shd CG13478
Links: FlyBase, BDGP InSitu, DOT, Permalink
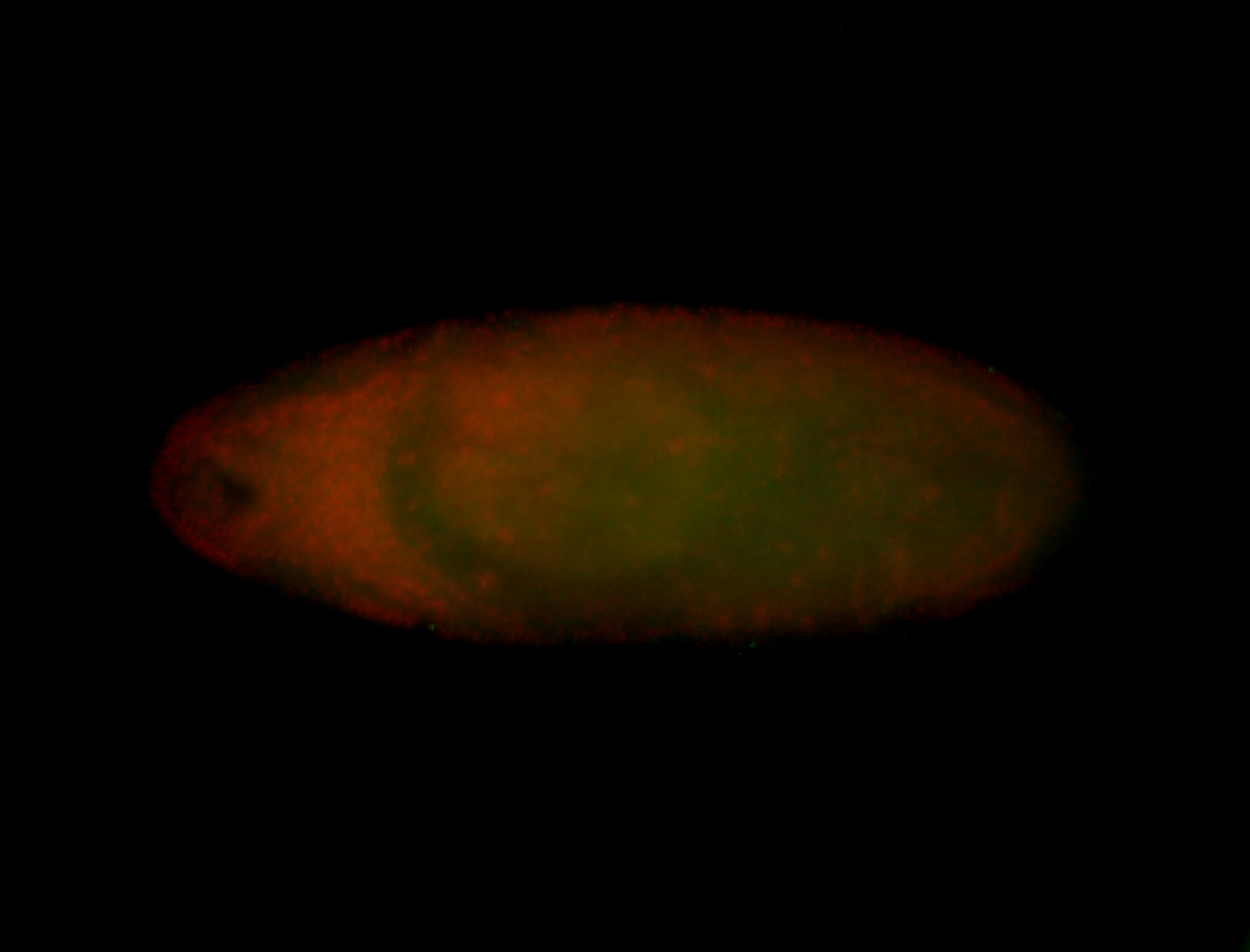
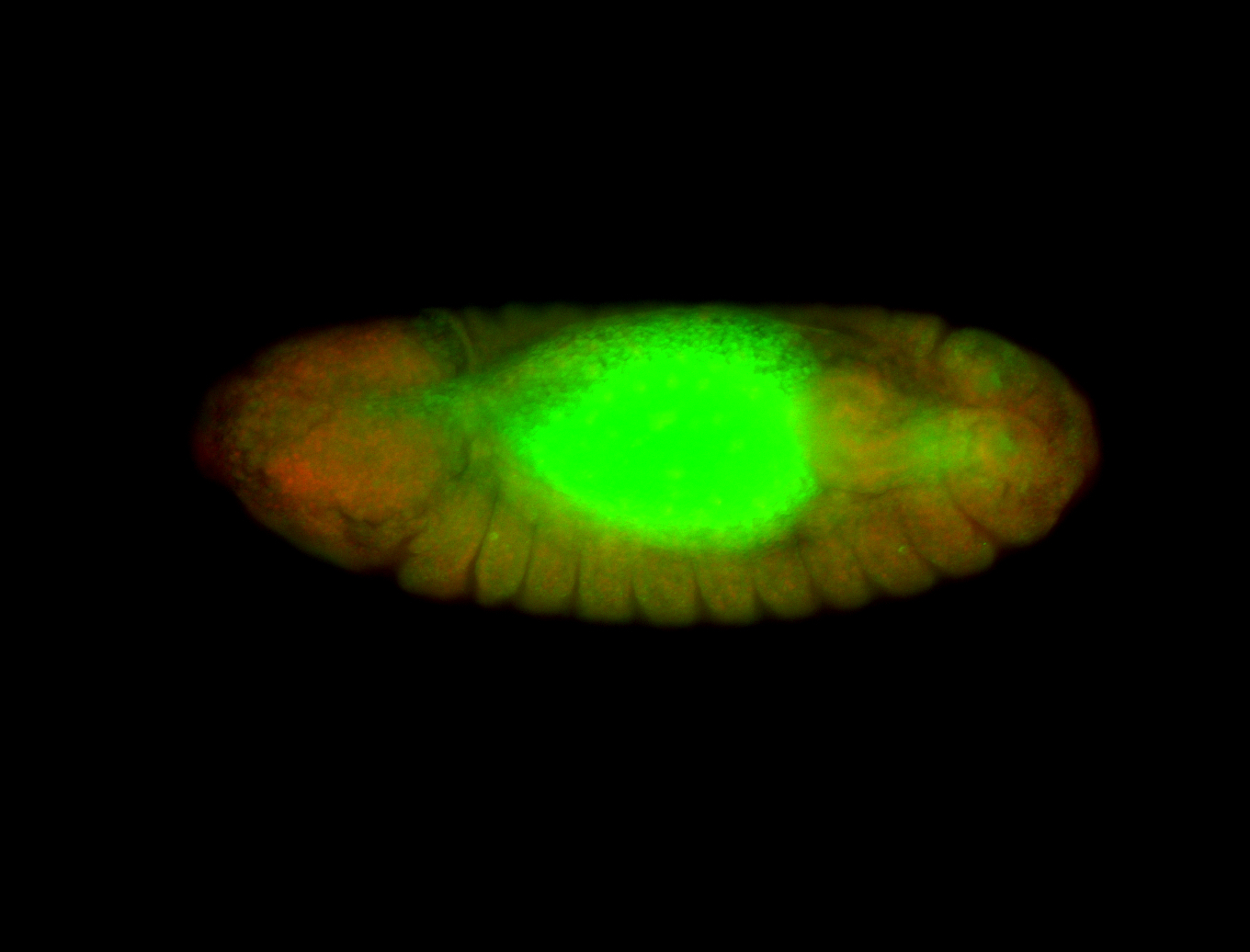
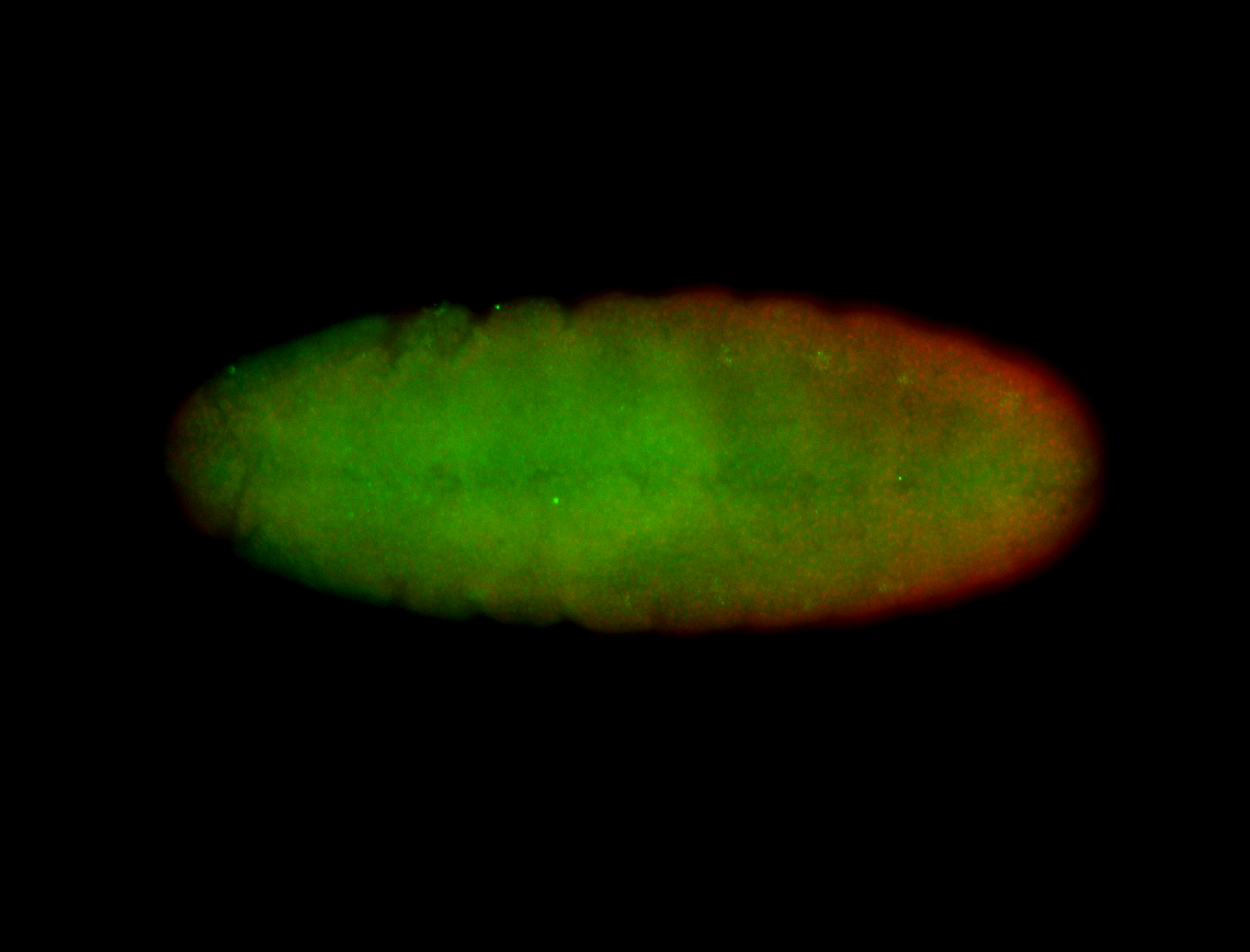
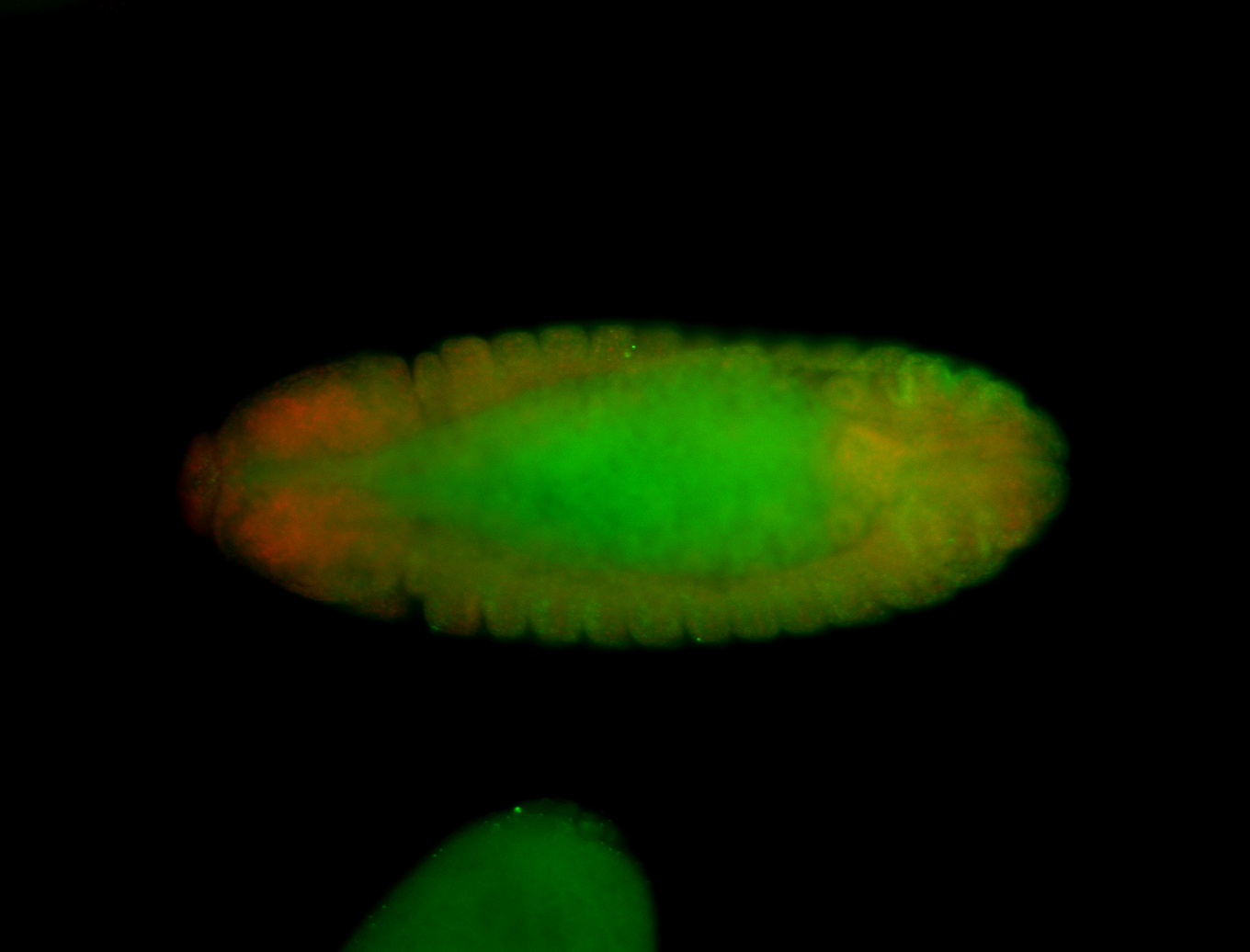
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| Re-annotated. Need more data. |
Embryo
Larva
| Area | Images | Terms |
|---|
Adult
| Area | Images | Terms |
|---|
Comments
| -performed via HCR-FISH using 6 probe sets targeting the RD isoform transcript (previously tested with 3 and was not detected) |
| -probes are from two different plates (3 sets from each plate); SH-HCR-Plate-1 and SH-HCR-Plate-4 |
| -this gene is very weakly expressed and more probe sets (if using HCR-FISH) should be considered for future retests |