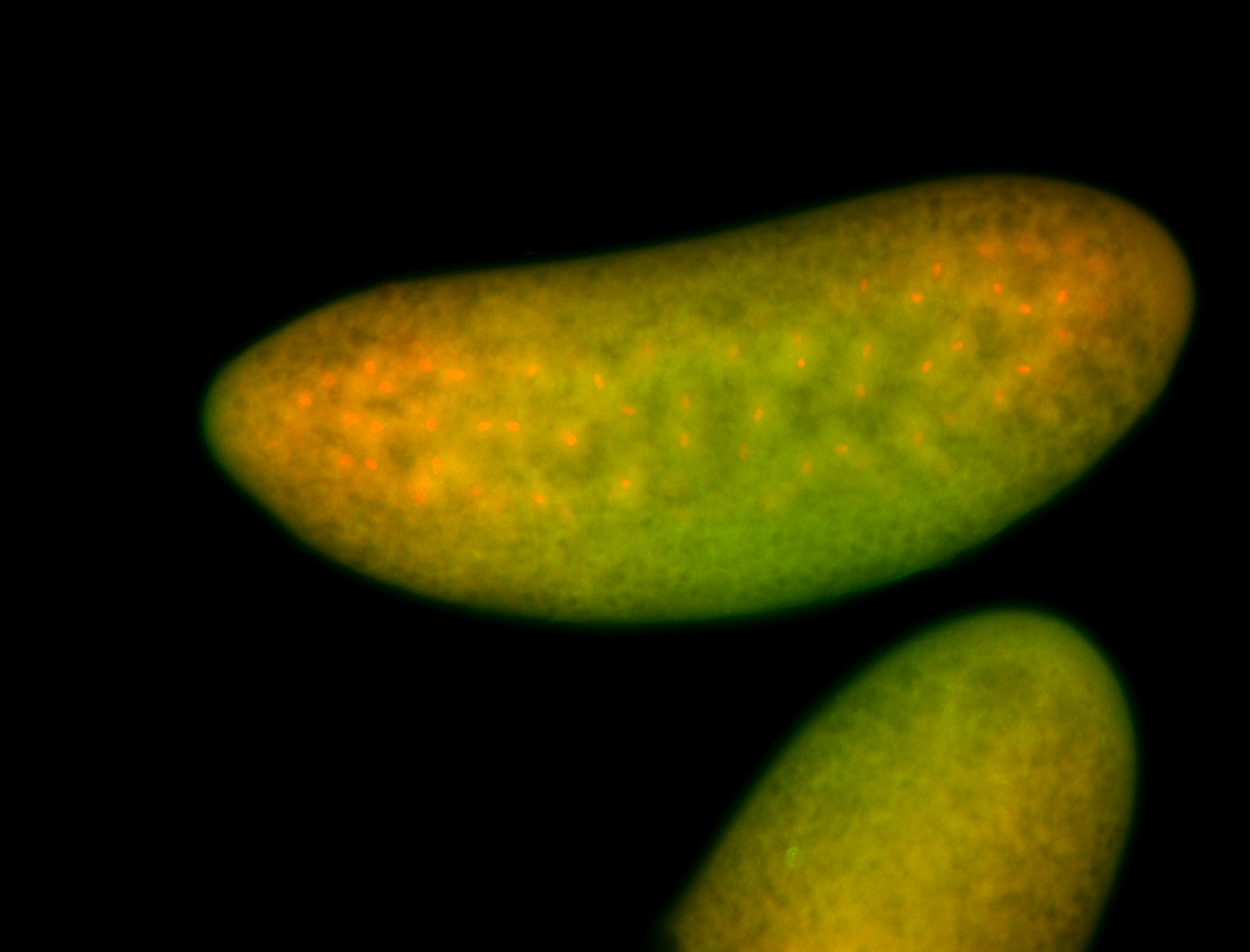
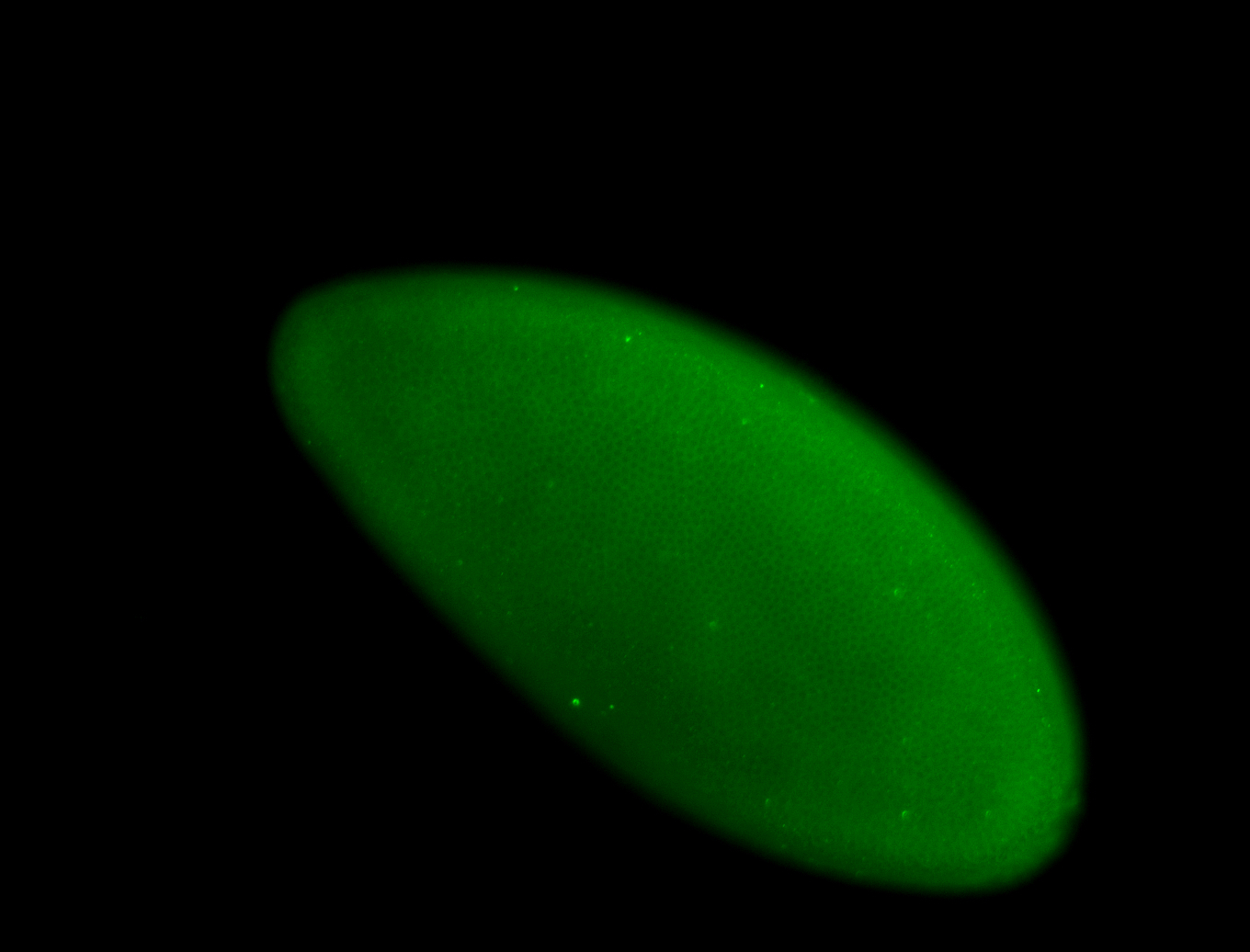
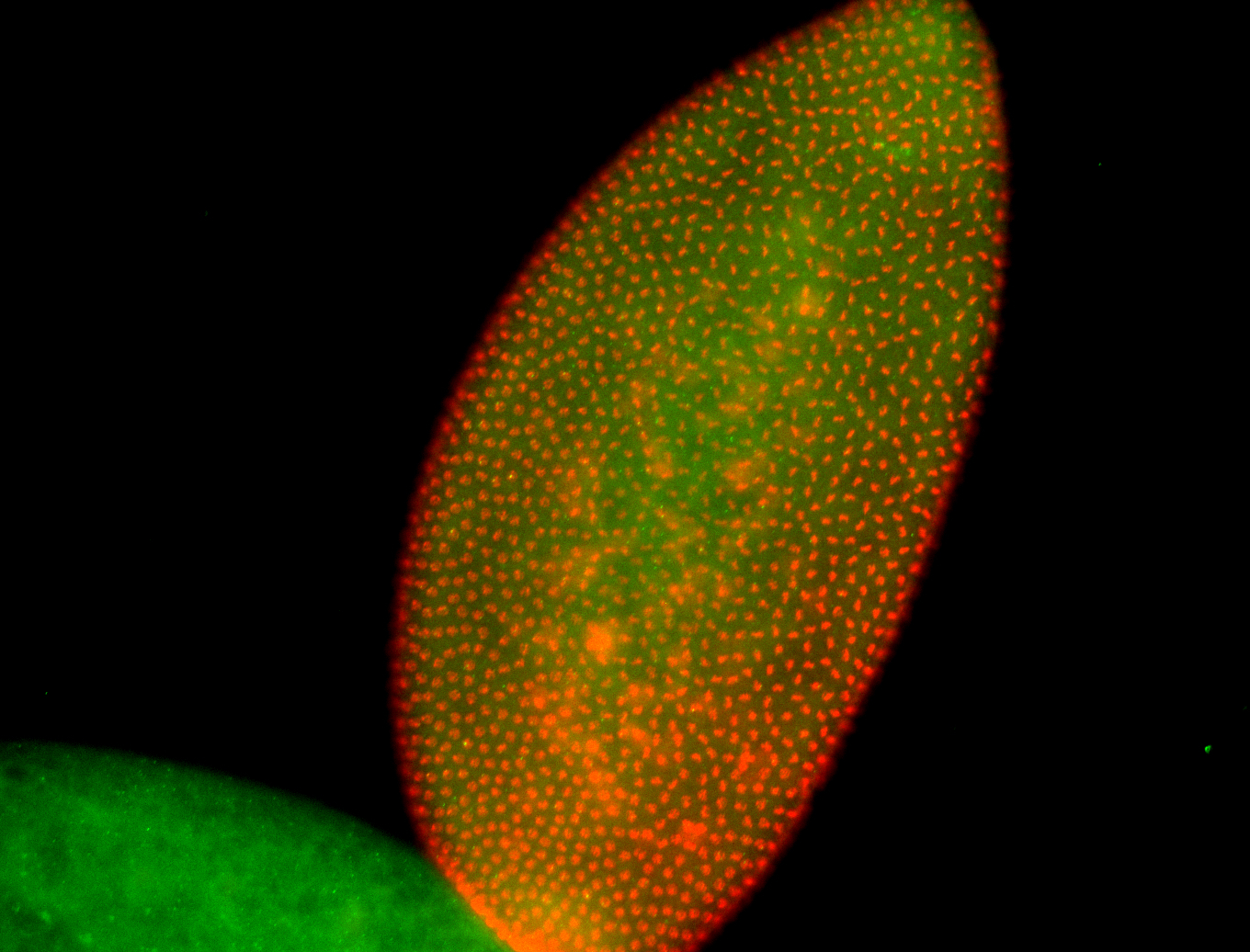
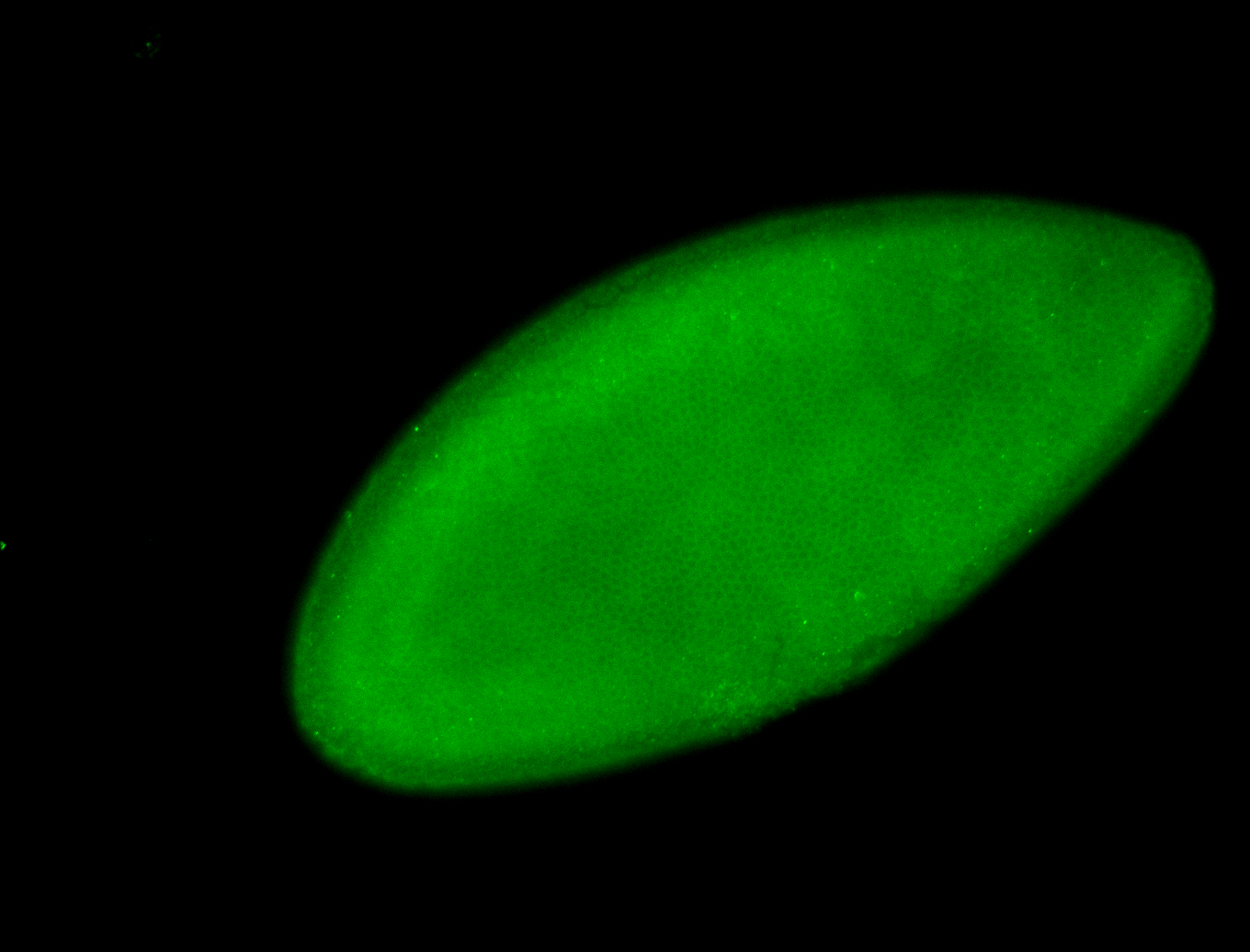
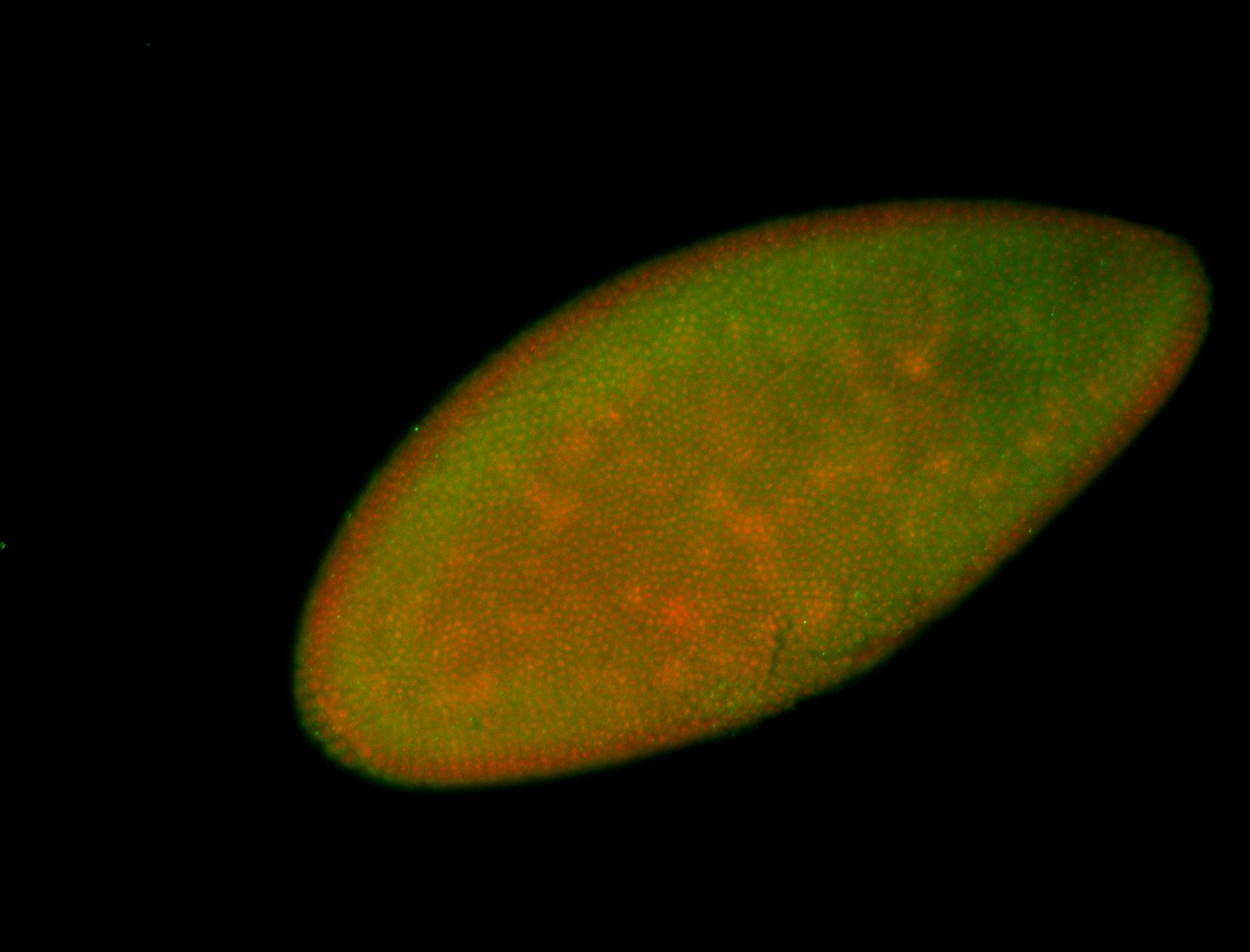
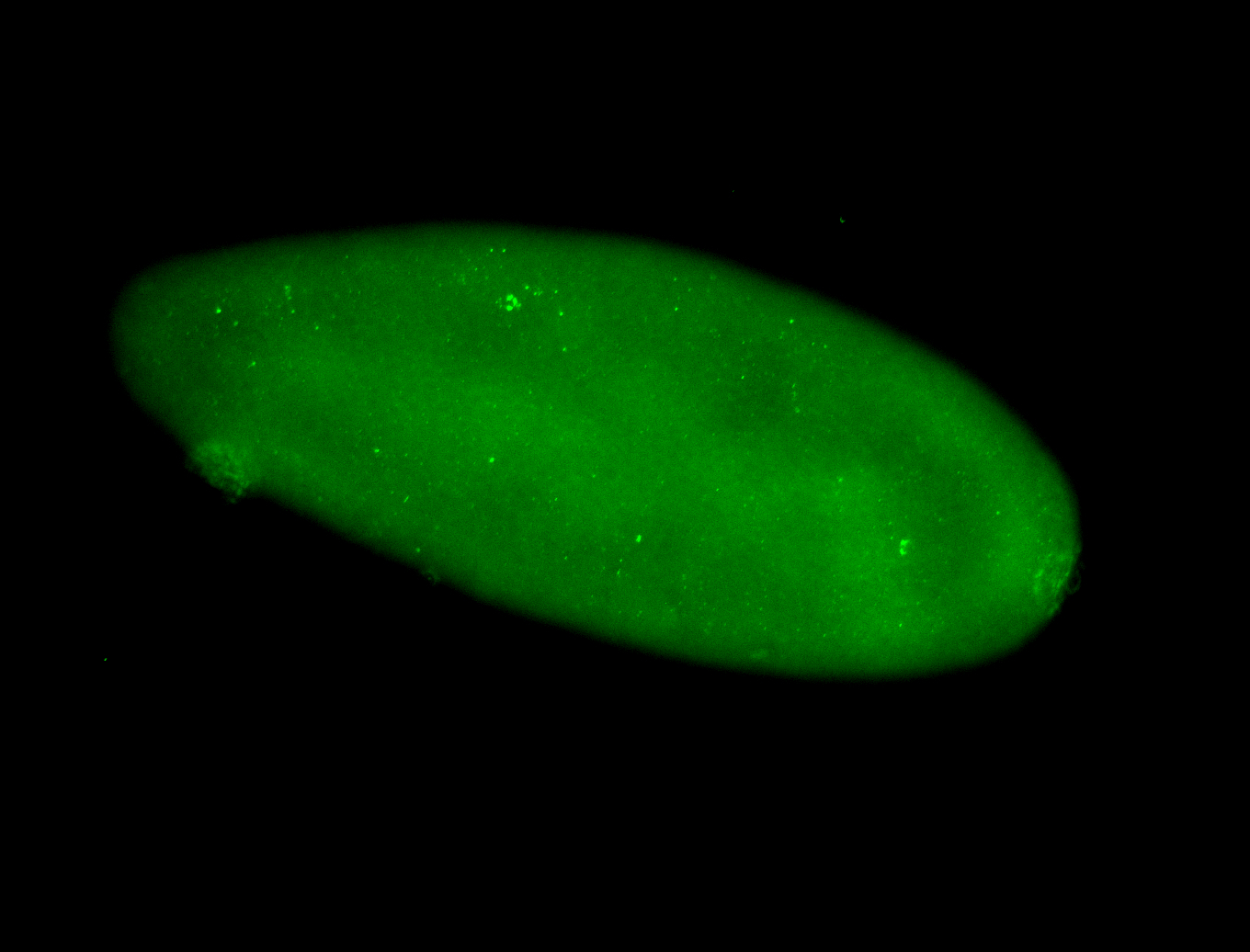
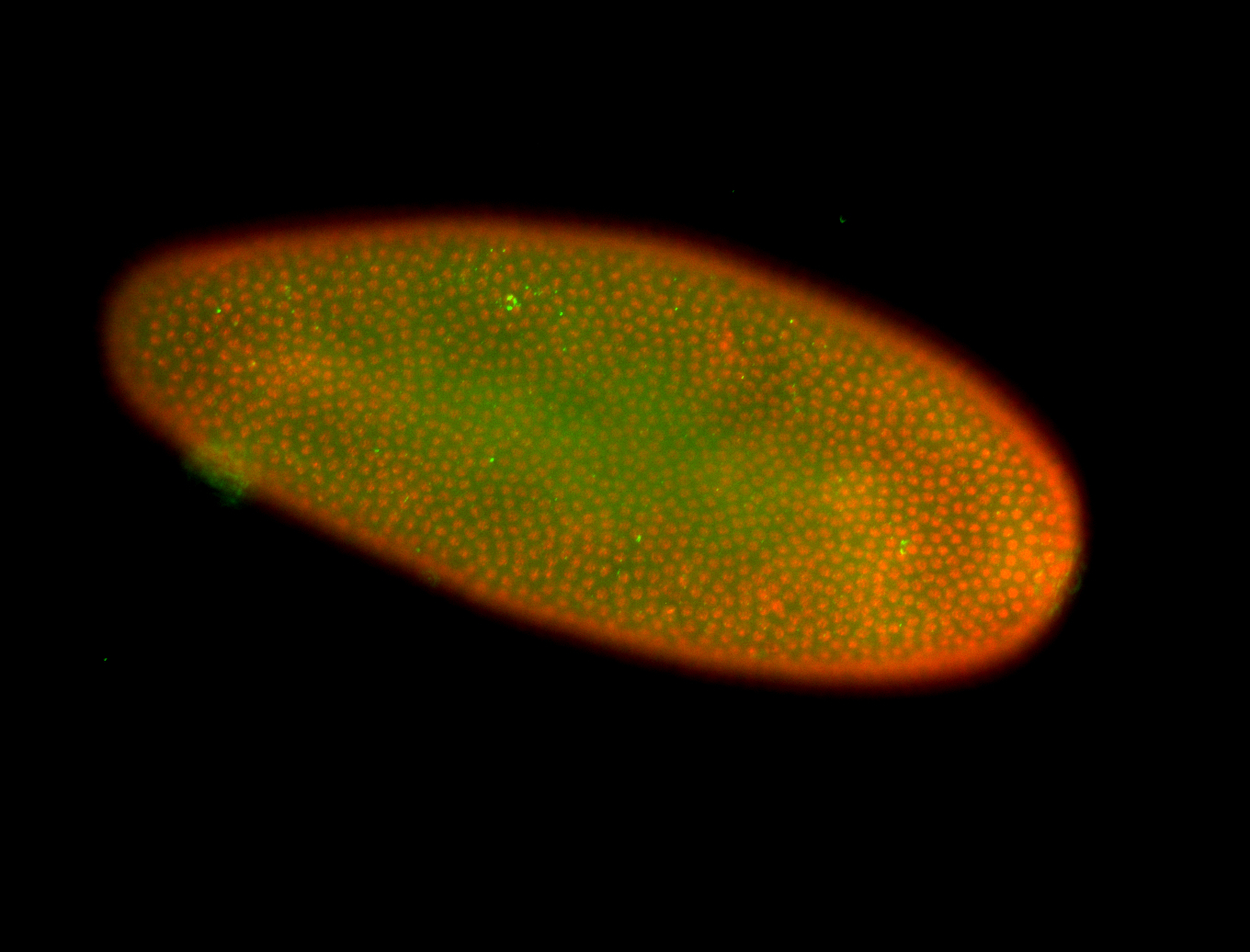
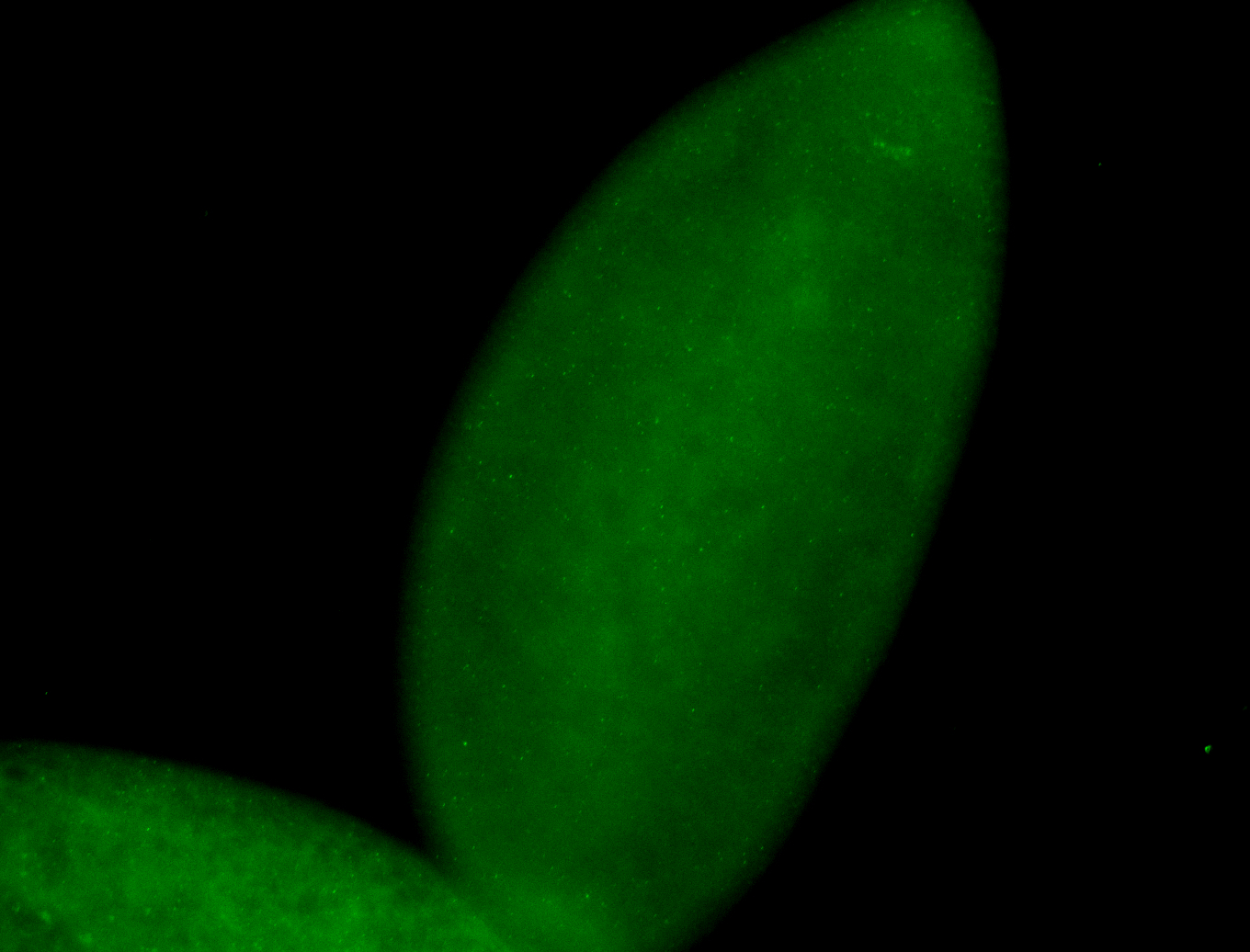
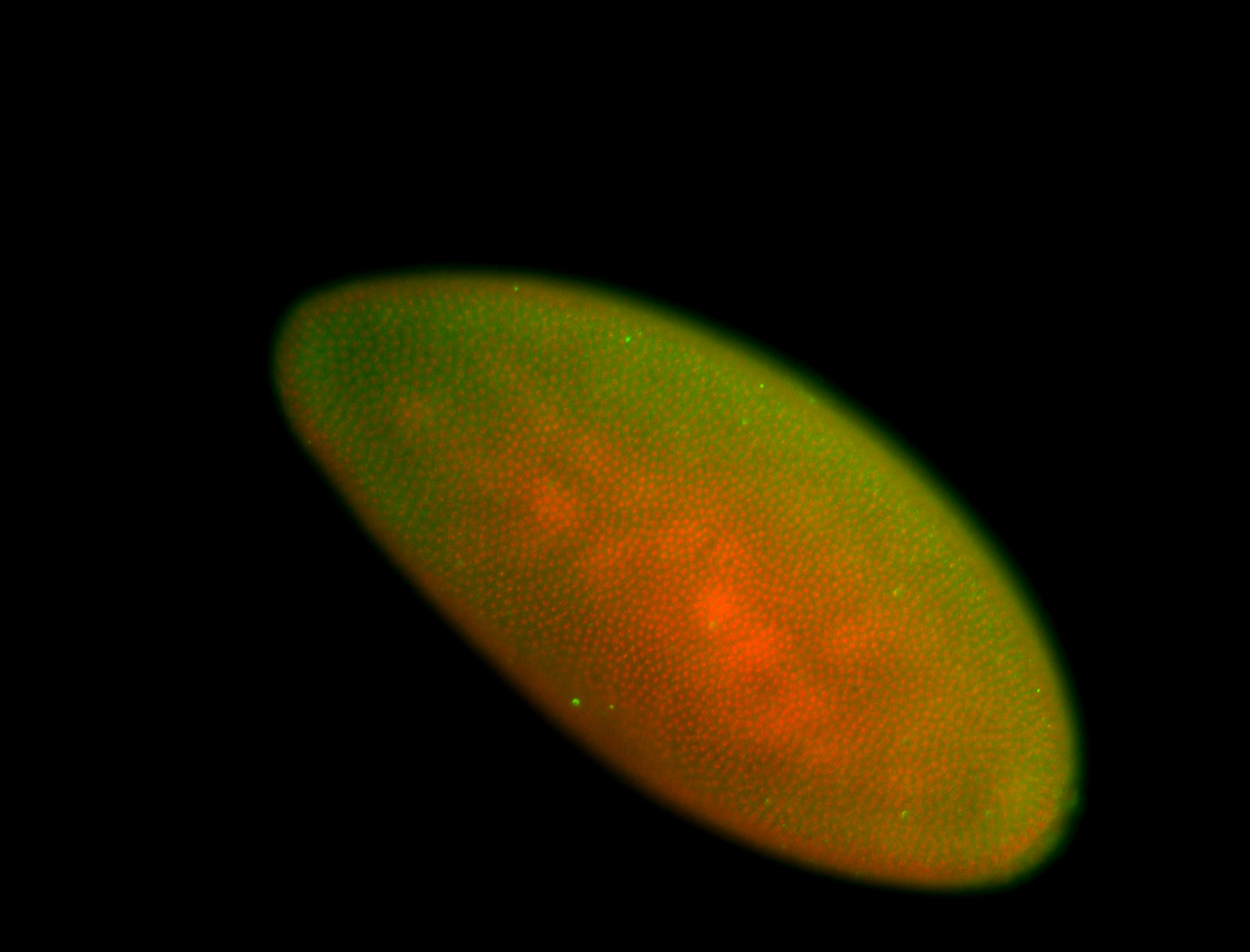
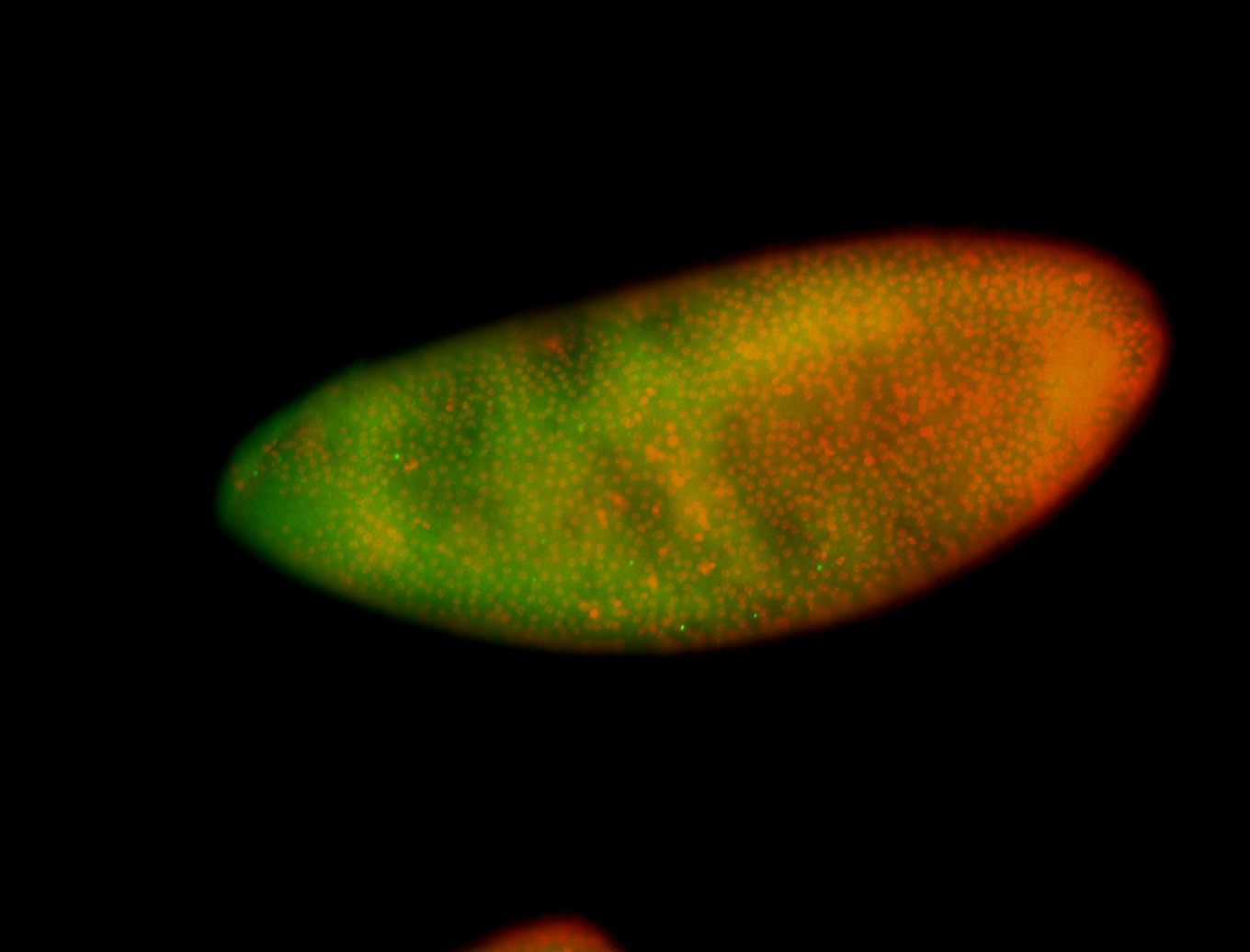
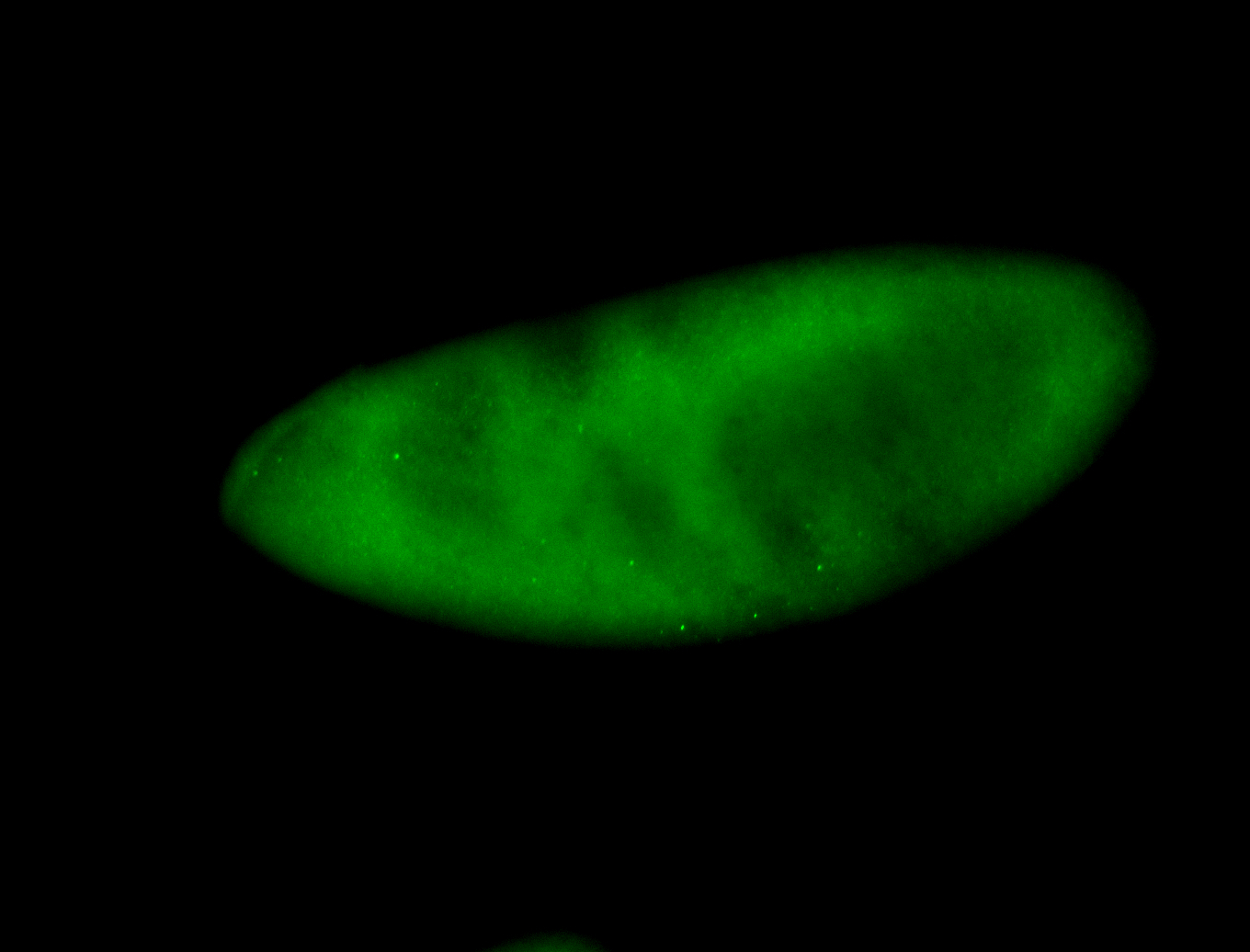
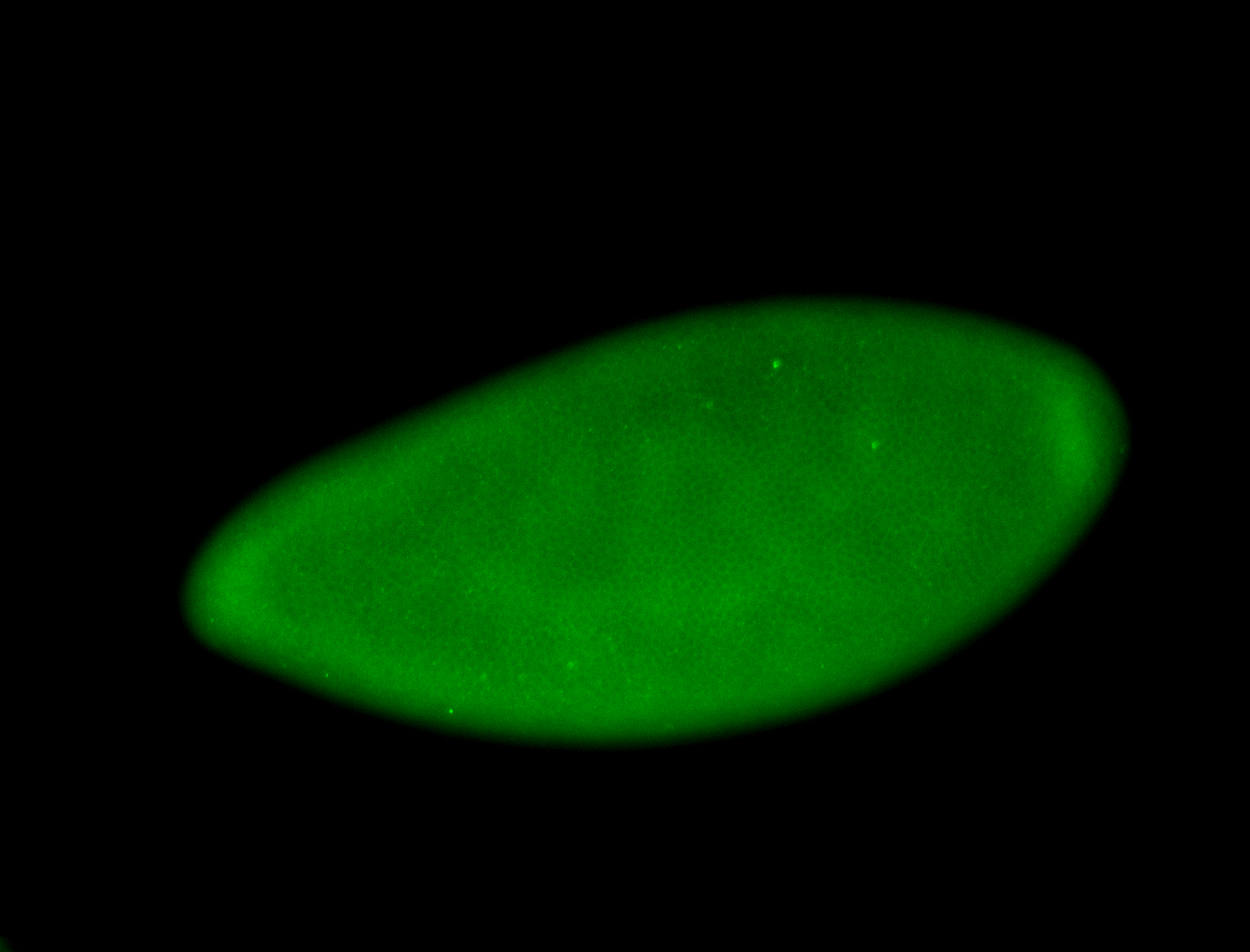
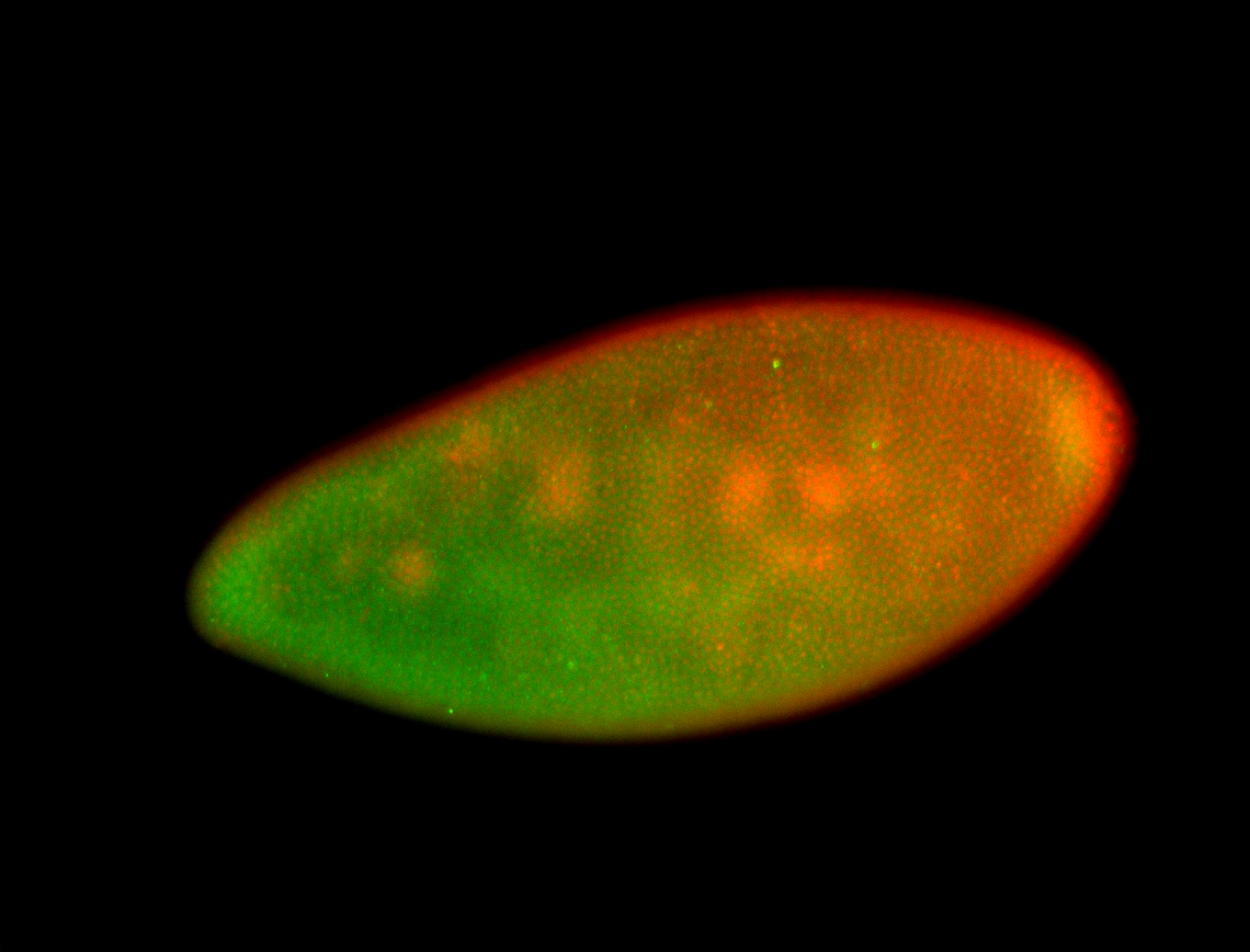
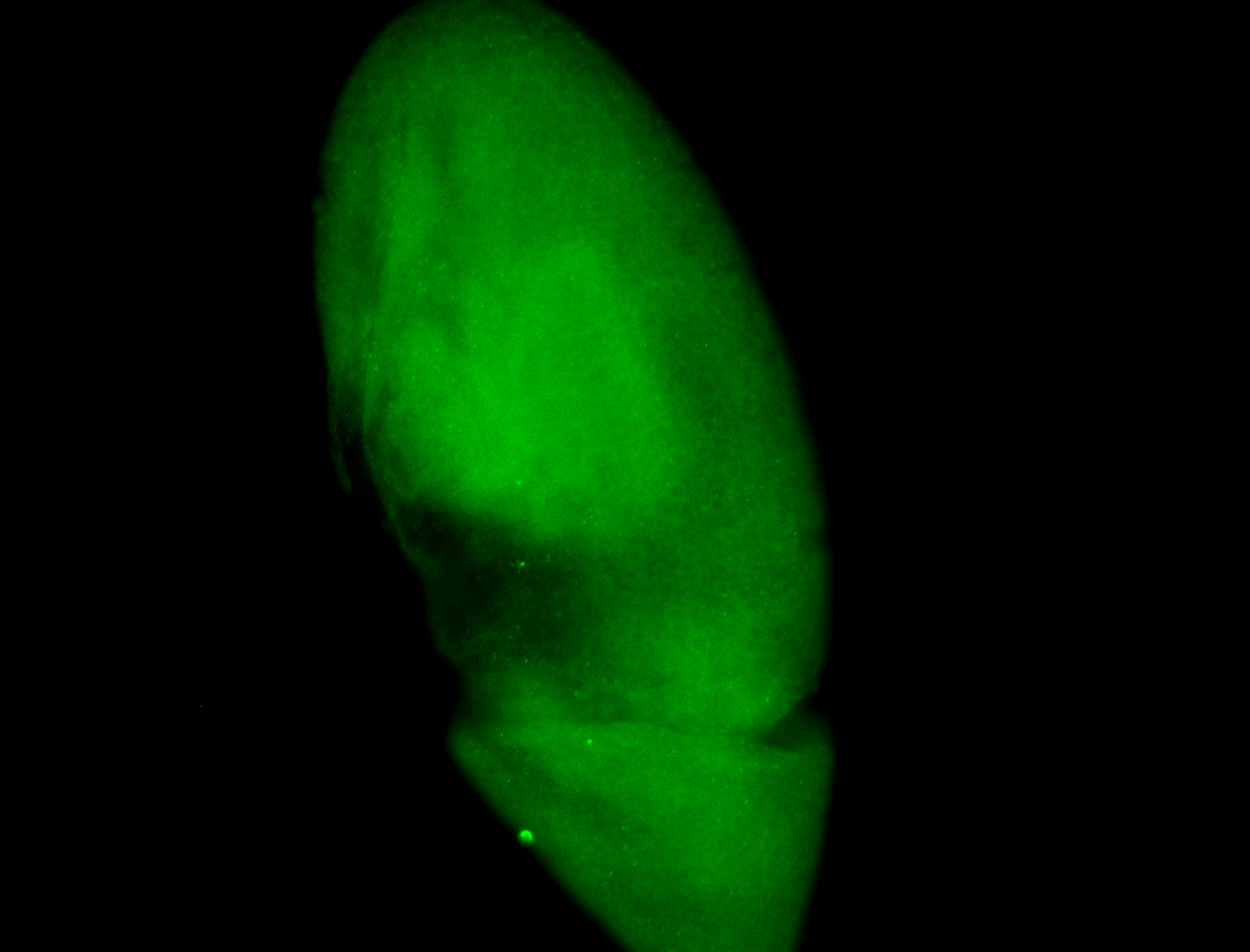
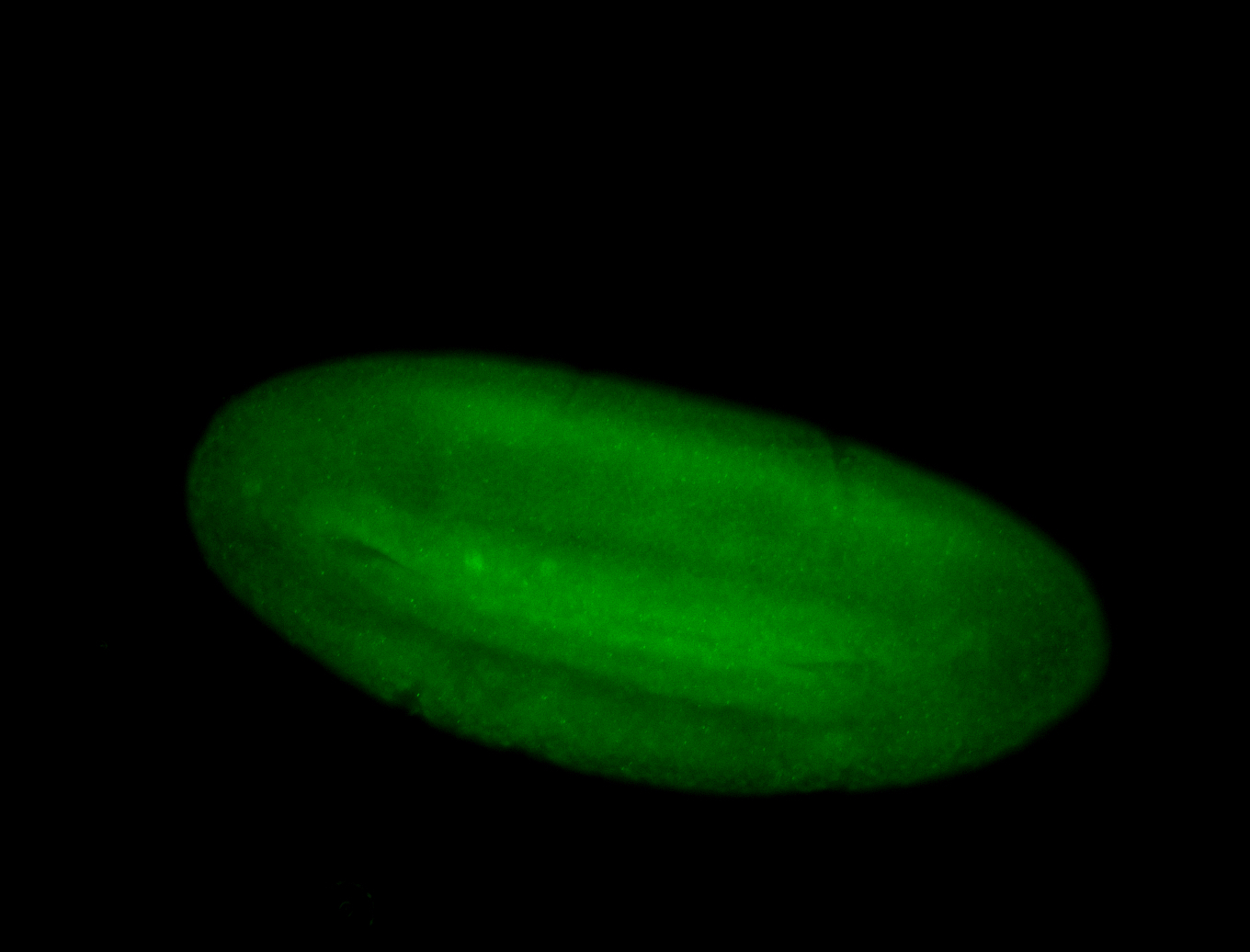
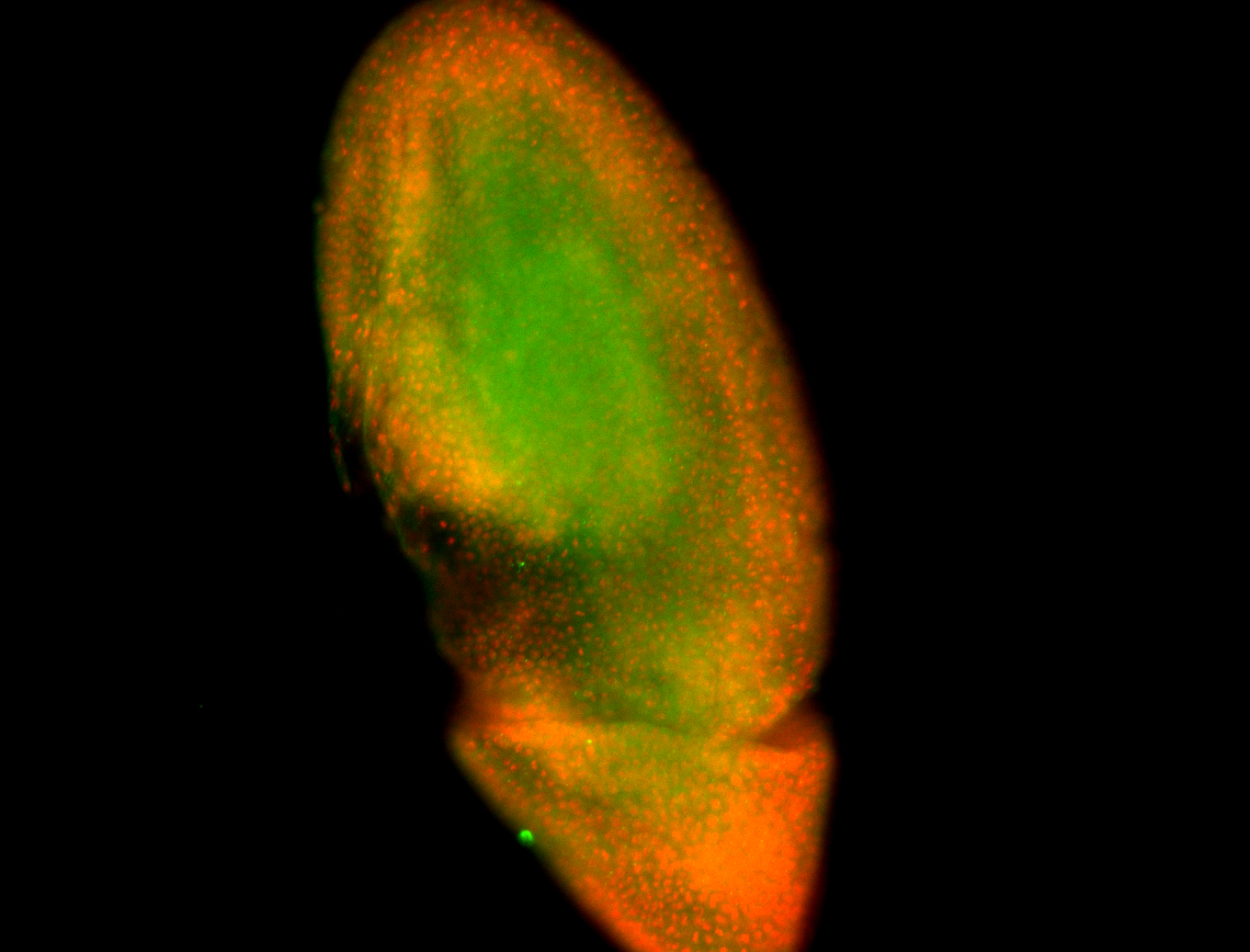
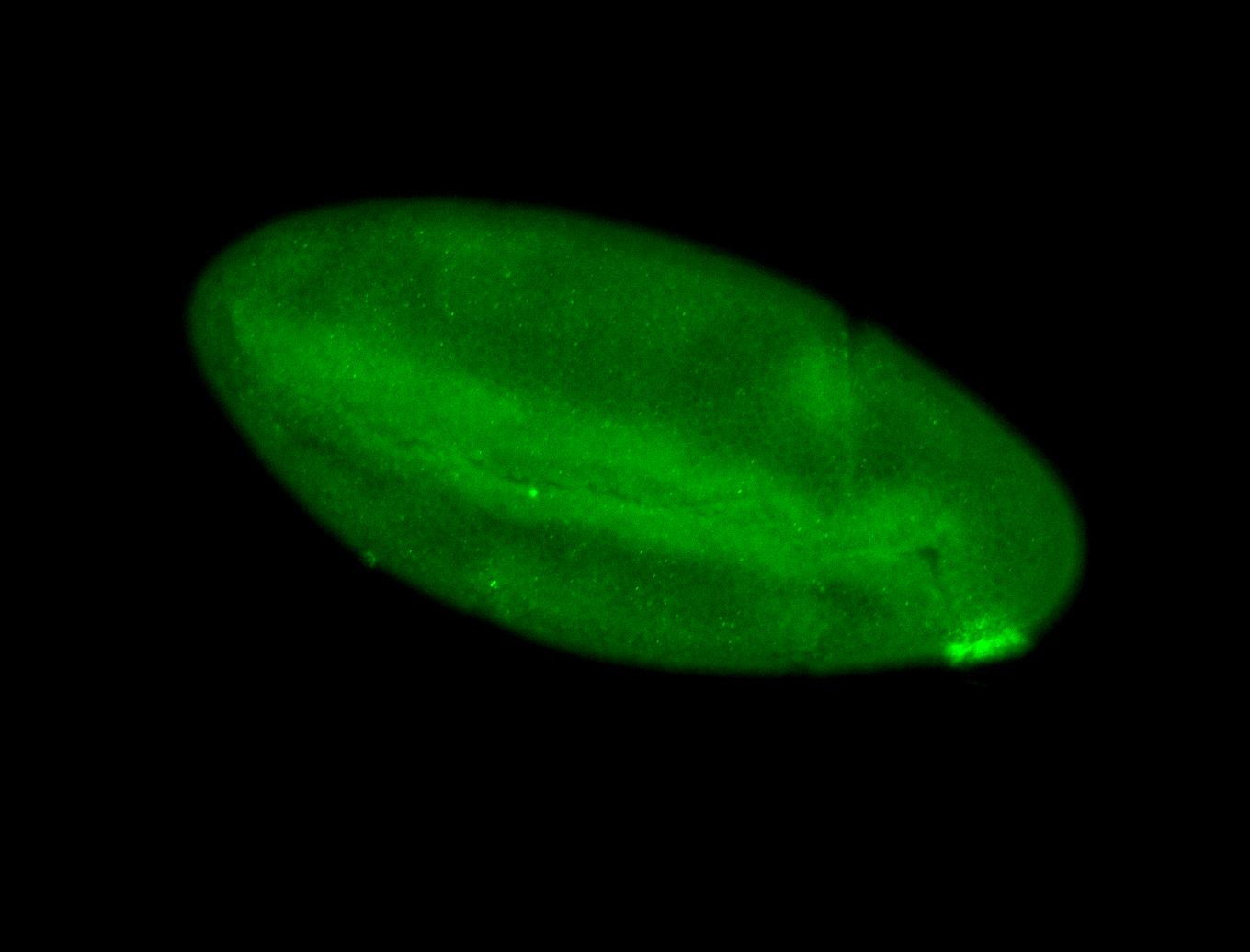
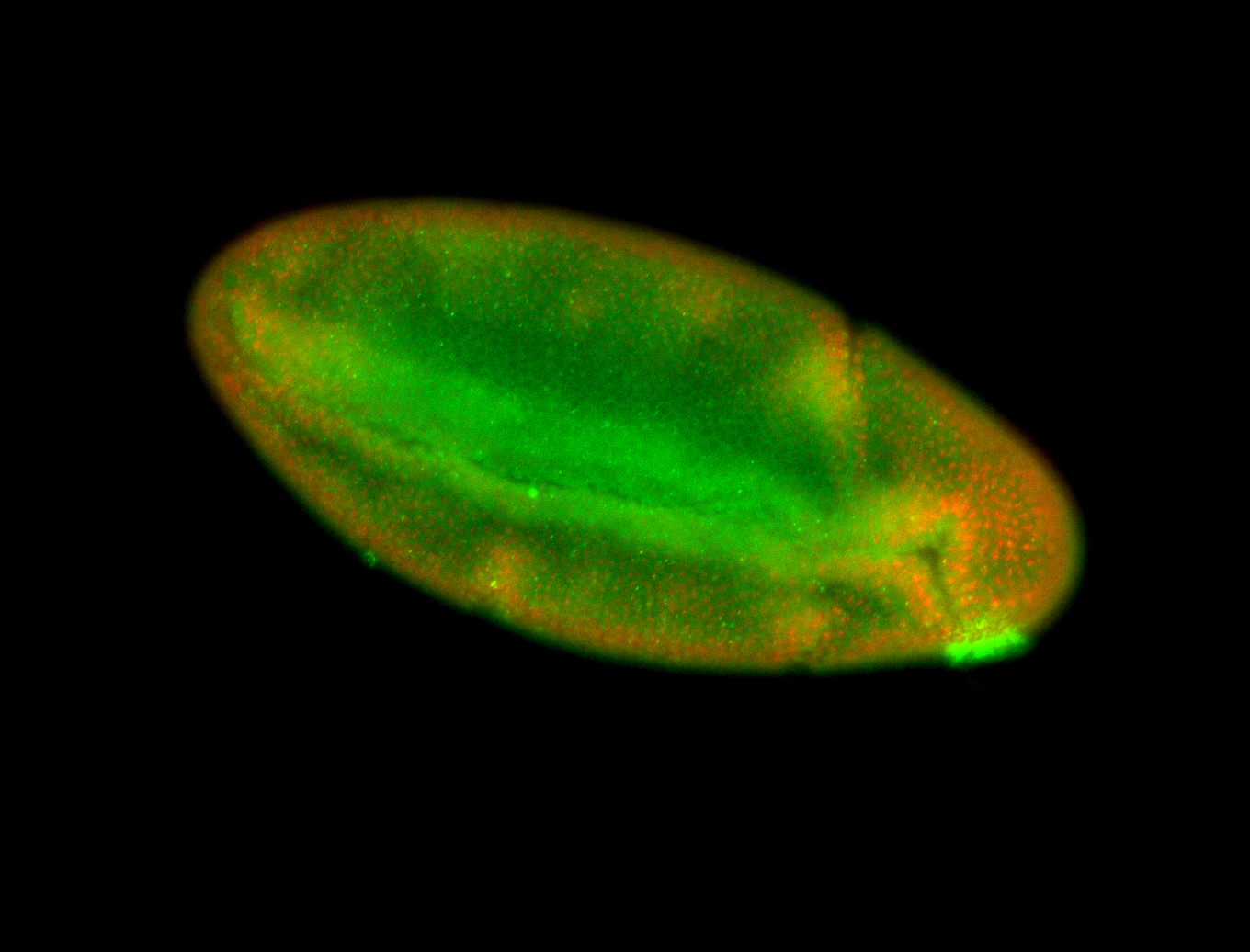
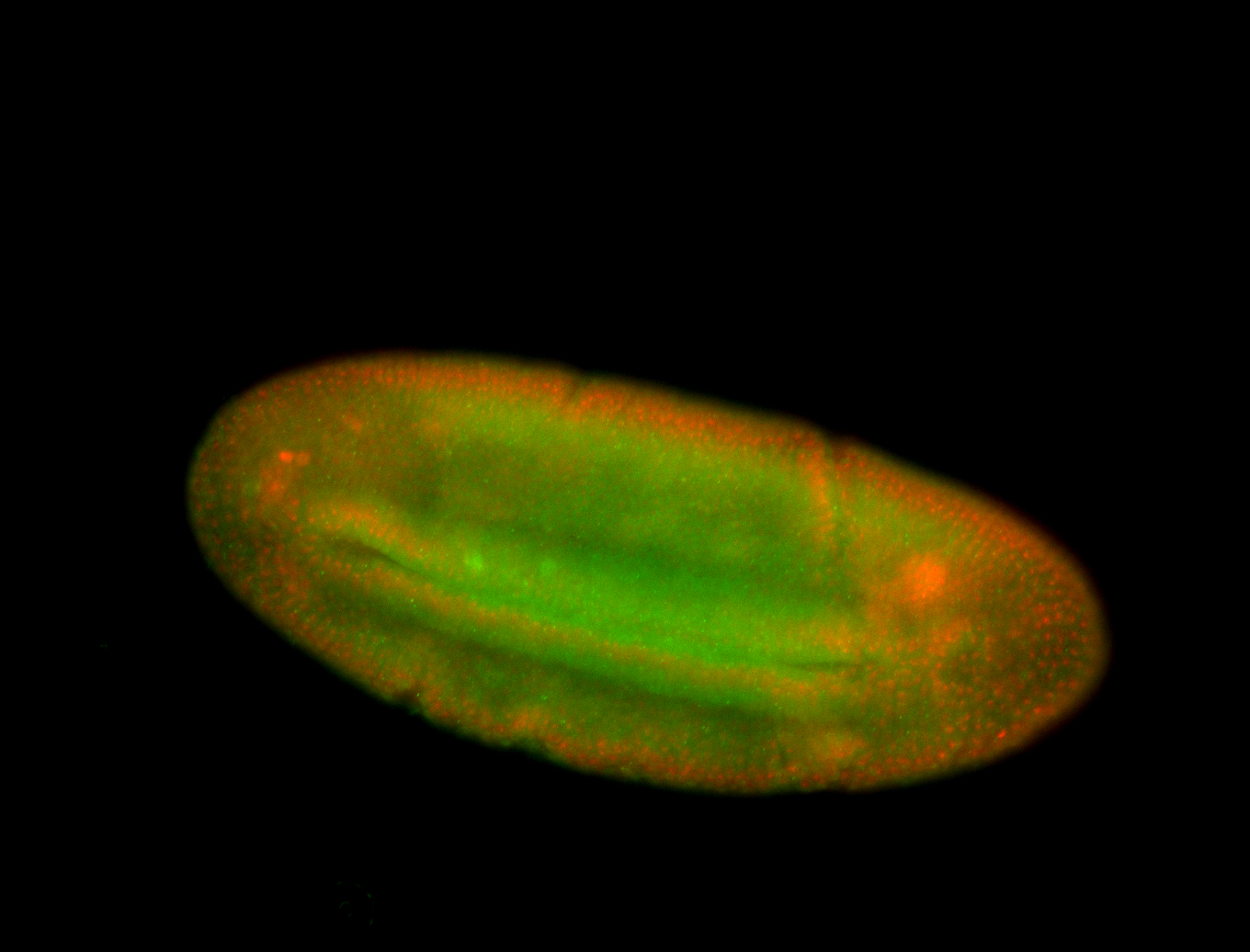
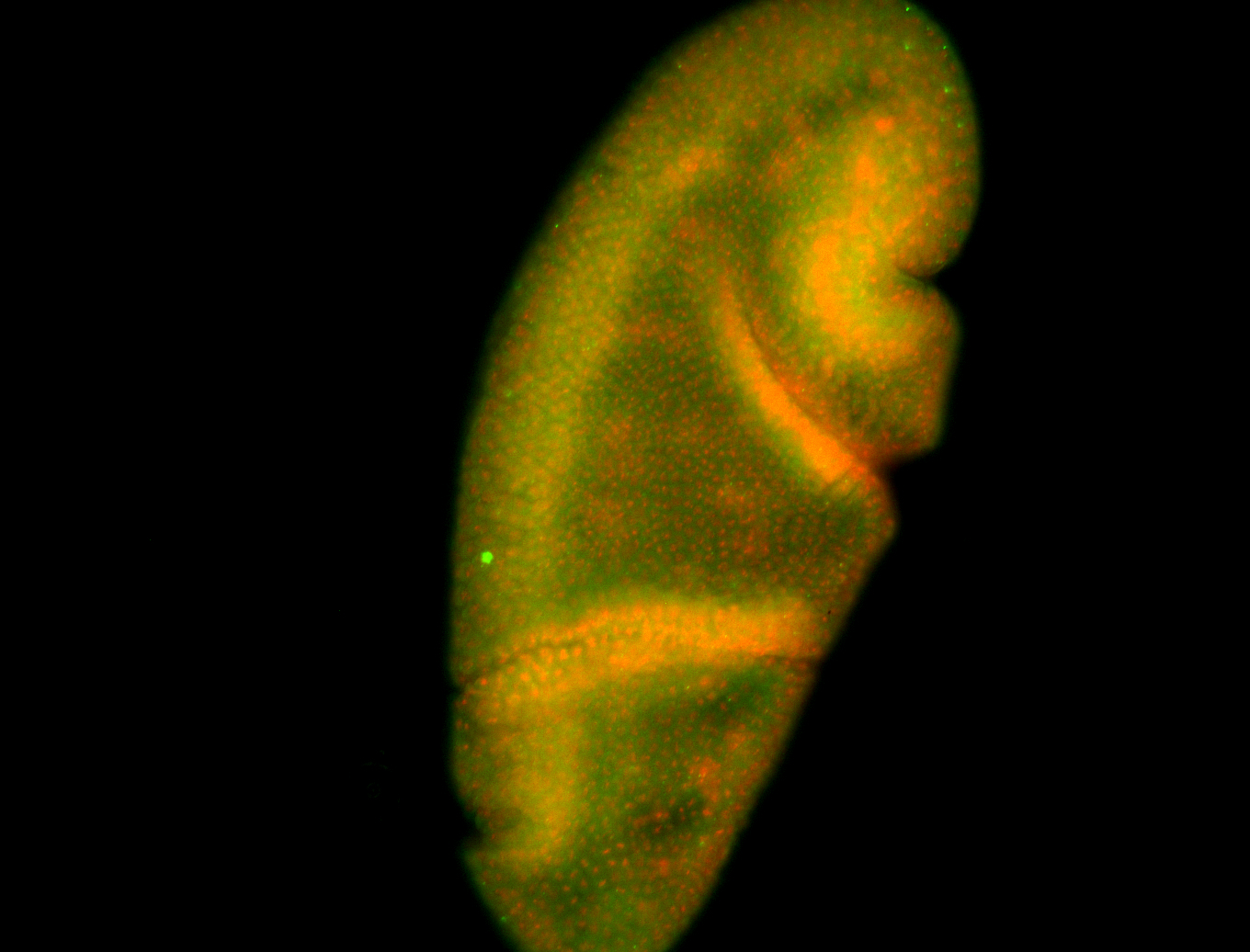
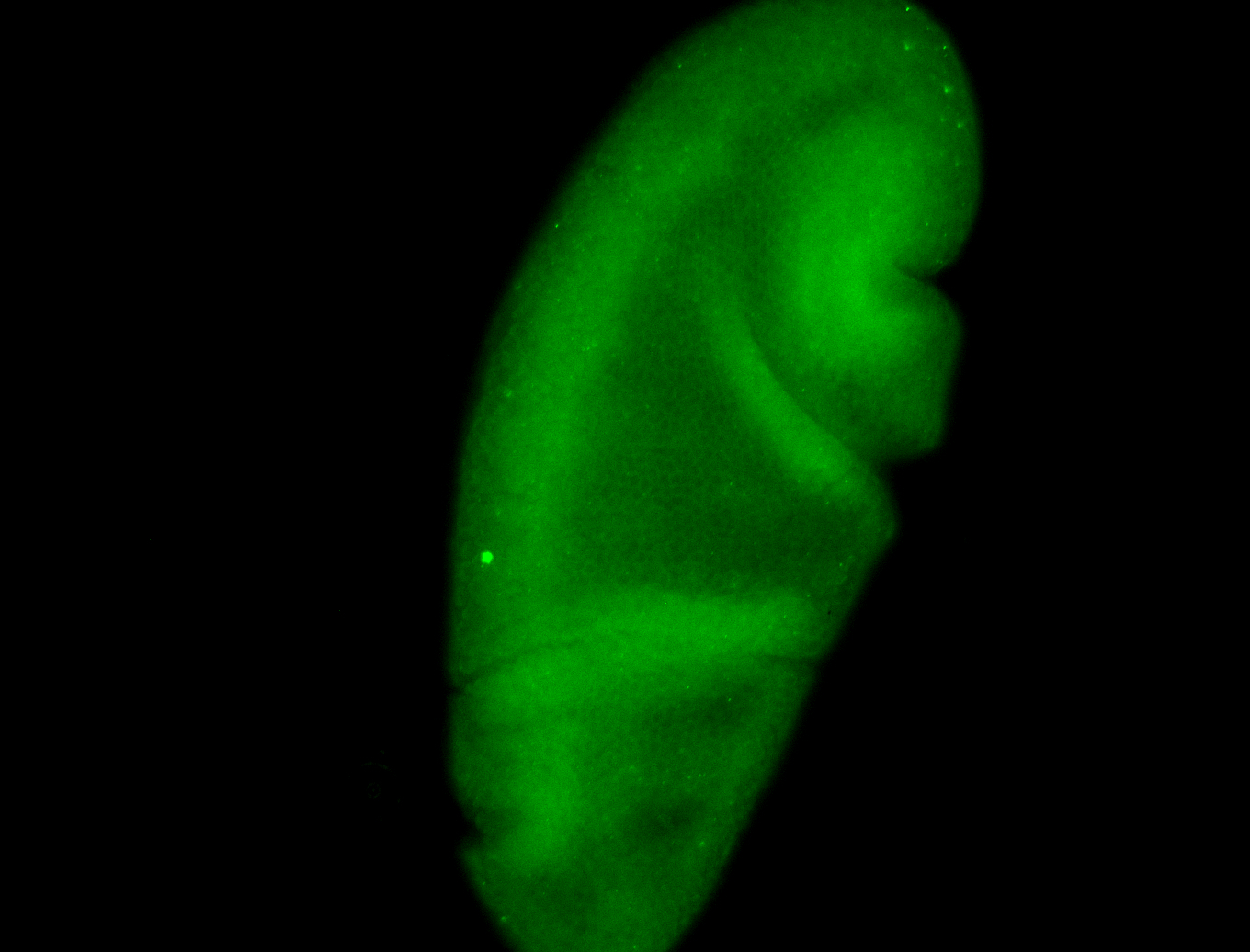
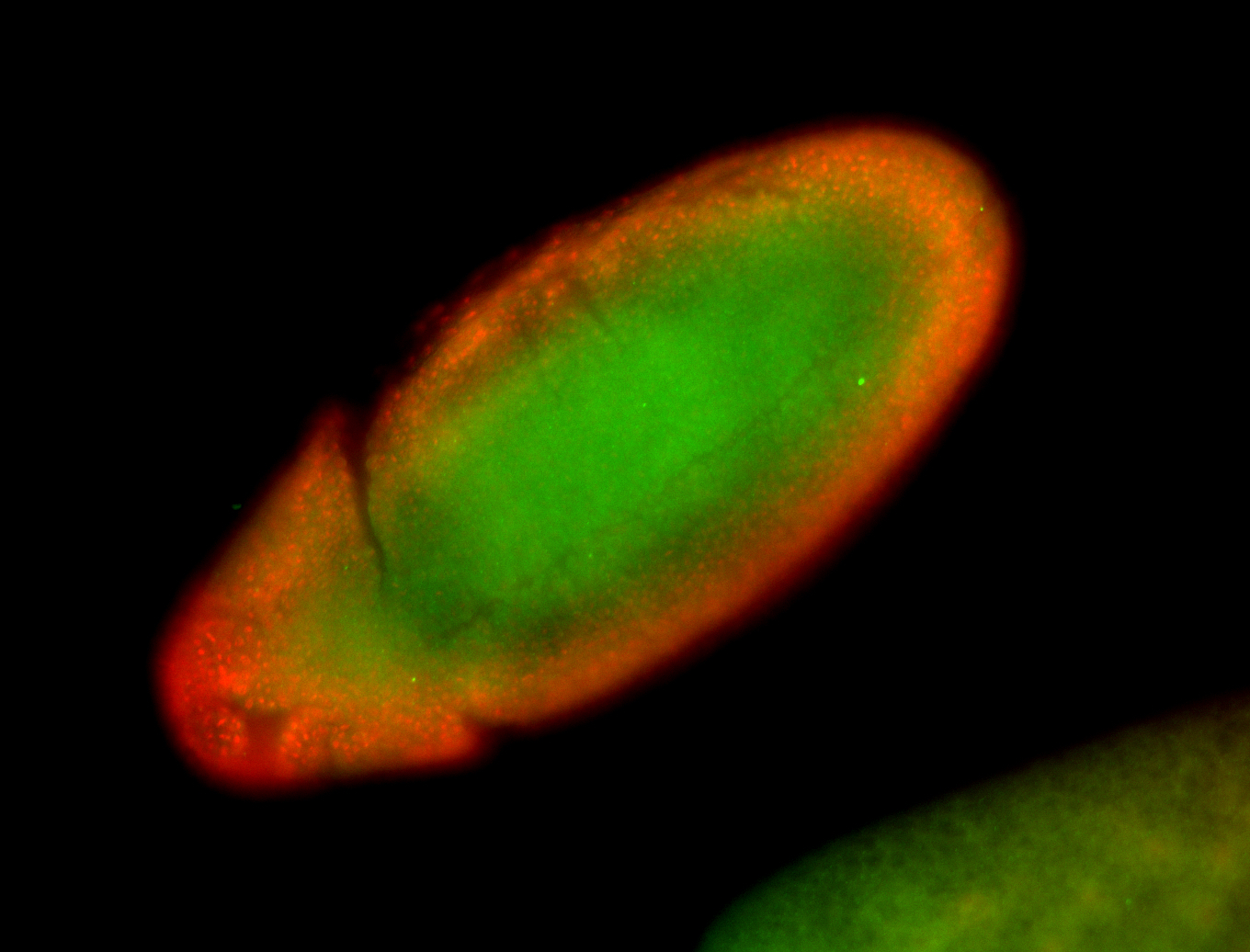
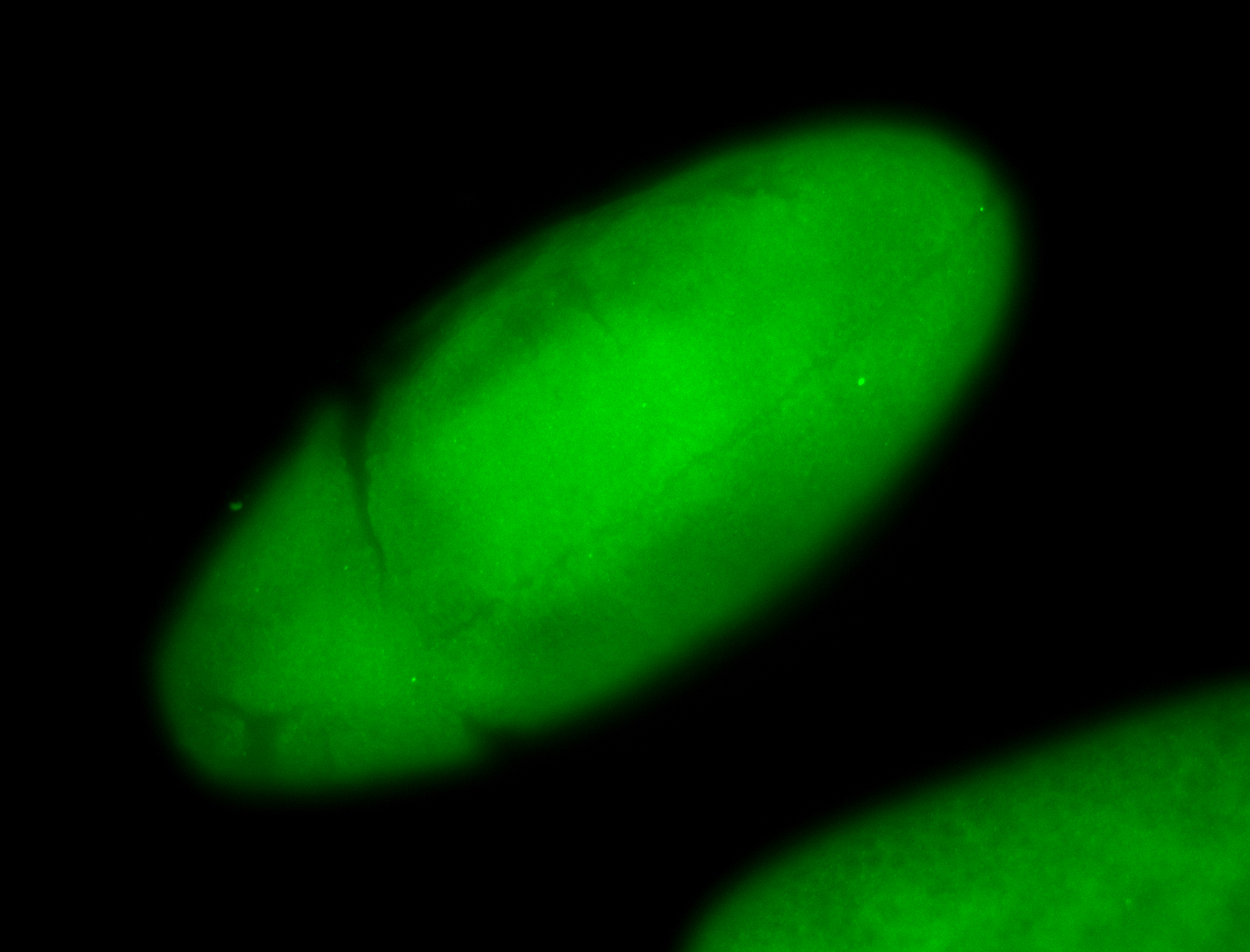
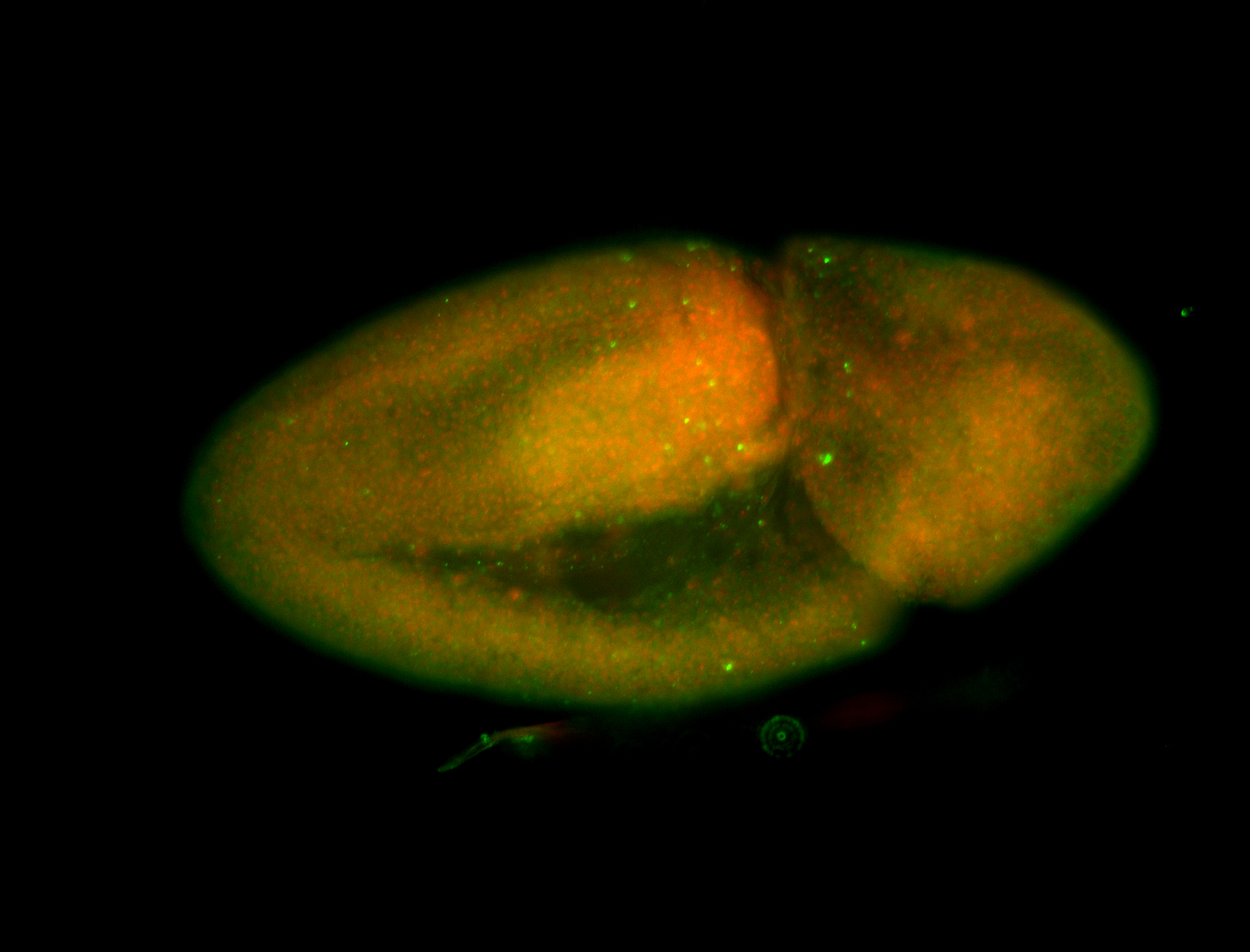
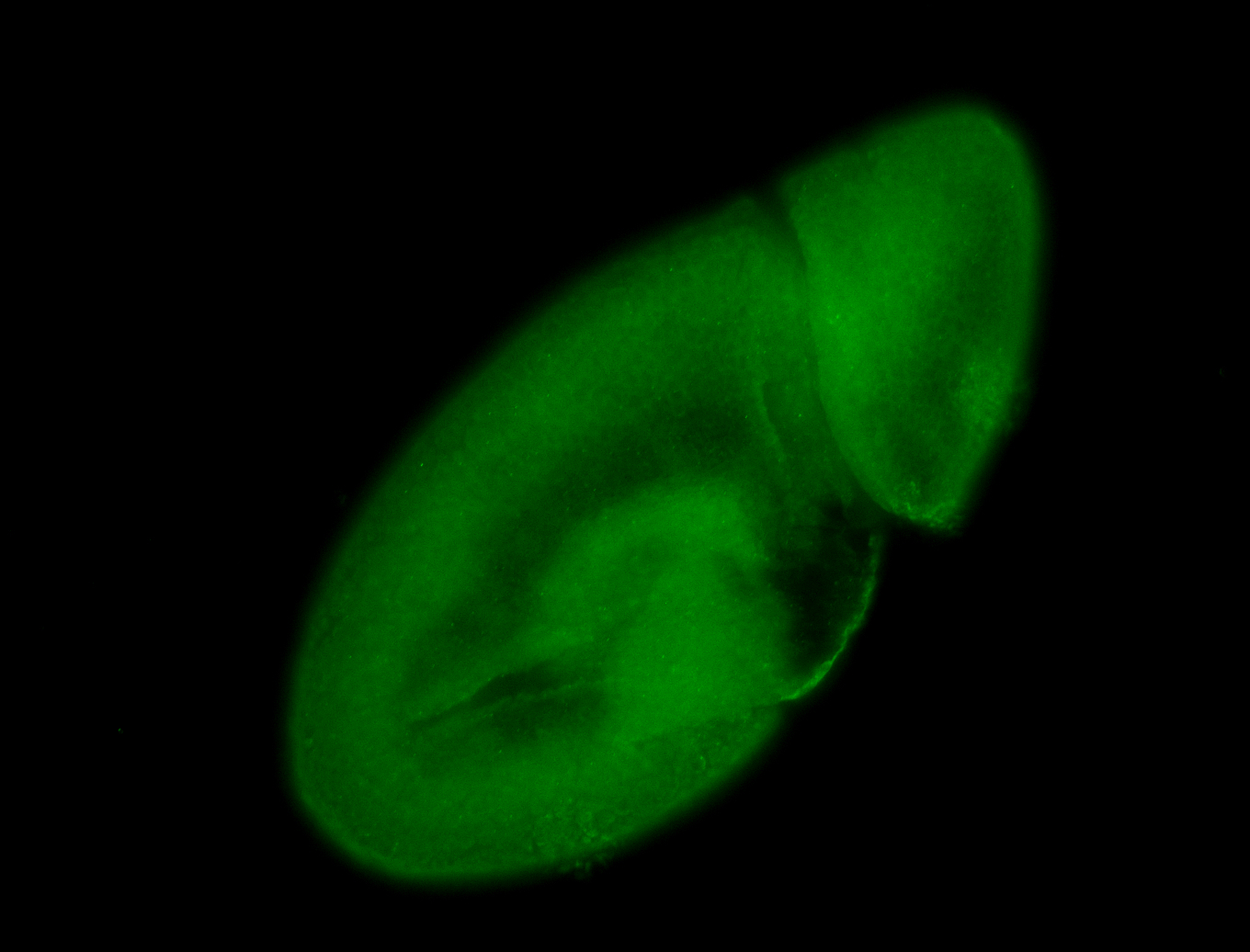
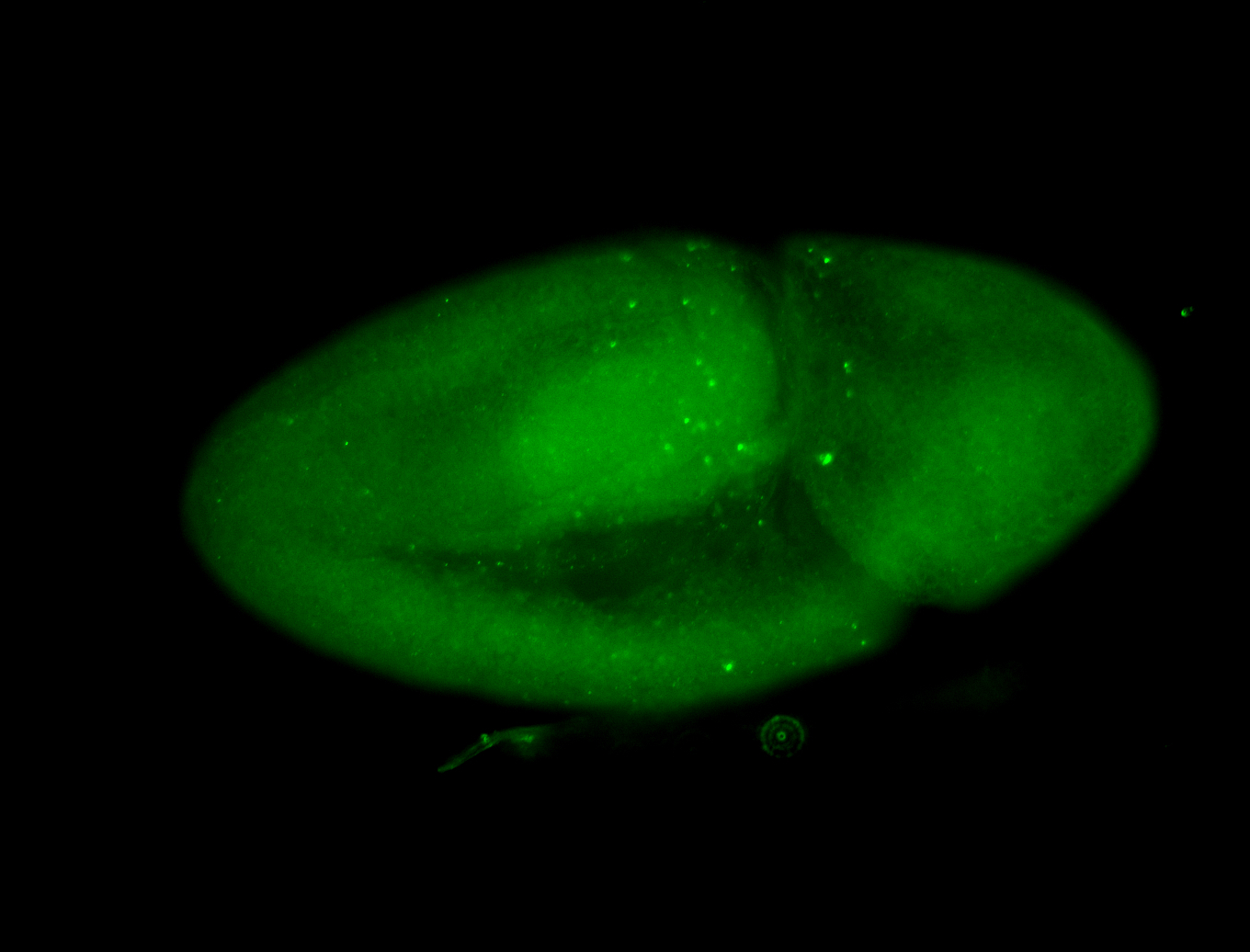
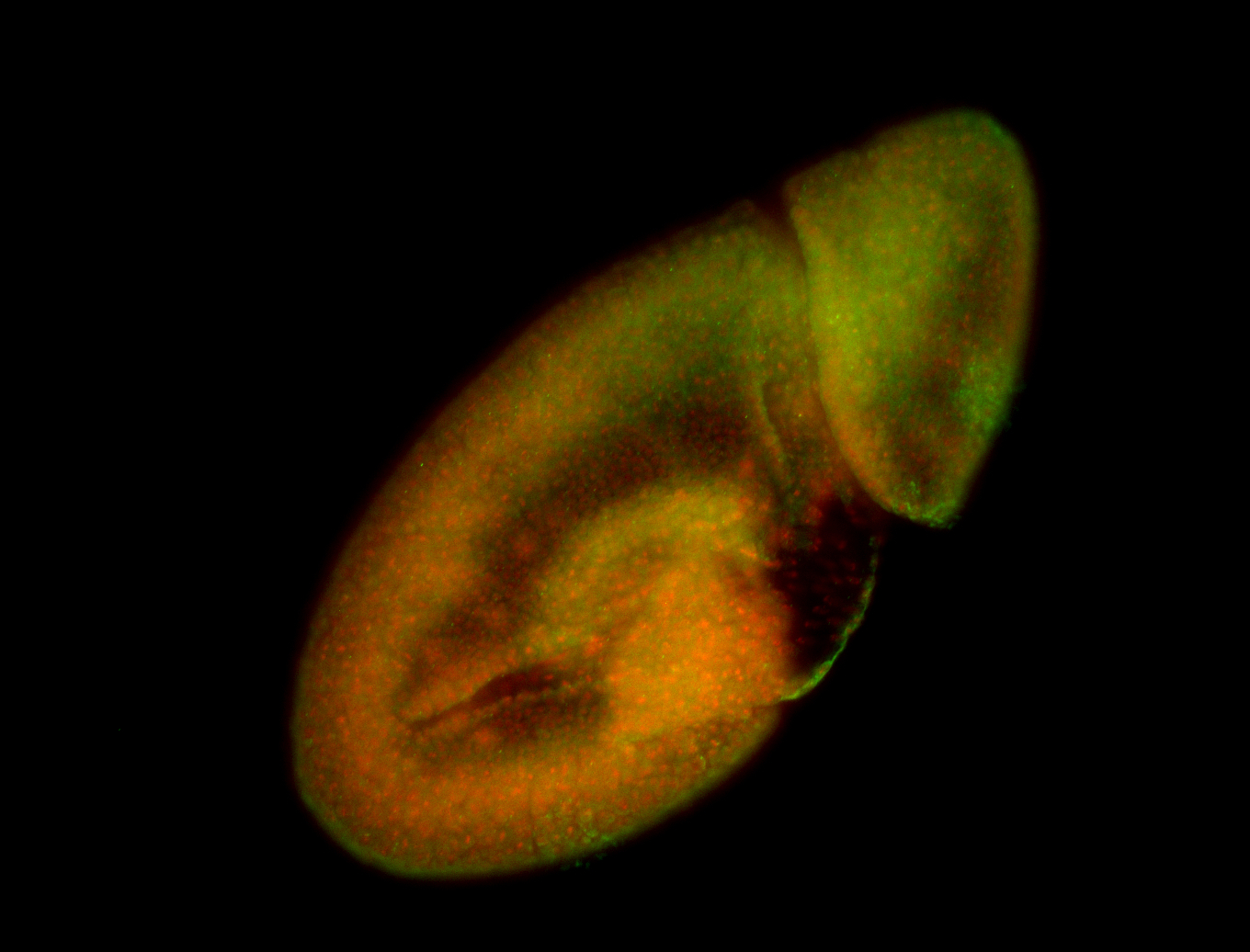
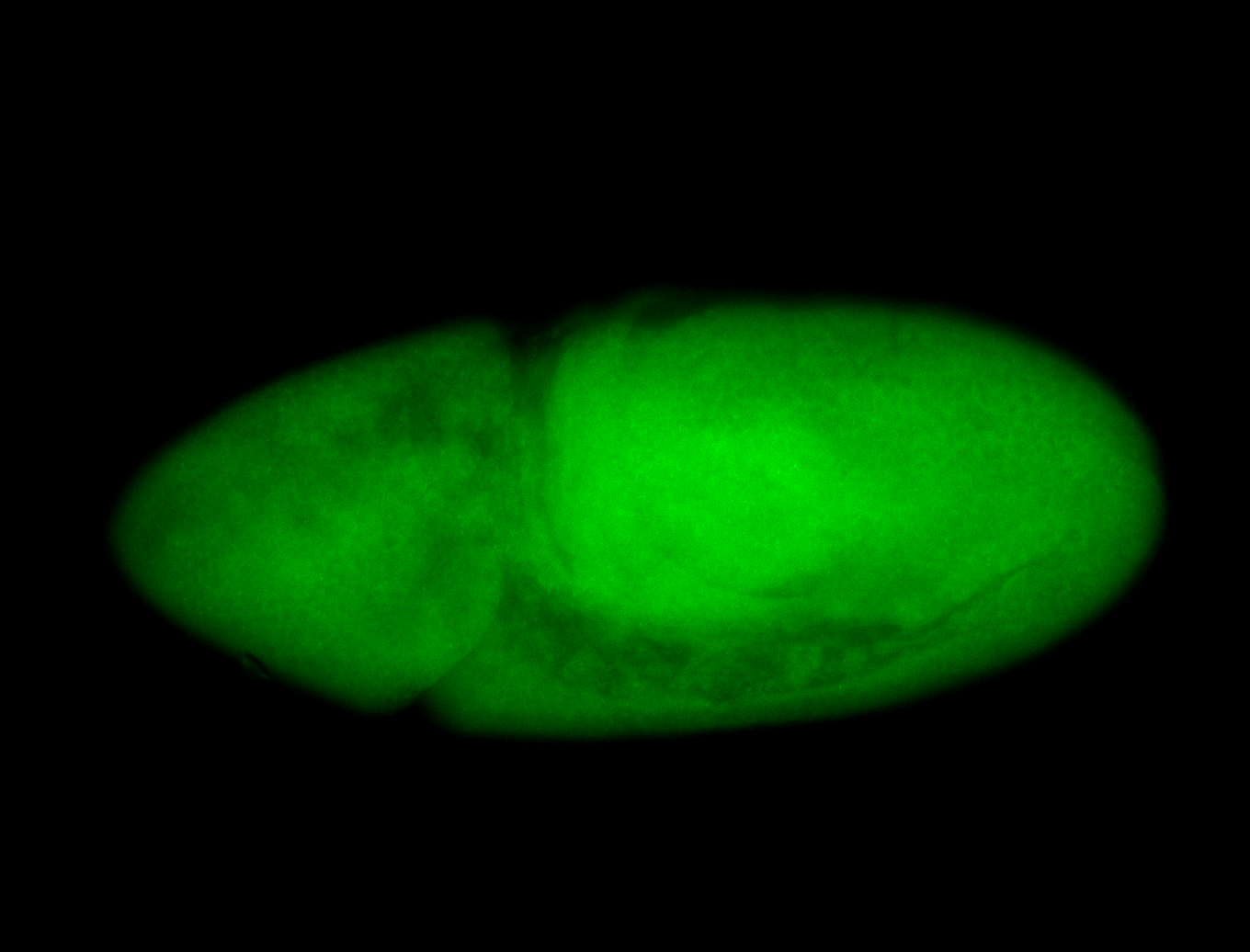
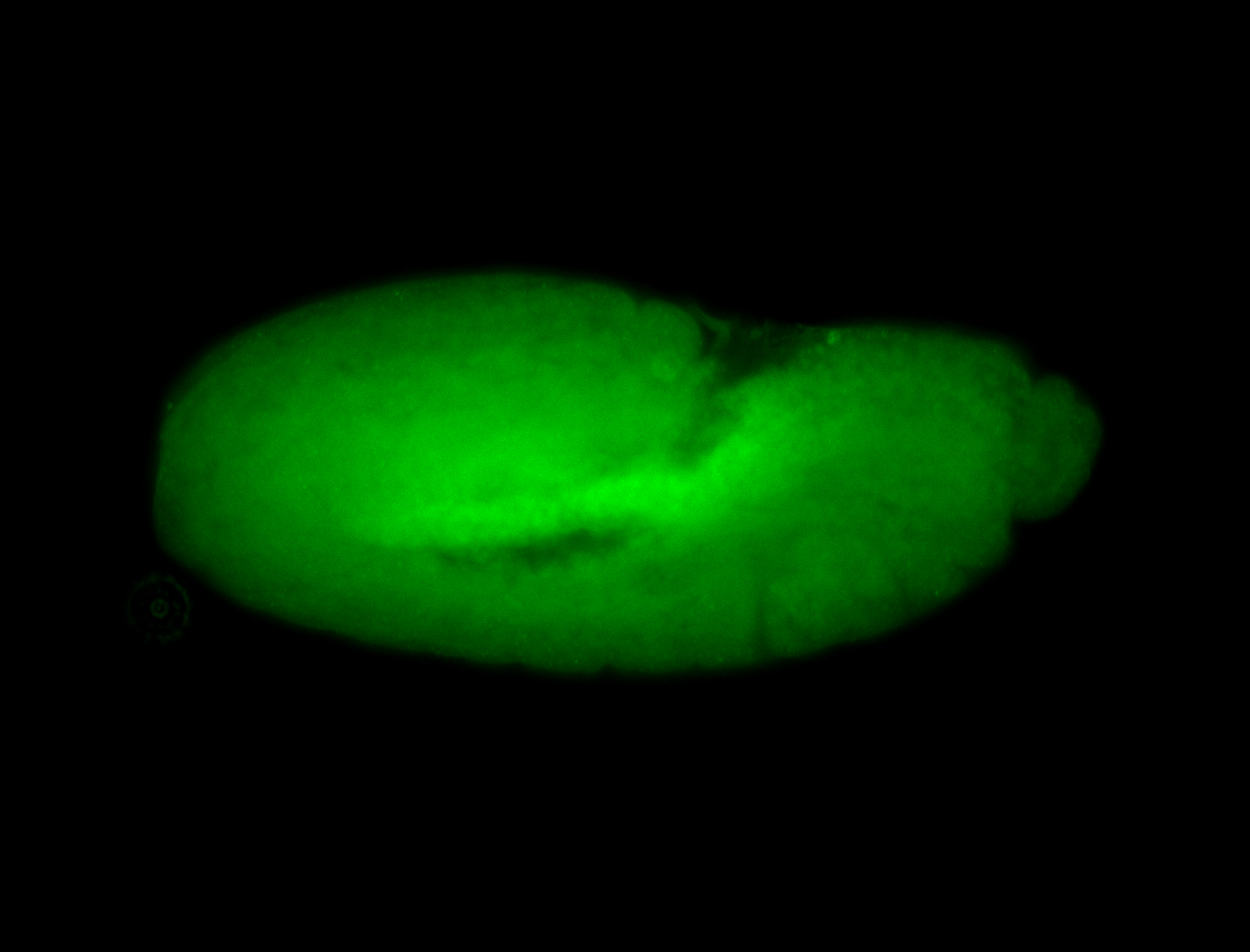
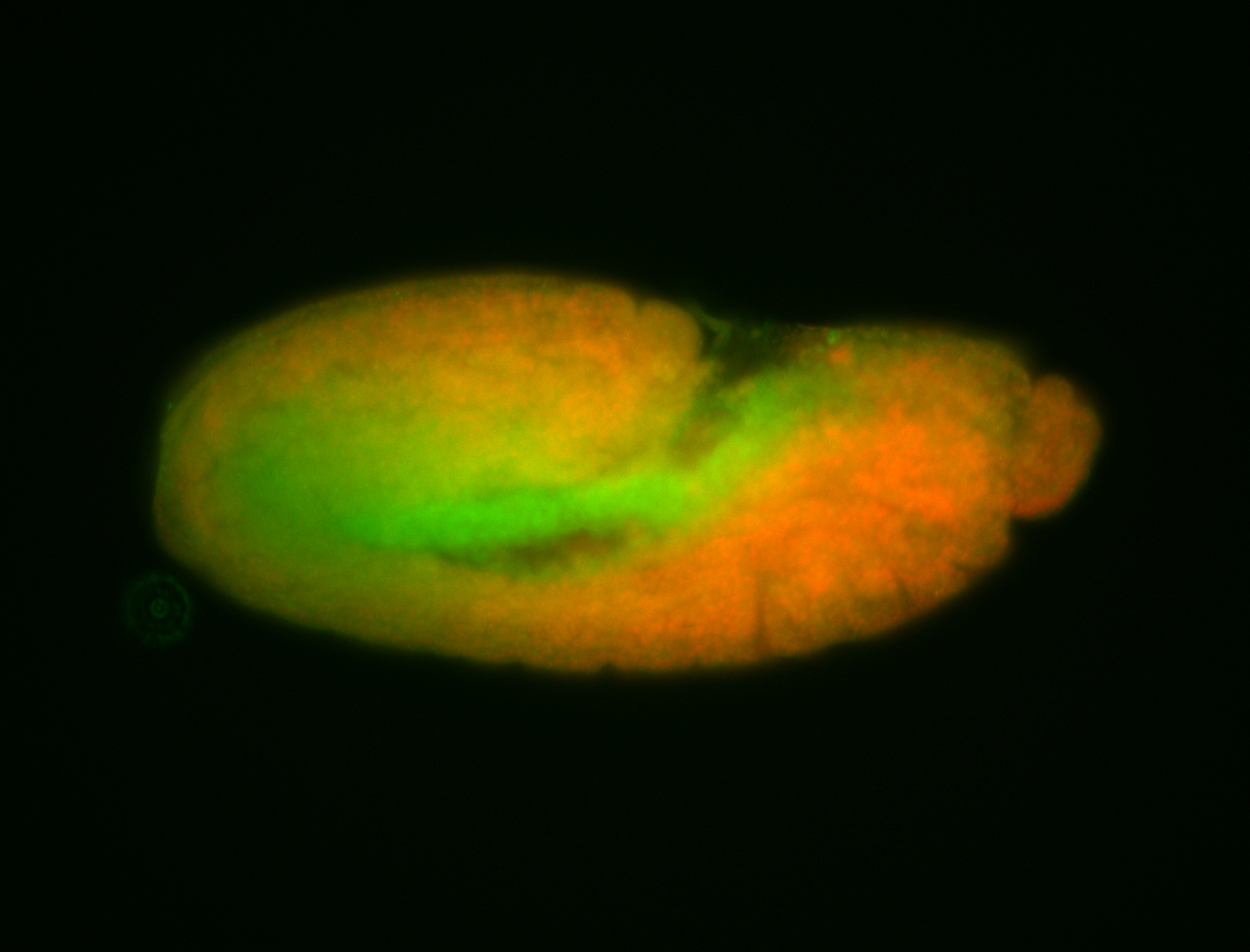
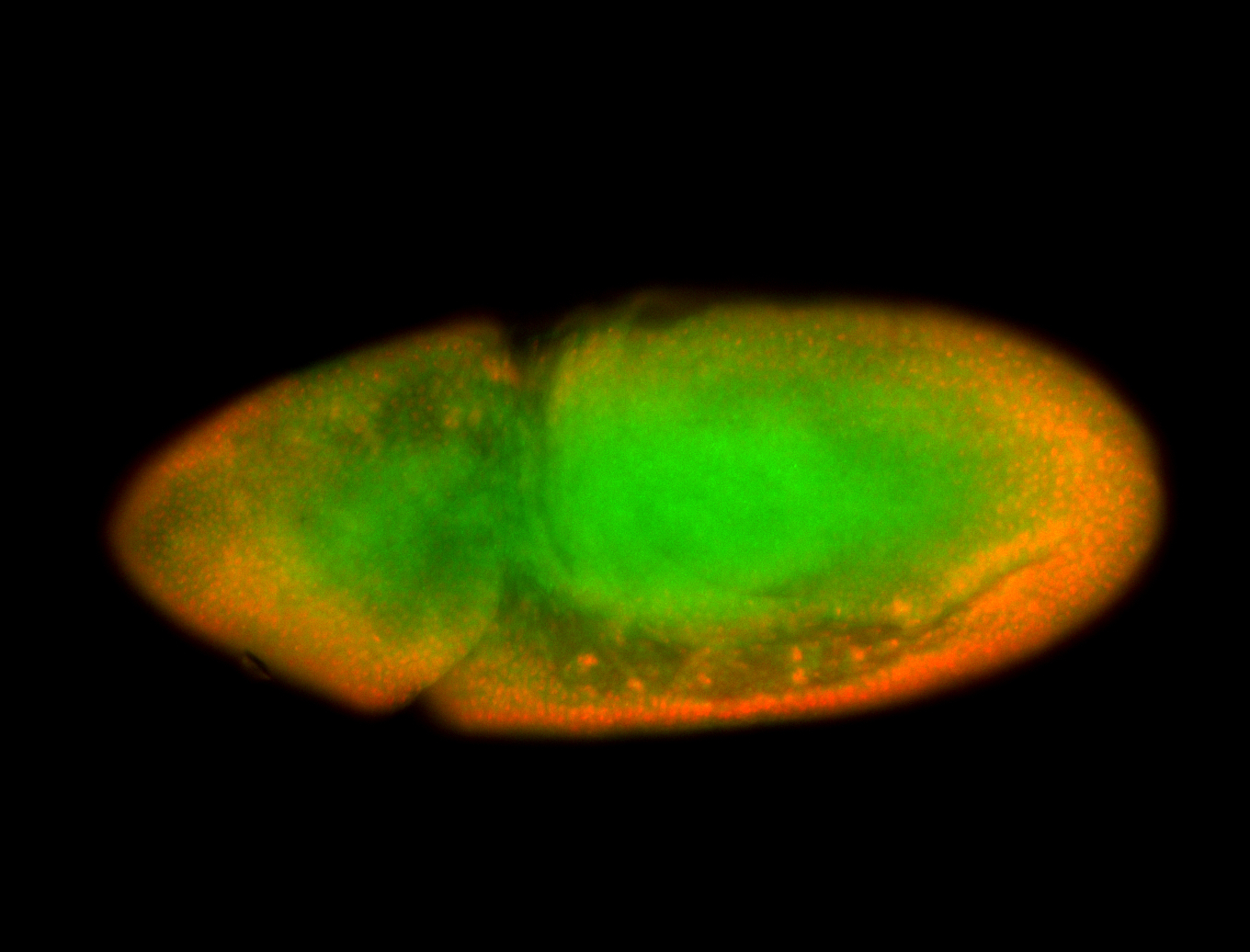
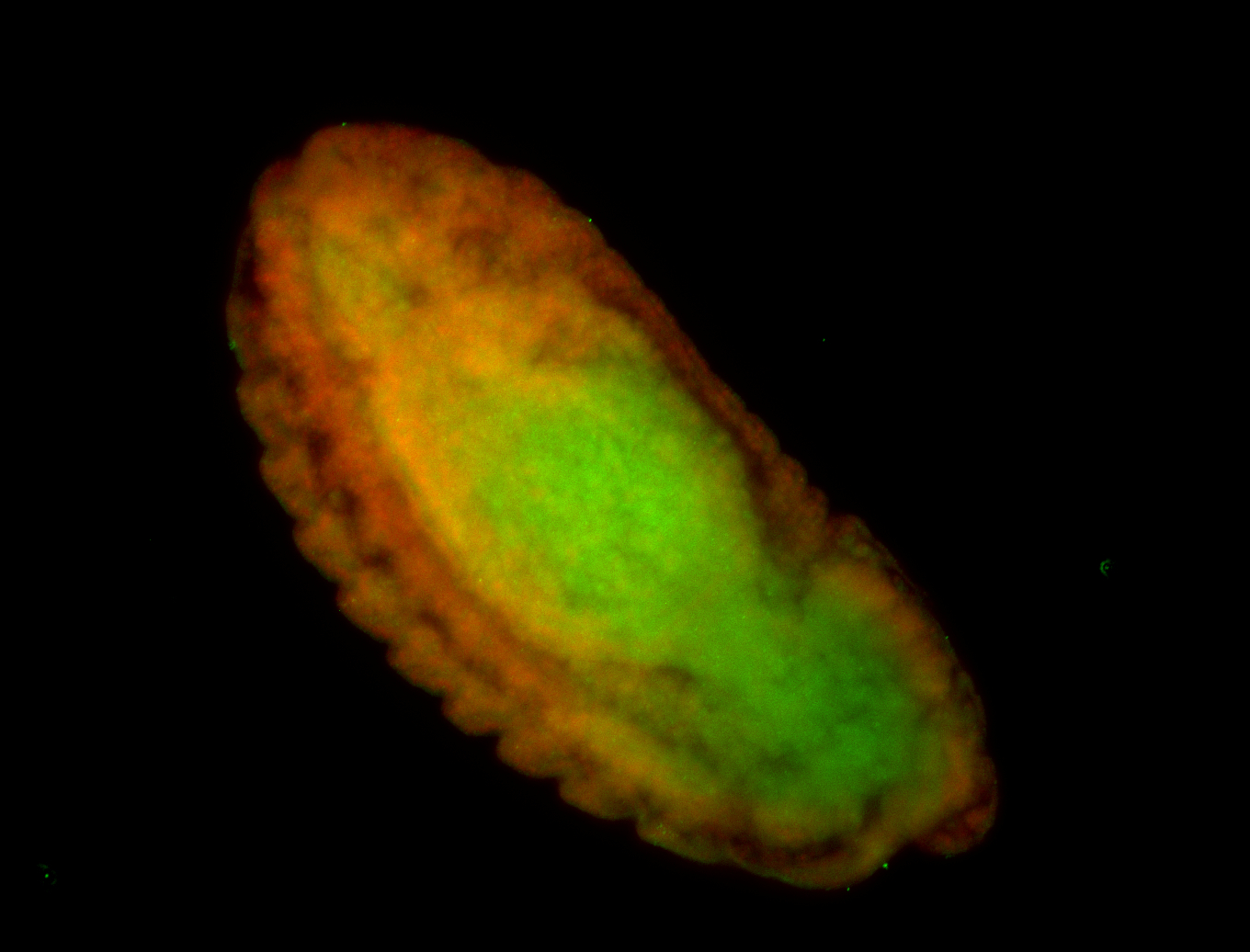
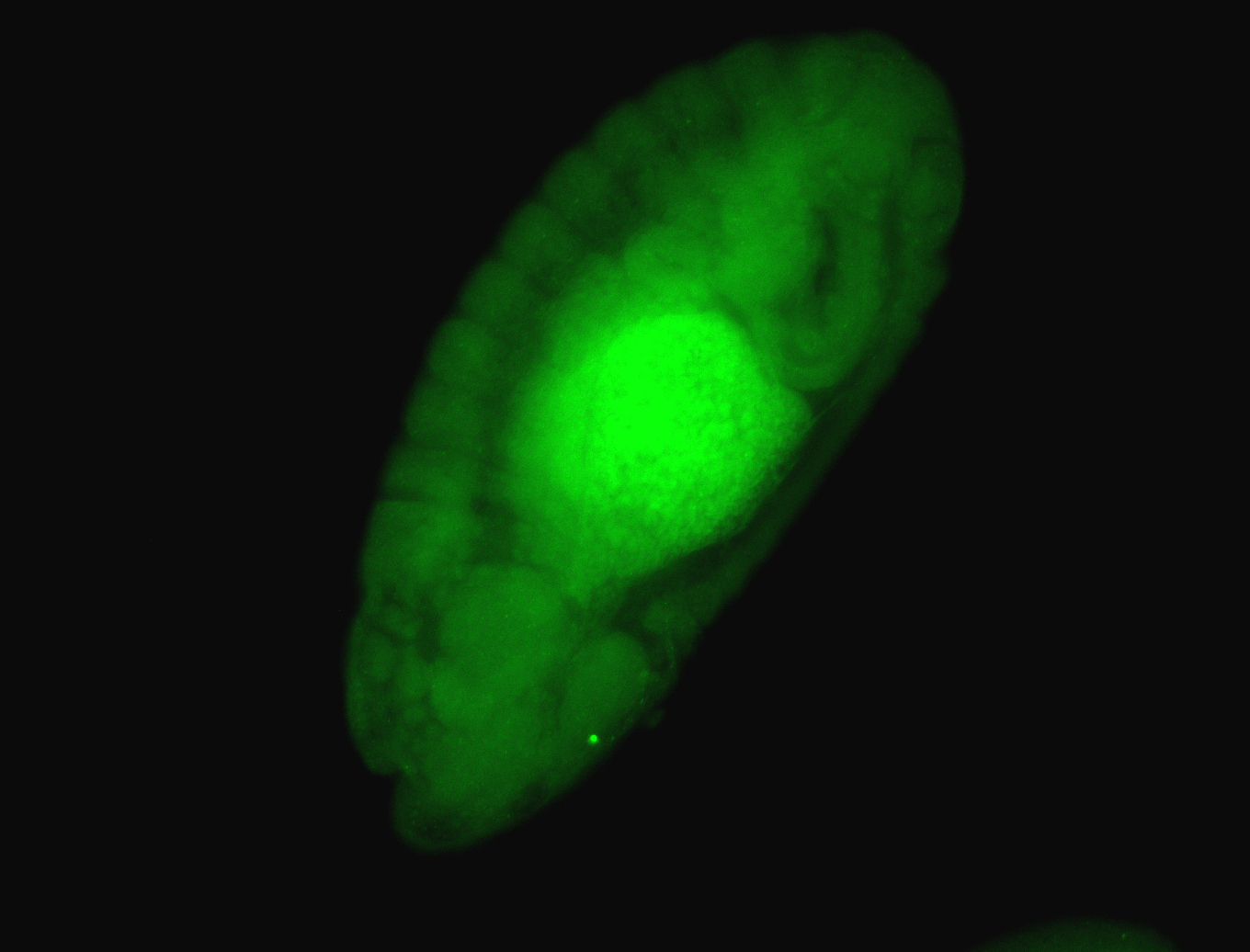
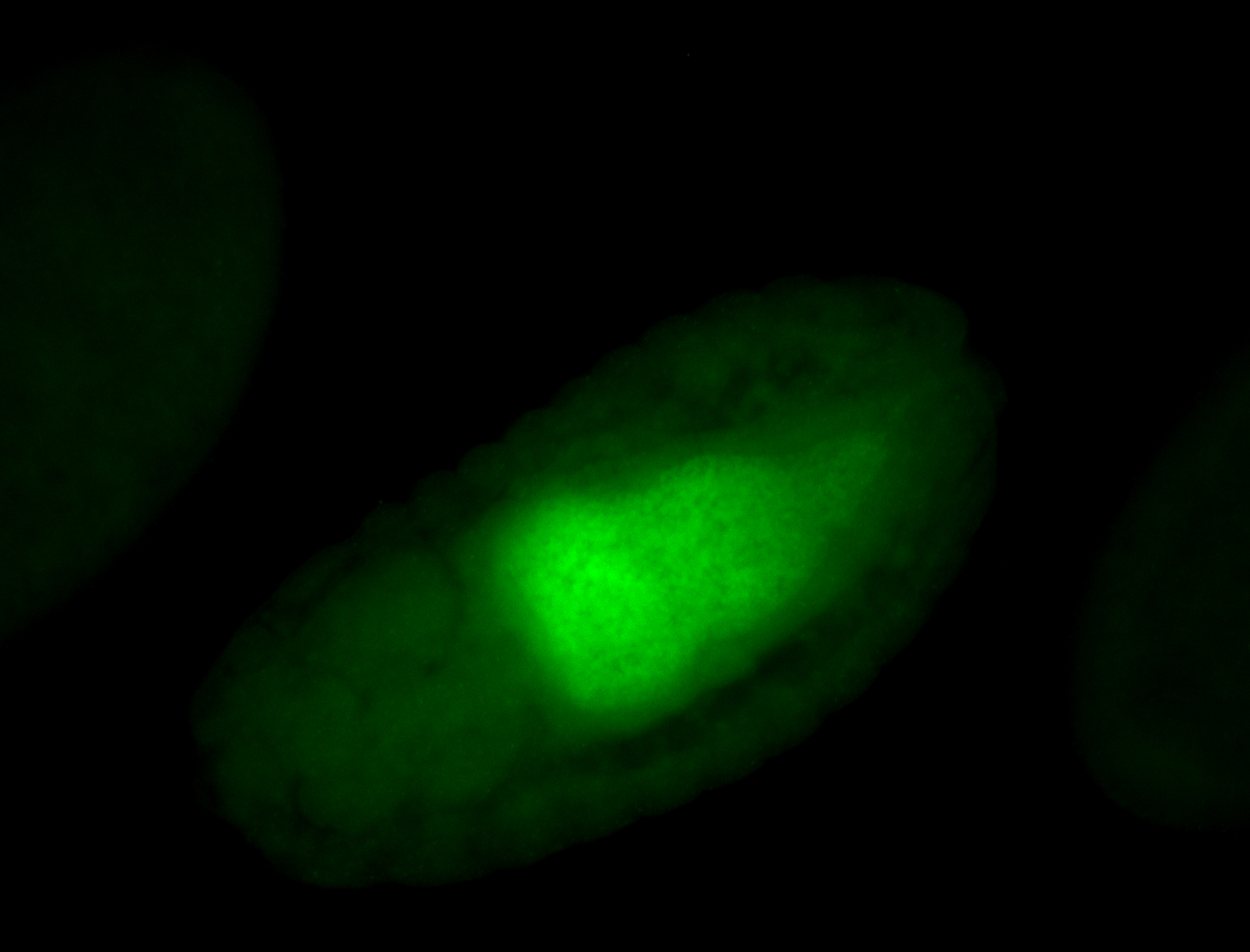
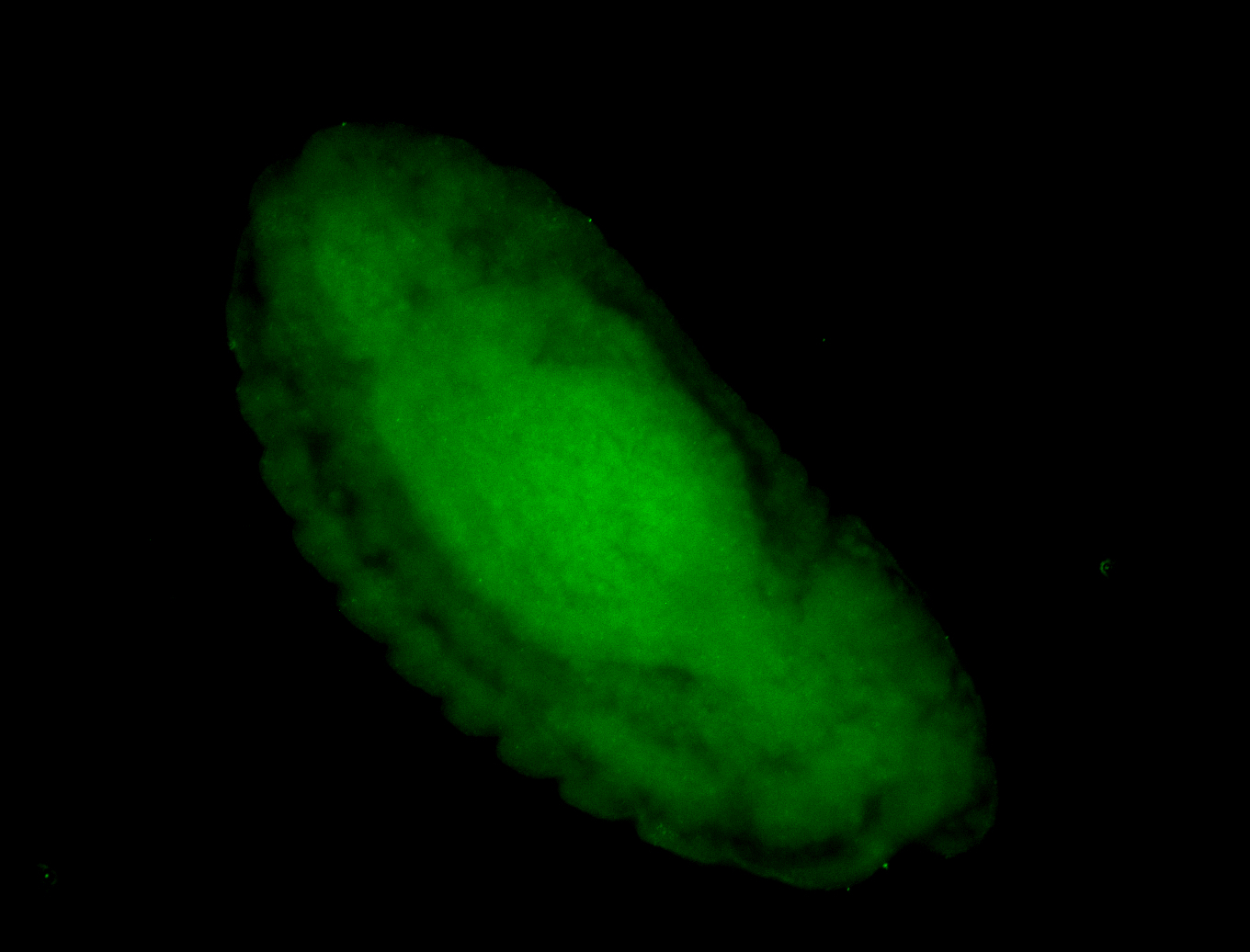
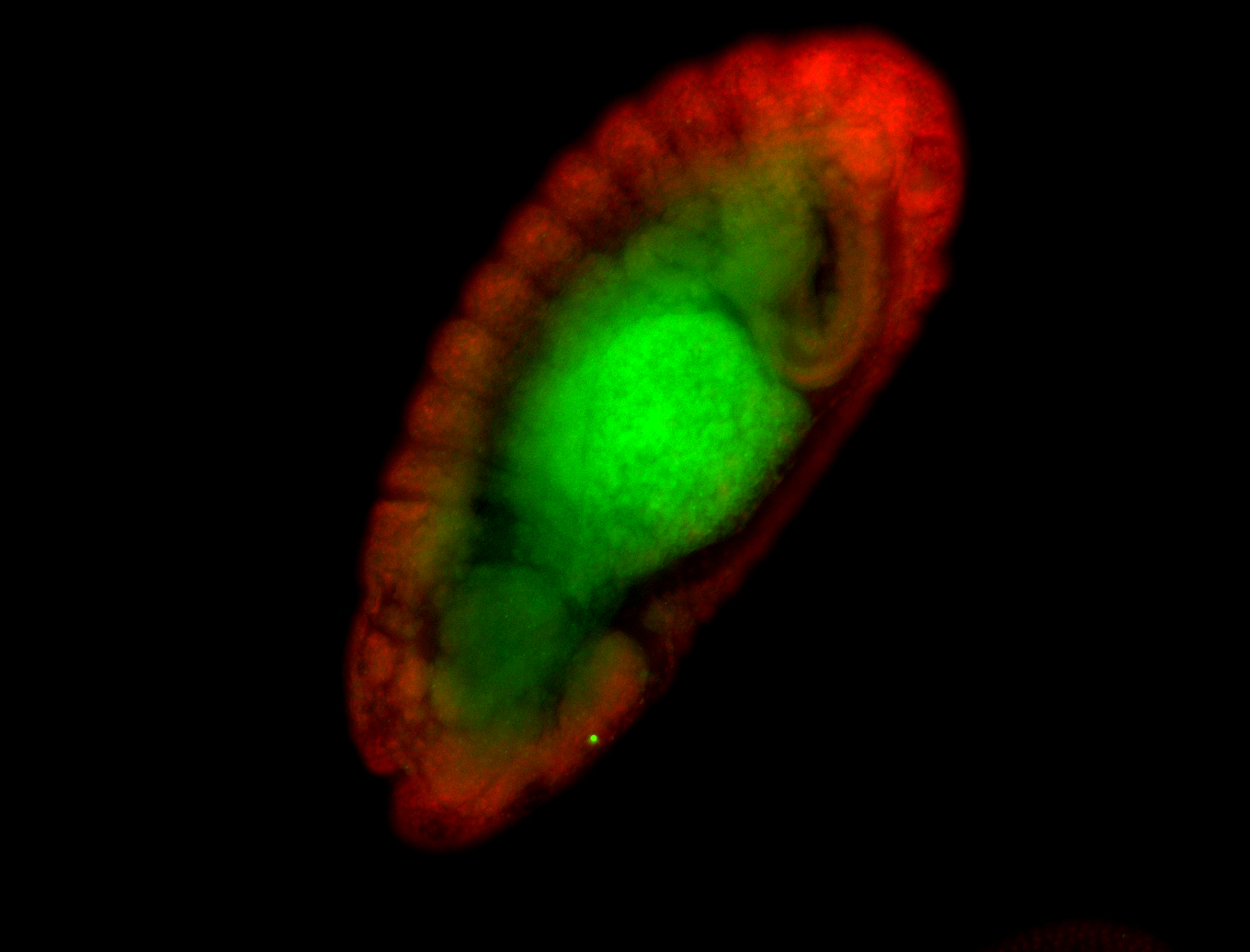
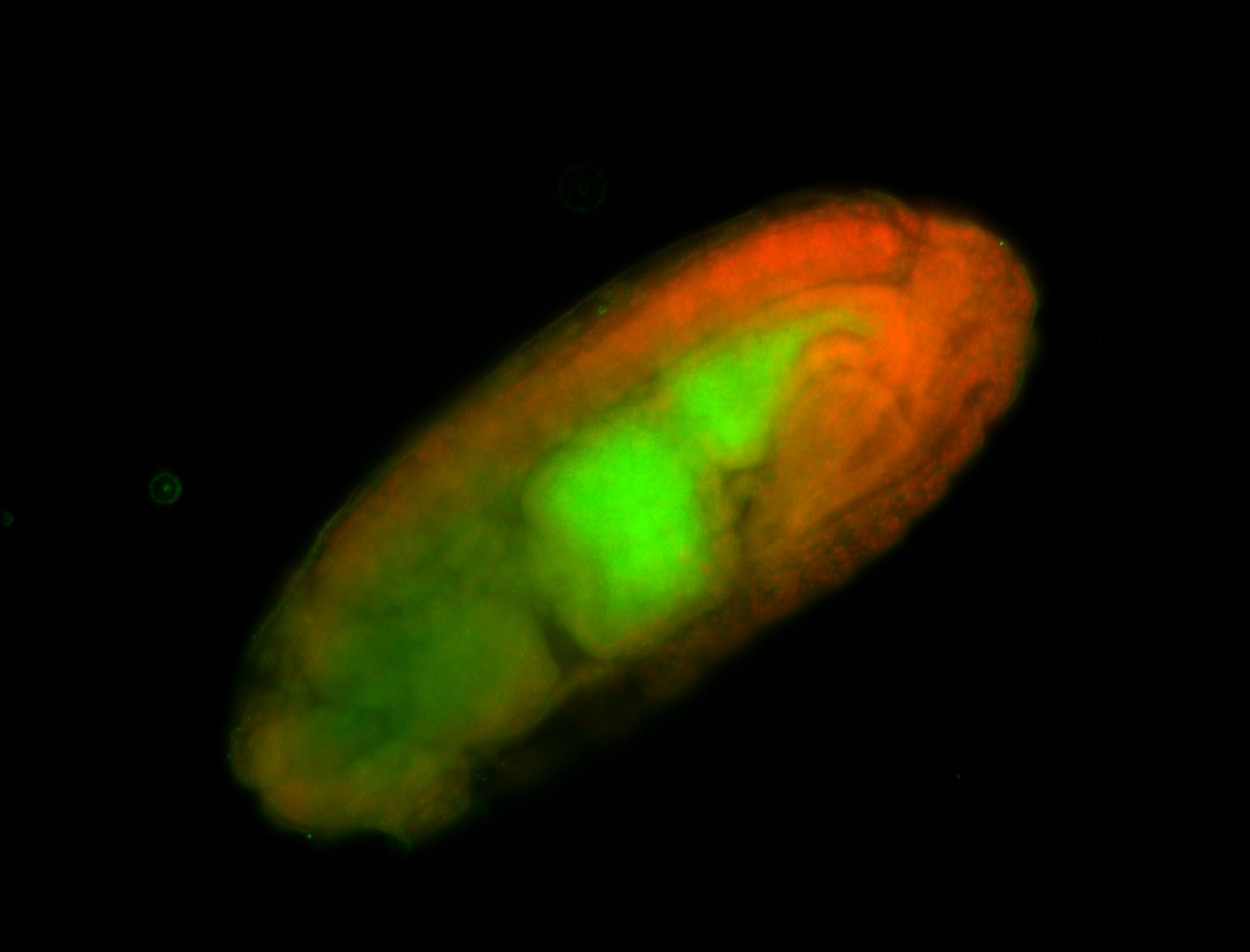
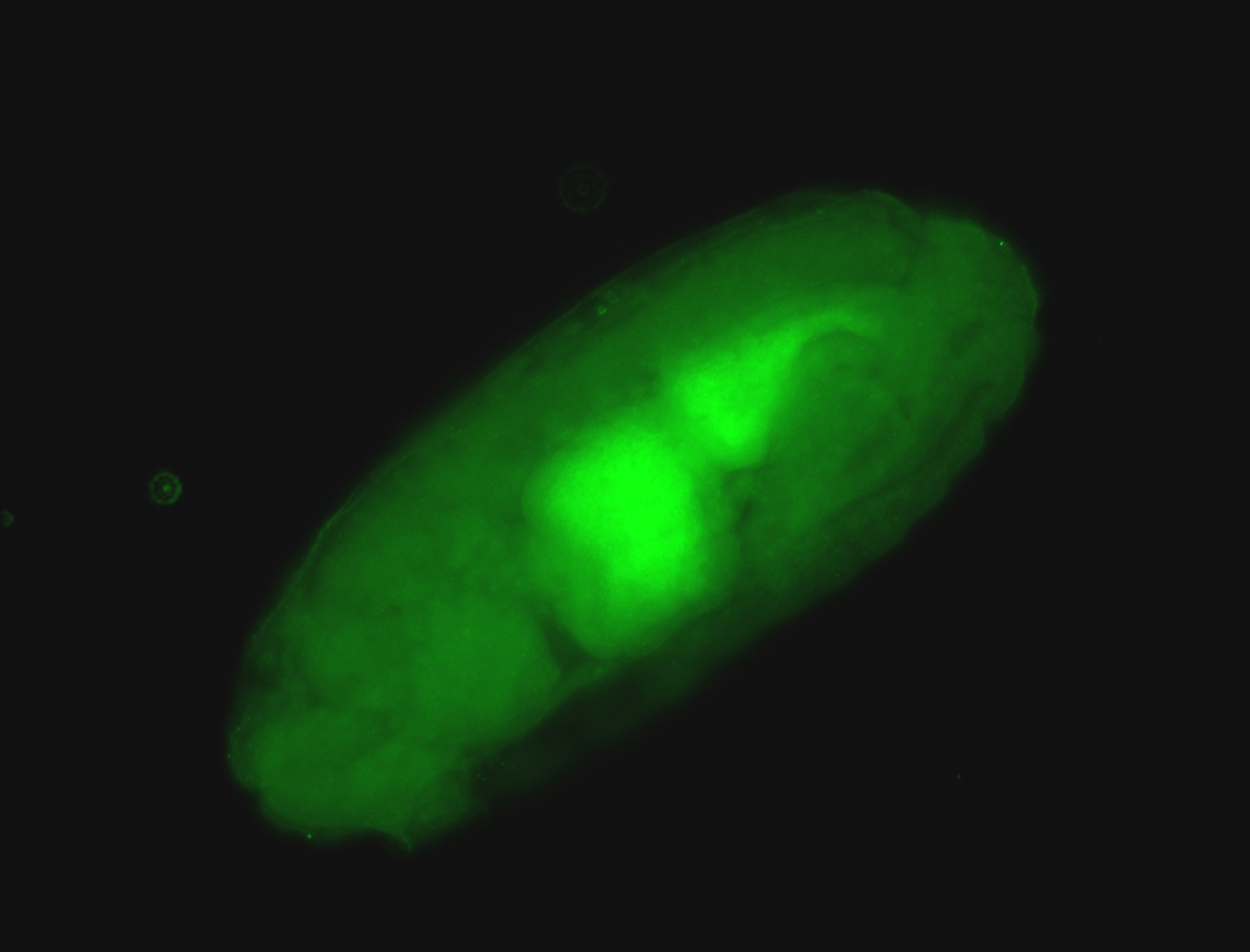
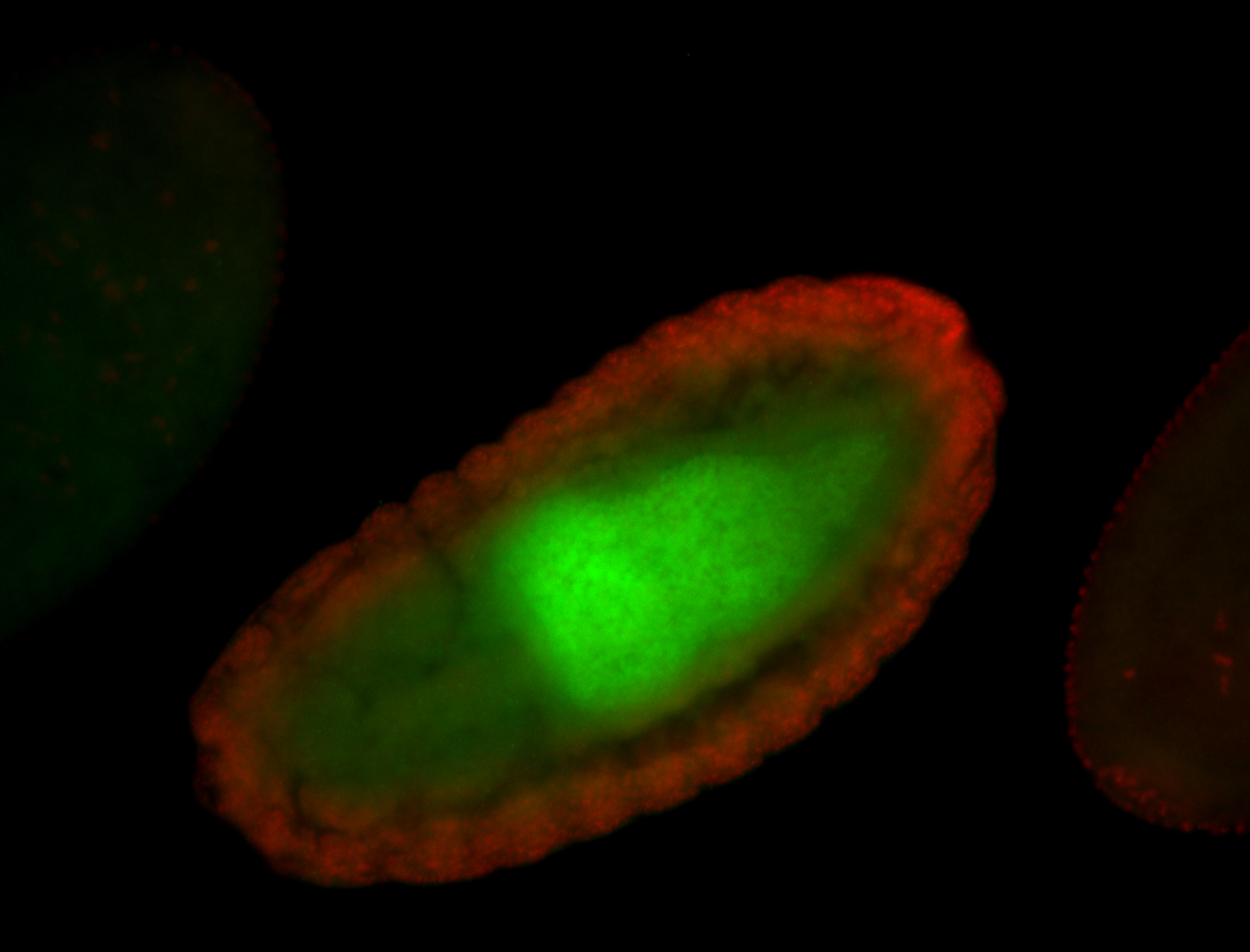
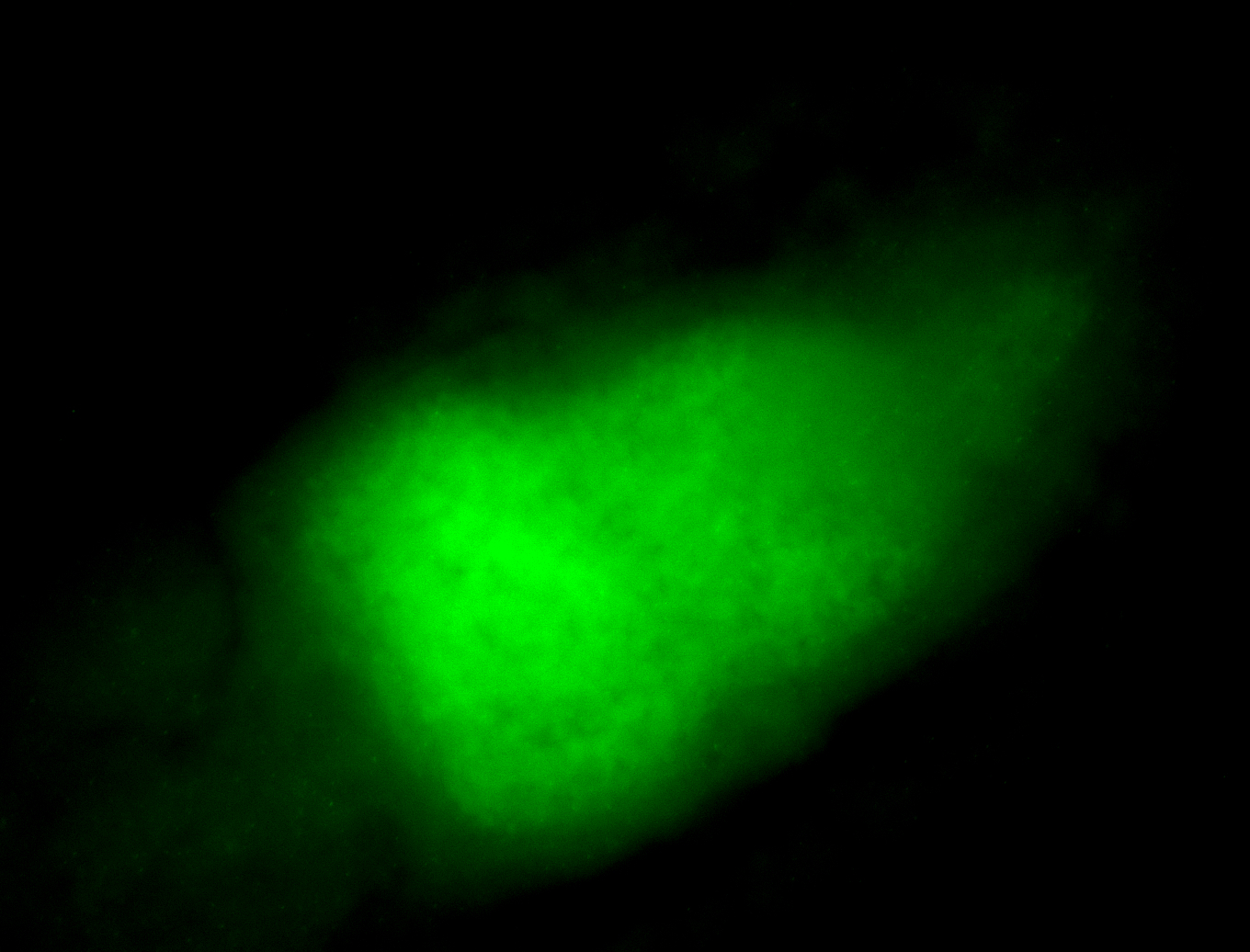
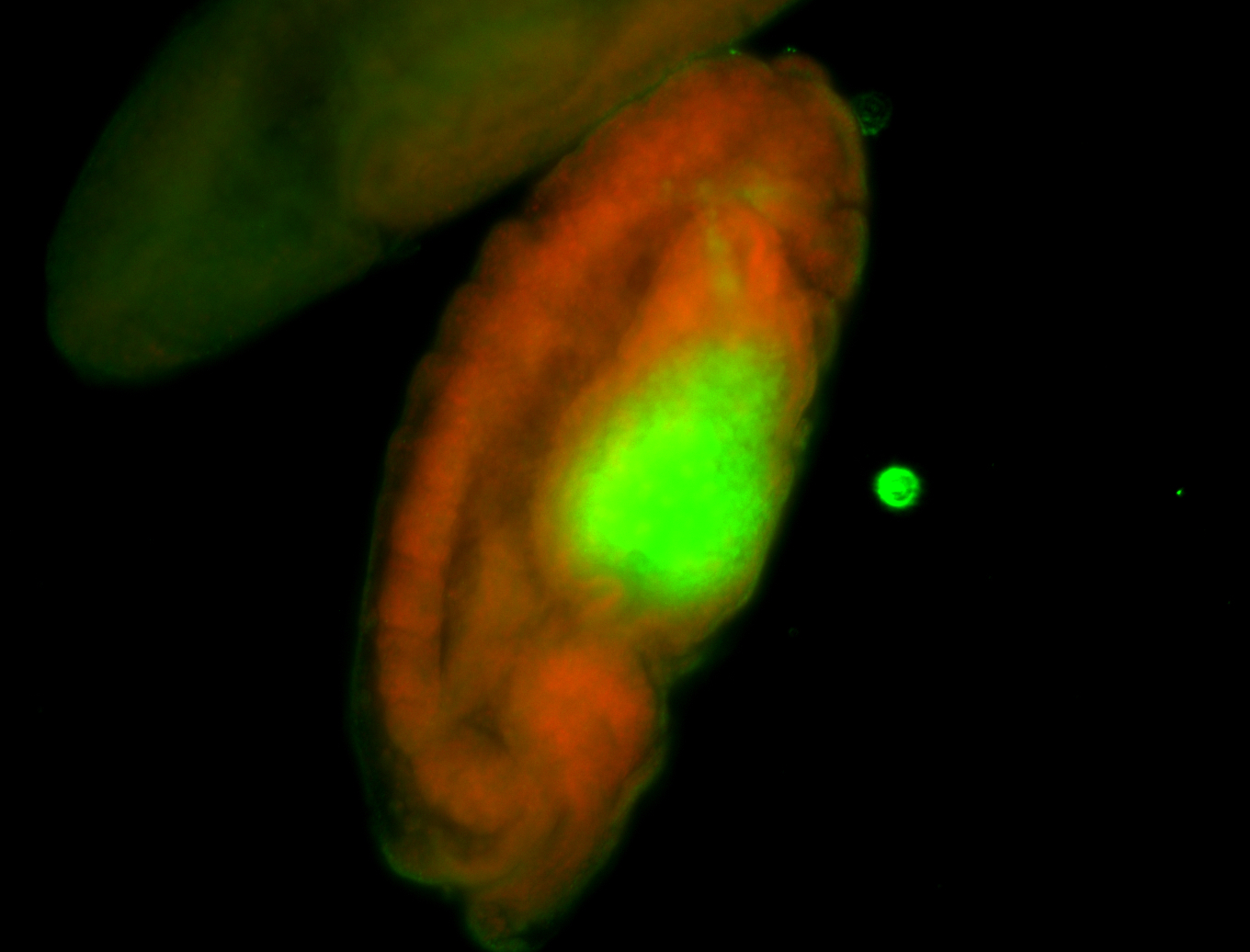
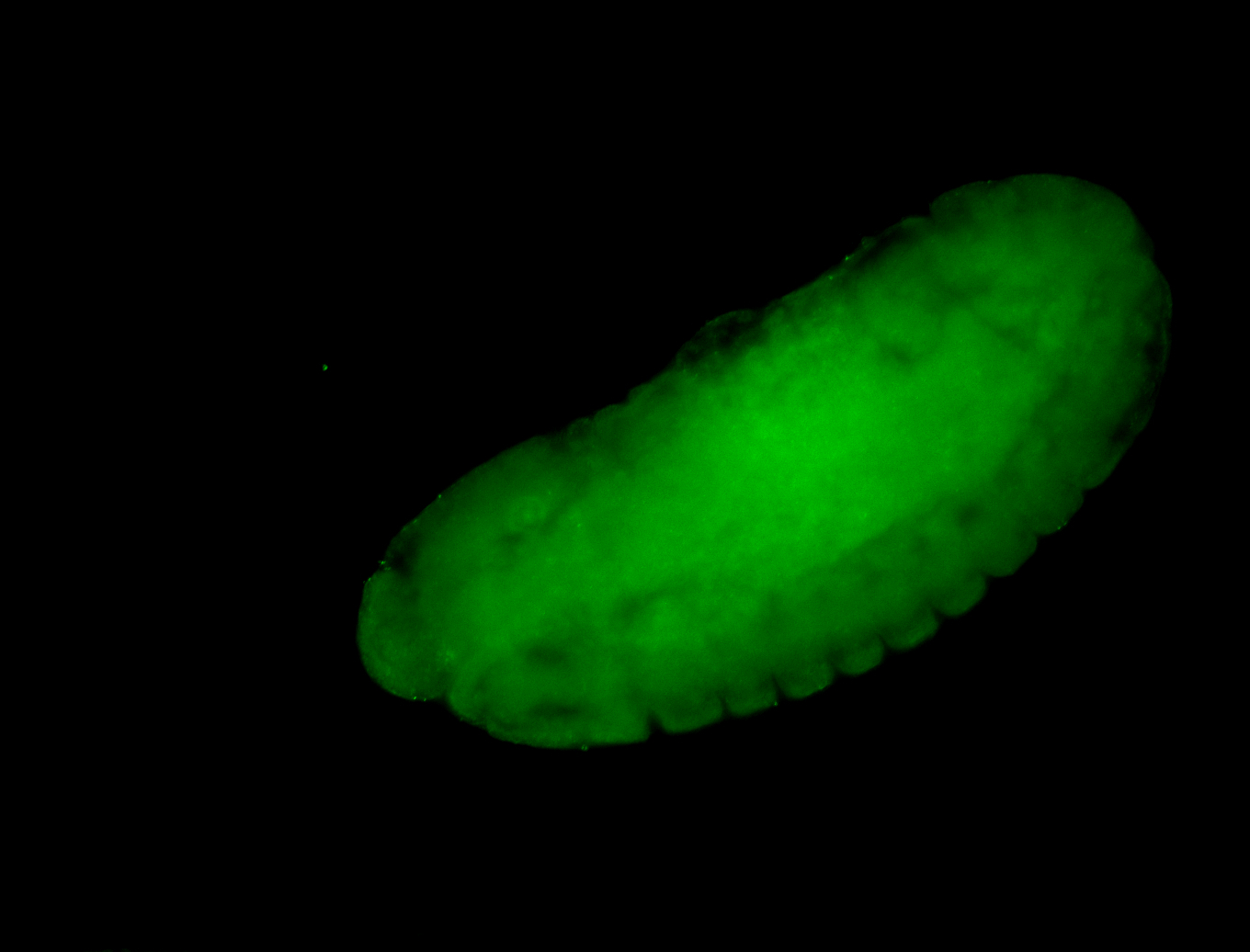
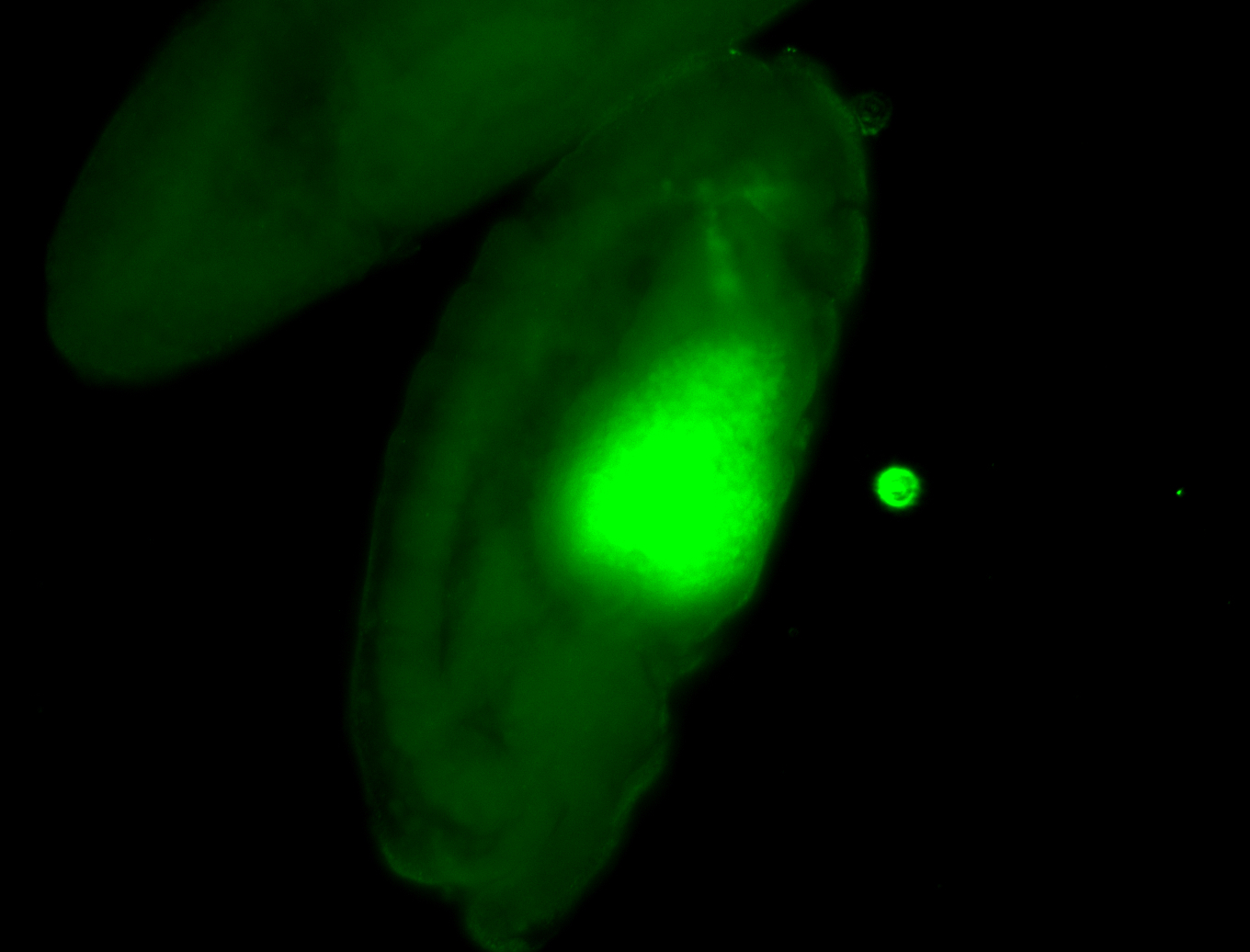
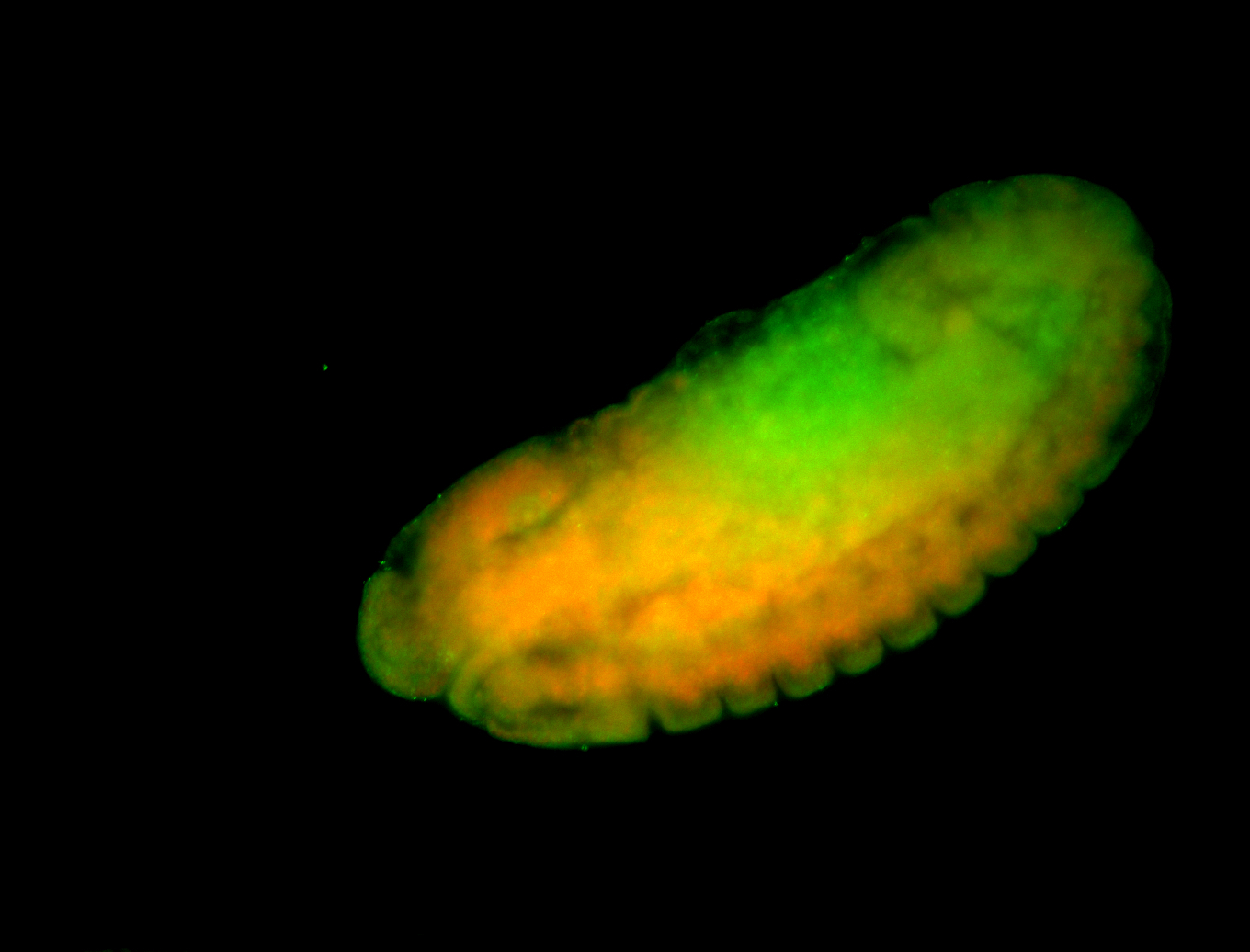
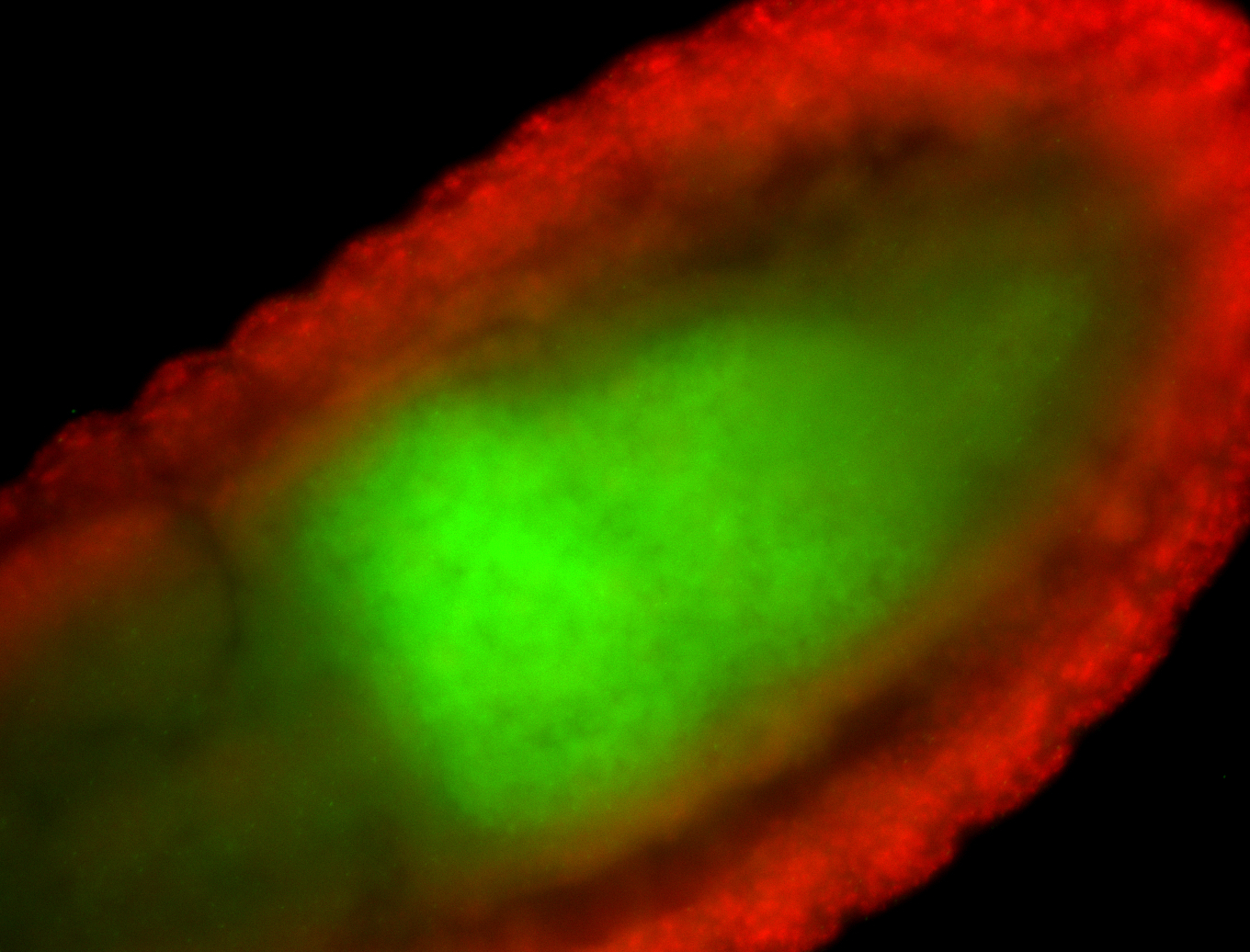
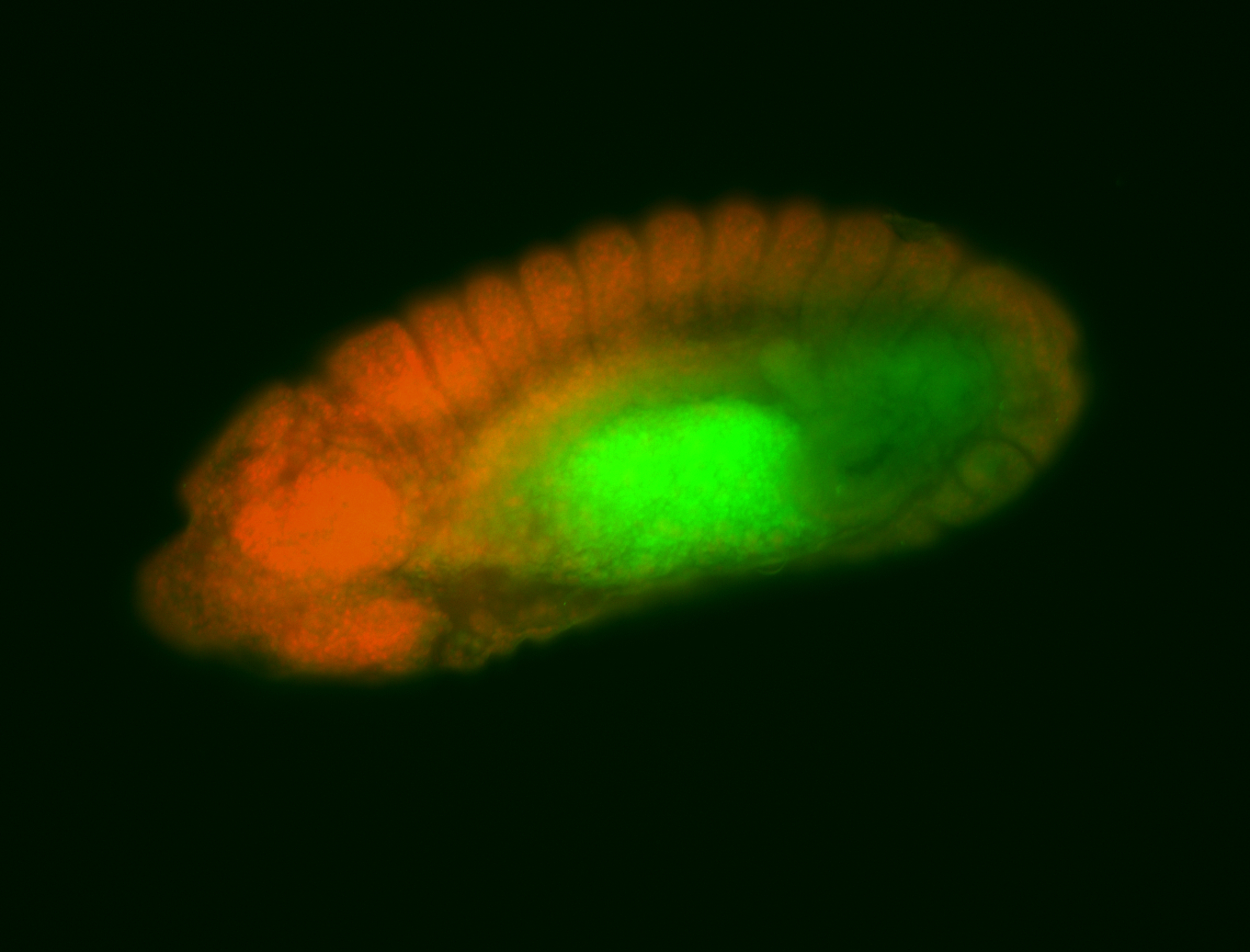
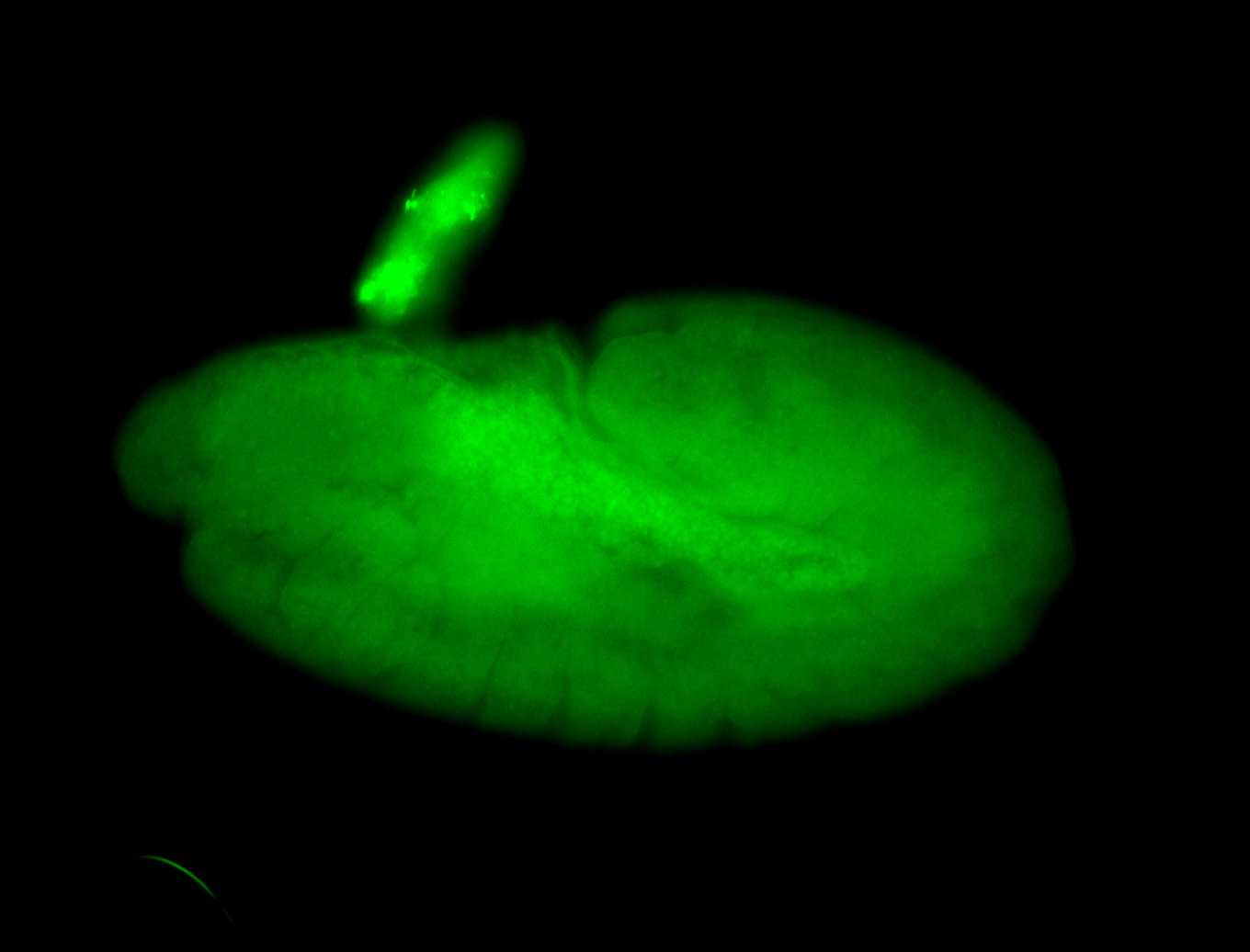
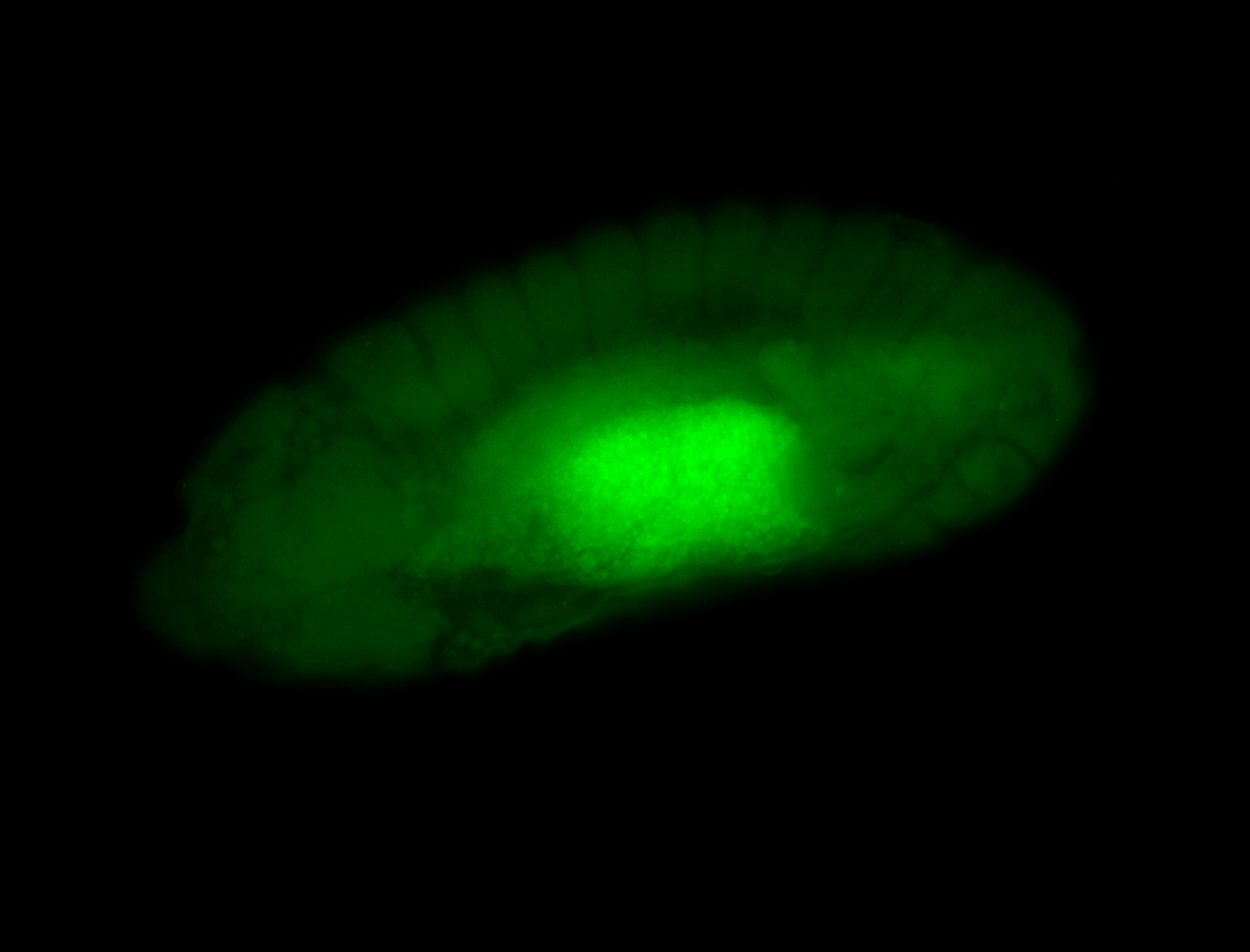
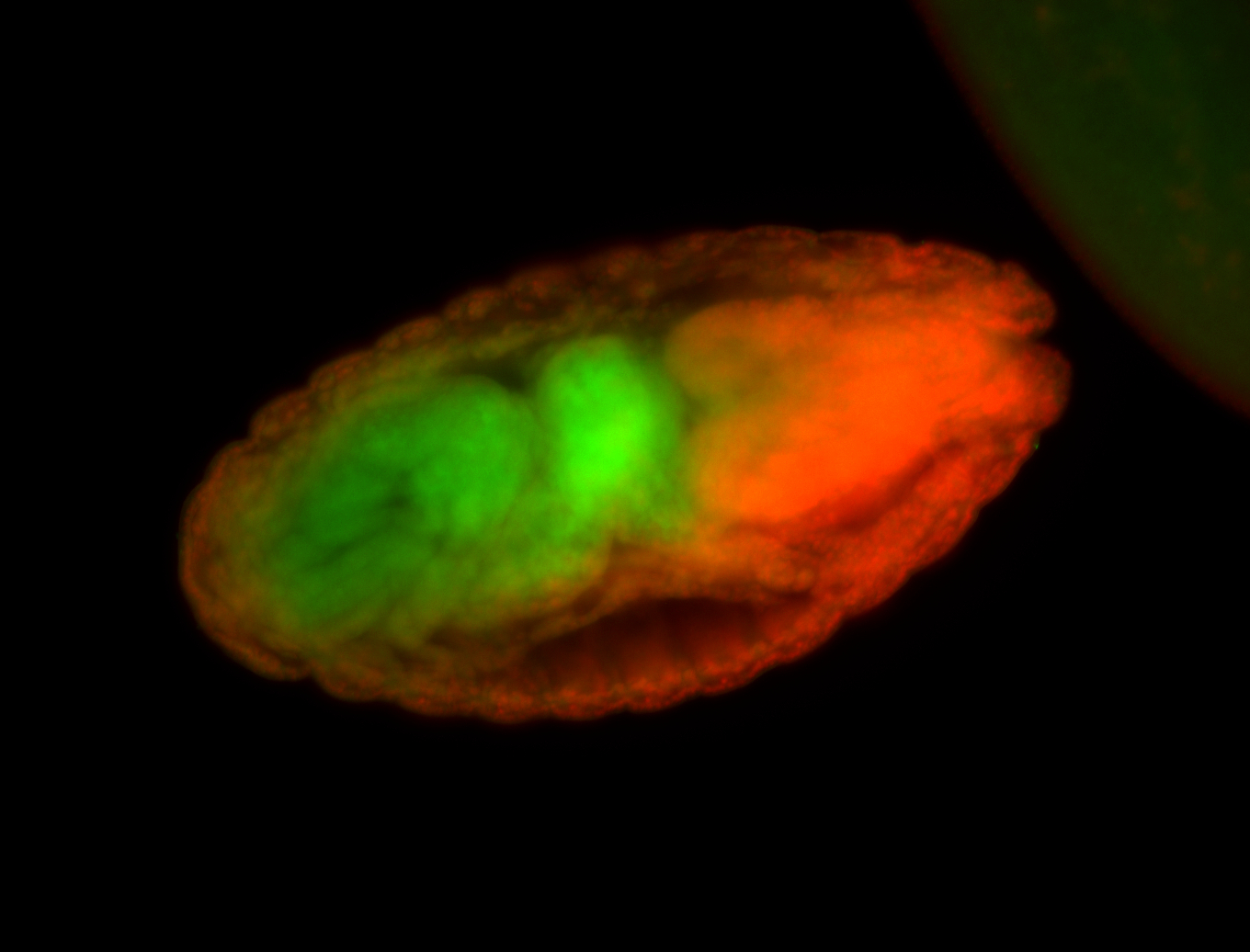
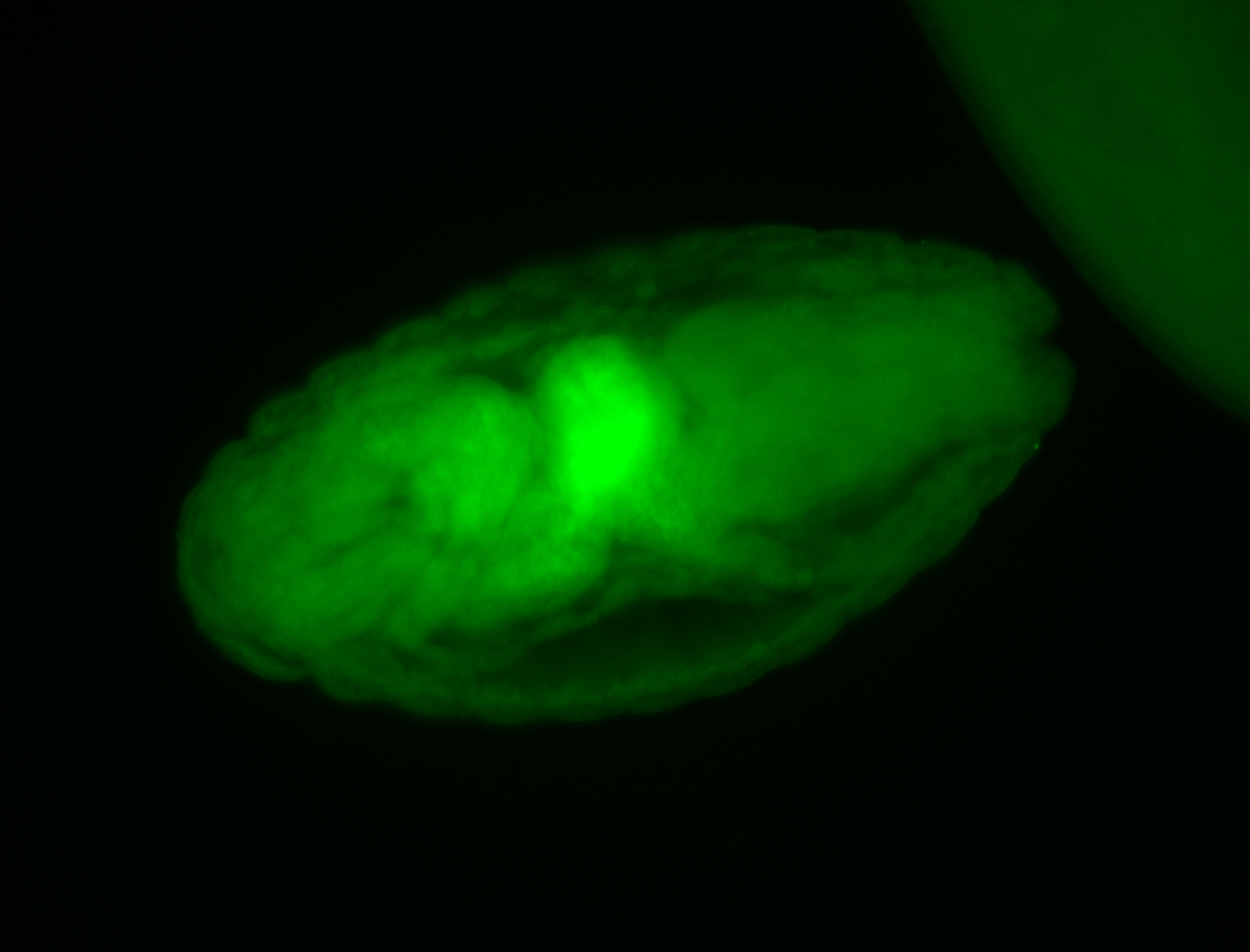
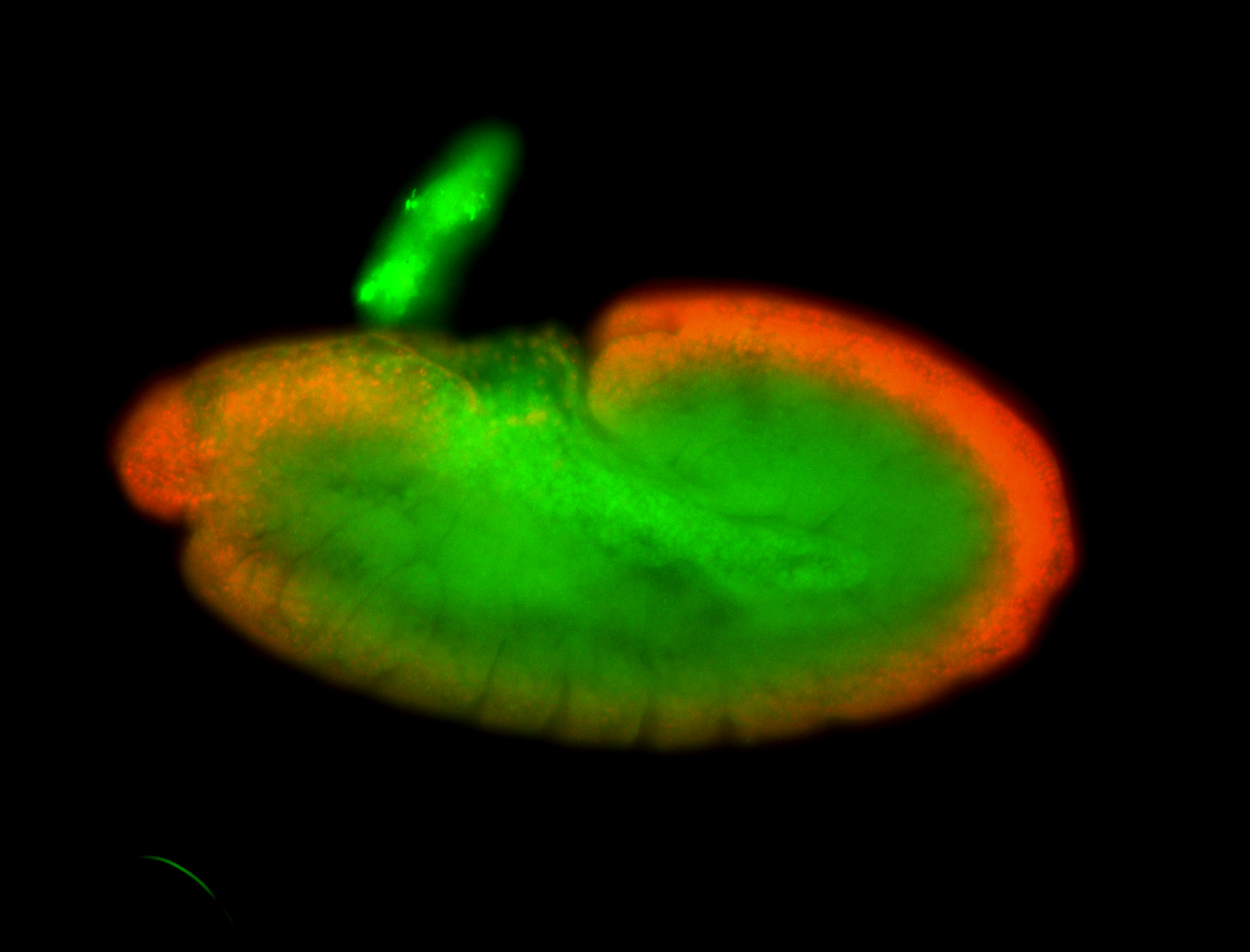
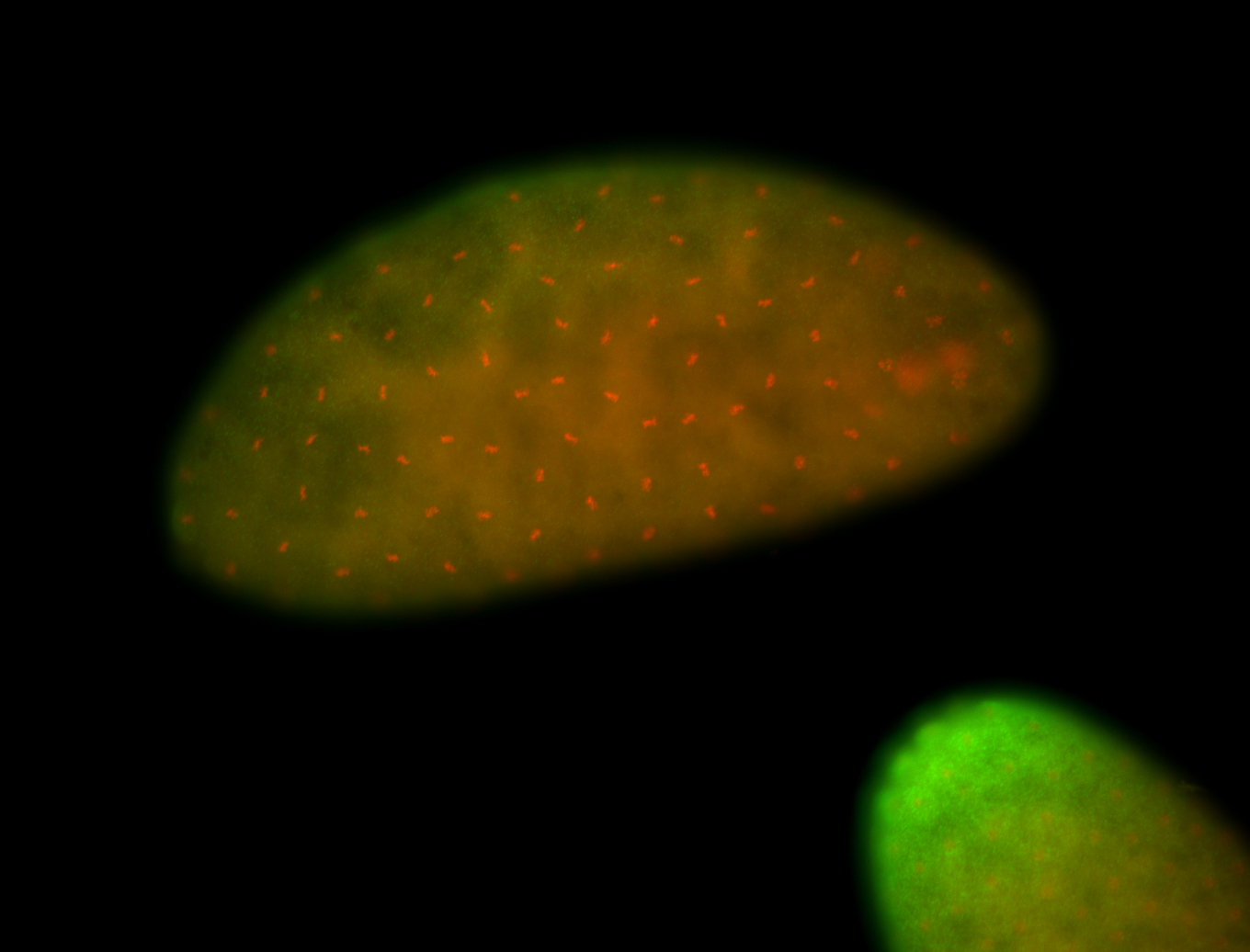
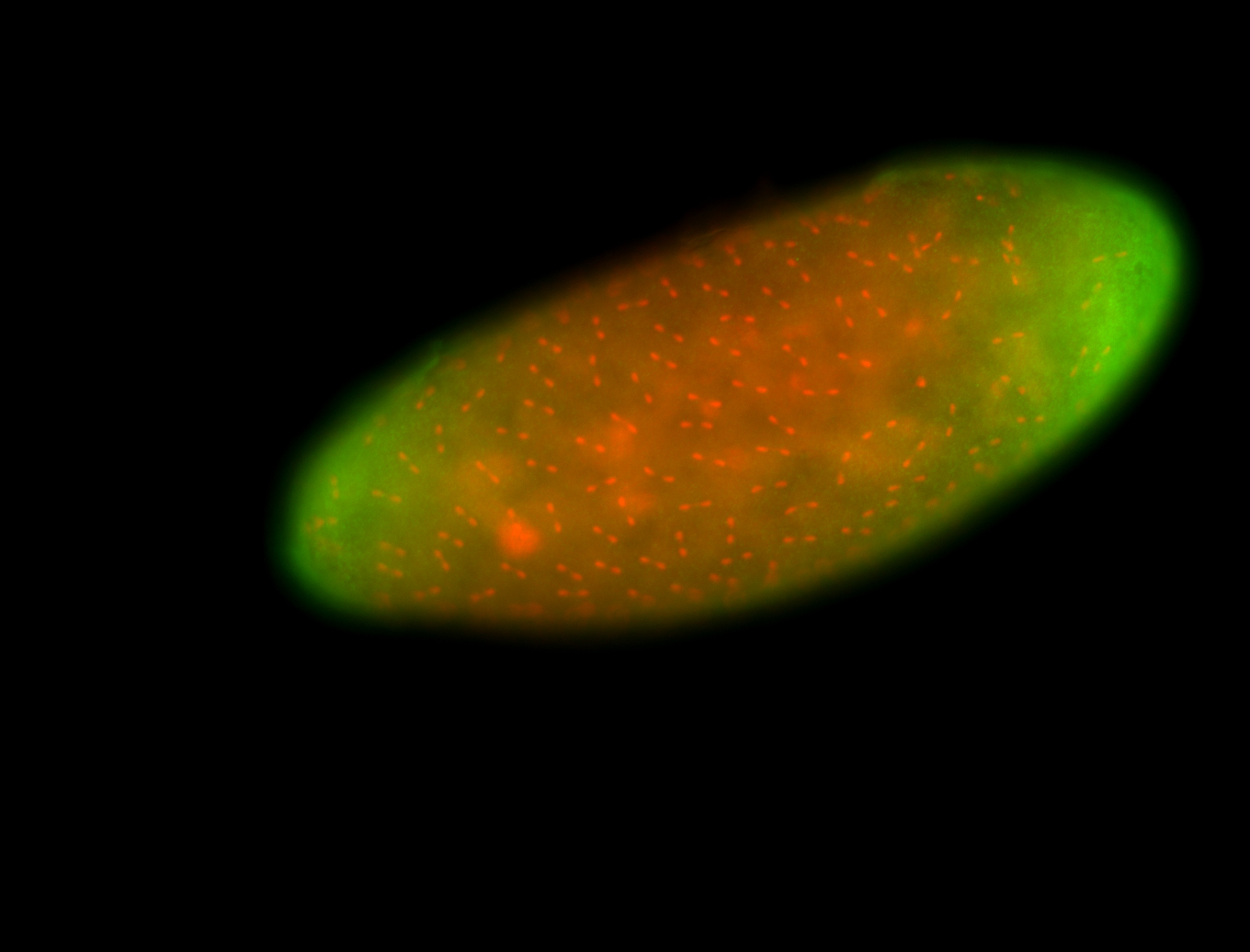
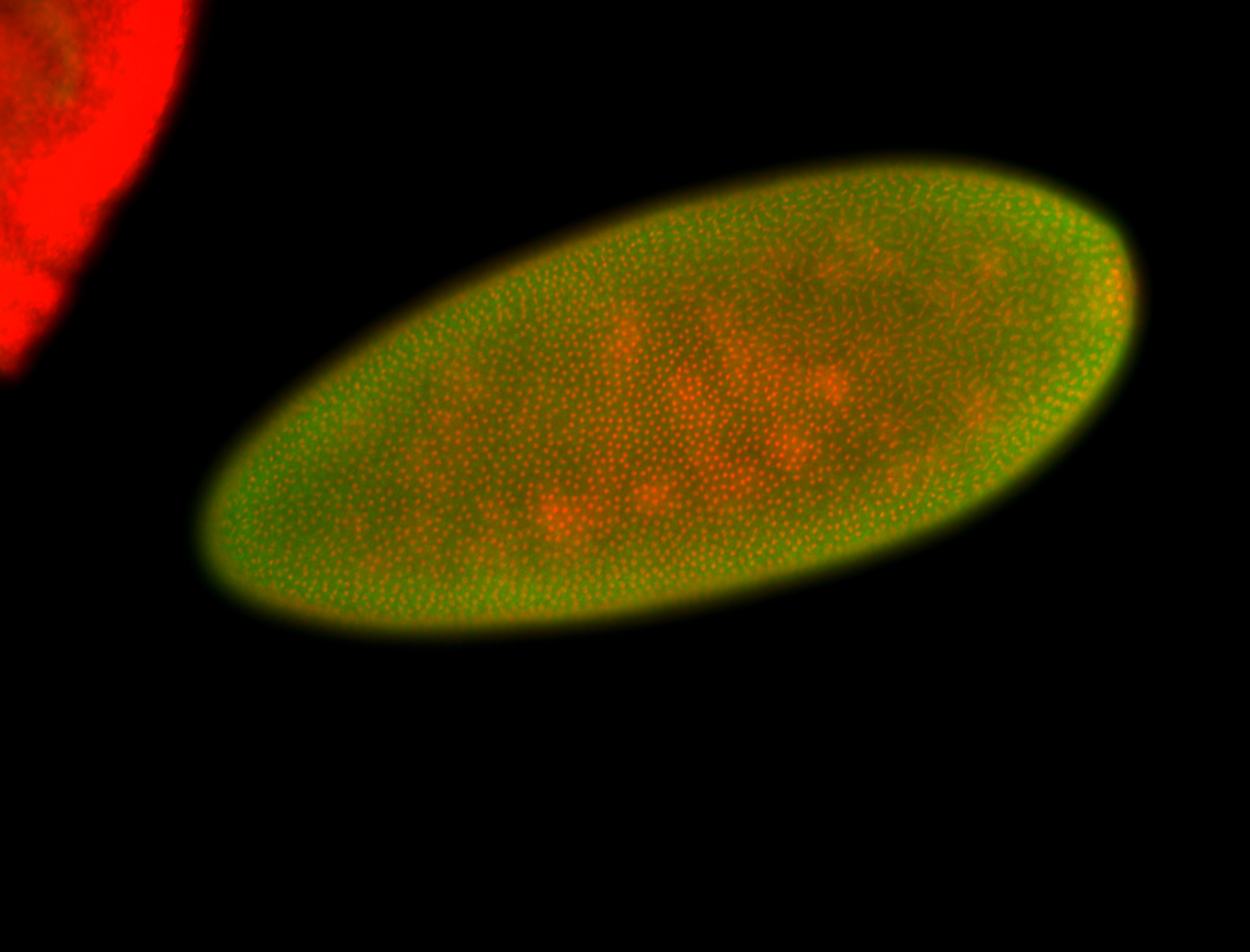
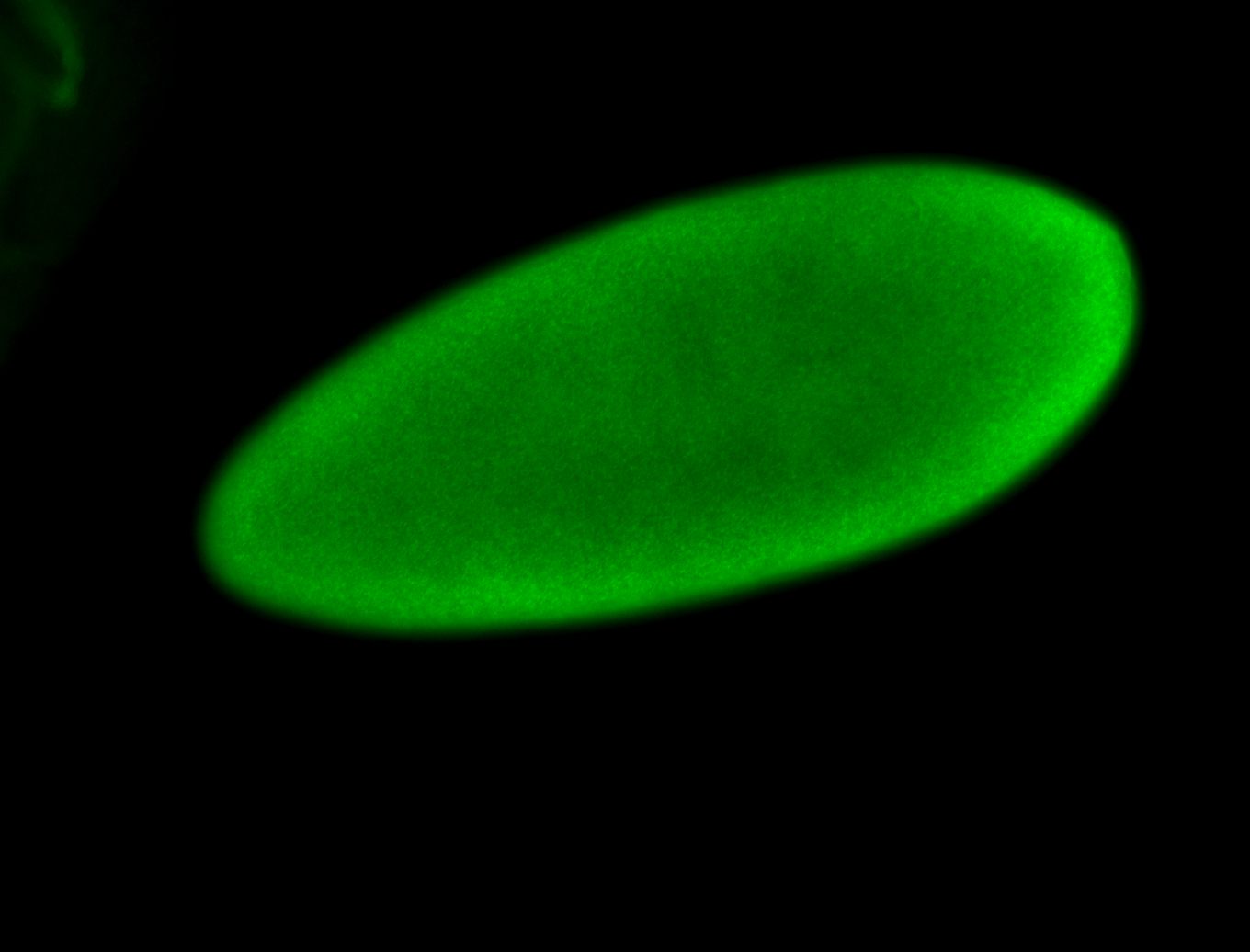
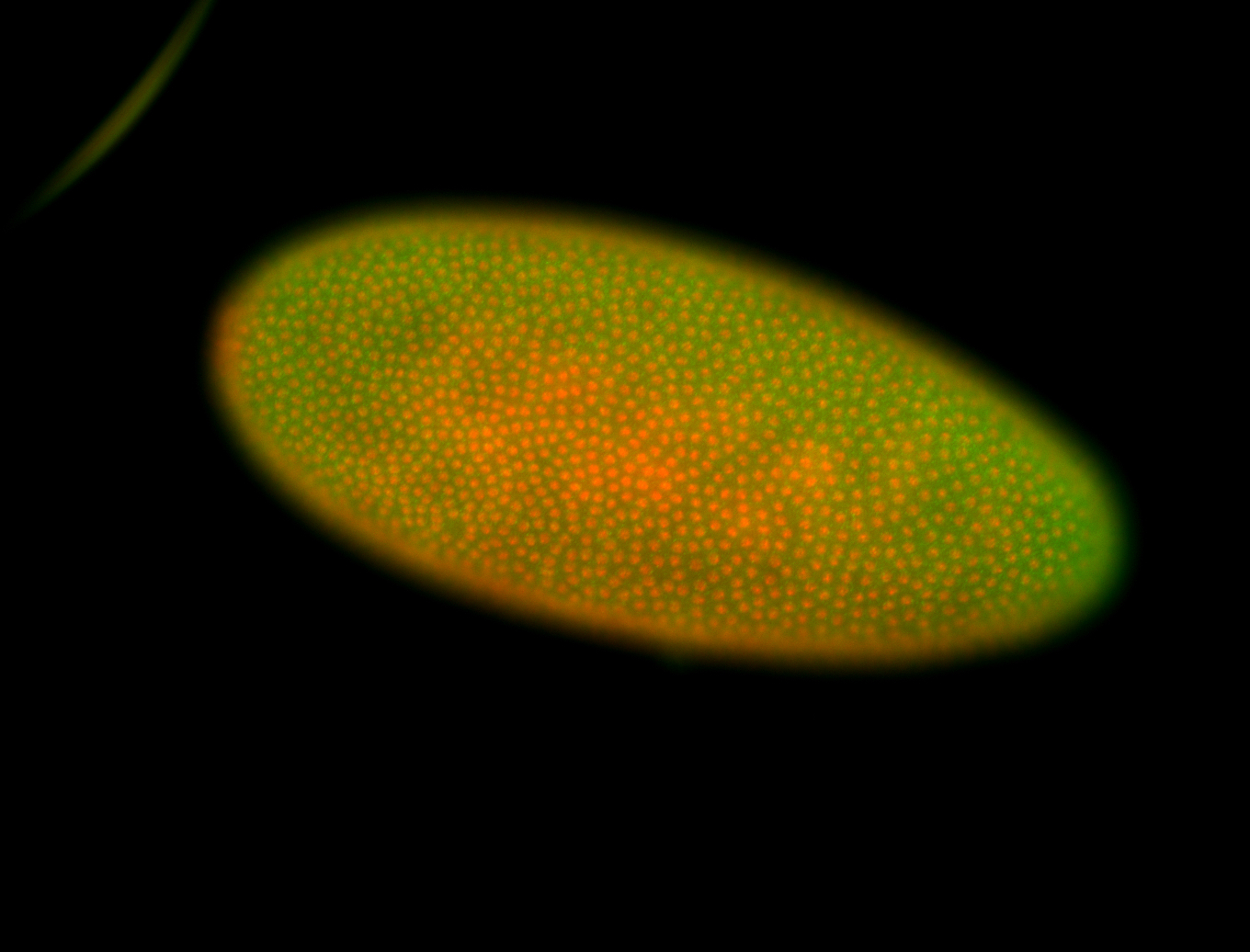
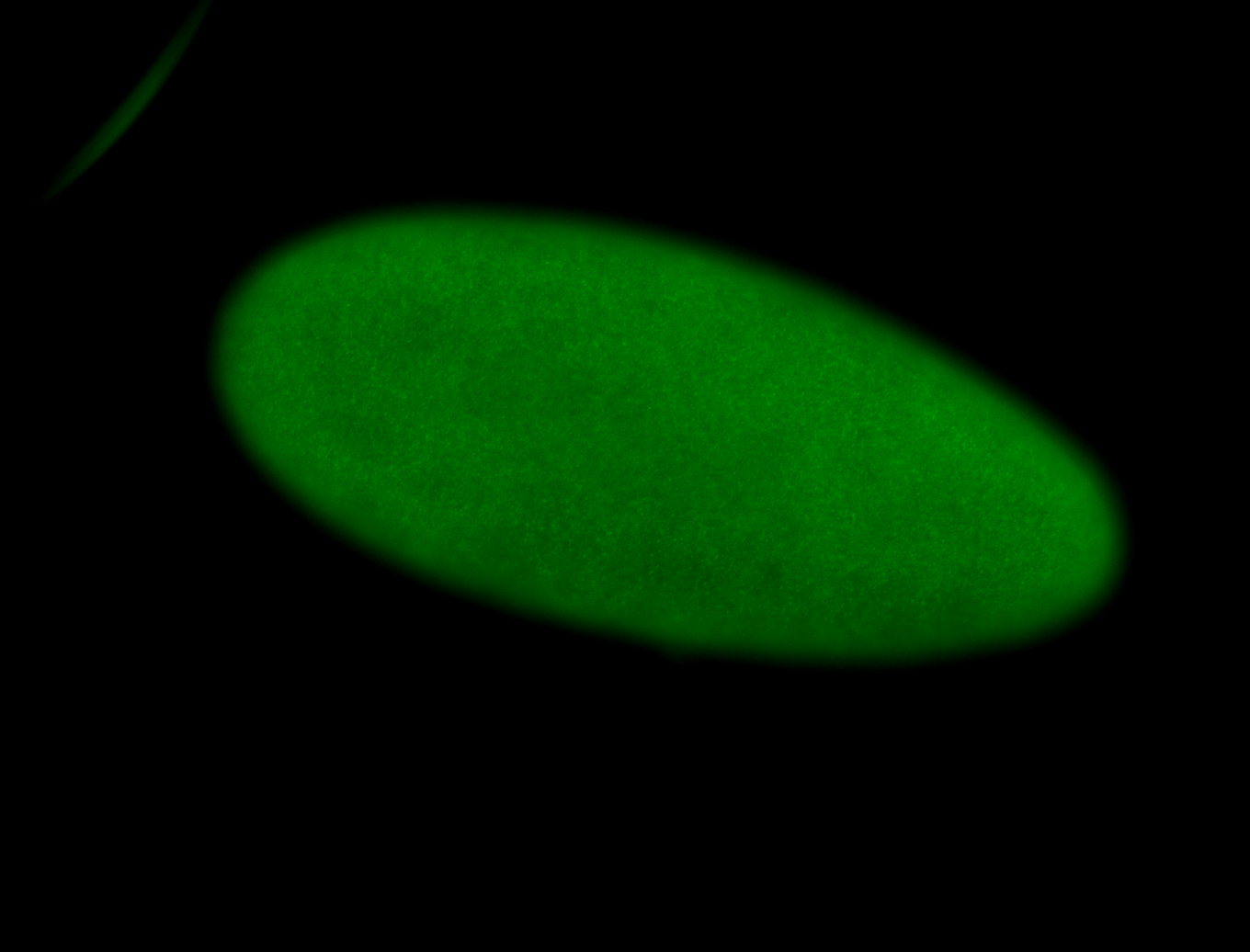
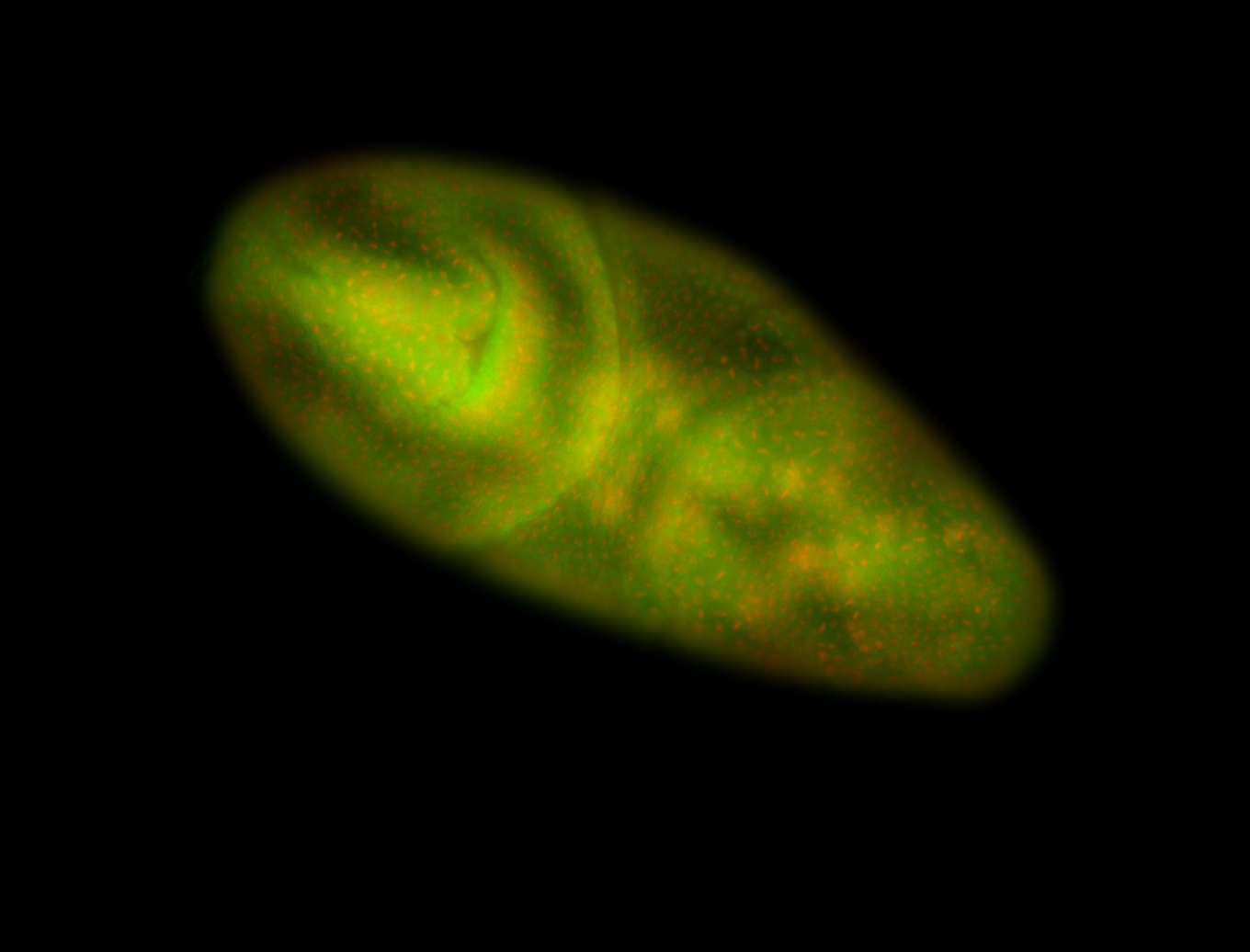
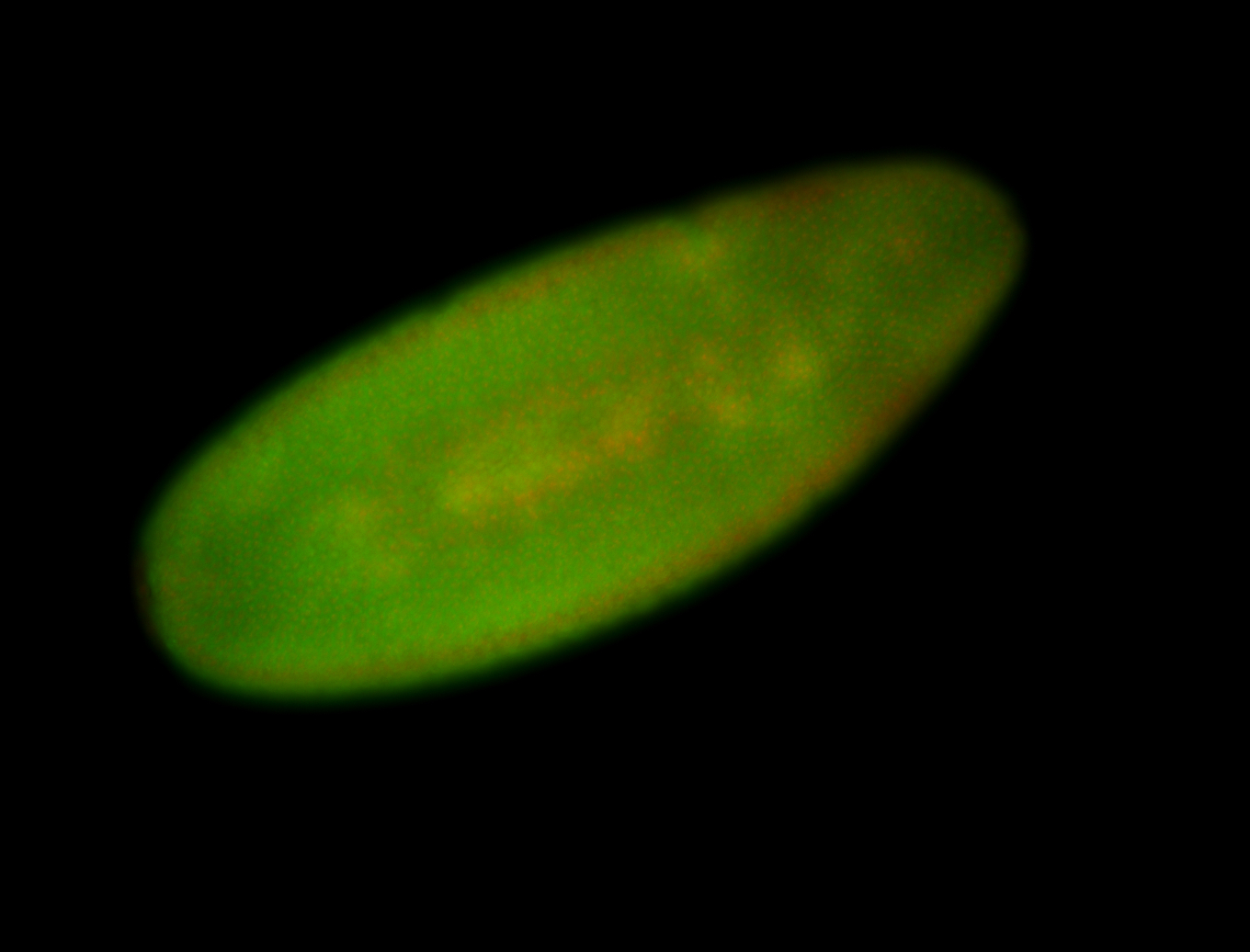
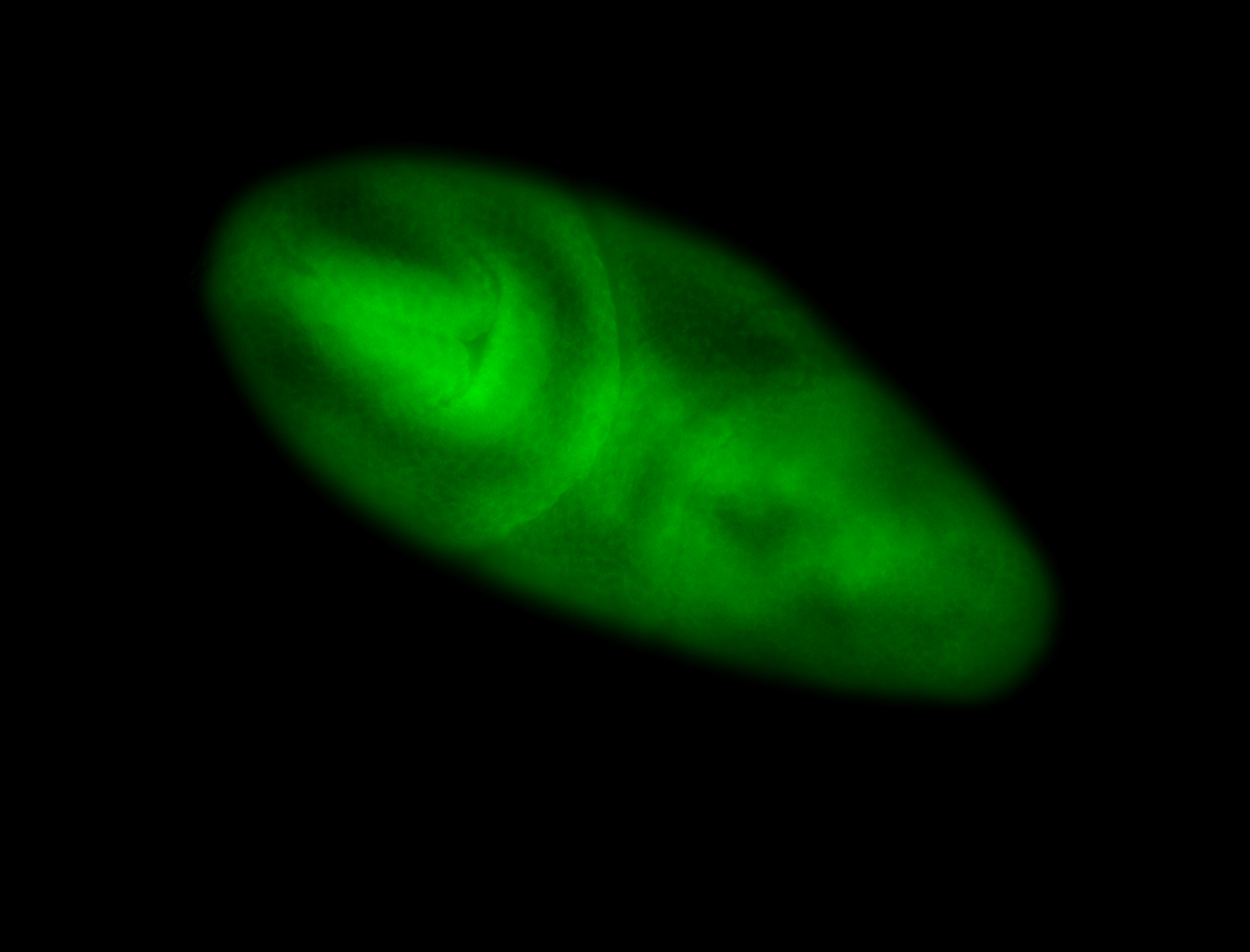
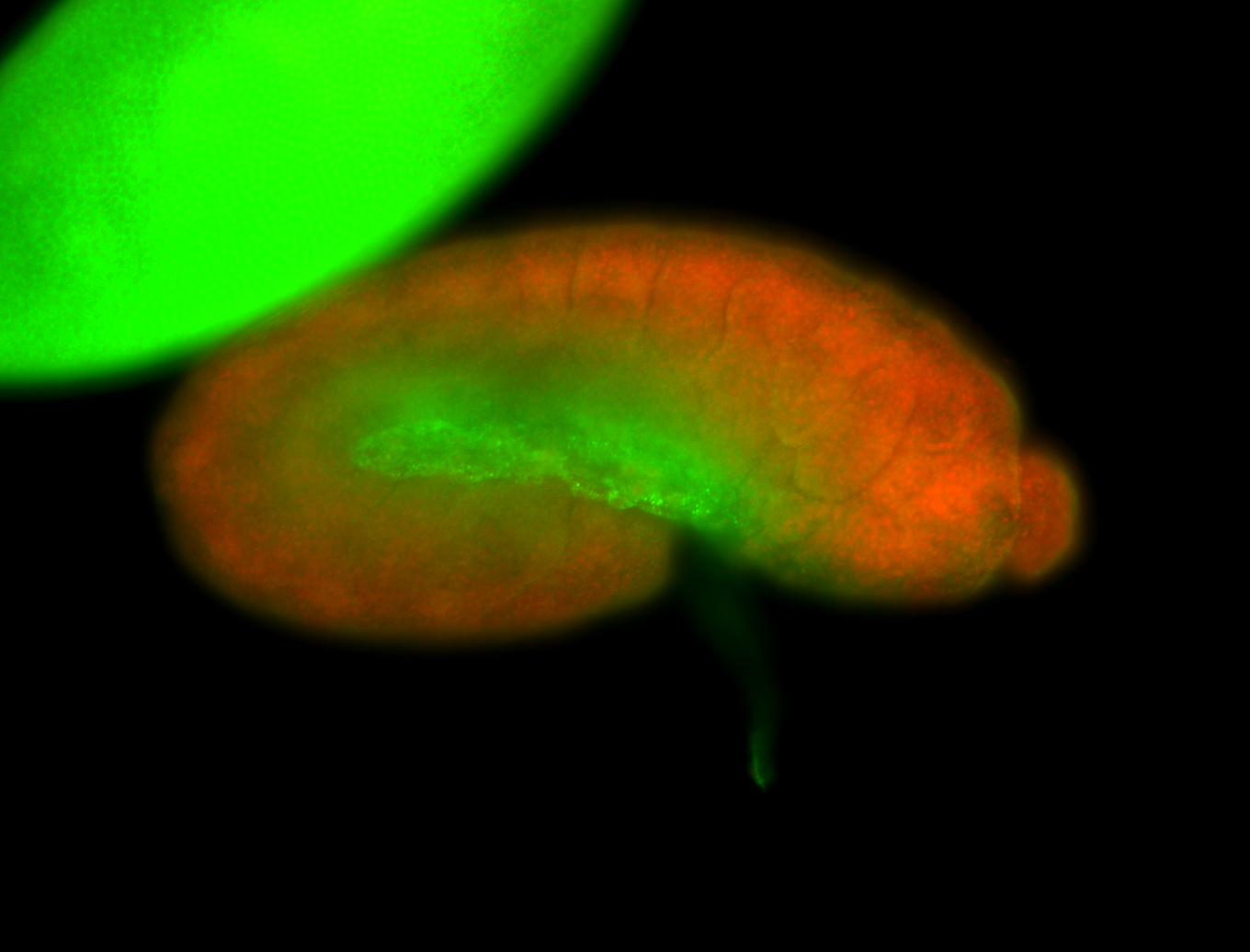
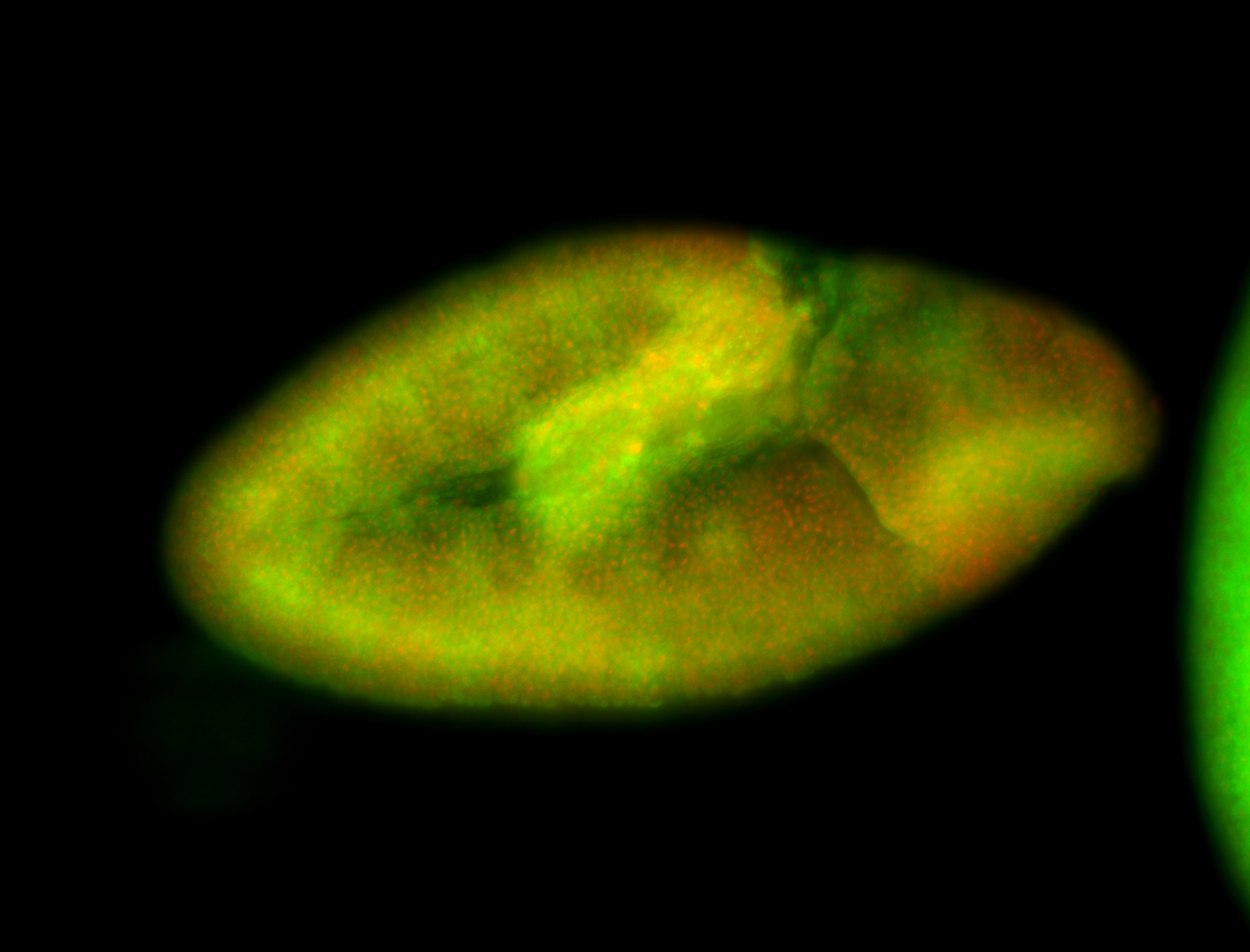
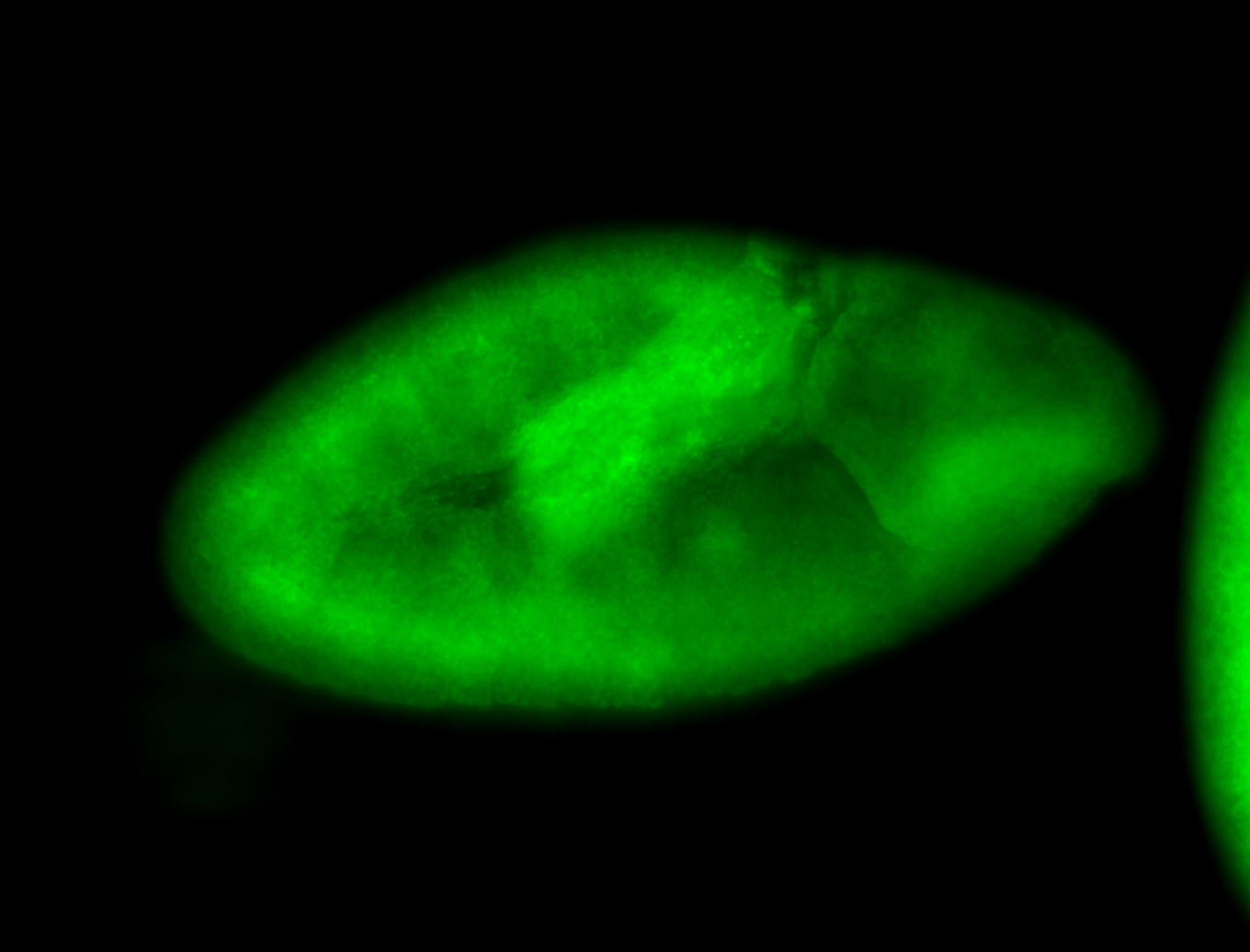
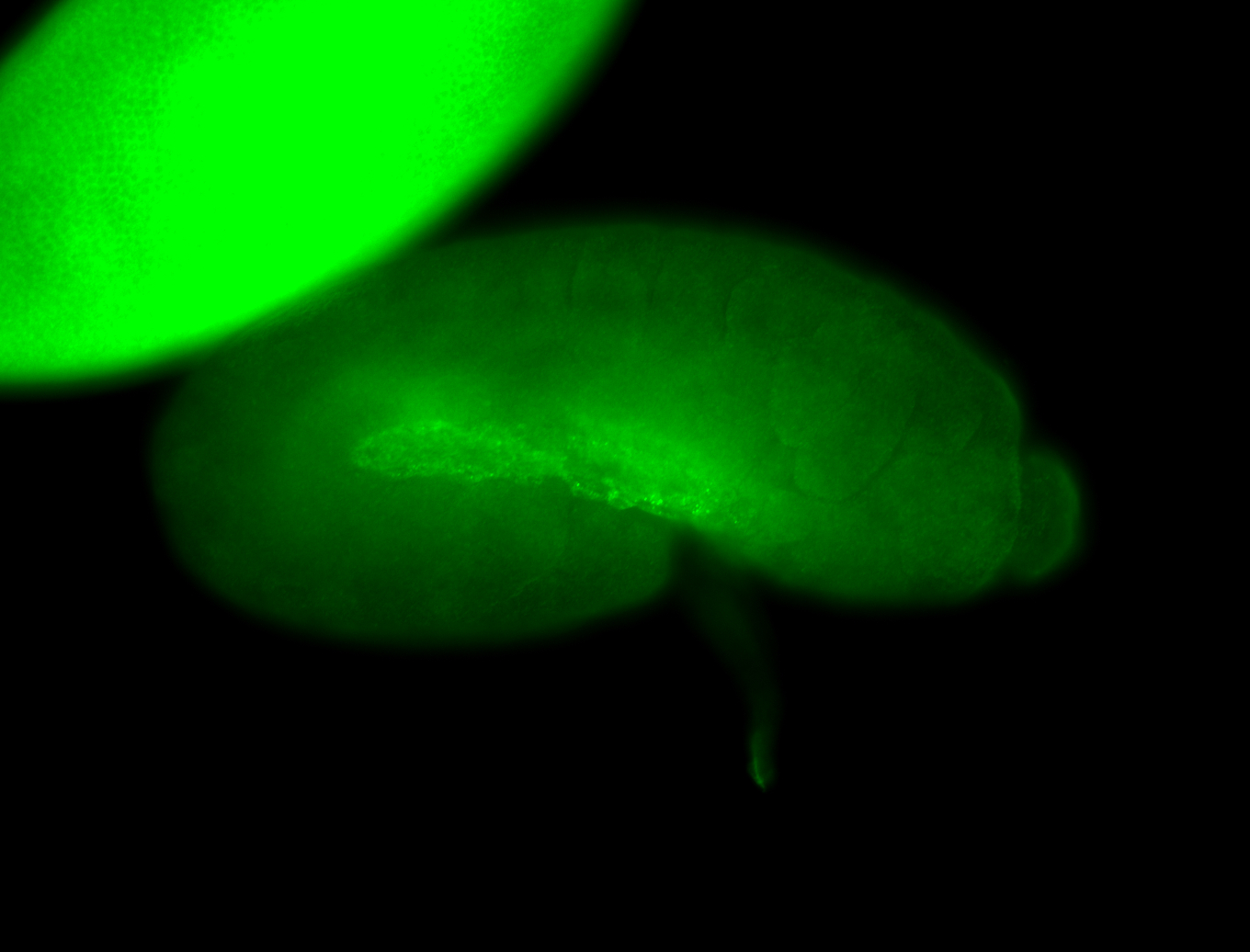
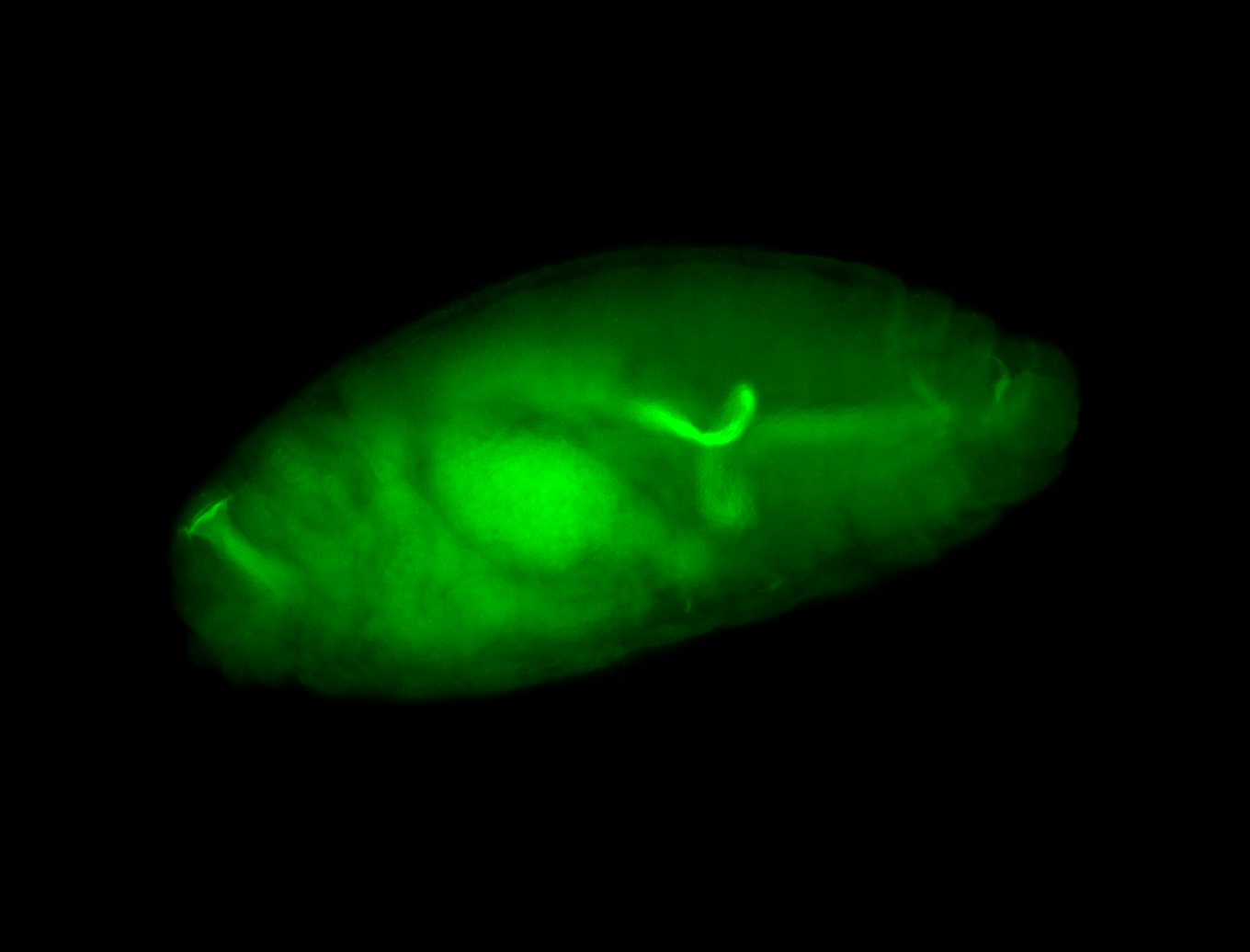
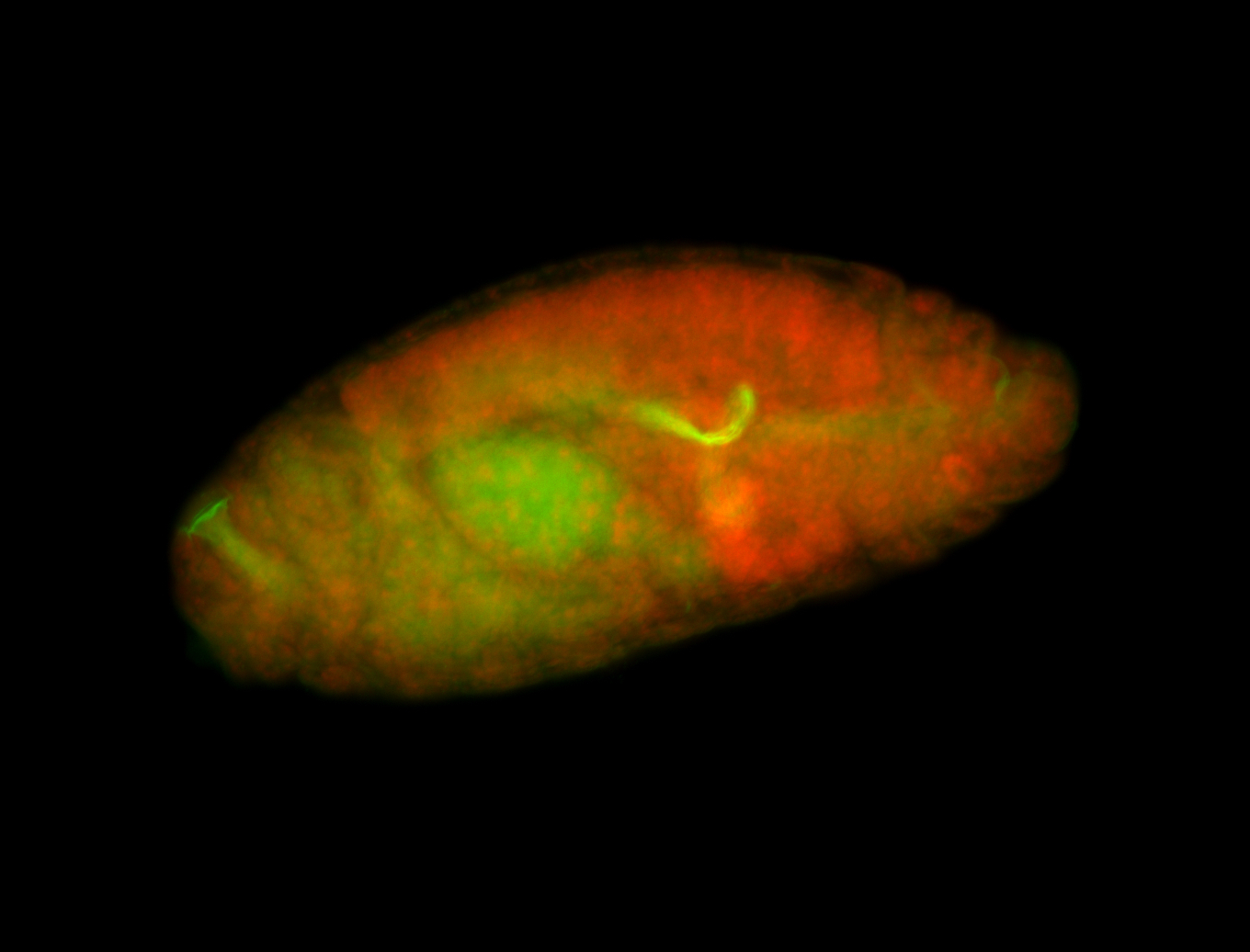
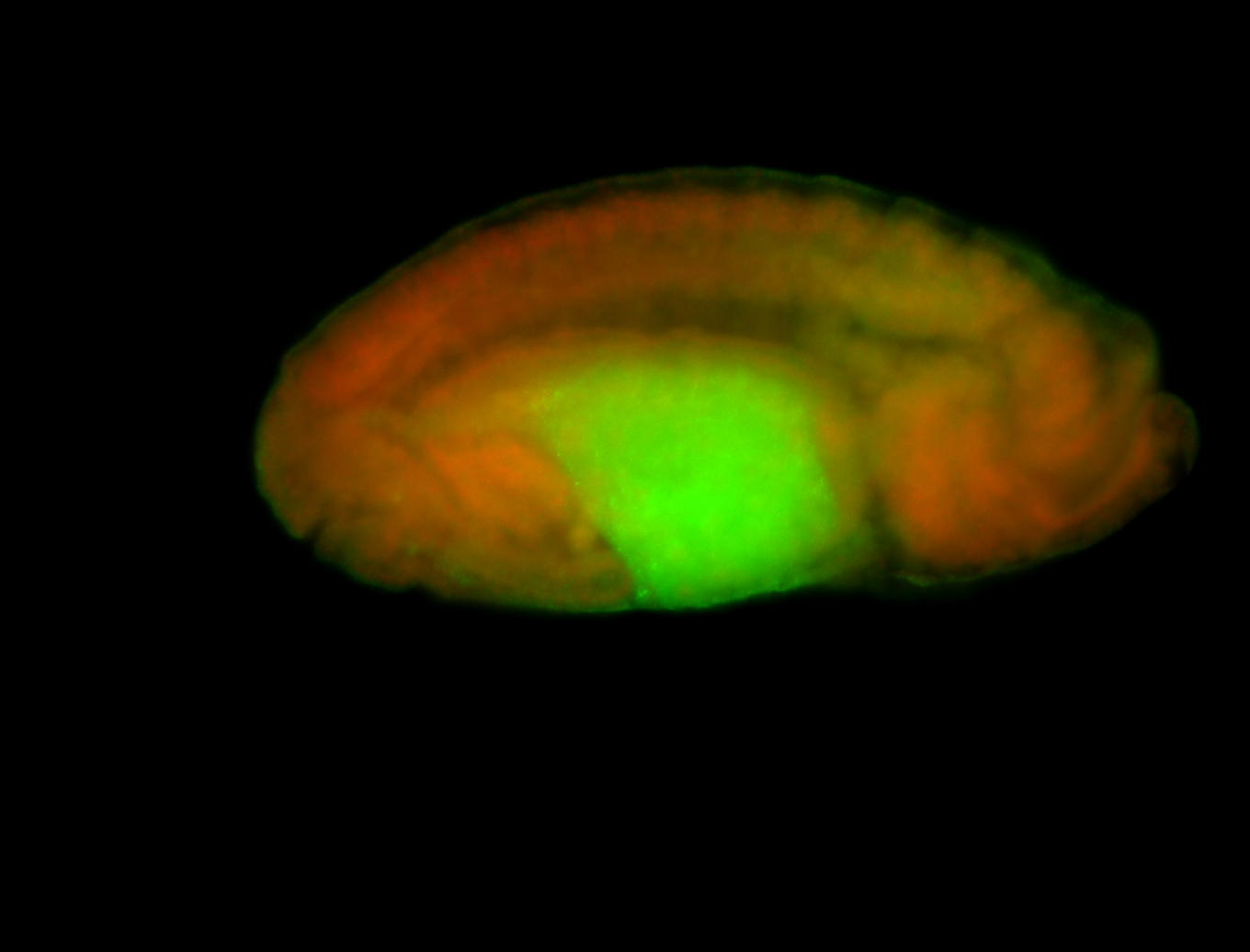
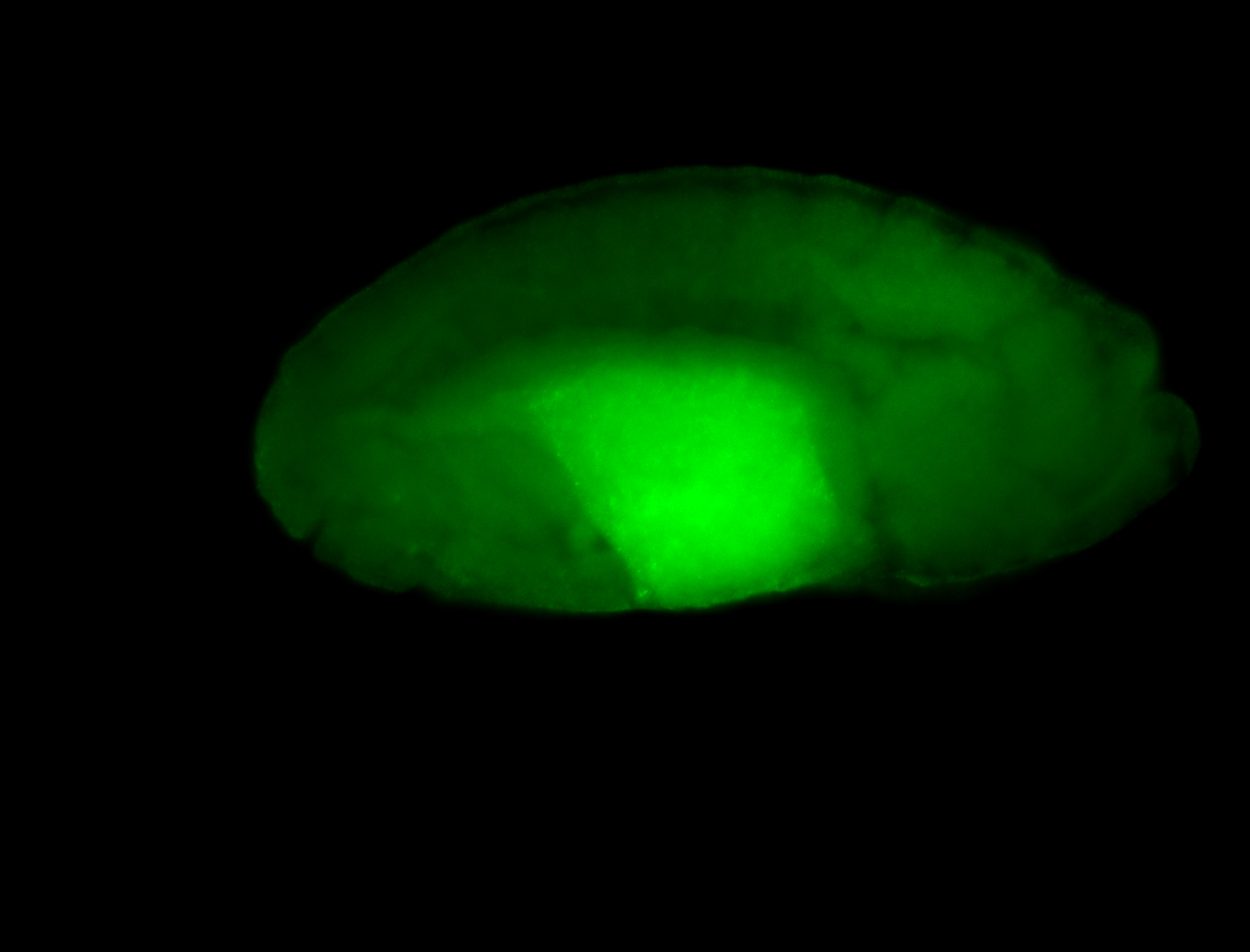
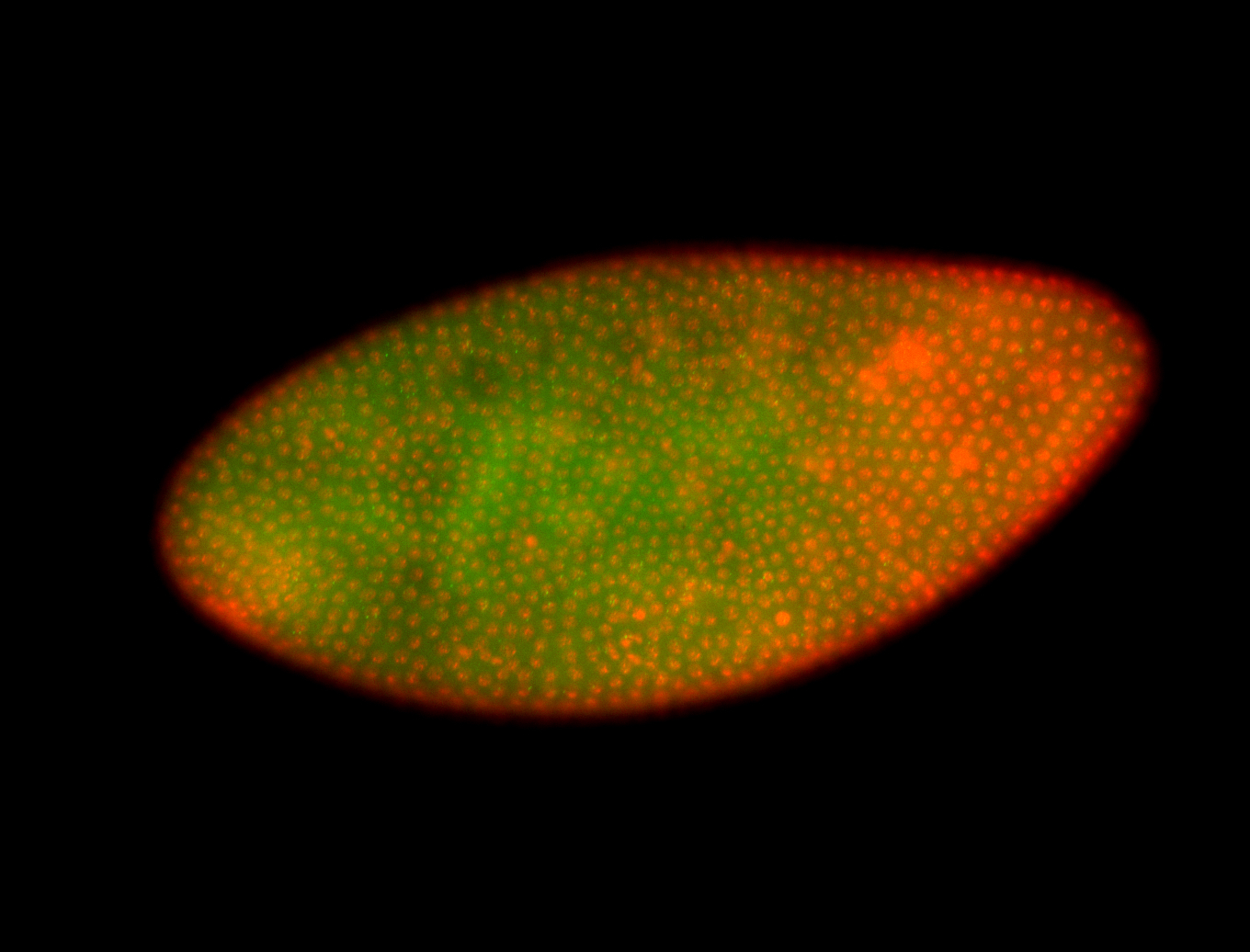
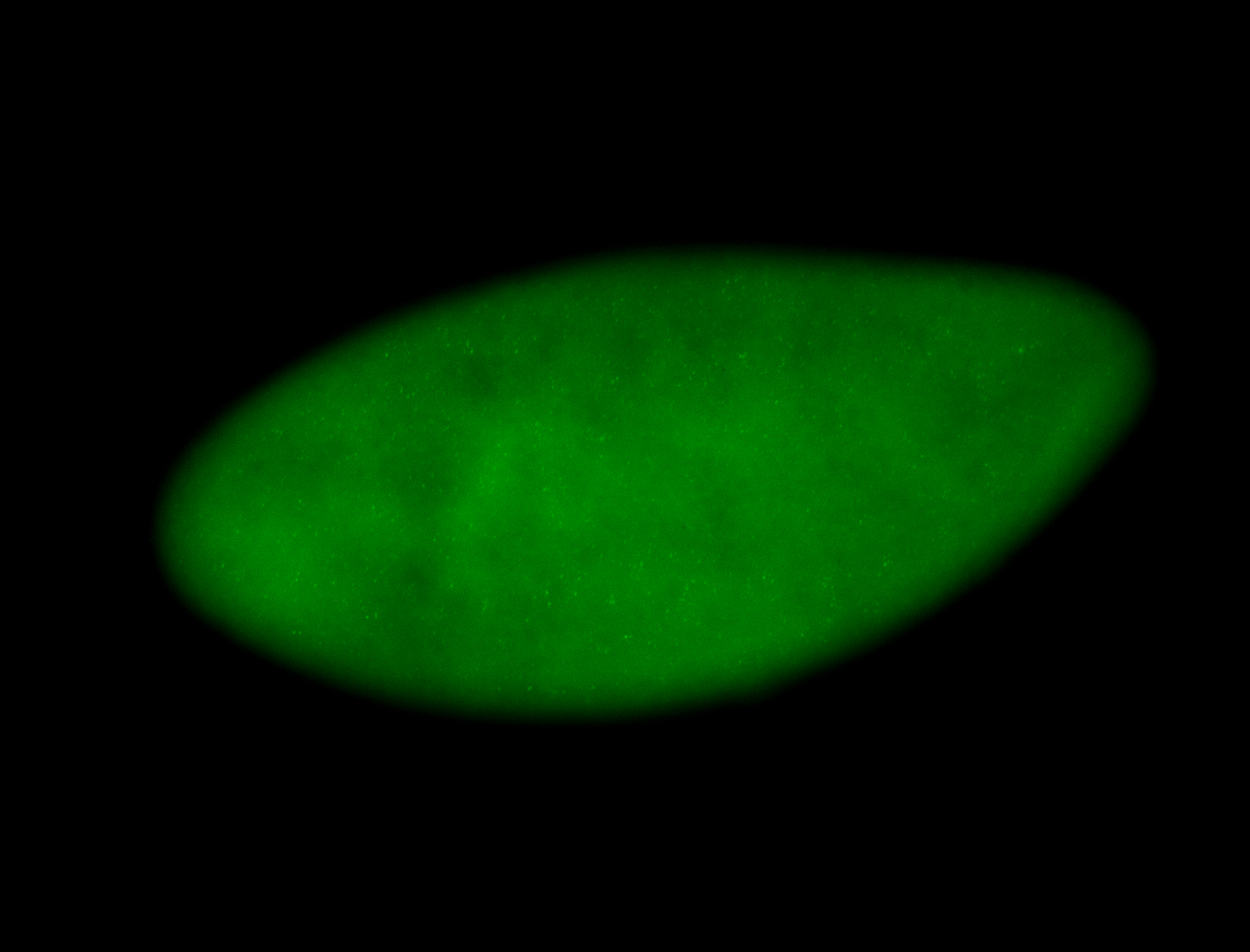
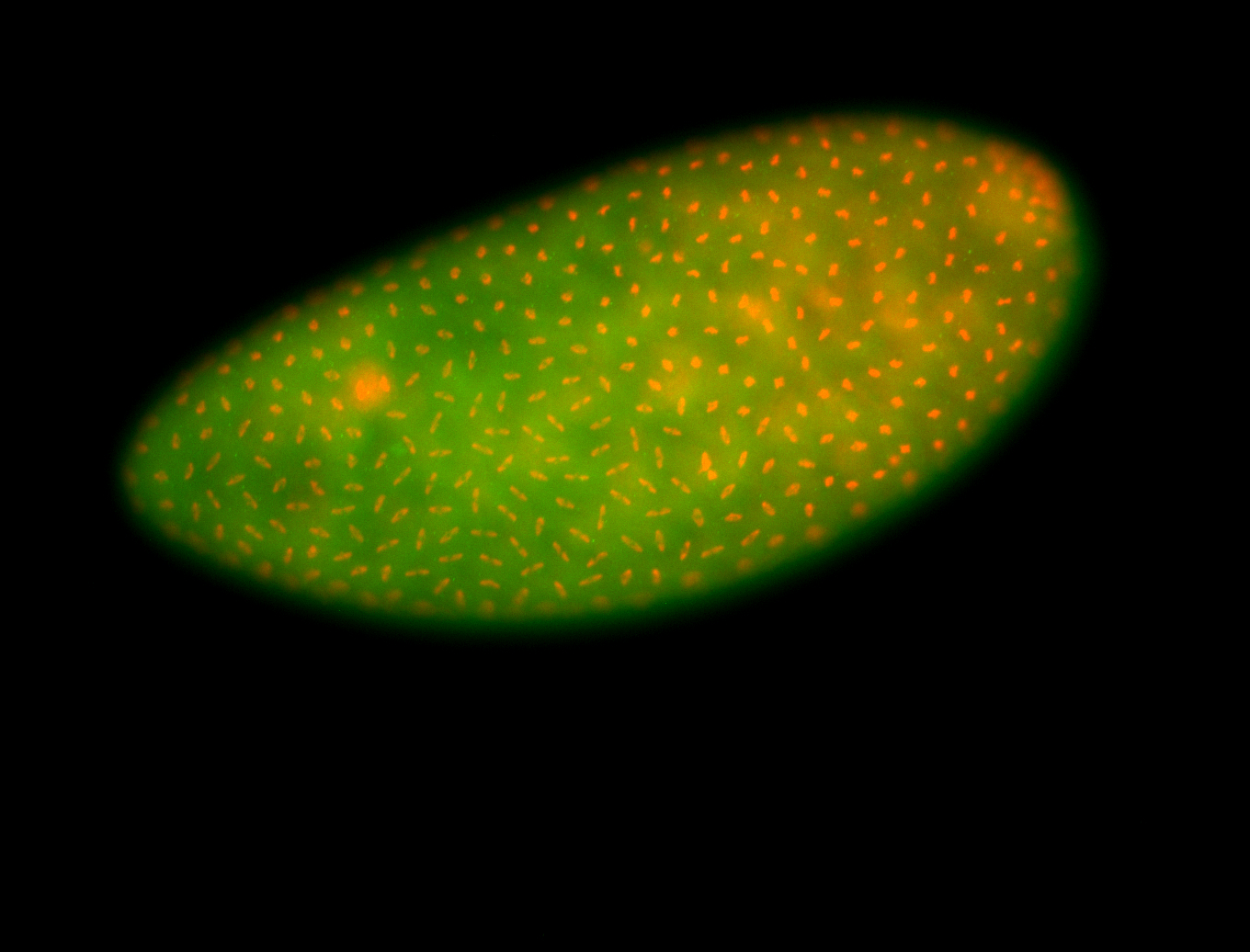
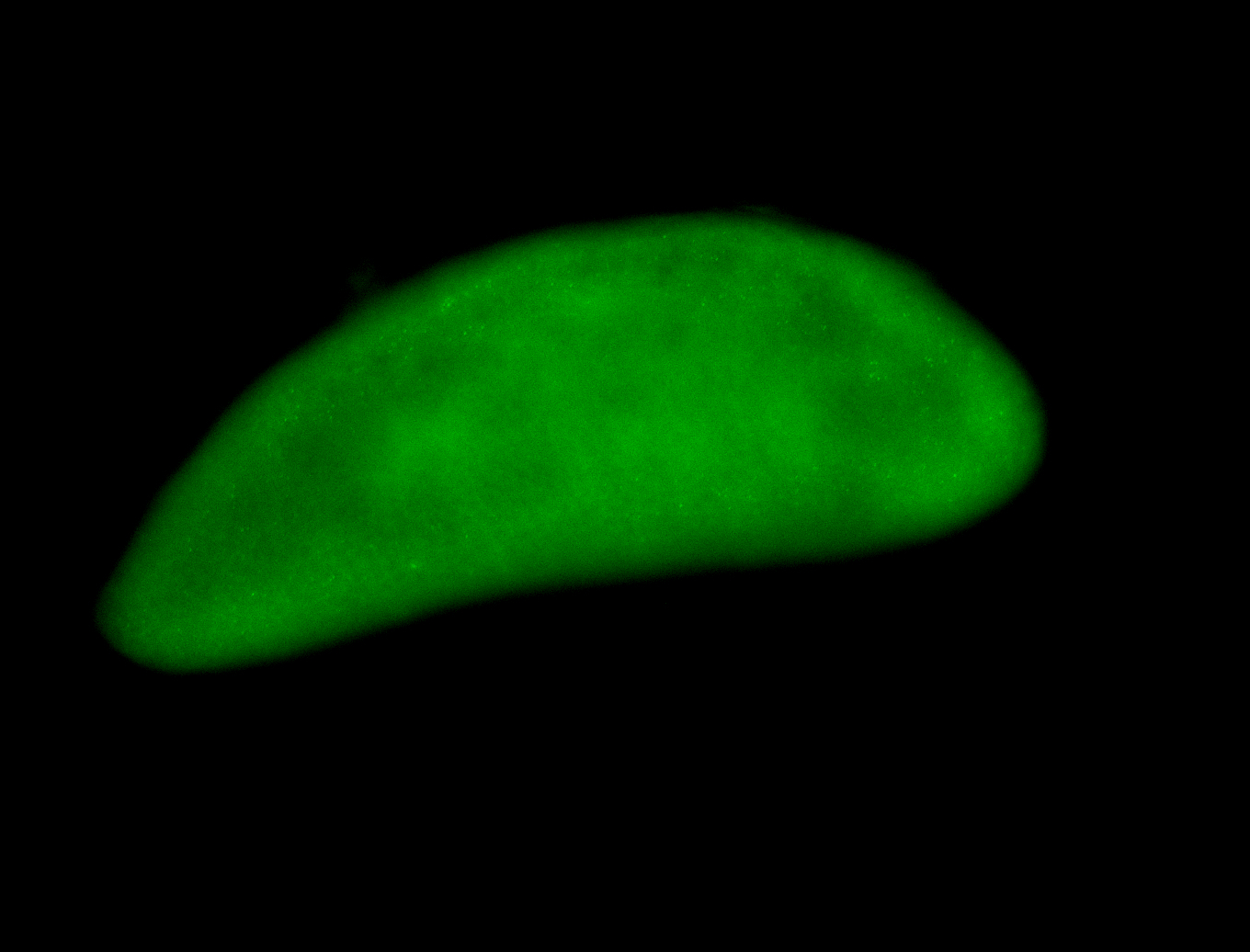
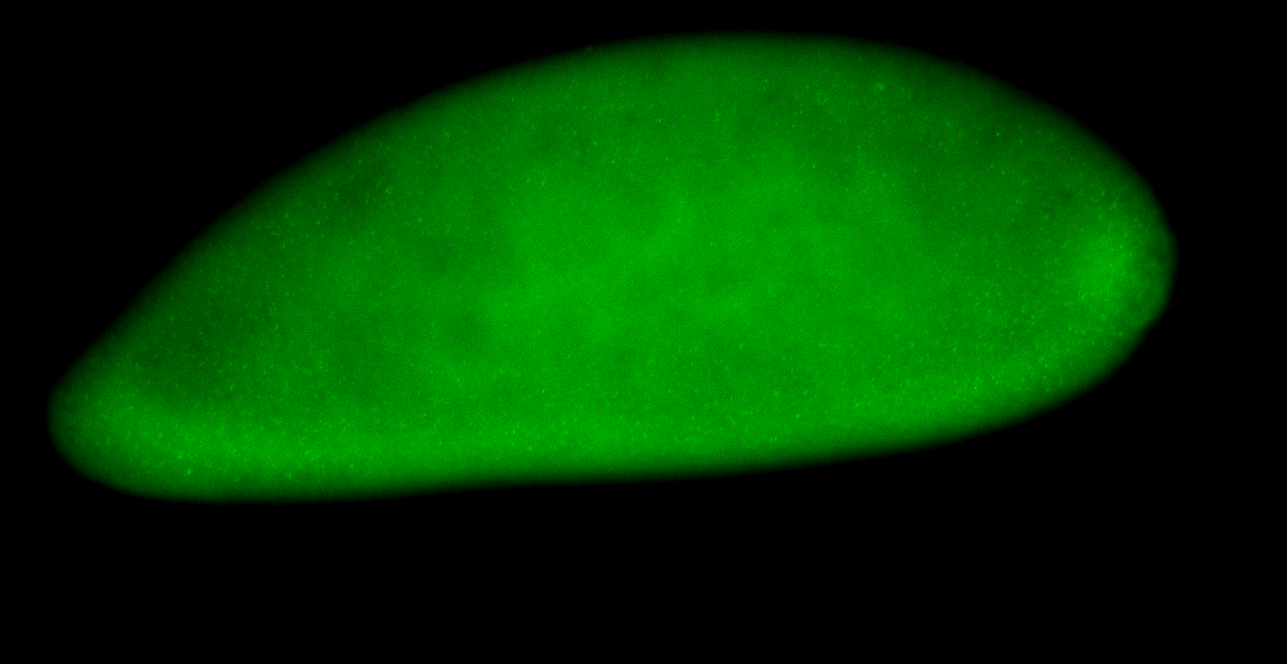
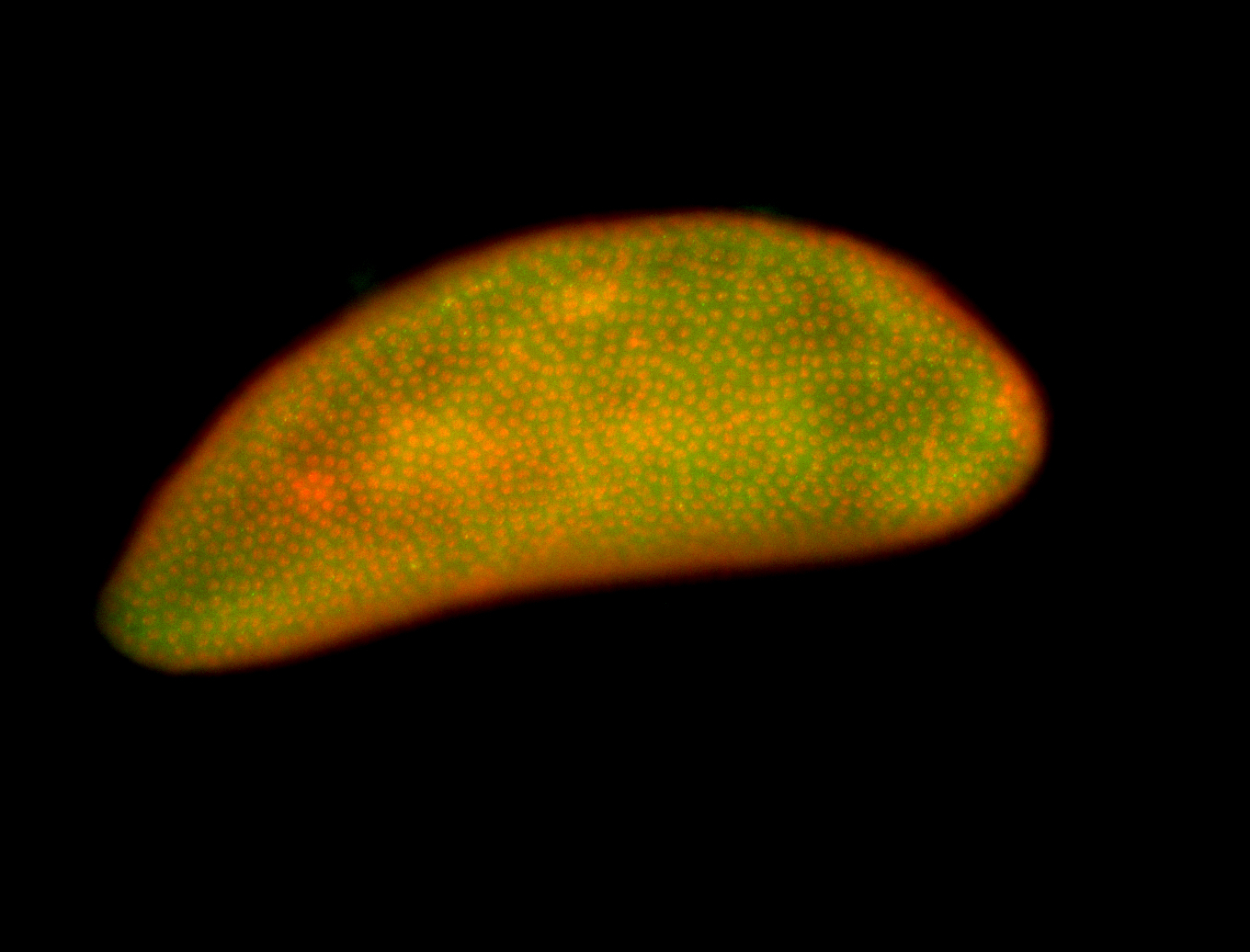
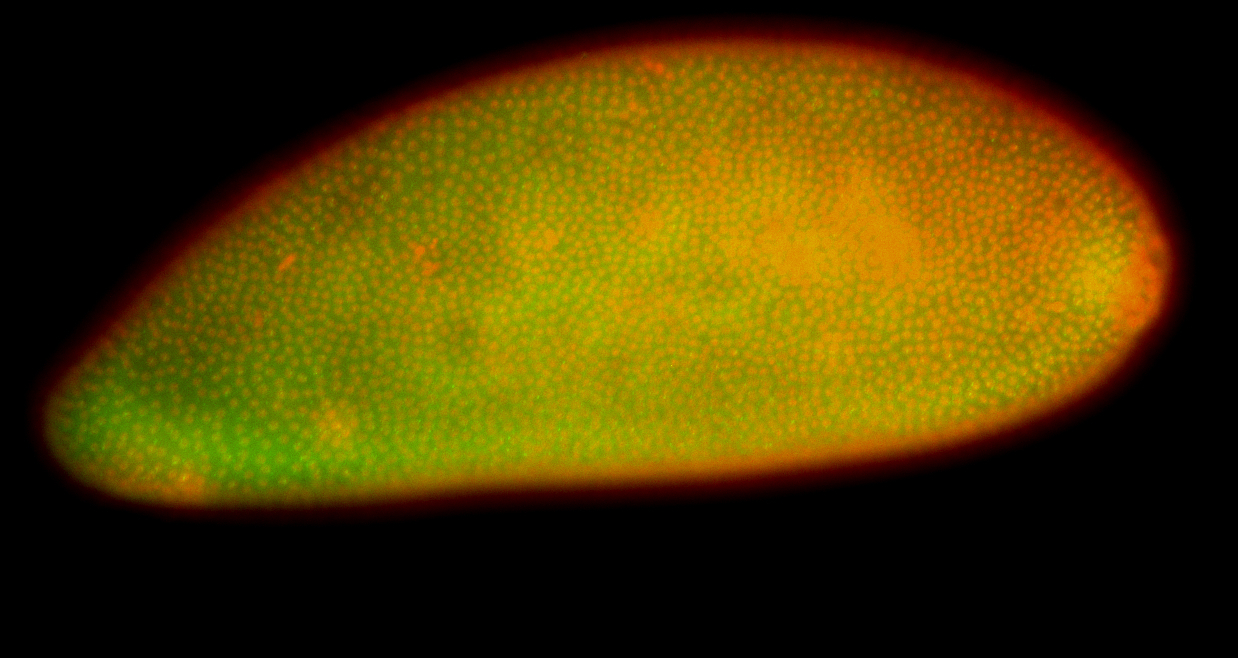
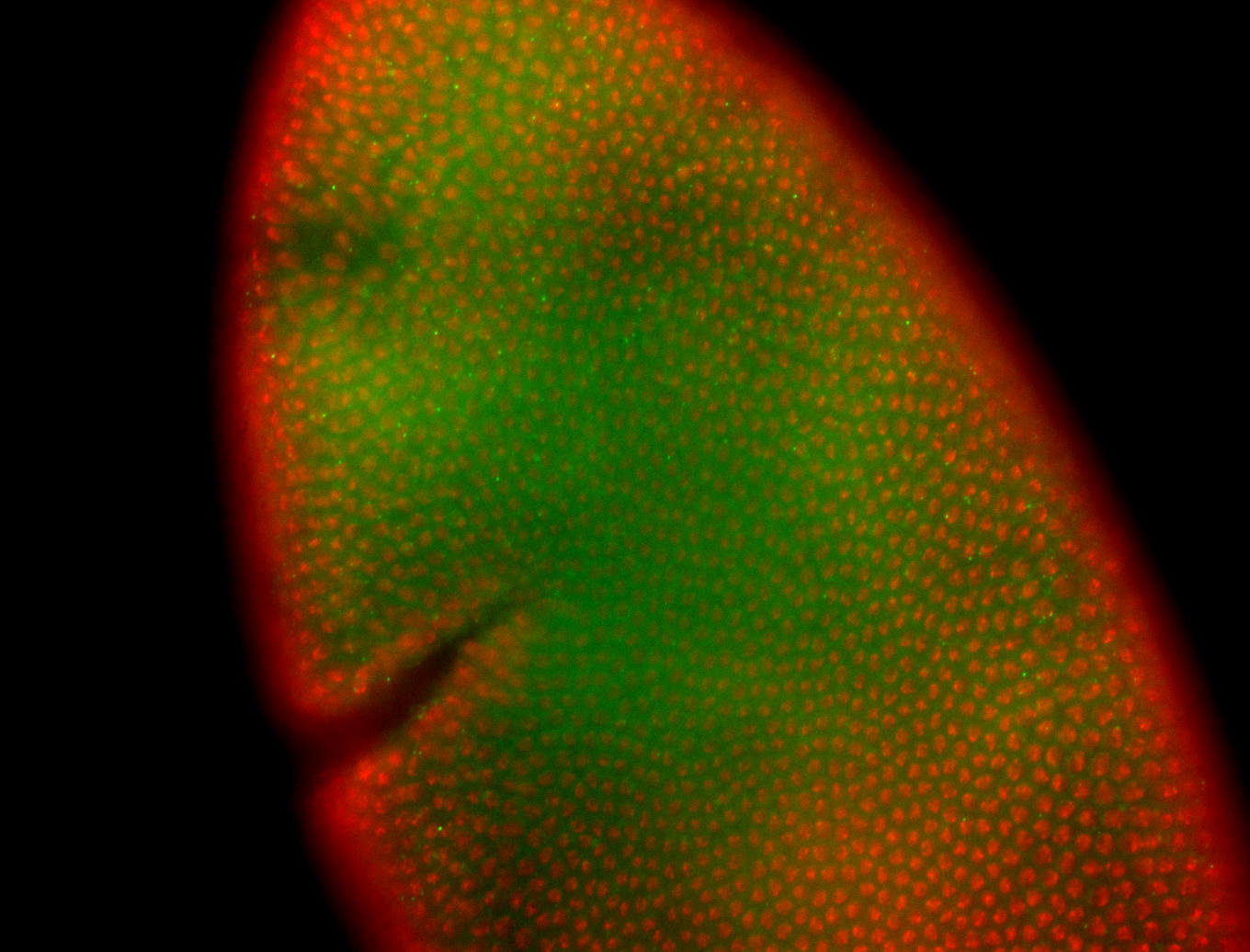
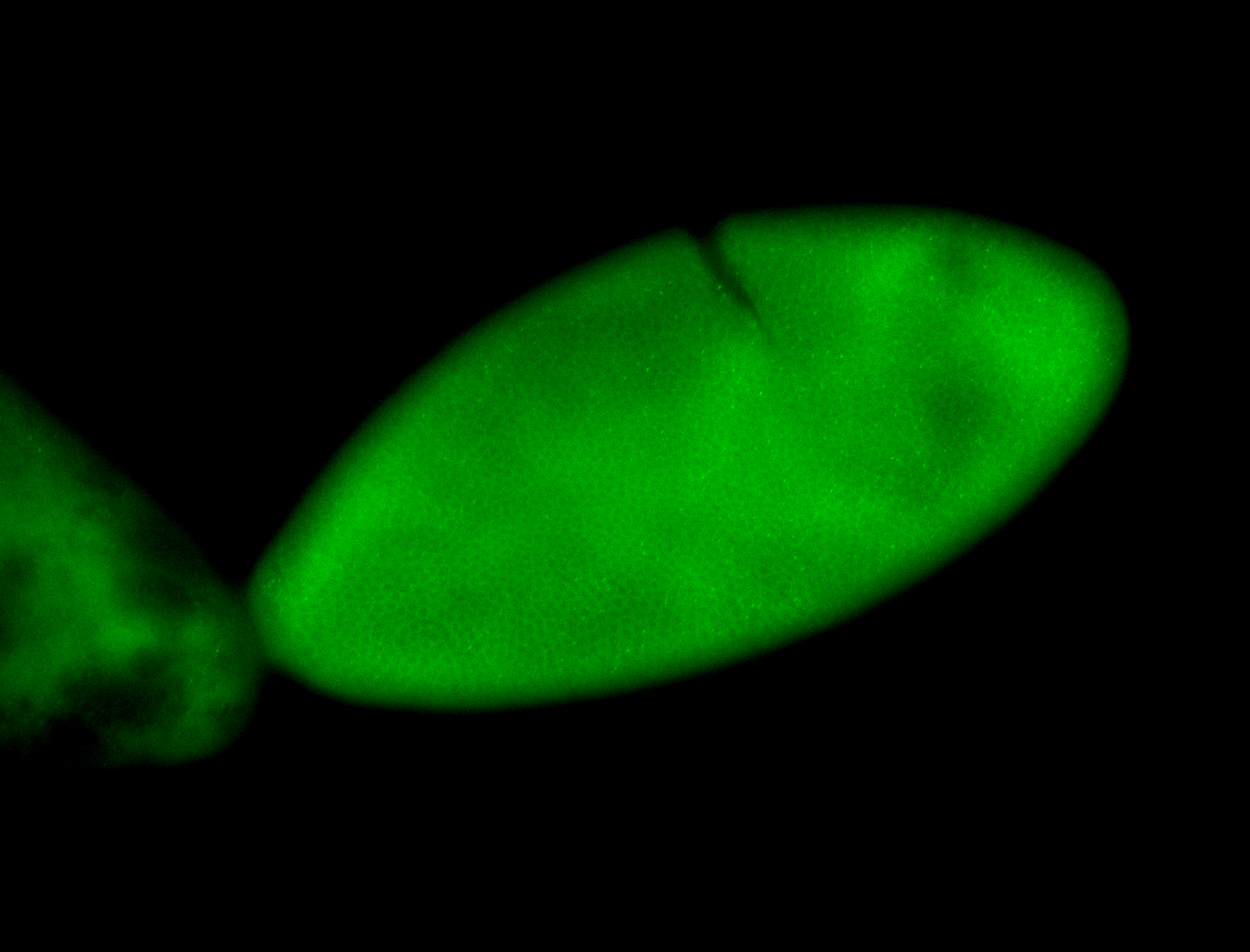
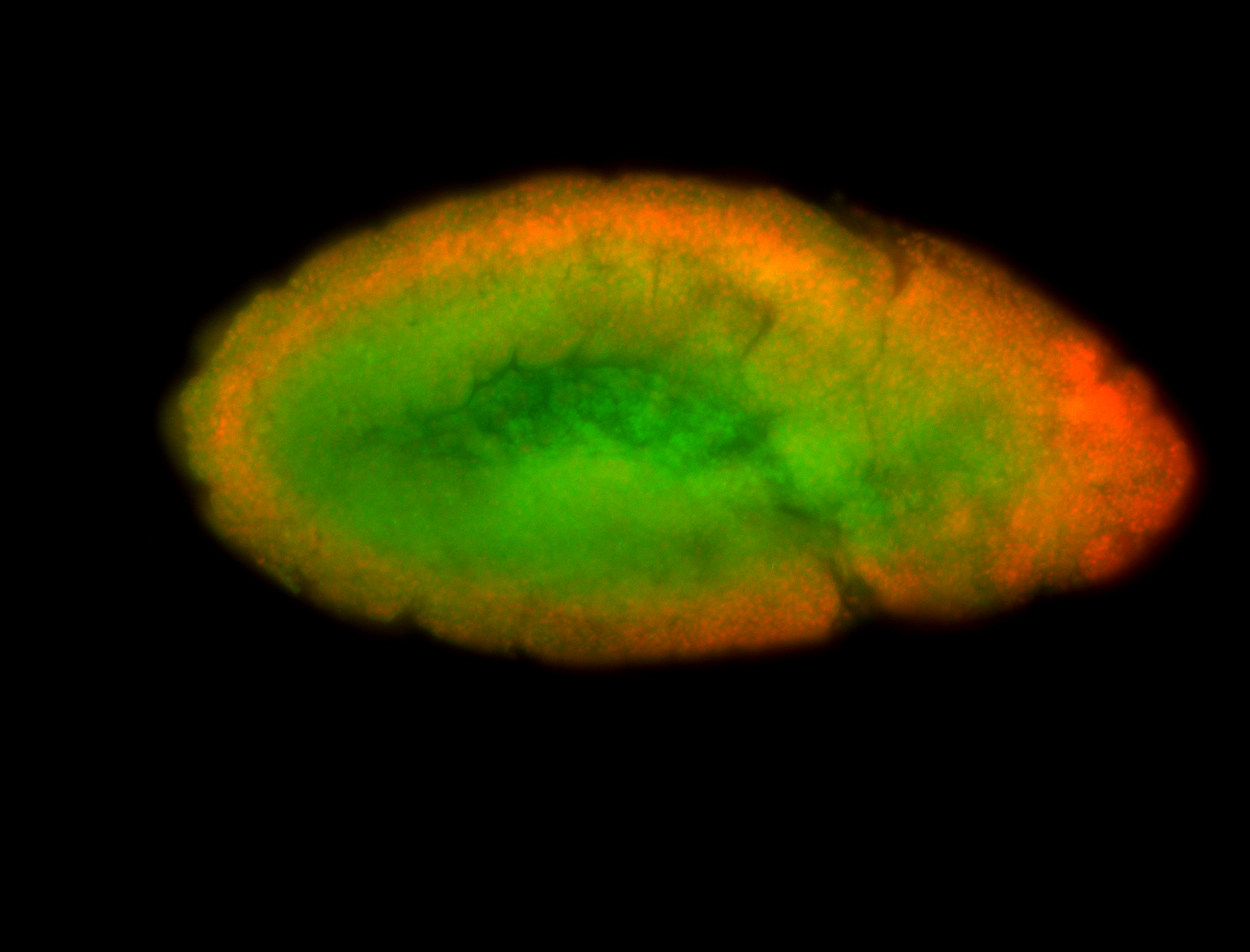
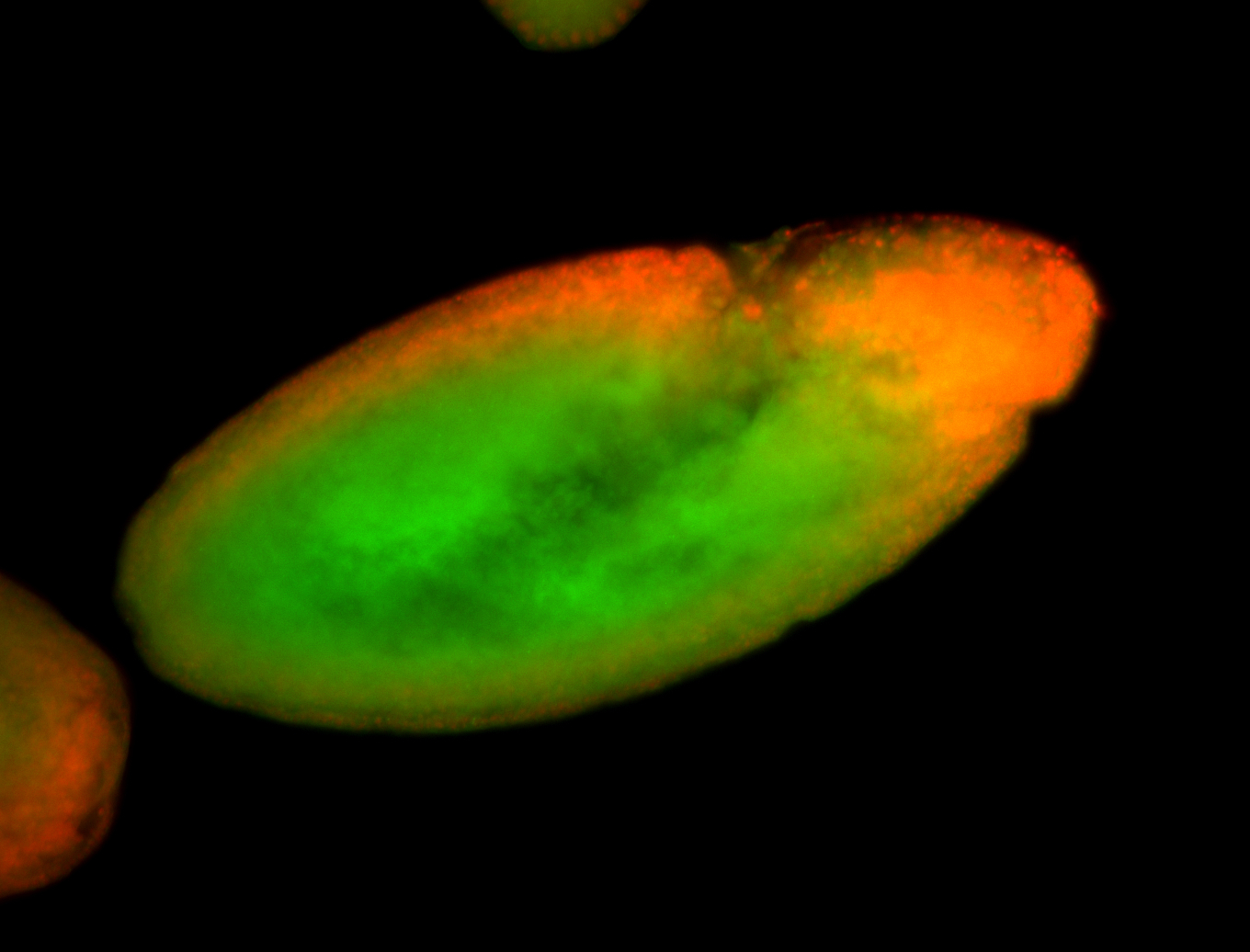
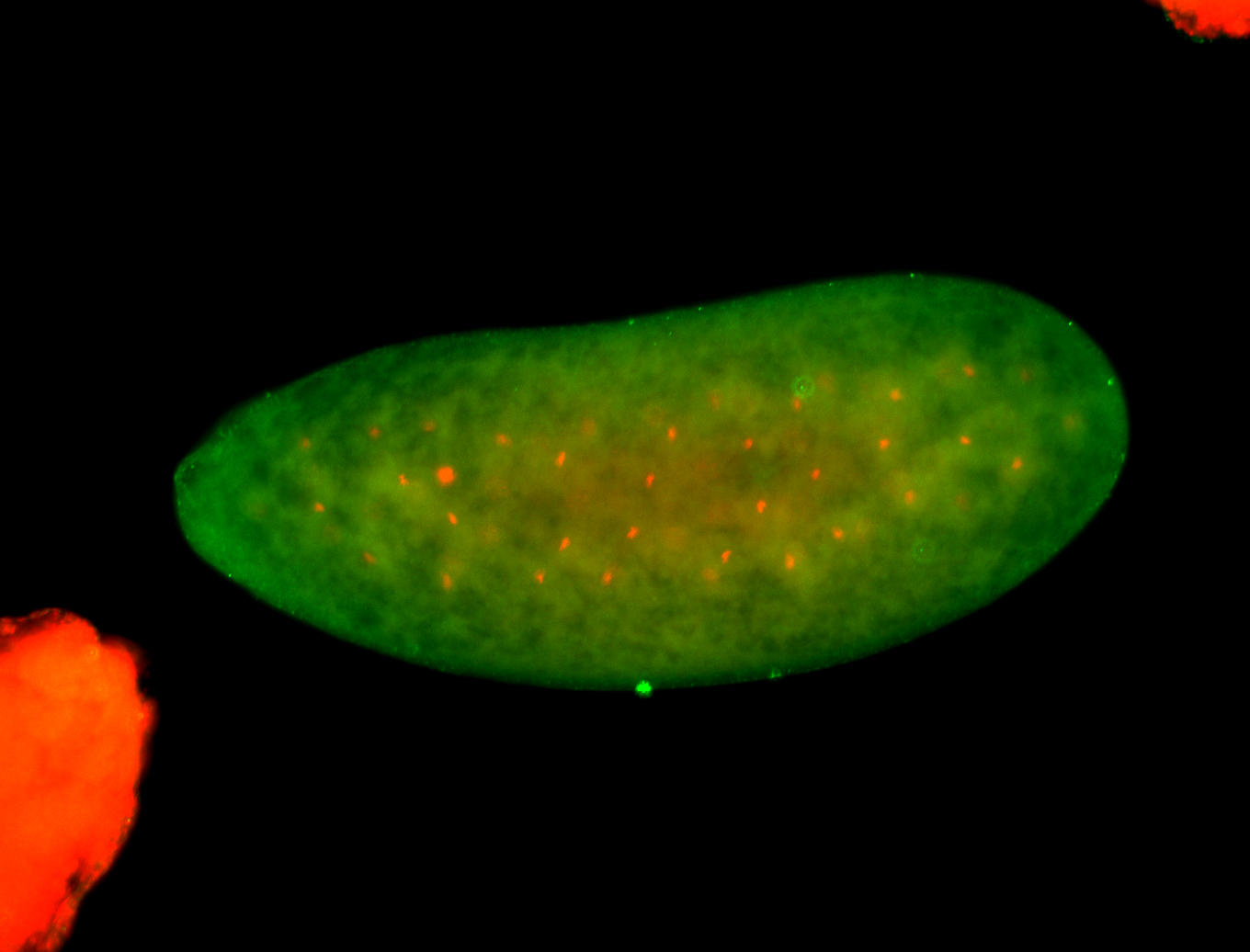
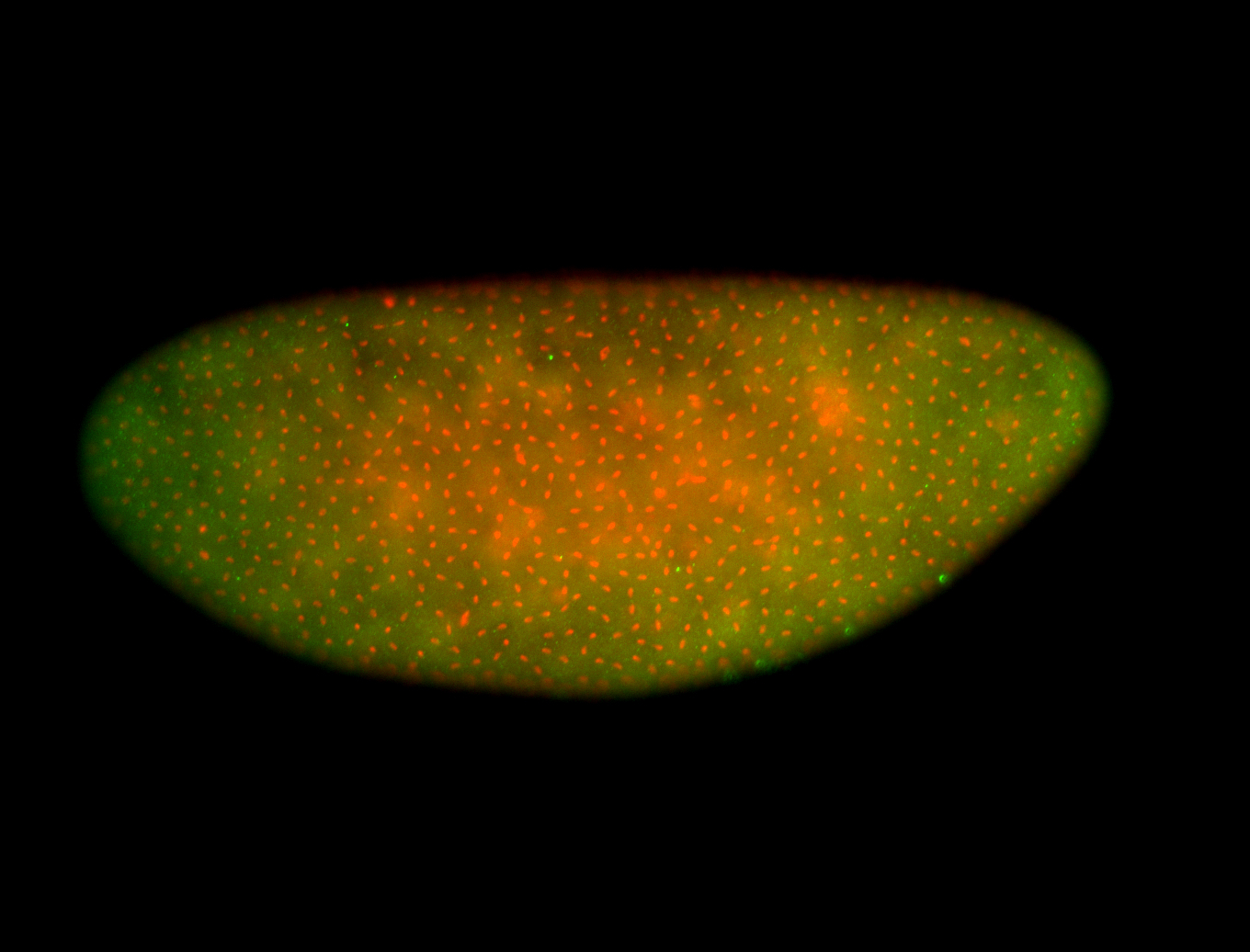
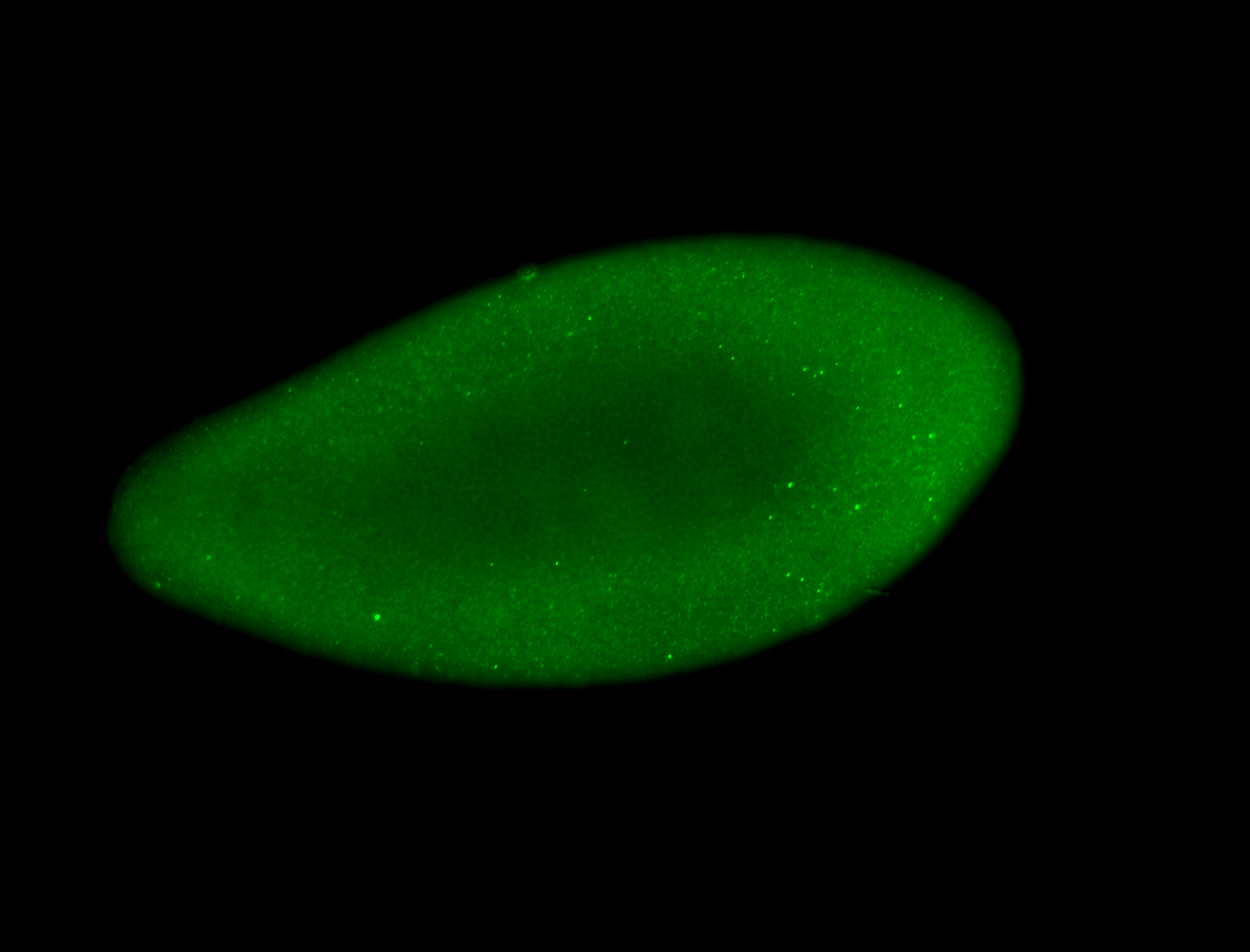
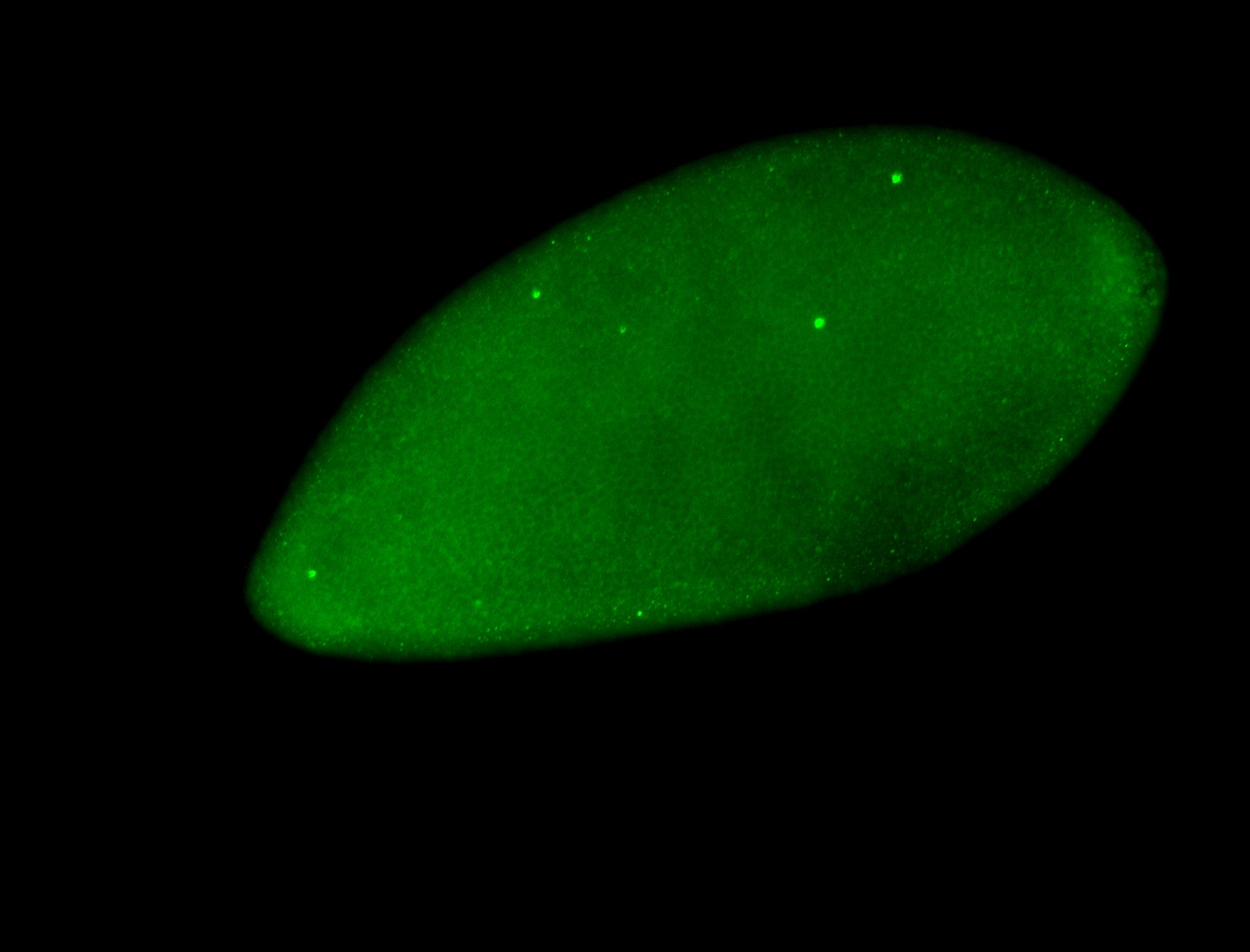
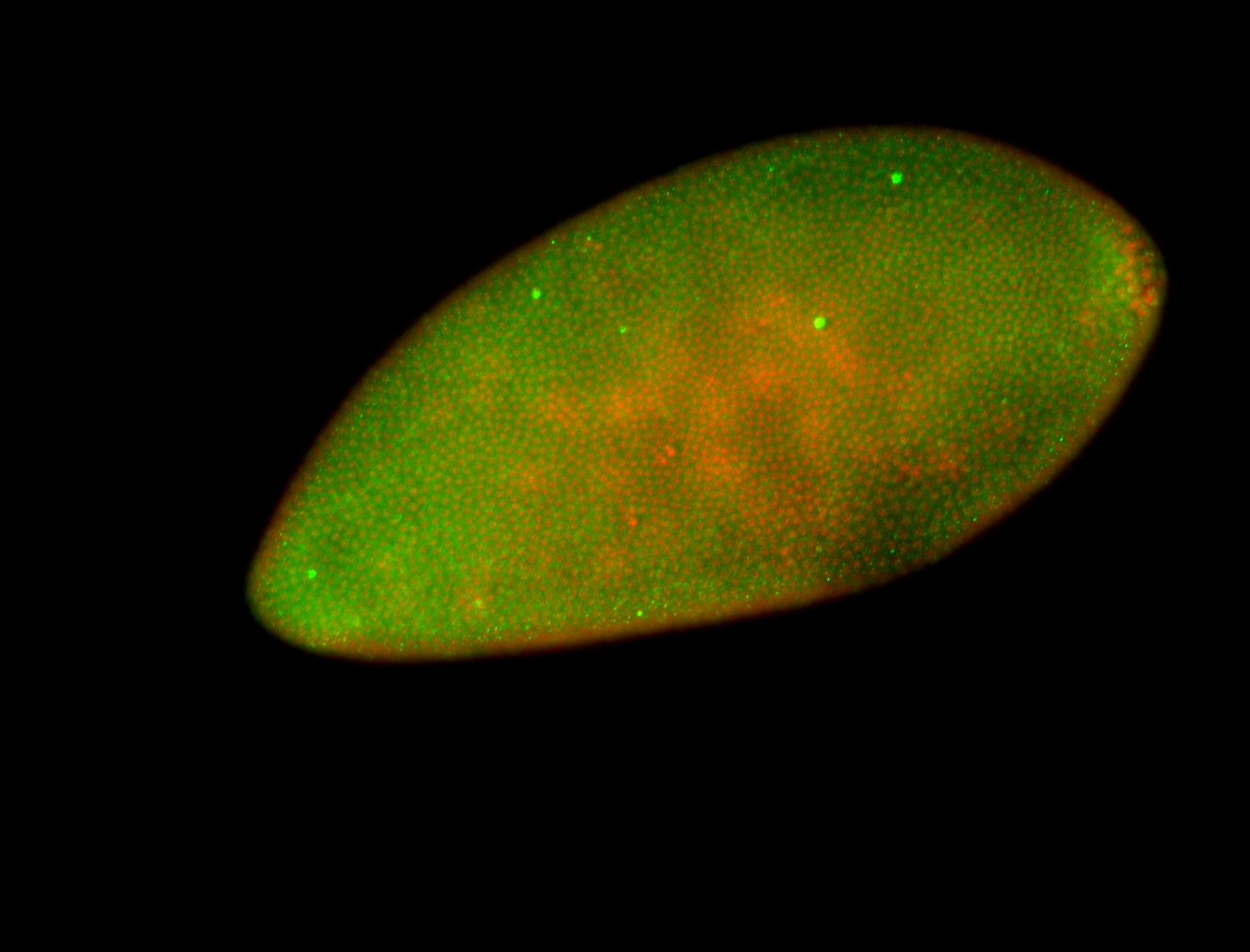
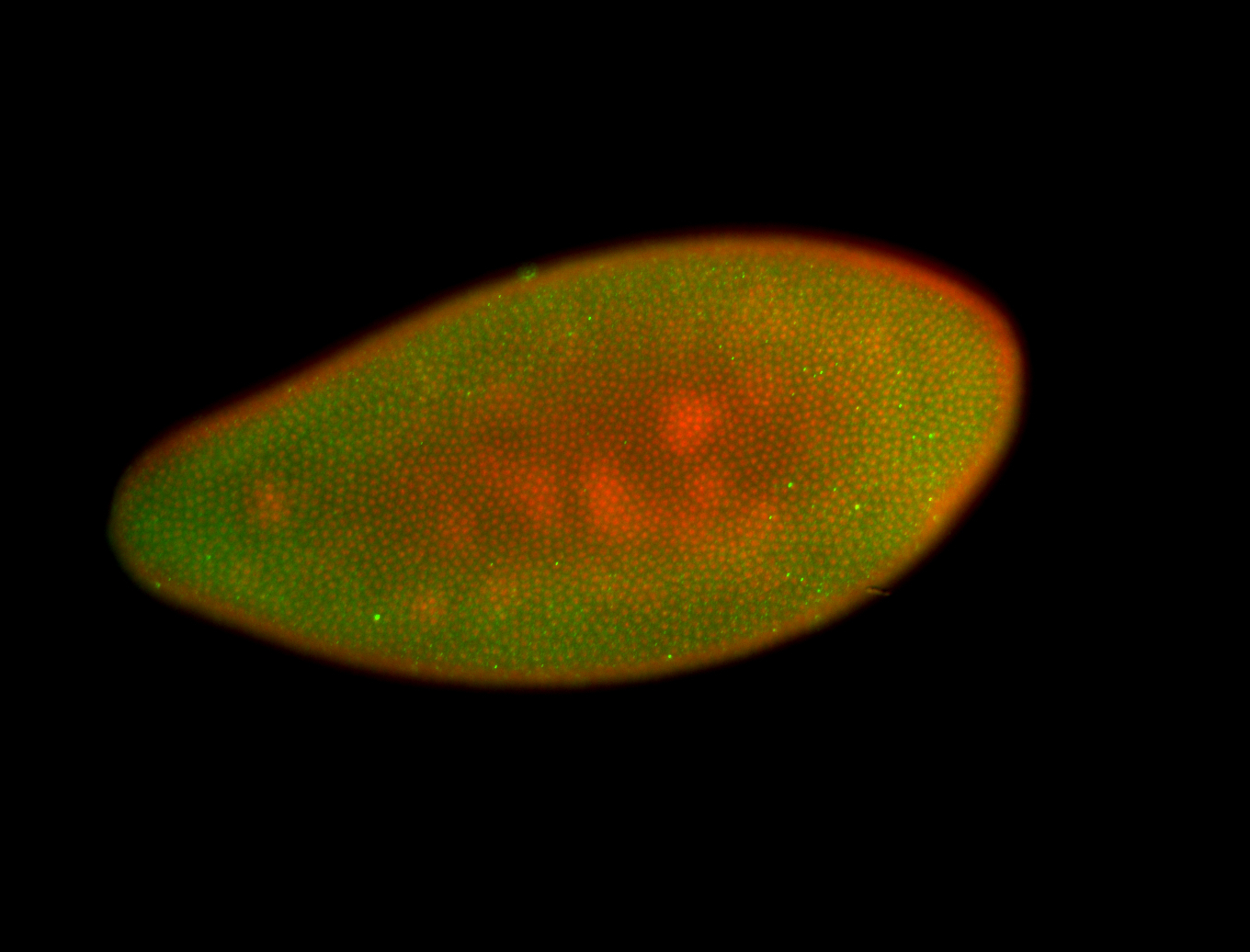
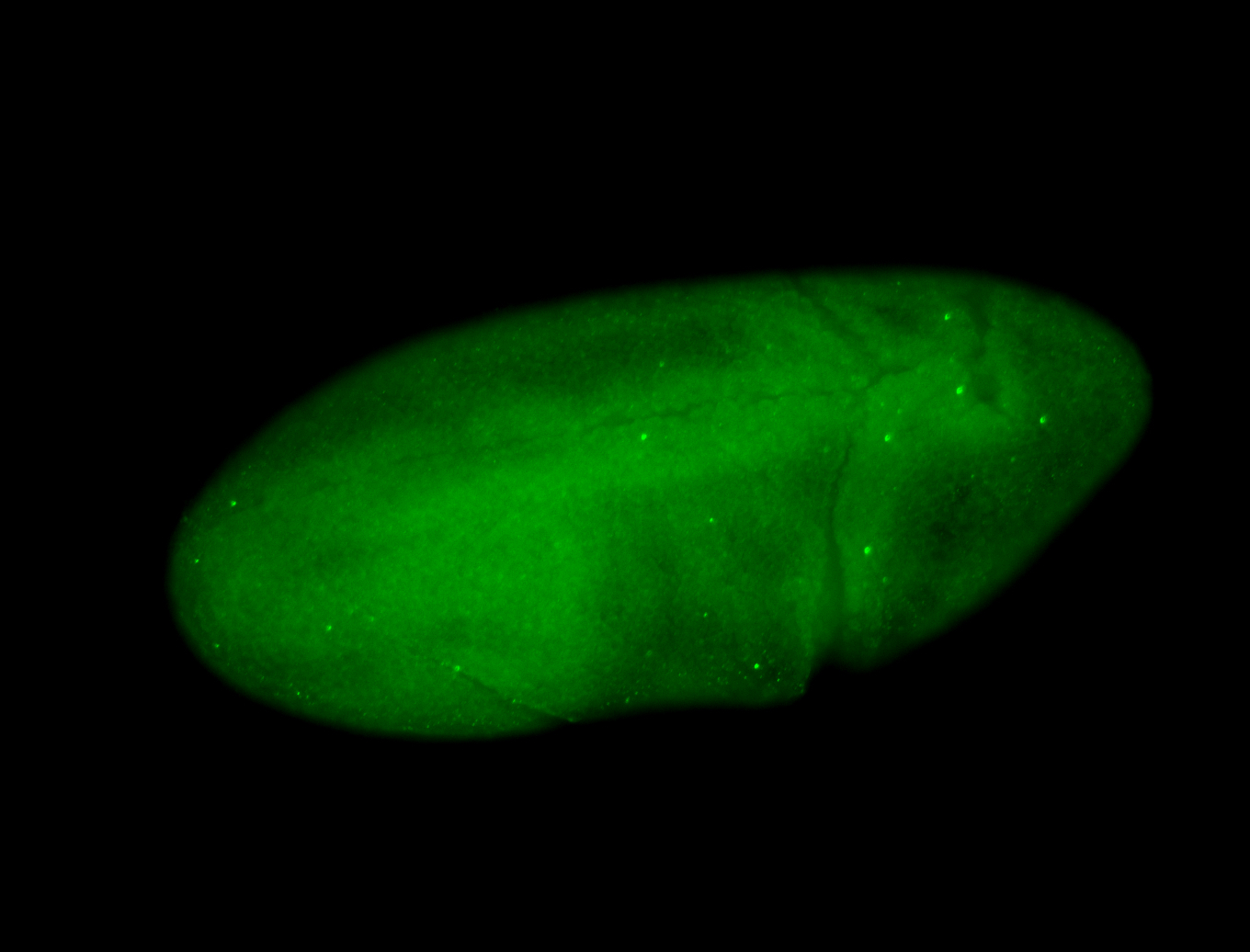
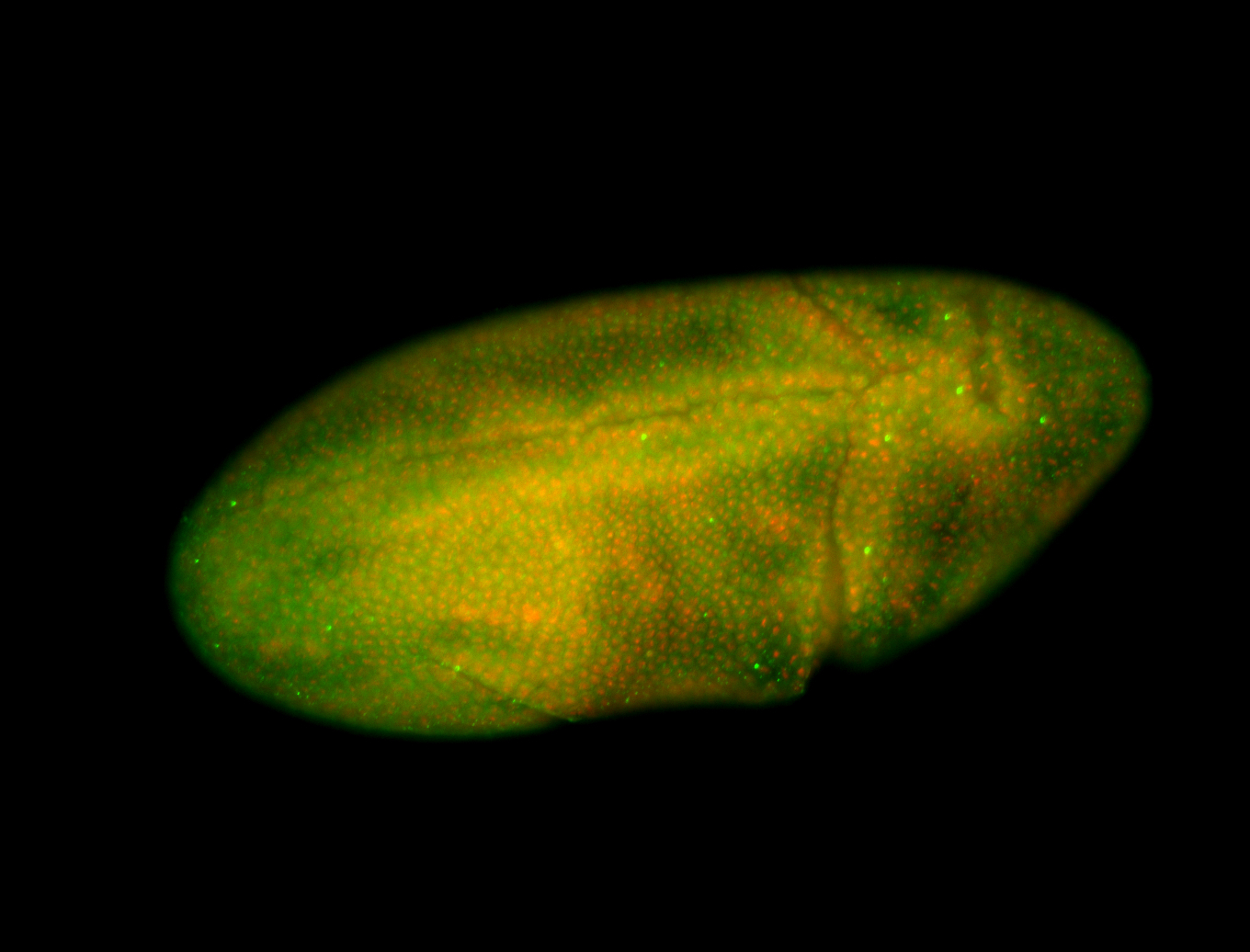
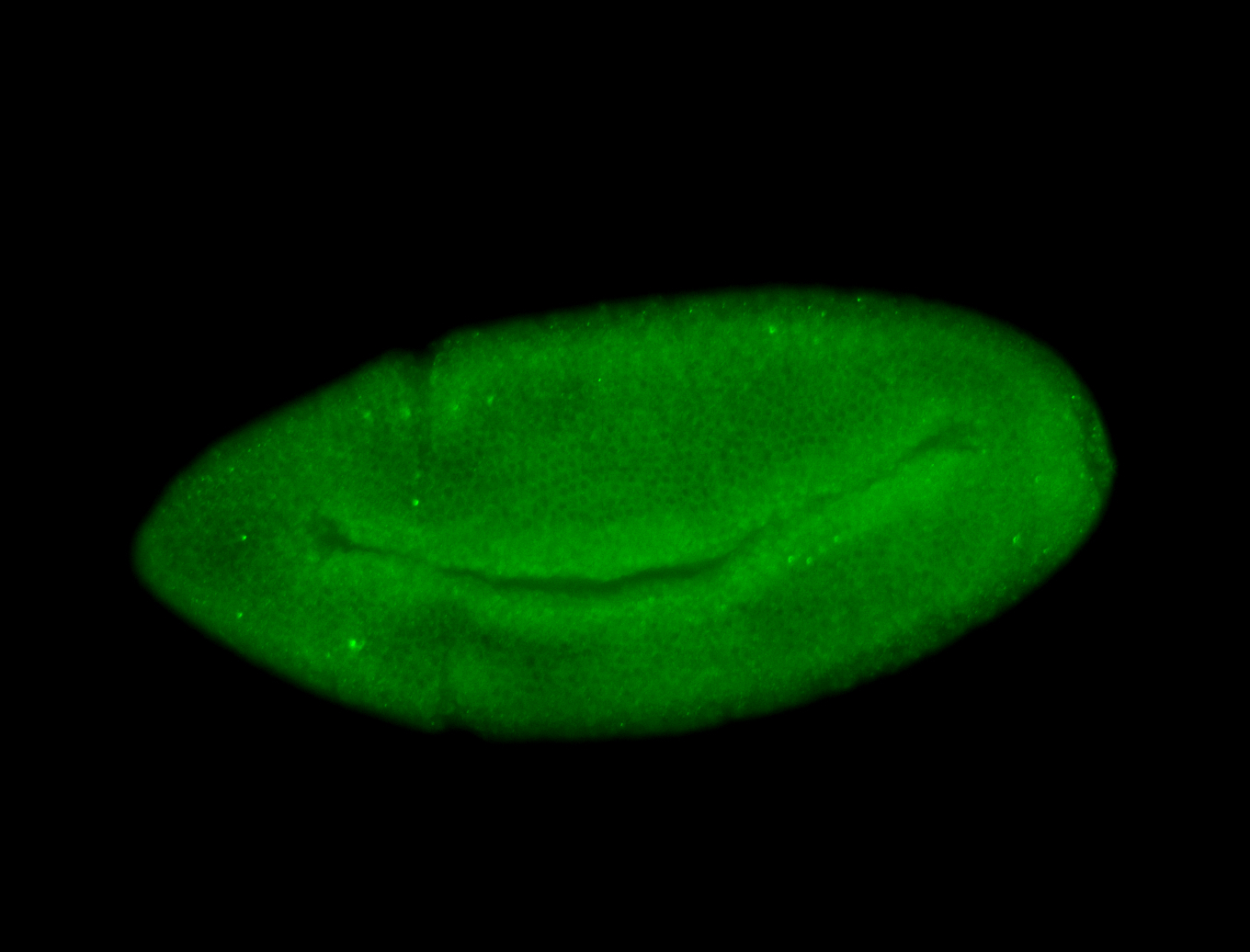
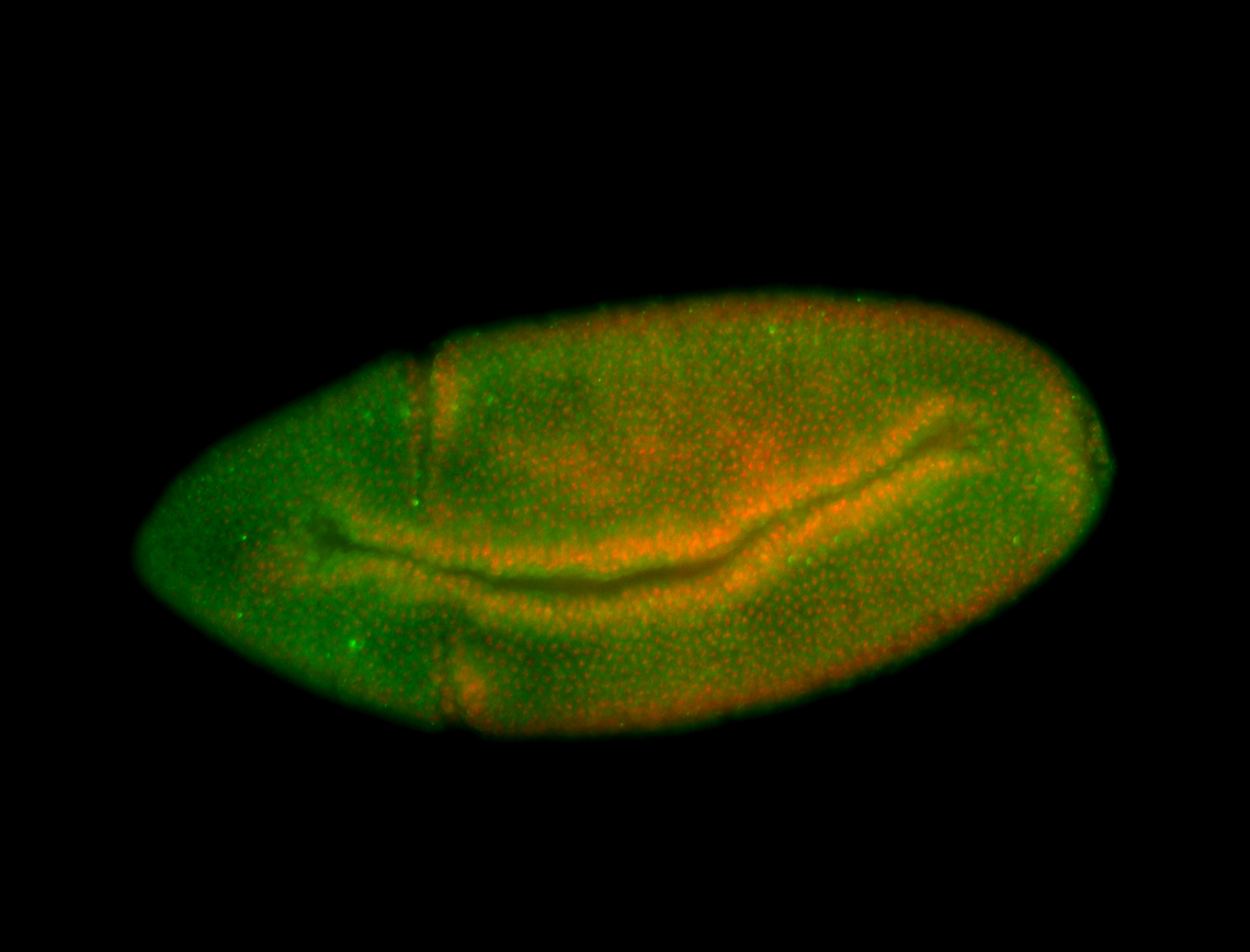
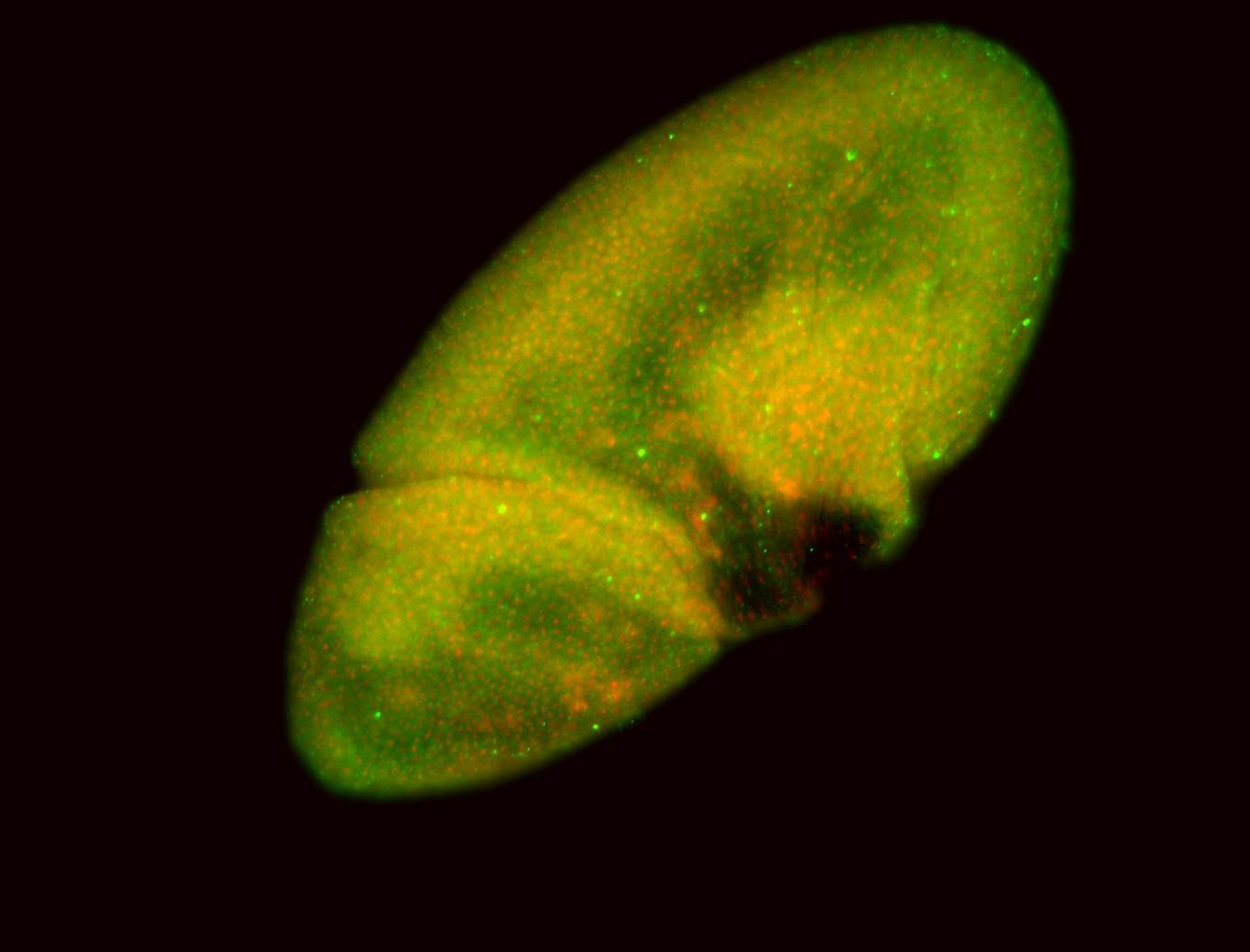
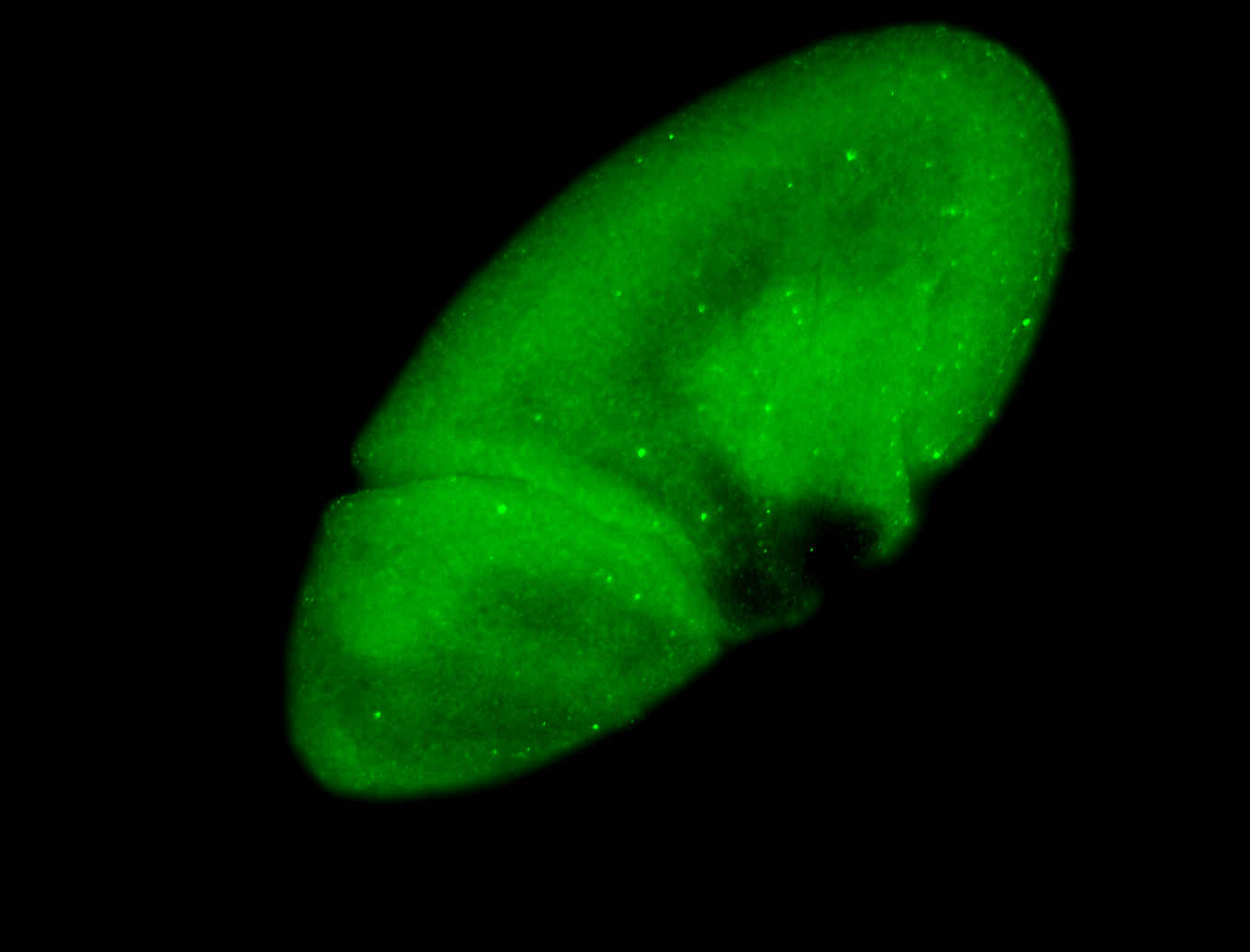
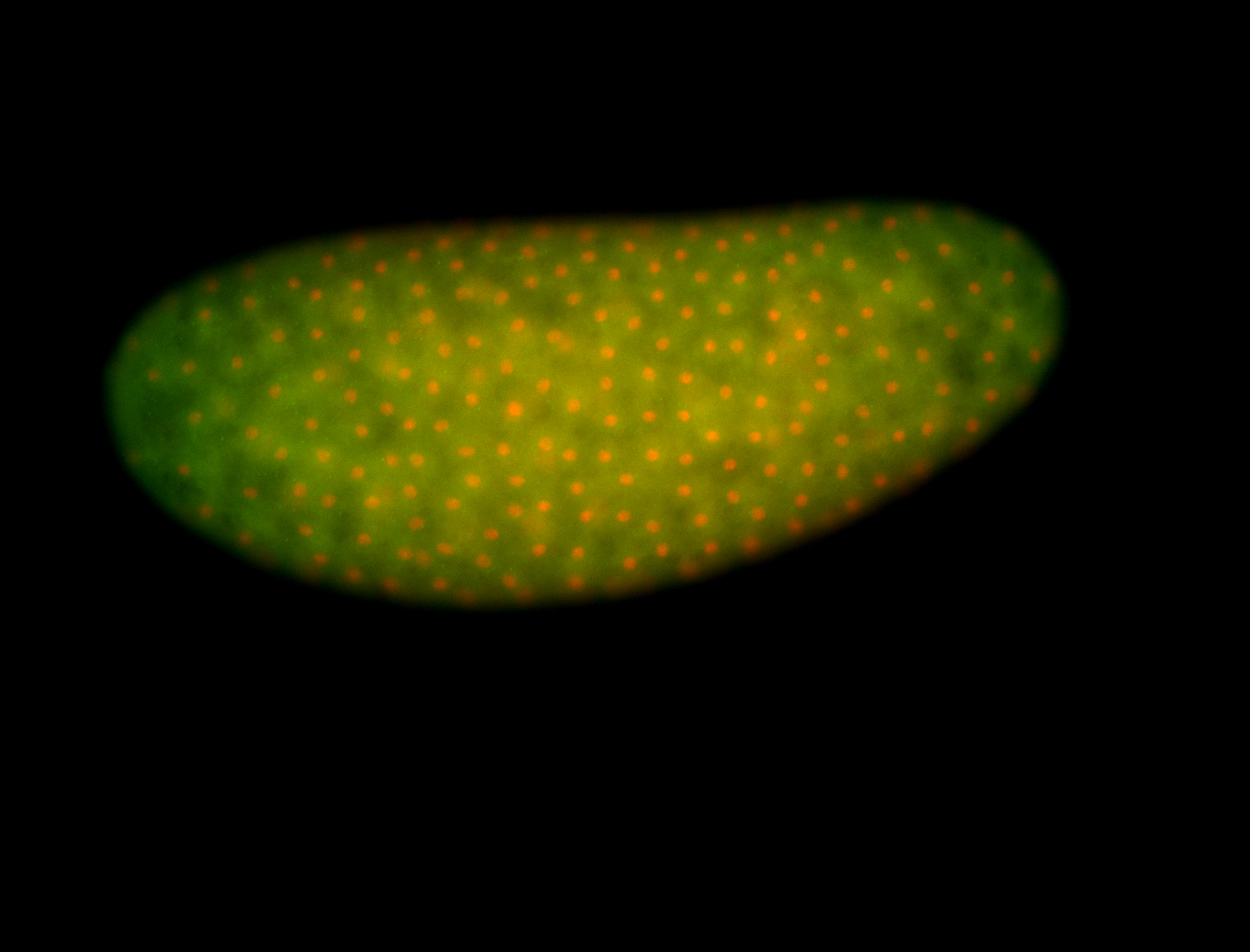
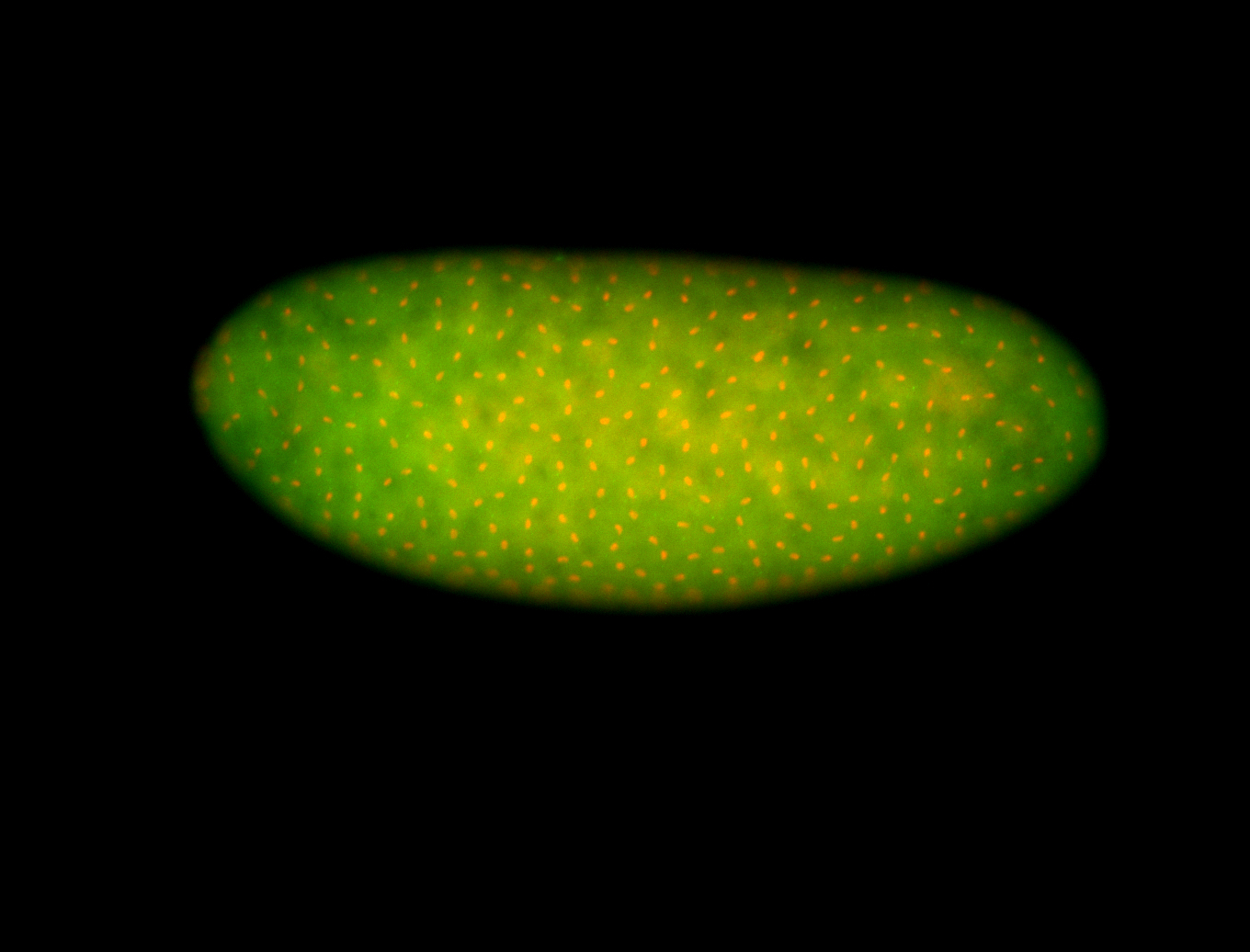
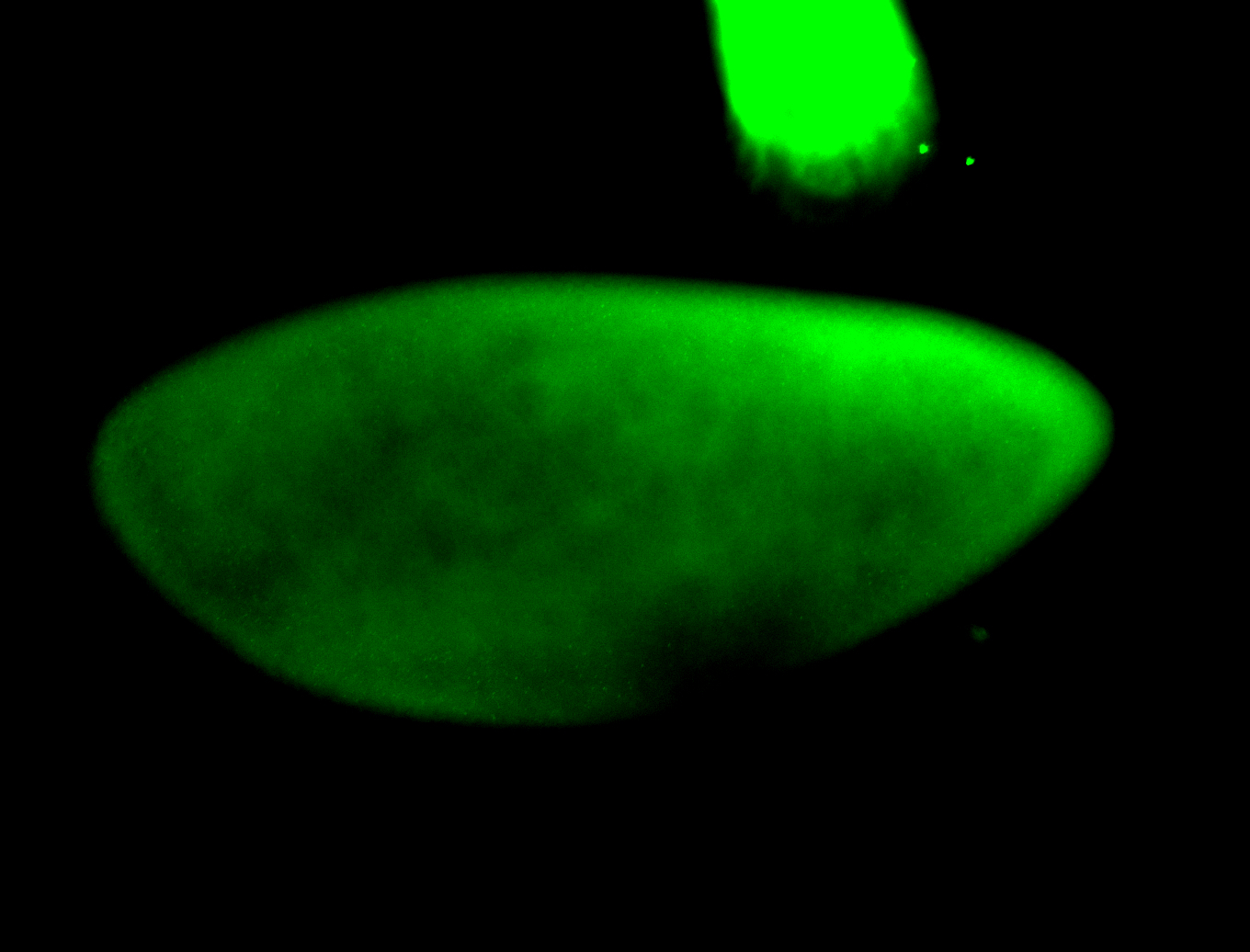
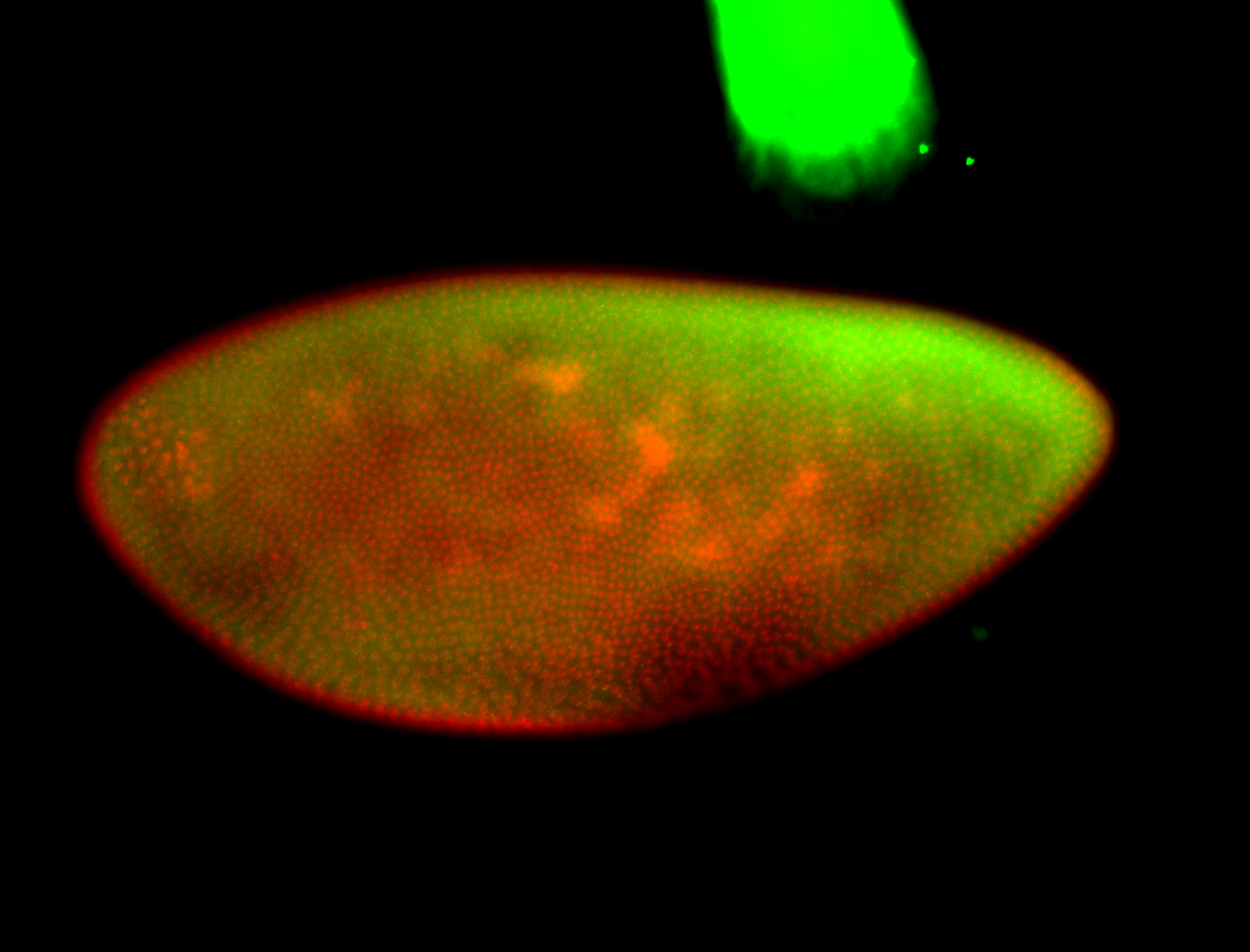
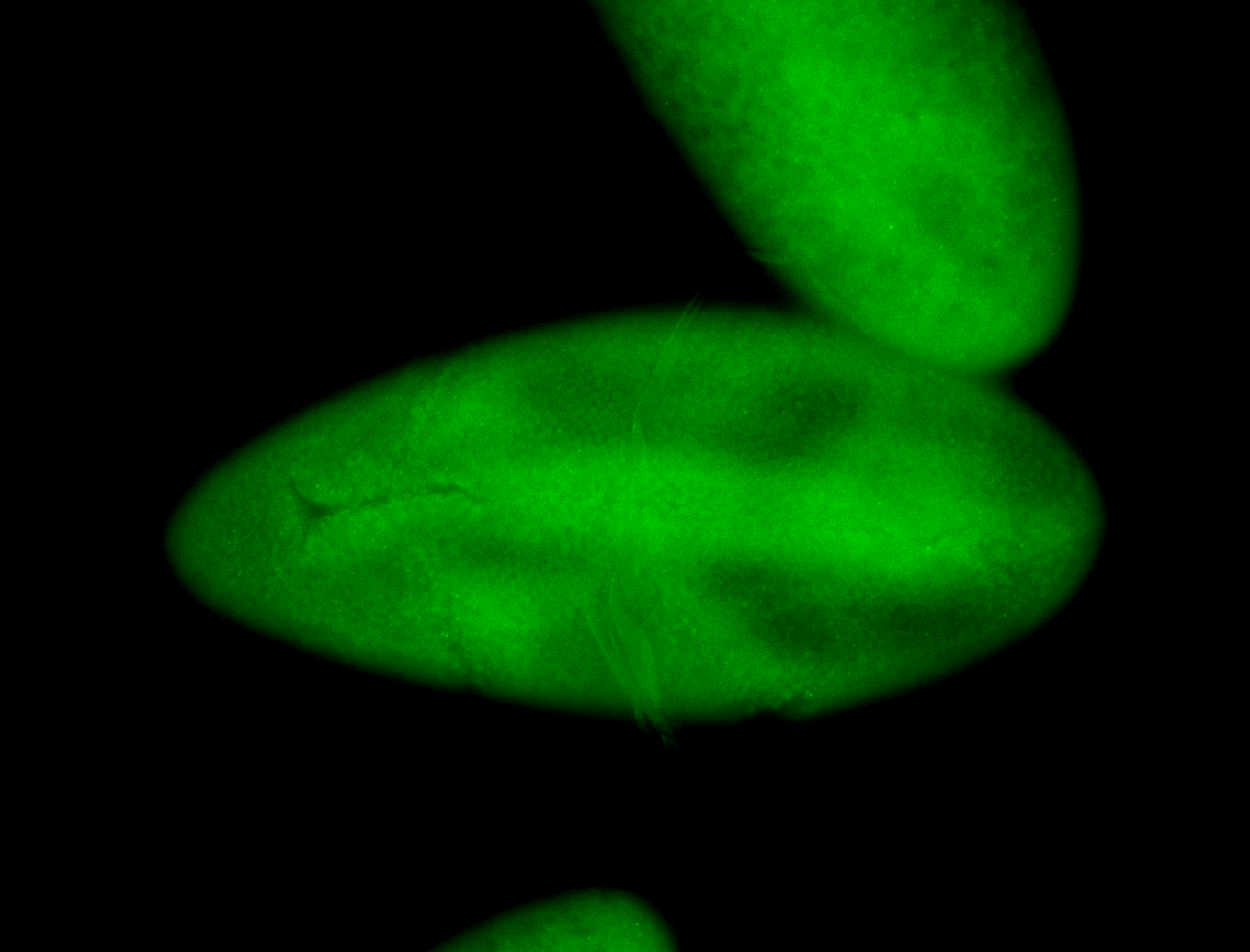
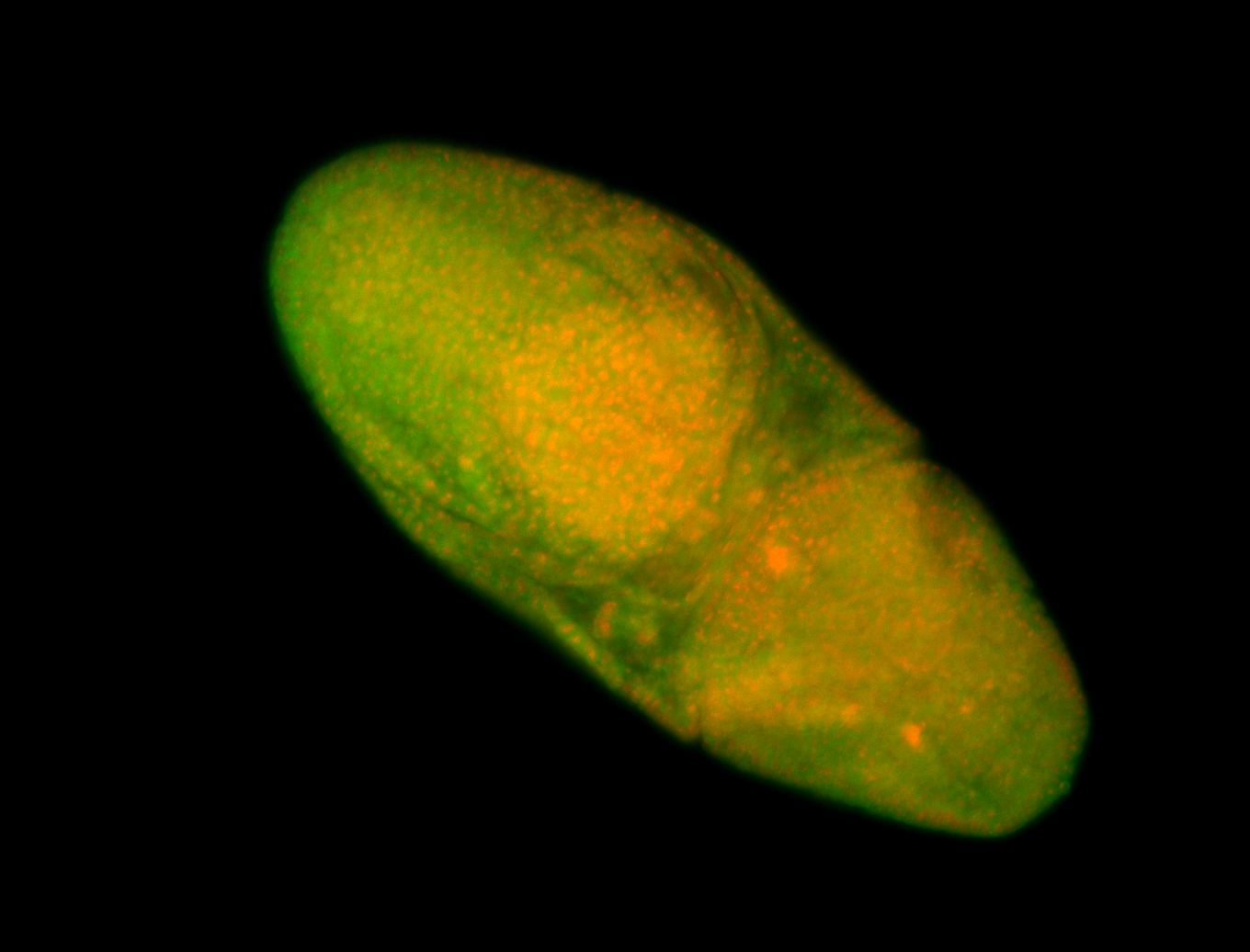
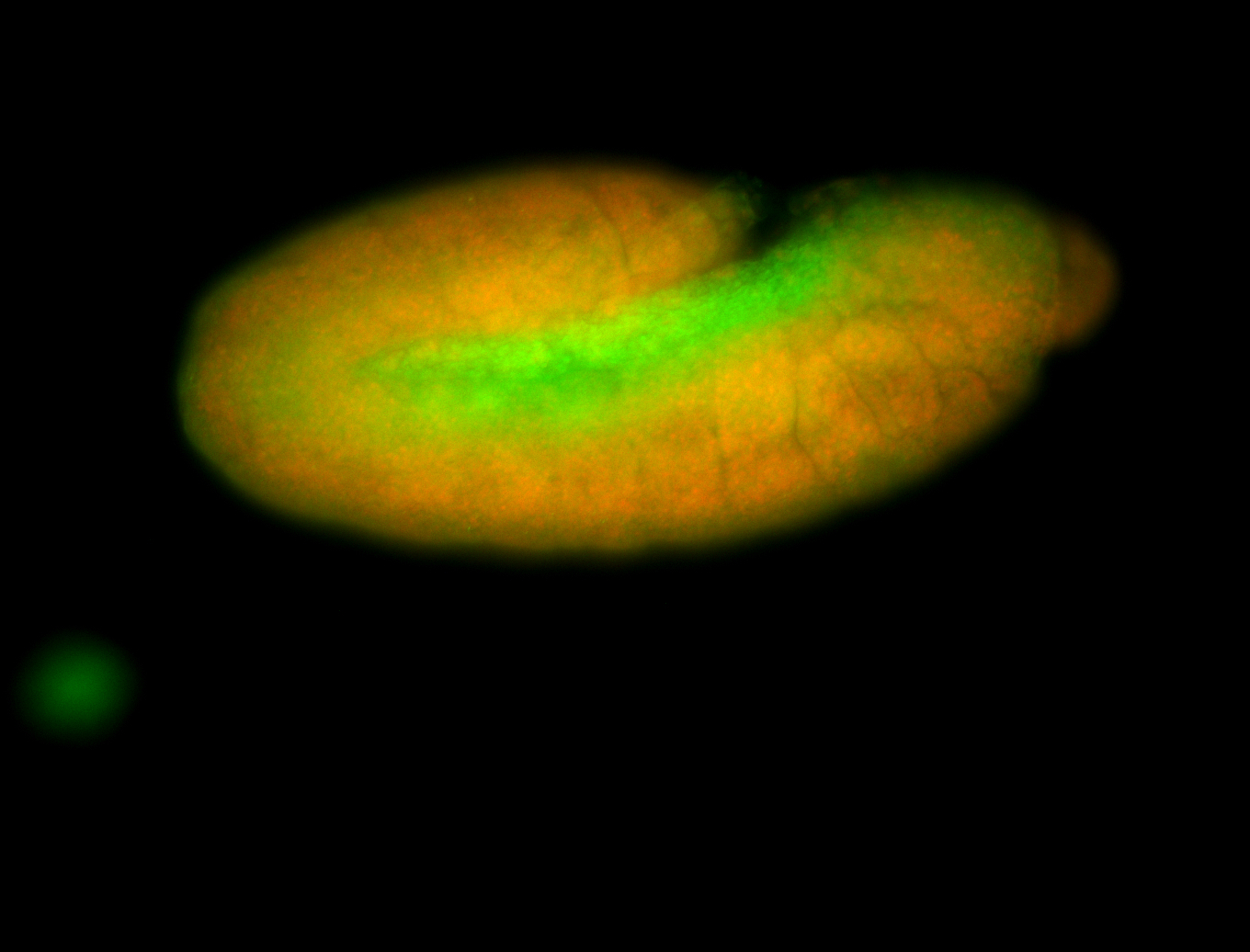
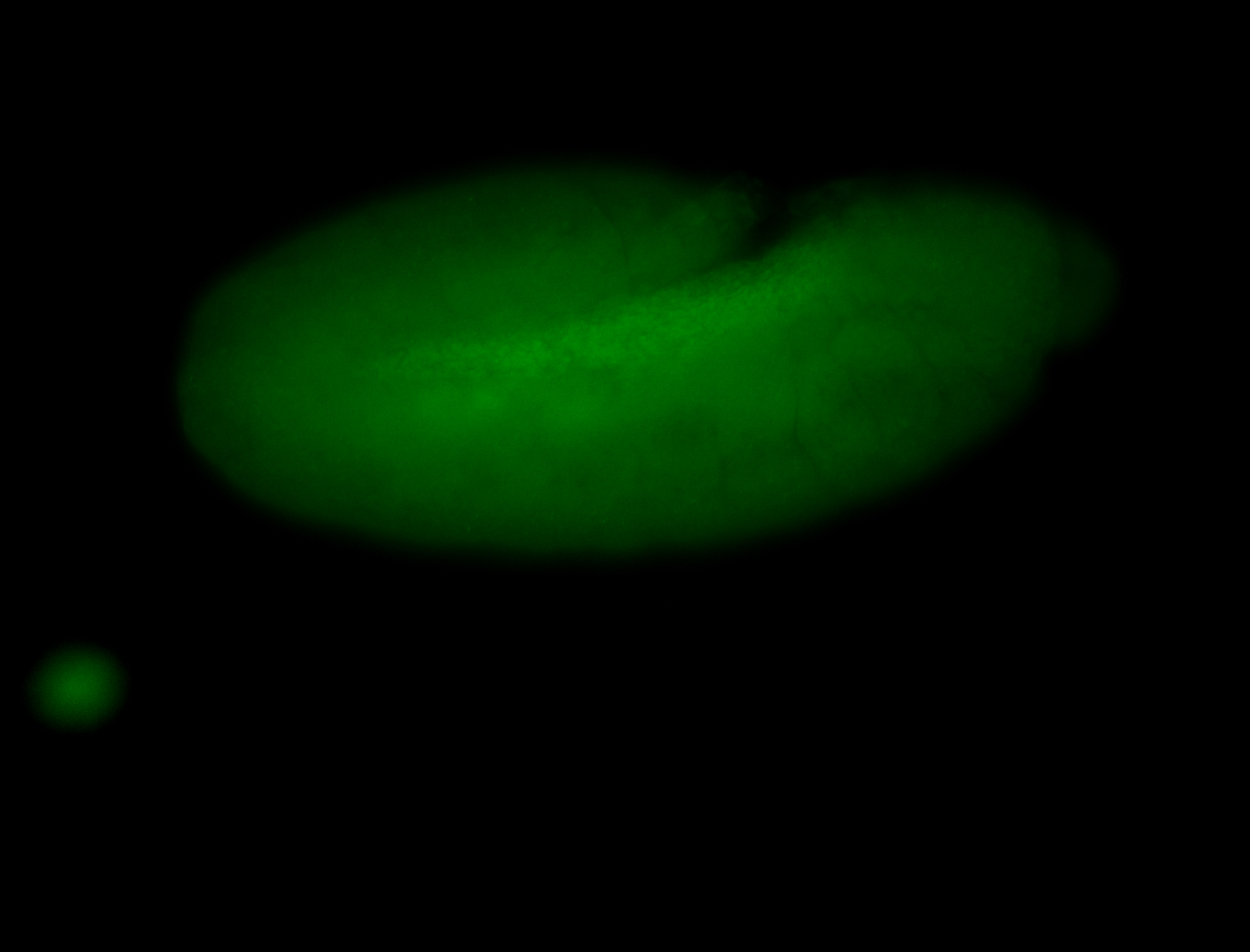
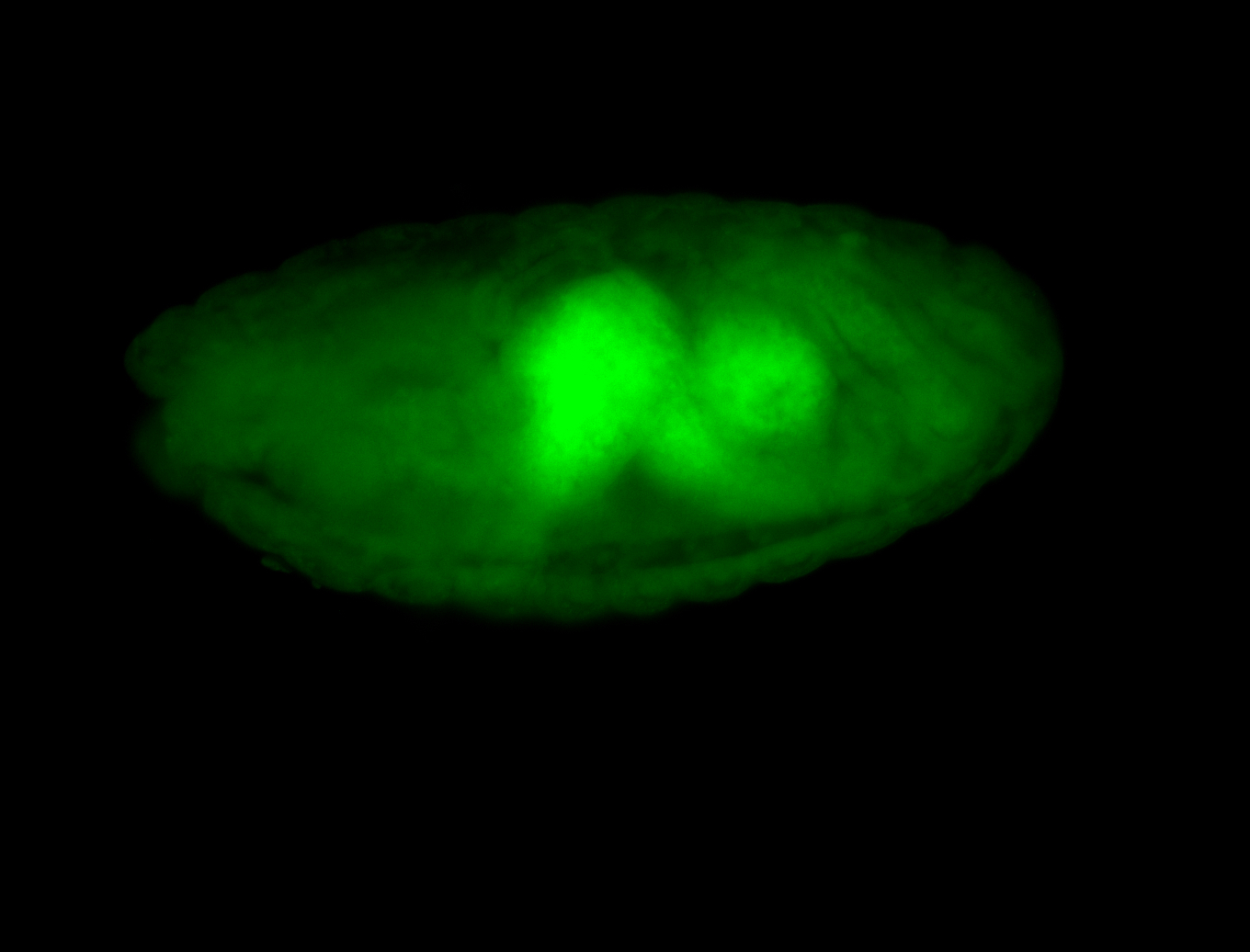
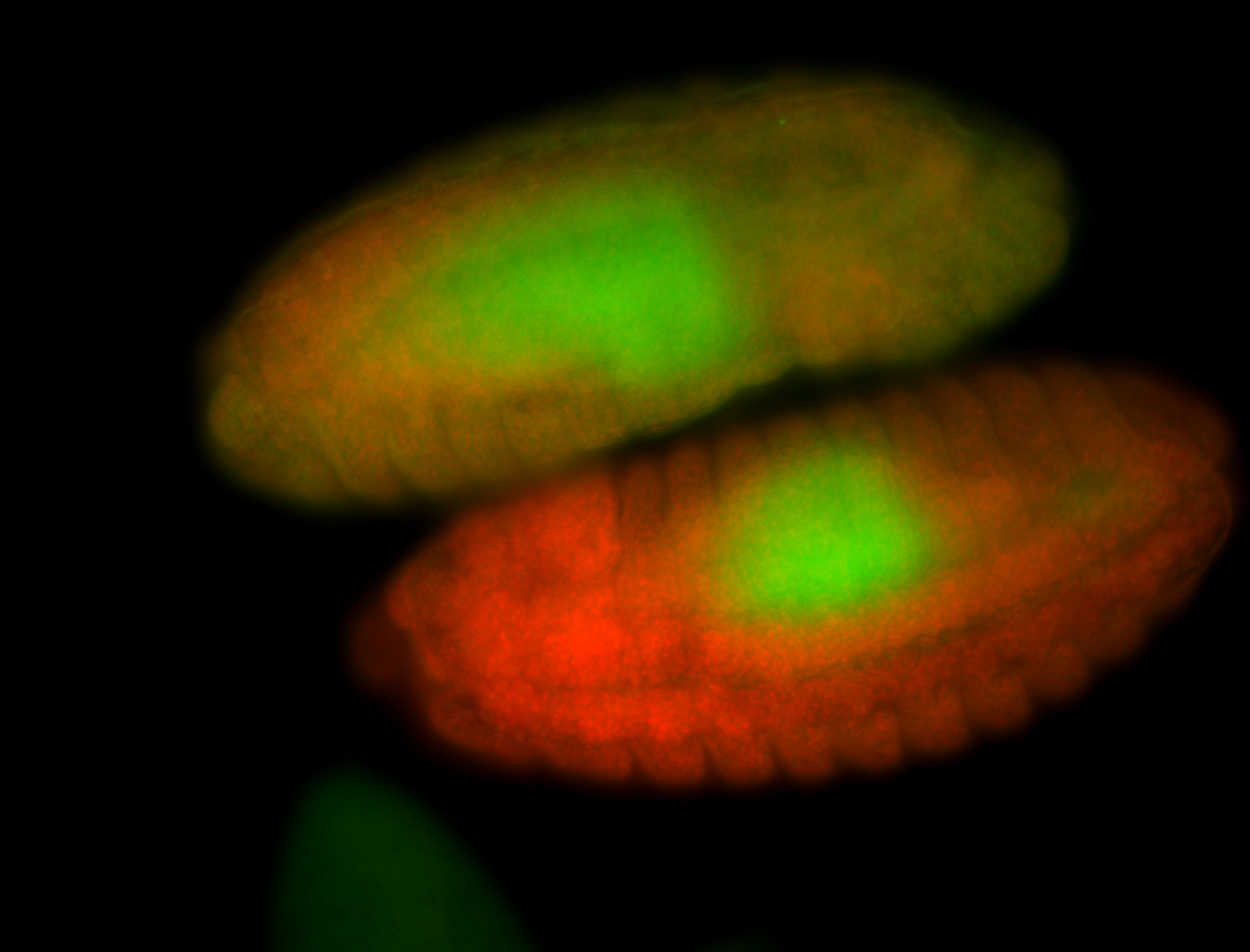
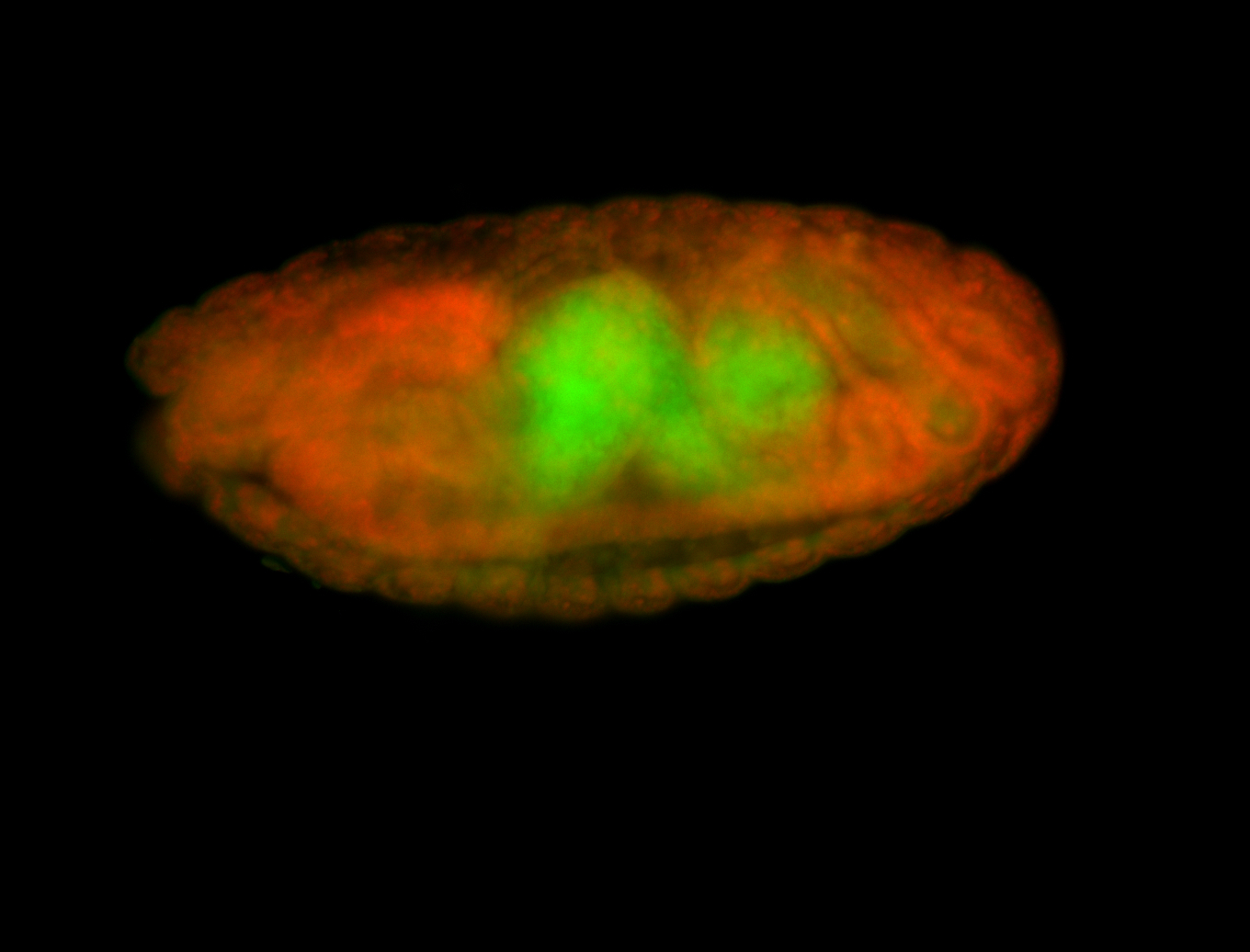
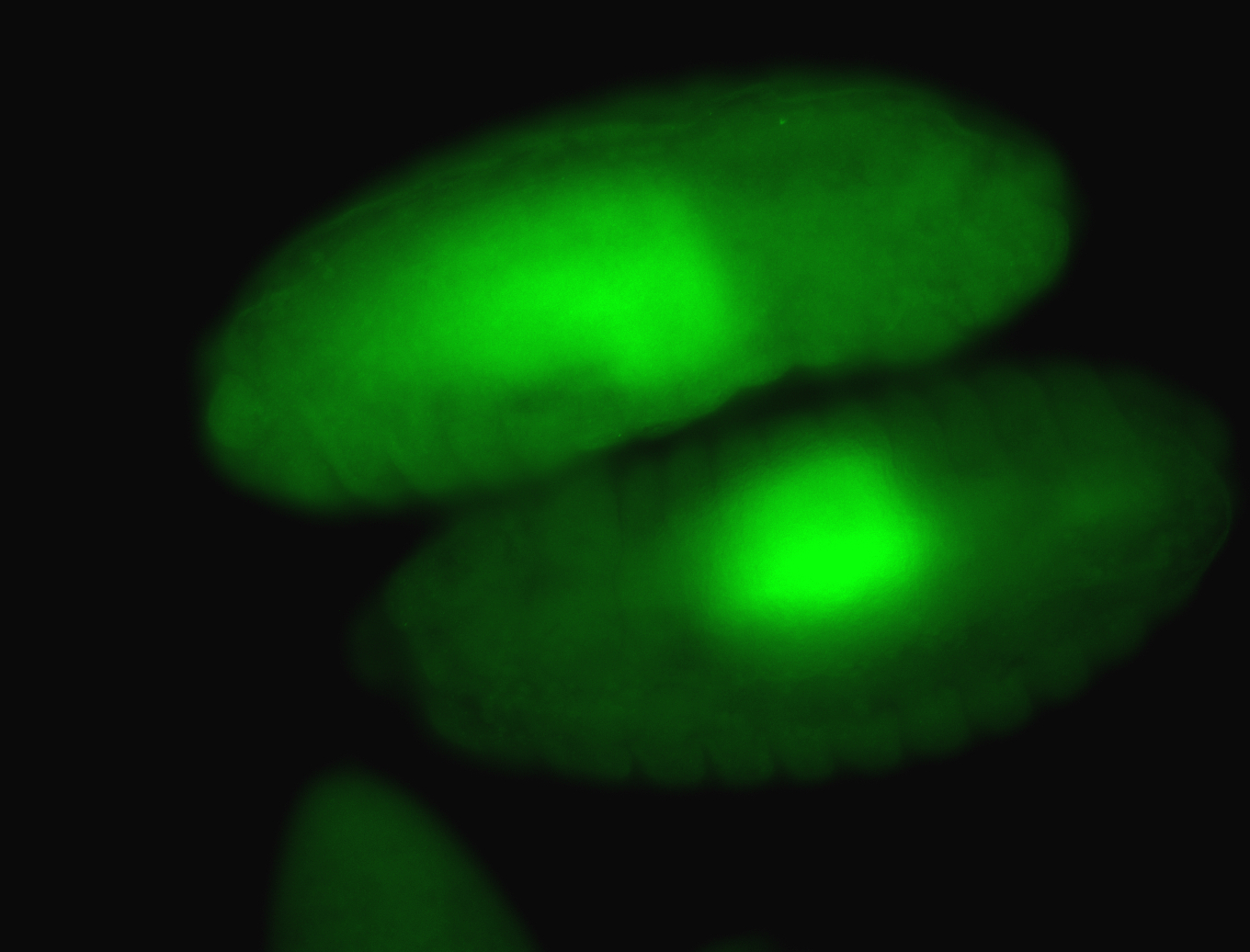
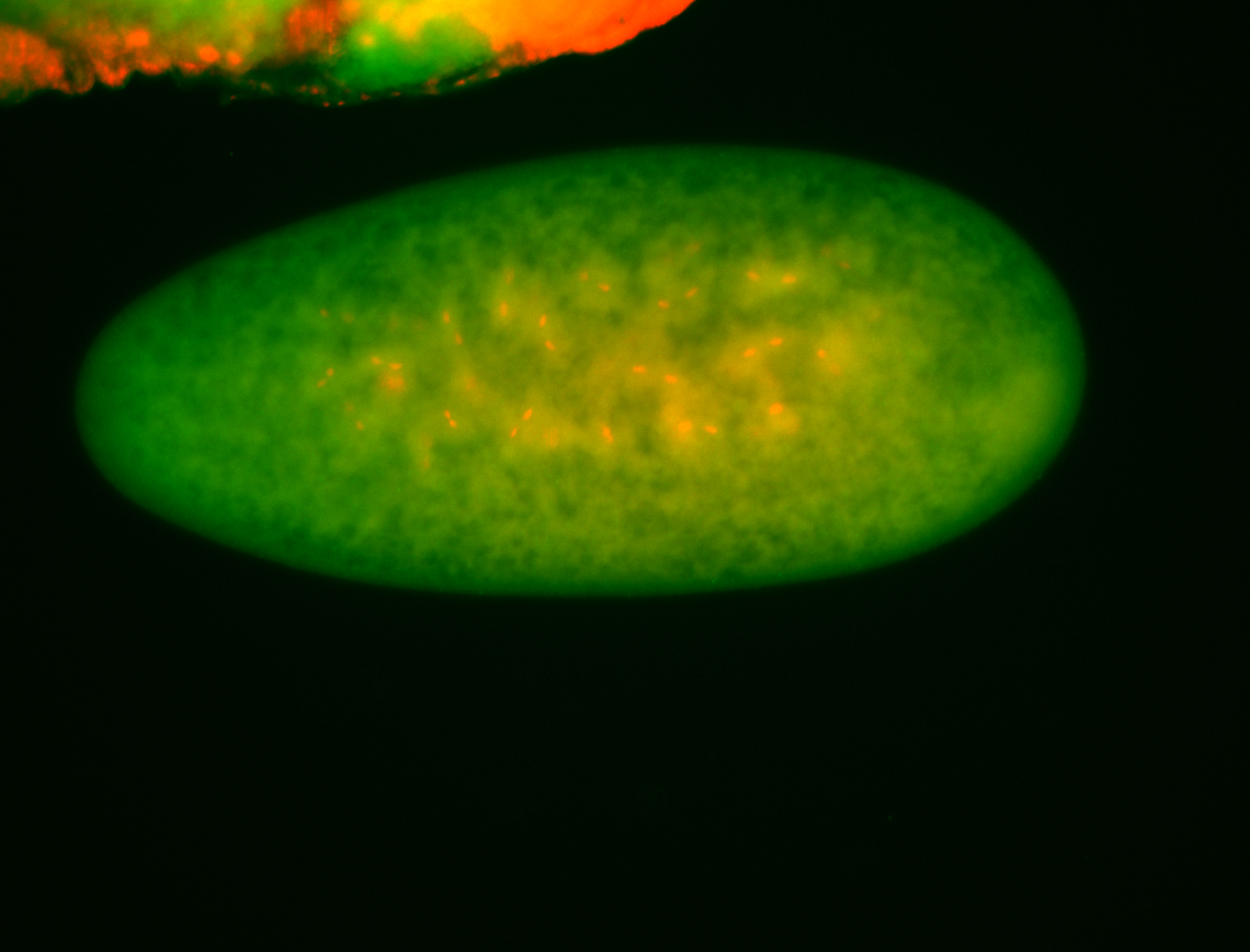
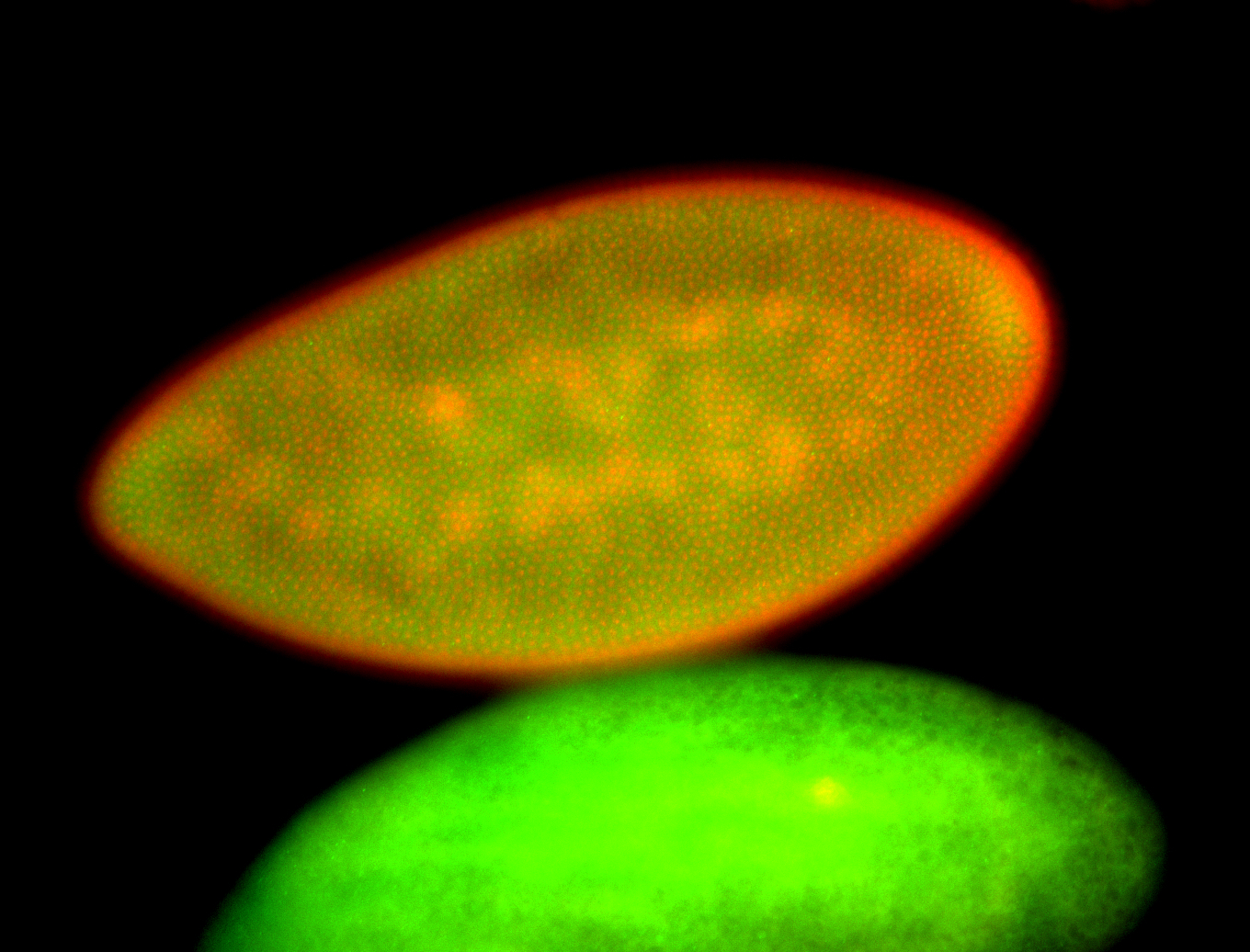
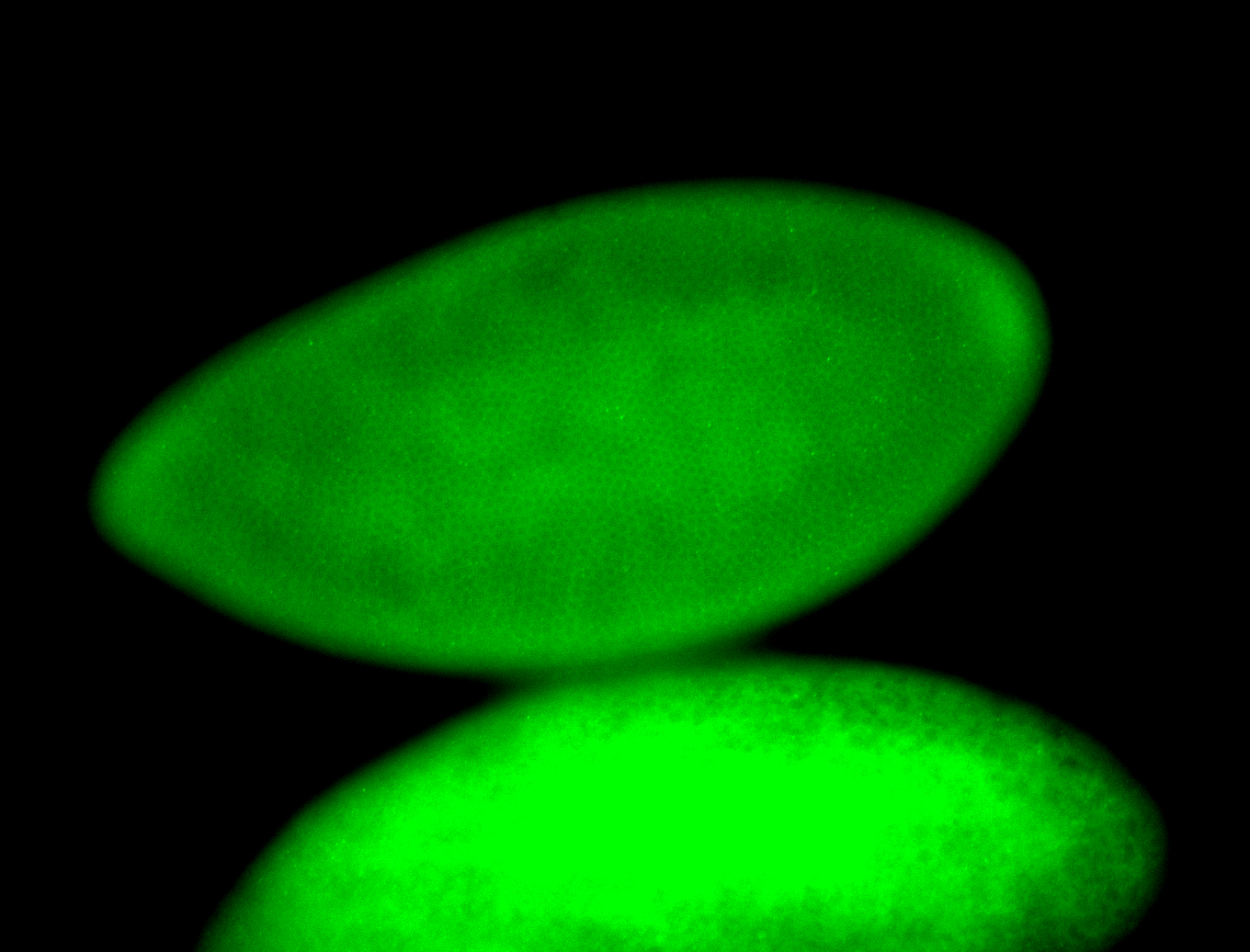
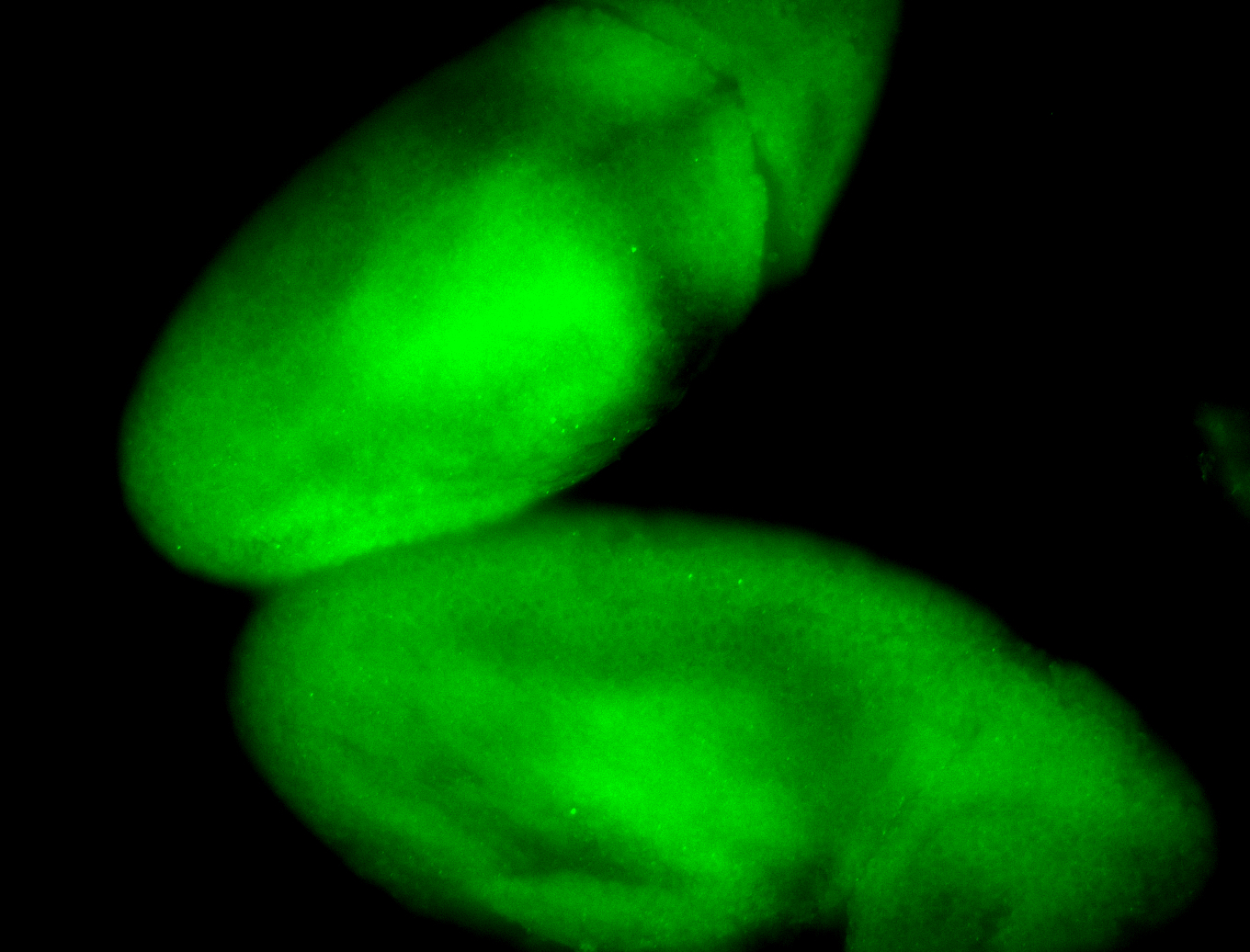
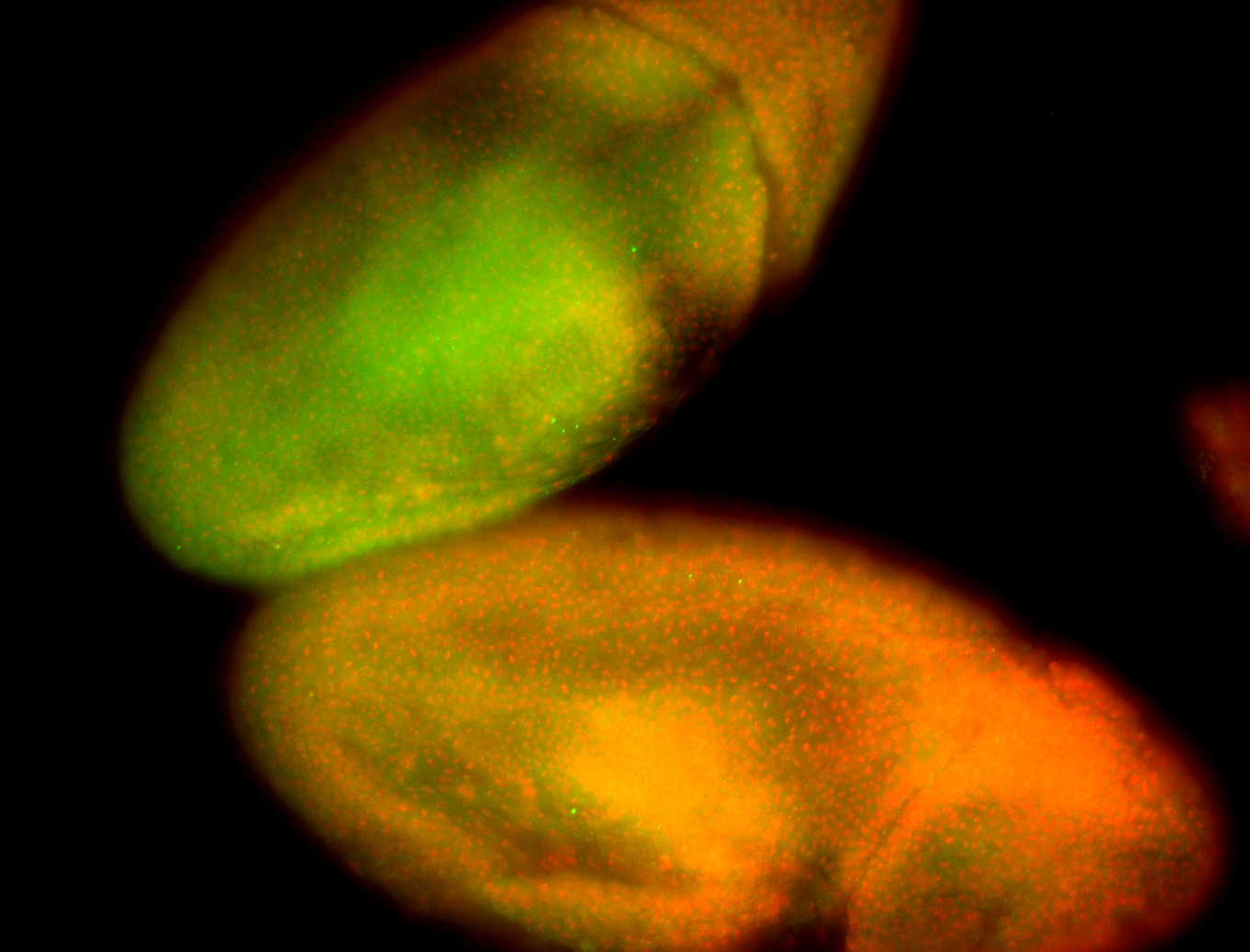
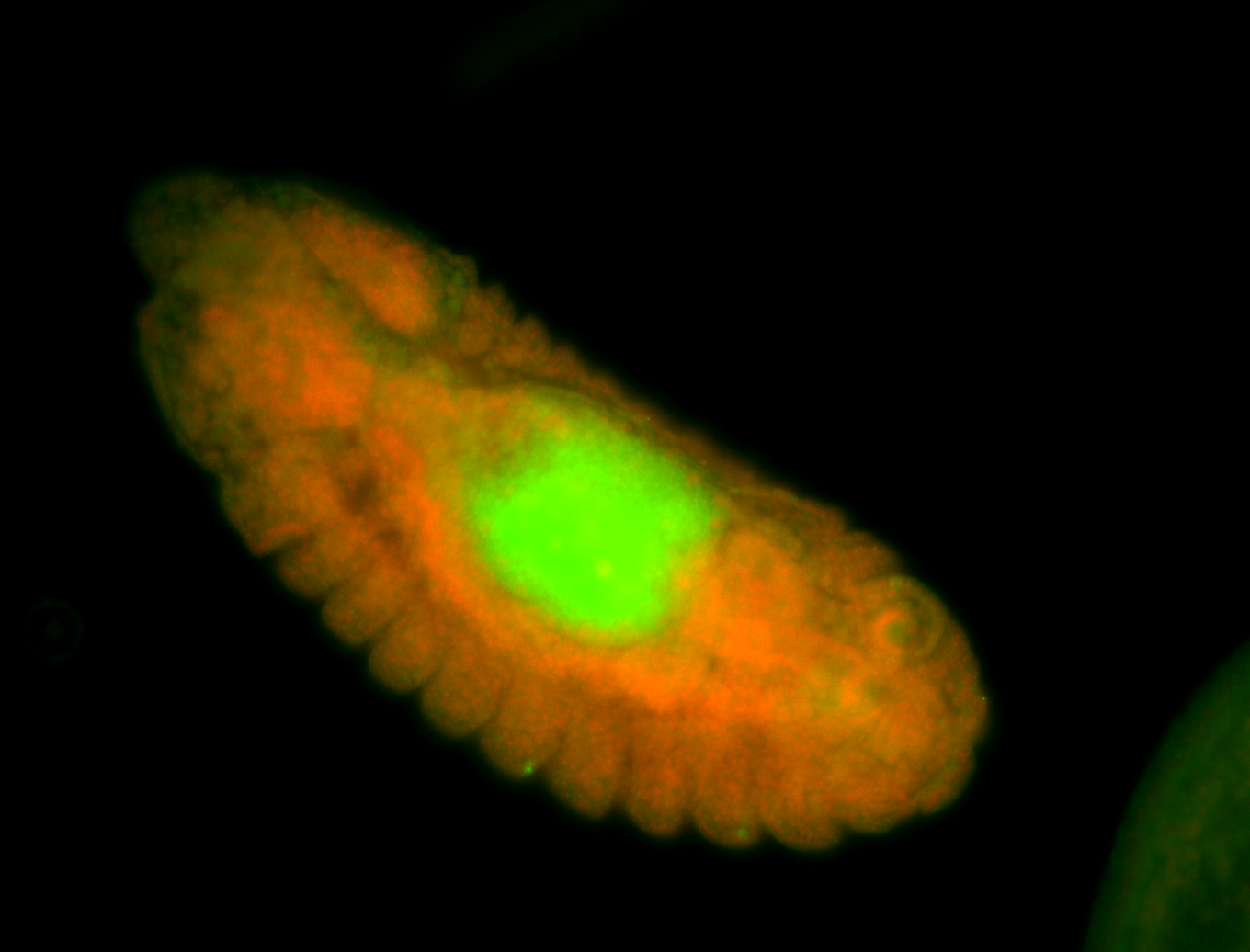
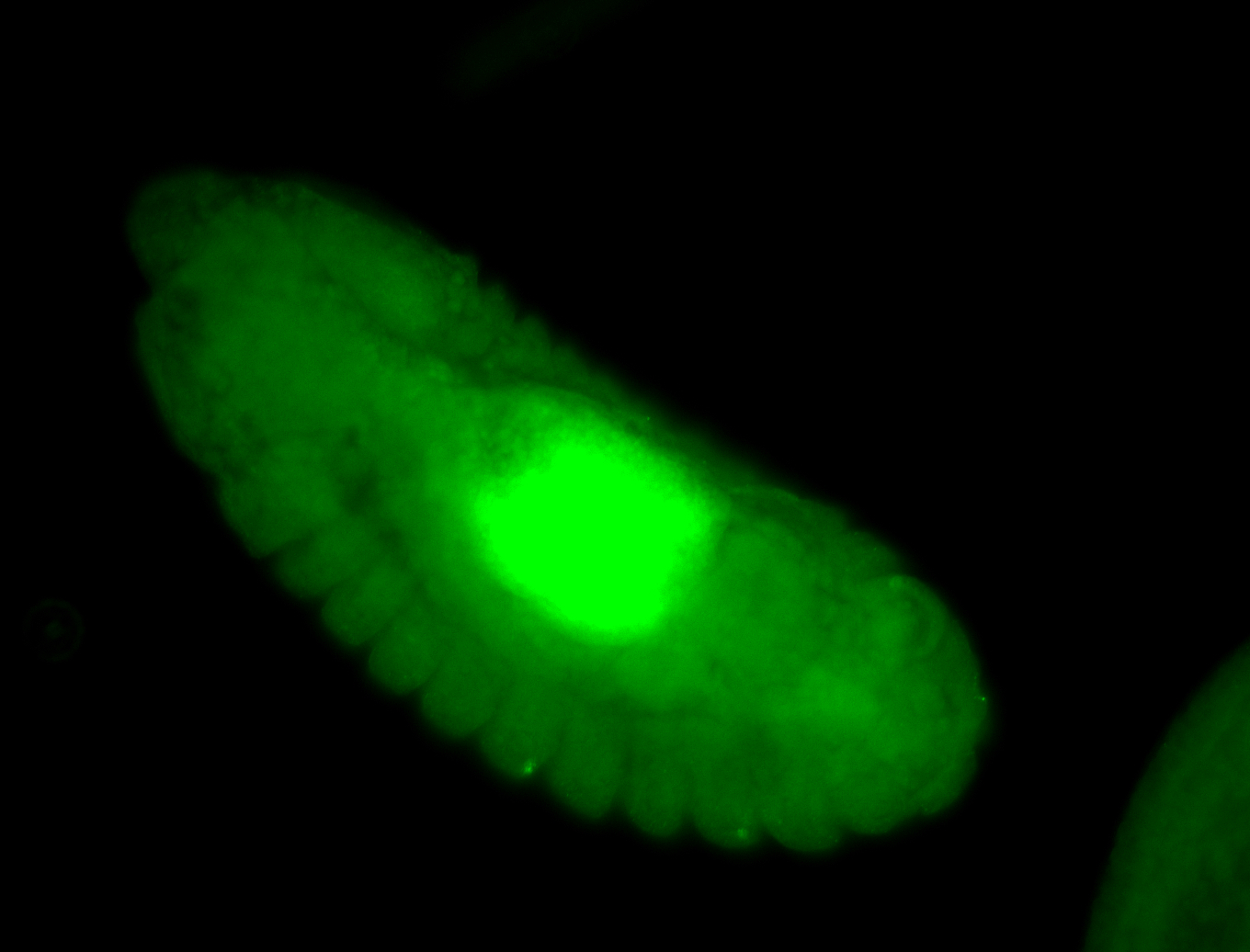
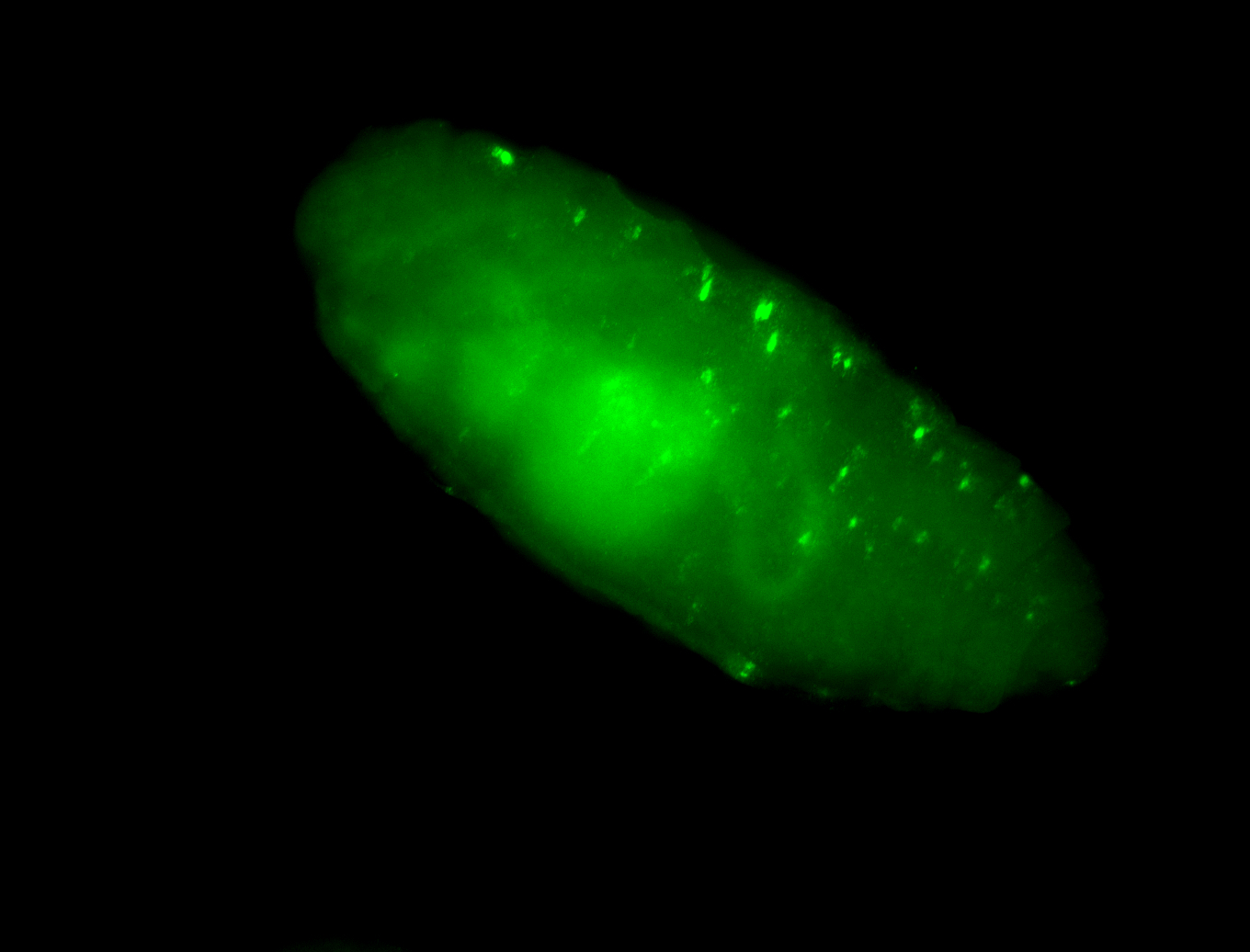
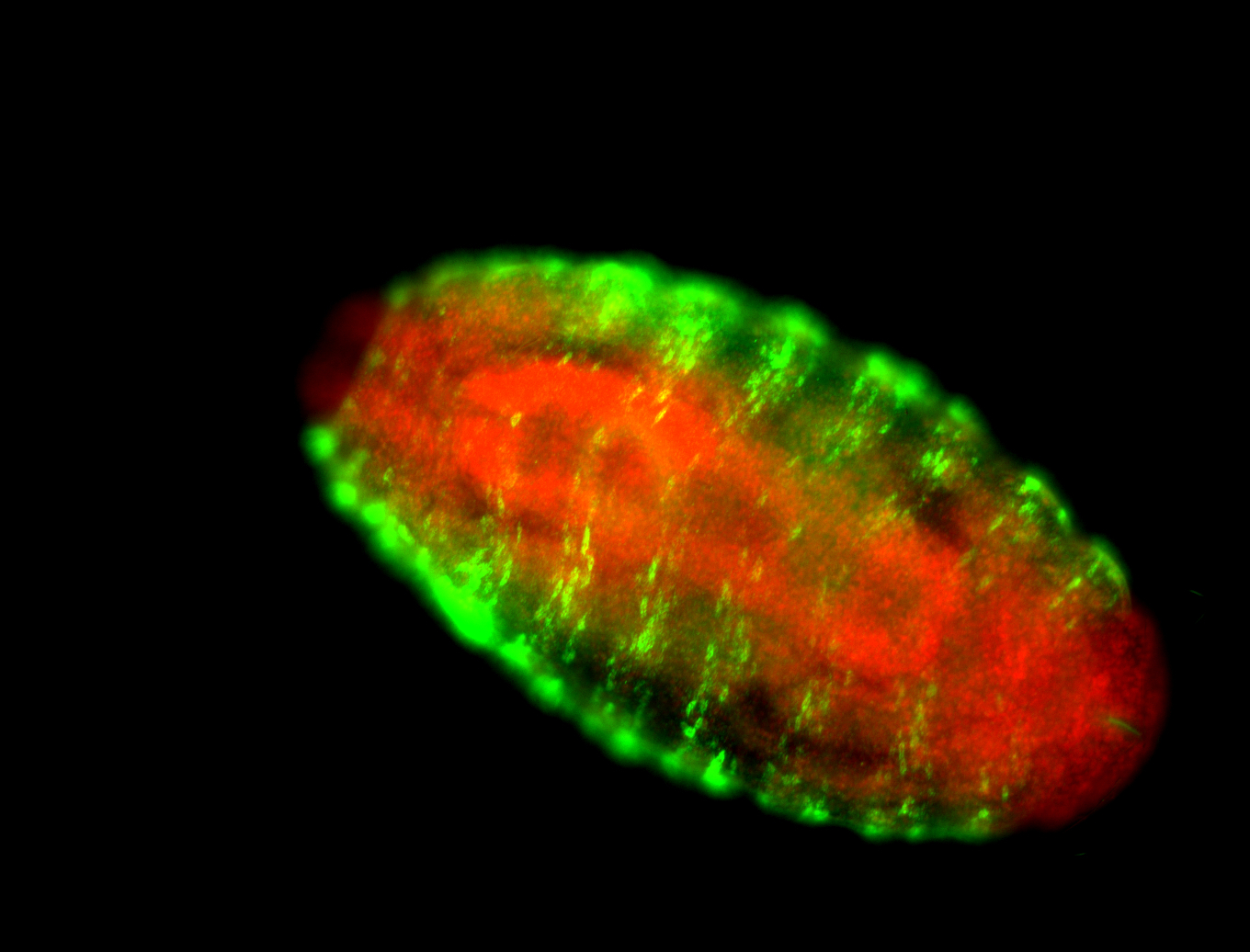
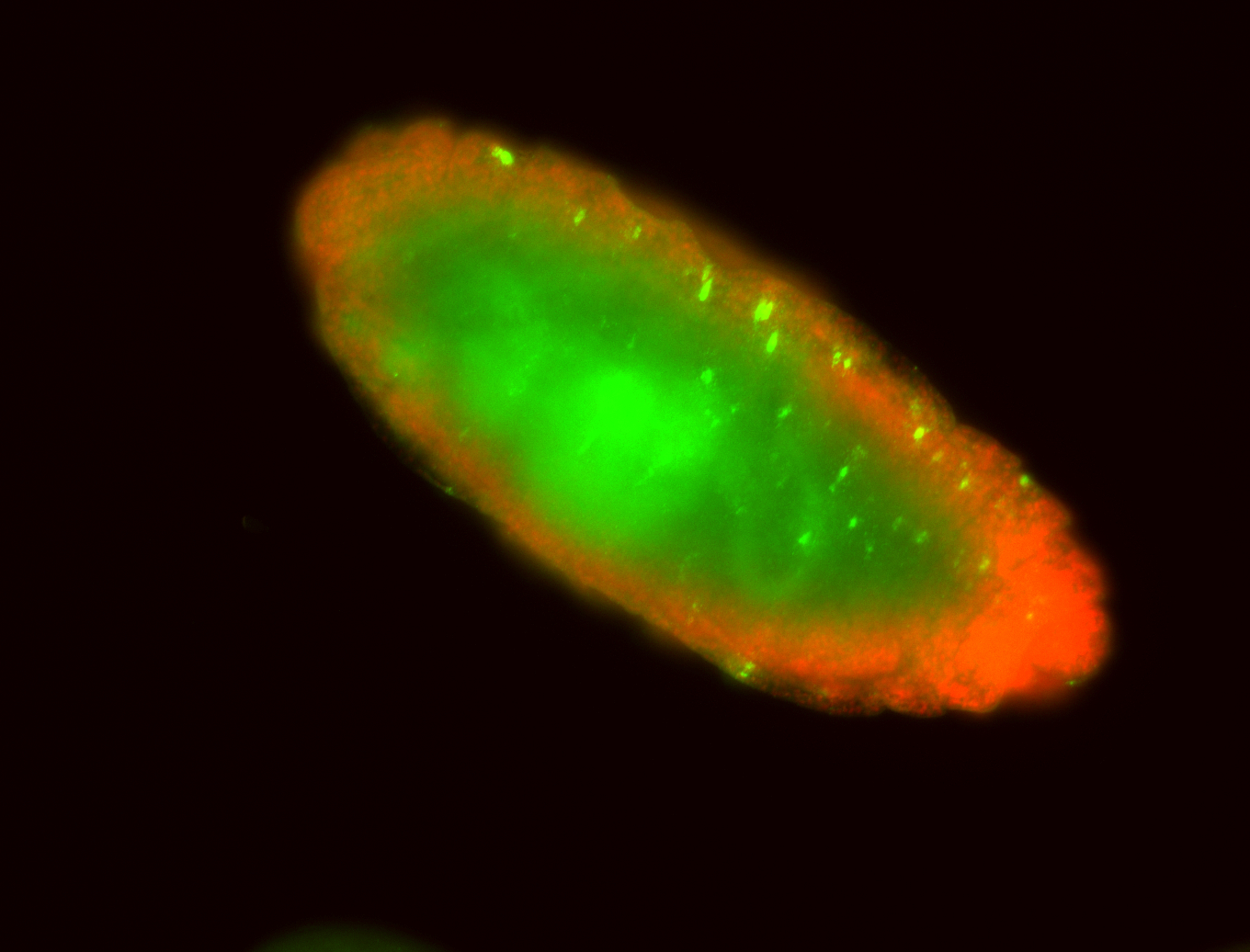
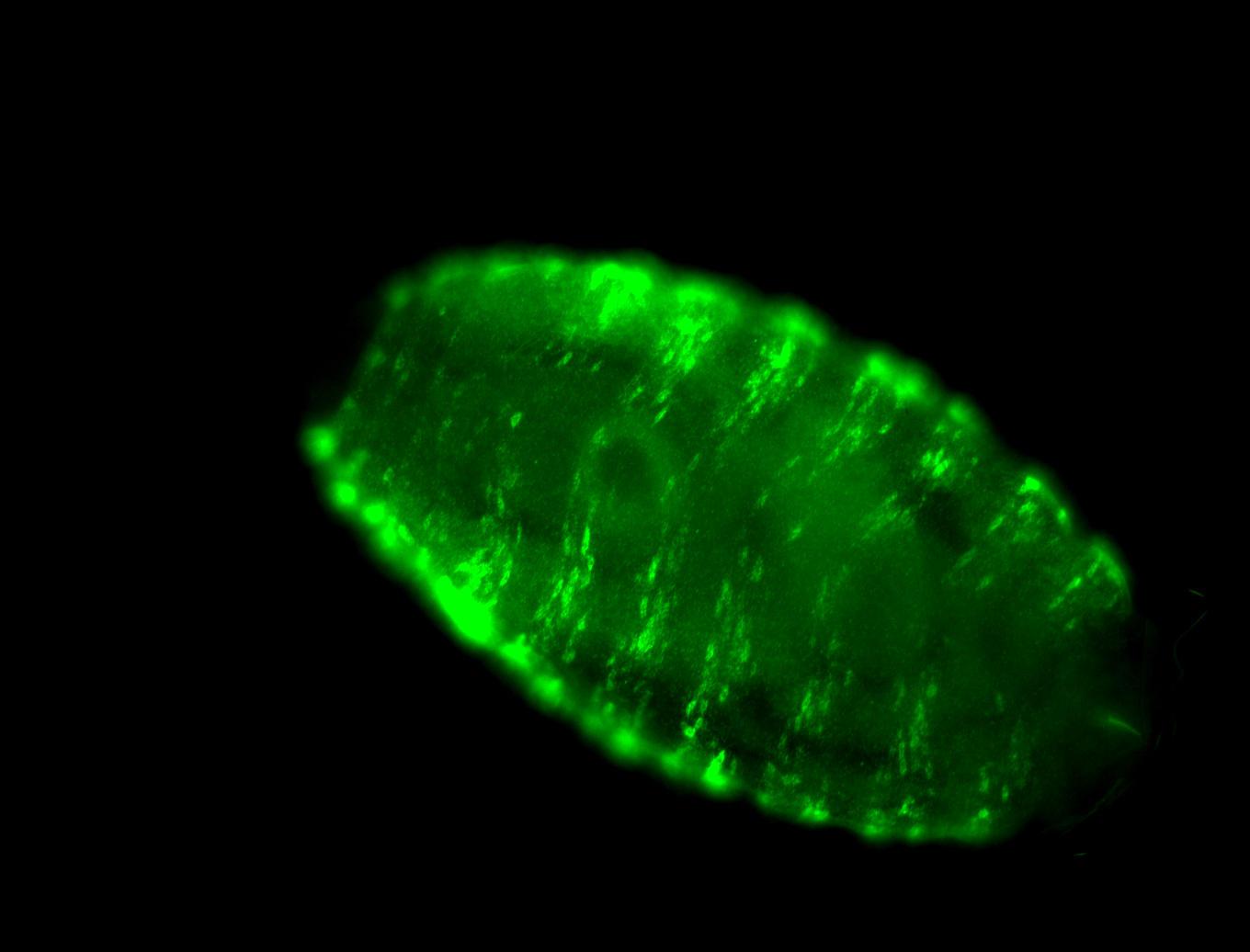
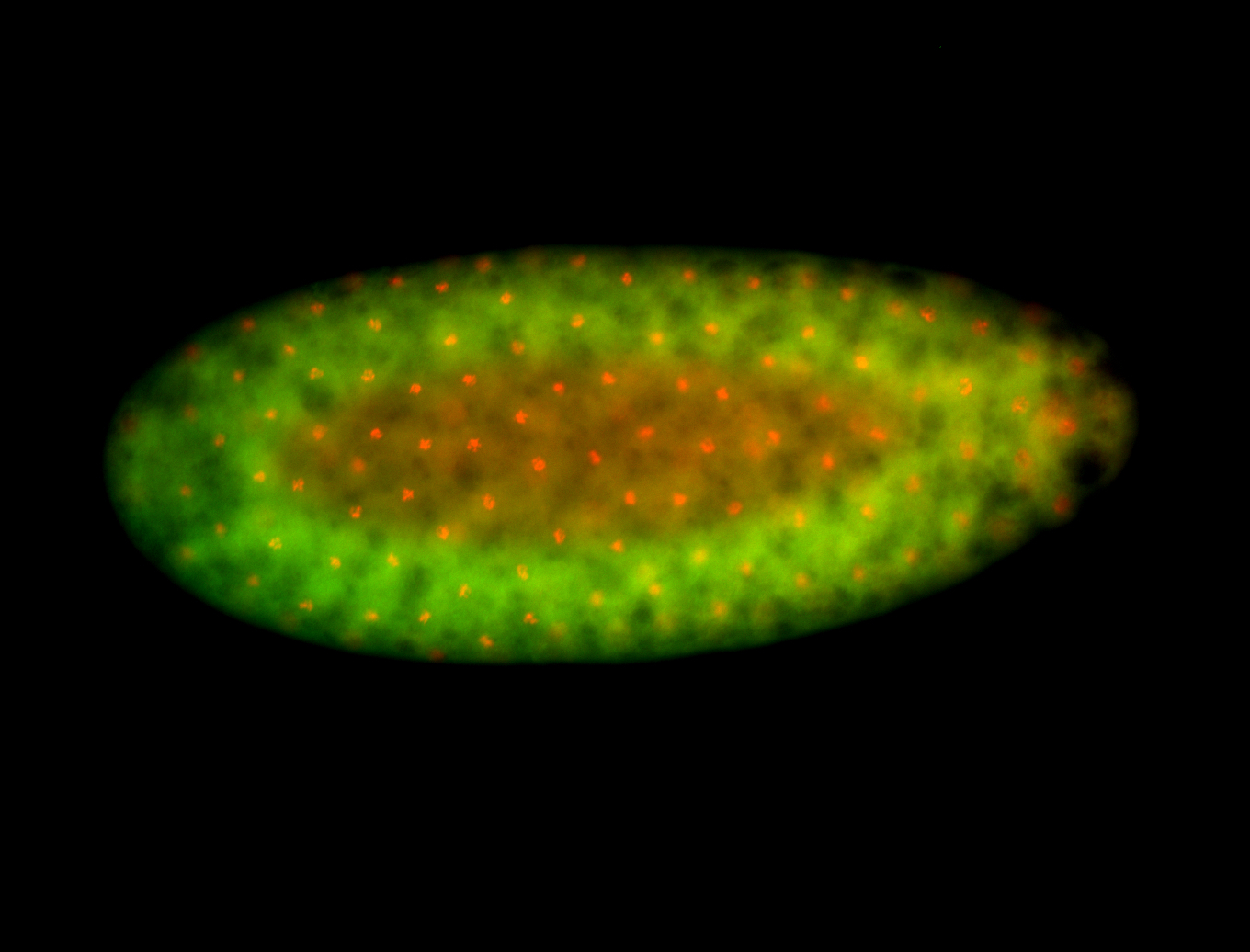
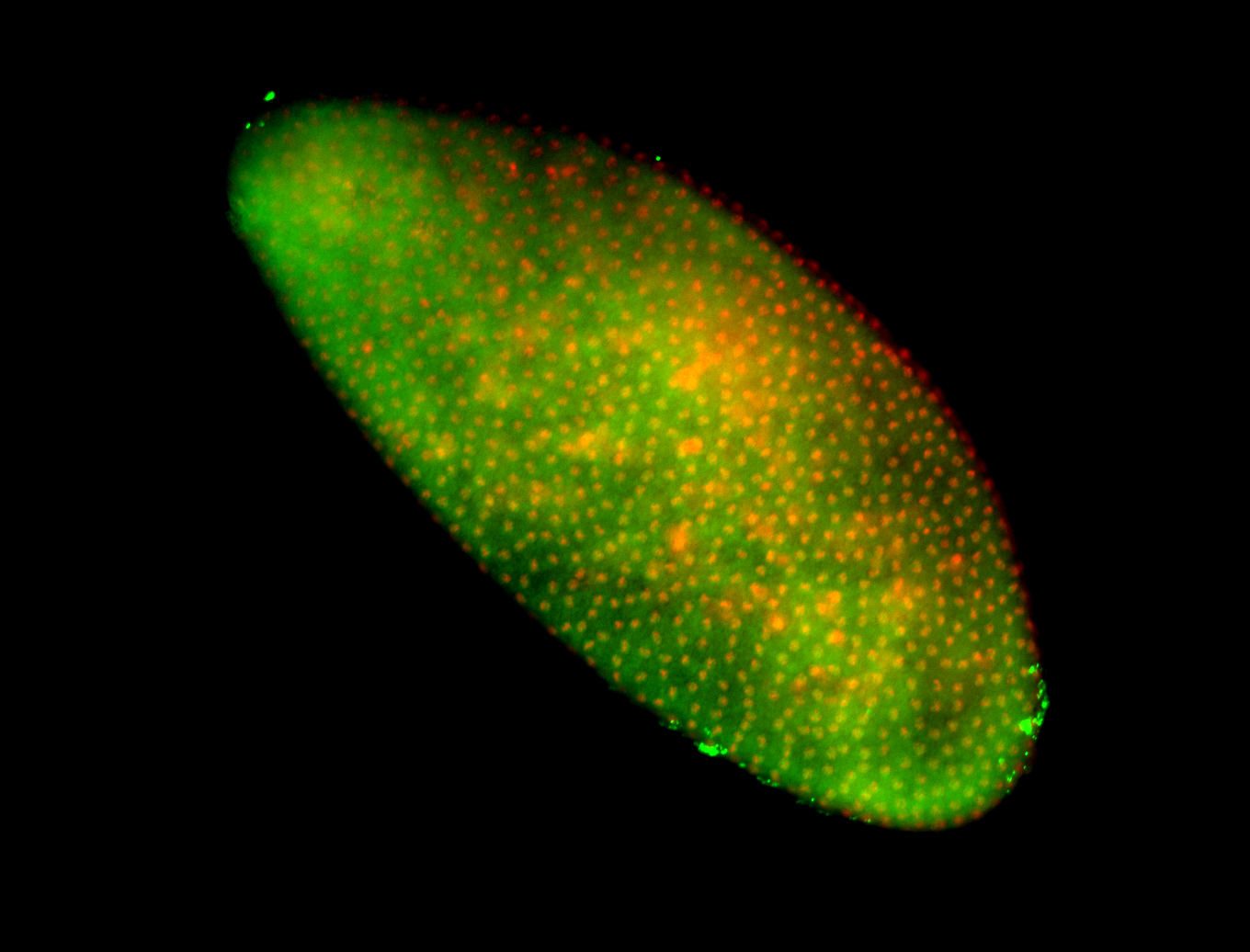
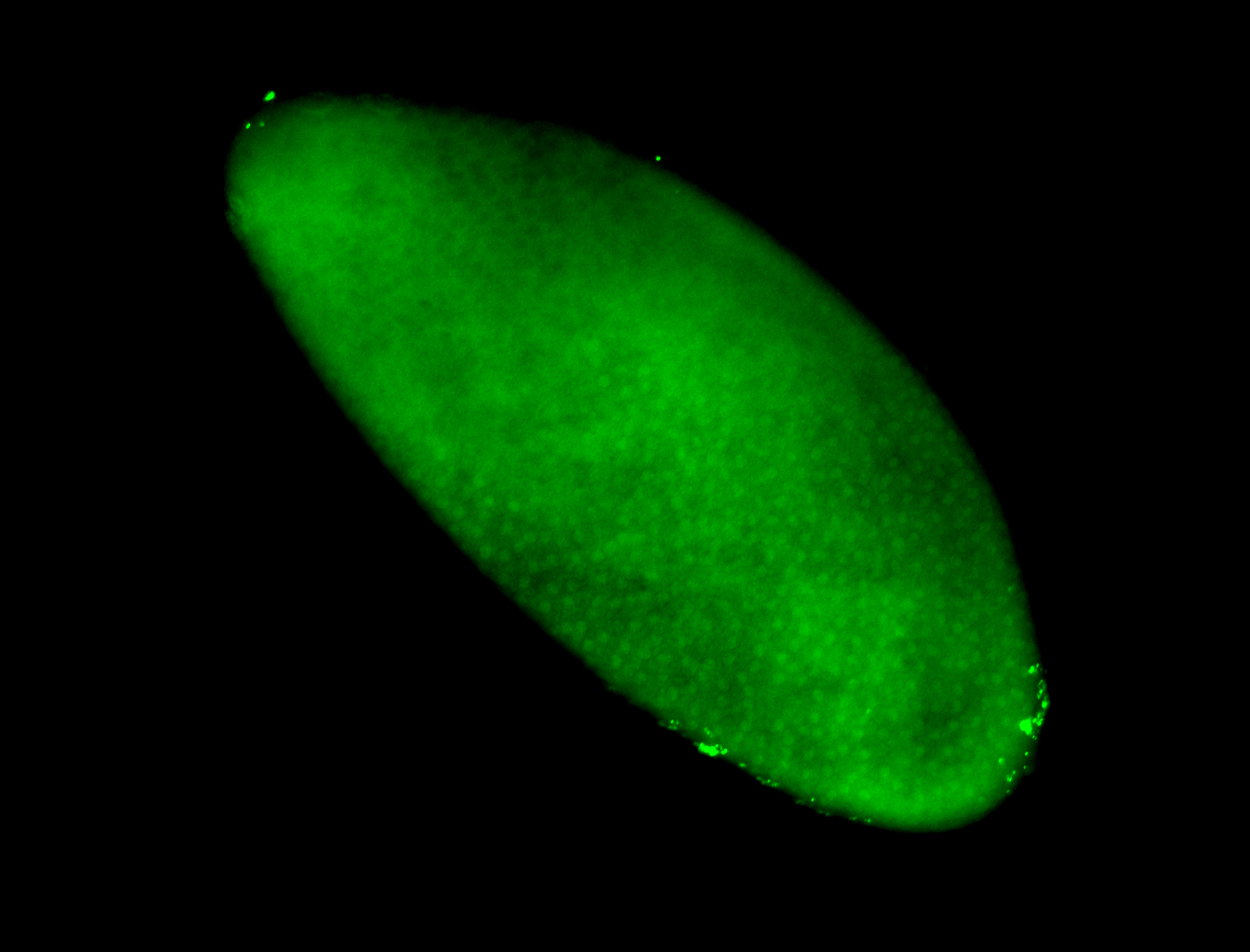
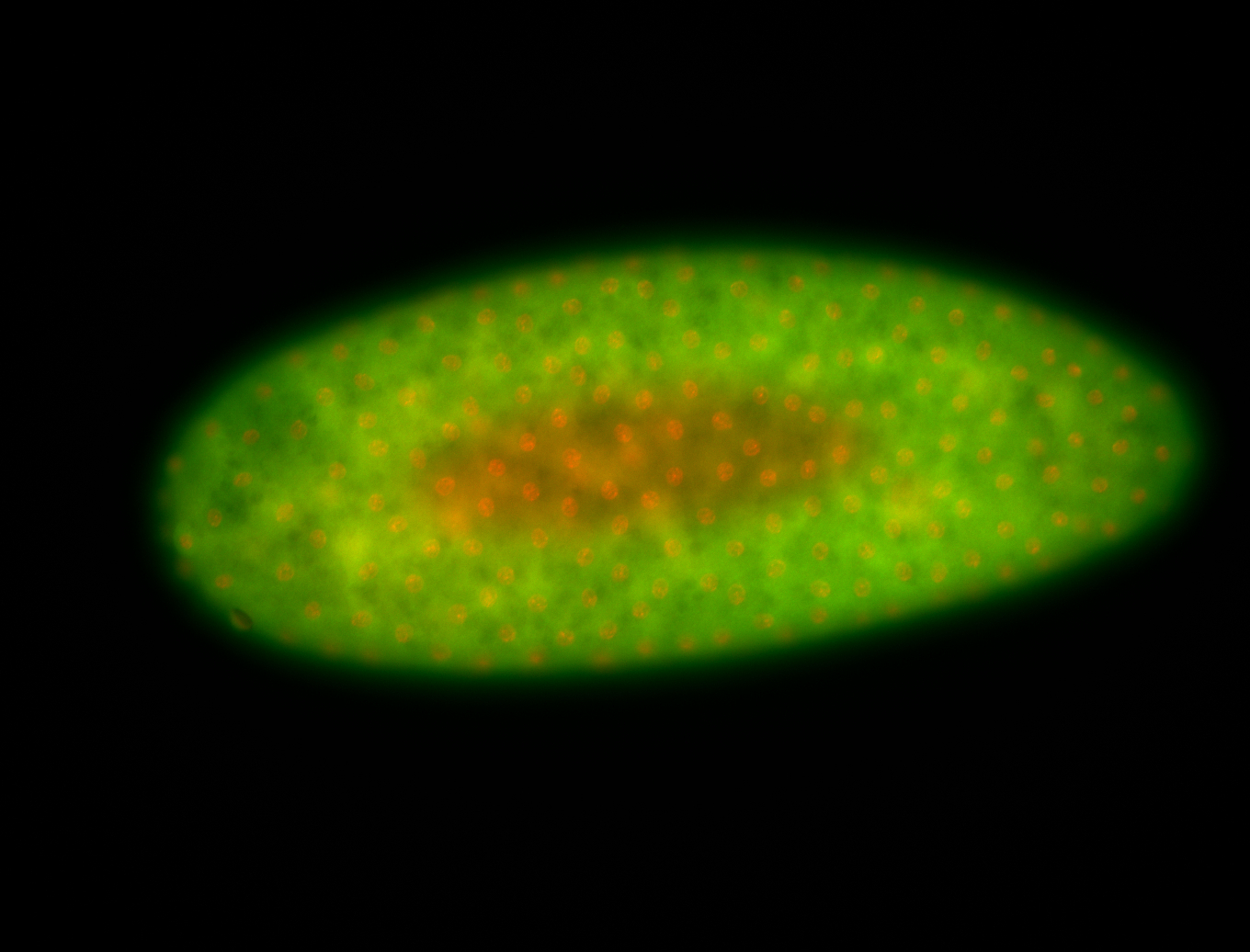
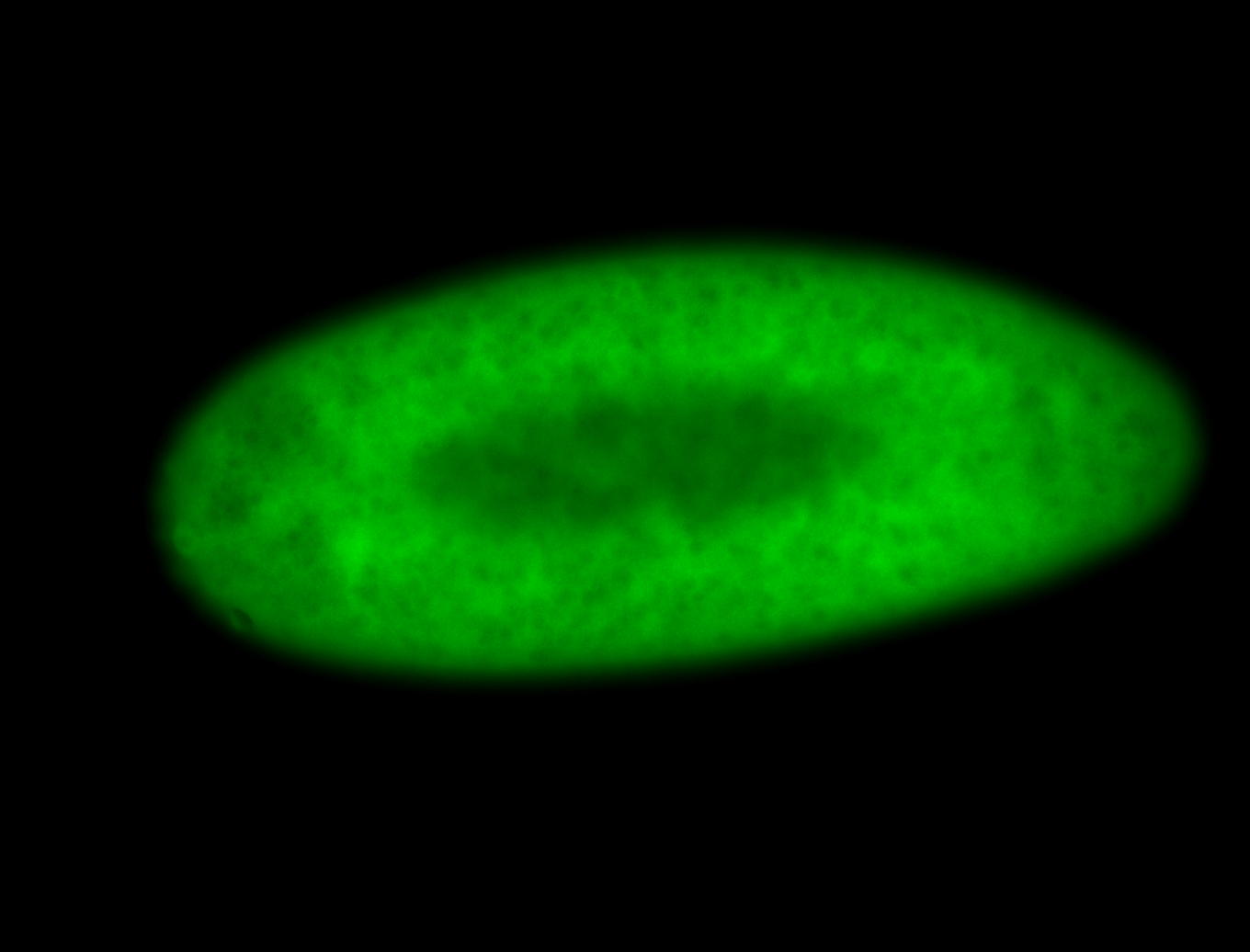
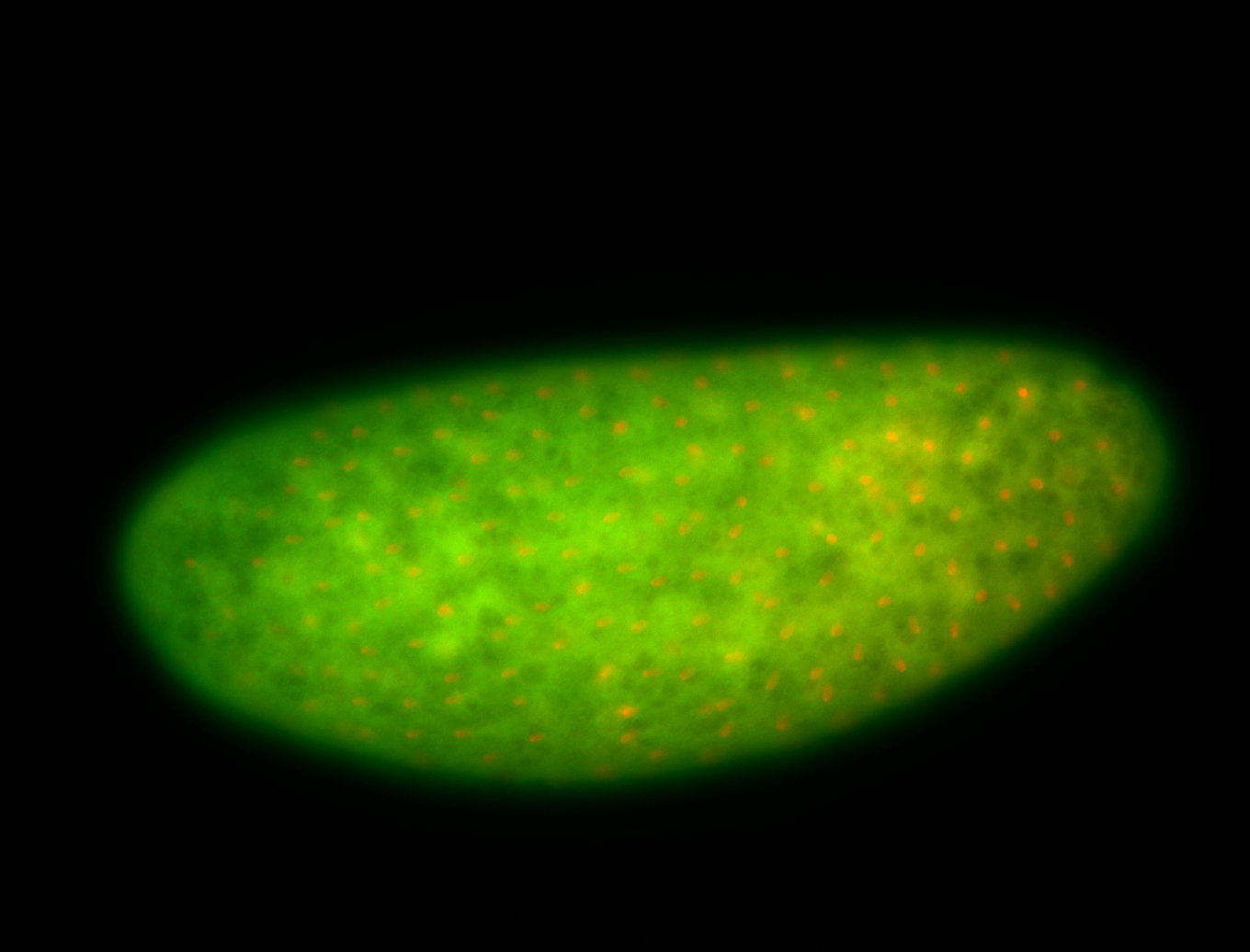
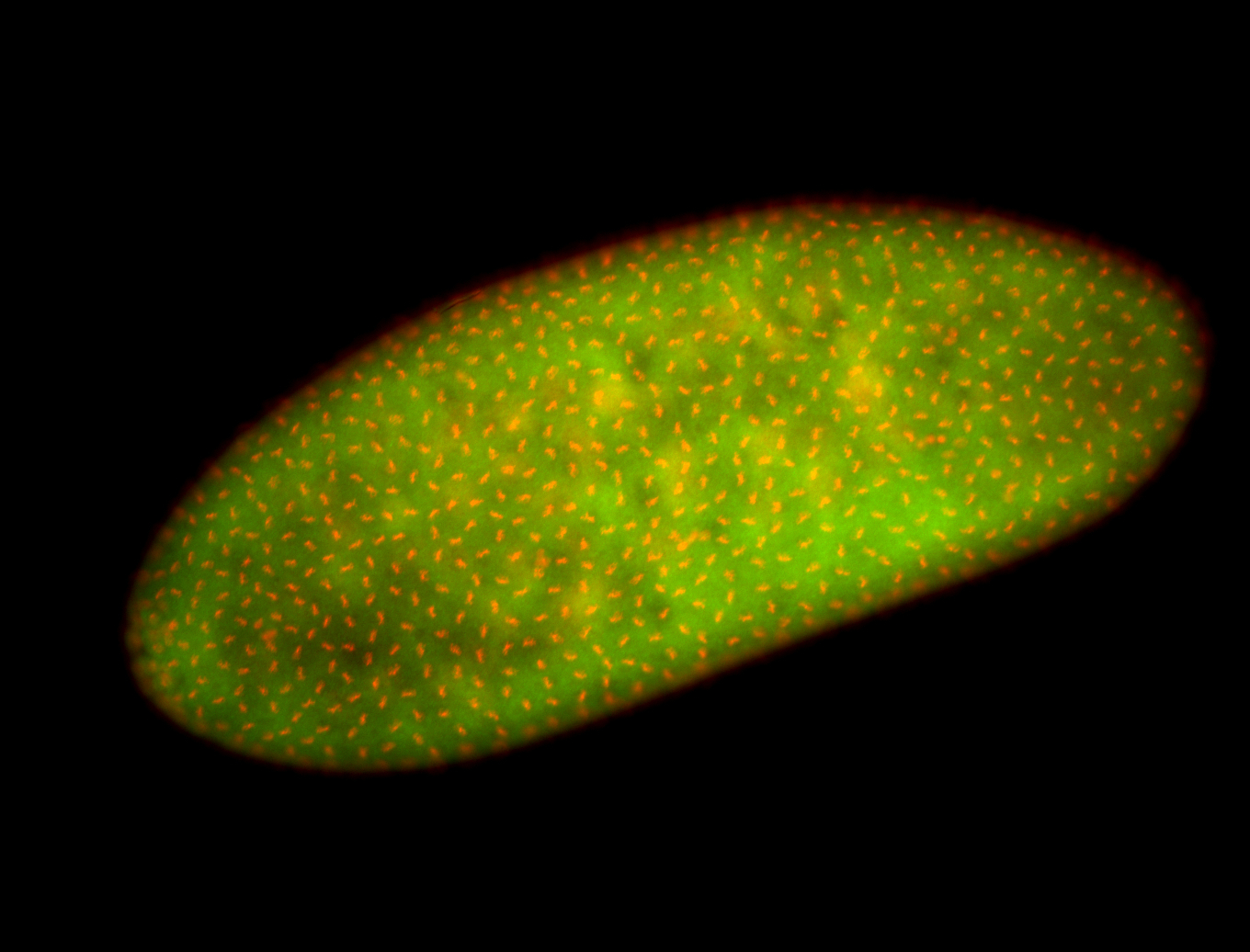
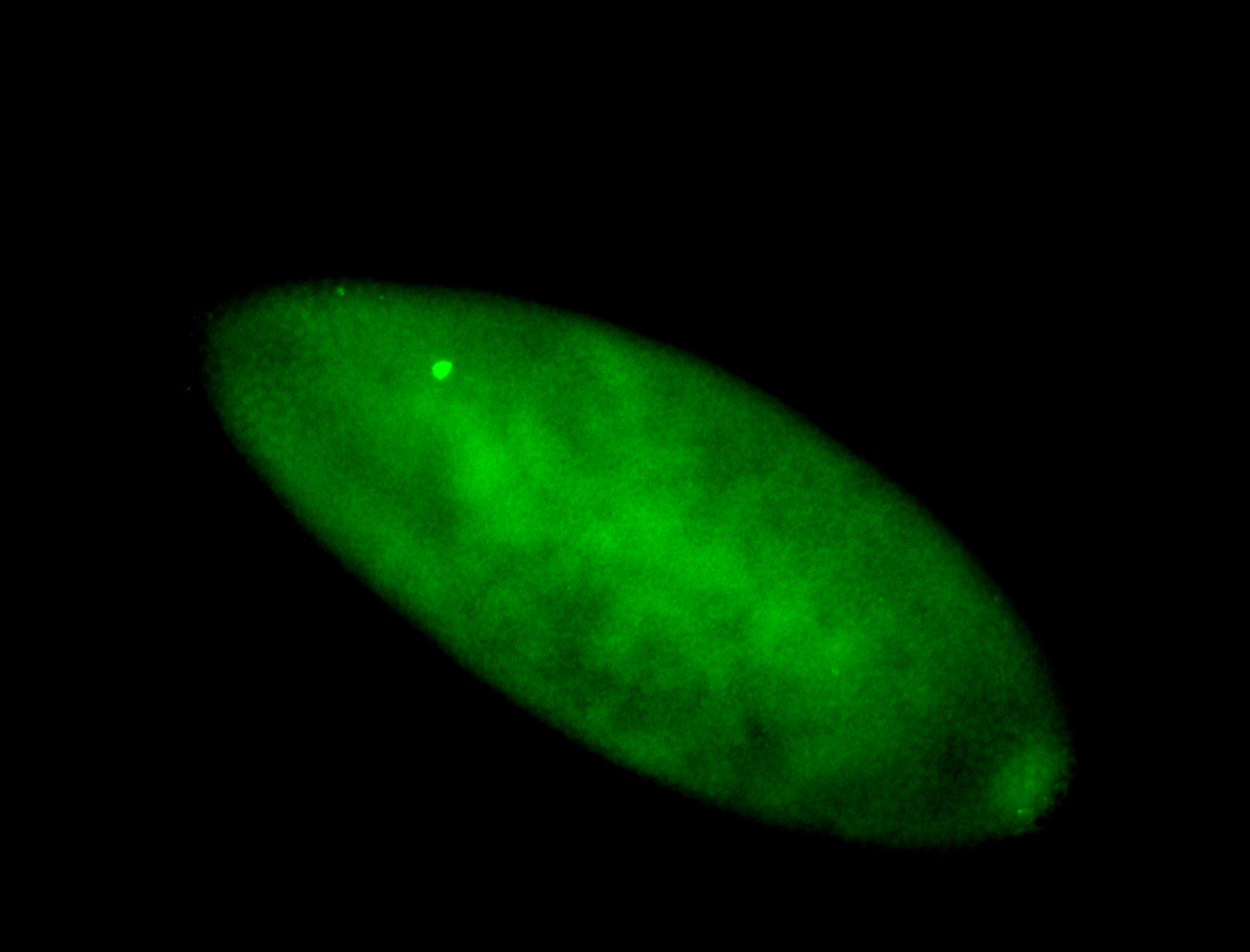
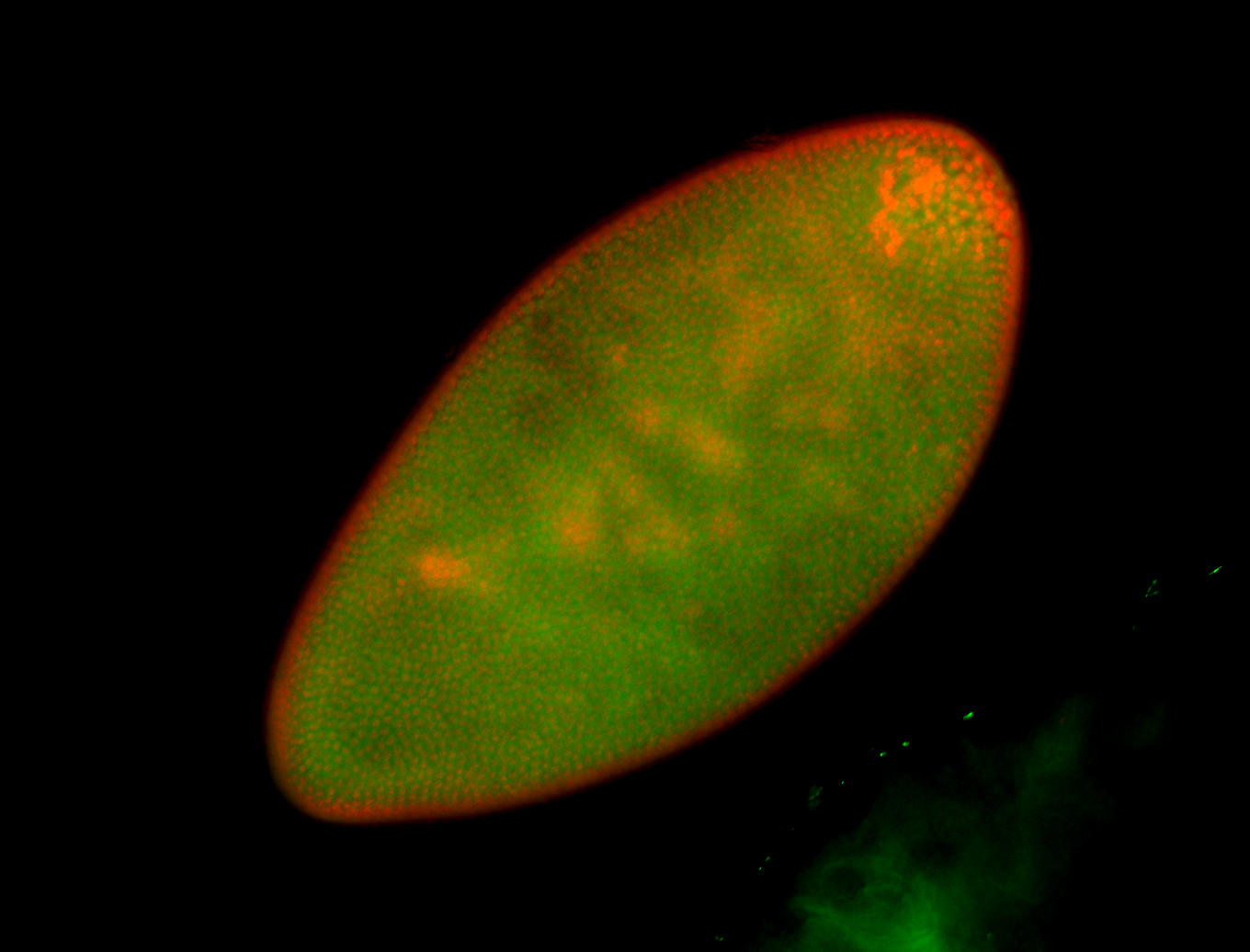
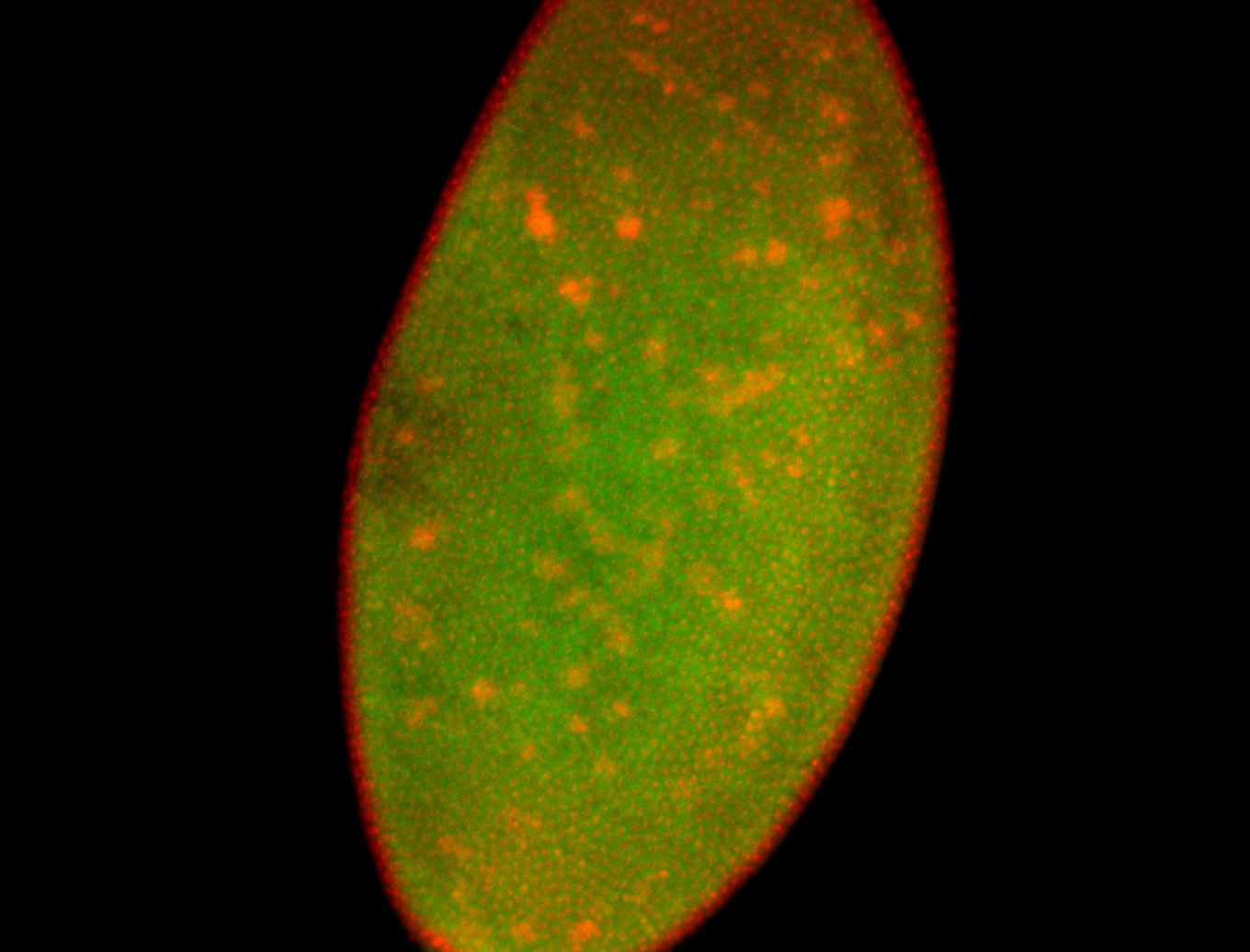
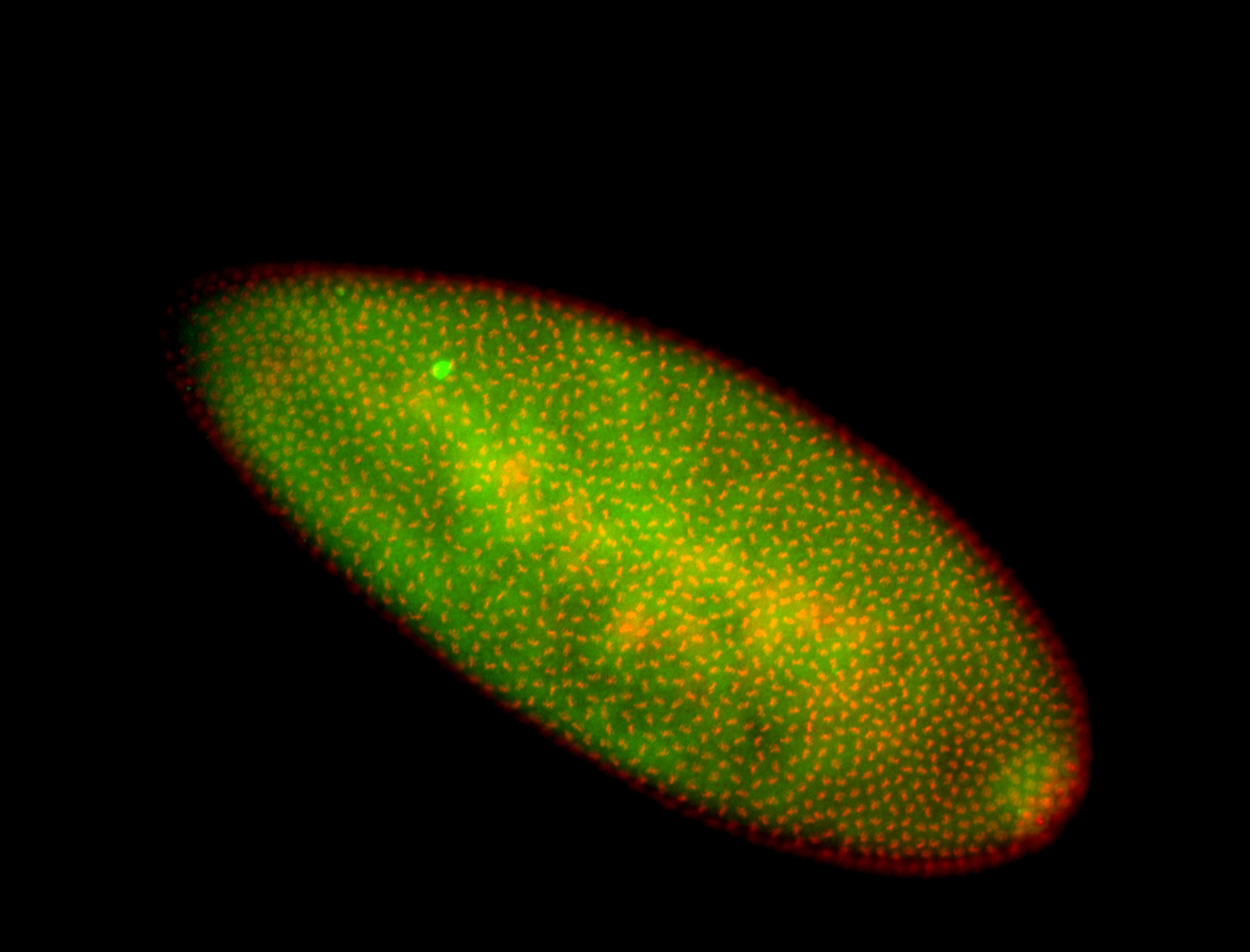
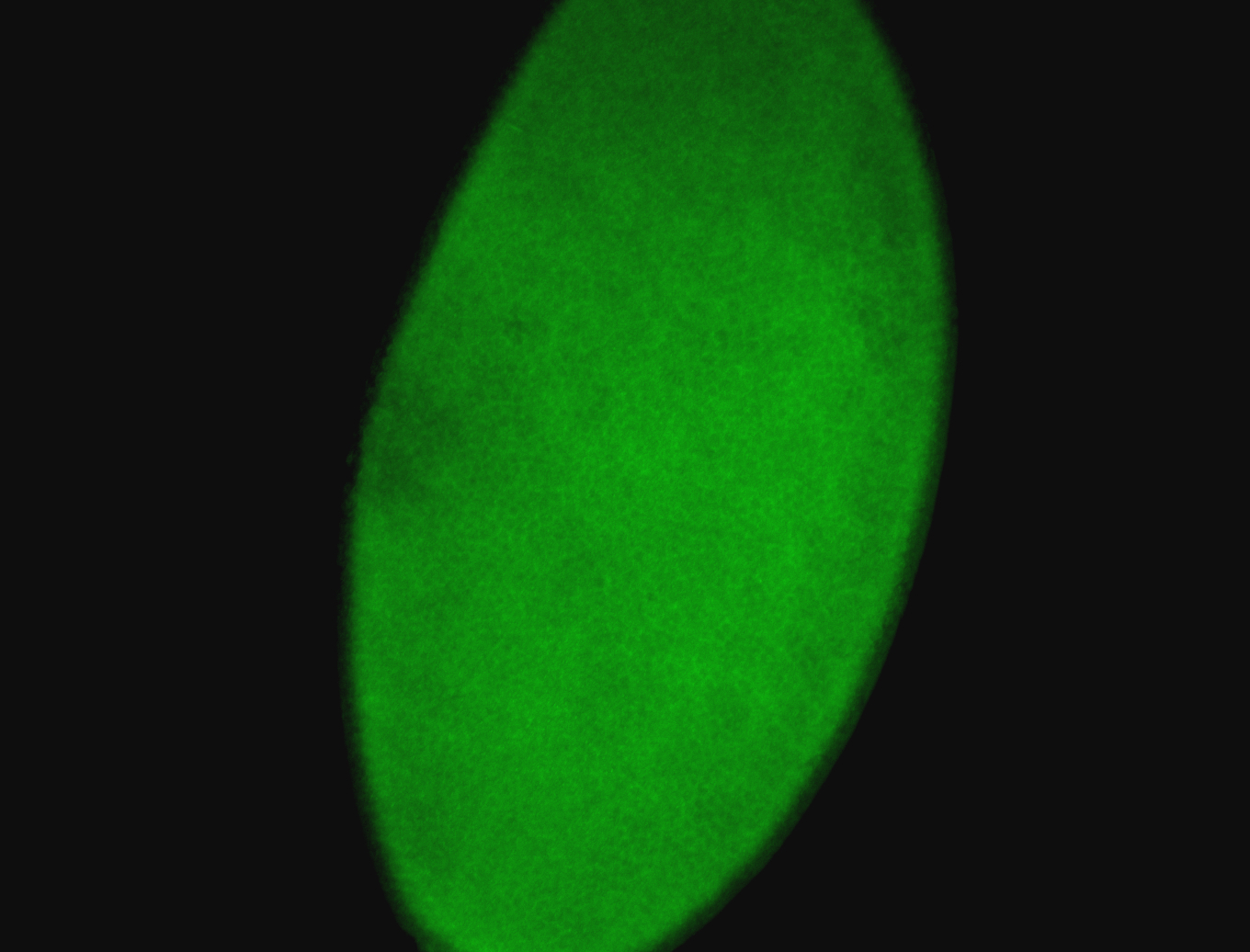
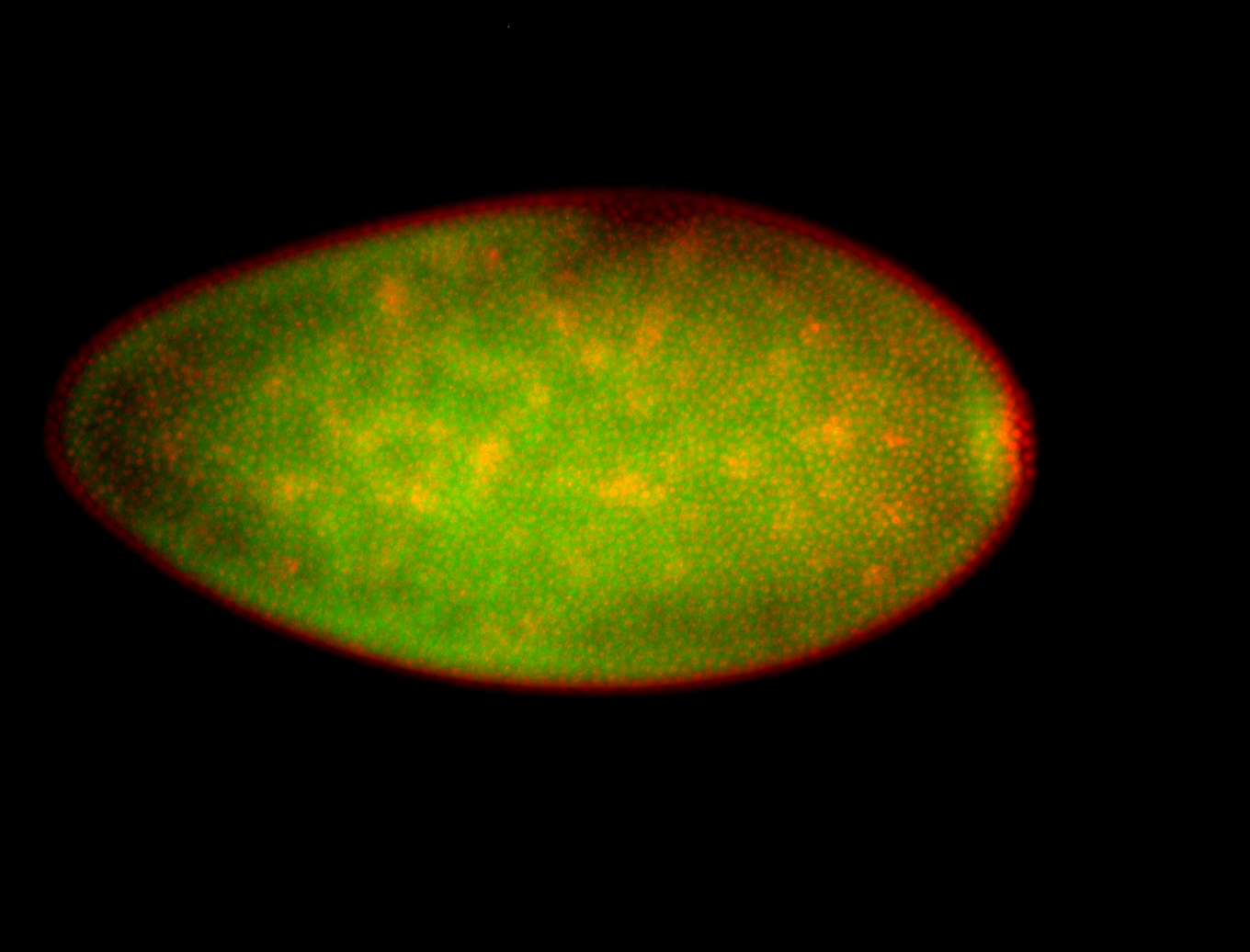
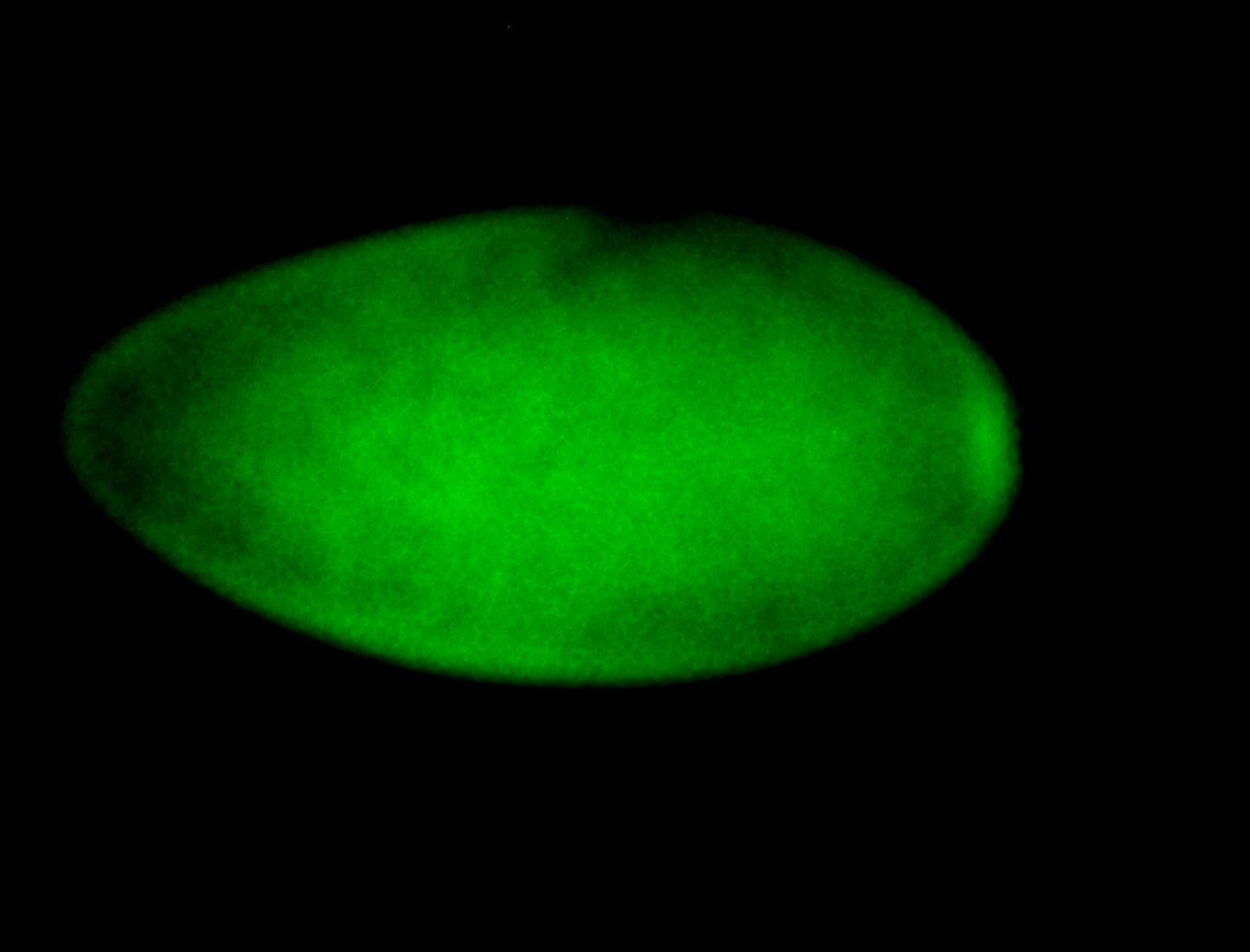
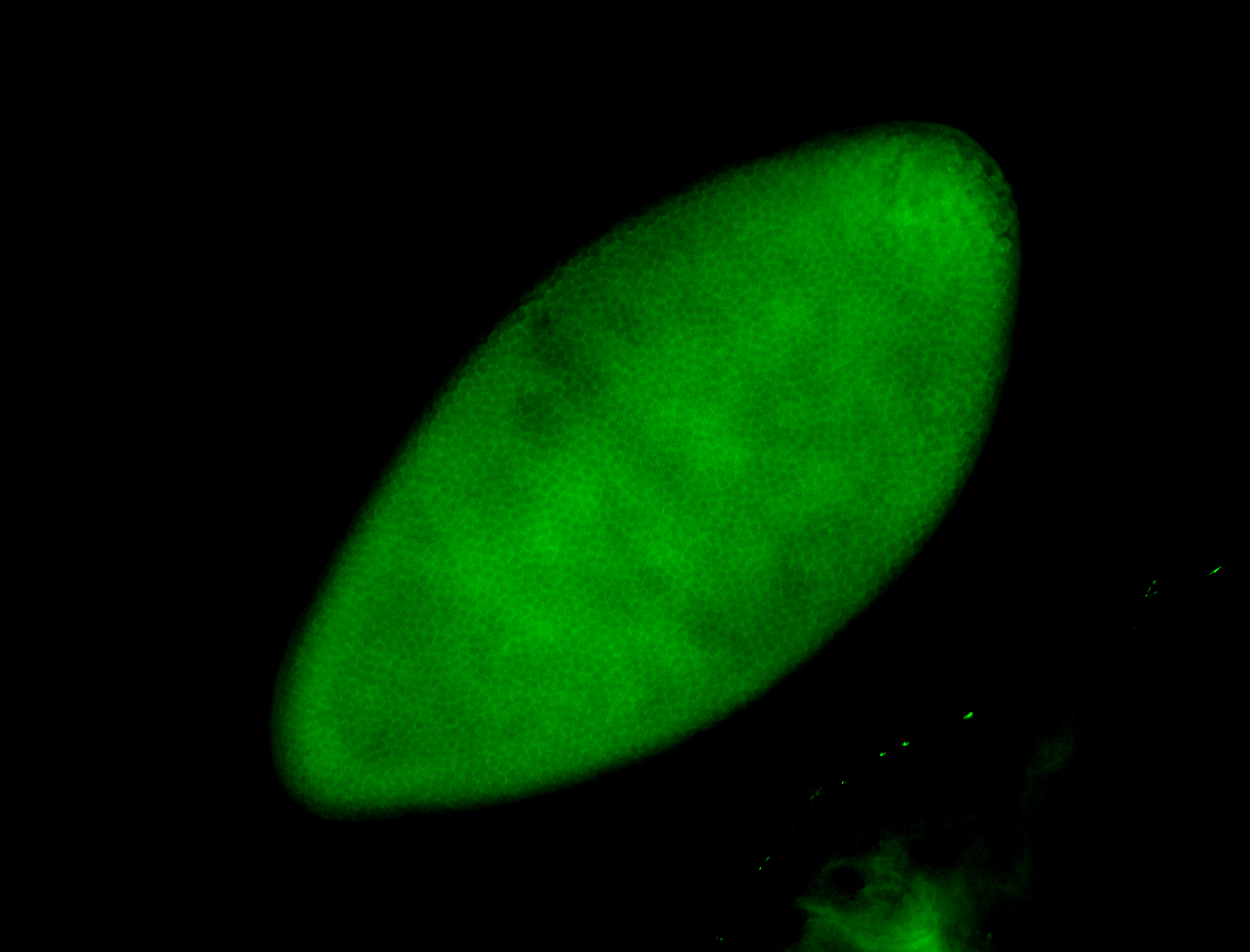
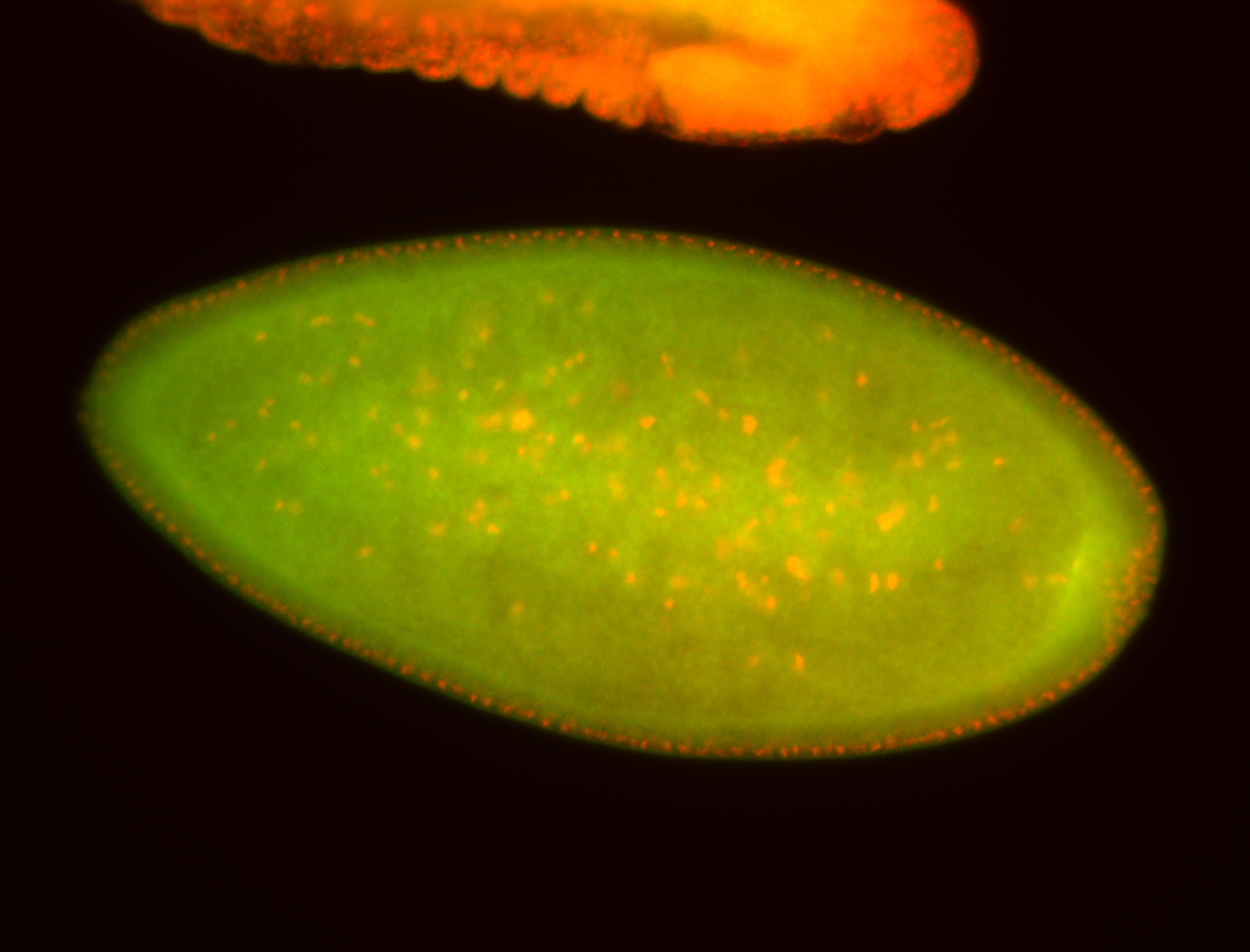
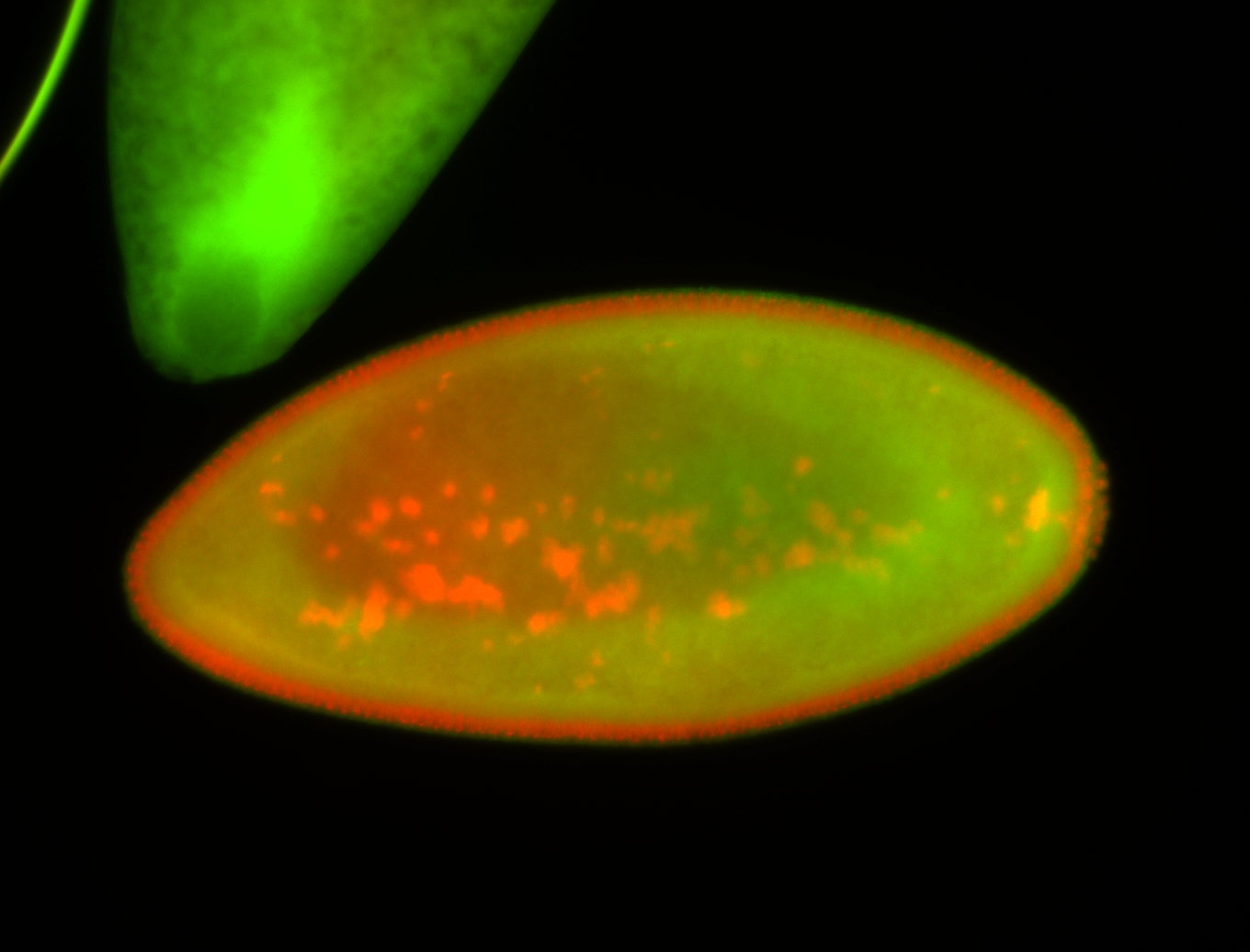
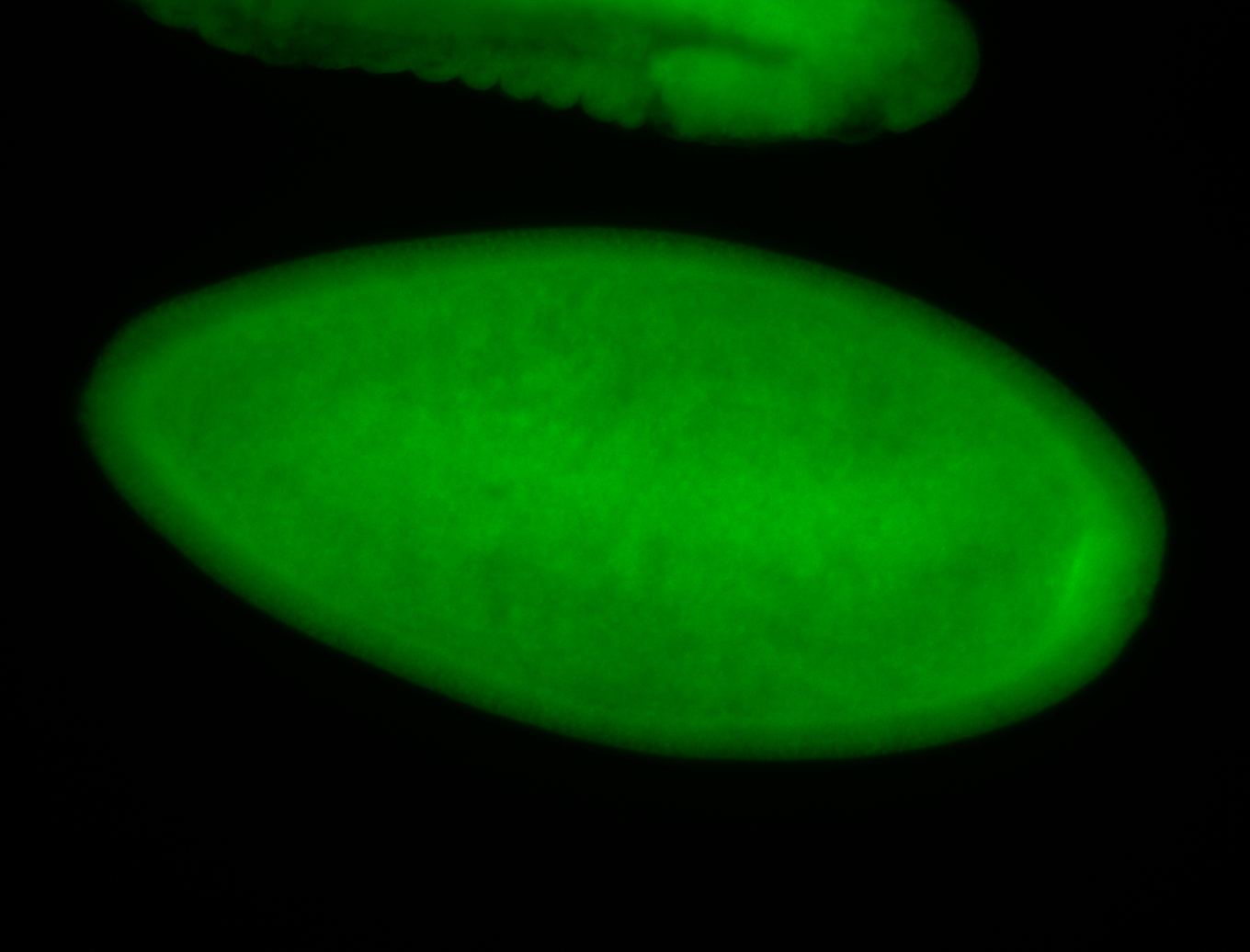
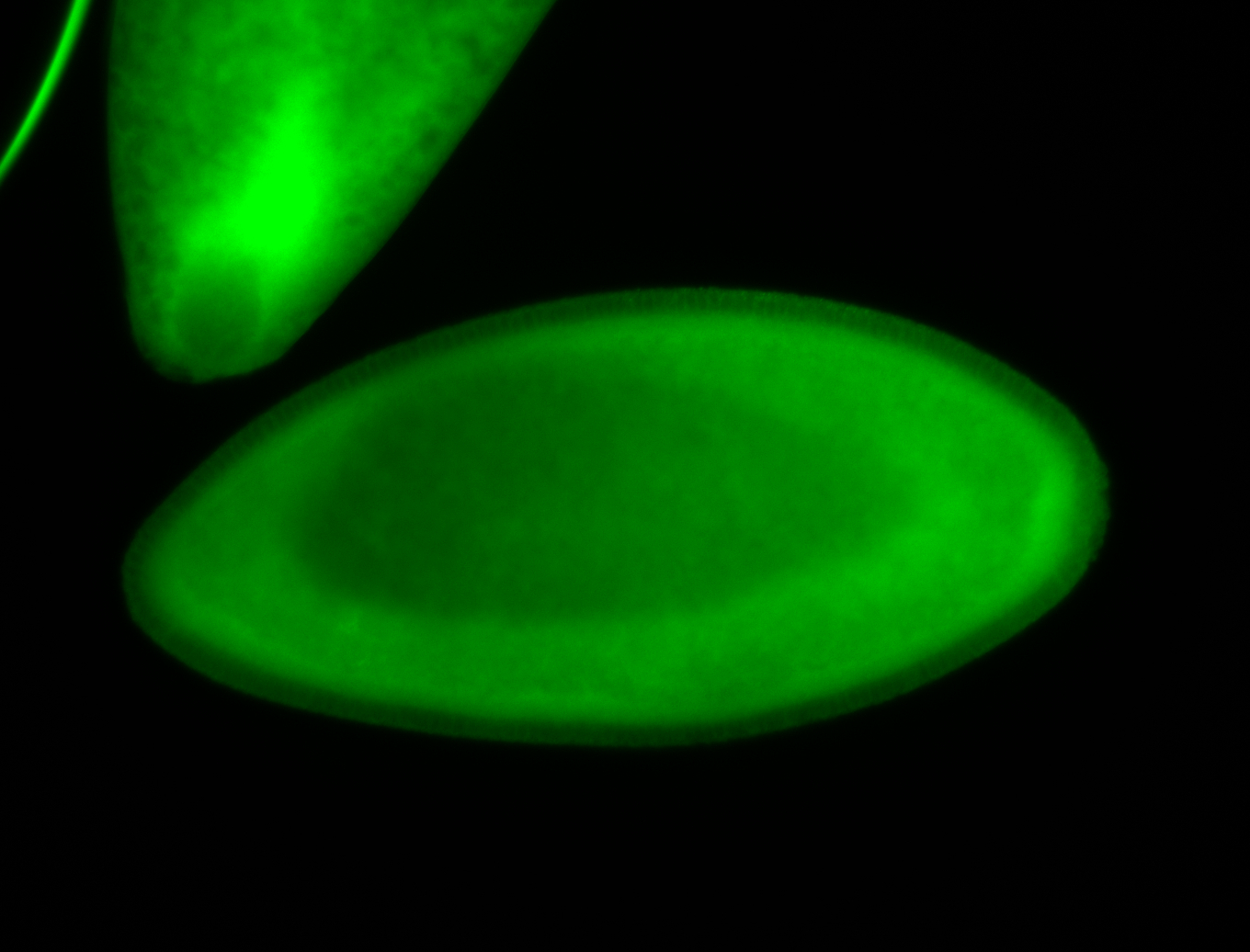
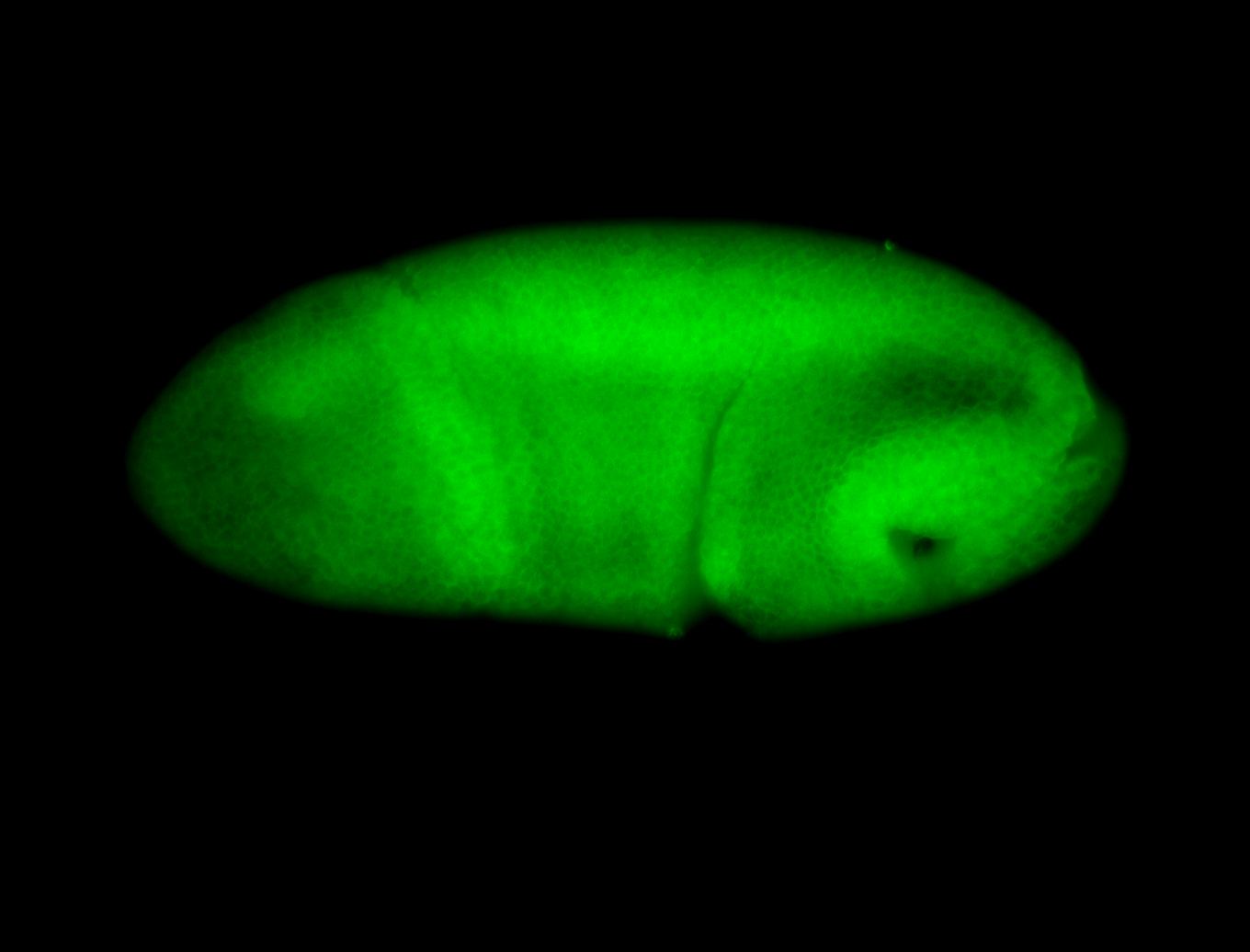
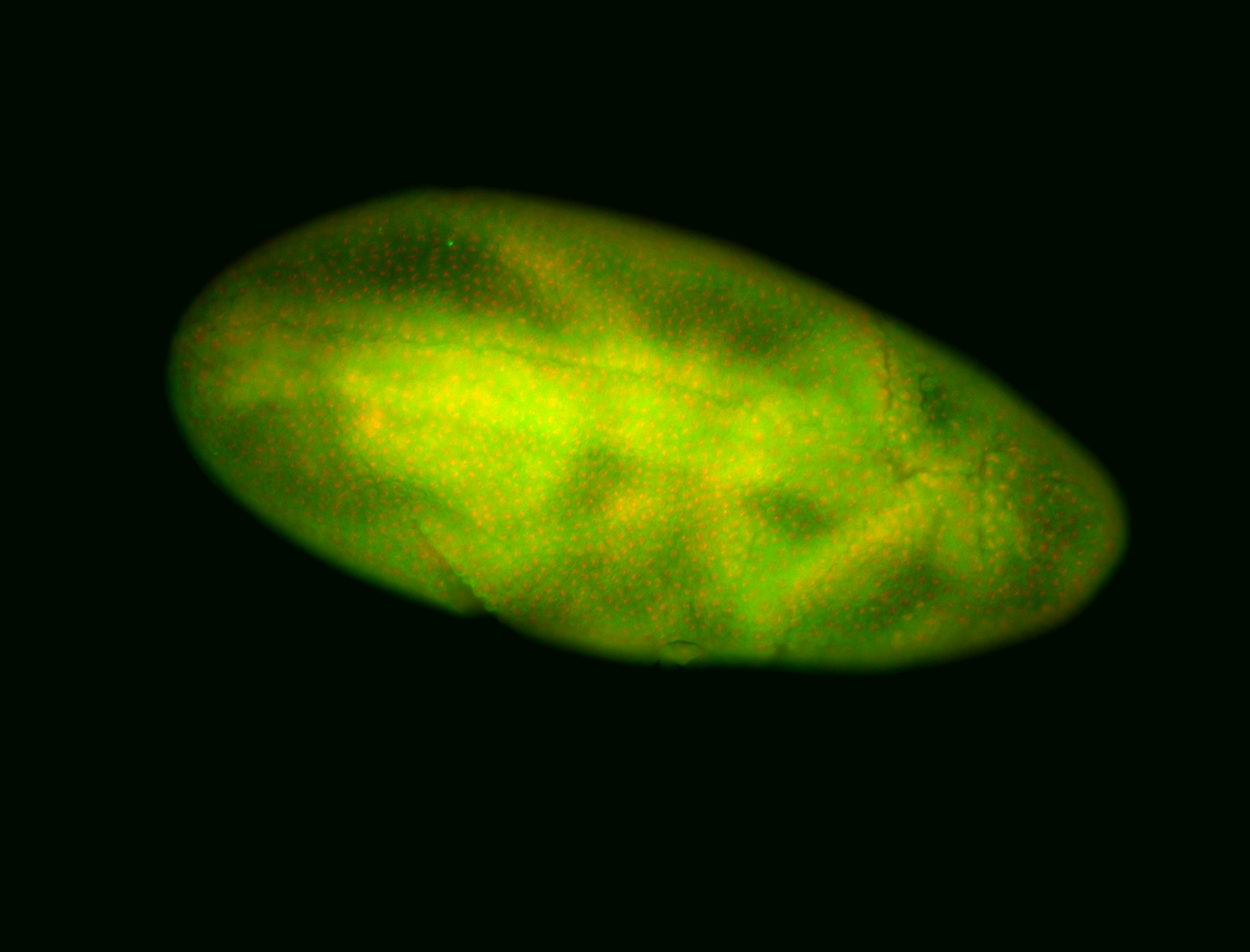
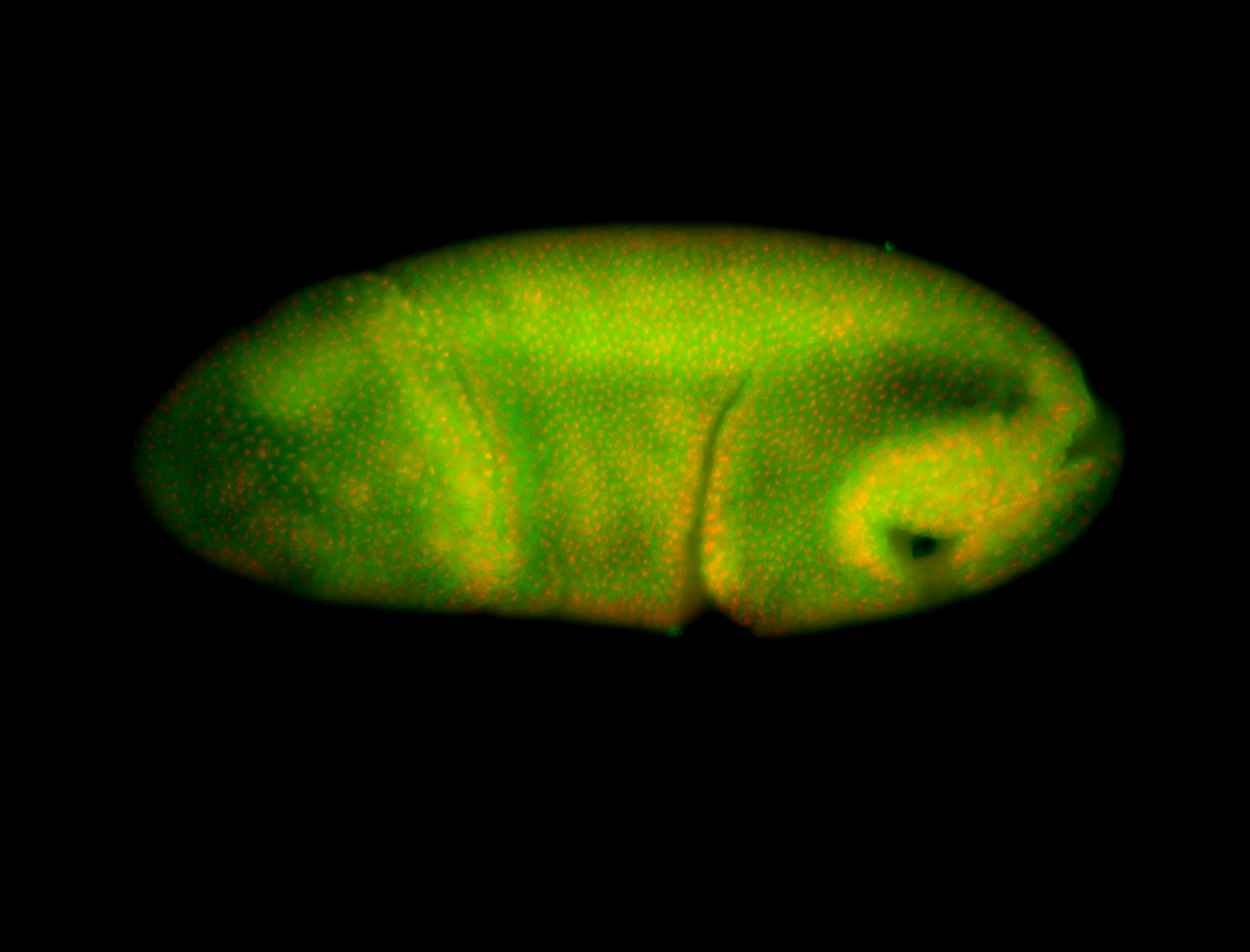
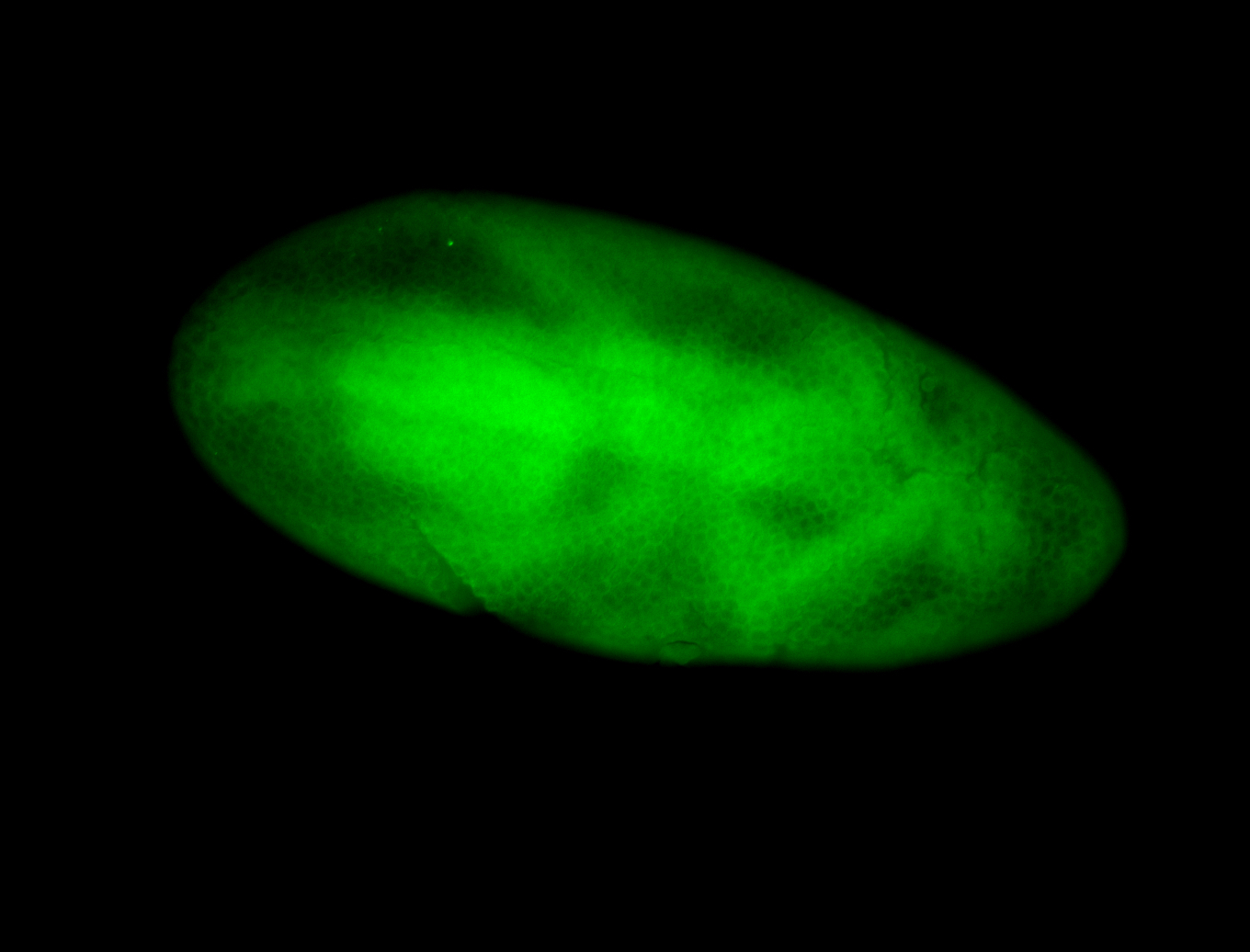
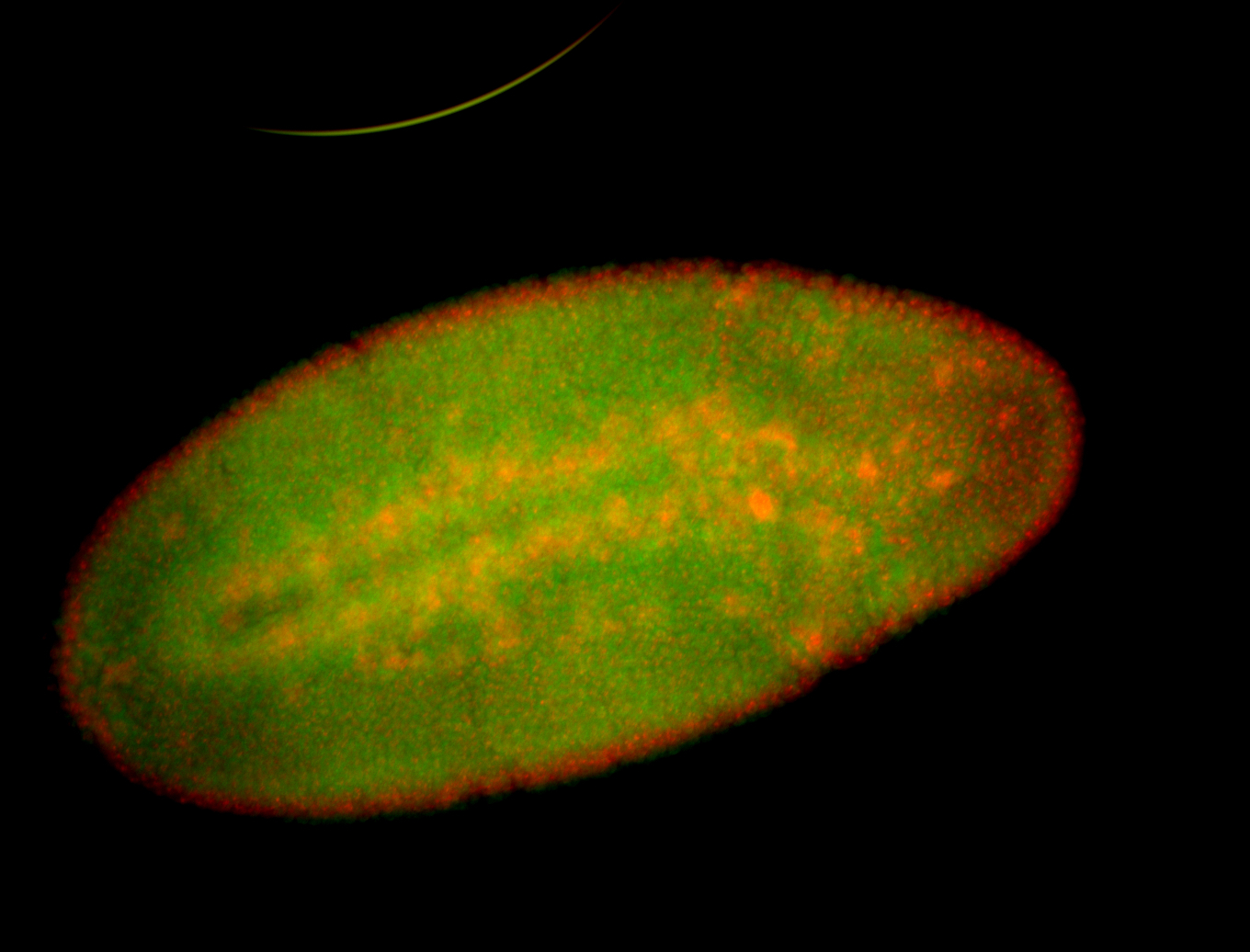
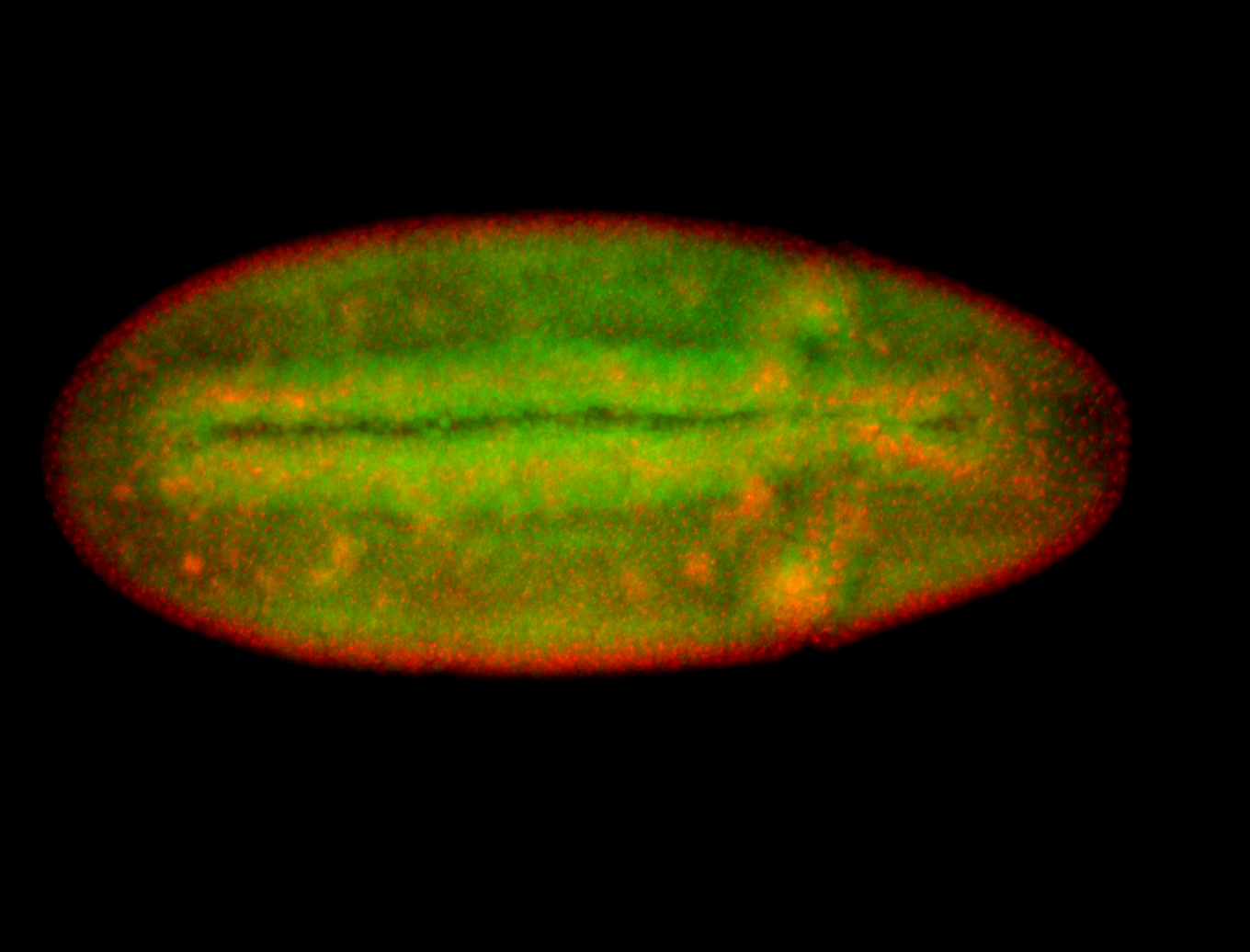
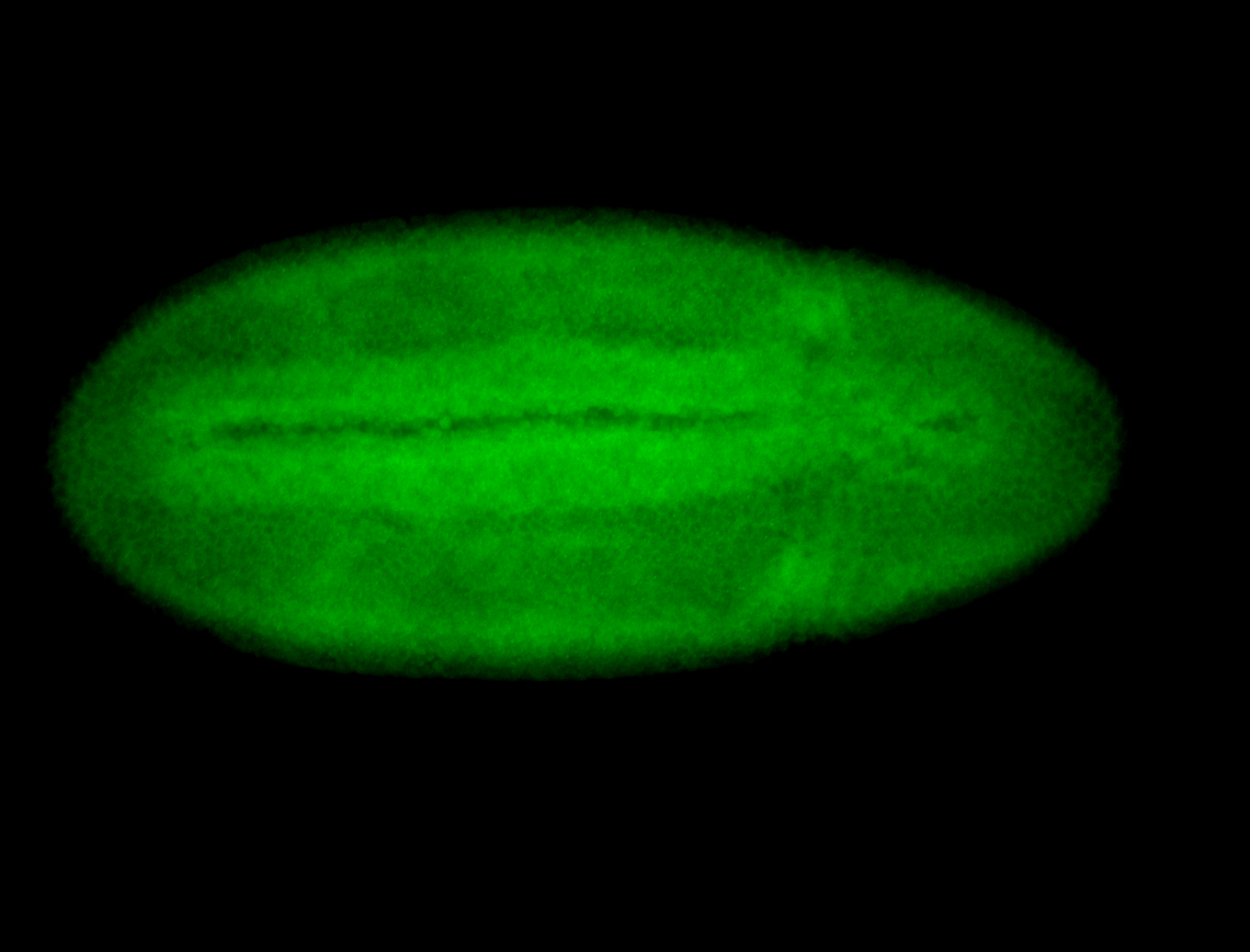
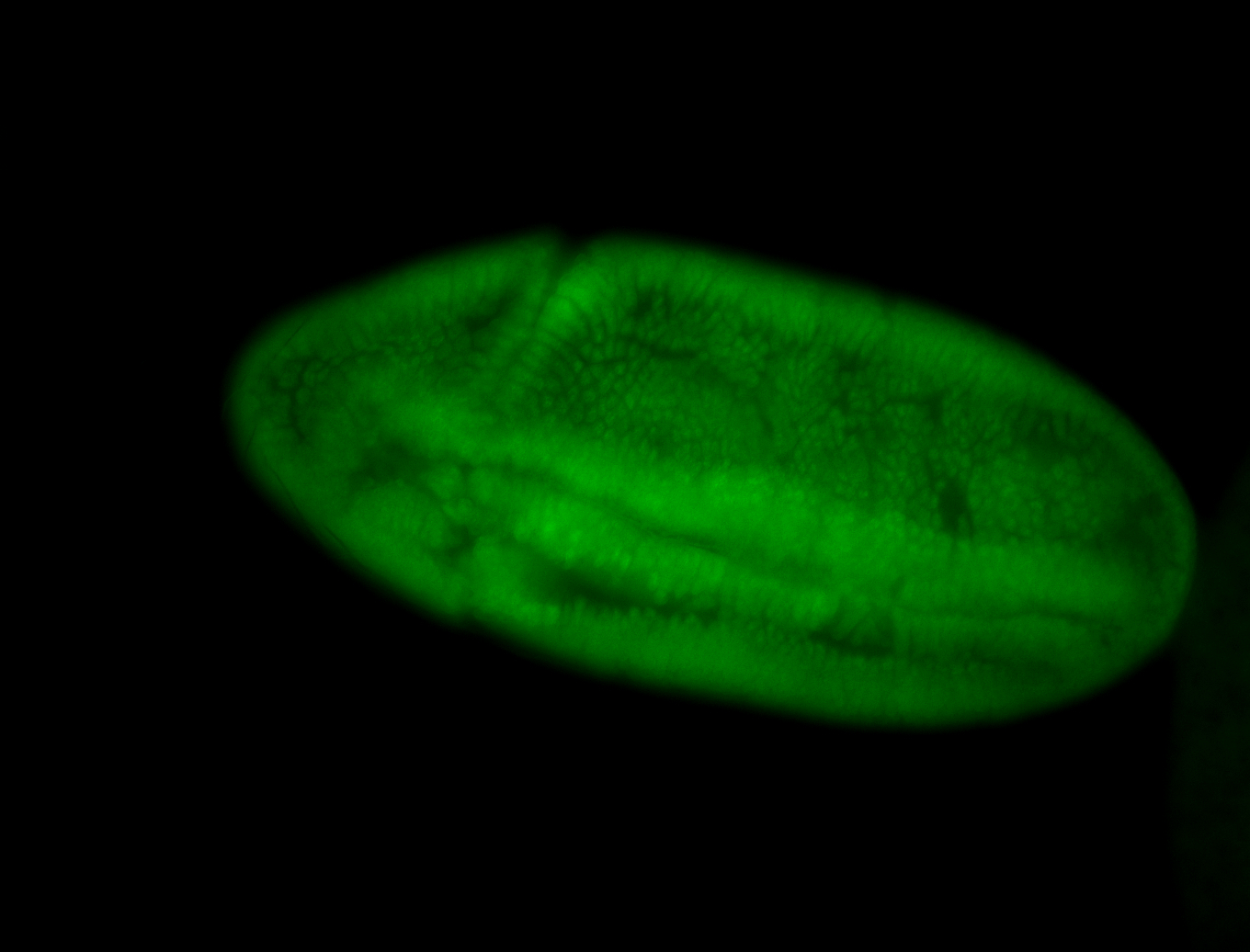
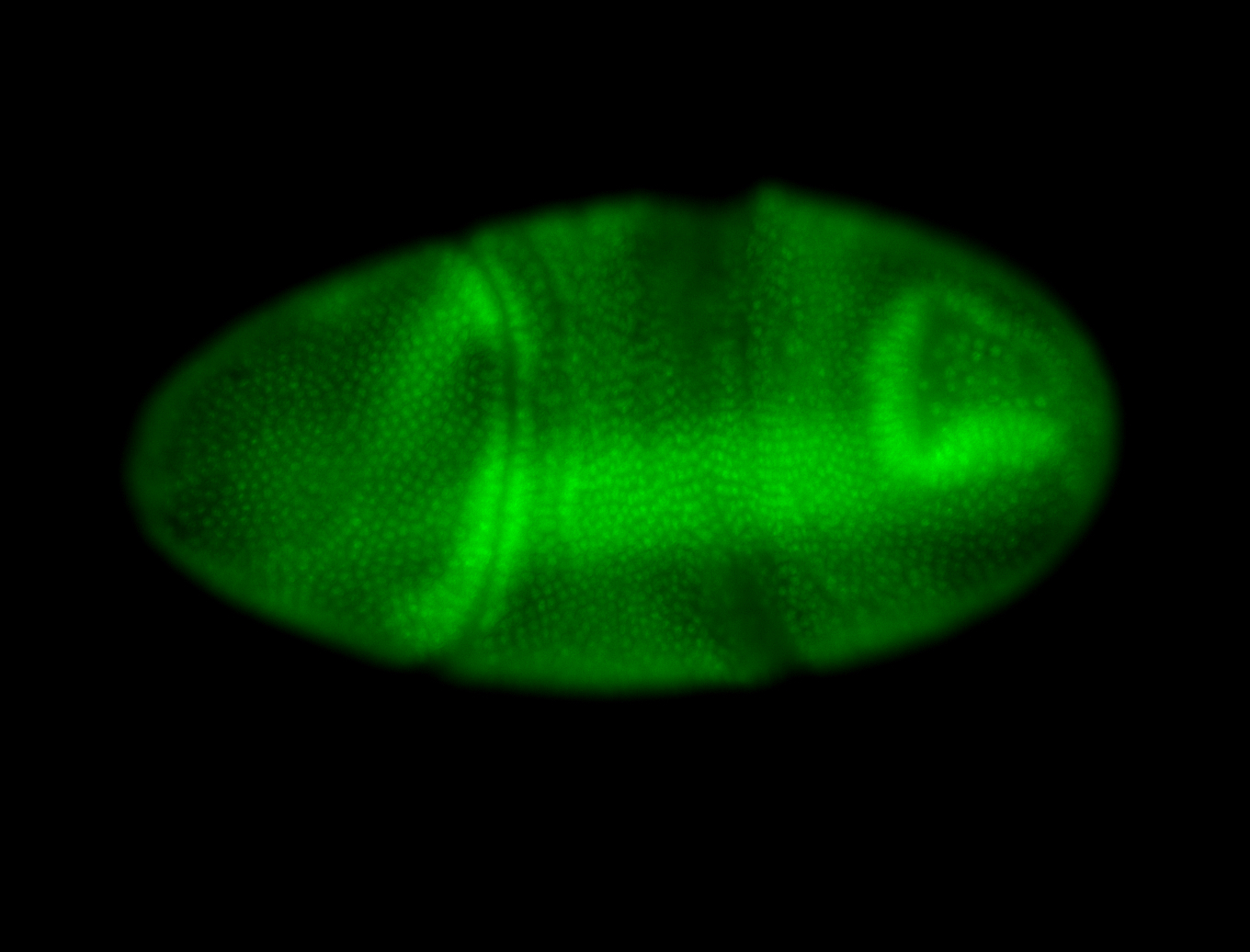
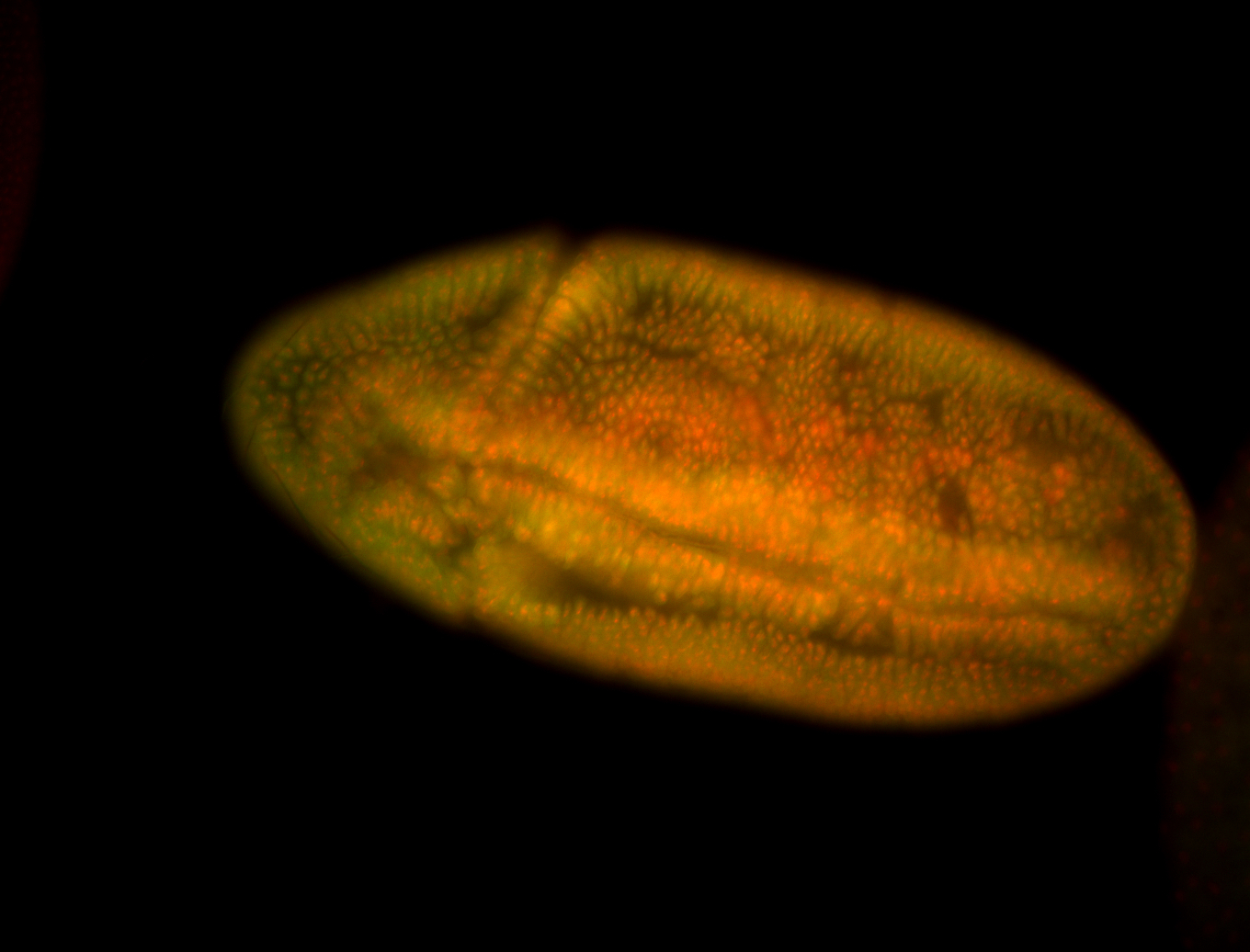
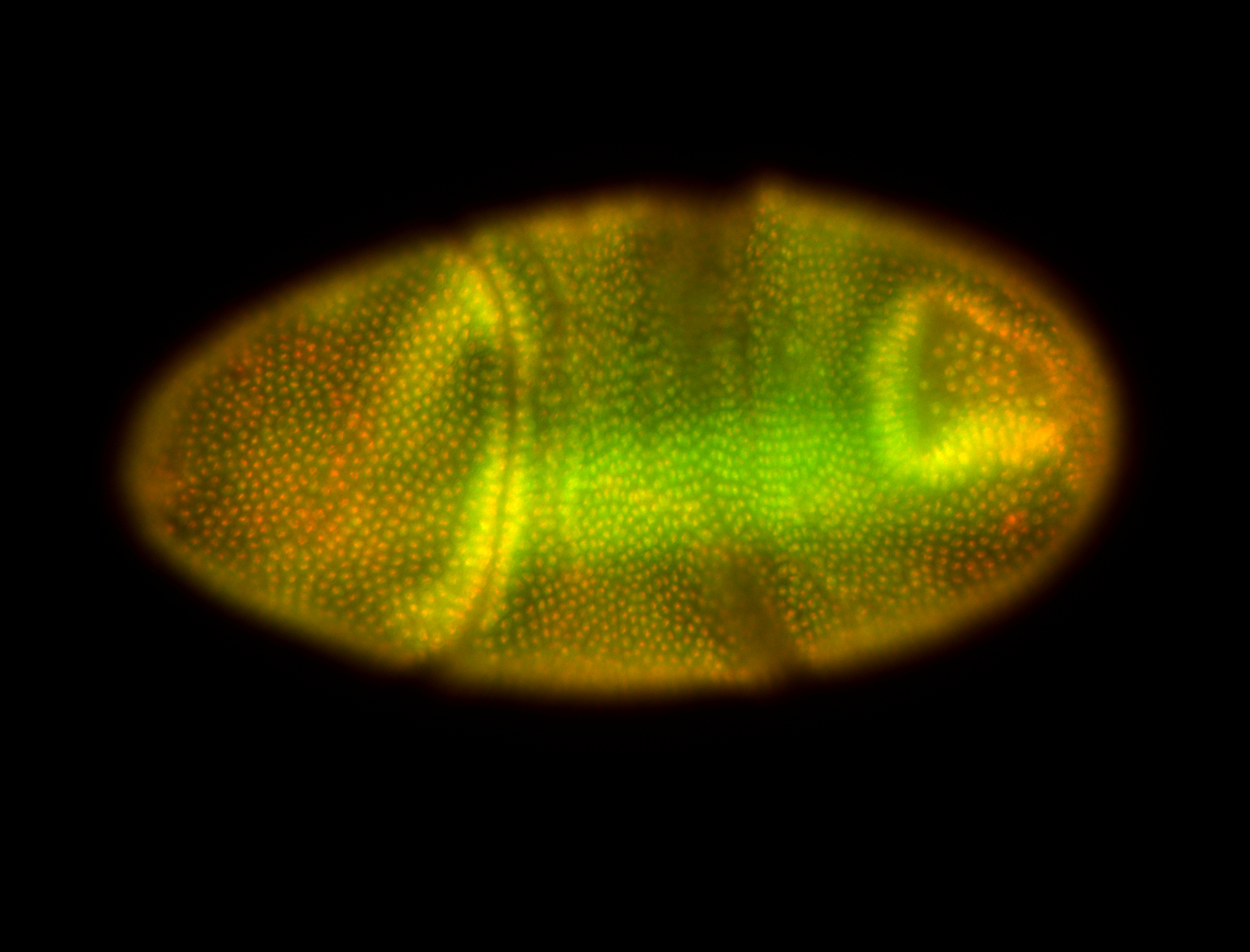
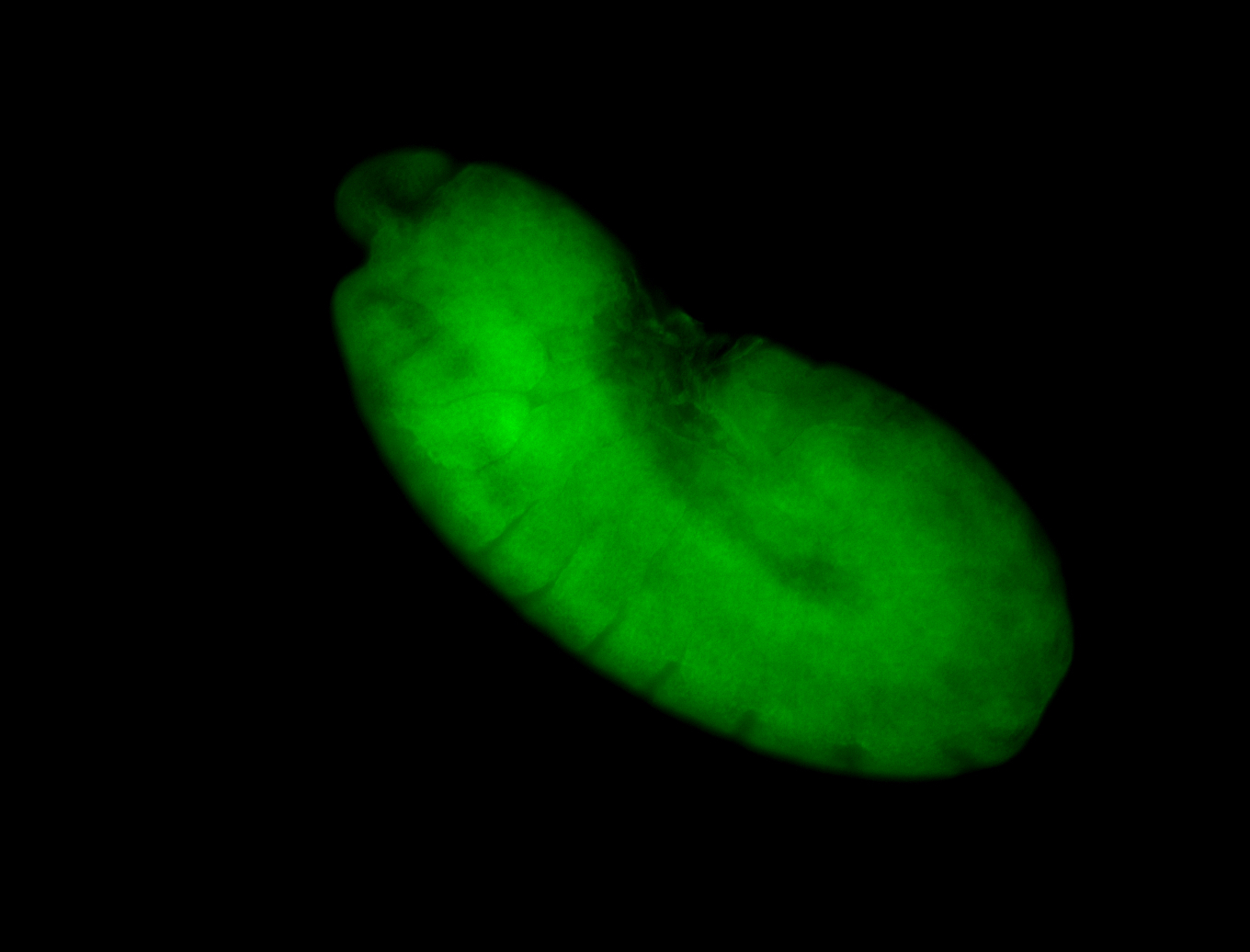
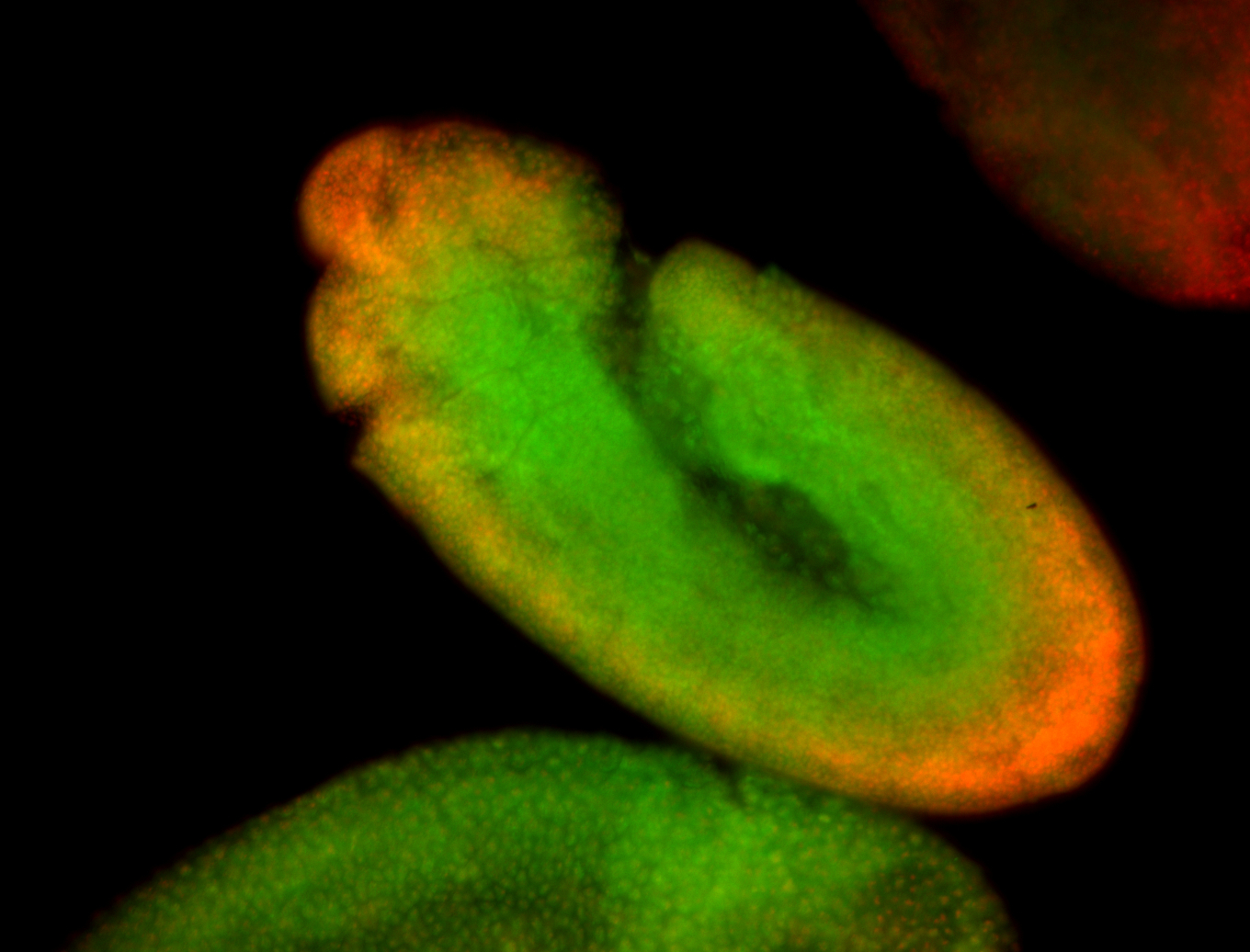
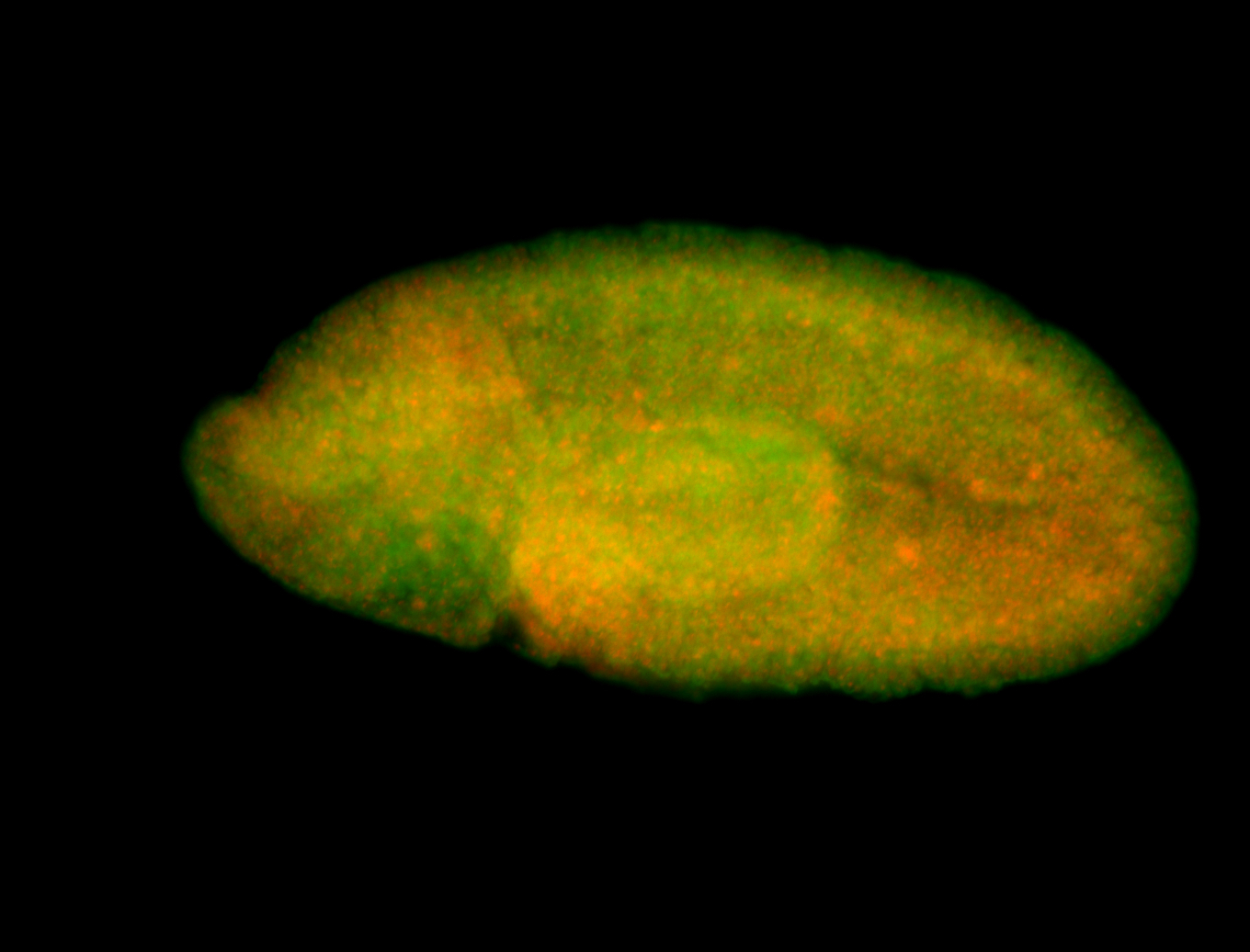
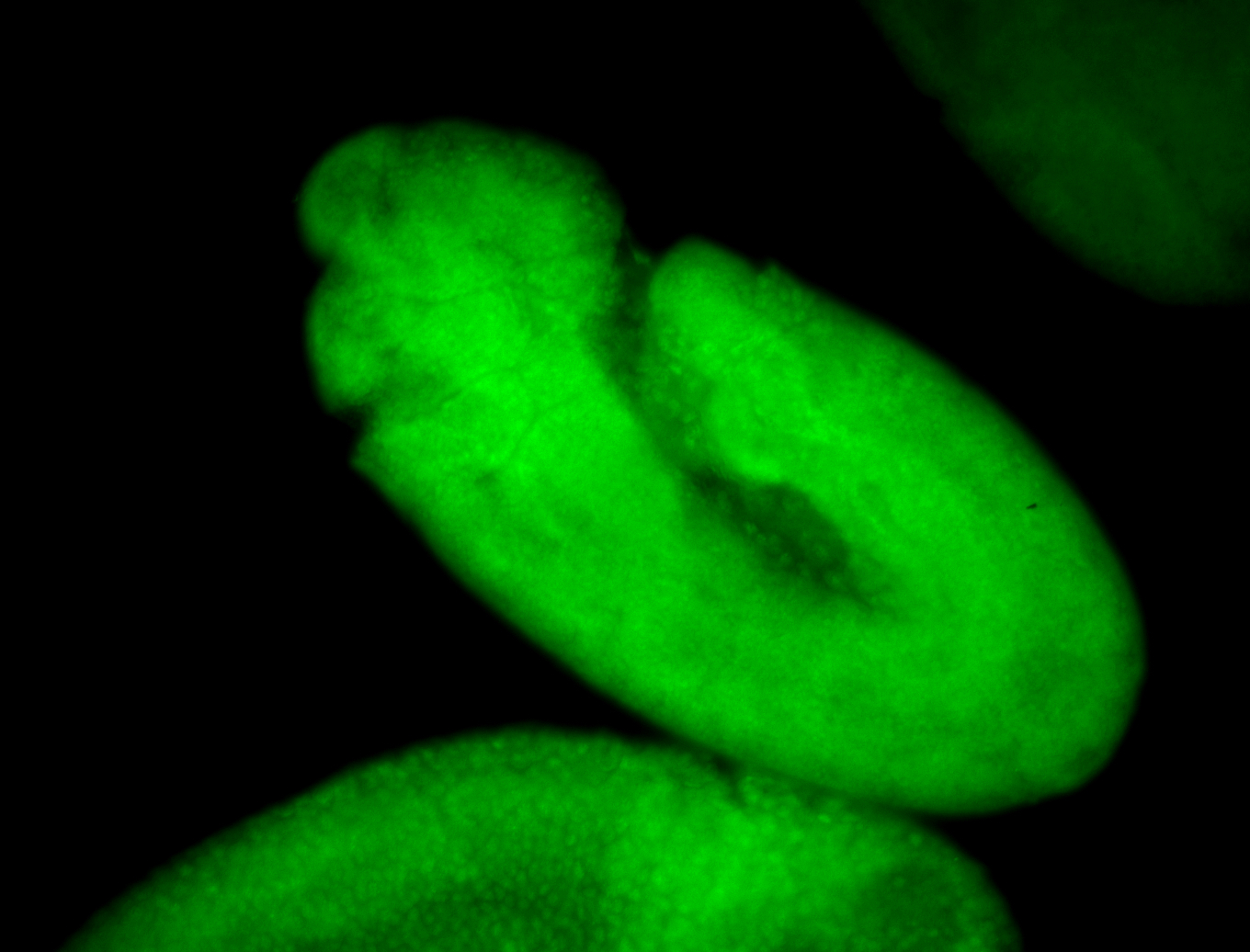
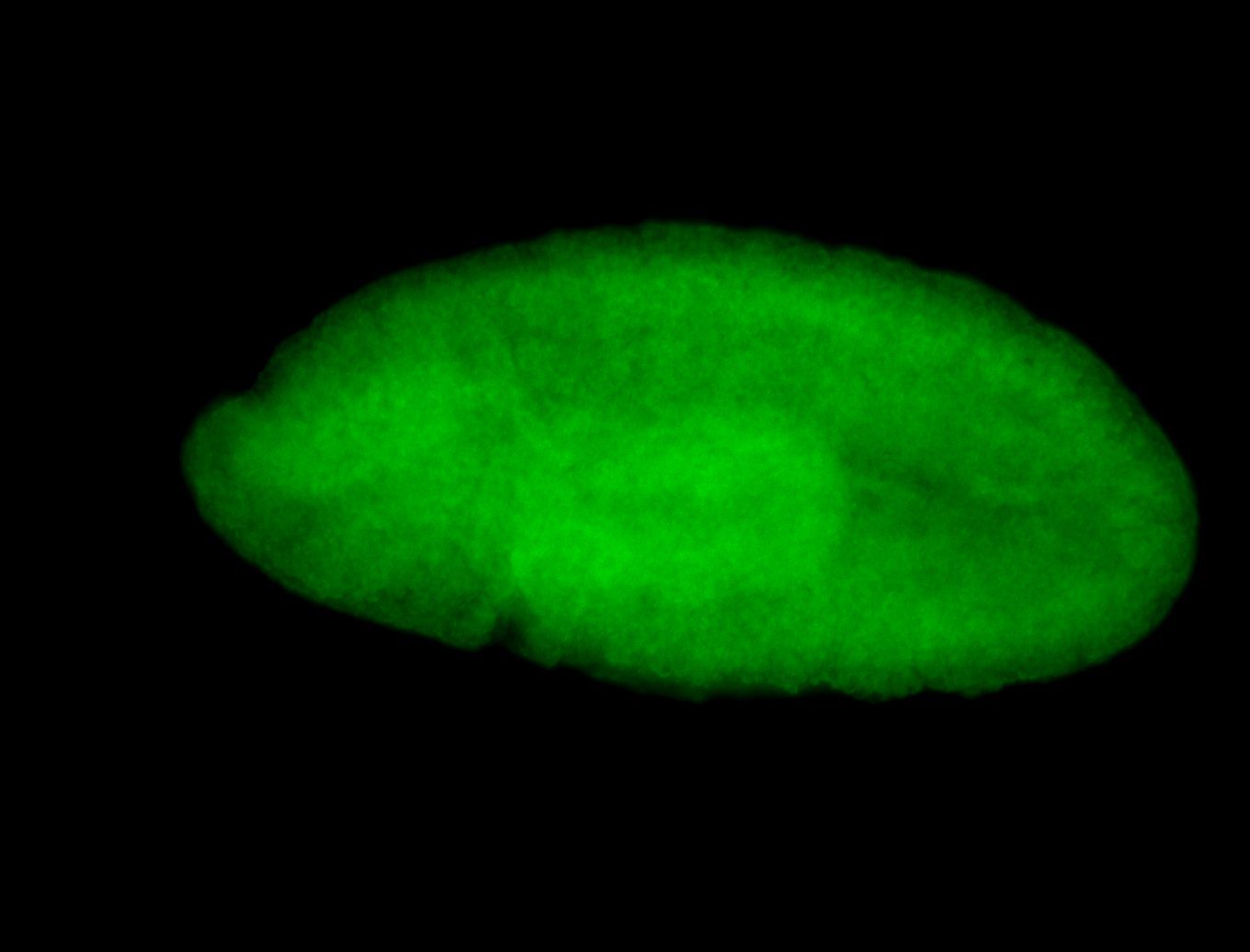
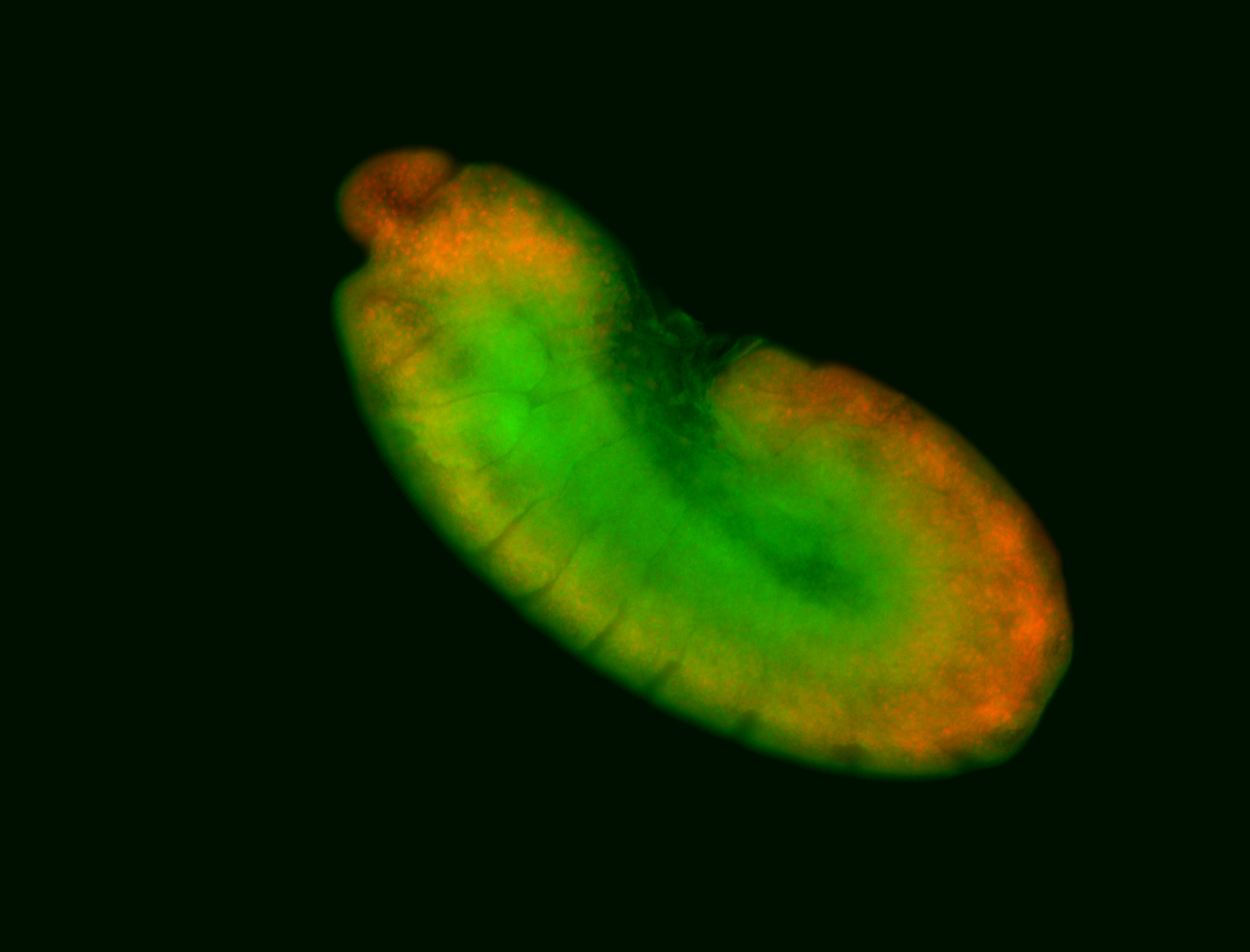
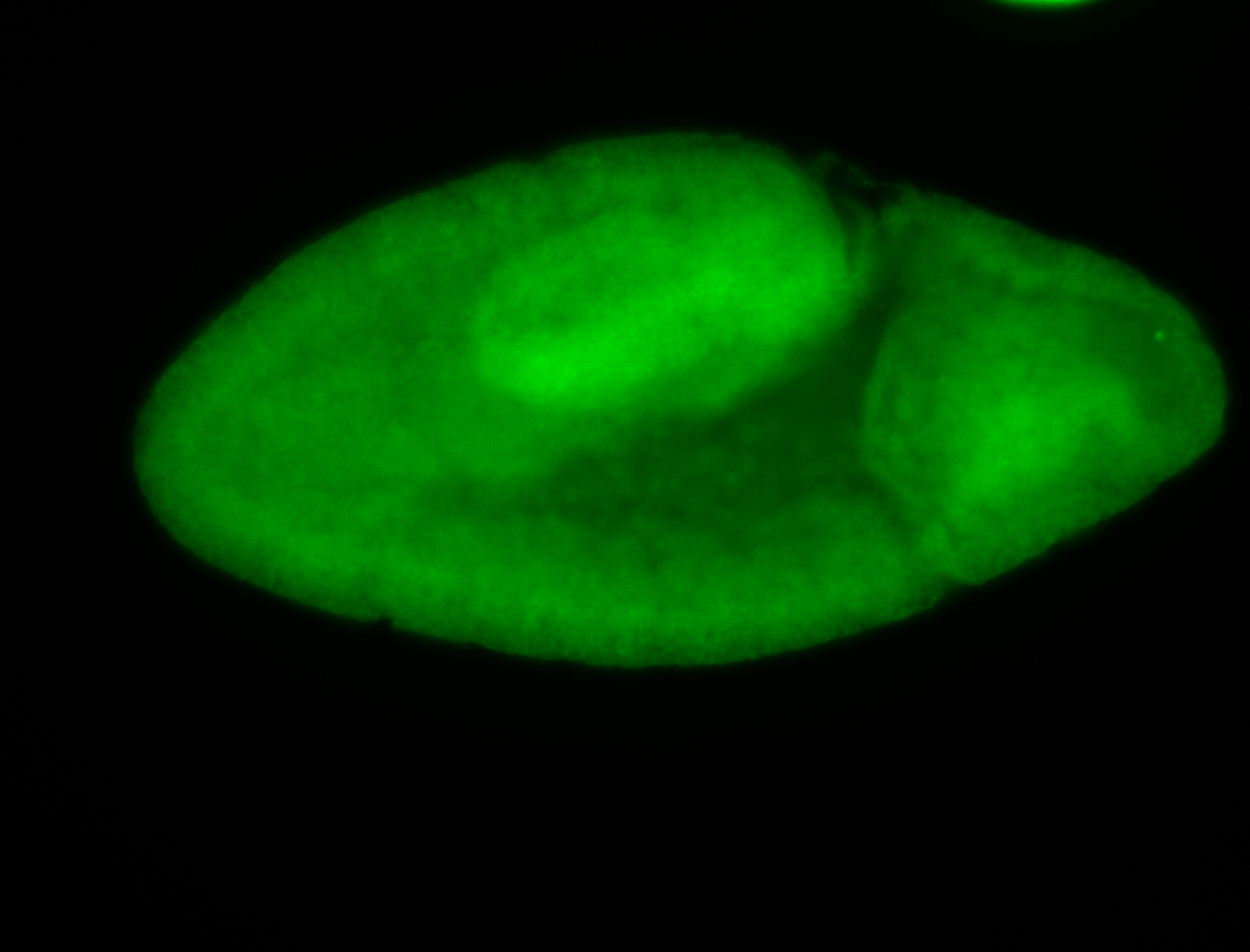
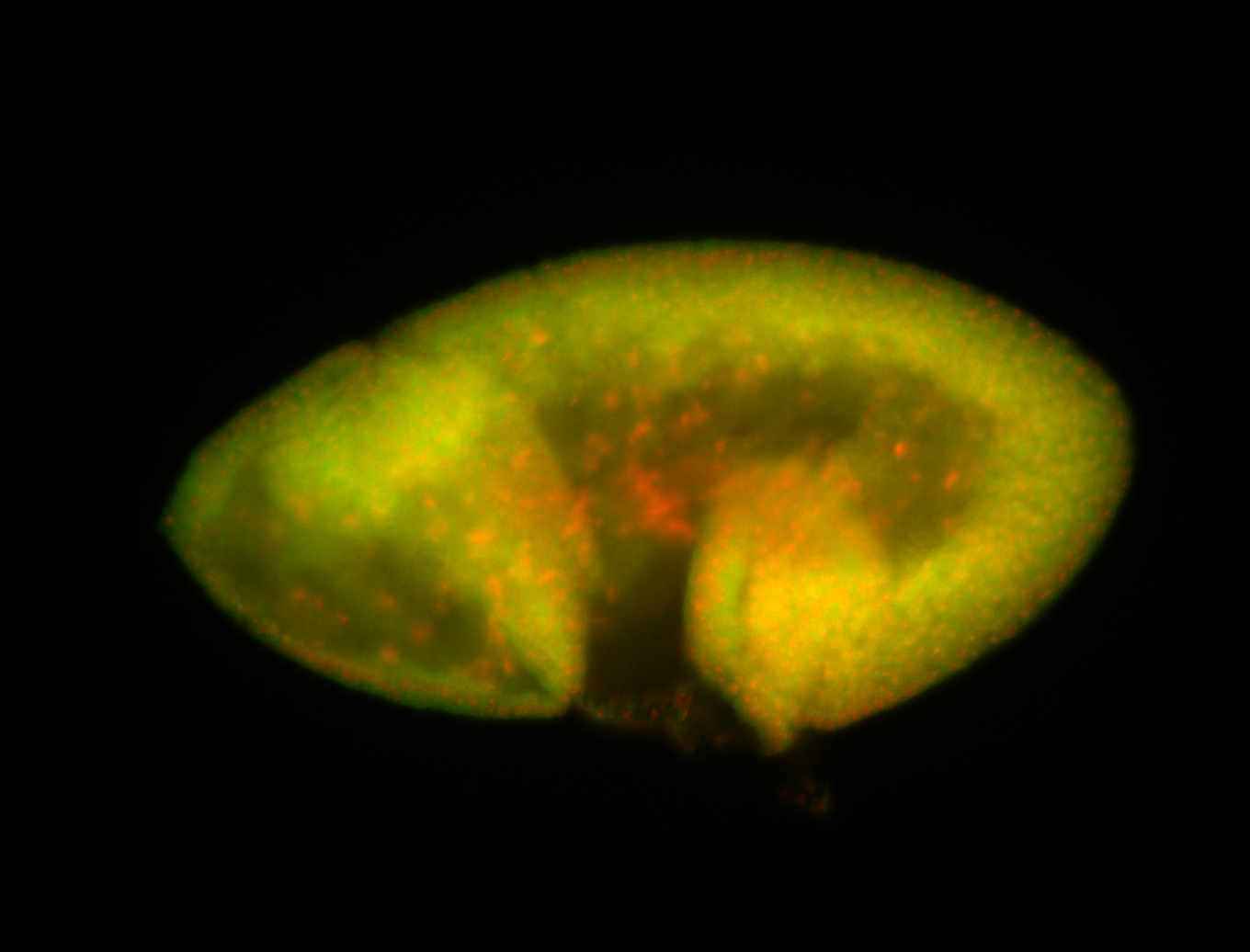
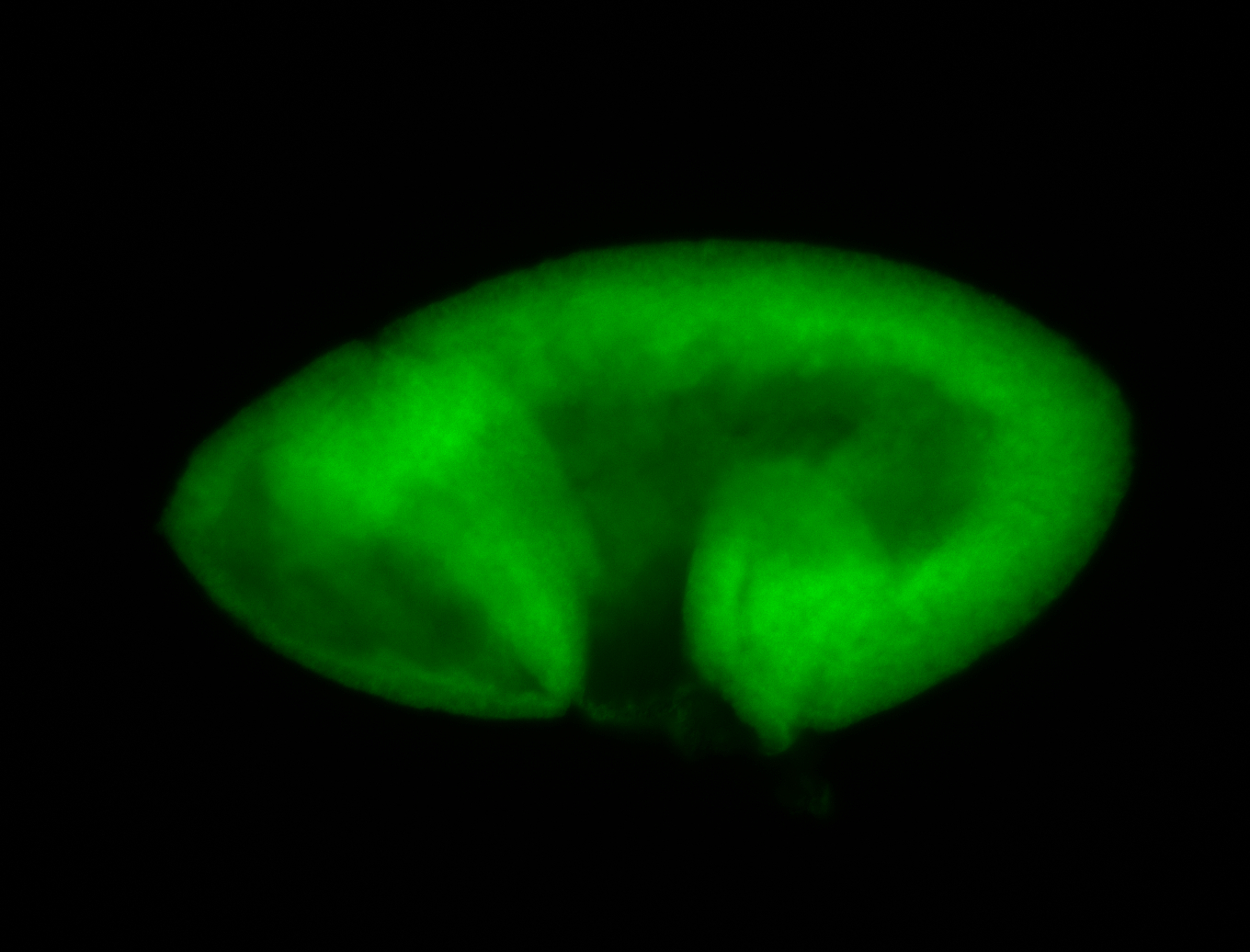
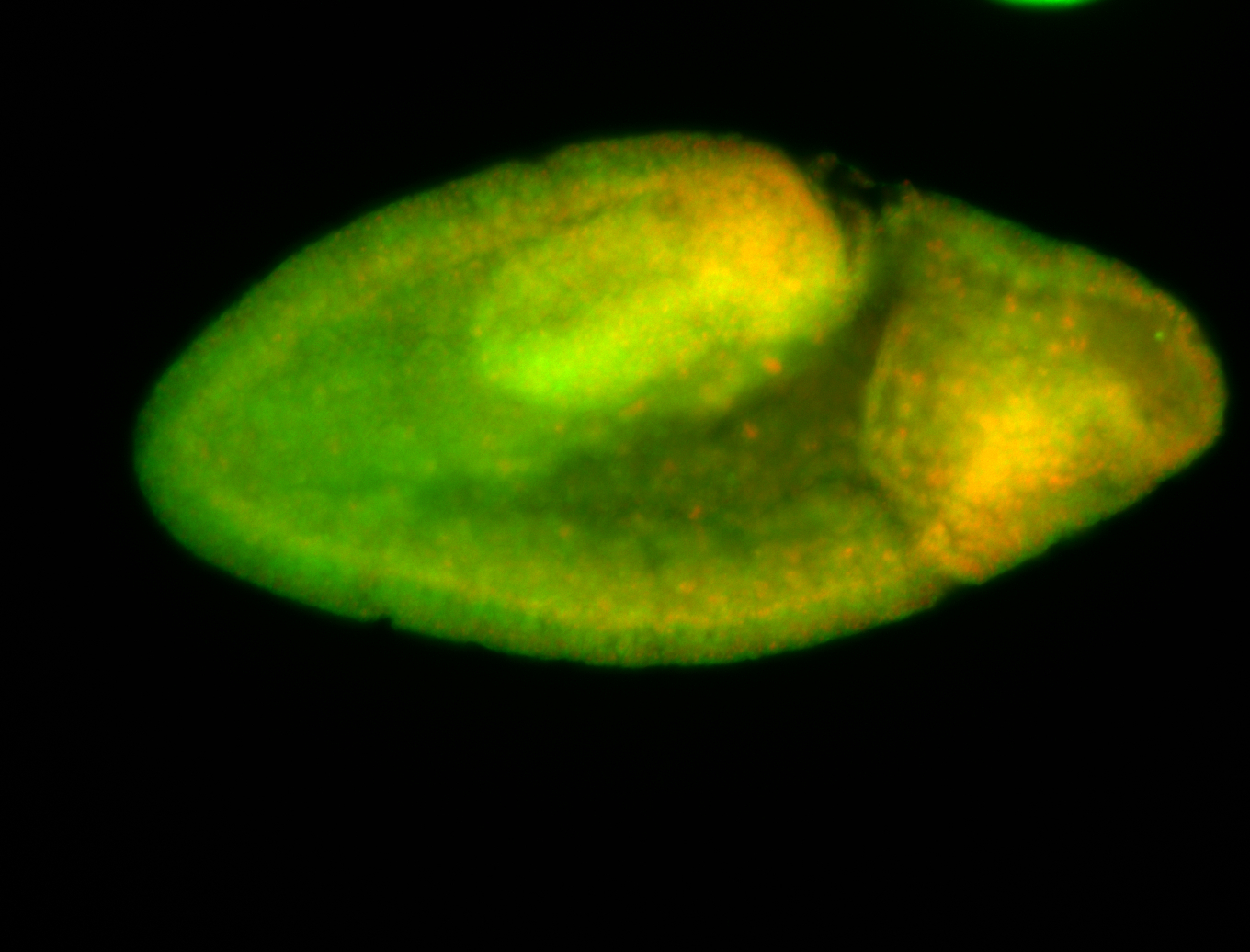
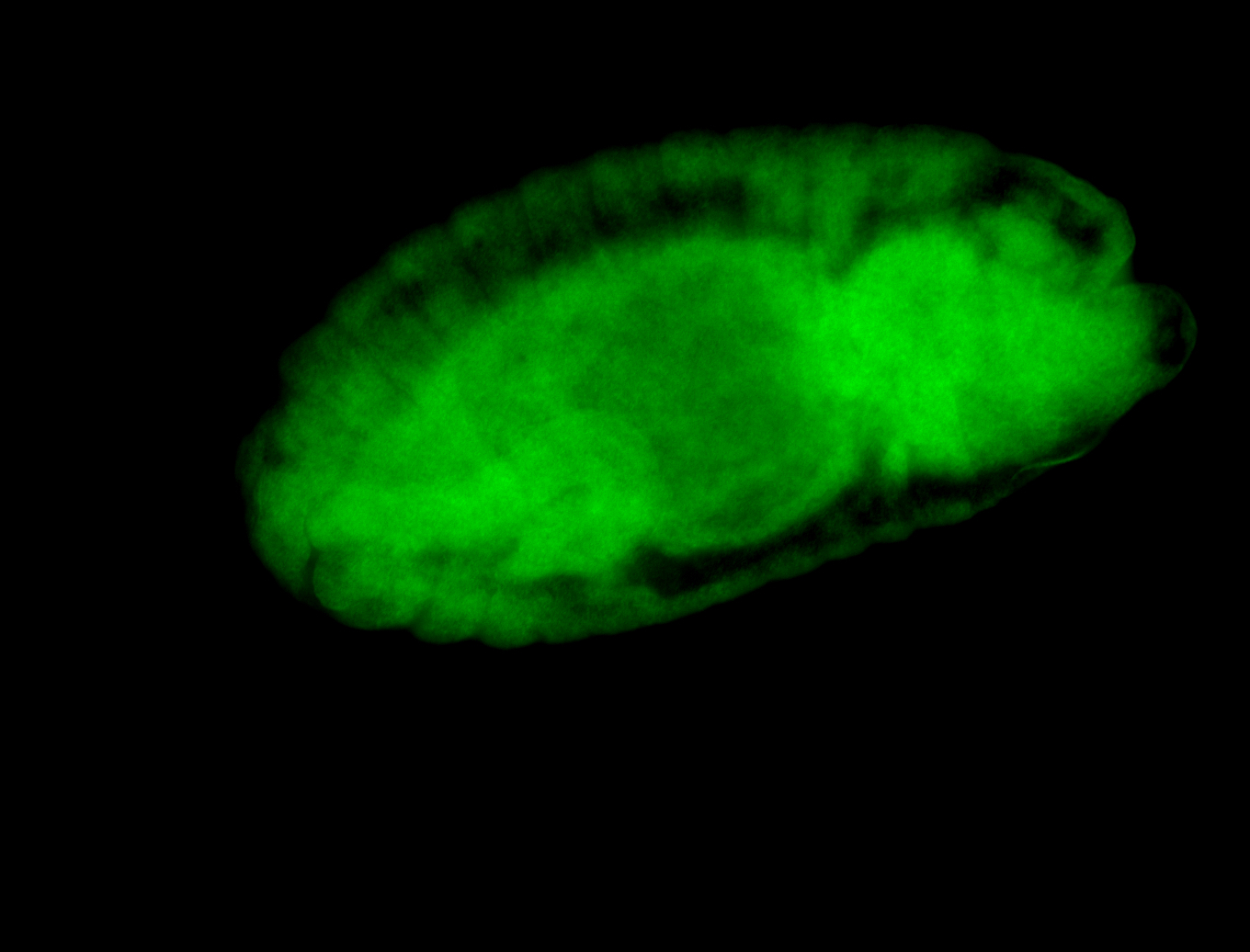
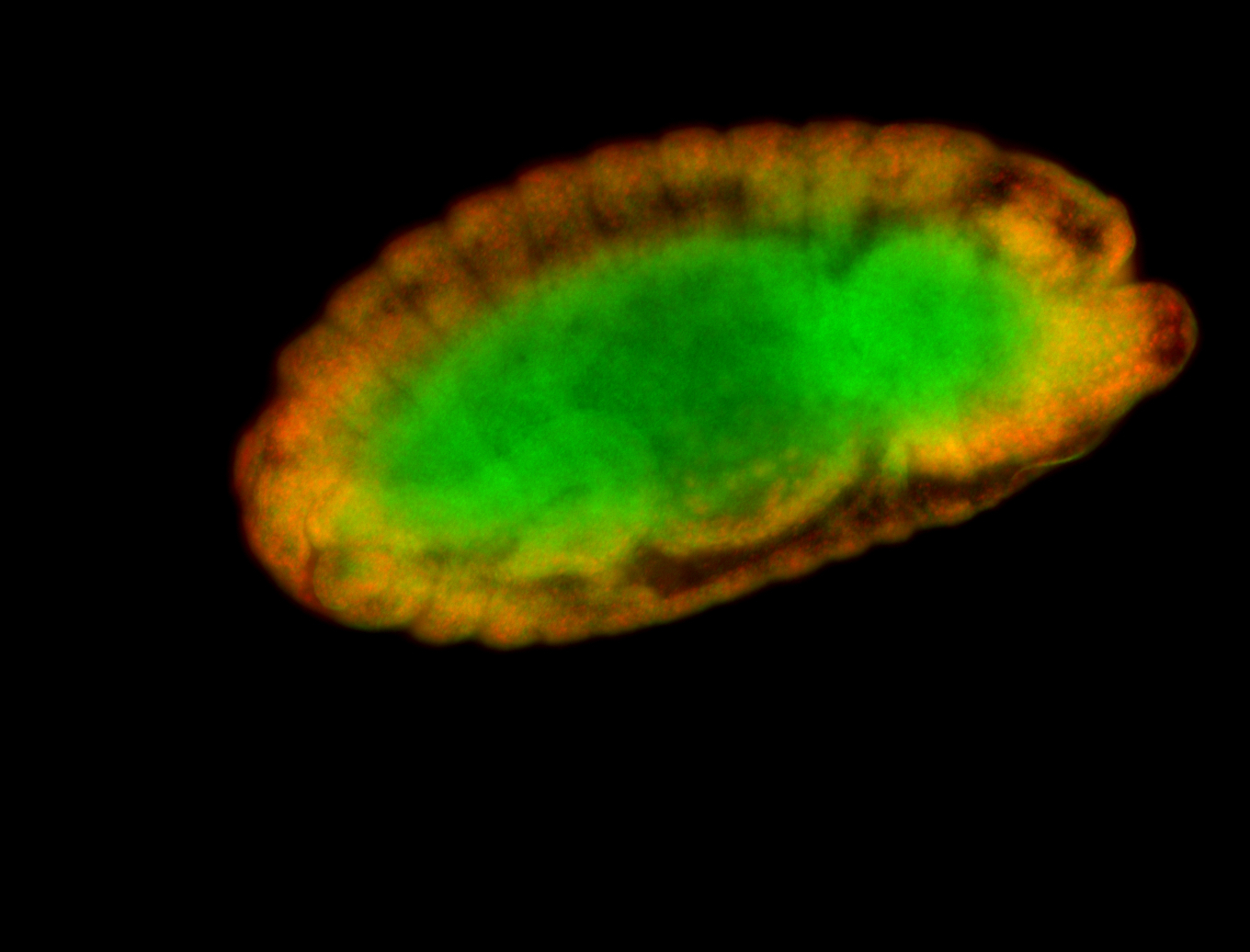
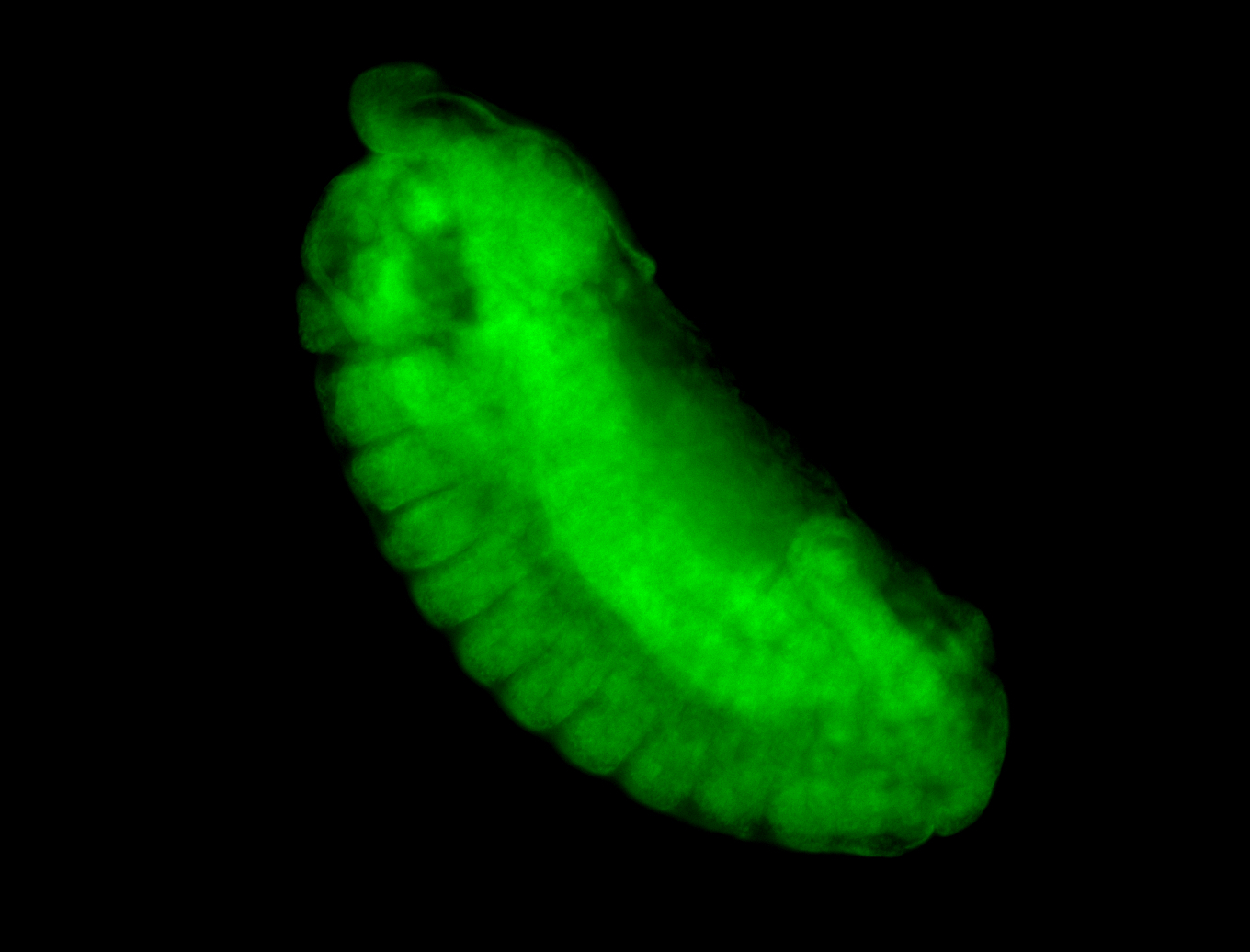
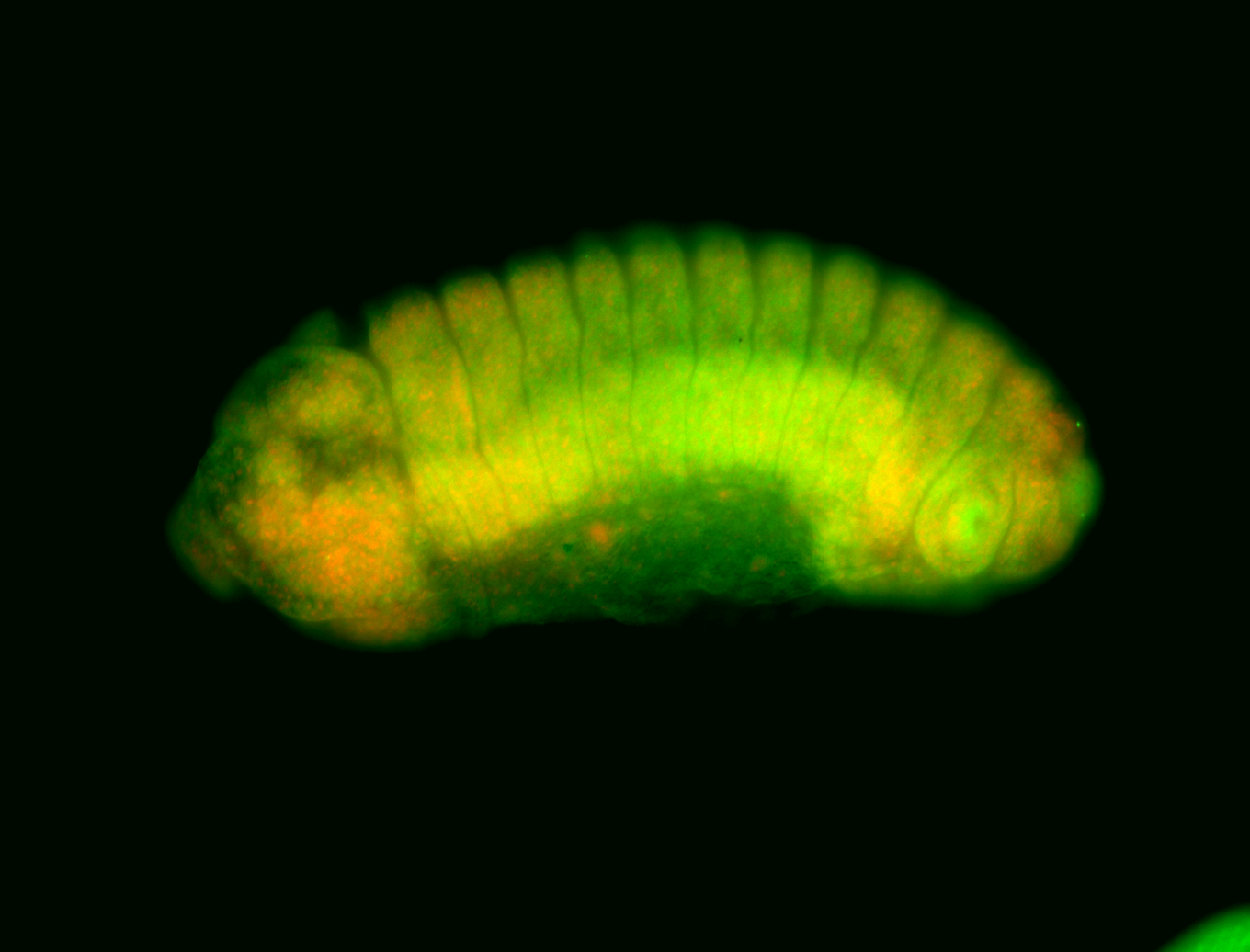
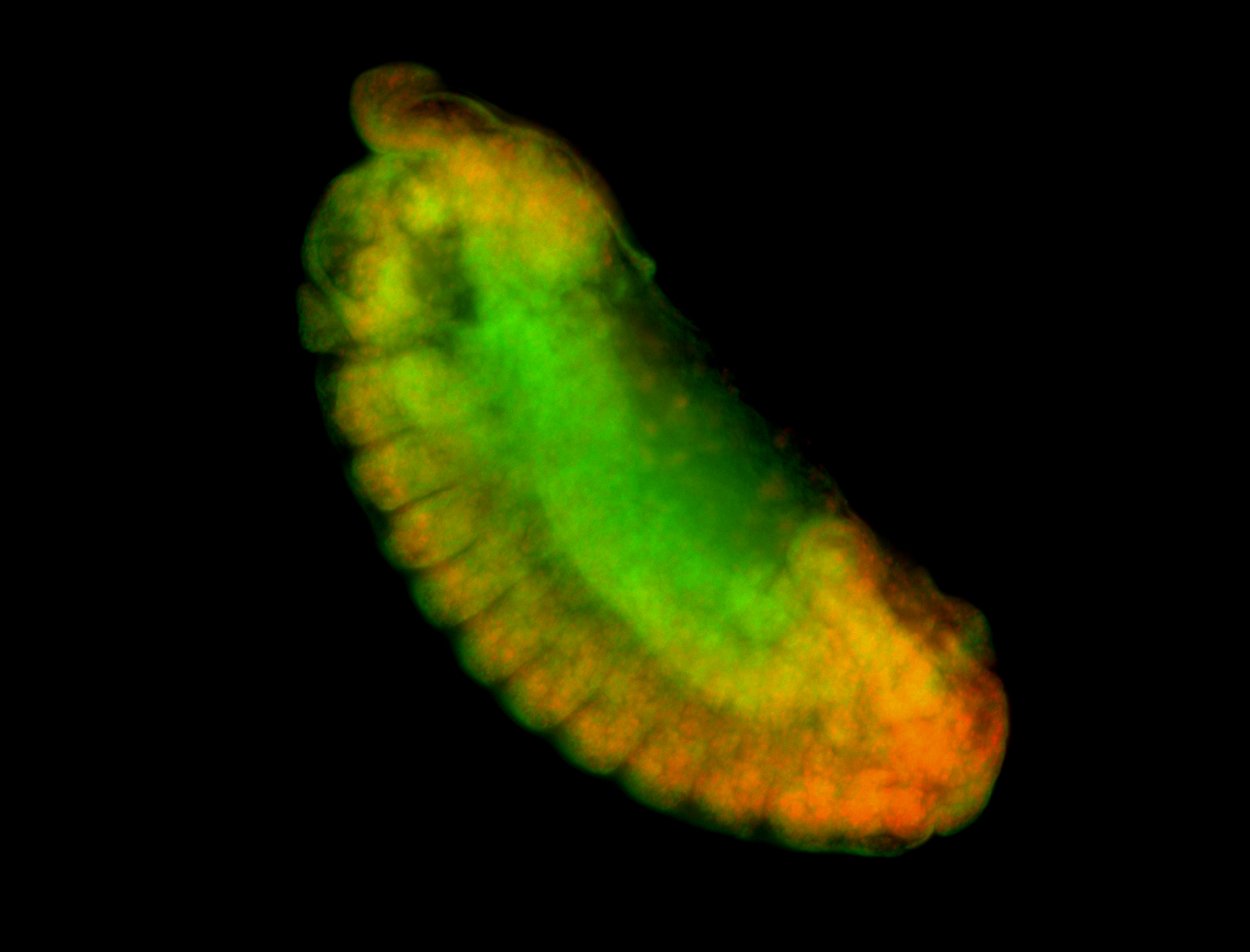
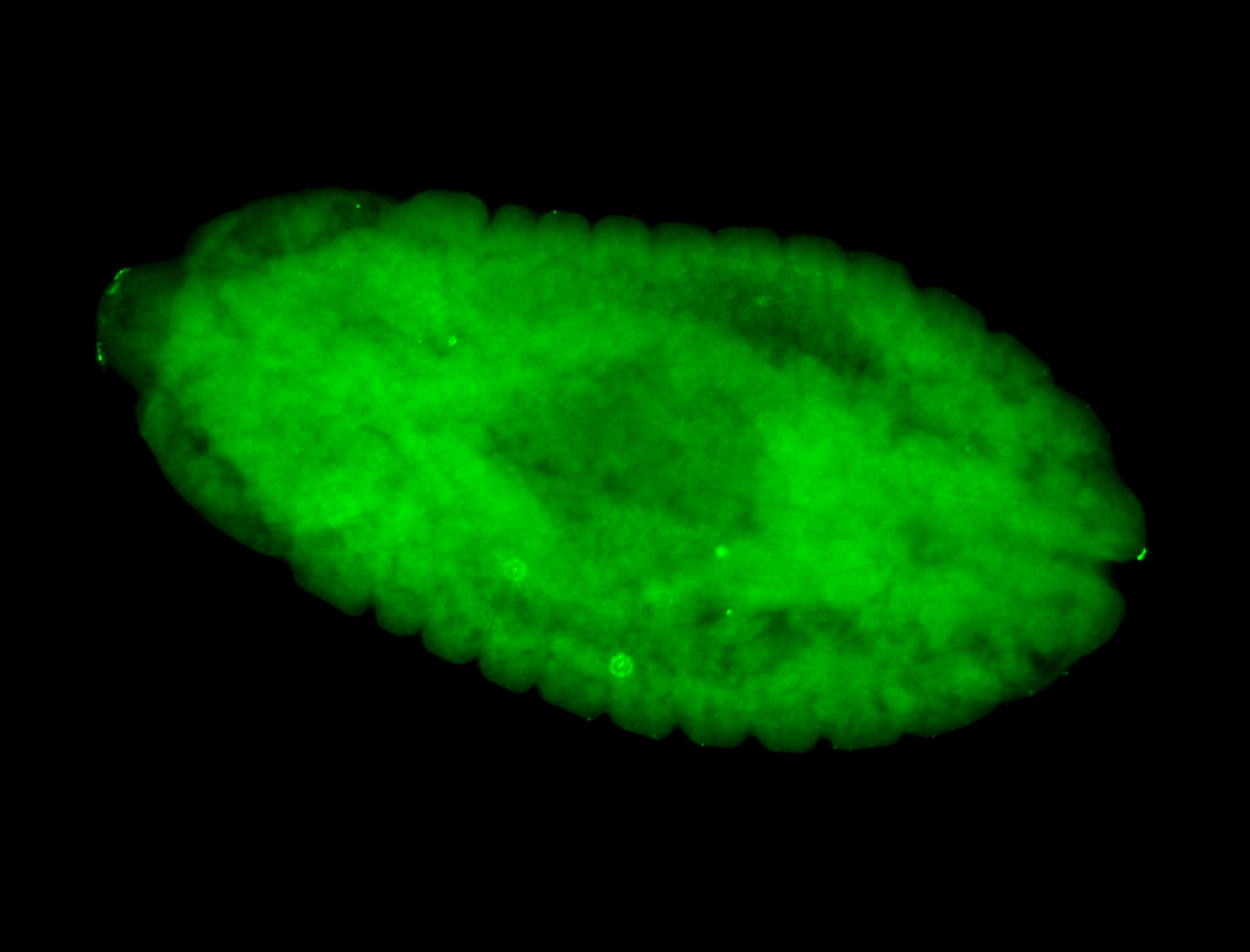
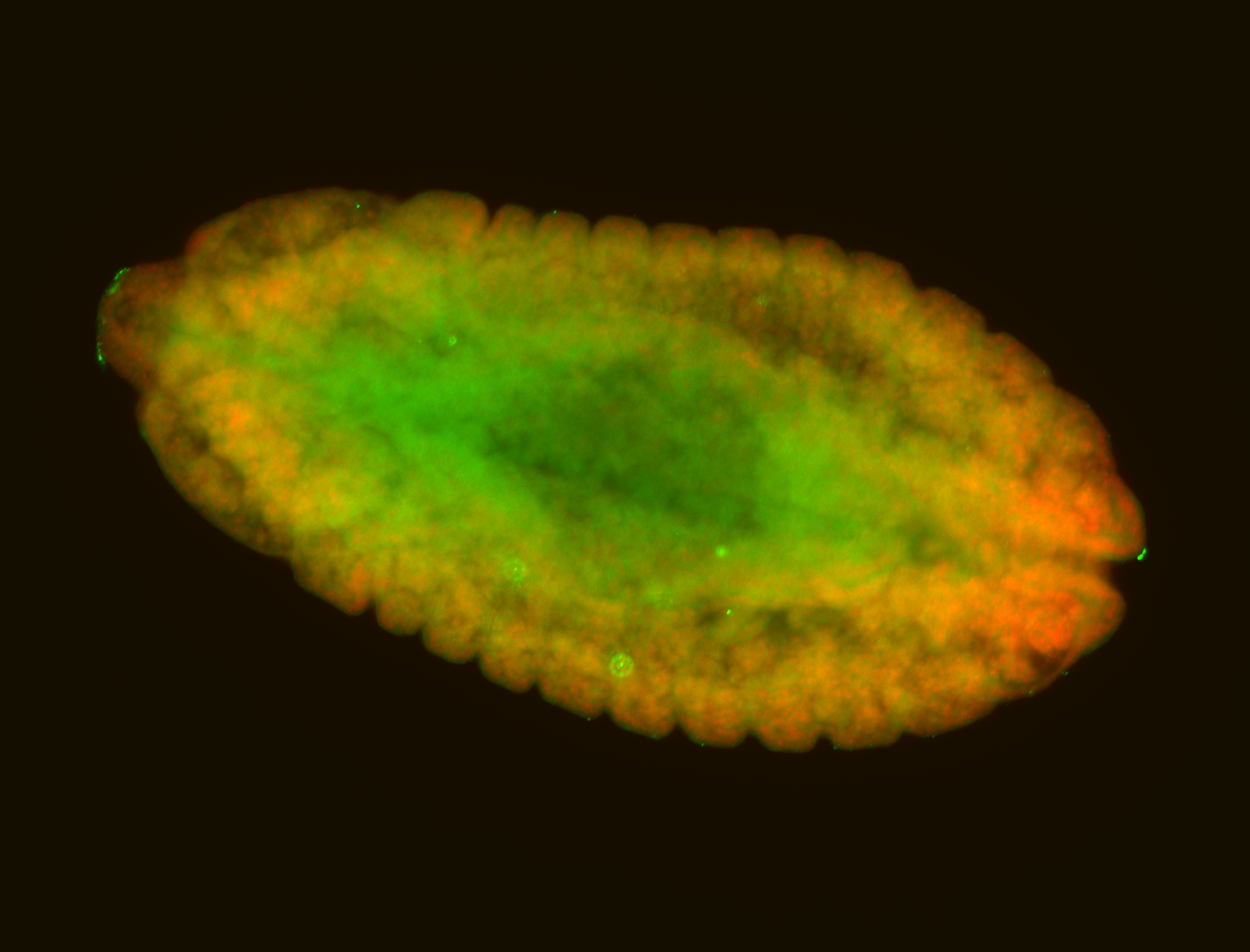
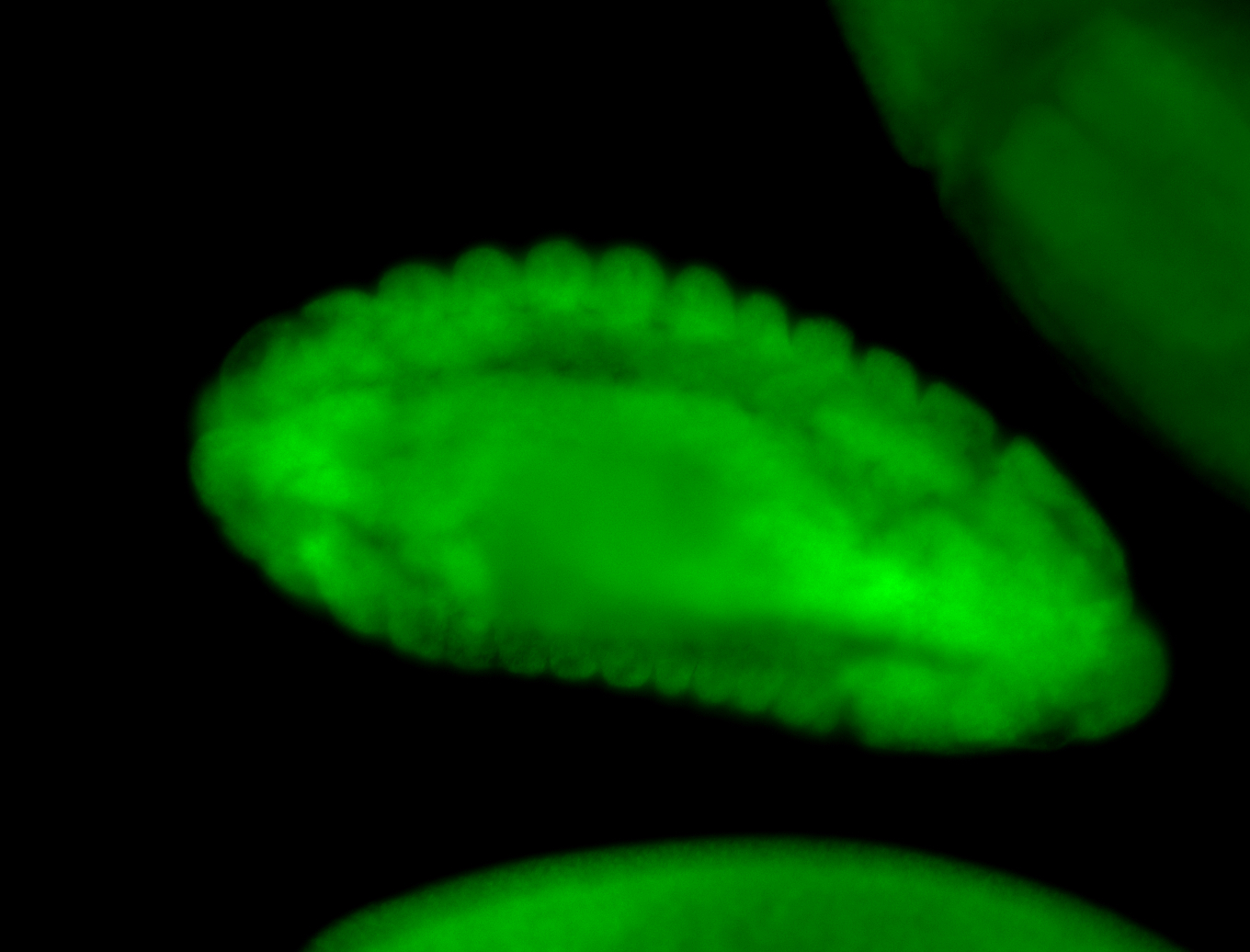
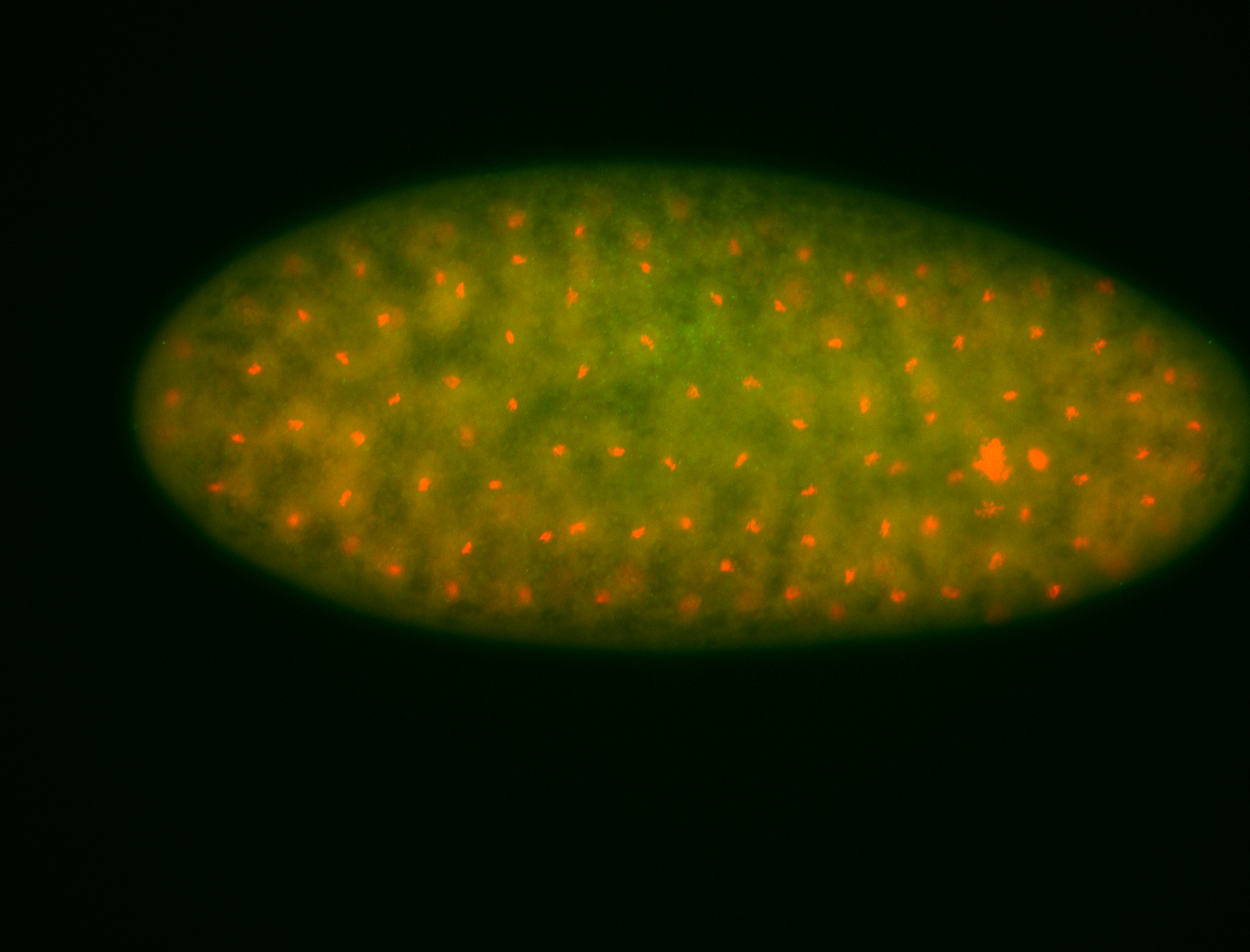
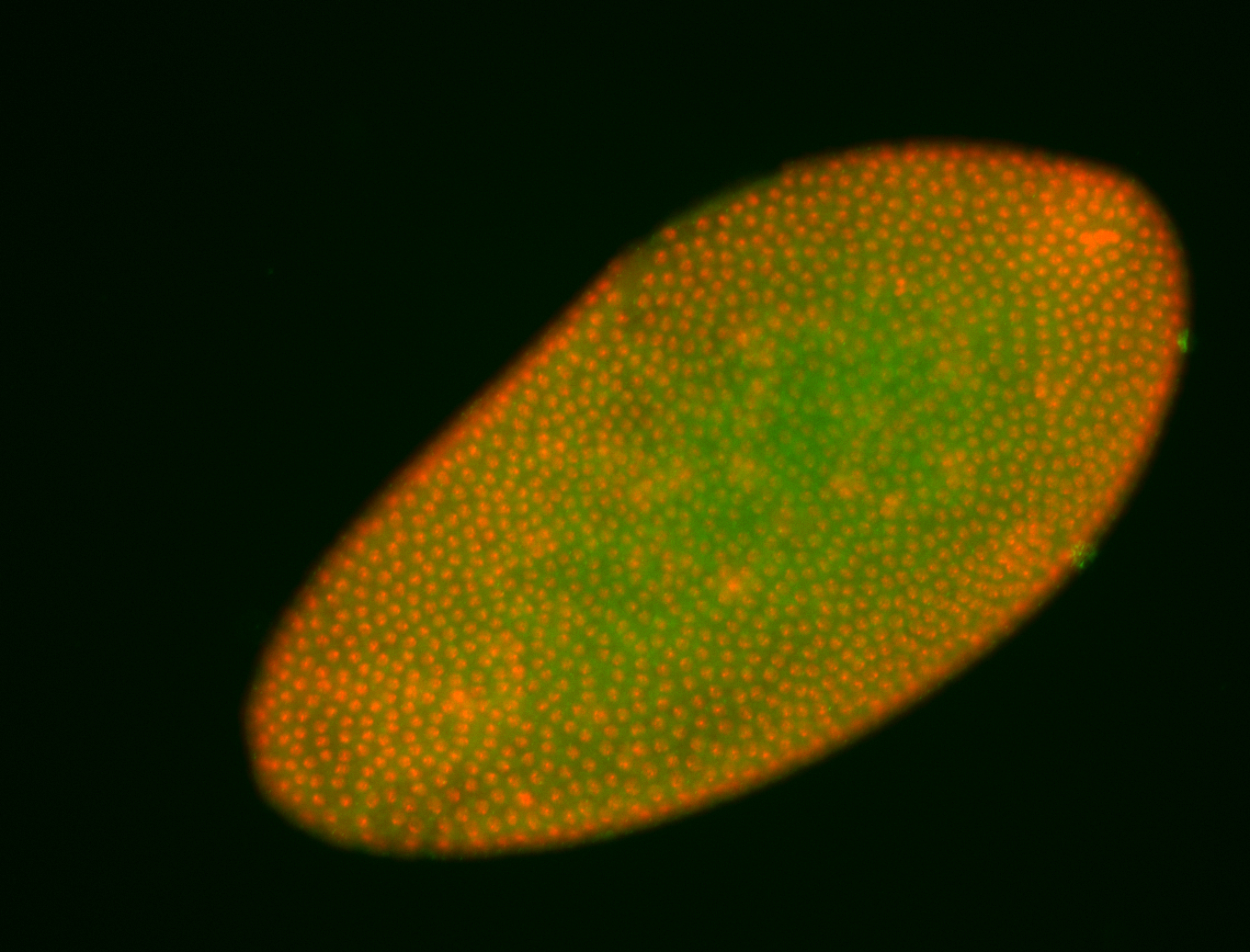
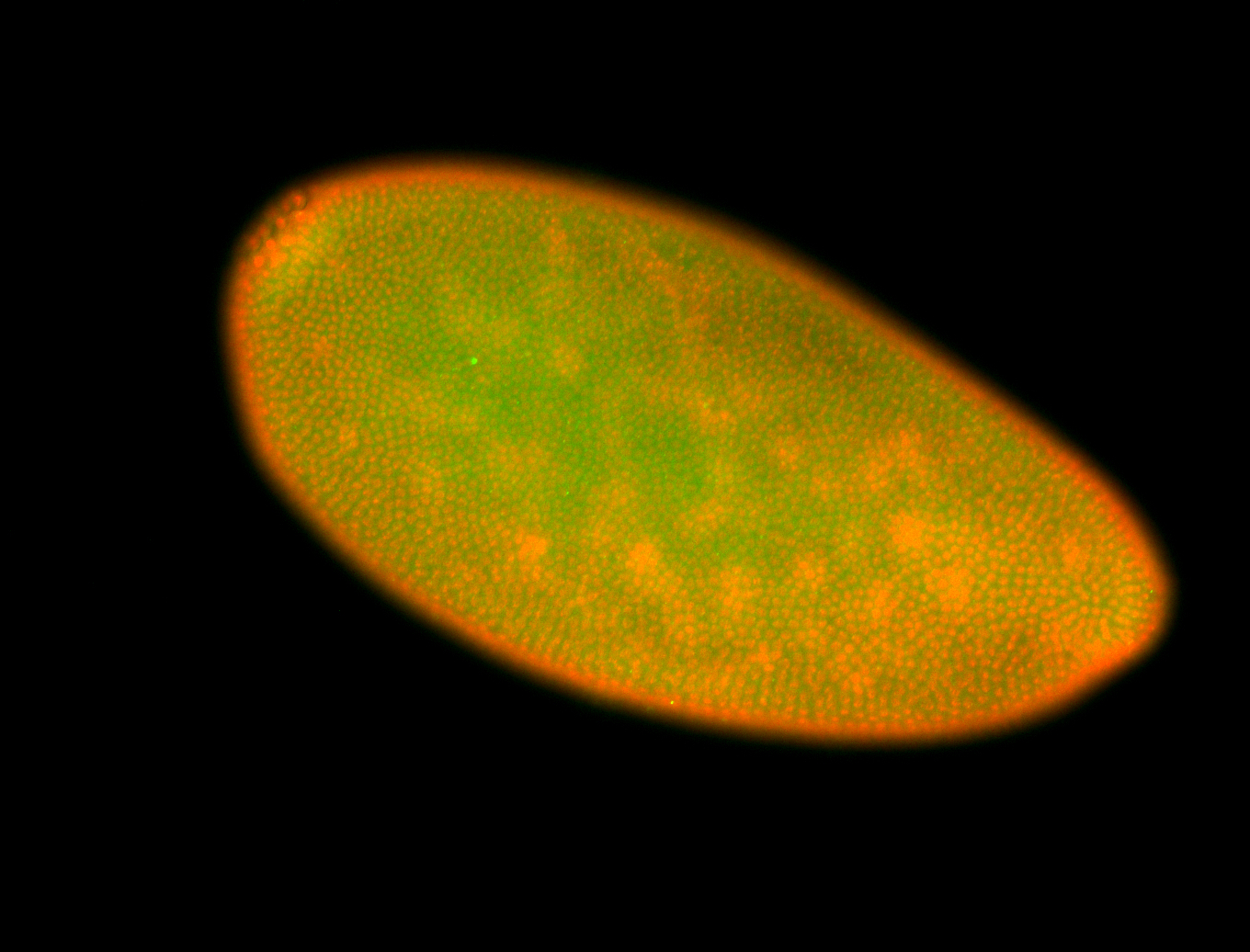
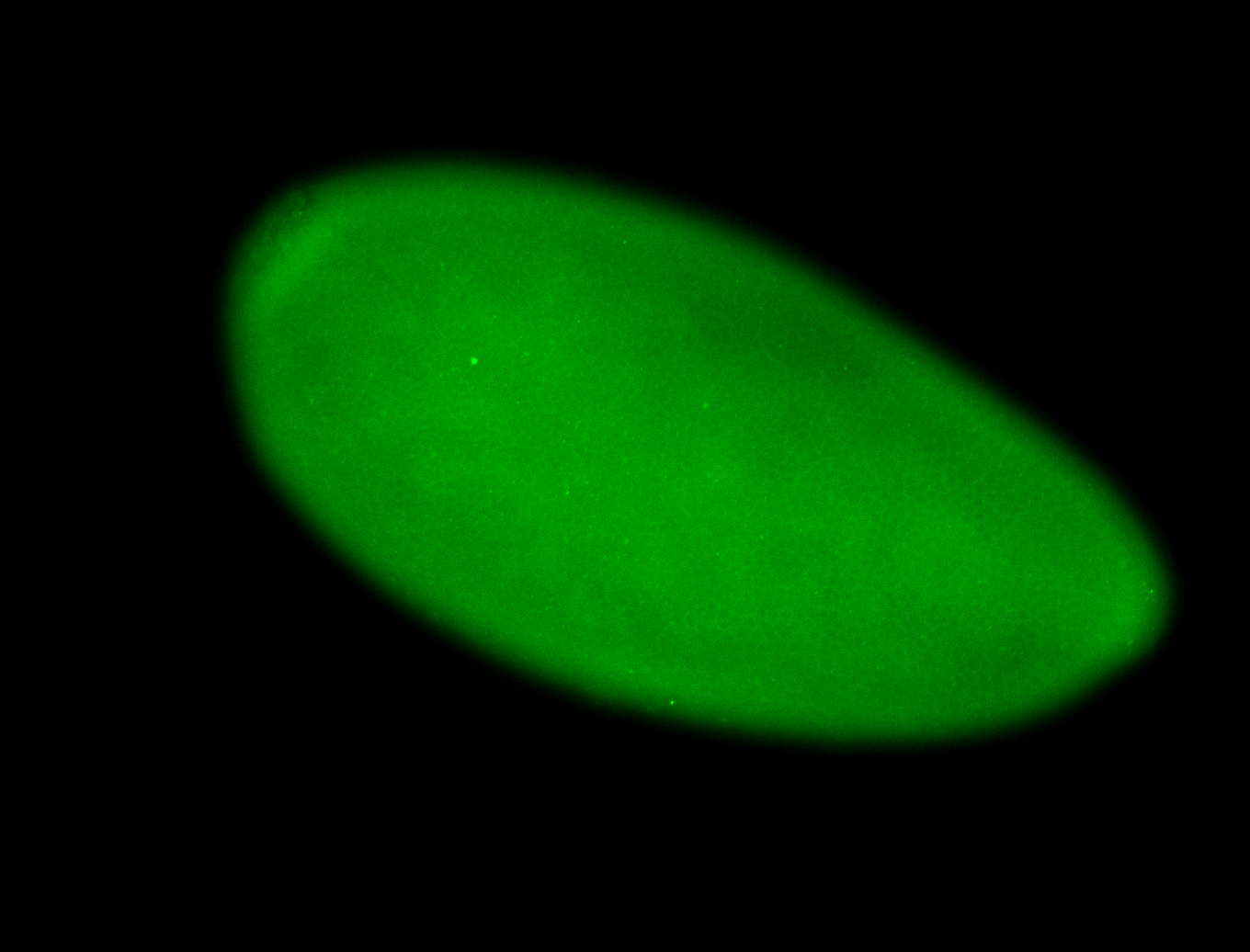
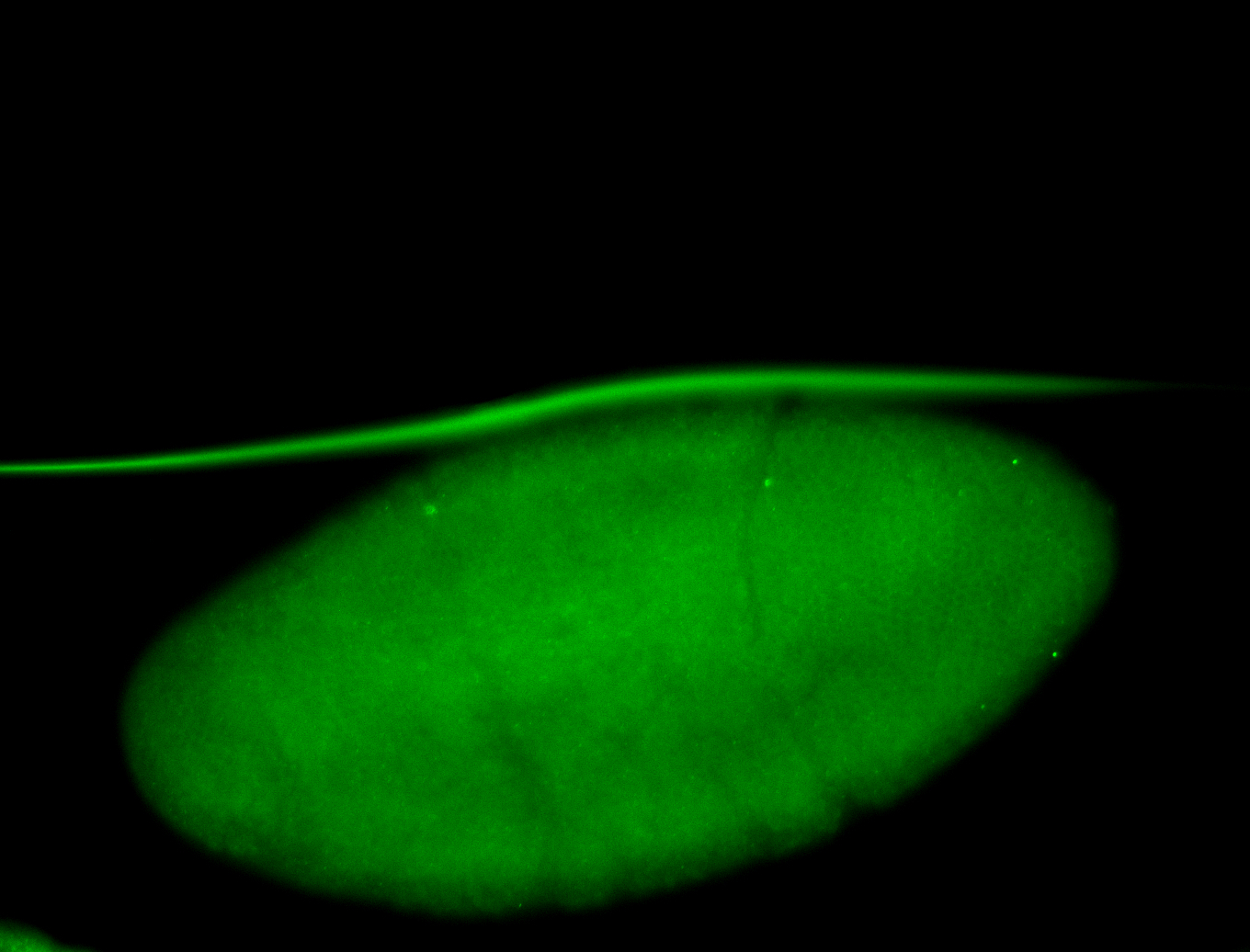
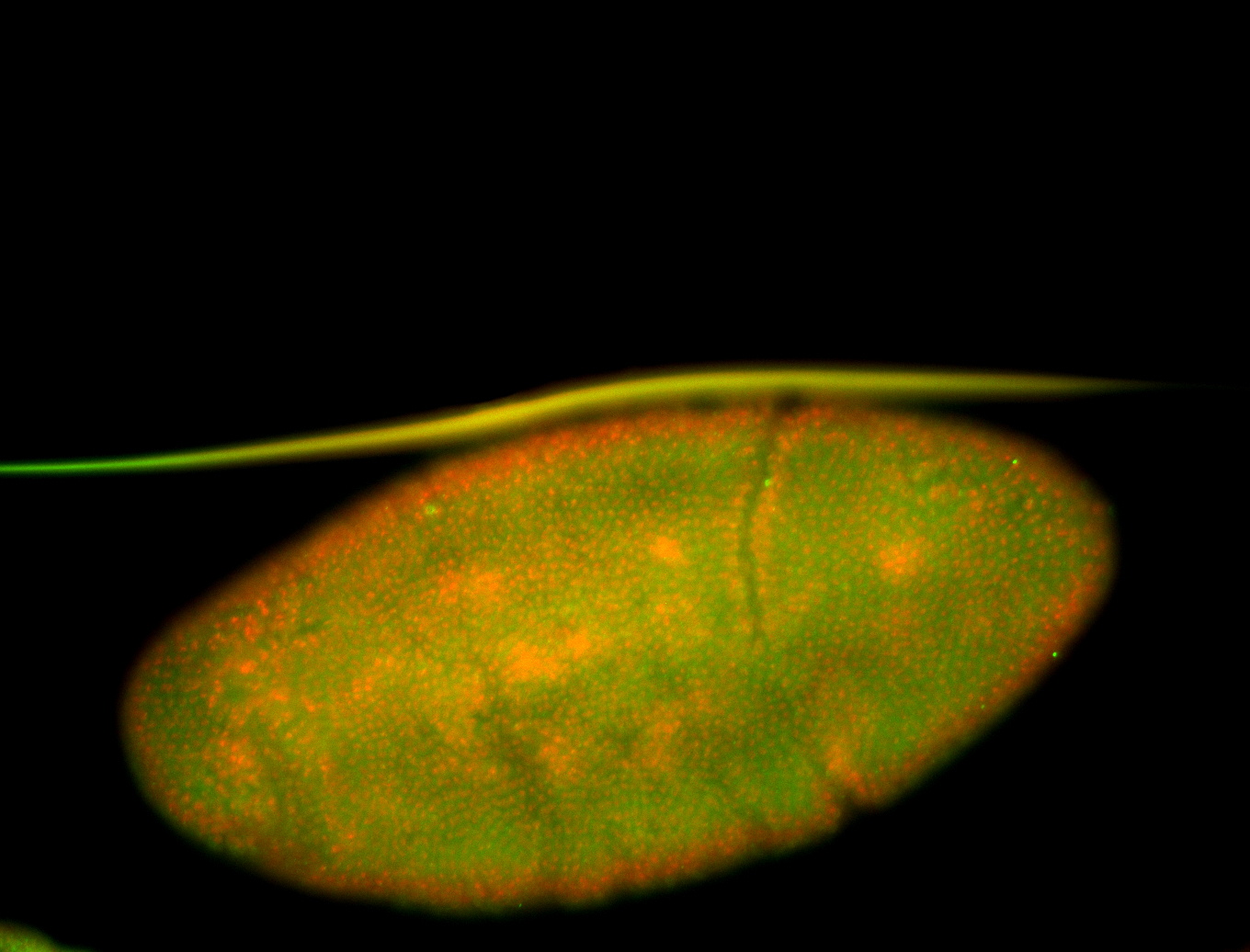
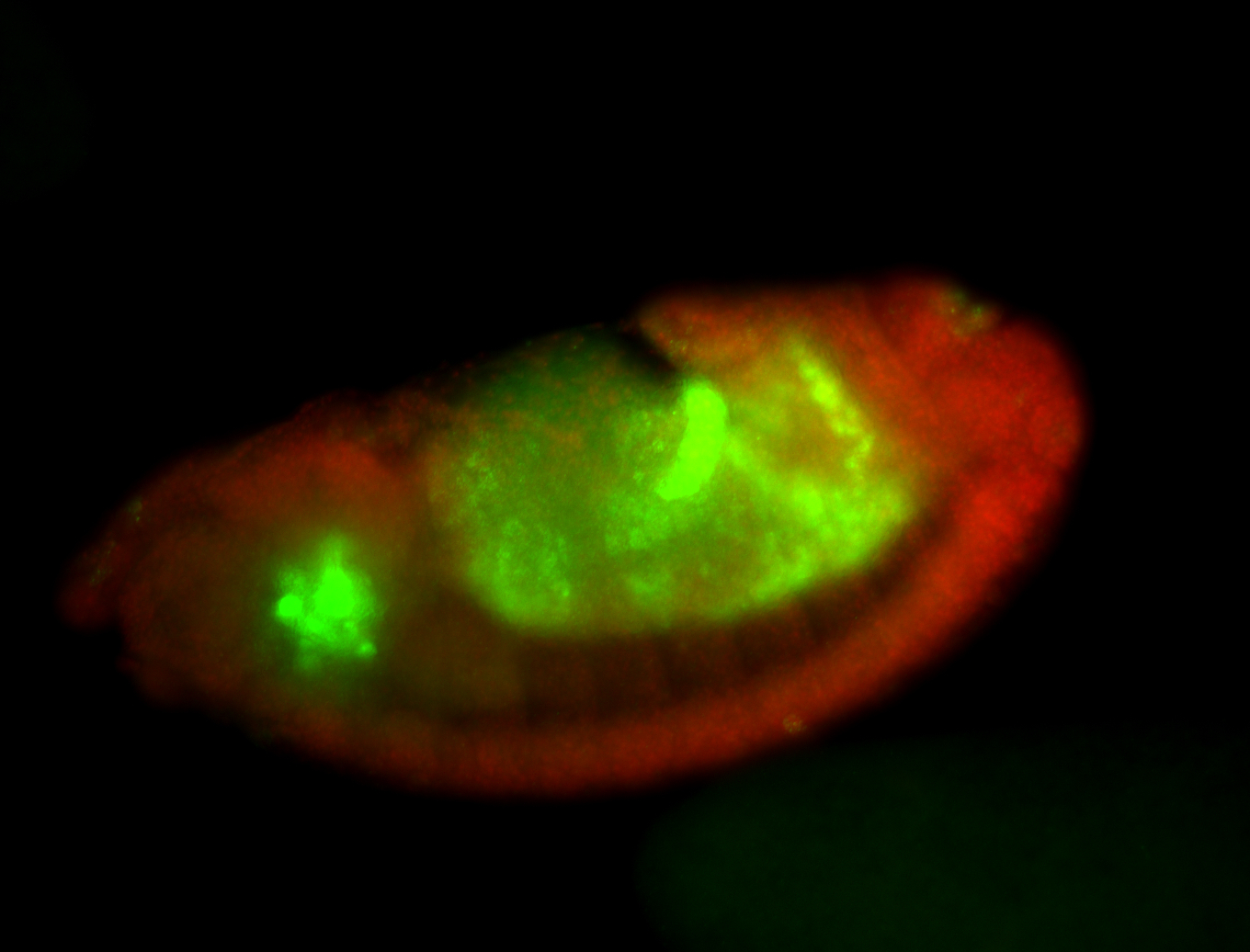
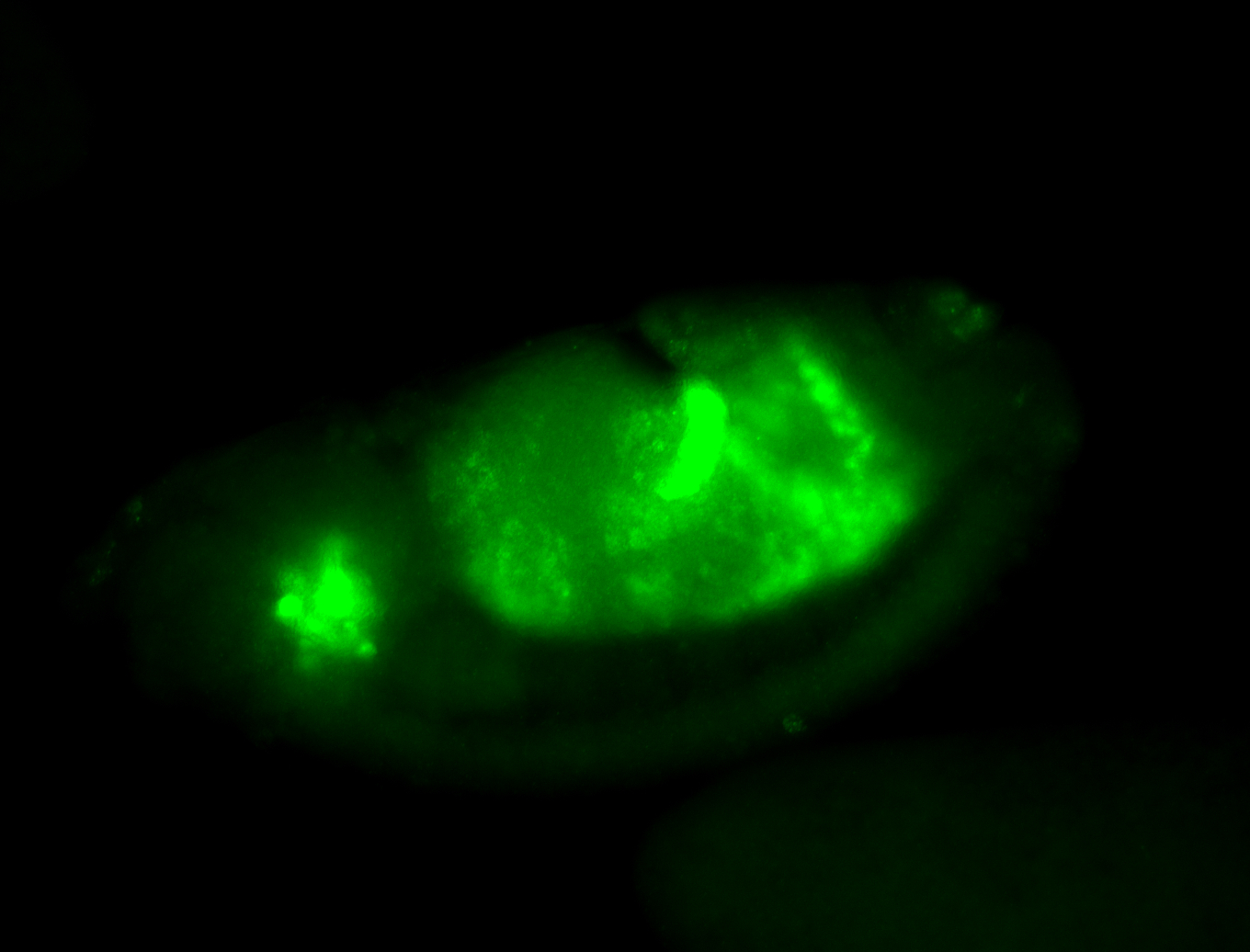
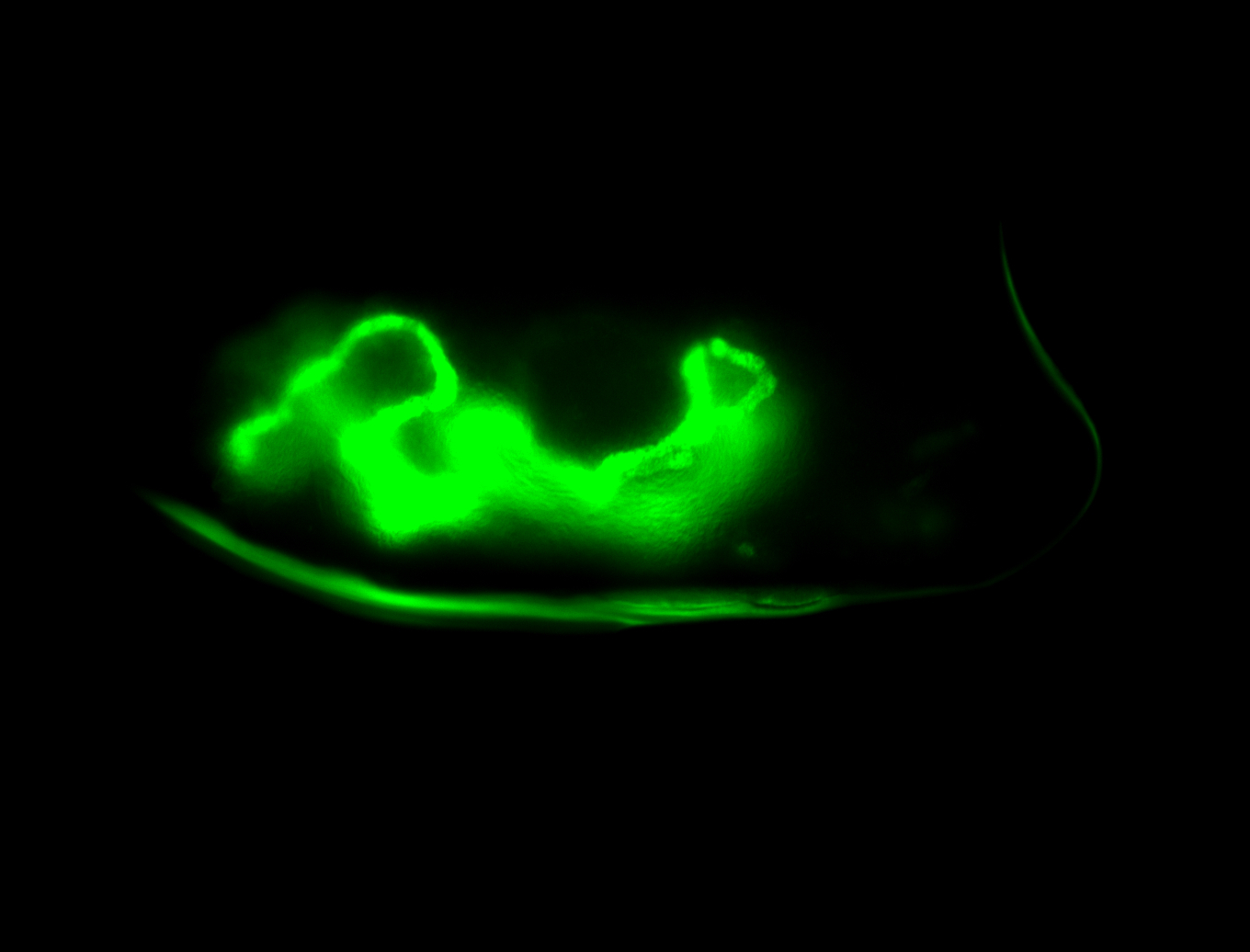
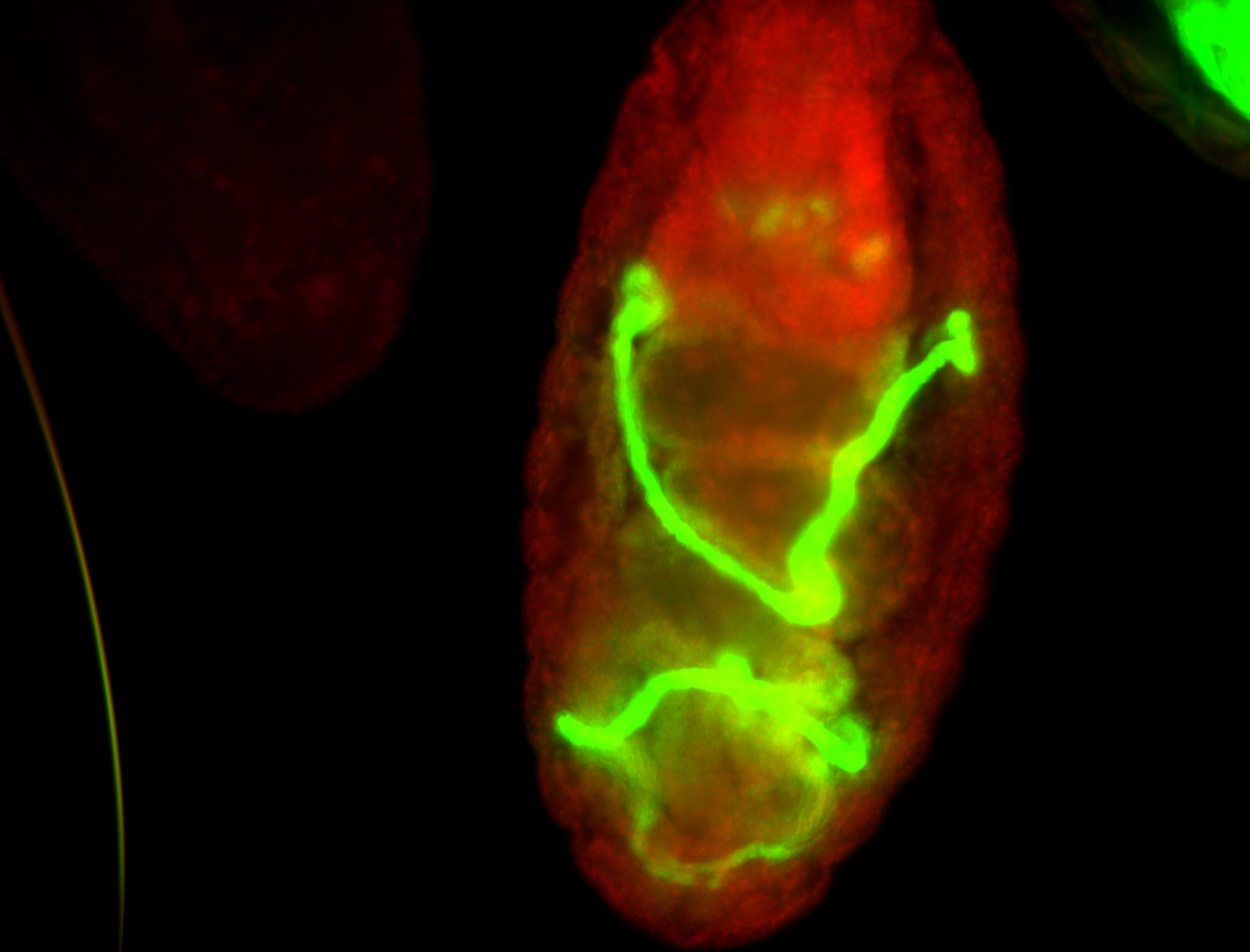
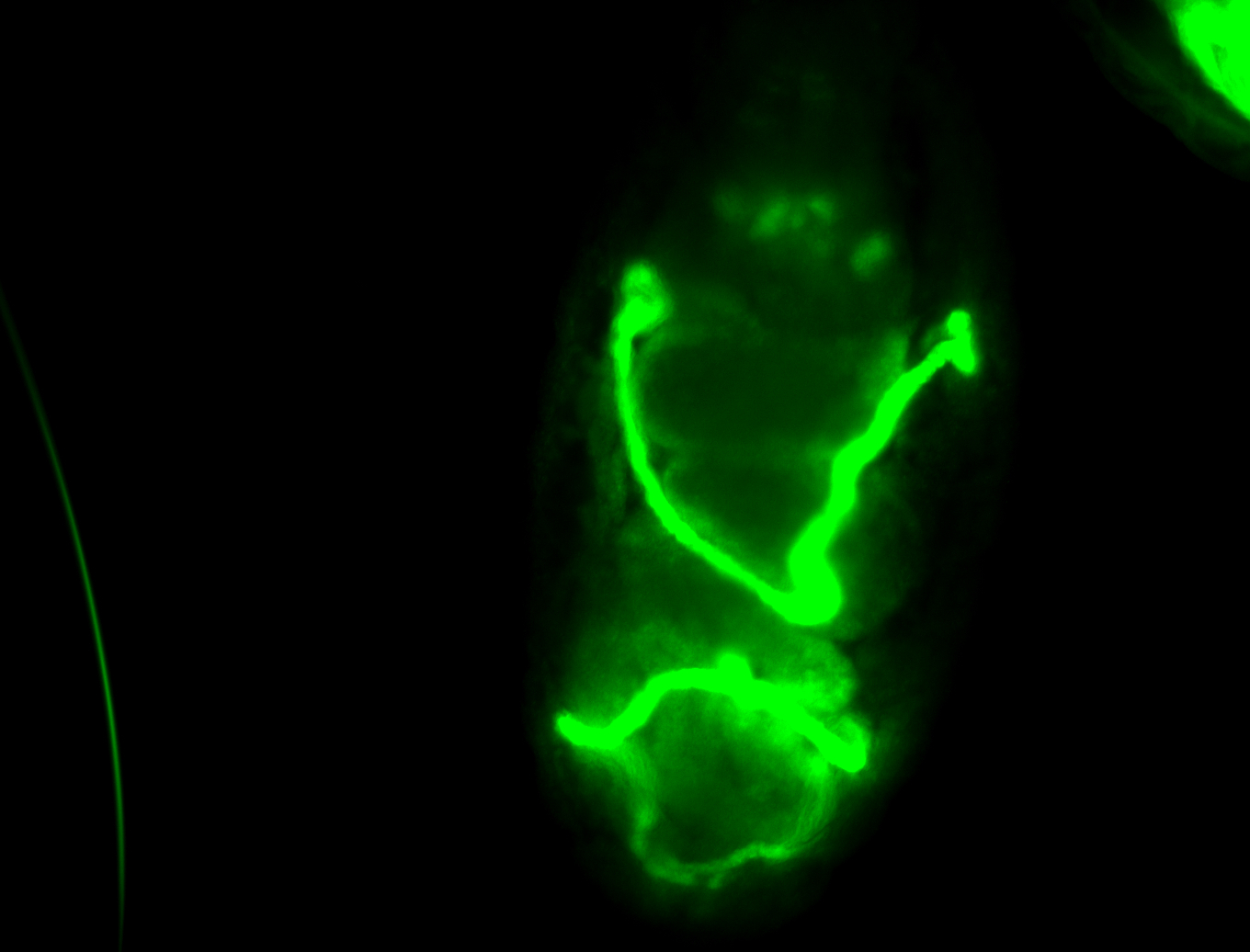
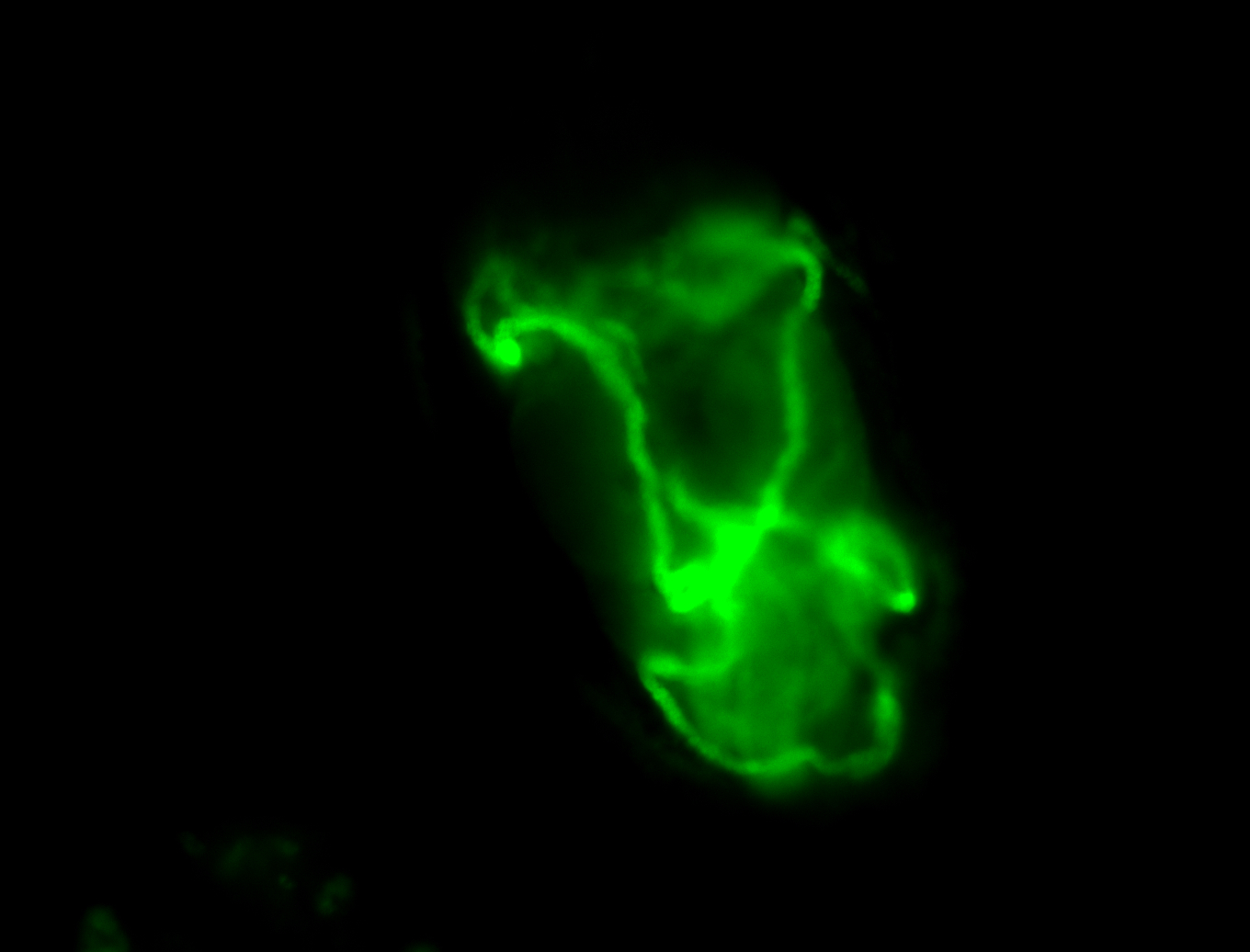
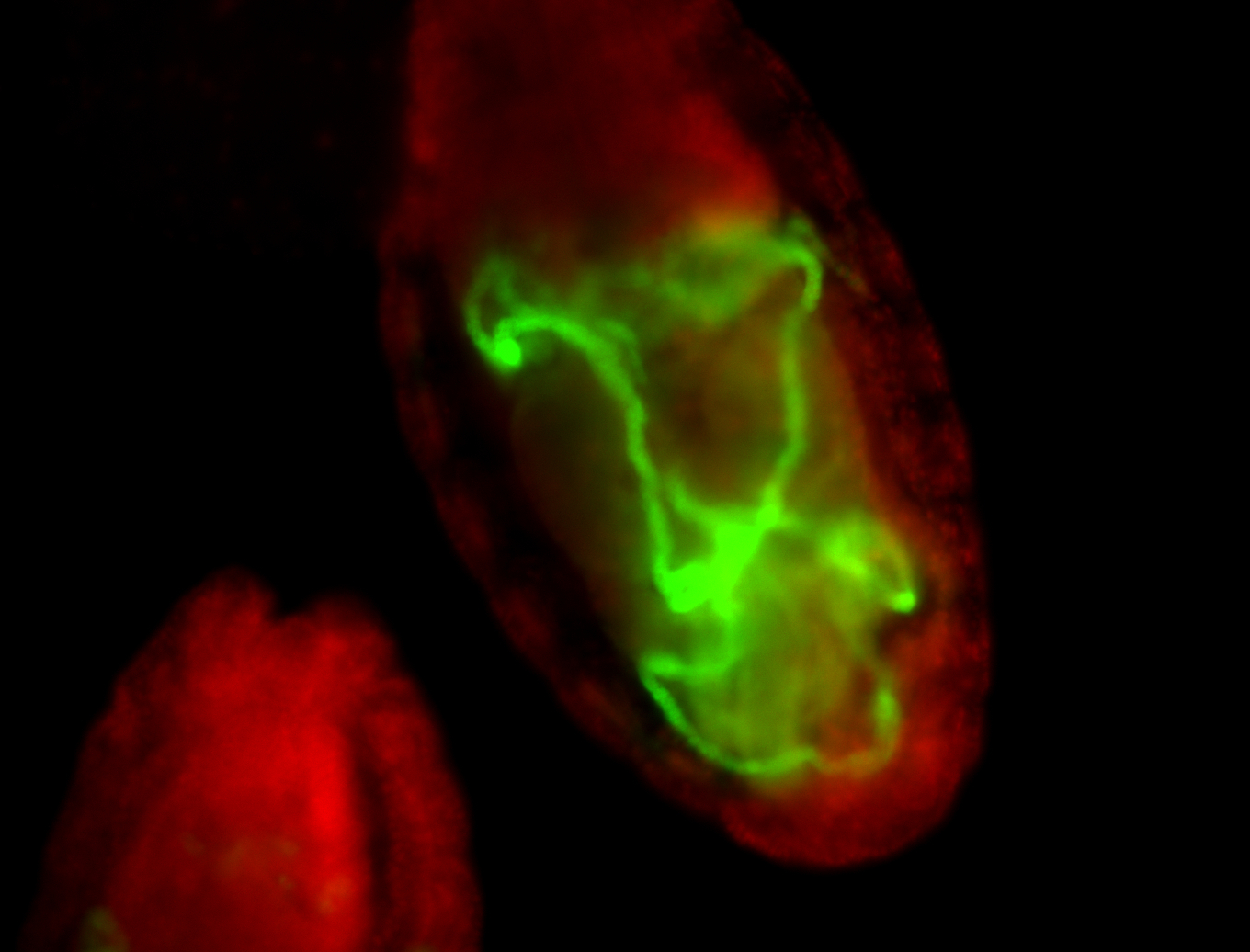
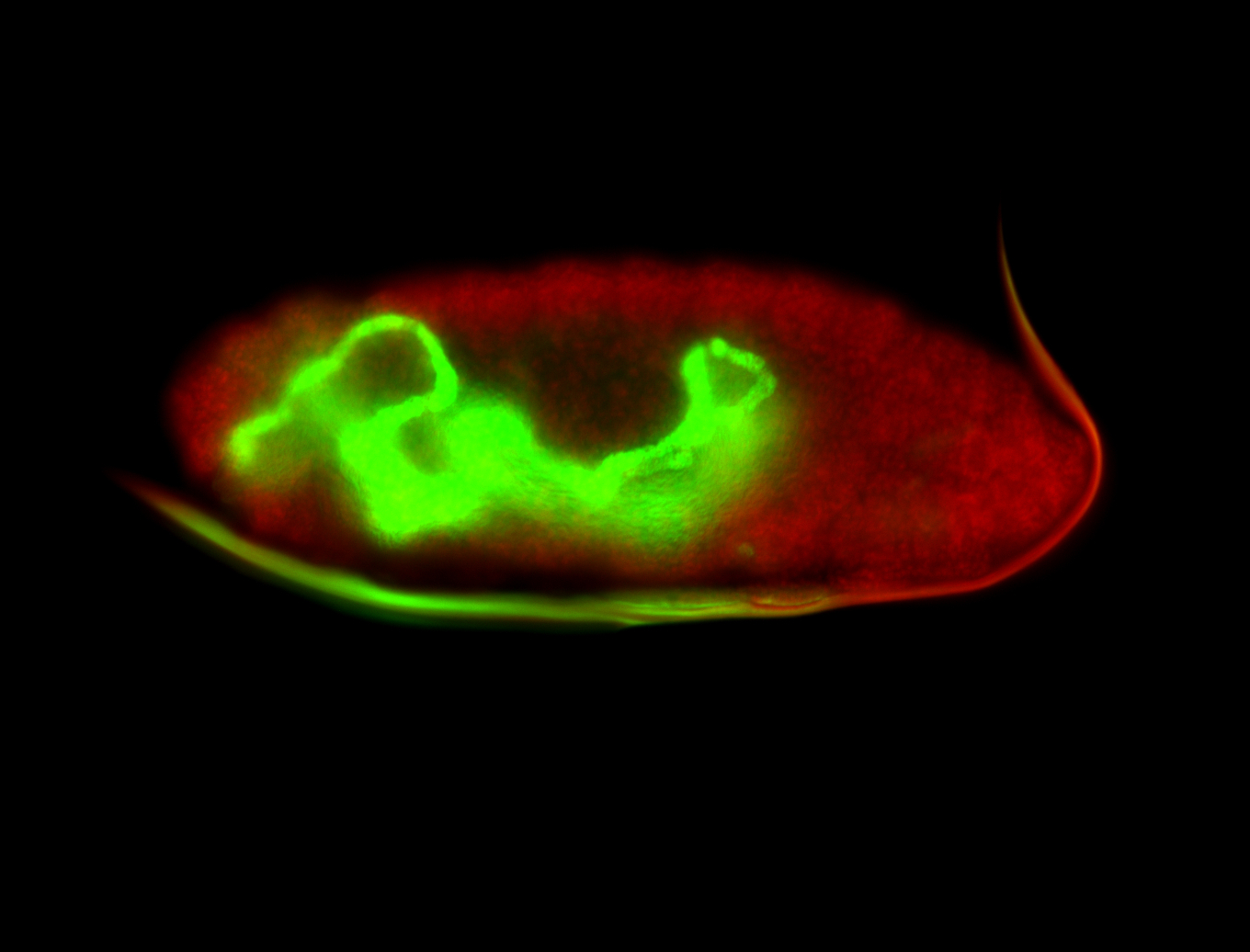
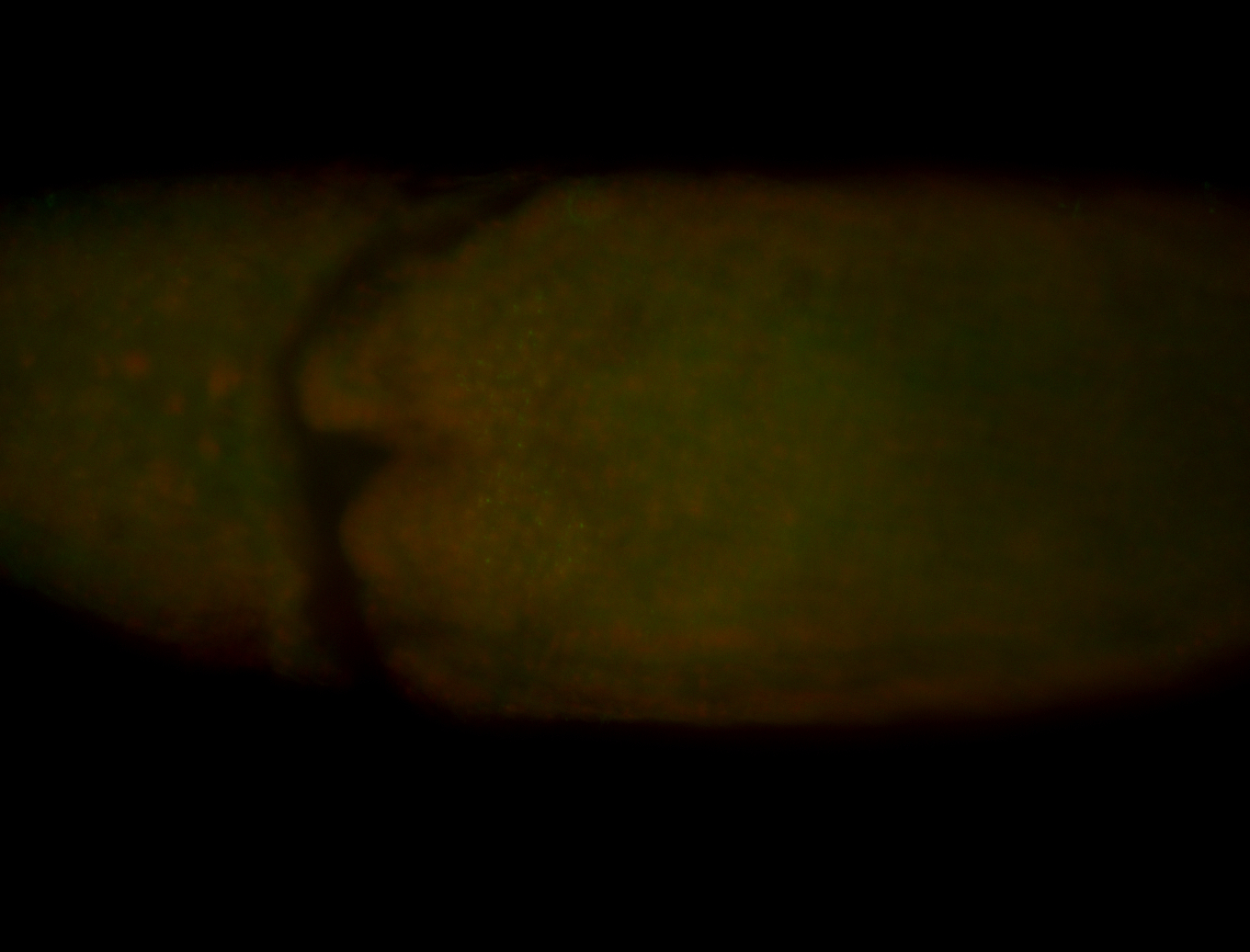
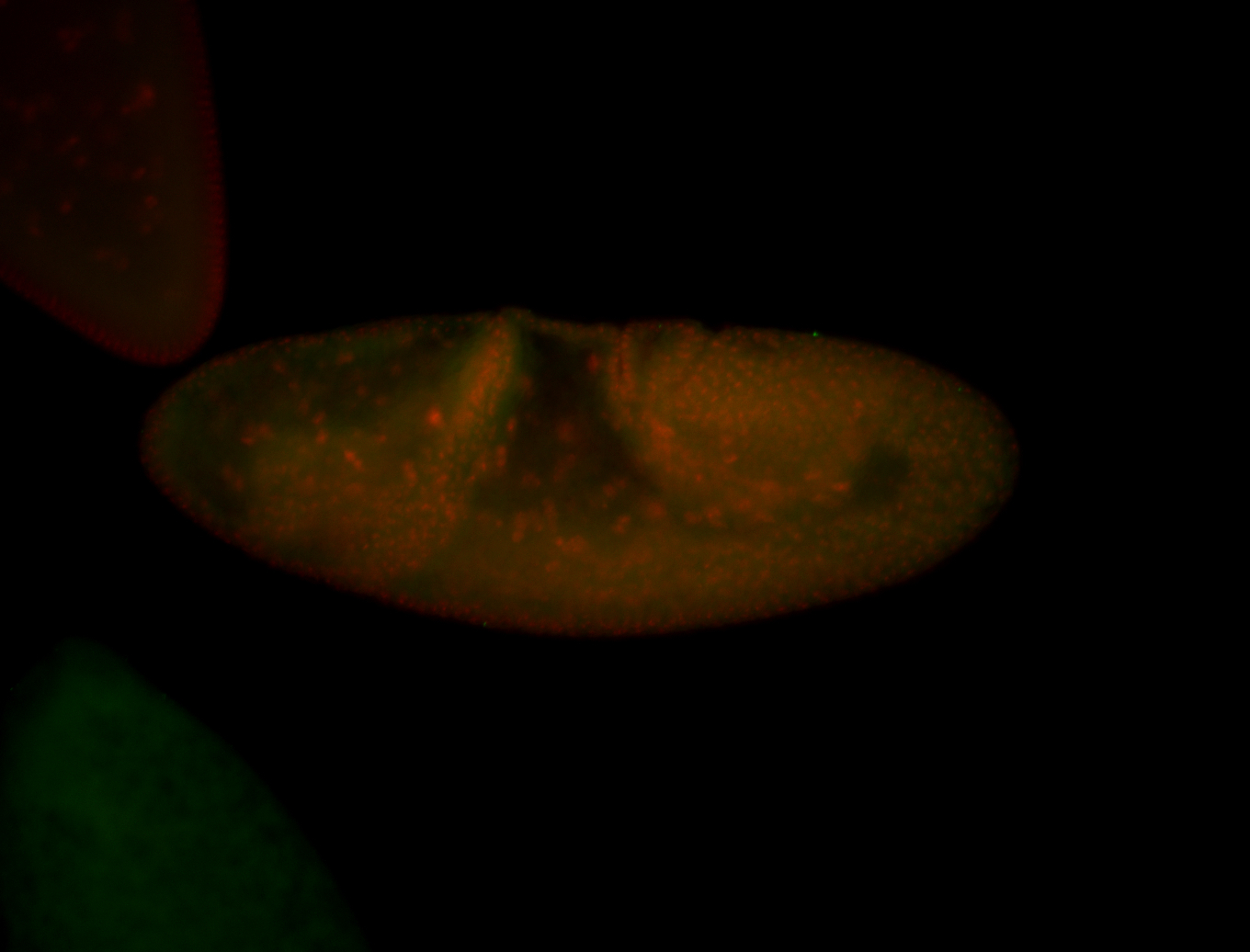
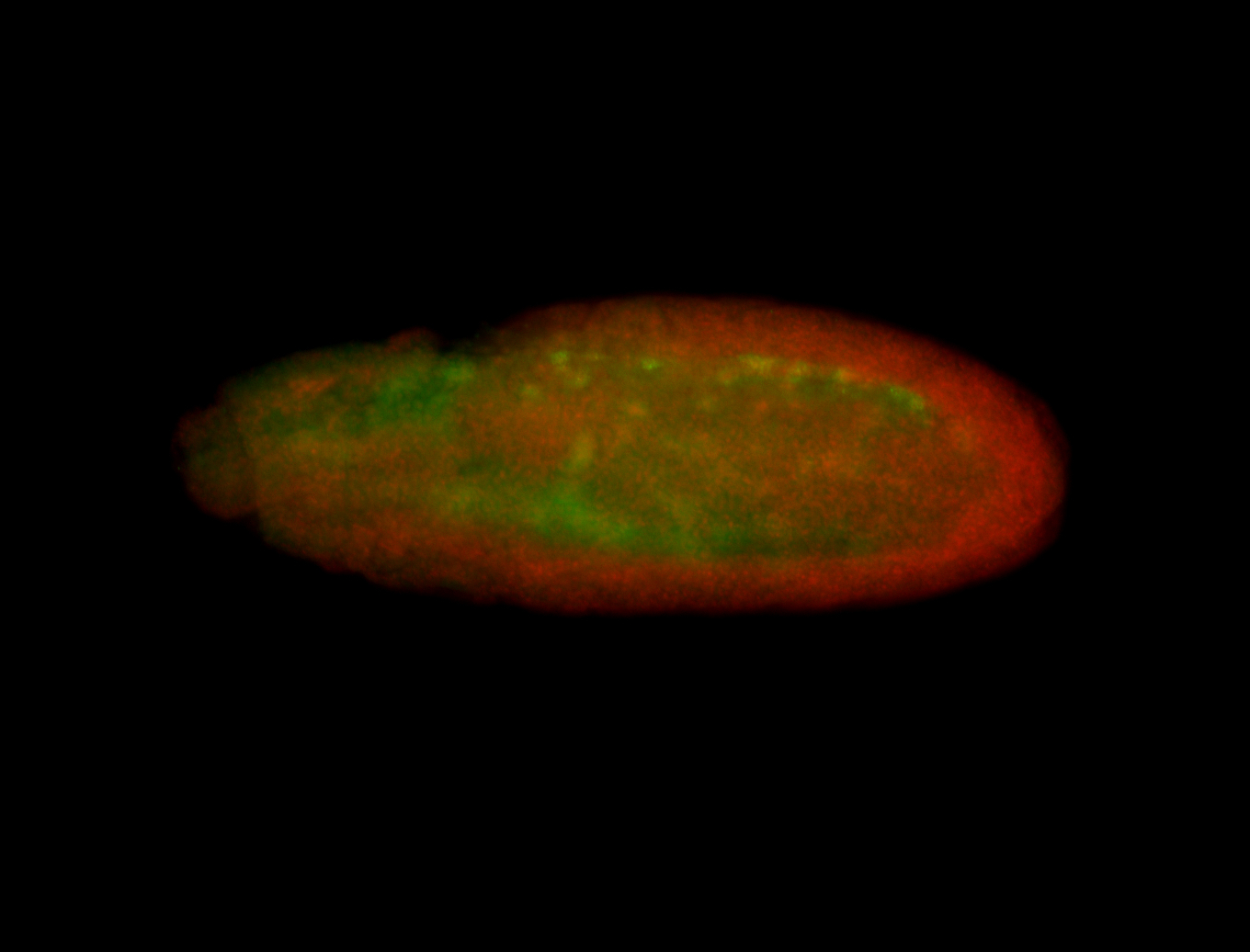
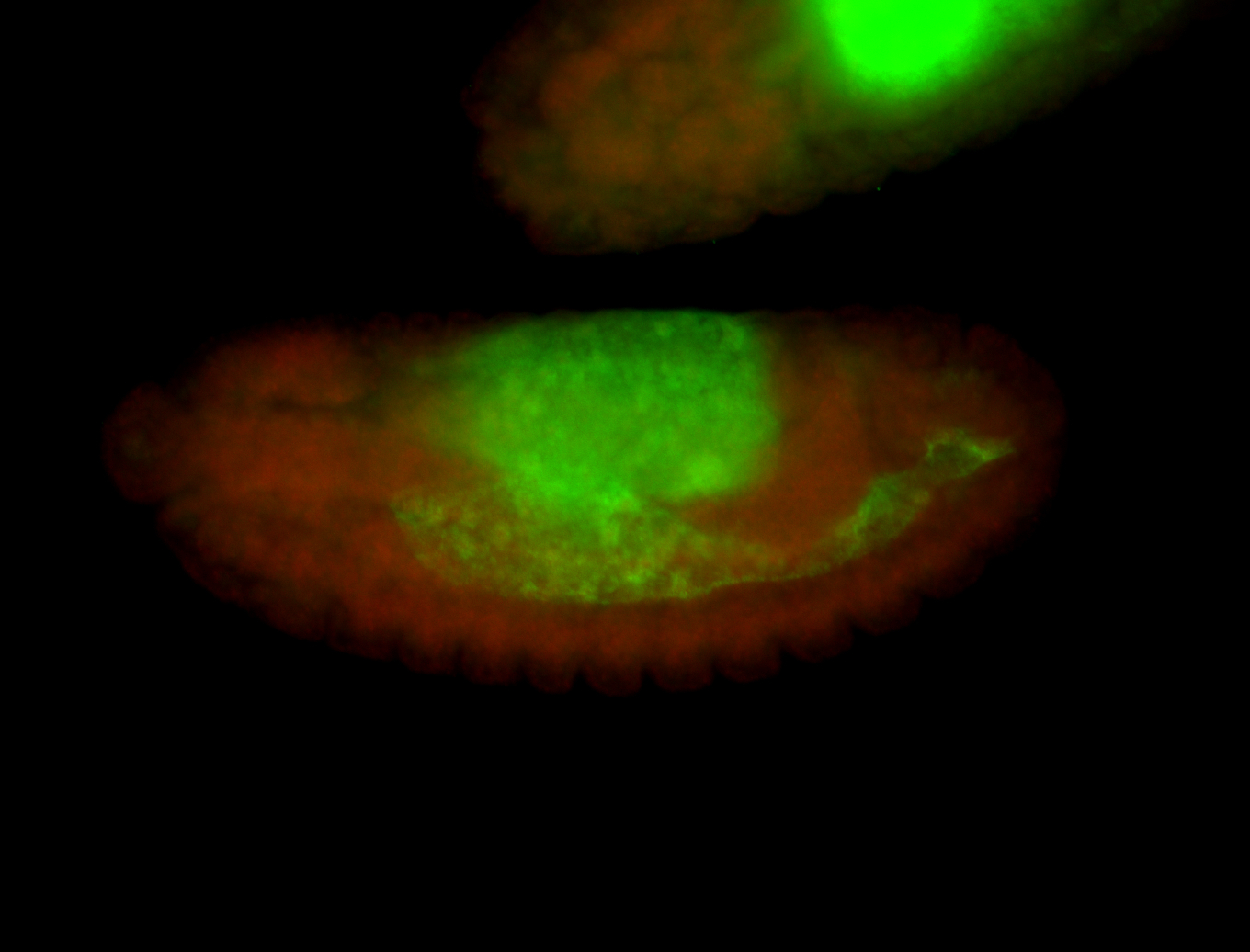
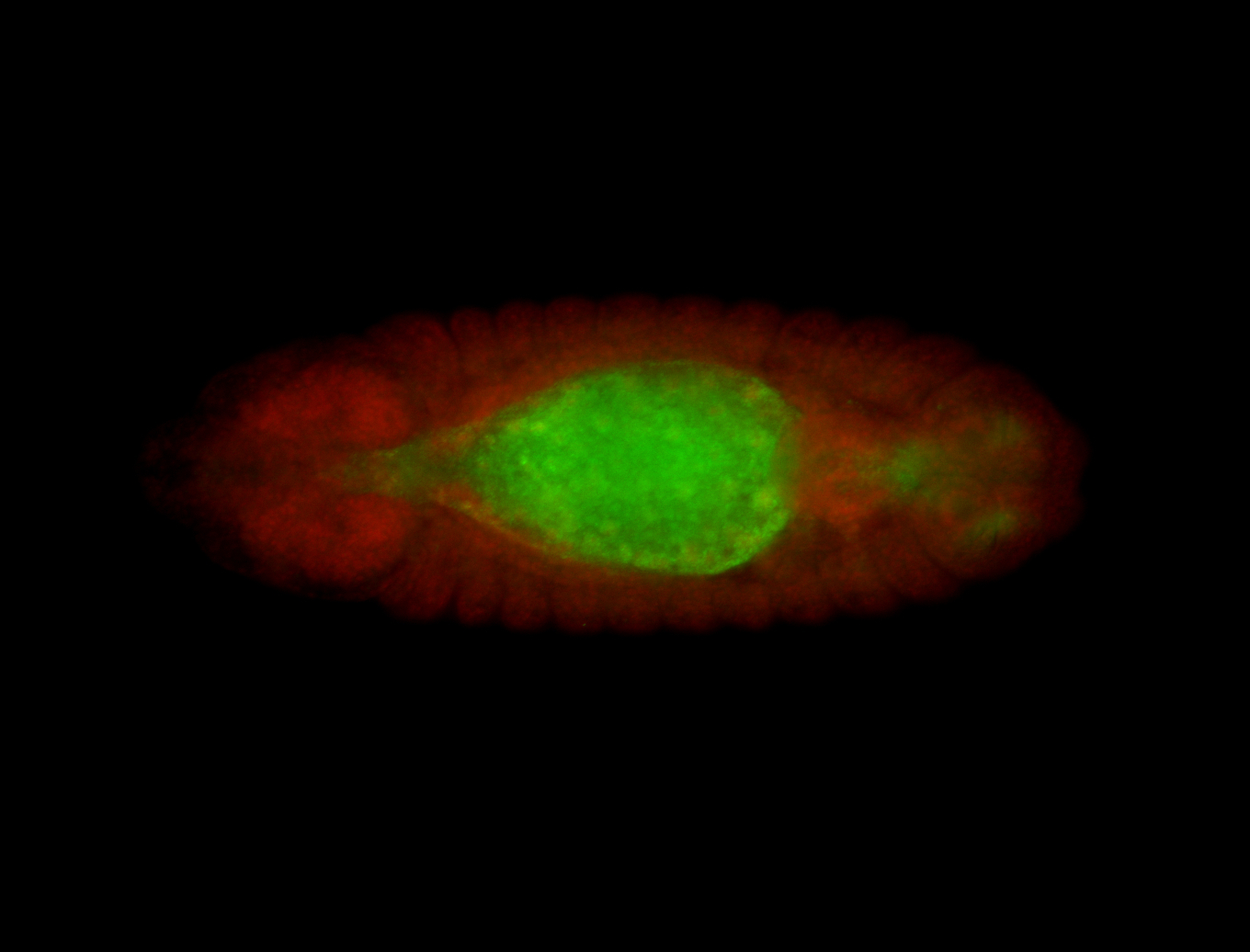
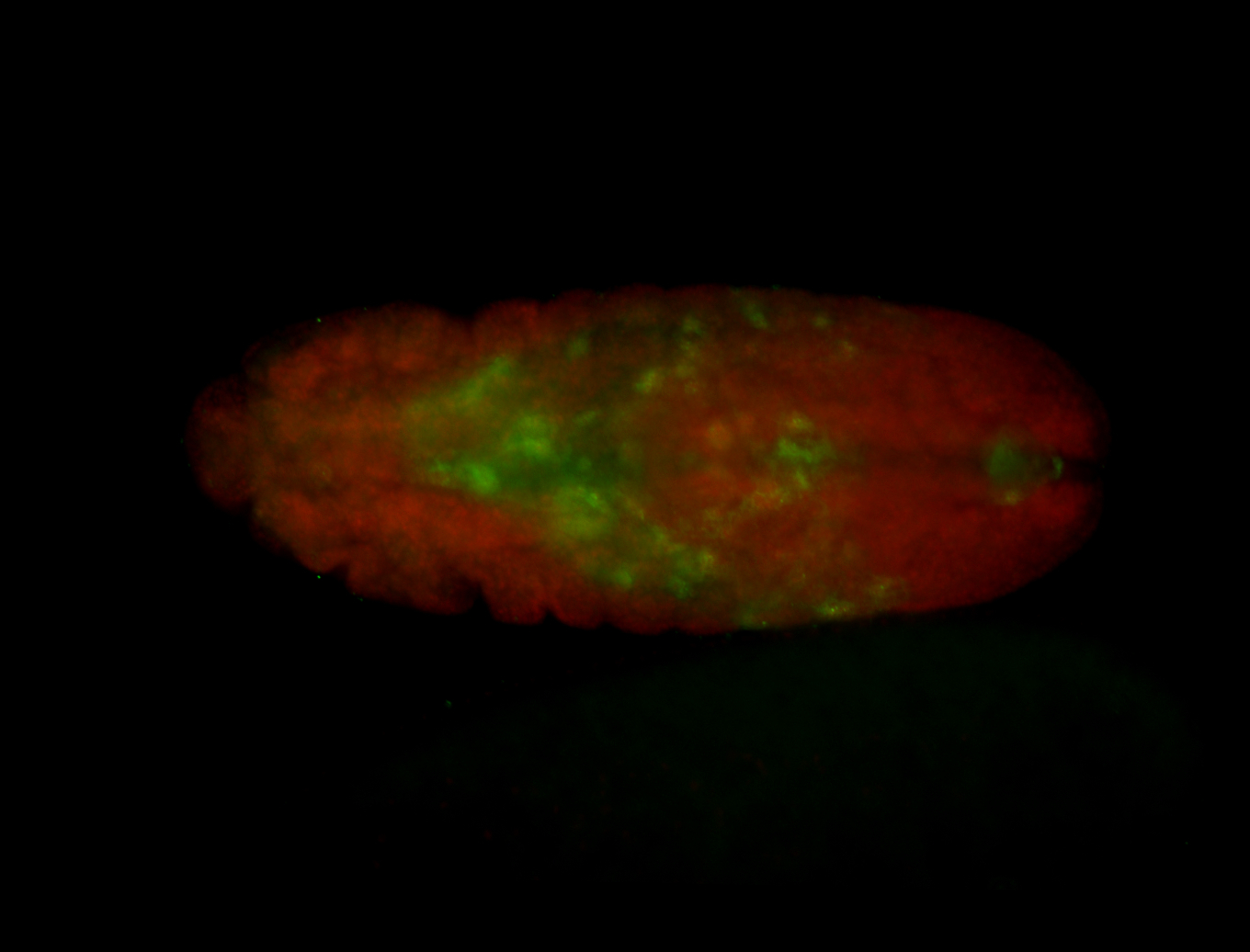
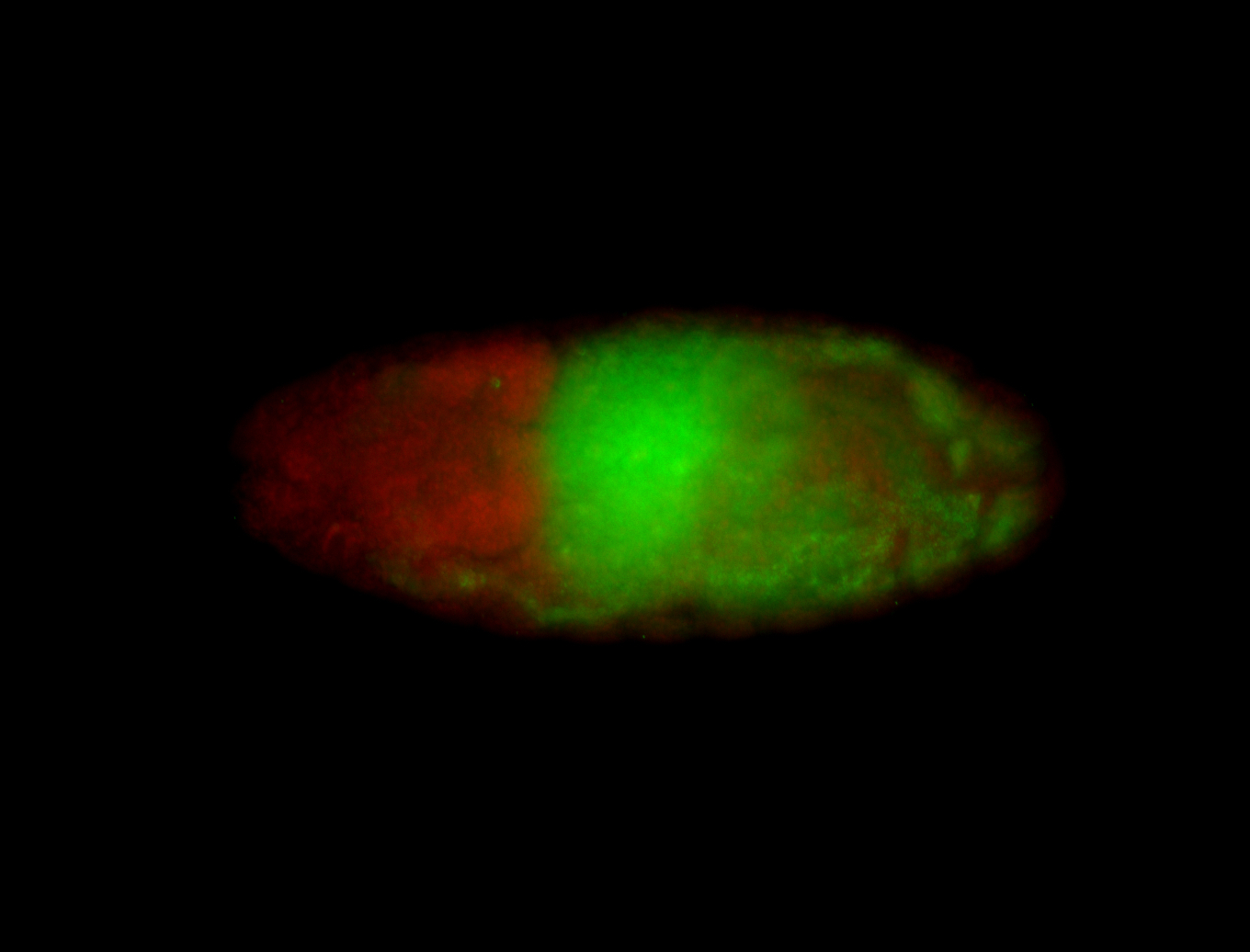
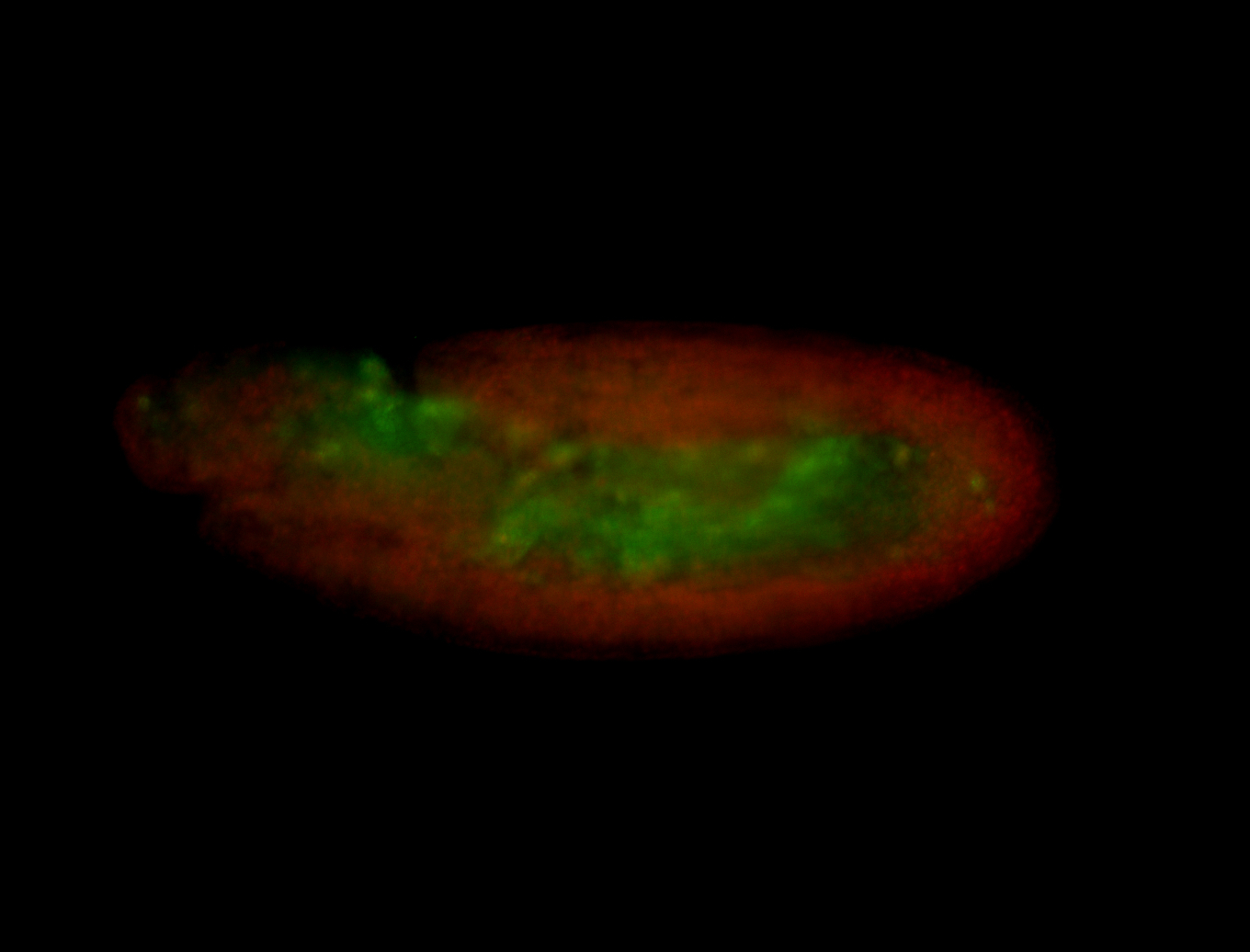
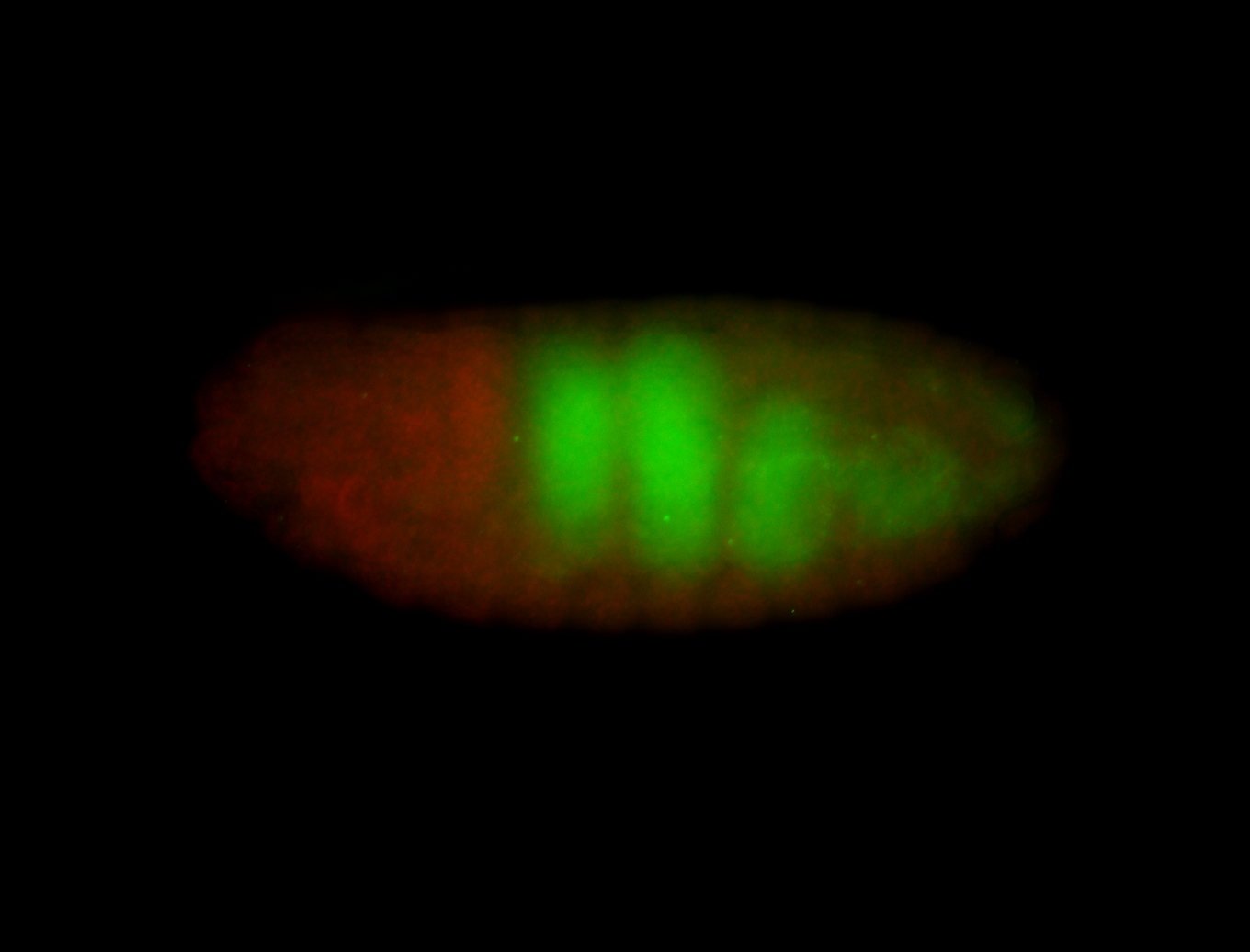
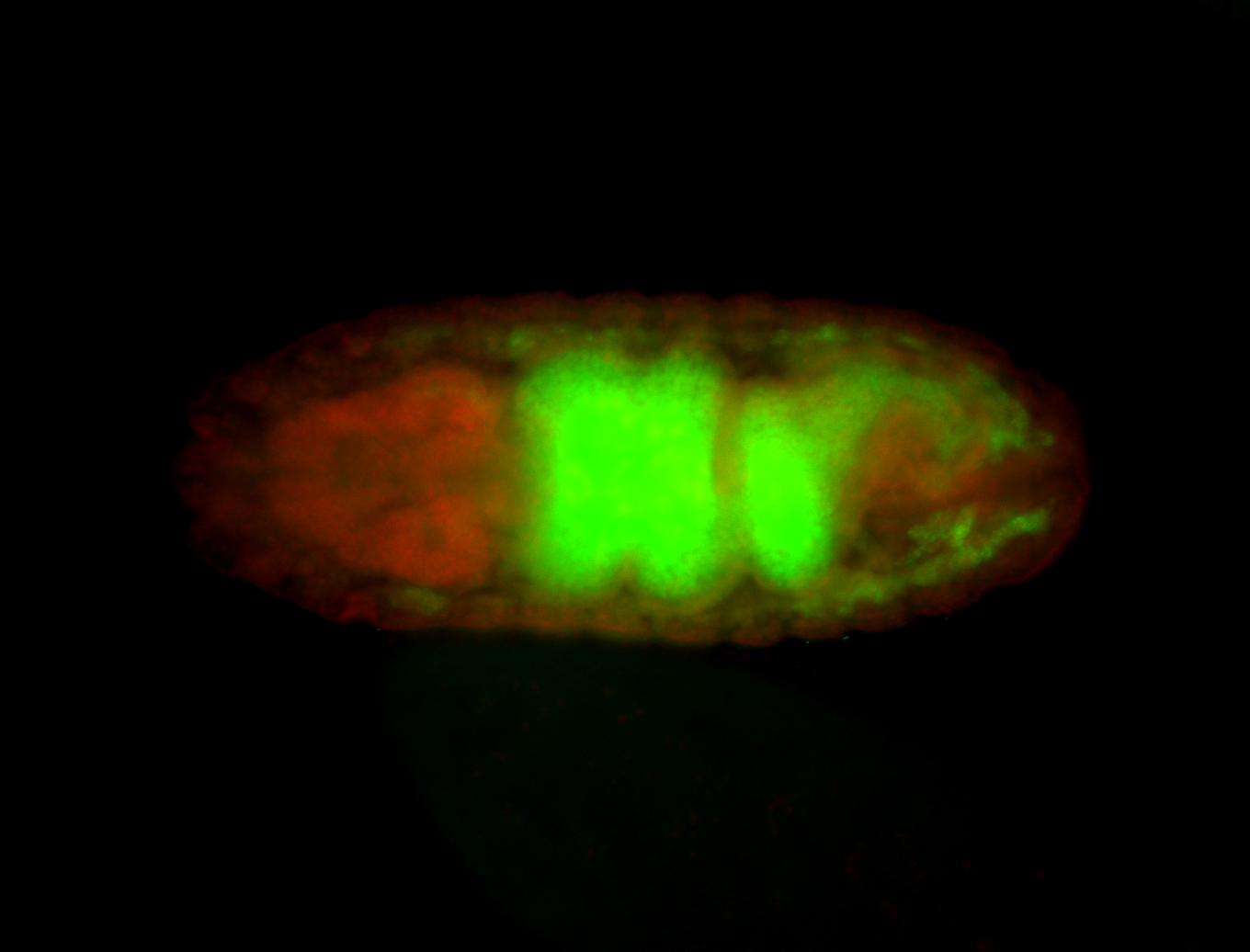
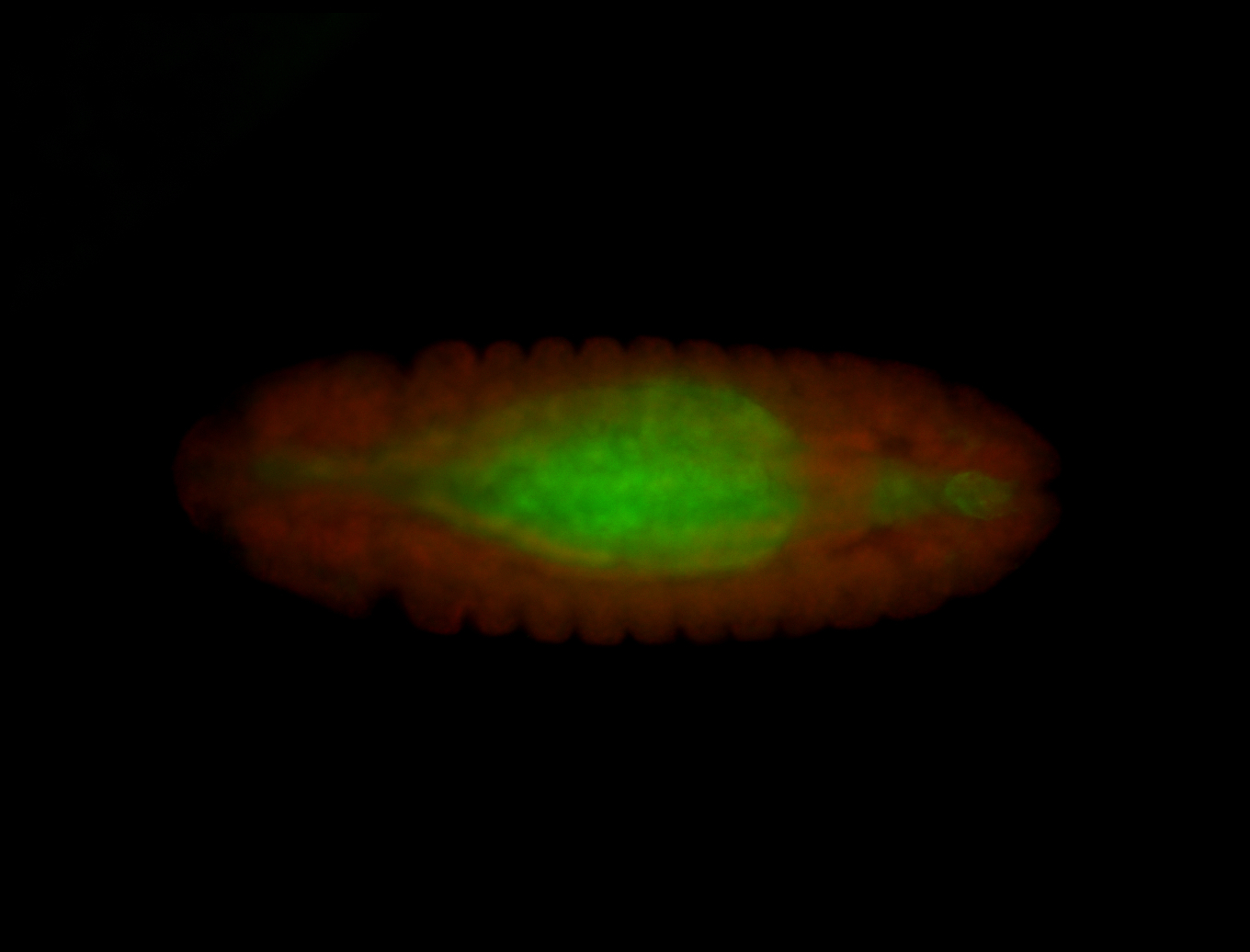
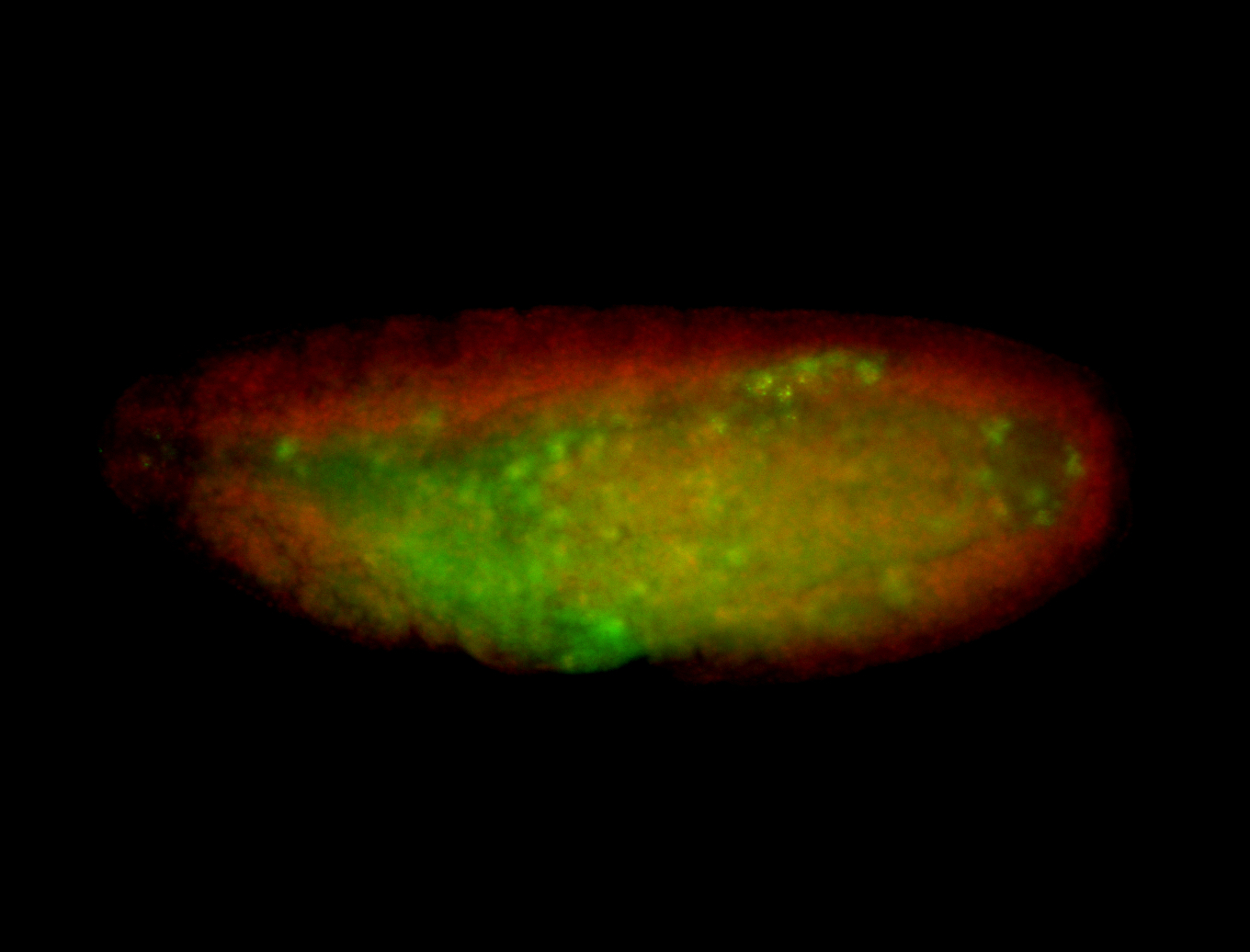
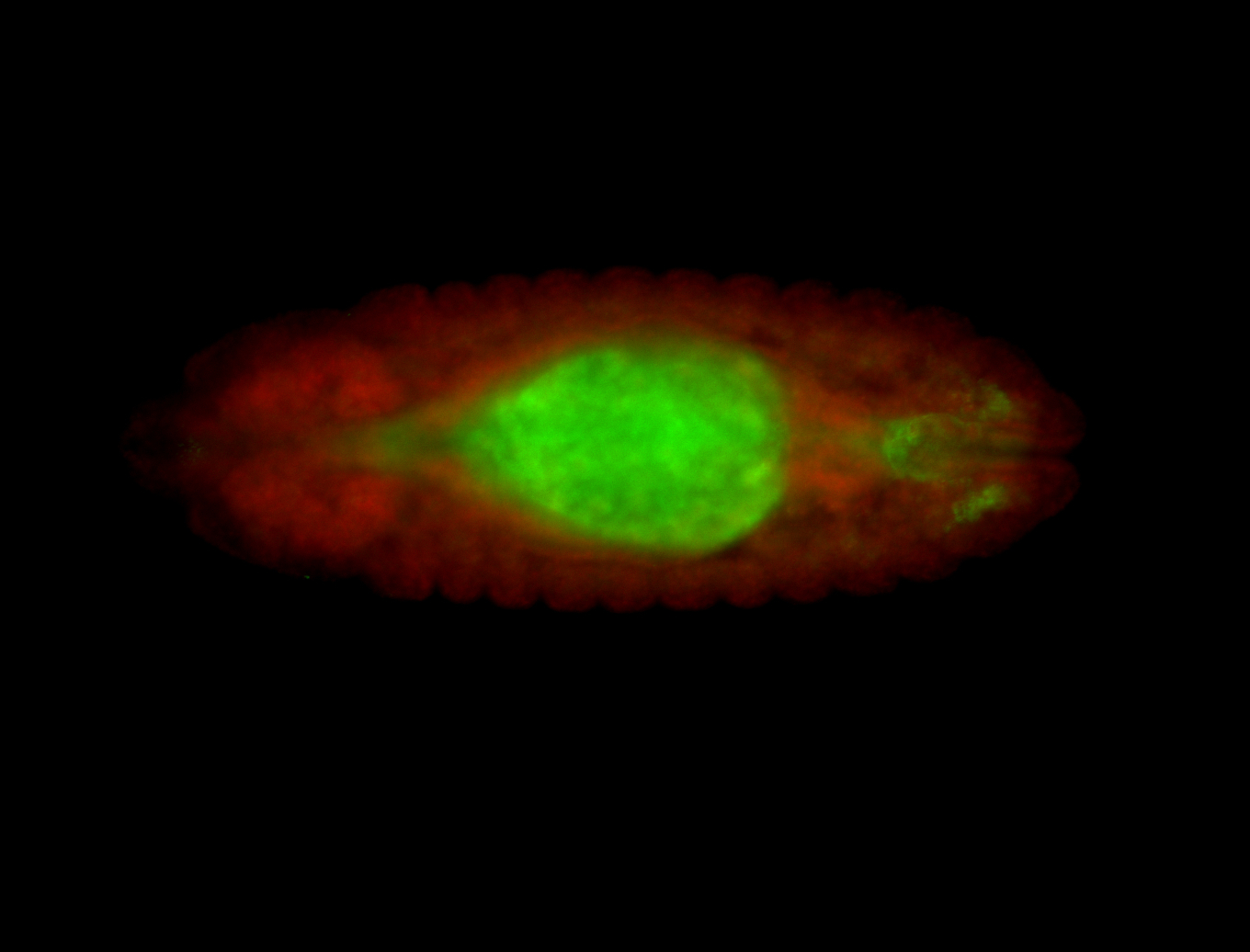
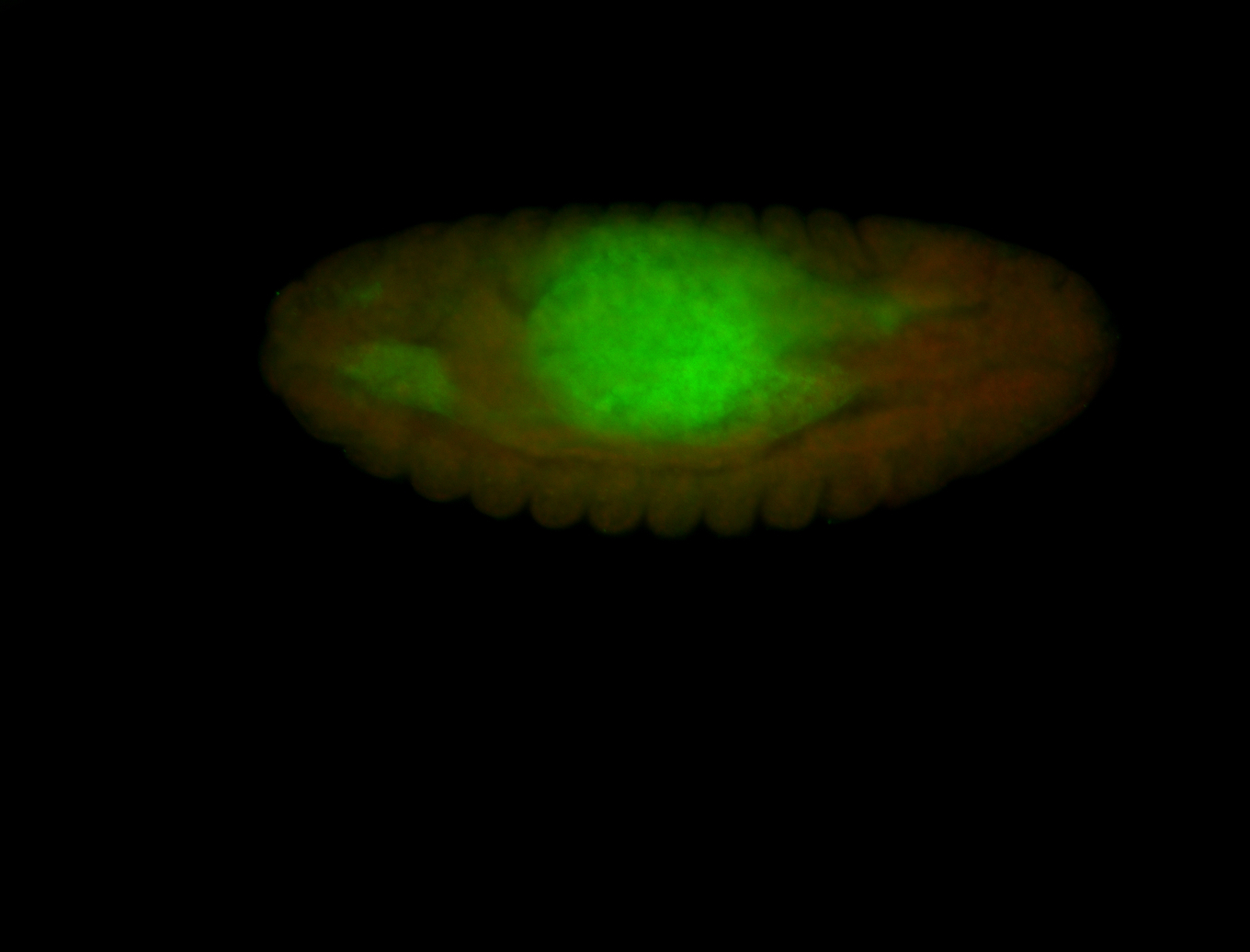
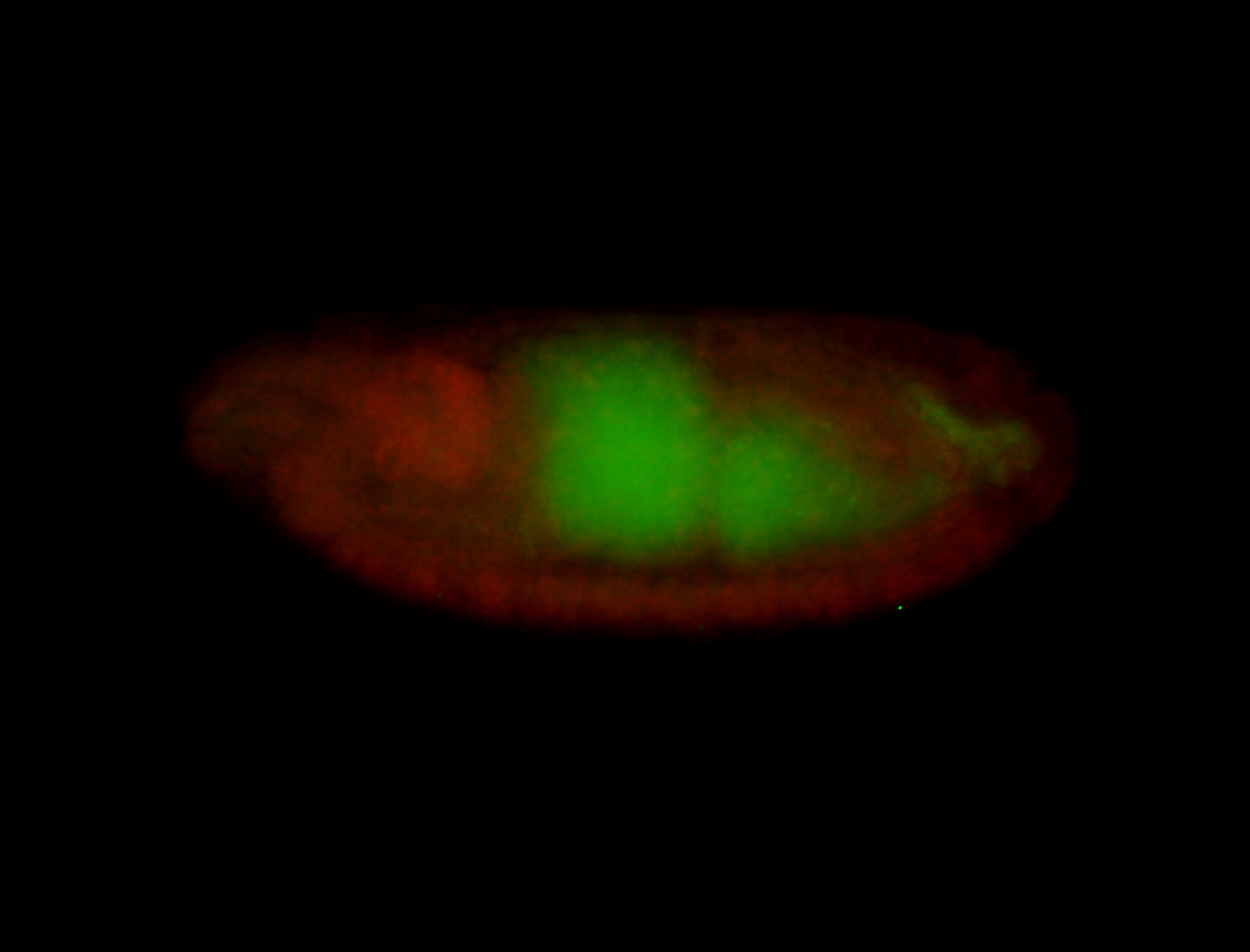
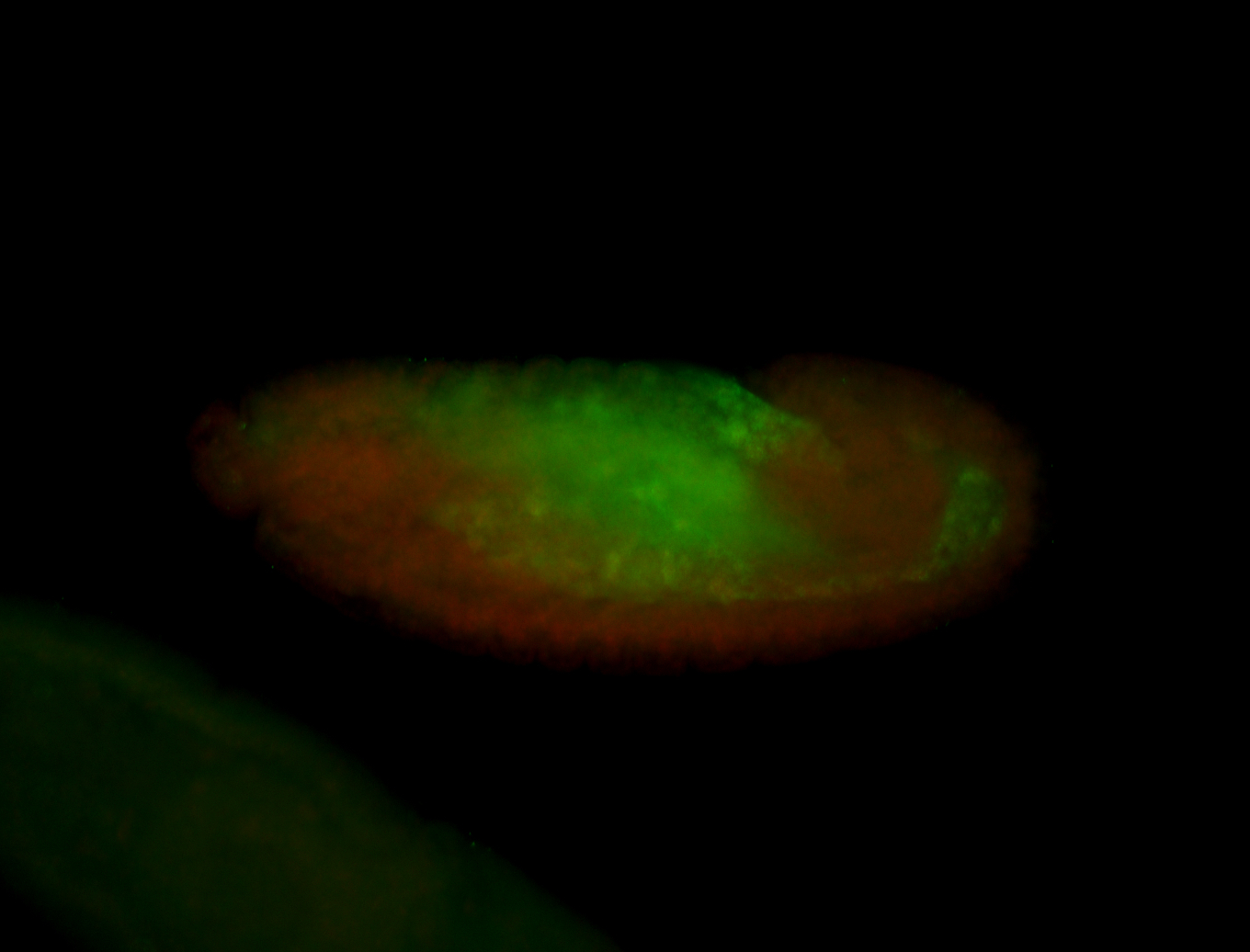
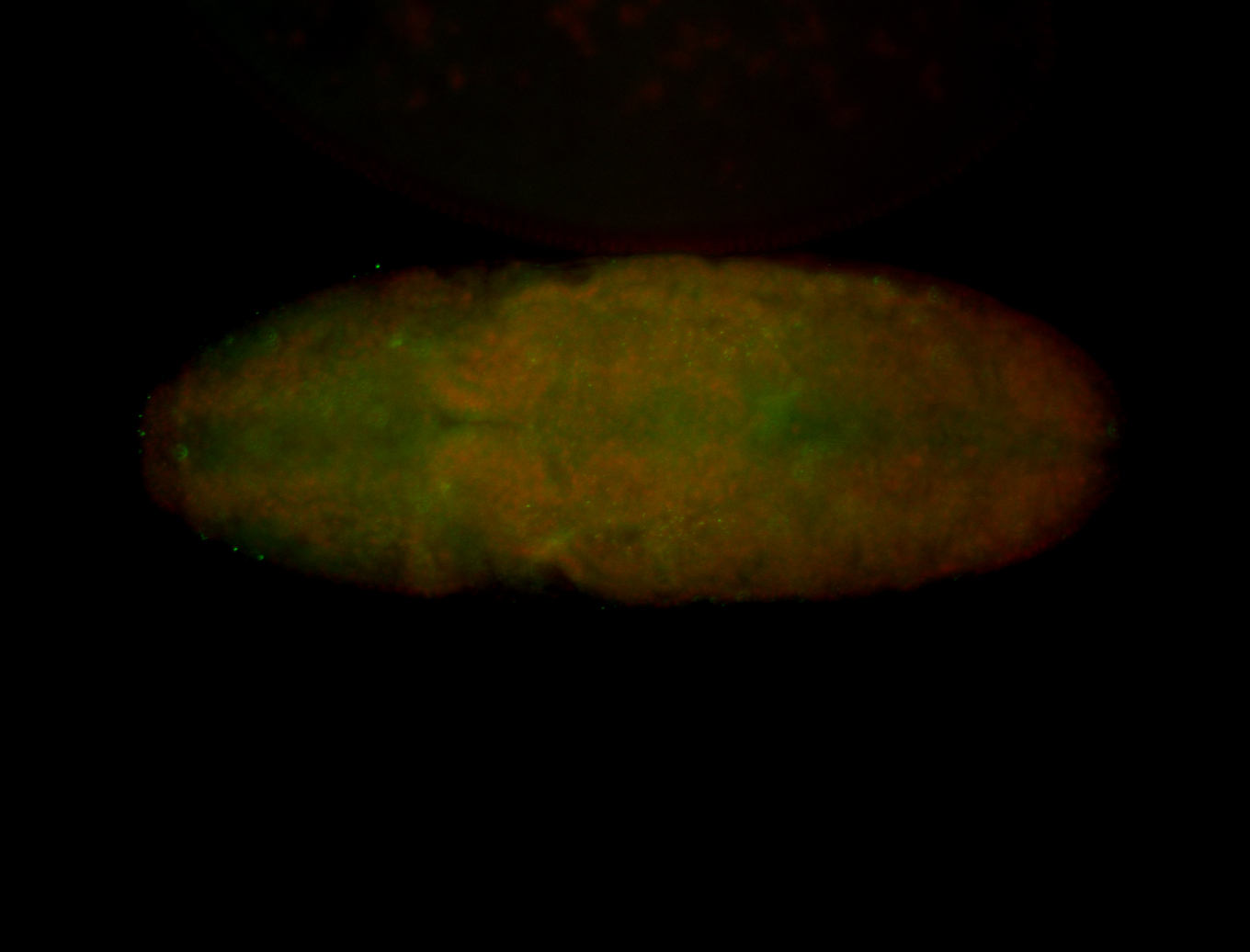
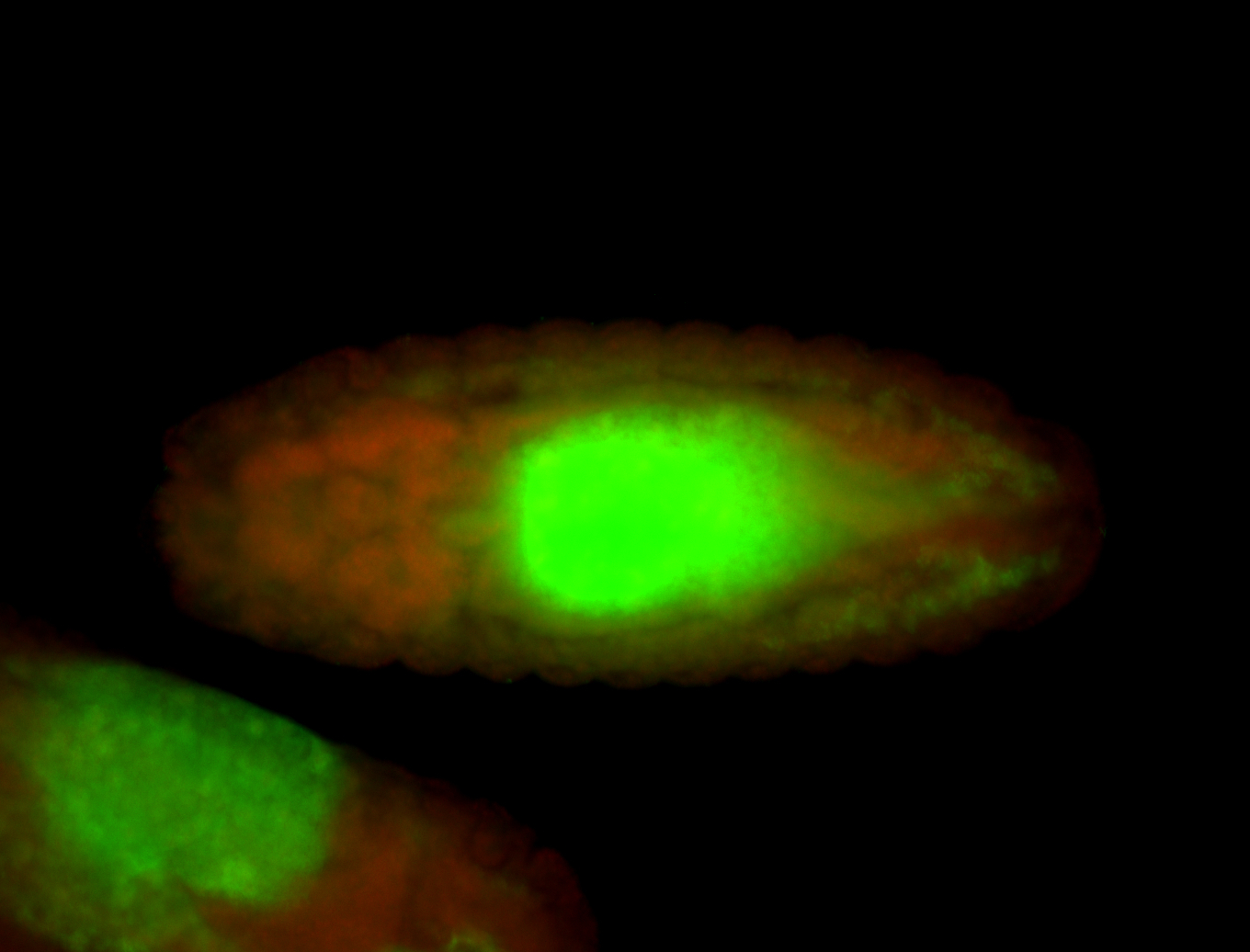
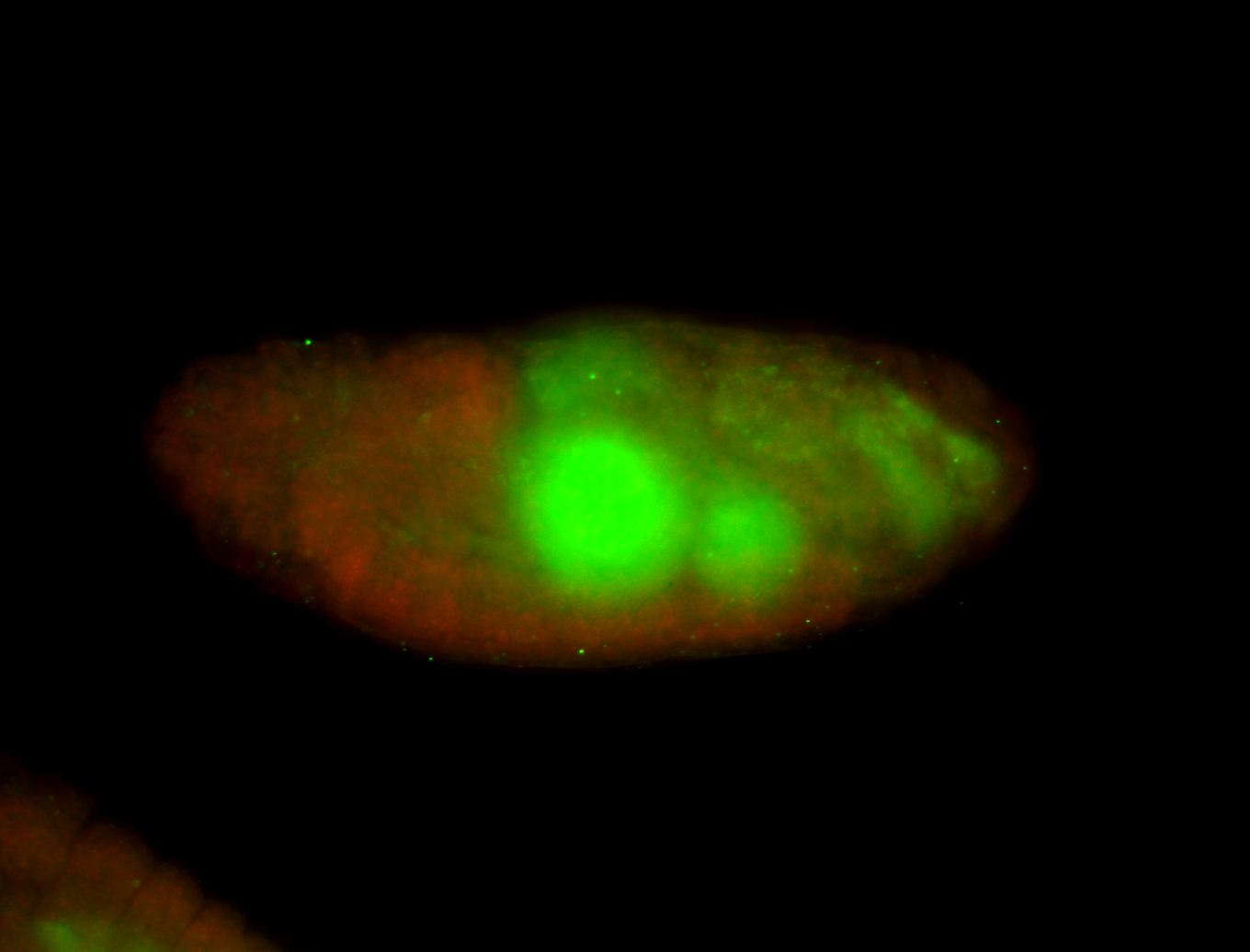
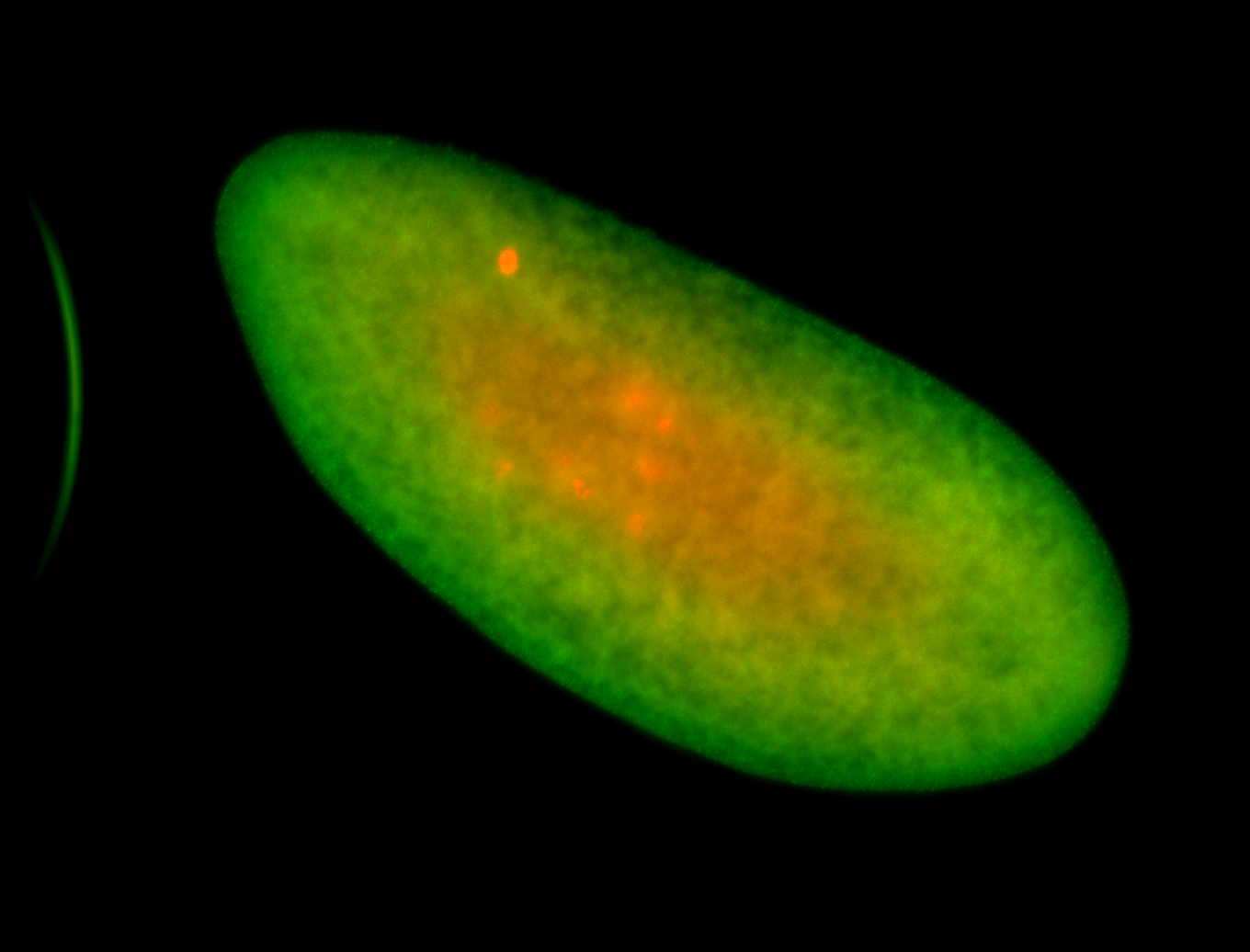
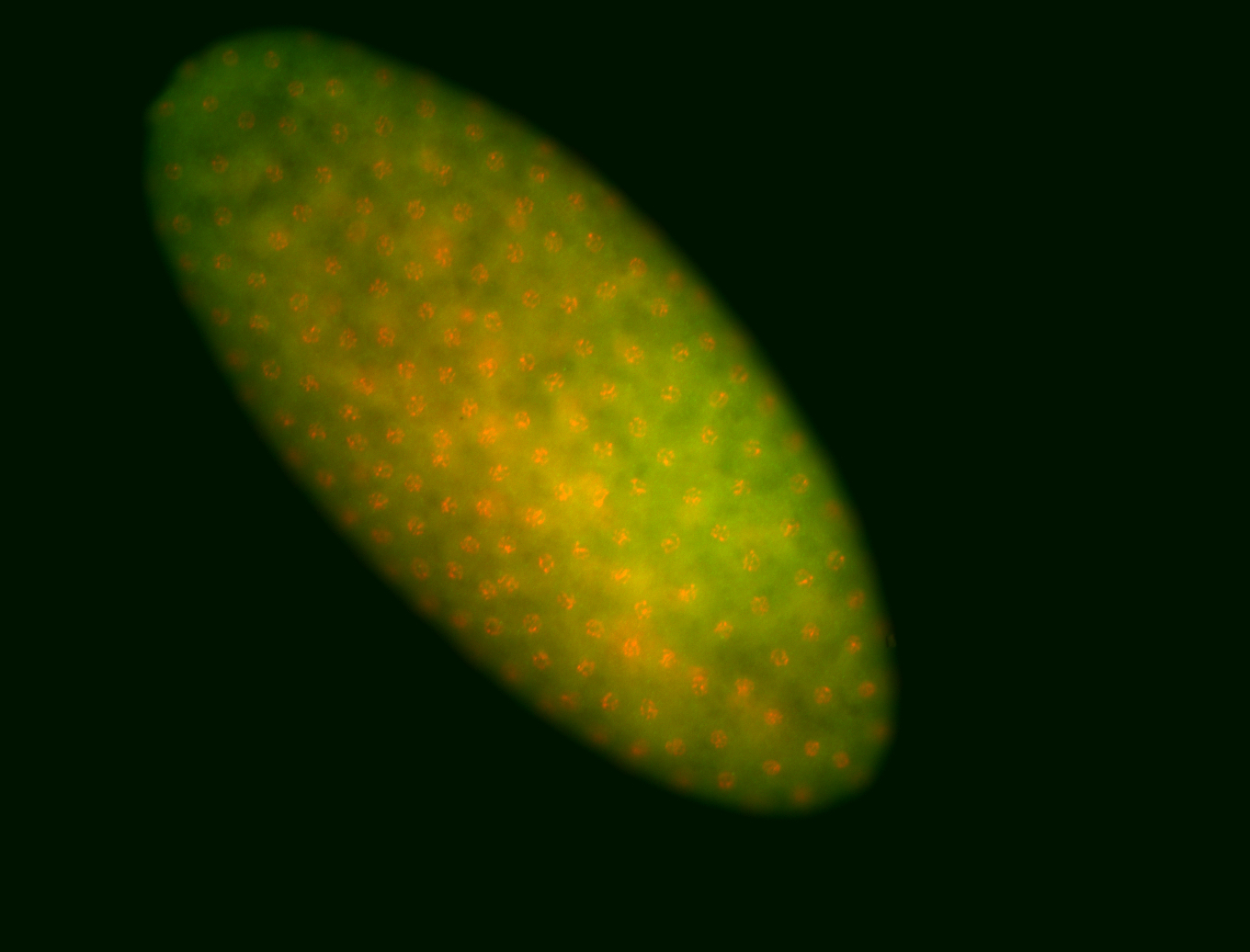
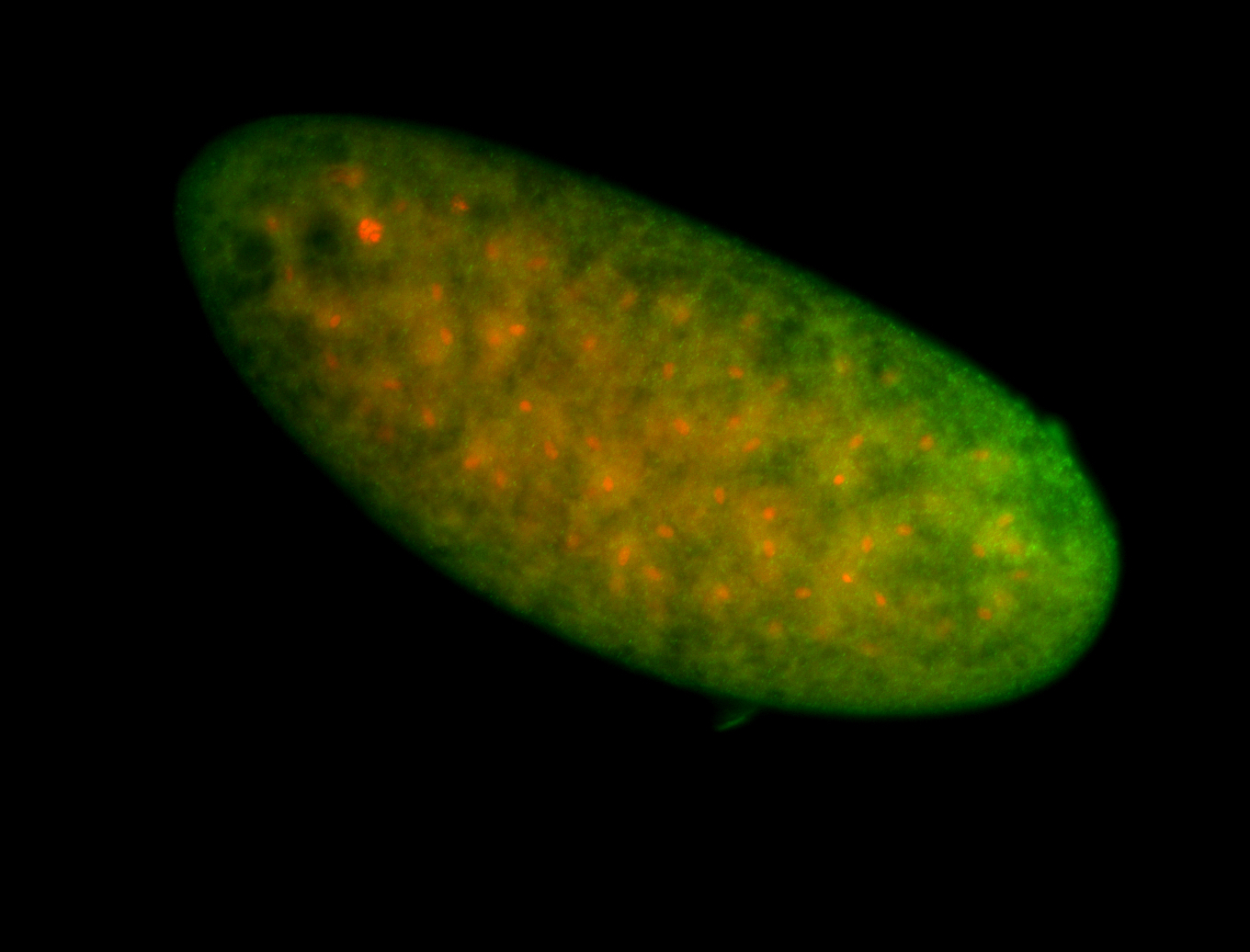
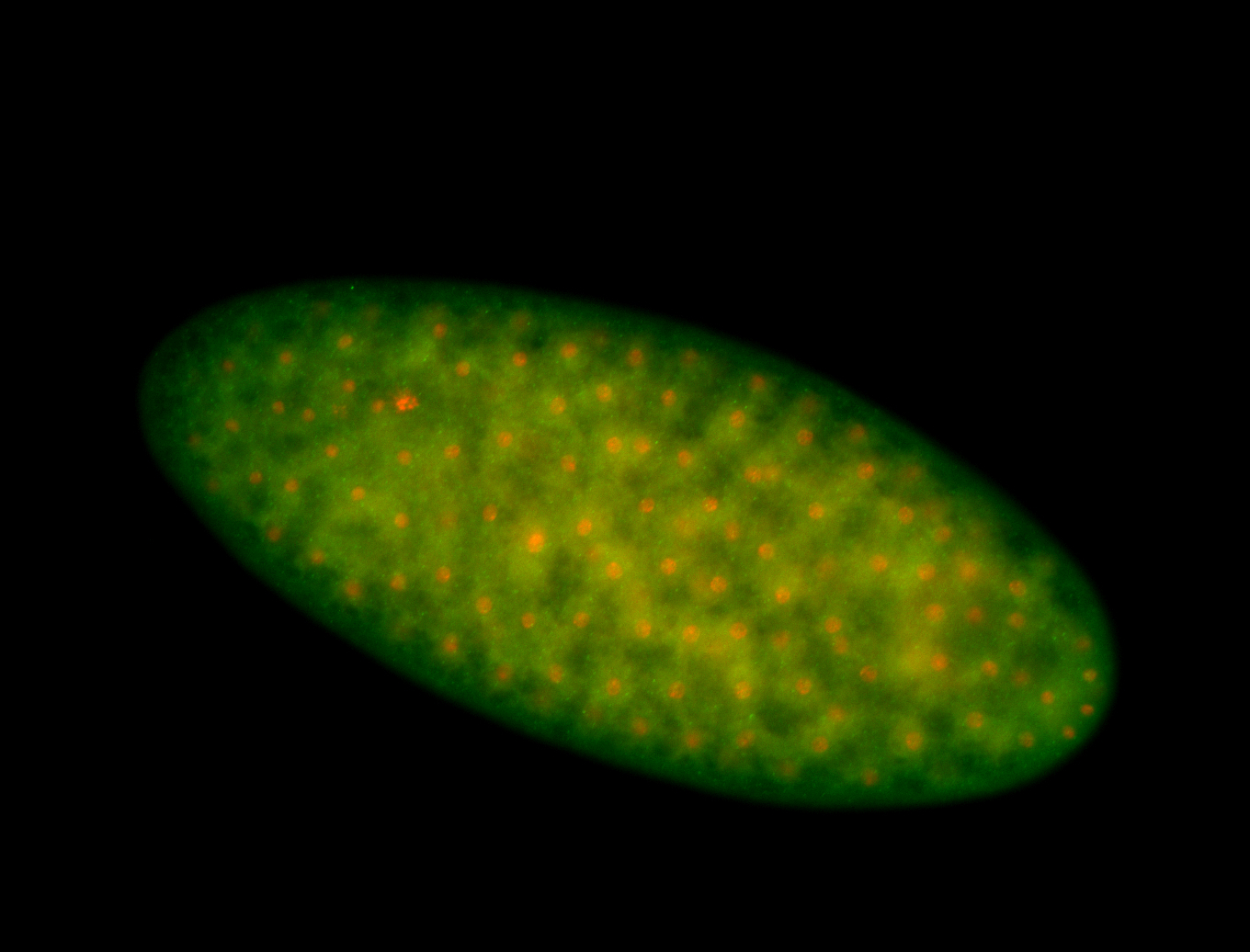
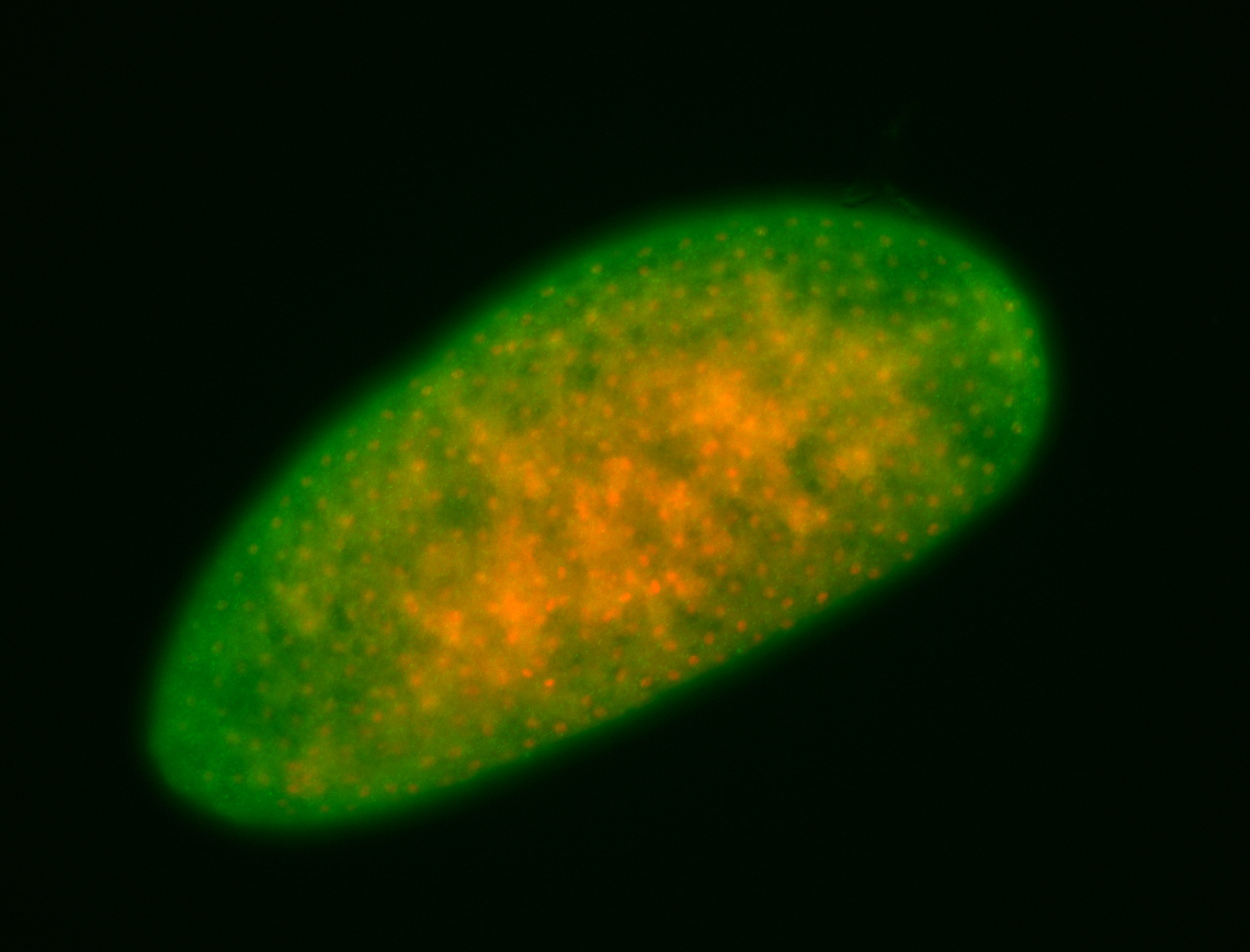
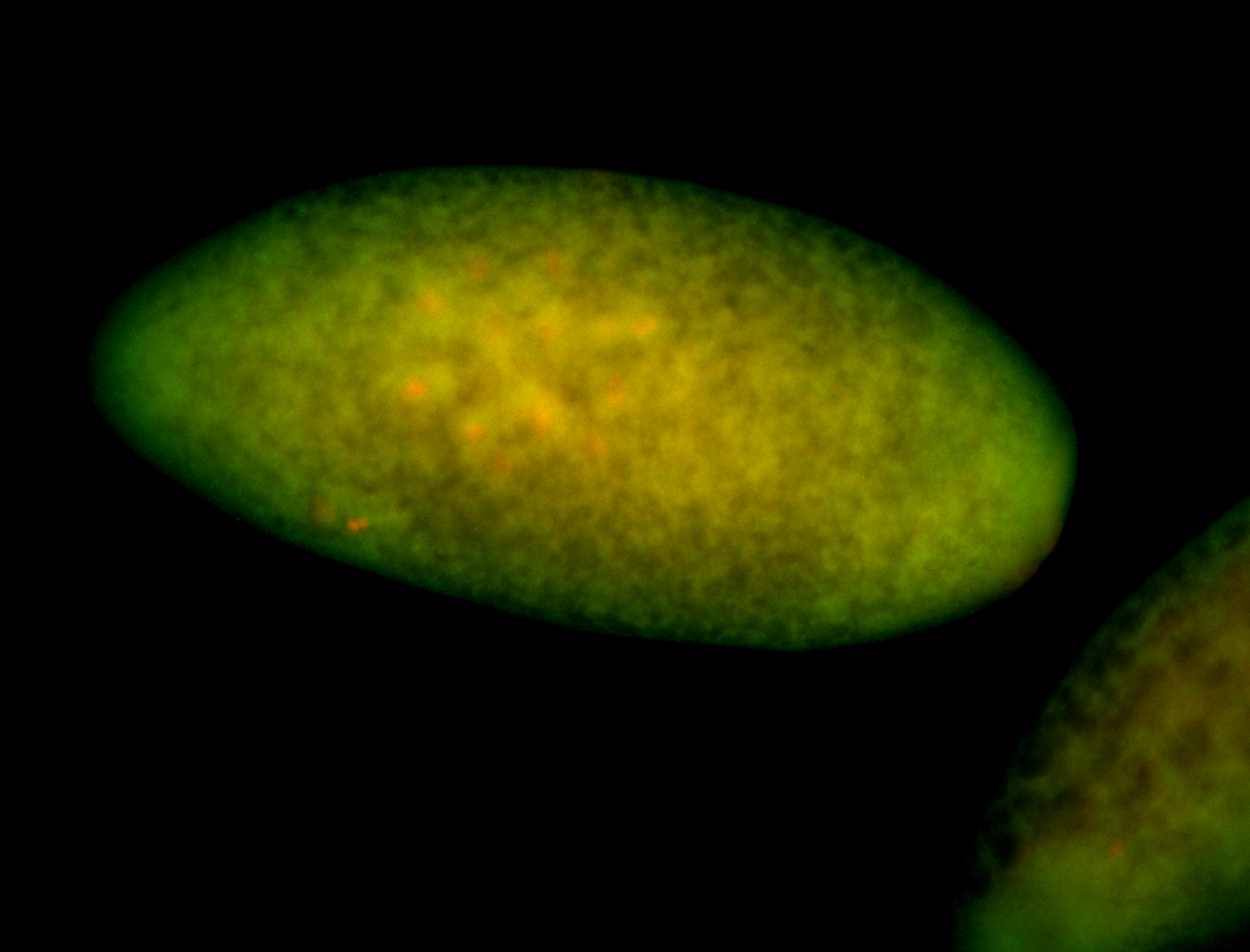
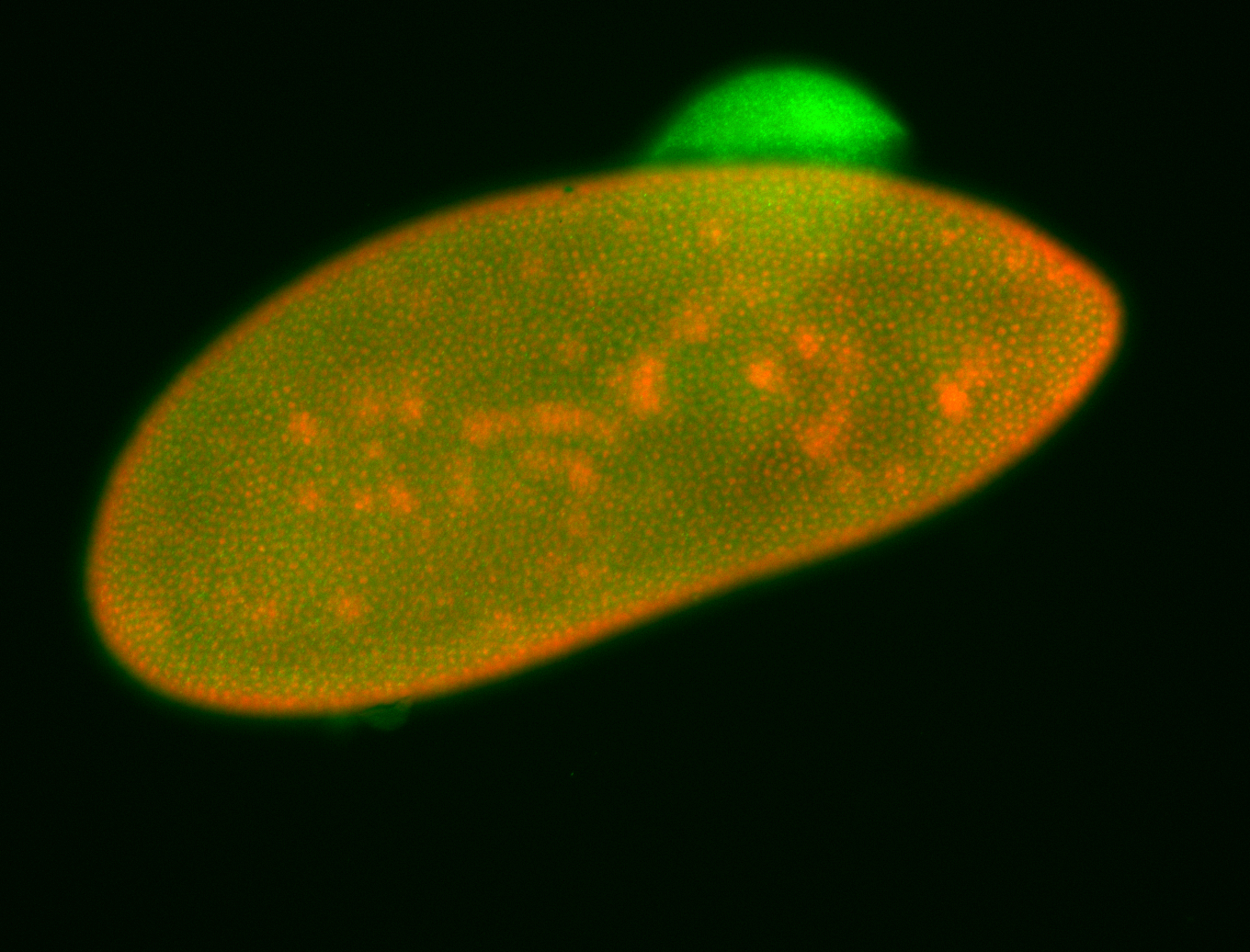
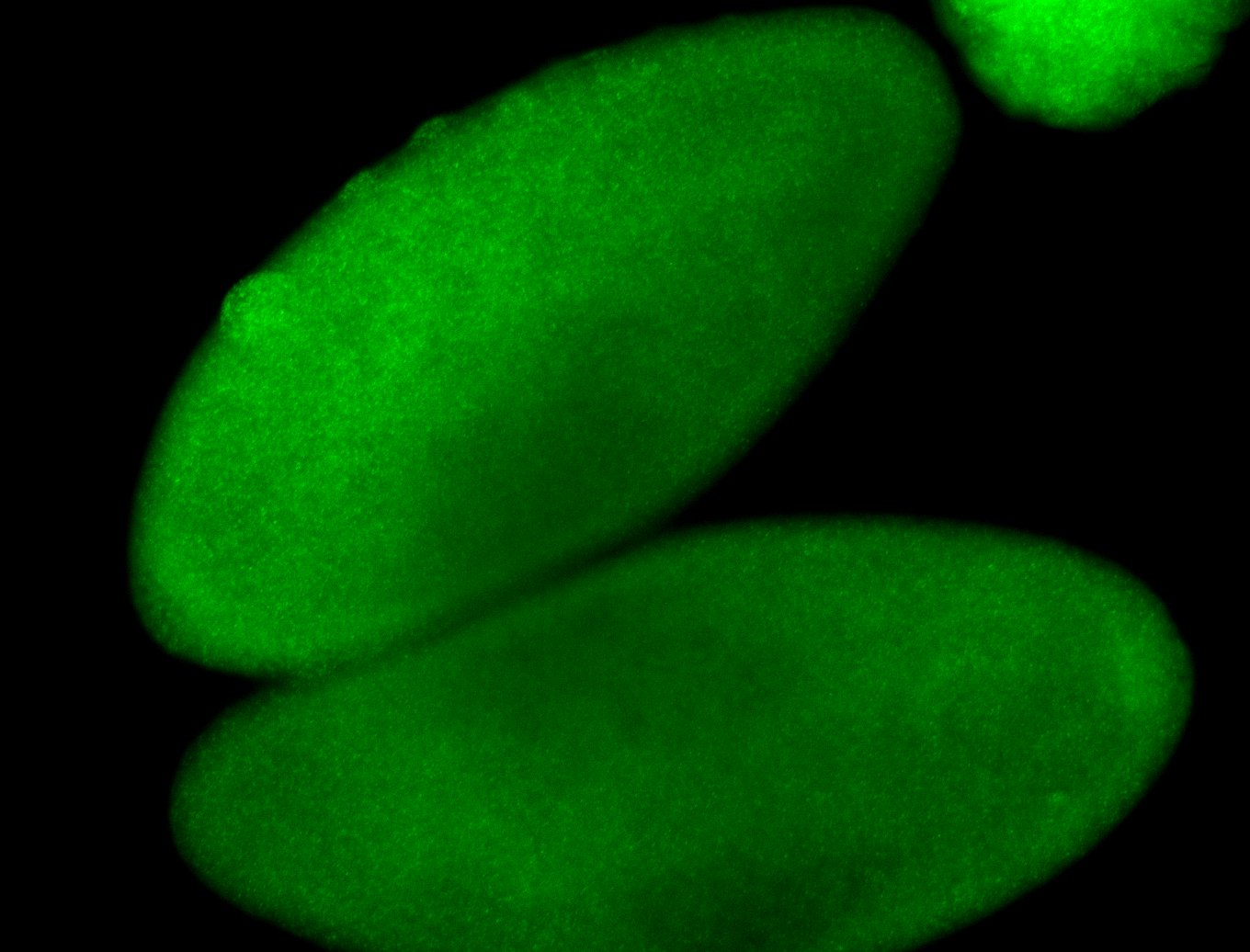
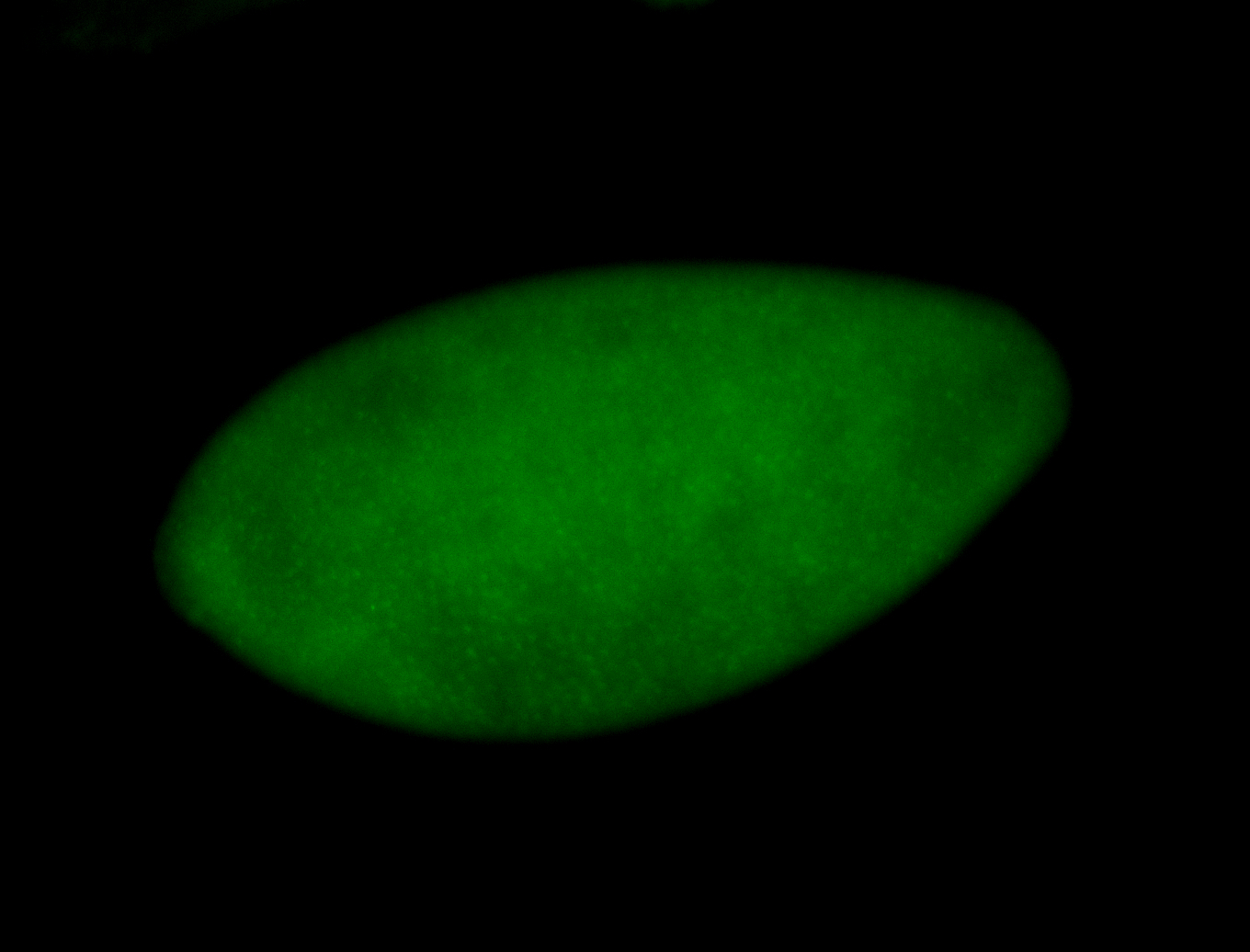
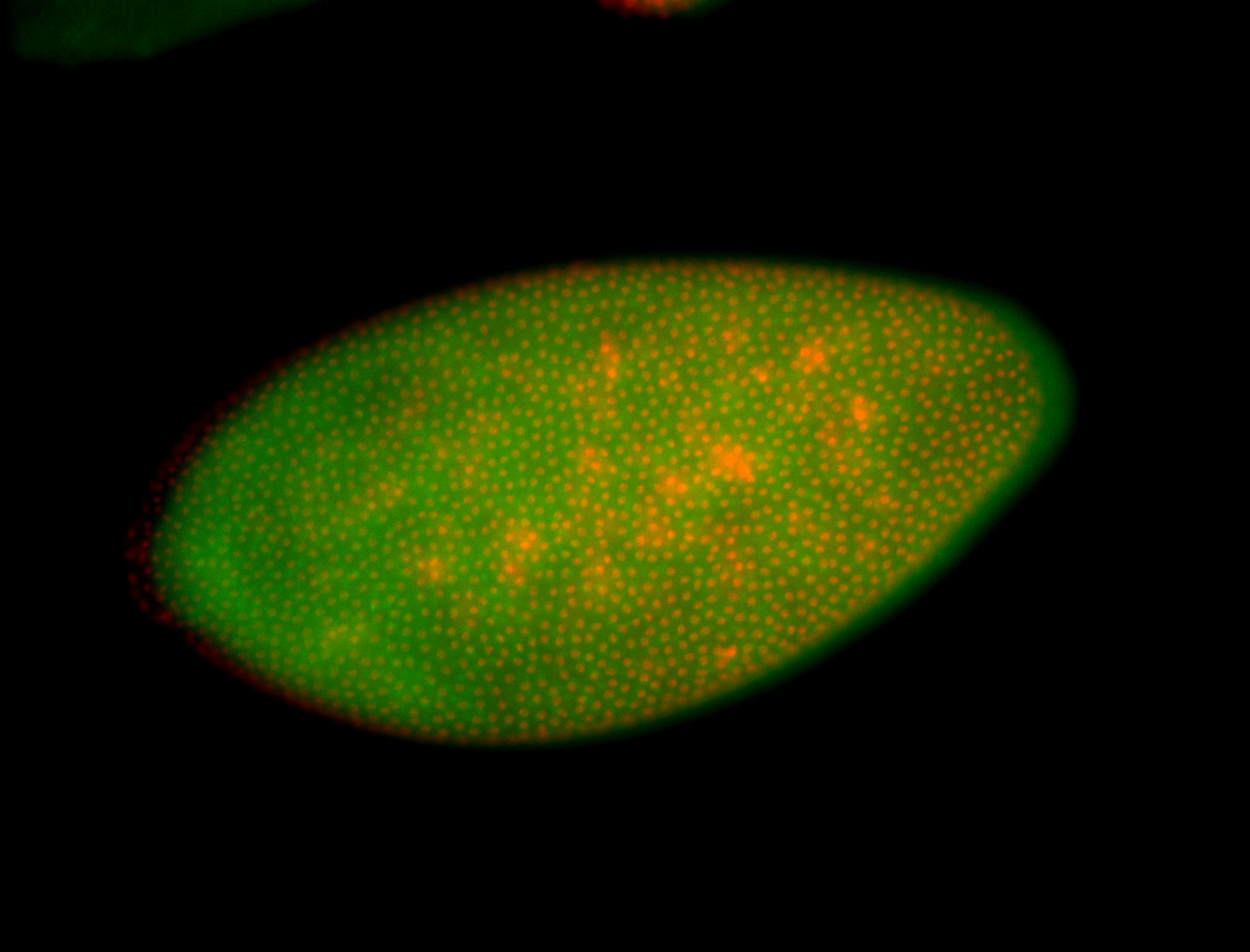
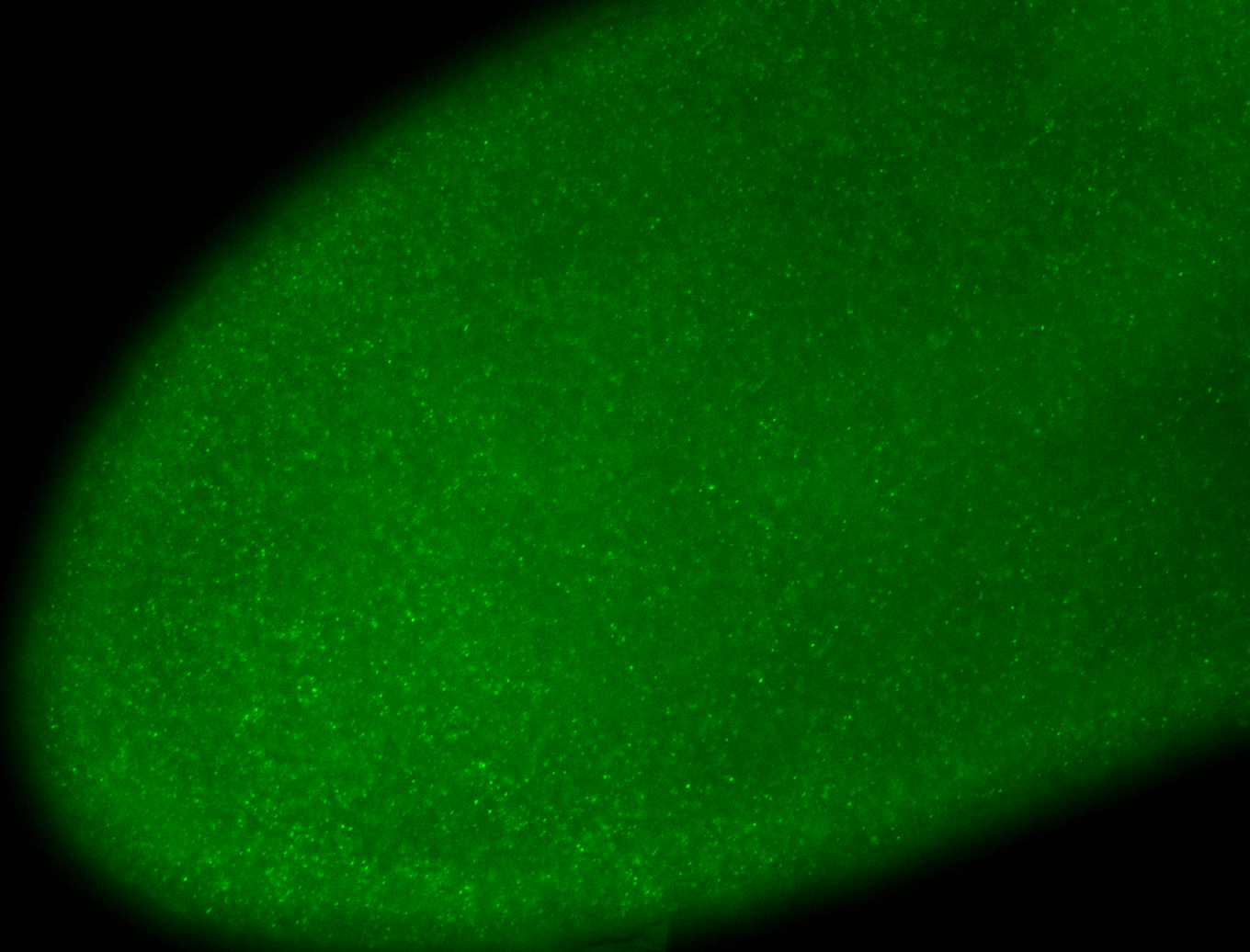
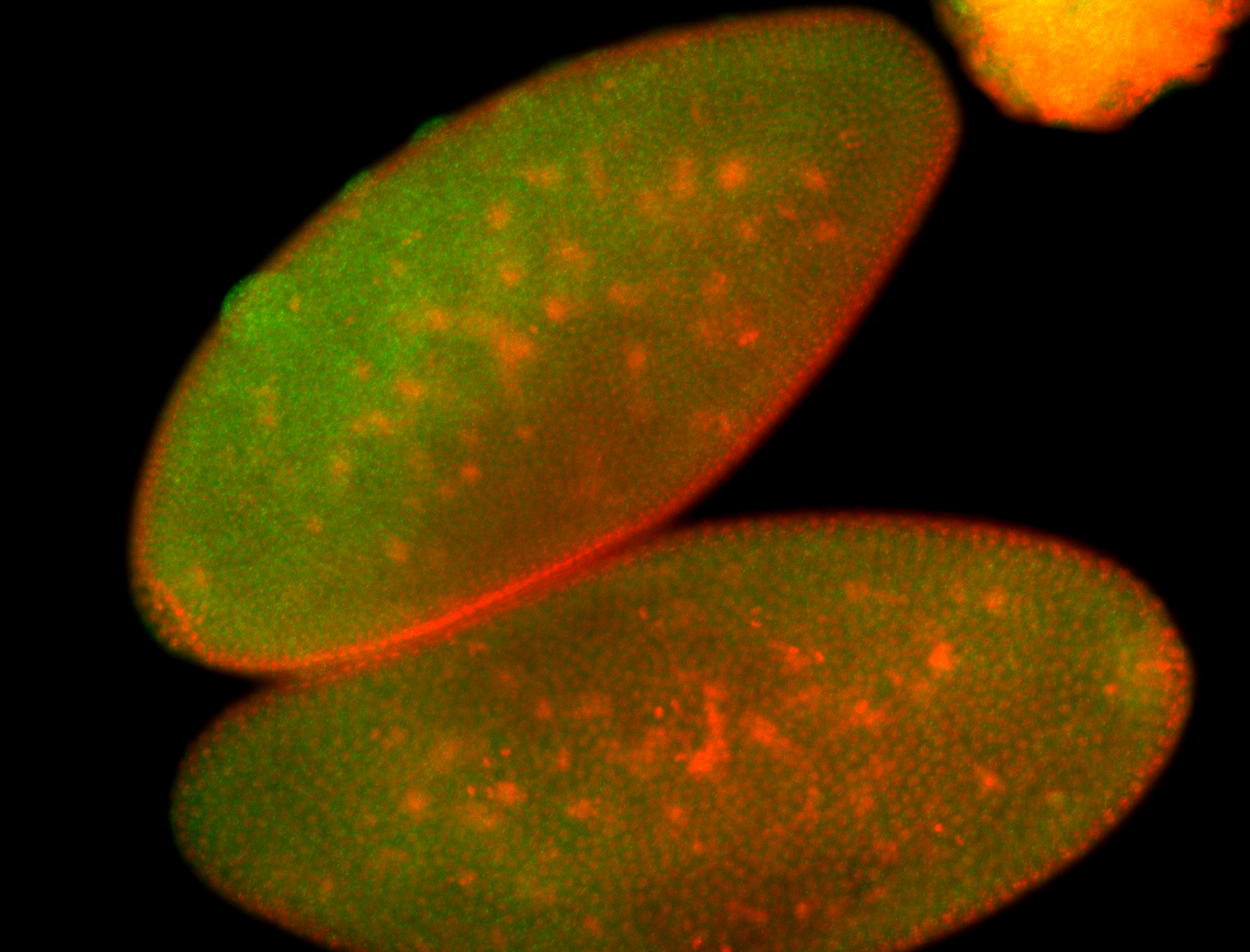
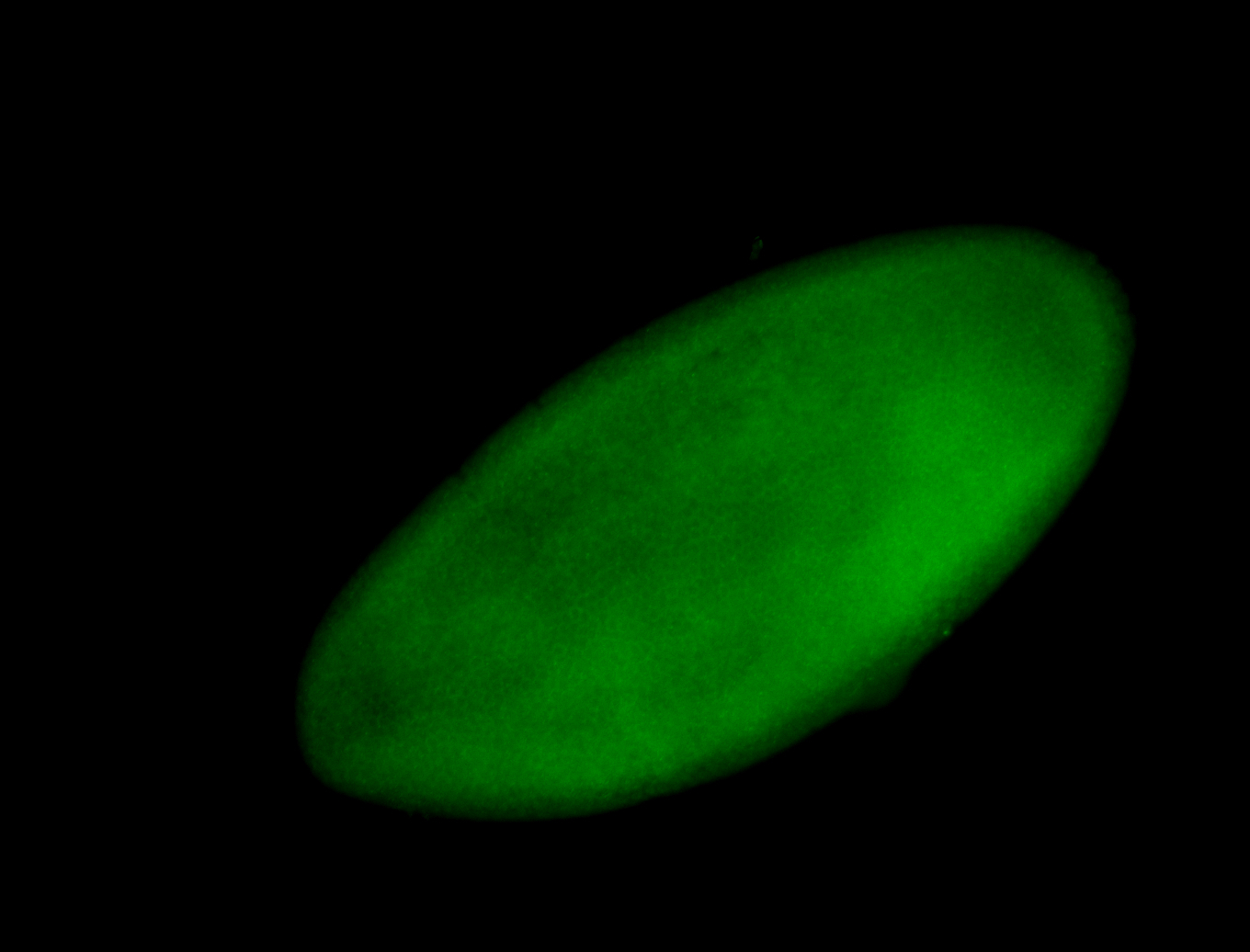
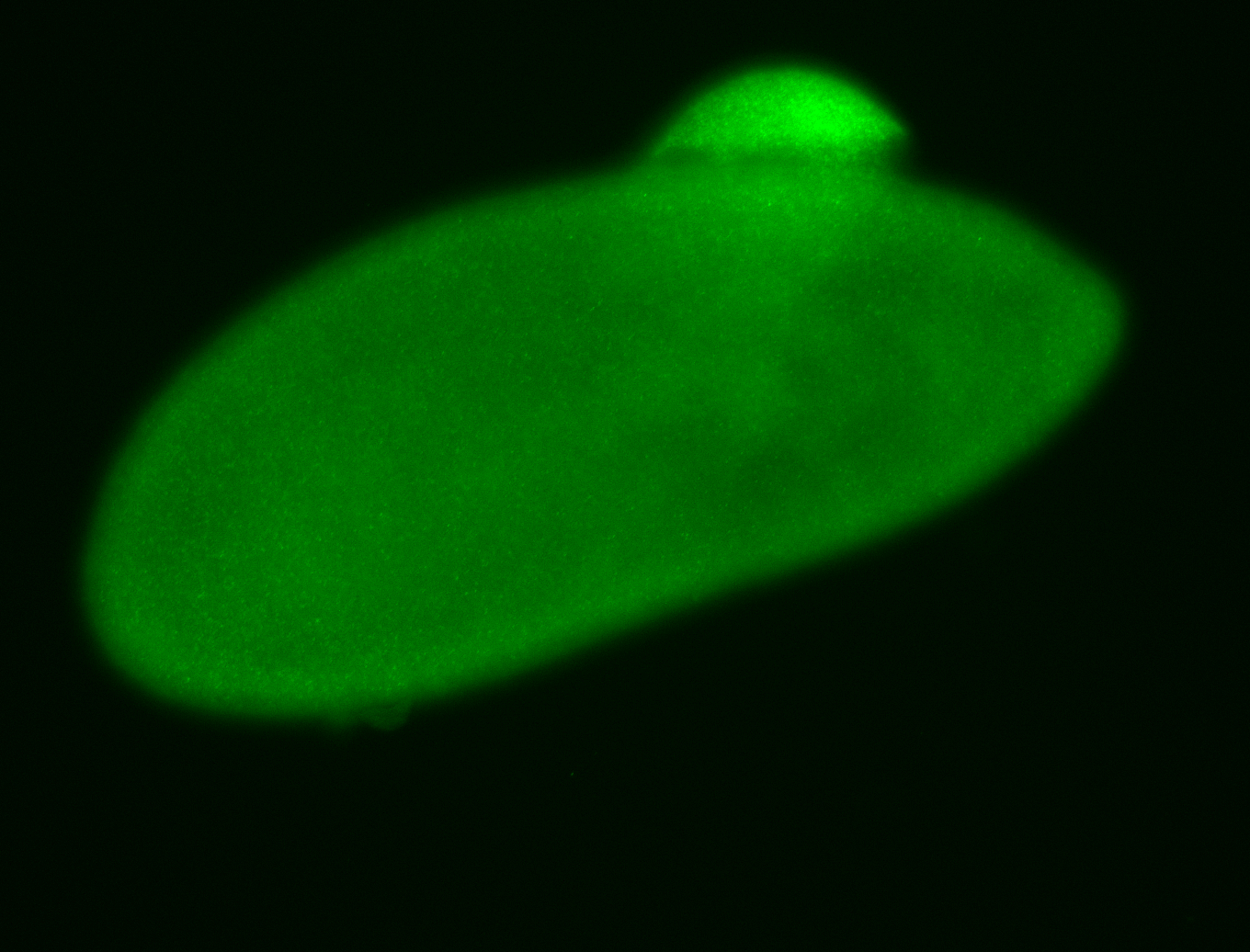
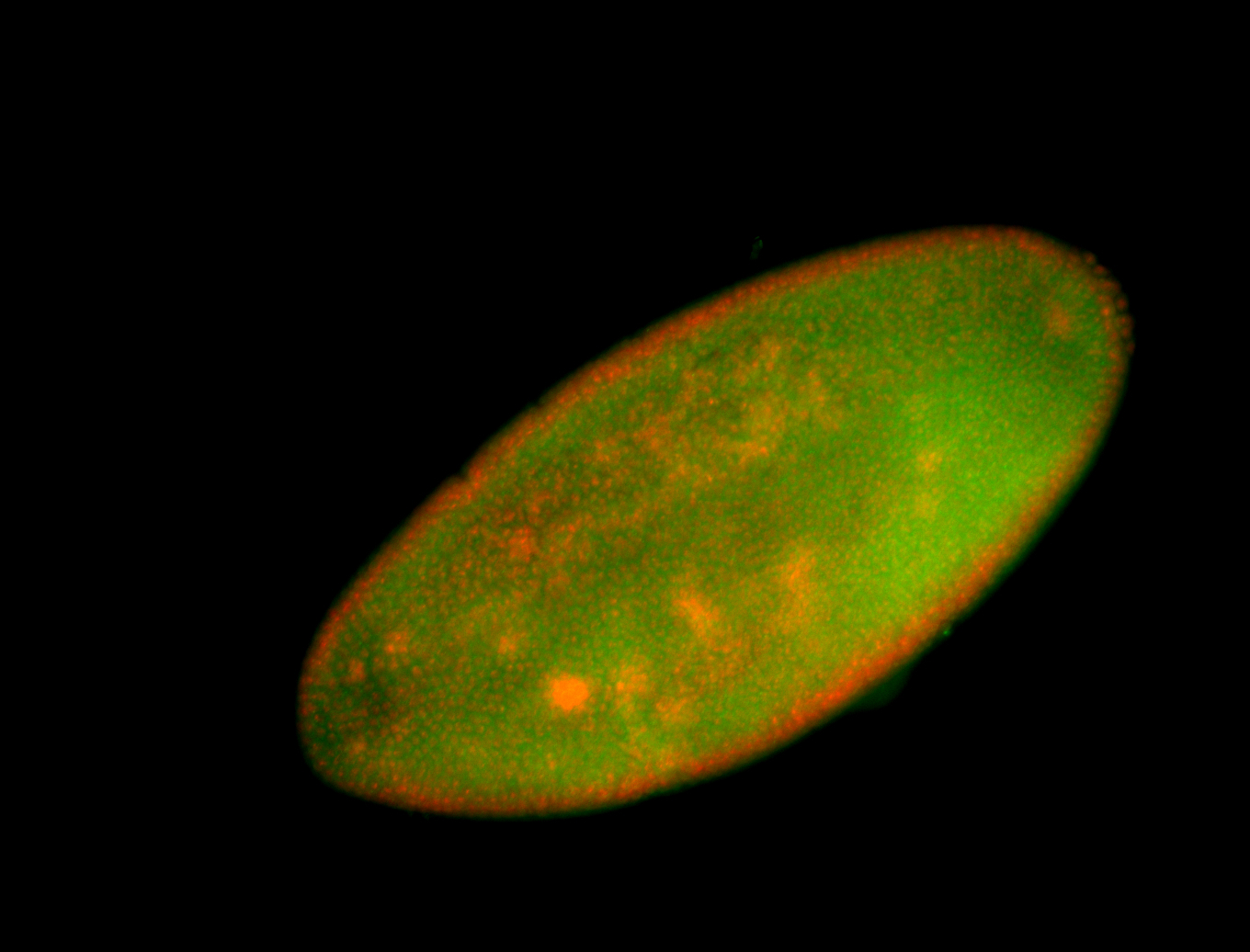
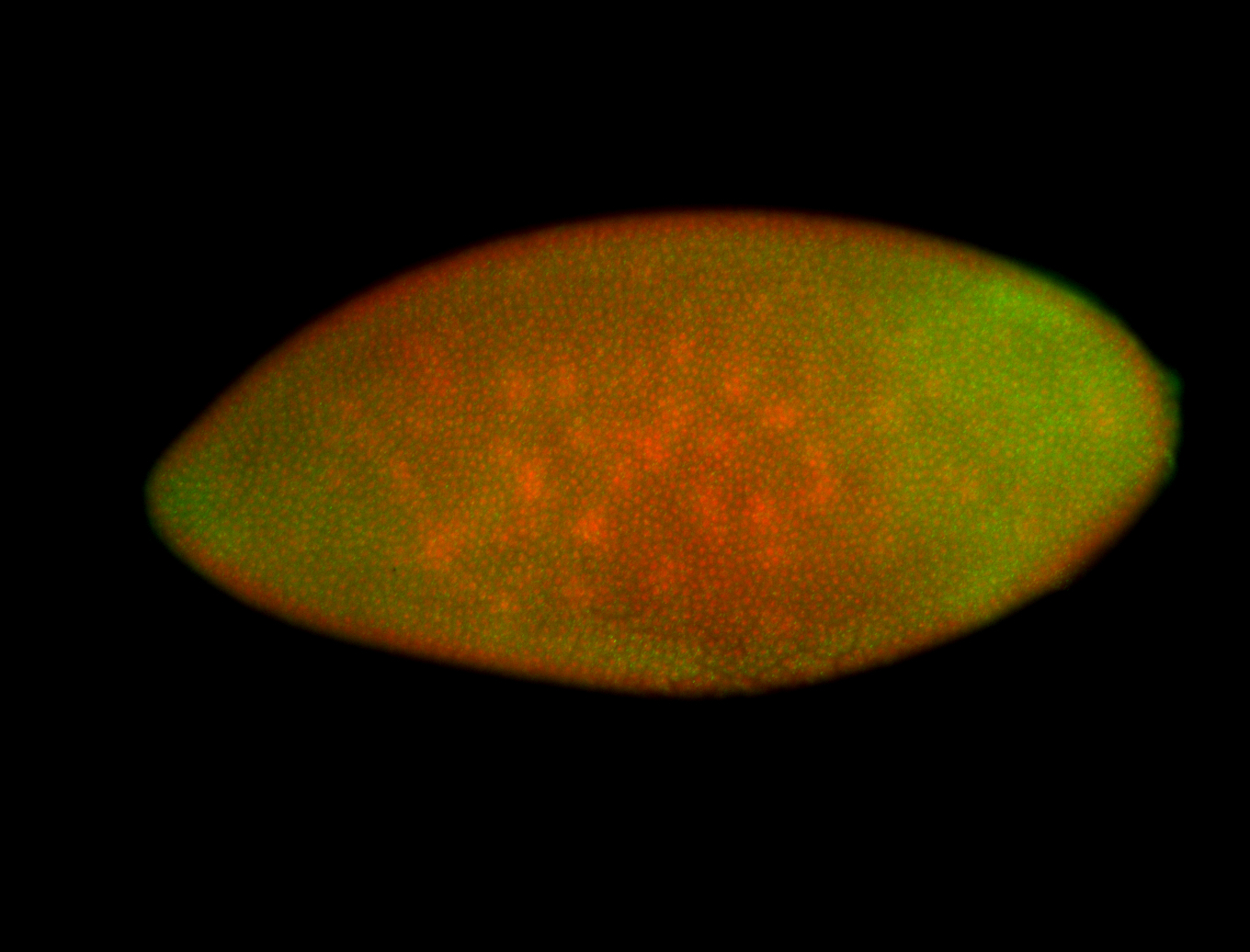
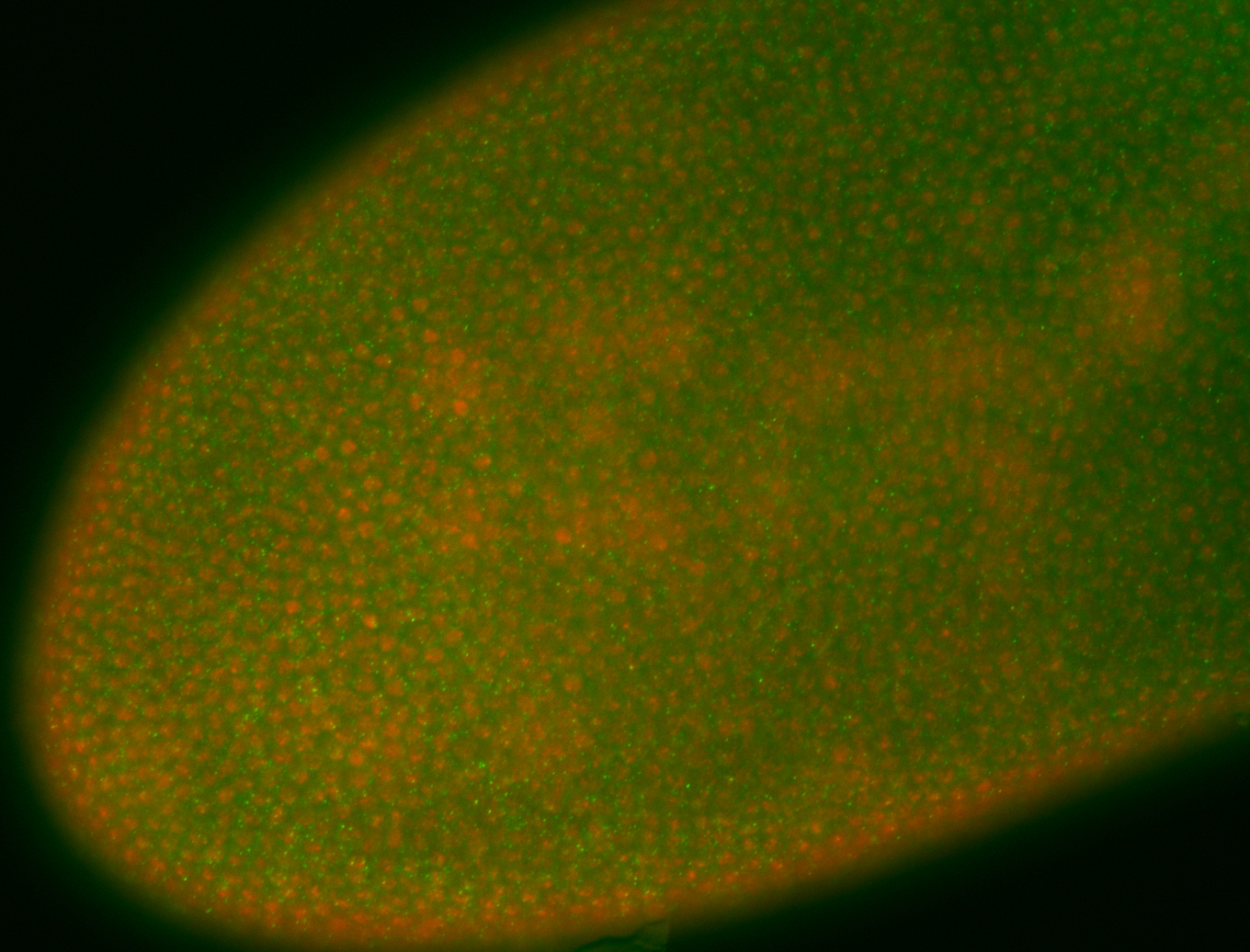
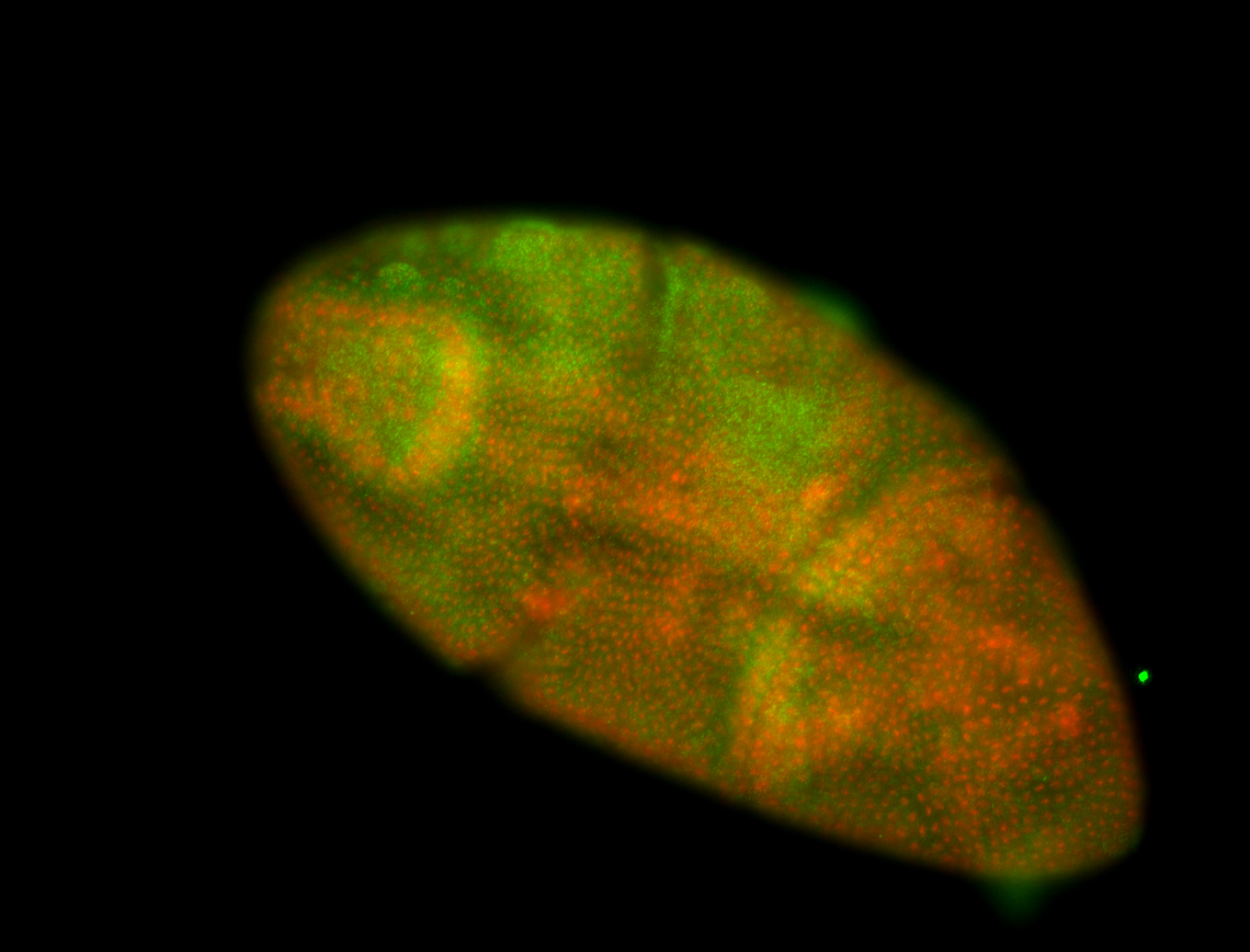
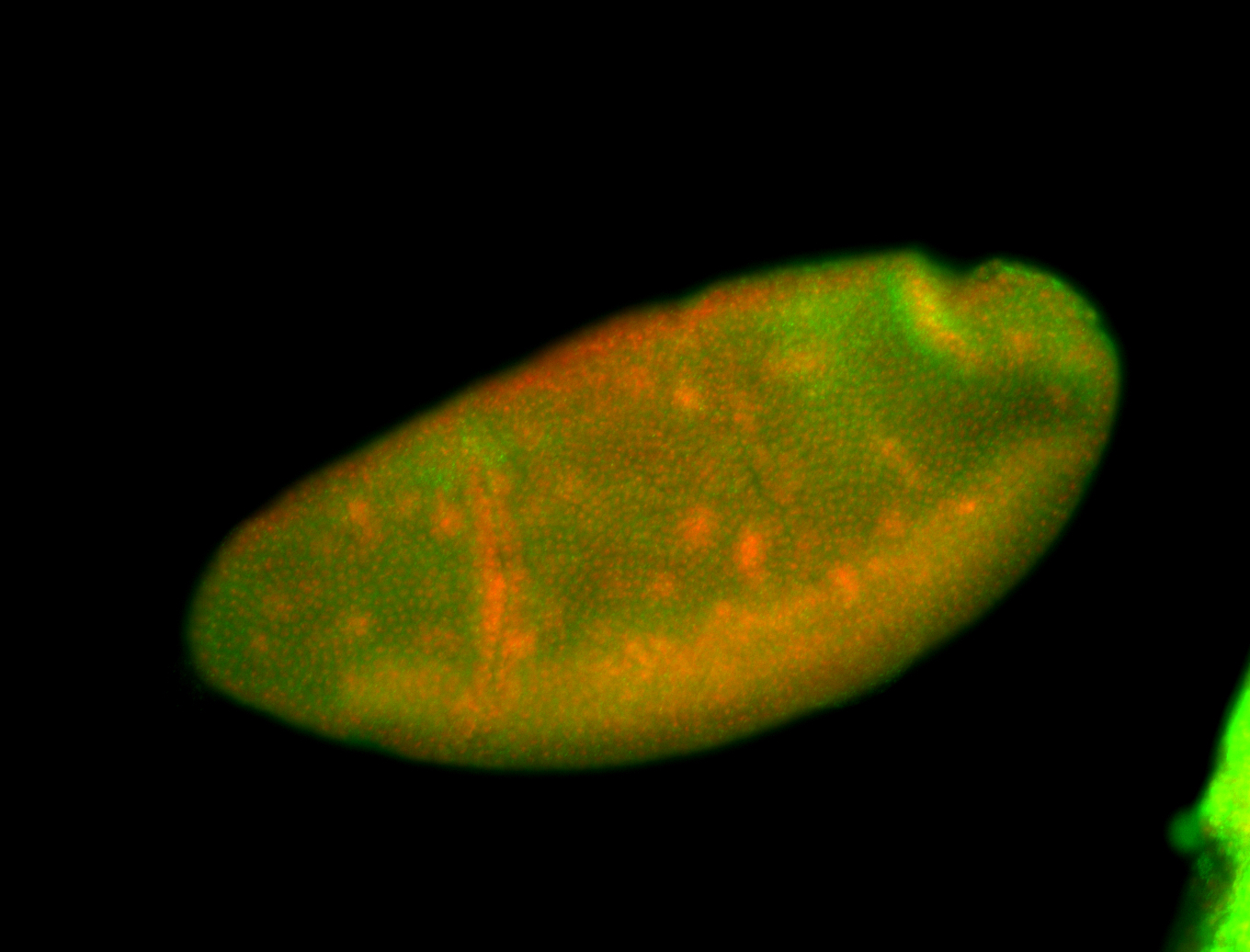
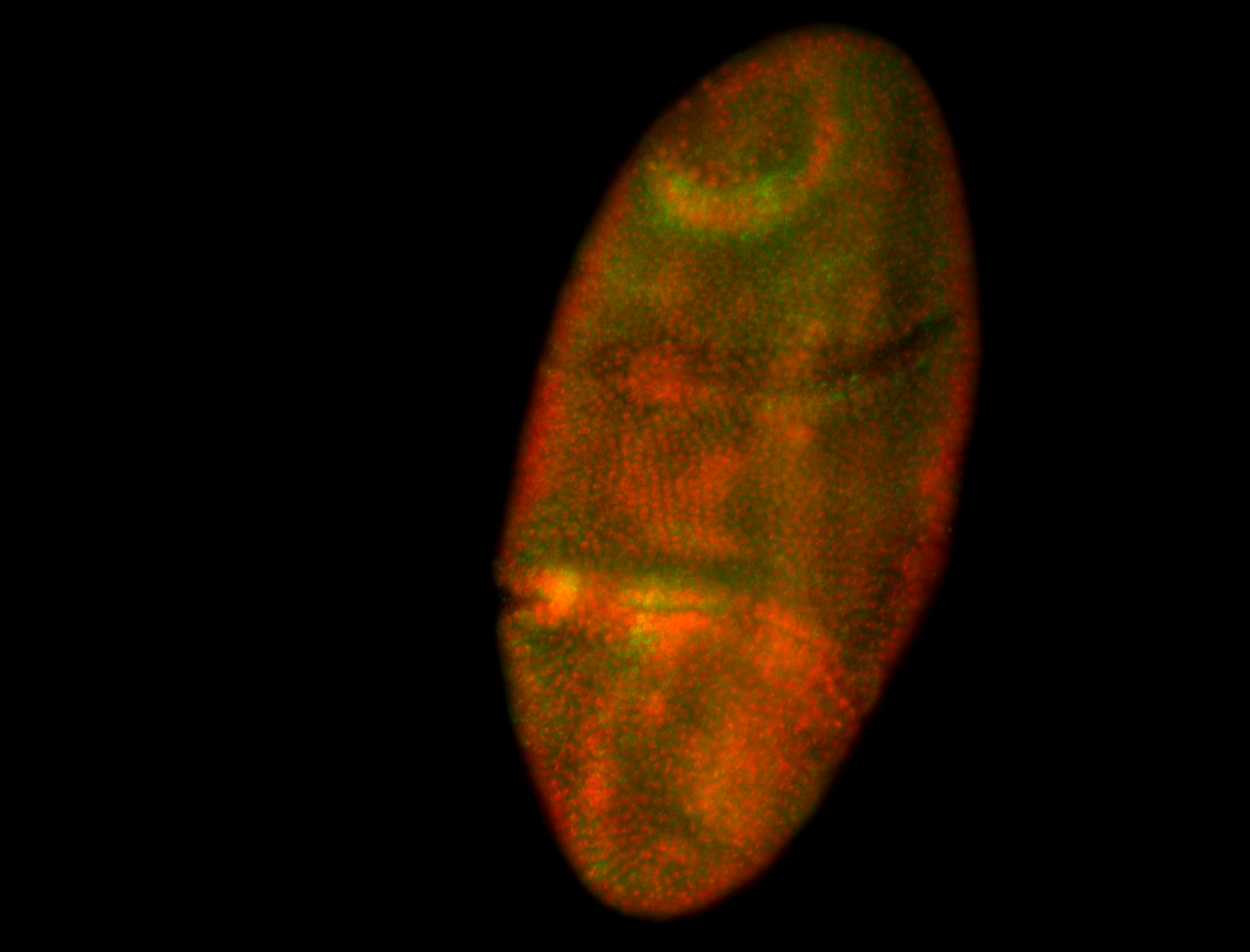
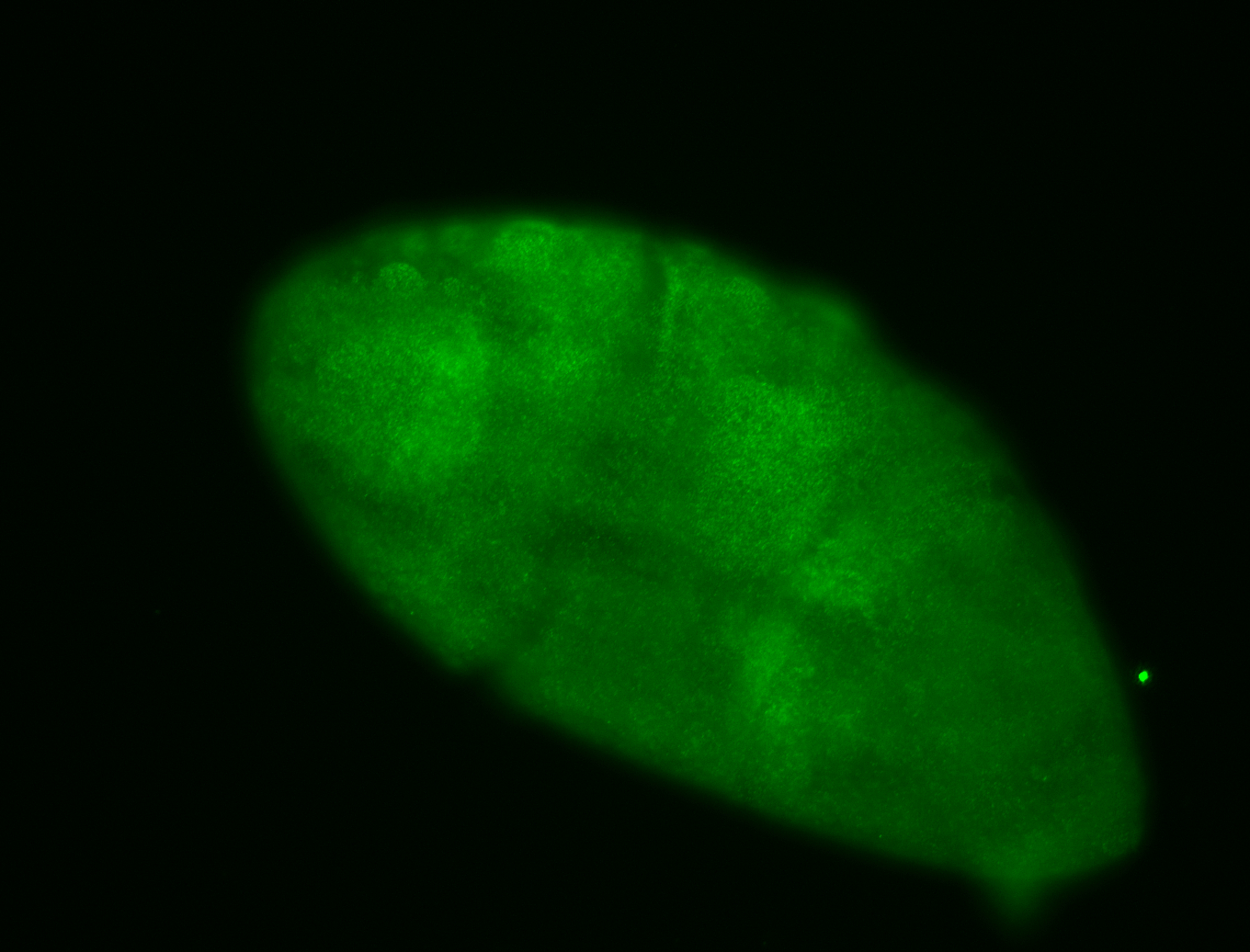
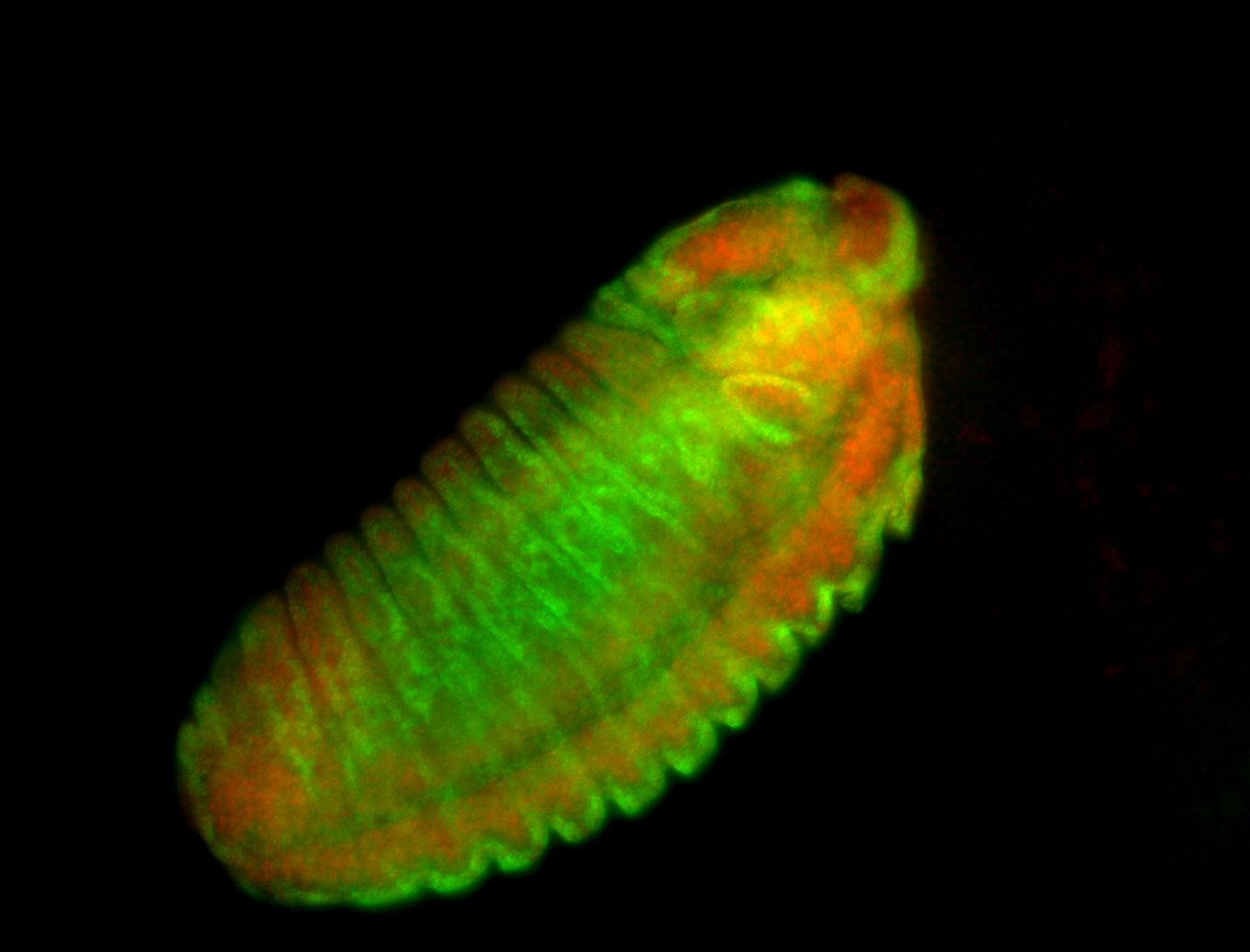
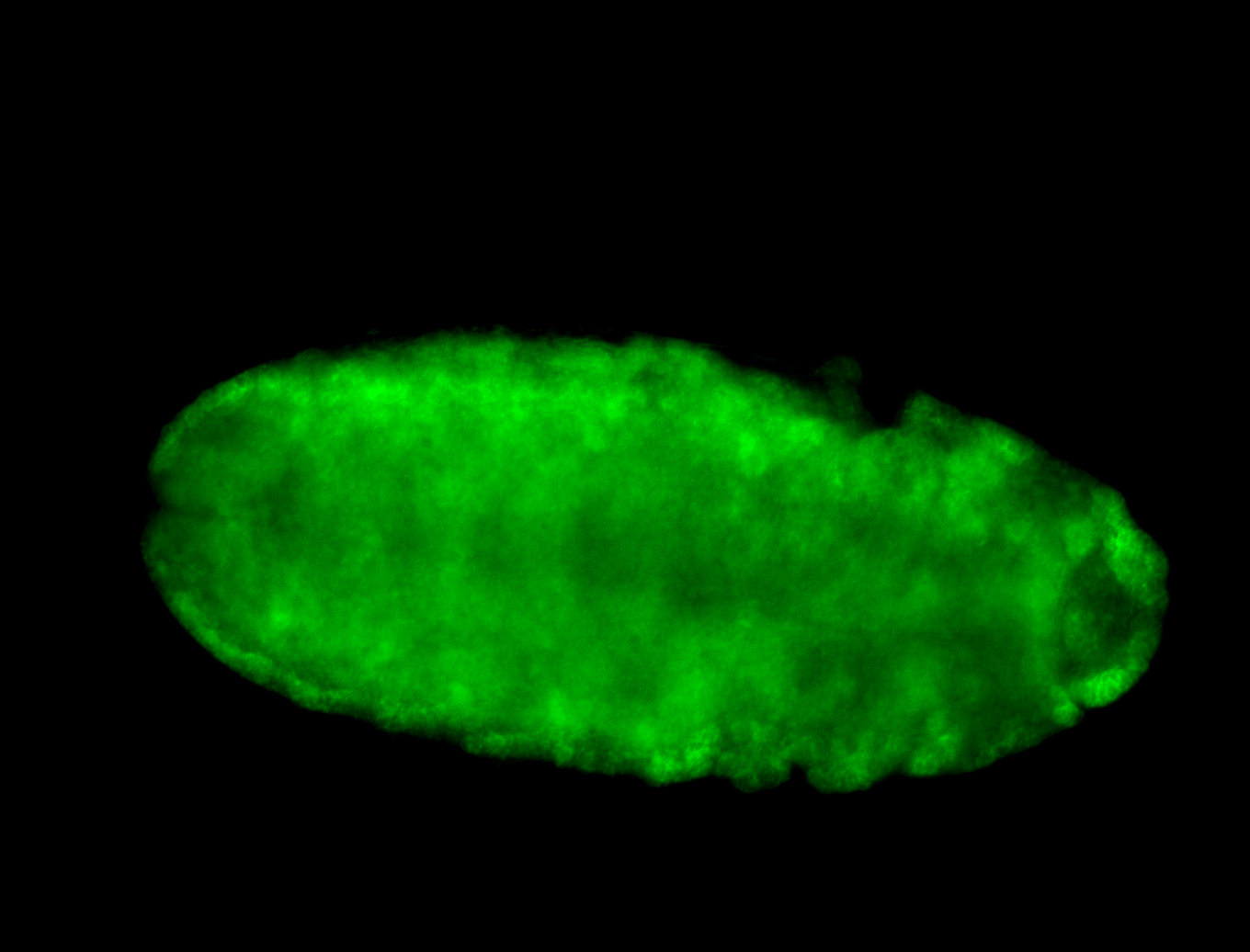
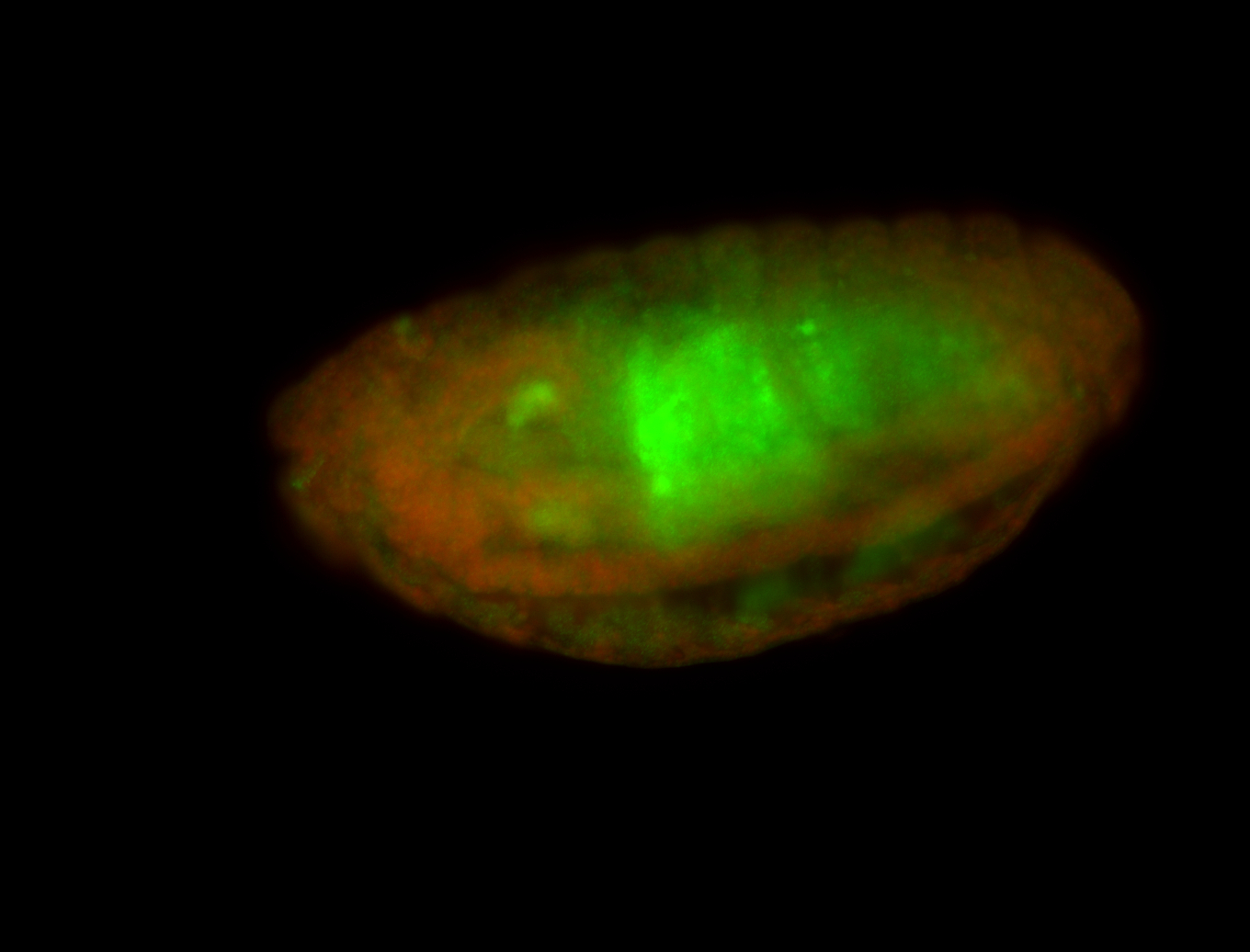
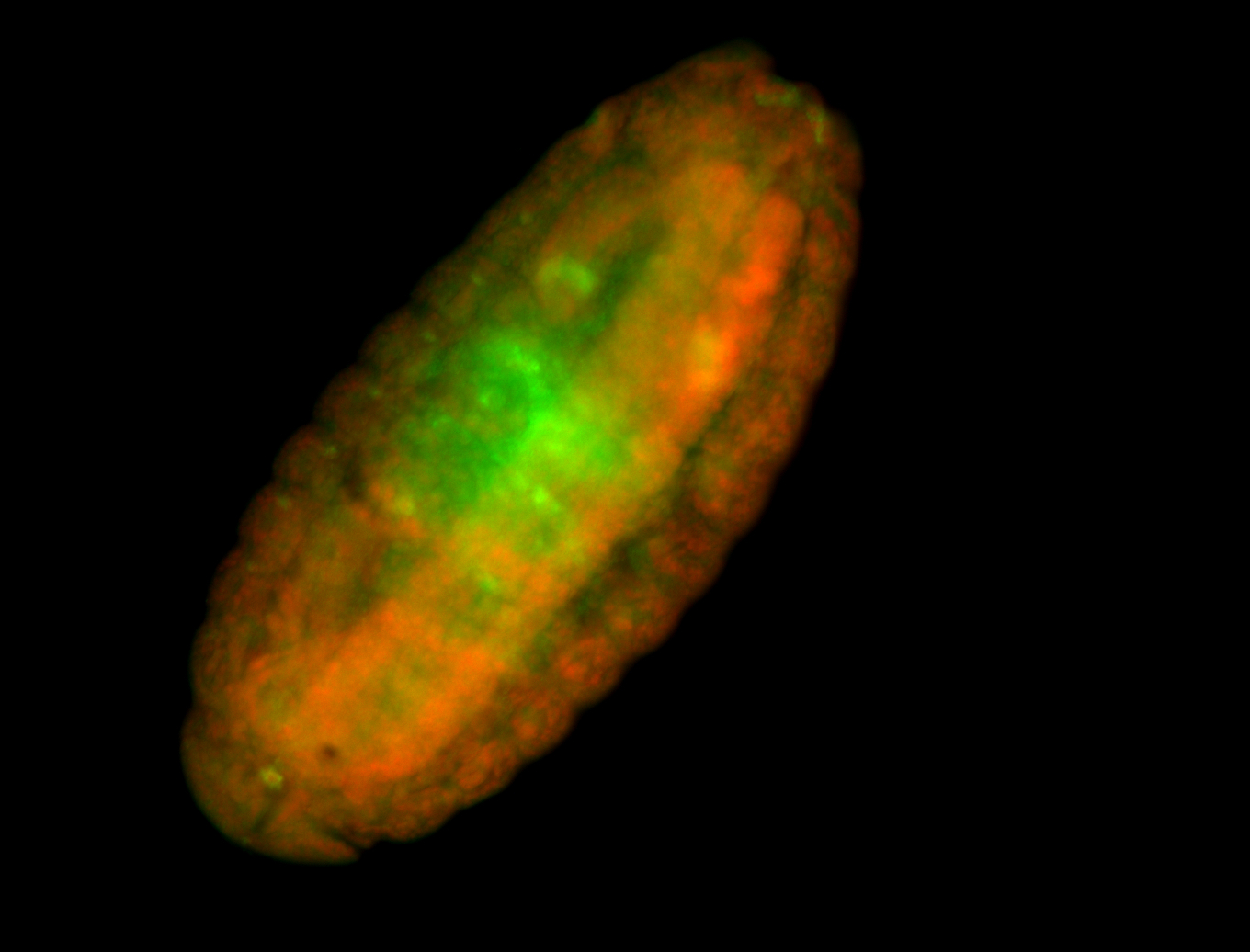
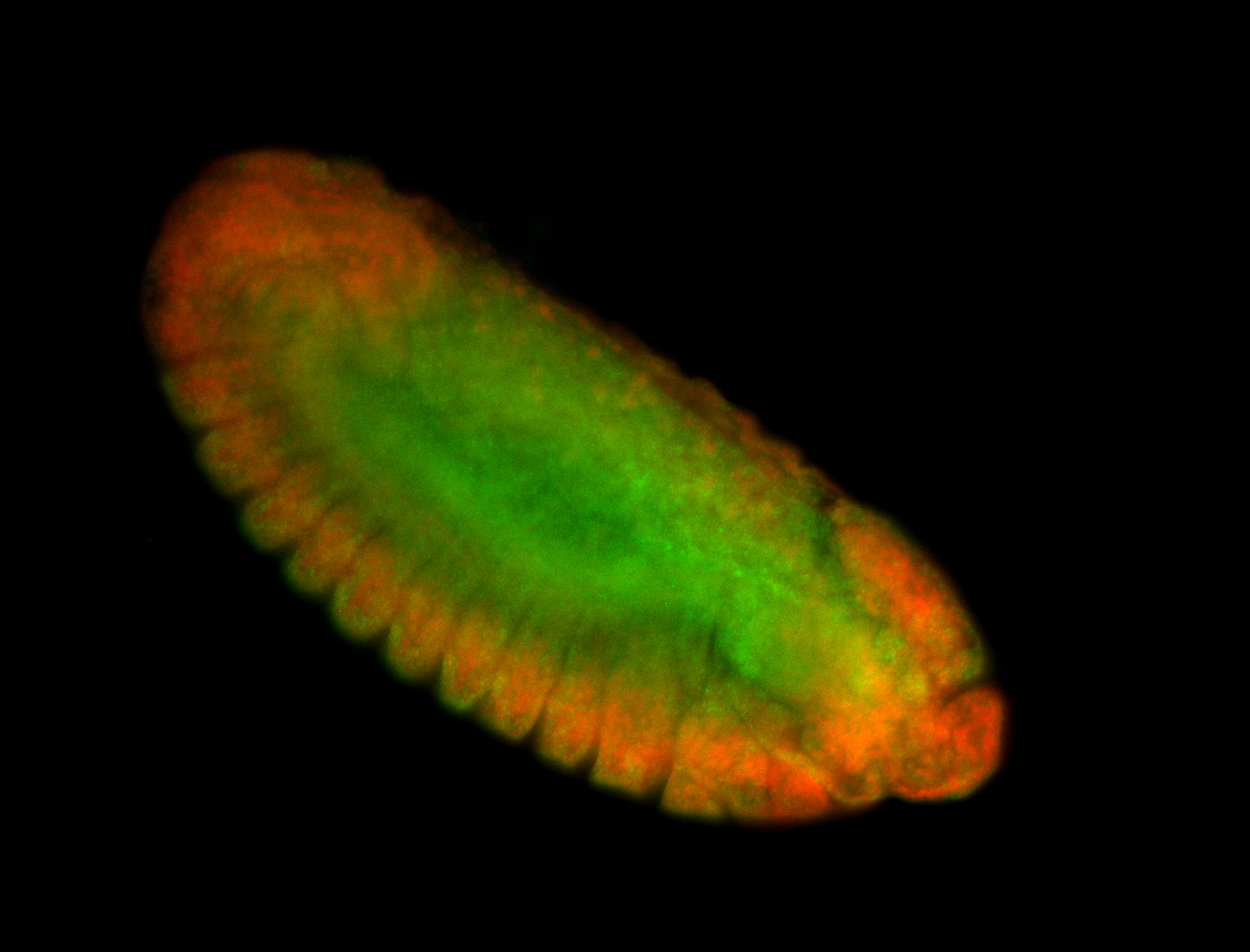
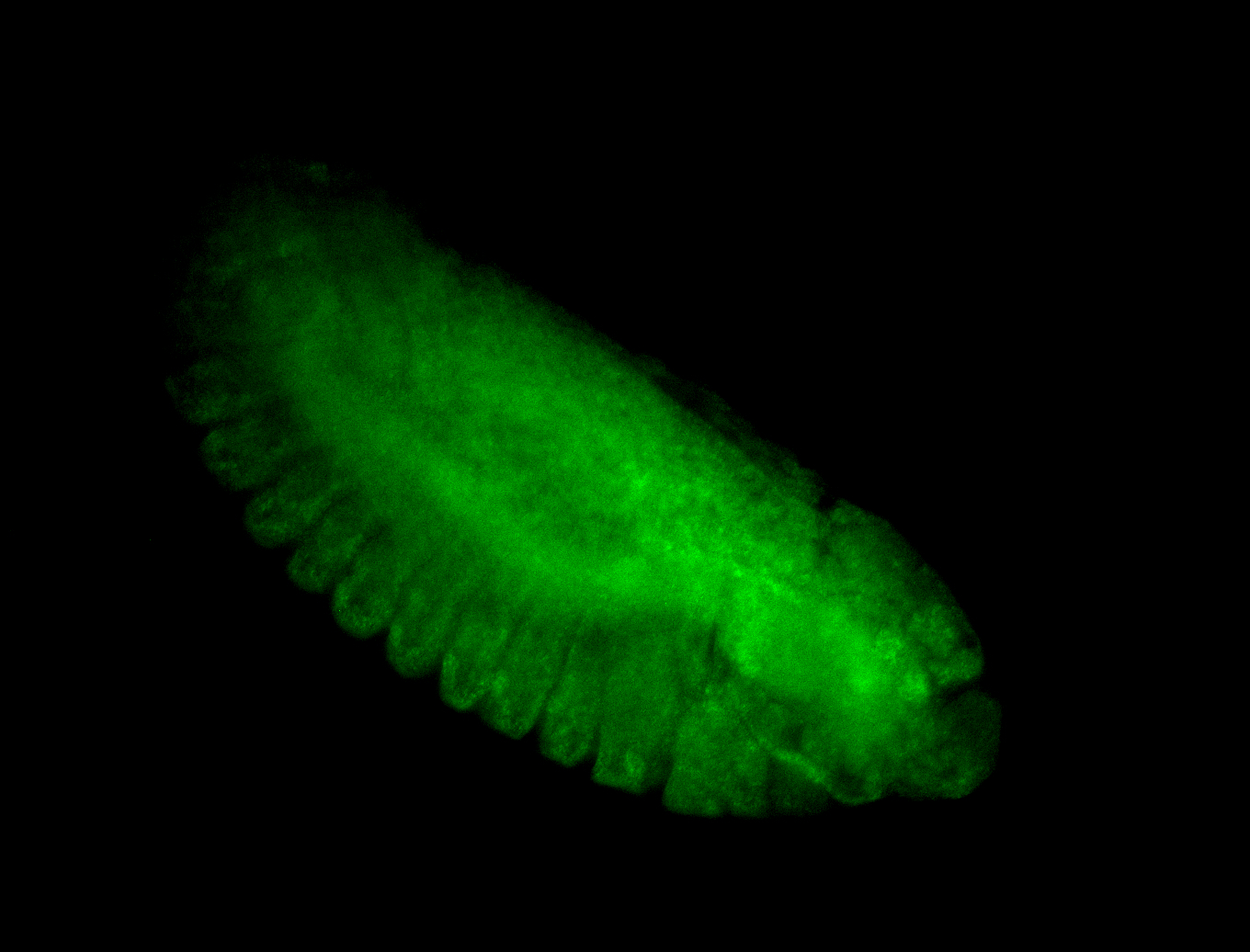
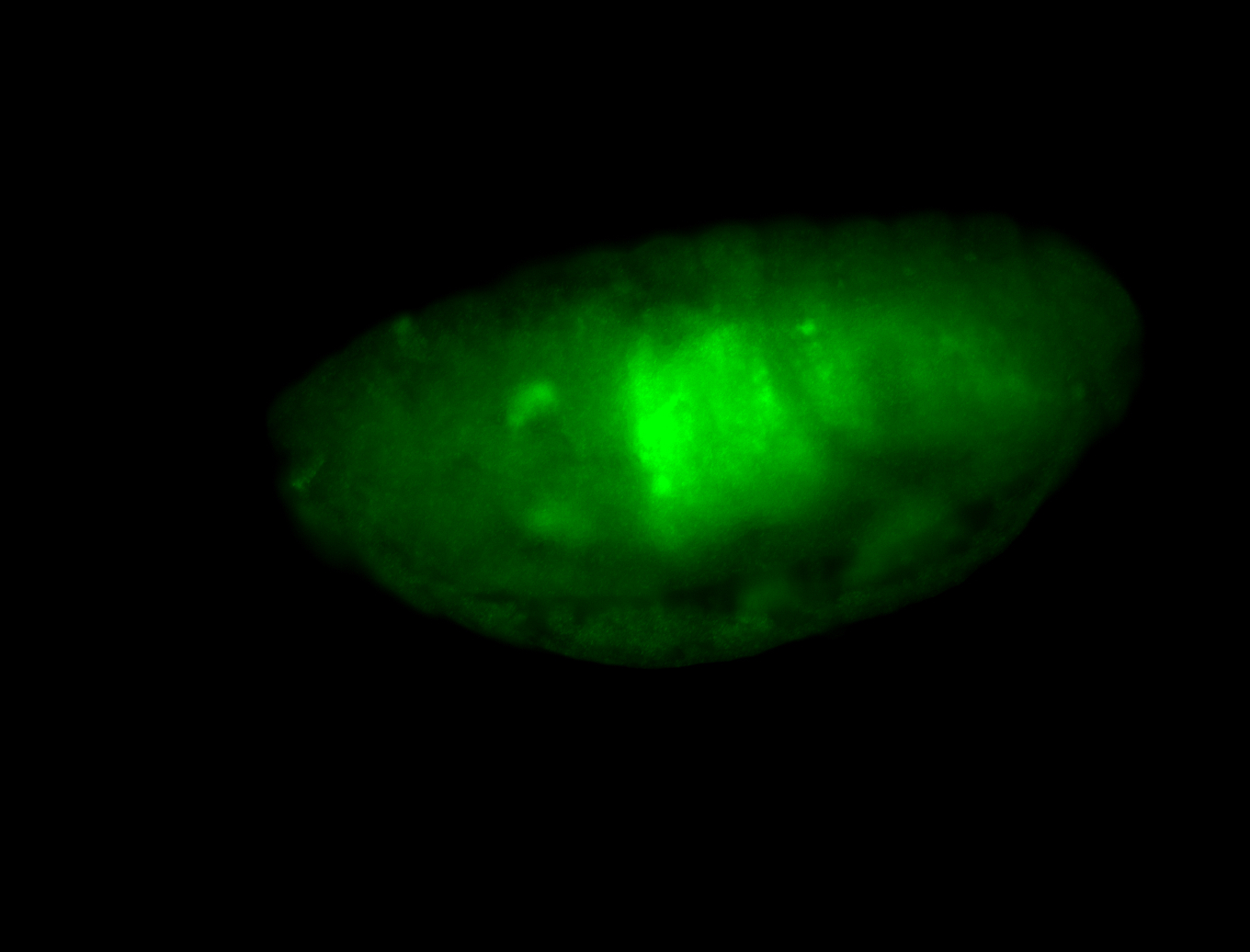
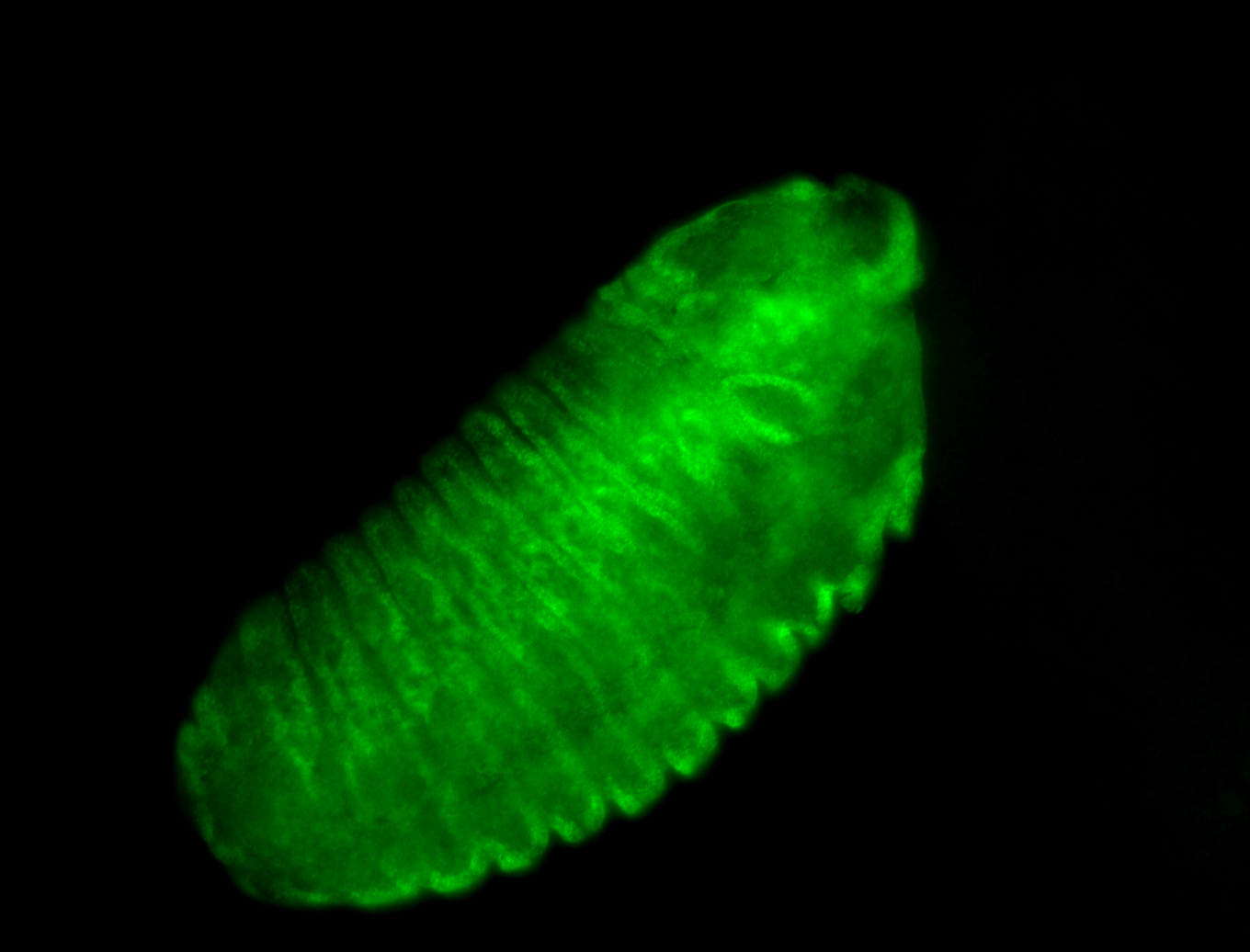
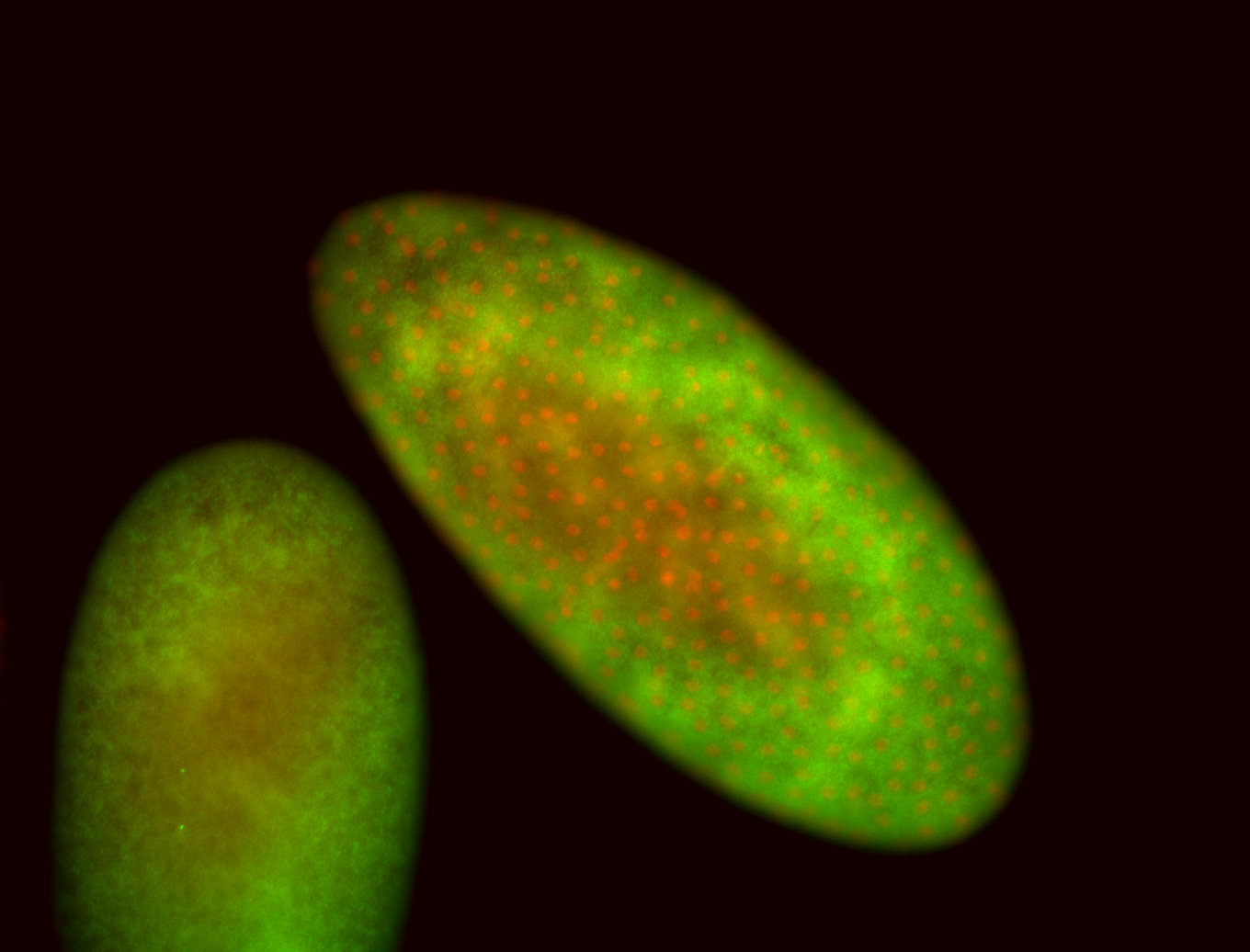
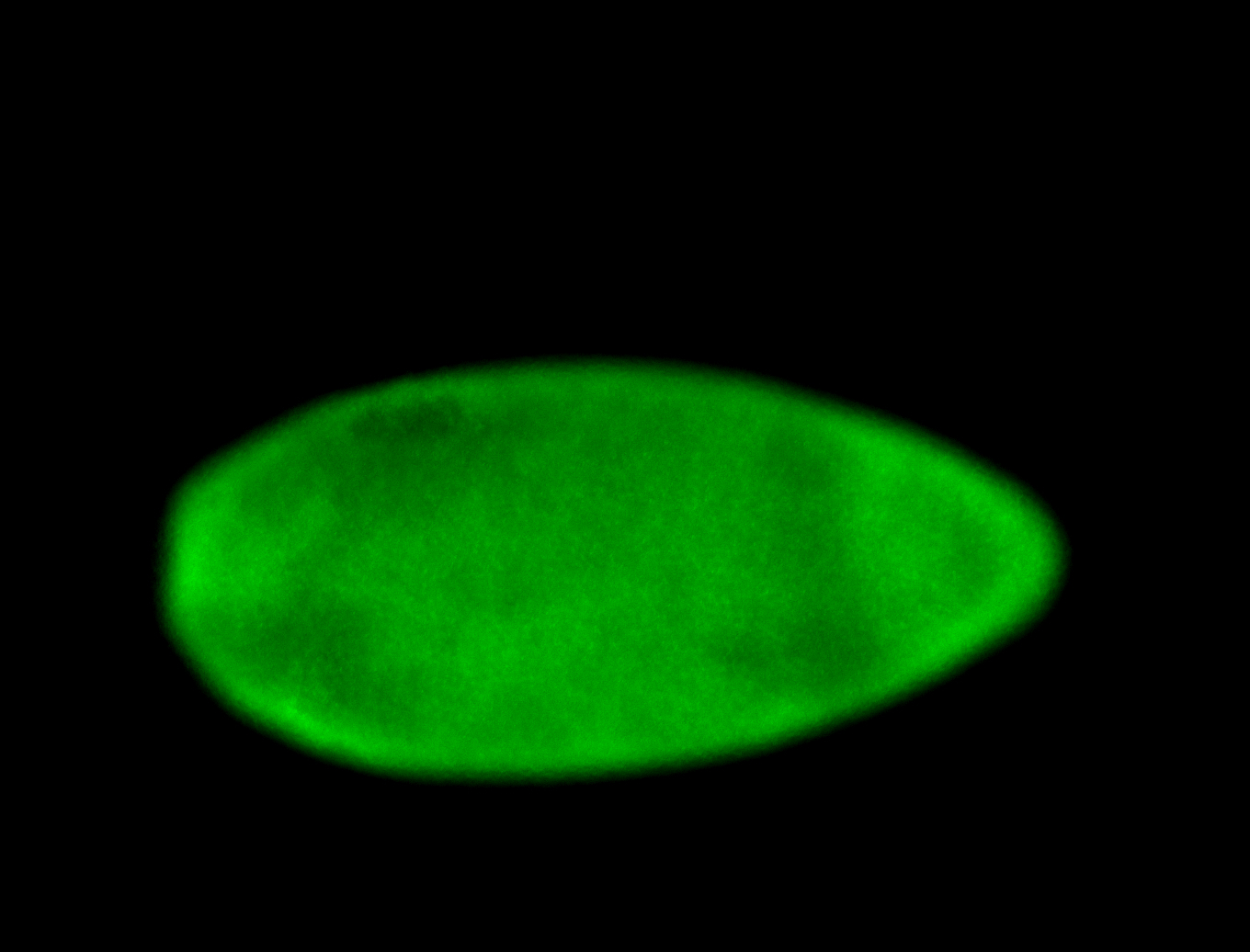
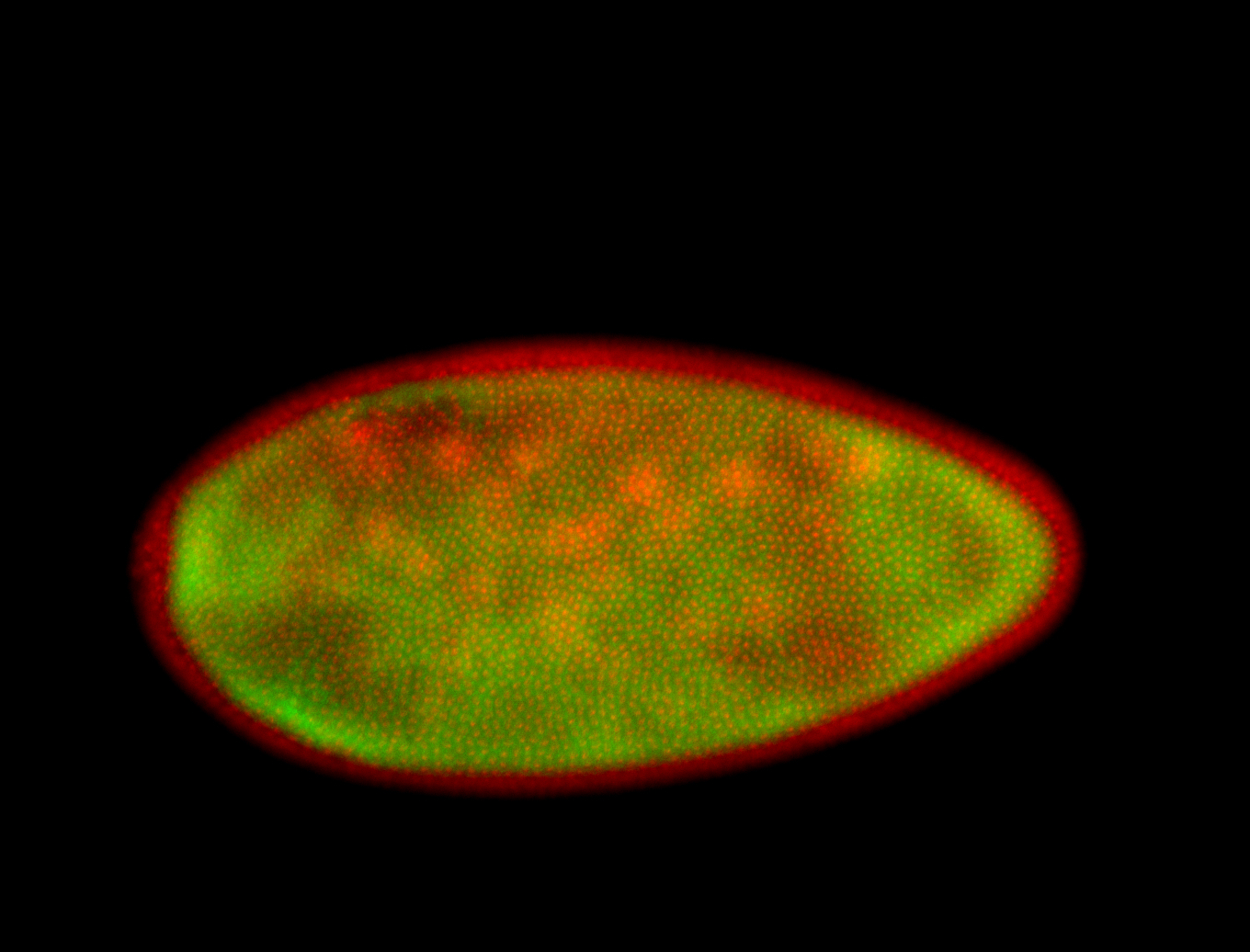
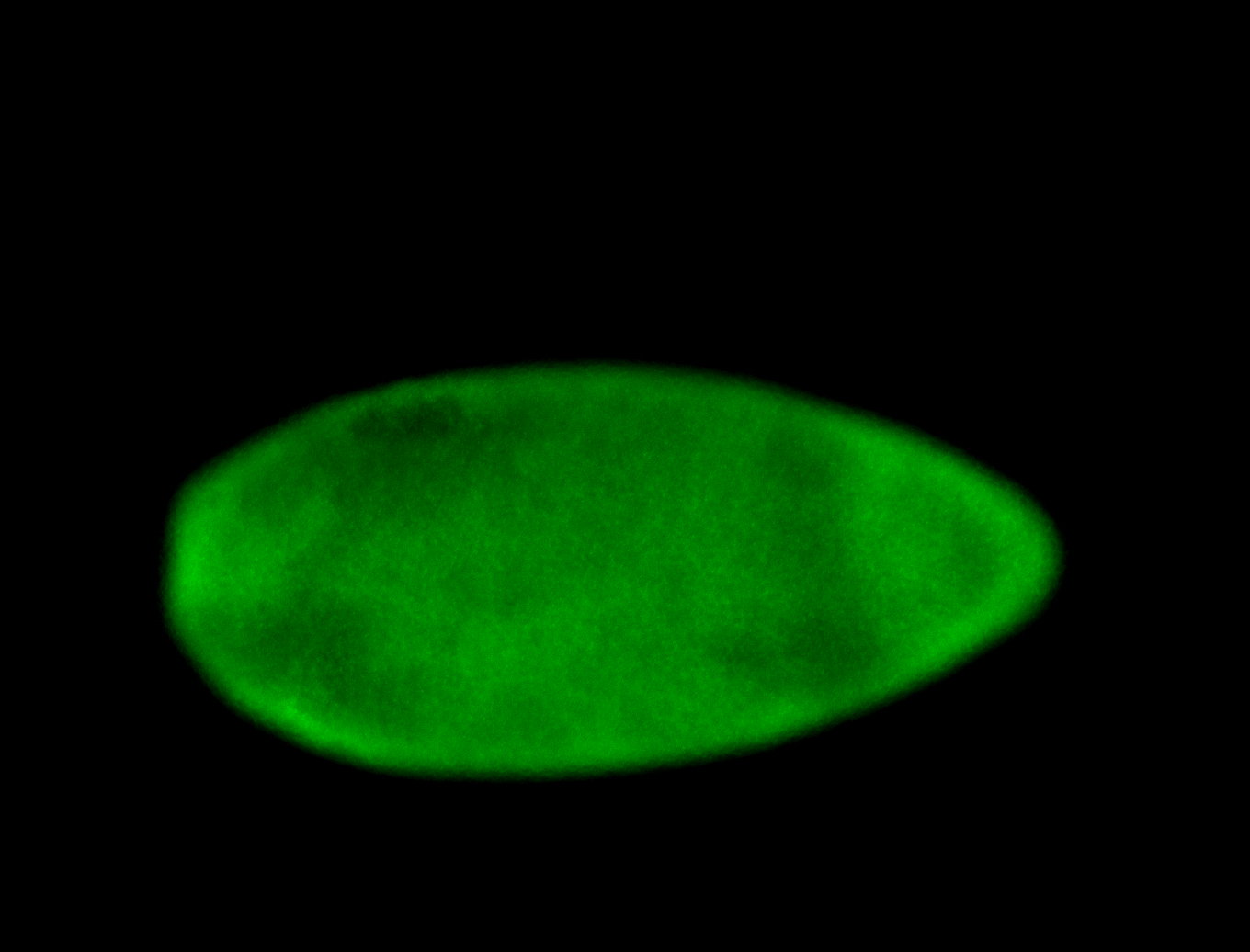
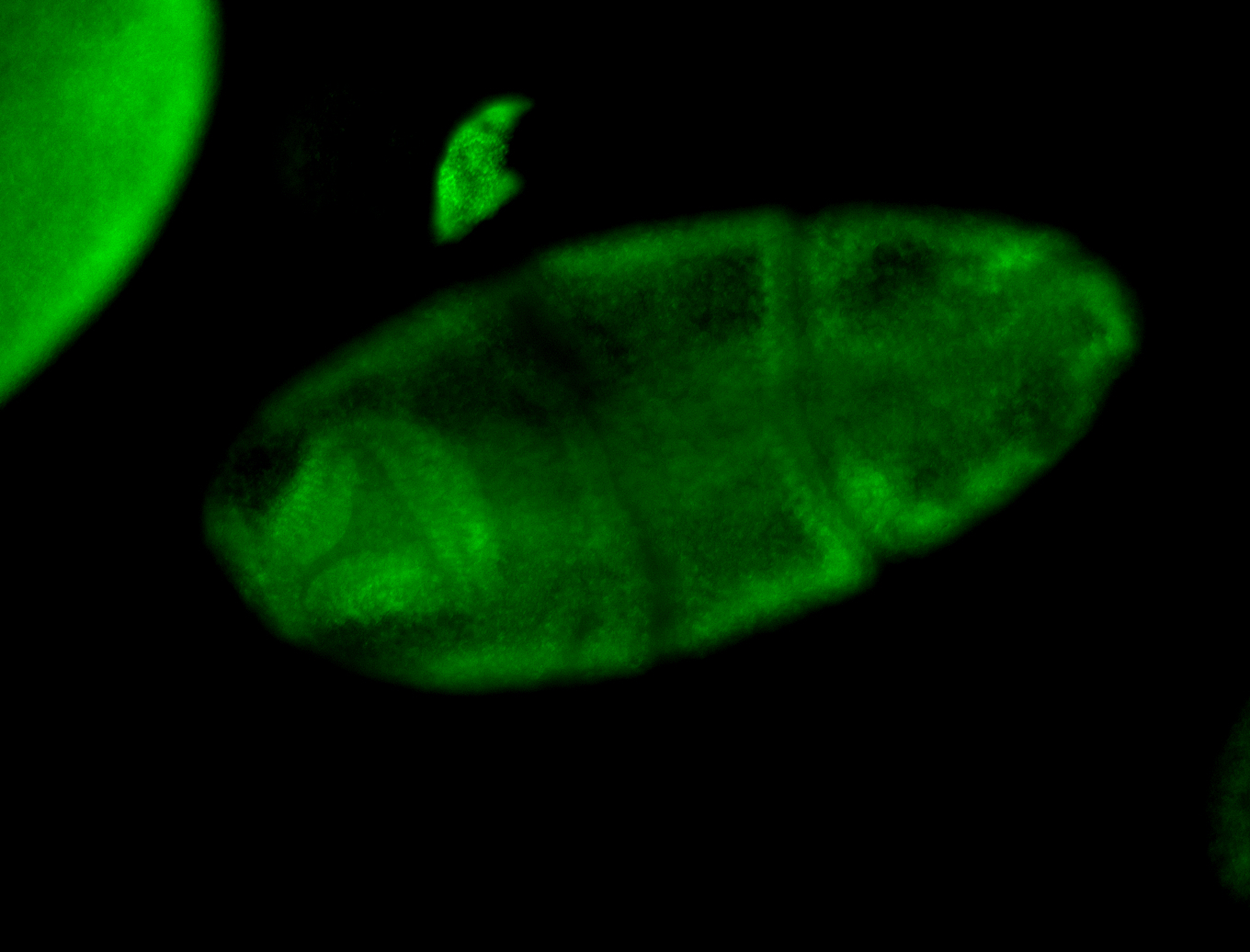
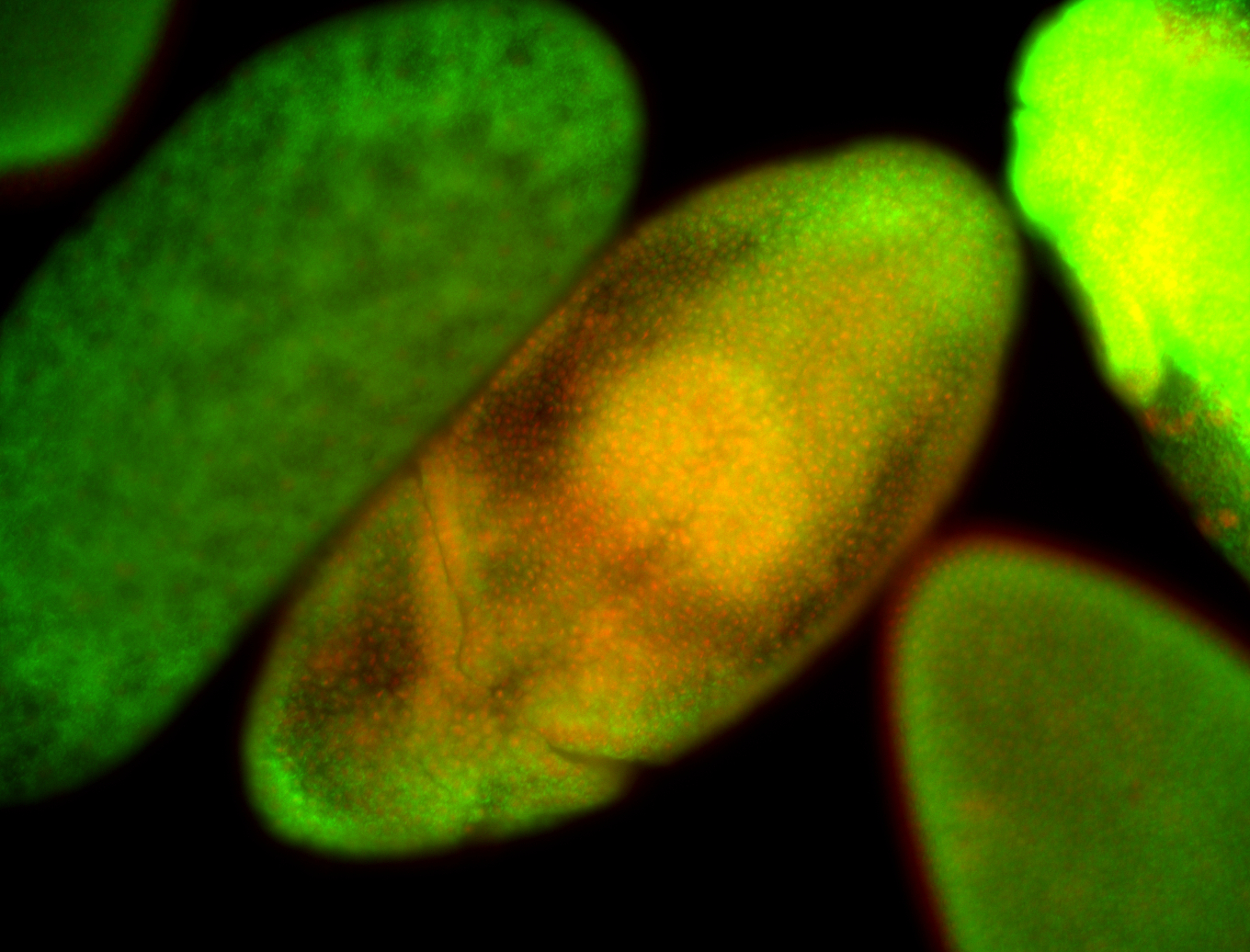
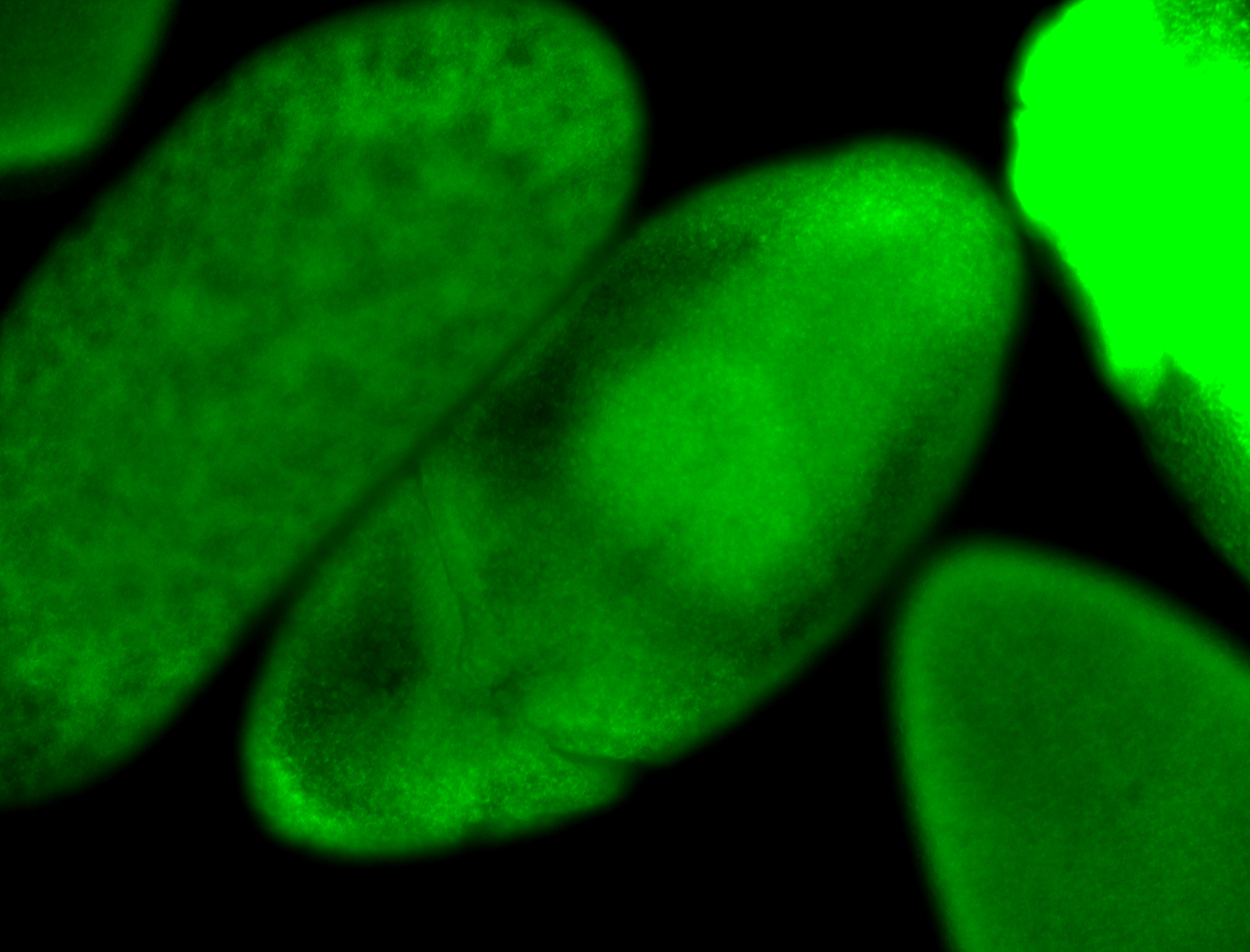
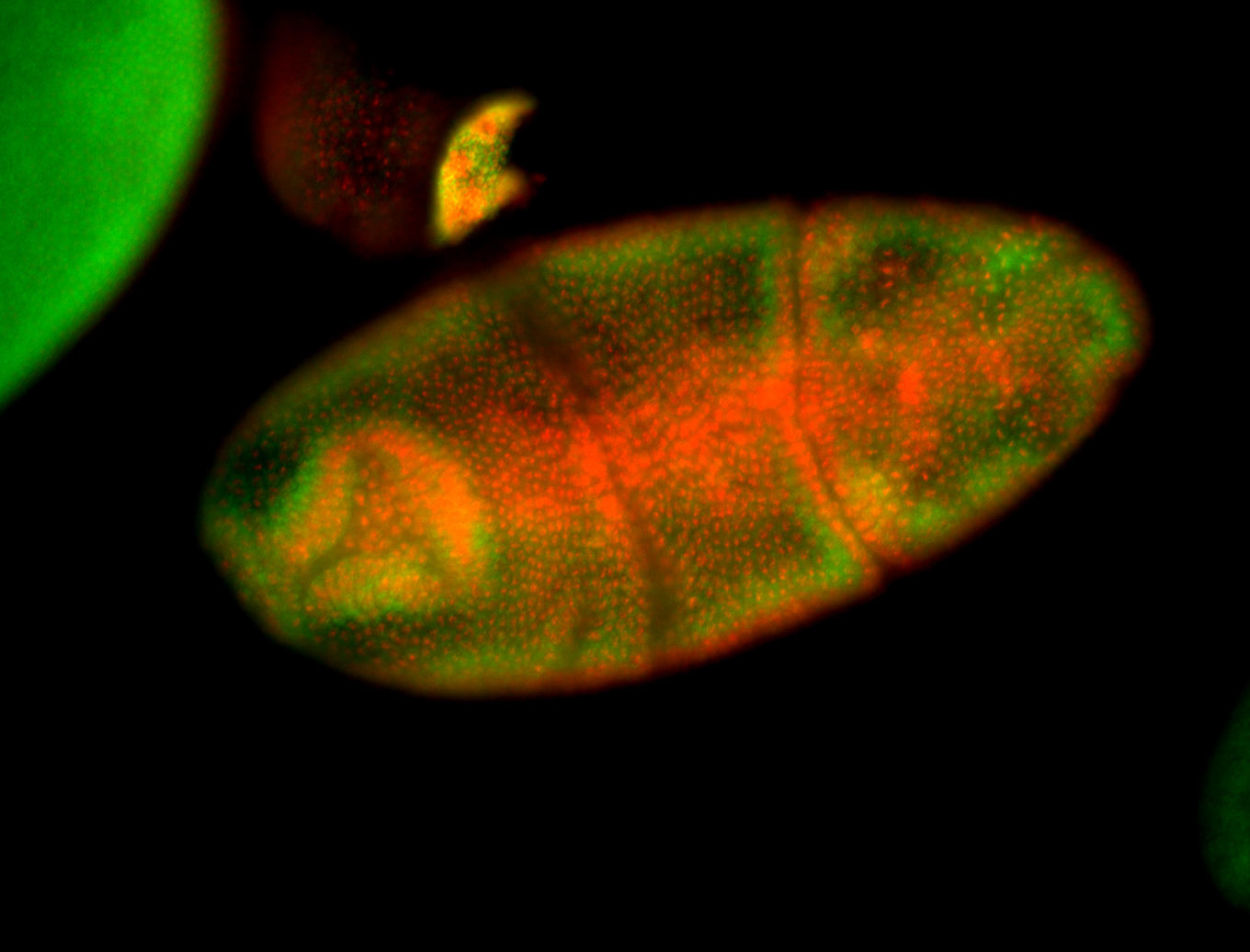
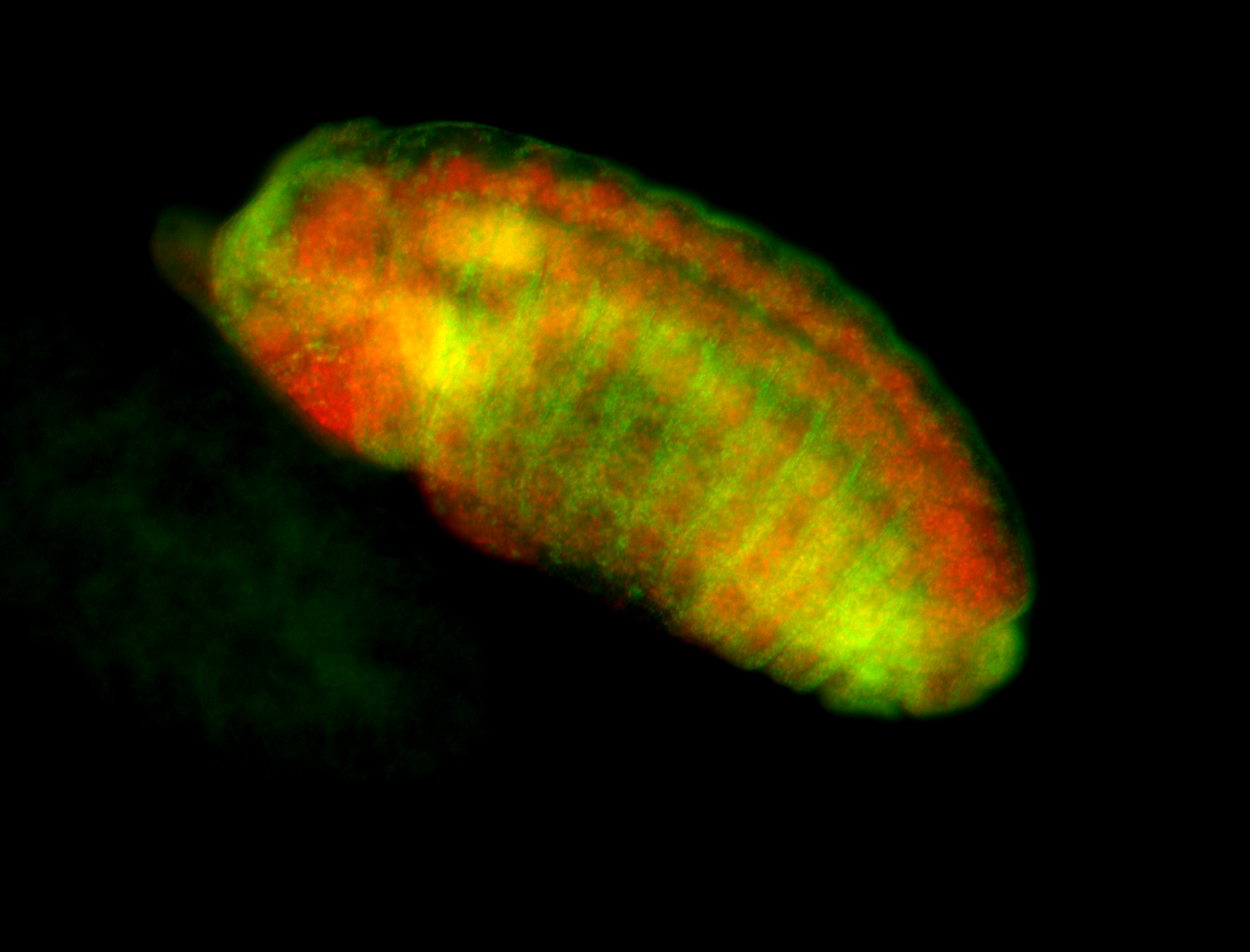
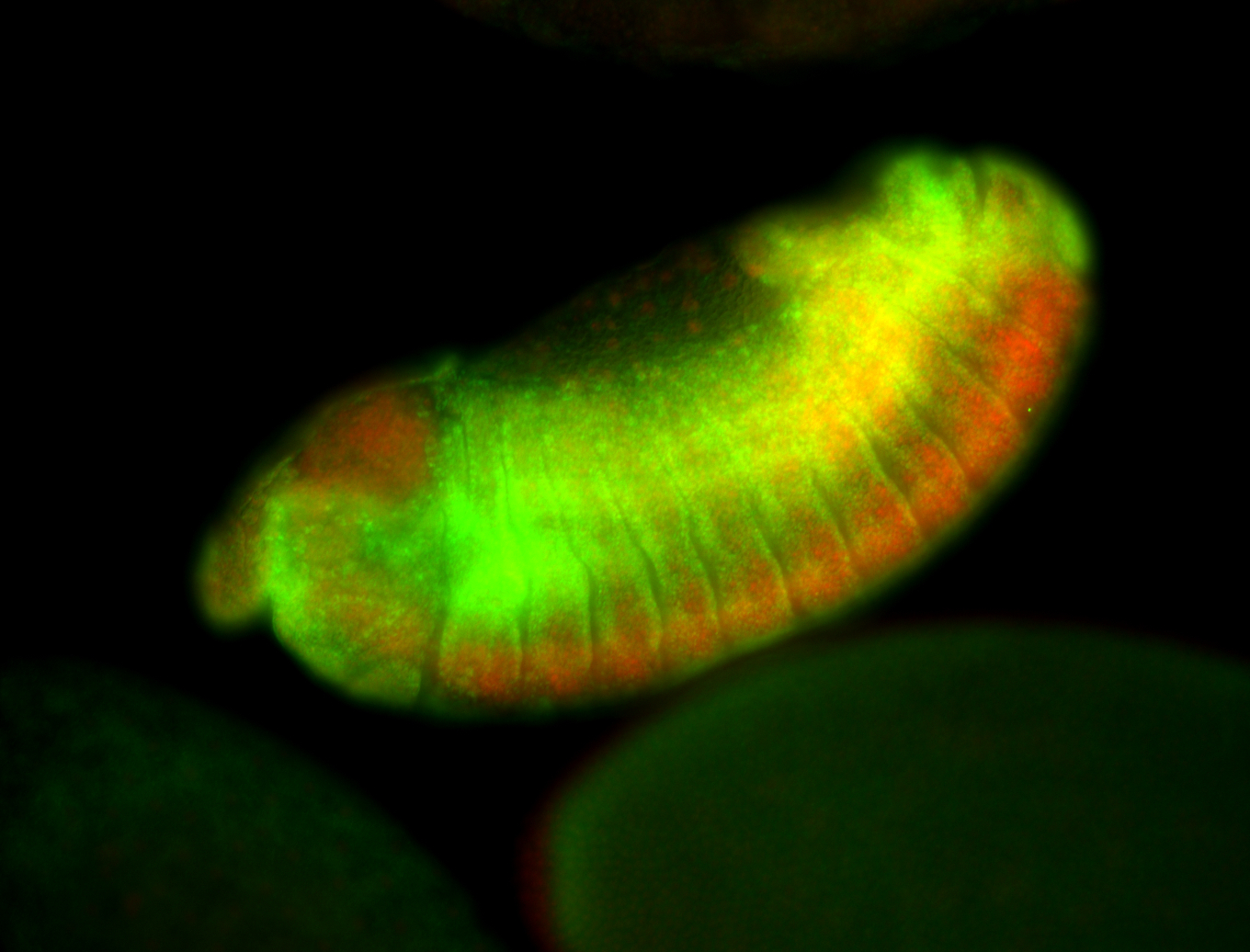
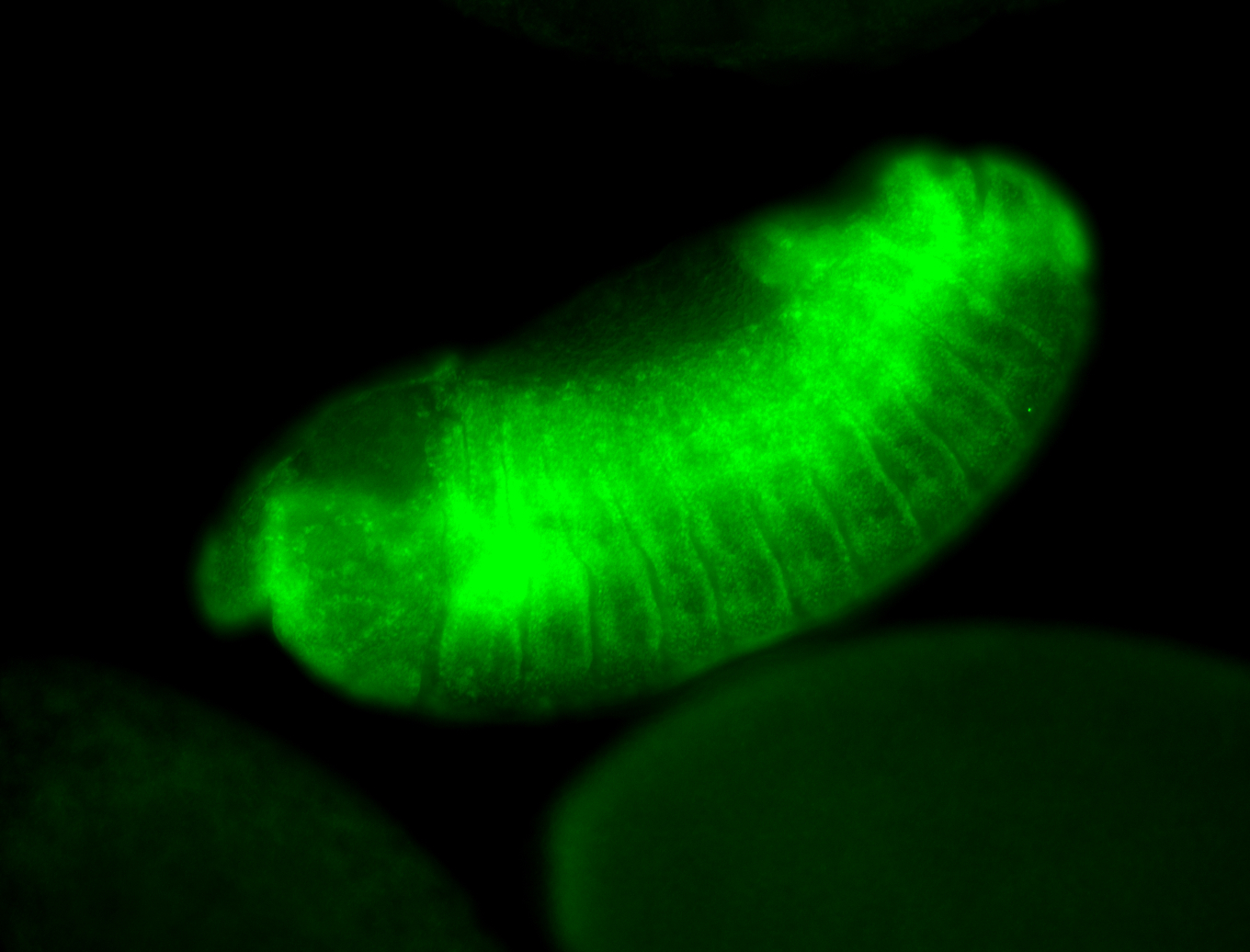
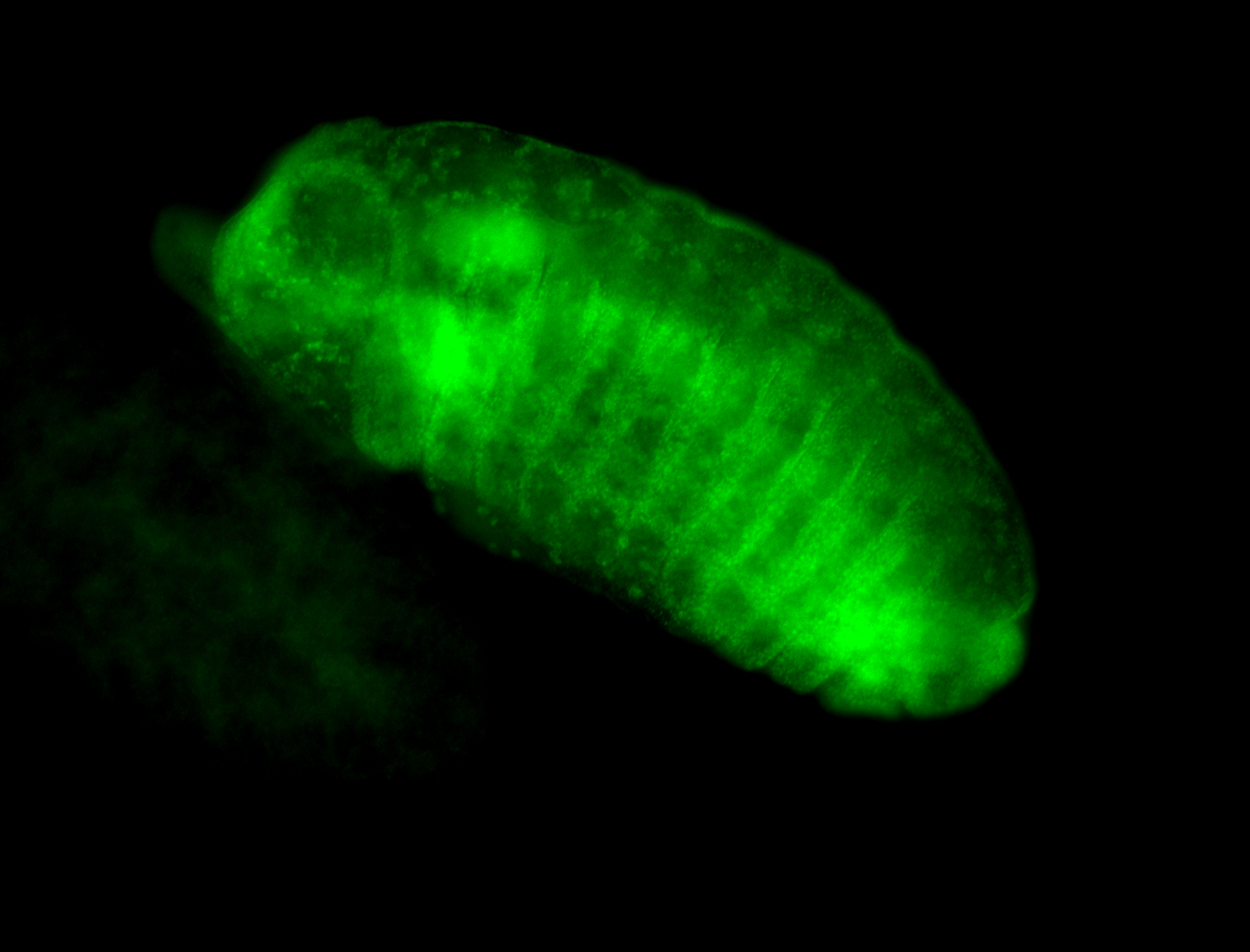
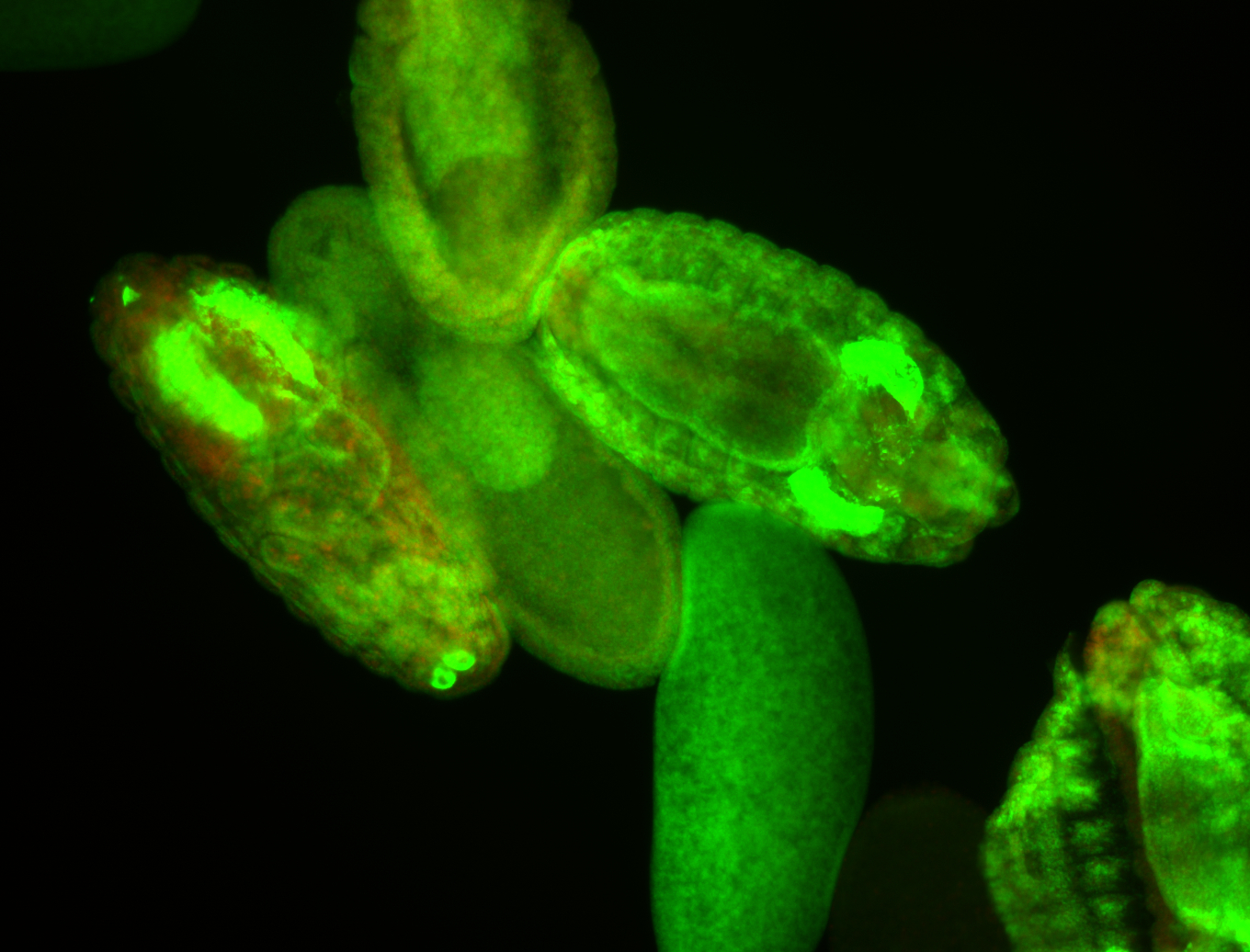
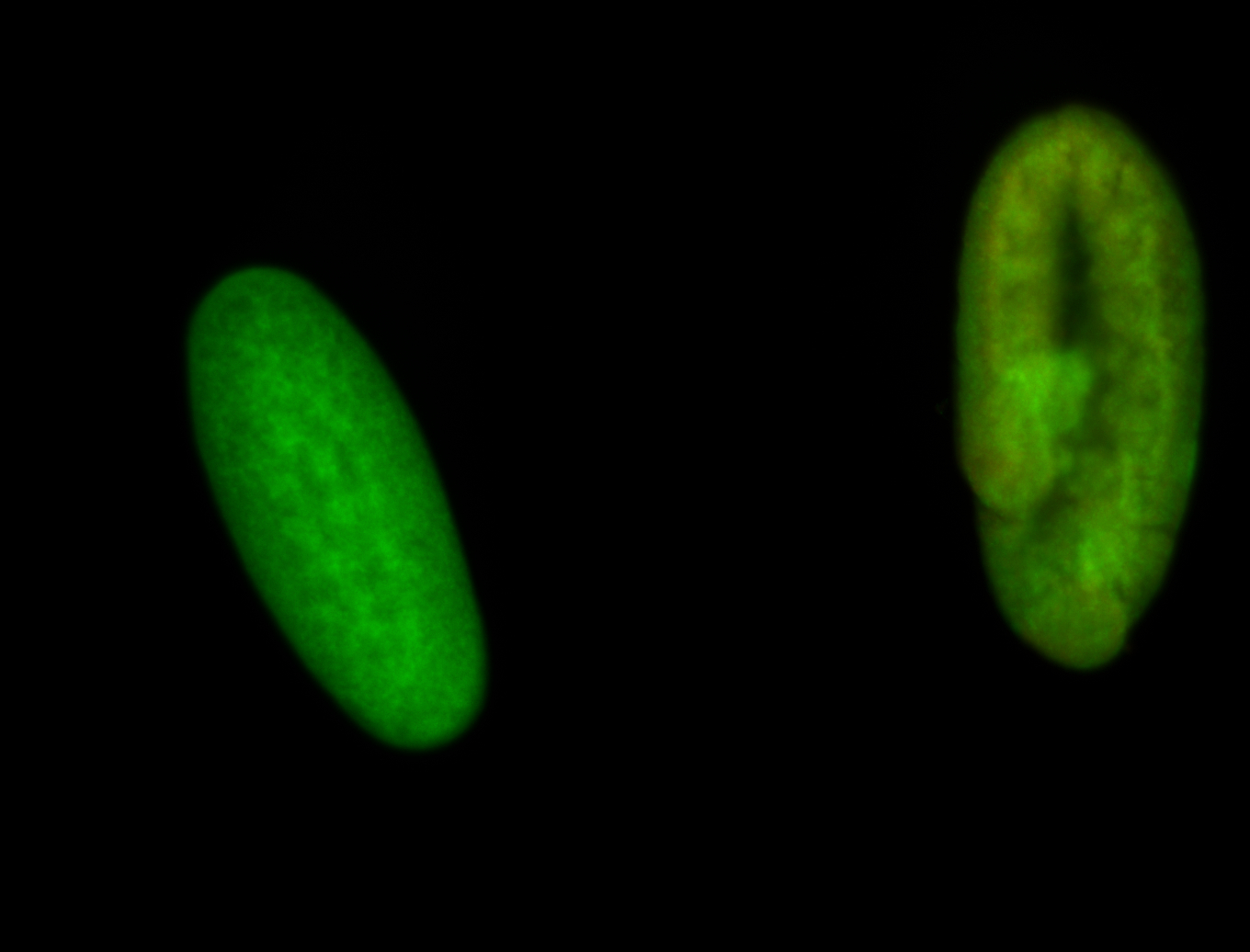
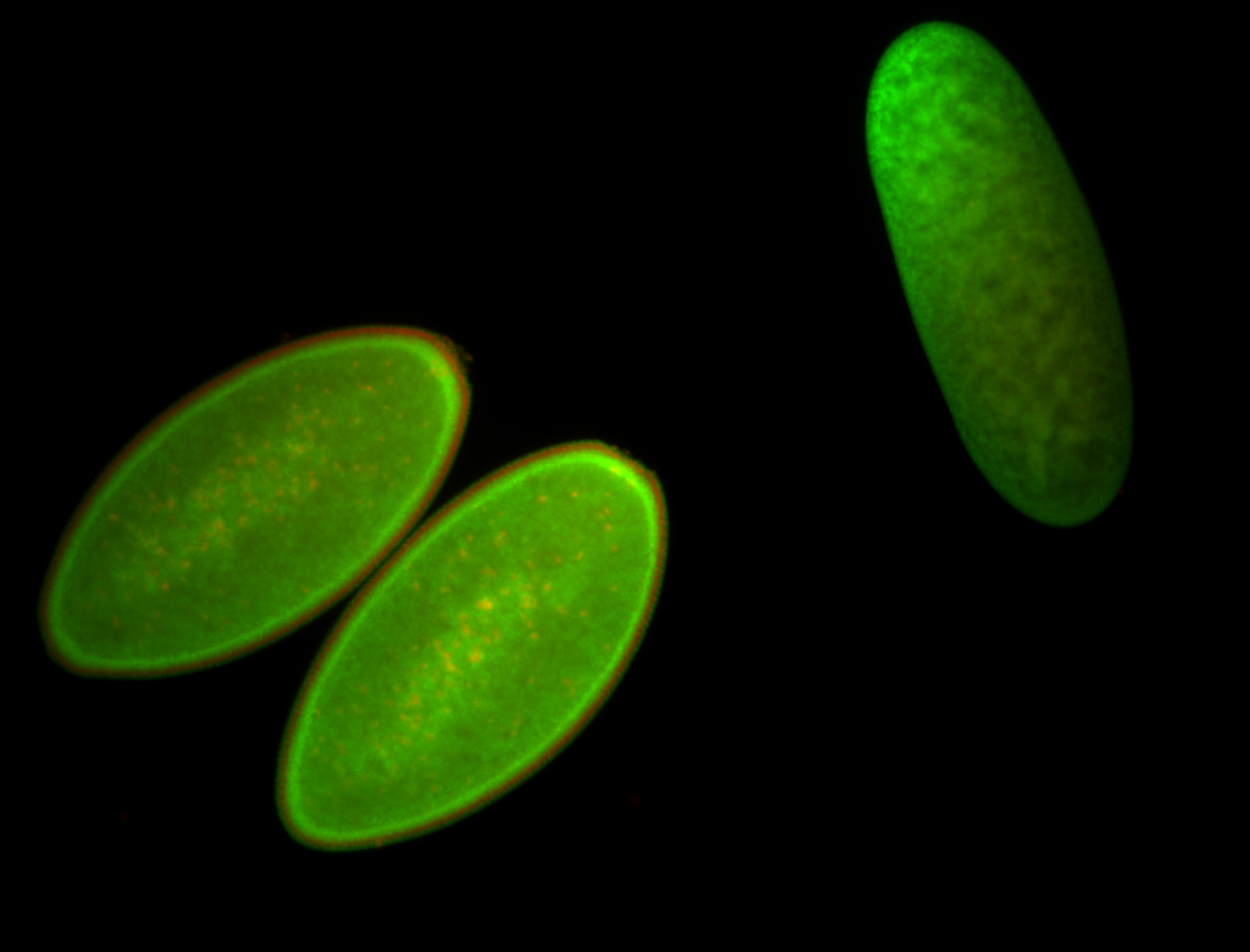
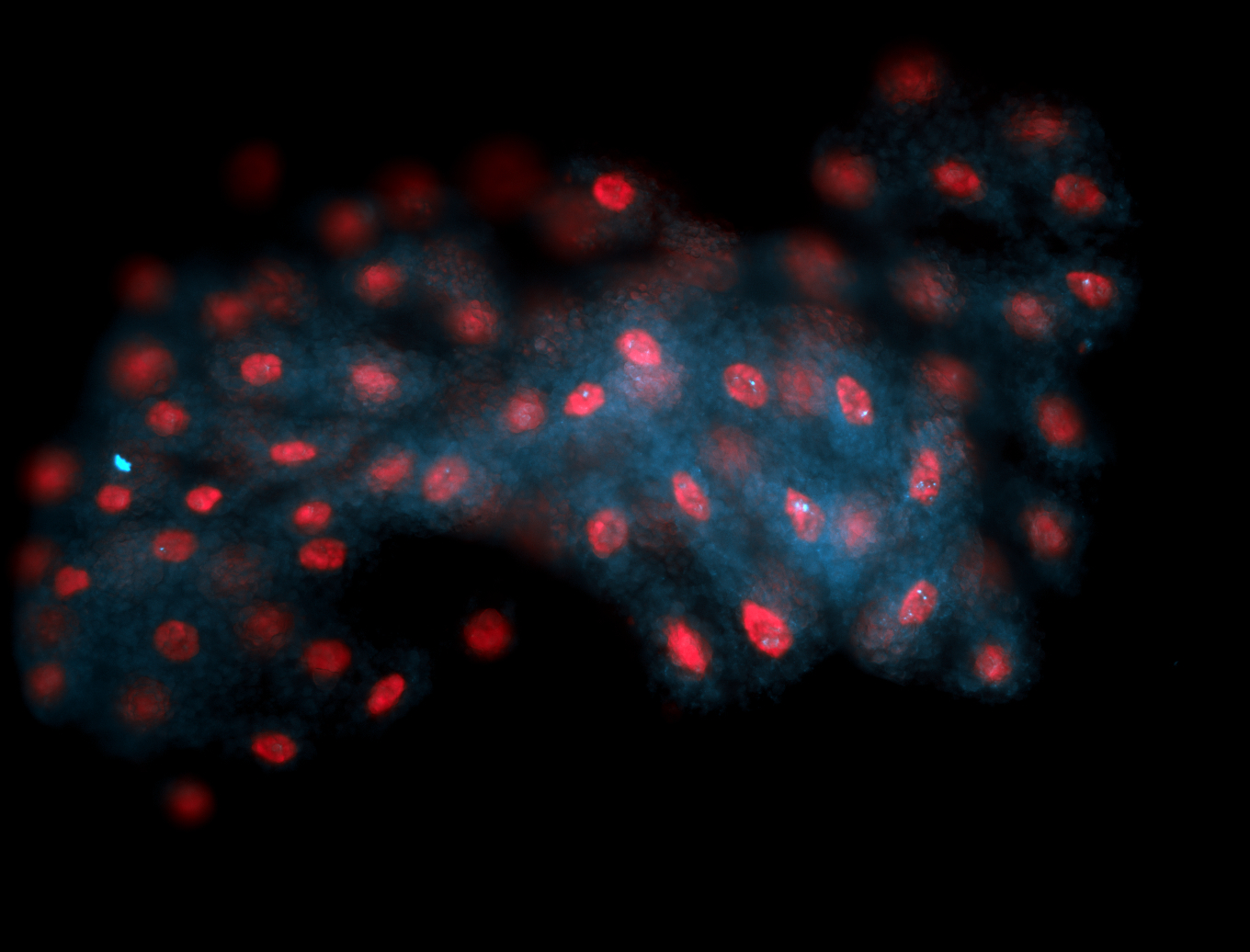
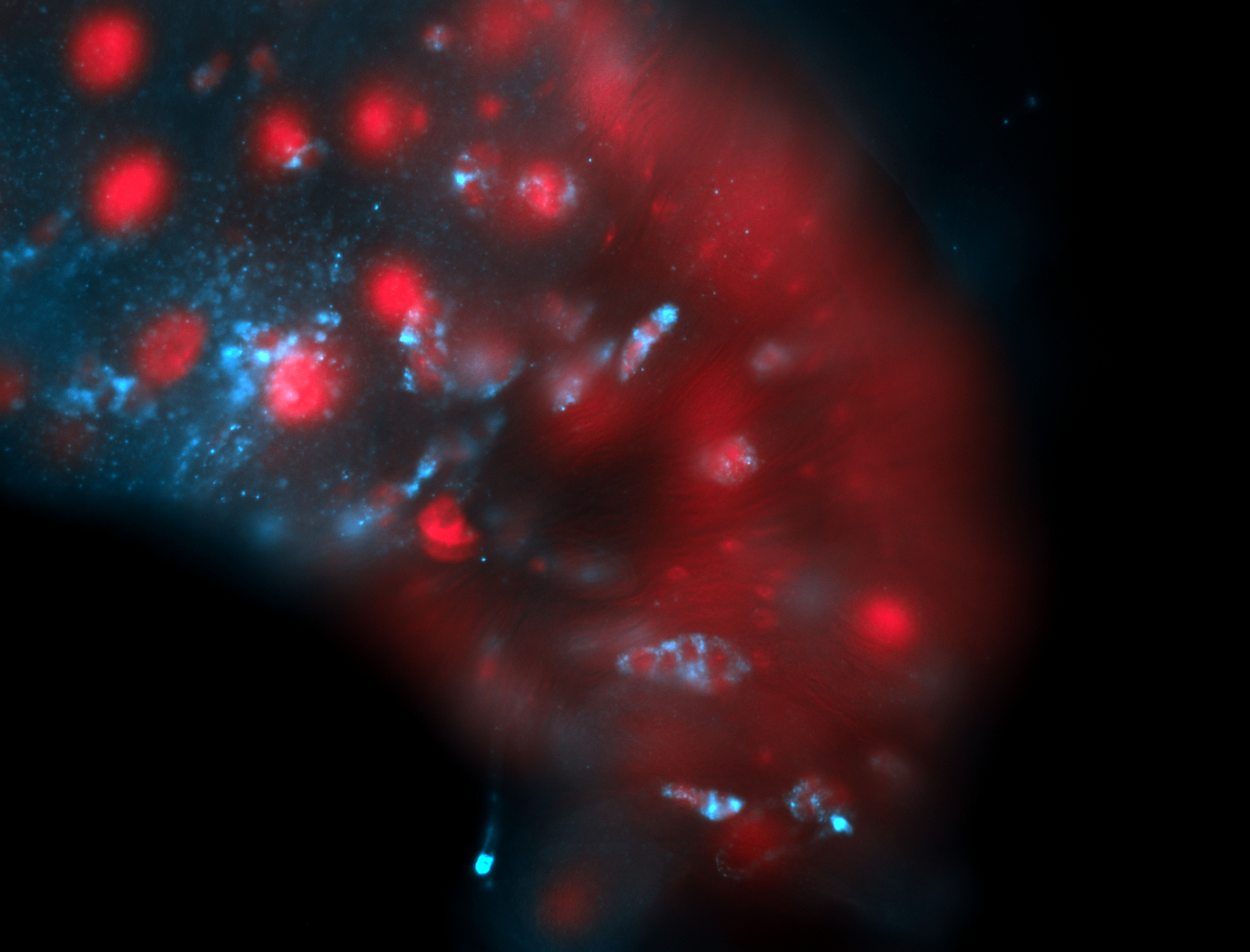
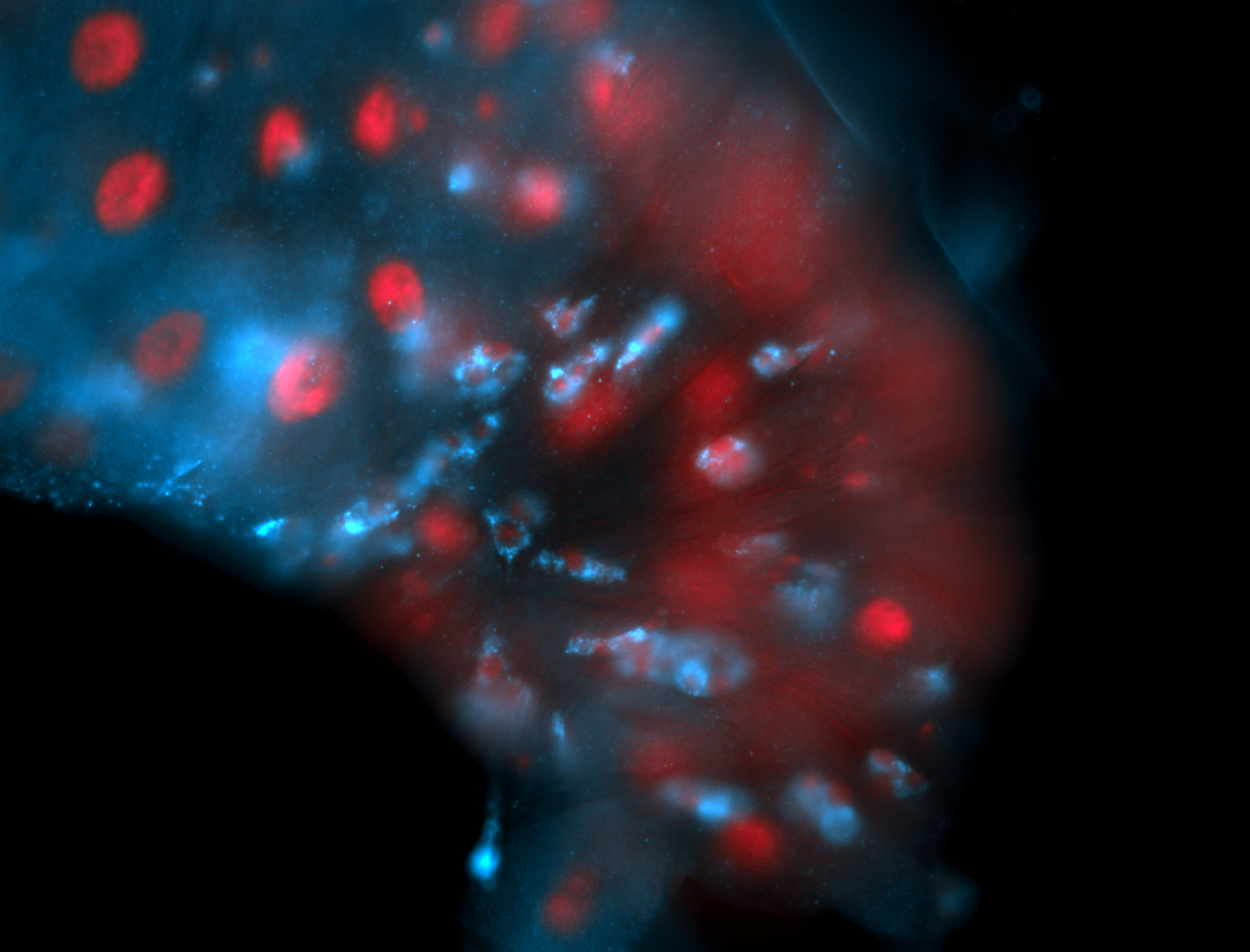
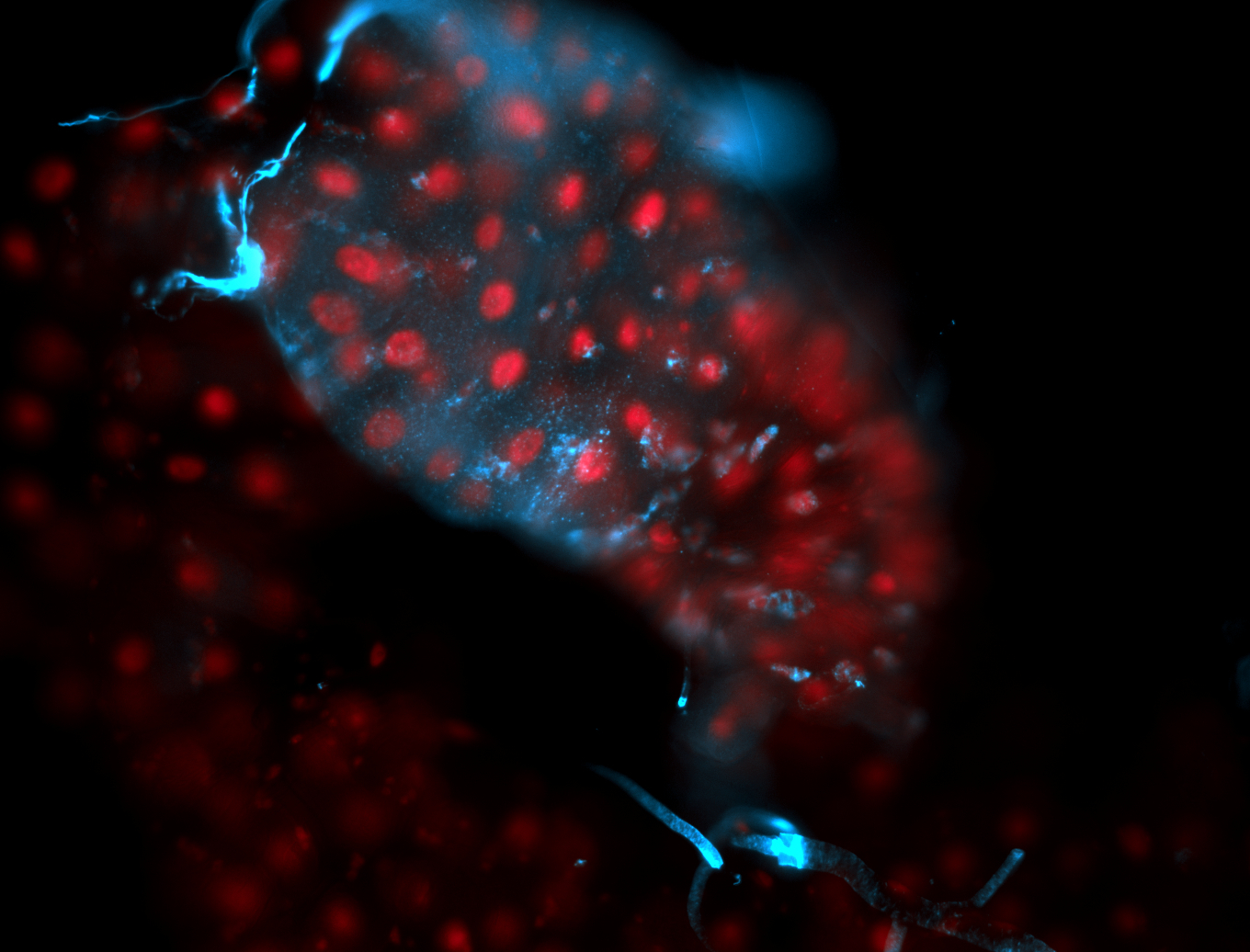
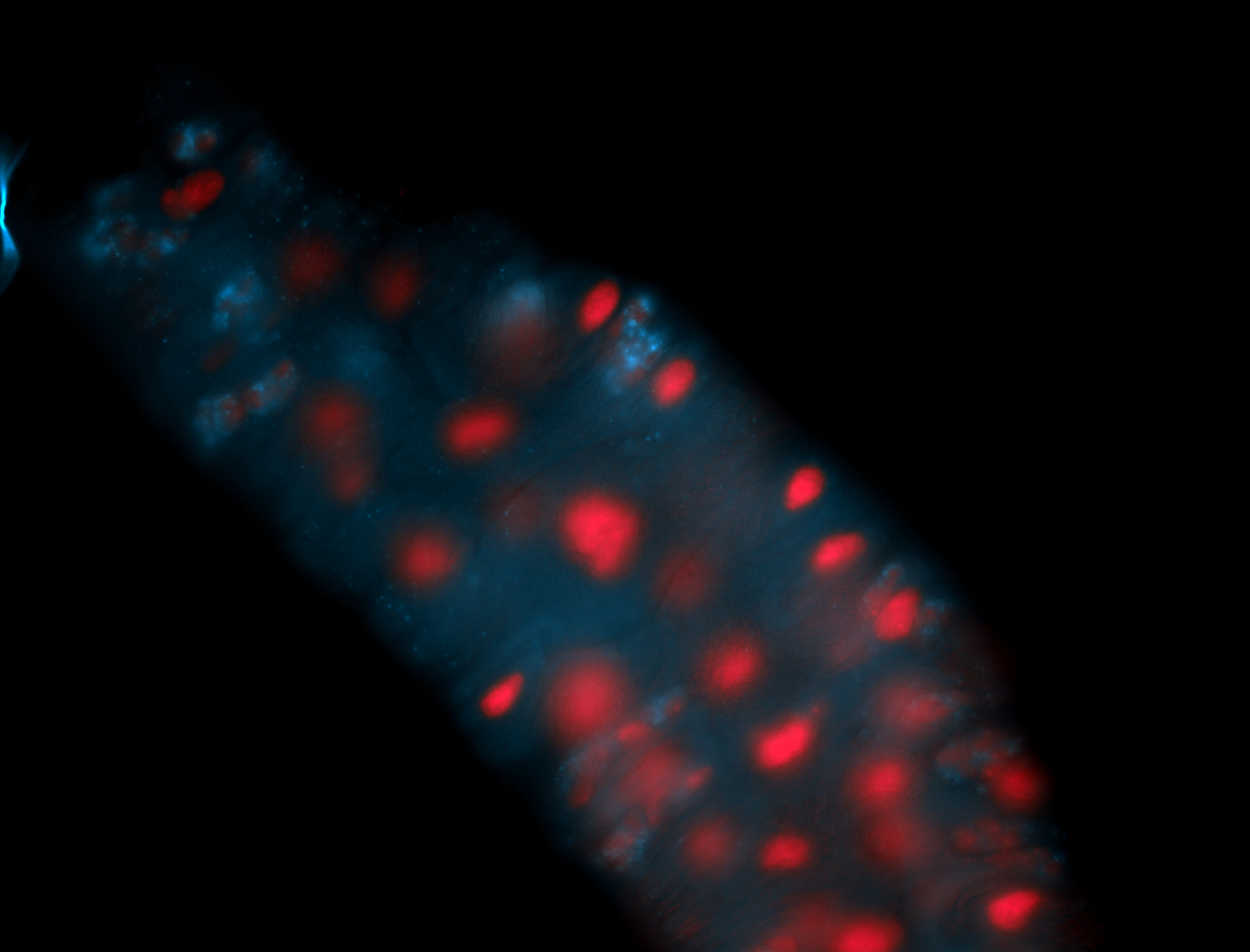
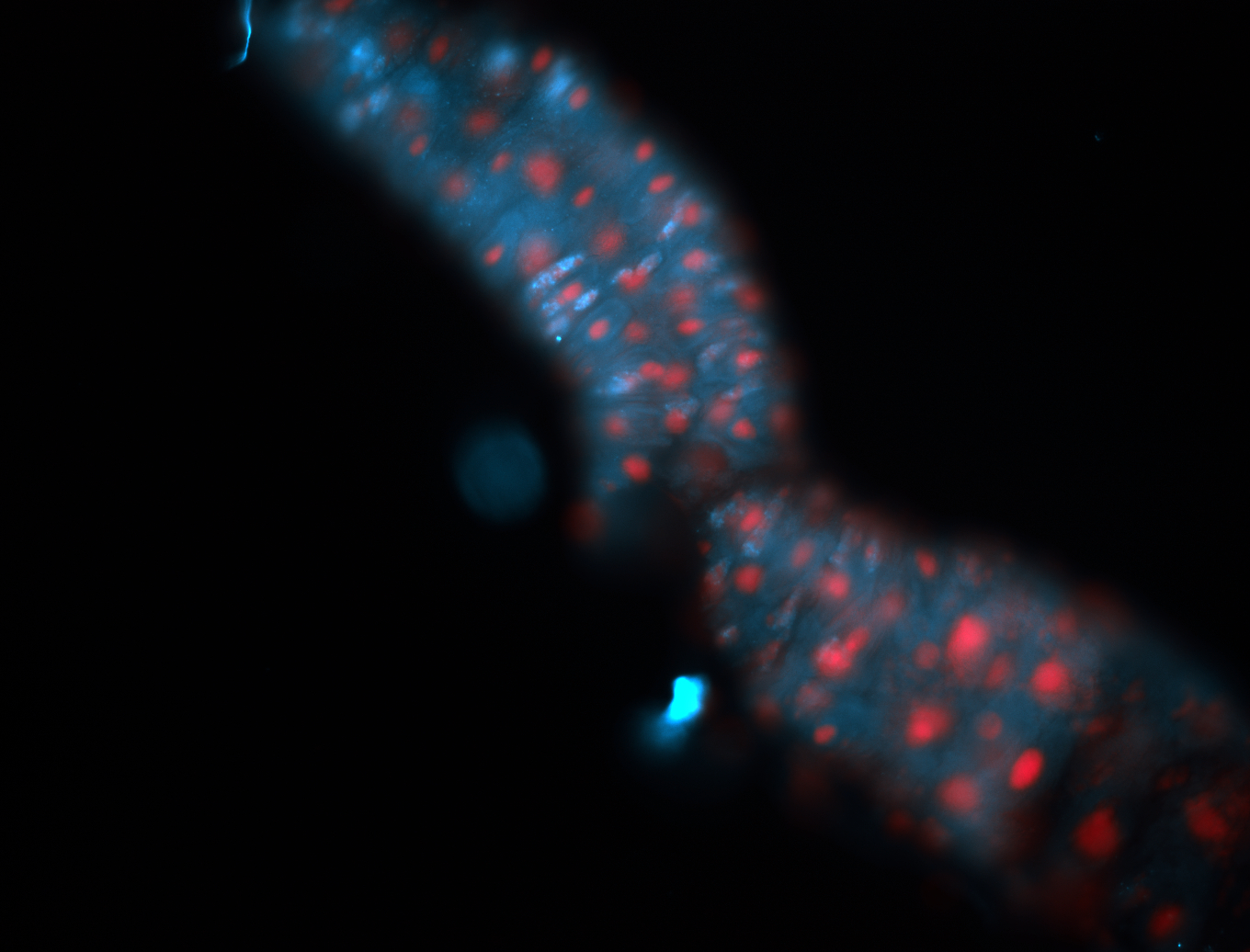
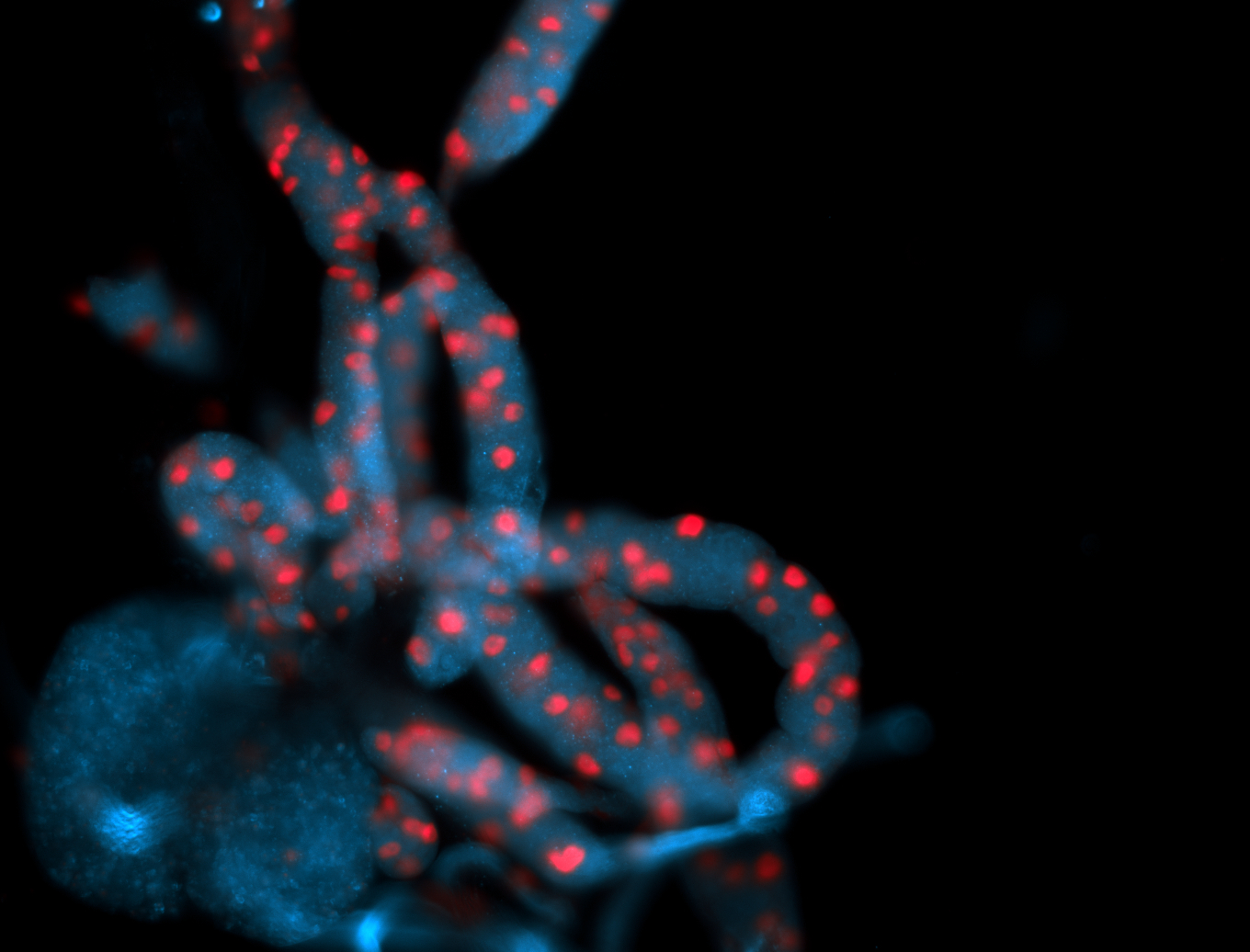
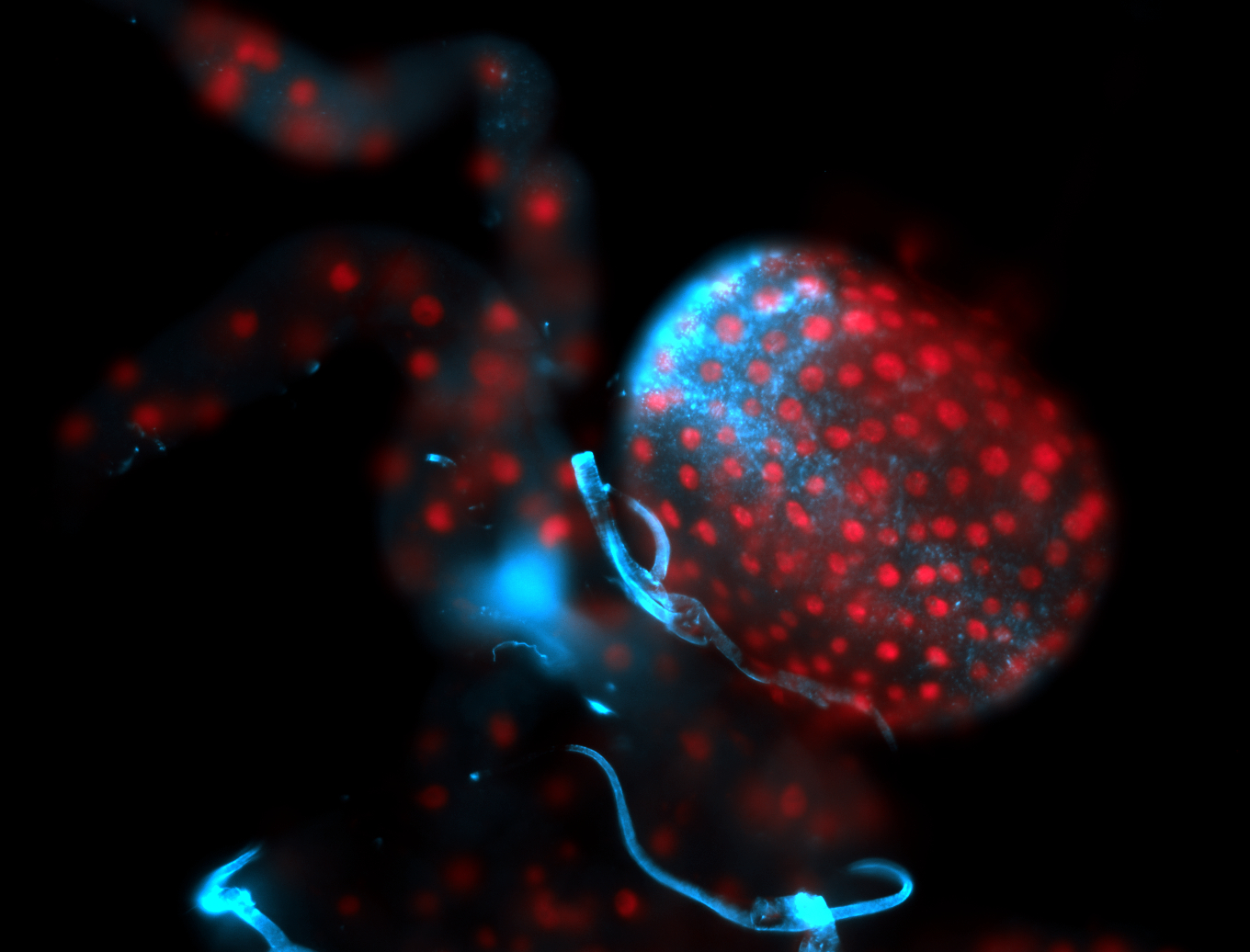
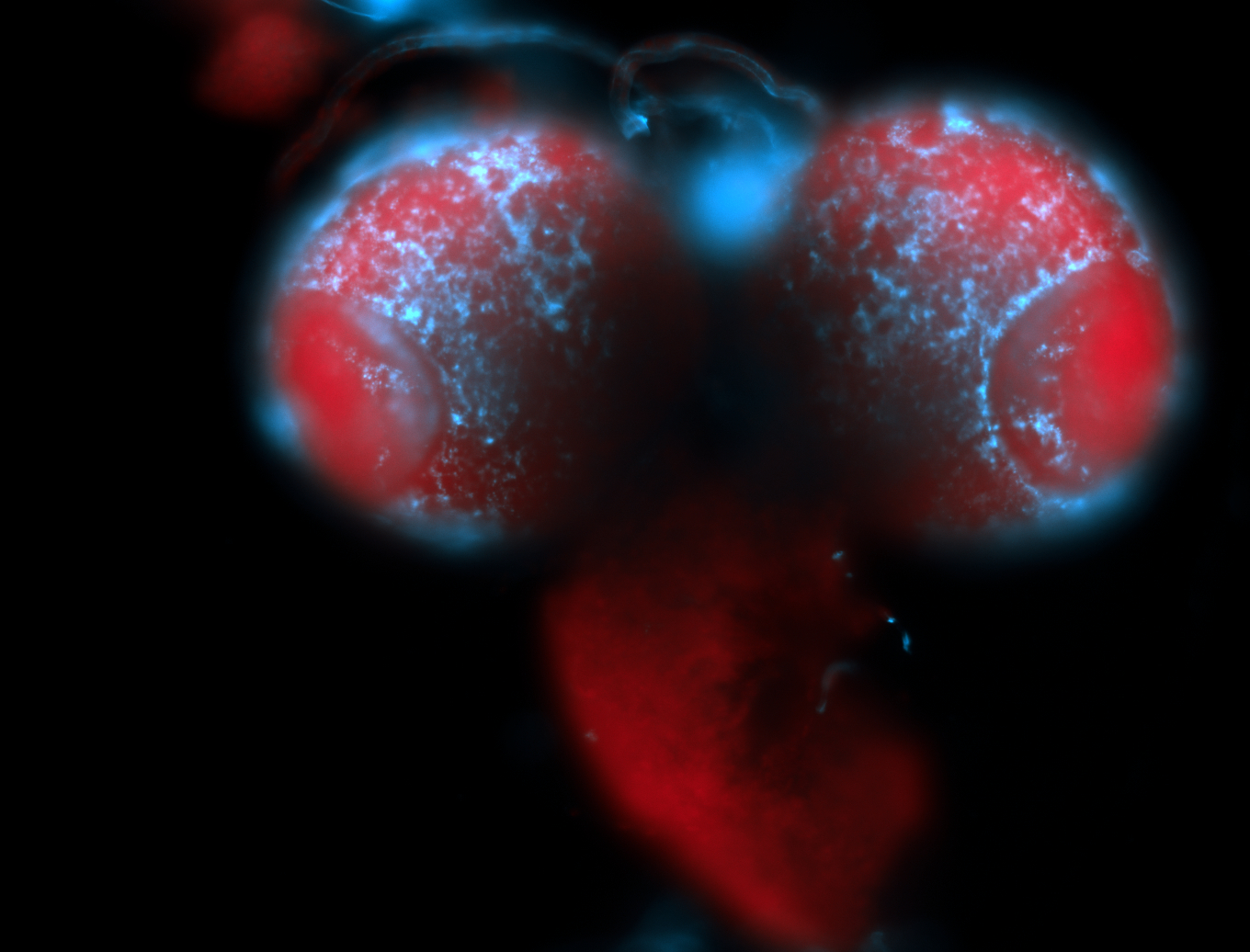
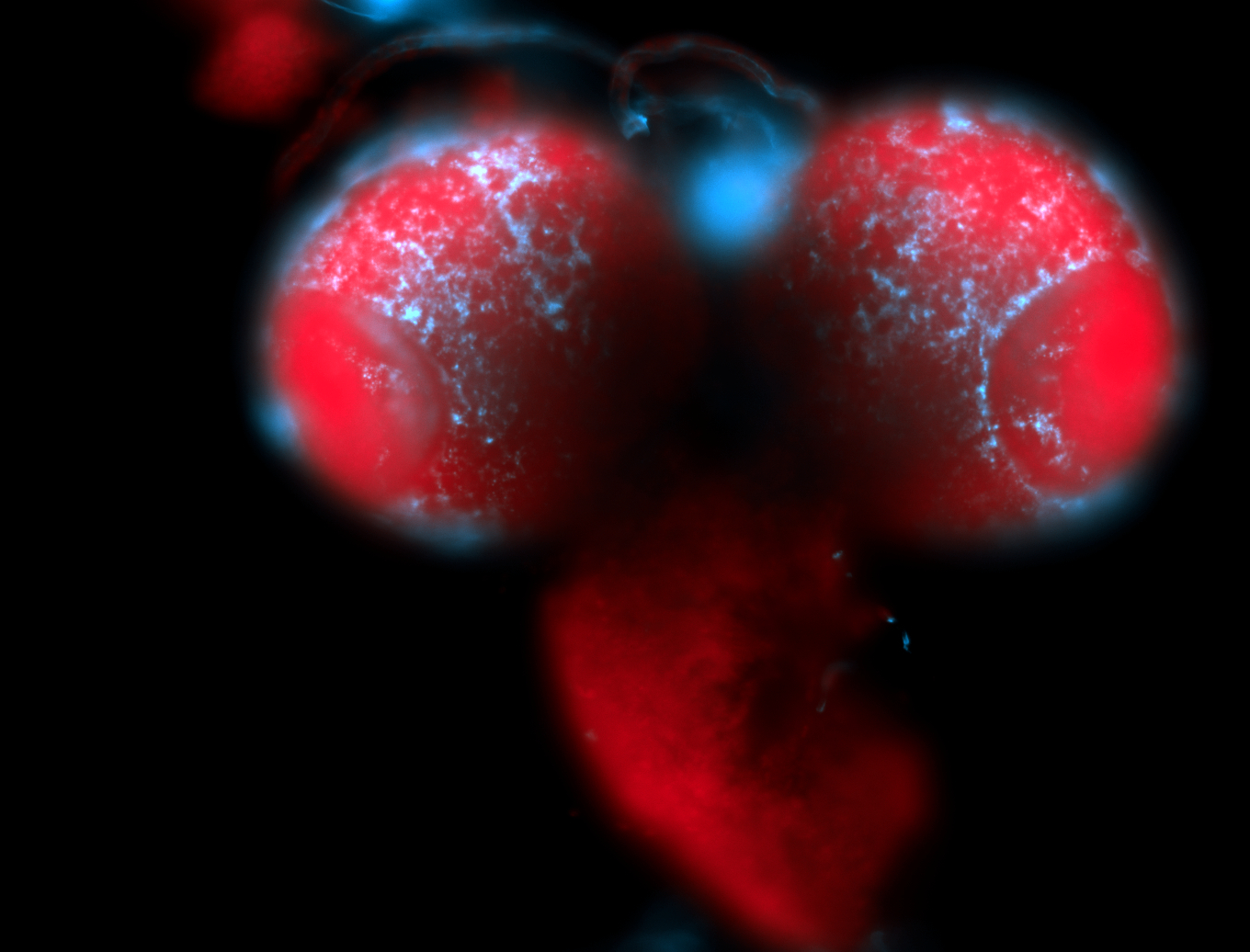
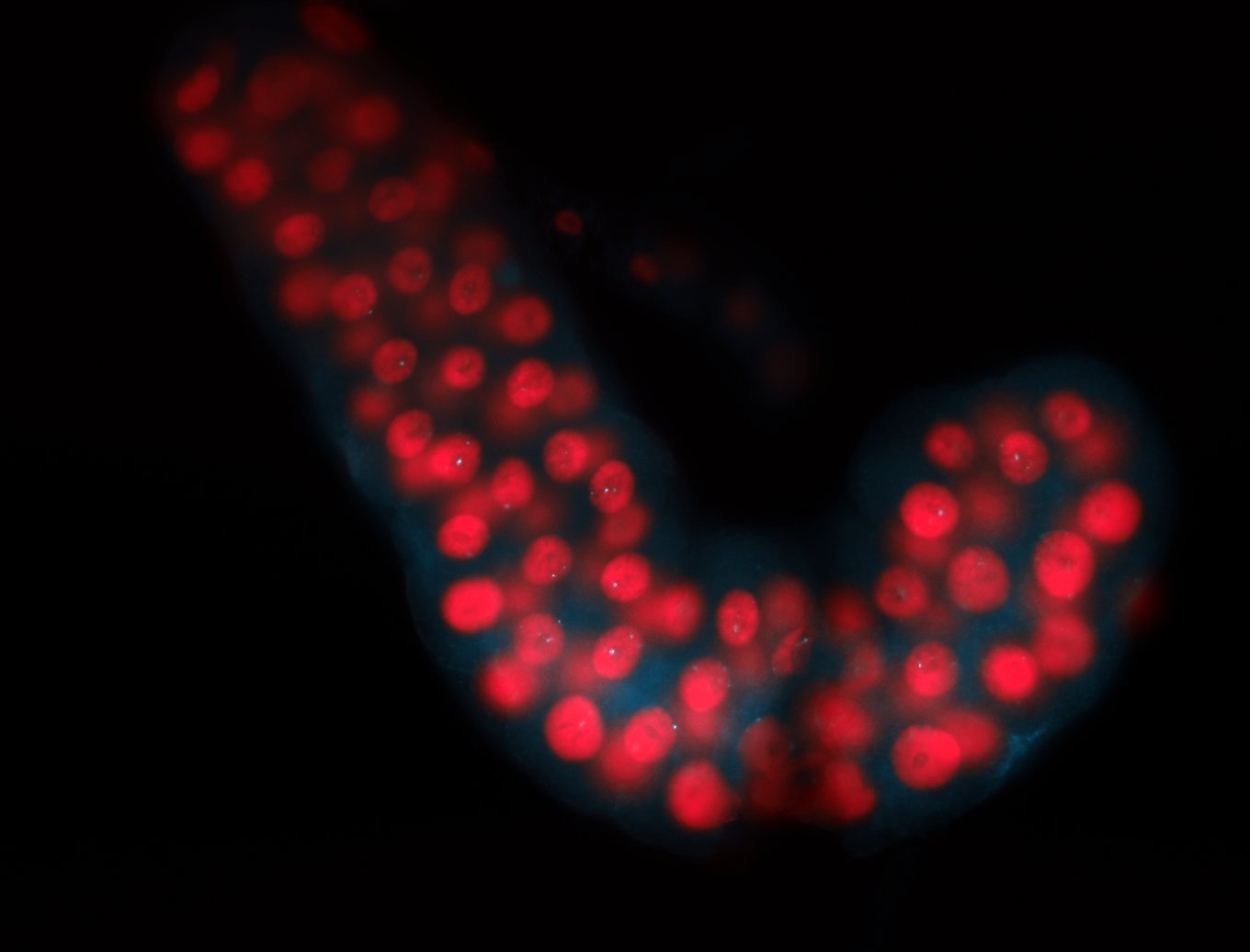
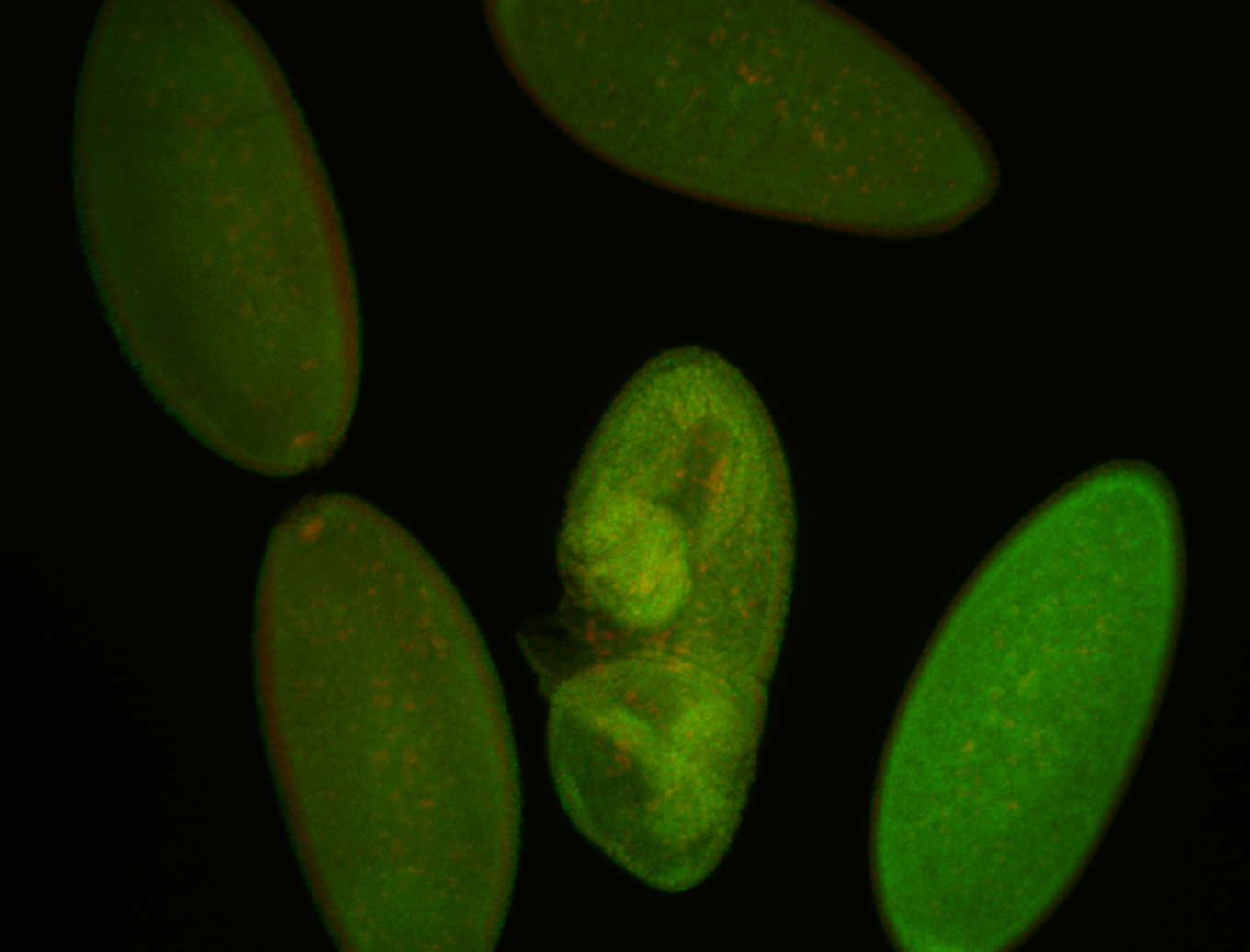
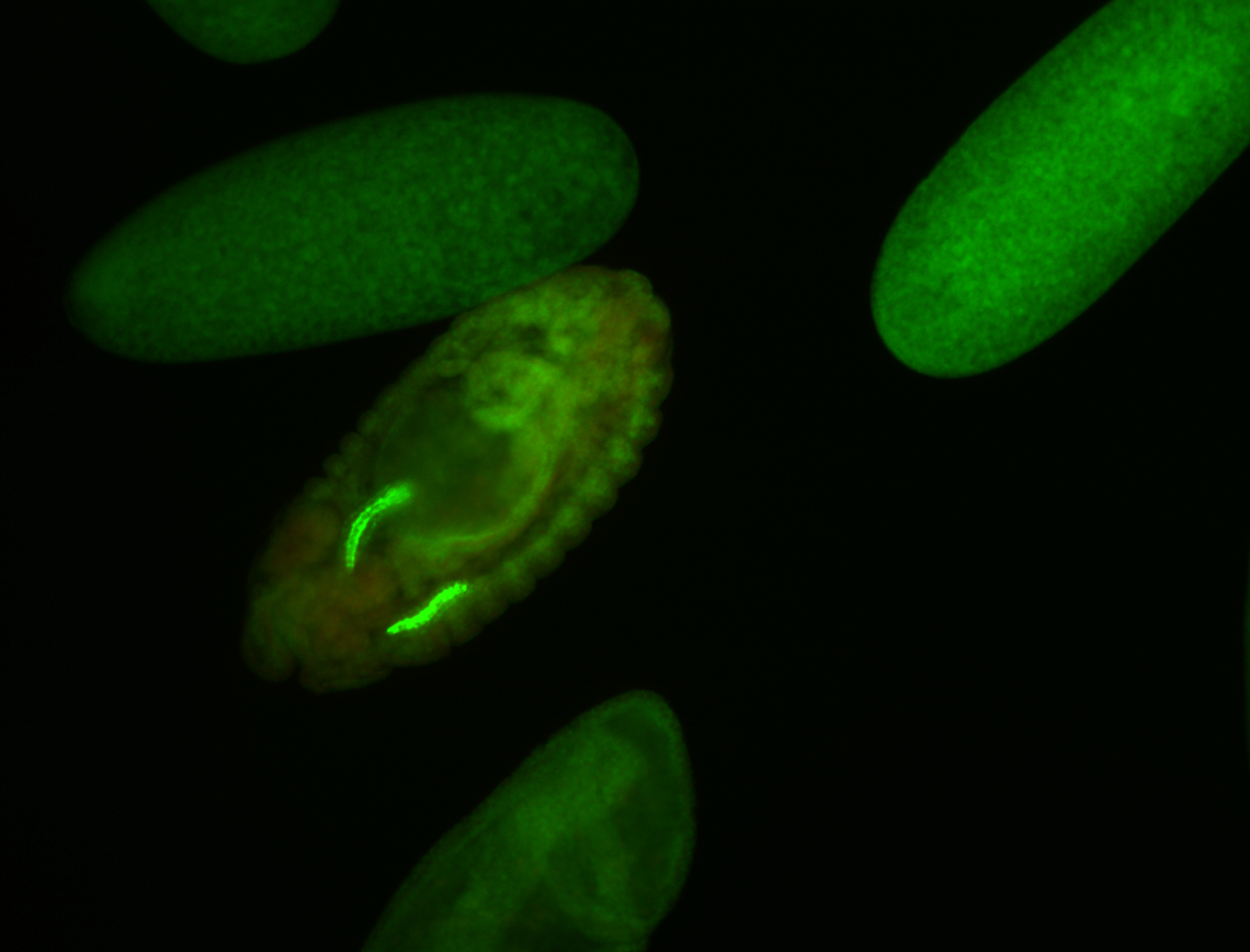
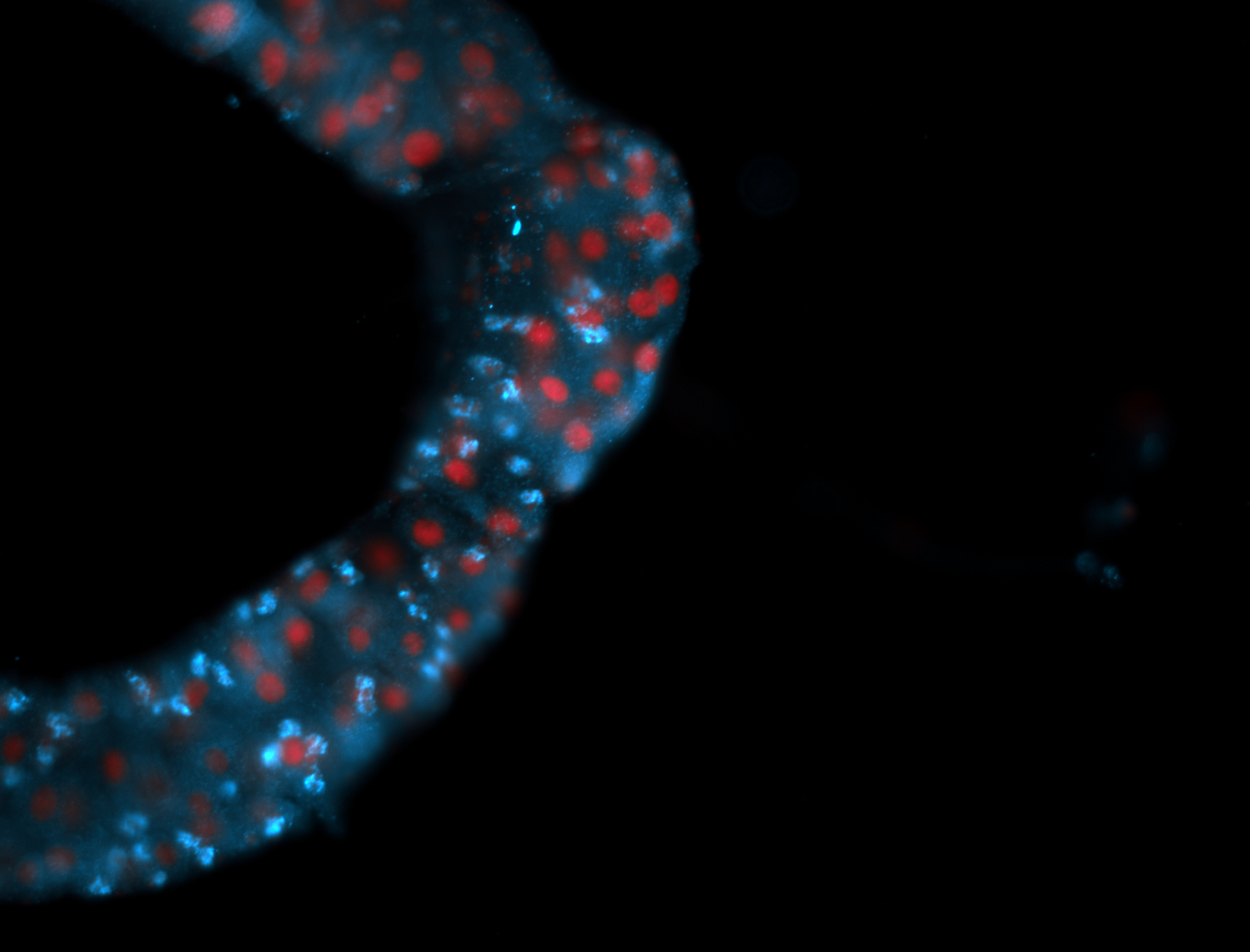
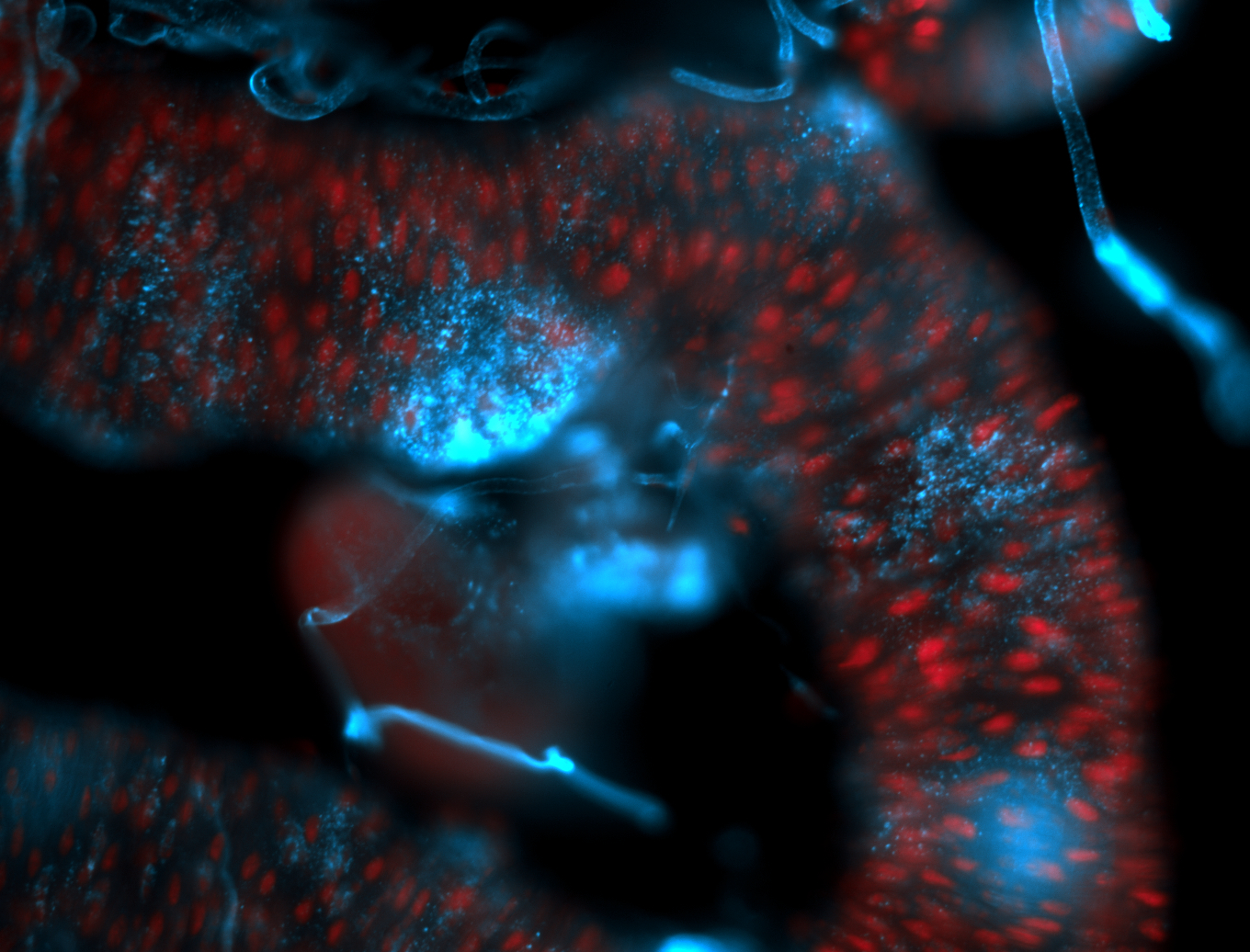
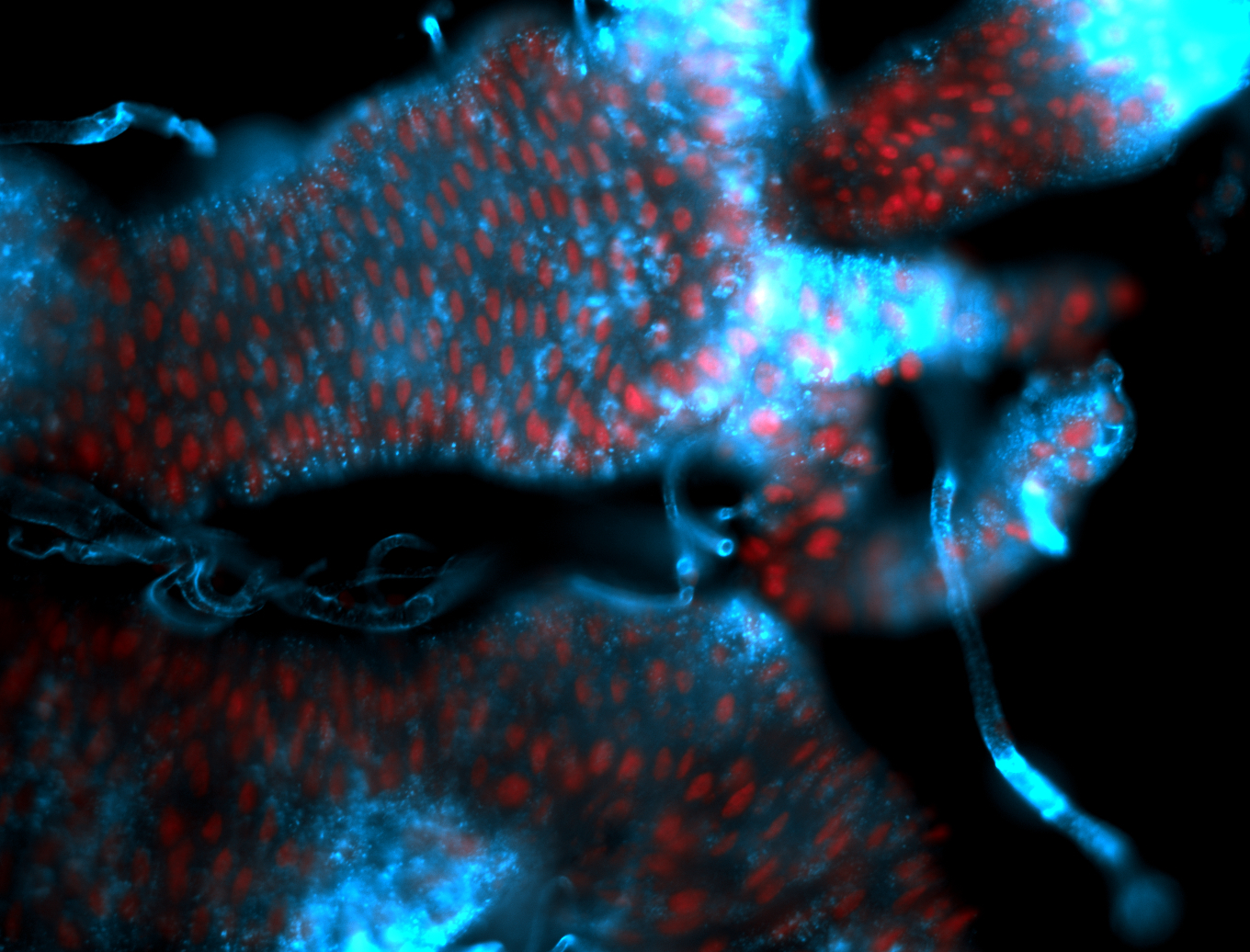
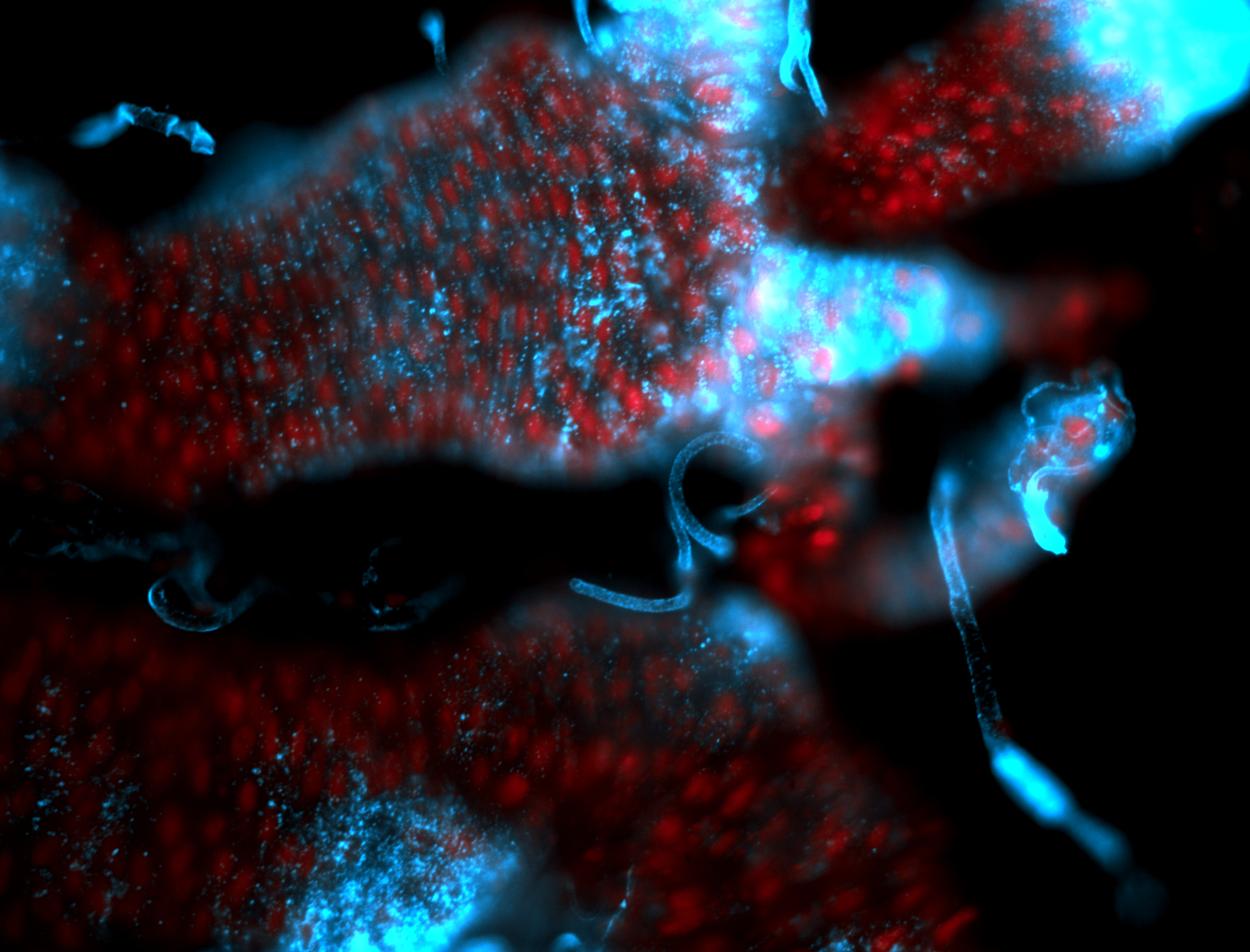
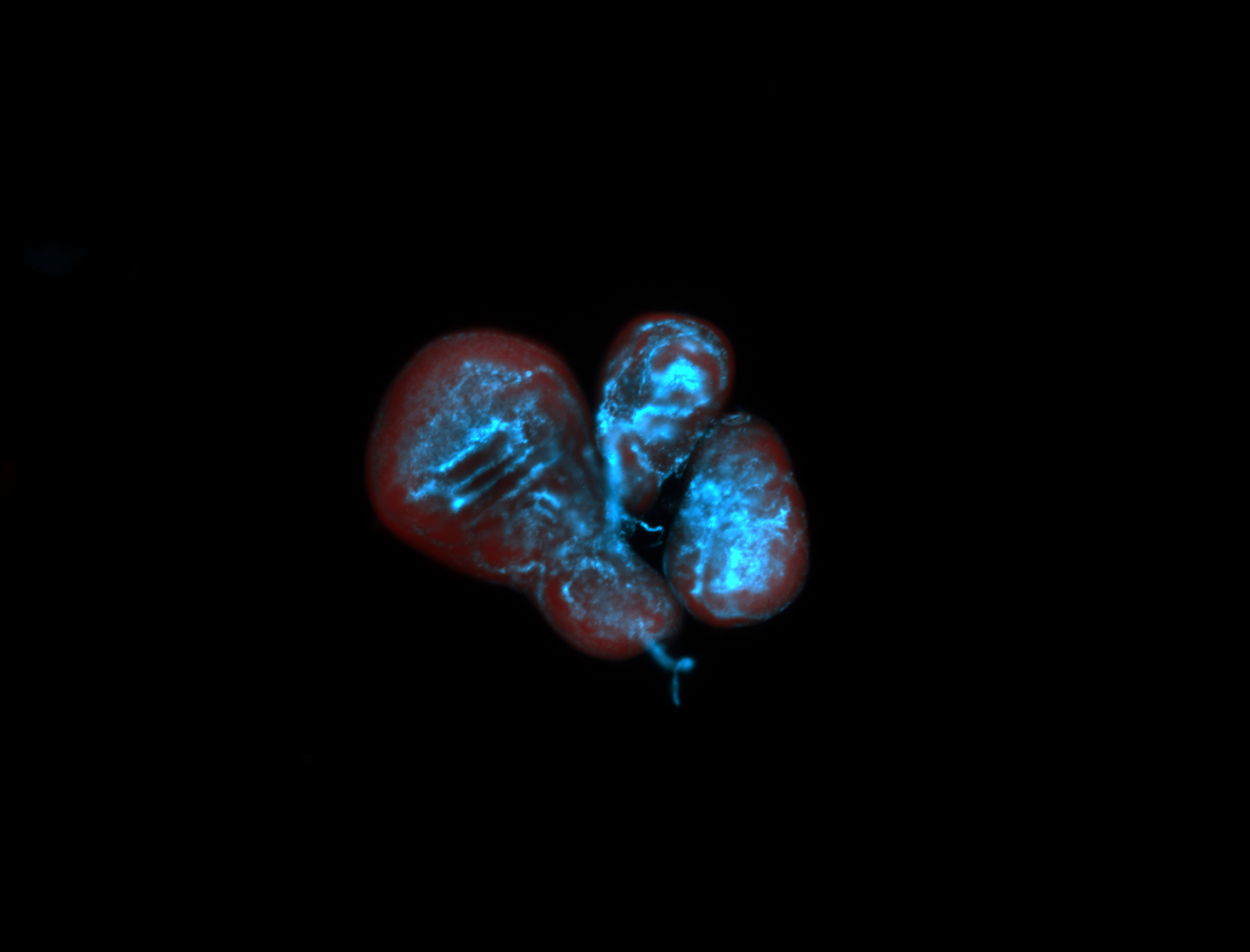
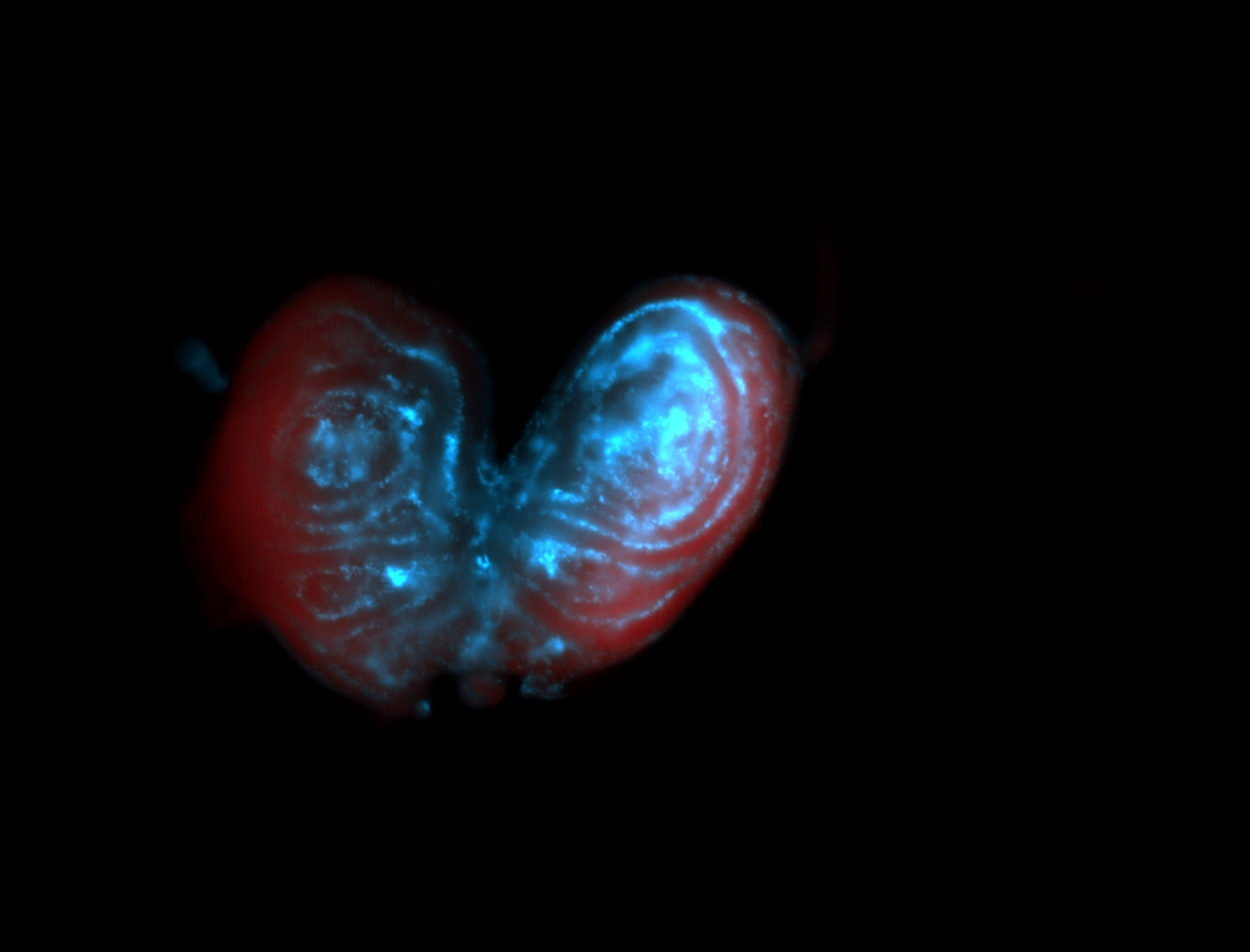
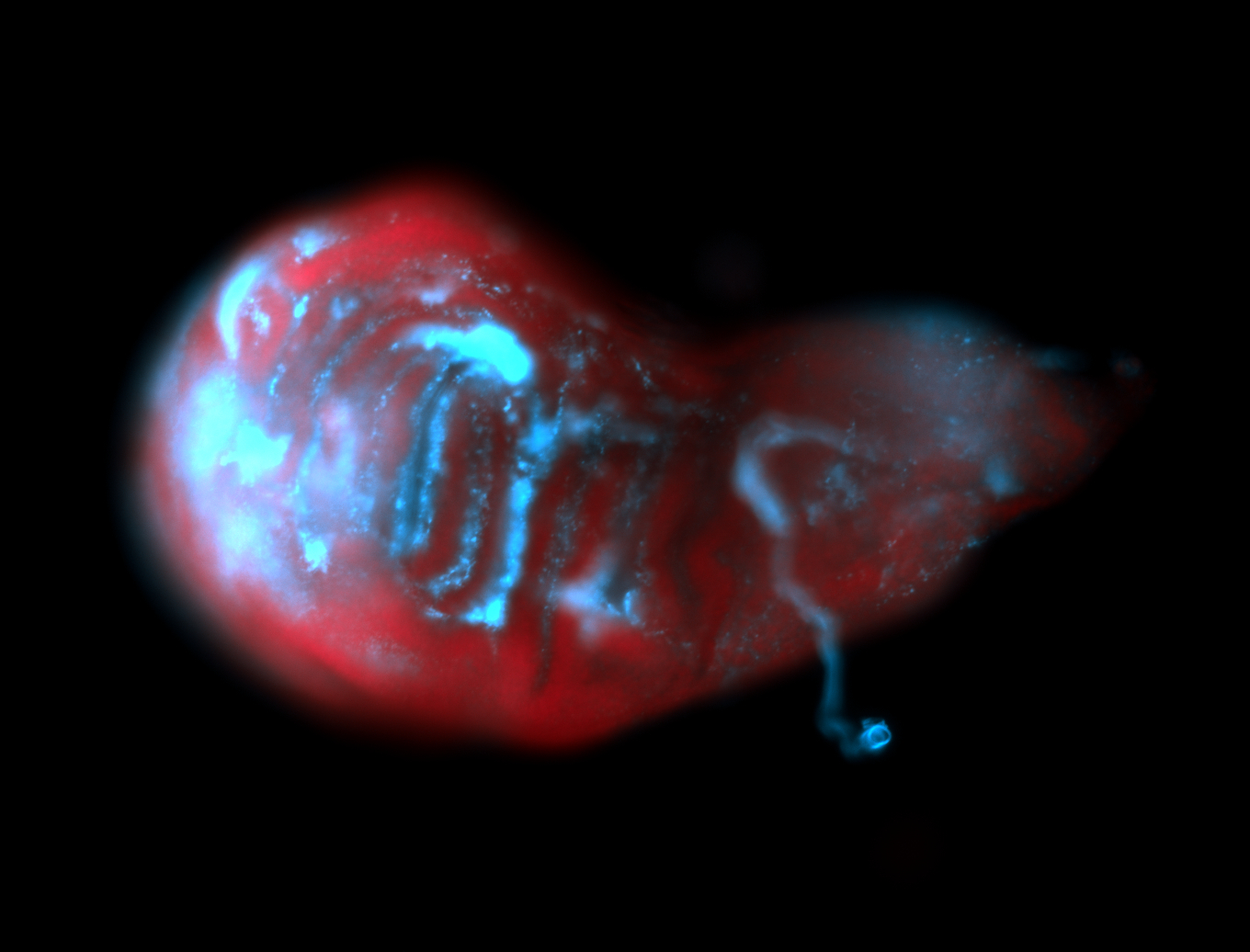
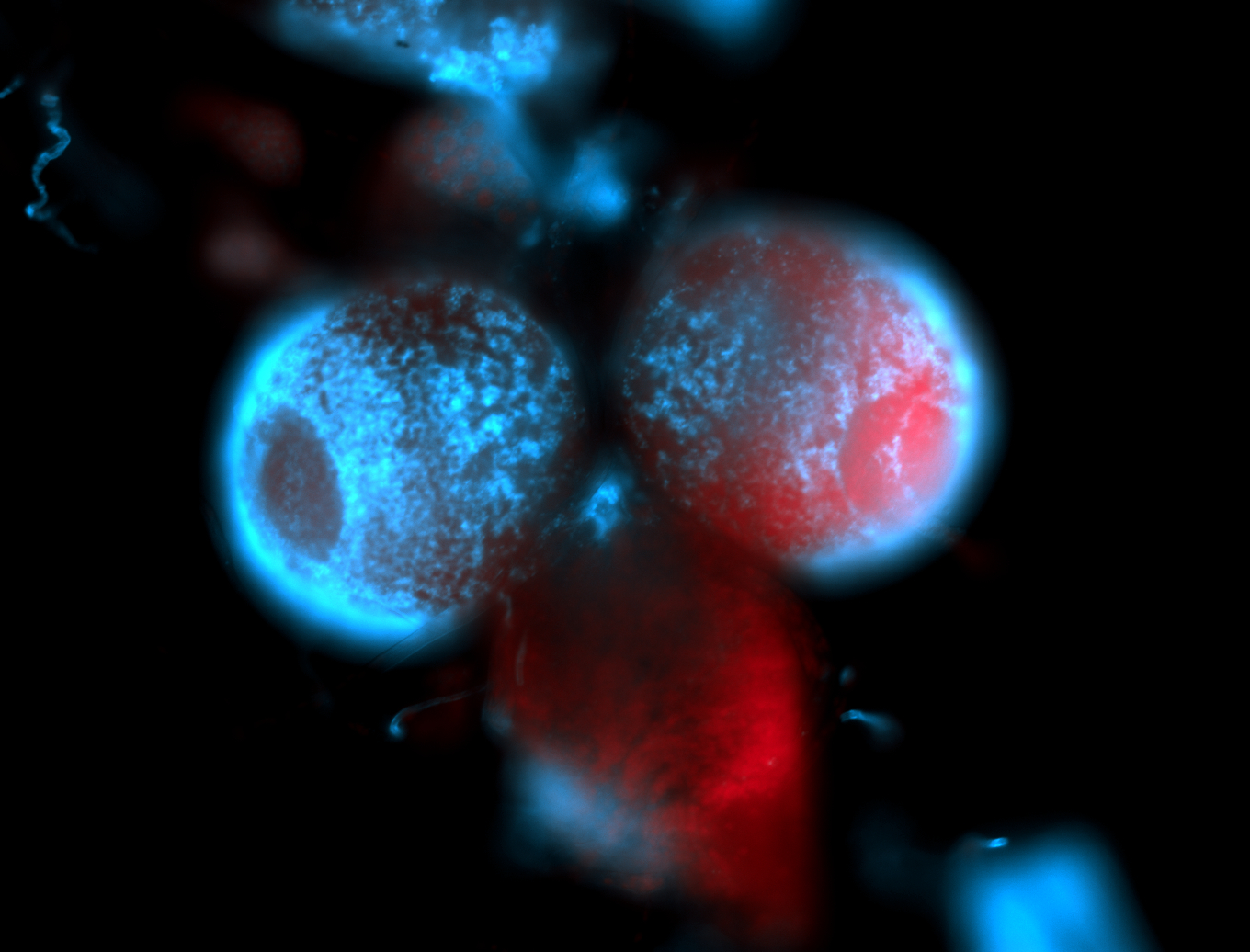
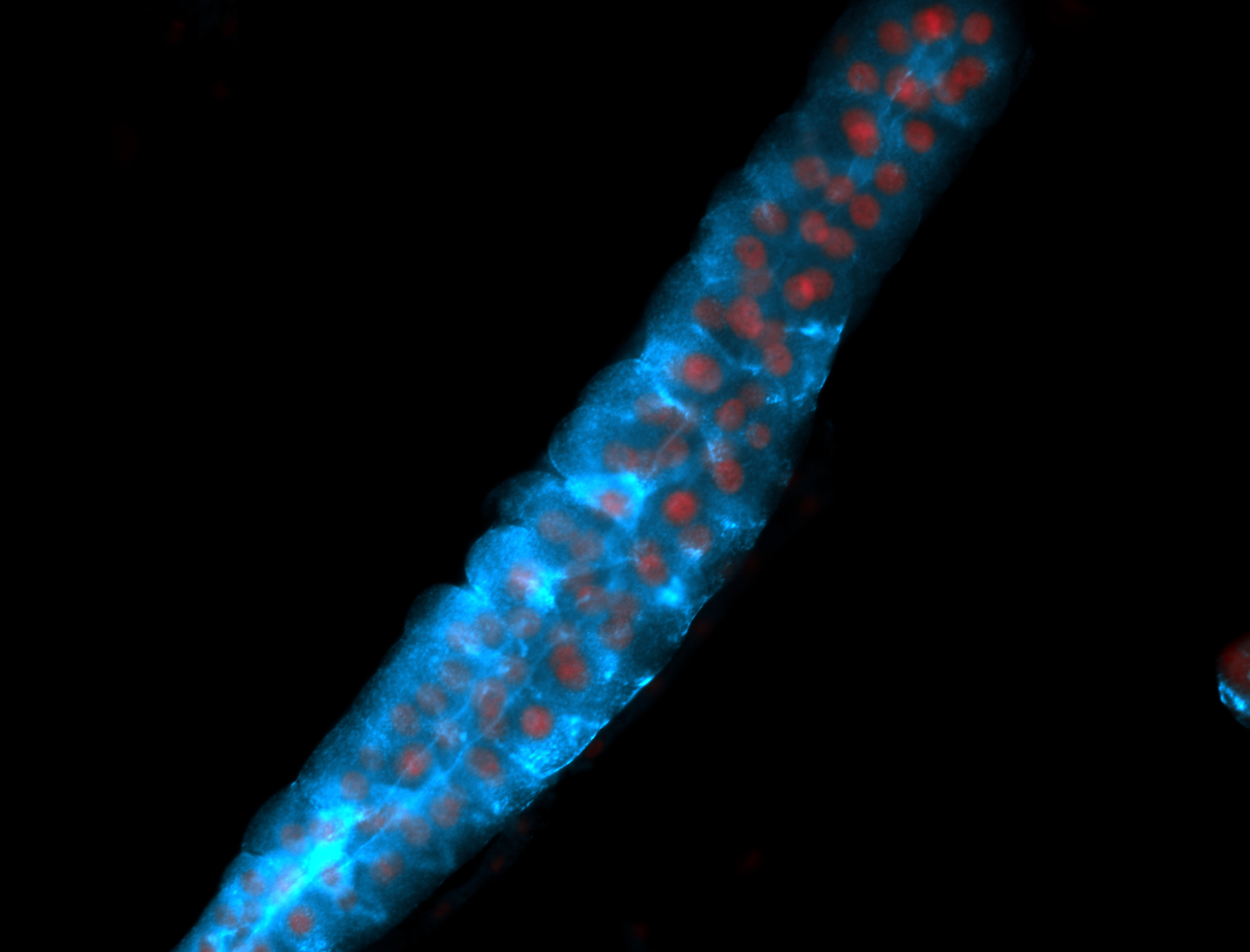
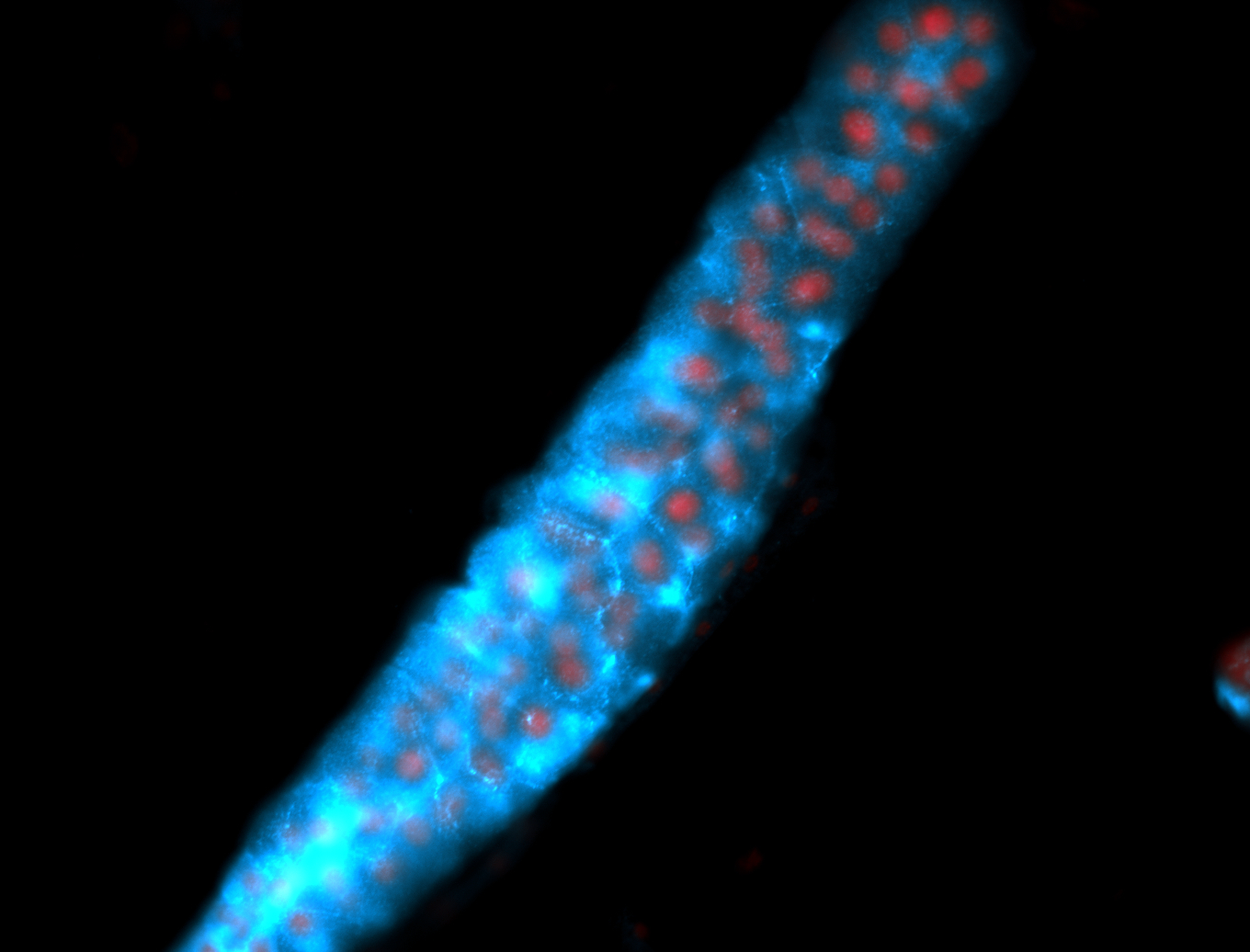
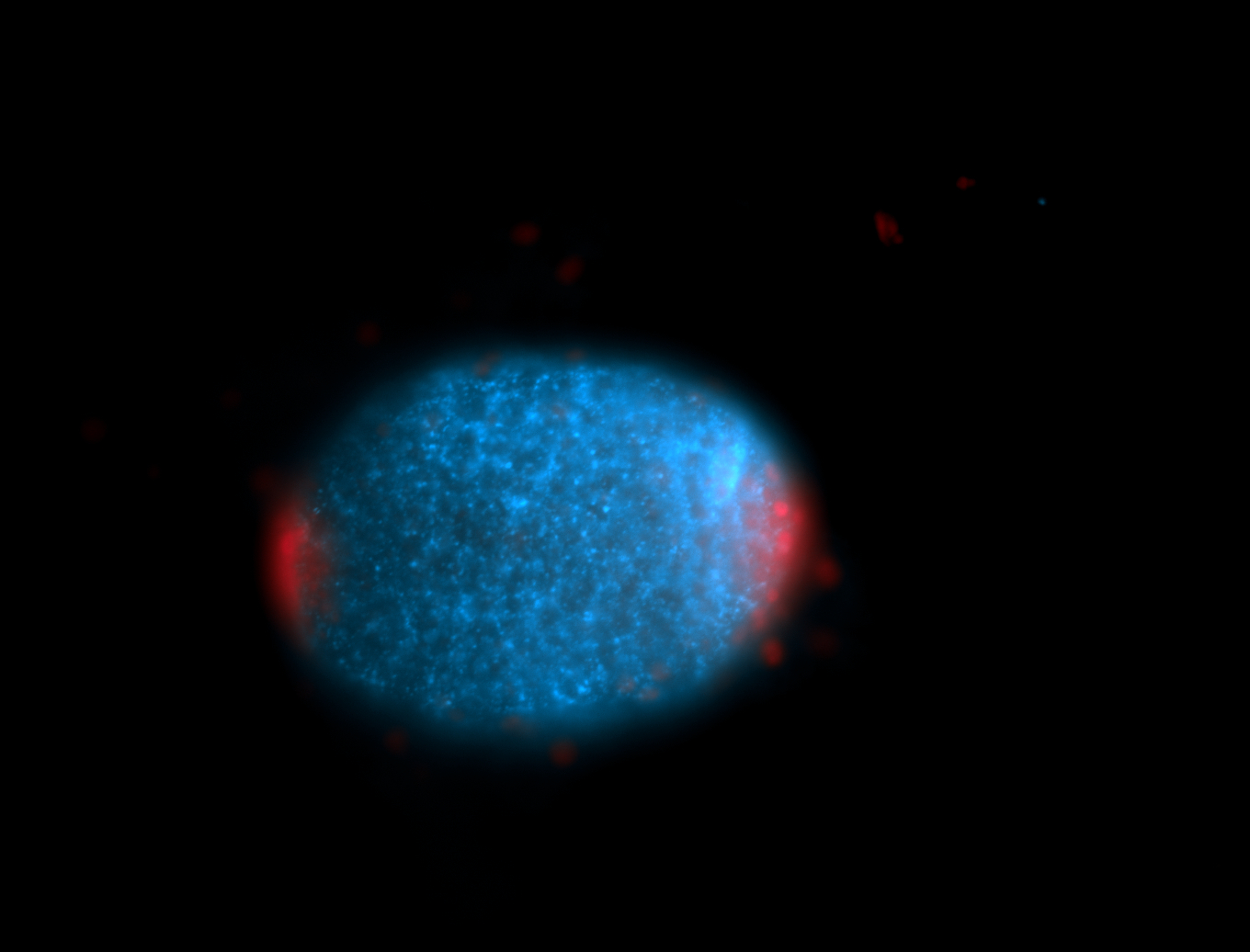
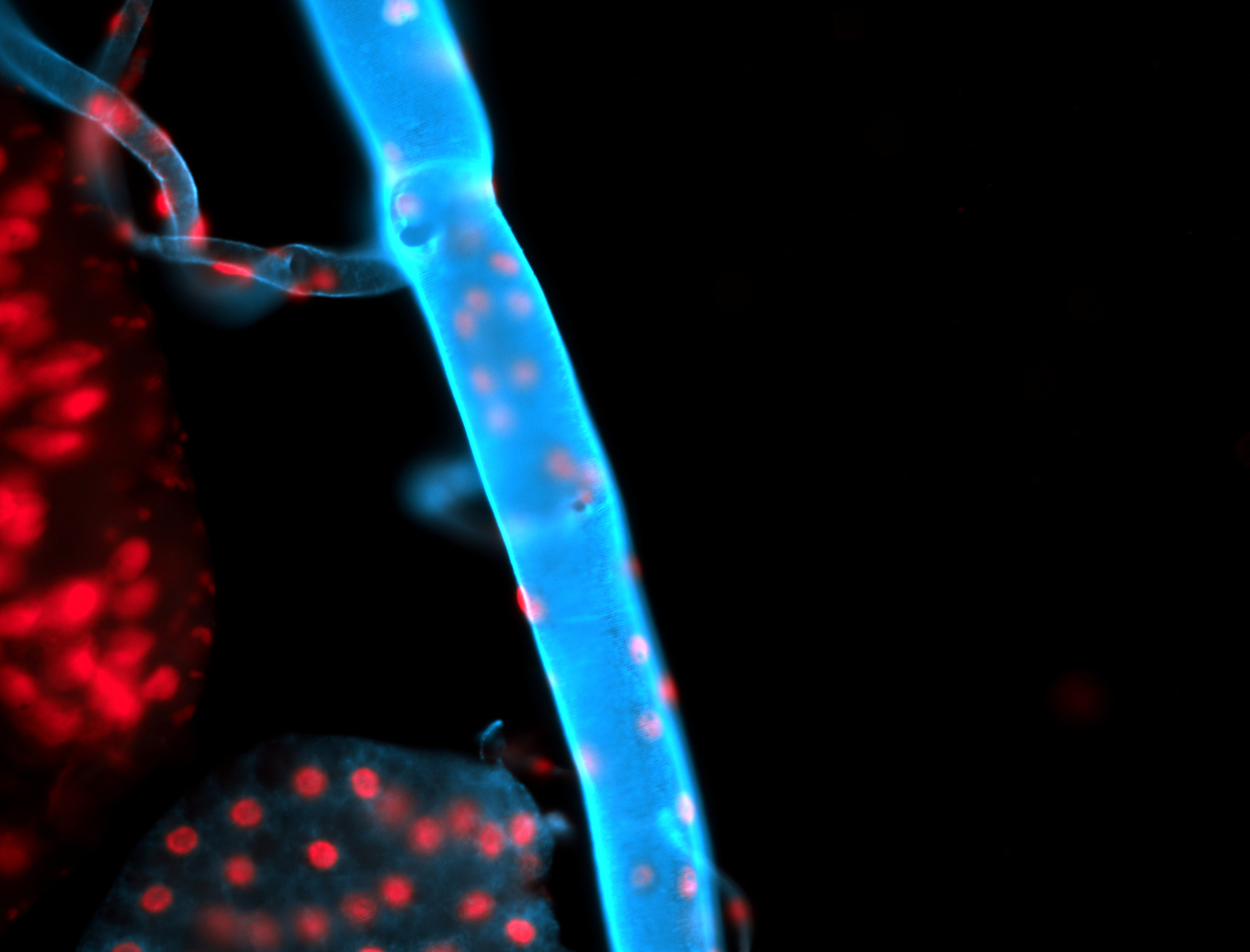
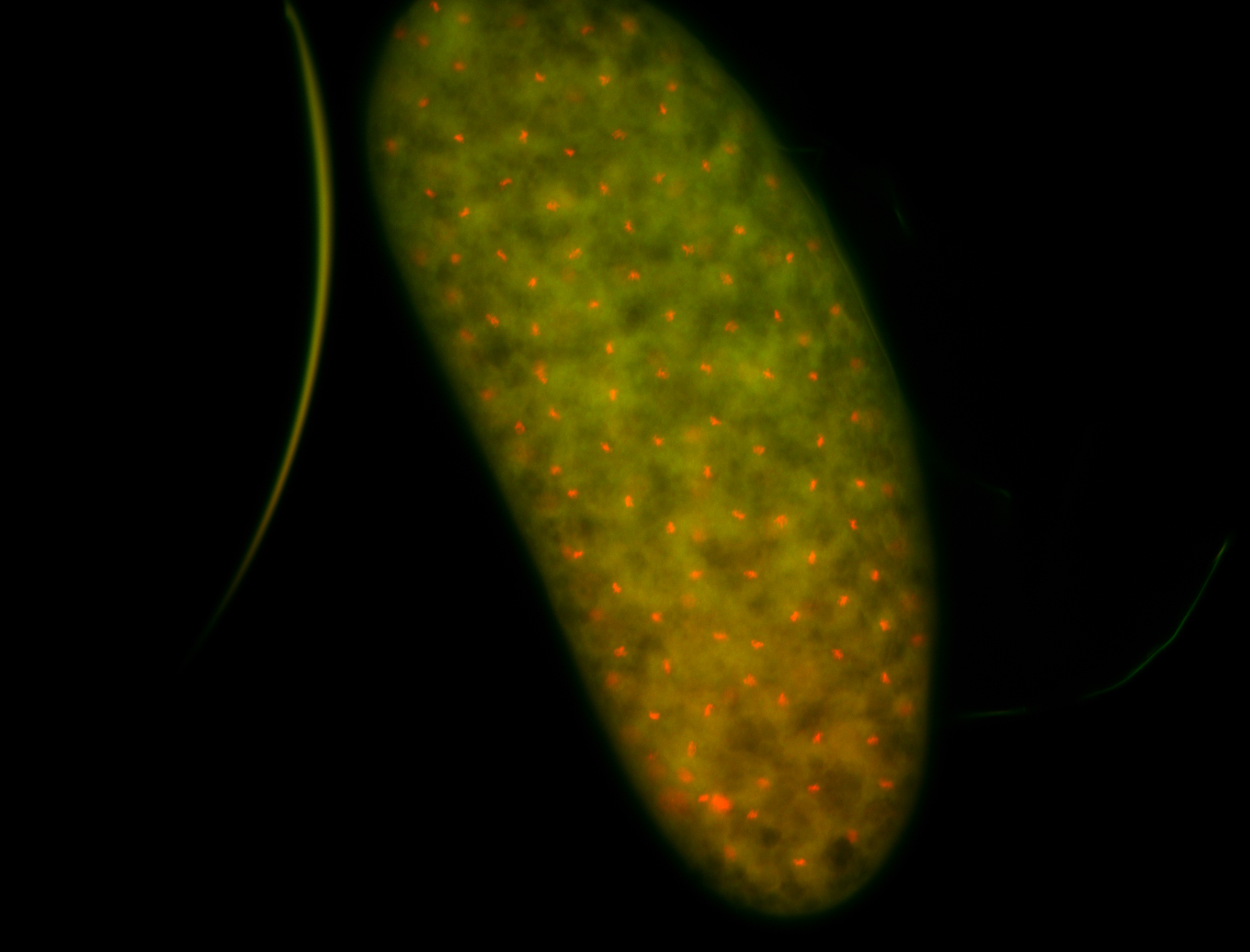
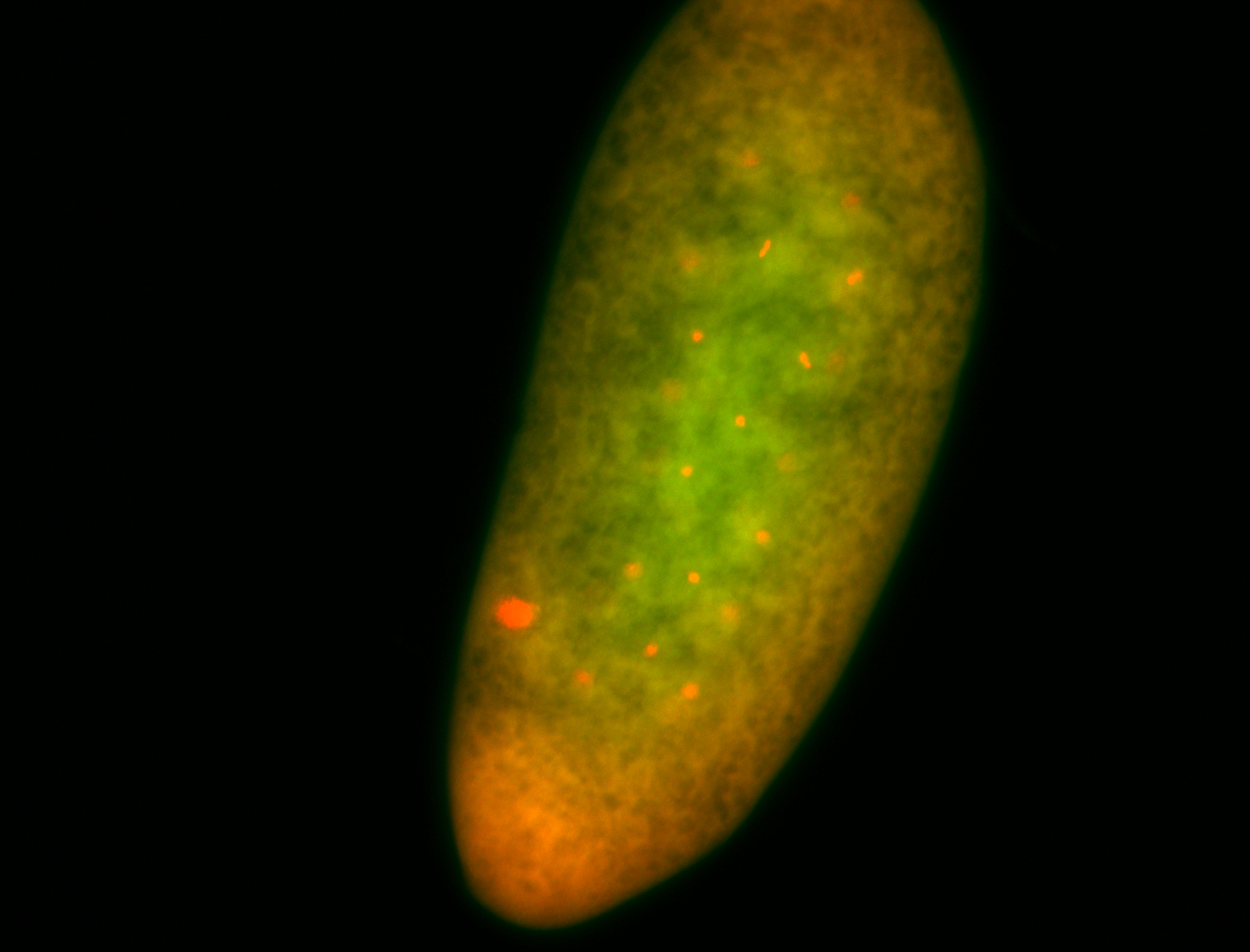
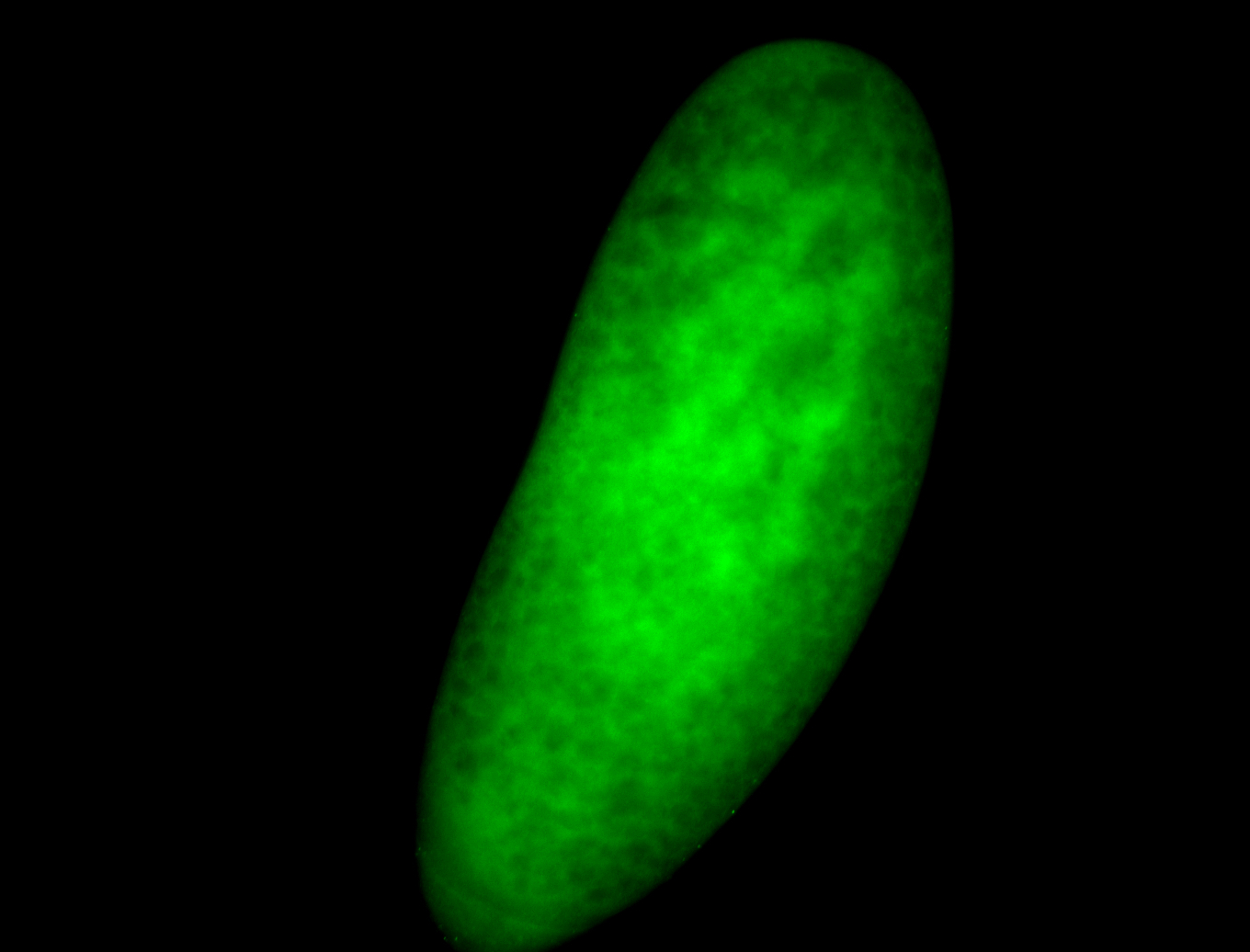
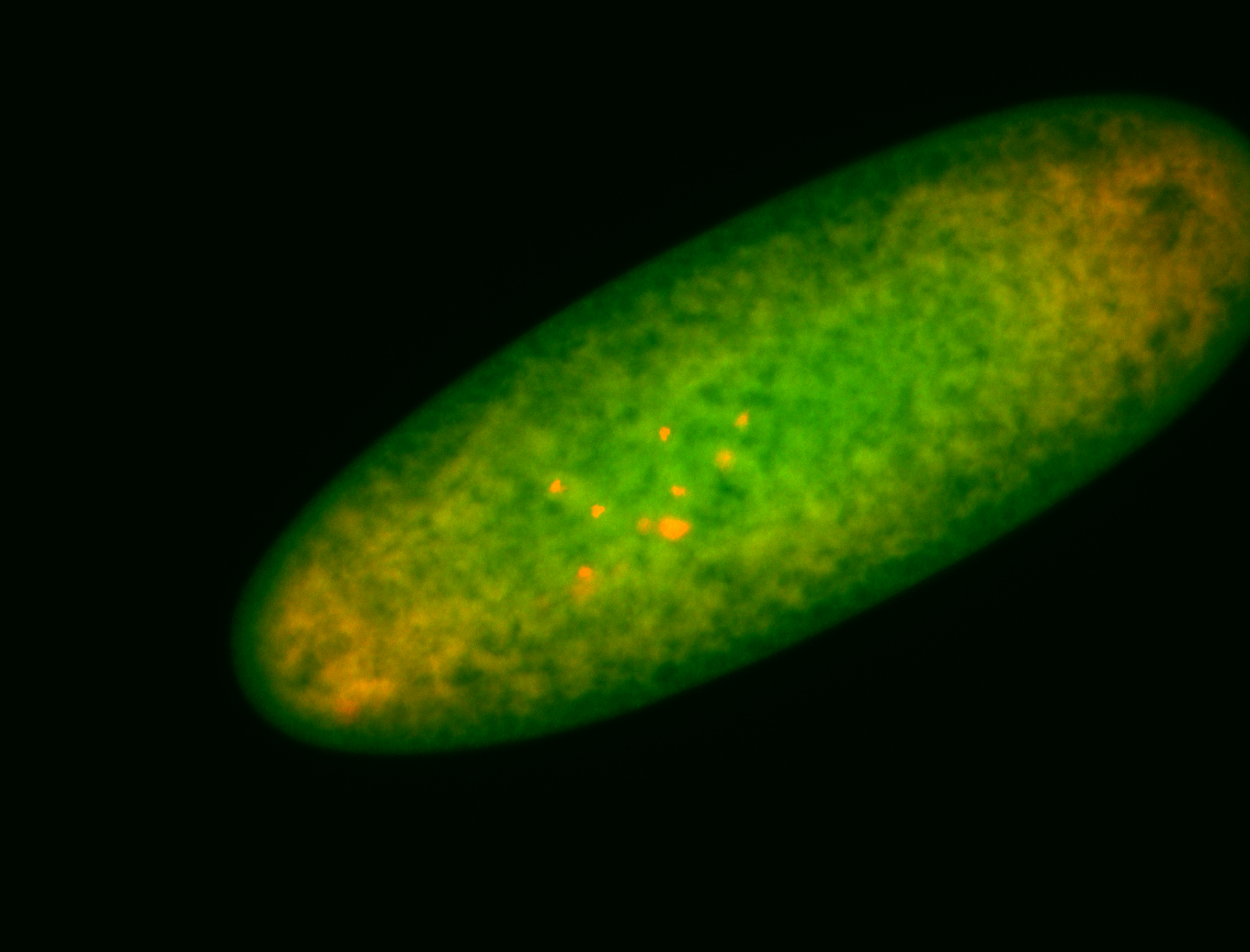
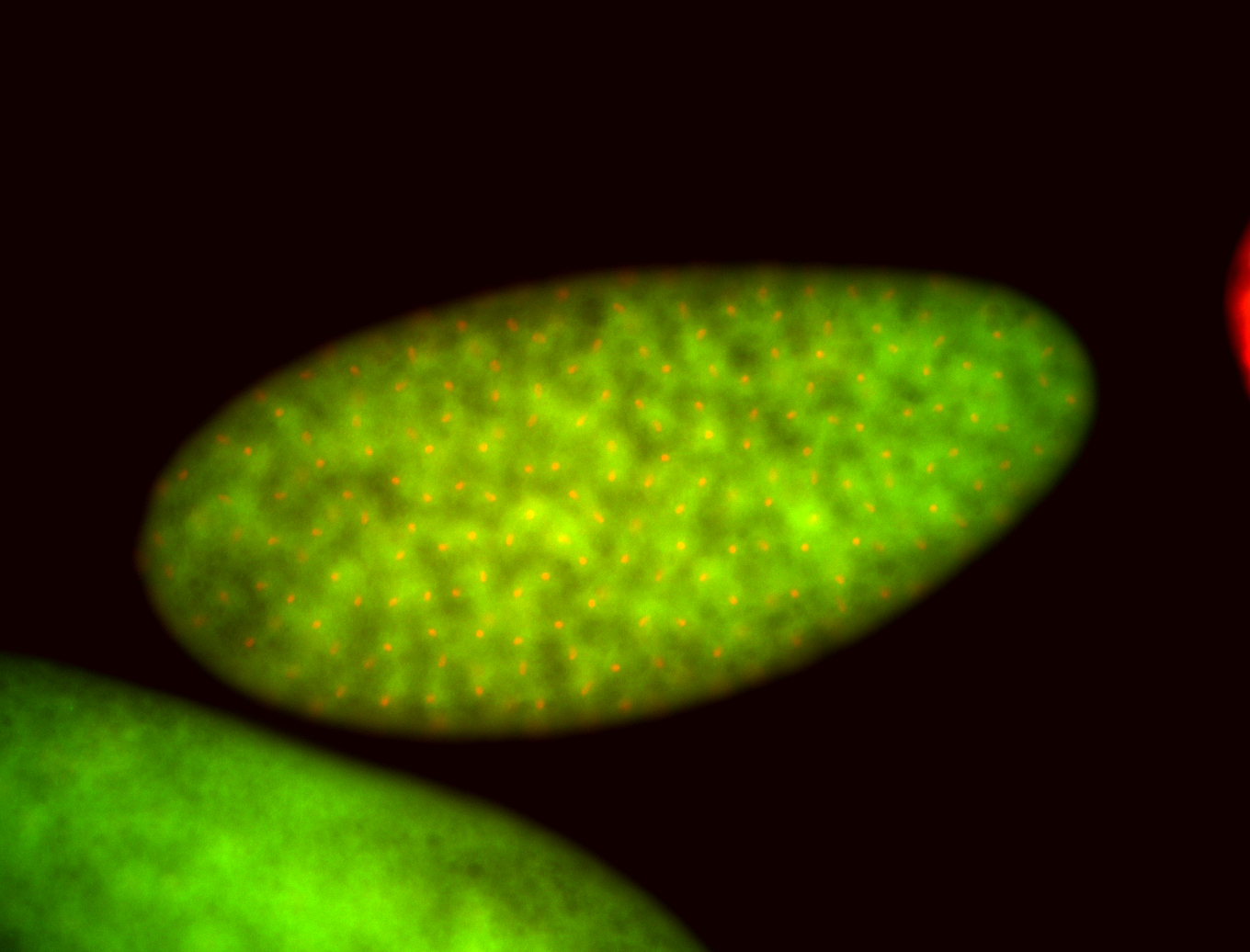
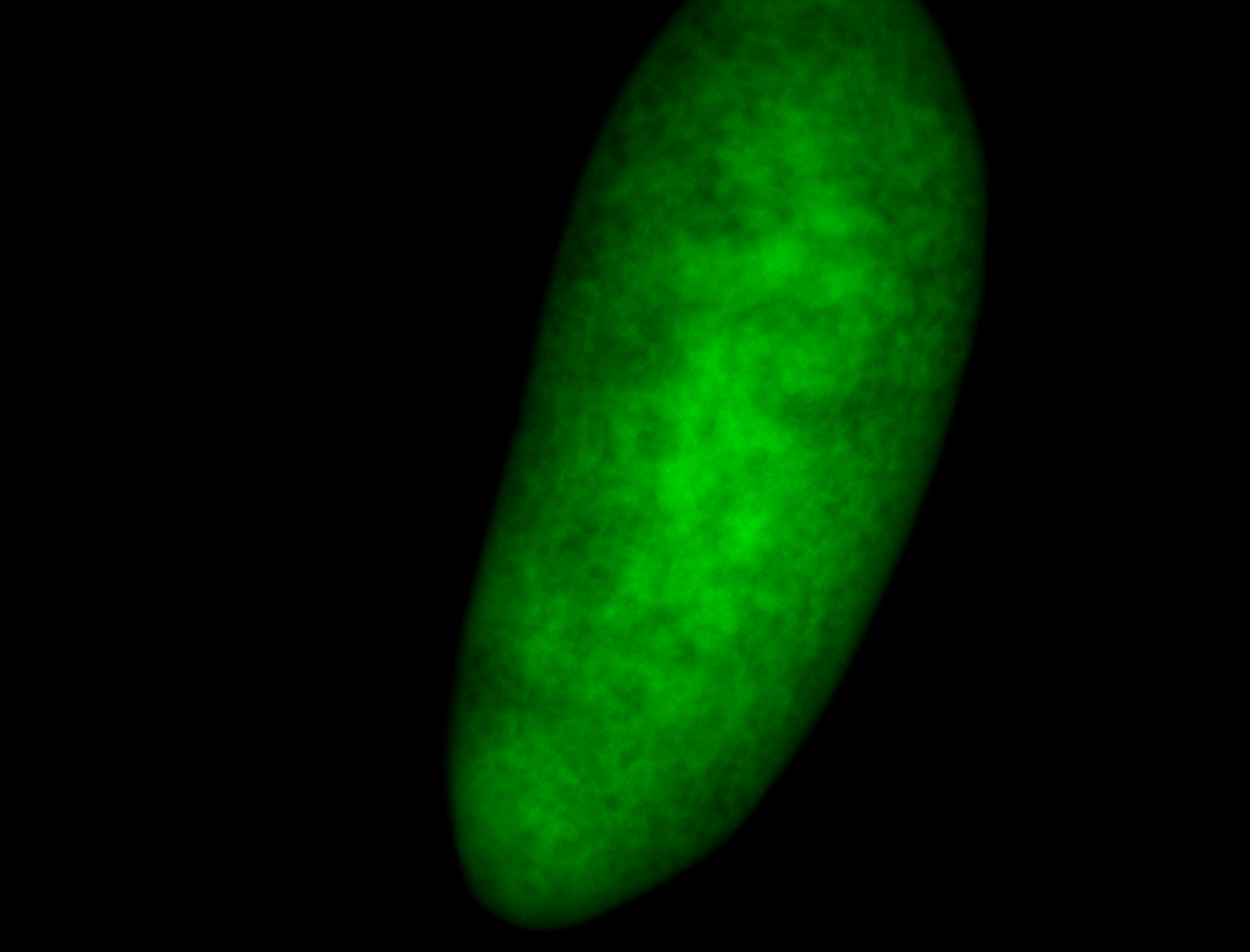
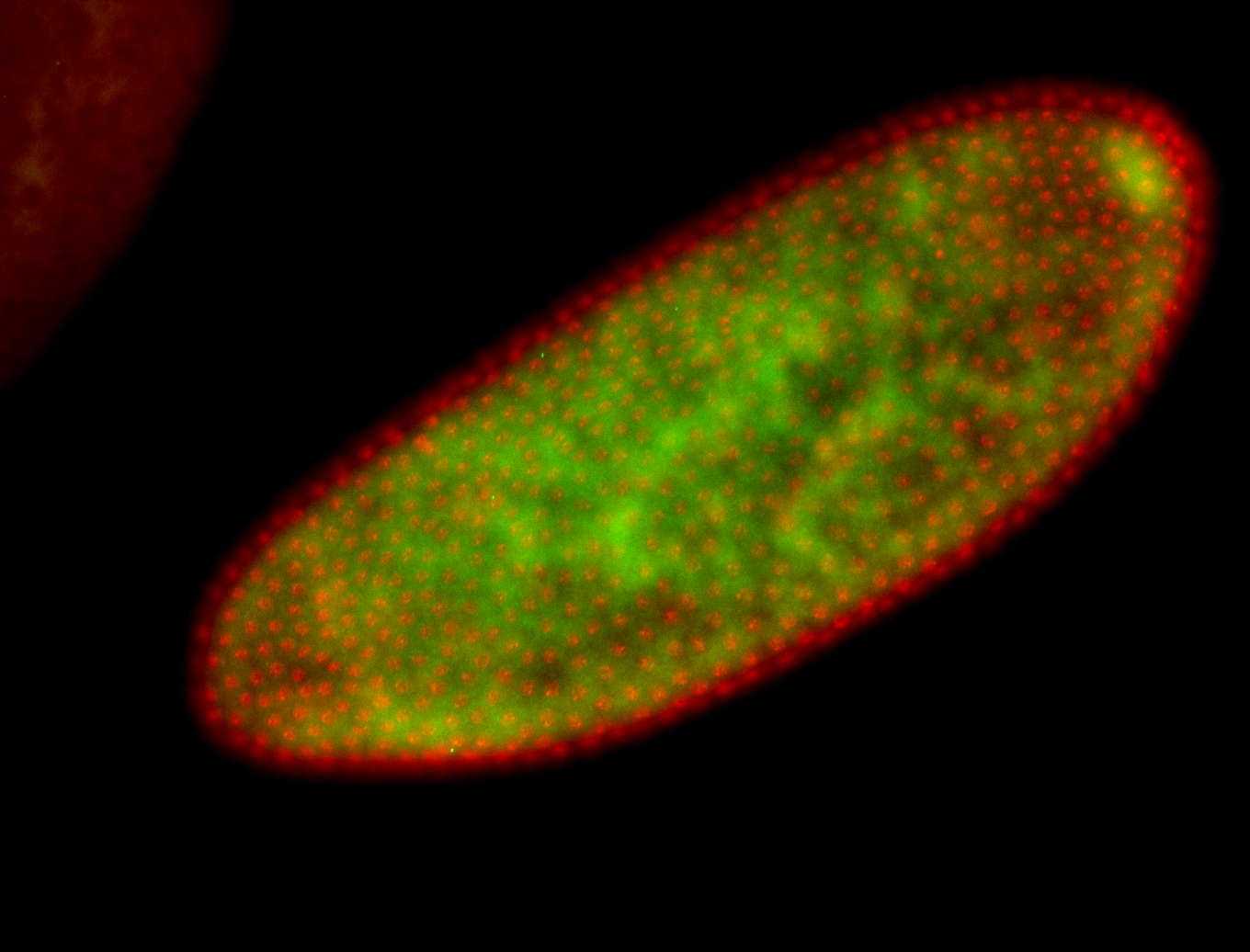
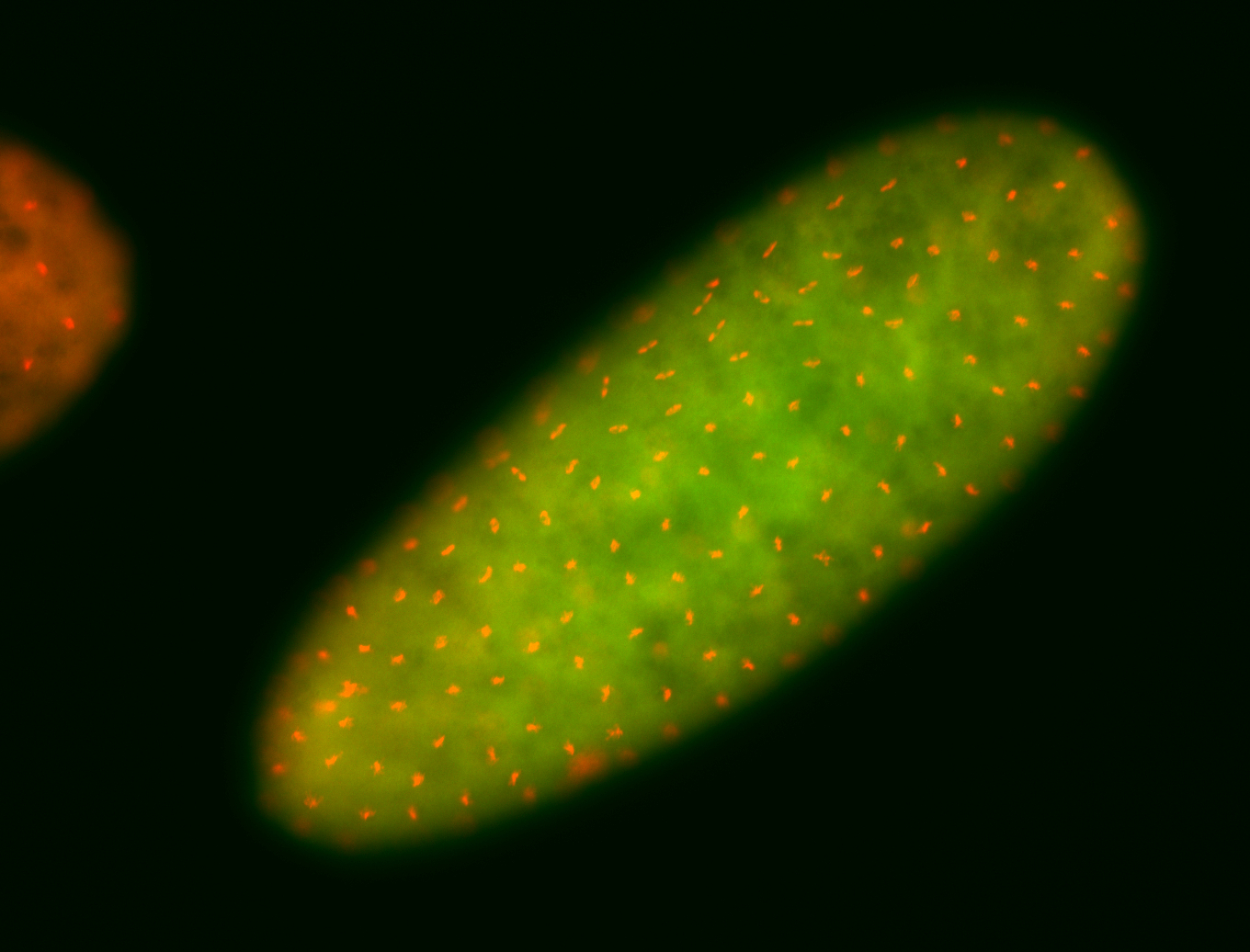
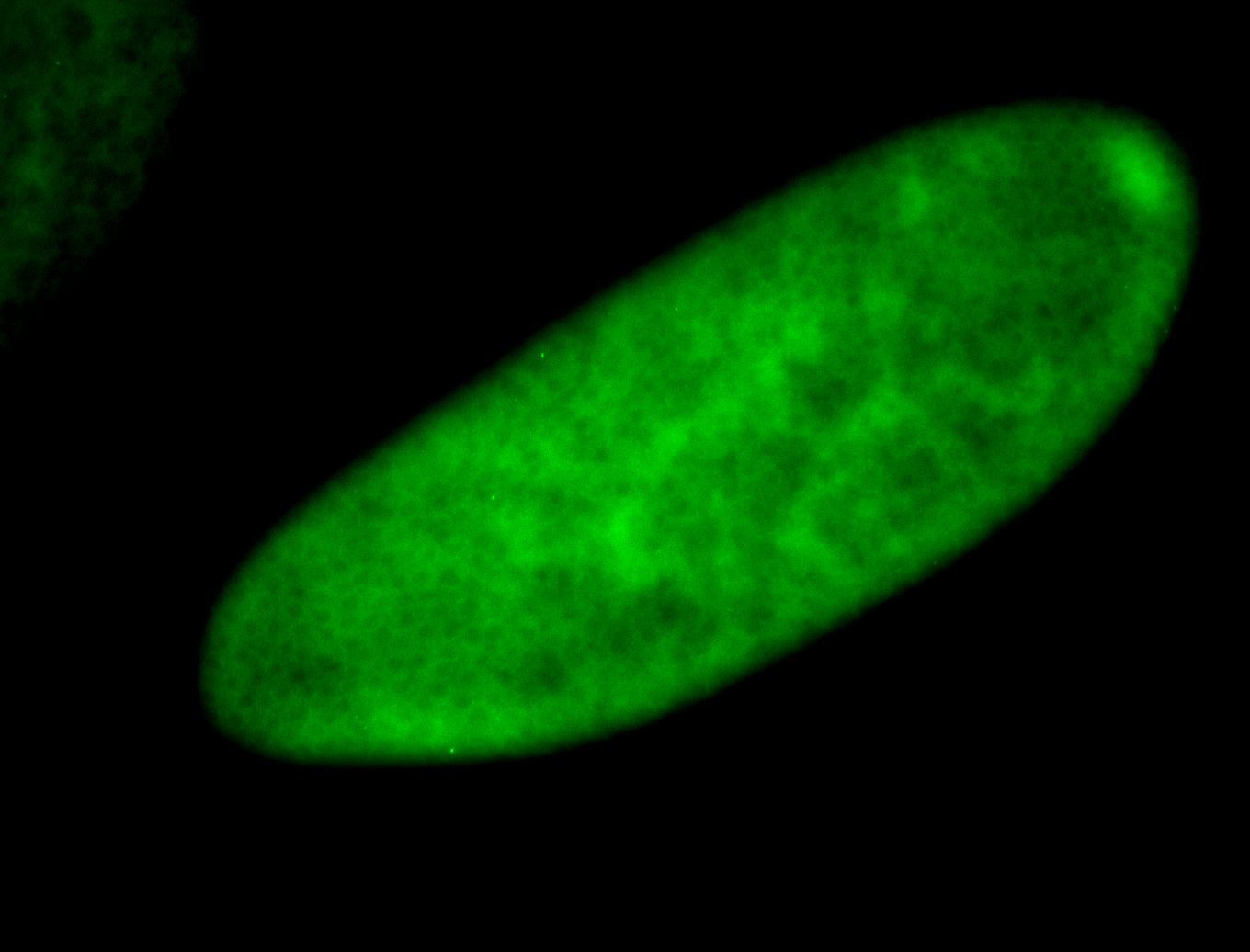
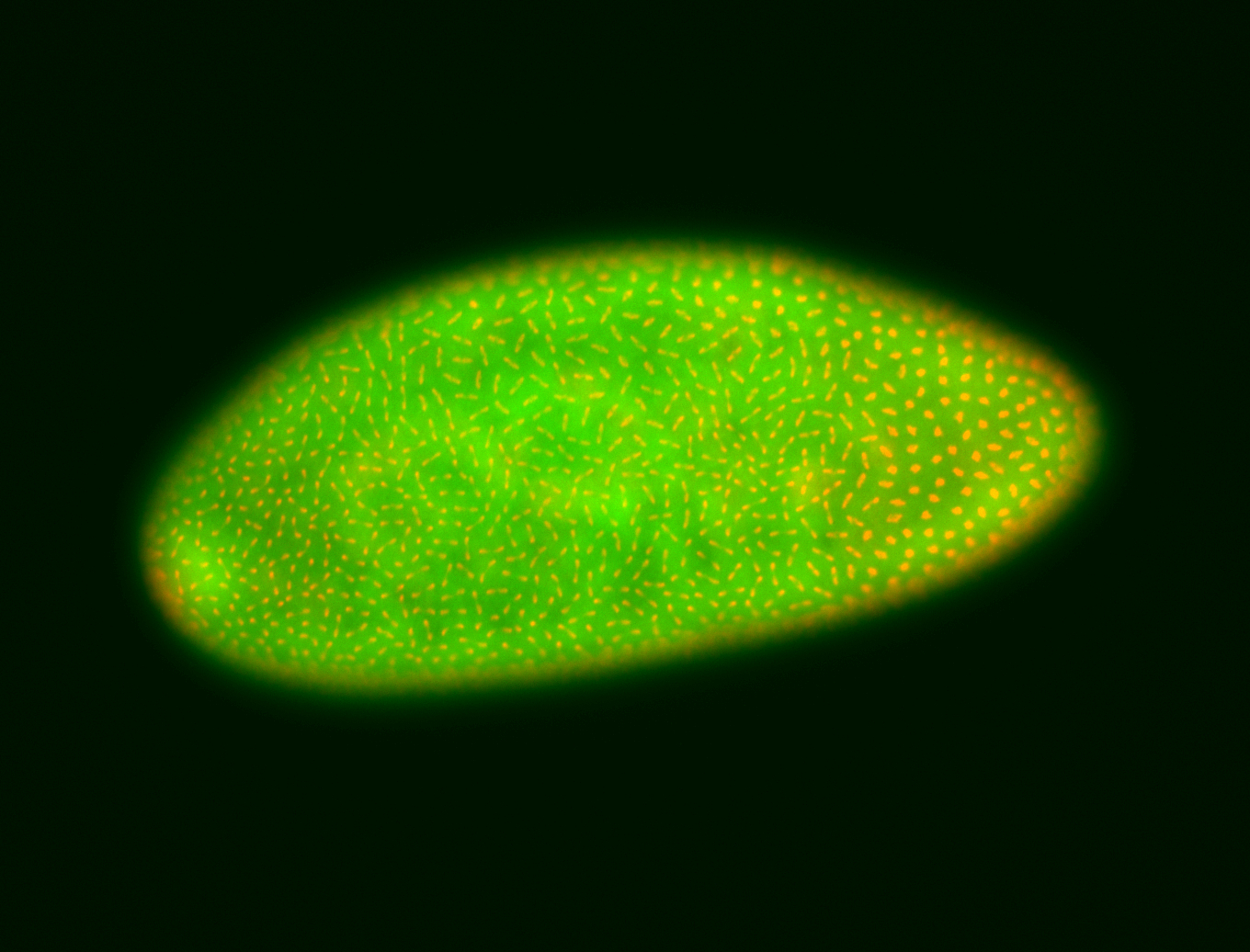
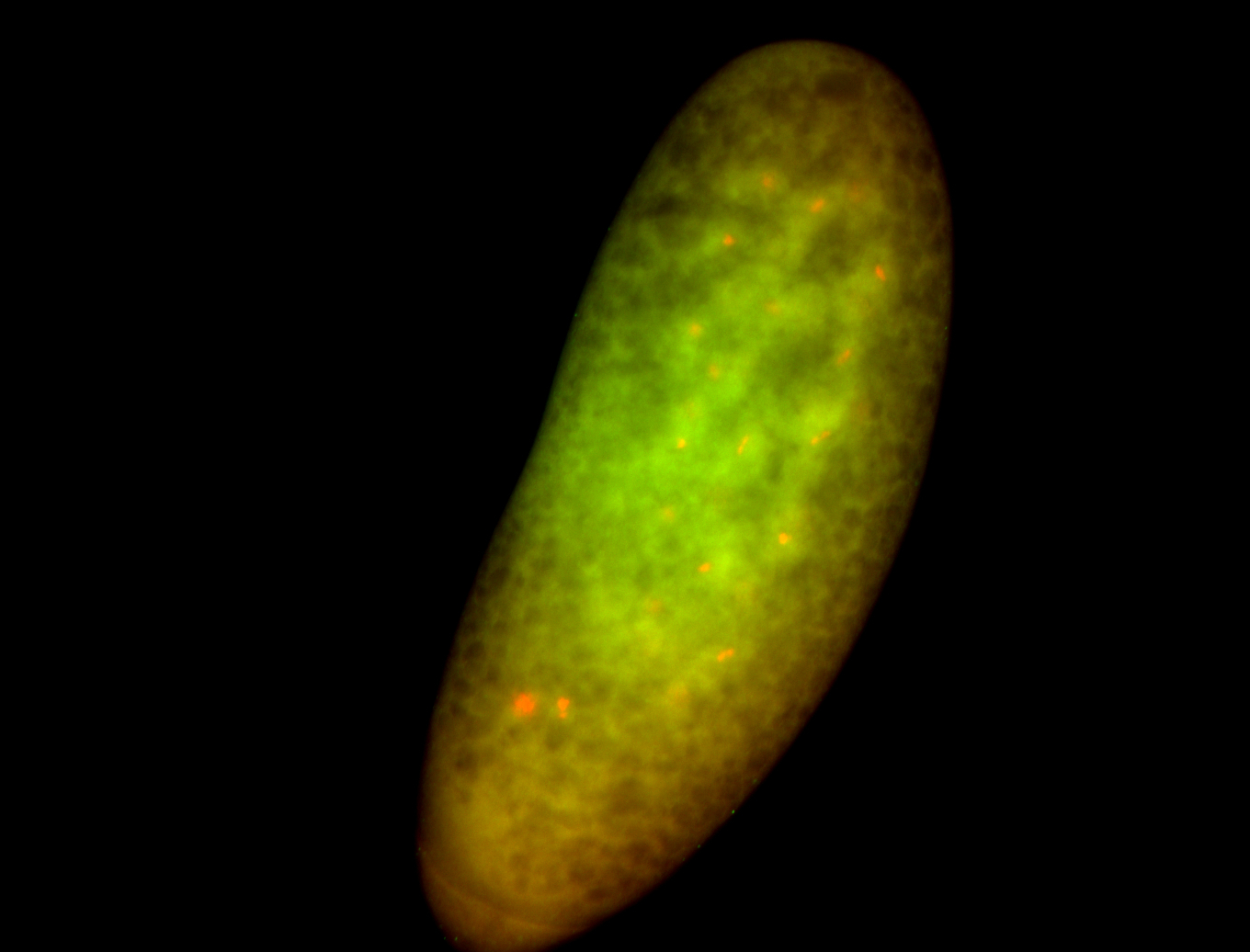
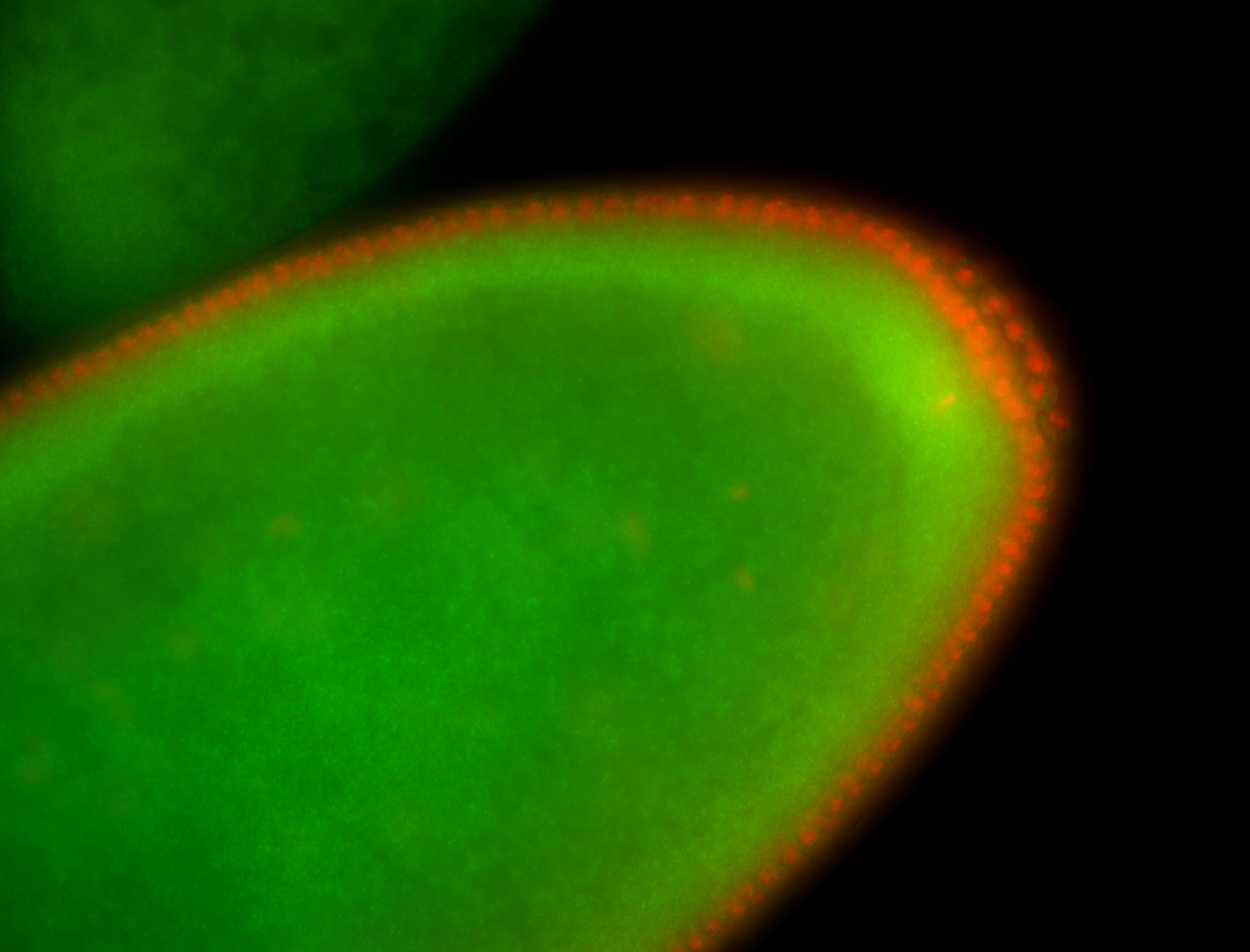
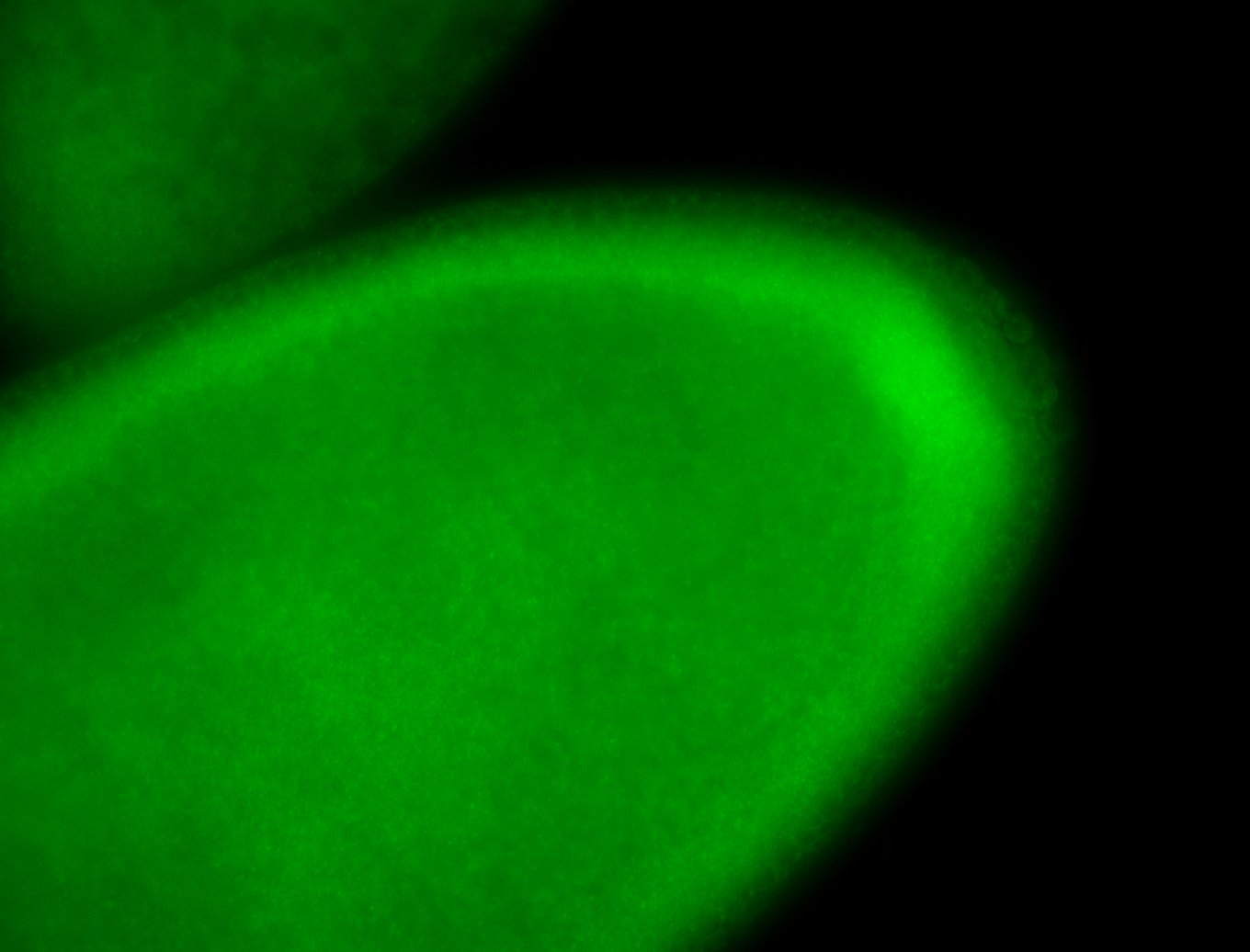
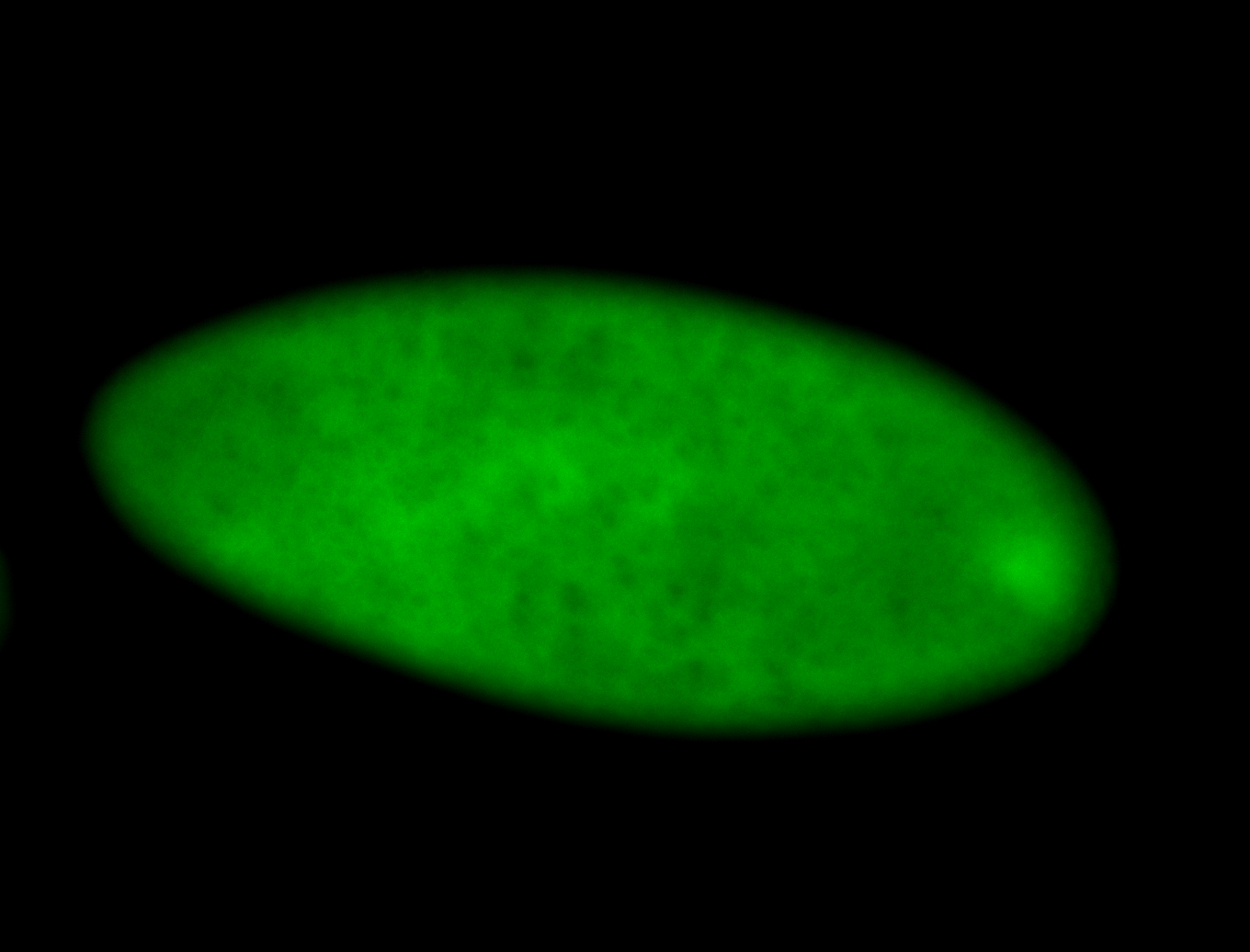
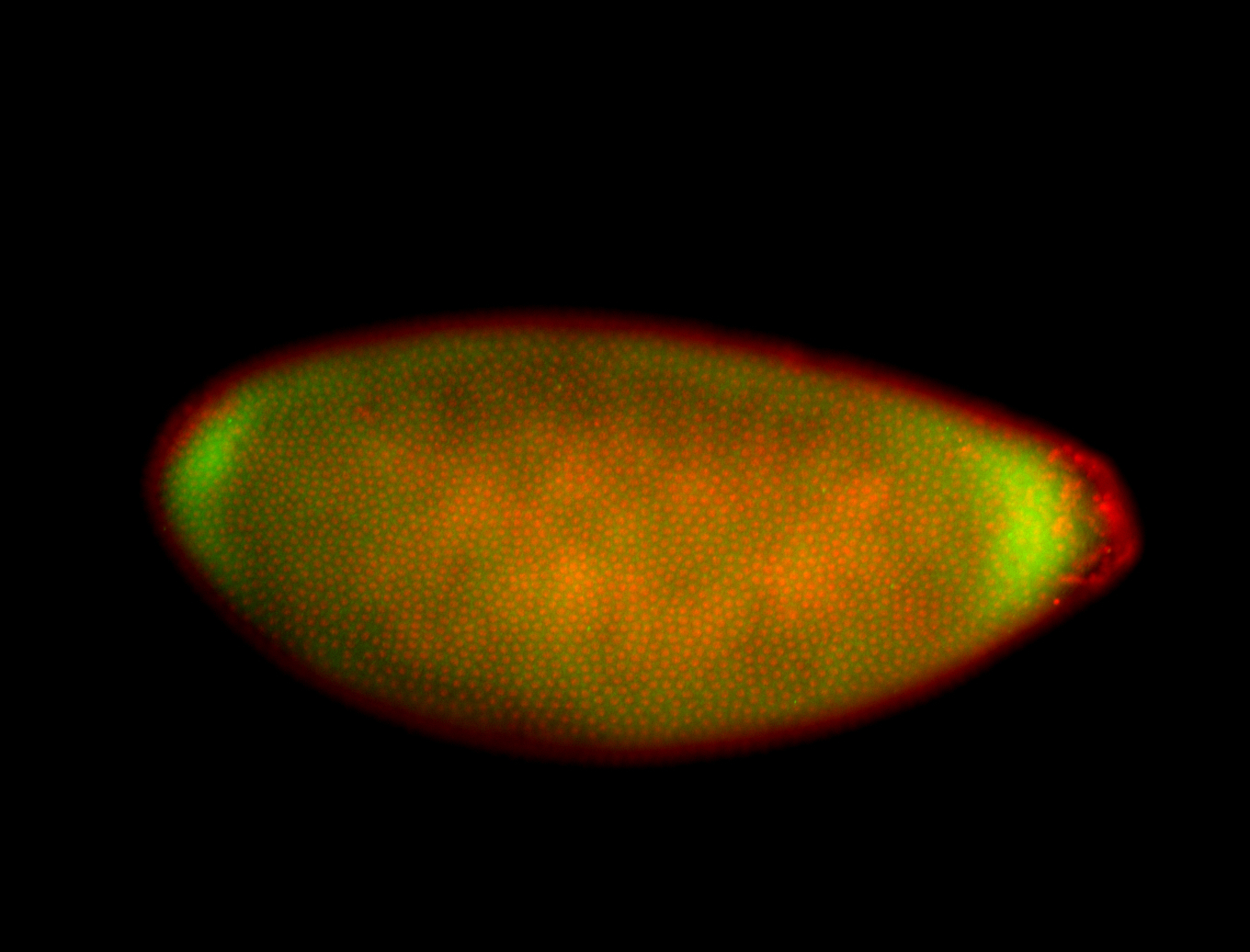
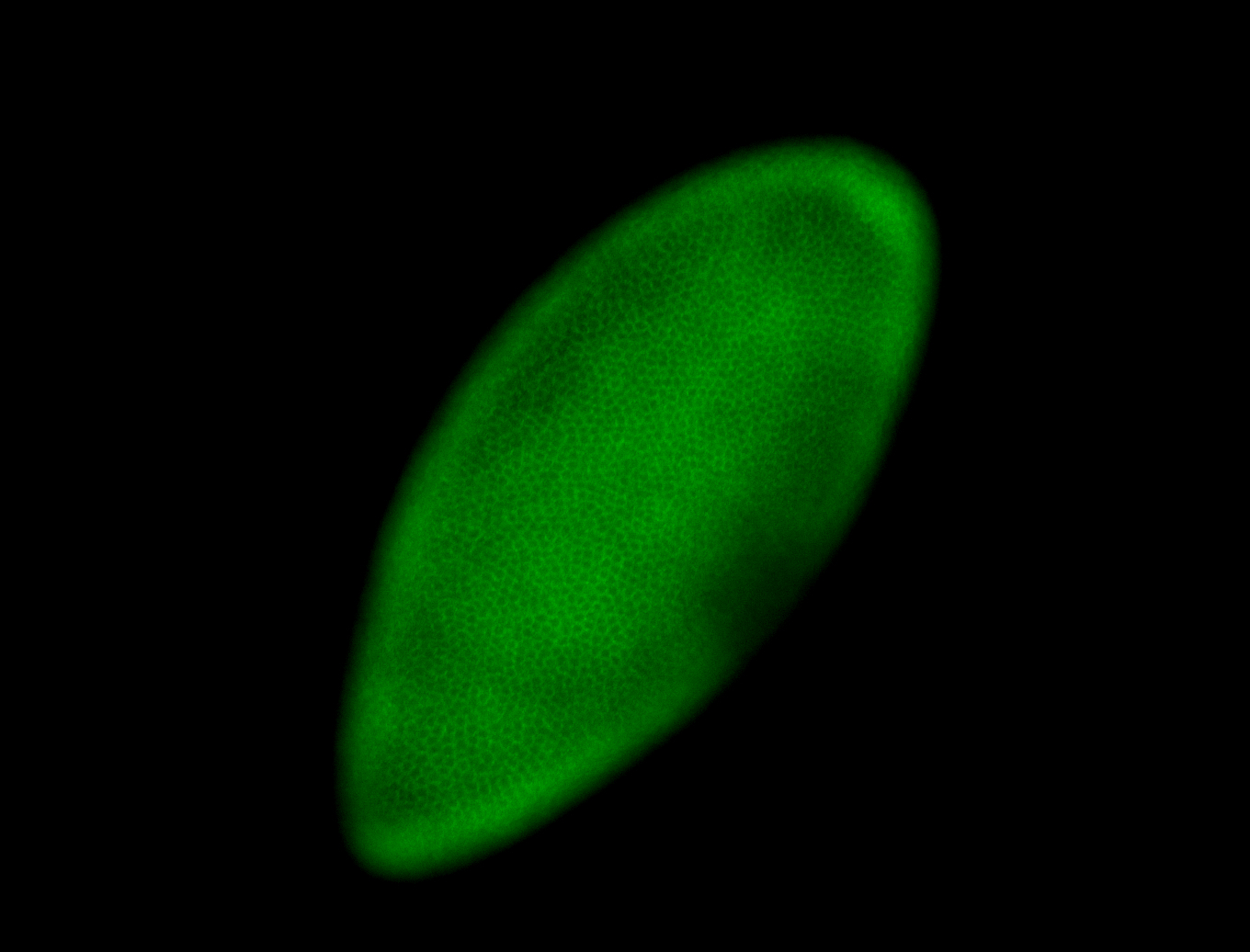
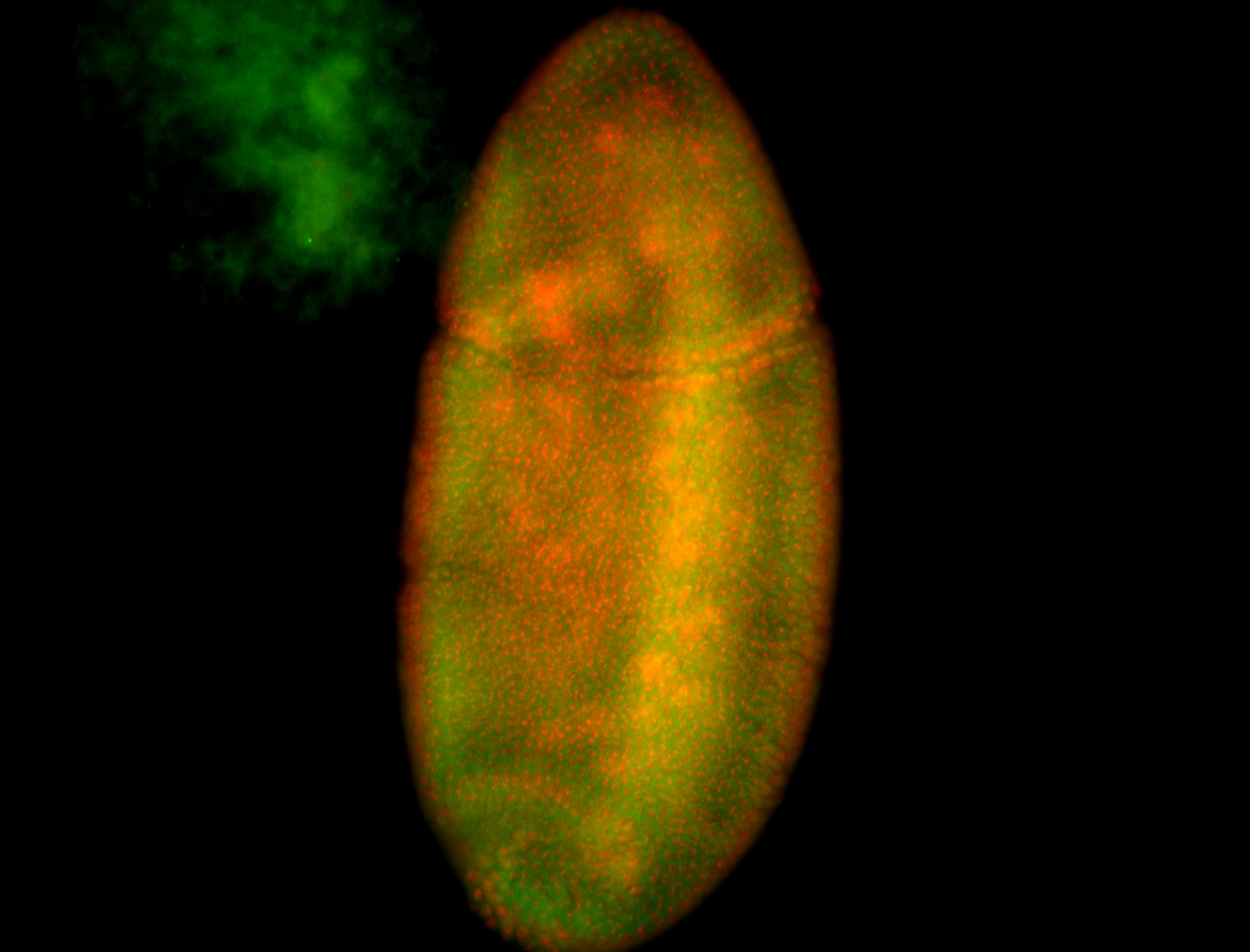
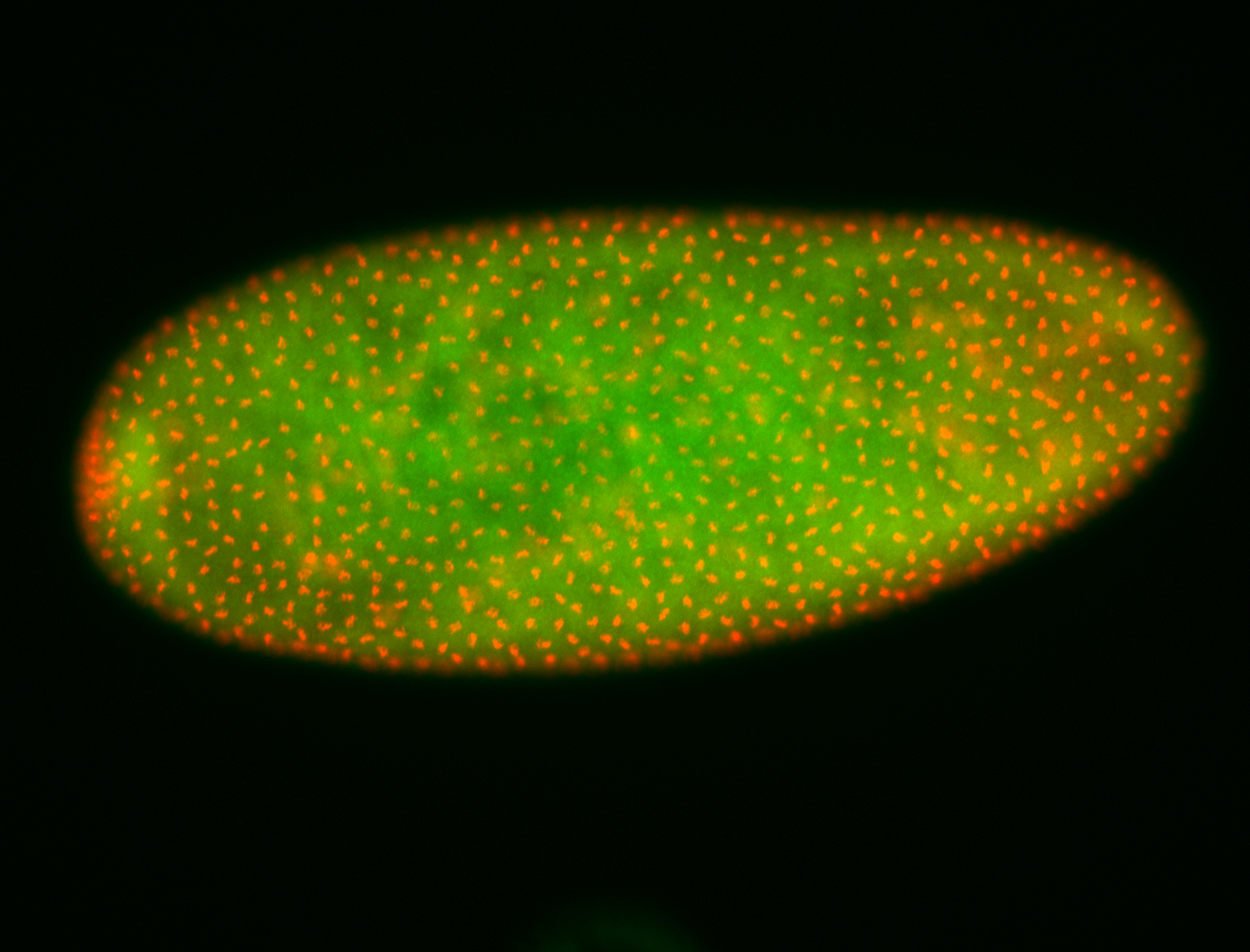
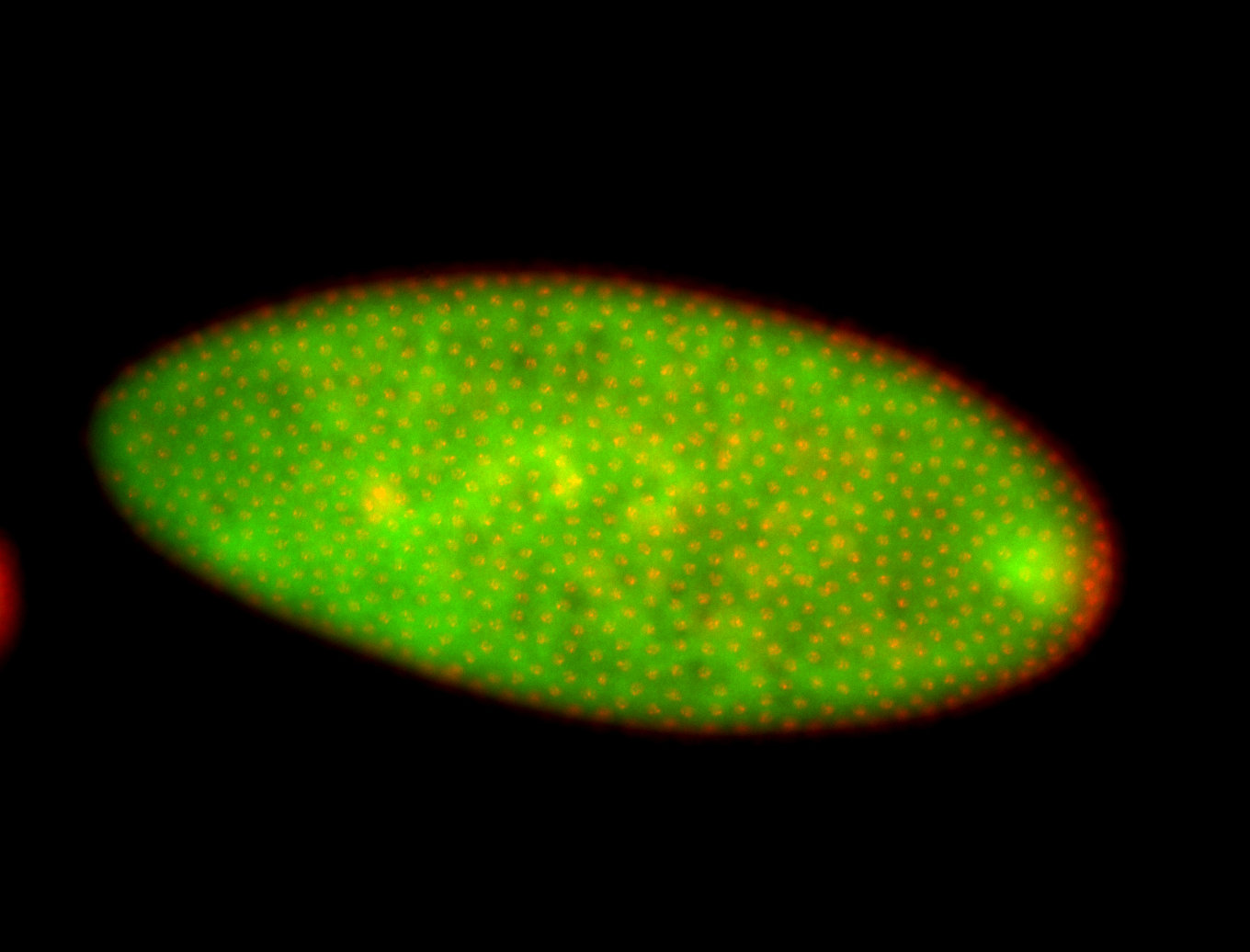
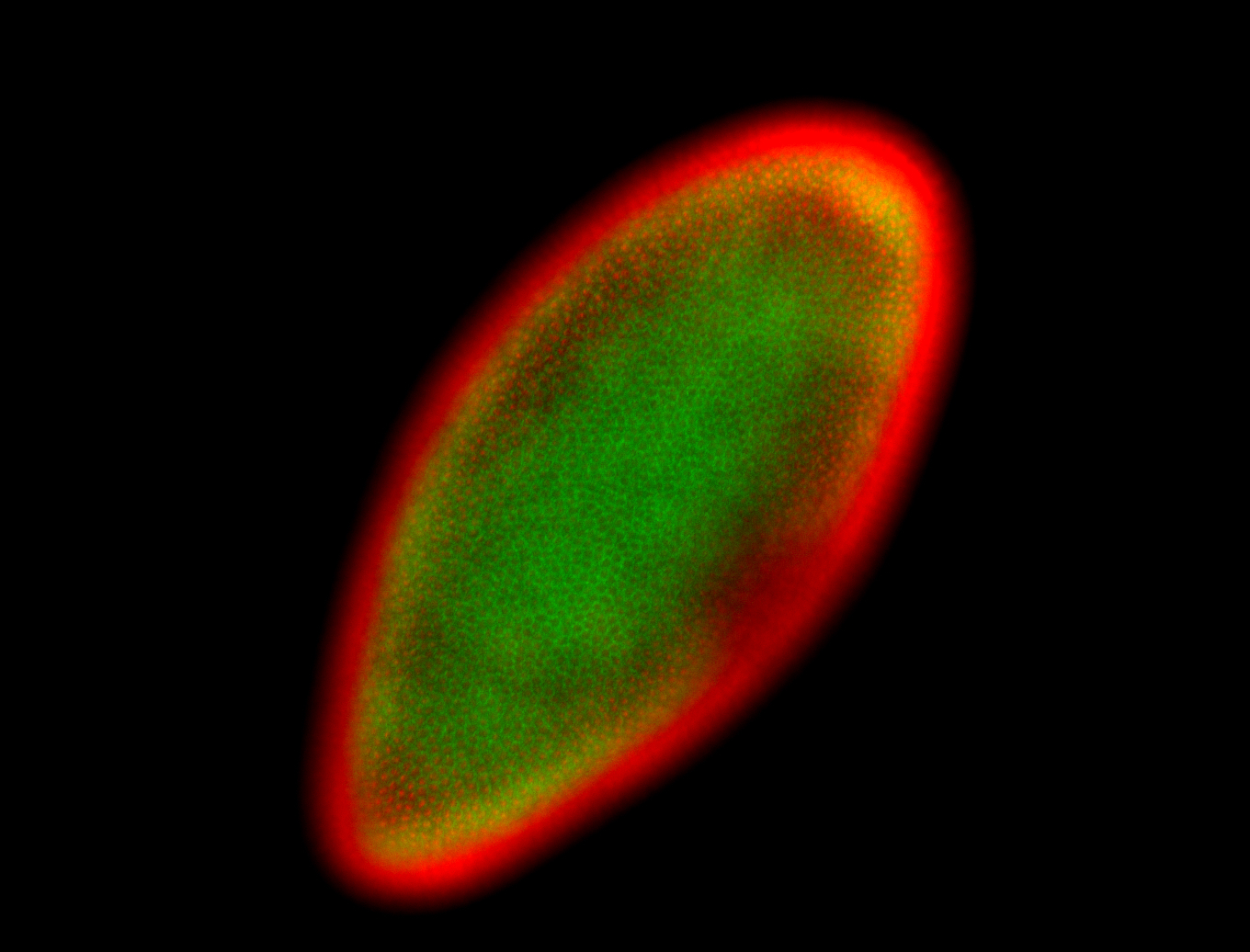
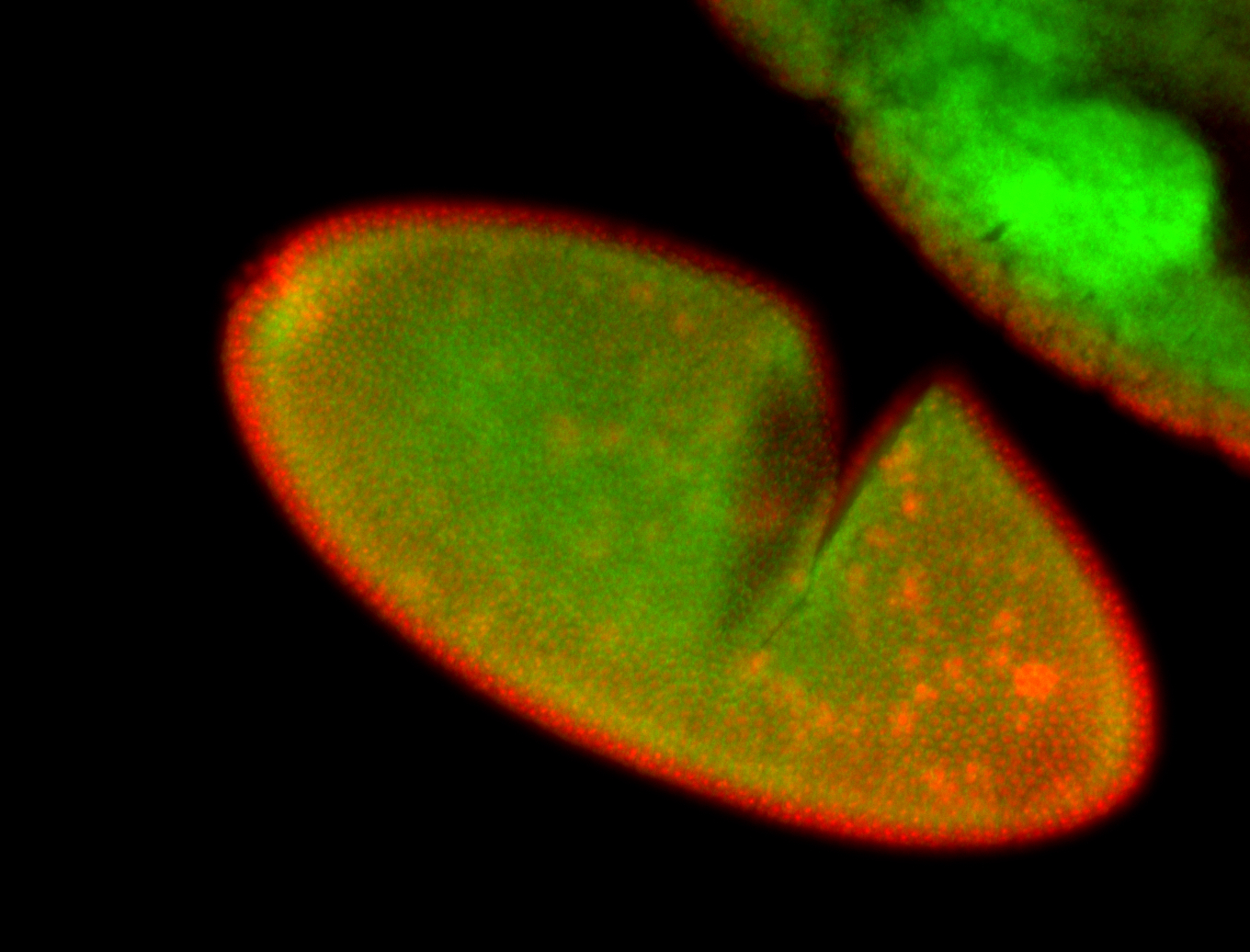
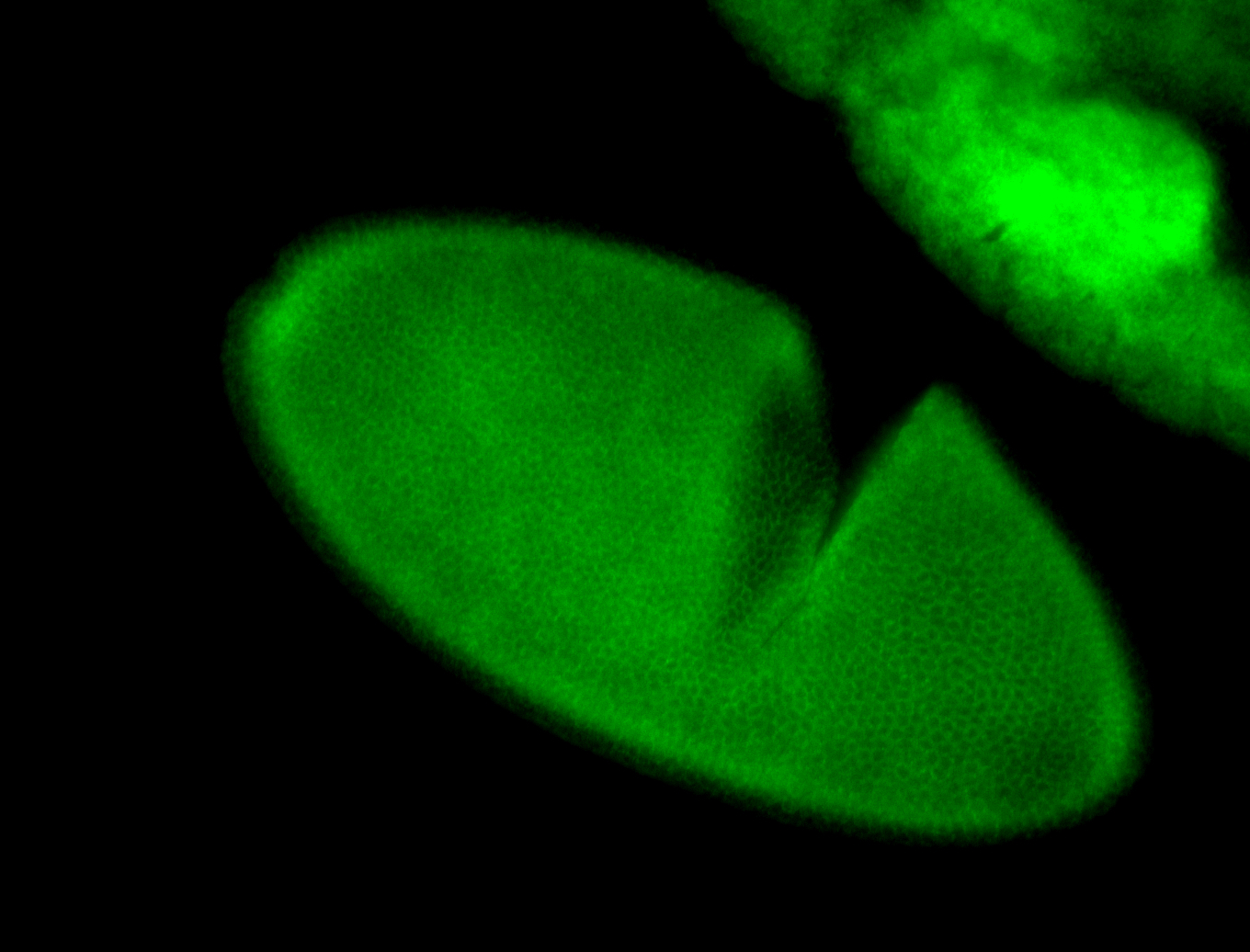
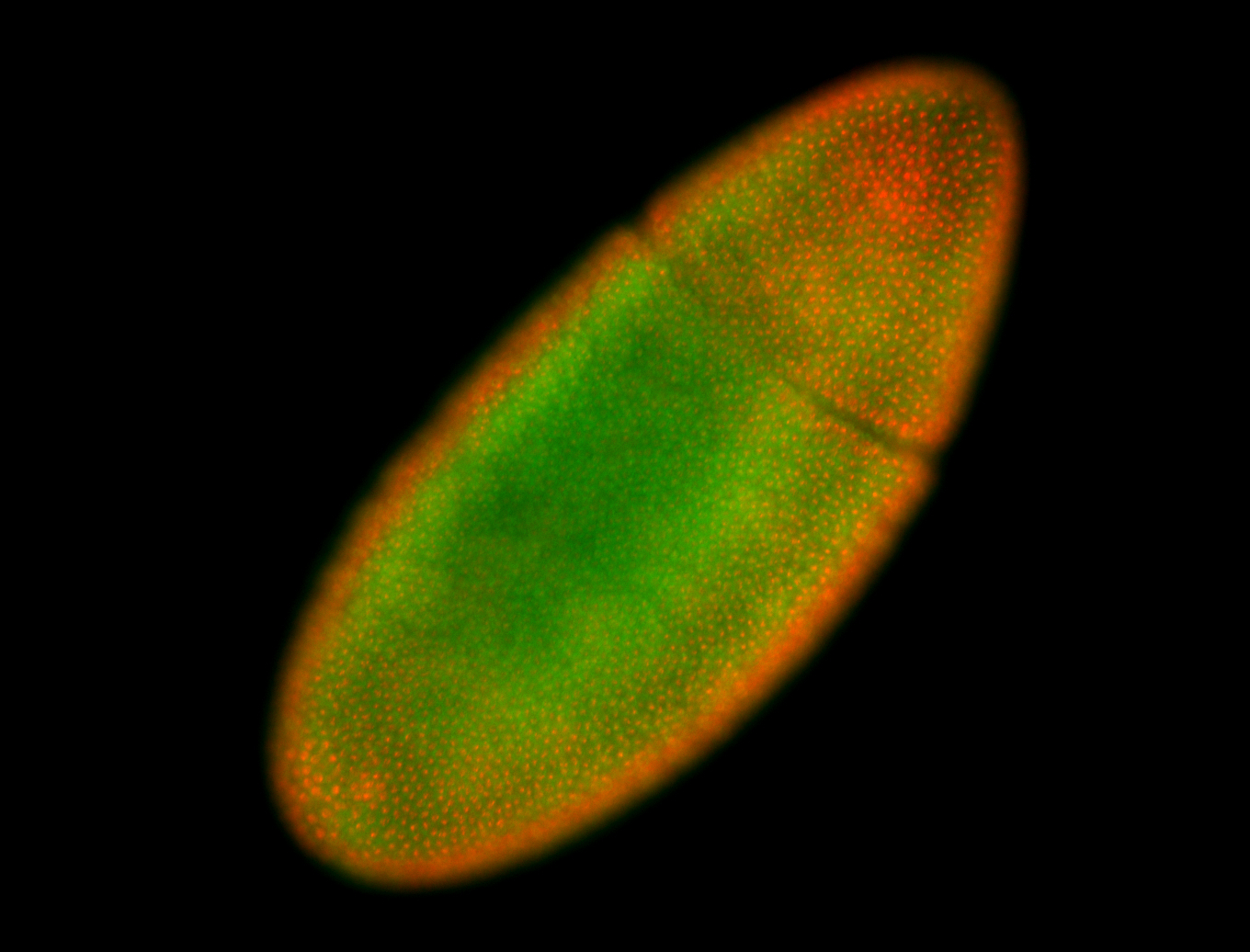
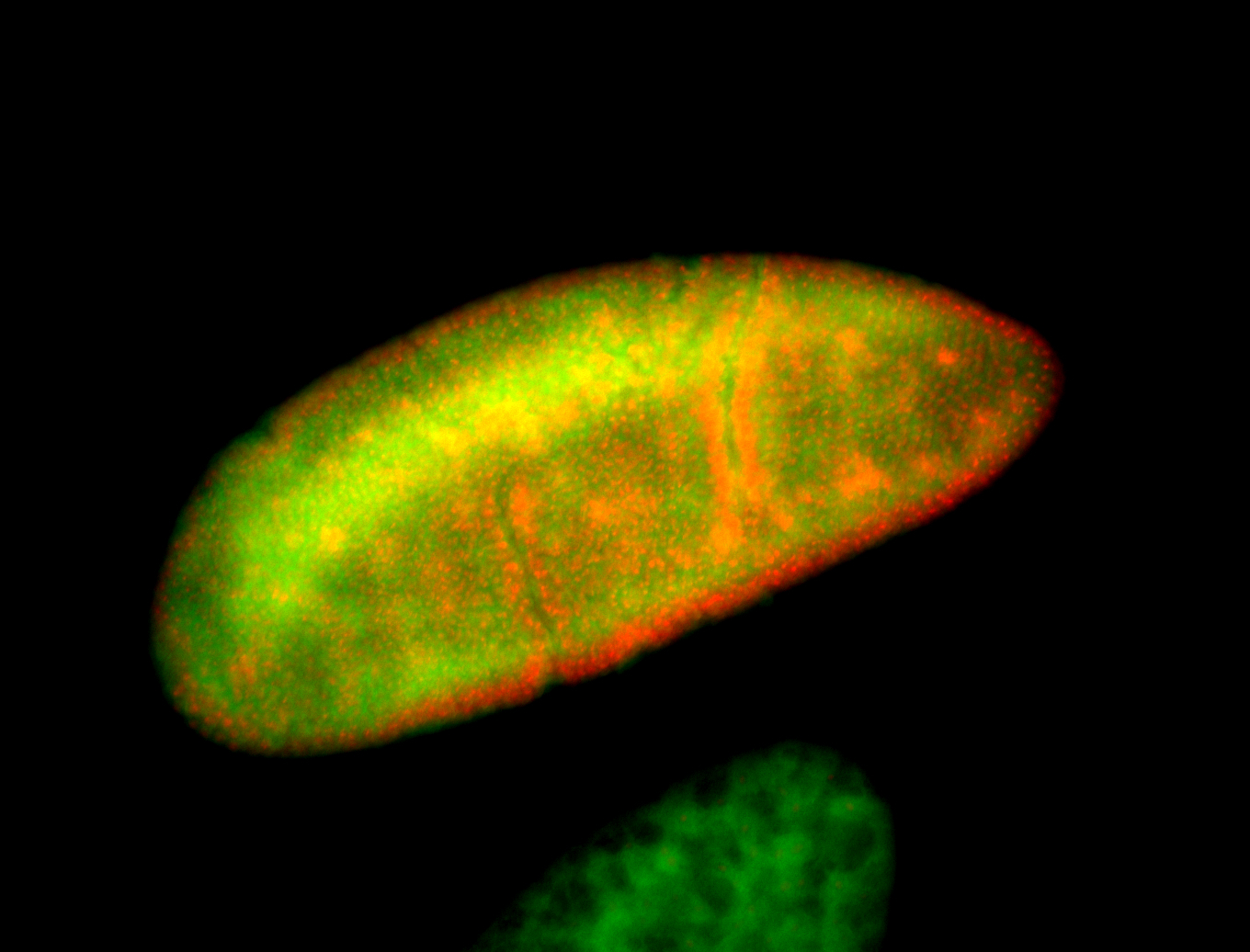
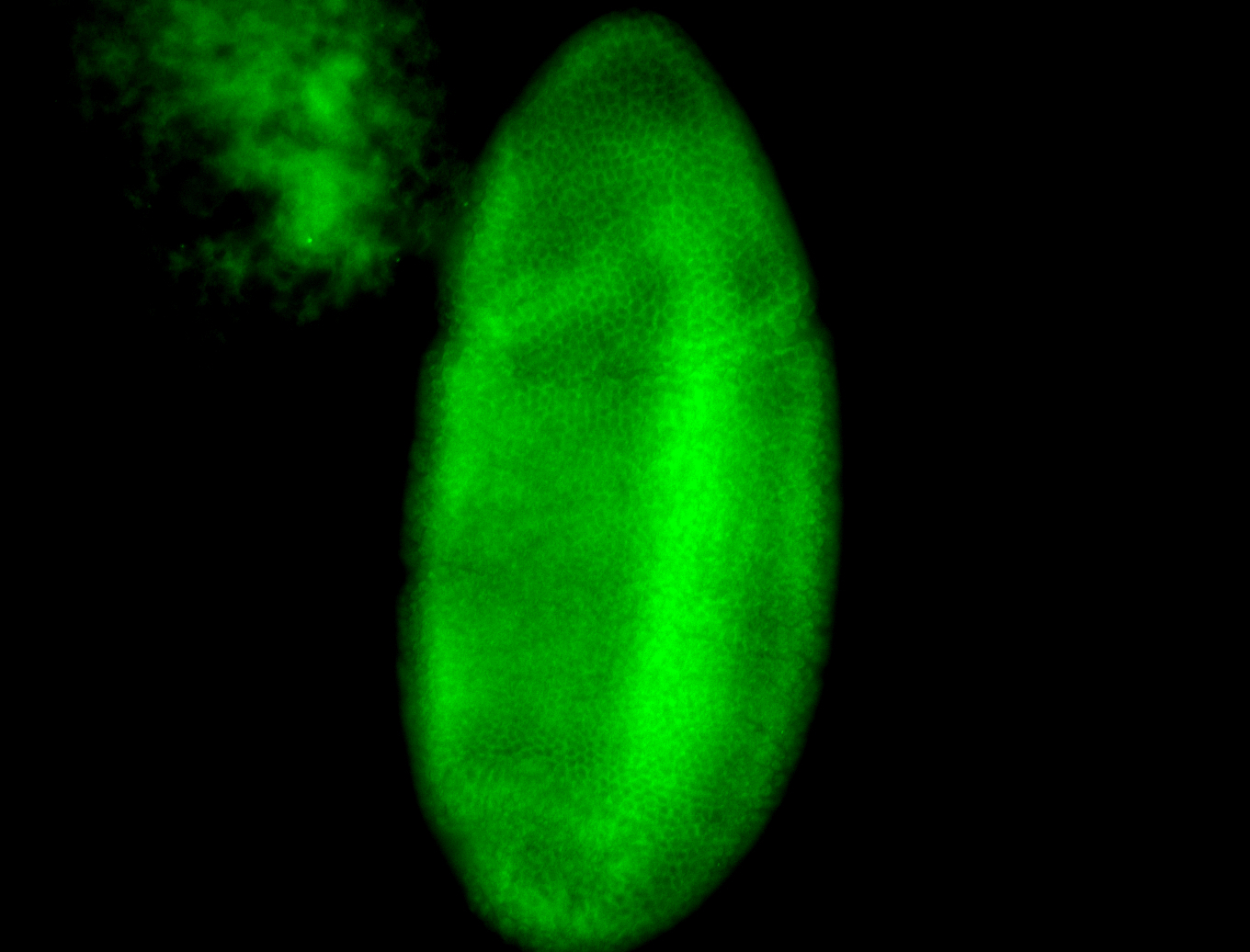
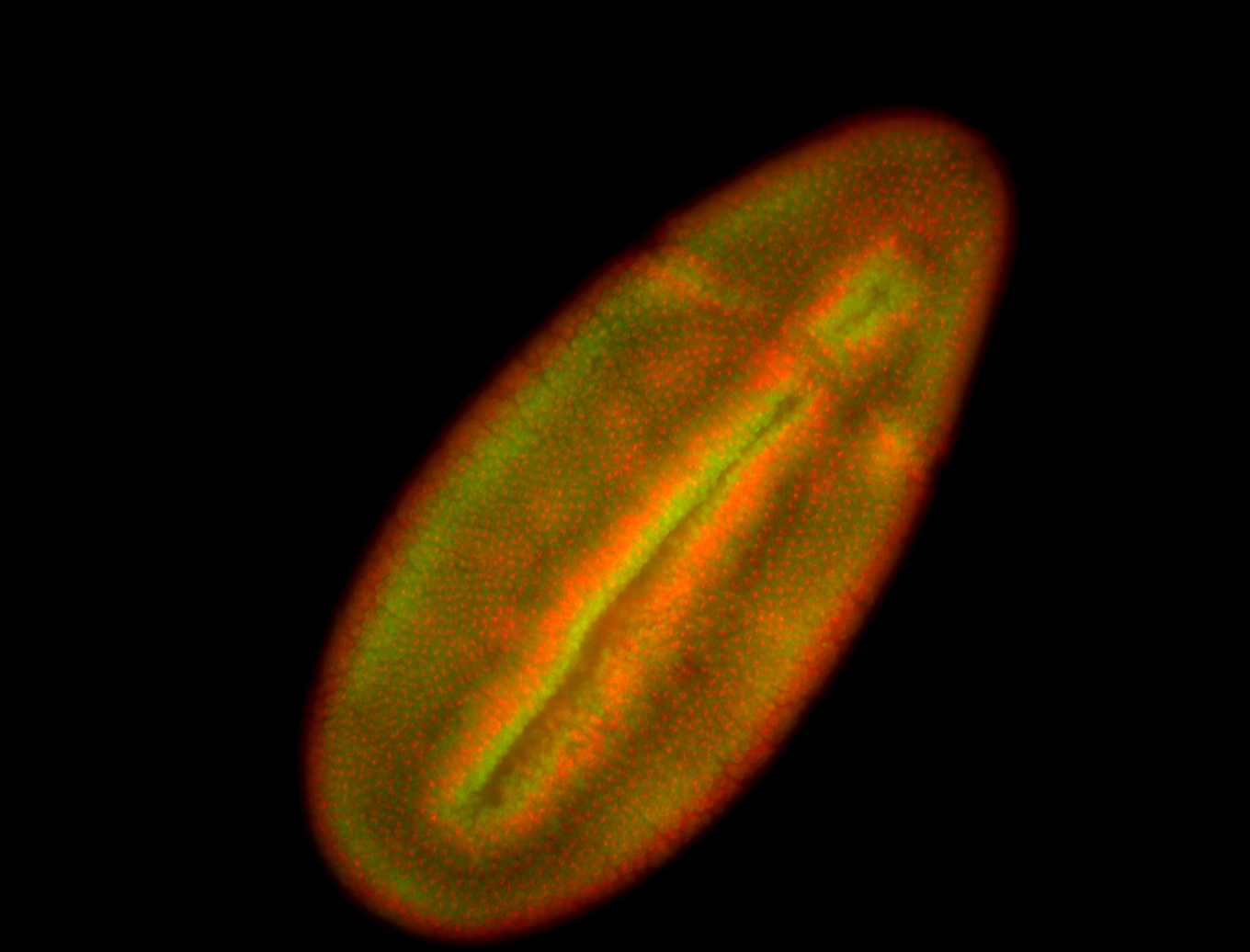
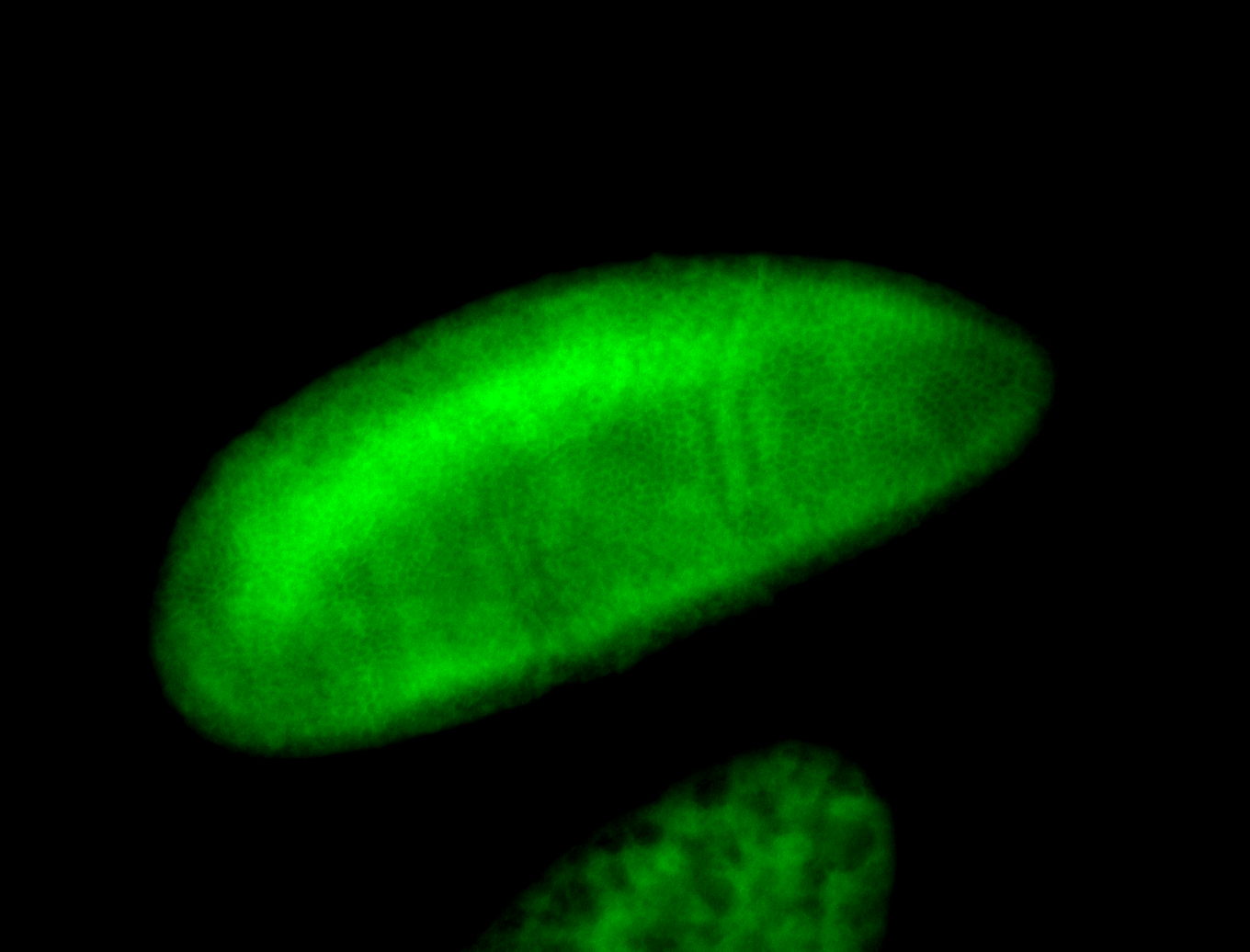
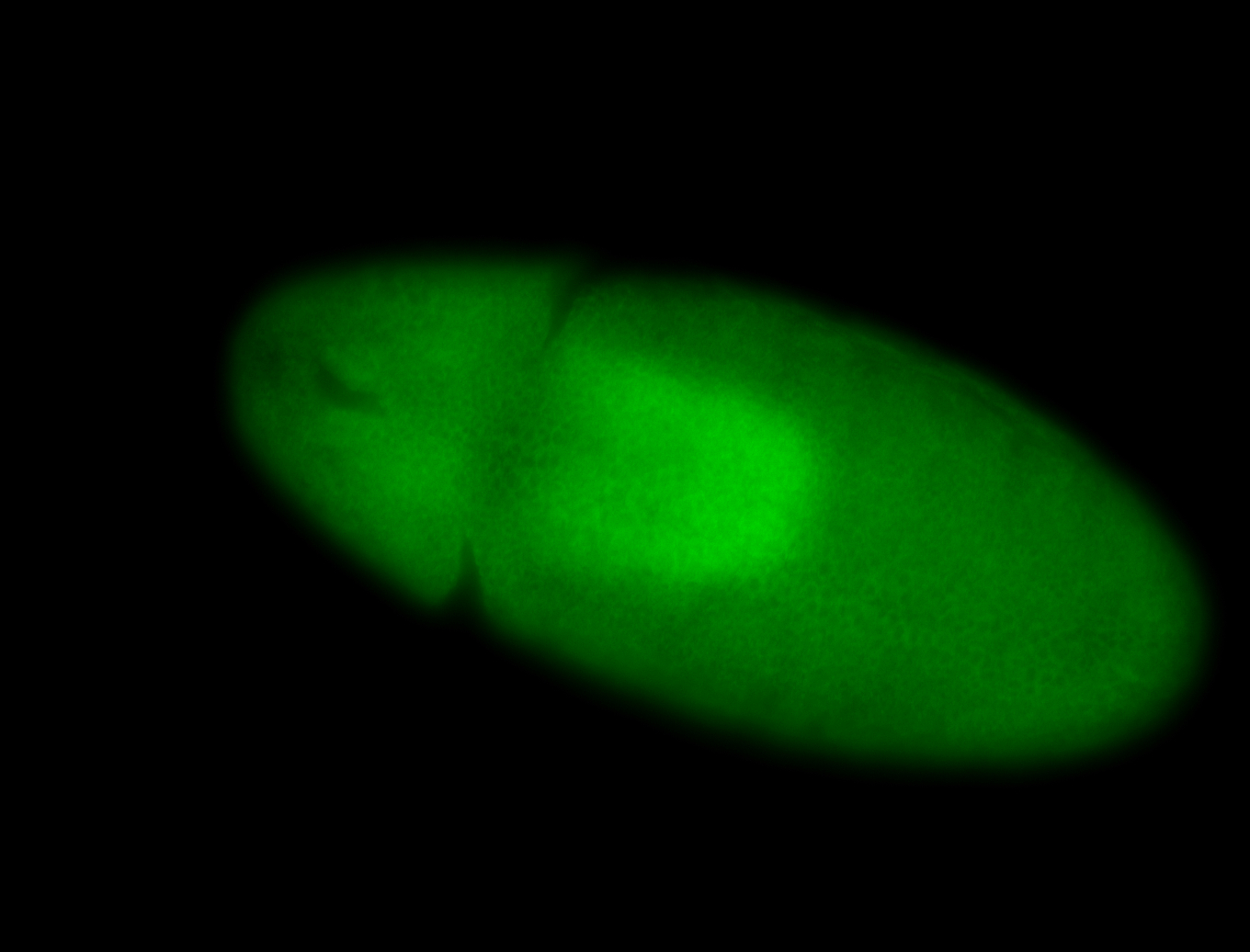
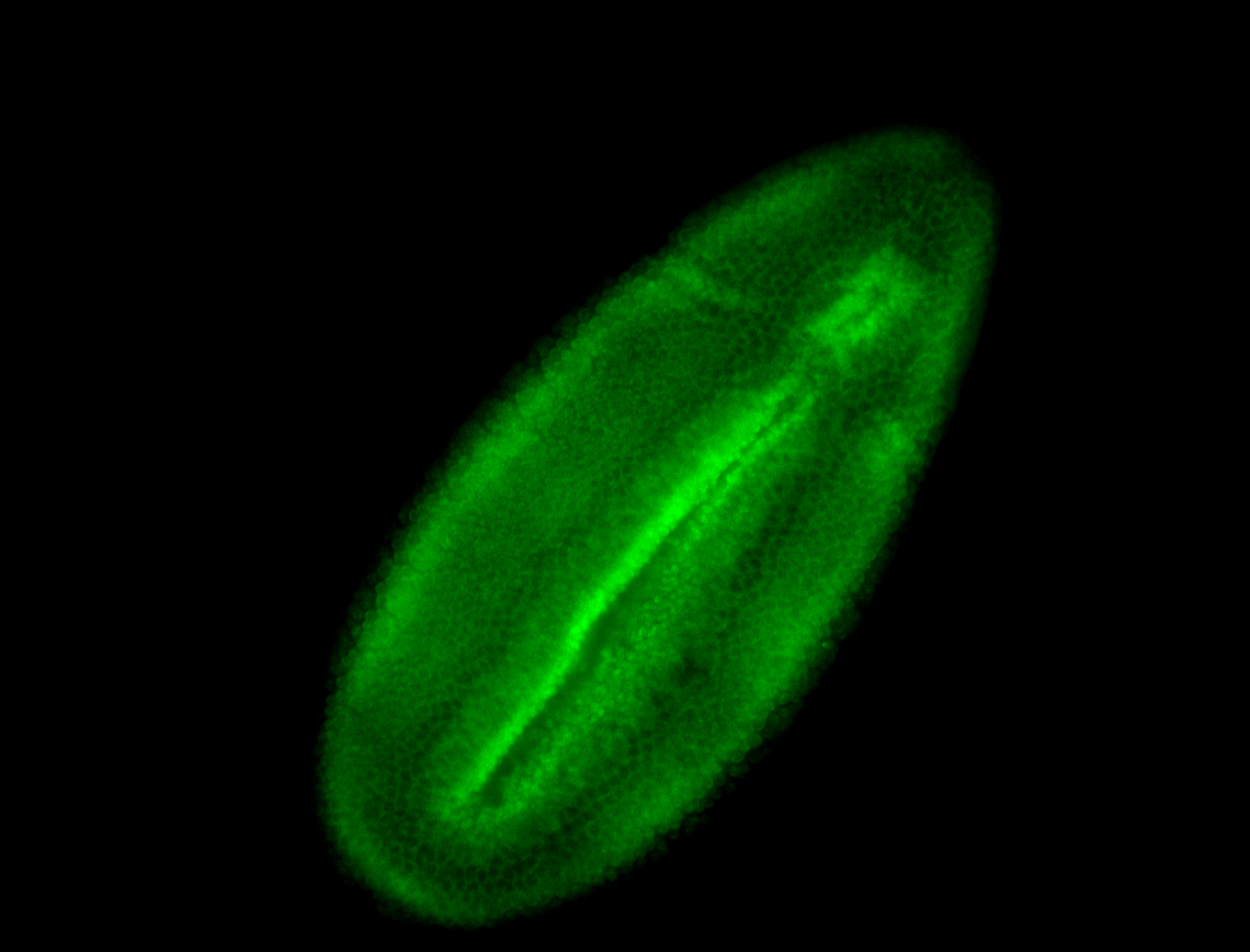
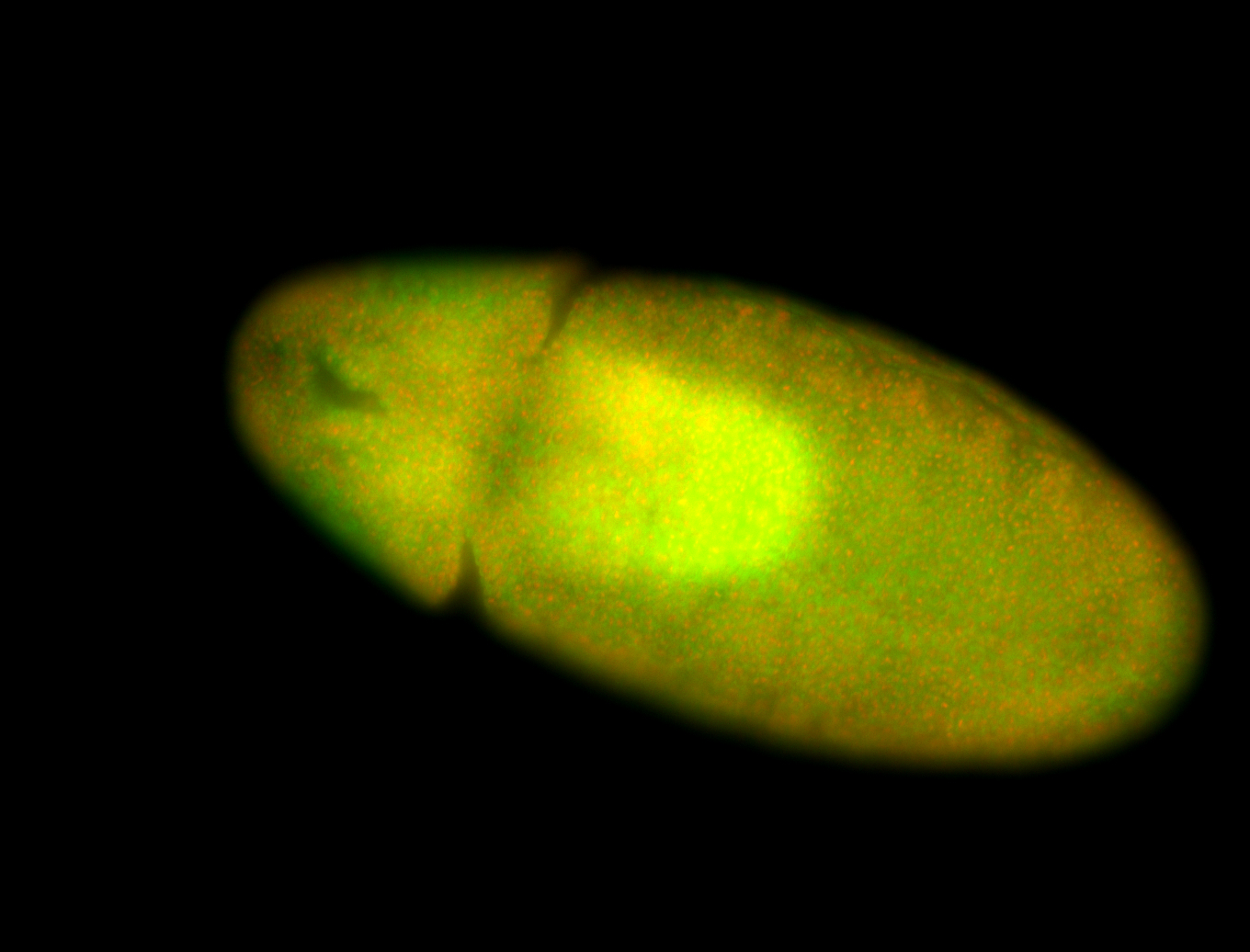
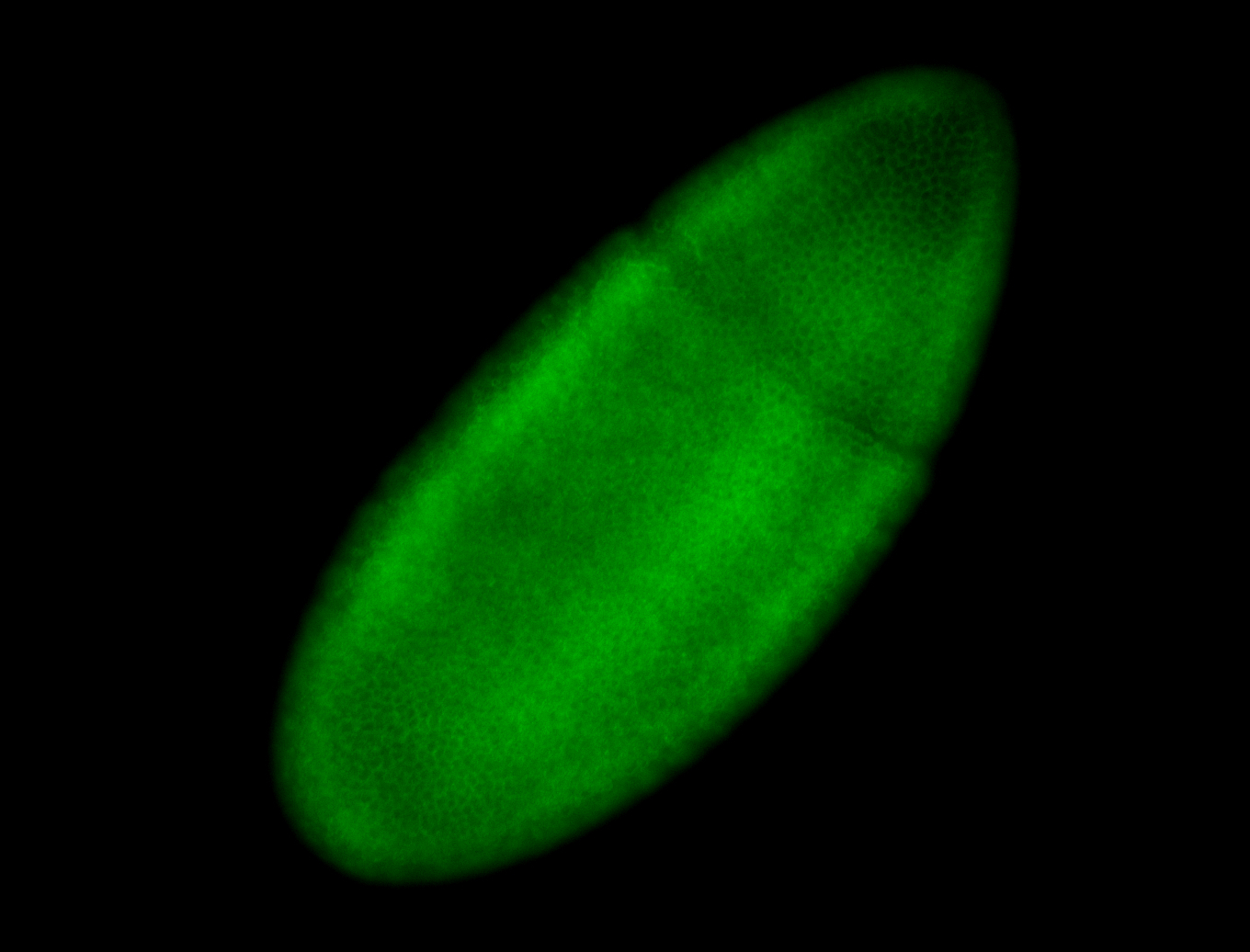
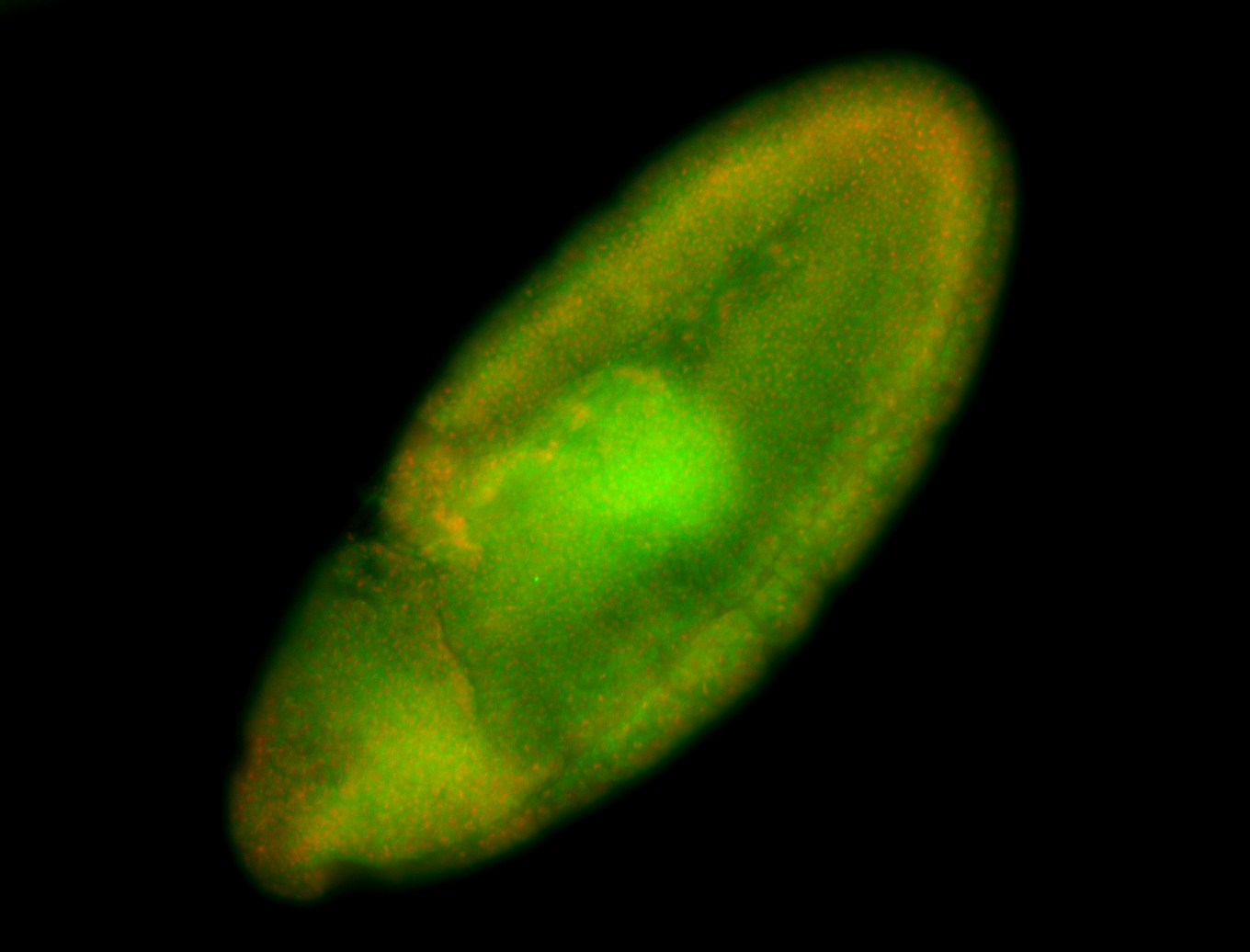
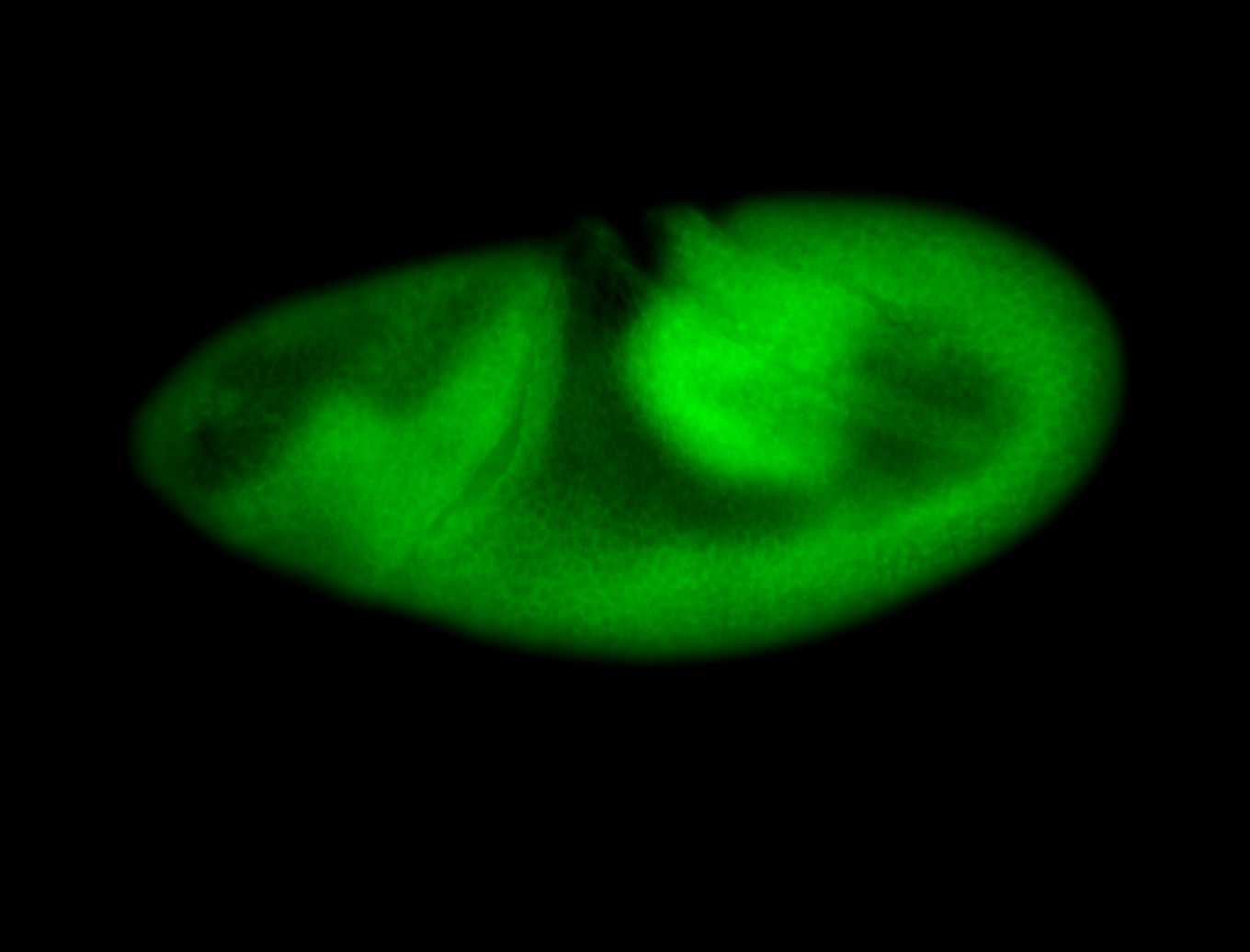
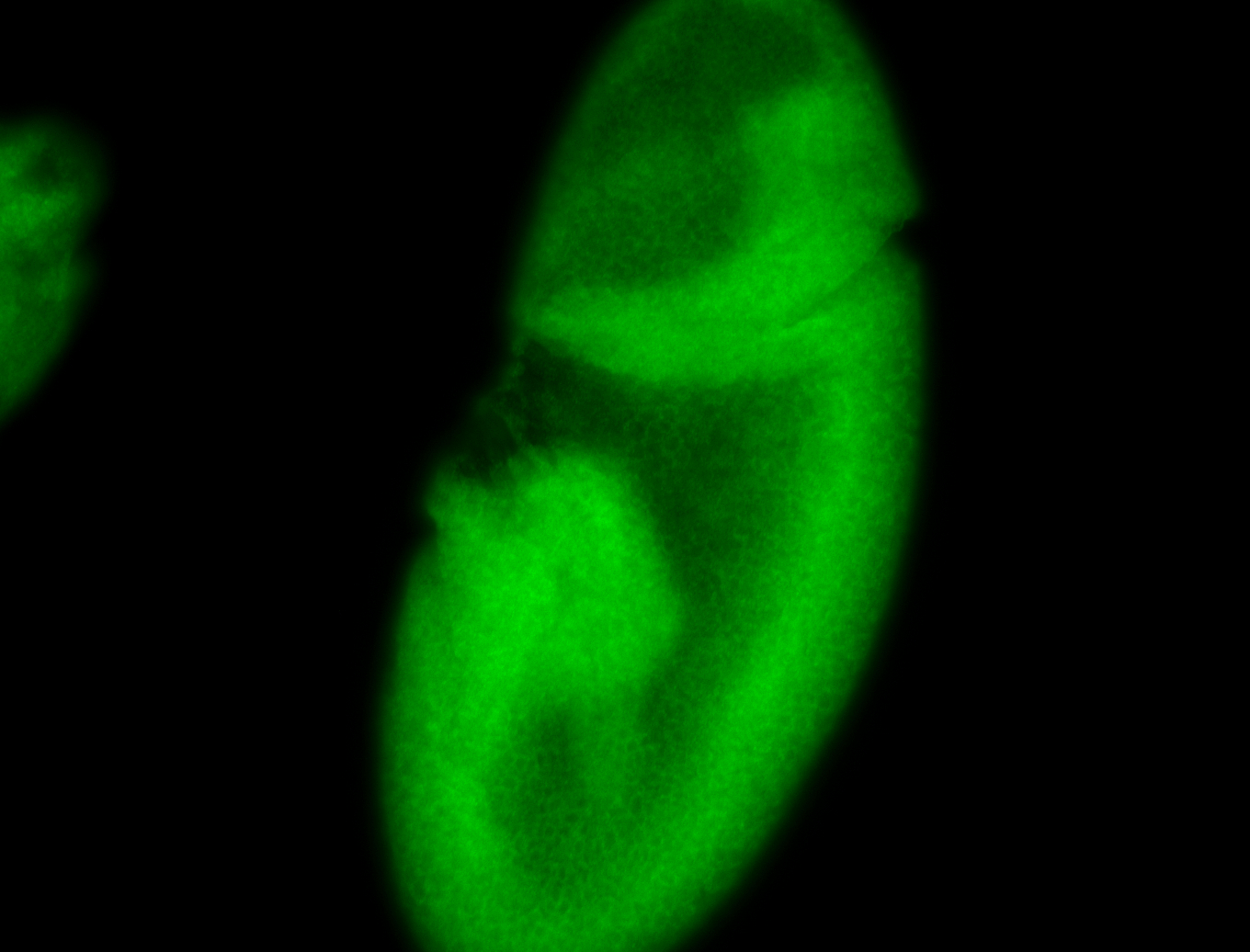
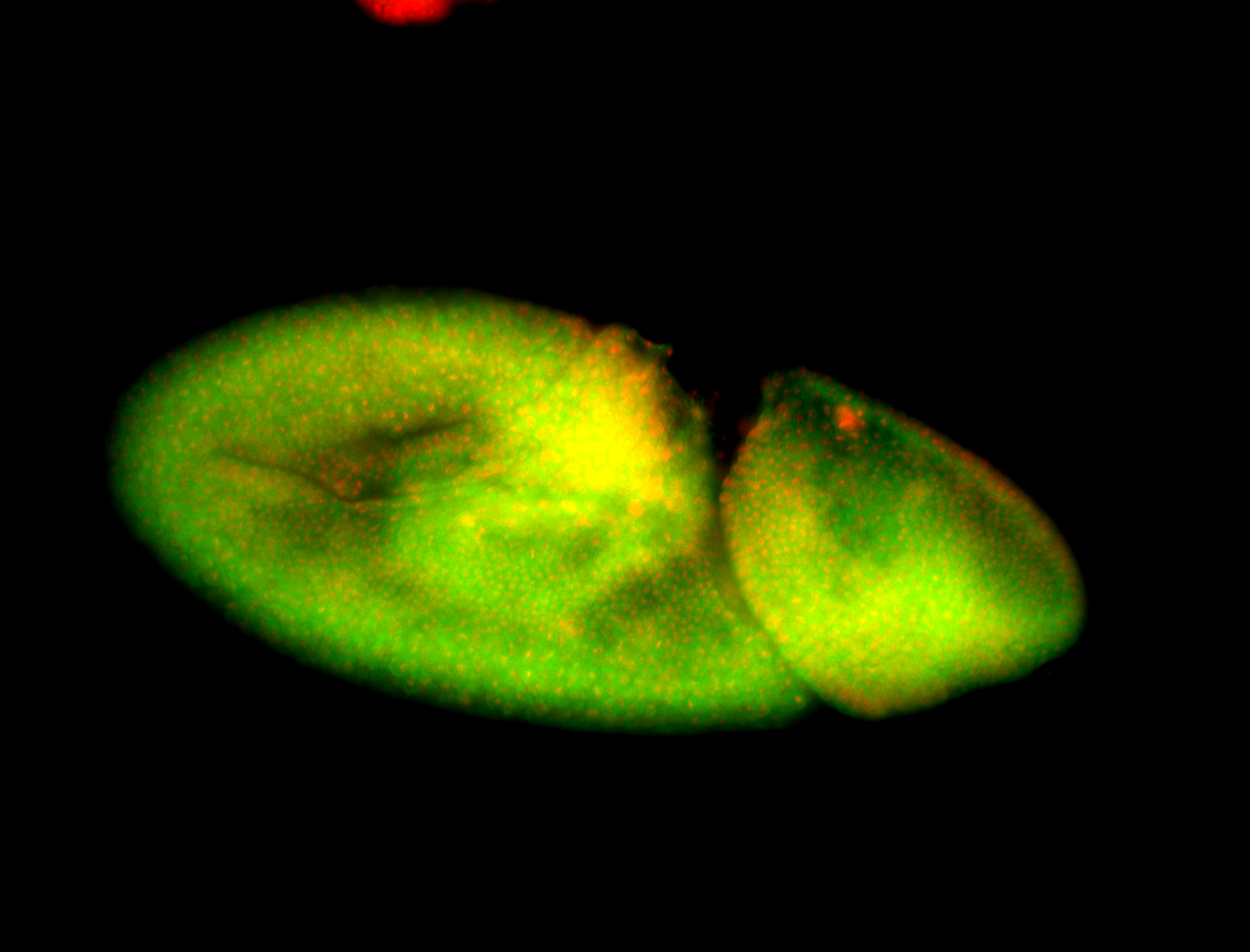
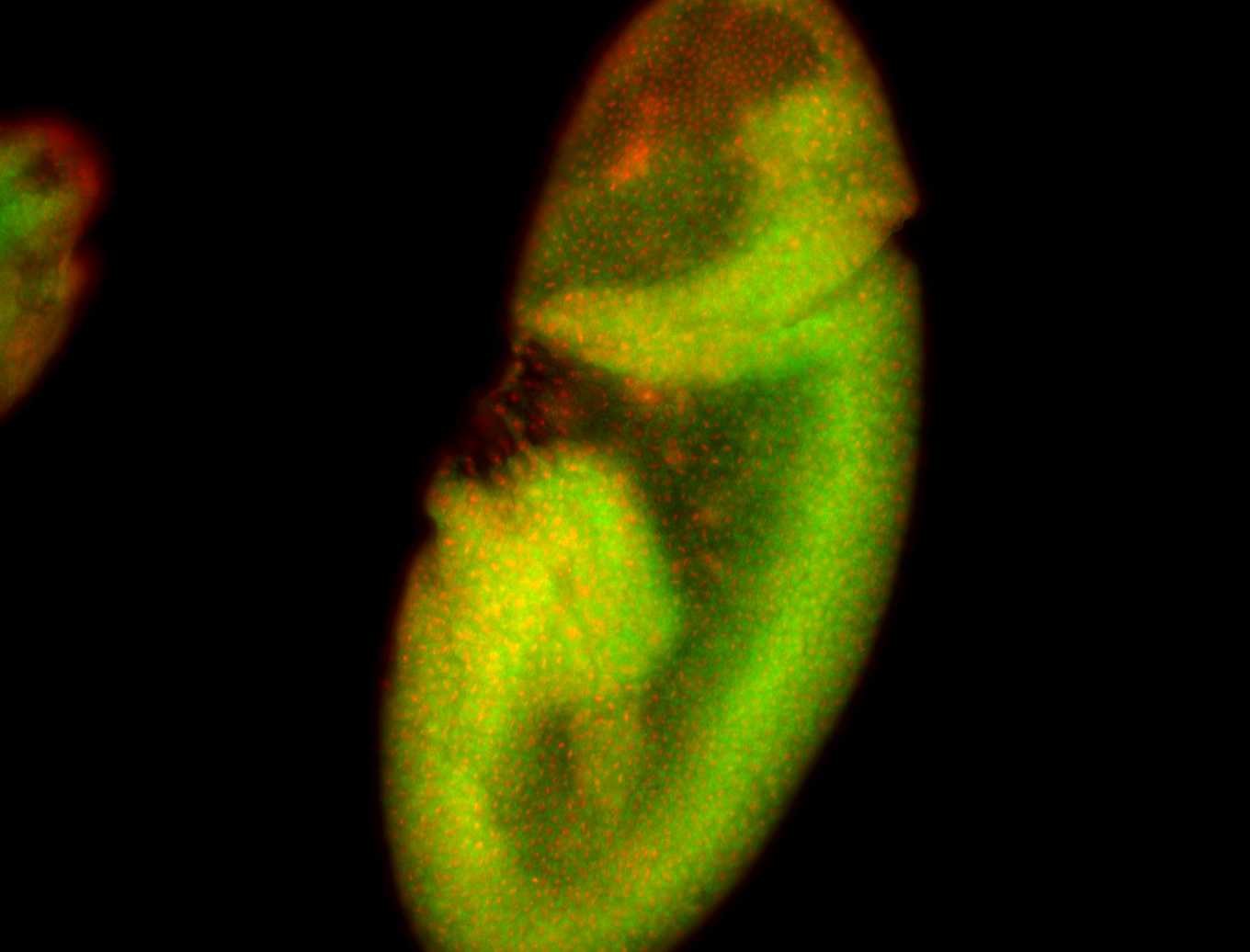
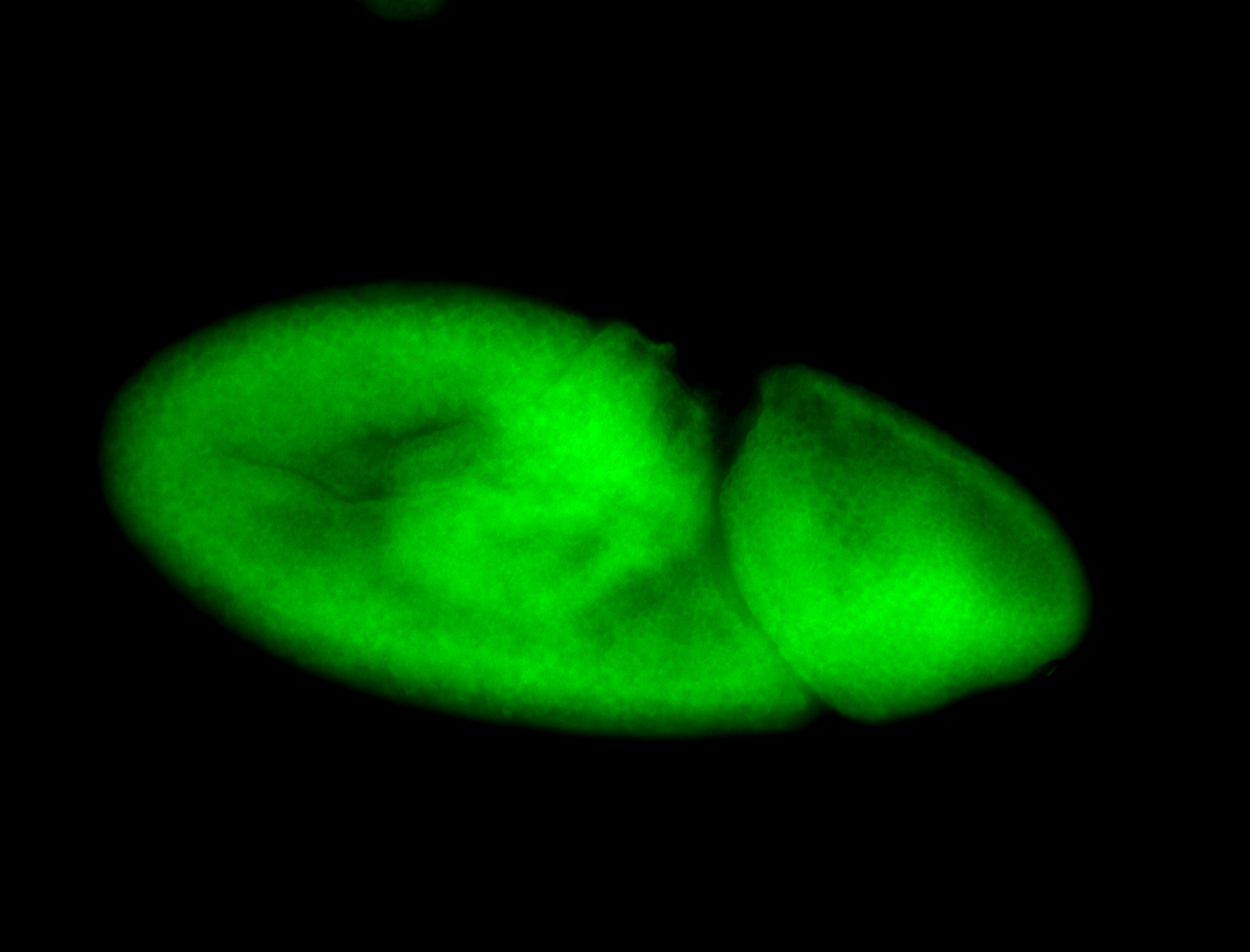
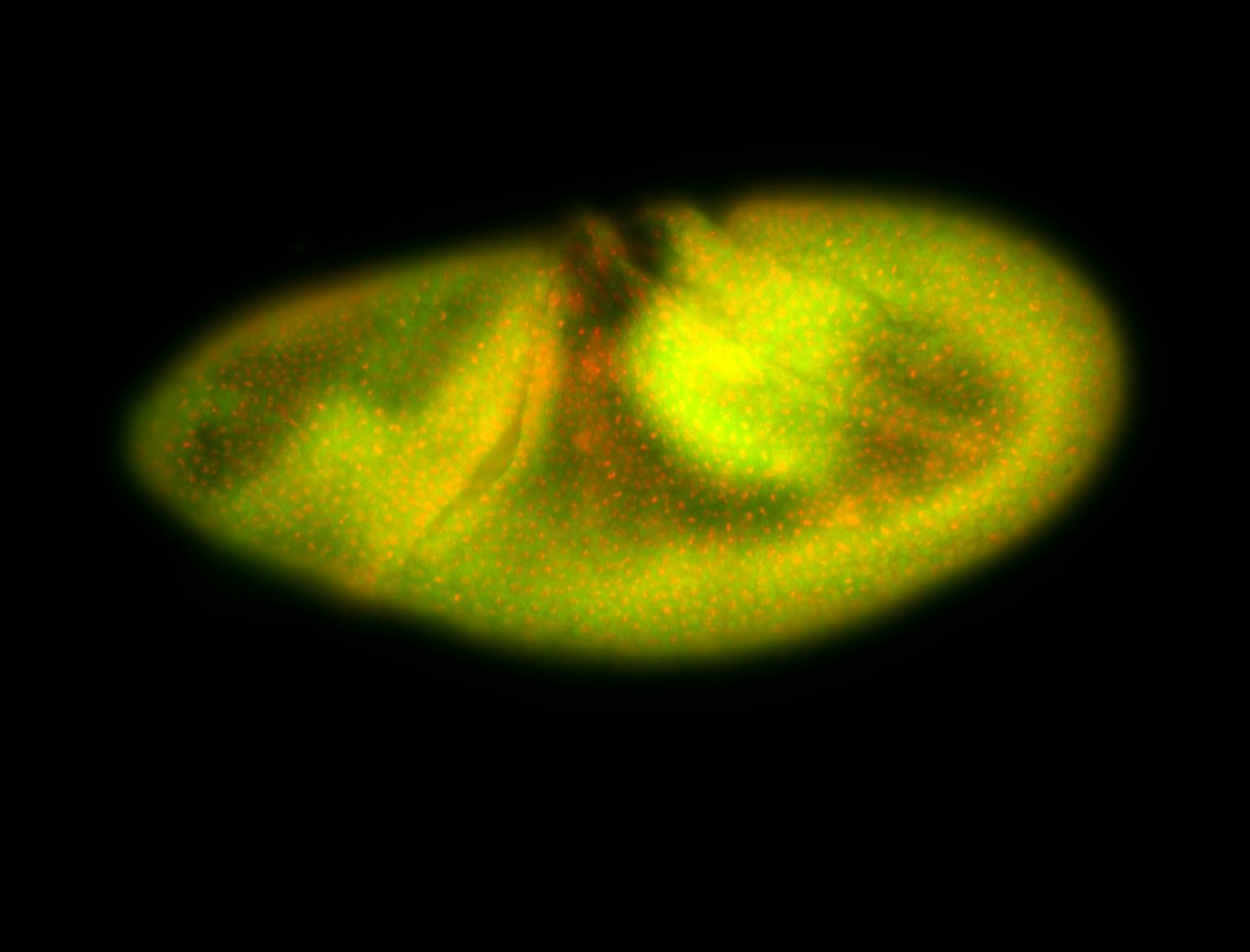
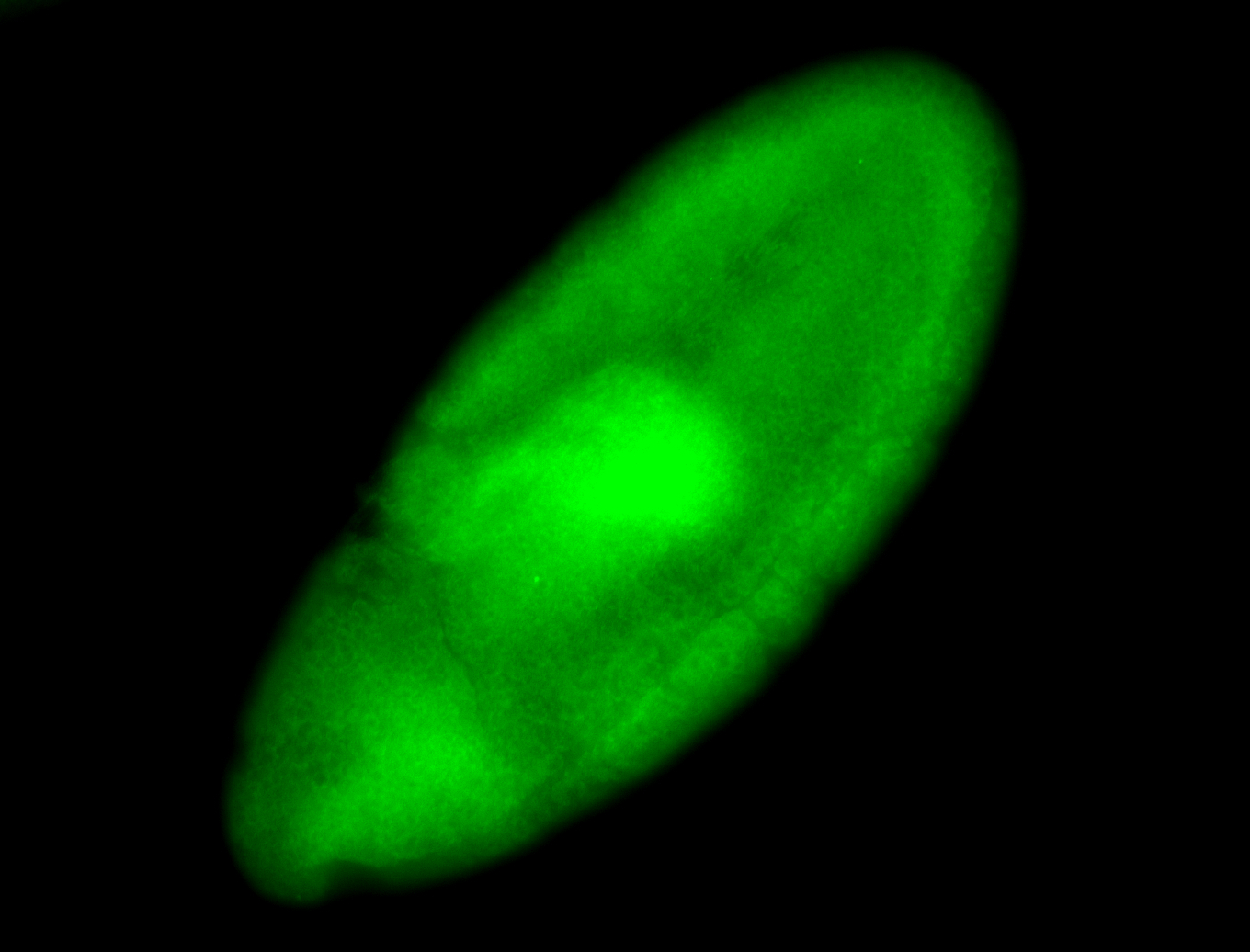
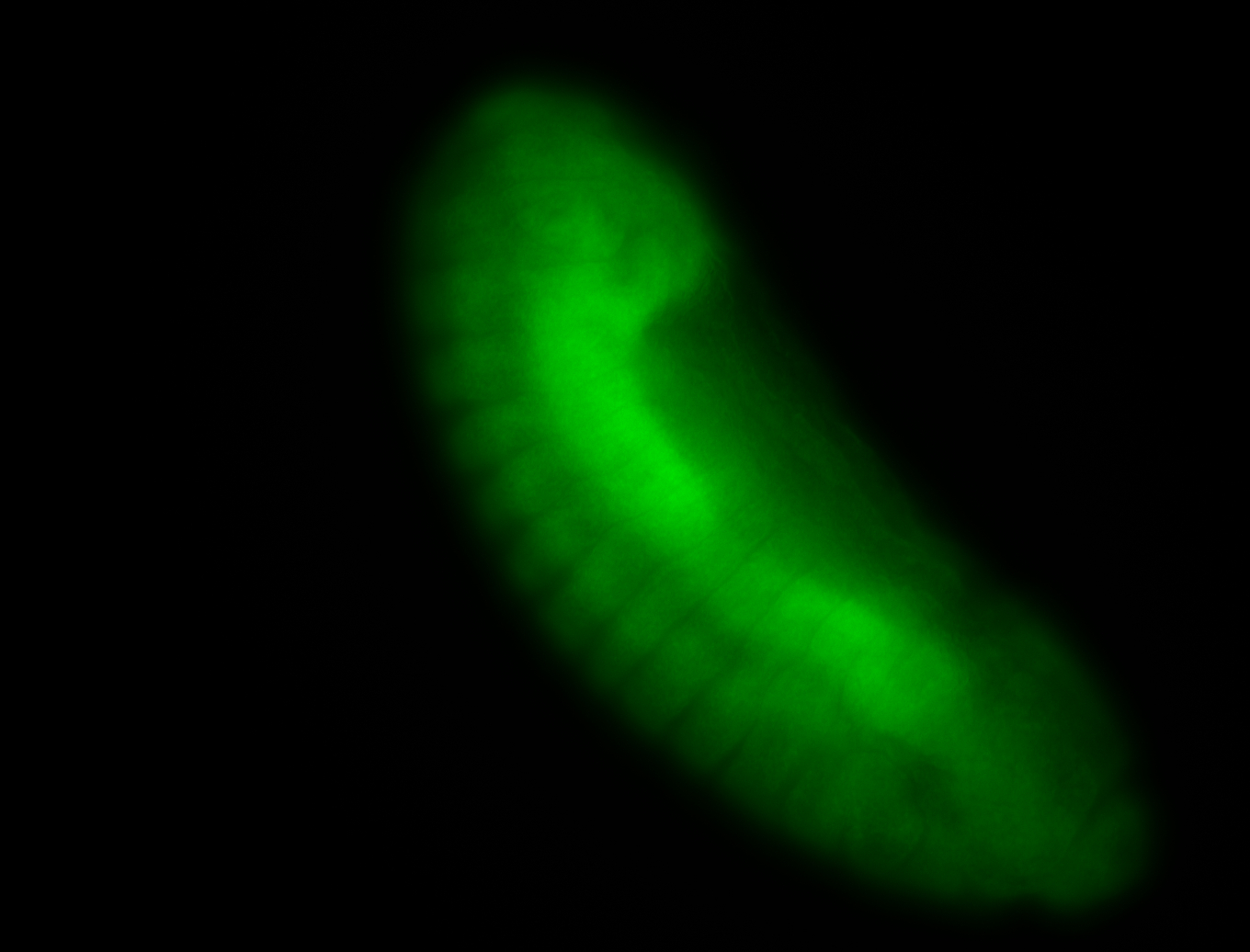
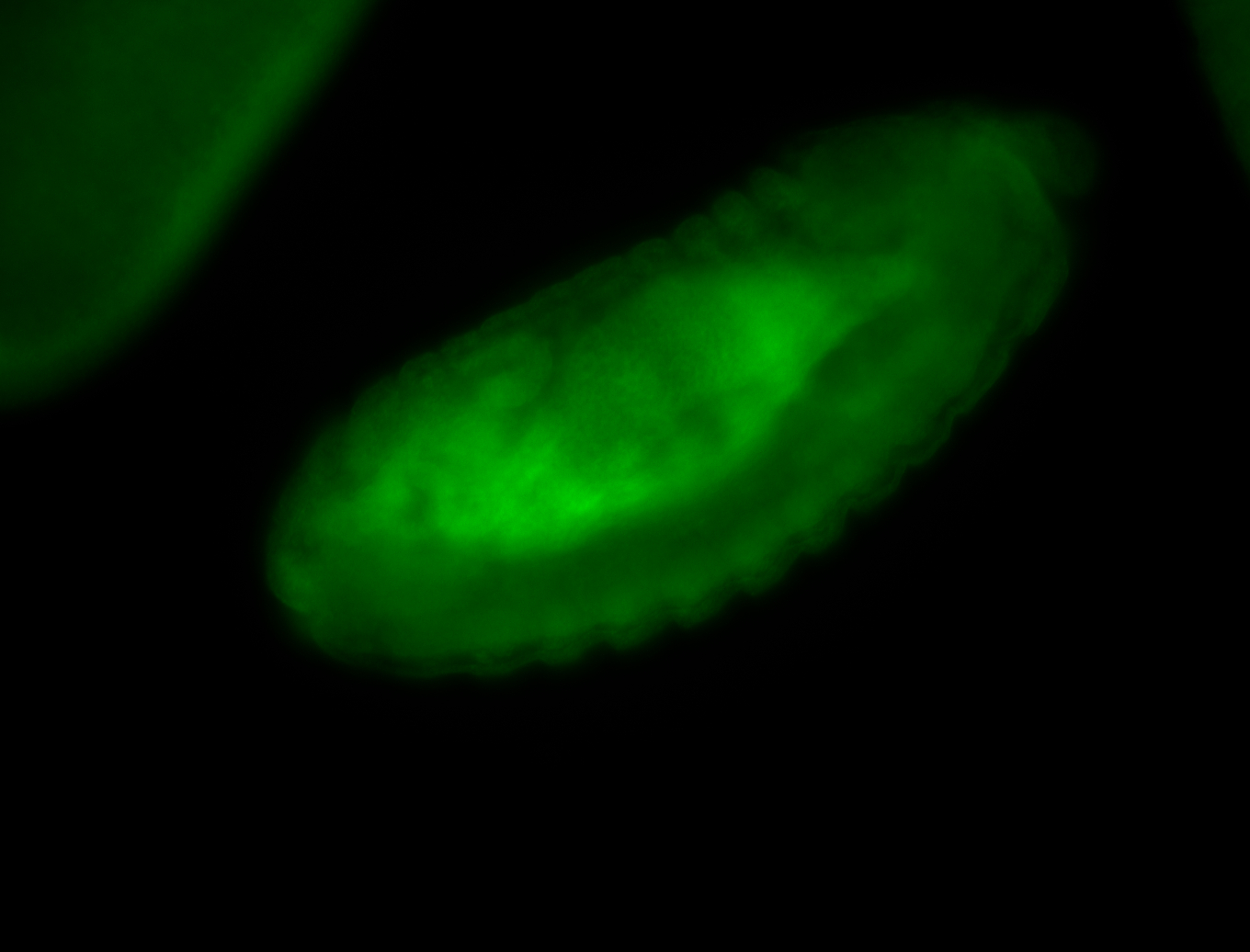
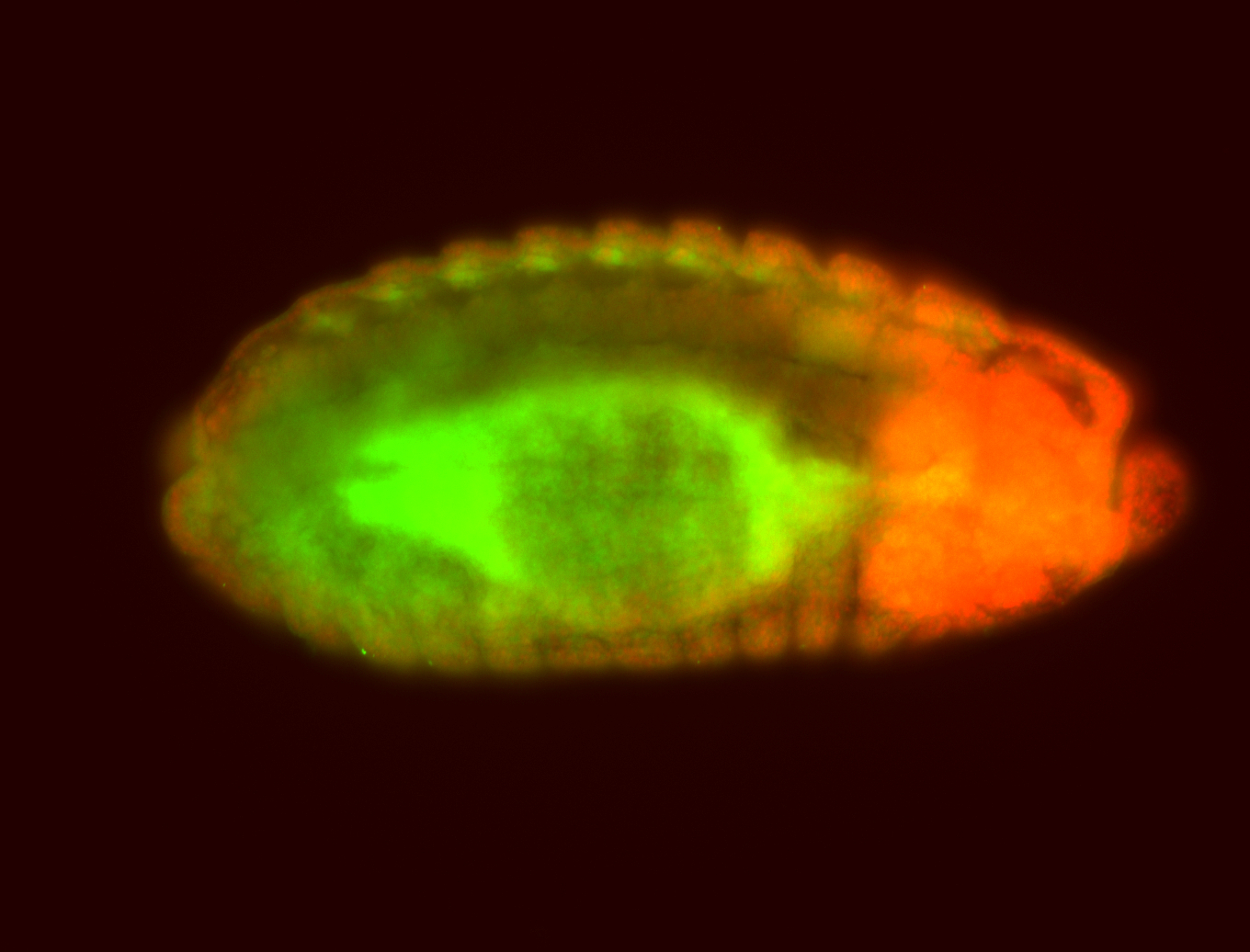
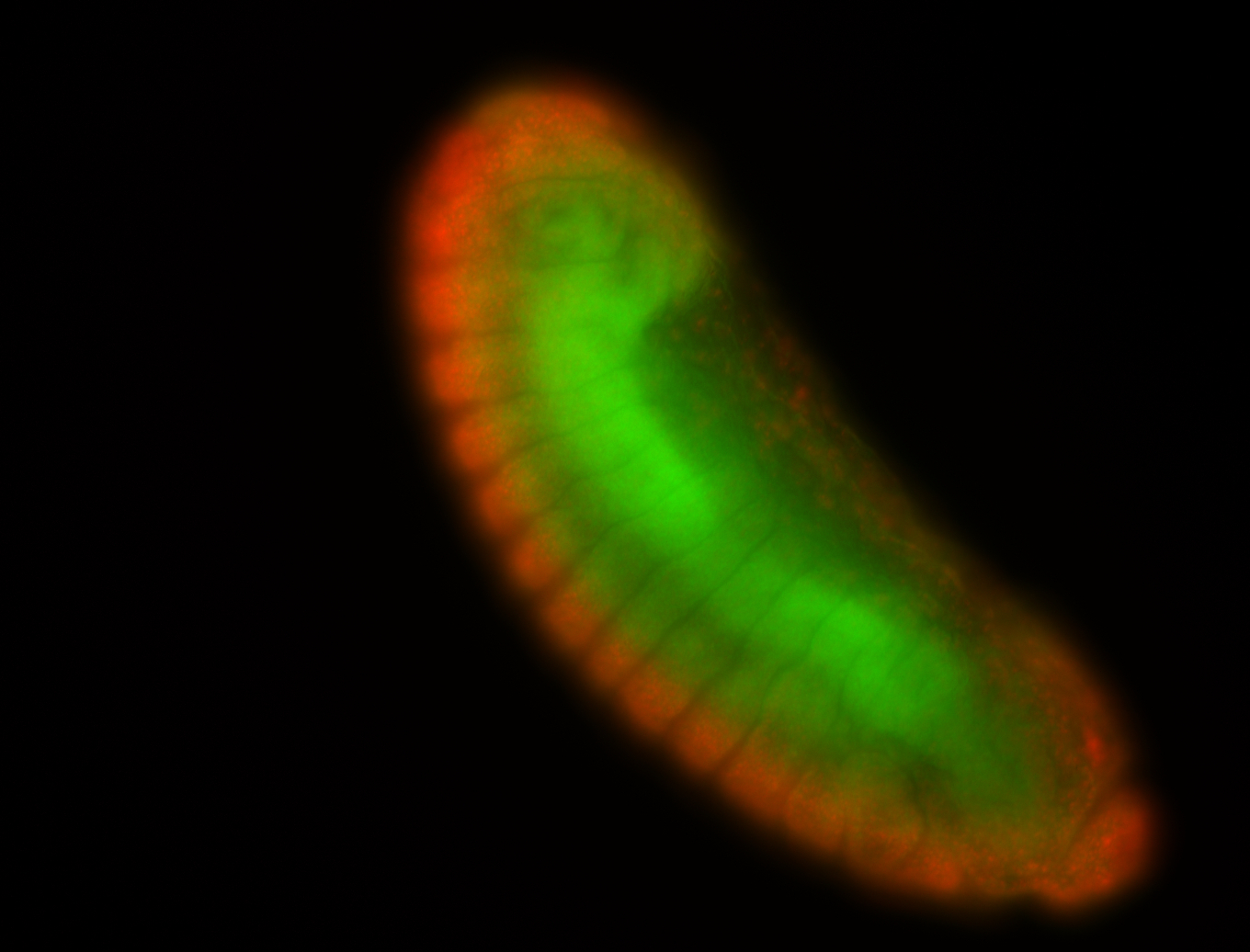
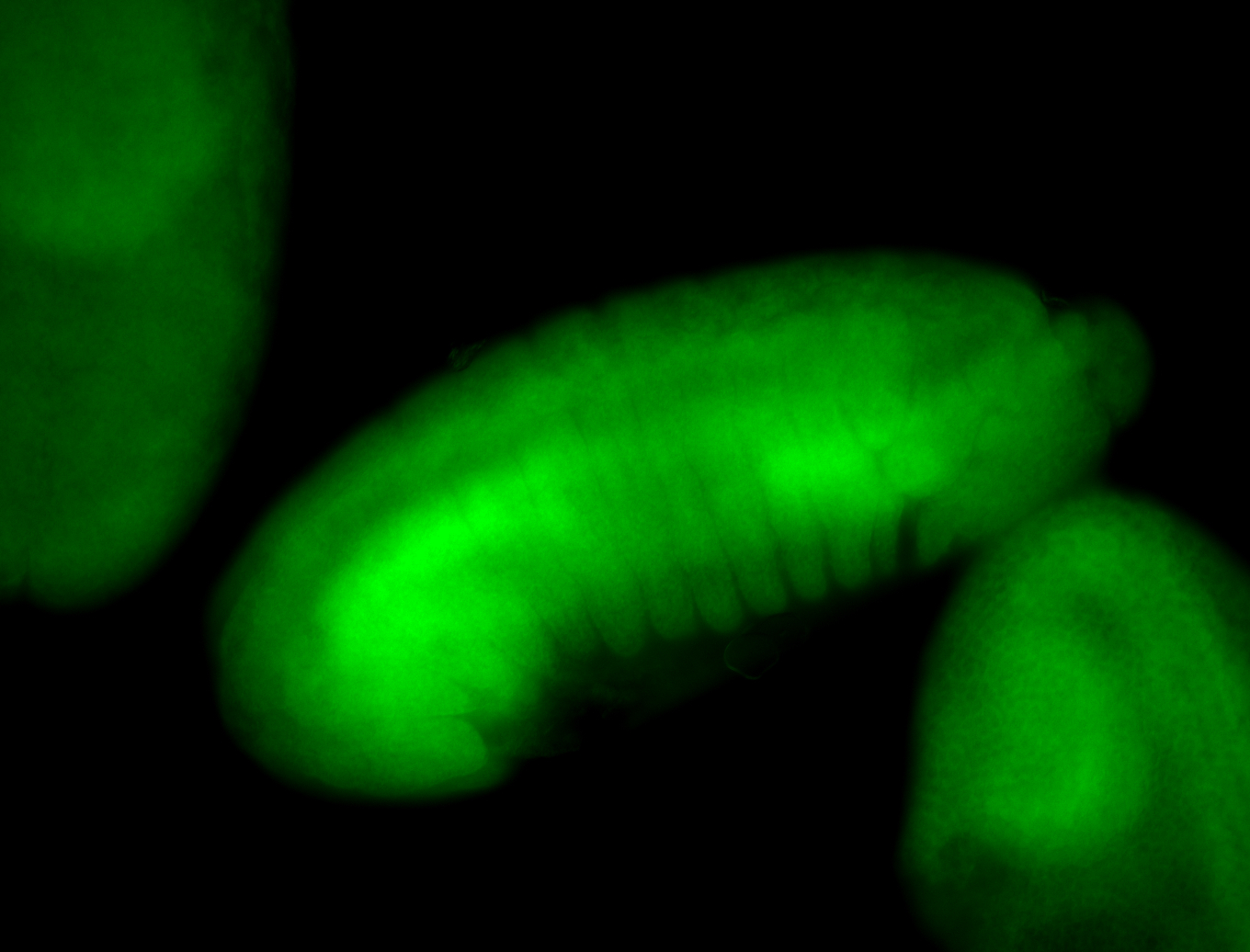
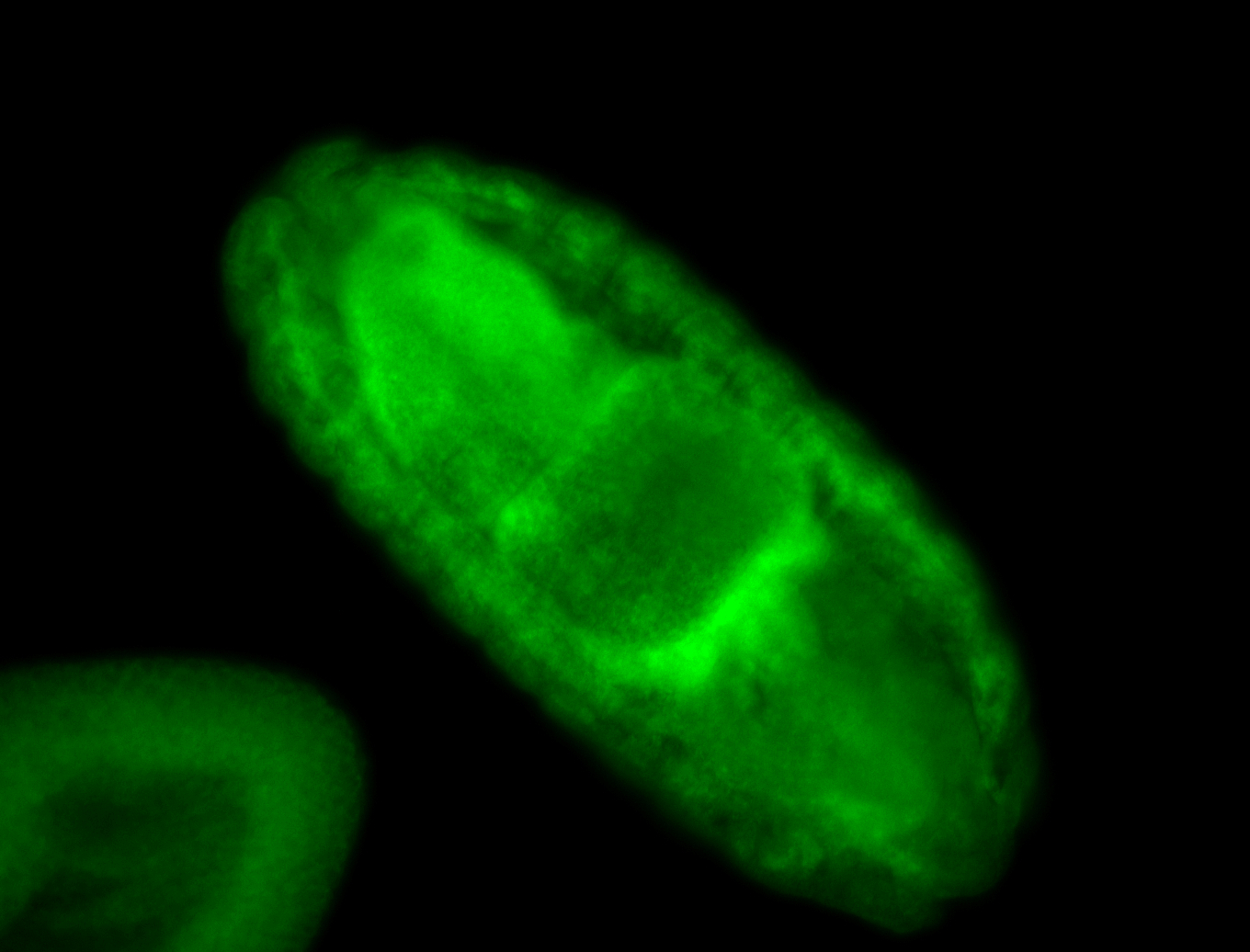
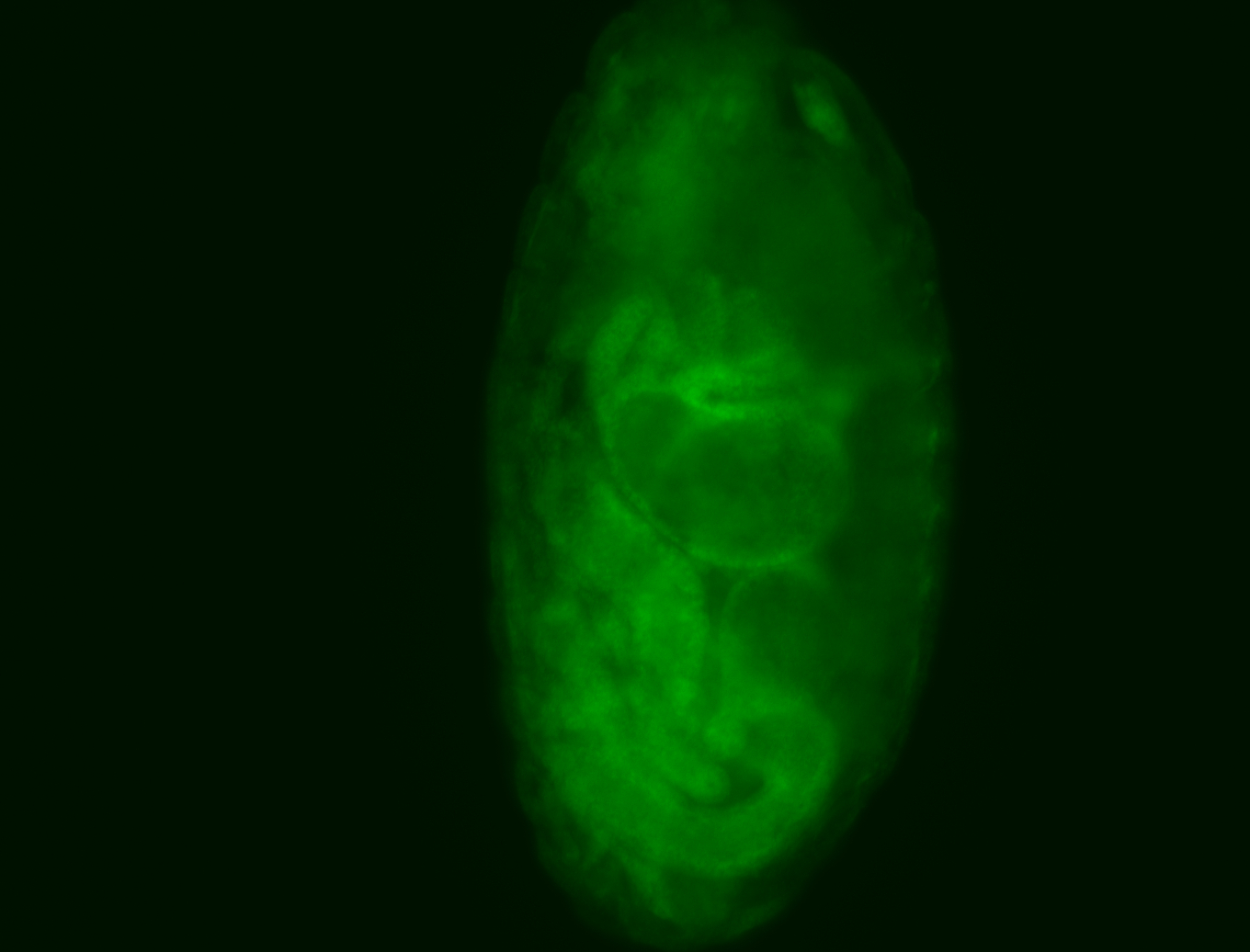
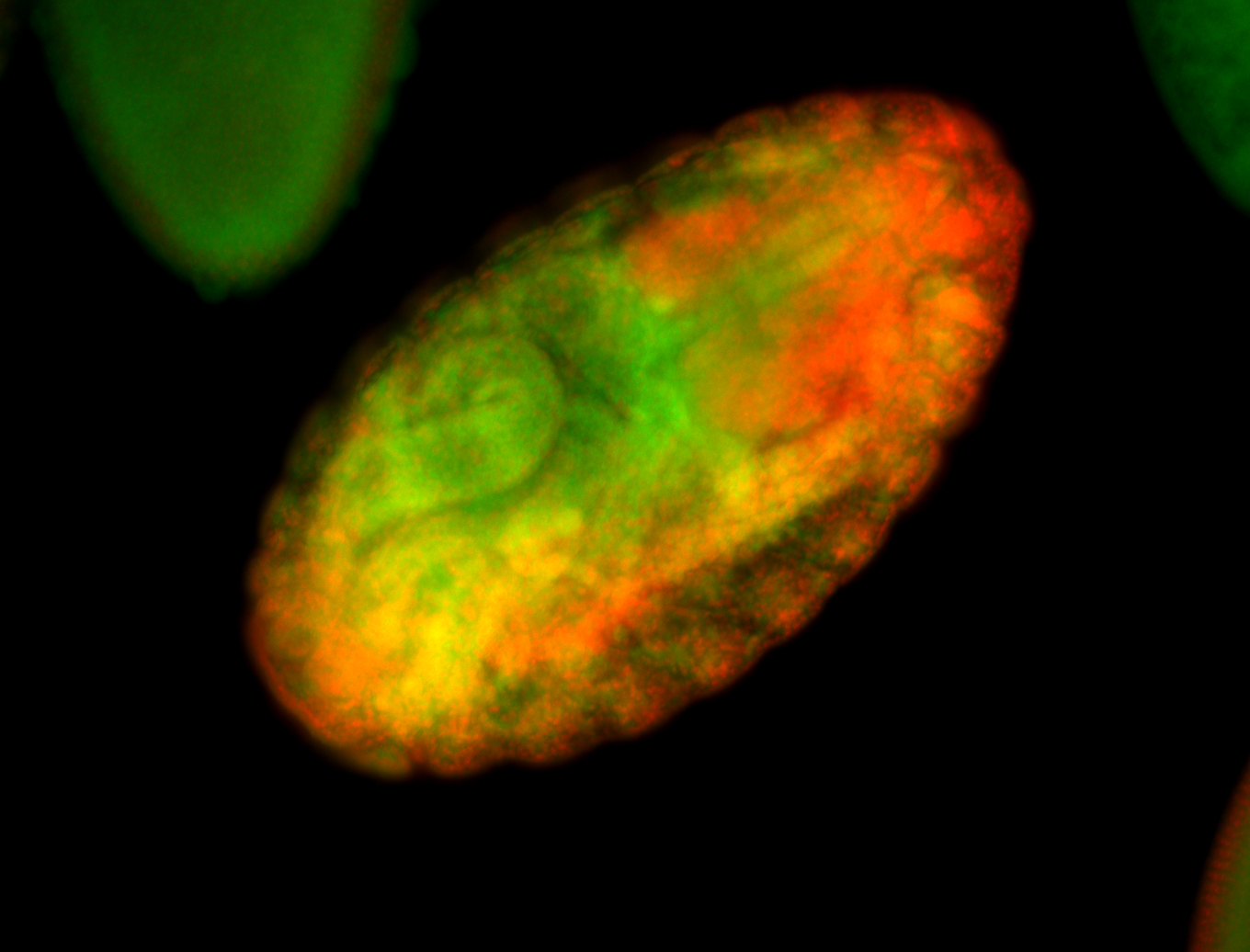
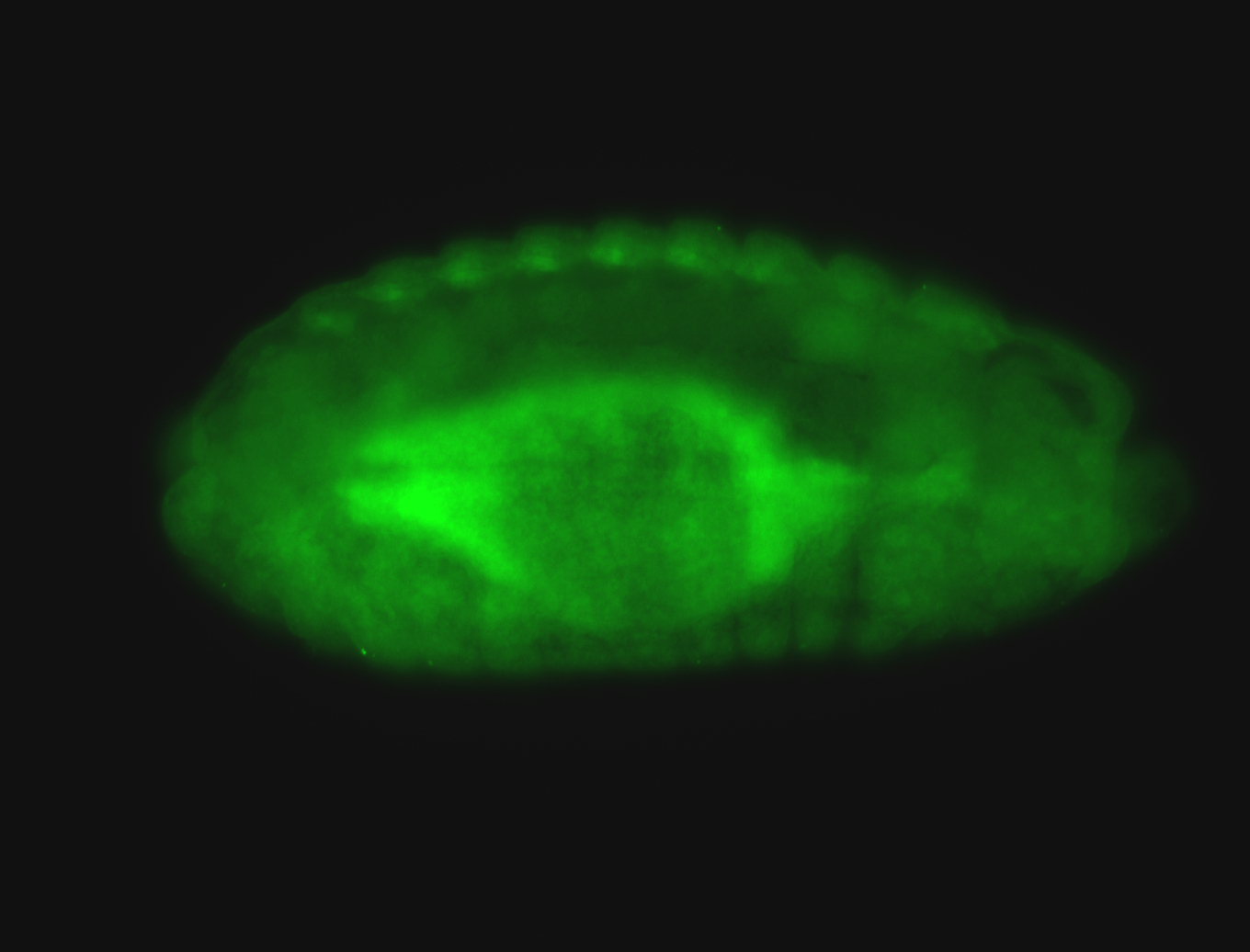
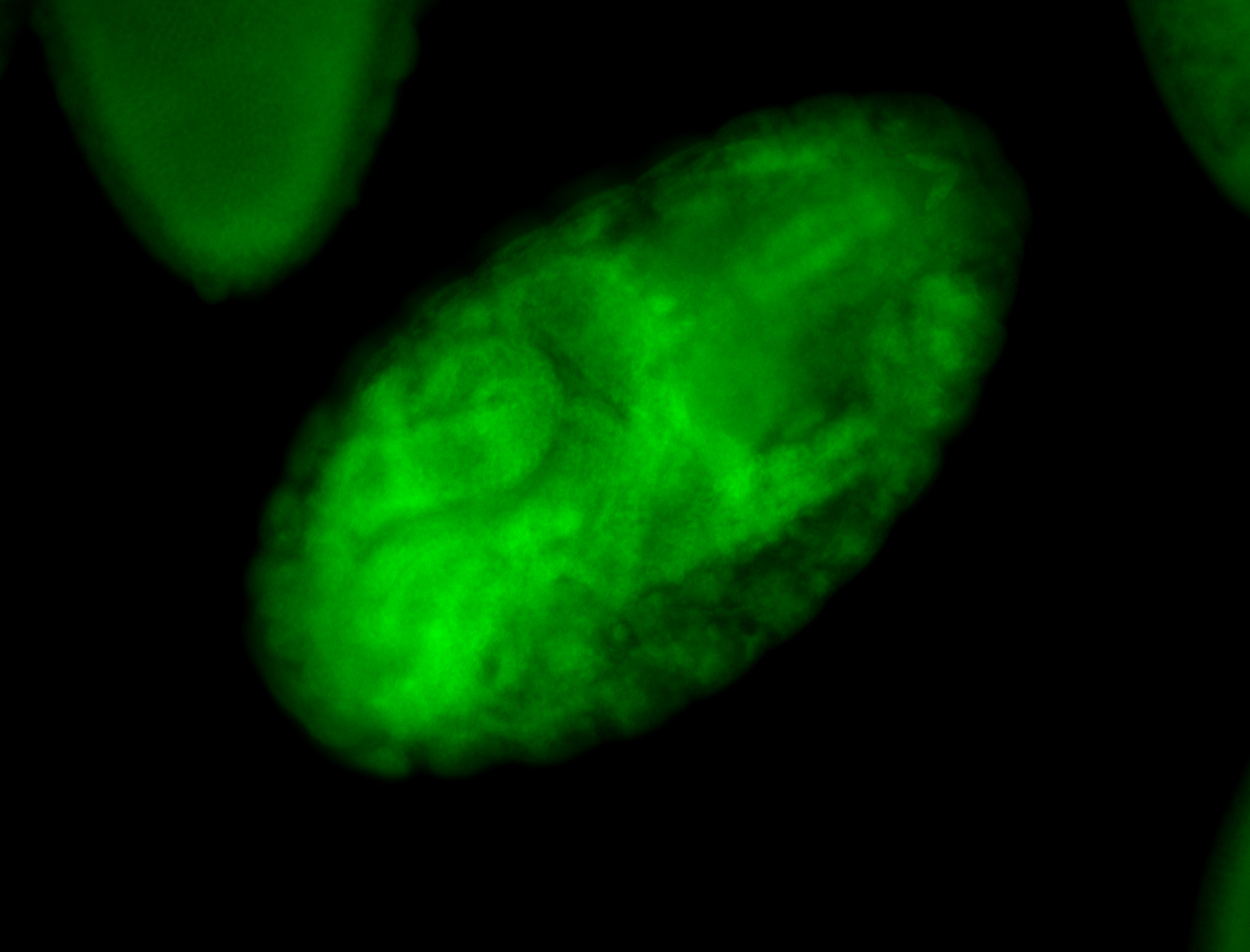
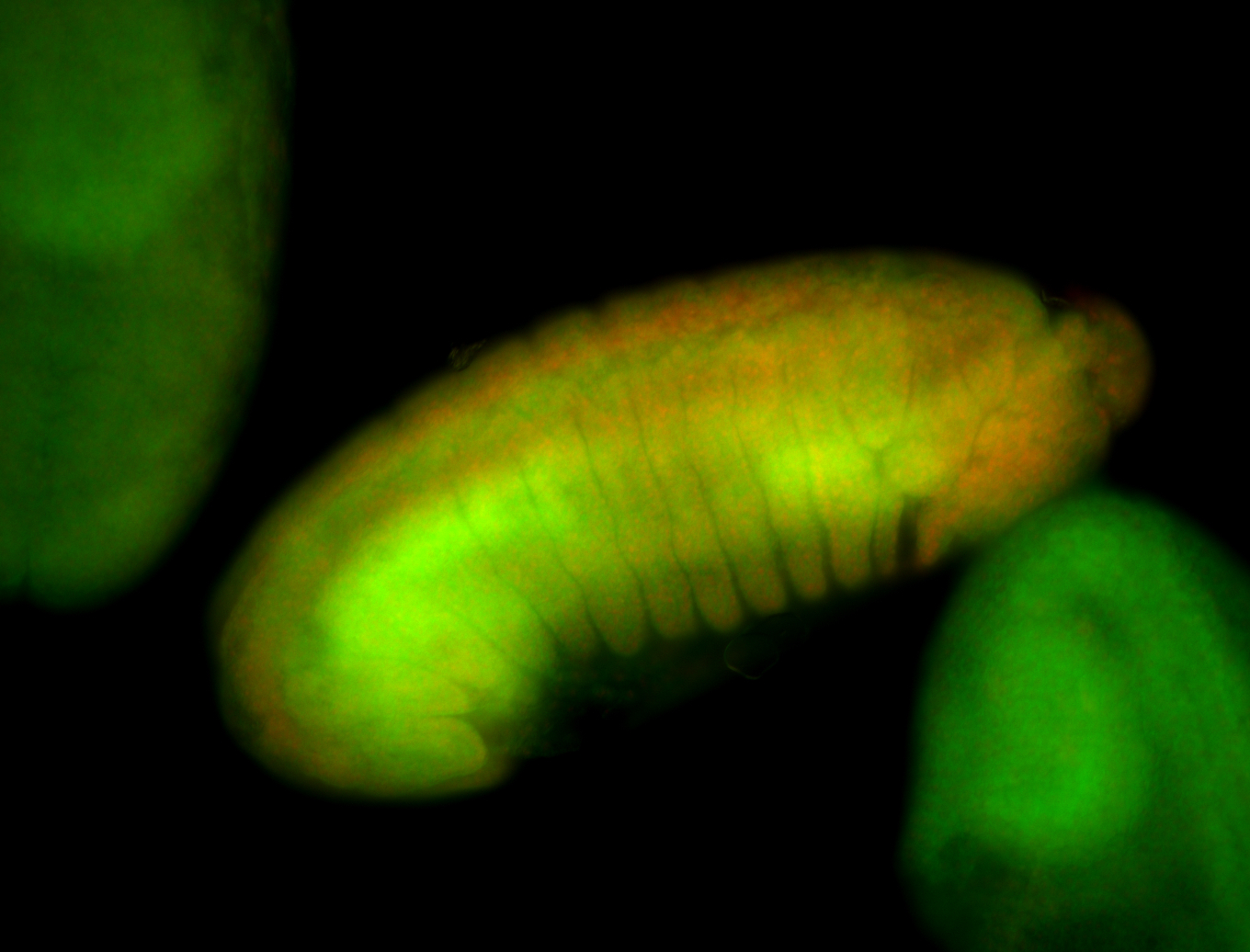
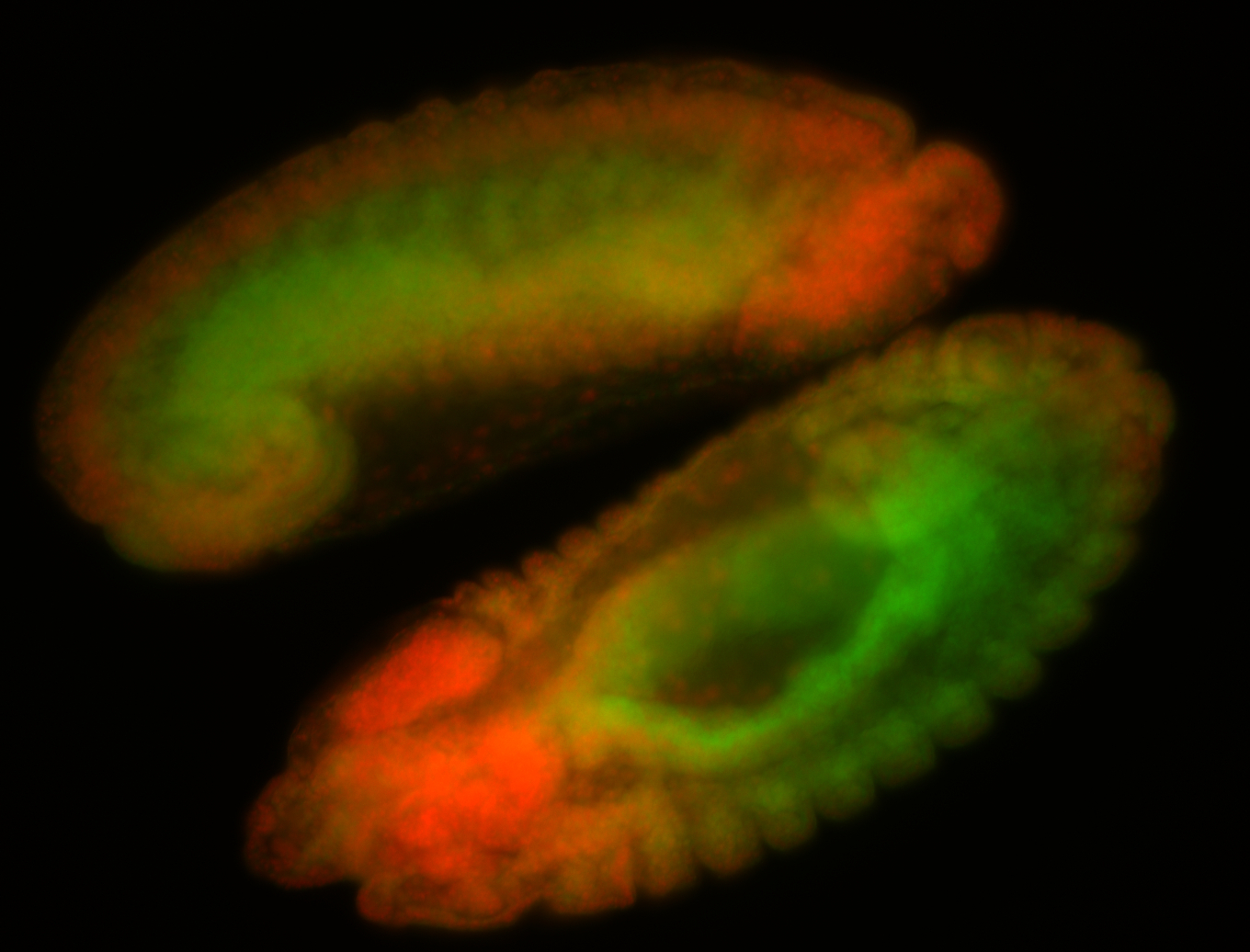
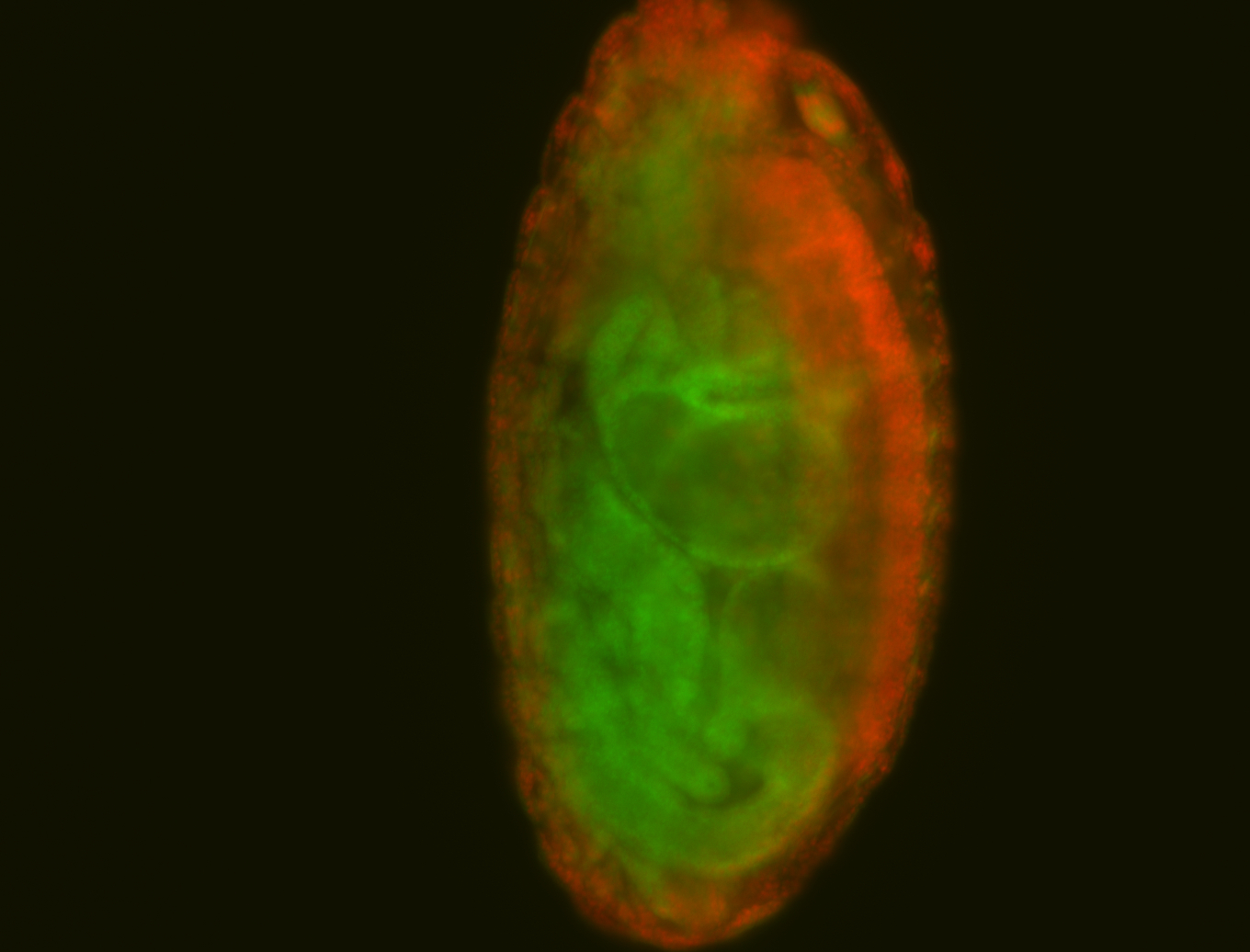
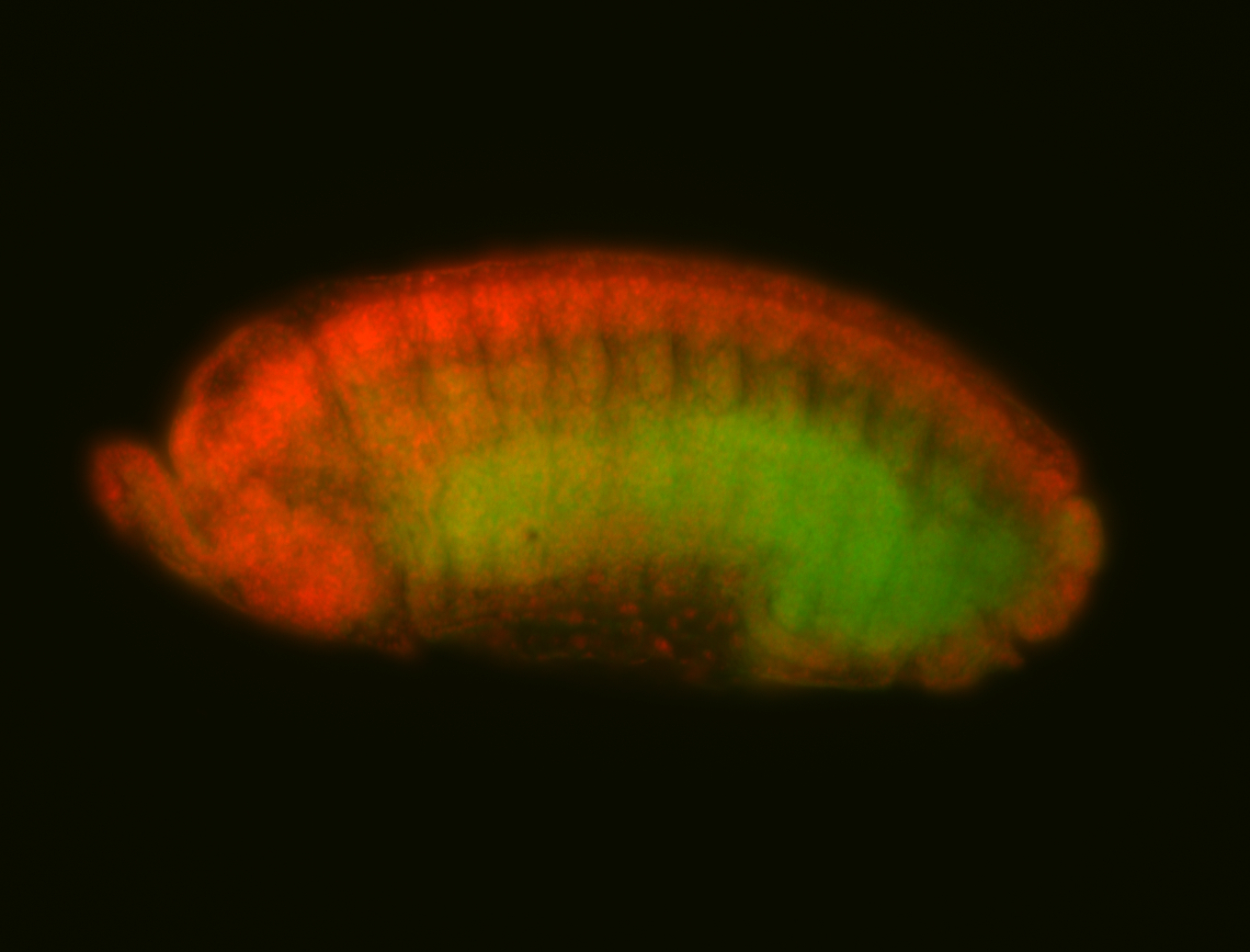
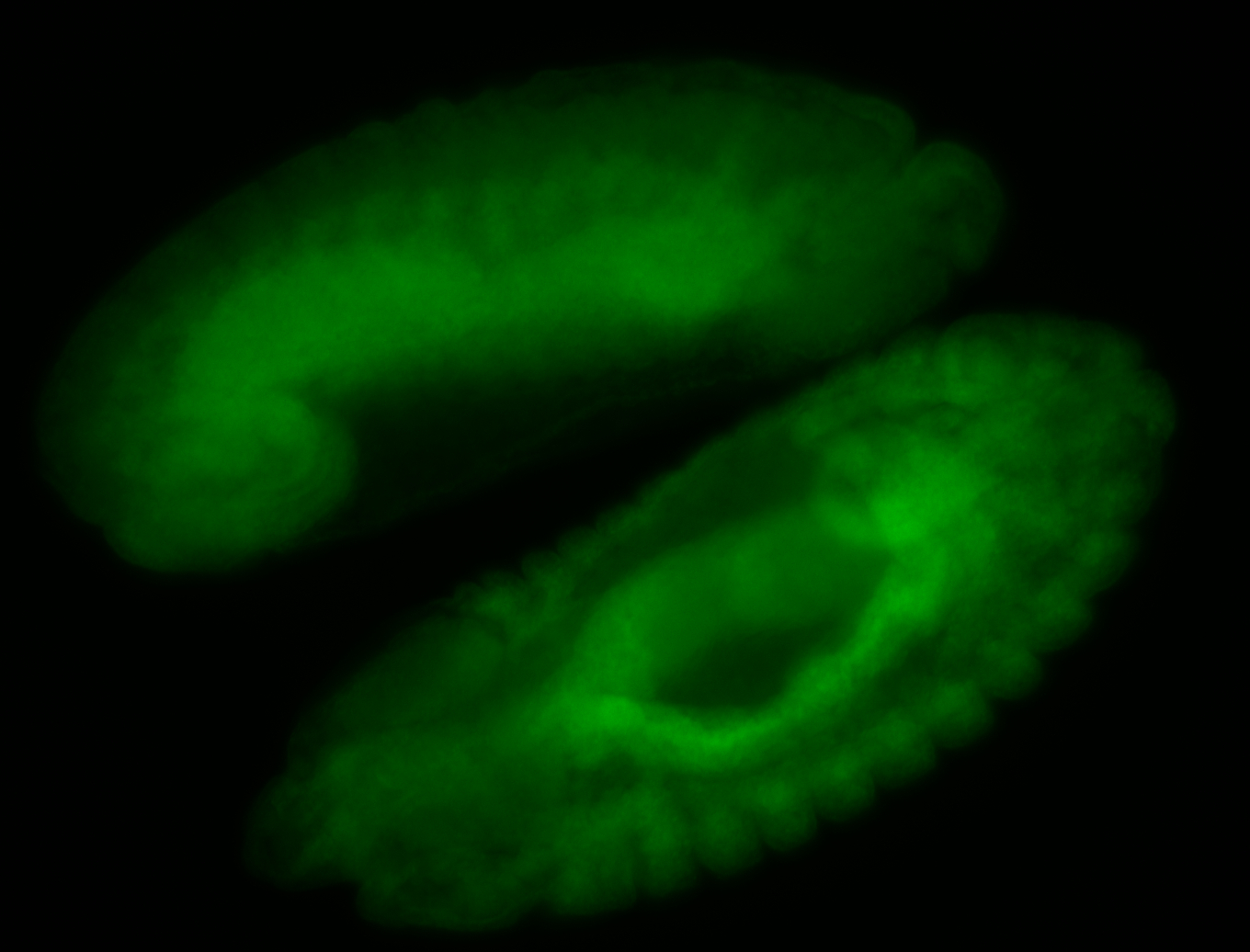
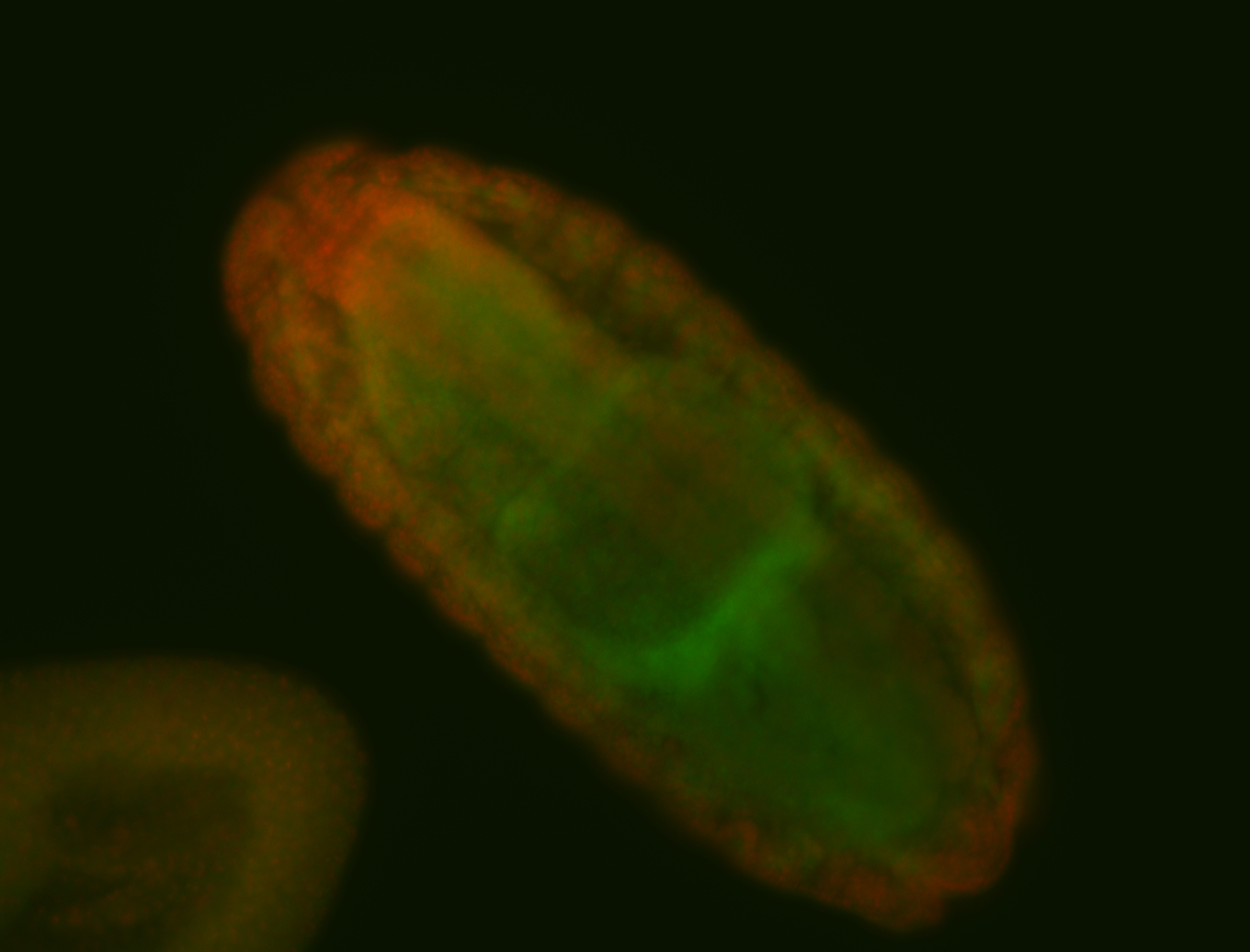
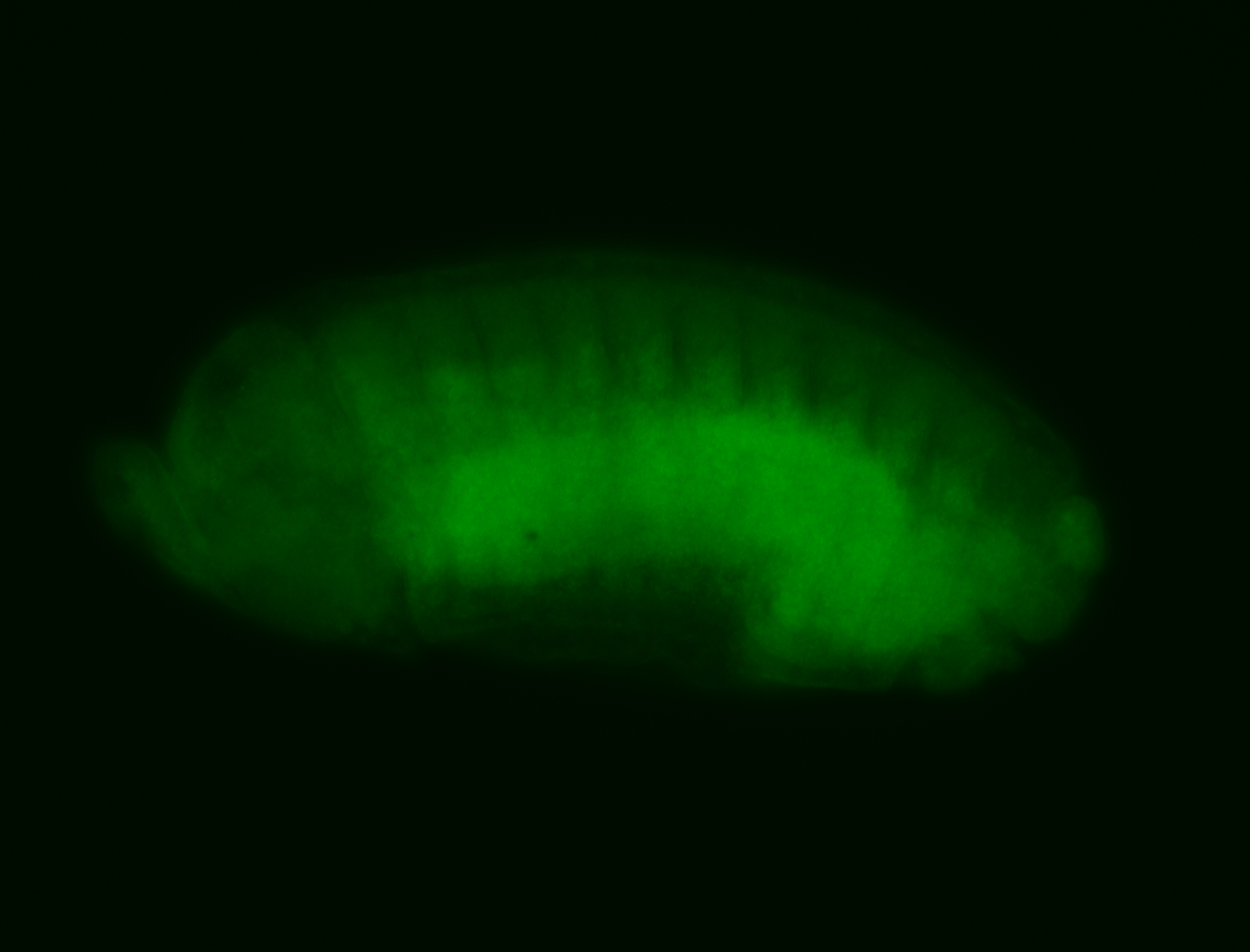
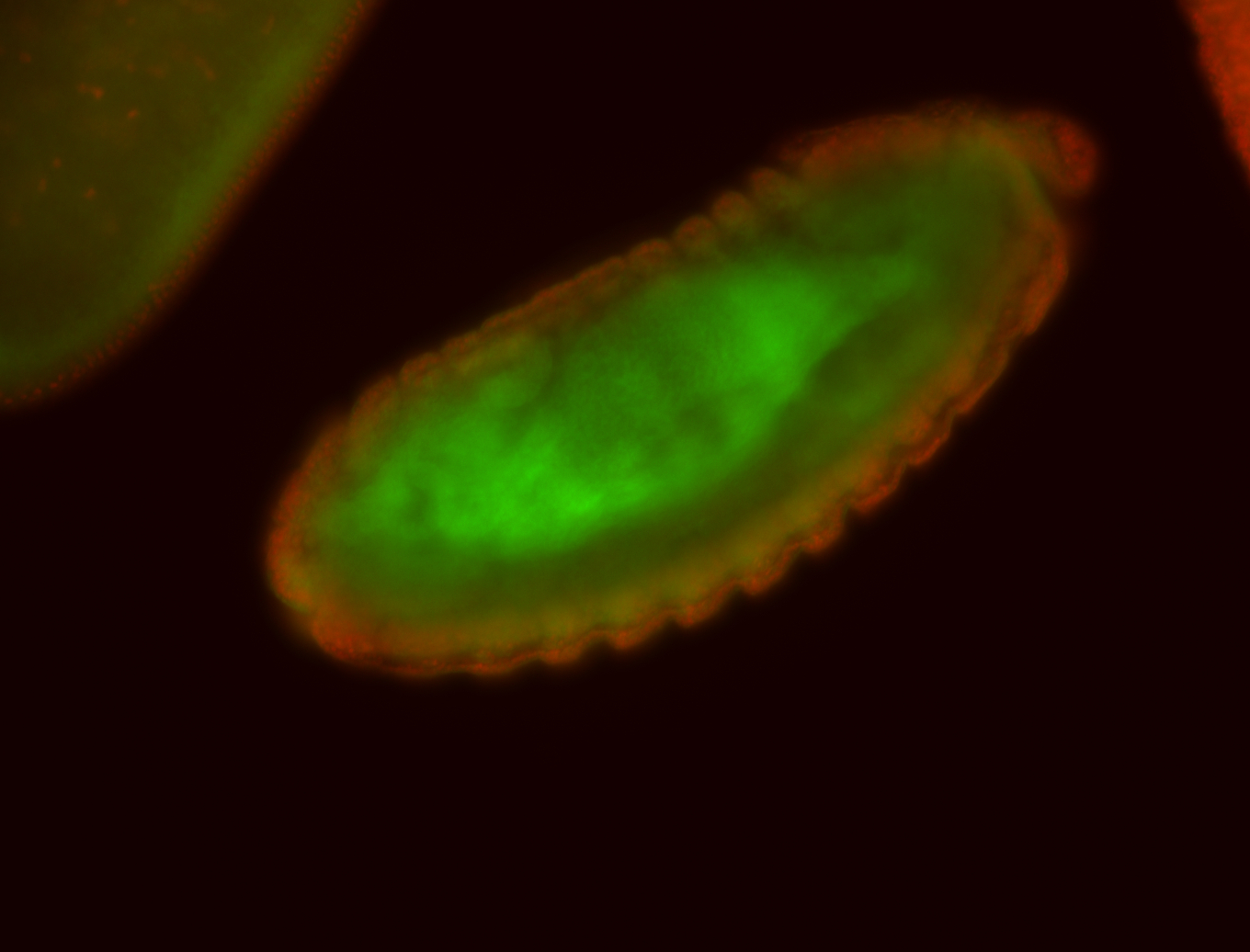
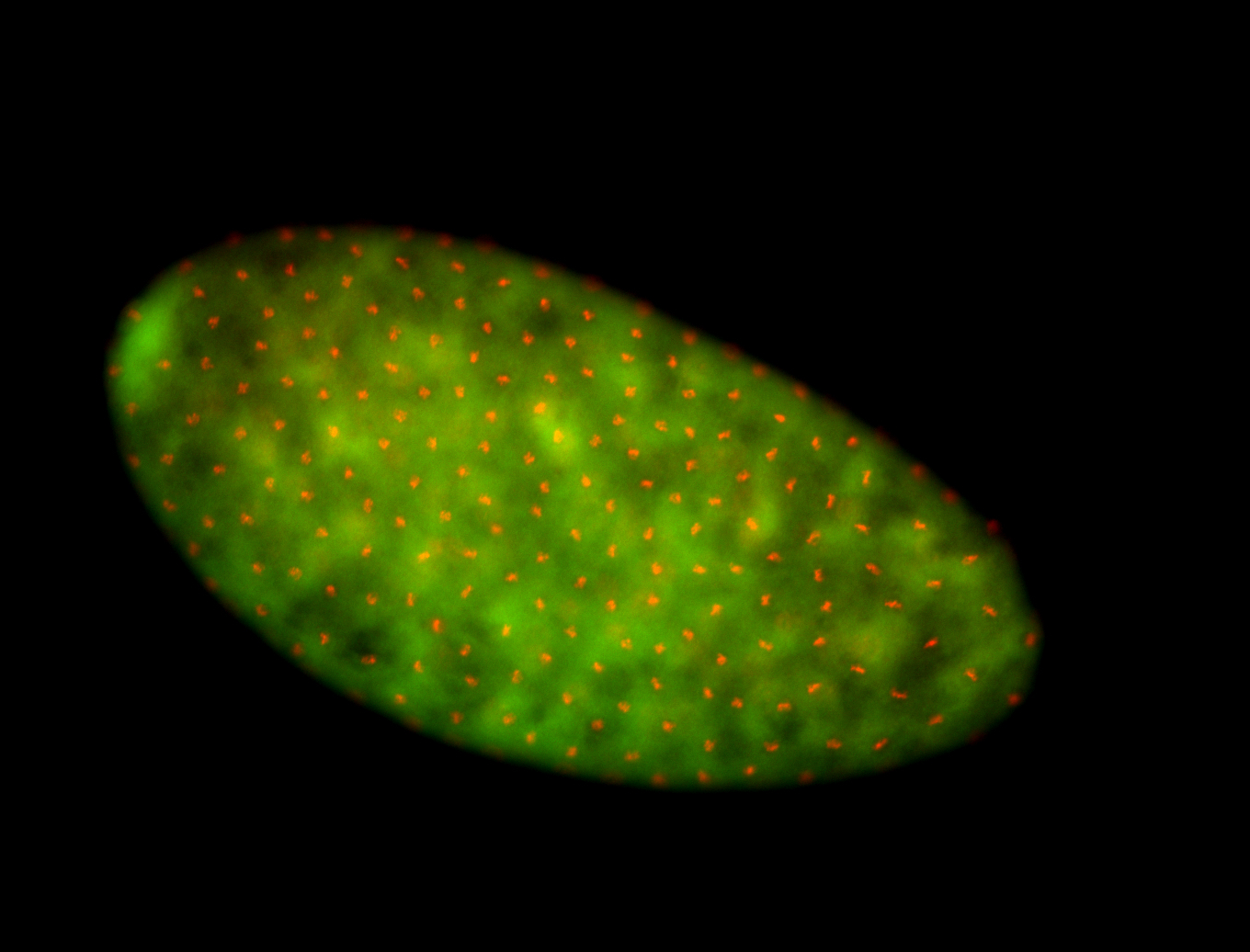
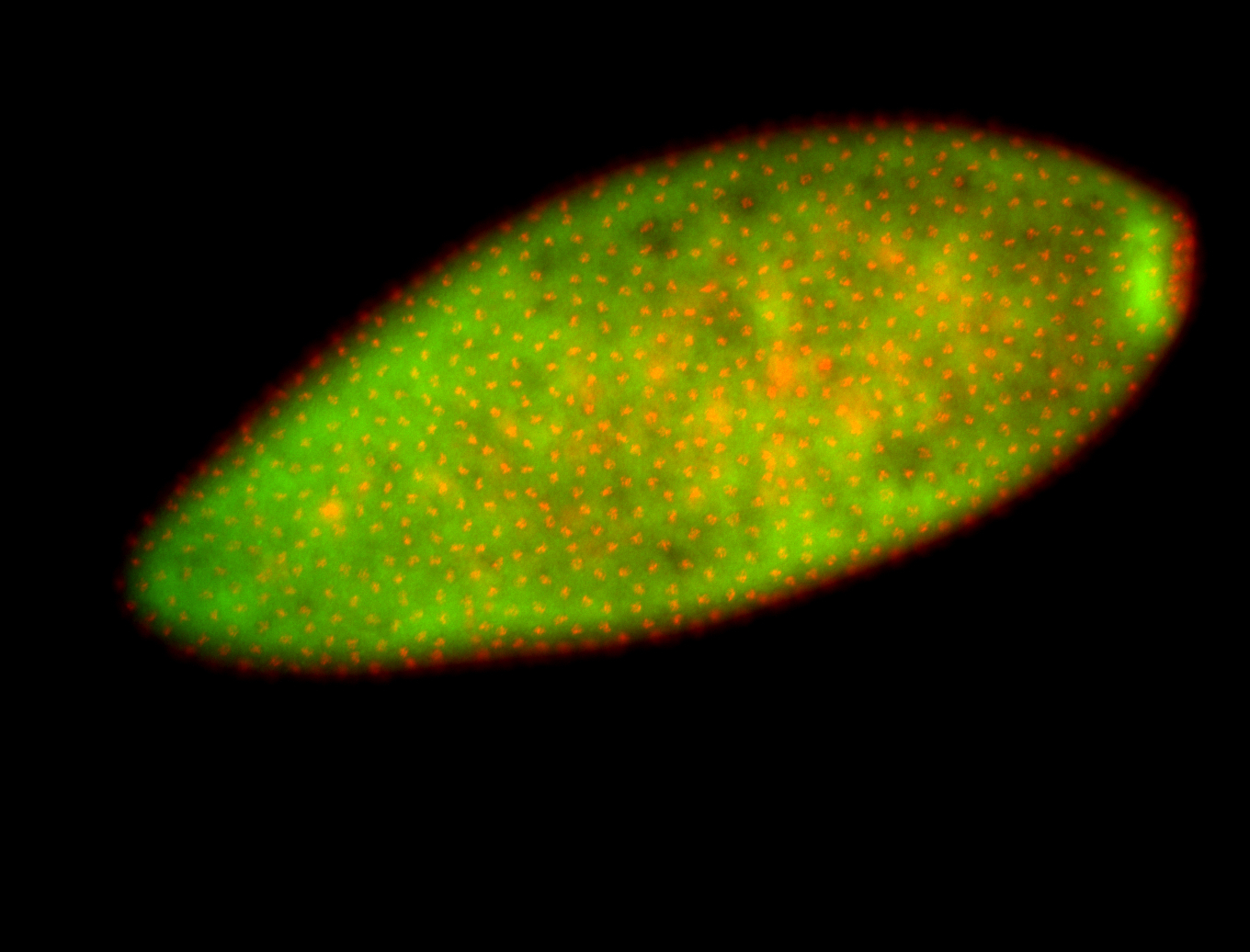
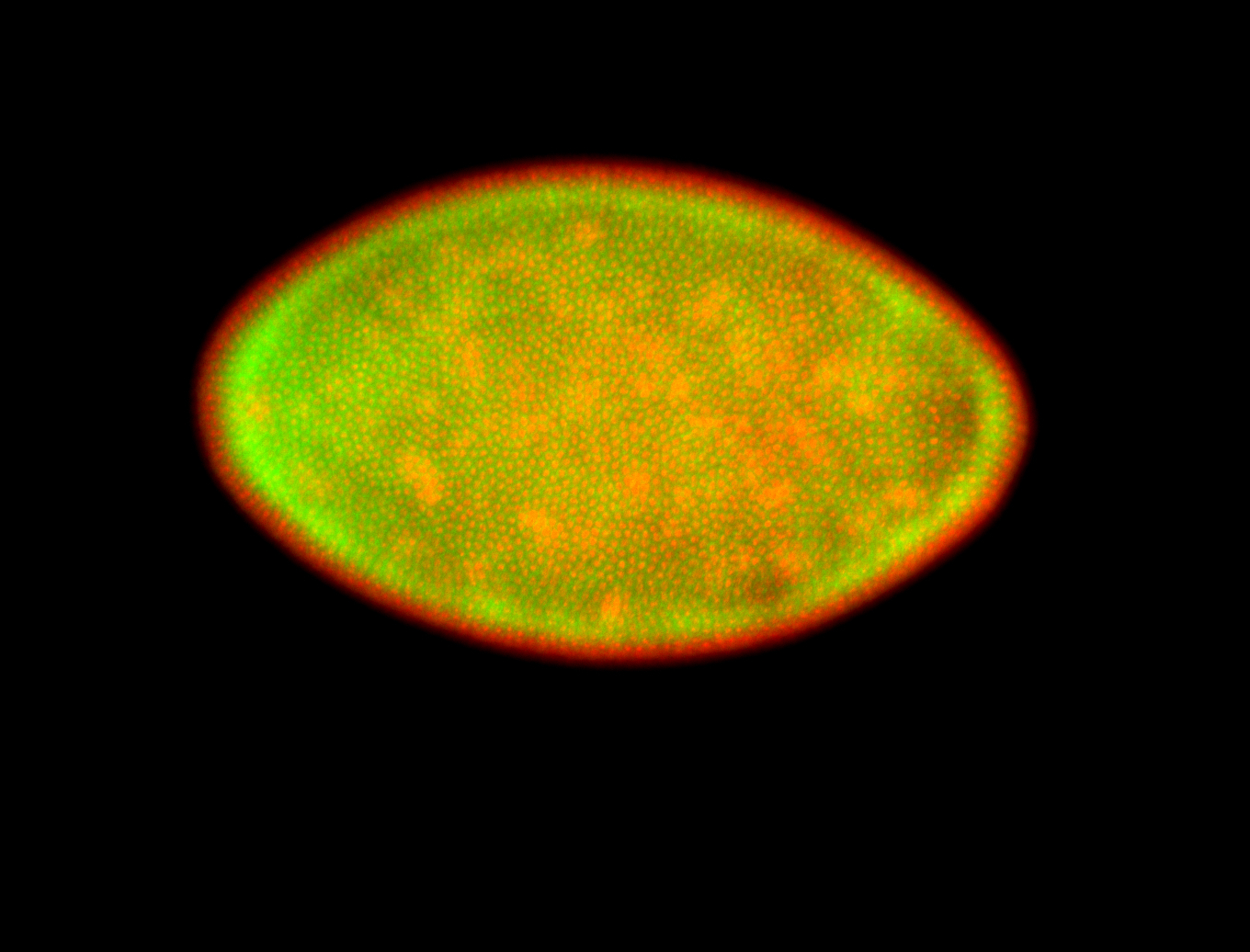
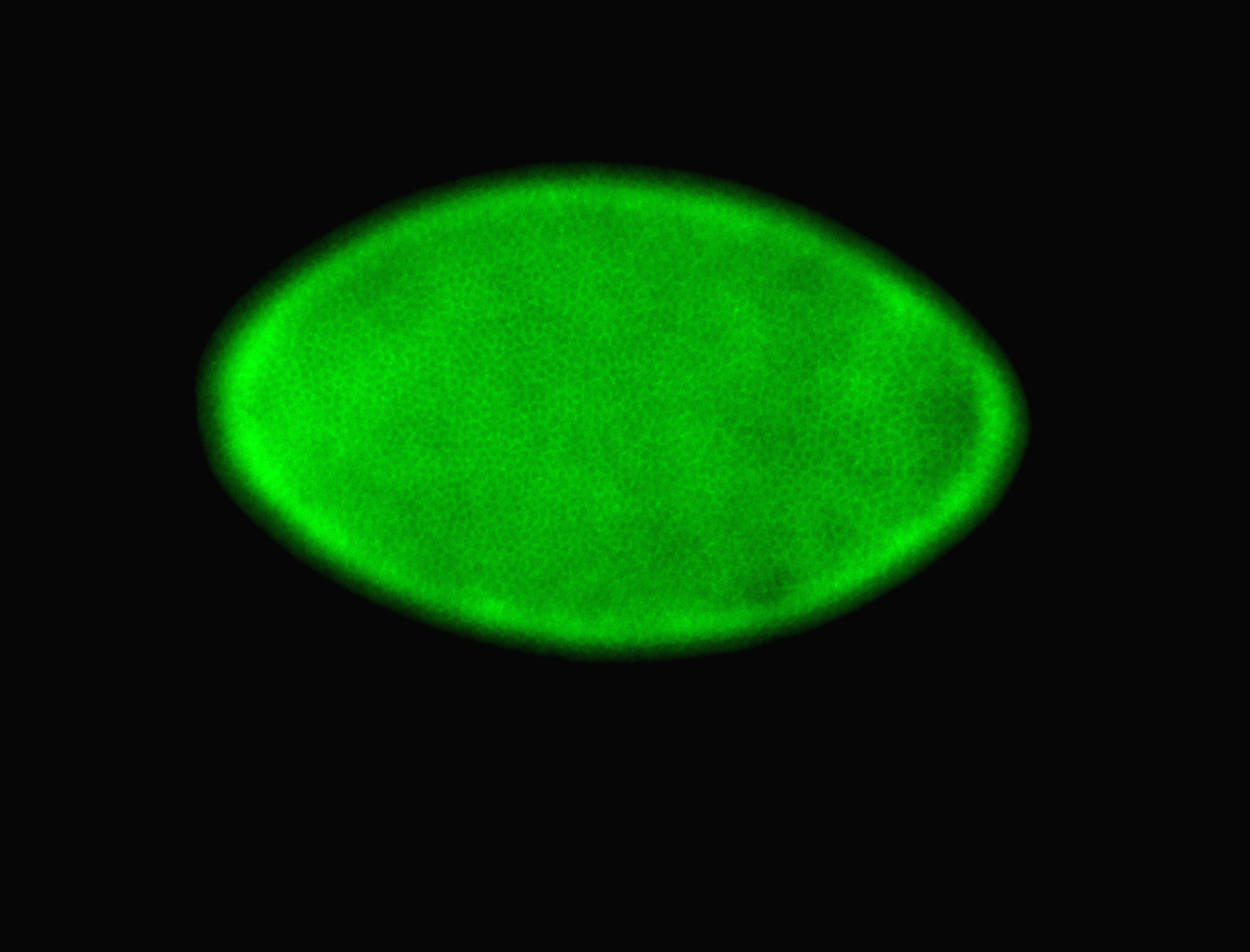
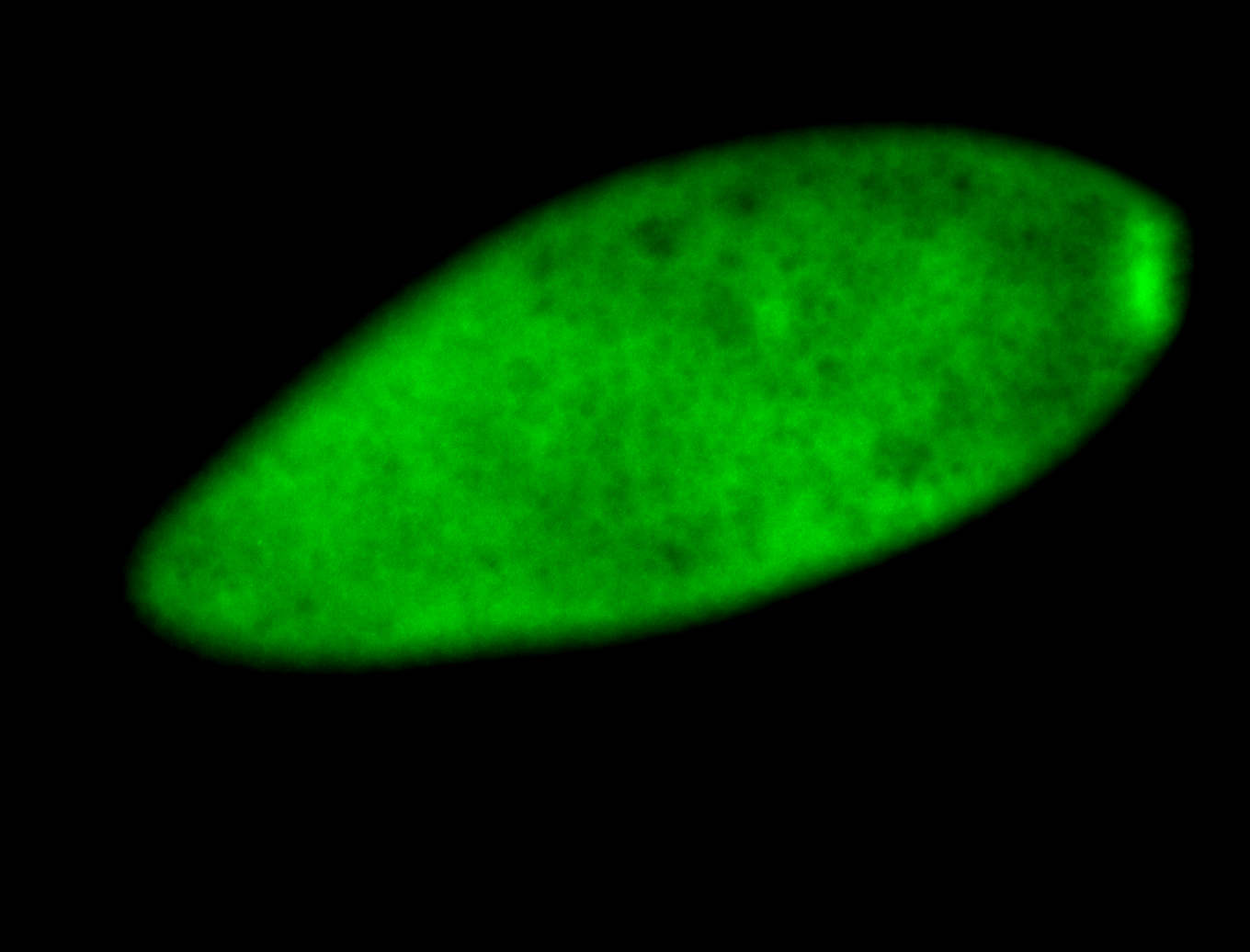
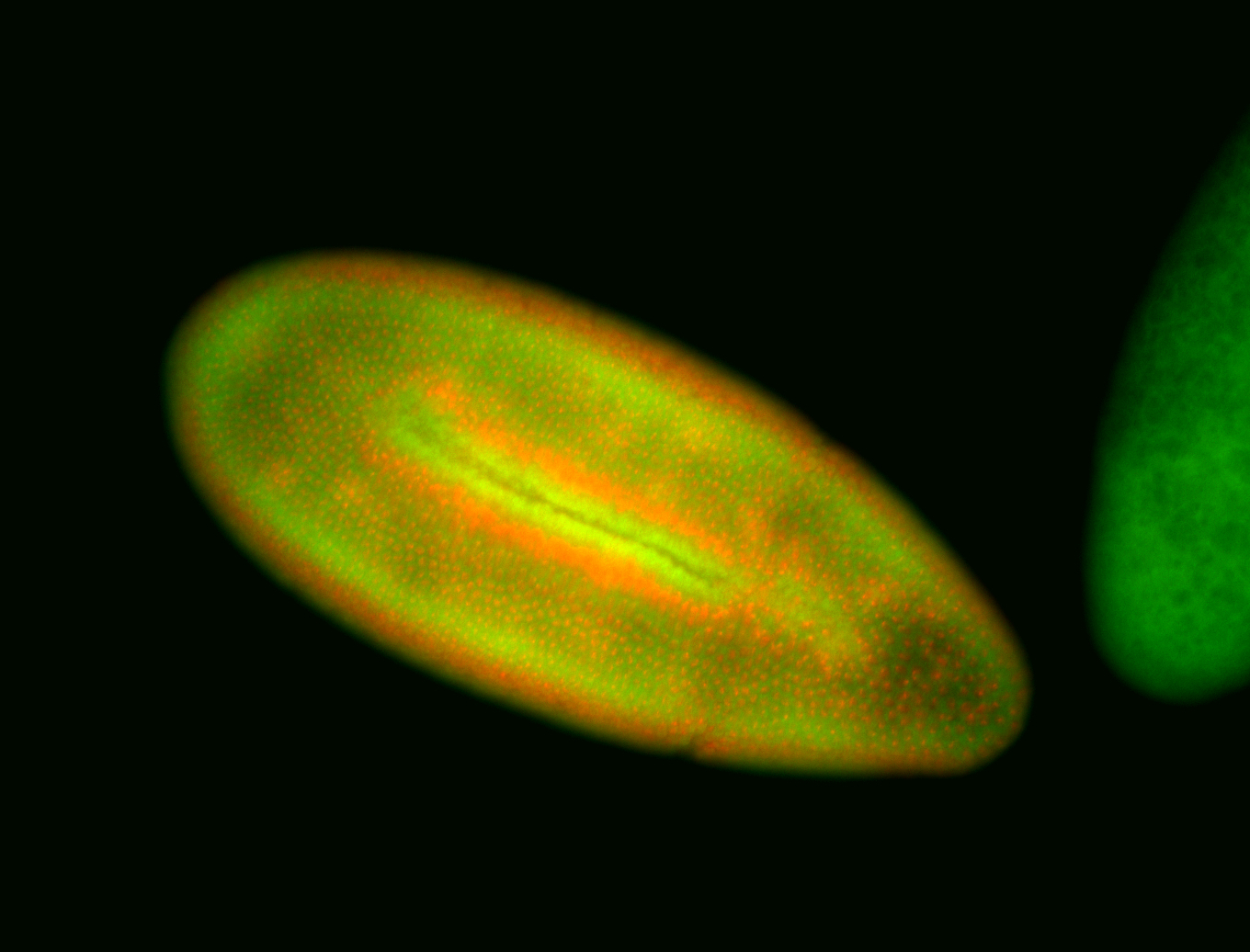
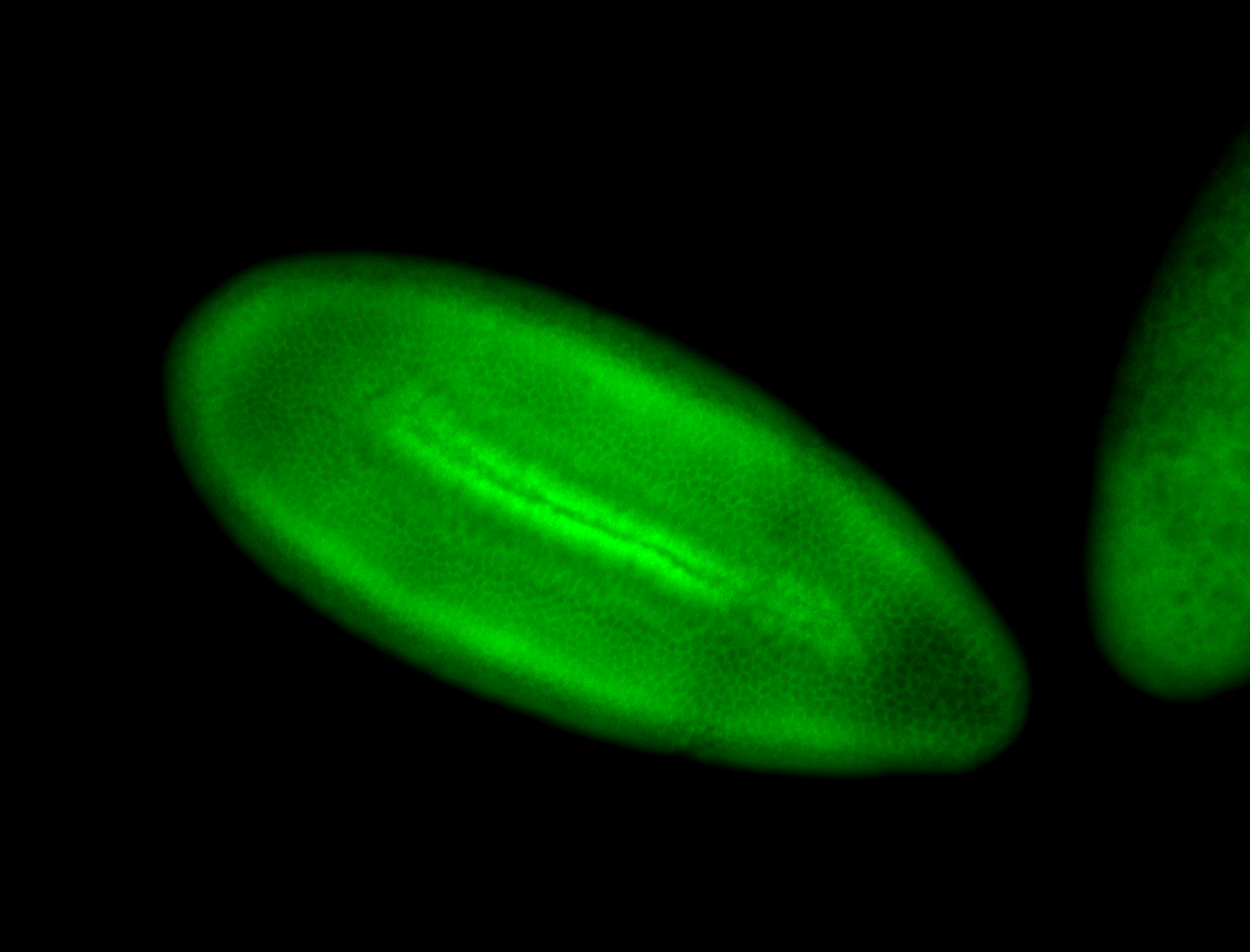
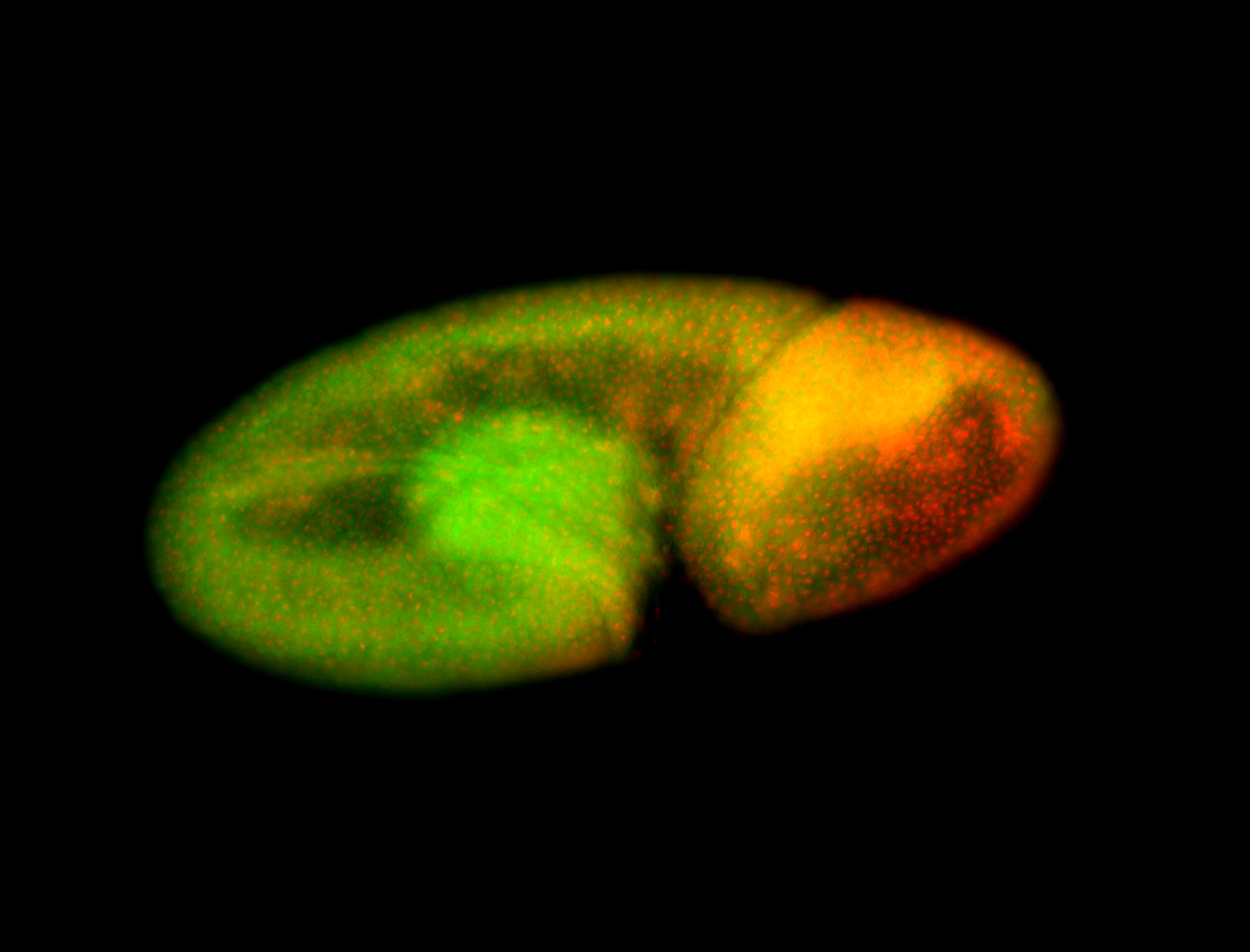
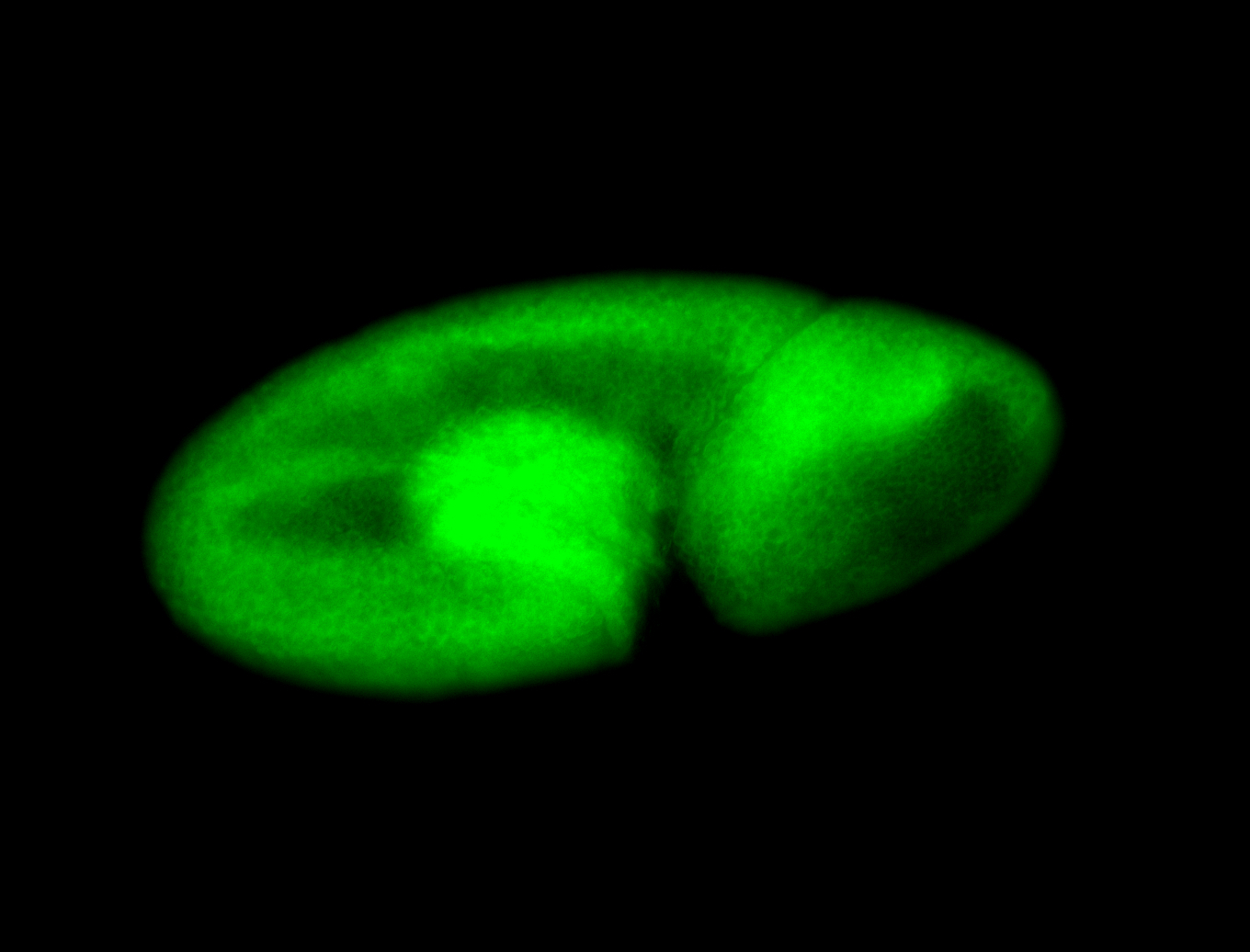
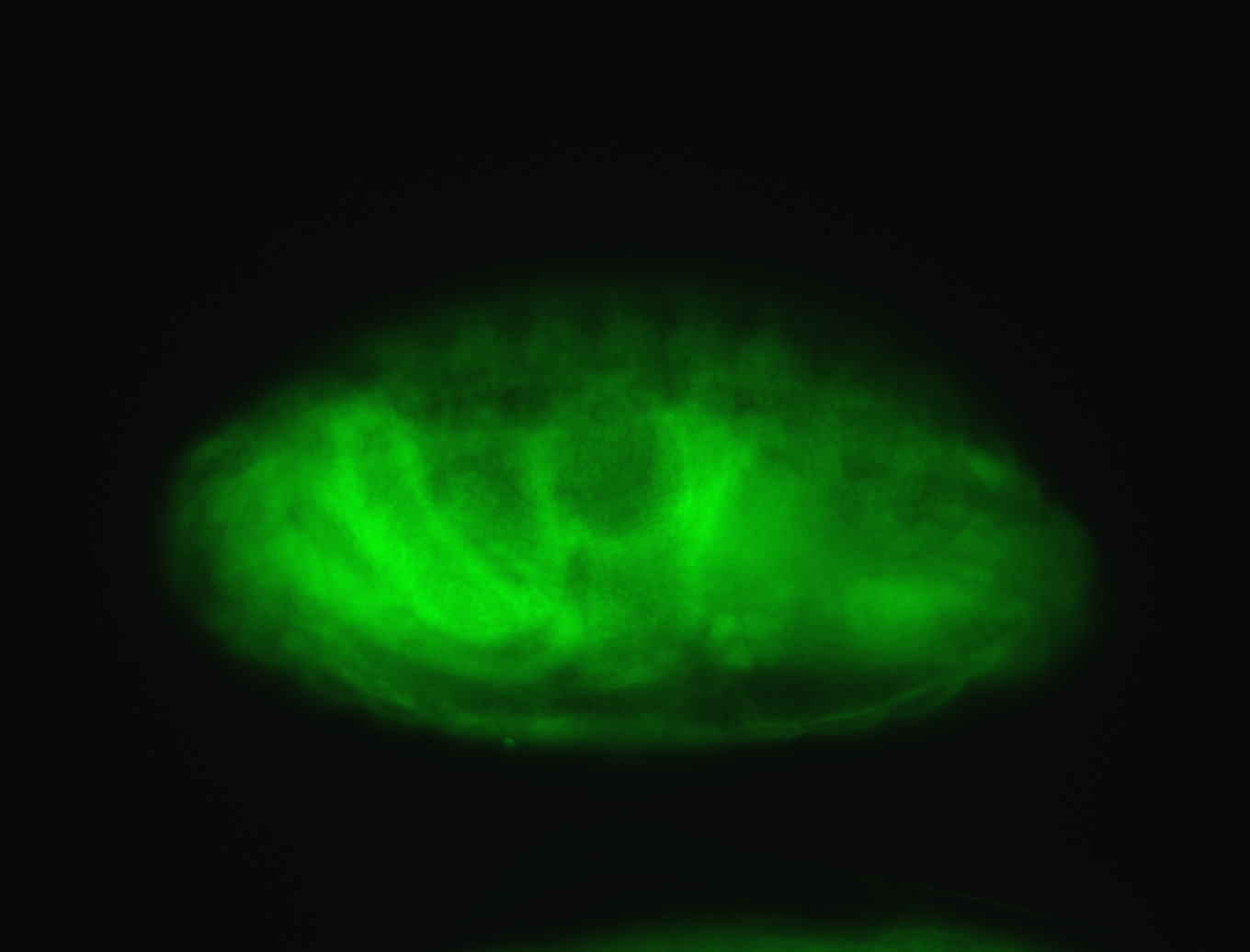
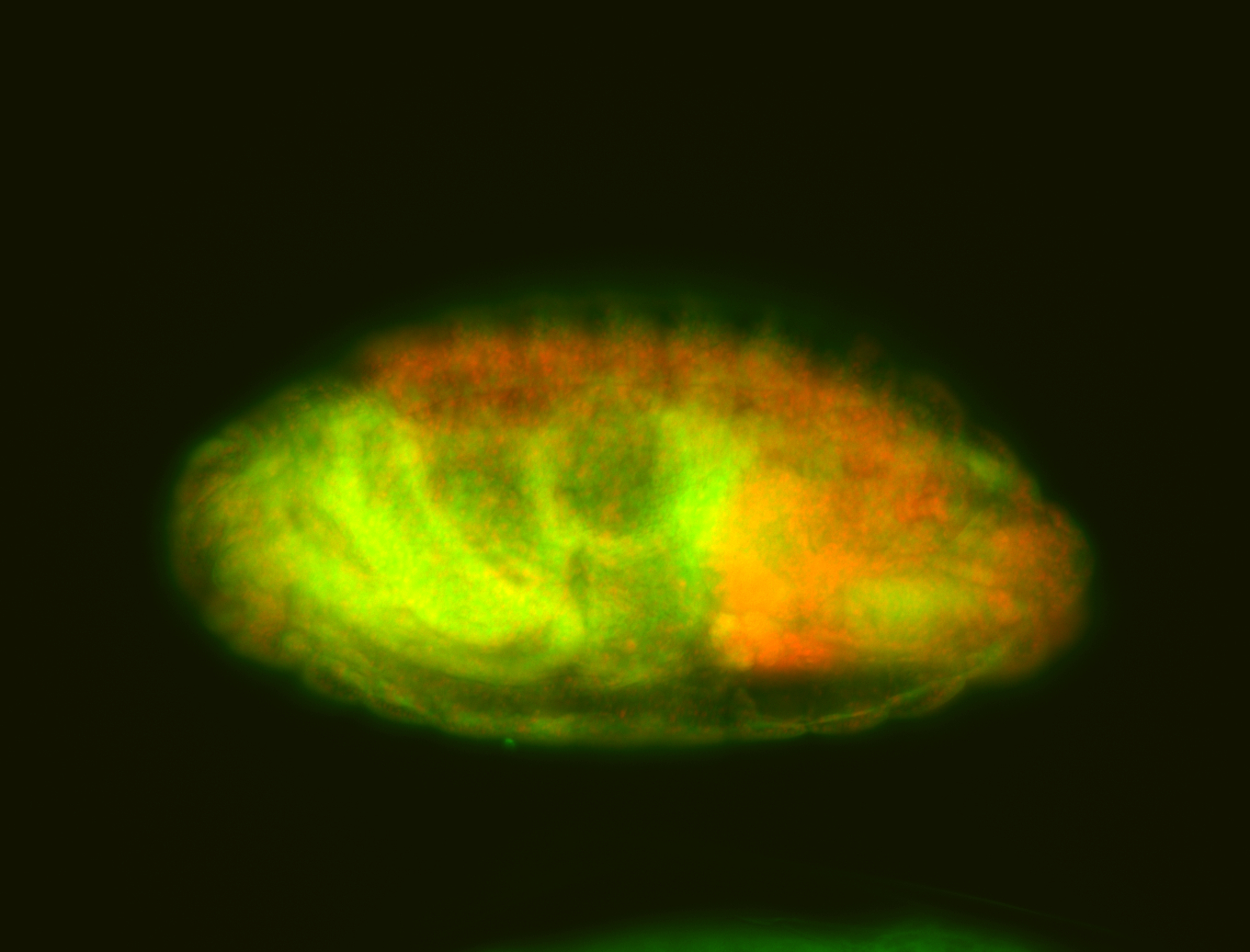
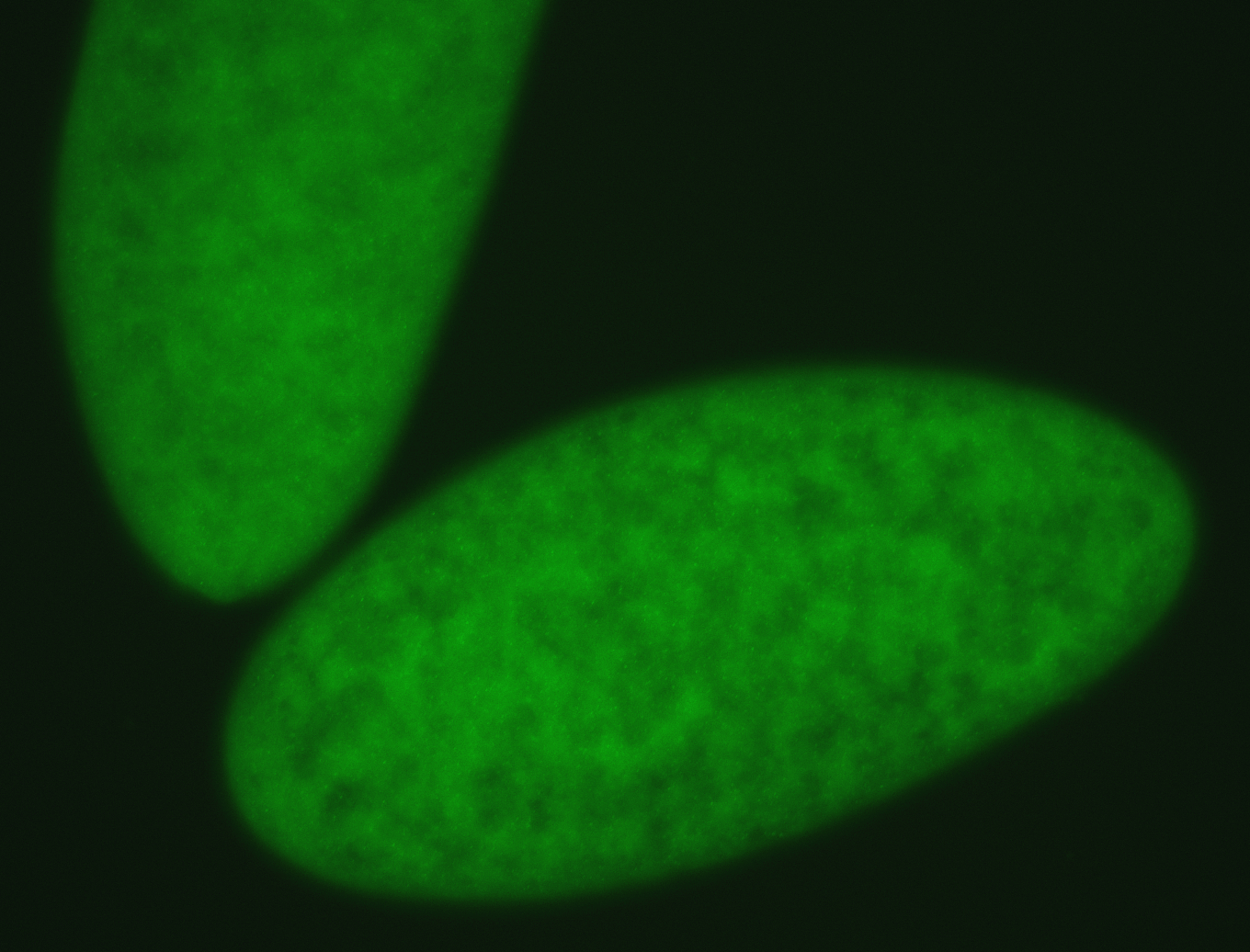
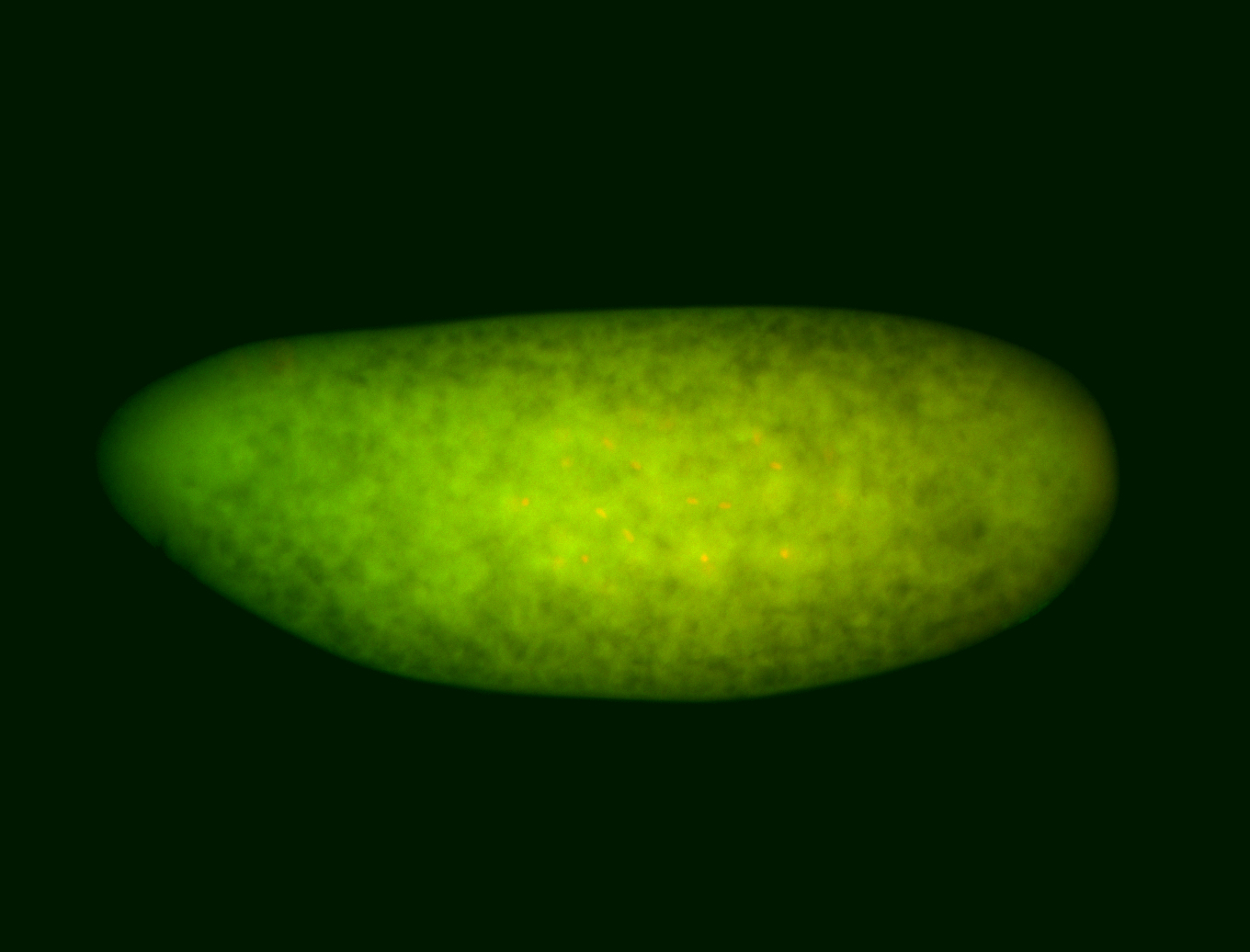
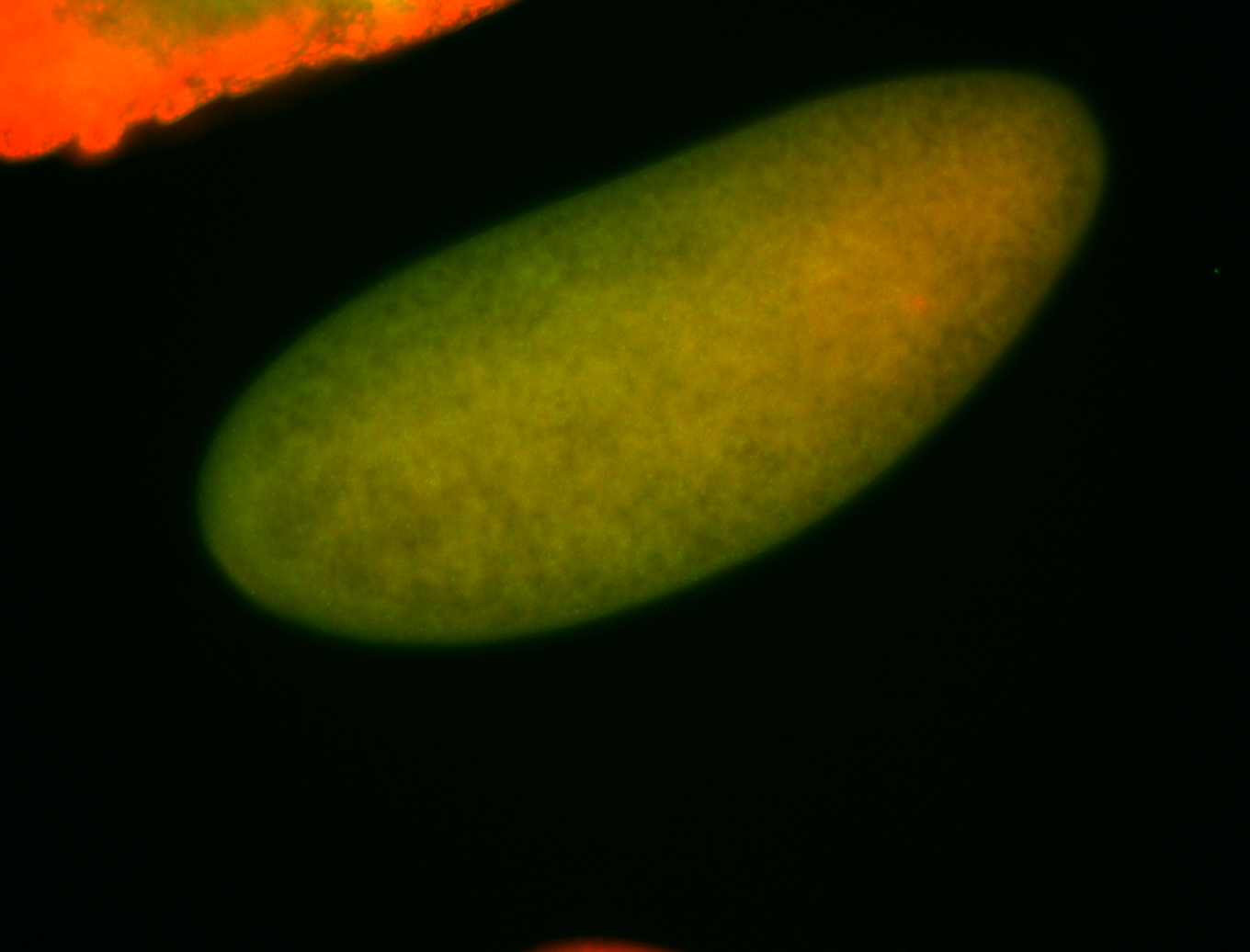
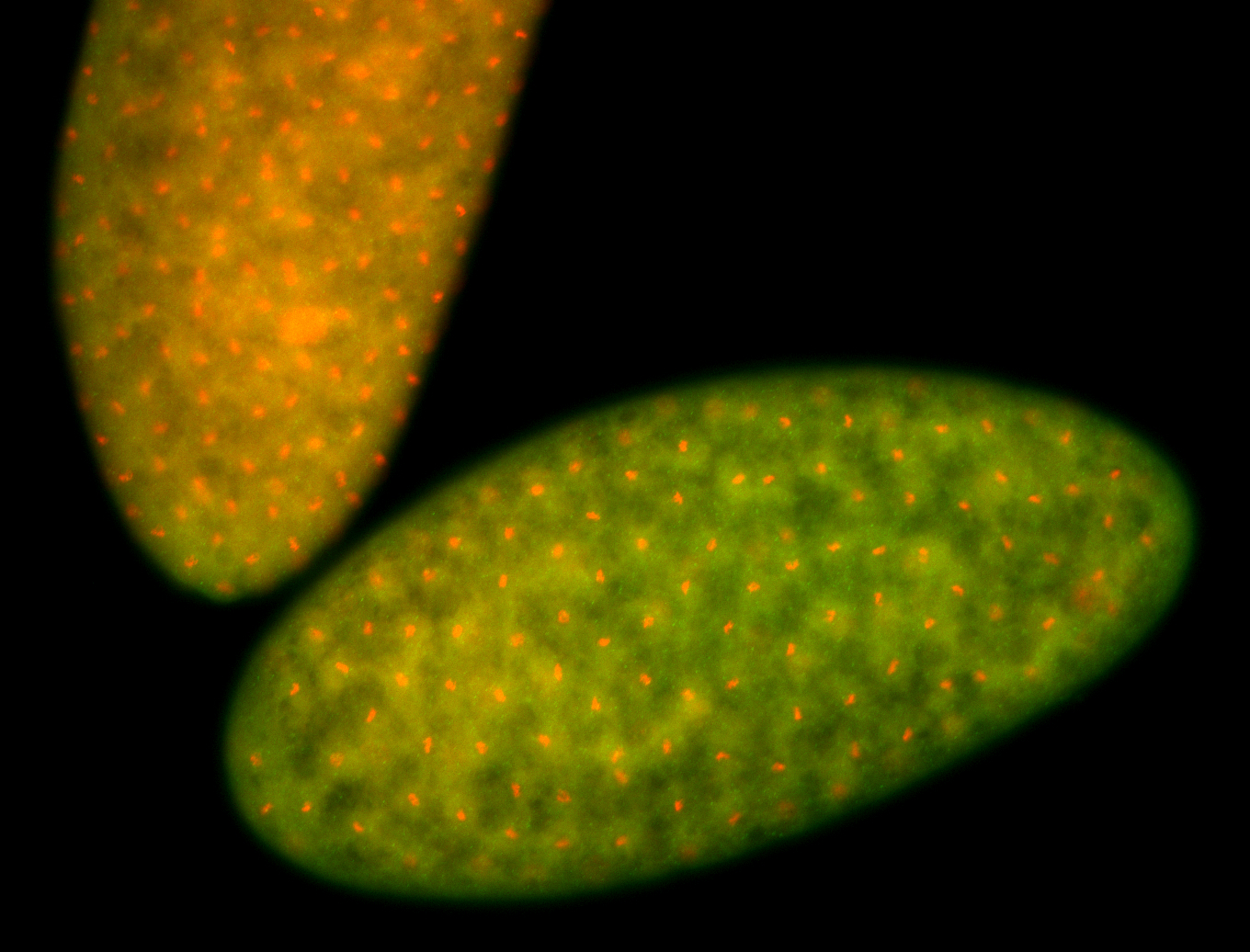
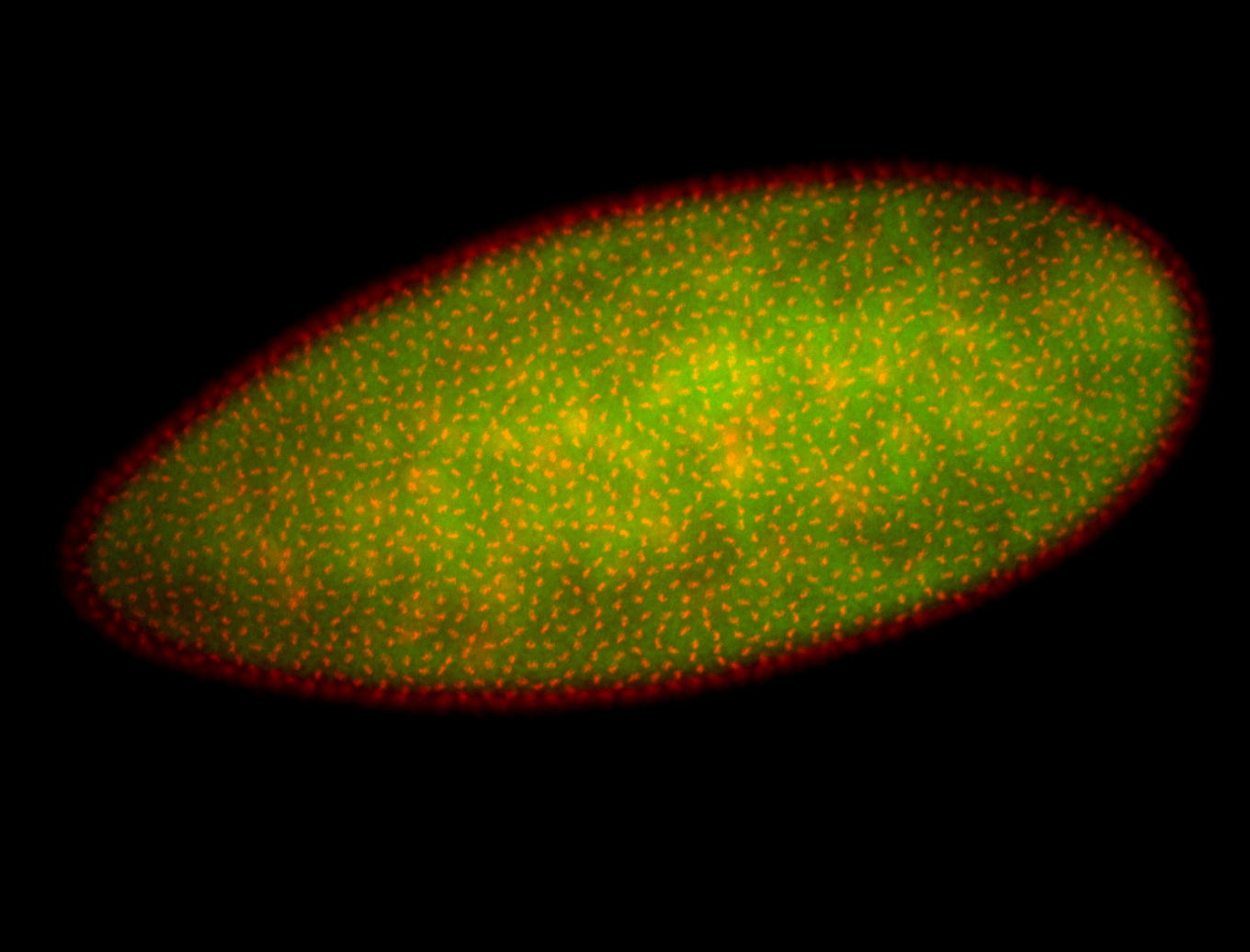
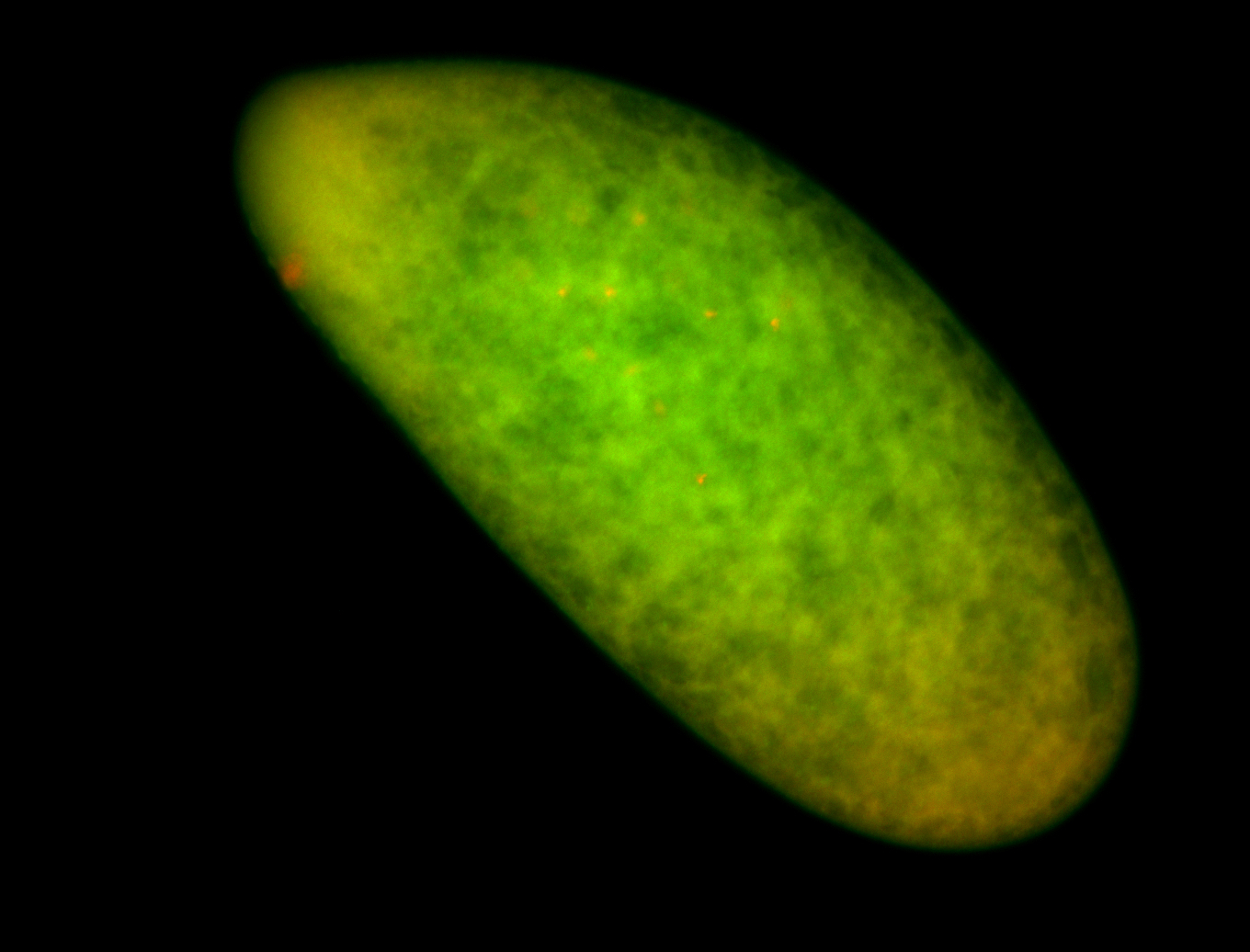
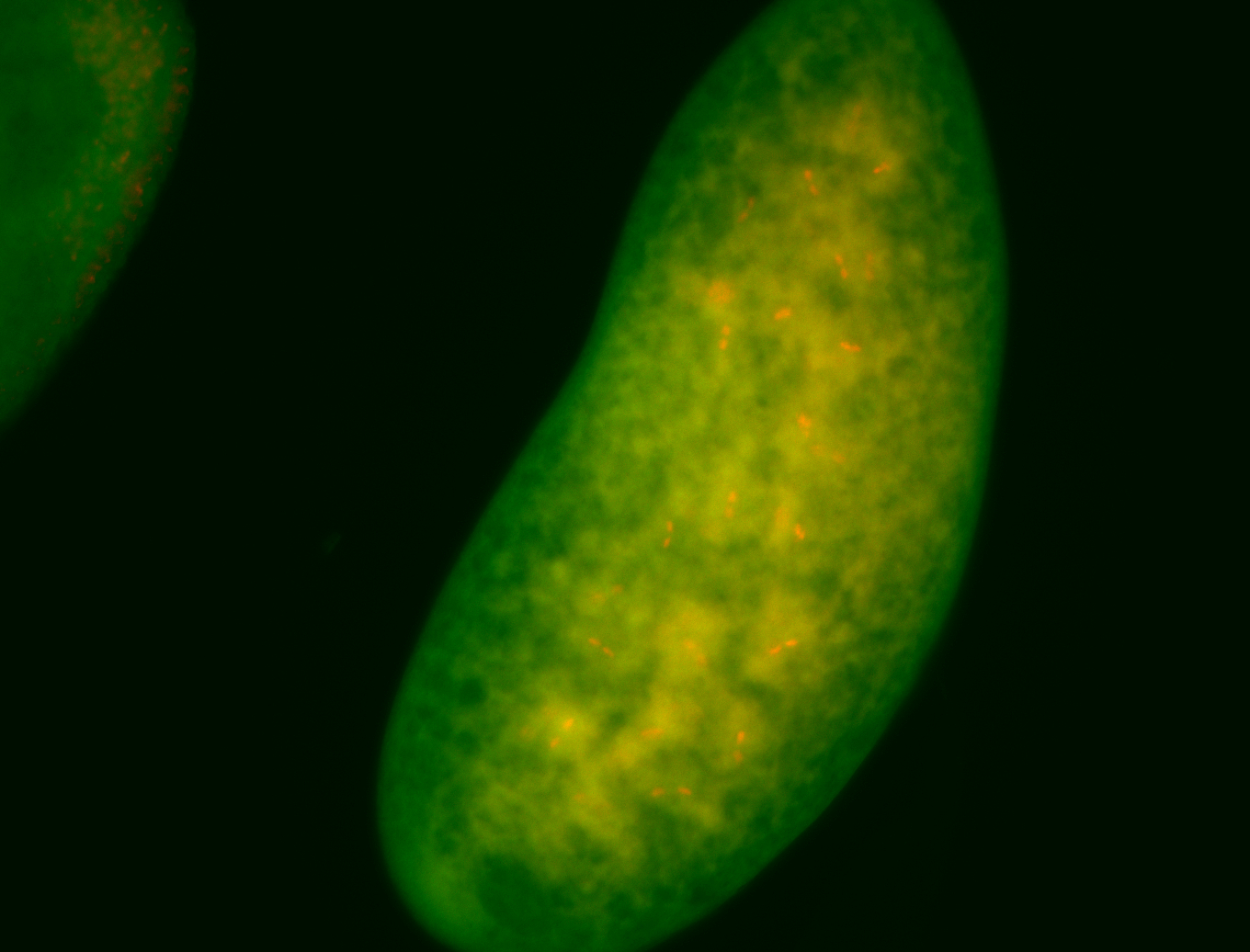
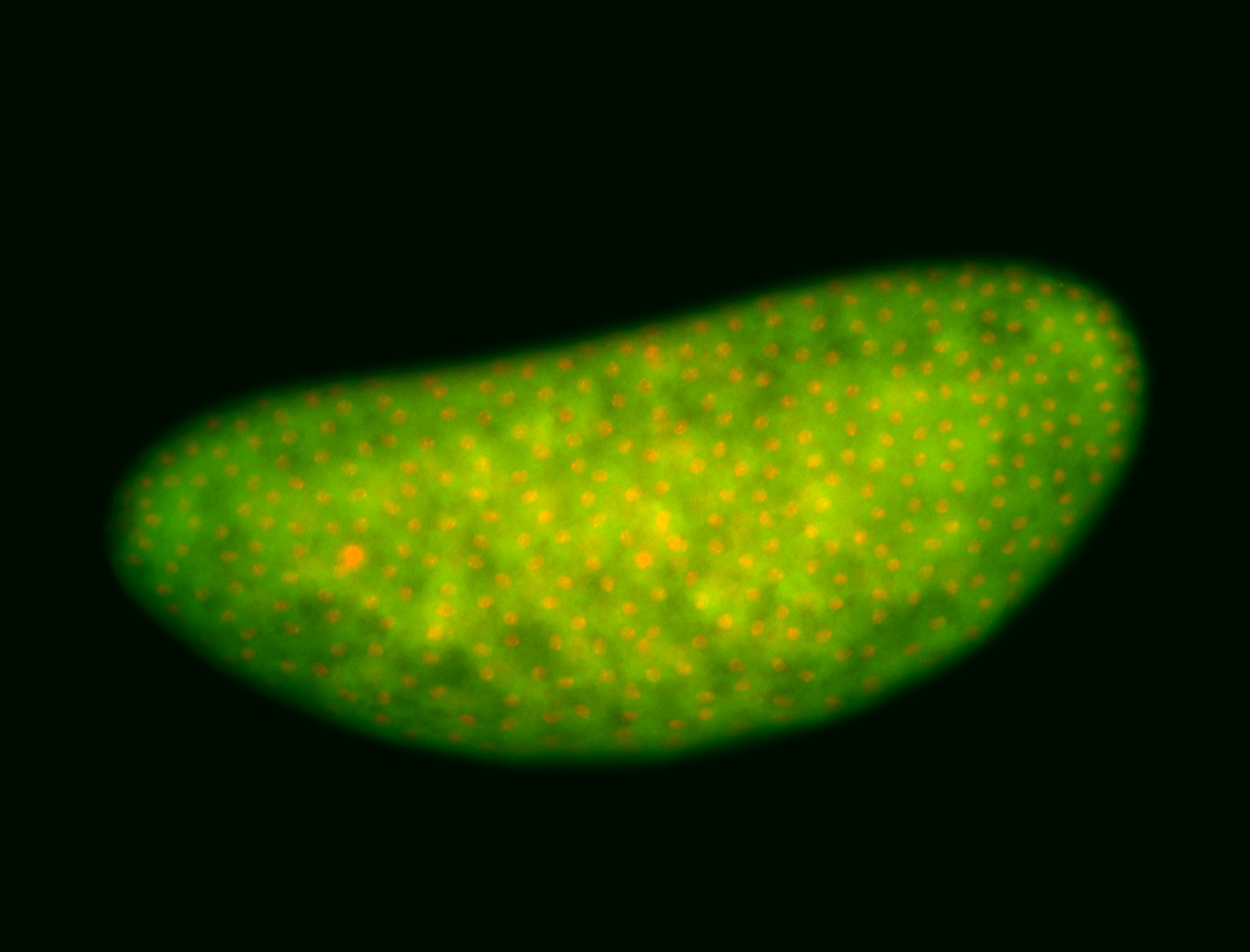
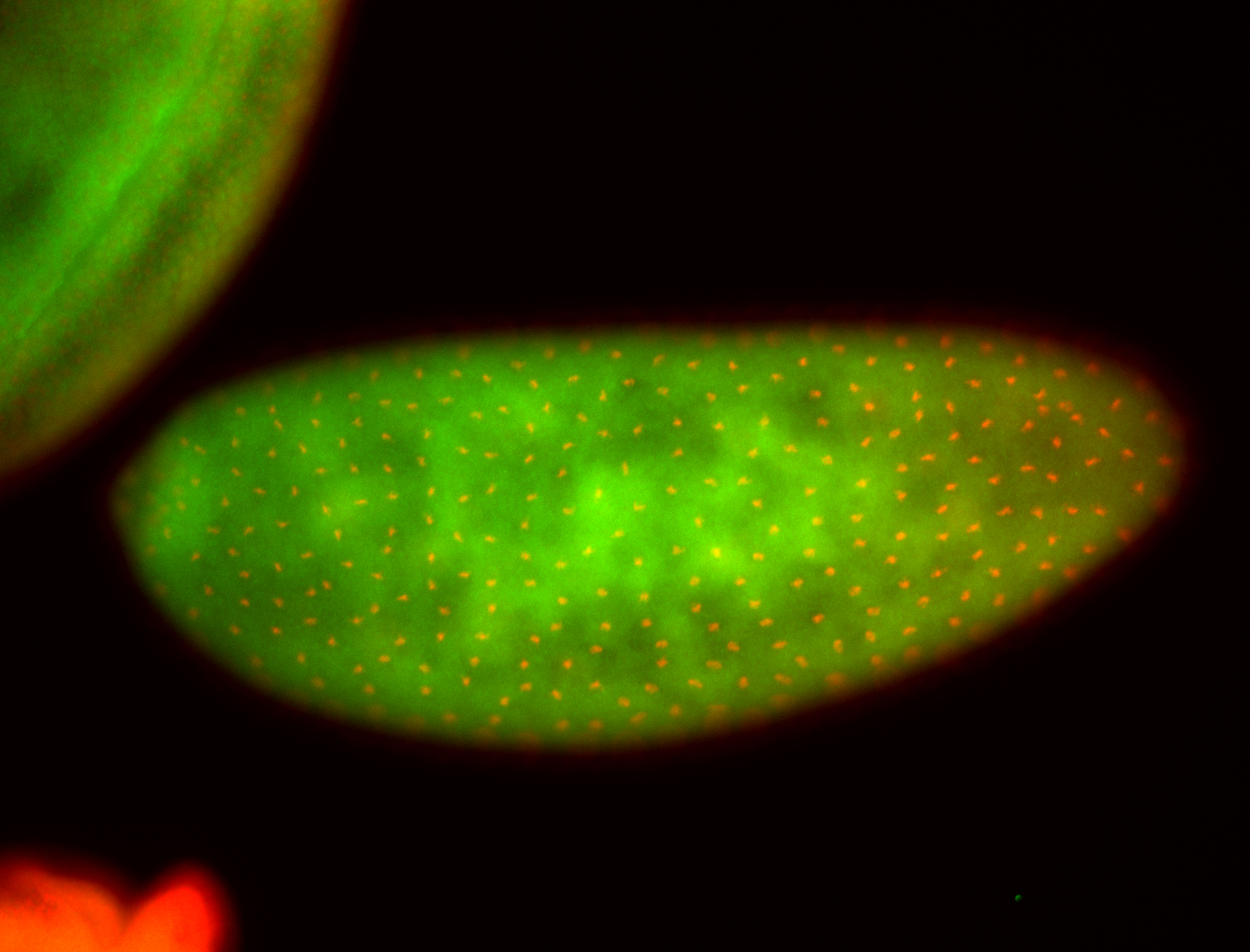
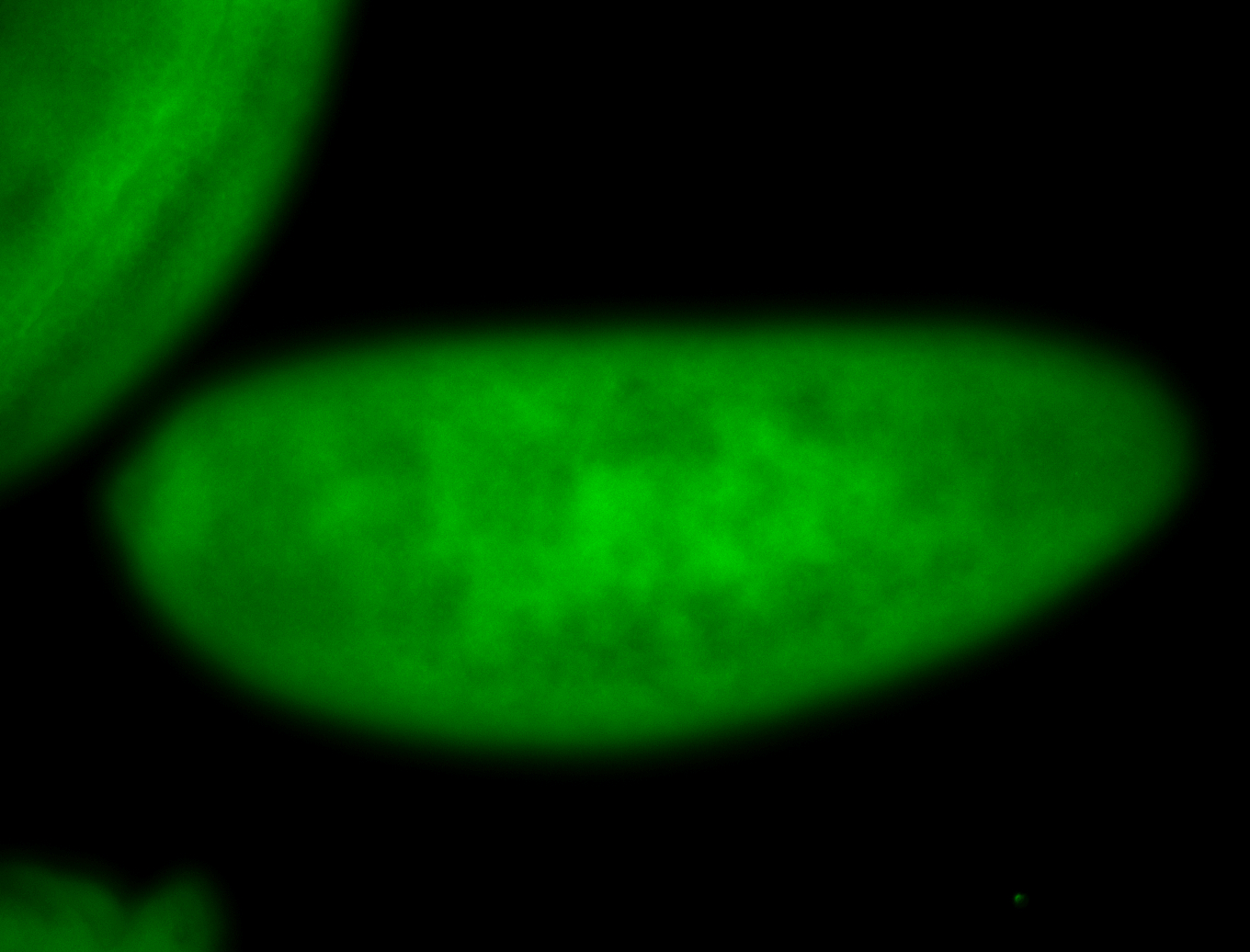
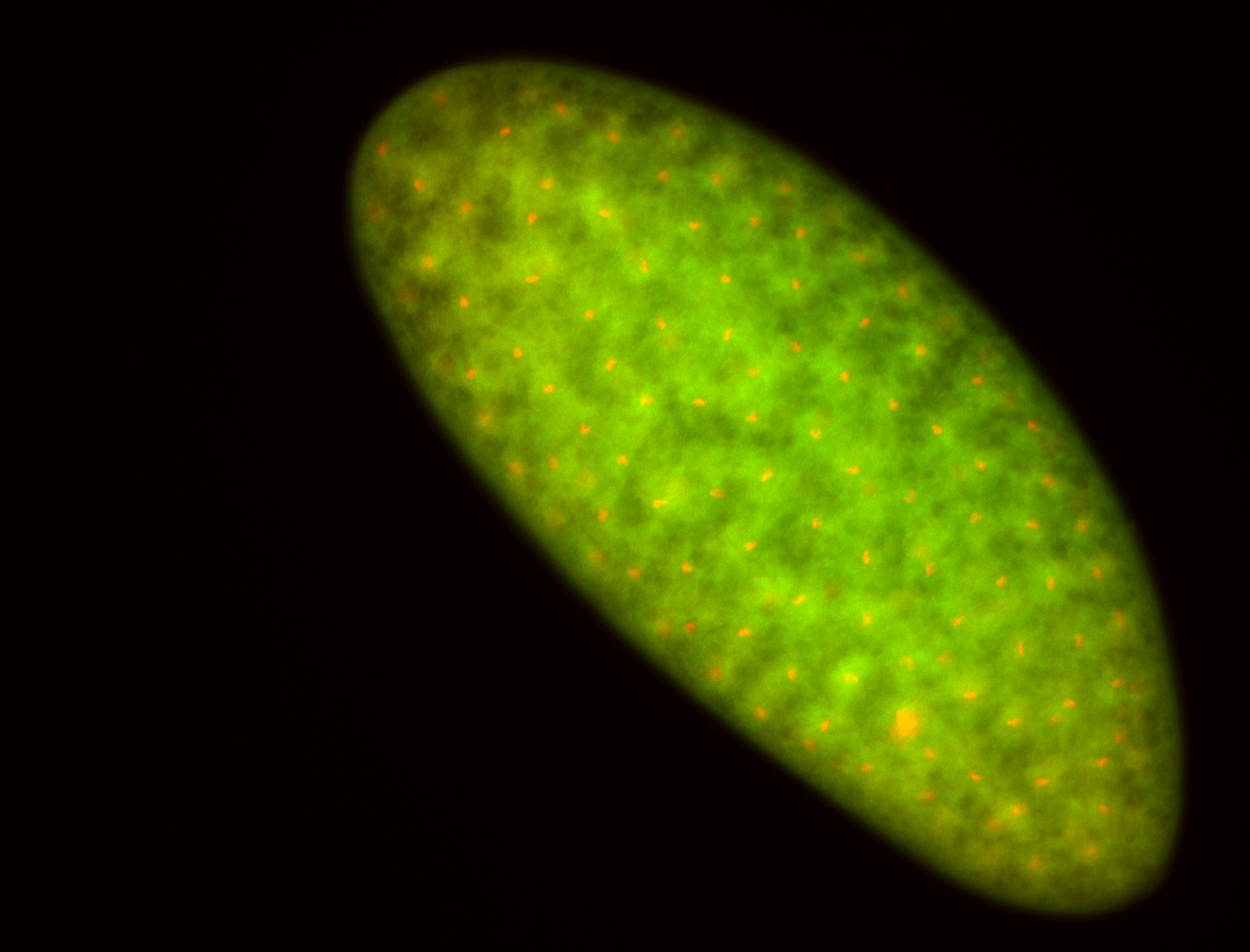
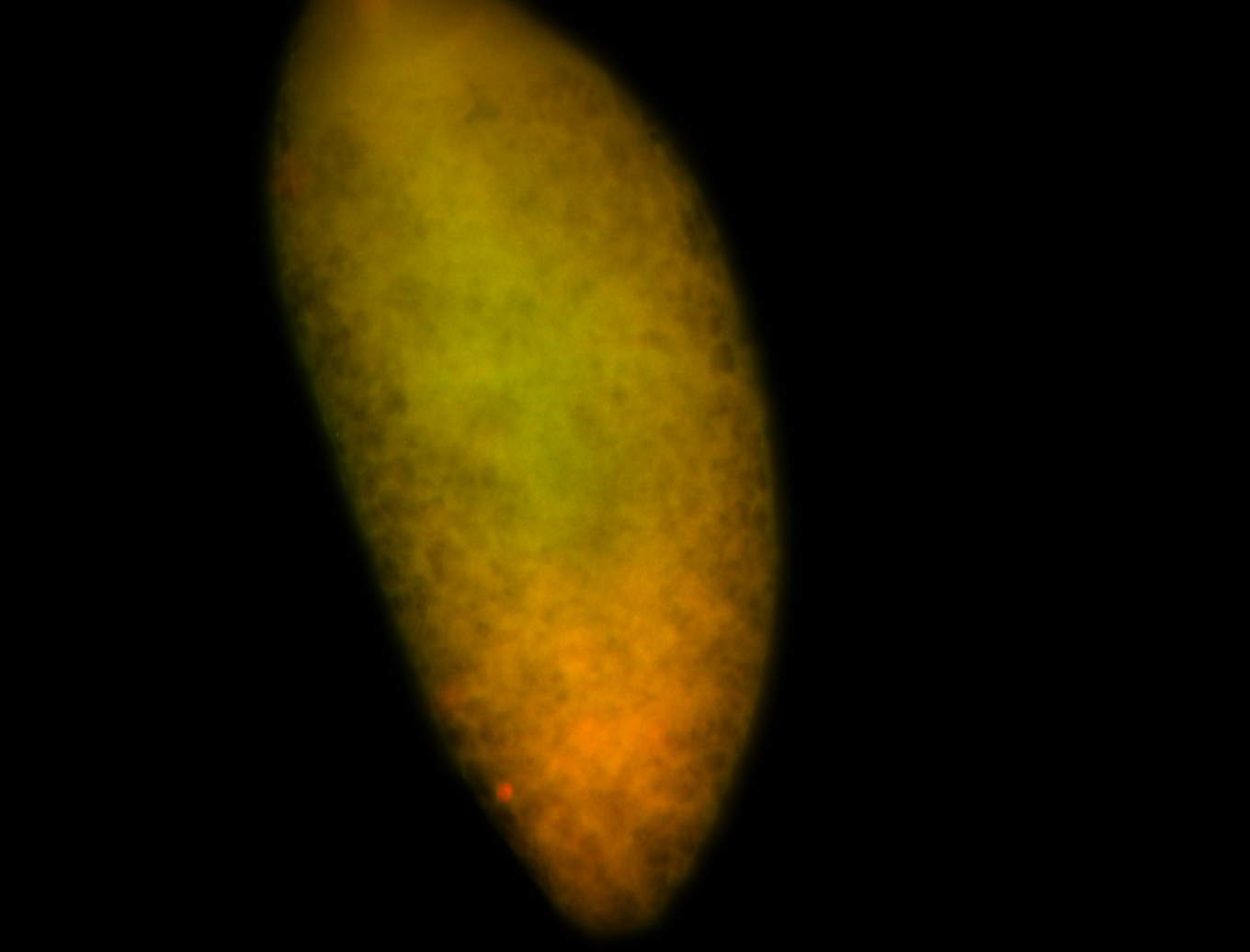
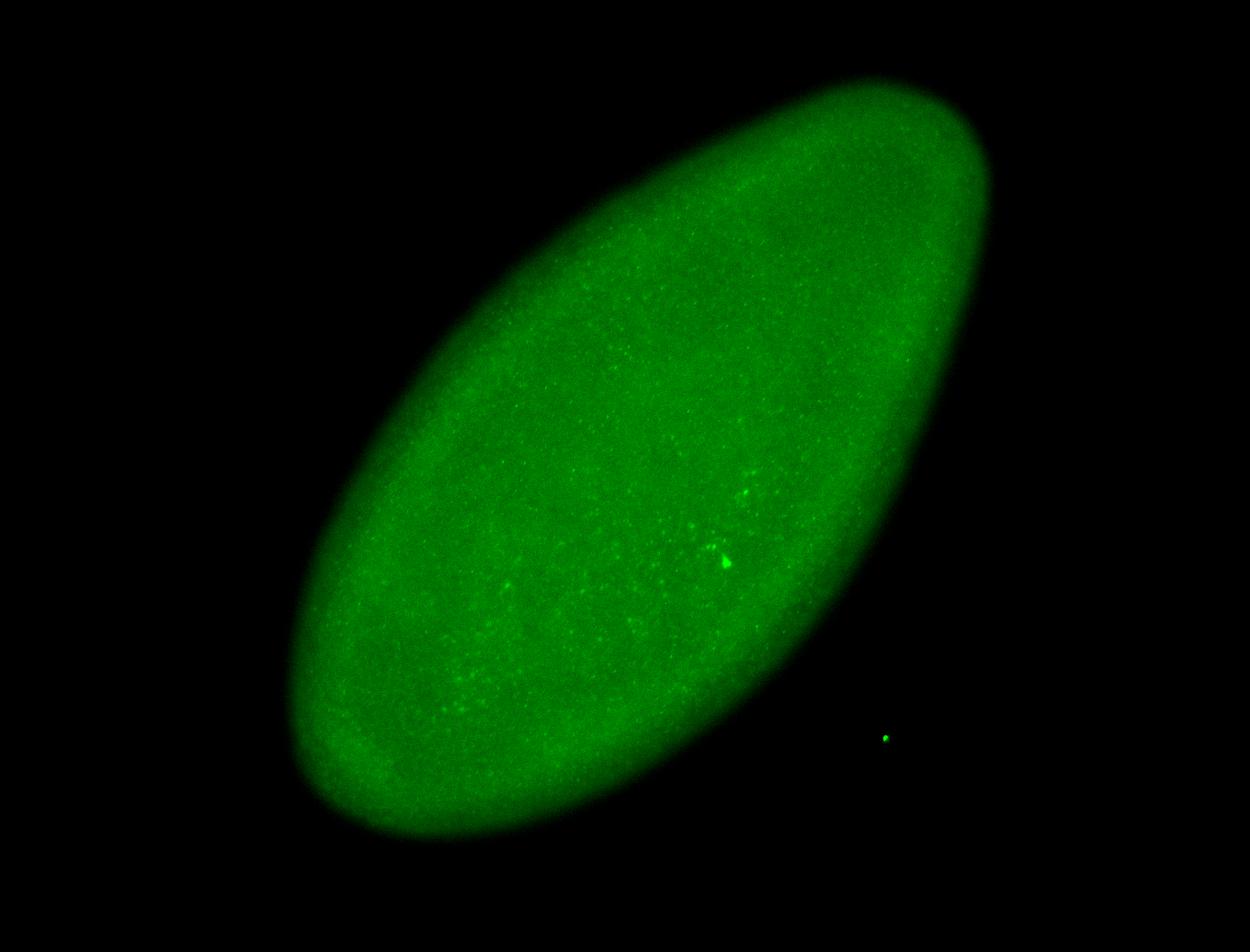
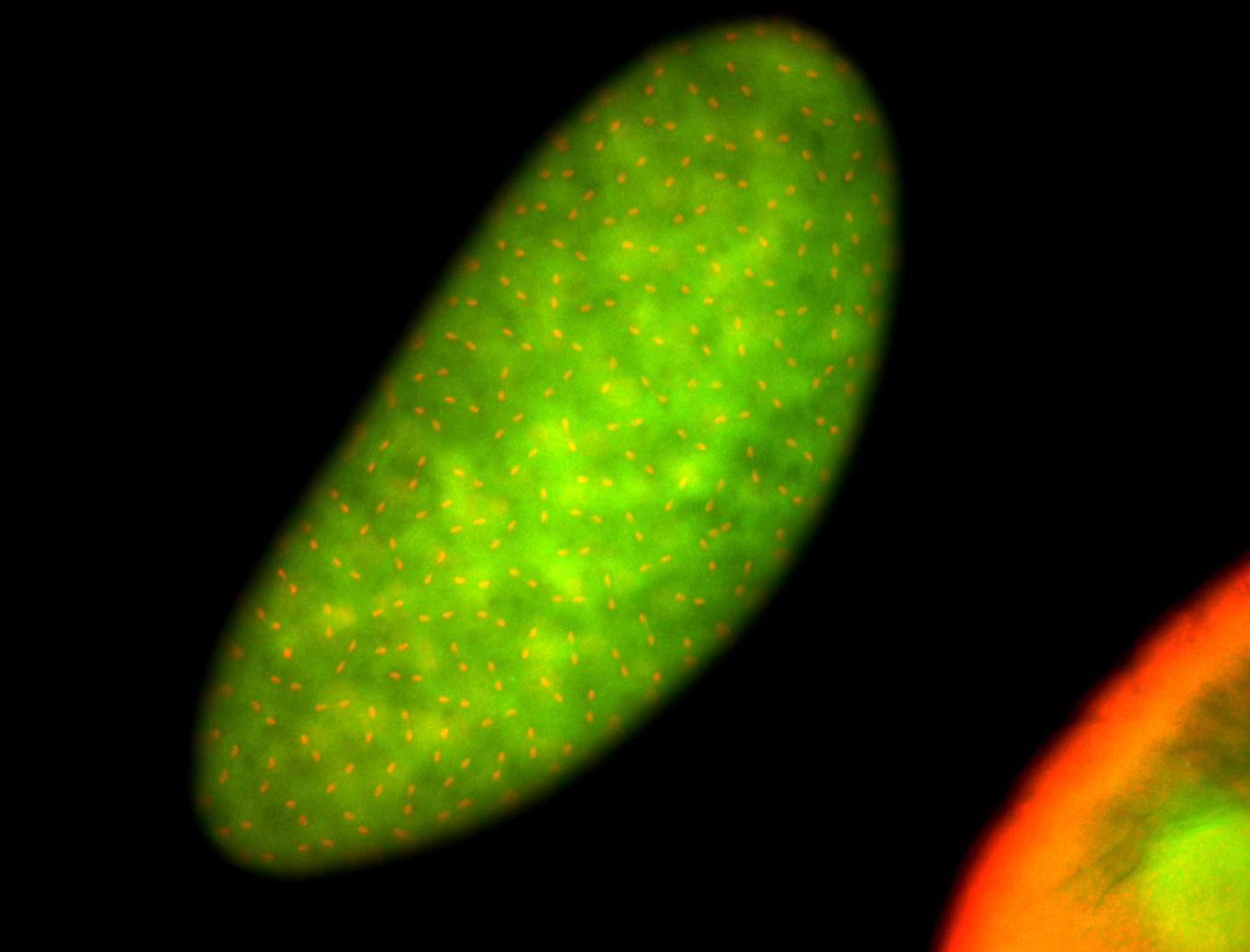
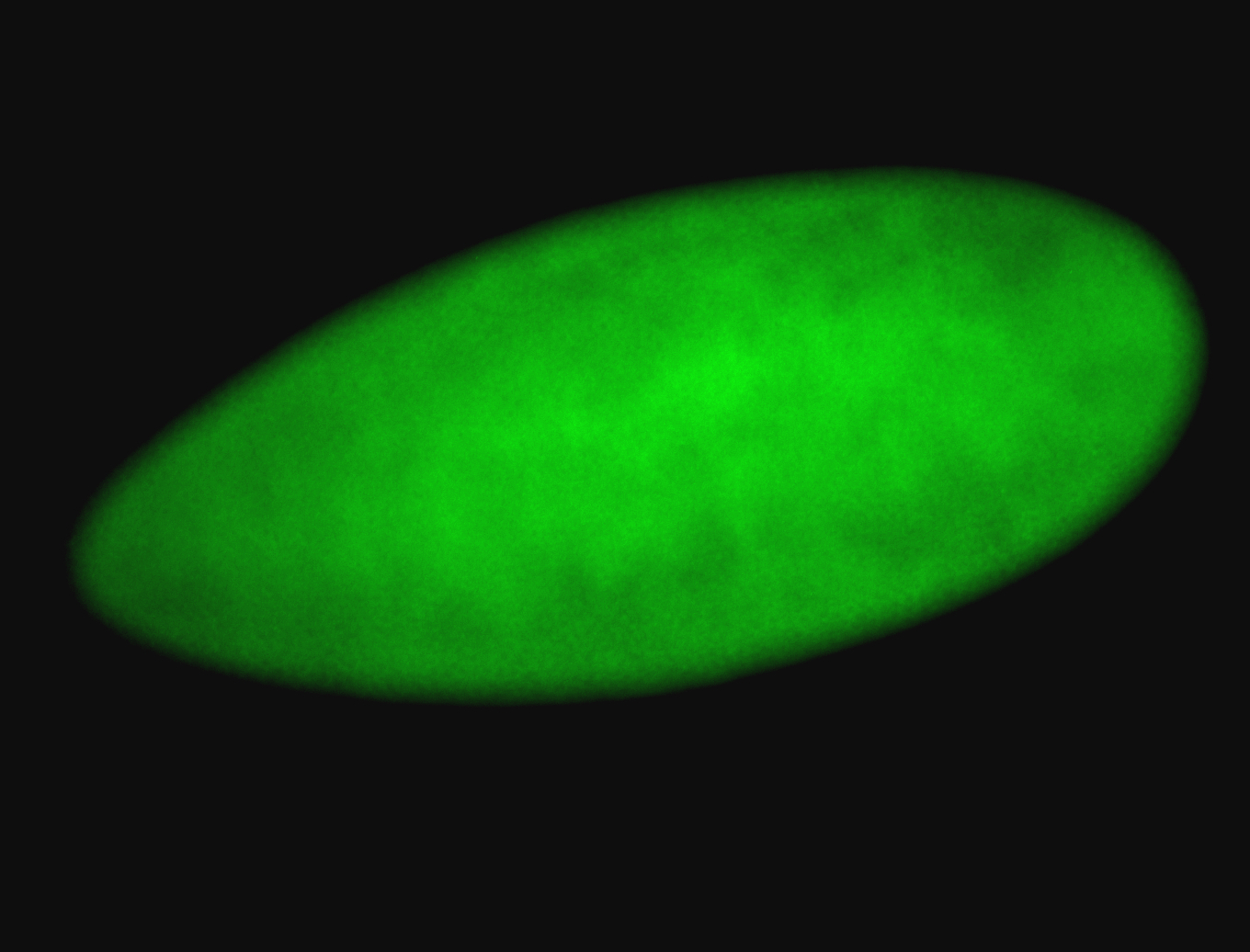
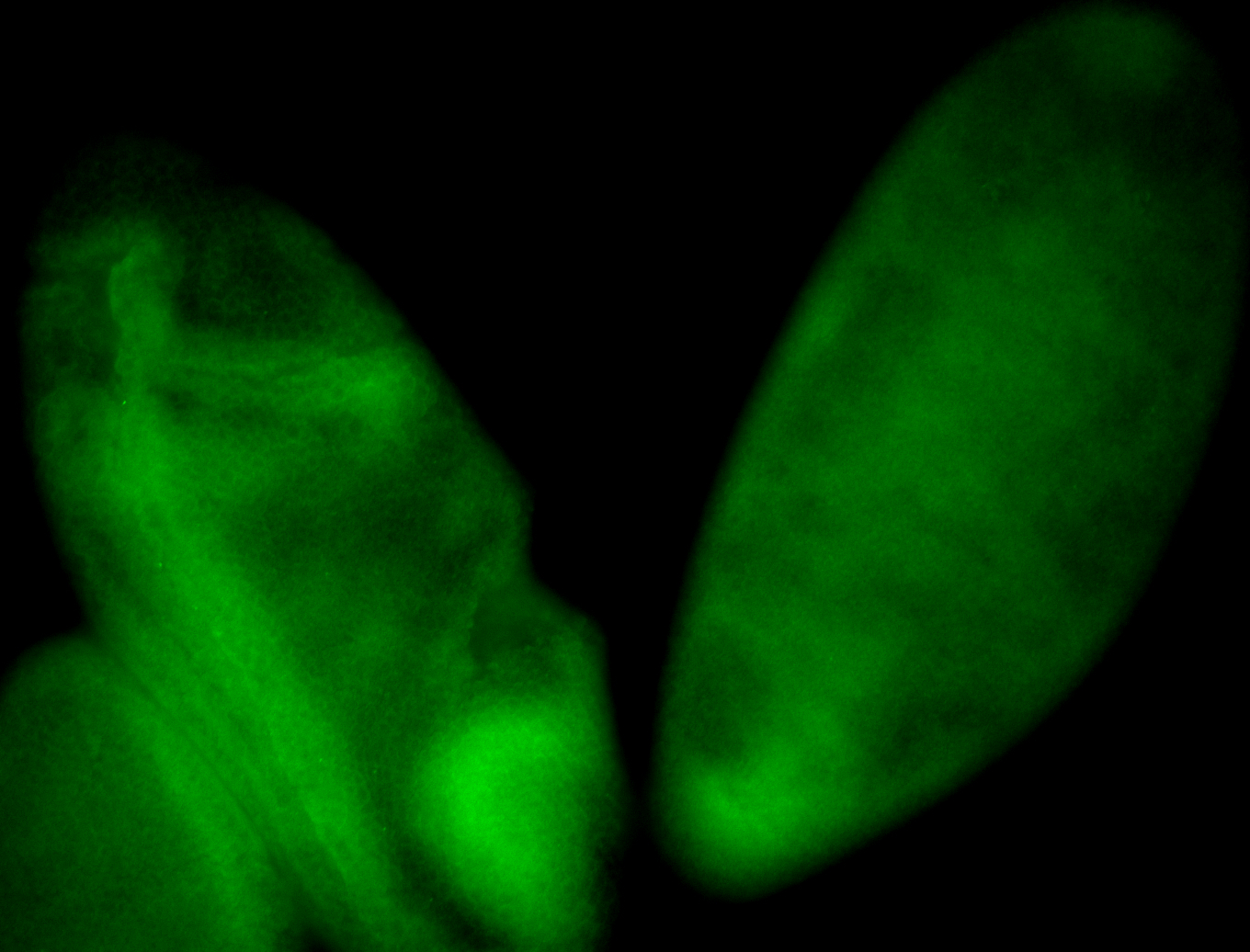
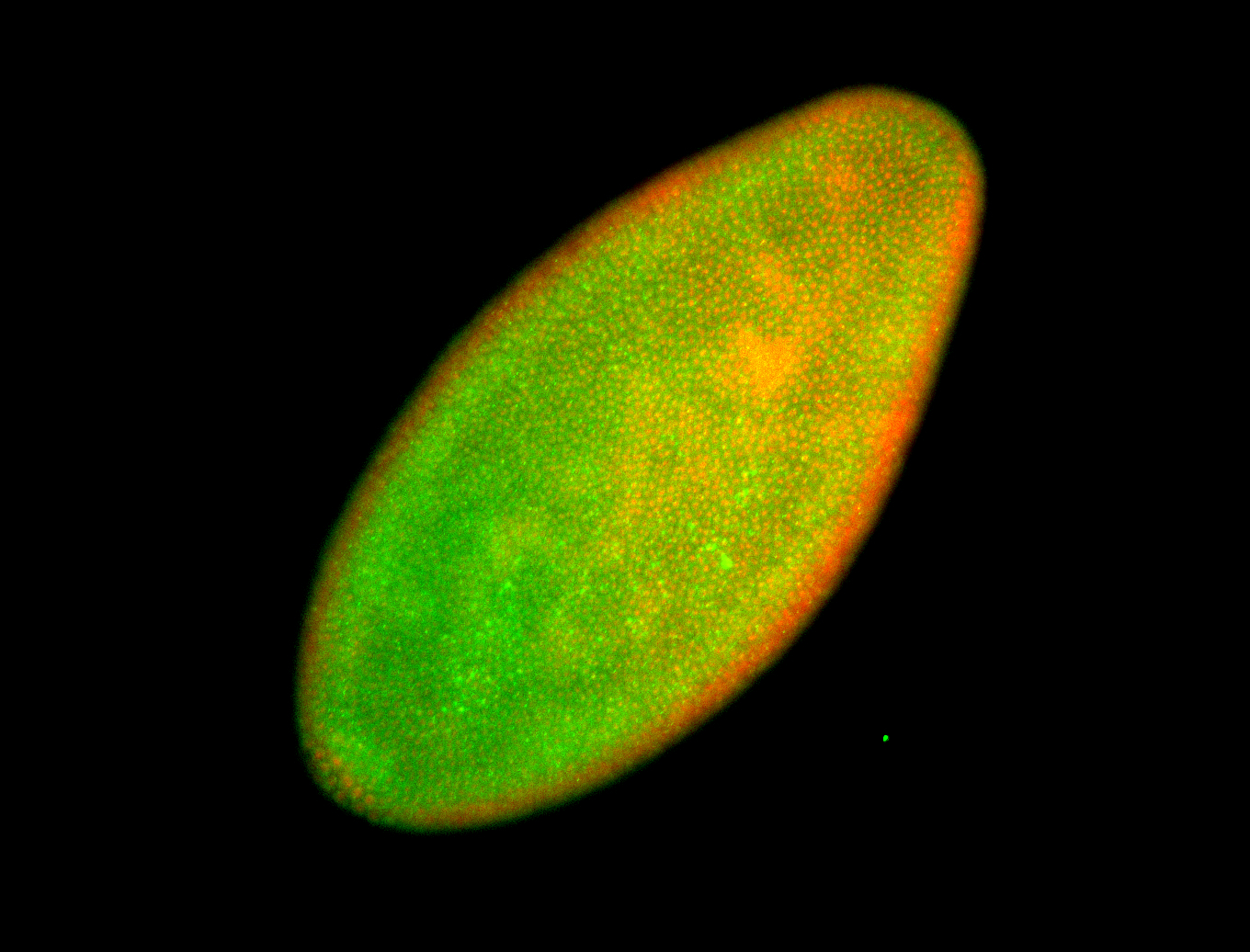
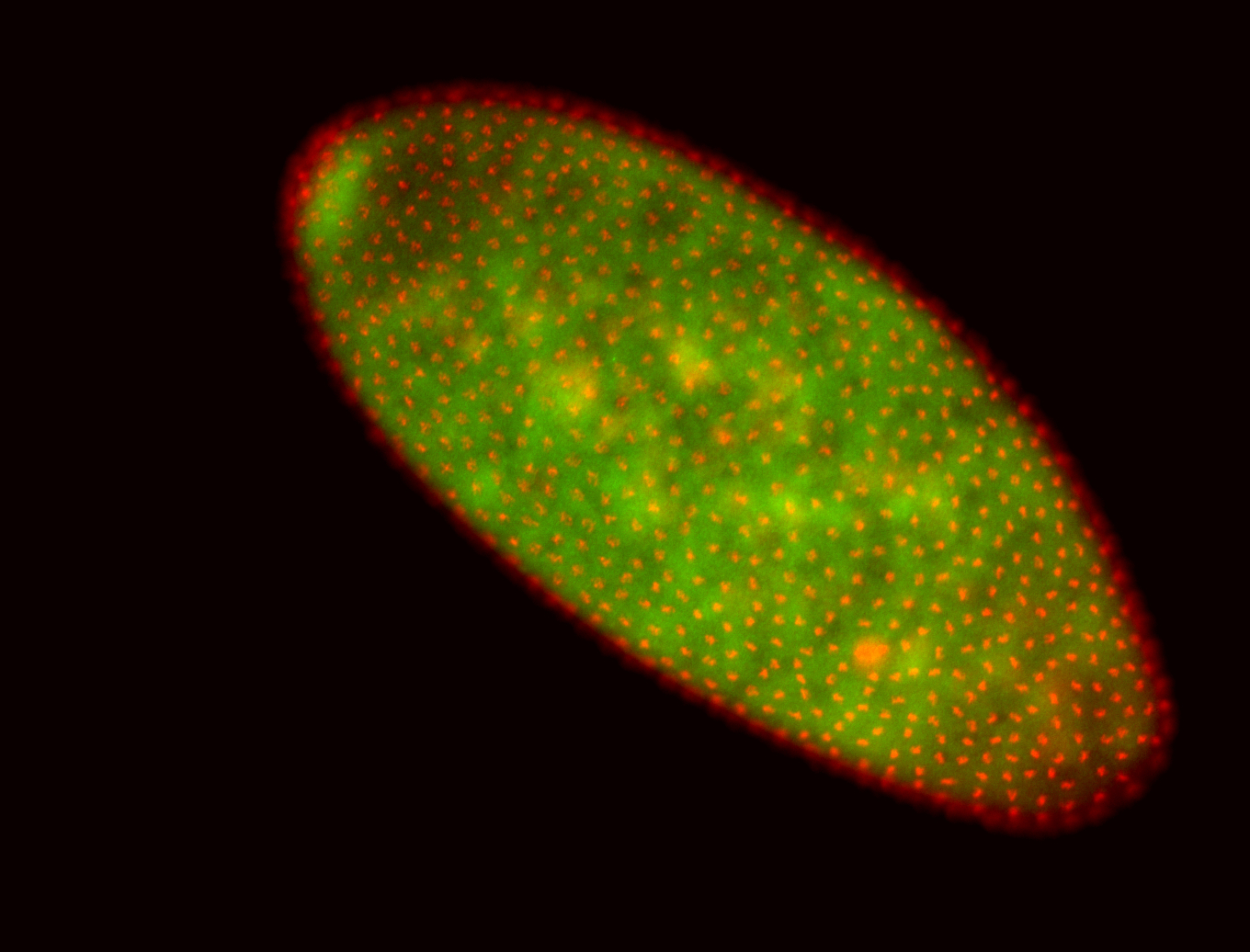
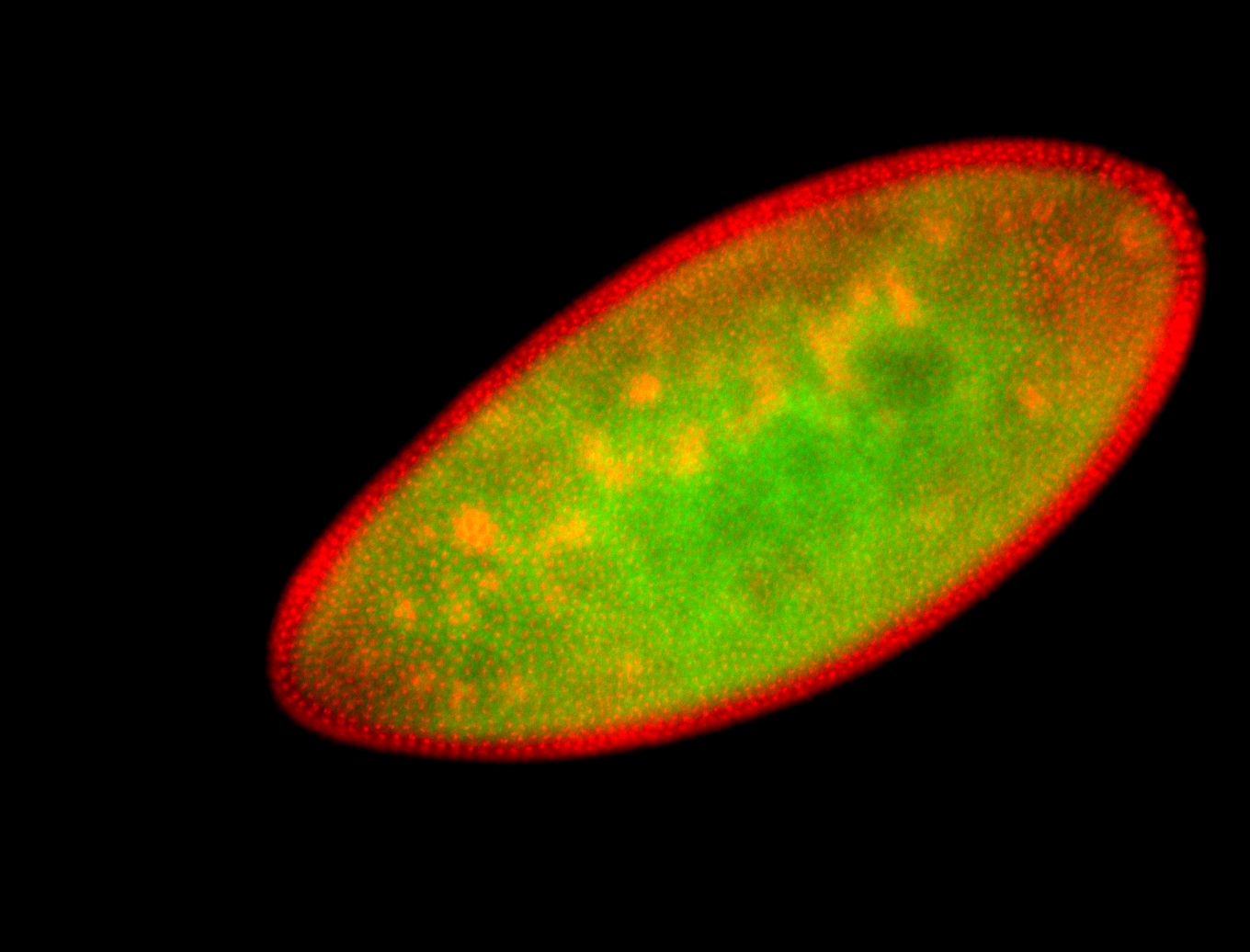
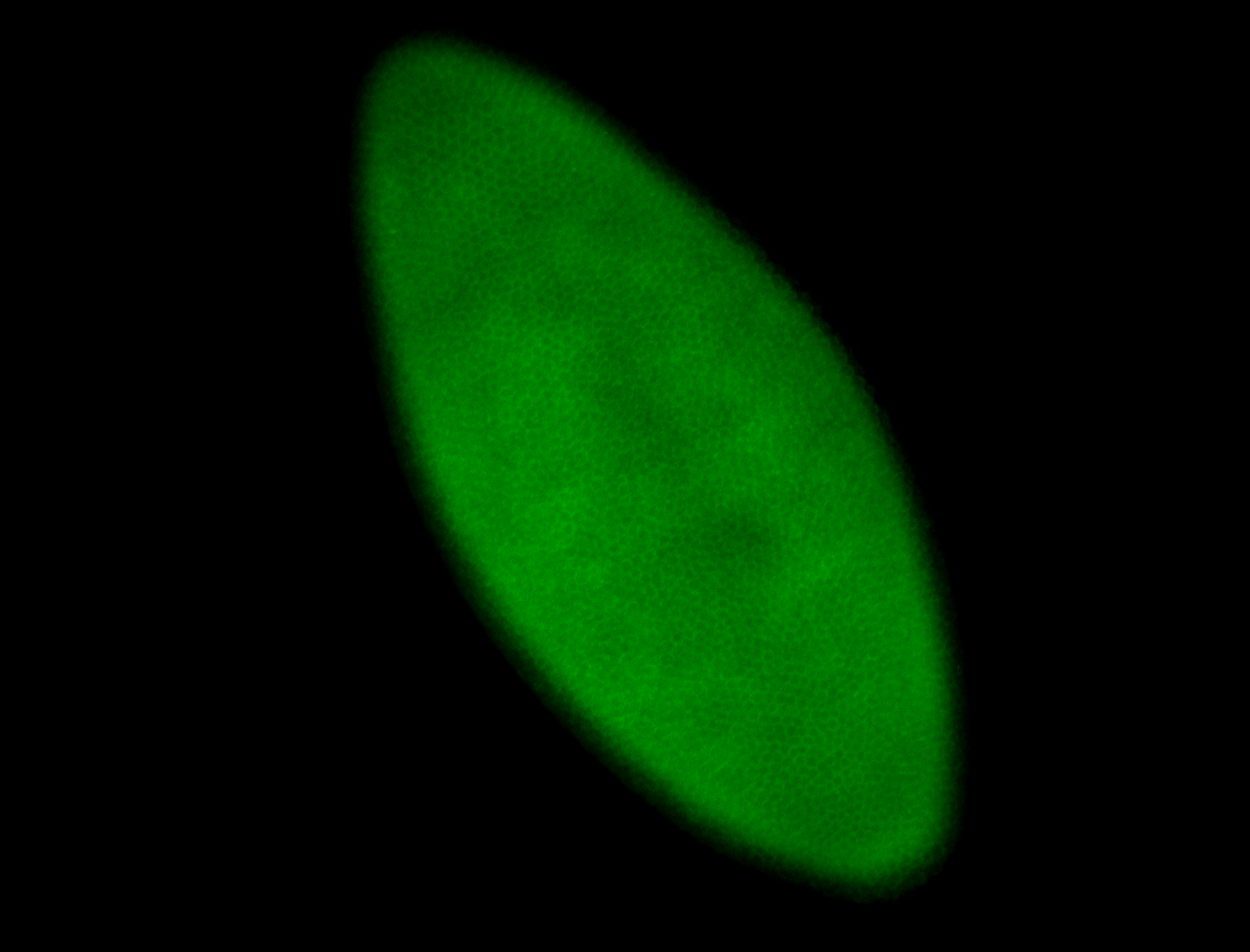
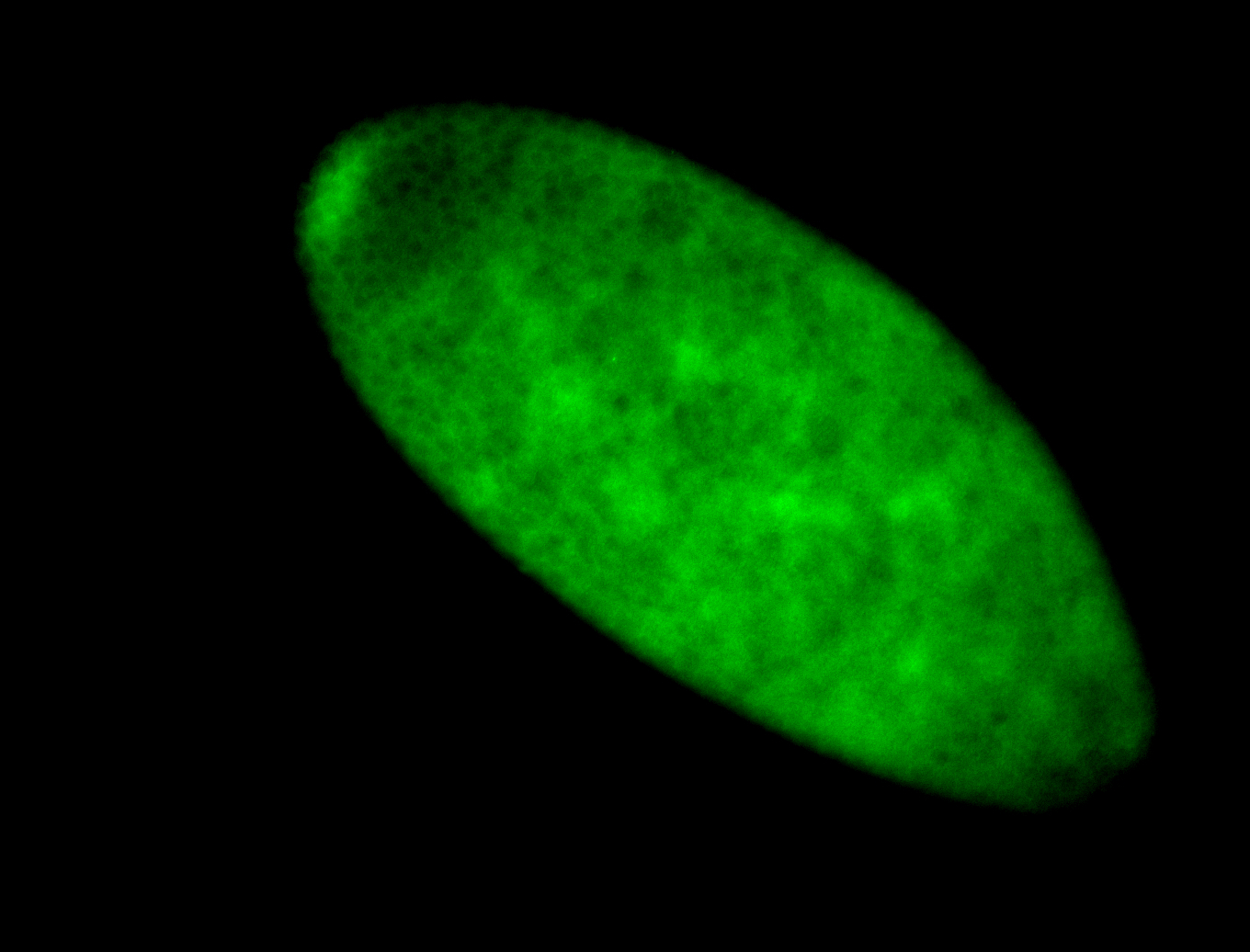
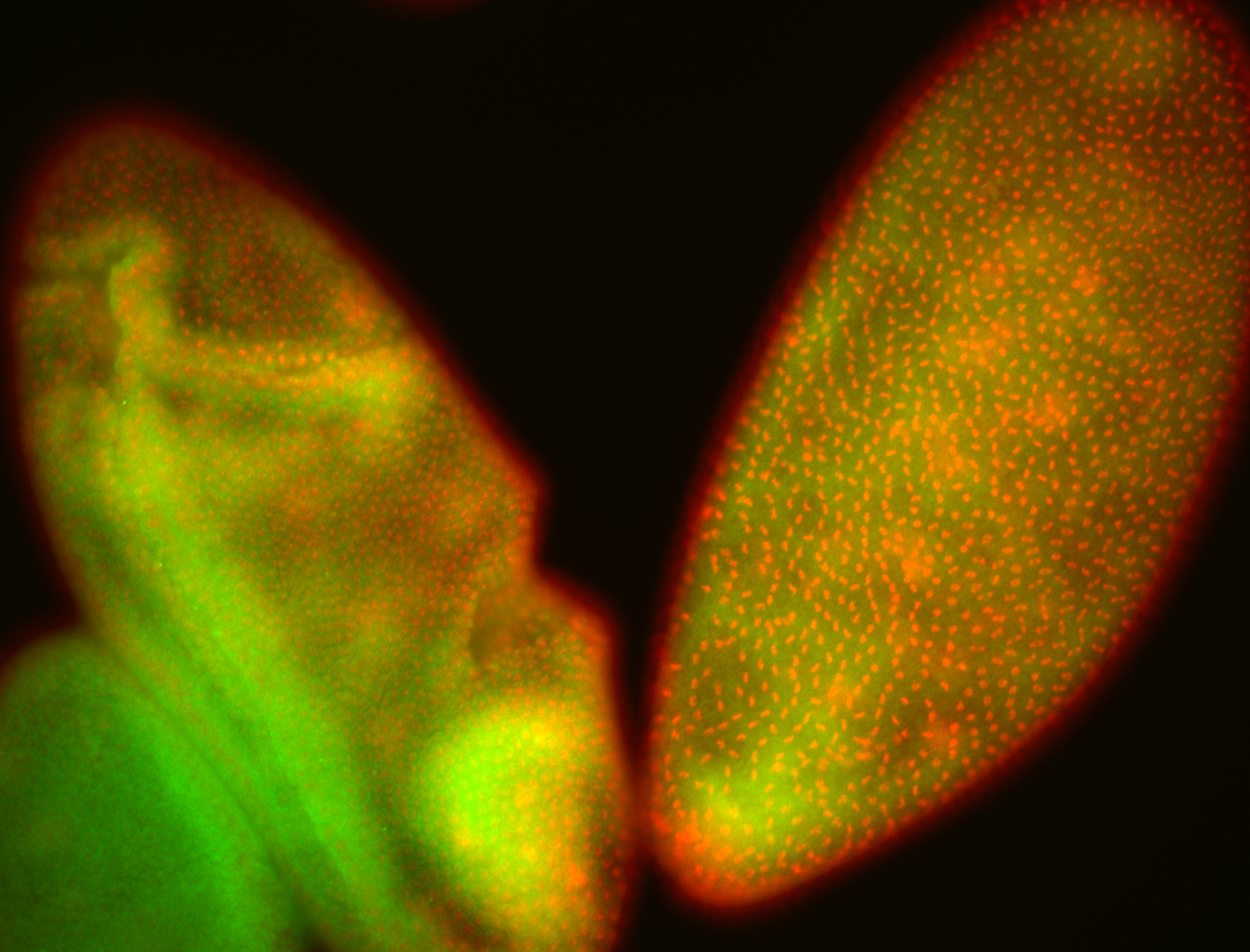
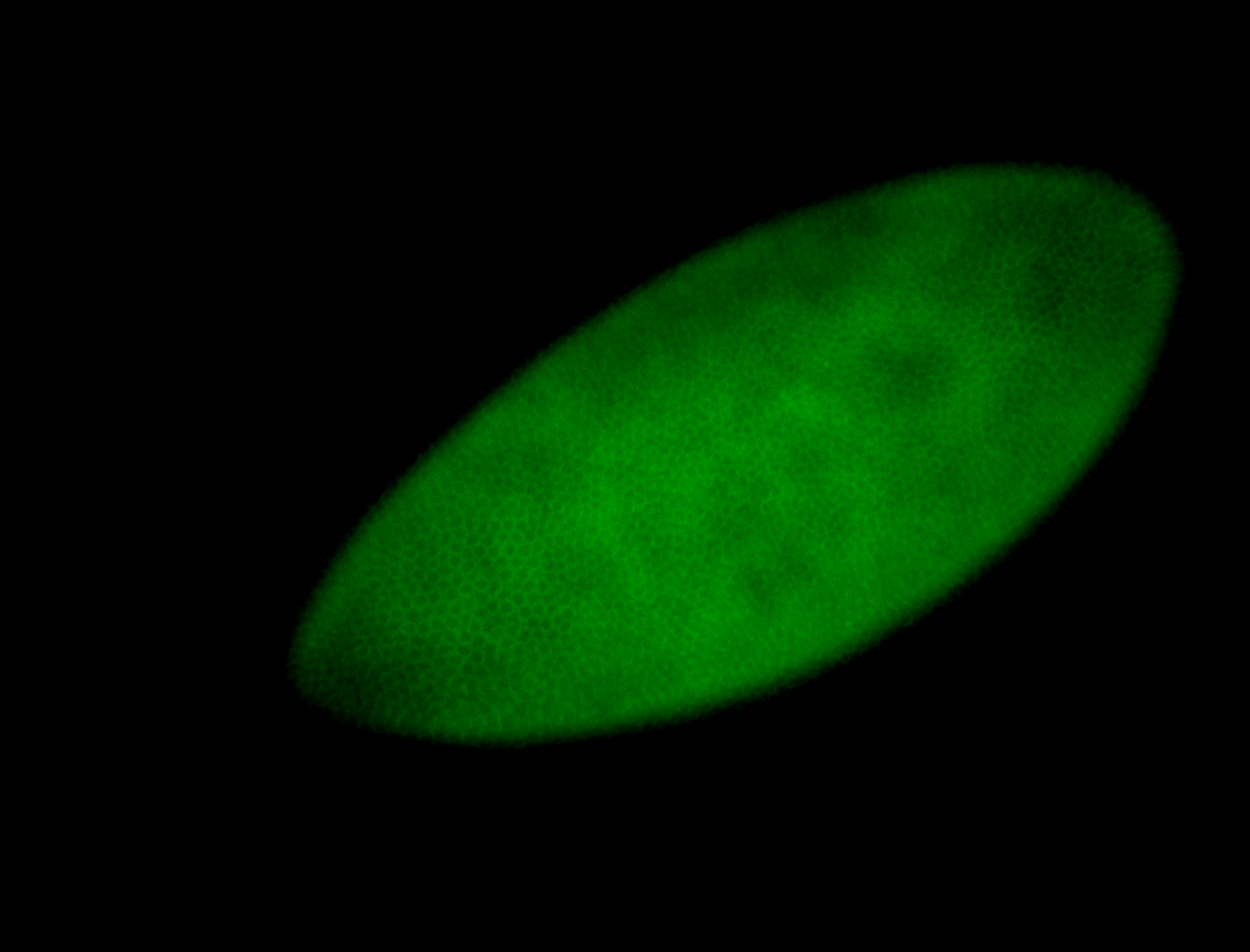
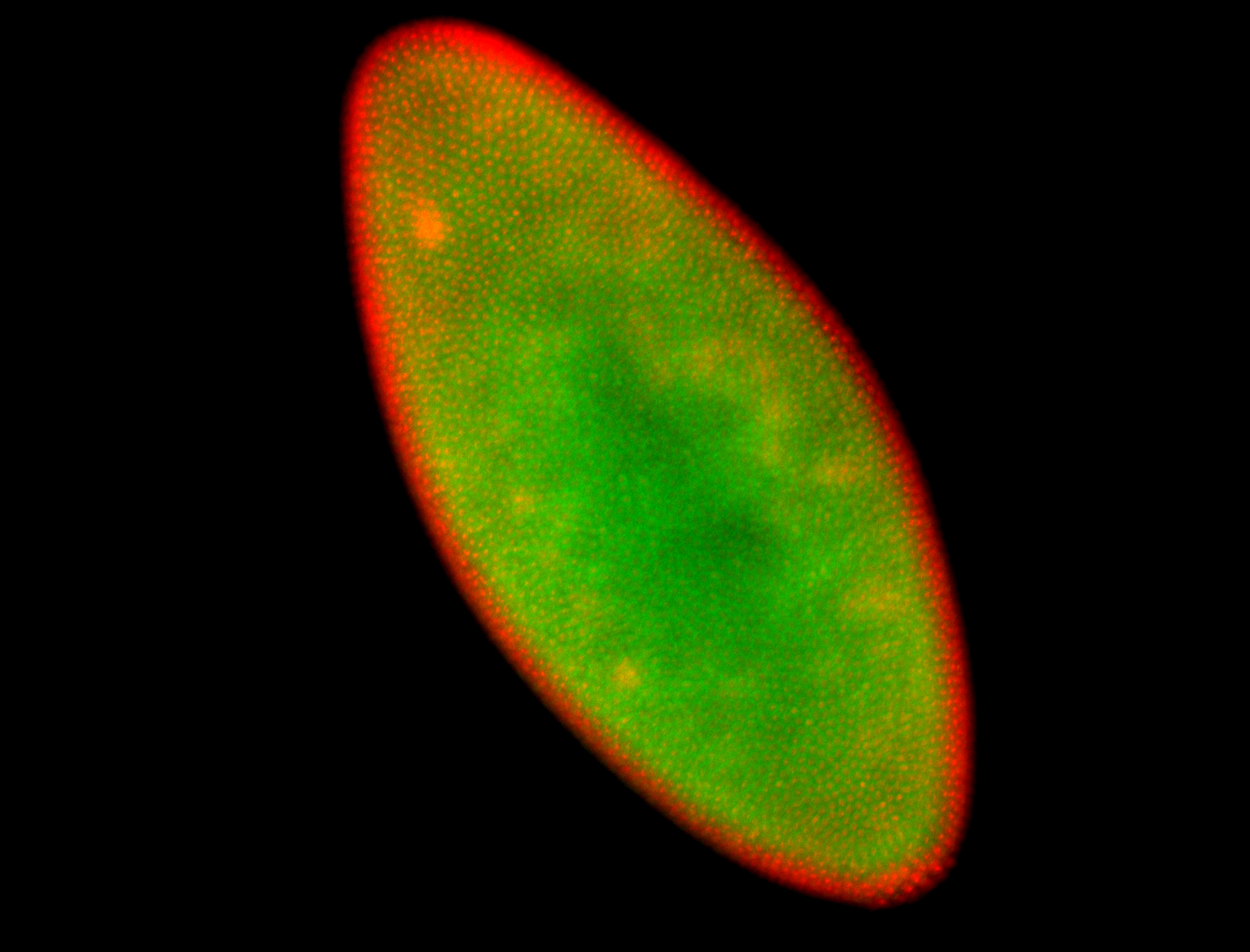
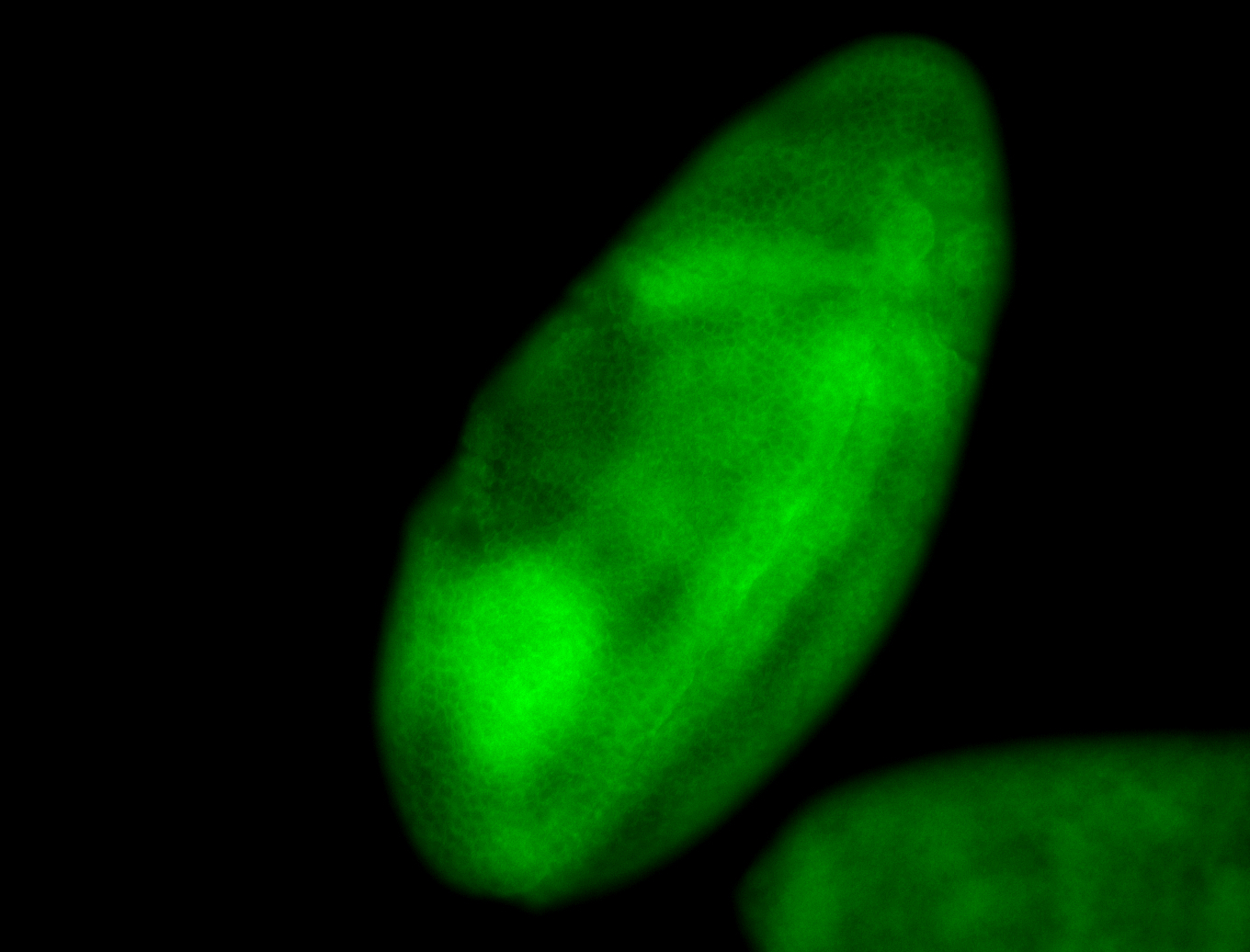
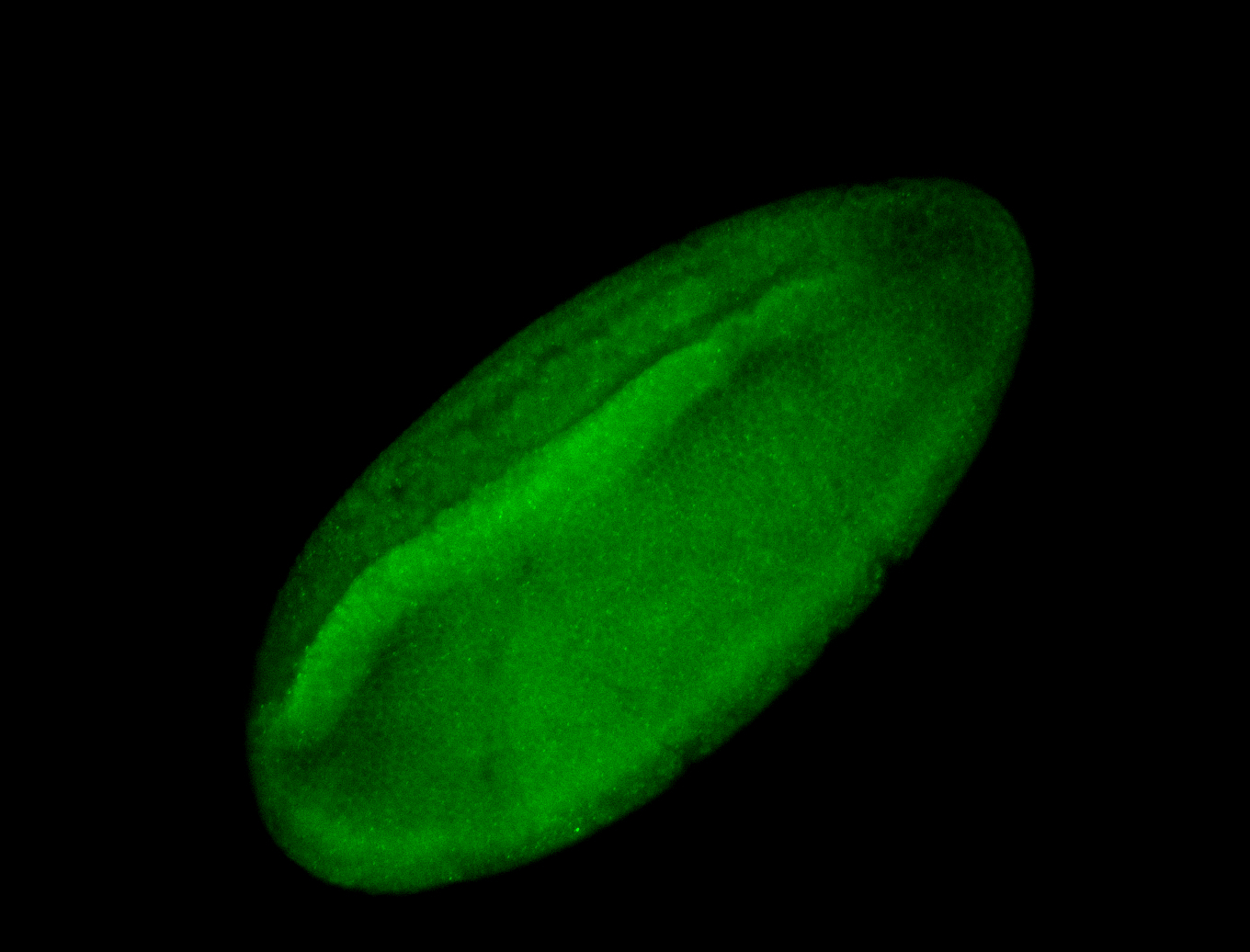
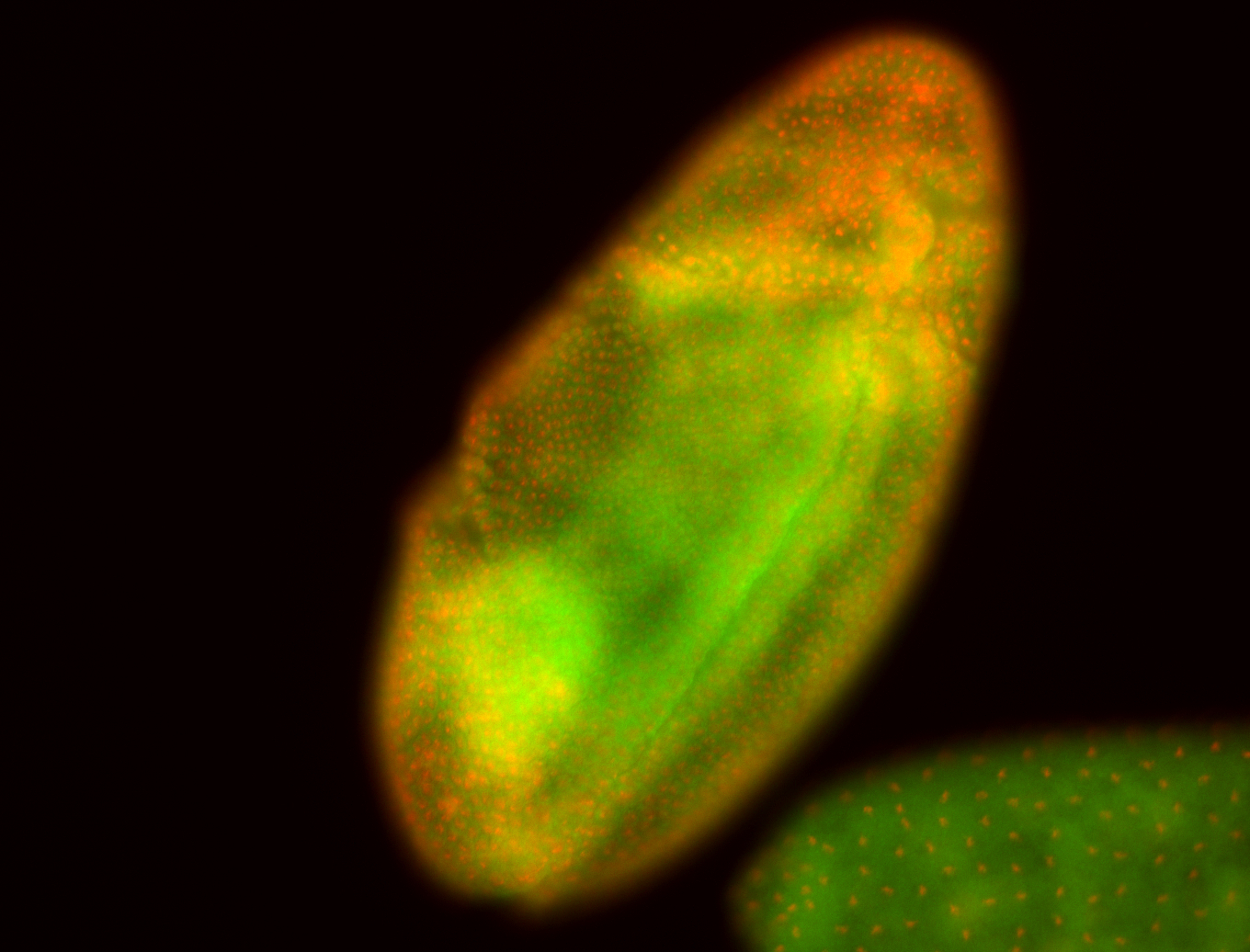
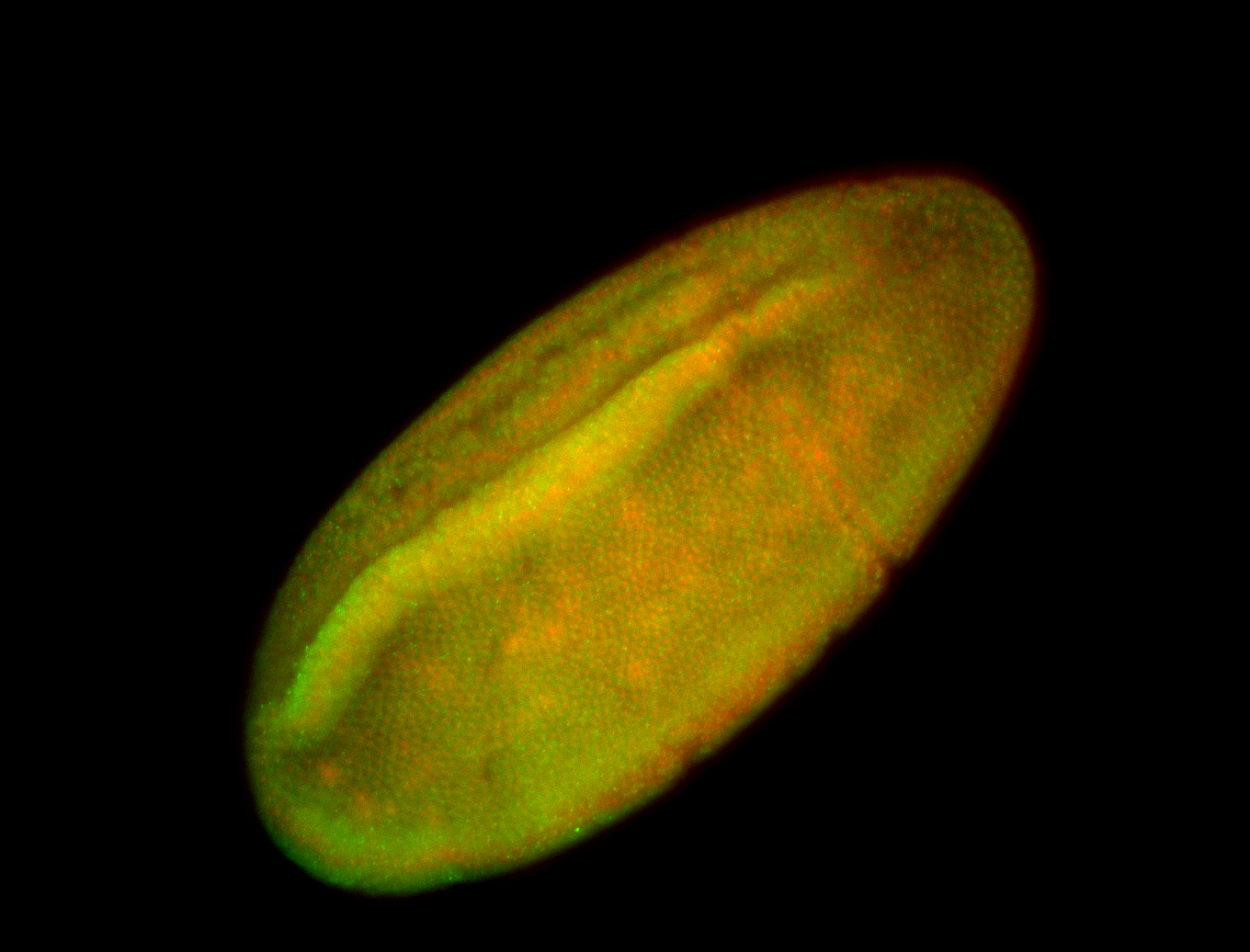
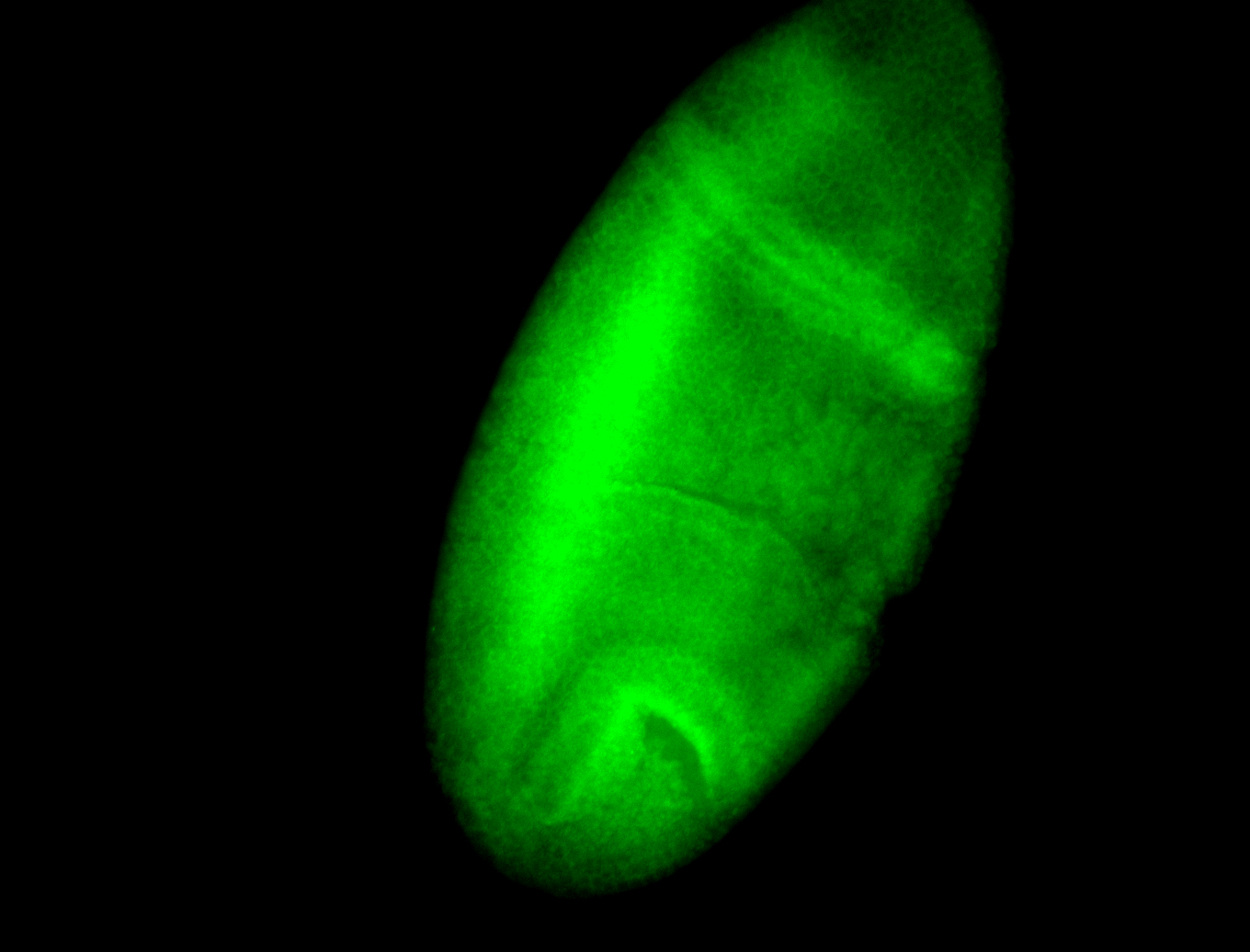
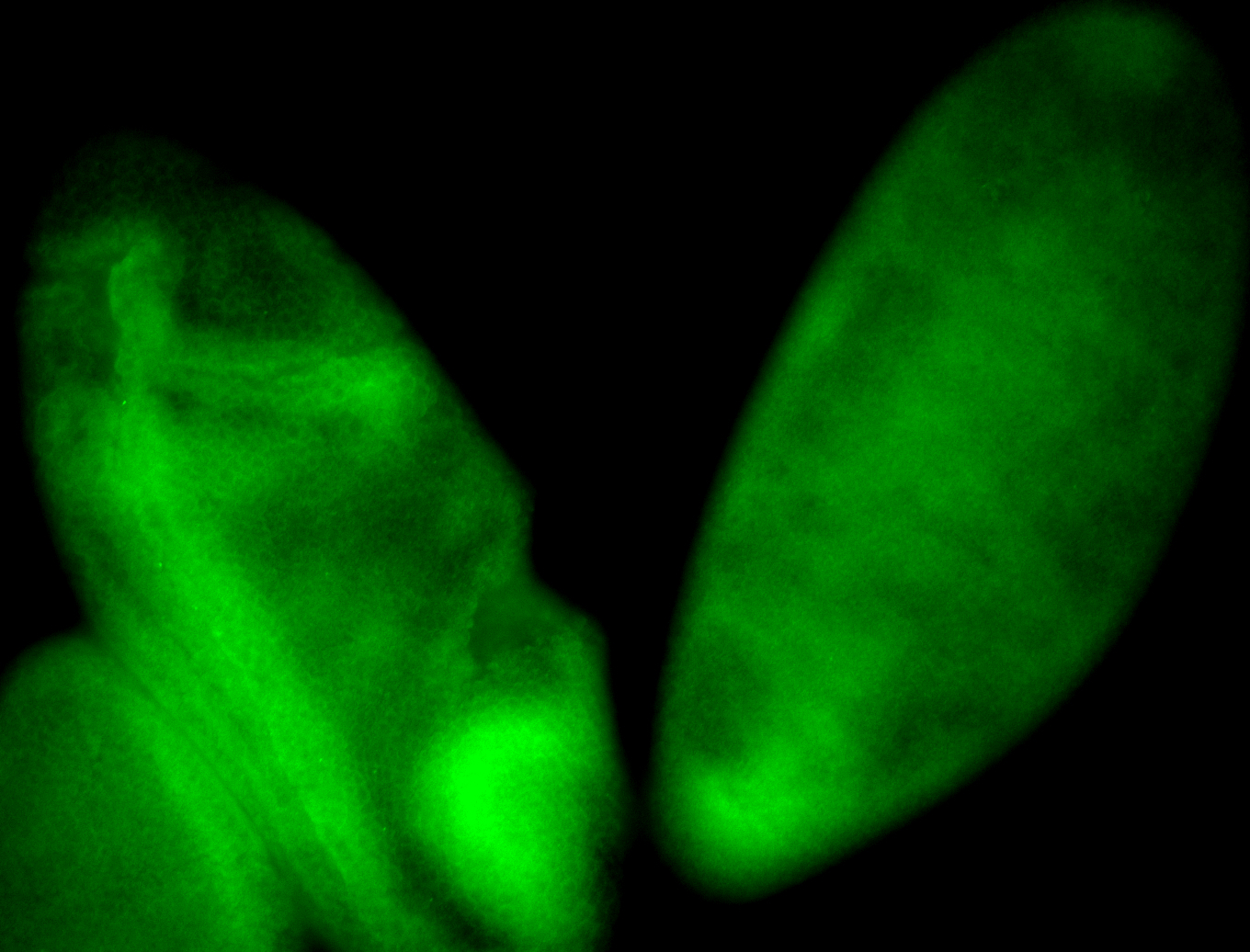
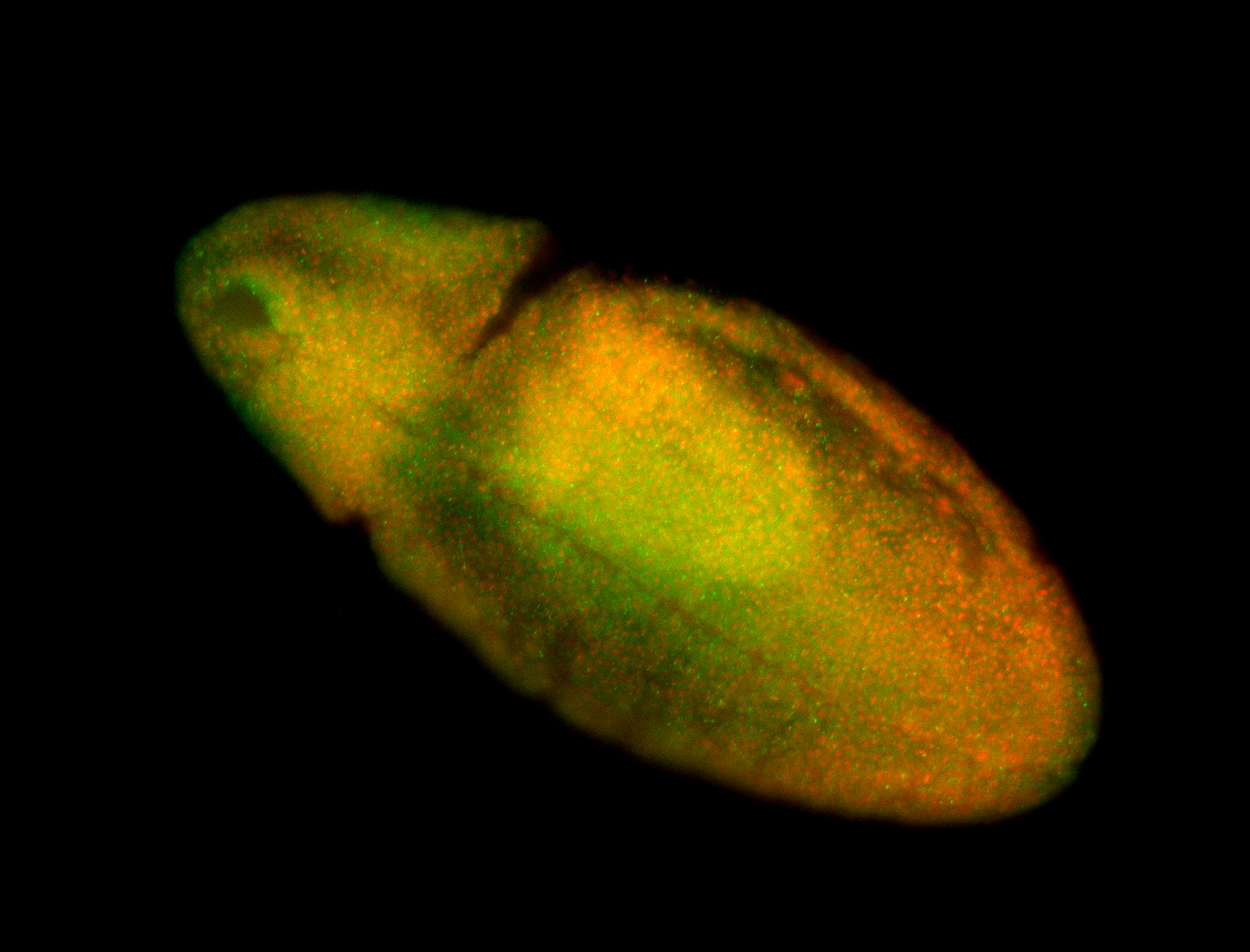
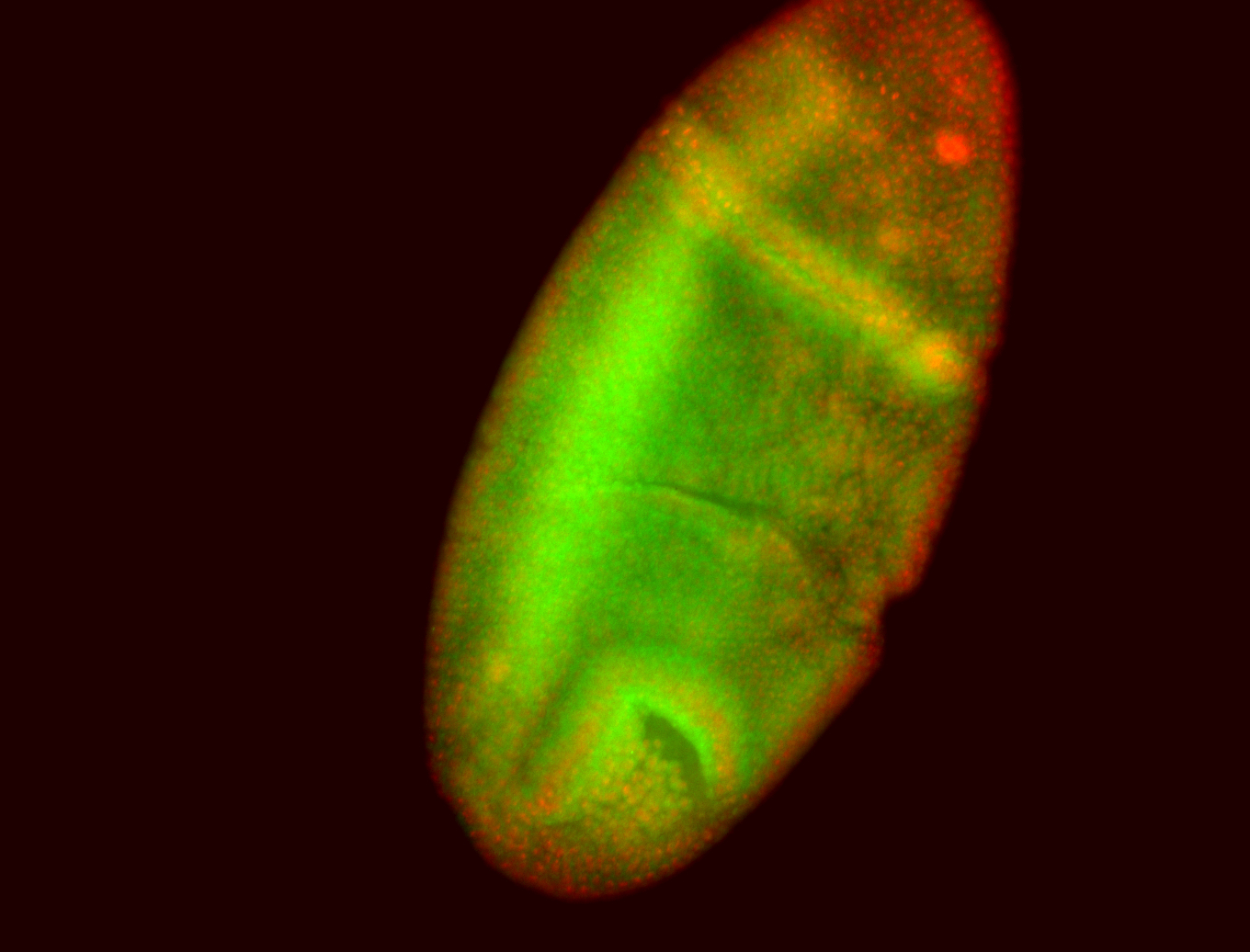
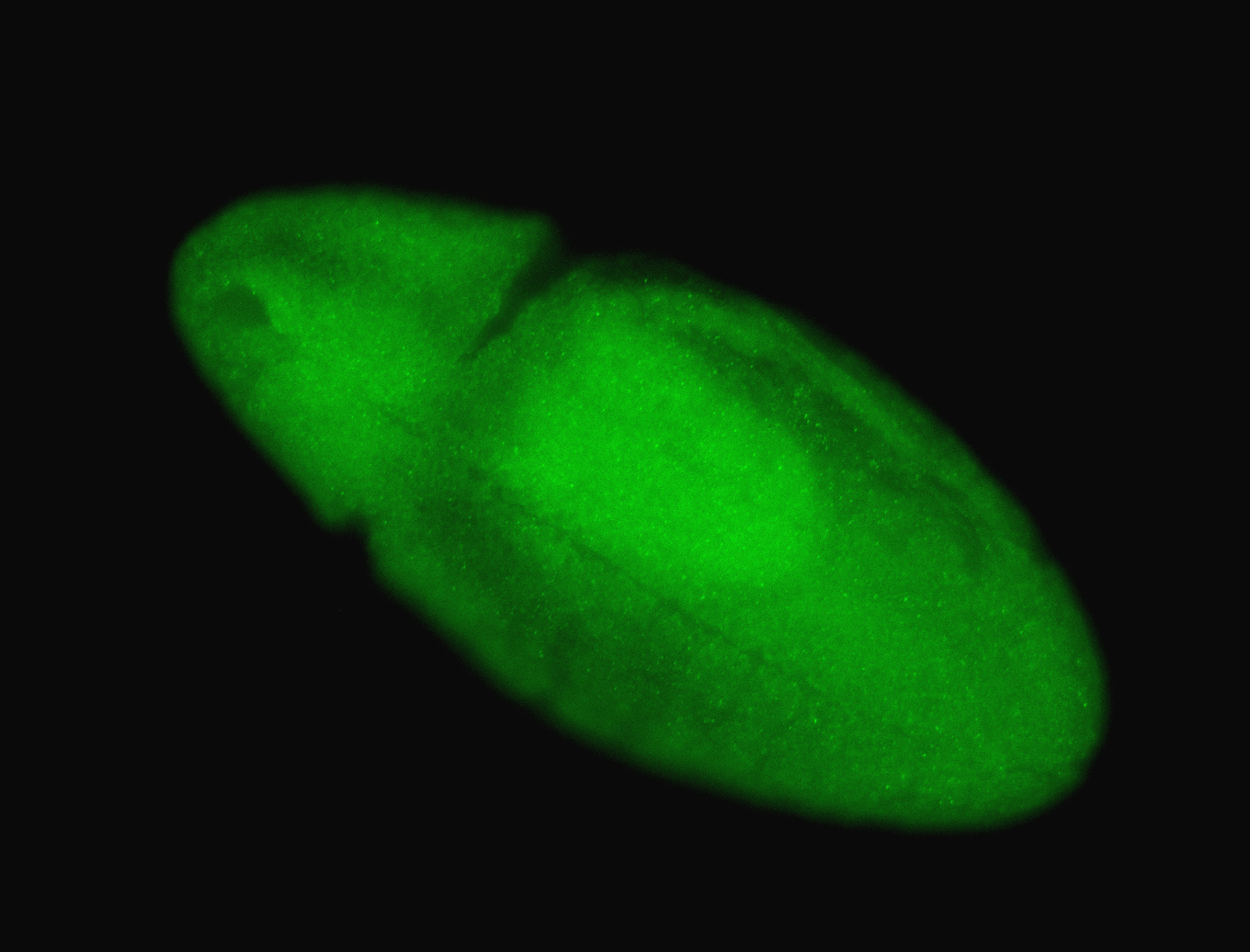
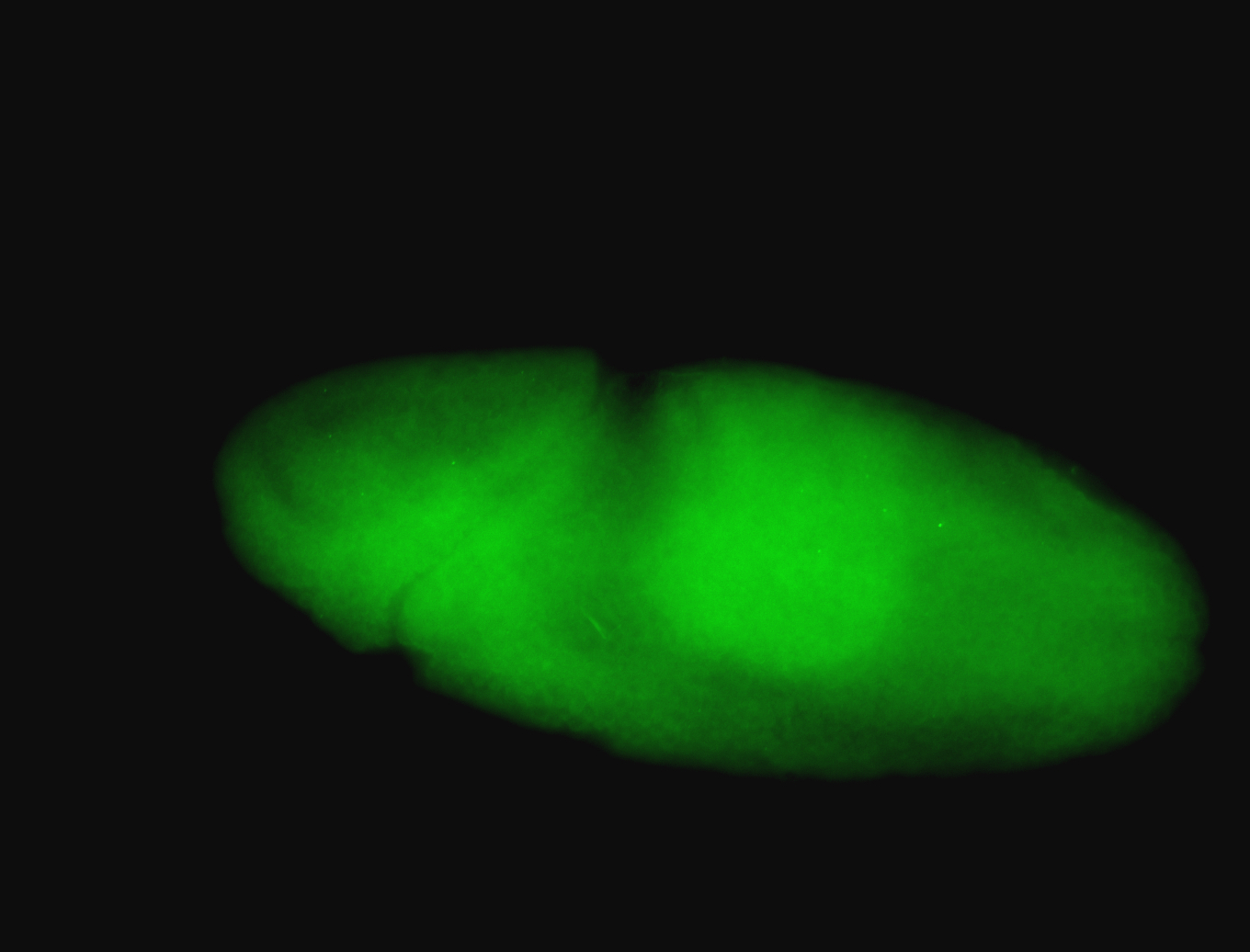
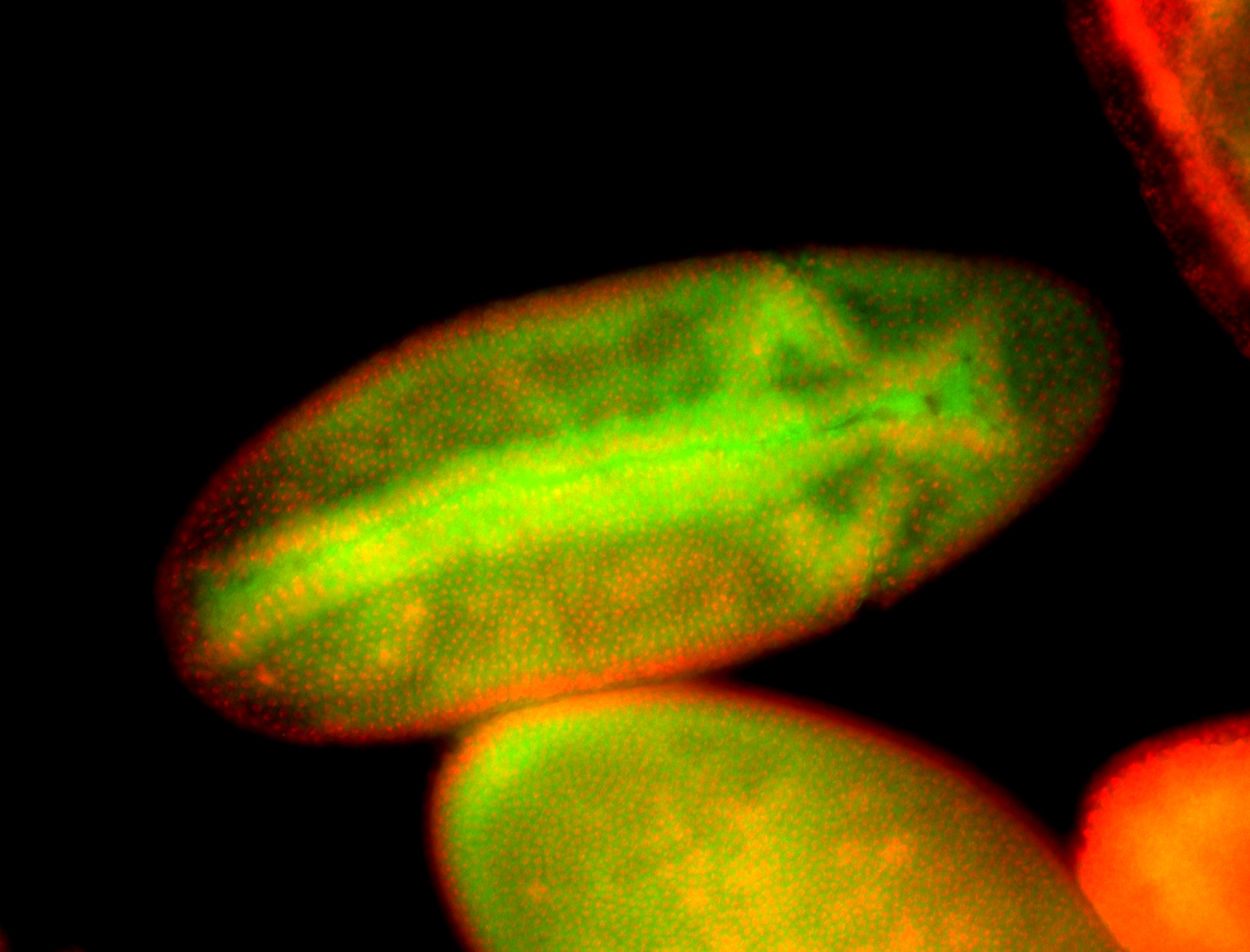
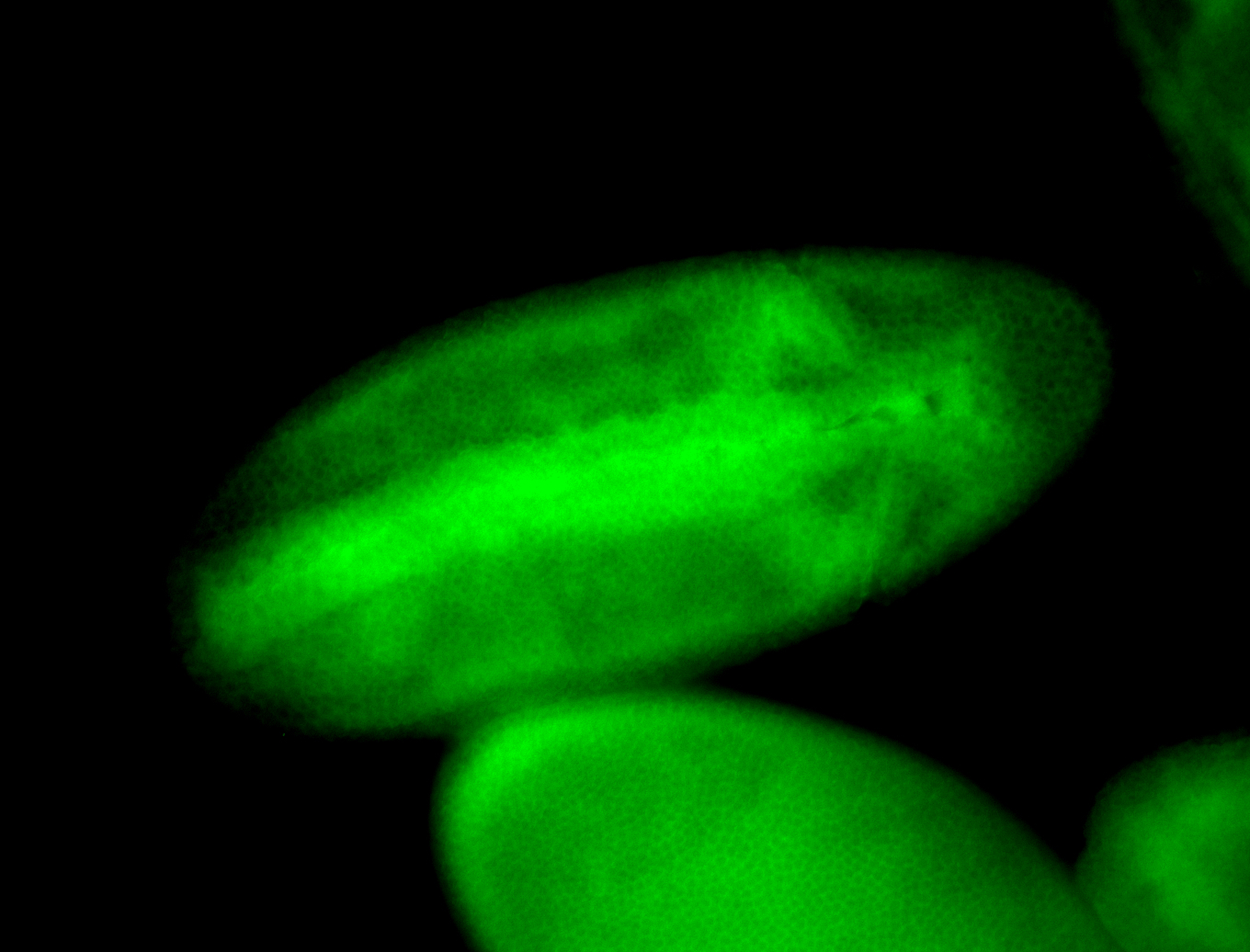
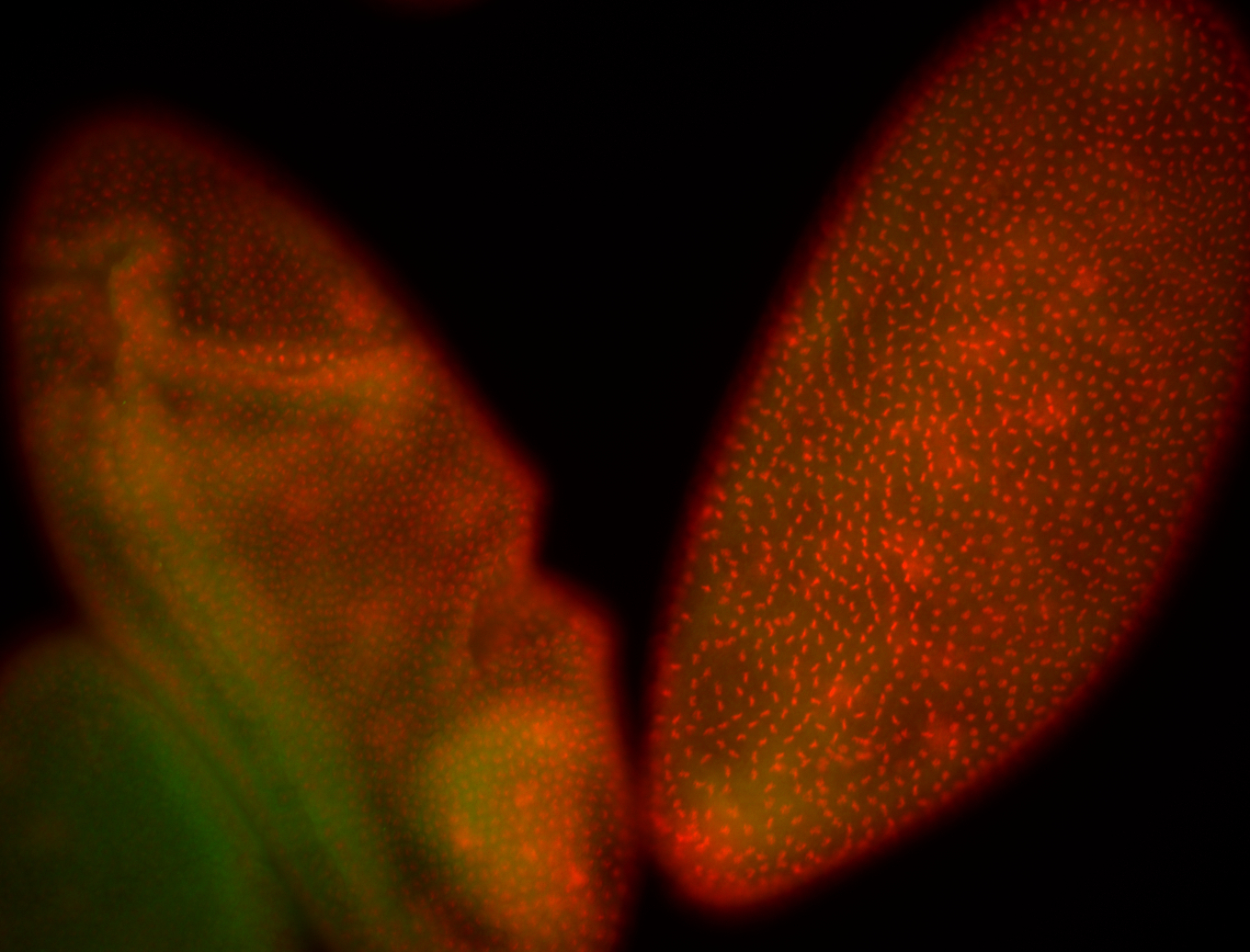
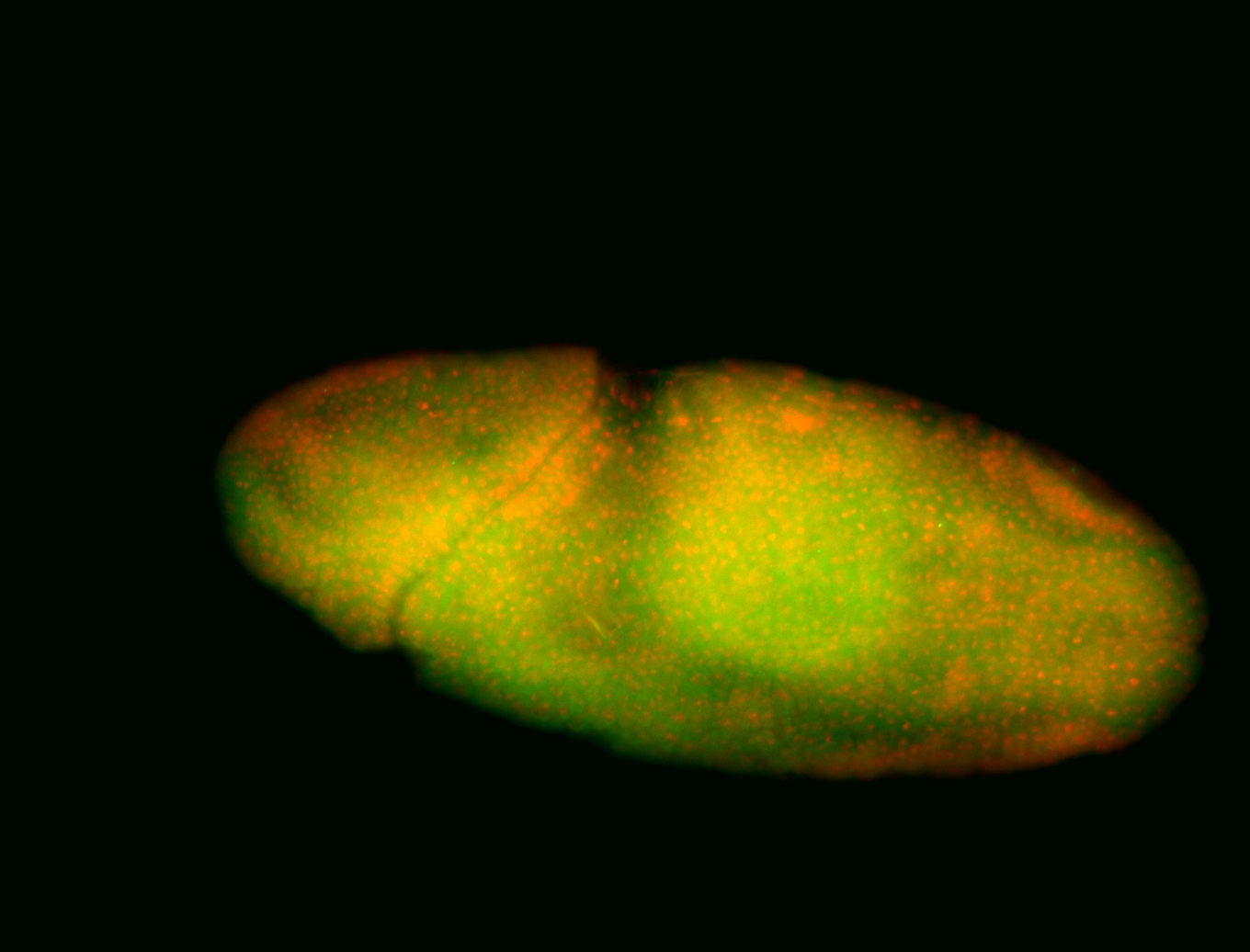
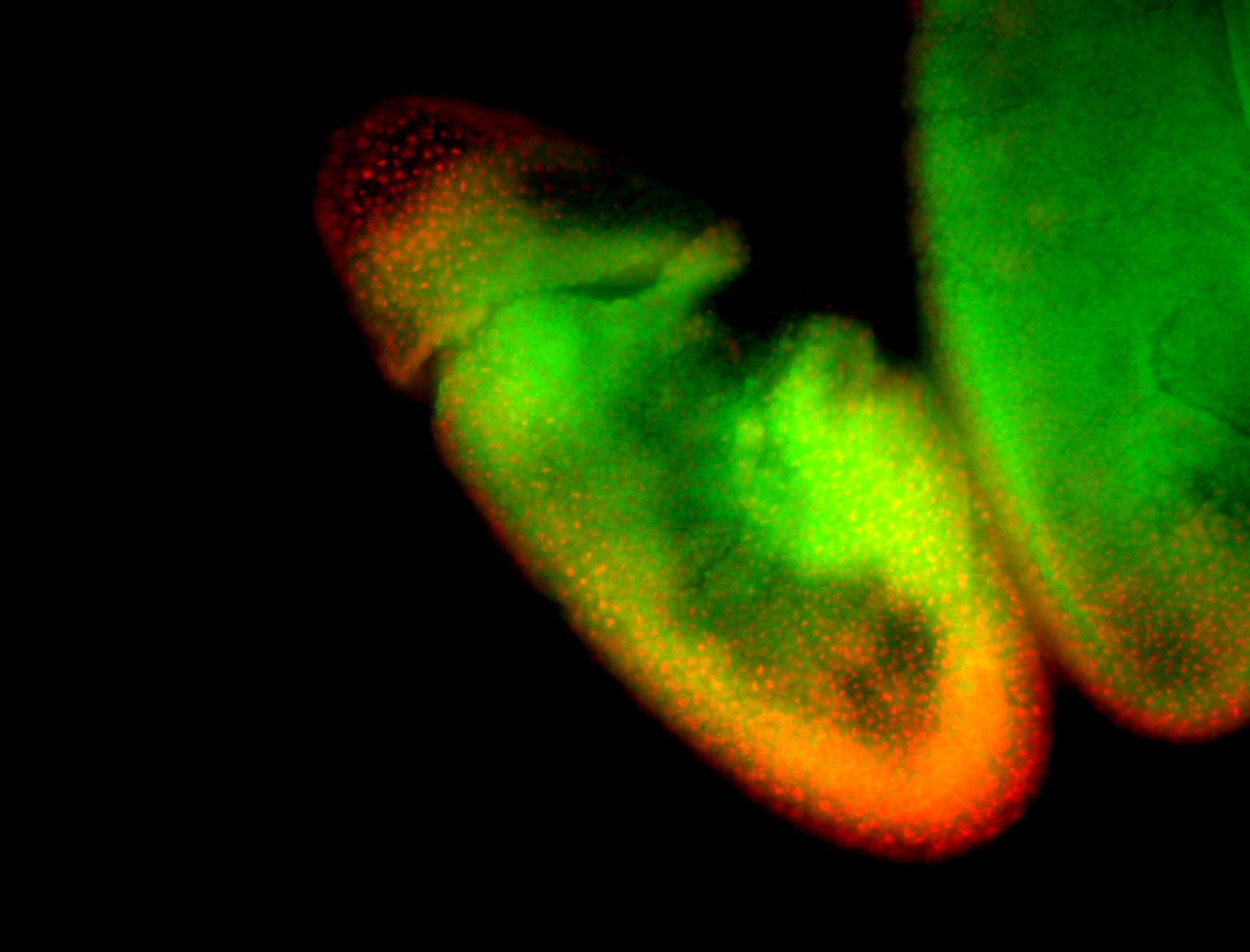
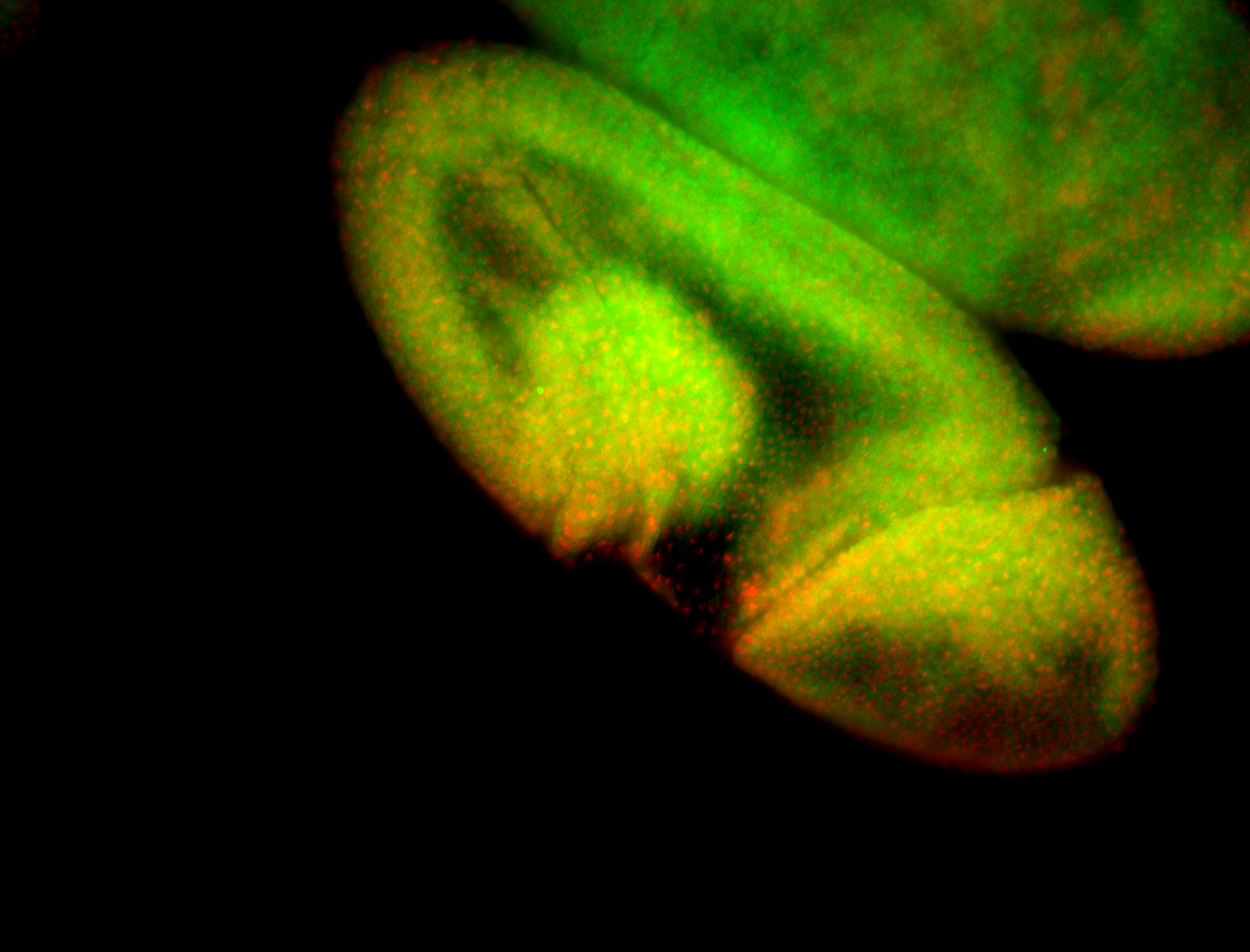
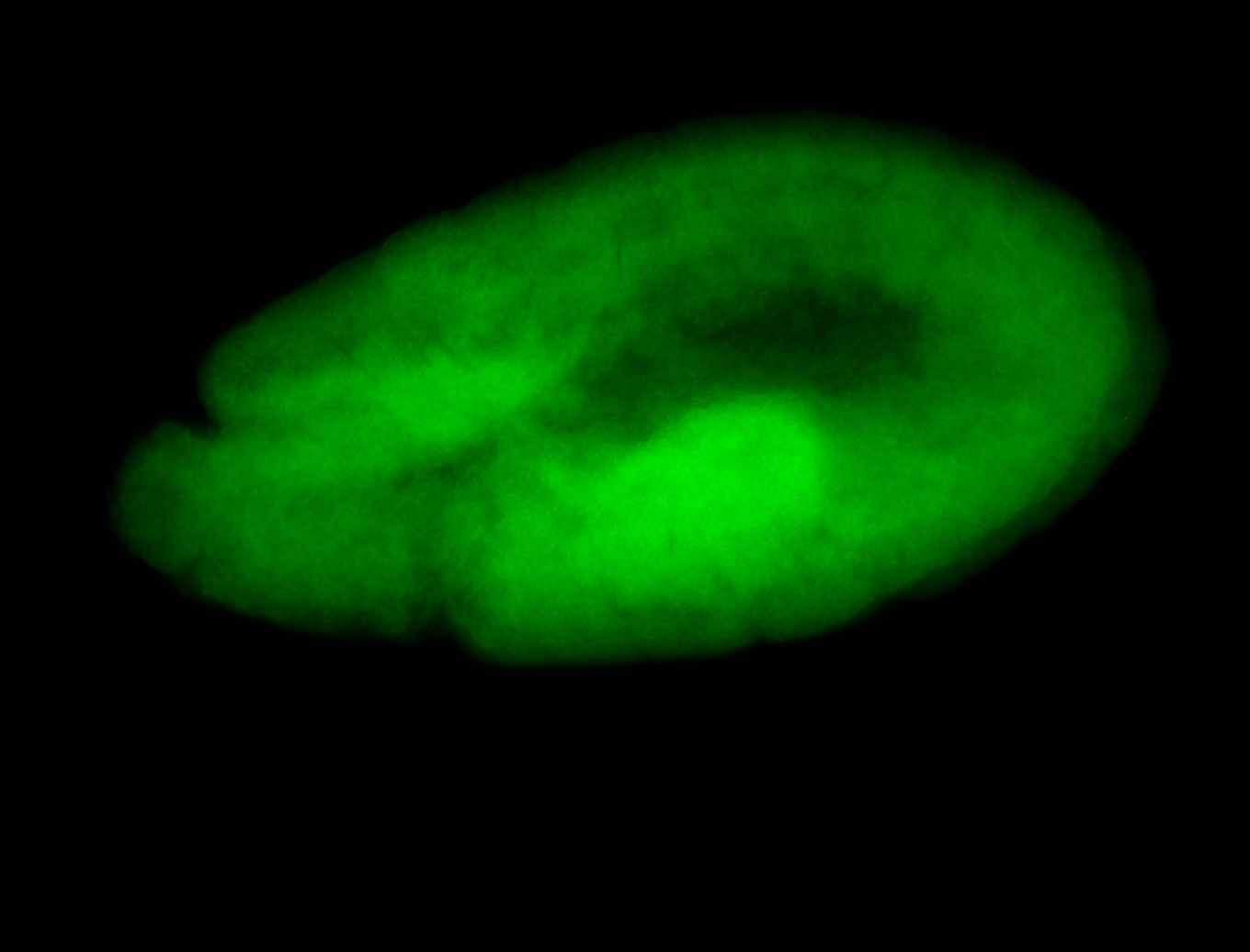
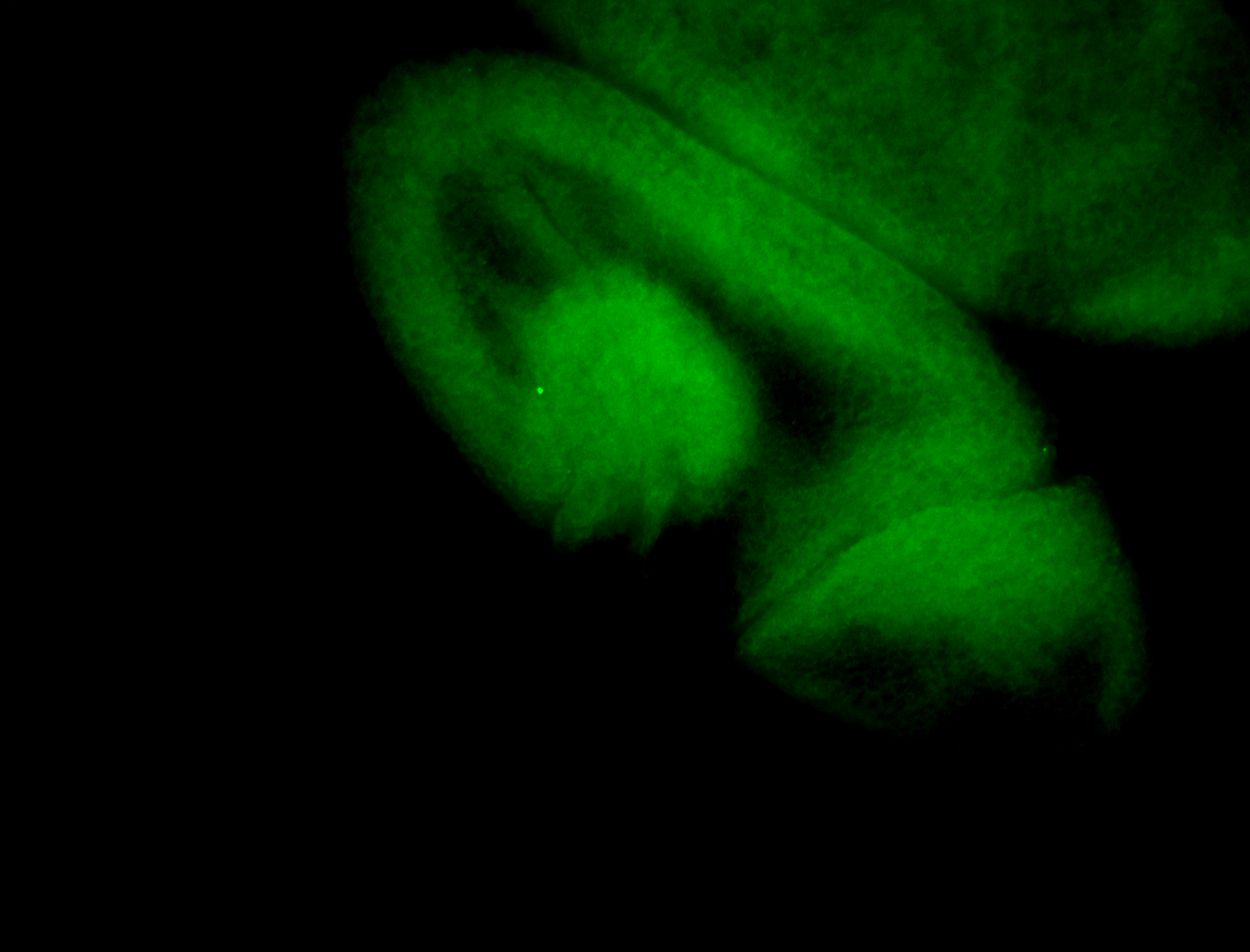
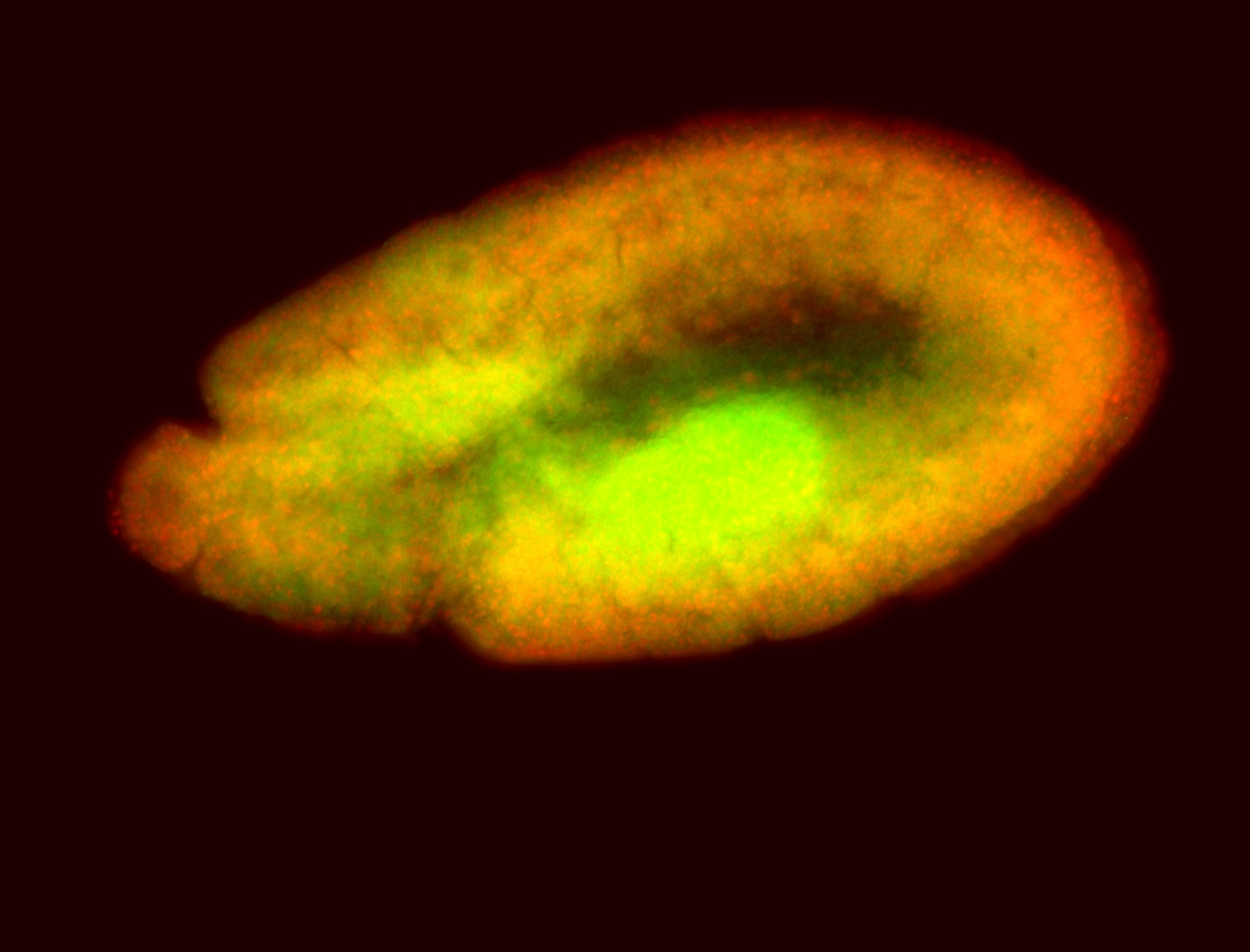
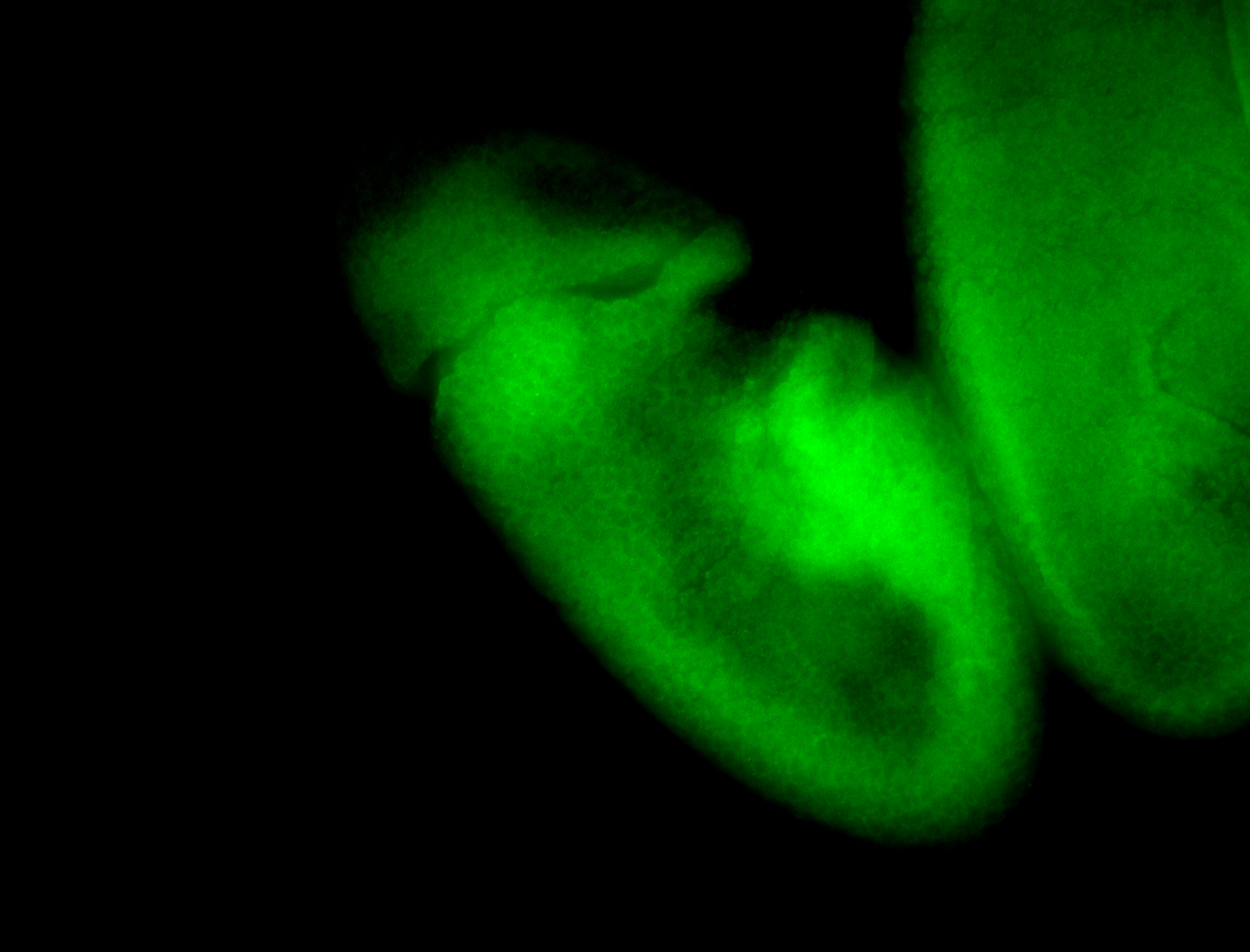
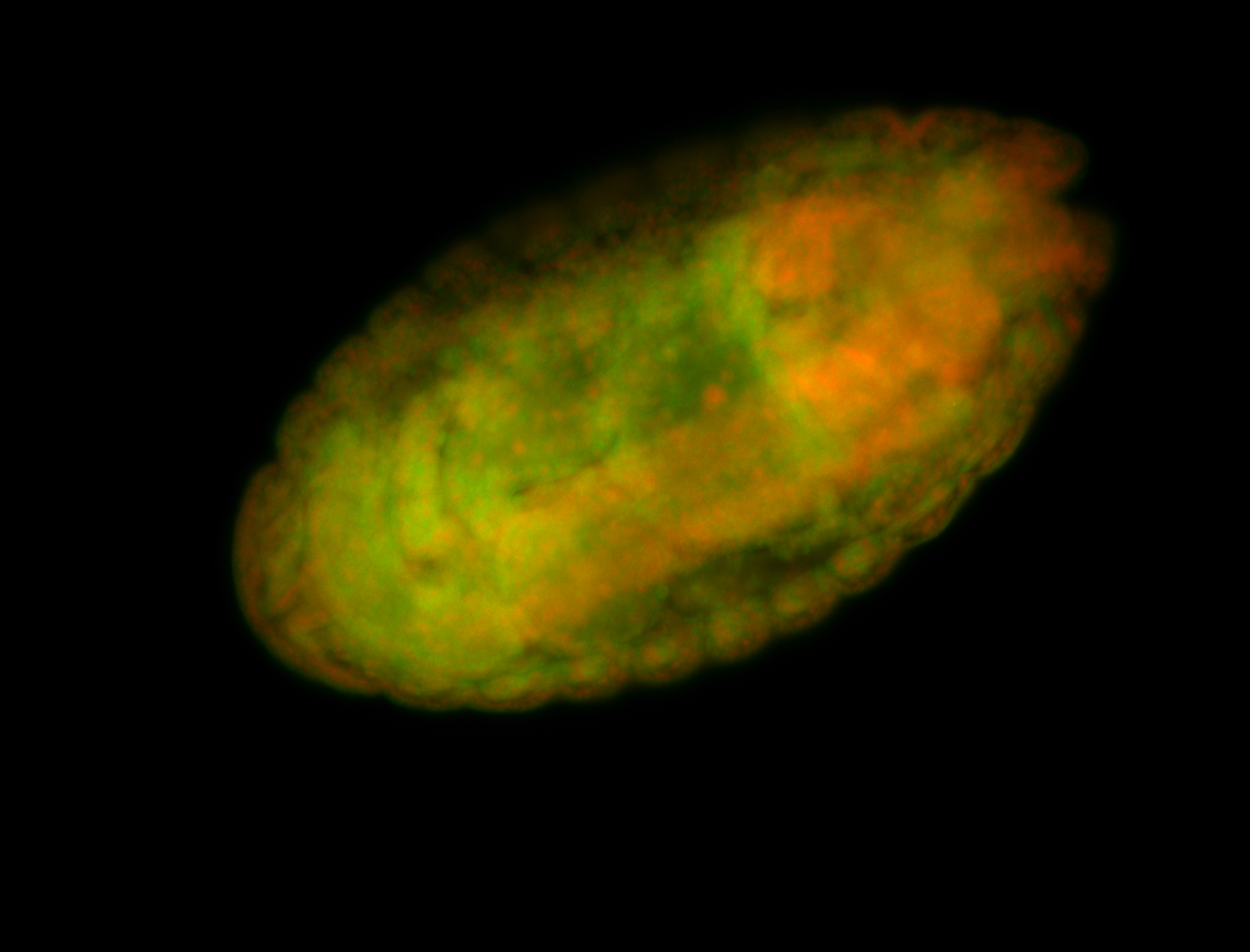
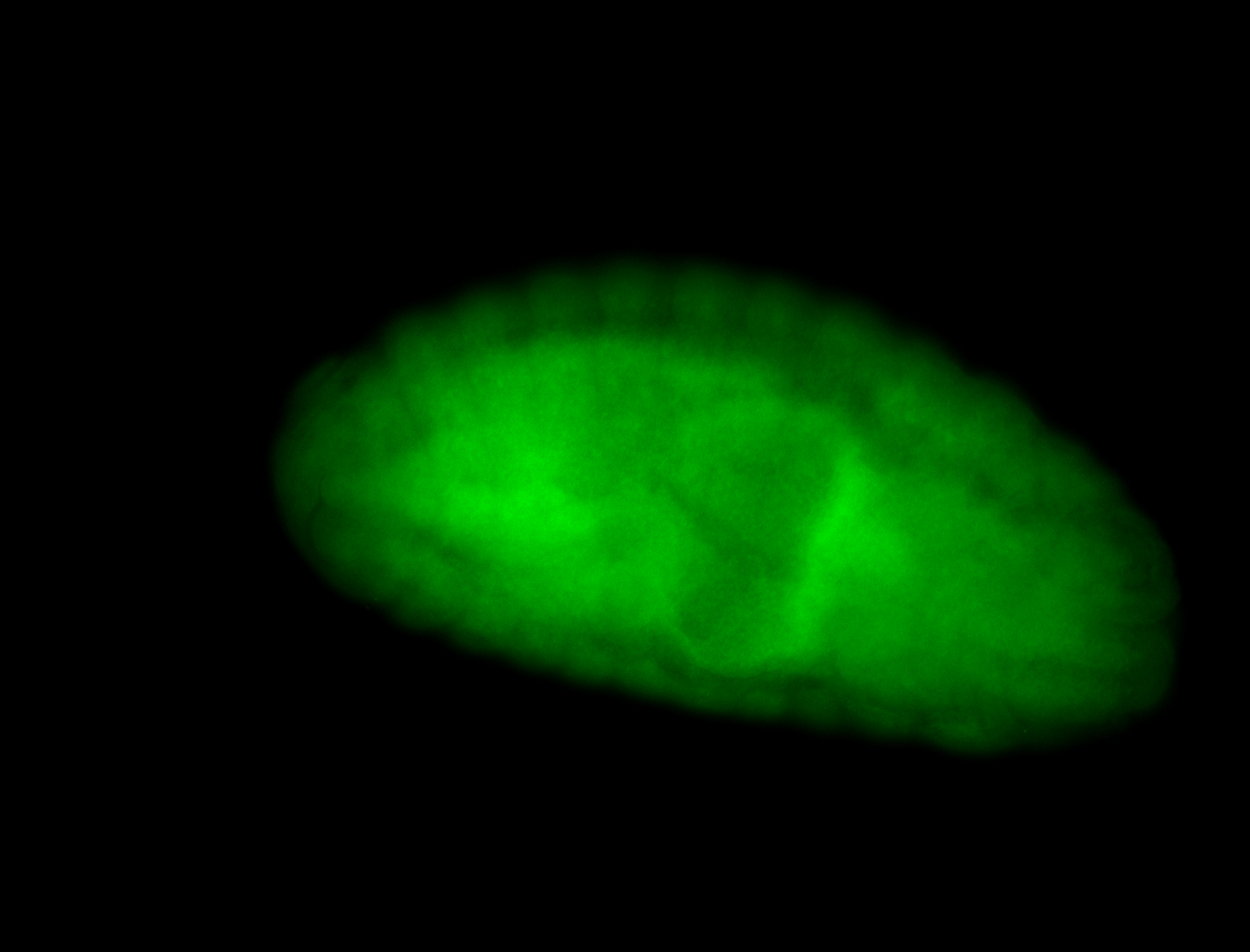
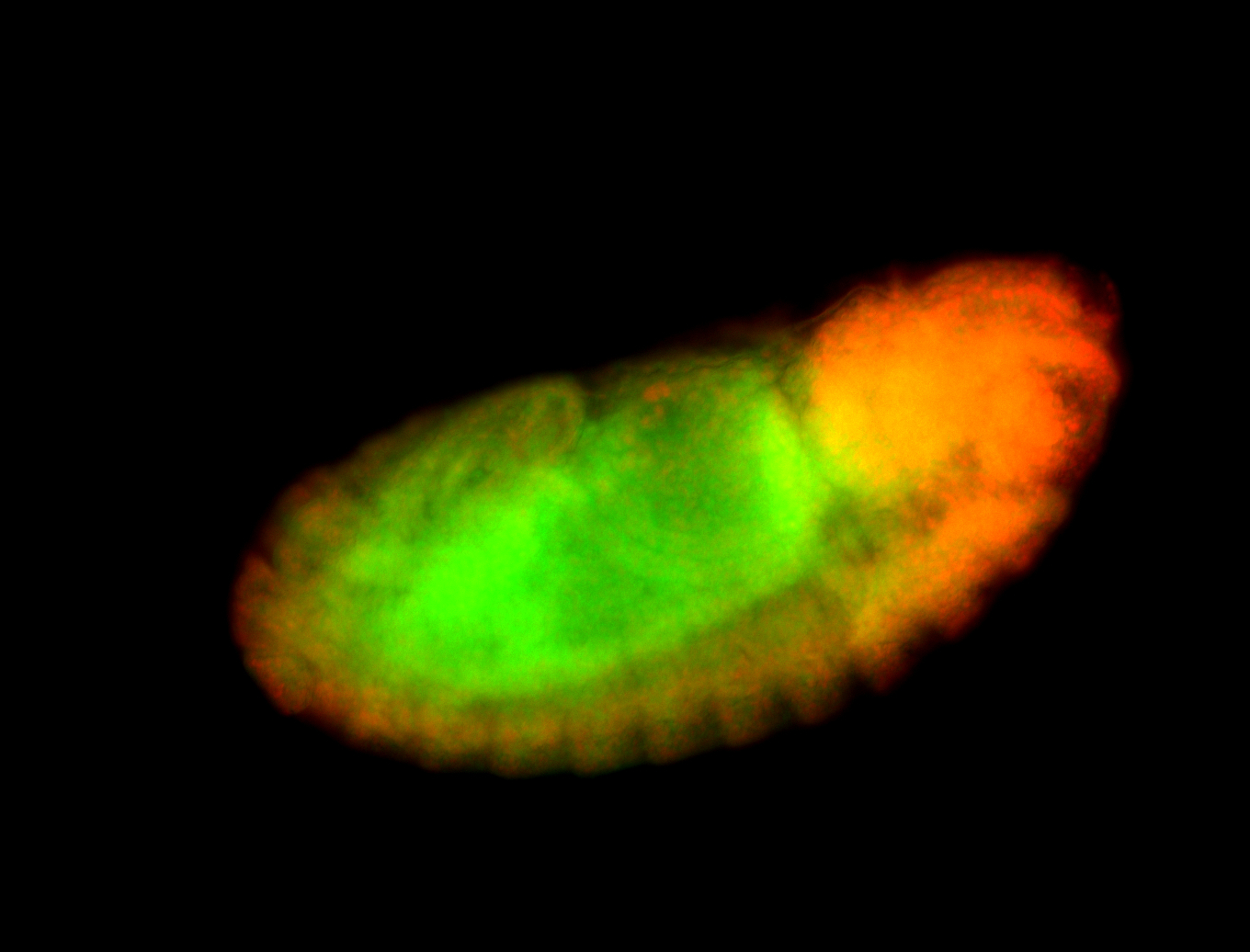
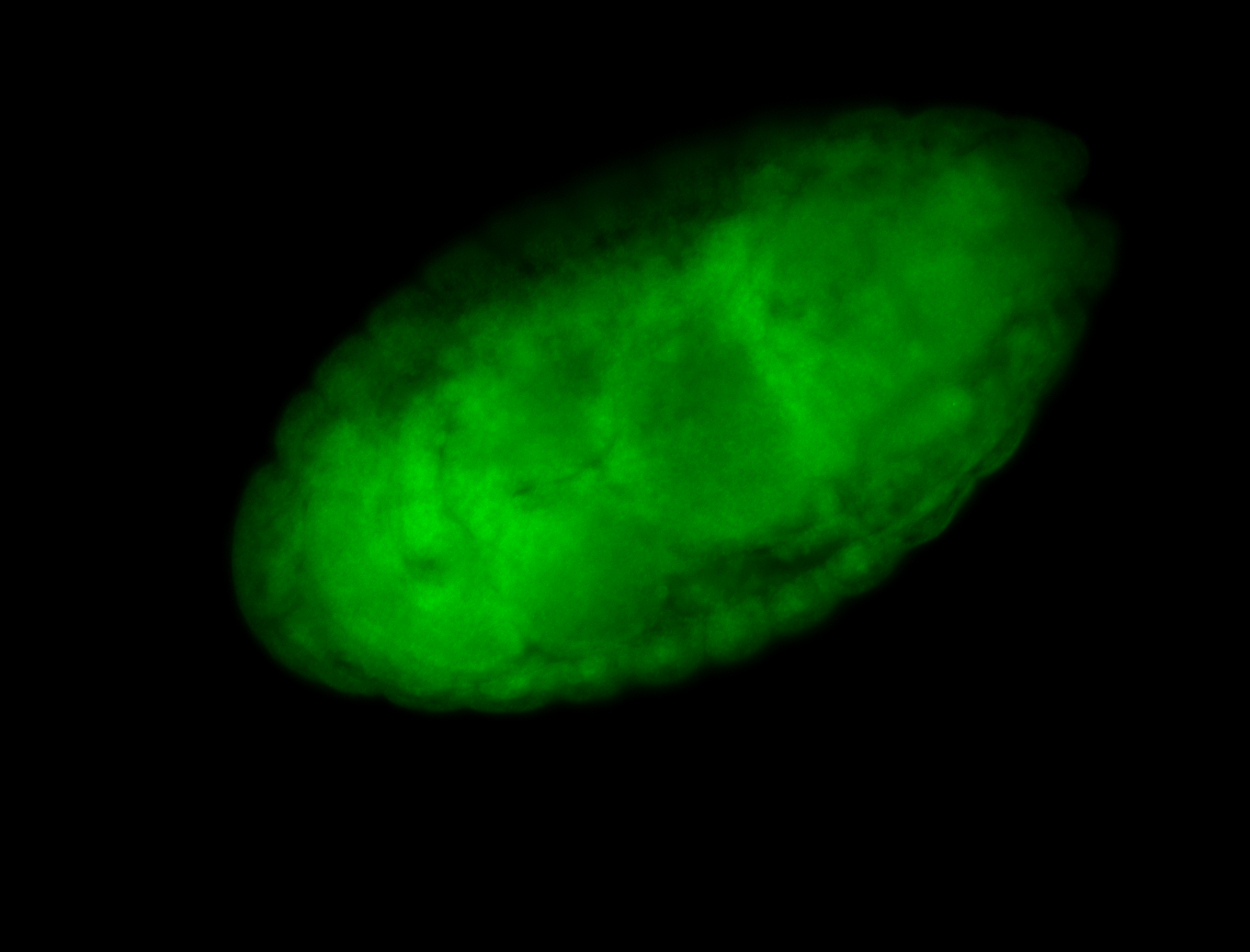
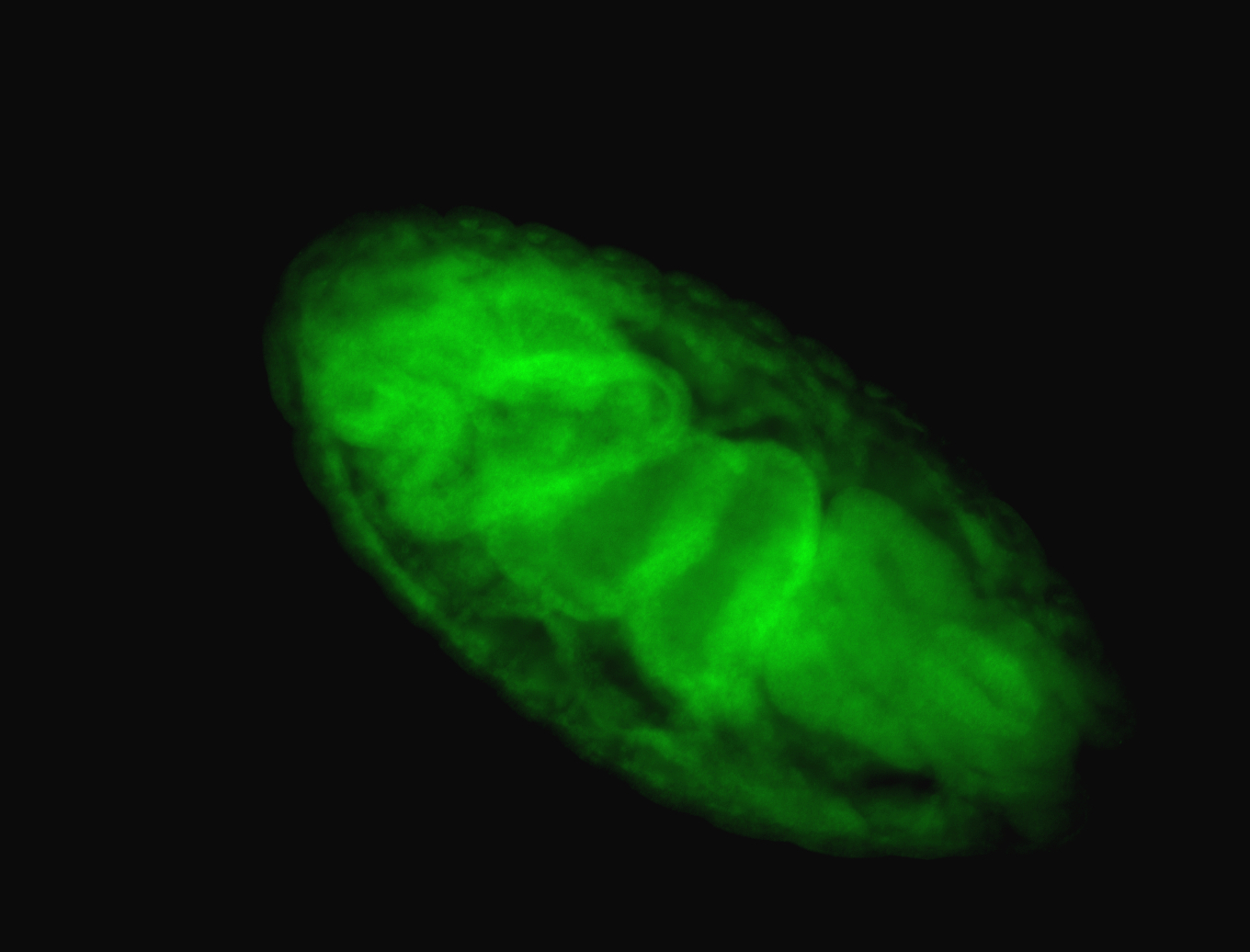
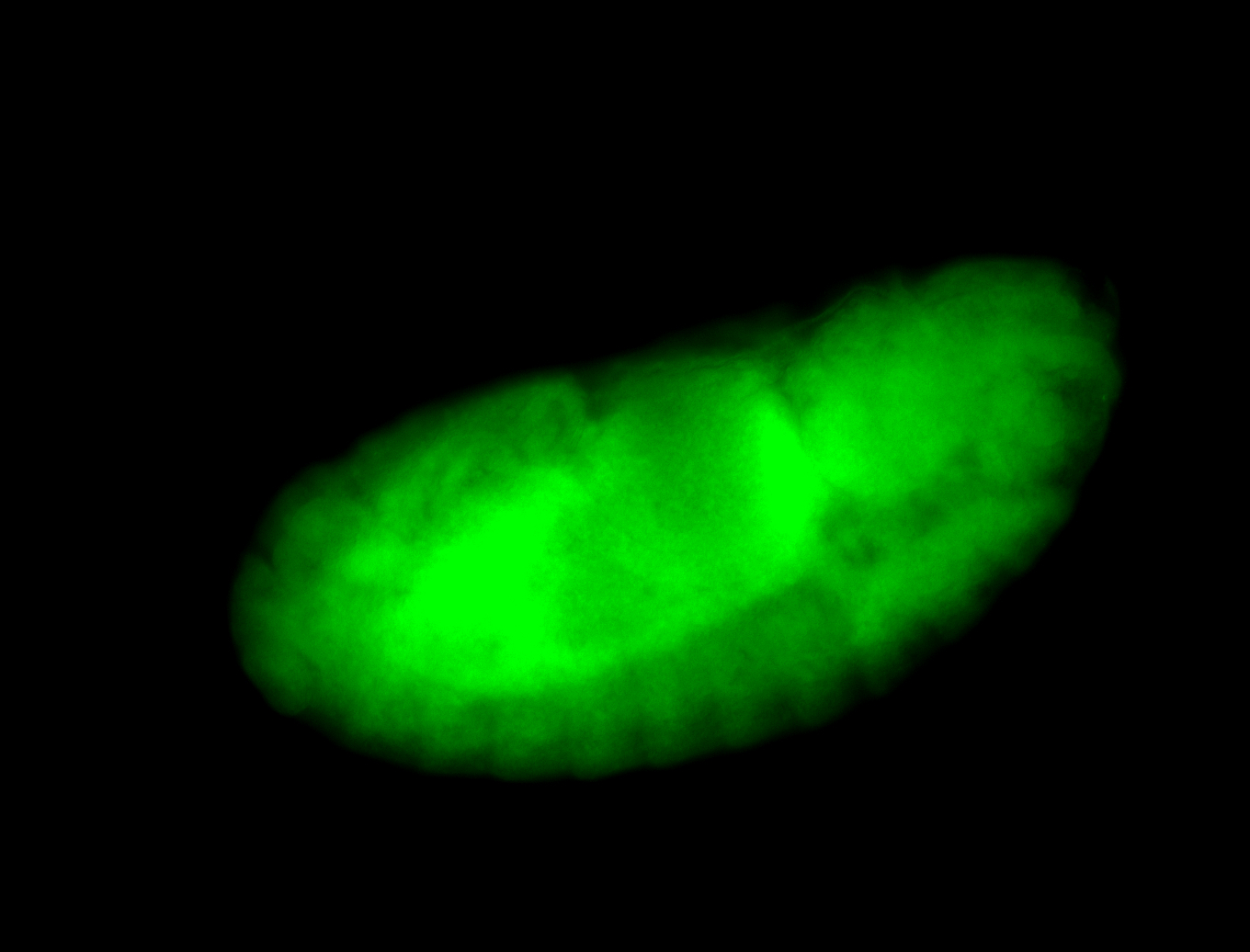
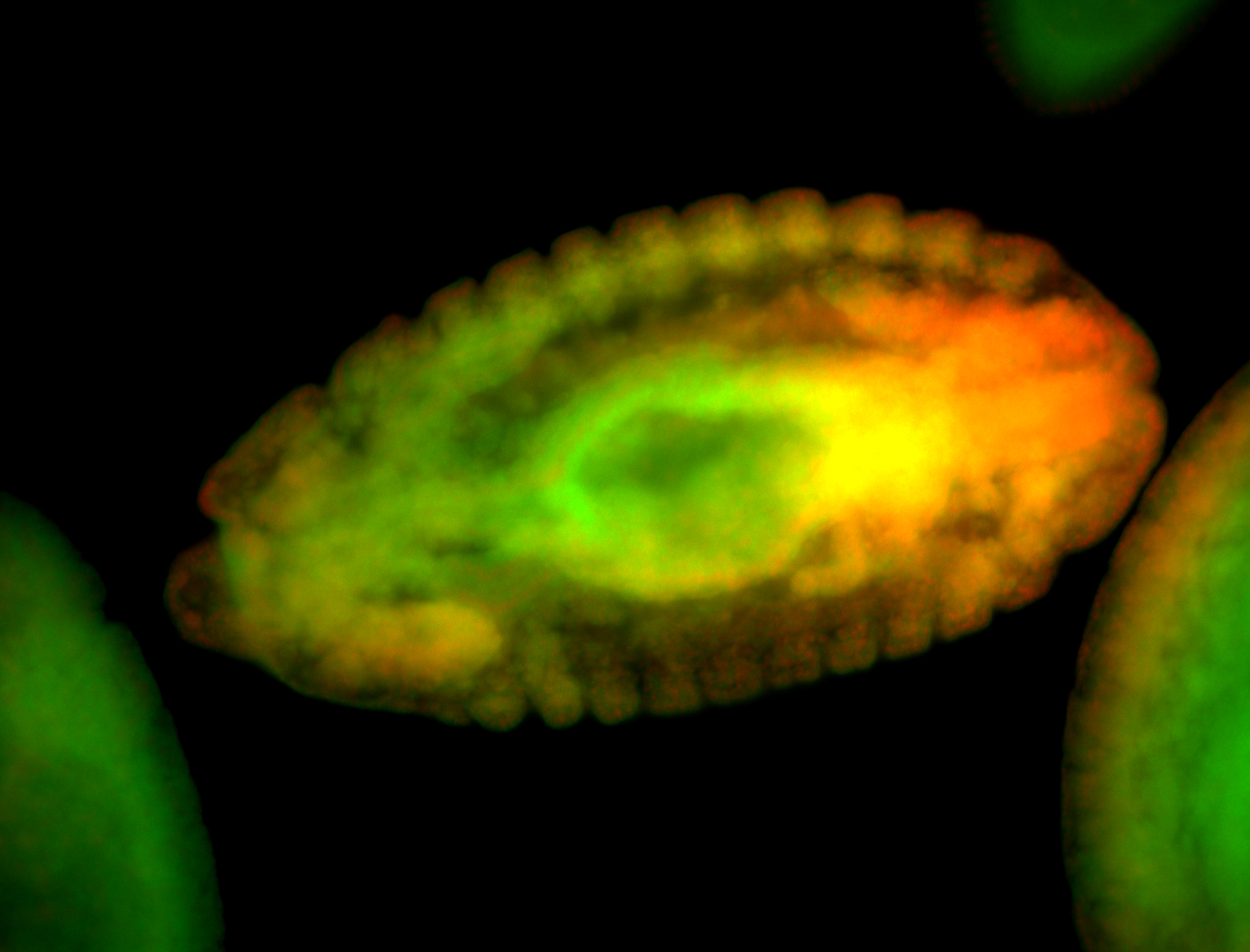
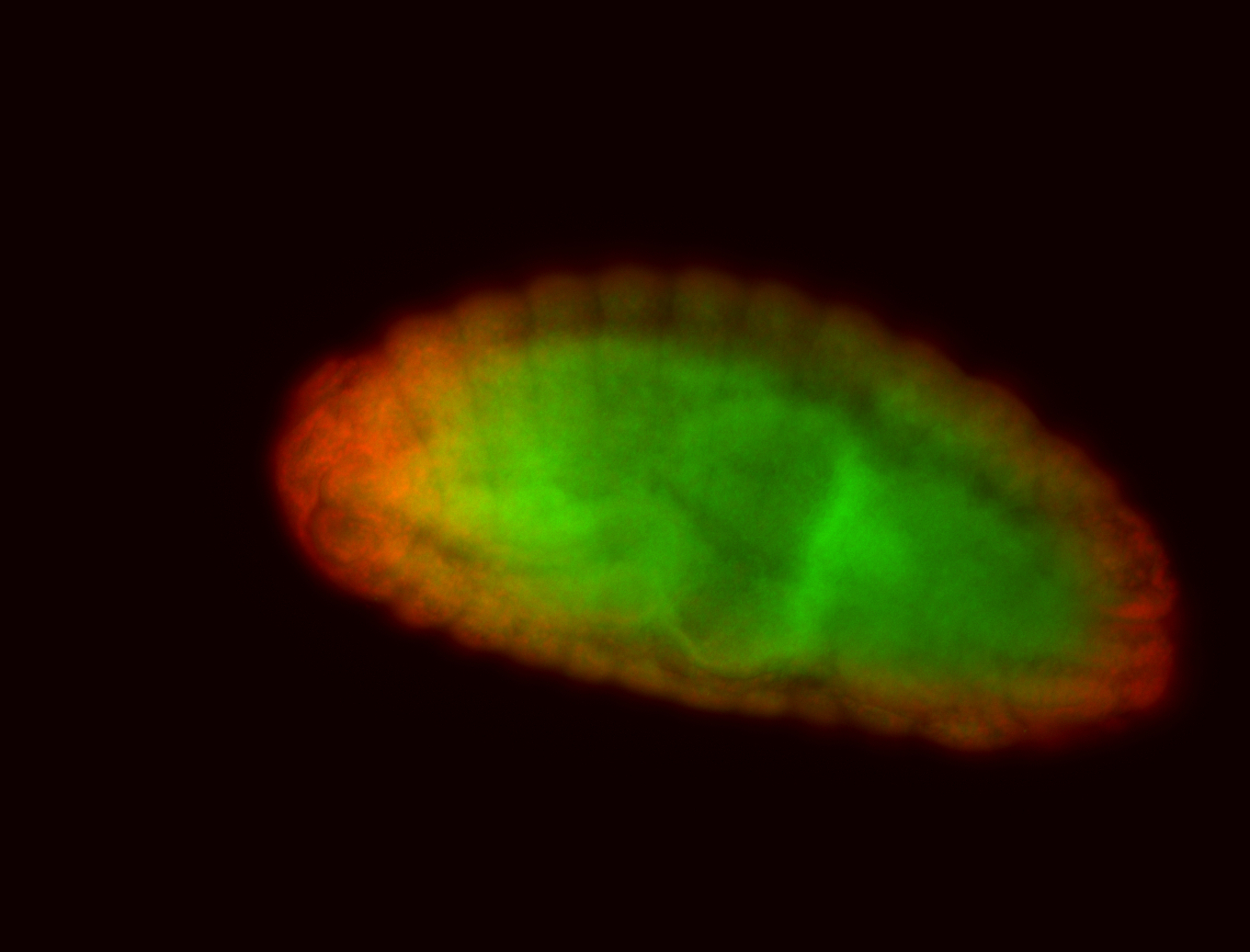
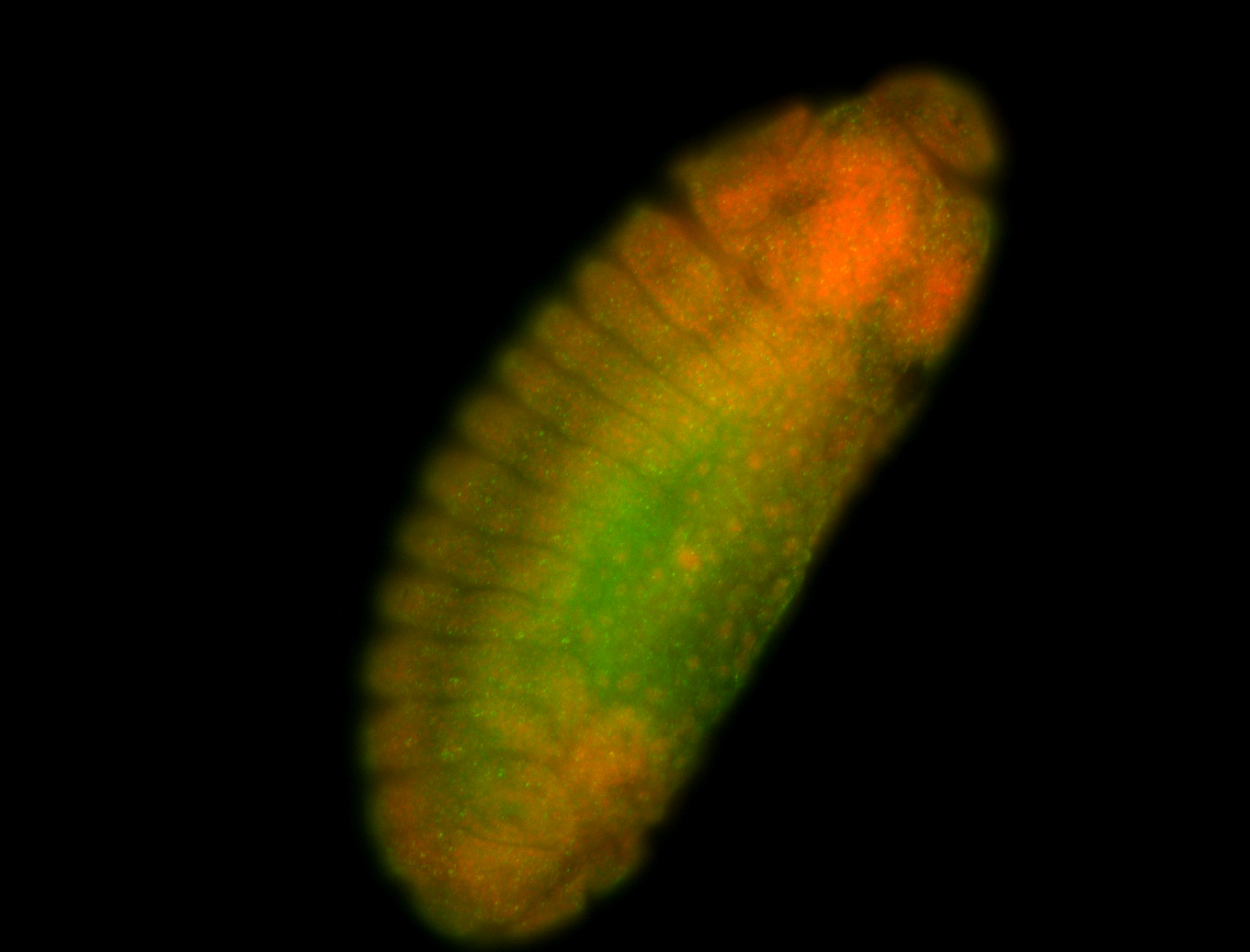
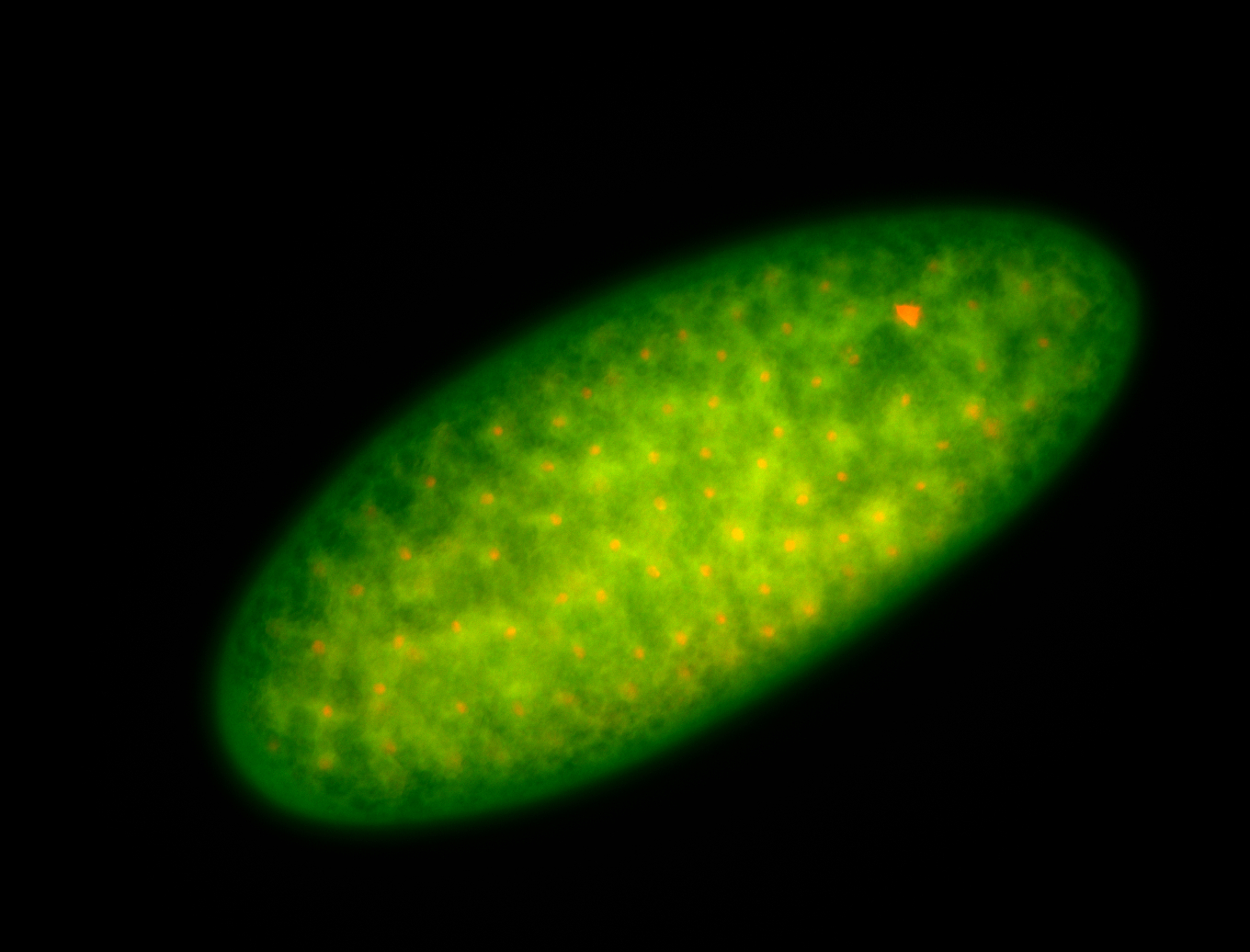
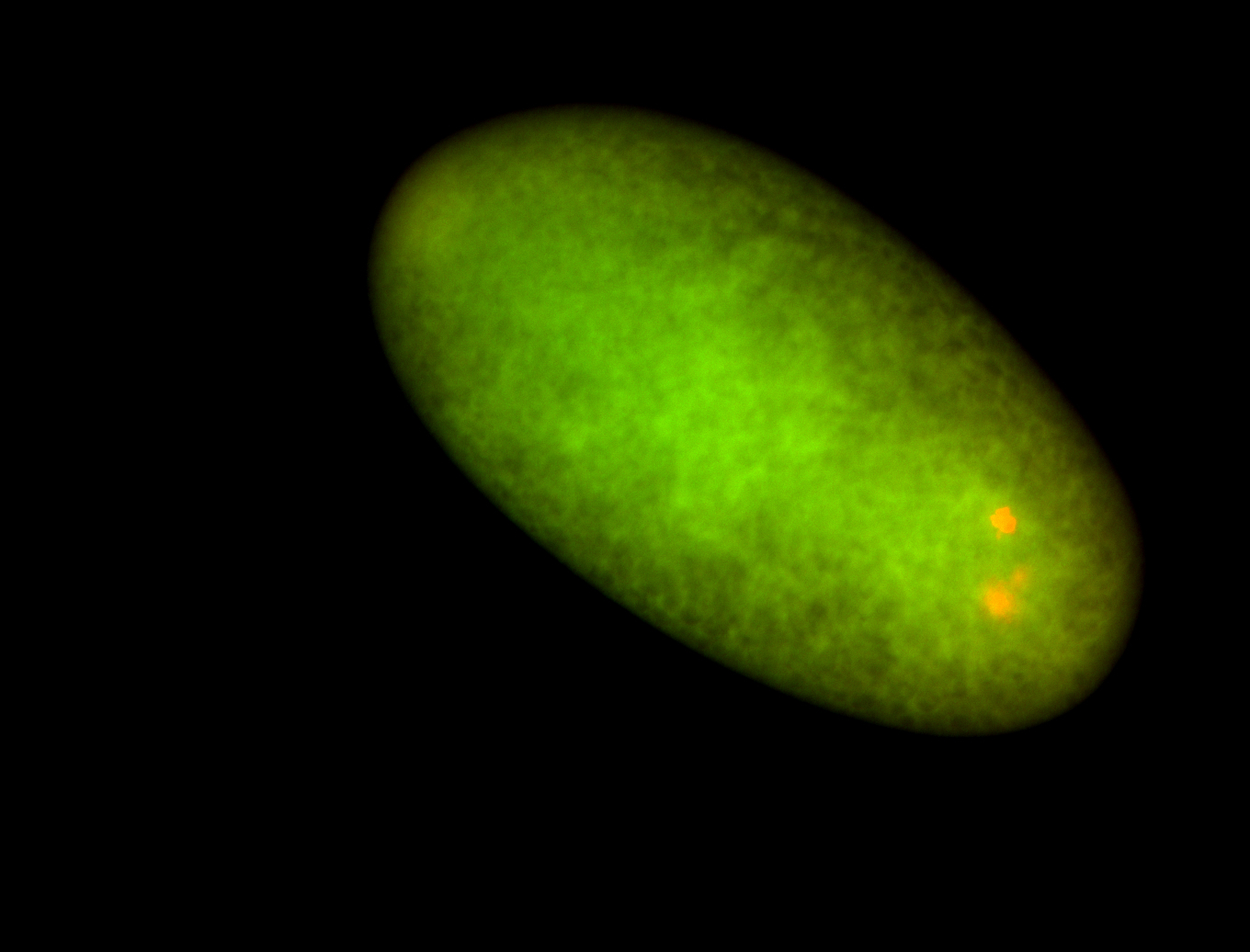
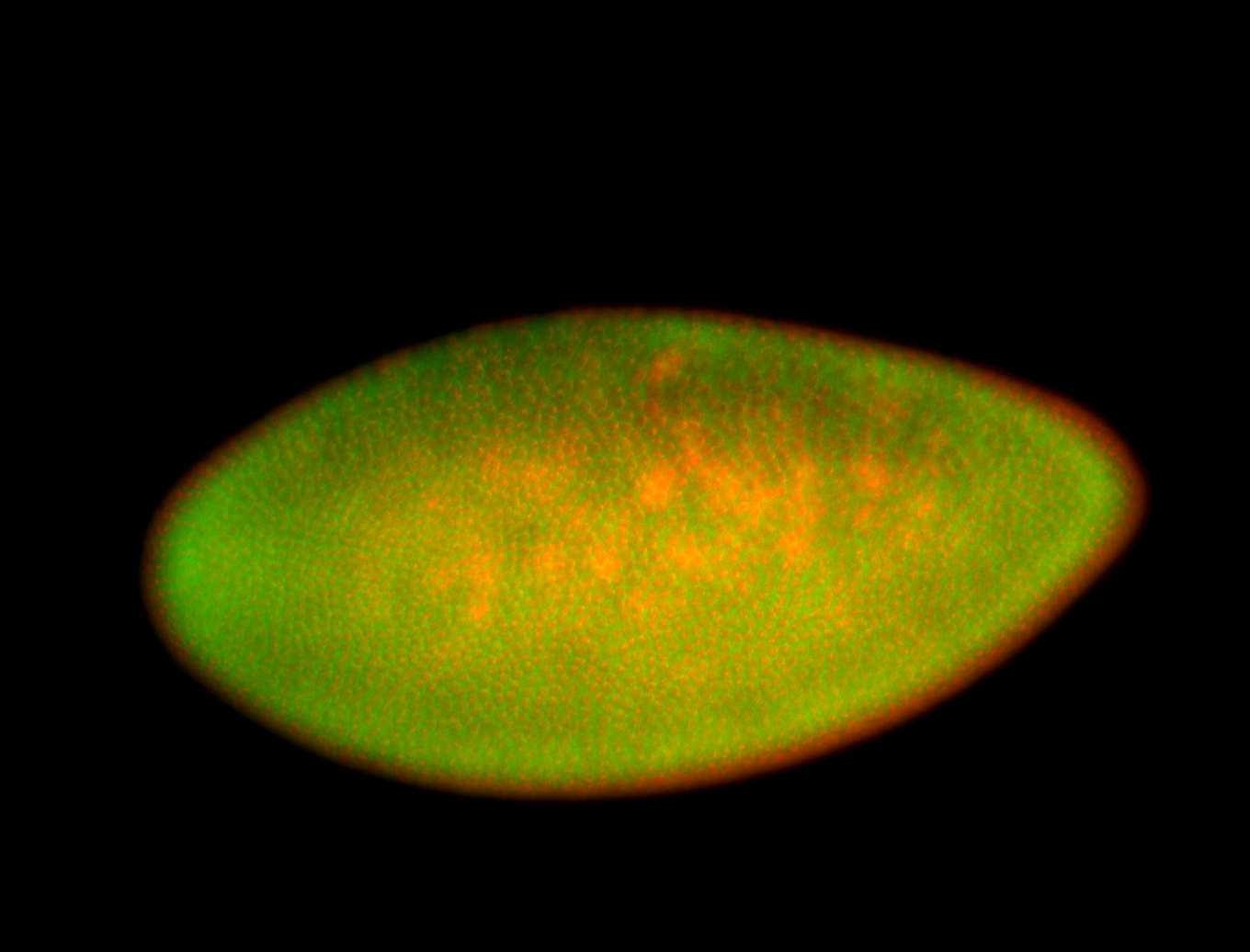
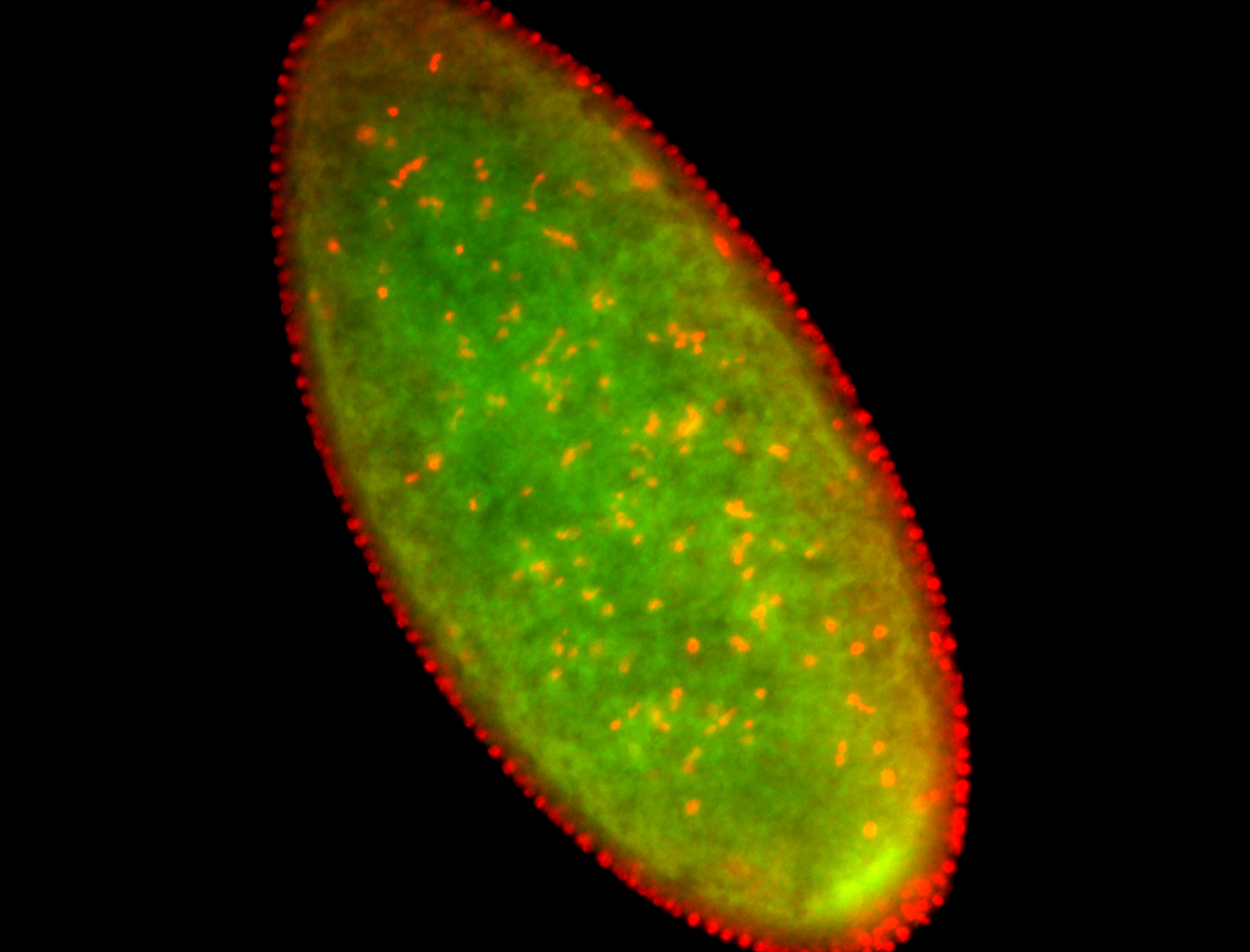
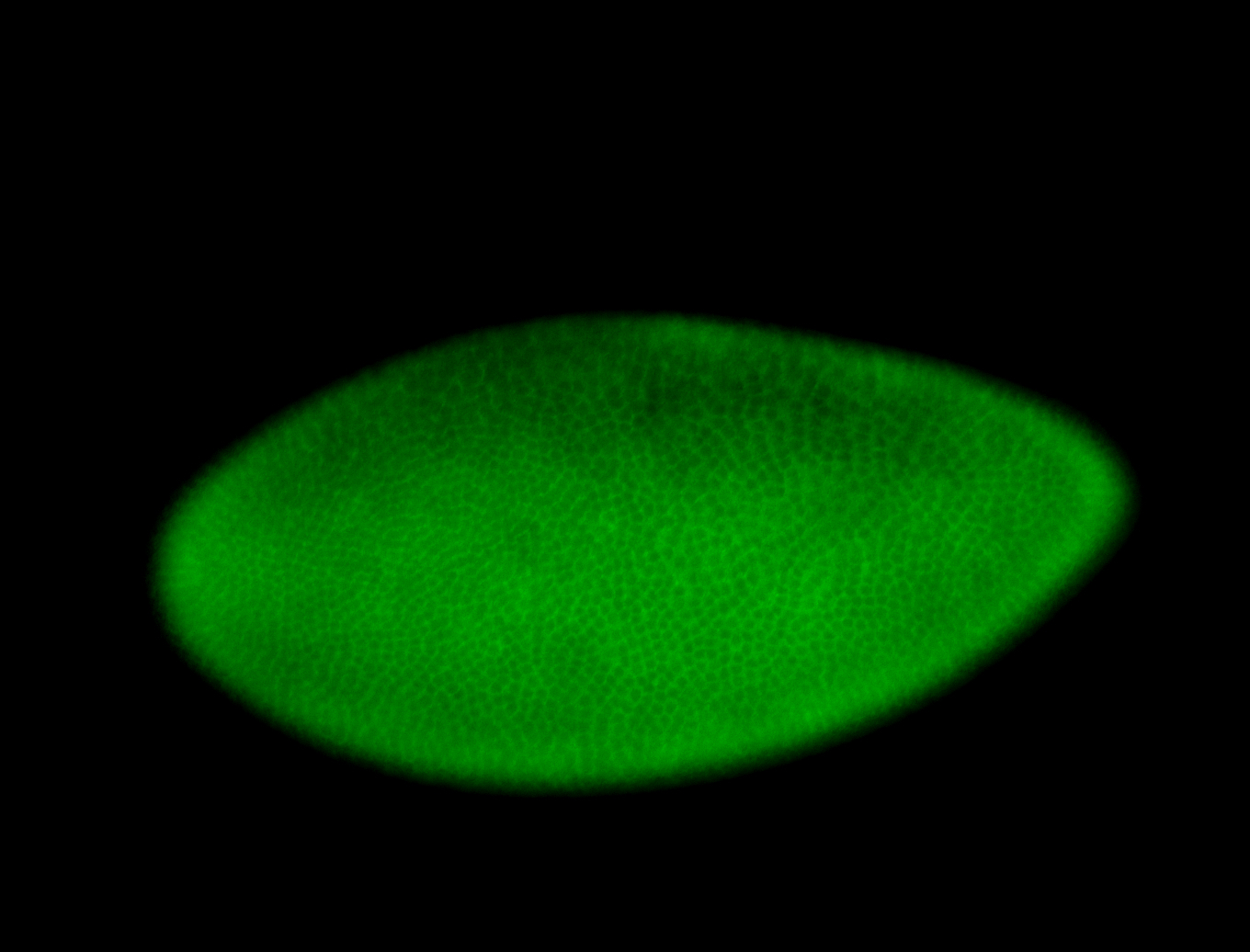
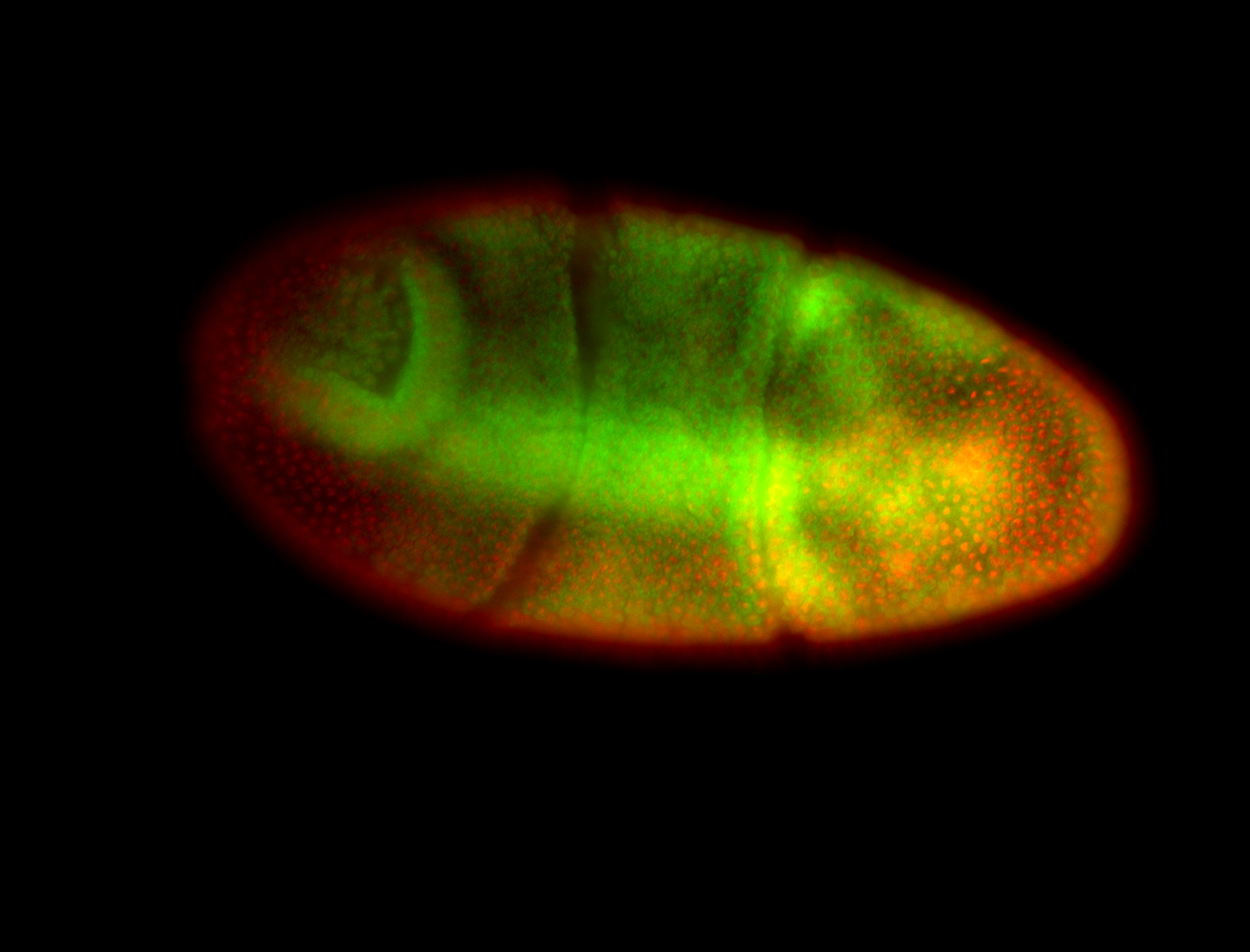
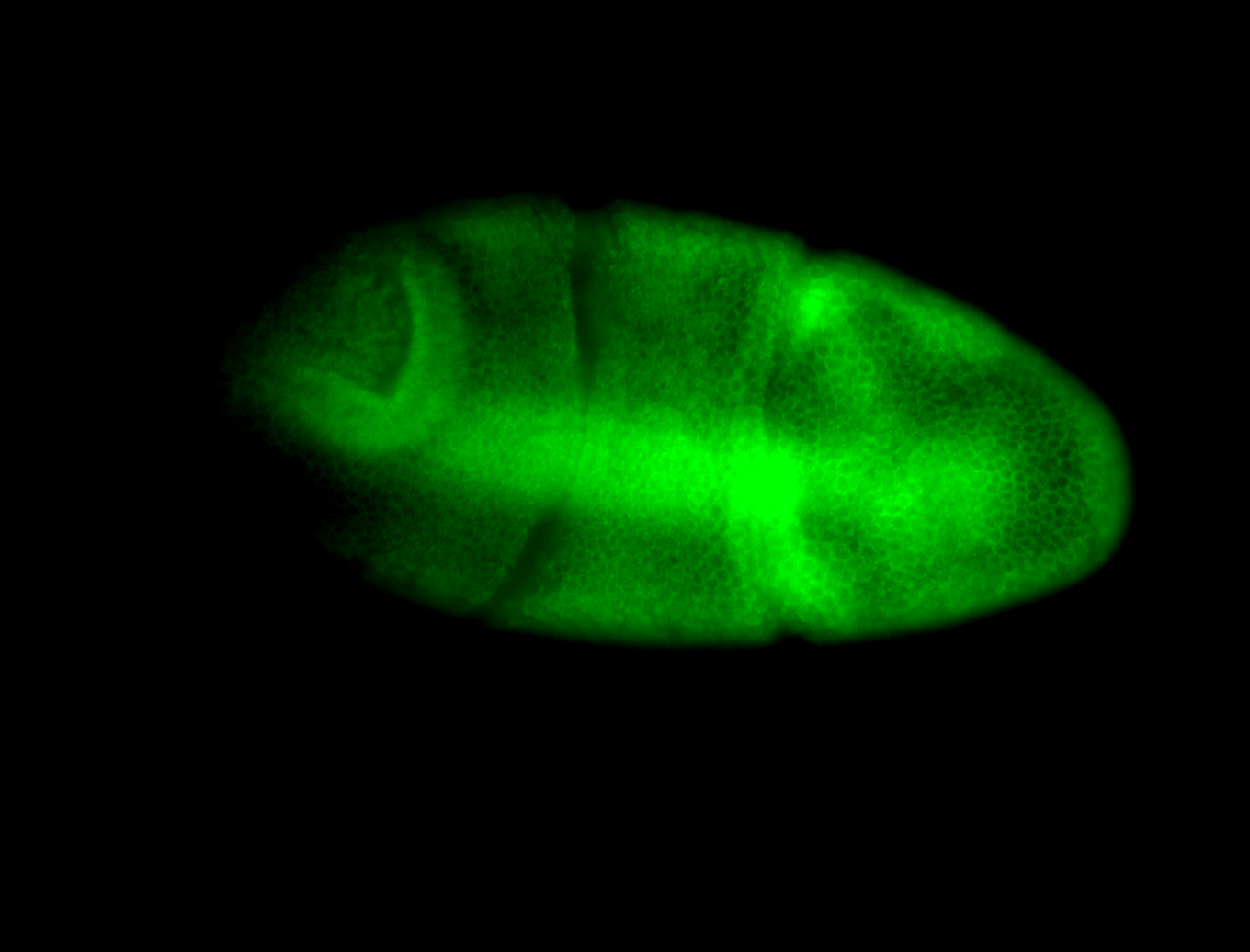
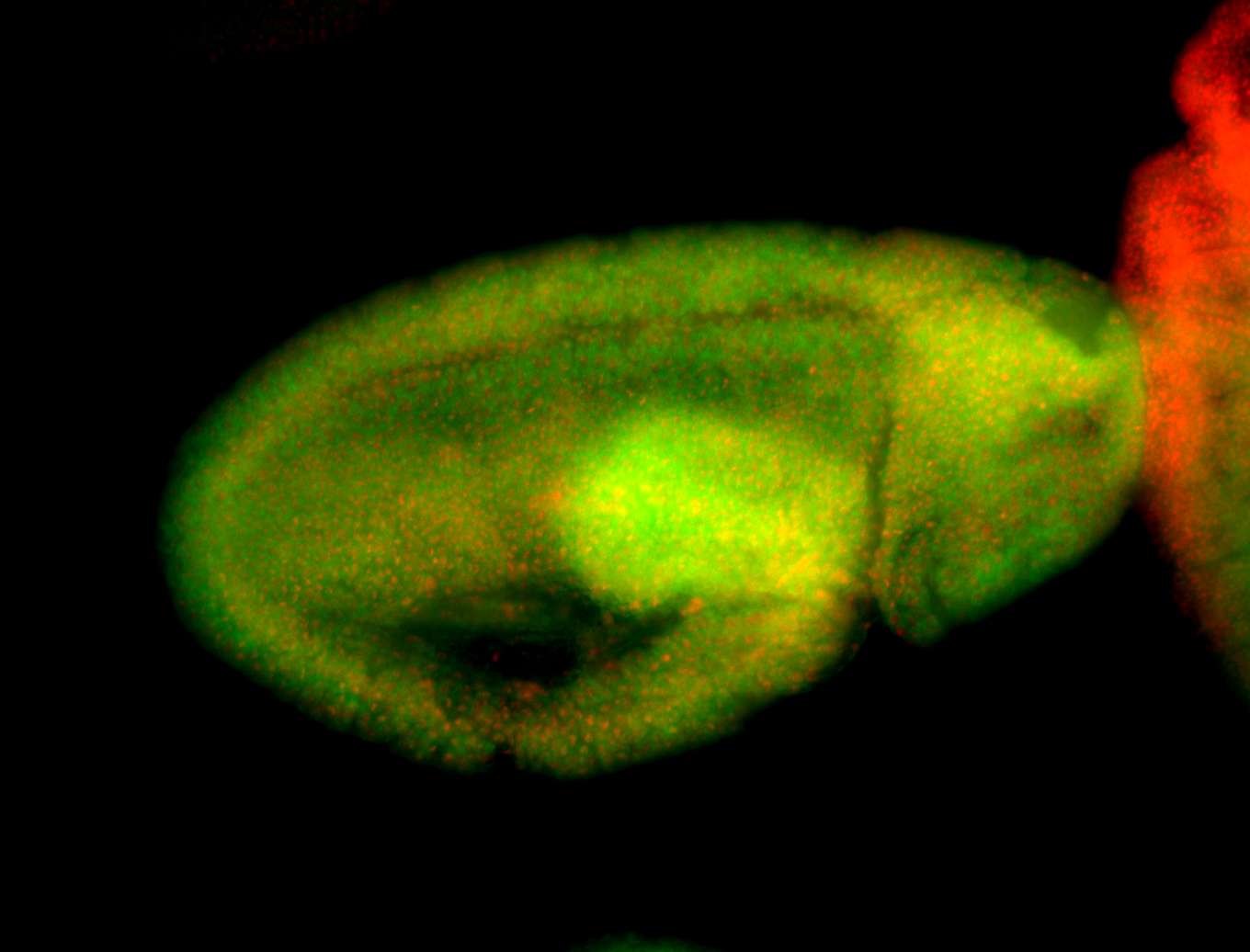
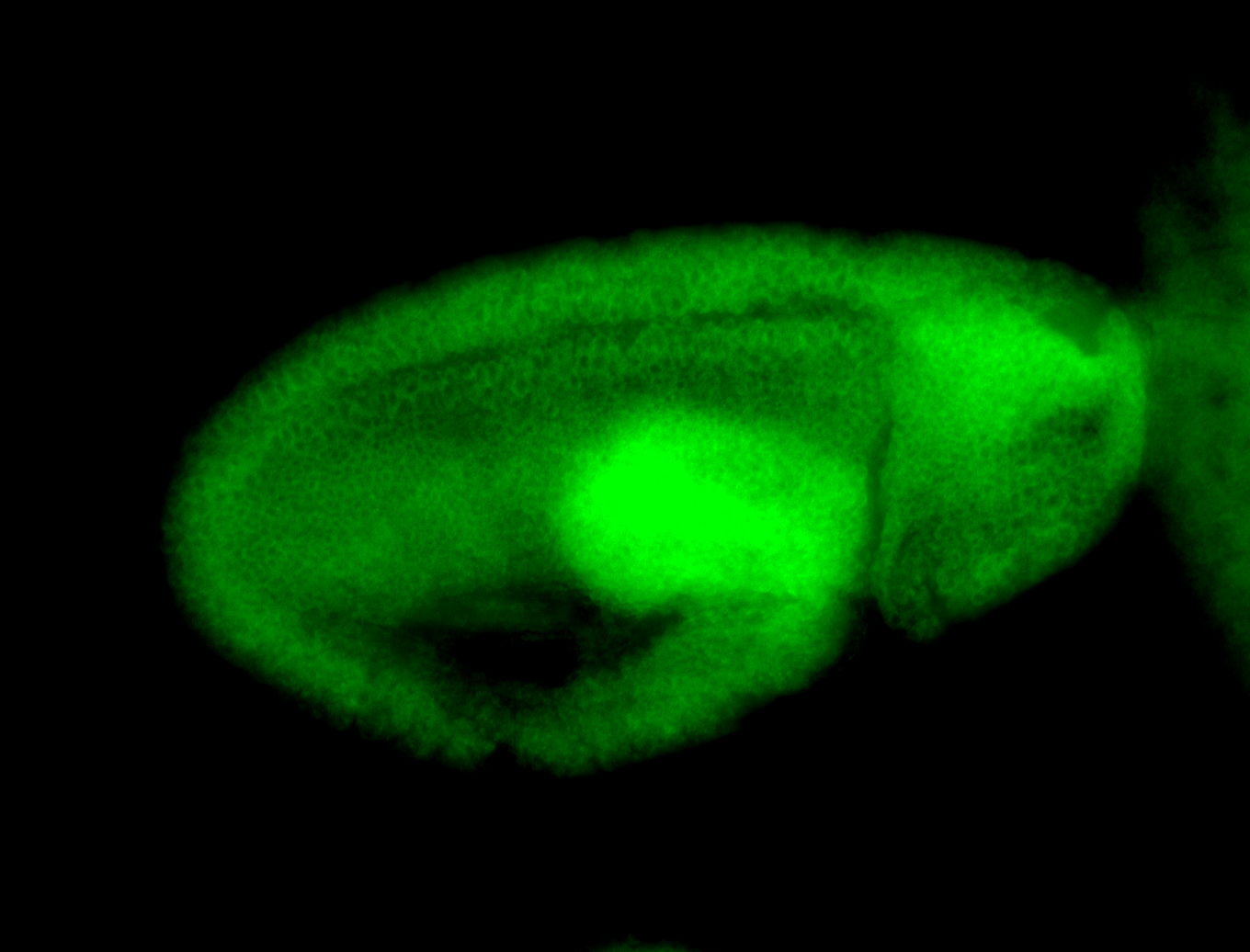
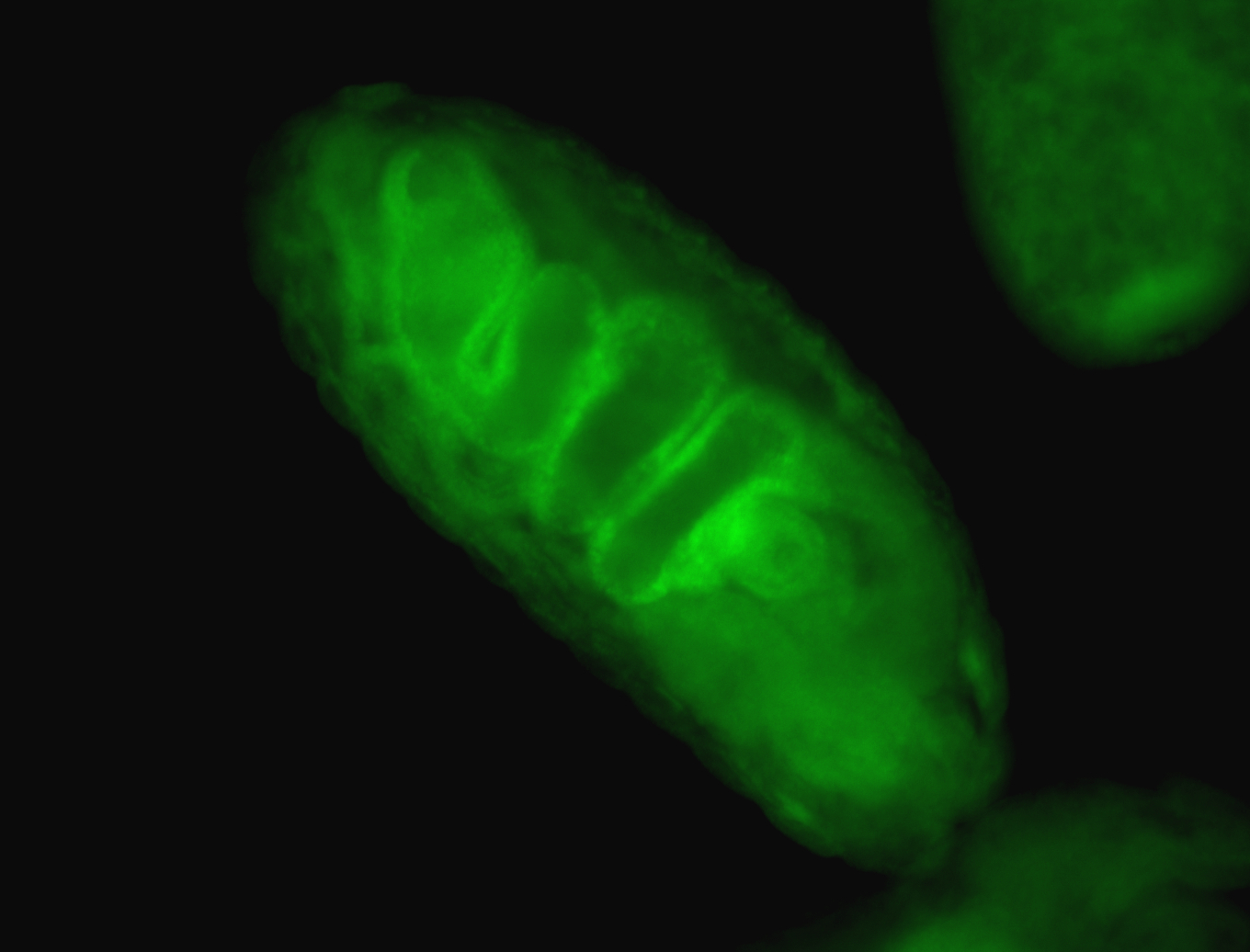
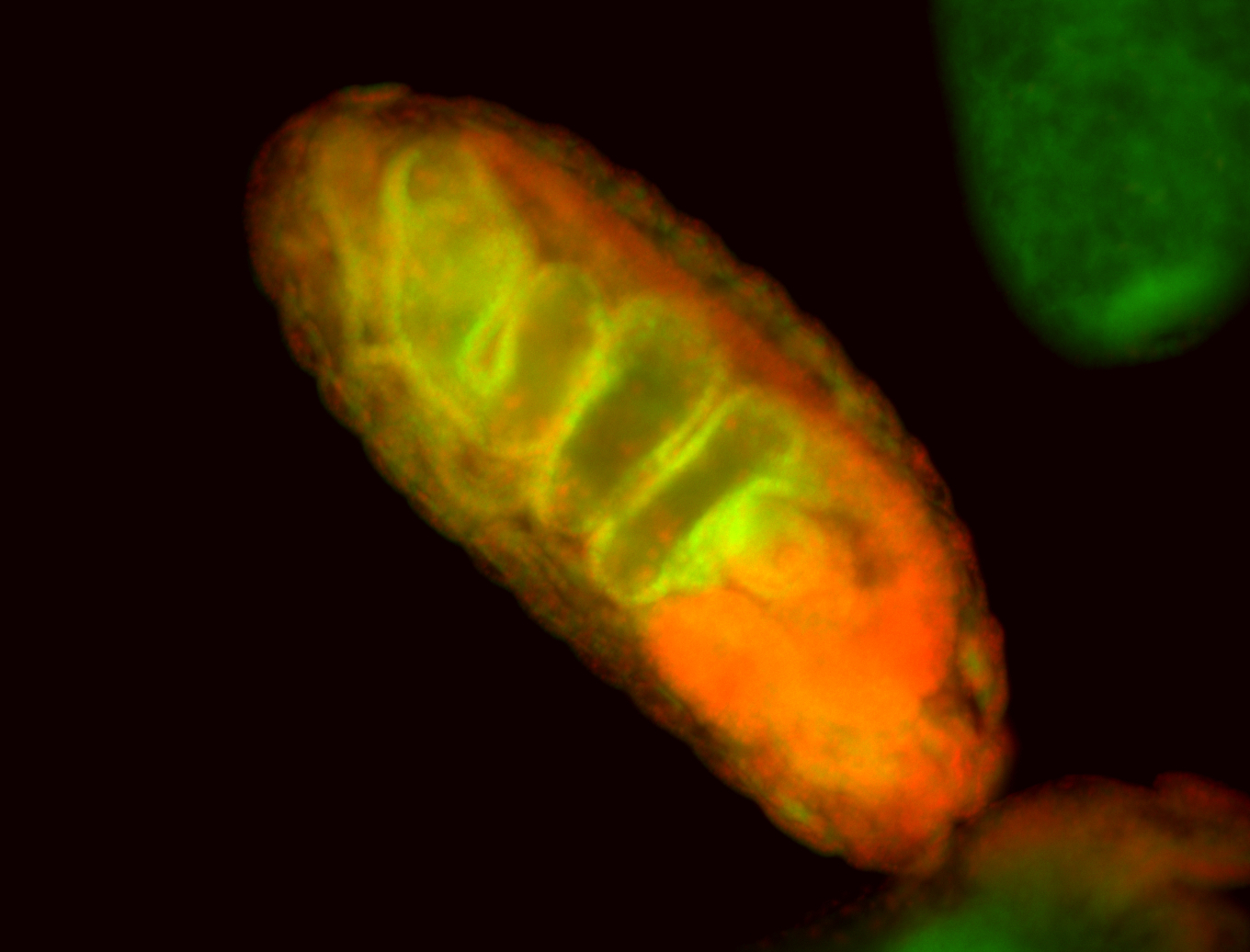
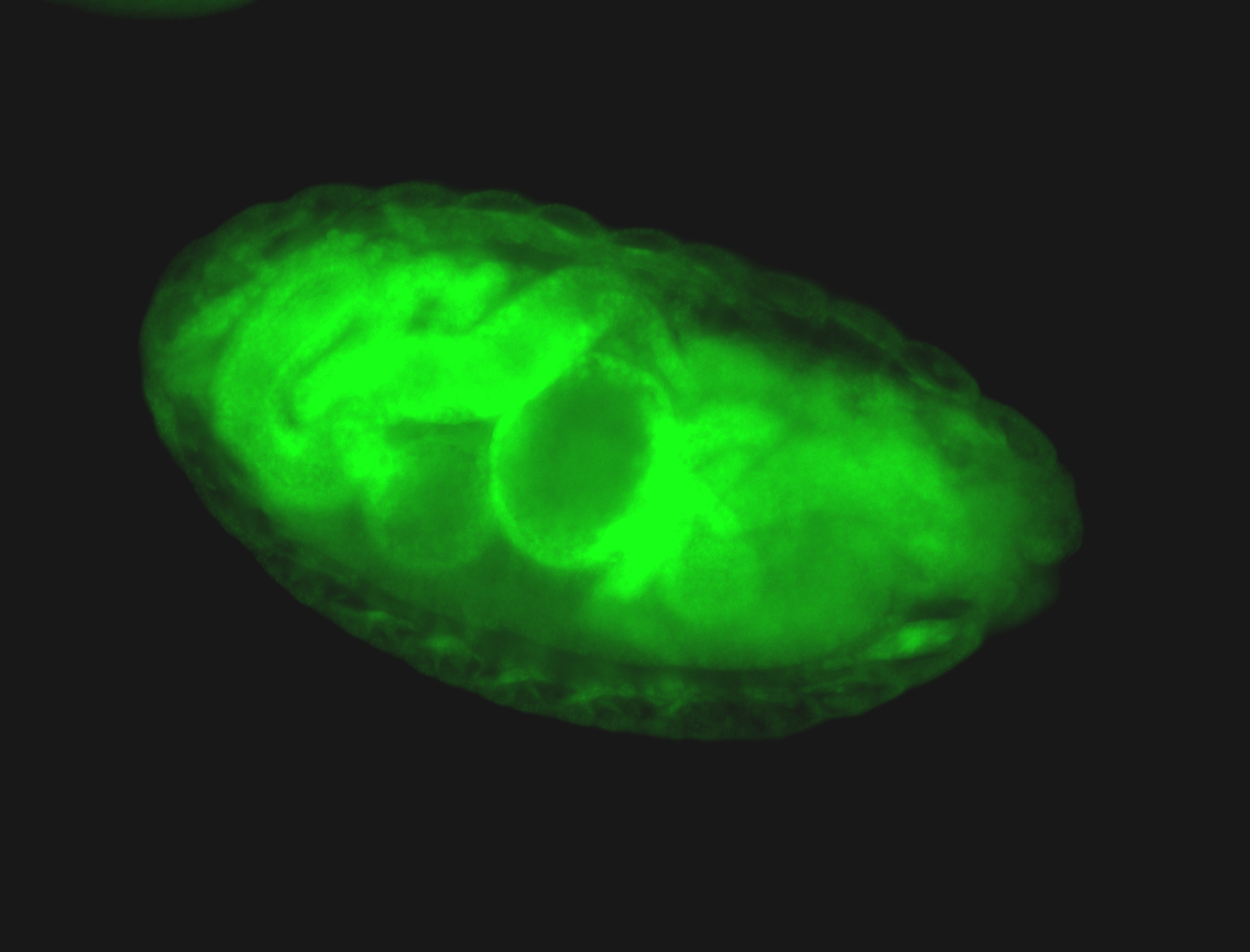
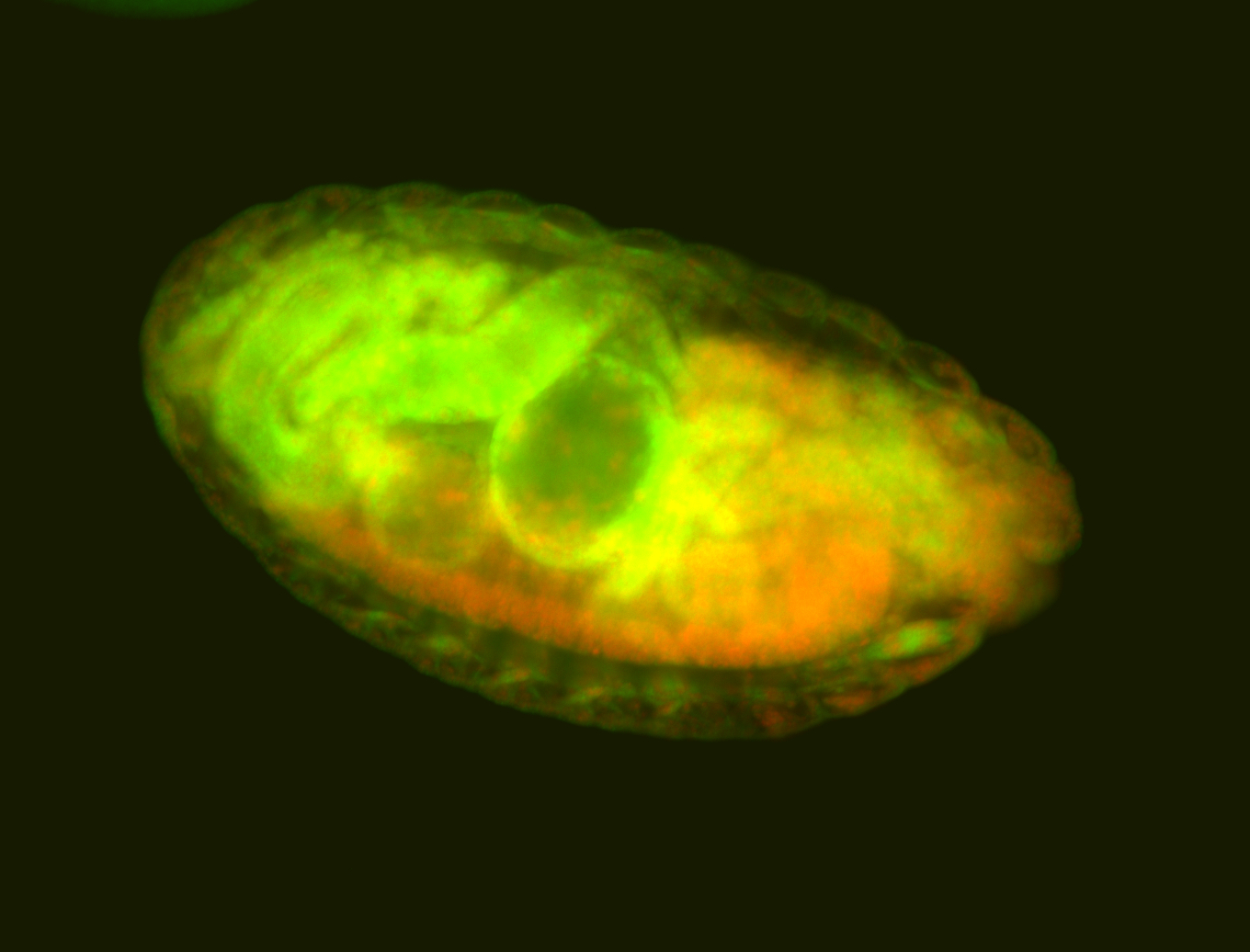
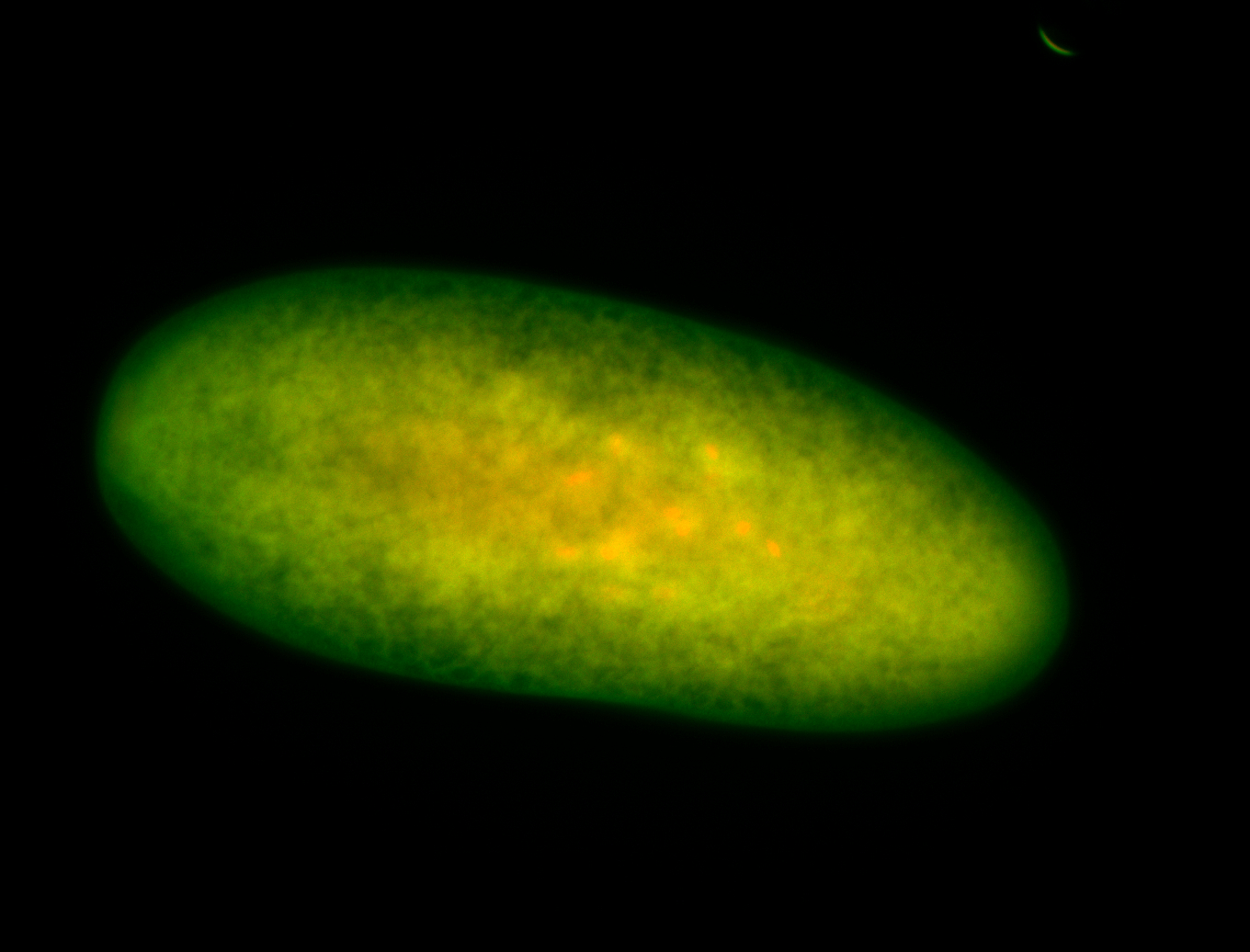
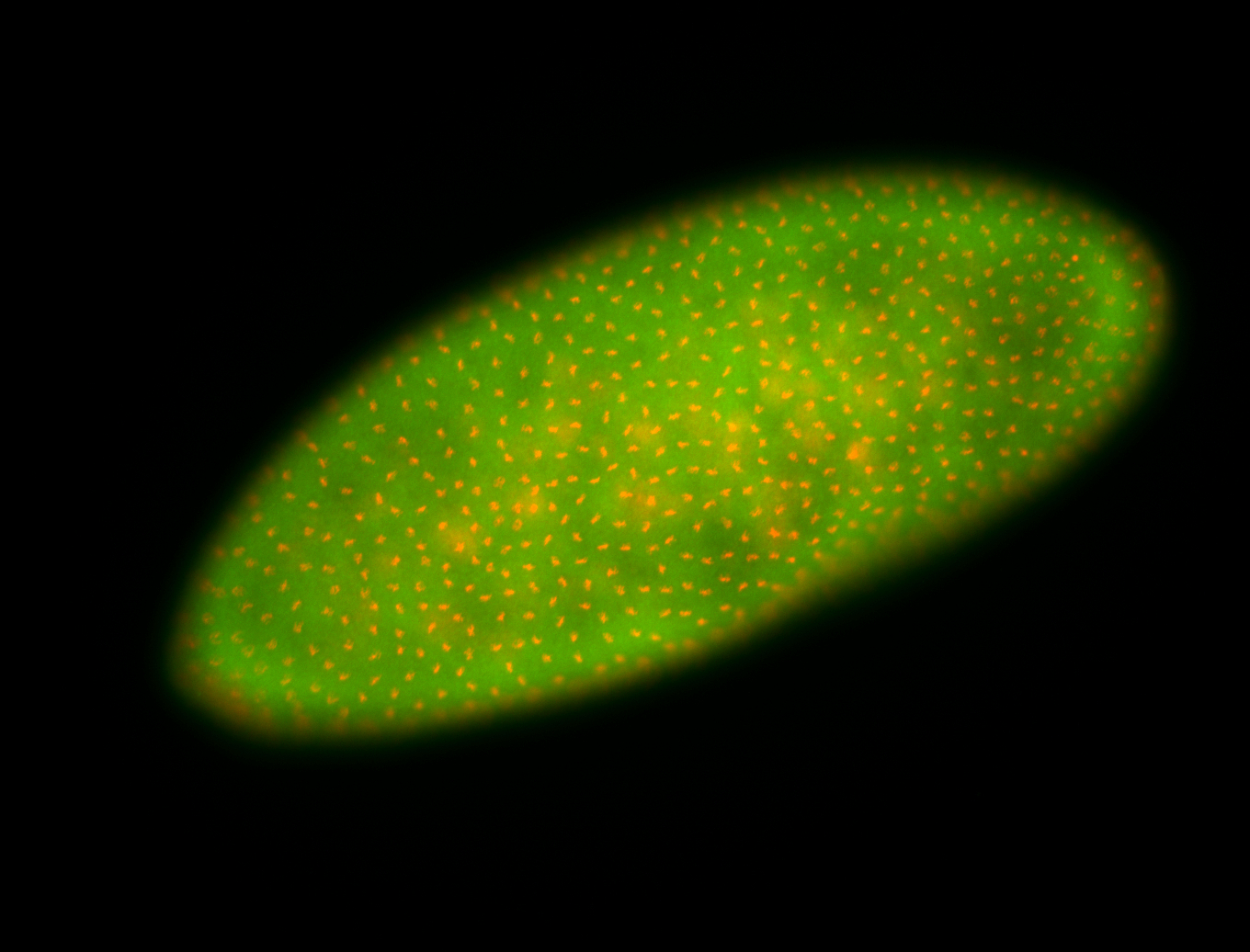
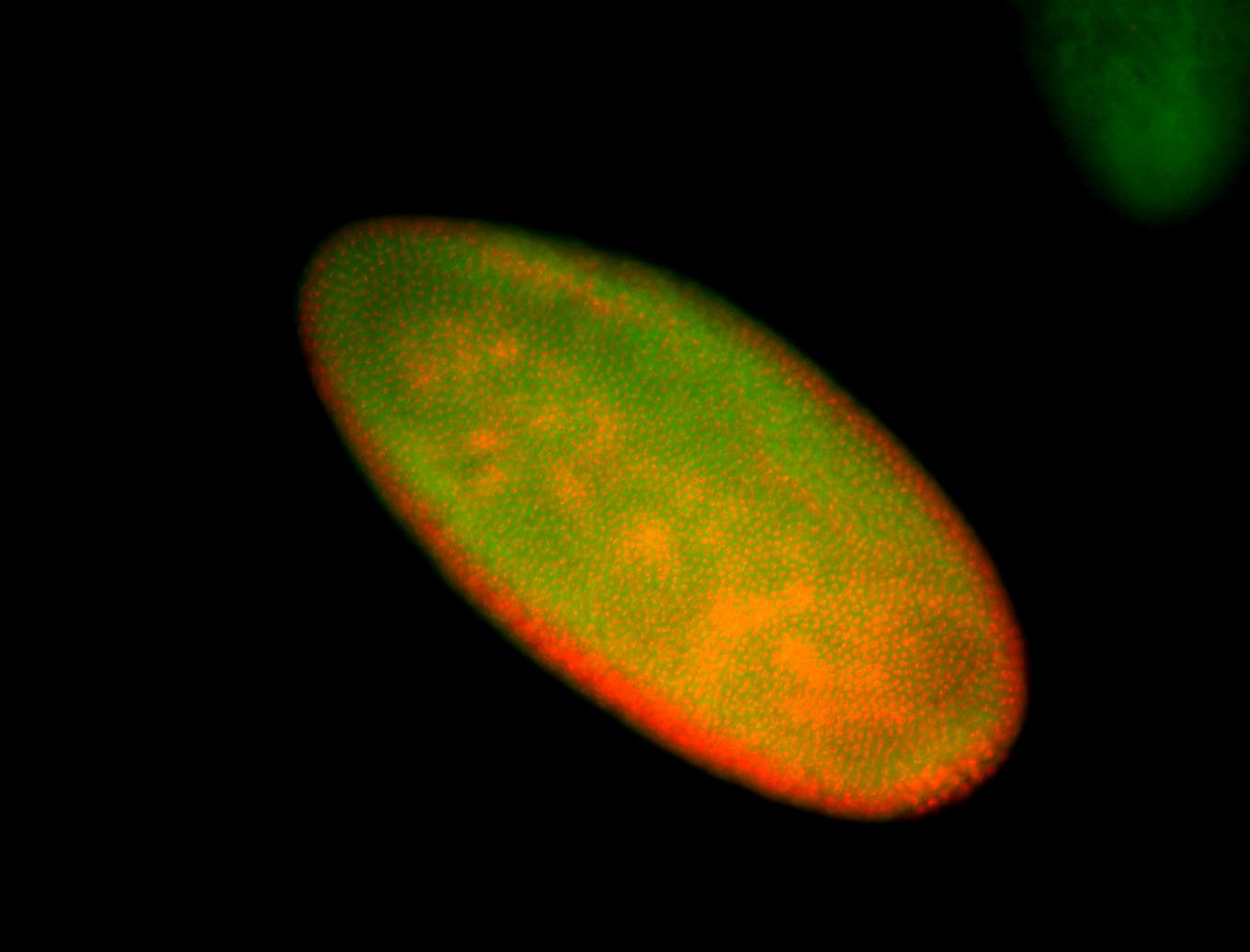
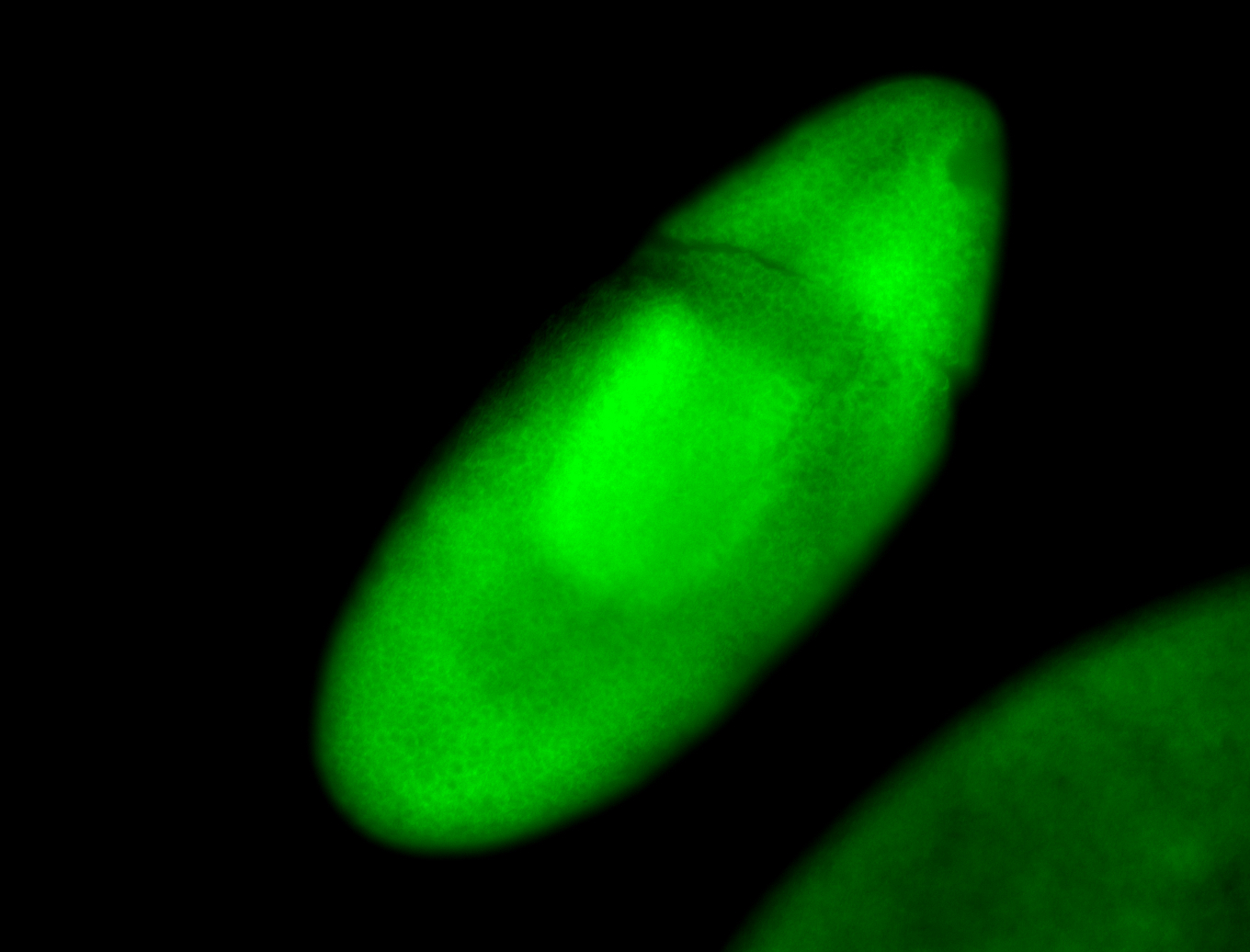
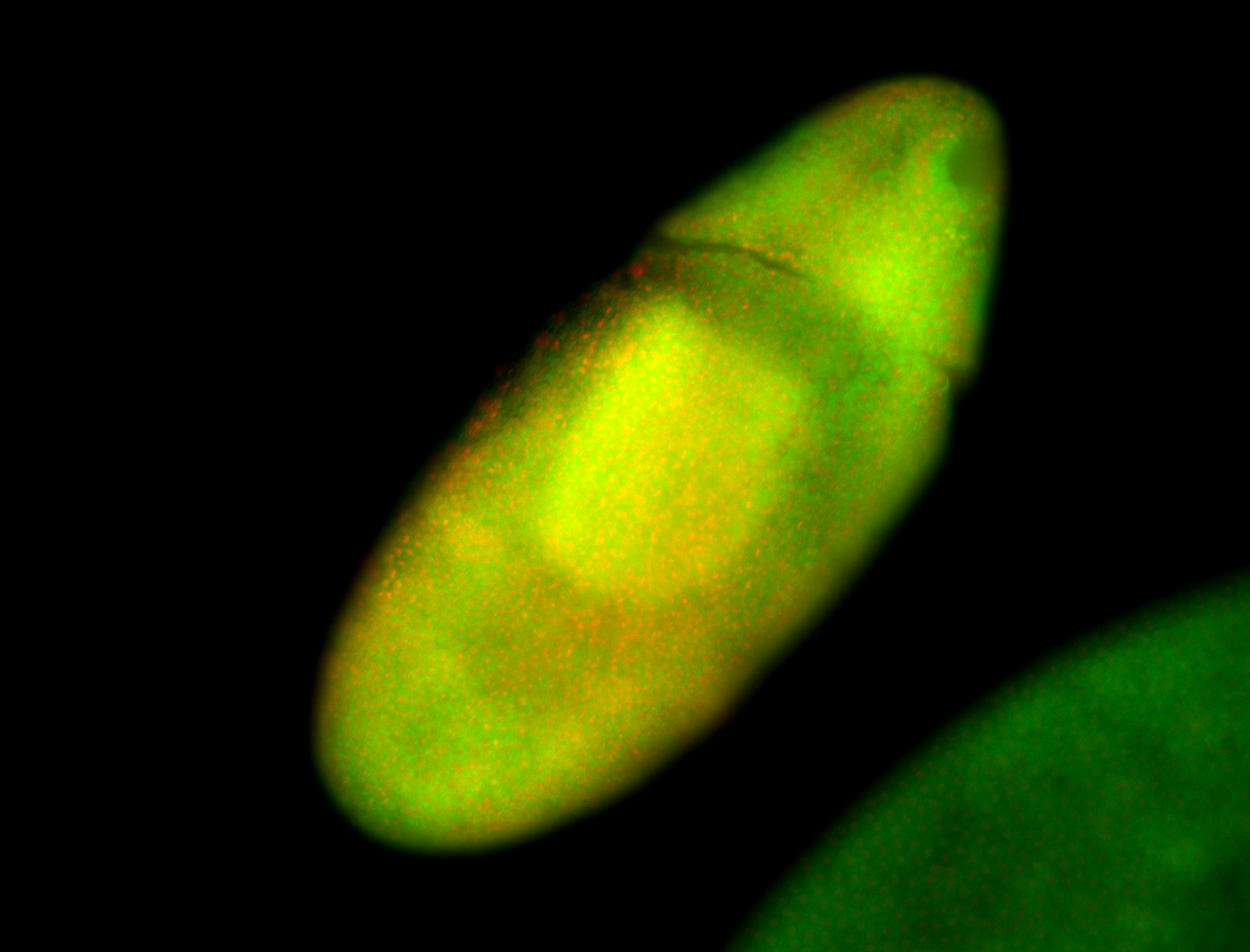
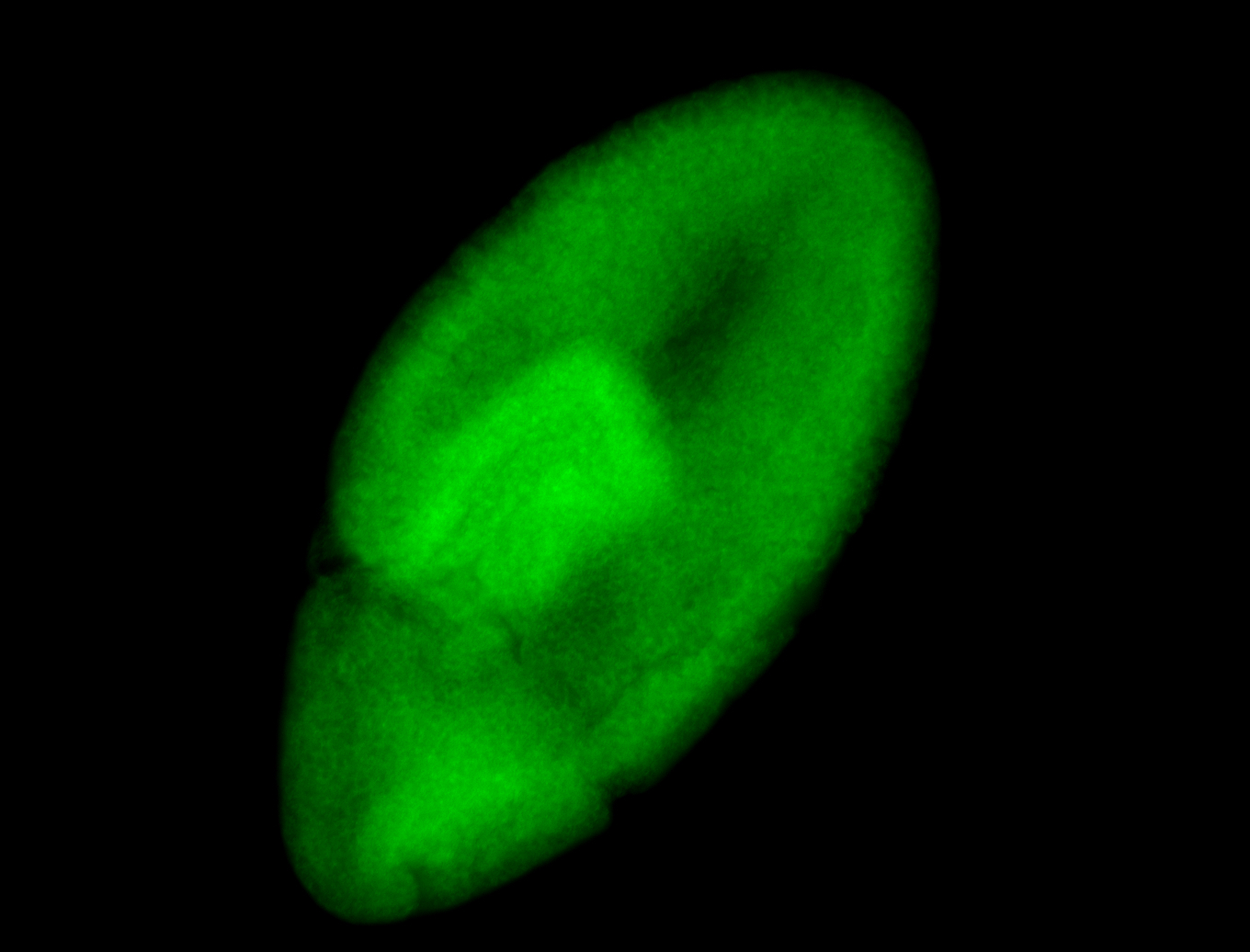
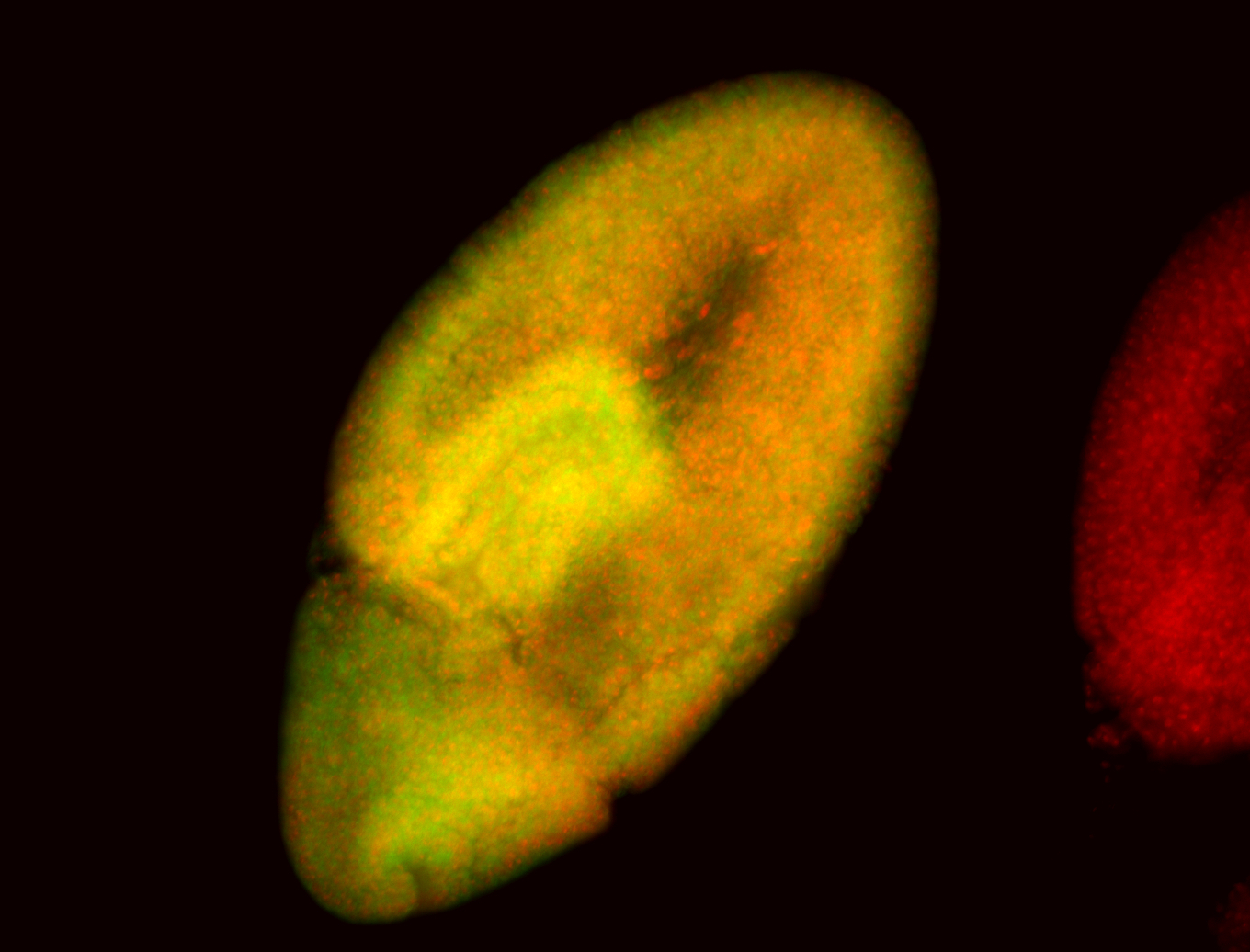
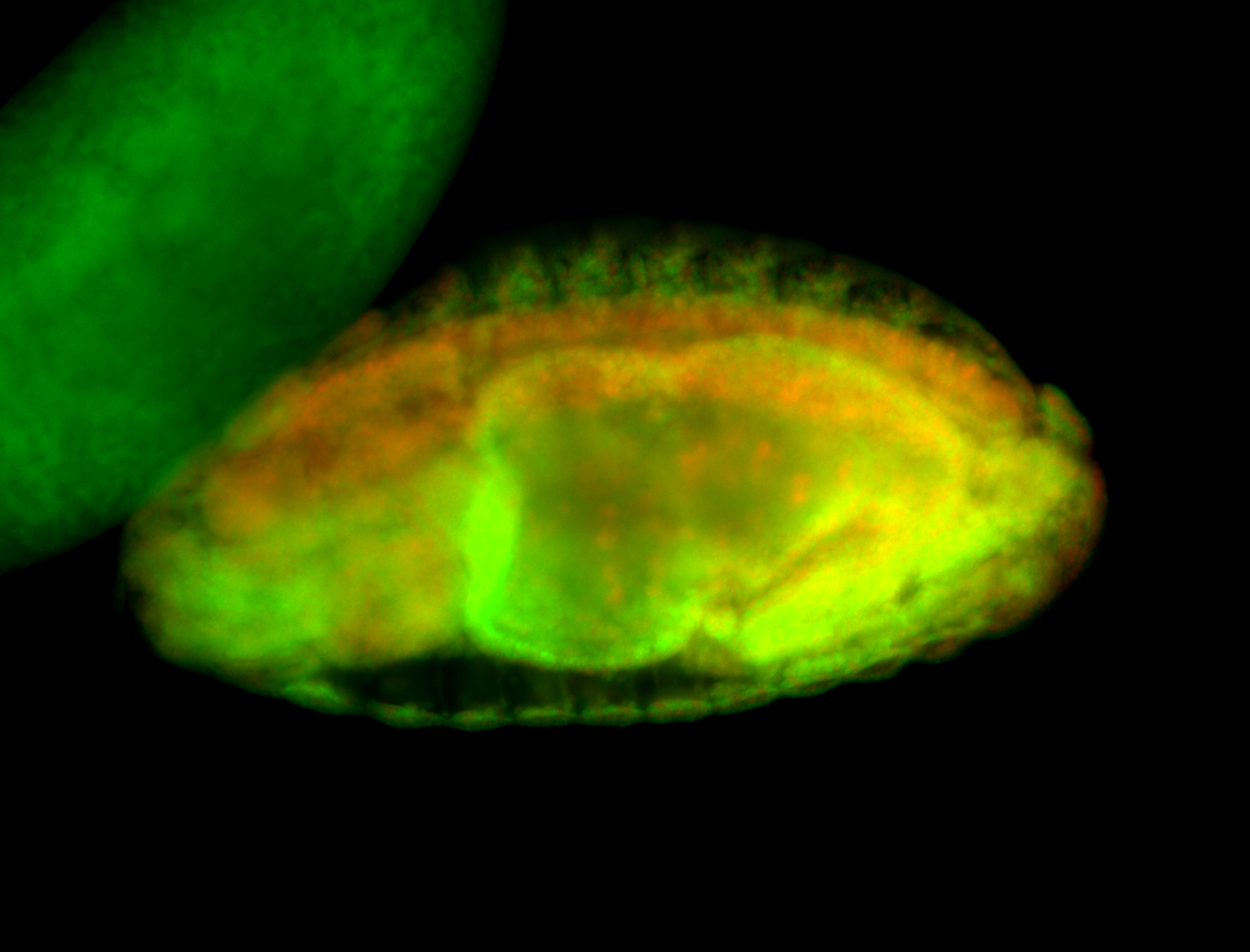
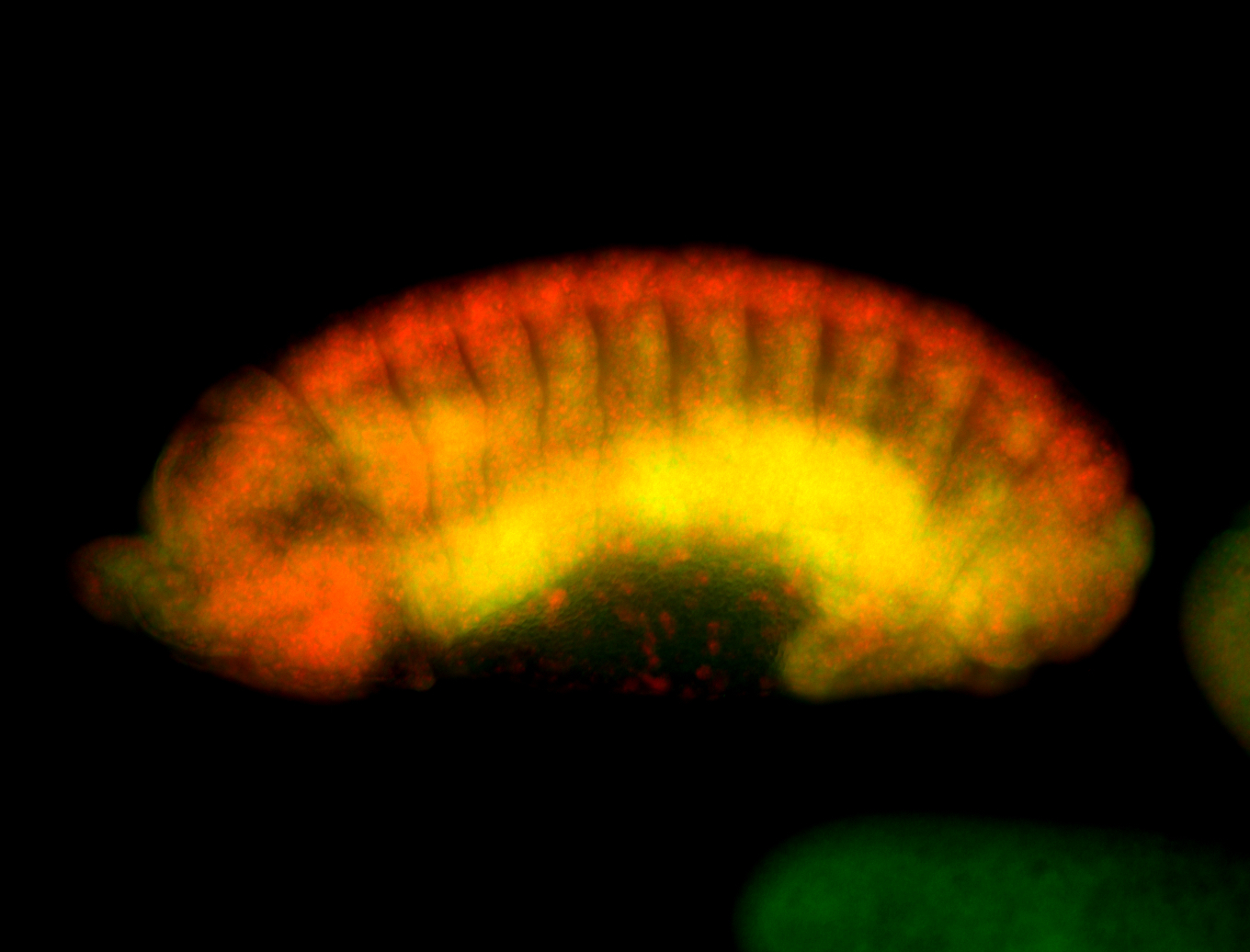
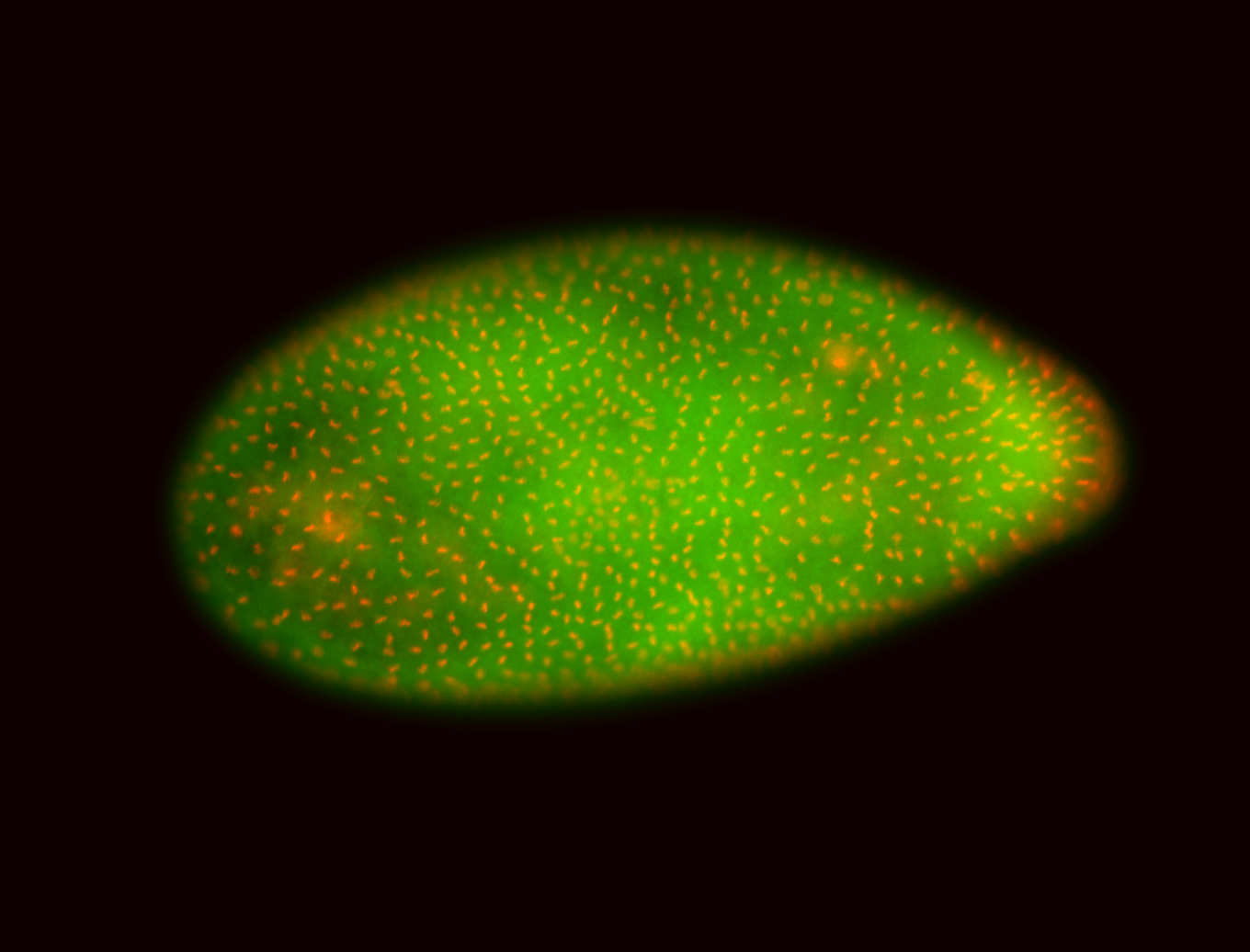
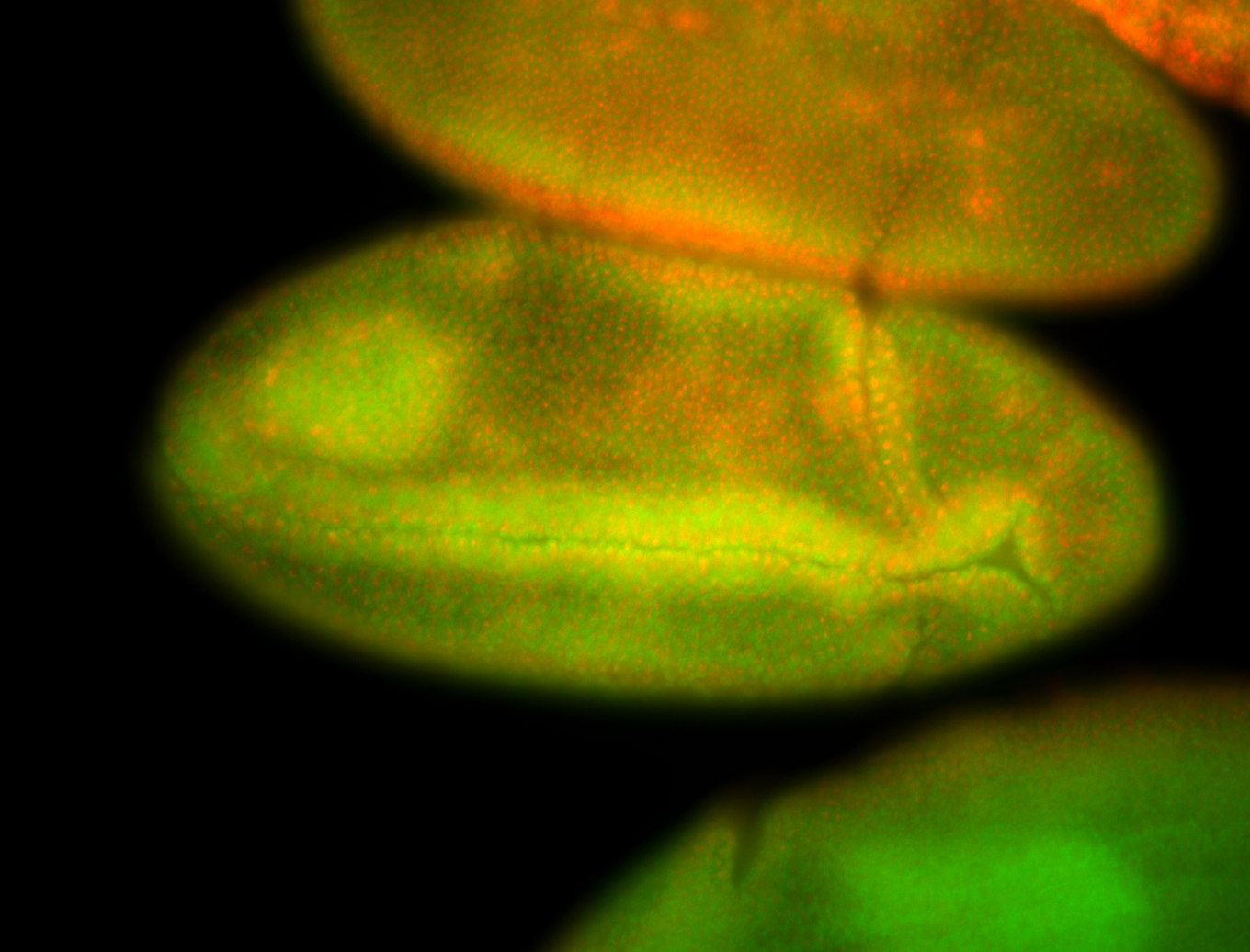
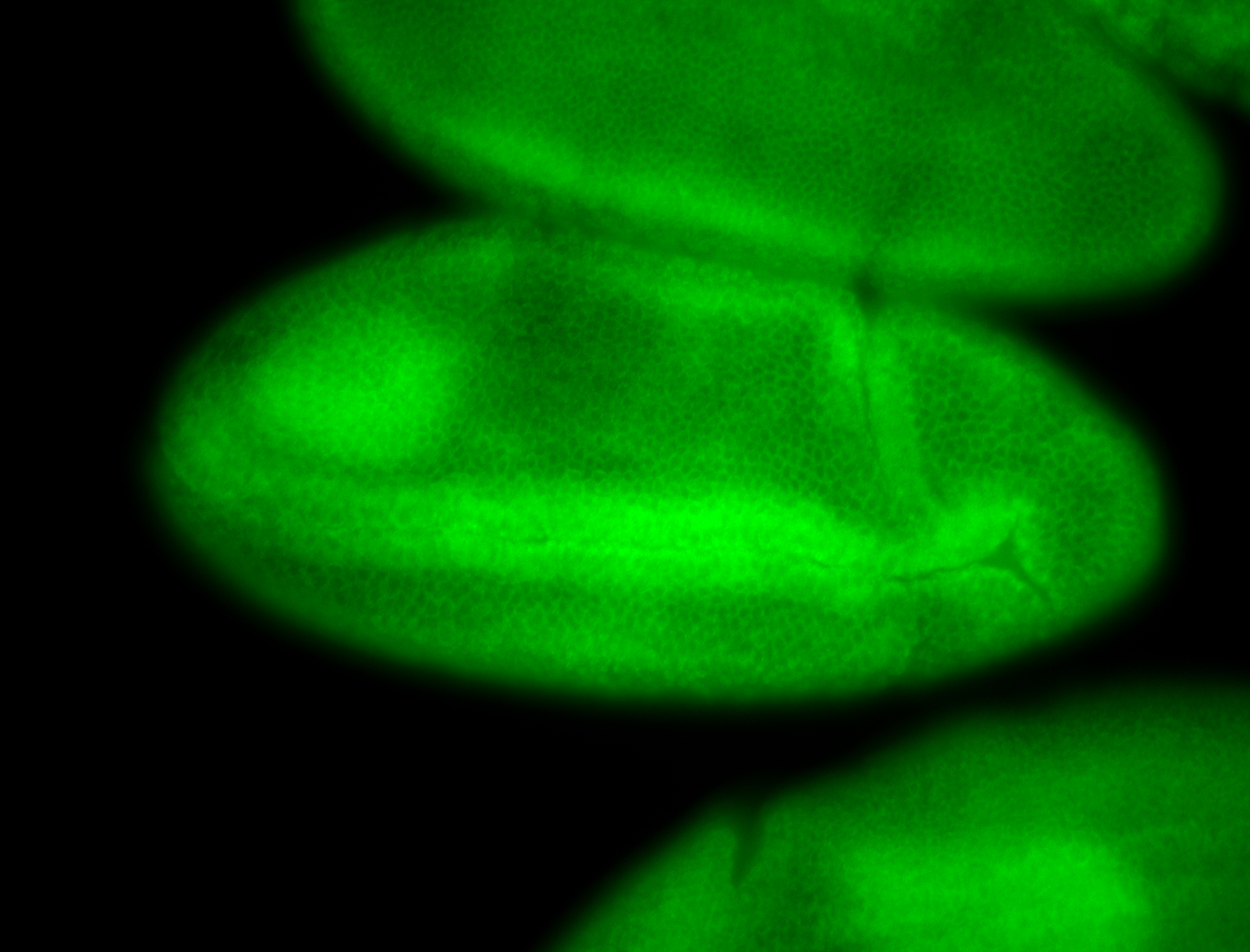
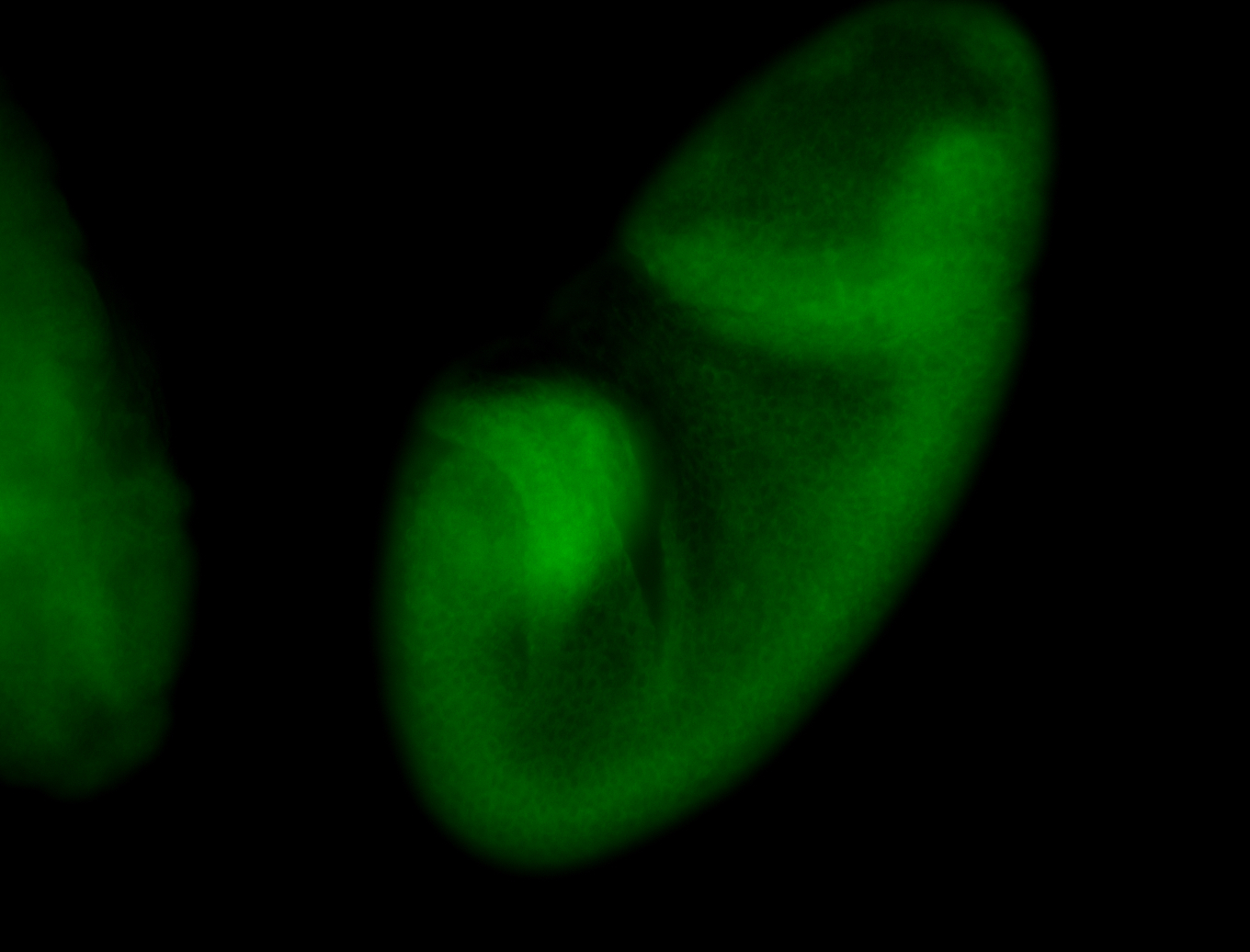
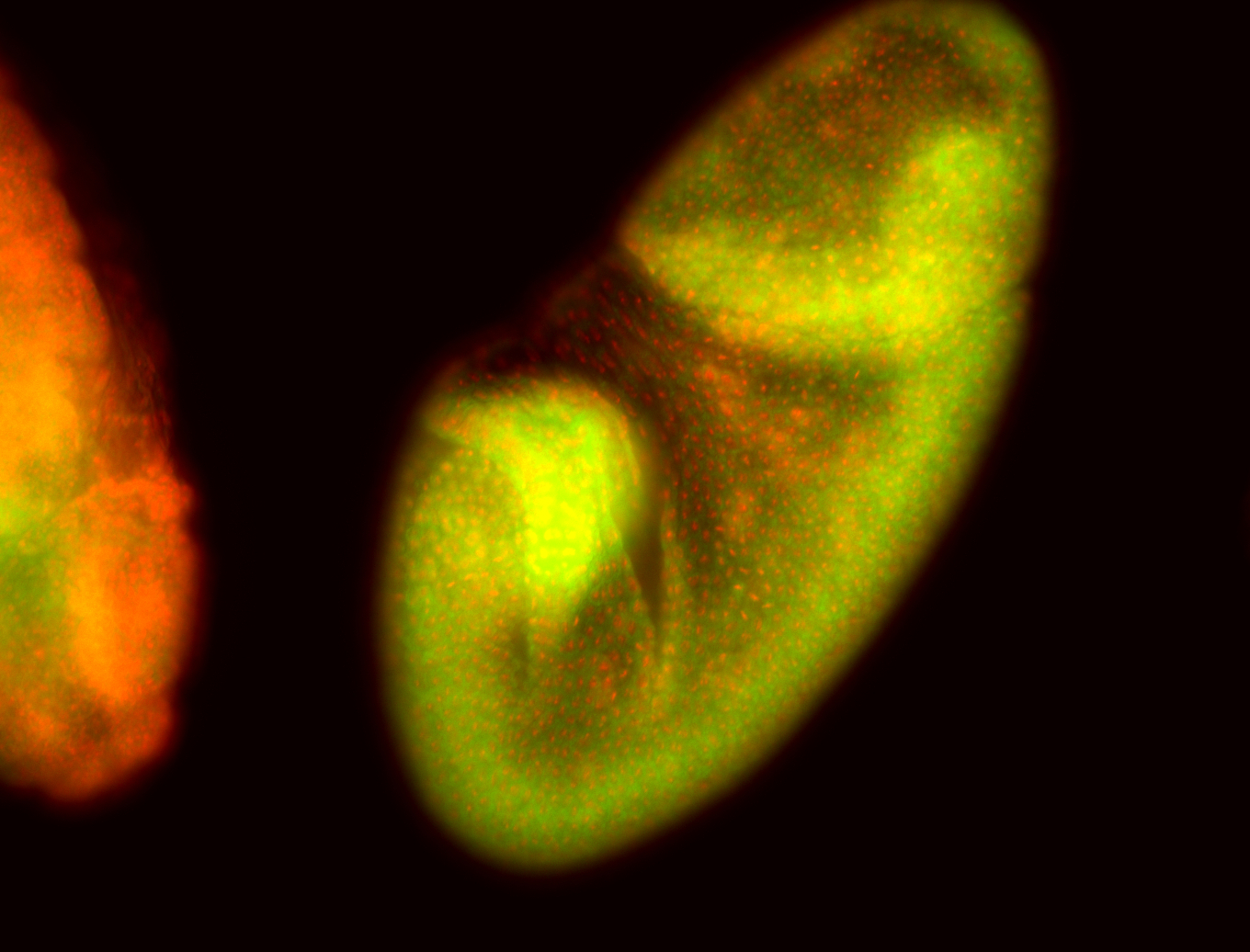
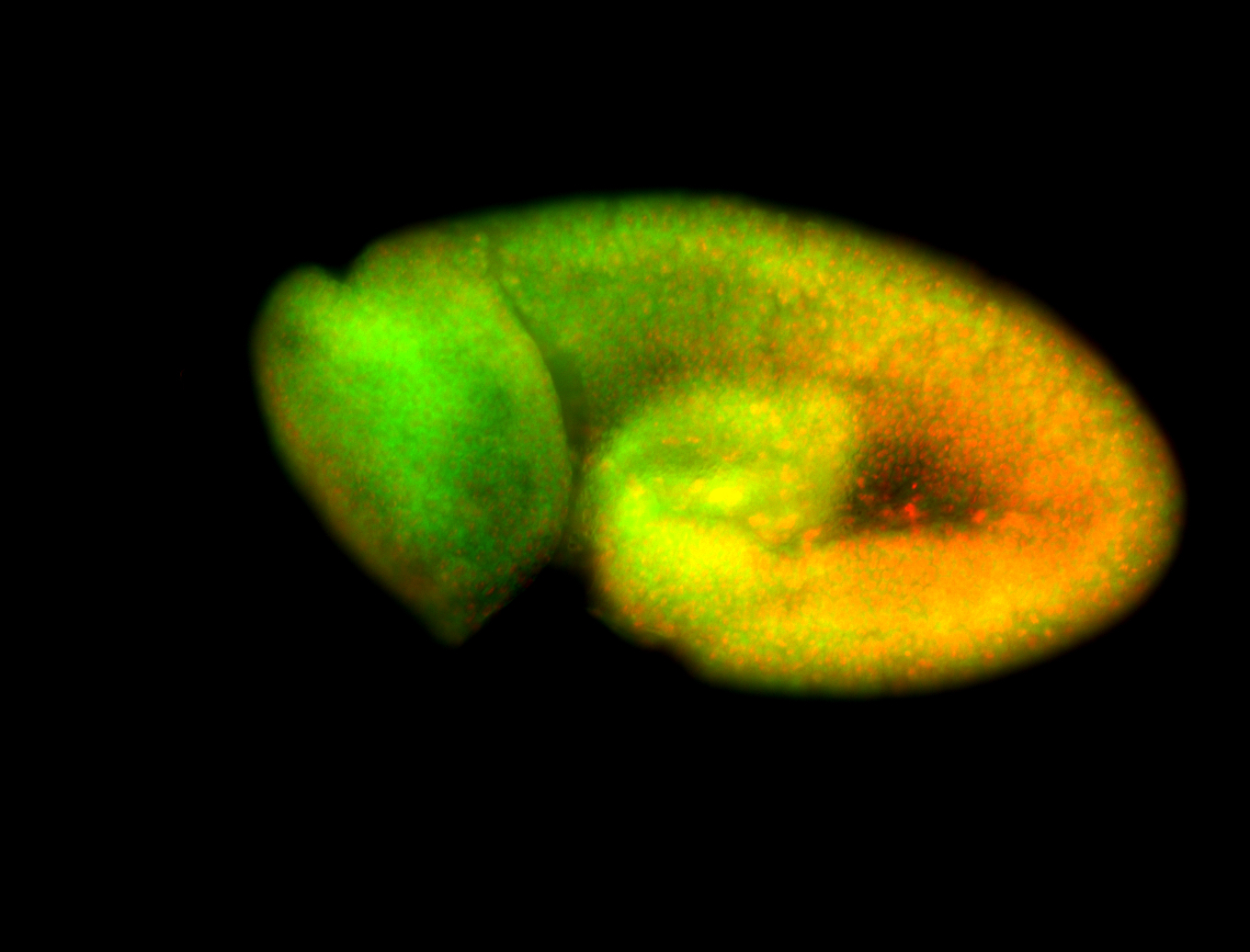
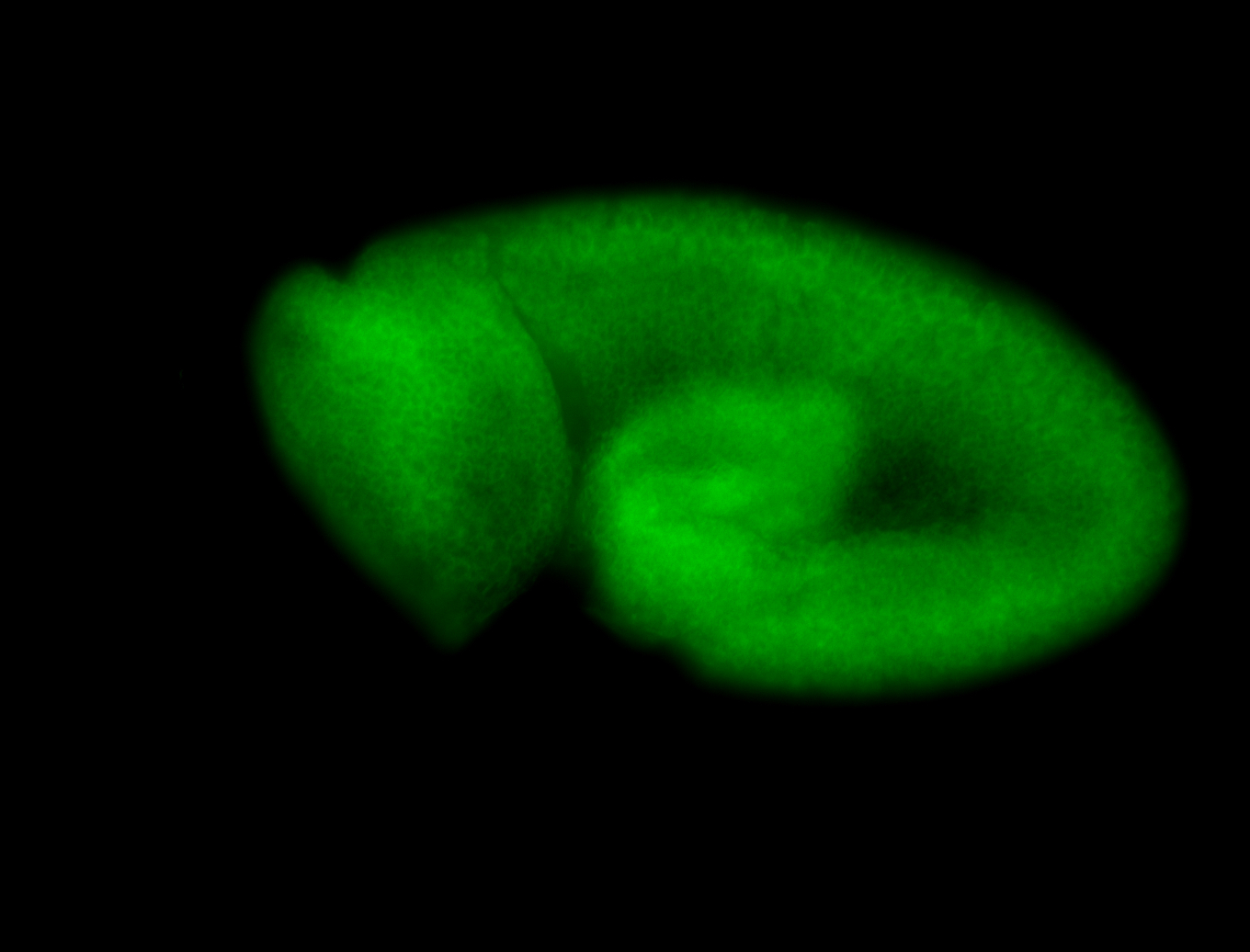
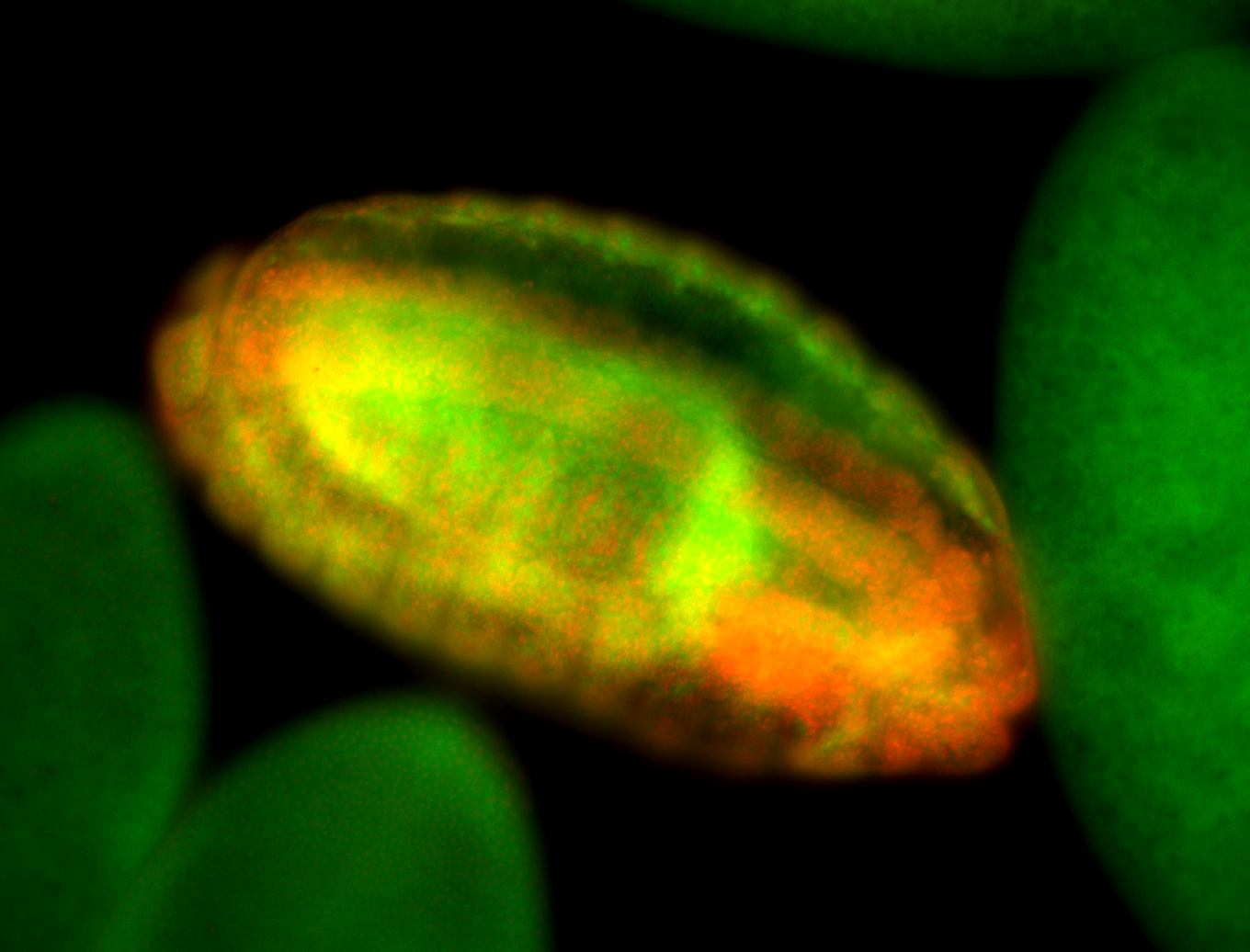
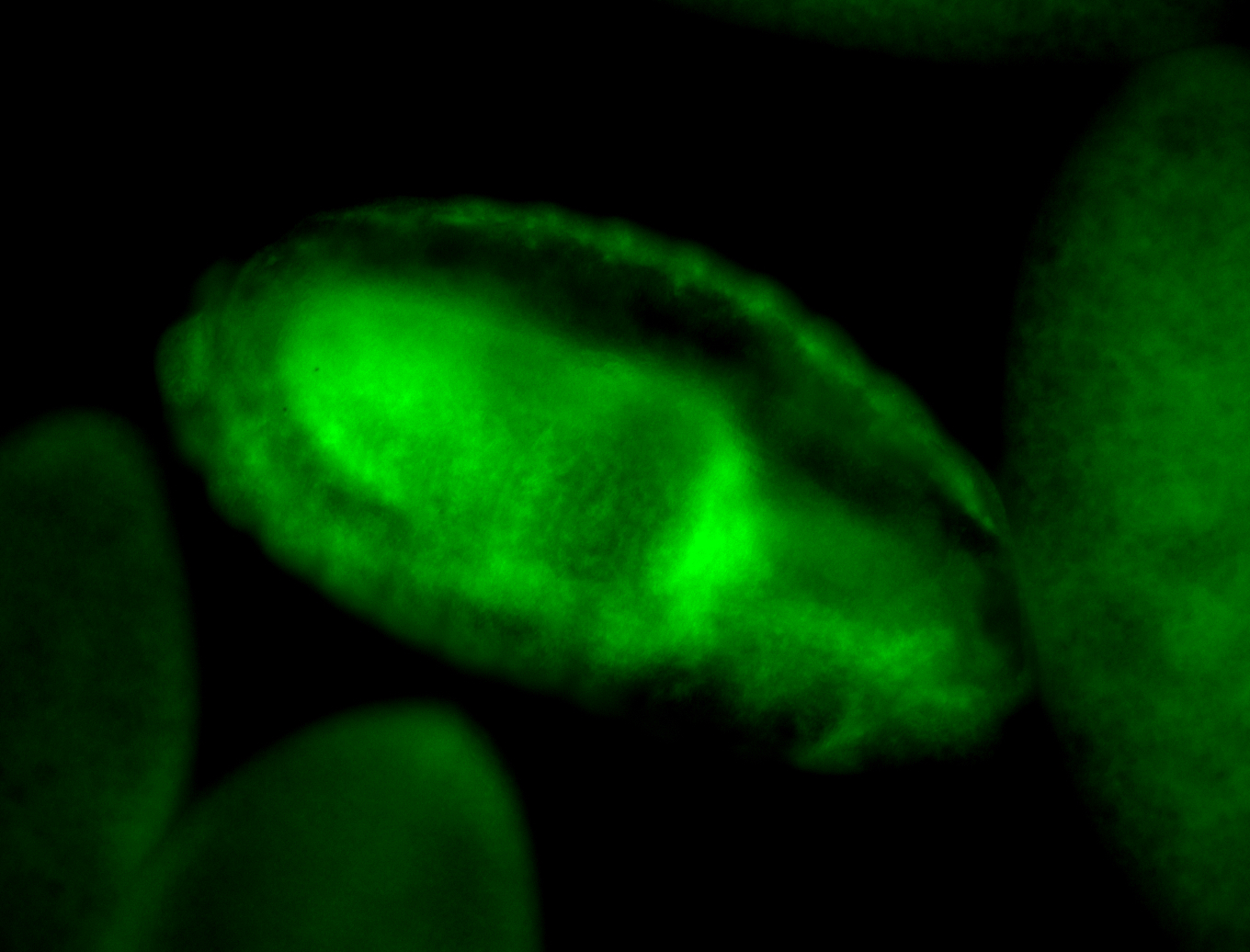
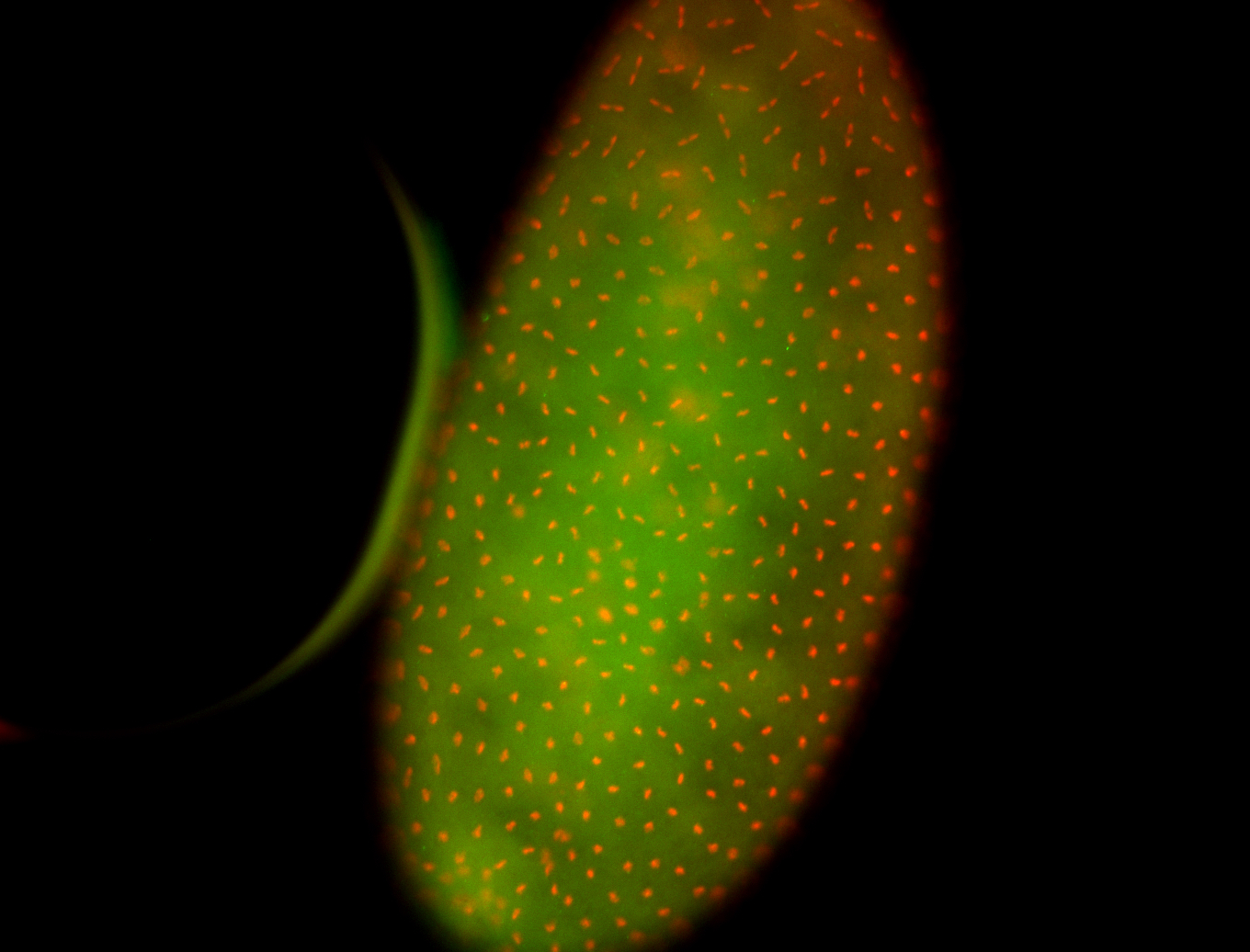
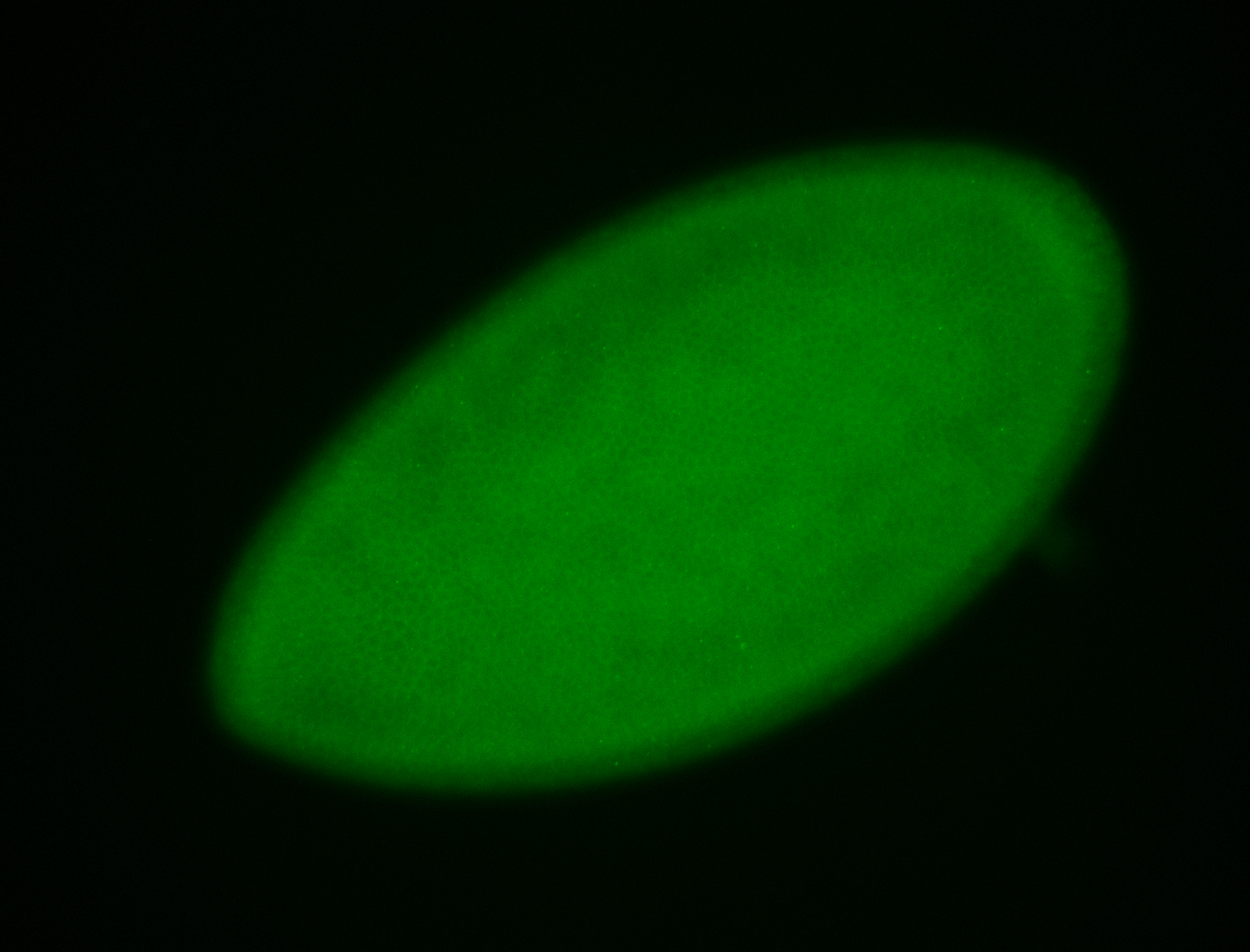
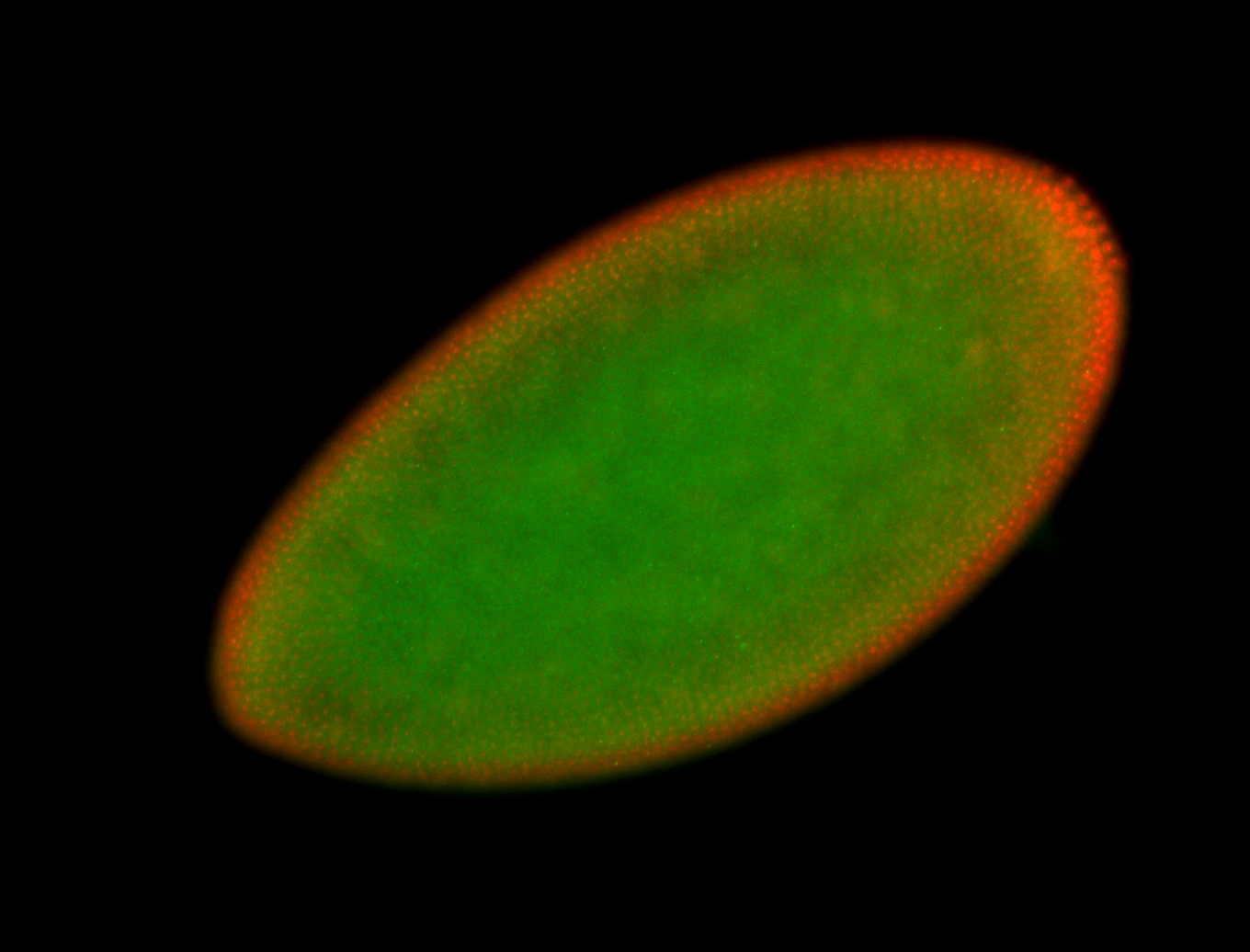
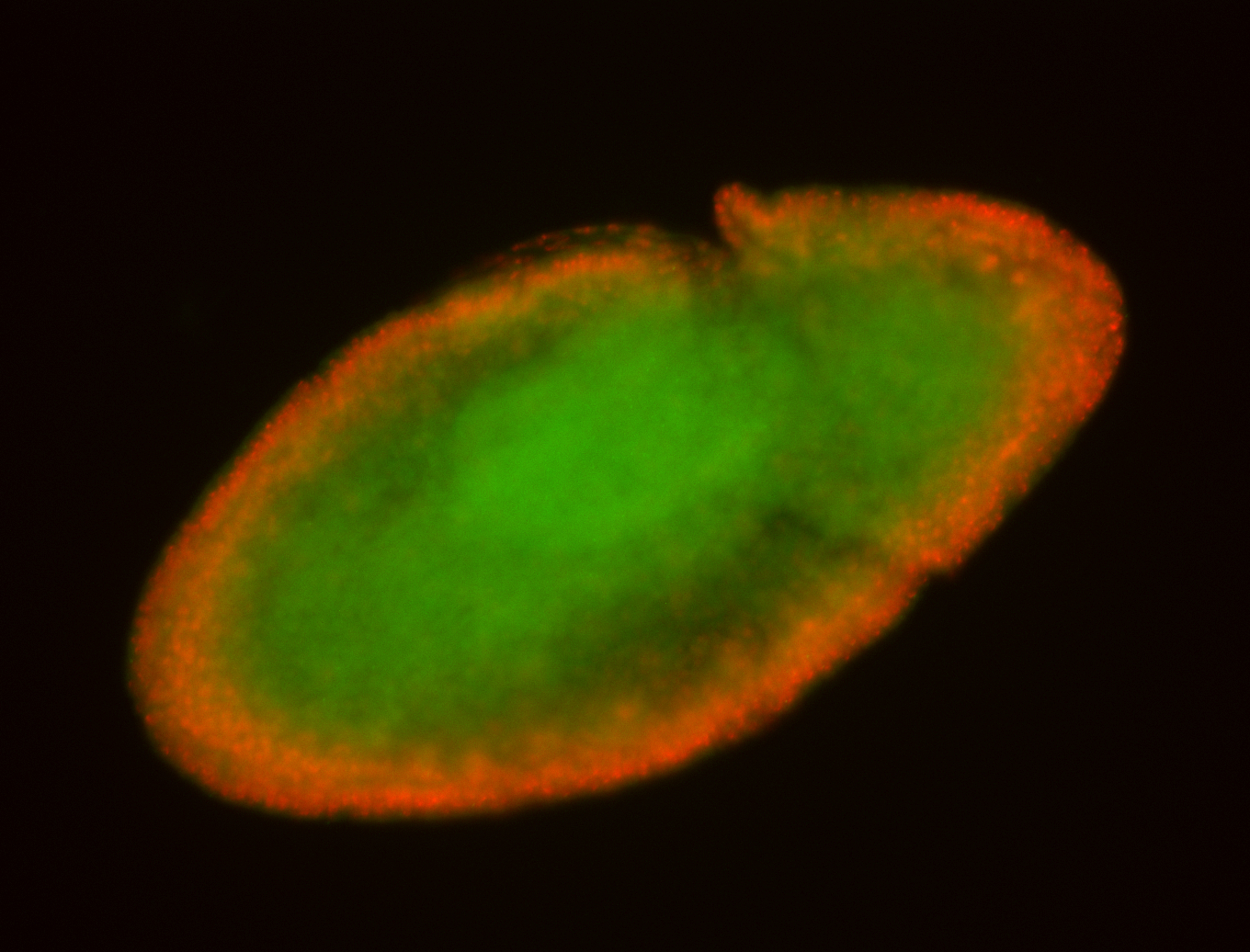
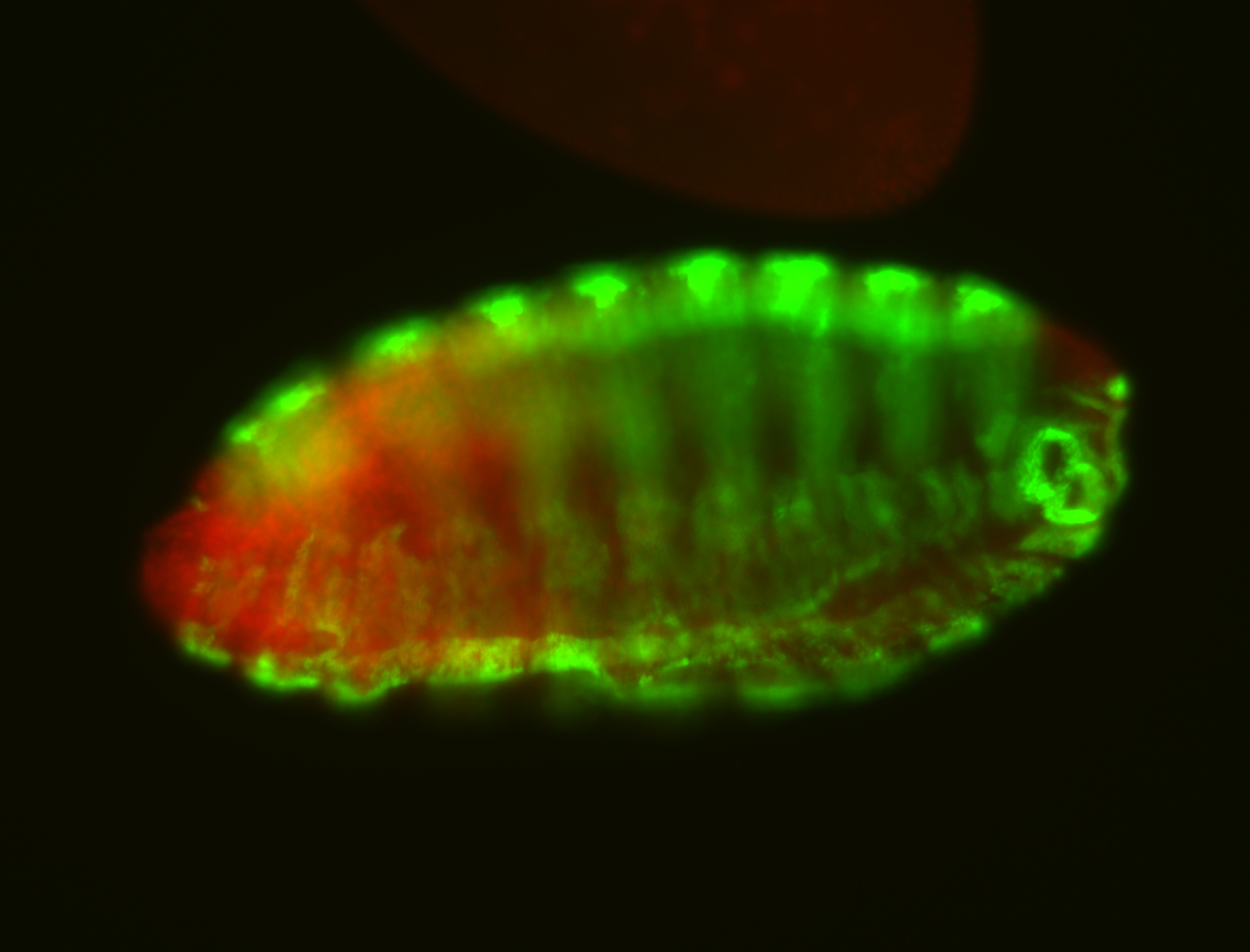
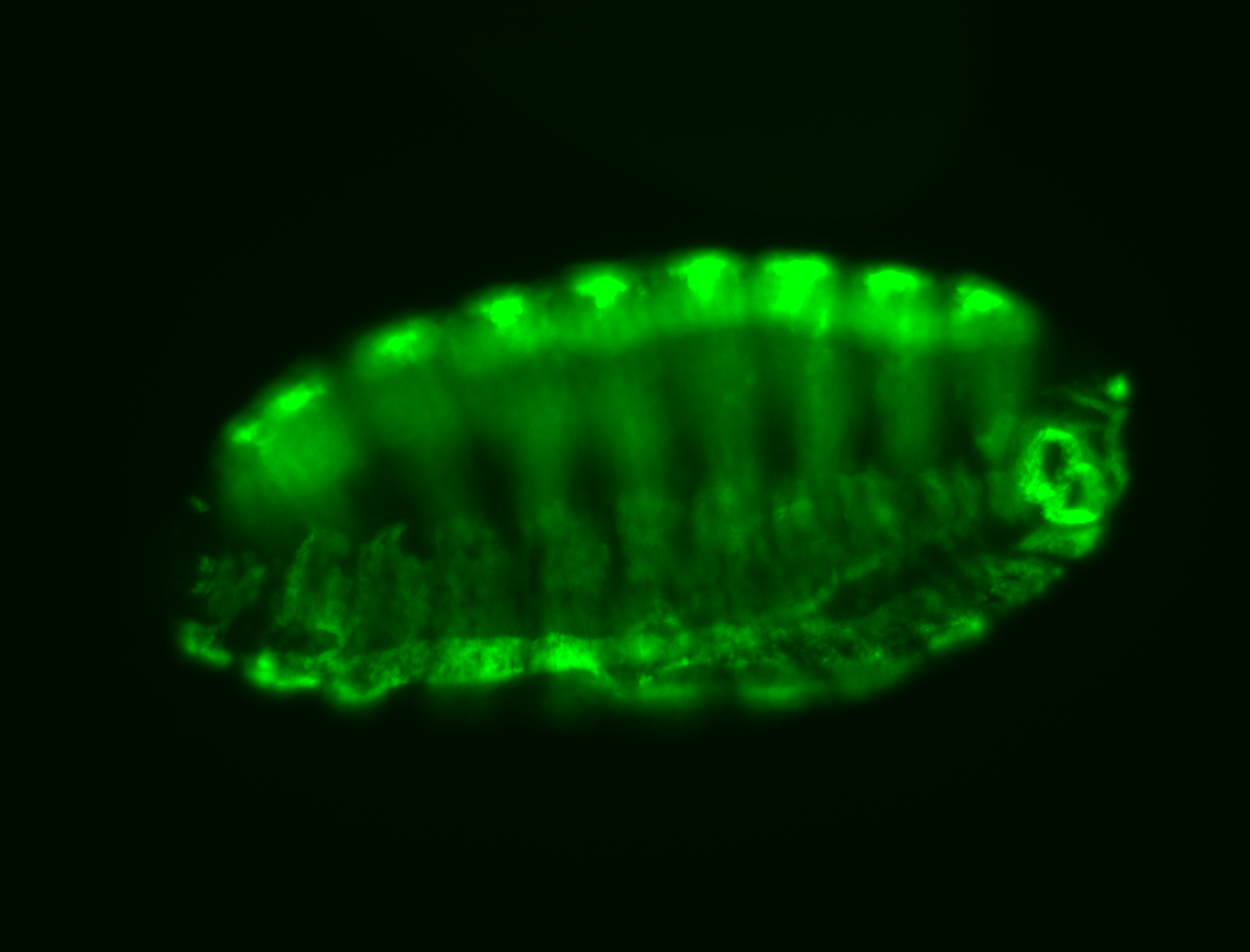
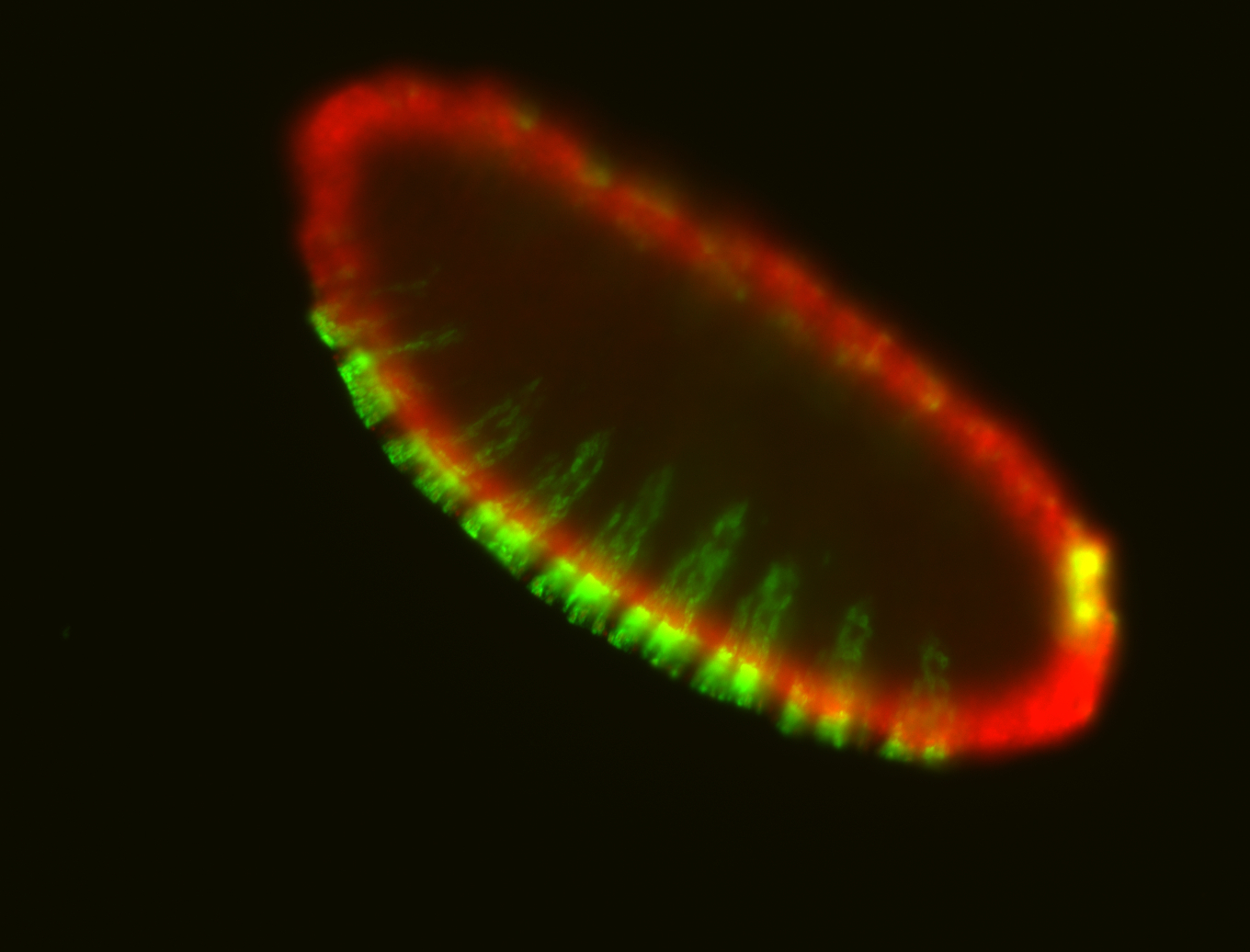
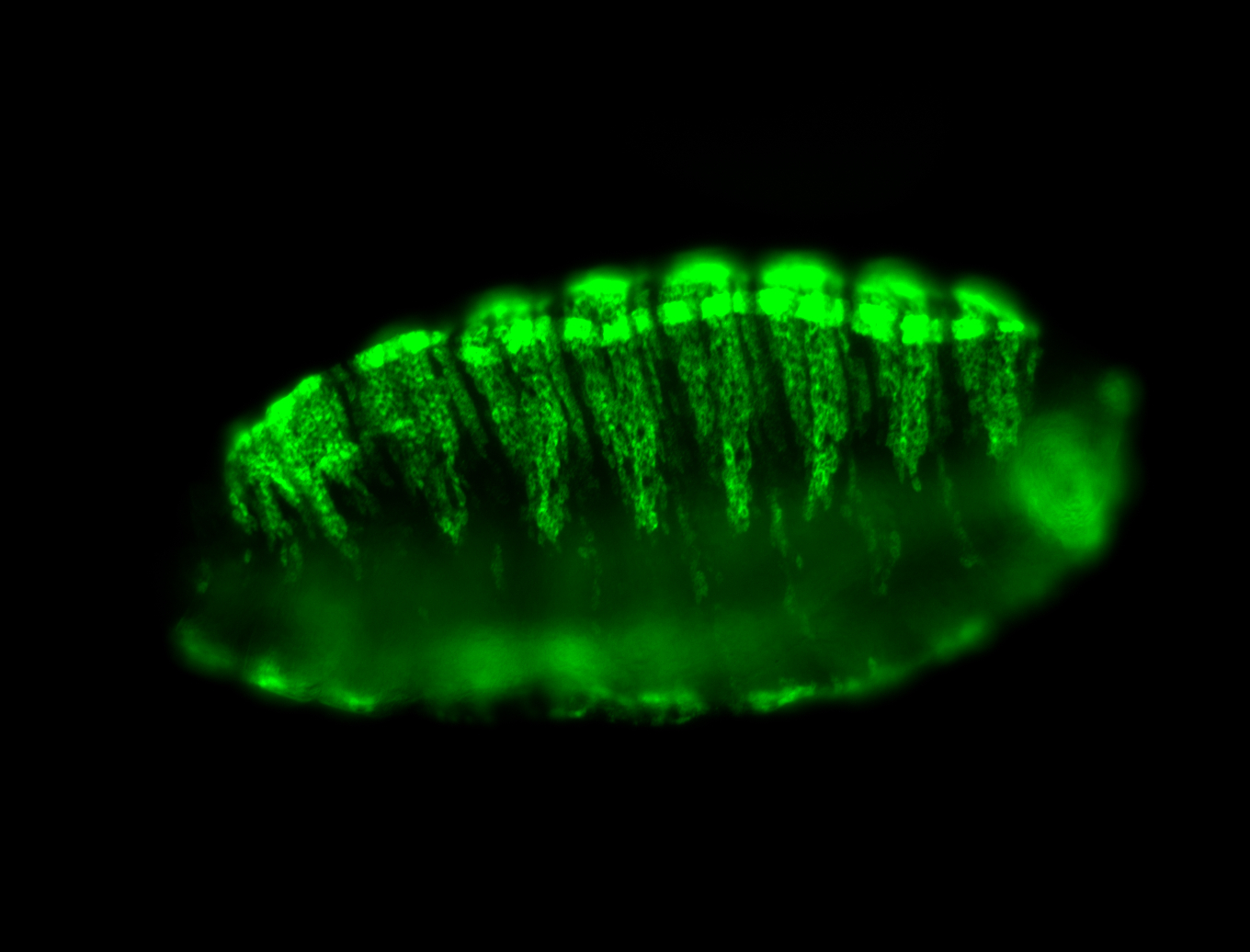
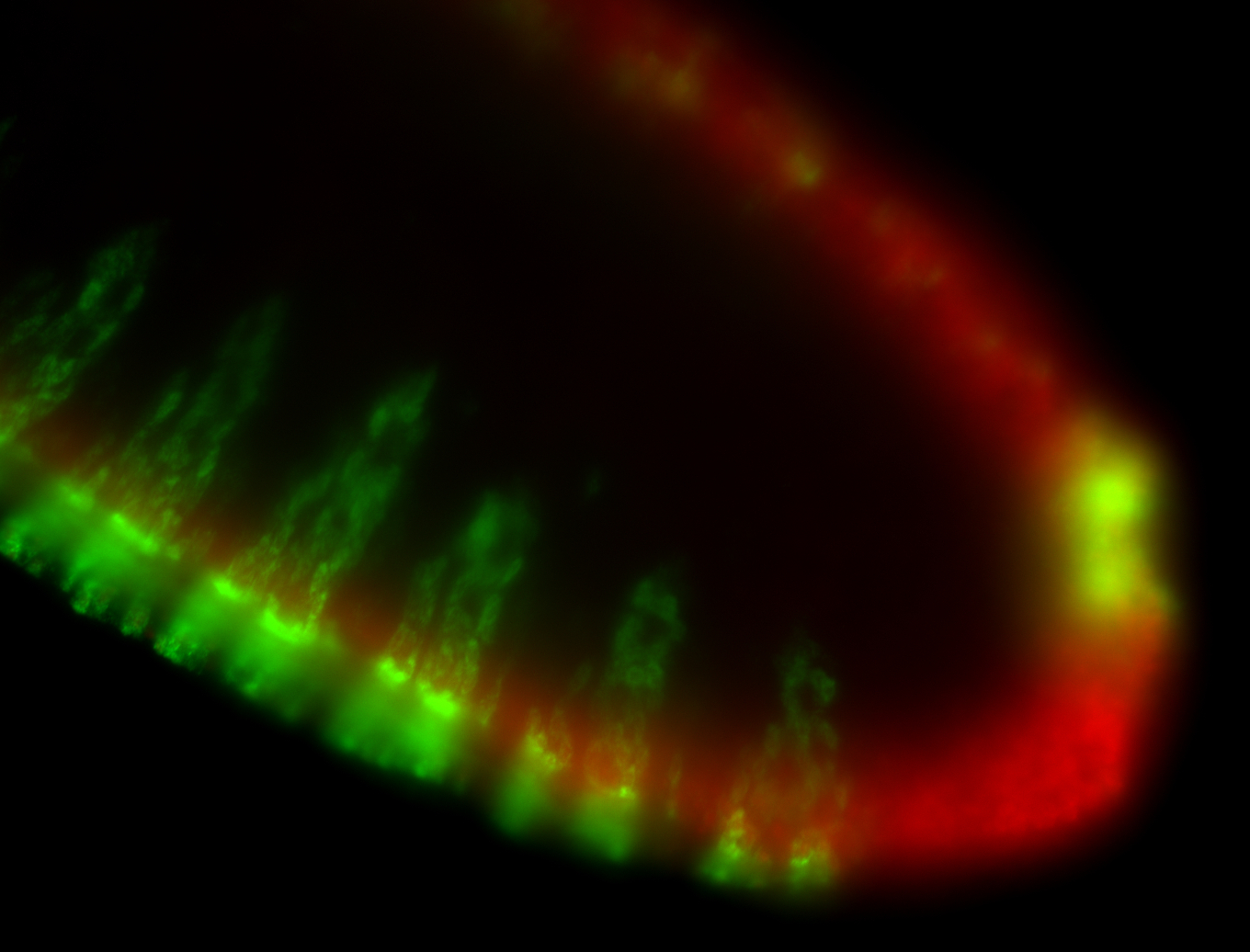
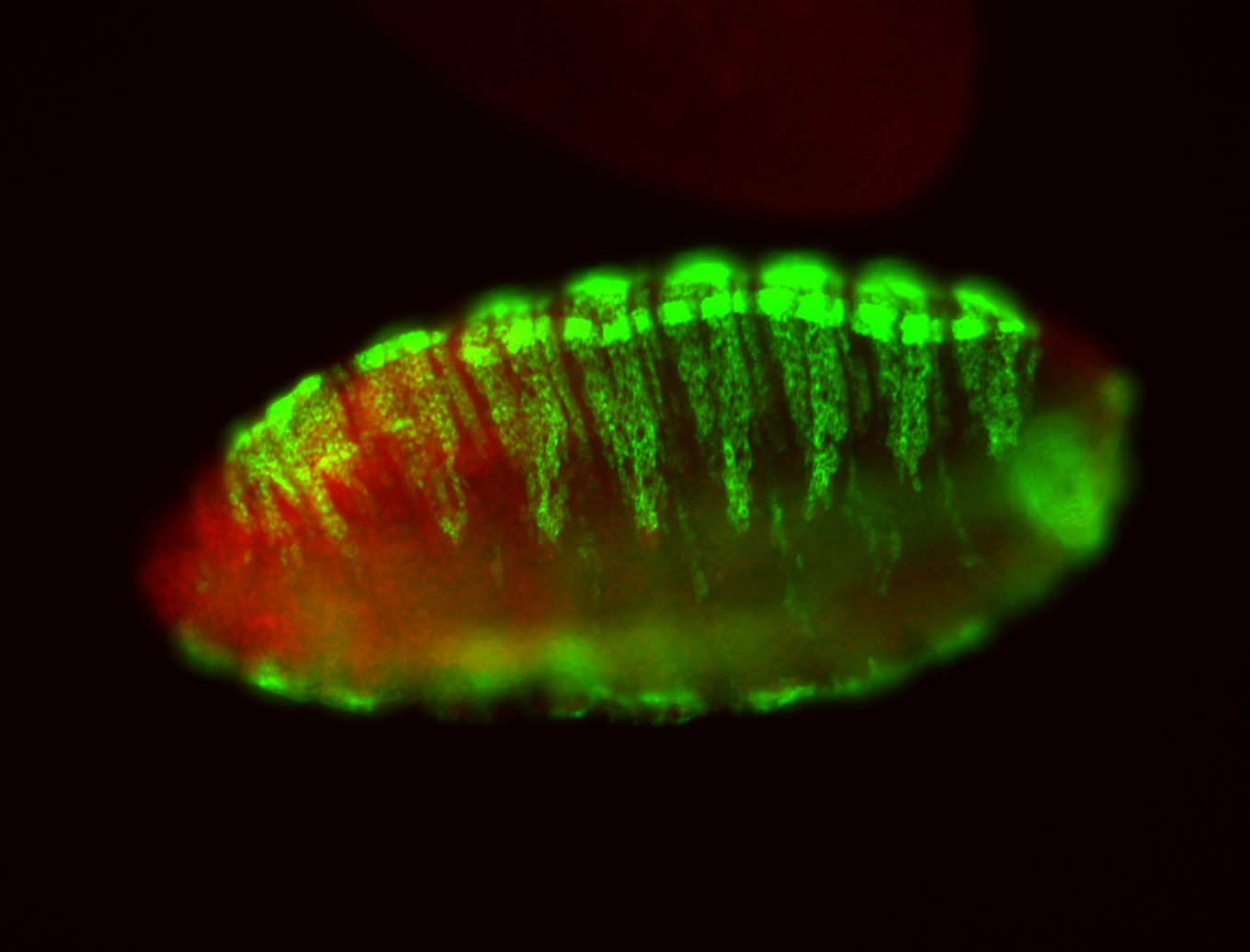
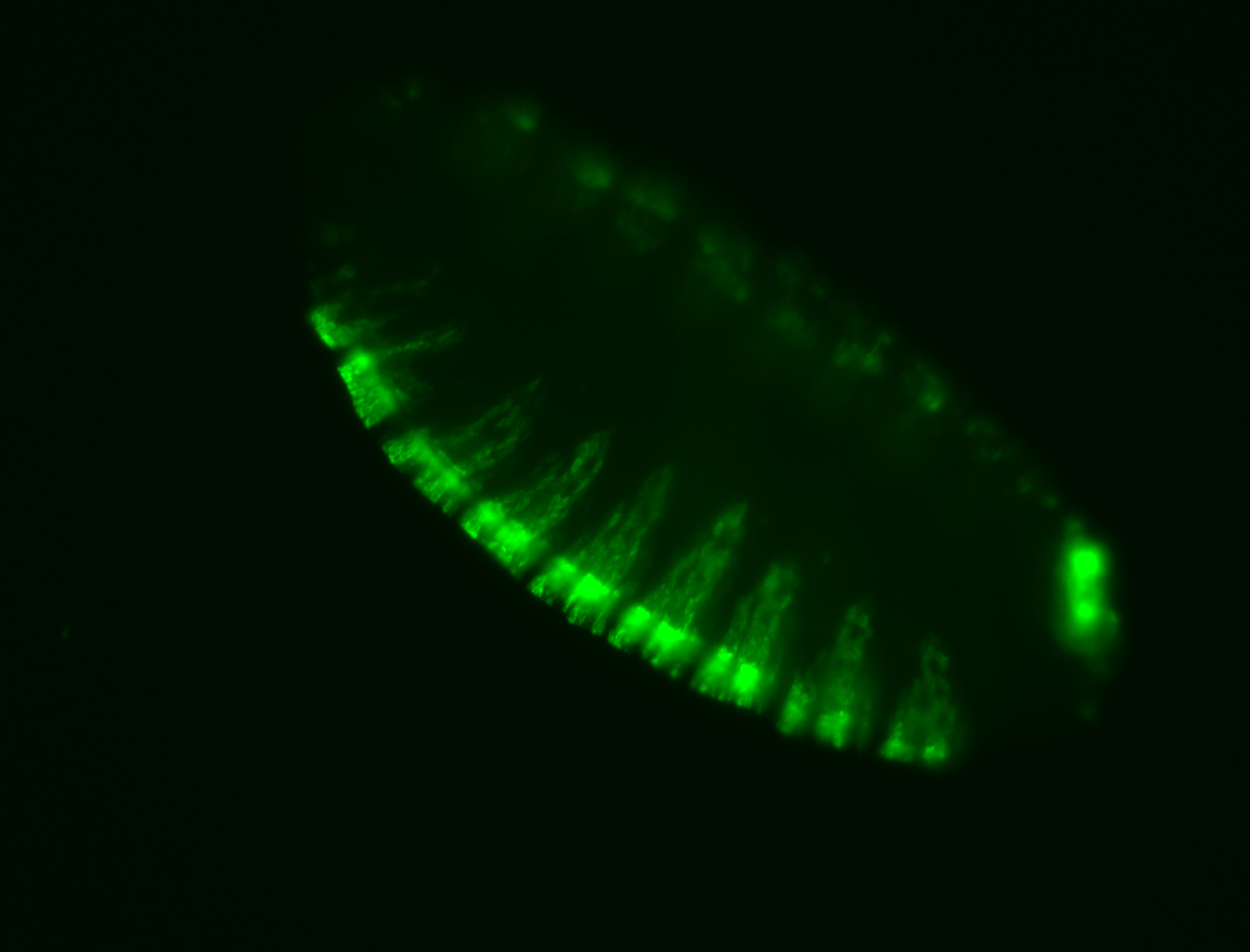
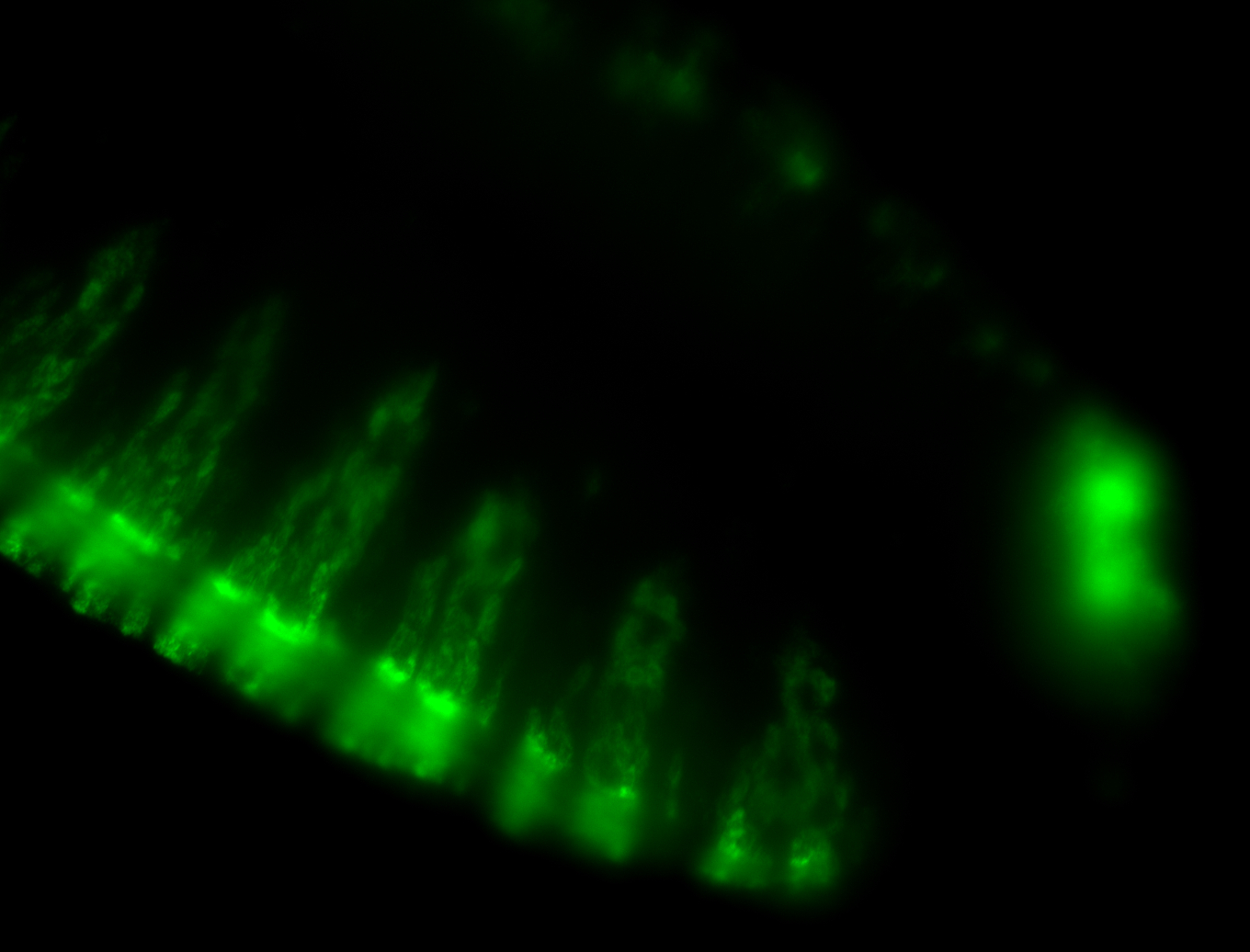
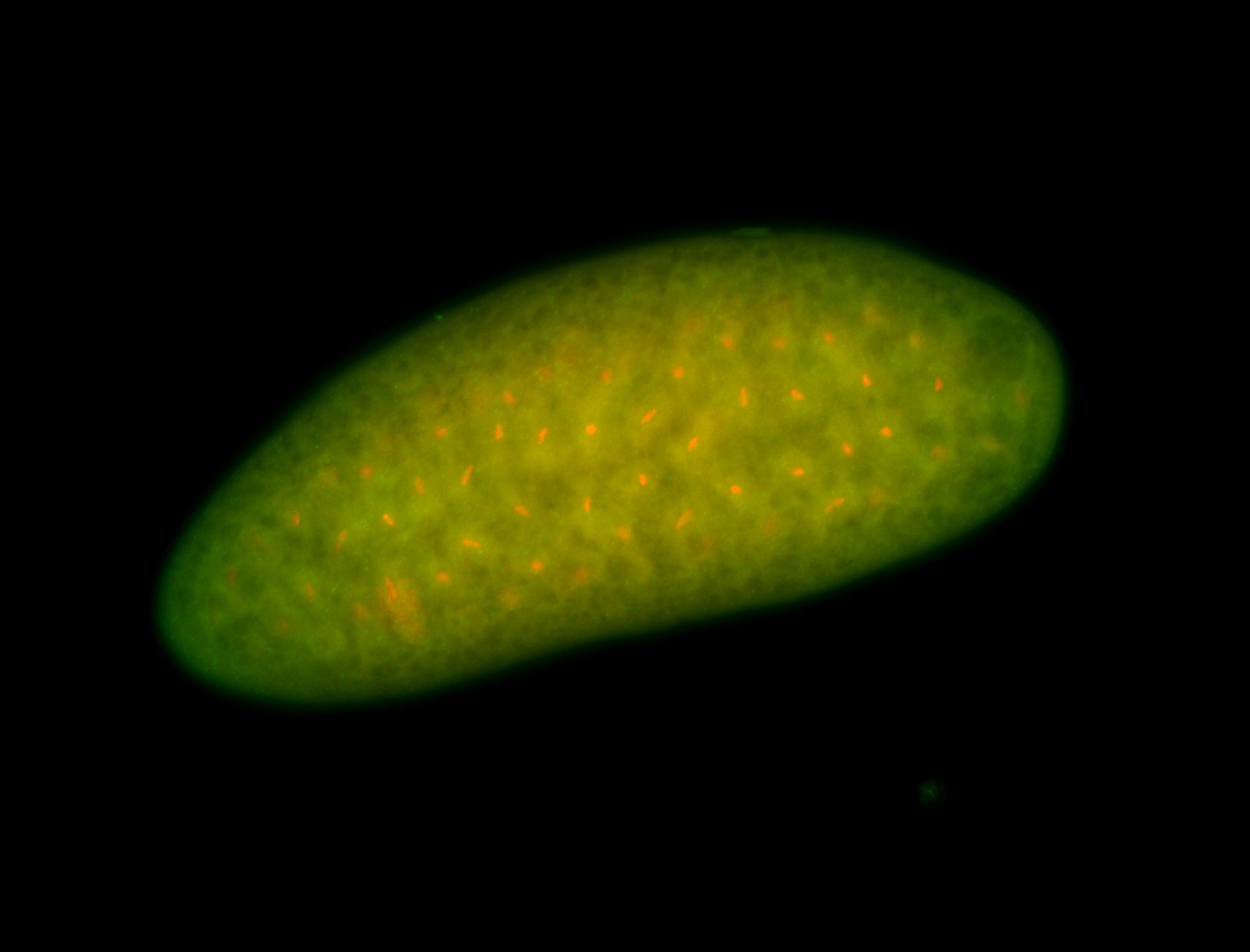
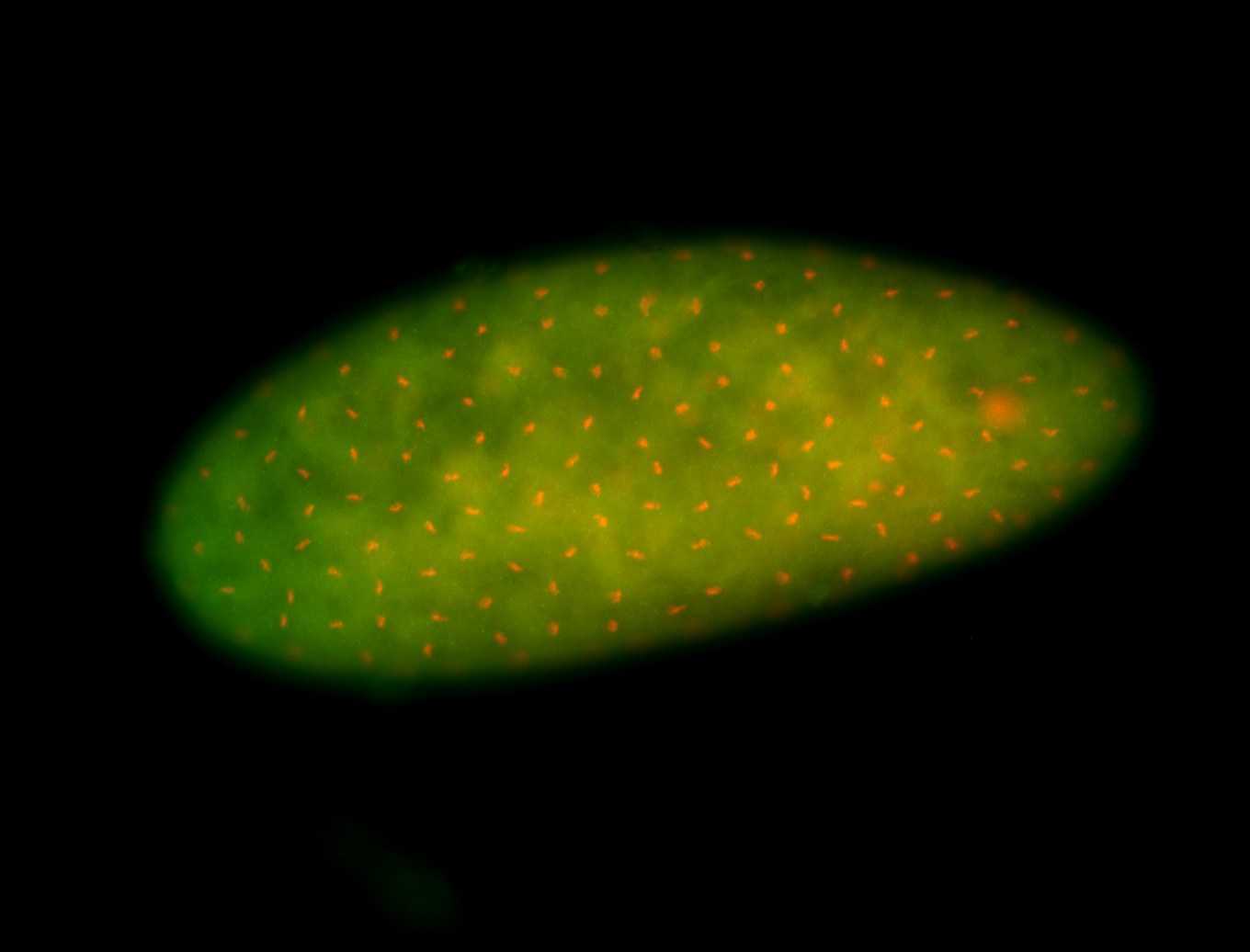
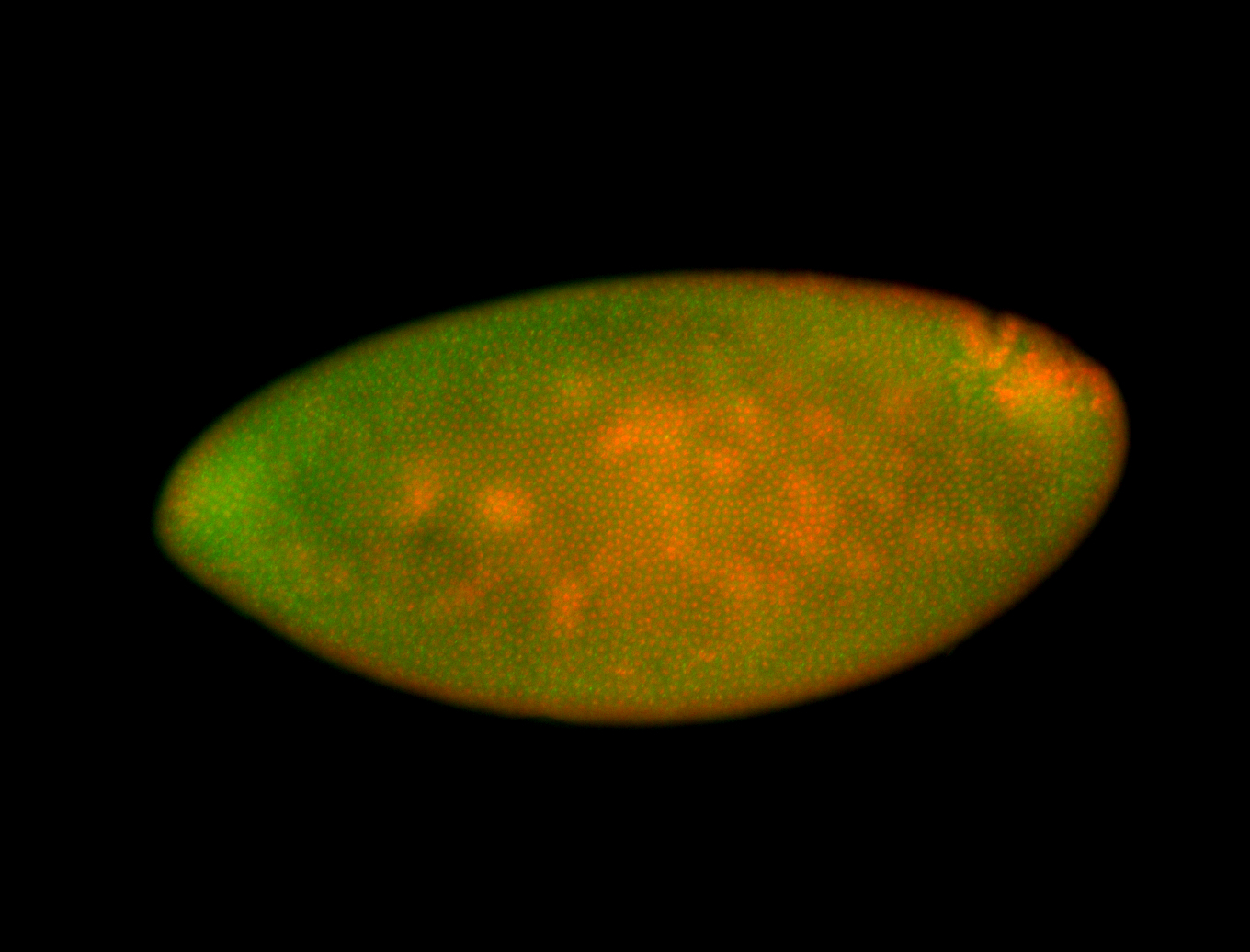
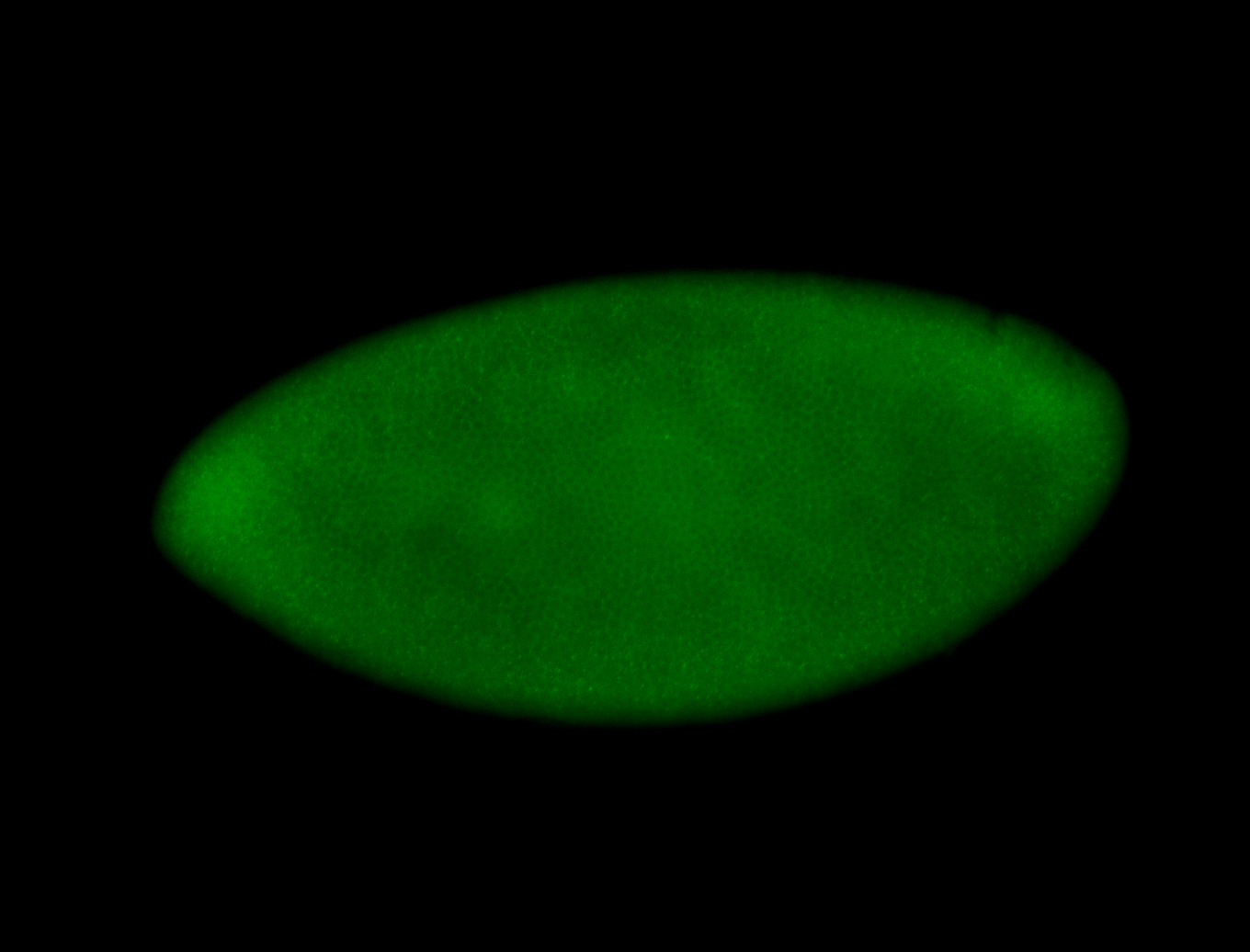
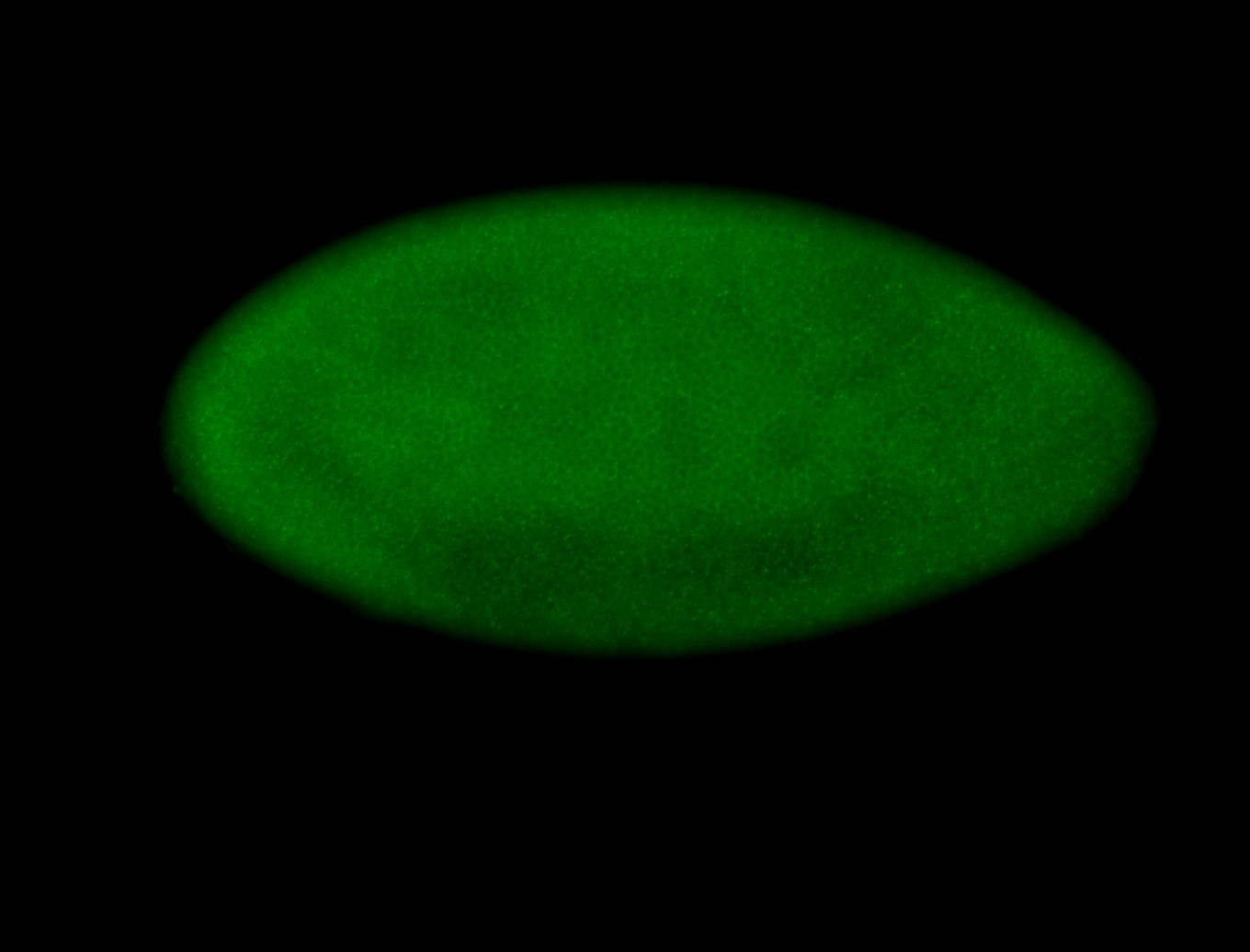
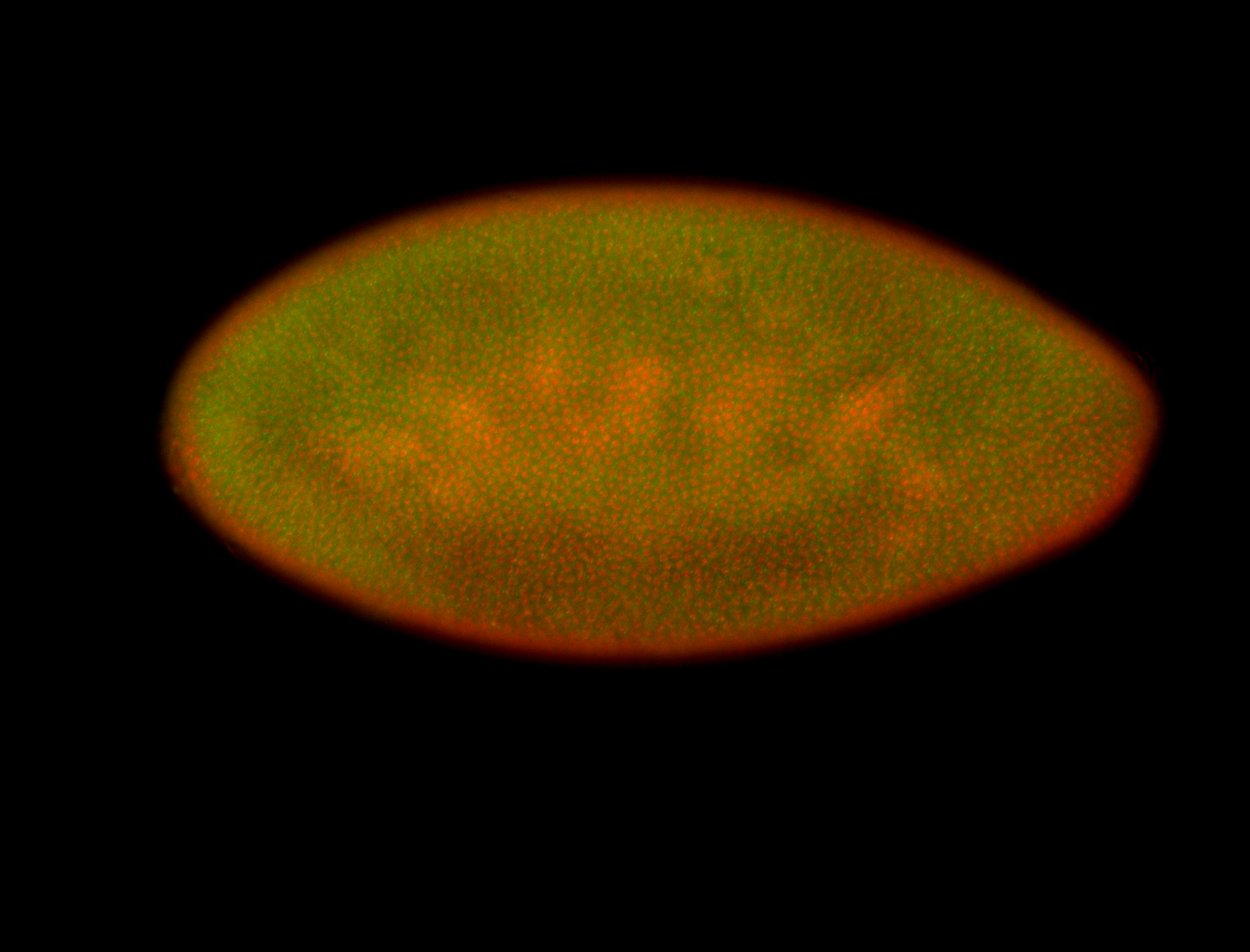
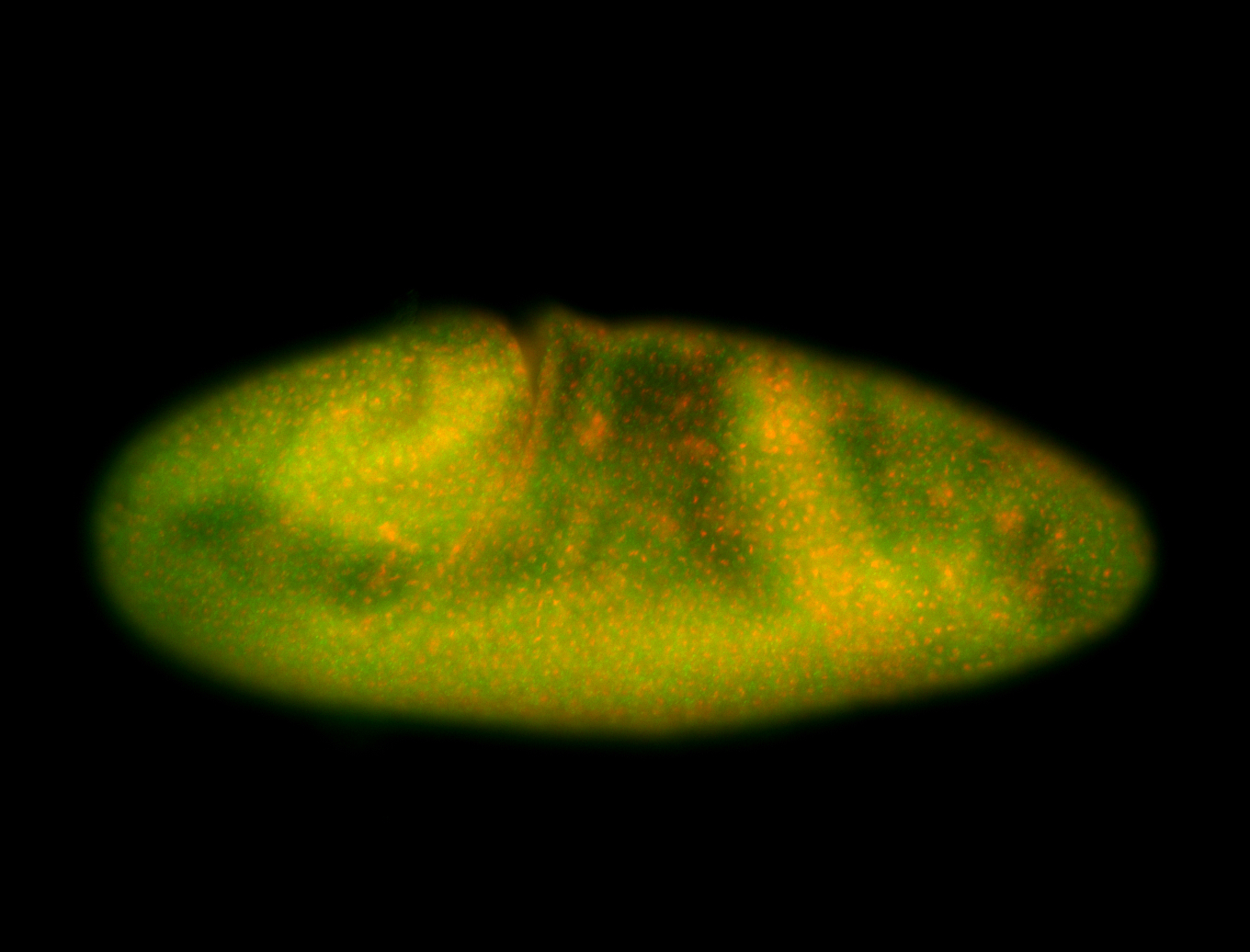
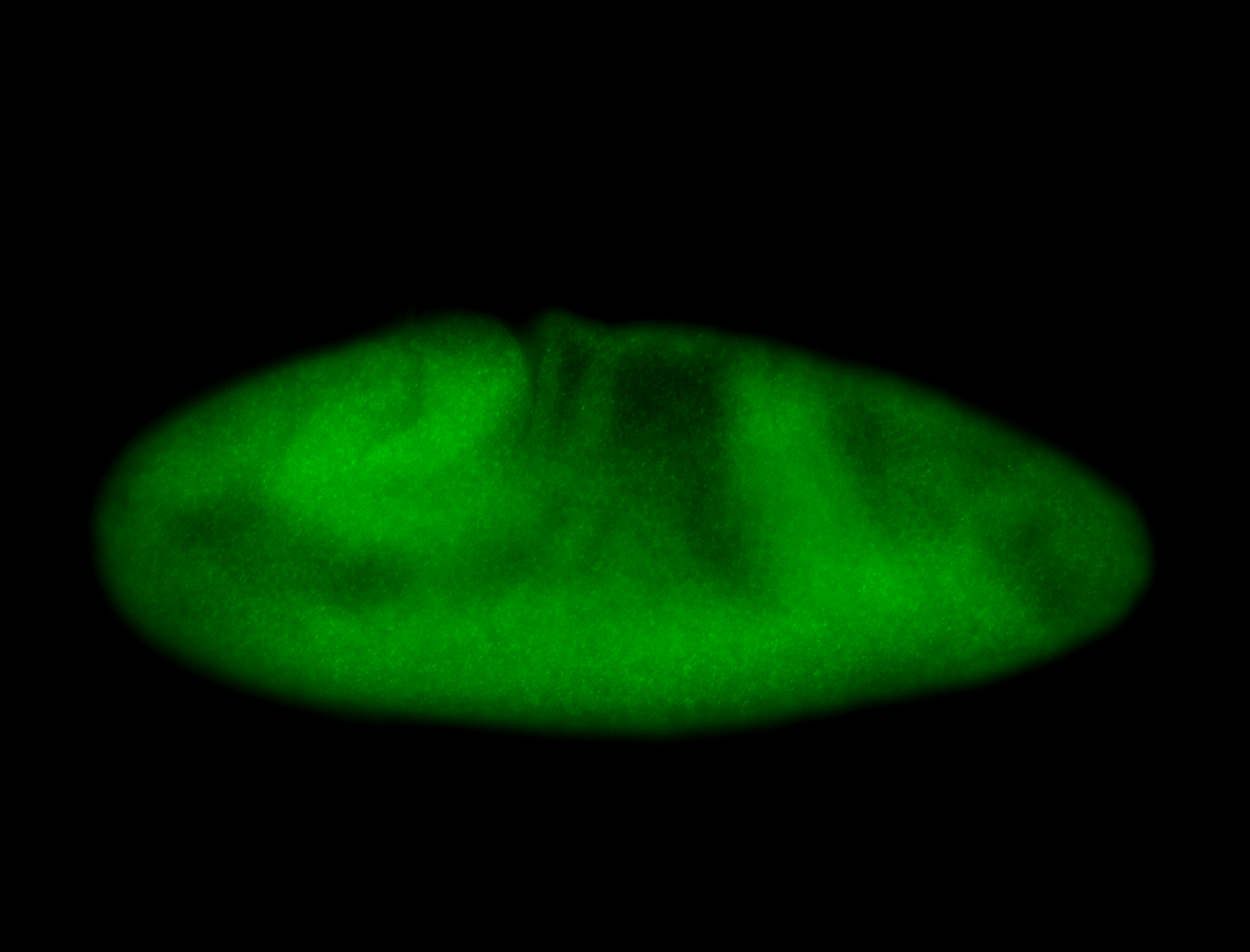
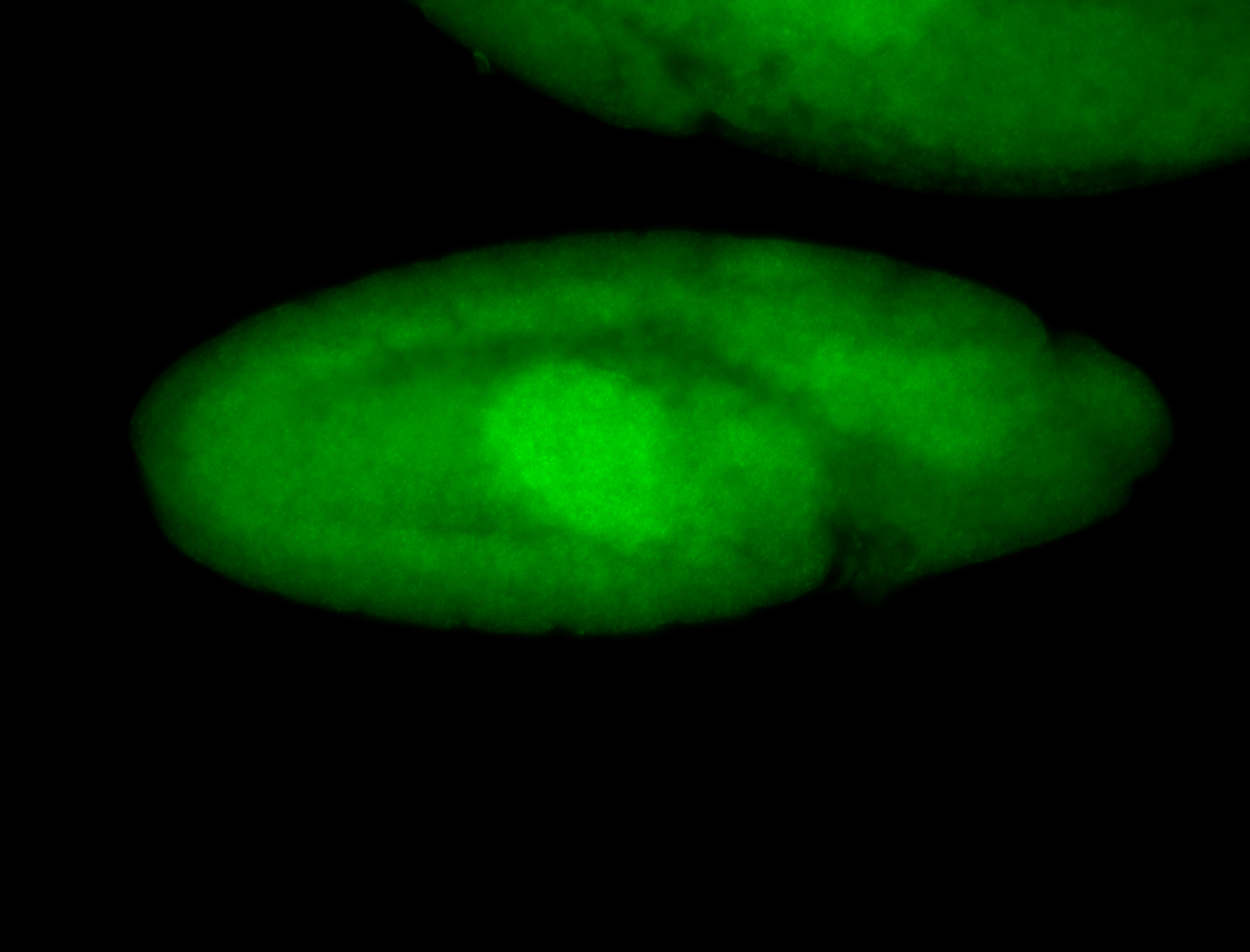
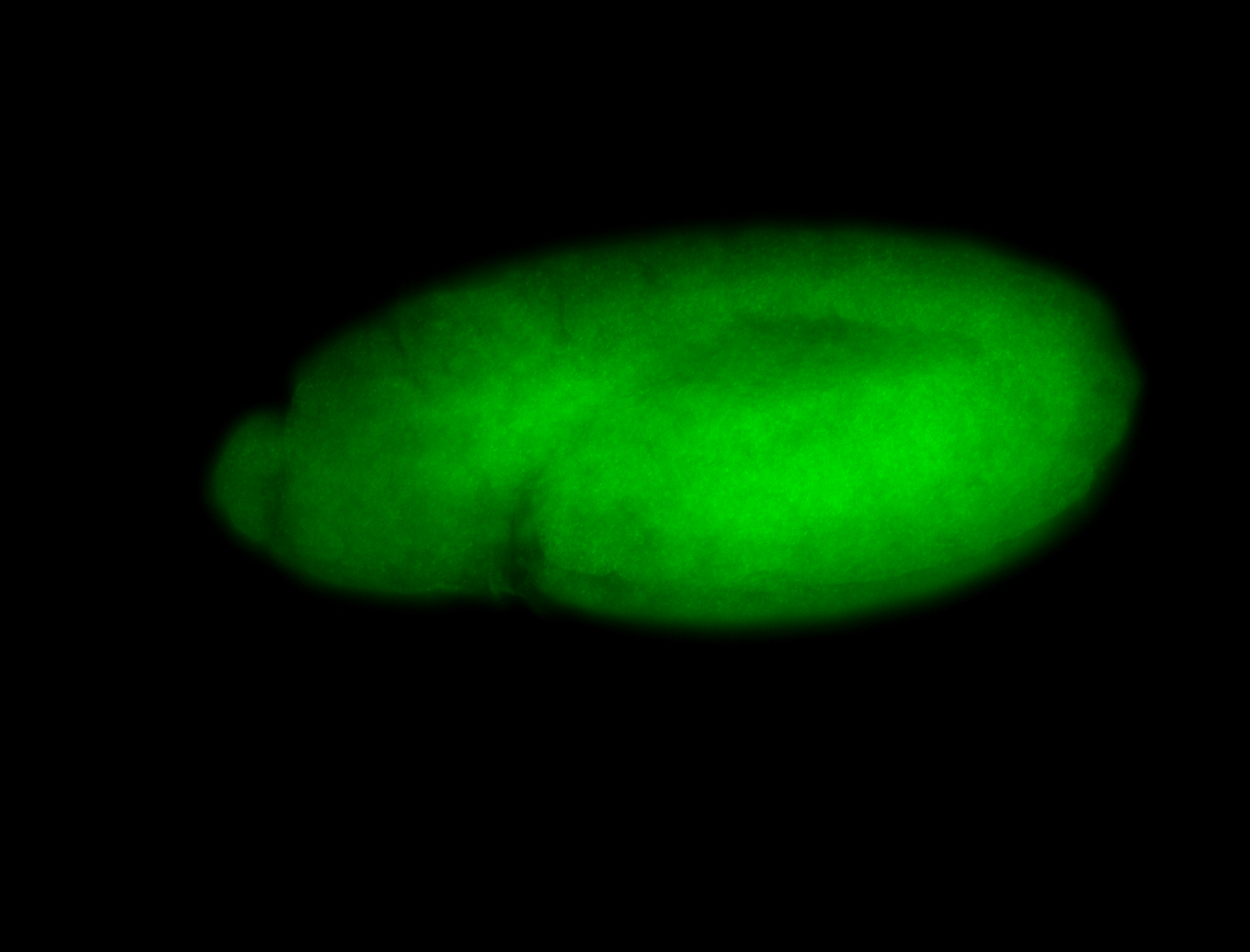
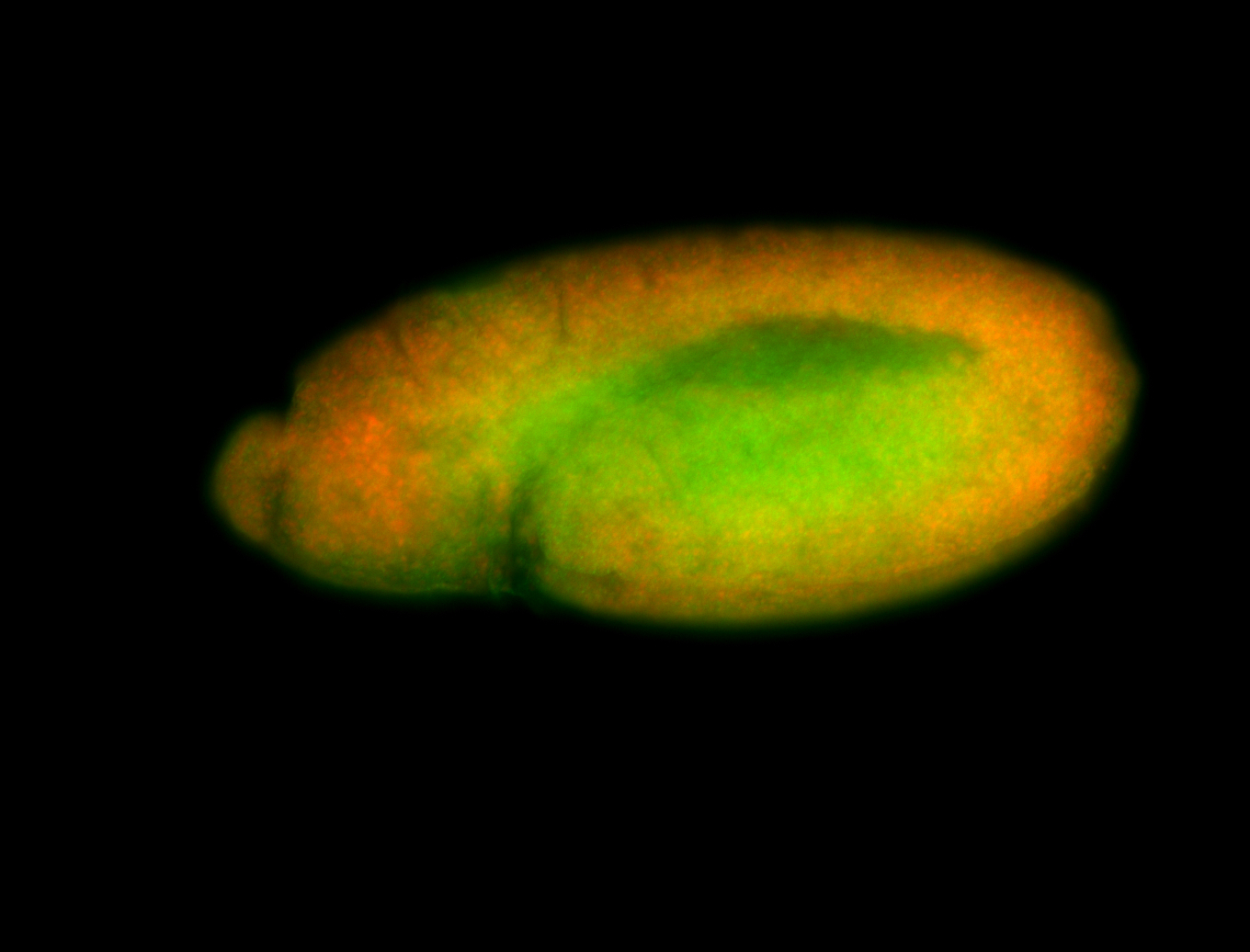
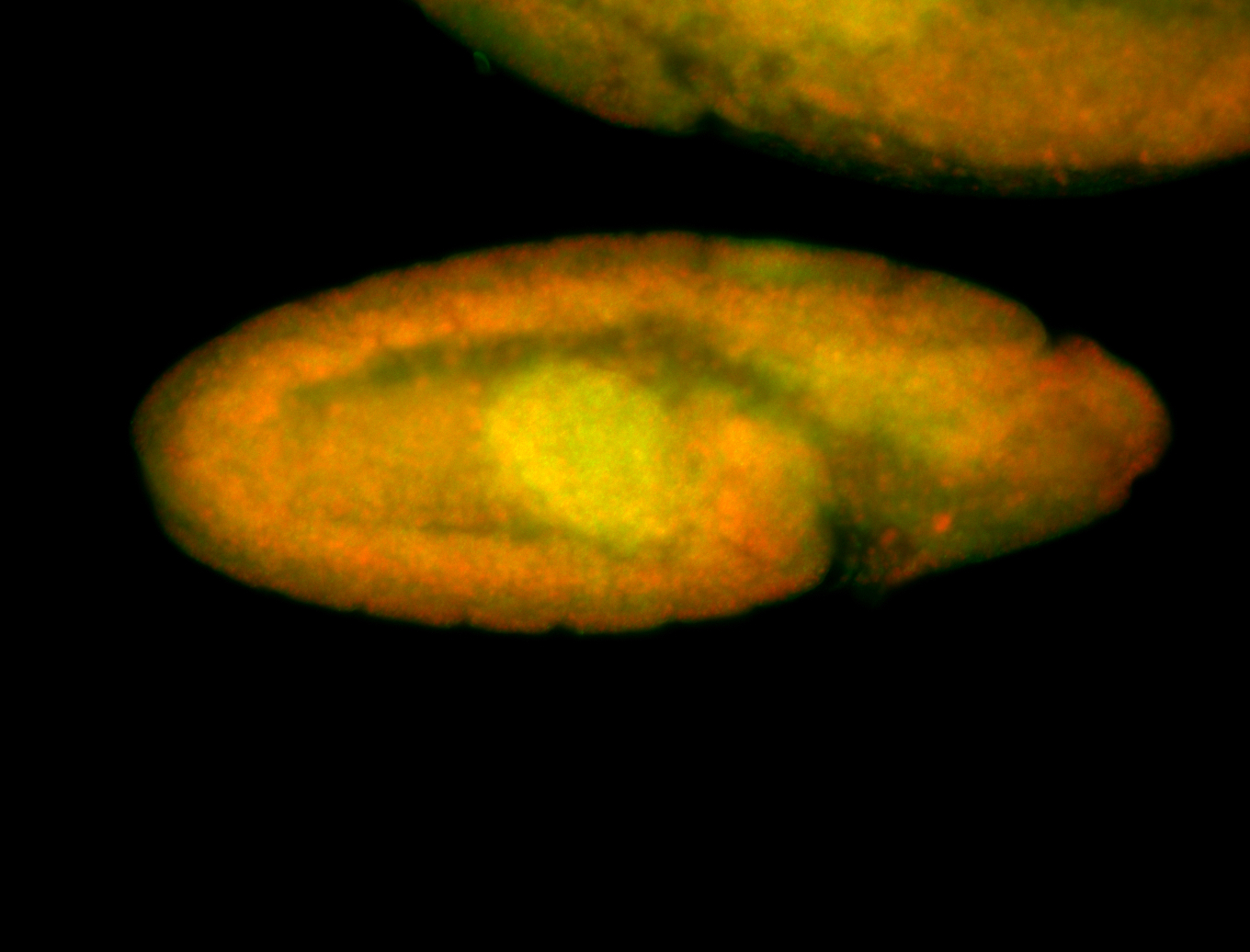
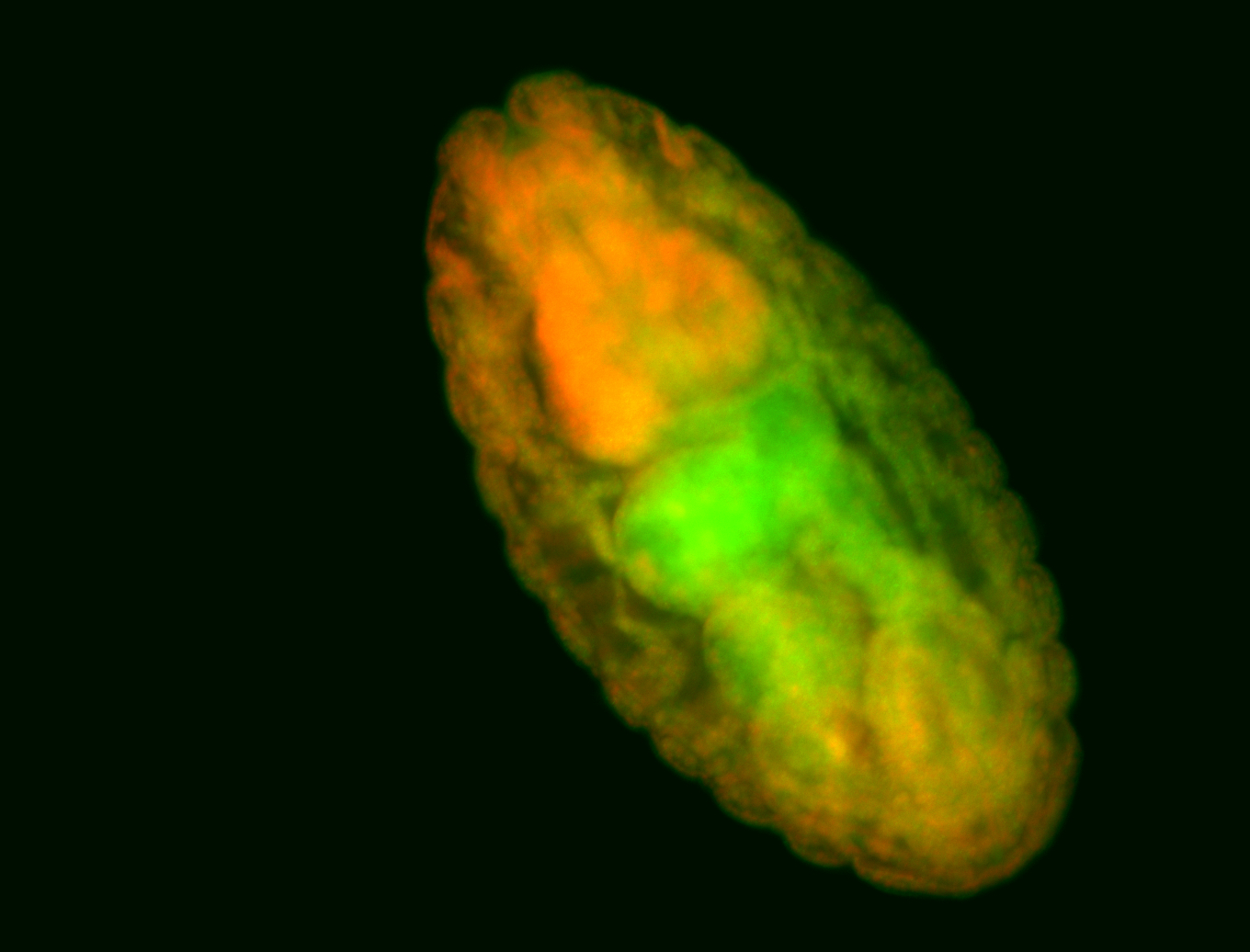
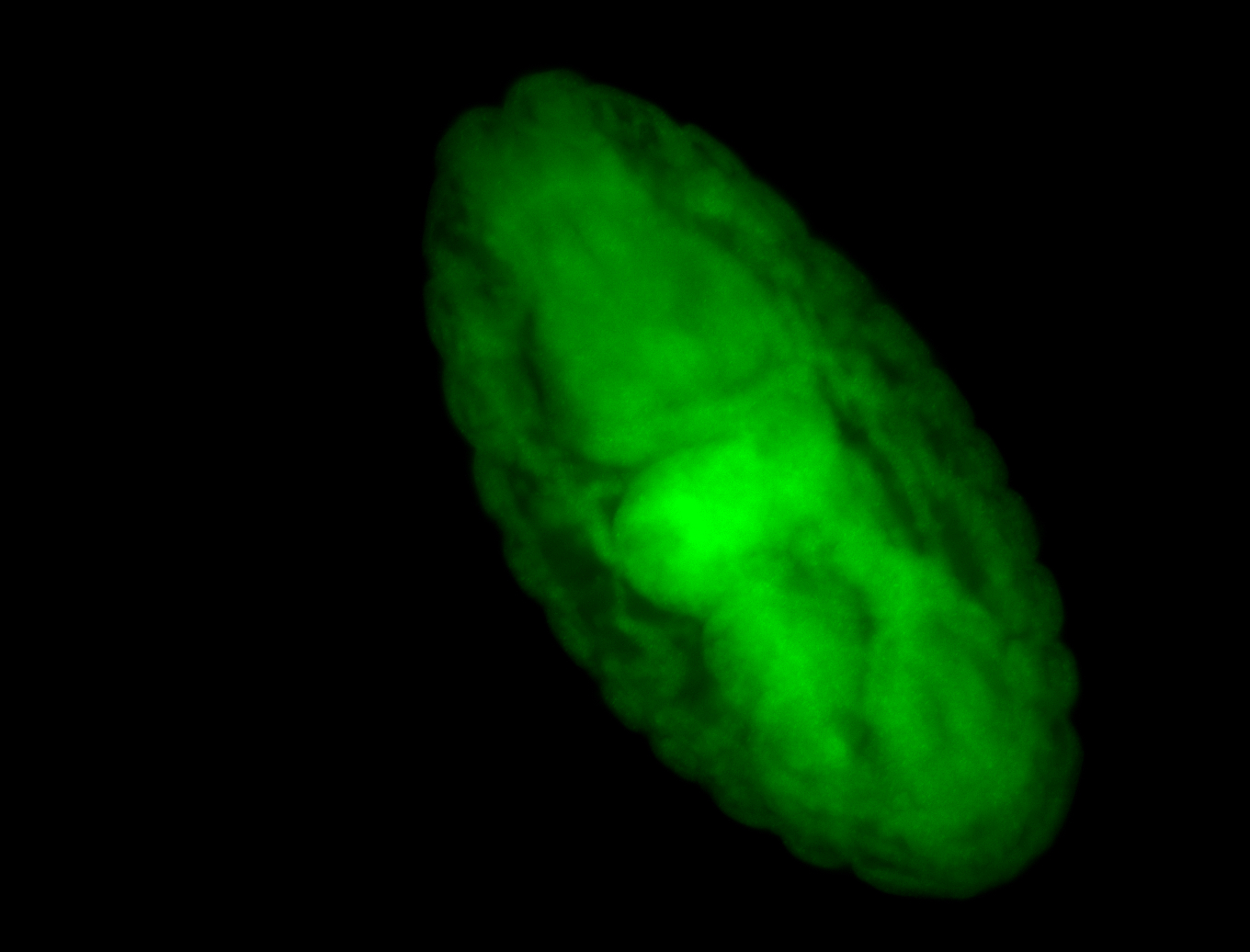
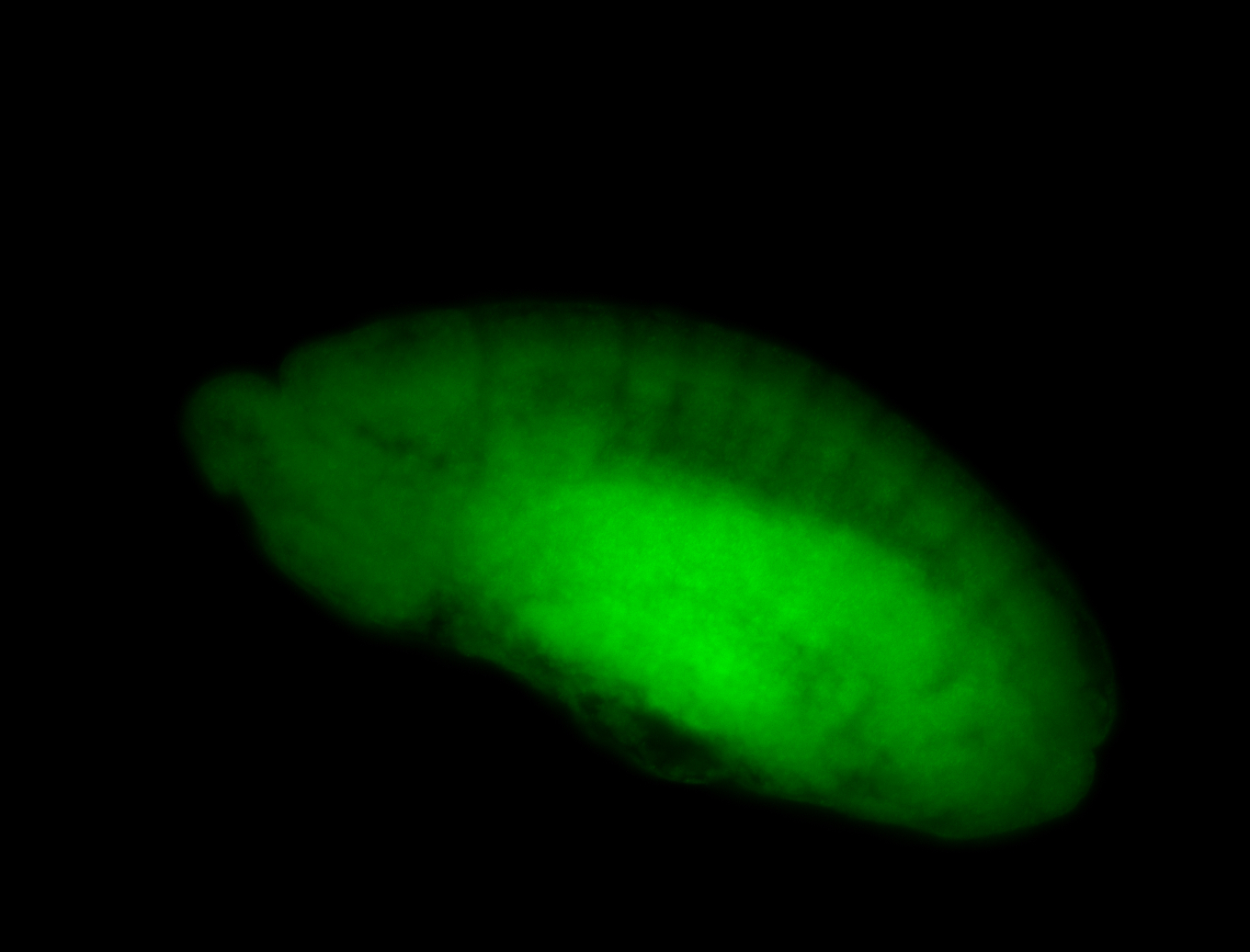
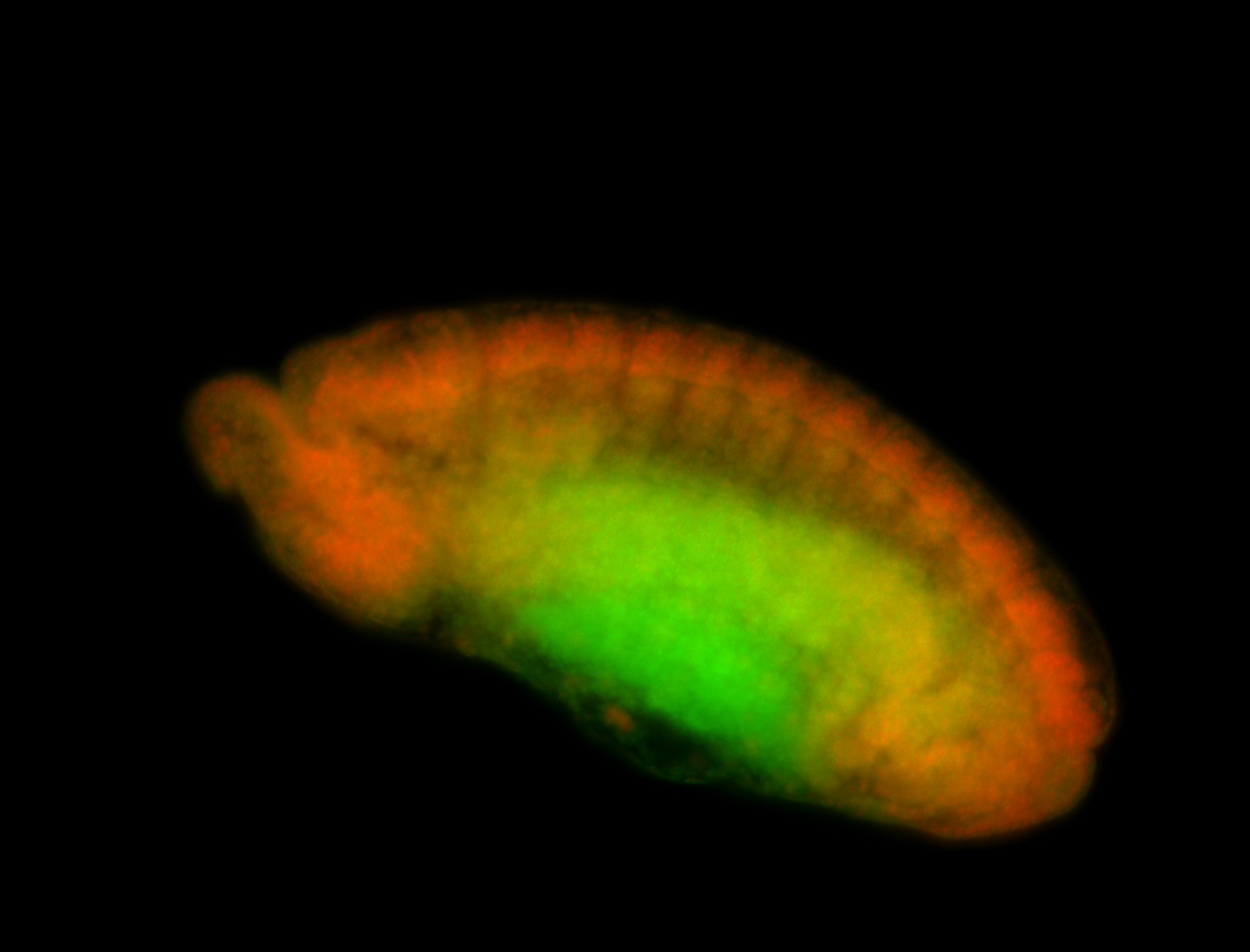
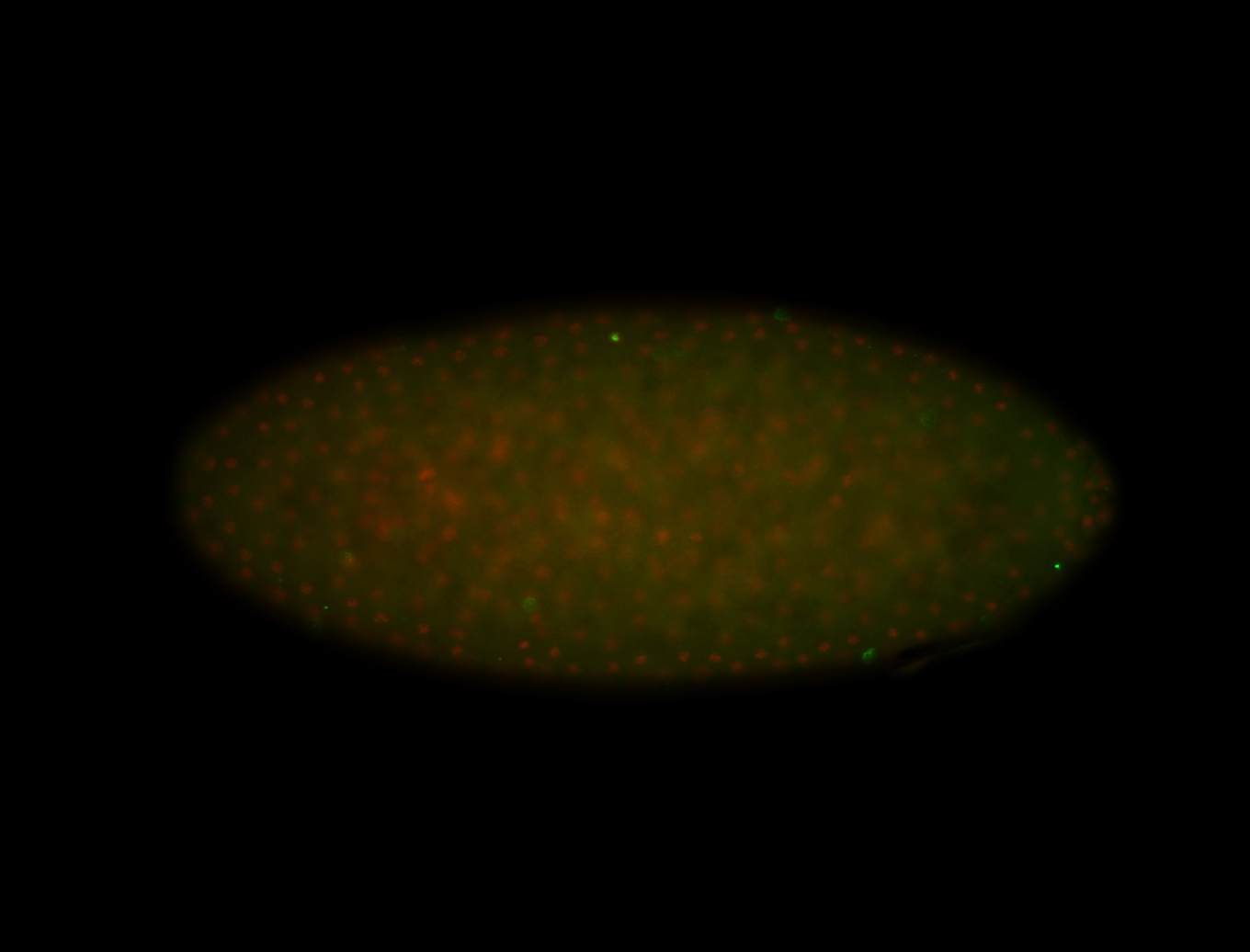
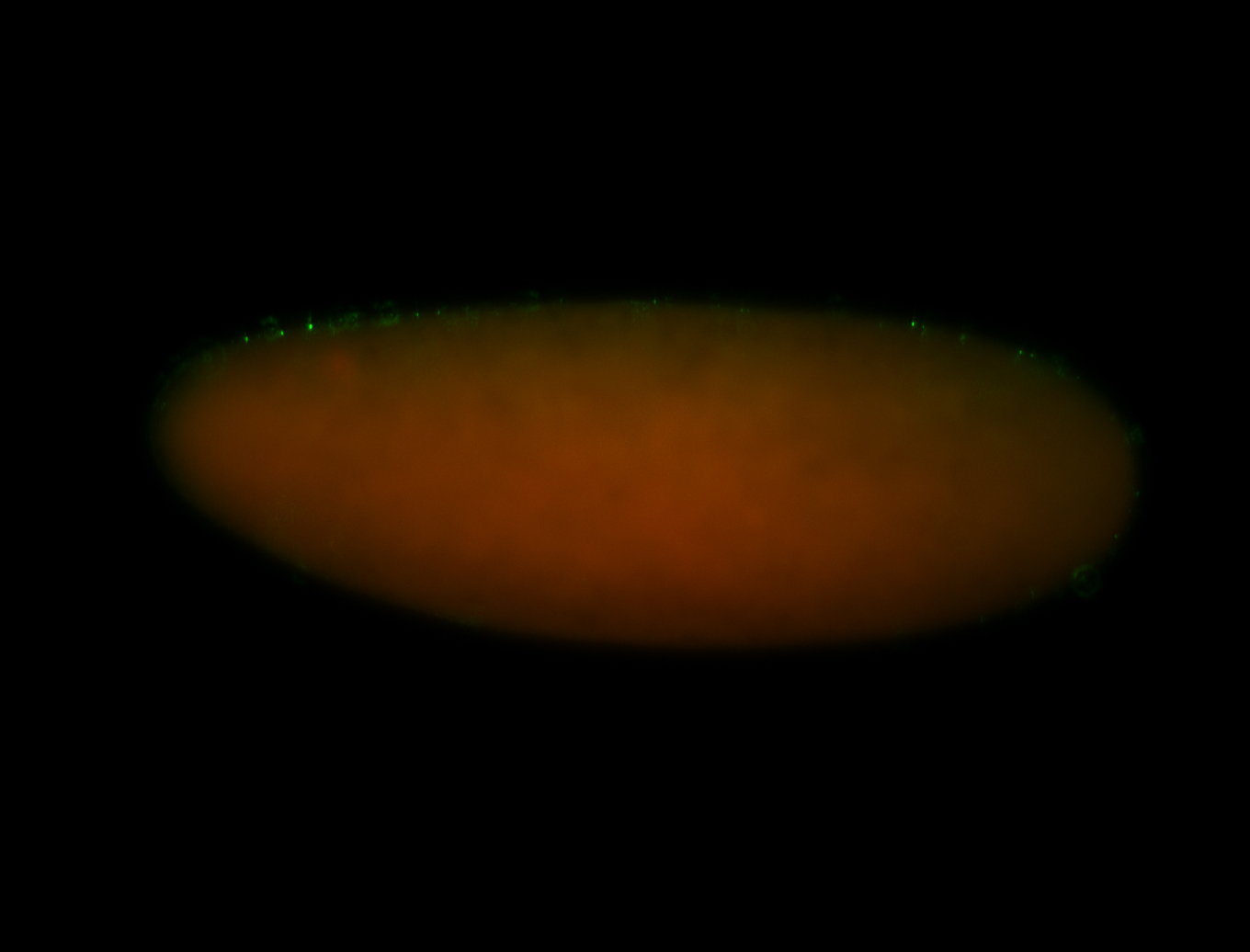
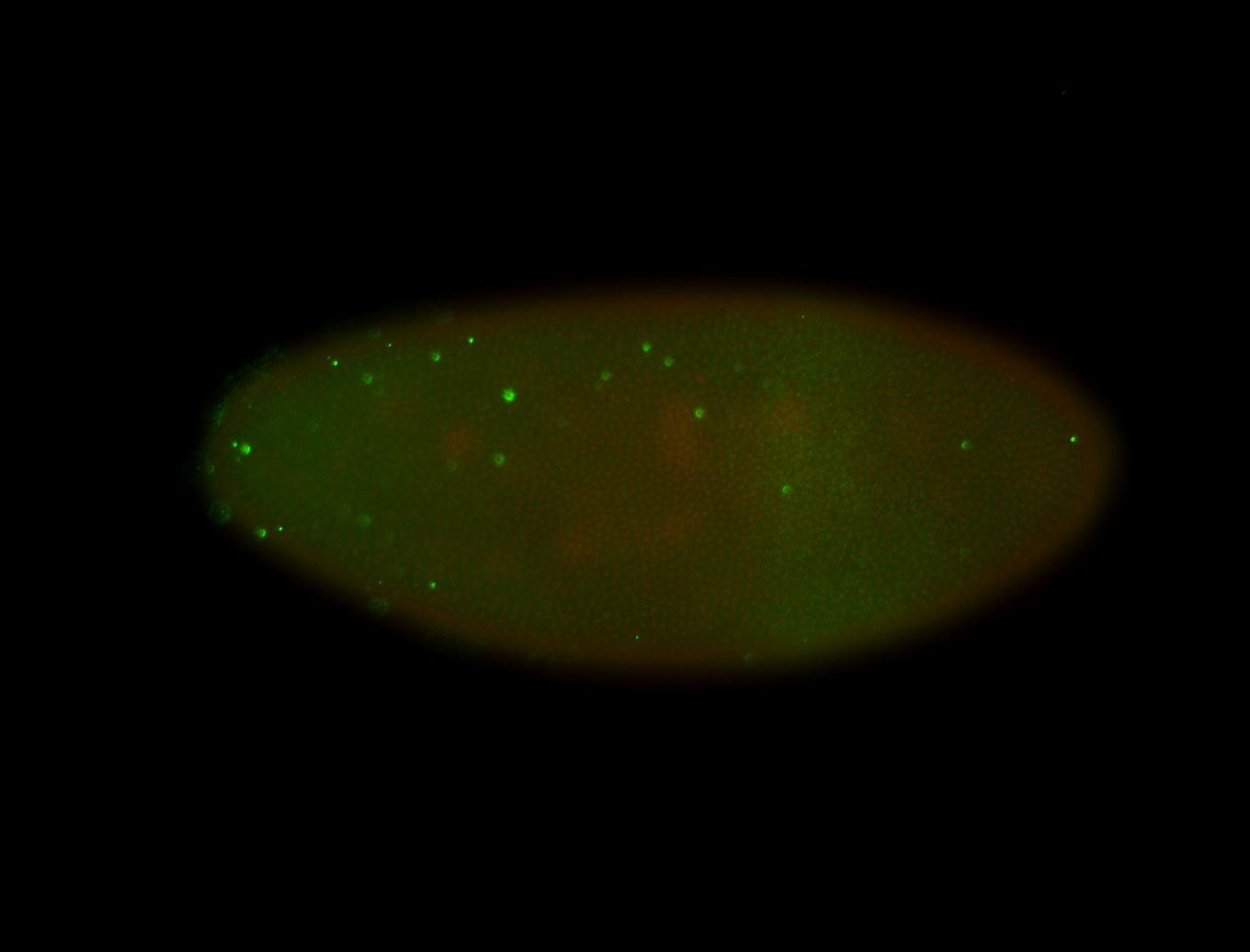
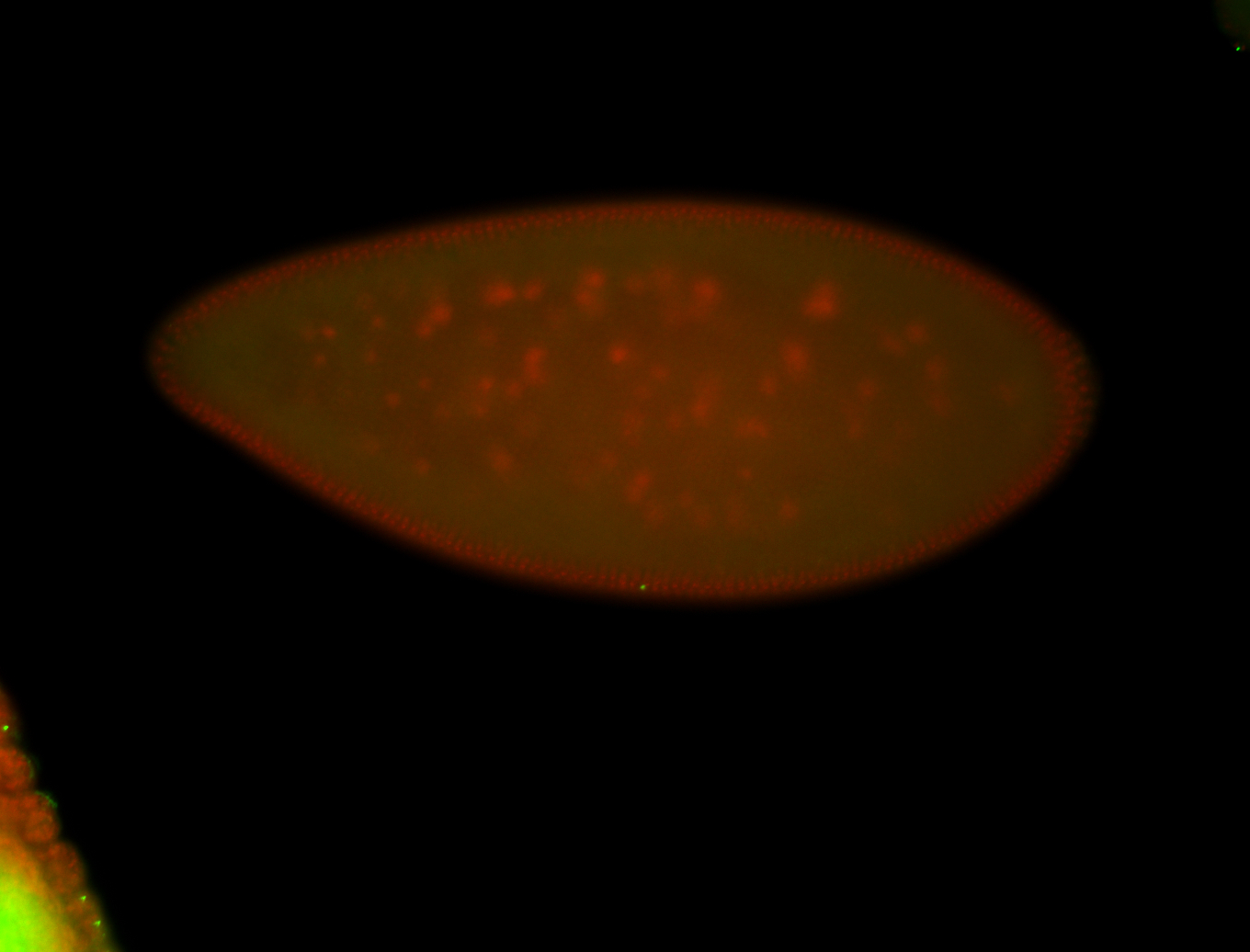
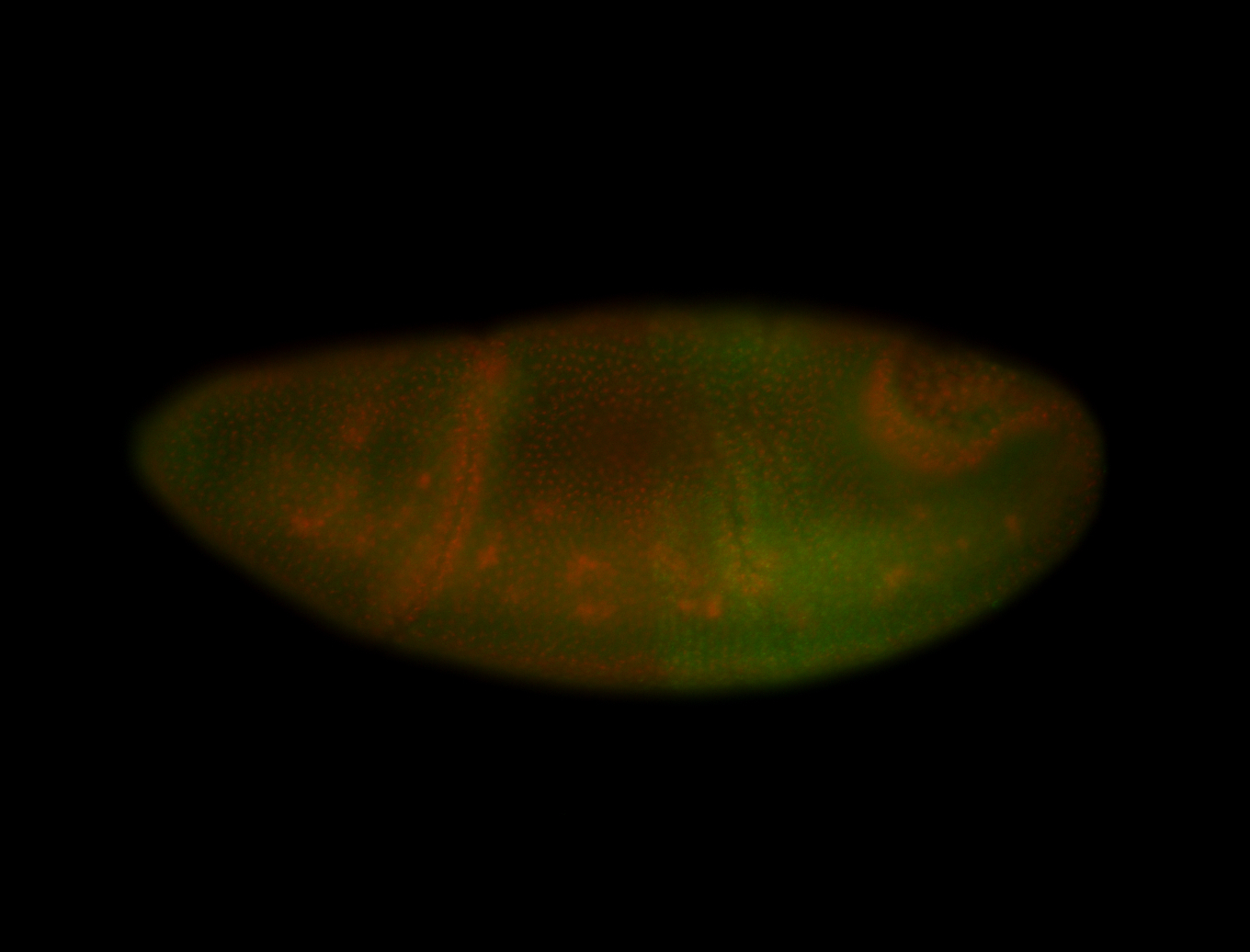
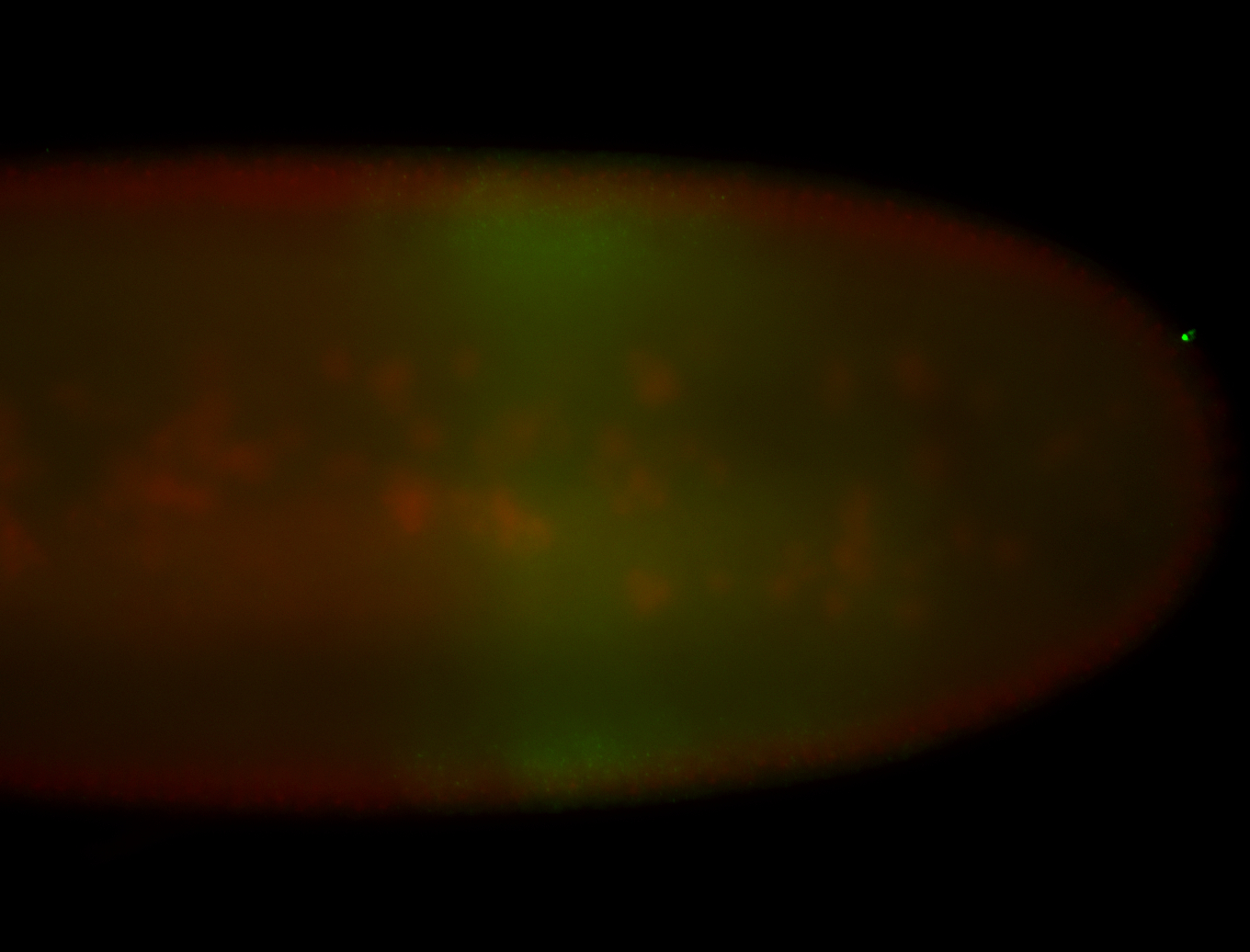
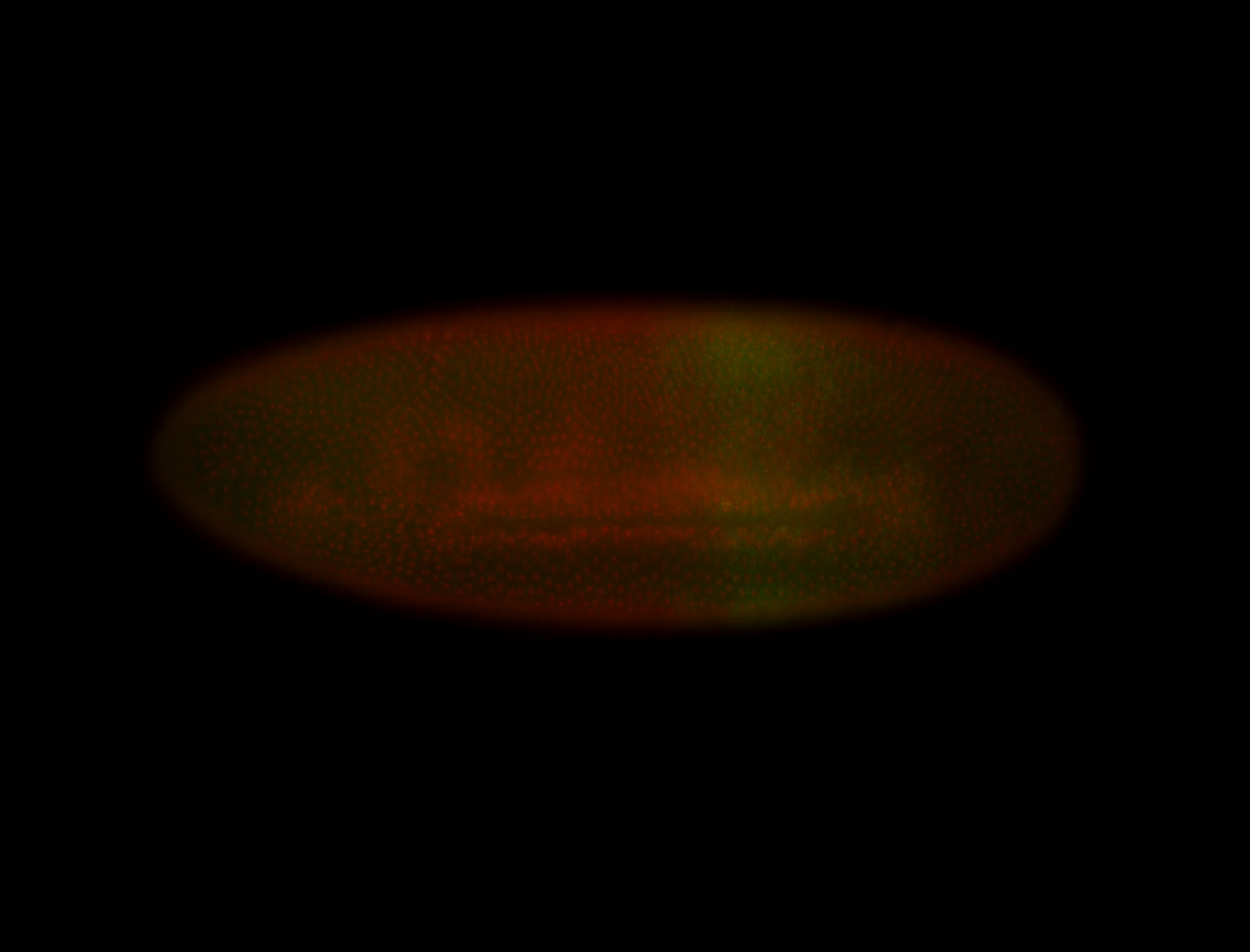
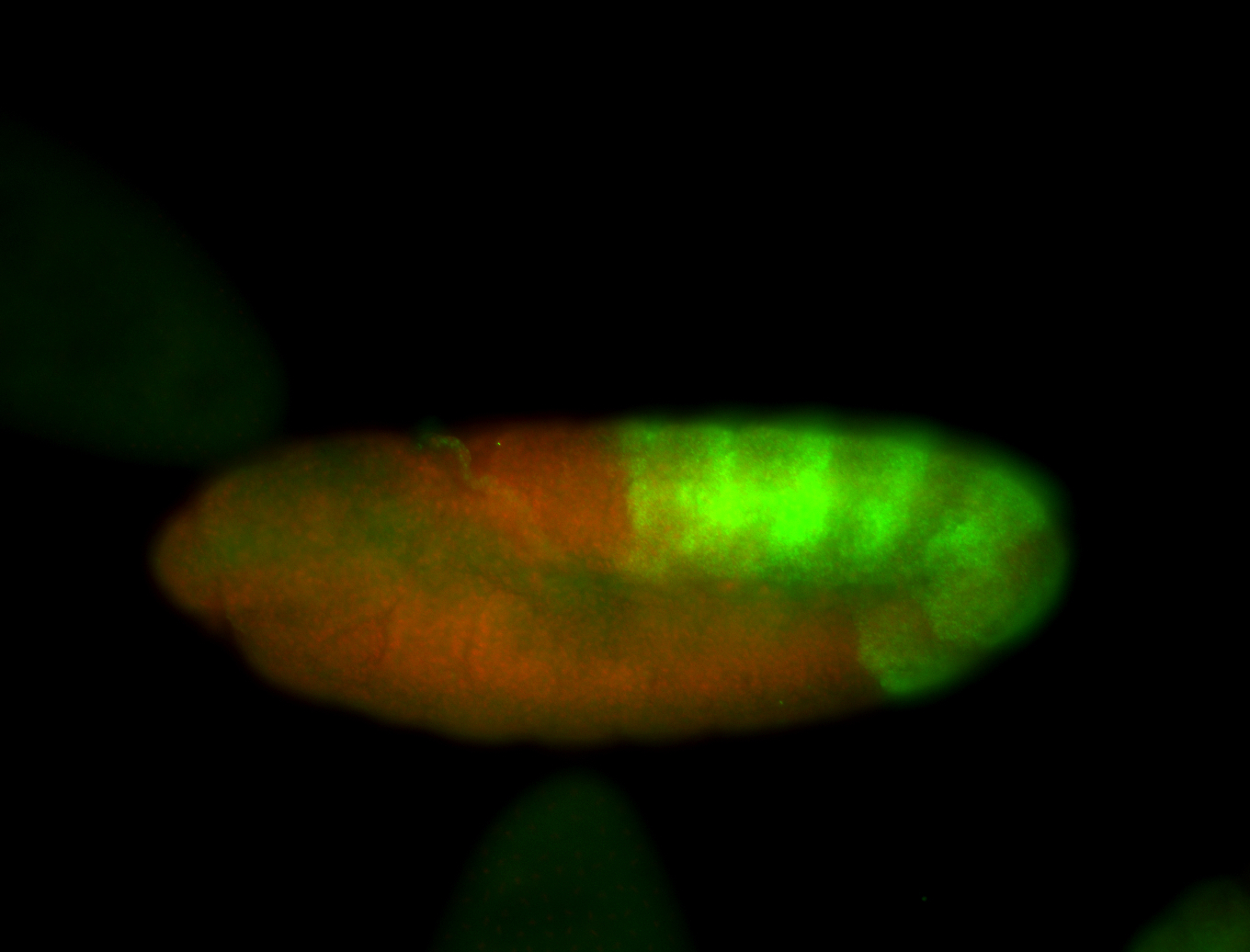
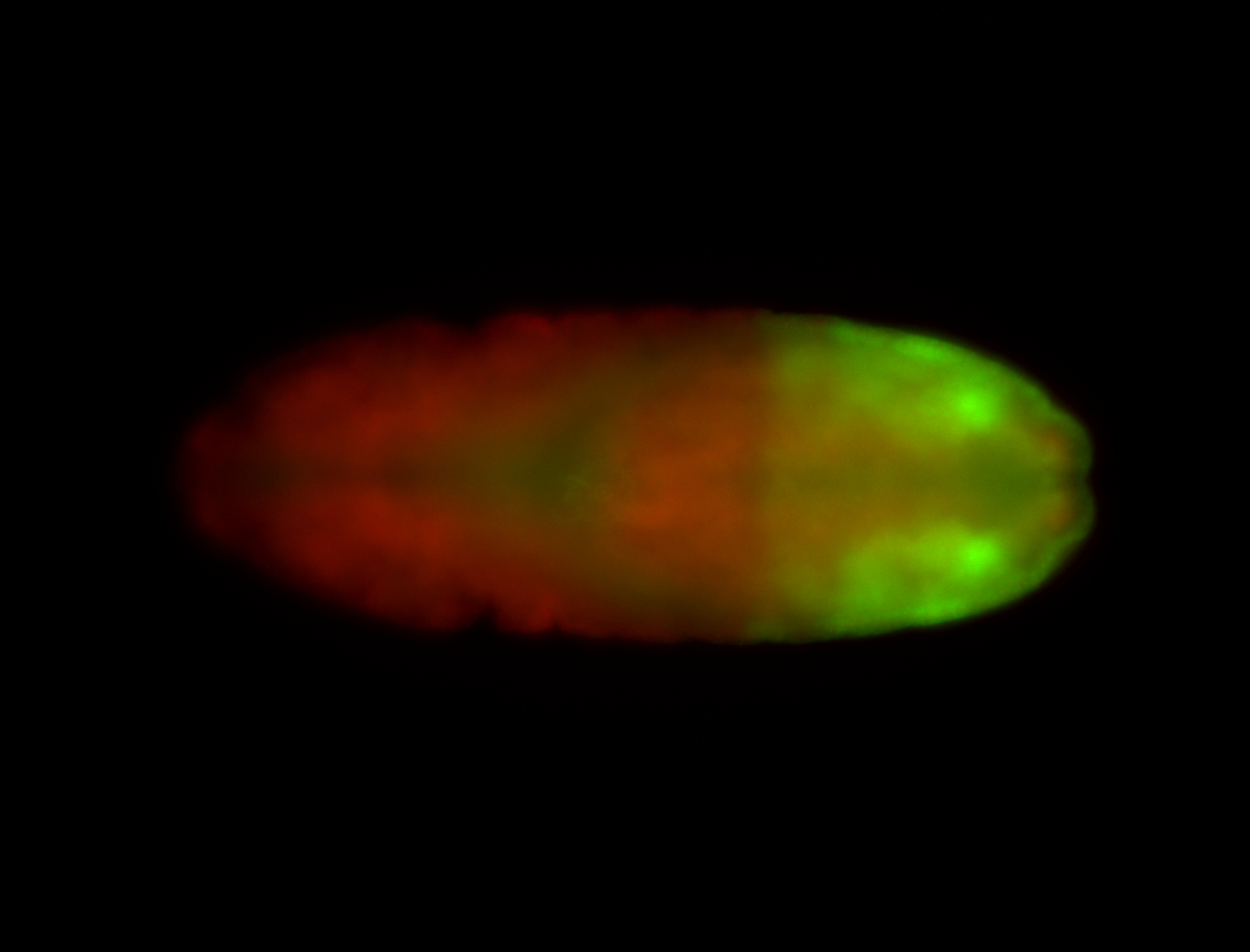
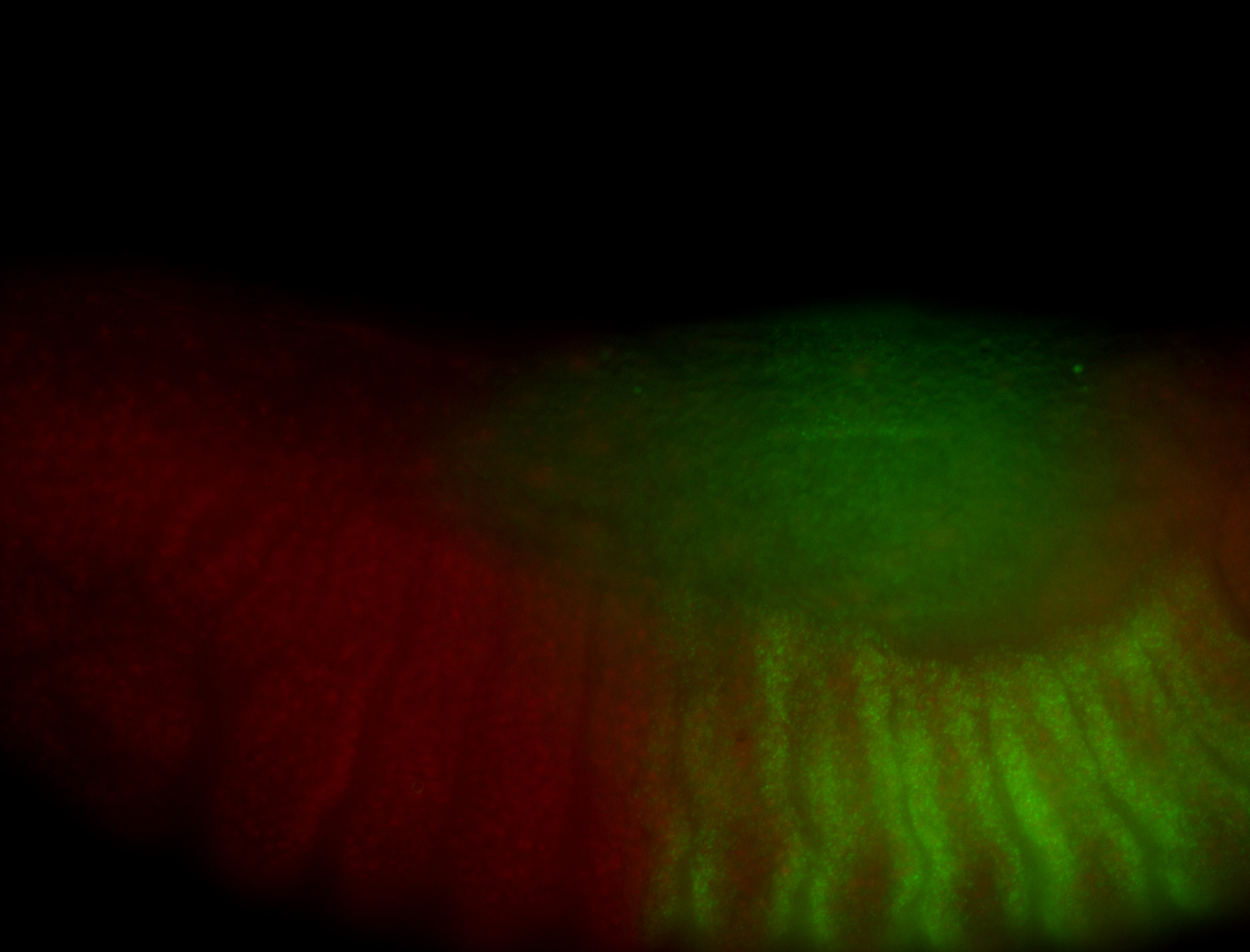
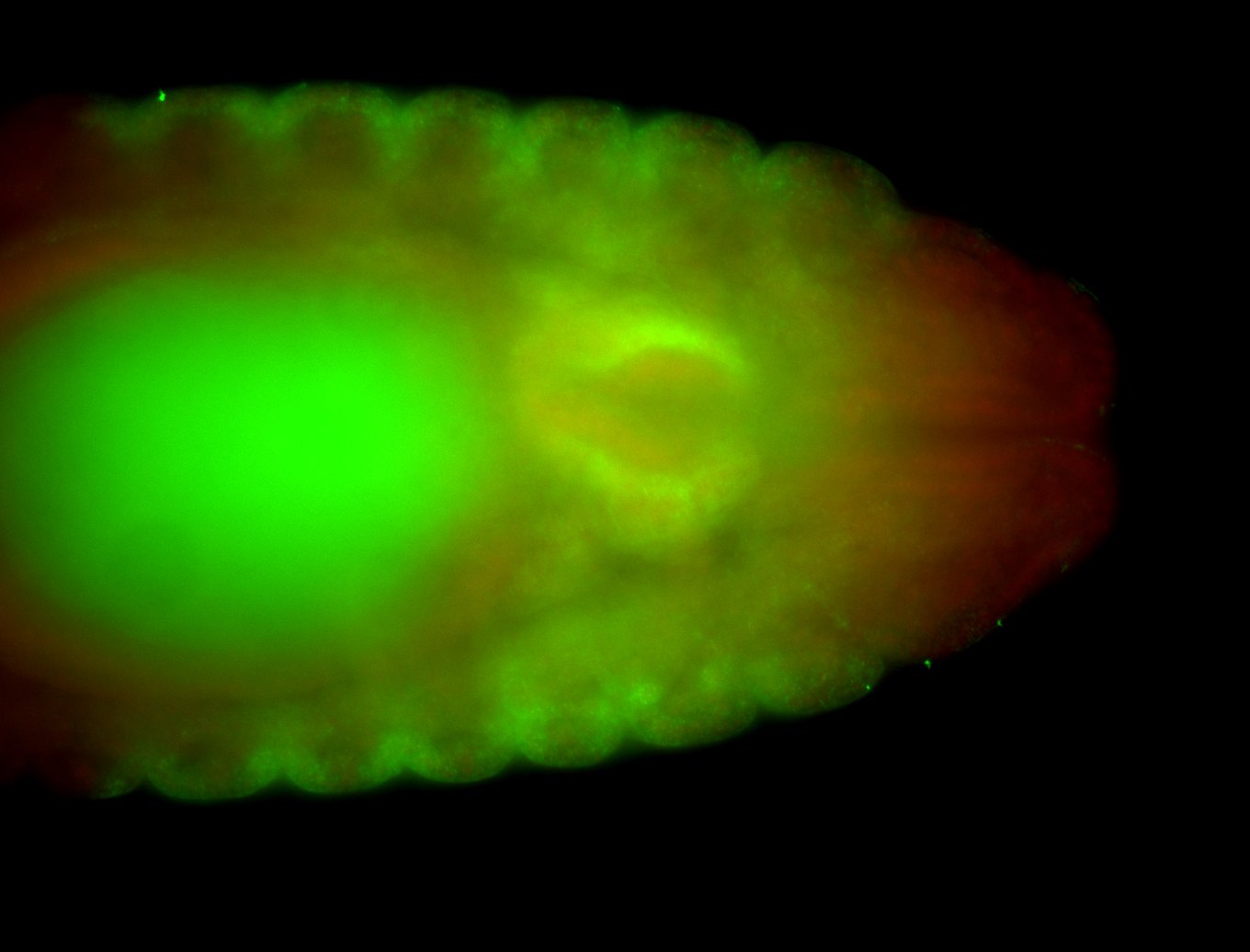
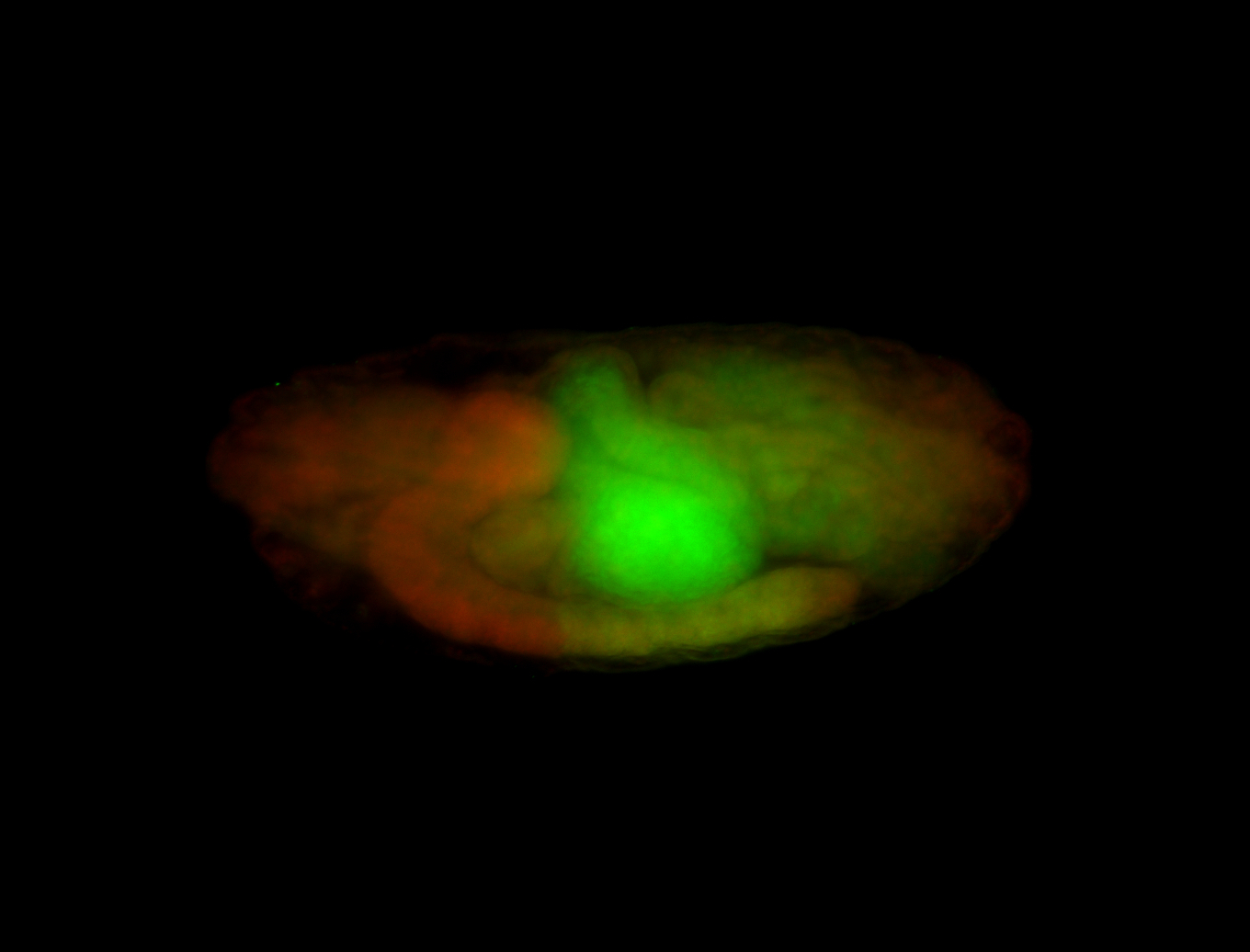
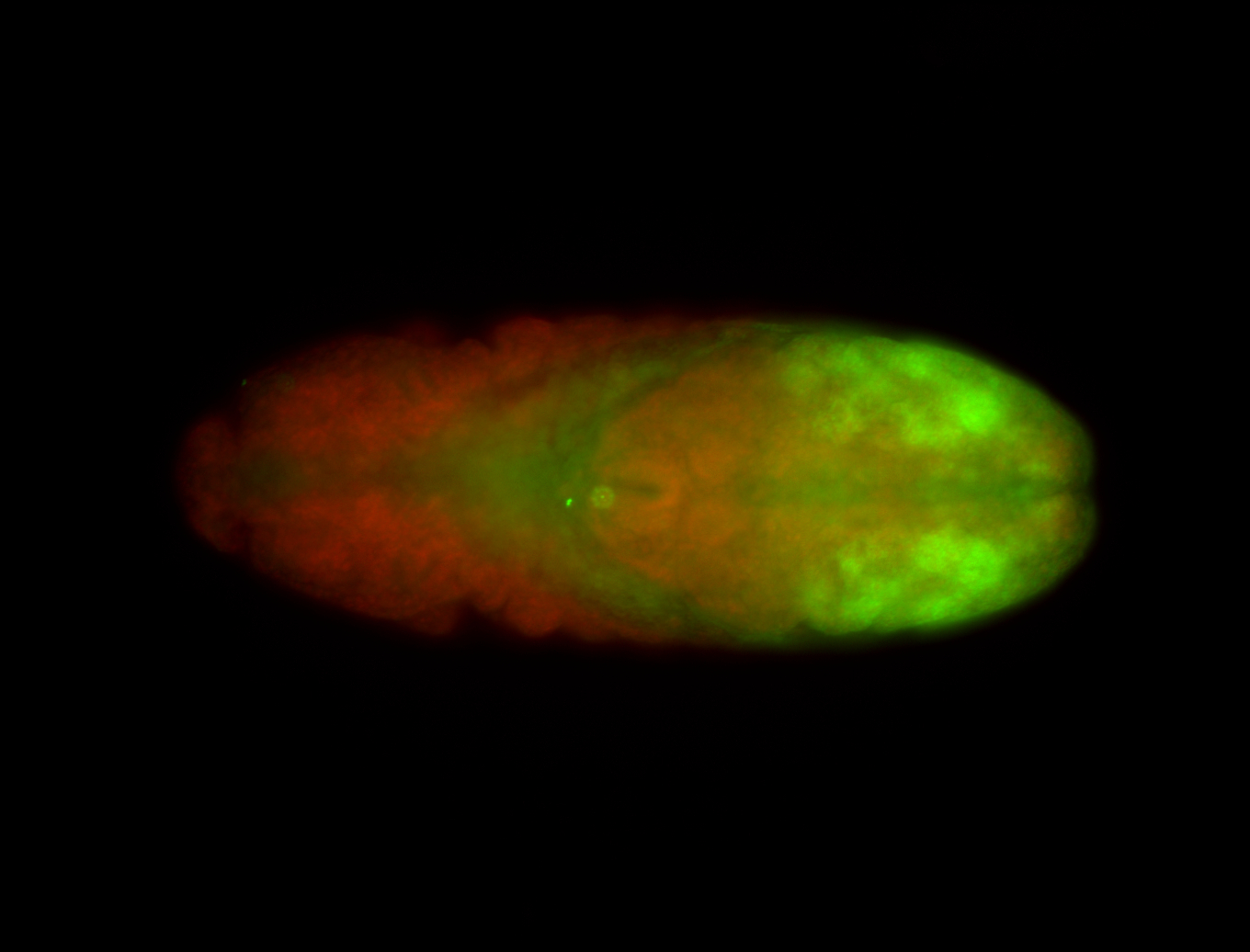
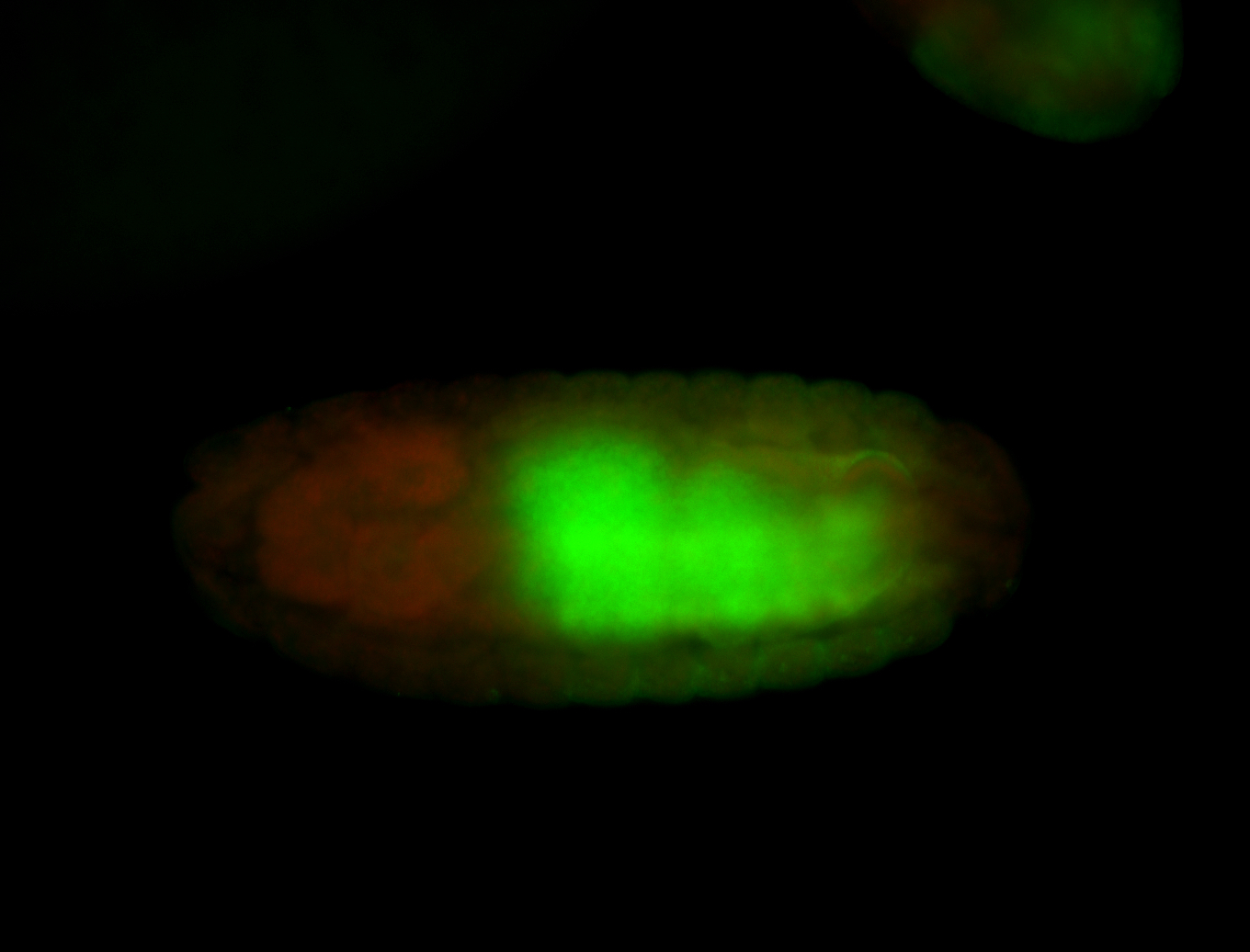
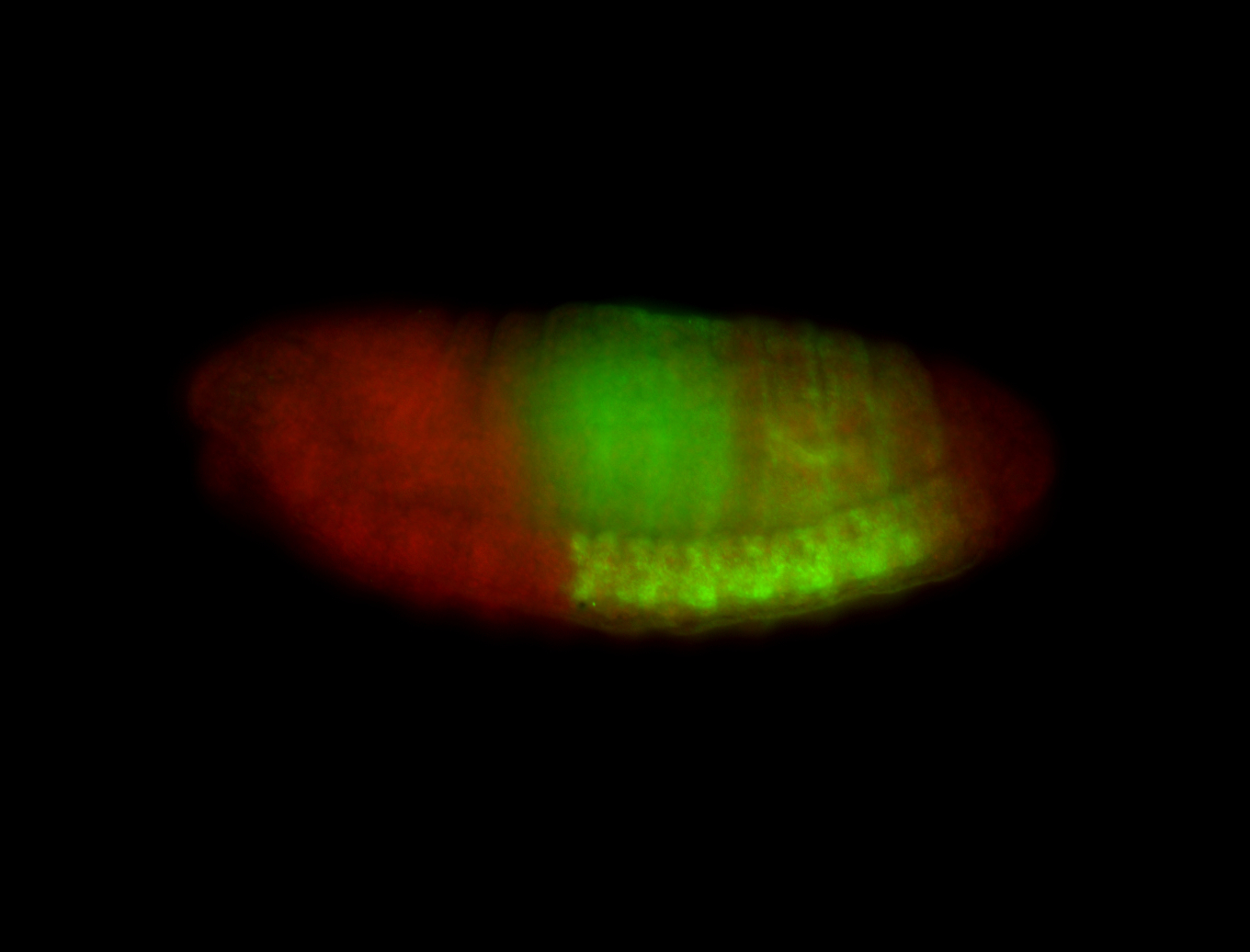
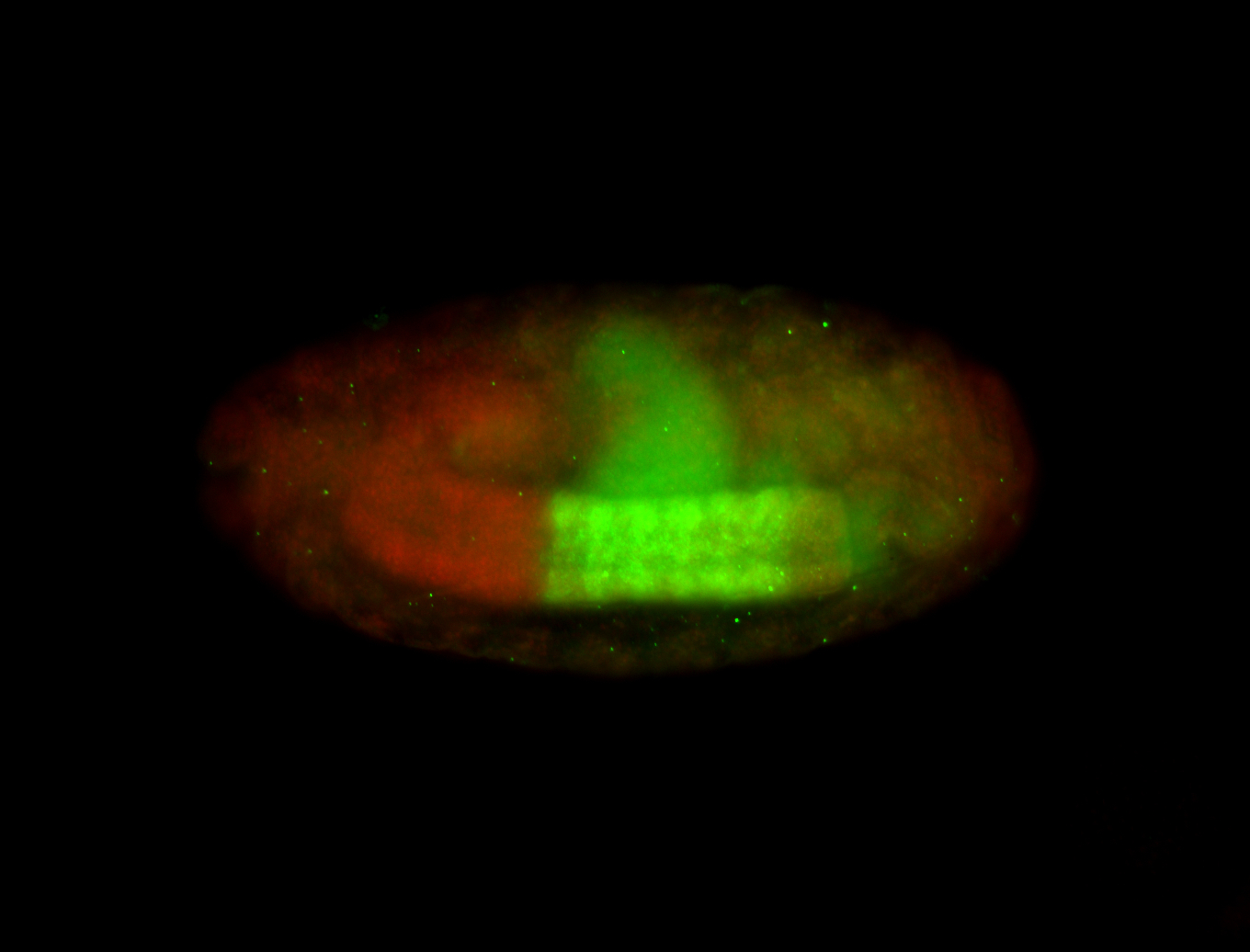
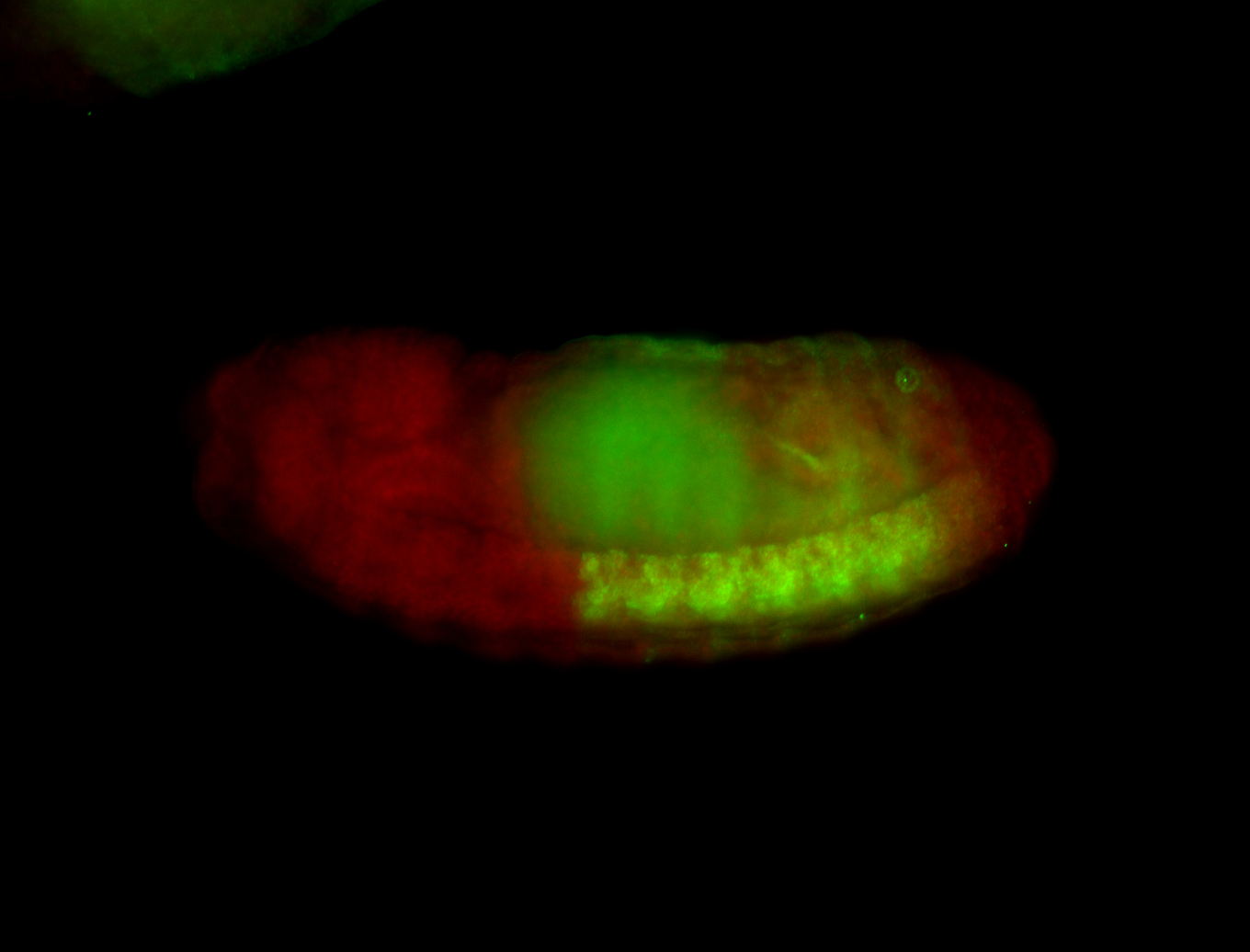
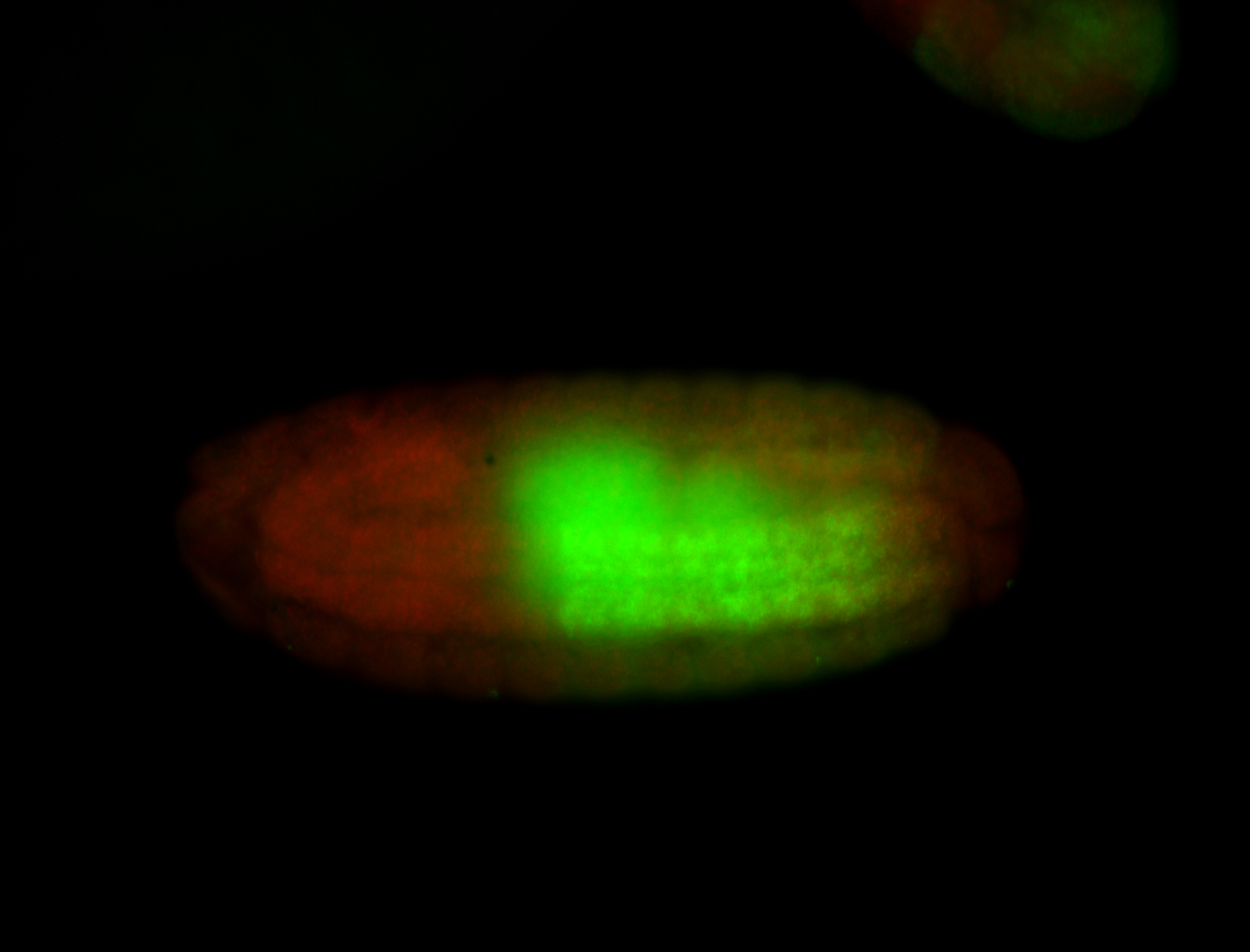
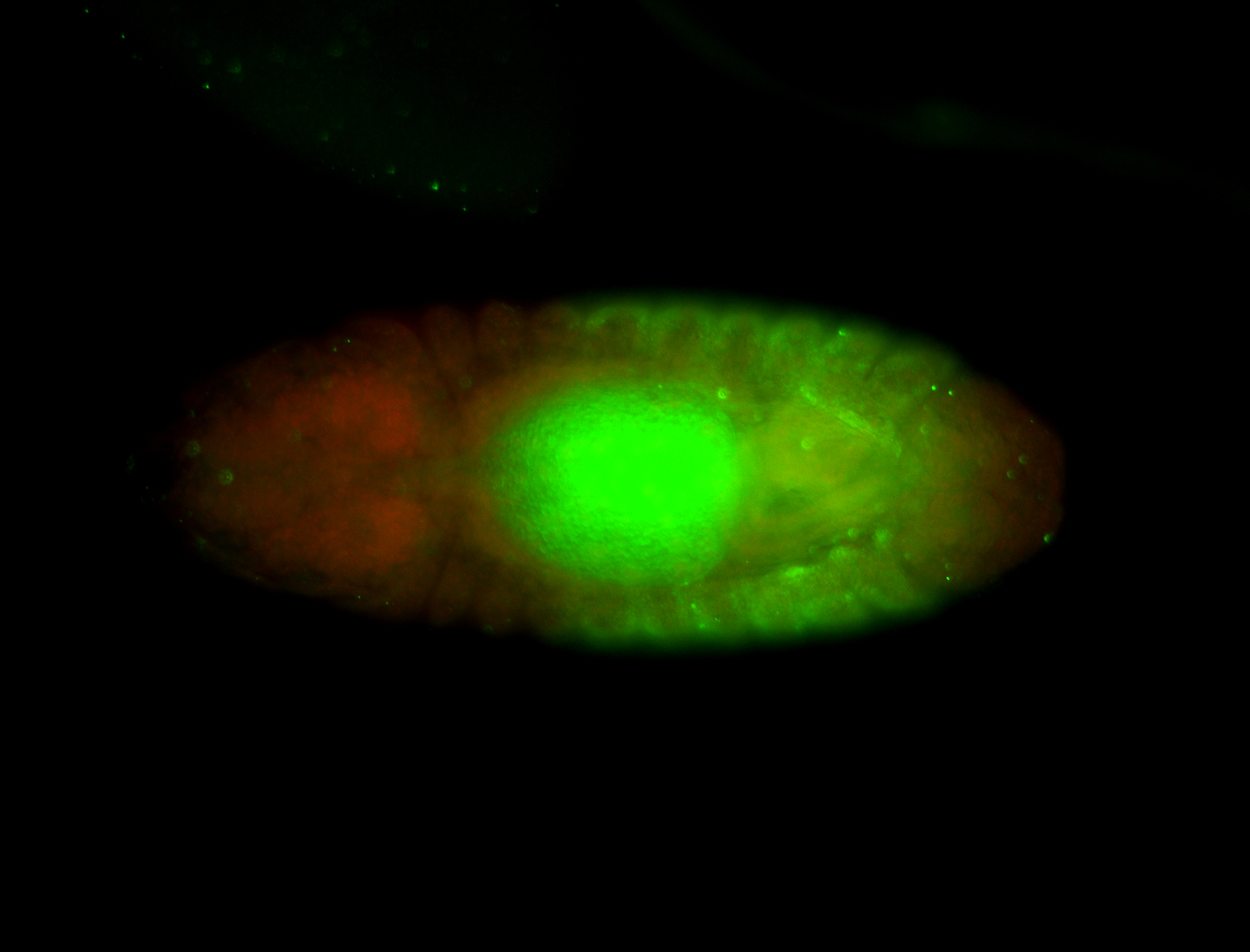
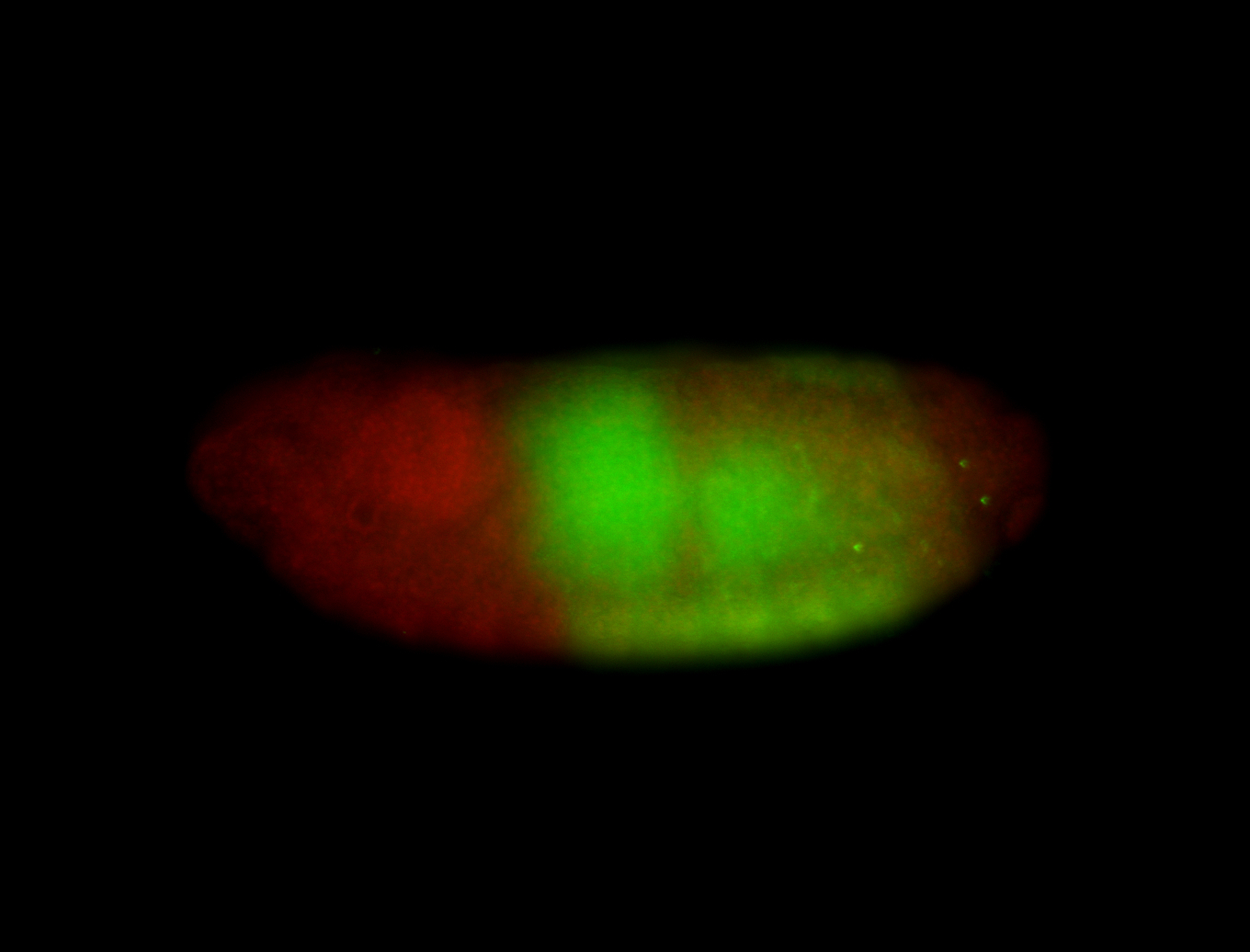
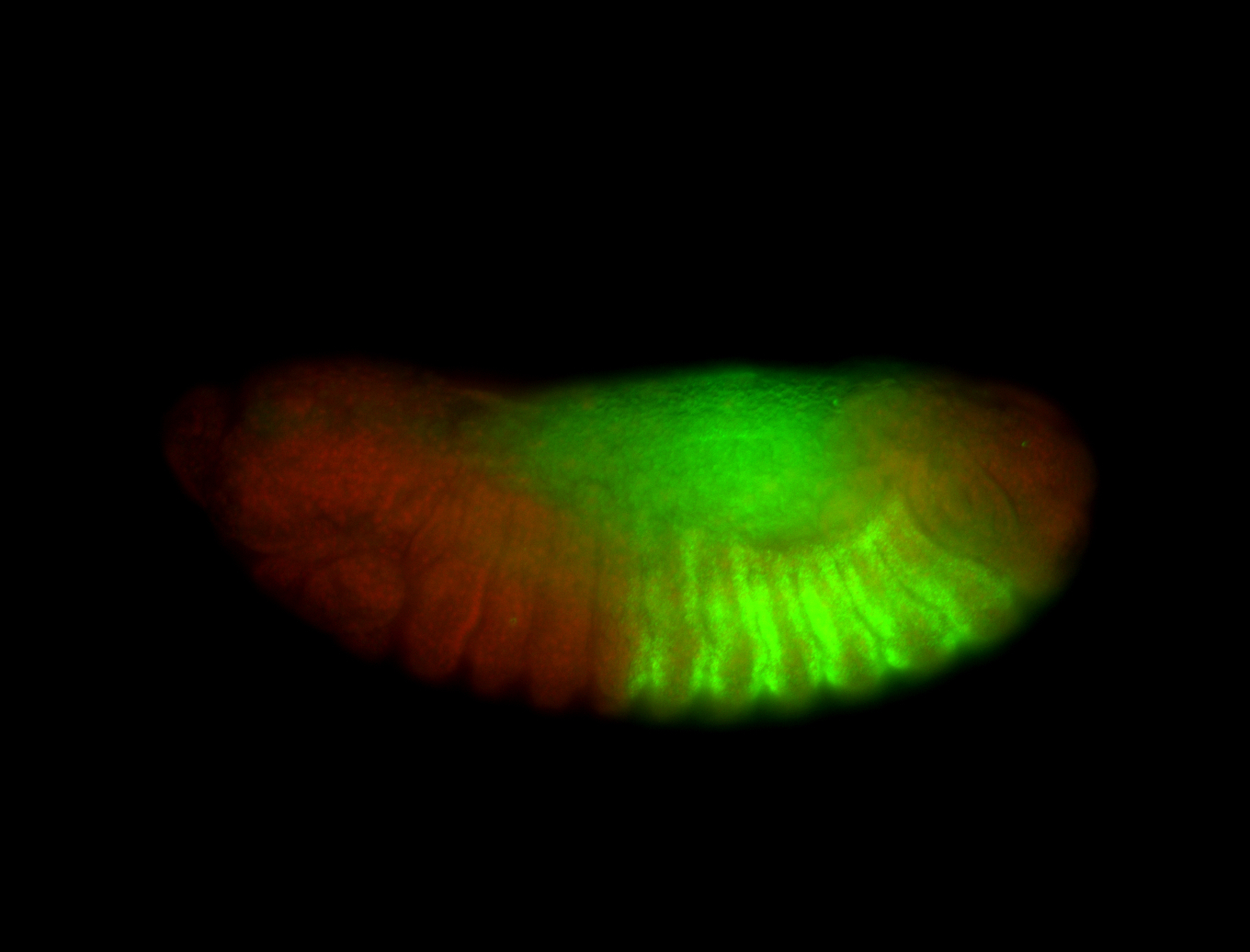
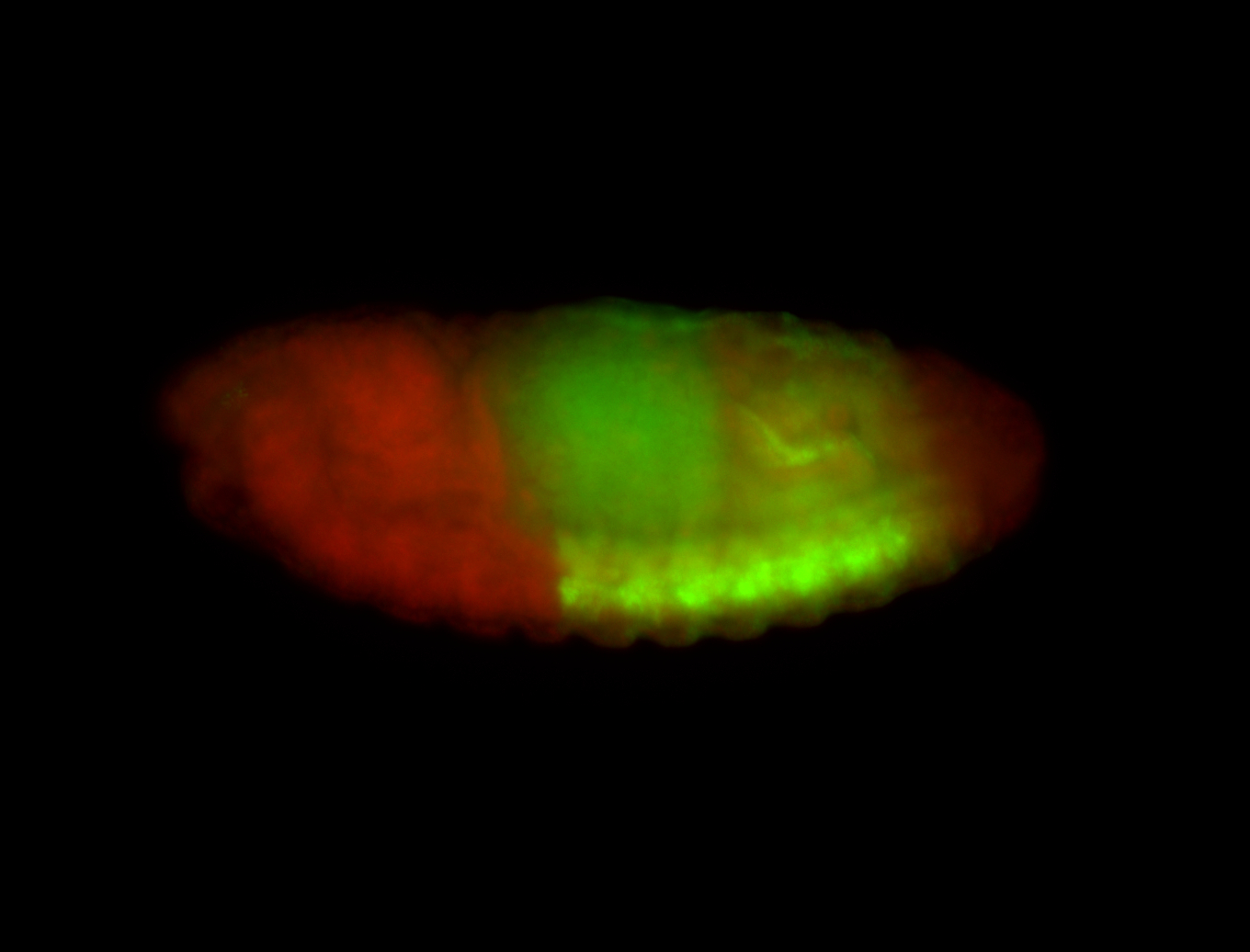
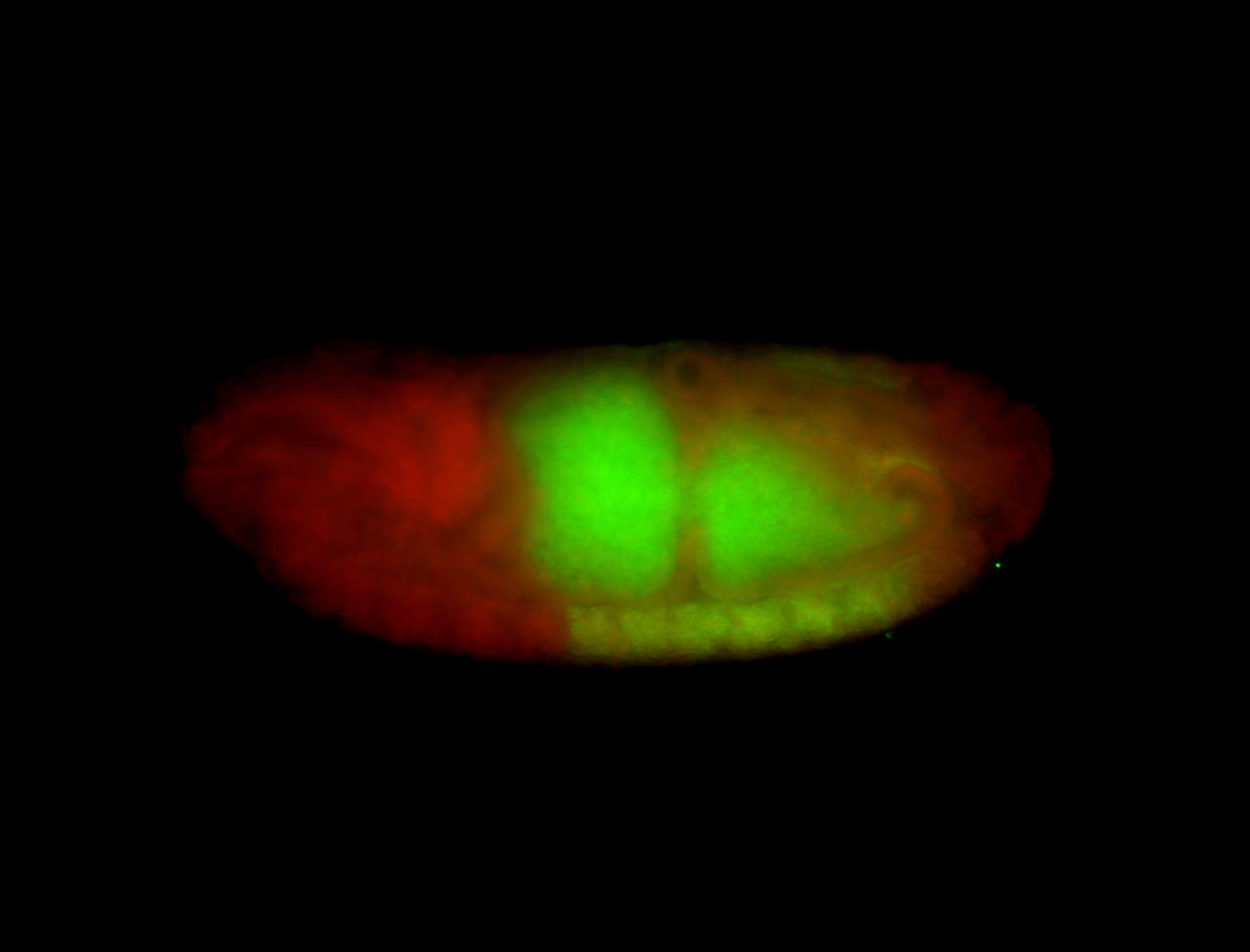
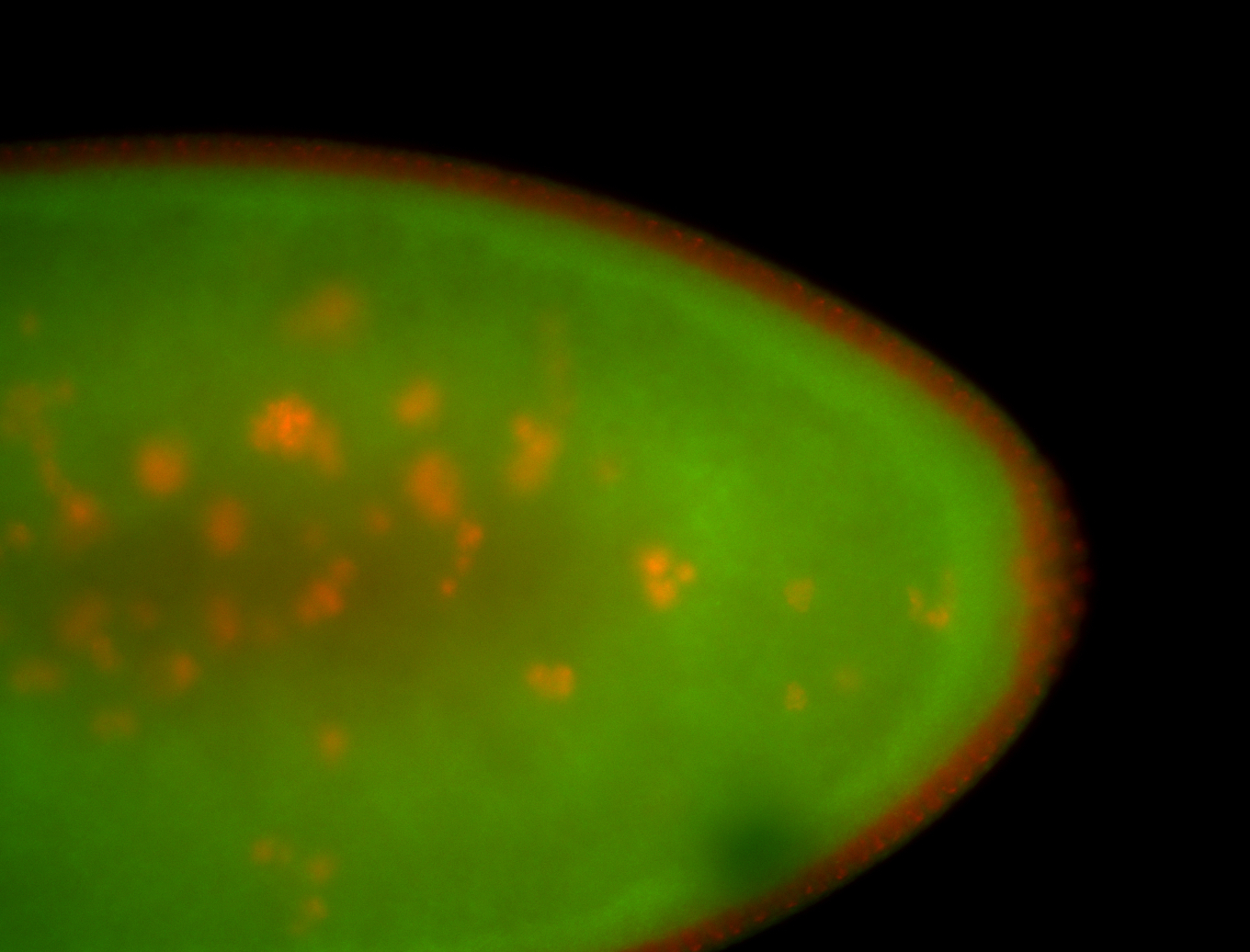
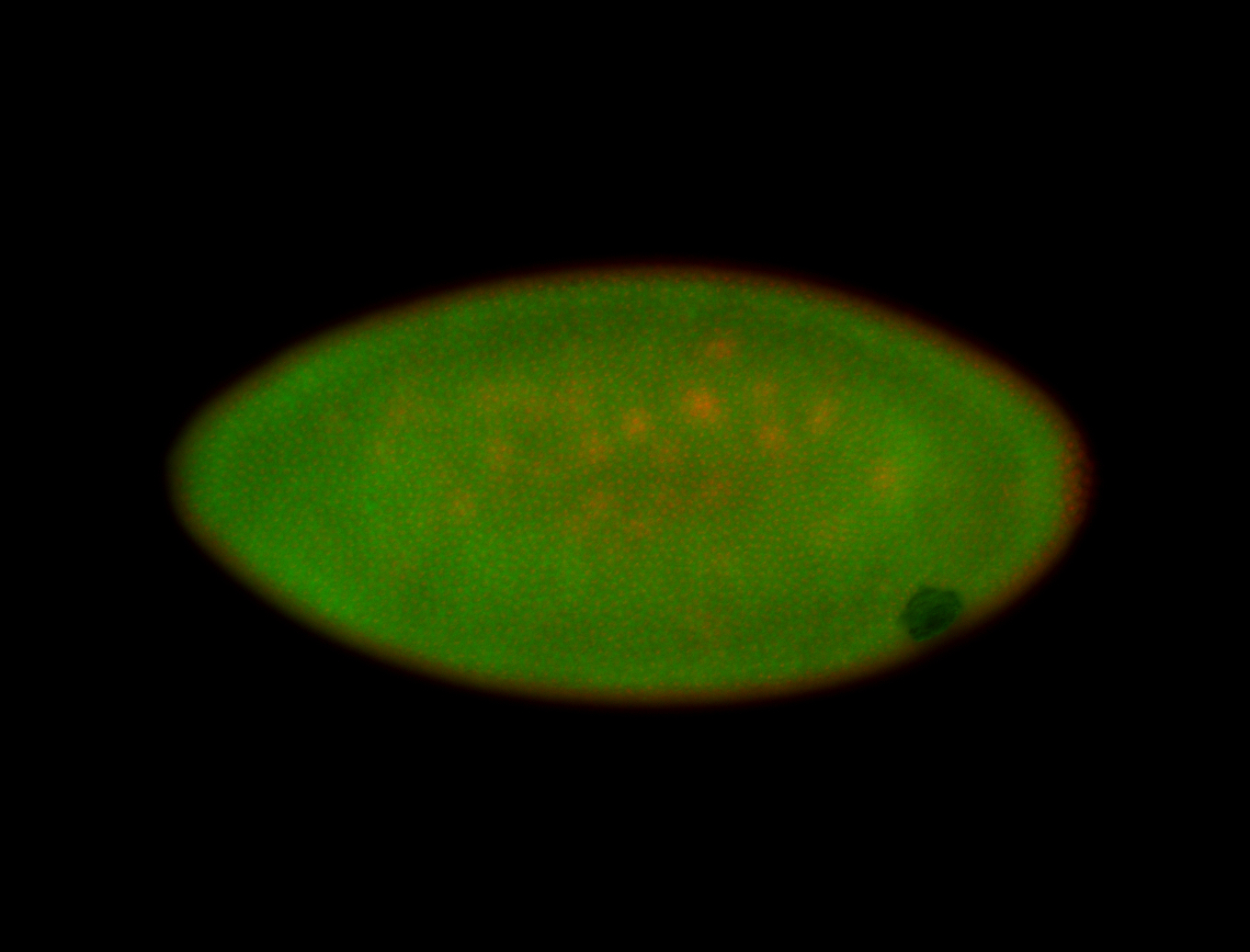
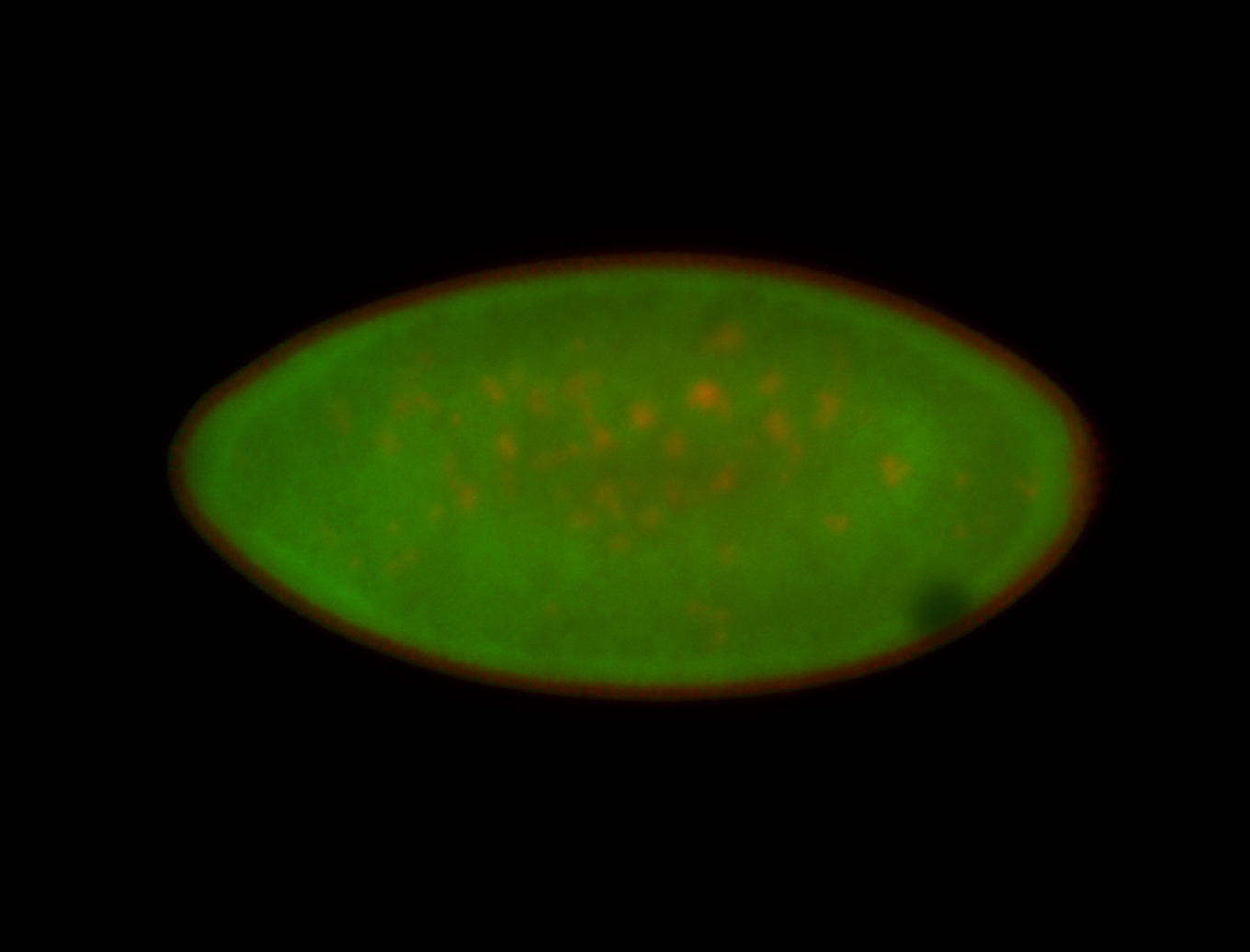
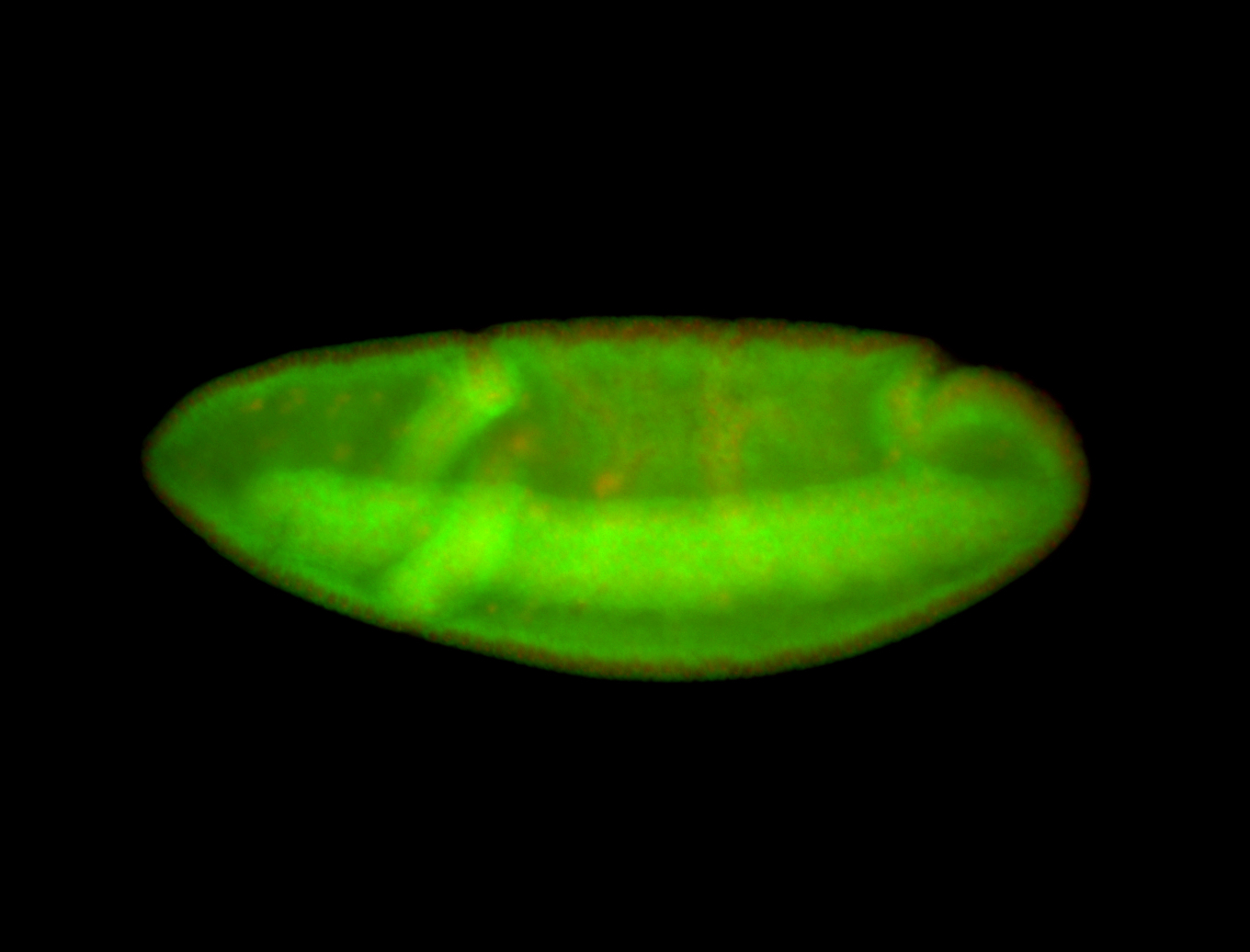
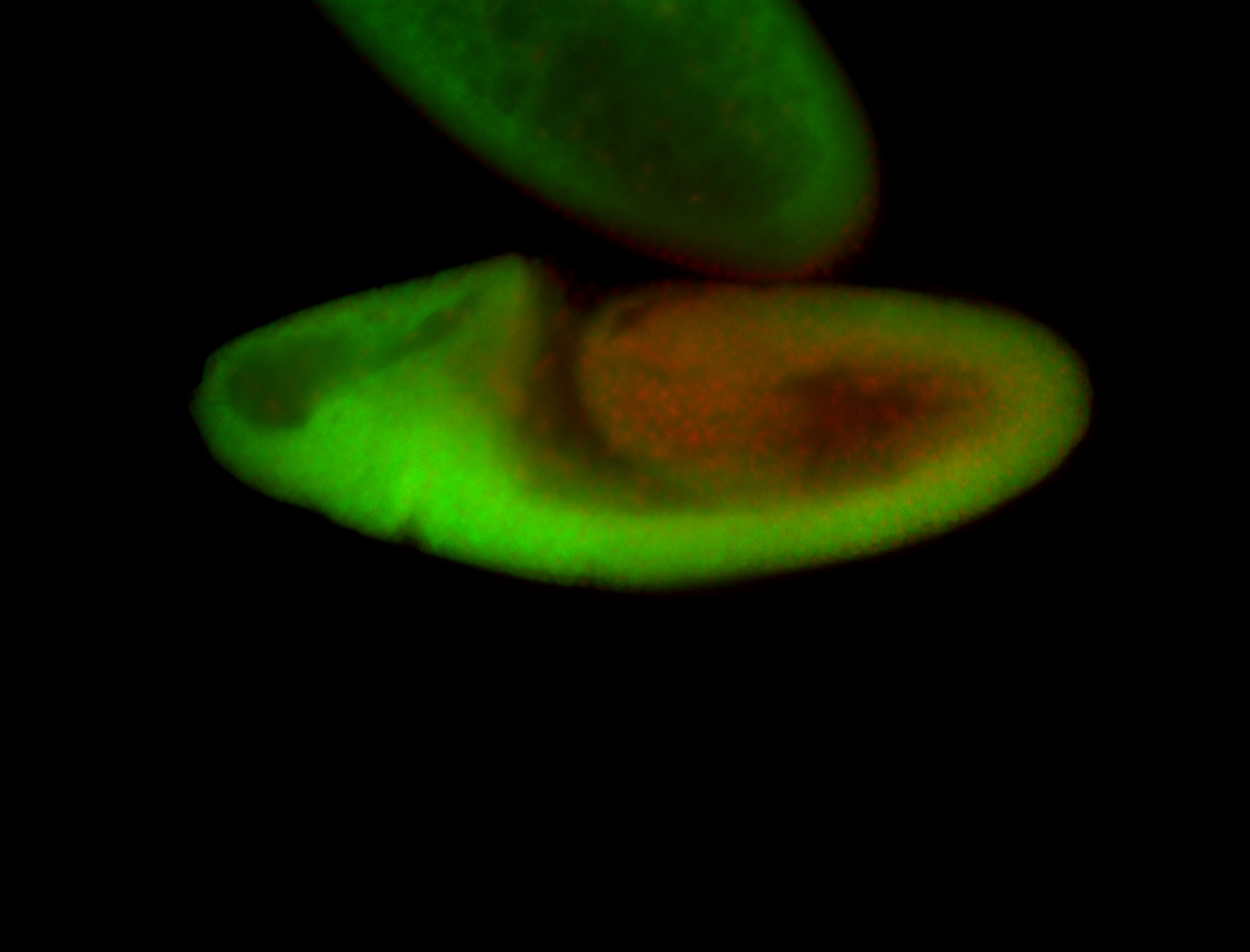
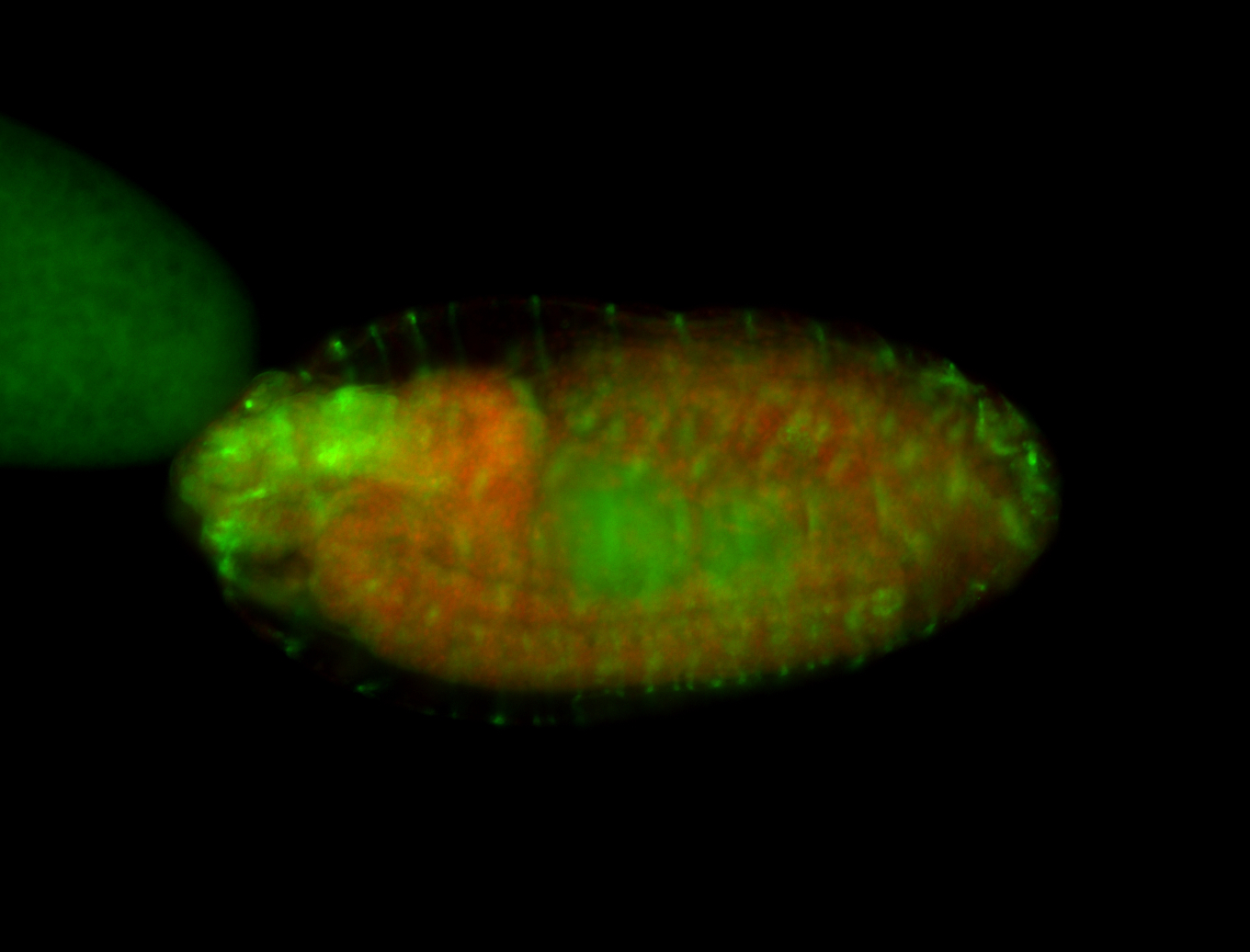
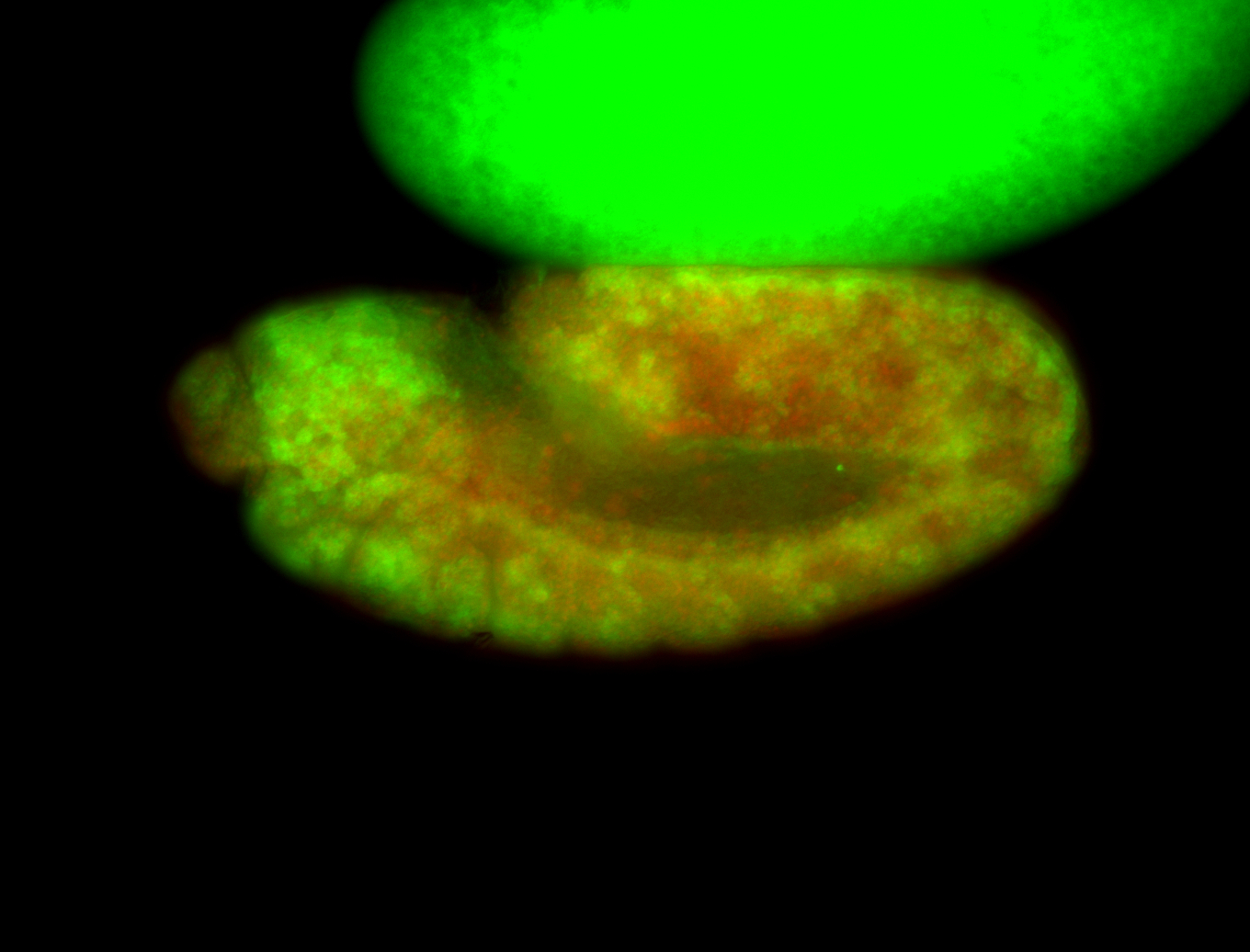
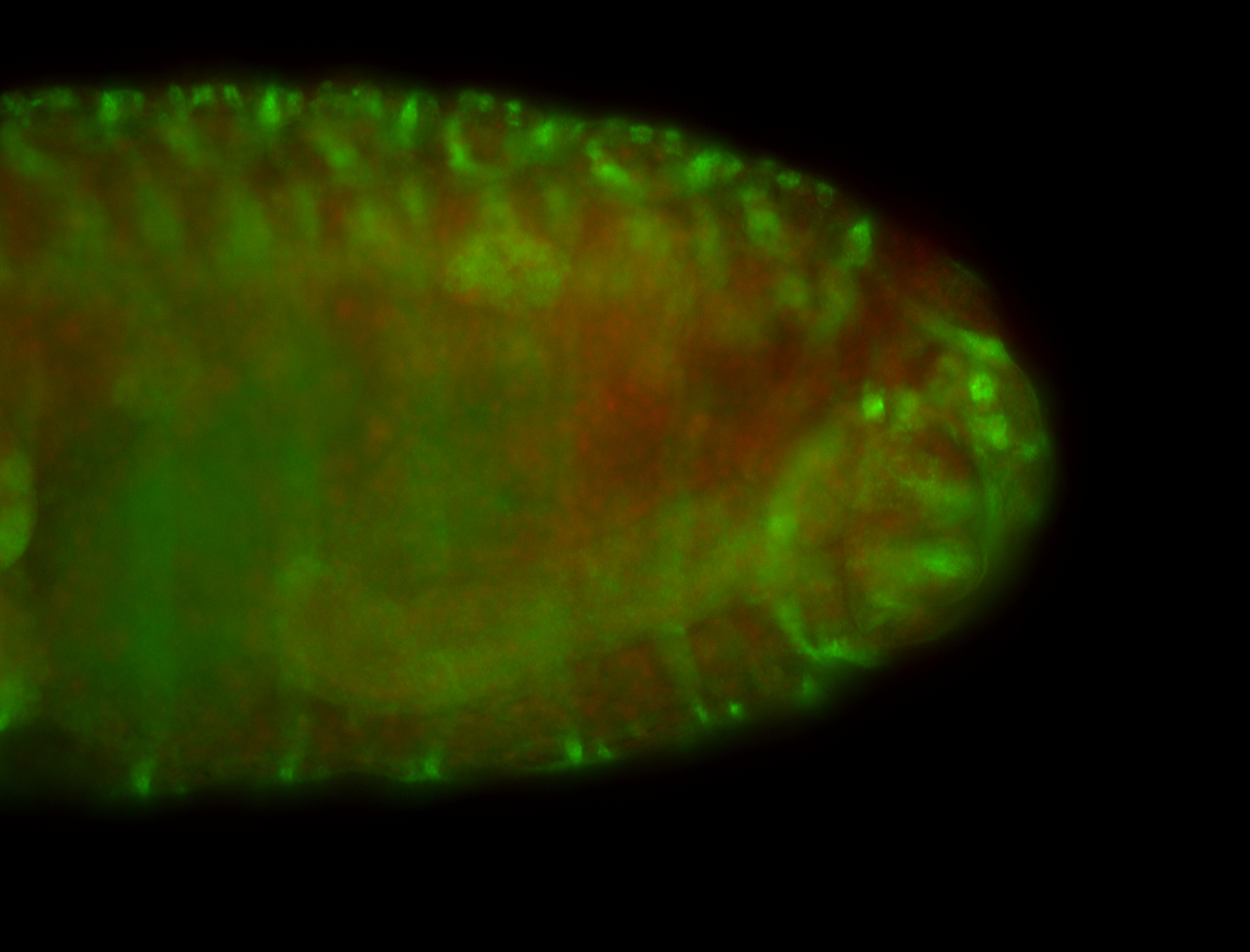
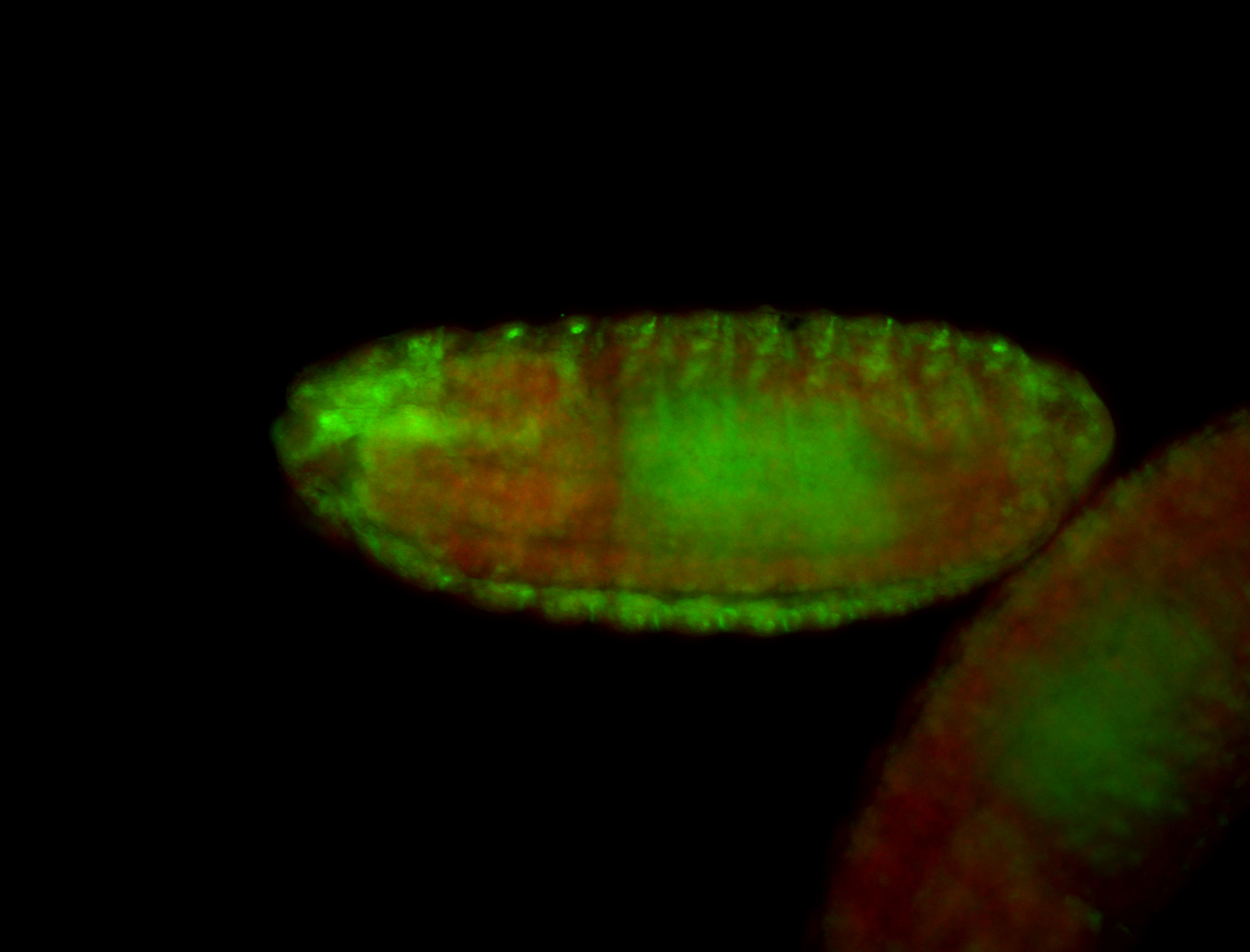
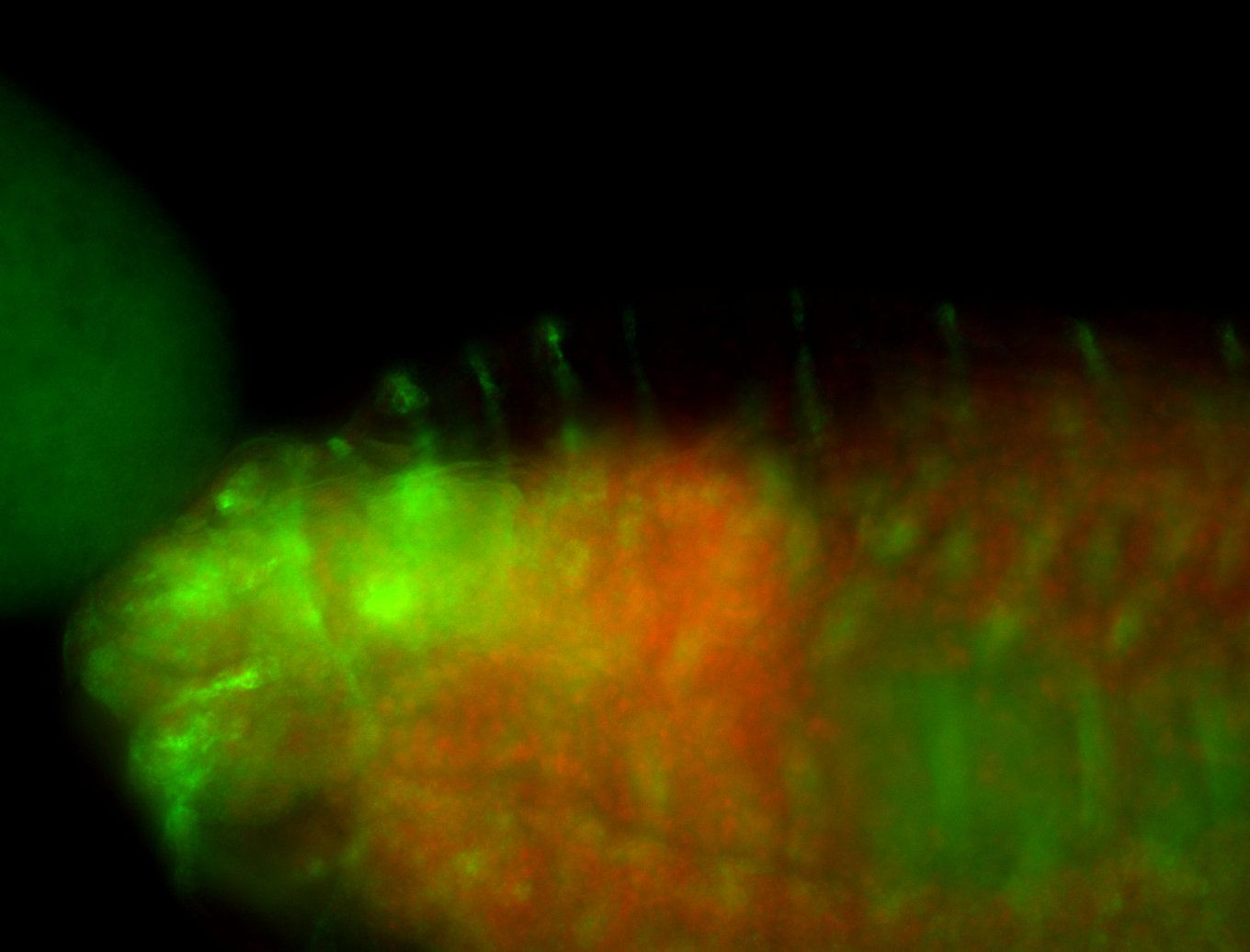
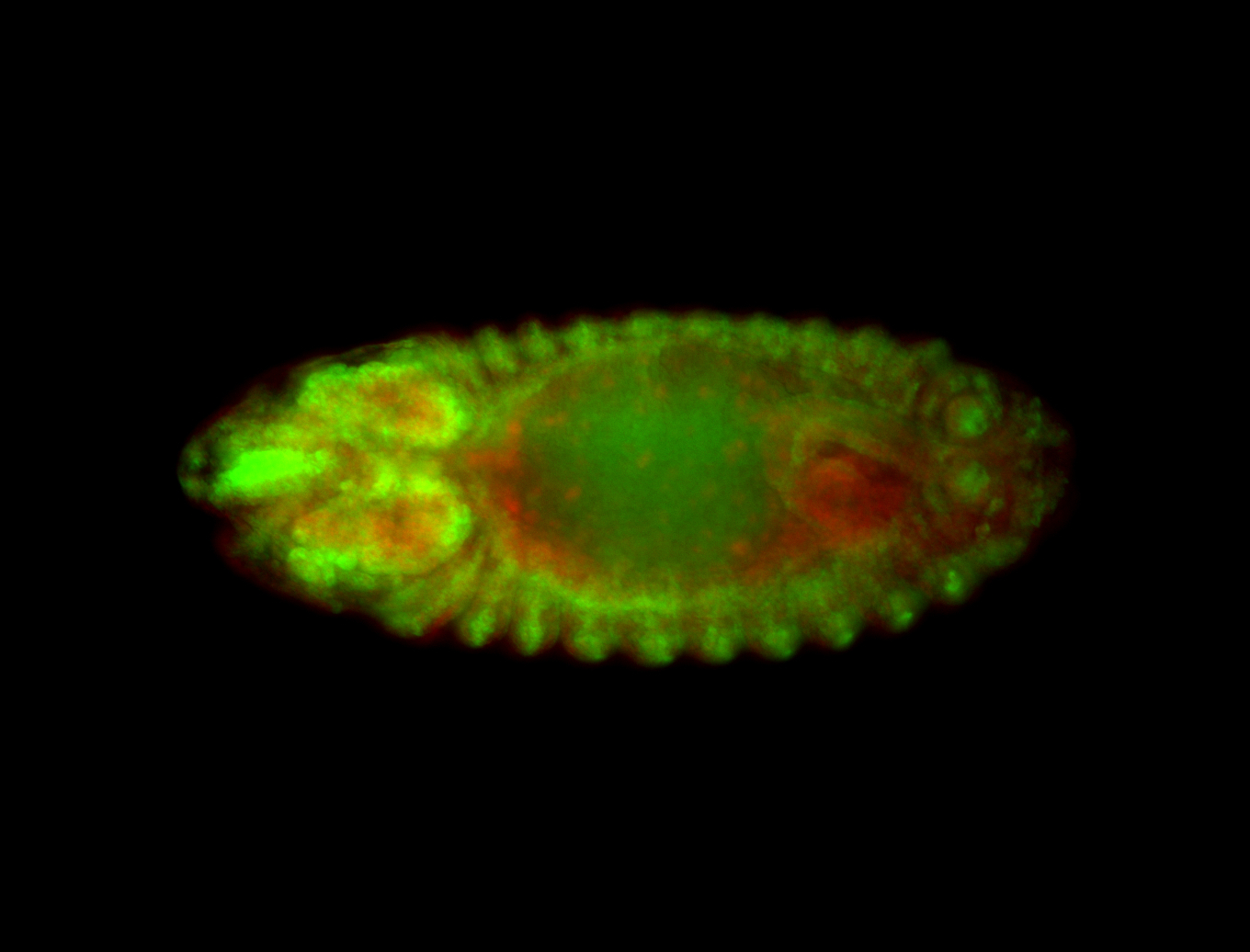
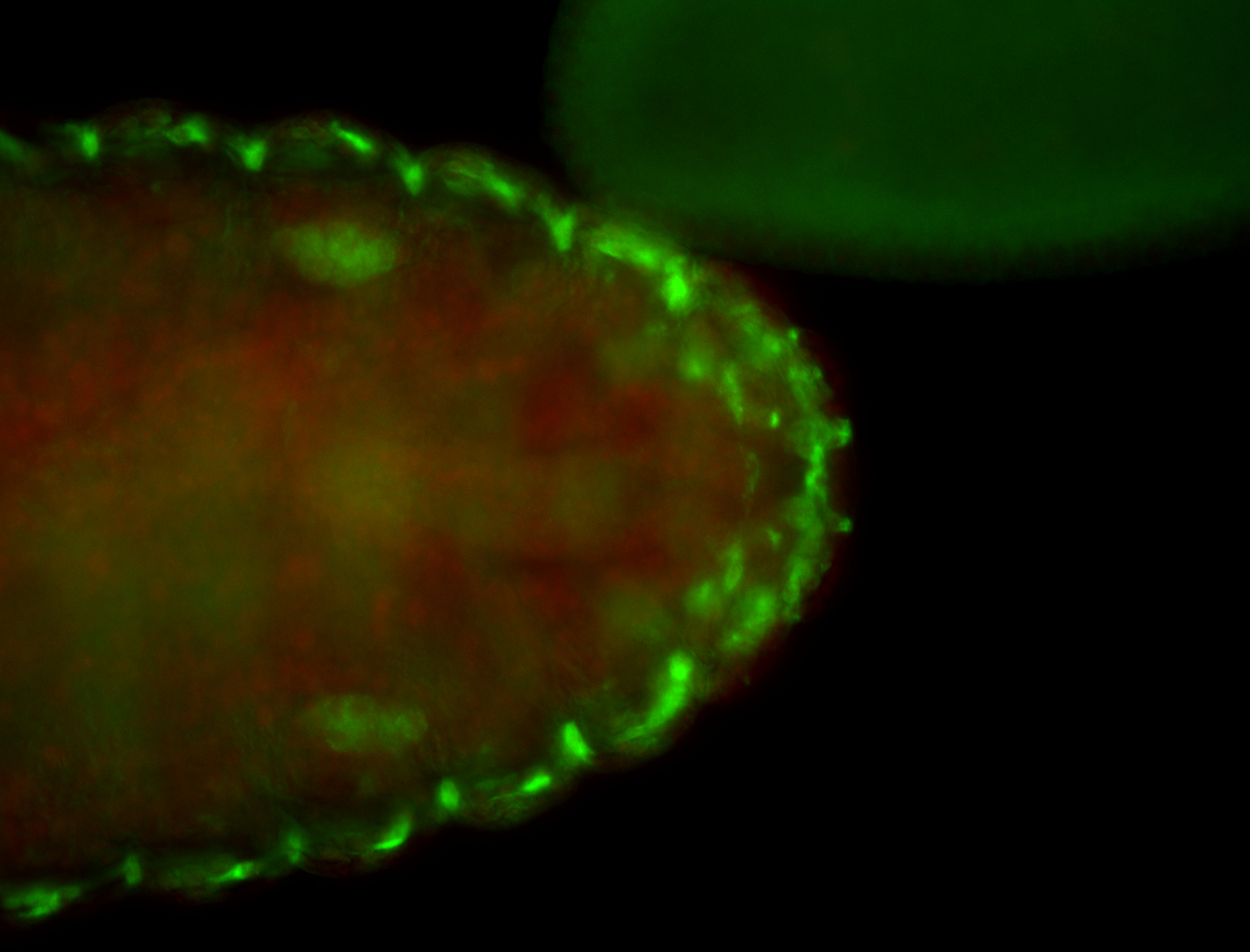
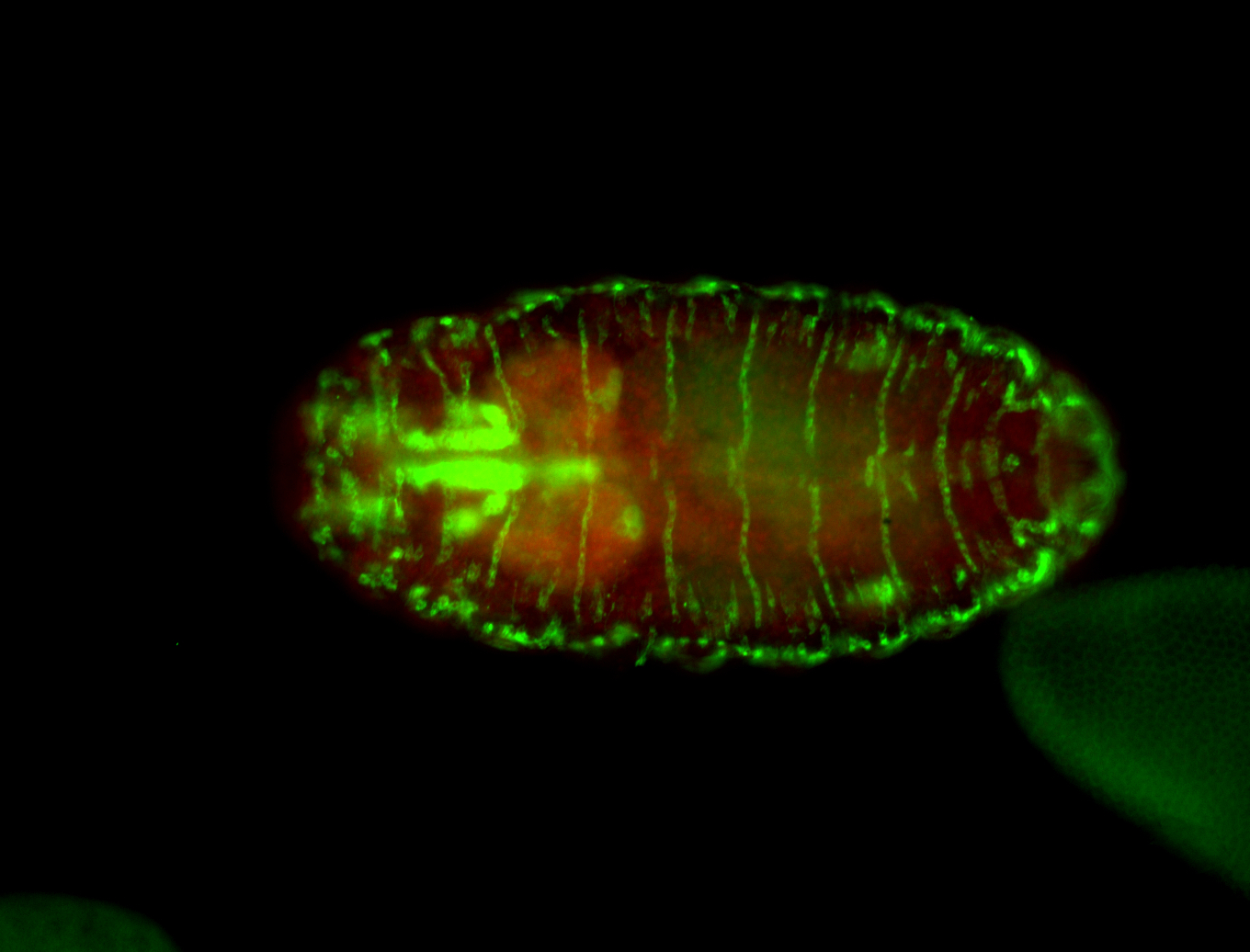
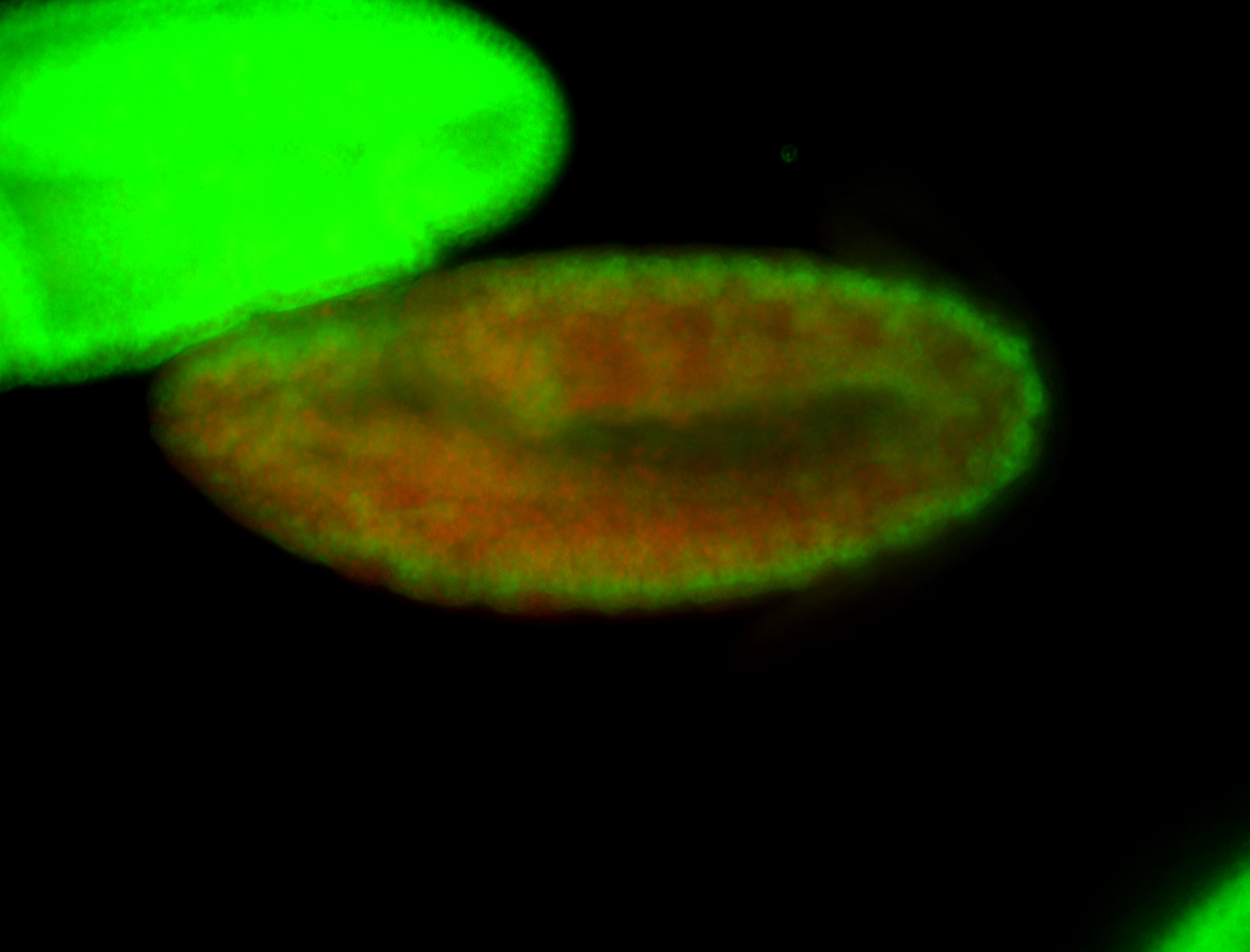
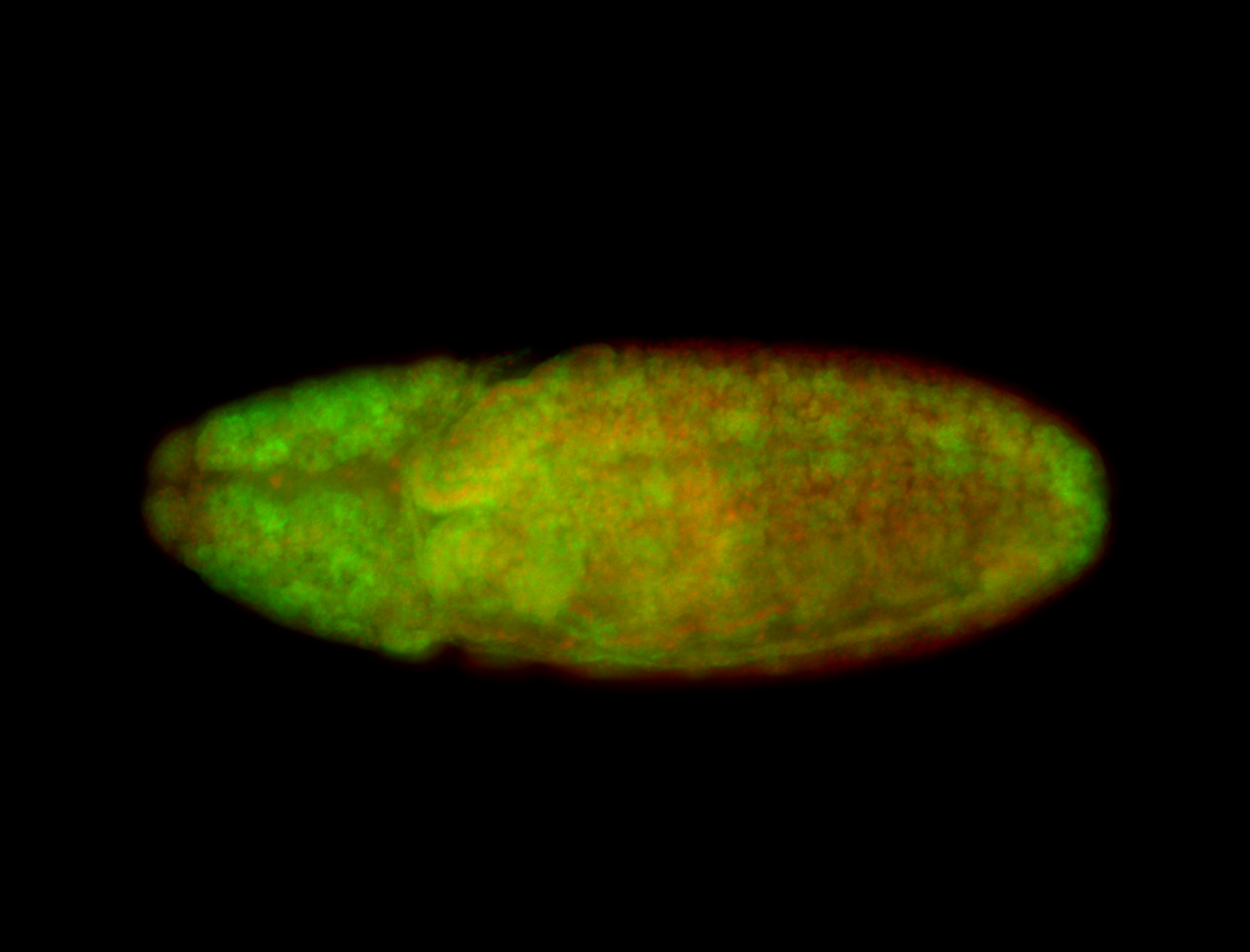
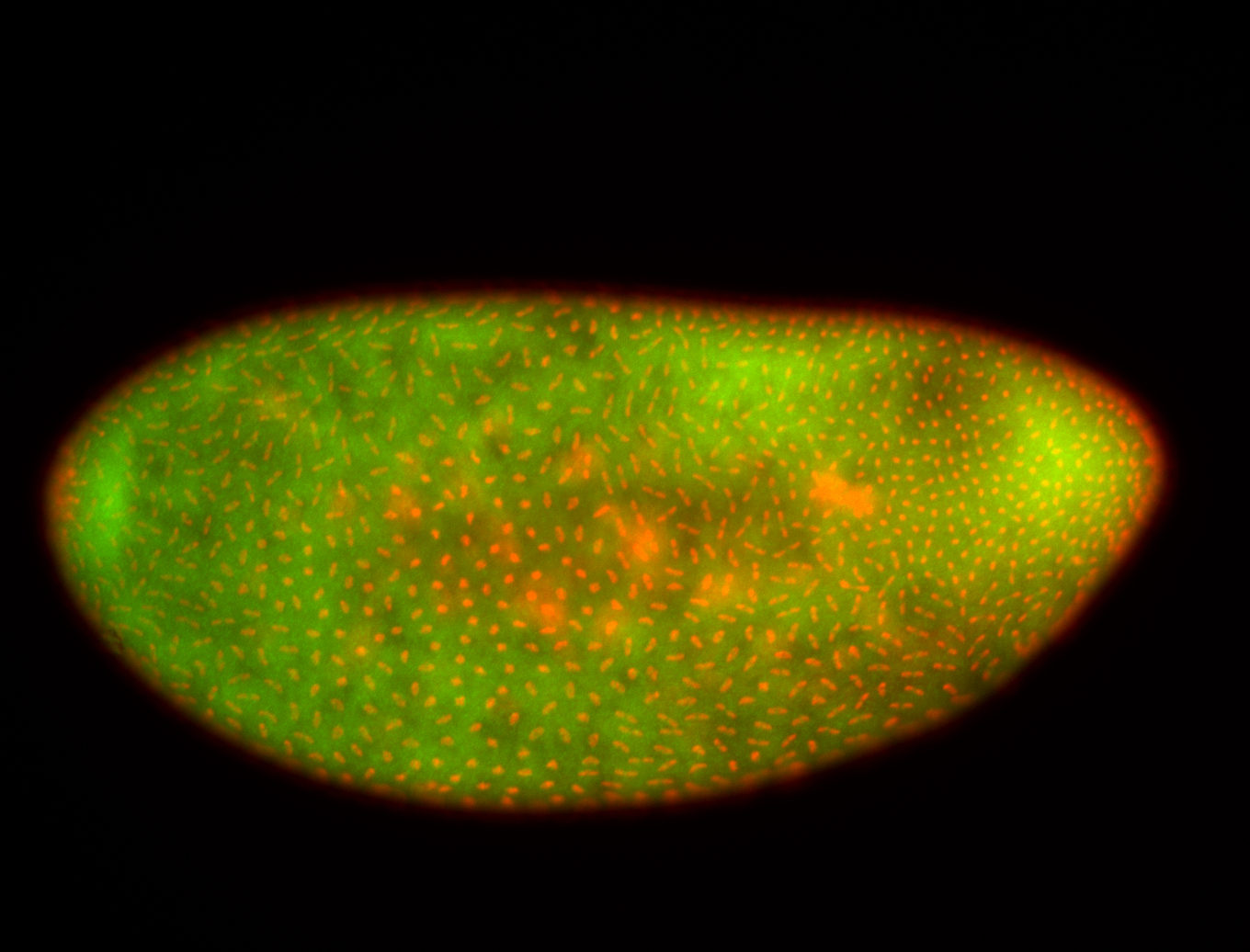
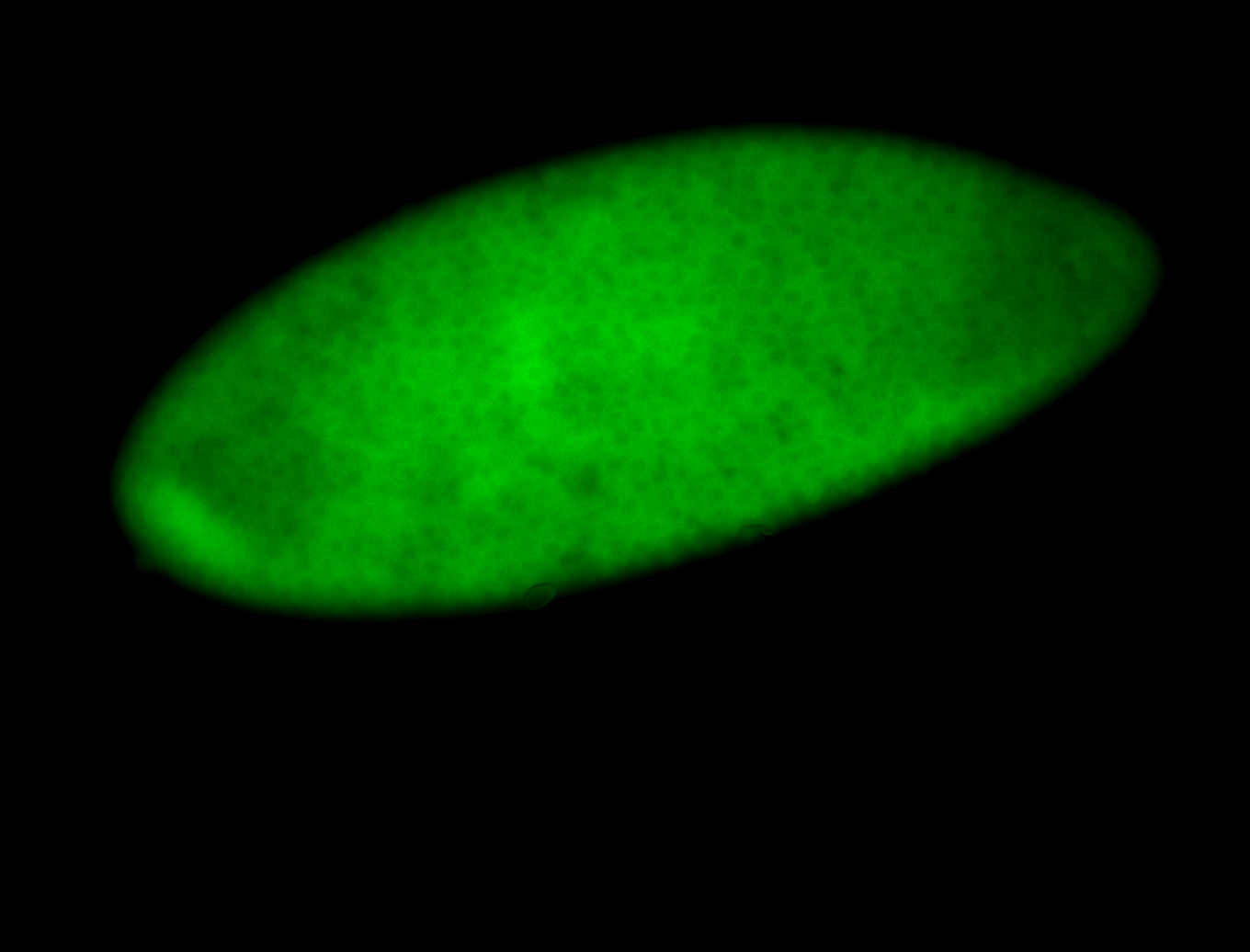
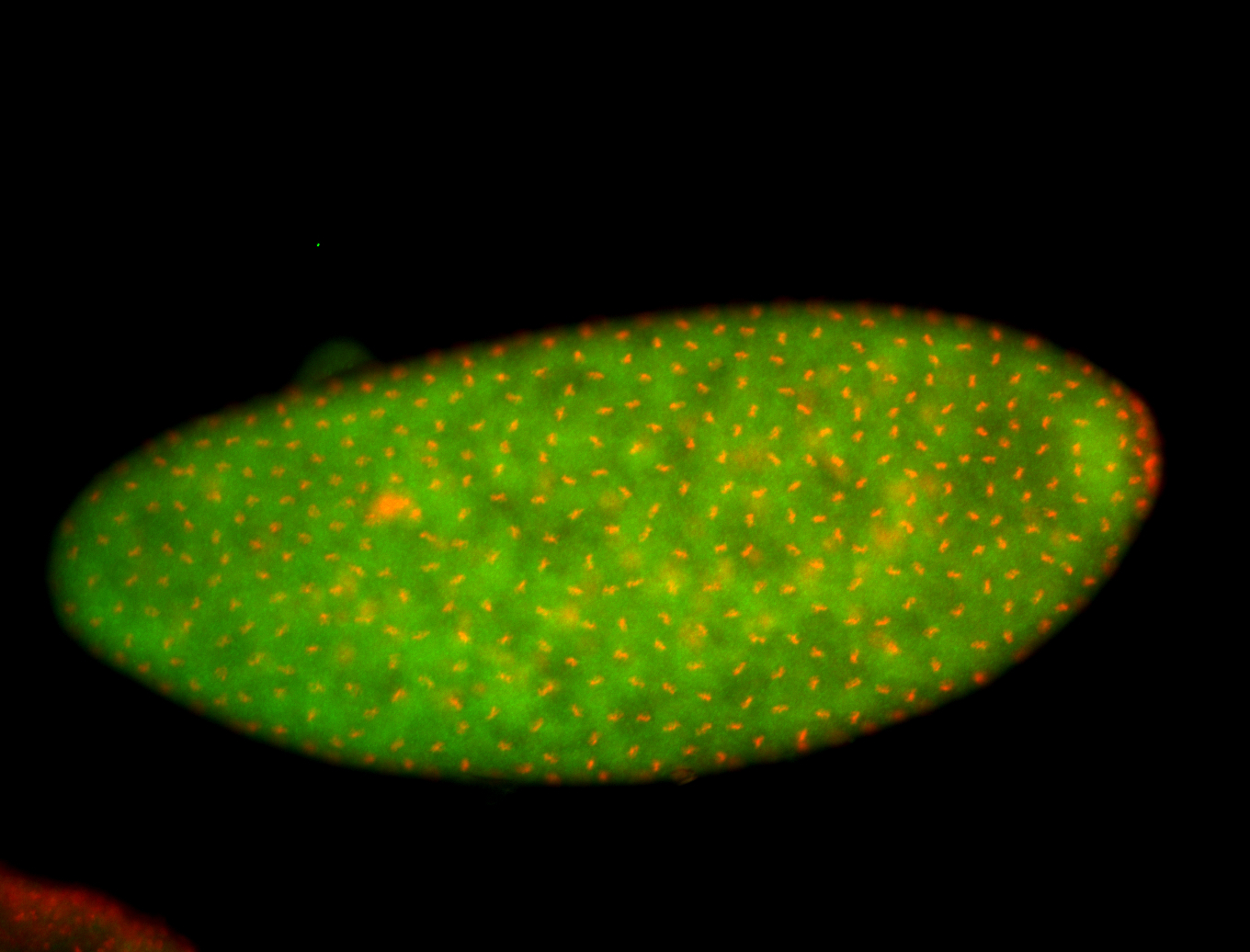
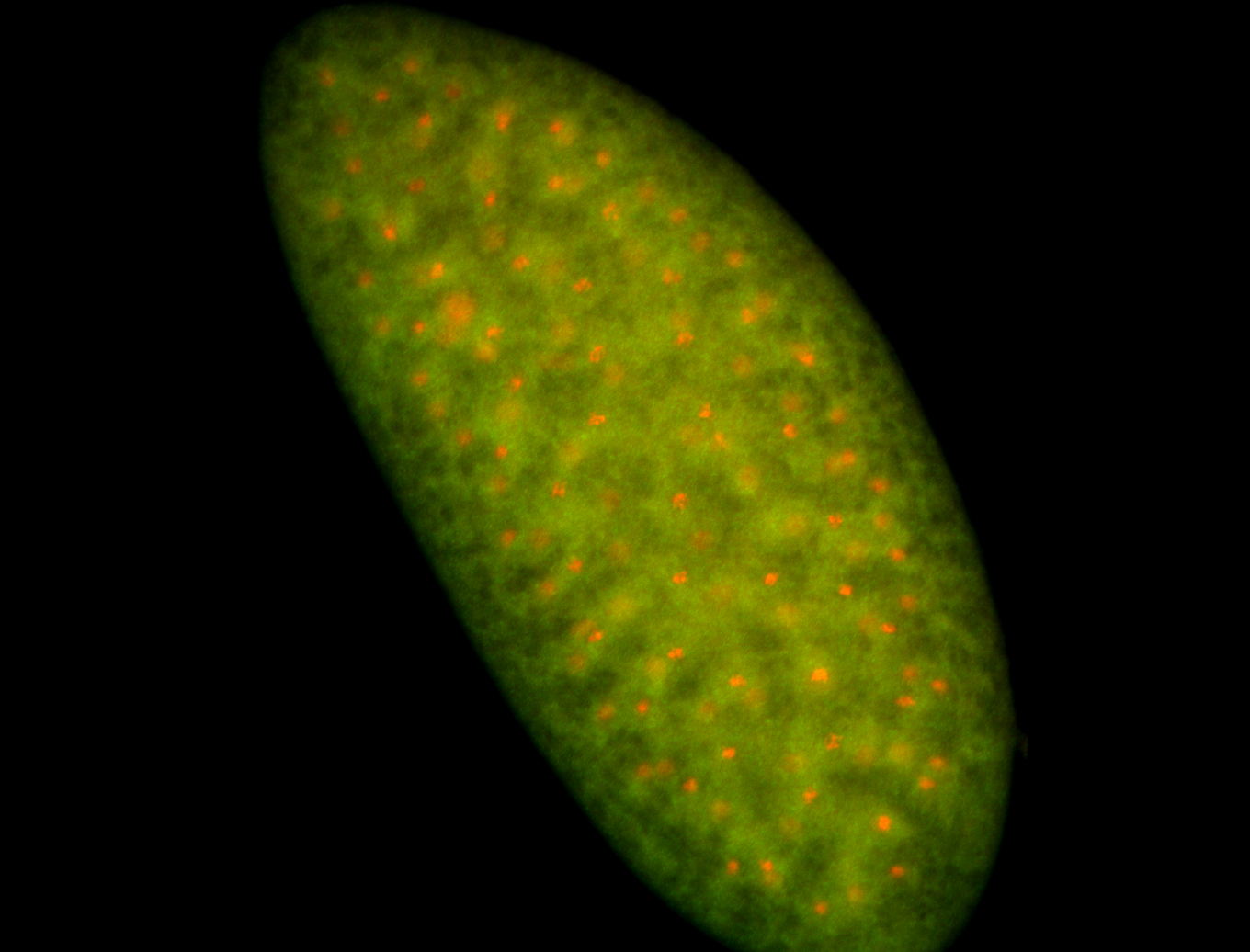
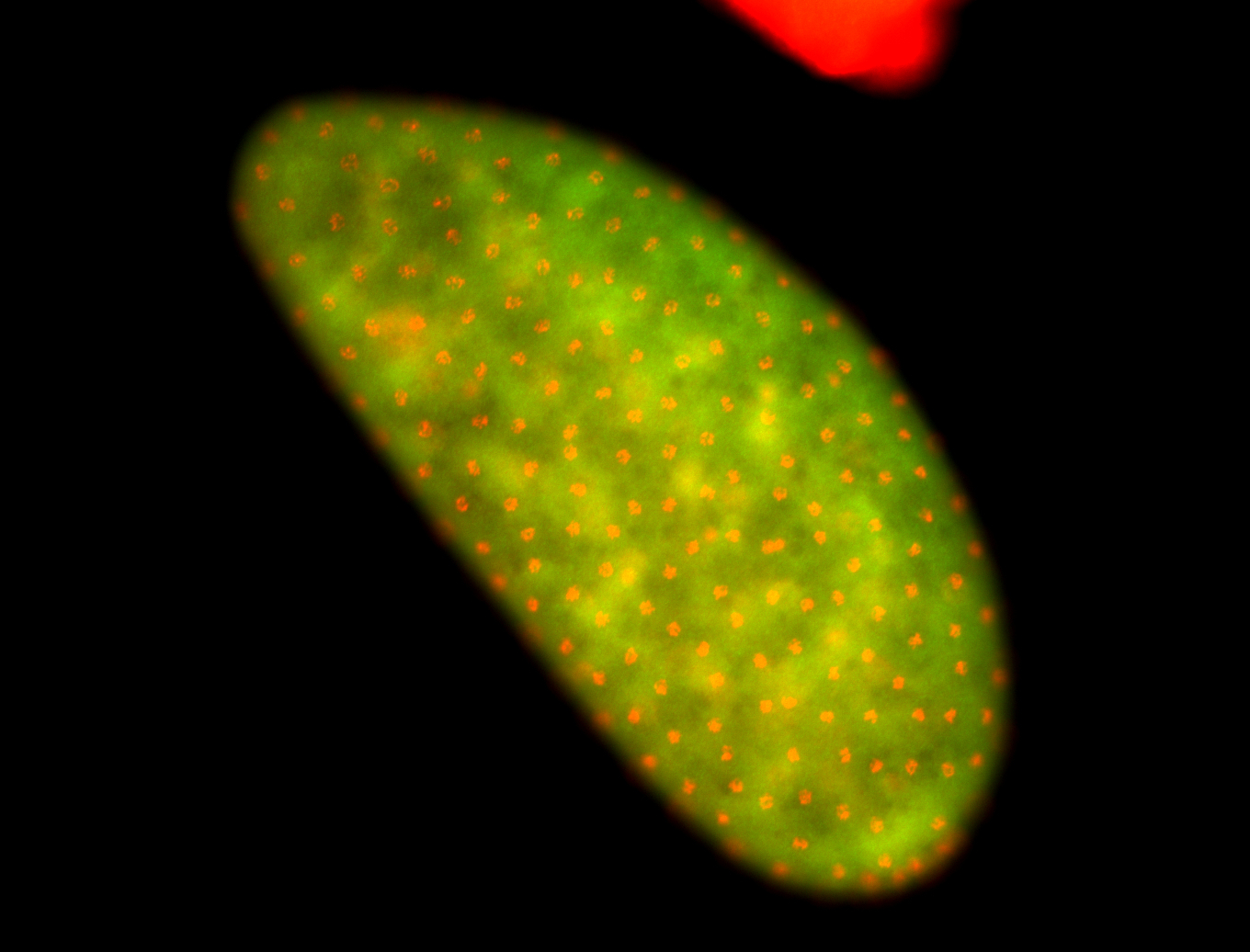
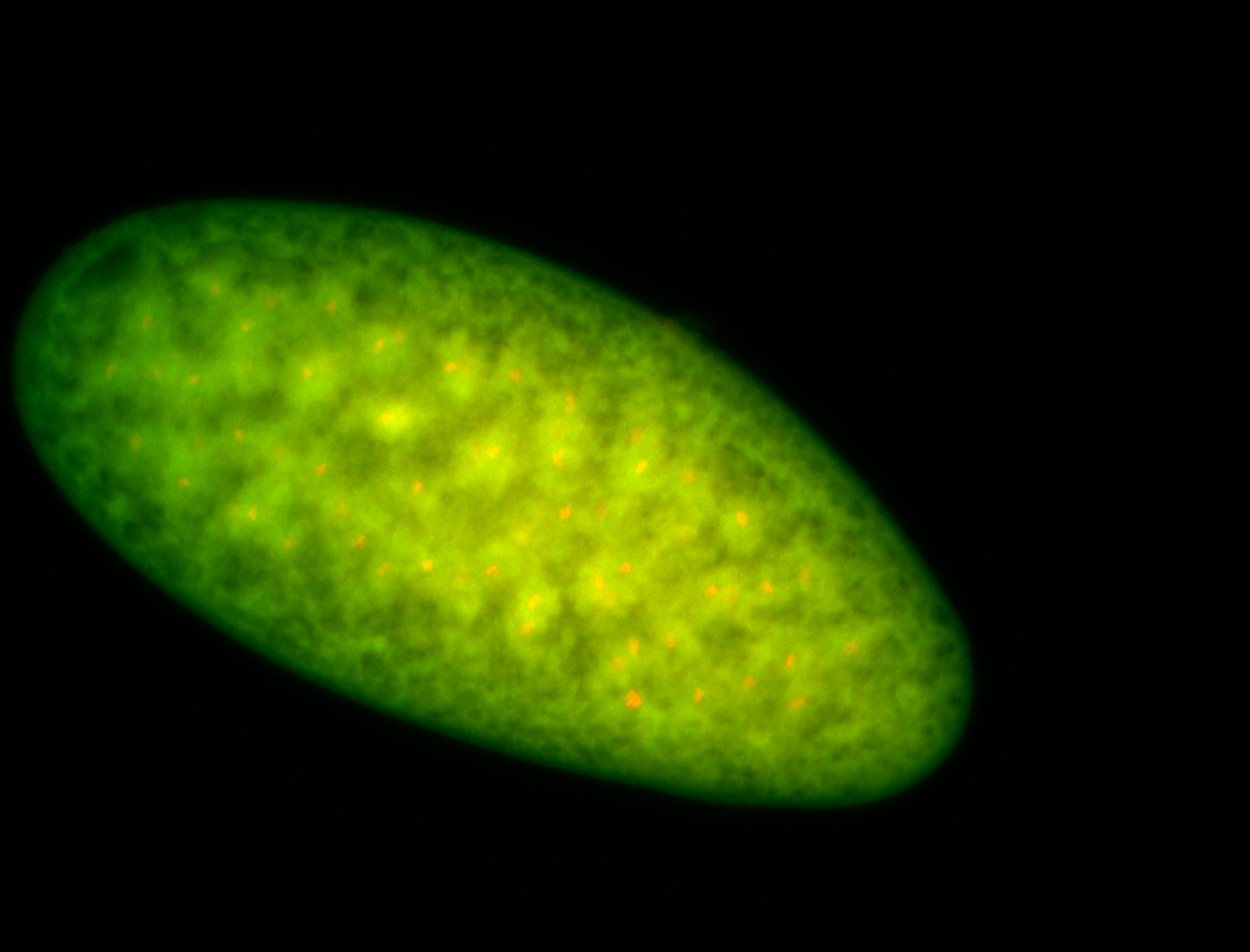
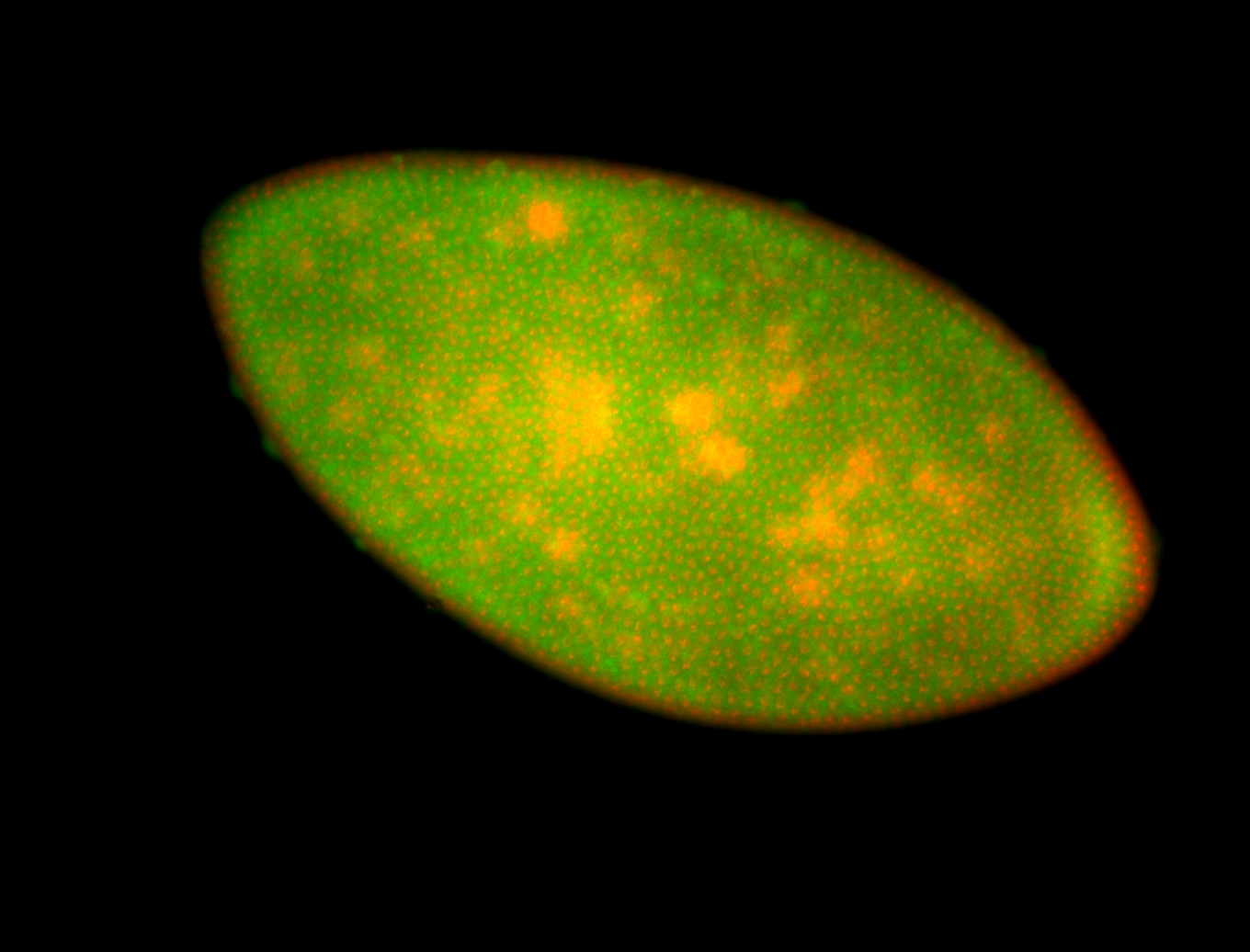
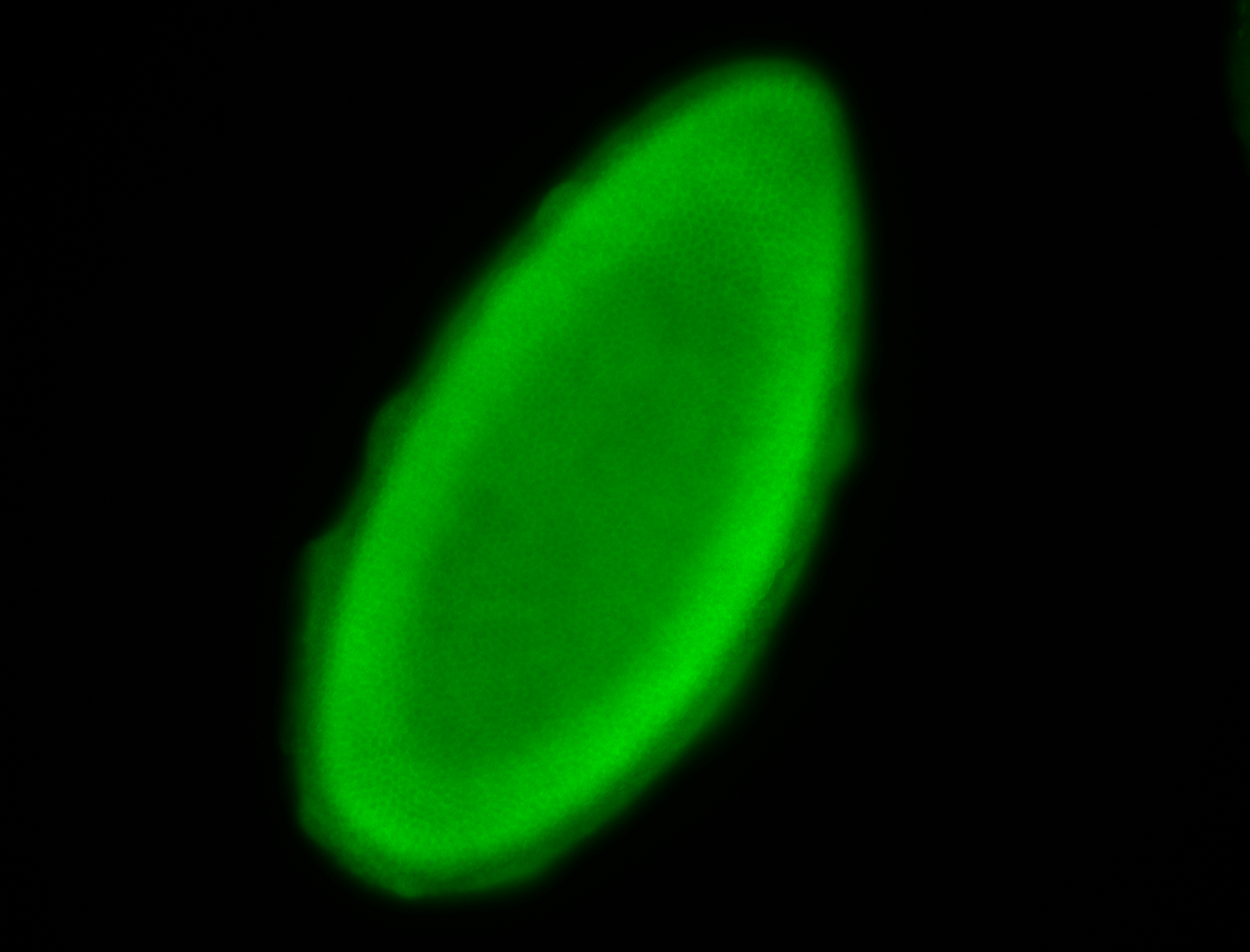
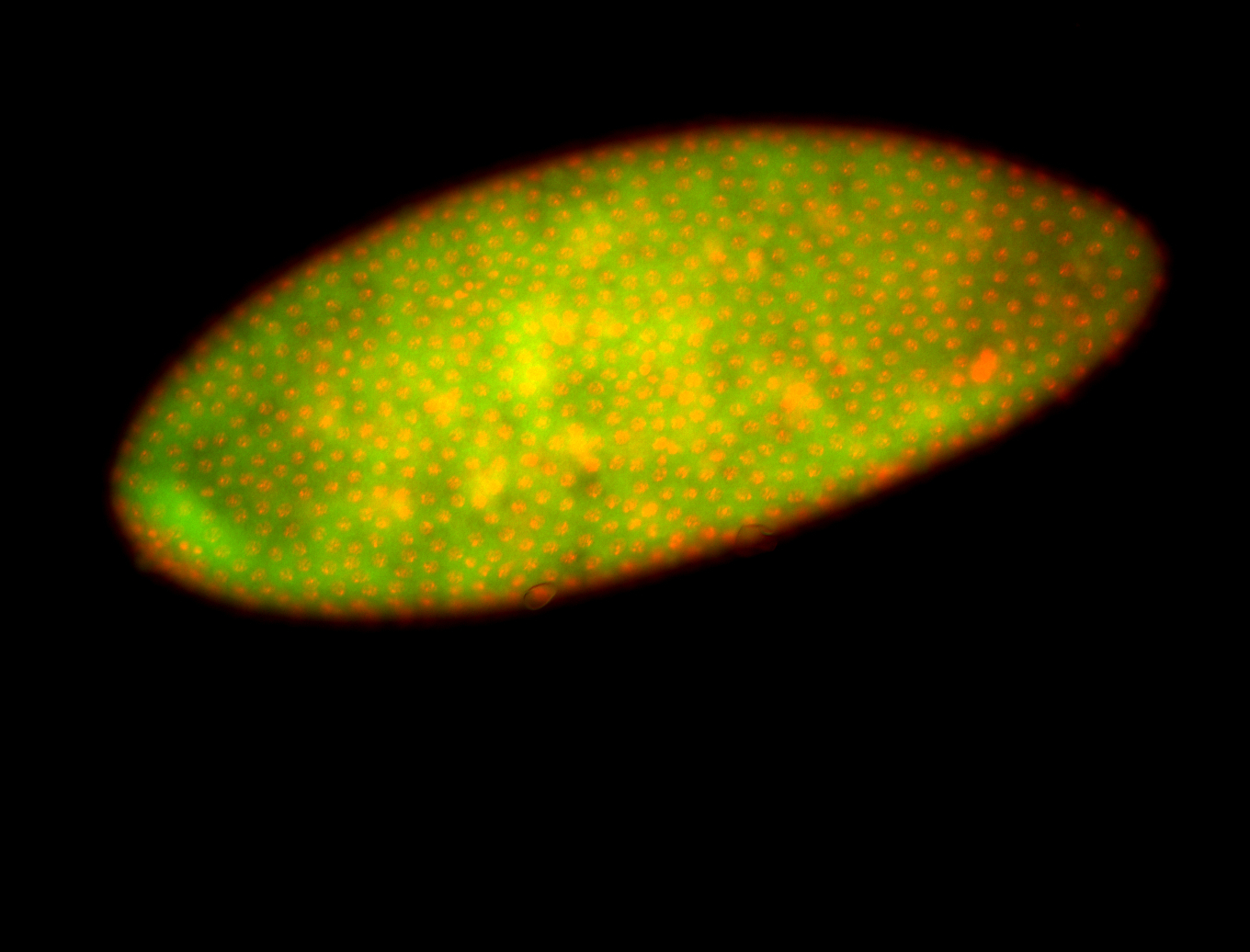
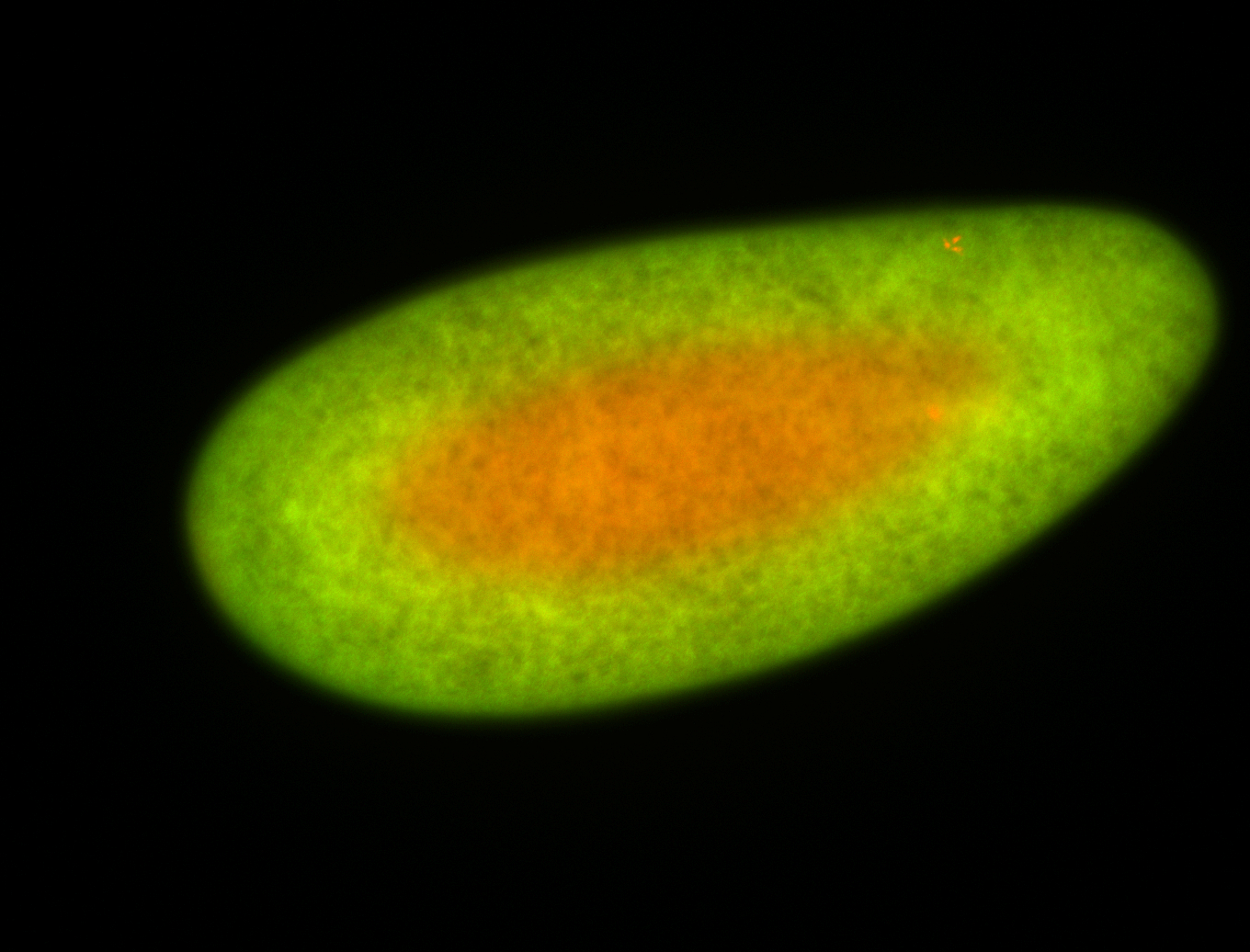
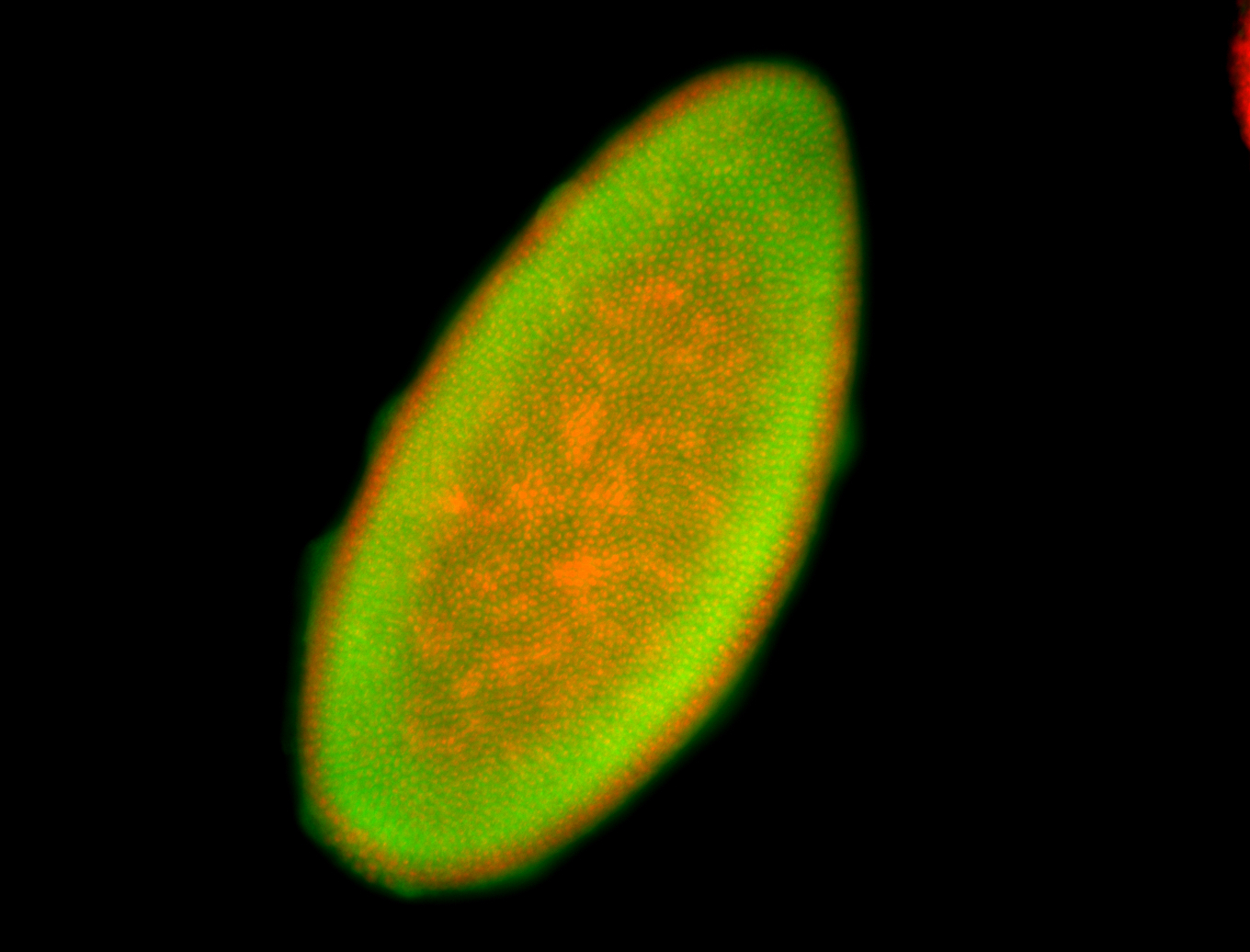
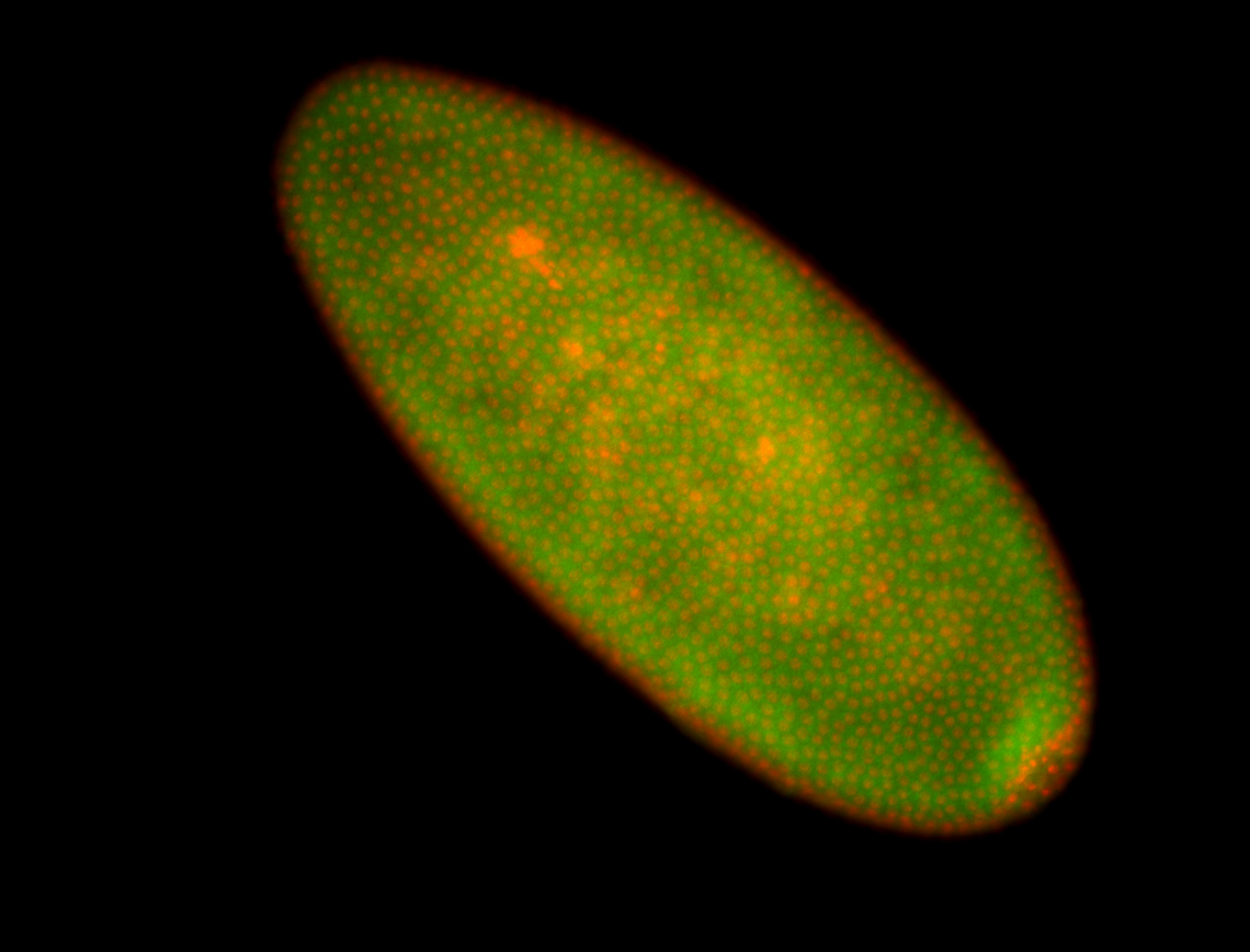
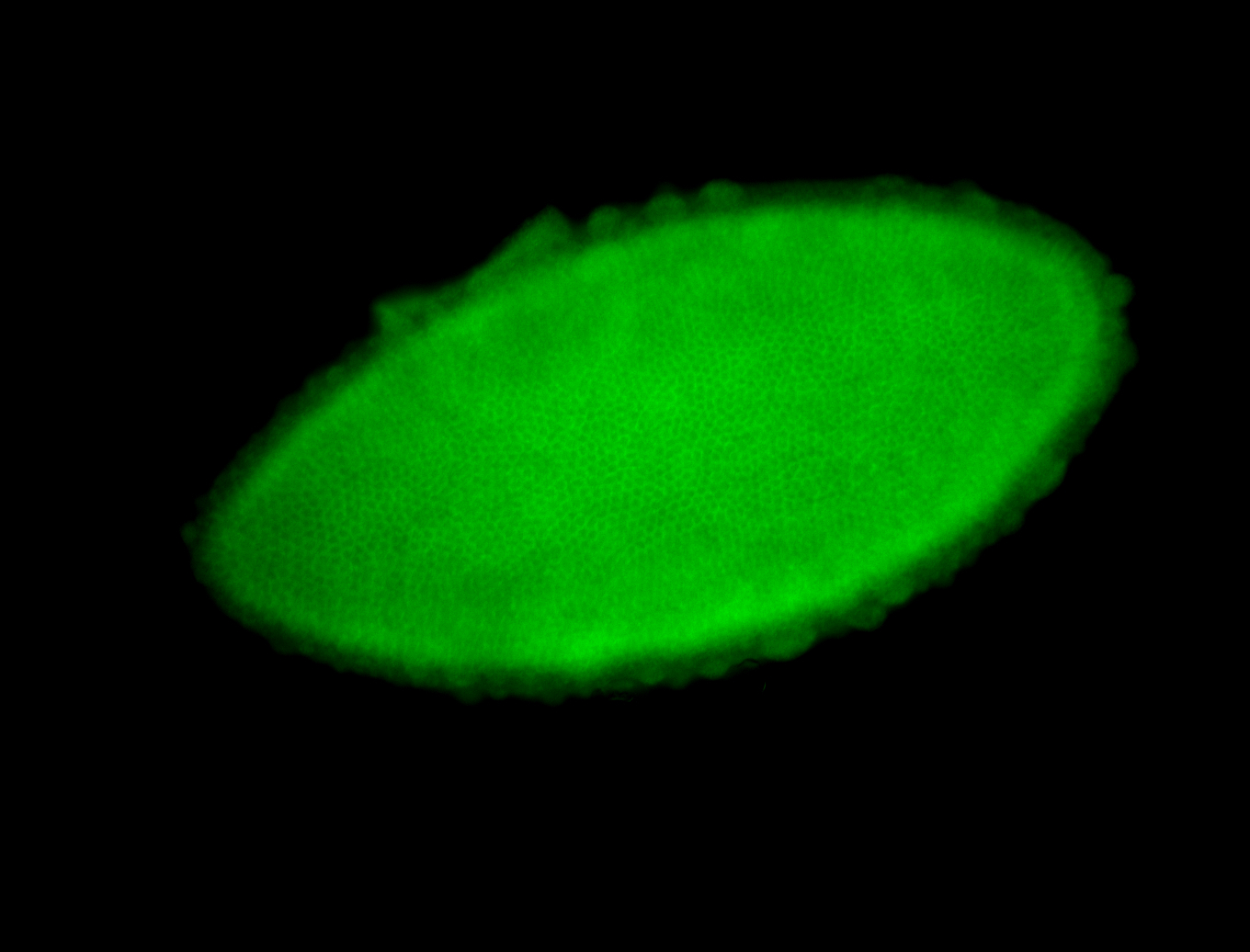
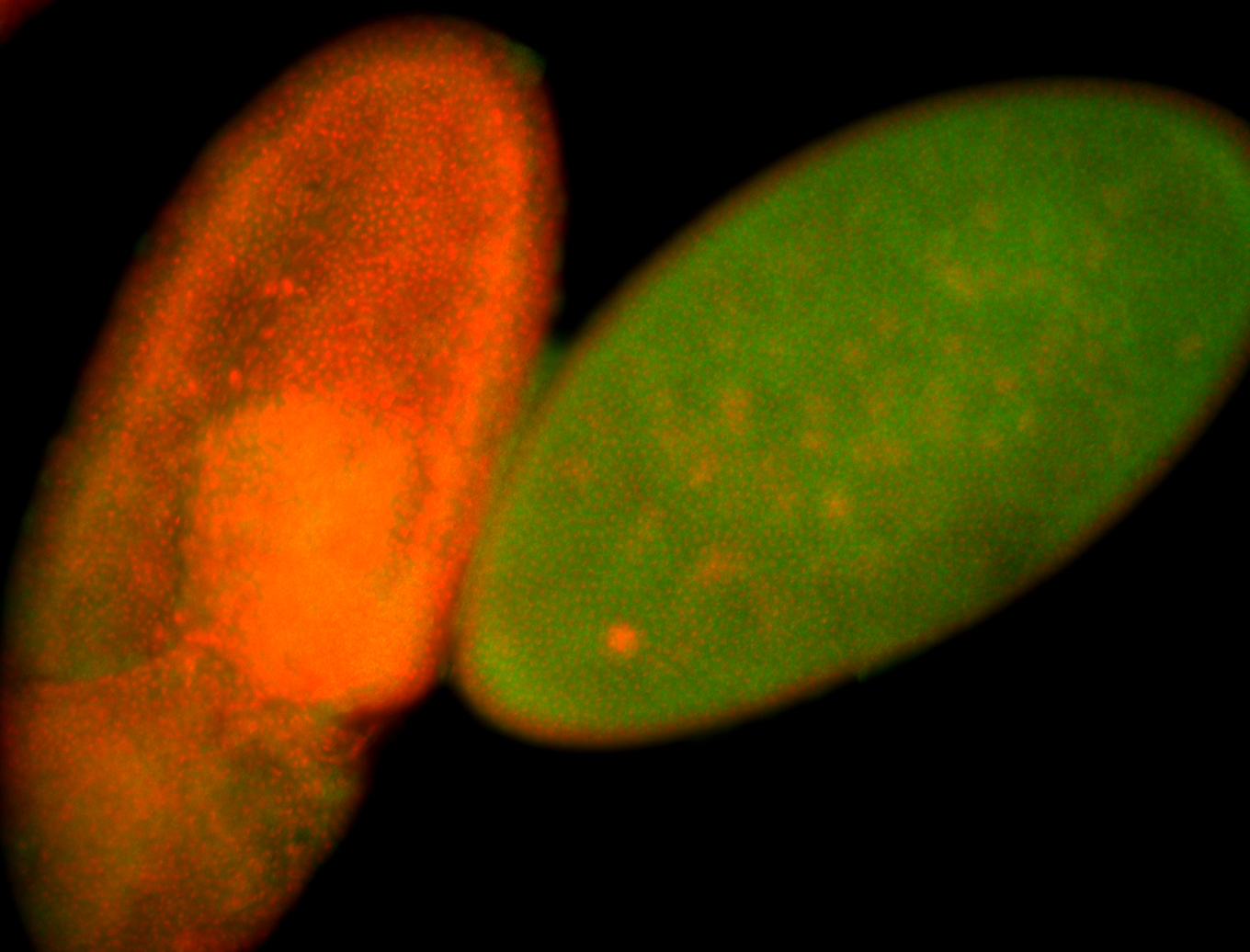
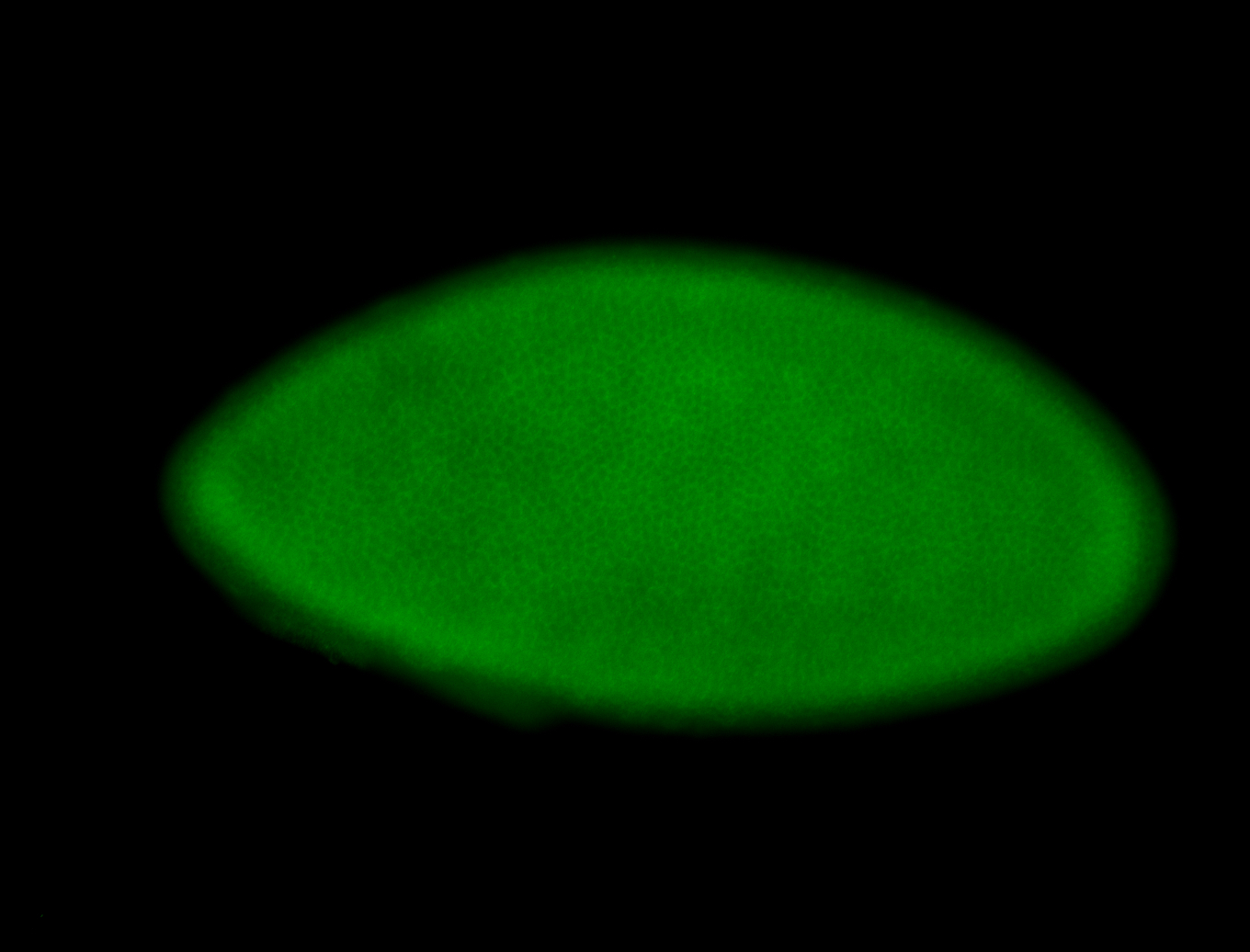
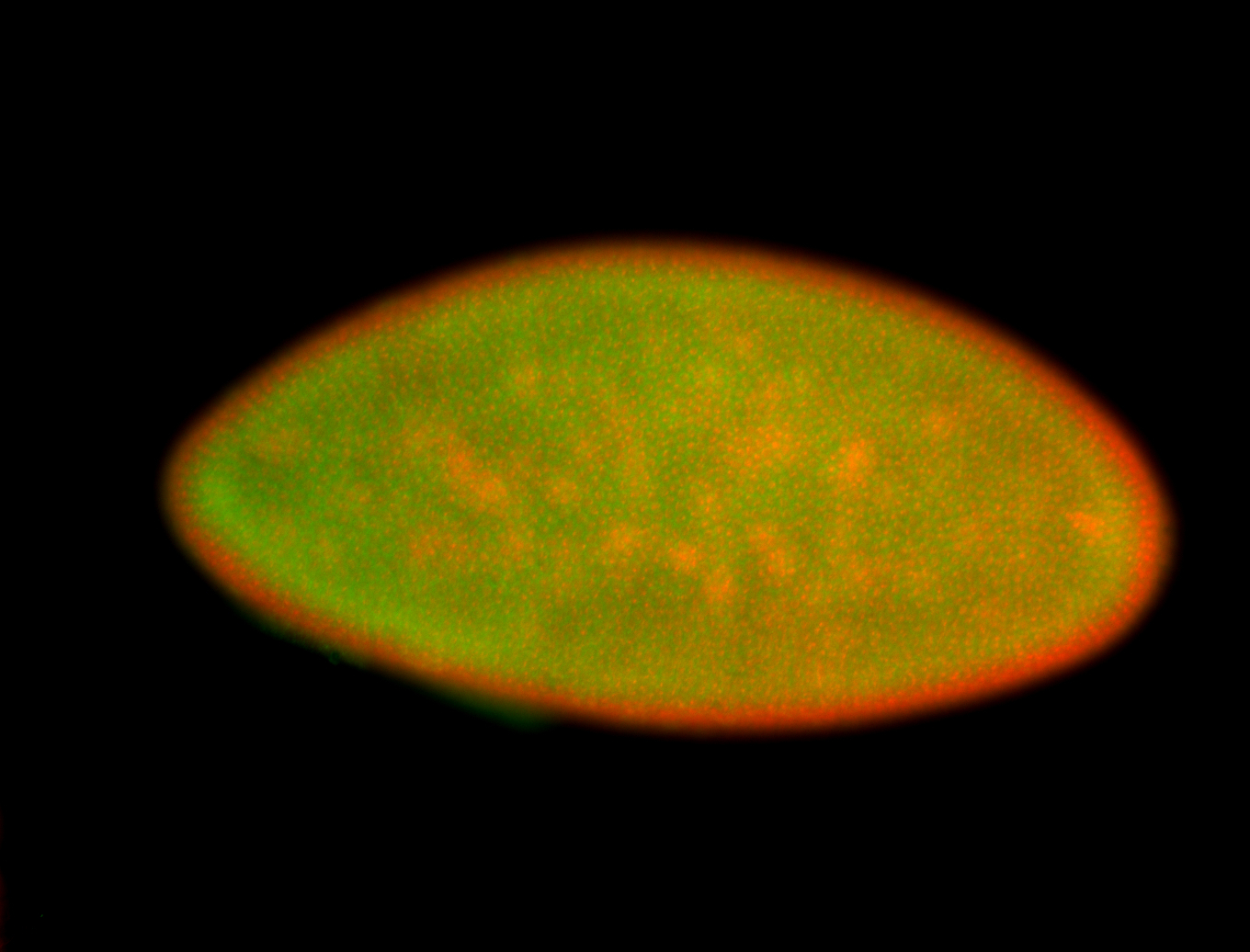
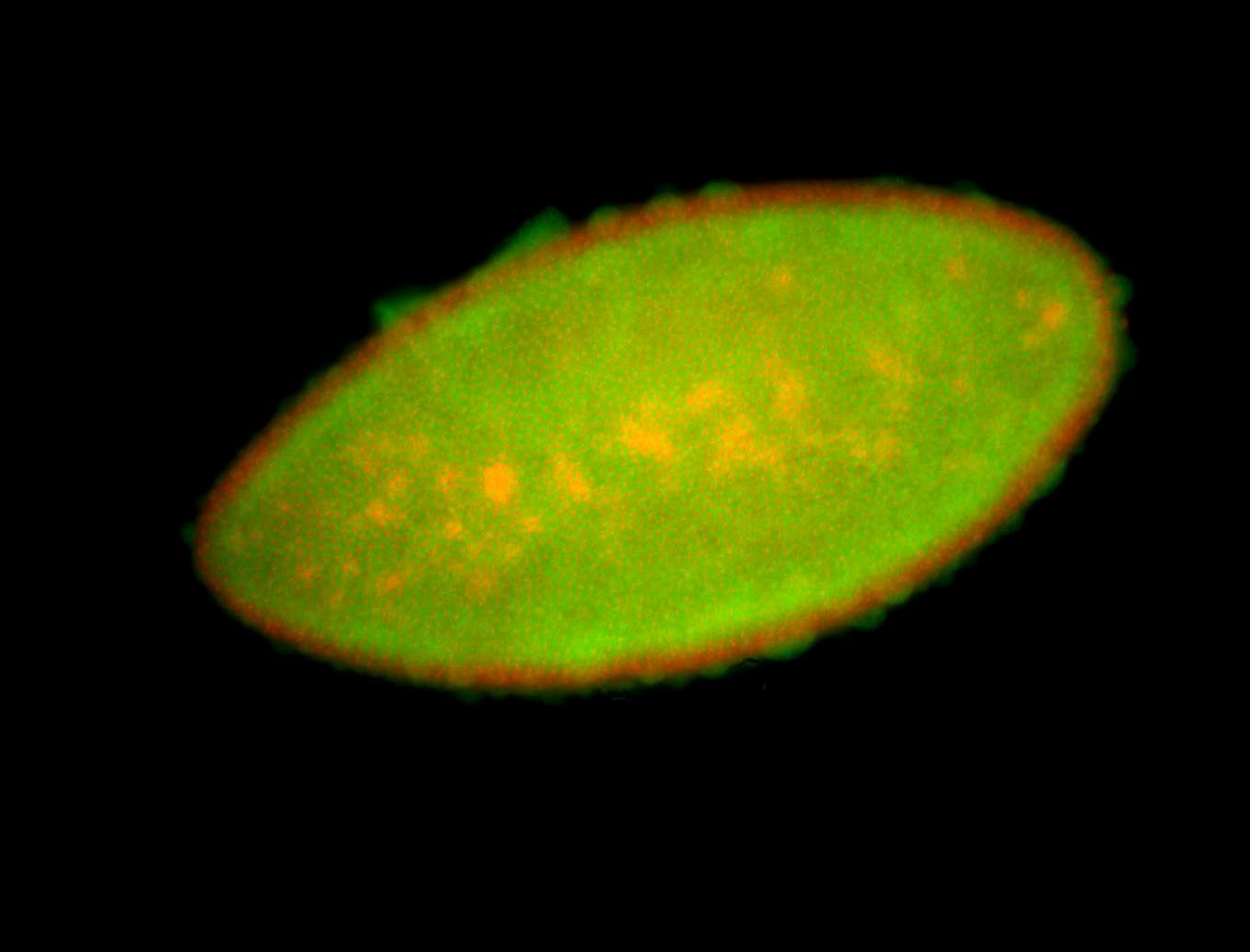
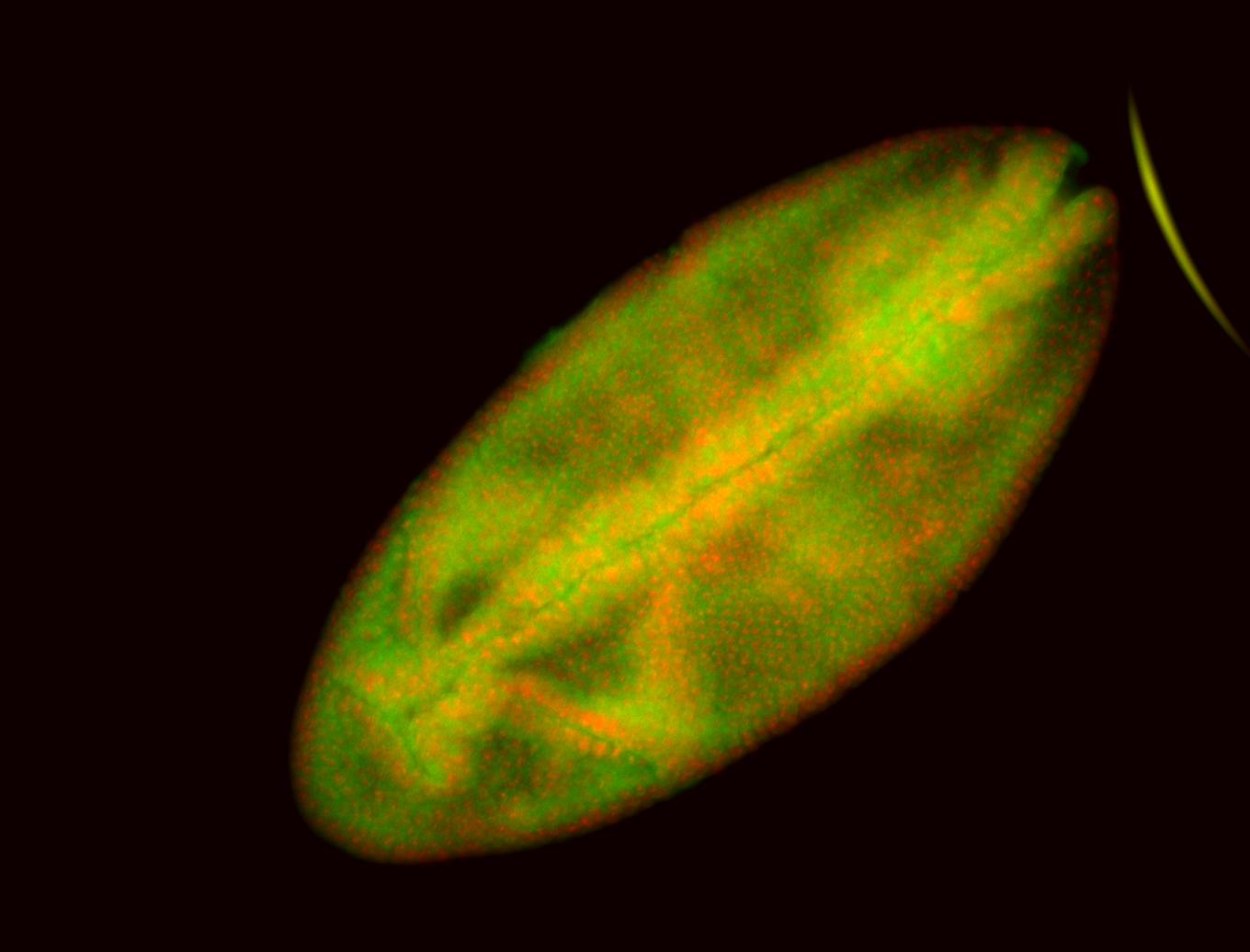
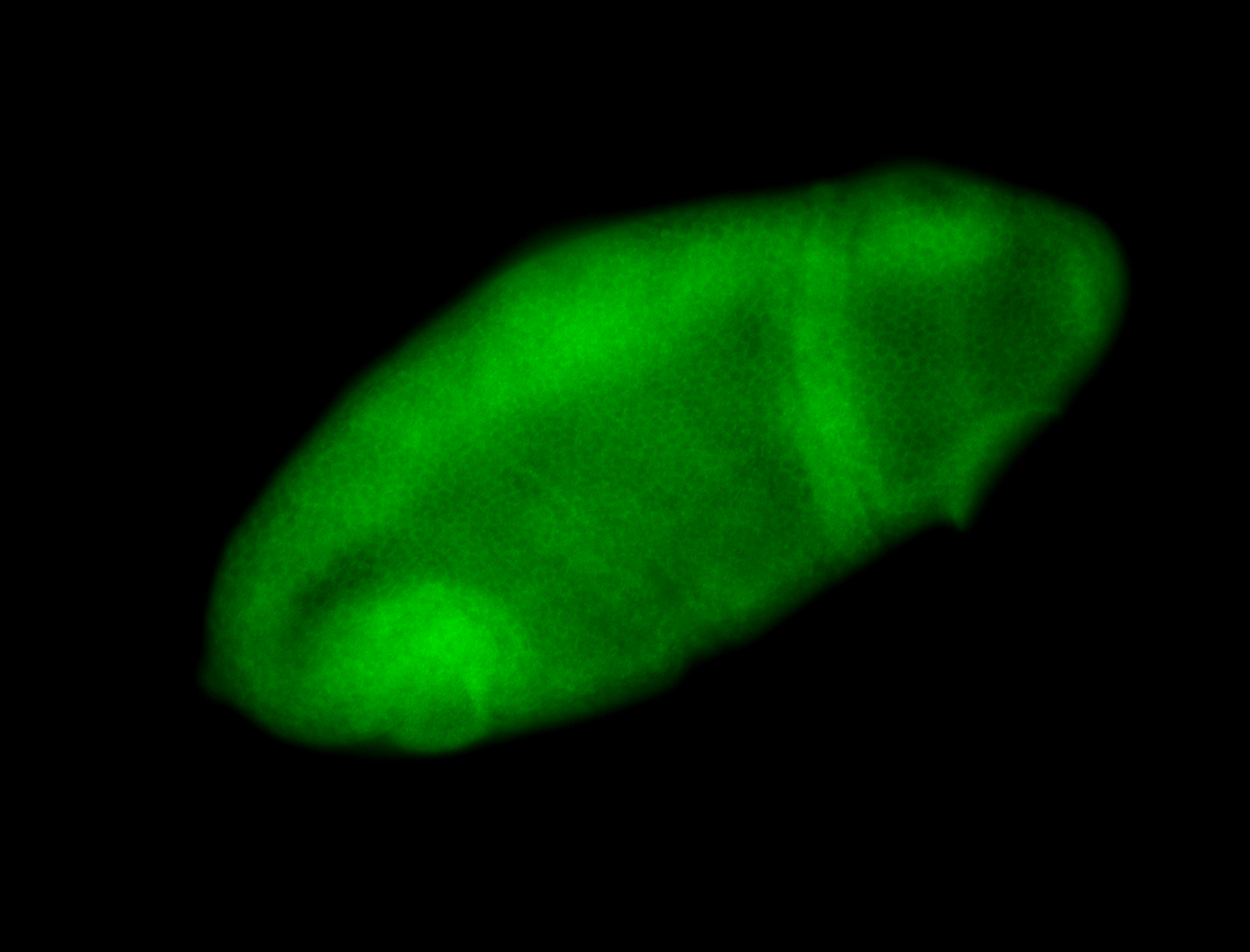
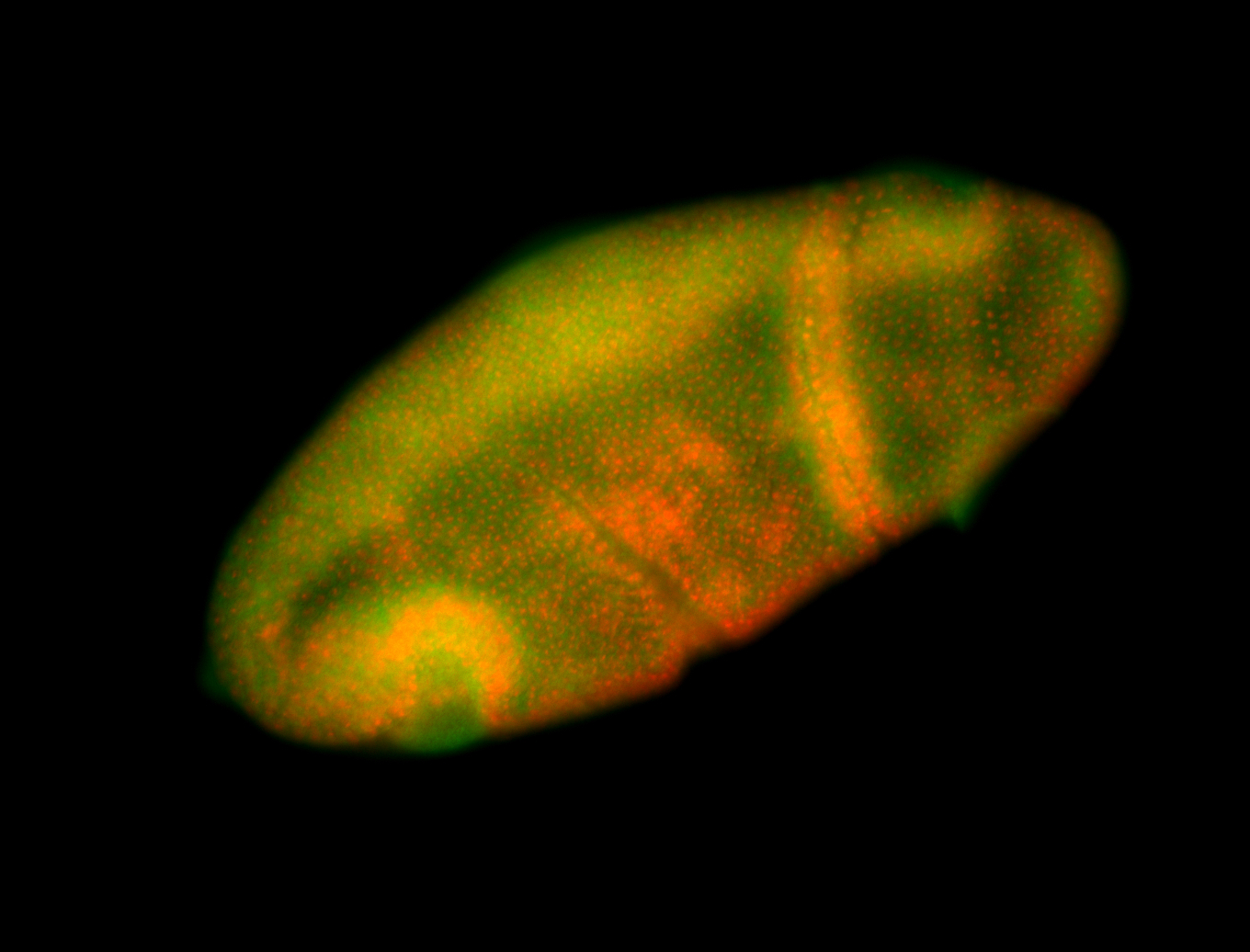
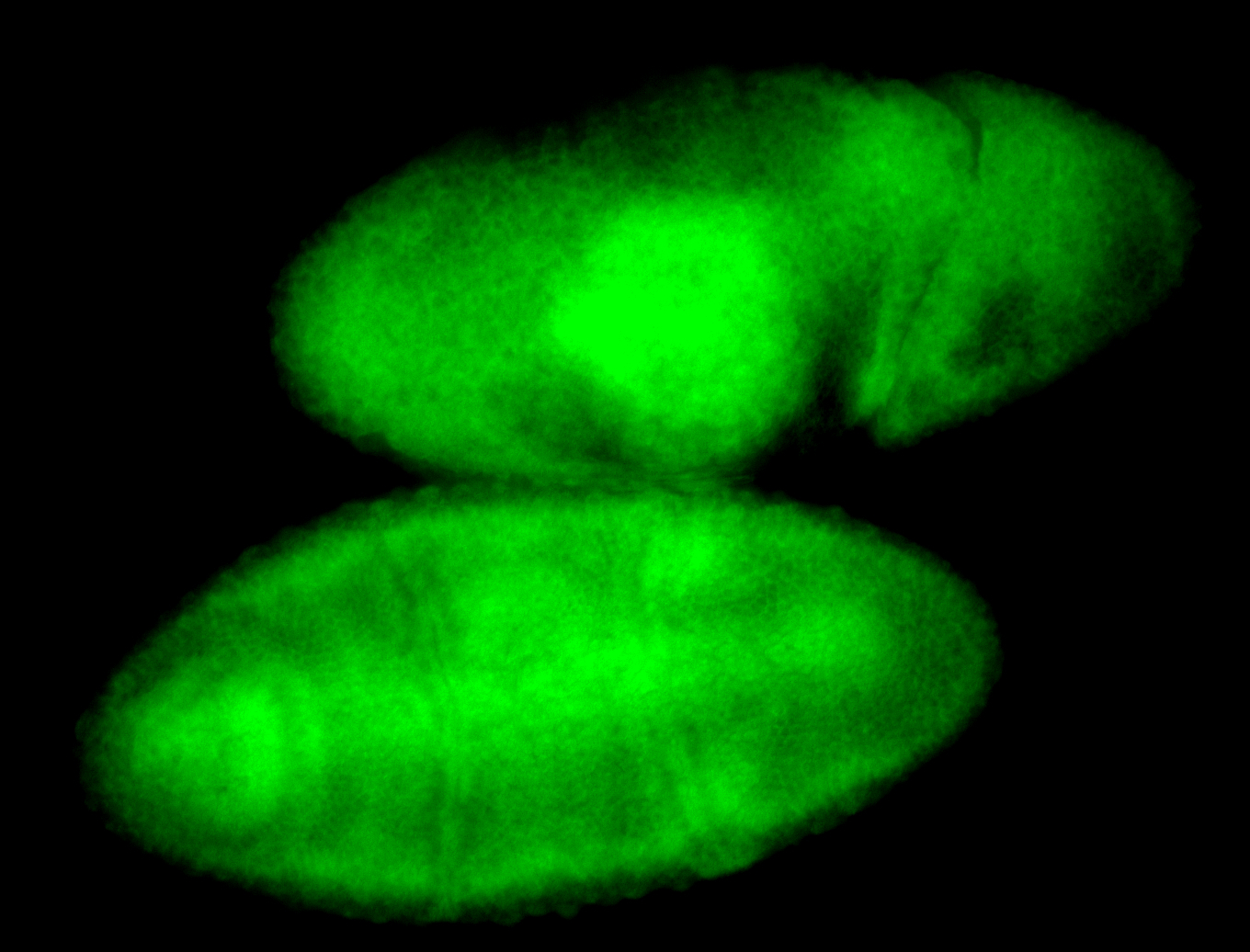
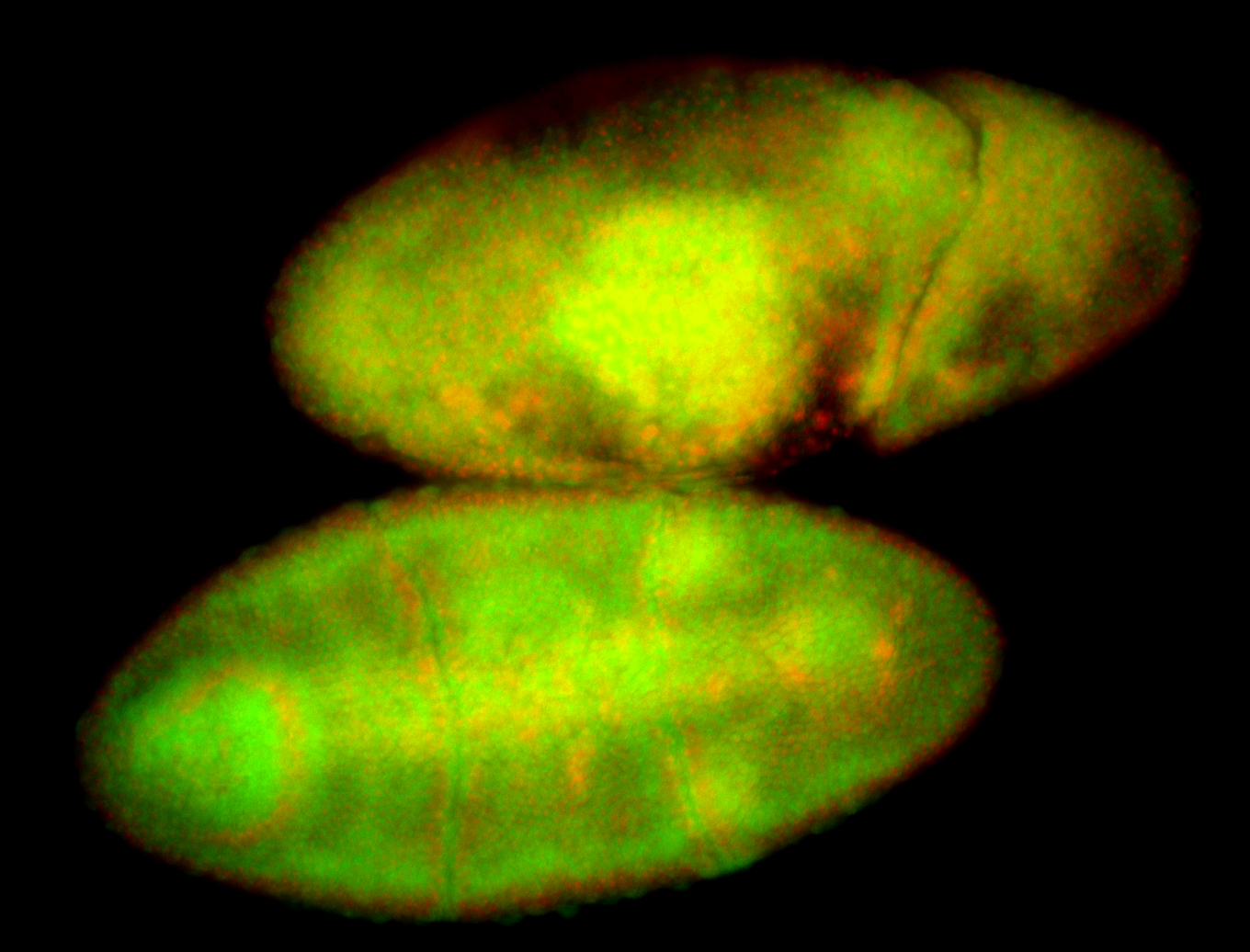
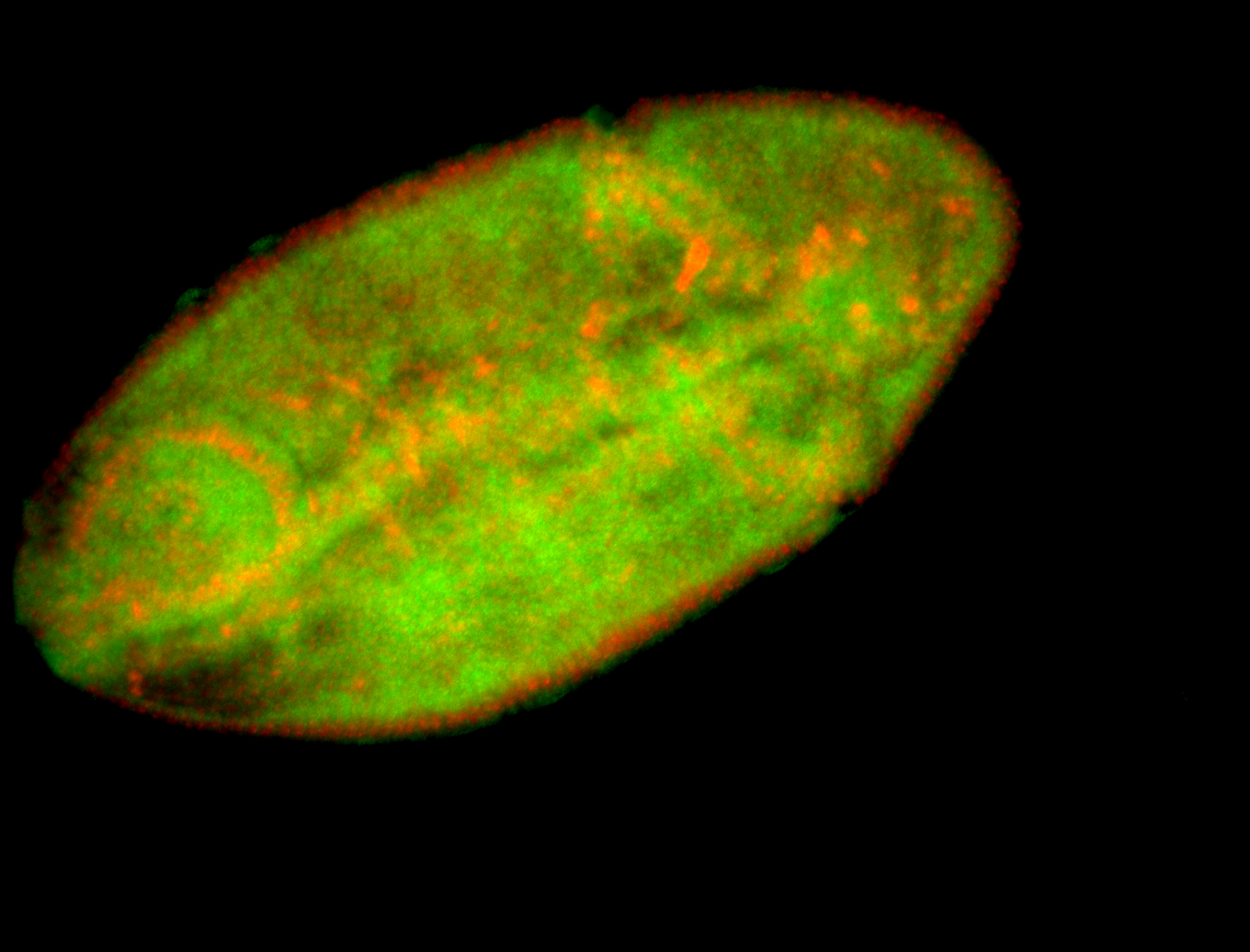
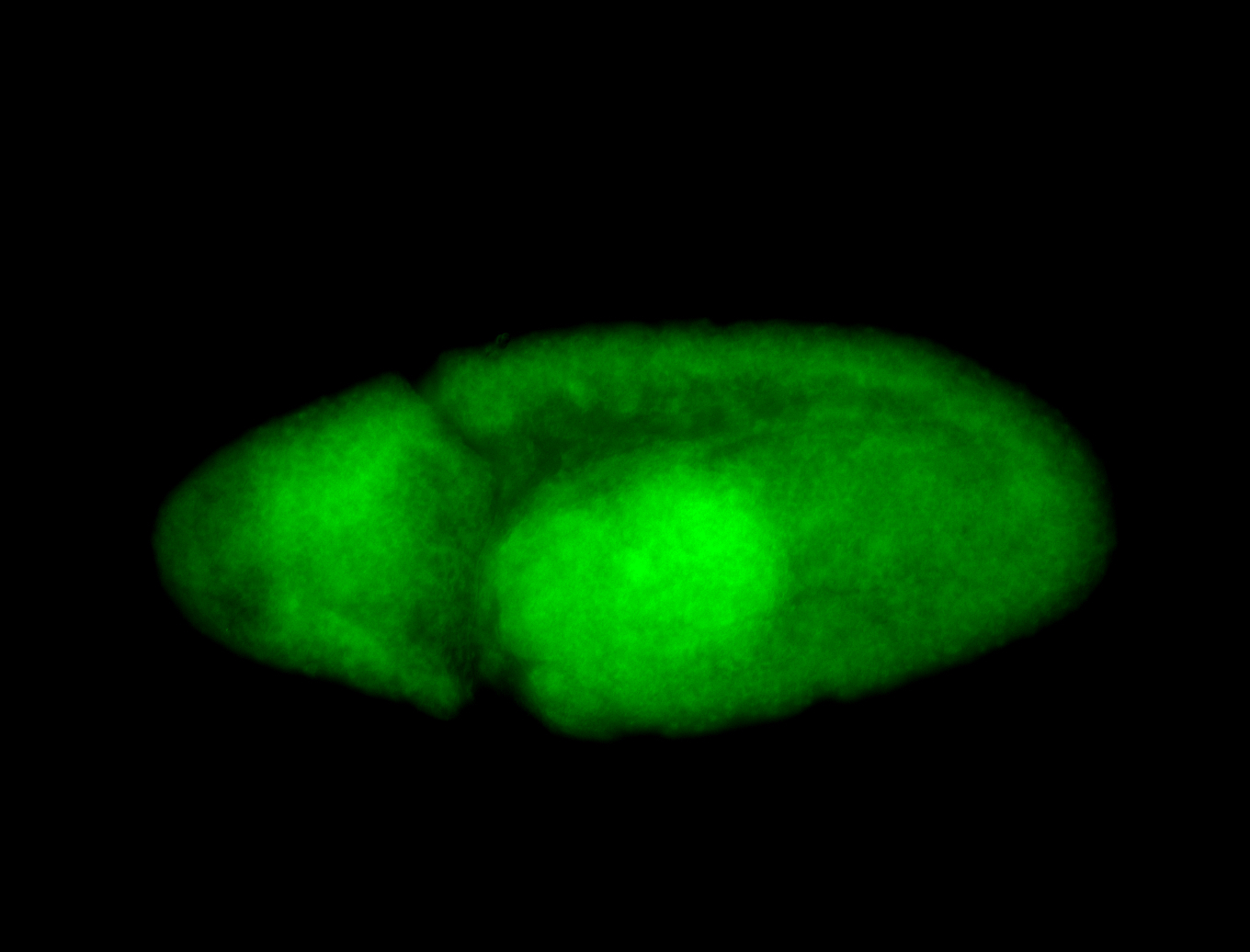
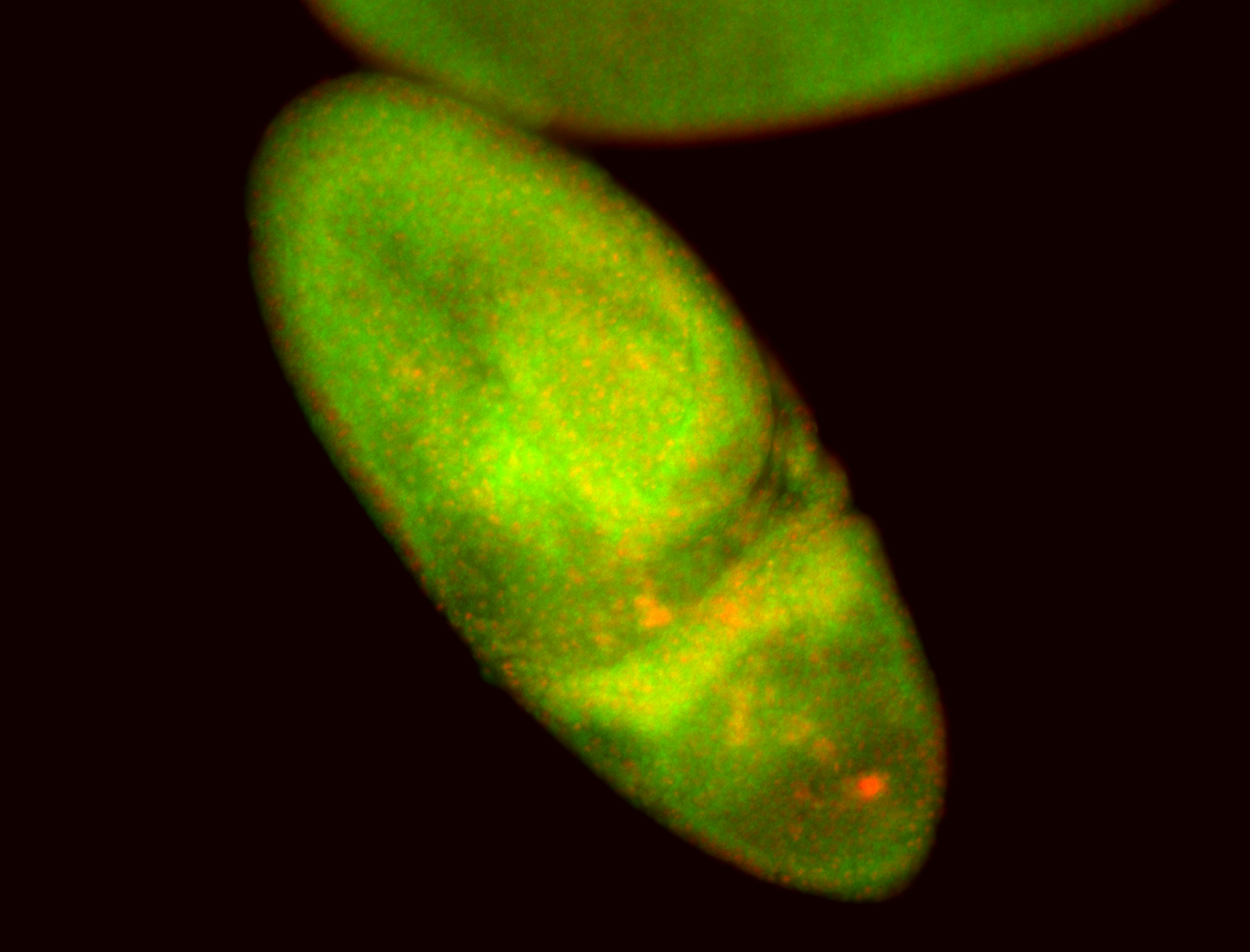
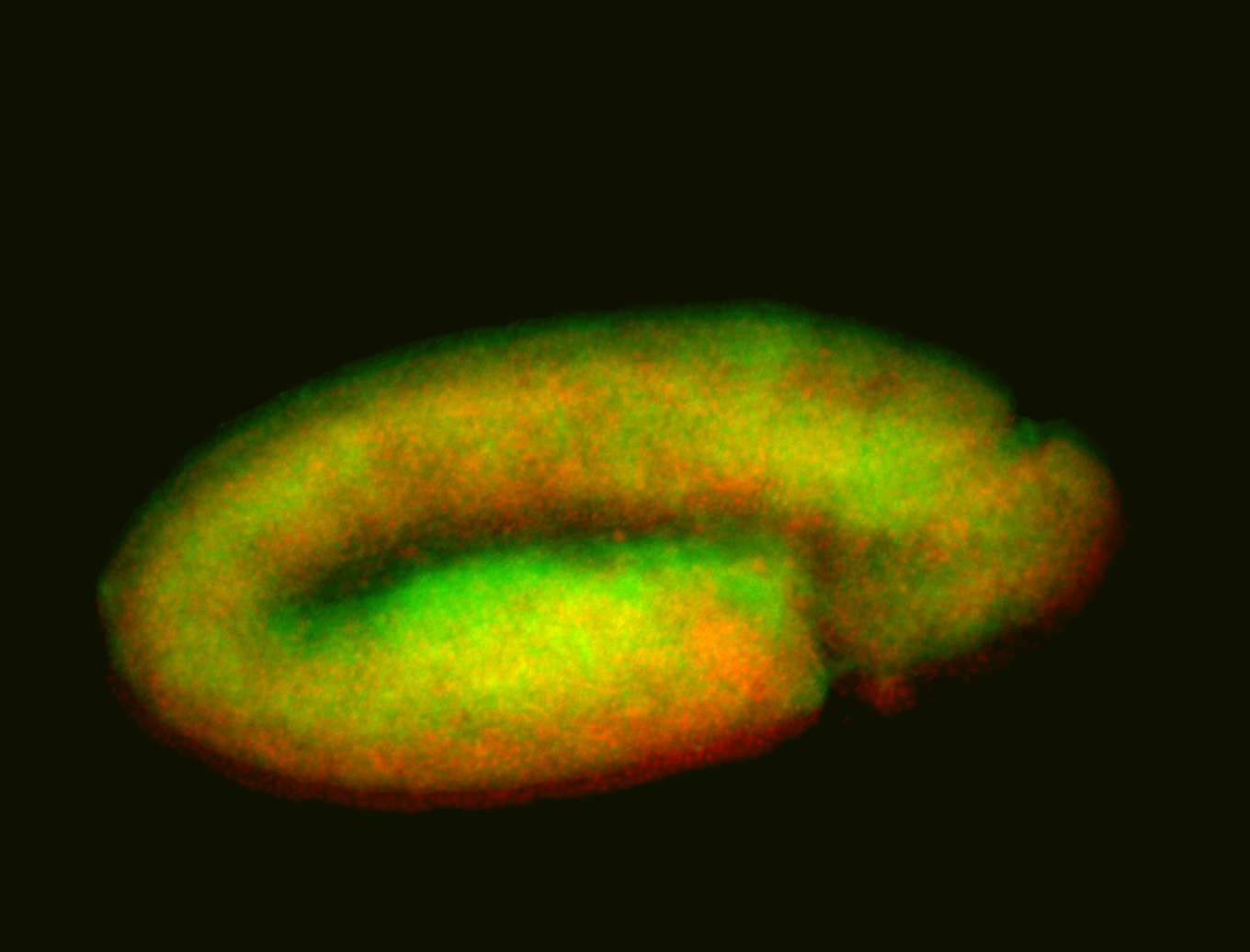
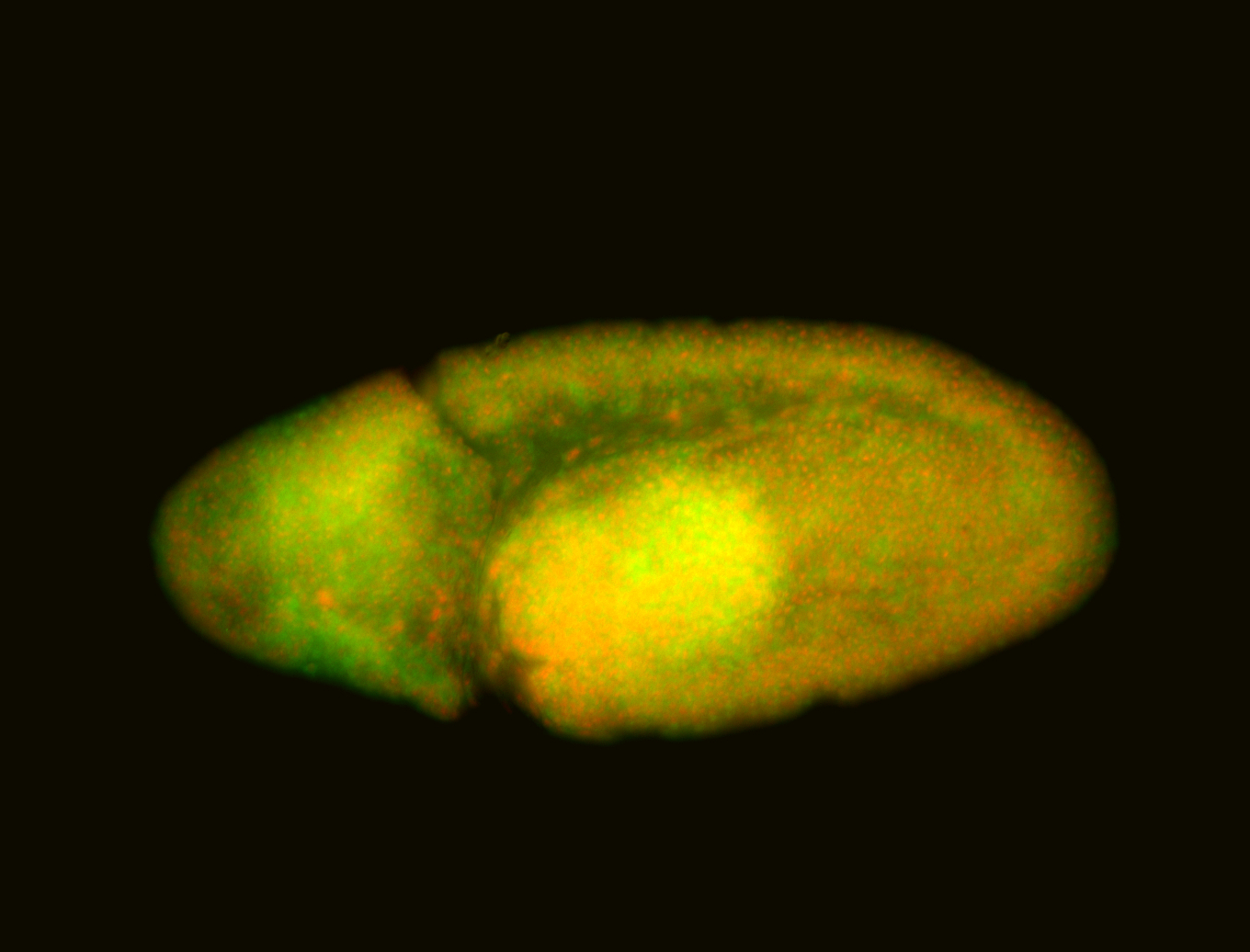
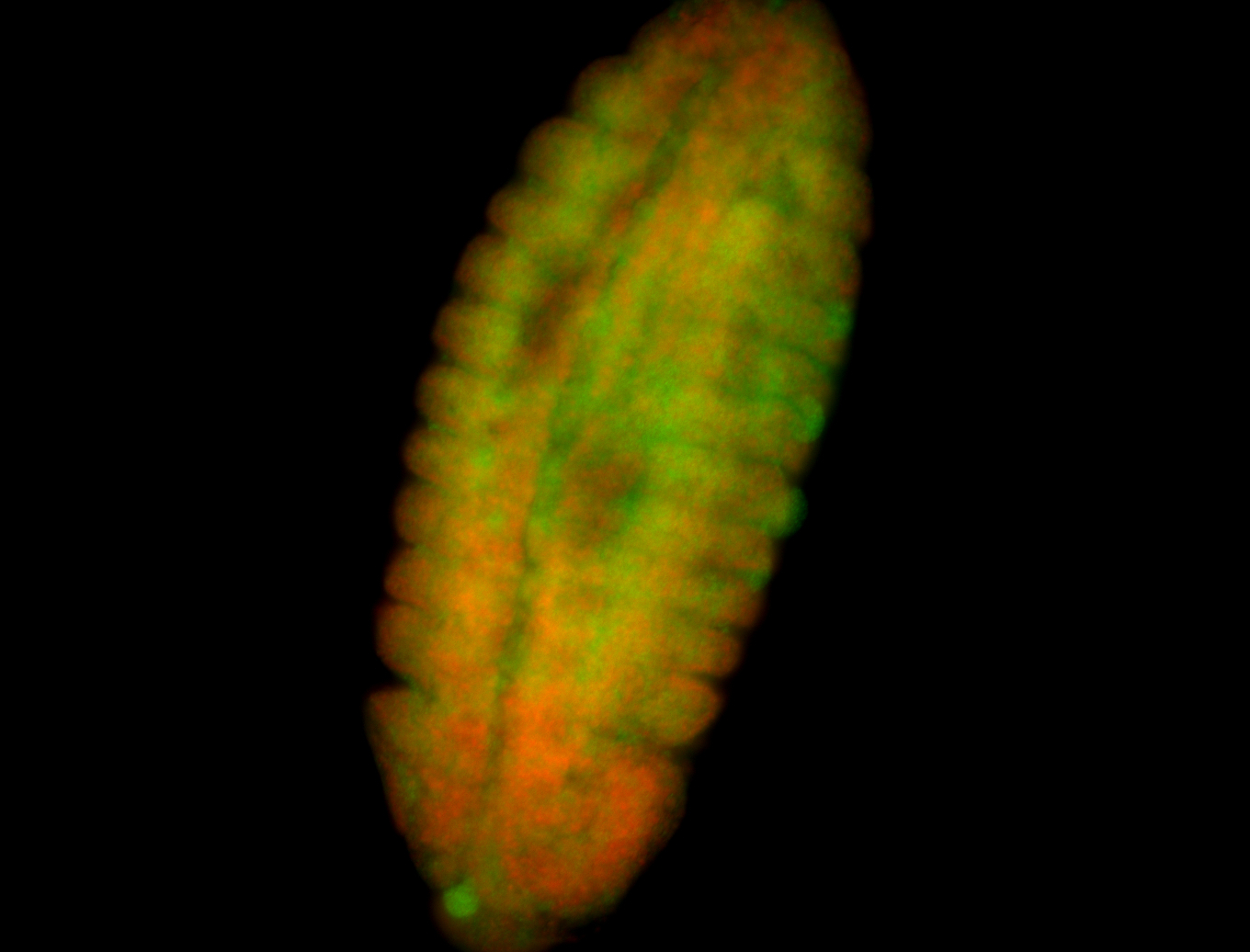
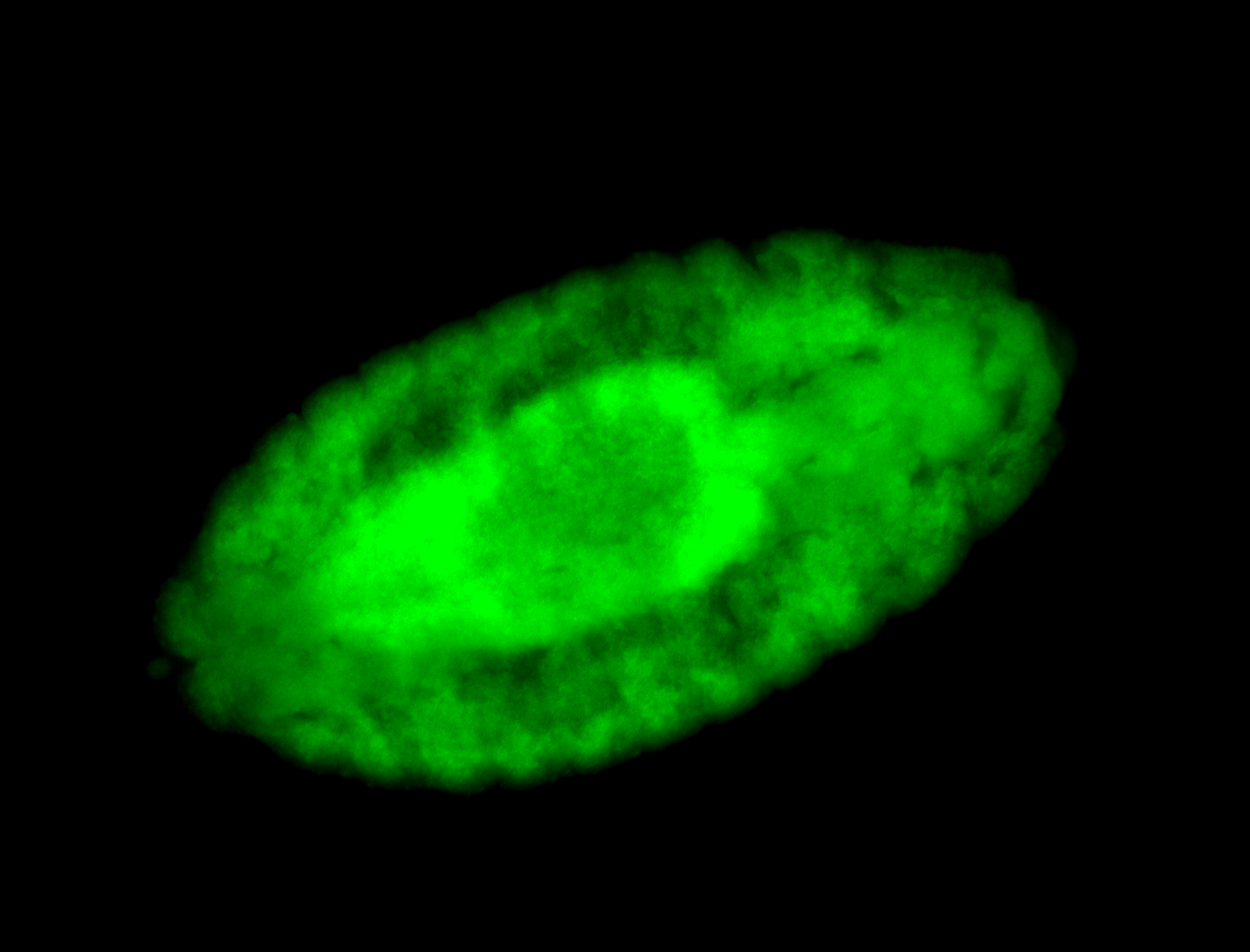
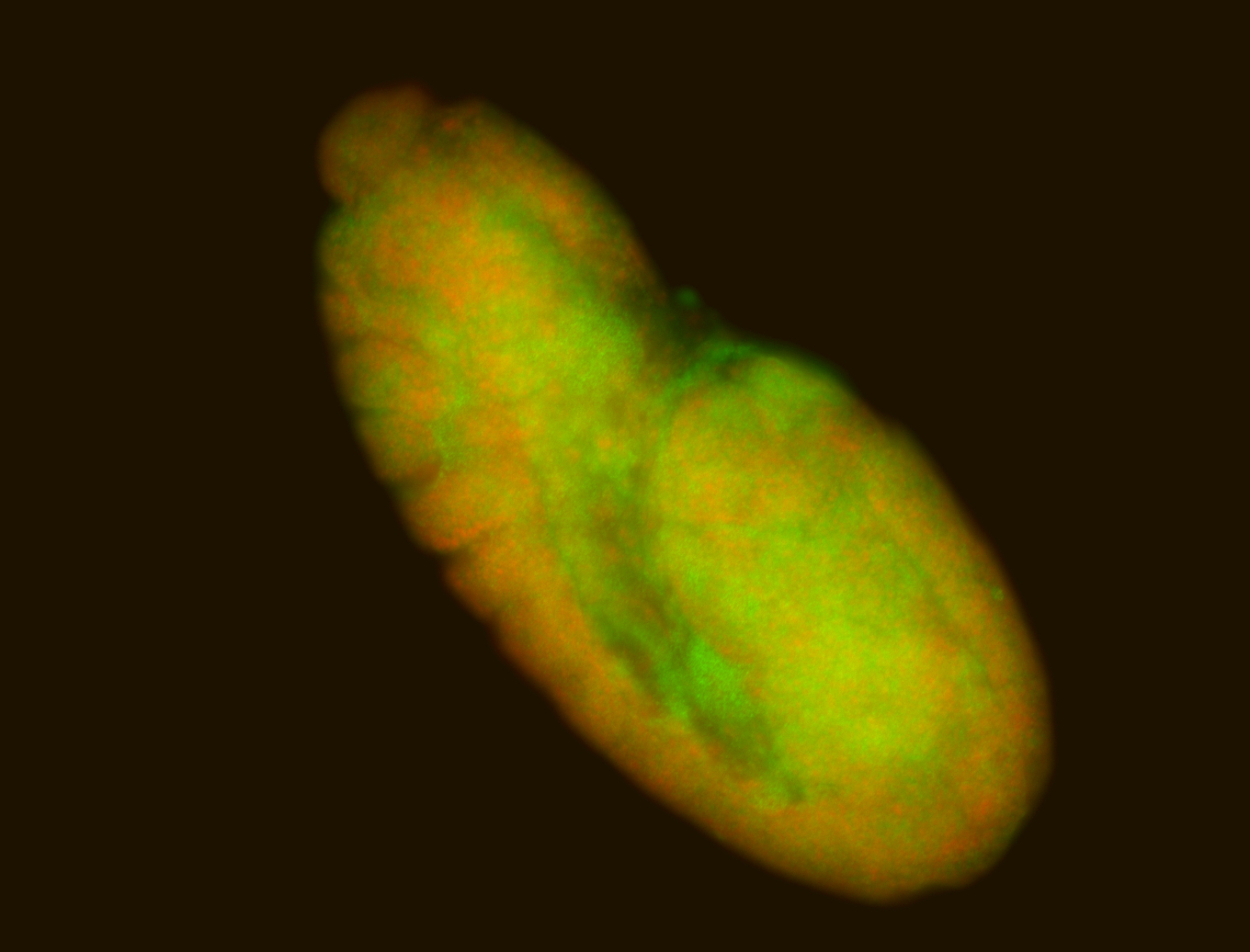
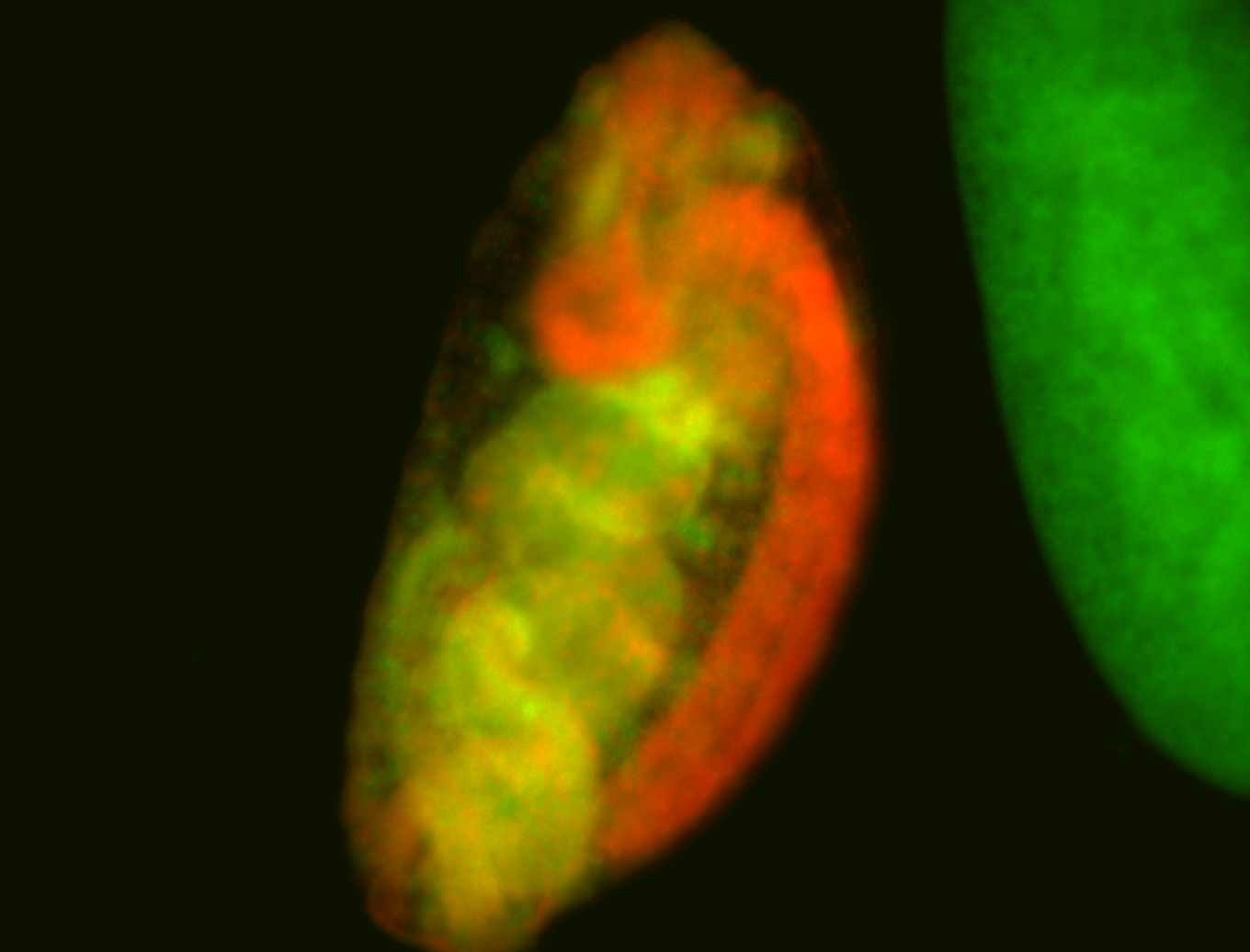
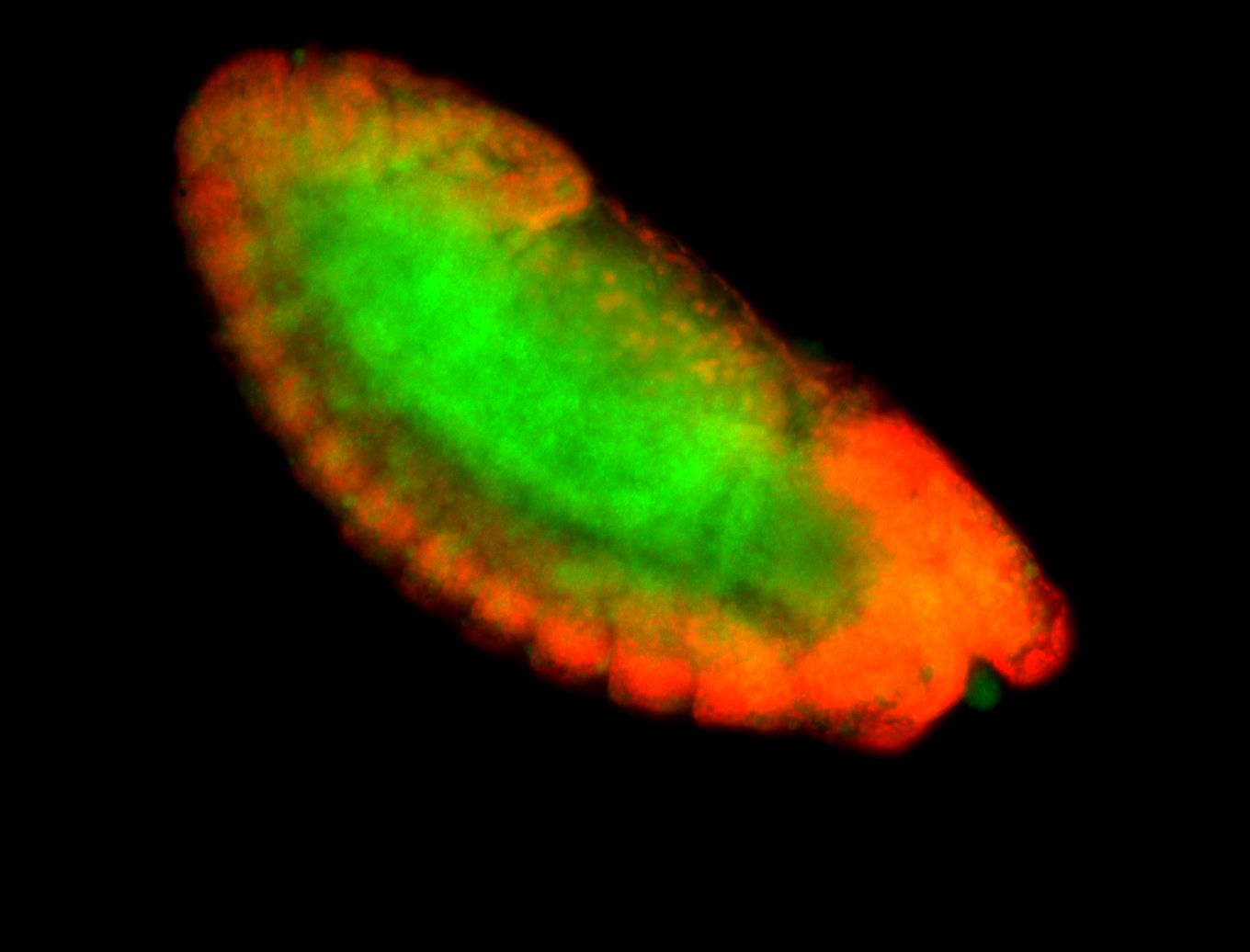
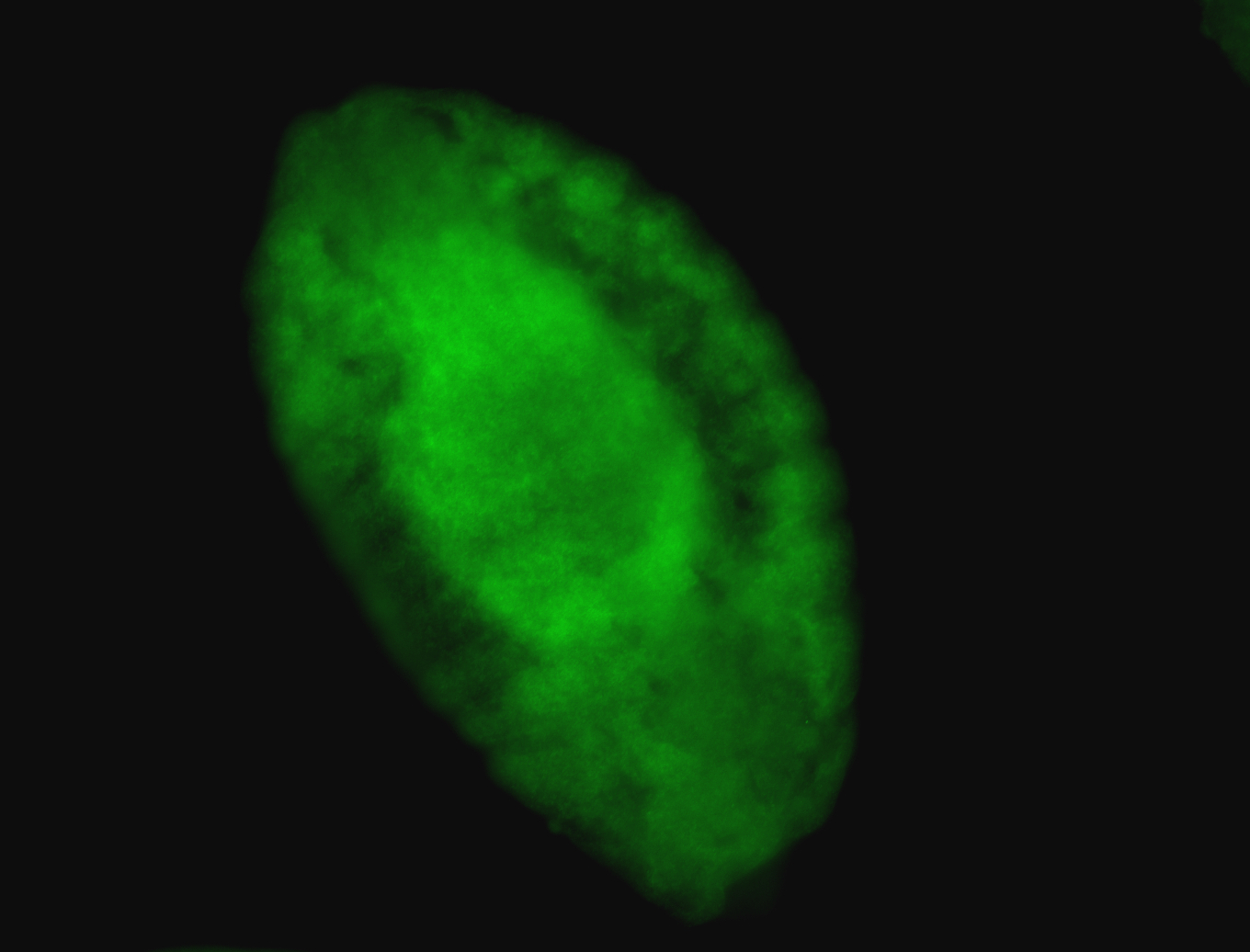
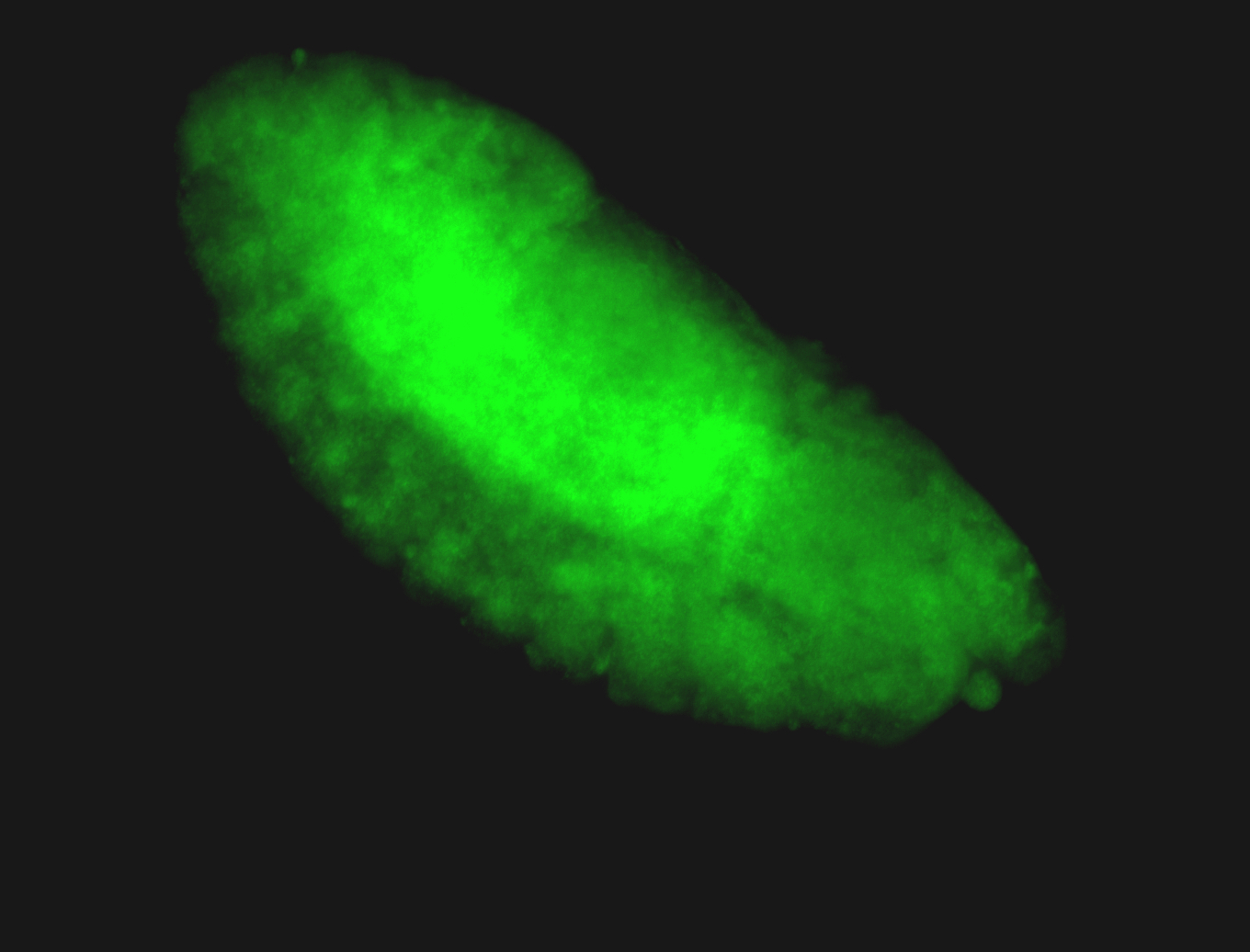
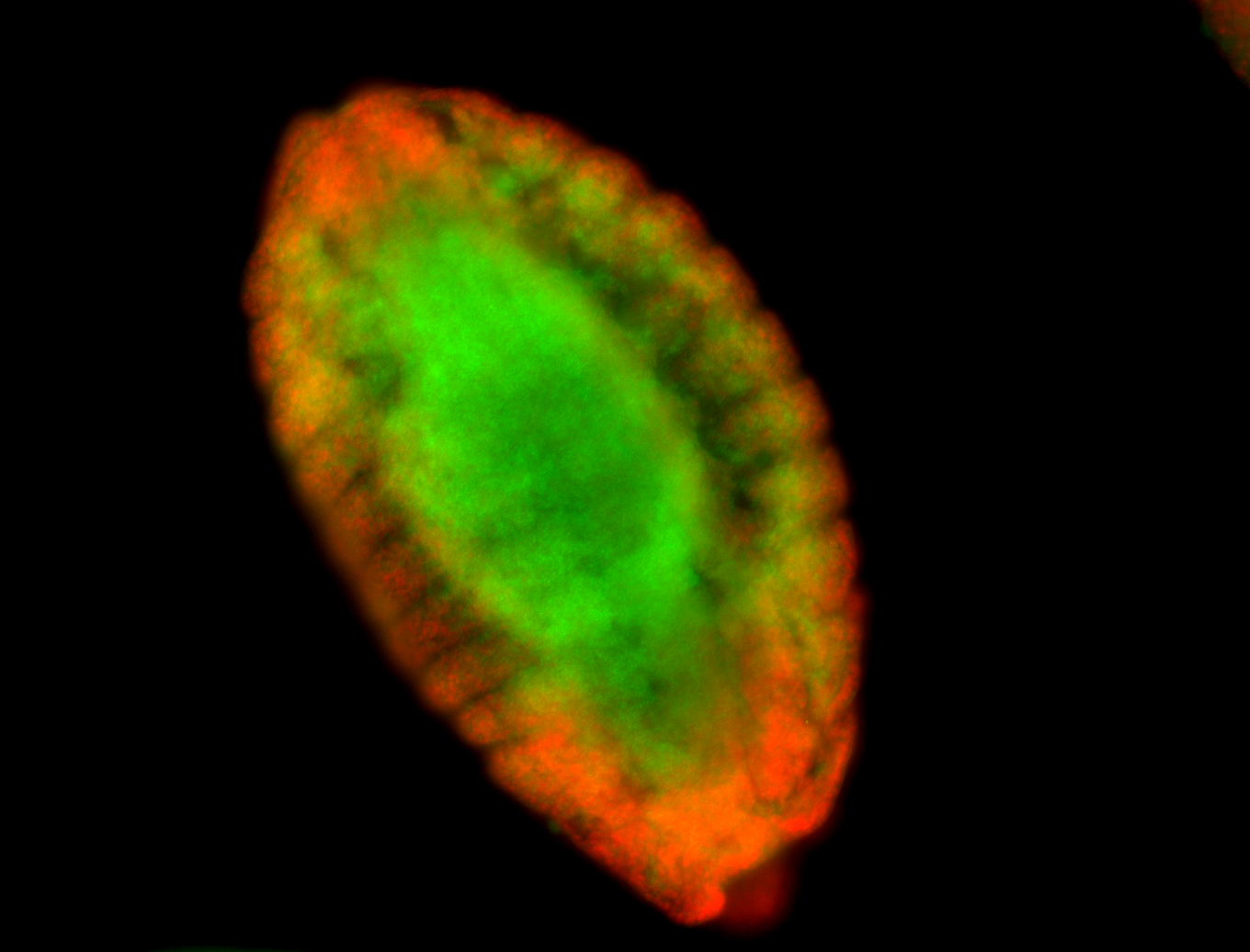
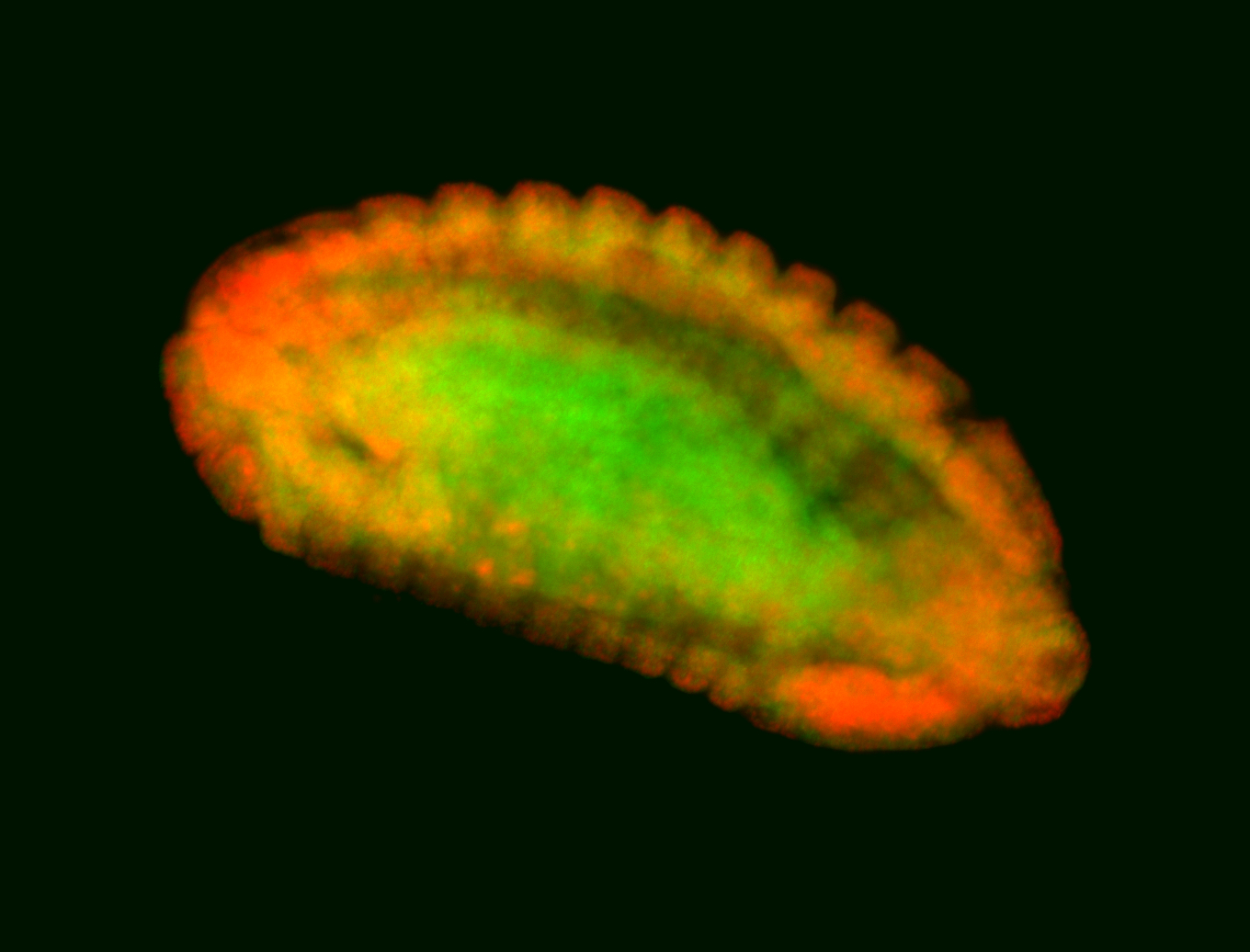
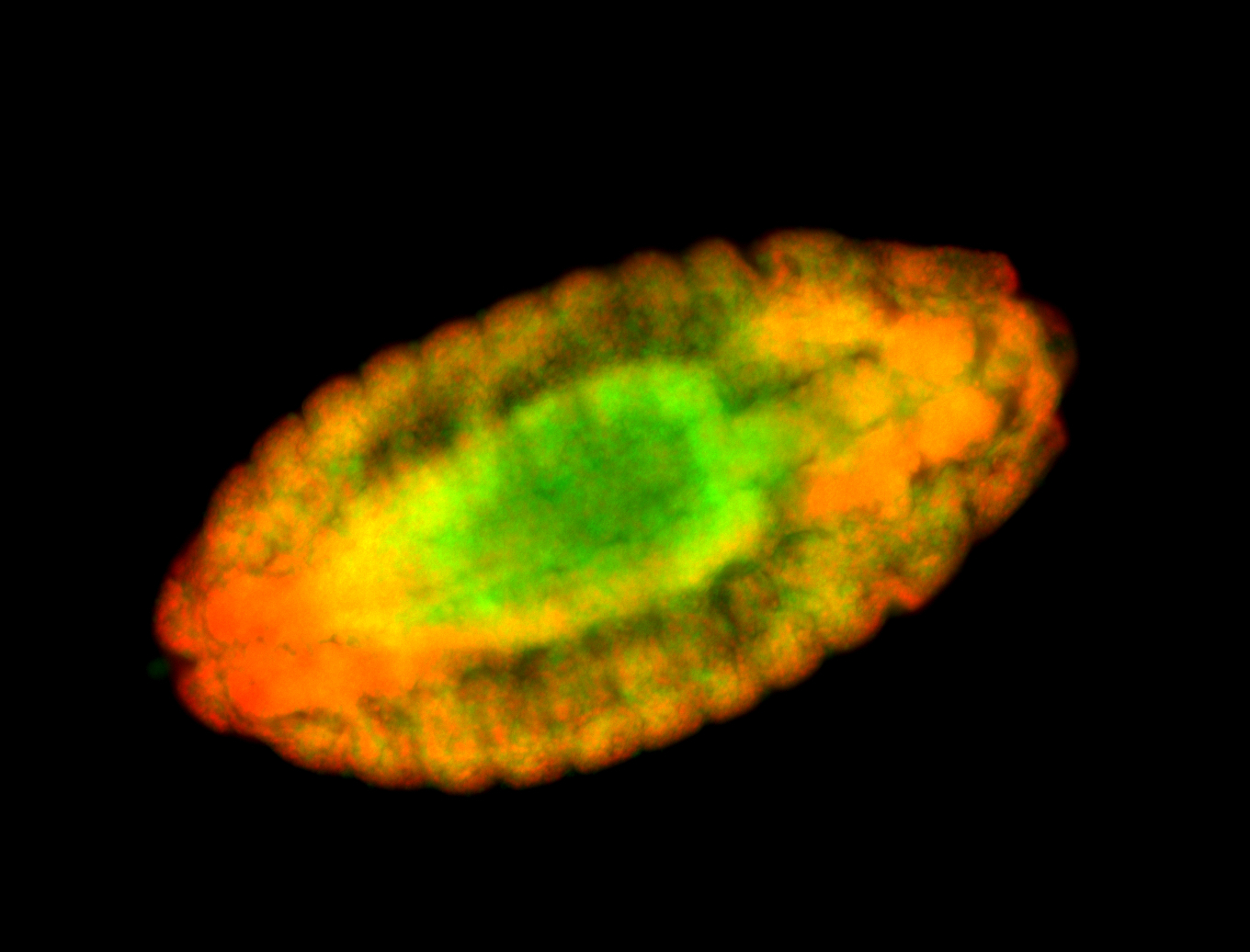
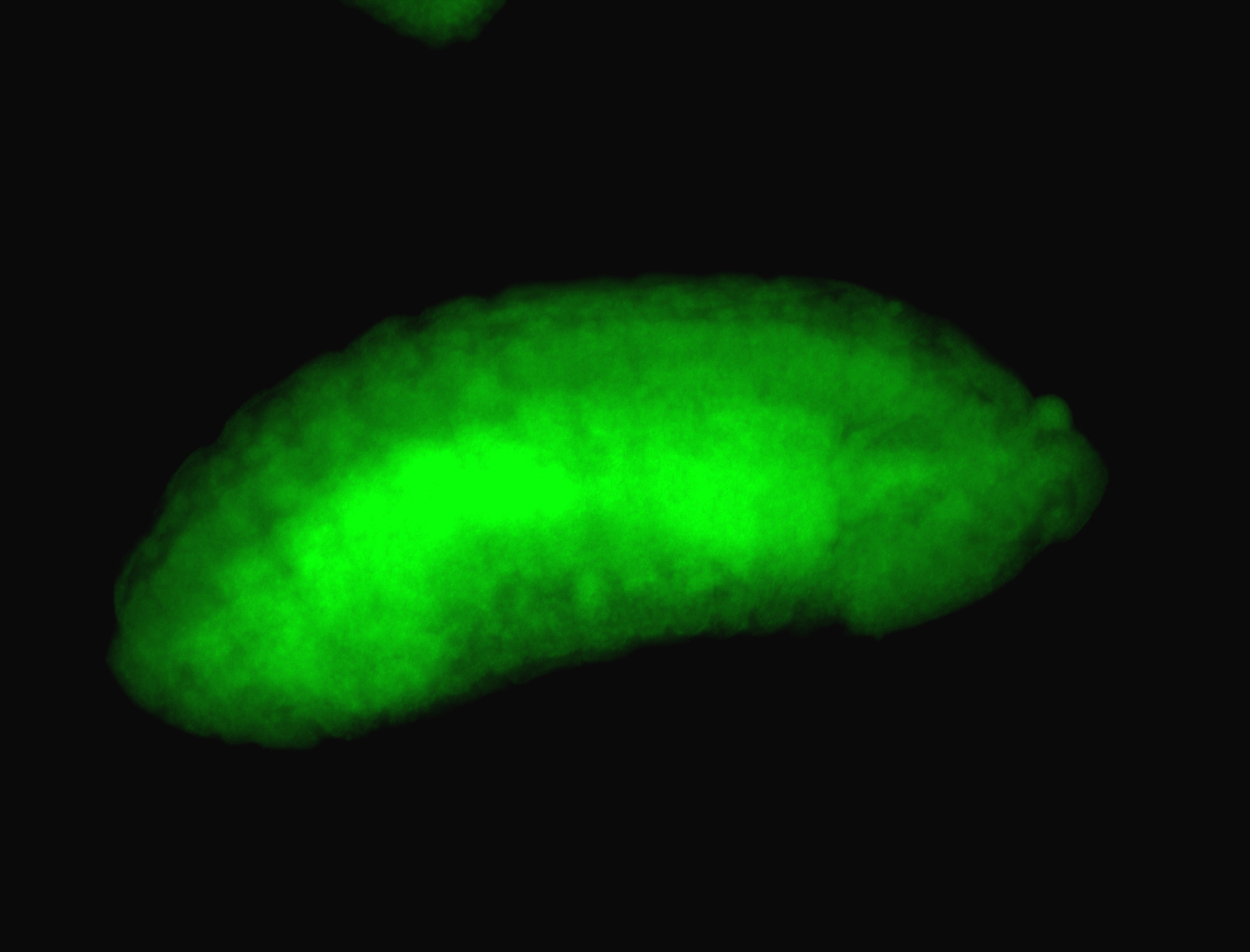
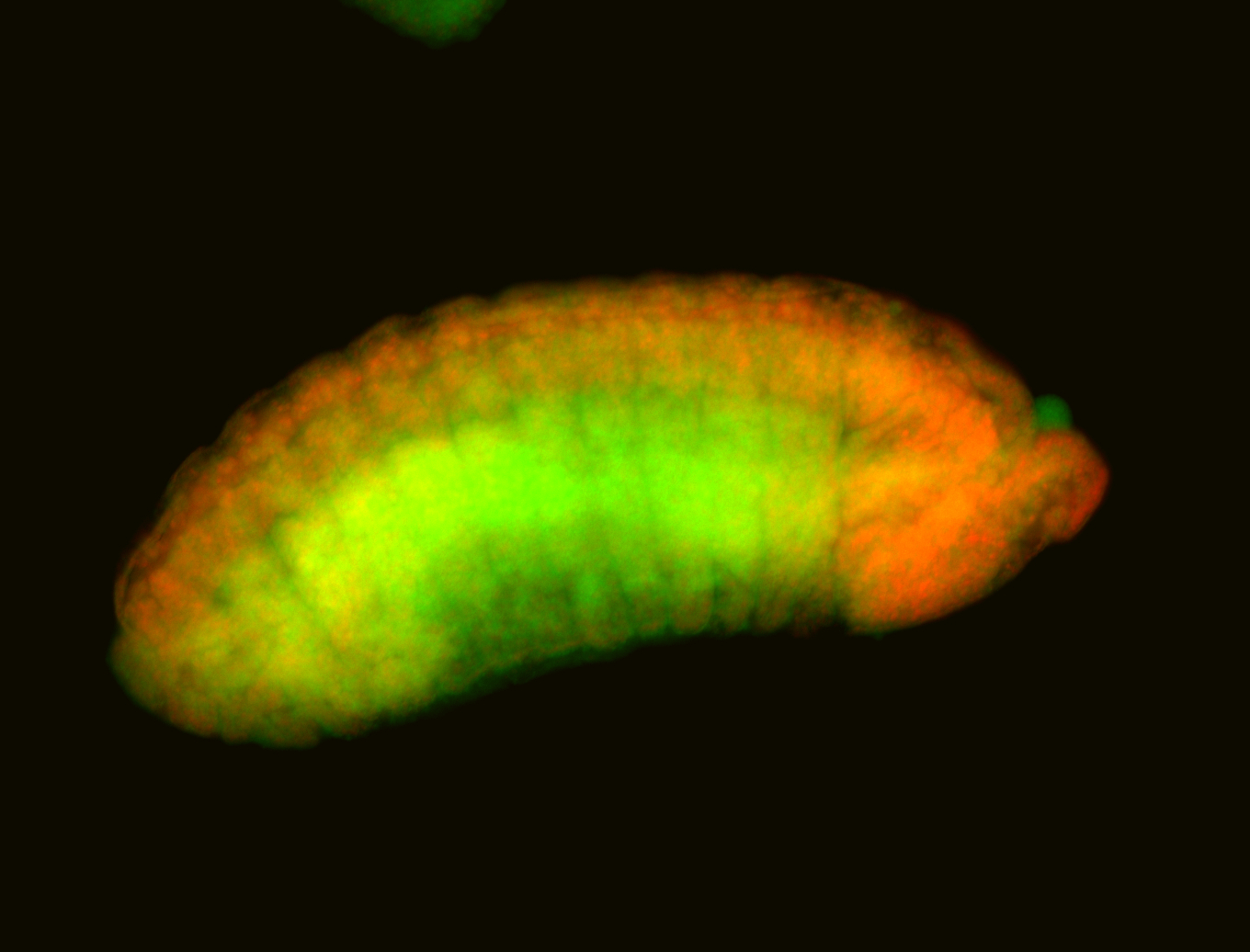
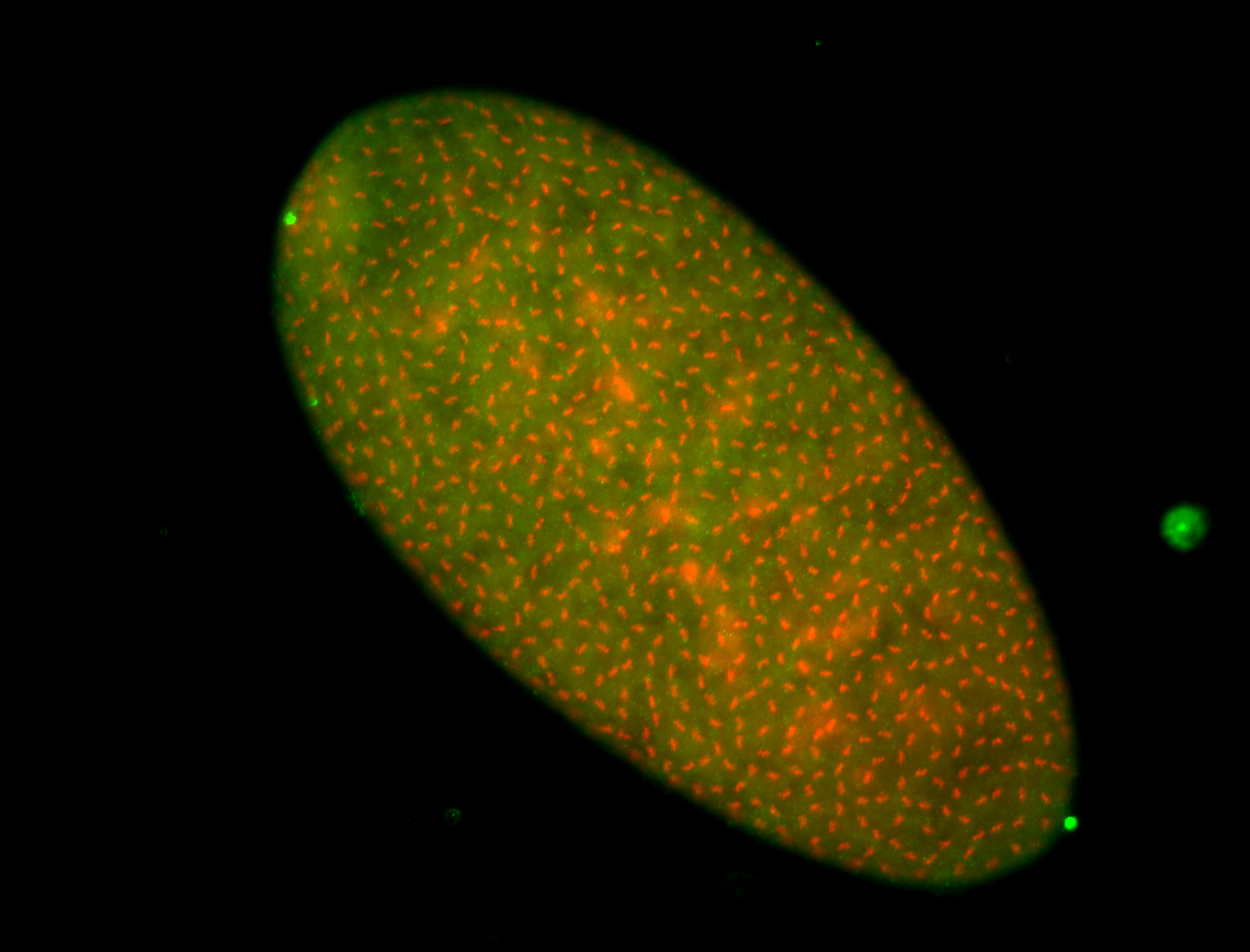
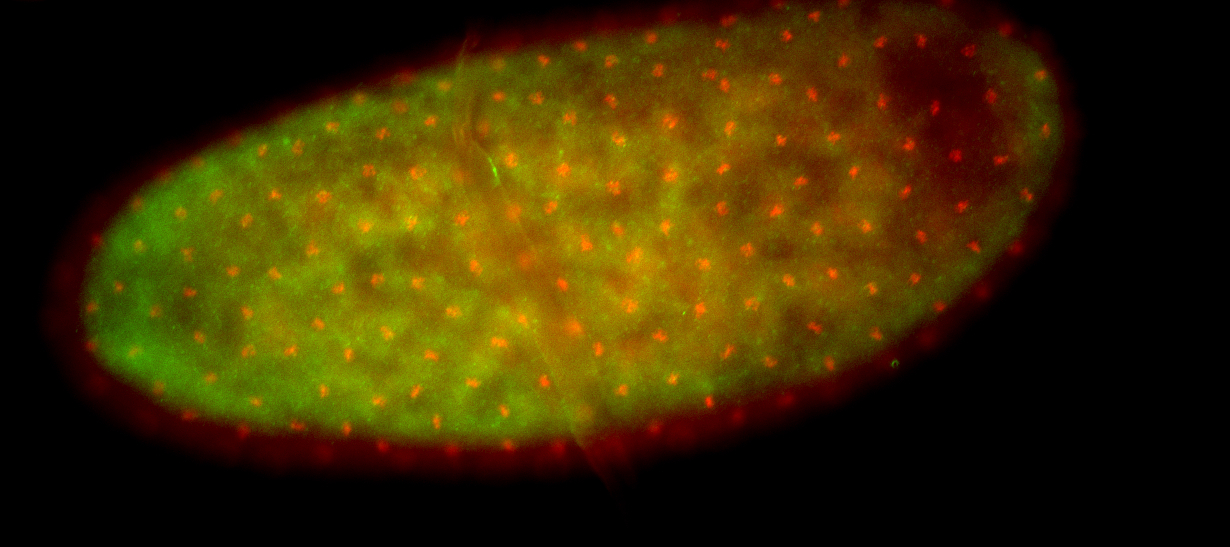
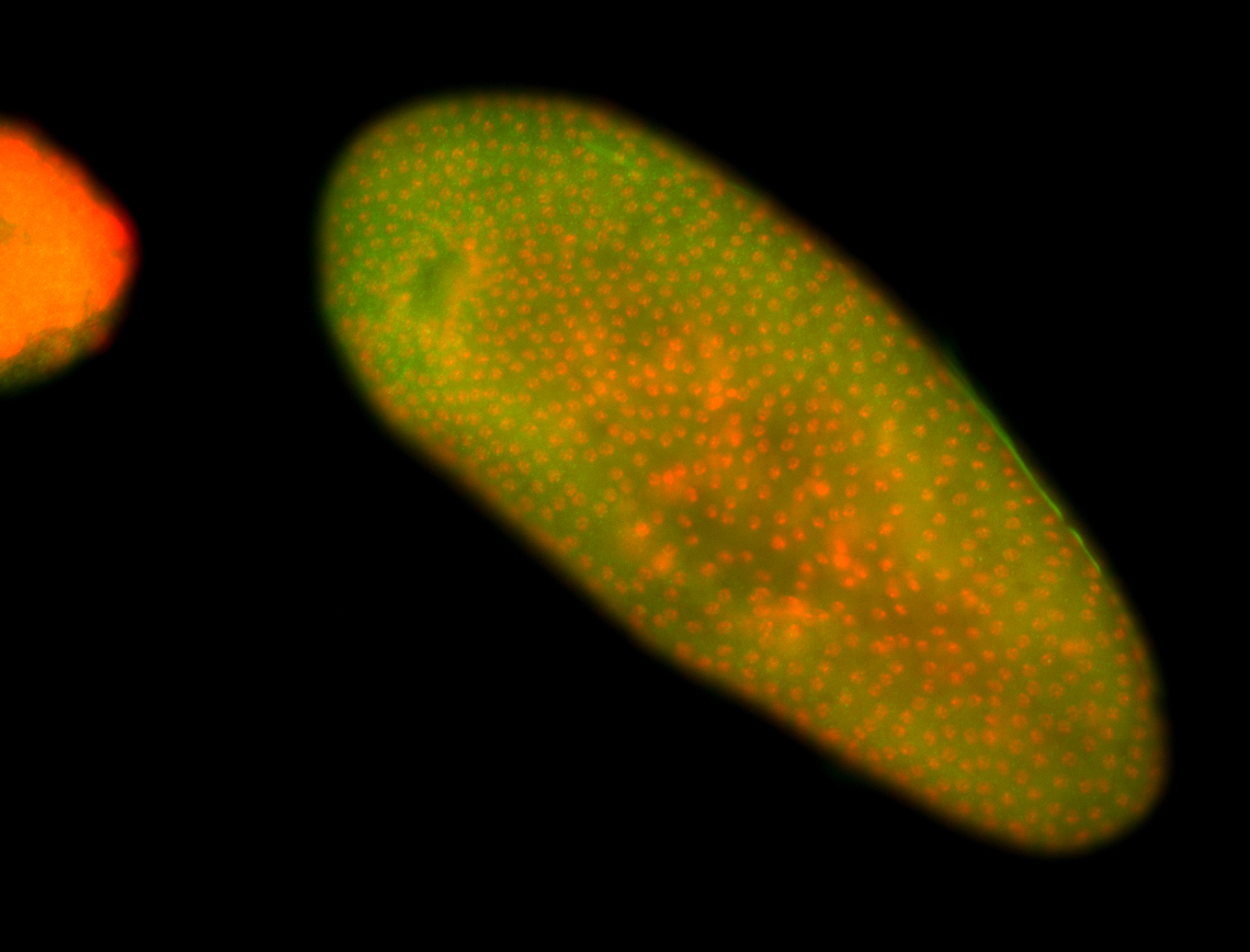
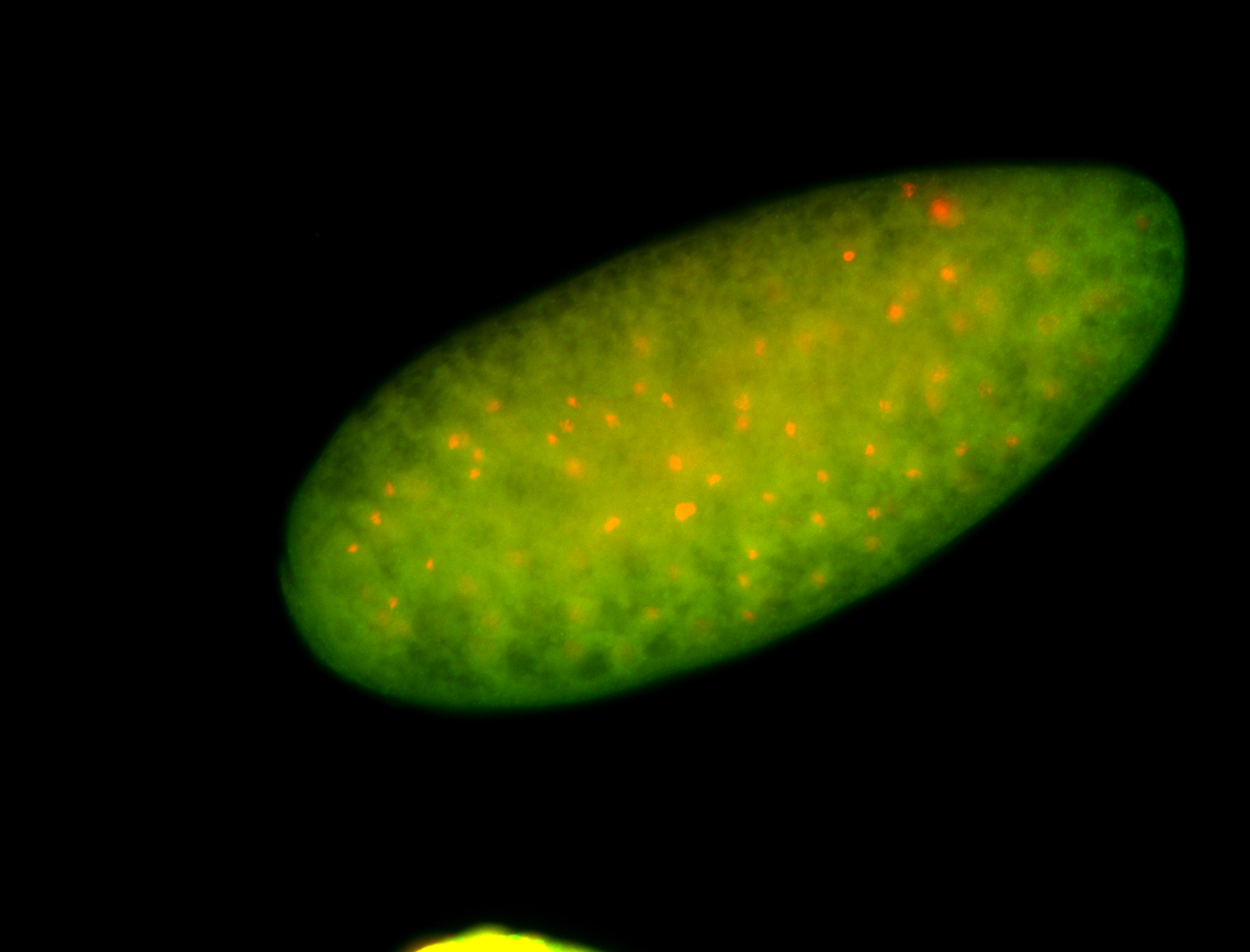
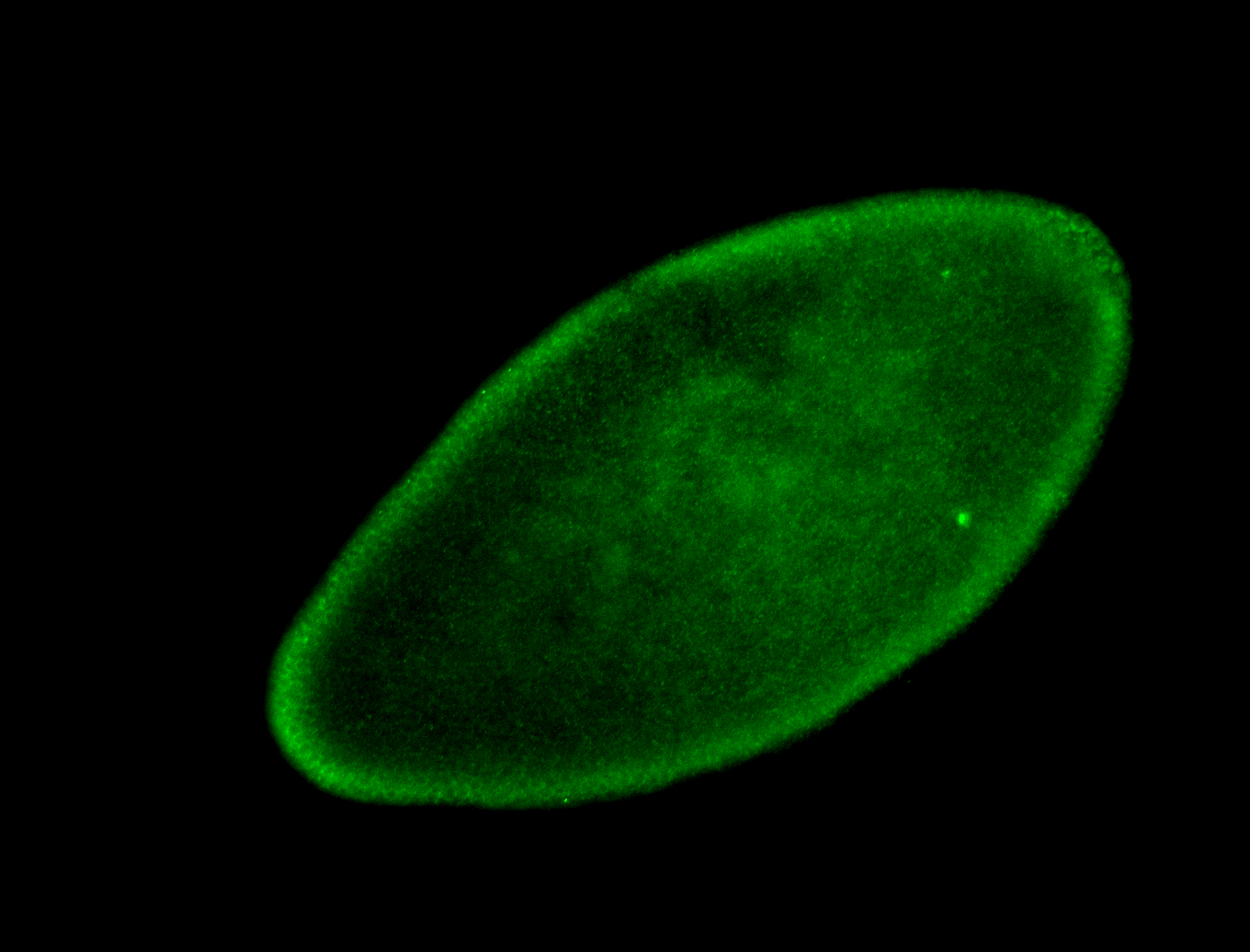
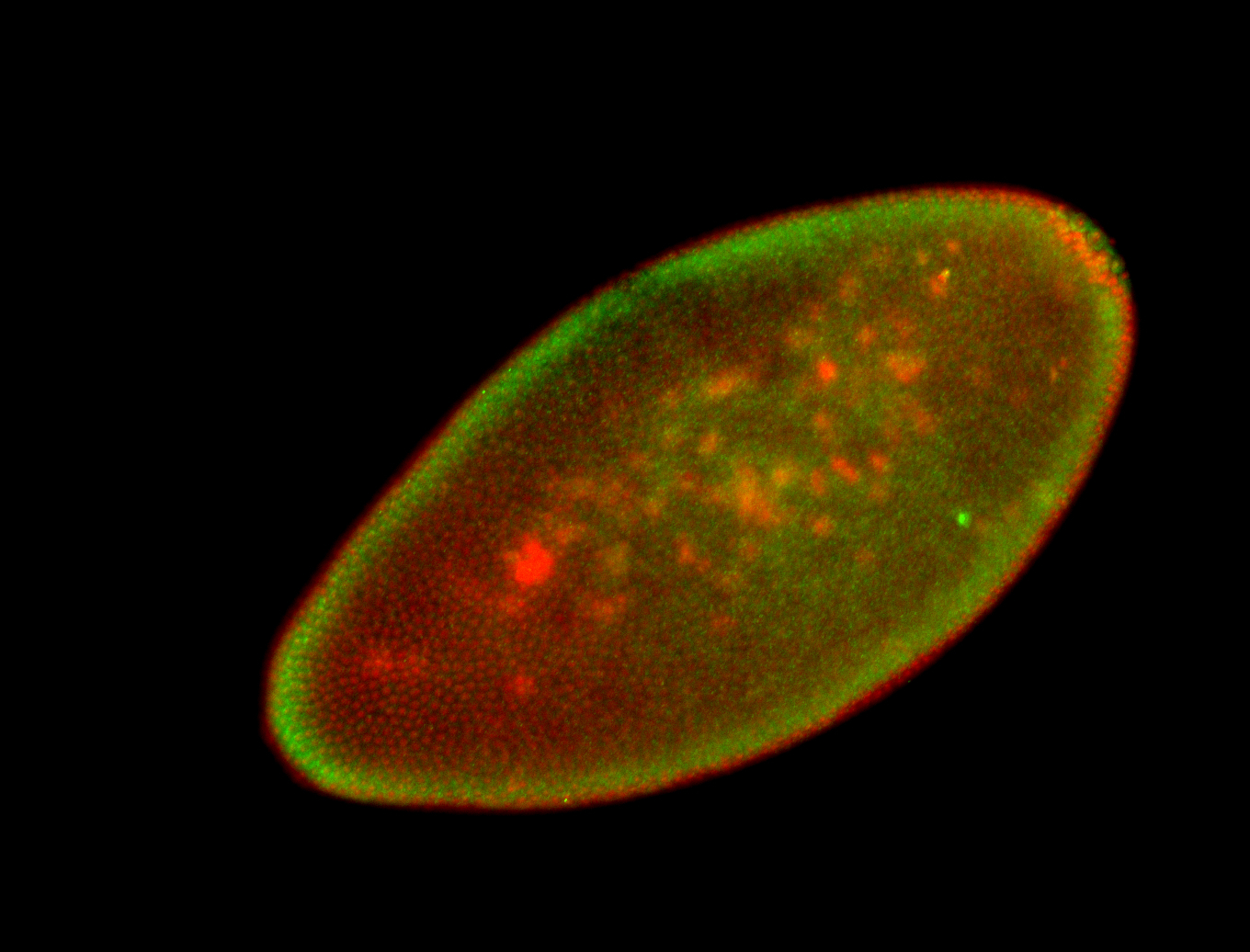
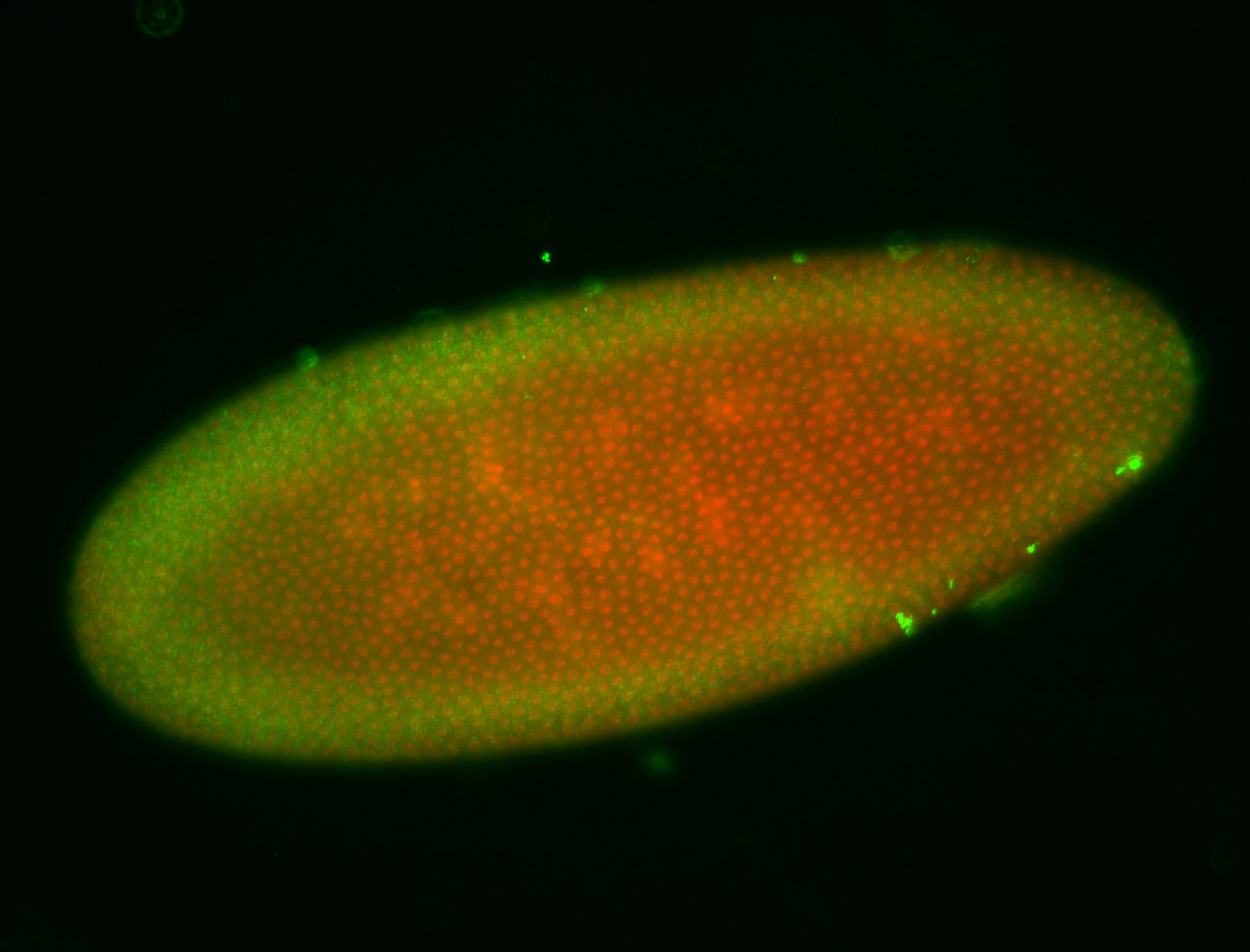
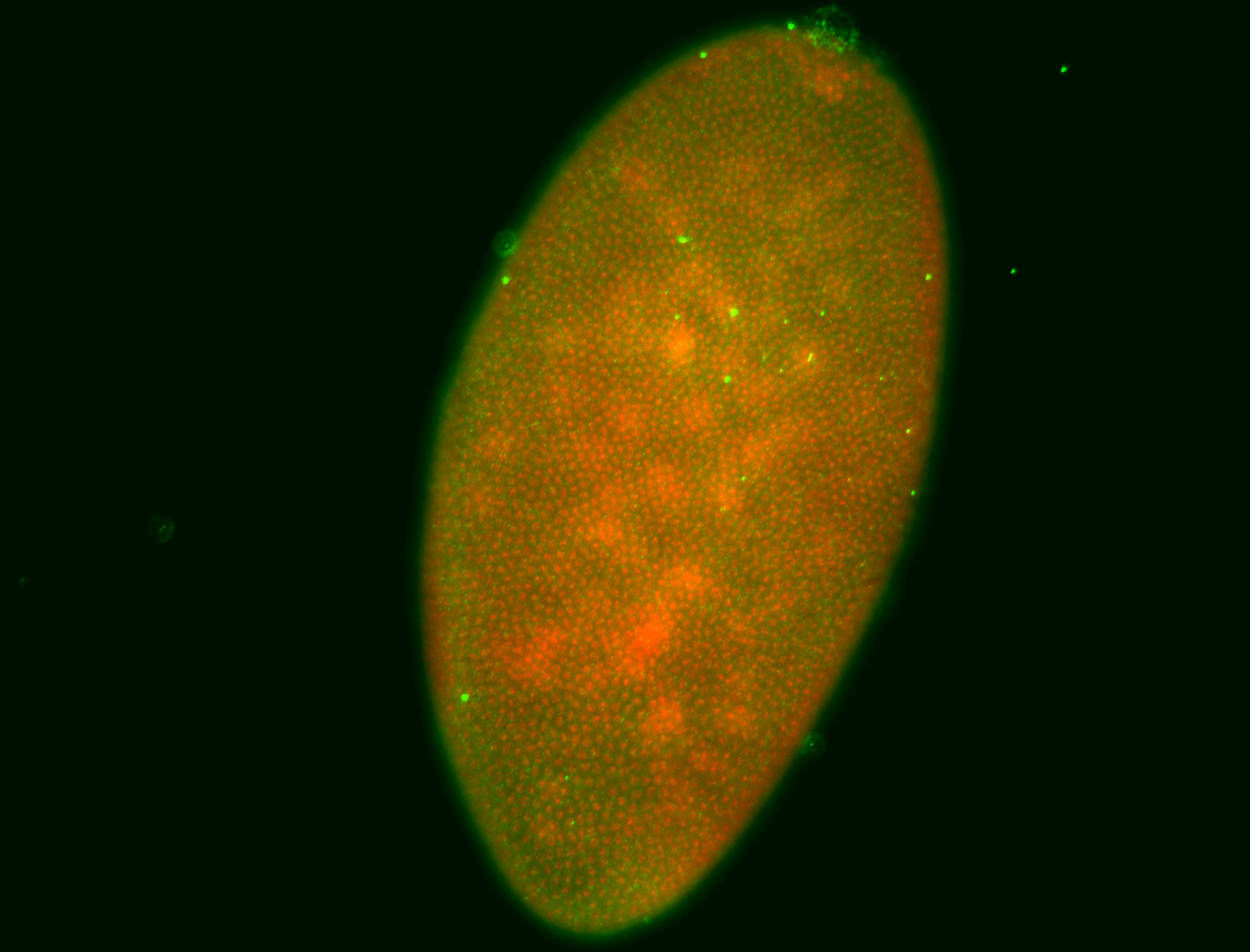
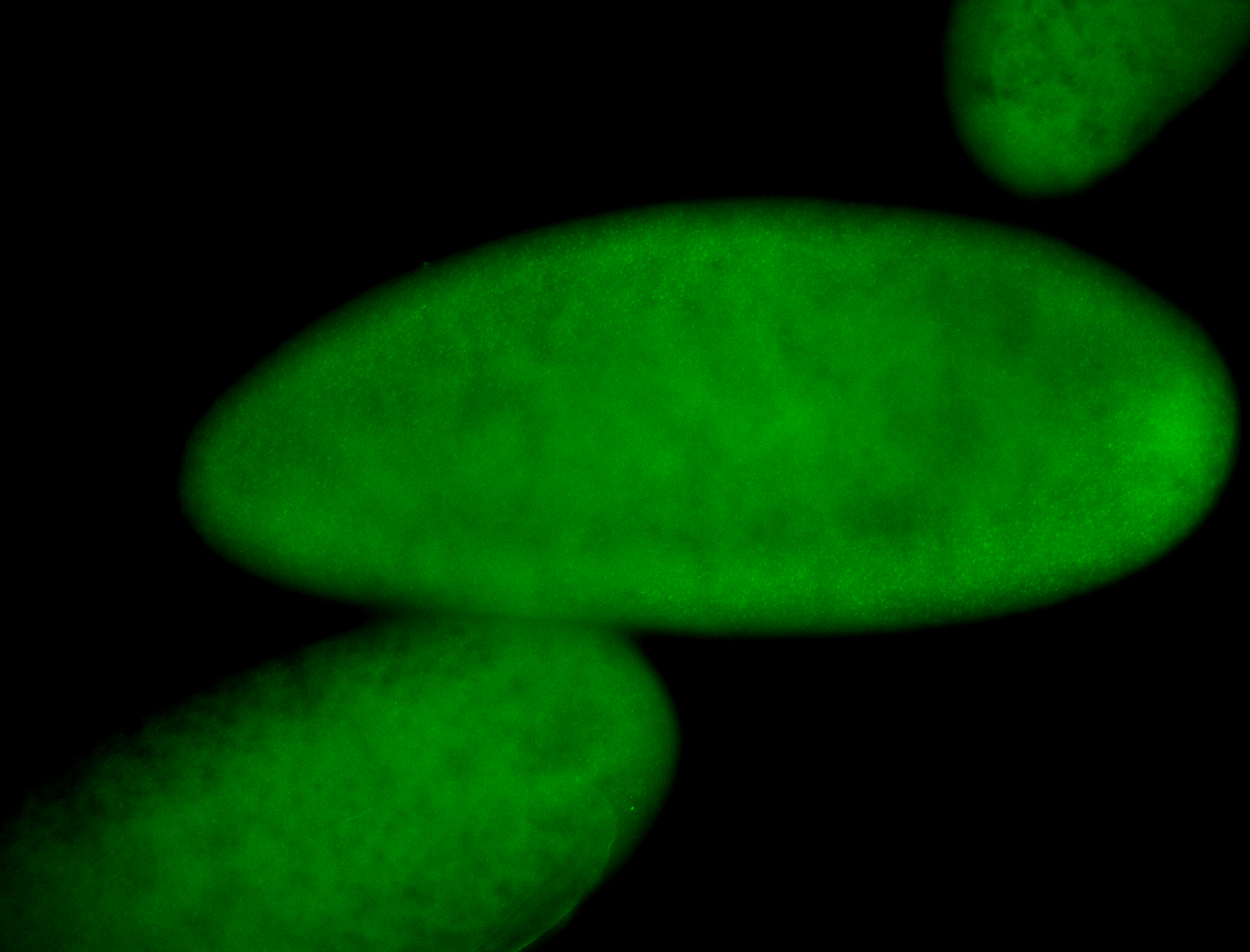
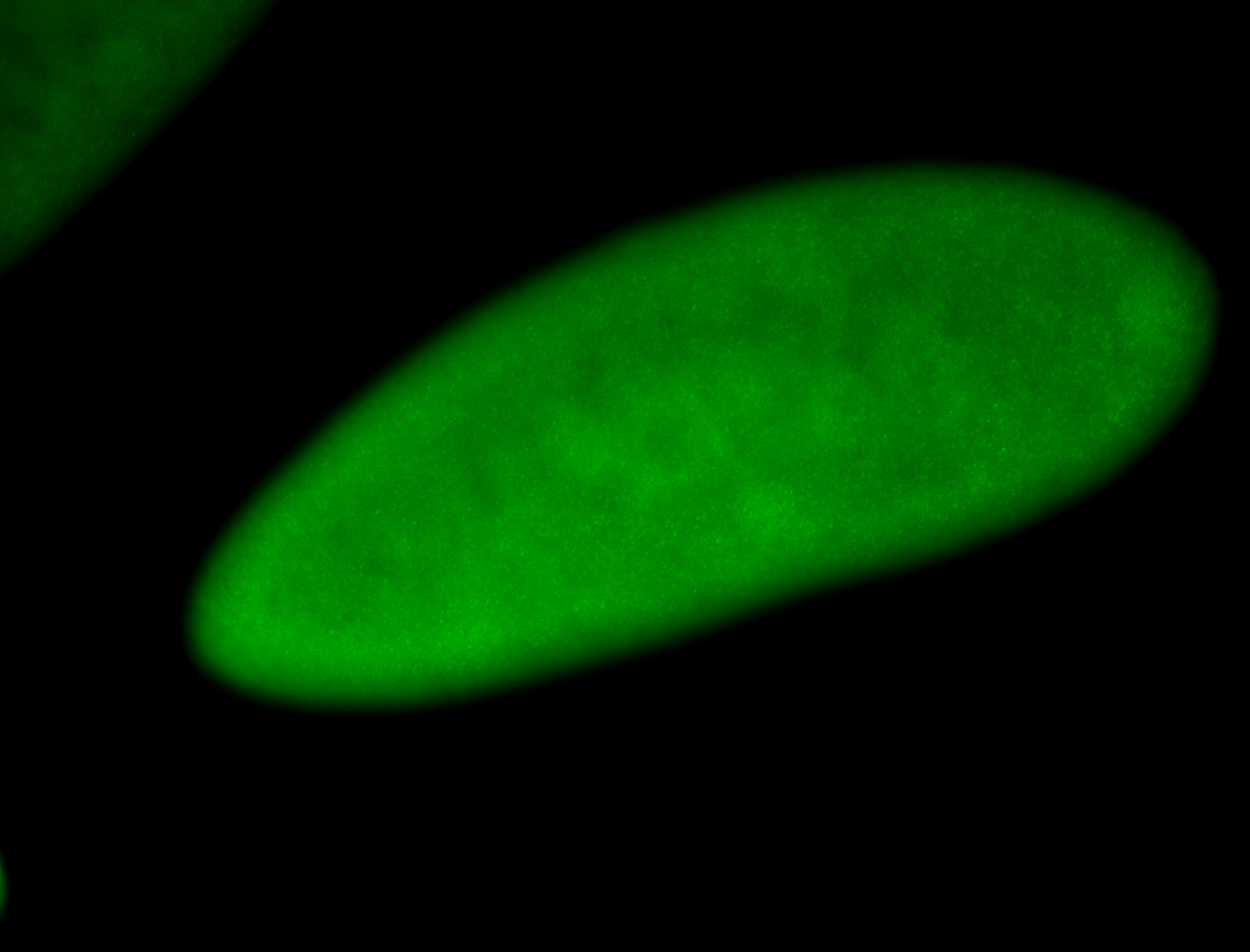
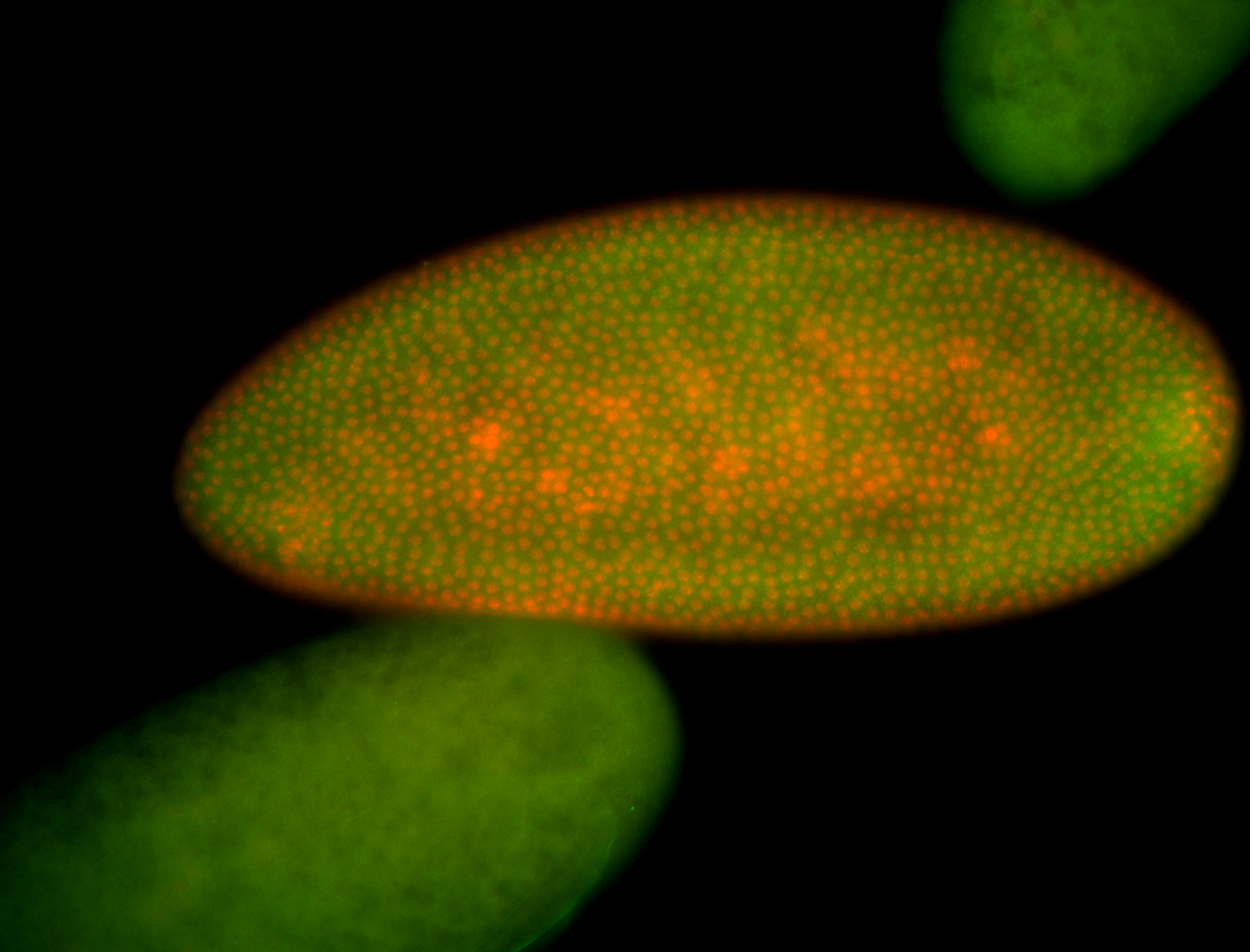
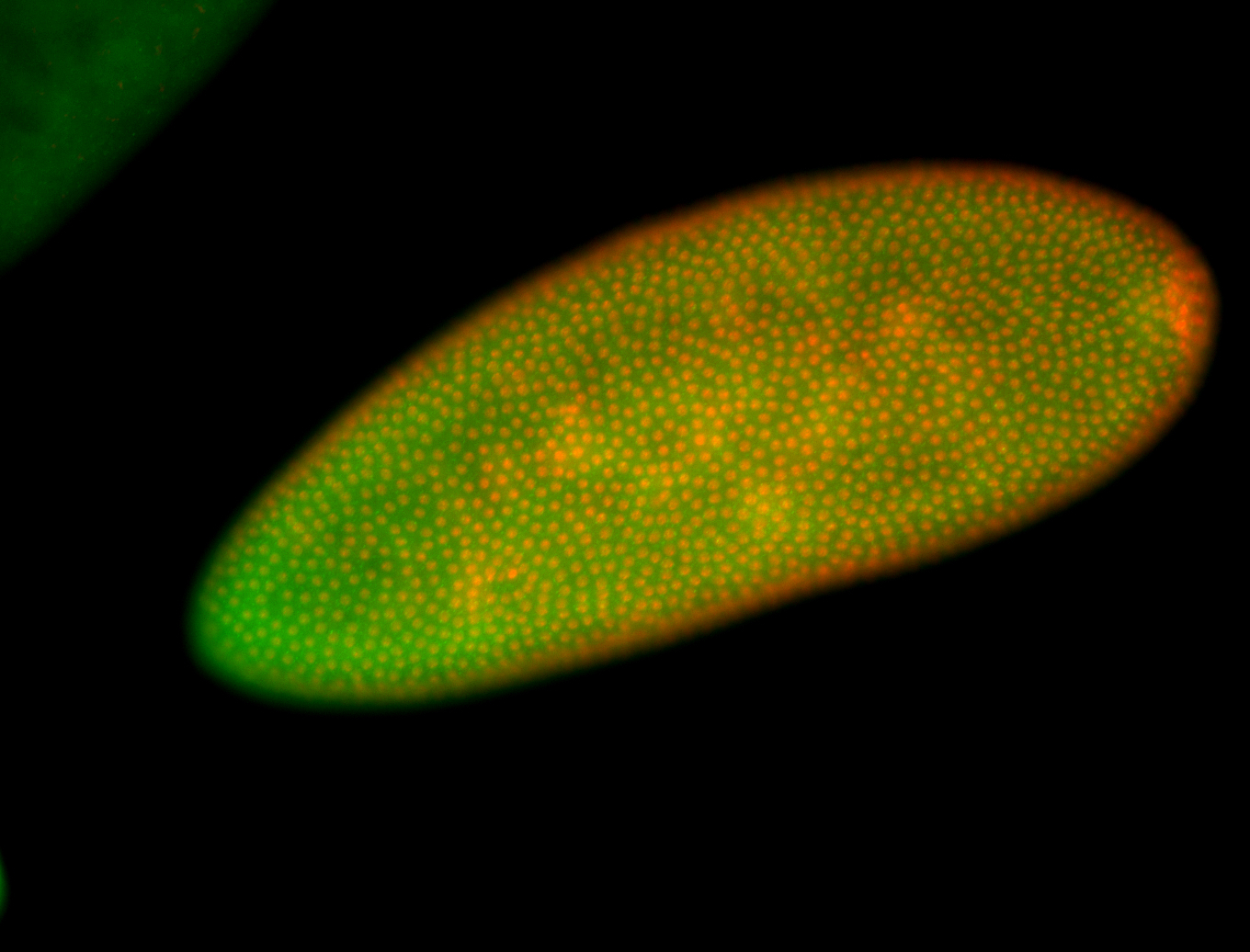
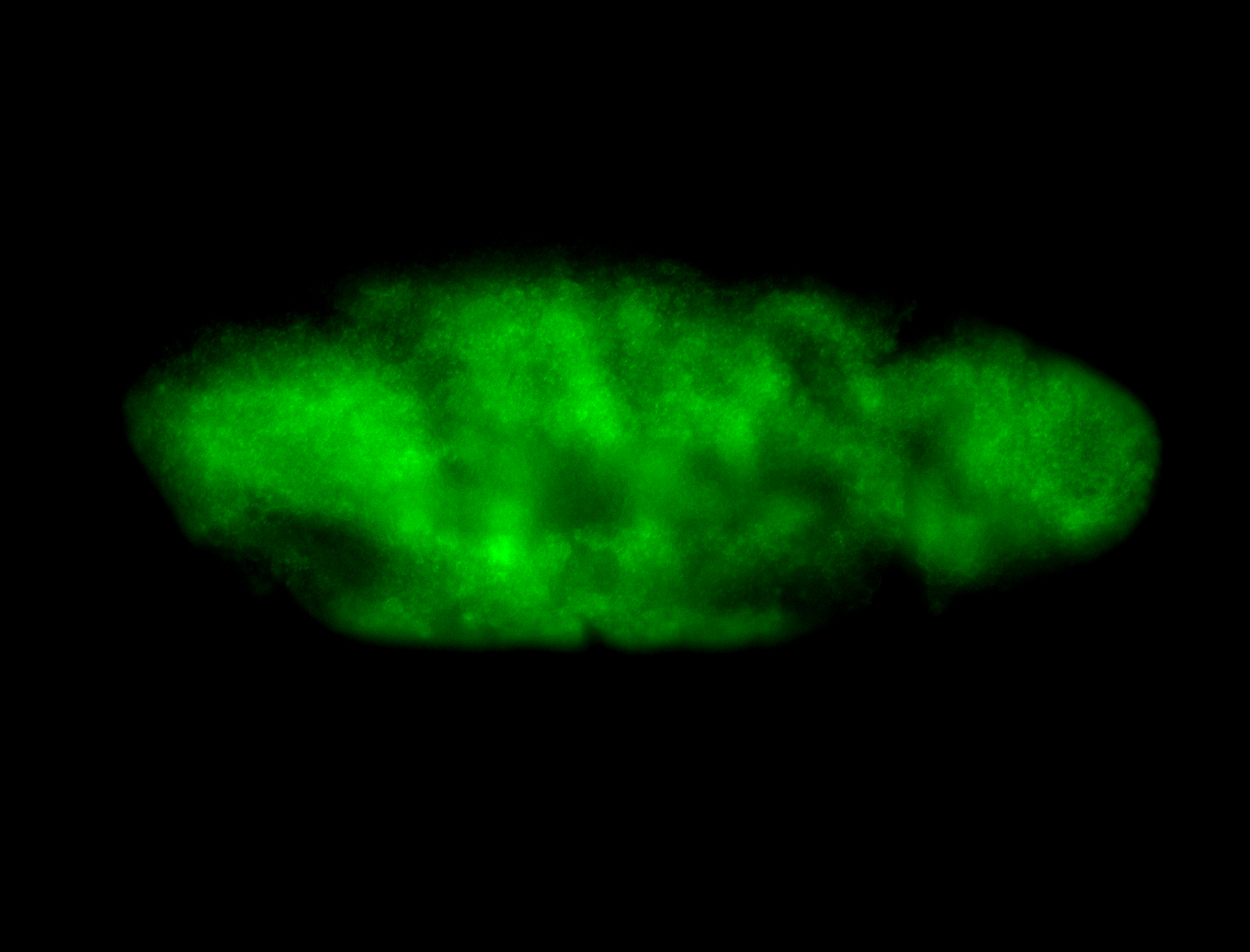
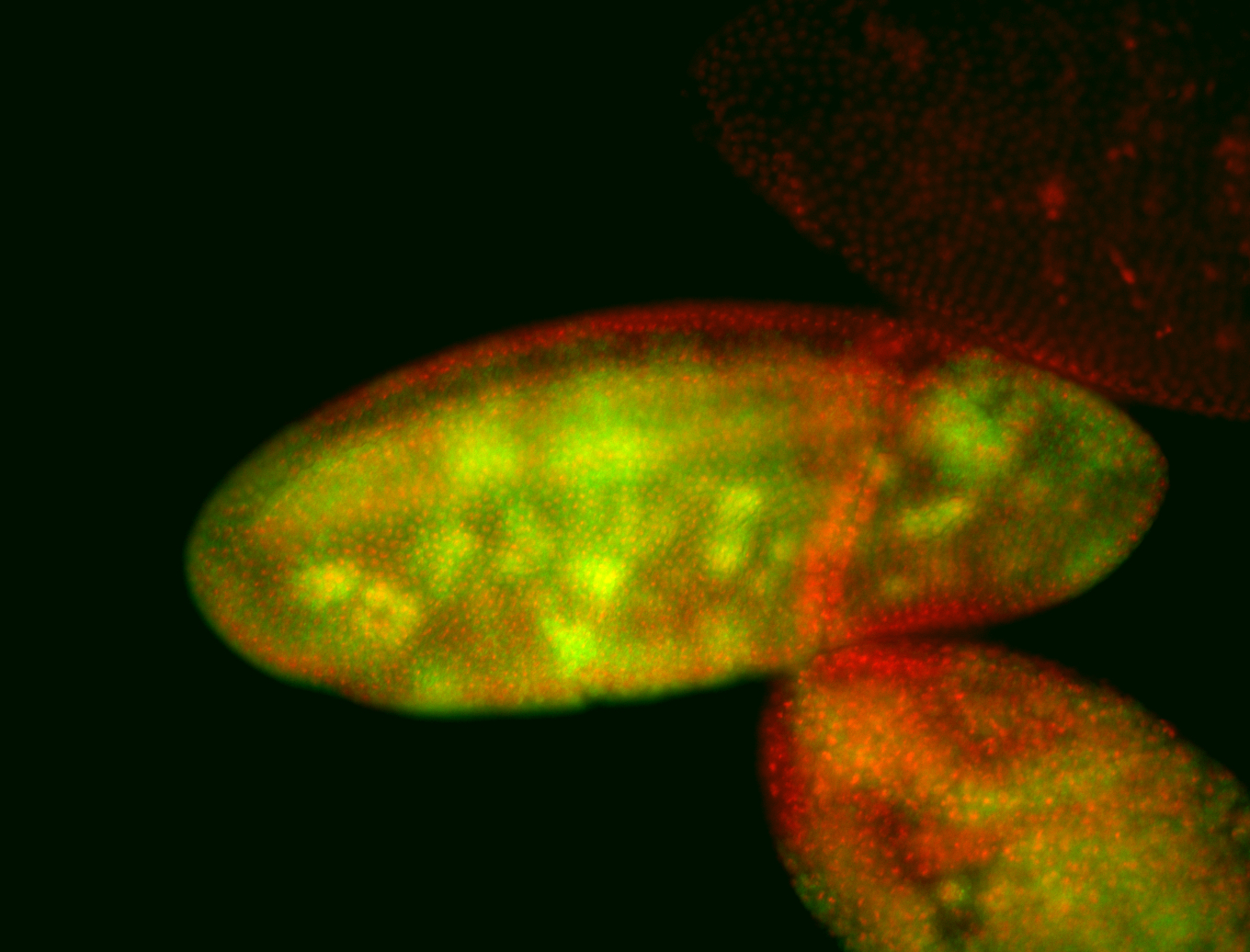
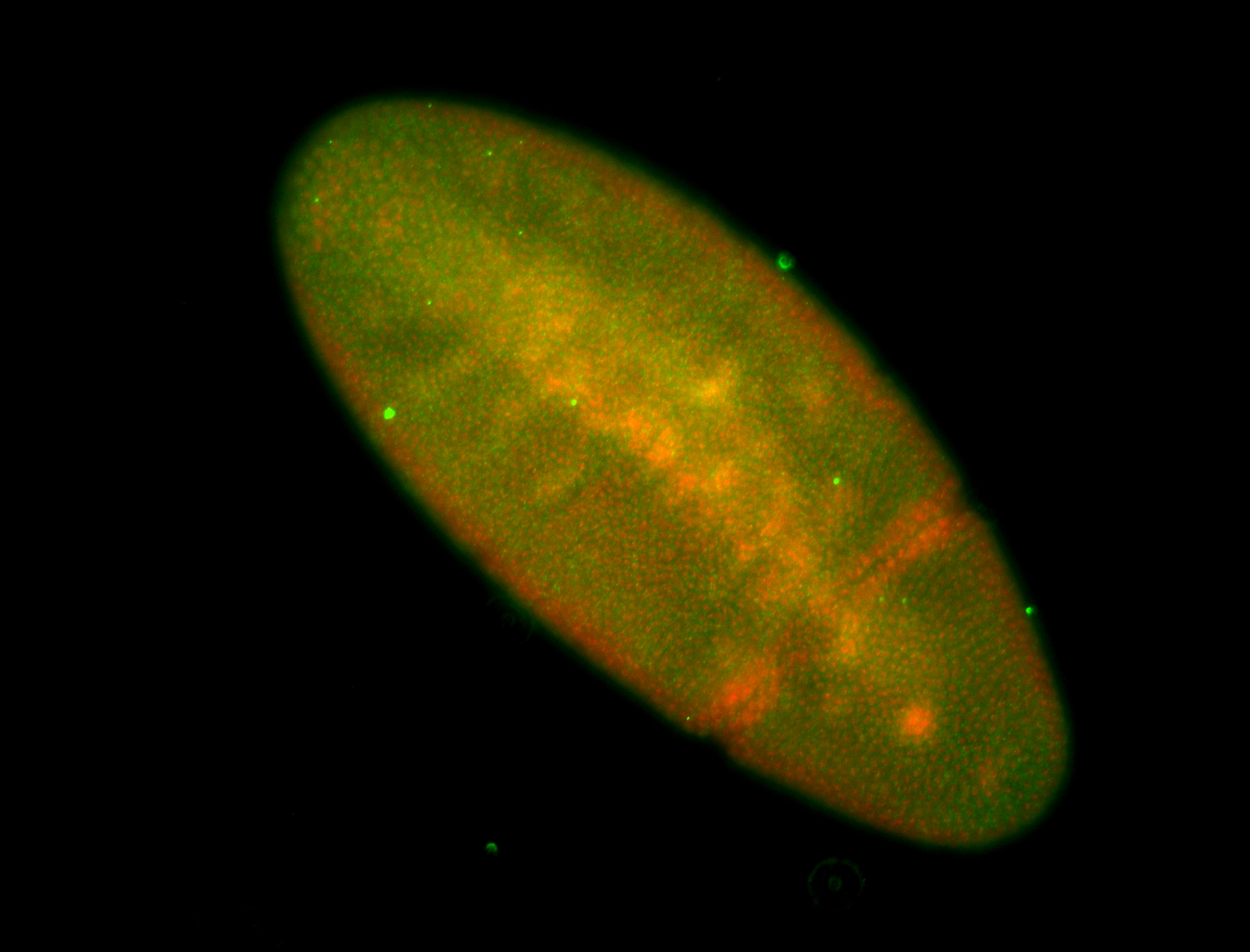
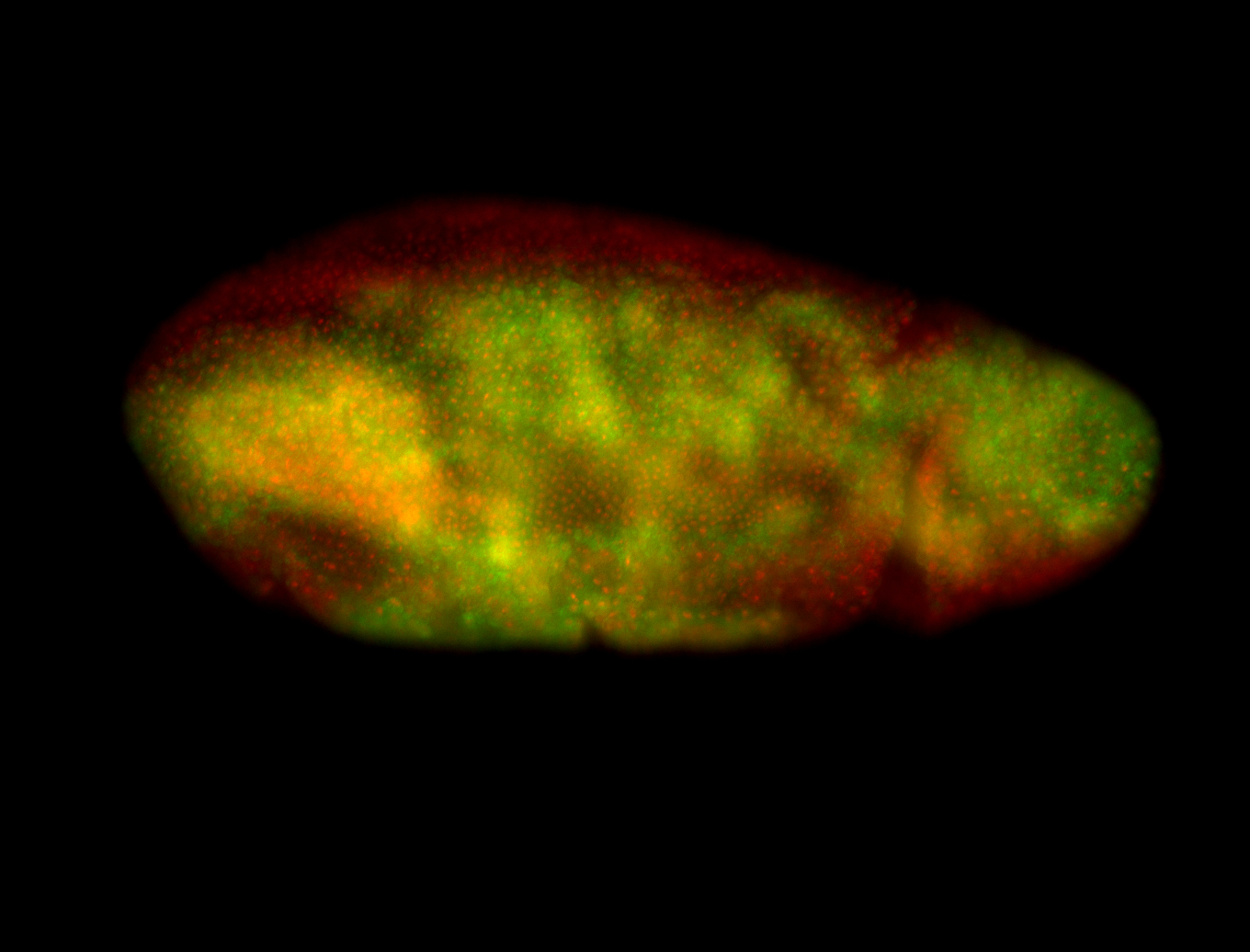
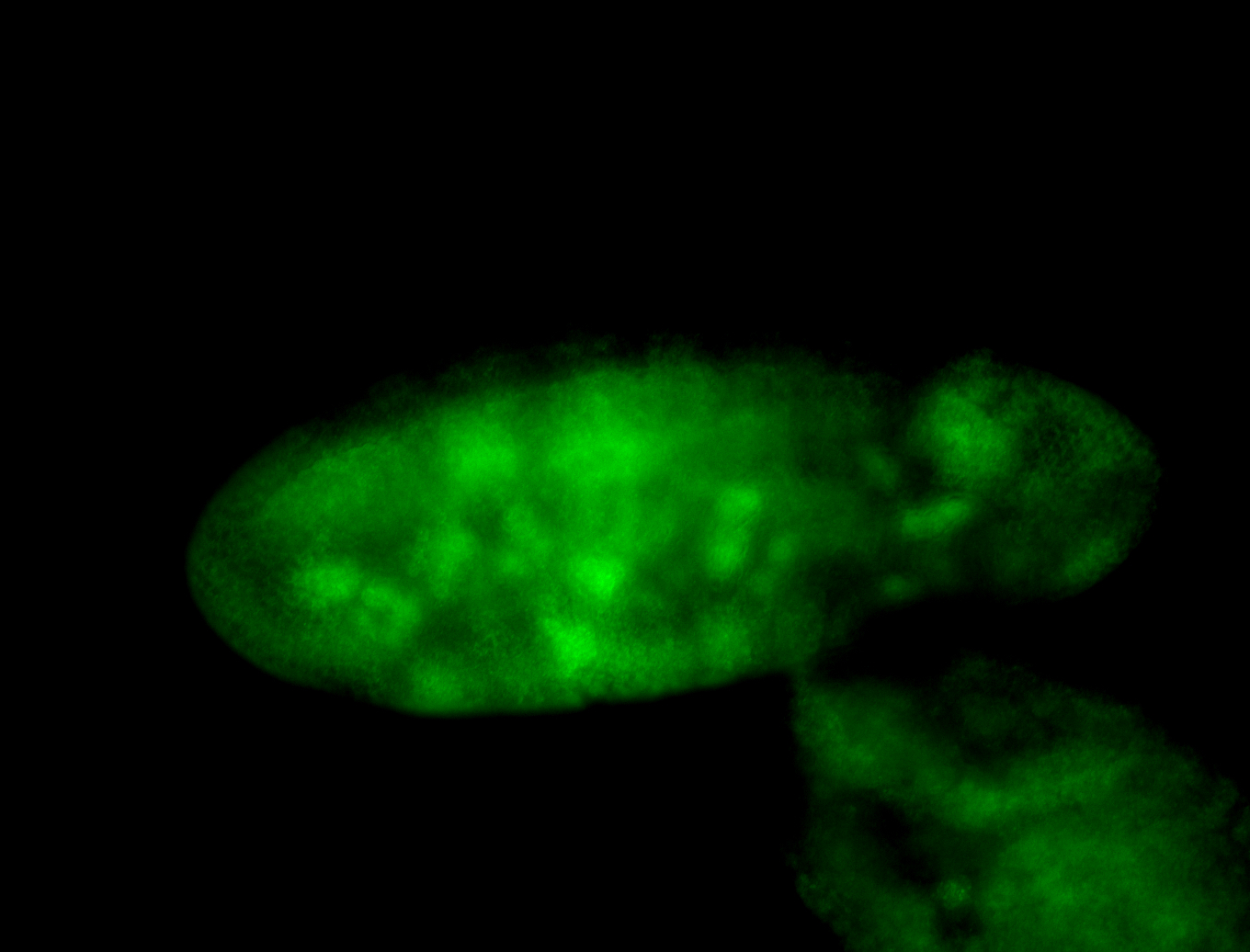
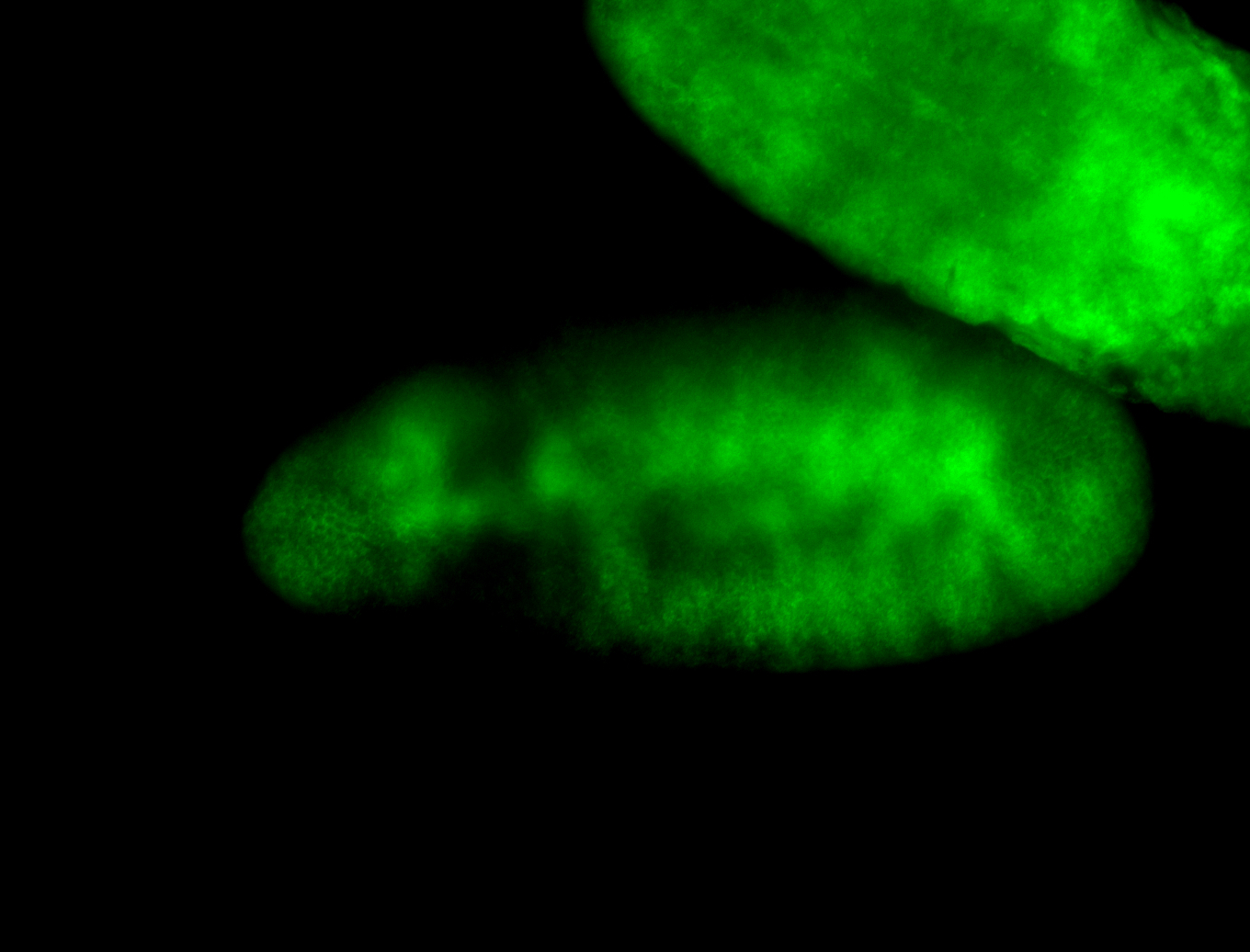
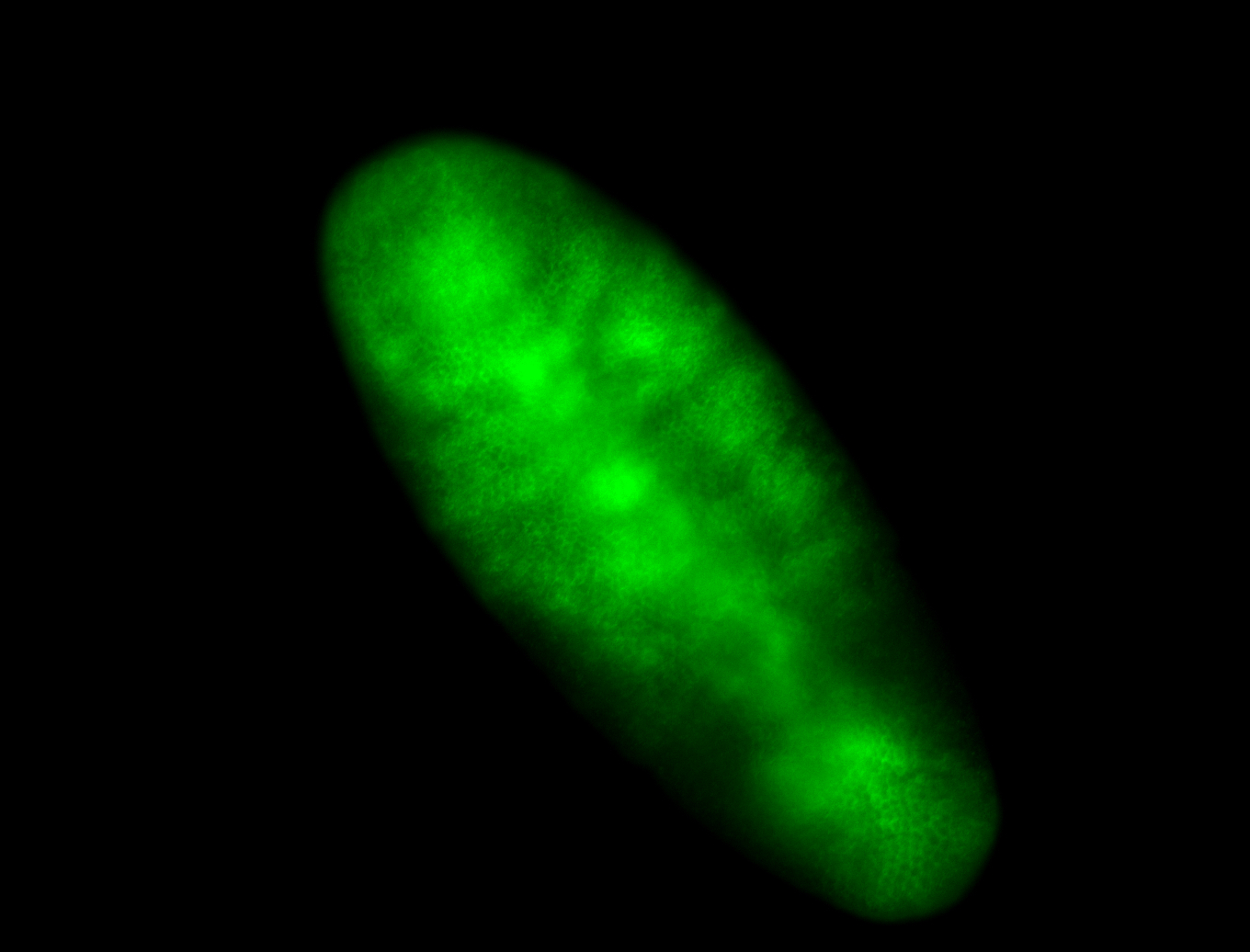
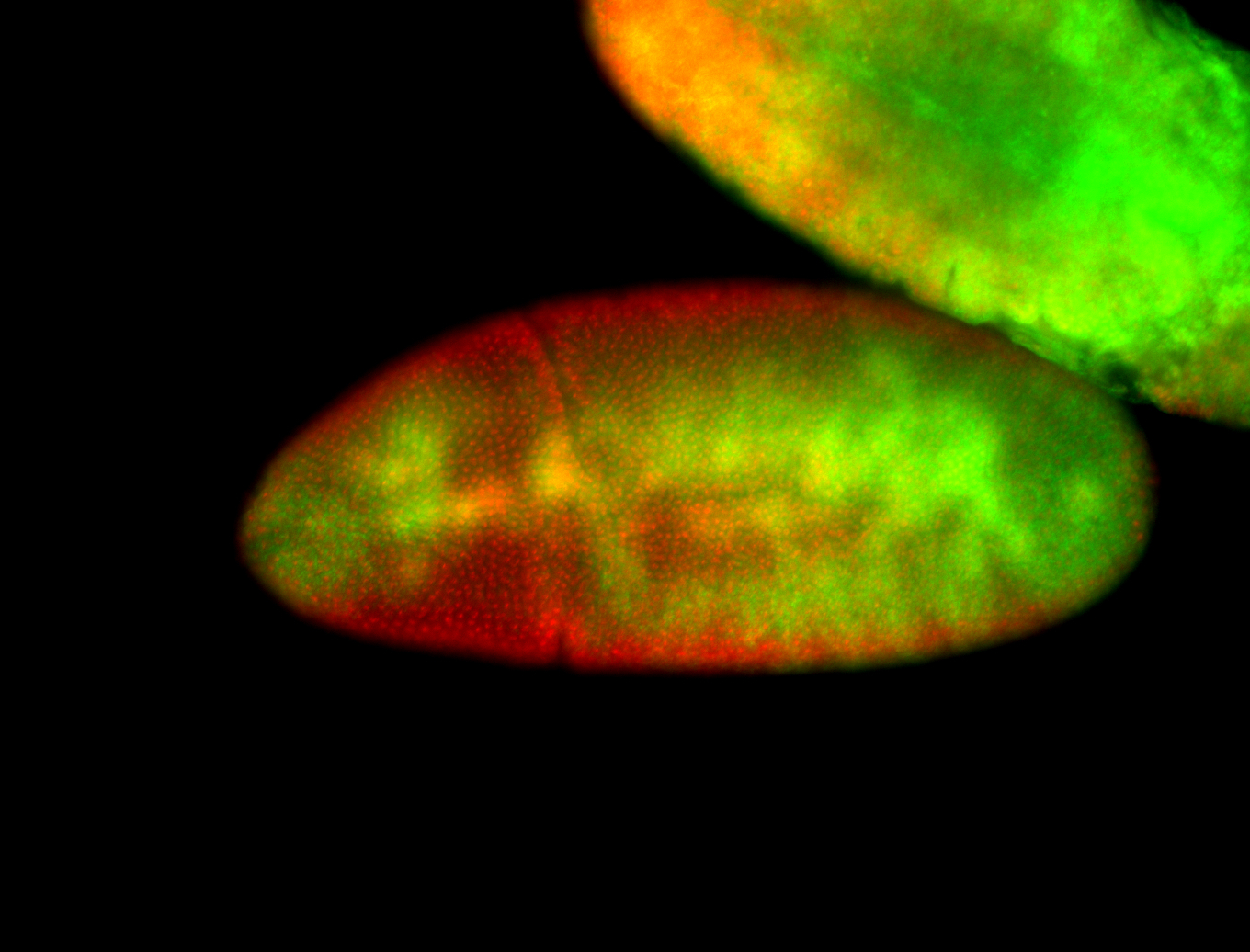
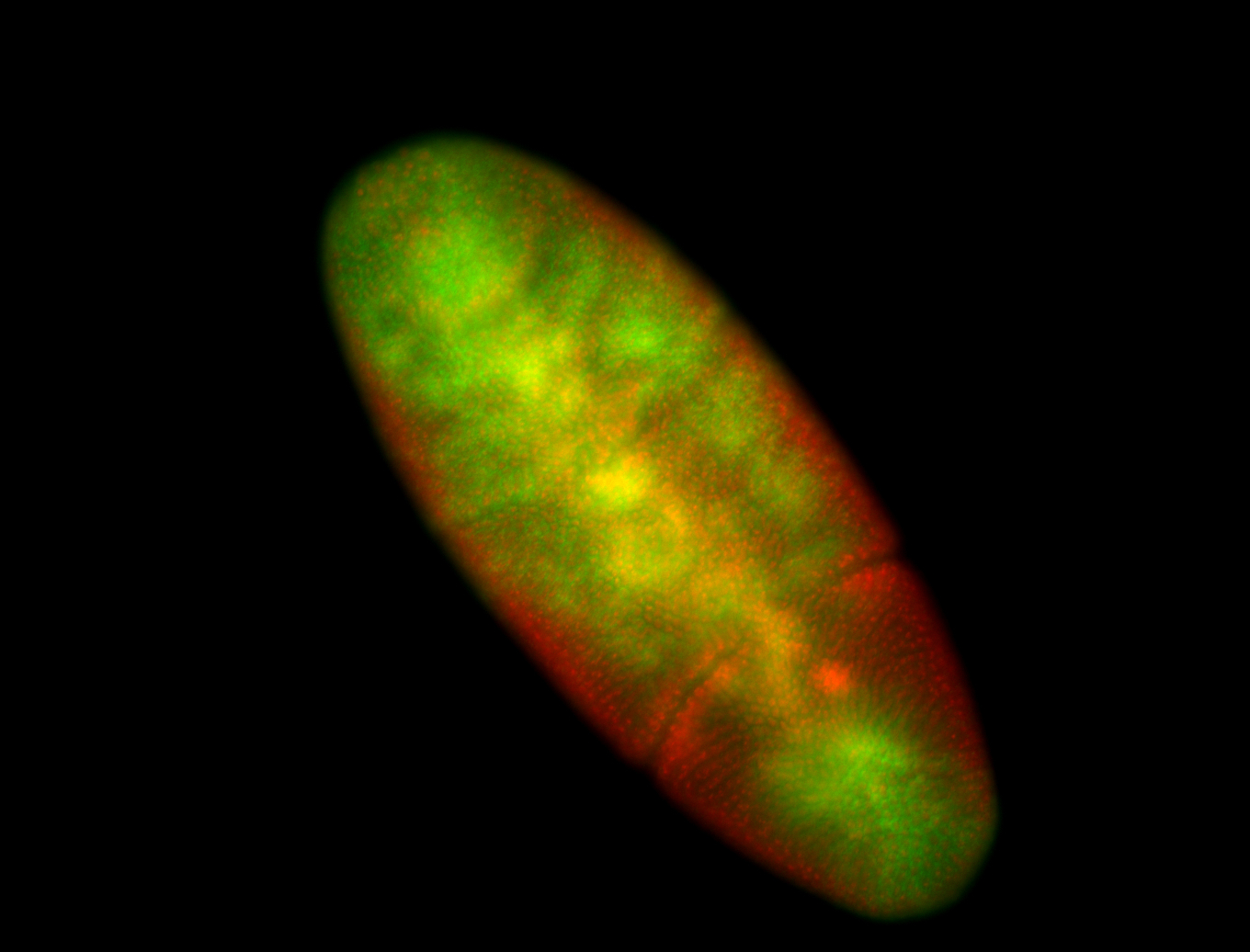
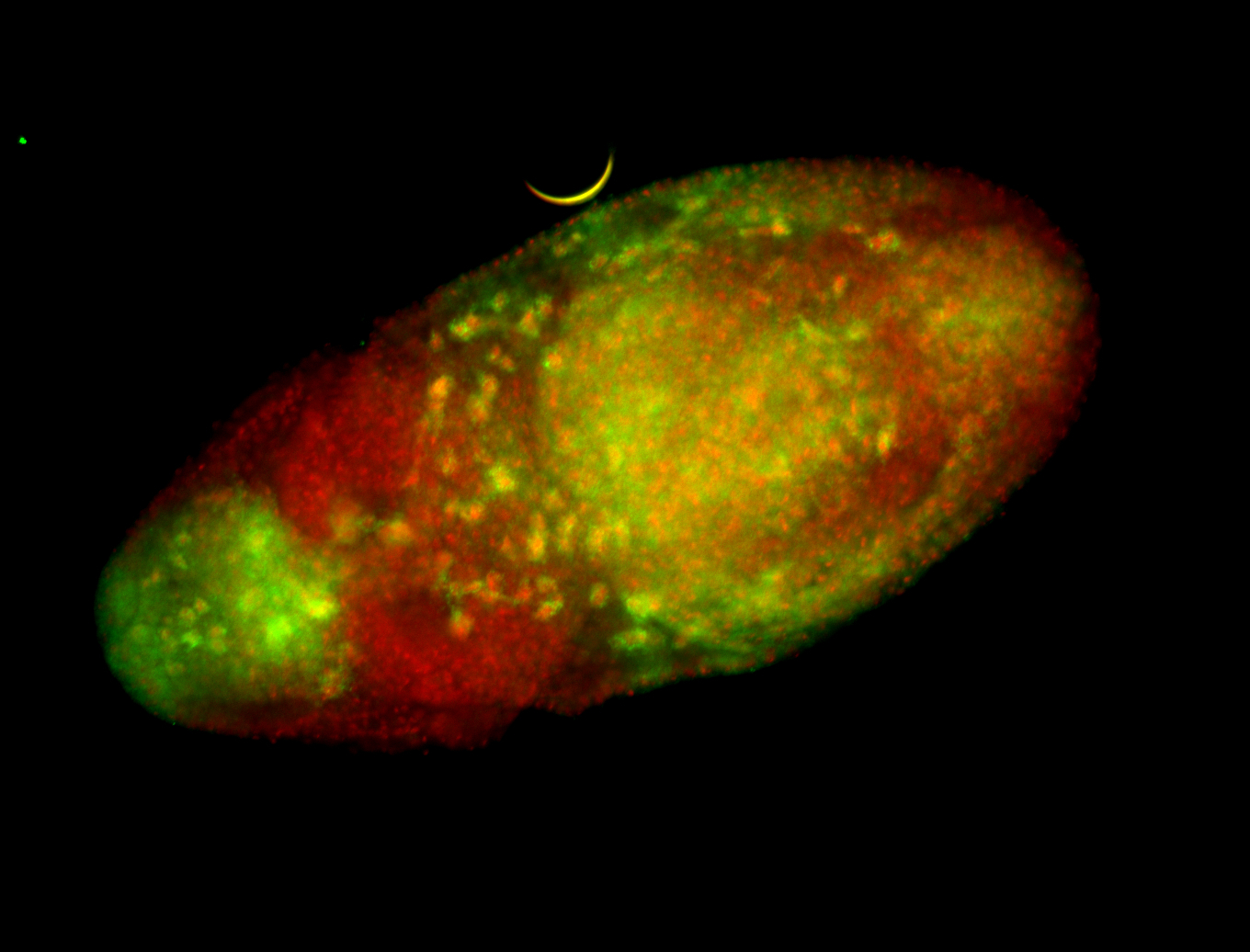
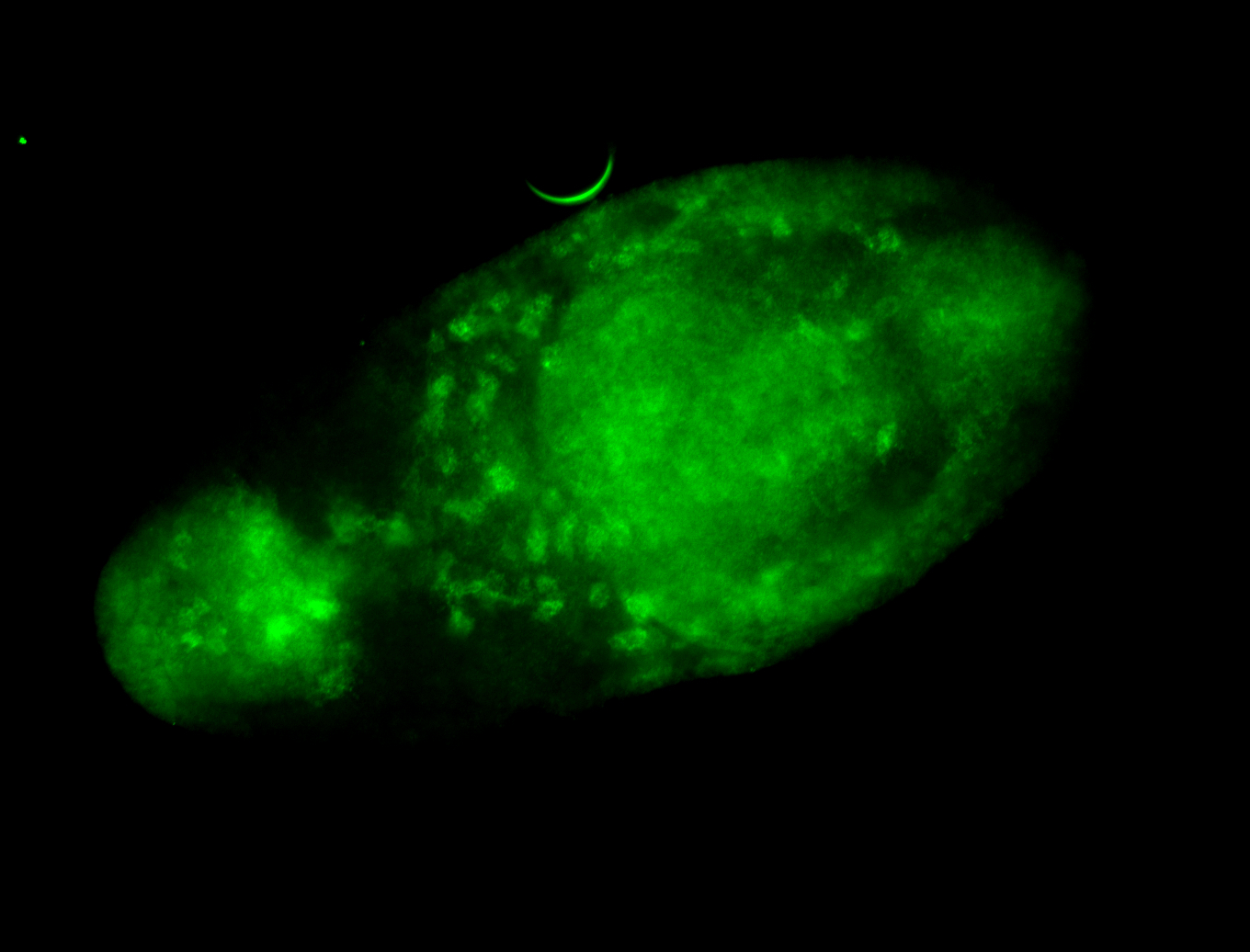
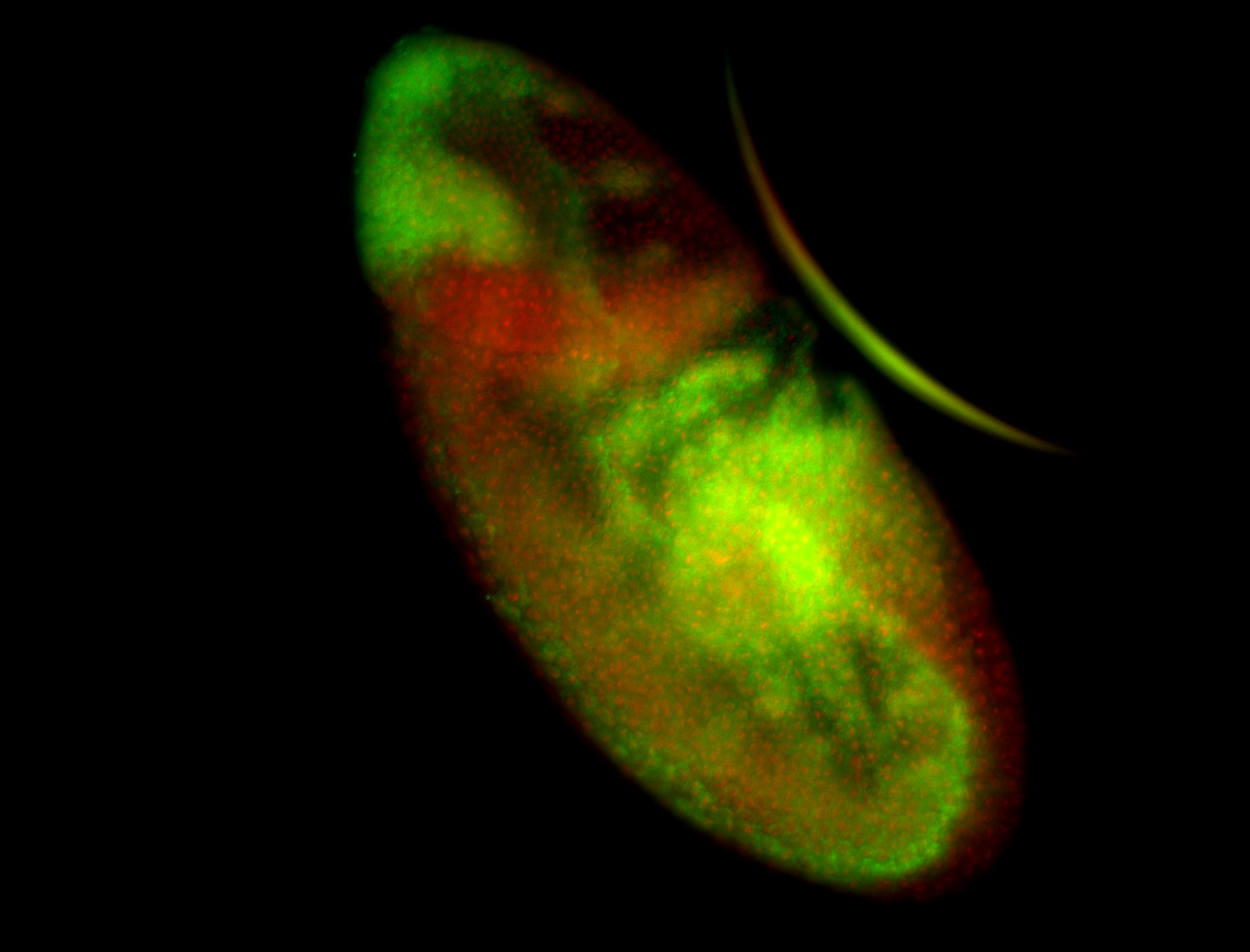
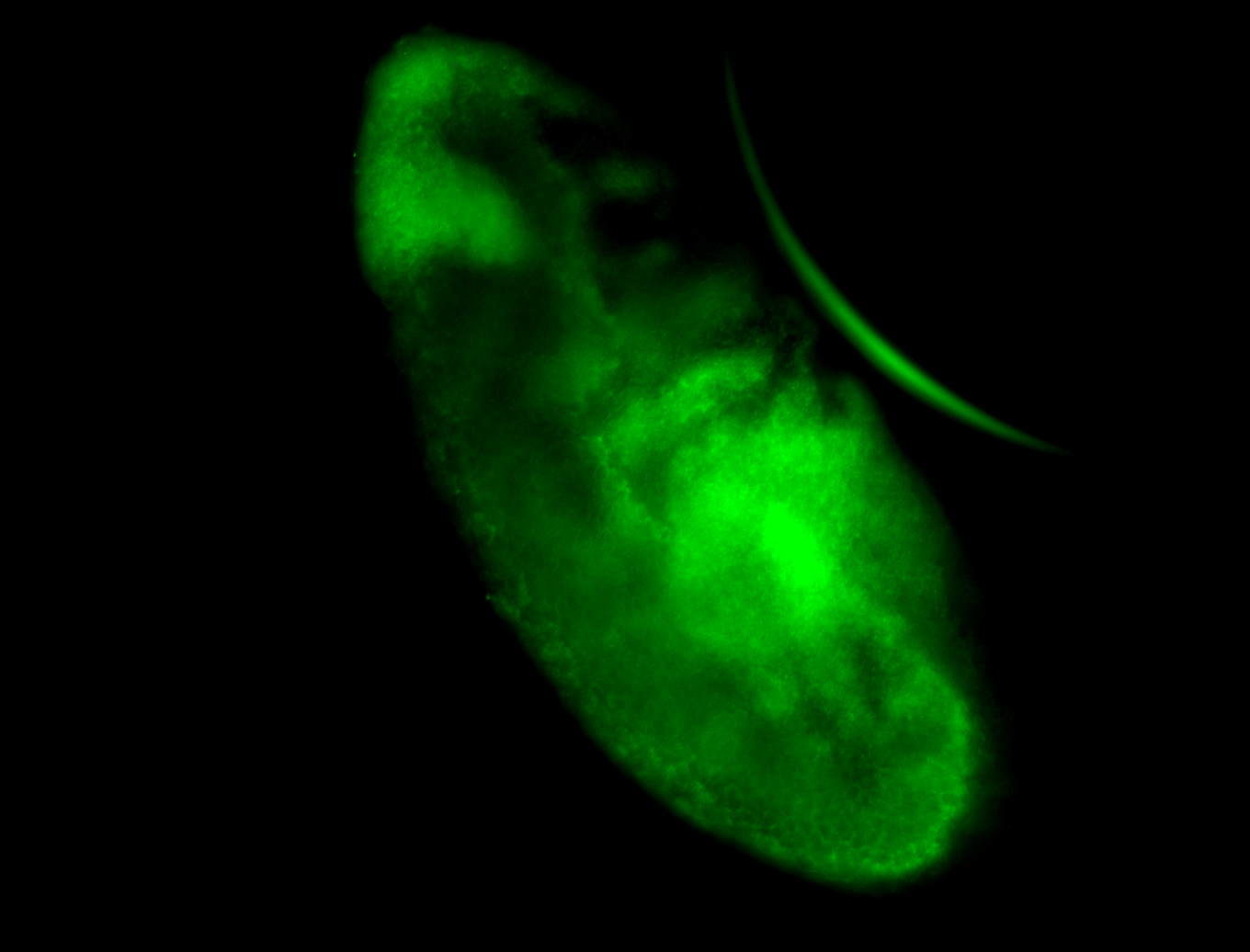
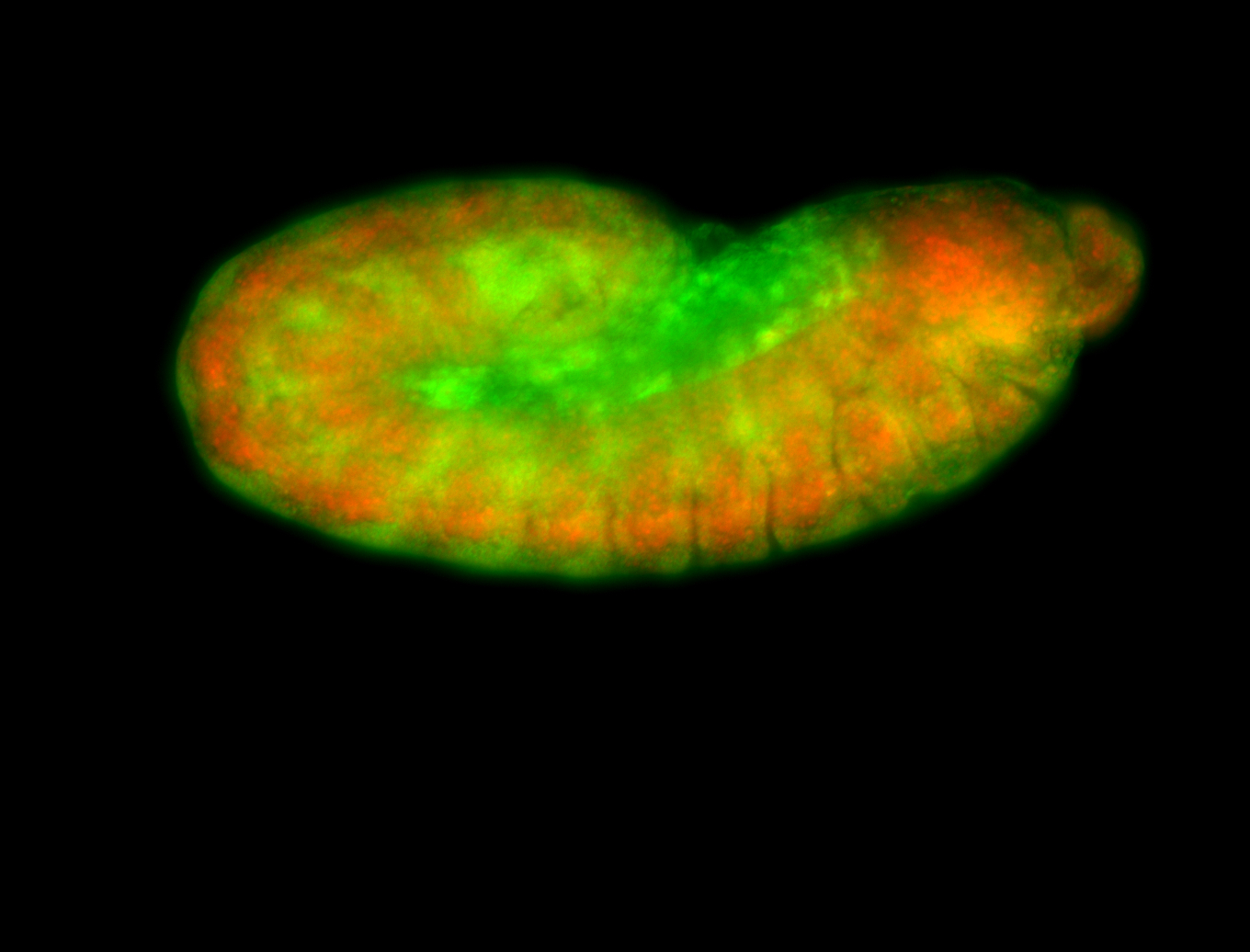
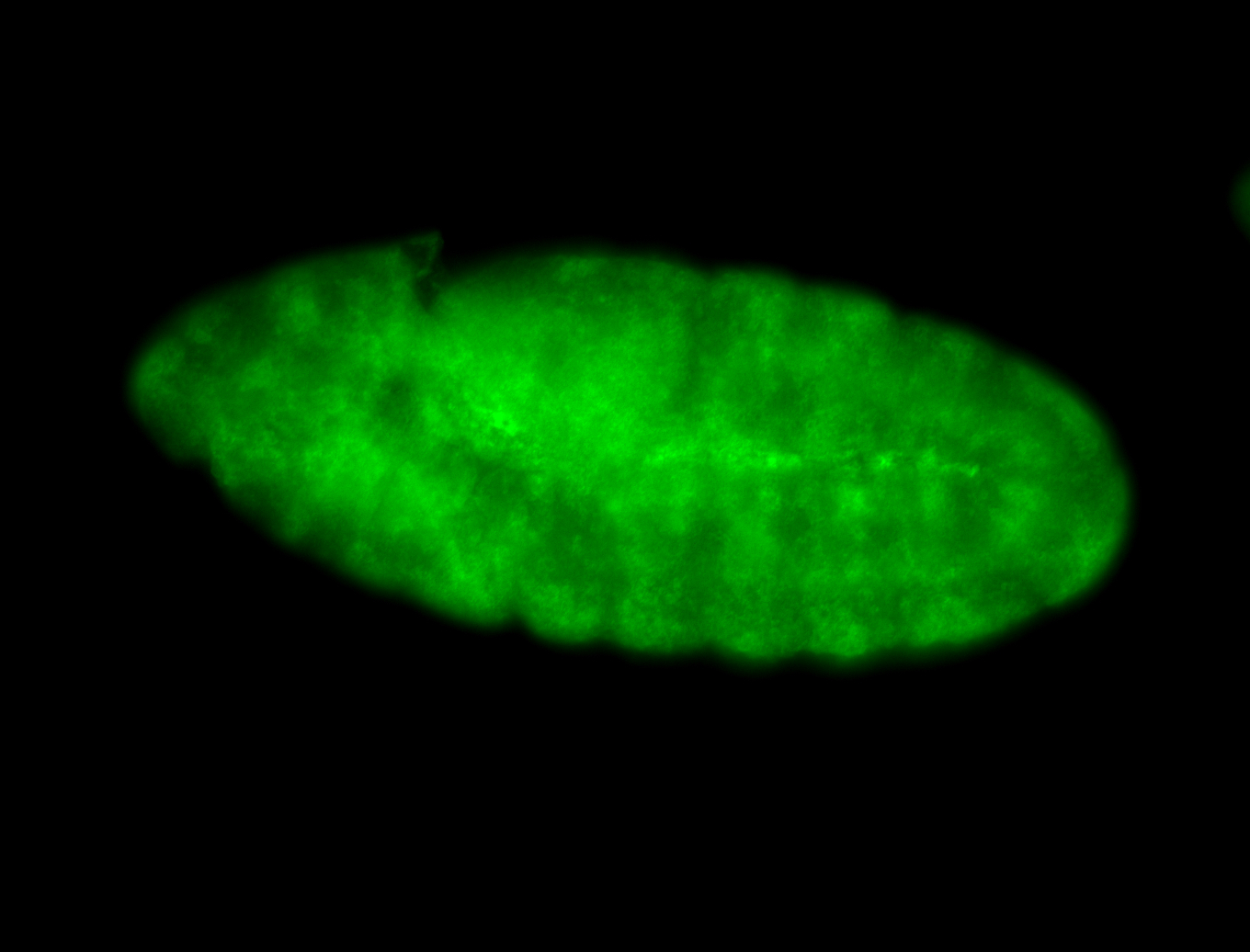
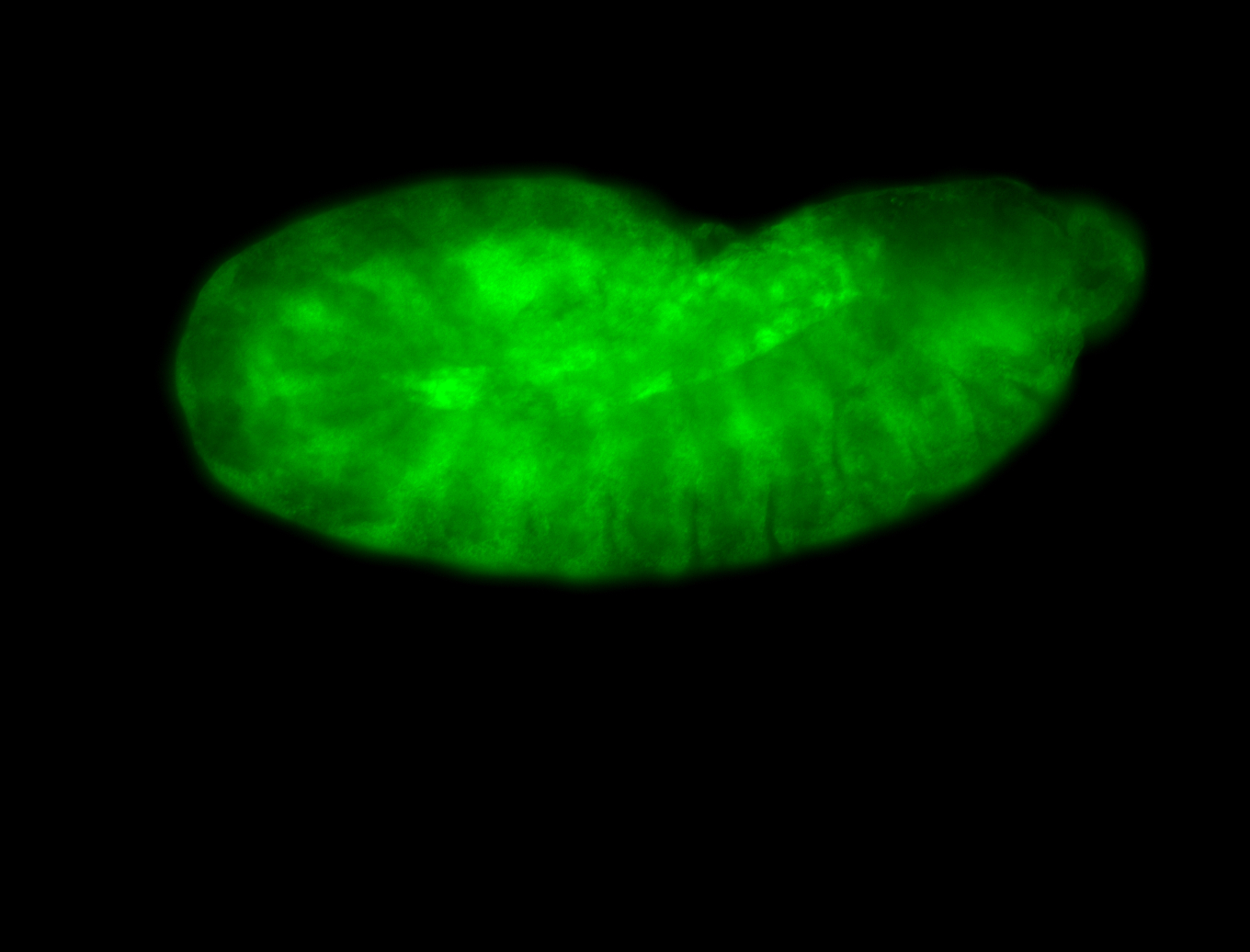
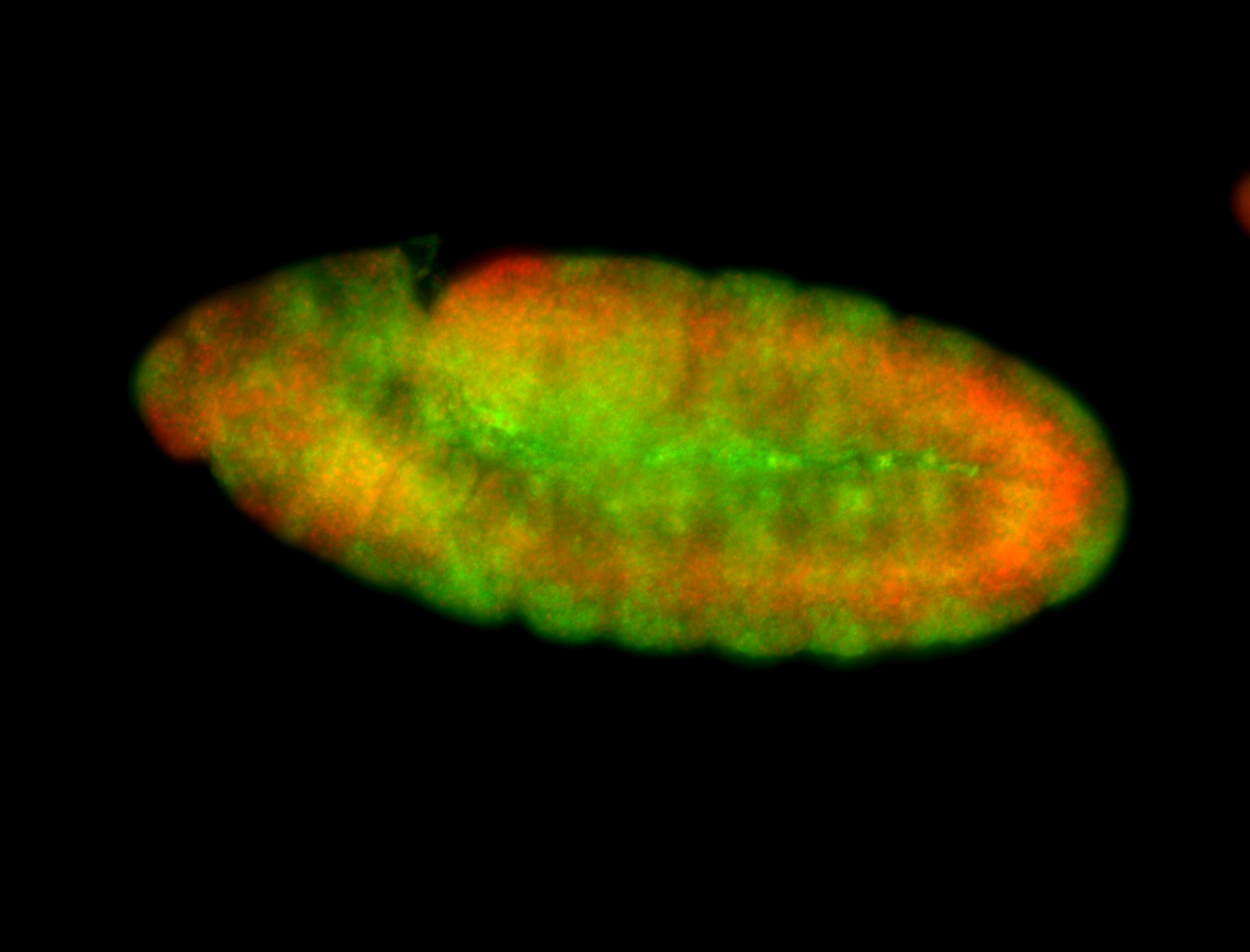
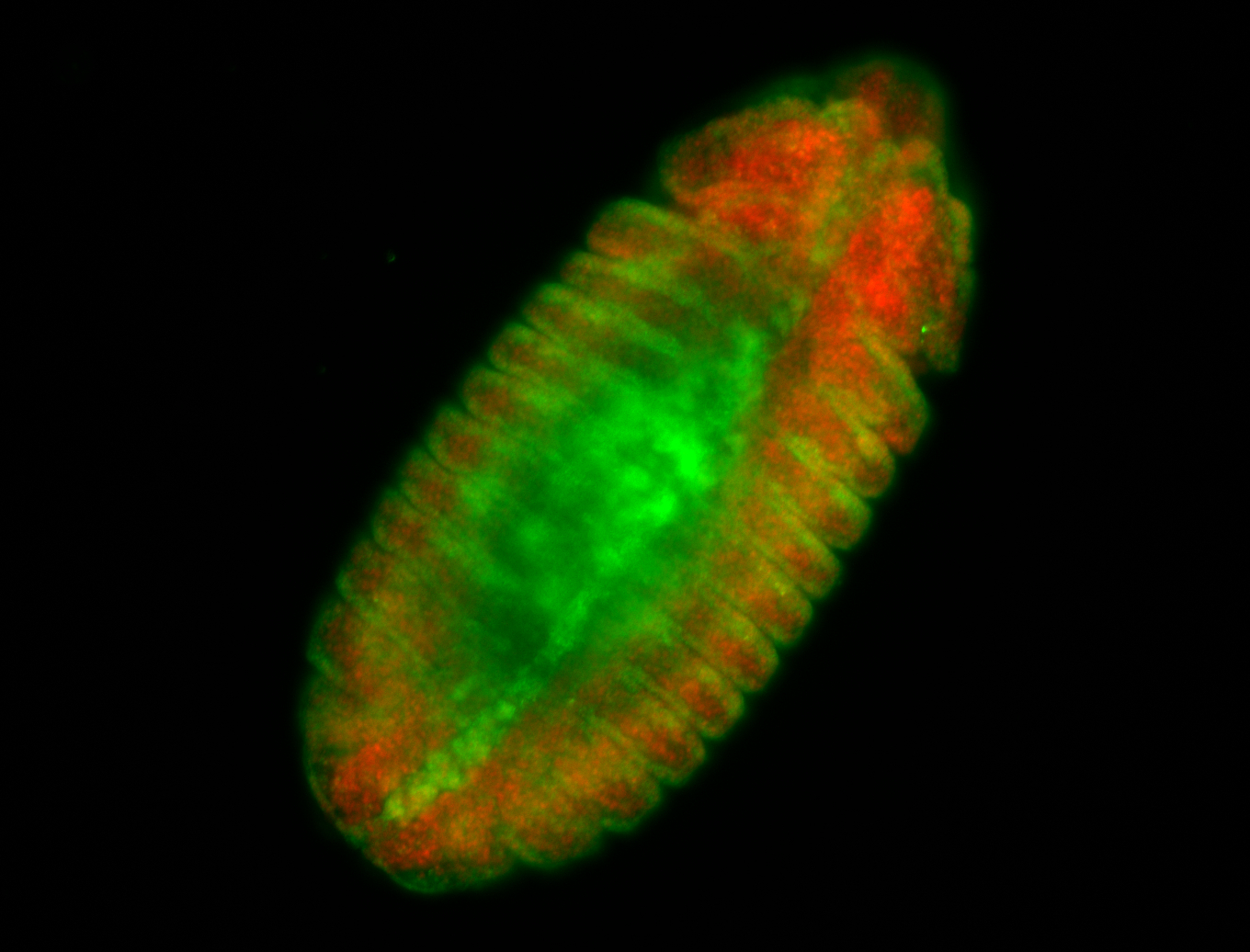
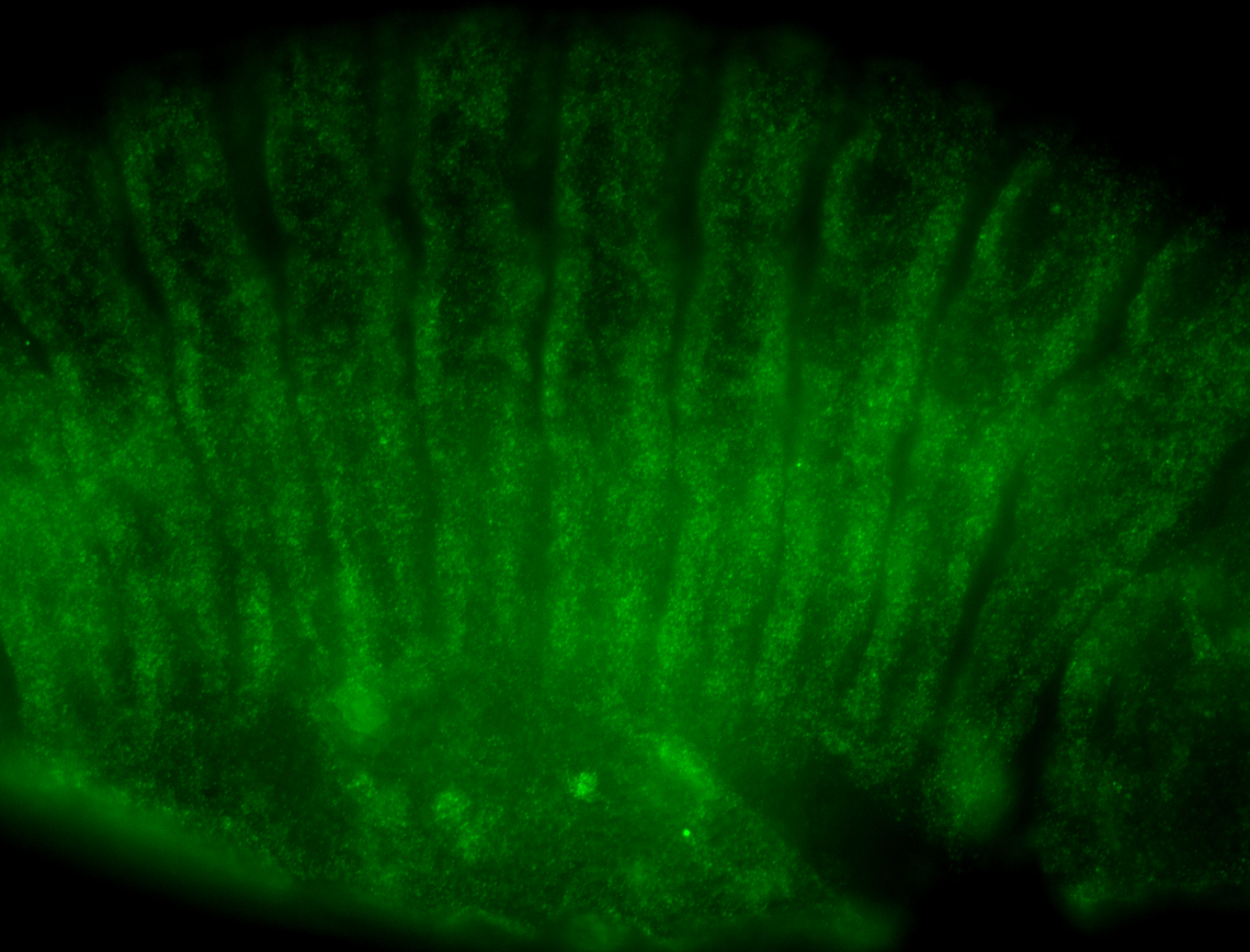
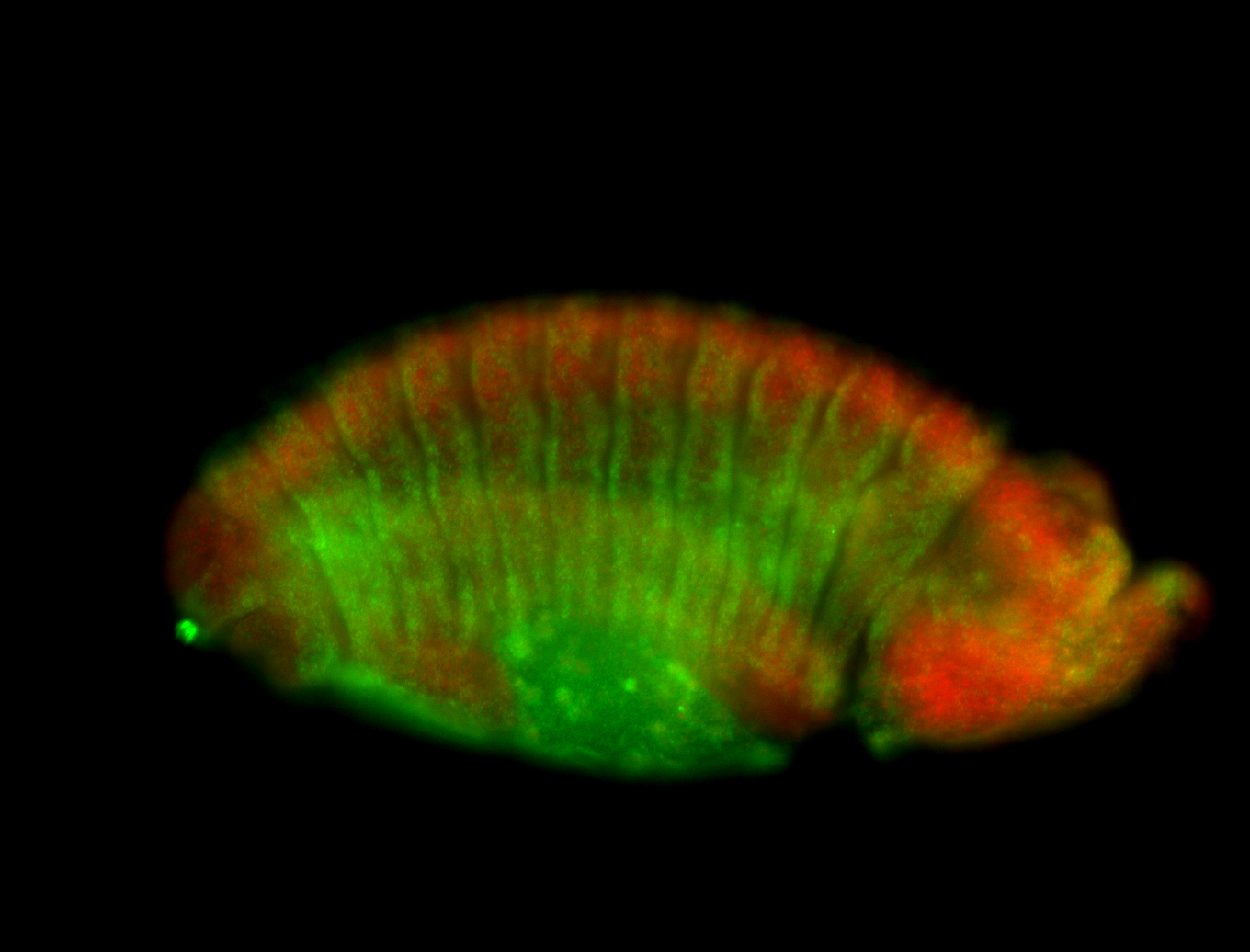
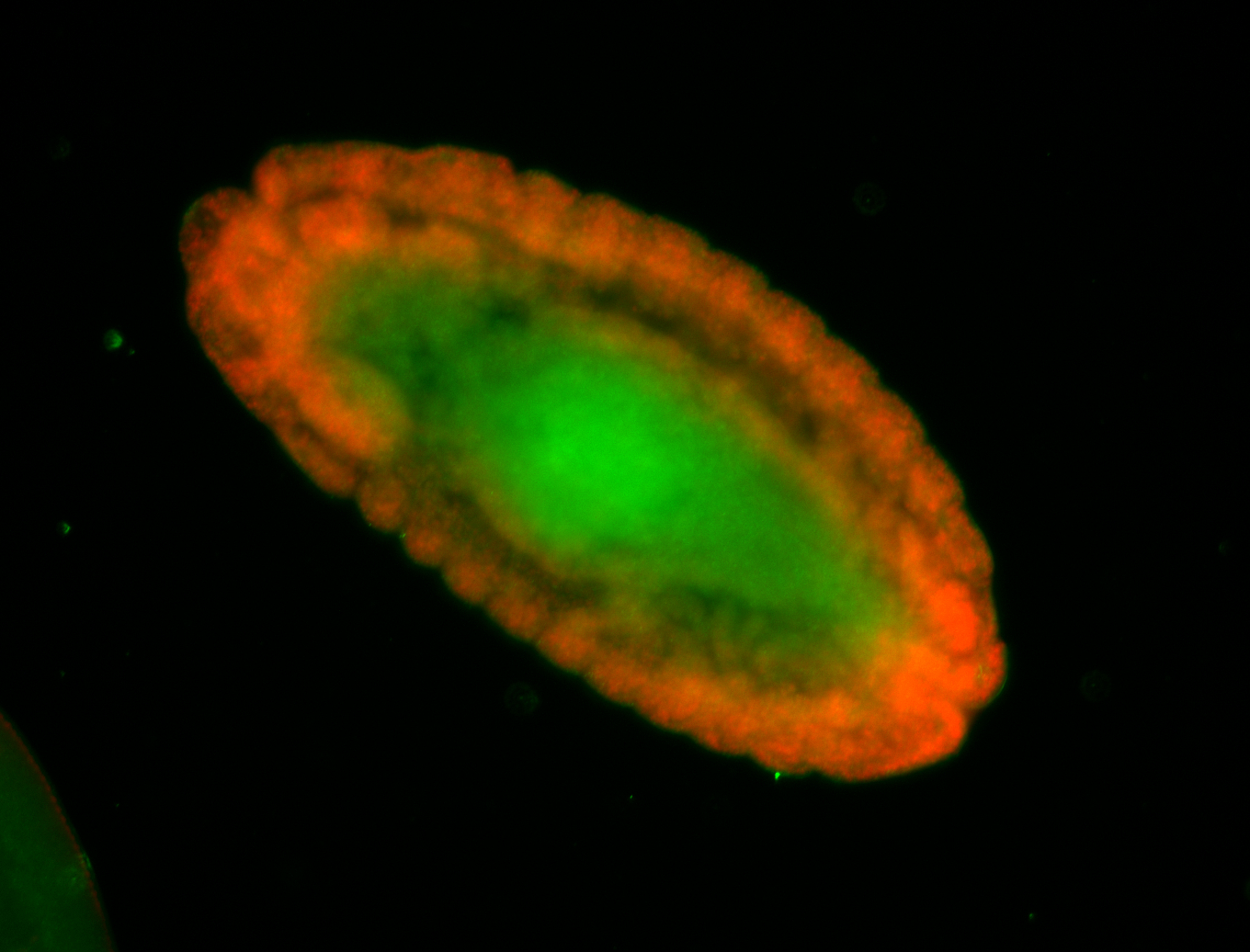
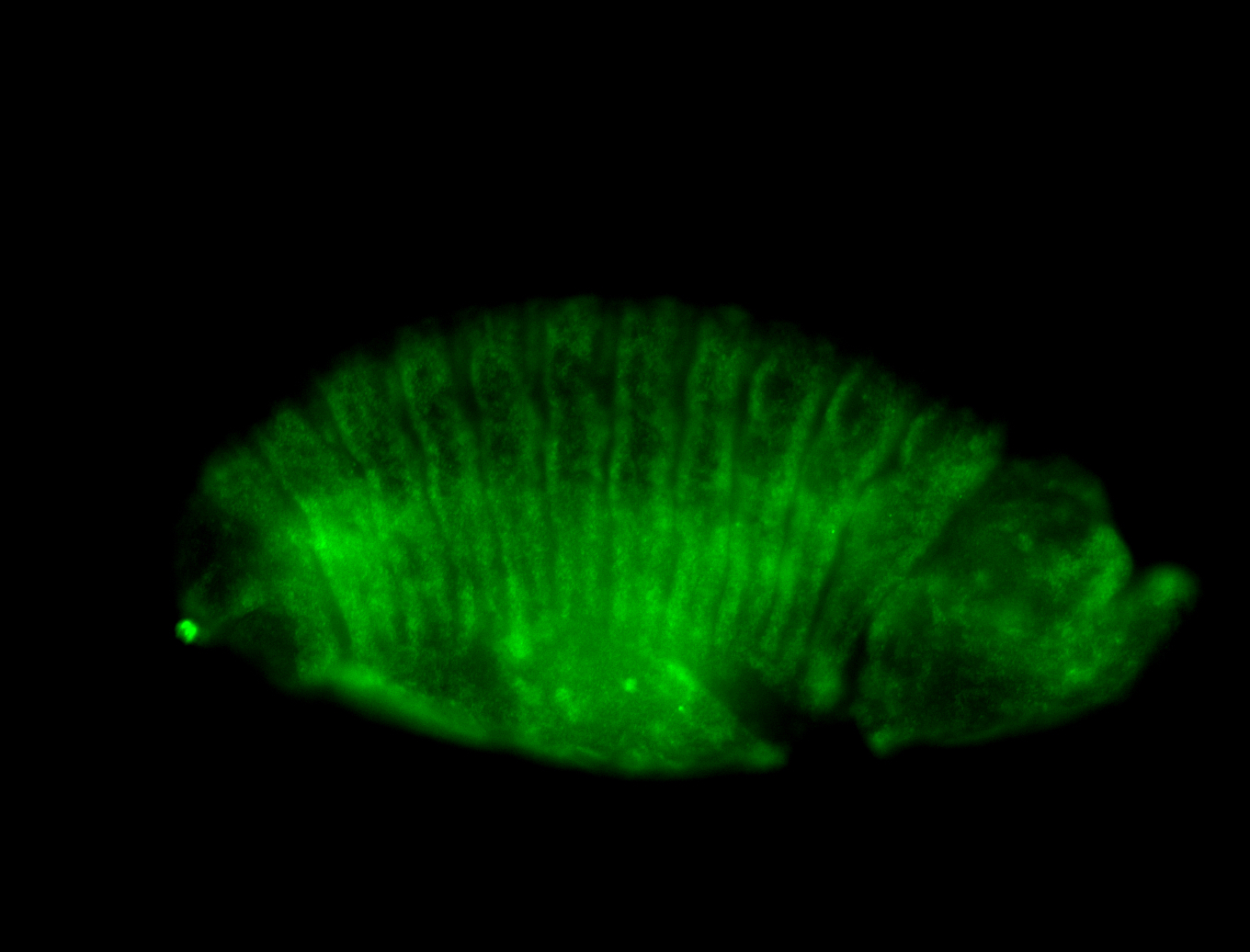
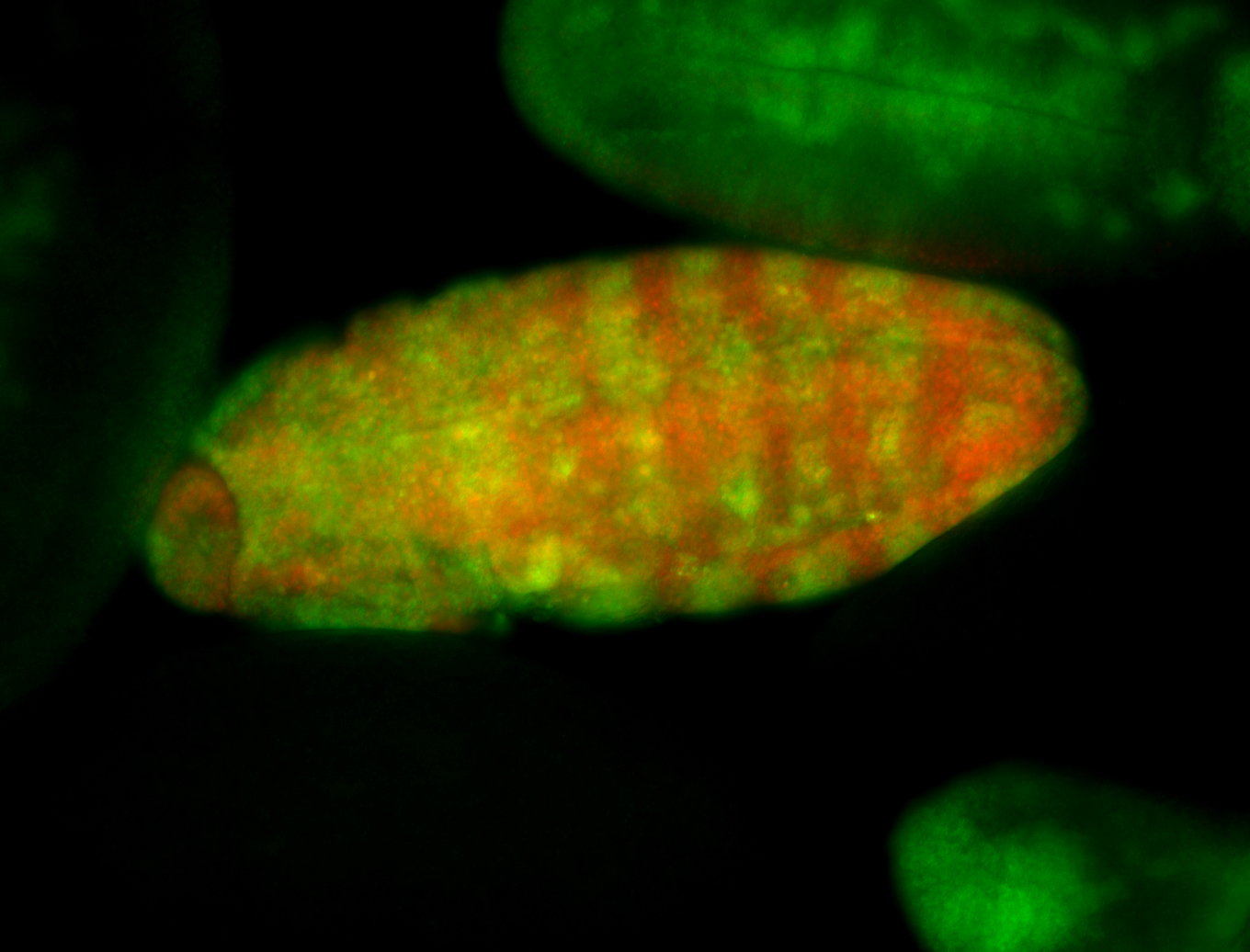
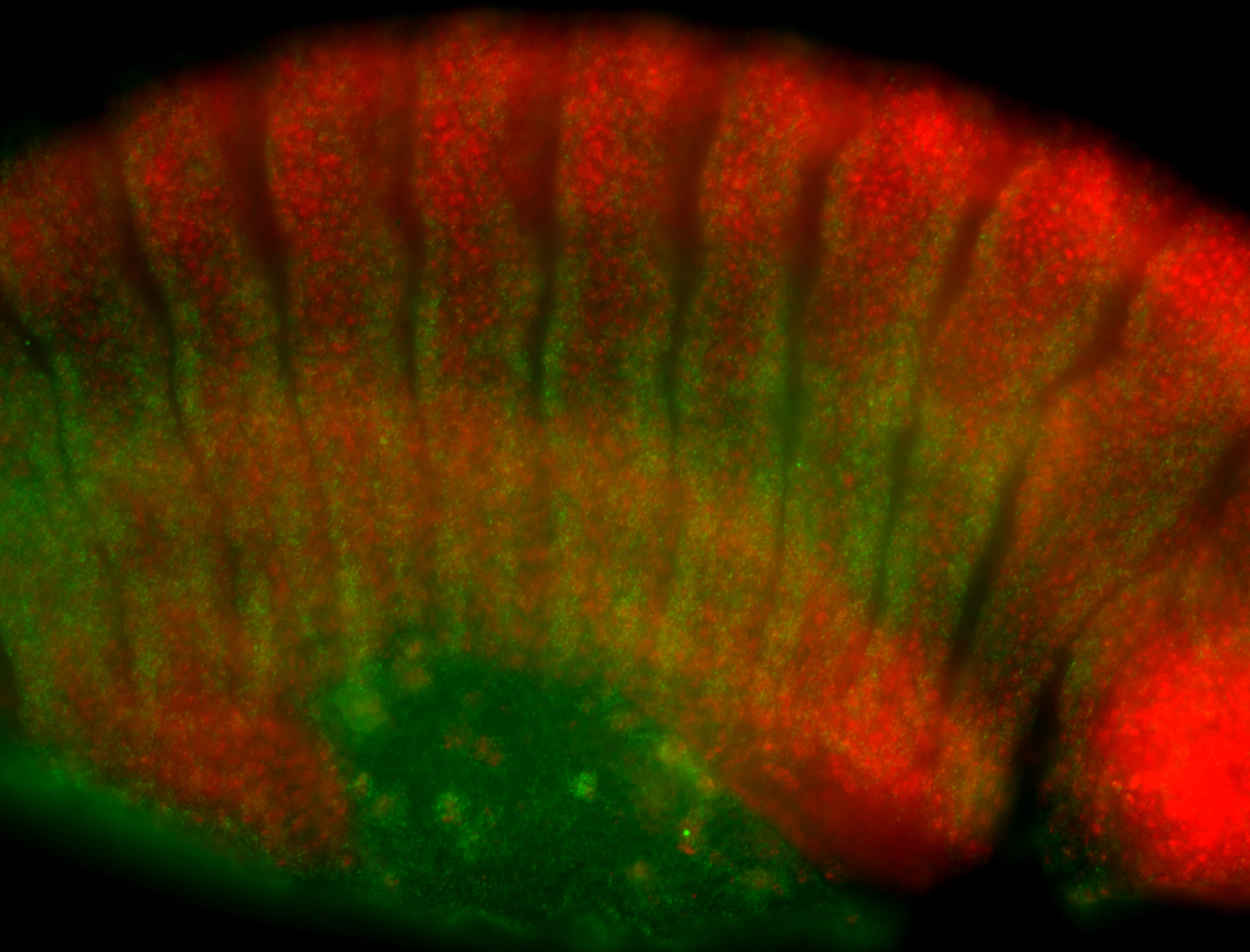
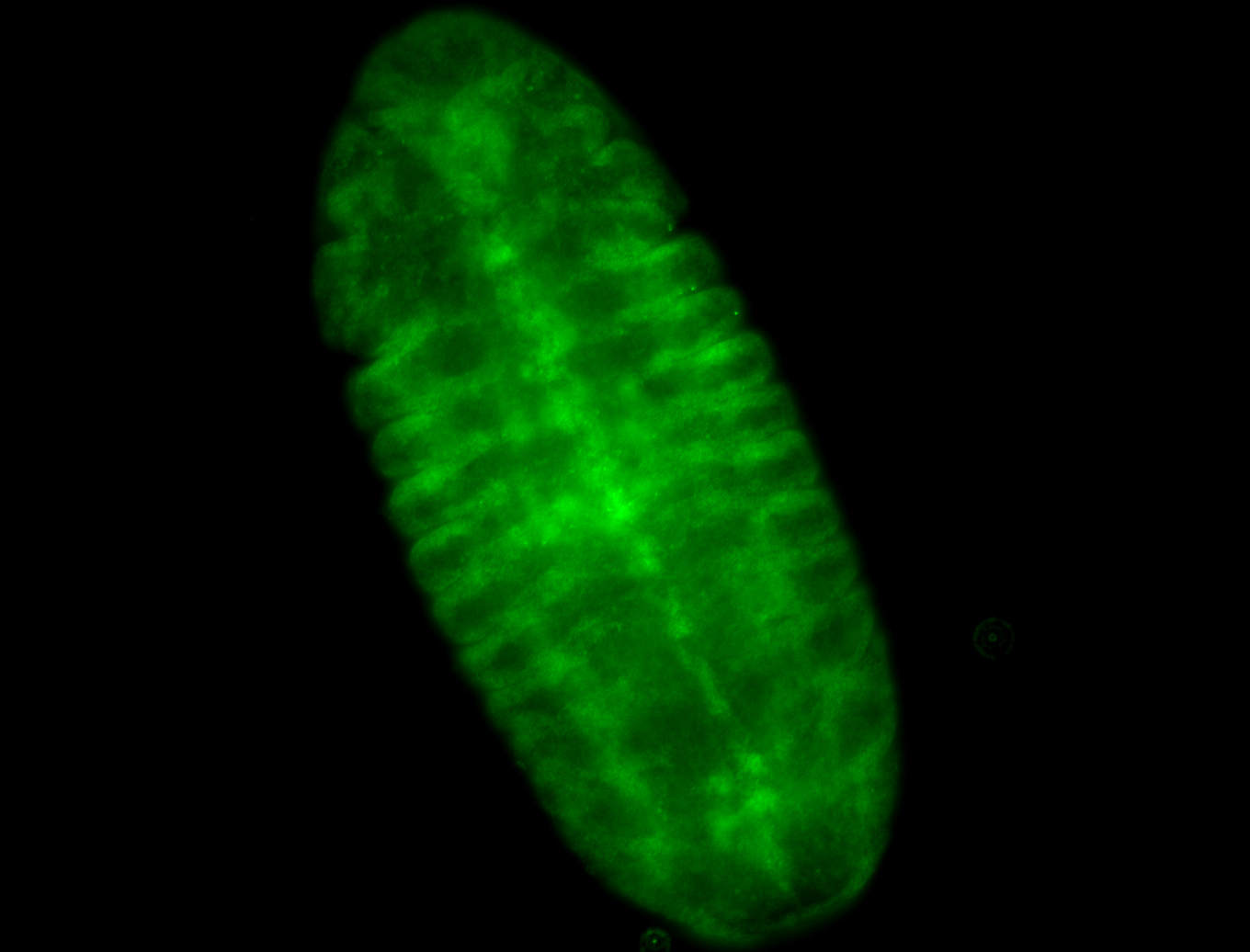
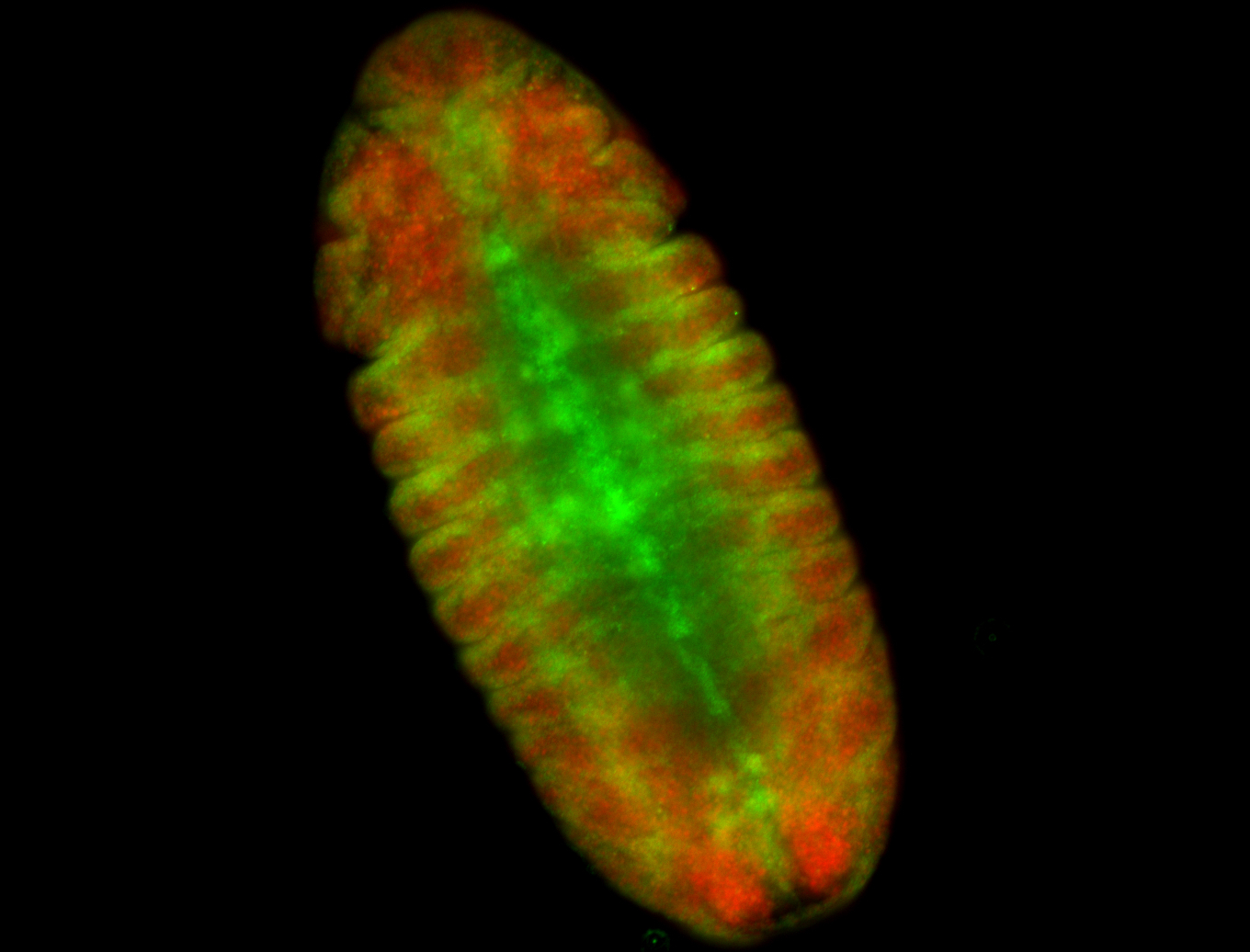
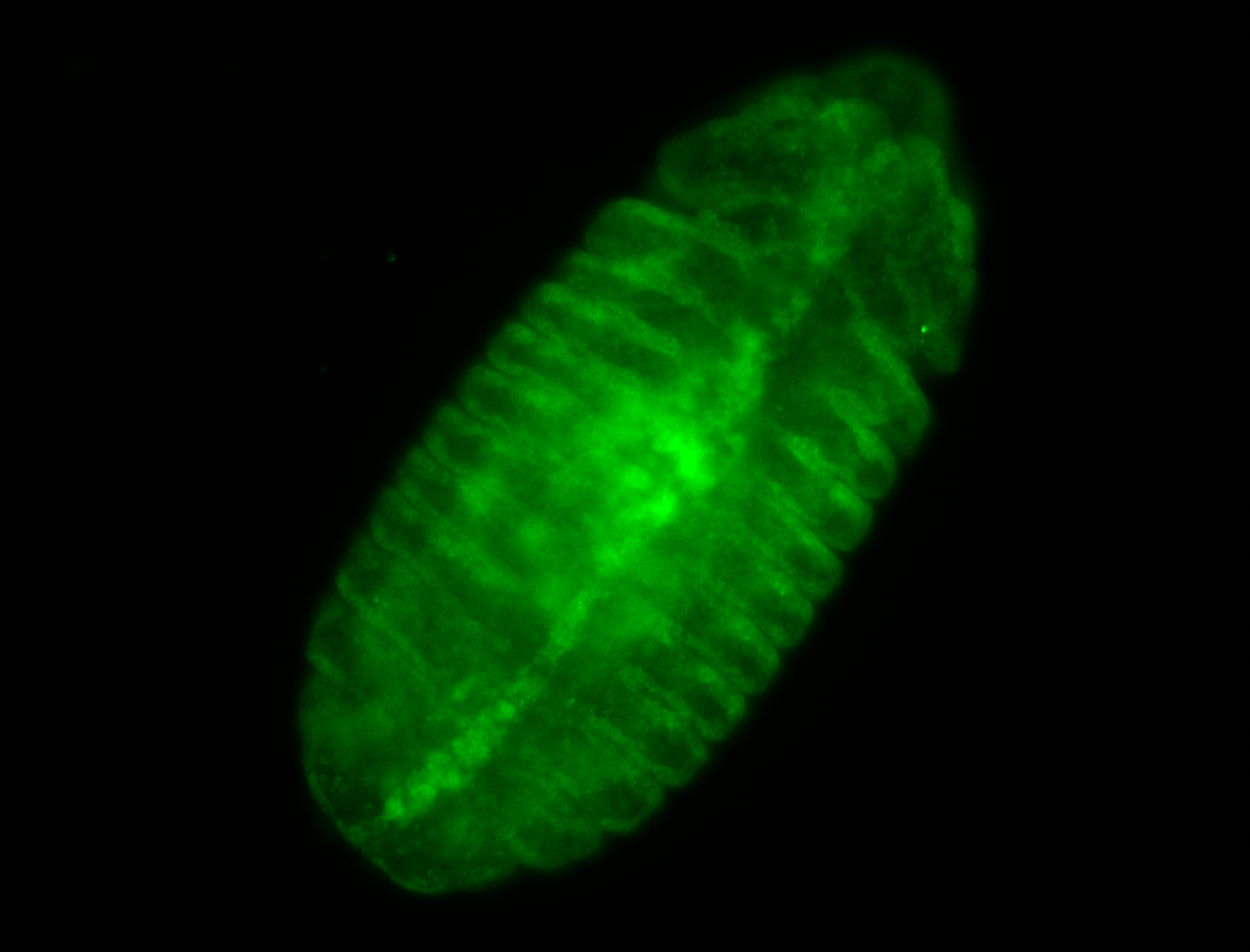
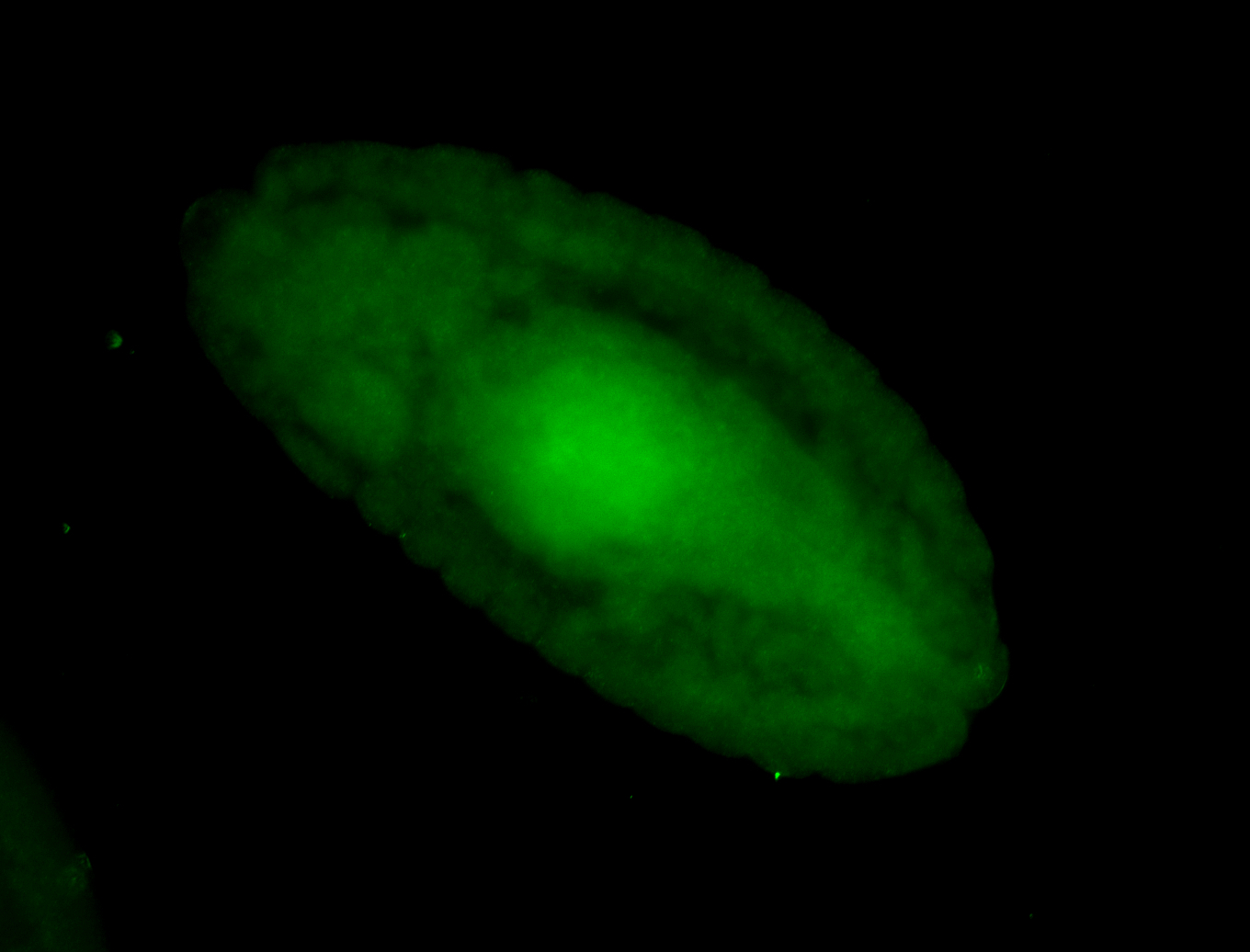
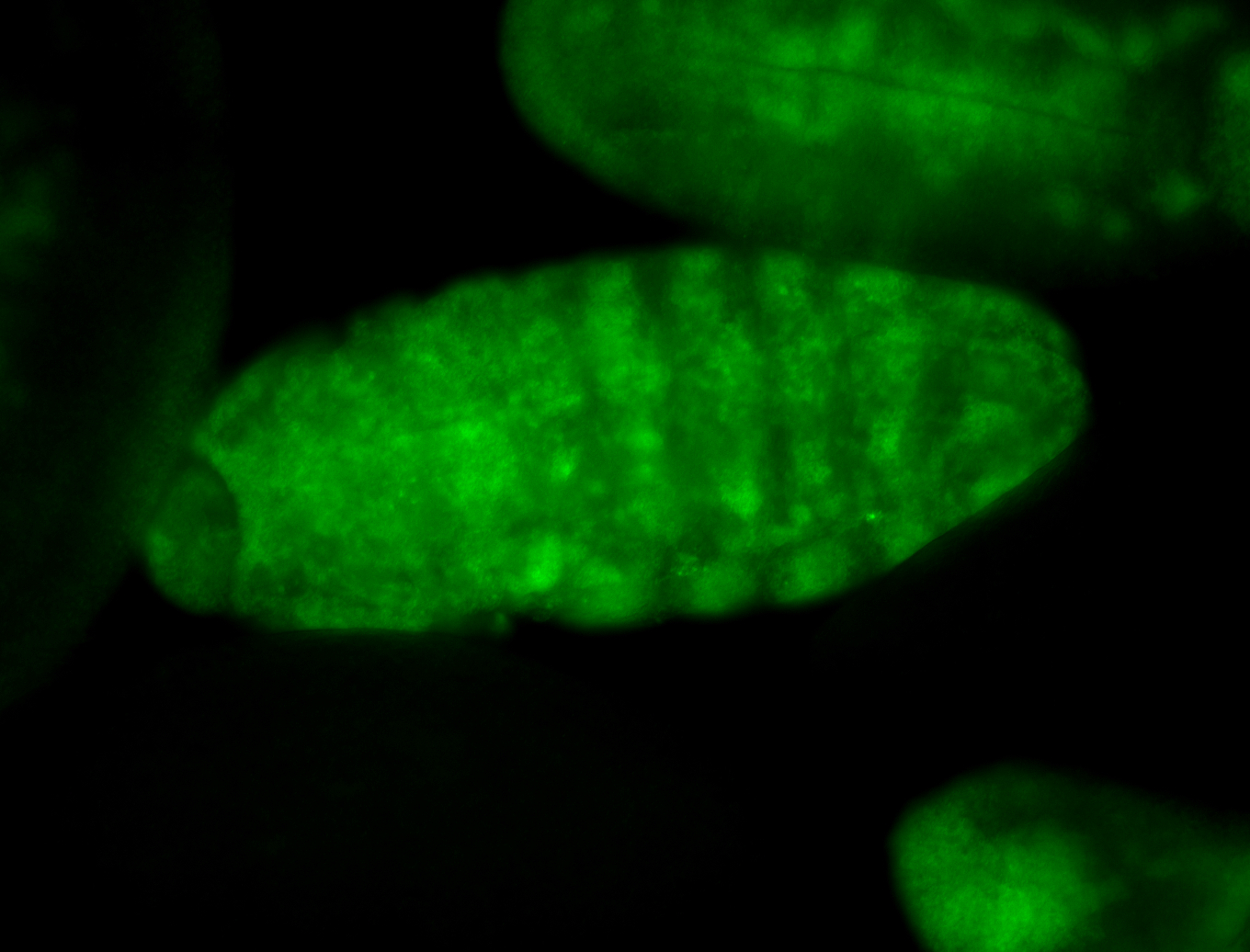
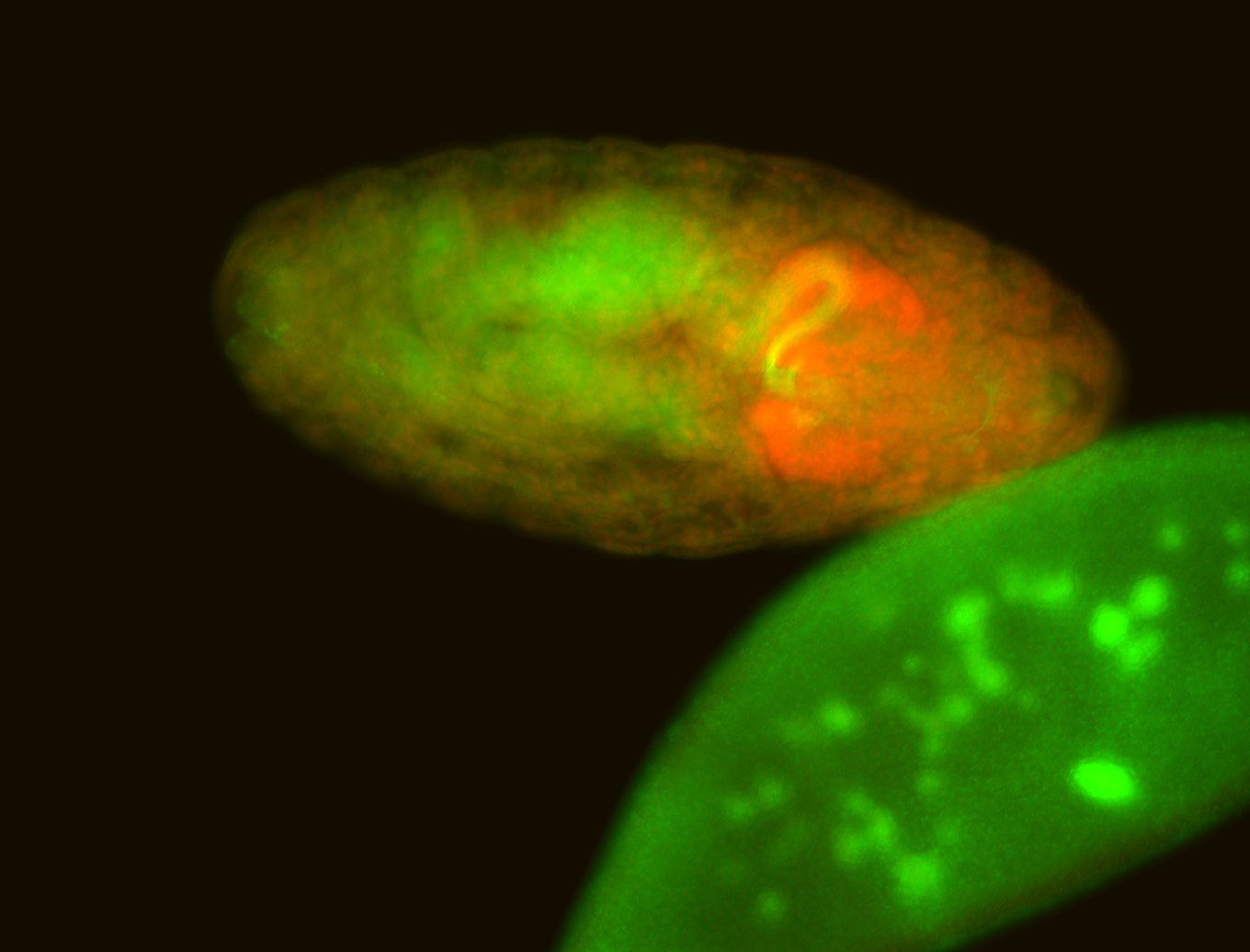
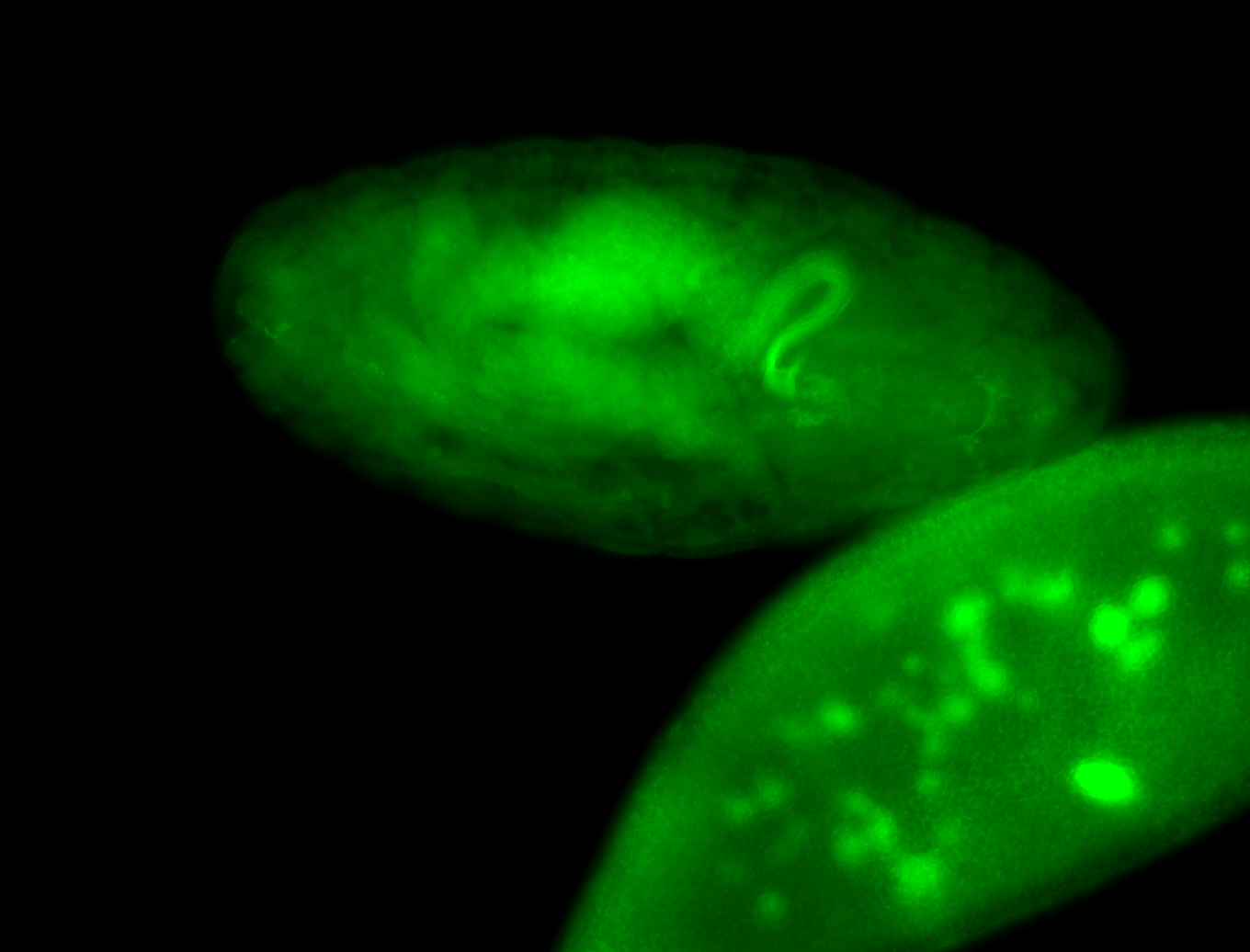
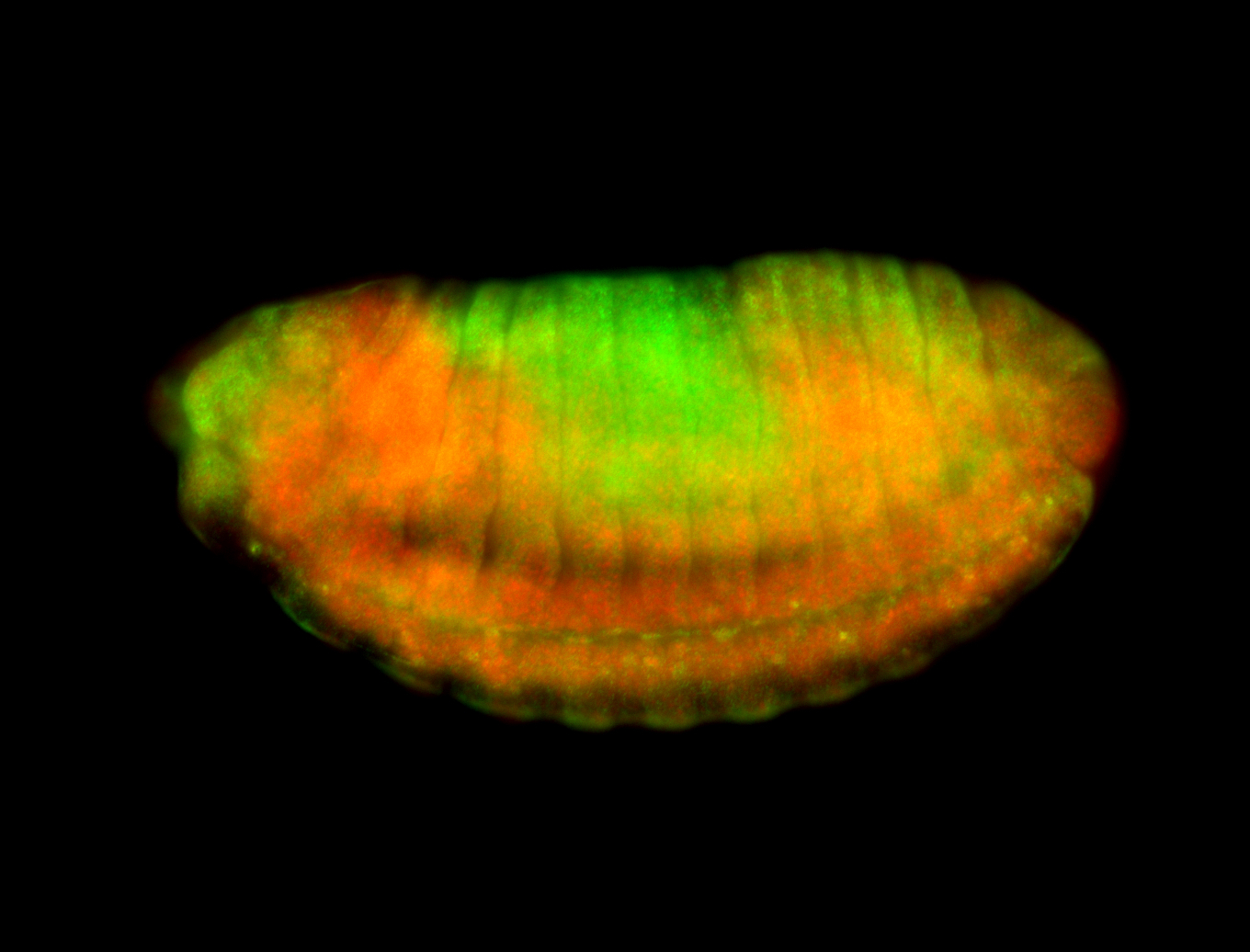
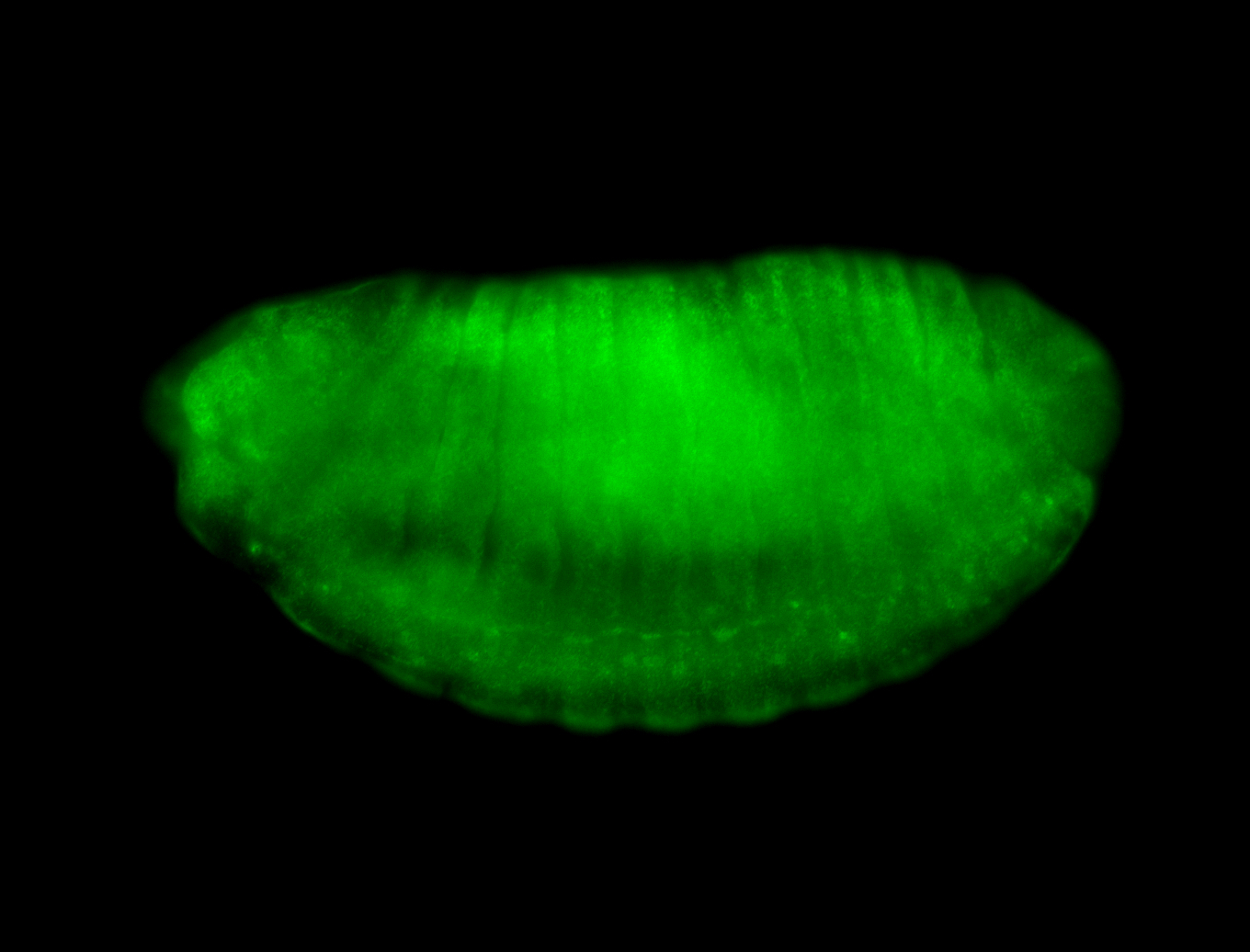
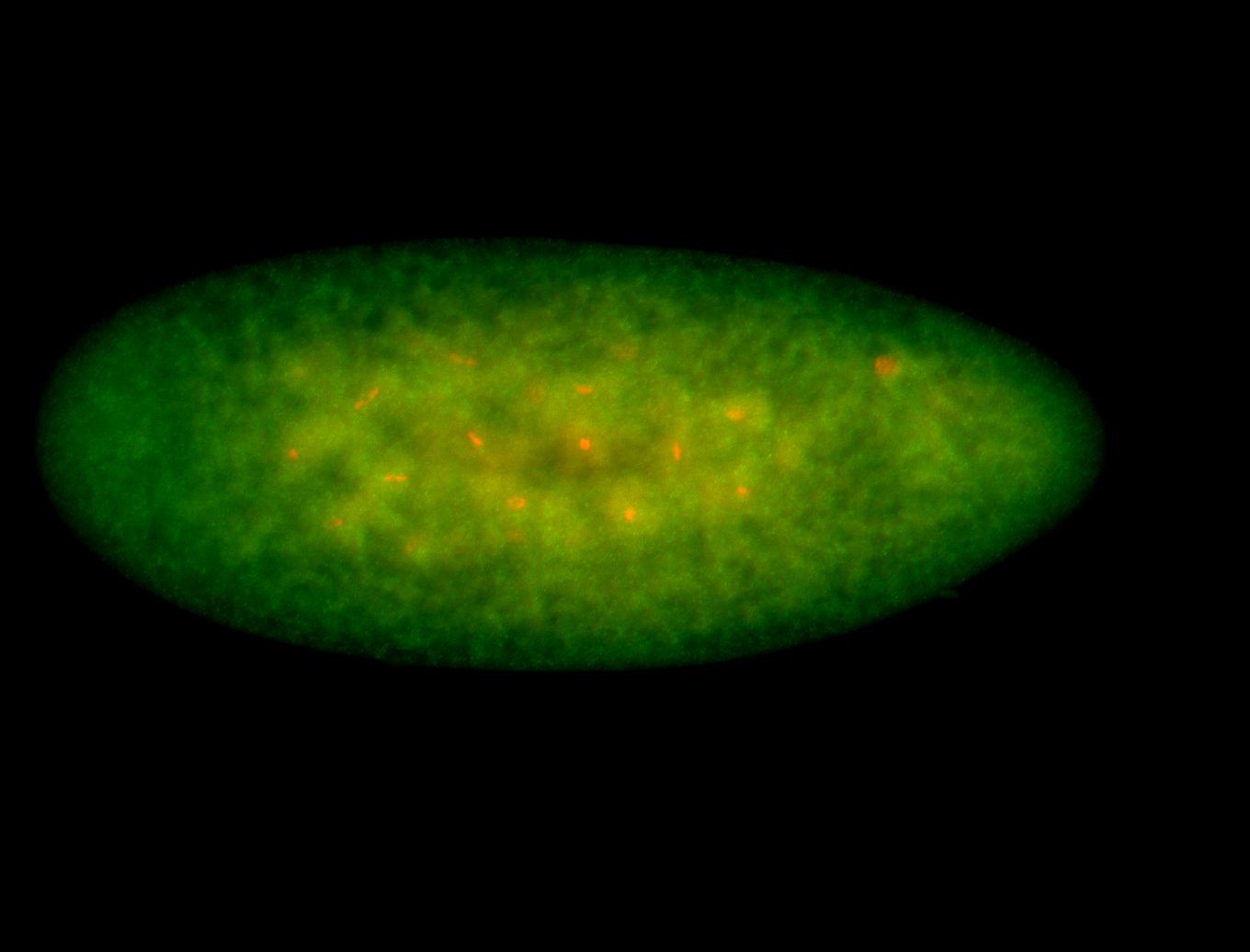
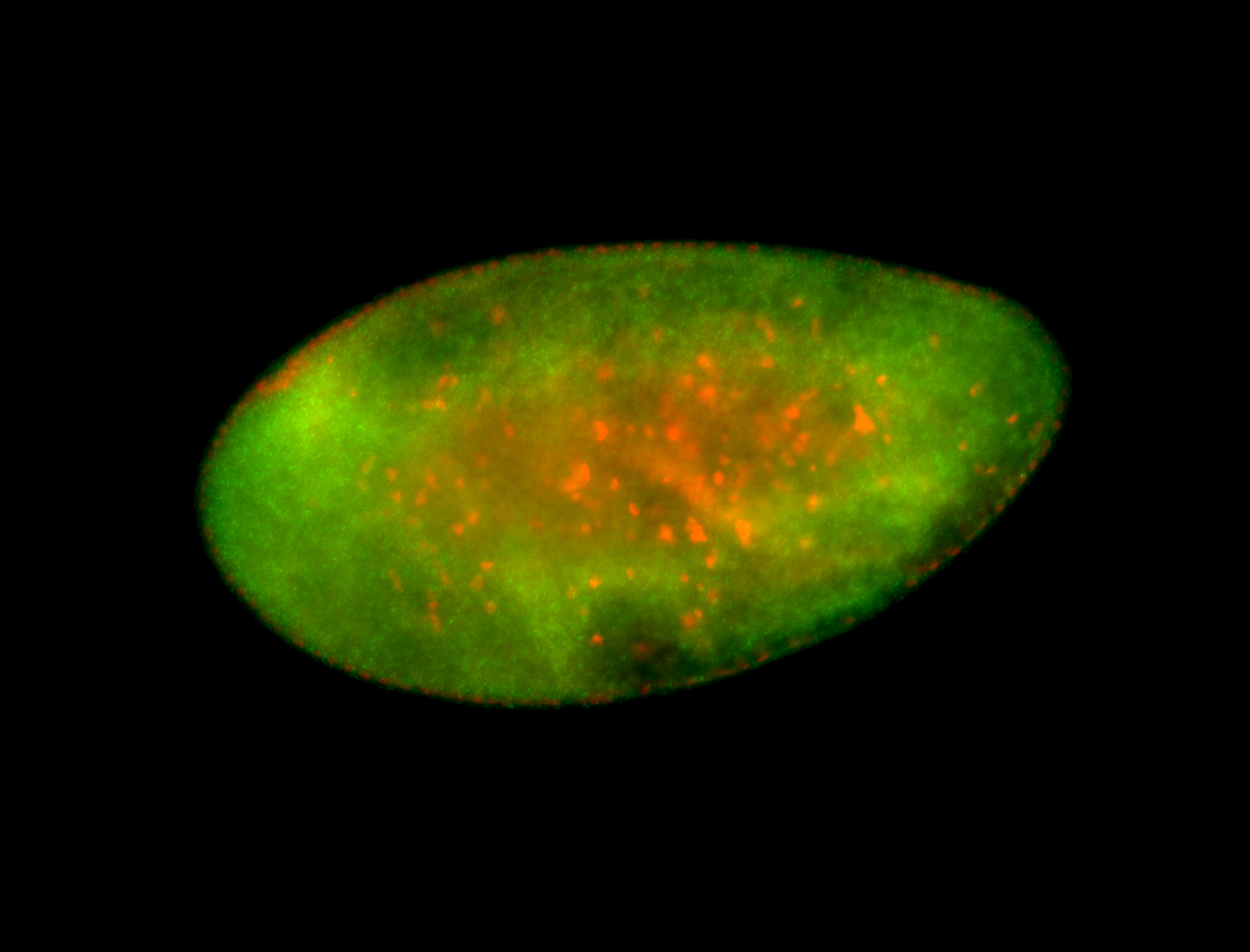
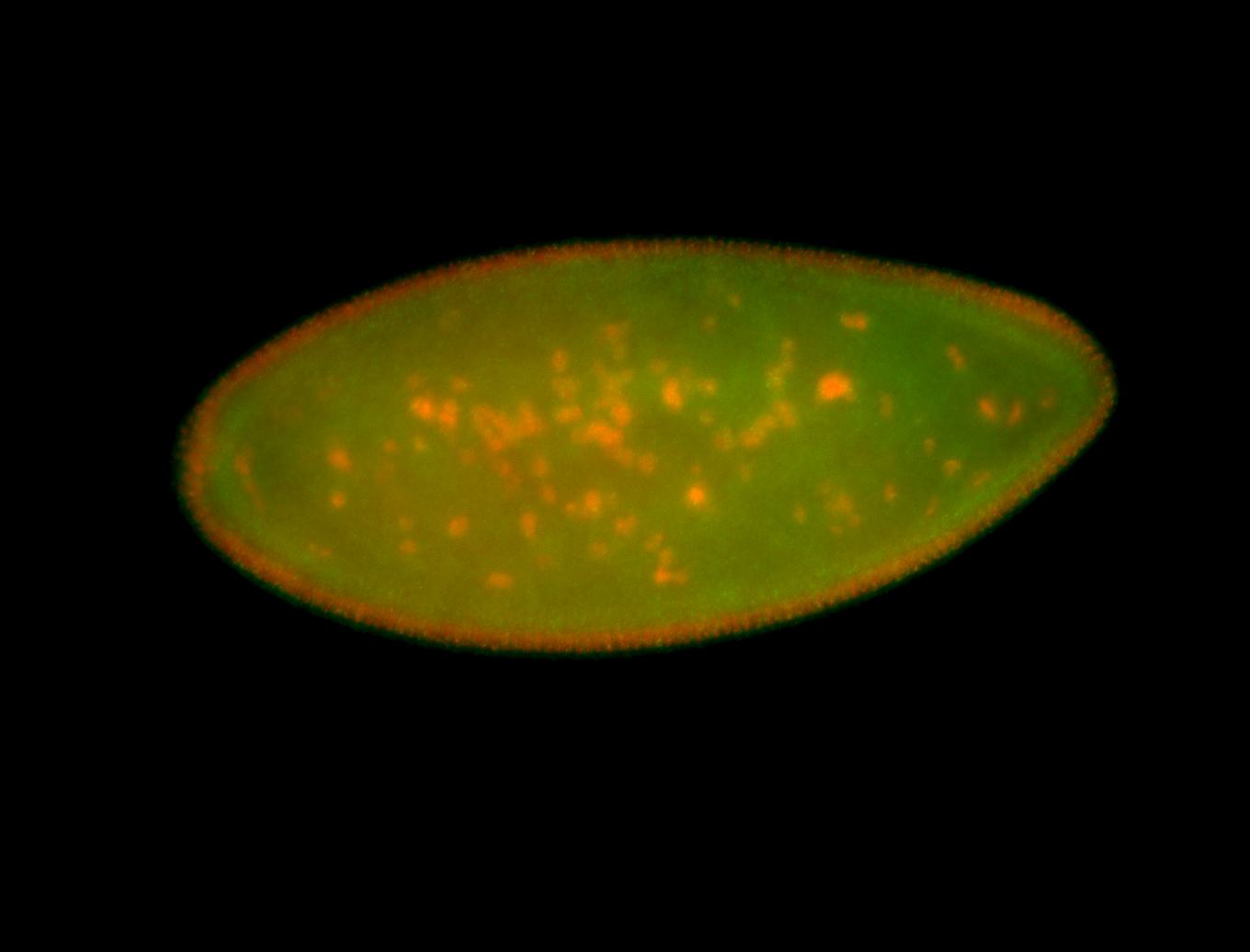
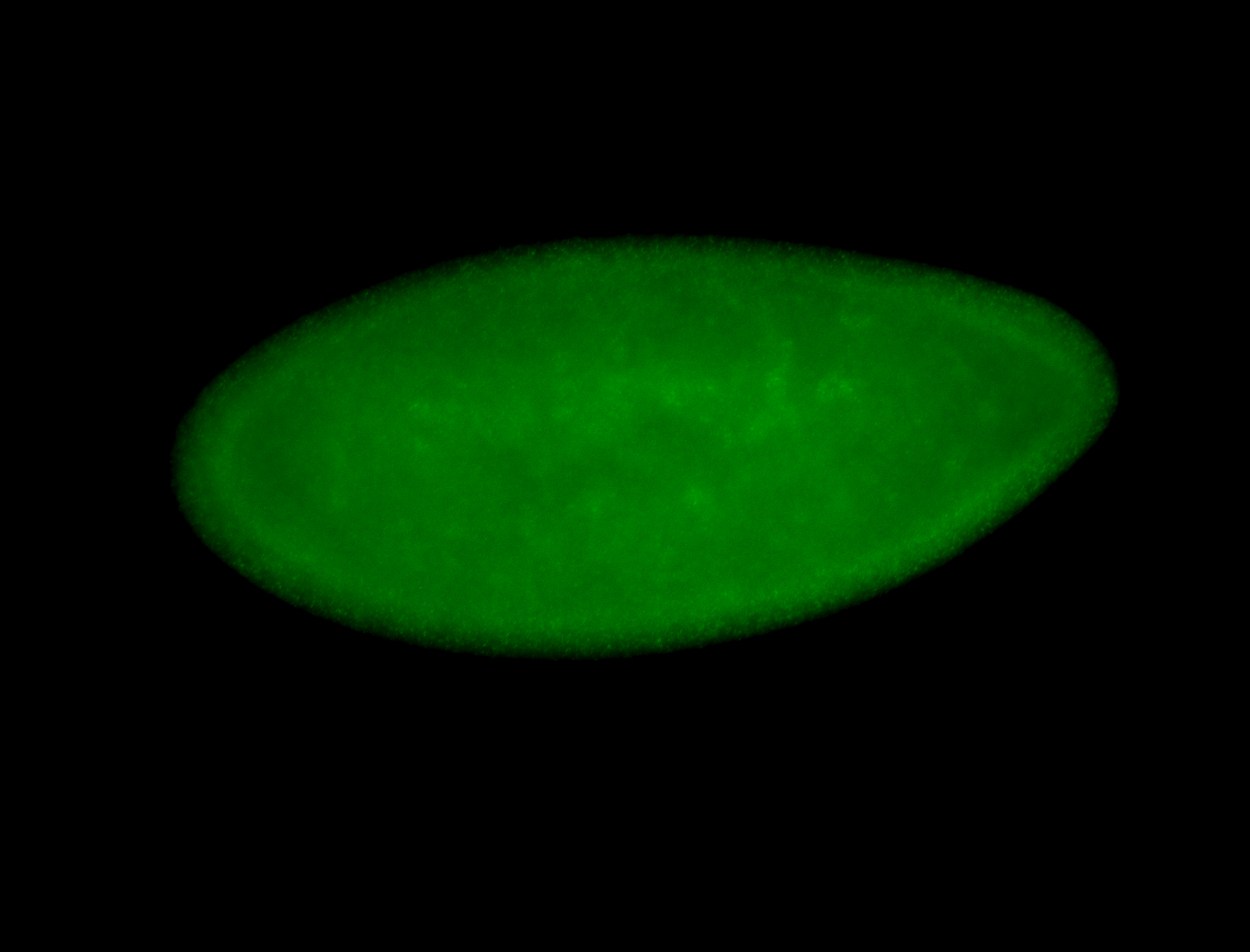
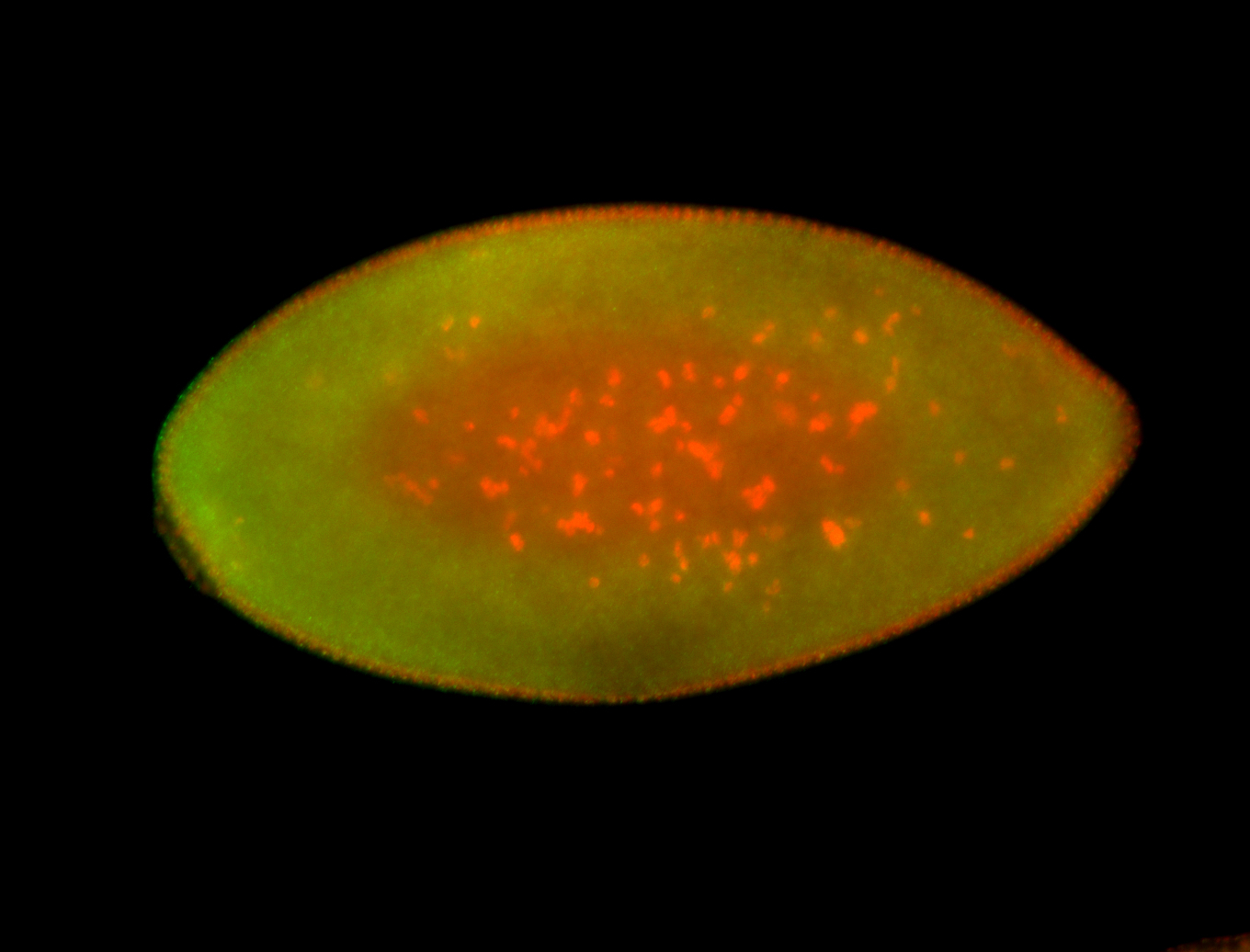
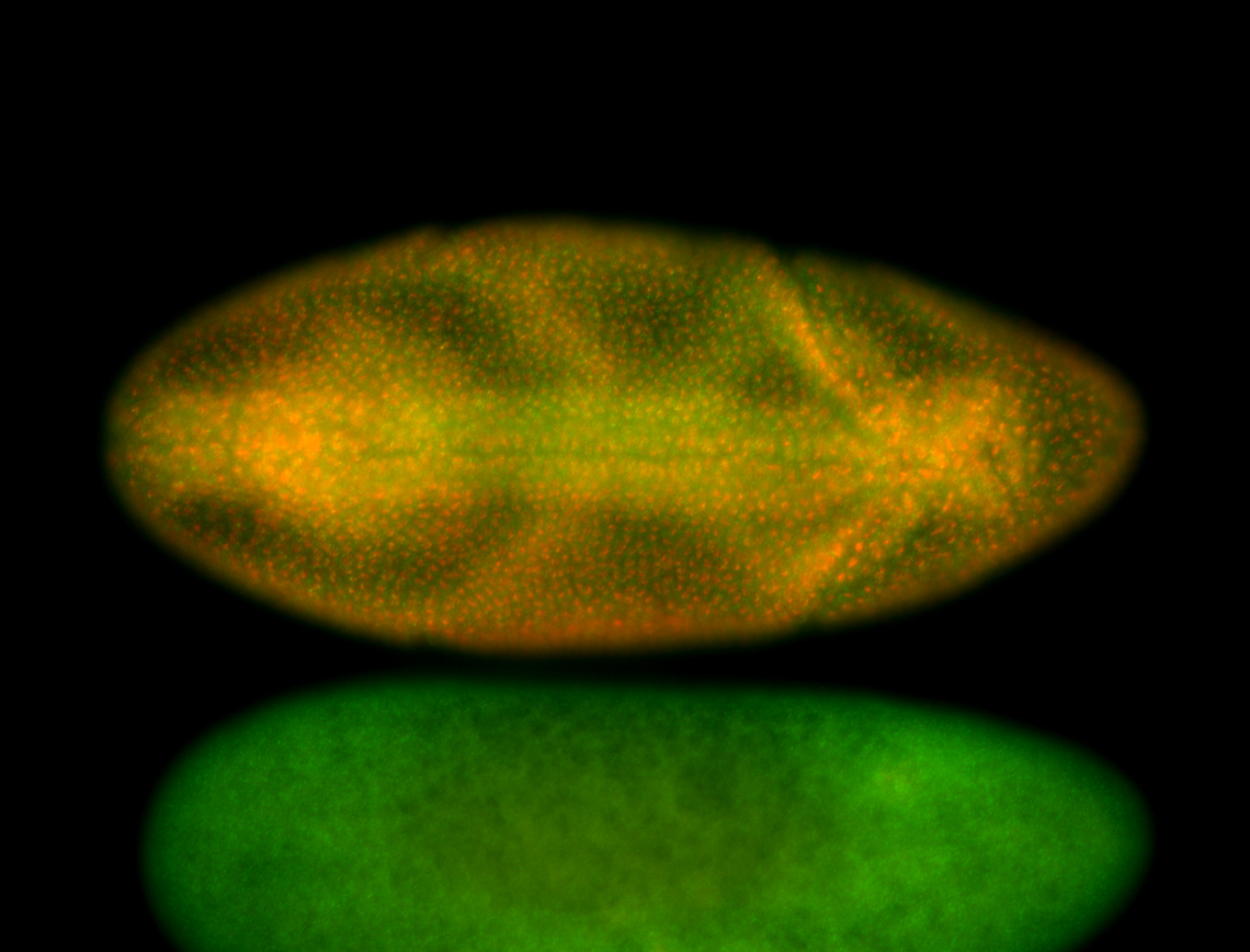
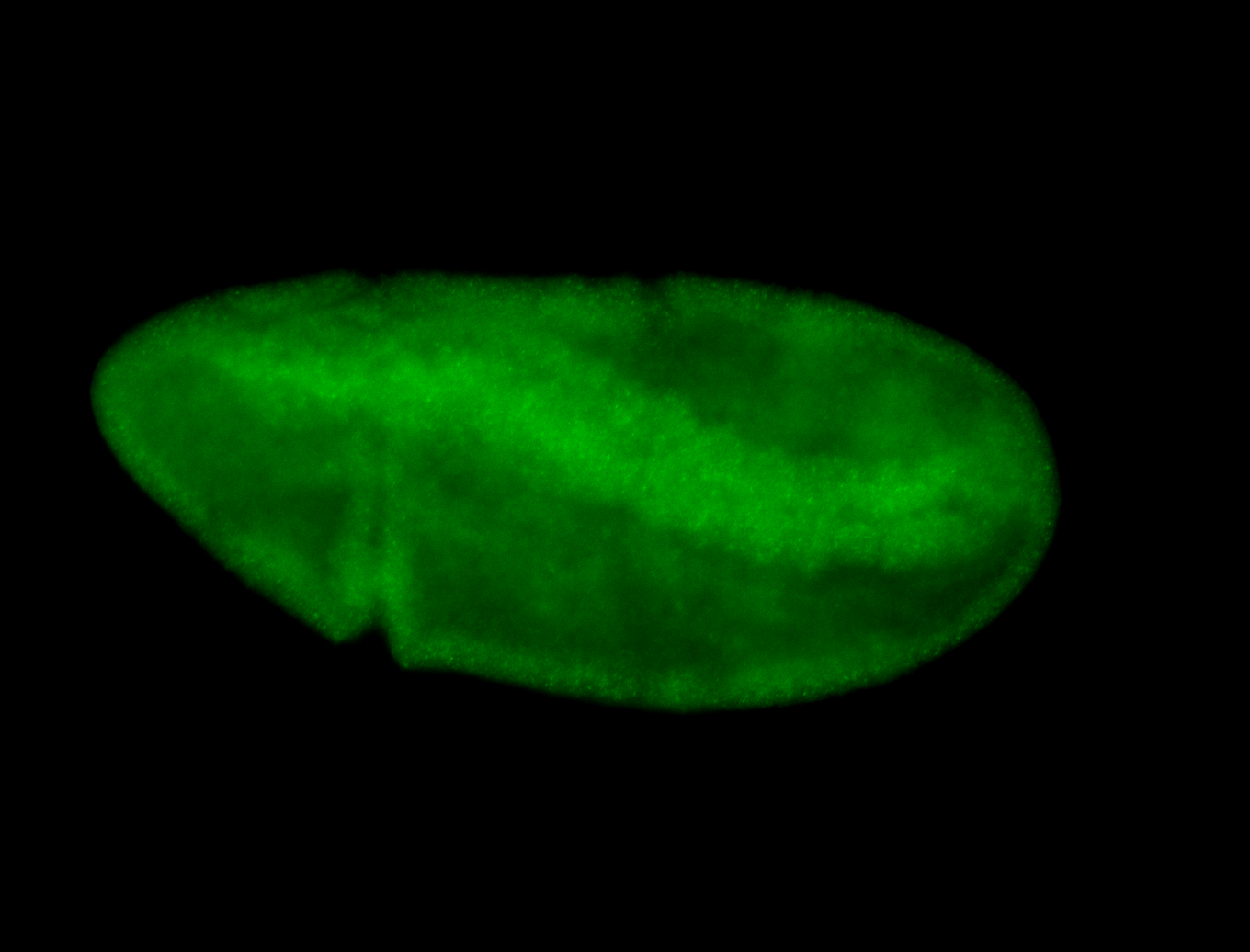
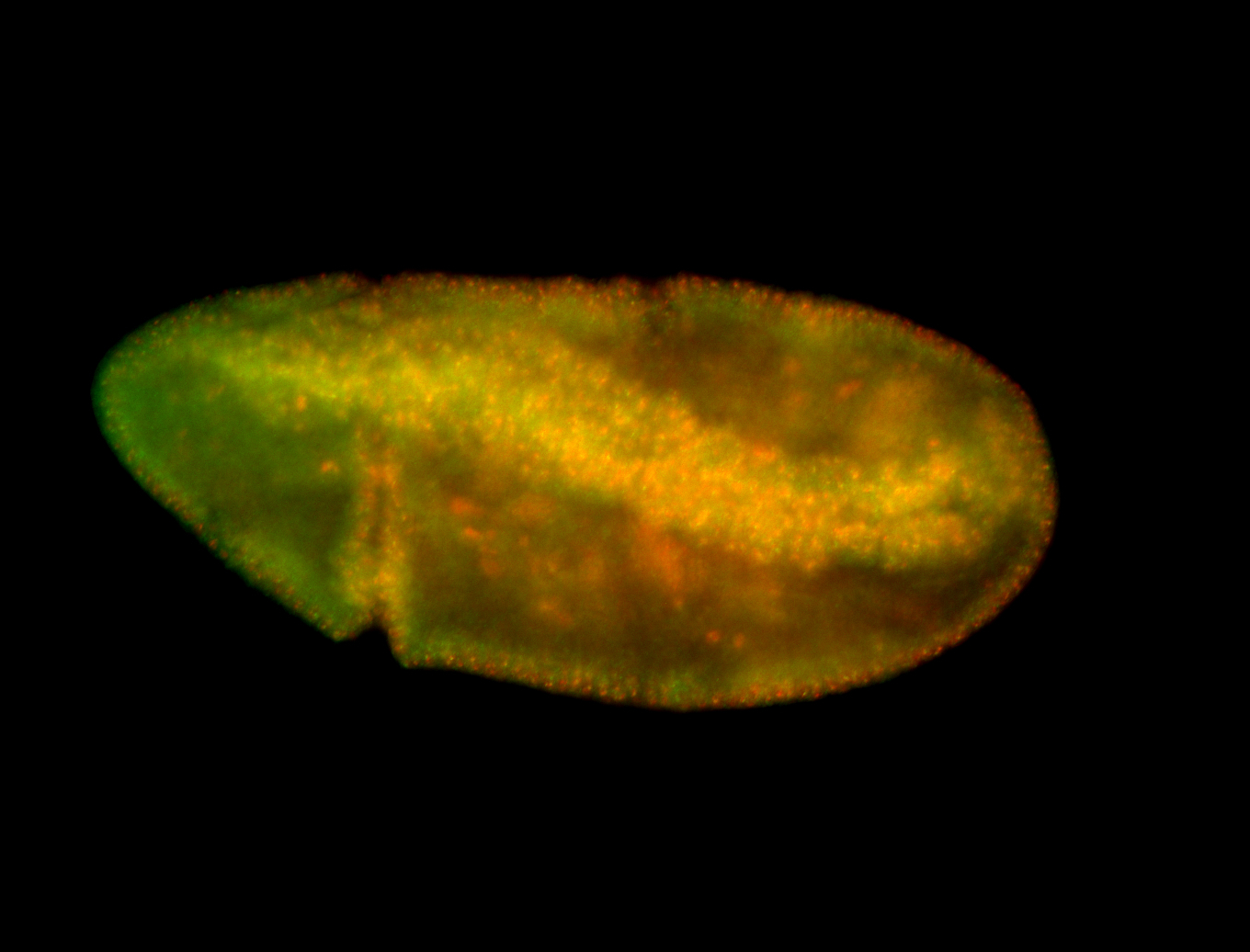
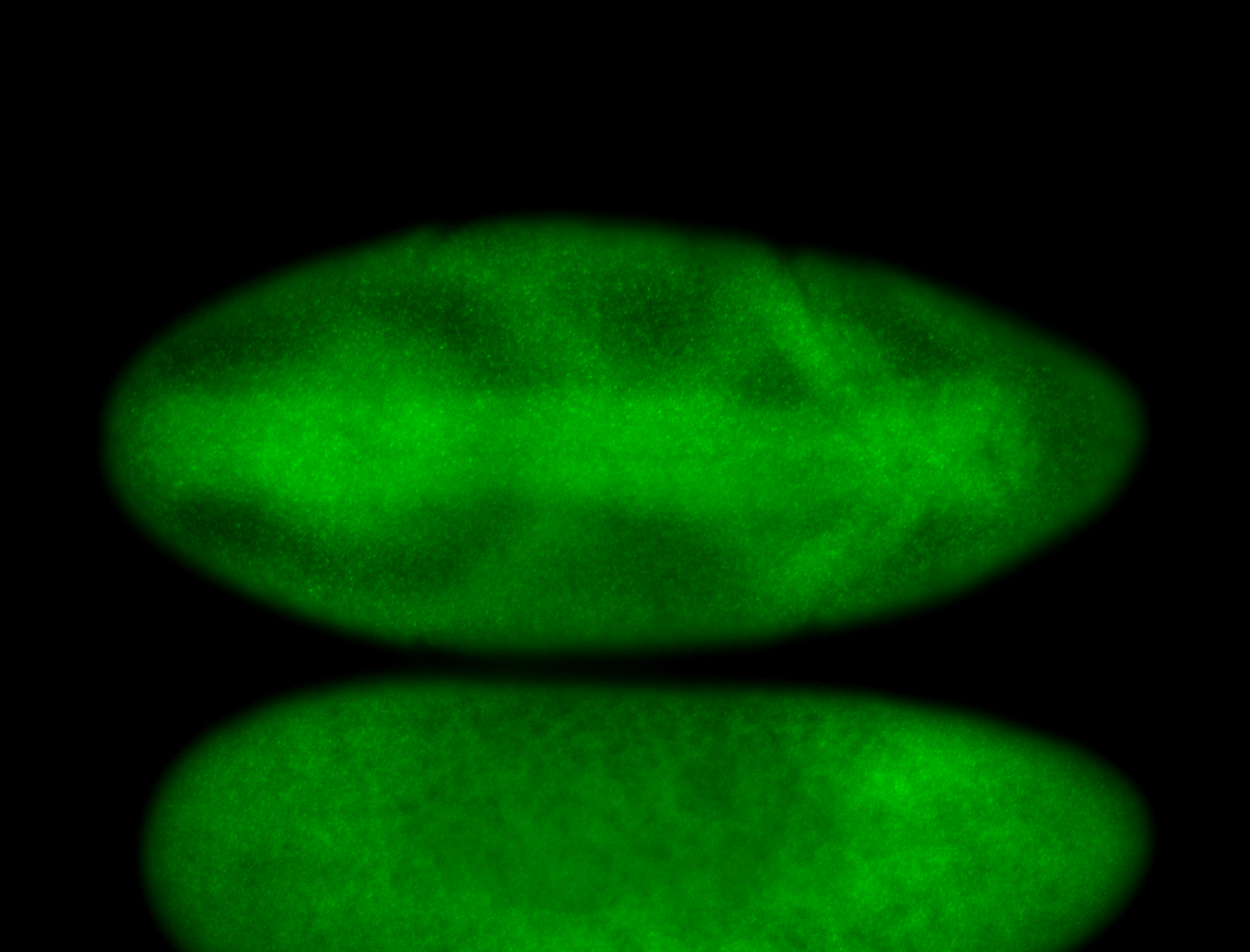
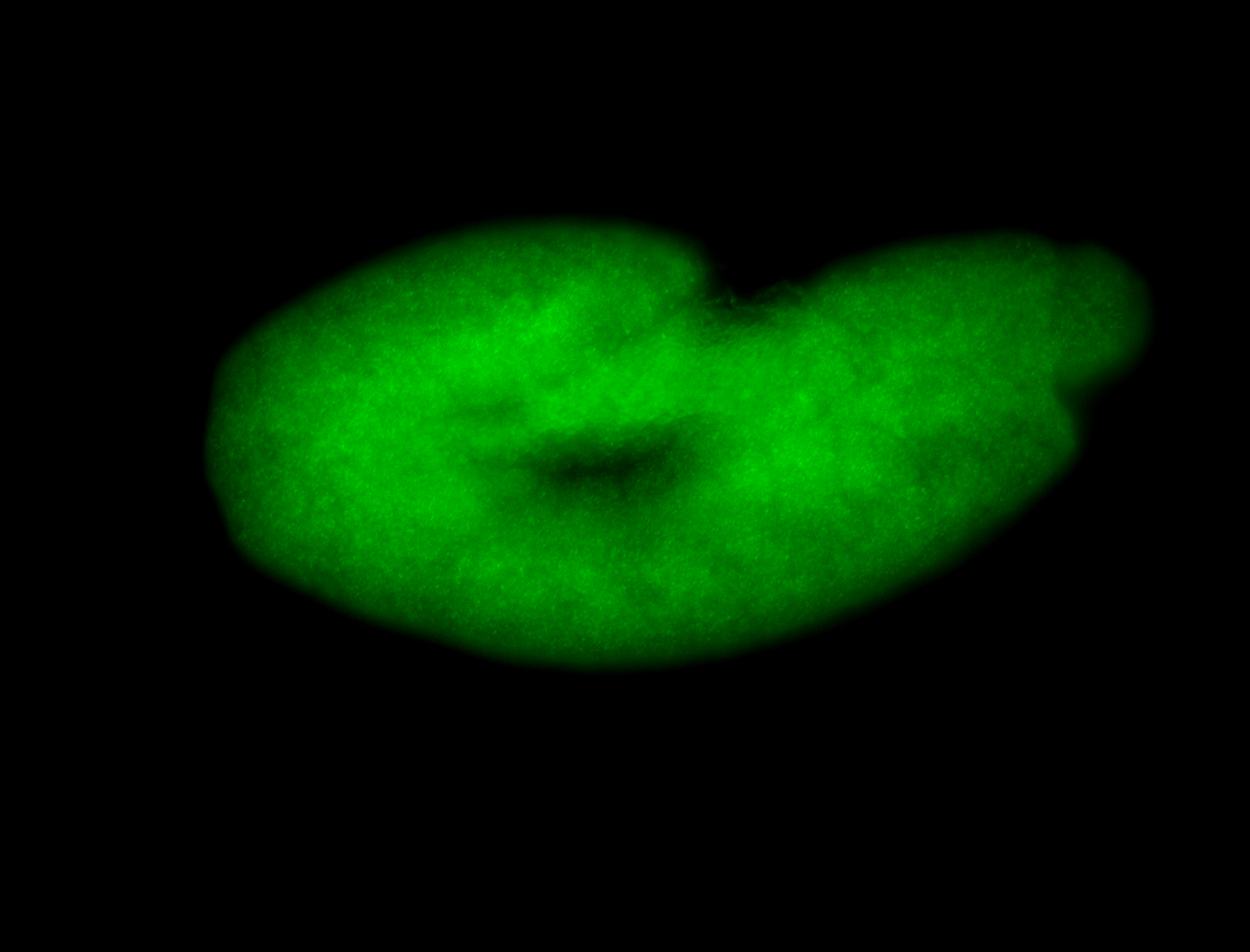
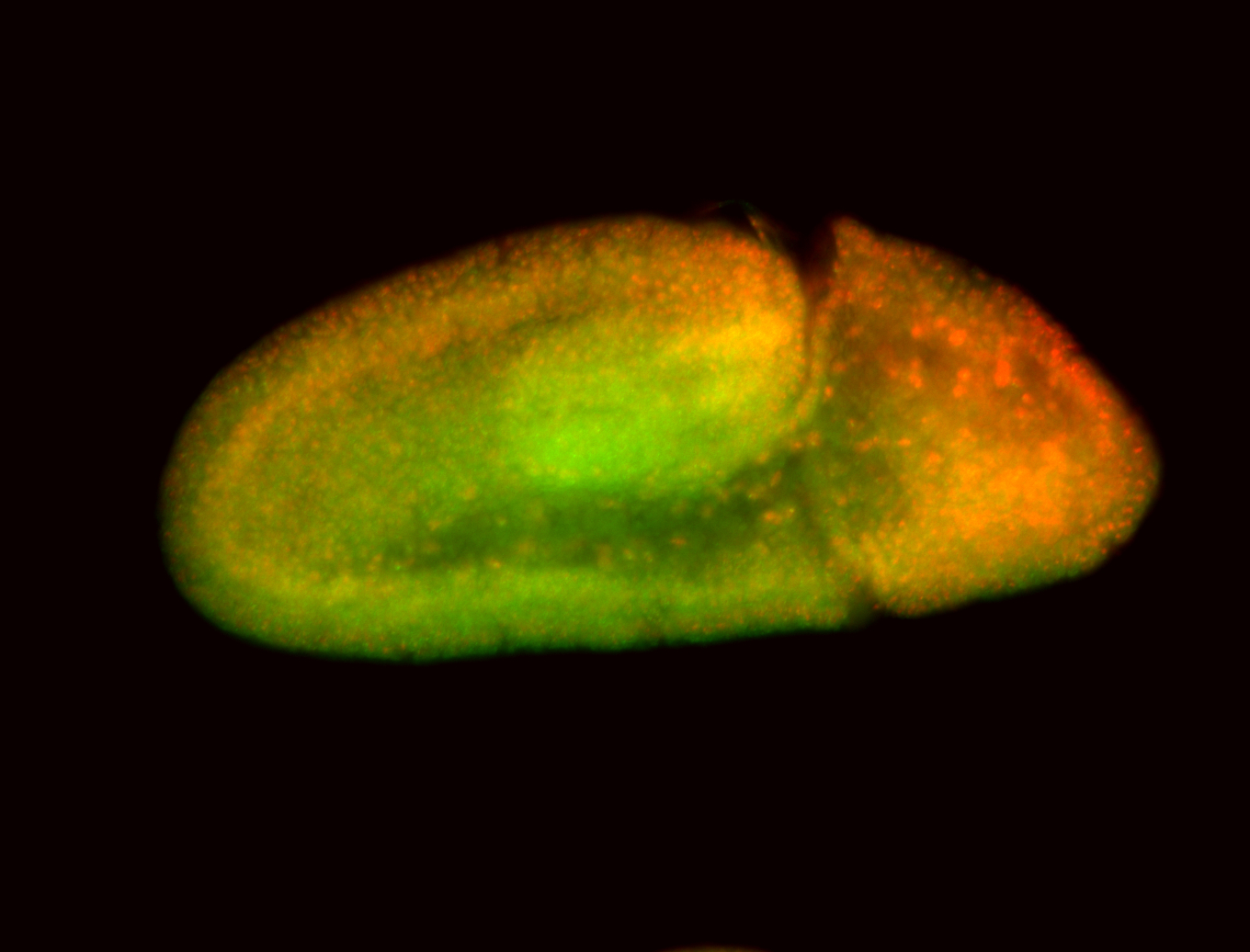
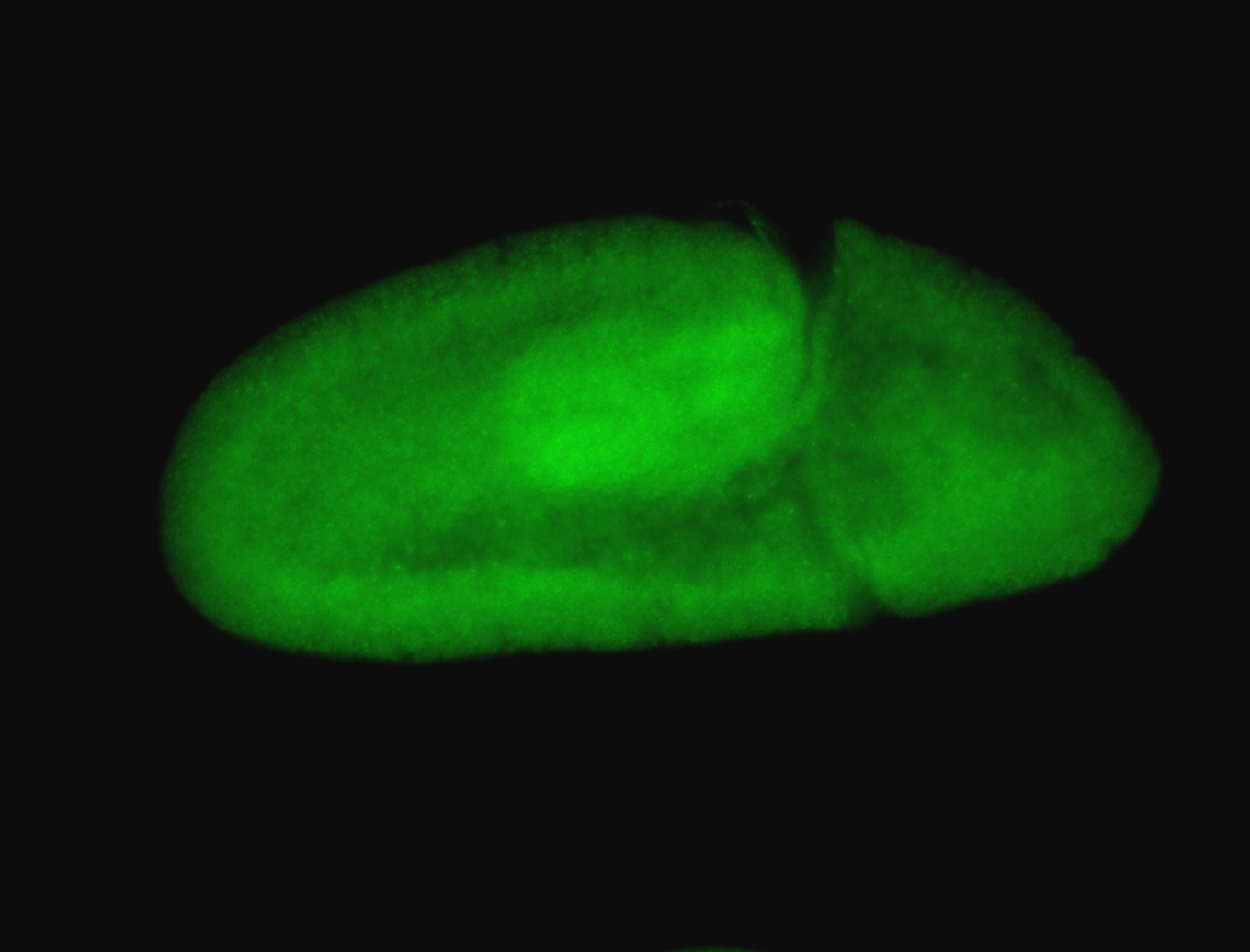
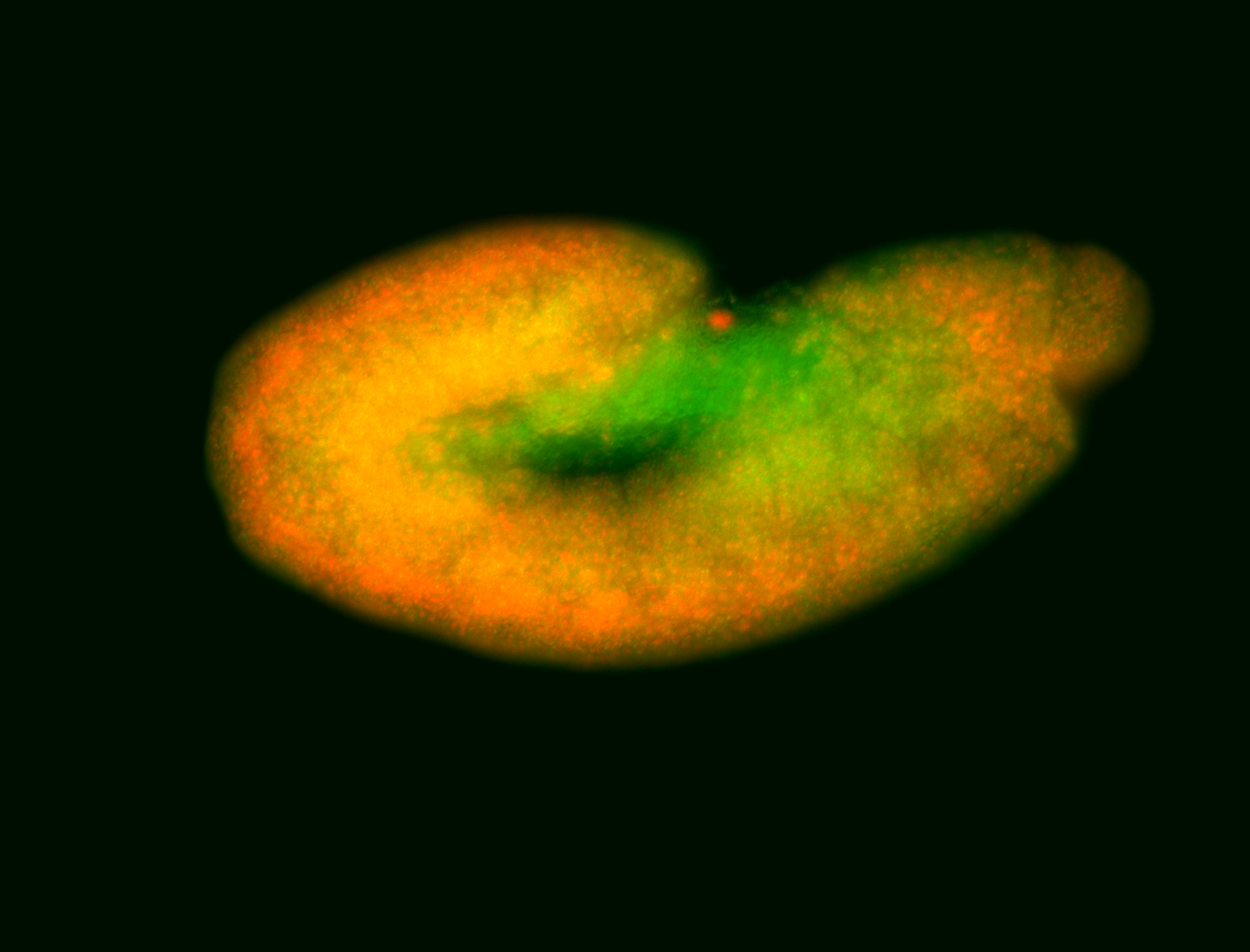
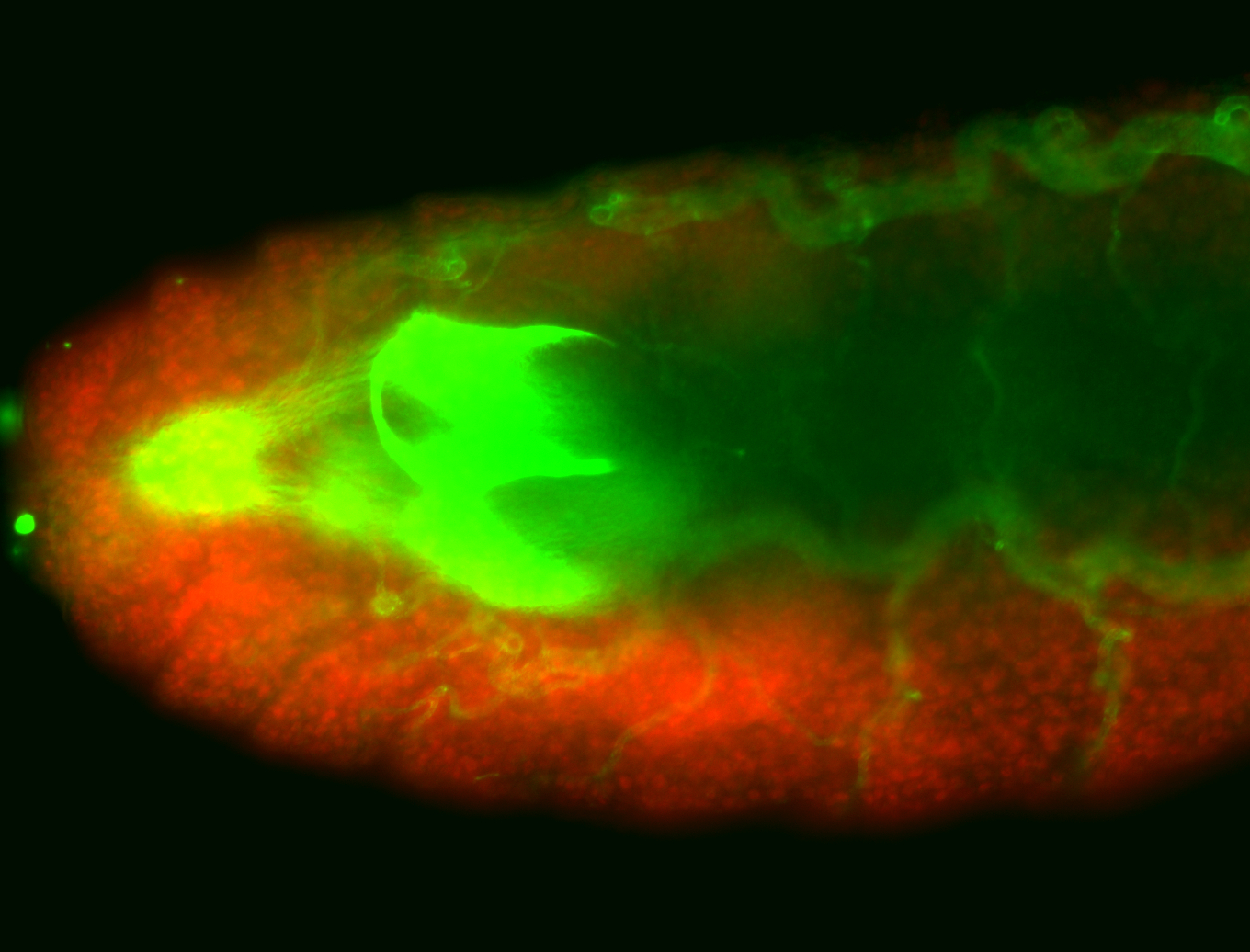
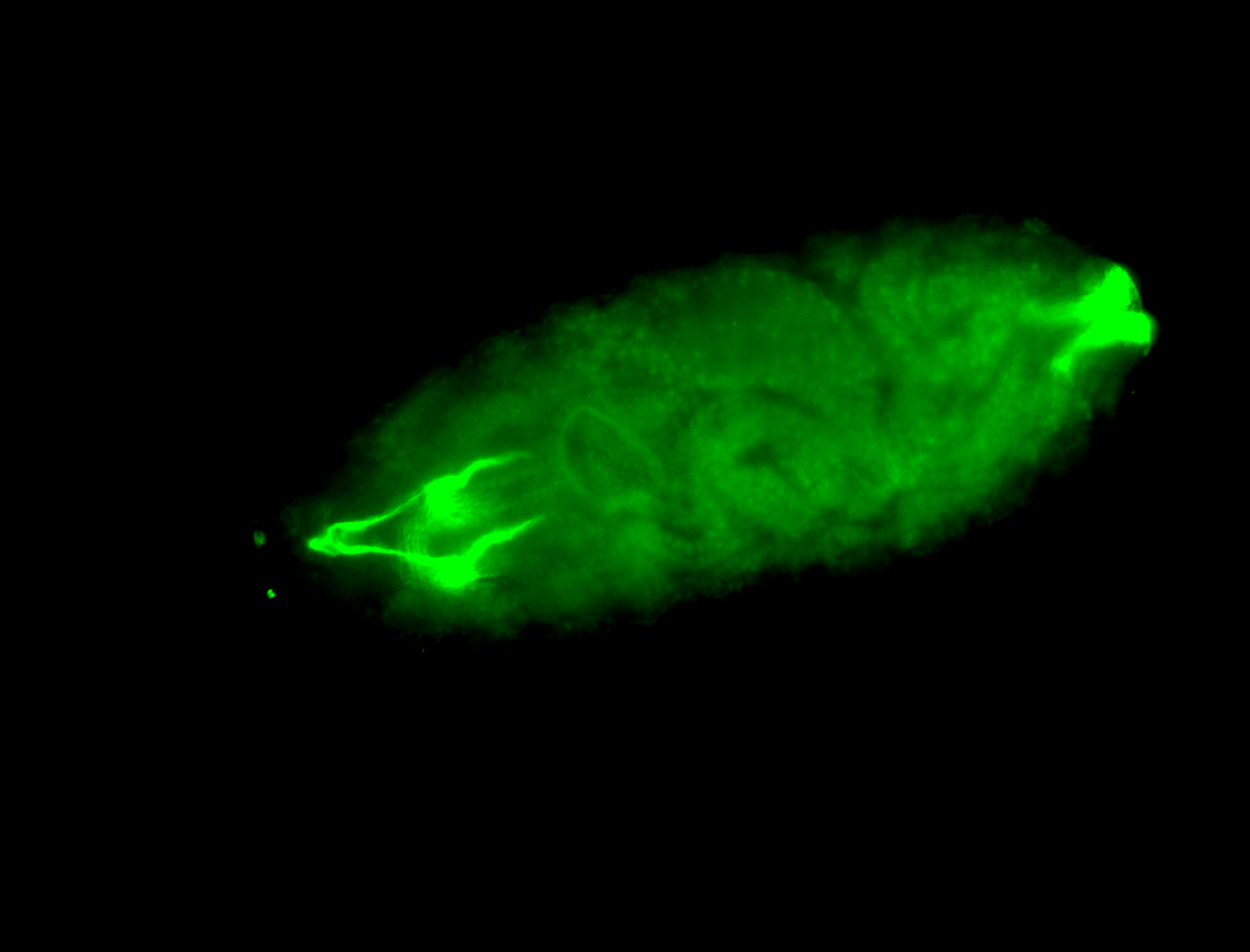
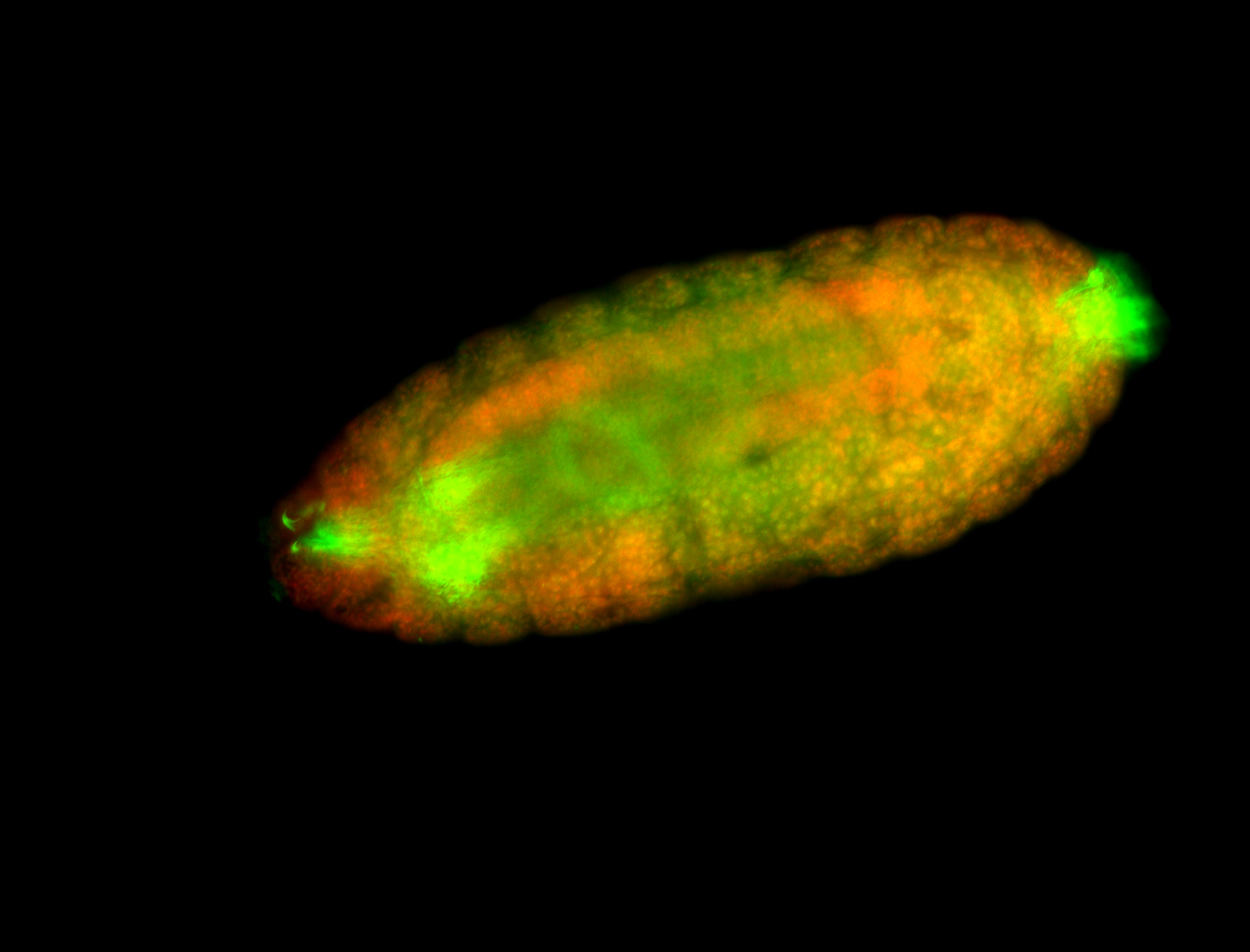
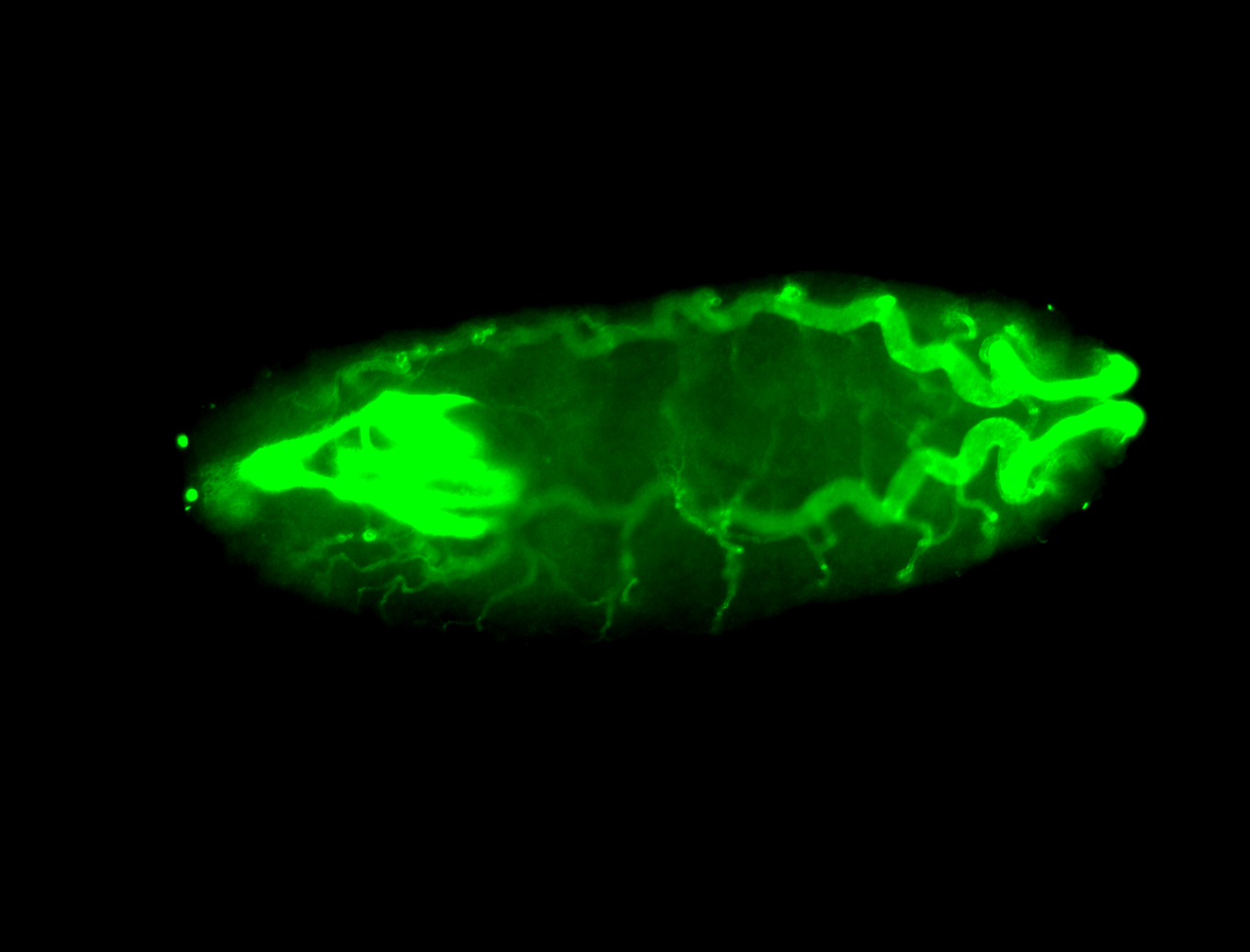
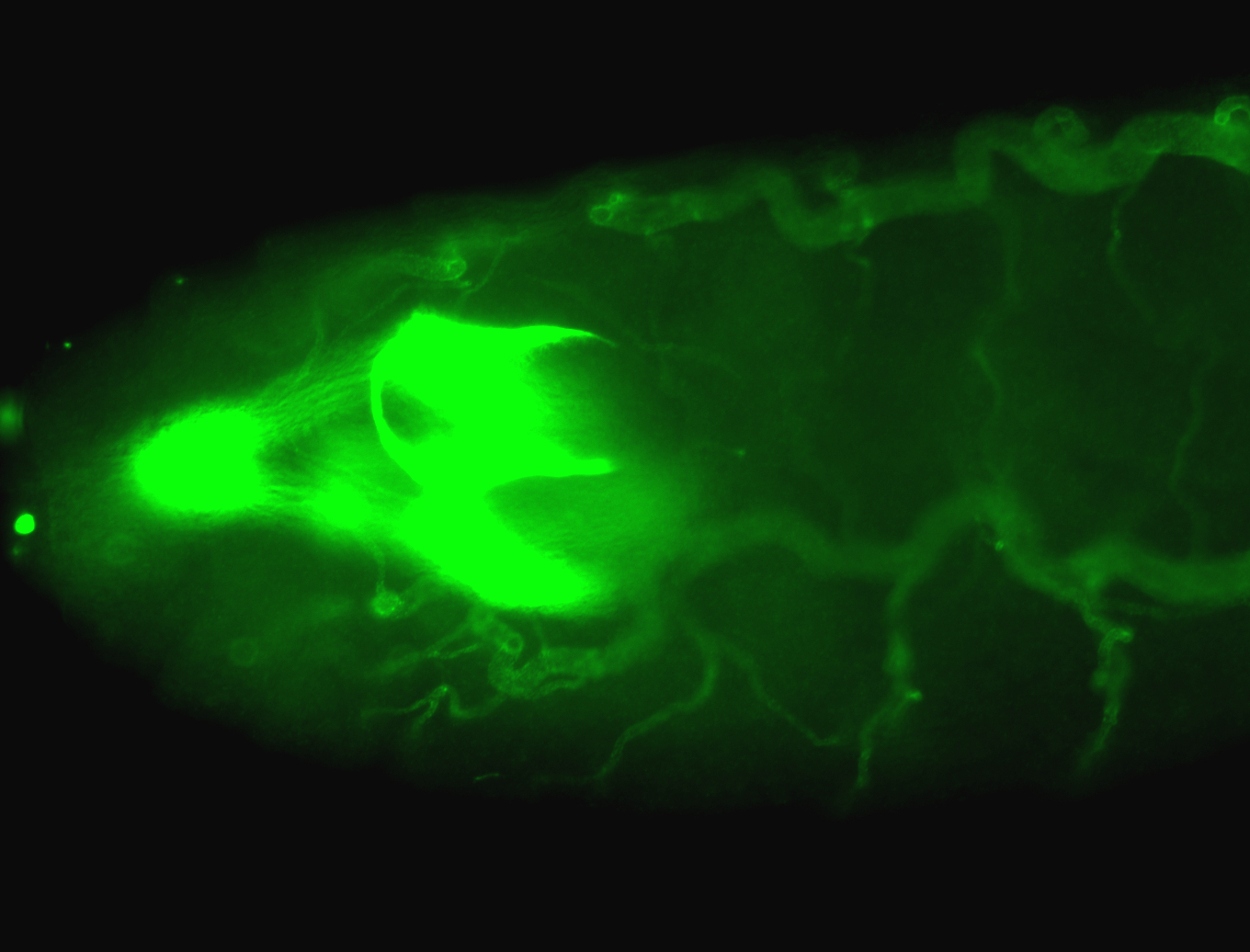
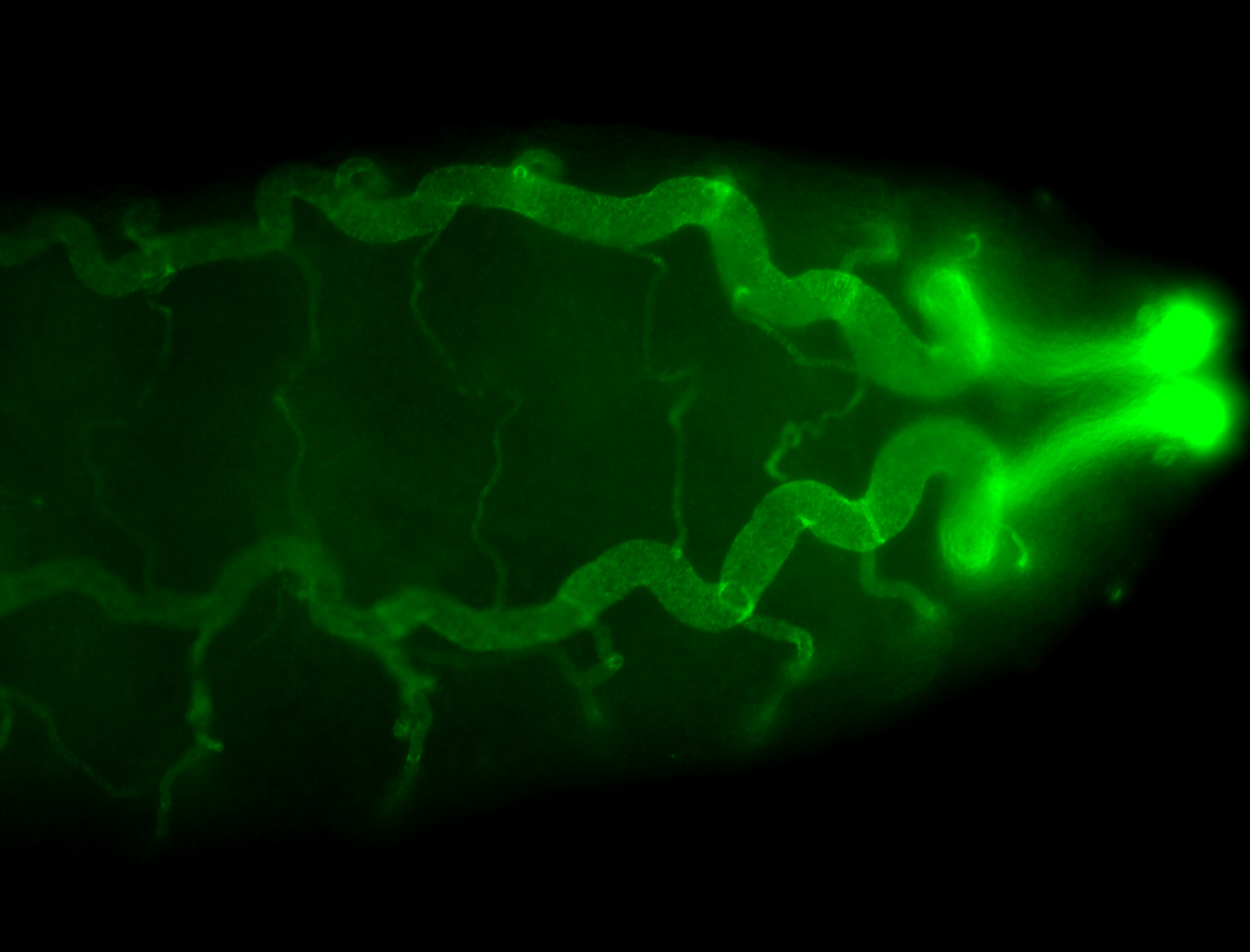
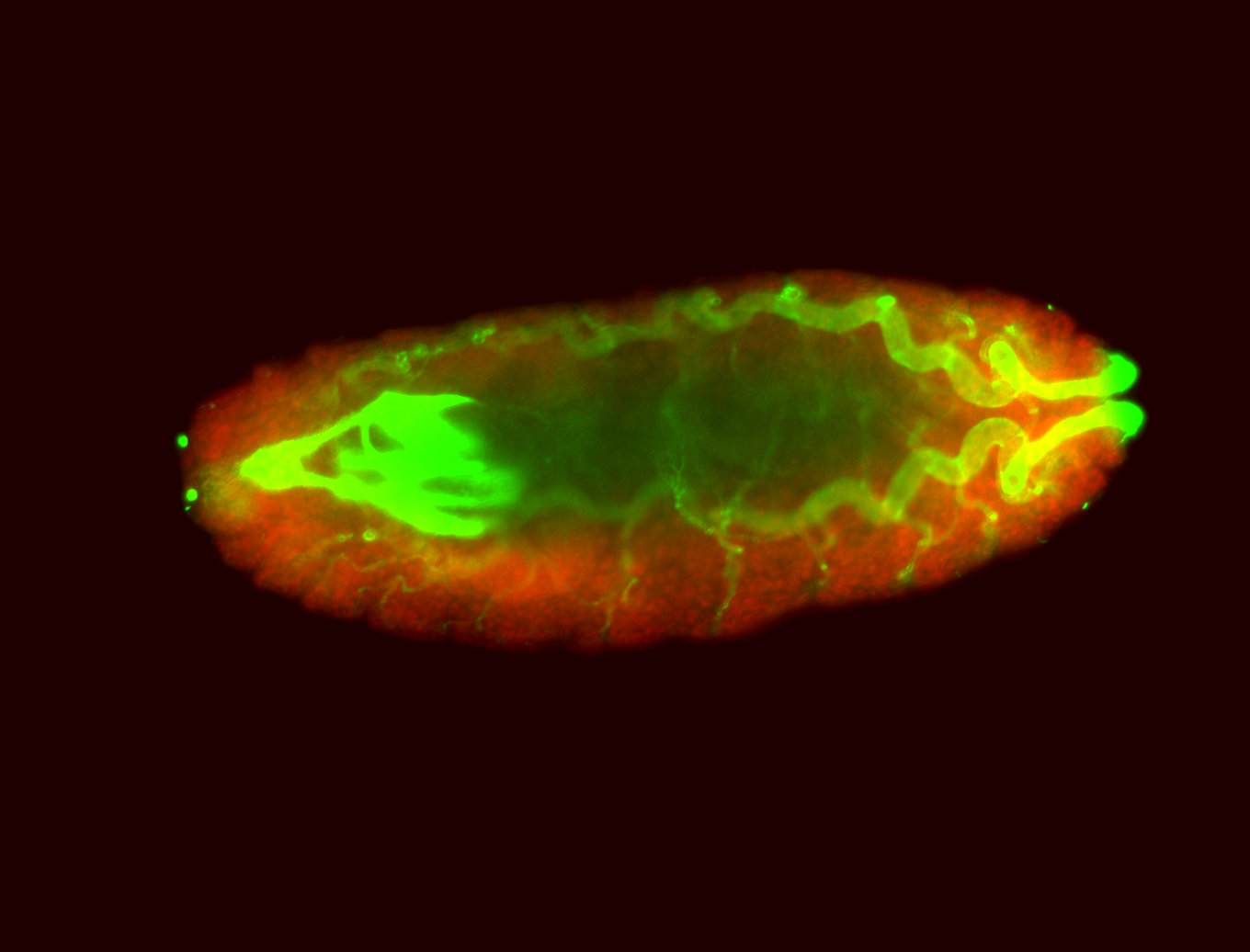
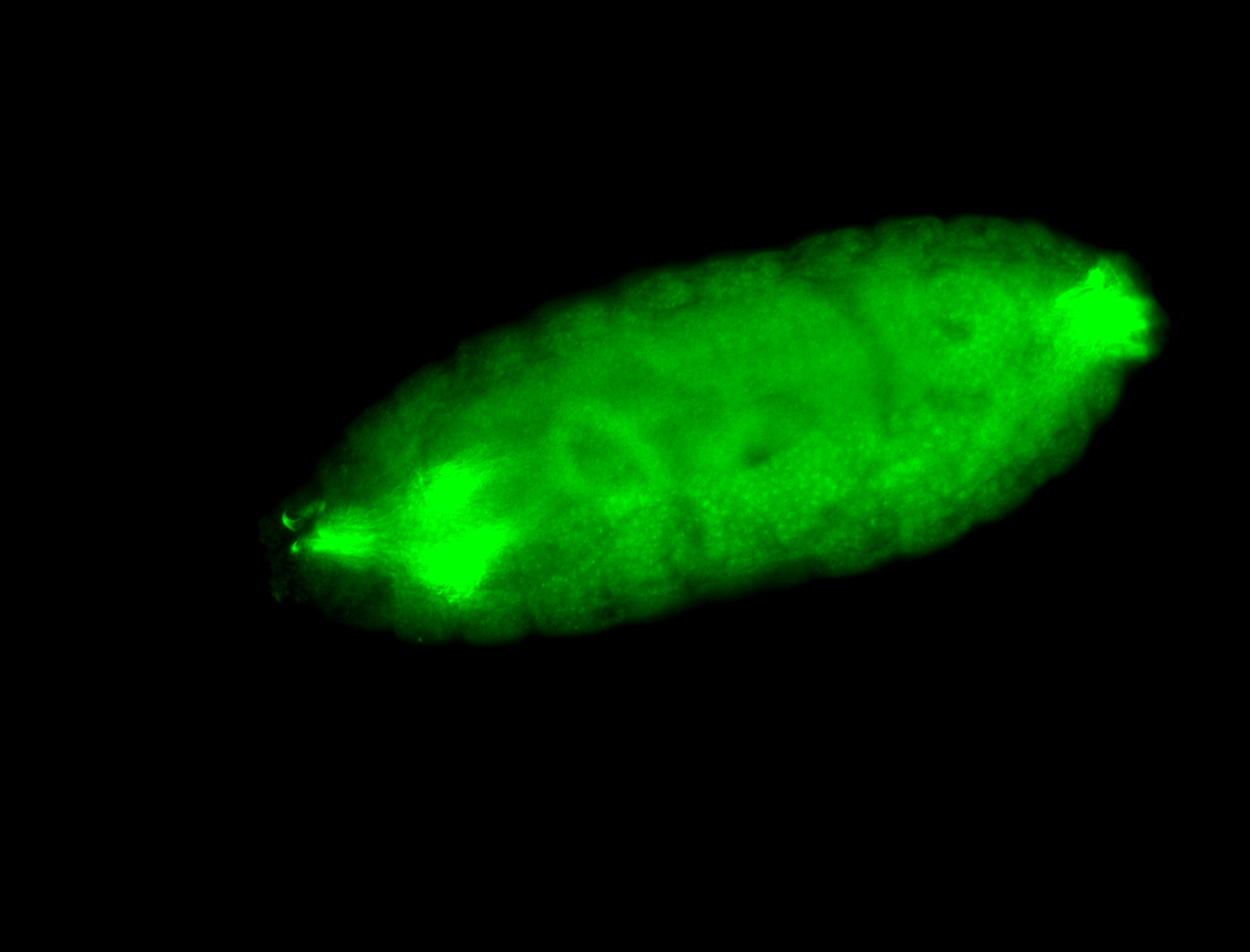
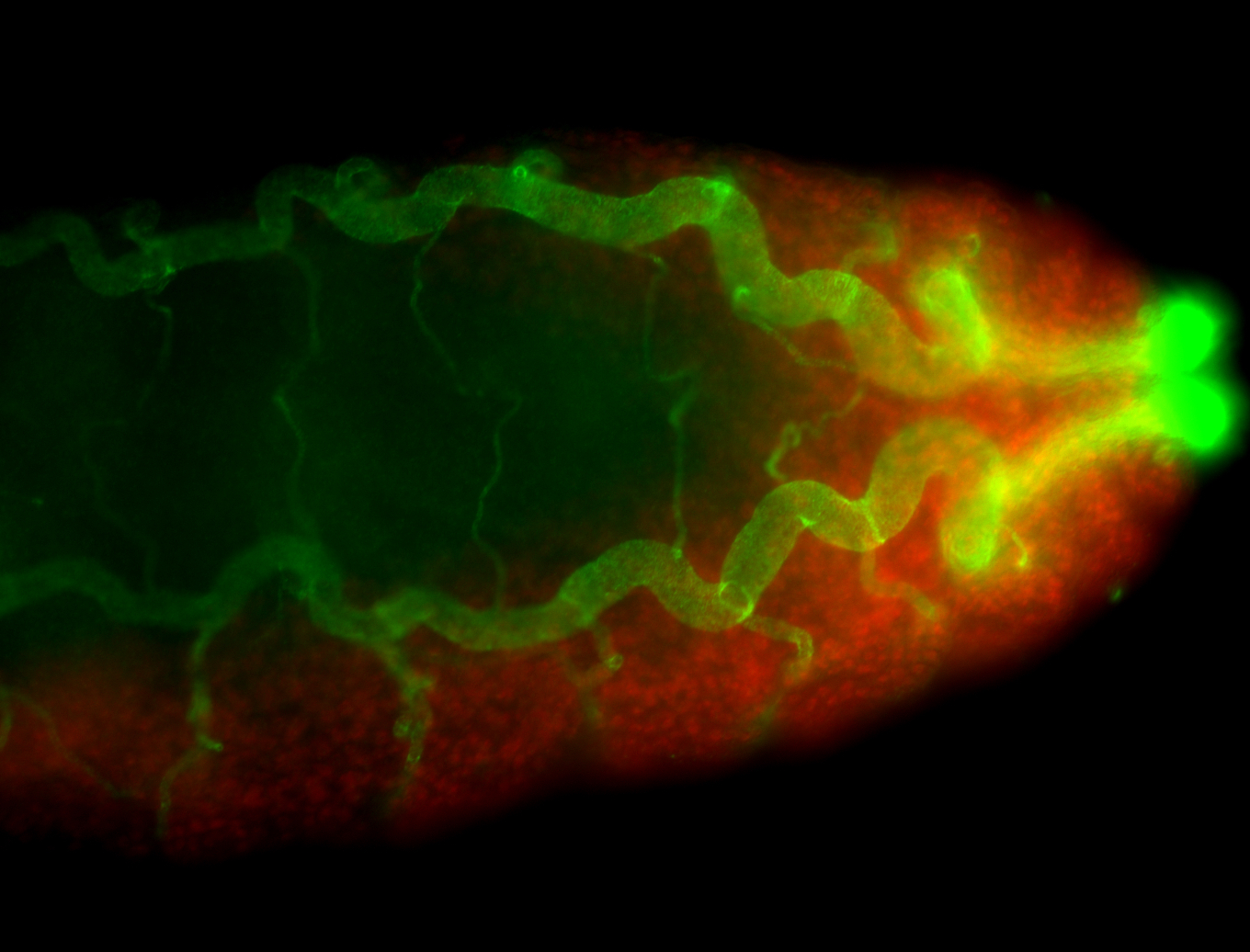
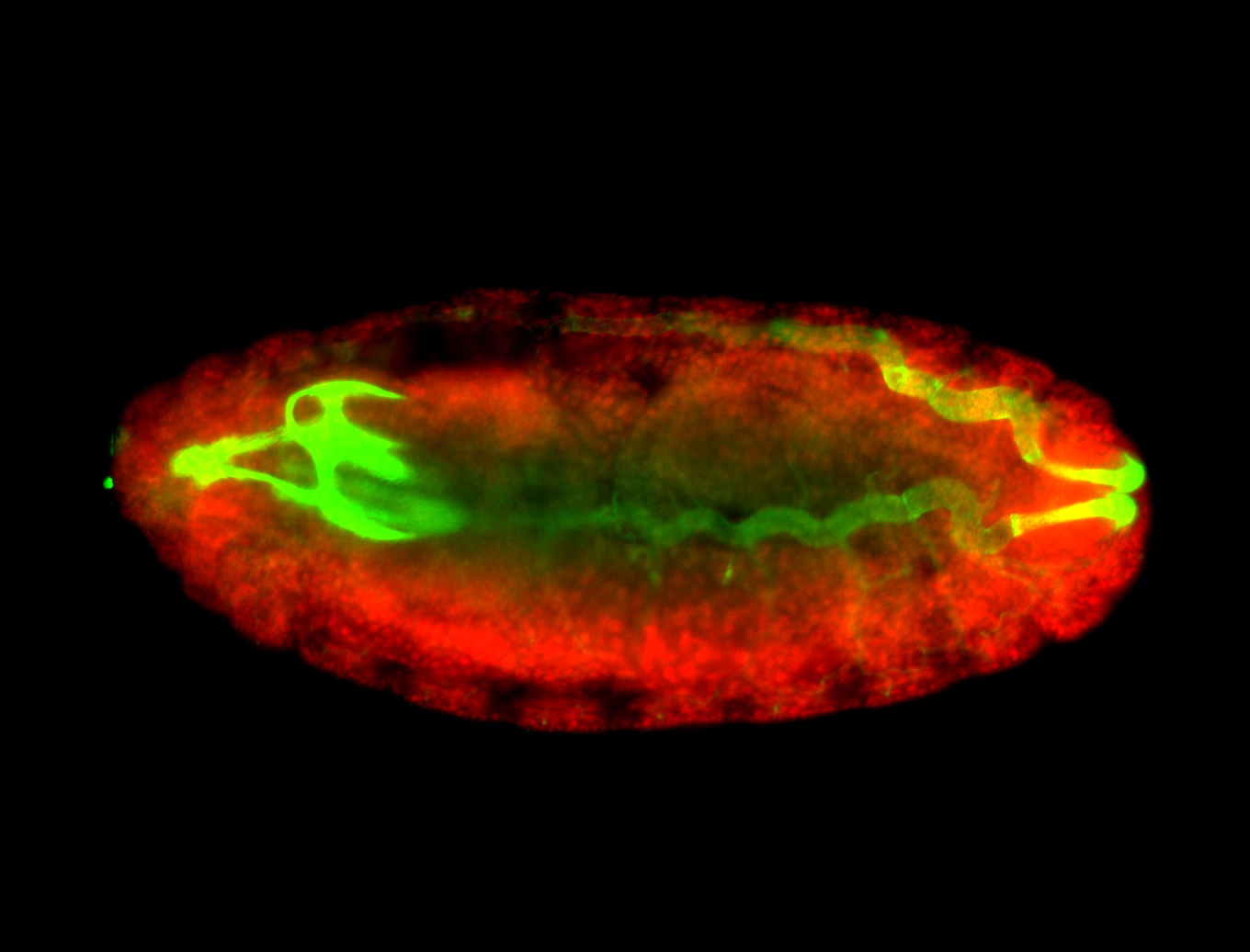
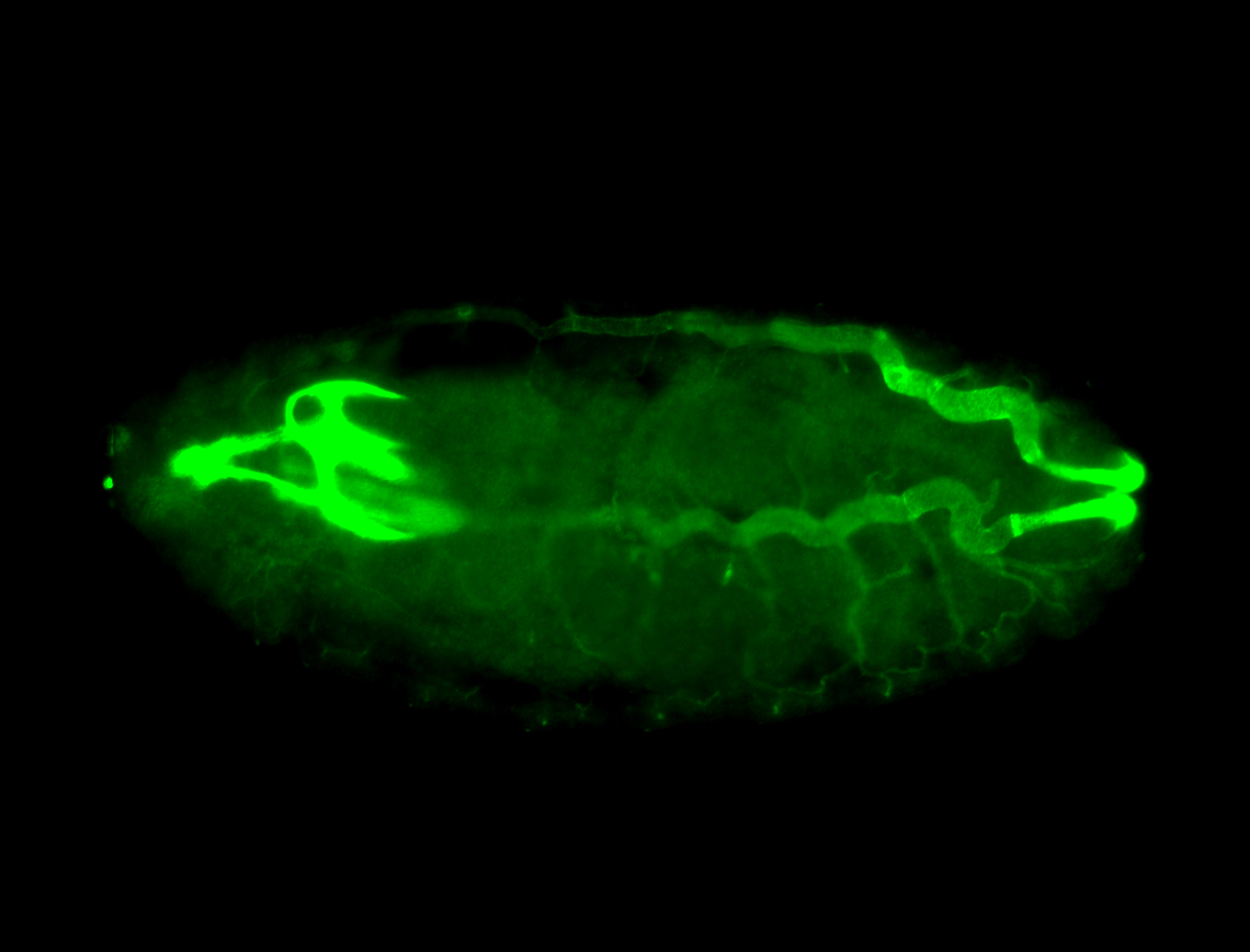
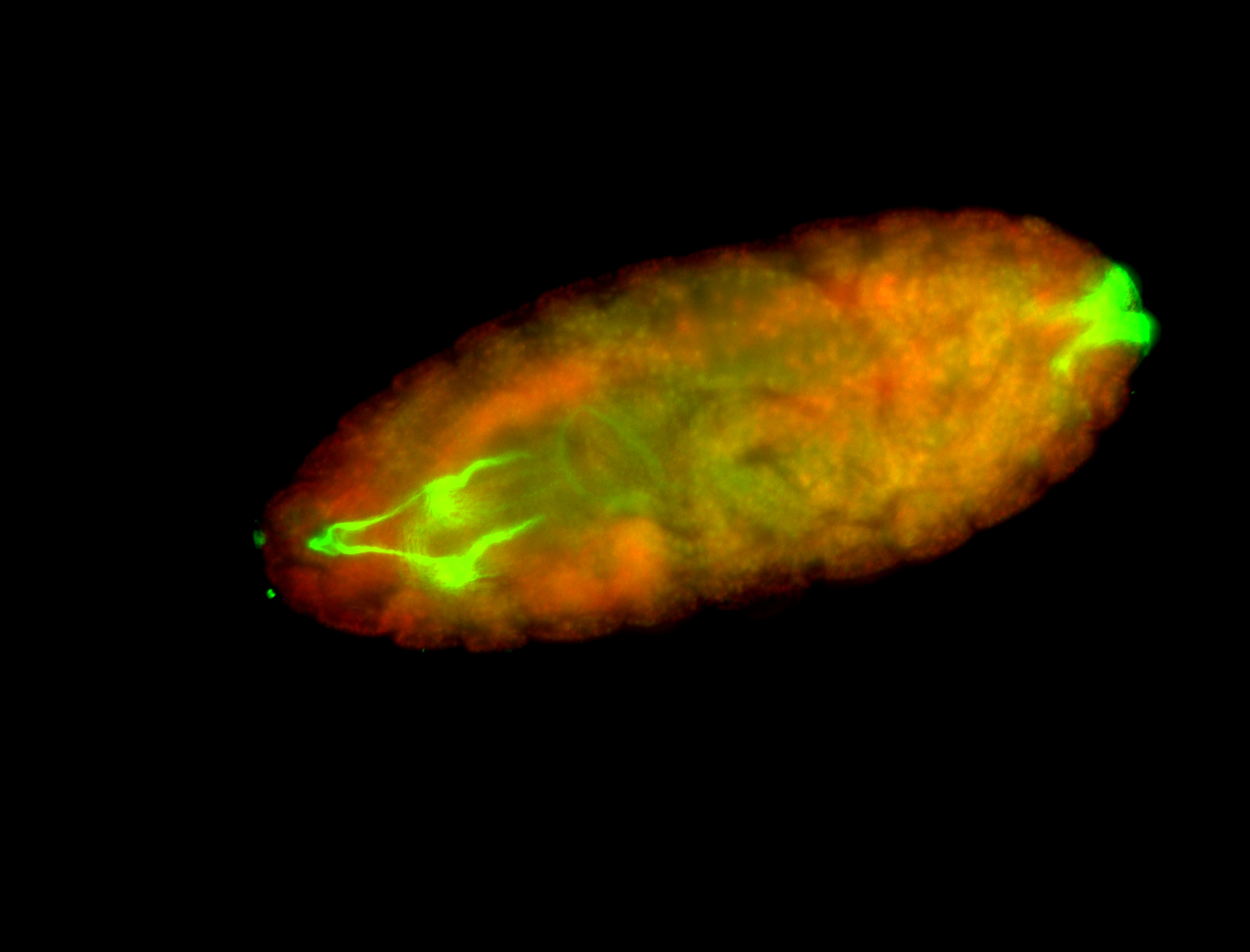
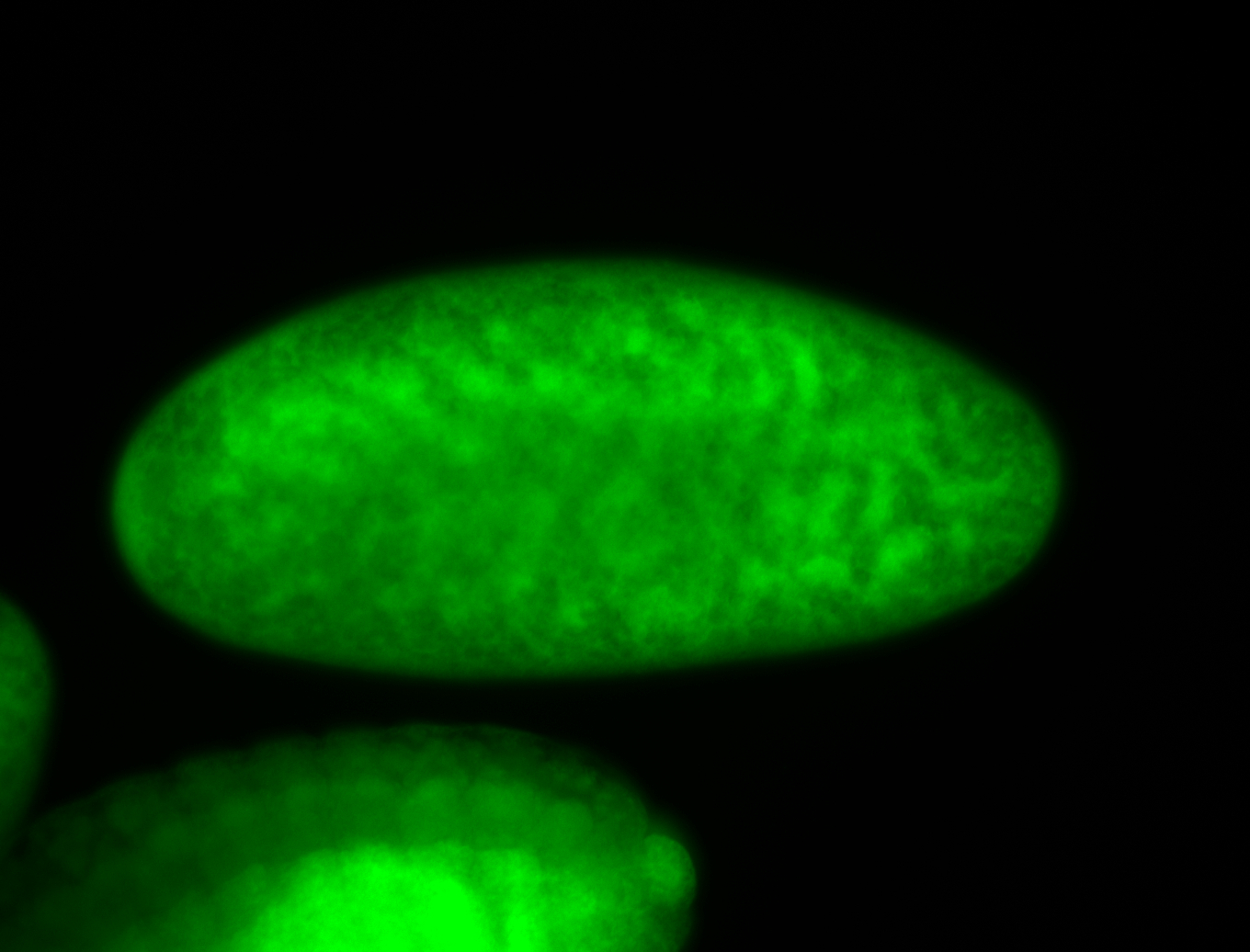
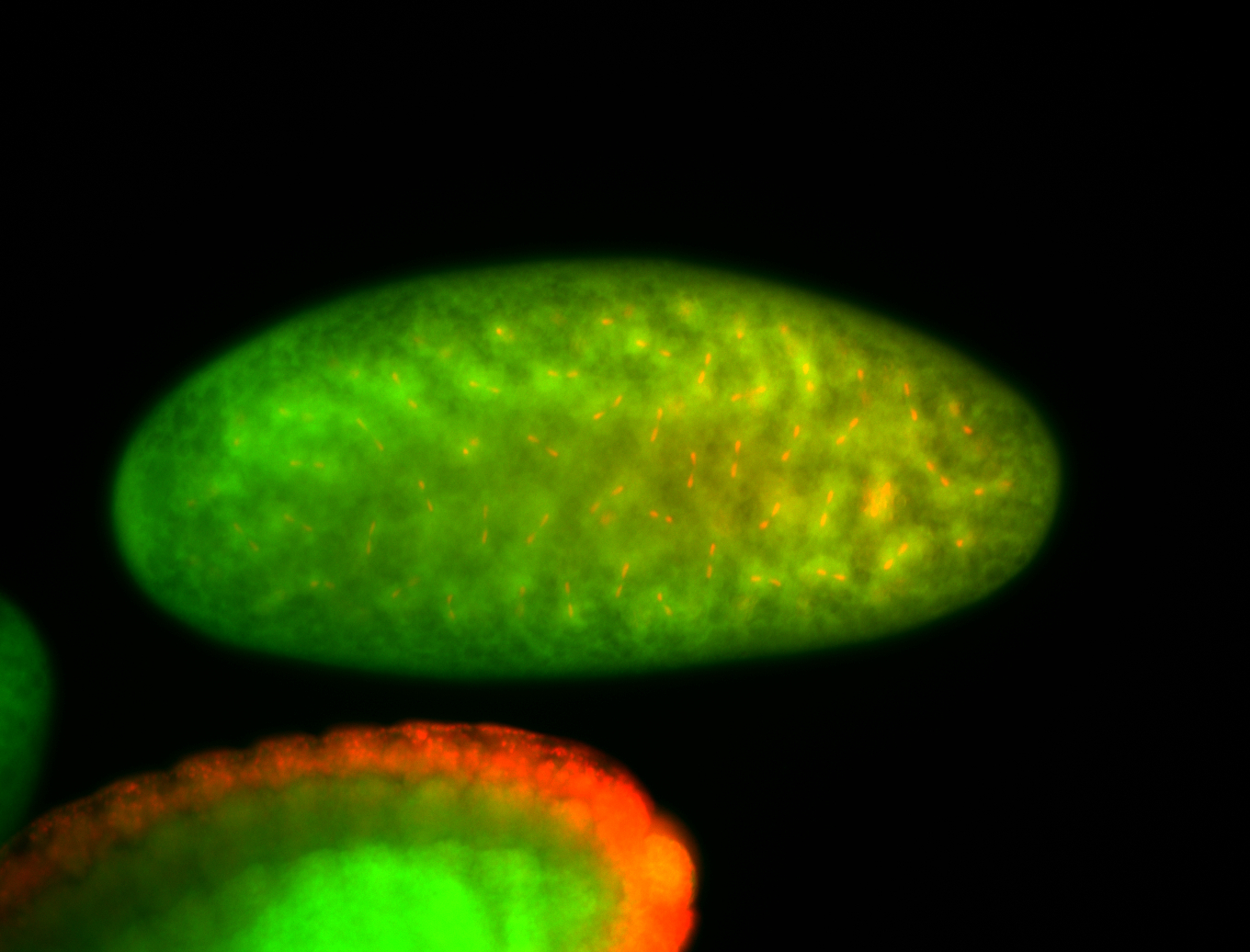
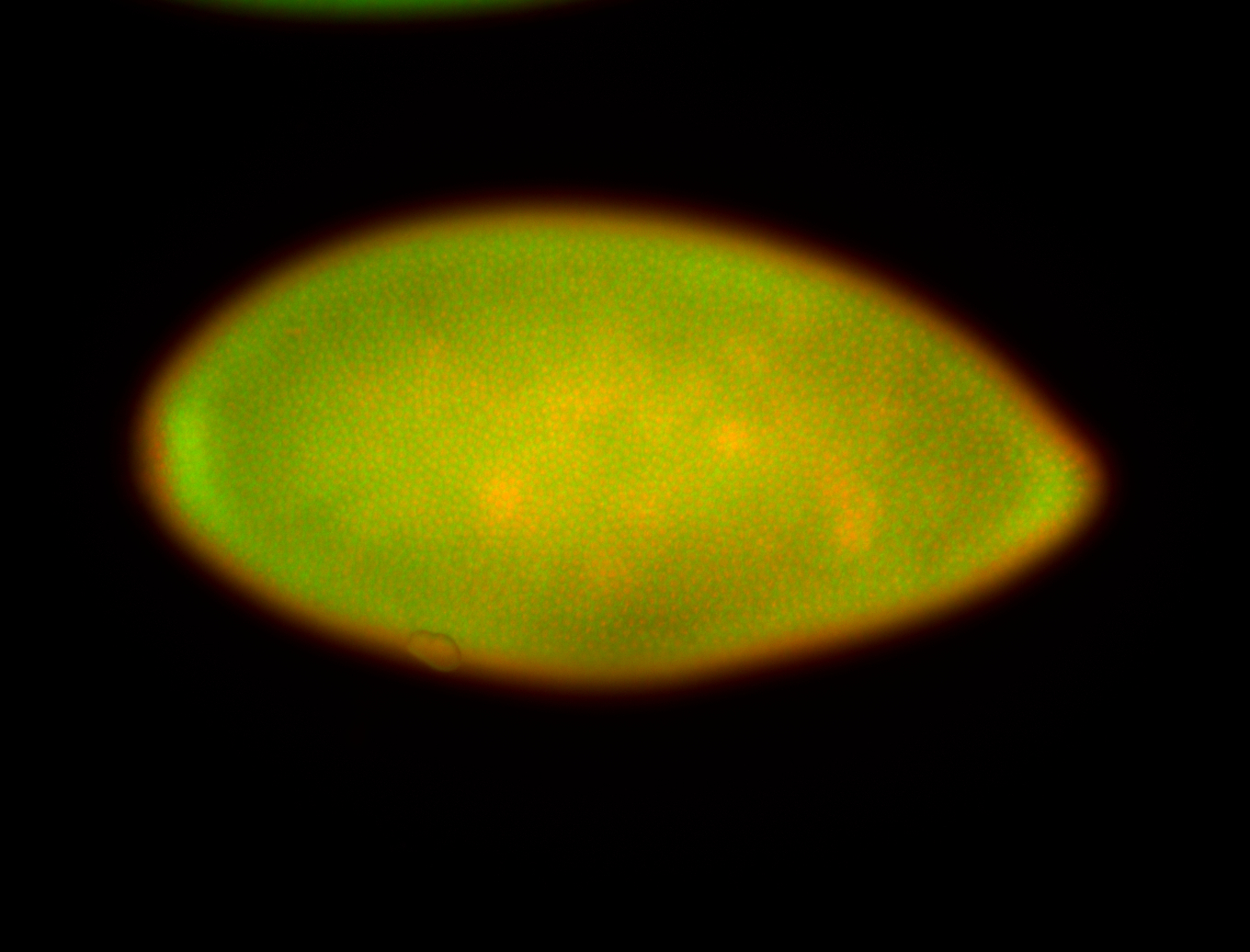
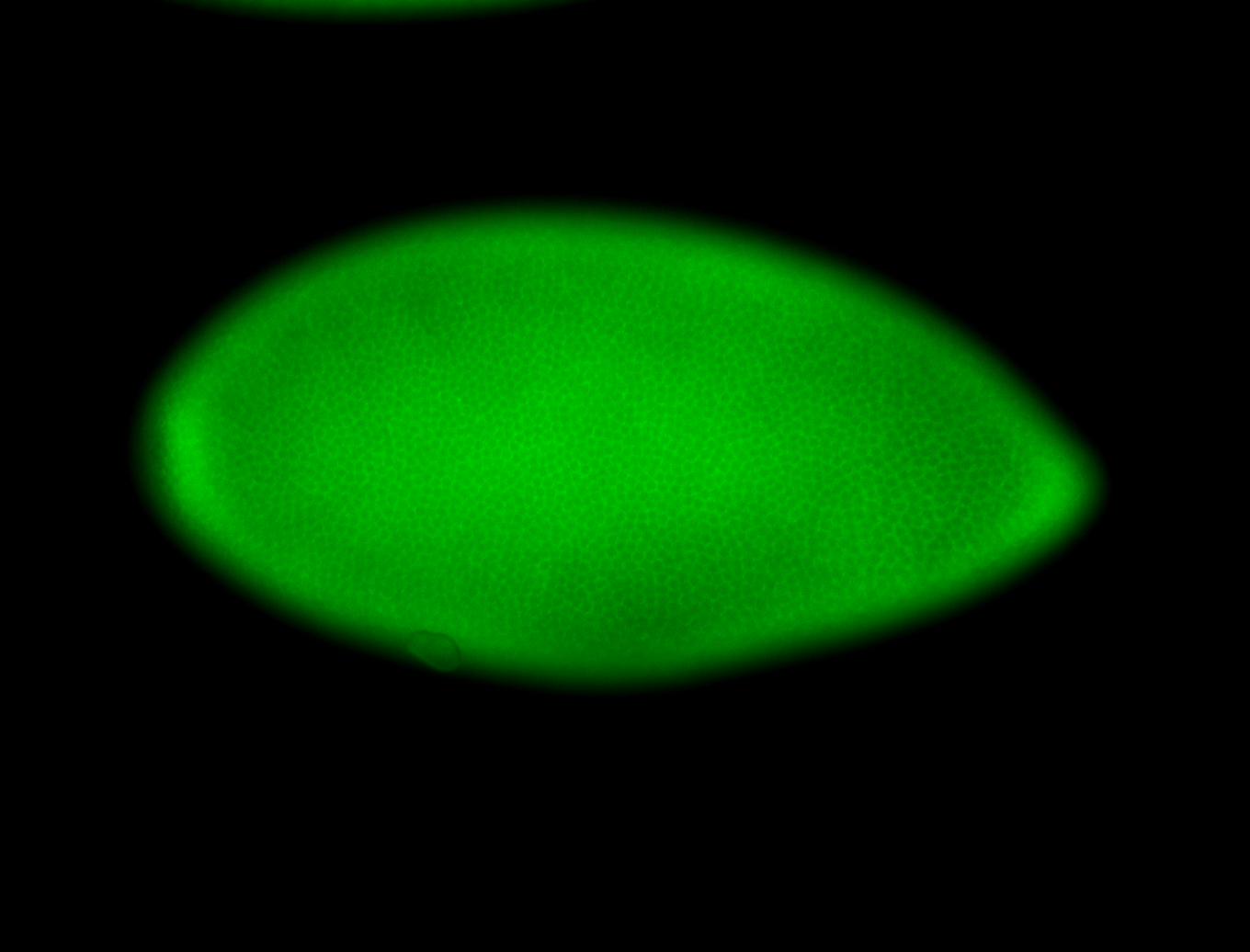
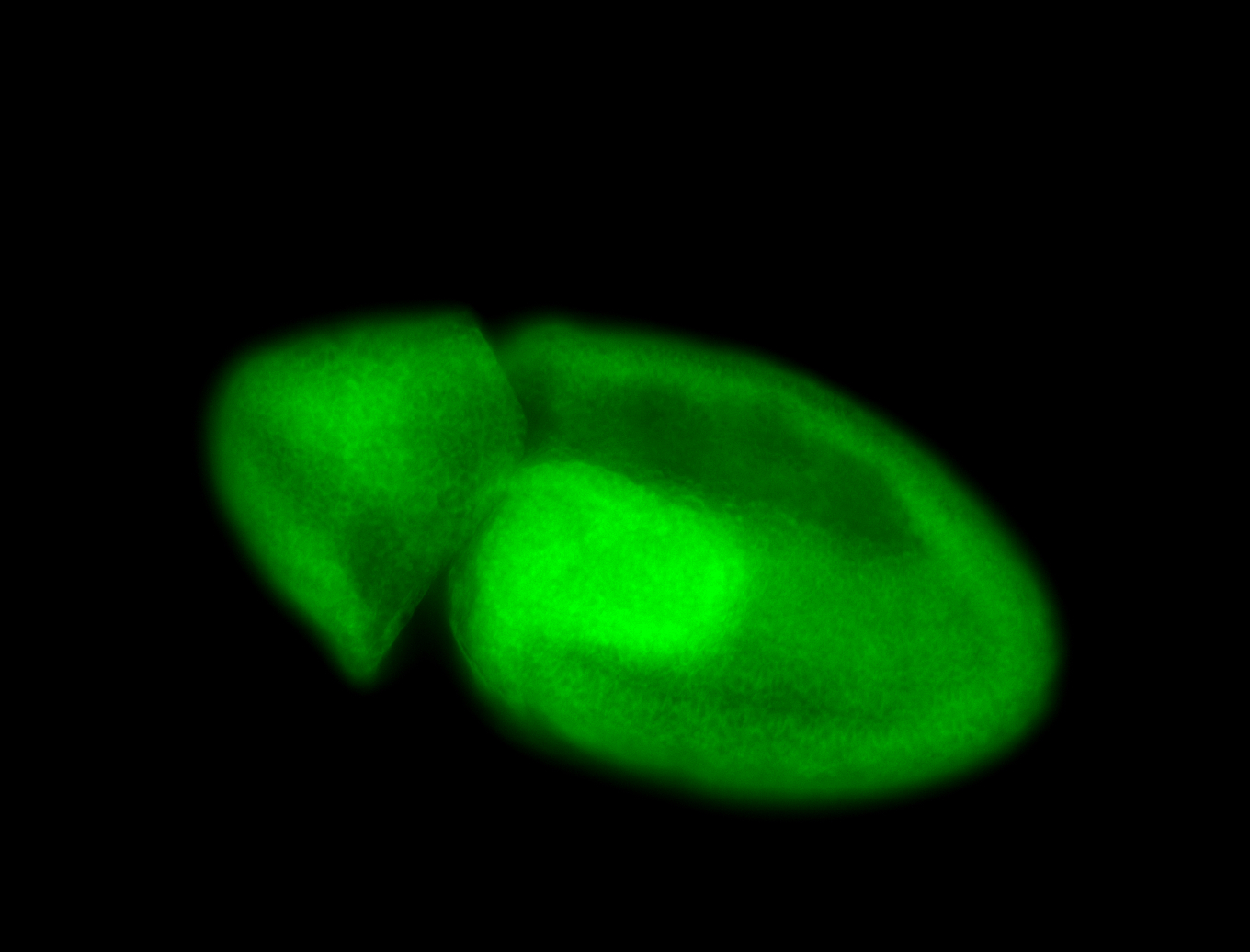
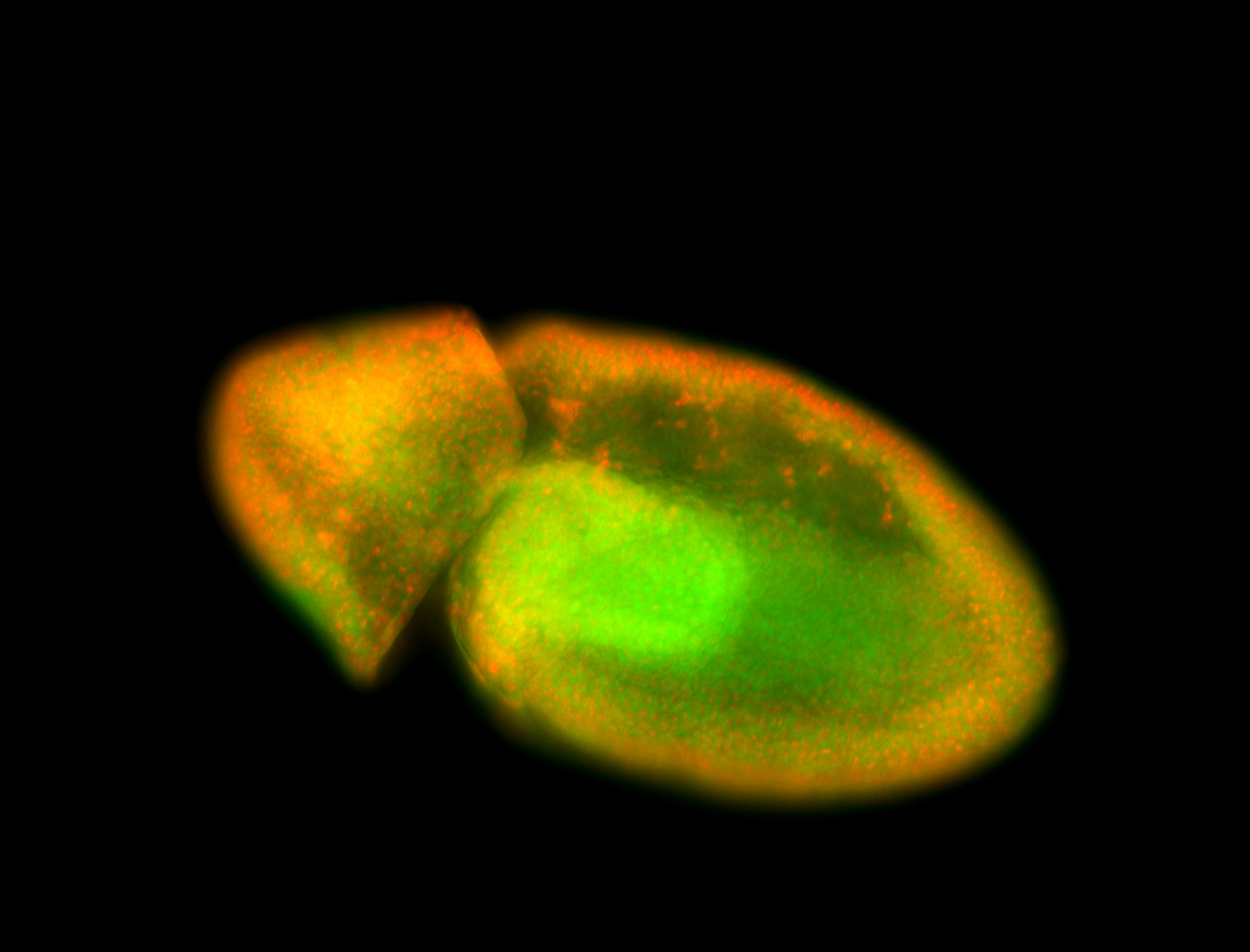
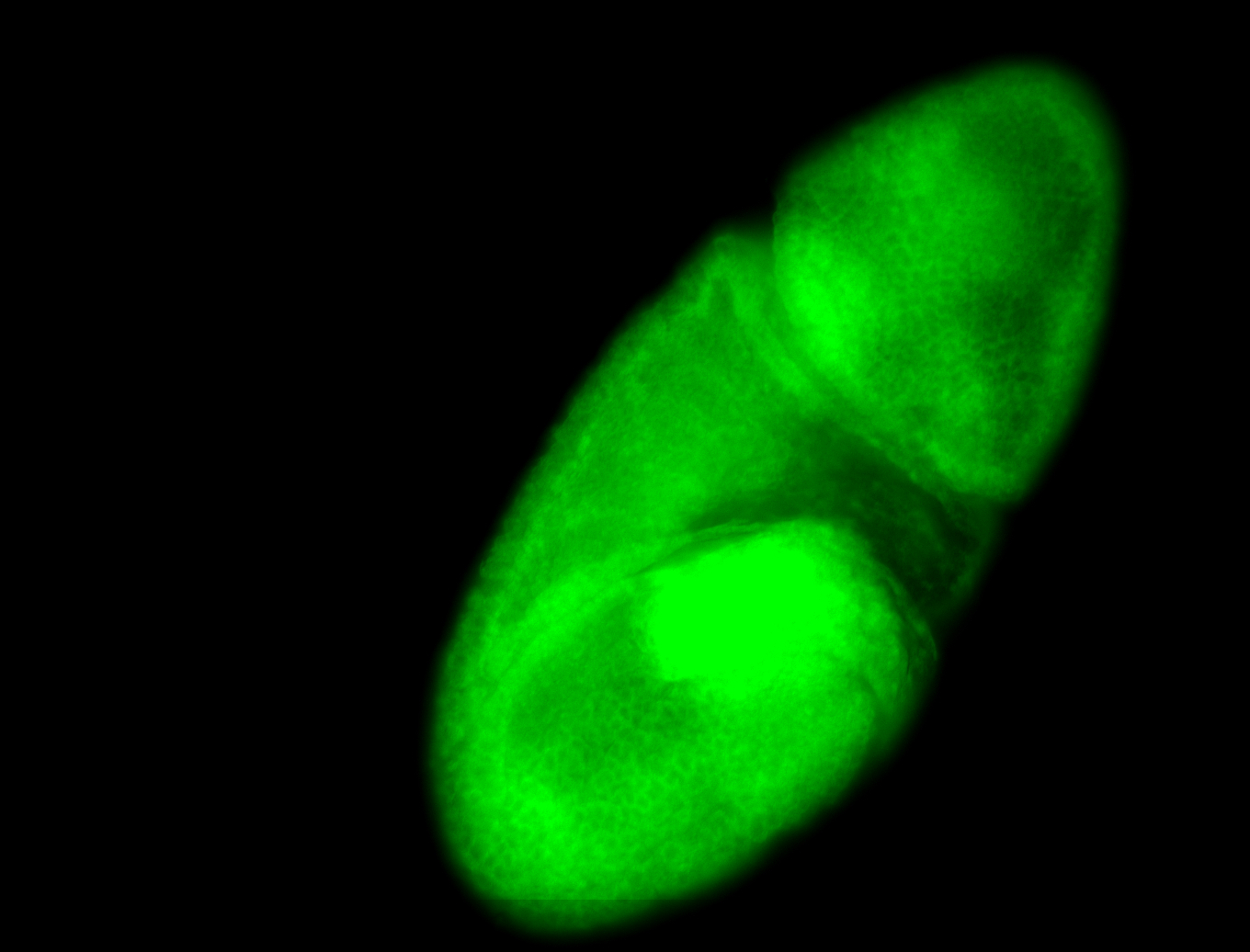
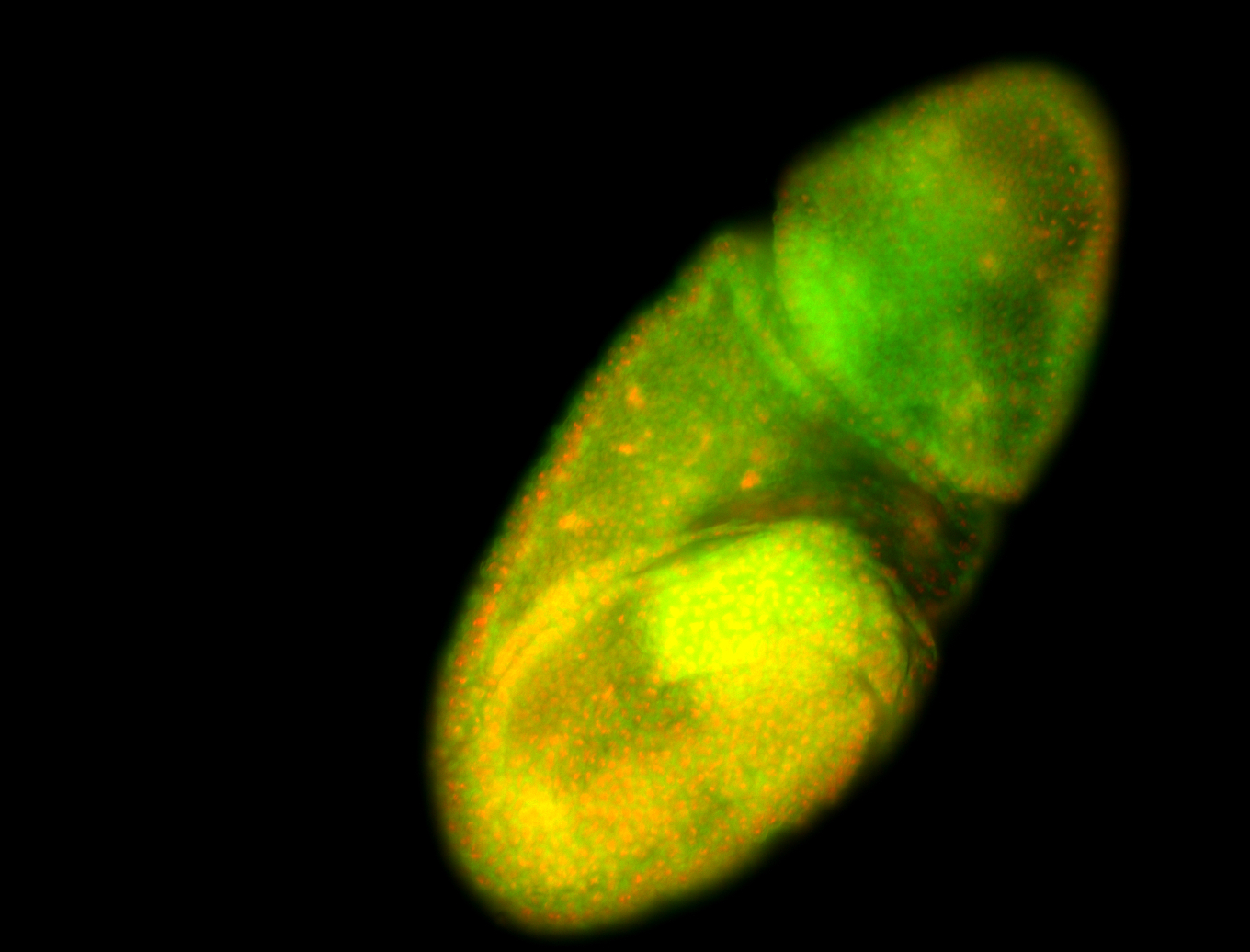
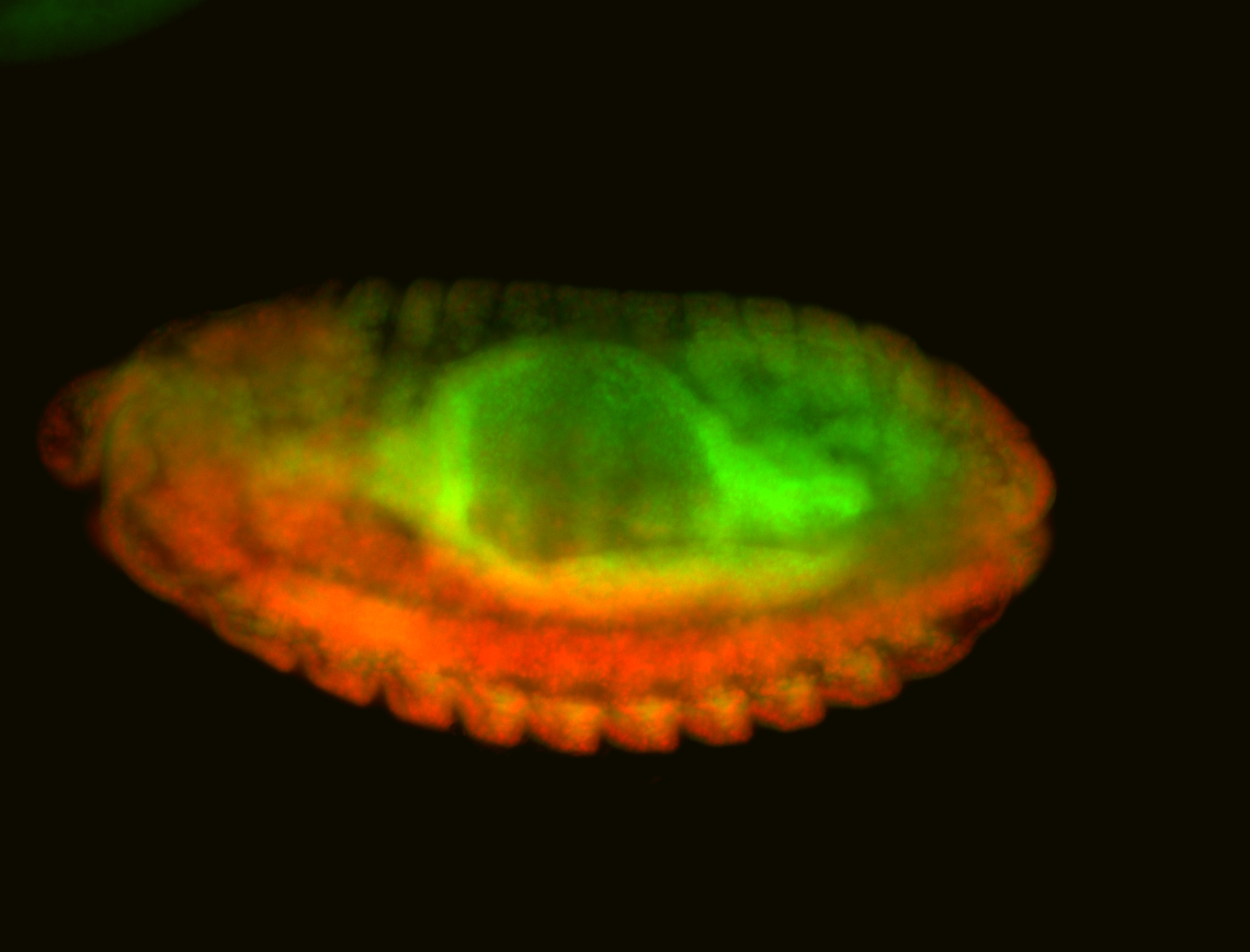
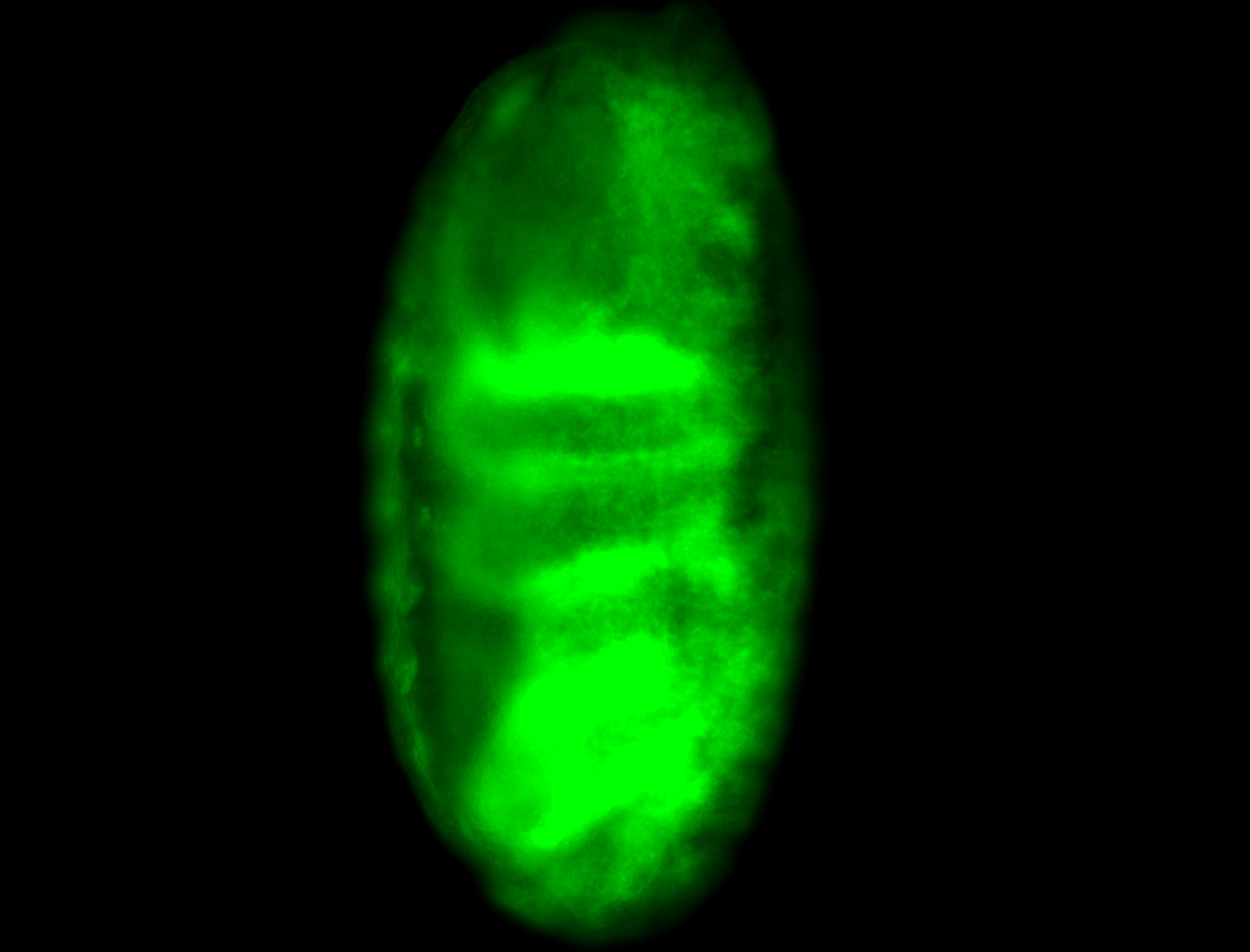
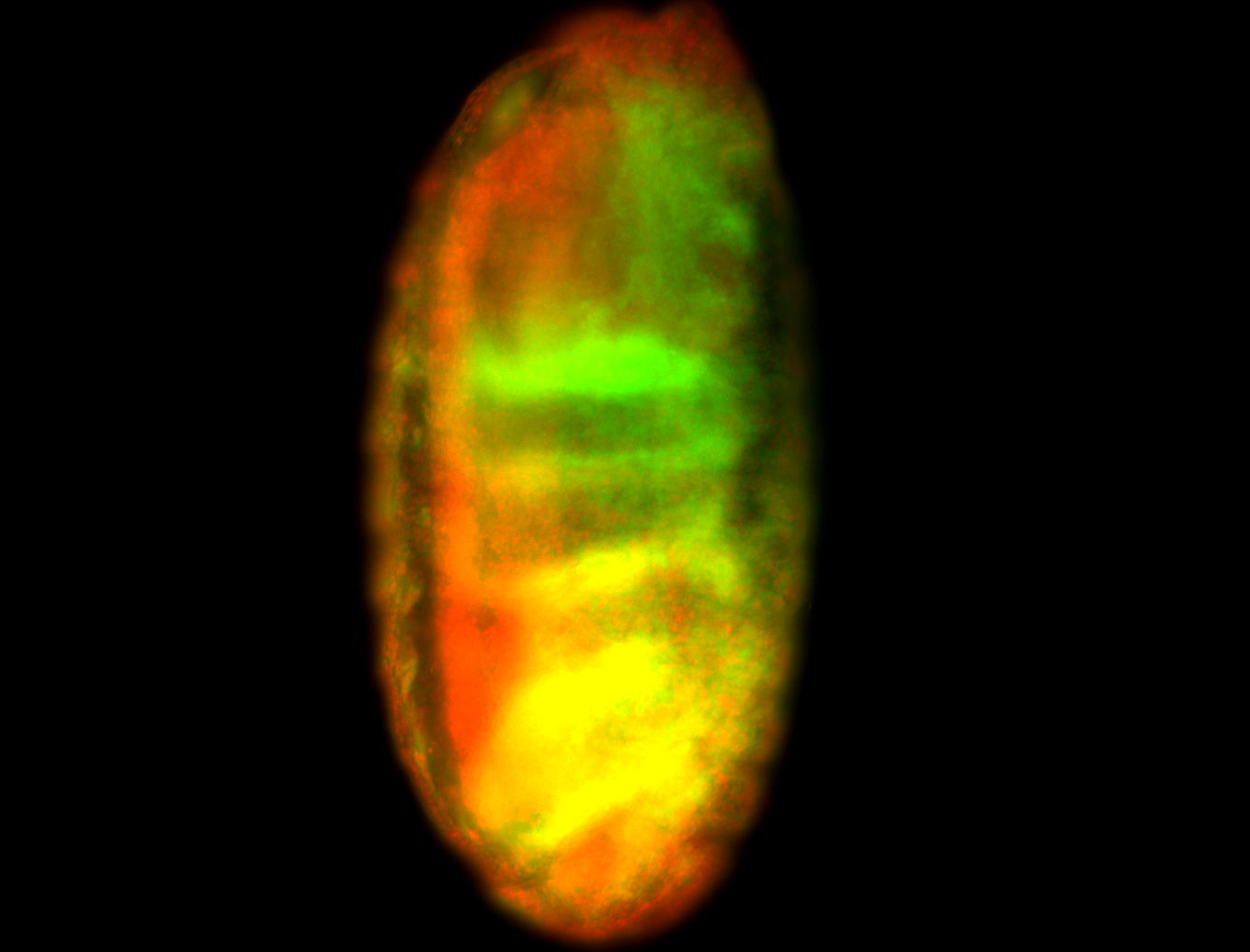
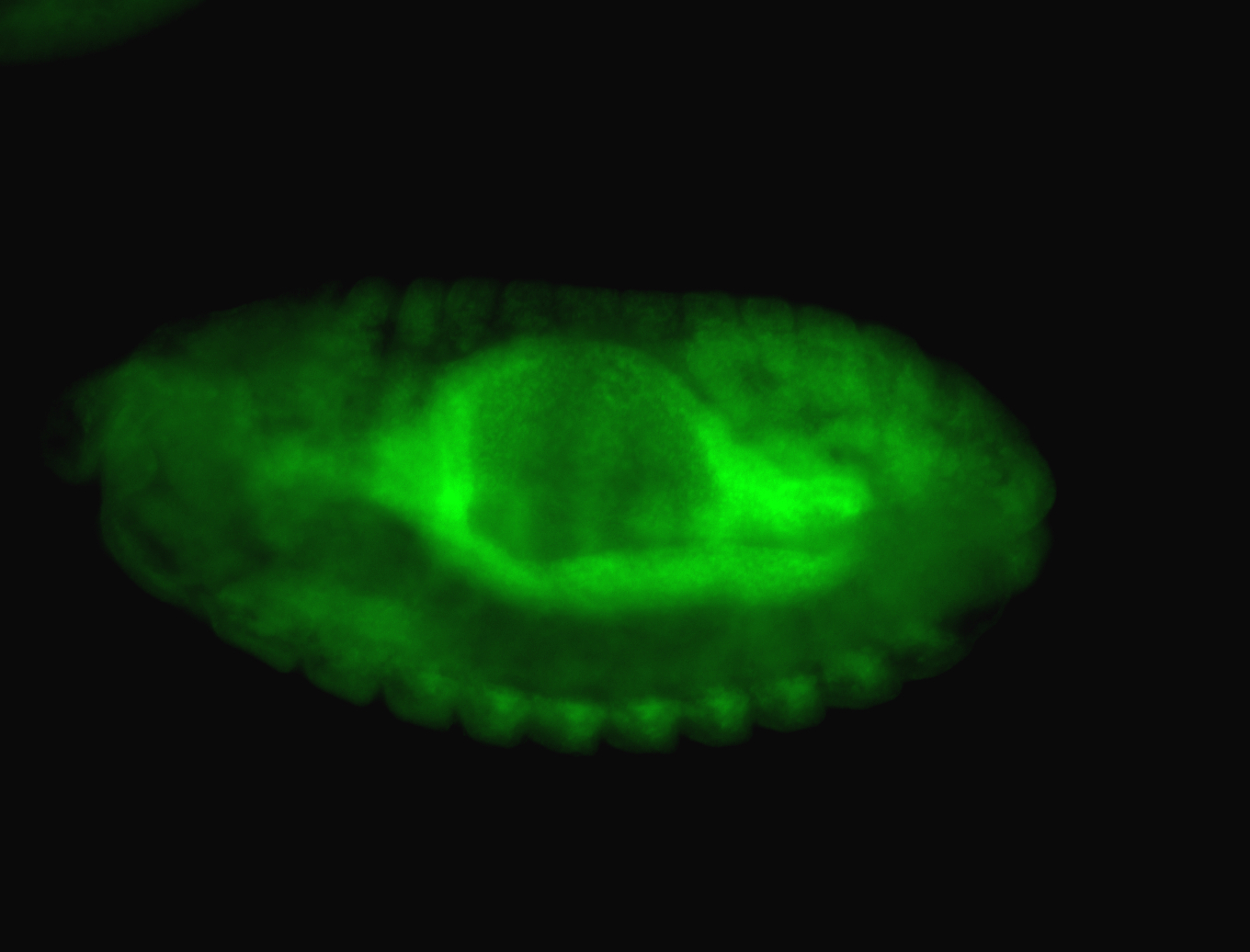
For a full list of searchable terms, please see the term page.
NOTE: It takes a few seconds for the term suggestions to load.
Result Summary
| Number of genes | 42 | |
| Export | ||
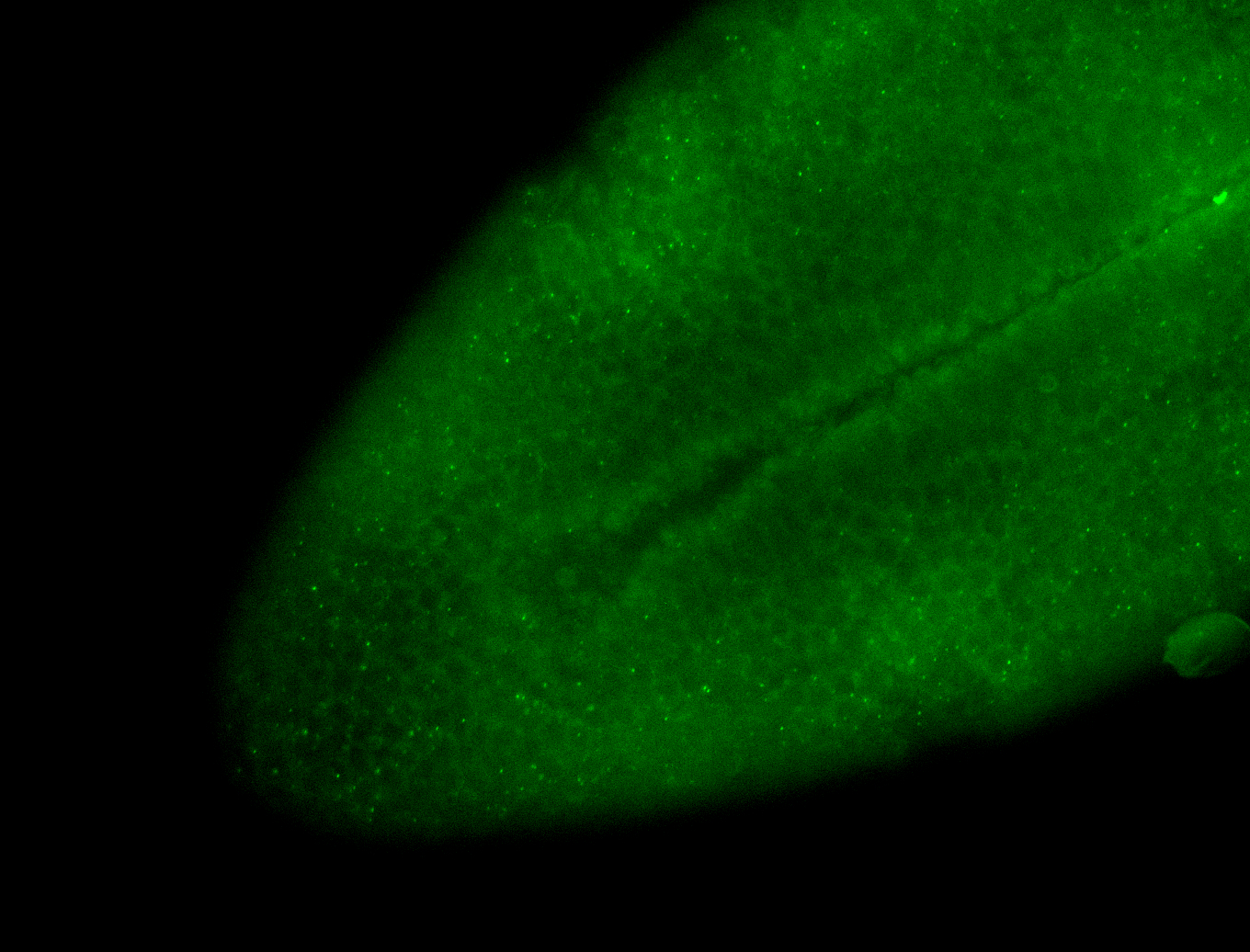
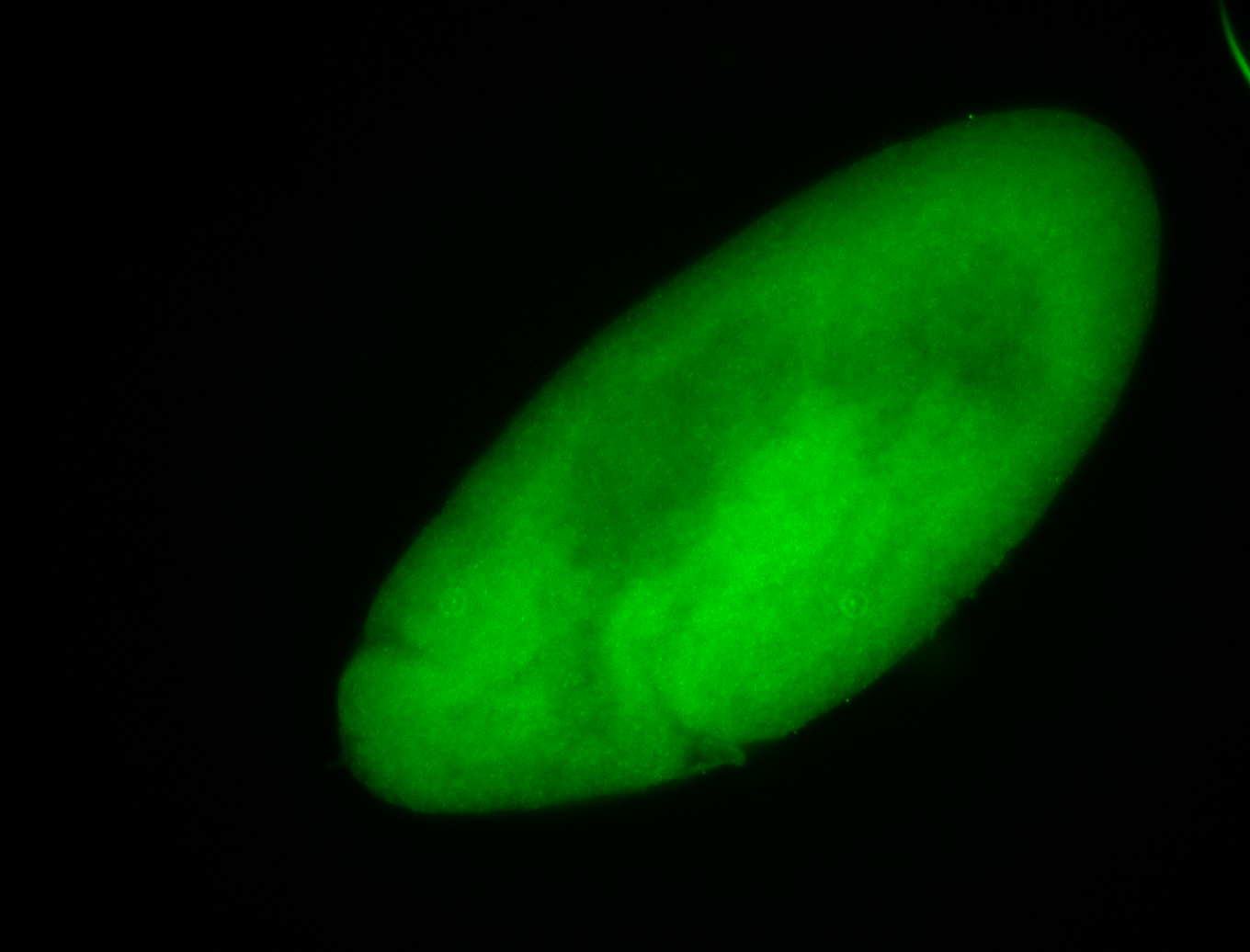
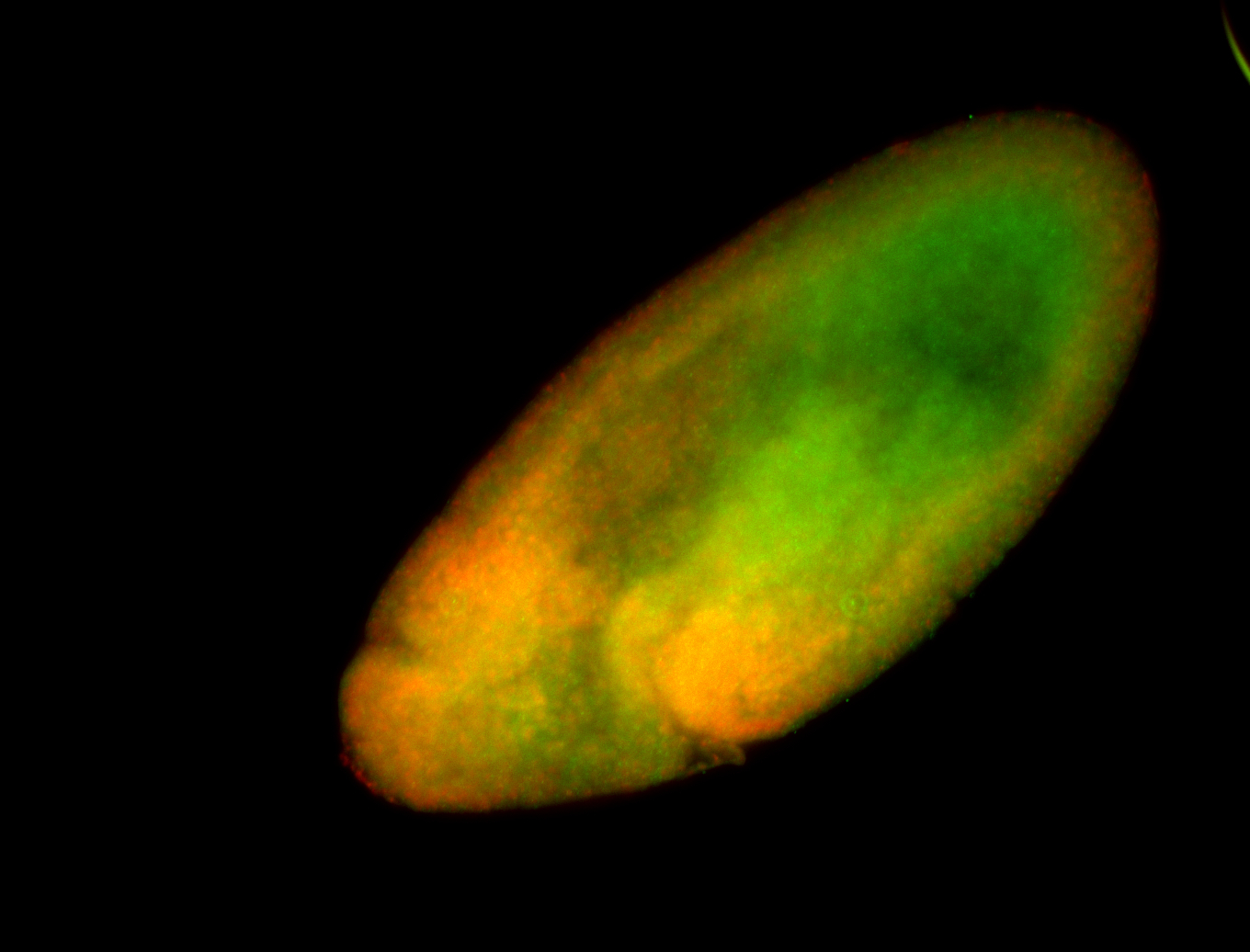
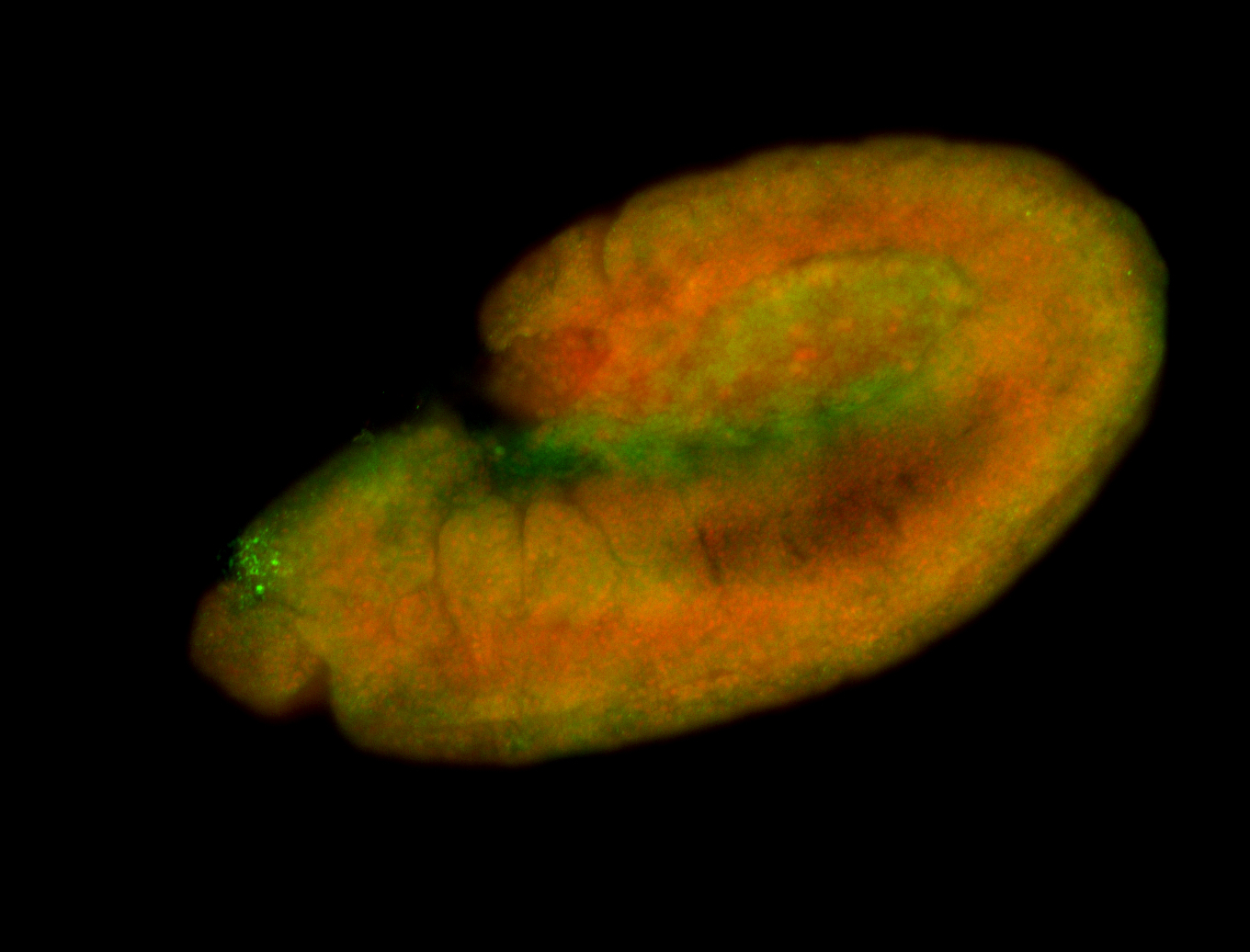
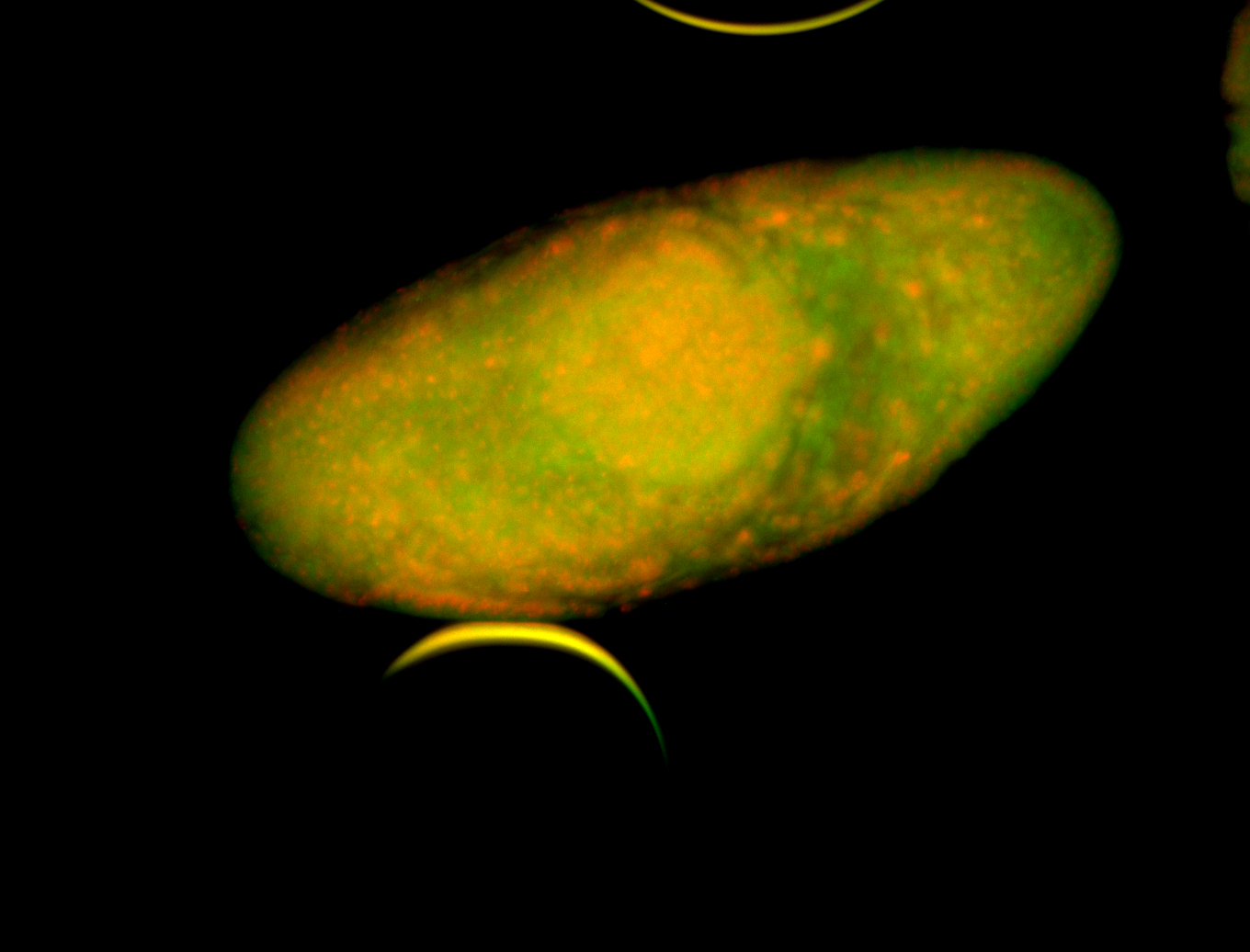
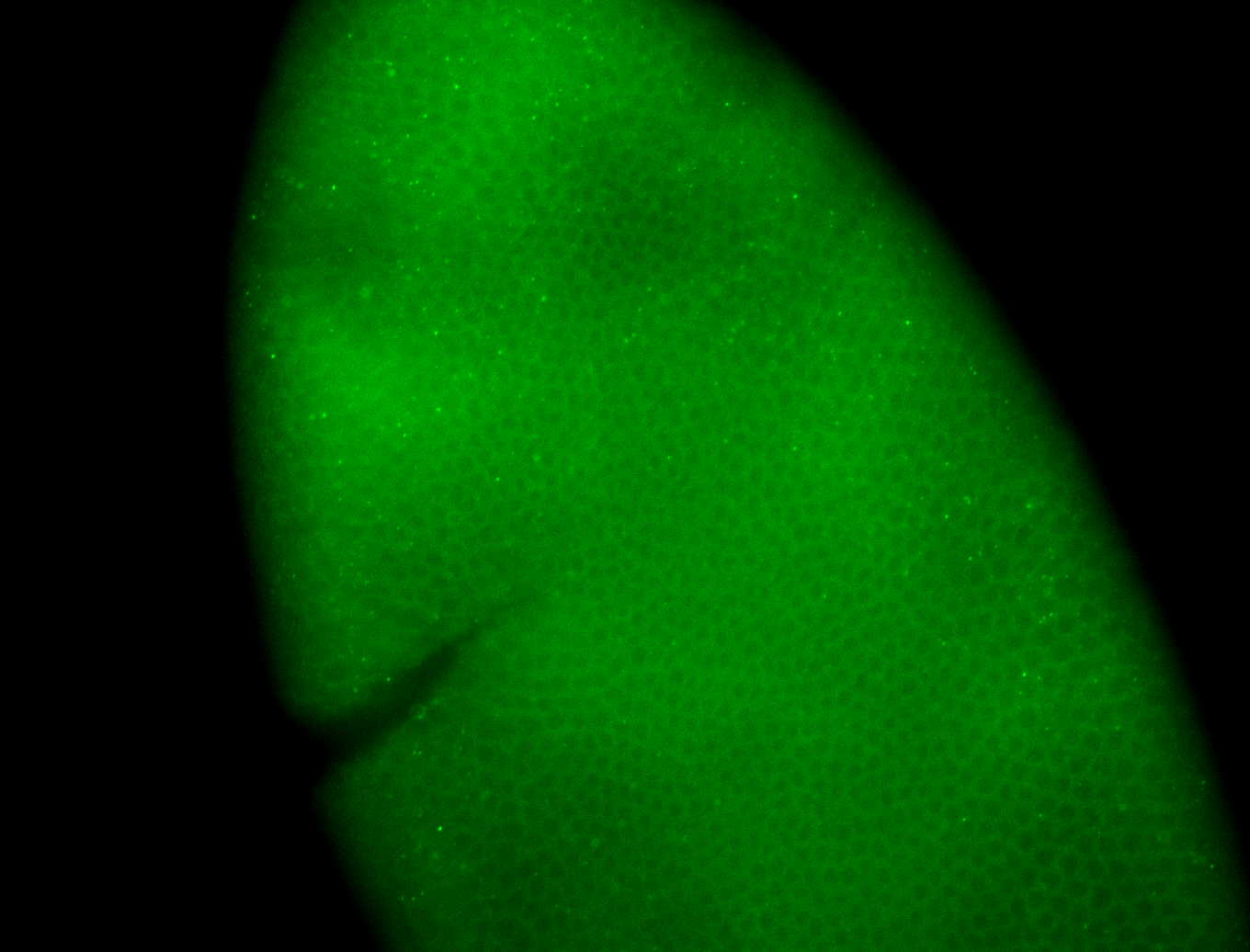
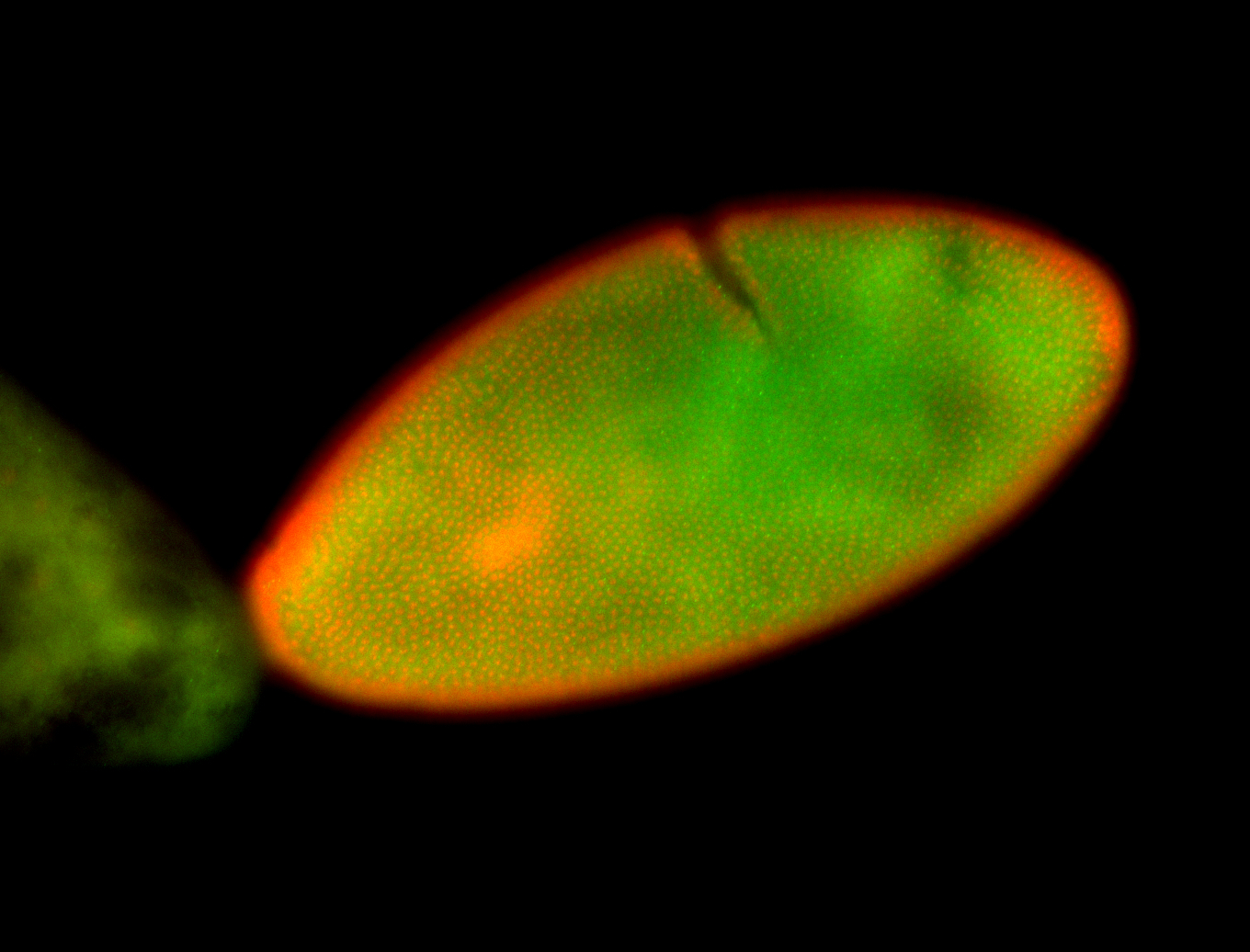
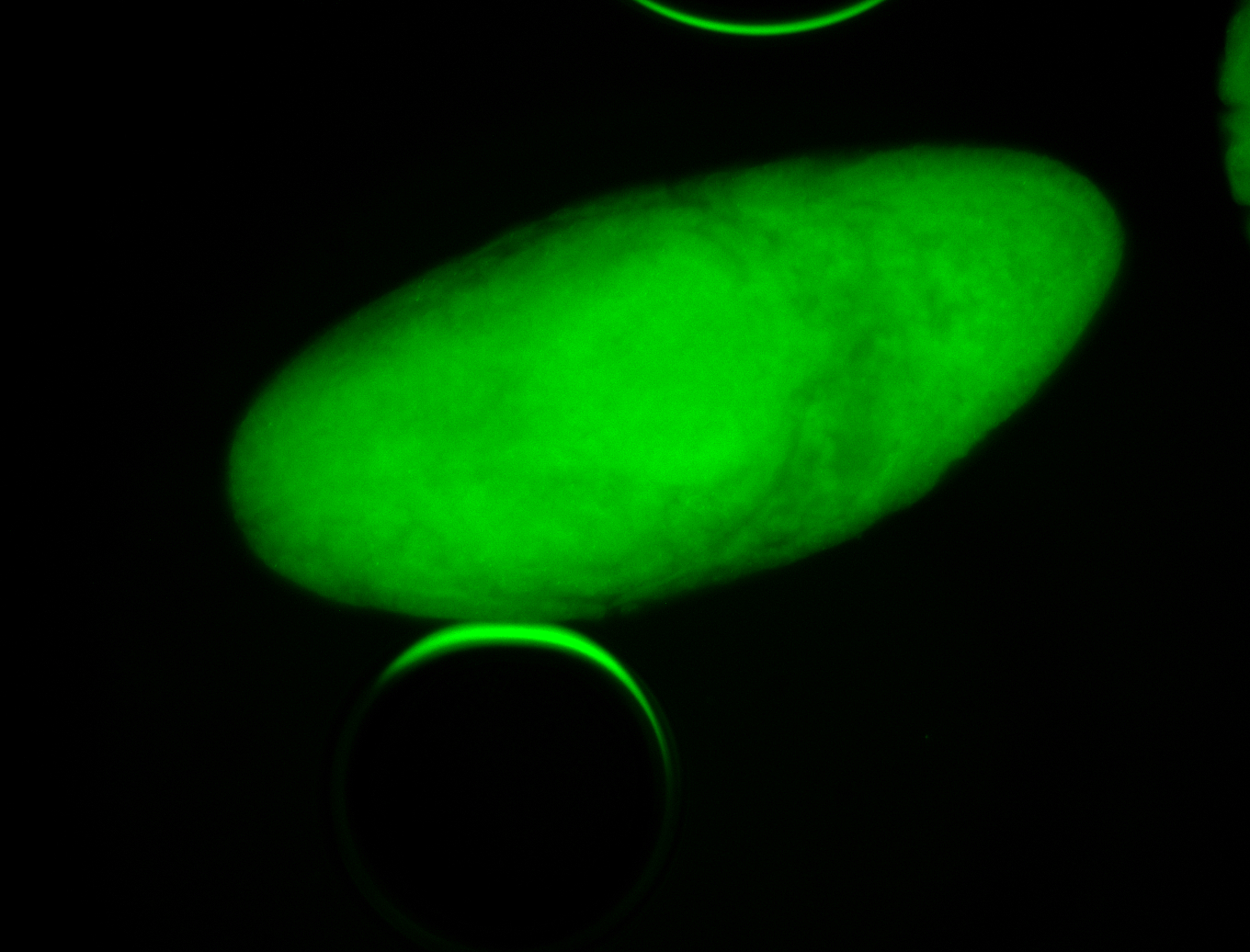
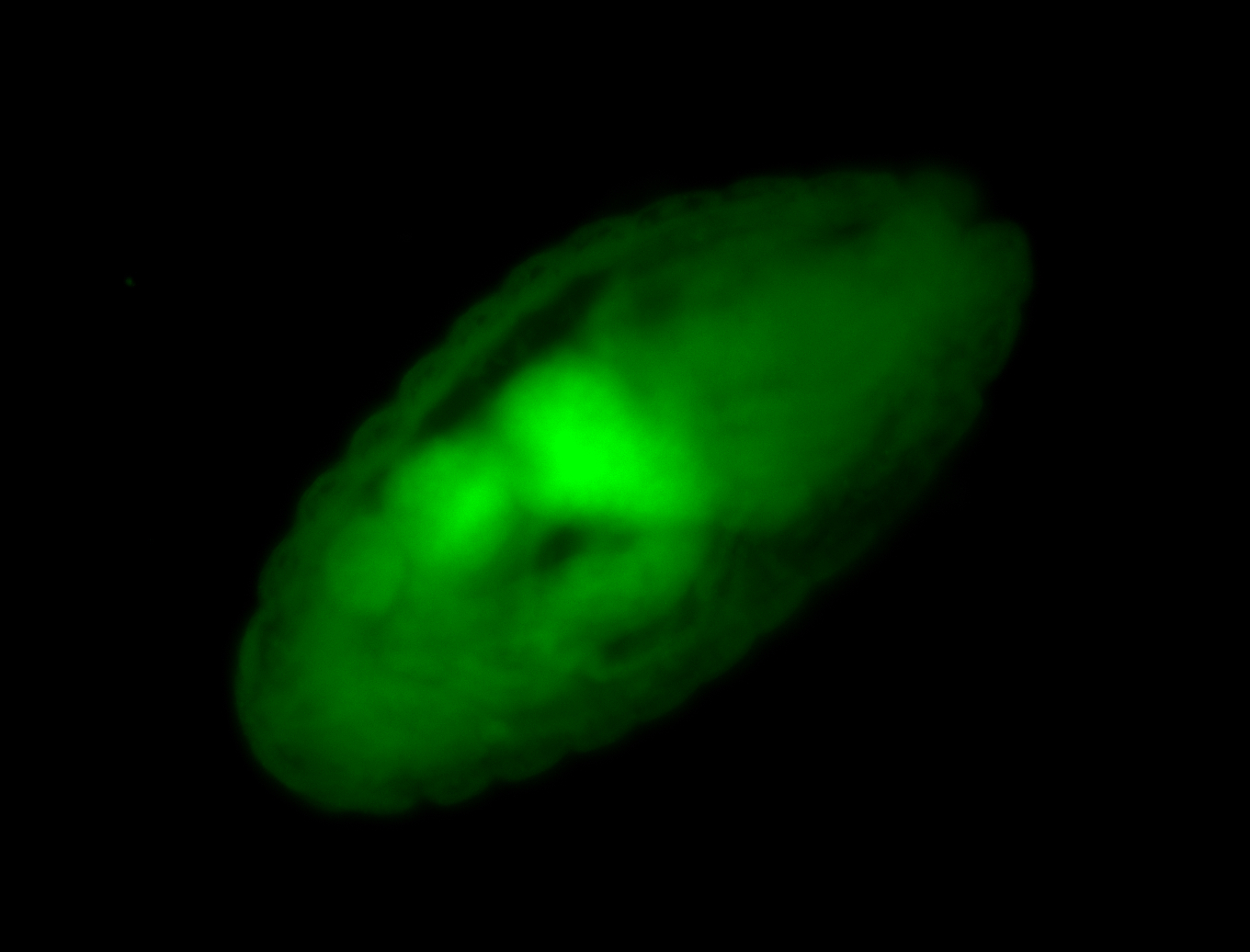
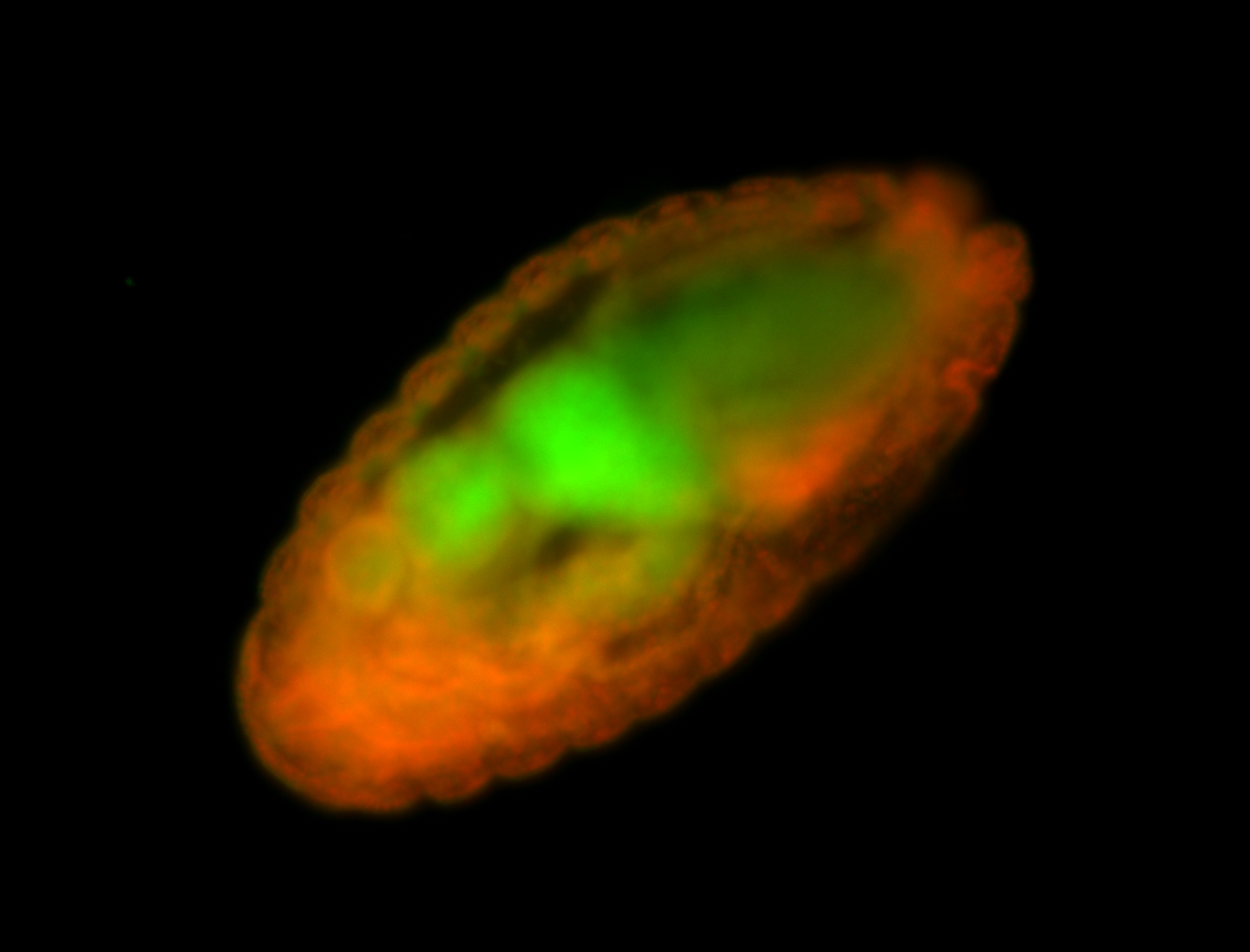
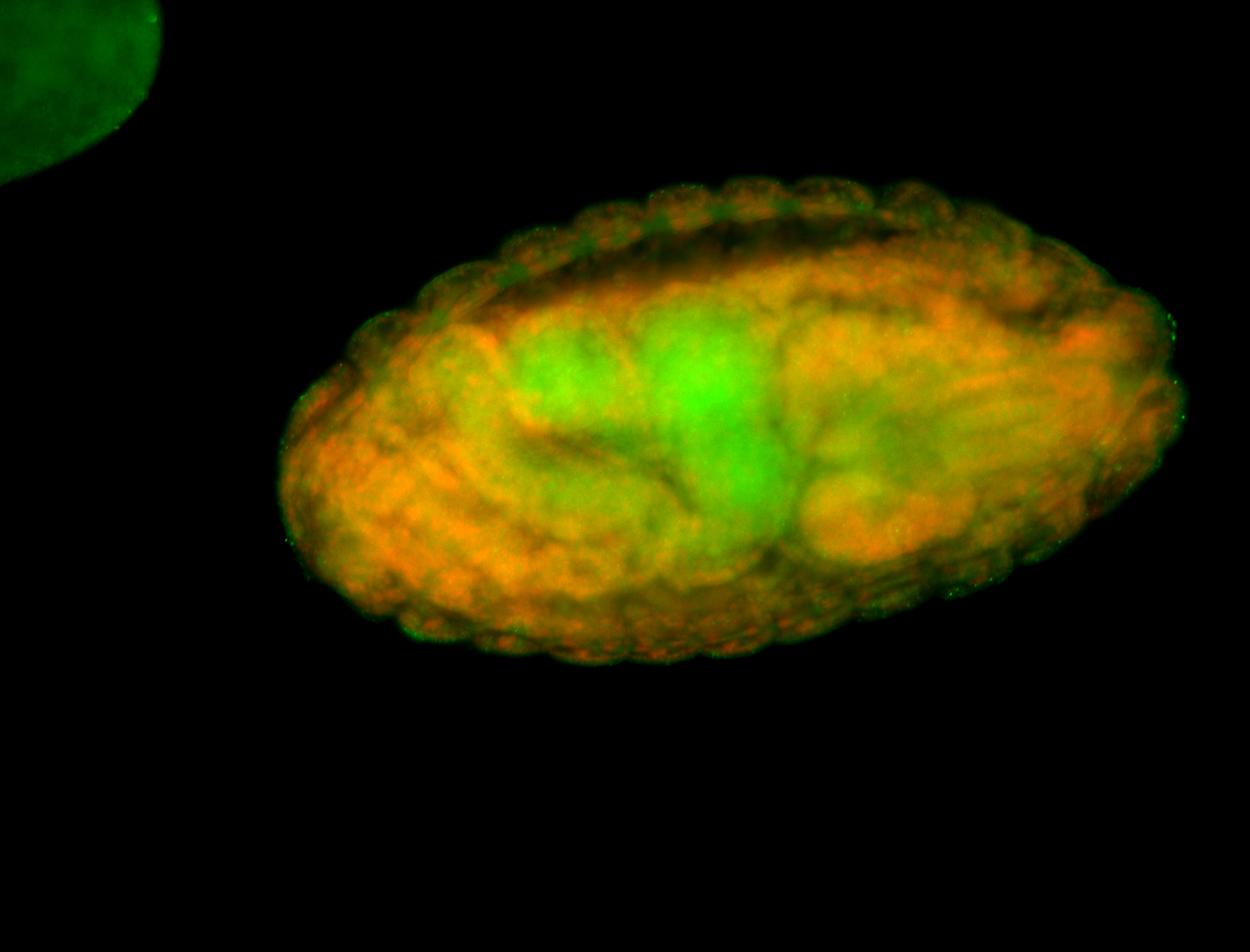
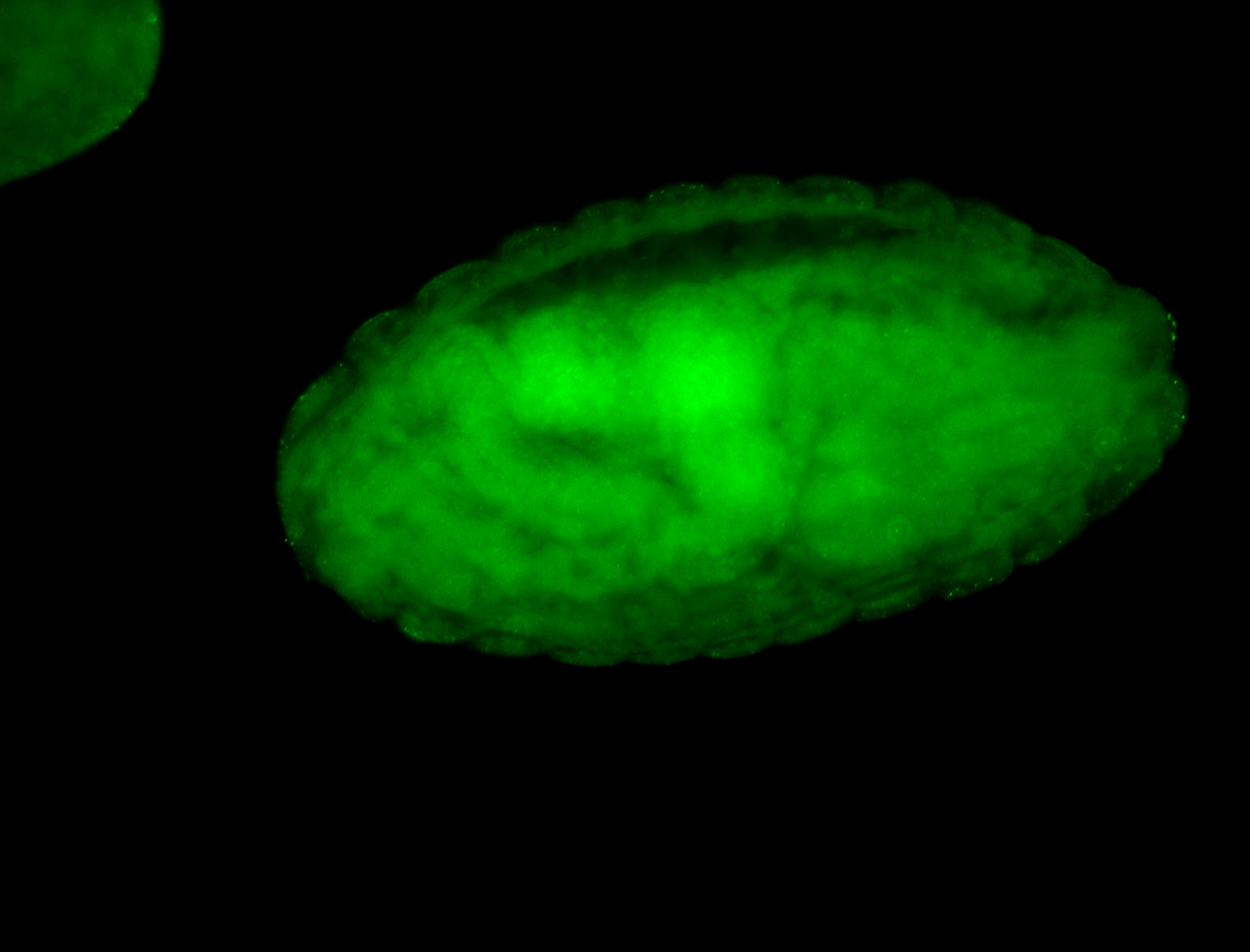
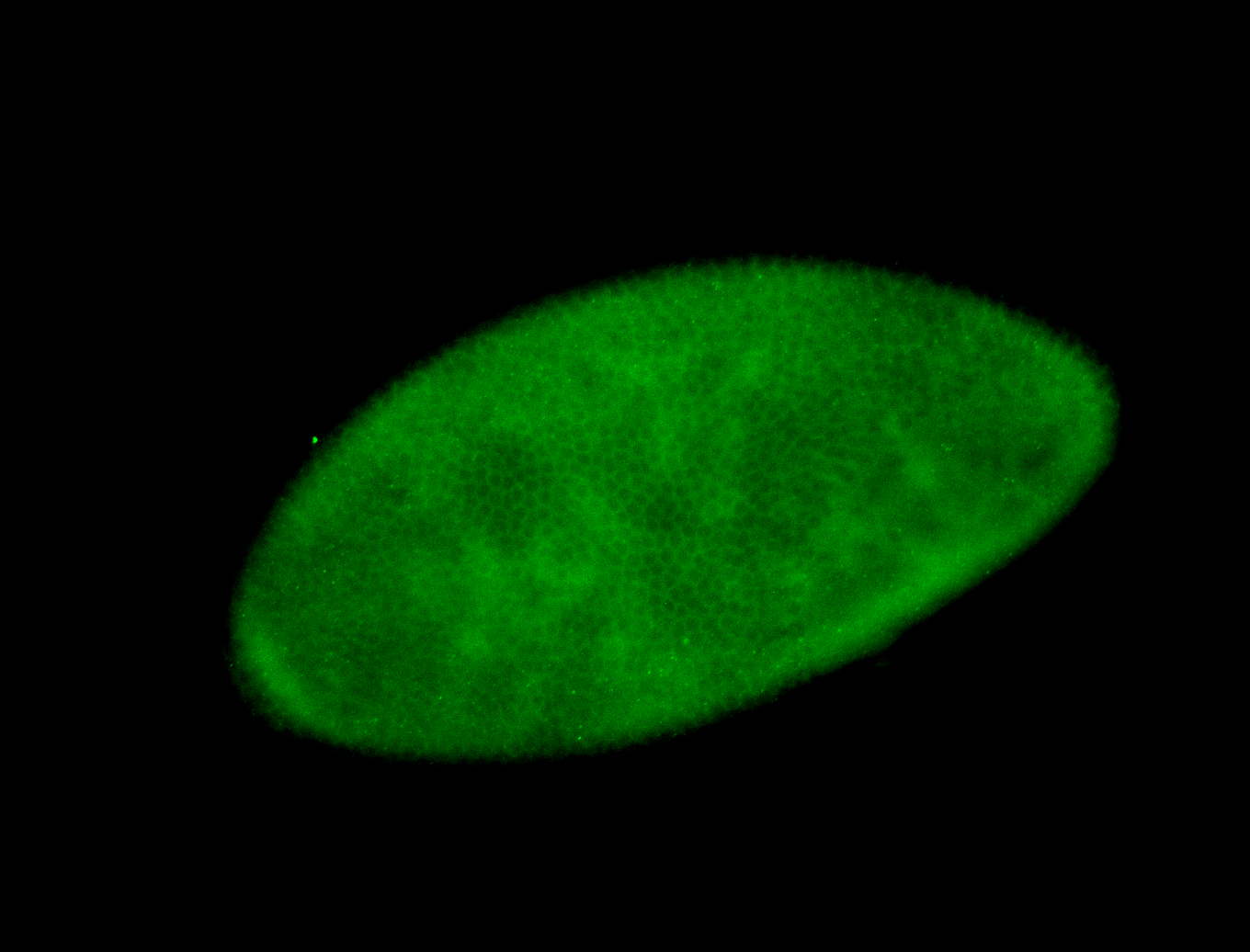
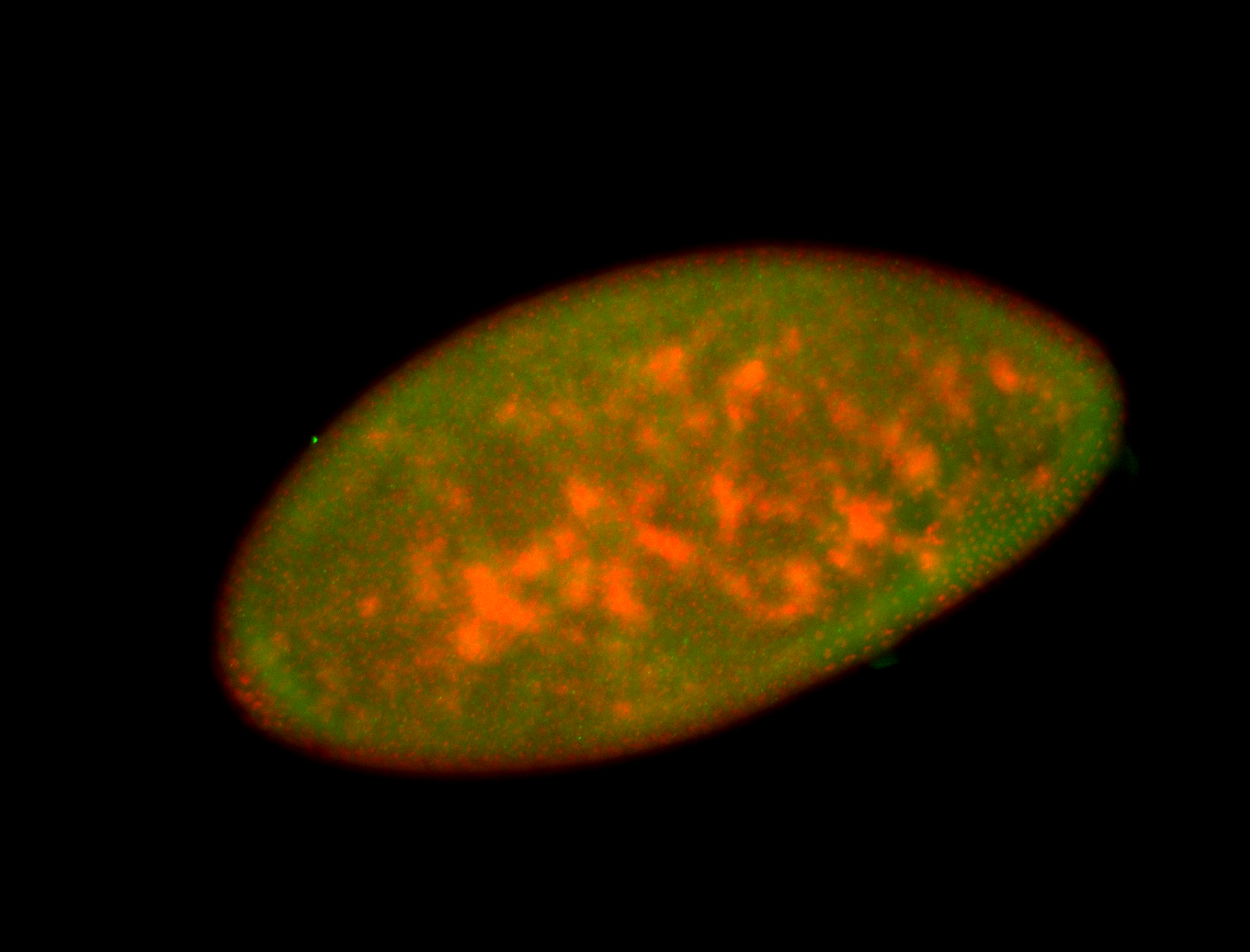
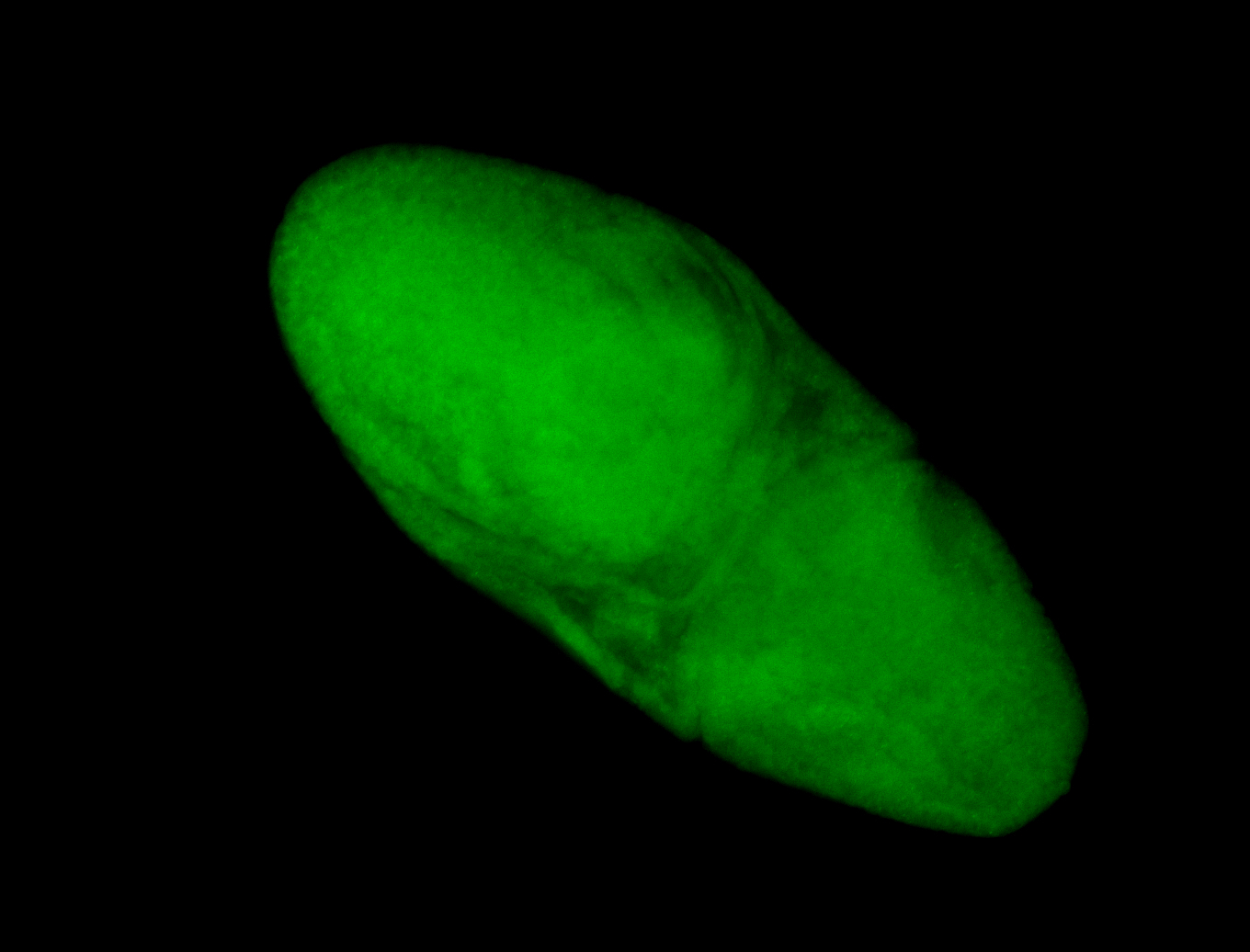
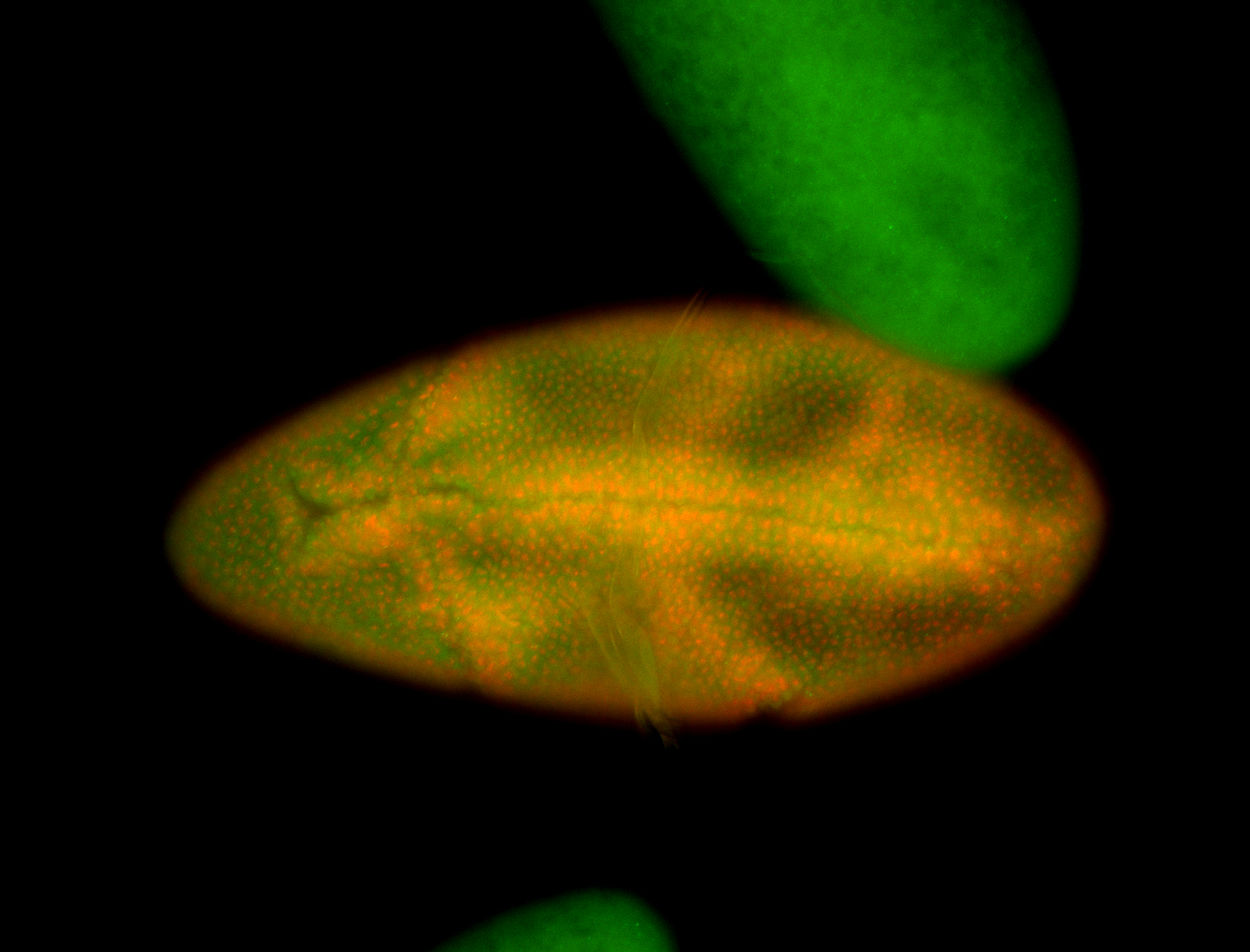
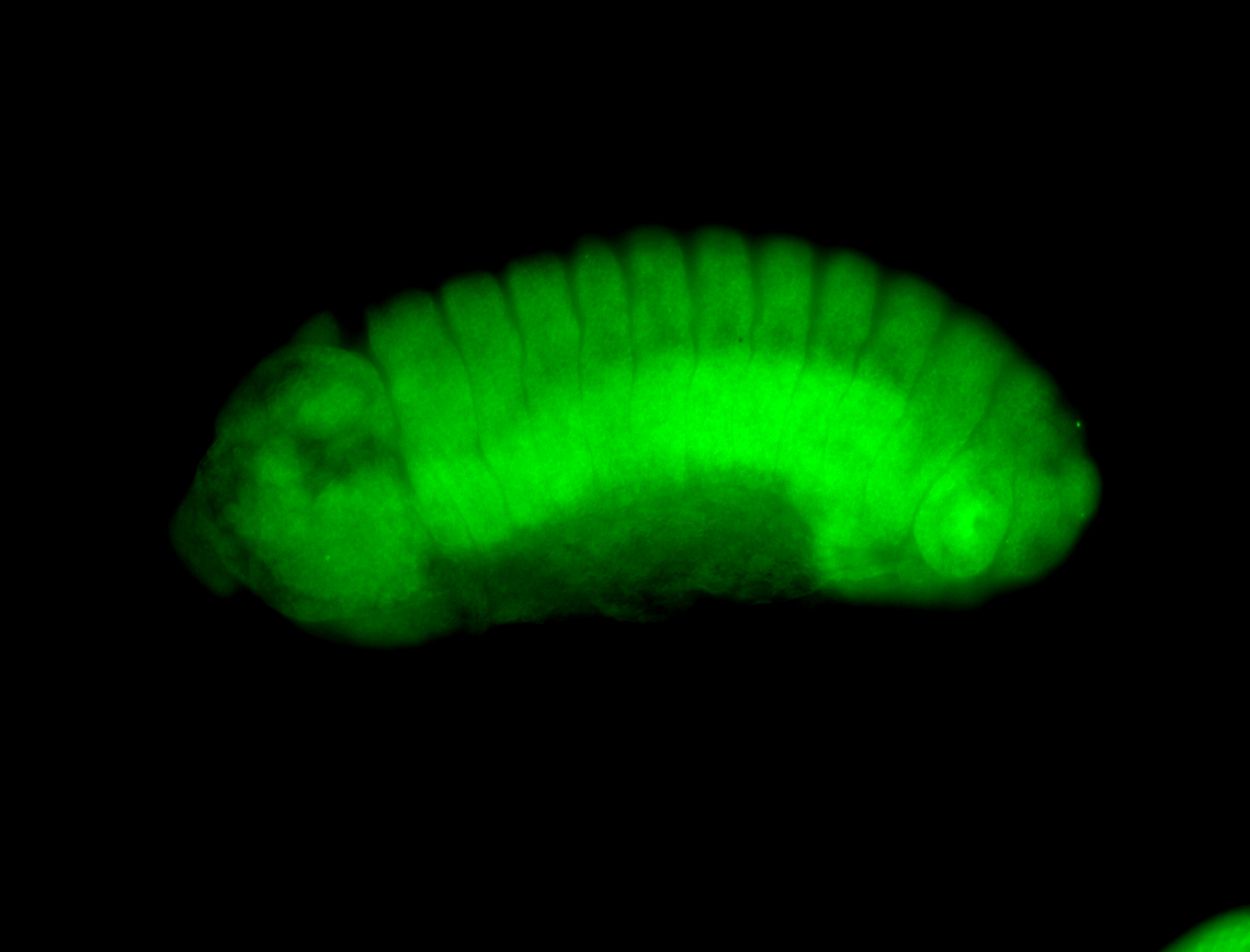
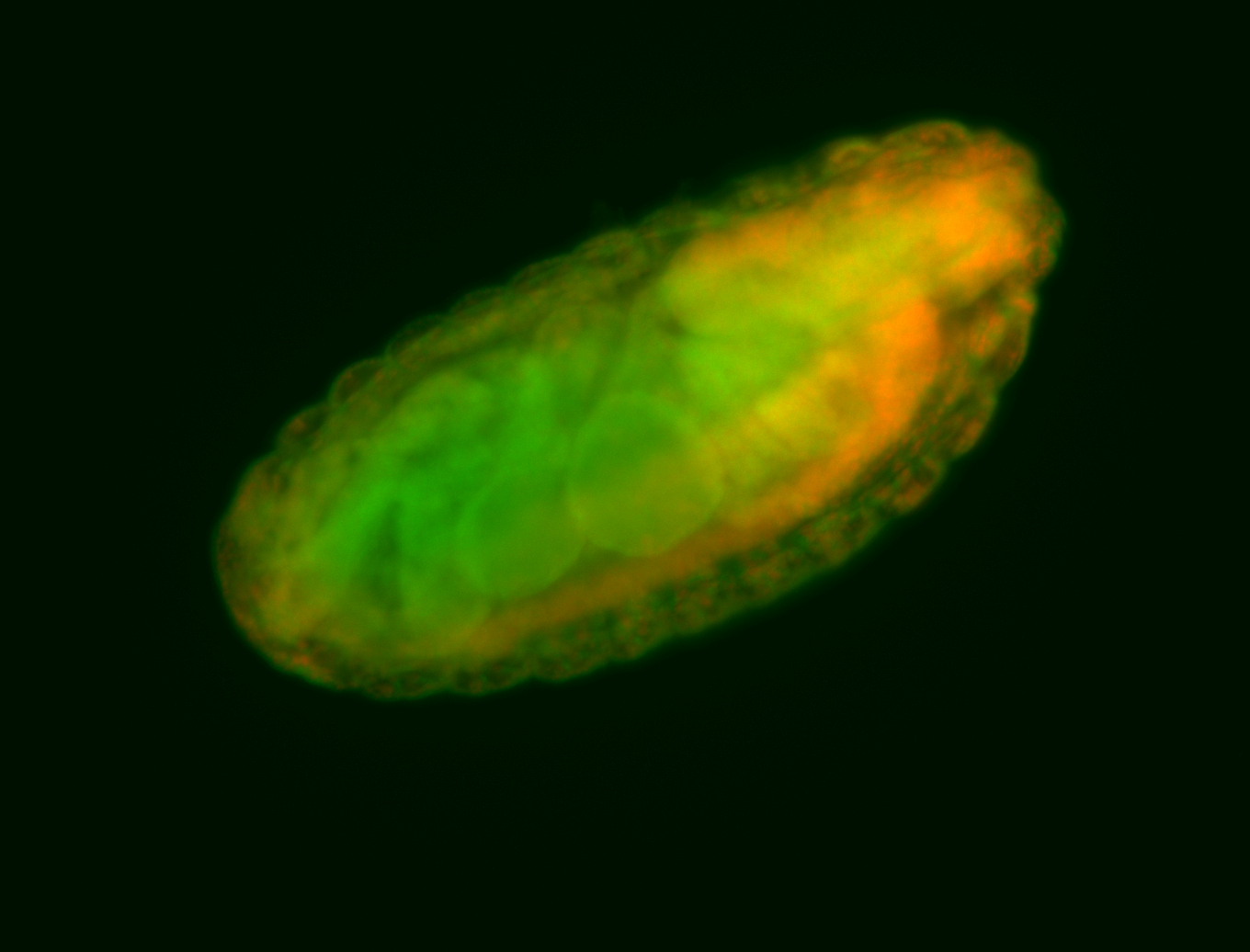
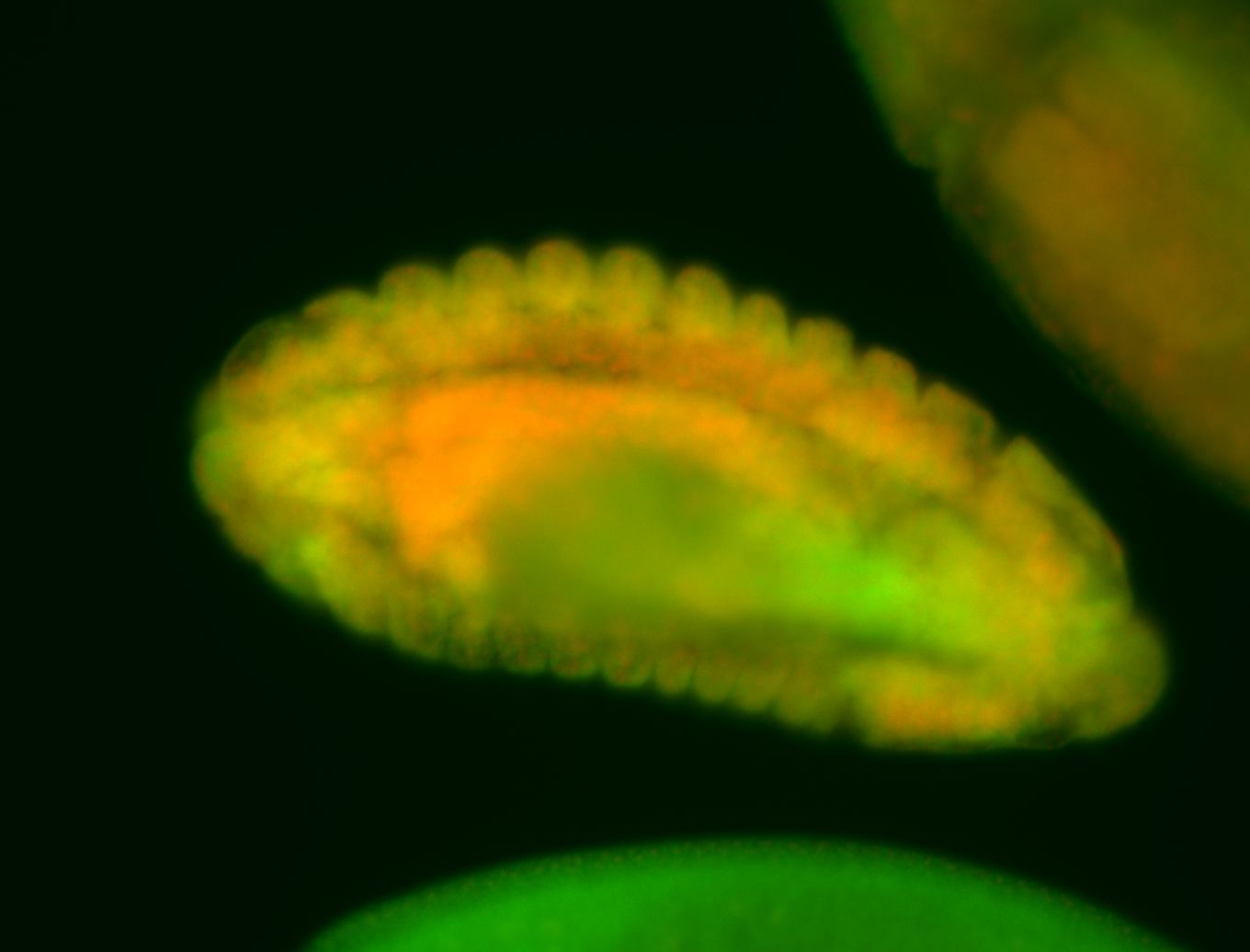
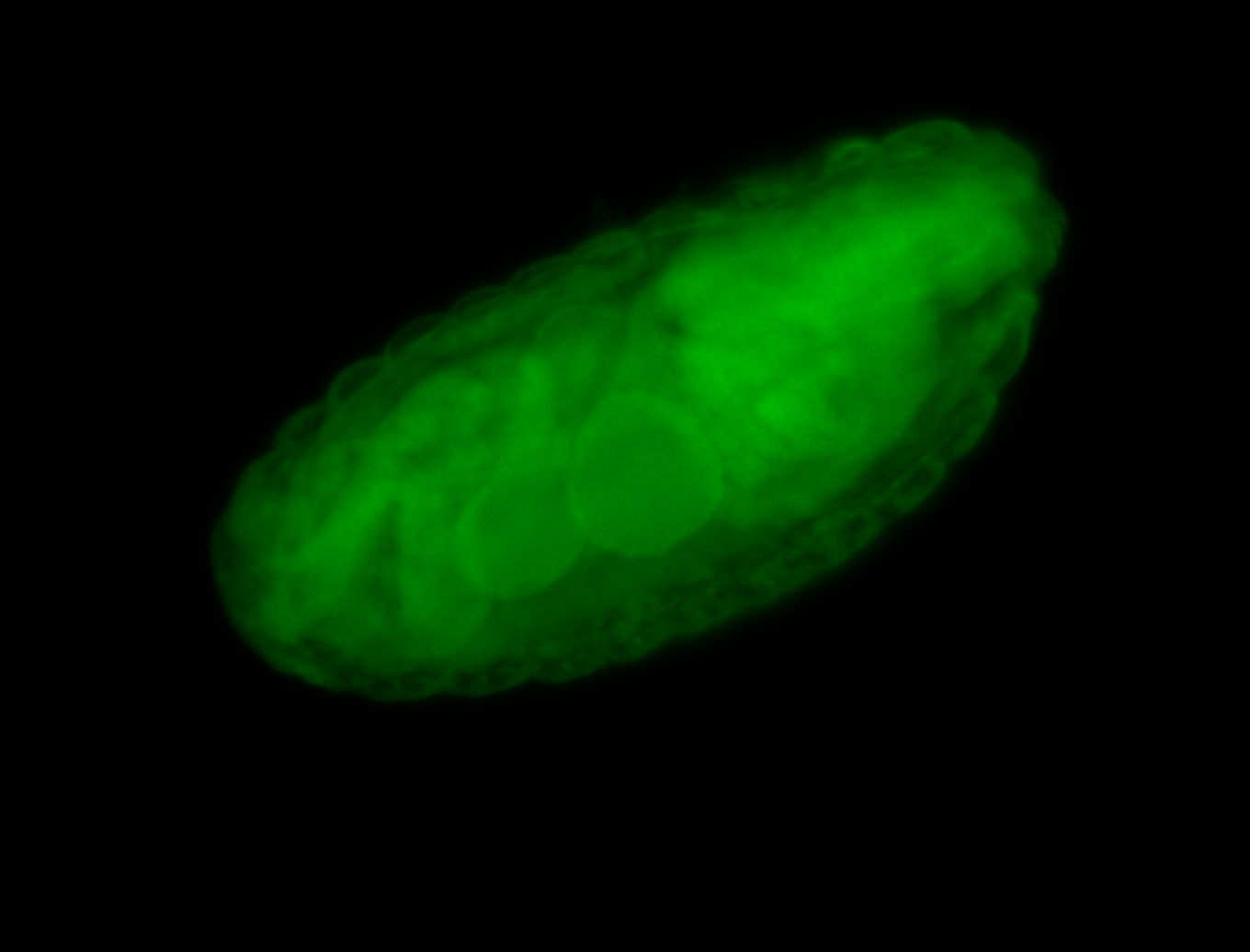
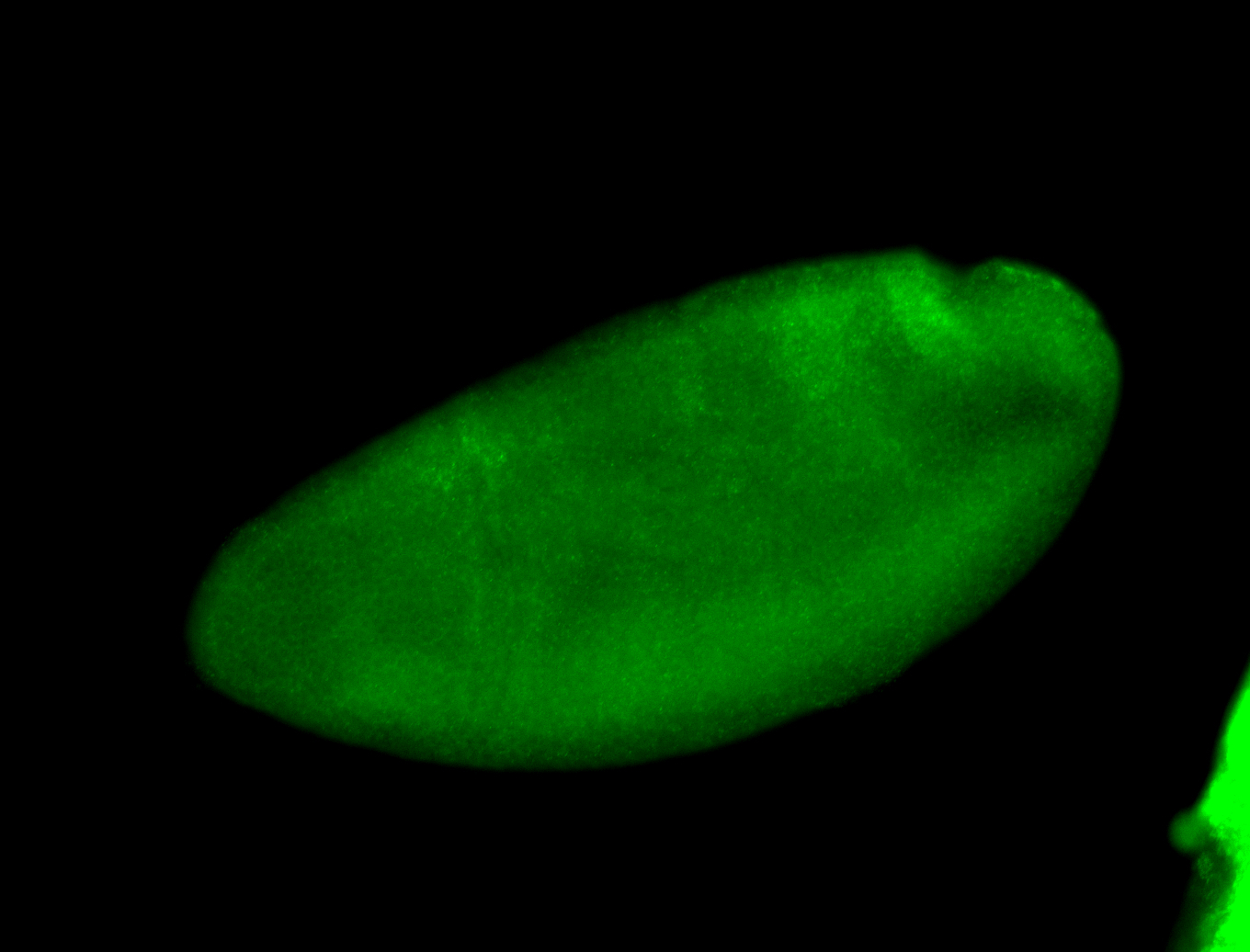
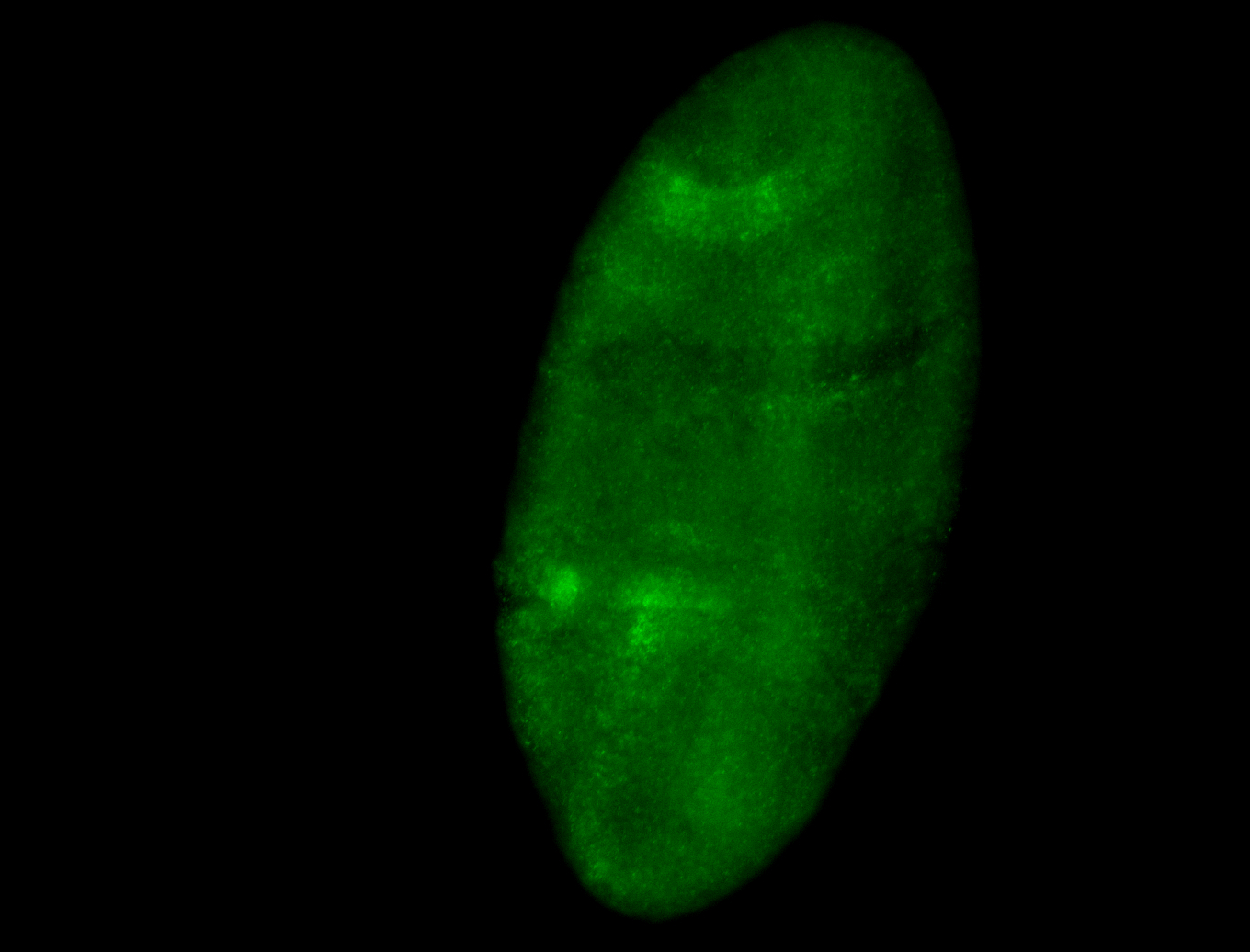
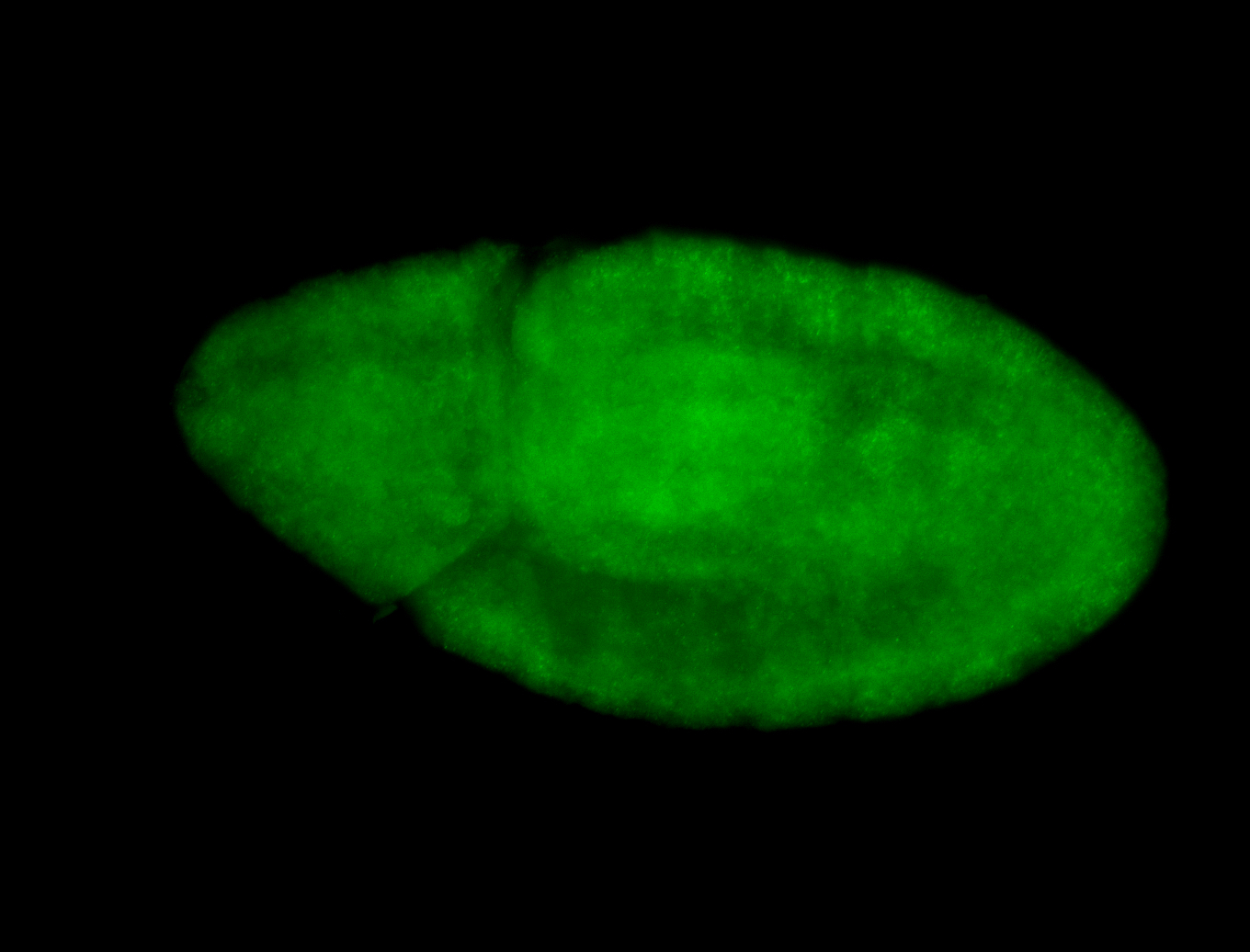
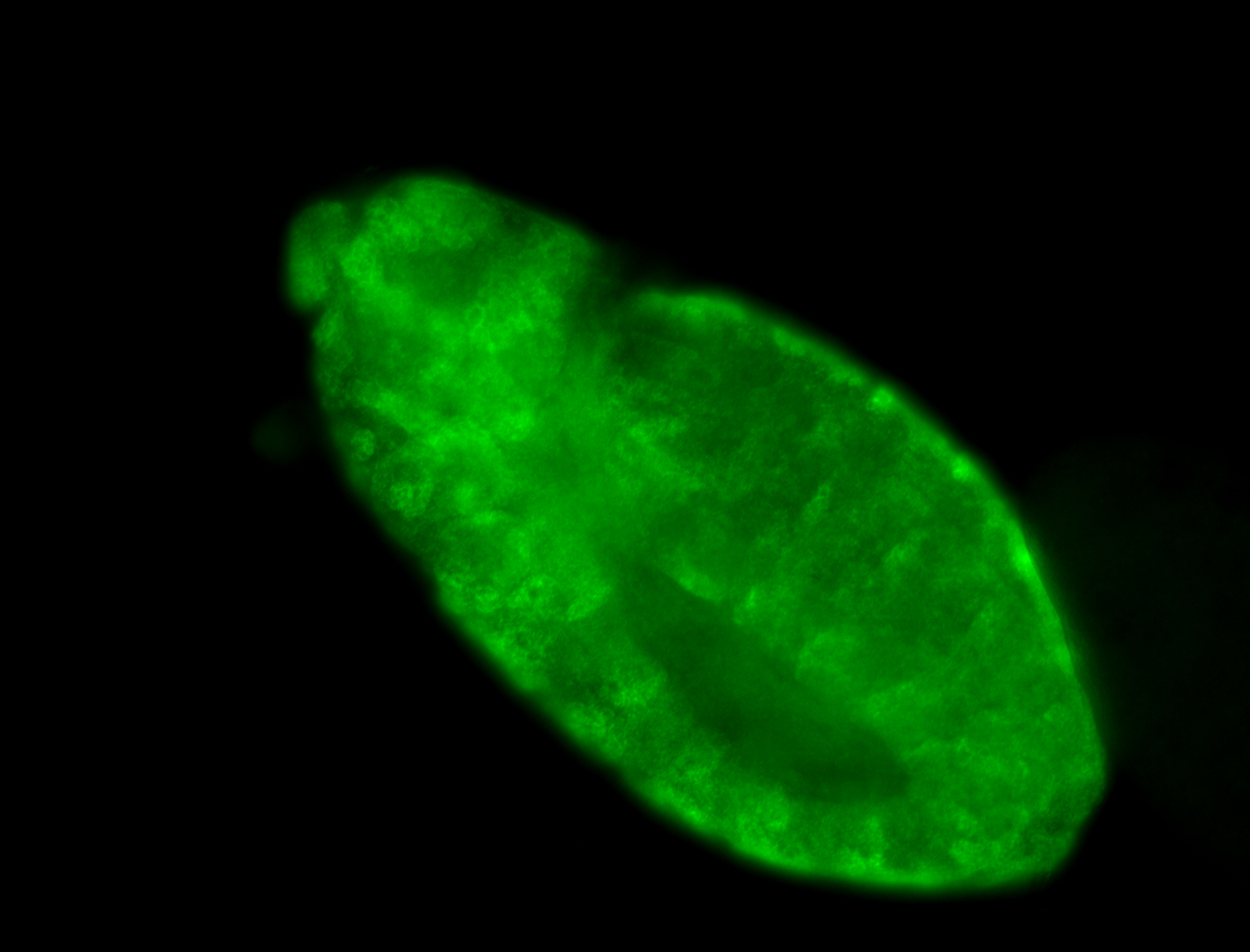
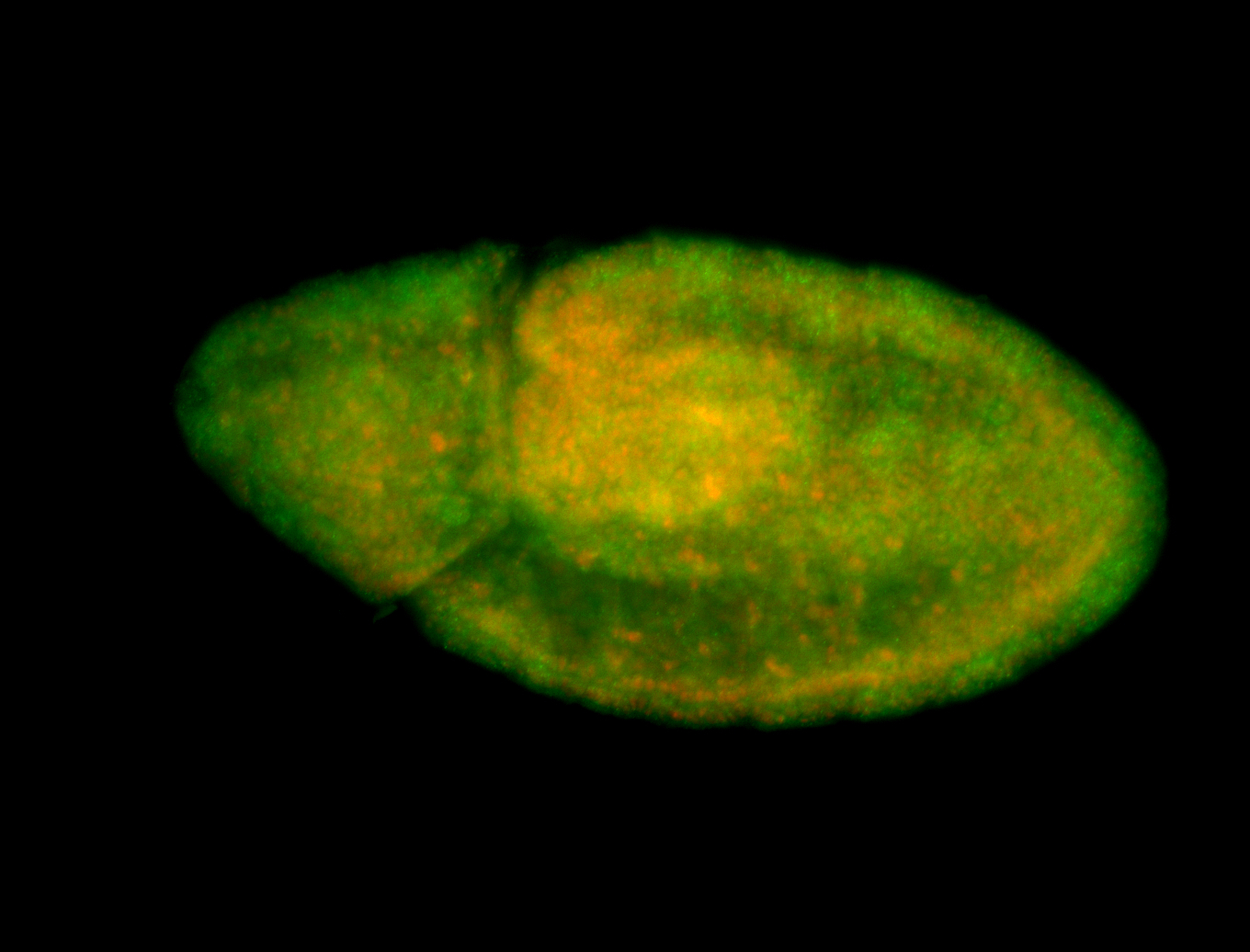
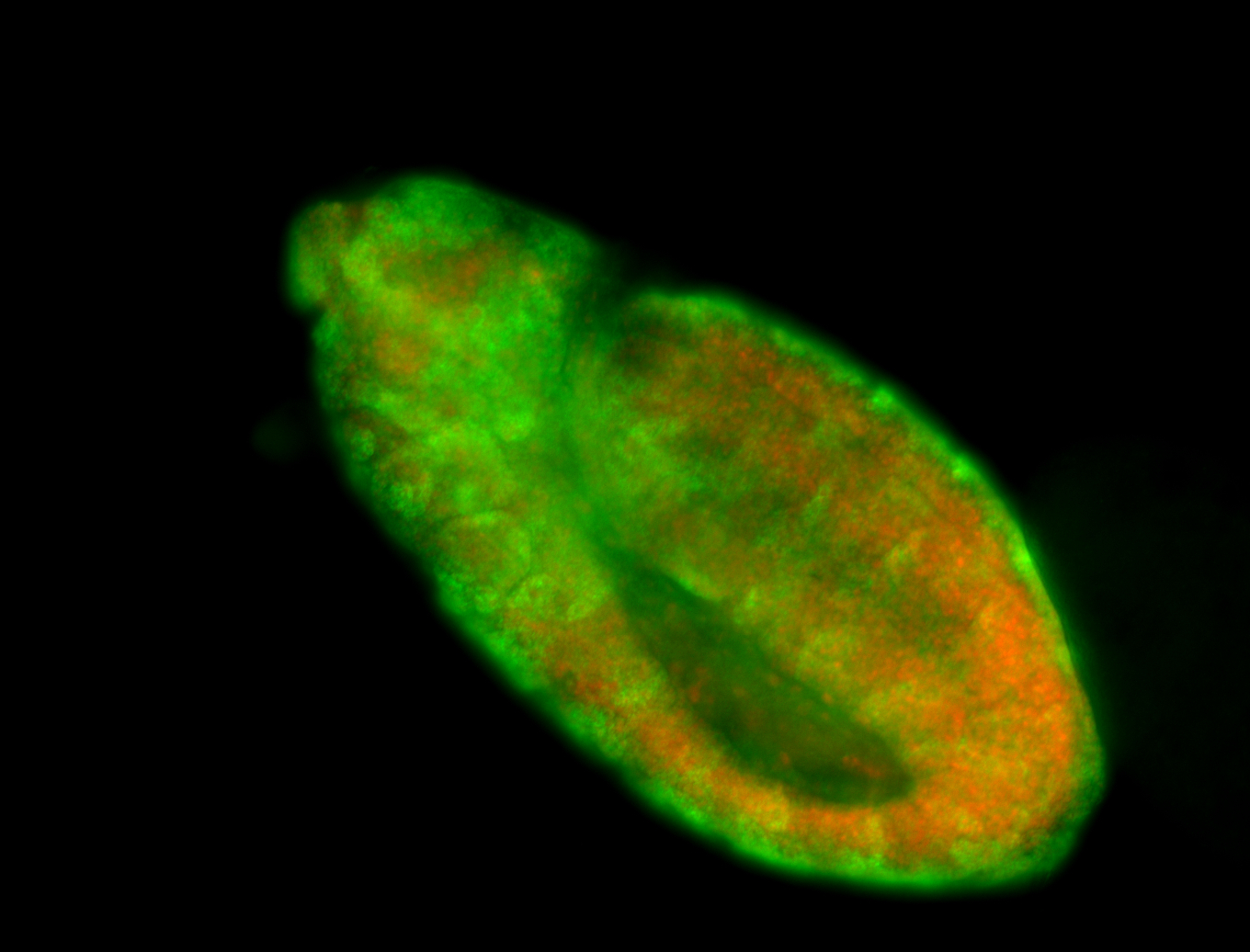
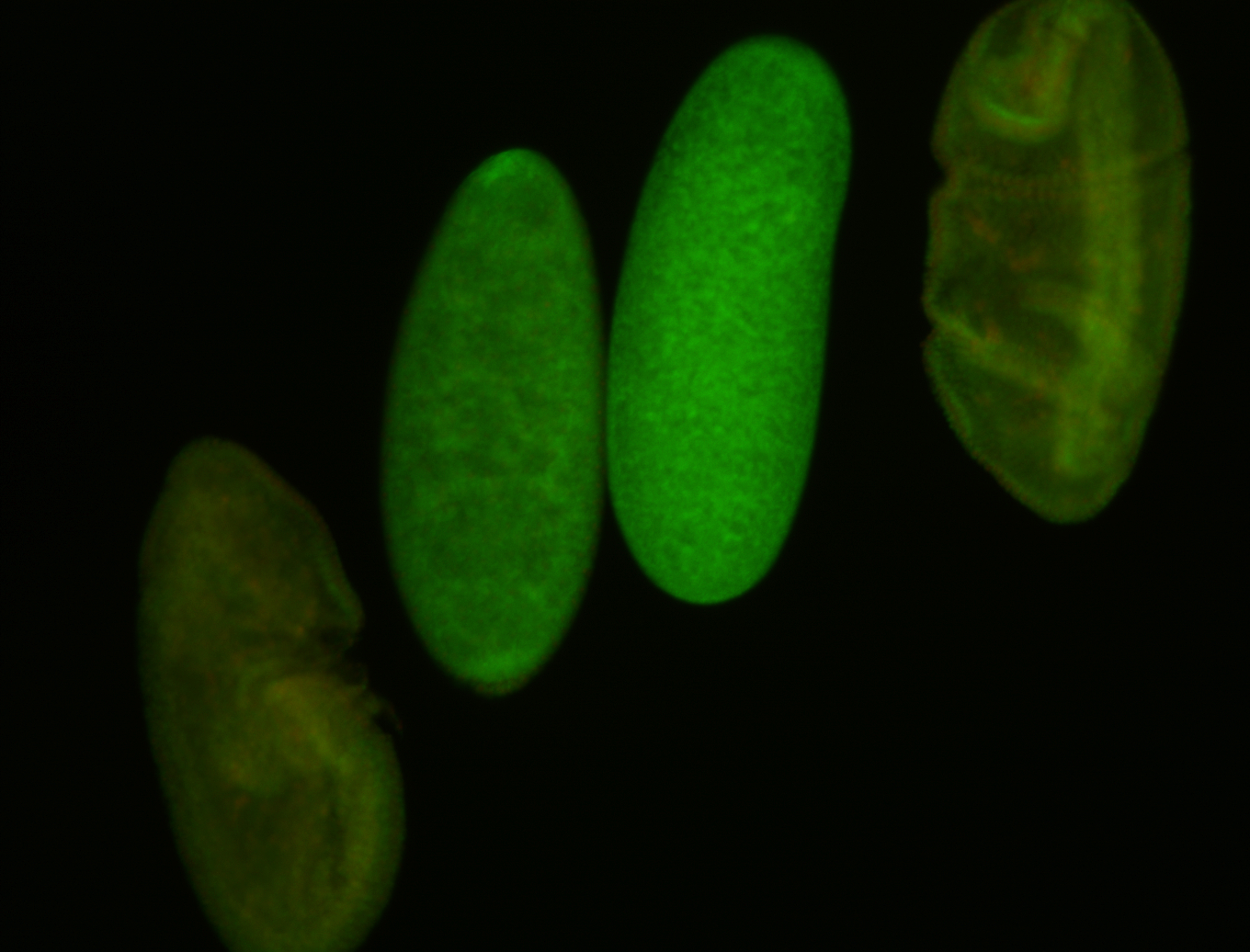
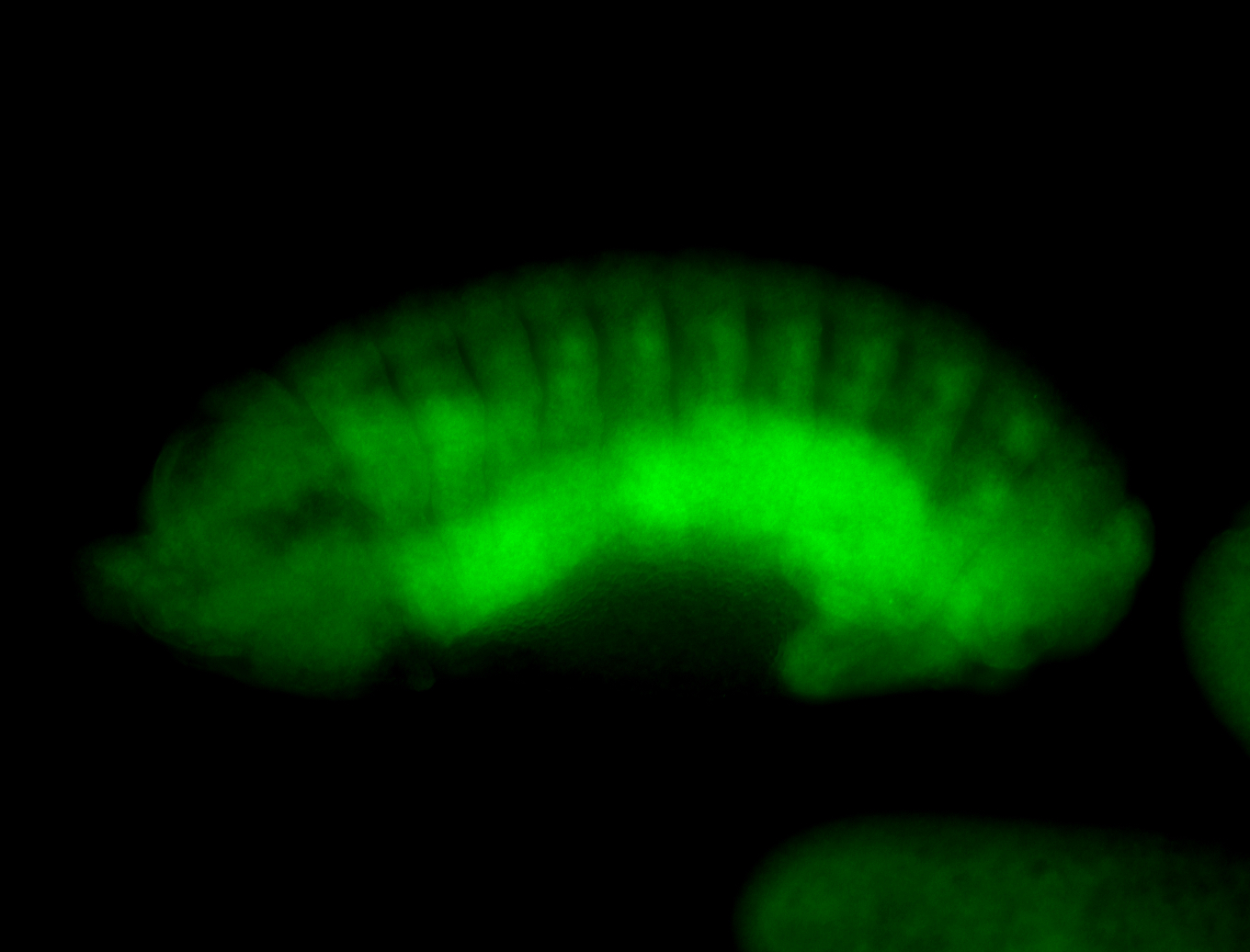
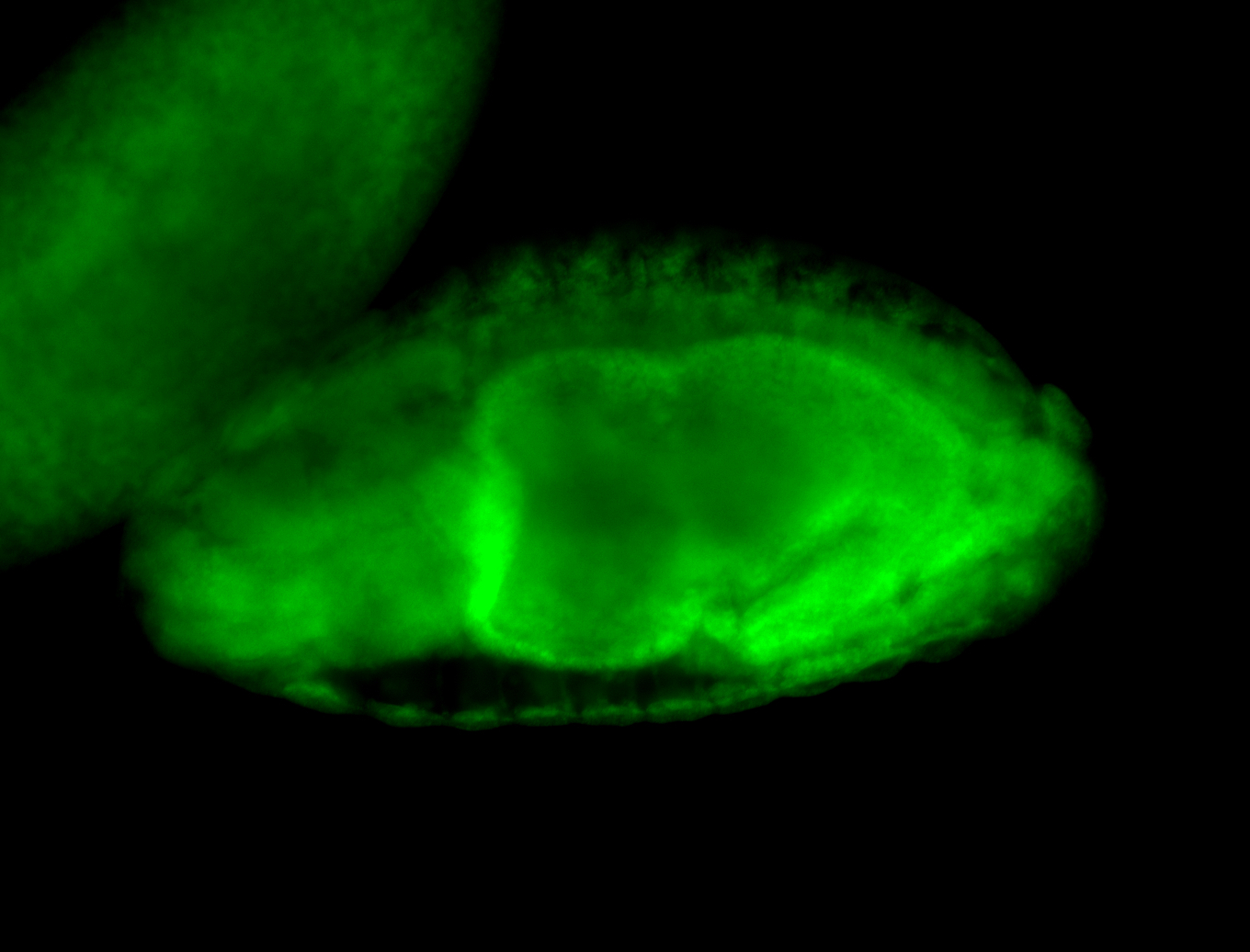
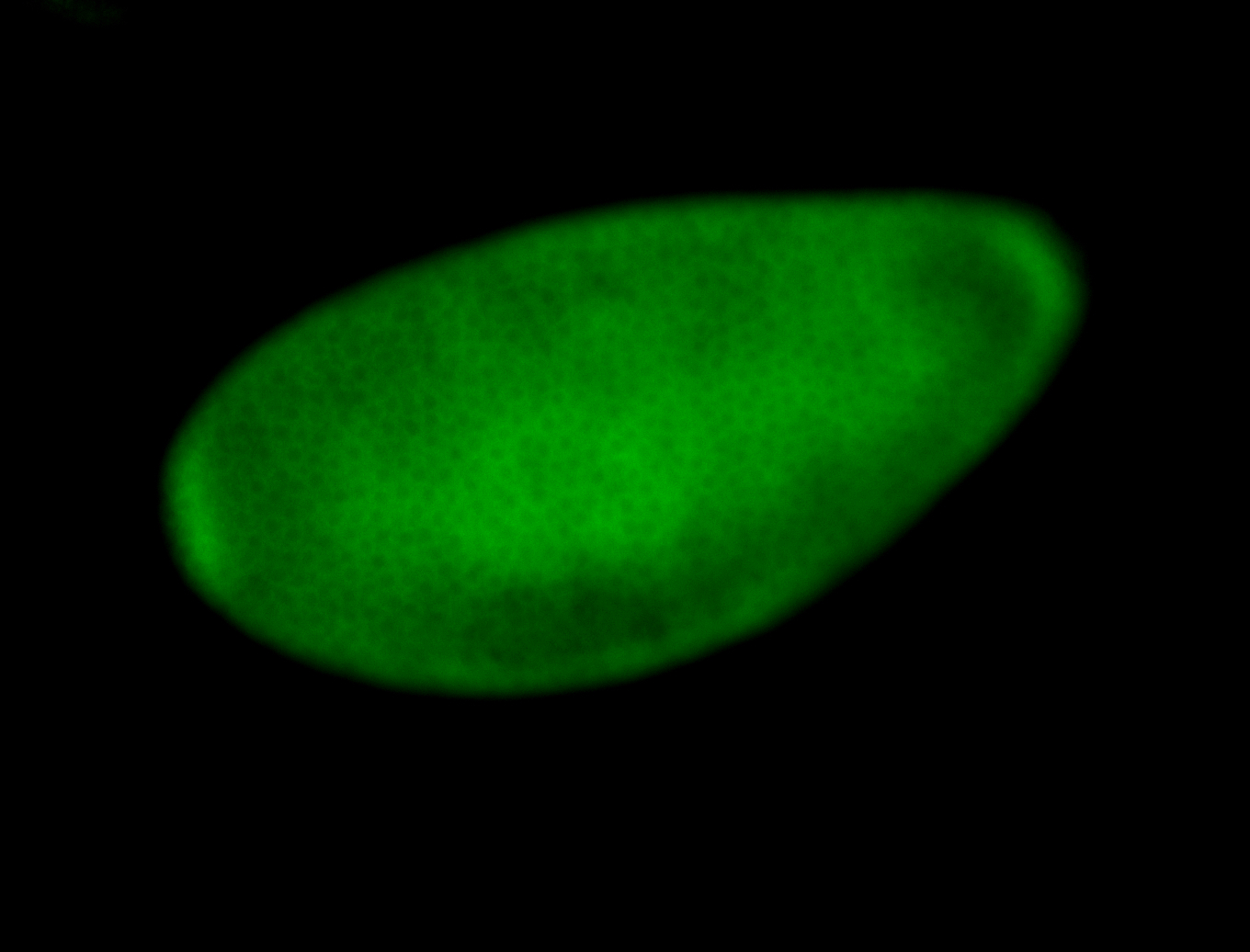
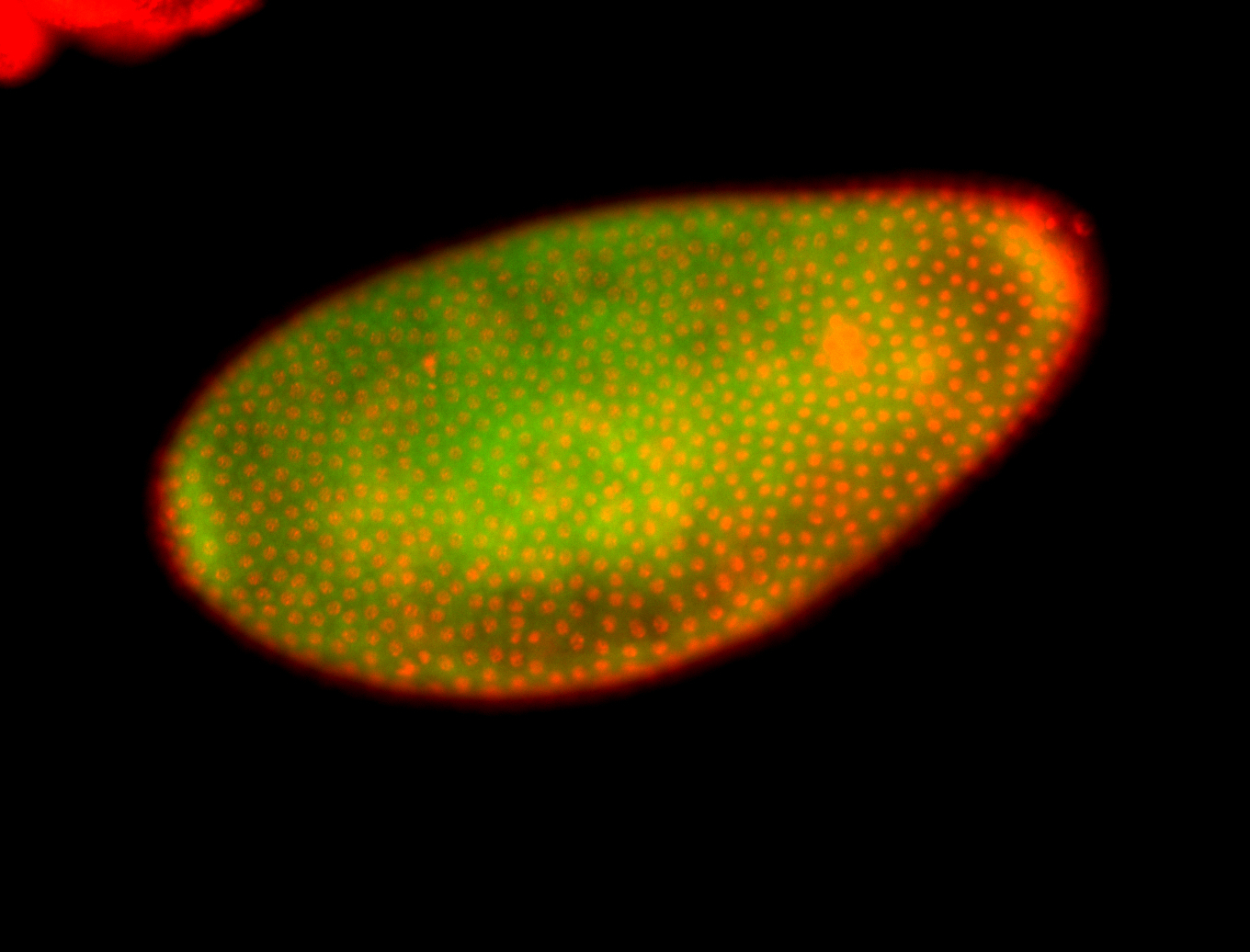
Embryo
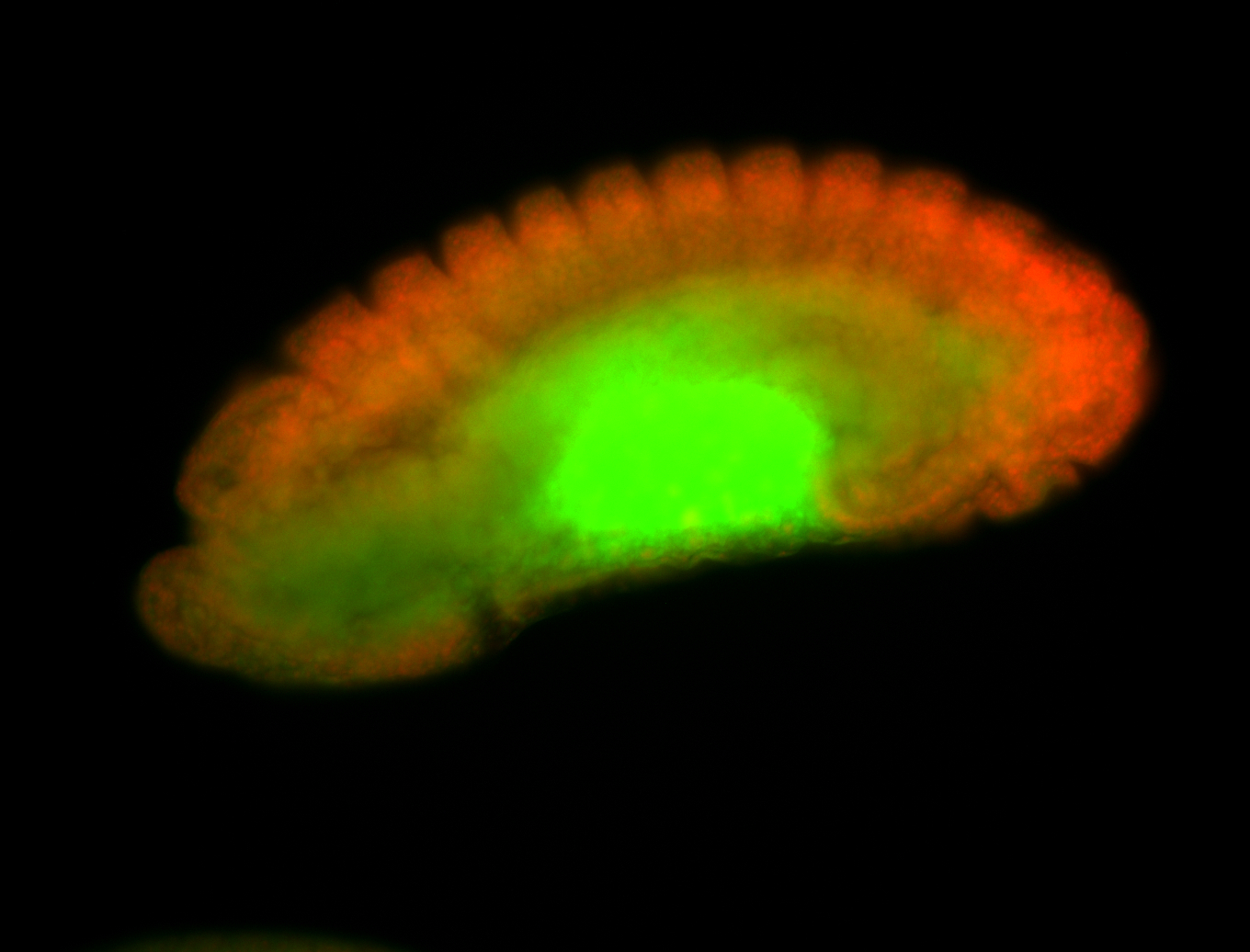
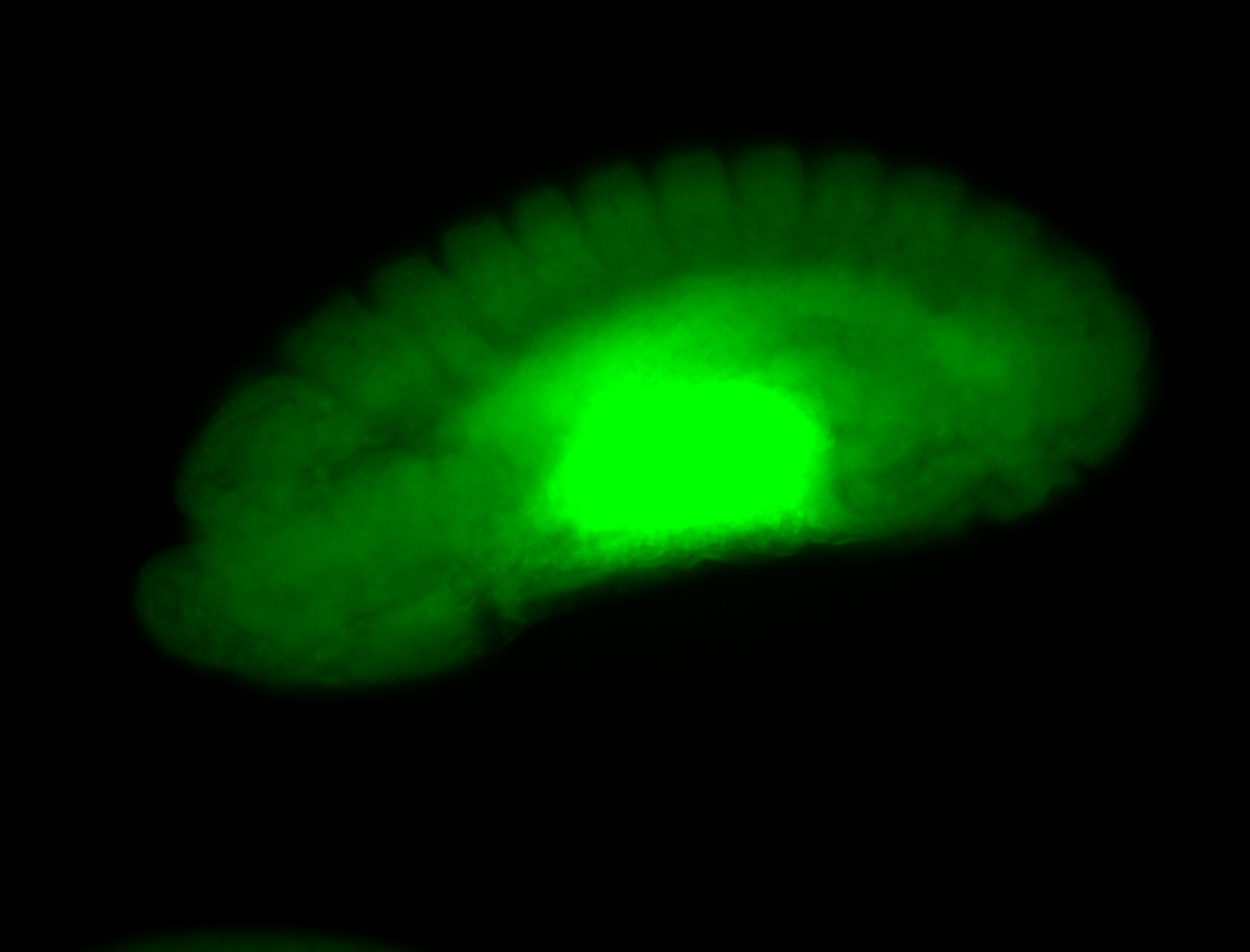
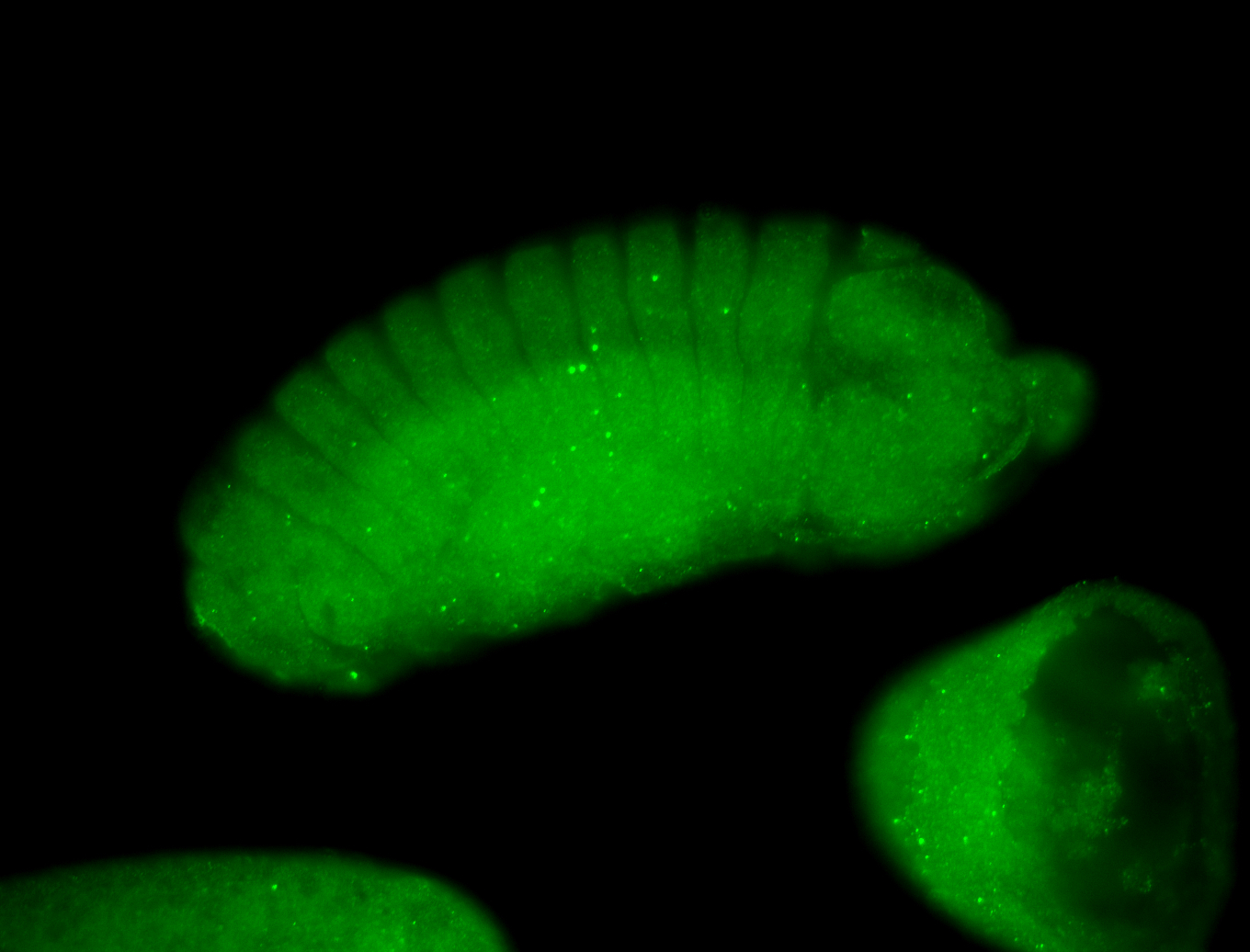
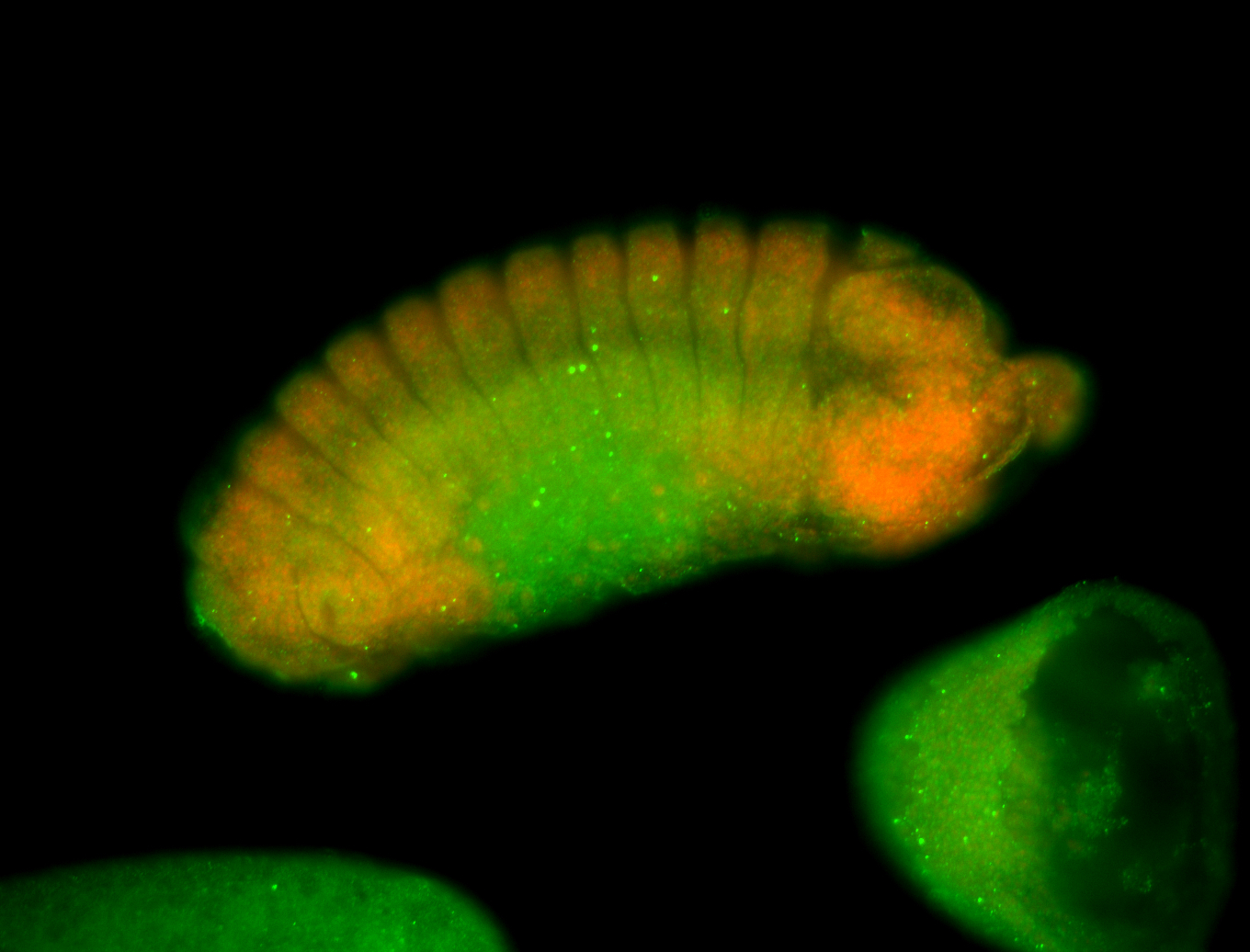
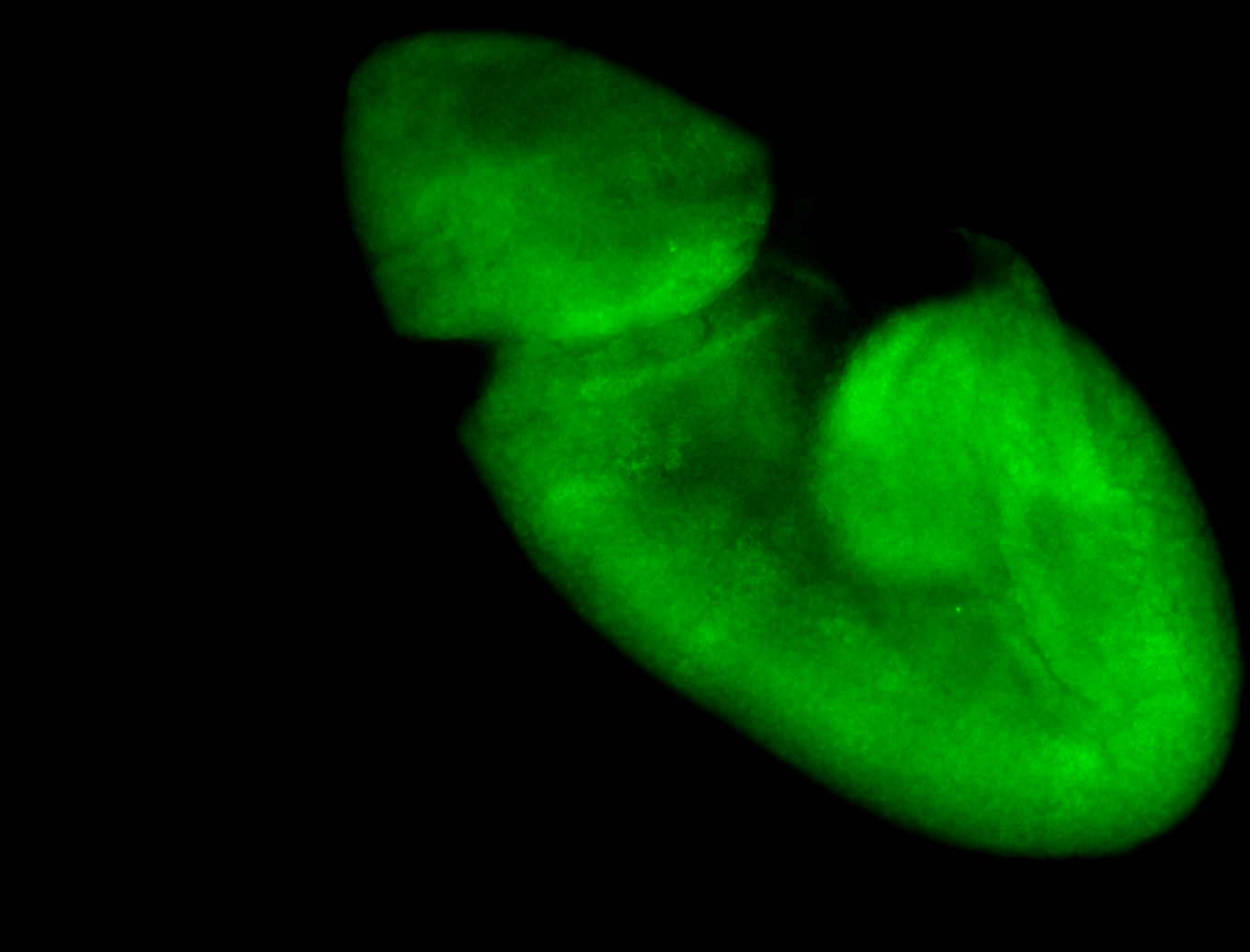
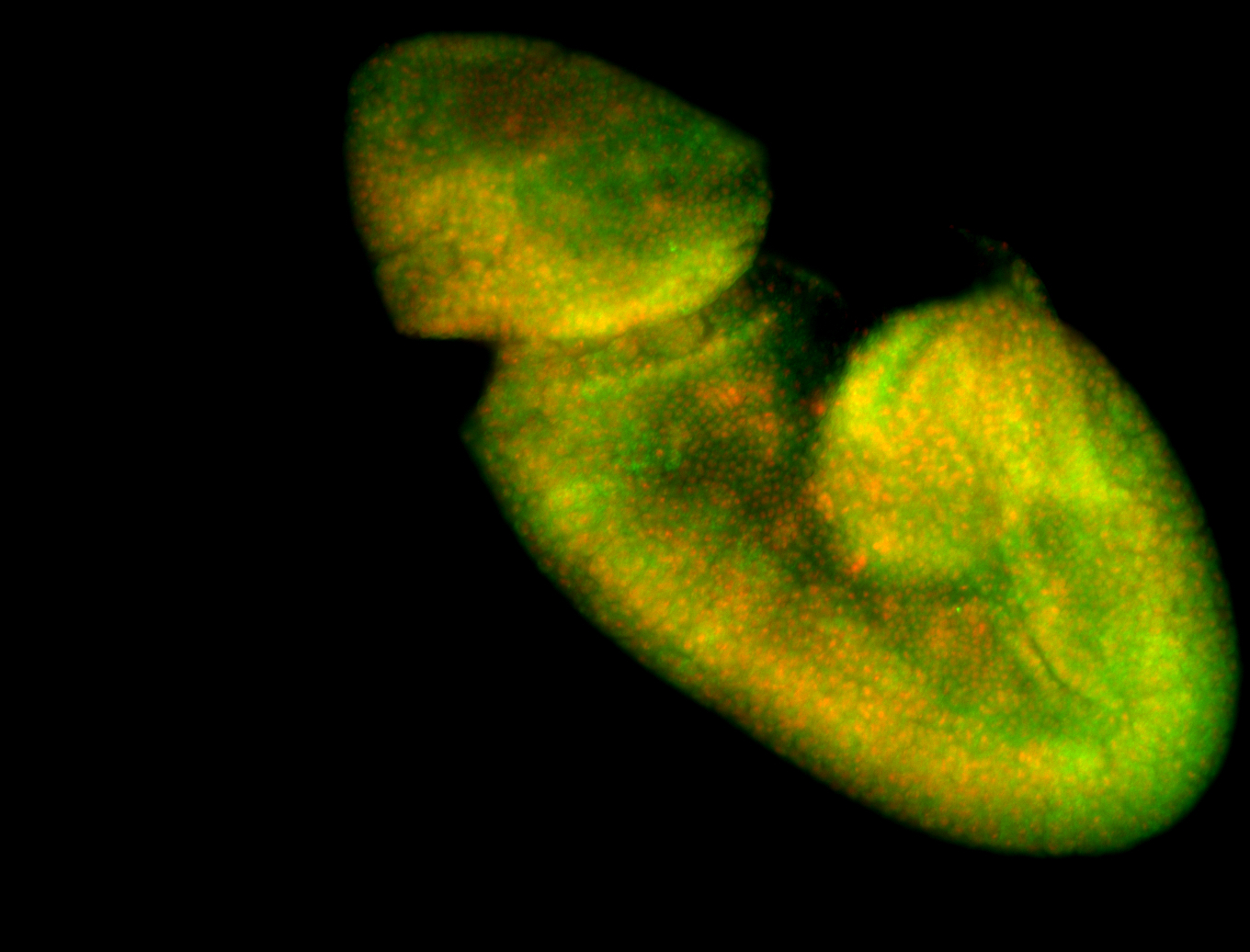
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -sample background seems to high and does not match current data - candidate for retest (2025) |
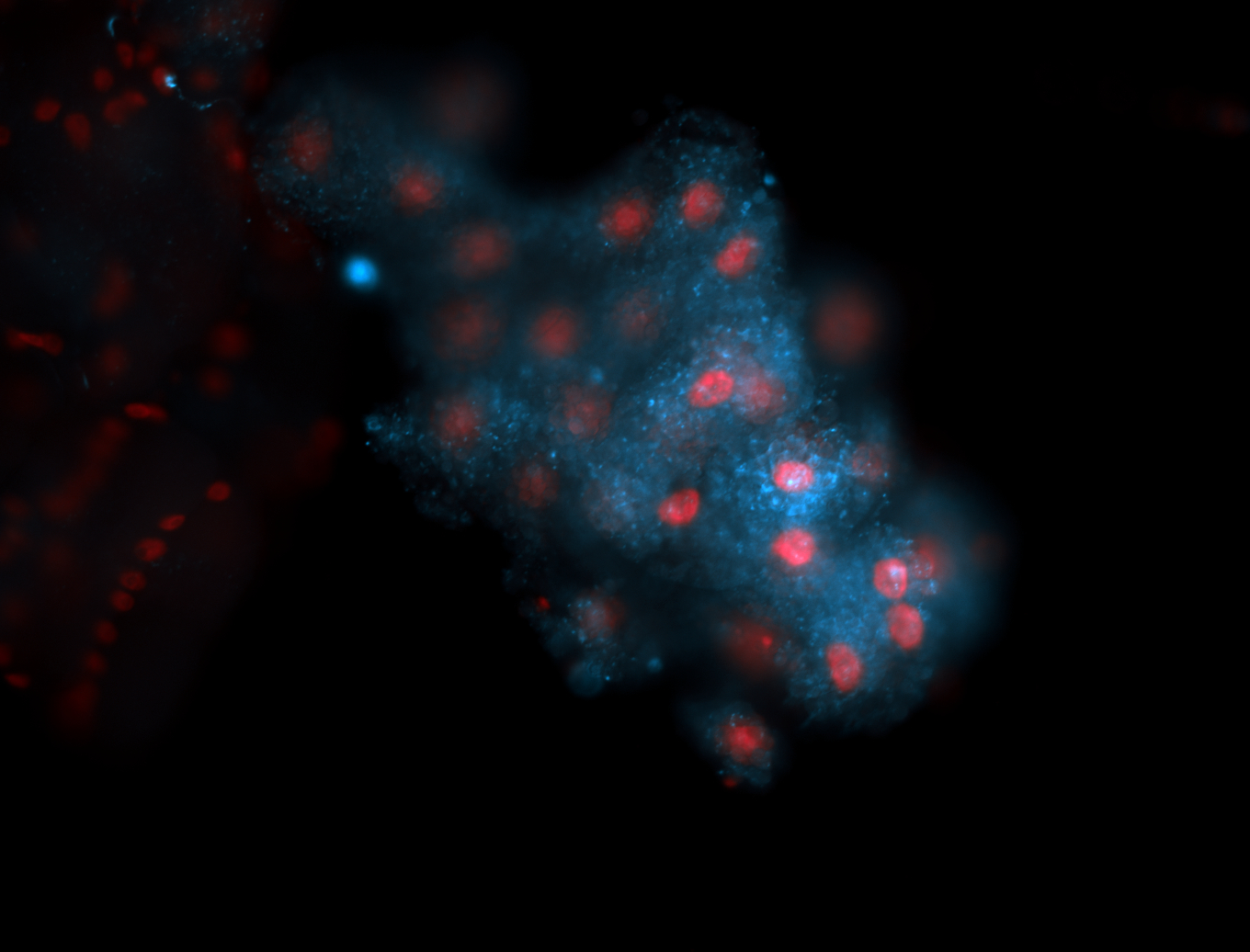
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
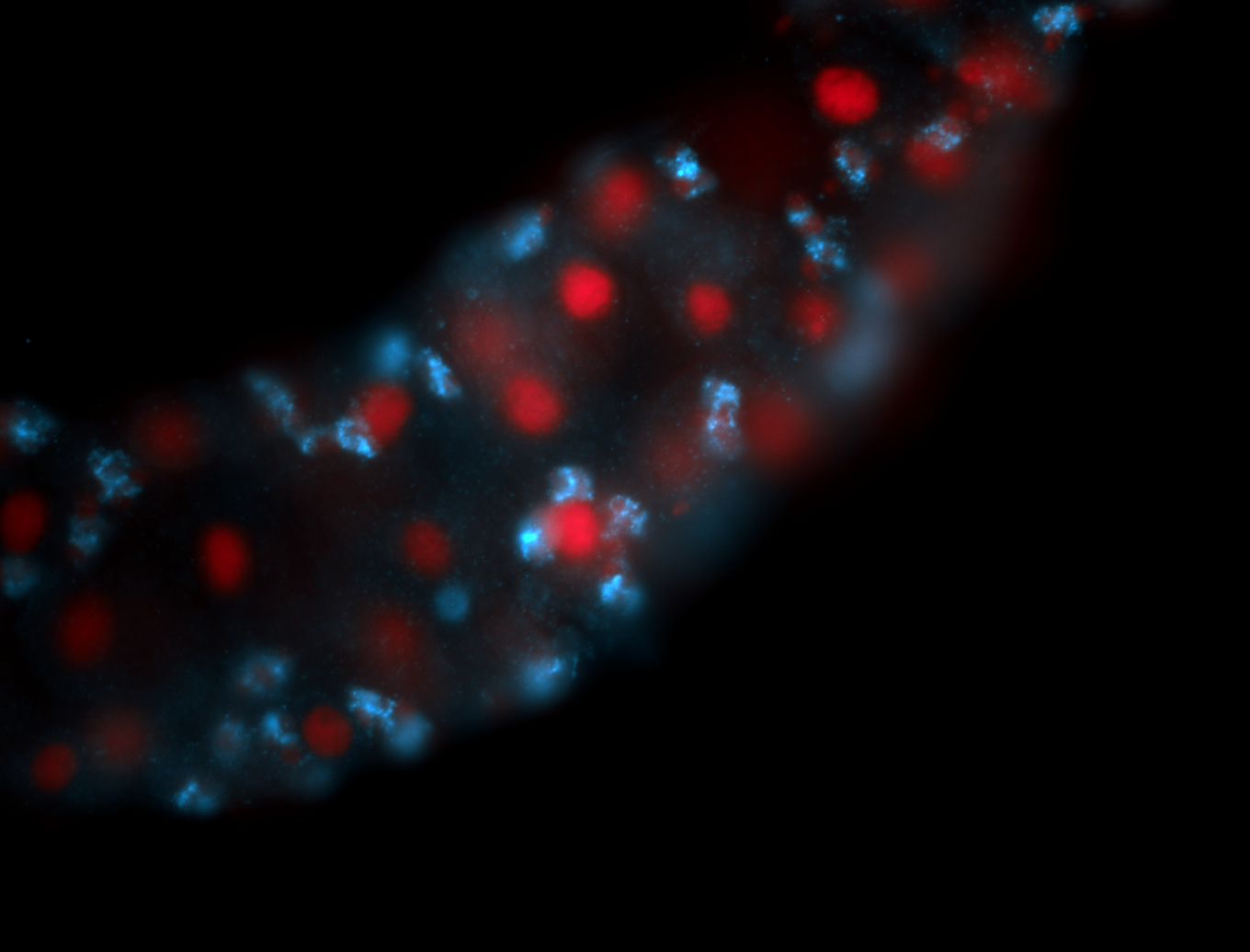
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
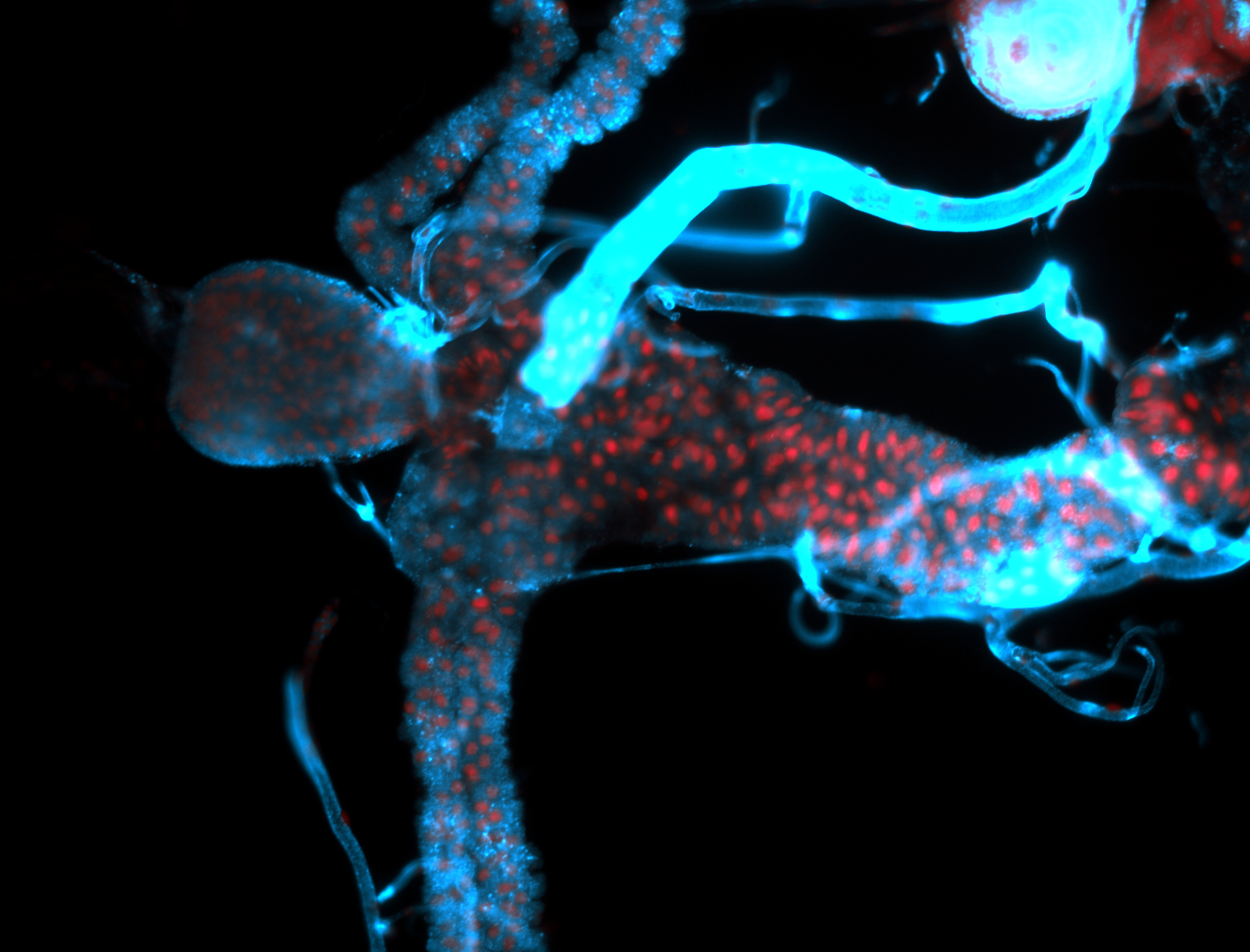
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -retested 2025 for better late stage coverage of patterns |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe on gel. |
Embryo
Larva
Adult
| Area | Images | Terms |
|---|
Comments
| Good probe on gel. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RE isoform transcript |
| -BDGP observes diffuse patterns, which in our experiment appears ubiquitous with some possible enrichment patterns in the later stages as outlined in the annotations |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -The results of our experiments do not align with those of BDGP, which observed no signal. They also do not align with modENCODE RNAseq data on FlyBase, which shows no expression throughout embryogenesis. No other genes overlap with TotA. |
| -Possible explanations for these discrepancies could be the design of probes by BDGP or ourselves, incorrect RNAseq data on FlyBase, or stress of flies during sample collection as TotA belongs to a family of stress-inducible genes. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -The results of our experiments do not align with those of BDGP, which observed no signal. They also do not align with modENCODE RNAseq data on FlyBase, which shows no expression throughout embryogenesis. No other genes overlap with TotC. |
| -Possible explanations for these discrepancies could be the design of probes by BDGP or ourselves, incorrect RNAseq data on FlyBase, or stress of flies during sample collection as TotC belongs to a family of stress-inducible genes. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -U2 us encoded at at least 6 different loci which this probe tries to target -U2: 34ABa 2L CR31850, U2: 34ABb 2L CR31854, U2: 34ABc 2L CR33788, U2: 38ABb 2L CR32878, U2: 38ABa 2L CR32882, and U2: 14B X CR32913 |
| -testing via HCR-FISH using a single probe set common to all listed loci |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
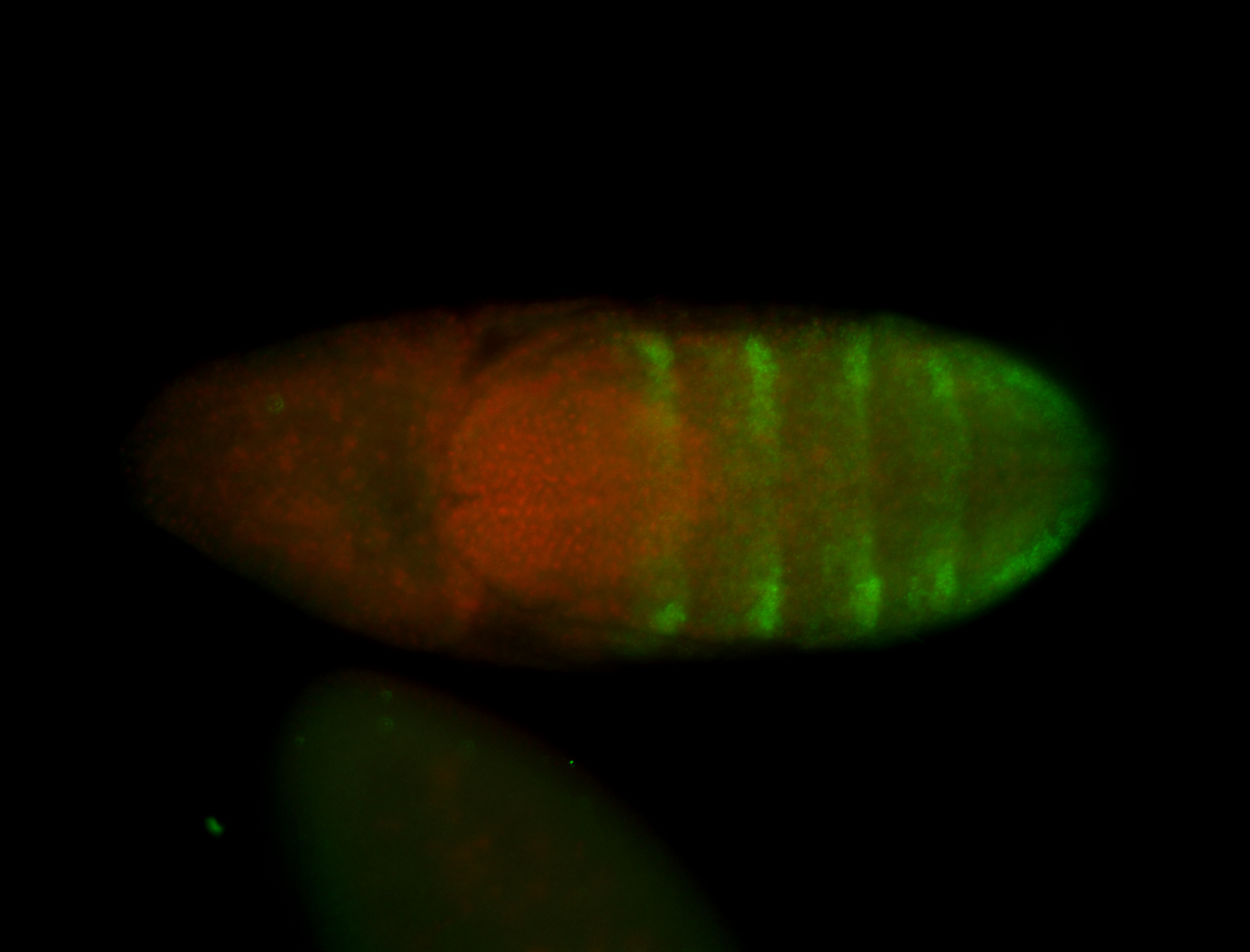
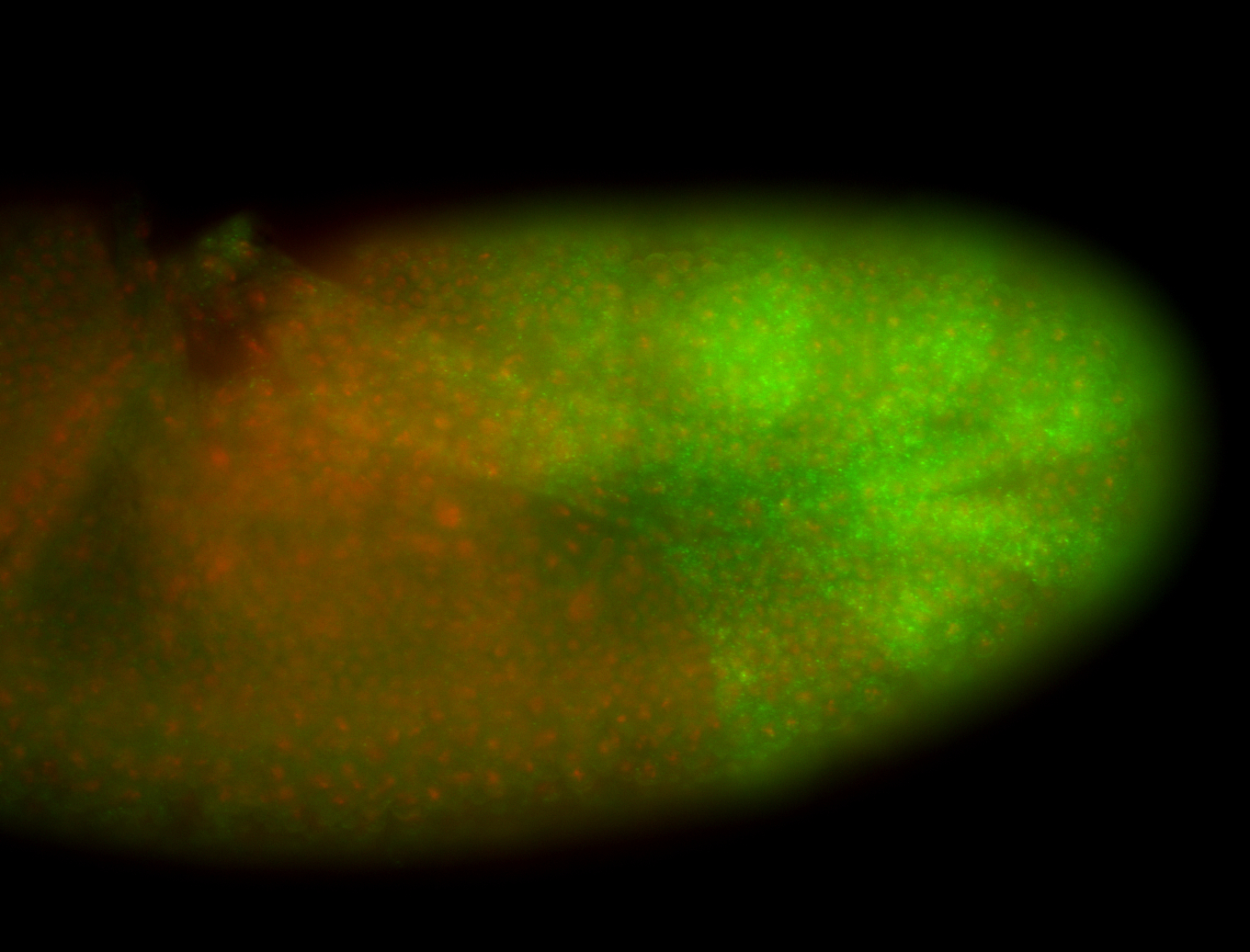
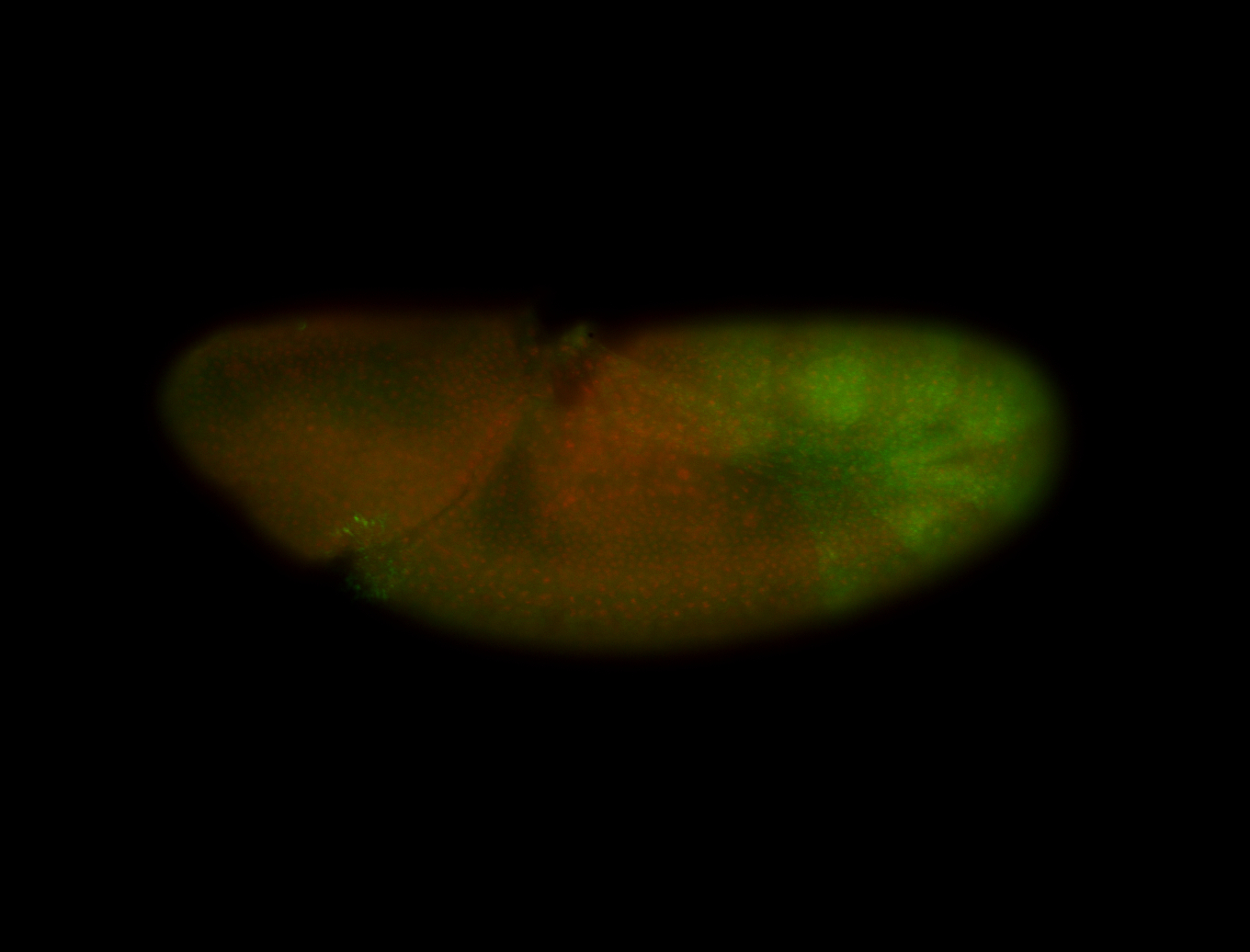
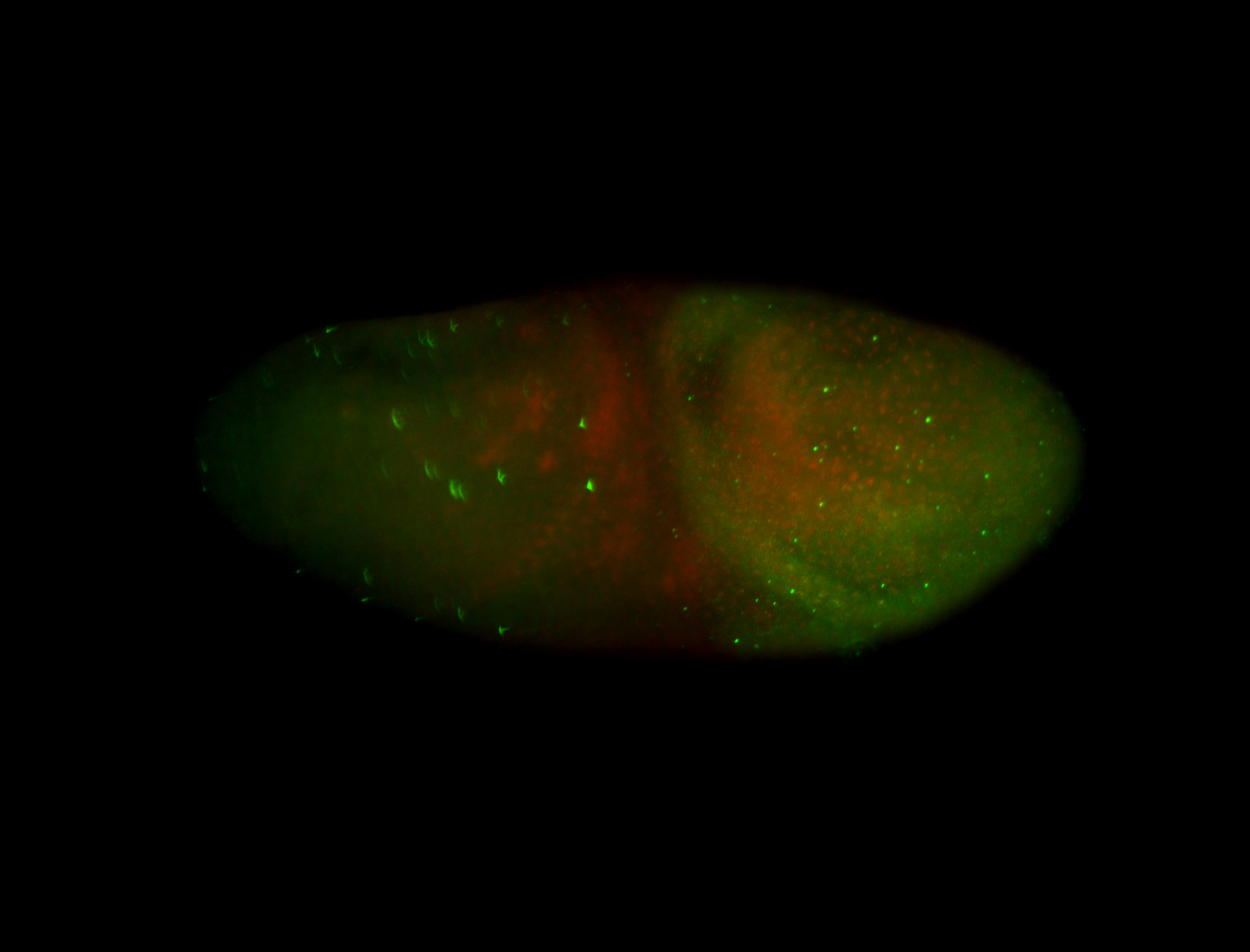
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -possesses a striking segmented pattern in the epidermis, mesoderm, and VNC; reports of expression in the cardiac primordium and dorsal vessel were not clearly observed and could possibly be amnioserosa expression |
| -a classic homeobox gene |
Probe betaTub56D-RA Links: Permalink
Genes: betaTub56D
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
| -gorgeous epidermal patterns, also expressed in CNS, certain muscles, and gonads |
Probe CG11901 Links: Permalink
Genes: eEF1 gamma
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting common exons |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting common exons |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting common exons |
| -Strong midgut signal was not observed by BDGP, likely a result of differing probe design |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 3 probe sets targeting the RA isoform transcript |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Probe CR34590 Links: Permalink
Genes: snoRNA:(psi)18S-531
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|